Nature Medicine: doi: /nm.4439
|
|
|
- Isabella Wiggins
- 5 years ago
- Views:
Transcription
1 Figure S1. Overview of the variant calling and verification process. This figure expands on Fig. 1c with details of verified variants identification in 547 additional validation samples. Somatic variants (SNVs, indels, focal and chromosome-armlevel CNVs, and fusion products) were first called in 197 diagnostic samples with remission DNA (for germline) using a Complete Genomics custom Whole Genome Sequencing (WGS) variant calling pipeline. Complete Genomics calls were optimized at the start of the TARGET project using 100 independentlyverified variants in WGS samples. Matched tumor and remission samples in 153 cases were used for somatic variant calling by both WGS and targeted capture sequencing (TCS) of genes recurrently impacted in the WGS samples. 72% of WGS SNVs, and 76% of WGS indels were confirmed by TCS (red & green text in figures). For focal copy number (CN) alterations spanning fewer than 7 genes, 75% of recurrent WGS deletion/loss and 85% gain/amplification calls matched recurrent alterations discovered by SNP6 arrays in 96 matching samples. For chromosomal junctions, we integrated WGS, clinical and RNA-seq data by majority vote, and confirmed 89% of WGS calls. An additional 29 samples from the WGS discovery cohort were verified by TCS of diagnostic cases only, as part of 146 tumors without matched remission (see top portion of the figure). The remainder of these 146 cases were not used for discovery or validation purposes, rather, we simply identified recurrence of variants that were observed and verified in other samples.
2 Figure S2. Cellular processes and pathways commonly impacted in pediatric AML. The height of each bar indicates the percentage of samples with verified fusions (green), SNVs/indels (grey), or focal CNVs (gold) in recurrently impacted genes within 684 pediatric AML samples. See Table S2b for a list of the impacted genes.
3 Figure S3. Data type overlap for TARGET and TCGA diagnostic samples. UpSet plots ( showing the set overlaps for whole genome sequencing (WGS), whole exome (WXS), mrna sequencing, DNA methylation arrays (CpGmeth), mirna sequencing and targeted capture sequencing (TCS) in the TARGET and TCGA cohorts. The numbers of assays analyzed for each type are indicated by the horizontal bar graphs and number in the set intersection is illustrated in vertical bar graphs. The Clinical category includes samples comprising the entire TARGET AML dataset, including those in TARGET AML subprojects (e.g. previously reported WXS analysis 6 ). Data from these samples are included in the chromosomal arm level and karyotype based assessments of copy loss and fusions. (a) All TARGET AML project samples available. (b) All TCGA samples used for comparisons to TARGET. (c) Assay type overlaps for TCGA and TARGET data combined. a TARGET AML assay overlap (n=1023) * Clinical annotations include ISCN karyotype b TCGA assay overlap (n=177) c Combined TARGET & TCGA assay data overlap (n=1200)
4 Figure S4. Clonality estimates are consistent by age across cohorts. Both TCGA and TARGET AML cohorts contain affected individuals between the ages of 15 and 39 (adolescent and young adult, or AYA). Mutational and karyotypic clonality were assessed in AYA patients with whole-genome or whole-exome sequencing from either cohort, resulting in estimates from 40 TARGET AML subjects and 22 TCGA AML subjects in this age group. No significant association between cohort and mutational clonality estimate (p = , Fisher s exact test) or karyotypic clonality (p= , Fisher s exact test) is observed (TCGA AYA cases are older and more likely to have normal karyotype, though not significantly so). A multivariate Poisson model similarly shows little evidence for a significant cohort-wise effect. The strongest predictor of (decreasing) mutational clonality in AYAs is age at diagnosis (p=0.28).
5 Figure S5. The context of genome-wide mutation burden in pediatric AML. The mutational burden of SNV and indels is low in pediatric AML (blue), with a median of 10 mutations/case across the 197 sample WGS cohort. This places pediatric AML, along with other pediatric malignancies (rhabdoid tumor, Ewing sarcoma, medulloblastoma) and adult AML (red) among the least mutated of human cancers. Figure reproduced from the raw data reported by Lawrence and colleagues 51 updated to reflect TARGET AML results, plotted using the ggplot2 package in the R statistical environment.
6 Figure S6. A simplified visualization of common genomic variants in TARGET and TCGA AML data. Selected small variants are grouped by those that appear distinctive from core binding factor (CBF; t(8;21) and inv(16)) and KMT2A (aka MLL) fusions (grp1: mutations of WT1, NPM1, PTPN11, GATA2, CEBPA) and those that frequently cooccur with CBF alterations (grp2: mutations of KIT or ASXL2, loss of chr X). C, chromosomal alteration; J, junction/translocation; M, mutation; I, ITD. Pediatric Adult CBF grp1.var KMT2A FLT3 grp2.var NRAS KRAS ZEB2 MBNL1 grp1.var DNMT3A IDH2 IDH1 CBF TET2 TP53 NRAS grp2.var KMT2A KRAS
7 Figure S7. Adult-Pediatric mutational contrasts in AML. Lollipop plots generated with ProteinPaint ( highlight differences in frequency, type, and location of sequence variants in pediatric and adult AML. The plotted data reflect all somatic coding variants identified at presentation in 177 TCGA cases and 815 TARGET AML cases (WGS + TCS). Mutations are coded by functional class: blue, missense; brown, insertion; gray, deletion; red, frameshifting; orange, stop-gain; green, tandem duplication. a MYC b GATA2 TARGET TARGET c KRAS d FLT3 TARGET TCGA TARGET TCGA e NRAS f KIT TARGET TCGA TARGET TCGA
8 Figure S8. The impact of pediatric gene fusions on clinical outcome. (a) 199 patients evaluated for CBFA2T3-GLIS2 fusion had clinical outcome data available for analysis. Those with the fusion (n=9) had significantly worse overall survival than patients without the fusion (n=190) (p=0.0101). (b) 824 patients were evaluated for fusions involving ETS family transcription factors (ETV6, FUS, or ERG) through karyotype and/or transcriptome sequencing and had clinical outcome data available for analysis. Those with fusions (n=20) had significantly worse event-free survival than patients without a fusion (n=804) (p=0.0060). (c) 824 patients were evaluated for fusions involving KAT6A through karyotype and/or transcriptome sequencing and had clinical outcome data available for analysis. Those with fusions (n=8) had significantly worse overall survival than patients without a fusion (n=816) (p=0.0195). Differences in outcome were assessed by log-rank test. EFS, event free survival; OS, overall survival.
9 Figure S9. Pediatric CBL Exonic Deletions Detected by cdna Fragment Length Analysis. Representative examples of CBL wild-type and deletion transcripts detected by capillary electrophoresis of cdna. Horizontal axis depicts size of the PCR fragment (bp), while vertical axis indicates strength of signal. WT size (full-length transcript) is 685bp, exon 8 deletion only is 563bp, and deletions of exons 8 and 9 is 354bp.
10 Figure S10. Mutational frequency differences in key myeloid genes. (a) ECOG comparison 4. (b) TCGA comparison, balanced by cytogenetic subtypes (see online Methods). Error bars indicate the empirical SD from the resampling procedure. a b TARGET ECOG TARGET TCGA
11 Figure S11. Mutational co-occurrence in KMT2A rearranged childhood AML. We identified single copy segmental deletions of ZEB2 and/or MBNL1 in 14 patients, 6 of whom had concurrent KMT2A fusions (p=0.035, Fisher s exact test). The row entitled KMT2A (clinical) shows the manually-curated classification of the tumor primary cytogenetic type by combining results from clinical, genomic and RNA-seq assays. By this measure, all samples are classified as belonging to the KMT2A fusion cytogenetic group. The row entitled KMT2A (WGS) shows KMT2A variants found by WGS alone. Note 2 samples have copy number alterations as well as fusions impacting KMT2A. C, copy number alteration; J, junction/translocation; M, mutation; I, ITD. KMT2A (clinical) KMT2A (WGS) MLLT3 NRAS FLT3 KRAS MLLT10 MBNL1 TMEM14E ZEB2
12 Figure S12. Clonality at presentation in pediatric AML. (a) Mutation-based inference of clonality in 197 TARGET AML cases with WGS and 177 TCGA AML cases identifies 2 or more detectable clones in the majority of patients across age ranges. (b) A similar pattern with overall fewer detectable clones was observed by karyotypic inference of clonal relationships at presentation. a Infants (age <3) Children (age 3-15) AYA (age 15-40) Adults (age >40) b Mutational clones detected at diagnosis Karyotypic clones detected at diagnosis
13 Figure S13. Gene variants alone and in combination impact pediatric AML outcomes. (a) 963 patients from the TARGET dataset with clinical results for FLT3 internal tandem duplication (ITD), NPM1, WT1, NUP98-NSD1 fusion had clinical outcome data for analysis. Patients with a combination of FLT3 ITD and WT1 or NUP98-NSD1 versus FLT3 ITD alone or in combination with NPM1 mutation exhibit significantly decreased overall survival (p<0.001). (b) Similar results were found for COG trial AAML0531 (b), COG trial CCG-2961 (c), and the Dutch Childhood Oncology Group (DCOG) (d). In each trial those with FLT3 ITD plus WT1 and/or NUP98-NSD1 fusion exhibit significantly worse overall survival. The exact numbers of patients in each subgroup are indicated in the table below the figures. The total numbers of evaluable patients is indicated in the table below. ITD, FLT3-ITD. Cohort ITD - ITD - NPM1 + ITD - WT1 + ITD - NPM1 + WT1 + ITD - WT1 + NUP98-NSD1 + ITD - NUP98-NSD1 + ITD + ITD + NPM1 + ITD + WT1 + ITD + NPM1 + WT1 + ITD + WT1 + NUP98-NSD1 + ITD + NUP98-NSD1 + ITD - NPM1 + NUP98-NSD1 + TARGET AAML CCG DCOG Total
14 Figure S14. Remission rates vary for pediatric AML with FLT3-ITD according to cooperating mutations. The CCG-2961, AAML0531 and DCOG cohorts were combined to compare complete remission (CR) rates after one cycle of induction therapy for groups with FLT3-ITD cooperating mutations, as shown. CR rates are consistent with the survival outcomes (Figs. 3c and S13) among these studies: the poorest outcome group containing FLT3-ITD and a cooperating WT1 and/or NUP98-NSD1 fusion had the lowest CR rate, at 54.8%. The most favorable group, FLT3-ITD positive, NPM1 positive at 93.0% (groupwise p<0.0001, Kruskal-Wallis).
