Supplementary Figure 1
|
|
|
- Bernadette McKinney
- 5 years ago
- Views:
Transcription
1 Supplementary Figure 1 Constitutive EGFR signaling does not activate canonical EGFR signals (a) U251EGFRInd cells with or without tetracycline exposure (24h, 1µg/ml) were treated with EGF for 15 minutes followed by preparation of lysates and Western blot with the indicated antibodies. (b) U87V or U87EGFR cells were treated with EGF for 15 minutes followed by preparation of lysates and Western blot with the indicated antibodies. (c) Tyrosine phosphorylation of the EGFR at residues 845 and 1173 was examined in U251V cells and U251EGFR cells in the absence of EGF. (d) In this experiment we compared tyrosine phosphorylation of the EGFR at residue 1068 in U251V cells compared to U251EGFR cells. Equal amounts of protein were loaded. (e) shows the same experiment as (d). except that that less protein (15%) was loaded in the U251EGFR lane in order to load equal amounts of EGFR. Tyrosine phosphorylation of the EGFR is still detectable only in U251EGFR cells. Densitometry was performed for A-E for pegfr/egfr and the values are shown. Cells were serum starved overnight for all experiments.
2 Supplementary Figure 2 Regulation of gene-transcription by EGFRwt and mutants (a) An experiment examining mrna levels of EGR2 in U251EGFR cells. In contradistinction to IFIT1 and IFI27, EGR2 is low in the absence of EGF and increased when cells are exposed to EGF (p<0.001). (b) Cetuximab fails to inhibit EGFR mediated induction of IFI27 mrna as detected by real time quantitative PCR. Cells were incubated overnight with either control IgG or Cetuximab (100ug/ml). (c) U251 cells conditionally expressing the mutant EGFRvIII in response to tetracycline were incubated with tetracycline (or no tetracycline) for 24h followed by RNA extraction and qpcr for IFI27. (d) U251 cells were transfected with EGFRwt, a kinase inactive EGFR mutant, or empty vector followed by extraction of RNA and qpcr for IFI27. Cells were serum starved and cultured in serum free DMEM overnight for all experiments. The EGF concentration used was 50ng/ml. Error bars represent the means±standard deviations of three independentexperiments.data were analyzed by 1way ANOVA followed by Tukey s multiple comparison test. (p values and asterisks: 0.05 (*) 0.01 (**), (***).
3 Supplementary Figure 3 EGFR regulates IFIT1 and IFI27 in MDAMB468 cells (a) Quantitative real time PCR examining IFIT1 mrna levels in MDAMB468 cells. In the absence of EGF, IFIT1 level is high and diminishes in response to EGF exposure (p<0.0001). In addition, use oferlotinib also diminishes IFIT1 mrna suggesting that the high level is EGFR driven (p<0.0001). (b) The same experiment examining IFI27 mrna. (c) A Western blot showing IFIT1 protein levels in MDAMB468 cells which is decreased in cells exposed to EGF for 6h. (d) ELISA for IFI27 levels in MDAMB468 cells shows a high level of IFI27 in the absence of EGF. When EGF is added IFI27 levels are decreased. Exposing cells to Erlotinib also results in a decrease in IFI27 levels. Cells were serum starved and cultured in serum free DMEM overnight for all experiments. The EGF concentration used was 50ng/ml. Error bars represent the means±standard deviations of three independent experiments.data were analyzed by 1way ANOVA followed by Tukey s multiple comparison test. (p values and asterisks: 0.05 (*) 0.01 (**), (***).
4 Supplementary Figure 4 EGFR regulates IFIT1 and IFI27 in glioma cell lines (a) In this experiment we used U251EGFRind cells that express high levels of EGFR conditionally in response to tetracycline and examined the level of IFIT1mRNA by quantitative real time PCR. In the absence of tetracycline IFI27 mrna levels are low with or without EGF. When tetracycline is added (1µg/ml for 24h), there is substantial increase in IFIT1 mrna. When EGF is added to tetracycline treated cells, IFIT1 mrna levels are downregulated (p<0.0001). (b) The same experiment examining IFI27 mrna levels. (c) Quantitative real time PCR examining IFIT1 mrna levels in U87EGFR cells and in empty vector transfected U87V cells. In U87EGFR cells IFIT1 level is high in the absence of EGF and diminishes in response to EGF exposure (p<0.0001). (d) The same experiment was conducted examining IFI27 mrna levels. (e) Western blot showing that the IFIT1 level is high in U87EGFR cells and decreased when cells are exposed to EGF for 6h. Cells were serum starved and cultured in serum free DMEM overnight for all experiments. The EGF concentration used was 50ng/ml. Error bars represent the means±standard deviations of three independent experiments.data were analyzed by 1way ANOVA followed by Tukey s multiple comparison test. (p values and asterisks: 0.05 (*) 0.01 (**), (***).
5 . Supplementary Figure 5 EGFR regulated IFIT1, IFI27 and TRAIL expression (a) An experiment examining IFIT1 mrna levels in GICs (glioma initiating cells) cultured as neurospheres derived from GBM429 (NS: neurosphere). In the 429NS condition, cells are grown continuously in medium containing EGF and show low levels of IFIT1 mrna. In 429NS-EGF condition, cells are grown in EGF free medium for 24h and show a high level of IFIT1 mrna. In 429NS+EGF, cells are grown in EGF free medium for 24h, then EGF is added for 4h, resulting in a decrease in IFIT1 mrna (p<0.0001). In 429NS+Erlotinib, cells are grown in EGF free medium for 24h in the presence of Erlotinib (10µM) again resulting in a decrease in IFIT1 mrna (p<0.0001). In the absence of EGF, IFIT1 level is high in the absence of EGF and diminishes in response to EGF exposure (p<0.0001). In addition, use of Erlotinib also diminishes IFIT1 mrna suggesting that the high level is EGFR driven (p<0.0001). (b) The same experiment examining IFI27 mrna in 429NS cells. (c) Quantitative real time PCR examining TRAIL mrna levels in U251EGFR cells. In the absence of EGF, TRAIL level is high in U251EGFR cells but not in U251V (vector transfected) cells and diminishes in response to EGF exposure in U251EGFR cells (p<0.0001). (d) The same experiment in U87EGFR cells (p<0.0001). (e) The same experiment in MDAMB468 cells. Since there is no vector control here, we used Erlotinib to demonstrate that TRAIL levels are EGFR driven. Addition of EGF or ErlotinibdownregulatesTRAIL levels in MDAMB468 cells (p<0.0001).error bars represent the means±standard deviations of three independent experiments.data were analyzed by 1way ANOVA followed by Tukey s multiple comparison test. (p values and asterisks: 0.05 (*) 0.01 (**), (***).
6 Supplementary Figure 6 EGFR induced IFIT1 and IFI27 requires IRF3 (a, b) U87EGFR cells were transfected with control (scrambled) sirna or with sirna directed against IRF3 followed by quantitative real time PCR for IFIT1 or IFI27. The mrna levels for both genes are decreased when IRF3 is silenced compared to control sirna. (c) shows silencing of IRF3 by Western blot. (d) The transcriptional activity of IRF3 was examined by using a synthetic pisre-luc reporter in U87EGFR cells with a dual luciferase assay. The activity of IRF3 is high in U87EGFR cells compared to U87V (vector transfected cells). Addition of EGF leads to a statistically significant decrease in reporter activity. Furthermore, Erlotinib also downregulates reporter activity, indicating that the high IRF3 transcriptional activity in the absence of EGF is driven by EGFR. Cells were serum starved and cultured in serum free DMEM overnight for all experiments. (e) The same experiment as (d) was conducted in U87EGFR cell using IFI27-Luc plasmidto examine IRF3 transcriptional activity. Error bars represent the means±standard deviations of three independent experiments.data were analyzed by 1way ANOVA followed by Tukey s multiple comparison test. (p values and asterisks: 0.05 (*) 0.01 (**), (***).
7 Supplementary Figure 7 Constitutive EGFR signaling activates IRF3 transcriptional activity in MDAMB468 cells. The transcriptional activity of IRF3 was examined by using a synthetic pisre-luc reporter in MDAMB468 cells with a dual luciferase assay. (a) The activity of IRF3 is high in MDAMB468 cells. Addition of EGF leads to a statistically significant decrease in reporter activity. Furthermore, Erlotinib also downregulates reporter activity, indicating that the high IRF3 transcriptional activity in -these cells is driven by EGFR. (b) The same experiment as (a) was conducted in MDAMB468 cells using IFI27-Luc plasmid to examine IRF3 transcriptional activity. Cells were serum starved and cultured in serum free DMEM overnight for all experiments. Error bars represent the means±standard deviations of three independent experiments.data were analyzed by 1way ANOVA followed by Tukey s multiple comparison test. (p values and asterisks: 0.05 (*) 0.01 (**), (***).
