Unique roles of the unfolded protein response pathway in fungal. development and differentiation. Kwang Woo Jung, Yee Seul So, & Yong Sun Bahn *
|
|
|
- Derrick Craig
- 5 years ago
- Views:
Transcription
1 Supplementry Informtion Unique roles of the unfolded protein response pthwy in fungl development nd differentition Kwng Woo Jung, Yee Seul So, & Yong Sun Bhn * Contents Supplementry Figure S1 Supplementry Figure S2 Supplementry Figure S3 Supplementry Figure S4 Supplementry Figure S5 Supplementry Figure S6 Supplementry Figure S7 Supplementry Figure S8 Supplementry Figure S9 Supplementry Tble S1 Supplementry Tble S2
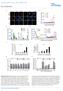
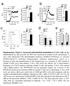
2 α α+h s α+h us α ire1 α ire1 +H s α ire1 +H us α hxl1 α hxl1 +H s α hxl1 +H us Supplementry Figure S1. Overctivtion of Hxl1 does not ffect sexul differentition in C. neoformns. Ech Cryptococcus strin ws cultured in the liquid YPD medium t 30 C overnight. Next, equl concentrtions of MATα nd MAT strins were mixed nd spotted onto V8 mting medium (ph 5). Serotype A strins were co cultured on the V8 medium (ph 5.0) for 10 d. Strin informtion: α (H99), (KN99), α ire1 mutnt (), α hxl1 mutnt (YSB723), α+h s (YSB742), α+h us (YSB737), α ire1 +H s (YSB1127), α ire1 +H us (YSB1126), α hxl1 +H s (YSB1225), nd α hxl1 +H us (YSB762).
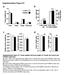
3 α α ire1 α RIM101oe ire1 α ADA2oe ire1 α GCN5oe ire1 15 dy Supplementry Figure S2. Overexpression of ADA2, RIM101, or GCN5 did not suppress the mting defect of the ire1 mutnt. Serotype A MATα nd MAT strins were grown in the liquid YPD medium t 30 C for 16 h. After cell counting, MATα nd MAT strins were co cultured on the V8 medium (ph 5.0) for 15 d t room temperture in the drk: α (H99) (KN99), α ire1 () ire1 (YSB550), α RIM101oe ire1 (YSB3308) ire1 (YSB550), α ADA2oe ire1 (YSB3376) ire1 (YSB550), nd α GCN5oe ire1 (YSB3372) ire1 (YSB550). Representtive edges of the mting ptches were photogrphed t 100 mgnifiction.
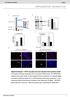
4 b c d E (6130) e h 3.5 kb 1.9 kb E (2918) ire1 ::NEO KW308* KW309* BmHI digestion B(0) B (1922) hxl1 ::NEO H kb f i KN99 YSB550* EcoRI digestion YSB550 E (4213) E (4213) ire1 ::NEO KW350* E (6785) ire1 ::NAT KW351* B(0) B (1922) H B (0) B (3495) E (12174) H ::NEO g j 8.5 kb 4.2 kb 2.5 kb H (0) crg1δ YSB1010* YSB1011* YSB kb 1.9 kb α crg1δ EcoRV digestion H (1217) YSB1007 KN99 YSB850* YSB851 BmHI digestion YSB1008* YSB1009* YSB849 H (4391) hxl1 ::NEO H ire1 ::NAT H B (0) B (3237) 3.2 kb hxl1 ::NAT 1.9 kb H CPK1 H (0) H (2863) H ::NAT cpk1 ::NEO BmHI digestion k l m YSB2505* YSB2506 YSB2567 E (2695) YSB2507* YSB2508 YSB kb cpk1 ::NEO RAS1 E(0) E (810) 2.6 kb RAS1 1.2 kb HindIII digestion CPK1 rs1 ::NEO 0.8 kb EcoRV digestion rs1 ::NEO Supplementry Figure S3. Disruption of C. neoformns nd H in serotype A MAT KN99, ire1 mutnts nd crg1 strins nd disruption of CPK1 nd RAS1 in the ire1 mutnts. (,c,f, h, j, nd l) A digrm of disruption of the gene, H gene, CPK1 gene, nd RAS1 gene in the serotype A (MAT) KN99 strin, ire1 mutnts ( nd YSB550), crg1 mutnts (H99 crg1 nd PPW196). (b, d, e, g, i, k, nd m) The correct gene disruption ws verified by Southern blot nlysis using genomic DNAs digested with the indicted restriction enzyme. Strins mrked with sterisk were used in this study.
5 b c d E (5231) JEC21 YSB2883 YSB2884 YSB2885 YSB2886* YSB2887 YSB2888 P (2377) JEC21 YSB2030* E (2591) 5.2 kb 2.5 kb ire1 ::NAT H P (3077) 3kb H H ::NAT hxl1 ::NAT EcoRI digestion ire1 ::NAT e f g h E (5231) JEC20 YSB2026* YSB2027 YSB2028 YSB2029 P (2377) 5.2 kb H PstI digestion JEC20 YSB1985* E (1191) 1. ire1 ::NEO P (1397) H ire1 ::NEO EcoRI digestion hxl1 ::NEO 1.3 kb PstI digestion H ::NEO Supplementry Figure S4. Disruption of C. neoformns nd H in serotype D JEC21 nd JEC20 strins. (, c, e, nd g) A digrm of disruption of the gene nd H gene in serotype D JEC21 (MATα) strin nd JEC20 (MAT). (b, d, f, nd h) The correct gene disruption ws verified by Southern blot nlysis using genomic DNAs digested with the indicted restriction enzyme. Strins mrked with sterisk were used in this study.
6 b 80 YSB2144* YSB2145* E (5231) 5.2 kb E (2591) 2.5 kb ire1 ::NAT ire1 ::NAT EcoRI digestion c P (2377) d 80 YSB2147* YSB2148* H P (3077) hxl1 ::NAT 3 kb H hxl1 ::NAT e P (2377) f PstI digestion YSB2145 KW345* KW346* H P (1397) 1.3 kb H hxl1 ::NEO hxl1 ::NEO PstI digestion Supplementry Figure S5. Deletion of C. neoformns nd H in the 80 strin nd ire1 mutnt. (, c, nd e) A digrm of disruption of the gene nd H gene in the 80 strin. (b, d, nd f) The correct gene disruption ws verified by Southern blot nlysis using genomic DNAs digested with the indicted restriction enzyme. Strins mrked with sterisk were used in this study.
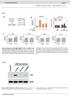
7 b c B(0) B (4772) YSB550 YSB3193 YSB3194* YSB550 YSB3193 YSB3194* B(0) NAT B (4592) E (6902) PH3 4.7 kb 4.5 kb BmHI digestion KA PH3:KA KA rrna Supplementry Figure S6. Construction of the constitutively KA overexpressing strins in C. neoformns. () The strtegy for construction of the PH3:KA strin contining the NAT resistnce mrker (NAT R ) nd the histone H3 gene promoter (PH3). (b) The correct genotype of the PH3:KA strin in the bckground of the ire1 mutnt ws confirmed by Southern blot nlysis using genomic DNAs digested with the restriction enzyme BmHI. The membrne ws hybridised with KA specific probe, wshed, nd developed. (c) Northern blot nlysis for mesurement of KA expression in PH3:KA strins. Dt from ethidium bromide stining of rrna ws used for loding control. Strins mrked with sterisk were used in this study.
8 E (4230) E (3861) E (6568) b 80 YSB3492* YSB3493* NEO PH3 c YSB2145 YSB3496 YSB3497 YSB3498 YSB3499* YSB3500* 4.2 kb 3.8 kb 2.5 kb KA PH3:KA 4.2 kb 3.8 kb 2.5 kb KA PH3:KA d EcoRI digestion 80 YSB3492 * YSB3493 * e YSB2145 YSB3496 EcoRI digestion YSB3497 YSB3498 YSB3499* YSB3500* KA KA rrna rrna Supplementry Figure S7. Construction of the constitutively KA overexpressing strins from the 80 nd ire1 mutnt.() A digrm of construction of the constitutively KA overexpressing strins using H3 promoter. (b nd c) The correct genotype of the PH3:KA strin in ech bckground strin (80: YSB3492 nd YSB3493 nd ire1δ mutnt: YSB3499 nd YSB3500) ws confirmed by Southern blot nlysis using genomic DNAs digested with the restriction enzyme EcoRI. The membrne ws hybridised with KA specific probe, wshed, nd developed. (d nd e) Northern blot nlysis for mesurement of KA expression in PH3:KA strins. Dt from ethidium bromide stining of rrna ws used for loding control. Strins mrked with sterisk were used in this study.
9 b c H (2611) H(0) PH3:ADA2 YSB3375 YSB3376* YSB3377 YSB3378* YSB3379 ADA2 H (0) H (4969) 4.9 kb PH3:ADA2 ADA2 PH3:ADA2 YSB3375 YSB3376* YSB3377 YSB3378* YSB3379 NEO PH3 2.6 kb ADA2 PH3:ADA2 HindIII digestion d e f H(0) H (2224) PH3:GCN5 YSB3372* YSB3373* rrna PH3:GCN5 YSB3372* YSB3373* H (0) H (4582) GCN5 4.5 kb PH3:GCN5 GCN5 NEO PH3 g h i K (2052) K (3115) K (0) RIM101 K(0) K (4410) K (5473) NEO PH3 PH3:RIM101 PH3:GCN5 GCN5 2.2 kb HindIII digestion PH3:RIM101 YSB3308* YSB3309* 4.4 kb RIM101 2 kb PH3:RIM101 KpnI digestion rrna RIM101 rrna PH3:RIM101 YSB3308* YSB3309* Supplementry Figure S8. Construction of the constitutively ADA2, GCN5, nd RIM101 overexpressing strins from the ire1 mutnt. (, d, nd g) A digrm of construction of the constitutively ADA2, GCN5, nd RIM101 overexpressing strins using the H3 promoter. (b, e, nd h) The correct genotype of the PH3:ADA2, PH3:GCN5, nd PH3:RIM101 strins in the strin (PH3:ADA2: YSB3376 nd YSB3378, PH3:GCN5: YSB3372 nd YSB3373, nd PH3:RIM101:YSB3308 nd YSB3309) ws confirmed by Southern blot nlysis using genomic DNAs digested with the indicted restriction enzyme. (c, f, nd i) Northern blot nlysis for mesurement of ADA2, GCN5, nd RIM101 expression levels. Ech membrne ws hybridised with ADA2, GCN5, nd RIM101 specific probes, wshed, nd developed. Dt from ethidium bromide stining of rrna ws used for loding control. Strins mrked with sterisk were used in this study.
10 P (7112) b c H99 YSB2618 YSB2619* YSB2619 YSB2744* STE6 P (9967) 7.9 kb P (7929) 7. STE6 STE6:GFP STE6:GFP NEO 2 kb PstI digestion d e f E(0) E (504) E (2216) STE3/CPRα E(0) E (504) 4.5 kb 1.7 kb E (5071) 0.5 kb H99 YSB2863 YSB2864* YSB2865 STE3:GFP STE3 4 Kb 4 Kb EcoRV, HindIII digestion YSB2864 YSB3019 ire1 ::NAT YSB3020* YSB3021 YSB3022 ire1 ::NAT STE3/CP:GFP NEO EcoRV digestion g h i E(0) E (2311) CP E (5617) E(0) E (2311) E (8472) kb H99 YSB3300* YSB3301 CP:GFP CP 4 Kb EcoRV, HindIII digestion YSB3300 YSB3311* ire1 ::NAT CP:GFP NEO EcoRV digestion EcoRV, HindIII digestion Supplementry Figure S9. Construction of the STE6:GFP, STE3/CPRα:GFP, nd CP:GFP strins nd ire1 mutnt in ech Gfp strin bckground. (, d, nd g) The scheme for construction of the STE6:GFP, STE3/CPRα:GFP, nd CP:GFP strins. (b, e, nd h) The correct genotype of the STE6:GFP, STE3/CPRα:GFP, nd CP:GFP strins in the strin (STE6:GFP: YSB2618 nd YSB2619, STE3/CPRα:GFP: YSB2864 nd YSB2865, nd CP:GFP: YSB3300 nd YSB3301) ws confirmed by Southern blot nlysis using genomic DNAs digested with the indicted restriction enzyme. (c, f, nd i) Southern blot nlysis for verifiction of the ire1 mutnt in ech Gfp strin bckground. Strins mrked with sterisk were used in this study.