15 Figure S15. Novel ZEB2 and MBNL1 Deletions. (a-b) show short (<500 Kbp) deletion segments along chromosomes 2 (panel a, ZEB2) and chromosome 3 (panel b, MBNL1) in TARGET discovery cohort samples (n=197). (c) With the exception of one ZEB2-deleted sample (red point at top right of panel c), samples with ZEB2 and MBNL1 deletions are not impacted by large numbers of other CNVs. a b c
16 Expression Value ELF1 expression Figure S16. Novel ELF1 focal deletions in the TARGET discovery cohort. (a) Genome browser view of segmental deletions covering the ELF1 locus. Patients (n=197) are in rows, blue bars indicate length of deletion in that genomic region. (b) Genomic deletions were confirmed in a secondary assay using the ncounter CNV assay (Nanostring Techologies), with verification (boxed specimens with low probes signals as identified by green signals in the heatmap below) of all ELF1 deletions initially identified by WGS. (c) Expression values (RPKM) of ELF1 differ between those with the deletion and those with wild-type copy number (p=0.0077). (d) Unsupervised clustering of 63 differentially expressed genes (p<0.01) between patients with and without ELF1 deletion shows many genes are upregulated in the samples with ELF1 deletions. Orange labels on the y axis indicate patients with an ELF1 deletion. a b c d Expression of ELF del ELF1 deletion WT ELF1 WT
17 Figure S17. Summary view of the key fusion classes in pediatric AML. Each colored region represents a fusion family. Descriptive labels are written adjacent to each family. The fusion partner genes for each family are indicated by their HGNC symbols. The lines connecting gene symbols indicate fusion partners. The thickness of each line reflects the frequency of the observed fusion.
18 Figure S18. Varying the age cutoff for infants (< 3 years in Figure 4b) vs. children, to < 2 or even < 1, does not substantially alter conclusions about fusion prevalence. Panel c is the same as Fig. 4b (reproduced here for comparison). Panels a and b show how samples shift between age groups if the infant-child threshold is reduced to <2 years (b), or <1 year (a). Fusions are listed in the same order as in 4b and used the same color scheme. a b c Infants Children AYA Adults Infants Children AYA Adults < < Infants <3 Children 3-15 AYA Adults
19 Figure S19. Co-occurring mutations with CEBPA. (a) Oncoprint ( showing all TARGET samples with functionally-validated CSF3R mutations 20. Green indicates samples with mutations. (b) CEBPA and GATA2 mutations combinatorially impact Event-Free Survival. a b Percent survival CEBPA and/or GATA2 in Normal Cyto EFS 100 GATA2 +, CEBPA - (N=7) 80 CEBPA +, GATA2 + (N=16) CEBPA +, GATA2 - (N=13) 60 Wildtype (N=143) 40 P= EFS (Days)
20 Figure S20. Patterns of mutual co-occurrence and mutual exclusion among somatic pediatric AML variants. (a) Patterns of co-occurrence and (b) mutual exclusion among variants in the TARGET cohort were evaluated using CoMEt (see online methods). Line thickness represents log(p-value) for the observed co-occurrence rates. Orange boxes indicate cytogenetic groups. Except for copy number alterations at the top-right, which were only evaluated within the 197 samples with WGS, all other relations are among 684 samples with TCS. (c) An alternative derivation of conditional gene-gene relationships using a penalized Ising model yields similar conditional dependencies. a b
21 c
22 Figure S21. Anti-correlated DNA methylation and reduced transcription potential. By scanning 2000 bp upstream and 200 bp downstream of the transcription start site (TSS) for all known ENSEMBL isoforms of ~8000 expressed genes in AML, we fit segmented regression models of DNA methylation (X axis) against asinh (transcripts per million, TPM, Y axis) of each transcript or gene. Hyperbolic arcsine (asinh) is similar to log transformation but is defined at all points along the real number line. Since large batch effects confound the biological differences between TARGET pediatric AML and TCGA adult AML mrna data, we opted to take the within-cohort median expression for samples with 10% or less methylation at a CpG locus, and the silencing threshold at the locus corresponding to the gene of interest was then defined as the methylation fraction beyond which no sample in a cohort exceeded the median unmethylated expression level (from samples with <= 10% methylation) within its cohort. Any locus where healthy progenitors or myeloid cells showed >= 10% methylation was omitted from consideration. After these filtering steps, the most significantly associated locus (ideally correlated with r > 0.8 against its neighboring loci) was then selected as a tag CpG for the downstream transcript(s). A tag CpG for HumanMethylation450 arrays and either the same locus or (if not present) the best surrogate locus for HumanMethylation27 arrays passing the filters was retained for silencing calls. If no suitable HumanMethylation27 locus could be found, only samples with HumanMethylation450 data were assayed for silencing of a given gene. This method identified 119 genes with recurrent silencing by promoter hypermethylation within the TARGET and TCGA datasets. Examples below include THRB and WDR35 (components of NMF signature 2 and 13 signals, respectively), CDKN2B, and ULBP1, ULBP2 and ULBP3 (NK ligands). The red line marks the empirically determined silencing threshold (% methylation).
23 Figure S22. Integrative analysis of gene mutations, deletion, and transcriptional silencing by promoter methylation. Silencing (gold) or mutation/deletion events (gray) for each gene (rows) are displayed for all assayed patients (columns), with marginal total of events per patient illustrated in the upper histogram. The plotted data reflects 172 TCGA cases and 284 TARGET cases at 119 genes and are outlined in Tables S8-S9. These data represent a complete illustration of the subset shown in Fig. 5a with differences in row/column ordering based on differing clustering solutions for greater numbers of samples and genes. Status silenced mutated Cohort
24 Figure S23. NMF Deconvolution of genome-wide methylation patterns. DNA methylation signatures derived by non-negative matrix factorization (NMF) and in silico purification. Samples are ordered by hierarchical clustering of signatures (labeled at right) and demonstrate the relative similarity of methylation features from samples within cytogenetic categories (top ribbon). The plotted data are outlined in Table S10 and represent a complete illustration of those shown in Fig 5b. Associations Cohort
25 Figure S24. Two DNA methylation signatures mark poor prognosis. Kaplan-Meier plots for signatures 2 and 13. After stratifying by cohort and adjusting for both TP53 mutation status and white blood cell count, these two signatures predict significantly (p < 0.05) poorer event-free survival in both pediatric and adult patients with above-median scores. DNA methylation signature #2 DNA methylation signature #13
26 Consensus matrix Figure S25. Unsupervised Nonnegative Matrix Factorization (NMF) Clustering of mirna Expression. This figure is a fully annotated version of Fig. 6A in the main text. Unsupervised NMF clustering of mirna expression patterns in pediatric AML samples revealed 4 discrete pediatric subgroups (marked by the numbered colored rectangles at the top) that were correlated with specific genomic alterations (indicated by blue bars in the gray annotation rows below the race and FAB category annotations, near the top) Consensus matrix Expression z-
27 Figure S26. Kaplan-Meier plots for samples expressing low and high levels of mirs let-7a-3p, let-7b-5p and 30a-3p. The expression (RPM) cut point between high and low expression groups for each mirna was defined using the X-tile method 77, where all separation points between patients are considered and the selected cut point is the one that provided the optimal (lowest) EFS log rank p-value. OS, overall survival. P=< P= P=<0.0001
28 Figure S27. Kaplan-Meier plots for samples expressing low and high levels of mirs 155-5p, p, p and 26a-2-3p. The expression (RPM) cut point between high and low expression groups for each mirna was defined using the X-tile method 77. OS, overall survival.
