Mouse Clec9a ORF sequence
|
|
|
- Lisa Page
- 5 years ago
- Views:
Transcription
1 Mouse Clec9a gene LOCUS NC_ bp DNA linear CON 1-JUL-27 DEFINITION Mus musculus chromosome 6, reference assembly (C57BL/6J). ACCESSION NC_72 REGION: Mouse Clec9a ORF sequence Exon 1 ATGCATGCGGAAGAAATATATACCTCTCTTCAGTGGGACATTCCTACCTCAGAGGCCTCTCAGAAGTGCCAATCCCCTAGCAAA TGTTCAG Exon2 GAGCATGGTGTGTTGTGACGATGATTTCCTGTGTGGTCTGTATGGGCTTGTTAGCAACGTCCATTTTCTTGGGCATCAAGT Exon 3 TCTTCCAGGTATCCTCTCTTGTCTTGGAGCAGCAGGAAAGACTCATCCAACAGGACACAGCATTGGTGAACCTTACACAGTGG CAGAGGAAATACACACTGGAATACTGCCAAGCCTTACTGCAGAGATCTCTCCATTCAG Exon 4 GCACAGATGCTTCTACTGGACCAGTTCTTCTGACCTCTCCACAGATGGTTCCACAGACCCTGGACAGCAAGGAAACAG Exon 5 GTAGTGACTGCAGCCCTTGTCCACACAACTGGATTCAGAATGGAAAAAGTTGTTACTATGTCTTTGAACGCTGGGAAATG Exon 6 TGGAACATCAGTAAGAAGAGCTGTTTAAAAGAGGGCGCTAGTCTCTTTCAAATAGACAGCAAAGAAGAAATGGAGTTCATCAGC AGTATAGGGAAACTCAAAGGAGGAAATAAATATTGGGTGGGAGTGTTTCAAGATGGAATCAGTGGATCTTGGTTCTGGGAAGA TGGCTCTTCTCCTCTCTCTGACTT Exon 7 GTTGCCTGCAGAAAGACAGCGATCAGCCGGCCAGATCTGTGGATACCTCAAAGATTCTACTCTCATCTCAGATAAGTGCGATA GCTGGAAATATTTTATCTGTGAGAAGAAGGCATTTGGATCCTGCATCTGA Mouse CLEC9a protein sequence. /Exon boundaries/ Cytoplasmic tail-transmembrane-neck-ctld
2 MHAEEIYTSLQWDIPTSEASQKCQSPSKCS/GAWCVVTMISCVVCMGLLATSIFLGIK/FFQVSSLVLEQQERLIQQDTALVNLTQW QRKYTLEYCQALLQRSLHS/GTDASTGPVLLTSPQMVPQTLDSKET/GSDCSPCPHNWIQNGKSCYYVFERWEM/WNISKKSCLK EGASLFQIDSKEEMEFISSIGKLKGGNKYWVGVFQDGISGSWFWEDGSSPLSD/LLPAERQRSAGQICGYLKDSTLISDKCDSWKY FICEKKAFGSCI
3 Human CLEC9A gene LOCUS NC_ bp DNA linear CON 3-AUG-26 DEFINITION Homo sapiens chromosome 12, reference assembly, complete sequence. ACCESSION NC_12 REGION: Human CLEC9a ORF sequence Exon 4 ATGCACGAGGAAGAAATATACACCTCTCTTCAGTGGGATAGCCCAGCACCAGACACTTACCAGAAATGTCTGTCTTC CAACAAATGTTCAG Exon 5 GAGCATGCTGTCTTGTGATGGTGATTTCATGTGTTTTCTGCATGGGATTATTAACAGCATCCATTTTCTTGGGCGTCAAGT Exon 6 TGTTGCAGGTGTCCACCATTGCGATGCAGCAGCAAGAAAAACTCATCCAACAAGAGAGGGCACTGCTAAACTTTACAGAATGG AAGAGAAGCTGTGCCCTTCAGATGAAATATTGCCAAGCCTTCATGCAAAACTCATTAAGTTCAG Exon 7 CCCATAACAGCAGTCCTTGTCCAAACAATTGGATTCAGAACAGAGAAAGTTGTTACTATGTCTCTGAAATTTGGAGCATTTGGC ACACCAGTCAAGAGAATTGTTTAAAGGAAGGTTCCACGCTGCTACAAATAGAGAGCAAAGAAGAAATG Exon 8 GATTTTATCACTGGCAGCTTGAGGAAGATTAAAGGAAGCTATGATTACTGGGTGGGGTTGTCTCAGGATGGACACAGCGGACG CTGGCTTTGGCAAGATGGCTCCTCTCCTTCTCCTGGCCT Exon 9 GTTGCCAGCAGAGAGATCCCAGTCAGCTAACCAAGTCTGTGGATACGTGAAAAGCAATTCCCTTCTTTCGTCTAACTGCAGCA CGTGGAAGTATTTTATCTGTGAGAAGTATGCGTTGAGATCCTCTGTCTGA Mouse CLEC9a protein sequence. /Exon boundaries/ Cytoplasmic tail-transmembrane-neck-ctld MHEEEIYTSLQWDSPAPDTYQKCLSSNKCS/GACCLVMVISCVFCMGLLTASIFLGVK/LLQVSTIAMQQQEKLIQQERALLNFTEW KRSCALQMKYCQAFMQNSLSS/AHNSSPCPNNWIQNRESCYYVSEIWSI/WHTSQENCLKEGSTLLQIESKEEMDFITGSLRKIKGS
4 YDYWVGLSQDGHSGRWLWQDGSSPSPG/LLPAERSQSANQVCGYVKSNSLLSSNCSTWKYFICEKYALRSSV
5 Fig. S2. Sequence alignment Mus m. 1 MHAEEIYTSLQWDIPTSEASQKCQSPSKCSGAWCVVTMISCVVCMGLLAT 5 Homo s. 1 MHEEEIYTSLQWDSPAPDTYQKCLSSNKCSGACCLVMVISCVFCMGLLTA 5 Rattus n. 1 MHEEEIYTSLQWDIPTSEASQKCPSLSKCPGTWCIVTVISCVVCVGLLAA 5 Macaca m. 1 MHEEEIYTSLQWDSPAPNTYQKCLSSNKCSGAWCLVMAISCIFCMGLLTA 5 Pan t. 1 MHEEEIYTSLQWDSPAPDTYQKCLSSNKCSGACCLVMVISCVFCMGLLTA 5 Canis f. 1 MQEEETYTSLRWDSPTPSFYQKHLSSTKYSGAWCLVTVITCILCVGSIAT 5 *. ** ****.** *. ** * * *. *.* *.*. *.*... Mus m. 51 SIFLGIKFFQVSSLVLEQQERLIQQDTALVNLTQWQRKYTLEY--CQALL 98 Homo s. 51 SIFLGVKLLQVSTIAMQQQEKLIQQERALLNFTEWKRSCALQMKYCQAFM 1 Rattus n. 51 SIFLGIKFSQVSSLVMEQRERLIRQDTALLNLTEWQRNHTLQLKSCQASL 1 Macaca m. 51 SIFLGVKLLQVSTIAMQQQEKLIQQERALLNFTEWKRSHVLQMKFCQTFM 1 Pan t. 51 SIFLGVKLLQVSTIAMQQQEKLIQQERALLNFTEWKRSCALQMKYCQAFM 1 Canis f. 51 SVFLGLKLFQVSTIAMKQREKLILQDRALLNFTQWERNHNLQMKYCQTLM 1 *.***.* ***... *.*.** *. **.* *.* * *. **.. Mus m. 99 QRSLHSGTDASTGPVLLTSPQMVPQTLDSKETGSDCSPCPHNWIQNGKSC 148 Homo s. 11 QNSLSS AHNSSPCPNNWIQNRESC 124 Rattus n. 11 QRSLRS GSNCNPCPPNWIQNGKSC 124 Macaca m. 11 QSSFSS AHNCSPCPNNWIQNRESC 124 Pan t. 11 QNSLSS AHNSSPCPNSWIQNRESC 124 Canis f. 11 QNSFSS AHNCSPCPDNWIQNGESC 124 * * *. *** **** ** Mus m. 149 YYVFERWEMWNISKKSCLKEGASLFQIDSKEEMEFISS-IGKLKGGNKYW 197 Homo s. 125 YYVSEIWSIWHTSQENCLKEGSTLLQIESKEEMDFITGSLRKIKGSYDYW 174 Rattus n. 125 YYAFDRWETWNNSKKSCLKEGDSLLQIDSKEEMEFINLSIWKLKGGYEYW 174 Macaca m. 125 YYVSEHWKIWHTSQENCLKEGSTLLQIESEEEMDFITGSLRKIRGSYDYW 174 Pan t. 125 YYVSEIWSIWHTSQENCLKEGSTLLQIESKEEMDFITGSLRKIKGSYDYW 174 Canis f. 125 YHVFENWKIWHTSKEDCLKEGSNLLQIDSKEEMDFITGSLKKVKSGFDYW 174 *. * *. *. ***** * **.* ***.**. *.. ** Mus m. 198 VGVFQDGISGSWFWEDGSSPLSDLLPAERQRSAGQICGYLKDSTLISDKC 247 Homo s. 175 VGLSQDGHSGRWLWQDGSSPSPGLLPAERSQSANQVCGYVKSNSLLSSNC 224 Rattus n. 175 VGVFQDGPSGSWFWEDGSSPLSDLLPTDRQLSASQICGYLKDHTLISDNC 224 Macaca m. 175 VGLSQDGHSGRWLWQDGSSPSPGLLPVEISQSTNQVCGYIKNSSLLSSNC 224 Pan t. 175 VGLSQDGHSGRWLWQDGSSPSPGLLPVERSQSANQVCGYMKSNSLLSSNC 224 Canis f. 175 VGLSQDGLSKPWLWQDGSSPSPDLSPVQTLQSTNQLCGYLKDKFLSSANC 224 **. *** * * *.***** * * *. *.***.* * * * Mus m. 248 DSWKYFICEKKAFGSCI 264 Homo s. 225 STWKYFICEKYALRSSV 241 Rattus n. 225 SNWKYFICEKKAFGSCI 241 Macaca m. 225 STWKYFICEKYALRSSV 241 Pan t. 225 STWKYFICEKYALRSSV 241 Canis f. 225 SIWKYFICEKYALRSSN 241 ******** * *.
