Genes methylated by DNA methyltransferase 3b are similar in mouse intestine and human colon cancer
|
|
|
- Lawrence Erick Shepherd
- 6 years ago
- Views:
Transcription
1 Genes methylated by DNA methyltransferase 3b are similar in mouse intestine and human colon cancer The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation As Published Publisher Steine, Eveline J. et al. Genes Methylated by DNA Methyltransferase 3b Are Similar in Mouse Intestine and Human Colon Cancer. Journal of Clinical Investigation (2011): Web. American Society for Clinical Investigation Version Final published version Accessed Sat Apr 21 20:01:45 EDT 2018 Citable Link Terms of Use Detailed Terms Article is made available in accordance with the publisher's policy and may be subject to US copyright law. Please refer to the publisher's site for terms of use.
2 Brief report Genes methylated by DNA methyltransferase 3b are similar in mouse intestine and human colon cancer Eveline J. Steine, 1 Mathias Ehrich, 2 George W. Bell, 1 Arjun Raj, 3 Seshamma Reddy, 1 Alexander van Oudenaarden, 3 Rudolf Jaenisch, 1 and Heinz G. Linhart 1,4,5 1 Whitehead Institute for Biomedical Research and Department of Biology, Massachusetts Institute of Technology (MIT), Cambridge, Massachusetts, USA. 2 Sequenom Inc., San Diego, California, USA. 3 Department of Physics and Department of Biology, Koch Institute for Integrative Cancer Research, MIT, Cambridge, Massachusetts, USA. 4 Department of Gastroenterology and Hepatology, University Hospital Freiburg, Freiburg, Germany. 5 Department of Translational Oncology and Department of Epigenetics, National Center for Tumor Diseases/DKFZ, Heidelberg, Germany. Human cancer cells frequently have regions of their DNA hypermethylated, which results in transcriptional silencing of affected genes and promotion of tumor formation. However, it is still unknown whether cancerassociated aberrant DNA methylation is targeted to specific genomic regions, whether this methylation also occurs in noncancerous cells, and whether these epigenetic events are maintained in the absence of the initiating cause. Here we have addressed some of these issues by demonstrating that transgenic expression of DNA methyltransferase 3b (Dnmt3b) in the mouse colon initiates de novo DNA methylation of genes that are similar to genes that become methylated in human colon cancer. This is consistent with the notion that aberrant methylation in cancer may be attributable to targeting of specific sequences by Dnmt3b rather than to random methylation followed by clonal selection. We also showed that Dnmt3b-induced aberrant DNA methylation was maintained in regenerating tissue, even in the absence of continuous Dnmt3b expression. This supports the concept that transient stressors can cause permanent epigenetic changes in somatic stem cells and that these accumulate over the lifetime of an organism in analogy to DNA mutations. Introduction Regional hypermethylation of DNA is frequently found in human cancer, causes transcriptional silencing of affected genes, and promotes tumor formation (1, 2). An unresolved question is whether aberrant DNA methylation of cancer is caused by stochastic DNA methylation followed by clonal selection, or whether specific sequences are targeted by the de novo methyltransferases (3). Furthermore, it is not known whether cancer-specific DNA methylation only occurs in neoplastic cells or whether it can also occur in normal cells. Another important question is whether aberrations in DNA methylation are maintained in the absence of the initiating cause. Abundant evidence indicates that DNA methyltransferase 3b (Dnmt3b) is involved in de novo methylation of mammalian cells during development, is frequently activated in human tumors, and promotes tumor development in APC Min/+ mice (4, 5). To address whether Dnmt3b targets specific sequences for de novo methylation, we used transgenic mice that allow tetracycline-inducible expression of Dnmt3b in WT mice and in APC Min/+ mice, a model of intestinal tumorigenesis. Results and Discussion To determine whether aberrant de novo methylation is a stochastic or a targeted process, we compared DNA methylation of colon tumors, representing a mono-/oligoclonal cell population, with normal colon epithelial cells of APC Min/+ mice induced to express Authorship note: Eveline J. Steine and Mathias Ehrich contributed equally to this work. Conflict of interest: M. Ehrich is employed by Sequenom Inc. S. Reddy is employed by Oral Cancer Prevention International. Citation for this article: J Clin Invest. 2011;121(5): doi: /jci the Dnmt3b transgene by exposure to doxycycline (dox). We analyzed DNA methylation of 23 candidate genes that either are frequently methylated in human colon cancer or represent classical tumor suppressor genes (6). As control regions, 3 imprinted loci were included, and control samples included ES cells deficient for Dnmt1; triple-knockout ES cells deficient for Dnmt1, Dnmt3a, and Dnmt3b; and WT ES cells. For methylation analysis, we used gene-specific amplification of bisulfite-treated DNA followed by reverse transcription and MALDI-TOF analysis (7). An almost identical methylation pattern was seen in all tumor samples and nontumor cell samples from Dnmt3b-induced mice (Figure 1A). A significant increase in DNA methylation (defined as methylation increase of at least 0.1 and P 0.05 by t test) was detected in 11 of 26 candidate regions (including imprinted loci) in both tumor samples and nontumor samples, and pairwise analysis of methylation data confirmed a strong correlation of de novo methylation patterns among all Dnmt3b-expressing samples (correlation between induced sample pairs, ; mean r, 0.90; Figure 1B). The observation that clonal and nonclonal cell populations showed the same de novo methylation pattern across multiple samples strongly suggests that de novo methylation by Dnmt3b is not random, but targets specific genetic loci. The data in Figure 1A also support the hypothesis that de novo methylation of targets such as Abpa2 (also known as Mint2), Hic1, Gata4, Gata5, Cacna1g, and Neurog1, considered to be hallmarks of methylated genes in cancer (6), can occur prior to tumor formation in normal cells (see also Supplemental Table 1; supplemental material available online with this article; doi: /jci43169ds1). To further compare DNA methylation in the mouse model with human colon cancer, we analyzed additional candidate genes that were previously reported to be methylated or unmethylated 1748 The Journal of Clinical Investigation Volume 121 Number 5 May 2011
3 Figure 1 DNA methylation analysis of colon tumors and colon epithelial cells with or without Dnmt3b transgene expression. 4 mo dox, Dnmt3b induction; no dox, no Dnmt3b induction; ES c/c, Dnmt1- deficient ES cells; ES tko, Dnmt1/ Dnmt3a/Dnmt3b triple-knockout ES cells; gray, no data. Heat map data were generated by mass array analysis and subjected to sample-based unsupervised hierarchical clustering. (A) Tumors and mucosa samples with Dnmt3b induction showed reproducible de novo DNA methylation of specific genomic loci. Imprinted regions (Nespas-, Peg3-, and H19-DMR) were partially methylated; ES tko and ES c/c cells showed loss of DNA methylation. (B) Pairwise correlation of methylation data confirmed similar de novo methylation in all samples with Dnmt3b induction (r, ; mean r, 0.90). (C) Dnmt3b-mediated DNA methylation in colon mucosa of APC WT mice closely resembled de novo methylation reported for human colon cancer (Supplemental Table 1), which suggests that Dnmt3b can induce cancer-specific de novo methylation in normal cells (red, methylated; blue, unmethylated; black, control/imprinted regions). in human colon cancer (6). To confirm that de novo methylation of cancer-specific targets can occur in normal cells, the expanded candidate gene analysis was conducted using APC WT mice as opposed to the previously studied mutant APC Min/+ mice. Indeed, the Dnmt3b-induced DNA methylation pattern in WT mice fully reproduced the DNA methylation pattern detected in APC Min/+ mice (Figure 1C). This showed that the de novo methylation patterns obtained in Figure 1A do not require the presence of a mutant APC allele. Further, the expanded candidate gene analysis demonstrated that human cancer related methylation targets were consistently methylated in mice with Dnmt3b expression: the target list (Figure 1C) included 31 genes that are frequently methylated in human colon cancer (6). Of these 31 human cancer methylation targets, 26 were also significantly methylated when Dnmt3b was induced in the colonic mucosa of WT mice (0.1 or greater absolute increase in DNA methylation and P 0.05 by t test; Supplemental Table 1). Conversely, 8 of 8 candidate genes reportedly unmethylated in human colon cancer also remained unmethylated in the Dnmt3b mouse model (0.1 or less de novo methylation and P 0.05; Supplemental Table 1). To complement this candidate gene approach and to further explore the similarities between de novo methylation in our transgenic model and aberrant DNA methylation of colon cancer, we next conducted genome-wide analysis of Dnmt3b-mediated de novo methylation using reduced representation bisulphite sequencing (RRBS; ref. 8). Using 3 colon mucosa samples with 3 months of Dnmt3b induction and 3 samples from age-matched control mice, we scored Dnmt3b-mediated de novo methylation in 13,361 promoter regions. Analogous to the above findings, we observed almost identical de novo methylation patterns among all 3 induced samples (pairwise correlation between induced sample pairs, r > 0.95; Supplemental Figure 2). This further sup- The Journal of Clinical Investigation Volume 121 Number 5 May