29 Figure S28. High expression levels of mirs 133a-3p, 212-3p, and 29c-5p have deleterious effects on event free survival (EFS). The expression (RPM) cut point between high and low expression groups for each mirna was defined using the X-tile method 77. EFS, event-free survival.
Molecular Markers in Acute Leukemia. Dr Muhd Zanapiah Zakaria Hospital Ampang
 Molecular Markers in Acute Leukemia Dr Muhd Zanapiah Zakaria Hospital Ampang Molecular Markers Useful at diagnosis Classify groups and prognosis Development of more specific therapies Application of risk-adjusted
Molecular Markers in Acute Leukemia Dr Muhd Zanapiah Zakaria Hospital Ampang Molecular Markers Useful at diagnosis Classify groups and prognosis Development of more specific therapies Application of risk-adjusted
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Patel JP, Gönen M, Figueroa ME, et al. Prognostic relevance
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Patel JP, Gönen M, Figueroa ME, et al. Prognostic relevance
Examining Genetics and Genomics of Acute Myeloid Leukemia in 2017
 Examining Genetics and Genomics of Acute Myeloid Leukemia in 2017 Elli Papaemmanuil, PhD Memorial Sloan Kettering Cancer Center New York, New York, United States Today s Talk Cancer genome introduction
Examining Genetics and Genomics of Acute Myeloid Leukemia in 2017 Elli Papaemmanuil, PhD Memorial Sloan Kettering Cancer Center New York, New York, United States Today s Talk Cancer genome introduction
New drugs in Acute Leukemia. Cristina Papayannidis, MD, PhD University of Bologna
 New drugs in Acute Leukemia Cristina Papayannidis, MD, PhD University of Bologna Challenges to targeted therapy in AML Multiple subtypes based upon mutations/cytogenetic aberrations No known uniform genomic
New drugs in Acute Leukemia Cristina Papayannidis, MD, PhD University of Bologna Challenges to targeted therapy in AML Multiple subtypes based upon mutations/cytogenetic aberrations No known uniform genomic
Concomitant WT1 mutations predicted poor prognosis in CEBPA double-mutated acute myeloid leukemia
 Concomitant WT1 mutations predicted poor prognosis in CEBPA double-mutated acute myeloid leukemia Feng-Ming Tien, Hsin-An Hou, Jih-Luh Tang, Yuan-Yeh Kuo, Chien-Yuan Chen, Cheng-Hong Tsai, Ming Yao, Chi-Cheng
Concomitant WT1 mutations predicted poor prognosis in CEBPA double-mutated acute myeloid leukemia Feng-Ming Tien, Hsin-An Hou, Jih-Luh Tang, Yuan-Yeh Kuo, Chien-Yuan Chen, Cheng-Hong Tsai, Ming Yao, Chi-Cheng
Acute leukemia and myelodysplastic syndromes
 11/01/2012 Post-ASH meeting 1 Acute leukemia and myelodysplastic syndromes Peter Vandenberghe Centrum Menselijke Erfelijkheid & Afdeling Hematologie, UZ Leuven 11/01/2012 Post-ASH meeting 2 1. Acute myeloid
11/01/2012 Post-ASH meeting 1 Acute leukemia and myelodysplastic syndromes Peter Vandenberghe Centrum Menselijke Erfelijkheid & Afdeling Hematologie, UZ Leuven 11/01/2012 Post-ASH meeting 2 1. Acute myeloid
Nature Methods: doi: /nmeth.3115
 Supplementary Figure 1 Analysis of DNA methylation in a cancer cohort based on Infinium 450K data. RnBeads was used to rediscover a clinically distinct subgroup of glioblastoma patients characterized by
Supplementary Figure 1 Analysis of DNA methylation in a cancer cohort based on Infinium 450K data. RnBeads was used to rediscover a clinically distinct subgroup of glioblastoma patients characterized by
Cancer Informatics Lecture
 Cancer Informatics Lecture Mayo-UIUC Computational Genomics Course June 22, 2018 Krishna Rani Kalari Ph.D. Associate Professor 2017 MFMER 3702274-1 Outline The Cancer Genome Atlas (TCGA) Genomic Data Commons
Cancer Informatics Lecture Mayo-UIUC Computational Genomics Course June 22, 2018 Krishna Rani Kalari Ph.D. Associate Professor 2017 MFMER 3702274-1 Outline The Cancer Genome Atlas (TCGA) Genomic Data Commons
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10866 a b 1 2 3 4 5 6 7 Match No Match 1 2 3 4 5 6 7 Turcan et al. Supplementary Fig.1 Concepts mapping H3K27 targets in EF CBX8 targets in EF H3K27 targets in ES SUZ12 targets in ES
doi:10.1038/nature10866 a b 1 2 3 4 5 6 7 Match No Match 1 2 3 4 5 6 7 Turcan et al. Supplementary Fig.1 Concepts mapping H3K27 targets in EF CBX8 targets in EF H3K27 targets in ES SUZ12 targets in ES
Nature Genetics: doi: /ng Supplementary Figure 1. HOX fusions enhance self-renewal capacity.
 Supplementary Figure 1 HOX fusions enhance self-renewal capacity. Mouse bone marrow was transduced with a retrovirus carrying one of three HOX fusion genes or the empty mcherry reporter construct as described
Supplementary Figure 1 HOX fusions enhance self-renewal capacity. Mouse bone marrow was transduced with a retrovirus carrying one of three HOX fusion genes or the empty mcherry reporter construct as described
Whole Genome and Transcriptome Analysis of Anaplastic Meningioma. Patrick Tarpey Cancer Genome Project Wellcome Trust Sanger Institute
 Whole Genome and Transcriptome Analysis of Anaplastic Meningioma Patrick Tarpey Cancer Genome Project Wellcome Trust Sanger Institute Outline Anaplastic meningioma compared to other cancers Whole genomes
Whole Genome and Transcriptome Analysis of Anaplastic Meningioma Patrick Tarpey Cancer Genome Project Wellcome Trust Sanger Institute Outline Anaplastic meningioma compared to other cancers Whole genomes
Mutational Impact on Diagnostic and Prognostic Evaluation of MDS
 Mutational Impact on Diagnostic and Prognostic Evaluation of MDS Elsa Bernard, PhD Papaemmanuil Lab, Computational Oncology, MSKCC MDS Foundation ASH 2018 Symposium Disclosure Research funds provided by
Mutational Impact on Diagnostic and Prognostic Evaluation of MDS Elsa Bernard, PhD Papaemmanuil Lab, Computational Oncology, MSKCC MDS Foundation ASH 2018 Symposium Disclosure Research funds provided by
Nature Genetics: doi: /ng Supplementary Figure 1. Somatic coding mutations identified by WES/WGS for 83 ATL cases.
 Supplementary Figure 1 Somatic coding mutations identified by WES/WGS for 83 ATL cases. (a) The percentage of targeted bases covered by at least 2, 10, 20 and 30 sequencing reads (top) and average read
Supplementary Figure 1 Somatic coding mutations identified by WES/WGS for 83 ATL cases. (a) The percentage of targeted bases covered by at least 2, 10, 20 and 30 sequencing reads (top) and average read
Reporting cytogenetics Can it make sense? Daniel Weisdorf MD University of Minnesota
 Reporting cytogenetics Can it make sense? Daniel Weisdorf MD University of Minnesota Reporting cytogenetics What is it? Terminology Clinical value What details are important Diagnostic Tools for Leukemia
Reporting cytogenetics Can it make sense? Daniel Weisdorf MD University of Minnesota Reporting cytogenetics What is it? Terminology Clinical value What details are important Diagnostic Tools for Leukemia
BCR ABL1 like ALL: molekuliniai mechanizmai ir klinikinė reikšmė. IKAROS delecija: molekulinė biologija, prognostinė reikšmė. ASH 2015 naujienos
 BCR ABL1 like ALL: molekuliniai mechanizmai ir klinikinė reikšmė. IKAROS delecija: molekulinė biologija, prognostinė reikšmė. ASH 2015 naujienos Ph like ALL BCR ABL1 like acute lymphoblastic leukemia (ALL)
BCR ABL1 like ALL: molekuliniai mechanizmai ir klinikinė reikšmė. IKAROS delecija: molekulinė biologija, prognostinė reikšmė. ASH 2015 naujienos Ph like ALL BCR ABL1 like acute lymphoblastic leukemia (ALL)
Relationship between genomic features and distributions of RS1 and RS3 rearrangements in breast cancer genomes.
