A Blueprint for Drug/Diagnostic Development: Facilitating Development and Use of Curated Genetic Databases
|
|
|
- Cornelius Gordon
- 5 years ago
- Views:
Transcription
1 A Blueprint for Drug/Diagnostic Development: Facilitating Development and Use of Curated Genetic Databases TABLE OF CONTENTS Forum Goals Summary Terminology and Considerations Proposals o Proposal #1: Define the minimum core data elements required for the interpretation of clinical significance of variants o Proposal #2: Establish a framework to evaluate the strength of evidence for genotype/phenotype associations o Proposal #3: Determine the context of use of a database that encompasses information outlined in proposal #1 and supported with the levels of evidence captured in proposal #2 o Proposal #4: Incentivize data sharing into publicly available research- and regulatorygrade databases 1
2 Forum Goals Expanding on the 2014 conference that explored regulatory considerations and strategies for FDA approval of an NGS platform, the objectives for this forum are to: Define strategies for developing and using curated genetic databases to establish clinical relevance for various intended uses Develop a method for evaluating the quality of such databases for different stakeholders, including (for example) researchers, regulators and payers Standardize the interpretation and reporting of genetic alterations to databases in order to facilitate interoperability Summary High-throughput genomic technologies, including next generation sequencing (NGS), allow for rapid assessment of many analytes, enabling diagnosis, and/or prognosis, of disease and helping to predict a patient s risk for developing certain conditions or response to therapies. There are many advantages to high throughput sequencing tests over those that detect a single analyte; however, with multiple tests and platforms under development, demonstrating adequate analytical and clinical performance for a set of variants with differing intended uses is challenging. There is a particular tension between the need to provide reasonable assurance of the analytical and clinical validity of variants and practical limitations for submitting such data for every possible variant the test could identify. Thus, consensus, evidence-based guidelines are needed at multiple levels, including: sample collection, preparation, and analysis; clinical reporting (including variant calling, interpretation, and report content), and data storage and protection. With the increased use of genomic profiling in cancer therapy and the development of more powerful technologies, there is great interest in developing new regulatory pathways to facilitate the rapid approval of therapeutics and the accompanying diagnostics that enable precision medicine. Despite recent successes of drug-diagnostic combinations 1, there exists an opportunity to address some of the possible inefficiencies in regulatory review and coverage and reimbursement of these diagnostics. The FDA, aligned with the Precision Medicine Initiative, proposed 2 minimizing the burden of the traditional regulatory requirement for submitting clinical data by each sponsor through permitting the use of existing databases to support the associations between genetic variants and clinical outcomes (i.e., clinical validity ). A precedent for such an approach was the 2013 FDA-clearance of the Illumina MiSeqDx system and accompanying assays (Cystic Fibrosis 139 Variant and Cystic Fibrosis Sequencing Assays) for diagnosing cystic fibrosis, which relied on a curated database of mutations associated with symptoms of cystic fibrosis (the Clinical and Functional TRanslation of CFTR database). Other efforts are underway to determine whether existing databases, such as ClinGen and ClinVar, could be leveraged to support the clinical significance of variants detected by NGS tests. A number of these databases are publicly accessible, and continually updated, with many additional resources available or in development. Identifying strategies to improve and connect existing private or public systems should facilitate the creation and sharing of critical knowledge and limit potential isolation of current genomic data collection efforts. A working group, comprised of representatives from the academic, corporate, and government sectors with expertise in pharmaceutical and diagnostic development and regulatory affairs, explored strategies for using curated databases containing up-to-date clinical evidence and correlates of genomic alterations to support clinical validity of sequencing-based diagnostics. The working group focused on the proposals outlined below to 1 DR Parkinson et al. Evidence of Clinical Utility: An Unmet Need in Molecular Diagnostics for Patients with Cancer Clin Cancer Res; 20(6) 2 Optimizing FDA s Regulatory Oversight of Next Generation Sequencing Diagnostic Tests Preliminary Discussion Paper
3 address the challenges associated with the integration of evidence from databases into NGS-based companion diagnostic (CDx) and complementary diagnostic development. Successful implementation of such an effort could facilitate making diagnostic tests available to patients more rapidly, thus increasing patient access to needed therapeutics by, o Defining the clinical relevance of rare variants and building on existing knowledgebases o Promoting sharing of data, standard mechanisms for querying shared data, and development of consistent, transparent data sources to support clinical validity o o Introducing efficiencies into biomarker development and validation Improving the design of novel biomarker-driven clinical trials and clinical decision-making using existing biomarkers While the main focus of this document is addressing the regulatory challenges, the strategies that evolve may also be applicable for addressing coverage and reimbursement needs for payers. It will be important to consider the implications of these proposals on the payer community. Terminology and Considerations: With the multiple terminologies in use for genetic alterations (also markers, or variants), we will use the terms interchangeably in this document, preferring the term most commonly applied in a given context. Additionally, the following definitions will be used in this document: Actionable variant (or group of variants): Variants with supporting data that allow for a benefit-risk assessment of treatment choice, link patients to an FDA approved drug (on- or off-label) or an investigational drug enrolled in a registrational trial, or that are prognostic or predictive of outcome. Some markers may be specific (e.g., BRAF V600E), while others may represent a functional group (e.g., alterations in Exon 19 of the EGFR gene or loss-of-function mutations in a tumor suppressor gene such as TP53) Regulatory-grade database: Refers to a database that contains genetic information and related data (i.e., clinical) about a disease condition and is recognized by the FDA, or other health authorities, as able to provide evidence of clinical relevance of the variants detected for the intended use of such a test. Such a database would be reflected in the Intended Use statement and/or in the product label of a test Research-grade database: Refers to a database that contains genetic and clinical information about a disease condition from one or more research studies that may be supportive of clinical validity, but not recognized by the FDA as sufficient for regulatory approval For the purposes of this paper, both targeted sequencing and whole genome or whole exome sequencing may be considered; while technical differences need to be addressed between the various sequencing technologies (such as the need for confirmation with orthogonal methods), the link between variant and phenotype should be independent of the tool used to identify the variant. Thus, this document aims to not be limited in scope by the current technologies used in clinical practice and also to be forward looking to those technologies in development. While the effort to develop potential regulatory-grade databases could facilitate diagnostic development, there are significant challenges to overcome. First, the literature is not comprehensively or consistently curated. Second, even entries within the same database may not be equally valid (e.g., the clinical validity of variants may differ based on intended use). Additionally, the level of evidence supporting a link between a genetic variant and a clinical phenotype will fall into a continuum and investigational or preliminary findings may not be well described. To begin addressing some of the challenges, the working group makes the following proposals: 3
4 Proposals Pre-Meeting DRAFT Proposal 1: Define the minimum core data elements required for the interpretation of clinical significance of variants Broad utility of genetic data within the clinical and research settings requires the establishment of standards of how information is collected and processed, capturing details on the functional consequences of variants and relevant clinical details. Such a set of standards should begin by considering the following aspects for inclusion: 1. Functional classification of variants: a. Hereditary cancer risk (germ-line) variants vs. somatic variants arising in tumors. Somatic variants are typically targets of cancer therapy, although some germ-line variants are likewise targetable (e.g., PARP inhibitors for BRCA1/2). i. Hereditary cancer risk variants are classified similarly to risk variants for other diseases: 1. Benign, 2. Likely benign, 3. Unknown, 4. Likely pathogenic, and 5. Pathogenic. Given the focus of this paper on variants relevant to the care of patients with established cancer and extensive ongoing efforts in hereditary disease risk field, cancer risk assessment is not further addressed here 3. ii. Somatic variants may be classified as cancer driver or passenger mutations or variants whose function is unknown. Somatic driver variants and germ-line cancer risk variants relevant to cancer care (e.g. BRCA1/2) should be prioritized for database inclusion and are the focus of these proposals iii. Driver mutation clinical implications may be impacted by additional factors which should be captured including: 1. Clonal vs. sub-clonal status 2. Level of copy number amplification 2. Grouping of variants into classes ( alteration groups ) that may be interpreted equivalently on the basis of either clinical or pre-clinical data (e.g., all EGFR exon 19 deletions) 3. Association of variant groups with clinically relevant phenotypes and the indication for which this association would be pertinent, including a. Diagnosis i. Tumor type (i.e. lung, multiple) ii. Stage of cancer iii. Prior therapies, etc. b. Prognosis c. Therapy response prediction, including detail on evidence type (e.g., trial in biomarker selected population vs. all-comers) and type of response data i. Responsive (e.g., RECIST, PFS, OS) ii. Resistant d. Adverse events, safety signals, and patient reported outcomes e. Demographic group i. Age ii. Gender iii. Ethnicity 4. Levels of supporting evidence for the association (see proposal 2) 5. Data and technology source to ensure the validity and comparability of the data, including: 3 Richards S. et al., Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in Medicine 17,
