Supporting information
|
|
|
- Kelley Weaver
- 5 years ago
- Views:
Transcription
1 Supporting information Supplementary Figure 1 Gating strategies for cells depicted in Table 2. Supplementary Figure 2 Viral load in humans measured at TP comparing (A) mild and severe disease in all ph1n1 patients and (B) comparing mild and severe NRF patients. (C)-(D) Viral load in the same groups as (A)-(B) shown in relation to days from illness onset. p values calculated by Mann-Whitney test in (A) and (B); and with Spearman s rank correlation in (C) and (D). Supplementary Figure 3 (A)-(I) Circulating immune profile as described in Table 2, comparing mild and severe disease in NRF and WRF separately (Mann Whitney Rank Sum test if not normal distribution or Student t Test if normal distribution). (J) Correlation between CD14 monocytes:cd3 T cell ratio and IFN-γ Elispot SFU to ph1n1 HA [as in (D)] for NRF severe patients. Supplementary Figure 4 (A)-(B) Circulating CD14 + monocytes and CD15 + LDG expressed as % of live cells for NRF (n=18) and WRF (n=14) patients. (C)-(D) Monocytes and LDGs at TP1 and 4-6 weeks later (TP2) for 9 patients from NRF severe group expressed as % of live cells. (E)-(H) Relationship between CD14 + monocytes and CD15 + LDG with BMI for all patients. (I) Difference in BMI between patients with severe and mild disease for all patients. (J)-(K) Data for Figure 1G-H when patients with BMI>3 are removed. All values are mean ± SEM for normally distributed sets or median ± interquartile range for non-normal distribution. p values calculated using Mann-Whitney test in non-parametric analysis and Student s t-test for parametric. Wilcoxon matched-pairs signed rank test was used for TP1 vs TP2 in (C). Correlation and signficance calculated using Spearman s rank test for (E) and
2 (F). p values calculated using Kuskall-Wallis test and Dunn s multiple comparison test for (A)-(B), and (J)-(K). Supplementary Figure 5 (A) Representative TNF-α and IL-1 ICS for Figure 2H and (C)-(D) in this figure. (B) IL-1 expressing monocytes as % of total monocytes for NRF severe patients compared to healthy controls. (C)-(D) TNF-α and IL-1 expression for monocytes from patients from NRF severe group. ICS of cytokines measured following 6 hours of LPS stimulation of PBMCs and represented as proportion of CD14 + monocytes. Paired t test used. Supplementary Figure 6 (A) Weight change relative to initial weight in C57BL/6 mice following infection with pathogenic A/PR/8/34/H1N1 (PR8) or low pathogenic NYMC-X-179A (X-179A) influenza viruses. n=6 mice per group from 2 experiments. Difference between groups analyzed using two-way ANOVA with repeated measures on different days. Error bars are not visible for Days 2 and 3 of PR8 mice. (B) Viral titers in lungs of PR8- and X179A-infected mice on day 3 and 5 after inoculation. Viral titers measured by qpcr using primers for Matrix (M) gene, and normalized to b2m house keeping gene. PR8-infected mice reached limits of severity before day 5 and were culled. (C) Weight change following infection of 9 week old female C57BL/6 mice with 1 4 EID5/ml PR8 that were either naïve ( PR8 ), or had been prechallenged with X179A 14 days previously ( X179A+PR8 ); both groups of mice were of the same age and weight. Mice were culled at 2% weight loss limit. n=3 per group. (D)-(E) Number of transferred cells (identifiable as CD cells) at 6 and 24 hours after transfer in BAL and perfused lung digest. Supplementary Figure 7 (A) Percentage weight change relative to initial weight following infection with either 1 5 EID5/ml of X179A or H5N1 and uninfected mice. n=6 in each group. Difference between groups analyzed using two-way ANOVA with
3 repeated measures on different days. **p<.1; ****p<.1. (B)-(C). Viral titers in lungs of H5N1- and X179A-infected mice on day 3 and 5 after inoculation with 1 5 EID5/ml of H5N1 or X179A. Viral titers measured by qpcr using primers for Matrix (M) gene, and normalized to b2m house keeping gene. For (B), single value is combined RNA for 6 mice, which was then averaged. This was to maximize efficiency and reduce possibility of contamination in Category 4 conditions. Supplementary Figure 8 (A-D) GSEA summaries from GSE3269, GSE53321, GSE63245 and GSE51466 showing the normalized enrichment scores of M1 or M2 gene sets in H5N1 or X179A infection on day 3 and day 5. n.s - gene set was not significantly enriched in sample. Tables show all details from GSEA analysis for all 4 gene sets analyzed. ES= enrichment score (degree to which the gene set is overrepresented at the top or bottom of the ranked input list of genes), NES= normalized enrichment score (adjusted for gene set size or multiple hypothesis testing), NOM P= nominal p value, FDR q= false discovery rate, FWER Q= family wise-error rate, size= number of genes in analysis after filtering out those not in expression dataset. Supplementary Table 1. Characterization of previously healthy ph1n1-infected patients with no risk factors (NRF) for severe disease. Resp score respiratory score as defined in Methods. Supplementary Table 2 Gene ontology terms that were enriched in H5N1 or X179A infection on day 3 and day 5, analyzed by GOrilla. Only GO terms with at least 5 genes ( b ) were used in the analysis. Enrichment of these terms was tested against gene lists ranked in order of highest to lowest expression in infected mice (H5N1 or X-179A) relative to uninfected controls. Enrichment = (b/n) / (B/N): N- the total number of genes; B- the total number of genes associated with a specific GO
4 term; n- the number of genes in the top of the user's input list; b- the number of genes in the intersection. Supplementary Table 3 Gene sets used to identify M1 or M2 macrophage genes for use in GSEA for GSEA (maximum input). Supplementary Table 4 Antibodies used in mouse flow cytometry analysis. Supplementary Table 5 Antibodies used in human flow cytometry analysis. Supplementary Table 6 qpcr primers used for human monocyte M1-M2 gene expression studies and mouse viral load assessment. Supplementary Table 7 Top genes significantly upregulated in M1 or M2 macrophages at adjusted p<.5 and logfc>1.5. Gene sets from published work were analysed using GEO2R and used for input into GSEA. MOSAIC data and clinical samples were provided by the MOSAIC consortium.**mosaic investigators: Chelsea and Westminster NHS Foundation Trust: B.G. Gazzard. Francis Crick Institute, Mill Hill Laboratory: A. Hay, J. McCauley, A. O GGarra. Imperial College London, UK: P. Aylin, D. Ashby, W.S. Barclay, S.J. Brett, W.O. Cookson, M.J. Cox, J. Dunning, L.N. Drumright, R.A. Elderfield, L. Garcia-Alvarez, M.J. Griffiths, M.S. Habibi, T.T. Hansel, J.A. Herberg, A.H. Holmes, S.L. Johnston, O.M. Kon, M. Levin, M.F. Moffatt, S. Nadel, J.O. Warner. Liverpool School of Tropical Medicine, UK: S.J. Aston, S.B. Gordon. Manchester Collaborative Centre for Inflammation Research (MCCIR) T. Hussell. Public Health England (formerly Health Protection Agency), UK: C.
5 Thompson, M.C. Zambon. The Roslin Institute, University of Edinburgh: D.A. Hume. University College London, UK: A. Hayward. UCL Institute of Child Health: R.L. Smyth; University of Edinburgh, UK: J.K. Baillie, P. Simmonds University of Liverpool, UK: P.S. McNamara; M.G. Semple; University of Nottingham, UK: J.S. Nguyen-Van-Tam; University of Oxford, UK: L-P. Ho, A. J. McMichael Wellcome Trust Sanger Institute, UK: P. Kellam West of Scotland Specialist Virology Centre, Glasgow, UK: W.E. Adamson, W.F. Carman.
