UC Irvine UC Irvine Previously Published Works
|
|
|
- Chad Gregory
- 6 years ago
- Views:
Transcription
1 UC Irvine UC Irvine Previously Published Works Title Lipidomic analysis of endocannabinoid metabolism in biological samples Permalink Journal Journal of Chromatography B: Analytical Technologies in the Biomedical and Life Sciences, 877(26) ISSN Authors Astarita, G Piomelli, D Publication Date DOI /j.jchromb License CC BY 4.0 Peer reviewed escholarship.org Powered by the California Digital Library University of California
2 NIH Public Access Author Manuscript J Chromatogr B Analyt Technol Biomed Life Sci. Author manuscript; available in PMC 2010 September 15. Published in final edited form as: J Chromatogr B Analyt Technol Biomed Life Sci September 15; 877(26): doi: / j.jchromb Lipidomic analysis of endocannabinoid metabolism in biological samples Giuseppe Astarita and Daniele Piomelli Department of Pharmacology, University of California, Irvine, CA Abstract The endocannabinoids are signaling lipids present in many living organisms. They activate G proteincoupled cannabinoid receptors to modulate a broad range of biological processes that include emotion, cognition, inflammation and reproduction. The endocannabinoids are embedded in an interconnected network of cellular lipid pathways, the regulation of which is likely to control the strength and duration of endocannabinoid signals. Therefore, physiopathological or pharmacological perturbations of these pathways may indirectly affect endocannabinoid activity and, vice versa, endocannabinoid activity may influence lipid pathways involved in other metabolic and signaling events. Recent progress in liquid chromatography and mass spectrometry has fueled the development of targeted lipidomic approaches, which allow researchers to examine complex lipid interactions in cells and gain a broader view of the endocannabinoid system. Here, we review these new developments from the perspective of our laboratory s experience in the field. Keywords lipidomics; anandamide; 2-arachidonoylglycerol (2-AG); N-acyl phosphatidylethanolamine (NAPE); fatty acid amide hydrolase (FAAH); diacylglycerol (DAG); triacylglycerol (TAG); arachidonic acid; prostaglandin 1. Introduction The endocannabinoid signaling system is part of a network of cellular lipid pathways. Perturbations of such pathways, induced by physiological or pathological events, may strongly affect endocannabinoid function. Recent advances liquid chromatography (LC) and mass spectrometry (MS) have fueled the development of lipidomics, a branch of metabolomics that has enormous potential for the study of endocannabinoid metabolism. Indeed, lipidomic approaches can be combined with genetic and pharmacological strategies to address many unsolved questions on the roles and regulation of endocannabinoid signaling. Considerable progress has been made in understanding the physiological and pathological roles of endocannabinoids and several recent reviews discuss these topics in detail [1,2]. Here, we will focus on recent advances in the development and application of lipidomics to the study of the endocannabinoid system. Correspondence to: Daniele Piomelli, PhD, Department of Pharmacology, University of California, Irvine; Irvine, CA ; Phone: (949) ; Fax: (949) ; piomelli@uci.edu. Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
3 Astarita and Piomelli Page Endocannabinoid metabolism: a complex picture The endocannabinoids are endogenous lipids derived from polyunsaturated fatty acids. The two best known endocannabinoids, arachidonoylethanolamide (anandamide) and 2- arachidonoyl-sn-glycerol (2-AG), are both derivatives of arachidonic acid. These molecules are found in all mammalian tissues and provide a classic example of signaling lipids. They are produced by cells on demand and bind to G protein-coupled receptors, called cannabinoid receptors, which mediate their action. Cannabinoid receptors participate in the regulation of many physiological processes, such as emotion, reward, cognition, inflammation and reproduction. In the last two decades, much research has been devoted to studying the formation and deactivation of individual endocannabinoid lipids in mammalian tissues, with the intent of elucidating their biological regulation. These analyses have initially relied on techniques such as thin-layer chromatography (TLC) and gas chromatography-mass spectrometry (GC/ MS). Recent progress in LC/MS has greatly facilitated these efforts, allowing researchers to place individual endocannabinoid molecules in the context of the interconnected network of their substrates and products. Indeed, not only are the endocannabinoids produced through biochemical pathways that yield a sequence of functionally distinct signaling lipids, but also different biochemical pathways are used to generate the same endocannabinoid molecule [3]. Thus, the same enzymes and lipid substrates can be often shared between endocannabinoid and other metabolic pathways. The development of analytical approaches that consider the ensemble of the endocannabinoid system, rather than its individual components in isolation, could be invaluable to elucidate the physiopathological roles of this signaling system LC/MS of endocannabinoids The beginning of the endocannabinoid field was marked by the discovery in 1992 of the first endogenous cannabinoid receptor ligand, the fatty acid ethanolamide anandamide [4]. Initially, endocannabinoids analysis relied heavily on TLC, LC and GC/MS [5-7]. However, these techniques often require multiple analytical steps, including chemical derivatization and serial enzymatic hydrolysis, which render analyses very laborious and time-consuming. The advent of LC/MS has completely changed this scenario. Using LC/MS we can identify and quantify individual endocannabinoid molecules in a single step using analytical platforms, such as tandem mass spectrometry (MS/MS), which provide the detailed structural information necessary for the characterization of lipids in complex biological matrices [8]. Furthermore, the invention of ionization techniques such as electrospray ionization (ESI) and atmospheric pressure chemical ionization (APCI), which permit the direct coupling of LC to MS, has allowed researchers to identify and quantify the endocannabinoids with greater sensitivity then ever before possible [9-13]. Therefore, the combination of LC and MS has emerged as the technique of choice for endocannabinoid analyses. 