SAP1, SAP3, SAP4, SAP7, SAP8,
|
|
|
- Iris Spencer
- 6 years ago
- Views:
Transcription
1 MAJOR ARTICLE Differential Expression of Candida albicans Secreted Aspartyl Proteinase and Phospholipase B Genes in Humans Correlates with Active Oral and Vaginal Infections Julian R. Naglik, 1 Catherine A. Rodgers, 2 Penelope J. Shirlaw, 1 Jennifer L. Dobbie, 2 Lynette L. Fernandes-Naglik, 1 Deborah Greenspan, 3 Nina Agabian, 3 and Stephen J. Challacombe 1 1 Department of Oral Medicine, Pathology, and Immunology, Guy s, King s, and St. Thomas Dental Institute, Kings College London (Guy s Campus), and 2 Department of Genitourinary and HIV Medicine, Guy s and St Thomas Hospital Trust, St Thomas Hospital, London, United Kingdom; 3 Department of Stomatology, University of California, San Francisco The in vivo expression of Candida albicans secreted aspartyl proteinase (SAP1 SAP8) and phospholipase B (PLB1 and PLB2) genes was analyzed in 137 human subjects with oral and vaginal candidiasis or carriage. Total RNA was isolated from whole unstimulated saliva or vaginal swabs, and the expression of SAP1 8 and PLB1 2 was evaluated by reverse-transcriptase polymerase chain reaction using specific primer sets. A spectrum of SAP gene expression profiles was obtained from different C. albicans strains during symptomatic disease and asymptomatic carriage. SAP2 and SAP5 were the most common genes expressed during both infection and carriage. SAP1, SAP3, SAP4, SAP7, SAP8, and PLB1 expression was correlated with oral disease, whereas SAP1, SAP3, and SAP6 SAP8 expression was correlated with vaginal disease. Furthermore, SAP1, SAP3, and SAP8 were preferentially expressed in vaginal, rather than oral, infections. This study demonstrates the differential expression of the hydrolytic enzyme genes in humans and correlates the expression of specific Candida species virulence genes with active disease and anatomical location. Candida species infections of mucosal surfaces are common, debilitating, and often recurring diseases. Approximately three-quarters of all healthy women of fertile age suffer from vaginal candidiasis, with significant associated physical and psychological morbidity [1, 2]. Candida albicans, the most virulent of the Candida species, can cause severe oral infections in even mildly immunocompromised hosts. Attributes that putatively contribute to Received 17 December 2002; accepted 15 February 2003; electronically published 14 July Presented in part: EURESCO Conference on Fungal Virulence Factors and Disease, 8 13 September 2001, Seefeld, Austria; 6th American Society for Microbiology Conference on Candida and Candidiasis, January 2002, Tampa, Florida (abstract 48). Financial support: Dunhill Medical Trust; National Institutes of Health (grants AI and PO1-DE07946). Reprints and correspondence: Dr. Julian R. Naglik, Dept. of Oral Medicine, Pathology, and Immunology, Floor 28, Guy s Tower, GKT Dental Institute, Kings College London (Guy s Campus), St. Thomas St., London SE1 9RT, United Kingdom (julian.naglik@kcl.ac.uk). The Journal of Infectious Diseases 2003; 188: by the Infectious Diseases Society of America. All rights reserved /2003/ $15.00 C. albicans virulence include adhesion, hyphal formation, phenotypic switching, and extracellular hydrolytic enzyme production (reviewed in [3 7]). The extracellular hydrolytic enzymes, including the secreted aspartyl proteinase (SAP) and phospholipase (PLB) genes, are among the few gene products that have been shown to directly contribute to C. albicans virulence. Four types of phospholipases have been reported in C. albicans phospholipase A [8], B [9, 10], C [11], and D [12] but only the PLB1 and PLB2 gene products have been detected extracellularly. Although PLB1 is thought to account for most of the secreted phospholipase B activity in C. albicans, PLB2 contributes in a minor way, because a PLB1-deficient strain still produces residual amounts of phospholipase B activity [13]. Phospholipase B1 is secreted during invasion of the gastrointestinal tract of mice [13], and C. albicans mutants lacking PLB1 are significantly attenuated in virulence in both an intragastric infant-mouse model [13] and an intravenous model of murine candidiasis C. albicans SAP and PLB Analysis In Vivo JID 2003:188 (1 August) 469
2 [14]. Serum samples from patients with invasive Candida species infections reacted with purified phospholipase B, which indicates that phospholipase B is produced in humans [13] and that blood isolates of C. albicans produce more phospholipase B activity in vitro than oral isolates from carriers of Candida species [15]. Although these studies implicate the phospholipase B enzyme family in C. albicans virulence, no studies have yet analyzed the expression of the PLB1 and PLB2 genes in human mucosal infections. The C. albicans Saps are, by far, the best characterized members of the hydrolytic enzymes secreted by C. albicans and are encoded by 10 SAP genes (reviewed in [16 18]). Two distinct groups are clustered within the family; Sap1 Sap3 are 67% identical, and Sap4 Sap6 are 89% identical, with Sap7 being the most divergent, with only 20% 27% sequence homology to the other Saps [19]. The proteinase family is differentially regulated and expressed under a variety of laboratory growth conditions [20, 21], during experimental C. albicans infections using reconstituted human oral epithelium (RHE) [22], and in vivo [23]. Different SAP genes appear to be essential for mucosal (SAP1 SAP3) [24 27] and systemic (SAP4 SAP6) [28] infections and also are involved in C. albicans adherence [29], tissue damage [24, 30], and evasion of host immune responses [31]. The proteinases possess distinct differences in ph optima, with Sap1 Sap3 (yeast associated) having optimum activity at lower ph values, and Sap4 Sap6 (hyphal associated) having optimum activity at higher ph values [31]. C. albicans SAP gene expression in oral or vaginal infections has been studied in vitro and in vivo in animal models and in patient isolates. Using infected oral RHE, Schaller et al. [22] detected a specific order of SAP gene expression by reversetranscriptase polymerase chain reaction (RT-PCR); SAP1 and SAP3 were expressed in the initial stages (possibly reflecting colonization), followed by SAP6 (concomitant with germ tube formation and severe lesions), and SAP2 and SAP8 were expressed in late phases of infection. Of interest, a different SAP expression profile was observed using an RHE model of vaginal candidiasis; initially SAP2, SAP9, and SAP10 transcripts were detected; followed by SAP1, SAP4, and SAP5 (concomitant with lesions); and, finally, by SAP6 and SAP7 (M. Schaller, M. Bein, H. Korting, et al., personal communication). By use of SAPdeficient mutants, the SAP1 SAP3 subfamily was shown to contribute to tissue damage in both oral and vaginal RHE models [24] (M. Schaller, M. Bein, H. Korting, et al., personal communication). Together, these 2 studies support a role for SAP1 SAP3 in establishing C. albicans infections at human mucosal surfaces. In a murine model of oropharyngeal candidiasis, using both normal and transgenic mice expressing human immunodeficiency virus (HIV) type 1, SAP1 SAP6 and SAP9 transcripts were detected continuously throughout the course of infection, with SAP5 and SAP9 being the most highly expressed; SAP7 and SAP8 were the only genes expressed transiently during infection [32]. Finally, by use of in vivo expression technology (IVET), as well as in a mouse model of esophageal candidiasis, transcription of SAP5 and SAP6 predominated [23], indicating a possible role for the SAP4 SAP6 subfamily in mouse mucosal infections. We have been interested in determining whether SAP gene expression patterns observed in vitro and in animal models are representative of those expressed in the context of human disease. We previously have described a radiation-based RT-PCR protocol that allowed us to assay the expression of individual SAP genes directly in the oral cavity without intermediate laboratory cultivation of the clinical isolates [33]. Using RT-PCR, we were able to amplify SAP mrna from as little as 1 pg of total RNA and from!10 cfu of C. albicans in a patient sample. We analyzed the expression of C. albicans SAP1 SAP7 in a small number of patients presenting with oral disease, as well as in asymptomatic carriers, and obtained data that suggested differential expression of individual members of the SAP gene family in humans [33]. In the present study, to determine whether the expression of individual extracellular hydrolytic enzyme genes correlates with oral or vaginal candidiasis, as opposed to carriage, and to discover whether certain SAP and PLB genes are specifically or selectively expressed during oral or vaginal disease, SAP1 SAP8 and PLB1 and PLB2 gene expression were analyzed in 137 clinical samples obtained from patients colonized or infected either orally or vaginally with C. albicans. MATERIALS AND METHODS Strains, media, and culture conditions. The following Candida species and strains were used in this study: C. albicans NCPF 3156, C. dubliniensis NCPF 3949, C. tropicalis ATCC 750, C. parapsilosis ATCC 22019, C. guilliermondii NCPF 3099, and C. glabrata ATCC Strains were maintained on Sabouraud (SAB) dextrose agar (Oxoid) stab cultures at room temperature. For DNA isolation, all cultures were grown in 10 ml of SAB dextrose broth (Oxoid) for 2 days at 27 C onan orbital incubator. C. albicans NCPF 3156 was used as a positive control for all optimization experiments. Selection of primers and specificity of RT-PCR. In our initial study [33], a single set of PCR primers was used to amplify SAP4 SAP6 together, which are highly homologous in nucleotide sequence [19]. However, in the present study, individual primer sets were designed to amplify SAP4 SAP6 separately. One primer pair each for ACT1 (actin), SAP1 SAP8, and PLB1 and PLB2 were designed (table 1). None of the primer sets amplified regions containing introns. The primer sets were tested against C. albicans DNA to check for accurate amplifi- 470 JID 2003:188 (1 August) Naglik et al.
