Single-Molecule Analysis of Gene Expression Using Two-Color RNA- Labeling in Live Yeast
|
|
|
- Jason Watkins
- 5 years ago
- Views:
Transcription
1 Supplemental Figures, Tables and Results Single-Molecule Analysis of Gene Expression Using Two-Color RNA- Labeling in Live Yeast Sami Hocine 1, Pascal Raymond 2, Daniel Zenklusen 2, Jeffrey A. Chao 1 & Robert H. Singer 1 1 Department of Anatomy and Structural Biology, Albert Einstein College of Medicine, Bronx, NY 2 Department of Biochemistry, Université de Montréal, Montreal, Canada Correspondance should be addressed to R.H.S. (robert.singer@einstein.yu.edu).
2 a 24 PP7 KAN R loxp loxp MDN1 UTR MDN1 24 PP7 KAN R loxp loxp UTR MDN1 24 PP7 loxp UTR (tagged mrna) b MATa MDN1-24PP7 MET25 promoter PCP 2yEGFP HIS3 c MAT a (labeled mrna) MAT α Supplementary Figure 1. Schematic for tagging of yeast genes (a) Targeted integration of the 24PP7 cassette including the kanamycin selectable marker is performed by transformation and homologous recombination. The marker is removed by inducing expression of the CRE recombinase yielding mrnas that are precisely tagged in the 3 UTR. (b) PP7 tagged strains are then transformed with a plasmid encoding PCP-2yEGFP and mrnas become fluorescently labeled, allowing visualization of single molecules. (c) Two-color strains were generated by mating one-color haploids, generating yeast that express green or red MDN1 mrnas depending upon which allele they were transcribed from.
3 bps a 24MS2-GAL7 24PP7-GAL b Frequency of Loss of Repeats (%) 24MS2-GAL 24PP7-GAL 24MS2 bps c 24PP7 bps d Supplementary Figure 2. Assessing PP7 and MS2 hairpin copy number by PCR (a) GAL gene tagged in 5 UTR by 24MS2 and 24PP7. Hairpin copy number is determined by PCR product size (24x = 2039bps) for colonies resulting from independent integration events (24x = blue arrowhead; 1-23x = yellow arrowhead, 0x = red arrowhead). (b) 5 Taggings of GAL1, GAL7 and GAL10 indicates the occurrence of hairpin loss as 53 ± 7% for MS2 vs. 27 ± 7% PP7 (n = 15 colonies). (c,d) Single colonies confirmed to have the full complement of MS2 or PP7 hairpins were repeatedly diluted and grown in log-phase for approximately 80 generations. Cultured cells were plated and colonies were again screened for hairpin copy number. Both MS2 and PP7 hairpins remain stable (retain 24 stem loops) in culture following integration. Error bars indicate s.e.m.
4 a + TAG TAG MDN1-24PP7 b WT MDN1 + PCP-FP c MDN1-24MS2 d WT MDN1 + MCP-FP Supplementary Figure 3. Fluorescent spots require RNA tags (a-d) Maximum intensity projections highlight single MDN1 mrnas within the cell volume. MDN1 mrnas are labeled by co-expression of CP-FP driven off the MET25 promoter. Fluorescent spots require the mrna tag and do not result from CP-FP aggregation alone. Scale bars, 3 µm.
5 a z-sweep Acquisition Exposure (ms) b z-sweep c z-sections MDN1-24PP7 Stage Position (um) EMCCD Read-out Intervals Acquisition time: < 700ms 4000+ms Supplementary Figure 4. Visualization of single MDN1 mrnas (a) Detection of diffusing mrnas depends on rapid acquisition, such that the fluorescent signal is not spatially distributed during acquisition. This is achieved by a continuous z-sweep in which the stage continually moves along the z-axis (green lines). The sample receives constant 488 nm laser illumination (1.9 mw measured from back focal plane) and the EMCCD camera maintains a read-out interval of 0.75 µm (red arrows) over a total range of 4.5 µm. (b) Maximum intensity projection of MDN1-24PP7 mrnas acquired as z-sweep. Optimized conditions: 40% 488nm laser power; 90 ms exposure every 0.75 µm. (c) Maximum intensity projection of MDN1-24PP7 mrnas acquired as z-stack. Optimized conditions: 60% 488nm laser power; 50 ms exposure every 0.25 µm. Acquisition by z-sweep is ~6 times faster than standard z-sections, providing a snapshot of single mrnas within the cell volume. Scale bars, 3 µm.
6 ] a WT MDN1 MDN1-24MS2 MDN1-24PP7 b MDN1-24MS2 c g h ]FISHLive-cell MDN1-24PP7 Average MDN1 mrnas per cell + CP-FP CP-FP d e f i STEADY-STATE mrna FISH ( CP-FP) FISH (+ CP-FP) j NASCENT mrna Live-cell Average MDN1 nascent chains per cell WT MDN1 MDN1-24MS2 MDN1-24PP7 WT MDN1 MDN1-24MS2 MDN1-24PP7 MDN1-24MS2 MDN1-24PP7 WT MDN1 MDN1-24MS2 MDN1-24PP7 Supplementary Figure 5. MS2 and PP7 3 UTR tagging does not disrupt MDN1 expression Single-molecule FISH performed on haploid yeast using Cy3 labeled MDN1 probes (red) with nuclei labeled by DAPI (blue). Cells lacking CP-GFP expression are grown in YPD rich media while those expressing CP-GFP are grown in the appropriate synthetic minimal media. (a-c) WT MDN1 mrnas average 5.94 ± 0.16 per cell (n = 269 cells) whereas MDN1-24MS2 mrnas average 5.59 ± 0.17 per cell (n = 305 cells) and MDN1-24PP7 mrnas average 5.41 ± 0.15 per cell (n = 319 cells). (d-f) When expressing CP-GFP, WT MDN1 mrnas average 5.71 ± 0.19 per cell (n = 248 cells) whereas MDN1-24MS2 mrnas average 5.25 ± 0.16 per cell (n = 259 cells) and MDN1-24PP7 mrnas average 5.56 ± 0.16 per cell (n = 292 cells). (g,h) Live cell mrna counting yields an average of 5.59 ± 0.46 per cell (n = 85 cells) for MDN1-24MS2 and 5.54 ± 0.42 per cell (n = 92 cells) for MDN1-24PP7. (i) Quantification of mean MDN1 steady-state expression levels for all conditions. (j) WT MDN1 shows an average of 1.82 ± 0.16 nascent mrnas per cell (n = 61 cells), as compared to 1.85 ± 0.15 nascent mrnas for MDN1-24MS2 (n = 59 cells) and 1.78 ± 0.15 nascent mrnas for MDN1-24PP7 (n = 65 cells). Scale bars, 3 µm. Error bars indicate s.e.m.
7 MDN1-24PP7 + PCP-FP + MCP-FP a b MDN1-24MS2 c d Supplementary Figure 6. PP7 and MS2 are orthogonal (a-d) PCP does not bind to MS2 hairpins and MCP does not bind to PP7 hairpins. MDN1-24PP7 mrnas are only visualized by co-expressing the appropriate fluorescent coat protein, PCP-2yEGFP, and MDN1-24MS2 mrnas are only visualized by co-expressing the appropriate fluorescent coat protein, MCP-2yEGFP. Scale bars, 3 µm.
8 S G2 Pre-Mitotis Late-Mitotis G1 a b c d e 0 min 20 min 40 min 60 min 80 min Mitotic Progression f 0 min g 5 min h 10 min i 15 min j 20 min Supplementary Figure 7. Assessment of cell cycle stage MDN1-24PP7 mrnas are labeled with PCP-2yEGFP and nuclei are visualized by H2A2-yECFP. Cell cycle stage is determined by nuclear signal and cell morphology. (a-e) Cells can be imaged every 20 minutes in order to assess MDN1-24PP7 expression as the cell progresses through the cell cycle. (f-j) Cells can also be imaged at shorter timescales (5 min) in order to assess MDN1-24PP7 expression at different stages in the mitotic cycle. Scale bars, 3 µm.
9 MDN1-24PP7 + PCP-2yEGFP MDN1-24MS2 + MCP-mCherry H2A2-yECFP Merge a b c d e f g 0 min 20 min 40 min h i j 60 min 80 min 100 min Supplementary Figure 8. Two-color system paired with a nuclear marker (a) MDN1-24PP7 mrnas labeled with PCP-2yEGFP. (b) MDN1-24MS2 mrnas labeled with MCP-mCherry. (c) yecfp is fused to endogenous histone H2A2 such that cell nuclei can be easily visualized by low-power excitation with 436 nm light. (d) Merge. (e-j) Cells are imaged every 20 minutes and MDN1 expression can be quantified for each allele within the context of cell cycle stage. Scale bars, 3 µm.
10 a b transcript abundance transcript abundance time (min) time (min) Supplementary Figure 9. Cells exhibit different expression profiles over time Transcript counts for two cells (MDN1-24PP7 in green and MDN1-24MS2 in red) over 80 minutes. (a) Cell in which both alleles fluctuate around the mean. (b) Cell in which one allele (MDN1-24PP7 in green) exhibits persistently high expression while the other allele (MDN1-24MS2 in red) exhibits persistently low expression.
11 a MDN1 24 PP7 b c PP7-Cy3 24 MS2 d MS2-Cy5 e Merge DIC Supplementary Figure 10. Intramolecular tagging of MDN1 (a) Schematic showing MDN1 being 5 -tagged with 24PP7 and 3 -tagged with 24MS2. (b-e) PP7 probes in red colocalize with MS2 probes in green, demonstrating that both tags are present within the same molecules. DAPI signal (blue) highlights the nucleus and DIC images show cell morphology.