6 Fig. S3 mdngr-1 L S VS CD4 + DC CD8α + DC pdc DN DC β-actin
7 Fig. S4A. Phylogenetic tree of Clec9a CTLD.5 Clec9a Clec7a Clec12a Clec12b Clec1a Clec1b Klrc2 Klrc1 Klrc3 Klrk1 Klrd1 Klre1 Gm156 Klri2 Klri1 Olr1 Klrg1 Klrg2 Clec2l Clec5a Klrb1f Klrb6 Klrb1d Klrb1a Klrb1c Klrb1b Cd D21Rik LOC67744 Clec2f Clec2i Clec2d Clec2g Clec2e Clec2h
8 Fig S4B. Percentage identity and similarity of Clec9a CTLD. Protein % id % sim Clec9a 1 1 Klrk Clec7a Clec12b Klre Gm Klrc Klrc Klrc Klri Clec1b Clec2e Klrb1d Klrb1f Klri Clec2h Clec2l Klrb1b Klrb1c Klrb1a Klrb Klrg Klrg Clec1a Klrd Olr LOC Clec2g Clec2d Clec2f Cd D21Rik Clec12a Clec5a Clec2i
9 Fig. S5 MFI rat IgG F H EGFP
10 Fig. S6. CD11c - CD4 + cdc CD8α + cdc DN cdc pdc % of Max CD11c rat IgG1 / DNGR-1 cdc.4.4 skin DC % of Max skin-dc CD8α - cdc CD8α + cdc 1 CD4 rat IgG1 / DNGR-1
11 Fig S CD11c IgG1-A αdngr-1-a αdec25-a488
12 A B H2Kb SIINFEKL % Specific lysis CD8 p <.1 2 nm 2nM Peptide (nm) S1 peptide + αcd IgG1 S1 + αcd4 peptide S1 + αcd4 IgG1 S1 + αcd4 DNGR-1 S1 + αcd αdngr-1 S1 + αcd % Tetramer + CD S1 peptide + αcd4 p <.5 IgG1 S1 αdngr-1 S1 + αcd4 + αcd4 Fig. S8
13 p =.3 Tumors per mouse IgG1-Endo + Adj αdngr-1-endo + Adj Fig. S9
14 Supplementary figure legends Figure S1. Mouse Clec9a and human CLEC9A sequences. Gene localization is indicated using Genbank nomenclature. The open reading frame (ORF) and predicted protein sequence is depicted. The exon boundaries are mapped onto the protein sequence and the protein domains are highlighted: cytoplasmic tail (red), transmembrane region (green), stalk region (black) and CTLD (blue). Figure S2. Sequence alignment of CLEC9 proteins in different species. CLEC9A sequences from Mus musculus, Homo sapiens, Rattus norvegicus, Macacca mulata, Pan troglodytes and Canis familiaris were aligned using Clustal W. Conserved features are highlighted: cytoplasmic tyrosine (red); transmembrane domain (grey); Asn81, putative site for N-glycosylation (green); putative Cys involved in stabilization of dimer (blue); Ser 14-16, putative site for glycosaminoglycan binding (green); conserved CTLD residues, including six Cys involved in intramolecular disulphide bonds (yellow). Identity is represented by an asterisk (*) and similarity with a dot (.). Figure S3. Distribution of mouse DNGR-1 transcripts. mrna from subsets of spleen DC were subjected to RT-PCR using mouse DNGR-1 specific primers (upper lanes) or β- actin primers (lower lanes). Arrows indicate the long (L), short (S) and very short (VS) isoforms detected for mouse DNGR-1. Figure S4. Phylogenetic analysis of mouse CLEC9A. (A) Phylogenetic tree of mouse CLEC9A. NK receptor-like lectins were identified by searching the mouse proteome with
15 the domain alignment sourced from NCBI's Conserved Domain Database (CDD).The identified proteins were aligned to Clec9a and the tree was generated in Clustal W using the CDD domain as a guide. The distance data represents the minimum number of substitutions required to convert one sequence into another. (B) Percentage identity and similarity was calculated from the alignment using MacBoxshade. Figure S5. Generation of rat anti-mouse DNGR-1 mabs. mab were generated as indicated in the Materials and Methods and were used for staining a mixture of DNGR-1- expressing B3Z cells (GFP positive) and parental cells (GFP negative). MFI of the GFP positive (DNGR-1 + ) and GFP negative (parental) is indicated for the rat IgG irrelevant control or the anti-dngr-1 1F6, 397 and 7H11 mabs. Figure S6. Expression of DNGR-1 in peripheral lymph nodes cells. Analysis of DNGR-1 expression (blue) compared to the isotype control (red) in the indicated subsets of peripheral lymph nodes cells. Upper and lower panels represent different experiments comparing DNGR-1 expression in blood-derived DC subsets and skin-derived DC subsets (defined as shown on the dot plot), respectively. Figure S7. Comparison of in vivo labeling with anti-dngr-1 and anti-dec-25 mabs. (A) Mice were injected i.v. with 1 µg of Alexa-488 conjugated anti-dngr-1 (7H11), anti-dec-25 (NLDC-145) or isotype-matched control (rat IgG1) and lymph nodes were analyzed one day later. Dot plots show CD11c versus Alexa488 in anti-dec-
16 25 (right panel), anti-dngr-1 (middle panel) or rat IgG1 (left panel) injected mice. Numbers represent % events in the indicated quadrant. Figure S8. Comparison of anti-ova CTL priming by free OVA peptide versus peptide targeted via anti-dngr-1. 2µg S1 conjugated anti-dngr-1 or rat IgG1 isotype-matched control mab or 2µg of free S1 peptide (1 fold excess compared to the amount present in 2µg of the antibody conjugates) were injected s.c. with anti-cd4 (25 µg). Target cells were injected five days later and mice analyzed on day 6 as in Figure 6A. (A) In vivo CTL activity as measured by target cell elimination. Graph shows mean ± SEM of % specific lysis in one experiment of two (n=3 mice/group). All groups are shown but the only one in which killing was detectable was that receiving anti-dngr-1- S1 + anti-cd4. (B) H-2K b -SIINFEKL tetramer staining of splenocytes. Left panel: representative dot plots of tetramer staining vs. CD8 in gated CD8 + CD3 + T cells. Right panel: frequency of tetramer + CD8 + T cells in one experiment of two (n=3 mice/group). p values were calculated using Student s t test. Figure S9. Immunoprophylaxis of B16 melanoma via targeting of tumor antigens to DNGR-1. Experiments were carried out as in Figure 8 except that the vaccine was given one day prior to infusion of B16 cells. Data show the number of lung tumors per mouse. Data are pooled from two independent experiments (n=7 mice/group) and each point represents one mouse. p values were calculated using the Mann Whitney U test.
SUPPLEMENTARY INFORMATION
 doi: 1.138/nature775 4 O.D. (595-655) 3 1 -ζ no antibody isotype ctrl Plated Soluble 1F6 397 7H11 Supplementary Figure 1 Soluble and plated anti- Abs induce -! signalling. B3Z cells stably expressing!
doi: 1.138/nature775 4 O.D. (595-655) 3 1 -ζ no antibody isotype ctrl Plated Soluble 1F6 397 7H11 Supplementary Figure 1 Soluble and plated anti- Abs induce -! signalling. B3Z cells stably expressing!
Nature Immunology: doi: /ni Supplementary Figure 1. Gene expression profile of CD4 + T cells and CTL responses in Bcl6-deficient mice.