3 Table 1. Primer sets detecting Candida albicans secreted aspartyl proteinase genes SAP1 SAP8, phospholipase B genes PLB1 and PLB2, and ACT1 genes. Gene [reference], primer Sequence Product, bp SAP1 [34] Forward 5 -TCA ATC AAT TTA CTC TTC CAT TTC TAA CA Reverse 5 -CCA GTA GCA TTA ACA GGA GTT TTA ATG ACA-3 SAP2 [35] Forward 5 -AAC AAC AAC CCA CTA GAC ATC ACC Reverse 5 -TGA CCA TTA GTA ACT GGG AAT GCT TTA GGA-3 SAP3 [36] Forward 5 -CCT TCT CTA AAA TTA TGG ATT GGA AC Reverse 5 -TTG ATT TCA CCT TGG GGA CCA GTA ACA TTT-3 SAP4 [37] Forward 5 -TTA TTT TTA GAT ATT GAG CCC ACA GAA A Reverse 5 -GCC AGT GTC AAC AAT AAC GCT AAG TT-3 SAP5 [19] Forward 5 -AGA ATT TCC CGT CGA TGA GAC TGG T Reverse 5 -CAA ATT TTG GGA AGT GCG GGA AGA-3 SAP6 [19] Forward 5 -CCC GTT TTG AAA TTA AAT ATG CTG ATG G Reverse 5 -GTC GTA AGG AGT TCT GGT AGC TTC G-3 SAP7 [19] Forward 5 -GAA ATG CAA AGA GTA TTA GAG TTA TTA C Reverse 5 -GAA TGA TTT GGT TTA CAT CAT CTT CAA CTG-3 SAP8 [38] Forward 5 -GCC GTT GGT GCC AAA TGG AAT AGT TA Reverse 5 -ATT TGA CTT GAG CCA ACA GAA TGG T-3 PLB1 [10] Forward 5 -CCT ATT GCC AAA CAA GCA TTG TC Reverse 5 -CCA AGC TAC TGA TTT CAC CTG CTC C-3 PLB2 [39] Forward 5 -GTG GGA TCT TGC AGA GTT CAA GC Reverse 5 -CTC AAA GCT CTC CCA TAG ACA TCT G-3 ACT1 [40] Forward 5 -GAT TTT GTC TGA ACG TGG TAA CAG Reverse 5 -GGA GTT GAA AGT GGT TTG GTC AAT AC-3 cation of the correct genes, as well as against a panel of genomic DNA isolated from different Candida species to test for crossreactivity. All the amplified gene products were purified by use of the QIAquick PCR purification system, according to the manufacturer s instructions (Qiagen), and were sequenced at the Advanced Biotechnology Centre (London, UK) to confirm the specificity of the reaction. Study populations and type of Candida species infection. Patients were selected as they presented to the clinic by a single experienced clinician at each of the Oral Medicine and Genitourinary Medicine clinics. Two populations were used to obtain clinical samples for the analysis of SAP and PLB expression. The first population included patients with active Candida species infection, with clinical signs and symptoms of oral or vaginal candidiasis. Patients presenting with oral candidiasis were 3 infected with C. albicans cfu/ml of saliva ( n p 40; 25 men and 15 women; mean age, years). Of these 40 patients, 24 with oral candidiasis were HIV infected, and 16 were HIV uninfected. None of these 40 patients had been treated with anti-hiv proteinase drugs before or at the time of sample collection. The majority of patients presented with a single clinical form of C. albicans infection that is, pseudomembranous (thrush) candidiasis (PC; n p 14) or erythema- tous candidiasis (EC; n p 9) although some presented with C. albicans SAP and PLB Analysis In Vivo JID 2003:188 (1 August) 471
4 both types ( n p 4). Six patients suffered from chronic atrophic candidiasis (CAC), which also can result from wearing dentures, and 7 patients with Sjögren s Syndrome, an autoimmune disease characterized by xerostomia (dry mouth), suffered from chronic candidiasis (CC). Vaginal infection was diagnosed primarily on clinical signs and symptoms and cultures positive for C. albicans ( n p 40; mean age, years; Candida species colony-forming unit count range, 2 to cfu). Of the 40 patients, the majority presented with PC or EC, although the clinical diagnosis of PC or EC for vaginal infection is not as explicit as it is for oral infection, probably due, in part, to physical, mechanical, and immunological [41, 42] differences between the vaginal lumen and the oral cavity. Positive diagnosis of vaginal infection met the following criteria: culture positive and the presence of at least 1 sign and 1 symptom of infection. Signs included edema (swelling), erythema, pseudomembranous plaques, and discharge; and symptoms included pruritus (itch), pain, and soreness. None of the patients presenting with vaginal disease was tested for HIV infection. In addition, none of the patients with oral or vaginal infection was treated with either antibiotic or antimycotic drugs for at least 1 month before sampling. The second population included subjects who were asymptomatic carriers (Candida species carrier group) that is, without any clinical signs or symptoms of infection. Oral Candida species carriers harbored C. albicans cfu/ml of saliva ( n p 29; 2 men and 27 women; mean age, years), and vaginal carriage status was based on cultures positive for C. albicans, regardless of colony-forming unit count (range, cfu/swab; n p 28; mean age, 26 5 years). All Candida carriers were either laboratory volunteers or individuals attending the Oral Medicine clinic at Guy s Hospital or the Genitourinary Medicine Clinic at St Thomas Hospital. None of the individuals who comprised the Candida species carrier population was tested for HIV infection. Informed consent was obtained from all patients regarding the nature of the study. The collection of clinical samples was conducted according to the rules of the Guy s and St Thomas Hospital Trust ethical review board. Sample collection and preparation. Specimens were collected as either whole unstimulated saliva or vaginal swabs, because these provided reliable and consistent mrna detection (data not shown). Duplicate samples of whole unstimulated saliva or vaginal swabs were collected at the same visit from each subject. The vaginal swabs were taken from the lateral vaginal wall; for patients presenting with infection, swabs were taken directly from areas of infection. One of the whole saliva samples was immediately frozen on dry ice at 70 C, and one of the vaginal swabs was immediately suspended in 1 ml of RNAlater (Ambion) and then frozen on dry ice. The purpose of each procedure was to preserve the integrity of RNA for the subsequent analysis of SAP and PLB gene expression in the clinical samples. The second saliva and vaginal swab samples were used to determine Candida colony-forming unit counts and for yeast identification, using the API 32C AUX system (BioMerieux), CHROMagar (CHROMagar), germ tube formation, and growth at 42 C [33]. Two hundred five individuals were enrolled in the oral study during a 2-year period. After Candida species identification, 64 matched the criteria of having clinical signs and symptoms of infection and of harboring C. albicans cfu/ml of saliva, and 38 were designated as asymptomatic Candida carriers harboring C. albicans cfu/ml of saliva. For the vaginal study, 202 patients were enrolled during an 18-month period. Of these, 42 matched the criteria of having symptomatic C. albicans vaginal infection, and 34 were carriers. Total RNA was isolated from all qualifying samples. RT-PCR and PCR (without avian myeloblastosis virus [AMV] reverse transcriptase [RT]), using ACT1 primers, were performed on each sample, to determine whether sufficient C. albicans total RNA was isolated for mrna detection and whether the total RNA samples contained contaminating DNA, respectively. ACT1 mrna was successfully and reproducibly amplified from samples obtained from 40 patients with oral infections, 29 oral carriers, 40 patients with vaginal infections, and 28 vaginal carriers. Transcription of SAP1 8 and PLB1 2 genes was assayed in each of these samples by RT-PCR. DNA and RNA isolation. Genomic DNA from Candida species was isolated by use of a phenol/chloroform-based protocol [33]. Total RNA from clinical samples was prepared using the RNeasy RNA isolation system (Qiagen), incorporating glass beads ( mmol, Sigma). The QIAshredder unit (Qiagen) was used to homogenize the cell lysates, and, at the end of the procedure, the total RNA was treated overnight with DNAse (Qiagen), to allow the complete digestion of any contaminating DNA. Purified RNA from each sample was confirmed to be DNA-free by the absence of an amplified product after PCR (without RT) by use of ACT1 (actin) primers. RT-PCR analysis of C. albicans SAP and PLB mrna expression. Thirteen RT-PCRs were performed for each RNA sample prepared from each clinical isolate. These included the 10 experimental reactions (SAP1 8 and PLB1 2), and 3 different control reactions: an ACT1 control to demonstrate the presence or absence of Candida species, a negative (water) control, and a positive control using genomic DNA isolated from C. albicans NCPF 3156 cells. Each RNA sample was analyzed in duplicate and, in many cases, in triplicate to verify the hydrolytic gene expression data. Complete congruence in gene expression was required in 2 separate analyses using the same RNA sample. RT-PCR conditions were optimized by use of modified Taguchi methods [43] and included a touchdown and 472 JID 2003:188 (1 August) Naglik et al.
5 RESULTS Figure 1. Reactivity of secreted aspartyl proteinase genes SAP1 SAP8 and phospholipase B genes PLB1 and PLB2 primer sets with genomic DNA isolated from different Candida species: C. albicans NCPF 3156 (1), C. dubliniensis NCPF 3949 (2), C. tropicalis ATCC 750 (3), C. parapsilosis ATCC (4), C. guilliermondii NCPF 3099 (5), and C. glabrata ATCC (6). The expected SAP and PLB gene products were amplified from C. albicans. SAP1 SAP7 and PLB2 primers were specific for C. albicans, whereas the SAP8 and PLB1 primers amplified a polymerase chain reaction (PCR) product from C. dubliniensis, which was identical in size to C. albicans. hot start protocol, as described elsewhere [33]. RT-PCR experiments using the ACT1, SAP, and PLB primers were performed by use of the Access RT-PCR system (Promega). Template RNA was added to the RT-PCR mix containing 1 AMV/ Tfl buffer (Promega; proprietary information), 1 mmol MgSO 4, 0.1 mmol dntps, 0.6 mmol primers, 3.75 U AMV RT, and 1 mci 32 P-dCTP (ICN Pharmaceutical). Radioactive-labeling was used to maximize sensitivity. After RT (48 C for 45 min), the sample was denatured at 94 C for 3 min, and 2.5 U of Tfl DNA polymerase was added to the reaction (hot start). Cycling times were as follows: denaturation at 94 C, annealing at 60 C, and extension at 72 C, each for 30 s. A final extension at 72 C for 10 min followed cycling. All radiolabeled RT-PCR products were electrophoresed through a 7% denaturing 7-mol urea polyacrylamide gel, exposed to autoradiography film at 70 C, and developed [33]. Statistical analysis. Fisher s exact test (2-tailed) was used to determine statistical differences in hydrolytic enzyme gene expression between patients with oral or vaginal infection and carrier subjects. The number of individuals used in this study was based on power calculations from our previous study, which showed a difference in proportions of the indicator variable of 0.0 and 0.6 [33]. Eighty percent power to show a statistically significant difference between proportions of subjects with an indicator variable of 0.2 versus 0.6 can be achieved with 28 subjects in each of our 4 main groups. Therefore, between 28 and 40 subjects were recruited in each group. ACT1, SAP1 8, and PLB1 2 primers. To control for the presence of Candida species in the absence of SAP gene expression, primers were designed to detect Candida actin (ACT1) gene expression [33]. ACT1 could be amplified from Candida species samples obtained from carriers that contained!10 cfu. The SAP1 SAP8 and PLB1 and PLB2 primers amplified small PCR fragments, ranging in size from 161 to 277 bp (table 1). The specificity of each primer pair was determined by PCR using C. albicans genomic DNA, as described elsewhere [33], and the identity of the SAP1 SAP8 and PLB1 and PLB2 PCR products was confirmed by DNA sequencing at the Advanced Biotechnology Centre (London; data not shown). The SAP1 SAP7 and PLB2 primer sets were C. albicans specific and did not amplify any gene products from C. dubliniensis, C. tropicalis, C. parapsilosis, C. guilliermondii, or C. glabrata (figure 1). However, the SAP8 and PLB1 primers did amplify gene products identical in size in both C. albicans and its closely related species C. dubliniensis (figure 1), indicating that C. dubliniensis may possess homologues of SAP8 and PLB1. However, we were able to ascertain that the expression of SAP8 and PLB1 detected in our experiments was attributable to C. albicans, because each of the clinical samples were plated and identified as monocultures of C. albicans. To control for cross-reactivity with microbial flora or host RNA present in each sample, RT-PCR was performed using SAP and PLB primer sets and RNA isolated from oral (n p 7) and vaginal ( n p 7) samples that were culture negative for Candida species, as described elsewhere [33]. No RT-PCR amplification products were detected under any of these conditions, demonstrating the specificity of the primer sets for C. albicans gene expression (data not shown). RNA preparation from patient samples. The RNA and Candida colony-forming unit concentration means and ranges for the 4 study populations are displayed in table 2. It should be noted that only a small fraction of the total RNA is attributable to C. albicans, because the majority of RNA present in the entire sample is derived from variable amounts of host and bacterial RNA. RNA isolated from patient samples with as few Table 2. Total RNA and Candida albicans colony-forming unit means and ranges in the 4 study populations. Study population RNA concentration mean SEM (range), mg/ml Total C. albicans count, cfu Oral infection (n p 40) (13 414) 2880 to Oral carriage (n p 29) (15 352) Vaginal infection (n p 40) (8 274) 2 to Vaginal carriage (n p 28) (11 298) C. albicans SAP and PLB Analysis In Vivo JID 2003:188 (1 August) 473
6 Figure 2. Detection of Candida albicans ACT1 (actin), secreted aspartyl proteinase genes SAP1 SAP8, and phospholipase B genes PLB1 and PLB2 mrna expression from clinical saliva and vaginal wash samples. Five (1 5) representative samples are presented for each study group: patients with oral infection, patients with vaginal infection, asymptomatic oral Candida carriers, and asymptomatic vaginal Candida carriers. The presence of actin signals (ACT1) indicates the presence of C. albicans in the clinical samples. The exclusion of avian myelobastosis virus reverse transcriptase ( RT) resulted in the absence of any polymerase chain reaction (PCR) signals using ACT1 primers, confirming the absence of C. albicans DNA in the clinical samples. A spectrum of SAP and PLB gene expression profiles was obtained from different C. albicans clinical strains during symptomatic disease and asymptomatic carriage. Two patients with oral infection and 2 patients with vaginal infection (1 and 2) harbored C. albicans expressing the full repertoire of all 10 hydrolytic enzyme genes tested, and 3 patients (3 5) possessed C. albicans expressing fewer genes. The 5 oral and vaginal asymptomatic carriers harbor C. albicans expressing fewer hydrolytic enzyme genes than the oral and vaginal infected groups. SAP2 and SAP5 are the most common genes expressed during both infection and carriage. as 50 cfu/ml in saliva or 2 cfu/ml in vaginal swabs was sufficient to detect C. albicans specific gene expression. There was no observable difference in the colony-forming unit counts between patients with different types of infection (i.e., PC or EC). All the total RNA preparations were confirmed to be Candida DNA free by the lack of an ACT1 amplification product by PCR (figure 2). In our previous study [33], we showed that differences in SAP gene expression profiles between the oral infection and asymptomatic carrier groups were unlikely to be due to the greater numbers of C. albicans cells present in the infected patient group (i.e., that detection of SAP genes was independent of C. albicans RNA in the sample). This study confirms our initial assertion, because many samples from patients with oral and vaginal disease with greater C. albicans colony-forming unit counts expressed fewer hydrolytic enzyme genes than many oral and vaginal carriers who harbored lower C. albicans colonyforming unit counts. Furthermore, the detection of C. albicans actin (ACT1) from!10 C. albicans cfu in vaginal swab samples indicated that the methods employed were sufficiently sensitive to also detect SAP and PLB expression. The sensitivity of detection of SAP and PLB expression (independent of initial RNA concentration) also was assayed by diluting samples containing high numbers of C. albicans colony-forming units to levels of colony-forming units typically found in samples obtained from carriers ( cfu/ml) and assaying gene expression in the diluted and undiluted samples. In samples obtained from 5 different patients, we found identical gene expression profiles in both the diluted and undiluted samples (data not shown), indicating that any differences in gene expression profiles between the disease and carrier populations were unlikely to be due to the greater numbers of C. albicans cells present in the infected patient groups. C. albicans SAP and PLB expression in samples from patients with oral candidiasis. The pattern of SAP1 SAP8 and PLB1 and PLB2 expression obtained from RT-PCR assay of patient saliva samples is shown in table 3 (and representative samples are illustrated in figure 2). SAP2 and SAP5 are the most common transcripts in both C. albicans infected patients (90% and 95%, respectively) and carriers (79% and 93%, respectively). SAP3 is expressed with the least frequency; only 35% of samples from infected patients and 14% of samples from carriers were positive for SAP3 transcripts. Although none of the samples obtained from carriers expressed all of the genes assayed, 10 samples obtained from C. albicans infected patients expressed the full repertoire of SAP1 SAP8 and PLB1 and PLB2 genes (representative samples in figure 2). In general, the autoradiographic profiles indicate that C. albicans from patients with oral infection express a greater repertoire of hydrolytic enzyme genes (6 7 genes/patient) than did those from oral 474 JID 2003:188 (1 August) Naglik et al.
