SUPPLEMENTARY INFORMATION
|
|
|
- Roderick Waters
- 5 years ago
- Views:
Transcription
1 -. -. SUPPLEMENTARY INFORMATION DOI: 1.1/ncb86 a WAT-1 WAT- BAT-1 BAT- sk-muscle-1 sk-muscle- mir-133b mir-133a mir-6 mir-378 mir-1 mir-85 mir-378 mir-6a mir-18 mir-133a mir- mir- mir-341 mir-196a mir-17 mir-434-3p mir- mir-99 mir-486 mir-361 mir- mir-155 mir-379 mir-181b mir-14 mir-185 mir-15b-5p mir-199a-3p mir-181a mir-199a-5p mir-76 mir-14 mir-13 mir-17 mir-1a mir-78 mir-19 mir-148a mir-3d mir-93 mir-451 mir-191 mir-3 mir-35 mir-15 mir-45 let-7i mir-146a mir-34-3p mir-99b mir- mir-1b mir-34a mir-195 mir-8 mir-18 mir-15a-5p mir-139-5p mir-15b mir-1 mir-335-5p mir-146b mir-13a mir-4 mir-143 mir-1 mir-99a mir-1 mir-c mir-b mir-151-5p mir-18 mir-455 mir-16 mir-16-3p mir-3c mir-16b mir-3 mir-9a mir-3a mir-3 mir-15 let-7g mir-a mir-193 mir-6b mir-15a mir-17 mir-9b b c Non-RT control d P1 mir-193b mir k 134k 1345k 13451k 1345k 13453k 13454k 13455k P mir-193b P3 P4 P1 P P3 P4 P5 P6 P7 P8 P9 P1 P P3 P4 f P5 P5 P6 Wt ob/ob P6 e P7 P7 P8 P9 4.kb P8 P9 P1 Basal 4 O C P11 P1 mir P13 P14 P1 P11 P1 P13 P14 P1 P11 P1 P13 Basal camp Dio P14 log(sample/mean) Supplemental Fig 1 Figure S1 MiR-193b-365 is a brown fat-enriched mirna cluster. (a). Detailed heat map showing the expression levels of 91 mirnas that are expressed in at least one sample at an intensity > 5 arbitrary units and are differentially expressed between the three tissues (P <.1, ANOVA). Red denotes higher and green denotes lower level relative to the mean of the six samples for each mirna. Tissues from 4-5 mice were pooled for each RNA sample preparation. (b). Cap-analysis gene expression (CAGE) Basic and Analysis Databases store original results produced by CAGE-seq, which measures expression levels of transcription start sites by sequencing large numbers of 5 transcript ends, termed CAGE tags. CAGE tag distribution across the genomic region around -365 is shown here. A black arrow represents a single 5 transcript tag, and a red arrow represents a cluster of 5 transcript tags. There is no CAGE tag between and -1 in the direction of transcription, supporting the idea that and are co-transcribed. (c). RT- PCR analysis of the primary transcript of Random primers were used to generate cdna from primary brown adipocytes. 14 pairs of primers (P1 to P14) depicted in the diagram above were then used to amplify segments of the putative primary transcript that includes both and. The amplified fragments covered the entire region between and, indicating that -365 is a bona fide cotranscribed mirna cluster. (d). Real-time PCR analysis of and mir- 365 expression in BAT of wild- type and ob/ob mice (1 week old). n=5. (e). 8 week old male mice were exposed to 4 o C for 4 hrs to induce thermogenesis in BAT. RNAs of interscapular BAT were harvested and real-time PCR was performed to examine the expression of and, and two markers genes upregulated during thermogenesis, type II iodothyronine deiodinase (Dio) and uncoupling protein 1 (). n=5. (f). Brown fat SVF cells were cultured and differentiated to day 6 and stimulated with 5uM dibutyrul camp for 4h. The expression of, and was examined by RT-PCR. n=3. P < 5, Student s t-test; Means ± SEM Macmillan Publishers Limited. All rights reserved.
2 SUPPLEMENTARY INFORMATION a 1 5 d 1μm LNA-miR-1 LNA- LNA-miR-193a+b FITC - CD LNA-miR-1 LNA-miR-193a LNA- LNA-miR-193a+b b 65.8% APC - CD11b 87.9% PE - Sca1 LNA mir-193a+b LNA 6 8 e f g.. LNA- Gapdh Cebpa mir-1 LNA- LNA Pgc-1a Pparg Prdm16 LNA- LNA- LNA- LNA- LNA-+365 LNA-+365 c Relative Expression Relative Expression Cidea LNA-193a+b LNA-365 Adipogenesis markers AdipoQ Dio CebpA Pgc1a Brown fat markers Ppara Pparg Prdm16 i h LNA-miR-193a+b LNA- Fig S AdipoQ Cebpa LNA-miR-193a+b LNA- 5μm Glut4 Pparg Figure S MiR-193b-365 is required for brown fat adipogenesis. (a). Brown fat SVF cells were FACS-sorted to enrich brown preadipocyte population (CD45 -, CD11b -, Sca1 + ). Sorted cells were transfected with LNA inhibitors for mir-193a+b and, then induced to differentiation for 4 day. (b). ORO staining and (c). Real time-pcr were used to examine the effects of blocking these mirnas on brown fat adipogenesis. n=3. (d). SVF cells were transfected with LNA mirna inhibitors (1nM) one day before differentiation. 4 days after differentiation, representative micrographs were taken and are shown here. (e). Western blot to examine the expression of in response to blocking and/or during brown fat adipogenesis. (f, g). Primary brown preadipocytes were cultured and differentiated to day 6. These cells were transfected by LNA mirna inhibitors as indicated in figure. 3 days after, RNAs were extracted and RT-PCR was performed to examine the expression of (f) mirnas and (g) brown fat markers. (h). Subcutaneous white fat SVF cells were transfected with LNA mirna inhibitors for mir-193a+b and, and then were induced to differentiation for 4 day. ORO staining and Real time-pcr (i) were used to examine the effects of blocking these mirnas on white fat adipogenesis. n=3. P < 5, Student s t-test; Means ± SEM Macmillan Publishers Limited. All rights reserved.
3 SUPPLEMENTARY INFORMATION c e a D D6 Runx1t1-P1 Runx1t1-P1 Runx1t1-P Runx1t1-P b GFP Runx1T1 d Runx1t1 AdipoQ 5μm Adipogenesis markers Cebpα Pparγ Runx1t1 Cidea Dio Brown fat markers Pgc1α Pparα Prdm16 f LNA-miR-193a+b LNA-miR-193a+b Runx1t Gapdh 8 Figure S3 MiR-193b regulates adipogenesis by targeting Runx1t1. (a). Real-time PCR to examine the change of Run1t1 during brown fat adipogenesis. (b-e). Brown fat SVF cells were infected by retrovius expressing RUNX1T1, and then differentiated for 4 days. (b) ORO staining and (c-e) Real-time PCR were performed to examine the effects of ectopic Runx1t1 on brown fat adipogenesis. n=3. (f). Brown fat SVF cells were transfected with LNA mirna inhibitors for mir-193a and mir- 193b and then were induced to differentiate. Western was performed to examine the level of Runx1t1. P < 5, Student s t-test; Means ± SEM Macmillan Publishers Limited. All rights reserved.
4 SUPPLEMENTARY INFORMATION a Primary brown adipocytes Primary brown adipocytes b mir-193a AACUGGCCUACAAAGUCCCAGU AACUGGCCCACAAAGUCCCGCU mir-193b mir c mir-13 d mir-193a CC1 mir-17 Myf5 Myf6 MyoG Ckm Tnni Tnnt3 Casq1 e f Myosin GAPDH Cdon Gapdh Igfbp5 Figure S4 MiR-193b-365 represses myogenesis of CC1. (a). Realtime- PCR analysis of and in differentiated CC1 myoblasts retrovirally expressing these two mirnas. Primary brown adipocytes directly isolated from animals were used as a positive control. Ectopic expression resulted in a mirna level about ~3 and ~6.5-fold higher than in primary brown adipocytes for and, respectively, indicating that the mirna expression levels were in a physiological range. n=3. (b). Diagram to show the nucleotide sequences of mir-193a (upper), and mir- 193b (lower) and their relative positions in the genome. (c). Representative micrographs of cells (Day 6) differentiated from CC1 myoblasts expressing retroviral mir-193a or the control vector. (d). Real-time PCR analysis of cells depicted in (c) for their expression of muscle markers, Myf5, Myf6, Myog, muscle creatine kinase [Ckm], troponin I type [Tnni], troponin T type 3 [Tnnt3], calsequestrin 1 [Casq1]. n=3. (e). Western blot with triplicate biological repeats to examine the expression of myosin upon ectopic expression of and/or. n=3. (f). Western analysis for predicted targets, cell adhesion molecule-related/down-regulated by oncogenes (Cdon) and insulin-like growth factor binding protein 5 (Igfbp5), in CC1 cells expressing or a control vector. n=3. P < 5, Student s t-test; Means ± SEM Macmillan Publishers Limited. All rights reserved.