 Supplementary Figure 1 Relationship between genomic features and distributions of RS1 and RS3 rearrangements in breast cancer genomes. (a,b) Values of coefficients associated with genomic features, separately
Supplementary Figure 1 Relationship between genomic features and distributions of RS1 and RS3 rearrangements in breast cancer genomes. (a,b) Values of coefficients associated with genomic features, separately
Corporate Medical Policy. Policy Effective February 23, 2018
 Corporate Medical Policy Genetic Testing for FLT3, NPM1 and CEBPA Mutations in Acute File Name: Origination: Last CAP Review: Next CAP Review: Last Review: genetic_testing_for_flt3_npm1_and_cebpa_mutations_in_acute_myeloid_leukemia
Corporate Medical Policy Genetic Testing for FLT3, NPM1 and CEBPA Mutations in Acute File Name: Origination: Last CAP Review: Next CAP Review: Last Review: genetic_testing_for_flt3_npm1_and_cebpa_mutations_in_acute_myeloid_leukemia
Supplementary Figure 1. Estimation of tumour content
 Supplementary Figure 1. Estimation of tumour content a, Approach used to estimate the tumour content in S13T1/T2, S6T1/T2, S3T1/T2 and S12T1/T2. Tissue and tumour areas were evaluated by two independent
Supplementary Figure 1. Estimation of tumour content a, Approach used to estimate the tumour content in S13T1/T2, S6T1/T2, S3T1/T2 and S12T1/T2. Tissue and tumour areas were evaluated by two independent
Illumina Trusight Myeloid Panel validation A R FHAN R A FIQ
 Illumina Trusight Myeloid Panel validation A R FHAN R A FIQ G E NETIC T E CHNOLOGIST MEDICAL G E NETICS, CARDIFF To Cover Background to the project Choice of panel Validation process Genes on panel, Protocol
Illumina Trusight Myeloid Panel validation A R FHAN R A FIQ G E NETIC T E CHNOLOGIST MEDICAL G E NETICS, CARDIFF To Cover Background to the project Choice of panel Validation process Genes on panel, Protocol
Supplementary Figure 1: Features of IGLL5 Mutations in CLL: a) Representative IGV screenshot of first
 Supplementary Figure 1: Features of IGLL5 Mutations in CLL: a) Representative IGV screenshot of first intron IGLL5 mutation depicting biallelic mutations. Red arrows highlight the presence of out of phase
Supplementary Figure 1: Features of IGLL5 Mutations in CLL: a) Representative IGV screenshot of first intron IGLL5 mutation depicting biallelic mutations. Red arrows highlight the presence of out of phase
SUPPLEMENTARY FIGURES: Supplementary Figure 1
 SUPPLEMENTARY FIGURES: Supplementary Figure 1 Supplementary Figure 1. Glioblastoma 5hmC quantified by paired BS and oxbs treated DNA hybridized to Infinium DNA methylation arrays. Workflow depicts analytic
SUPPLEMENTARY FIGURES: Supplementary Figure 1 Supplementary Figure 1. Glioblastoma 5hmC quantified by paired BS and oxbs treated DNA hybridized to Infinium DNA methylation arrays. Workflow depicts analytic
Computational Analysis of UHT Sequences Histone modifications, CAGE, RNA-Seq
 Computational Analysis of UHT Sequences Histone modifications, CAGE, RNA-Seq Philipp Bucher Wednesday January 21, 2009 SIB graduate school course EPFL, Lausanne ChIP-seq against histone variants: Biological
Computational Analysis of UHT Sequences Histone modifications, CAGE, RNA-Seq Philipp Bucher Wednesday January 21, 2009 SIB graduate school course EPFL, Lausanne ChIP-seq against histone variants: Biological
Impact of Biomarkers in the Management of Patients with Acute Myeloid Leukemia
 Impact of Biomarkers in the Management of Patients with Acute Myeloid Leukemia Hartmut Döhner Medical Director, Department of Internal Medicine III Director, Comprehensive Cancer Center Ulm Ulm University,
Impact of Biomarkers in the Management of Patients with Acute Myeloid Leukemia Hartmut Döhner Medical Director, Department of Internal Medicine III Director, Comprehensive Cancer Center Ulm Ulm University,
Nature Structural & Molecular Biology: doi: /nsmb.2419
 Supplementary Figure 1 Mapped sequence reads and nucleosome occupancies. (a) Distribution of sequencing reads on the mouse reference genome for chromosome 14 as an example. The number of reads in a 1 Mb
Supplementary Figure 1 Mapped sequence reads and nucleosome occupancies. (a) Distribution of sequencing reads on the mouse reference genome for chromosome 14 as an example. The number of reads in a 1 Mb
Session 4 Rebecca Poulos
 The Cancer Genome Atlas (TCGA) & International Cancer Genome Consortium (ICGC) Session 4 Rebecca Poulos Prince of Wales Clinical School Introductory bioinformatics for human genomics workshop, UNSW 20
The Cancer Genome Atlas (TCGA) & International Cancer Genome Consortium (ICGC) Session 4 Rebecca Poulos Prince of Wales Clinical School Introductory bioinformatics for human genomics workshop, UNSW 20
About OMICS Group Conferences
 About OMICS Group OMICS Group International is an amalgamation of Open Access publications and worldwide international science conferences and events. Established in the year 2007 with the sole aim of
About OMICS Group OMICS Group International is an amalgamation of Open Access publications and worldwide international science conferences and events. Established in the year 2007 with the sole aim of
BWA alignment to reference transcriptome and genome. Convert transcriptome mappings back to genome space
 Whole genome sequencing Whole exome sequencing BWA alignment to reference transcriptome and genome Convert transcriptome mappings back to genome space genomes Filter on MQ, distance, Cigar string Annotate
Whole genome sequencing Whole exome sequencing BWA alignment to reference transcriptome and genome Convert transcriptome mappings back to genome space genomes Filter on MQ, distance, Cigar string Annotate
Molecular Genetics of Paediatric Tumours. Gino Somers MBBS, BMedSci, PhD, FRCPA Pathologist-in-Chief Hospital for Sick Children, Toronto, ON, CANADA
 Molecular Genetics of Paediatric Tumours Gino Somers MBBS, BMedSci, PhD, FRCPA Pathologist-in-Chief Hospital for Sick Children, Toronto, ON, CANADA Financial Disclosure NanoString - conference costs for
Molecular Genetics of Paediatric Tumours Gino Somers MBBS, BMedSci, PhD, FRCPA Pathologist-in-Chief Hospital for Sick Children, Toronto, ON, CANADA Financial Disclosure NanoString - conference costs for
ADRL Advanced Diagnostics Research Laboratory
 ADRL Advanced Diagnostics Research Laboratory John DeCoteau, MD FRCP Department of Pathology, Division of Hematopathology University of Saskatchewan Saskatchewan Cancer Agency ADRL Project Objectives New
ADRL Advanced Diagnostics Research Laboratory John DeCoteau, MD FRCP Department of Pathology, Division of Hematopathology University of Saskatchewan Saskatchewan Cancer Agency ADRL Project Objectives New
Please Silence Your Cell Phones. Thank You
 Please Silence Your Cell Phones Thank You Utility of NGS and Comprehensive Genomic Profiling in Hematopathology Practice Maria E. Arcila M.D. Memorial Sloan Kettering Cancer Center New York, NY Disclosure
Please Silence Your Cell Phones Thank You Utility of NGS and Comprehensive Genomic Profiling in Hematopathology Practice Maria E. Arcila M.D. Memorial Sloan Kettering Cancer Center New York, NY Disclosure
Blastic Plasmacytoid Dendritic Cell Neoplasm with DNMT3A and TET2 mutations (SH )
 Blastic Plasmacytoid Dendritic Cell Neoplasm with DNMT3A and TET2 mutations (SH2017-0314) Habibe Kurt, Joseph D. Khoury, Carlos E. Bueso-Ramos, Jeffrey L. Jorgensen, Guilin Tang, L. Jeffrey Medeiros, and
Blastic Plasmacytoid Dendritic Cell Neoplasm with DNMT3A and TET2 mutations (SH2017-0314) Habibe Kurt, Joseph D. Khoury, Carlos E. Bueso-Ramos, Jeffrey L. Jorgensen, Guilin Tang, L. Jeffrey Medeiros, and
The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions
Test Name Results Units Bio. Ref. Interval. Positive
 LL - LL-ROHINI (NATIONAL REFERENCE 135091534 Age 36 Years Gender Female 1/9/2017 120000AM 1/9/2017 105316AM 2/9/2017 104147AM Ref By Final LEUKEMIA GENETIC ROFILE ANY SIX MARKERS, CR QUALITATIVE AML ETO
LL - LL-ROHINI (NATIONAL REFERENCE 135091534 Age 36 Years Gender Female 1/9/2017 120000AM 1/9/2017 105316AM 2/9/2017 104147AM Ref By Final LEUKEMIA GENETIC ROFILE ANY SIX MARKERS, CR QUALITATIVE AML ETO
Journal: Nature Methods
 Journal: Nature Methods Article Title: Network-based stratification of tumor mutations Corresponding Author: Trey Ideker Supplementary Item Supplementary Figure 1 Supplementary Figure 2 Supplementary Figure
Journal: Nature Methods Article Title: Network-based stratification of tumor mutations Corresponding Author: Trey Ideker Supplementary Item Supplementary Figure 1 Supplementary Figure 2 Supplementary Figure
GENETIC TESTING FOR FLT3, NPM1 AND CEBPA VARIANTS IN CYTOGENETICALLY NORMAL ACUTE MYELOID LEUKEMIA