12 Supplementary Table 1. Sequences for mrna tagging PP7V3 TAAGGTACCTAATTGCCTAGAAAGGAGCAGACGATATGGCGTCGCTCCCTGCAGGTCGACTCTAGAAACCAG CAGAGCATATGGGCTCGCTGGCTGCAGTATTCCCGGGTTCATT MS2ORF TACGGTACTTATTGCCAAGAAAGCACGAGCATCAGCCGTGCCTCCAGGTCGAATCTTCA AACGACGACGATCACGCGTCGCTCCAGTA TTCCAGGGTTCATC *bold regions indicate hairpin sequences
13 Supplementary Table 2. PCR primer pairs Forward Primer Reverse Primer a TTCACTGATTTTGCGTCAATACTTTACAGACCTGGCA TCCAGCTAAGCCGCTCTAGAACTAGTGGATCC CCTTTGATTCGTGTAGTAAACCTCCTCTTCTTGGTTTTCA CGATATAGCATAGGCCACTAGTGGATCTG b CCTCGGTGAGTTTTCTCCTTCA ATATATTTAATTAACCTATAGAACTGAATGGGAAACT c GCGTCAATACTTTACAGACCTGGC ATATAT TTAATTAACCTATAGAACTGAATGGGAAACT d ACTCCACTTCAAGTAAGAGTTTG AGTCACATCAAGATCGTTTATGG e GCACGGAATATGGGACTACTTCG AGTCACATCAAGATCGTTTATGG
14 Supplementary Table 3. FISH probes MDN1 794 TTT GTC GTG GAT AGT GTG GAC CTT AGG GAC GAT AAC GCC ACA GAT TGA CG MDN1 860 CTC CCG AGT TGA CGA AGA GAG GAA ACC GTT TTA TGA GTA GGG ACA AAG GTT MDN CTA TAA GTA CCC ATC TCC CTT CTT TGA CCG CGG TAG CGA GAA CAC CAG CTC MDN TTT GCA GCC TTT ACA GTC TCT CCT CTG GAT GGA ATG GTT AGT TCG CGC TT PP7 GTA CCT TAG GAT CTA ATG AAC CCG GGA ATA CTG CAG CCA GCG AGC CCA TA MS2 TCT TGG CAA TAA GTA CCG TAG GAT CTG ATG AAC CCT GGA ATA CTG GAG CG *bold nucleotides indicate dye conjugation sites (Cy3, Cy3.5 or Cy5)
15 Supplemental Results Strain preparation for visualization of MDN1 mrnas Strains were prepared by site-specific genomic integration of 24PP7 or 24MS2 into the 3 UTR of one MDN1 allele (Supplementary Fig. 1a). Each stem loop binds two viral coat proteins, so RNA molecules become labeled following transformation with a plasmid that encodes fluorescent coat protein (Supplementary Fig. 1b). Two-color strains were obtained by mating one-color haploids, each expressing a different CP-FP (Supplementary Fig. 1c). MS2 and PP7 hairpin sequences were also optimized to minimize loss of hairpins during propagation and cloning in bacteria, a phenomenon observed with the use of prior MS2 versions. In addition to the hairpin array, the integration cassettes include a kanamycin selectable marker flanked by LoxP sites. The cassette was integrated into the 3 UTR of MDN1 immediately downstream of the stop codon, leaving the 3 UTR fully intact. Tagged strains were selected for by kanamycin resistance and integration sites were confirmed by PCR. Interestingly, fluorescent mrnas were not visible in all kanamycin-resistant colonies. Further PCR analysis revealed that during genomic integration by homologous recombination, varying numbers of hairpins could be lost (Supplementary Fig. 2a). Because hairpin copy number is directly proportional to the fluorescent signal of single mrnas, it was necessary to use only those strains that retained the full complement of hairpins for imaging experiments. MS2 hairpins were found to be twice as likely to undergo loss of hairpins compared to PP7 hairpins during homologous recombination in yeast (Supplementary Fig. 2b), however both were found to be stable once integrated (Supplementary Fig. 2c,d). In vivo imaging of single MDN1 mrnas We visualized fluorescent MDN1 mrnas for both the MDN1-24PP7 and MDN1-MS2 haploid strains by epifluorescence. Fluorescent spots that could be observed in yeast cells were shown to require the RNA tags, and did not result from CP-FP aggregation (Supplementary Fig. 3a-d).
16 Achieving the appropriate signal-to-noise levels in vivo depends on several factors. Yeast cells exhibit auto-fluorescence in the green and red channels and we reduced auto-fluorescence by culturing cells at very low log-phase growth for hours. Rapid diffusion of single mrnas in the x, y and z directions is also a concern, as it results in blurred fluorescence signal due to photons being spatially distributed on the EMCCD chip during exposure. Diffusion of individual mrnas in and out of z focal planes during image acquisition can also lead to oversampling of the total number of cellular mrnas. Therefore, a balance of high-intensity laser excitation and rapid acquisition through the cell volume is critical for accurately counting MDN1 steady-state levels. Cells were illuminated as the stage moved continuously in the z-axis, with the EMCCD camera reading out at 90ms intervals (Supplementary Fig. 4a). We have achieved ~6-fold faster acquisition using this z-sweeps acquisition as compared to traditional z-sections, minimizing blurring by providing a snap-shot of diffusing mrnas (Supplementary Fig. 4b,c). Tagging does not disrupt MDN1 expression Theoretically, PP7 and MS2 systems can be used to tag yeast genes at any position, however the addition of each cassette (approximately 1.3 kb) and the binding of fluorescent coat proteins to a transcript has the potential to alter a gene s expression level. Since MDN1 is an essential gene, we created MDN1-24PP7 and MDN1-24MS2 haploid strains by first tagging diploid yeast and then dissecting and screening tetrads. We observed that sporulation always resulted in four tetrads indicating that tagging did not render cells inviable. Haploid strains containing either tag also exhibited wild type doubling times in liquid media, indicating that tagging did not confer any growth defect. There have been mixed reports regarding how exactly the use of these tags may affect gene expression 1,2. In order to more closely examine the effects of tagging on MDN1 expression levels and whether such effects might differ based on which tag was used, we performed single-molecule FISH to
17 quantify MDN1 expression for WT and tagged strains (Supplementary Fig. 5a-c). WT MDN1 was expressed on average at 5.94 ± 0.16 mrnas per cell compared to 5.59 ± 0.17 mrnas per cell for MDN1-24MS2 and 5.41 ± 0.15 mrnas per cell for MDN1-24PP7. We performed the same experiments on WT and tagged strains expressing either PCP-2yEGFP or MCP-2yEGFP in order to determine if labeling of the mrna by the appropriate CP-FP affected transcript numbers. (Supplementary Fig. 5d-f). Under these conditions, FISH experiments revealed that WT MDN1 was expressed on average at 5.71 ± 0.19 mrnas per cell compared to 5.25 ± 0.16 mrnas per cell for MDN1-24MS2 + MCP-2yEGFP and 5.56 ± 0.16 mrnas per cell for MDN1-24PP7 + PCP-2yEGFP. We used the same analysis to quantify MDN1 expression in living cells (Supplementary Fig. 5g,h) and counted an average of 5.59 ± 0.46 mrnas per cell for MDN1-24MS2 + MCP-2yEGFP and 5.54 ± 0.42 mrnas per cell for MDN1-24PP7 + PCP-2yEGFP.. Taken together, it appears that tagging of the 3 UTR of MDN1 did not perturb steady-state expression levels and that live cell experiments had detection sensitivities similar to those of FISH (Supplementary Fig. 5i). Furthermore, we see by FISH that MDN1 remains transcriptionally active in nearly all cells regardless of whether it is tagged or not. More specifically, WT MDN1 shows an average of 1.82 ± 0.16 nascent mrnas per cell, as compared to 1.85 ± 0.15 nascent mrnas for MDN1-24MS2 and 1.78 ± 0.15 nascent mrnas for MDN1-24PP7 (Supplementary Fig. 5j), suggesting that transcriptional activity is also unaffected. These experiments provided a foundation for two-color quantitative analysis of gene expression with physiological relevance. MS2 and PP7 labeling systems are orthogonal The prospect of a two-color imaging utilizing both PP7 and MS2 tags requires that the individual imaging systems be orthogonal. The RNA-binding surfaces of PCP and MCP have evolved to recognize distinct RNA hairpins and have been shown in vitro to discriminately bind their own hairpin with ~1000- fold difference in affinity 3-5. In order to confirm this specificity in living yeast cells, we expressed MCP-
18 2yEGFP in MDN1-24PP7 haploids and PCP-2yECFP in MDN1-24MS2 haploids (Supplementary Fig 6a-d). We did not observe fluorescently-labeled mrnas in these strains, indicating that each CP maintains high specificity for its cognate hairpin sequence at concentrations required for live cell imaging. Determining transcript counts within the context of cell cycle As mentioned, we were interested in applying this two-color system to correlate the mrna expression for two identical MDN1 alleles. However, partitioning of mrnas during cell division can complicate the evaluation of MDN1 allelic correlation, as the division and partitioning of transcripts will automatically translate to a decrease in steady-state numbers for both alleles within the parental cell. We therefore fused yecfp to endogenous histone H2A2 in order to visualize nuclei and used cell and nuclear morphology to determine cell cycle stage (Supplementary Fig. 7a-e). Imaging at a faster acquisition rate, we can observe different time points during progression through mitosis (Supplementary Fig. 7f-j). We applied this to generate three-color strains (Supplementary Fig. 8a-d) in which we quantified expression of each allele at 20 minute time points up until mitosis (Supplementary Fig. 8e-j, Supplementary Movie 1-4). Single-cell resolution provides a window into different expression behaviors This system also provides a window into the different behaviors that cells exhibit in terms of allelic coordination over time. While single-molecule FISH has demonstrated cell-to-cell variability in expression levels, it was not previously possible to determine whether this variation resulted from subpopulations of cells that vary in expression levels that persist or whether expression levels fluctuate rapidly and equally in all cells over time. While most cells within a population exhibit MDN1 expression that fluctuates about the mean through time (Supplementary Fig. 9a), we also observe cells that exhibit persistently high or low expression as compared to the mean for the population. In fact, certain cells show one allele exhibiting persistently high expression and the other allele exhibiting persistently low
19 expression (Supplementary Fig. 9b). These expression regimes are not likely to result from changes in transcription factor binding, and may instead point to inherent differences in chromatin states at these two loci. Dual-tagged MDN1 is transcribed into mrnas that contain both tags within the same molecule Before determining elongation rates using dual-tagged MDN1, it was necessary to first confirm that both tags were indeed present within the same molecule. We prepared this strain by integrating 24PP7 into the 5 UTR of a strain in which MDN1 was already tagged in the 3 UTR with 24MS2 (Supplementary Fig. 10a). Because it was a diploid, it was necessary to verify that this 5 tag was integrated into the appropriate allele. We used single-molecule FISH to confirm dual-tagging of MDN1, showing that PP7 and MS2 probes colocalize to single transcripts in this intramolecular strain (Supplementary Fig. 10b-e). We induced transcription in confirmed strains by adding galactose, and acquired at 2 minute intervals to visualize 3 and 5 signals (Supplementary Movie 5-8).
