Supplementary Figure 1
|
|
|
- Stephanie Blake
- 5 years ago
- Views:
Transcription
1 Count Count Supplementary Figure 1 Coverage per amplicon for error-corrected sequencing experiments. Errorcorrected consensus sequence (ECCS) coverage was calculated for each of the 568 amplicons in the capture panel. a, Histogram of ECCS coverage for all amplicons. b, Histogram of ECCS coverage in amplicons in which a variant was detected. a 2 Coverage Per Amplicon b Coverage per Amplicon Coverage per Amplicon with Called Variant Coverage per Amplicon
2 Tareget Space (bp) Target Space (bp) Supplementary Figure 2 Number of mutations detected per gene in exons (top panel) and introns (bottom panel) relative to the capture space (bp = base pairs) targeting that gene in the panel. 125 Detected Exonic SNVs Relative to Capture Space per Gene CUX KDM6A STAG2 BCOR BCORL1 TET2 ATRX DNMT3A 5 25 ASXL1 RAD21 NOTCH1 TP53 IKZF1 ETV6 SMC3 MYD88 CBL KRAS Detected SNVs 125 Detected Intronic SNVs Relative to Capture Space per Gene CUX1 1 TET2 STAG2 KDM6A 75 ATRX DNMT3A 5 EZH2 25 IKZF1 ETV6 FBXW7 MYD88 KRAS 2 4 Detected SNVs 6
3 probe fluorescence (A.U.) Supplementary Figure 3 Representative droplet digital PCR (ddpcr) results from NHS participant 12 for the detected DNMT3A G543A clonal variant. The wild-type probe intensity in arbitrary units (A.U.) was plotted relative to the DNMT3A G543A (mutant) probe intensity for each droplet. a-d, Variant quantification at the first time point for a, all cells; b, B lymphocytes; c, T lymphocytes; and d, myeloid cells. e-h, Variant quantification at the second time point for e, all cells; f, B lymphocytes; g, T lymphocytes; and h, myeloid cells. i, The DNMT3A G543A variant was not detected in the negative control sample from participant 2, time point 2. j, DNMT3A G543A positive (or empty) droplets were detected in the gblock positive control. a 11,79 e 92,45 i 96, , , , b 885 f 2,3 j , , ,75 c ,196 g , , ,864 d ,24 h , , , probe fluorescence (A.U.)
4 VAF by ddpcr Supplementary Figure 4. Several mutations identified by error-corrected sequencing (ECS) were verified using droplet digital PCR (ddpcr). The variant allele fractions (VAFs) identified ECS and ddpcr were highly correlated (R 2 =.98) VAF by ECS
5 Supplementary Figure 5 Heat map depicting the number of exonic single nucleotide variants (SNVs) detected in each gene per study participant. Gene DNMT3A TET2 STAG2 BCORL1 TP53 KRAS ASXL1 SMC3 RAD21 CBL BCOR NOTCH1 MYD88 KDM6A IKZF1 ETV6 CUX1 ATRX Detected Clonal Exonic SNVs per Individual SNVs Participant ID
6 K67E S129X Q231X Q249X E283X V296M R31W G32C W314X R32X S337X R379H E482X C494F G498V L58P C537F G543S G543A A572D R635W R635Q L637P L639F E667X T691I I75T S714C F731S F732S H739P A741E R749H S775_E2splice P799S H821R T834A T835A S839P W86R R882H R899G P94L Supplementary Figure 6 Detected exonic clonal single nucleotide variants (SNVs) in DNMT3A. The detected SNVs were predominantly nonsense mutations (blue) in the first half of the gene or missense mutations (red) in the three functional domains a proline-tryptophan-tryptophan-proline (PWWP) chromatin targeting domain, a zinc finger nuclease (ZFN) domain and a S-adenosylmethionine (SAM) dependent methyltransferase (MTase) domain. Exons AA Residue DNMT3A PWWP domain ZFN domain SAM-dependent MTase Missense Nonsense Splice
7 L446X W954X E1137_E3splice R1216G A1217A G1288S F139L Y1337X R1359H S1486X E1513_E1splice Y1589X T1793I Supplementary Figure 7 Detected exonic clonal single nucleotide variants in TET2. Exons AA Residue TET2 Oxygenase domain Silent Missense Nonsense Splice
8 DNMT3A KDM6A ATRX STAG2 CUX1 ETV6 EZH2 FBXW7 IKZF1 KRAS MYD88 TET2 Detected Clonal Intronic SNVs Supplementary Figure 8 Number of intronic clonal single nucleotide variants (SNVs) detected by gene. 6 Detected Intronic SNVs by Gene 4 2 Gene
9 Gene Supplementary Figure 9 Heat map depicting the number of intronic single nucleotide variants (SNVs) detected in each gene per individual. DNMT3A KDM6A STAG2 Detected Clonal Intronic SNVs per Individual ATRX TET2 MYD88 KRAS IKZF1 FBXW7 EZH2 ETV6 CUX1 Count Participant ID
10 CD19 SSC SSC SSC Supplementary Figure 1 Representative flow cytometry gating strategy from NHS participant 5, time point 1 to isolate B lymphocytes, T lymphocytes and myeloid cells. Myeloid Cells FSC CD45 CD33 B Cells T Cells CD3
11 Supplementary Table 1 Sequenced reads and error-corrected consensus sequences generated for each library. Collection 1 Replicate 1 Collection 1 Replicate 2 Collection 2 Replicate 1 Collection 2 Replicate 2 Participant ID Raw Reads ECCS Raw Reads ECCS Raw Reads ECCS Raw Reads ECCS 1 38,483,255 3,333,626 33,448,218 2,84,567 35,427,539 2,987,274 33,392,978 2,784, ,268,72 3,318,173 41,984,812 3,558,516 35,157,811 3,27,67 33,657,29 2,86, ,63,819 2,581,39 36,17,671 2,959,152 48,998,142 3,584,46 4,599,238 3,291, ,932,163 2,212,764 3,623,846 2,51,632 39,579,433 3,254,544 48,529,452 3,53, ,11,143 2,727,3 34,151,27 2,411,821 52,32,285 3,759,16 55,49,72 4,17, ,27,169 2,863,69 35,84,657 2,946,669 5,852,817 3,682,33 48,351,486 3,514, ,658,678 2,663,917 42,58,68 2,714,869 45,885,262 3,233,3 44,468,353 3,548, ,771,597 2,734,288 41,632,517 2,528,357 5,72,31 3,399,553 5,378,471 3,698, ,449,116 2,531,229 41,67,14 2,599,127 6,14,462 4,197,532 5,347,145 3,993,77 1 4,492,765 2,554,6 38,729,489 2,4,5 59,87,612 4,34,423 58,962,293 3,996, ,94,33 3,684,38 44,34,692 3,456,949 64,52,183 4,96,893 56,51,287 3,797, ,115,177 4,245,185 48,446,875 3,692,857 61,813,583 4,322,748 59,452,7 4,11, ,368,839 3,59,66 41,269,631 3,343,48 59,327,495 4,213,628 52,689,35 4,173, ,837,743 3,76,61 37,36,17 2,976,419 6,366,37 4,53,326 58,19,532 4,51, ,65,75 3,47,283 44,547,457 2,49,226 58,11,11 3,98,542 51,539,869 4,14, ,226,742 2,986,829 6,744,391 3,97,372 45,532,881 3,414,68 6,632,143 4,27, ,985,852 4,79,429 51,835,232 3,373,746 55,355,241 3,721,52 59,51,527 4,96, ,185,217 3,337,38 54,495,83 3,388,54 57,117,288 4,147,18 58,64,485 3,999, ,213,946 2,947,31 51,28,264 3,343,682 53,983,868 3,723,161 48,268,843 3,796, ,521,626 3,376,26 41,417,619 2,86,99 58,651,16 4,92,482 56,692,728 3,793,691