5 SUPPLEMENTARY INFORMATION a Pparγ +365 Cebpα b AdipoQ BAT_D6 CC1 D5 Cebpα Pparγ c Pax MyoD Gapdh 8 d f 5μm Myogenic condition Ckm Myf5 Myf6 Myod Myog Tnnt3 Tnni e Adipogenic condition g AdipoQ Cebpa Ckm Myf5 Myf6 Myod Myog Tnni Tnnt3 Pparg Lpl Pgc1a Figure S5 induces adipogenesis in myoblasts. (a). CC1 cells were infected with retrovirus expressing mir-193 and/or and then exposed to myogenic condition for 6 days. Real-time PCR analysis to examine the expression of Pparγ and Cebpα. n=3. (b). CC1 myoblasts were infected by retrovirus and exposed to proadipogenic differentiation conditions for 5 days. RNAs were harvested, and Real-time PCR was used to compare the levels of Pparg, Cebpa, AdipoQ and in these cells and their levels in primary brown adipocyte cultures. Values of BAT cells are set at. n=3, (c). Western blot for myogenic markers Pax3, MyoD and brown fat marker. n=3. (d-g). Primary human myoblasts were infected with virus expressing or vector and then exposed to myogenic condition (d, e) or adipogenic condition (f, g) for 6 days. Representative micrographs of cells were shown in (d). Realtime PCR was performed to examine the expression of myogenic markers (e, f) and adipogenic markers (g). n=3. P < 5, Student s t-test; Means ± SEM Macmillan Publishers Limited. All rights reserved.
6 SUPPLEMENTARY INFORMATION a b c d WAT Cox5b CC1 Pparγ Cidea Pparα 3 1 Pparγ WAT CC1 Myf5 Myod Cebpα Myog mir-193b mir-365 f Amplicon 1345k 13451k 1345k 13453k 13454k WAT Amplicon Amplicon BAT sh- m e g CC1 Amplicon sinc sippara Amplicon i k l h j Retroviral AdipoQ Amplicon WT PPARα KO Day (-3) Day (-1) Day () Day (3) LNA mir-193a+b LNA Adipogenesis markers Cebpa Inhibitor Pparg Brown fat markers Adipogenic induction Cidea Dio Ppara Prdm16 ORO staining RNA extraction LNA mir-193a+b LNA LNA mir-193a+b LNA PPARα -365 Runx1t1 Myf5+ Cdon Igfbp5 Brown adipocytes Figure S6 Figure S6-365 is regulated by. (a) Real-time PCR analysis of differentiated primary white adipocytes and (b) differentiated CC1 myoblasts that retrovirally express. White preadipocytes and CC1 myoblasts were cultured to about 5% confluence and infected by retroviruses expressing Prdm16. Six days after differentiation, cells were harvested and analyzed by real-time PCR. n=3. P < 5. (c). RT-PCR primers were designed to amplify four fragments in the primary transcript between and. (d). Subcutaneous white pre-adipocytes and (e). CC1 myoblasts were infected by retrovirus expressing Prdm16 two days before differentiation. 5 days after differentiation, real-time PCR were performed to examine the expression of pri n=3. (f), Primary brown preadipocytes were infected by retrovirus expressing shrna targeting two days before differentiation. By D5 of differentiation, real-time PCR was used to examine the mrna level of pri n=3. (g), Muscle Brown fat SVF cells were transfected with sirnas targeting PPARα, and differentiated for 3 days. RT-PCR was performed to examine the expression of pri n=3. (h), RT-PCR was performed to examine the expression of pri--365 in brown adipose tissue isolated from PPARα knockout mice (8 week old, Male). Age-matched wild- type mice were used as control. n=6. (i). The diagram of experimental design for (i-l). (j). ORO staining and (k, l) Real-time PCR were performed to examine the effects of LNA mirna inhibitors on brown fat adipogenesis in the presence of ectopic Prdm16. (m) Brown adipocytes arise from a Myf5-positive, myoblastic lineage. During this process, Prdm16 is strongly induced. Probably through interactions with Ppara, Prdm16 promotes the expression of the mir- 193b-365 cluster, which is required for brown fat adipogenesis. At the same time, -365 blocks the otherwise myogenic potential of brown preadipocytes. P < 5, Student s t-test; Means ± SEM Macmillan Publishers Limited. All rights reserved.
7 Supplementary Table 1. Real-time PCR primers FORWARD REVERSE 18S GTAACCCGTTGAACCCCATT CCATCCAATCGGTAGTAGC AdipoQ CGATTGTCAGTGGATCTGACG CAACAGTAGCATCCTGAGCCCT Casq1 GGGGGCTCATGGTAGAGGAG CTGGCACTGCTGTTTGTACTG Cdon ATCCAGACCTCGGACCCTTAT GGCAGTAACAGGTTTAGCAGAAC Cebpa TGCGCAAGAGCCGAGATAAA CCTTCTGTTGCGTCTCCACG Cidea TGCTCTTCTGTATCGCCCAGT GCCGTGTTAAGGAATCTGCTG Ckm GCAAGCACCCCAAGTTTGA ACCTGTGCCGCGCTTCT Cox8b GAACCATGAAGCCAACGACT GCGAAGTTCACAGTGGTTCC Dio CAGTGTGGTGCACGTCTCCAATC TGAACCAAAGTTGACCACCAG Elovl3 TCCGCGTTCTCATGTAGGTCT GGACCTGATGCAACCCTATGA Eva1 GTCCCAACCAGACCATCAAC CTCCATCTTGCTCTGGAAGC ACAAGCTGGTGGTGGAATGTG CCTTTGGCTCATGCCCTTT Glut4 CTGTCGCTGGTTTCTCCAACT CCCATAGCATCCGCAACATA Igfbp5 CCCTGCGACGAGAAAGCTC GCTCTTTTCGTTGAGGCAAACC Myf5 AAGGCTCCTGTATCCCCTCAC TGACCTTCTTCAGGCGTCTAC Myf6 ATCAGCTACATTGAGCGTCTACA CCTGGAATGATCCGAAACACTTG MyoD CGCCACTCCGGGACATAG GAAGTCGTCTGCTGTCTCAAAGG MyoG AGCGCAGGCTCAAGAAAGTGAATG CTGTAGGCGCTCAATGTACTGGAT Nrip1 GGCTGTCTGATTCCATCGTGA CCAGCAATGTGCTATCCTGTT Pgc1a CCCTGCCATTGTTAAGACC TGCTGCTGTTCCTGTTTTC Ppara AGAGCCCCATCTGTCCTCTC ACTGGTAGTCTGCAAAACCAAA Pparg GTGCCAGTTTCGATCCGTAGA GGCCAGCATCGTGTAGATGA Prdm16 CAGCACGGTGAAGCCATTC GCGTGCATCCGCTTGTG Retnla TCCACTCTGGATCTCCCAAGA TCCCTCCACTGTAACGAAGACTC Tnni AGAGTGTGATGCTCCAGATAGC AGCAACGTCGATCTTCGCA Tnnt3 GGAACGCCAGAACAGATTGG TGGAGGACAGAGCCTTTTTCTT 11 Macmillan Publishers Limited. All rights reserved.
8 ACTGCCACACCTCCAGTCATT CTTTGCCTCACTCAGGATTGG Casq1 (human) CACCCAAGTCAGGGGTACAG GTGCCAGCACCTCATACTTCT myog (human) GCTGTATGAGACATCCCCCTA CGACTTCCTCTTACACACCTTAC CEBPA (human) ACTGCGATGGGGGTCACCAG CTCCGGTCCCTCTGGGATGG FABP4 (human) AGCACCATAACCTTAGATGGGG CGTGGAAGTGACGCCTTTCA Tnni (human) ATCTGCGGGGCAAGTTCAAG AGGACTCGGACTCAAACATCT CKM (human) ATGCCATTCGGTAACACCCAC TCTTGTAGAGTTCAAGGGTCAGT PPARg (human) CCTATTGACCCAGAAAGCGATT CATTACGGAGAGATCCACGGA MyoD1 (human) CTTTGCCACAACGGACGACT GTGCTCTTCGGGTTTCAGGAG Myf6 (human) GGAGCGCCATCAGCTATATTG ATCCGCACCCTCAAGATTTTC Myf5 (human) GTCCCCCGAGGGTGAATTTG GCCCACATGAGGCAGTGAC LPL (human) TCATTCCCGGAGTAGCAGAGT GGCCACAAGTTTTGGCACC Tnnt3 (human) TCCAGAAGAAGCGTCAGAACA TCTCTGCACGAATCCTCTGCT PGC1 (human) CCAGGAGGCAGAAGAGCCGT GCTCCGAGCAGGGACGTCTT Runx1t1-P1 ATGCCTGATCGTACCGAGAAG GTCGTTGGCGTAAATGAGCTG Runx1t1-P ATTTACGCCAACGACATTAACGA CTGAGTTGCCTAGCACCACA Pri--365 Pri--365 Amplicon Pri--365 Pri--365 AGGGAGACGGTTGCAGGGTA GCTGGCTGTCACATGATAA GCTTCCCTCCCCTGCCAATA TCACTCCGTGCCAGTCCAGA TGTGACAGCCAGCAACTCAGC GGGAAGGATAACTGAGTGTCT CCTTTCCTAGGTAAGACTCCACCAC AGCACTGATGCCAACAGGCC 11 Macmillan Publishers Limited. All rights reserved.