11 Supplementry Tble S1. Strins used in this study Strin Genotype Prent Reference H99 MAT ref. 1 KN99 MAT ref. 2 JEC21 MAT ref. 3 JEC20 MAT ref MAT ref. 4 MAT ire1 ::NAT STM#224 H99 ref. 5 YSB1000 MAT ire1 ::NAT STM#224 NEO ref. 5 YSB723 MAT hxl1 ::NAT STM#295 H99 ref. 5 YSB762 MAT hxl1 ::NAT STM#295 H NEO YSB723 ref. 5 YSB737 MAT H us NEO H99 ref. 5 YSB742 MAT H s NEO H99 ref. 5 YSB1126 MAT ire1 ::NAT STM#224 H us NEO ref. 5 YSB1127 MAT ire1 ::NAT STM#224 H s NEO ref. 5 YSB1225 MAT hxl1 ::NAT STM#295 H s NEO YSB723 ref. 5 YSB119 MAT c1 ::NAT STM#43 ur5 ACA1 URA5 YSB108 ref. 6 YSB121 MAT c1 ::NEO ur5 ACA1 URA5 YSB109 ref. 6 H99 crg1 MAT ur5 crg1 ::URA5 F99 ref. 7 PPW196 MAT ur5 crg1 ::URA5 F99 ref. 7 YSB53 MAT rs1 ::NAT STM#150 H99 ref. 6 YSB127 MAT cpk1 ::NAT STM#184 H99 ref. 8 YSB1366 MAT rim101 ::NAT STM#208 H99 ref. 9 YSB550 MAT ire1 ::NEO KN99 This study YSB850 MAT hxl1 ::NEO KN99 This study KW308 MAT ire1 ::NAT STM#224 hxl1 ::NEO This study KW309 MAT ire1 ::NAT STM#224 hxl1 ::NEO This study KW350 MAT ire1 ::NEO hxl1 ::NAT STM#56 YSB550 This study KW351 MAT ire1 ::NEO hxl1 ::NAT STM#56 YSB550 This study YSB1741 MAT PH3:KA NEO ire1 ::NAT STM#224 ref. 10 YSB3194 MAT PH3:KA NAT ire1 ::NEO YSB550 This study YSB3308 MAT PH3:RIM101 NEO ire1 ::NAT STM#224 This study YSB3309 MAT PH3:RIM101 NEO ire1 ::NAT STM#224 This study YSB3372 MAT PH3:GCN5 NEO ire1 ::NAT STM#224 This study YSB3373 MAT PH3:GCN5 NEO ire1 ::NAT STM#224 This study YSB3376 MAT PH3:ADA2 NEO ire1 ::NAT STM#224 This study YSB3378 MAT PH3:ADA2 NEO ire1 ::NAT STM#224 This study YSB1008 MAT ur5 crg1 ::URA5 ire1 ::NAT STM#125 H99 crg1 This study YSB1009 MAT ur5 crg1 ::URA5 ire1 ::NAT STM#125 H99 crg1 This study YSB1010 MAT ur5 crg1 ::URA5 ire1 ::NAT STM#125 PPW196 This study YSB1011 MAT ur5 crg1 ::URA5 ire1 ::NAT STM#125 PPW196 This study YSB2505 MAT ire1 ::NAT STM#224 cpk1 ::NEO This study YSB2507 MAT ire1 ::NAT STM#224 rs1 ::NEO This study YSB2619 MAT STE6:GFP NEO H99 This study YSB2744 MAT STE6:GFP NEO ire1 ::NAT STM#169 YSB2619 This study YSB2864 MAT STE3:GFP NEO H99 This study YSB3020 MAT STE3:GFP NEO ire1 ::NAT STM#169 YSB2864 This study YSB3000 MAT CP:GFP NEO H99 This study YSB3311 MAT CP:GFP NEO ire1 ::NAT STM#169 YSB3000 This study YSB2886 MAT ire1 ::NAT STM#273 JEC21 This study YSB2026 MAT ire1 ::NEO JEC20 This study YSB2030 MAT hxl1 ::NAT STM#58 JEC21 This study YSB1985 MAT hxl1 ::NEO JEC20 This study YSB2144 MAT ire1 ::NAT STM# This study YSB2145 MAT ire1 ::NAT STM# This study YSB2147 MAT hxl1 ::NAT STM#43 80 This study YSB2148 MAT hxl1 ::NAT STM#43 80 This study
12 KW345 MAT ire1 ::NAT STM#273 hxl1 ::NEO YSB2145 This study KW346 MAT ire1 ::NAT STM#273 hxl1 ::NEO YSB2145 This study YSB3596 MAT ire1 ::NAT STM#273 NEO YSB2144 This study YSB3492 MAT PH3:KA NEO 80 This study YSB3493 MAT PH3:KA NEO 80 This study YSB3499 MAT PH3:KA NEO ire1 ::NAT STM#273 YSB2145 This study YSB3500 MAT PH3:KA NEO ire1 ::NAT STM#273 YSB2145 This study Ech NAT STM# indictes the Nt r mrker with unique signture tg.
13 Supplementry Tble S2. Primers used in this study Primer Nme Sequence (5 3 ) Comment B79 TGTGGATGCTGGCGGAGGATA Screening primer on ACT promoter B1026 GTAAAACGACGGCCAGTGAGC M13 forwrd (extended) B1027 CAGGAAACAGCTATGACCATG M13 reverse (extended) B1454 AAGGTGTTCCCCGACGACGAATCG NSL B1455 AACTCCGTCGCGAGCCCCATCAAC NSR B1886 TGGAAGAGATGGATGTGC NSL NEO B1887 ATTGTCTGTTGTGCCCAG NSR NEO B4017 GCATGCAGGATTCGAGTG H3 promoter left flnking primer 1 B4018 GTGATAGATGTGTTGTGGTG H3 promoter right flnking primer 2 B354 GCATGCAGGATTCGAGTG GFPht NEO left flnking primer 1 B5665 GGTGGCGGTGGCTCTGTGAGC GFPht NEO left flnking primer 2 B1648 AGGAATACGAGGTTTATCGG screening primer (for serotype A) B1644 GCCCCATCATCATAATCAC left flnking primer 1 (for serotype A) B1645 GCTCACTGGCCGTCGTTTTACACTATGTGTCCATCTGAGGC left flnking primer 2 (for serotype A) B1646 CATGGTCATAGCTGTTTCCTGAGTGAGTTGAGGGAGGAAAG right flnking primer 1 (for serotype A) B1647 GAAGAAGAGCGTCAAGAAGG right flnking primer 2 (for serotype A) B1683 AGCATTAGGGGTGTAGGTG Southern blot probe primer 1 (for serotype A) B1880 AACTCTTCTCAGCCTTCGG H screening primer (for serotype A) B1881 GTTTGAGGCTGGTAAAAAGG H left flnking primer 1 (for serotype A) B1882 GCTCACTGGCCGTCGTTTTACATGGGGAATGAAAGCGTG H left flnking primer 2 (for serotype A) B1883 CATGGTCATAGCTGTTTCCTGAAGGGGCGAGAGTAGTTCAG H right flnking primer 1 (for serotype A) B1884 GACTGTAAAGGAGGGCATAAG H right flnking primer 2 (for serotype A) B1885 CGTTCTCCGTCTTGATAGC H Southern blot probe primer 1 (for serotype A) B2167 CAAGGTTGACGAGATGAGTATG screening primer (for serotype D nd 80) B2168 CTTTCTTTTTCCGCCTACC left flnking primer 1 (for serotype D nd 80) B2169 TCACTGGCCGTCGTTTTAC GTCGCCGAGAGAATAAAATC left flnking primer 2 (for serotype D nd 80) B2170 CATGGTCATAGCTGTTTCCTG TGAGTTGAGGGAGGAAAGTC right flnking primer 1 (for serotype D nd 80) B2171 AAGGACACTATCCGTTCCG right flnking primer 2 (for serotype D nd 80) B4948 GATTTGGGTCGGAGATTC Southern blot probe primer 1 (for serotype D nd 80) B4769 GGAGTGAAAGCAGGAGTTG H screening primer (for serotype D nd 80) B4770 CCATCGTTCGGTATGCTAC H left flnking primer 1 (for serotype D nd 80) B4771 TCACTGGCCGTCGTTTTACAGTGGAAATAGGTGCGATG H left flnking primer 2 (for serotype D nd 80) B4772 CATGGTCATAGCTGTTTCCTGTTGGAAGGGAGGAAATGC H right flnking primer 1 (for serotype D nd 80) B4773 GGGTTAGTAGGAAGAAGTAGGC H right flnking primer 2 (for serotype D nd 80) B4774 CGTTGGCGAAGGACAATAC H Southern blot probe primer 1 (for serotype D nd 80) B3550 TCCCAATCTACTGACCTATCG KA 5 screening primer (H3 promoter replcement for serotype A) B3551 CGTAGGGTATGTCTCTGATGAG KA left flnking primer 1 (H3 promoter replcement for serotype A) B4270 CACTCGAATCCTGCATGCGGTGGCAAAAGTCTTGAGGA KA left flnking primer 2 (H3 promoter replcement for serotype A) B4264 CAAGACCTCAAAGACACCG KA right flnking primer 1 (H3 promoter replcement for serotype A) B4271 ACCACAACACATCTATCACATGGCATACCCTTCAAGAAT KA right flnking primer 2 (H3 promoter replcement for serotype A) B3555 CAAGCAGGGACAGTAACAAC KA Southern blot probe primer 1 (for serotype A) B4978 AGGCAGTCTGGAGTGTCATC KA Northern blot probe primer 1 (for serotype A) B2182 TCAGCAGCAGCAGGTAAAC ADA2 5 screening primer (H3 promoter replcement for serotype A) B2183 GGATGATGGAATCGTATGC ADA2 left flnking primer 1 (H3 promoter replcement for serotype A) B6399 CACTCGAATCCTGCATGCACTCGTTTGTTGATGCCTTT ADA2 left flnking primer 2 (H3 promoter replcement for serotype A) B6400 ACCACAACACATCTATCACATGACTGTCACGCAGAGGAA ADA2 right flnking primer 1 (H3 promoter replcement for serotype A)