12 Supplementary Table 2 Clonal SNVs detected by error-corrected sequencing. Participant ID Chr Start End Ref Alt Gene Amino Acid COSMIC VAF1.1 VAF1.2 VAF2.1 VAF T C IKZF1 intronic X G T STAG2 V376L G T DNMT3A A741E T G DNMT3A H739P A G DNMT3A L637P A G DNMT3A L58P G A DNMT3A R31W C G TET2 R1216G T A TET2 A1217A G C ETV6 intronic G A DNMT3A R32X G A NOTCH1 Q2338X X G T STAG2 R35L C T DNMT3A R635Q C G DNMT3A intronic C T MYD88 I192I T G TET2 F139L T G CUX1 intronic A G CBL I383M G A DNMT3A P94L T C DNMT3A T834A G A DNMT3A P799S G A DNMT3A intronic G A DNMT3A intronic C G DNMT3A intronic C G DNMT3A intronic G A DNMT3A intronic G A DNMT3A T691I C A DNMT3A E667X G A DNMT3A L639F C T DNMT3A G543S G A TET2 splicing T C FBXW7 intronic C T IKZF1 S21S X A T ATRX intronic X G T STAG2 intronic C A DNMT3A E482X G A TET2 G1288S T C DNMT3A H821R T C DNMT3A splicing C T DNMT3A R749H G C DNMT3A S714C G A DNMT3A R635W G T DNMT3A A572D C T DNMT3A R379H C T DNMT3A V296M G A DNMT3A Q249X C G TET2 S1486X A G ASXL1 N859S X T G KDM6A intronic G C KRAS intronic X C G BCORL1 H1376Q A G DNMT3A W86R C A DNMT3A E283X G C DNMT3A S129X T A MYD88 intronic A G TP53 F27S A T ASXL1 K131X X G A BCOR A414V X G T KDM6A intronic X A T ATRX F2165Y X A G BCORL1 L47L C G DNMT3A G543A C G TET2 intronic C T SMC3 S243F G C DNMT3A R899G C T DNMT3A R882H A G DNMT3A S839P A G DNMT3A F732S C A DNMT3A C537F C A DNMT3A G498V C A DNMT3A C494F C T DNMT3A W314X C A DNMT3A G32C T C RAD21 Y598C G C DNMT3A S337X T G KRAS A66A A G KRAS S65S X C T KDM6A A239V T C DNMT3A T835A T C SMC3 L242P G T TP53 P278H G T ASXL1 R119S X T A STAG2 UTR A G DNMT3A I75T A G DNMT3A F731S T G TET2 L446X C T TET2 T1793I X G A BCOR P652P X A T STAG2 K113I X G A BCORL1 G1522S G A DNMT3A Q231X G A TET2 W954X T A TET2 Y1337X G A CUX1 R62H A G EZH2 intronic X A G KDM6A intronic X C T BCORL1 R784X T G TET2 Y1589X G A CBL C381Y T C TP53 H168R X A G ATRX intronic X C T STAG2 intronic T C DNMT3A K67E G A TET2 R1359H G A TET2 splicing C A RAD21 V32V C A ETV6 V32V C T KRAS G12D
13 Supplementary Table 3 Clonal insertion/deletion variants detected by error-corrected sequencing. Participant ID Chr Start End Ref Alt Gene AA_Change VAF1.1 VAF1.2 VAF2.1 VAF GTG DNMT3A intronic CAGT - TET2 N253fs A TET2 Y1255_G1256delinsX A - DNMT3A I75fs AGCAGCGGGAAGGGTCAGAA - DNMT3A intronic T DNMT3A T53fs TCTC - RAD21 E528fs X ATTAATTTT - STAG2 intronic C - TET2 A1381fs
14 Supplementary Table 4 Summary of droplet digital PCR validation experiments. Participant ID Gene AA_Change Cosmic Wells ECS VAF Replicate 1 Collection 1 Collection 2 ECS VAF ddpcr ECS VAF Replicate VAF Replicate 1 2 ECS VAF Replicate 2 ddpcr VAF ddpcr Control UPN.Collection 4 DNMT3A R32X STAG2 R35L DNMT3A R635Q DNMT3A P94L TET2 splice TET2 G1288S A DNMT3A S714C A DNMT3A R635W TET2 S1486X TP53 F27S A DNMT3A W86R DNMT3A G543A A DNMT3A R882H TP53 P278H DNMT3A I75T TET2 W954X TET2 Y1337X BCORL1 R784X CBL C381Y TP53 H168R KRAS G12D TET2 A1381fs Control VAF
15 Supplementary Table 5 Summary of variant VAFs detected by droplet digital PCR in sorted hematopoietic compartments. Participant ID Gene Amino Acid COSMIC Collection Bulk B T M 4 DNMT3A R32X DNMT3A R635Q DNMT3A P94L TET2 G1288S DNMT3A S714C DNMT3A W86R DNMT3A G543A DNMT3A R882H TP53 P278H DNMT3A I75T TET2 W954X TP53 H168R KRAS G12D
16 Supplementary Table 6 Primer sequences for library preparation. Primer Name Sequence i5 16N Random AATGATACGGCGACCACCGAGATCTACACNNNNNNNNNNNNNNNNACACTCTTTCCCTACACGACGCTCTTCCGATCT P5 AATGATACGGCGACCACCGA P7 CAAGCAGAAGACGGCATACGA
Illumina Trusight Myeloid Panel validation A R FHAN R A FIQ
 Illumina Trusight Myeloid Panel validation A R FHAN R A FIQ G E NETIC T E CHNOLOGIST MEDICAL G E NETICS, CARDIFF To Cover Background to the project Choice of panel Validation process Genes on panel, Protocol
Illumina Trusight Myeloid Panel validation A R FHAN R A FIQ G E NETIC T E CHNOLOGIST MEDICAL G E NETICS, CARDIFF To Cover Background to the project Choice of panel Validation process Genes on panel, Protocol
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature13898 Supplementary Information Table 1 Kras mutation status of carcinogen-induced mouse lung adenomas Tumour Treatment Strain Grade Genotype Kras status (WES)* Kras status (Sanger) 32T1
doi:10.1038/nature13898 Supplementary Information Table 1 Kras mutation status of carcinogen-induced mouse lung adenomas Tumour Treatment Strain Grade Genotype Kras status (WES)* Kras status (Sanger) 32T1
A Comprehensive Study of TP53 Mutations in Chronic Lymphocytic Leukemia: Analysis of 1,287 Diagnostic CLL Samples
 A Comprehensive Study of TP53 Mutations in Chronic Lymphocytic Leukemia: Analysis of 1,287 Diagnostic CLL Samples Sona Pekova, MD., PhD. Chambon Ltd., Laboratory for molecular diagnostics, Prague, Czech
A Comprehensive Study of TP53 Mutations in Chronic Lymphocytic Leukemia: Analysis of 1,287 Diagnostic CLL Samples Sona Pekova, MD., PhD. Chambon Ltd., Laboratory for molecular diagnostics, Prague, Czech
SUPPLEMENTARY INFORMATION. Rare independent mutations in renal salt handling genes contribute to blood pressure variation
 SUPPLEMENTARY INFORMATION Rare independent mutations in renal salt handling genes contribute to blood pressure variation Weizhen Ji, Jia Nee Foo, Brian J. O Roak, Hongyu Zhao, Martin G. Larson, David B.
SUPPLEMENTARY INFORMATION Rare independent mutations in renal salt handling genes contribute to blood pressure variation Weizhen Ji, Jia Nee Foo, Brian J. O Roak, Hongyu Zhao, Martin G. Larson, David B.