6 SSc Supplemental Figure 1 FSc (gated on lymphocytes).52 (gated on CD3+) 61.2 (gated on CD3+ CD8+) SEVERE CD CD CD CD3 CD8 HLA-DR MILD CD CD CD CD3 CD8 HLA-DR
7 Relative viral burden (relative pful/ml equivalent) A All ph1n1 B NRF C All ph1n1 D MIld p=.3 Severe Relative viral burden (relative pful/ml equivalent) NRF Mild p=.2 NRF Severe Relative viral burden (relative pful/ml equivalent) r=-.2 p= Days from first symptoms Relative viral burden (relative pful/ml equivalent) NRF Days from first symptoms r=-.4 p=.1 Supplemental Figure 2
8 CD3 + T cells (per ml blood) IFN-γ elispot SFU to ph1n1 NA A B C D p=.6 NK cells (per ml blood) IFN-γ elispot SFU to ph1n1 NP p= IFN-γ elispot SFU to ph1n1 PA IFN-γ elispot SFU to ph1n1 HA E F G H CD8 + et cells (per ml blood) 5 IFN-γ elispot SFU to ph1n1 PB p=.16 I IFN-γ elispot SFU to ph1n1 M1/M IFNγ Elispot to HA J r=.6 p= CD14 hi to CD3+ ratio Supplemental Figure 3
9 A Monocytes (as % of live cells) *p=.5 B CD15 + LDG (as % of live cells) *p=.5 C Monocytes (% of live cells) D p=.1 LDG (% of live cells) p<.1 Still hospitalized at TP2 TP1 TP2 TP1 TP2 Supplemental Figure 4
10 Supplemental Figure 4 E F G H Monocytes x1^6 (per ml of blood) BMI BMI I p=.15 Mild Severe CD15 + LDG x1 6 (per ml of blood) 3. n.s p=.9 p=.58 Monocytes x 1 6 (per ml blood) BMI J * p=.5 Monocytes x1^6 (per ml of blood) BMI < 3 CD15 + LDG x 1 6 (per ml blood) K BMI >3 CD15 + LDG x1 6 (per ml of blood) p=.8 BMI < 3 p=.49 BMI >3
11 A B IL1 + monocytes (% of total monocytes) NRF severe *p=.1 HC C TNFα ICS (% of CD14 + monocytes) D IL-1 ICS (% of CD14 + monocytes) TP1 TP2 TP1 TP2 Supplemental Figure 5
12 A % weight change p<.1 **** **** Day post infection Uninfected X179A PR8 B Relative expression (normalised to β2m) IAV matrix gene expression PR8 day 3 X-179A day 3 X-179A day 5 C % weight change X179A + PR8 ** **** ******** Day post infection PR8 Supplemental Figure 6
13 D Number of transferred cells ( x1 6 per ml of BAL) M1 BMDM +M2 BMDM 6hr 24hr Time after transfer E Number of transferred cells (x1 6 per lung digest) M1 BMDM +M2 BMDM 6hr 24hr Time after transfer Supplemental Figure 6
14 A % weight change p<.1 ** **** **** **** **** -3 X179A -4 H5N1 Uninfected Day post infection B Relative expression (normalised to β2m) H5N1 IAV matrix gene expression H5N1 day 1 H5N1 day 3 H5N1 day 5 C Relative expression (normalised to β2m) X-179A IAV matrix gene expression X-179A day 1 X-179A day 3 X-179A day 5 Supplemental figure 7
15 A GSE3269 B GSE53321 C GSE63245 D GSE51466 M1 M2 H5N1 d3 H5N1 d5 X179A d3 X179A d5 H5N1 d3 H5N1 d5 X179A d3 X179A d Normalised enrichment score M1 M2 H5N1 d3 H5N1 d5 X179A d3 X179A d5 H5N1 d3 H5N1 d5 X179A d3 X179A d Normalised enrichment score M1 M2 H5N1 d3 H5N1 d5 X179A d3 X179A d5 H5N1 d3 H5N1 d5 X179A d3 X179A d Normalised enrichment score M1 M2 H5N1 d3 H5N1 d5 X179A d3 X179A d5 H5N1 d3 H5N1 d5 X179A d3 X179A d Normalised enrichment score E M1 enrichment F M2 enrichment H5N1 H5N1 X179A X179A ES NES NOM P FDR Q FWER Q SIZE GSE <.1 <.1 < GSE <.1 <.1 < GSE <.1 <.1 <.1 35 day 3 GSE <.1 <.1 < GSE <.1 <.1 < GSE <.1 <.1 < GSE <.1 <.1 <.1 35 day 5 GSE <.1 <.1 < GSE <.1 <.1 < GSE <.1 <.1 < GSE <.1 <.1 <.1 35 day 3 GSE <.1 <.1 < GSE <.1 <.1 < GSE <.1 <.1 < GSE <.1 <.1 <.1 35 day 5 GSE <.1 <.1 < H5N1 H5N1 X179A X179A ES NES NOM P FDR Q FWER Q SIZE GSE <.1 <.1 < GSE GSE <.1 <.1 <.1 23 day 3 GSE <.1 < GSE <.1 <.1 < GSE GSE <.1 <.1 <.1 23 day 5 GSE <.1 < GSE GSE GSE day 3 GSE <.1 <.1 < GSE GSE GSE day 5 GSE <.1 <.1 < Supplemental figure 8
16 Patient number (NRF) TP2 Resp score BMI Mild (M) or severe (S) Comorbidities Days from first symptom 1 yes 1 33 M NONE 5 5 F S NONE 9 33 F 3 yes 1 >3 M NONE 8 43 M M NONE F S NONE 12 4 M S NONE F S NONE 8 36 F S NONE 1 33 F 9 yes 3 24 S NONE M M NONE 1 23 M 11 yes 3 39 S NONE 8 44 M S NONE M 13 yes 3 25 S NONE M S NONE M M NONE 12 5 F 16 yes 3 33 S NONE M 17 yes 3 33 S NONE F 18 yes 1 23 M NONE 1 44 F Age Gender Supplemental table 1
17 H5N1 day 3 H5N1 day 5 P-value FDR q-value Enrichment (N, B, n, b) P-value FDR q-value Enrichment (N, B, n, b) GO:926 positive regulation of monocyte chemotaxis 4.22E E (16186,11,258,6) 6.18E E (16186,11,369,7) GO:925 regulation of monocyte chemotaxis 9.4E E (16186,12,258,6) 1.5E-8 6.5E (16186,12,369,7) GO:42116 macrophage activation 2.53E-8 1.4E (16186,24,967,12) 5.4E-8 2.2E (16186,24,6,1) GO:1758 regulation of macrophage chemotaxis 3.56E E (16186,16,1651,1) 8.92E-5 1.8E (16186,16,1328,8) GO:45649 regulation of macrophage differentiation 3.37E E (16186,13,19,6) 1.54E-4 2.9E (16186,13,869,6) GO:71621 granulocyte chemotaxis 3.33E E (16186,41,396,18) 3.3E E (16186,41,596,21) GO:71622 regulation of granulocyte chemotaxis 1.34E E (16186,32,172,18) 5.45E E (16186,32,58,12) GO:9753 granulocyte migration 1.1E E (16186,43,396,18) 1.16E E (16186,43,596,21) GO:587 positive regulation of T cell activation 2.35E-8 9.8E (16186,12,685,21) 1.75E E (16186,12,194,28) GO:4211 T cell activation 5.34E E (16186,16,1449,44) 2.3E E (16186,16,1657,48) GO:42129 regulation of T cell proliferation 8.8E E (16186,16,618,21) 1.7E-9 5.2E (16186,16,552,21) GO:7489 T cell aggregation 6.62E E (16186,161,1449,44) 2.75E E (16186,161,1657,48) GO:46631 alpha-beta T cell activation 4.55E E (16186,39,1591,17) 4.74E E (16186,39,1174,16) GO:4212 positive regulation of T cell proliferation 3.1E E (16186,6,59,14) 4.8E E (16186,6,5,14) GO:1914 regulation of T cell mediated cytotoxicity 7.E E (16186,16,1284,8) 5.62E-5 1.2E (16186,16,863,7) GO:244 regulation of T cell migration 4.71E E (16186,23,165,13) 1.11E-8 4.6E (16186,23,728,11) GO:72678 T cell migration 1.82E E (16186,11,1176,9) 2.35E E (16186,11,137,8) GO:4558 regulation of T cell differentiation 1.74E E (16186,87,685,14) 5.46E E (16186,87,1425,2) GO:45582 positive regulation of T cell differentiation 1.37E E (16186,52,685,12) 1.11E E (16186,52,337,8) GO:46635 positive regulation of alpha-beta T cell activation 9.28E-4 1.4E (16186,37,685,8) 8.65E E (16186,37,5,7) GO:5863 regulation of T cell activation 1.83E E (16186,181,685,31) 5.43E E (16186,181,1428,52) GO:19724 B cell mediated immunity 5.8E E (16186,21,1562,13) 1.15E-6 3.4E (16186,21,13,11) GO:2712 regulation of B cell mediated immunity 1.18E E (16186,3,1494,15) 2.54E E (16186,3,374,8) GO:2714 positive regulation of B cell mediated immunity 1.55E E (16186,24,1416,13) 6.56E E (16186,24,374,7) GO:2715 regulation of natural killer cell mediated immunity 8.36E E (16186,24,1657,12) 1.2E E (16186,24,863,11) GO:42269 regulation of natural killer cell mediated cytotoxicity 8.36E E (16186,24,1657,12) 1.2E E (16186,24,863,11) Supplemental table 2
18 X179A day 3 X179A day 5 P-value FDR q-value Enrichment (N, B, n, b) P-value FDR q-value Enrichment (N, B, n, b) GO:926 positive regulation of monocyte chemotaxis 1.17E E (16186,11,676,6) 2.18E E (16186,11,314,7) GO:925 regulation of monocyte chemotaxis 2.29E E (16186,12,676,6) 5.1E E (16186,12,314,7) GO:42116 macrophage activation 4.51E-5 2.8E (16186,24,729,8) 5.16E E (16186,24,537,8) GO:1758 regulation of macrophage chemotaxis 4.9E E (16186,16,478,5) 7.34E-5 1.7E (16186,16,577,6) GO:45649 regulation of macrophage differentiation 2.45E-4 8.5E (16186,13,948,6) 2.9E E (16186,13,577,5) GO:71621 granulocyte chemotaxis 3.29E-4 1.2E (16186,41,516,8) 3.33E E (16186,41,536,14) GO:71622 regulation of granulocyte chemotaxis 4.E E (16186,32,211,5) 3.27E E (16186,32,665,11) GO:9753 granulocyte migration 4.53E E (16186,43,516,8) 7.47E E (16186,43,536,14) GO:587 positive regulation of T cell activation 3.21E E (16186,12,16,35) 9.5E E (16186,12,14,33) GO:4211 T cell activation 3.4E-4 1.4E (16186,16,651,18) 6.95E E (16186,16,15,57) GO:42129 regulation of T cell proliferation 6.66E E (16186,16,1588,32) 1.41E E (16186,16,852,28) GO:7489 T cell aggregation 3.67E E (16186,161,651,18) 9.87E E (16186,161,15,57) GO:46631 alpha-beta T cell activation 9.99E-4 2.4E (16186,39,651,8) 3.2E E (16186,39,15,21) GO:4212 positive regulation of T cell proliferation 1.67E E (16186,6,141,21) 4.36E E (16186,6,14,21) GO:1914 regulation of T cell mediated cytotoxicity 3.2E-6 2.2E (16186,16,55,7) 6.17E E (16186,16,677,8) GO:244 regulation of T cell migration 7.13E-5 3.1E (16186,23,211,5) 1.23E E (16186,23,179,6) GO:72678 T cell migration 3.2E E (16186,11,82,6) 2.14E E (16186,11,12,9) GO:4558 regulation of T cell differentiation 7.26E-5 3.4E (16186,87,63,14) 4.19E E (16186,87,966,21) GO:45582 positive regulation of T cell differentiation 2.36E E (16186,52,724,12) 1.31E-6 5.2E (16186,52,121,17) GO:46635 positive regulation of alpha-beta T cell activation 1.9E E (16186,37,538,8) 6.15E E (16186,37,1175,12) GO:5863 regulation of T cell activation 8.45E E (16186,181,16,46) 1.46E E (16186,181,14,46) GO:19724 B cell mediated immunity 2.72E E (16186,21,581,8) 1.55E-6 5.8E (16186,21,536,8) GO:2712 regulation of B cell mediated immunity 1.71E-5 9.5E (16186,3,168,13) 2.24E-7 1.1E (16186,3,1321,14) GO:2714 positive regulation of B cell mediated immunity 3.54E E (16186,24,488,7) 7.34E-6 2.4E (16186,24,1321,11) GO:2715 regulation of natural killer cell mediated immunity 7.78E E (16186,24,1689,11) 3.E-4 5.6E (16186,24,687,7) GO:42269 regulation of natural killer cell mediated cytotoxicity 7.78E E (16186,24,1689,11) 3.E-4 5.6E (16186,24,687,7) Supplemental table 2 cont