 Supplementary Figure 1 Gene expression profile of CD4 + T cells and CTL responses in Bcl6-deficient mice. (a) Gene expression profile in the resting CD4 + T cells were analyzed by an Affymetrix microarray
Supplementary Figure 1 Gene expression profile of CD4 + T cells and CTL responses in Bcl6-deficient mice. (a) Gene expression profile in the resting CD4 + T cells were analyzed by an Affymetrix microarray
Dendritic cells in cancer immunotherapy Aimin Jiang
 Dendritic cells in cancer immunotherapy Aimin Jiang Feb. 11, 2014 Dendritic cells at the interface of innate and adaptive immune responses Dendritic cells: initiators of adaptive immune responses Dendritic
Dendritic cells in cancer immunotherapy Aimin Jiang Feb. 11, 2014 Dendritic cells at the interface of innate and adaptive immune responses Dendritic cells: initiators of adaptive immune responses Dendritic
Supplemental Figure 1
 Supplemental Figure 1 1a 1c PD-1 MFI fold change 6 5 4 3 2 1 IL-1α IL-2 IL-4 IL-6 IL-1 IL-12 IL-13 IL-15 IL-17 IL-18 IL-21 IL-23 IFN-α Mut Human PD-1 promoter SBE-D 5 -GTCTG- -1.2kb SBE-P -CAGAC- -1.kb
Supplemental Figure 1 1a 1c PD-1 MFI fold change 6 5 4 3 2 1 IL-1α IL-2 IL-4 IL-6 IL-1 IL-12 IL-13 IL-15 IL-17 IL-18 IL-21 IL-23 IFN-α Mut Human PD-1 promoter SBE-D 5 -GTCTG- -1.2kb SBE-P -CAGAC- -1.kb
Supplementary Figure S1. PTPN2 levels are not altered in proliferating CD8+ T cells. Lymph node (LN) CD8+ T cells from C57BL/6 mice were stained with
 Supplementary Figure S1. PTPN2 levels are not altered in proliferating CD8+ T cells. Lymph node (LN) CD8+ T cells from C57BL/6 mice were stained with CFSE and stimulated with plate-bound α-cd3ε (10µg/ml)
Supplementary Figure S1. PTPN2 levels are not altered in proliferating CD8+ T cells. Lymph node (LN) CD8+ T cells from C57BL/6 mice were stained with CFSE and stimulated with plate-bound α-cd3ε (10µg/ml)
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Supplementary Figure 1: Chemokine receptor expression profiles of CCR6 + and CCR6 - CD4 + IL-17A +/ex and Treg cells. Quantitative PCR analysis of chemokine receptor transcript abundance
SUPPLEMENTARY FIGURES Supplementary Figure 1: Chemokine receptor expression profiles of CCR6 + and CCR6 - CD4 + IL-17A +/ex and Treg cells. Quantitative PCR analysis of chemokine receptor transcript abundance
Supporting Information Table of Contents
 Supporting Information Table of Contents Supporting Information Figure 1 Page 2 Supporting Information Figure 2 Page 4 Supporting Information Figure 3 Page 5 Supporting Information Figure 4 Page 6 Supporting
Supporting Information Table of Contents Supporting Information Figure 1 Page 2 Supporting Information Figure 2 Page 4 Supporting Information Figure 3 Page 5 Supporting Information Figure 4 Page 6 Supporting
Supplemental Figure 1. Cell-bound Cetuximab reduces EGFR staining intensity. Blood
 Antibody-mediated depletion of CD19-CAR T cells Supplemental 1 Supplemental Materials Supplemental Figure 1. Supplemental Figure 1. Cell-bound Cetuximab reduces EGFR staining intensity. Blood cells were
Antibody-mediated depletion of CD19-CAR T cells Supplemental 1 Supplemental Materials Supplemental Figure 1. Supplemental Figure 1. Cell-bound Cetuximab reduces EGFR staining intensity. Blood cells were
activation with anti-cd3/cd28 beads and 3d following transduction. Supplemental Figure 2 shows
 Supplemental Data Supplemental Figure 1 compares CXCR4 expression in untreated CD8 + T cells, following activation with anti-cd3/cd28 beads and 3d following transduction. Supplemental Figure 2 shows the
Supplemental Data Supplemental Figure 1 compares CXCR4 expression in untreated CD8 + T cells, following activation with anti-cd3/cd28 beads and 3d following transduction. Supplemental Figure 2 shows the
SUPPLEMENTARY INFORMATION
 doi:1.138/nature1554 a TNF-α + in CD4 + cells [%] 1 GF SPF 6 b IL-1 + in CD4 + cells [%] 5 4 3 2 1 Supplementary Figure 1. Effect of microbiota on cytokine profiles of T cells in GALT. Frequencies of TNF-α
doi:1.138/nature1554 a TNF-α + in CD4 + cells [%] 1 GF SPF 6 b IL-1 + in CD4 + cells [%] 5 4 3 2 1 Supplementary Figure 1. Effect of microbiota on cytokine profiles of T cells in GALT. Frequencies of TNF-α
Supplementary Figure 1. Using DNA barcode-labeled MHC multimers to generate TCR fingerprints
 Supplementary Figure 1 Using DNA barcode-labeled MHC multimers to generate TCR fingerprints (a) Schematic overview of the workflow behind a TCR fingerprint. Each peptide position of the original peptide
Supplementary Figure 1 Using DNA barcode-labeled MHC multimers to generate TCR fingerprints (a) Schematic overview of the workflow behind a TCR fingerprint. Each peptide position of the original peptide
SUPPLEMENTARY INFORMATION
 doi: 1.138/nature89 IFN- (ng ml ) 5 4 3 1 Splenocytes NS IFN- (ng ml ) 6 4 Lymph node cells NS Nfkbiz / Nfkbiz / Nfkbiz / Nfkbiz / IL- (ng ml ) 3 1 Splenocytes IL- (ng ml ) 1 8 6 4 *** ** Lymph node cells
doi: 1.138/nature89 IFN- (ng ml ) 5 4 3 1 Splenocytes NS IFN- (ng ml ) 6 4 Lymph node cells NS Nfkbiz / Nfkbiz / Nfkbiz / Nfkbiz / IL- (ng ml ) 3 1 Splenocytes IL- (ng ml ) 1 8 6 4 *** ** Lymph node cells
Supplementary Figure 1. Generation of knockin mice expressing L-selectinN138G. (a) Schematics of the Sellg allele (top), the targeting vector, the
 Supplementary Figure 1. Generation of knockin mice expressing L-selectinN138G. (a) Schematics of the Sellg allele (top), the targeting vector, the targeted allele in ES cells, and the mutant allele in
Supplementary Figure 1. Generation of knockin mice expressing L-selectinN138G. (a) Schematics of the Sellg allele (top), the targeting vector, the targeted allele in ES cells, and the mutant allele in
Supplemental Materials for. Effects of sphingosine-1-phosphate receptor 1 phosphorylation in response to. FTY720 during neuroinflammation
 Supplemental Materials for Effects of sphingosine-1-phosphate receptor 1 phosphorylation in response to FTY7 during neuroinflammation This file includes: Supplemental Table 1. EAE clinical parameters of
Supplemental Materials for Effects of sphingosine-1-phosphate receptor 1 phosphorylation in response to FTY7 during neuroinflammation This file includes: Supplemental Table 1. EAE clinical parameters of
Supplementary figure legends
 Supplementary figure legends SUPPLEMENTRY FIGURE S1. Lentiviral construct. Schematic representation of the PCR fragment encompassing the genomic locus of mir-33a that was introduced in the lentiviral construct.
Supplementary figure legends SUPPLEMENTRY FIGURE S1. Lentiviral construct. Schematic representation of the PCR fragment encompassing the genomic locus of mir-33a that was introduced in the lentiviral construct.
Supplementary Figure 1.
 Supplementary Figure 1. Female Pro-ins2 -/- mice at 5-6 weeks of age were either inoculated i.p. with a single dose of CVB4 (1x10 5 PFU/mouse) or PBS and treated with αgalcer or control vehicle. On day
Supplementary Figure 1. Female Pro-ins2 -/- mice at 5-6 weeks of age were either inoculated i.p. with a single dose of CVB4 (1x10 5 PFU/mouse) or PBS and treated with αgalcer or control vehicle. On day
Eosinophils are required. for the maintenance of plasma cells in the bone marrow
 Eosinophils are required for the maintenance of plasma cells in the bone marrow Van Trung Chu, Anja Fröhlich, Gudrun Steinhauser, Tobias Scheel, Toralf Roch, Simon Fillatreau, James J. Lee, Max Löhning
Eosinophils are required for the maintenance of plasma cells in the bone marrow Van Trung Chu, Anja Fröhlich, Gudrun Steinhauser, Tobias Scheel, Toralf Roch, Simon Fillatreau, James J. Lee, Max Löhning
Nature Immunology: doi: /ni Supplementary Figure 1. Id2 and Id3 define polyclonal T H 1 and T FH cell subsets.