14 B2187 TTCATCTGGAGGACGAGTG ADA2 right flnking primer 2 (H3 promoter replcement for serotype A) B4269 CCCAGACCGTTTTGAATG ADA2 Southern blot probe primer 1 (for serotype A) B2183 GGATGATGGAATCGTATGC ADA2 Southern blot probe primer 2 (for serotype A) B2187 TTCATCTGGAGGACGAGTG ADA2 Northern blot probe primer 1 (for serotype A) B6400 ACCACAACACATCTATCACATGACTGTCACGCAGAGGAA ADA2 Northern blot probe primer 2 (for serotype A) B2986 CACGGCAACTTATGCTCTC RIM101 5 screening primer (H3 promoter replcement) B2982 CATCAGTCTTGCTTCTTCTGC RIM101 left flnking primer 1 (H3 promoter replcement) B6201 CACTCGAATCCTGCATGCCTTGGCCTTGCTGTTAACTT RIM101 left flnking primer 2 (H3 promoter replcement) B6202 ACCACAACACATCTATCACATGGCTTACCCAATTCTCCC RIM101 right flnking primer 1 (H3 promoter replcement) B6203 AGCACCAAAAGGTTCAGC RIM101 right flnking primer 2 (H3 promoter replcement) B4980 ACTCGGTGTTGGTGAAACGG RIM101 Southern blot probe primer 1 B4980 ACTCGGTGTTGGTGAAACGG RIM101 Northern blot probe primer 1 B6170 GCCAAATCTTCTCAGCACTC RIM101 Northern blot probe primer 2 B6401 CAAAAATGGCGTCAGGTC GCN5 5 screening primer (H3 promoter replcement) B6402 CAATGATGAATGACCACGAC GCN5 left flnking primer 1 (H3 promoter replcement) B6403 CACTCGAATCCTGCATGCAGCAGAAACGGAAAGGCTTA GCN5 left flnking primer 2 (H3 promoter replcement) B6404 ACCACAACACATCTATCACATGGCGCCAAAACCACGTCGT GCN5 right flnking primer 1 (H3 promoter replcement) B6405 TCAGAGTGGGTAGATTCGC GCN5 right flnking primer 2 (H3 promoter replcement) B6406 CGTCTTCGTCTTCATTGC GCN5 Southern blot probe primer 1 B6402 CAATGATGAATGACCACGAC GCN5 Southern blot probe primer 2 B6405 TCAGAGTGGGTAGATTCGC GCN5 Northern blot probe primer 1 B6406 CGTCTTCGTCTTCATTGC GCN5 Northern blot probe primer 2 B6632 TGGCGGAGAAACAGTAGAG KA 5 screening primer (H3 promoter replcement for 80) B6633 GGCAACCCTCATCTACTGAC KA left flnking primer 1 (H3 promoter replcement for 80) B6634 CACTCGAATCCTGCATGCCCCGGGCCTTTTTACCCTTT KA left flnking primer 2 (H3 promoter replcement for 80) B6635 ACCACAACACATCTATCACATGAAATGGCGAGTGAGGCG KA right flnking primer 1 (H3 promoter replcement for 80) B6644 CACTATTACCCACAAGCGG KA right flnking primer 2 (H3 promoter replcement for 80) B6637 AAGTCGTTCCTCCTCAGTG KA Southern blot probe primer 1 (for 80) B5661 CAGTCTAACTTTTGGCAGTGG STE6:GFP tgging left flnking primer 1 B5662 GCTCACAGAGCCACCGCCACCCTCCCATTCTCCCGTCTTCATCA STR6:GFP tgging left flnking primer 2 B5663 GCCACTCGAATCCTGCATGCATTATAGAGGTTGTCTTGC STE6:GFP tgging right flnking primer 1 B5664 GCTCGGGTAAGATACTGAGG STE6:GFP tgging right flnking primer 2 B5660 GAGGGTTGCCTTTGTTTG STE6:GFP tgging screening primer B5741 TTGGTTTCATAGCCCTGC STE6:GFP tgging Southern blot probe primer B5897 TTTTCATCGGAGTCCCTG STE3/CPRα:GFP tgging left flnking primer 1 B5898 GCTCACAGAGCCACCGCCACCAACGACAGCACGCTCGACCGA STE3/CPRα:GFP tgging left flnking primer 2 B5899 GCCACTCGAATCCTGCATGCTTCCACGATGGTTGATGTAAA STE3/CPRα:GFP tgging right flnking primer 1 B5900 GACAAGAATGCGATGTGG STE3/CPRα:GFP tgging right flnking primer 2 B4790 CTTCTACCTCTGCCTCTTCAC STE3/CPRα:GFP tgging screening primer B5959 CGTCTCCCATCTCACTTTG STE3/CPRα:GFP tgging Southern blot probe primer B5943 TATGATTCTTCCGCTCGC CP:GFP tgging left flnking primer 1 B5902 GCTCACAGAGCCACCGCCACCAACCATTGATGAGCTAGTCTT CP:GFP tgging left flnking primer 2 B5903 GCCACTCGAATCCTGCATGCAATGTCCATTTGTTGGGACCG CP:GFP tgging right flnking primer 1 B5904 GACCAAGCAAGAAGAAGATG CP:GFP tgging right flnking primer 2 B4792 CCATCTGTCATCGCTTTG CP:GFP tgging dignostic primer B4793 CAGTGGTAGCAGAGGAATAGG CP:GFP tgging Southern blot probe primer B1894 TTTTACGCTTTTTGCAGATTCCGCCAAA MFα1 primer 1 for Northern blot B1895 GACCACTGTTTCTTTCGTTCT MFα1 primer 2 for Northern blot J94 CGCCTTCACTGCCATCTTC MFα1 primer 1 for qrt nlysis J95 ACAAAGGGTCATGCCACCGG MFα1 primer 2 for qrt nlysis B679 CGCCCTTGCTCCTTCTTCTATG ACT1 primer 1 for qrt nlysis B680 GACTCGTCGTATTCGCTCTTCG ACT1 primer 2 for qrt nlysis
15 JOHE12039 CTGTAGAAGATGTGAGTTTGGG CPK1 left flnking primer 1 JOHE12040 CTGGCCGTCGTTTTACTGATTGATGAGAGATACGGG CPK1 left flnking primer 2 JOHE12041 GTCATAGCTGTTTCCTGGGCGGAGAAATAGAGGTTG CPK1 right flnking primer 1 JOHE12042 CGCACAAGAAGTAAGAGGTG CPK1 right flnking primer 2 JOHE12043 GGCTATGGACCGTATTCAC CPK1 screening primer JOHE12045 TATCTCACAAGCCACTCCC CPK1 Southern blot probe primer 1 JOHE12046 ATGCTGCTCACCGTTAGTC CPK1 Southern blot probe primer 2 JOHE11600 AAGACTGGTTCAGCAGAGC RAS1 left flnking primer 1 JOHE11601 CTGGCCGTCGTTTTACCTAAATGGGGATGGTTCG RAS1 left flnking primer 2 JOHE11602 GTCATAGCTGTTTCCTGAAACATCCGCCAAGCAAC RAS1 right flnking primer 1 JOHE11603 GAGCAAAATGAGGAACTTGG RAS1 right flnking primer 2 JOHE11604 TGTGCTTTACCAGGCAGTCG RAS1 screening primer JOHE11606 CACACTCTTTGTCCTTCCG RAS1 Southern blot probe primer 1 JOHE11607 ACCGTATTCTTCTTG ACC G RAS1 Southern blot probe primer 2
16 References 1. Perfect, J. R., Ketbchi, N., Cox, G. M., Ingrm, C. W. & Beiser, C. L. Kryotyping of Cryptococcus neoformns s n epidemiologicl tool. J. Clin. Microbiol. 31, (1993). 2. Nielsen, K. et l. Sexul cycle of Cryptococcus neoformns vr. grubii nd virulence of congenic nd isoltes. Infect. Immun. 71, (2003). 3. Kwon Chung, K. J., Edmn, J. C. & Wickes, B. L. Genetic ssocition of mting types nd virulence in Cryptococcus neoformns. Infect. Immun. 60, (1992). 4. Lin,., Hull, C. M. & Heitmn, J. Sexul reproduction between prtners of the sme mting type in Cryptococcus neoformns. Nture 434, (2005). 5. Cheon, S. A. et l. Unique evolution of the UPR pthwy with novel bzip trnscription fctor, Hxl1, for controlling pthogenicity of Cryptococcus neoformns. PLoS Pthog. 7, e (2011). 6. Bhn, Y. S., Hicks, J. K., Giles, S. S., Cox, G. M. & Heitmn, J. Adenylyl cyclse ssocited protein Ac1 regultes virulence nd differentition of Cryptococcus neoformns vi the cyclic AMP protein kinse A cscde. Eukryot. Cell 3, (2004). 7. Wng, P., Cutler, J., King, J. & Plmer, D. Muttion of the regultor of G protein signling Crg1 increses virulence in Cryptococcus neoformns. Eukryot. Cell 3, (2004). 8. Bhn, Y. S., Geunes Boyer, S. & Heitmn, J. Ssk2 mitogen ctivted protein kinse kinse kinse governs divergent ptterns of the stress ctivted Hog1 signling pthwy in Cryptococcus neoformns. Eukryot. Cell 6, (2007). 9. Jung, K. W. et l. Systemtic functionl profiling of trnscription fctor networks in Cryptococcus neoformns. Nt. Commun. 6, 6757 (2015)
17 10. Jung, K. W., Kng, H. A. & Bhn, Y. S. Essentil roles of the Kr2/BiP moleculr chperone downstrem of the UPR pthwy in Cryptococcus neoformns. PLoS ONE 8, e58956 (2013).
Supplementary figure 1
 Supplementry figure 1 Dy 8 post LCMV infection Vsculr Assoc. Prenchym Dy 3 post LCMV infection 1 5 6.7.29 1 4 1 3 1 2 88.9 4.16 1 2 1 3 1 4 1 5 1 5 1.59 5.97 1 4 1 3 1 2 21.4 71 1 2 1 3 1 4 1 5 1 5.59.22
Supplementry figure 1 Dy 8 post LCMV infection Vsculr Assoc. Prenchym Dy 3 post LCMV infection 1 5 6.7.29 1 4 1 3 1 2 88.9 4.16 1 2 1 3 1 4 1 5 1 5 1.59 5.97 1 4 1 3 1 2 21.4 71 1 2 1 3 1 4 1 5 1 5.59.22
SUPPLEMENTARY INFORMATION
 doi:1.138/nture1794 BR EPFs BRI1? ERECTA TMM BSKs YDA PP2A BSU1 BIN2 pbzr1/2 BZR1/2 MKK4/5/7/9 MPK3/6 SPCH Cell growth Stomtl production Supplementry Figure 1. The model of BR nd stomtl signling pthwys.
doi:1.138/nture1794 BR EPFs BRI1? ERECTA TMM BSKs YDA PP2A BSU1 BIN2 pbzr1/2 BZR1/2 MKK4/5/7/9 MPK3/6 SPCH Cell growth Stomtl production Supplementry Figure 1. The model of BR nd stomtl signling pthwys.
2018 American Diabetes Association. Published online at
 Supplementry Figure S1. Ft-1 mice exhibit reduced diposity when fed n HFHS diet. WT nd ft-1 mice were fed either control or n HFHS diet for 18 weeks. A: Representtive photogrphs for side-by-side comprison
Supplementry Figure S1. Ft-1 mice exhibit reduced diposity when fed n HFHS diet. WT nd ft-1 mice were fed either control or n HFHS diet for 18 weeks. A: Representtive photogrphs for side-by-side comprison
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nture07679 Emryonic Stem (ES) cell Hemngiolst Flk1 + Blst Colony 3 to 3.5 Dys 3-4 Dys ES differentition Sort of Flk1 + cells Supplementry Figure 1. Chrcteristion of lst colony development.
doi: 10.1038/nture07679 Emryonic Stem (ES) cell Hemngiolst Flk1 + Blst Colony 3 to 3.5 Dys 3-4 Dys ES differentition Sort of Flk1 + cells Supplementry Figure 1. Chrcteristion of lst colony development.
