Lung Met 1 Lung Met 2 Lung Met Lung Met H3K4me1. Lung Met H3K27ac Primary H3K4me1
|
|
|
- Ferdinand Shelton
- 5 years ago
- Views:
Transcription
1 a Gained Met-VELs Lung Met 1 Lung Met Lung Met 3 1. Lung Met H3K4me1 Lung Met H3K4me1 1 Lung Met H3K4me1 Lung Met H3K7ac 1.5 Lung Met H3K7ac Lung Met H3K7ac.8 Primary H3K4me1 Primary H3K7ac 1 Primary H3K4me1 Primary H3K7ac.6 Primary H3K4me1 Primary H3K7ac Lost Met-VELs Lung Met H3K4me1 Lung Met H3K7ac Primary H3K4me1 Primary H3K7ac Lung Met H3K4me1 Lung Met H3K7ac Primary H3K4me1 Primary H3K7ac Lung Met H3K4me1 Lung Met H3K7ac Primary H3K4me1 Primary H3K7ac Gained Met-VELs Lost Met-VELs Gained Met-VELs b 1.! 1!.8!.6!.4!.!! -.! -.4! Lung Met MNNG H3K4me1! MNNG H3K7ac! MNNG DNase! HOS H3K4me1! HOS H3K7ac! HOS DNase! 1.8! 1.6! 1.4! 1.! 1!.8!.6!.4!.!! -.! -.4! Lung Met H3K4me1 Lung Met H3K7ac Primary H3K4me1 Primary H3K7ac B H3K4me1! 143B H3K7ac! 143B DNase! HOS H3K4me1! HOS H3K7ac! HOS DNase! 1.6! 1.4! 1.! 1!.8!.6!.4!.!! -.! -.4! Lung Met H3K4me1 Lung Met H3K7ac Primary H3K4me1 Primary H3K7ac MNNG 143B LM7 LM7 H3K4me1! LM7 H3K7ac! LM7 DNase! SaOS H3K4me1! SaOS H3K7ac! SaOS DNase! Lost Met-VELs 1.! 1!.8!.6!.4!.!! -.! MNNG H3K4me1! MNNG H3K7ac! MNNG DNase! HOS H3K4me1! HOS H3K7ac! HOS DNase! 1.! 1!.8!.6!.4!.!! -.! 143B H3K4me1! 143B H3K7ac! 143B DNase! HOS H3K4me1! HOS H3K7ac! HOS DNase! 1.4! 1.! 1!.8!.6!.4!.!! -.! -.4! LM7 H3K4me1! LM7 H3K7ac! LM7 DNase! SaOS H3K4me1! SaOS H3K7ac! SaOS DNase! M Gained Met-VELs M11 H3K4me1 M11 H3K7ac M11 DNase Hu9 H3K4me1 Hu9 H3K7ac Hu9 DNase Lost Met-VELs M11 H3K4me1 M11 H3K7ac M11 DNase Hu9 H3K4me1 Hu9 H3K7ac Hu9 DNase Nature Medicine: doi:1.138/nm.4475
2 Supplementary Figure 1: Met-VEL profiles of osteosarcoma patient lung metastases and human osteosarcoma cell lines. a, Aggregate plots showing H3K4me1 ChIP-seq and H3K7ac ChIP-seq signal +/- 3Kb from midpoints of gained and lost Met-VELs in paired patient lung metastases and primary tumors. b, Aggregate plots showing H3K4me1 ChIP-seq, H3K7ac ChIP-seq and DNase-seq signal +/- 3Kb from mid-points of gained and lost Met-VELs in metastatic/parental human osteosarcoma cell lines. Nature Medicine: doi:1.138/nm.4475
3 a Gained Met-VEL Cluster Lost Met-VEL Cluster.5 1. H3K4me1 H3K4me1 GHR COL3A1 MG63 (Parental) MG63.3 (Metastatic) Gained Met-VEL 1kb MG63 (Parental) MG63.3 (Metastatic) Gained Met-VEL 1kb b MG63.3 Lost Met-VEL Clusters LHFP Scaled Coordinates WASF3 COL3A1 Chromosomes 16 Lost Met-VELs in kb window c Number of Clusters Lung Met 1 Lung Met Lung Met 3 Lung Met 4 Lung Met 5 MG63.3 LM7 M11 MNNG 143B Gained Met-VELs Lost Met-VELs d % Total Met-VELs 1 5 Gained Met-VELs outside clusters within clusters Lung Met 1 Lung Met Lung Met 3 Lung Met 4 Lung Met 5 MG63.3 LM7 M11 MNNG 143B % Total Met-VELs 1 5 Lung Met 1 Lung Met Lost Met-VELs outside clusters within clusters Lung Met 3 Lung Met 4 Lung Met 5 MG63.3 LM7 M11 MNNG 143B Nature Medicine: doi:1.138/nm.4475
4 Supplementary Figure : Met-VEL clusters occur across metastatic cancers. a. UCSC browser view of H3K4me1 profiles in MG63.3 (metastatic) and MG63 (parental) cell lines illustrating an example of a gained (left) and lost (right) Met-VEL cluster. Met-VELs identified by black bars. kb Met-VEL clusters highlighted in gray. b. Genome-wide lost Met-VEL landscape for MG63.3 cell line. Rows represent scaled chromosomal coordinates. Peaks represent maximum gained Met-VEL counts in kb sliding windows. Predicted target genes for selected peaks are labeled. c. Gained and lost Met-VEL cluster counts in patient lung metastases/primary tumors and metastatic/parental cell line pairs. d. Percentage of total gained (top) and lost (bottom) Met-VELs within and outside of clusters in patient lung metastases/primary tumors and metastatic/parental cell line pairs. Nature Medicine: doi:1.138/nm.4475
5 a Parental Metastatic b 1 MNNG Pair Gained Gained Met-VEL Met-VEL Genes Cluster Genes Lost Met-VEL Genes Lost Met-VEL Cluster Genes 4hrs 4hrs 14 days log(fpkm) *** *** *** *** ** *** * RNA-seq d Lost Met-VEL Motifs MG63.3 MNNG 143B FOSL1 JUNB JUND UBP1 FOS JUN JDP BACH1 MAF NFEL TFAPA NFIX ZIC1 CTCF TFAP4 ZBTB7B TFAPC EF6 EF1 ZIC SMAD4 TCF1 ATF4 NFIB MYC ZNF711 TFCP ATF MAFA MAFF log(fpkm) B Pair Gained Gained Met-VEL Met-VEL Genes Cluster Genes *** Lost Met-VEL Genes *** *** * Lost Met-VEL Cluster Genes Parental line 4hrs Metastatic line 4hrs Metastatic line day 14 *** * 3-1*log(p-value) G B Color Key e Lost Met-VEL Genes MG63.3 MNNG 143B embryonic morphogenesis extracellular matrix proteinaceous extracellular matrix transcription factor activity tube development ECM-receptor interaction cell motion -1*log p-value <3-7 <7-1 <1-13 <13-15 >15 enzyme linked receptor protein signaling pathway extracellular matrix part phosphoprotein nucleus embryonic skeletal system development acetylation 1 6 c MNNG 143B Early Constitutive Late Early Constitutive Late Normalized Expression -3 3 Met Day 1 Met Day 14 Parental Day 1 Met Day 1 Met Day 14 Parental Day 1 Nature Medicine: doi:1.138/nm.4475
6 Supplementary Figure 3: Assessment of Met-VEL associated gene expression during metastatic colonization of the lung. a. Schematic of experimental design for assessment of Met-VEL gene expression in parental and metastatic cell lines in ex vivo lung metastasis model. Image adapted from 9. b. Log quantile-normalized FPKM values for gained (left) and lost (right) Met-VEL and Met-VEL cluster genes in HOS/MNNG (top) and HOS/143B (bottom) cell line pairs. Asterisks indicate significant differences in FPKM distributions between parental and metastatic cell lines (* P<.5; ** P<1E-3; *** P<1E-4). P-values calculated by Mann-Whitney Test. c. Heatmap of up-regulated gained Met-VEL genes in HOS/MNNG (top) and HOS/143B (bottom) cell line pairs. d. Expressed transcription factors with enriched motifs in lost Met-VELs in three metastatic/parental cell line pairs and corresponding motif enrichment p-values. e. Gene Ontology (GO) terms for lost Met-VEL genes in three metastatic/parental cell line pairs and corresponding p-values. Nature Medicine: doi:1.138/nm.4475
7 a Gained Met-VEL Genes b Lost Met-VEL Genes MG63.3 MNNG 143B MG63.3 MNNG 143B 4 p=8.5e-4 4 p<.e-16 p<.e-16 p=.17 p=9.33e-5 p=4.1e-1 p=.76 p=.57 4 p=.15 p=1.74e-5 p=1.1e-4 p=1.1e-5 log(fold change expression) Met vs. Parental in vitro Met 4hrs lung vs. Parental 4hrs lung Met 14 days lung vs. Parental 4hrs lung Met vs. Parental in vitro Met 4hrs lung vs. Parental 4hrs lung Met 14 days lung vs. Parental 4hrs lung c log(fold-change expression met. vs. non-met) MG63.3 Pair MNNG Pair 143B Pair All Expressed Genes Gained Met-VEL Genes Lost Met-VEL Genes d Gained Met-VELs 5 5 Lost Met-VELs Number of Genes 15 1 Number of Genes MG63.3 MNNG 143B MG63.3 MNNG 143B Top up-regulated genes Overlap Gained Met-VEL Genes Top down-regulated genes Overlap Lost Met-VEL Genes Nature Medicine: doi:1.138/nm.4475