5 a. Sequencing technology, sample type (blood, tissue, etc.), and key validation including analytical performance parameters (i.e., analytical performance characteristics with consideration of small or large deletions, insertions, rearrangements, and genomic context)that indicate accuracy, to ensure adequate coverage b. Establishment of assay/platform-specific standards to ensure adequate analytical performance for various types of variants, such as small or large deletions, insertions, rearrangements, and genomic context Ensuring the above parameters are captured when a variant is submitted should introduce transparency and consistency to the database. However, while some variants fall into clear categories of driver or passenger mutations, others will have conflicting classifications among researchers, clinicians and laboratories. This may be due in part to the specific disease setting, differences in bioinformatic analyses, and interpretation of the literature. Thus, it may be important to create a mechanism to adjudicate those variants, particularly those that may have clinical implications on regulatory and reimbursement decisions and ultimately treatment or prognosis. One mechanism is to establish a panel of subject matter experts who would arbitrate inconsistent results and finalize the variant assignment based on the most current evidence. Such a panel would need to be structured, transparent in their methods, and composed of a variety of community experts both broad and deep in scope. Such efforts are already underway for germ-line variant classification, where expert working groups have been organized to apply a set of standards developed in the context of particular diseases. These panels would develop a master classification for an individual variant or group of variants, would facilitate review of the pertinent literature, and ensure the classification system is regularly updated. The standards would then need to be applied broadly among the databases used to support clinical performance. While this system would help minimize variability and provide verifiable content within a database, independent review of the raw data would still be needed. In fact, studies to evaluate the concordance among publicly available databases note significant levels of discrepancies in the classification of germ-line variants 4,5. Further, with the standardization of content, it may be possible to query and generate automatic reports to disclose the level of inconsistency within a database (i.e., dbgap), thus providing transparency to the degree of inconsistency within a database and among databases. Proposal 1: Questions for discussion: Should additional data elements be considered? Are any proposed data elements not relevant and should be de-prioritized? Should raw data should be deposited into the database? o If so, through what mechanism? Is inconsistency, as long as it is transparent, acceptable in variant classification? To what degree should the community strive to resolve it? Can independent expert panel review help resolve database inconsistencies? If so, how would it be structured and governed? What other mechanisms can be put in place? Is it appropriate to include patient reported outcomes or other data in databases of these types? Could data be submitted from international sources? Does this pose any barriers? Proposal 2: Establish a framework to evaluate the strength of evidence for genotype/phenotype associations 4 Vail PJ, et al Comparison of locus-specific databases for BRCA1 and BRCA2 variants reveals disparity in variant classification within and among databases. J Community Genet. 5 Thompson BA et al Application of a 5-tiered scheme for standardized classification of 2,300 unique mismatch repair gene variants in the insight locus-specific database. Nat Genet 46:
6 Assessing levels of evidence for particular genotype/phenotype association will be essential to the appropriate application of any proposed database. The following table 6 describes a proposed grading system for the level of evidence supporting an association, ranging from the weakest association based upon pre-clinical evidence, at level 3B, increasing in strength to FDA approval (i.e. inclusion on drug label). Classification for a given variant is not intended to be static and will change over time as new evidence becomes available. Thus, to ensure robust classification of the variants, continued evaluation will be needed. This could be accomplished by ensuring that along with every new submission into a database, information is captured regarding: 1) the variant detected and phenotype reported according to the framework presented in proposal 1; 2) the level of concordance with the current classification; 3) descriptions of any differences or changes; and 4) the scientific rationale for these changes. Additionally, while intra- and inter-database discordances cannot be eliminated, the system could also facilitate the ranking of aggregate evidence with conflicting results. Level Decreasing strength of evidence 1A 1B 2A 2B 3A 3B Supporting Evidence Drug is FDA-approved for the patient s tumor type and indication and outcomes are associated with a specific biomarker (i.e., on-label) An adequately powered, prospective study with biomarker selection/stratification or multi-study meta-analysis is available, demonstrating that a biomarker predicts tumor response (or resistance) to a drug or that the drug is clinically more (or less) effective in a biomarker-selected cohort in the patient s tumor type compared to an unselected cohort Robust study demonstrating a biomarker is associated with tumor response or resistance to the drug in the patient s tumor type that may not meet statistical criteria for 1B. Single or few unusual responder(s), or case studies, show a biomarker is associated with response or resistance to drug in the patient s tumor type, supported by scientific rationale Clinical data is available that demonstrates that the biomarker predicts tumor response to drug in a different tumor type Preclinical data (in vitro or in vivo models or functional genomics) demonstrates that a biomarker predicts cell-based response to drug treatment *Adapted from Meric-Bernstam et al., 2015 Example 1: EGFR mutation in metastatic non-small cell lung cancer (mnsclc) a. An illustrative example includes kinase domain mutations in the EGFR gene in mnsclc. The EGFR tyrosine kinase inhibitors (TKIs) afatinib, erlotinib, and gefitinib are FDA-approved for the treatment of patients with mnsclc whose tumors harbor exon 19 deletion or the exon 21 L858R substitution mutation based on randomized controlled trials (RCTs) demonstrating improved progression-free survival (PFS), objective response rate (ORR) and favorable benefit-risk in this patient population compared to standard chemotherapy. Detection of exon 19 deletion or exon 21 L858R in mnsclc could be considered level 1A evidence supporting biomarker patient selection for use of a drug b. Other, less common EGFR kinase domain mutations are thought to be activating and confer sensitivity to EGFR TKIs such as afatinib and erlotinib. For example, exon 21 substitution mutation L861Q and exon 18 substitution mutation G719X are thought to be activating in vitro and are thought to confer sensitivity (though less so than L858R and exon 19 deletion) to EGFR TKIs. Patients with mnsclc whose tumors harbor L861Q and G719X may be administered EGFR TKI in the community. While strictly speaking this may be considered as off-label use, pre-clinical and case report data suggest that 6 Meric-Bernstam F. et al., A Decision Support Framework for Genomically Informed Investigational Cancer Therapy JNCI J Natl Cancer Inst 107(7) 6
7 patients with these mutations may be responsive to EGFR TKI, though a prospective RCT is not feasible. Detection of EGFR L861Q or G719X in mnsclc could be considered level 2B evidence supporting the biomarker for patient selection for use of an EGFR TKI c. In addition, there are activating mutations that are thought to confer resistance to EGFR TKI, such as exon 20 insertion mutations, or the T790M resistance mutation. Pre-clinical and clinical data suggest that first and second generation EGFR TKIs may not be active in patients whose tumors harbor exon 20 insertion or T790M. In the afatinib product label, the forest plots of a subgroup of patients with uncommon mutations (predominantly resistance mutations such as T790M and exon 20 insertions) show that PFS and OS tend to favor chemotherapy over afatinib. Detection of T790M and insertions in exon 20 in mnsclc could be considered level 2B evidence arguing against the use of erlotinib and afatinib in these patients Example 2: ROS1 rearrangement in mnsclc a. Crizotinib is approved for ALK-rearranged mnsclc (~5% incidence) based on improved PFS, ORR, and favorable risk-benefit compared to conventional chemotherapy in RCTs. A recent robust single arm trial in patients with ROS1-rearranged NSCLC (~1% incidence) published in NEJM (Shaw et. al 7 ) indicated that crizotinib in ROS1 fusion-positive mnsclc has a large and durable ORR in this molecular subgroup, and current NCCN guidelines recommend crizotinib for ROS1 fusion-positive mnsclc. Detection of ROS1 rearrangement in mnsclc could be considered level 2A supporting use of crizotinib for these patients Example 3: BRAF mutation a. Dabrafenib and vemurafenib are approved for V600E metastatic melanoma (MM), and dabrafenib in combination with trametinib is approved for V600E and V600K MM. The basis for these approvals included improvements in ORR, PFS, and in certain circumstances overall survival (OS) as compared to conventional chemotherapy in RCTs. Detection of BRAF V600E or K in MM could be considered level 1A evidence supporting these biomarkers for patient selection for use of a drug or combination of drugs b. At ASCO 2015, Planchard et al 8 reported high and potentially durable ORR with dabrafenib in combination with trametinib in patients with V600E positive mnsclc based on a single arm trial. Detection of BRAF V600E in mnsclc could be considered level 2A supporting use of dabrafenib with trametinib for these patients c. Data from single arm trials along with anecdotal reports suggest that patients with V600E positive metastatic colorectal cancer (mcrc) have a low response to vemurafenib. Detection of BRAF V600E in mcrc could be considered level 2A evidence not to use vemurafenib in this setting Example 4: RAS in mcrc a. Prior to treatment of metastatic colorectal cancer patients with the anti-egfr monoclonal antibodies cetuximab and panitumumab it must be determined if the patient s tumor harbors somatic mutations in exon 2 (codons 12 and 13), exon 3 (codons 59 and 61) and exon 4 (codons 117 and 146) of either KRAS or NRAS (RAS) since mutations in RAS have been shown to be predictive of resistance. A recent metaanalysis describes results from retrospective subset analyses of RAS-mutant or wild-type populations across ninercts conducted to investigate the role of RAS mutations on the clinical effects of anti-egfrdirected monoclonal antibodies [Sorich reference]. Treatment with cetuximab or panitumumab was shown to provide prolonged survival over best supportive care and in combination with different chemotherapies over chemotherapies alone for mcrc patients having tumors that are RAS wild type. 7 Shaw AT, et al Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med. 371(21): Planchard D., et al. Interim results of a phase II study of the BRAF inhibitor (BRAFi) dabrafenib (D) in combination with the MEK inhibitor trametinib (T) in patients (pts) with BRAF V600E mutated (mut) metastatic non-small cell lung cancer (NSCLC). Presented at ASCO J Clin Oncol 33, 2015 (suppl; abstr 8006). 7