9 Supplementary Table. Cloning primers Name Forward Reverse mmu- mir- 193 mmu- mir- 193b mmu- mir- 365 CDON -3UTR IGFBP 5-3UTR CDON - 3UTR- Mut IGFBP 5-3UTR- Mut RUNX 1T1-3UTR RUNX 1T1_ Mut ccgctcgagccccgtcgtgtaaaccttcga G ccgctcgagcccctacagcaggaagaggt AGG ccgctcgagcccgttgctgtacagatgctg AG CCGCTCGAGGAGGAGTGTGAGCAGA CCT CCGCTCGAGCAGTTGGCTGGGAAGG AA TCTTCGTGTTCCCACTGCAAGGATCC AACTGTACCACACAGGCCTGG CCCAACTCAAACTTCAATATATCTTAG GGATCCATCCTAGACCTAAGTCTCTC CTTTCTGC CCGCTCGAGCTGTCAGAGGAAAGACA CCAC TGAGGCGATAGTAGAACACAGAGGGA TCCAACGGGTCGTAATGACTTACTGT G ggaattcttaattaaaaactttccagtcgcttc TATCC ggaattcttaattaaaaacttccctttgacgga ATGAG ggaattcttaattaaaaatattcaccactggct TTGAG ATAAGAATGCGGCCGCTCCCCTGTTTT CCAGATGA ATAAGAATGCGGCCGCTGTGGCTCAC AGCGAAGA CCAGGCCTGTGTGGTACAGTTGGATC CTTGCAGTGGGAACACGAAGA GCAGAAAGGAGAGACTTAGGTCTAGG ATGGATCCCTAAGATATATTGAAGTTTG AGTTGGG ATAAGAATGCGGCCGCTTAGCATTTTT GCTTTCAACAG CACAGTAAGTCATTACGACCCGTTGGA TCCCTCTGTGTTCTACTATCGCCTCA 11 Macmillan Publishers Limited. All rights reserved.
10 Supplementary Table 3. Primers to detect the co-transcript of -365 mmu--365 transcript; P1-F mmu--365 transcript; P1-R mmu--365 transcript; P-F mmu--365 transcript; P-R mmu--365 transcript; P3-F mmu--365 transcript; P3-R mmu--365 transcript; P4-F mmu--365 transcript; P4-R mmu--365 transcript; P5-F mmu--365 transcript; P5-R mmu--365 transcript; P6-F mmu--365 transcript; P6-R mmu--365 transcript; P7-F mmu--365 transcript; P7-R mmu--365 transcript; P8-F mmu--365 transcript; P8-R mmu--365 transcript; P9-F mmu--365 transcript; P9-R mmu--365 transcript; P1-F mmu--365 transcript; P1-R mmu--365 transcript; P11-F mmu--365 transcript; P11-R mmu--365 transcript; P1-F mmu--365 transcript; P1-R mmu--365 transcript; P13-F mmu--365 transcript; P13-R CGAGAGGGGAGGTAGCG AGTAAGCTGCTTCAGACTCTCG CAGCTTACTACCTTCCCCTAA CAATCCAGATTGGGAGAA TTCACGTCTTCTGAGGACACATCC AGGGAGGTTCATCTGCACCC GAACCTCCCTACAGCAGGAAGAG CGGAGCTAGATAAGAATACCCTCCC GTGCCAAATTCTCGGAACAAAC TGTCTGTTGTCATTCAGGTGACTGA CACACACATTCCATATATGCACCC GCTCGTTAATTCACAGAAAGCACTC GCCAGATCCCTCTACTCTGCTAGA TATTGGCAGGGGAGGGAAGC GCTTCCCTCCCCTGCCAATA CCTTTCCTAGGTAAGACTCCACCAC GAAAGGCCCTGGGTTCATCC TGGGAAGGGACCACAAAGCG TCACTCCGTGCCAGTCCAGA AGCACTGATGCCAACAGGCC TGCTCAAGACAGCCTGGACC TGCTTCTAGGCCCATGGCTT AGGGAGACGGTTGCAGGGTA TGTGACAGCCAGCAACTCAGC GCTGGCTGTCACATGATAA GGGAAGGATAACTGAGTGTCT 11 Macmillan Publishers Limited. All rights reserved.
11 mmu--365 transcript; P14-F mmu--365 transcript; P14-R TCCTTCCCTGAGGCTTCACC AACCAATCAGCTGGCGGGAG 11 Macmillan Publishers Limited. All rights reserved.
12 Supplementary Table 4. ChIP primers ChIP 193b 1F ChIP 193b 1R ChIP 193b F ChIP 193b R ChIP 193b 3F ChIP 193b 3R GTGTGCTTGCAGCGGTGTCC CCCCCAAAGCCAGGGAGTCA GGTGGATGGGGTGGGGTGTT CCCCTCTGGGGTTCGCAGTT GTTAGGGGACCCCCTCCCTC GCACATTCCTCCGCTCCCCT 11 Macmillan Publishers Limited. All rights reserved.
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
SUPPLEMENTARY INFORMATION
 doi: 1.138/nature7221 Brown fat selective genes 12 1 Control Q-RT-PCR (% of Control) 8 6 4 2 Ntrk3 Cox7a1 Cox8b Cox5b ATPase b2 ATPase f1a1 Sirt3 ERRα Elovl3/Cig3 PPARα Zic1 Supplementary Figure S1. stimulates
doi: 1.138/nature7221 Brown fat selective genes 12 1 Control Q-RT-PCR (% of Control) 8 6 4 2 Ntrk3 Cox7a1 Cox8b Cox5b ATPase b2 ATPase f1a1 Sirt3 ERRα Elovl3/Cig3 PPARα Zic1 Supplementary Figure S1. stimulates
Supplemental Information. Brown Adipogenic Reprogramming. Induced by a Small Molecule
 Cell Reports, Volume 18 Supplemental Information rown dipogenic Reprogramming Induced by a Small Molecule aoming Nie, Tao Nie, Xiaoyan Hui, Ping Gu, Liufeng Mao, Kuai Li, Ran Yuan, Jiashun Zheng, Haixia
Cell Reports, Volume 18 Supplemental Information rown dipogenic Reprogramming Induced by a Small Molecule aoming Nie, Tao Nie, Xiaoyan Hui, Ping Gu, Liufeng Mao, Kuai Li, Ran Yuan, Jiashun Zheng, Haixia
GPR120 *** * * Liver BAT iwat ewat mwat Ileum Colon. UCP1 mrna ***
 a GPR120 GPR120 mrna/ppia mrna Arbitrary Units 150 100 50 Liver BAT iwat ewat mwat Ileum Colon b UCP1 mrna Fold induction 20 15 10 5 - camp camp SB202190 - - - H89 - - - - - GW7647 Supplementary Figure
a GPR120 GPR120 mrna/ppia mrna Arbitrary Units 150 100 50 Liver BAT iwat ewat mwat Ileum Colon b UCP1 mrna Fold induction 20 15 10 5 - camp camp SB202190 - - - H89 - - - - - GW7647 Supplementary Figure
A synergistic anti-obesity effect by a combination of capsinoids and cold temperature through the promotion of beige adipocyte biogenesis
 A synergistic anti-obesity effect by a combination of capsinoids and cold temperature through the promotion of beige adipocyte biogenesis Kana Ohyama, 1,2 Yoshihito Nogusa, 1 Kosaku Shinoda, 2 Katsuya
A synergistic anti-obesity effect by a combination of capsinoids and cold temperature through the promotion of beige adipocyte biogenesis Kana Ohyama, 1,2 Yoshihito Nogusa, 1 Kosaku Shinoda, 2 Katsuya
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2211 a! mir-143! b! mir-103/107! let-7a! mir-144! mir-122a! mir-126-3p! mir-194! mir-27a! mir-30c! Figure S1 Northern blot analysis of mir-143 expression dependent on feeding conditions.
DOI: 10.1038/ncb2211 a! mir-143! b! mir-103/107! let-7a! mir-144! mir-122a! mir-126-3p! mir-194! mir-27a! mir-30c! Figure S1 Northern blot analysis of mir-143 expression dependent on feeding conditions.