8 Supplementary Figure 4: Assessment of Met-VEL associated gene expression in vitro and in ex vivo lung culture. a. Log fold-change in quantile-normalized FPKM values of gained Met-VEL genes in metastatic cell lines versus parental cell lines in various conditions. P-values calculated by Mann-Whitney Test. b. Log fold-change in quantile-normalized FPKM values of lost Met-VEL genes in metastatic cell lines versus parental cell lines in various conditions. P-values calculated by Mann-Whitney Test. c. Violin plots of log(fold-change quantile-normalized FPKM values) distributions for gene sets in three metastatic cell lines relative to parental non-metastatic lines. Fold-change values for all expressed genes and gained Met-VEL gene sets represent comparisons of expression in metastatic lines at day 14 in ex vivo lung culture to non-metastatic lines at 4hrs. Fold-change values for lost Met-VEL gene set represent comparison of expression in metastatic lines at 4hrs in ex vivo lung culture to non-metastatic lines at 4hrs. d. Bar charts indicating overlap between gained (left) and lost (right) Met-VEL gene sets and top 1 up- and down-regulated genes in corresponding conditions, respectively. Nature Medicine: doi:1.138/nm.4475
9 Regulation of cell migration Angiogenesis and Endothelial cell migration Endothelial Developmental Programs Embyronic Epithelial tube Neg. regulation of cell migration Growth Urothelial Adherens junction Epithelial cell migration Actin cytoskeleton organization Epithelial cell proliferation Pathways in cancer Signaling Pathways Ossification Rap1 Regulation of cell shape WNT ERBB Hippo IL-6 response Node Color = FDR Protein dephosphorylation Protein localization Negative regulation to plasma membrane of protein phosphorylation.5.1 <.5 Nature Medicine: doi:1.138/nm.4475
10 Supplementary Figure 5: Assessment of enriched functions of gained Met-VEL genes in osteosarcoma patient metastases. Enriched Map representation of all Gene Ontology (GO) terms for gained Met-VEL genes calculated by aggregating gene lists from two patient metastases (Lung Met 4 and Lung Met 5). Nature Medicine: doi:1.138/nm.4475
11 a Normalized Metastatic Burden Vehicle 5nM JQ1 MNNG p=7.45e-8 b Mouse 1 Mouse Vehicle MNNG 5nM JQ1 c Day 15 Vehicle Time (days) 143B Vehicle 143B 5nM JQ1 Day 15 5nM JQ1 Normalized Metastatic Burden Vehicle 5nM JQ1 p=1.73e-5 Mouse 1 Mouse Time (days) d MNNG Activated Gained Met-VEL Genes. 143B Activated Gained Met-VEL Genes ES -.6 NES = FDR <.5 ES NES = -1.7 FDR < f 4 * * MNNG * 1 143B * * Vehicle Treated JQ1 Treated Vehicle Treated JQ1 Treated 3 5 e MNNG Activated Met-VEL Gene Expression Log(fold-change vs. parental line) p<.e Vehicle 5nM JQ1 Log(fold-change vs. parental line) 143B Activated Gained Met-VEL Genes 4 - p=5.78e-7 % Reduction in Expression 1-1 All Activated Genes Lost Met-VEL Genes Activated Gained Met-VEL Genes Activated Gained Met-VEL Genes no SEs SE Genes % Reduction in Expression All Activated Genes Lost Met-VEL Genes Activated Gained Met-VEL Genes Activated Gained Met-VEL Genes no SEs SE Genes Nature Medicine: doi:1.138/nm.4475
12 Supplementary Figure 6: Analysis of anti-metastatic and gene expression effects of BET inhibition with JQ1. a. Kinetics of metastatic outgrowth of MNNG (top) and 143B (bottom) metastatic cell lines in ex vivo lung culture with 5nM JQ1 or vehicle (DMSO) treatment. Metastatic burden measured as total GFP+ area per lung section normalized to GFP+ area on day. Values represent averages of 8 lung sections (4 sections per mouse x mice) +/-SEM. P-value calculated by Mann-Whitney Test. Dashed line indicates time points chosen for RNA-seq studies. b. Representative.5x images of vehicle (left) and 5nM JQ1 treated (right) lung sections at day 15. Lung sections outlined with dashed white line. Scale bar = 5µm. c. X image of hematoxylin and eosin stained section of lung slice after 15 days in ex vivo culture treated with DMSO (top) and 5nM JQ1 (bottom) illustrating viable lung cells and architecture. Scale bar = 1µm. d. GSEA plots of -fold up-regulated gained Met-VEL gene sets in vehicle versus JQ1-treated MNNG (left) and 143B (right) cells isolated from ex vivo lung culture. Cells isolated at time points indicated by dashed lines in Supplemental Figure -5a. e. Log fold-change expression -fold up-regulated gained Met-VEL gene sets in vehicle versus JQ1-treated MNNG (left) and 143B (right) cells isolated from ex vivo lung culture at time points indicated by dashed lines in Extended Data Figure 7a relative to parental cell line. f. Percent reduction in gene expression with 5nM JQ1 treatment of all genes up-regulated -fold in metastatic cell lines relative to parental cell lines at sorting time points, lost Met-VEL genes, gained Met-VEL genes -fold up-regulated relative to parental cell lines at sorting time points, upregulated gained Met-VEL genes without SE genes, and all SE genes in MNNG and 143B cells growing in ex vivo lung culture sorted at time points indicated in Extended Data Figure 7a. Nature Medicine: doi:1.138/nm.4475
13 Nature Medicine: doi:1.138/nm.4475
14 Supplementary Figure 7: Assessment of LT3REPIR shrna construct induction and leakiness. a. Schematic of doxycycline-inducible LT3REPIR shrna construct. Modified from (Fellmann et al., 13). b. Cytometric analysis of GFP and DsRed expression in GFP+ MG63.3 cells transduced with LT3REPIR construct at baseline (left) and 4hrs after exposure to 5ug/ml doxycycline (right). Nature Medicine: doi:1.138/nm.4475
15 Nature Medicine: doi:1.138/nm.4475
16 Supplementary Figure 8: Assessment of Tissue Factor (F3) dysregulation in metastatic osteosarcoma. a. IGV browser view of H3K7ac, H3K4me1, and DNase profiles at F3 gained Met-VEL cluster locus in MG63.3 (metastatic) and MG63 (parental) cell lines. Top of figure shows local contact profile analysis of F3 locus in MG63.3. In the top panel (main trend), the contact intensity (black line) is calculated by using a running mean analysis of normalized read counts with a 1kb sliding window. The th and 8th percentile are visualized as a gray trend graph. In the bottom panel, contact intensities are computed using linearly increasing sliding windows (scaled -5 kb) and are displayed as a color-coded heatmap of positive 4C signals (maximum of interaction set to 1). Local color changes are log-scaled to indicate changes of statistical enrichment of captured sequences, corresponding to the enhancer-promoter interaction. Areas of significant contact highlighted. b. Fold-change quantile normalized F3 FPKM values in 3 metastatic cell lines at 4hrs and 14 days of metastatic outgrowth in ex vivo lung culture relative to parental line at 4hrs. c. Tissue Factor (F3) relative expression in human patient primary tumors and lung metastases normalized to expression in normal osteoblasts. d. IGV browser view of H3K7ac, H3K4me1, and DNase profiles at F3 gained Met-VEL cluster locus in MG63.3, MNNG, and 143B metastatic cell lines. Nature Medicine: doi:1.138/nm.4475
17 Nature Medicine: doi:1.138/nm.4475
18 Supplementary Figure 9: F3 expression in human osteosarcoma tumors. Immunohistochemical staining of F3 in human osteosarcoma lung metastases and primary tumors. Scale bars = µm. Nature Medicine: doi:1.138/nm.4475
19 a. MG63.3 DHS chr1 p36.3 p36. p36.13 p36.11 p35.1 p34. p33 p3.3 p3.1 p31. p31.1 p.3 p.1 p1. p13.3 p13.1 p11.1 q1 q1.1 q1.3 q3.1 q3.3 q4. q5.1 q5.3 q31.1 q31.3 q3.1 q3. q41 q4.11 q4.mg63 q43h3k7ac q kb MG63.3 H3K7ac Patient Lung Mets F3 ESC TADs IMR9 TADs b. SEs lost in Mets SEs gained in Mets log1(wilcox p_val) F mean(met) mean(non_met) Nature Medicine: doi:1.138/nm.4475
20 Supplementary Figure 1: Assessment of Tissue Factor (F3) Met-VELs in patient lung metastases. a. IGV browser views of the F3 locus showing H3K7ac ChIP-seq tracks of lung metastases from 1 osteosarcoma patients. Blue bars below each track correspond to super enhancers defined using the ROSE script. Hi-C defined topologically associating domains (TADs) are displayed below. b. Volcano plot of 5571 total super enhancers detected in all osteosarcoma patient samples and cell lines used in this study. Points marked in red denote super enhancers meeting the threshold of significance (P <.5) for being gained or lost in metastatic samples. Points in grey denote super enhancers below the significance threshold. The F3 super enhancer is indicated by the arrow. Nature Medicine: doi:1.138/nm.4475