7 Table 3. Candida albicans secreted aspartyl proteinase (SAP) and phospholipase B (PLB) gene expression in oral infection and carriage. Group SAP1 SAP2 SAP3 SAP4 SAP5 SAP6 SAP7 SAP8 PLB1 PLB2 Oral infection (n p 40) 28 (70) 36 (90) 14 (35) 28 (70) 38 (95) 32 (80) 31 (78) 19 (48) 23 (58) 19 (48) Oral carrier (n p 29) 9 (31) 23 (79) 2 (14) 12 (41) 27 (93) 20 (69) 15 (48) 5 (17) 4 (14) 10 (34) P a ! NOTE. Data are no. subjects (%) who were positive for the expression of the particular gene. a Probability values between the no. of subjects who were positive for the expression of a particular gene in the oral infection and oral carrier groups were determined by use of the 2-tailed Fisher s exact test. Significant differences in the frequency of detection are in boldface type. carriers (4 5 genes/carrier). However, there were 4 instances when only 3 genes were expressed in samples from infected individuals and 9 instances when 7 genes were expressed in samples from carriers. When comparing SAP and PLB expression between patients with oral infection and carriers, we found that SAP1, SAP3, SAP4, SAP7, SAP8, and PLB1, are preferentially expressed in C. albicans infection (table 3). C. albicans SAP and PLB expression in samples from patients with vaginal candidiasis. SAP1 SAP8 and PLB1 and PLB2 expression by C. albicans in swab samples from vaginal infection and carriage is shown in table 4 (and representative samples in figure 2). SAP2, SAP4, and SAP5 are the most common transcripts detected in both vaginal infections (100%, 88%, and 98%, respectively) and carrier states (89%, 75%, and 89%, respectively). A high proportion of samples from patients with vaginal infections ( n p 17), compared with 1 sample from the carriers, harbored C. albicans expressing the full repertoire of all 10 genes (SAP1 SAP8 and PLB1 and PLB2; representative samples in figure 2). In general, C. albicans from vaginal infections express more hydrolytic enzyme genes (7 8 genes/patient) than did the vaginal carriers ( 5 genes/carrier). When comparing C. albicans SAP and PLB expression between patients with vaginal infection and carriers, SAP1, SAP3, and SAP6 SAP8 were preferentially expressed to a significant level in C. albicans infection (table 4). Comparison of C. albicans SAP and PLB gene expression patterns from oral and vaginal samples. Similarities in the overall pattern of SAP gene expression are evident between C. albicans assayed in oral and vaginal sites. SAP2 and SAP5 are expressed with the greatest frequency in both the oral and vaginal infection and carrier groups; notably, 1 gene each from the SAP1 SAP3 and SAP4 SAP6 subfamilies (tables 3 and 4). Four genes, SAP1, SAP3, SAP7, and SAP8 are preferentially expressed to a significant level during infection rather than carriage in both the oral and vaginal study groups (tables 3 and 4). The main differences between the oral and vaginal populations is that both SAP4 ( P p.026) and PLB1 ( P.001) are preferentially expressed during oral infection versus oral carriage (table 3), but not in vaginal infection versus vaginal carriage (table 4). In addition, SAP6 ( P.001) is preferentially expressed during vaginal infection versus vaginal carriage, but not in oral infection versus oral carriage. A comparison of the oral and vaginal C. albicans infected groups alone reveals that SAP1, SAP3, and SAP8 are expressed significantly more frequently during vaginal infection than during oral infection (table 5). SAP and PLB expression in HIV-infected and HIV-uninfected patients with oral candidiasis and in different clinical forms of C. albicans infection. No statistical differences in the SAP or PLB expression profiles between HIV-infected and HIV-uninfected patients (table 6) or between the various clinical forms of disease (data not shown) were evident. DISCUSSION This study analyzed C. albicans SAP1 SAP8 and PLB1 and PLB2 gene expression during oral and vaginal infection and colonization in 137 samples from separate individuals, and the results provide insight with regard to patterns of extracellular Table 4. Candida albicans secreted aspartyl proteinase (SAP) and phospholipase B (PLB) gene expression in vaginal infection and carriage. Group SAP1 SAP2 SAP3 SAP4 SAP5 SAP6 SAP7 SAP8 PLB1 PLB2 Vaginal infection (n p 40) 36 (90) 40 (100) 27 (68) 35 (88) 39 (98) 31 (78) 24 (60) 30 (75) 25 (63) 25 (63) Vaginal carrier (n p 28) 12 (43) 25 (89) 7 (25) 21 (75) 25 (89) 9 (32) 8 (29) 9 (32) 12 (43) 14 (50) P a! !.001! NOTE. Data are no. subjects (%) who were positive for the expression of the particular gene. a Probability values between the no. of subjects who were positive for the expression of a particular gene in the vaginal infected and carrier groups were determined by use of the 2-tailed Fisher s exact test. Significant differences in the frequency of detection are in boldface type. C. albicans SAP and PLB Analysis In Vivo JID 2003:188 (1 August) 475
8 Table 5. Candida albicans secreted aspartyl proteinase (SAP) and phospholipase B (PLB) gene expression in oral and vaginal infection. Group SAP1 SAP2 SAP3 SAP4 SAP5 SAP6 SAP7 SAP8 PLB1 PLB2 Oral infection (n p 40) Vaginal infection (n p 40) P a NOTE. Data are no. of subjects who were positive for the expression of the particular gene. a Probability values between the no. of subjects who were positive for the expression of a particular gene in the oral and vaginal infection groups were determined by use of the 2-tailed Fisher s exact test. Significant differences in the frequency of detection are in boldface type. hydrolytic enzyme gene expression in the context of human disease. The data show that all members of the SAP gene family can be expressed by C. albicans strains in vivo during the course of the same disease presentation (i.e., oral or vaginal, PC or EC); not only are individual SAP and PLB gene transcripts more frequently detected during active C. albicans infection compared with carriage (tables 3 and 4; figure 2), but there also is differential expression of certain hydrolytic enzymes during infection in the oral cavity and vaginal lumen (table 5). This study expands our initial analysis [33] by comparing PLB1 and PLB2, as well as SAP1 SAP8 gene expression, and by inclusion of C. albicans obtained not only from oral infections and carriage but also from vaginal infections and carriage. This study did not analyze the expression of SAP9 and SAP10, because there is good evidence to suggest that Sap9 and Sap10 both have C-terminal consensus sequences typical for glycophosphotidylinositol (GPI) anchored proteins and, therefore, may not be secreted from the cell. In addition, Sap9 and Sap10 might be regulatory proteinases that may play a role in the cell surface integrity of the cell, which is in contrast to the putative functions of the other Saps (A. Albrecht, I. Pichova, M. Monod, and B. Hube [principal investigator], personal communication). Many laboratory experiments have been performed to assess the expression patterns of genes in the SAP gene family. In general, Sap2 is the predominant C. albicans proteinase secreted in protein containing medium [20, 21, 35], a finding that agrees well with our own that SAP2 is a predominant proteinase gene expressed by C. albicans isolated from colonization and infection of the oral cavity (100%) and vagina (90%) (table 3 and 4; figure 2). SAP5 (like SAP2) is also universally expressed in samples obtained from oral and vaginal infections and colonization (tables 3 and 4; figure 2). This dominant expression of SAP5 also has been observed in murine models of oropharyngeal candidiasis using IVET [23] and RT-PCR [32]. However, SAP5 gene expression was not detected in an RHE model of oral candidiasis [22], a finding which contrasts with our data showing that SAP5 is the most commonly expressed SAP gene at human mucosal surfaces. Because we find both SAP2 and SAP5 expressed in nearly every patient sample, whether from active disease or colonization, this suggests that individual members of the 2 main subfamilies (SAP1 3 and SAP4 6) of the SAP gene superfamily are required by C. albicans to fulfill basic functions in relation to survival and proliferation in the human host. In support of this conclusion is the broad substrate specificity of Sap2 (which can degrade many proteins found in the oral cavity and vaginal lumen [17]), and its role in causing tissue damage as assessed in an RHE oral model [24] and virulence in a rat vaginitis model [25]. In comparisons of symptomatic disease and asymptomatic carriage, SAP1 and SAP3 are preferentially expressed in active oral and vaginal candidiasis (tables 3 and 4) but are more frequently found in association with vaginal infections (table 5). In laboratory settings, SAP1 and SAP3 gene expression are tightly coupled and appear to be regulated by phenotypic switching [20, 21, 44], a phenomenon that is thought to allow C. albicans to adapt to different host environments during the Table 6. Candida albicans secreted aspartyl proteinase (SAP) and phospholipase B (PLB) gene expression in human immunodeficiency virus (HIV) infected and HIV-uninfected patients with oral candidiasis. Status SAP1 SAP2 SAP3 SAP4 SAP5 SAP6 SAP7 SAP8 PLB1 PLB2 HIV infected (n p 24) HIV uninfected (n p 16) P a NOTE. Data are no. of subjects who were positive for the expression of a particular gene. a Probability values between the no. of subjects who were positive for the expression of a particular gene in the HIV-infected and HIV-uninfected groups with oral candidiasis were determined by use of the 2-tailed Fisher s exact test. 476 JID 2003:188 (1 August) Naglik et al.