4 Figure 2 RRBS analysis of colon epithelial cells with or without Dnmt3b transgene expression in vivo. RRBS data from colon epithelial cells with or without Dnmt3b transgene expression (n = 3 each) were used to calculate de novo methylation in 13,361 annotated promoter regions. (A) Scatter plot demonstrating Dnmt3b-mediated hypermethylation of a distinct subpopulation of promoter regions (11.7% of promoters showed >0.5 methylation gain in Dnmt3bexpressing tissue; lines parallel to the diagonal indicate the position of 0.1 methylation gain or loss). (B) Histogram further illustrating the appearance of a discrete subpopulation of hypermethylated promoters in Dnmt3b-expressing samples (blue), whereas the control samples (brown) contained few promoter regions with methylation scores >0.5. ports the concept that Dnmt3b directly targets certain promoter regions. Interestingly, a discrete subset of genes (11.7%; 1,564 of 13,361) showed a strong methylation gain upon Dnmt3b expression (absolute methylation gain >0.5, false discovery rate [FDR] P < 0.01, complete methylation assigned as 1), whereas the majority of promoter regions remained unmethylated in both test and control DNA (Figure 2, A and B). This suggests that a subgroup of genes in the genome is inherently methylation sensitive. Importantly, in agreement with the results of the candidate gene analysis, we again found a strong concordance when comparing the murine Dnmt3b-mediated de novo methylation data with published data from human colon cancer: for the human cancer/mouse comparison, we selected human genes from the study of Widschwendter et al. (6) that were classified either as cancer-related methylation targets (absolute methylation gain >0.1 in tumors, PMR gain in tumors >10) or as nontargets (tumor methylation gain <0.1, excluding genes with baseline methylation in human control tissue >100% [PMR >100]). Mouse RRBS data were available for 44 human colon cancer methylation targets and for 71 nontargets (115 total). As shown in Supplemental Tables 2 and 3, 84% of genes (37 of 44) methylated in human colon cancer were also methylated by Dnmt3b (methylation gain >0.1, FDR P < 0.01); conversely, 92% of genes (65 of 71) not methylated in human colon cancer were also not methylated by Dnmt3b (methylation gain <0.1). This supports the concept that Dnmt3b targets the very same set of genes that are frequently methylated in human cancer. In addition, the similarity between murine and human data suggests conservation of DNA methylation sensitivity across species. Previous studies had shown a strong correlation between genes methylated in human cancer and genes targeted by the polycomb repressive complex 2 (PRC2) in ES cells (6, 9). We therefore used published data on mouse ES cells (10) to compare the PRC2 occupancy of genes targeted by Dnmt3b (methylation gain >0.5) with that of genes not targeted by Dnmt3b (methylation gain <0.1). Analogous to previous reports, we defined genes associated with either Suz12, Eed, or H3K27 methylation in mouse ES cells as PRC2 targets. Interestingly, ES cell PRC2 occupancy was 71% (721 of 1,015) for genes targeted by Dnmt3b and only 13% (716 of 5,564) for genes not targeted by Dnmt3b (Supplemental Tables 4 and 5). This strong enrichment of PRC2 targets (in ES cells) among genes methylated by Dnmt3b is in good agreement with the previously reported enrichment of PRC2 targets in genes methylated in human cancer (6) and further supports the concept that Dnmt3b targets the same genes that are methylated in cancer. Taken together, our results indicate that colon epithelial cell methylation in the Dnmt3b model predicts DNA methylation of human colon cancer with high confidence. Having established that Dnmt3b can induce a cancer-specific signature of de novo methylation in healthy tissue, we next analyzed the stability of such alterations. Such information would help resolve the question whether altered DNA methylation is a potential initiating cause of disease, rather than only an effect of disease. We induced de novo methylation in the colon of transgenic mice by adding dox to the drinking water for 5 months and analyzed maintenance of these methylation marks after 4 months of dox withdrawal and silencing of transgene expression. The epithelial cell layer of the colon fully renews every 5 to 1750 The Journal of Clinical Investigation Volume 121 Number 5 May 2011
5 Figure 3 Transient expression of Dnmt3b in vivo causes permanent epigenetic alterations of colonic mucosa. (A) Dnmt3b transgene expression in colon epithelial cells in vivo was activated within 2 days of dox administration and returned to baseline within 1 week of dox withdrawal (n = 2 each; quantitative PCR analysis, mean ± SD). (B) Dnmt3b was induced in transgenic mice for 5 months (On, n = 5), and a subset was followed up for an additional period of 4 months after dox withdrawal (On/Off, n = 5). Controls were agematched to the On/Off group (n = 4). Almost all genes that underwent significant de novo methylation (P < 0.01, t test) were equally methylated in the On and On/Off groups. (C) Dnmt3b-mediated de novo methylation was analyzed in dividing (prolif, n = 5) and nondividing mouse embryonic fibroblasts (mit, mitomycin C, n = 3; irrad, γ-irradiated, n = 2) in the absence of dox or after 14 days of dox-induced Dnmt3b transgene expression. Box plot shows combined methylation data of genes with significant de novo methylation in proliferating cells (P < 0.05, paired t test; n = 25). Box denotes interquartile range; line within box denotes median; whiskers denote 1.5 interquartile range; symbols denote outliers. Significant Dnmt3b-mediated de novo methylation was detected in both proliferating and growth-arrested cells, which suggests that cell division is not required for this process. P values between groups are shown by brackets. 7 days (11) and is therefore ideal to test maintenance of epimutations in vivo. Dnmt3b expression in transgenic mice was rapidly activated by dox in the drinking water and quickly returned to baseline within 1 week of dox withdrawal (Figure 3A). Importantly, de novo methylation of individual genes was almost identical in mice on dox compared with mice that were maintained in the absence of dox for 4 months (Figure 3B). This supports the hypothesis that aberrant de novo DNA methylation is maintained in regenerating healthy tissue, even in the absence of the initiating cause. To test whether this may be caused by de novo methylation of the intestinal stem cell compartment, we used RNA FISH analysis to measure Dnmt3b transgene expression in colonic epithelial cells that express the intestinal stem cell marker Lgr5 (Supplemental Figure 1 and refs. 11, 12). Indeed, Dnmt3b expression in Lgr5-positive cells of a dox-induced transgenic mouse was approximately twice that of a littermate control (control, ± Dnmt3b particles/μm 3 ; Dnmt3b transgene induction, ± Dnmt3b particles/μm 3 ; P 0.001), supporting the concept that transgene-mediated de novo methylation of intestinal stem cells contributes to irreversible epigenetic alteration of the colonic mucosa. Several previous studies have shown a strong correlation between cell proliferation and acquisition of aberrant de novo DNA methylation (8, 13), which suggests that infrequently dividing cells, such as somatic stem cells, are possibly less sensitive to Dnmt3b-mediated de novo methylation than more rapidly dividing cells. To address this question, we analyzed Dnmt3b-mediated de novo methylation of proliferating and growth-arrested (mitomycin C treated or irradiated) mouse embryonic fibroblasts after 14 days of dox-induced transgene expression. We did not detect a significant difference between de novo methylation of actively dividing cells and growth-arrested cells (mitomycin C, P = 0.23; γ-irradiated, P = 0.41; paired t test versus proliferating cells; Figure 3C). This further supports the concept that Dnmt3b activation in the slow-cycling stem cell compartment can contribute to permanent epigenetic alterations in regenerating tissues. In summary, our results show that Dnmt3b induction in the mouse colon generates de novo methylation that closely resembles aberrant DNA methylation of human colon cancer. We demonstrated that cancer-specific DNA methylation can be a targeted process and may not be the result of stochastic methylation followed by clonal selection in the incipient tumor cell population. Importantly, our results showed that de novo methylation can occur in normal cells, which suggests that regional hypermethylation may contribute to the earliest stages of tumor formation. Finally, we demonstrated that transient induction of aberrant DNA methylation, most likely in intestinal stem cells, caused permanent epigenetic alterations of the intestinal mucosa. These observations support the concept that transient stressors can cause permanent epigenetic changes in somatic stem cells that accumulate over the lifetime of an organism, analogous to DNA mutations. Methods Transgenic mice. All mice used in this study and the genotyping protocol were described previously (5). Transgene induction was achieved by admin- The Journal of Clinical Investigation Volume 121 Number 5 May