1.5 ASK1KO fed. fasted 16 hrs w/o water. Fed. 4th. 4th WT ASK1KO N=29, 11(WT), ,5(ASK1KO) ASK1KO ASK1KO **** Time [h]
![1.5 ASK1KO fed. fasted 16 hrs w/o water. Fed. 4th. 4th WT ASK1KO N=29, 11(WT), ,5(ASK1KO) ASK1KO ASK1KO **** Time [h] 1.5 ASK1KO fed. fasted 16 hrs w/o water. Fed. 4th. 4th WT ASK1KO N=29, 11(WT), ,5(ASK1KO) ASK1KO ASK1KO **** Time [h]](/thumbs/87/97258189.jpg) 7: 13: 19: 1: 7: 151117 a 151117 4th 4th b c RQ.95 KO.9.85.8.75.7 light dark light dark.65 7: 19: 7: 19: 7: Means ± SEM, N=6 RQ 1..9.8.7.6.6 KO CL (-) CL (+) ibat weight ratio (/body weight) [%].5.4.3.2.1
7: 13: 19: 1: 7: 151117 a 151117 4th 4th b c RQ.95 KO.9.85.8.75.7 light dark light dark.65 7: 19: 7: 19: 7: Means ± SEM, N=6 RQ 1..9.8.7.6.6 KO CL (-) CL (+) ibat weight ratio (/body weight) [%].5.4.3.2.1
SUPPLEMENTARY DATA. Supplementary Table 1. Primer sequences for qrt-pcr
 Supplementary Table 1. Primer sequences for qrt-pcr Gene PRDM16 UCP1 PGC1α Dio2 Elovl3 Cidea Cox8b PPARγ AP2 mttfam CyCs Nampt NRF1 16s-rRNA Hexokinase 2, intron 9 β-actin Primer Sequences 5'-CCA CCA GCG
Supplementary Table 1. Primer sequences for qrt-pcr Gene PRDM16 UCP1 PGC1α Dio2 Elovl3 Cidea Cox8b PPARγ AP2 mttfam CyCs Nampt NRF1 16s-rRNA Hexokinase 2, intron 9 β-actin Primer Sequences 5'-CCA CCA GCG
control kda ATGL ATGLi HSL 82 GAPDH * ** *** WT/cTg WT/cTg ATGLi AKO/cTg AKO/cTg ATGLi WT/cTg WT/cTg ATGLi AKO/cTg AKO/cTg ATGLi iwat gwat ibat
 body weight (g) tissue weights (mg) ATGL protein expression (relative to GAPDH) HSL protein expression (relative to GAPDH) ### # # kda ATGL 55 HSL 82 GAPDH 37 2.5 2. 1.5 1..5 2. 1.5 1..5.. Supplementary
body weight (g) tissue weights (mg) ATGL protein expression (relative to GAPDH) HSL protein expression (relative to GAPDH) ### # # kda ATGL 55 HSL 82 GAPDH 37 2.5 2. 1.5 1..5 2. 1.5 1..5.. Supplementary
Nature Immunology: doi: /ni Supplementary Figure 1. Huwe1 has high expression in HSCs and is necessary for quiescence.
 Supplementary Figure 1 Huwe1 has high expression in HSCs and is necessary for quiescence. (a) Heat map visualizing expression of genes with a known function in ubiquitin-mediated proteolysis (KEGG: Ubiquitin
Supplementary Figure 1 Huwe1 has high expression in HSCs and is necessary for quiescence. (a) Heat map visualizing expression of genes with a known function in ubiquitin-mediated proteolysis (KEGG: Ubiquitin
Fig. S1. Validation of ChIP-seq binding sites by single gene ChIP-PCR Fig. S2. Transactivation potential of PPAR
 Fig. S1. Validation of ChIP-seq binding sites by single gene ChIP-PCR ChIP-PCR was performed on PPARγ and RXR-enriched chromatin harvested during adipocyte differentiation at day and day 6 as described
Fig. S1. Validation of ChIP-seq binding sites by single gene ChIP-PCR ChIP-PCR was performed on PPARγ and RXR-enriched chromatin harvested during adipocyte differentiation at day and day 6 as described
SUPPLEMENTARY INFORMATION. Supplemental Figure 1. Body weight and blood glucose parameters of chow-diet (CD)
 SUPPLEMENTARY INFORMATION LEGENDS Supplemental Figure. Body weight and blood glucose parameters of chow-diet (CD) fed and high-fat diet (HFD) fed mice. (A) Body weight was measured at the beginning of
SUPPLEMENTARY INFORMATION LEGENDS Supplemental Figure. Body weight and blood glucose parameters of chow-diet (CD) fed and high-fat diet (HFD) fed mice. (A) Body weight was measured at the beginning of
Supplementary Figure 1. DJ-1 modulates ROS concentration in mouse skeletal muscle.
 Supplementary Figure 1. DJ-1 modulates ROS concentration in mouse skeletal muscle. (a) mrna levels of Dj1 measured by quantitative RT-PCR in soleus, gastrocnemius (Gastroc.) and extensor digitorum longus
Supplementary Figure 1. DJ-1 modulates ROS concentration in mouse skeletal muscle. (a) mrna levels of Dj1 measured by quantitative RT-PCR in soleus, gastrocnemius (Gastroc.) and extensor digitorum longus
Metabolic ER stress and inflammation in white adipose tissue (WAT) of mice with dietary obesity.
 Supplementary Figure 1 Metabolic ER stress and inflammation in white adipose tissue (WAT) of mice with dietary obesity. Male C57BL/6J mice were fed a normal chow (NC, 10% fat) or a high-fat diet (HFD,
Supplementary Figure 1 Metabolic ER stress and inflammation in white adipose tissue (WAT) of mice with dietary obesity. Male C57BL/6J mice were fed a normal chow (NC, 10% fat) or a high-fat diet (HFD,
Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v)
 SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
Supplementary Information
 Supplementary Information GADD34-deficient mice develop obesity, nonalcoholic fatty liver disease, hepatic carcinoma and insulin resistance Naomi Nishio and Ken-ichi Isobe Department of Immunology, Nagoya
Supplementary Information GADD34-deficient mice develop obesity, nonalcoholic fatty liver disease, hepatic carcinoma and insulin resistance Naomi Nishio and Ken-ichi Isobe Department of Immunology, Nagoya
Supplementary Table 1.
 Supplementary Table 1. Expression of genes involved in brown fat differentiation in WAT of db/db mice treated with HDAC inhibitors. Data are expressed as fold change (FC) versus control. symbol FC SAHA
Supplementary Table 1. Expression of genes involved in brown fat differentiation in WAT of db/db mice treated with HDAC inhibitors. Data are expressed as fold change (FC) versus control. symbol FC SAHA
Supplementary Table 2. Plasma lipid profiles in wild type and mutant female mice submitted to a HFD for 12 weeks wt ERα -/- AF-1 0 AF-2 0
 Supplementary Table 1. List of specific primers used for gene expression analysis. Genes Primer forward Primer reverse Hprt GCAGTACAGCCCCAAAATGG AACAAAGTCTGGCCTGTATCCA Srebp-1c GGAAGCTGTCGGGGTAGCGTC CATGTCTTCAAATGTGCAATCCAT
Supplementary Table 1. List of specific primers used for gene expression analysis. Genes Primer forward Primer reverse Hprt GCAGTACAGCCCCAAAATGG AACAAAGTCTGGCCTGTATCCA Srebp-1c GGAAGCTGTCGGGGTAGCGTC CATGTCTTCAAATGTGCAATCCAT
Supplementary Figure 1
 Supplementary Figure 1 a Percent of body weight! (%) 4! 3! 1! Epididymal fat Subcutaneous fat Liver SD Percent of body weight! (%) ** 3! 1! SD Percent of body weight! (%) 6! 4! SD ** b Blood glucose (mg/dl)!
Supplementary Figure 1 a Percent of body weight! (%) 4! 3! 1! Epididymal fat Subcutaneous fat Liver SD Percent of body weight! (%) ** 3! 1! SD Percent of body weight! (%) 6! 4! SD ** b Blood glucose (mg/dl)!
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1. Differential expression of mirnas from the pri-mir-17-92a locus.
 Supplementary Figure 1 Differential expression of mirnas from the pri-mir-17-92a locus. (a) The mir-17-92a expression unit in the third intron of the host mir-17hg transcript. (b,c) Impact of knockdown
Supplementary Figure 1 Differential expression of mirnas from the pri-mir-17-92a locus. (a) The mir-17-92a expression unit in the third intron of the host mir-17hg transcript. (b,c) Impact of knockdown
Histone demethylase JMJD1A coordinates acute and chronic adaptation to cold stress via thermogenic phospho-switch. Abe et al.
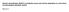 Histone demethylase JMJD1A coordinates acute and chronic adaptation to cold stress via thermogenic phospho-switch Abe et al. a BAT 1 kb Ucp1 H3Kme3 H3K7ac (., ) (., ) -13 kb -. kb -. kb b scwat Chronic
Histone demethylase JMJD1A coordinates acute and chronic adaptation to cold stress via thermogenic phospho-switch Abe et al. a BAT 1 kb Ucp1 H3Kme3 H3K7ac (., ) (., ) -13 kb -. kb -. kb b scwat Chronic
A microrna-34a/fgf21 Regulatory Axis and Browning of White Fat
 A microrna-34a/fgf21 Regulatory Axis and Browning of White Fat Jongsook Kim Kemper, Ph.D Department of Molecular and Integrative Physiology, University of Illinois at Urbana-Champaign, USA 213 International
A microrna-34a/fgf21 Regulatory Axis and Browning of White Fat Jongsook Kim Kemper, Ph.D Department of Molecular and Integrative Physiology, University of Illinois at Urbana-Champaign, USA 213 International
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/2/1/ra81/dc1 Supplementary Materials for Delivery of MicroRNA-126 by Apoptotic Bodies Induces CXCL12- Dependent Vascular Protection Alma Zernecke,* Kiril Bidzhekov,
www.sciencesignaling.org/cgi/content/full/2/1/ra81/dc1 Supplementary Materials for Delivery of MicroRNA-126 by Apoptotic Bodies Induces CXCL12- Dependent Vascular Protection Alma Zernecke,* Kiril Bidzhekov,
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2638 Figure S1 Morphological characteristics of fetal testes and ovaries from 6.5-20 developmental weeks. Representative images of Hematoxylin and Eosin staining of testes and ovaries over
DOI: 10.1038/ncb2638 Figure S1 Morphological characteristics of fetal testes and ovaries from 6.5-20 developmental weeks. Representative images of Hematoxylin and Eosin staining of testes and ovaries over
Figure S1. Reduction in glomerular mir-146a levels correlate with progression to higher albuminuria in diabetic patients.