 Supplementary Figure 1 Id2 and Id3 define polyclonal T H 1 and T FH cell subsets. Id2 YFP/+ (a) or Id3 GFP/+ (b) mice were analyzed 7 days after LCMV infection. T H 1 (SLAM + CXCR5 or CXCR5 PD-1 ), T FH
Supplementary Figure 1 Id2 and Id3 define polyclonal T H 1 and T FH cell subsets. Id2 YFP/+ (a) or Id3 GFP/+ (b) mice were analyzed 7 days after LCMV infection. T H 1 (SLAM + CXCR5 or CXCR5 PD-1 ), T FH
Supplementary Fig. 1 p38 MAPK negatively regulates DC differentiation. (a) Western blot analysis of p38 isoform expression in BM cells, immature DCs
 Supplementary Fig. 1 p38 MAPK negatively regulates DC differentiation. (a) Western blot analysis of p38 isoform expression in BM cells, immature DCs (idcs) and mature DCs (mdcs). A myeloma cell line expressing
Supplementary Fig. 1 p38 MAPK negatively regulates DC differentiation. (a) Western blot analysis of p38 isoform expression in BM cells, immature DCs (idcs) and mature DCs (mdcs). A myeloma cell line expressing
Supplementary Figure 1
 S U P P L E M E N TA R Y I N F O R M AT I O N DOI: 10.1038/ncb2896 Supplementary Figure 1 Supplementary Figure 1. Sequence alignment of TERB1 homologs in vertebrates. M. musculus TERB1 was derived from
S U P P L E M E N TA R Y I N F O R M AT I O N DOI: 10.1038/ncb2896 Supplementary Figure 1 Supplementary Figure 1. Sequence alignment of TERB1 homologs in vertebrates. M. musculus TERB1 was derived from
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/9/430/ra57/dc1 Supplementary Materials for The 4E-BP eif4e axis promotes rapamycinsensitive growth and proliferation in lymphocytes Lomon So, Jongdae Lee, Miguel
www.sciencesignaling.org/cgi/content/full/9/430/ra57/dc1 Supplementary Materials for The 4E-BP eif4e axis promotes rapamycinsensitive growth and proliferation in lymphocytes Lomon So, Jongdae Lee, Miguel
Supporting Information
 Supporting Information McCullough et al. 10.1073/pnas.0801567105 A α10 α8 α9 N α7 α6 α5 C β2 β1 α4 α3 α2 α1 C B N C Fig. S1. ALIX Bro1 in complex with the C-terminal CHMP4A helix. (A) Ribbon diagram showing
Supporting Information McCullough et al. 10.1073/pnas.0801567105 A α10 α8 α9 N α7 α6 α5 C β2 β1 α4 α3 α2 α1 C B N C Fig. S1. ALIX Bro1 in complex with the C-terminal CHMP4A helix. (A) Ribbon diagram showing
Hands-On Ten The BRCA1 Gene and Protein
 Hands-On Ten The BRCA1 Gene and Protein Objective: To review transcription, translation, reading frames, mutations, and reading files from GenBank, and to review some of the bioinformatics tools, such
Hands-On Ten The BRCA1 Gene and Protein Objective: To review transcription, translation, reading frames, mutations, and reading files from GenBank, and to review some of the bioinformatics tools, such
The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep
 SUPPLEMENTARY INFORMATION The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep degradation associated with lymphocyte and dendritic cell hyperresponsiveness Jinyi Zhang, Naima
SUPPLEMENTARY INFORMATION The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep degradation associated with lymphocyte and dendritic cell hyperresponsiveness Jinyi Zhang, Naima
Spleen. mlns. E Spleen 4.1. mlns. Spleen. mlns. Mock 17. Mock CD8 HIV-1 CD38 HLA-DR. Ki67. Spleen. Spleen. mlns. Cheng et al. Fig.
 C D E F Mock 17 Mock 4.1 CD38 57 CD8 23.7 HLA-DR Ki67 G H I Cheng et al. Fig.S1 Supplementary Figure 1. persistent infection leads to human T cell depletion and hyper-immune activation. Humanized mice
C D E F Mock 17 Mock 4.1 CD38 57 CD8 23.7 HLA-DR Ki67 G H I Cheng et al. Fig.S1 Supplementary Figure 1. persistent infection leads to human T cell depletion and hyper-immune activation. Humanized mice
SUPPLEMENTARY INFORMATION. Supp. Fig. 1. Autoimmunity. Tolerance APC APC. T cell. T cell. doi: /nature06253 ICOS ICOS TCR CD28 TCR CD28
 Supp. Fig. 1 a APC b APC ICOS ICOS TCR CD28 mir P TCR CD28 P T cell Tolerance Roquin WT SG Icos mrna T cell Autoimmunity Roquin M199R SG Icos mrna www.nature.com/nature 1 Supp. Fig. 2 CD4 + CD44 low CD4
Supp. Fig. 1 a APC b APC ICOS ICOS TCR CD28 mir P TCR CD28 P T cell Tolerance Roquin WT SG Icos mrna T cell Autoimmunity Roquin M199R SG Icos mrna www.nature.com/nature 1 Supp. Fig. 2 CD4 + CD44 low CD4
Supplementary Figure 1. mrna expression of chitinase and chitinase-like protein in splenic immune cells. Each splenic immune cell population was
 Supplementary Figure 1. mrna expression of chitinase and chitinase-like protein in splenic immune cells. Each splenic immune cell population was sorted by FACS. Surface markers for sorting were CD11c +
Supplementary Figure 1. mrna expression of chitinase and chitinase-like protein in splenic immune cells. Each splenic immune cell population was sorted by FACS. Surface markers for sorting were CD11c +
Supplementary Table 1 Clinicopathological characteristics of 35 patients with CRCs
 Supplementary Table Clinicopathological characteristics of 35 patients with CRCs Characteristics Type-A CRC Type-B CRC P value Sex Male / Female 9 / / 8.5 Age (years) Median (range) 6. (9 86) 6.5 (9 76).95
Supplementary Table Clinicopathological characteristics of 35 patients with CRCs Characteristics Type-A CRC Type-B CRC P value Sex Male / Female 9 / / 8.5 Age (years) Median (range) 6. (9 86) 6.5 (9 76).95
well for 2 h at rt. Each dot represents an individual mouse and bar is the mean ±
 Supplementary data: Control DC Blimp-1 ko DC 8 6 4 2-2 IL-1β p=.5 medium 8 6 4 2 IL-2 Medium p=.16 8 6 4 2 IL-6 medium p=.3 5 4 3 2 1-1 medium IL-1 n.s. 25 2 15 1 5 IL-12(p7) p=.15 5 IFNγ p=.65 4 3 2 1
Supplementary data: Control DC Blimp-1 ko DC 8 6 4 2-2 IL-1β p=.5 medium 8 6 4 2 IL-2 Medium p=.16 8 6 4 2 IL-6 medium p=.3 5 4 3 2 1-1 medium IL-1 n.s. 25 2 15 1 5 IL-12(p7) p=.15 5 IFNγ p=.65 4 3 2 1
Supplementary Figure 1. CFTR protein structure and domain architecture.
 A Plasma Membrane NH ₂ COOH Supplementary Figure. CFT protein structure and domain architecture. (A) Open state CFT homology model, ribbon representation from Serohijos et al. 8 PNAS 5:356. CFT domains
A Plasma Membrane NH ₂ COOH Supplementary Figure. CFT protein structure and domain architecture. (A) Open state CFT homology model, ribbon representation from Serohijos et al. 8 PNAS 5:356. CFT domains
Pearson r = P (one-tailed) = n = 9
 8F4-Specific Lysis, % 1 UPN1 UPN3 8 UPN7 6 Pearson r =.69 UPN2 UPN5 P (one-tailed) =.192 4 UPN8 n = 9 2 UPN9 UPN4 UPN6 5 1 15 2 25 8 8F4, % Max MFI Supplementary Figure S1. AML samples UPN1-UPN9 show variable
8F4-Specific Lysis, % 1 UPN1 UPN3 8 UPN7 6 Pearson r =.69 UPN2 UPN5 P (one-tailed) =.192 4 UPN8 n = 9 2 UPN9 UPN4 UPN6 5 1 15 2 25 8 8F4, % Max MFI Supplementary Figure S1. AML samples UPN1-UPN9 show variable
and follicular helper T cells is Egr2-dependent. (a) Diagrammatic representation of the
 Supplementary Figure 1. LAG3 + Treg-mediated regulation of germinal center B cells and follicular helper T cells is Egr2-dependent. (a) Diagrammatic representation of the experimental protocol for the
Supplementary Figure 1. LAG3 + Treg-mediated regulation of germinal center B cells and follicular helper T cells is Egr2-dependent. (a) Diagrammatic representation of the experimental protocol for the
SUPPLEMENTARY MATERIAL
 SYPPLEMENTARY FIGURE LEGENDS SUPPLEMENTARY MATERIAL Figure S1. Phylogenic studies of the mir-183/96/182 cluster and 3 -UTR of Casp2. (A) Genomic arrangement of the mir-183/96/182 cluster in vertebrates.
SYPPLEMENTARY FIGURE LEGENDS SUPPLEMENTARY MATERIAL Figure S1. Phylogenic studies of the mir-183/96/182 cluster and 3 -UTR of Casp2. (A) Genomic arrangement of the mir-183/96/182 cluster in vertebrates.