6 istering 0.5 g/l dox in the drinking water. All animal studies were reviewed and approved by the Committee on Animal Care (protocol no ; Department of Comparative Medicine, MIT). Tissue harvesting. Colon tumors from APC Min/+ mice were harvested under the dissecting microscope. Colon epithelial cells from tumor-free colon samples of APC Min/+ mice and WT mice were harvested as described previously (14). Mouse embryonic fibroblasts. Double-homozygous embryos (Dnmt3b +/+ and Rosa-rtTA +/+ ) were harvested 12.5 days after conception. Growth arrest of cells was achieved by 24 Gy irradiation or mitomycin C treatment and verified using BrdU staining. After 14 days of dox treatment, cells were harvested for DNA methylation analysis. See Supplemental Methods for details. Lgr5 and Dnmt3b mrna FISH. Frozen sections of colon samples were hybridized with labeled probes targeting mouse Lgr5 and Dnmt3b mrna (12). Individual mrna molecules were located using the semiautomated method described previously (12). Lgr5-positive cells were manually identified to calculate Dmnt3b mrna density in intestinal stem cells. See Supplemental Methods for details and probe sequences. cdna and quantitative PCR. cdna production and quantitative PCR was conducted as described previously (5). See Supplemental Methods for details and primer sequences. DNA methylation analysis (mass array platform). Genomic DNA was sodium bisulfite converted and PCR amplified. When feasible, amplicons were designed to cover CpG islands in the same region as the 5 untranslated region. MassCLEAVE biochemistry, mass spectra acquisition, and methylation ratio analysis was performed as previously described (7). Primers are listed in Supplemental Methods. DNA methylation analysis (reduced representation bisulfite sequencing). RRBS methylation analysis was conducted as described previously (8). Briefly, mouse genomic DNA was digested with MspI, and resulting fragments between 40 and 220 bp were subjected to sequencing (Illumina genome Analyzer II) (8). Sequence reads that mapped to a promoter region (defined as 2 kb up- and downstream of the transcription start site) were then used to calculate promoter methylation. Raw data and processed RRBS data were deposited in GEO ( accession no. GSE26758). See Supplemental Methods for details. Statistics. All correlations were performed using the Pearson correlation. Significant difference in methylation fraction for the RRBS data was assayed with the moderated 2-tailed t test, corrected for FDR, with significance defined as FDR P value less than Significant difference for colon mass array methylation data was assayed by 2-tailed t test and for cell culture mass array data by 2-tailed paired t test. For RNA FISH, we obtained errors by bootstrap resampling; for P values, we computed the fraction of events that the null hypothesis would be satisfied given a random resampling of the data set. In all cases, P values less than 0.05 were considered significant. Acknowledgments We thank Jessica Dausman, Ruth Flannery, and Qing Gao for help with maintenance of the mouse colony and Tom DiCesare for support in preparation of figures. E.J. Steine is supported by Prins Bernhard Cultuurfonds Banning-de Jong Fonds ; H.G. Linhart is supported by a fellowship from the Fritz-Thyssen Stiftung. Research described in this article was supported by grants to R. Jaenisch by Philip Morris International and by NIH grant RO1-CA A. Raj was supported by a Burroughs-Wellcome Fund Career Award at the Scientific Interface. A. Raj was supported by a Burroughs- Wellcome Fund Career Award at the Scientific Interface. A. van Oudenaarden is supported by an NIH Director s Pioneer award and award U54CA from the National Cancer Institute. Received for publication March 31, 2010, and accepted in revised form January 26, Address correspondence to: Rudolf Jaenisch, Whitehead Institute and Department of Biology, MIT, 9 Cambridge Center, Cambridge, Massachusetts 02142, USA. Phone: ; Fax: ; jaenisch@wi.mit.edu. Or to: Heinz Linhart, DKFZ, Im Neuenheimer Feld 581 (TP4), Heidelberg, Germany. Phone: ; Fax: ; heinz. linhart@nct-heidelberg.de. 1. Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128(4): Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447(7143): Opavsky R, et al. CpG island methylation in a mouse model of lymphoma is driven by the genetic configuration of tumor cells. PLoS Genet. 2007;3(9): Nosho K, et al. DNMT3B expression might contribute to CpG island methylator phenotype in colorectal cancer. Clin Cancer Res. 2009;15(11): Linhart HG, et al. Dnmt3b promotes tumorigenesis in vivo by gene-specific de novo methylation and transcriptional silencing. Genes Dev. 2007; 21(23): Widschwendter M, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39(2): Ehrich M, et al. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci U S A. 2005;102(44): Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454(7205): Schlesinger Y, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007; 39(2): Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441(7091): Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007; 449(7165): Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mrna molecules using multiple singly labeled probes. Nat Methods. 2008;5(10): Velicescu M, Weisenberger DJ, Gonzales FA, Tsai YC, Nguyen CT, Jones PA. Cell division is required for de novo methylation of CpG islands in bladder cancer cells. Cancer Res. 2002;62(8): Fujimoto K, Beauchamp RD, Whitehead RH. Identification and isolation of candidate human colonic clonogenic cells based on cell surface integrin expression. Gastroenterology. 2002;123(6): The Journal of Clinical Investigation Volume 121 Number 5 May 2011
Whitehead Institute for Biomedical Research and Department of Biology, Massachusetts Institute of Technology (MIT), Cambridge, Massachusetts, USA.
 Eveline J. Steine, 1 Mathias Ehrich, 2 George W. Bell, 1 Arjun Raj, 3 Seshamma Reddy, 1 Alexander van Oudenaarden, 3 Rudolf Jaenisch, 1 and Heinz G. Linhart 1,4,5 1 Whitehead Institute for Biomedical Research
Eveline J. Steine, 1 Mathias Ehrich, 2 George W. Bell, 1 Arjun Raj, 3 Seshamma Reddy, 1 Alexander van Oudenaarden, 3 Rudolf Jaenisch, 1 and Heinz G. Linhart 1,4,5 1 Whitehead Institute for Biomedical Research
Stem Cell Epigenetics
 Stem Cell Epigenetics Philippe Collas University of Oslo Institute of Basic Medical Sciences Norwegian Center for Stem Cell Research www.collaslab.com Source of stem cells in the body Somatic ( adult )
Stem Cell Epigenetics Philippe Collas University of Oslo Institute of Basic Medical Sciences Norwegian Center for Stem Cell Research www.collaslab.com Source of stem cells in the body Somatic ( adult )
FOLLICULAR LYMPHOMA- ILLUMINA METHYLATION. Jude Fitzgibbon
 FOLLICULAR LYMPHOMA- ILLUMINA METHYLATION Jude Fitzgibbon j.fitzgibbon@qmul.ac.uk Molecular Predictors of Clinical outcome Responders Alive Non-responders Dead TUMOUR GENETICS Targeted therapy, Prognostic
FOLLICULAR LYMPHOMA- ILLUMINA METHYLATION Jude Fitzgibbon j.fitzgibbon@qmul.ac.uk Molecular Predictors of Clinical outcome Responders Alive Non-responders Dead TUMOUR GENETICS Targeted therapy, Prognostic
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10866 a b 1 2 3 4 5 6 7 Match No Match 1 2 3 4 5 6 7 Turcan et al. Supplementary Fig.1 Concepts mapping H3K27 targets in EF CBX8 targets in EF H3K27 targets in ES SUZ12 targets in ES
doi:10.1038/nature10866 a b 1 2 3 4 5 6 7 Match No Match 1 2 3 4 5 6 7 Turcan et al. Supplementary Fig.1 Concepts mapping H3K27 targets in EF CBX8 targets in EF H3K27 targets in ES SUZ12 targets in ES
Nature Methods: doi: /nmeth.3115
 Supplementary Figure 1 Analysis of DNA methylation in a cancer cohort based on Infinium 450K data. RnBeads was used to rediscover a clinically distinct subgroup of glioblastoma patients characterized by
Supplementary Figure 1 Analysis of DNA methylation in a cancer cohort based on Infinium 450K data. RnBeads was used to rediscover a clinically distinct subgroup of glioblastoma patients characterized by
Transcriptional repression of Xi
 Transcriptional repression of Xi Xist Transcription of Xist Xist RNA Spreading of Xist Recruitment of repression factors. Stable repression Translocated Xic cannot efficiently silence autosome regions.
Transcriptional repression of Xi Xist Transcription of Xist Xist RNA Spreading of Xist Recruitment of repression factors. Stable repression Translocated Xic cannot efficiently silence autosome regions.
The silence of the genes: clinical applications of (colorectal) cancer epigenetics
 The silence of the genes: clinical applications of (colorectal) cancer epigenetics Manon van Engeland, PhD Dept. of Pathology GROW - School for Oncology & Developmental Biology Maastricht University Medical
The silence of the genes: clinical applications of (colorectal) cancer epigenetics Manon van Engeland, PhD Dept. of Pathology GROW - School for Oncology & Developmental Biology Maastricht University Medical
Repressive Transcription
 Repressive Transcription The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation As Published Publisher Guenther, M. G., and R. A.
Repressive Transcription The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation As Published Publisher Guenther, M. G., and R. A.
R. Piazza (MD, PhD), Dept. of Medicine and Surgery, University of Milano-Bicocca EPIGENETICS
 R. Piazza (MD, PhD), Dept. of Medicine and Surgery, University of Milano-Bicocca EPIGENETICS EPIGENETICS THE STUDY OF CHANGES IN GENE EXPRESSION THAT ARE POTENTIALLY HERITABLE AND THAT DO NOT ENTAIL A
R. Piazza (MD, PhD), Dept. of Medicine and Surgery, University of Milano-Bicocca EPIGENETICS EPIGENETICS THE STUDY OF CHANGES IN GENE EXPRESSION THAT ARE POTENTIALLY HERITABLE AND THAT DO NOT ENTAIL A
SUPPLEMENTARY FIGURES: Supplementary Figure 1
 SUPPLEMENTARY FIGURES: Supplementary Figure 1 Supplementary Figure 1. Glioblastoma 5hmC quantified by paired BS and oxbs treated DNA hybridized to Infinium DNA methylation arrays. Workflow depicts analytic
SUPPLEMENTARY FIGURES: Supplementary Figure 1 Supplementary Figure 1. Glioblastoma 5hmC quantified by paired BS and oxbs treated DNA hybridized to Infinium DNA methylation arrays. Workflow depicts analytic
Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types.