8 Supplementary Figure 8 EGFR mediated affects on IRF3 promoter occupancy (a) The effect of Cetuximab on EGFR driven transcriptional activity of IRF3 was examined by using a synthetic pisre-luc reporter in MDAMB468 cells using a dual luciferase assay. The EGFR induced transcriptional activity of IRF3 is not blocked by Cetuximab compared to control IgG in U251EGFR cells. (b) A CHIP assay examining IFIT1 promoter occupancy by IRF3 in MDAMB468 cells with or without EGF. IRF3 promoter binding can be detected in the absence of EGF and decreases by 6h of EGF stimulation and remains decreased 24h after EGF treatment. (c) A CHIP assay examining IFI27 promoter occupancy by IRF3 in MDAMB468 cells with or without EGF. IRF3 promoter binding can be detected in the absence of EGF and decreases by 6h of EGF stimulation coming back to baseline at 24h. (d) A CHIP assay examining IFIT1 promoter occupancy by IRF3 in U87EGFR cells with or without EGF. IRF3 promoter binding can be detected in the absence of EGF and decreases by 6h of EGF stimulation and remains decreased 24h after EGF treatment. (e) A CHIP assay examining IFI27 promoter occupancy by IRF3 in U87EGFR cells with or without EGF. IRF3 promoter binding can be detected in the absence of EGF and decreases by 6h of EGF stimulation and remains decreased 24h after EGF treatment. (f, g) Overexpression of the LDL receptor fails to upregulate IFIT1 or IFI27 mrna in U251MG cells as measured by qpcr. EGFR was used as a positive control. (h) Overexpression of the LDL receptor in U251MG cells detected by Western blot. Error bars represent the means±standard deviations of three independent experiments.data were analyzed by 1way ANOVA followed by Tukey s multiple comparison test. (p values and asterisks: 0.05 (*) 0.01 (**), (***).
9 Supplementary Figure 9 Constitutive EGFR signaling does not result in increased ER stress (a) U87V cells (transfected with empty vector) and U87EGFR cells were examined for phosphorylation of EIF2α. EGF or Erlotinib were added as indicated followed by lysate preparation and Western blot with phospo-eif2α antibodies. Densitometry was performed to quantitate the peif2α signal. (b) Splicing of XBP-1 was examined by Western blot in U251EGFR cells. Splicing of XBP-1 can only be detected in EGF treated cells. (c) Splicing of XBP-1 was examined at an RNA level by treating U251EGFR or MDAMB468 cells with EGF as indicated followed by RNA extraction and reverse transcriptase PCR. Thapsigargin was used as a positive control. (d) U251EGFR cells were treated with a PERK inhibitor, GSK (1μM) overnight followed by RNA extraction and qpcr for IFI27. GSK fails to prevent EGFR mediated IFI27 induction, (e) GSK fails to prevent EGFR induced activation of IRF3 mediated transcriptional activity assessed using a synthetic pisre-luc reporter. (f, g) MDAMB468 cells were treated with a PERK inhibitor, GSK (1μM) overnight followed by RNA extraction and qpcr for IFIT1 or IFI27. GSK fails to prevent EGFR mediated IFIT1 or IFI27 induction. (h) U251EGFR cells were treated with an ATF6 inhibitor, AEBSF (300μM) overnight followed by RNA extraction and qpcr for IFI27. AEBSF fails to prevent EGFR mediated IFI27 induction.error bars represent the means±standard deviations of three independent experiments.data were analyzed by 1way ANOVA followed by Tukey s multiple comparison test. (p values and asterisks: 0.05 (*) 0.01 (**), (***).
10 Supplementary Figure 10 Inhibition of ER stress fails to block constitutive EGFR signaling (a) GSK fails to prevent EGFR induced activation of IRF3 mediated transcriptional activity in U251EGFR cells assessed using a synthetic pisre-luc reporter. (b) AEBSF fails to prevent EGFR induced activation of IRF3 mediated transcriptional activity in U251EGFR assessed using a synthetic pisre-luc reporter. (c, d) MDAMB468 cells were treated with an ATF6 inhibitor, AEBSF, followed by RNA extraction and qpcr for IFIT1 and IFI27. AEBSF fails to prevent EGFR mediated IFI27 induction. (e) AEBSF fails to prevent EGFR induced activation of IRF3 mediated transcriptional activity in MDAMB468 cells assessed using a synthetic pisre-luc reporter. (f) Silencing XBP-1 fails to prevent EGFR induced induction of IFI27 in U251EGFR cells. (g) Silencing XBP-1 fails to prevent EGFR induced activation of IRF3 mediated transcriptional activity assessed using a synthetic pisre-luc reporter in MDAMB468 cells. (h) Effective silencing of XBP-1 is demonstrated in MDAMB468 cells by Western blot. Error bars represent the means±standard deviations of three independent experiments.data were analyzed by 1way ANOVA followed by Tukey s multiple comparison test. (p values and asterisks: 0.05 (*) 0.01 (**), (***).
11 Supplementary Figure 11 EGFR associated signaling complexes (a) EGFR co-immunoprecipitates with IRF3 in U87EGFR cells in the absence of EGF. When EGF is added there is a rapid loss of the EGFR-IRF3 association. (b) Similarly, the EGFR coimmunoprecipitates with TBK1 in U87EGFR cells. When EGF is added there is a rapid loss of the EGFR-TBK1 association. (c) EGFR drives TBK1-IRF3 association in U87EGFR cells. TBK1 coimmunoprecipitates with IRF3 in the absence of EGF. When EGF or Erlotinib is added the TBK1-IRF3 association is disrupted. (d) A coimmunoprecipitation experiment examining the EGFR-Shc association in U87EGFR cells. In the absence of EGF there is no association between Shc and EGFR. Shc becomes associated with the EGFR only when EGF is added to U87EGFR cells. For (a-d). Cells were serum starved and cultured in serum free DMEM overnight for all experiments. (e) IRF3 localizes to the cytosolic compartment in U251V cells. Very little IRF3 can be detected in the membrane or nuclear fractions. (f) IKKε is not expressed in U251EGFR cells or U87EGFR cells. MCF7 cells were used as a positive control for IKKε expression. Three independent experiments were done.
12 Supplementary Figure 12 EGFR activates IFN signaling (a) There is a high level of IFNβ mrna in MDAMB468 cells. Addition of EGF or Erlotinib leads to a loss of this increase in IFNβ. (b) There is a high level of IFNα mrna in MDAMB468 cells. Addition of EGF or Erlotinib leads to a loss of this increase in IFNα. (c) High level of IFNβ in supernatants from U251EGFR and MDAMB468 cells. Both cell lines express high levels of EGFR. (d) U251EGFR and U87EGFR cells do not express IRF7 as determined by Western blot. Jurkat cells are used as a positive control. (e, f) Ruxolitinib blocks EGFR induced upregulation of IFIT1 and IFI27 in MDAMB468 cells. Cells were incubated with Ruxolinitib (1uM) overnight followed by extraction of RNA and qpcr for IFI27 or IFIT1. Error bars represent the means±standard deviations of three independent experiments.data were analyzed by 1way ANOVA followed by Tukey s multiple comparison test. (p values and asterisks: 0.05 (*) 0.01 (**), (***).
13 Supplementary Figure 13 JAK inhibitors block EGFR mediated IFIT1 and IFI27 induction (a) U251EGFR cells were incubated with Tofacitinib (300nM) overnight followed by extraction of RNA and qpcr for IFI27. Tofacitinib blocks EGFR mediated induction of IFI27. (b, c) MDAMB468 cells were incubated with Tofacitinib (300nM) overnight followed by extraction of RNA and qpcr for IFIT1 or IFI27. Tofacitinib blocks EGFR mediated induction of IFIT1 and IFI27. (d, e) JAK inhibitors fail to inhibit ERK and Akt activation induced by EGF in U251EGFR cells. Cells were serum starved and treated overnight with JAK inhibitors. EGF was used for 15 min at a concentration of 50ng/ml. Error bars represent the means±standard deviations of three independent experiments.data were analyzed by 1way ANOVA followed by Tukey s multiple comparison test. (p values and asterisks: 0.05 (*) 0.01 (**), (***).
14 Supplementary Figure 14 TLR3 in not expressed in glioma cell lines TLR3 is not expressed in U251EGFR and U87EGFR cells as determined by Western blot. MCF10A cells were used as a positive control. The experiment was repeated once.