8 Average overall survival and progression free survival were significantly improved for the different anti- EGFR drugs regardless of line of therapy or therapy combination with hazard ratios of 0.87 (95% CI ) and 0.62 (95% CI ) respectively. Detection of RAS mutation in exon 2 (codons 12 and 13), exon 3 (codons 59 and 61) and exon 4 (codons 117 and 146) could be considered level 1B evidence arguing against use of cetuximab or panitumumab for these patients and level 1A if clinical utility is validated for a RAS companion diagnostic in conjunction with a RCT for one of these anti-egfr agents. Ongoing evidence assessment according to the framework proposed above will be required to ensure that the best available data and evidence are utilized at any given time to determine which variants are most relevant to patient care. Proposal #2: Questions for discussion: Are the levels of supporting evidence suggested appropriate? Are they consistent with the work that others in the field are undertaking? How should database consistency be captured and presented? Can retrospective studies be sufficient? Or are prospective confirmatory trials always required? What level of data should be reported to physicians and patients? What other level-of-evidence information should be retained and accessible in the database? o Data may become outdated with time, or determined to be unreliable. Is there any way to minimize the ability of this data to skew information? Should this type of categorization include prognostic as well as predictive markers? Proposal 3: Determine the context of use of a database that encompasses information outlined in proposal #1 and supported with the levels of evidence captured in proposal #2 After the challenges associated with database creation are addressed, it is possible to specify how such data may be used. To begin addressing utility, the working group considered several applications, including 1) Research-grade (i.e., for use in scientific research and publications), 2) Regulatory-grade (i.e., for use in regulatory approvals), and 3) Reimbursement-grade (i.e., for use in the clinical setting). Provided database characteristics outlined above, i.e. capturing the core data elements as well as the strength of evidence for variant/phenotype association, the working group proposed an approach, summarized in the figure below and adapted from the current clinical trial paradigm for developing companion diagnostics (Appendix 1), for determining the application of associations in the database. Briefly, associations with level 3 evidence, reflecting pre-clinical or clinical data from other tumor types, would be appropriate for further investigation and early clinical trials to determine more clearly the appropriateness of matching variant with therapy. There may be a possibility that associations with stronger evidence, level 2, could be used to support clinical validity, for example as part of a PMA review process for a CDx drug-diagnostic pair. Associations with level 1 evidence may possibly not require additional FDA review, only analytical validation of variant detection. Labeling for such a test would need to reflect the database used, the scientific rationale supporting the association, whether there was any discordance between the diagnostic test and database for the specific variant or grouping of variants and rationale for providing the result of such variant, etc. Thus, elaborating on selected case studies above: Example 3b: A diagnostic test that detects the BRAF V600E mutation in mnsclc (level 2A), for use of the dabrafenib/trametinib combination therapy, may use a database capturing the therapeutic association between BRAF V600E and durable ORR in mnsclc patients treated with dabrafenib in combination with trametinib to support clinical significance for FDA test approval. 8
9 Example 4a: Alternatively, a diagnostic detecting RAS-mutation in exon 2 (codons 12 and 13), exon 3 (codons 59 and 61) and exon 4 (codons 117 and 146), considered level 1B, may only require analytical validation of variant detection to guide against the use of cetuximab or panitumumab for these patients with mcrc tumors. Capture minimum core data elements (proposal 1) Establish the strength of supporting evidence for variant phenotype association: (proposal 2) Level 1 Level 2 Level 3 Sufficient to meet definition of clinical validity for CDx and reimbursement purposes FDA review of clinical validity required to assess adequacy for CDx claims and need/feasibility of additional studies Further investigation via genotype-selected clinical trials Proposal #3: Questions for discussion: Is this level-based framework appropriate to guide database use for regulatory approval? How could this framework translate to other contexts, e.g., payer community? If there are multiple databases that meet the requirements of a FDA-grade database, but contain discordant results, how will these be resolved? How would products that relied on these data be affected? How would the FDA resolve the discrepancy? What is meant by analytical data only? (What is required to establish acceptable analytical performance?) When specific claims are made as to variants identified by a test, are accuracy studies required? How should data from databases be analyzed? E.g., Statistical analysis for hypothesis generation versus validation? Proposal 4: Incentivize data sharing into publicly available research- and regulatory-grade databases A significant fraction of data being generated from genetic testing is not publicly accessible. While there are multiple contributing factors, two key data sharing issues are: 1. clinical laboratories have limited resources to collate and upload their genetic findings into publicly accessible databases and 2. sponsors building internal databases are reluctant to make information publicly available due to competitive considerations Given that accurate interpretation of genomic data is essential to patient health, developing incentives to facilitate all laboratories (small and large, public and private) to share data should ultimately help all players develop and perform more effective diagnostic tests. Following are suggestions to address these issues. Data Sharing Issue #1: Many laboratories, particularly small labs, do not have the resources or the technical expertise required to gather and format data in a manner that is conducive to inclusion in clinical databases. Further, organizations 9
10 that are aggregating data may not have funding to reformat data from laboratories or create sophisticated pipelines that make data sharing easier. In order to address these issues, the working group suggests: Developing proposals for either establishing or upgrading databases to include the costs of creating easy-to-use pipelines to facilitate data transfer, or sufficient funds to allow for manual upload of data from smaller laboratories Creating mechanisms for industry stakeholders to either participate more broadly in the creation of public databases (without establishing ownership) or provide non-restricted funding to aid in their creation Data Sharing Issue #2: Competitive incentives often do not align with data sharing. For example, for FDA-cleared or -approved diagnostics, sponsors may be required to perform studies to establish the clinical validity of the marker the test detects. While this process sets a high standard for ensuring test efficacy and safety, it is also often lengthy and resource intensive for sponsors with limited value for sharing these data as it may lower the bar for market entry by subsequent product developers. For LDTs, the aggregated variant database is often proprietary, providing a critical competitive barrier needed to be successful in a highly competitive marketplace. In order to address these issues, the working group offers the following suggestions for discussion: Provide sponsors that contribute data to public databases access to priority review paths that provide opportunities for intensive guidance on efficient Dx development and accelerated review timelines for FDA-cleared or approved products Similar to FDA approved pharmaceutical products, provide competitive protection (i.e., similar to patenting) that gives an organization that has made the investment to aggregate data a time-bound competitive advantage in the marketplace If data from sponsors are published in peer-reviewed literature the data must also be shared with at least one publicly available database (if available) Finally, the development in recent years of FDA programs intended to facilitate and expedite the development and review of new drugs to address unmet medical need (e.g. fast track designation, breakthrough therapy designation, accelerated approval, and priority review), have greatly accelerated the approval of therapies for the treatment of serious or life-threatening conditions. In some disease settings such as oncology where targeted therapy development is highly dependent on the co-development of a CDx, the accelerated approval provisions have created unrealistic timeline requirements for the development of CDx. In a prior whitepaper published by this working group 9, it was proposed that an expedited review path be created to ensure that both drug and CDx can reach the market within an optimal timeframe. The use of aggregated publicly available databases to support the clinical relevance of variants in the CDx genes may enable cleared or approved products to reach the market expeditiously. Simultaneous inclusion of data submission requirements as a qualifying criteria for expedited review may help to establish a virtuous feedback loop and provide sponsors with the framework needed to share proprietary data sources. Proposal #4: Questions for discussion: Would establishments of database standards alone incentivize data sharing? If so, would focusing on implementation strategies result in improved data sharing? I.e., development of bioinformatics support and data formatting, to facilitate submission and curation Would development of an alternate pathway to submit data for a diagnostic test that requires database submission and community/expert panel review to be a compelling path for regulatory approval? 9 A Blueprint for Drug/Diagnostic Co-Development: Next-Generation Sequencing (NGS) in Oncology
11 Is it possible to begin with promoting sharing into research-grade databases, using the established metrics, such that these could be elevated to regulatory-grade with improved evidence base? What would such a proposal look like? What legal mechanisms exist for expediting regulatory approval of CDx? What would competitive protection for diagnostics look like? Are these incentives sufficient to promote data sharing? What are the drawbacks of these suggestions? 11
12 Appendix 1: Using the current clinical trial paradigm, this figure illustrates the strength of evidence used to support CDx approval. Extrapolating from this process of approval, the working group suggests applying this model to broader utilization of genetic databases. Investigational Drug Investigational Device Clinical trial Variants Detected Drug Response genotype phenotype Variants with sufficient evidence No database needed Variants with insufficient evidence Use database for clinical validity Variants not detected but in database Further evaluation needed drug response Drug and Device Approval 12
NGS IN ONCOLOGY: FDA S PERSPECTIVE
 NGS IN ONCOLOGY: FDA S PERSPECTIVE ASQ Biomed/Biotech SIG Event April 26, 2018 Gaithersburg, MD You Li, Ph.D. Division of Molecular Genetics and Pathology Food and Drug Administration (FDA) Center for
NGS IN ONCOLOGY: FDA S PERSPECTIVE ASQ Biomed/Biotech SIG Event April 26, 2018 Gaithersburg, MD You Li, Ph.D. Division of Molecular Genetics and Pathology Food and Drug Administration (FDA) Center for
NGS ONCOPANELS: FDA S PERSPECTIVE
 NGS ONCOPANELS: FDA S PERSPECTIVE CBA Workshop: Biomarker and Application in Drug Development August 11, 2018 Rockville, MD You Li, Ph.D. Division of Molecular Genetics and Pathology Food and Drug Administration
NGS ONCOPANELS: FDA S PERSPECTIVE CBA Workshop: Biomarker and Application in Drug Development August 11, 2018 Rockville, MD You Li, Ph.D. Division of Molecular Genetics and Pathology Food and Drug Administration
Transform genomic data into real-life results
 CLINICAL SUMMARY Transform genomic data into real-life results Biomarker testing and targeted therapies can drive improved outcomes in clinical practice New FDA-Approved Broad Companion Diagnostic for
CLINICAL SUMMARY Transform genomic data into real-life results Biomarker testing and targeted therapies can drive improved outcomes in clinical practice New FDA-Approved Broad Companion Diagnostic for
33 rd Annual J.P. Morgan Healthcare Conference. January 2015
 33 rd Annual J.P. Morgan Healthcare Conference January 2015 Forward-looking Statements This presentation contains forward-looking statements, which express the current beliefs and expectations of management.
33 rd Annual J.P. Morgan Healthcare Conference January 2015 Forward-looking Statements This presentation contains forward-looking statements, which express the current beliefs and expectations of management.