19 Title Macrophage treatment for M1 or M2 (n=) Citation GSE53321 Expression data from polarized macrophages: effect of p53 Bone marrow derived macrophages polarised with E.coli LPS+IFN-γ (M1) or IL4+IL13 (M2) Li L, Ng DS, Mah WC, Almeida FF et al. A unique role for p53 in the regulation of M2 macrophage polarization. Cell Death Differ 215 Jul;22(7): PMID: GSE3269 IFN-γ-induced inos expression in regulatory macrophages prolongs allograft graft survival in fully immunocompetent recipients FACS sorted L6C+ Ly6G- CD11b+ monocytes from bone marrow cultured for 6 days in 1% FCS + 2ng/ml M- CSF, then for 24 hours with 25ng/ml IFN-g + 1ug/ml LPS (M1) or 2ng/ml IL-4 (M2) Riquelme P, Tomiuk S, Kammler A, Fändrich F et al. IFN-γ-induced inos expression in mouse regulatory macrophages prolongs allograft survival in fully immunocompetent recipients. Mol Ther 213 Feb;21(2): PMID: GSE63245 Gene expression data from murine M1 and M2 macrophages Bone marrow derived cells cultured with G-MCSF (M1) or M-CSF (M2) for 7 days n/a GSE51466 Polarization profiling of renal macrophages in stone model mice Bone marrow derived macrophages grown in L- conditioned medium over 7 days. BMM were stimulated for 2 hours with 1ng/ml LPS and 2ng/ml GM-CSF (M1) or 5ng/ml IL-4 and 1ng/ml M-CSF (M2) Taguchi K, Okada A, Kitamura H, Yasui T et al. Colonystimulating factor-1 signaling suppresses renal crystal formation. J Am Soc Nephrol 214 Aug;25(8): PMID: Supplemental table 3
20 Antibody Conjugate Clone Host & Supplier Cat. Number Isotype CCR2 APC Rat IgG2B R&D FAB5538A CCR7 efluor45 4B12 Rat IgG2a, κ ebioscience CD11c PE N418 Hamster IgG Miltenyi CD26 PE-cy7 C68C2 Rat IgG2a, κ BioLegend CD3 APCeFluor C11 Armenian ebioscience Hamster IgG CD86 FITC GL1 Rat IgG2a, κ ebioscience Dectin-1 PE RH1 Rat IgG1, κ BioLegend F4/8 APC-cy7 BM8 Rat IgG2a, κ BioLegend Ly6C efluor45 HK1.4 Rat IgG2c, κ ebioscience Ly6G FITC 1A8 Rat IgG2A Miltenyi Ly6G APC 1A8 Rat IgG2a, κ BioLegend Zombie Aqua TM BV51 n/a n/a BioLegend Supplementary table 4
21 Antibody Conjugate Clone Host & Isotype Supplier Cat. Number CCR7 PB G43H7 Mouse IgG2a, κ BioLegend CD11b APCe-fluor78 ICRF44 Mouse IgG1, κ ebioscience CD14 PEcy7 61D3 Mouse IgG1, κ ebioscience CD15 APC HI98 Mouse IgM, κ BioLegend 3197 CD16 efluor45 CB16 Mouse IgG1, κ ebioscience CD16 PE B73.1 Mouse IgG1, κ BioLegend 3674 CD163 PE GHI/61 Mouse IgG1, κ BioLegend CD3 APCeFluor78 UCHT1 Mouse IgG1, κ ebioscience CD3 FITC UCHT1 Mouse IgG1, κ ebioscience CD38 APC HIT2 Mouse IgG1, κ ebioscience CD4 PB RPA-T4 Mouse IgG1, κ BioLegend 3521 CD4 PEcy7 RPA-T4 Mouse IgG1, κ ebioscience CD56 PEcy7 HCD56 Mouse IgG1, κ BioLegend CD8a efluor45 RPA-T8 Mouse IgG1, κ ebioscience HLA-DR FITC L243 Mouse IgG2a, κ BioLegend 3763 Vα24 Jα18 FITC 6B11 Mouse IgG1, κ BD IL-1 APC JES3-9D7 Rat IgG1, κ BioLegend 5141 IL-6 FITC MQ2-13A5 Rat IgG1, κ BioLegend 5114 TNFα efluor45 Mab11 Mouse IgG1, κ ebioscience Fixable viability dye efluor78 n/a n/a ebioscience Supplementary table 5
22 Gene symbol Gene Description Forward primer Reverse primer ACTB Actin, beta (ACTB) CCTGGCACCCAGCACAAT GCCGATCCACACGGAGTACT CD163 CD163 molecule TTTGTCAACTTGAGTCCCTTCAC TCCCGCTACACTTGTTTTCAC CD2R1 CD2 receptor 1 GACCAGAGAGGGTCTCACCA TTGAAGCGGCCACTAAGAAG GBP1 Guanylate binding protein 1, interferon-inducible AGGAGTTCCTTCAAAGATGTGGA GCAACTGGACCCTGTCGTT IDO1 Indoleamine 2,3-dioxygenase 1 (IDO1) GCCAGCTTCGAGAAAGAGTTG ATCCCAGAACTAGACGTGCAA IL-8 Interleukin 8 TGCTAAAGAACTTAGATGTCAGTGCAT TGGTCCACTCTCAATCACTCTCA IL1 Interleukin 1 TACGGCGCTGTCATCGATT GGCTTTGTAGATGCCTTTCTCTTG IL12b Interleukin 12B (p4) AGAAGATGGTATCACCTGGACC GAACCTCGCCTCCTTTGTGAC IL6 Interleukin 6 (interferon, beta 2) AGTAACATGTGTGAAAGCAGCAAAG GGCAAGTCTCCTCATTGAATCC MRC1 mannose receptor, C type 1 (MRC1) AAGGCGGTGACCTCACAAG AAAGTCCAATTCCTCGATGGTG TGM2 transglutaminase 2 (TGM2) GCCACTTCATTTTGCTCTTCAA TCCTCTTCCGAGTCCAGGTACA TNFalpha Tumor necrosis factor GGCCAAGCCCTGGTATGAG TAGTCGGGCCGATTGATCTC Primer Forward primer Reverse primer Influenza Matrix CTTCTAACCGAGGTCGAAACGTA GGTGACAGGATTGGTCTTGTCTTTA Supplementary table 6
Online supplement. Phenotypic, functional and plasticity features of classical and alternatively activated
 Online supplement Phenotypic, functional and plasticity features of classical and alternatively activated human macrophages Abdullah Al Tarique*, Jayden Logan *, Emma Thomas *, Patrick G Holt *, Peter
Online supplement Phenotypic, functional and plasticity features of classical and alternatively activated human macrophages Abdullah Al Tarique*, Jayden Logan *, Emma Thomas *, Patrick G Holt *, Peter
IL-6Rα IL-6RαT-KO KO. IL-6Rα f/f bp. f/f 628 bp deleted 368 bp. 500 bp
 STD H 2 O WT KO IL-6Rα f/f IL-6Rα IL-6RαT-KO KO 1000 bp 500 bp f/f 628 bp deleted 368 bp Supplementary Figure 1 Confirmation of T-cell IL-6Rα deficiency. (a) Representative histograms and (b) quantification
STD H 2 O WT KO IL-6Rα f/f IL-6Rα IL-6RαT-KO KO 1000 bp 500 bp f/f 628 bp deleted 368 bp Supplementary Figure 1 Confirmation of T-cell IL-6Rα deficiency. (a) Representative histograms and (b) quantification
Bezzi et al., Supplementary Figure 1 *** Nature Medicine: doi: /nm Pten pc-/- ;Zbtb7a pc-/- Pten pc-/- ;Pml pc-/- Pten pc-/- ;Trp53 pc-/-
 Gr-1 Gr-1 Gr-1 Bezzi et al., Supplementary Figure 1 a Gr1-CD11b 3 months Spleen T cells 3 months Spleen B cells 3 months Spleen Macrophages 3 months Spleen 15 4 8 6 c CD11b+/Gr1+ cells [%] 1 5 b T cells
Gr-1 Gr-1 Gr-1 Bezzi et al., Supplementary Figure 1 a Gr1-CD11b 3 months Spleen T cells 3 months Spleen B cells 3 months Spleen Macrophages 3 months Spleen 15 4 8 6 c CD11b+/Gr1+ cells [%] 1 5 b T cells
Supplementary Figure 1. NAFL enhanced immunity of other vaccines (a) An over-the-counter, hand-held non-ablative fractional laser (NAFL).
 Supplementary Figure 1. NAFL enhanced immunity of other vaccines (a) An over-the-counter, hand-held non-ablative fractional laser (NAFL). (b) Depiction of a MTZ array generated by NAFL. (c-e) IgG production
Supplementary Figure 1. NAFL enhanced immunity of other vaccines (a) An over-the-counter, hand-held non-ablative fractional laser (NAFL). (b) Depiction of a MTZ array generated by NAFL. (c-e) IgG production
Supplemental Figure 1. IL-3 blockade with Fab CSL362 depletes plasmacytoid dendritic cells (pdcs), but not basophils, at higher doses.
 Supplemental Figure 1. IL-3 blockade with Fab CSL362 depletes plasmacytoid dendritic cells (pdcs), but not basophils, at higher doses. Percentage of viable (A) pdcs (Sytox Blue-, Lin1-, HLADR+, BDCA2++)
Supplemental Figure 1. IL-3 blockade with Fab CSL362 depletes plasmacytoid dendritic cells (pdcs), but not basophils, at higher doses. Percentage of viable (A) pdcs (Sytox Blue-, Lin1-, HLADR+, BDCA2++)
Supplementary Materials
 Supplementary Materials 43 Figure S1. CD123 in acute lymphoblastic leukemia and leukemia-initiating cells. A. CD123 (histograms) is highly and homogenously expressed in B-ALL blasts (as defined by live,
Supplementary Materials 43 Figure S1. CD123 in acute lymphoblastic leukemia and leukemia-initiating cells. A. CD123 (histograms) is highly and homogenously expressed in B-ALL blasts (as defined by live,
Supporting Information
 Supporting Information Idoyaga et al. 10.1073/pnas.0812247106 SSC a) Single cell suspension 99 Aqua b) Live cells 96 -W c) Singlets 92 -A CD19+ER119 d) CD19 ER119 cells 97 CD3 e) CD3 cells 27 f) DX5 cells
Supporting Information Idoyaga et al. 10.1073/pnas.0812247106 SSC a) Single cell suspension 99 Aqua b) Live cells 96 -W c) Singlets 92 -A CD19+ER119 d) CD19 ER119 cells 97 CD3 e) CD3 cells 27 f) DX5 cells
Supplementary Figures
 Inhibition of Pulmonary Anti Bacterial Defense by IFN γ During Recovery from Influenza Infection By Keer Sun and Dennis W. Metzger Supplementary Figures d a Ly6G Percentage survival f 1 75 5 1 25 1 5 1
Inhibition of Pulmonary Anti Bacterial Defense by IFN γ During Recovery from Influenza Infection By Keer Sun and Dennis W. Metzger Supplementary Figures d a Ly6G Percentage survival f 1 75 5 1 25 1 5 1
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Supplementary Figure 1: Chemokine receptor expression profiles of CCR6 + and CCR6 - CD4 + IL-17A +/ex and Treg cells. Quantitative PCR analysis of chemokine receptor transcript abundance
SUPPLEMENTARY FIGURES Supplementary Figure 1: Chemokine receptor expression profiles of CCR6 + and CCR6 - CD4 + IL-17A +/ex and Treg cells. Quantitative PCR analysis of chemokine receptor transcript abundance
Nature Immunology: doi: /ni Supplementary Figure 1. Cytokine pattern in skin in response to urushiol.