9 course of an infection. Therefore, we did not expect to observe an uncoupling of SAP1 and SAP3 gene expression; SAP1 was expressed in 70% and 90% of the samples obtained from patients with oral and vaginal infections, respectively, whereas SAP3 was expressed in only 35% and 68% of samples, respectively (tables 3 and 4). The association of these switch phenotype-related members of the SAP gene family with active disease at both oral and vaginal sites is consistent with a role for phenotypic switching in the disease process [33] and supports observations that C. albicans isolated from patients with active oral candidiasis exhibit higher rates of phenotypic switching than similarly localized commensal strains [45]. Although SAP3 is expressed at higher overall frequency in active mucosal infections, compared with asymptomatic colonization, it is not one of the more frequently detected members of the SAP gene family (tables 3 and 4). Within that context, however, SAP3 expression is 2-fold more prevalent in both carriers and patients suffering from vaginal rather than oral carriage and disease, raising the possibility that SAP3 expression could facilitate C. albicans colonization and infection of the vaginal mucosa. Our data from C. albicans infections in humans also are consistent with those of Ripeau et al. [32], who showed that SAP3 expression in the oral cavity of mice is down-regulated during persistent colonization. In summary, the universal expression of SAP2 during colonization and infection and the preferential expression of SAP1, in particular, during active human disease supports the current evidence that the Sap1 Sap3 subfamily contribute to the pathogenesis and virulence of C. albicans at mucosal surfaces. Perhaps one of our more surprising results was the detection of transcripts from genes in the SAP4 SAP6 subfamily in all but 1oftheC. albicans infected patients ( n p 79) and all but 5 carriers ( n p 52). Most studies using either SAP-deficient mu- tants or proteinase inhibitors indicate that SAP4 SAP6 do not contribute to tissue damage and pathogenesis at mucosal surfaces [24, 25]. Rather, SAP4 SAP6 are thought to promote systemic disease, because SAP4-, SAP5-, and SAP6-deficient mutants are more attenuated in virulence in systemic models of C. albicans infections [28, 31, 46, 47]. Our results that SAP4 and SAP6 are expressed more frequently during oral and vaginal infections, compared with carriage ( P p.026 and P.001, respectively; tables 3 and 4) are the first to indicate that the SAP4 SAP6 subfamily also is involved in human mucosal disease. The expression of C. albicans SAP7 mrna in samples from both oral and vaginal infections extends the findings of our initial study, where the expression of this gene was first reported [33]. SAP7 transcripts have since been detected during late stages of in vitro vaginal infections in an RHE model (M. Schaller, M. Bein, H. Korting, et al., personal communication) and transiently expressed in a murine model of oropharyngeal candidiasis [32]. Unexpectedly, SAP7 expression correlates with both oral and vaginal infection rather than carriage (P p.038 and P.010, respectively; tables 3 and 4), which is particularly noteworthy, because SAP7 is the most divergent of all the SAP genes [19]. Moreover, SAP7 transcripts are not detected in a systemic murine model of intraperitoneal infection [47], leading to the possibility that SAP7 may be expressed only when C. albicans is located at mucosal surfaces. Although the functions of Sap7 are completely unknown, its apparent preferential expression during oral and vaginal infections tentatively supports some role for this proteinase during human mucosal infections. Similarly, SAP8 is expressed in C. albicans isolated from active oral and vaginal infections rather than carriage ( P p.011 and P p.001, respectively; tables 3 and 4). This is the first study to associate SAP8 expression with human mucosal infections and corroborates the work of Ripeau et al. [32], who showed transient expression of SAP8 in a murine model of oropharyngeal candidiasis. Little is known about Sap8 except that it is most similar to the SAP1 SAP3 subfamily ( 50%) and is temperature regulated in vitro [38]. Thus, the mechanism by which Sap8 contributes to human mucosal infections is still unclear and requires more functional data based on SAP8 deficient mutants and biochemical characterization of the Sap8 protein. PLB1 expression correlates with oral infection ( P.001; table 3) but not vaginal infection. PLB1 transcripts also have been detected in the oral cavity of mice [32], and using PLB1-deficient mutants PLB1 has been shown to contribute to an intragastric model [13] and an intravenous model of murine candidiasis [14]. However, our study is the first, to our knowledge, to specifically correlate PLB1 expression with human oral infections, rather than carriage. Because PLB1 transcripts were detected in only 14% of oral carriers (table 3), our results are in agreement with a previous study that demonstrated little phospholipase activity in C. albicans strains isolated from the oral cavity of carriers [15]. It is possible that PLB1 expression may be regulated by factors that also regulate the expression of hyphal morphology [10]; however, since hyphae are probably present in the vast majority of the samples examined (concluded from the widespread expression of SAP4 SAP6, which are coordinately expressed with hyphal development in laboratory settings [20, 21]), the infrequent detection of PLB1 transcripts in comparison with SAP4 SAP6 transcripts suggests that PLB1 expression in vivo does not correlate with the presence of hyphae. PLB2 was expressed in a similar number of subjects as PLB1 but was not associated with either oral or vaginal infections (tables 3 and 4). Although PLB2 transcripts are found in many samples in which PLB1 transcripts are absent, any major functional role for PLB2 during colonization and infection in humans may be limited considering that disruption of the PLB1 gene results in a loss of most of the detectable extracellular phospholipase activity of C. albicans [13]. C. albicans SAP and PLB Analysis In Vivo JID 2003:188 (1 August) 477
10 No correlations between the expression of specific SAP or PLB genes with either the HIV status (table 6) or the different clinical forms of candidiasis were evident. With respect to the different clinical forms of infection, the number of samples in each group (see Materials and Methods) was not sufficiently large to draw any firm conclusions. However, the former observation was perhaps surprising because there is a selection of C. albicans strains in HIV infection that appears to be associated with increased Sap2 production [48, 49]. Although no qualitative differences in SAP gene expression were found in this study to support those conclusions, it is possible that quantitative differences between these groups existed, which could be addressed by real-time RT-PCR. In summary, the present study together with our initial study [33] are the only reports to address the expression of the C. albicans hydrolytic enzyme genes in humans, and have demonstrated differences in SAP1 SAP8 and PLB1 and PLB2 expression during colonization and active disease in both the oral cavity and vaginal lumen. However, because this is a point prevalence study, it remains to be confirmed whether repeated samples from the same subject would consistently give the same profile of gene expression. In addition, it is likely that SAP or PLB expression varies with time and with different stages and severity of diseases in the same subject, regardless of whether being colonized or infected with C. albicans. It is also possible that some of the described differences in SAP and PLB expression that did not quite reach statistical significance might reach the predefined significance level ( P p.05) if more subjects are analyzed. Furthermore, it cannot be ruled out that some hydrolytic enzymes might be expressed at a very low level under the detection limit for the assay, which may nonetheless be biologically relevant or may contribute to the regulation of expression of other genes. Finally, SAP gene expression and protein secretion are tightly coupled for Sap2 [50], although there is no solid experimental evidence to demonstrate a similar phenomenon for the other Saps, or whether all SAP or PLB mrnas are equally translated with the same efficiency. Nevertheless, although there is an absence of proteomic tools to discriminate fully among the SAP genes, the differential expression data described in this study are suggestive for a differential virulence role of the respective enzymes at the oral and vaginal sites and between colonization and infection. Because Candida species are able to colonize and infect essentially every tissue in the human host, it may be crucial for the fungus to possess a number of similar but independently regulated, functionally distinct hydrolytic enzyme genes (which can be expressed under a variety of environmental conditions) to provide sufficient flexibility to survive and promote infection at different niche sites [17]. However, the precise regulatory factors that control or influence these expression profiles in vivo remain to be elucidated. With access to the recently completed genome sequence of C. albicans ( and with the development of DNA microarrays specifically for Candida research, our understanding of C. albicans SAP gene regulation and their interactions with the human host should soon rapidly increase. Acknowledgments We thank Sue Howell, for kindly supplying the Candida reference strains used in this study, and Ron Wilson, for assistance with the statistical analysis. References 1. Sobel JD. Pathogenesis and treatment of recurrent vulvovaginal candidiasis. Clin Infect Dis 1992; 14 (Suppl 1):S Sobel JD. Pathogenesis and epidemiology of vulvovaginal candidiasis. Ann NY Acad Sci 1988; 544: Monod M, Borg-von Zepelin M. Secreted proteinases and other virulence mechanisms of Candida albicans. Chem Immunol 2002; 81: Calderone RA, Fonzi WA. Virulence factors of Candida albicans. Trends Microbiol 2001; 9: Agabian N, Odds FC, Poulain D, Soll DR, White TC. Pathogenesis of invasive candidiasis. J Med Vet Mycol 1994; 32: Odds FC. Candida species and virulence. ASM News 1994; 60: Cutler JE. Putative virulence factors of Candida albicans. Annu Rev Microbiol 1991; 45: Banno Y, Yamada T, Nozawa Y. Secreted phospholipases of the dimorphic fungus, Candida albicans; separation of three enzymes and some biological properties. Sabouraudia 1985; 23: Barrett-Bee K, Hayes Y, Wilson RG, Ryley JF. A comparison of phospholipase activity, cellular adherence and pathogenicity of yeasts. J Gen Microbiol 1985; 131: Hoover CI, Jantapour MJ, Newport G, Agabian N, Fisher SJ. Cloning and regulated expression of the Candida albicans phospholipase B (PLB1) gene. FEMS Microbiol Lett 1998; 167: Pugh D, Cawson RA. The cytochemical localization of phospholipase in Candida albicans infecting the chick chorio-allantoic membrane. Sabouraudia 1977; 15: Hube B, Hess D, Baker CA, Schaller M, Schafer W, Dolan JW. The role and relevance of phospholipase D1 during growth and dimorphism of Candida albicans. Microbiology 2001; 147: Ghannoum MA. Potential role of phospholipases in virulence and fungal pathogenesis. Clin Microbiol Rev 2000; 13: Leidich SD, Ibrahim AS, Fu Y, et al. Cloning and disruption of caplb1, a phospholipase B gene involved in the pathogenicity of Candida albicans. J Biol Chem 1998; 273: Ibrahim AS, Mirbod F, Filler SG, et al. Evidence implicatingphospholipase as a virulence factor of Candida albicans. Infect Immun 1995; 63: Hube B, Naglik JR. Extracellular hydrolases. In: Calderone RA, ed. Candida and candidiasis. Washington, DC: American Society for Microbiology Press, 2002: Hube B, Naglik J. Candida albicans proteinases: resolving the mystery of a gene family. Microbiology 2001; 147: Hube B. Extracellular proteinases of human pathogenic fungi. In: Ernst JF, Schmidt A, eds. Contributions to microbiology: dimorphisminhuman pathogenic and apathogenic yeasts. Vol. 5. Basel: S. Karger AG, 2000: Monod M, Togni G, Hube B, Sanglard D. Multiplicity of genes encoding secreted aspartic proteinases in Candida species. Mol Microbiol 1994; 13: White TC, Agabian N. Candida albicans secreted aspartyl proteinases: 478 JID 2003:188 (1 August) Naglik et al.
Supplementary Table 3. 3 UTR primer sequences. Primer sequences used to amplify and clone the 3 UTR of each indicated gene are listed.
 Supplemental Figure 1. DLKI-DIO3 mirna/mrna complementarity. Complementarity between the indicated DLK1-DIO3 cluster mirnas and the UTR of SOX2, SOX9, HIF1A, ZEB1, ZEB2, STAT3 and CDH1with mirsvr and PhastCons
Supplemental Figure 1. DLKI-DIO3 mirna/mrna complementarity. Complementarity between the indicated DLK1-DIO3 cluster mirnas and the UTR of SOX2, SOX9, HIF1A, ZEB1, ZEB2, STAT3 and CDH1with mirsvr and PhastCons
c Tuj1(-) apoptotic live 1 DIV 2 DIV 1 DIV 2 DIV Tuj1(+) Tuj1/GFP/DAPI Tuj1 DAPI GFP
 Supplementary Figure 1 Establishment of the gain- and loss-of-function experiments and cell survival assays. a Relative expression of mature mir-484 30 20 10 0 **** **** NCP mir- 484P NCP mir- 484P b Relative
Supplementary Figure 1 Establishment of the gain- and loss-of-function experiments and cell survival assays. a Relative expression of mature mir-484 30 20 10 0 **** **** NCP mir- 484P NCP mir- 484P b Relative
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Sherman SI, Wirth LJ, Droz J-P, et al. Motesanib diphosphate
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Sherman SI, Wirth LJ, Droz J-P, et al. Motesanib diphosphate
Supplemental Data. Shin et al. Plant Cell. (2012) /tpc YFP N
 MYC YFP N PIF5 YFP C N-TIC TIC Supplemental Data. Shin et al. Plant Cell. ()..5/tpc..95 Supplemental Figure. TIC interacts with MYC in the nucleus. Bimolecular fluorescence complementation assay using
MYC YFP N PIF5 YFP C N-TIC TIC Supplemental Data. Shin et al. Plant Cell. ()..5/tpc..95 Supplemental Figure. TIC interacts with MYC in the nucleus. Bimolecular fluorescence complementation assay using
a) Primary cultures derived from the pancreas of an 11-week-old Pdx1-Cre; K-MADM-p53
 1 2 3 4 5 6 7 8 9 10 Supplementary Figure 1. Induction of p53 LOH by MADM. a) Primary cultures derived from the pancreas of an 11-week-old Pdx1-Cre; K-MADM-p53 mouse revealed increased p53 KO/KO (green,
1 2 3 4 5 6 7 8 9 10 Supplementary Figure 1. Induction of p53 LOH by MADM. a) Primary cultures derived from the pancreas of an 11-week-old Pdx1-Cre; K-MADM-p53 mouse revealed increased p53 KO/KO (green,
Supplementary Document
 Supplementary Document 1. Supplementary Table legends 2. Supplementary Figure legends 3. Supplementary Tables 4. Supplementary Figures 5. Supplementary References 1. Supplementary Table legends Suppl.
Supplementary Document 1. Supplementary Table legends 2. Supplementary Figure legends 3. Supplementary Tables 4. Supplementary Figures 5. Supplementary References 1. Supplementary Table legends Suppl.
Supplementary Materials
 Supplementary Materials 1 Supplementary Table 1. List of primers used for quantitative PCR analysis. Gene name Gene symbol Accession IDs Sequence range Product Primer sequences size (bp) β-actin Actb gi
Supplementary Materials 1 Supplementary Table 1. List of primers used for quantitative PCR analysis. Gene name Gene symbol Accession IDs Sequence range Product Primer sequences size (bp) β-actin Actb gi
Supplementary Figure 1 a
 Supplementary Figure a Normalized expression/tbp (A.U.).6... Trip-br transcripts Trans Trans Trans b..5. Trip-br Ctrl LPS Normalized expression/tbp (A.U.) c Trip-br transcripts. adipocytes.... Trans Trans
Supplementary Figure a Normalized expression/tbp (A.U.).6... Trip-br transcripts Trans Trans Trans b..5. Trip-br Ctrl LPS Normalized expression/tbp (A.U.) c Trip-br transcripts. adipocytes.... Trans Trans
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 U1 inhibition causes a shift of RNA-seq reads from exons to introns. (a) Evidence for the high purity of 4-shU-labeled RNAs used for RNA-seq. HeLa cells transfected with control
Supplementary Figure 1 U1 inhibition causes a shift of RNA-seq reads from exons to introns. (a) Evidence for the high purity of 4-shU-labeled RNAs used for RNA-seq. HeLa cells transfected with control
Abbreviations: P- paraffin-embedded section; C, cryosection; Bio-SA, biotin-streptavidin-conjugated fluorescein amplification.