 Supplementary Figure 1 Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types. (a) Pearson correlation heatmap among open chromatin profiles of different
Supplementary Figure 1 Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types. (a) Pearson correlation heatmap among open chromatin profiles of different
Dnmt3b promotes tumorigenesis in vivo by gene-specific de novo methylation and transcriptional silencing
 Dnmt3b promotes tumorigenesis in vivo by gene-specific de novo methylation and transcriptional silencing Heinz G. Linhart, 1 Haijiang Lin, 1,6 Yasuhiro Yamada, 2 Eva Moran, 1 Eveline J. Steine, 1 Sumita
Dnmt3b promotes tumorigenesis in vivo by gene-specific de novo methylation and transcriptional silencing Heinz G. Linhart, 1 Haijiang Lin, 1,6 Yasuhiro Yamada, 2 Eva Moran, 1 Eveline J. Steine, 1 Sumita
Epigenetics DNA methylation. Biosciences 741: Genomics Fall, 2013 Week 13. DNA Methylation
 Epigenetics DNA methylation Biosciences 741: Genomics Fall, 2013 Week 13 DNA Methylation Most methylated cytosines are found in the dinucleotide sequence CG, denoted mcpg. The restriction enzyme HpaII
Epigenetics DNA methylation Biosciences 741: Genomics Fall, 2013 Week 13 DNA Methylation Most methylated cytosines are found in the dinucleotide sequence CG, denoted mcpg. The restriction enzyme HpaII
DNA methylation & demethylation
 DNA methylation & demethylation Lars Schomacher (Group Christof Niehrs) What is Epigenetics? Epigenetics is the study of heritable changes in gene expression (active versus inactive genes) that do not
DNA methylation & demethylation Lars Schomacher (Group Christof Niehrs) What is Epigenetics? Epigenetics is the study of heritable changes in gene expression (active versus inactive genes) that do not
SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer.
 Supplementary Figure 1 SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer. Scatter plots comparing expression profiles of matched pretreatment
Supplementary Figure 1 SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer. Scatter plots comparing expression profiles of matched pretreatment
SUPPLEMENTAL INFORMATION
 SUPPLEMENTAL INFORMATION GO term analysis of differentially methylated SUMIs. GO term analysis of the 458 SUMIs with the largest differential methylation between human and chimp shows that they are more
SUPPLEMENTAL INFORMATION GO term analysis of differentially methylated SUMIs. GO term analysis of the 458 SUMIs with the largest differential methylation between human and chimp shows that they are more
Biochemical Determinants Governing Redox Regulated Changes in Gene Expression and Chromatin Structure
 Biochemical Determinants Governing Redox Regulated Changes in Gene Expression and Chromatin Structure Frederick E. Domann, Ph.D. Associate Professor of Radiation Oncology The University of Iowa Iowa City,
Biochemical Determinants Governing Redox Regulated Changes in Gene Expression and Chromatin Structure Frederick E. Domann, Ph.D. Associate Professor of Radiation Oncology The University of Iowa Iowa City,
Not IN Our Genes - A Different Kind of Inheritance.! Christopher Phiel, Ph.D. University of Colorado Denver Mini-STEM School February 4, 2014
 Not IN Our Genes - A Different Kind of Inheritance! Christopher Phiel, Ph.D. University of Colorado Denver Mini-STEM School February 4, 2014 Epigenetics in Mainstream Media Epigenetics *Current definition:
Not IN Our Genes - A Different Kind of Inheritance! Christopher Phiel, Ph.D. University of Colorado Denver Mini-STEM School February 4, 2014 Epigenetics in Mainstream Media Epigenetics *Current definition:
Epigenetic programming in chronic lymphocytic leukemia
 Epigenetic programming in chronic lymphocytic leukemia Christopher Oakes 10 th Canadian CLL Research Meeting September 18-19 th, 2014 Epigenetics and DNA methylation programming in normal and tumor cells:
Epigenetic programming in chronic lymphocytic leukemia Christopher Oakes 10 th Canadian CLL Research Meeting September 18-19 th, 2014 Epigenetics and DNA methylation programming in normal and tumor cells:
Genetics and Genomics in Medicine Chapter 6 Questions
 Genetics and Genomics in Medicine Chapter 6 Questions Multiple Choice Questions Question 6.1 With respect to the interconversion between open and condensed chromatin shown below: Which of the directions
Genetics and Genomics in Medicine Chapter 6 Questions Multiple Choice Questions Question 6.1 With respect to the interconversion between open and condensed chromatin shown below: Which of the directions
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers
 Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers Gordon Blackshields Senior Bioinformatician Source BioScience 1 To Cancer Genetics Studies
Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers Gordon Blackshields Senior Bioinformatician Source BioScience 1 To Cancer Genetics Studies
Jayanti Tokas 1, Puneet Tokas 2, Shailini Jain 3 and Hariom Yadav 3
 Jayanti Tokas 1, Puneet Tokas 2, Shailini Jain 3 and Hariom Yadav 3 1 Department of Biotechnology, JMIT, Radaur, Haryana, India 2 KITM, Kurukshetra, Haryana, India 3 NIDDK, National Institute of Health,
Jayanti Tokas 1, Puneet Tokas 2, Shailini Jain 3 and Hariom Yadav 3 1 Department of Biotechnology, JMIT, Radaur, Haryana, India 2 KITM, Kurukshetra, Haryana, India 3 NIDDK, National Institute of Health,
Tissue of origin determines cancer-associated CpG island promoter hypermethylation patterns
 RESEARCH Open Access Tissue of origin determines cancer-associated CpG island promoter hypermethylation patterns Duncan Sproul 1,2, Robert R Kitchen 1,3, Colm E Nestor 1,2, J Michael Dixon 1, Andrew H
RESEARCH Open Access Tissue of origin determines cancer-associated CpG island promoter hypermethylation patterns Duncan Sproul 1,2, Robert R Kitchen 1,3, Colm E Nestor 1,2, J Michael Dixon 1, Andrew H
Epigenetics. Lyle Armstrong. UJ Taylor & Francis Group. f'ci Garland Science NEW YORK AND LONDON
 ... Epigenetics Lyle Armstrong f'ci Garland Science UJ Taylor & Francis Group NEW YORK AND LONDON Contents CHAPTER 1 INTRODUCTION TO 3.2 CHROMATIN ARCHITECTURE 21 THE STUDY OF EPIGENETICS 1.1 THE CORE
... Epigenetics Lyle Armstrong f'ci Garland Science UJ Taylor & Francis Group NEW YORK AND LONDON Contents CHAPTER 1 INTRODUCTION TO 3.2 CHROMATIN ARCHITECTURE 21 THE STUDY OF EPIGENETICS 1.1 THE CORE
Liposarcoma*Genome*Project*
 LiposarcomaGenomeProject July2015! Submittedby: JohnMullen,MD EdwinChoy,MD,PhD GregoryCote,MD,PhD G.PeturNielsen,MD BradBernstein,MD,PhD Liposarcoma Background Liposarcoma is the most common soft tissue
LiposarcomaGenomeProject July2015! Submittedby: JohnMullen,MD EdwinChoy,MD,PhD GregoryCote,MD,PhD G.PeturNielsen,MD BradBernstein,MD,PhD Liposarcoma Background Liposarcoma is the most common soft tissue
Fragile X Syndrome. Genetics, Epigenetics & the Role of Unprogrammed Events in the expression of a Phenotype
 Fragile X Syndrome Genetics, Epigenetics & the Role of Unprogrammed Events in the expression of a Phenotype A loss of function of the FMR-1 gene results in severe learning problems, intellectual disability
Fragile X Syndrome Genetics, Epigenetics & the Role of Unprogrammed Events in the expression of a Phenotype A loss of function of the FMR-1 gene results in severe learning problems, intellectual disability
SUPPLEMENTARY INFORMATION
 doi:.38/nature8975 SUPPLEMENTAL TEXT Unique association of HOTAIR with patient outcome To determine whether the expression of other HOX lincrnas in addition to HOTAIR can predict patient outcome, we measured
doi:.38/nature8975 SUPPLEMENTAL TEXT Unique association of HOTAIR with patient outcome To determine whether the expression of other HOX lincrnas in addition to HOTAIR can predict patient outcome, we measured
Nature Immunology: doi: /ni Supplementary Figure 1. DNA-methylation machinery is essential for silencing of Cd4 in cytotoxic T cells.