Supplementary Figure 1
 Roles of endoplsmic reticulum stress-medited poptosis in -polrized mcrophges during mycocteril infections Supplementry informtion Yun-Ji Lim, Min-Hee Yi, Ji-Ae Choi, Jung-hwn Lee, Ji-Ye Hn, Sung-Hee Jo,
Roles of endoplsmic reticulum stress-medited poptosis in -polrized mcrophges during mycocteril infections Supplementry informtion Yun-Ji Lim, Min-Hee Yi, Ji-Ae Choi, Jung-hwn Lee, Ji-Ye Hn, Sung-Hee Jo,
TNF-α (pg/ml) IL-6 (ng/ml)
 Xio, et l., Supplementry Figure 1 IL-6 (ng/ml) TNF-α (pg/ml) 16 12 8 4 1,4 1,2 1, 8 6 4 2 med Cl / Pm3CSK4 zymosn curdln Poly (I:C) LPS flgelin MALP-2 imiquimod R848 CpG TNF-α (pg/ml) IL-6 (ng/ml) 2 1.6
Xio, et l., Supplementry Figure 1 IL-6 (ng/ml) TNF-α (pg/ml) 16 12 8 4 1,4 1,2 1, 8 6 4 2 med Cl / Pm3CSK4 zymosn curdln Poly (I:C) LPS flgelin MALP-2 imiquimod R848 CpG TNF-α (pg/ml) IL-6 (ng/ml) 2 1.6
Ulk λ PPase. 32 P-Ulk1 32 P-GST-TSC2. Ulk1 GST (TSC2) : Ha-Ulk1 : AMPK. WB: Ha (Ulk1) : Glu. h CON - Glu - A.A WB: LC3 AMPK-WT AMPK-DKO
 DOI: 10.1038/ncb2152 C.C + - + - : Glu b Ulk1 - - + λ PPse c AMPK + - + + : ATP P-GST-TSC2 WB: Flg (Ulk1) WB Ulk1 WB: H (Ulk1) GST (TSC2) C.C d e WT K46R - + - + : H-Ulk1 : AMPK - + - + + + AMPK H-Ulk1
DOI: 10.1038/ncb2152 C.C + - + - : Glu b Ulk1 - - + λ PPse c AMPK + - + + : ATP P-GST-TSC2 WB: Flg (Ulk1) WB Ulk1 WB: H (Ulk1) GST (TSC2) C.C d e WT K46R - + - + : H-Ulk1 : AMPK - + - + + + AMPK H-Ulk1
NappHS. rrna. transcript abundance. NappHS relative con W+W 0.8. nicotine [µg mg -1 FM]
![NappHS. rrna. transcript abundance. NappHS relative con W+W 0.8. nicotine [µg mg -1 FM] NappHS. rrna. transcript abundance. NappHS relative con W+W 0.8. nicotine [µg mg -1 FM]](/thumbs/92/110177253.jpg) (A) W+OS 3 min 6 min con L S L S RNA loding control NppHS rrna (B) (C) 8 1 k NppHS reltive trnscript undnce 6 4.5 *** *** *** *** 3 k. + + + line 1 line (D) nicotine [µg mg -1 FM] 1..8.4. con W+W Supplementl
(A) W+OS 3 min 6 min con L S L S RNA loding control NppHS rrna (B) (C) 8 1 k NppHS reltive trnscript undnce 6 4.5 *** *** *** *** 3 k. + + + line 1 line (D) nicotine [µg mg -1 FM] 1..8.4. con W+W Supplementl
Supplementary information for: Low bone mass and changes in the osteocyte network in mice lacking autophagy in the osteoblast lineage
 Supplementry informtion for: Low one mss nd chnges in the osteocyte network in mice lcking utophgy in the osteolst linege Mrilin Piemontese, Meld Onl, Jinhu Xiong, Li Hn, Jeff D. Thostenson, Mri Almeid,
Supplementry informtion for: Low one mss nd chnges in the osteocyte network in mice lcking utophgy in the osteolst linege Mrilin Piemontese, Meld Onl, Jinhu Xiong, Li Hn, Jeff D. Thostenson, Mri Almeid,
Received 16 July 2004/Accepted 21 August 2004
 EUKARYOTIC CELL, Dec. 2004, p. 1476 1491 Vol. 3, No. 6 1535-9778/04/$08.00 0 DOI: 10.1128/EC.3.6.1476 1491.2004 Copyright 2004, American Society for Microbiology. All Rights Reserved. Adenylyl Cyclase-Associated
EUKARYOTIC CELL, Dec. 2004, p. 1476 1491 Vol. 3, No. 6 1535-9778/04/$08.00 0 DOI: 10.1128/EC.3.6.1476 1491.2004 Copyright 2004, American Society for Microbiology. All Rights Reserved. Adenylyl Cyclase-Associated
SUPPLEMENTARY INFORMATION
 DOI:.38/nc393 Nnog DAPI Nnog/DAPI c-jun DAPI c-jun/dapi c e Reltive expression to Gpdh mescs ( Feeder free) mescs ( Feeder) MEFs.5 MEFs ipscs ESCs..5 p=.24 p=.483 p=.22. JunB JunD Fos Fr Fr2 ATF2 ATF3
DOI:.38/nc393 Nnog DAPI Nnog/DAPI c-jun DAPI c-jun/dapi c e Reltive expression to Gpdh mescs ( Feeder free) mescs ( Feeder) MEFs.5 MEFs ipscs ESCs..5 p=.24 p=.483 p=.22. JunB JunD Fos Fr Fr2 ATF2 ATF3
Acute and gradual increases in BDNF concentration elicit distinct signaling and functions in neurons
 nd grdul increses in BDNF concentrtion elicit distinct signling nd functions in neurons Yunyun Ji,, Yun Lu, Feng Yng, Wnhu Shen, Tin Tze-Tsng Tng,, Linyin Feng, Shumin Dun, nd Bi Lu,.. - Grdul (normlized
nd grdul increses in BDNF concentrtion elicit distinct signling nd functions in neurons Yunyun Ji,, Yun Lu, Feng Yng, Wnhu Shen, Tin Tze-Tsng Tng,, Linyin Feng, Shumin Dun, nd Bi Lu,.. - Grdul (normlized
Received 24 September 2007/Accepted 8 October 2007
 EUKARYOTIC CELL, Dec. 2007, p. 2278 2289 Vol. 6, No. 12 1535-9778/07/$08.00 0 doi:10.1128/ec.00349-07 Copyright 2007, American Society for Microbiology. All Rights Reserved. Ssk2 Mitogen-Activated Protein
EUKARYOTIC CELL, Dec. 2007, p. 2278 2289 Vol. 6, No. 12 1535-9778/07/$08.00 0 doi:10.1128/ec.00349-07 Copyright 2007, American Society for Microbiology. All Rights Reserved. Ssk2 Mitogen-Activated Protein
SUPPLEMENTARY INFORMATION
 doi:1.138/nture1228 Totl Cell Numer (cells/μl of lood) 12 1 8 6 4 2 d Peripherl Blood 2 4 7 Time (d) fter nti-cd3 i.p. + TCRβ + IL17A + cells (%) 7 6 5 4 3 2 1 Totl Cell Numer (x1 3 ) 8 7 6 5 4 3 2 1 %
doi:1.138/nture1228 Totl Cell Numer (cells/μl of lood) 12 1 8 6 4 2 d Peripherl Blood 2 4 7 Time (d) fter nti-cd3 i.p. + TCRβ + IL17A + cells (%) 7 6 5 4 3 2 1 Totl Cell Numer (x1 3 ) 8 7 6 5 4 3 2 1 %
SUPPLEMENTARY INFORMATION
 SUPPLEMEARY IFORMAIO doi:./nture correction to Supplementry Informtion Adenom-linked rrier defects nd microil products drive IL-/IL-7-medited tumour growth Sergei I. Grivennikov, Kepeng Wng, Dniel Mucid,
SUPPLEMEARY IFORMAIO doi:./nture correction to Supplementry Informtion Adenom-linked rrier defects nd microil products drive IL-/IL-7-medited tumour growth Sergei I. Grivennikov, Kepeng Wng, Dniel Mucid,
Supplementary Information
 Supplementry Informtion Cutneous immuno-surveillnce nd regultion of inflmmtion y group 2 innte lymphoid cells Ben Roediger, Ryn Kyle, Kwok Ho Yip, Nitl Sumri, Thoms V. Guy, Brin S. Kim, Andrew J. Mitchell,
Supplementry Informtion Cutneous immuno-surveillnce nd regultion of inflmmtion y group 2 innte lymphoid cells Ben Roediger, Ryn Kyle, Kwok Ho Yip, Nitl Sumri, Thoms V. Guy, Brin S. Kim, Andrew J. Mitchell,
EFFECTS OF AN ACUTE ENTERIC DISEASE CHALLENGE ON IGF-1 AND IGFBP-3 GENE EXPRESSION IN PORCINE SKELETAL MUSCLE
 Swine Dy 22 Contents EFFECTS OF AN ACUTE ENTERIC DISEASE CHALLENGE ON IGF-1 AND IGFBP-3 GENE EXPRESSION IN PORCINE SKELETAL MUSCLE B. J. Johnson, J. P. Kyser, J. D. Dunn, A. T. Wyln, S. S. Dritz 1, J.
Swine Dy 22 Contents EFFECTS OF AN ACUTE ENTERIC DISEASE CHALLENGE ON IGF-1 AND IGFBP-3 GENE EXPRESSION IN PORCINE SKELETAL MUSCLE B. J. Johnson, J. P. Kyser, J. D. Dunn, A. T. Wyln, S. S. Dritz 1, J.
Alimonti_Supplementary Figure 1. Pten +/- Pten + Pten. Pten hy. β-actin. Pten - wt hy/+ +/- wt hy/+ +/- Pten. Pten. Relative Protein level (% )
 Alimonti_Supplementry Figure 1 hy 3 4 5 3 Neo 4 5 5 Proe 5 Proe hy/ hy/ /- - 3 6 Neo β-tin d Reltive Protein level (% ) 15 1 5 hy/ /- Reltive Gene Expr. (% ) 15 1 5 hy/ /- Supplementry Figure 1 Chrteriztion
Alimonti_Supplementry Figure 1 hy 3 4 5 3 Neo 4 5 5 Proe 5 Proe hy/ hy/ /- - 3 6 Neo β-tin d Reltive Protein level (% ) 15 1 5 hy/ /- Reltive Gene Expr. (% ) 15 1 5 hy/ /- Supplementry Figure 1 Chrteriztion
SUPPLEMENTARY INFORMATION
 doi:0.08/nture0987 SUPPLEMENTARY FIGURE Structure of rbbit Xist gene. Exons re shown in boxes with romn numbers, introns in thin lines. Arrows indicte the locliztion of primers used for mplifiction. WWW.NATURE.COM/NATURE
doi:0.08/nture0987 SUPPLEMENTARY FIGURE Structure of rbbit Xist gene. Exons re shown in boxes with romn numbers, introns in thin lines. Arrows indicte the locliztion of primers used for mplifiction. WWW.NATURE.COM/NATURE
Supplementary Information Titles
 Supplementry Informtion Titles Journl: Nture Medicine Article Title: Corresponding Author: Modelling colorectl cncer using CRISPR-Cs9-medited engineering of humn intestinl orgnoids Toshiro Sto Supplementry
Supplementry Informtion Titles Journl: Nture Medicine Article Title: Corresponding Author: Modelling colorectl cncer using CRISPR-Cs9-medited engineering of humn intestinl orgnoids Toshiro Sto Supplementry
SUPPLEMENTARY INFORMATION
 Supplementry Figure 1. Genertion of N- nd C-tgged cyclin D1 knock-in mice., N-tgged cyclin D1 gene trgeting construct, cyclin D1 genomic locus, cyclin D1 locus following homologous recomintion (trgeted
Supplementry Figure 1. Genertion of N- nd C-tgged cyclin D1 knock-in mice., N-tgged cyclin D1 gene trgeting construct, cyclin D1 genomic locus, cyclin D1 locus following homologous recomintion (trgeted
Supplementary Information
 Supplementary Information Overexpression of Fto leads to increased food intake and results in obesity Chris Church, Lee Moir, Fiona McMurray, Christophe Girard, Gareth T Banks, Lydia Teboul, Sara Wells,
Supplementary Information Overexpression of Fto leads to increased food intake and results in obesity Chris Church, Lee Moir, Fiona McMurray, Christophe Girard, Gareth T Banks, Lydia Teboul, Sara Wells,
Interaction between genetic background and the mating type locus in Cryptococcus neoformans virulence potential
 Genetics: Published Articles Ahead of Print, published on June 18, 2005 as 10.1534/genetics.105.045039 Interaction between genetic background and the mating type locus in Cryptococcus neoformans virulence
Genetics: Published Articles Ahead of Print, published on June 18, 2005 as 10.1534/genetics.105.045039 Interaction between genetic background and the mating type locus in Cryptococcus neoformans virulence
SUPPLEMENTARY INFORMATION
 Supplementry Tble 1. Sttistics of dt sets nd structure refinement PYL1 po PYL2/ABA PYL2 po PYL2/ABA/HAB1 PDB code 3KAY 3KAZ 3KB0 3KB3 Dt collection APS bem line 21-ID 21-ID 21-ID 21-ID Spce group P6 5
Supplementry Tble 1. Sttistics of dt sets nd structure refinement PYL1 po PYL2/ABA PYL2 po PYL2/ABA/HAB1 PDB code 3KAY 3KAZ 3KB0 3KB3 Dt collection APS bem line 21-ID 21-ID 21-ID 21-ID Spce group P6 5
Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome
 Supplementry Informtion Microtuule-driven sptil rrngement of mitochondri promotes ctivtion of the NLRP3 inflmmsome Tkum Misw 1,2, Michihiro Tkhm 1,2, Ttsuy Kozki 1,2, Hnn Lee 1,2, Jin Zou 1,2, Ttsuy Sitoh
Supplementry Informtion Microtuule-driven sptil rrngement of mitochondri promotes ctivtion of the NLRP3 inflmmsome Tkum Misw 1,2, Michihiro Tkhm 1,2, Ttsuy Kozki 1,2, Hnn Lee 1,2, Jin Zou 1,2, Ttsuy Sitoh
Copy Number ID2 MYCN ID2 MYCN. Copy Number MYCN DDX1 ID2 KIDINS220 MBOAT2 ID2
 Copy Numer Copy Numer Copy Numer Copy Numer DIPG38 DIPG49 ID2 MYCN ID2 MYCN c DIPG01 d DIPG29 ID2 MYCN ID2 MYCN e STNG2 f MYCN DIPG01 Chr. 2 DIPG29 Chr. 1 MYCN DDX1 Chr. 2 ID2 KIDINS220 MBOAT2 ID2 Supplementry
Copy Numer Copy Numer Copy Numer Copy Numer DIPG38 DIPG49 ID2 MYCN ID2 MYCN c DIPG01 d DIPG29 ID2 MYCN ID2 MYCN e STNG2 f MYCN DIPG01 Chr. 2 DIPG29 Chr. 1 MYCN DDX1 Chr. 2 ID2 KIDINS220 MBOAT2 ID2 Supplementry
Mating-Type-Specific and Nonspecific PAK Kinases Play Shared and Divergent Roles in Cryptococcus neoformans
 EUKARYOTIC CELL, Apr. 2002, p. 257 272 Vol. 1, No. 2 1535-9778/02/$04.00 0 DOI: 10.1128/EC.1.2.257 272.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Mating-Type-Specific
EUKARYOTIC CELL, Apr. 2002, p. 257 272 Vol. 1, No. 2 1535-9778/02/$04.00 0 DOI: 10.1128/EC.1.2.257 272.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Mating-Type-Specific
Supplementary Information
 Supplementry Informtion Non-nonil prevents skeletl ging n inflmmtion y inhiiting NF-κB Bo Yu, Ji Chng, Yunsong Liu, Jiong Li, Kreen Kevork, Khli Al Hezimi, Dn T. Grves, No-Hee Prk, Cun-Yu Wng Supplementry
Supplementry Informtion Non-nonil prevents skeletl ging n inflmmtion y inhiiting NF-κB Bo Yu, Ji Chng, Yunsong Liu, Jiong Li, Kreen Kevork, Khli Al Hezimi, Dn T. Grves, No-Hee Prk, Cun-Yu Wng Supplementry
Supplementary Figure S1
 Supplementry Figure S Tissue weights (g).... Liver Hert Brin Pncres Len mss (g) 8 6 -% +% 8 6 Len mss Len mss (g) (% ody weight) Len mss (% ody weight) c Tiilis nterior weight (g).6...... Qudriceps weight
Supplementry Figure S Tissue weights (g).... Liver Hert Brin Pncres Len mss (g) 8 6 -% +% 8 6 Len mss Len mss (g) (% ody weight) Len mss (% ody weight) c Tiilis nterior weight (g).6...... Qudriceps weight
SUPPLEMENTARY INFORMATION
 doi:0.08/nture078 RNse VifHA VifHA βctin 6 Cell lyste IP: ntiha MG VifHA VifHA β ctin 6 7 Cell lyste IP: ntiha Supplementry Figure. Effect of RNse nd MG tretment on the Vif interction., RNse tretment does
doi:0.08/nture078 RNse VifHA VifHA βctin 6 Cell lyste IP: ntiha MG VifHA VifHA β ctin 6 7 Cell lyste IP: ntiha Supplementry Figure. Effect of RNse nd MG tretment on the Vif interction., RNse tretment does
* * * * * liver kidney ileum. Supplementary Fig.S1
 Supplementry Fig.S1 liver kidney ileum Fig.S1. Orlly delivered Fexrmine is intestinlly-restricted Mice received vehicle or Fexrmine (100mg/kg) vi per os (PO) or intrperitonel (IP) injection for 5 dys (n=3/group).
Supplementry Fig.S1 liver kidney ileum Fig.S1. Orlly delivered Fexrmine is intestinlly-restricted Mice received vehicle or Fexrmine (100mg/kg) vi per os (PO) or intrperitonel (IP) injection for 5 dys (n=3/group).
SUPPLEMENTARY INFORMATION
 doi: 1.138/nture862 humn hr. 21q MRPL39 murine Chr.16 Mrpl39 Dyrk1A Runx1 murine Chr. 17 ZNF295 Ets2 Znf295 murine Chr. 1 COL18A1 -/- lot: nti-dscr1 IgG hevy hin DSCR1 DSCR1 expression reltive to hevy
doi: 1.138/nture862 humn hr. 21q MRPL39 murine Chr.16 Mrpl39 Dyrk1A Runx1 murine Chr. 17 ZNF295 Ets2 Znf295 murine Chr. 1 COL18A1 -/- lot: nti-dscr1 IgG hevy hin DSCR1 DSCR1 expression reltive to hevy
Impact of Mating Type, Serotype, and Ploidy on the Virulence of Cryptococcus neoformans
 INFECTION AND IMMUNITY, July 2008, p. 2923 2938 Vol. 76, No. 7 0019-9567/08/$08.00 0 doi:10.1128/iai.00168-08 Copyright 2008, American Society for Microbiology. All Rights Reserved. Impact of Mating Type,
INFECTION AND IMMUNITY, July 2008, p. 2923 2938 Vol. 76, No. 7 0019-9567/08/$08.00 0 doi:10.1128/iai.00168-08 Copyright 2008, American Society for Microbiology. All Rights Reserved. Impact of Mating Type,
Conservation of morphogenesis and virulence in Cryptococcus neoformans. Xiaorong Lin Texas A&M University
 Conservation of morphogenesis and virulence in Cryptococcus neoformans Xiaorong Lin Texas A&M University Cryptococcus infection Lymphatic and hematogenous dissemination Pulmonary infection Cryptococcal
Conservation of morphogenesis and virulence in Cryptococcus neoformans Xiaorong Lin Texas A&M University Cryptococcus infection Lymphatic and hematogenous dissemination Pulmonary infection Cryptococcal
Overview: The Cellular Internet. General Biology. 7. Cell Communication & Signalling
 Course o: BG00 Credits:.00 Generl Biology Overview: The Cellulr Internet Cell-to-cell communiction is bsolutely essentil for multicellulr orgnisms Biologists hve discovered some universl mechnisms of cellulr
Course o: BG00 Credits:.00 Generl Biology Overview: The Cellulr Internet Cell-to-cell communiction is bsolutely essentil for multicellulr orgnisms Biologists hve discovered some universl mechnisms of cellulr
SUPPLEMENTARY INFORMATION
 SUPPEMENTARY INFORMATION DOI: 1.138/ncb956 Norml CIS Invsive crcinom 4 months months b Bldder #1 Bldder # Bldder #3 6 months (Invsive crcinom) Supplementry Figure 1 Mouse model of bldder cncer. () Schemtic
SUPPEMENTARY INFORMATION DOI: 1.138/ncb956 Norml CIS Invsive crcinom 4 months months b Bldder #1 Bldder # Bldder #3 6 months (Invsive crcinom) Supplementry Figure 1 Mouse model of bldder cncer. () Schemtic
Specialization of the HOG pathway and its impact on differentiation and virulence. of Cryptococcus neoformans
 Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans Yong-Sun Bahn, Kaihei Kojima, Gary M. Cox, and Joseph Heitman,,,, Departments of Molecular Genetics
Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans Yong-Sun Bahn, Kaihei Kojima, Gary M. Cox, and Joseph Heitman,,,, Departments of Molecular Genetics
SUPPLEMENTARY INFORMATION
 Prentl doi:.8/nture57 Figure S HPMECs LM Cells Cell lines VEGF (ng/ml) Prentl 7. +/-. LM 7. +/-.99 LM 7. +/-.99 Fold COX induction 5 VEGF: - + + + Bevcizum: - - 5 (µg/ml) Reltive MMP LM mock COX MMP LM+
Prentl doi:.8/nture57 Figure S HPMECs LM Cells Cell lines VEGF (ng/ml) Prentl 7. +/-. LM 7. +/-.99 LM 7. +/-.99 Fold COX induction 5 VEGF: - + + + Bevcizum: - - 5 (µg/ml) Reltive MMP LM mock COX MMP LM+
SUPPLEMENTARY INFORMATION
 % ells with ili (mrke y A-Tu) Reltive Luiferse % ells with ili (mrke y Arl13) % ells with ili DOI: 1.138/n2259 A-Tuulin Hoehst % Cilite Non-ilite -Serum 9% 8% 7% 1 6% % 4% +Serum 1 3% 2% 1% % Serum: -
% ells with ili (mrke y A-Tu) Reltive Luiferse % ells with ili (mrke y Arl13) % ells with ili DOI: 1.138/n2259 A-Tuulin Hoehst % Cilite Non-ilite -Serum 9% 8% 7% 1 6% % 4% +Serum 1 3% 2% 1% % Serum: -
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nture11225 Numer of OTUs sed on 3% distnce Numer of 16s rrna-sed V2-V4 tg sequences LF MF PUFA Supplementry Figure 1. High-ft diets decrese the richness nd diversity
SUPPLEMENTARY INFORMATION doi:10.1038/nture11225 Numer of OTUs sed on 3% distnce Numer of 16s rrna-sed V2-V4 tg sequences LF MF PUFA Supplementry Figure 1. High-ft diets decrese the richness nd diversity
Identification of ENA1 as a Virulence Gene of the Human Pathogenic Fungus Cryptococcus neoformans through Signature-Tagged Insertional Mutagenesis
 EUKARYOTIC CELL, Mar. 2009, p. 315 326 Vol. 8, No. 3 1535-9778/09/$08.00 0 doi:10.1128/ec.00375-08 Copyright 2009, American Society for Microbiology. All Rights Reserved. Identification of ENA1 as a Virulence
EUKARYOTIC CELL, Mar. 2009, p. 315 326 Vol. 8, No. 3 1535-9778/09/$08.00 0 doi:10.1128/ec.00375-08 Copyright 2009, American Society for Microbiology. All Rights Reserved. Identification of ENA1 as a Virulence
Supplementary Fig. 1. Aortic micrornas are differentially expressed in PFM v. GFM.
 Reltive expression 5 1 15 2-5 5 (n = 3) (n = 3) GFM nd PFM PFM GFM mmu-mir-145 mmu-mir-143 mmu-mir-72 mmu-mir-22 mmu-mir-27 mmu-mir-125-5p mmu-mir-23 mmu-mir-29 mmu-mir-126-3p mmu-let-7d mmu-let-7c mmu-mir-199-3p
Reltive expression 5 1 15 2-5 5 (n = 3) (n = 3) GFM nd PFM PFM GFM mmu-mir-145 mmu-mir-143 mmu-mir-72 mmu-mir-22 mmu-mir-27 mmu-mir-125-5p mmu-mir-23 mmu-mir-29 mmu-mir-126-3p mmu-let-7d mmu-let-7c mmu-mir-199-3p
Effect of fungicide timing and wheat varietal resistance on Mycosphaerella graminicola and its sterol 14 α-demethylation-inhibitorresistant
 Effect of fungicide timing nd whet vrietl resistnce on Mycospherell grminicol nd its sterol 14 α-demethyltion-inhiitorresistnt genotypes Didierlurent L., Roisin-Fichter C., Snssené J., Selim S. Pltform
Effect of fungicide timing nd whet vrietl resistnce on Mycospherell grminicol nd its sterol 14 α-demethyltion-inhiitorresistnt genotypes Didierlurent L., Roisin-Fichter C., Snssené J., Selim S. Pltform
Primers used for real time qpcr
 Supplementry Tble 1. Primers used for rel time qpcr Gene Accession number Forwrd/reverse primers Tgfα Tgfβ1 Hgf Cyclin A2 Cyclin B1 Cyclin D1 Cyclin E1 FoxM1 p21 Lrt Cyp26A1 CrbpI Rrβ Bcmo1 Bcmo2 NM_31199
Supplementry Tble 1. Primers used for rel time qpcr Gene Accession number Forwrd/reverse primers Tgfα Tgfβ1 Hgf Cyclin A2 Cyclin B1 Cyclin D1 Cyclin E1 FoxM1 p21 Lrt Cyp26A1 CrbpI Rrβ Bcmo1 Bcmo2 NM_31199
A. Kinoshita 1, L. Locher 2, R. Tienken 3, U. Meyer 3, S. Dänicke 3, J. Rehage 4, K. Huber 5
 Effects of dietry nicin supplementtion on heptic expression of FoxO nd genes involved in glucose production in diry cows during the trnsition period A. Kinoshit, L. Locher, R. Tienken 3, U. Meyer 3, S.