 Supplementary Table 1. Sequence of primers for real time PCR. Gene Forward primer Reverse primer S25 5 -GTG GTC CAC ACT ACT CTC TGA GTT TC-3 5 - GAC TTT CCG GCA TCC TTC TTC-3 Mafa cds 5 -CTT CAG CAA GGA
Supplementary Table 1. Sequence of primers for real time PCR. Gene Forward primer Reverse primer S25 5 -GTG GTC CAC ACT ACT CTC TGA GTT TC-3 5 - GAC TTT CCG GCA TCC TTC TTC-3 Mafa cds 5 -CTT CAG CAA GGA
Figure S1. Analysis of genomic and cdna sequences of the targeted regions in WT-KI and
 Figure S1. Analysis of genomic and sequences of the targeted regions in and indicated mutant KI cells, with WT and corresponding mutant sequences underlined. (A) cells; (B) K21E-KI cells; (C) D33A-KI cells;
Figure S1. Analysis of genomic and sequences of the targeted regions in and indicated mutant KI cells, with WT and corresponding mutant sequences underlined. (A) cells; (B) K21E-KI cells; (C) D33A-KI cells;
Supplementary Figure 1. ROS induces rapid Sod1 nuclear localization in a dosagedependent manner. WT yeast cells (SZy1051) were treated with 4NQO at
 Supplementary Figure 1. ROS induces rapid Sod1 nuclear localization in a dosagedependent manner. WT yeast cells (SZy1051) were treated with 4NQO at different concentrations for 30 min and analyzed for
Supplementary Figure 1. ROS induces rapid Sod1 nuclear localization in a dosagedependent manner. WT yeast cells (SZy1051) were treated with 4NQO at different concentrations for 30 min and analyzed for
CD31 5'-AGA GAC GGT CTT GTC GCA GT-3' 5 ' -TAC TGG GCT TCG AGA GCA GT-3'
 Table S1. The primer sets used for real-time RT-PCR analysis. Gene Forward Reverse VEGF PDGFB TGF-β MCP-1 5'-GTT GCA GCA TGA ATC TGA GG-3' 5'-GGA GAC TCT TCG AGG AGC ACT T-3' 5'-GAA TCA GGC ATC GAG AGA
Table S1. The primer sets used for real-time RT-PCR analysis. Gene Forward Reverse VEGF PDGFB TGF-β MCP-1 5'-GTT GCA GCA TGA ATC TGA GG-3' 5'-GGA GAC TCT TCG AGG AGC ACT T-3' 5'-GAA TCA GGC ATC GAG AGA
Supplementary Figure 1 MicroRNA expression in human synovial fibroblasts from different locations. MicroRNA, which were identified by RNAseq as most
 Supplementary Figure 1 MicroRNA expression in human synovial fibroblasts from different locations. MicroRNA, which were identified by RNAseq as most differentially expressed between human synovial fibroblasts
Supplementary Figure 1 MicroRNA expression in human synovial fibroblasts from different locations. MicroRNA, which were identified by RNAseq as most differentially expressed between human synovial fibroblasts
Toluidin-Staining of mast cells Ear tissue was fixed with Carnoy (60% ethanol, 30% chloroform, 10% acetic acid) overnight at 4 C, afterwards
 Toluidin-Staining of mast cells Ear tissue was fixed with Carnoy (60% ethanol, 30% chloroform, 10% acetic acid) overnight at 4 C, afterwards incubated in 100 % ethanol overnight at 4 C and embedded in
Toluidin-Staining of mast cells Ear tissue was fixed with Carnoy (60% ethanol, 30% chloroform, 10% acetic acid) overnight at 4 C, afterwards incubated in 100 % ethanol overnight at 4 C and embedded in
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. H3F3B expression in lung cancer. a. Comparison of H3F3B expression in relapsed and non-relapsed lung cancer patients. b. Prognosis of two groups of lung cancer
Supplementary Figures Supplementary Figure 1. H3F3B expression in lung cancer. a. Comparison of H3F3B expression in relapsed and non-relapsed lung cancer patients. b. Prognosis of two groups of lung cancer
Supplementary Table 2. Conserved regulatory elements in the promoters of CD36.
 Supplementary Table 1. RT-qPCR primers for CD3, PPARg and CEBP. Assay Forward Primer Reverse Primer 1A CAT TTG TGG CCT TGT GCT CTT TGA TGA GTC ACA GAA AGA ATC AAT TC 1B AGG AAA TGA ACT GAT GAG TCA CAG
Supplementary Table 1. RT-qPCR primers for CD3, PPARg and CEBP. Assay Forward Primer Reverse Primer 1A CAT TTG TGG CCT TGT GCT CTT TGA TGA GTC ACA GAA AGA ATC AAT TC 1B AGG AAA TGA ACT GAT GAG TCA CAG
BIOLOGY 621 Identification of the Snorks
 Name: Date: Block: BIOLOGY 621 Identification of the Snorks INTRODUCTION: In this simulation activity, you will examine the DNA sequence of a fictitious organism - the Snork. Snorks were discovered on
Name: Date: Block: BIOLOGY 621 Identification of the Snorks INTRODUCTION: In this simulation activity, you will examine the DNA sequence of a fictitious organism - the Snork. Snorks were discovered on
Table S1. Oligonucleotides used for the in-house RT-PCR assays targeting the M, H7 or N9. Assay (s) Target Name Sequence (5 3 ) Comments
 SUPPLEMENTAL INFORMATION 2 3 Table S. Oligonucleotides used for the in-house RT-PCR assays targeting the M, H7 or N9 genes. Assay (s) Target Name Sequence (5 3 ) Comments CDC M InfA Forward (NS), CDC M
SUPPLEMENTAL INFORMATION 2 3 Table S. Oligonucleotides used for the in-house RT-PCR assays targeting the M, H7 or N9 genes. Assay (s) Target Name Sequence (5 3 ) Comments CDC M InfA Forward (NS), CDC M
Culture Density (OD600) 0.1. Culture Density (OD600) Culture Density (OD600) Culture Density (OD600) Culture Density (OD600)
 A. B. C. D. E. PA JSRI JSRI 2 PA DSAM DSAM 2 DSAM 3 PA LNAP LNAP 2 LNAP 3 PAO Fcor Fcor 2 Fcor 3 PAO Wtho Wtho 2 Wtho 3 Wtho 4 DTSB Low Iron 2 4 6 8 2 4 6 8 2 22 DTSB Low Iron 2 4 6 8 2 4 6 8 2 22 DTSB
A. B. C. D. E. PA JSRI JSRI 2 PA DSAM DSAM 2 DSAM 3 PA LNAP LNAP 2 LNAP 3 PAO Fcor Fcor 2 Fcor 3 PAO Wtho Wtho 2 Wtho 3 Wtho 4 DTSB Low Iron 2 4 6 8 2 4 6 8 2 22 DTSB Low Iron 2 4 6 8 2 4 6 8 2 22 DTSB
Plasmids Western blot analysis and immunostaining Flow Cytometry Cell surface biotinylation RNA isolation and cdna synthesis
 Plasmids psuper-retro-s100a10 shrna1 was constructed by cloning the dsdna oligo 5 -GAT CCC CGT GGG CTT CCA GAG CTT CTT TCA AGA GAA GAA GCT CTG GAA GCC CAC TTT TTA-3 and 5 -AGC TTA AAA AGT GGG CTT CCA GAG
Plasmids psuper-retro-s100a10 shrna1 was constructed by cloning the dsdna oligo 5 -GAT CCC CGT GGG CTT CCA GAG CTT CTT TCA AGA GAA GAA GCT CTG GAA GCC CAC TTT TTA-3 and 5 -AGC TTA AAA AGT GGG CTT CCA GAG
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature05883 SUPPLEMENTARY INFORMATION Supplemental Figure 1 Prostaglandin agonists and antagonists alter runx1/cmyb expression. a-e, Embryos were exposed to (b) PGE2 and (c) PGI2 (20μM) and
doi: 10.1038/nature05883 SUPPLEMENTARY INFORMATION Supplemental Figure 1 Prostaglandin agonists and antagonists alter runx1/cmyb expression. a-e, Embryos were exposed to (b) PGE2 and (c) PGI2 (20μM) and
Citation for published version (APA): Oosterveer, M. H. (2009). Control of metabolic flux by nutrient sensors Groningen: s.n.
 University of Groningen Control of metabolic flux by nutrient sensors Oosterveer, Maaike IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from it.
University of Groningen Control of metabolic flux by nutrient sensors Oosterveer, Maaike IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from it.
A smart acid nanosystem for ultrasensitive. live cell mrna imaging by the target-triggered intracellular self-assembly
 Electronic Supplementary Material (ESI) for Chemical Science. This journal is The Royal Society of Chemistry 2017 A smart ZnO@polydopamine-nucleic acid nanosystem for ultrasensitive live cell mrna imaging
Electronic Supplementary Material (ESI) for Chemical Science. This journal is The Royal Society of Chemistry 2017 A smart ZnO@polydopamine-nucleic acid nanosystem for ultrasensitive live cell mrna imaging
Beta Thalassemia Case Study Introduction to Bioinformatics
 Beta Thalassemia Case Study Sami Khuri Department of Computer Science San José State University San José, California, USA sami.khuri@sjsu.edu www.cs.sjsu.edu/faculty/khuri Outline v Hemoglobin v Alpha
Beta Thalassemia Case Study Sami Khuri Department of Computer Science San José State University San José, California, USA sami.khuri@sjsu.edu www.cs.sjsu.edu/faculty/khuri Outline v Hemoglobin v Alpha
Nature Immunology: doi: /ni.3836
 Supplementary Figure 1 Recombinant LIGHT-VTP induces pericyte contractility and endothelial cell activation. (a) Western blot showing purification steps for full length murine LIGHT-VTP (CGKRK) protein:
Supplementary Figure 1 Recombinant LIGHT-VTP induces pericyte contractility and endothelial cell activation. (a) Western blot showing purification steps for full length murine LIGHT-VTP (CGKRK) protein:
SUPPLEMENTARY DATA. Supplementary Table 1. Primer sequences for qrt-pcr
 Supplementary Table 1. Primer sequences for qrt-pcr Gene PRDM16 UCP1 PGC1α Dio2 Elovl3 Cidea Cox8b PPARγ AP2 mttfam CyCs Nampt NRF1 16s-rRNA Hexokinase 2, intron 9 β-actin Primer Sequences 5'-CCA CCA GCG
Supplementary Table 1. Primer sequences for qrt-pcr Gene PRDM16 UCP1 PGC1α Dio2 Elovl3 Cidea Cox8b PPARγ AP2 mttfam CyCs Nampt NRF1 16s-rRNA Hexokinase 2, intron 9 β-actin Primer Sequences 5'-CCA CCA GCG
Phylogenetic analysis of human and chicken importins. Only five of six importins were studied because
 Supplementary Figure S1 Phylogenetic analysis of human and chicken importins. Only five of six importins were studied because importin-α6 was shown to be testis-specific. Human and chicken importin protein
Supplementary Figure S1 Phylogenetic analysis of human and chicken importins. Only five of six importins were studied because importin-α6 was shown to be testis-specific. Human and chicken importin protein
Beta Thalassemia Sami Khuri Department of Computer Science San José State University Spring 2015
 Bioinformatics in Medical Product Development SMPD 287 Three Beta Thalassemia Sami Khuri Department of Computer Science San José State University Hemoglobin Outline Anatomy of a gene Hemoglobinopathies
Bioinformatics in Medical Product Development SMPD 287 Three Beta Thalassemia Sami Khuri Department of Computer Science San José State University Hemoglobin Outline Anatomy of a gene Hemoglobinopathies
Supplementary Figure 1
 Supplementary Figure 1 3 3 3 1 1 Bregma -1.6mm 3 : Bregma Ref) Http://www.mbl.org/atlas165/atlas165_start.html Bregma -.18mm Supplementary Figure 1 Schematic representation of the utilized brain slice
Supplementary Figure 1 3 3 3 1 1 Bregma -1.6mm 3 : Bregma Ref) Http://www.mbl.org/atlas165/atlas165_start.html Bregma -.18mm Supplementary Figure 1 Schematic representation of the utilized brain slice
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1: Cryopreservation alters CD62L expression by CD4 T cells. Freshly isolated (left) or cryopreserved PBMCs (right) were stained with the mix of antibodies described
Supplementary Figure 1 Supplementary Figure 1: Cryopreservation alters CD62L expression by CD4 T cells. Freshly isolated (left) or cryopreserved PBMCs (right) were stained with the mix of antibodies described
BHP 2-7 and Nthy-ori 3-1 cells were grown in RPMI1640 medium (Hyclone) supplemented with 10% fetal bovine serum (Gibco), 2mM L-glutamine, and 100 U/mL
 1 2 3 4 Materials and Methods Cell culture BHP 2-7 and Nthy-ori 3-1 cells were grown in RPMI1640 medium (Hyclone) 5 supplemented with 10% fetal bovine serum (Gibco), 2mM L-glutamine, and 100 U/mL 6 penicillin-streptomycin.