 Supplementary Figure 1 DNA-methylation machinery is essential for silencing of Cd4 in cytotoxic T cells. (a) Scheme for the retroviral shrna screen. (b) Histogram showing CD4 expression (MFI) in WT cytotoxic
Supplementary Figure 1 DNA-methylation machinery is essential for silencing of Cd4 in cytotoxic T cells. (a) Scheme for the retroviral shrna screen. (b) Histogram showing CD4 expression (MFI) in WT cytotoxic
Epigenetics. Jenny van Dongen Vrije Universiteit (VU) Amsterdam Boulder, Friday march 10, 2017
 Epigenetics Jenny van Dongen Vrije Universiteit (VU) Amsterdam j.van.dongen@vu.nl Boulder, Friday march 10, 2017 Epigenetics Epigenetics= The study of molecular mechanisms that influence the activity of
Epigenetics Jenny van Dongen Vrije Universiteit (VU) Amsterdam j.van.dongen@vu.nl Boulder, Friday march 10, 2017 Epigenetics Epigenetics= The study of molecular mechanisms that influence the activity of
Histones modifications and variants
 Histones modifications and variants Dr. Institute of Molecular Biology, Johannes Gutenberg University, Mainz www.imb.de Lecture Objectives 1. Chromatin structure and function Chromatin and cell state Nucleosome
Histones modifications and variants Dr. Institute of Molecular Biology, Johannes Gutenberg University, Mainz www.imb.de Lecture Objectives 1. Chromatin structure and function Chromatin and cell state Nucleosome
p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs
 p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs 2013, Katerina I. Leonova et al. Kolmogorov Mikhail Noncoding DNA Mammalian
p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs 2013, Katerina I. Leonova et al. Kolmogorov Mikhail Noncoding DNA Mammalian
Alpha thalassemia mental retardation X-linked. Acquired alpha-thalassemia myelodysplastic syndrome
 Alpha thalassemia mental retardation X-linked Acquired alpha-thalassemia myelodysplastic syndrome (Alpha thalassemia mental retardation X-linked) Acquired alpha-thalassemia myelodysplastic syndrome Schematic
Alpha thalassemia mental retardation X-linked Acquired alpha-thalassemia myelodysplastic syndrome (Alpha thalassemia mental retardation X-linked) Acquired alpha-thalassemia myelodysplastic syndrome Schematic
Dominic J Smiraglia, PhD Department of Cancer Genetics. DNA methylation in prostate cancer
 Dominic J Smiraglia, PhD Department of Cancer Genetics DNA methylation in prostate cancer Overarching theme Epigenetic regulation allows the genome to be responsive to the environment Sets the tone for
Dominic J Smiraglia, PhD Department of Cancer Genetics DNA methylation in prostate cancer Overarching theme Epigenetic regulation allows the genome to be responsive to the environment Sets the tone for
mirna Dr. S Hosseini-Asl
 mirna Dr. S Hosseini-Asl 1 2 MicroRNAs (mirnas) are small noncoding RNAs which enhance the cleavage or translational repression of specific mrna with recognition site(s) in the 3 - untranslated region
mirna Dr. S Hosseini-Asl 1 2 MicroRNAs (mirnas) are small noncoding RNAs which enhance the cleavage or translational repression of specific mrna with recognition site(s) in the 3 - untranslated region
A Genome-Wide Study of DNA Methylation Patterns and Gene Expression Levels in Multiple Human and Chimpanzee Tissues
 A Genome-Wide Study of DNA Methylation Patterns and Gene Expression Levels in Multiple Human and Chimpanzee Tissues Athma A. Pai 1 *, Jordana T. Bell 1 a, John C. Marioni 1 b, Jonathan K. Pritchard 1,2
A Genome-Wide Study of DNA Methylation Patterns and Gene Expression Levels in Multiple Human and Chimpanzee Tissues Athma A. Pai 1 *, Jordana T. Bell 1 a, John C. Marioni 1 b, Jonathan K. Pritchard 1,2
Results. Abstract. Introduc4on. Conclusions. Methods. Funding
 . expression that plays a role in many cellular processes affecting a variety of traits. In this study DNA methylation was assessed in neuronal tissue from three pigs (frontal lobe) and one great tit (whole
. expression that plays a role in many cellular processes affecting a variety of traits. In this study DNA methylation was assessed in neuronal tissue from three pigs (frontal lobe) and one great tit (whole
Generating Spontaneous Copy Number Variants (CNVs) Jennifer Freeman Assistant Professor of Toxicology School of Health Sciences Purdue University
 Role of Chemical lexposure in Generating Spontaneous Copy Number Variants (CNVs) Jennifer Freeman Assistant Professor of Toxicology School of Health Sciences Purdue University CNV Discovery Reference Genetic
Role of Chemical lexposure in Generating Spontaneous Copy Number Variants (CNVs) Jennifer Freeman Assistant Professor of Toxicology School of Health Sciences Purdue University CNV Discovery Reference Genetic
Breeding scheme, transgenes, histological analysis and site distribution of SB-mutagenized osteosarcoma.
 Supplementary Figure 1 Breeding scheme, transgenes, histological analysis and site distribution of SB-mutagenized osteosarcoma. (a) Breeding scheme. R26-LSL-SB11 homozygous mice were bred to Trp53 LSL-R270H/+
Supplementary Figure 1 Breeding scheme, transgenes, histological analysis and site distribution of SB-mutagenized osteosarcoma. (a) Breeding scheme. R26-LSL-SB11 homozygous mice were bred to Trp53 LSL-R270H/+
he micrornas of Caenorhabditis elegans (Lim et al. Genes & Development 2003)
 MicroRNAs: Genomics, Biogenesis, Mechanism, and Function (D. Bartel Cell 2004) he micrornas of Caenorhabditis elegans (Lim et al. Genes & Development 2003) Vertebrate MicroRNA Genes (Lim et al. Science
MicroRNAs: Genomics, Biogenesis, Mechanism, and Function (D. Bartel Cell 2004) he micrornas of Caenorhabditis elegans (Lim et al. Genes & Development 2003) Vertebrate MicroRNA Genes (Lim et al. Science
Supplementary Figure 1: High-throughput profiling of survival after exposure to - radiation. (a) Cells were plated in at least 7 wells in a 384-well
 Supplementary Figure 1: High-throughput profiling of survival after exposure to - radiation. (a) Cells were plated in at least 7 wells in a 384-well plate at cell densities ranging from 25-225 cells in
Supplementary Figure 1: High-throughput profiling of survival after exposure to - radiation. (a) Cells were plated in at least 7 wells in a 384-well plate at cell densities ranging from 25-225 cells in
The role of DNMT3A and HOXA9 hypomethylation in acute myeloid leukemia (AML)
 Campbell Drohan BIOL 463 December 2016 The role of DNMT3A and HOXA9 hypomethylation in acute myeloid leukemia (AML) Introduction Epigenetic modifications of DNA and histones are key to gene regulation
Campbell Drohan BIOL 463 December 2016 The role of DNMT3A and HOXA9 hypomethylation in acute myeloid leukemia (AML) Introduction Epigenetic modifications of DNA and histones are key to gene regulation
Chromatin-Based Regulation of Gene Expression
 Chromatin-Based Regulation of Gene Expression.George J. Quellhorst, Jr., PhD.Associate Director, R&D.Biological Content Development Topics to be Discussed Importance of Chromatin-Based Regulation Mechanism
Chromatin-Based Regulation of Gene Expression.George J. Quellhorst, Jr., PhD.Associate Director, R&D.Biological Content Development Topics to be Discussed Importance of Chromatin-Based Regulation Mechanism
BIOL2005 WORKSHEET 2008
 BIOL2005 WORKSHEET 2008 Answer all 6 questions in the space provided using additional sheets where necessary. Hand your completed answers in to the Biology office by 3 p.m. Friday 8th February. 1. Your
BIOL2005 WORKSHEET 2008 Answer all 6 questions in the space provided using additional sheets where necessary. Hand your completed answers in to the Biology office by 3 p.m. Friday 8th February. 1. Your
Nature Getetics: doi: /ng.3471
 Supplementary Figure 1 Summary of exome sequencing data. ( a ) Exome tumor normal sample sizes for bladder cancer (BLCA), breast cancer (BRCA), carcinoid (CARC), chronic lymphocytic leukemia (CLLX), colorectal
Supplementary Figure 1 Summary of exome sequencing data. ( a ) Exome tumor normal sample sizes for bladder cancer (BLCA), breast cancer (BRCA), carcinoid (CARC), chronic lymphocytic leukemia (CLLX), colorectal
Supplementary Information Titles Journal: Nature Medicine
 Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
Computational Analysis of UHT Sequences Histone modifications, CAGE, RNA-Seq
 Computational Analysis of UHT Sequences Histone modifications, CAGE, RNA-Seq Philipp Bucher Wednesday January 21, 2009 SIB graduate school course EPFL, Lausanne ChIP-seq against histone variants: Biological
Computational Analysis of UHT Sequences Histone modifications, CAGE, RNA-Seq Philipp Bucher Wednesday January 21, 2009 SIB graduate school course EPFL, Lausanne ChIP-seq against histone variants: Biological
LGR5. Dnmt3b induced. Dnmt3b. Supplemental Figure 1
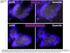 Supplemental Figure 1 LGR5 Control Dnmt3b LGR5 Dnmt3b induced Dnmt3b Supplemental Figure 1: Illustration of RNA FISH analysis of Lgr5 (left panel) and Dmnt3b expression (right panel) in colon crypts isolated
Supplemental Figure 1 LGR5 Control Dnmt3b LGR5 Dnmt3b induced Dnmt3b Supplemental Figure 1: Illustration of RNA FISH analysis of Lgr5 (left panel) and Dmnt3b expression (right panel) in colon crypts isolated
Today. Genomic Imprinting & X-Inactivation
 Today 1. Quiz (~12 min) 2. Genomic imprinting in mammals 3. X-chromosome inactivation in mammals Note that readings on Dosage Compensation and Genomic Imprinting in Mammals are on our web site. Genomic
Today 1. Quiz (~12 min) 2. Genomic imprinting in mammals 3. X-chromosome inactivation in mammals Note that readings on Dosage Compensation and Genomic Imprinting in Mammals are on our web site. Genomic
The Epigenome Tools 2: ChIP-Seq and Data Analysis
 The Epigenome Tools 2: ChIP-Seq and Data Analysis Chongzhi Zang zang@virginia.edu http://zanglab.com PHS5705: Public Health Genomics March 20, 2017 1 Outline Epigenome: basics review ChIP-seq overview
The Epigenome Tools 2: ChIP-Seq and Data Analysis Chongzhi Zang zang@virginia.edu http://zanglab.com PHS5705: Public Health Genomics March 20, 2017 1 Outline Epigenome: basics review ChIP-seq overview
Neurotrophic factor GDNF and camp suppress glucocorticoid-inducible PNMT expression in a mouse pheochromocytoma model.