Effects of dietry nicin supplementtion on heptic expression of FoxO nd genes involved in glucose production in diry cows during the trnsition period A. Kinoshit, L. Locher, R. Tienken 3, U. Meyer 3, S.
*** *** *** *** T-cells T-ALL. T-cells T-ALL. T-cells T-ALL. T-cells T-ALL. T-cells T-ALL. Relative ATP content. Relative ATP content RLU RLU
 RLU Events 1 1 1 Luciferin (μm) T-cells T-ALL 1 1 Time (min) T-cells T-ALL 1 1 1 1 DCF-DA Reltive ATP content....1.1.. T-cells T-ALL RLU 1 1 T-cells T-ALL Luciferin (μm) 1 1 Time (min) c d Control e DCFH-DA
RLU Events 1 1 1 Luciferin (μm) T-cells T-ALL 1 1 Time (min) T-cells T-ALL 1 1 1 1 DCF-DA Reltive ATP content....1.1.. T-cells T-ALL RLU 1 1 T-cells T-ALL Luciferin (μm) 1 1 Time (min) c d Control e DCFH-DA
Supplementary Figure S1
 Supplementry Figure S1 d MAP2 GFAP e MAP2 GFAP GFAP c f Clindin GFAP Supplementry Figure S1. Neuronl deth nd ltered strocytes in the rin of n ffected child. Neuron specific MAP2 ntiody stining in the hippocmpus
Supplementry Figure S1 d MAP2 GFAP e MAP2 GFAP GFAP c f Clindin GFAP Supplementry Figure S1. Neuronl deth nd ltered strocytes in the rin of n ffected child. Neuron specific MAP2 ntiody stining in the hippocmpus
Pde1 Phosphodiesterase Modulates Cyclic AMP Levels through a Protein Kinase A-Mediated Negative Feedback Loop in Cryptococcus neoformans
 EUKARYOTIC CELL, Dec. 2005, p. 1971 1981 Vol. 4, No. 12 1535-9778/05/$08.00 0 doi:10.1128/ec.4.12.1971 1981.2005 Copyright 2005, American Society for Microbiology. All Rights Reserved. Pde1 Phosphodiesterase
EUKARYOTIC CELL, Dec. 2005, p. 1971 1981 Vol. 4, No. 12 1535-9778/05/$08.00 0 doi:10.1128/ec.4.12.1971 1981.2005 Copyright 2005, American Society for Microbiology. All Rights Reserved. Pde1 Phosphodiesterase
Supplementary Information. SAMHD1 Restricts HIV-1 Infection in Resting CD4 + T Cells
 Supplementry Informtion SAMHD Restricts HIV- Infection in Resting CD T Cells Hnn-Mri Blduf,2,, Xioyu Pn,, Elin Erikson,2, Srh Schmidt, Wqo Dddch 3, Mnj Burggrf, Kristin Schenkov, In Amiel,2, Guido Wnitz
Supplementry Informtion SAMHD Restricts HIV- Infection in Resting CD T Cells Hnn-Mri Blduf,2,, Xioyu Pn,, Elin Erikson,2, Srh Schmidt, Wqo Dddch 3, Mnj Burggrf, Kristin Schenkov, In Amiel,2, Guido Wnitz
Heparanase promotes tumor infiltration and antitumor activity of CAR-redirected T- lymphocytes
 Supporting Online Mteril for Heprnse promotes tumor infiltrtion nd ntitumor ctivity of -redirected T- lymphocytes IgnzioCrun, Brr Svoldo, VlentinHoyos, Gerrit Weer, Ho Liu, Eugene S. Kim, Michel M. Ittmnn,
Supporting Online Mteril for Heprnse promotes tumor infiltrtion nd ntitumor ctivity of -redirected T- lymphocytes IgnzioCrun, Brr Svoldo, VlentinHoyos, Gerrit Weer, Ho Liu, Eugene S. Kim, Michel M. Ittmnn,
SUPPLEMENTARY INFORMATION
 DOI: 1.13/n7 Reltive Pprg mrna 3 1 1 Time (weeks) Interspulr Inguinl Epididyml Reltive undne..1.5. - 5 5-51 51-1 1-7 7 - - 1 1-1 Lipid droplet size ( m ) 1-3 3 - - - 1 1-1 1-1 1-175 175-3 3-31 31-5 >5
DOI: 1.13/n7 Reltive Pprg mrna 3 1 1 Time (weeks) Interspulr Inguinl Epididyml Reltive undne..1.5. - 5 5-51 51-1 1-7 7 - - 1 1-1 Lipid droplet size ( m ) 1-3 3 - - - 1 1-1 1-1 1-175 175-3 3-31 31-5 >5
FoxP3 + regulatory CD4 T cells control the generation of functional CD8 memory
 Received Fe Accepted 6 Jul Pulished 7 Aug DOI:.8/ncomms99 FoxP + regultory CD T cells control the genertion of functionl memory M.G. de Goër de Herve,,, S. Jfour,,, M. Vllée, & Y. Toufik, During the primry
Received Fe Accepted 6 Jul Pulished 7 Aug DOI:.8/ncomms99 FoxP + regultory CD T cells control the genertion of functionl memory M.G. de Goër de Herve,,, S. Jfour,,, M. Vllée, & Y. Toufik, During the primry
Laboratory of Clinical Investigation, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD 20892
 The second STE12 homologue of Cryptococcus neoformans is MATa-specific and plays an important role in virulence Y. C. Chang, L. A. Penoyer, and K. J. Kwon-Chung* Laboratory of Clinical Investigation, National
The second STE12 homologue of Cryptococcus neoformans is MATa-specific and plays an important role in virulence Y. C. Chang, L. A. Penoyer, and K. J. Kwon-Chung* Laboratory of Clinical Investigation, National
Supplementary Figure 1
 doi: 1.138/nture6188 SUPPLEMENTARY INFORMATION Supplementry Figure 1 c CFU-F colonies per 1 5 stroml cells 14 12 1 8 6 4 2 Mtrigel plug Neg. MCF7/Rs MDA-MB-231 * * MCF7/Rs-Lung MDA-MB-231-Lung MCF7/Rs-Kidney
doi: 1.138/nture6188 SUPPLEMENTARY INFORMATION Supplementry Figure 1 c CFU-F colonies per 1 5 stroml cells 14 12 1 8 6 4 2 Mtrigel plug Neg. MCF7/Rs MDA-MB-231 * * MCF7/Rs-Lung MDA-MB-231-Lung MCF7/Rs-Kidney
TO produce organs predictably, meristems tightly
 Copyright Ó 2009 by the Genetics Society of Americ DOI: 10.1534/genetics.108.098350 Note The Interction of knotted1 nd thick tssel dwrf1 in Vegettive nd Reproductive Meristems of Mize Chin Lunde nd Srh
Copyright Ó 2009 by the Genetics Society of Americ DOI: 10.1534/genetics.108.098350 Note The Interction of knotted1 nd thick tssel dwrf1 in Vegettive nd Reproductive Meristems of Mize Chin Lunde nd Srh
Serotype AD Strains of Cryptococcus neoformans Are Diploid or Aneuploid and Are Heterozygous at the Mating-Type Locus
 INFECTION AND IMMUNITY, Jan. 2001, p. 115 122 Vol. 69, No. 1 0019-9567/01/$04.00 0 DOI: 10.1128/IAI.69.1.115 122.2001 Copyright 2001, American Society for Microbiology. All Rights Reserved. Serotype AD
INFECTION AND IMMUNITY, Jan. 2001, p. 115 122 Vol. 69, No. 1 0019-9567/01/$04.00 0 DOI: 10.1128/IAI.69.1.115 122.2001 Copyright 2001, American Society for Microbiology. All Rights Reserved. Serotype AD
SUPPLEMENTARY INFORMATION
 X p -lu c ct ivi ty doi:.8/nture8 S CsA - THA + DAPI Merge FSK THA TUN Supplementry Figure : A. Ad-Xp luc ctivity in primry heptocytes exposed to FSK, THA, or TUN s indicted. Luciferse ctivity normlized
X p -lu c ct ivi ty doi:.8/nture8 S CsA - THA + DAPI Merge FSK THA TUN Supplementry Figure : A. Ad-Xp luc ctivity in primry heptocytes exposed to FSK, THA, or TUN s indicted. Luciferse ctivity normlized
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nture1188 1mM CCl 2 (min) 3 4 6 CCl 2 (mm) for 4min.1. 1 (mm) Pro- d WT GdCl 3 R-68 -/- P2x7r -/- -/- Csp1 -/- WT -/- P2x7r -/- -/- Csp1 -/- Csp1 (p2) (p17) Pro-Csp1
SUPPLEMENTARY INFORMATION doi:1.138/nture1188 1mM CCl 2 (min) 3 4 6 CCl 2 (mm) for 4min.1. 1 (mm) Pro- d WT GdCl 3 R-68 -/- P2x7r -/- -/- Csp1 -/- WT -/- P2x7r -/- -/- Csp1 -/- Csp1 (p2) (p17) Pro-Csp1
Supporting Information
 Supporting Informtion Ethylbromoisobutyrte (EBIB), tin (II) 2-ethylhexnote (Sn(EH) 2 ), copper (II) bromide (CuBr 2 ), toluene, sodium zide, dimethylformmide (DMF), nd N, N, N, N, N pentmethyltriethylenedimine
Supporting Informtion Ethylbromoisobutyrte (EBIB), tin (II) 2-ethylhexnote (Sn(EH) 2 ), copper (II) bromide (CuBr 2 ), toluene, sodium zide, dimethylformmide (DMF), nd N, N, N, N, N pentmethyltriethylenedimine
The β-globin nuclear compartment in development and erythroid differentiation
 The β-gloin nucler comprtment in development nd erythroid differentition Roert-Jn Plstr 1,2, Bs Tolhuis 1,2, Erik Splinter 1, Rin Nijmeijer 1, Frnk Grosveld 1 & Wouter de Lt 1 Efficient trnscription of
The β-gloin nucler comprtment in development nd erythroid differentition Roert-Jn Plstr 1,2, Bs Tolhuis 1,2, Erik Splinter 1, Rin Nijmeijer 1, Frnk Grosveld 1 & Wouter de Lt 1 Efficient trnscription of
SUPPLEMENTARY INFORMATION
 doi:.3/nture93 d 5 Rttlesnke DRG (reds) Rttlesnke TG (reds) c 3 TRPV1 other TRPs 1 1 3 Non-pit snke TG (reds) SFig. 1 5 5 3 other TRPs TRPV1 1 1 3 Non-pit snke DRG (reds) 5 Antomy of the pit orgn nd comprison
doi:.3/nture93 d 5 Rttlesnke DRG (reds) Rttlesnke TG (reds) c 3 TRPV1 other TRPs 1 1 3 Non-pit snke TG (reds) SFig. 1 5 5 3 other TRPs TRPV1 1 1 3 Non-pit snke DRG (reds) 5 Antomy of the pit orgn nd comprison
Supplementary Figure S1
 Supplementry Figure S Connexin4 TroponinI Merge Plsm memrne Met Intrcellulr Met Supplementry Figure S H9c rt crdiomyolsts cell line. () Immunofluorescence of crdic mrkers: Connexin4 (green) nd TroponinI
Supplementry Figure S Connexin4 TroponinI Merge Plsm memrne Met Intrcellulr Met Supplementry Figure S H9c rt crdiomyolsts cell line. () Immunofluorescence of crdic mrkers: Connexin4 (green) nd TroponinI
Study on the association between PI3K/AKT/mTOR signaling pathway gene polymorphism and susceptibility to gastric
 JBUON 2017; 22(6): 1488-1493 ISSN: 1107-0625, online ISSN: 2241-6293 www.jbuon.com E-mil: editoril_office@jbuon.com ORIGINAL ARTICLE Study on the ssocition between PI3K/AKT/mTOR signling pthwy gene polymorphism
JBUON 2017; 22(6): 1488-1493 ISSN: 1107-0625, online ISSN: 2241-6293 www.jbuon.com E-mil: editoril_office@jbuon.com ORIGINAL ARTICLE Study on the ssocition between PI3K/AKT/mTOR signling pthwy gene polymorphism
DR. MARC PAGÈS Project Manager R&D Biologicals - Coccidia Projects, HIPRA
 DR. MARC PAGÈS Project Mnger R&D Biologicls - Coccidi Projects, HIPRA Dr. Mrc Pgès Bosch otined Microiology nd Genetics degree t the University of Brcelon in 1998. He otined his PhD working on the synptoneml
DR. MARC PAGÈS Project Mnger R&D Biologicls - Coccidi Projects, HIPRA Dr. Mrc Pgès Bosch otined Microiology nd Genetics degree t the University of Brcelon in 1998. He otined his PhD working on the synptoneml
Pheromones Stimulate Mating and Differentiation via Paracrine and Autocrine Signaling in Cryptococcus neoformans
 EUKARYOTIC CELL, June 2002, p. 366 377 Vol. 1, No. 3 1535-9778/02/$04.00 0 DOI: 10.1128/EC.1.3.366 377.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Pheromones Stimulate
EUKARYOTIC CELL, June 2002, p. 366 377 Vol. 1, No. 3 1535-9778/02/$04.00 0 DOI: 10.1128/EC.1.3.366 377.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Pheromones Stimulate
SUPPLEMENTARY INFORMATION
 DOI: 1.138/nc286 Figure S1 e f Medium DMSO AktVIII PP242 Rp S6K1-I Gr1 + + + + + + Strvtion + + + + + IB: Akt-pT38 IB: Akt K-pT389 K IB: Rptor Gr1 shs6k1-a shs6k1-b shs6k1-c shrictor shrptor Gr1 c IB:
DOI: 1.138/nc286 Figure S1 e f Medium DMSO AktVIII PP242 Rp S6K1-I Gr1 + + + + + + Strvtion + + + + + IB: Akt-pT38 IB: Akt K-pT389 K IB: Rptor Gr1 shs6k1-a shs6k1-b shs6k1-c shrictor shrptor Gr1 c IB:
Type II monocytes modulate T cell-mediated central nervous system autoimmunity
 Type II monocytes modulte T cell-medited centrl nervous system utoimmunity Mrtin S. Weer, Thoms Prod homme, Swsn Youssef, Shnnon E. Dunn, Cynthi D. Rundle, Lind Lee, Jun C. Ptrroyo, Olf Stüve, Rymond A.