21 a GAPDH b F3 d.5 c Relative GAPDH Expression -Dox vs. +Dox MG63.3 shf3a MG63.3 shf3b Normalized F3 Expression Dox +Dox -Dox +Dox shf3-a shf3-b Nature Medicine: doi:1.138/nm.4475
22 Supplementary Figure 11: Assessment of F3 knockdown by RT-qPCR. a, Amplification plots and standard curves of GAPDH RT-qPCR using.ng, ng, ng, and ng of template cdna demonstrating efficiency value of 1% and R value of 1.. All Cq values used for quantification of GAPDH were confirmed to be within linear range of standards. b, Amplification plots and standard curves of F3 RT-qPCR using.ng, ng, ng, and ng of template cdna demonstrating efficiency value of 97.% and R value of.999. All Cq values used for quantification of F3 were confirmed to be within linear range of standards. c, Relative GAPDH expression in DsRed+ (induced) cells transduced with F3 shrna constructs after 4hrs treatment with 5ug/ml doxycycline relative to uninduced controls (average of 3 replicates +/-SEM). d, Relative F3 expression normalized to GAPDH in DsRed+ (induced) cells transduced with F3 shrna constructs after 4hrs treatment with 5ug/ml doxycycline relative to uninduced controls (average of 3 replicates +/-SEM). Nature Medicine: doi:1.138/nm.4475
23 +Dox (F3 kd) -Dox a 8 MG63.3 shf3a 1 MG63.3 shf3b Fold-Change Confluence 6 4 MG63.3 shf3a untreated MG63.3 shf3a 5ug/ml dox Fold-Change Confluence MG63.3 shf3b untreated MG63.3 shf3b 5ug/ml dox 5 1 Time (hrs) 5 1 Time (hrs) MNNG shf3a MNNG shf3b 5 5 Fold-Change Confluence MNNG shf3a untreated MNNG shf3a 5ug/ml dox Fold-Change Confluence MNNG shf3b untreated MNNG shf3b 5ug/ml dox 5 1 Time (hrs) d b Inducible shf3a MG63.3 Inducible shf3b Inducible shf3a MNNG Inducible shf3b Mouse 1 -Dox +Dox (F3 kd) Mouse 5 1 Time (hrs) c 1 MG63.3 MNNG 15 p =.3 p =. p =.13 p =.3 P=.3 P=. P=.13 P=.3 Mouse 3 Normalized Metastatic Burden Dox +Dox Inducible shf3a x x -Dox +Dox Inducible shf3b x x Burden Normalized Metastatic Burden 1 5 -Dox +Dox Inducible shf3a x x -Dox +Dox Inducible shf3b x x e log[total area per.5x field(pixels)] P<.1 6 n=5 mice per condition -Dox +Dox (F3 kd) Nature Medicine: doi:1.138/nm.4475
24 Supplementary Figure 1: Assessment of F3 knockdown on metastatic osteosarcoma cell in vitro growth and lung colonization. a. Change in confluence relative to day of MG63.3 (top) and MNNG (bottom) cells transduced with F3 shrna constructs grown in standard culture conditions over 18hrs in the presence or absence of 5ug/ml doxycycline. Values represent averages from 6 plates +/- SD. b. Representative.5x images of from day 1 of ex vivo lung culture sections of GFP+ MG63.3 (left) and MNNG (right) cells transduced with F3 shrna constructs untreated (top) or treated with 5ug/ml doxycycline (bottom). Lung sections outlined with dashed white line. Scale bar = 5µm. c. Quantification of metastatic burden at day 1 of ex vivo lung culture sections of GFP+ MG63.3 (left) and MNNG (right) cells transduced with F3 shrna constructs untreated (red) or treated (blue) with 5ug/ml doxycycline. Values represent averages +/-SEM from 8 sections per condition (4 sections per mouse x mice) normalized to the same section at day. P-Value calculated by Mann-Whitney Test. d. Representative.5x images of in vivo metastatic burden in untreated (left) or doxycyclinetreated (right) mice receiving tail vein injection of 5x1 5 GFP+ MG63.3 cells transduced with shf3b construct. Scale bar = 5µm. e. Quantification of in vivo metastatic burden in untreated (red) or doxycycline-treated (blue) mice receiving tail vein injection of 5x1 5 GFP+ MG63.3 cells transduced with shf3b construct. Values represent log of total GFP+ area per.5x field, black bars represent average +/- SEM (N= 5 mice per condition, 5 images per mouse). P-Value calculated by Mann-Whitney Test. Nature Medicine: doi:1.138/nm.4475
25 a Field 1 Field Field 3 Field 4 Field 5 Mouse 5 Mouse 4 Mouse 3 Mouse Mouse 1 Mouse 5 -Dox (Uninduced) Mouse 4 Mouse 3 Mouse Mouse 1 b Field 1 Field Field 3 Field 4 Field 5 +Dox (F3 knockdown) Nature Medicine: doi:1.138/nm.4475
26 Supplementary Figure 13: Assessment of lung metastatic burden at experimental end point of orthotopic spontaneous metastasis experiment with F3 knockdown. a. All images used for quantification of in vivo metastatic lesions in lungs 1 days after measureable tumor formation in untreated mice receiving orthotopic injection of 8x1 5 MG63.3 cells transduced with shf3b construct (5 mice per condition, 5 images per mouse). Scale bar = 5µm. b. All images used for quantification of in vivo metastatic lesions in lungs 1 days after measureable tumor formation in doxycycline-treated mice receiving orthotopic injection of 8x1 5 MG63.3 cells transduced with shf3b construct (5 mice per condition, 5 images per mouse). Scale bar = 5µm. Nature Medicine: doi:1.138/nm.4475
27 Gene Inclusion Criteria Metastasis Specific? BACH Enriched Motif N DOCK4 Common Gained Met-VEL N Gained Met-VEL Cluster (MG63.3) F3 Gained Met-VEL Cluster (MG63.3) Y FBXO4 Gained Met-VEL Cluster (MNNG/143B) Y FLNA Common Gained Met-VEL Y Gained Met-VEL Cluster (MNNG/143B) FOS Enriched Motif Y FOSL1 Enriched Motif Y FOXO3 Gained Met-VEL Cluster (MNNG/143B) Y GHR Gained Met-VEL Cluster (MG63.3) N JUN Enriched Motif N JUNB Enriched Motif N RASSF Common Gained Met-VEL N TMEM3 Common Gained Met-VEL N Nature Medicine: doi:1.138/nm.4475
28 Supplementary Table 1: Candidate metastasis dependency genes identified by in vivo highthroughput RNAi functional assay. Nature Medicine: doi:1.138/nm.4475
7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans.
 Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
Nature Immunology: doi: /ni Supplementary Figure 1. Transcriptional program of the TE and MP CD8 + T cell subsets.
 Supplementary Figure 1 Transcriptional program of the TE and MP CD8 + T cell subsets. (a) Comparison of gene expression of TE and MP CD8 + T cell subsets by microarray. Genes that are 1.5-fold upregulated
Supplementary Figure 1 Transcriptional program of the TE and MP CD8 + T cell subsets. (a) Comparison of gene expression of TE and MP CD8 + T cell subsets by microarray. Genes that are 1.5-fold upregulated
Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types.
 Supplementary Figure 1 Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types. (a) Pearson correlation heatmap among open chromatin profiles of different
Supplementary Figure 1 Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types. (a) Pearson correlation heatmap among open chromatin profiles of different
Supplementary Figure S1. Gene expression analysis of epidermal marker genes and TP63.
 Supplementary Figure Legends Supplementary Figure S1. Gene expression analysis of epidermal marker genes and TP63. A. Screenshot of the UCSC genome browser from normalized RNAPII and RNA-seq ChIP-seq data
Supplementary Figure Legends Supplementary Figure S1. Gene expression analysis of epidermal marker genes and TP63. A. Screenshot of the UCSC genome browser from normalized RNAPII and RNA-seq ChIP-seq data
Supplementary Figure 1. Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Nature Immunology: doi: /ni.