 161 Neurotrophic factor GDNF and camp suppress glucocorticoid-inducible PNMT expression in a mouse pheochromocytoma model. Marian J. Evinger a, James F. Powers b and Arthur S. Tischler b a. Department
161 Neurotrophic factor GDNF and camp suppress glucocorticoid-inducible PNMT expression in a mouse pheochromocytoma model. Marian J. Evinger a, James F. Powers b and Arthur S. Tischler b a. Department
Cellecta Overview. Started Operations in 2007 Headquarters: Mountain View, CA
 Cellecta Overview Started Operations in 2007 Headquarters: Mountain View, CA Focus: Development of flexible, scalable, and broadly parallel genetic screening assays to expedite the discovery and characterization
Cellecta Overview Started Operations in 2007 Headquarters: Mountain View, CA Focus: Development of flexible, scalable, and broadly parallel genetic screening assays to expedite the discovery and characterization
Barrett s Esophagus and Esophageal Adenocarcinoma Epigenetic Biomarker Discovery Using Infinium Methylation
 February 2008 Application Note arrett s Esophagus and Esophageal Adenocarcinoma Epigenetic iomarker Discovery Using Infinium Methylation Contributed by Patricia C. Galipeau, Dennis L. Chao, Xiaohong Li,
February 2008 Application Note arrett s Esophagus and Esophageal Adenocarcinoma Epigenetic iomarker Discovery Using Infinium Methylation Contributed by Patricia C. Galipeau, Dennis L. Chao, Xiaohong Li,
DNA methylation: a potential clinical biomarker for the detection of human cancers
 DNA methylation: a potential clinical biomarker for the detection of human cancers Name: Tong Samuel Supervisor: Zigui CHEN Date: 1 st December 2016 Department: Microbiology Source: cited from Jakubowski,
DNA methylation: a potential clinical biomarker for the detection of human cancers Name: Tong Samuel Supervisor: Zigui CHEN Date: 1 st December 2016 Department: Microbiology Source: cited from Jakubowski,
Epigenetics and Chromatin Remodeling
 Epigenetics and Chromatin Remodeling Bradford Coffee, PhD, FACMG Emory University Atlanta, GA Speaker Disclosure Information Grant/Research Support: none Salary/Consultant Fees: none Board/Committee/Advisory
Epigenetics and Chromatin Remodeling Bradford Coffee, PhD, FACMG Emory University Atlanta, GA Speaker Disclosure Information Grant/Research Support: none Salary/Consultant Fees: none Board/Committee/Advisory
Analysis of shotgun bisulfite sequencing of cancer samples
 Analysis of shotgun bisulfite sequencing of cancer samples Kasper Daniel Hansen Postdoc with Rafael Irizarry Johns Hopkins Bloomberg School of Public Health Brixen, July 1st, 2011 The
Analysis of shotgun bisulfite sequencing of cancer samples Kasper Daniel Hansen Postdoc with Rafael Irizarry Johns Hopkins Bloomberg School of Public Health Brixen, July 1st, 2011 The
Clinical and Pathological Significance of Epigenomic Changes in Colorectal Cancer
 Clinical and Pathological Significance of Epigenomic Changes in Colorectal Cancer Shuji Ogino, M.D., Ph.D. Associate Professor of Pathology Harvard Medical School Brigham and Women s Hospital Dana-Farber
Clinical and Pathological Significance of Epigenomic Changes in Colorectal Cancer Shuji Ogino, M.D., Ph.D. Associate Professor of Pathology Harvard Medical School Brigham and Women s Hospital Dana-Farber
Back to the Basics: Methyl-Seq 101
 Back to the Basics: Methyl-Seq 101 Presented By: Alex Siebold, Ph.D. October 9, 2013 Field Applications Scientist Agilent Technologies Life Sciences & Diagnostics Group Life Sciences & Diagnostics Group
Back to the Basics: Methyl-Seq 101 Presented By: Alex Siebold, Ph.D. October 9, 2013 Field Applications Scientist Agilent Technologies Life Sciences & Diagnostics Group Life Sciences & Diagnostics Group
Epigenetic Pathways Linking Parental Effects to Offspring Development. Dr. Frances A. Champagne Department of Psychology, Columbia University
 Epigenetic Pathways Linking Parental Effects to Offspring Development Dr. Frances A. Champagne Department of Psychology, Columbia University Prenatal & Postnatal Experiences Individual differences in brain
Epigenetic Pathways Linking Parental Effects to Offspring Development Dr. Frances A. Champagne Department of Psychology, Columbia University Prenatal & Postnatal Experiences Individual differences in brain
Epigenetics: Basic Principals and role in health and disease
 Epigenetics: Basic Principals and role in health and disease Cambridge Masterclass Workshop on Epigenetics in GI Health and Disease 3 rd September 2013 Matt Zilbauer Overview Basic principals of Epigenetics
Epigenetics: Basic Principals and role in health and disease Cambridge Masterclass Workshop on Epigenetics in GI Health and Disease 3 rd September 2013 Matt Zilbauer Overview Basic principals of Epigenetics
Copyright 2014 The Authors.
 n Lund, K., Cole, J., VanderKraats, N. D., McBryan, T., Pchelintsev, N. A., Clark, W., Copland, M., Edwards, J. R., and Adams, P.(214) DNMT inhibitors reverse a specific signature of aberrant promoter
n Lund, K., Cole, J., VanderKraats, N. D., McBryan, T., Pchelintsev, N. A., Clark, W., Copland, M., Edwards, J. R., and Adams, P.(214) DNMT inhibitors reverse a specific signature of aberrant promoter
Cluster Dendrogram. dist(cor(na.omit(tss.exprs.chip[, c(1:10, 24, 27, 30, 48:50, dist(cor(na.omit(tss.exprs.chip[, c(1:99, 103, 104, 109, 110,
 A Transcriptome (RNA-seq) Transcriptome (RNA-seq) 3. 2.5 2..5..5...5..5 2. 2.5 3. 2.5 2..5..5...5..5 2. 2.5 Cluster Dendrogram RS_ES3.2 RS_ES3. RS_SHS5.2 RS_SHS5. PS_SHS5.2 PS_SHS5. RS_LJ3 PS_LJ3..4 _SHS5.2
A Transcriptome (RNA-seq) Transcriptome (RNA-seq) 3. 2.5 2..5..5...5..5 2. 2.5 3. 2.5 2..5..5...5..5 2. 2.5 Cluster Dendrogram RS_ES3.2 RS_ES3. RS_SHS5.2 RS_SHS5. PS_SHS5.2 PS_SHS5. RS_LJ3 PS_LJ3..4 _SHS5.2
Epigenetics: A historical overview Dr. Robin Holliday
 Epigenetics 1 Rival hypotheses Epigenisis - The embryo is initially undifferentiated. As development proceeds, increasing levels of complexity emerge giving rise to the larval stage or to the adult organism.
Epigenetics 1 Rival hypotheses Epigenisis - The embryo is initially undifferentiated. As development proceeds, increasing levels of complexity emerge giving rise to the larval stage or to the adult organism.