Next Generation Sequencing in Haematological Malignancy: A European Perspective. Wolfgang Kern, Munich Leukemia Laboratory
 Next Generation Sequencing in Haematological Malignancy: A European Perspective Wolfgang Kern, Munich Leukemia Laboratory Diagnostic Methods Cytomorphology Cytogenetics Immunophenotype Histology FISH Molecular
Next Generation Sequencing in Haematological Malignancy: A European Perspective Wolfgang Kern, Munich Leukemia Laboratory Diagnostic Methods Cytomorphology Cytogenetics Immunophenotype Histology FISH Molecular
Vertical Magnetic Separation of Circulating Tumor Cells and Somatic Genomic-Alteration Analysis in Lung Cancer Patients
 Vertical Magnetic Separation of Circulating Cells and Somatic Genomic-Alteration Analysis in Lung Cancer Patients Chang Eun Yoo 1,2#, Jong-Myeon Park 3#, Hui-Sung Moon 1,2, Je-Gun Joung 2, Dae-Soon Son
Vertical Magnetic Separation of Circulating Cells and Somatic Genomic-Alteration Analysis in Lung Cancer Patients Chang Eun Yoo 1,2#, Jong-Myeon Park 3#, Hui-Sung Moon 1,2, Je-Gun Joung 2, Dae-Soon Son
Supplementary Figure 1. Cytoscape bioinformatics toolset was used to create the network of protein-protein interactions between the product of each
 Supplementary Figure 1. Cytoscape bioinformatics toolset was used to create the network of protein-protein interactions between the product of each mutated gene and the panel of 125 cancer-driving genes
Supplementary Figure 1. Cytoscape bioinformatics toolset was used to create the network of protein-protein interactions between the product of each mutated gene and the panel of 125 cancer-driving genes
To test the possible source of the HBV infection outside the study family, we searched the Genbank
 Supplementary Discussion The source of hepatitis B virus infection To test the possible source of the HBV infection outside the study family, we searched the Genbank and HBV Database (http://hbvdb.ibcp.fr),
Supplementary Discussion The source of hepatitis B virus infection To test the possible source of the HBV infection outside the study family, we searched the Genbank and HBV Database (http://hbvdb.ibcp.fr),
NeoTYPE Cancer Profiles
 NeoTYPE Cancer Profiles Multimethod Analysis of 25+ Hematologic Diseases and Solid Tumors Anatomic Pathology FISH Molecular The next generation of diagnostic, prognostic, and therapeutic assessment NeoTYPE
NeoTYPE Cancer Profiles Multimethod Analysis of 25+ Hematologic Diseases and Solid Tumors Anatomic Pathology FISH Molecular The next generation of diagnostic, prognostic, and therapeutic assessment NeoTYPE
Next generation sequencing analysis - A UK perspective. Nicholas Lea
 Next generation sequencing analysis - A UK perspective Nicholas Lea King s HMDC LMH is part of an integrated pathology service at King s Haematological Malignancy Diagnostic Centre (HMDC) HMDC serves population
Next generation sequencing analysis - A UK perspective Nicholas Lea King s HMDC LMH is part of an integrated pathology service at King s Haematological Malignancy Diagnostic Centre (HMDC) HMDC serves population
Accel-Amplicon Panels
 Accel-Amplicon Panels Amplicon sequencing has emerged as a reliable, cost-effective method for ultra-deep targeted sequencing. This highly adaptable approach is especially applicable for in-depth interrogation
Accel-Amplicon Panels Amplicon sequencing has emerged as a reliable, cost-effective method for ultra-deep targeted sequencing. This highly adaptable approach is especially applicable for in-depth interrogation
Laboratory Service Report
 Client C7028846-DLP Rochester Rochester, N 55901 Specimen Type Peripheral blood CR PDF Report available at: https://test.mmlaccess.com/reports/c7028846-zwselwql7p.ashx Indication for Test DS CR Pathogenic
Client C7028846-DLP Rochester Rochester, N 55901 Specimen Type Peripheral blood CR PDF Report available at: https://test.mmlaccess.com/reports/c7028846-zwselwql7p.ashx Indication for Test DS CR Pathogenic
NeoTYPE Cancer Profiles
 NeoTYPE Cancer Profiles 30+ Multimethod Assays for Hematologic Diseases and Solid Tumors Molecular FISH Anatomic Pathology The next generation of diagnostic, prognostic, and therapeutic assessment What
NeoTYPE Cancer Profiles 30+ Multimethod Assays for Hematologic Diseases and Solid Tumors Molecular FISH Anatomic Pathology The next generation of diagnostic, prognostic, and therapeutic assessment What
Susanne Schnittger. Workflow of molecular investigations in JAK2-negative MPNs - the Munich experience
 Susanne Schnittger Workflow of molecular investigations in JAK2negative MPNs the Munich experience Cohort single centre experience to apply new markers in a daily diagnostic work flow total: 20,547 cases
Susanne Schnittger Workflow of molecular investigations in JAK2negative MPNs the Munich experience Cohort single centre experience to apply new markers in a daily diagnostic work flow total: 20,547 cases
Supplementary Figure 1. FACS analysis of cells infected with TY93/H5N1 GFP-627E,
 Supplementary Figure 1. FACS analysis of cells infected with TY93/H5N1 GFP-627E, TY93/H5N1 GFP-627K, or the TY93/H5N1 PB2(588-759) virus library. To establish our GFP- FACS screening platform, we compared
Supplementary Figure 1. FACS analysis of cells infected with TY93/H5N1 GFP-627E, TY93/H5N1 GFP-627K, or the TY93/H5N1 PB2(588-759) virus library. To establish our GFP- FACS screening platform, we compared
Please Silence Your Cell Phones. Thank You
 Please Silence Your Cell Phones Thank You Utility of NGS and Comprehensive Genomic Profiling in Hematopathology Practice Maria E. Arcila M.D. Memorial Sloan Kettering Cancer Center New York, NY Disclosure
Please Silence Your Cell Phones Thank You Utility of NGS and Comprehensive Genomic Profiling in Hematopathology Practice Maria E. Arcila M.D. Memorial Sloan Kettering Cancer Center New York, NY Disclosure
Introduction of an NGS gene panel into the Haemato-Oncology MPN service
 Introduction of an NGS gene panel into the Haemato-Oncology MPN service Dr. Anna Skowronska, Dr Jane Bryon, Dr Samuel Clokie, Dr Yvonne Wallis and Professor Mike Griffiths West Midlands Regional Genetics
Introduction of an NGS gene panel into the Haemato-Oncology MPN service Dr. Anna Skowronska, Dr Jane Bryon, Dr Samuel Clokie, Dr Yvonne Wallis and Professor Mike Griffiths West Midlands Regional Genetics
NGS in tissue and liquid biopsy
 NGS in tissue and liquid biopsy Ana Vivancos, PhD Referencias So, why NGS in the clinics? 2000 Sanger Sequencing (1977-) 2016 NGS (2006-) ABIPrism (Applied Biosystems) Up to 2304 per day (96 sequences
NGS in tissue and liquid biopsy Ana Vivancos, PhD Referencias So, why NGS in the clinics? 2000 Sanger Sequencing (1977-) 2016 NGS (2006-) ABIPrism (Applied Biosystems) Up to 2304 per day (96 sequences
TP53 mutational profile in CLL : A retrospective study of the FILO group.
 TP53 mutational profile in CLL : A retrospective study of the FILO group. Fanny Baran-Marszak Hopital Avicenne Bobigny France 2nd ERIC workshop on TP53 analysis in CLL, Stresa 2017 TP53 abnormalities :
TP53 mutational profile in CLL : A retrospective study of the FILO group. Fanny Baran-Marszak Hopital Avicenne Bobigny France 2nd ERIC workshop on TP53 analysis in CLL, Stresa 2017 TP53 abnormalities :
Laboratory Service Report
 Specimen Type Peripheral blood CR PDF Report available at: https://test.mmlaccess.com/reports/c7028846-ih2xuglwpq.ashx Indication for Test DS CR Pathogenic utations Detected CR 1. JAK2: c.1849g>t;p.val617phe
Specimen Type Peripheral blood CR PDF Report available at: https://test.mmlaccess.com/reports/c7028846-ih2xuglwpq.ashx Indication for Test DS CR Pathogenic utations Detected CR 1. JAK2: c.1849g>t;p.val617phe
modified dye uptake assay including formazan test EC 90 not tested plaque reduction assay
 Sauerbrei A, Bohn-Wippert K, Kaspar M, Krumbholz A, Karrasch M, Zell R. 2015. Database on natural polymorphisms and resistance-related non-synonymous mutations in thymidine kinase and DNA polymerase genes
Sauerbrei A, Bohn-Wippert K, Kaspar M, Krumbholz A, Karrasch M, Zell R. 2015. Database on natural polymorphisms and resistance-related non-synonymous mutations in thymidine kinase and DNA polymerase genes
Clinical Utility of Droplet ddpcr, moving to diagnostics. Koen De Gelas, PhD, CRIG ddpcr mini symposium, 15/05/2018
 1 Clinical Utility of Droplet ddpcr, moving to diagnostics 2 Koen De Gelas, PhD, CRIG ddpcr mini symposium, 15/05/2018 Disclaimer Disclaimer: all consumables, instruments, applications and software covered
1 Clinical Utility of Droplet ddpcr, moving to diagnostics 2 Koen De Gelas, PhD, CRIG ddpcr mini symposium, 15/05/2018 Disclaimer Disclaimer: all consumables, instruments, applications and software covered
Sample Metrics. Allele Frequency (%) Read Depth Ploidy. Gene CDS Effect Protein Effect. LN Metastasis Tumor Purity Computational Pathology 80% 60%
 Supplemental Table 1: Estimated tumor purity, allele frequency, and independent read depth for all gene mutations classified as either potentially pathogenic or VUS in the metatastic and primary tumor
Supplemental Table 1: Estimated tumor purity, allele frequency, and independent read depth for all gene mutations classified as either potentially pathogenic or VUS in the metatastic and primary tumor
Frequency(%) KRAS G12 KRAS G13 KRAS A146 KRAS Q61 KRAS K117N PIK3CA H1047 PIK3CA E545 PIK3CA E542K PIK3CA Q546. EGFR exon19 NFS-indel EGFR L858R
 Frequency(%) 1 a b ALK FS-indel ALK R1Q HRAS Q61R HRAS G13R IDH R17K IDH R14Q MET exon14 SS-indel KIT D8Y KIT L76P KIT exon11 NFS-indel SMAD4 R361 IDH1 R13 CTNNB1 S37 CTNNB1 S4 AKT1 E17K ERBB D769H ERBB
Frequency(%) 1 a b ALK FS-indel ALK R1Q HRAS Q61R HRAS G13R IDH R17K IDH R14Q MET exon14 SS-indel KIT D8Y KIT L76P KIT exon11 NFS-indel SMAD4 R361 IDH1 R13 CTNNB1 S37 CTNNB1 S4 AKT1 E17K ERBB D769H ERBB
Supplemental Table 1. The list of variants with their respective scores for each variant classifier Gene DNA Protein Align-GVGD Polyphen-2 CADD MAPP
 Supplemental Table 1. The list of variants with their respective scores for each variant classifier Gene DNA Protein Align-GVGD Polyphen-2 CADD MAPP Frequency Domain Mammals a 3 S/P b Mammals a 3 S/P b
Supplemental Table 1. The list of variants with their respective scores for each variant classifier Gene DNA Protein Align-GVGD Polyphen-2 CADD MAPP Frequency Domain Mammals a 3 S/P b Mammals a 3 S/P b
Importance of minor TP53 mutated clones in the clinic
 Importance of minor TP53 mutated clones in the clinic Davide Rossi, M.D., Ph.D. Hematology IOSI - Oncology Institute of Southern Switzerland IOR - Institute of Oncology Reserach Bellinzona - Switzerland
Importance of minor TP53 mutated clones in the clinic Davide Rossi, M.D., Ph.D. Hematology IOSI - Oncology Institute of Southern Switzerland IOR - Institute of Oncology Reserach Bellinzona - Switzerland
Supplementary Information
 Supplementary Information Table of Contents Supplementary methods... 2 Figure S1 - Variable DNA yield proportional to bone marrow aspirate cellularity.... 3 Figure S2 - Mutations by clinical ontogeny group....