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12005 a S Ψ ΨΨ ΨΨ ΨΨ Ψ Ψ Ψ Ψ Ψ ΨΨ Ψ Ψ ΨΨΨΨΨΨ S1 (a.a. 1 751) S2 (a.a. 752 1353) S1 Fc S1 (a.a. 1-747) Fc b HCoV-EMC-S1-Fc SARS-CoV-S1-Fc HCoV-EMC-S1-Fc SARS-CoV-S1-Fc kda - 170 - - 130
doi:10.1038/nature12005 a S Ψ ΨΨ ΨΨ ΨΨ Ψ Ψ Ψ Ψ Ψ ΨΨ Ψ Ψ ΨΨΨΨΨΨ S1 (a.a. 1 751) S2 (a.a. 752 1353) S1 Fc S1 (a.a. 1-747) Fc b HCoV-EMC-S1-Fc SARS-CoV-S1-Fc HCoV-EMC-S1-Fc SARS-CoV-S1-Fc kda - 170 - - 130
Supplementary Information
 Supplementary Information Supplementary Figure 1! a! b! Nfatc1!! Nfatc1"! P1! P2! pa1! pa2! ex1! ex2! exons 3-9! ex1! ex11!!" #" Nfatc1A!!" Nfatc1B! #"!" Nfatc1C! #" DN1! DN2! DN1!!A! #A!!B! #B!!C! #C!!A!
Supplementary Information Supplementary Figure 1! a! b! Nfatc1!! Nfatc1"! P1! P2! pa1! pa2! ex1! ex2! exons 3-9! ex1! ex11!!" #" Nfatc1A!!" Nfatc1B! #"!" Nfatc1C! #" DN1! DN2! DN1!!A! #A!!B! #B!!C! #C!!A!
Supplementary Materials for
 www.sciencemag.org/content/348/6241/aaa825/suppl/dc1 Supplementary Materials for A mucosal vaccine against Chlamydia trachomatis generates two waves of protective memory T cells Georg Stary,* Andrew Olive,
www.sciencemag.org/content/348/6241/aaa825/suppl/dc1 Supplementary Materials for A mucosal vaccine against Chlamydia trachomatis generates two waves of protective memory T cells Georg Stary,* Andrew Olive,
(A) RT-PCR for components of the Shh/Gli pathway in normal fetus cell (MRC-5) and a
 Supplementary figure legends Supplementary Figure 1. Expression of Shh signaling components in a panel of gastric cancer. (A) RT-PCR for components of the Shh/Gli pathway in normal fetus cell (MRC-5) and
Supplementary figure legends Supplementary Figure 1. Expression of Shh signaling components in a panel of gastric cancer. (A) RT-PCR for components of the Shh/Gli pathway in normal fetus cell (MRC-5) and
SUPPORTING INFORMATIONS
 SUPPORTING INFORMATIONS Mice MT/ret RetCD3ε KO α-cd25 treated MT/ret Age 1 month 3 mnths 6 months 1 month 3 months 6 months 1 month 3 months 6 months 2/87 Survival 87/87 incidence of 17/87 1 ary tumor
SUPPORTING INFORMATIONS Mice MT/ret RetCD3ε KO α-cd25 treated MT/ret Age 1 month 3 mnths 6 months 1 month 3 months 6 months 1 month 3 months 6 months 2/87 Survival 87/87 incidence of 17/87 1 ary tumor
Supplemental Figure 1. Activated splenocytes upregulate Serpina3g and Serpina3f expression.
 Relative Serpin expression 25 2 15 1 5 Serpina3f 1 2 3 4 5 6 8 6 4 2 Serpina3g 1 2 3 4 5 6 C57BL/6 DBA/2 Supplemental Figure 1. Activated splenocytes upregulate Serpina3g and Serpina3f expression. Splenocytes
Relative Serpin expression 25 2 15 1 5 Serpina3f 1 2 3 4 5 6 8 6 4 2 Serpina3g 1 2 3 4 5 6 C57BL/6 DBA/2 Supplemental Figure 1. Activated splenocytes upregulate Serpina3g and Serpina3f expression. Splenocytes
Figure S1. (A) SDS-PAGE separation of GST-fusion proteins purified from E.coli BL21 strain is shown. An equal amount of GST-tag control, LRRK2 LRR
 Figure S1. (A) SDS-PAGE separation of GST-fusion proteins purified from E.coli BL21 strain is shown. An equal amount of GST-tag control, LRRK2 LRR and LRRK2 WD40 GST fusion proteins (5 µg) were loaded
Figure S1. (A) SDS-PAGE separation of GST-fusion proteins purified from E.coli BL21 strain is shown. An equal amount of GST-tag control, LRRK2 LRR and LRRK2 WD40 GST fusion proteins (5 µg) were loaded
% of live splenocytes. STAT5 deletion. (open shapes) % ROSA + % floxed
 Supp. Figure 1. a 14 1 1 8 6 spleen cells (x1 6 ) 16 % of live splenocytes 5 4 3 1 % of live splenocytes 8 6 4 b 1 1 c % of CD11c + splenocytes (closed shapes) 8 6 4 8 6 4 % ROSA + (open shapes) % floxed
Supp. Figure 1. a 14 1 1 8 6 spleen cells (x1 6 ) 16 % of live splenocytes 5 4 3 1 % of live splenocytes 8 6 4 b 1 1 c % of CD11c + splenocytes (closed shapes) 8 6 4 8 6 4 % ROSA + (open shapes) % floxed
Nature Medicine: doi: /nm.3922
 Title: Glucocorticoid-induced tumor necrosis factor receptor-related protein co-stimulation facilitates tumor regression by inducing IL-9-producing helper T cells Authors: Il-Kyu Kim, Byung-Seok Kim, Choong-Hyun
Title: Glucocorticoid-induced tumor necrosis factor receptor-related protein co-stimulation facilitates tumor regression by inducing IL-9-producing helper T cells Authors: Il-Kyu Kim, Byung-Seok Kim, Choong-Hyun
CD80 and PD-L2 define functionally distinct memory B cell subsets that are. Griselda V Zuccarino-Catania, Saheli Sadanand, Florian J Weisel, Mary M
 Supplementary Figures CD8 and PD-L define functionally distinct memory B cell subsets that are independent of antibody isotype Running title: Memory B Cell Subset Function Griselda V Zuccarino-Catania,
Supplementary Figures CD8 and PD-L define functionally distinct memory B cell subsets that are independent of antibody isotype Running title: Memory B Cell Subset Function Griselda V Zuccarino-Catania,
SUPPLEMENTARY MATERIAL
 SUPPLEMENTARY MATERIAL IL-1 signaling modulates activation of STAT transcription factors to antagonize retinoic acid signaling and control the T H 17 cell it reg cell balance Rajatava Basu 1,5, Sarah K.
SUPPLEMENTARY MATERIAL IL-1 signaling modulates activation of STAT transcription factors to antagonize retinoic acid signaling and control the T H 17 cell it reg cell balance Rajatava Basu 1,5, Sarah K.
pplementary Figur Supplementary Figure 1. a.
 pplementary Figur Supplementary Figure 1. a. Quantification by RT-qPCR of YFV-17D and YFV-17D pol- (+) RNA in the supernatant of cultured Huh7.5 cells following viral RNA electroporation of respective
pplementary Figur Supplementary Figure 1. a. Quantification by RT-qPCR of YFV-17D and YFV-17D pol- (+) RNA in the supernatant of cultured Huh7.5 cells following viral RNA electroporation of respective
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Supplementary Figure 1. NAFL enhanced immunity of other vaccines (a) An over-the-counter, hand-held non-ablative fractional laser (NAFL).