Epigenetic Regulation of Health and Disease Nutritional and environmental effects on epigenetic regulation
 Epigenetic Regulation of Health and Disease Nutritional and environmental effects on epigenetic regulation Robert FEIL Director of Research CNRS & University of Montpellier, Montpellier, France. E-mail:
Epigenetic Regulation of Health and Disease Nutritional and environmental effects on epigenetic regulation Robert FEIL Director of Research CNRS & University of Montpellier, Montpellier, France. E-mail:
Iso-Seq Method Updates and Target Enrichment Without Amplification for SMRT Sequencing
 Iso-Seq Method Updates and Target Enrichment Without Amplification for SMRT Sequencing PacBio Americas User Group Meeting Sample Prep Workshop June.27.2017 Tyson Clark, Ph.D. For Research Use Only. Not
Iso-Seq Method Updates and Target Enrichment Without Amplification for SMRT Sequencing PacBio Americas User Group Meeting Sample Prep Workshop June.27.2017 Tyson Clark, Ph.D. For Research Use Only. Not
Carcinogenesis in IBD
 Oxford Inflammatory Bowel Disease MasterClass Carcinogenesis in IBD Dr Simon Leedham, Oxford, UK Oxford Inflammatory Bowel Disease MasterClass Carcinogenesis in Inflammatory Bowel Disease Dr Simon Leedham
Oxford Inflammatory Bowel Disease MasterClass Carcinogenesis in IBD Dr Simon Leedham, Oxford, UK Oxford Inflammatory Bowel Disease MasterClass Carcinogenesis in Inflammatory Bowel Disease Dr Simon Leedham
International Graduate Research Programme in Cardiovascular Science
 1 International Graduate Research Programme in Cardiovascular Science This work has been supported by the European Community s Sixth Framework Programme under grant agreement n LSHM-CT-2005-01883 EUGeneHeart.
1 International Graduate Research Programme in Cardiovascular Science This work has been supported by the European Community s Sixth Framework Programme under grant agreement n LSHM-CT-2005-01883 EUGeneHeart.
Imprinting. Joyce Ohm Cancer Genetics and Genomics CGP-L2-319 x8821
 Imprinting Joyce Ohm Cancer Genetics and Genomics CGP-L2-319 x8821 Learning Objectives 1. To understand the basic concepts of genomic imprinting Genomic imprinting is an epigenetic phenomenon that causes
Imprinting Joyce Ohm Cancer Genetics and Genomics CGP-L2-319 x8821 Learning Objectives 1. To understand the basic concepts of genomic imprinting Genomic imprinting is an epigenetic phenomenon that causes
PRC2 crystal clear. Matthieu Schapira
 PRC2 crystal clear Matthieu Schapira Epigenetic mechanisms control the combination of genes that are switched on and off in any given cell. In turn, this combination, called the transcriptional program,
PRC2 crystal clear Matthieu Schapira Epigenetic mechanisms control the combination of genes that are switched on and off in any given cell. In turn, this combination, called the transcriptional program,
iplex genotyping IDH1 and IDH2 assays utilized the following primer sets (forward and reverse primers along with extension primers).
 Supplementary Materials Supplementary Methods iplex genotyping IDH1 and IDH2 assays utilized the following primer sets (forward and reverse primers along with extension primers). IDH1 R132H and R132L Forward:
Supplementary Materials Supplementary Methods iplex genotyping IDH1 and IDH2 assays utilized the following primer sets (forward and reverse primers along with extension primers). IDH1 R132H and R132L Forward:
DOES THE BRCAX GENE EXIST? FUTURE OUTLOOK
 CHAPTER 6 DOES THE BRCAX GENE EXIST? FUTURE OUTLOOK Genetic research aimed at the identification of new breast cancer susceptibility genes is at an interesting crossroad. On the one hand, the existence
CHAPTER 6 DOES THE BRCAX GENE EXIST? FUTURE OUTLOOK Genetic research aimed at the identification of new breast cancer susceptibility genes is at an interesting crossroad. On the one hand, the existence
Studying The Role Of DNA Mismatch Repair In Brain Cancer Malignancy
 Kavya Puchhalapalli CALS Honors Project Report Spring 2017 Studying The Role Of DNA Mismatch Repair In Brain Cancer Malignancy Abstract Malignant brain tumors including medulloblastomas and primitive neuroectodermal
Kavya Puchhalapalli CALS Honors Project Report Spring 2017 Studying The Role Of DNA Mismatch Repair In Brain Cancer Malignancy Abstract Malignant brain tumors including medulloblastomas and primitive neuroectodermal
An epigenetic approach to understanding (and predicting?) environmental effects on gene expression
 www.collaslab.com An epigenetic approach to understanding (and predicting?) environmental effects on gene expression Philippe Collas University of Oslo Institute of Basic Medical Sciences Stem Cell Epigenetics
www.collaslab.com An epigenetic approach to understanding (and predicting?) environmental effects on gene expression Philippe Collas University of Oslo Institute of Basic Medical Sciences Stem Cell Epigenetics
Supplementary Figure 1: Hsp60 / IEC mice are embryonically lethal (A) Light microscopic pictures show mouse embryos at developmental stage E12.
 Supplementary Figure 1: Hsp60 / IEC mice are embryonically lethal (A) Light microscopic pictures show mouse embryos at developmental stage E12.5 and E13.5 prepared from uteri of dams and subsequently genotyped.
Supplementary Figure 1: Hsp60 / IEC mice are embryonically lethal (A) Light microscopic pictures show mouse embryos at developmental stage E12.5 and E13.5 prepared from uteri of dams and subsequently genotyped.
Accessing and Using ENCODE Data Dr. Peggy J. Farnham
 1 William M Keck Professor of Biochemistry Keck School of Medicine University of Southern California How many human genes are encoded in our 3x10 9 bp? C. elegans (worm) 959 cells and 1x10 8 bp 20,000
1 William M Keck Professor of Biochemistry Keck School of Medicine University of Southern California How many human genes are encoded in our 3x10 9 bp? C. elegans (worm) 959 cells and 1x10 8 bp 20,000
CANCER. Inherited Cancer Syndromes. Affects 25% of US population. Kills 19% of US population (2nd largest killer after heart disease)
 CANCER Affects 25% of US population Kills 19% of US population (2nd largest killer after heart disease) NOT one disease but 200-300 different defects Etiologic Factors In Cancer: Relative contributions
CANCER Affects 25% of US population Kills 19% of US population (2nd largest killer after heart disease) NOT one disease but 200-300 different defects Etiologic Factors In Cancer: Relative contributions
Loss of Dnmt3b function upregulates the tumor modifier Ment and accelerates mouse lymphomagenesis
 Research article Loss of Dnmt3b function upregulates the tumor modifier Ment and accelerates mouse lymphomagenesis Ryan A. Hlady, 1 Slavomira Novakova, 1 Jana Opavska, 1 David Klinkebiel, 2 Staci L. Peters,
Research article Loss of Dnmt3b function upregulates the tumor modifier Ment and accelerates mouse lymphomagenesis Ryan A. Hlady, 1 Slavomira Novakova, 1 Jana Opavska, 1 David Klinkebiel, 2 Staci L. Peters,
GCATCCATCTTGGGGCGTCCCAATTGCTGAGTAACAAATGAGACGC TGTGGCCAAACTCAGTCATAACTAATGACATTTCTAGACAAAGTGAC TTCAGATTTTCAAAGCGTACCCTGTTTACATCATTTTGCCAATTTCG
 Lecture 6 GCATCCATCTTGGGGCGTCCCAATTGCTGAGTAACAAATGAGACGC TGTGGCCAAACTCAGTCATAACTAATGACATTTCTAGACAAAGTGAC TTCAGATTTTCAAAGCGTACCCTGTTTACATCATTTTGCCAATTTCG CGTACTGCAACCGGCGGGCCACGCCCCCGTGAAAAGAAGGTTGTT TTCTCCACATTTCGGGGTTCTGGACGTTTCCCGGCTGCGGGGCGG
Lecture 6 GCATCCATCTTGGGGCGTCCCAATTGCTGAGTAACAAATGAGACGC TGTGGCCAAACTCAGTCATAACTAATGACATTTCTAGACAAAGTGAC TTCAGATTTTCAAAGCGTACCCTGTTTACATCATTTTGCCAATTTCG CGTACTGCAACCGGCGGGCCACGCCCCCGTGAAAAGAAGGTTGTT TTCTCCACATTTCGGGGTTCTGGACGTTTCCCGGCTGCGGGGCGG
Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression.
 Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Supplementary Figure 1. Estimation of tumour content
 Supplementary Figure 1. Estimation of tumour content a, Approach used to estimate the tumour content in S13T1/T2, S6T1/T2, S3T1/T2 and S12T1/T2. Tissue and tumour areas were evaluated by two independent
Supplementary Figure 1. Estimation of tumour content a, Approach used to estimate the tumour content in S13T1/T2, S6T1/T2, S3T1/T2 and S12T1/T2. Tissue and tumour areas were evaluated by two independent
Nature Immunology: doi: /ni Supplementary Figure 1. Characteristics of SEs in T reg and T conv cells.
 Supplementary Figure 1 Characteristics of SEs in T reg and T conv cells. (a) Patterns of indicated transcription factor-binding at SEs and surrounding regions in T reg and T conv cells. Average normalized
Supplementary Figure 1 Characteristics of SEs in T reg and T conv cells. (a) Patterns of indicated transcription factor-binding at SEs and surrounding regions in T reg and T conv cells. Average normalized
EPIGENOMICS PROFILING SERVICES
 EPIGENOMICS PROFILING SERVICES Chromatin analysis DNA methylation analysis RNA-seq analysis Diagenode helps you uncover the mysteries of epigenetics PAGE 3 Integrative epigenomics analysis DNA methylation
EPIGENOMICS PROFILING SERVICES Chromatin analysis DNA methylation analysis RNA-seq analysis Diagenode helps you uncover the mysteries of epigenetics PAGE 3 Integrative epigenomics analysis DNA methylation
Nature Genetics: doi: /ng Supplementary Figure 1. Assessment of sample purity and quality.