Corporate Medical Policy
 Corporate Medical Policy Molecular Analysis for Targeted Therapy for Non-Small Cell Lung File Name: Origination: Last CAP Review: Next CAP Review: Last Review: molecular_analysis_for_targeted_therapy_for_non_small_cell_lung_cancer
Corporate Medical Policy Molecular Analysis for Targeted Therapy for Non-Small Cell Lung File Name: Origination: Last CAP Review: Next CAP Review: Last Review: molecular_analysis_for_targeted_therapy_for_non_small_cell_lung_cancer
Precision Genetic Testing in Cancer Treatment and Prognosis
 Precision Genetic Testing in Cancer Treatment and Prognosis Deborah Cragun, PhD, MS, CGC Genetic Counseling Graduate Program Director University of South Florida Case #1 Diana is a 47 year old cancer patient
Precision Genetic Testing in Cancer Treatment and Prognosis Deborah Cragun, PhD, MS, CGC Genetic Counseling Graduate Program Director University of South Florida Case #1 Diana is a 47 year old cancer patient
Regulatory Landscape for Precision Medicine
 Regulatory Landscape for Precision Medicine Adam C. Berger, Ph.D. Office of In Vitro Diagnostics and Radiological Health, FDA FOCR-Alexandria A Blueprint for Breakthrough Meeting September 13, 2017 1 What
Regulatory Landscape for Precision Medicine Adam C. Berger, Ph.D. Office of In Vitro Diagnostics and Radiological Health, FDA FOCR-Alexandria A Blueprint for Breakthrough Meeting September 13, 2017 1 What
NCCN Non-Small Cell Lung Cancer V Meeting June 15, 2018
 Guideline Page and Request Illumina Inc. requesting to replace Testing should be conducted as part of broad molecular profiling with Consider NGS-based assays that include EGFR, ALK, ROS1, and BRAF as
Guideline Page and Request Illumina Inc. requesting to replace Testing should be conducted as part of broad molecular profiling with Consider NGS-based assays that include EGFR, ALK, ROS1, and BRAF as
Big data vs. the individual liver from a regulatory perspective
 Big data vs. the individual liver from a regulatory perspective Robert Schuck, Pharm.D., Ph.D. Genomics and Targeted Therapy Office of Clinical Pharmacology Center for Drug Evaluation and Research Food
Big data vs. the individual liver from a regulatory perspective Robert Schuck, Pharm.D., Ph.D. Genomics and Targeted Therapy Office of Clinical Pharmacology Center for Drug Evaluation and Research Food
GENETIC TESTING FOR TARGETED THERAPY FOR NON-SMALL CELL LUNG CANCER (NSCLC)
 CANCER (NSCLC) Non-Discrimination Statement and Multi-Language Interpreter Services information are located at the end of this document. Coverage for services, procedures, medical devices and drugs are
CANCER (NSCLC) Non-Discrimination Statement and Multi-Language Interpreter Services information are located at the end of this document. Coverage for services, procedures, medical devices and drugs are
Molecular Testing Updates. Karen Rasmussen, PhD, FACMG Clinical Molecular Genetics Spectrum Medical Group, Pathology Division Portland, Maine
 Molecular Testing Updates Karen Rasmussen, PhD, FACMG Clinical Molecular Genetics Spectrum Medical Group, Pathology Division Portland, Maine Keeping Up with Predictive Molecular Testing in Oncology: Technical
Molecular Testing Updates Karen Rasmussen, PhD, FACMG Clinical Molecular Genetics Spectrum Medical Group, Pathology Division Portland, Maine Keeping Up with Predictive Molecular Testing in Oncology: Technical
PLENARY SESSION 1: CLINICAL TRIAL DESIGN IN AN ERA OF HORIZONTAL DRUG DEVELOPMENT Industry Perspective
 PLENARY SESSION 1: CLINICAL TRIAL DESIGN IN AN ERA OF HORIZONTAL DRUG DEVELOPMENT Industry Perspective Davy Chiodin, VP - Regulatory Science, QA and Compliance, Acerta Pharma (A Member of the AstraZeneca
PLENARY SESSION 1: CLINICAL TRIAL DESIGN IN AN ERA OF HORIZONTAL DRUG DEVELOPMENT Industry Perspective Davy Chiodin, VP - Regulatory Science, QA and Compliance, Acerta Pharma (A Member of the AstraZeneca
Patient Leader Education Summit. Precision Medicine: Today and Tomorrow March 31, 2017
 Patient Leader Education Summit Precision Medicine: Today and Tomorrow March 31, 2017 Precision Medicine: Presentation Outline Agenda What is a Precision Medicine What is its clinical value Overview of
Patient Leader Education Summit Precision Medicine: Today and Tomorrow March 31, 2017 Precision Medicine: Presentation Outline Agenda What is a Precision Medicine What is its clinical value Overview of
Molecular Testing in Lung Cancer
 Molecular Testing in Lung Cancer Pimpin Incharoen, M.D. Assistant Professor, Thoracic Pathology Department of Pathology, Ramathibodi Hospital Genetic alterations in lung cancer Source: Khono et al, Trans
Molecular Testing in Lung Cancer Pimpin Incharoen, M.D. Assistant Professor, Thoracic Pathology Department of Pathology, Ramathibodi Hospital Genetic alterations in lung cancer Source: Khono et al, Trans
1. Q: What has changed from the draft recommendations posted for public comment in November/December 2011?
 Frequently Asked Questions (FAQs) in regard to Molecular Testing Guideline for Selection of Lung Cancer Patients for EGFR and ALK Tyrosine Kinase Inhibitors 1. Q: What has changed from the draft recommendations
Frequently Asked Questions (FAQs) in regard to Molecular Testing Guideline for Selection of Lung Cancer Patients for EGFR and ALK Tyrosine Kinase Inhibitors 1. Q: What has changed from the draft recommendations
Delivering Value Through Personalized Medicine: An Industry Perspective
 Delivering Value Through Personalized Medicine: An Industry Perspective Josephine A. Sollano, Dr.PH Head, Global HEOR and Medical Communications Pfizer Oncology, NY, USA josephine.sollano@pfizer.com What
Delivering Value Through Personalized Medicine: An Industry Perspective Josephine A. Sollano, Dr.PH Head, Global HEOR and Medical Communications Pfizer Oncology, NY, USA josephine.sollano@pfizer.com What
Basket Trials: Features, Examples, and Challenges
 : Features, s, and Challenges Lindsay A. Renfro, Ph.D. Associate Professor of Research Division of Biostatistics University of Southern California ASA Biopharm / Regulatory / Industry Statistics Workshop
: Features, s, and Challenges Lindsay A. Renfro, Ph.D. Associate Professor of Research Division of Biostatistics University of Southern California ASA Biopharm / Regulatory / Industry Statistics Workshop
MATCHMAKING IN ONCOLOGY CHALLENGES AND COMBINATION STRATEGY FOR NOVEL TARGETED AGENTS
 MATCHMAKING IN ONCOLOGY CHALLENGES AND COMBINATION STRATEGY FOR NOVEL TARGETED AGENTS DR. RACHEL E. LAING*, OLIVIER LESUEUR*, STAN HOPKINS AND DR. SEAN X. HU MAY 2012 Recent advances in oncology, such
MATCHMAKING IN ONCOLOGY CHALLENGES AND COMBINATION STRATEGY FOR NOVEL TARGETED AGENTS DR. RACHEL E. LAING*, OLIVIER LESUEUR*, STAN HOPKINS AND DR. SEAN X. HU MAY 2012 Recent advances in oncology, such
Personalised Healthcare (PHC) with Foundation Medicine (FMI) Fatma Elçin KINIKLI, FMI Turkey, Science Leader
 Personalised Healthcare (PHC) with Foundation Medicine (FMI) Fatma Elçin KINIKLI, FMI Turkey, Science Leader Agenda PHC Approach Provides Better Patient Outcome FMI offers Comprehensive Genomic Profiling,
Personalised Healthcare (PHC) with Foundation Medicine (FMI) Fatma Elçin KINIKLI, FMI Turkey, Science Leader Agenda PHC Approach Provides Better Patient Outcome FMI offers Comprehensive Genomic Profiling,
Implementation of nation-wide molecular testing in oncology in the French Health care system : quality assurance issues & challenges
 Implementation of nation-wide molecular testing in oncology in the French Health care system : quality assurance issues & challenges Frédérique Nowak - 21 october 2015 "Putting Science into Standards event:
Implementation of nation-wide molecular testing in oncology in the French Health care system : quality assurance issues & challenges Frédérique Nowak - 21 october 2015 "Putting Science into Standards event:
DENOMINATOR: Adult patients with metastatic colorectal cancer who have a RAS (KRAS or NRAS) gene mutation
 Quality ID #452 (NQF 1860): Patients with Metastatic Colorectal Cancer and RAS (KRAS or NRAS) 4 with Anti-epidermal Growth Factor Receptor (EGFR) Monoclonal Antibodies National Quality Strategy Domain:
Quality ID #452 (NQF 1860): Patients with Metastatic Colorectal Cancer and RAS (KRAS or NRAS) 4 with Anti-epidermal Growth Factor Receptor (EGFR) Monoclonal Antibodies National Quality Strategy Domain:
Targeted therapies for advanced non-small cell lung cancer. Tom Stinchcombe Duke Cancer Insitute
 Targeted therapies for advanced non-small cell lung cancer Tom Stinchcombe Duke Cancer Insitute Topics ALK rearranged NSCLC ROS1 rearranged NSCLC EGFR mutation: exon 19/exon 21 L858R and uncommon mutations
Targeted therapies for advanced non-small cell lung cancer Tom Stinchcombe Duke Cancer Insitute Topics ALK rearranged NSCLC ROS1 rearranged NSCLC EGFR mutation: exon 19/exon 21 L858R and uncommon mutations
VeriStrat Poor Patients Show Encouraging Overall Survival and Progression Free Survival Signal; Confirmatory Phase 2 Study Planned by Year-End
 AVEO and Biodesix Announce Exploratory Analysis of VeriStrat-Selected Patients with Non-Small Cell Lung Cancer in Phase 2 Study of Ficlatuzumab Presented at ESMO 2014 Congress VeriStrat Poor Patients Show
AVEO and Biodesix Announce Exploratory Analysis of VeriStrat-Selected Patients with Non-Small Cell Lung Cancer in Phase 2 Study of Ficlatuzumab Presented at ESMO 2014 Congress VeriStrat Poor Patients Show
Data Sharing Consortiums and Large Datasets to Inform Cancer Diagnosis
 February, 13th 2018 Data Sharing Consortiums and Large Datasets to Inform Cancer Diagnosis Improving Cancer Diagnosis and Care: Patient Access to Oncologic Imaging and Pathology Expertise and Technologies:
February, 13th 2018 Data Sharing Consortiums and Large Datasets to Inform Cancer Diagnosis Improving Cancer Diagnosis and Care: Patient Access to Oncologic Imaging and Pathology Expertise and Technologies:
How Personalized Medicine is Changing the Biopharmaceutical Marketplace
 How Personalized Medicine is Changing the Biopharmaceutical Marketplace Marc Chioda, PharmD Associate Medical Director, Pfizer Oncology. Presentation to the Cancer Action Coalition of Virginia January
How Personalized Medicine is Changing the Biopharmaceutical Marketplace Marc Chioda, PharmD Associate Medical Director, Pfizer Oncology. Presentation to the Cancer Action Coalition of Virginia January
Therapeutic Options for Patients with BRAF-mutant Metastatic Colorectal Cancer
 Therapeutic Options for Patients with BRAF-mutant Metastatic Colorectal Cancer Axel Grothey, M.D., Professor of Oncology, Clinical and Translational Science Division of Medical Oncology Mayo Clinic, Rochester,
Therapeutic Options for Patients with BRAF-mutant Metastatic Colorectal Cancer Axel Grothey, M.D., Professor of Oncology, Clinical and Translational Science Division of Medical Oncology Mayo Clinic, Rochester,
MP Circulating Tumor DNA Management of Non-Small-Cell Lung Cancer (Liquid Biopsy)
 Medical Policy BCBSA Ref. Policy: 2.04.143 Last Review: 10/18/2018 Effective Date: 10/18/2018 Section: Medicine Related Policies 2.04.121 Miscellaneous Genetic and Molecular Diagnostic Tests 2.04.45 Molecular
Medical Policy BCBSA Ref. Policy: 2.04.143 Last Review: 10/18/2018 Effective Date: 10/18/2018 Section: Medicine Related Policies 2.04.121 Miscellaneous Genetic and Molecular Diagnostic Tests 2.04.45 Molecular
Molecular Targets in Lung Cancer
 Molecular Targets in Lung Cancer Robert Ramirez, DO, FACP Thoracic and Neuroendocrine Oncology November 18 th, 2016 Disclosures Consulting and speaker fees for Ipsen Pharmaceuticals, AstraZeneca and Merck
Molecular Targets in Lung Cancer Robert Ramirez, DO, FACP Thoracic and Neuroendocrine Oncology November 18 th, 2016 Disclosures Consulting and speaker fees for Ipsen Pharmaceuticals, AstraZeneca and Merck
Circulating Tumor DNA for Management of Non-Small-Cell Lung Cancer (Liquid Biopsy)
 Circulating Tumor DNA for Management of Non-Small-Cell Lung Cancer (Liquid Biopsy) Policy Number: 2.04.143 Last Review: 1/2019 Origination: 1/2018 Next Review: 1/2020 Policy Blue Cross and Blue Shield
Circulating Tumor DNA for Management of Non-Small-Cell Lung Cancer (Liquid Biopsy) Policy Number: 2.04.143 Last Review: 1/2019 Origination: 1/2018 Next Review: 1/2020 Policy Blue Cross and Blue Shield
Spectrum Pharmaceuticals
 Spectrum Pharmaceuticals Joe Turgeon President and CEO June 2018 Investor Presentation 1 Safe Harbor Statement This presentation contains forward looking statements regarding future events and the future
Spectrum Pharmaceuticals Joe Turgeon President and CEO June 2018 Investor Presentation 1 Safe Harbor Statement This presentation contains forward looking statements regarding future events and the future
Giorgio V. Scagliotti Università di Torino Dipartimento di Oncologia
 Giorgio V. Scagliotti Università di Torino Dipartimento di Oncologia giorgio.scagliotti@unito.it Politi K & Herbst R. Clin. Cancer Res. 2015; 21:2213 Breast Colorectal Gastric/GE Junction Tumor Type Head
Giorgio V. Scagliotti Università di Torino Dipartimento di Oncologia giorgio.scagliotti@unito.it Politi K & Herbst R. Clin. Cancer Res. 2015; 21:2213 Breast Colorectal Gastric/GE Junction Tumor Type Head
Comprehensive genomic profiling for various solid tumors
 Content Highlight Test Specification Test Content Performance Validation Test Report I II III IV V The NovoPM TM comprehensive cancer genomic profiling test Comprehensive genomic profiling for various
Content Highlight Test Specification Test Content Performance Validation Test Report I II III IV V The NovoPM TM comprehensive cancer genomic profiling test Comprehensive genomic profiling for various
Corporate Medical Policy
 Corporate Medical Policy Proteomic Testing for Targeted Therapy in Non-Small Cell Lung File Name: Origination: Last CAP Review: Next CAP Review: Last Review: proteomic_testing_for_targeted_therapy_in_non_small_cell_lung_cancer
Corporate Medical Policy Proteomic Testing for Targeted Therapy in Non-Small Cell Lung File Name: Origination: Last CAP Review: Next CAP Review: Last Review: proteomic_testing_for_targeted_therapy_in_non_small_cell_lung_cancer
Lung Cancer Genetics: Common Mutations and How to Treat Them David J. Kwiatkowski, MD, PhD. Mount Carrigain 2/4/17
 Lung Cancer Genetics: Common Mutations and How to Treat Them David J. Kwiatkowski, MD, PhD Mount Carrigain 2/4/17 Histology Adenocarcinoma: Mixed subtype, acinar, papillary, solid, micropapillary, lepidic
Lung Cancer Genetics: Common Mutations and How to Treat Them David J. Kwiatkowski, MD, PhD Mount Carrigain 2/4/17 Histology Adenocarcinoma: Mixed subtype, acinar, papillary, solid, micropapillary, lepidic
CURRENT STANDARD OF CARE OF COLORECTAL CANCER: THE EVOLUTION OF ESMO CLINICAL PRACTICE GUIDELINES
 CURRENT STANDARD OF CARE OF COLORECTAL CANCER: THE EVOLUTION OF ESMO CLINICAL PRACTICE GUIDELINES Fortunato Ciardiello ESMO Past-President 2018-2019 Dipartimento di Medicina di Precisione Università degli
CURRENT STANDARD OF CARE OF COLORECTAL CANCER: THE EVOLUTION OF ESMO CLINICAL PRACTICE GUIDELINES Fortunato Ciardiello ESMO Past-President 2018-2019 Dipartimento di Medicina di Precisione Università degli
Use of Single-Arm Cohorts/Trials to Demonstrate Clinical Benefit for Breakthrough Therapies. Eric H. Rubin, MD Merck Research Laboratories
 Use of Single-Arm Cohorts/Trials to Demonstrate Clinical Benefit for Breakthrough Therapies Eric H. Rubin, MD Merck Research Laboratories Outline Pembrolizumab P001 study - example of multiple expansion
Use of Single-Arm Cohorts/Trials to Demonstrate Clinical Benefit for Breakthrough Therapies Eric H. Rubin, MD Merck Research Laboratories Outline Pembrolizumab P001 study - example of multiple expansion
If multiple KRAS mutation tests have been performed, refer to the most recent test results.
 Measure #452 (NQF 1860): Patients with Metastatic Colorectal Cancer and KRAS Gene Mutation Spared Treatment with Anti-epidermal Growth Factor Receptor (EGFR) Monoclonal Antibodies National Quality Strategy
Measure #452 (NQF 1860): Patients with Metastatic Colorectal Cancer and KRAS Gene Mutation Spared Treatment with Anti-epidermal Growth Factor Receptor (EGFR) Monoclonal Antibodies National Quality Strategy
Health Systems Adoption of Personalized Medicine: Promise and Obstacles. Scott Ramsey Fred Hutchinson Cancer Research Center Seattle, WA
 Health Systems Adoption of Personalized Medicine: Promise and Obstacles Scott Ramsey Fred Hutchinson Cancer Research Center Seattle, WA Cancer Pharmaceutical Pricing Bach P. NEJM 2009;360:626 Opportunities
Health Systems Adoption of Personalized Medicine: Promise and Obstacles Scott Ramsey Fred Hutchinson Cancer Research Center Seattle, WA Cancer Pharmaceutical Pricing Bach P. NEJM 2009;360:626 Opportunities
Clinical Policy: Dabrafenib (Tafinlar) Reference Number: CP.PHAR.239 Effective Date: 07/16 Last Review Date: 07/17 Line of Business: Medicaid
 Clinical Policy: (Tafinlar) Reference Number: CP.PHAR.239 Effective Date: 07/16 Last Review Date: 07/17 Line of Business: Medicaid Coding Implications Revision Log See Important Reminder at the end of
Clinical Policy: (Tafinlar) Reference Number: CP.PHAR.239 Effective Date: 07/16 Last Review Date: 07/17 Line of Business: Medicaid Coding Implications Revision Log See Important Reminder at the end of
Medical Policy An independent licensee of the Blue Cross Blue Shield Association
 Circulating Tumor DNA Management of Non-Small-Cell Lung Cancer (Liquid Biopsy) Page 1 of 36 Medical Policy An independent licensee of the Blue Cross Blue Shield Association Title: Circulating Tumor DNA
Circulating Tumor DNA Management of Non-Small-Cell Lung Cancer (Liquid Biopsy) Page 1 of 36 Medical Policy An independent licensee of the Blue Cross Blue Shield Association Title: Circulating Tumor DNA
Personalized medecine Biomarker
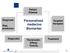 Patient Disease Diagnosis Risk prediction Personalized medecine Biomarker Targeted therapies Diagnostic Theranostic: Efficay Toxicity Treatment 1 THE CASE FOR PERSONALIZED MEDICINE IS COMPELLING.. Toxicity
Patient Disease Diagnosis Risk prediction Personalized medecine Biomarker Targeted therapies Diagnostic Theranostic: Efficay Toxicity Treatment 1 THE CASE FOR PERSONALIZED MEDICINE IS COMPELLING.. Toxicity
Page: 1 of 27. Molecular Analysis for Targeted Therapy of Non-Small-Cell Lung Cancer
 Last Review Status/Date: December 2014 Page: 1 of 27 Non-Small-Cell Lung Cancer Description Over half of patients with non-small-cell lung cancer (NSCLC) present with advanced and therefore incurable disease,
Last Review Status/Date: December 2014 Page: 1 of 27 Non-Small-Cell Lung Cancer Description Over half of patients with non-small-cell lung cancer (NSCLC) present with advanced and therefore incurable disease,
Opportunities and Challenges in the Development of Companion Diagnostics
 Opportunities and Challenges in the Development of Companion Diagnostics E. Patrick Groody, Ph.D. Divisional Vice President Research and Development Abbott Molecular Agenda Value of Personalized Medicine
Opportunities and Challenges in the Development of Companion Diagnostics E. Patrick Groody, Ph.D. Divisional Vice President Research and Development Abbott Molecular Agenda Value of Personalized Medicine
See Important Reminder at the end of this policy for important regulatory and legal information.