20 1. A. B. Muthukrishnan, M. Kandhavelu, J. Lloyd-Price et al., Nucleic Acids Res 40 (17), 8472 (2012). 2. J. R. Chubb, T. Trcek, S. M. Shenoy et al., Curr Biol 16 (10), 1018 (2006). 3. J. Carey, V. Cameron, P. L. de Haseth et al., Biochemistry 22 (11), 2601 (1983). 4. F. Lim, T. P. Downey, and D. S. Peabody, J Biol Chem 276 (25), (2001). 5. J. A. Chao, Y. Patskovsky, S. C. Almo et al., Nat Struct Mol Biol 15 (1), 103 (2008).
c Tuj1(-) apoptotic live 1 DIV 2 DIV 1 DIV 2 DIV Tuj1(+) Tuj1/GFP/DAPI Tuj1 DAPI GFP
 Supplementary Figure 1 Establishment of the gain- and loss-of-function experiments and cell survival assays. a Relative expression of mature mir-484 30 20 10 0 **** **** NCP mir- 484P NCP mir- 484P b Relative
Supplementary Figure 1 Establishment of the gain- and loss-of-function experiments and cell survival assays. a Relative expression of mature mir-484 30 20 10 0 **** **** NCP mir- 484P NCP mir- 484P b Relative
a) Primary cultures derived from the pancreas of an 11-week-old Pdx1-Cre; K-MADM-p53
 1 2 3 4 5 6 7 8 9 10 Supplementary Figure 1. Induction of p53 LOH by MADM. a) Primary cultures derived from the pancreas of an 11-week-old Pdx1-Cre; K-MADM-p53 mouse revealed increased p53 KO/KO (green,
1 2 3 4 5 6 7 8 9 10 Supplementary Figure 1. Induction of p53 LOH by MADM. a) Primary cultures derived from the pancreas of an 11-week-old Pdx1-Cre; K-MADM-p53 mouse revealed increased p53 KO/KO (green,
Supplementary Table 3. 3 UTR primer sequences. Primer sequences used to amplify and clone the 3 UTR of each indicated gene are listed.
 Supplemental Figure 1. DLKI-DIO3 mirna/mrna complementarity. Complementarity between the indicated DLK1-DIO3 cluster mirnas and the UTR of SOX2, SOX9, HIF1A, ZEB1, ZEB2, STAT3 and CDH1with mirsvr and PhastCons
Supplemental Figure 1. DLKI-DIO3 mirna/mrna complementarity. Complementarity between the indicated DLK1-DIO3 cluster mirnas and the UTR of SOX2, SOX9, HIF1A, ZEB1, ZEB2, STAT3 and CDH1with mirsvr and PhastCons
Supplemental Data. Shin et al. Plant Cell. (2012) /tpc YFP N
 MYC YFP N PIF5 YFP C N-TIC TIC Supplemental Data. Shin et al. Plant Cell. ()..5/tpc..95 Supplemental Figure. TIC interacts with MYC in the nucleus. Bimolecular fluorescence complementation assay using
MYC YFP N PIF5 YFP C N-TIC TIC Supplemental Data. Shin et al. Plant Cell. ()..5/tpc..95 Supplemental Figure. TIC interacts with MYC in the nucleus. Bimolecular fluorescence complementation assay using
Supplementary Document
 Supplementary Document 1. Supplementary Table legends 2. Supplementary Figure legends 3. Supplementary Tables 4. Supplementary Figures 5. Supplementary References 1. Supplementary Table legends Suppl.
Supplementary Document 1. Supplementary Table legends 2. Supplementary Figure legends 3. Supplementary Tables 4. Supplementary Figures 5. Supplementary References 1. Supplementary Table legends Suppl.
Supplementary Figure 1. ROS induces rapid Sod1 nuclear localization in a dosagedependent manner. WT yeast cells (SZy1051) were treated with 4NQO at
 Supplementary Figure 1. ROS induces rapid Sod1 nuclear localization in a dosagedependent manner. WT yeast cells (SZy1051) were treated with 4NQO at different concentrations for 30 min and analyzed for
Supplementary Figure 1. ROS induces rapid Sod1 nuclear localization in a dosagedependent manner. WT yeast cells (SZy1051) were treated with 4NQO at different concentrations for 30 min and analyzed for
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 U1 inhibition causes a shift of RNA-seq reads from exons to introns. (a) Evidence for the high purity of 4-shU-labeled RNAs used for RNA-seq. HeLa cells transfected with control
Supplementary Figure 1 U1 inhibition causes a shift of RNA-seq reads from exons to introns. (a) Evidence for the high purity of 4-shU-labeled RNAs used for RNA-seq. HeLa cells transfected with control
Supplementary Figure 1 a
 Supplementary Figure a Normalized expression/tbp (A.U.).6... Trip-br transcripts Trans Trans Trans b..5. Trip-br Ctrl LPS Normalized expression/tbp (A.U.) c Trip-br transcripts. adipocytes.... Trans Trans
Supplementary Figure a Normalized expression/tbp (A.U.).6... Trip-br transcripts Trans Trans Trans b..5. Trip-br Ctrl LPS Normalized expression/tbp (A.U.) c Trip-br transcripts. adipocytes.... Trans Trans
Abbreviations: P- paraffin-embedded section; C, cryosection; Bio-SA, biotin-streptavidin-conjugated fluorescein amplification.