 Supplementary Figure 1. NAFL enhanced immunity of other vaccines (a) An over-the-counter, hand-held non-ablative fractional laser (NAFL). (b) Depiction of a MTZ array generated by NAFL. (c-e) IgG production
Supplementary Figure 1. NAFL enhanced immunity of other vaccines (a) An over-the-counter, hand-held non-ablative fractional laser (NAFL). (b) Depiction of a MTZ array generated by NAFL. (c-e) IgG production
B220 CD4 CD8. Figure 1. Confocal Image of Sensitized HLN. Representative image of a sensitized HLN
 B220 CD4 CD8 Natarajan et al., unpublished data Figure 1. Confocal Image of Sensitized HLN. Representative image of a sensitized HLN showing B cell follicles and T cell areas. 20 µm thick. Image of magnification
B220 CD4 CD8 Natarajan et al., unpublished data Figure 1. Confocal Image of Sensitized HLN. Representative image of a sensitized HLN showing B cell follicles and T cell areas. 20 µm thick. Image of magnification
SUPPLEMENTARY INFORMATION
 Supplemental Figure 1. Furin is efficiently deleted in CD4 + and CD8 + T cells. a, Western blot for furin and actin proteins in CD4cre-fur f/f and fur f/f Th1 cells. Wild-type and furin-deficient CD4 +
Supplemental Figure 1. Furin is efficiently deleted in CD4 + and CD8 + T cells. a, Western blot for furin and actin proteins in CD4cre-fur f/f and fur f/f Th1 cells. Wild-type and furin-deficient CD4 +
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/4/3/eaaq0762/dc1 Supplementary Materials for Structures of monomeric and oligomeric forms of the Toxoplasma gondii perforin-like protein 1 Tao Ni, Sophie I. Williams,
advances.sciencemag.org/cgi/content/full/4/3/eaaq0762/dc1 Supplementary Materials for Structures of monomeric and oligomeric forms of the Toxoplasma gondii perforin-like protein 1 Tao Ni, Sophie I. Williams,
Hua Tang, Weiping Cao, Sudhir Pai Kasturi, Rajesh Ravindran, Helder I Nakaya, Kousik
 SUPPLEMENTARY FIGURES 1-19 T H 2 response to cysteine-proteases requires dendritic cell-basophil cooperation via ROS mediated signaling Hua Tang, Weiping Cao, Sudhir Pai Kasturi, Rajesh Ravindran, Helder
SUPPLEMENTARY FIGURES 1-19 T H 2 response to cysteine-proteases requires dendritic cell-basophil cooperation via ROS mediated signaling Hua Tang, Weiping Cao, Sudhir Pai Kasturi, Rajesh Ravindran, Helder
Bioinformatics Laboratory Exercise
 Bioinformatics Laboratory Exercise Biology is in the midst of the genomics revolution, the application of robotic technology to generate huge amounts of molecular biology data. Genomics has led to an explosion
Bioinformatics Laboratory Exercise Biology is in the midst of the genomics revolution, the application of robotic technology to generate huge amounts of molecular biology data. Genomics has led to an explosion
Kerdiles et al - Figure S1
 Kerdiles et al - Figure S1 a b Homo sapiens T B ce ce l ls c l M ls ac r PM oph N ag es Mus musculus Foxo1 PLCγ Supplementary Figure 1 Foxo1 expression pattern is conserved between mouse and human. (a)
Kerdiles et al - Figure S1 a b Homo sapiens T B ce ce l ls c l M ls ac r PM oph N ag es Mus musculus Foxo1 PLCγ Supplementary Figure 1 Foxo1 expression pattern is conserved between mouse and human. (a)
HD1 (FLU) HD2 (EBV) HD2 (FLU)
 ramer staining + anti-pe beads ramer staining a HD1 (FLU) HD2 (EBV) HD2 (FLU).73.11.56.46.24 1.12 b CD127 + c CD127 + d CD127 - e CD127 - PD1 - PD1 + PD1 + PD1-1 1 1 1 %CD127 + PD1-8 6 4 2 + anti-pe %CD127
ramer staining + anti-pe beads ramer staining a HD1 (FLU) HD2 (EBV) HD2 (FLU).73.11.56.46.24 1.12 b CD127 + c CD127 + d CD127 - e CD127 - PD1 - PD1 + PD1 + PD1-1 1 1 1 %CD127 + PD1-8 6 4 2 + anti-pe %CD127
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3073 LATS2 Binding ability to SIAH2 Degradation by SIAH2 1-160 161-402 403-480 -/ 481-666 1-666 - - 667-1088 -/ 1-0 Supplementary Figure 1 Schematic drawing of LATS2 deletion mutants and
DOI: 10.1038/ncb3073 LATS2 Binding ability to SIAH2 Degradation by SIAH2 1-160 161-402 403-480 -/ 481-666 1-666 - - 667-1088 -/ 1-0 Supplementary Figure 1 Schematic drawing of LATS2 deletion mutants and
CD44
 MR1-5-OP-RU CD24 CD24 CD44 MAIT cells 2.78 11.2 WT RORγt- GFP reporter 1 5 1 4 1 3 2.28 1 5 1 4 1 3 4.8 1.6 8.1 1 5 1 4 1 3 1 5 1 4 1 3 3.7 3.21 8.5 61.7 1 2 1 3 1 4 1 5 TCRβ 2 1 1 3 1 4 1 5 CD44 1 2 GFP
MR1-5-OP-RU CD24 CD24 CD44 MAIT cells 2.78 11.2 WT RORγt- GFP reporter 1 5 1 4 1 3 2.28 1 5 1 4 1 3 4.8 1.6 8.1 1 5 1 4 1 3 1 5 1 4 1 3 3.7 3.21 8.5 61.7 1 2 1 3 1 4 1 5 TCRβ 2 1 1 3 1 4 1 5 CD44 1 2 GFP
Supplementary Figure 1 IL-27 IL
 Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
sequences of a styx mutant reveals a T to A transversion in the donor splice site of intron 5
 sfigure 1 Styx mutant mice recapitulate the phenotype of SHIP -/- mice. (A) Analysis of the genomic sequences of a styx mutant reveals a T to A transversion in the donor splice site of intron 5 (GTAAC
sfigure 1 Styx mutant mice recapitulate the phenotype of SHIP -/- mice. (A) Analysis of the genomic sequences of a styx mutant reveals a T to A transversion in the donor splice site of intron 5 (GTAAC
Supplementary Figure 1 Protease allergens induce IgE and IgG1 production. (a-c)
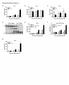 1 Supplementary Figure 1 Protease allergens induce IgE and IgG1 production. (a-c) Serum IgG1 (a), IgM (b) and IgG2 (c) concentrations in response to papain immediately before primary immunization (day
1 Supplementary Figure 1 Protease allergens induce IgE and IgG1 production. (a-c) Serum IgG1 (a), IgM (b) and IgG2 (c) concentrations in response to papain immediately before primary immunization (day
Fig. S1. Upregulation of K18 and K14 mrna levels during ectoderm specification of hescs. Quantitative real-time PCR analysis of mrna levels of OCT4
 Fig. S1. Upregulation of K18 and K14 mrna levels during ectoderm specification of hescs. Quantitative real-time PCR analysis of mrna levels of OCT4 (n=3 independent differentiation experiments for each
Fig. S1. Upregulation of K18 and K14 mrna levels during ectoderm specification of hescs. Quantitative real-time PCR analysis of mrna levels of OCT4 (n=3 independent differentiation experiments for each
Supplemental Information. Checkpoint Blockade Immunotherapy. Induces Dynamic Changes. in PD-1 CD8 + Tumor-Infiltrating T Cells
 Immunity, Volume 50 Supplemental Information Checkpoint Blockade Immunotherapy Induces Dynamic Changes in PD-1 CD8 + Tumor-Infiltrating T Cells Sema Kurtulus, Asaf Madi, Giulia Escobar, Max Klapholz, Jackson
Immunity, Volume 50 Supplemental Information Checkpoint Blockade Immunotherapy Induces Dynamic Changes in PD-1 CD8 + Tumor-Infiltrating T Cells Sema Kurtulus, Asaf Madi, Giulia Escobar, Max Klapholz, Jackson
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Choi YL, Soda M, Yamashita Y, et al. EML4-ALK mutations in
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Choi YL, Soda M, Yamashita Y, et al. EML4-ALK mutations in
Generation of ST2-GFP reporter mice and characterization of ILC1 cells following infection
 Supplementary Figure 1 Generation of ST2-GFP reporter mice and characterization of ILC1 cells following infection with influenza virus. (a) ST2-GFP reporter mice were generated as described in Methods.
Supplementary Figure 1 Generation of ST2-GFP reporter mice and characterization of ILC1 cells following infection with influenza virus. (a) ST2-GFP reporter mice were generated as described in Methods.
a Beckman Coulter Life Sciences: White Paper
 a Beckman Coulter Life Sciences: White Paper An 8-color DuraClone IM panel for detection of Human blood dendritic cells by flow cytometry Nathalie Dupas 1, Snehita Sattiraju 2, Neha Girish 2, Murthy Pendyala
a Beckman Coulter Life Sciences: White Paper An 8-color DuraClone IM panel for detection of Human blood dendritic cells by flow cytometry Nathalie Dupas 1, Snehita Sattiraju 2, Neha Girish 2, Murthy Pendyala
Atypical Natural Killer T-cell receptor recognition of CD1d-lipid antigens supplementary Information.
 Atypical Natural Killer T-cell receptor recognition of CD1d-lipid antigens supplementary Information. Supplementary Figure 1. Phenotypic analysis of TRBV25-1 + and TRBV25-1 - CD1d-α-GalCerreactive cells.
Atypical Natural Killer T-cell receptor recognition of CD1d-lipid antigens supplementary Information. Supplementary Figure 1. Phenotypic analysis of TRBV25-1 + and TRBV25-1 - CD1d-α-GalCerreactive cells.
Cell isolation. Spleen and lymph nodes (axillary, inguinal) were removed from mice
 Supplementary Methods: Cell isolation. Spleen and lymph nodes (axillary, inguinal) were removed from mice and gently meshed in DMEM containing 10% FBS to prepare for single cell suspensions. CD4 + CD25
Supplementary Methods: Cell isolation. Spleen and lymph nodes (axillary, inguinal) were removed from mice and gently meshed in DMEM containing 10% FBS to prepare for single cell suspensions. CD4 + CD25
Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2)
 Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
Supplementary Figure 1. Deletion of Smad3 prevents B16F10 melanoma invasion and metastasis in a mouse s.c. tumor model.