 Supplementary Figure 1 Cytokine pattern in skin in response to urushiol. Wild-type (WT) and CD1a-tg mice (n = 3 per group) were sensitized and challenged with urushiol (uru) or vehicle (veh). Quantitative
Supplementary Figure 1 Cytokine pattern in skin in response to urushiol. Wild-type (WT) and CD1a-tg mice (n = 3 per group) were sensitized and challenged with urushiol (uru) or vehicle (veh). Quantitative
Supplementary Materials for
 www.sciencetranslationalmedicine.org/cgi/content/full/8/352/352ra110/dc1 Supplementary Materials for Spatially selective depletion of tumor-associated regulatory T cells with near-infrared photoimmunotherapy
www.sciencetranslationalmedicine.org/cgi/content/full/8/352/352ra110/dc1 Supplementary Materials for Spatially selective depletion of tumor-associated regulatory T cells with near-infrared photoimmunotherapy
Fluorochrome Panel 1 Panel 2 Panel 3 Panel 4 Panel 5 CTLA-4 CTLA-4 CD15 CD3 FITC. Bio) PD-1 (MIH4, BD) ICOS (C398.4A, Biolegend) PD-L1 (MIH1, BD)
 Additional file : Table S. Antibodies used for panel stain to identify peripheral immune cell subsets. Panel : PD- signaling; Panel : CD + T cells, CD + T cells, B cells; Panel : Tregs; Panel :, -T, cdc,
Additional file : Table S. Antibodies used for panel stain to identify peripheral immune cell subsets. Panel : PD- signaling; Panel : CD + T cells, CD + T cells, B cells; Panel : Tregs; Panel :, -T, cdc,
Blocking antibodies and peptides. Rat anti-mouse PD-1 (29F.1A12, rat IgG2a, k), PD-
 Supplementary Methods Blocking antibodies and peptides. Rat anti-mouse PD-1 (29F.1A12, rat IgG2a, k), PD- L1 (10F.9G2, rat IgG2b, k), and PD-L2 (3.2, mouse IgG1) have been described (24). Anti-CTLA-4 (clone
Supplementary Methods Blocking antibodies and peptides. Rat anti-mouse PD-1 (29F.1A12, rat IgG2a, k), PD- L1 (10F.9G2, rat IgG2b, k), and PD-L2 (3.2, mouse IgG1) have been described (24). Anti-CTLA-4 (clone
Supplementary Figure 1. Characterization of basophils after reconstitution of SCID mice
 Supplementary figure legends Supplementary Figure 1. Characterization of after reconstitution of SCID mice with CD4 + CD62L + T cells. (A-C) SCID mice (n = 6 / group) were reconstituted with 2 x 1 6 CD4
Supplementary figure legends Supplementary Figure 1. Characterization of after reconstitution of SCID mice with CD4 + CD62L + T cells. (A-C) SCID mice (n = 6 / group) were reconstituted with 2 x 1 6 CD4
Supporting Information
 Supporting Information Desnues et al. 10.1073/pnas.1314121111 SI Materials and Methods Mice. Toll-like receptor (TLR)8 / and TLR9 / mice were generated as described previously (1, 2). TLR9 / mice were
Supporting Information Desnues et al. 10.1073/pnas.1314121111 SI Materials and Methods Mice. Toll-like receptor (TLR)8 / and TLR9 / mice were generated as described previously (1, 2). TLR9 / mice were
SUPPLEMENTARY INFORMATION
 doi:1.138/nature1554 a TNF-α + in CD4 + cells [%] 1 GF SPF 6 b IL-1 + in CD4 + cells [%] 5 4 3 2 1 Supplementary Figure 1. Effect of microbiota on cytokine profiles of T cells in GALT. Frequencies of TNF-α
doi:1.138/nature1554 a TNF-α + in CD4 + cells [%] 1 GF SPF 6 b IL-1 + in CD4 + cells [%] 5 4 3 2 1 Supplementary Figure 1. Effect of microbiota on cytokine profiles of T cells in GALT. Frequencies of TNF-α
Table S1. Viral load and CD4 count of HIV-infected patient population
 Table S1. Viral load and CD4 count of HIV-infected patient population Subject ID Viral load (No. of copies per ml of plasma) CD4 count (No. of cells/µl of blood) 28 7, 14 29 7, 23 21 361,99 94 217 7, 11
Table S1. Viral load and CD4 count of HIV-infected patient population Subject ID Viral load (No. of copies per ml of plasma) CD4 count (No. of cells/µl of blood) 28 7, 14 29 7, 23 21 361,99 94 217 7, 11
sequences of a styx mutant reveals a T to A transversion in the donor splice site of intron 5
 sfigure 1 Styx mutant mice recapitulate the phenotype of SHIP -/- mice. (A) Analysis of the genomic sequences of a styx mutant reveals a T to A transversion in the donor splice site of intron 5 (GTAAC
sfigure 1 Styx mutant mice recapitulate the phenotype of SHIP -/- mice. (A) Analysis of the genomic sequences of a styx mutant reveals a T to A transversion in the donor splice site of intron 5 (GTAAC
Supplementary Information. Tissue-wide immunity against Leishmania. through collective production of nitric oxide
 Supplementary Information Tissue-wide immunity against Leishmania through collective production of nitric oxide Romain Olekhnovitch, Bernhard Ryffel, Andreas J. Müller and Philippe Bousso Supplementary
Supplementary Information Tissue-wide immunity against Leishmania through collective production of nitric oxide Romain Olekhnovitch, Bernhard Ryffel, Andreas J. Müller and Philippe Bousso Supplementary
Supplementary Table e-1. Flow cytometry reagents and staining combinations
 Supplementary data Supplementary Table e-1. Flow cytometry reagents and staining combinations Reagents Antibody Fluorochrome Clone Source conjugation CD3 FITC UCHT1 BD Biosciences CD3 PerCP-Cy5.5 SK7 Biolegend
Supplementary data Supplementary Table e-1. Flow cytometry reagents and staining combinations Reagents Antibody Fluorochrome Clone Source conjugation CD3 FITC UCHT1 BD Biosciences CD3 PerCP-Cy5.5 SK7 Biolegend
L-selectin Is Essential for Delivery of Activated CD8 + T Cells to Virus-Infected Organs for Protective Immunity
 Cell Reports Supplemental Information L-selectin Is Essential for Delivery of Activated CD8 + T Cells to Virus-Infected Organs for Protective Immunity Rebar N. Mohammed, H. Angharad Watson, Miriam Vigar,
Cell Reports Supplemental Information L-selectin Is Essential for Delivery of Activated CD8 + T Cells to Virus-Infected Organs for Protective Immunity Rebar N. Mohammed, H. Angharad Watson, Miriam Vigar,
Supporting Information
 Supporting Information Aldridge et al. 10.1073/pnas.0900655106 Fig. S1. Flow diagram of sublethal (a) and lethal (b) influenza virus infections. (a) Infection of lung epithelial cells by influenza virus
Supporting Information Aldridge et al. 10.1073/pnas.0900655106 Fig. S1. Flow diagram of sublethal (a) and lethal (b) influenza virus infections. (a) Infection of lung epithelial cells by influenza virus
Supplementary Fig. 1: Ex vivo tetramer enrichment with anti-c-myc beads
 Supplementary Fig. 1: Ex vivo tetramer enrichment with anti-c-myc beads Representative example of comparative ex vivo tetramer enrichment performed in three independent experiments with either conventional
Supplementary Fig. 1: Ex vivo tetramer enrichment with anti-c-myc beads Representative example of comparative ex vivo tetramer enrichment performed in three independent experiments with either conventional
Interferon γ regulates idiopathic pneumonia syndrome, a. Th17 + CD4 + T-cell-mediated GvH disease
 Interferon γ regulates idiopathic pneumonia syndrome, a Th17 + CD4 + T-cell-mediated GvH disease Nora Mauermann, Julia Burian, Christophe von Garnier, Stefan Dirnhofer, Davide Germano, Christine Schuett,
Interferon γ regulates idiopathic pneumonia syndrome, a Th17 + CD4 + T-cell-mediated GvH disease Nora Mauermann, Julia Burian, Christophe von Garnier, Stefan Dirnhofer, Davide Germano, Christine Schuett,
SUPPLEMENTARY METHODS
 SUPPLEMENTARY METHODS Histological analysis. Colonic tissues were collected from 5 parts of the middle colon on day 7 after the start of DSS treatment, and then were cut into segments, fixed with 4% paraformaldehyde,
SUPPLEMENTARY METHODS Histological analysis. Colonic tissues were collected from 5 parts of the middle colon on day 7 after the start of DSS treatment, and then were cut into segments, fixed with 4% paraformaldehyde,
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10134 Supplementary Figure 1. Anti-inflammatory activity of sfc. a, Autoantibody immune complexes crosslink activating Fc receptors, promoting activation of macrophages, and WWW.NATURE.COM/NATURE
doi:10.1038/nature10134 Supplementary Figure 1. Anti-inflammatory activity of sfc. a, Autoantibody immune complexes crosslink activating Fc receptors, promoting activation of macrophages, and WWW.NATURE.COM/NATURE
CD44
 MR1-5-OP-RU CD24 CD24 CD44 MAIT cells 2.78 11.2 WT RORγt- GFP reporter 1 5 1 4 1 3 2.28 1 5 1 4 1 3 4.8 1.6 8.1 1 5 1 4 1 3 1 5 1 4 1 3 3.7 3.21 8.5 61.7 1 2 1 3 1 4 1 5 TCRβ 2 1 1 3 1 4 1 5 CD44 1 2 GFP
MR1-5-OP-RU CD24 CD24 CD44 MAIT cells 2.78 11.2 WT RORγt- GFP reporter 1 5 1 4 1 3 2.28 1 5 1 4 1 3 4.8 1.6 8.1 1 5 1 4 1 3 1 5 1 4 1 3 3.7 3.21 8.5 61.7 1 2 1 3 1 4 1 5 TCRβ 2 1 1 3 1 4 1 5 CD44 1 2 GFP
Suppl Video: Tumor cells (green) and monocytes (white) are seeded on a confluent endothelial
 Supplementary Information Häuselmann et al. Monocyte induction of E-selectin-mediated endothelial activation releases VE-cadherin junctions to promote tumor cell extravasation in the metastasis cascade