 Supplementary Table 1. Sequence of primers for real time PCR. Gene Forward primer Reverse primer S25 5 -GTG GTC CAC ACT ACT CTC TGA GTT TC-3 5 - GAC TTT CCG GCA TCC TTC TTC-3 Mafa cds 5 -CTT CAG CAA GGA
Supplementary Table 1. Sequence of primers for real time PCR. Gene Forward primer Reverse primer S25 5 -GTG GTC CAC ACT ACT CTC TGA GTT TC-3 5 - GAC TTT CCG GCA TCC TTC TTC-3 Mafa cds 5 -CTT CAG CAA GGA
Figure S1. Analysis of genomic and cdna sequences of the targeted regions in WT-KI and
 Figure S1. Analysis of genomic and sequences of the targeted regions in and indicated mutant KI cells, with WT and corresponding mutant sequences underlined. (A) cells; (B) K21E-KI cells; (C) D33A-KI cells;
Figure S1. Analysis of genomic and sequences of the targeted regions in and indicated mutant KI cells, with WT and corresponding mutant sequences underlined. (A) cells; (B) K21E-KI cells; (C) D33A-KI cells;
Supplementary Figure 1 MicroRNA expression in human synovial fibroblasts from different locations. MicroRNA, which were identified by RNAseq as most
 Supplementary Figure 1 MicroRNA expression in human synovial fibroblasts from different locations. MicroRNA, which were identified by RNAseq as most differentially expressed between human synovial fibroblasts
Supplementary Figure 1 MicroRNA expression in human synovial fibroblasts from different locations. MicroRNA, which were identified by RNAseq as most differentially expressed between human synovial fibroblasts
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Sherman SI, Wirth LJ, Droz J-P, et al. Motesanib diphosphate
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Sherman SI, Wirth LJ, Droz J-P, et al. Motesanib diphosphate
CD31 5'-AGA GAC GGT CTT GTC GCA GT-3' 5 ' -TAC TGG GCT TCG AGA GCA GT-3'
 Table S1. The primer sets used for real-time RT-PCR analysis. Gene Forward Reverse VEGF PDGFB TGF-β MCP-1 5'-GTT GCA GCA TGA ATC TGA GG-3' 5'-GGA GAC TCT TCG AGG AGC ACT T-3' 5'-GAA TCA GGC ATC GAG AGA
Table S1. The primer sets used for real-time RT-PCR analysis. Gene Forward Reverse VEGF PDGFB TGF-β MCP-1 5'-GTT GCA GCA TGA ATC TGA GG-3' 5'-GGA GAC TCT TCG AGG AGC ACT T-3' 5'-GAA TCA GGC ATC GAG AGA
Supplementary Materials
 Supplementary Materials 1 Supplementary Table 1. List of primers used for quantitative PCR analysis. Gene name Gene symbol Accession IDs Sequence range Product Primer sequences size (bp) β-actin Actb gi
Supplementary Materials 1 Supplementary Table 1. List of primers used for quantitative PCR analysis. Gene name Gene symbol Accession IDs Sequence range Product Primer sequences size (bp) β-actin Actb gi
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. H3F3B expression in lung cancer. a. Comparison of H3F3B expression in relapsed and non-relapsed lung cancer patients. b. Prognosis of two groups of lung cancer
Supplementary Figures Supplementary Figure 1. H3F3B expression in lung cancer. a. Comparison of H3F3B expression in relapsed and non-relapsed lung cancer patients. b. Prognosis of two groups of lung cancer
Culture Density (OD600) 0.1. Culture Density (OD600) Culture Density (OD600) Culture Density (OD600) Culture Density (OD600)
 A. B. C. D. E. PA JSRI JSRI 2 PA DSAM DSAM 2 DSAM 3 PA LNAP LNAP 2 LNAP 3 PAO Fcor Fcor 2 Fcor 3 PAO Wtho Wtho 2 Wtho 3 Wtho 4 DTSB Low Iron 2 4 6 8 2 4 6 8 2 22 DTSB Low Iron 2 4 6 8 2 4 6 8 2 22 DTSB
A. B. C. D. E. PA JSRI JSRI 2 PA DSAM DSAM 2 DSAM 3 PA LNAP LNAP 2 LNAP 3 PAO Fcor Fcor 2 Fcor 3 PAO Wtho Wtho 2 Wtho 3 Wtho 4 DTSB Low Iron 2 4 6 8 2 4 6 8 2 22 DTSB Low Iron 2 4 6 8 2 4 6 8 2 22 DTSB
Table S1. Oligonucleotides used for the in-house RT-PCR assays targeting the M, H7 or N9. Assay (s) Target Name Sequence (5 3 ) Comments
 SUPPLEMENTAL INFORMATION 2 3 Table S. Oligonucleotides used for the in-house RT-PCR assays targeting the M, H7 or N9 genes. Assay (s) Target Name Sequence (5 3 ) Comments CDC M InfA Forward (NS), CDC M
SUPPLEMENTAL INFORMATION 2 3 Table S. Oligonucleotides used for the in-house RT-PCR assays targeting the M, H7 or N9 genes. Assay (s) Target Name Sequence (5 3 ) Comments CDC M InfA Forward (NS), CDC M
BIOLOGY 621 Identification of the Snorks
 Name: Date: Block: BIOLOGY 621 Identification of the Snorks INTRODUCTION: In this simulation activity, you will examine the DNA sequence of a fictitious organism - the Snork. Snorks were discovered on
Name: Date: Block: BIOLOGY 621 Identification of the Snorks INTRODUCTION: In this simulation activity, you will examine the DNA sequence of a fictitious organism - the Snork. Snorks were discovered on
Toluidin-Staining of mast cells Ear tissue was fixed with Carnoy (60% ethanol, 30% chloroform, 10% acetic acid) overnight at 4 C, afterwards
 Toluidin-Staining of mast cells Ear tissue was fixed with Carnoy (60% ethanol, 30% chloroform, 10% acetic acid) overnight at 4 C, afterwards incubated in 100 % ethanol overnight at 4 C and embedded in
Toluidin-Staining of mast cells Ear tissue was fixed with Carnoy (60% ethanol, 30% chloroform, 10% acetic acid) overnight at 4 C, afterwards incubated in 100 % ethanol overnight at 4 C and embedded in
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature05883 SUPPLEMENTARY INFORMATION Supplemental Figure 1 Prostaglandin agonists and antagonists alter runx1/cmyb expression. a-e, Embryos were exposed to (b) PGE2 and (c) PGI2 (20μM) and
doi: 10.1038/nature05883 SUPPLEMENTARY INFORMATION Supplemental Figure 1 Prostaglandin agonists and antagonists alter runx1/cmyb expression. a-e, Embryos were exposed to (b) PGE2 and (c) PGI2 (20μM) and
Citation for published version (APA): Oosterveer, M. H. (2009). Control of metabolic flux by nutrient sensors Groningen: s.n.
 University of Groningen Control of metabolic flux by nutrient sensors Oosterveer, Maaike IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from it.
University of Groningen Control of metabolic flux by nutrient sensors Oosterveer, Maaike IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from it.
A basic helix loop helix transcription factor controls cell growth
 A basic helix loop helix transcription factor controls cell growth and size in root hairs Keke Yi 1,2, Benoît Menand 1,3, Elizabeth Bell 1, Liam Dolan 1,4 Supplementary note Low soil phosphate availability
A basic helix loop helix transcription factor controls cell growth and size in root hairs Keke Yi 1,2, Benoît Menand 1,3, Elizabeth Bell 1, Liam Dolan 1,4 Supplementary note Low soil phosphate availability
Supplementary Table 2. Conserved regulatory elements in the promoters of CD36.
 Supplementary Table 1. RT-qPCR primers for CD3, PPARg and CEBP. Assay Forward Primer Reverse Primer 1A CAT TTG TGG CCT TGT GCT CTT TGA TGA GTC ACA GAA AGA ATC AAT TC 1B AGG AAA TGA ACT GAT GAG TCA CAG
Supplementary Table 1. RT-qPCR primers for CD3, PPARg and CEBP. Assay Forward Primer Reverse Primer 1A CAT TTG TGG CCT TGT GCT CTT TGA TGA GTC ACA GAA AGA ATC AAT TC 1B AGG AAA TGA ACT GAT GAG TCA CAG
Supplementary Materials and Methods
 DD2 suppresses tumorigenicity of ovarian cancer cells by limiting cancer stem cell population Chunhua Han et al. Supplementary Materials and Methods Analysis of publicly available datasets: To analyze
DD2 suppresses tumorigenicity of ovarian cancer cells by limiting cancer stem cell population Chunhua Han et al. Supplementary Materials and Methods Analysis of publicly available datasets: To analyze
Phylogenetic analysis of human and chicken importins. Only five of six importins were studied because
 Supplementary Figure S1 Phylogenetic analysis of human and chicken importins. Only five of six importins were studied because importin-α6 was shown to be testis-specific. Human and chicken importin protein
Supplementary Figure S1 Phylogenetic analysis of human and chicken importins. Only five of six importins were studied because importin-α6 was shown to be testis-specific. Human and chicken importin protein
A smart acid nanosystem for ultrasensitive. live cell mrna imaging by the target-triggered intracellular self-assembly
 Electronic Supplementary Material (ESI) for Chemical Science. This journal is The Royal Society of Chemistry 2017 A smart ZnO@polydopamine-nucleic acid nanosystem for ultrasensitive live cell mrna imaging
Electronic Supplementary Material (ESI) for Chemical Science. This journal is The Royal Society of Chemistry 2017 A smart ZnO@polydopamine-nucleic acid nanosystem for ultrasensitive live cell mrna imaging
Beta Thalassemia Case Study Introduction to Bioinformatics
 Beta Thalassemia Case Study Sami Khuri Department of Computer Science San José State University San José, California, USA sami.khuri@sjsu.edu www.cs.sjsu.edu/faculty/khuri Outline v Hemoglobin v Alpha
Beta Thalassemia Case Study Sami Khuri Department of Computer Science San José State University San José, California, USA sami.khuri@sjsu.edu www.cs.sjsu.edu/faculty/khuri Outline v Hemoglobin v Alpha