15 Supplementary Figure 15 Ligand-activated EGFR is more sensitive to virus-induced cell death An MTT conversion assay was undertaken to determine whether EGFR overexpression alters sensitivity to adenovirus infection. (a) Comparison of the effects of adenovirus on U251V cells and U251EGFR cells, with or without EGF. U251EGFR cells are more resistant to adenovirus compared to U251V cells, and this resistance is lost when U251EGFR cells are exposed to EGF. (b) The same experiment performed in U87V and U87EGFR cells. EGF was added for 24h in serum free conditions. Error bars represent the means±standard deviations of three independent experiments.data were analyzed by 1way ANOVA followed by Tukey s multiple comparison test. (p values and asterisks: 0.05 (*) 0.01 (**), (***).
16 Supplementary Figure 16 Ligand-activated EGFR is more sensitive to chemotherapy An MTT conversion assay was undertaken to determine whether EGFR overexpression alters sensitivity to temozolomide. (a) Comparison of the effects of temozolomide on U251V cells and U251EGFR cells, with or without EGF. U251EGFR cells are more resistant to temozolomide compared to U251V cells, and this resistance is lost when U251EGFR cells are exposed to EGF. (b) The same experiment performed in U87V and U87EGFR cells. (c) The same experiment was performed in MDAMB468 cells transfected with EGFR sirna or control sirna. In all cell lines EGFR overexpression confers resistance to temozolomide and this resistance is lost if EGF is added. EGF was added for 24h in serum free conditions. Error bars represent the means±standarddeviations of three independent experiments.data were analyzed by 1way ANOVA followed by Tukey s multiple comparison test. (p values and asterisks: 0.05 (*) 0.01 (**), (***).
17 Supplementary Figure 17 Constitutive EGFR signaling in GBM. (a) shows the inverse relationship between TGFα levels and IFIT1 in GBM tumors overexpressing EGFR. (b) shows the inverse relationship between TGFα levels and IFI27 in GBM tumors overexpressing EGFR. (c) levels of TGFα, IFIT1 and IFI27 in tumors expressing a low level of EGFR as determined by real time quantitative PCR. (d) EGFR levels were examined in the 27 GBM tumors used in this study by Western blot. Error bars represent the means±standarddeviations of three independent experiments.data were analyzed by 1way ANOVA followed by Tukey s multiple comparison test. (p values and asterisks: 0.05 (*) 0.01 (**), (***).
18 Supplementary Figure 18 Uncropped blots Key uncropped western blot images are shown above.
NIH Public Access Author Manuscript Nat Commun. Author manuscript; available in PMC 2015 June 15.
 NIH Public Access Author Manuscript Published in final edited form as: Nat Commun. ; 5: 5811. doi:10.1038/ncomms6811. Constitutive and ligand-induced EGFR signaling triggers distinct and mutually exclusive
NIH Public Access Author Manuscript Published in final edited form as: Nat Commun. ; 5: 5811. doi:10.1038/ncomms6811. Constitutive and ligand-induced EGFR signaling triggers distinct and mutually exclusive
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 EGFR inhibition activates signaling pathways (a-b) EGFR inhibition activates signaling pathways (a) U251EGFR cells were treated with erlotinib (1µM) for the indicated times followed
Supplementary Figure 1 EGFR inhibition activates signaling pathways (a-b) EGFR inhibition activates signaling pathways (a) U251EGFR cells were treated with erlotinib (1µM) for the indicated times followed
HEK293FT cells were transiently transfected with reporters, N3-ICD construct and
 Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
Table S1. Primer sequences used for qrt-pcr. CACCATTGGCAATGAGCGGTTC AGGTCTTTGCGGATGTCCACGT ACTB AAGTCCATGTGCTGGCAGCACT ATCACCACTCCGAAGTCCGTCT LCOR
 Table S1. Primer sequences used for qrt-pcr. ACTB LCOR KLF6 CTBP1 CDKN1A CDH1 ATF3 PLAU MMP9 TFPI2 CACCATTGGCAATGAGCGGTTC AGGTCTTTGCGGATGTCCACGT AAGTCCATGTGCTGGCAGCACT ATCACCACTCCGAAGTCCGTCT CGGCTGCAGGAAAGTTTACA
Table S1. Primer sequences used for qrt-pcr. ACTB LCOR KLF6 CTBP1 CDKN1A CDH1 ATF3 PLAU MMP9 TFPI2 CACCATTGGCAATGAGCGGTTC AGGTCTTTGCGGATGTCCACGT AAGTCCATGTGCTGGCAGCACT ATCACCACTCCGAAGTCCGTCT CGGCTGCAGGAAAGTTTACA
SUPPLEMENTARY FIGURES AND TABLE
 SUPPLEMENTARY FIGURES AND TABLE Supplementary Figure S1: Characterization of IRE1α mutants. A. U87-LUC cells were transduced with the lentiviral vector containing the GFP sequence (U87-LUC Tet-ON GFP).
SUPPLEMENTARY FIGURES AND TABLE Supplementary Figure S1: Characterization of IRE1α mutants. A. U87-LUC cells were transduced with the lentiviral vector containing the GFP sequence (U87-LUC Tet-ON GFP).
RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh-
 1 a b Supplementary Figure 1. Effects of GSK3b knockdown on poly I:C-induced cytokine production. RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh- GSK3b) were stimulated
1 a b Supplementary Figure 1. Effects of GSK3b knockdown on poly I:C-induced cytokine production. RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh- GSK3b) were stimulated
Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus
 Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus changes in corresponding proteins between wild type and Gprc5a-/-
Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus changes in corresponding proteins between wild type and Gprc5a-/-
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells
 Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Intracellular MHC class II molecules promote TLR-triggered innate. immune responses by maintaining Btk activation
 Intracellular MHC class II molecules promote TLR-triggered innate immune responses by maintaining Btk activation Xingguang Liu, Zhenzhen Zhan, Dong Li, Li Xu, Feng Ma, Peng Zhang, Hangping Yao and Xuetao
Intracellular MHC class II molecules promote TLR-triggered innate immune responses by maintaining Btk activation Xingguang Liu, Zhenzhen Zhan, Dong Li, Li Xu, Feng Ma, Peng Zhang, Hangping Yao and Xuetao
p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO
 Supplementary Information p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO Yuri Shibata, Masaaki Oyama, Hiroko Kozuka-Hata, Xiao Han, Yuetsu Tanaka,
Supplementary Information p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO Yuri Shibata, Masaaki Oyama, Hiroko Kozuka-Hata, Xiao Han, Yuetsu Tanaka,
Supplemental Figure 1
 Supplemental Figure 1 1a 1c PD-1 MFI fold change 6 5 4 3 2 1 IL-1α IL-2 IL-4 IL-6 IL-1 IL-12 IL-13 IL-15 IL-17 IL-18 IL-21 IL-23 IFN-α Mut Human PD-1 promoter SBE-D 5 -GTCTG- -1.2kb SBE-P -CAGAC- -1.kb
Supplemental Figure 1 1a 1c PD-1 MFI fold change 6 5 4 3 2 1 IL-1α IL-2 IL-4 IL-6 IL-1 IL-12 IL-13 IL-15 IL-17 IL-18 IL-21 IL-23 IFN-α Mut Human PD-1 promoter SBE-D 5 -GTCTG- -1.2kb SBE-P -CAGAC- -1.kb
Supplementary Information
 Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
p = formed with HCI-001 p = Relative # of blood vessels that formed with HCI-002 Control Bevacizumab + 17AAG Bevacizumab 17AAG
 A.. Relative # of ECs associated with HCI-001 1.4 1.2 1.0 0.8 0.6 0.4 0.2 0.0 ol b p < 0.001 Relative # of blood vessels that formed with HCI-001 1.4 1.2 1.0 0.8 0.6 0.4 0.2 0.0 l b p = 0.002 Control IHC:
A.. Relative # of ECs associated with HCI-001 1.4 1.2 1.0 0.8 0.6 0.4 0.2 0.0 ol b p < 0.001 Relative # of blood vessels that formed with HCI-001 1.4 1.2 1.0 0.8 0.6 0.4 0.2 0.0 l b p = 0.002 Control IHC:
Tbk1-TKO! DN cells (%)! 15! 10!