 CYTOGENETICALLY NORMAL ACUTE MYELOID LEUKEMIA Non-Discrimination Statement and Multi-Language Interpreter Services information are located at the end of this document. Coverage for services, procedures,
CYTOGENETICALLY NORMAL ACUTE MYELOID LEUKEMIA Non-Discrimination Statement and Multi-Language Interpreter Services information are located at the end of this document. Coverage for services, procedures,
Objectives. Morphology and IHC. Flow and Cyto FISH. Testing for Heme Malignancies 3/20/2013
 Molecular Markers in Hematologic Malignancy: Ways to locate the needle in the haystack. Objectives Review the types of testing for hematologic malignancies Understand rationale for molecular testing Marcie
Molecular Markers in Hematologic Malignancy: Ways to locate the needle in the haystack. Objectives Review the types of testing for hematologic malignancies Understand rationale for molecular testing Marcie
Molecular Markers. Marcie Riches, MD, MS Associate Professor University of North Carolina Scientific Director, Infection and Immune Reconstitution WC
 Molecular Markers Marcie Riches, MD, MS Associate Professor University of North Carolina Scientific Director, Infection and Immune Reconstitution WC Overview Testing methods Rationale for molecular testing
Molecular Markers Marcie Riches, MD, MS Associate Professor University of North Carolina Scientific Director, Infection and Immune Reconstitution WC Overview Testing methods Rationale for molecular testing
Supplemental Material. The new provisional WHO entity RUNX1 mutated AML shows specific genetics without prognostic influence of dysplasia
 Supplemental Material The new provisional WHO entity RUNX1 mutated AML shows specific genetics without prognostic influence of dysplasia Torsten Haferlach, 1 Anna Stengel, 1 Sandra Eckstein, 1 Karolína
Supplemental Material The new provisional WHO entity RUNX1 mutated AML shows specific genetics without prognostic influence of dysplasia Torsten Haferlach, 1 Anna Stengel, 1 Sandra Eckstein, 1 Karolína
Deep Learning Analytics for Predicting Prognosis of Acute Myeloid Leukemia with Cytogenetics, Age, and Mutations
 Deep Learning Analytics for Predicting Prognosis of Acute Myeloid Leukemia with Cytogenetics, Age, and Mutations Andy Nguyen, M.D., M.S. Medical Director, Hematopathology, Hematology and Coagulation Laboratory,
Deep Learning Analytics for Predicting Prognosis of Acute Myeloid Leukemia with Cytogenetics, Age, and Mutations Andy Nguyen, M.D., M.S. Medical Director, Hematopathology, Hematology and Coagulation Laboratory,
A pediatric patient with acute leukemia of ambiguous lineage with a NUP98-NSD1 rearrangement SH
 A pediatric patient with acute leukemia of ambiguous lineage with a NUP98NSD1 rearrangement SH20170203 Rebecca LeemanNeill, Ronald Rice, Anita Malek, Patricia Raciti, Susan Hsiao, Mahesh Mansukhani, Bachir
A pediatric patient with acute leukemia of ambiguous lineage with a NUP98NSD1 rearrangement SH20170203 Rebecca LeemanNeill, Ronald Rice, Anita Malek, Patricia Raciti, Susan Hsiao, Mahesh Mansukhani, Bachir
Nature Genetics: doi: /ng Supplementary Figure 1. SEER data for male and female cancer incidence from
 Supplementary Figure 1 SEER data for male and female cancer incidence from 1975 2013. (a,b) Incidence rates of oral cavity and pharynx cancer (a) and leukemia (b) are plotted, grouped by males (blue),
Supplementary Figure 1 SEER data for male and female cancer incidence from 1975 2013. (a,b) Incidence rates of oral cavity and pharynx cancer (a) and leukemia (b) are plotted, grouped by males (blue),
DNA-seq Bioinformatics Analysis: Copy Number Variation
 DNA-seq Bioinformatics Analysis: Copy Number Variation Elodie Girard elodie.girard@curie.fr U900 institut Curie, INSERM, Mines ParisTech, PSL Research University Paris, France NGS Applications 5C HiC DNA-seq
DNA-seq Bioinformatics Analysis: Copy Number Variation Elodie Girard elodie.girard@curie.fr U900 institut Curie, INSERM, Mines ParisTech, PSL Research University Paris, France NGS Applications 5C HiC DNA-seq
Supplementary Figure S1. Gene expression analysis of epidermal marker genes and TP63.
 Supplementary Figure Legends Supplementary Figure S1. Gene expression analysis of epidermal marker genes and TP63. A. Screenshot of the UCSC genome browser from normalized RNAPII and RNA-seq ChIP-seq data
Supplementary Figure Legends Supplementary Figure S1. Gene expression analysis of epidermal marker genes and TP63. A. Screenshot of the UCSC genome browser from normalized RNAPII and RNA-seq ChIP-seq data
Frequency(%) KRAS G12 KRAS G13 KRAS A146 KRAS Q61 KRAS K117N PIK3CA H1047 PIK3CA E545 PIK3CA E542K PIK3CA Q546. EGFR exon19 NFS-indel EGFR L858R
 Frequency(%) 1 a b ALK FS-indel ALK R1Q HRAS Q61R HRAS G13R IDH R17K IDH R14Q MET exon14 SS-indel KIT D8Y KIT L76P KIT exon11 NFS-indel SMAD4 R361 IDH1 R13 CTNNB1 S37 CTNNB1 S4 AKT1 E17K ERBB D769H ERBB
Frequency(%) 1 a b ALK FS-indel ALK R1Q HRAS Q61R HRAS G13R IDH R17K IDH R14Q MET exon14 SS-indel KIT D8Y KIT L76P KIT exon11 NFS-indel SMAD4 R361 IDH1 R13 CTNNB1 S37 CTNNB1 S4 AKT1 E17K ERBB D769H ERBB
Supplemental Information. Molecular, Pathological, Radiological, and Immune. Profiling of Non-brainstem Pediatric High-Grade
 Cancer Cell, Volume 33 Supplemental Information Molecular, Pathological, Radiological, and Immune Profiling of Non-brainstem Pediatric High-Grade Glioma from the HERBY Phase II Randomized Trial Alan Mackay,
Cancer Cell, Volume 33 Supplemental Information Molecular, Pathological, Radiological, and Immune Profiling of Non-brainstem Pediatric High-Grade Glioma from the HERBY Phase II Randomized Trial Alan Mackay,
Out-Patient Billing CPT Codes
 Out-Patient Billing CPT Codes Updated Date: August 3, 08 Client Billed Molecular Tests HPV DNA Tissue Testing 8764 No Medicare Billed - Molecular Tests NeoARRAY NeoARRAY SNP/Cytogenetic No 89 NeoLAB NeoLAB
Out-Patient Billing CPT Codes Updated Date: August 3, 08 Client Billed Molecular Tests HPV DNA Tissue Testing 8764 No Medicare Billed - Molecular Tests NeoARRAY NeoARRAY SNP/Cytogenetic No 89 NeoLAB NeoLAB
Molecular and genetic alterations associated with therapy resistance and relapse of acute myeloid leukemia
 Hackl et al. Journal of Hematology & Oncology (2017) 10:51 DOI 10.1186/s13045-017-0416-0 REVIEW Open Access Molecular and genetic alterations associated with therapy resistance and relapse of acute myeloid
Hackl et al. Journal of Hematology & Oncology (2017) 10:51 DOI 10.1186/s13045-017-0416-0 REVIEW Open Access Molecular and genetic alterations associated with therapy resistance and relapse of acute myeloid
ARTICLE RESEARCH. Macmillan Publishers Limited. All rights reserved
 Extended Data Figure 6 Annotation of drivers based on clinical characteristics and co-occurrence patterns. a, Putative drivers affecting greater than 10 patients were assessed for enrichment in IGHV mutated
Extended Data Figure 6 Annotation of drivers based on clinical characteristics and co-occurrence patterns. a, Putative drivers affecting greater than 10 patients were assessed for enrichment in IGHV mutated
Next Generation Sequencing in Haematological Malignancy: A European Perspective. Wolfgang Kern, Munich Leukemia Laboratory
 Next Generation Sequencing in Haematological Malignancy: A European Perspective Wolfgang Kern, Munich Leukemia Laboratory Diagnostic Methods Cytomorphology Cytogenetics Immunophenotype Histology FISH Molecular
Next Generation Sequencing in Haematological Malignancy: A European Perspective Wolfgang Kern, Munich Leukemia Laboratory Diagnostic Methods Cytomorphology Cytogenetics Immunophenotype Histology FISH Molecular
Expert-guided Visual Exploration (EVE) for patient stratification. Hamid Bolouri, Lue-Ping Zhao, Eric C. Holland
 Expert-guided Visual Exploration (EVE) for patient stratification Hamid Bolouri, Lue-Ping Zhao, Eric C. Holland Oncoscape.sttrcancer.org Paul Lisa Ken Jenny Desert Eric The challenge Given - patient clinical
Expert-guided Visual Exploration (EVE) for patient stratification Hamid Bolouri, Lue-Ping Zhao, Eric C. Holland Oncoscape.sttrcancer.org Paul Lisa Ken Jenny Desert Eric The challenge Given - patient clinical
Session 4 Rebecca Poulos
 The Cancer Genome Atlas (TCGA) & International Cancer Genome Consortium (ICGC) Session 4 Rebecca Poulos Prince of Wales Clinical School Introductory bioinformatics for human genomics workshop, UNSW 28
The Cancer Genome Atlas (TCGA) & International Cancer Genome Consortium (ICGC) Session 4 Rebecca Poulos Prince of Wales Clinical School Introductory bioinformatics for human genomics workshop, UNSW 28
Application of Whole Genome Microarrays in Cancer: You should be doing this test!!