Supplementary Information Häuselmann et al. Monocyte induction of E-selectin-mediated endothelial activation releases VE-cadherin junctions to promote tumor cell extravasation in the metastasis cascade
Supplementary Information:
 Supplementary Information: Follicular regulatory T cells with Bcl6 expression suppress germinal center reactions by Yeonseok Chung, Shinya Tanaka, Fuliang Chu, Roza Nurieva, Gustavo J. Martinez, Seema
Supplementary Information: Follicular regulatory T cells with Bcl6 expression suppress germinal center reactions by Yeonseok Chung, Shinya Tanaka, Fuliang Chu, Roza Nurieva, Gustavo J. Martinez, Seema
BD Flow Cytometry Reagents Multicolor Panels Designed for Optimal Resolution with the BD LSRFortessa X-20 Cell Analyzer
 Multicolor Panels Designed for Optimal Resolution with the BD LSRFortessa X-2 Cell Analyzer Proper multicolor panel design takes into account fluorochrome brightness, antigen density, co-expression, and
Multicolor Panels Designed for Optimal Resolution with the BD LSRFortessa X-2 Cell Analyzer Proper multicolor panel design takes into account fluorochrome brightness, antigen density, co-expression, and
well for 2 h at rt. Each dot represents an individual mouse and bar is the mean ±
 Supplementary data: Control DC Blimp-1 ko DC 8 6 4 2-2 IL-1β p=.5 medium 8 6 4 2 IL-2 Medium p=.16 8 6 4 2 IL-6 medium p=.3 5 4 3 2 1-1 medium IL-1 n.s. 25 2 15 1 5 IL-12(p7) p=.15 5 IFNγ p=.65 4 3 2 1
Supplementary data: Control DC Blimp-1 ko DC 8 6 4 2-2 IL-1β p=.5 medium 8 6 4 2 IL-2 Medium p=.16 8 6 4 2 IL-6 medium p=.3 5 4 3 2 1-1 medium IL-1 n.s. 25 2 15 1 5 IL-12(p7) p=.15 5 IFNγ p=.65 4 3 2 1
Supplemental Information. Aryl Hydrocarbon Receptor Controls. Monocyte Differentiation. into Dendritic Cells versus Macrophages
 Immunity, Volume 47 Supplemental Information Aryl Hydrocarbon Receptor Controls Monocyte Differentiation into Dendritic Cells versus Macrophages Christel Goudot, Alice Coillard, Alexandra-Chloé Villani,
Immunity, Volume 47 Supplemental Information Aryl Hydrocarbon Receptor Controls Monocyte Differentiation into Dendritic Cells versus Macrophages Christel Goudot, Alice Coillard, Alexandra-Chloé Villani,
Title. CitationCancer science, 109(4): Issue Date Doc URL. Rights(URL)
 Title Toll-like receptor 3 signal augments radiation-induc Yoshida, Sumito; Shime, Hiroaki; Takeda, Yohei; Nam, Author(s) Hiroki; Kasahara, Masanori; Seya, Tsukasa CitationCancer science, 19(): 956-965
Title Toll-like receptor 3 signal augments radiation-induc Yoshida, Sumito; Shime, Hiroaki; Takeda, Yohei; Nam, Author(s) Hiroki; Kasahara, Masanori; Seya, Tsukasa CitationCancer science, 19(): 956-965
Nature Immunology: doi: /ni Supplementary Figure 1. Examples of staining for each antibody used for the mass cytometry analysis.
 Supplementary Figure 1 Examples of staining for each antibody used for the mass cytometry analysis. To illustrate the functionality of each antibody probe, representative plots illustrating the expected
Supplementary Figure 1 Examples of staining for each antibody used for the mass cytometry analysis. To illustrate the functionality of each antibody probe, representative plots illustrating the expected
Supplementary Figure 1. mtor LysM and Rictor LysM mice have normal cellularity and percentages of hematopoe>c cells. a. Cell numbers of lung, liver,
 a. b. c. Supplementary Figure 1. mtor LysM and Rictor LysM mice have normal cellularity and percentages of hematopoe>c cells. a. Cell numbers of lung, liver, and spleen. b. Cell numbers of bone marrow
a. b. c. Supplementary Figure 1. mtor LysM and Rictor LysM mice have normal cellularity and percentages of hematopoe>c cells. a. Cell numbers of lung, liver, and spleen. b. Cell numbers of bone marrow
Commercially available HLA Class II tetramers (Beckman Coulter) conjugated to
 Class II tetramer staining Commercially available HLA Class II tetramers (Beckman Coulter) conjugated to PE were combined with dominant HIV epitopes (DRB1*0101-DRFYKTLRAEQASQEV, DRB1*0301- PEKEVLVWKFDSRLAFHH,
Class II tetramer staining Commercially available HLA Class II tetramers (Beckman Coulter) conjugated to PE were combined with dominant HIV epitopes (DRB1*0101-DRFYKTLRAEQASQEV, DRB1*0301- PEKEVLVWKFDSRLAFHH,
Optimizing Intracellular Flow Cytometry:
 Optimizing Intracellular Flow Cytometry: Simultaneous Detection of Cytokines and Transcription Factors An encore presentation by Jurg Rohrer, PhD, BD Biosciences 10.26.10 Outline Introduction Cytokines
Optimizing Intracellular Flow Cytometry: Simultaneous Detection of Cytokines and Transcription Factors An encore presentation by Jurg Rohrer, PhD, BD Biosciences 10.26.10 Outline Introduction Cytokines
Supplementary Figure S1. Flow cytometric analysis of the expression of Thy1 in NH cells. Flow cytometric analysis of the expression of T1/ST2 and
 Supplementary Figure S1. Flow cytometric analysis of the expression of Thy1 in NH cells. Flow cytometric analysis of the expression of T1/ST2 and Thy1 in NH cells derived from the lungs of naïve mice.
Supplementary Figure S1. Flow cytometric analysis of the expression of Thy1 in NH cells. Flow cytometric analysis of the expression of T1/ST2 and Thy1 in NH cells derived from the lungs of naïve mice.
Synergy of radiotherapy and PD-1 blockade in Kras-mutant lung cancer
 Supplementary Information Synergy of radiotherapy and PD-1 blockade in Kras-mutant lung cancer Grit S. Herter-Sprie, Shohei Koyama, Houari Korideck, Josephine Hai, Jiehui Deng, Yvonne Y. Li, Kevin A. Buczkowski,
Supplementary Information Synergy of radiotherapy and PD-1 blockade in Kras-mutant lung cancer Grit S. Herter-Sprie, Shohei Koyama, Houari Korideck, Josephine Hai, Jiehui Deng, Yvonne Y. Li, Kevin A. Buczkowski,
CD14 + S100A9 + Monocytic Myeloid-Derived Suppressor Cells and Their Clinical Relevance in Non-Small Cell Lung Cancer
 CD14 + S1A9 + Monocytic Myeloid-Derived Suppressor Cells and Their Clinical Relevance in Non-Small Cell Lung Cancer Po-Hao, Feng M.D., Kang-Yun, Lee, M.D. Ph.D., Ya-Ling Chang, Yao-Fei Chan, Lu- Wei, Kuo,Ting-Yu
CD14 + S1A9 + Monocytic Myeloid-Derived Suppressor Cells and Their Clinical Relevance in Non-Small Cell Lung Cancer Po-Hao, Feng M.D., Kang-Yun, Lee, M.D. Ph.D., Ya-Ling Chang, Yao-Fei Chan, Lu- Wei, Kuo,Ting-Yu
% of live splenocytes. STAT5 deletion. (open shapes) % ROSA + % floxed
 Supp. Figure 1. a 14 1 1 8 6 spleen cells (x1 6 ) 16 % of live splenocytes 5 4 3 1 % of live splenocytes 8 6 4 b 1 1 c % of CD11c + splenocytes (closed shapes) 8 6 4 8 6 4 % ROSA + (open shapes) % floxed
Supp. Figure 1. a 14 1 1 8 6 spleen cells (x1 6 ) 16 % of live splenocytes 5 4 3 1 % of live splenocytes 8 6 4 b 1 1 c % of CD11c + splenocytes (closed shapes) 8 6 4 8 6 4 % ROSA + (open shapes) % floxed
CD25-PE (BD Biosciences) and labeled with anti-pe-microbeads (Miltenyi Biotec) for depletion of CD25 +
 Supplements Supplemental Materials and Methods Depletion of CD25 + T-cells from PBMC. Fresh or HD precultured PBMC were stained with the conjugate CD25-PE (BD Biosciences) and labeled with anti-pe-microbeads
Supplements Supplemental Materials and Methods Depletion of CD25 + T-cells from PBMC. Fresh or HD precultured PBMC were stained with the conjugate CD25-PE (BD Biosciences) and labeled with anti-pe-microbeads
Supplementary Figure 1
 Supplementary Figure 1 Identification of IFN-γ-producing CD8 + and CD4 + T cells with naive phenotype by alternative gating and sample-processing strategies. a. Contour 5% probability plots show definition
Supplementary Figure 1 Identification of IFN-γ-producing CD8 + and CD4 + T cells with naive phenotype by alternative gating and sample-processing strategies. a. Contour 5% probability plots show definition
Supplementary Figure 1. Efficient DC depletion in CD11c.DOG transgenic mice
 Supplementary Figure 1. Efficient DC depletion in CD11c.DOG transgenic mice (a) CD11c.DOG transgenic mice (tg) were treated with 8 ng/g body weight (b.w.) diphtheria toxin (DT) i.p. on day -1 and every
Supplementary Figure 1. Efficient DC depletion in CD11c.DOG transgenic mice (a) CD11c.DOG transgenic mice (tg) were treated with 8 ng/g body weight (b.w.) diphtheria toxin (DT) i.p. on day -1 and every
Supplementary information. The proton-sensing G protein-coupled receptor T-cell death-associated gene 8
 1 Supplementary information 2 3 The proton-sensing G protein-coupled receptor T-cell death-associated gene 8 4 (TDAG8) shows cardioprotective effects against myocardial infarction 5 Akiomi Nagasaka 1+,
1 Supplementary information 2 3 The proton-sensing G protein-coupled receptor T-cell death-associated gene 8 4 (TDAG8) shows cardioprotective effects against myocardial infarction 5 Akiomi Nagasaka 1+,
Metabolic ER stress and inflammation in white adipose tissue (WAT) of mice with dietary obesity.