 Clinical Policy: (Iressa) Reference Number: CP.PHAR.68 Effective Date: 11.16.16 Last Review Date: 11.18 Line of Business: Commercial, Medicaid Revision Log See Important Reminder at the end of this policy
Clinical Policy: (Iressa) Reference Number: CP.PHAR.68 Effective Date: 11.16.16 Last Review Date: 11.18 Line of Business: Commercial, Medicaid Revision Log See Important Reminder at the end of this policy
American Society of Clinical Oncology All rights reserved.
 Extended RAS Gene Mutation Testing in Metastatic Colorectal Carcinoma to Predict Response to Anti-EGFR Monoclonal Antibody Therapy: American Society of Clinical Oncology Provisional Clinical Opinion Update
Extended RAS Gene Mutation Testing in Metastatic Colorectal Carcinoma to Predict Response to Anti-EGFR Monoclonal Antibody Therapy: American Society of Clinical Oncology Provisional Clinical Opinion Update
Personalised cancer care Information for Medical Specialists. A new way to unlock treatment options for your patients
 Personalised cancer care Information for Medical Specialists A new way to unlock treatment options for your patients Contents Optimised for clinical benefit 4 Development history 4 Full FIND IT panel vs
Personalised cancer care Information for Medical Specialists A new way to unlock treatment options for your patients Contents Optimised for clinical benefit 4 Development history 4 Full FIND IT panel vs
Related Policies None
 Medical Policy MP 2.04.53 BCBSA Ref. Policy: 2.04.53 Last Review: 07/25/2018 Effective Date: 07/25/2018 Section: Medicine Related Policies None DISCLAIMER Our medical policies are designed for informational
Medical Policy MP 2.04.53 BCBSA Ref. Policy: 2.04.53 Last Review: 07/25/2018 Effective Date: 07/25/2018 Section: Medicine Related Policies None DISCLAIMER Our medical policies are designed for informational
Description of Procedure or Service. Policy. Benefits Application
 Corporate Medical Policy KRAS, NRAS, BRAF Mutation Analysis and Related File Name: Origination: Last CAP Review: Next CAP Review: Last Review: kras_nras_braf_mutation_analysis_and_related_treatment_in_metastatic_colorectal_cancer
Corporate Medical Policy KRAS, NRAS, BRAF Mutation Analysis and Related File Name: Origination: Last CAP Review: Next CAP Review: Last Review: kras_nras_braf_mutation_analysis_and_related_treatment_in_metastatic_colorectal_cancer
Circulating Tumor DNA in GIST and its Implications on Treatment
 Circulating Tumor DNA in GIST and its Implications on Treatment October 2 nd 2017 Dr. Ciara Kelly Assistant Attending Physician Sarcoma Medical Oncology Service Objectives Background Liquid biopsy & ctdna
Circulating Tumor DNA in GIST and its Implications on Treatment October 2 nd 2017 Dr. Ciara Kelly Assistant Attending Physician Sarcoma Medical Oncology Service Objectives Background Liquid biopsy & ctdna
Medical Policy An independent licensee of the Blue Cross Blue Shield Association
 KRAS, NRAS, and BRAF Variant Analysis in Metastatic Colorectal Cancer Page 1 of 25 Medical Policy An independent licensee of the Blue Cross Blue Shield Association Title: KRAS, NRAS, and BRAF Variant Analysis
KRAS, NRAS, and BRAF Variant Analysis in Metastatic Colorectal Cancer Page 1 of 25 Medical Policy An independent licensee of the Blue Cross Blue Shield Association Title: KRAS, NRAS, and BRAF Variant Analysis
Targeted therapy in NSCLC: do we progress? Prof. Dr. V. Surmont. Masterclass 27 september 2018
 Targeted therapy in NSCLC: do we progress? Prof. Dr. V. Surmont Masterclass 27 september 2018 Outline Introduction EGFR TKI ALK TKI TKI for uncommon driver mutations Take home messages The promise of
Targeted therapy in NSCLC: do we progress? Prof. Dr. V. Surmont Masterclass 27 september 2018 Outline Introduction EGFR TKI ALK TKI TKI for uncommon driver mutations Take home messages The promise of
September 23, The Role of In Vitro Diagnostic Tests in Pediatric Master Protocol Development
 The Role of In Vitro Diagnostic Tests in Pediatric Master Protocol Development September 23, 2016 Anand Pathak, MD, PhD, MPH Medical Officer Molecular Genetics Branch Division of Molecular Genetics and
The Role of In Vitro Diagnostic Tests in Pediatric Master Protocol Development September 23, 2016 Anand Pathak, MD, PhD, MPH Medical Officer Molecular Genetics Branch Division of Molecular Genetics and
Molecular Analysis for Targeted Therapy for Non- Small-Cell Lung Cancer Section 2.0 Medicine Subsection 2.04 Pathology/Laboratory
 2.04.45 Molecular Analysis for Targeted Therapy for Non- Small-Cell Lung Cancer Section 2.0 Medicine Subsection 2.04 Pathology/Laboratory Effective Date November 26, 2014 Original Policy Date November
2.04.45 Molecular Analysis for Targeted Therapy for Non- Small-Cell Lung Cancer Section 2.0 Medicine Subsection 2.04 Pathology/Laboratory Effective Date November 26, 2014 Original Policy Date November
2018 Edition The Current Landscape of Genetic Testing
 2018 Edition The Current Landscape of Genetic Testing Market growth, reimbursement trends, challenges and opportunities November April 20182017 EXECUTIVE SUMMARY Concert Genetics is a software and managed
2018 Edition The Current Landscape of Genetic Testing Market growth, reimbursement trends, challenges and opportunities November April 20182017 EXECUTIVE SUMMARY Concert Genetics is a software and managed
Clinical Policy: Pembrolizumab (Keytruda) Reference Number: CP.PHAR.322
 Clinical Policy: (Keytruda) Reference Number: CP.PHAR.322 Effective Date: 03/17 Last Review Date: 03/17 Coding Implications Revision Log See Important Reminder at the end of this policy for important regulatory
Clinical Policy: (Keytruda) Reference Number: CP.PHAR.322 Effective Date: 03/17 Last Review Date: 03/17 Coding Implications Revision Log See Important Reminder at the end of this policy for important regulatory
Biomarkers of Response to EGFR-TKIs EORTC-NCI-ASCO Meeting on Molecular Markers in Cancer November 17, 2007
 Biomarkers of Response to EGFR-TKIs EORTC-NCI-ASCO Meeting on Molecular Markers in Cancer November 17, 2007 Bruce E. Johnson, MD Dana-Farber Cancer Institute, Brigham and Women s Hospital, and Harvard
Biomarkers of Response to EGFR-TKIs EORTC-NCI-ASCO Meeting on Molecular Markers in Cancer November 17, 2007 Bruce E. Johnson, MD Dana-Farber Cancer Institute, Brigham and Women s Hospital, and Harvard
Personalised Healthcare. will it deliver? Bob Holland Head of Personalised Healthcare & Biomarkers Innovative Medicines AstraZeneca R&D
 Personalised Healthcare x will it deliver? Bob Holland Head of Personalised Healthcare & Biomarkers Innovative Medicines AstraZeneca R&D Personalised Healthcare It is delivering! Bob Holland Head of Personalised
Personalised Healthcare x will it deliver? Bob Holland Head of Personalised Healthcare & Biomarkers Innovative Medicines AstraZeneca R&D Personalised Healthcare It is delivering! Bob Holland Head of Personalised
Design considerations for Phase II trials incorporating biomarkers
 Design considerations for Phase II trials incorporating biomarkers Sumithra J. Mandrekar Professor of Biostatistics, Mayo Clinic Pre-Meeting Workshop Enhancing the Design and Conduct of Phase II Studies
Design considerations for Phase II trials incorporating biomarkers Sumithra J. Mandrekar Professor of Biostatistics, Mayo Clinic Pre-Meeting Workshop Enhancing the Design and Conduct of Phase II Studies
Delivering on the Promise of Personalised Healthcare
 Delivering on the Promise of Personalised Healthcare Dr John S Mills Diagnostic Team Director, Personalised Healthcare and Biomarkers Function, AstraZeneca, UK EBF 6 th Open Symposium, Barcelona, 20 Nov
Delivering on the Promise of Personalised Healthcare Dr John S Mills Diagnostic Team Director, Personalised Healthcare and Biomarkers Function, AstraZeneca, UK EBF 6 th Open Symposium, Barcelona, 20 Nov
Integration of Genomics Into Clinical Pathways. Precision Medicine and Decision Support
 Integration of Genomics Into Clinical Pathways Precision Medicine and Decision Support Faculty Andrew Hertler, MD, FACP Chief Medical Officer New Century Health Andrew Hertler, MD, FACP is employed by
Integration of Genomics Into Clinical Pathways Precision Medicine and Decision Support Faculty Andrew Hertler, MD, FACP Chief Medical Officer New Century Health Andrew Hertler, MD, FACP is employed by
KRAS, NRAS, and BRAF Variant Analysis in Metastatic Colorectal Cancer
 KRAS, NRAS, and BRAF Variant Analysis in Metastatic Colorectal Cancer Policy Number: 2.04.53 Last Review: 5/2018 Origination: 1/2011 Next Review: 5/2019 Policy Blue Cross and Blue Shield of Kansas City
KRAS, NRAS, and BRAF Variant Analysis in Metastatic Colorectal Cancer Policy Number: 2.04.53 Last Review: 5/2018 Origination: 1/2011 Next Review: 5/2019 Policy Blue Cross and Blue Shield of Kansas City
Companion & Complementary Diagnostics: Clinical and Regulatory Perspectives
 Companion & Complementary Diagnostics: Clinical and Regulatory Perspectives Workshop on Companion Diagnostics January 31, 2017 Jan Trøst Jørgensen, M.Sc.Pharm., Ph.D. Dx-Rx Institute Fredensborg, Denmark
Companion & Complementary Diagnostics: Clinical and Regulatory Perspectives Workshop on Companion Diagnostics January 31, 2017 Jan Trøst Jørgensen, M.Sc.Pharm., Ph.D. Dx-Rx Institute Fredensborg, Denmark
Personalized Healthcare Update
 Dr. Kai - Oliver Wesche Market Development Manager, Personalized Healthcare QIAGEN Personalized Healthcare Update Pioneering Personalized Medicine through Partnering TOMTOVOK BKM120 Zelboraf QIAGEN partners:
Dr. Kai - Oliver Wesche Market Development Manager, Personalized Healthcare QIAGEN Personalized Healthcare Update Pioneering Personalized Medicine through Partnering TOMTOVOK BKM120 Zelboraf QIAGEN partners:
K-Ras signalling in NSCLC
 Targeting the Ras-Raf-Mek-Erk pathway Egbert F. Smit MD PhD Dept. Pulmonary Diseases Vrije Universiteit VU Medical Centre Amsterdam, The Netherlands K-Ras signalling in NSCLC Sun et al. Nature Rev. Cancer
Targeting the Ras-Raf-Mek-Erk pathway Egbert F. Smit MD PhD Dept. Pulmonary Diseases Vrije Universiteit VU Medical Centre Amsterdam, The Netherlands K-Ras signalling in NSCLC Sun et al. Nature Rev. Cancer
Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment with Targeted Tyrosine Kinase Inhibitors
 Q: How is the strength of recommendation determined in the new molecular testing guideline? A: The strength of recommendation is determined by the strength of the available data (evidence). Strong Recommendation:
Q: How is the strength of recommendation determined in the new molecular testing guideline? A: The strength of recommendation is determined by the strength of the available data (evidence). Strong Recommendation:
Optimum Sequencing of EGFR targeted therapy in NSCLC. Dr. Sema SEZGİN GÖKSU Akdeniz Univercity, Antalya, Turkey
 Optimum Sequencing of EGFR targeted therapy in NSCLC Dr. Sema SEZGİN GÖKSU Akdeniz Univercity, Antalya, Turkey Lung cancer NSCLC SCLC adeno squamous EGFR ALK ROS1 BRAF HER2 KRAS EGFR Transl Lung Cancer
Optimum Sequencing of EGFR targeted therapy in NSCLC Dr. Sema SEZGİN GÖKSU Akdeniz Univercity, Antalya, Turkey Lung cancer NSCLC SCLC adeno squamous EGFR ALK ROS1 BRAF HER2 KRAS EGFR Transl Lung Cancer
July, ArQule, Inc.