 Application of Whole Genome Microarrays in Cancer: You should be doing this test!! Daynna Wolff, Ph.D. Director, Cytogenetics and Genomics Disclosures Clinical Laboratory Director and Employee, Medical
Application of Whole Genome Microarrays in Cancer: You should be doing this test!! Daynna Wolff, Ph.D. Director, Cytogenetics and Genomics Disclosures Clinical Laboratory Director and Employee, Medical
Supplementary Materials for
 www.sciencemag.org/content/355/6332/eaai8478/suppl/dc1 Supplementary Materials for Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq Andrew S. Venteicher,
www.sciencemag.org/content/355/6332/eaai8478/suppl/dc1 Supplementary Materials for Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq Andrew S. Venteicher,
Computer Science, Biology, and Biomedical Informatics (CoSBBI) Outline. Molecular Biology of Cancer AND. Goals/Expectations. David Boone 7/1/2015
 Goals/Expectations Computer Science, Biology, and Biomedical (CoSBBI) We want to excite you about the world of computer science, biology, and biomedical informatics. Experience what it is like to be a
Goals/Expectations Computer Science, Biology, and Biomedical (CoSBBI) We want to excite you about the world of computer science, biology, and biomedical informatics. Experience what it is like to be a
HEMATOLOGIC MALIGNANCIES BIOLOGY
 HEMATOLOGIC MALIGNANCIES BIOLOGY Failure of terminal differentiation Failure of differentiated cells to undergo apoptosis Failure to control growth Neoplastic stem cell FAILURE OF TERMINAL DIFFERENTIATION
HEMATOLOGIC MALIGNANCIES BIOLOGY Failure of terminal differentiation Failure of differentiated cells to undergo apoptosis Failure to control growth Neoplastic stem cell FAILURE OF TERMINAL DIFFERENTIATION
Genomic Medicine: What every pathologist needs to know
 Genomic Medicine: What every pathologist needs to know Stephen P. Ethier, Ph.D. Professor, Department of Pathology and Laboratory Medicine, MUSC Director, MUSC Center for Genomic Medicine Genomics and
Genomic Medicine: What every pathologist needs to know Stephen P. Ethier, Ph.D. Professor, Department of Pathology and Laboratory Medicine, MUSC Director, MUSC Center for Genomic Medicine Genomics and
Test Name Results Units Bio. Ref. Interval. Positive
 LL - LL-ROHINI (NATIONAL REFERENCE 135091533 Age 28 Years Gender Male 1/9/2017 120000AM 1/9/2017 105415AM 4/9/2017 23858M Ref By Final LEUKEMIA DIAGNOSTIC COMREHENSIVE ROFILE, ANY 6 MARKERS t (1;19) (q23
LL - LL-ROHINI (NATIONAL REFERENCE 135091533 Age 28 Years Gender Male 1/9/2017 120000AM 1/9/2017 105415AM 4/9/2017 23858M Ref By Final LEUKEMIA DIAGNOSTIC COMREHENSIVE ROFILE, ANY 6 MARKERS t (1;19) (q23
Nature Biotechnology: doi: /nbt.1904
 Supplementary Information Comparison between assembly-based SV calls and array CGH results Genome-wide array assessment of copy number changes, such as array comparative genomic hybridization (acgh), is
Supplementary Information Comparison between assembly-based SV calls and array CGH results Genome-wide array assessment of copy number changes, such as array comparative genomic hybridization (acgh), is
Plasma-Seq conducted with blood from male individuals without cancer.
 Supplementary Figures Supplementary Figure 1 Plasma-Seq conducted with blood from male individuals without cancer. Copy number patterns established from plasma samples of male individuals without cancer
Supplementary Figures Supplementary Figure 1 Plasma-Seq conducted with blood from male individuals without cancer. Copy number patterns established from plasma samples of male individuals without cancer
Abstract. Optimization strategy of Copy Number Variant calling using Multiplicom solutions APPLICATION NOTE. Introduction
 Optimization strategy of Copy Number Variant calling using Multiplicom solutions Michael Vyverman, PhD; Laura Standaert, PhD and Wouter Bossuyt, PhD Abstract Copy number variations (CNVs) represent a significant
Optimization strategy of Copy Number Variant calling using Multiplicom solutions Michael Vyverman, PhD; Laura Standaert, PhD and Wouter Bossuyt, PhD Abstract Copy number variations (CNVs) represent a significant
All patients with FLT3 mutant AML should receive midostaurin-based induction therapy. Not so fast!