Supplementary Information Table of Contents Supplementary methods... 2 Figure S1 - Variable DNA yield proportional to bone marrow aspirate cellularity.... 3 Figure S2 - Mutations by clinical ontogeny group....
Variant interpretation exercise. ACGS Somatic Variant Interpretation Workshop Joanne Mason 21/09/18
 Variant interpretation exercise ACGS Somatic Variant Interpretation Workshop Joanne Mason 21/09/18 Format of exercise Compile a list of tricky variants across solid cancer and haematological malignancy.
Variant interpretation exercise ACGS Somatic Variant Interpretation Workshop Joanne Mason 21/09/18 Format of exercise Compile a list of tricky variants across solid cancer and haematological malignancy.
Blastic Plasmacytoid Dendritic Cell Neoplasm with DNMT3A and TET2 mutations (SH )
 Blastic Plasmacytoid Dendritic Cell Neoplasm with DNMT3A and TET2 mutations (SH2017-0314) Habibe Kurt, Joseph D. Khoury, Carlos E. Bueso-Ramos, Jeffrey L. Jorgensen, Guilin Tang, L. Jeffrey Medeiros, and
Blastic Plasmacytoid Dendritic Cell Neoplasm with DNMT3A and TET2 mutations (SH2017-0314) Habibe Kurt, Joseph D. Khoury, Carlos E. Bueso-Ramos, Jeffrey L. Jorgensen, Guilin Tang, L. Jeffrey Medeiros, and
Supplementary Figure 1 Weight and body temperature of ferrets inoculated with
 Supplementary Figure 1 Weight and body temperature of ferrets inoculated with A/Anhui/1/2013 (H7N9) influenza virus. (a) Body temperature and (b) weight change of ferrets after intranasal inoculation with
Supplementary Figure 1 Weight and body temperature of ferrets inoculated with A/Anhui/1/2013 (H7N9) influenza virus. (a) Body temperature and (b) weight change of ferrets after intranasal inoculation with
The Center for PERSONALIZED DIAGNOSTICS
 The Center for PERSONALIZED DIAGNOSTICS Precision Diagnostics for Personalized Medicine A joint initiative between The Department of Pathology and Laboratory Medicine & The Abramson Cancer Center The (CPD)
The Center for PERSONALIZED DIAGNOSTICS Precision Diagnostics for Personalized Medicine A joint initiative between The Department of Pathology and Laboratory Medicine & The Abramson Cancer Center The (CPD)
Supplementary Figure 1. Schematic diagram of o2n-seq. Double-stranded DNA was sheared, end-repaired, and underwent A-tailing by standard protocols.
 Supplementary Figure 1. Schematic diagram of o2n-seq. Double-stranded DNA was sheared, end-repaired, and underwent A-tailing by standard protocols. A-tailed DNA was ligated to T-tailed dutp adapters, circularized
Supplementary Figure 1. Schematic diagram of o2n-seq. Double-stranded DNA was sheared, end-repaired, and underwent A-tailing by standard protocols. A-tailed DNA was ligated to T-tailed dutp adapters, circularized
Supplementary Figure 1. BMS enhances human T cell activation in vitro in a
 Supplementary Figure 1. BMS98662 enhances human T cell activation in vitro in a concentration-dependent manner. Jurkat T cells were activated with anti-cd3 and anti-cd28 antibody in the presence of titrated
Supplementary Figure 1. BMS98662 enhances human T cell activation in vitro in a concentration-dependent manner. Jurkat T cells were activated with anti-cd3 and anti-cd28 antibody in the presence of titrated
Reporting TP53 gene analysis results in CLL
 Reporting TP53 gene analysis results in CLL Mutations in TP53 - From discovery to clinical practice in CLL Discovery Validation Clinical practice Variant diversity *Leroy at al, Cancer Research Review
Reporting TP53 gene analysis results in CLL Mutations in TP53 - From discovery to clinical practice in CLL Discovery Validation Clinical practice Variant diversity *Leroy at al, Cancer Research Review
Relative activity (%) SC35M
 a 125 Bat (H17N) b 125 A/WSN (H1N1) Relative activity (%) 0 75 50 25 Relative activity (%) 0 75 50 25 0 Pos. Neg. PA PB1 Pos. Neg. NP PA PB1 PB2 0 Pos. Neg. NP PA PB1 PB2 SC35M Bat Supplementary Figure
a 125 Bat (H17N) b 125 A/WSN (H1N1) Relative activity (%) 0 75 50 25 Relative activity (%) 0 75 50 25 0 Pos. Neg. PA PB1 Pos. Neg. NP PA PB1 PB2 0 Pos. Neg. NP PA PB1 PB2 SC35M Bat Supplementary Figure
Supporting Information
 Supporting Information Rampal et al. 10.1073/pnas.1407792111 Fig. S1. Genetic events in leukemic transformation of chronic-phase MPNs. (A) Survival of post-mpn AML patients according to mutational status
Supporting Information Rampal et al. 10.1073/pnas.1407792111 Fig. S1. Genetic events in leukemic transformation of chronic-phase MPNs. (A) Survival of post-mpn AML patients according to mutational status
Identification and clinical detection of genetic alterations of pre-neoplastic lesions Time for the PML ome? David Sidransky MD Johns Hopkins
 Identification and clinical detection of genetic alterations of pre-neoplastic lesions Time for the PML ome? David Sidransky MD Johns Hopkins February 3-5, 2016 Lansdowne Resort, Leesburg, VA Molecular
Identification and clinical detection of genetic alterations of pre-neoplastic lesions Time for the PML ome? David Sidransky MD Johns Hopkins February 3-5, 2016 Lansdowne Resort, Leesburg, VA Molecular
NC bp. b 1481 bp
 Kcna3 NC 11 178 p 1481 p 346 p c *** *** d relative expression..4.3.2.1 CD4 T cells Kcna1 Kcna2 Kcna3 Kcna4 Kcna Kcna6 Kcna7 Kcnn4 relative expression..4.3.2.1 CD8 T cells Kcna1 Kcna2 Kcna3 Kcna4 Kcna
Kcna3 NC 11 178 p 1481 p 346 p c *** *** d relative expression..4.3.2.1 CD4 T cells Kcna1 Kcna2 Kcna3 Kcna4 Kcna Kcna6 Kcna7 Kcnn4 relative expression..4.3.2.1 CD8 T cells Kcna1 Kcna2 Kcna3 Kcna4 Kcna
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Fig. 1: Quality assessment of formalin-fixed paraffin-embedded (FFPE)-derived DNA and nuclei. (a) Multiplex PCR analysis of unrepaired and repaired bulk FFPE gdna from
Supplementary Figure 1 Supplementary Fig. 1: Quality assessment of formalin-fixed paraffin-embedded (FFPE)-derived DNA and nuclei. (a) Multiplex PCR analysis of unrepaired and repaired bulk FFPE gdna from
Nature Genetics: doi: /ng Supplementary Figure 1. Somatic coding mutations identified by WES/WGS for 83 ATL cases.