 Supplementary Figure 1 Metabolic ER stress and inflammation in white adipose tissue (WAT) of mice with dietary obesity. Male C57BL/6J mice were fed a normal chow (NC, 10% fat) or a high-fat diet (HFD,
Supplementary Figure 1 Metabolic ER stress and inflammation in white adipose tissue (WAT) of mice with dietary obesity. Male C57BL/6J mice were fed a normal chow (NC, 10% fat) or a high-fat diet (HFD,
Supplementary Figures for TSC1 controls macrophage polarization to prevent inflammatory disorder by Linnan Zhu et al
 Supplementary Figures for TSC1 controls macrophage polarization to prevent inflammatory disorder by Linnan Zhu et al Suppl. Fig. 1 Tissue DN C Proteins kd TSC1-17 TSC 1 loxp bp -48-285 ctin PEMs Neutrophils
Supplementary Figures for TSC1 controls macrophage polarization to prevent inflammatory disorder by Linnan Zhu et al Suppl. Fig. 1 Tissue DN C Proteins kd TSC1-17 TSC 1 loxp bp -48-285 ctin PEMs Neutrophils
Supplemental Table I.
 Supplemental Table I Male / Mean ± SEM n Mean ± SEM n Body weight, g 29.2±0.4 17 29.7±0.5 17 Total cholesterol, mg/dl 534.0±30.8 17 561.6±26.1 17 HDL-cholesterol, mg/dl 9.6±0.8 17 10.1±0.7 17 Triglycerides,
Supplemental Table I Male / Mean ± SEM n Mean ± SEM n Body weight, g 29.2±0.4 17 29.7±0.5 17 Total cholesterol, mg/dl 534.0±30.8 17 561.6±26.1 17 HDL-cholesterol, mg/dl 9.6±0.8 17 10.1±0.7 17 Triglycerides,
NK cell flow cytometric assay In vivo DC viability and migration assay
 NK cell flow cytometric assay 6 NK cells were purified, by negative selection with the NK Cell Isolation Kit (Miltenyi iotec), from spleen and lymph nodes of 6 RAG1KO mice, injected the day before with
NK cell flow cytometric assay 6 NK cells were purified, by negative selection with the NK Cell Isolation Kit (Miltenyi iotec), from spleen and lymph nodes of 6 RAG1KO mice, injected the day before with
D CD8 T cell number (x10 6 )
 IFNγ Supplemental Figure 1. CD T cell number (x1 6 ) 18 15 1 9 6 3 CD CD T cells CD6L C CD5 CD T cells CD6L D CD8 T cell number (x1 6 ) 1 8 6 E CD CD8 T cells CD6L F Log(1)CFU/g Feces 1 8 6 p
IFNγ Supplemental Figure 1. CD T cell number (x1 6 ) 18 15 1 9 6 3 CD CD T cells CD6L C CD5 CD T cells CD6L D CD8 T cell number (x1 6 ) 1 8 6 E CD CD8 T cells CD6L F Log(1)CFU/g Feces 1 8 6 p
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. NKT ligand-loaded tumour antigen-presenting B cell- and monocyte-based vaccine induces NKT, NK and CD8 T cell responses. (A) The cytokine profiles of liver
Supplementary Figures Supplementary Figure 1. NKT ligand-loaded tumour antigen-presenting B cell- and monocyte-based vaccine induces NKT, NK and CD8 T cell responses. (A) The cytokine profiles of liver
Targeting tumour associated macrophages in anti-cancer therapies. Annamaria Gal Seminar Series on Drug Discovery Budapest 5 January 2018
 Targeting tumour associated macrophages in anti-cancer therapies Annamaria Gal Seminar Series on Drug Discovery Budapest 5 January 2018 Macrophages: Professional phagocytes of the myeloid lineage APC,
Targeting tumour associated macrophages in anti-cancer therapies Annamaria Gal Seminar Series on Drug Discovery Budapest 5 January 2018 Macrophages: Professional phagocytes of the myeloid lineage APC,
Supplementary Figure 1. mrna expression of chitinase and chitinase-like protein in splenic immune cells. Each splenic immune cell population was
 Supplementary Figure 1. mrna expression of chitinase and chitinase-like protein in splenic immune cells. Each splenic immune cell population was sorted by FACS. Surface markers for sorting were CD11c +
Supplementary Figure 1. mrna expression of chitinase and chitinase-like protein in splenic immune cells. Each splenic immune cell population was sorted by FACS. Surface markers for sorting were CD11c +
Supplementary Figure 1: TSLP receptor skin expression in dcssc. A: Healthy control (HC) skin with TSLP receptor expression in brown (10x
 Supplementary Figure 1: TSLP receptor skin expression in dcssc. A: Healthy control (HC) skin with TSLP receptor expression in brown (10x magnification). B: Second HC skin stained for TSLP receptor in brown
Supplementary Figure 1: TSLP receptor skin expression in dcssc. A: Healthy control (HC) skin with TSLP receptor expression in brown (10x magnification). B: Second HC skin stained for TSLP receptor in brown
Human Innate Lymphoid Cells: crosstalk with CD4 + regulatory T cells and role in Type 1 Diabetes
 Joint Lab Meeting 03/03/2015 Human Innate Lymphoid Cells: crosstalk with CD4 + regulatory T cells and role in Type 1 Diabetes Caroline Raffin Bluestone Lab ILCs: Definition - Common lymphoid progenitor
Joint Lab Meeting 03/03/2015 Human Innate Lymphoid Cells: crosstalk with CD4 + regulatory T cells and role in Type 1 Diabetes Caroline Raffin Bluestone Lab ILCs: Definition - Common lymphoid progenitor
ACTIVATION AND EFFECTOR FUNCTIONS OF CELL-MEDIATED IMMUNITY AND NK CELLS. Choompone Sakonwasun, MD (Hons), FRCPT
 ACTIVATION AND EFFECTOR FUNCTIONS OF CELL-MEDIATED IMMUNITY AND NK CELLS Choompone Sakonwasun, MD (Hons), FRCPT Types of Adaptive Immunity Types of T Cell-mediated Immune Reactions CTLs = cytotoxic T lymphocytes
ACTIVATION AND EFFECTOR FUNCTIONS OF CELL-MEDIATED IMMUNITY AND NK CELLS Choompone Sakonwasun, MD (Hons), FRCPT Types of Adaptive Immunity Types of T Cell-mediated Immune Reactions CTLs = cytotoxic T lymphocytes
x Lymphocyte count /µl CD8+ count/µl 800 Calculated
 % Lymphocyte in CBC A. 50 40 30 20 10 Lymphocyte count /µl B. x10 3 2.5 1.5 C. 50 D. 1000 % CD3+CD8+ Cells 40 30 20 Calculated CD8+ count/µl 800 600 400 200 10 0 #61 #63 #64 #65 #68 #71 #72 #75 Figure
% Lymphocyte in CBC A. 50 40 30 20 10 Lymphocyte count /µl B. x10 3 2.5 1.5 C. 50 D. 1000 % CD3+CD8+ Cells 40 30 20 Calculated CD8+ count/µl 800 600 400 200 10 0 #61 #63 #64 #65 #68 #71 #72 #75 Figure
pplementary Figur Supplementary Figure 1. a.
 pplementary Figur Supplementary Figure 1. a. Quantification by RT-qPCR of YFV-17D and YFV-17D pol- (+) RNA in the supernatant of cultured Huh7.5 cells following viral RNA electroporation of respective
pplementary Figur Supplementary Figure 1. a. Quantification by RT-qPCR of YFV-17D and YFV-17D pol- (+) RNA in the supernatant of cultured Huh7.5 cells following viral RNA electroporation of respective
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
Supplementary Figure 1. Antibiotic partially rescues mice from sepsis. (ab) BALB/c mice under CLP were treated with antibiotic or PBS.