Nature Immunology: doi: /ni.3836
 Supplementary Figure 1 Recombinant LIGHT-VTP induces pericyte contractility and endothelial cell activation. (a) Western blot showing purification steps for full length murine LIGHT-VTP (CGKRK) protein:
Supplementary Figure 1 Recombinant LIGHT-VTP induces pericyte contractility and endothelial cell activation. (a) Western blot showing purification steps for full length murine LIGHT-VTP (CGKRK) protein:
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1. Lats1/2 deleted ihbs and ihps showed decreased transcripts of hepatocyte related genes (a and b) Western blots (a) and recombination PCR (b) of control and
Supplementary Figure 1 Supplementary Figure 1. Lats1/2 deleted ihbs and ihps showed decreased transcripts of hepatocyte related genes (a and b) Western blots (a) and recombination PCR (b) of control and
 www.lessonplansinc.com Topic: Protein Synthesis - Sentence Activity Summary: Students will simulate transcription and translation by building a sentence/polypeptide from words/amino acids. Goals & Objectives:
www.lessonplansinc.com Topic: Protein Synthesis - Sentence Activity Summary: Students will simulate transcription and translation by building a sentence/polypeptide from words/amino acids. Goals & Objectives:
Beta Thalassemia Sami Khuri Department of Computer Science San José State University Spring 2015
 Bioinformatics in Medical Product Development SMPD 287 Three Beta Thalassemia Sami Khuri Department of Computer Science San José State University Hemoglobin Outline Anatomy of a gene Hemoglobinopathies
Bioinformatics in Medical Product Development SMPD 287 Three Beta Thalassemia Sami Khuri Department of Computer Science San José State University Hemoglobin Outline Anatomy of a gene Hemoglobinopathies
Supplemental Information. Cancer-Associated Fibroblasts Neutralize. the Anti-tumor Effect of CSF1 Receptor Blockade
 Cancer Cell, Volume 32 Supplemental Information Cancer-Associated Fibroblasts Neutralize the Anti-tumor Effect of CSF1 Receptor Blockade by Inducing PMN-MDSC Infiltration of Tumors Vinit Kumar, Laxminarasimha
Cancer Cell, Volume 32 Supplemental Information Cancer-Associated Fibroblasts Neutralize the Anti-tumor Effect of CSF1 Receptor Blockade by Inducing PMN-MDSC Infiltration of Tumors Vinit Kumar, Laxminarasimha
SUPPLEMENTARY DATA. Supplementary Table 1. Primer sequences for qrt-pcr
 Supplementary Table 1. Primer sequences for qrt-pcr Gene PRDM16 UCP1 PGC1α Dio2 Elovl3 Cidea Cox8b PPARγ AP2 mttfam CyCs Nampt NRF1 16s-rRNA Hexokinase 2, intron 9 β-actin Primer Sequences 5'-CCA CCA GCG
Supplementary Table 1. Primer sequences for qrt-pcr Gene PRDM16 UCP1 PGC1α Dio2 Elovl3 Cidea Cox8b PPARγ AP2 mttfam CyCs Nampt NRF1 16s-rRNA Hexokinase 2, intron 9 β-actin Primer Sequences 5'-CCA CCA GCG
Supplementary Figure 1a
 Supplementary Figure 1a Hours: E-cadherin TGF-β On TGF-β Off 0 12 24 36 48 24 48 72 Vimentin βactin Fig. S1a. Treatment of AML12 cells with TGF-β induces EMT. Treatment of AML12 cells with TGF-β results
Supplementary Figure 1a Hours: E-cadherin TGF-β On TGF-β Off 0 12 24 36 48 24 48 72 Vimentin βactin Fig. S1a. Treatment of AML12 cells with TGF-β induces EMT. Treatment of AML12 cells with TGF-β results
Astaxanthin prevents and reverses diet-induced insulin resistance and. steatohepatitis in mice: A comparison with vitamin E
 Supplementary Information Astaxanthin prevents and reverses diet-induced insulin resistance and steatohepatitis in mice: A comparison with vitamin E Yinhua Ni, 1,2 Mayumi Nagashimada, 1 Fen Zhuge, 1 Lili
Supplementary Information Astaxanthin prevents and reverses diet-induced insulin resistance and steatohepatitis in mice: A comparison with vitamin E Yinhua Ni, 1,2 Mayumi Nagashimada, 1 Fen Zhuge, 1 Lili
Supplementary Figure 1
 Supplementary Figure 1 3 3 3 1 1 Bregma -1.6mm 3 : Bregma Ref) Http://www.mbl.org/atlas165/atlas165_start.html Bregma -.18mm Supplementary Figure 1 Schematic representation of the utilized brain slice
Supplementary Figure 1 3 3 3 1 1 Bregma -1.6mm 3 : Bregma Ref) Http://www.mbl.org/atlas165/atlas165_start.html Bregma -.18mm Supplementary Figure 1 Schematic representation of the utilized brain slice
Supplemental Information. Th17 Lymphocytes Induce Neuronal. Cell Death in a Human ipsc-based. Model of Parkinson's Disease
 Cell Stem Cell, Volume 23 Supplemental Information Th17 Lymphocytes Induce Neuronal Cell Death in a Human ipsc-based Model of Parkinson's Disease Annika Sommer, Franz Maxreiter, Florian Krach, Tanja Fadler,
Cell Stem Cell, Volume 23 Supplemental Information Th17 Lymphocytes Induce Neuronal Cell Death in a Human ipsc-based Model of Parkinson's Disease Annika Sommer, Franz Maxreiter, Florian Krach, Tanja Fadler,
SUPPORTING INFORMATION
 SUPPORTING INFORMATION Biology is different in small volumes: endogenous signals shape phenotype of primary hepatocytes cultured in microfluidic channels Amranul Haque, Pantea Gheibi, Yandong Gao, Elena
SUPPORTING INFORMATION Biology is different in small volumes: endogenous signals shape phenotype of primary hepatocytes cultured in microfluidic channels Amranul Haque, Pantea Gheibi, Yandong Gao, Elena
TetR repressor-based bioreporters for the detection of doxycycline using Escherichia
 Supplementary materials TetR repressor-based bioreporters for the detection of doxycycline using Escherichia coli and Acinetobacter oleivorans Hyerim Hong and Woojun Park * Department of Environmental
Supplementary materials TetR repressor-based bioreporters for the detection of doxycycline using Escherichia coli and Acinetobacter oleivorans Hyerim Hong and Woojun Park * Department of Environmental
Nucleotide Sequence of the Australian Bluetongue Virus Serotype 1 RNA Segment 10
 J. gen. Virol. (1988), 69, 945-949. Printed in Great Britain 945 Key words: BTV/genome segment lo/nucleotide sequence Nucleotide Sequence of the Australian Bluetongue Virus Serotype 1 RNA Segment 10 By
J. gen. Virol. (1988), 69, 945-949. Printed in Great Britain 945 Key words: BTV/genome segment lo/nucleotide sequence Nucleotide Sequence of the Australian Bluetongue Virus Serotype 1 RNA Segment 10 By
Supplementary Information. Bamboo shoot fiber prevents obesity in mice by. modulating the gut microbiota
 Supplementary Information Bamboo shoot fiber prevents obesity in mice by modulating the gut microbiota Xiufen Li 1,2, Juan Guo 1, Kailong Ji 1,2, and Ping Zhang 1,* 1 Key Laboratory of Tropical Plant Resources
Supplementary Information Bamboo shoot fiber prevents obesity in mice by modulating the gut microbiota Xiufen Li 1,2, Juan Guo 1, Kailong Ji 1,2, and Ping Zhang 1,* 1 Key Laboratory of Tropical Plant Resources
Lezione 10. Sommario. Bioinformatica. Lezione 10: Sintesi proteica Synthesis of proteins Central dogma: DNA makes RNA makes proteins Genetic code
 Lezione 10 Bioinformatica Mauro Ceccanti e Alberto Paoluzzi Lezione 10: Sintesi proteica Synthesis of proteins Dip. Informatica e Automazione Università Roma Tre Dip. Medicina Clinica Università La Sapienza
Lezione 10 Bioinformatica Mauro Ceccanti e Alberto Paoluzzi Lezione 10: Sintesi proteica Synthesis of proteins Dip. Informatica e Automazione Università Roma Tre Dip. Medicina Clinica Università La Sapienza
Supporting Information
 Supporting Information Malapeira et al. 10.1073/pnas.1217022110 SI Materials and Methods Plant Material and Growth Conditions. A. thaliana seedlings were stratified at 4 C in the dark for 3 d on Murashige
Supporting Information Malapeira et al. 10.1073/pnas.1217022110 SI Materials and Methods Plant Material and Growth Conditions. A. thaliana seedlings were stratified at 4 C in the dark for 3 d on Murashige
Journal of Cell Science Supplementary information. Arl8b +/- Arl8b -/- Inset B. electron density. genotype
 J. Cell Sci. : doi:.4/jcs.59: Supplementary information E9. A Arl8b /- Arl8b -/- Arl8b Arl8b non-specific band Gapdh Tbp E7.5 HE Inset B D Control al am hf C E Arl8b -/- al am hf E8.5 F low middle high
J. Cell Sci. : doi:.4/jcs.59: Supplementary information E9. A Arl8b /- Arl8b -/- Arl8b Arl8b non-specific band Gapdh Tbp E7.5 HE Inset B D Control al am hf C E Arl8b -/- al am hf E8.5 F low middle high
Integration Solutions
 Integration Solutions (1) a) With no active glycosyltransferase of either type, an ii individual would not be able to add any sugars to the O form of the lipopolysaccharide. Thus, the only lipopolysaccharide
Integration Solutions (1) a) With no active glycosyltransferase of either type, an ii individual would not be able to add any sugars to the O form of the lipopolysaccharide. Thus, the only lipopolysaccharide
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1: Cryopreservation alters CD62L expression by CD4 T cells. Freshly isolated (left) or cryopreserved PBMCs (right) were stained with the mix of antibodies described