 Supplementary Figure 1 Somatic coding mutations identified by WES/WGS for 83 ATL cases. (a) The percentage of targeted bases covered by at least 2, 10, 20 and 30 sequencing reads (top) and average read
Supplementary Figure 1 Somatic coding mutations identified by WES/WGS for 83 ATL cases. (a) The percentage of targeted bases covered by at least 2, 10, 20 and 30 sequencing reads (top) and average read
p.arg119gly p.arg119his p.ala179thr c.540+1g>a c.617_633+6del Prediction basis structure
 a Missense ATG p.arg119gly p.arg119his p.ala179thr p.ala189val p.gly206trp p.gly206arg p.arg251his p.ala257thr TGA 5 UTR 1 2 3 4 5 6 7 3 UTR Splice Site, Frameshift b Mutation p.gly206trp p.gly4fsx50 c.138+1g>a
a Missense ATG p.arg119gly p.arg119his p.ala179thr p.ala189val p.gly206trp p.gly206arg p.arg251his p.ala257thr TGA 5 UTR 1 2 3 4 5 6 7 3 UTR Splice Site, Frameshift b Mutation p.gly206trp p.gly4fsx50 c.138+1g>a
SureSelect Cancer All-In-One Custom and Catalog NGS Assays
 SureSelect Cancer All-In-One Custom and Catalog NGS Assays Detect all cancer-relevant variants in a single SureSelect assay SNV Indel TL SNV Indel TL Single DNA input Single AIO assay Single data analysis
SureSelect Cancer All-In-One Custom and Catalog NGS Assays Detect all cancer-relevant variants in a single SureSelect assay SNV Indel TL SNV Indel TL Single DNA input Single AIO assay Single data analysis
Supplementary information to:
 Supplementary information to: Digital Sorting of Pure Cell Populations Enables Unambiguous Genetic Analysis of Heterogeneous Formalin-Fixed Paraffin Embedded Tumors by Next Generation Sequencing Authors
Supplementary information to: Digital Sorting of Pure Cell Populations Enables Unambiguous Genetic Analysis of Heterogeneous Formalin-Fixed Paraffin Embedded Tumors by Next Generation Sequencing Authors
Aplastic Anaemia Genomics: Current data and interpretation
 An Academic Health Sciences Centre at the heart of a world city... Aplastic Anaemia Genomics: Current data and interpretation Austin G Kulasekararaj Consultant Haematologist King s College Hospital, London
An Academic Health Sciences Centre at the heart of a world city... Aplastic Anaemia Genomics: Current data and interpretation Austin G Kulasekararaj Consultant Haematologist King s College Hospital, London
MEDICAL GENOMICS LABORATORY. Next-Gen Sequencing and Deletion/Duplication Analysis of NF1 Only (NF1-NG)
 Next-Gen Sequencing and Deletion/Duplication Analysis of NF1 Only (NF1-NG) Ordering Information Acceptable specimen types: Fresh blood sample (3-6 ml EDTA; no time limitations associated with receipt)
Next-Gen Sequencing and Deletion/Duplication Analysis of NF1 Only (NF1-NG) Ordering Information Acceptable specimen types: Fresh blood sample (3-6 ml EDTA; no time limitations associated with receipt)
Nature Genetics: doi: /ng Supplementary Figure 1. Mutational signatures in BCC compared to melanoma.
 Supplementary Figure 1 Mutational signatures in BCC compared to melanoma. (a) The effect of transcription-coupled repair as a function of gene expression in BCC. Tumor type specific gene expression levels
Supplementary Figure 1 Mutational signatures in BCC compared to melanoma. (a) The effect of transcription-coupled repair as a function of gene expression in BCC. Tumor type specific gene expression levels
Supplementary. presence of the. (c) mrna expression. Error. in naive or
 Figure 1. (a) Naive CD4 + T cells were activated in the presence of the indicated cytokines for 3 days. Enpp2 mrna expression was measured by qrt-pcrhr, infected with (b, c) Naive CD4 + T cells were activated
Figure 1. (a) Naive CD4 + T cells were activated in the presence of the indicated cytokines for 3 days. Enpp2 mrna expression was measured by qrt-pcrhr, infected with (b, c) Naive CD4 + T cells were activated
7/12/2016 TESTING. Objectives. New Directions in Aplastic Anemia: What's on the Horizon? Better way to evaluate clonal evolution?
 New Directions in Aplastic Anemia: What's on the Horizon? Amy E. DeZern, MD; MHS Assistant Professor Hematologic Malignancies Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins Baltimore, MD Objectives
New Directions in Aplastic Anemia: What's on the Horizon? Amy E. DeZern, MD; MHS Assistant Professor Hematologic Malignancies Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins Baltimore, MD Objectives
Supplementary Tables. Supplementary Figures
 Supplementary Files for Zehir, Benayed et al. Mutational Landscape of Metastatic Cancer Revealed from Prospective Clinical Sequencing of 10,000 Patients Supplementary Tables Supplementary Table 1: Sample
Supplementary Files for Zehir, Benayed et al. Mutational Landscape of Metastatic Cancer Revealed from Prospective Clinical Sequencing of 10,000 Patients Supplementary Tables Supplementary Table 1: Sample
Nature Immunology: doi: /ni.3412
 Supplementary Figure 1 Gata1 expression in heamatopoietic stem and progenitor populations. (a) Unsupervised clustering according to 100 top variable genes across single pre-gm cells. The two main cell
Supplementary Figure 1 Gata1 expression in heamatopoietic stem and progenitor populations. (a) Unsupervised clustering according to 100 top variable genes across single pre-gm cells. The two main cell
HD1 (FLU) HD2 (EBV) HD2 (FLU)
 ramer staining + anti-pe beads ramer staining a HD1 (FLU) HD2 (EBV) HD2 (FLU).73.11.56.46.24 1.12 b CD127 + c CD127 + d CD127 - e CD127 - PD1 - PD1 + PD1 + PD1-1 1 1 1 %CD127 + PD1-8 6 4 2 + anti-pe %CD127
ramer staining + anti-pe beads ramer staining a HD1 (FLU) HD2 (EBV) HD2 (FLU).73.11.56.46.24 1.12 b CD127 + c CD127 + d CD127 - e CD127 - PD1 - PD1 + PD1 + PD1-1 1 1 1 %CD127 + PD1-8 6 4 2 + anti-pe %CD127
p.r623c p.p976l p.d2847fs p.t2671 p.d2847fs p.r2922w p.r2370h p.c1201y p.a868v p.s952* RING_C BP PHD Cbp HAT_KAT11
 ARID2 p.r623c KMT2D p.v650fs p.p976l p.r2922w p.l1212r p.d1400h DNA binding RFX DNA binding Zinc finger KMT2C p.a51s p.d372v p.c1103* p.d2847fs p.t2671 p.d2847fs p.r4586h PHD/ RING DHHC/ PHD PHD FYR N
ARID2 p.r623c KMT2D p.v650fs p.p976l p.r2922w p.l1212r p.d1400h DNA binding RFX DNA binding Zinc finger KMT2C p.a51s p.d372v p.c1103* p.d2847fs p.t2671 p.d2847fs p.r4586h PHD/ RING DHHC/ PHD PHD FYR N
Mutational Impact on Diagnostic and Prognostic Evaluation of MDS
 Mutational Impact on Diagnostic and Prognostic Evaluation of MDS Elsa Bernard, PhD Papaemmanuil Lab, Computational Oncology, MSKCC MDS Foundation ASH 2018 Symposium Disclosure Research funds provided by
Mutational Impact on Diagnostic and Prognostic Evaluation of MDS Elsa Bernard, PhD Papaemmanuil Lab, Computational Oncology, MSKCC MDS Foundation ASH 2018 Symposium Disclosure Research funds provided by
Clonal hematopoiesis of indeterminate potential and MDS. Siddhartha Jaiswal AAMDS Meeting 3/17/16
 Clonal hematopoiesis of indeterminate potential and MDS Siddhartha Jaiswal AAMDS Meeting 3/17/16 Clonal evolution from birth to death Might pre-malignant clones, bearing only the initiating lesion, be
Clonal hematopoiesis of indeterminate potential and MDS Siddhartha Jaiswal AAMDS Meeting 3/17/16 Clonal evolution from birth to death Might pre-malignant clones, bearing only the initiating lesion, be
Nature Immunology: doi: /ni Supplementary Figure 1. Cellularity of leukocytes and their progenitors in naive wild-type and Spp1 / mice.