1 2 3 4 Materials and Methods Cell culture BHP 2-7 and Nthy-ori 3-1 cells were grown in RPMI1640 medium (Hyclone) 5 supplemented with 10% fetal bovine serum (Gibco), 2mM L-glutamine, and 100 U/mL 6 penicillin-streptomycin.
Supplemental Information. Th17 Lymphocytes Induce Neuronal. Cell Death in a Human ipsc-based. Model of Parkinson's Disease
 Cell Stem Cell, Volume 23 Supplemental Information Th17 Lymphocytes Induce Neuronal Cell Death in a Human ipsc-based Model of Parkinson's Disease Annika Sommer, Franz Maxreiter, Florian Krach, Tanja Fadler,
Cell Stem Cell, Volume 23 Supplemental Information Th17 Lymphocytes Induce Neuronal Cell Death in a Human ipsc-based Model of Parkinson's Disease Annika Sommer, Franz Maxreiter, Florian Krach, Tanja Fadler,
Supplementary Figure 1a
 Supplementary Figure 1a Hours: E-cadherin TGF-β On TGF-β Off 0 12 24 36 48 24 48 72 Vimentin βactin Fig. S1a. Treatment of AML12 cells with TGF-β induces EMT. Treatment of AML12 cells with TGF-β results
Supplementary Figure 1a Hours: E-cadherin TGF-β On TGF-β Off 0 12 24 36 48 24 48 72 Vimentin βactin Fig. S1a. Treatment of AML12 cells with TGF-β induces EMT. Treatment of AML12 cells with TGF-β results
 www.lessonplansinc.com Topic: Protein Synthesis - Sentence Activity Summary: Students will simulate transcription and translation by building a sentence/polypeptide from words/amino acids. Goals & Objectives:
www.lessonplansinc.com Topic: Protein Synthesis - Sentence Activity Summary: Students will simulate transcription and translation by building a sentence/polypeptide from words/amino acids. Goals & Objectives:
Supplemental Information. Cancer-Associated Fibroblasts Neutralize. the Anti-tumor Effect of CSF1 Receptor Blockade
 Cancer Cell, Volume 32 Supplemental Information Cancer-Associated Fibroblasts Neutralize the Anti-tumor Effect of CSF1 Receptor Blockade by Inducing PMN-MDSC Infiltration of Tumors Vinit Kumar, Laxminarasimha
Cancer Cell, Volume 32 Supplemental Information Cancer-Associated Fibroblasts Neutralize the Anti-tumor Effect of CSF1 Receptor Blockade by Inducing PMN-MDSC Infiltration of Tumors Vinit Kumar, Laxminarasimha
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1. Lats1/2 deleted ihbs and ihps showed decreased transcripts of hepatocyte related genes (a and b) Western blots (a) and recombination PCR (b) of control and
Supplementary Figure 1 Supplementary Figure 1. Lats1/2 deleted ihbs and ihps showed decreased transcripts of hepatocyte related genes (a and b) Western blots (a) and recombination PCR (b) of control and
Astaxanthin prevents and reverses diet-induced insulin resistance and. steatohepatitis in mice: A comparison with vitamin E
 Supplementary Information Astaxanthin prevents and reverses diet-induced insulin resistance and steatohepatitis in mice: A comparison with vitamin E Yinhua Ni, 1,2 Mayumi Nagashimada, 1 Fen Zhuge, 1 Lili
Supplementary Information Astaxanthin prevents and reverses diet-induced insulin resistance and steatohepatitis in mice: A comparison with vitamin E Yinhua Ni, 1,2 Mayumi Nagashimada, 1 Fen Zhuge, 1 Lili
A basic helix loop helix transcription factor controls cell growth
 A basic helix loop helix transcription factor controls cell growth and size in root hairs Keke Yi 1,2, Benoît Menand 1,3, Elizabeth Bell 1, Liam Dolan 1,4 Supplementary note Low soil phosphate availability
A basic helix loop helix transcription factor controls cell growth and size in root hairs Keke Yi 1,2, Benoît Menand 1,3, Elizabeth Bell 1, Liam Dolan 1,4 Supplementary note Low soil phosphate availability
*To whom correspondence should be addressed. This PDF file includes:
 www.sciencemag.org/cgi/content/full/science.1212182/dc1 Supporting Online Material for Partial Retraction to Detection of an Infectious Retrovirus, XMRV, in Blood Cells of Patients with Chronic Fatigue
www.sciencemag.org/cgi/content/full/science.1212182/dc1 Supporting Online Material for Partial Retraction to Detection of an Infectious Retrovirus, XMRV, in Blood Cells of Patients with Chronic Fatigue
Supplemental Figures: Supplemental Figure 1
 Supplemental Figures: Supplemental Figure 1 Suppl. Figure 1. BM-DC infection with H. pylori does not induce cytotoxicity and treatment of BM-DCs with H. pylori sonicate, but not heat-inactivated bacteria,
Supplemental Figures: Supplemental Figure 1 Suppl. Figure 1. BM-DC infection with H. pylori does not induce cytotoxicity and treatment of BM-DCs with H. pylori sonicate, but not heat-inactivated bacteria,
Resistance to Tetracycline Antibiotics by Wangrong Yang, Ian F. Moore, Kalinka P. Koteva, Donald W. Hughes, David C. Bareich and Gerard D. Wright.
 Supplementary Data for TetX is a Flavin-Dependent Monooxygenase Conferring Resistance to Tetracycline Antibiotics by Wangrong Yang, Ian F. Moore, Kalinka P. Koteva, Donald W. Hughes, David C. Bareich and
Supplementary Data for TetX is a Flavin-Dependent Monooxygenase Conferring Resistance to Tetracycline Antibiotics by Wangrong Yang, Ian F. Moore, Kalinka P. Koteva, Donald W. Hughes, David C. Bareich and
Cross-talk between mineralocorticoid and angiotensin II signaling for cardiac
 ONLINE SUPPLEMENT TO Crosstalk between mineralocorticoid and angiotensin II signaling for cardiac remodeling An Di ZHANG,,3, Aurelie NGUYEN DINH CAT*,,3, Christelle SOUKASEUM *,,3, Brigitte ESCOUBET, 4,
ONLINE SUPPLEMENT TO Crosstalk between mineralocorticoid and angiotensin II signaling for cardiac remodeling An Di ZHANG,,3, Aurelie NGUYEN DINH CAT*,,3, Christelle SOUKASEUM *,,3, Brigitte ESCOUBET, 4,
Supplementary Materials and Methods
 DD2 suppresses tumorigenicity of ovarian cancer cells by limiting cancer stem cell population Chunhua Han et al. Supplementary Materials and Methods Analysis of publicly available datasets: To analyze
DD2 suppresses tumorigenicity of ovarian cancer cells by limiting cancer stem cell population Chunhua Han et al. Supplementary Materials and Methods Analysis of publicly available datasets: To analyze
TetR repressor-based bioreporters for the detection of doxycycline using Escherichia
 Supplementary materials TetR repressor-based bioreporters for the detection of doxycycline using Escherichia coli and Acinetobacter oleivorans Hyerim Hong and Woojun Park * Department of Environmental
Supplementary materials TetR repressor-based bioreporters for the detection of doxycycline using Escherichia coli and Acinetobacter oleivorans Hyerim Hong and Woojun Park * Department of Environmental
Supporting Information
 Supporting Information Malapeira et al. 10.1073/pnas.1217022110 SI Materials and Methods Plant Material and Growth Conditions. A. thaliana seedlings were stratified at 4 C in the dark for 3 d on Murashige
Supporting Information Malapeira et al. 10.1073/pnas.1217022110 SI Materials and Methods Plant Material and Growth Conditions. A. thaliana seedlings were stratified at 4 C in the dark for 3 d on Murashige
Supplementary Information. Bamboo shoot fiber prevents obesity in mice by. modulating the gut microbiota
 Supplementary Information Bamboo shoot fiber prevents obesity in mice by modulating the gut microbiota Xiufen Li 1,2, Juan Guo 1, Kailong Ji 1,2, and Ping Zhang 1,* 1 Key Laboratory of Tropical Plant Resources
Supplementary Information Bamboo shoot fiber prevents obesity in mice by modulating the gut microbiota Xiufen Li 1,2, Juan Guo 1, Kailong Ji 1,2, and Ping Zhang 1,* 1 Key Laboratory of Tropical Plant Resources
ice-cold 70% ethanol with gentle vortexing, incubated at -20 C for 4 hours, and washed with PBS.
 Cell cycle analysis For cell cycle analysis, single cell suspensions of E12.5 fetal liver cells were suspended in 4 ml ice-cold 7% ethanol with gentle vortexing, incubated at -2 C for 4 hours, and washed
Cell cycle analysis For cell cycle analysis, single cell suspensions of E12.5 fetal liver cells were suspended in 4 ml ice-cold 7% ethanol with gentle vortexing, incubated at -2 C for 4 hours, and washed
CIRCRESAHA/2004/098145/R1 - ONLINE 1. Validation by Semi-quantitative Real-Time Reverse Transcription PCR
 CIRCRESAHA/2004/098145/R1 - ONLINE 1 Expanded Materials and Methods Validation by Semi-quantitative Real-Time Reverse Transcription PCR Expression patterns of 13 genes (Online Table 2), selected with respect
CIRCRESAHA/2004/098145/R1 - ONLINE 1 Expanded Materials and Methods Validation by Semi-quantitative Real-Time Reverse Transcription PCR Expression patterns of 13 genes (Online Table 2), selected with respect
Description of Supplementary Files. File Name: Supplementary Information Description: Supplementary Figures and Supplementary Tables
 Description of Supplementary Files File Name: Supplementary Information Description: Supplementary Figures and Supplementary Tables Supplementary Figure 1: (A), HCT116 IDH1-WT and IDH1-R132H cells were
Description of Supplementary Files File Name: Supplementary Information Description: Supplementary Figures and Supplementary Tables Supplementary Figure 1: (A), HCT116 IDH1-WT and IDH1-R132H cells were
Journal of Cell Science Supplementary information. Arl8b +/- Arl8b -/- Inset B. electron density. genotype
 J. Cell Sci. : doi:.4/jcs.59: Supplementary information E9. A Arl8b /- Arl8b -/- Arl8b Arl8b non-specific band Gapdh Tbp E7.5 HE Inset B D Control al am hf C E Arl8b -/- al am hf E8.5 F low middle high
J. Cell Sci. : doi:.4/jcs.59: Supplementary information E9. A Arl8b /- Arl8b -/- Arl8b Arl8b non-specific band Gapdh Tbp E7.5 HE Inset B D Control al am hf C E Arl8b -/- al am hf E8.5 F low middle high
Supplementary Figure 1
 Metastatic melanoma Primary melanoma Healthy human skin Supplementary Figure 1 CD22 IgG4 Supplementary Figure 1: Immunohisochemical analysis of CD22+ (left) and IgG4 (right), cells (shown in red and indicated
Metastatic melanoma Primary melanoma Healthy human skin Supplementary Figure 1 CD22 IgG4 Supplementary Figure 1: Immunohisochemical analysis of CD22+ (left) and IgG4 (right), cells (shown in red and indicated
Nucleotide Sequence of the Australian Bluetongue Virus Serotype 1 RNA Segment 10
 J. gen. Virol. (1988), 69, 945-949. Printed in Great Britain 945 Key words: BTV/genome segment lo/nucleotide sequence Nucleotide Sequence of the Australian Bluetongue Virus Serotype 1 RNA Segment 10 By
J. gen. Virol. (1988), 69, 945-949. Printed in Great Britain 945 Key words: BTV/genome segment lo/nucleotide sequence Nucleotide Sequence of the Australian Bluetongue Virus Serotype 1 RNA Segment 10 By
Lezione 10. Sommario. Bioinformatica. Lezione 10: Sintesi proteica Synthesis of proteins Central dogma: DNA makes RNA makes proteins Genetic code
 Lezione 10 Bioinformatica Mauro Ceccanti e Alberto Paoluzzi Lezione 10: Sintesi proteica Synthesis of proteins Dip. Informatica e Automazione Università Roma Tre Dip. Medicina Clinica Università La Sapienza
Lezione 10 Bioinformatica Mauro Ceccanti e Alberto Paoluzzi Lezione 10: Sintesi proteica Synthesis of proteins Dip. Informatica e Automazione Università Roma Tre Dip. Medicina Clinica Università La Sapienza