 All patients with FLT3 mutant AML should receive midostaurin-based induction therapy Not so fast! Harry P. Erba, M.D., Ph.D. Professor, Internal Medicine Director, Hematologic Malignancy Program University
All patients with FLT3 mutant AML should receive midostaurin-based induction therapy Not so fast! Harry P. Erba, M.D., Ph.D. Professor, Internal Medicine Director, Hematologic Malignancy Program University
Nature Medicine: doi: /nm.3967
 Supplementary Figure 1. Network clustering. (a) Clustering performance as a function of inflation factor. The grey curve shows the median weighted Silhouette widths for varying inflation factors (f [1.6,
Supplementary Figure 1. Network clustering. (a) Clustering performance as a function of inflation factor. The grey curve shows the median weighted Silhouette widths for varying inflation factors (f [1.6,
Introduction. Introduction
 Introduction We are leveraging genome sequencing data from The Cancer Genome Atlas (TCGA) to more accurately define mutated and stable genes and dysregulated metabolic pathways in solid tumors. These efforts
Introduction We are leveraging genome sequencing data from The Cancer Genome Atlas (TCGA) to more accurately define mutated and stable genes and dysregulated metabolic pathways in solid tumors. These efforts
Changing AML Outcomes via Personalized Medicine: Transforming Cancer Management with Genetic Insight
 Changing AML Outcomes via Personalized Medicine: Transforming Cancer Management with Genetic Insight Co-Moderators: Rick Winneker, PhD, Senior Vice President, Research, Leukemia & Lymphoma Society Mike
Changing AML Outcomes via Personalized Medicine: Transforming Cancer Management with Genetic Insight Co-Moderators: Rick Winneker, PhD, Senior Vice President, Research, Leukemia & Lymphoma Society Mike
SUPPLEMENTAL APPENDIX METZELER ET AL.: SPECTRUM AND PROGNOSTIC RELEVANCE OF DRIVER GENE MUTATIONS IN ACUTE MYELOID LEUKEMIA
 SUPPLEMENTAL APPENDIX TO METZELER ET AL.: SPECTRUM AND PROGNOSTIC RELEVANCE OF DRIVER GENE MUTATIONS IN ACUTE MYELOID LEUKEMIA SUPPLEMENTAL METHODS Treatment protocols In the AMLCG-1999 trial 1-3 (clinicaltrials.gov
SUPPLEMENTAL APPENDIX TO METZELER ET AL.: SPECTRUM AND PROGNOSTIC RELEVANCE OF DRIVER GENE MUTATIONS IN ACUTE MYELOID LEUKEMIA SUPPLEMENTAL METHODS Treatment protocols In the AMLCG-1999 trial 1-3 (clinicaltrials.gov
NGS in tissue and liquid biopsy
 NGS in tissue and liquid biopsy Ana Vivancos, PhD Referencias So, why NGS in the clinics? 2000 Sanger Sequencing (1977-) 2016 NGS (2006-) ABIPrism (Applied Biosystems) Up to 2304 per day (96 sequences
NGS in tissue and liquid biopsy Ana Vivancos, PhD Referencias So, why NGS in the clinics? 2000 Sanger Sequencing (1977-) 2016 NGS (2006-) ABIPrism (Applied Biosystems) Up to 2304 per day (96 sequences
SUPPLEMENTARY APPENDIX
 SUPPLEMENTARY APPENDIX 1) Supplemental Figure 1. Histopathologic Characteristics of the Tumors in the Discovery Cohort 2) Supplemental Figure 2. Incorporation of Normal Epidermal Melanocytic Signature
SUPPLEMENTARY APPENDIX 1) Supplemental Figure 1. Histopathologic Characteristics of the Tumors in the Discovery Cohort 2) Supplemental Figure 2. Incorporation of Normal Epidermal Melanocytic Signature
Nature Genetics: doi: /ng.2995
 Supplementary Figure 1 Kaplan-Meier survival curves of patients with brainstem tumors. (a) Comparison of patients with PPM1D mutation versus wild-type PPM1D. (b) Comparison of patients with PPM1D mutation
Supplementary Figure 1 Kaplan-Meier survival curves of patients with brainstem tumors. (a) Comparison of patients with PPM1D mutation versus wild-type PPM1D. (b) Comparison of patients with PPM1D mutation
Genetic alterations of histone lysine methyltransferases and their significance in breast cancer
 Genetic alterations of histone lysine methyltransferases and their significance in breast cancer Supplementary Materials and Methods Phylogenetic tree of the HMT superfamily The phylogeny outlined in the
Genetic alterations of histone lysine methyltransferases and their significance in breast cancer Supplementary Materials and Methods Phylogenetic tree of the HMT superfamily The phylogeny outlined in the
The Center for PERSONALIZED DIAGNOSTICS
 The Center for PERSONALIZED DIAGNOSTICS Precision Diagnostics for Personalized Medicine A joint initiative between The Department of Pathology and Laboratory Medicine & The Abramson Cancer Center The (CPD)
The Center for PERSONALIZED DIAGNOSTICS Precision Diagnostics for Personalized Medicine A joint initiative between The Department of Pathology and Laboratory Medicine & The Abramson Cancer Center The (CPD)
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Heatmap of GO terms for differentially expressed genes. The terms were hierarchically clustered using the GO term enrichment beta. Darker red, higher positive
Supplementary Figures Supplementary Figure 1. Heatmap of GO terms for differentially expressed genes. The terms were hierarchically clustered using the GO term enrichment beta. Darker red, higher positive
Case Studies on High Throughput Gene Expression Data Kun Huang, PhD Raghu Machiraju, PhD
 Case Studies on High Throughput Gene Expression Data Kun Huang, PhD Raghu Machiraju, PhD Department of Biomedical Informatics Department of Computer Science and Engineering The Ohio State University Review
Case Studies on High Throughput Gene Expression Data Kun Huang, PhD Raghu Machiraju, PhD Department of Biomedical Informatics Department of Computer Science and Engineering The Ohio State University Review
Nature Genetics: doi: /ng Supplementary Figure 1
 Supplementary Figure 1 Expression deviation of the genes mapped to gene-wise recurrent mutations in the TCGA breast cancer cohort (top) and the TCGA lung cancer cohort (bottom). For each gene (each pair
Supplementary Figure 1 Expression deviation of the genes mapped to gene-wise recurrent mutations in the TCGA breast cancer cohort (top) and the TCGA lung cancer cohort (bottom). For each gene (each pair
Clasificación Molecular del Cáncer de Próstata. JM Piulats
 Clasificación Molecular del Cáncer de Próstata JM Piulats Introduction The Gleason score is the major method for prostate cancer tissue grading and the most important prognostic factor in this disease.
Clasificación Molecular del Cáncer de Próstata JM Piulats Introduction The Gleason score is the major method for prostate cancer tissue grading and the most important prognostic factor in this disease.
Variant Classification. Author: Mike Thiesen, Golden Helix, Inc.
 Variant Classification Author: Mike Thiesen, Golden Helix, Inc. Overview Sequencing pipelines are able to identify rare variants not found in catalogs such as dbsnp. As a result, variants in these datasets
Variant Classification Author: Mike Thiesen, Golden Helix, Inc. Overview Sequencing pipelines are able to identify rare variants not found in catalogs such as dbsnp. As a result, variants in these datasets
Global variation in copy number in the human genome
 Global variation in copy number in the human genome Redon et. al. Nature 444:444-454 (2006) 12.03.2007 Tarmo Puurand Study 270 individuals (HapMap collection) Affymetrix 500K Whole Genome TilePath (WGTP)
Global variation in copy number in the human genome Redon et. al. Nature 444:444-454 (2006) 12.03.2007 Tarmo Puurand Study 270 individuals (HapMap collection) Affymetrix 500K Whole Genome TilePath (WGTP)
Laboratory Service Report
 Client C7028846-DLP Rochester Rochester, N 55901 Specimen Type Peripheral blood CR PDF Report available at: https://test.mmlaccess.com/reports/c7028846-zwselwql7p.ashx Indication for Test DS CR Pathogenic
Client C7028846-DLP Rochester Rochester, N 55901 Specimen Type Peripheral blood CR PDF Report available at: https://test.mmlaccess.com/reports/c7028846-zwselwql7p.ashx Indication for Test DS CR Pathogenic
Molecular Genetic Testing for the Diagnosis of Haematological Malignancies
 Molecular Genetic Testing for the Diagnosis of Haematological Malignancies Dr Anthony Bench Haemto-Oncology Diagnostic Service Cambrıdge Unıversıty Hospitals NHS Foundatıon Trust Cambridge UK Molecular
Molecular Genetic Testing for the Diagnosis of Haematological Malignancies Dr Anthony Bench Haemto-Oncology Diagnostic Service Cambrıdge Unıversıty Hospitals NHS Foundatıon Trust Cambridge UK Molecular
Supplementary Information. Supplementary Figures
 Supplementary Information Supplementary Figures.8 57 essential gene density 2 1.5 LTR insert frequency diversity DEL.5 DUP.5 INV.5 TRA 1 2 3 4 5 1 2 3 4 1 2 Supplementary Figure 1. Locations and minor
Supplementary Information Supplementary Figures.8 57 essential gene density 2 1.5 LTR insert frequency diversity DEL.5 DUP.5 INV.5 TRA 1 2 3 4 5 1 2 3 4 1 2 Supplementary Figure 1. Locations and minor
Supplementary Figure 1: Comparison of acgh-based and expression-based CNA analysis of tumors from breast cancer GEMMs.
 Supplementary Figure 1: Comparison of acgh-based and expression-based CNA analysis of tumors from breast cancer GEMMs. (a) CNA analysis of expression microarray data obtained from 15 tumors in the SV40Tag
Supplementary Figure 1: Comparison of acgh-based and expression-based CNA analysis of tumors from breast cancer GEMMs. (a) CNA analysis of expression microarray data obtained from 15 tumors in the SV40Tag
Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells.
 SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
The 16th KJC Bioinformatics Symposium Integrative analysis identifies potential DNA methylation biomarkers for pan-cancer diagnosis and prognosis
 The 16th KJC Bioinformatics Symposium Integrative analysis identifies potential DNA methylation biomarkers for pan-cancer diagnosis and prognosis Tieliu Shi tlshi@bio.ecnu.edu.cn The Center for bioinformatics
The 16th KJC Bioinformatics Symposium Integrative analysis identifies potential DNA methylation biomarkers for pan-cancer diagnosis and prognosis Tieliu Shi tlshi@bio.ecnu.edu.cn The Center for bioinformatics
The Cancer Genome Atlas & International Cancer Genome Consortium
 The Cancer Genome Atlas & International Cancer Genome Consortium Session 3 Dr Jason Wong Prince of Wales Clinical School Introductory bioinformatics for human genomics workshop, UNSW 31 st July 2014 1
The Cancer Genome Atlas & International Cancer Genome Consortium Session 3 Dr Jason Wong Prince of Wales Clinical School Introductory bioinformatics for human genomics workshop, UNSW 31 st July 2014 1
Acute Myeloid Leukemia Progress at last
 Acute Myeloid Leukemia Progress at last Bruno C. Medeiros, MD September 9, 217 Introduction Mechanisms of leukemogenesis Emerging therapies in AML Previously untreated AML Relapsed and refractory patients
Acute Myeloid Leukemia Progress at last Bruno C. Medeiros, MD September 9, 217 Introduction Mechanisms of leukemogenesis Emerging therapies in AML Previously untreated AML Relapsed and refractory patients
Supplementary Figure 1. Copy Number Alterations TP53 Mutation Type. C-class TP53 WT. TP53 mut. Nature Genetics: doi: /ng.