 Supplementary Figure 1 Cellularity of leukocytes and their progenitors in naive wild-type and Spp1 / mice. (a, b) Gating strategies for differentiated cells including PMN (CD11b + Ly6G hi and CD11b + Ly6G
Supplementary Figure 1 Cellularity of leukocytes and their progenitors in naive wild-type and Spp1 / mice. (a, b) Gating strategies for differentiated cells including PMN (CD11b + Ly6G hi and CD11b + Ly6G
Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A.
 Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A. Upper part, three-primer PCR strategy at the Mcm3 locus yielding
Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A. Upper part, three-primer PCR strategy at the Mcm3 locus yielding
Performance Characteristics BRCA MASTR Plus Dx
 Performance Characteristics BRCA MASTR Plus Dx with drmid Dx for Illumina NGS systems Manufacturer Multiplicom N.V. Galileïlaan 18 2845 Niel Belgium Table of Contents 1. Workflow... 4 2. Performance Characteristics
Performance Characteristics BRCA MASTR Plus Dx with drmid Dx for Illumina NGS systems Manufacturer Multiplicom N.V. Galileïlaan 18 2845 Niel Belgium Table of Contents 1. Workflow... 4 2. Performance Characteristics
Comprehensive Analyses of Circulating Cell- Free Tumor DNA
 Comprehensive Analyses of Circulating Cell- Free Tumor DNA Boston, MA June 28th, 2016 Derek Murphy, Ph.D. Scientist, Research and Development Personal Genome Diagnostics Acquisition of Somatic Alterations
Comprehensive Analyses of Circulating Cell- Free Tumor DNA Boston, MA June 28th, 2016 Derek Murphy, Ph.D. Scientist, Research and Development Personal Genome Diagnostics Acquisition of Somatic Alterations
SUPPLEMENTAL MATERIAL
 SUPPLEMENTAL MATERIAL SUPPLEMENTAL MATERIAL Supplemental Methods Phenotype Prediction Analyses In order to assess the phylogenetic properties of nssnvs, sequence conservation analysis was conducted using
SUPPLEMENTAL MATERIAL SUPPLEMENTAL MATERIAL Supplemental Methods Phenotype Prediction Analyses In order to assess the phylogenetic properties of nssnvs, sequence conservation analysis was conducted using
Nature Genetics: doi: /ng Supplementary Figure 1. Clinical timeline for the discovery WES cases.
 Supplementary Figure 1 Clinical timeline for the discovery WES cases. This illustrates the timeline of the disease events during the clinical course of each patient s disease, further indicating the available
Supplementary Figure 1 Clinical timeline for the discovery WES cases. This illustrates the timeline of the disease events during the clinical course of each patient s disease, further indicating the available
SUPPLEMENTARY INFORMATION. Supp. Fig. 1. Autoimmunity. Tolerance APC APC. T cell. T cell. doi: /nature06253 ICOS ICOS TCR CD28 TCR CD28
 Supp. Fig. 1 a APC b APC ICOS ICOS TCR CD28 mir P TCR CD28 P T cell Tolerance Roquin WT SG Icos mrna T cell Autoimmunity Roquin M199R SG Icos mrna www.nature.com/nature 1 Supp. Fig. 2 CD4 + CD44 low CD4
Supp. Fig. 1 a APC b APC ICOS ICOS TCR CD28 mir P TCR CD28 P T cell Tolerance Roquin WT SG Icos mrna T cell Autoimmunity Roquin M199R SG Icos mrna www.nature.com/nature 1 Supp. Fig. 2 CD4 + CD44 low CD4
Supplementary Fig. 1. Delivery of mirnas via Red Fluorescent Protein.
 prfp-vector RFP Exon1 Intron RFP Exon2 prfp-mir-124 mir-93/124 RFP Exon1 Intron RFP Exon2 Untransfected prfp-vector prfp-mir-93 prfp-mir-124 Supplementary Fig. 1. Delivery of mirnas via Red Fluorescent
prfp-vector RFP Exon1 Intron RFP Exon2 prfp-mir-124 mir-93/124 RFP Exon1 Intron RFP Exon2 Untransfected prfp-vector prfp-mir-93 prfp-mir-124 Supplementary Fig. 1. Delivery of mirnas via Red Fluorescent
Fig S1. ZNF697-NOTCH2 rearrangement: Diagram of the ZNF697-NOTCH2 fusion and
 SUPPLEMENTAL FIGURES: Fig S1. ZNF697-NOTCH2 rearrangement: Diagram of the ZNF697-NOTCH2 fusion and breakpoint. The 5 untranslated region of ZNF697 is joined out of frame with exon 26 of NOTCH2, with the
SUPPLEMENTAL FIGURES: Fig S1. ZNF697-NOTCH2 rearrangement: Diagram of the ZNF697-NOTCH2 fusion and breakpoint. The 5 untranslated region of ZNF697 is joined out of frame with exon 26 of NOTCH2, with the
MET skipping mutation, EGFR
 New NSCLC biomarkers in clinical research: detection of MET skipping mutation, EGFR T790M, and other important biomarkers Fernando López-Ríos Laboratorio de Dianas Terapéuticas Hospital Universitario HM
New NSCLC biomarkers in clinical research: detection of MET skipping mutation, EGFR T790M, and other important biomarkers Fernando López-Ríos Laboratorio de Dianas Terapéuticas Hospital Universitario HM
CHAPTER 8 ONCOGENIC MARKER DETECTION FROM P53 MUTANT AMINO-ACID SUBSTITUTIONS
 134 CHAPTER 8 ONCOGENIC MARKER DETECTION FROM P53 MUTANT AMINO-ACID SUBSTITUTIONS The recent past has witnessed a rapid rise in the utilization of computational techniques to aid and accelerate biological
134 CHAPTER 8 ONCOGENIC MARKER DETECTION FROM P53 MUTANT AMINO-ACID SUBSTITUTIONS The recent past has witnessed a rapid rise in the utilization of computational techniques to aid and accelerate biological
SUPPLEMENTARY INFORMATION. Intron retention is a widespread mechanism of tumor suppressor inactivation.