 Supplementary Figure 1 Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Expression of Mll4 floxed alleles (16-19) in naive CD4 + T cells isolated from lymph nodes and
Supplementary Figure 1 Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Expression of Mll4 floxed alleles (16-19) in naive CD4 + T cells isolated from lymph nodes and
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/375/ra41/dc1 Supplementary Materials for Actin cytoskeletal remodeling with protrusion formation is essential for heart regeneration in Hippo-deficient mice
www.sciencesignaling.org/cgi/content/full/8/375/ra41/dc1 Supplementary Materials for Actin cytoskeletal remodeling with protrusion formation is essential for heart regeneration in Hippo-deficient mice
Supplementary Figure 1. Characterization of NMuMG-ErbB2 and NIC breast cancer cells expressing shrnas targeting LPP. NMuMG-ErbB2 cells (a) and NIC
 Supplementary Figure 1. Characterization of NMuMG-ErbB2 and NIC breast cancer cells expressing shrnas targeting LPP. NMuMG-ErbB2 cells (a) and NIC cells (b) were engineered to stably express either a LucA-shRNA
Supplementary Figure 1. Characterization of NMuMG-ErbB2 and NIC breast cancer cells expressing shrnas targeting LPP. NMuMG-ErbB2 cells (a) and NIC cells (b) were engineered to stably express either a LucA-shRNA
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Effect of HSP90 inhibition on expression of endogenous retroviruses. (a) Inducible shrna-mediated Hsp90 silencing in mouse ESCs. Immunoblots of total cell extract expressing the
Supplementary Figure 1 Effect of HSP90 inhibition on expression of endogenous retroviruses. (a) Inducible shrna-mediated Hsp90 silencing in mouse ESCs. Immunoblots of total cell extract expressing the
SHREE ET AL, SUPPLEMENTAL MATERIALS. (A) Workflow for tumor cell line derivation and orthotopic implantation.
 SHREE ET AL, SUPPLEMENTAL MATERIALS SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure 1. Derivation and characterization of TS1-TGL and TS2-TGL PyMT cell lines and development of an orthotopic
SHREE ET AL, SUPPLEMENTAL MATERIALS SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure 1. Derivation and characterization of TS1-TGL and TS2-TGL PyMT cell lines and development of an orthotopic
Nature Structural & Molecular Biology: doi: /nsmb.2419
 Supplementary Figure 1 Mapped sequence reads and nucleosome occupancies. (a) Distribution of sequencing reads on the mouse reference genome for chromosome 14 as an example. The number of reads in a 1 Mb
Supplementary Figure 1 Mapped sequence reads and nucleosome occupancies. (a) Distribution of sequencing reads on the mouse reference genome for chromosome 14 as an example. The number of reads in a 1 Mb
(a) Significant biological processes (upper panel) and disease biomarkers (lower panel)
 Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES 1 Supplementary Figure 1, Adult hippocampal QNPs and TAPs uniformly express REST a-b) Confocal images of adult hippocampal mouse sections showing GFAP (green), Sox2 (red), and REST
SUPPLEMENTARY FIGURES 1 Supplementary Figure 1, Adult hippocampal QNPs and TAPs uniformly express REST a-b) Confocal images of adult hippocampal mouse sections showing GFAP (green), Sox2 (red), and REST
EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH
 EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH Supplementary Figure 1. Supplementary Figure 1. Characterization of KP and KPH2 autochthonous UPS tumors. a) Genotyping of KPH2
EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH Supplementary Figure 1. Supplementary Figure 1. Characterization of KP and KPH2 autochthonous UPS tumors. a) Genotyping of KPH2
Nature Immunology: doi: /ni Supplementary Figure 1. Characteristics of SEs in T reg and T conv cells.
 Supplementary Figure 1 Characteristics of SEs in T reg and T conv cells. (a) Patterns of indicated transcription factor-binding at SEs and surrounding regions in T reg and T conv cells. Average normalized
Supplementary Figure 1 Characteristics of SEs in T reg and T conv cells. (a) Patterns of indicated transcription factor-binding at SEs and surrounding regions in T reg and T conv cells. Average normalized
a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation,
 Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
fl/+ KRas;Atg5 fl/+ KRas;Atg5 fl/fl KRas;Atg5 fl/fl KRas;Atg5 Supplementary Figure 1. Gene set enrichment analyses. (a) (b)
 KRas;At KRas;At KRas;At KRas;At a b Supplementary Figure 1. Gene set enrichment analyses. (a) GO gene sets (MSigDB v3. c5) enriched in KRas;Atg5 fl/+ as compared to KRas;Atg5 fl/fl tumors using gene set
KRas;At KRas;At KRas;At KRas;At a b Supplementary Figure 1. Gene set enrichment analyses. (a) GO gene sets (MSigDB v3. c5) enriched in KRas;Atg5 fl/+ as compared to KRas;Atg5 fl/fl tumors using gene set
Nature Genetics: doi: /ng Supplementary Figure 1. Assessment of sample purity and quality.
 Supplementary Figure 1 Assessment of sample purity and quality. (a) Hematoxylin and eosin staining of formaldehyde-fixed, paraffin-embedded sections from a human testis biopsy collected concurrently with
Supplementary Figure 1 Assessment of sample purity and quality. (a) Hematoxylin and eosin staining of formaldehyde-fixed, paraffin-embedded sections from a human testis biopsy collected concurrently with
Supplementary information
 Supplementary information High fat diet-induced changes of mouse hepatic transcription and enhancer activity can be reversed by subsequent weight loss Majken Siersbæk, Lyuba Varticovski, Shutong Yang,
Supplementary information High fat diet-induced changes of mouse hepatic transcription and enhancer activity can be reversed by subsequent weight loss Majken Siersbæk, Lyuba Varticovski, Shutong Yang,
Supplementary Figure 1. Metabolic landscape of cancer discovery pipeline. RNAseq raw counts data of cancer and healthy tissue samples were downloaded
 Supplementary Figure 1. Metabolic landscape of cancer discovery pipeline. RNAseq raw counts data of cancer and healthy tissue samples were downloaded from TCGA and differentially expressed metabolic genes
Supplementary Figure 1. Metabolic landscape of cancer discovery pipeline. RNAseq raw counts data of cancer and healthy tissue samples were downloaded from TCGA and differentially expressed metabolic genes
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/7/308/ra4/dc1 Supplementary Materials for Antipsychotics Activate mtorc1-dependent Translation to Enhance Neuronal Morphological Complexity Heather Bowling, Guoan
www.sciencesignaling.org/cgi/content/full/7/308/ra4/dc1 Supplementary Materials for Antipsychotics Activate mtorc1-dependent Translation to Enhance Neuronal Morphological Complexity Heather Bowling, Guoan
Breeding scheme, transgenes, histological analysis and site distribution of SB-mutagenized osteosarcoma.
 Supplementary Figure 1 Breeding scheme, transgenes, histological analysis and site distribution of SB-mutagenized osteosarcoma. (a) Breeding scheme. R26-LSL-SB11 homozygous mice were bred to Trp53 LSL-R270H/+
Supplementary Figure 1 Breeding scheme, transgenes, histological analysis and site distribution of SB-mutagenized osteosarcoma. (a) Breeding scheme. R26-LSL-SB11 homozygous mice were bred to Trp53 LSL-R270H/+
Supplementary Figure 1 IL-27 IL
 Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
SUPPLEMENTARY INFORMATION
 doi:.38/nature8975 SUPPLEMENTAL TEXT Unique association of HOTAIR with patient outcome To determine whether the expression of other HOX lincrnas in addition to HOTAIR can predict patient outcome, we measured
doi:.38/nature8975 SUPPLEMENTAL TEXT Unique association of HOTAIR with patient outcome To determine whether the expression of other HOX lincrnas in addition to HOTAIR can predict patient outcome, we measured
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Heatmap of GO terms for differentially expressed genes. The terms were hierarchically clustered using the GO term enrichment beta. Darker red, higher positive
Supplementary Figures Supplementary Figure 1. Heatmap of GO terms for differentially expressed genes. The terms were hierarchically clustered using the GO term enrichment beta. Darker red, higher positive
Nature Genetics: doi: /ng Supplementary Figure 1. Phenotypic characterization of MES- and ADRN-type cells.