 Supplementary Materials Supplementary Figures Figure S1. Reduction in glomerular mir-146a levels correlate with progression to higher albuminuria in diabetic patients. Figure S2. Expression level of podocyte
Supplementary Materials Supplementary Figures Figure S1. Reduction in glomerular mir-146a levels correlate with progression to higher albuminuria in diabetic patients. Figure S2. Expression level of podocyte
MicroRNA networks regulate development of brown adipocytes
 MicroRNA networks regulate development of brown adipocytes The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation As Published Publisher
MicroRNA networks regulate development of brown adipocytes The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation As Published Publisher
RNA interference induced hepatotoxicity results from loss of the first synthesized isoform of microrna-122 in mice
 SUPPLEMENTARY INFORMATION RNA interference induced hepatotoxicity results from loss of the first synthesized isoform of microrna-122 in mice Paul N Valdmanis, Shuo Gu, Kirk Chu, Lan Jin, Feijie Zhang,
SUPPLEMENTARY INFORMATION RNA interference induced hepatotoxicity results from loss of the first synthesized isoform of microrna-122 in mice Paul N Valdmanis, Shuo Gu, Kirk Chu, Lan Jin, Feijie Zhang,
SUPPLEMENTARY INFORMATION. Supp. Fig. 1. Autoimmunity. Tolerance APC APC. T cell. T cell. doi: /nature06253 ICOS ICOS TCR CD28 TCR CD28
 Supp. Fig. 1 a APC b APC ICOS ICOS TCR CD28 mir P TCR CD28 P T cell Tolerance Roquin WT SG Icos mrna T cell Autoimmunity Roquin M199R SG Icos mrna www.nature.com/nature 1 Supp. Fig. 2 CD4 + CD44 low CD4
Supp. Fig. 1 a APC b APC ICOS ICOS TCR CD28 mir P TCR CD28 P T cell Tolerance Roquin WT SG Icos mrna T cell Autoimmunity Roquin M199R SG Icos mrna www.nature.com/nature 1 Supp. Fig. 2 CD4 + CD44 low CD4
Figure SⅠ: Expression of mir-155, mir-122 and mir-196a in allografts compared with
 Figure SⅠ: Expression of mir-155, mir-122 and mir-196a in allografts compared with isografts (control) at the 2nd week, 4th and 8th week by RT-PCR. At the advanced stage, the expression of these three
Figure SⅠ: Expression of mir-155, mir-122 and mir-196a in allografts compared with isografts (control) at the 2nd week, 4th and 8th week by RT-PCR. At the advanced stage, the expression of these three
Supplementary Material
 Supplementary Material Summary: The supplementary information includes 1 table (Table S1) and 4 figures (Figure S1 to S4). Supplementary Figure Legends Figure S1 RTL-bearing nude mouse model. (A) Tumor
Supplementary Material Summary: The supplementary information includes 1 table (Table S1) and 4 figures (Figure S1 to S4). Supplementary Figure Legends Figure S1 RTL-bearing nude mouse model. (A) Tumor
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Supplementary Figure 1 IL-27 IL
 Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression.
 Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/407/ra127/dc1 Supplementary Materials for Loss of FTO in adipose tissue decreases Angptl4 translation and alters triglyceride metabolism Chao-Yung Wang,* Shian-Sen
www.sciencesignaling.org/cgi/content/full/8/407/ra127/dc1 Supplementary Materials for Loss of FTO in adipose tissue decreases Angptl4 translation and alters triglyceride metabolism Chao-Yung Wang,* Shian-Sen
Supplementary Figure 1 a
 Supplementary Figure a Normalized expression/tbp (A.U.).6... Trip-br transcripts Trans Trans Trans b..5. Trip-br Ctrl LPS Normalized expression/tbp (A.U.) c Trip-br transcripts. adipocytes.... Trans Trans
Supplementary Figure a Normalized expression/tbp (A.U.).6... Trip-br transcripts Trans Trans Trans b..5. Trip-br Ctrl LPS Normalized expression/tbp (A.U.) c Trip-br transcripts. adipocytes.... Trans Trans
Supplementary Information. Induction of p53-independent apoptosis by ectopic expression of HOXA5
 Supplementary Information Induction of p53-independent apoptosis by ectopic expression of in human liposarcomas Dhong Hyun Lee 1, *, Charles Forscher 1, Dolores Di Vizio 2, 3, and H. Phillip Koeffler 1,
Supplementary Information Induction of p53-independent apoptosis by ectopic expression of in human liposarcomas Dhong Hyun Lee 1, *, Charles Forscher 1, Dolores Di Vizio 2, 3, and H. Phillip Koeffler 1,
Supplementary Fig. S1. Schematic diagram of minigenome segments.
 open reading frame 1565 (segment 5) 47 (-) 3 5 (+) 76 101 125 149 173 197 221 246 287 open reading frame 890 (segment 8) 60 (-) 3 5 (+) 172 Supplementary Fig. S1. Schematic diagram of minigenome segments.
open reading frame 1565 (segment 5) 47 (-) 3 5 (+) 76 101 125 149 173 197 221 246 287 open reading frame 890 (segment 8) 60 (-) 3 5 (+) 172 Supplementary Fig. S1. Schematic diagram of minigenome segments.
HSP72 HSP90. Quadriceps Muscle. MEF2c MyoD1 MyoG Myf5 Hsf1 Hsp GLUT4/GAPDH (AU)
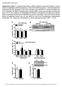 Supplementary Figure 1. Impaired insulin action in HSP72 deficient muscle and myotubes in culture cannot be explained by altered myogenesis or reduced total GLUT4 expression. Genes associated with myogenesis
Supplementary Figure 1. Impaired insulin action in HSP72 deficient muscle and myotubes in culture cannot be explained by altered myogenesis or reduced total GLUT4 expression. Genes associated with myogenesis
PAX8-PPARγ Fusion Protein in thyroid carcinoma
 Sidney H. Ingbar Lecture, ATA 10/17/2013: PAX8-PPARγ Fusion Protein in thyroid carcinoma Ronald J. Koenig, MD, PhD Division of Metabolism, Endocrinology & Diabetes University of Michigan Learning Objectives
Sidney H. Ingbar Lecture, ATA 10/17/2013: PAX8-PPARγ Fusion Protein in thyroid carcinoma Ronald J. Koenig, MD, PhD Division of Metabolism, Endocrinology & Diabetes University of Michigan Learning Objectives
Supplemental Information. Metabolic Maturation during Muscle Stem Cell. Differentiation Is Achieved by mir-1/133a-mediated
 Cell Metabolism, Volume 27 Supplemental Information Metabolic Maturation during Muscle Stem Cell Differentiation Is Achieved by mir-1/133a-mediated Inhibition of the Dlk1-Dio3 Mega Gene Cluster Stas Wüst,
Cell Metabolism, Volume 27 Supplemental Information Metabolic Maturation during Muscle Stem Cell Differentiation Is Achieved by mir-1/133a-mediated Inhibition of the Dlk1-Dio3 Mega Gene Cluster Stas Wüst,
(A) RT-PCR for components of the Shh/Gli pathway in normal fetus cell (MRC-5) and a
 Supplementary figure legends Supplementary Figure 1. Expression of Shh signaling components in a panel of gastric cancer. (A) RT-PCR for components of the Shh/Gli pathway in normal fetus cell (MRC-5) and
Supplementary figure legends Supplementary Figure 1. Expression of Shh signaling components in a panel of gastric cancer. (A) RT-PCR for components of the Shh/Gli pathway in normal fetus cell (MRC-5) and
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12215 Supplementary Figure 1. The effects of full and dissociated GR agonists in supporting BFU-E self-renewal divisions. BFU-Es were cultured in self-renewal medium with indicated GR
doi:10.1038/nature12215 Supplementary Figure 1. The effects of full and dissociated GR agonists in supporting BFU-E self-renewal divisions. BFU-Es were cultured in self-renewal medium with indicated GR
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells
 MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
Supplementary Fig. 1. Delivery of mirnas via Red Fluorescent Protein.
 prfp-vector RFP Exon1 Intron RFP Exon2 prfp-mir-124 mir-93/124 RFP Exon1 Intron RFP Exon2 Untransfected prfp-vector prfp-mir-93 prfp-mir-124 Supplementary Fig. 1. Delivery of mirnas via Red Fluorescent
prfp-vector RFP Exon1 Intron RFP Exon2 prfp-mir-124 mir-93/124 RFP Exon1 Intron RFP Exon2 Untransfected prfp-vector prfp-mir-93 prfp-mir-124 Supplementary Fig. 1. Delivery of mirnas via Red Fluorescent
Supplemental Material:
 Supplemental Material: MATERIALS AND METHODS RNA interference Mouse CHOP sirna (ON-TARGETplus SMARTpool Cat# L-062068-00) and control sirna (ON-TARGETplus Control) were purchased from Dharmacon. Transfection
Supplemental Material: MATERIALS AND METHODS RNA interference Mouse CHOP sirna (ON-TARGETplus SMARTpool Cat# L-062068-00) and control sirna (ON-TARGETplus Control) were purchased from Dharmacon. Transfection
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS)
 Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Figure 1. Quantile-quantile (Q-Q) plots. (Panel A) Q-Q plot graphical
 Supplementary Figure 1. Quantile-quantile (Q-Q) plots. (Panel A) Q-Q plot graphical representation using all SNPs (n= 13,515,798) including the region on chromosome 1 including SORT1 which was previously
Supplementary Figure 1. Quantile-quantile (Q-Q) plots. (Panel A) Q-Q plot graphical representation using all SNPs (n= 13,515,798) including the region on chromosome 1 including SORT1 which was previously
Supplemental Fig. 1. Relative mrna Expression. Relative mrna Expression WT KO WT KO RT 4 0 C
 Supplemental Fig. 1 A 1.5 1..5 Hdac11 (ibat) n=4 n=4 n=4 n=4 n=4 n=4 n=4 n=4 WT KO WT KO WT KO WT KO RT 4 C RT 4 C Supplemental Figure 1. Hdac11 mrna is undetectable in KO adipose tissue. Quantitative
Supplemental Fig. 1 A 1.5 1..5 Hdac11 (ibat) n=4 n=4 n=4 n=4 n=4 n=4 n=4 n=4 WT KO WT KO WT KO WT KO RT 4 C RT 4 C Supplemental Figure 1. Hdac11 mrna is undetectable in KO adipose tissue. Quantitative
Nature Immunology: doi: /ni Supplementary Figure 1. DNA-methylation machinery is essential for silencing of Cd4 in cytotoxic T cells.