 Supplementary Figure a Copy Number Alterations in M-class b TP53 Mutation Type Recurrent Copy Number Alterations 8 6 4 2 TP53 WT TP53 mut TP53-mutated samples (%) 7 6 5 4 3 2 Missense Truncating M-class
Supplementary Figure a Copy Number Alterations in M-class b TP53 Mutation Type Recurrent Copy Number Alterations 8 6 4 2 TP53 WT TP53 mut TP53-mutated samples (%) 7 6 5 4 3 2 Missense Truncating M-class
ncounter Assay Automated Process Immobilize and align reporter for image collecting and barcode counting ncounter Prep Station
 ncounter Assay ncounter Prep Station Automated Process Hybridize Reporter to RNA Remove excess reporters Bind reporter to surface Immobilize and align reporter Image surface Count codes Immobilize and
ncounter Assay ncounter Prep Station Automated Process Hybridize Reporter to RNA Remove excess reporters Bind reporter to surface Immobilize and align reporter Image surface Count codes Immobilize and
August 17, Dear Valued Client:
 August 7, 08 Re: CMS Announces 6-Month Period of Enforcement Discretion for Laboratory Date of Service Exception Policy Under the Medicare Clinical Laboratory Fee Schedule (the 4 Day Rule ) Dear Valued
August 7, 08 Re: CMS Announces 6-Month Period of Enforcement Discretion for Laboratory Date of Service Exception Policy Under the Medicare Clinical Laboratory Fee Schedule (the 4 Day Rule ) Dear Valued
Expanded View Figures
 Solip Park & Ben Lehner Epistasis is cancer type specific Molecular Systems Biology Expanded View Figures A B G C D E F H Figure EV1. Epistatic interactions detected in a pan-cancer analysis and saturation
Solip Park & Ben Lehner Epistasis is cancer type specific Molecular Systems Biology Expanded View Figures A B G C D E F H Figure EV1. Epistatic interactions detected in a pan-cancer analysis and saturation
Nature Genetics: doi: /ng Supplementary Figure 1. PCA for ancestry in SNV data.
 Supplementary Figure 1 PCA for ancestry in SNV data. (a) EIGENSTRAT principal-component analysis (PCA) of SNV genotype data on all samples. (b) PCA of only proband SNV genotype data. (c) PCA of SNV genotype
Supplementary Figure 1 PCA for ancestry in SNV data. (a) EIGENSTRAT principal-component analysis (PCA) of SNV genotype data on all samples. (b) PCA of only proband SNV genotype data. (c) PCA of SNV genotype
Nature Immunology: doi: /ni Supplementary Figure 1. Characteristics of SEs in T reg and T conv cells.
 Supplementary Figure 1 Characteristics of SEs in T reg and T conv cells. (a) Patterns of indicated transcription factor-binding at SEs and surrounding regions in T reg and T conv cells. Average normalized
Supplementary Figure 1 Characteristics of SEs in T reg and T conv cells. (a) Patterns of indicated transcription factor-binding at SEs and surrounding regions in T reg and T conv cells. Average normalized
TCF3 breakpoints of TCF3-PBX1 (patients 1a 5a) and TCF3-HLF (patients 6a 9a and11a) translocations.
 Supplementary Figure 1 TCF3 breakpoints of TCF3-PBX1 (patients 1a 5a) and TCF3-HLF (patients 6a 9a and11a) translocations. The CpG motifs closest to the breakpoints are highlighted in red boxes and the
Supplementary Figure 1 TCF3 breakpoints of TCF3-PBX1 (patients 1a 5a) and TCF3-HLF (patients 6a 9a and11a) translocations. The CpG motifs closest to the breakpoints are highlighted in red boxes and the
TEST MENU TEST CPT CODES TAT. Chromosome Analysis Bone Marrow x 2, 88264, x 3, Days
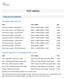 TEST MENU CANCER/LEUKEMIA CHROMOSOME ANALYSIS Chromosome Analysis Bone Marrow 88237 x 2, 88264, 88280 x 3, 88291 4 Days Chromosome Analysis Bone Marrow Core 88237 x 2, 88264, 88280 x 3, 88291 4 Days Chromosome
TEST MENU CANCER/LEUKEMIA CHROMOSOME ANALYSIS Chromosome Analysis Bone Marrow 88237 x 2, 88264, 88280 x 3, 88291 4 Days Chromosome Analysis Bone Marrow Core 88237 x 2, 88264, 88280 x 3, 88291 4 Days Chromosome
Supplemental Figure legends
 Supplemental Figure legends Supplemental Figure S1 Frequently mutated genes. Frequently mutated genes (mutated in at least four patients) with information about mutation frequency, RNA-expression and copy-number.
Supplemental Figure legends Supplemental Figure S1 Frequently mutated genes. Frequently mutated genes (mutated in at least four patients) with information about mutation frequency, RNA-expression and copy-number.
Predicting clinical outcomes in neuroblastoma with genomic data integration
 Predicting clinical outcomes in neuroblastoma with genomic data integration Ilyes Baali, 1 Alp Emre Acar 1, Tunde Aderinwale 2, Saber HafezQorani 3, Hilal Kazan 4 1 Department of Electric-Electronics Engineering,
Predicting clinical outcomes in neuroblastoma with genomic data integration Ilyes Baali, 1 Alp Emre Acar 1, Tunde Aderinwale 2, Saber HafezQorani 3, Hilal Kazan 4 1 Department of Electric-Electronics Engineering,
p.r623c p.p976l p.d2847fs p.t2671 p.d2847fs p.r2922w p.r2370h p.c1201y p.a868v p.s952* RING_C BP PHD Cbp HAT_KAT11
 ARID2 p.r623c KMT2D p.v650fs p.p976l p.r2922w p.l1212r p.d1400h DNA binding RFX DNA binding Zinc finger KMT2C p.a51s p.d372v p.c1103* p.d2847fs p.t2671 p.d2847fs p.r4586h PHD/ RING DHHC/ PHD PHD FYR N
ARID2 p.r623c KMT2D p.v650fs p.p976l p.r2922w p.l1212r p.d1400h DNA binding RFX DNA binding Zinc finger KMT2C p.a51s p.d372v p.c1103* p.d2847fs p.t2671 p.d2847fs p.r4586h PHD/ RING DHHC/ PHD PHD FYR N
a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation,
 Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
Molecular. Oncology & Pathology. Diagnostic, Prognostic, Therapeutic, and Predisposition Tests in Precision Medicine. Liquid Biopsy.
 Molecular Oncology & Pathology Hereditary Cancer Somatic Cancer Liquid Biopsy Next-Gen Sequencing qpcr Sanger Sequencing Diagnostic, Prognostic, Therapeutic, and Predisposition Tests in Precision Medicine
Molecular Oncology & Pathology Hereditary Cancer Somatic Cancer Liquid Biopsy Next-Gen Sequencing qpcr Sanger Sequencing Diagnostic, Prognostic, Therapeutic, and Predisposition Tests in Precision Medicine
AVENIO family of NGS oncology assays ctdna and Tumor Tissue Analysis Kits
 AVENIO family of NGS oncology assays ctdna and Tumor Tissue Analysis Kits Accelerating clinical research Next-generation sequencing (NGS) has the ability to interrogate many different genes and detect
AVENIO family of NGS oncology assays ctdna and Tumor Tissue Analysis Kits Accelerating clinical research Next-generation sequencing (NGS) has the ability to interrogate many different genes and detect
Future Targets for Acute Myeloid Leukemia
 Future Targets for Acute Myeloid Leukemia E. Anders Kolb, M.D. Director, Nemours Center for Cancer and Blood Disorders Chair, Children s Oncology Group Myeloid Disease Committee Future Targets for Acute
Future Targets for Acute Myeloid Leukemia E. Anders Kolb, M.D. Director, Nemours Center for Cancer and Blood Disorders Chair, Children s Oncology Group Myeloid Disease Committee Future Targets for Acute