 SUPPLEMENTARY INFORMATION Intron retention is a widespread mechanism of tumor suppressor inactivation. Hyunchul Jung 1,2,3, Donghoon Lee 1,4, Jongkeun Lee 1,5, Donghyun Park 2,6, Yeon Jeong Kim 2,6, Woong-Yang
SUPPLEMENTARY INFORMATION Intron retention is a widespread mechanism of tumor suppressor inactivation. Hyunchul Jung 1,2,3, Donghoon Lee 1,4, Jongkeun Lee 1,5, Donghyun Park 2,6, Yeon Jeong Kim 2,6, Woong-Yang
Supplemental material Mutant DNMT3A: A Marker of Poor Prognosis in Acute Myeloid Leukemia Ana Flávia Tibúrcio Ribeiro, Marta Pratcorona, Claudia
 Supplemental material Mutant DNMT3A: A Marker of Poor Prognosis in Acute Myeloid Leukemia Ana Flávia Tibúrcio Ribeiro, Marta Pratcorona, Claudia Erpelinck-Verschueren, Veronika Rockova, Mathijs Sanders,
Supplemental material Mutant DNMT3A: A Marker of Poor Prognosis in Acute Myeloid Leukemia Ana Flávia Tibúrcio Ribeiro, Marta Pratcorona, Claudia Erpelinck-Verschueren, Veronika Rockova, Mathijs Sanders,
Characterisation of structural variation in breast. cancer genomes using paired-end sequencing on. the Illumina Genome Analyser
 Characterisation of structural variation in breast cancer genomes using paired-end sequencing on the Illumina Genome Analyser Phil Stephens Cancer Genome Project Why is it important to study cancer? Why
Characterisation of structural variation in breast cancer genomes using paired-end sequencing on the Illumina Genome Analyser Phil Stephens Cancer Genome Project Why is it important to study cancer? Why
Next generation histopathological diagnosis for precision medicine in solid cancers
 Next generation histopathological diagnosis for precision medicine in solid cancers from genomics to clinical application Aldo Scarpa ARC-NET Applied Research on Cancer Department of Pathology and Diagnostics
Next generation histopathological diagnosis for precision medicine in solid cancers from genomics to clinical application Aldo Scarpa ARC-NET Applied Research on Cancer Department of Pathology and Diagnostics
Clinical molecular profiling of archival tumor tissue and cell-free DNA from patients with Erdheim-Chester disease
 ECD International Medical Symposium New York City October 26, 2017 Clinical molecular profiling of archival tumor tissue and cell-free DNA from patients with Erdheim-Chester disease Filip Janku, Eli Diamond,
ECD International Medical Symposium New York City October 26, 2017 Clinical molecular profiling of archival tumor tissue and cell-free DNA from patients with Erdheim-Chester disease Filip Janku, Eli Diamond,
Abstract. Optimization strategy of Copy Number Variant calling using Multiplicom solutions APPLICATION NOTE. Introduction
 Optimization strategy of Copy Number Variant calling using Multiplicom solutions Michael Vyverman, PhD; Laura Standaert, PhD and Wouter Bossuyt, PhD Abstract Copy number variations (CNVs) represent a significant
Optimization strategy of Copy Number Variant calling using Multiplicom solutions Michael Vyverman, PhD; Laura Standaert, PhD and Wouter Bossuyt, PhD Abstract Copy number variations (CNVs) represent a significant
Clinical Genetics. Functional validation in a diagnostic context. Robert Hofstra. Leading the way in genetic issues
 Clinical Genetics Leading the way in genetic issues Functional validation in a diagnostic context Robert Hofstra Clinical Genetics Leading the way in genetic issues Future application of exome sequencing
Clinical Genetics Leading the way in genetic issues Functional validation in a diagnostic context Robert Hofstra Clinical Genetics Leading the way in genetic issues Future application of exome sequencing
Discovering Rare Hematopoietic Clones Harboring Leukemia-Associated Mutations Using Error-Corrected Sequencing
 Washington University in St. Louis Washington University Open Scholarship Arts & Sciences Electronic Theses and Dissertations Arts & Sciences Spring 5-15-2018 Discovering Rare Hematopoietic Clones Harboring
Washington University in St. Louis Washington University Open Scholarship Arts & Sciences Electronic Theses and Dissertations Arts & Sciences Spring 5-15-2018 Discovering Rare Hematopoietic Clones Harboring
Acute leukemia and myelodysplastic syndromes
 11/01/2012 Post-ASH meeting 1 Acute leukemia and myelodysplastic syndromes Peter Vandenberghe Centrum Menselijke Erfelijkheid & Afdeling Hematologie, UZ Leuven 11/01/2012 Post-ASH meeting 2 1. Acute myeloid
11/01/2012 Post-ASH meeting 1 Acute leukemia and myelodysplastic syndromes Peter Vandenberghe Centrum Menselijke Erfelijkheid & Afdeling Hematologie, UZ Leuven 11/01/2012 Post-ASH meeting 2 1. Acute myeloid
LIPOPROTEINE ATEROGENE E ANTI-ATEROGENE ATEROGENE
 LIPOPROTEINE ATEROGENE E ANTI-ATEROGENE ATEROGENE Sebastiano Calandra Dipartimento di Scienze Biomediche Università di Modena e Reggio Emilia Incidence Rate/1000 200-150 - 100-50 - Women 0 Men
LIPOPROTEINE ATEROGENE E ANTI-ATEROGENE ATEROGENE Sebastiano Calandra Dipartimento di Scienze Biomediche Università di Modena e Reggio Emilia Incidence Rate/1000 200-150 - 100-50 - Women 0 Men
MEDICAL GENOMICS LABORATORY. Peripheral Nerve Sheath Tumor Panel by Next-Gen Sequencing (PNT-NG)
 Peripheral Nerve Sheath Tumor Panel by Next-Gen Sequencing (PNT-NG) Ordering Information Acceptable specimen types: Blood (3-6ml EDTA; no time limitations associated with receipt) Saliva (OGR-575 DNA Genotek;
Peripheral Nerve Sheath Tumor Panel by Next-Gen Sequencing (PNT-NG) Ordering Information Acceptable specimen types: Blood (3-6ml EDTA; no time limitations associated with receipt) Saliva (OGR-575 DNA Genotek;
Fabry Disease X-linked genetic, multi-organ disorder. Fabry disease screening program in Hypertrophic p Cardiomyopathy: preliminary results.
 Fabry Disease X-linked genetic, multi-organ disorder Fabry disease screening program in Hypertrophic p Cardiomyopathy: y preliminary results. Globotriaosylceramide, GL3 Brain -galactosidase A Eyes Lactosylceramide
Fabry Disease X-linked genetic, multi-organ disorder Fabry disease screening program in Hypertrophic p Cardiomyopathy: y preliminary results. Globotriaosylceramide, GL3 Brain -galactosidase A Eyes Lactosylceramide
iplex genotyping IDH1 and IDH2 assays utilized the following primer sets (forward and reverse primers along with extension primers).
 Supplementary Materials Supplementary Methods iplex genotyping IDH1 and IDH2 assays utilized the following primer sets (forward and reverse primers along with extension primers). IDH1 R132H and R132L Forward:
Supplementary Materials Supplementary Methods iplex genotyping IDH1 and IDH2 assays utilized the following primer sets (forward and reverse primers along with extension primers). IDH1 R132H and R132L Forward:
Case Presentation. Pei Lin, M. D.
 Case Presentation Pei Lin, M. D. History A 26 yr man reports a history of numerous skin and upper respiratory infections as a child, including lymphadenitis and meningitis. In March 2013 during a preoperative
Case Presentation Pei Lin, M. D. History A 26 yr man reports a history of numerous skin and upper respiratory infections as a child, including lymphadenitis and meningitis. In March 2013 during a preoperative
Supplementary Figure 1: Features of IGLL5 Mutations in CLL: a) Representative IGV screenshot of first
 Supplementary Figure 1: Features of IGLL5 Mutations in CLL: a) Representative IGV screenshot of first intron IGLL5 mutation depicting biallelic mutations. Red arrows highlight the presence of out of phase
Supplementary Figure 1: Features of IGLL5 Mutations in CLL: a) Representative IGV screenshot of first intron IGLL5 mutation depicting biallelic mutations. Red arrows highlight the presence of out of phase
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description:
 File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. Schematic of Ras biochemical coupled assay.
File name: Supplementary Information Description: Supplementary figures and supplementary tables. File name: Peer review file Description: Supplementary Figure 1. Schematic of Ras biochemical coupled assay.
The preclinical efficacy of a novel telomerase inhibitor, imetelstat, in AML: A randomized trial in patient-derived xenografts
 The preclinical efficacy of a novel telomerase inhibitor, imetelstat, in AML: A randomized trial in patient-derived xenografts Claudia Bruedigam, Ph.D Gordon and Jessie Gilmour Leukaemia Research Laboratory
The preclinical efficacy of a novel telomerase inhibitor, imetelstat, in AML: A randomized trial in patient-derived xenografts Claudia Bruedigam, Ph.D Gordon and Jessie Gilmour Leukaemia Research Laboratory
Pharmacogenomics in Rare Diseases: Development Strategy for Ivacaftor as a Therapy for Cystic Fibrosis
 Pharmacogenomics in Rare Diseases: Development Strategy for Ivacaftor as a Therapy for Cystic Fibrosis Federico Goodsaid Vice President Strategic Regulatory Intelligence Vertex Pharmaceuticals Is there
Pharmacogenomics in Rare Diseases: Development Strategy for Ivacaftor as a Therapy for Cystic Fibrosis Federico Goodsaid Vice President Strategic Regulatory Intelligence Vertex Pharmaceuticals Is there
Molecular profiling in confirming the diagnosis of early myelodysplastic syndrome
 Molecular profiling of early MDS Hematopathology - March 2016 Article Molecular profiling in confirming the diagnosis of early myelodysplastic syndrome Maya Thangavelu 1,*, Ryan Olson 2, Li Li 2, Wanlong
Molecular profiling of early MDS Hematopathology - March 2016 Article Molecular profiling in confirming the diagnosis of early myelodysplastic syndrome Maya Thangavelu 1,*, Ryan Olson 2, Li Li 2, Wanlong
Pediatric Oncology & Pathology Services
 Pediatric Oncology & Pathology Services Anatomic Pathology Flow Cytometry Cytogenetics Pharma Services Diagnostic, Prognostic, Predictive, and Predisposition Testing Pediatric Oncology & Pathology Services
Pediatric Oncology & Pathology Services Anatomic Pathology Flow Cytometry Cytogenetics Pharma Services Diagnostic, Prognostic, Predictive, and Predisposition Testing Pediatric Oncology & Pathology Services
Spectrum of somatically acquired mutations identified by combining WES and genome-wide DNA array analysis in the discovery cohort of 30 JMML cases.