Characterizing intra-host influenza virus populations to predict emergence
 Characterizing intra-host influenza virus populations to predict emergence June 12, 2012 Forum on Microbial Threats Washington, DC Elodie Ghedin Center for Vaccine Research Dept. Computational & Systems
Characterizing intra-host influenza virus populations to predict emergence June 12, 2012 Forum on Microbial Threats Washington, DC Elodie Ghedin Center for Vaccine Research Dept. Computational & Systems
SUPPORTING INFORMATION
 SUPPORTING INFORMATION Biology is different in small volumes: endogenous signals shape phenotype of primary hepatocytes cultured in microfluidic channels Amranul Haque, Pantea Gheibi, Yandong Gao, Elena
SUPPORTING INFORMATION Biology is different in small volumes: endogenous signals shape phenotype of primary hepatocytes cultured in microfluidic channels Amranul Haque, Pantea Gheibi, Yandong Gao, Elena
Relationship of the APOA5/A4/C3/A1 gene cluster and APOB gene polymorphisms with dyslipidemia
 elationship of the APOA5/A4/C3/A1 gene cluster and APOB gene polymorphisms with dyslipidemia H.J. Ou 1, G. Huang 2, W. Liu 3, X.L. Ma 2, Y. Wei 4, T. Zhou 5 and Z.M. Pan 3 1 Department of Neurology, The
elationship of the APOA5/A4/C3/A1 gene cluster and APOB gene polymorphisms with dyslipidemia H.J. Ou 1, G. Huang 2, W. Liu 3, X.L. Ma 2, Y. Wei 4, T. Zhou 5 and Z.M. Pan 3 1 Department of Neurology, The
Formylpeptide receptor2 contributes to colon epithelial homeostasis, inflammation, and tumorigenesis
 Supplementary Data Formylpeptide receptor2 contributes to colon epithelial homeostasis, inflammation, and tumorigenesis Keqiang Chen, Mingyong Liu, Ying Liu, Teizo Yoshimura, Wei Shen, Yingying Le, Scott
Supplementary Data Formylpeptide receptor2 contributes to colon epithelial homeostasis, inflammation, and tumorigenesis Keqiang Chen, Mingyong Liu, Ying Liu, Teizo Yoshimura, Wei Shen, Yingying Le, Scott
PATHOGENIC CHARACTERISTICS OF Candida albicans ISOLATED FROM ORAL
 PATHOGENIC CHARACTERISTICS OF Candida albicans ISOLATED FROM ORAL CAVITIES OF DENTURE WEARERS AND CANCER PATIENTS WEARING ORAL PROSTHESES Junior Vivian Mothibe A dissertation submitted to the Faculty of
PATHOGENIC CHARACTERISTICS OF Candida albicans ISOLATED FROM ORAL CAVITIES OF DENTURE WEARERS AND CANCER PATIENTS WEARING ORAL PROSTHESES Junior Vivian Mothibe A dissertation submitted to the Faculty of
Candida albicans Secreted Aspartyl Proteinases in Virulence and Pathogenesis
 MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS, Sept. 2003, p. 400 428 Vol. 67, No. 3 1092-2172/03/$08.00 0 DOI: 10.1128/MMBR.67.3.400 428.2003 Copyright 2003, American Society for Microbiology. All Rights
MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS, Sept. 2003, p. 400 428 Vol. 67, No. 3 1092-2172/03/$08.00 0 DOI: 10.1128/MMBR.67.3.400 428.2003 Copyright 2003, American Society for Microbiology. All Rights
Expression of Selected Inflammatory Cytokine Genes in Bladder Biopsies
 Borneo Journal of Resource Science and Technology (2013) 3(2): 15-20 Expression of Selected Inflammatory Cytokine Genes in Bladder Biopsies EDMUND UI-HANG SIM *1, NUR DIANA ANUAR 2, TENG-AIK ONG 3, GUAN-
Borneo Journal of Resource Science and Technology (2013) 3(2): 15-20 Expression of Selected Inflammatory Cytokine Genes in Bladder Biopsies EDMUND UI-HANG SIM *1, NUR DIANA ANUAR 2, TENG-AIK ONG 3, GUAN-
Baseline clinical characteristics for the 81 CMML patients Routine diagnostic testing and statistical analyses... 3
 Next-Generation Sequencing Technology Reveals a Characteristic Pattern of Molecular Mutations in 72.8% of Chronic Myelomonocytic Leukemia (CMML) by Detecting Frequent Alterations in TET2, CBL, RAS, and
Next-Generation Sequencing Technology Reveals a Characteristic Pattern of Molecular Mutations in 72.8% of Chronic Myelomonocytic Leukemia (CMML) by Detecting Frequent Alterations in TET2, CBL, RAS, and
Integration Solutions
 Integration Solutions (1) a) With no active glycosyltransferase of either type, an ii individual would not be able to add any sugars to the O form of the lipopolysaccharide. Thus, the only lipopolysaccharide
Integration Solutions (1) a) With no active glycosyltransferase of either type, an ii individual would not be able to add any sugars to the O form of the lipopolysaccharide. Thus, the only lipopolysaccharide
Mutation Screening and Association Studies of the Human UCP 3 Gene in Normoglycemic and NIDDM Morbidly Obese Patients
 Mutation Screening and Association Studies of the Human UCP 3 Gene in Normoglycemic and NIDDM Morbidly Obese Patients Shuichi OTABE, Karine CLEMENT, Séverine DUBOIS, Frederic LEPRETRE, Veronique PELLOUX,
Mutation Screening and Association Studies of the Human UCP 3 Gene in Normoglycemic and NIDDM Morbidly Obese Patients Shuichi OTABE, Karine CLEMENT, Séverine DUBOIS, Frederic LEPRETRE, Veronique PELLOUX,
SUPPLEMENTAL METHODS Cell culture RNA extraction and analysis Immunohistochemical analysis and laser capture microdissection (LCM)
 SUPPLEMENTAL METHODS Cell culture Human peripheral blood mononuclear cells were isolated from healthy donors by Ficoll density gradient centrifugation. Monocyte differentiation to resting macrophages ()
SUPPLEMENTAL METHODS Cell culture Human peripheral blood mononuclear cells were isolated from healthy donors by Ficoll density gradient centrifugation. Monocyte differentiation to resting macrophages ()
in the Gastrointestinal and Reproductive Tracts of Quarter Horse Mares
 Influence of Probiotics on Microflora in the Gastrointestinal and Reproductive Tracts of Quarter Horse Mares Katie Barnhart Research Advisors: Dr. Kimberly Cole and Dr. John Mark Reddish Department of
Influence of Probiotics on Microflora in the Gastrointestinal and Reproductive Tracts of Quarter Horse Mares Katie Barnhart Research Advisors: Dr. Kimberly Cole and Dr. John Mark Reddish Department of
Single-Molecule Analysis of Gene Expression Using Two-Color RNA- Labeling in Live Yeast
 Supplemental Figures, Tables and Results Single-Molecule Analysis of Gene Expression Using Two-Color RNA- Labeling in Live Yeast Sami Hocine 1, Pascal Raymond 2, Daniel Zenklusen 2, Jeffrey A. Chao 1 &
Supplemental Figures, Tables and Results Single-Molecule Analysis of Gene Expression Using Two-Color RNA- Labeling in Live Yeast Sami Hocine 1, Pascal Raymond 2, Daniel Zenklusen 2, Jeffrey A. Chao 1 &
Cancer Genetics 204 (2011) 45e52
 Cancer Genetics 204 (2011) 45e52 Exon scanning by reverse transcriptaseepolymerase chain reaction for detection of known and novel EML4eALK fusion variants in nonesmall cell lung cancer Heather R. Sanders
Cancer Genetics 204 (2011) 45e52 Exon scanning by reverse transcriptaseepolymerase chain reaction for detection of known and novel EML4eALK fusion variants in nonesmall cell lung cancer Heather R. Sanders
Advanced Subsidiary Unit 1: Lifestyle, Transport, Genes and Health
 Write your name here Surname Other names Edexcel GCE Centre Number Candidate Number Biology Advanced Subsidiary Unit 1: Lifestyle, Transport, Genes and Health Thursday 8 January 2009 Morning Time: 1 hour
Write your name here Surname Other names Edexcel GCE Centre Number Candidate Number Biology Advanced Subsidiary Unit 1: Lifestyle, Transport, Genes and Health Thursday 8 January 2009 Morning Time: 1 hour
University of Groningen. Vasoregression in incipient diabetic retinopathy Pfister, Frederick
 University of Groningen Vasoregression in incipient diabetic retinopathy Pfister, Frederick IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from
University of Groningen Vasoregression in incipient diabetic retinopathy Pfister, Frederick IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from
Isolate Sexual Idiomorph Species
 SUPLEMENTARY TABLE 1. Isolate identification, sexual idiomorph and species of each isolate used for MAT locus distribution in Paracoccidioides species. Isolate Sexual Idiomorph Species Pb01 MAT1-1 P. lutzii
SUPLEMENTARY TABLE 1. Isolate identification, sexual idiomorph and species of each isolate used for MAT locus distribution in Paracoccidioides species. Isolate Sexual Idiomorph Species Pb01 MAT1-1 P. lutzii
Supplementary information
 Supplementary information Unique polypharmacology nuclear receptor modulator blocks inflammatory signaling pathways Mi Ra Chang 1, Anthony Ciesla 1, Timothy S. Strutzenberg 1, Scott J. Novick 1, Yuanjun
Supplementary information Unique polypharmacology nuclear receptor modulator blocks inflammatory signaling pathways Mi Ra Chang 1, Anthony Ciesla 1, Timothy S. Strutzenberg 1, Scott J. Novick 1, Yuanjun
Supplementary Information
 Supplementary Information Remodeling of heterochromatin structure slows neuropathological progression and prolongs survival in an animal model of Huntington s disease Junghee Lee, Yu Jin Hwang, Yunha Kim,
Supplementary Information Remodeling of heterochromatin structure slows neuropathological progression and prolongs survival in an animal model of Huntington s disease Junghee Lee, Yu Jin Hwang, Yunha Kim,
Enhanced detection and serotyping of Streptococcus pneumoniae using multiplex polymerase chain reaction
 Original article http://dx.doi.org/10.3345/kjp.2012.55.11.424 Korean J Pediatr 2012;55(11):424-429 eissn 1738-1061 pissn 2092-7258 Enhanced detection and serotyping of Streptococcus pneumoniae using multiplex
Original article http://dx.doi.org/10.3345/kjp.2012.55.11.424 Korean J Pediatr 2012;55(11):424-429 eissn 1738-1061 pissn 2092-7258 Enhanced detection and serotyping of Streptococcus pneumoniae using multiplex
L I F E S C I E N C E S
 1a L I F E S C I E N C E S 5 -UUA AUA UUC GAA AGC UGC AUC GAA AAC UGU GAA UCA-3 5 -TTA ATA TTC GAA AGC TGC ATC GAA AAC TGT GAA TCA-3 3 -AAT TAT AAG CTT TCG ACG TAG CTT TTG ACA CTT AGT-5 OCTOBER 31, 2006
1a L I F E S C I E N C E S 5 -UUA AUA UUC GAA AGC UGC AUC GAA AAC UGU GAA UCA-3 5 -TTA ATA TTC GAA AGC TGC ATC GAA AAC TGT GAA TCA-3 3 -AAT TAT AAG CTT TCG ACG TAG CTT TTG ACA CTT AGT-5 OCTOBER 31, 2006
PATIENTS AND METHODS. Subjects
 PATIENTS AND METHODS Subjects Twenty-nine morbidly obese subjects involved in a gastric surgery program were enrolled in the study between October 25 and March 21. Bariatric surgery was performed in patients
PATIENTS AND METHODS Subjects Twenty-nine morbidly obese subjects involved in a gastric surgery program were enrolled in the study between October 25 and March 21. Bariatric surgery was performed in patients
Isolation and Genetic Characterization of New Reassortant H3N1 Swine Influenza Virus from Pigs in the Midwestern United States
 JOURNAL OF VIROLOGY, May 2006, p. 5092 5096 Vol. 80, No. 10 0022-538X/06/$08.00 0 doi:10.1128/jvi.80.10.5092 5096.2006 Copyright 2006, American Society for Microbiology. All Rights Reserved. Isolation
JOURNAL OF VIROLOGY, May 2006, p. 5092 5096 Vol. 80, No. 10 0022-538X/06/$08.00 0 doi:10.1128/jvi.80.10.5092 5096.2006 Copyright 2006, American Society for Microbiology. All Rights Reserved. Isolation
Europe PMC Funders Group Author Manuscript Int J Antimicrob Agents. Author manuscript; available in PMC 2010 November 15.