Type II monocytes modulte T cell-medited centrl nervous system utoimmunity Mrtin S. Weer, Thoms Prod homme, Swsn Youssef, Shnnon E. Dunn, Cynthi D. Rundle, Lind Lee, Jun C. Ptrroyo, Olf Stüve, Rymond A.
Supporting information
 Supporting informtion Multiple Univrite Dt Anlysis Revels the Inulin Effects on the High-ft-diet Induced Metolic Altertions in Rt Myocrdium nd Testicles in the Pre-oesity Stte Yixun Dun #, Ynpeng An #,
Supporting informtion Multiple Univrite Dt Anlysis Revels the Inulin Effects on the High-ft-diet Induced Metolic Altertions in Rt Myocrdium nd Testicles in the Pre-oesity Stte Yixun Dun #, Ynpeng An #,
Increased expression of receptor for advanced glycation end-products worsens focal brain ischemia in diabetic rats*
 NEURAL REGENERATION RESEARCH Volume 7, Issue 13, My 2012 www.nrronline.org Cite this rticle s: Neurl Regen Res. 2012;7(13):1000-1005. Incresed expression of receptor for dvnced glyction end-products worsens
NEURAL REGENERATION RESEARCH Volume 7, Issue 13, My 2012 www.nrronline.org Cite this rticle s: Neurl Regen Res. 2012;7(13):1000-1005. Incresed expression of receptor for dvnced glyction end-products worsens
Patterns of Proviral Insertion and Deletion in Avian Leukosis Virus- Induced Lymphomas
 JOURNAL OF VIROLOGY, Jn. 1986, p. 28-36 0022-538X/86/010028-09$02.00/0 Copyright C 1986, Americn Society for Microbiology Vol. 57, No. 1 Ptterns of Provirl Insertion nd Deletion in Avin Leukosis Virus-
JOURNAL OF VIROLOGY, Jn. 1986, p. 28-36 0022-538X/86/010028-09$02.00/0 Copyright C 1986, Americn Society for Microbiology Vol. 57, No. 1 Ptterns of Provirl Insertion nd Deletion in Avin Leukosis Virus-
Received 27 June 2003/Accepted 26 July 2003
 EUKARYOTIC CELL, Oct. 2003, p. 1036 1045 Vol. 2, No. 5 1535-9778/03/$08.00 0 DOI: 10.1128/EC.2.5.1036 1045.2003 Copyright 2003, American Society for Microbiology. All Rights Reserved. Recapitulation of
EUKARYOTIC CELL, Oct. 2003, p. 1036 1045 Vol. 2, No. 5 1535-9778/03/$08.00 0 DOI: 10.1128/EC.2.5.1036 1045.2003 Copyright 2003, American Society for Microbiology. All Rights Reserved. Recapitulation of
Arabidopsis downy mildew effector HaRxL106 suppresses plant immunity by binding to RADICAL-INDUCED CELL DEATH1
 Reserch Arbidopsis downy mildew effector HRxL106 suppresses plnt immunity by binding to RADICAL-INDUCED CELL DEATH1 Lennrt Wirthmueller 1,2, Shut Asi 1, Ghnsym Rllplli 1, Jn Sklenr 1, Georgin Fbro 1, De
Reserch Arbidopsis downy mildew effector HRxL106 suppresses plnt immunity by binding to RADICAL-INDUCED CELL DEATH1 Lennrt Wirthmueller 1,2, Shut Asi 1, Ghnsym Rllplli 1, Jn Sklenr 1, Georgin Fbro 1, De
A rt i c l e s. a Events (% of max)
 Continuous requirement for the TCR in regultory T cell function Andrew G Levine 1,, Aron Arvey 1,,4, Wei Jin 1,,4 & Alexnder Y Rudensky 1 3 14 Nture Americ, Inc. All rights reserved. Foxp3 + regultory
Continuous requirement for the TCR in regultory T cell function Andrew G Levine 1,, Aron Arvey 1,,4, Wei Jin 1,,4 & Alexnder Y Rudensky 1 3 14 Nture Americ, Inc. All rights reserved. Foxp3 + regultory
T cell intrinsic role of Nod2 in promoting type 1 immunity to Toxoplasma gondii
 T cell intrinsic role of Nod in promoting type 1 immunity to Toxoplsm gondii Michel H Shw 1, Thornik Reimer 1, Crmen Sánchez-Vldepeñs, Neil Wrner 1, Yun-Gi Kim 1, Mnuel Fresno & Griel Nuñez 1 Nod elongs
T cell intrinsic role of Nod in promoting type 1 immunity to Toxoplsm gondii Michel H Shw 1, Thornik Reimer 1, Crmen Sánchez-Vldepeñs, Neil Wrner 1, Yun-Gi Kim 1, Mnuel Fresno & Griel Nuñez 1 Nod elongs
Feeding state and age dependent changes in melaninconcentrating hormone expression in the hypothalamus of broiler chickens
 Supplementry Mterils Epub: No 2017_23 Vol. 65, 2018 https://doi.org/10.183/bp.2017_23 Regulr pper Feeding stte nd ge dependent chnges in melninconcentrting hormone expression in the hypothlmus of broiler
Supplementry Mterils Epub: No 2017_23 Vol. 65, 2018 https://doi.org/10.183/bp.2017_23 Regulr pper Feeding stte nd ge dependent chnges in melninconcentrting hormone expression in the hypothlmus of broiler
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/nc2824 Hcn4 Tx5 Mlc2 c Hcn4- ISH d Tx5- ISH e Mlc2-ISH Hcn4-ISH f e Tx5-ISH f -ISH Figure S1 Section in situ hyridistion nlysis of crescent stge mouse emryos (E7.5). () More nterior section
DOI: 10.1038/nc2824 Hcn4 Tx5 Mlc2 c Hcn4- ISH d Tx5- ISH e Mlc2-ISH Hcn4-ISH f e Tx5-ISH f -ISH Figure S1 Section in situ hyridistion nlysis of crescent stge mouse emryos (E7.5). () More nterior section
Transcription factor Foxo3 controls the magnitude of T cell immune responses by modulating the function of dendritic cells
 Trnscription fctor Foxo3 controls the mgnitude of T cell immune responses by modulting the function of dendritic cells 9 Nture Americ, Inc. All rights reserved. Anne S Dejen, Dniel R Beisner,4, Irene L
Trnscription fctor Foxo3 controls the mgnitude of T cell immune responses by modulting the function of dendritic cells 9 Nture Americ, Inc. All rights reserved. Anne S Dejen, Dniel R Beisner,4, Irene L
Pleiotropic roles of the Msi1-like protein, Msl1, in Cryptococcus neoformans
 EC Accepts, published online ahead of print on 5 October 2012 Eukaryotic Cell doi:10.1128/ec.00261-12 Copyright 2012, American Society for Microbiology. All Rights Reserved. 1 2 3 4 5 6 7 8 9 10 11 12
EC Accepts, published online ahead of print on 5 October 2012 Eukaryotic Cell doi:10.1128/ec.00261-12 Copyright 2012, American Society for Microbiology. All Rights Reserved. 1 2 3 4 5 6 7 8 9 10 11 12
Defective Wnt-dependent cerebellar midline fusion in a mouse model of Joubert syndrome
 correction notice Nt. Med. 17, 726 731 (2011) Defective Wnt-dependent cereellr midline fusion in mouse model of Jouert syndrome Mdeline A Lncster, Dipik J Gopl, Joon Kim, Shr N Sleem, Jennifer L Silhvy,
correction notice Nt. Med. 17, 726 731 (2011) Defective Wnt-dependent cereellr midline fusion in mouse model of Jouert syndrome Mdeline A Lncster, Dipik J Gopl, Joon Kim, Shr N Sleem, Jennifer L Silhvy,
THE EFFECT OF DIFFERENT STIMULI ON MEAGRE (Argyrosomus regius) FEEDING BEHAVIOUR.