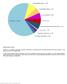 Supplementary Figure 1 Spectrum of somatically acquired mutations identified by combining WES and genome-wide DNA array analysis in the discovery cohort of 30 JMML cases. A total of 85 somatically acquired
Supplementary Figure 1 Spectrum of somatically acquired mutations identified by combining WES and genome-wide DNA array analysis in the discovery cohort of 30 JMML cases. A total of 85 somatically acquired
Click to edit Master /tle style
 Click to edit Master /tle style Tel: (314) 747-7337 Toll Free: (866) 450-7697 Fax: (314) 747-7336 Email: gps@wustl.edu Website: gps.wustl.edu GENETIC TESTING IN CANCER Ka/nka Vigh-Conrad, PhD Genomics
Click to edit Master /tle style Tel: (314) 747-7337 Toll Free: (866) 450-7697 Fax: (314) 747-7336 Email: gps@wustl.edu Website: gps.wustl.edu GENETIC TESTING IN CANCER Ka/nka Vigh-Conrad, PhD Genomics
Nature Immunology: doi: /ni Supplementary Figure 1. Huwe1 has high expression in HSCs and is necessary for quiescence.
 Supplementary Figure 1 Huwe1 has high expression in HSCs and is necessary for quiescence. (a) Heat map visualizing expression of genes with a known function in ubiquitin-mediated proteolysis (KEGG: Ubiquitin
Supplementary Figure 1 Huwe1 has high expression in HSCs and is necessary for quiescence. (a) Heat map visualizing expression of genes with a known function in ubiquitin-mediated proteolysis (KEGG: Ubiquitin
Molecular Genetic Testing for the Diagnosis of Haematological Malignancies
 Molecular Genetic Testing for the Diagnosis of Haematological Malignancies Dr Anthony Bench Haemto-Oncology Diagnostic Service Cambrıdge Unıversıty Hospitals NHS Foundatıon Trust Cambridge UK Molecular
Molecular Genetic Testing for the Diagnosis of Haematological Malignancies Dr Anthony Bench Haemto-Oncology Diagnostic Service Cambrıdge Unıversıty Hospitals NHS Foundatıon Trust Cambridge UK Molecular
The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep
 SUPPLEMENTARY INFORMATION The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep degradation associated with lymphocyte and dendritic cell hyperresponsiveness Jinyi Zhang, Naima
SUPPLEMENTARY INFORMATION The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep degradation associated with lymphocyte and dendritic cell hyperresponsiveness Jinyi Zhang, Naima
CHR POS REF OBS ALLELE BUILD CLINICAL_SIGNIFICANCE
 CHR POS REF OBS ALLELE BUILD CLINICAL_SIGNIFICANCE is_clinical dbsnp MITO GENE chr1 13273 G C heterozygous - - -. - DDX11L1 chr1 949654 A G Homozygous 52 - - rs8997 - ISG15 chr1 1021346 A G heterozygous
CHR POS REF OBS ALLELE BUILD CLINICAL_SIGNIFICANCE is_clinical dbsnp MITO GENE chr1 13273 G C heterozygous - - -. - DDX11L1 chr1 949654 A G Homozygous 52 - - rs8997 - ISG15 chr1 1021346 A G heterozygous
Supplementary Figure 1
 A B D Relative TAp73 mrna p73 Supplementary Figure 1 25 2 15 1 5 p63 _-tub. MDA-468 HCC1143 HCC38 SUM149 MDA-468 HCC1143 HCC38 SUM149 HCC-1937 MDA-MB-468 ΔNp63_ TAp73_ TAp73β E C Relative ΔNp63 mrna TAp73
A B D Relative TAp73 mrna p73 Supplementary Figure 1 25 2 15 1 5 p63 _-tub. MDA-468 HCC1143 HCC38 SUM149 MDA-468 HCC1143 HCC38 SUM149 HCC-1937 MDA-MB-468 ΔNp63_ TAp73_ TAp73β E C Relative ΔNp63 mrna TAp73
Nature Immunology: doi: /ni Supplementary Figure 1. Gene expression profile of CD4 + T cells and CTL responses in Bcl6-deficient mice.
 Supplementary Figure 1 Gene expression profile of CD4 + T cells and CTL responses in Bcl6-deficient mice. (a) Gene expression profile in the resting CD4 + T cells were analyzed by an Affymetrix microarray
Supplementary Figure 1 Gene expression profile of CD4 + T cells and CTL responses in Bcl6-deficient mice. (a) Gene expression profile in the resting CD4 + T cells were analyzed by an Affymetrix microarray
Bio 111 Study Guide Chapter 17 From Gene to Protein
 Bio 111 Study Guide Chapter 17 From Gene to Protein BEFORE CLASS: Reading: Read the introduction on p. 333, skip the beginning of Concept 17.1 from p. 334 to the bottom of the first column on p. 336, and
Bio 111 Study Guide Chapter 17 From Gene to Protein BEFORE CLASS: Reading: Read the introduction on p. 333, skip the beginning of Concept 17.1 from p. 334 to the bottom of the first column on p. 336, and
Supplementary Table e1. Clinical and genetic data on the 37 participants from the WUSM
 Supplementary Data Supplementary Table e1. Clinical and genetic data on the 37 participants from the WUSM cohort. Supplementary Table e2. Specificity, sensitivity and unadjusted ORs for glioma in participants
Supplementary Data Supplementary Table e1. Clinical and genetic data on the 37 participants from the WUSM cohort. Supplementary Table e2. Specificity, sensitivity and unadjusted ORs for glioma in participants
SUPPLEMENTARY FIGURE S1: nlp-22 is expressed in the RIA interneurons and is secreted. (a) An animal expressing both the RIA specific reporter
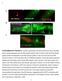 1 SUPPLEMENTARY FIGURE S1: nlp-22 is expressed in the RIA interneurons and is secreted. (a) An animal expressing both the RIA specific reporter Pglr-3:mCherry (red) and Pnlp-22:gfp (green) shows co-localization
1 SUPPLEMENTARY FIGURE S1: nlp-22 is expressed in the RIA interneurons and is secreted. (a) An animal expressing both the RIA specific reporter Pglr-3:mCherry (red) and Pnlp-22:gfp (green) shows co-localization
Juvenile and Chronic Myelo-Monocytic Leukemia
 Juvenile and Chronic Myelo-Monocytic Leukemia Haematopoietic stem cell Lympho-myeloid progenitor cell MEP CFU-GM lymphoid progenitor cell BFU-E CFU-MK CFU-E erythro CFU-M CFU-G CFU-T CFU-B MGK red blood
Juvenile and Chronic Myelo-Monocytic Leukemia Haematopoietic stem cell Lympho-myeloid progenitor cell MEP CFU-GM lymphoid progenitor cell BFU-E CFU-MK CFU-E erythro CFU-M CFU-G CFU-T CFU-B MGK red blood
Deliverable 2.1 List of relevant genetic variants for pre-emptive PGx testing
 GA N 668353 H2020 Research and Innovation Deliverable 2.1 List of relevant genetic variants for pre-emptive PGx testing WP N and Title: WP2 - Towards shared European Guidelines for PGx Lead beneficiary:
GA N 668353 H2020 Research and Innovation Deliverable 2.1 List of relevant genetic variants for pre-emptive PGx testing WP N and Title: WP2 - Towards shared European Guidelines for PGx Lead beneficiary:
Nature Getetics: doi: /ng.3471
 Supplementary Figure 1 Summary of exome sequencing data. ( a ) Exome tumor normal sample sizes for bladder cancer (BLCA), breast cancer (BRCA), carcinoid (CARC), chronic lymphocytic leukemia (CLLX), colorectal
Supplementary Figure 1 Summary of exome sequencing data. ( a ) Exome tumor normal sample sizes for bladder cancer (BLCA), breast cancer (BRCA), carcinoid (CARC), chronic lymphocytic leukemia (CLLX), colorectal
La biopsia liquida. Aldo Scarpa. Anatomia Patologica e ARC-NET Centro di Ricerca Applicata sul Cancro
 La biopsia liquida Aldo Scarpa Anatomia Patologica e ARC-NET Centro di Ricerca Applicata sul Cancro Azienda Ospedaliera Universitaria Integrata di Verona Obstacles to precision oncology Genomic heterogeneity
La biopsia liquida Aldo Scarpa Anatomia Patologica e ARC-NET Centro di Ricerca Applicata sul Cancro Azienda Ospedaliera Universitaria Integrata di Verona Obstacles to precision oncology Genomic heterogeneity