 a! T Cells! TKO! B Cells! TKO! b! CD4! 8.9 85.2 3.4 2.88 CD8! Tbk1-TKO! 1.1 84.8 2.51 2.54 c! DN cells (%)! 4 3 2 1 DP cells (%)! 9 8 7 6 CD4 + SP cells (%)! 5 4 3 2 1 5 TKO! TKO! TKO! TKO! 15 1 5 CD8
a! T Cells! TKO! B Cells! TKO! b! CD4! 8.9 85.2 3.4 2.88 CD8! Tbk1-TKO! 1.1 84.8 2.51 2.54 c! DN cells (%)! 4 3 2 1 DP cells (%)! 9 8 7 6 CD4 + SP cells (%)! 5 4 3 2 1 5 TKO! TKO! TKO! TKO! 15 1 5 CD8
S1a S1b S1c. S1d. S1f S1g S1h SUPPLEMENTARY FIGURE 1. - si sc Il17rd Il17ra bp. rig/s IL-17RD (ng) -100 IL-17RD
 SUPPLEMENTARY FIGURE 1 0 20 50 80 100 IL-17RD (ng) S1a S1b S1c IL-17RD β-actin kda S1d - si sc Il17rd Il17ra rig/s15-574 - 458-361 bp S1f S1g S1h S1i S1j Supplementary Figure 1. Knockdown of IL-17RD enhances
SUPPLEMENTARY FIGURE 1 0 20 50 80 100 IL-17RD (ng) S1a S1b S1c IL-17RD β-actin kda S1d - si sc Il17rd Il17ra rig/s15-574 - 458-361 bp S1f S1g S1h S1i S1j Supplementary Figure 1. Knockdown of IL-17RD enhances
Supplementary Figure 1. Establishment of prostacyclin-secreting hmscs. (a) PCR showed the integration of the COX-1-10aa-PGIS transgene into the
 Supplementary Figure 1. Establishment of prostacyclin-secreting hmscs. (a) PCR showed the integration of the COX-1-10aa-PGIS transgene into the genomic DNA of hmscs (PGI2- hmscs). Native hmscs and plasmid
Supplementary Figure 1. Establishment of prostacyclin-secreting hmscs. (a) PCR showed the integration of the COX-1-10aa-PGIS transgene into the genomic DNA of hmscs (PGI2- hmscs). Native hmscs and plasmid
Supplementary Figure 1
 Supplementary Figure 1 YAP negatively regulates IFN- signaling. (a) Immunoblot analysis of Yap knockdown efficiency with sh-yap (#1 to #4 independent constructs) in Raw264.7 cells. (b) IFN- -Luc and PRDs
Supplementary Figure 1 YAP negatively regulates IFN- signaling. (a) Immunoblot analysis of Yap knockdown efficiency with sh-yap (#1 to #4 independent constructs) in Raw264.7 cells. (b) IFN- -Luc and PRDs
Title: Cytosolic DNA-mediated, STING-dependent pro-inflammatory gene. Fig. S1. STING ligands-mediated signaling response in MEFs. (A) Primary MEFs (1
 1 Supporting Information 2 3 4 Title: Cytosolic DNA-mediated, STING-dependent pro-inflammatory gene induction necessitates canonical NF-κB activation through TBK1 5 6 Authors: Abe et al. 7 8 9 Supporting
1 Supporting Information 2 3 4 Title: Cytosolic DNA-mediated, STING-dependent pro-inflammatory gene induction necessitates canonical NF-κB activation through TBK1 5 6 Authors: Abe et al. 7 8 9 Supporting
PID1 increases chemotherapy-induced apoptosis in medulloblastoma and glioblastoma cells in a manner that involves NFκB
 SUPPLEMENTARY FIGURES: PID1 increases chemotherapy-induced apoptosis in medulloblastoma and glioblastoma cells in a manner that involves NFκB Jingying Xu, Xiuhai Ren, Anup Singh Pathania, G. Esteban Fernandez,
SUPPLEMENTARY FIGURES: PID1 increases chemotherapy-induced apoptosis in medulloblastoma and glioblastoma cells in a manner that involves NFκB Jingying Xu, Xiuhai Ren, Anup Singh Pathania, G. Esteban Fernandez,
Relative Rates. SUM159 CB- 839-Resistant *** n.s Intracellular % Labeled by U- 13 C-Asn 0.
 A Relative Growth Rates 1.2 1.8.6.4.2 B Relative Rates 1.6 1.4 1.2 1.8.6.4.2 LPS2 Parental LPS2 Q-Independent SUM159 Parental SUM159 CB-839-Resistant LPS2 Parental LPS2 Q- Independent SUM159 Parental SUM159
A Relative Growth Rates 1.2 1.8.6.4.2 B Relative Rates 1.6 1.4 1.2 1.8.6.4.2 LPS2 Parental LPS2 Q-Independent SUM159 Parental SUM159 CB-839-Resistant LPS2 Parental LPS2 Q- Independent SUM159 Parental SUM159
Xenoestrogen-induced Regulation of EZH2 and Histone Methylation via Non-Genomic Estrogen
 Xenoestrogen-induced Regulation of EZH2 and Histone Methylation via Non-Genomic Estrogen Receptor Signaling to PI3K/AKT Tiffany G. Bredfeldt, Kristen L. Greathouse, Stephen H. Safe, Mien-Chie Hung, Mark
Xenoestrogen-induced Regulation of EZH2 and Histone Methylation via Non-Genomic Estrogen Receptor Signaling to PI3K/AKT Tiffany G. Bredfeldt, Kristen L. Greathouse, Stephen H. Safe, Mien-Chie Hung, Mark
microrna-200b and microrna-200c promote colorectal cancer cell proliferation via
 Supplementary Materials microrna-200b and microrna-200c promote colorectal cancer cell proliferation via targeting the reversion-inducing cysteine-rich protein with Kazal motifs Supplementary Table 1.
Supplementary Materials microrna-200b and microrna-200c promote colorectal cancer cell proliferation via targeting the reversion-inducing cysteine-rich protein with Kazal motifs Supplementary Table 1.
Integrin CD11b negatively regulates TLR-triggered inflammatory responses by. activating Syk and promoting MyD88 and TRIF degradation via cbl-b
 Integrin CD11b negatively regulates TLR-triggered inflammatory responses by activating Syk and promoting MyD88 and TRIF degradation via cbl-b Chaofeng Han, Jing Jin, Sheng Xu, Haibo Liu, Nan Li, and Xuetao
Integrin CD11b negatively regulates TLR-triggered inflammatory responses by activating Syk and promoting MyD88 and TRIF degradation via cbl-b Chaofeng Han, Jing Jin, Sheng Xu, Haibo Liu, Nan Li, and Xuetao
Figures S1-S5, Figure Legends, Table S1 List of primers used in the study
 Insulin receptor alternative splicing is regulated by insulin signaling and modulates beta cell survival Pushkar Malakar,4, Lital Chartarifsky,4, Ayat Hija, Gil Leibowitz 3, Benjamin Glaser 3, Yuval Dor,
Insulin receptor alternative splicing is regulated by insulin signaling and modulates beta cell survival Pushkar Malakar,4, Lital Chartarifsky,4, Ayat Hija, Gil Leibowitz 3, Benjamin Glaser 3, Yuval Dor,
Stewart et al. CD36 ligands promote sterile inflammation through assembly of a TLR 4 and 6 heterodimer
 NFκB (fold induction) Stewart et al. ligands promote sterile inflammation through assembly of a TLR 4 and 6 heterodimer a. mrna (fold induction) 5 4 3 2 1 LDL oxldl Gro1a MIP-2 RANTES mrna (fold induction)
NFκB (fold induction) Stewart et al. ligands promote sterile inflammation through assembly of a TLR 4 and 6 heterodimer a. mrna (fold induction) 5 4 3 2 1 LDL oxldl Gro1a MIP-2 RANTES mrna (fold induction)
Supplemental Figure 1. Western blot analysis indicated that MIF was detected in the fractions of
 Supplemental Figure Legends Supplemental Figure 1. Western blot analysis indicated that was detected in the fractions of plasma membrane and cytosol but not in nuclear fraction isolated from Pkd1 null
Supplemental Figure Legends Supplemental Figure 1. Western blot analysis indicated that was detected in the fractions of plasma membrane and cytosol but not in nuclear fraction isolated from Pkd1 null
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Chairoungdua et al., http://www.jcb.org/cgi/content/full/jcb.201002049/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Expression of CD9 and CD82 inhibits Wnt/ -catenin
Supplemental material Chairoungdua et al., http://www.jcb.org/cgi/content/full/jcb.201002049/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Expression of CD9 and CD82 inhibits Wnt/ -catenin
Supplementary data Supplementary Figure 1 Supplementary Figure 2
 Supplementary data Supplementary Figure 1 SPHK1 sirna increases RANKL-induced osteoclastogenesis in RAW264.7 cell culture. (A) RAW264.7 cells were transfected with oligocassettes containing SPHK1 sirna
Supplementary data Supplementary Figure 1 SPHK1 sirna increases RANKL-induced osteoclastogenesis in RAW264.7 cell culture. (A) RAW264.7 cells were transfected with oligocassettes containing SPHK1 sirna
Supplemental information
 Carcinoemryonic antigen-related cell adhesion molecule 6 (CEACAM6) promotes EGF receptor signaling of oral squamous cell carcinoma metastasis via the complex N-glycosylation y Chiang et al. Supplemental
Carcinoemryonic antigen-related cell adhesion molecule 6 (CEACAM6) promotes EGF receptor signaling of oral squamous cell carcinoma metastasis via the complex N-glycosylation y Chiang et al. Supplemental