 THE EFFECT OF DIFFERENT STIMULI ON MEGRE (rgyrosomus regius) FEEDING EHVIOUR. Ionnis E. Ppdkis, Nikos Ppndroulkis, lkioni Sfendourki, Veronic Cmporesi 3, Mnolis Vsilkis, Constntinos C. Mylons Institute
THE EFFECT OF DIFFERENT STIMULI ON MEGRE (rgyrosomus regius) FEEDING EHVIOUR. Ionnis E. Ppdkis, Nikos Ppndroulkis, lkioni Sfendourki, Veronic Cmporesi 3, Mnolis Vsilkis, Constntinos C. Mylons Institute
SUPPLEMENTARY INFORMATION
 TM TM tip link horizontl top connectors 1 leucine-rich (21 %) otoncorin-like 1809 ntigenic peptides B D signl peptide hydrophoic segment proline/threonine-rich (79 %) Supplementry Figure 1. () The outer
TM TM tip link horizontl top connectors 1 leucine-rich (21 %) otoncorin-like 1809 ntigenic peptides B D signl peptide hydrophoic segment proline/threonine-rich (79 %) Supplementry Figure 1. () The outer
Critical role of c-kit in beta cell function: increased insulin secretion and protection against diabetes in a mouse model
 Dietologi (1) 55:14 5 DOI 1.17/s15-1-5-5 ARTICLE Criticl role of c-kit in et cell function: incresed insulin secretion nd protection ginst dietes in mouse model Z. C. Feng & J. Li & B. A. Turco & M. Riopel
Dietologi (1) 55:14 5 DOI 1.17/s15-1-5-5 ARTICLE Criticl role of c-kit in et cell function: incresed insulin secretion nd protection ginst dietes in mouse model Z. C. Feng & J. Li & B. A. Turco & M. Riopel
Supplementary Information. Title: RAS-MAPK dependence underlies a rational polytherapy strategy in EML4-
 Supplementry Informtion Title: RAS-MAPK dependence underlies rtionl polytherpy strtegy in EML4- ALK positive lung cncer Authors: Gorjn Hrustnovic, Victor Olivs, Evngelos Pzrentzos, Asmin Tulpule, Surh
Supplementry Informtion Title: RAS-MAPK dependence underlies rtionl polytherpy strtegy in EML4- ALK positive lung cncer Authors: Gorjn Hrustnovic, Victor Olivs, Evngelos Pzrentzos, Asmin Tulpule, Surh
Expression of Three Cell Cycle Inhibitors during Development of Adipose Tissue
 Expression of Three Cell Cycle Inhiitors during Development of Adipose Tissue Jiin Zhng Deprtment of Animl Sciences Advisor: Michel E. Dvis Co-dvisor: Kichoon Lee Development of niml dipose tissue Hypertrophy
Expression of Three Cell Cycle Inhiitors during Development of Adipose Tissue Jiin Zhng Deprtment of Animl Sciences Advisor: Michel E. Dvis Co-dvisor: Kichoon Lee Development of niml dipose tissue Hypertrophy
Supplementary Figure 1
 Supplementry Figure 1 c d Wistr SHR Wistr AF-353 SHR AF-353 n = 6 n = 6 n = 28 n = 3 n = 12 n = 12 Supplementry Figure 1 Neurophysiologicl properties of petrosl chemoreceptive neurones in Wistr nd SH rts.
Supplementry Figure 1 c d Wistr SHR Wistr AF-353 SHR AF-353 n = 6 n = 6 n = 28 n = 3 n = 12 n = 12 Supplementry Figure 1 Neurophysiologicl properties of petrosl chemoreceptive neurones in Wistr nd SH rts.
AMPK maintains energy homeostasis and survival in cancer cells via. regulating p38/pgc-1α-mediated mitochondrial biogenesis
 SUPPLEMENTARY INFORMATION AMPK mintins energy homeostsis nd survivl in cncer cells vi regulting p38/pgc-1α-medited mitochondril iogenesis Blkrishn Chue 1, Prmnnd Mlvi 1, Shivendr Vikrm Singh 1, Noshd Mohmmd
SUPPLEMENTARY INFORMATION AMPK mintins energy homeostsis nd survivl in cncer cells vi regulting p38/pgc-1α-medited mitochondril iogenesis Blkrishn Chue 1, Prmnnd Mlvi 1, Shivendr Vikrm Singh 1, Noshd Mohmmd
Journal of Hainan Medical University.
 132 Journl of Hinn Medicl University 2017; 23(11): 132-136 Journl of Hinn Medicl University http://www.hnykdxxb.com Assessment of the efficcy nd sfety of bronchil rtery perfusion chemotherpy combined with
132 Journl of Hinn Medicl University 2017; 23(11): 132-136 Journl of Hinn Medicl University http://www.hnykdxxb.com Assessment of the efficcy nd sfety of bronchil rtery perfusion chemotherpy combined with
Goal: Evaluate plant health effects while suppressing dollar spot and brown patch
 Newer Fungicide Products Alone nd In Rottion on Chicgo Golf Green Reserchers: Chicgo District Golf Assoc. Derek Settle, Tim Sibicky, nd Nick DeVries Gol: Evlute plnt helth effects while suppressing dollr
Newer Fungicide Products Alone nd In Rottion on Chicgo Golf Green Reserchers: Chicgo District Golf Assoc. Derek Settle, Tim Sibicky, nd Nick DeVries Gol: Evlute plnt helth effects while suppressing dollr
Cyanidin-3-O-glucoside ameliorates lipid and glucose accumulation in C57BL/6J mice via activation of PPAR-α and AMPK
 3 rd Interntionl Conference nd Exhiition on Nutrition & Food Sciences Septemer 23-25, 214 Vlenci, Spin Cynidin-3-O-glucoside meliortes lipid nd glucose ccumultion in C57BL/6J mice vi ctivtion of PPAR-α
3 rd Interntionl Conference nd Exhiition on Nutrition & Food Sciences Septemer 23-25, 214 Vlenci, Spin Cynidin-3-O-glucoside meliortes lipid nd glucose ccumultion in C57BL/6J mice vi ctivtion of PPAR-α
Northern blot analysis
 Northern blot nlysis RNA SCD RNA SCD FAS C c-9 t-1 C c-9 t-1 PRE PI PDMI PRE PI PDMI PRE PDMI PIM An W c-9, t-11 t-1, c-12 C 5 2 4 1 um C 5 2 4 1 um Angus dipocytes expressed SCD higher thn Wgyu dipocyte
Northern blot nlysis RNA SCD RNA SCD FAS C c-9 t-1 C c-9 t-1 PRE PI PDMI PRE PI PDMI PRE PDMI PIM An W c-9, t-11 t-1, c-12 C 5 2 4 1 um C 5 2 4 1 um Angus dipocytes expressed SCD higher thn Wgyu dipocyte
PDGF-BB secreted by preosteoclasts induces angiogenesis during coupling with osteogenesis
 Supplementry Informtion PDGF-BB secreted y preosteoclsts induces ngiogenesis during coupling with osteogenesis Hui Xie, Zhung Cui, Long Wng, Zhuying Xi, Yin Hu, Lingling Xin, Chngjun Li, Ling Xie, Jnet
Supplementry Informtion PDGF-BB secreted y preosteoclsts induces ngiogenesis during coupling with osteogenesis Hui Xie, Zhung Cui, Long Wng, Zhuying Xi, Yin Hu, Lingling Xin, Chngjun Li, Ling Xie, Jnet
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Stelter et al., http://www.jcb.org/cgi/content/full/jcb.201105042/dc1 S1 Figure S1. Dyn2 recruitment to edid-labeled FG domain Nups.
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Stelter et al., http://www.jcb.org/cgi/content/full/jcb.201105042/dc1 S1 Figure S1. Dyn2 recruitment to edid-labeled FG domain Nups.
Single-Molecule Studies of Unlabelled Full-Length p53 Protein Binding to DNA
 Single-Molecule Studies of Unlbelled Full-Length p53 Protein Binding to DNA Philipp Nuttll, 1 Kidn Lee, 2 Pietro Ciccrell, 3 Mrco Crminti, 3 Giorgio Ferrri, 3 Ki- Bum Kim, 2 Tim Albrecht 1* 1 Imperil College
Single-Molecule Studies of Unlbelled Full-Length p53 Protein Binding to DNA Philipp Nuttll, 1 Kidn Lee, 2 Pietro Ciccrell, 3 Mrco Crminti, 3 Giorgio Ferrri, 3 Ki- Bum Kim, 2 Tim Albrecht 1* 1 Imperil College
Supplementary Figure 1
 Supplementry Figure 1 Ncor1 Expression 2. 1.5 1..5. Muscle Liver Lmi Ppropi Kindey Pncres Lung Testis Bone Mrrow Thymus Spleen Peripherl Lymph nods Smll Intestine Ncor1 Expression 1.5 1..5. DN DP SP CD8
Supplementry Figure 1 Ncor1 Expression 2. 1.5 1..5. Muscle Liver Lmi Ppropi Kindey Pncres Lung Testis Bone Mrrow Thymus Spleen Peripherl Lymph nods Smll Intestine Ncor1 Expression 1.5 1..5. DN DP SP CD8
Transcriptional reprogramming of mature CD4 + helper T cells generates distinct MHC class II restricted cytotoxic T lymphocytes
 Trnscriptionl reprogrmming of mture + helper T cells genertes distinct MHC clss II restricted cytotoxic T lymphocytes npg 213 Nture Americ, Inc. All rights reserved. Dniel Mucid 1,8,9, Mohmmd Mushtq Husin
Trnscriptionl reprogrmming of mture + helper T cells genertes distinct MHC clss II restricted cytotoxic T lymphocytes npg 213 Nture Americ, Inc. All rights reserved. Dniel Mucid 1,8,9, Mohmmd Mushtq Husin
TLR7 induces anergy in human CD4 + T cells
 TLR7 induces nergy in humn CD T cells Mrgrit Dominguez-Villr 1, Anne-Sophie Gutron 1, Mrine de Mrcken 1, Mrl J Keller & Dvid A Hfler 1 The recognition of microil ptterns y Toll-like receptors (TLRs) is
TLR7 induces nergy in humn CD T cells Mrgrit Dominguez-Villr 1, Anne-Sophie Gutron 1, Mrine de Mrcken 1, Mrl J Keller & Dvid A Hfler 1 The recognition of microil ptterns y Toll-like receptors (TLRs) is
SUPPLEMENTARY INFORMATION. Cytochrome P450-2E1 promotes fast food-mediated hepatic fibrosis
 SUPPLEMENTARY INRMATION Cytochrome P-E1 promotes fst food-medited heptic fibrosis Mohmed A. Abdelmegeed, Youngshim Choi, Grzegorz Godlewski b, Seung-Kwon H, Atryee Bnerjee, Sehwn Jng, nd Byoung-Joon Song
SUPPLEMENTARY INRMATION Cytochrome P-E1 promotes fst food-medited heptic fibrosis Mohmed A. Abdelmegeed, Youngshim Choi, Grzegorz Godlewski b, Seung-Kwon H, Atryee Bnerjee, Sehwn Jng, nd Byoung-Joon Song
The effect of encapsulated butyric acid and zinc on performance, gut integrity and meat quality in male broiler chickens 1
 The effect of encpsulted utyric cid nd zinc on performnce, gut integrity nd met qulity in mle roiler chickens 1 Astrct This study evluted the impct of encpsulted utyric cid nd zinc (ButiPEARL Z) on performnce
The effect of encpsulted utyric cid nd zinc on performnce, gut integrity nd met qulity in mle roiler chickens 1 Astrct This study evluted the impct of encpsulted utyric cid nd zinc (ButiPEARL Z) on performnce
R-α-Lipoic acid and acetyl-l-carnitine complementarily promote mitochondrial biogenesis in murine 3T3-L1 adipocytes
 Dietologi (28) 51:165 174 DOI 1.17/s125-7-852-4 ARTICLE R-α-Lipoic cid nd cetyl-l-crnitine complementrily promote mitochondril iogenesis in murine 3T3-L1 dipocytes W. Shen & K. Liu & C. Tin & L. Yng &
Dietologi (28) 51:165 174 DOI 1.17/s125-7-852-4 ARTICLE R-α-Lipoic cid nd cetyl-l-crnitine complementrily promote mitochondril iogenesis in murine 3T3-L1 dipocytes W. Shen & K. Liu & C. Tin & L. Yng &
N α -Acetylation of yeast ribosomal proteins and its effect on protein synthesis
 JOURNAL OF PROTEOMICS 74 (2011) 431 441 available at www.sciencedirect.com www.elsevier.com/locate/jprot N α -Acetylation of yeast ribosomal proteins and its effect on protein synthesis Masahiro Kamita
JOURNAL OF PROTEOMICS 74 (2011) 431 441 available at www.sciencedirect.com www.elsevier.com/locate/jprot N α -Acetylation of yeast ribosomal proteins and its effect on protein synthesis Masahiro Kamita