 Europe PMC Funders Group Author Manuscript Published in final edited form as: Int J Antimicrob Agents. 2005 October ; 26(4): 335 337. doi:10.1016/j.ijantimicag.2005.07.006. Interaction of serotonin with
Europe PMC Funders Group Author Manuscript Published in final edited form as: Int J Antimicrob Agents. 2005 October ; 26(4): 335 337. doi:10.1016/j.ijantimicag.2005.07.006. Interaction of serotonin with
Supporting Information. Mutational analysis of a phenazine biosynthetic gene cluster in
 Supporting Information for Mutational analysis of a phenazine biosynthetic gene cluster in Streptomyces anulatus 9663 Orwah Saleh 1, Katrin Flinspach 1, Lucia Westrich 1, Andreas Kulik 2, Bertolt Gust
Supporting Information for Mutational analysis of a phenazine biosynthetic gene cluster in Streptomyces anulatus 9663 Orwah Saleh 1, Katrin Flinspach 1, Lucia Westrich 1, Andreas Kulik 2, Bertolt Gust
Trichophyton Microsporum Epidermophyton. dermatomycosis. Dematiaceous(pigmented fungi ) Dimorphic fungi Yeast and yeast like saprophyte
 Cutaneous candidiasis dermatophytosis Trichophyton Microsporum Epidermophyton dermatomycosis Dematiaceous(pigmented fungi ) Dimorphic fungi Yeast and yeast like saprophyte dermatomycosis Yeast & yeast
Cutaneous candidiasis dermatophytosis Trichophyton Microsporum Epidermophyton dermatomycosis Dematiaceous(pigmented fungi ) Dimorphic fungi Yeast and yeast like saprophyte dermatomycosis Yeast & yeast
The Clinical Performance of Primary HPV Screening, Primary HPV Screening Plus Cytology Cotesting, and Cytology Alone at a Tertiary Care Hospital
 The Clinical Performance of Primary HPV Screening, Primary HPV Screening Plus Cytology Cotesting, and Cytology Alone at a Tertiary Care Hospital Jung-Woo Choi MD, PhD; Younghye Kim MD, PhD; Ju-Han Lee
The Clinical Performance of Primary HPV Screening, Primary HPV Screening Plus Cytology Cotesting, and Cytology Alone at a Tertiary Care Hospital Jung-Woo Choi MD, PhD; Younghye Kim MD, PhD; Ju-Han Lee
Recommended laboratory tests to identify influenza A/H5 virus in specimens from patients with an influenza-like illness
 World Health Organization Recommended laboratory tests to identify influenza A/H5 virus in specimens from patients with an influenza-like illness General information Highly pathogenic avian influenza (HPAI)
World Health Organization Recommended laboratory tests to identify influenza A/H5 virus in specimens from patients with an influenza-like illness General information Highly pathogenic avian influenza (HPAI)
HCV Persistence and Immune Evasion in the Absence of Memory T Cell Help.
 SOM Text HCV Persistence and Immune Evasion in the Absence of Memory T Cell Help. Arash Grakoui 1, Naglaa H. Shoukry 2, David J. Woollard 2, Jin-Hwan Han 1, Holly L. Hanson 1, John Ghrayeb 3, Krishna K.
SOM Text HCV Persistence and Immune Evasion in the Absence of Memory T Cell Help. Arash Grakoui 1, Naglaa H. Shoukry 2, David J. Woollard 2, Jin-Hwan Han 1, Holly L. Hanson 1, John Ghrayeb 3, Krishna K.
Loyer, et al. microrna-21 contributes to NASH Suppl 1/15
 Loyer, et al. microrna-21 contributes to NASH Suppl 1/15 SUPPLEMENTARY MATERIAL: Liver MicroRNA-21 is Overexpressed in Non Alcoholic Steatohepatitis and Contributes to the Disease in Experimental Models
Loyer, et al. microrna-21 contributes to NASH Suppl 1/15 SUPPLEMENTARY MATERIAL: Liver MicroRNA-21 is Overexpressed in Non Alcoholic Steatohepatitis and Contributes to the Disease in Experimental Models
Patterns of hemagglutinin evolution and the epidemiology of influenza
 2 8 US Annual Mortality Rate All causes Infectious Disease Patterns of hemagglutinin evolution and the epidemiology of influenza DIMACS Working Group on Genetics and Evolution of Pathogens, 25 Nov 3 Deaths
2 8 US Annual Mortality Rate All causes Infectious Disease Patterns of hemagglutinin evolution and the epidemiology of influenza DIMACS Working Group on Genetics and Evolution of Pathogens, 25 Nov 3 Deaths
Detection of 549 new HLA alleles in potential stem cell donors from the United States, Poland and Germany
 HLA ISSN 2059-2302 BRIEF COMMUNICATION Detection of 549 new HLA alleles in potential stem cell donors from the United States, Poland and Germany C. J. Hernández-Frederick 1, N. Cereb 2,A.S.Giani 1, J.
HLA ISSN 2059-2302 BRIEF COMMUNICATION Detection of 549 new HLA alleles in potential stem cell donors from the United States, Poland and Germany C. J. Hernández-Frederick 1, N. Cereb 2,A.S.Giani 1, J.
Mutation analysis of a Chinese family with oculocutaneous albinism
 /, 2016, Vol. 7, (No. 51), pp: 84981-84988 Mutation analysis of a Chinese family with oculocutaneous albinism Xiong Wang 1, Yaowu Zhu 1, Na Shen 1, Jing Peng 1, Chunyu Wang 1, Haiyi Liu 2, Yanjun Lu 1
/, 2016, Vol. 7, (No. 51), pp: 84981-84988 Mutation analysis of a Chinese family with oculocutaneous albinism Xiong Wang 1, Yaowu Zhu 1, Na Shen 1, Jing Peng 1, Chunyu Wang 1, Haiyi Liu 2, Yanjun Lu 1
CHAPTER 4 RESULTS. showed that all three replicates had similar growth trends (Figure 4.1) (p<0.05; p=0.0000)
 CHAPTER 4 RESULTS 4.1 Growth Characterization of C. vulgaris 4.1.1 Optical Density Growth study of Chlorella vulgaris based on optical density at 620 nm (OD 620 ) showed that all three replicates had similar
CHAPTER 4 RESULTS 4.1 Growth Characterization of C. vulgaris 4.1.1 Optical Density Growth study of Chlorella vulgaris based on optical density at 620 nm (OD 620 ) showed that all three replicates had similar
Malignant Amelanotic Melanoma of the Pleura without Primary Skin Lesion: An Autopsy Case Report. a a*
 2009 63 6 379 384 Malignant Amelanotic Melanoma of the Pleura without Primary Skin Lesion: An Autopsy Case Report a b a a a* a b 380 63 6 Chest x ray and computed tomography (CT). A, Chest x ray on admission
2009 63 6 379 384 Malignant Amelanotic Melanoma of the Pleura without Primary Skin Lesion: An Autopsy Case Report a b a a a* a b 380 63 6 Chest x ray and computed tomography (CT). A, Chest x ray on admission
Ho Young Jung Hye Seung Han Hyo Bin Kim 1 Seo Young Oh 1 Sun-Joo Lee 2 Wook Youn Kim
 Journal of Pathology and Translational Medicine 2016; 50: 138-146 ORIGINAL ARTICLE Comparison of Analytical and Clinical Performance of HPV 9G DNA Chip, PANArray HPV Genotyping Chip, and Hybrid-Capture
Journal of Pathology and Translational Medicine 2016; 50: 138-146 ORIGINAL ARTICLE Comparison of Analytical and Clinical Performance of HPV 9G DNA Chip, PANArray HPV Genotyping Chip, and Hybrid-Capture
Journal of Microbes and Infection,June 2007,Vol 2,No. 2. (HBsAg)2 , (PCR) 1762/ 1764
 68 2007 6 2 2 Journal of Microbes and Infection,June 2007,Vol 2,No. 2 2 S 1 1 1 2 2 3 1 (HBsAg)2 ( YIC) S 5 30g 60g YIC ( HBV) DNA > 2 log10 e (HBeAg), 6 DNA, 1 YIC 1, (PCR) (0 ) (44 ) HBV DNA S 2, S a
68 2007 6 2 2 Journal of Microbes and Infection,June 2007,Vol 2,No. 2 2 S 1 1 1 2 2 3 1 (HBsAg)2 ( YIC) S 5 30g 60g YIC ( HBV) DNA > 2 log10 e (HBeAg), 6 DNA, 1 YIC 1, (PCR) (0 ) (44 ) HBV DNA S 2, S a
Mechanistic and functional insights into fatty acid activation in Mycobacterium tuberculosis SUPPLEMENTARY INFORMATION
 Mechanistic and functional insights into fatty acid activation in Mycobacterium tuberculosis Pooja Arora 1, Aneesh Goyal 2, Vivek T atarajan 1, Eerappa Rajakumara 2, Priyanka Verma 1, Radhika Gupta 3,
Mechanistic and functional insights into fatty acid activation in Mycobacterium tuberculosis Pooja Arora 1, Aneesh Goyal 2, Vivek T atarajan 1, Eerappa Rajakumara 2, Priyanka Verma 1, Radhika Gupta 3,
without LOI phenotype by breeding female wild-type C57BL/6J and male H19 +/.
 Sakatani et al. 1 Supporting Online Material Materials and methods Mice and genotyping: H19 mutant mice with C57BL/6J background carrying a deletion in the structural H19 gene (3 kb) and 10 kb of 5 flanking
Sakatani et al. 1 Supporting Online Material Materials and methods Mice and genotyping: H19 mutant mice with C57BL/6J background carrying a deletion in the structural H19 gene (3 kb) and 10 kb of 5 flanking
SUPPLEMENTARY INFORMATION
 BASELINE ISCHAEMIA a b Phd2 +/- c d Collateral growth and maintenance SMC recruitment SMC proliferation Phd2 +/- NF- B off NF- B on NF- B on NF- B on Endothelial cell Smooth muscle cell Pro-arteriogenic
BASELINE ISCHAEMIA a b Phd2 +/- c d Collateral growth and maintenance SMC recruitment SMC proliferation Phd2 +/- NF- B off NF- B on NF- B on NF- B on Endothelial cell Smooth muscle cell Pro-arteriogenic
Bacterial Gene Finding CMSC 423
 Bacterial Gene Finding CMSC 423 Finding Signals in DNA We just have a long string of A, C, G, Ts. How can we find the signals encoded in it? Suppose you encountered a language you didn t know. How would
Bacterial Gene Finding CMSC 423 Finding Signals in DNA We just have a long string of A, C, G, Ts. How can we find the signals encoded in it? Suppose you encountered a language you didn t know. How would
IMMUNOLOGY, HEALTH, AND DISEASE
 IMMUNOLOGY, HEALTH, AND DISEASE Multiplex polymerase chain reaction for the detection and differentiation of avian influenza viruses and other poultry respiratory pathogens S. Rashid,* K. Naeem, 1 Z. Ahmed,
IMMUNOLOGY, HEALTH, AND DISEASE Multiplex polymerase chain reaction for the detection and differentiation of avian influenza viruses and other poultry respiratory pathogens S. Rashid,* K. Naeem, 1 Z. Ahmed,
Frequency of mosaicism points towards mutation prone early cleavage cell divisions.
 Frequency of mosaicism points towards mutation prone early cleavage cell divisions. Chad Harland, Wouter Coppieters, Latifa Karim, Carole Charlier, Michel Georges Germ-line de novo mutations Definition:
Frequency of mosaicism points towards mutation prone early cleavage cell divisions. Chad Harland, Wouter Coppieters, Latifa Karim, Carole Charlier, Michel Georges Germ-line de novo mutations Definition:
Supplementary Figure S1
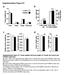 Supplementry Figure S Tissue weights (g).... Liver Hert Brin Pncres Len mss (g) 8 6 -% +% 8 6 Len mss Len mss (g) (% ody weight) Len mss (% ody weight) c Tiilis nterior weight (g).6...... Qudriceps weight
Supplementry Figure S Tissue weights (g).... Liver Hert Brin Pncres Len mss (g) 8 6 -% +% 8 6 Len mss Len mss (g) (% ody weight) Len mss (% ody weight) c Tiilis nterior weight (g).6...... Qudriceps weight
Finding protein sites where resistance has evolved
 Finding protein sites where resistance has evolved The amino acid (Ka) and synonymous (Ks) substitution rates Please sit in row K or forward The Berlin patient: first person cured of HIV Contracted HIV
Finding protein sites where resistance has evolved The amino acid (Ka) and synonymous (Ks) substitution rates Please sit in row K or forward The Berlin patient: first person cured of HIV Contracted HIV
Viral hepatitis, which affects half a billion people
 GASTROENTEROLOGY 2006;130:435 452 BASIC LIVER, PANCREAS, AND BILIARY TRACT Natural Killer Cells Ameliorate Liver Fibrosis by Killing Activated Stellate Cells in NKG2D-Dependent and Tumor Necrosis Factor
GASTROENTEROLOGY 2006;130:435 452 BASIC LIVER, PANCREAS, AND BILIARY TRACT Natural Killer Cells Ameliorate Liver Fibrosis by Killing Activated Stellate Cells in NKG2D-Dependent and Tumor Necrosis Factor