Supplementary Figure 1 Supplementary Figure 1: Cryopreservation alters CD62L expression by CD4 T cells. Freshly isolated (left) or cryopreserved PBMCs (right) were stained with the mix of antibodies described
Supplementary Information
 Supplementary Information Remodeling of heterochromatin structure slows neuropathological progression and prolongs survival in an animal model of Huntington s disease Junghee Lee, Yu Jin Hwang, Yunha Kim,
Supplementary Information Remodeling of heterochromatin structure slows neuropathological progression and prolongs survival in an animal model of Huntington s disease Junghee Lee, Yu Jin Hwang, Yunha Kim,
Bacterial Gene Finding CMSC 423
 Bacterial Gene Finding CMSC 423 Finding Signals in DNA We just have a long string of A, C, G, Ts. How can we find the signals encoded in it? Suppose you encountered a language you didn t know. How would
Bacterial Gene Finding CMSC 423 Finding Signals in DNA We just have a long string of A, C, G, Ts. How can we find the signals encoded in it? Suppose you encountered a language you didn t know. How would
Supplementary Figure 1
 Metastatic melanoma Primary melanoma Healthy human skin Supplementary Figure 1 CD22 IgG4 Supplementary Figure 1: Immunohisochemical analysis of CD22+ (left) and IgG4 (right), cells (shown in red and indicated
Metastatic melanoma Primary melanoma Healthy human skin Supplementary Figure 1 CD22 IgG4 Supplementary Figure 1: Immunohisochemical analysis of CD22+ (left) and IgG4 (right), cells (shown in red and indicated
Description of Supplementary Files. File Name: Supplementary Information Description: Supplementary Figures and Supplementary Tables
 Description of Supplementary Files File Name: Supplementary Information Description: Supplementary Figures and Supplementary Tables Supplementary Figure 1: (A), HCT116 IDH1-WT and IDH1-R132H cells were
Description of Supplementary Files File Name: Supplementary Information Description: Supplementary Figures and Supplementary Tables Supplementary Figure 1: (A), HCT116 IDH1-WT and IDH1-R132H cells were
Rapid blue-light mediated induction of protein interactions in living cells
 Nature Methods Rapid blue-light mediated induction of protein interactions in living cells Matthew J Kennedy, Robert M Hughes, Leslie A Peteya, Joel W Schwartz, Michael D Ehlers & Chandra L Tucker Supplementary
Nature Methods Rapid blue-light mediated induction of protein interactions in living cells Matthew J Kennedy, Robert M Hughes, Leslie A Peteya, Joel W Schwartz, Michael D Ehlers & Chandra L Tucker Supplementary
Cross-talk between mineralocorticoid and angiotensin II signaling for cardiac
 ONLINE SUPPLEMENT TO Crosstalk between mineralocorticoid and angiotensin II signaling for cardiac remodeling An Di ZHANG,,3, Aurelie NGUYEN DINH CAT*,,3, Christelle SOUKASEUM *,,3, Brigitte ESCOUBET, 4,
ONLINE SUPPLEMENT TO Crosstalk between mineralocorticoid and angiotensin II signaling for cardiac remodeling An Di ZHANG,,3, Aurelie NGUYEN DINH CAT*,,3, Christelle SOUKASEUM *,,3, Brigitte ESCOUBET, 4,
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/10/473/eaai7696/dc1 Supplementary Materials for Astrocyte-shed extracellular vesicles regulate the peripheral leukocyte response to inflammatory brain lesions
www.sciencesignaling.org/cgi/content/full/10/473/eaai7696/dc1 Supplementary Materials for Astrocyte-shed extracellular vesicles regulate the peripheral leukocyte response to inflammatory brain lesions
Isolate Sexual Idiomorph Species
 SUPLEMENTARY TABLE 1. Isolate identification, sexual idiomorph and species of each isolate used for MAT locus distribution in Paracoccidioides species. Isolate Sexual Idiomorph Species Pb01 MAT1-1 P. lutzii
SUPLEMENTARY TABLE 1. Isolate identification, sexual idiomorph and species of each isolate used for MAT locus distribution in Paracoccidioides species. Isolate Sexual Idiomorph Species Pb01 MAT1-1 P. lutzii
Plasmids Western blot analysis and immunostaining Flow Cytometry Cell surface biotinylation RNA isolation and cdna synthesis
 Plasmids psuper-retro-s100a10 shrna1 was constructed by cloning the dsdna oligo 5 -GAT CCC CGT GGG CTT CCA GAG CTT CTT TCA AGA GAA GAA GCT CTG GAA GCC CAC TTT TTA-3 and 5 -AGC TTA AAA AGT GGG CTT CCA GAG
Plasmids psuper-retro-s100a10 shrna1 was constructed by cloning the dsdna oligo 5 -GAT CCC CGT GGG CTT CCA GAG CTT CTT TCA AGA GAA GAA GCT CTG GAA GCC CAC TTT TTA-3 and 5 -AGC TTA AAA AGT GGG CTT CCA GAG
Loyer, et al. microrna-21 contributes to NASH Suppl 1/15
 Loyer, et al. microrna-21 contributes to NASH Suppl 1/15 SUPPLEMENTARY MATERIAL: Liver MicroRNA-21 is Overexpressed in Non Alcoholic Steatohepatitis and Contributes to the Disease in Experimental Models
Loyer, et al. microrna-21 contributes to NASH Suppl 1/15 SUPPLEMENTARY MATERIAL: Liver MicroRNA-21 is Overexpressed in Non Alcoholic Steatohepatitis and Contributes to the Disease in Experimental Models
BHP 2-7 and Nthy-ori 3-1 cells were grown in RPMI1640 medium (Hyclone) supplemented with 10% fetal bovine serum (Gibco), 2mM L-glutamine, and 100 U/mL
 1 2 3 4 Materials and Methods Cell culture BHP 2-7 and Nthy-ori 3-1 cells were grown in RPMI1640 medium (Hyclone) 5 supplemented with 10% fetal bovine serum (Gibco), 2mM L-glutamine, and 100 U/mL 6 penicillin-streptomycin.
1 2 3 4 Materials and Methods Cell culture BHP 2-7 and Nthy-ori 3-1 cells were grown in RPMI1640 medium (Hyclone) 5 supplemented with 10% fetal bovine serum (Gibco), 2mM L-glutamine, and 100 U/mL 6 penicillin-streptomycin.
SUPPLEMENTARY INFORMATION
 BASELINE ISCHAEMIA a b Phd2 +/- c d Collateral growth and maintenance SMC recruitment SMC proliferation Phd2 +/- NF- B off NF- B on NF- B on NF- B on Endothelial cell Smooth muscle cell Pro-arteriogenic
BASELINE ISCHAEMIA a b Phd2 +/- c d Collateral growth and maintenance SMC recruitment SMC proliferation Phd2 +/- NF- B off NF- B on NF- B on NF- B on Endothelial cell Smooth muscle cell Pro-arteriogenic
*To whom correspondence should be addressed. This PDF file includes:
 www.sciencemag.org/cgi/content/full/science.1212182/dc1 Supporting Online Material for Partial Retraction to Detection of an Infectious Retrovirus, XMRV, in Blood Cells of Patients with Chronic Fatigue
www.sciencemag.org/cgi/content/full/science.1212182/dc1 Supporting Online Material for Partial Retraction to Detection of an Infectious Retrovirus, XMRV, in Blood Cells of Patients with Chronic Fatigue
CIRCRESAHA/2004/098145/R1 - ONLINE 1. Validation by Semi-quantitative Real-Time Reverse Transcription PCR
 CIRCRESAHA/2004/098145/R1 - ONLINE 1 Expanded Materials and Methods Validation by Semi-quantitative Real-Time Reverse Transcription PCR Expression patterns of 13 genes (Online Table 2), selected with respect
CIRCRESAHA/2004/098145/R1 - ONLINE 1 Expanded Materials and Methods Validation by Semi-quantitative Real-Time Reverse Transcription PCR Expression patterns of 13 genes (Online Table 2), selected with respect
Oligo Sequence* bp %GC Tm Hair Hm Ht Position Size Ref. HIVrt-F 5 -CTA-gAA-CTT-TRA-ATg-CAT-ggg-TAA-AAg-TA
 Human immunodeficiency virus (HIV) detection & quantitation by qrt-pcr (Taqman). Created on: Oct 26, 2010; Last modified by: Jul 17, 2017; Version: 3.0 This protocol describes the qrt-pcr taqman based
Human immunodeficiency virus (HIV) detection & quantitation by qrt-pcr (Taqman). Created on: Oct 26, 2010; Last modified by: Jul 17, 2017; Version: 3.0 This protocol describes the qrt-pcr taqman based
without LOI phenotype by breeding female wild-type C57BL/6J and male H19 +/.
 Sakatani et al. 1 Supporting Online Material Materials and methods Mice and genotyping: H19 mutant mice with C57BL/6J background carrying a deletion in the structural H19 gene (3 kb) and 10 kb of 5 flanking
Sakatani et al. 1 Supporting Online Material Materials and methods Mice and genotyping: H19 mutant mice with C57BL/6J background carrying a deletion in the structural H19 gene (3 kb) and 10 kb of 5 flanking
Supporting Information. Mutational analysis of a phenazine biosynthetic gene cluster in
 Supporting Information for Mutational analysis of a phenazine biosynthetic gene cluster in Streptomyces anulatus 9663 Orwah Saleh 1, Katrin Flinspach 1, Lucia Westrich 1, Andreas Kulik 2, Bertolt Gust
Supporting Information for Mutational analysis of a phenazine biosynthetic gene cluster in Streptomyces anulatus 9663 Orwah Saleh 1, Katrin Flinspach 1, Lucia Westrich 1, Andreas Kulik 2, Bertolt Gust
Detection of 549 new HLA alleles in potential stem cell donors from the United States, Poland and Germany
 HLA ISSN 2059-2302 BRIEF COMMUNICATION Detection of 549 new HLA alleles in potential stem cell donors from the United States, Poland and Germany C. J. Hernández-Frederick 1, N. Cereb 2,A.S.Giani 1, J.
HLA ISSN 2059-2302 BRIEF COMMUNICATION Detection of 549 new HLA alleles in potential stem cell donors from the United States, Poland and Germany C. J. Hernández-Frederick 1, N. Cereb 2,A.S.Giani 1, J.