 Supplementary Figure 1 Assessment of sample purity and quality. (a) Hematoxylin and eosin staining of formaldehyde-fixed, paraffin-embedded sections from a human testis biopsy collected concurrently with
Supplementary Figure 1 Assessment of sample purity and quality. (a) Hematoxylin and eosin staining of formaldehyde-fixed, paraffin-embedded sections from a human testis biopsy collected concurrently with
September 20, Submitted electronically to: Cc: To Whom It May Concern:
 History Study (NOT-HL-12-147), p. 1 September 20, 2012 Re: Request for Information (RFI): Building a National Resource to Study Myelodysplastic Syndromes (MDS) The MDS Cohort Natural History Study (NOT-HL-12-147).
History Study (NOT-HL-12-147), p. 1 September 20, 2012 Re: Request for Information (RFI): Building a National Resource to Study Myelodysplastic Syndromes (MDS) The MDS Cohort Natural History Study (NOT-HL-12-147).
Circular RNAs (circrnas) act a stable mirna sponges
 Circular RNAs (circrnas) act a stable mirna sponges cernas compete for mirnas Ancestal mrna (+3 UTR) Pseudogene RNA (+3 UTR homolgy region) The model holds true for all RNAs that share a mirna binding
Circular RNAs (circrnas) act a stable mirna sponges cernas compete for mirnas Ancestal mrna (+3 UTR) Pseudogene RNA (+3 UTR homolgy region) The model holds true for all RNAs that share a mirna binding
Chromatin marks identify critical cell-types for fine-mapping complex trait variants
 Chromatin marks identify critical cell-types for fine-mapping complex trait variants Gosia Trynka 1-4 *, Cynthia Sandor 1-4 *, Buhm Han 1-4, Han Xu 5, Barbara E Stranger 1,4#, X Shirley Liu 5, and Soumya
Chromatin marks identify critical cell-types for fine-mapping complex trait variants Gosia Trynka 1-4 *, Cynthia Sandor 1-4 *, Buhm Han 1-4, Han Xu 5, Barbara E Stranger 1,4#, X Shirley Liu 5, and Soumya
Alan G. Casson FRCSC Professor of Surgery & VP Integrated Health Services University of Saskatchewan & Saskatoon Health Region
 Identification and Characterization of Stem-like Cells in Human Esophageal Adenocarcinoma and Normal Epithelial Cell Lines No disclosures / conflict of interest Alan G. Casson FRCSC Professor of Surgery
Identification and Characterization of Stem-like Cells in Human Esophageal Adenocarcinoma and Normal Epithelial Cell Lines No disclosures / conflict of interest Alan G. Casson FRCSC Professor of Surgery
Epigenetics Armstrong_Prelims.indd 1 04/11/2013 3:28 pm
 Epigenetics Epigenetics Lyle Armstrong vi Online resources Accessible from www.garlandscience.com, the Student and Instructor Resource Websites provide learning and teaching tools created for Epigenetics.
Epigenetics Epigenetics Lyle Armstrong vi Online resources Accessible from www.garlandscience.com, the Student and Instructor Resource Websites provide learning and teaching tools created for Epigenetics.
Towards an automated procedure for annotation of gene products through SAS Methods. Henrik Tveit Norwegian University of Science and Technology
 Towards an automated procedure for annotation of gene products through SAS Methods Henrik Tveit Norwegian University of Science and Technology Overview! Biology: the Structure of Life! Functional genomics!
Towards an automated procedure for annotation of gene products through SAS Methods Henrik Tveit Norwegian University of Science and Technology Overview! Biology: the Structure of Life! Functional genomics!
Comparative DNA methylome analysis of endometrial carcinoma reveals complex and distinct deregulation of cancer promoters and enhancers
 Zhang et al. BMC Genomics 2014, 15:868 RESEARCH ARTICLE Open Access Comparative DNA methylome analysis of endometrial carcinoma reveals complex and distinct deregulation of cancer promoters and enhancers
Zhang et al. BMC Genomics 2014, 15:868 RESEARCH ARTICLE Open Access Comparative DNA methylome analysis of endometrial carcinoma reveals complex and distinct deregulation of cancer promoters and enhancers
Epigenetics 101. Kevin Sweet, MS, CGC Division of Human Genetics
 Epigenetics 101 Kevin Sweet, MS, CGC Division of Human Genetics Learning Objectives 1. Evaluate the genetic code and the role epigenetic modification plays in common complex disease 2. Evaluate the effects
Epigenetics 101 Kevin Sweet, MS, CGC Division of Human Genetics Learning Objectives 1. Evaluate the genetic code and the role epigenetic modification plays in common complex disease 2. Evaluate the effects
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1171320/dc1 Supporting Online Material for A Frazzled/DCC-Dependent Transcriptional Switch Regulates Midline Axon Guidance Long Yang, David S. Garbe, Greg J. Bashaw*
www.sciencemag.org/cgi/content/full/1171320/dc1 Supporting Online Material for A Frazzled/DCC-Dependent Transcriptional Switch Regulates Midline Axon Guidance Long Yang, David S. Garbe, Greg J. Bashaw*
MicroRNA and Male Infertility: A Potential for Diagnosis
 Review Article MicroRNA and Male Infertility: A Potential for Diagnosis * Abstract MicroRNAs (mirnas) are small non-coding single stranded RNA molecules that are physiologically produced in eukaryotic
Review Article MicroRNA and Male Infertility: A Potential for Diagnosis * Abstract MicroRNAs (mirnas) are small non-coding single stranded RNA molecules that are physiologically produced in eukaryotic
Integrated genomic analysis of human osteosarcomas
 Integrated genomic analysis of human osteosarcomas Leonardo A. Meza-Zepeda Project Leader Genomic Section Department of Tumor Biology The Norwegian Radium Hospital Head Microarray Core Facility Norwegian
Integrated genomic analysis of human osteosarcomas Leonardo A. Meza-Zepeda Project Leader Genomic Section Department of Tumor Biology The Norwegian Radium Hospital Head Microarray Core Facility Norwegian
Aberrant Promoter CpG Methylation is a Mechanism for Lack of Hypoxic Induction of
 Aberrant Promoter CpG Methylation is a Mechanism for Lack of Hypoxic Induction of PHD3 in a Diverse Set of Malignant Cells Abstract The prolyl-hydroxylase domain family of enzymes (PHD1-3) plays an important
Aberrant Promoter CpG Methylation is a Mechanism for Lack of Hypoxic Induction of PHD3 in a Diverse Set of Malignant Cells Abstract The prolyl-hydroxylase domain family of enzymes (PHD1-3) plays an important
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/7/310/ra11/dc1 Supplementary Materials for STAT3 Induction of mir-146b Forms a Feedback Loop to Inhibit the NF-κB to IL-6 Signaling Axis and STAT3-Driven Cancer
www.sciencesignaling.org/cgi/content/full/7/310/ra11/dc1 Supplementary Materials for STAT3 Induction of mir-146b Forms a Feedback Loop to Inhibit the NF-κB to IL-6 Signaling Axis and STAT3-Driven Cancer
Profiles of gene expression & diagnosis/prognosis of cancer. MCs in Advanced Genetics Ainoa Planas Riverola
 Profiles of gene expression & diagnosis/prognosis of cancer MCs in Advanced Genetics Ainoa Planas Riverola Gene expression profiles Gene expression profiling Used in molecular biology, it measures the
Profiles of gene expression & diagnosis/prognosis of cancer MCs in Advanced Genetics Ainoa Planas Riverola Gene expression profiles Gene expression profiling Used in molecular biology, it measures the
AP VP DLP H&E. p-akt DLP
 A B AP VP DLP H&E AP AP VP DLP p-akt wild-type prostate PTEN-null prostate Supplementary Fig. 1. Targeted deletion of PTEN in prostate epithelium resulted in HG-PIN in all three lobes. (A) The anatomy
A B AP VP DLP H&E AP AP VP DLP p-akt wild-type prostate PTEN-null prostate Supplementary Fig. 1. Targeted deletion of PTEN in prostate epithelium resulted in HG-PIN in all three lobes. (A) The anatomy
DNA and Histone Methylation in Learning and Memory
 EXPERIMENTAL BIOLOGY ANNUAL MEETING April 2010 DNA and Histone Methylation in Learning and Memory J. David Sweatt Dept of Neurobiology McKnight Brain Institute UAB School of Medicine The Molecular Basis
EXPERIMENTAL BIOLOGY ANNUAL MEETING April 2010 DNA and Histone Methylation in Learning and Memory J. David Sweatt Dept of Neurobiology McKnight Brain Institute UAB School of Medicine The Molecular Basis
Lecture 27. Epigenetic regulation of gene expression during development
 Lecture 27 Epigenetic regulation of gene expression during development Development of a multicellular organism is not only determined by the DNA sequence but also epigenetically through DNA methylation
Lecture 27 Epigenetic regulation of gene expression during development Development of a multicellular organism is not only determined by the DNA sequence but also epigenetically through DNA methylation