2.5. AMPK activity
 Supplement Fig. A 3 B phos-ampk 2.5 * Control AICAR AMPK AMPK activity (Absorbance at 45 nm) 2.5.5 Control AICAR Supplement Fig. Effects of AICAR on AMPK activation in macrophages. J774. macrophages were
Supplement Fig. A 3 B phos-ampk 2.5 * Control AICAR AMPK AMPK activity (Absorbance at 45 nm) 2.5.5 Control AICAR Supplement Fig. Effects of AICAR on AMPK activation in macrophages. J774. macrophages were
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk
 Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Plasma exposure levels from individual mice 4 hours post IP administration at the
 Supplemental Figure Legends Figure S1. Plasma exposure levels of MKC-3946 in mice. Plasma exposure levels from individual mice 4 hours post IP administration at the indicated dose mg/kg. Data represent
Supplemental Figure Legends Figure S1. Plasma exposure levels of MKC-3946 in mice. Plasma exposure levels from individual mice 4 hours post IP administration at the indicated dose mg/kg. Data represent
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12215 Supplementary Figure 1. The effects of full and dissociated GR agonists in supporting BFU-E self-renewal divisions. BFU-Es were cultured in self-renewal medium with indicated GR
doi:10.1038/nature12215 Supplementary Figure 1. The effects of full and dissociated GR agonists in supporting BFU-E self-renewal divisions. BFU-Es were cultured in self-renewal medium with indicated GR
Supplementary Material
 Supplementary Material The Androgen Receptor is a negative regulator of eif4e Phosphorylation at S209: Implications for the use of mtor inhibitors in advanced prostate cancer Supplementary Figures Supplemental
Supplementary Material The Androgen Receptor is a negative regulator of eif4e Phosphorylation at S209: Implications for the use of mtor inhibitors in advanced prostate cancer Supplementary Figures Supplemental
Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis
 Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis of PD-L1 in ovarian cancer cells. (c) Western blot analysis
Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis of PD-L1 in ovarian cancer cells. (c) Western blot analysis
Pro-apoptotic signalling through Toll-like receptor 3 involves TRIF-dependent
 Pro-apoptotic signalling through Toll-like receptor 3 involves TRIF-dependent activation of caspase-8 and is under the control of inhibitor of apoptosis proteins in melanoma cells Arnim Weber, Zofia Kirejczyk,
Pro-apoptotic signalling through Toll-like receptor 3 involves TRIF-dependent activation of caspase-8 and is under the control of inhibitor of apoptosis proteins in melanoma cells Arnim Weber, Zofia Kirejczyk,
Nature Immunology doi: /ni Supplementary Figure 1. Raf-1 inhibition does not affect TLR4-induced type I IFN responses.
 Supplementary Figure 1 Raf-1 inhibition does not affect TLR4-induced type I IFN responses. Real-time PCR analyses of IFNB, ISG15, TRIM5, TRIM22 and APOBEC3G mrna in modcs 6 h after stimulation with TLR4
Supplementary Figure 1 Raf-1 inhibition does not affect TLR4-induced type I IFN responses. Real-time PCR analyses of IFNB, ISG15, TRIM5, TRIM22 and APOBEC3G mrna in modcs 6 h after stimulation with TLR4
Supplemental Material:
 Supplemental Material: MATERIALS AND METHODS RNA interference Mouse CHOP sirna (ON-TARGETplus SMARTpool Cat# L-062068-00) and control sirna (ON-TARGETplus Control) were purchased from Dharmacon. Transfection
Supplemental Material: MATERIALS AND METHODS RNA interference Mouse CHOP sirna (ON-TARGETplus SMARTpool Cat# L-062068-00) and control sirna (ON-TARGETplus Control) were purchased from Dharmacon. Transfection
Supplementary information. MARCH8 inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins
 Supplementary information inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins Takuya Tada, Yanzhao Zhang, Takayoshi Koyama, Minoru Tobiume, Yasuko Tsunetsugu-Yokota, Shoji
Supplementary information inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins Takuya Tada, Yanzhao Zhang, Takayoshi Koyama, Minoru Tobiume, Yasuko Tsunetsugu-Yokota, Shoji
Supplementary Information and Figure legends
 Supplementary Information and Figure legends Table S1. Primers for quantitative RT-PCR Target Sequence (5 -> 3 ) Target Sequence (5 -> 3 ) DAB2IP F:TGGACGATGTGCTCTATGCC R:GGATGGTGATGGTTTGGTAG Snail F:CCTCCCTGTCAGATGAGGAC
Supplementary Information and Figure legends Table S1. Primers for quantitative RT-PCR Target Sequence (5 -> 3 ) Target Sequence (5 -> 3 ) DAB2IP F:TGGACGATGTGCTCTATGCC R:GGATGGTGATGGTTTGGTAG Snail F:CCTCCCTGTCAGATGAGGAC
supplementary information
 DOI: 1.138/ncb1 Control Atg7 / NAC 1 1 1 1 (mm) Control Atg7 / NAC 1 1 1 1 (mm) Lamin B Gstm1 Figure S1 Neither the translocation of into the nucleus nor the induction of antioxidant proteins in autophagydeficient
DOI: 1.138/ncb1 Control Atg7 / NAC 1 1 1 1 (mm) Control Atg7 / NAC 1 1 1 1 (mm) Lamin B Gstm1 Figure S1 Neither the translocation of into the nucleus nor the induction of antioxidant proteins in autophagydeficient
Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was
 Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was painted on the shaved back skin of CBL/J and BALB/c mice for consecutive days. (a, b) Phenotypic presentation of mouse back skin
Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was painted on the shaved back skin of CBL/J and BALB/c mice for consecutive days. (a, b) Phenotypic presentation of mouse back skin
(A) RT-PCR for components of the Shh/Gli pathway in normal fetus cell (MRC-5) and a
 Supplementary figure legends Supplementary Figure 1. Expression of Shh signaling components in a panel of gastric cancer. (A) RT-PCR for components of the Shh/Gli pathway in normal fetus cell (MRC-5) and
Supplementary figure legends Supplementary Figure 1. Expression of Shh signaling components in a panel of gastric cancer. (A) RT-PCR for components of the Shh/Gli pathway in normal fetus cell (MRC-5) and
(A) Dose response curves of HMLE_shGFP (blue circle), HMLE_shEcad (red square),
 Supplementary Figures and Tables Figure S1. Validation of EMT-selective small molecules (A) Dose response curves of HMLE_shGFP (blue circle), HMLE_shEcad (red square), and HMLE_Twist (black diamond) cells
Supplementary Figures and Tables Figure S1. Validation of EMT-selective small molecules (A) Dose response curves of HMLE_shGFP (blue circle), HMLE_shEcad (red square), and HMLE_Twist (black diamond) cells
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3311 A B TSC2 -/- MEFs C Rapa Hours WCL 0 6 12 24 36 pakt.s473 AKT ps6k S6K CM IGF-1 Recipient WCL - + - + - + pigf-1r IGF-1R pakt ps6 AKT D 1 st SILAC 2 nd SILAC E GAPDH FGF21 ALKPGVIQILGVK
DOI: 10.1038/ncb3311 A B TSC2 -/- MEFs C Rapa Hours WCL 0 6 12 24 36 pakt.s473 AKT ps6k S6K CM IGF-1 Recipient WCL - + - + - + pigf-1r IGF-1R pakt ps6 AKT D 1 st SILAC 2 nd SILAC E GAPDH FGF21 ALKPGVIQILGVK
Supplementary Figure 1. Characterization of ALDH-positive cell population in MCF-7 cells. (a) Expression level of stem cell markers in MCF-7 cells or
 Supplementary Figure 1. Characterization of ALDH-positive cell population in MCF-7 cells. (a) Expression level of stem cell markers in MCF-7 cells or ALDH-positive cell population by qpcr. Data represent
Supplementary Figure 1. Characterization of ALDH-positive cell population in MCF-7 cells. (a) Expression level of stem cell markers in MCF-7 cells or ALDH-positive cell population by qpcr. Data represent
A Normal Exencephaly Craniora- Spina bifida Microcephaly chischisis. Midbrain Forebrain/ Forebrain/ Hindbrain Spinal cord Hindbrain Hindbrain
 A Normal Exencephaly Craniora- Spina bifida Microcephaly chischisis NTD Number of embryos % among NTD Embryos Exencephaly 52 74.3% Craniorachischisis 6 8.6% Spina bifida 5 7.1% Microcephaly 7 1% B Normal