 Supplementary Figure 1 DNA-methylation machinery is essential for silencing of Cd4 in cytotoxic T cells. (a) Scheme for the retroviral shrna screen. (b) Histogram showing CD4 expression (MFI) in WT cytotoxic
Supplementary Figure 1 DNA-methylation machinery is essential for silencing of Cd4 in cytotoxic T cells. (a) Scheme for the retroviral shrna screen. (b) Histogram showing CD4 expression (MFI) in WT cytotoxic
Nature Genetics: doi: /ng.3731
 Supplementary Figure 1 Circadian profiles of Adarb1 transcript and ADARB1 protein in mouse tissues. (a) Overlap of rhythmic transcripts identified in the previous transcriptome analyses. The mouse liver
Supplementary Figure 1 Circadian profiles of Adarb1 transcript and ADARB1 protein in mouse tissues. (a) Overlap of rhythmic transcripts identified in the previous transcriptome analyses. The mouse liver
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb3399 a b c d FSP DAPI 5mm mm 5mm 5mm e Correspond to melanoma in-situ Figure a DCT FSP- f MITF mm mm MlanaA melanoma in-situ DCT 5mm FSP- mm mm mm mm mm g melanoma in-situ MITF MlanaA mm mm
DOI:.38/ncb3399 a b c d FSP DAPI 5mm mm 5mm 5mm e Correspond to melanoma in-situ Figure a DCT FSP- f MITF mm mm MlanaA melanoma in-situ DCT 5mm FSP- mm mm mm mm mm g melanoma in-situ MITF MlanaA mm mm
Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid.
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
 DOI: 10.1038/ncb2210 b. ICAM1 ng ml -1 P = 0.0001 Small RNA (15-30nts) ng ml -1 Cell Lysate Exosome HDL Plasma HDL Normal Human HDL mirnas R = 0.45 P < 0.0001 Normal Human Exosome mirnas Figure S1. Characterization
DOI: 10.1038/ncb2210 b. ICAM1 ng ml -1 P = 0.0001 Small RNA (15-30nts) ng ml -1 Cell Lysate Exosome HDL Plasma HDL Normal Human HDL mirnas R = 0.45 P < 0.0001 Normal Human Exosome mirnas Figure S1. Characterization
ECM1 controls T H 2 cell egress from lymph nodes through re-expression of S1P 1
 ZH, Li et al, page 1 ECM1 controls T H 2 cell egress from lymph nodes through re-expression of S1P 1 Zhenhu Li 1,4,Yuan Zhang 1,4, Zhiduo Liu 1, Xiaodong Wu 1, Yuhan Zheng 1, Zhiyun Tao 1, Kairui Mao 1,
ZH, Li et al, page 1 ECM1 controls T H 2 cell egress from lymph nodes through re-expression of S1P 1 Zhenhu Li 1,4,Yuan Zhang 1,4, Zhiduo Liu 1, Xiaodong Wu 1, Yuhan Zheng 1, Zhiyun Tao 1, Kairui Mao 1,
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11095 Supplementary Table 1. Summary of the binding between Angptls and various Igdomain containing receptors as determined by flow cytometry analysis. The results were summarized from
doi:10.1038/nature11095 Supplementary Table 1. Summary of the binding between Angptls and various Igdomain containing receptors as determined by flow cytometry analysis. The results were summarized from
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Effect of HSP90 inhibition on expression of endogenous retroviruses. (a) Inducible shrna-mediated Hsp90 silencing in mouse ESCs. Immunoblots of total cell extract expressing the
Supplementary Figure 1 Effect of HSP90 inhibition on expression of endogenous retroviruses. (a) Inducible shrna-mediated Hsp90 silencing in mouse ESCs. Immunoblots of total cell extract expressing the
HIF-inducible mir-191 promotes migration in breast cancer through complex regulation of TGFβ-signaling in hypoxic microenvironment.
 HIF-inducible mir-9 promotes migration in breast cancer through complex regulation of TGFβ-signaling in hypoxic microenvironment. Neha Nagpal, Hafiz M Ahmad, Shibu Chameettachal3, Durai Sundar, Sourabh
HIF-inducible mir-9 promotes migration in breast cancer through complex regulation of TGFβ-signaling in hypoxic microenvironment. Neha Nagpal, Hafiz M Ahmad, Shibu Chameettachal3, Durai Sundar, Sourabh
York criteria, 6 RA patients and 10 age- and gender-matched healthy controls (HCs).
 MATERIALS AND METHODS Study population Blood samples were obtained from 15 patients with AS fulfilling the modified New York criteria, 6 RA patients and 10 age- and gender-matched healthy controls (HCs).
MATERIALS AND METHODS Study population Blood samples were obtained from 15 patients with AS fulfilling the modified New York criteria, 6 RA patients and 10 age- and gender-matched healthy controls (HCs).
Supplementary. presence of the. (c) mrna expression. Error. in naive or
 Figure 1. (a) Naive CD4 + T cells were activated in the presence of the indicated cytokines for 3 days. Enpp2 mrna expression was measured by qrt-pcrhr, infected with (b, c) Naive CD4 + T cells were activated
Figure 1. (a) Naive CD4 + T cells were activated in the presence of the indicated cytokines for 3 days. Enpp2 mrna expression was measured by qrt-pcrhr, infected with (b, c) Naive CD4 + T cells were activated
Supplementary Figure 1. mtor LysM and Rictor LysM mice have normal cellularity and percentages of hematopoe>c cells. a. Cell numbers of lung, liver,
 a. b. c. Supplementary Figure 1. mtor LysM and Rictor LysM mice have normal cellularity and percentages of hematopoe>c cells. a. Cell numbers of lung, liver, and spleen. b. Cell numbers of bone marrow
a. b. c. Supplementary Figure 1. mtor LysM and Rictor LysM mice have normal cellularity and percentages of hematopoe>c cells. a. Cell numbers of lung, liver, and spleen. b. Cell numbers of bone marrow
Supplementary Figure 1. DNA methylation of the adiponectin promoter R1, Pparg2, and Tnfa promoter in adipocytes is not affected by obesity.