 1 Supplementary Figure 1. Antibiotic partially rescues mice from sepsis. (ab) BALB/c mice under CLP were treated with antibiotic or PBS. (a) Survival curves. WT Sham (n=5), WT CLP or WT CLP antibiotic
1 Supplementary Figure 1. Antibiotic partially rescues mice from sepsis. (ab) BALB/c mice under CLP were treated with antibiotic or PBS. (a) Survival curves. WT Sham (n=5), WT CLP or WT CLP antibiotic
Application Information Bulletin: Human NK Cells Phenotypic characterizing of human Natural Killer (NK) cell populations in peripheral blood
 Application Information Bulletin: Human NK Cells Phenotypic characterizing of human Natural Killer (NK) cell populations in peripheral blood Christopher A Fraker, Ph.D., University of Miami - Miami, Florida
Application Information Bulletin: Human NK Cells Phenotypic characterizing of human Natural Killer (NK) cell populations in peripheral blood Christopher A Fraker, Ph.D., University of Miami - Miami, Florida
Supporting Information
 Supporting Information Bellora et al. 10.1073/pnas.1007654108 SI Materials and Methods Cells. NK cells purified from peripheral blood mononuclear cells (PBMC) of healthy donors (Human NK Cell Isolation
Supporting Information Bellora et al. 10.1073/pnas.1007654108 SI Materials and Methods Cells. NK cells purified from peripheral blood mononuclear cells (PBMC) of healthy donors (Human NK Cell Isolation
Supplemental Materials for. Effects of sphingosine-1-phosphate receptor 1 phosphorylation in response to. FTY720 during neuroinflammation
 Supplemental Materials for Effects of sphingosine-1-phosphate receptor 1 phosphorylation in response to FTY7 during neuroinflammation This file includes: Supplemental Table 1. EAE clinical parameters of
Supplemental Materials for Effects of sphingosine-1-phosphate receptor 1 phosphorylation in response to FTY7 during neuroinflammation This file includes: Supplemental Table 1. EAE clinical parameters of
Pearson r = P (one-tailed) = n = 9
 8F4-Specific Lysis, % 1 UPN1 UPN3 8 UPN7 6 Pearson r =.69 UPN2 UPN5 P (one-tailed) =.192 4 UPN8 n = 9 2 UPN9 UPN4 UPN6 5 1 15 2 25 8 8F4, % Max MFI Supplementary Figure S1. AML samples UPN1-UPN9 show variable
8F4-Specific Lysis, % 1 UPN1 UPN3 8 UPN7 6 Pearson r =.69 UPN2 UPN5 P (one-tailed) =.192 4 UPN8 n = 9 2 UPN9 UPN4 UPN6 5 1 15 2 25 8 8F4, % Max MFI Supplementary Figure S1. AML samples UPN1-UPN9 show variable
Supplemental Table 1. Primer sequences for transcript analysis
 Supplemental Table 1. Primer sequences for transcript analysis Primer Sequence (5 3 ) Primer Sequence (5 3 ) Mmp2 Forward CCCGTGTGGCCCTC Mmp15 Forward CGGGGCTGGCT Reverse GCTCTCCCGGTTTC Reverse CCTGGTGTGCCTGCTC
Supplemental Table 1. Primer sequences for transcript analysis Primer Sequence (5 3 ) Primer Sequence (5 3 ) Mmp2 Forward CCCGTGTGGCCCTC Mmp15 Forward CGGGGCTGGCT Reverse GCTCTCCCGGTTTC Reverse CCTGGTGTGCCTGCTC
Nature Medicine: doi: /nm.3922
 Title: Glucocorticoid-induced tumor necrosis factor receptor-related protein co-stimulation facilitates tumor regression by inducing IL-9-producing helper T cells Authors: Il-Kyu Kim, Byung-Seok Kim, Choong-Hyun
Title: Glucocorticoid-induced tumor necrosis factor receptor-related protein co-stimulation facilitates tumor regression by inducing IL-9-producing helper T cells Authors: Il-Kyu Kim, Byung-Seok Kim, Choong-Hyun
Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Table
 Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Table Title of file for HTML: Peer Review File Description: Innate Scavenger Receptor-A regulates
Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Table Title of file for HTML: Peer Review File Description: Innate Scavenger Receptor-A regulates
Supporting Information
 Supporting Information Valkenburg et al. 10.1073/pnas.1403684111 SI Materials and Methods ELISA and Microneutralization. Sera were treated with Receptor Destroying Enzyme II (RDE II, Accurate) before ELISA
Supporting Information Valkenburg et al. 10.1073/pnas.1403684111 SI Materials and Methods ELISA and Microneutralization. Sera were treated with Receptor Destroying Enzyme II (RDE II, Accurate) before ELISA
CELLULAR IMMUNE CORRELATES OF PROTECTION AGAINST
 CELLULAR IMMUNE CORRELATES OF PROTECTION AGAINST SYMPTOMATIC PANDEMIC INFLUENZA Saranya Sridhar 1, Shaima Begom 1, Alison Bermingham 2, Katja Hoschler 2, Walt Adamson 3, William Carman 3, Thomas Bean 4,
CELLULAR IMMUNE CORRELATES OF PROTECTION AGAINST SYMPTOMATIC PANDEMIC INFLUENZA Saranya Sridhar 1, Shaima Begom 1, Alison Bermingham 2, Katja Hoschler 2, Walt Adamson 3, William Carman 3, Thomas Bean 4,
Categorical analysis of human T cell heterogeneity with One-SENSE
 1 2 3 4 Supplementary Information Categorical analysis of human T cell heterogeneity with One-SENSE 5 6 Running title: T cell Analysis by One-SENSE 7 8 9 1 11 12 13 14 15 16 17 18 19 2 21 22 23 24 25 26
1 2 3 4 Supplementary Information Categorical analysis of human T cell heterogeneity with One-SENSE 5 6 Running title: T cell Analysis by One-SENSE 7 8 9 1 11 12 13 14 15 16 17 18 19 2 21 22 23 24 25 26
The encephalitogenicity of TH17 cells is dependent on IL-1- and IL-23- induced production of the cytokine GM-CSF
 CORRECTION NOTICE Nat.Immunol. 12, 568 575 (2011) The encephalitogenicity of TH17 cells is dependent on IL-1- and IL-23- induced production of the cytokine GM-CSF Mohamed El-Behi, Bogoljub Ciric, Hong
CORRECTION NOTICE Nat.Immunol. 12, 568 575 (2011) The encephalitogenicity of TH17 cells is dependent on IL-1- and IL-23- induced production of the cytokine GM-CSF Mohamed El-Behi, Bogoljub Ciric, Hong
Nature Immunology: doi: /ni Supplementary Figure 1. Gene expression profile of CD4 + T cells and CTL responses in Bcl6-deficient mice.
 Supplementary Figure 1 Gene expression profile of CD4 + T cells and CTL responses in Bcl6-deficient mice. (a) Gene expression profile in the resting CD4 + T cells were analyzed by an Affymetrix microarray
Supplementary Figure 1 Gene expression profile of CD4 + T cells and CTL responses in Bcl6-deficient mice. (a) Gene expression profile in the resting CD4 + T cells were analyzed by an Affymetrix microarray
Supplementary Figure 1. Immune profiles of untreated and PD-1 blockade resistant EGFR and Kras mouse lung tumors (a) Total lung weight of untreated
 1 Supplementary Figure 1. Immune profiles of untreated and PD-1 blockade resistant EGFR and Kras mouse lung tumors (a) Total lung weight of untreated (U) EGFR TL mice (n=7), Kras mice (n=7), PD-1 blockade
1 Supplementary Figure 1. Immune profiles of untreated and PD-1 blockade resistant EGFR and Kras mouse lung tumors (a) Total lung weight of untreated (U) EGFR TL mice (n=7), Kras mice (n=7), PD-1 blockade
ndln NK Cells (x10 3 ) Days post-infection (A/PR/8) *** *** *** Liver NK Cells (x10 4 ) Days post-infection (MCMV)
 A mln NK Cells(x ) 6 1 * ndln NK Cells (x ) ns C Lung NK Cells(x ) 1 1 7 * D LN NK Cells (x ) 1 7 1 7 Days post-infection (A/PR/8) * * E Liver NK Cells (x ) 1 7 Days post-infection (A/PR/8) * * * 1 7 Days
A mln NK Cells(x ) 6 1 * ndln NK Cells (x ) ns C Lung NK Cells(x ) 1 1 7 * D LN NK Cells (x ) 1 7 1 7 Days post-infection (A/PR/8) * * E Liver NK Cells (x ) 1 7 Days post-infection (A/PR/8) * * * 1 7 Days
Human and mouse T cell regulation mediated by soluble CD52 interaction with Siglec-10. Esther Bandala-Sanchez, Yuxia Zhang, Simone Reinwald,
 Human and mouse T cell regulation mediated by soluble CD52 interaction with Siglec-1 Esther Bandala-Sanchez, Yuxia Zhang, Simone Reinwald, James A. Dromey, Bo Han Lee, Junyan Qian, Ralph M Böhmer and Leonard
Human and mouse T cell regulation mediated by soluble CD52 interaction with Siglec-1 Esther Bandala-Sanchez, Yuxia Zhang, Simone Reinwald, James A. Dromey, Bo Han Lee, Junyan Qian, Ralph M Böhmer and Leonard
Supplementary Materials for
 www.sciencemag.org/content/348/6241/aaa825/suppl/dc1 Supplementary Materials for A mucosal vaccine against Chlamydia trachomatis generates two waves of protective memory T cells Georg Stary,* Andrew Olive,
www.sciencemag.org/content/348/6241/aaa825/suppl/dc1 Supplementary Materials for A mucosal vaccine against Chlamydia trachomatis generates two waves of protective memory T cells Georg Stary,* Andrew Olive,
Supplemental Figure 1
 Supplemental Figure 1 1a 1c PD-1 MFI fold change 6 5 4 3 2 1 IL-1α IL-2 IL-4 IL-6 IL-1 IL-12 IL-13 IL-15 IL-17 IL-18 IL-21 IL-23 IFN-α Mut Human PD-1 promoter SBE-D 5 -GTCTG- -1.2kb SBE-P -CAGAC- -1.kb
Supplemental Figure 1 1a 1c PD-1 MFI fold change 6 5 4 3 2 1 IL-1α IL-2 IL-4 IL-6 IL-1 IL-12 IL-13 IL-15 IL-17 IL-18 IL-21 IL-23 IFN-α Mut Human PD-1 promoter SBE-D 5 -GTCTG- -1.2kb SBE-P -CAGAC- -1.kb
MAIT cell function is modulated by PD-1 signaling in patients with active
 MAIT cell function is modulated by PD-1 signaling in patients with active tuberculosis Jing Jiang, M.D., Xinjing Wang, M.D., Hongjuan An, M.Sc., Bingfen Yang, Ph.D., Zhihong Cao, M.Sc., Yanhua Liu, Ph.D.,
MAIT cell function is modulated by PD-1 signaling in patients with active tuberculosis Jing Jiang, M.D., Xinjing Wang, M.D., Hongjuan An, M.Sc., Bingfen Yang, Ph.D., Zhihong Cao, M.Sc., Yanhua Liu, Ph.D.,
SUPPLEMENTAL INFORMATIONS
 1 SUPPLEMENTAL INFORMATIONS Figure S1 Cumulative ZIKV production by testis explants over a 9 day-culture period. Viral titer values presented in Figure 1B (viral release over a 3 day-culture period measured
1 SUPPLEMENTAL INFORMATIONS Figure S1 Cumulative ZIKV production by testis explants over a 9 day-culture period. Viral titer values presented in Figure 1B (viral release over a 3 day-culture period measured
Measuring Dendritic Cells
 Measuring Dendritic Cells A.D. Donnenberg, V.S. Donnenberg UNIVERSITY of PITTSBURGH CANCER INSTITUTE CCS Longbeach 10_04 Measuring DC Rare event detection The basics of DC measurement Applications Cancer
Measuring Dendritic Cells A.D. Donnenberg, V.S. Donnenberg UNIVERSITY of PITTSBURGH CANCER INSTITUTE CCS Longbeach 10_04 Measuring DC Rare event detection The basics of DC measurement Applications Cancer
Supplementary Figure 1. Enhanced detection of CTLA-4 on the surface of HIV-specific
 SUPPLEMENTARY FIGURE LEGEND Supplementary Figure 1. Enhanced detection of CTLA-4 on the surface of HIV-specific CD4 + T cells correlates with intracellular CTLA-4 levels. (a) Comparative CTLA-4 levels
SUPPLEMENTARY FIGURE LEGEND Supplementary Figure 1. Enhanced detection of CTLA-4 on the surface of HIV-specific CD4 + T cells correlates with intracellular CTLA-4 levels. (a) Comparative CTLA-4 levels
Human Immunodeficiency Virus Type-1 Myeloid Derived Suppressor Cells Inhibit Cytomegalovirus Inflammation through Interleukin-27 and B7-H4
 Human Immunodeficiency Virus Type-1 Myeloid Derived Suppressor Cells Inhibit Cytomegalovirus Inflammation through Interleukin-27 and B7-H4 Ankita Garg, Rodney Trout and Stephen A. Spector,,* Department
Human Immunodeficiency Virus Type-1 Myeloid Derived Suppressor Cells Inhibit Cytomegalovirus Inflammation through Interleukin-27 and B7-H4 Ankita Garg, Rodney Trout and Stephen A. Spector,,* Department
HD1 (FLU) HD2 (EBV) HD2 (FLU)
 ramer staining + anti-pe beads ramer staining a HD1 (FLU) HD2 (EBV) HD2 (FLU).73.11.56.46.24 1.12 b CD127 + c CD127 + d CD127 - e CD127 - PD1 - PD1 + PD1 + PD1-1 1 1 1 %CD127 + PD1-8 6 4 2 + anti-pe %CD127
ramer staining + anti-pe beads ramer staining a HD1 (FLU) HD2 (EBV) HD2 (FLU).73.11.56.46.24 1.12 b CD127 + c CD127 + d CD127 - e CD127 - PD1 - PD1 + PD1 + PD1-1 1 1 1 %CD127 + PD1-8 6 4 2 + anti-pe %CD127
Nature Immunology: doi: /ni Supplementary Figure 1. Huwe1 has high expression in HSCs and is necessary for quiescence.