 Supplementary Figure 1 Phenotypic characterization of MES- and ADRN-type cells. (a) Bright-field images showing cellular morphology of MES-type (691-MES, 700-MES, 717-MES) and ADRN-type (691-ADRN, 700-
Supplementary Figure 1 Phenotypic characterization of MES- and ADRN-type cells. (a) Bright-field images showing cellular morphology of MES-type (691-MES, 700-MES, 717-MES) and ADRN-type (691-ADRN, 700-
Nature Genetics: doi: /ng Supplementary Figure 1
 Supplementary Figure 1 Expression deviation of the genes mapped to gene-wise recurrent mutations in the TCGA breast cancer cohort (top) and the TCGA lung cancer cohort (bottom). For each gene (each pair
Supplementary Figure 1 Expression deviation of the genes mapped to gene-wise recurrent mutations in the TCGA breast cancer cohort (top) and the TCGA lung cancer cohort (bottom). For each gene (each pair
Supplementary Figure 1. IHC and proliferation analysis of pten-deficient mammary tumors
 Wang et al LEGENDS TO SUPPLEMENTARY INFORMATION Supplementary Figure 1. IHC and proliferation analysis of pten-deficient mammary tumors A. Induced expression of estrogen receptor α (ERα) in AME vs PDA
Wang et al LEGENDS TO SUPPLEMENTARY INFORMATION Supplementary Figure 1. IHC and proliferation analysis of pten-deficient mammary tumors A. Induced expression of estrogen receptor α (ERα) in AME vs PDA
Sirt1 Hmg20b Gm (0.17) 24 (17.3) 877 (857)
 3 (0.17) 24 (17.3) Sirt1 Hmg20 Gm4763 877 (857) c d Suppl. Figure 1. Screen validation for top candidate antagonists of Dot1L (a) Numer of genes with one (gray), two (cyan) or three (red) shrna scored
3 (0.17) 24 (17.3) Sirt1 Hmg20 Gm4763 877 (857) c d Suppl. Figure 1. Screen validation for top candidate antagonists of Dot1L (a) Numer of genes with one (gray), two (cyan) or three (red) shrna scored
SUPPLEMENTARY FIGURES AND TABLES
 SUPPLEMENTARY FIGURES AND TABLES Supplementary Figure S1: CaSR expression in neuroblastoma models. A. Proteins were isolated from three neuroblastoma cell lines and from the liver metastasis of a MYCN-non
SUPPLEMENTARY FIGURES AND TABLES Supplementary Figure S1: CaSR expression in neuroblastoma models. A. Proteins were isolated from three neuroblastoma cell lines and from the liver metastasis of a MYCN-non
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
Nature Immunology: doi: /ni Supplementary Figure 1. DNA-methylation machinery is essential for silencing of Cd4 in cytotoxic T cells.
 Supplementary Figure 1 DNA-methylation machinery is essential for silencing of Cd4 in cytotoxic T cells. (a) Scheme for the retroviral shrna screen. (b) Histogram showing CD4 expression (MFI) in WT cytotoxic
Supplementary Figure 1 DNA-methylation machinery is essential for silencing of Cd4 in cytotoxic T cells. (a) Scheme for the retroviral shrna screen. (b) Histogram showing CD4 expression (MFI) in WT cytotoxic
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 Characterization of stable expression of GlucB and sshbira in the CT26 cell line (a) Live cell imaging of stable CT26 cells expressing green fluorescent protein
Supplementary Figures Supplementary Figure 1 Characterization of stable expression of GlucB and sshbira in the CT26 cell line (a) Live cell imaging of stable CT26 cells expressing green fluorescent protein
Title: Epigenetic mechanisms underlying maternal diabetes-associated risk of congenital heart disease
 1 Supplemental Materials 2 3 Title: Epigenetic mechanisms underlying maternal diabetes-associated risk of congenital heart disease 4 5 6 Authors: Madhumita Basu, 1 Jun-Yi Zhu, 2 Stephanie LaHaye 1,3, Uddalak
1 Supplemental Materials 2 3 Title: Epigenetic mechanisms underlying maternal diabetes-associated risk of congenital heart disease 4 5 6 Authors: Madhumita Basu, 1 Jun-Yi Zhu, 2 Stephanie LaHaye 1,3, Uddalak
(A) Cells grown in monolayer were fixed and stained for surfactant protein-c (SPC,
 Supplemental Figure Legends Figure S1. Cell line characterization (A) Cells grown in monolayer were fixed and stained for surfactant protein-c (SPC, green) and co-stained with DAPI to visualize the nuclei.
Supplemental Figure Legends Figure S1. Cell line characterization (A) Cells grown in monolayer were fixed and stained for surfactant protein-c (SPC, green) and co-stained with DAPI to visualize the nuclei.
Supplementary Figure 1. Deletion of Smad3 prevents B16F10 melanoma invasion and metastasis in a mouse s.c. tumor model.
 A B16F1 s.c. Lung LN Distant lymph nodes Colon B B16F1 s.c. Supplementary Figure 1. Deletion of Smad3 prevents B16F1 melanoma invasion and metastasis in a mouse s.c. tumor model. Highly invasive growth
A B16F1 s.c. Lung LN Distant lymph nodes Colon B B16F1 s.c. Supplementary Figure 1. Deletion of Smad3 prevents B16F1 melanoma invasion and metastasis in a mouse s.c. tumor model. Highly invasive growth
Supplementary figure 1
 Supplementary figure 1 Nature Medicine: doi:1.138/nm.275 CLUSTER BY SELF-ORGANIZING MAPS SELECTED PATHWAY ANALISYS TERMS Cluster : up-regulated genes in acute patients Cell cycle/dna repair Fatty acid
Supplementary figure 1 Nature Medicine: doi:1.138/nm.275 CLUSTER BY SELF-ORGANIZING MAPS SELECTED PATHWAY ANALISYS TERMS Cluster : up-regulated genes in acute patients Cell cycle/dna repair Fatty acid
Nature Immunology: doi: /ni Supplementary Figure 1. Huwe1 has high expression in HSCs and is necessary for quiescence.
 Supplementary Figure 1 Huwe1 has high expression in HSCs and is necessary for quiescence. (a) Heat map visualizing expression of genes with a known function in ubiquitin-mediated proteolysis (KEGG: Ubiquitin
Supplementary Figure 1 Huwe1 has high expression in HSCs and is necessary for quiescence. (a) Heat map visualizing expression of genes with a known function in ubiquitin-mediated proteolysis (KEGG: Ubiquitin
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature19360 Supplementary Tables Supplementary Table 1. Number of monoclonal reads in each sample Sample Number of cells Total reads Aligned reads Monoclonal reads
SUPPLEMENTARY INFORMATION doi:10.1038/nature19360 Supplementary Tables Supplementary Table 1. Number of monoclonal reads in each sample Sample Number of cells Total reads Aligned reads Monoclonal reads
TEB. Id4 p63 DAPI Merge. Id4 CK8 DAPI Merge
 a Duct TEB b Id4 p63 DAPI Merge Id4 CK8 DAPI Merge c d e Supplementary Figure 1. Identification of Id4-positive MECs and characterization of the Comma-D model. (a) IHC analysis of ID4 expression in the
a Duct TEB b Id4 p63 DAPI Merge Id4 CK8 DAPI Merge c d e Supplementary Figure 1. Identification of Id4-positive MECs and characterization of the Comma-D model. (a) IHC analysis of ID4 expression in the
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3311 A B TSC2 -/- MEFs C Rapa Hours WCL 0 6 12 24 36 pakt.s473 AKT ps6k S6K CM IGF-1 Recipient WCL - + - + - + pigf-1r IGF-1R pakt ps6 AKT D 1 st SILAC 2 nd SILAC E GAPDH FGF21 ALKPGVIQILGVK
DOI: 10.1038/ncb3311 A B TSC2 -/- MEFs C Rapa Hours WCL 0 6 12 24 36 pakt.s473 AKT ps6k S6K CM IGF-1 Recipient WCL - + - + - + pigf-1r IGF-1R pakt ps6 AKT D 1 st SILAC 2 nd SILAC E GAPDH FGF21 ALKPGVIQILGVK
Supplementary methods:
 Supplementary methods: Primers sequences used in real-time PCR analyses: β-actin F: GACCTCTATGCCAACACAGT β-actin [11] R: AGTACTTGCGCTCAGGAGGA MMP13 F: TTCTGGTCTTCTGGCACACGCTTT MMP13 R: CCAAGCTCATGGGCAGCAACAATA
Supplementary methods: Primers sequences used in real-time PCR analyses: β-actin F: GACCTCTATGCCAACACAGT β-actin [11] R: AGTACTTGCGCTCAGGAGGA MMP13 F: TTCTGGTCTTCTGGCACACGCTTT MMP13 R: CCAAGCTCATGGGCAGCAACAATA
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb3399 a b c d FSP DAPI 5mm mm 5mm 5mm e Correspond to melanoma in-situ Figure a DCT FSP- f MITF mm mm MlanaA melanoma in-situ DCT 5mm FSP- mm mm mm mm mm g melanoma in-situ MITF MlanaA mm mm
DOI:.38/ncb3399 a b c d FSP DAPI 5mm mm 5mm 5mm e Correspond to melanoma in-situ Figure a DCT FSP- f MITF mm mm MlanaA melanoma in-situ DCT 5mm FSP- mm mm mm mm mm g melanoma in-situ MITF MlanaA mm mm
Supplemental Table S1
 Supplemental Table S. Tumorigenicity and metastatic potential of 44SQ cell subpopulations a Tumorigenicity b Average tumor volume (mm ) c Lung metastasis d CD high /4 8. 8/ CD low /4 6./ a Mice were injected
Supplemental Table S. Tumorigenicity and metastatic potential of 44SQ cell subpopulations a Tumorigenicity b Average tumor volume (mm ) c Lung metastasis d CD high /4 8. 8/ CD low /4 6./ a Mice were injected
Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells.
 SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
Supplementary Materials for
 www.sciencetranslationalmedicine.org/cgi/content/full/4/117/117ra8/dc1 Supplementary Materials for Notch4 Normalization Reduces Blood Vessel Size in Arteriovenous Malformations Patrick A. Murphy, Tyson
www.sciencetranslationalmedicine.org/cgi/content/full/4/117/117ra8/dc1 Supplementary Materials for Notch4 Normalization Reduces Blood Vessel Size in Arteriovenous Malformations Patrick A. Murphy, Tyson
Supplementary Table S1. List of PTPRK-RSPO3 gene fusions in TCGA's colon cancer cohort. Chr. # of Gene 2. Chr. # of Gene 1
 Supplementary Tale S1. List of PTPRK-RSPO3 gene fusions in TCGA's colon cancer cohort TCGA Case ID Gene-1 Gene-2 Chr. # of Gene 1 Chr. # of Gene 2 Genomic coordiante of Gene 1 at fusion junction Genomic
Supplementary Tale S1. List of PTPRK-RSPO3 gene fusions in TCGA's colon cancer cohort TCGA Case ID Gene-1 Gene-2 Chr. # of Gene 1 Chr. # of Gene 2 Genomic coordiante of Gene 1 at fusion junction Genomic
Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression.
 Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
* * * * Supplementary Figure 1. DS Lv CK HSA CK HSA. CK Col-3. CK Col-3. See overleaf for figure legend. Cancer cells
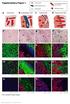 Supplementary Figure 1 Cancer cells Desmoplastic stroma Hepatocytes Pre-existing sinusoidal blood vessel New blood vessel a Normal liver b Desmoplastic HGP c Pushing HGP d Replacement HGP e f g h i DS
Supplementary Figure 1 Cancer cells Desmoplastic stroma Hepatocytes Pre-existing sinusoidal blood vessel New blood vessel a Normal liver b Desmoplastic HGP c Pushing HGP d Replacement HGP e f g h i DS
Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor suppressor genes
 Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor suppressor genes Kaifu Chen 1,2,3,4,5,10, Zhong Chen 6,10, Dayong Wu 6, Lili Zhang 7, Xueqiu Lin 1,2,8,
Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor suppressor genes Kaifu Chen 1,2,3,4,5,10, Zhong Chen 6,10, Dayong Wu 6, Lili Zhang 7, Xueqiu Lin 1,2,8,
Nature Immunology: doi: /ni Supplementary Figure 1. Id2 and Id3 define polyclonal T H 1 and T FH cell subsets.
 Supplementary Figure 1 Id2 and Id3 define polyclonal T H 1 and T FH cell subsets. Id2 YFP/+ (a) or Id3 GFP/+ (b) mice were analyzed 7 days after LCMV infection. T H 1 (SLAM + CXCR5 or CXCR5 PD-1 ), T FH
Supplementary Figure 1 Id2 and Id3 define polyclonal T H 1 and T FH cell subsets. Id2 YFP/+ (a) or Id3 GFP/+ (b) mice were analyzed 7 days after LCMV infection. T H 1 (SLAM + CXCR5 or CXCR5 PD-1 ), T FH
A263 A352 A204. Pan CK. pstat STAT3 pstat3 STAT3 pstat3. Columns Columns 1-6 Positive control. Omentum. Rectosigmoid A195.
 pstat3 75 Pan CK A A263 A352 A24 B Columns 1-6 Positive control A195 A22 A24 A183 Rectal Nodule STAT3 pstat3 STAT3 pstat3 Columns 7-12 Omentum Rectosigmoid Left Ovary Right Ovary Omentum Uterus Uterus
pstat3 75 Pan CK A A263 A352 A24 B Columns 1-6 Positive control A195 A22 A24 A183 Rectal Nodule STAT3 pstat3 STAT3 pstat3 Columns 7-12 Omentum Rectosigmoid Left Ovary Right Ovary Omentum Uterus Uterus
Supplemental Information. Genomic Characterization of Murine. Monocytes Reveals C/EBPb Transcription. Factor Dependence of Ly6C Cells
 Immunity, Volume 46 Supplemental Information Genomic Characterization of Murine Monocytes Reveals C/EBPb Transcription Factor Dependence of Ly6C Cells Alexander Mildner, Jörg Schönheit, Amir Giladi, Eyal
Immunity, Volume 46 Supplemental Information Genomic Characterization of Murine Monocytes Reveals C/EBPb Transcription Factor Dependence of Ly6C Cells Alexander Mildner, Jörg Schönheit, Amir Giladi, Eyal
SUPPLEMENTARY MATERIAL
 SUPPLEMENTARY MATERIAL IL-1 signaling modulates activation of STAT transcription factors to antagonize retinoic acid signaling and control the T H 17 cell it reg cell balance Rajatava Basu 1,5, Sarah K.
SUPPLEMENTARY MATERIAL IL-1 signaling modulates activation of STAT transcription factors to antagonize retinoic acid signaling and control the T H 17 cell it reg cell balance Rajatava Basu 1,5, Sarah K.
Suppl Video: Tumor cells (green) and monocytes (white) are seeded on a confluent endothelial
 Supplementary Information Häuselmann et al. Monocyte induction of E-selectin-mediated endothelial activation releases VE-cadherin junctions to promote tumor cell extravasation in the metastasis cascade
Supplementary Information Häuselmann et al. Monocyte induction of E-selectin-mediated endothelial activation releases VE-cadherin junctions to promote tumor cell extravasation in the metastasis cascade
Supplementary Figure 1: Features of IGLL5 Mutations in CLL: a) Representative IGV screenshot of first
 Supplementary Figure 1: Features of IGLL5 Mutations in CLL: a) Representative IGV screenshot of first intron IGLL5 mutation depicting biallelic mutations. Red arrows highlight the presence of out of phase
Supplementary Figure 1: Features of IGLL5 Mutations in CLL: a) Representative IGV screenshot of first intron IGLL5 mutation depicting biallelic mutations. Red arrows highlight the presence of out of phase
Nature Genetics: doi: /ng Supplementary Figure 1. SEER data for male and female cancer incidence from
 Supplementary Figure 1 SEER data for male and female cancer incidence from 1975 2013. (a,b) Incidence rates of oral cavity and pharynx cancer (a) and leukemia (b) are plotted, grouped by males (blue),
Supplementary Figure 1 SEER data for male and female cancer incidence from 1975 2013. (a,b) Incidence rates of oral cavity and pharynx cancer (a) and leukemia (b) are plotted, grouped by males (blue),
Supplemental Figure 1
 Supplemental Figure 1 1a 1c PD-1 MFI fold change 6 5 4 3 2 1 IL-1α IL-2 IL-4 IL-6 IL-1 IL-12 IL-13 IL-15 IL-17 IL-18 IL-21 IL-23 IFN-α Mut Human PD-1 promoter SBE-D 5 -GTCTG- -1.2kb SBE-P -CAGAC- -1.kb
Supplemental Figure 1 1a 1c PD-1 MFI fold change 6 5 4 3 2 1 IL-1α IL-2 IL-4 IL-6 IL-1 IL-12 IL-13 IL-15 IL-17 IL-18 IL-21 IL-23 IFN-α Mut Human PD-1 promoter SBE-D 5 -GTCTG- -1.2kb SBE-P -CAGAC- -1.kb
Supplementary Information
 Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
Nature Genetics: doi: /ng Supplementary Figure 1. HOX fusions enhance self-renewal capacity.
 Supplementary Figure 1 HOX fusions enhance self-renewal capacity. Mouse bone marrow was transduced with a retrovirus carrying one of three HOX fusion genes or the empty mcherry reporter construct as described
Supplementary Figure 1 HOX fusions enhance self-renewal capacity. Mouse bone marrow was transduced with a retrovirus carrying one of three HOX fusion genes or the empty mcherry reporter construct as described
Nature Neuroscience: doi: /nn Supplementary Figure 1. Confirmation that optogenetic inhibition of dopaminergic neurons affects choice
 Supplementary Figure 1 Confirmation that optogenetic inhibition of dopaminergic neurons affects choice (a) Sample behavioral trace as in Figure 1d, but with NpHR stimulation trials depicted as green blocks
Supplementary Figure 1 Confirmation that optogenetic inhibition of dopaminergic neurons affects choice (a) Sample behavioral trace as in Figure 1d, but with NpHR stimulation trials depicted as green blocks
Accessing and Using ENCODE Data Dr. Peggy J. Farnham
 1 William M Keck Professor of Biochemistry Keck School of Medicine University of Southern California How many human genes are encoded in our 3x10 9 bp? C. elegans (worm) 959 cells and 1x10 8 bp 20,000
1 William M Keck Professor of Biochemistry Keck School of Medicine University of Southern California How many human genes are encoded in our 3x10 9 bp? C. elegans (worm) 959 cells and 1x10 8 bp 20,000
Supplementary Figure 1 Cytokine receptors on developing thymocytes that can potentially signal Runx3d expression.