A Normal Exencephaly Craniora- Spina bifida Microcephaly chischisis NTD Number of embryos % among NTD Embryos Exencephaly 52 74.3% Craniorachischisis 6 8.6% Spina bifida 5 7.1% Microcephaly 7 1% B Normal
Egr-1 regulates RTA transcription through a cooperative involvement of transcriptional regulators
 /, 2017, Vol. 8, (No. 53), pp: 91425-91444 Egr-1 regulates RTA transcription through a cooperative involvement of transcriptional regulators Roni Sarkar 1 and Subhash C. Verma 1 1 Department of Microbiology
/, 2017, Vol. 8, (No. 53), pp: 91425-91444 Egr-1 regulates RTA transcription through a cooperative involvement of transcriptional regulators Roni Sarkar 1 and Subhash C. Verma 1 1 Department of Microbiology
Supplementary Information
 Supplementary Information Supplementary Figure 1. Effect of mir mimics and anti-mirs on DTPs a, Representative fluorescence microscopy images of GFP vector control or mir mimicexpressing parental and DTP
Supplementary Information Supplementary Figure 1. Effect of mir mimics and anti-mirs on DTPs a, Representative fluorescence microscopy images of GFP vector control or mir mimicexpressing parental and DTP
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 DOT1L regulates the expression of epithelial and mesenchymal markers. (a) The expression levels and cellular localizations of EMT markers were confirmed by
Supplementary Figures Supplementary Figure 1 DOT1L regulates the expression of epithelial and mesenchymal markers. (a) The expression levels and cellular localizations of EMT markers were confirmed by
Supplementary fig. 1. Crystals induce necroptosis does not involve caspases, TNF receptor or NLRP3. A. Mouse tubular epithelial cells were pretreated
 Supplementary fig. 1. Crystals induce necroptosis does not involve caspases, TNF receptor or NLRP3. A. Mouse tubular epithelial cells were pretreated with zvad-fmk (10µM) and exposed to calcium oxalate
Supplementary fig. 1. Crystals induce necroptosis does not involve caspases, TNF receptor or NLRP3. A. Mouse tubular epithelial cells were pretreated with zvad-fmk (10µM) and exposed to calcium oxalate
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb2822 a MTC02 FAO cells EEA1 b +/+ MEFs /DAPI -/- MEFs /DAPI -/- MEFs //DAPI c HEK 293 cells WCE N M C P AKT TBC1D7 Lamin A/C EEA1 VDAC d HeLa cells WCE N M C P AKT Lamin A/C EEA1 VDAC Figure
DOI:.38/ncb2822 a MTC02 FAO cells EEA1 b +/+ MEFs /DAPI -/- MEFs /DAPI -/- MEFs //DAPI c HEK 293 cells WCE N M C P AKT TBC1D7 Lamin A/C EEA1 VDAC d HeLa cells WCE N M C P AKT Lamin A/C EEA1 VDAC Figure
Supplementary Figure 1. The mir-182 binding site of SMAD7 3 UTR and the. mutated sequence.
 Supplementary Figure 1. The mir-182 binding site of SMAD7 3 UTR and the mutated sequence. 1 Supplementary Figure 2. Expression of mir-182 and SMAD7 in various cell lines. (A) Basal levels of mir-182 expression
Supplementary Figure 1. The mir-182 binding site of SMAD7 3 UTR and the mutated sequence. 1 Supplementary Figure 2. Expression of mir-182 and SMAD7 in various cell lines. (A) Basal levels of mir-182 expression
Doctoral Degree Program in Marine Biotechnology, College of Marine Sciences, Doctoral Degree Program in Marine Biotechnology, Academia Sinica, Taipei,
 Cyclooxygenase 2 facilitates dengue virus replication and serves as a potential target for developing antiviral agents Chun-Kuang Lin 1,2, Chin-Kai Tseng 3,4, Yu-Hsuan Wu 3,4, Chih-Chuang Liaw 1,5, Chun-
Cyclooxygenase 2 facilitates dengue virus replication and serves as a potential target for developing antiviral agents Chun-Kuang Lin 1,2, Chin-Kai Tseng 3,4, Yu-Hsuan Wu 3,4, Chih-Chuang Liaw 1,5, Chun-
FIG S1 Examination of eif4b expression after virus infection. (A) A549 cells
 Supplementary Figure Legends FIG S1 Examination of expression after virus infection. () 549 cells were infected with herpes simplex virus (HSV) (MOI = 1), and harvested at the indicated times, followed
Supplementary Figure Legends FIG S1 Examination of expression after virus infection. () 549 cells were infected with herpes simplex virus (HSV) (MOI = 1), and harvested at the indicated times, followed
Figure S1. Reduction in glomerular mir-146a levels correlate with progression to higher albuminuria in diabetic patients.
 Supplementary Materials Supplementary Figures Figure S1. Reduction in glomerular mir-146a levels correlate with progression to higher albuminuria in diabetic patients. Figure S2. Expression level of podocyte
Supplementary Materials Supplementary Figures Figure S1. Reduction in glomerular mir-146a levels correlate with progression to higher albuminuria in diabetic patients. Figure S2. Expression level of podocyte
Supplementary Figure 1: STAT3 suppresses Kras-induced lung tumorigenesis
 Supplementary Figure 1: STAT3 suppresses Kras-induced lung tumorigenesis (a) Immunohistochemical (IHC) analysis of tyrosine 705 phosphorylation status of STAT3 (P- STAT3) in tumors and stroma (all-time
Supplementary Figure 1: STAT3 suppresses Kras-induced lung tumorigenesis (a) Immunohistochemical (IHC) analysis of tyrosine 705 phosphorylation status of STAT3 (P- STAT3) in tumors and stroma (all-time
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Supplementary Figure 1. (A) Left, western blot analysis of ISGylated proteins in Jurkat T cells treated with 1000U ml -1 IFN for 16h (IFN) or left untreated (CONT); right, western
SUPPLEMENTARY FIGURES Supplementary Figure 1. (A) Left, western blot analysis of ISGylated proteins in Jurkat T cells treated with 1000U ml -1 IFN for 16h (IFN) or left untreated (CONT); right, western
a. b. c. d. e. f. g. h. i. j. k. l. m. n. o. p.
 a. b. c. d. e. f. g. h. i. j. k. l. 2.5 2 1.5 1.5 IL-1β 12 8 6 4 2 IL-1β 9 8 7 6 4 3 3 2.9 IL-1β m. n. o. p. 1.8 1.6 1.4 1.2 1.8.6.4.2 6h LPS 2 15 1 5 6h LPS 2 6h LPS 6 4 3 6h LPS Supplementary Figure
a. b. c. d. e. f. g. h. i. j. k. l. 2.5 2 1.5 1.5 IL-1β 12 8 6 4 2 IL-1β 9 8 7 6 4 3 3 2.9 IL-1β m. n. o. p. 1.8 1.6 1.4 1.2 1.8.6.4.2 6h LPS 2 15 1 5 6h LPS 2 6h LPS 6 4 3 6h LPS Supplementary Figure
Nature Medicine: doi: /nm.4078
 Supplementary Figure 1. Cetuximab induces ER stress response in DiFi cells. (a) Scheme of SILAC proteome. (b) MS-base read out of SILAC experiment. The histogram of log 2 -transformed normalized H/L ratios
Supplementary Figure 1. Cetuximab induces ER stress response in DiFi cells. (a) Scheme of SILAC proteome. (b) MS-base read out of SILAC experiment. The histogram of log 2 -transformed normalized H/L ratios
SUPPLEMENTARY INFORMATION
 Supplementary Table 1. Cell sphingolipids and S1P bound to endogenous TRAF2. Sphingolipid Cell pmol/mg TRAF2 immunoprecipitate pmol/mg Sphingomyelin 4200 ± 250 Not detected Monohexosylceramide 311 ± 18
Supplementary Table 1. Cell sphingolipids and S1P bound to endogenous TRAF2. Sphingolipid Cell pmol/mg TRAF2 immunoprecipitate pmol/mg Sphingomyelin 4200 ± 250 Not detected Monohexosylceramide 311 ± 18
Supplementary Figure 1. mir124 does not change neuron morphology and synaptic
 Supplementary Figure 1. mir124 does not change neuron morphology and synaptic density. Hippocampal neurons were transfected with mir124 (containing DsRed) or DsRed as a control. 2 d after transfection,
Supplementary Figure 1. mir124 does not change neuron morphology and synaptic density. Hippocampal neurons were transfected with mir124 (containing DsRed) or DsRed as a control. 2 d after transfection,
Supplementary Information
 Supplementary Information EGFRL858R mutant enhances lung adenocarcinoma cell invasive aility and promotes malignant pleural effusion formation through activation of the CXCL12CXCR4 pathway MengFeng Tsai
Supplementary Information EGFRL858R mutant enhances lung adenocarcinoma cell invasive aility and promotes malignant pleural effusion formation through activation of the CXCL12CXCR4 pathway MengFeng Tsai