 A B16F1 s.c. Lung LN Distant lymph nodes Colon B B16F1 s.c. Supplementary Figure 1. Deletion of Smad3 prevents B16F1 melanoma invasion and metastasis in a mouse s.c. tumor model. Highly invasive growth
A B16F1 s.c. Lung LN Distant lymph nodes Colon B B16F1 s.c. Supplementary Figure 1. Deletion of Smad3 prevents B16F1 melanoma invasion and metastasis in a mouse s.c. tumor model. Highly invasive growth
Supplementary Figure 1. ETBF activate Stat3 in B6 and Min mice colons
 Supplementary Figure 1 ETBF activate Stat3 in B6 and Min mice colons a pstat3 controls Pos Neg ETBF 1 2 3 4 b pstat1 pstat2 pstat3 pstat4 pstat5 pstat6 Actin Figure Legend: (a) ETBF induce predominantly
Supplementary Figure 1 ETBF activate Stat3 in B6 and Min mice colons a pstat3 controls Pos Neg ETBF 1 2 3 4 b pstat1 pstat2 pstat3 pstat4 pstat5 pstat6 Actin Figure Legend: (a) ETBF induce predominantly
Combined Rho-kinase inhibition and immunogenic cell death triggers and propagates immunity against cancer
 Supplementary Information Combined Rho-kinase inhibition and immunogenic cell death triggers and propagates immunity against cancer Gi-Hoon Nam, Eun-Jung Lee, Yoon Kyoung Kim, Yeonsun Hong, Yoonjeong Choi,
Supplementary Information Combined Rho-kinase inhibition and immunogenic cell death triggers and propagates immunity against cancer Gi-Hoon Nam, Eun-Jung Lee, Yoon Kyoung Kim, Yeonsun Hong, Yoonjeong Choi,
(a) Significant biological processes (upper panel) and disease biomarkers (lower panel)
 Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figures
 Supplementary Figures Supplementary Fig. 1. Surface thiol groups and reduction of activated T cells. (a) Activated CD8 + T-cells have high expression levels of free thiol groups on cell surface proteins.
Supplementary Figures Supplementary Fig. 1. Surface thiol groups and reduction of activated T cells. (a) Activated CD8 + T-cells have high expression levels of free thiol groups on cell surface proteins.
Supplementary methods:
 Supplementary methods: Primers sequences used in real-time PCR analyses: β-actin F: GACCTCTATGCCAACACAGT β-actin [11] R: AGTACTTGCGCTCAGGAGGA MMP13 F: TTCTGGTCTTCTGGCACACGCTTT MMP13 R: CCAAGCTCATGGGCAGCAACAATA
Supplementary methods: Primers sequences used in real-time PCR analyses: β-actin F: GACCTCTATGCCAACACAGT β-actin [11] R: AGTACTTGCGCTCAGGAGGA MMP13 F: TTCTGGTCTTCTGGCACACGCTTT MMP13 R: CCAAGCTCATGGGCAGCAACAATA
Supplemental Table 1. Primer sequences for transcript analysis
 Supplemental Table 1. Primer sequences for transcript analysis Primer Sequence (5 3 ) Primer Sequence (5 3 ) Mmp2 Forward CCCGTGTGGCCCTC Mmp15 Forward CGGGGCTGGCT Reverse GCTCTCCCGGTTTC Reverse CCTGGTGTGCCTGCTC
Supplemental Table 1. Primer sequences for transcript analysis Primer Sequence (5 3 ) Primer Sequence (5 3 ) Mmp2 Forward CCCGTGTGGCCCTC Mmp15 Forward CGGGGCTGGCT Reverse GCTCTCCCGGTTTC Reverse CCTGGTGTGCCTGCTC
Dual Targeting Nanoparticle Stimulates the Immune
 Dual Targeting Nanoparticle Stimulates the Immune System to Inhibit Tumor Growth Alyssa K. Kosmides, John-William Sidhom, Andrew Fraser, Catherine A. Bessell, Jonathan P. Schneck * Supplemental Figure
Dual Targeting Nanoparticle Stimulates the Immune System to Inhibit Tumor Growth Alyssa K. Kosmides, John-William Sidhom, Andrew Fraser, Catherine A. Bessell, Jonathan P. Schneck * Supplemental Figure
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/3/114/ra23/dc1 Supplementary Materials for Regulation of Zap70 Expression During Thymocyte Development Enables Temporal Separation of CD4 and CD8 Repertoire Selection
www.sciencesignaling.org/cgi/content/full/3/114/ra23/dc1 Supplementary Materials for Regulation of Zap70 Expression During Thymocyte Development Enables Temporal Separation of CD4 and CD8 Repertoire Selection
Phenylketonuria (PKU) Structure of Phenylalanine Hydroxylase. Biol 405 Molecular Medicine
 Phenylketonuria (PKU) Structure of Phenylalanine Hydroxylase Biol 405 Molecular Medicine 1998 Crystal structure of phenylalanine hydroxylase solved. The polypeptide consists of three regions: Regulatory
Phenylketonuria (PKU) Structure of Phenylalanine Hydroxylase Biol 405 Molecular Medicine 1998 Crystal structure of phenylalanine hydroxylase solved. The polypeptide consists of three regions: Regulatory
IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia
 Supplementary Figures IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia Yaming Wang, Kristy J. Szretter, William Vermi, Susan Gilfillan, Cristina
Supplementary Figures IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia Yaming Wang, Kristy J. Szretter, William Vermi, Susan Gilfillan, Cristina
The Emergence of Alternative 39 and 59 Splice Site Exons from Constitutive Exons
 The Emergence of Alternative 39 and 59 Splice Site Exons from Constitutive Exons Eli Koren, Galit Lev-Maor, Gil Ast * Department of Human Molecular Genetics, Sackler Faculty of Medicine, Tel Aviv University,
The Emergence of Alternative 39 and 59 Splice Site Exons from Constitutive Exons Eli Koren, Galit Lev-Maor, Gil Ast * Department of Human Molecular Genetics, Sackler Faculty of Medicine, Tel Aviv University,
Protein SD Units (P-value) Cluster order
 SUPPLEMENTAL TABLE AND FIGURES Table S1. Signature Phosphoproteome of CD22 E12 Transgenic Mouse BPL Cells. T-test vs. Other Protein SD Units (P-value) Cluster order ATPase (Ab-16) 1.41 0.000880 1 mtor
SUPPLEMENTAL TABLE AND FIGURES Table S1. Signature Phosphoproteome of CD22 E12 Transgenic Mouse BPL Cells. T-test vs. Other Protein SD Units (P-value) Cluster order ATPase (Ab-16) 1.41 0.000880 1 mtor
Akt and mtor pathways differentially regulate the development of natural and inducible. T H 17 cells
 Akt and mtor pathways differentially regulate the development of natural and inducible T H 17 cells Jiyeon S Kim, Tammarah Sklarz, Lauren Banks, Mercy Gohil, Adam T Waickman, Nicolas Skuli, Bryan L Krock,
Akt and mtor pathways differentially regulate the development of natural and inducible T H 17 cells Jiyeon S Kim, Tammarah Sklarz, Lauren Banks, Mercy Gohil, Adam T Waickman, Nicolas Skuli, Bryan L Krock,
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/2/4/e1500980/dc1 Supplementary Materials for The crystal structure of human dopamine -hydroxylase at 2.9 Å resolution Trine V. Vendelboe, Pernille Harris, Yuguang
advances.sciencemag.org/cgi/content/full/2/4/e1500980/dc1 Supplementary Materials for The crystal structure of human dopamine -hydroxylase at 2.9 Å resolution Trine V. Vendelboe, Pernille Harris, Yuguang
Supplemental information
 Supplemental information PI(3)K p11δ controls the sucellular compartmentalization of TLR4 signaling and protects from endotoxic shock Ezra Aksoy, Salma Taoui, David Torres, Sandrine Delauve, Aderrahman
Supplemental information PI(3)K p11δ controls the sucellular compartmentalization of TLR4 signaling and protects from endotoxic shock Ezra Aksoy, Salma Taoui, David Torres, Sandrine Delauve, Aderrahman
Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was
 Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was painted on the shaved back skin of CBL/J and BALB/c mice for consecutive days. (a, b) Phenotypic presentation of mouse back skin
Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was painted on the shaved back skin of CBL/J and BALB/c mice for consecutive days. (a, b) Phenotypic presentation of mouse back skin
Blocking antibodies and peptides. Rat anti-mouse PD-1 (29F.1A12, rat IgG2a, k), PD-
 Supplementary Methods Blocking antibodies and peptides. Rat anti-mouse PD-1 (29F.1A12, rat IgG2a, k), PD- L1 (10F.9G2, rat IgG2b, k), and PD-L2 (3.2, mouse IgG1) have been described (24). Anti-CTLA-4 (clone
Supplementary Methods Blocking antibodies and peptides. Rat anti-mouse PD-1 (29F.1A12, rat IgG2a, k), PD- L1 (10F.9G2, rat IgG2b, k), and PD-L2 (3.2, mouse IgG1) have been described (24). Anti-CTLA-4 (clone
Nature Immunology doi: /ni.2771
 Supplementary Figure 1. Lymphadenopathy, mitogen response, effector cells, and serum Ig assessment. (a) Computerized Tomography (CT) images demonstrating lymphadenopathy (arrows) in patient F.II.1 and
Supplementary Figure 1. Lymphadenopathy, mitogen response, effector cells, and serum Ig assessment. (a) Computerized Tomography (CT) images demonstrating lymphadenopathy (arrows) in patient F.II.1 and
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature11429 S1a 6 7 8 9 Nlrc4 allele S1b Nlrc4 +/+ Nlrc4 +/F Nlrc4 F/F 9 Targeting construct 422 bp 273 bp FRT-neo-gb-PGK-FRT 3x.STOP S1c Nlrc4 +/+ Nlrc4 F/F casp1
SUPPLEMENTARY INFORMATION doi:10.1038/nature11429 S1a 6 7 8 9 Nlrc4 allele S1b Nlrc4 +/+ Nlrc4 +/F Nlrc4 F/F 9 Targeting construct 422 bp 273 bp FRT-neo-gb-PGK-FRT 3x.STOP S1c Nlrc4 +/+ Nlrc4 F/F casp1
Nature Immunology: doi: /ni Supplementary Figure 1. DNA-methylation machinery is essential for silencing of Cd4 in cytotoxic T cells.