Supplementary information
 Supplementary information Unique polypharmacology nuclear receptor modulator blocks inflammatory signaling pathways Mi Ra Chang 1, Anthony Ciesla 1, Timothy S. Strutzenberg 1, Scott J. Novick 1, Yuanjun
Supplementary information Unique polypharmacology nuclear receptor modulator blocks inflammatory signaling pathways Mi Ra Chang 1, Anthony Ciesla 1, Timothy S. Strutzenberg 1, Scott J. Novick 1, Yuanjun
Cancer Genetics 204 (2011) 45e52
 Cancer Genetics 204 (2011) 45e52 Exon scanning by reverse transcriptaseepolymerase chain reaction for detection of known and novel EML4eALK fusion variants in nonesmall cell lung cancer Heather R. Sanders
Cancer Genetics 204 (2011) 45e52 Exon scanning by reverse transcriptaseepolymerase chain reaction for detection of known and novel EML4eALK fusion variants in nonesmall cell lung cancer Heather R. Sanders
SUPPLEMENTAL METHODS Cell culture RNA extraction and analysis Immunohistochemical analysis and laser capture microdissection (LCM)
 SUPPLEMENTAL METHODS Cell culture Human peripheral blood mononuclear cells were isolated from healthy donors by Ficoll density gradient centrifugation. Monocyte differentiation to resting macrophages ()
SUPPLEMENTAL METHODS Cell culture Human peripheral blood mononuclear cells were isolated from healthy donors by Ficoll density gradient centrifugation. Monocyte differentiation to resting macrophages ()
Mechanistic and functional insights into fatty acid activation in Mycobacterium tuberculosis SUPPLEMENTARY INFORMATION
 Mechanistic and functional insights into fatty acid activation in Mycobacterium tuberculosis Pooja Arora 1, Aneesh Goyal 2, Vivek T atarajan 1, Eerappa Rajakumara 2, Priyanka Verma 1, Radhika Gupta 3,
Mechanistic and functional insights into fatty acid activation in Mycobacterium tuberculosis Pooja Arora 1, Aneesh Goyal 2, Vivek T atarajan 1, Eerappa Rajakumara 2, Priyanka Verma 1, Radhika Gupta 3,
SUPPLEMENTARY RESULTS
 SUPPLEMENTARY RESULTS Supplementary Table 1. hfpr1- Flpln-CHO hfpr2-flpln-cho pec 50 E max (%) Log( /K A) Log( /K A) N pec 50 E max (%) Log( /K A) Log( /K A) n ERK1/2 phosphorylation fmlp 9.0±0.6 80±7
SUPPLEMENTARY RESULTS Supplementary Table 1. hfpr1- Flpln-CHO hfpr2-flpln-cho pec 50 E max (%) Log( /K A) Log( /K A) N pec 50 E max (%) Log( /K A) Log( /K A) n ERK1/2 phosphorylation fmlp 9.0±0.6 80±7
Resistance to Tetracycline Antibiotics by Wangrong Yang, Ian F. Moore, Kalinka P. Koteva, Donald W. Hughes, David C. Bareich and Gerard D. Wright.
 Supplementary Data for TetX is a Flavin-Dependent Monooxygenase Conferring Resistance to Tetracycline Antibiotics by Wangrong Yang, Ian F. Moore, Kalinka P. Koteva, Donald W. Hughes, David C. Bareich and
Supplementary Data for TetX is a Flavin-Dependent Monooxygenase Conferring Resistance to Tetracycline Antibiotics by Wangrong Yang, Ian F. Moore, Kalinka P. Koteva, Donald W. Hughes, David C. Bareich and
Supplementary Data. Clinical Setup. References. Program for Embryo Donation
 Supplementary Data Clinical Setup Notwithstanding their value in stem cell research, the source of supernumerary human blastocysts for the derivation of new stem cell lines and the modalities of their
Supplementary Data Clinical Setup Notwithstanding their value in stem cell research, the source of supernumerary human blastocysts for the derivation of new stem cell lines and the modalities of their
Baseline clinical characteristics for the 81 CMML patients Routine diagnostic testing and statistical analyses... 3
 Next-Generation Sequencing Technology Reveals a Characteristic Pattern of Molecular Mutations in 72.8% of Chronic Myelomonocytic Leukemia (CMML) by Detecting Frequent Alterations in TET2, CBL, RAS, and
Next-Generation Sequencing Technology Reveals a Characteristic Pattern of Molecular Mutations in 72.8% of Chronic Myelomonocytic Leukemia (CMML) by Detecting Frequent Alterations in TET2, CBL, RAS, and
Relationship of the APOA5/A4/C3/A1 gene cluster and APOB gene polymorphisms with dyslipidemia
 elationship of the APOA5/A4/C3/A1 gene cluster and APOB gene polymorphisms with dyslipidemia H.J. Ou 1, G. Huang 2, W. Liu 3, X.L. Ma 2, Y. Wei 4, T. Zhou 5 and Z.M. Pan 3 1 Department of Neurology, The
elationship of the APOA5/A4/C3/A1 gene cluster and APOB gene polymorphisms with dyslipidemia H.J. Ou 1, G. Huang 2, W. Liu 3, X.L. Ma 2, Y. Wei 4, T. Zhou 5 and Z.M. Pan 3 1 Department of Neurology, The
Malignant Amelanotic Melanoma of the Pleura without Primary Skin Lesion: An Autopsy Case Report. a a*
 2009 63 6 379 384 Malignant Amelanotic Melanoma of the Pleura without Primary Skin Lesion: An Autopsy Case Report a b a a a* a b 380 63 6 Chest x ray and computed tomography (CT). A, Chest x ray on admission
2009 63 6 379 384 Malignant Amelanotic Melanoma of the Pleura without Primary Skin Lesion: An Autopsy Case Report a b a a a* a b 380 63 6 Chest x ray and computed tomography (CT). A, Chest x ray on admission
University of Groningen. Vasoregression in incipient diabetic retinopathy Pfister, Frederick
 University of Groningen Vasoregression in incipient diabetic retinopathy Pfister, Frederick IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from
University of Groningen Vasoregression in incipient diabetic retinopathy Pfister, Frederick IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from
Mutation analysis of a Chinese family with oculocutaneous albinism
 /, 2016, Vol. 7, (No. 51), pp: 84981-84988 Mutation analysis of a Chinese family with oculocutaneous albinism Xiong Wang 1, Yaowu Zhu 1, Na Shen 1, Jing Peng 1, Chunyu Wang 1, Haiyi Liu 2, Yanjun Lu 1
/, 2016, Vol. 7, (No. 51), pp: 84981-84988 Mutation analysis of a Chinese family with oculocutaneous albinism Xiong Wang 1, Yaowu Zhu 1, Na Shen 1, Jing Peng 1, Chunyu Wang 1, Haiyi Liu 2, Yanjun Lu 1
The Effect of Epstein-Barr Virus Latent Membrane Protein 2 Expression on the Kinetics of Early B Cell Infection
 The Effect of Epstein-Barr Virus Latent Membrane Protein 2 Expression on the Kinetics of Early B Cell Infection Laura R. Wasil, Monica J. Tomaszewski, Aki Hoji, David T. Rowe* Department of Infectious
The Effect of Epstein-Barr Virus Latent Membrane Protein 2 Expression on the Kinetics of Early B Cell Infection Laura R. Wasil, Monica J. Tomaszewski, Aki Hoji, David T. Rowe* Department of Infectious
Mutation Screening and Association Studies of the Human UCP 3 Gene in Normoglycemic and NIDDM Morbidly Obese Patients
 Mutation Screening and Association Studies of the Human UCP 3 Gene in Normoglycemic and NIDDM Morbidly Obese Patients Shuichi OTABE, Karine CLEMENT, Séverine DUBOIS, Frederic LEPRETRE, Veronique PELLOUX,
Mutation Screening and Association Studies of the Human UCP 3 Gene in Normoglycemic and NIDDM Morbidly Obese Patients Shuichi OTABE, Karine CLEMENT, Séverine DUBOIS, Frederic LEPRETRE, Veronique PELLOUX,
Formylpeptide receptor2 contributes to colon epithelial homeostasis, inflammation, and tumorigenesis
 Supplementary Data Formylpeptide receptor2 contributes to colon epithelial homeostasis, inflammation, and tumorigenesis Keqiang Chen, Mingyong Liu, Ying Liu, Teizo Yoshimura, Wei Shen, Yingying Le, Scott
Supplementary Data Formylpeptide receptor2 contributes to colon epithelial homeostasis, inflammation, and tumorigenesis Keqiang Chen, Mingyong Liu, Ying Liu, Teizo Yoshimura, Wei Shen, Yingying Le, Scott
SUPPLEMENTAL FIGURE 1
 SUPPLEMENTL FIGURE 1 C Supplemental Figure 1. pproach for removal of snorns from Rpl13a gene. () Wild type Rpl13a exonintron structure is shown, with exo in black and intronic snorns in red rectangles.
SUPPLEMENTL FIGURE 1 C Supplemental Figure 1. pproach for removal of snorns from Rpl13a gene. () Wild type Rpl13a exonintron structure is shown, with exo in black and intronic snorns in red rectangles.
Advanced Subsidiary Unit 1: Lifestyle, Transport, Genes and Health
 Write your name here Surname Other names Edexcel GCE Centre Number Candidate Number Biology Advanced Subsidiary Unit 1: Lifestyle, Transport, Genes and Health Thursday 8 January 2009 Morning Time: 1 hour
Write your name here Surname Other names Edexcel GCE Centre Number Candidate Number Biology Advanced Subsidiary Unit 1: Lifestyle, Transport, Genes and Health Thursday 8 January 2009 Morning Time: 1 hour
ice-cold 70% ethanol with gentle vortexing, incubated at -20 C for 4 hours, and washed with PBS.
 Cell cycle analysis For cell cycle analysis, single cell suspensions of E12.5 fetal liver cells were suspended in 4 ml ice-cold 7% ethanol with gentle vortexing, incubated at -2 C for 4 hours, and washed
Cell cycle analysis For cell cycle analysis, single cell suspensions of E12.5 fetal liver cells were suspended in 4 ml ice-cold 7% ethanol with gentle vortexing, incubated at -2 C for 4 hours, and washed
Supplementary Figure S1
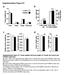 Supplementry Figure S Tissue weights (g).... Liver Hert Brin Pncres Len mss (g) 8 6 -% +% 8 6 Len mss Len mss (g) (% ody weight) Len mss (% ody weight) c Tiilis nterior weight (g).6...... Qudriceps weight
Supplementry Figure S Tissue weights (g).... Liver Hert Brin Pncres Len mss (g) 8 6 -% +% 8 6 Len mss Len mss (g) (% ody weight) Len mss (% ody weight) c Tiilis nterior weight (g).6...... Qudriceps weight
Nucleotide diversity of the TNF gene region in an African village
 (2001) 2, 343 348 2001 Nature Publishing Group All rights reserved 1466-4879/01 $15.00 www.nature.com/gene Nucleotide diversity of the TNF gene region in an African village A Richardson 1, F Sisay-Joof
(2001) 2, 343 348 2001 Nature Publishing Group All rights reserved 1466-4879/01 $15.00 www.nature.com/gene Nucleotide diversity of the TNF gene region in an African village A Richardson 1, F Sisay-Joof
Analysis of Human Cytomegalovirus orilyt Sequence Requirements in the Context of the Viral Genome
 JOURNAL OF VIROLOGY, Mar. 2005, p. 3615 3626 Vol. 79, No. 6 0022-538X/05/$08.00 0 doi:10.1128/jvi.79.6.3615 3626.2005 Copyright 2005, American Society for Microbiology. All Rights Reserved. Analysis of
JOURNAL OF VIROLOGY, Mar. 2005, p. 3615 3626 Vol. 79, No. 6 0022-538X/05/$08.00 0 doi:10.1128/jvi.79.6.3615 3626.2005 Copyright 2005, American Society for Microbiology. All Rights Reserved. Analysis of
A Rapid and Sensitive Chip-based Assay for Detection of rpob Gene Mutations Conferring Rifampicin Resistance in Mycobacterium tuberculosis (TB).