 Supplementary Figure 1. DNA methylation of the adiponectin promoter R1, Pparg2, and Tnfa promoter in adipocytes is not affected by obesity. (a) Relative amounts of adiponectin, Ppar 2, C/ebp, and Tnf mrna
Supplementary Figure 1. DNA methylation of the adiponectin promoter R1, Pparg2, and Tnfa promoter in adipocytes is not affected by obesity. (a) Relative amounts of adiponectin, Ppar 2, C/ebp, and Tnf mrna
p53 Plays a Role in Mesenchymal Differentiation Programs, in a Cell Fate Dependent Manner
 p53 Plays a Role in Mesenchymal Differentiation Programs, in a Cell Fate Dependent Manner Alina Molchadsky 1., Igor Shats 1., Naomi Goldfinger 1, Meirav Pevsner-Fischer 1, Melissa Olson 2, Ariel Rinon
p53 Plays a Role in Mesenchymal Differentiation Programs, in a Cell Fate Dependent Manner Alina Molchadsky 1., Igor Shats 1., Naomi Goldfinger 1, Meirav Pevsner-Fischer 1, Melissa Olson 2, Ariel Rinon
Supplemental Information Supplementary Table 1. Tph1+/+ Tph1 / Analyte Supplementary Table 2. Tissue Vehicle LP value
 Supplemental Information Supplementary Table. Urinary and adipose tissue catecholamines in Tph +/+ and Tph / mice fed a high fat diet for weeks. Tph +/+ Tph / Analyte ewat ibat ewat ibat Urine (ng/ml)
Supplemental Information Supplementary Table. Urinary and adipose tissue catecholamines in Tph +/+ and Tph / mice fed a high fat diet for weeks. Tph +/+ Tph / Analyte ewat ibat ewat ibat Urine (ng/ml)
Supplementary Fig. 1 eif6 +/- mice show a reduction in white adipose tissue, blood lipids and normal glycogen synthesis. The cohort of the original
 Supplementary Fig. 1 eif6 +/- mice show a reduction in white adipose tissue, blood lipids and normal glycogen synthesis. The cohort of the original phenotypic screening was n=40. For specific tests, the
Supplementary Fig. 1 eif6 +/- mice show a reduction in white adipose tissue, blood lipids and normal glycogen synthesis. The cohort of the original phenotypic screening was n=40. For specific tests, the
SUPPLEMENTARY DATA. Nature Medicine: doi: /nm.4171
 SUPPLEMENTARY DATA Supplementary Figure 1 a b c PF %Change - -4-6 Body weight Lean mass Body fat Tissue weight (g).4.3.2.1. PF GC iwat awat BAT PF d e f g week 2 week 3 NEFA (mmol/l) 1..5. PF phsl (Ser565)
SUPPLEMENTARY DATA Supplementary Figure 1 a b c PF %Change - -4-6 Body weight Lean mass Body fat Tissue weight (g).4.3.2.1. PF GC iwat awat BAT PF d e f g week 2 week 3 NEFA (mmol/l) 1..5. PF phsl (Ser565)
GW(g)/BW(g) GW(g)/BW(g) Con Dex Con Dex. GW(g)/BW(g) Relative mrna levels. Atrogin-1 Murf-1. Atrogin-1 Murf-1. Soleus
 a b c GW(g) GW(g) GW(g) 0.20 0.15 0.10 0.05 0.00 0.20 0.15 0.10 0.05 Den * ** GW(g)/BW(g) Den Dex Dex GW(g)/BW(g) d 0.00 Fasting GW(g) e 0.15 Cancer Cachexia f GW(g) 0.10 0.05 0.00 Young ** Old g GW(g)/BW(g)
a b c GW(g) GW(g) GW(g) 0.20 0.15 0.10 0.05 0.00 0.20 0.15 0.10 0.05 Den * ** GW(g)/BW(g) Den Dex Dex GW(g)/BW(g) d 0.00 Fasting GW(g) e 0.15 Cancer Cachexia f GW(g) 0.10 0.05 0.00 Young ** Old g GW(g)/BW(g)
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2535 Figure S1 SOX10 is expressed in human giant congenital nevi and its expression in human melanoma samples suggests that SOX10 functions in a MITF-independent manner. a, b, Representative
DOI: 10.1038/ncb2535 Figure S1 SOX10 is expressed in human giant congenital nevi and its expression in human melanoma samples suggests that SOX10 functions in a MITF-independent manner. a, b, Representative
SUPPLEMENTARY FIGURES AND TABLES
 SUPPLEMENTARY FIGURES AND TABLES Supplementary Figure S1: CaSR expression in neuroblastoma models. A. Proteins were isolated from three neuroblastoma cell lines and from the liver metastasis of a MYCN-non
SUPPLEMENTARY FIGURES AND TABLES Supplementary Figure S1: CaSR expression in neuroblastoma models. A. Proteins were isolated from three neuroblastoma cell lines and from the liver metastasis of a MYCN-non
Supplementary Figure 1 Chemokine and chemokine receptor expression during muscle regeneration (a) Analysis of CR3CR1 mrna expression by real time-pcr
 Supplementary Figure 1 Chemokine and chemokine receptor expression during muscle regeneration (a) Analysis of CR3CR1 mrna expression by real time-pcr at day 0, 1, 4, 10 and 21 post- muscle injury. (b)
Supplementary Figure 1 Chemokine and chemokine receptor expression during muscle regeneration (a) Analysis of CR3CR1 mrna expression by real time-pcr at day 0, 1, 4, 10 and 21 post- muscle injury. (b)
Journal of Cell Science Accepted manuscript
 2013. Published by The Company of Biologists Ltd. Heterogeneous lineage origin underlies phenotypic and molecular differences of white and beige adipocytes Weiyi Liu 1, Tizhong Shan 1, Xin Yang 1, Sandra
2013. Published by The Company of Biologists Ltd. Heterogeneous lineage origin underlies phenotypic and molecular differences of white and beige adipocytes Weiyi Liu 1, Tizhong Shan 1, Xin Yang 1, Sandra
A Normal Exencephaly Craniora- Spina bifida Microcephaly chischisis. Midbrain Forebrain/ Forebrain/ Hindbrain Spinal cord Hindbrain Hindbrain
 A Normal Exencephaly Craniora- Spina bifida Microcephaly chischisis NTD Number of embryos % among NTD Embryos Exencephaly 52 74.3% Craniorachischisis 6 8.6% Spina bifida 5 7.1% Microcephaly 7 1% B Normal
A Normal Exencephaly Craniora- Spina bifida Microcephaly chischisis NTD Number of embryos % among NTD Embryos Exencephaly 52 74.3% Craniorachischisis 6 8.6% Spina bifida 5 7.1% Microcephaly 7 1% B Normal
a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation,
 Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
ECM microenvironment unlocks brown adipogenic potential of adult human bone marrow-derived MSCs
 SUPPLEMENTARY INFORMATION ECM microenvironment unlocks brown adipogenic potential of adult human bone marrow-derived MSCs Michelle H. Lee 1,2,3,+, Anna G. Goralczyk 1,3,+, Rókus Kriszt 1,2,3, Xiu Min Ang
SUPPLEMENTARY INFORMATION ECM microenvironment unlocks brown adipogenic potential of adult human bone marrow-derived MSCs Michelle H. Lee 1,2,3,+, Anna G. Goralczyk 1,3,+, Rókus Kriszt 1,2,3, Xiu Min Ang
(a) Significant biological processes (upper panel) and disease biomarkers (lower panel)
 Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1 Schematic depiction of the tandem Fc GDF15. Supplementary Figure 2 Supplementary Figure 2 Gfral mrna levels in the brains of both wild-type and knockout Gfral
Supplementary Figure 1 Supplementary Figure 1 Schematic depiction of the tandem Fc GDF15. Supplementary Figure 2 Supplementary Figure 2 Gfral mrna levels in the brains of both wild-type and knockout Gfral
Lack of TRPV2 impairs thermogenesis in mouse brown adipose tissue
 Manuscript EMBO-2015-40819 Lack of TRPV2 impairs thermogenesis in mouse brown adipose tissue Wuping Sun, Kunitoshi Uchida, Yoshiro Suzuki, Yiming Zhou, Minji Kim, Yasunori Takayama, Nobuyuki Takahashi,
Manuscript EMBO-2015-40819 Lack of TRPV2 impairs thermogenesis in mouse brown adipose tissue Wuping Sun, Kunitoshi Uchida, Yoshiro Suzuki, Yiming Zhou, Minji Kim, Yasunori Takayama, Nobuyuki Takahashi,
mir-7a regulation of Pax6 in neural stem cells controls the spatial origin of forebrain dopaminergic neurons
 Supplemental Material mir-7a regulation of Pax6 in neural stem cells controls the spatial origin of forebrain dopaminergic neurons Antoine de Chevigny, Nathalie Coré, Philipp Follert, Marion Gaudin, Pascal
Supplemental Material mir-7a regulation of Pax6 in neural stem cells controls the spatial origin of forebrain dopaminergic neurons Antoine de Chevigny, Nathalie Coré, Philipp Follert, Marion Gaudin, Pascal
SUPPLEMENTARY DATA. Supplementary Table 1. Primers used in qpcr
 Supplementary Table 1. Primers used in qpcr Gene forward primer (5'-3') reverse primer (5'-3') β-actin AGAGGGAAATCGTGCGTGAC CAATAGTGATGACCTGGCCGT Hif-p4h-2 CTGGGCAACTACAGGATAAAC GCGTCCCAGTCTTTATTTAGATA
Supplementary Table 1. Primers used in qpcr Gene forward primer (5'-3') reverse primer (5'-3') β-actin AGAGGGAAATCGTGCGTGAC CAATAGTGATGACCTGGCCGT Hif-p4h-2 CTGGGCAACTACAGGATAAAC GCGTCCCAGTCTTTATTTAGATA
Supporting Information
 Supporting Information Charalambous et al. 10.1073/pnas.1406119111 SI Experimental Procedures Serum and Tissue Biochemistry. Enzymatic assay kits were used for determination of plasma FFAs (Roche), TAGs
Supporting Information Charalambous et al. 10.1073/pnas.1406119111 SI Experimental Procedures Serum and Tissue Biochemistry. Enzymatic assay kits were used for determination of plasma FFAs (Roche), TAGs
Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Table
 Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Table Title of file for HTML: Peer Review File Description: Innate Scavenger Receptor-A regulates
Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Table Title of file for HTML: Peer Review File Description: Innate Scavenger Receptor-A regulates
Up-Regulation of Mitochondrial Activity and Acquirement of Brown Adipose Tissue-Like Property in the White Adipose Tissue of Fsp27 Deficient Mice
 Up-Regulation of Mitochondrial Activity and Acquirement of Brown Adipose Tissue-Like Property in the White Adipose Tissue of Fsp27 Deficient Mice Shen Yon Toh 1,2,3., Jingyi Gong 2., Guoli Du 2., John
Up-Regulation of Mitochondrial Activity and Acquirement of Brown Adipose Tissue-Like Property in the White Adipose Tissue of Fsp27 Deficient Mice Shen Yon Toh 1,2,3., Jingyi Gong 2., Guoli Du 2., John
File Name: Supplementary Information Description: Supplementary Figures and Supplementary Tables. File Name: Peer Review File Description:
 File Name: Supplementary Information Description: Supplementary Figures and Supplementary Tables File Name: Peer Review File Description: Primer Name Sequence (5'-3') AT ( C) RT-PCR USP21 F 5'-TTCCCATGGCTCCTTCCACATGAT-3'
File Name: Supplementary Information Description: Supplementary Figures and Supplementary Tables File Name: Peer Review File Description: Primer Name Sequence (5'-3') AT ( C) RT-PCR USP21 F 5'-TTCCCATGGCTCCTTCCACATGAT-3'
Supplementary Figure 1
 Supplementary Figure 1 Asymmetrical function of 5p and 3p arms of mir-181 and mir-30 families and mir-142 and mir-154. (a) Control experiments using mirna sensor vector and empty pri-mirna overexpression
Supplementary Figure 1 Asymmetrical function of 5p and 3p arms of mir-181 and mir-30 families and mir-142 and mir-154. (a) Control experiments using mirna sensor vector and empty pri-mirna overexpression
Oestrogen signalling in white adipose progenitor cells inhibits differentiation into brown adipose and smooth muscle cells
 Received Feb 4 Accepted 8 Sep 4 Published Oct 4 DOI:.8/ncomms696 Oestrogen signalling in white adipose progenitor cells inhibits differentiation into brown adipose and smooth muscle cells Kfir Lapid,,
Received Feb 4 Accepted 8 Sep 4 Published Oct 4 DOI:.8/ncomms696 Oestrogen signalling in white adipose progenitor cells inhibits differentiation into brown adipose and smooth muscle cells Kfir Lapid,,
TISSUE-SPECIFIC STEM CELLS
 TISSUE-SPECIFIC STEM CELLS p107 Is a Crucial Regulator for Determining the Adipocyte Lineage Fate Choices of Stem Cells MARTINA DE SOUSA, a DEANNA P. PORRAS, a,b CHRISTOPHER G. R. PERRY, b PATRICK SEALE,
TISSUE-SPECIFIC STEM CELLS p107 Is a Crucial Regulator for Determining the Adipocyte Lineage Fate Choices of Stem Cells MARTINA DE SOUSA, a DEANNA P. PORRAS, a,b CHRISTOPHER G. R. PERRY, b PATRICK SEALE,
Supporting Information Table of content
 Supporting Information Table of content Supporting Information Fig. S1 Supporting Information Fig. S2 Supporting Information Fig. S3 Supporting Information Fig. S4 Supporting Information Fig. S5 Supporting
Supporting Information Table of content Supporting Information Fig. S1 Supporting Information Fig. S2 Supporting Information Fig. S3 Supporting Information Fig. S4 Supporting Information Fig. S5 Supporting
Supplementary Figure 1
 VO (ml kg - min - ) VCO (ml kg - min - ) Respiratory exchange ratio Energy expenditure (cal kg - min - ) Locomotor activity (x count) Body temperature ( C) Relative mrna expression TA Sol EDL PT Heart
VO (ml kg - min - ) VCO (ml kg - min - ) Respiratory exchange ratio Energy expenditure (cal kg - min - ) Locomotor activity (x count) Body temperature ( C) Relative mrna expression TA Sol EDL PT Heart
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2566 Figure S1 CDKL5 protein expression pattern and localization in mouse brain. (a) Multiple-tissue western blot from a postnatal day (P) 21 mouse probed with an antibody against CDKL5.
DOI: 10.1038/ncb2566 Figure S1 CDKL5 protein expression pattern and localization in mouse brain. (a) Multiple-tissue western blot from a postnatal day (P) 21 mouse probed with an antibody against CDKL5.
Supplementary Table 3. 3 UTR primer sequences. Primer sequences used to amplify and clone the 3 UTR of each indicated gene are listed.
 Supplemental Figure 1. DLKI-DIO3 mirna/mrna complementarity. Complementarity between the indicated DLK1-DIO3 cluster mirnas and the UTR of SOX2, SOX9, HIF1A, ZEB1, ZEB2, STAT3 and CDH1with mirsvr and PhastCons
Supplemental Figure 1. DLKI-DIO3 mirna/mrna complementarity. Complementarity between the indicated DLK1-DIO3 cluster mirnas and the UTR of SOX2, SOX9, HIF1A, ZEB1, ZEB2, STAT3 and CDH1with mirsvr and PhastCons
sequences of a styx mutant reveals a T to A transversion in the donor splice site of intron 5
 sfigure 1 Styx mutant mice recapitulate the phenotype of SHIP -/- mice. (A) Analysis of the genomic sequences of a styx mutant reveals a T to A transversion in the donor splice site of intron 5 (GTAAC
sfigure 1 Styx mutant mice recapitulate the phenotype of SHIP -/- mice. (A) Analysis of the genomic sequences of a styx mutant reveals a T to A transversion in the donor splice site of intron 5 (GTAAC
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3311 A B TSC2 -/- MEFs C Rapa Hours WCL 0 6 12 24 36 pakt.s473 AKT ps6k S6K CM IGF-1 Recipient WCL - + - + - + pigf-1r IGF-1R pakt ps6 AKT D 1 st SILAC 2 nd SILAC E GAPDH FGF21 ALKPGVIQILGVK
DOI: 10.1038/ncb3311 A B TSC2 -/- MEFs C Rapa Hours WCL 0 6 12 24 36 pakt.s473 AKT ps6k S6K CM IGF-1 Recipient WCL - + - + - + pigf-1r IGF-1R pakt ps6 AKT D 1 st SILAC 2 nd SILAC E GAPDH FGF21 ALKPGVIQILGVK
Targeting of the circadian clock via CK1δ/ε to improve glucose homeostasis in obesity
 Targeting of the circadian clock via CK1δ/ε to improve glucose homeostasis in obesity Peter S. Cunningham, Siobhán A. Ahern, Laura C. Smith, Carla S. da Silva Santos, Travis T. Wager and David A. Bechtold
Targeting of the circadian clock via CK1δ/ε to improve glucose homeostasis in obesity Peter S. Cunningham, Siobhán A. Ahern, Laura C. Smith, Carla S. da Silva Santos, Travis T. Wager and David A. Bechtold
Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein
 Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein content relative to GAPDH in two independent experiments.
Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein content relative to GAPDH in two independent experiments.
Table S1. Primer sequences used for qrt-pcr. CACCATTGGCAATGAGCGGTTC AGGTCTTTGCGGATGTCCACGT ACTB AAGTCCATGTGCTGGCAGCACT ATCACCACTCCGAAGTCCGTCT LCOR
 Table S1. Primer sequences used for qrt-pcr. ACTB LCOR KLF6 CTBP1 CDKN1A CDH1 ATF3 PLAU MMP9 TFPI2 CACCATTGGCAATGAGCGGTTC AGGTCTTTGCGGATGTCCACGT AAGTCCATGTGCTGGCAGCACT ATCACCACTCCGAAGTCCGTCT CGGCTGCAGGAAAGTTTACA
Table S1. Primer sequences used for qrt-pcr. ACTB LCOR KLF6 CTBP1 CDKN1A CDH1 ATF3 PLAU MMP9 TFPI2 CACCATTGGCAATGAGCGGTTC AGGTCTTTGCGGATGTCCACGT AAGTCCATGTGCTGGCAGCACT ATCACCACTCCGAAGTCCGTCT CGGCTGCAGGAAAGTTTACA
Supplementary methods:
 Supplementary methods: Primers sequences used in real-time PCR analyses: β-actin F: GACCTCTATGCCAACACAGT β-actin [11] R: AGTACTTGCGCTCAGGAGGA MMP13 F: TTCTGGTCTTCTGGCACACGCTTT MMP13 R: CCAAGCTCATGGGCAGCAACAATA
Supplementary methods: Primers sequences used in real-time PCR analyses: β-actin F: GACCTCTATGCCAACACAGT β-actin [11] R: AGTACTTGCGCTCAGGAGGA MMP13 F: TTCTGGTCTTCTGGCACACGCTTT MMP13 R: CCAAGCTCATGGGCAGCAACAATA
SUPPLEMENTARY MATERIAL
 SYPPLEMENTARY FIGURE LEGENDS SUPPLEMENTARY MATERIAL Figure S1. Phylogenic studies of the mir-183/96/182 cluster and 3 -UTR of Casp2. (A) Genomic arrangement of the mir-183/96/182 cluster in vertebrates.
SYPPLEMENTARY FIGURE LEGENDS SUPPLEMENTARY MATERIAL Figure S1. Phylogenic studies of the mir-183/96/182 cluster and 3 -UTR of Casp2. (A) Genomic arrangement of the mir-183/96/182 cluster in vertebrates.
Fig. S1. Weight of embryos during development. Embryos are collected at different time points (E12.5, E14.5, E16.5 and E18.5) from matings between
 Fig. S1. Weight of embryos during development. Embryos are collected at different time points (E12.5, E14.5, E16.5 and E18.5) from matings between Myod +/ or Myod / females and Myod +/ ;Igf2 +/ males and
Fig. S1. Weight of embryos during development. Embryos are collected at different time points (E12.5, E14.5, E16.5 and E18.5) from matings between Myod +/ or Myod / females and Myod +/ ;Igf2 +/ males and