A dual PI3 kinase/mtor inhibitor reveals emergent efficacy in glioma
 Supplemental data A dual PI3 kinase/mtor inhibitor reveals emergent efficacy in glioma Qi-Wen Fan, Zachary A. Knight, David D. Goldenberg, Wei Yu, Keith E. Mostov, David Stokoe, Kevan M. Shokat, and William
Supplemental data A dual PI3 kinase/mtor inhibitor reveals emergent efficacy in glioma Qi-Wen Fan, Zachary A. Knight, David D. Goldenberg, Wei Yu, Keith E. Mostov, David Stokoe, Kevan M. Shokat, and William
Title:Role of LPAR3, PKC and EGFR in LPA-induced cell migration in oral squamous carcinoma cells
 Author's response to reviews Title:Role of LPAR3, PKC and EGFR in LPA-induced cell migration in oral squamous carcinoma cells Authors: Ingvild J Brusevold (i.j.brusevold@medisin.uio.no) Ingun H Tveteraas
Author's response to reviews Title:Role of LPAR3, PKC and EGFR in LPA-induced cell migration in oral squamous carcinoma cells Authors: Ingvild J Brusevold (i.j.brusevold@medisin.uio.no) Ingun H Tveteraas
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/366/ra25/dc1 Supplementary Materials for Viral entry route determines how human plasmacytoid dendritic cells produce type I interferons Daniela Bruni, Maxime
www.sciencesignaling.org/cgi/content/full/8/366/ra25/dc1 Supplementary Materials for Viral entry route determines how human plasmacytoid dendritic cells produce type I interferons Daniela Bruni, Maxime
Predictive PP1Ca binding region in BIG3 : 1,228 1,232aa (-KAVSF-) HEK293T cells *** *** *** KPL-3C cells - E E2 treatment time (h)
 Relative expression ERE-luciferase activity activity (pmole/min) activity (pmole/min) activity (pmole/min) activity (pmole/min) MCF-7 KPL-3C ZR--1 BT-474 T47D HCC15 KPL-1 HBC4 activity (pmole/min) a d
Relative expression ERE-luciferase activity activity (pmole/min) activity (pmole/min) activity (pmole/min) activity (pmole/min) MCF-7 KPL-3C ZR--1 BT-474 T47D HCC15 KPL-1 HBC4 activity (pmole/min) a d
William C. Comb, Jessica E. Hutti, Patricia Cogswell, Lewis C. Cantley, and Albert S. Baldwin
 Molecular Cell, Volume 45 Supplemental Information p85 SH2 Domain Phosphorylation by IKK Promotes Feedback Inhibition of PI3K and Akt in Response to Cellular Starvation William C. Comb, Jessica E. Hutti,
Molecular Cell, Volume 45 Supplemental Information p85 SH2 Domain Phosphorylation by IKK Promotes Feedback Inhibition of PI3K and Akt in Response to Cellular Starvation William C. Comb, Jessica E. Hutti,
Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v)
 SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
Supplemental Information
 Supplemental Information Tobacco-specific Carcinogen Induces DNA Methyltransferases 1 Accumulation through AKT/GSK3β/βTrCP/hnRNP-U in Mice and Lung Cancer patients Ruo-Kai Lin, 1 Yi-Shuan Hsieh, 2 Pinpin
Supplemental Information Tobacco-specific Carcinogen Induces DNA Methyltransferases 1 Accumulation through AKT/GSK3β/βTrCP/hnRNP-U in Mice and Lung Cancer patients Ruo-Kai Lin, 1 Yi-Shuan Hsieh, 2 Pinpin
Supplemental Figure 1. (A) Western blot for the expression of RIPK1 in HK-2 cells treated with or without LPS (1 µg/ml) for indicated times.
 Supplemental Figure 1. (A) Western blot for the expression of RIPK1 in HK-2 cells treated with or without LPS (1 µg/ml) for indicated times. Western blots shown are representative results from 3 independent
Supplemental Figure 1. (A) Western blot for the expression of RIPK1 in HK-2 cells treated with or without LPS (1 µg/ml) for indicated times. Western blots shown are representative results from 3 independent
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein
 Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein content relative to GAPDH in two independent experiments.
Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein content relative to GAPDH in two independent experiments.
Supplementary Material
 Supplementary Material accompanying the manuscript Interleukin 37 is a fundamental inhibitor of innate immunity Marcel F Nold, Claudia A Nold-Petry, Jarod A Zepp, Brent E Palmer, Philip Bufler & Charles
Supplementary Material accompanying the manuscript Interleukin 37 is a fundamental inhibitor of innate immunity Marcel F Nold, Claudia A Nold-Petry, Jarod A Zepp, Brent E Palmer, Philip Bufler & Charles
Autocrine production of IL-11 mediates tumorigenicity in hypoxic cancer cells
 Research article Autocrine production of IL-11 mediates tumorigenicity in hypoxic cancer cells Barbara Onnis, 1 Nicole Fer, 2 Annamaria Rapisarda, 2 Victor S. Perez, 1 and Giovanni Melillo 2 1 Developmental
Research article Autocrine production of IL-11 mediates tumorigenicity in hypoxic cancer cells Barbara Onnis, 1 Nicole Fer, 2 Annamaria Rapisarda, 2 Victor S. Perez, 1 and Giovanni Melillo 2 1 Developmental
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS)
 Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Figure S I: Effects of D4F on body weight and serum lipids in apoe -/- mice.
 Supplementary Figures: Supplementary Figure S I: Effects of D4F on body weight and serum lipids in apoe -/- mice. Male apoe -/- mice were fed a high-fat diet for 8 weeks, and given PBS (model group) or
Supplementary Figures: Supplementary Figure S I: Effects of D4F on body weight and serum lipids in apoe -/- mice. Male apoe -/- mice were fed a high-fat diet for 8 weeks, and given PBS (model group) or
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation.
 Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
gliomas. Fetal brain expected who each low-
 Supplementary Figure S1. Grade-specificity aberrant expression of HOXA genes in gliomas. (A) Representative RT-PCR analyses of HOXA gene expression in human astrocytomas. Exemplified glioma samples include
Supplementary Figure S1. Grade-specificity aberrant expression of HOXA genes in gliomas. (A) Representative RT-PCR analyses of HOXA gene expression in human astrocytomas. Exemplified glioma samples include
Supplementary Fig. 1 eif6 +/- mice show a reduction in white adipose tissue, blood lipids and normal glycogen synthesis. The cohort of the original
 Supplementary Fig. 1 eif6 +/- mice show a reduction in white adipose tissue, blood lipids and normal glycogen synthesis. The cohort of the original phenotypic screening was n=40. For specific tests, the
Supplementary Fig. 1 eif6 +/- mice show a reduction in white adipose tissue, blood lipids and normal glycogen synthesis. The cohort of the original phenotypic screening was n=40. For specific tests, the
Interleukin-6 promotes pancreatic cancer cell migration by rapidly activating the small GTPase CDC42
 Interleukin-6 promotes pancreatic cancer cell migration by rapidly activating the small GTPase CDC42 Gina L. Razidlo, Kevin M. Burton, and Mark A. McNiven SUPPORTING INFORMATION Figure S1. IL-6 promotes
Interleukin-6 promotes pancreatic cancer cell migration by rapidly activating the small GTPase CDC42 Gina L. Razidlo, Kevin M. Burton, and Mark A. McNiven SUPPORTING INFORMATION Figure S1. IL-6 promotes
SUPPLEMENTARY INFORMATION
 sirna pool: Control Tetherin -HA-GFP HA-Tetherin -Tubulin Supplementary Figure S1. Knockdown of HA-tagged tetherin expression by tetherin specific sirnas. HeLa cells were cotransfected with plasmids expressing
sirna pool: Control Tetherin -HA-GFP HA-Tetherin -Tubulin Supplementary Figure S1. Knockdown of HA-tagged tetherin expression by tetherin specific sirnas. HeLa cells were cotransfected with plasmids expressing
Supplementary Figure 1. Characterization of NMuMG-ErbB2 and NIC breast cancer cells expressing shrnas targeting LPP. NMuMG-ErbB2 cells (a) and NIC
 Supplementary Figure 1. Characterization of NMuMG-ErbB2 and NIC breast cancer cells expressing shrnas targeting LPP. NMuMG-ErbB2 cells (a) and NIC cells (b) were engineered to stably express either a LucA-shRNA