 A Rapid and Sensitive Chip-based Assay for Detection of rpob Gene Mutations Conferring Rifampicin Resistance in Mycobacterium tuberculosis (TB). Wanyuan Ao, Steve Aldous, Evelyn Woodruff, Brian Hicke,
A Rapid and Sensitive Chip-based Assay for Detection of rpob Gene Mutations Conferring Rifampicin Resistance in Mycobacterium tuberculosis (TB). Wanyuan Ao, Steve Aldous, Evelyn Woodruff, Brian Hicke,
Characterizing intra-host influenza virus populations to predict emergence
 Characterizing intra-host influenza virus populations to predict emergence June 12, 2012 Forum on Microbial Threats Washington, DC Elodie Ghedin Center for Vaccine Research Dept. Computational & Systems
Characterizing intra-host influenza virus populations to predict emergence June 12, 2012 Forum on Microbial Threats Washington, DC Elodie Ghedin Center for Vaccine Research Dept. Computational & Systems
Patterns of hemagglutinin evolution and the epidemiology of influenza
 2 8 US Annual Mortality Rate All causes Infectious Disease Patterns of hemagglutinin evolution and the epidemiology of influenza DIMACS Working Group on Genetics and Evolution of Pathogens, 25 Nov 3 Deaths
2 8 US Annual Mortality Rate All causes Infectious Disease Patterns of hemagglutinin evolution and the epidemiology of influenza DIMACS Working Group on Genetics and Evolution of Pathogens, 25 Nov 3 Deaths
mir-1202: A Primate Specific and Brain Enriched mirna Involved in Major Depression and Antidepressant Treatment. Supplementary Information
 Title: mir-1202: A Primate Specific and Brain Enriched mirna Involved in Major Depression and Antidepressant Treatment. Authors: Juan Pablo Lopez 1, Raymond Lim 3, Cristiana Cruceanu 1, Liam Crapper 1,
Title: mir-1202: A Primate Specific and Brain Enriched mirna Involved in Major Depression and Antidepressant Treatment. Authors: Juan Pablo Lopez 1, Raymond Lim 3, Cristiana Cruceanu 1, Liam Crapper 1,
The Clinical Performance of Primary HPV Screening, Primary HPV Screening Plus Cytology Cotesting, and Cytology Alone at a Tertiary Care Hospital
 The Clinical Performance of Primary HPV Screening, Primary HPV Screening Plus Cytology Cotesting, and Cytology Alone at a Tertiary Care Hospital Jung-Woo Choi MD, PhD; Younghye Kim MD, PhD; Ju-Han Lee
The Clinical Performance of Primary HPV Screening, Primary HPV Screening Plus Cytology Cotesting, and Cytology Alone at a Tertiary Care Hospital Jung-Woo Choi MD, PhD; Younghye Kim MD, PhD; Ju-Han Lee
Enhanced detection and serotyping of Streptococcus pneumoniae using multiplex polymerase chain reaction
 Original article http://dx.doi.org/10.3345/kjp.2012.55.11.424 Korean J Pediatr 2012;55(11):424-429 eissn 1738-1061 pissn 2092-7258 Enhanced detection and serotyping of Streptococcus pneumoniae using multiplex
Original article http://dx.doi.org/10.3345/kjp.2012.55.11.424 Korean J Pediatr 2012;55(11):424-429 eissn 1738-1061 pissn 2092-7258 Enhanced detection and serotyping of Streptococcus pneumoniae using multiplex
Genome-wide identification of TCF7L2/TCF4 target mirnas reveals a role for mir-21 in Wnt-driven epithelial cancer
 INTERNATIONAL JOURNAL OF ONCOLOGY 40: 519-526, 2012 Genome-wide identification of TCF7L2/TCF4 target mirnas reveals a role for mir-21 in Wnt-driven epithelial cancer FENGMING LAN 1-3*, XIAO YUE 1-3,5*,
INTERNATIONAL JOURNAL OF ONCOLOGY 40: 519-526, 2012 Genome-wide identification of TCF7L2/TCF4 target mirnas reveals a role for mir-21 in Wnt-driven epithelial cancer FENGMING LAN 1-3*, XIAO YUE 1-3,5*,
Supplemental Figures: Supplemental Figure 1
 Supplemental Figures: Supplemental Figure 1 Suppl. Figure 1. BM-DC infection with H. pylori does not induce cytotoxicity and treatment of BM-DCs with H. pylori sonicate, but not heat-inactivated bacteria,
Supplemental Figures: Supplemental Figure 1 Suppl. Figure 1. BM-DC infection with H. pylori does not induce cytotoxicity and treatment of BM-DCs with H. pylori sonicate, but not heat-inactivated bacteria,
Isolation and Genetic Characterization of New Reassortant H3N1 Swine Influenza Virus from Pigs in the Midwestern United States
 JOURNAL OF VIROLOGY, May 2006, p. 5092 5096 Vol. 80, No. 10 0022-538X/06/$08.00 0 doi:10.1128/jvi.80.10.5092 5096.2006 Copyright 2006, American Society for Microbiology. All Rights Reserved. Isolation
JOURNAL OF VIROLOGY, May 2006, p. 5092 5096 Vol. 80, No. 10 0022-538X/06/$08.00 0 doi:10.1128/jvi.80.10.5092 5096.2006 Copyright 2006, American Society for Microbiology. All Rights Reserved. Isolation
HCV Persistence and Immune Evasion in the Absence of Memory T Cell Help.
 SOM Text HCV Persistence and Immune Evasion in the Absence of Memory T Cell Help. Arash Grakoui 1, Naglaa H. Shoukry 2, David J. Woollard 2, Jin-Hwan Han 1, Holly L. Hanson 1, John Ghrayeb 3, Krishna K.
SOM Text HCV Persistence and Immune Evasion in the Absence of Memory T Cell Help. Arash Grakoui 1, Naglaa H. Shoukry 2, David J. Woollard 2, Jin-Hwan Han 1, Holly L. Hanson 1, John Ghrayeb 3, Krishna K.
Development of RT-qPCR-based molecular diagnostic assays for therapeutic target selection of breast cancer patients
 Development of RT-qPCR-based molecular diagnostic assays for therapeutic target selection of breast cancer patients Sangjung Park The Graduate School Yonsei University Department of Biomedical Laboratory
Development of RT-qPCR-based molecular diagnostic assays for therapeutic target selection of breast cancer patients Sangjung Park The Graduate School Yonsei University Department of Biomedical Laboratory
McAlpine PERK-GSK3 regulates foam cell formation. Supplemental Material. Supplementary Table I. Sequences of real time PCR primers.
 Mclpine PERK-GSK3 regulates foam cell formation Supplemental Material Supplementary Table I. Sequences of real time PCR primers. Primer Name Primer Sequences (5-3 ) Product Size (bp) GRP78 (human) Fwd:
Mclpine PERK-GSK3 regulates foam cell formation Supplemental Material Supplementary Table I. Sequences of real time PCR primers. Primer Name Primer Sequences (5-3 ) Product Size (bp) GRP78 (human) Fwd:
SUPPLEMENTARY INFORMATION Glucosylceramide synthase (GlcT-1) in the fat body controls energy metabolism in Drosophila
 SUPPLEMENTARY INFORMATION Glucosylceramide synthase (GlcT-1) in the fat body controls energy metabolism in Drosophila Ayako Kohyama-Koganeya, 1,2 Takuji Nabetani, 1 Masayuki Miura, 2,3 Yoshio Hirabayashi
SUPPLEMENTARY INFORMATION Glucosylceramide synthase (GlcT-1) in the fat body controls energy metabolism in Drosophila Ayako Kohyama-Koganeya, 1,2 Takuji Nabetani, 1 Masayuki Miura, 2,3 Yoshio Hirabayashi
PATIENTS AND METHODS. Subjects
 PATIENTS AND METHODS Subjects Twenty-nine morbidly obese subjects involved in a gastric surgery program were enrolled in the study between October 25 and March 21. Bariatric surgery was performed in patients
PATIENTS AND METHODS Subjects Twenty-nine morbidly obese subjects involved in a gastric surgery program were enrolled in the study between October 25 and March 21. Bariatric surgery was performed in patients
Engineering a polarity-sensitive biosensor for time-lapse imaging of apoptotic processes and degeneration
 nature methods Engineering a polarity-sensitive biosensor for time-lapse imaging of apoptotic processes and degeneration Yujin E Kim, Jeannie Chen, Jonah R Chan & Ralf Langen Supplementary figures and
nature methods Engineering a polarity-sensitive biosensor for time-lapse imaging of apoptotic processes and degeneration Yujin E Kim, Jeannie Chen, Jonah R Chan & Ralf Langen Supplementary figures and