 July, 2012 Safe Harbor This presentation and other statements by ArQule may contain forward-looking statements within the meaning of the Private Securities Litigation Reform Act with respect to clinical
July, 2012 Safe Harbor This presentation and other statements by ArQule may contain forward-looking statements within the meaning of the Private Securities Litigation Reform Act with respect to clinical
SUBJECT: GENOTYPING - EPIDERMAL GROWTH
 MEDICAL POLICY SUBJECT: GENOTYPING - EPIDERMAL GROWTH Clinical criteria used to make utilization review decisions are based on credible scientific evidence published in peer reviewed medical literature
MEDICAL POLICY SUBJECT: GENOTYPING - EPIDERMAL GROWTH Clinical criteria used to make utilization review decisions are based on credible scientific evidence published in peer reviewed medical literature
Are Payers Getting Tougher? Essential Insights on How to Smooth Acceptance of New Genetic Tests
 Are Payers Getting Tougher? Essential Insights on How to Smooth Acceptance of New Genetic Tests Jacqueline Huang Senior Associate Reimbursement Policy and Government Affairs Objectives Learn how payers
Are Payers Getting Tougher? Essential Insights on How to Smooth Acceptance of New Genetic Tests Jacqueline Huang Senior Associate Reimbursement Policy and Government Affairs Objectives Learn how payers
GLOBAL REGISTRATION STRATEGIES:
 GLOBAL REGISTRATION STRATEGIES: Therapeutic Product (TP) and the Companion Diagnostic (CDx) Erin Pedalino Regulatory Affairs International MSD Unique Role as CDx Regulatory Liaison at a Therapeutic Product
GLOBAL REGISTRATION STRATEGIES: Therapeutic Product (TP) and the Companion Diagnostic (CDx) Erin Pedalino Regulatory Affairs International MSD Unique Role as CDx Regulatory Liaison at a Therapeutic Product
7/6/2015. Cancer Related Deaths: United States. Management of NSCLC TODAY. Emerging mutations as predictive biomarkers in lung cancer: Overview
 Emerging mutations as predictive biomarkers in lung cancer: Overview Kirtee Raparia, MD Assistant Professor of Pathology Cancer Related Deaths: United States Men Lung and bronchus 28% Prostate 10% Colon
Emerging mutations as predictive biomarkers in lung cancer: Overview Kirtee Raparia, MD Assistant Professor of Pathology Cancer Related Deaths: United States Men Lung and bronchus 28% Prostate 10% Colon
MEDICAL POLICY. SUBJECT: GENOTYPING - RAS MUTATION ANALYSIS IN METASTATIC COLORECTAL CANCER (KRAS/NRAS) POLICY NUMBER: CATEGORY: Laboratory
 MEDICAL POLICY Clinical criteria used to make utilization review decisions are based on credible scientific evidence published in peer reviewed medical literature generally recognized by the medical community.
MEDICAL POLICY Clinical criteria used to make utilization review decisions are based on credible scientific evidence published in peer reviewed medical literature generally recognized by the medical community.
AD (Leave blank) TITLE: Genomic Characterization of Brain Metastasis in Non-Small Cell Lung Cancer Patients
 AD (Leave blank) Award Number: W81XWH-12-1-0444 TITLE: Genomic Characterization of Brain Metastasis in Non-Small Cell Lung Cancer Patients PRINCIPAL INVESTIGATOR: Mark A. Watson, MD PhD CONTRACTING ORGANIZATION:
AD (Leave blank) Award Number: W81XWH-12-1-0444 TITLE: Genomic Characterization of Brain Metastasis in Non-Small Cell Lung Cancer Patients PRINCIPAL INVESTIGATOR: Mark A. Watson, MD PhD CONTRACTING ORGANIZATION:
Osamu Tetsu, MD, PhD Associate Professor Department of Otolaryngology-Head and Neck Surgery School of Medicine, University of California, San
 Osamu Tetsu, MD, PhD Associate Professor Department of Otolaryngology-Head and Neck Surgery School of Medicine, University of California, San Francisco Lung Cancer Classification Pathological Classification
Osamu Tetsu, MD, PhD Associate Professor Department of Otolaryngology-Head and Neck Surgery School of Medicine, University of California, San Francisco Lung Cancer Classification Pathological Classification
See Important Reminder at the end of this policy for important regulatory and legal information.
 Clinical Policy: (Erbitux) Reference Number: CP.PHAR.317 Effective Date: 02.01.17 Last Review Date: 11.18 Line of Business: Commercial, HIM, Medicaid Coding Implications Revision Log See Important Reminder
Clinical Policy: (Erbitux) Reference Number: CP.PHAR.317 Effective Date: 02.01.17 Last Review Date: 11.18 Line of Business: Commercial, HIM, Medicaid Coding Implications Revision Log See Important Reminder
Oncology Drug Development Using Molecular Pathology
 Oncology Drug Development Using Molecular Pathology Introduction PRESENTERS Lee Schacter, PhD, MD, FACP Executive Medical Director, Oncology Clinipace Worldwide Martha Bonino Director, Strategic Accounts
Oncology Drug Development Using Molecular Pathology Introduction PRESENTERS Lee Schacter, PhD, MD, FACP Executive Medical Director, Oncology Clinipace Worldwide Martha Bonino Director, Strategic Accounts
See Important Reminder at the end of this policy for important regulatory and legal information.
 Clinical Policy: (Tafinlar) Reference Number: CP.PHAR.239 Effective Date: 11.16.16 Last Review Date: 08.18 Line of Business: Commercial, HIM, Medicaid Revision Log See Important Reminder at the end of
Clinical Policy: (Tafinlar) Reference Number: CP.PHAR.239 Effective Date: 11.16.16 Last Review Date: 08.18 Line of Business: Commercial, HIM, Medicaid Revision Log See Important Reminder at the end of
Next Generation Sequencing in Clinical Practice: Impact on Therapeutic Decision Making
 Next Generation Sequencing in Clinical Practice: Impact on Therapeutic Decision Making November 20, 2014 Capturing Value in Next Generation Sequencing Symposium Douglas Johnson MD, MSCI Vanderbilt-Ingram
Next Generation Sequencing in Clinical Practice: Impact on Therapeutic Decision Making November 20, 2014 Capturing Value in Next Generation Sequencing Symposium Douglas Johnson MD, MSCI Vanderbilt-Ingram
pan-canadian Oncology Drug Review Final Clinical Guidance Report Dabrafenib (Tafinlar) and Trametinib (Mekinist) for Non-Small Cell Lung Cancer
 pan-canadian Oncology Drug Review Final Clinical Guidance Report Dabrafenib (Tafinlar) and Trametinib (Mekinist) for Non-Small Cell Lung Cancer November 2, 2017 DISCLAIMER Not a Substitute for Professional
pan-canadian Oncology Drug Review Final Clinical Guidance Report Dabrafenib (Tafinlar) and Trametinib (Mekinist) for Non-Small Cell Lung Cancer November 2, 2017 DISCLAIMER Not a Substitute for Professional
EVIDENCE IN BRIEF OVERALL CLINICAL BENEFIT
 information aligned with the expected toxicity profile of panitumumab, which is well-known as panitumumab is already used as a third-line therapy for patients with mcrc. perc also noted QoL did not deteriorate
information aligned with the expected toxicity profile of panitumumab, which is well-known as panitumumab is already used as a third-line therapy for patients with mcrc. perc also noted QoL did not deteriorate
Liquid biopsy: the experience of real life case studies
 Liquid biopsy: the experience of real life case studies 10 th September 2018 Beatriz Bellosillo Servicio de Anatomía Patológica Hospital del Mar, Barcelona Agenda Introduction Experience in colorectal
Liquid biopsy: the experience of real life case studies 10 th September 2018 Beatriz Bellosillo Servicio de Anatomía Patológica Hospital del Mar, Barcelona Agenda Introduction Experience in colorectal
Molecular Analysis for Targeted Therapy of Non-Small Cell Lung Cancer (NSCLC)
 Medical Policy Manual Genetic Testing, Policy No. 56 Molecular Analysis for Targeted Therapy of Non-Small Cell Lung Cancer (NSCLC) Next Review: November 2018 Last Review: April 2018 Effective: May 1, 2018
Medical Policy Manual Genetic Testing, Policy No. 56 Molecular Analysis for Targeted Therapy of Non-Small Cell Lung Cancer (NSCLC) Next Review: November 2018 Last Review: April 2018 Effective: May 1, 2018
How the Changing Landscape of Oncology Drug Development and Approval Will Affect Advanced Practitioners
 How the Changing Landscape of Oncology Drug Development and Approval Will Affect Advanced Practitioners Richard Pazdur, MD Director Oncology Center of Excellence US Food and Drug Administration Learning
How the Changing Landscape of Oncology Drug Development and Approval Will Affect Advanced Practitioners Richard Pazdur, MD Director Oncology Center of Excellence US Food and Drug Administration Learning
Overview of Biomarker Development for Immune PD-1/L1 Checkpoint Blockade
 Overview of Biomarker Development for Immune PD-1/L1 Checkpoint Blockade David L. Rimm MD-PhD Professor Departments of Pathology and Medicine (Oncology) Director, Yale Pathology Tissue Services Disclosures
Overview of Biomarker Development for Immune PD-1/L1 Checkpoint Blockade David L. Rimm MD-PhD Professor Departments of Pathology and Medicine (Oncology) Director, Yale Pathology Tissue Services Disclosures
See Important Reminder at the end of this policy for important regulatory and legal information.