 Supplementary Figure 1 DNA-methylation machinery is essential for silencing of Cd4 in cytotoxic T cells. (a) Scheme for the retroviral shrna screen. (b) Histogram showing CD4 expression (MFI) in WT cytotoxic
Supplementary Figure 1 DNA-methylation machinery is essential for silencing of Cd4 in cytotoxic T cells. (a) Scheme for the retroviral shrna screen. (b) Histogram showing CD4 expression (MFI) in WT cytotoxic
SUPPLEMENTARY INFORMATION
 1. Supplementary Figures and Legends Supplementary Fig. 1. S1P-mediated transcriptional regulation of integrins expressed in OP/monocytoid cells. Real-time quantitative PCR analyses of mrna for two integrins,
1. Supplementary Figures and Legends Supplementary Fig. 1. S1P-mediated transcriptional regulation of integrins expressed in OP/monocytoid cells. Real-time quantitative PCR analyses of mrna for two integrins,
Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A.
 Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A. Upper part, three-primer PCR strategy at the Mcm3 locus yielding
Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A. Upper part, three-primer PCR strategy at the Mcm3 locus yielding
Supplementary information
 Supplementary information Lipid peroxidation causes endosomal antigen release for cross-presentation Ilse Dingjan 1, Daniëlle RJ Verboogen 1, Laurent M Paardekooper 1, Natalia H Revelo 1, Simone P Sittig
Supplementary information Lipid peroxidation causes endosomal antigen release for cross-presentation Ilse Dingjan 1, Daniëlle RJ Verboogen 1, Laurent M Paardekooper 1, Natalia H Revelo 1, Simone P Sittig
COMPUTATIONAL OPTIMISATION OF TARGETED DNA SEQUENCING FOR CANCER DETECTION
 COMPUTATIONAL OPTIMISATION OF TARGETED DNA SEQUENCING FOR CANCER DETECTION Pierre Martinez, Nicholas McGranahan, Nicolai Juul Birkbak, Marco Gerlinger, Charles Swanton* SUPPLEMENTARY INFORMATION SUPPLEMENTARY
COMPUTATIONAL OPTIMISATION OF TARGETED DNA SEQUENCING FOR CANCER DETECTION Pierre Martinez, Nicholas McGranahan, Nicolai Juul Birkbak, Marco Gerlinger, Charles Swanton* SUPPLEMENTARY INFORMATION SUPPLEMENTARY
NK cell flow cytometric assay In vivo DC viability and migration assay
 NK cell flow cytometric assay 6 NK cells were purified, by negative selection with the NK Cell Isolation Kit (Miltenyi iotec), from spleen and lymph nodes of 6 RAG1KO mice, injected the day before with
NK cell flow cytometric assay 6 NK cells were purified, by negative selection with the NK Cell Isolation Kit (Miltenyi iotec), from spleen and lymph nodes of 6 RAG1KO mice, injected the day before with
SUPPLEMENTARY METHODS
 SUPPLEMENTARY METHODS Histological analysis. Colonic tissues were collected from 5 parts of the middle colon on day 7 after the start of DSS treatment, and then were cut into segments, fixed with 4% paraformaldehyde,
SUPPLEMENTARY METHODS Histological analysis. Colonic tissues were collected from 5 parts of the middle colon on day 7 after the start of DSS treatment, and then were cut into segments, fixed with 4% paraformaldehyde,
Supplemental Data. Integrating omics and alternative splicing i reveals insights i into grape response to high temperature
 Supplemental Data Integrating omics and alternative splicing i reveals insights i into grape response to high temperature Jianfu Jiang 1, Xinna Liu 1, Guotian Liu, Chonghuih Liu*, Shaohuah Li*, and Lijun
Supplemental Data Integrating omics and alternative splicing i reveals insights i into grape response to high temperature Jianfu Jiang 1, Xinna Liu 1, Guotian Liu, Chonghuih Liu*, Shaohuah Li*, and Lijun
Supplemental Table 1: Demographics and characteristics of study participants. Male, n (%) 3 (20%) 6 (50%) Age, years [mean ± SD] 33.3 ± ± 9.
![Supplemental Table 1: Demographics and characteristics of study participants. Male, n (%) 3 (20%) 6 (50%) Age, years [mean ± SD] 33.3 ± ± 9. Supplemental Table 1: Demographics and characteristics of study participants. Male, n (%) 3 (20%) 6 (50%) Age, years [mean ± SD] 33.3 ± ± 9.](/thumbs/90/103696086.jpg) SUPPLEMENTAL DATA Supplemental Table 1: Demographics and characteristics of study participants Lean (n=15) Obese (n=12) Male, n (%) 3 (20%) 6 (50%) Age, years [mean ± SD] 33.3 ± 9.5 44.8 ± 9.1 White, n
SUPPLEMENTAL DATA Supplemental Table 1: Demographics and characteristics of study participants Lean (n=15) Obese (n=12) Male, n (%) 3 (20%) 6 (50%) Age, years [mean ± SD] 33.3 ± 9.5 44.8 ± 9.1 White, n
Supplementary Figure 1 Chemokine and chemokine receptor expression during muscle regeneration (a) Analysis of CR3CR1 mrna expression by real time-pcr
 Supplementary Figure 1 Chemokine and chemokine receptor expression during muscle regeneration (a) Analysis of CR3CR1 mrna expression by real time-pcr at day 0, 1, 4, 10 and 21 post- muscle injury. (b)
Supplementary Figure 1 Chemokine and chemokine receptor expression during muscle regeneration (a) Analysis of CR3CR1 mrna expression by real time-pcr at day 0, 1, 4, 10 and 21 post- muscle injury. (b)
Uninformative BRCA Tests. Rebecca Sutphen, M.D. Professor, USF College of Medicine Chief Medical Officer, InformedDNA
 Uninformative RCA Tests Rebecca Sutphen, M.D. Professor, USF College of Medicine Chief Medical Officer, InformedDNA Uninformative RCA tests 1. R/OV cancer patient with normal results 2. Cancer-free person
Uninformative RCA Tests Rebecca Sutphen, M.D. Professor, USF College of Medicine Chief Medical Officer, InformedDNA Uninformative RCA tests 1. R/OV cancer patient with normal results 2. Cancer-free person
Dendritic cell subsets and CD4 T cell immunity in Melanoma. Ben Wylie 1 st year PhD Candidate
 Dendritic cell subsets and CD4 T cell immunity in Melanoma Ben Wylie 1 st year PhD Candidate Melanoma Melanoma is the 4 th most common cancer in Australia. Current treatment options are ineffective resulting
Dendritic cell subsets and CD4 T cell immunity in Melanoma Ben Wylie 1 st year PhD Candidate Melanoma Melanoma is the 4 th most common cancer in Australia. Current treatment options are ineffective resulting
Supplemental Figure 1. Signature gene expression in in vitro differentiated Th0, Th1, Th2, Th17 and Treg cells. (A) Naïve CD4 + T cells were cultured
 Supplemental Figure 1. Signature gene expression in in vitro differentiated Th0, Th1, Th2, Th17 and Treg cells. (A) Naïve CD4 + T cells were cultured under Th0, Th1, Th2, Th17, and Treg conditions. mrna
Supplemental Figure 1. Signature gene expression in in vitro differentiated Th0, Th1, Th2, Th17 and Treg cells. (A) Naïve CD4 + T cells were cultured under Th0, Th1, Th2, Th17, and Treg conditions. mrna