Supplementary Figure 1. Characterization of NMuMG-ErbB2 and NIC breast cancer cells expressing shrnas targeting LPP. NMuMG-ErbB2 cells (a) and NIC cells (b) were engineered to stably express either a LucA-shRNA
Supporting Information. FADD regulates NF-кB activation and promotes ubiquitination of cflip L to induce. apoptosis
 1 2 Supporting Information 3 4 5 FADD regulates NF-кB activation and promotes ubiquitination of cflip L to induce apoptosis 6 7 Kishu Ranjan and Chandramani Pathak* 8 9 Department of Cell Biology, School
1 2 Supporting Information 3 4 5 FADD regulates NF-кB activation and promotes ubiquitination of cflip L to induce apoptosis 6 7 Kishu Ranjan and Chandramani Pathak* 8 9 Department of Cell Biology, School
Supplementary Figure S1 Supplementary Figure S2
 Supplementary Figure S A) The blots shown in Figure B were qualified by using Gel-Pro analyzer software (Rockville, MD, USA). The ratio of LC3II/LC3I to actin was then calculated. The data are represented
Supplementary Figure S A) The blots shown in Figure B were qualified by using Gel-Pro analyzer software (Rockville, MD, USA). The ratio of LC3II/LC3I to actin was then calculated. The data are represented
The p53 protein induces apoptosis, cell cycle arrest, senescence,
 SCO2 Induces p53-mediated Apoptosis by Thr 845 Phosphorylation of ASK-1 and Dissociation of the ASK-1 Trx Complex Esha Madan, a,b Rajan Gogna, b Periannan Kuppusamy, c Madan Bhatt, d Abbas Ali Mahdi, a
SCO2 Induces p53-mediated Apoptosis by Thr 845 Phosphorylation of ASK-1 and Dissociation of the ASK-1 Trx Complex Esha Madan, a,b Rajan Gogna, b Periannan Kuppusamy, c Madan Bhatt, d Abbas Ali Mahdi, a
FOXO Reporter Kit PI3K/AKT Pathway Cat. #60643
 Data Sheet FOXO Reporter Kit PI3K/AKT Pathway Cat. #60643 Background The PI3K/AKT signaling pathway is essential for cell growth and survival. Disruption of this pathway or its regulation has been linked
Data Sheet FOXO Reporter Kit PI3K/AKT Pathway Cat. #60643 Background The PI3K/AKT signaling pathway is essential for cell growth and survival. Disruption of this pathway or its regulation has been linked
Organisms have evolved multiple ways to detect and respond
 Cyclin-dependent kinase activity is required for type I interferon production Oya Cingöz a,b and Stephen P. Goff a,b,c,1 a Department of Biochemistry and Molecular Biophysics, Columbia University Medical
Cyclin-dependent kinase activity is required for type I interferon production Oya Cingöz a,b and Stephen P. Goff a,b,c,1 a Department of Biochemistry and Molecular Biophysics, Columbia University Medical
Supplementary Figure 1: Tissue of Origin analysis on 152 cell lines. (a) Heatmap representation of the 30 Tissue scores for the 152 cell lines.
 Supplementary Figure 1: Tissue of Origin analysis on 152 cell lines. (a) Heatmap representation of the 30 Tissue scores for the 152 cell lines. The scores summarize the global expression of the tissue
Supplementary Figure 1: Tissue of Origin analysis on 152 cell lines. (a) Heatmap representation of the 30 Tissue scores for the 152 cell lines. The scores summarize the global expression of the tissue
EGFR kinase activity is required for TLR4 signaling and the septic shock response
 Manuscript EMBOR-2015-40337 EGFR kinase activity is required for TLR4 signaling and the septic shock response Saurabh Chattopadhyay, Manoj Veleeparambil, Darshana Poddar, Samar Abdulkhalek, Sudip K. Bandyopadhyay,
Manuscript EMBOR-2015-40337 EGFR kinase activity is required for TLR4 signaling and the septic shock response Saurabh Chattopadhyay, Manoj Veleeparambil, Darshana Poddar, Samar Abdulkhalek, Sudip K. Bandyopadhyay,
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/7/310/ra11/dc1 Supplementary Materials for STAT3 Induction of mir-146b Forms a Feedback Loop to Inhibit the NF-κB to IL-6 Signaling Axis and STAT3-Driven Cancer
www.sciencesignaling.org/cgi/content/full/7/310/ra11/dc1 Supplementary Materials for STAT3 Induction of mir-146b Forms a Feedback Loop to Inhibit the NF-κB to IL-6 Signaling Axis and STAT3-Driven Cancer
Supplementary Figure 1. Confocal immunofluorescence showing mitochondrial translocation of Drp1. Cardiomyocytes treated with H 2 O 2 were prestained
 Supplementary Figure 1. Confocal immunofluorescence showing mitochondrial translocation of Drp1. Cardiomyocytes treated with H 2 O 2 were prestained with MitoTracker (red), then were immunostained with
Supplementary Figure 1. Confocal immunofluorescence showing mitochondrial translocation of Drp1. Cardiomyocytes treated with H 2 O 2 were prestained with MitoTracker (red), then were immunostained with
Supplementary Figure 1
 Supplementary Figure 1 6 HE-50 HE-116 E-1 HE-108 Supplementary Figure 1. Targeted drug response curves of endometrial cancer cells. Endometrial cancer cell lines were incubated with serial dilutions of
Supplementary Figure 1 6 HE-50 HE-116 E-1 HE-108 Supplementary Figure 1. Targeted drug response curves of endometrial cancer cells. Endometrial cancer cell lines were incubated with serial dilutions of
4.2 RESULTS AND DISCUSSION
 phosphorylated proteins on treatment with Erlotinib.This chapter describes the finding of the SILAC experiment. 4.2 RESULTS AND DISCUSSION This is the first global report of its kind using dual strategies
phosphorylated proteins on treatment with Erlotinib.This chapter describes the finding of the SILAC experiment. 4.2 RESULTS AND DISCUSSION This is the first global report of its kind using dual strategies
Figure S1. ERBB3 mrna levels are elevated in Luminal A breast cancers harboring ERBB3
 Supplemental Figure Legends. Figure S1. ERBB3 mrna levels are elevated in Luminal A breast cancers harboring ERBB3 ErbB3 gene copy number gain. Supplemental Figure S1. ERBB3 mrna levels are elevated in
Supplemental Figure Legends. Figure S1. ERBB3 mrna levels are elevated in Luminal A breast cancers harboring ERBB3 ErbB3 gene copy number gain. Supplemental Figure S1. ERBB3 mrna levels are elevated in
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/4/199/ra75/dc1 Supplementary Materials for Signaling by the Matrix Proteoglycan Decorin Controls Inflammation and Cancer Through PDCD4 and MicroRNA-21 Rosetta
www.sciencesignaling.org/cgi/content/full/4/199/ra75/dc1 Supplementary Materials for Signaling by the Matrix Proteoglycan Decorin Controls Inflammation and Cancer Through PDCD4 and MicroRNA-21 Rosetta
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
Supplementary Information
 Supplementary Information An orally available, small-molecule interferon inhibits viral replication Hideyuki Konishi 1, Koichi Okamoto 1, Yusuke Ohmori 1, Hitoshi Yoshino 2, Hiroshi Ohmori 1, Motooki Ashihara
Supplementary Information An orally available, small-molecule interferon inhibits viral replication Hideyuki Konishi 1, Koichi Okamoto 1, Yusuke Ohmori 1, Hitoshi Yoshino 2, Hiroshi Ohmori 1, Motooki Ashihara
Inhibition of TGFβ enhances chemotherapy action against triple negative breast cancer by abrogation of
 SUPPLEMENTAL DATA Inhibition of TGFβ enhances chemotherapy action against triple negative breast cancer by abrogation of cancer stem cells and interleukin-8 Neil E. Bhola 1, Justin M. Balko 1, Teresa C.
SUPPLEMENTAL DATA Inhibition of TGFβ enhances chemotherapy action against triple negative breast cancer by abrogation of cancer stem cells and interleukin-8 Neil E. Bhola 1, Justin M. Balko 1, Teresa C.
Irf1 fold changes (D) 24h 48h. p-p65. t-p65. p-irf3. t-irf3. β-actin SKO TKO 100% 80% 60% 40% 20%
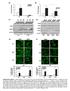 Irf7 Fold changes 3 1 Irf1 fold changes 3 1 8h h 8h 8h h 8h p-p6 p-p6 t-p6 p-irf3 β-actin p-irf3 t-irf3 β-actin TKO TKO STKO (E) (F) TKO TKO % of p6 nuclear translocation % % 1% 1% % % p6 TKO % of IRF3
Irf7 Fold changes 3 1 Irf1 fold changes 3 1 8h h 8h 8h h 8h p-p6 p-p6 t-p6 p-irf3 β-actin p-irf3 t-irf3 β-actin TKO TKO STKO (E) (F) TKO TKO % of p6 nuclear translocation % % 1% 1% % % p6 TKO % of IRF3