 Clinical Policy: (Tagrisso) Reference Number: CP.PHAR.294 Effective Date: 12.01.16 Last Review Date: 08.18 Line of Business: Commercial, Medicaid Revision Log See Important Reminder at the end of this
Clinical Policy: (Tagrisso) Reference Number: CP.PHAR.294 Effective Date: 12.01.16 Last Review Date: 08.18 Line of Business: Commercial, Medicaid Revision Log See Important Reminder at the end of this
Supplementary Online Content
 Supplementary Online Content Kris MG, Johnson BE, Berry LD, et al. Using Multiplexed Assays of Oncogenic Drivers in Lung Cancers to Select Targeted Drugs. JAMA. doi:10.1001/jama.2014.3741 etable 1. Trials
Supplementary Online Content Kris MG, Johnson BE, Berry LD, et al. Using Multiplexed Assays of Oncogenic Drivers in Lung Cancers to Select Targeted Drugs. JAMA. doi:10.1001/jama.2014.3741 etable 1. Trials
Clinical Policy: Nivolumab (Opdivo) Reference Number: CP.PHAR.121 Effective Date: Last Review Date: Line of Business: Medicaid
 Clinical Policy: (Opdivo) Reference Number: CP.PHAR.121 Effective Date: 07.15 Last Review Date: 01.18 Line of Business: Medicaid Revision Log See Important Reminder at the end of this policy for important
Clinical Policy: (Opdivo) Reference Number: CP.PHAR.121 Effective Date: 07.15 Last Review Date: 01.18 Line of Business: Medicaid Revision Log See Important Reminder at the end of this policy for important
Determined to realize a future in which people with cancer live longer and better than ever before
 Determined to realize a future in which people with cancer live longer and better than ever before 4Q 2016 EARNINGS PRESENTATION MARCH 2017 1 Forward-looking statements disclosure This presentation contains
Determined to realize a future in which people with cancer live longer and better than ever before 4Q 2016 EARNINGS PRESENTATION MARCH 2017 1 Forward-looking statements disclosure This presentation contains
PRECISION INSIGHTS. GPS Cancer. Molecular Insights You Can Rely On. Tumor-normal sequencing of DNA + RNA expression.
 PRECISION INSIGHTS GPS Cancer Molecular Insights You Can Rely On Tumor-normal sequencing of DNA + RNA expression www.nanthealth.com Cancer Care is Evolving Oncologists use all the information available
PRECISION INSIGHTS GPS Cancer Molecular Insights You Can Rely On Tumor-normal sequencing of DNA + RNA expression www.nanthealth.com Cancer Care is Evolving Oncologists use all the information available
Practical Implications of Next Generation Sequencing (NGS) in Breast Cancer: Promises and Challenges
 Practical Implications of Next Generation Sequencing (NGS) in Breast Cancer: Promises and Challenges Naoto T. Ueno, MD, PhD, FACP Professor of Medicine Executive Director, Morgan Welch Inflammatory Breast
Practical Implications of Next Generation Sequencing (NGS) in Breast Cancer: Promises and Challenges Naoto T. Ueno, MD, PhD, FACP Professor of Medicine Executive Director, Morgan Welch Inflammatory Breast
December 13, The Future Reimbursement Environment for NGS for Oncology
 WELCOME! Today s Speaker Mr. Mathews has worked on a variety of diagnostic reimbursement projects involving the analysis, development, and implementation of coding, coverage, and payment strategies for
WELCOME! Today s Speaker Mr. Mathews has worked on a variety of diagnostic reimbursement projects involving the analysis, development, and implementation of coding, coverage, and payment strategies for
Lung Cancer Update 2016 BAONS Oncology Care Update
 Lung Cancer Update 2016 BAONS Oncology Care Update Matthew Gubens, MD, MS Assistant Professor Chair, Thoracic Oncology Site Committee UCSF Helen Diller Family Comprehensive Cancer Center Disclosures Consulting
Lung Cancer Update 2016 BAONS Oncology Care Update Matthew Gubens, MD, MS Assistant Professor Chair, Thoracic Oncology Site Committee UCSF Helen Diller Family Comprehensive Cancer Center Disclosures Consulting
Erlotinib for the first-line treatment of EGFR-TK mutation positive non-small cell lung cancer
 ERRATUM Erlotinib for the first-line treatment of EGFR-TK mutation positive non-small cell lung cancer This report was commissioned by the NIHR HTA Programme as project number 11/08 Completed 6 th January
ERRATUM Erlotinib for the first-line treatment of EGFR-TK mutation positive non-small cell lung cancer This report was commissioned by the NIHR HTA Programme as project number 11/08 Completed 6 th January
Frequency(%) KRAS G12 KRAS G13 KRAS A146 KRAS Q61 KRAS K117N PIK3CA H1047 PIK3CA E545 PIK3CA E542K PIK3CA Q546. EGFR exon19 NFS-indel EGFR L858R
 Frequency(%) 1 a b ALK FS-indel ALK R1Q HRAS Q61R HRAS G13R IDH R17K IDH R14Q MET exon14 SS-indel KIT D8Y KIT L76P KIT exon11 NFS-indel SMAD4 R361 IDH1 R13 CTNNB1 S37 CTNNB1 S4 AKT1 E17K ERBB D769H ERBB
Frequency(%) 1 a b ALK FS-indel ALK R1Q HRAS Q61R HRAS G13R IDH R17K IDH R14Q MET exon14 SS-indel KIT D8Y KIT L76P KIT exon11 NFS-indel SMAD4 R361 IDH1 R13 CTNNB1 S37 CTNNB1 S4 AKT1 E17K ERBB D769H ERBB
EXAMPLE. - Potentially responsive to PI3K/mTOR and MEK combination therapy or mtor/mek and PKC combination therapy. ratio (%)
 Dr Kate Goodhealth Goodhealth Medical Clinic 123 Address Road SUBURBTOWN NSW 2000 Melanie Citizen Referring Doctor Your ref Address Dr John Medico 123 Main Street, SUBURBTOWN NSW 2000 Phone 02 9999 9999
Dr Kate Goodhealth Goodhealth Medical Clinic 123 Address Road SUBURBTOWN NSW 2000 Melanie Citizen Referring Doctor Your ref Address Dr John Medico 123 Main Street, SUBURBTOWN NSW 2000 Phone 02 9999 9999
1.Basis of resistance 2.Mechanisms of resistance 3.How to overcome resistance. 13/10/2017 Sara Redaelli
 Dott.ssa Sara Redaelli 13/10/2017 1.Basis of resistance 2.Mechanisms of resistance 3.How to overcome resistance Tumor Heterogeneity: Oncogenic Drivers in NSCLC The Promise of Genotype-Directed Therapy
Dott.ssa Sara Redaelli 13/10/2017 1.Basis of resistance 2.Mechanisms of resistance 3.How to overcome resistance Tumor Heterogeneity: Oncogenic Drivers in NSCLC The Promise of Genotype-Directed Therapy
Tissue or Liquid Biopsy? ~For Diagnosis, Monitoring and Early detection of Resistance~
 16 th Dec. 2016. ESMO Preceptorship Program Non-Small-Cell Lung Cancer @Singapore Tissue or Liquid Biopsy? ~For Diagnosis, Monitoring and Early detection of Resistance~ Research Institute for Disease of
16 th Dec. 2016. ESMO Preceptorship Program Non-Small-Cell Lung Cancer @Singapore Tissue or Liquid Biopsy? ~For Diagnosis, Monitoring and Early detection of Resistance~ Research Institute for Disease of
La biopsia liquida dei tumori: il viaggio. Paola Gazzaniga Liquid Biopsy Unit Dept. Molecular Medicine Sapienza University of Rome
 La biopsia liquida dei tumori: il viaggio Paola Gazzaniga Liquid Biopsy Unit Dept. Molecular Medicine Sapienza University of Rome Must Know Necessary Travel Tips There is lack of drugs to treat all the
La biopsia liquida dei tumori: il viaggio Paola Gazzaniga Liquid Biopsy Unit Dept. Molecular Medicine Sapienza University of Rome Must Know Necessary Travel Tips There is lack of drugs to treat all the