 Supplementary Figure 1 Huwe1 has high expression in HSCs and is necessary for quiescence. (a) Heat map visualizing expression of genes with a known function in ubiquitin-mediated proteolysis (KEGG: Ubiquitin
Supplementary Figure 1 Huwe1 has high expression in HSCs and is necessary for quiescence. (a) Heat map visualizing expression of genes with a known function in ubiquitin-mediated proteolysis (KEGG: Ubiquitin
Granulocyte macrophage colonystimulating factor receptor α expression and its targeting in antigen-induced arthritis and inflammation
 Cook et al. Arthritis Research & Therapy (2016) 18:287 DOI 10.1186/s13075-016-1185-9 RESEARCH ARTICLE Open Access Granulocyte macrophage colonystimulating factor receptor α expression and its targeting
Cook et al. Arthritis Research & Therapy (2016) 18:287 DOI 10.1186/s13075-016-1185-9 RESEARCH ARTICLE Open Access Granulocyte macrophage colonystimulating factor receptor α expression and its targeting
Supplementary Table 1 Clinicopathological characteristics of 35 patients with CRCs
 Supplementary Table Clinicopathological characteristics of 35 patients with CRCs Characteristics Type-A CRC Type-B CRC P value Sex Male / Female 9 / / 8.5 Age (years) Median (range) 6. (9 86) 6.5 (9 76).95
Supplementary Table Clinicopathological characteristics of 35 patients with CRCs Characteristics Type-A CRC Type-B CRC P value Sex Male / Female 9 / / 8.5 Age (years) Median (range) 6. (9 86) 6.5 (9 76).95
Supplementary Figure 1. Example of gating strategy
 Supplementary Figure 1. Example of gating strategy Legend Supplementary Figure 1: First, gating is performed to include only single cells (singlets) (A) and CD3+ cells (B). After gating on the lymphocyte
Supplementary Figure 1. Example of gating strategy Legend Supplementary Figure 1: First, gating is performed to include only single cells (singlets) (A) and CD3+ cells (B). After gating on the lymphocyte
Therapeutic effect of baicalin on experimental autoimmune encephalomyelitis. is mediated by SOCS3 regulatory pathway
 Therapeutic effect of baicalin on experimental autoimmune encephalomyelitis is mediated by SOCS3 regulatory pathway Yuan Zhang 1,2, Xing Li 1,2, Bogoljub Ciric 1, Cun-gen Ma 3, Bruno Gran 4, Abdolmohamad
Therapeutic effect of baicalin on experimental autoimmune encephalomyelitis is mediated by SOCS3 regulatory pathway Yuan Zhang 1,2, Xing Li 1,2, Bogoljub Ciric 1, Cun-gen Ma 3, Bruno Gran 4, Abdolmohamad
Combined Rho-kinase inhibition and immunogenic cell death triggers and propagates immunity against cancer
 Supplementary Information Combined Rho-kinase inhibition and immunogenic cell death triggers and propagates immunity against cancer Gi-Hoon Nam, Eun-Jung Lee, Yoon Kyoung Kim, Yeonsun Hong, Yoonjeong Choi,
Supplementary Information Combined Rho-kinase inhibition and immunogenic cell death triggers and propagates immunity against cancer Gi-Hoon Nam, Eun-Jung Lee, Yoon Kyoung Kim, Yeonsun Hong, Yoonjeong Choi,
Supplementalgfigureg1gSchematicgdiagramgofgtumor1modellingg
 SChinjectionh F:LuchLCLsh IVhinjectionh T:cellsh Monitorhforhtumorh growthhandhxeno: reactivehgvhd GVLgexperimentg kcbgvsgpbgt1cellse Xeno1reactiveg experimentg kcbgvsgpbgt1cellse IVhinjectionh 5xh,N^6
SChinjectionh F:LuchLCLsh IVhinjectionh T:cellsh Monitorhforhtumorh growthhandhxeno: reactivehgvhd GVLgexperimentg kcbgvsgpbgt1cellse Xeno1reactiveg experimentg kcbgvsgpbgt1cellse IVhinjectionh 5xh,N^6
Supplementary Figure S1. PTPN2 levels are not altered in proliferating CD8+ T cells. Lymph node (LN) CD8+ T cells from C57BL/6 mice were stained with
 Supplementary Figure S1. PTPN2 levels are not altered in proliferating CD8+ T cells. Lymph node (LN) CD8+ T cells from C57BL/6 mice were stained with CFSE and stimulated with plate-bound α-cd3ε (10µg/ml)
Supplementary Figure S1. PTPN2 levels are not altered in proliferating CD8+ T cells. Lymph node (LN) CD8+ T cells from C57BL/6 mice were stained with CFSE and stimulated with plate-bound α-cd3ε (10µg/ml)
January 25, 2017 Scientific Research Process Name of Journal: ESPS Manuscript NO: Manuscript Type: Title: Authors: Correspondence to
 January 25, 2017 Scientific Research Process Name of Journal: World Journal of Gastroenterology ESPS Manuscript NO: 31928 Manuscript Type: ORIGINAL ARTICLE Title: Thiopurine use associated with reduced
January 25, 2017 Scientific Research Process Name of Journal: World Journal of Gastroenterology ESPS Manuscript NO: 31928 Manuscript Type: ORIGINAL ARTICLE Title: Thiopurine use associated with reduced
PBMC from each patient were suspended in AIM V medium (Invitrogen) with 5% human
 Anti-CD19-CAR transduced T-cell preparation PBMC from each patient were suspended in AIM V medium (Invitrogen) with 5% human AB serum (Gemini) and 300 international units/ml IL-2 (Novartis). T cell proliferation
Anti-CD19-CAR transduced T-cell preparation PBMC from each patient were suspended in AIM V medium (Invitrogen) with 5% human AB serum (Gemini) and 300 international units/ml IL-2 (Novartis). T cell proliferation
Supplementary Figure 1. Deletion of Smad3 prevents B16F10 melanoma invasion and metastasis in a mouse s.c. tumor model.
 A B16F1 s.c. Lung LN Distant lymph nodes Colon B B16F1 s.c. Supplementary Figure 1. Deletion of Smad3 prevents B16F1 melanoma invasion and metastasis in a mouse s.c. tumor model. Highly invasive growth
A B16F1 s.c. Lung LN Distant lymph nodes Colon B B16F1 s.c. Supplementary Figure 1. Deletion of Smad3 prevents B16F1 melanoma invasion and metastasis in a mouse s.c. tumor model. Highly invasive growth
York criteria, 6 RA patients and 10 age- and gender-matched healthy controls (HCs).
 MATERIALS AND METHODS Study population Blood samples were obtained from 15 patients with AS fulfilling the modified New York criteria, 6 RA patients and 10 age- and gender-matched healthy controls (HCs).
MATERIALS AND METHODS Study population Blood samples were obtained from 15 patients with AS fulfilling the modified New York criteria, 6 RA patients and 10 age- and gender-matched healthy controls (HCs).
Supplementary Figure 1: Fn14 is upregulated in the epidermis and dermis of mice
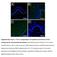 Supplementary Figure 1: Fn14 is upregulated in the epidermis and dermis of mice undergoing AD- and psoriasis-like disease. Immunofluorescence staining for Fn14 (green) and DAPI (blue) in skin of naïve
Supplementary Figure 1: Fn14 is upregulated in the epidermis and dermis of mice undergoing AD- and psoriasis-like disease. Immunofluorescence staining for Fn14 (green) and DAPI (blue) in skin of naïve
<10. IL-1β IL-6 TNF + _ TGF-β + IL-23
 3 ns 25 ns 2 IL-17 (pg/ml) 15 1 ns ns 5 IL-1β IL-6 TNF
3 ns 25 ns 2 IL-17 (pg/ml) 15 1 ns ns 5 IL-1β IL-6 TNF
Ex vivo Human Antigen-specific T Cell Proliferation and Degranulation Willemijn Hobo 1, Wieger Norde 1 and Harry Dolstra 2*
 Ex vivo Human Antigen-specific T Cell Proliferation and Degranulation Willemijn Hobo 1, Wieger Norde 1 and Harry Dolstra 2* 1 Department of Laboratory Medicine - Laboratory of Hematology, Radboud University
Ex vivo Human Antigen-specific T Cell Proliferation and Degranulation Willemijn Hobo 1, Wieger Norde 1 and Harry Dolstra 2* 1 Department of Laboratory Medicine - Laboratory of Hematology, Radboud University
Supplemental Figure S1. RANK expression on human lung cancer cells.
 Supplemental Figure S1. RANK expression on human lung cancer cells. (A) Incidence and H-Scores of RANK expression determined from IHC in the indicated primary lung cancer subgroups. The overall expression
Supplemental Figure S1. RANK expression on human lung cancer cells. (A) Incidence and H-Scores of RANK expression determined from IHC in the indicated primary lung cancer subgroups. The overall expression
Supplementary Figure 1. Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Nature Immunology: doi: /ni.
 Supplementary Figure 1 Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Expression of Mll4 floxed alleles (16-19) in naive CD4 + T cells isolated from lymph nodes and
Supplementary Figure 1 Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Expression of Mll4 floxed alleles (16-19) in naive CD4 + T cells isolated from lymph nodes and