 Supplementary Figure 1 Cytokine receptors on developing thymocytes that can potentially signal Runx3d expression. (a) Characterization of c-independent SP8 cells. Stainings for maturation markers (top)
Supplementary Figure 1 Cytokine receptors on developing thymocytes that can potentially signal Runx3d expression. (a) Characterization of c-independent SP8 cells. Stainings for maturation markers (top)
MIR retrotransposon sequences provide insulators to the human genome
 Supplementary Information: MIR retrotransposon sequences provide insulators to the human genome Jianrong Wang, Cristina Vicente-García, Davide Seruggia, Eduardo Moltó, Ana Fernandez- Miñán, Ana Neto, Elbert
Supplementary Information: MIR retrotransposon sequences provide insulators to the human genome Jianrong Wang, Cristina Vicente-García, Davide Seruggia, Eduardo Moltó, Ana Fernandez- Miñán, Ana Neto, Elbert
Supplemental Figures:
 Supplemental Figures: Figure 1: Intracellular distribution of VWF by electron microscopy in human endothelial cells. a) Immunogold labeling of LC3 demonstrating an LC3-positive autophagosome (white arrow)
Supplemental Figures: Figure 1: Intracellular distribution of VWF by electron microscopy in human endothelial cells. a) Immunogold labeling of LC3 demonstrating an LC3-positive autophagosome (white arrow)
Computational Analysis of UHT Sequences Histone modifications, CAGE, RNA-Seq
 Computational Analysis of UHT Sequences Histone modifications, CAGE, RNA-Seq Philipp Bucher Wednesday January 21, 2009 SIB graduate school course EPFL, Lausanne ChIP-seq against histone variants: Biological
Computational Analysis of UHT Sequences Histone modifications, CAGE, RNA-Seq Philipp Bucher Wednesday January 21, 2009 SIB graduate school course EPFL, Lausanne ChIP-seq against histone variants: Biological
Supplementary Materials. for Garmy-Susini, et al, Integrin 4 1 signaling is required for lymphangiogenesis and tumor metastasis
 Supplementary Materials for Garmy-Susini, et al, Integrin 4 1 signaling is required for lymphangiogenesis and tumor metastasis 1 Supplementary Figure Legends Supplementary Figure 1: Integrin expression
Supplementary Materials for Garmy-Susini, et al, Integrin 4 1 signaling is required for lymphangiogenesis and tumor metastasis 1 Supplementary Figure Legends Supplementary Figure 1: Integrin expression
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 Bidirectional optogenetic modulation of the tonic activity of CEA PKCδ + neurons in vitro. a, Top, Cell-attached voltage recording illustrating the blue light-induced increase in
Supplementary Figure 1 Bidirectional optogenetic modulation of the tonic activity of CEA PKCδ + neurons in vitro. a, Top, Cell-attached voltage recording illustrating the blue light-induced increase in
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 Atlas representations of the midcingulate (MCC) region targeted in this study compared against the anterior cingulate (ACC) region commonly reported. Coronal sections are shown on
Supplementary Figure 1 Atlas representations of the midcingulate (MCC) region targeted in this study compared against the anterior cingulate (ACC) region commonly reported. Coronal sections are shown on
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1. Differential expression of mirnas from the pri-mir-17-92a locus.
 Supplementary Figure 1 Differential expression of mirnas from the pri-mir-17-92a locus. (a) The mir-17-92a expression unit in the third intron of the host mir-17hg transcript. (b,c) Impact of knockdown
Supplementary Figure 1 Differential expression of mirnas from the pri-mir-17-92a locus. (a) The mir-17-92a expression unit in the third intron of the host mir-17hg transcript. (b,c) Impact of knockdown
Supplementary Figure 1: Attenuation of association signals after conditioning for the lead SNP. a) attenuation of association signal at the 9p22.
 Supplementary Figure 1: Attenuation of association signals after conditioning for the lead SNP. a) attenuation of association signal at the 9p22.32 PCOS locus after conditioning for the lead SNP rs10993397;
Supplementary Figure 1: Attenuation of association signals after conditioning for the lead SNP. a) attenuation of association signal at the 9p22.32 PCOS locus after conditioning for the lead SNP rs10993397;
Supplementary Information Titles Journal: Nature Medicine
 Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
Supplementary. presence of the. (c) mrna expression. Error. in naive or
 Figure 1. (a) Naive CD4 + T cells were activated in the presence of the indicated cytokines for 3 days. Enpp2 mrna expression was measured by qrt-pcrhr, infected with (b, c) Naive CD4 + T cells were activated
Figure 1. (a) Naive CD4 + T cells were activated in the presence of the indicated cytokines for 3 days. Enpp2 mrna expression was measured by qrt-pcrhr, infected with (b, c) Naive CD4 + T cells were activated
(a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable
 Supplementary Figure 1. Frameshift (FS) mutation in UVRAG. (a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable A 10 DNA repeat, generating a premature stop codon
Supplementary Figure 1. Frameshift (FS) mutation in UVRAG. (a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable A 10 DNA repeat, generating a premature stop codon
SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer.
 Supplementary Figure 1 SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer. Scatter plots comparing expression profiles of matched pretreatment
Supplementary Figure 1 SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer. Scatter plots comparing expression profiles of matched pretreatment
Nature Neuroscience: doi: /nn Supplementary Figure 1. Behavioral training.
 Supplementary Figure 1 Behavioral training. a, Mazes used for behavioral training. Asterisks indicate reward location. Only some example mazes are shown (for example, right choice and not left choice maze
Supplementary Figure 1 Behavioral training. a, Mazes used for behavioral training. Asterisks indicate reward location. Only some example mazes are shown (for example, right choice and not left choice maze
Supplementary Figure 1 (Related with Figure 4). Molecular consequences of Eed deletion. (a) ChIP analysis identifies 3925 genes that are associated
 Supplementary Figure 1 (Related with Figure 4). Molecular consequences of Eed deletion. (a) ChIP analysis identifies 3925 genes that are associated with the H3K27me3 mark in chondrocytes (see Table S1,
Supplementary Figure 1 (Related with Figure 4). Molecular consequences of Eed deletion. (a) ChIP analysis identifies 3925 genes that are associated with the H3K27me3 mark in chondrocytes (see Table S1,
Nature Immunology: doi: /ni Supplementary Figure 1. RNA-Seq analysis of CD8 + TILs and N-TILs.
 Supplementary Figure 1 RNA-Seq analysis of CD8 + TILs and N-TILs. (a) Schematic representation of the tumor and cell types used for the study. HNSCC, head and neck squamous cell cancer; NSCLC, non-small
Supplementary Figure 1 RNA-Seq analysis of CD8 + TILs and N-TILs. (a) Schematic representation of the tumor and cell types used for the study. HNSCC, head and neck squamous cell cancer; NSCLC, non-small
Supplementary Figure 1 ITGB1 and ITGA11 increase with evidence for heterodimers following HSC activation. (a) Time course of rat HSC activation
 Supplementary Figure 1 ITGB1 and ITGA11 increase with evidence for heterodimers following HSC activation. (a) Time course of rat HSC activation indicated by the detection of -SMA and COL1 (log scale).
Supplementary Figure 1 ITGB1 and ITGA11 increase with evidence for heterodimers following HSC activation. (a) Time course of rat HSC activation indicated by the detection of -SMA and COL1 (log scale).
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3021 Supplementary figure 1 Characterisation of TIMPless fibroblasts. a) Relative gene expression of TIMPs1-4 by real time quantitative PCR (RT-qPCR) in WT or ΔTimp fibroblasts (mean ±
DOI: 10.1038/ncb3021 Supplementary figure 1 Characterisation of TIMPless fibroblasts. a) Relative gene expression of TIMPs1-4 by real time quantitative PCR (RT-qPCR) in WT or ΔTimp fibroblasts (mean ±
Type of file: PDF Size of file: 0 KB Title of file for HTML: Supplementary Information Description: Supplementary Figures
 Type of file: PDF Size of file: 0 KB Title of file for HTML: Supplementary Information Description: Supplementary Figures Supplementary Figure 1 mir-128-3p is highly expressed in chemoresistant, metastatic
Type of file: PDF Size of file: 0 KB Title of file for HTML: Supplementary Information Description: Supplementary Figures Supplementary Figure 1 mir-128-3p is highly expressed in chemoresistant, metastatic
Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A.
 Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A. Upper part, three-primer PCR strategy at the Mcm3 locus yielding
Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A. Upper part, three-primer PCR strategy at the Mcm3 locus yielding
Supplementary Figure 1:
 Supplementary Figure 1: (A) Whole aortic cross-sections stained with Hematoxylin and Eosin (H&E), 7 days after porcine-pancreatic-elastase (PPE)-induced AAA compared to untreated, healthy control aortas
Supplementary Figure 1: (A) Whole aortic cross-sections stained with Hematoxylin and Eosin (H&E), 7 days after porcine-pancreatic-elastase (PPE)-induced AAA compared to untreated, healthy control aortas
Large conserved domains of low DNA methylation maintained by Dnmt3a
 Supplementary information Large conserved domains of low DNA methylation maintained by Dnmt3a Mira Jeong# 1, Deqiang Sun # 2, Min Luo# 1, Yun Huang 3, Grant A. Challen %1, Benjamin Rodriguez 2, Xiaotian
Supplementary information Large conserved domains of low DNA methylation maintained by Dnmt3a Mira Jeong# 1, Deqiang Sun # 2, Min Luo# 1, Yun Huang 3, Grant A. Challen %1, Benjamin Rodriguez 2, Xiaotian
Supplementary Figure 1. BMS enhances human T cell activation in vitro in a
 Supplementary Figure 1. BMS98662 enhances human T cell activation in vitro in a concentration-dependent manner. Jurkat T cells were activated with anti-cd3 and anti-cd28 antibody in the presence of titrated
Supplementary Figure 1. BMS98662 enhances human T cell activation in vitro in a concentration-dependent manner. Jurkat T cells were activated with anti-cd3 and anti-cd28 antibody in the presence of titrated
(A) Dose response curves of HMLE_shGFP (blue circle), HMLE_shEcad (red square),
 Supplementary Figures and Tables Figure S1. Validation of EMT-selective small molecules (A) Dose response curves of HMLE_shGFP (blue circle), HMLE_shEcad (red square), and HMLE_Twist (black diamond) cells
Supplementary Figures and Tables Figure S1. Validation of EMT-selective small molecules (A) Dose response curves of HMLE_shGFP (blue circle), HMLE_shEcad (red square), and HMLE_Twist (black diamond) cells
Supplementary Figure 1. Expression of phospho-sik3 in normal and osteoarthritic articular cartilage in the knee. (a) Semiserial histological sections
 Supplementary Figure 1. Expression of phospho-sik3 in normal and osteoarthritic articular cartilage in the knee. (a) Semiserial histological sections of normal cartilage were stained with safranin O-fast
Supplementary Figure 1. Expression of phospho-sik3 in normal and osteoarthritic articular cartilage in the knee. (a) Semiserial histological sections of normal cartilage were stained with safranin O-fast
p.r623c p.p976l p.d2847fs p.t2671 p.d2847fs p.r2922w p.r2370h p.c1201y p.a868v p.s952* RING_C BP PHD Cbp HAT_KAT11
 ARID2 p.r623c KMT2D p.v650fs p.p976l p.r2922w p.l1212r p.d1400h DNA binding RFX DNA binding Zinc finger KMT2C p.a51s p.d372v p.c1103* p.d2847fs p.t2671 p.d2847fs p.r4586h PHD/ RING DHHC/ PHD PHD FYR N
ARID2 p.r623c KMT2D p.v650fs p.p976l p.r2922w p.l1212r p.d1400h DNA binding RFX DNA binding Zinc finger KMT2C p.a51s p.d372v p.c1103* p.d2847fs p.t2671 p.d2847fs p.r4586h PHD/ RING DHHC/ PHD PHD FYR N
m 6 A mrna methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer
 SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41556-018-0174-4 In the format provided by the authors and unedited. m 6 A mrna methylation regulates AKT activity to promote the proliferation
SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41556-018-0174-4 In the format provided by the authors and unedited. m 6 A mrna methylation regulates AKT activity to promote the proliferation
Tcf21 MCM ; R26 mtmg Sham GFP Col 1/3 TAC 8W TAC 2W. Postn MCM ; R26 mtmg Sham GFP Col 1/3 TAC 8W TAC 2W
 A Tcf21 MCM ; R26 mtmg Sham GFP Col 1/3 Tcf21 MCM ; R26 mtmg TAC 2W Tcf21 MCM ; R26 mtmg TAC 8W B Postn MCM ; R26 mtmg Sham GFP Col 1/3 Postn MCM ; R26 mtmg TAC 2W Postn MCM ; R26 mtmg TAC 8W Supplementary
A Tcf21 MCM ; R26 mtmg Sham GFP Col 1/3 Tcf21 MCM ; R26 mtmg TAC 2W Tcf21 MCM ; R26 mtmg TAC 8W B Postn MCM ; R26 mtmg Sham GFP Col 1/3 Postn MCM ; R26 mtmg TAC 2W Postn MCM ; R26 mtmg TAC 8W Supplementary
Supplementary Figure 1: Comparison of acgh-based and expression-based CNA analysis of tumors from breast cancer GEMMs.
 Supplementary Figure 1: Comparison of acgh-based and expression-based CNA analysis of tumors from breast cancer GEMMs. (a) CNA analysis of expression microarray data obtained from 15 tumors in the SV40Tag
Supplementary Figure 1: Comparison of acgh-based and expression-based CNA analysis of tumors from breast cancer GEMMs. (a) CNA analysis of expression microarray data obtained from 15 tumors in the SV40Tag
Supplementary Information. Induction of human pancreatic beta cell replication by inhibitors of dual specificity tyrosine regulated kinase
 Journal: Nature Medicine Supplementary Information Induction of human pancreatic beta cell replication by inhibitors of dual specificity tyrosine regulated kinase 1,2 Peng Wang PhD, 1,2 Juan-Carlos Alvarez-Perez
Journal: Nature Medicine Supplementary Information Induction of human pancreatic beta cell replication by inhibitors of dual specificity tyrosine regulated kinase 1,2 Peng Wang PhD, 1,2 Juan-Carlos Alvarez-Perez
Supplementary Figures
 Supplementary Figures Supplementary Fig. 1. Galectin-3 is present within tumors. (A) mrna expression levels of Lgals3 (galectin-3) and Lgals8 (galectin-8) in the four classes of cell lines as determined
Supplementary Figures Supplementary Fig. 1. Galectin-3 is present within tumors. (A) mrna expression levels of Lgals3 (galectin-3) and Lgals8 (galectin-8) in the four classes of cell lines as determined
Supplementary Figure 1 Expression of Crb3 in mouse sciatic nerve: biochemical analysis (a) Schematic of Crb3 isoforms, ERLI and CLPI, indicating the
 Supplementary Figure 1 Expression of Crb3 in mouse sciatic nerve: biochemical analysis (a) Schematic of Crb3 isoforms, ERLI and CLPI, indicating the location of the transmembrane (TM), FRM binding (FB)
Supplementary Figure 1 Expression of Crb3 in mouse sciatic nerve: biochemical analysis (a) Schematic of Crb3 isoforms, ERLI and CLPI, indicating the location of the transmembrane (TM), FRM binding (FB)
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/7/322/ra38/dc1 Supplementary Materials for Dynamic Reprogramming of Signaling Upon Met Inhibition Reveals a Mechanism of Drug Resistance in Gastric Cancer Andrea
www.sciencesignaling.org/cgi/content/full/7/322/ra38/dc1 Supplementary Materials for Dynamic Reprogramming of Signaling Upon Met Inhibition Reveals a Mechanism of Drug Resistance in Gastric Cancer Andrea
Cluster Dendrogram. dist(cor(na.omit(tss.exprs.chip[, c(1:10, 24, 27, 30, 48:50, dist(cor(na.omit(tss.exprs.chip[, c(1:99, 103, 104, 109, 110,
 A Transcriptome (RNA-seq) Transcriptome (RNA-seq) 3. 2.5 2..5..5...5..5 2. 2.5 3. 2.5 2..5..5...5..5 2. 2.5 Cluster Dendrogram RS_ES3.2 RS_ES3. RS_SHS5.2 RS_SHS5. PS_SHS5.2 PS_SHS5. RS_LJ3 PS_LJ3..4 _SHS5.2
A Transcriptome (RNA-seq) Transcriptome (RNA-seq) 3. 2.5 2..5..5...5..5 2. 2.5 3. 2.5 2..5..5...5..5 2. 2.5 Cluster Dendrogram RS_ES3.2 RS_ES3. RS_SHS5.2 RS_SHS5. PS_SHS5.2 PS_SHS5. RS_LJ3 PS_LJ3..4 _SHS5.2
SUPPLEMENTARY FIGURES AND TABLE
 SUPPLEMENTARY FIGURES AND TABLE Supplementary Figure S1: Characterization of IRE1α mutants. A. U87-LUC cells were transduced with the lentiviral vector containing the GFP sequence (U87-LUC Tet-ON GFP).
SUPPLEMENTARY FIGURES AND TABLE Supplementary Figure S1: Characterization of IRE1α mutants. A. U87-LUC cells were transduced with the lentiviral vector containing the GFP sequence (U87-LUC Tet-ON GFP).
Figure S1, Beyer et al.
 Figure S1, eyer et al. Pax7 Myogenin si sitrl Hoechst T = 72h 14 1.8.6.4.2 12 1 8 6 4 2 24h 48h 96h diff. sitrl siset1 212 72h diff. b1 td r t Se km MyH Vinculin Myogenin β-ctin Vinculin MW b1 ka td r
Figure S1, eyer et al. Pax7 Myogenin si sitrl Hoechst T = 72h 14 1.8.6.4.2 12 1 8 6 4 2 24h 48h 96h diff. sitrl siset1 212 72h diff. b1 td r t Se km MyH Vinculin Myogenin β-ctin Vinculin MW b1 ka td r