1.3 Lipidomics to study endocannabinoid metabolism Many aspects of endocannabinoid metabolism are still incompletely understood. For example, it is unclear how changes in endocannabinoid signaling affect distal lipid pathways, and, vice versa, how alterations in cellular lipid metabolism influence endocannabinoid signaling. In addition, we do not understand the specific contributions of each of the biosynthetic and degradative enzymes that have been identified in endocannabinoid metabolism. Lipidomics can help answer these questions by offering a large-scale view of lipid alterations accompanying perturbations of endocannabinoid metabolism. For example, the analysis of lipid profiles associated with genetic or pharmacological manipulations of the endocannabinoid system might shed new light on the biochemical mechanisms underlying the regulation of endocannabinoid metabolism [12,14,15]. Furthermore, lipidomic information can be integrated into multidisciplinary datasets deriving from genomics, transcriptomics and
4 Astarita and Piomelli Page 3 proteomics. Such a system biology approach would offer novel insights on the effects of genetic or protein diversity on endocannabinoid metabolism. In the following sections, we illustrate the development and application of a comprehensive lipidomic approach to study endocannabinoid metabolism in biological samples. 2. Lipidomic analysis of endocannabinoids and related lipids The endocannabinoid system is constituted of three major lipid classes: glycerophospholipids, glycerolipids and fatty acyls. Because these lipids have a wide variety of chemical structures and dynamic range of concentrations, several approaches have been developed for their analysis [12,16-18]. A large-scale LC/MS lipidomic strategy for the identification and quantification of various classes of lipids in biological samples is presented in Fig. 1. Such strategy requires the combination of three analytical steps: 1) sample preparation by organic solvent extraction and lipid fractionation by open-bed silica-gel chromatography, which separates low-abundance lipids from molecules that might interfere or suppress their detection; 2) chromatographic separation using two different octadecyl (C18) reversed-phase LC columns, which can fractionate lipids based on their size, number of double bonds and diffusion properties; and 3) intrasource-ionization separation coupled to MS in either the positive or negative mode, which differentiates lipids based on their ionization properties Step 1. Sample preparation Lipid extraction Sample preparation includes lipid extraction from the biological matrix and removal of most non-lipid contaminants from the extract. Usually, lipid concentrations in biological samples are normalized by tissue weight or protein, total phospholipid or DNA concentration. Absolute quantification is made possible by using nonendogenous structural analogs (internal standards), which are added before the extraction procedure. A variety of mixtures of organic solvents have been utilized to extract lipid classes from biological samples [19-25]. The modified Folch procedure for lipid extraction utilized in our laboratory allows for greater than 90% recovery of most lipids [26]. In such procedure, snap-frozen tissues are weighed and homogenized in ice-cold methanol (1 ml per 100 mg tissue) to quench any possible enzymatic reaction that might interfere with the analysis. The methanol contains appropriate internal standards (see next section), which also act as carriers to reduce analyte loss during sample work-up. Samples from the homogenates are taken for protein measurements. Lipids are extracted with chloroform (2 vol) and washed with water (0.75 vol). Organic phases are collected and dried under a stream of N 2 (important to prevent lipid oxidation). Alternative extraction methods are recommended for the quantitative isolation of acidic phospholipids such as phosphoinositides [18,27] Internal standards Quantitation of lipid species in biological samples can be achieved by normalizing the individual molecular ion peak intensity with an internal standard for each lipid class. A mixture of non-endogenous molecules can be used as internal standards, which allow lipid levels to be normalized for both extraction efficiency and instrument response. A representative list of commercially available internal standards for each lipid class includes the following chemicals: 1) fatty-acid ethanolamides: deuterated and non-deuterated fatty acid ethanolamides can be synthesized as previously reported [12] or purchased from commercial sources such as Cayman Chemicals (Ann Arbor, MI, USA); 2) fatty acids: heptadecanoic acid from Nu-Chek Prep (Elysian, MN, USA) or d 8 -arachidonic acid (Cayman Chemicals); 3) N-acyl-phosphatidylethanolamine: 1,2-dipalmitoyl-sn-glycero-3- phosphoethanolamine-n-heptadecanoyl can be synthesized as previously described [12]; alternatively 1-O-hexadecyl-2-palmitoyl-sn-glycero-3-phosphoethanolamine-N-palmitoyl may be purchased from Sigma-Aldrich (St. Louis, MI, USA); 4) oxygenated
5 Astarita and Piomelli Page 4 endocannabinoids: d 4 -PGF 2α ethanolamide and prostaglandin: d 4 -prostaglandin-e 2 (Cayman Chemicals); 5) triacylglycerol: trinonadecenoin from Nu-Chek Prep; 6) diacylglycerol: dinonadienoin (Nu-Chek Prep); 7) monoacylglycerol: monoheptadecanoin (Nu-Chek Prep) or d 8-2-arachidonoyl-sn-glycerol (Cayman Chemicals); 8) phosphatidylethanolamine: 1,2- diheptadecanoyl-sn-glycero-3- phosphoethanolamine (Avanti Polar Lipids; Alabaster, AL, USA); 9) phosphatidylcholine: 1,2-diheptadecanoyl-sn-glycero-3-phosphocholine (Avanti); 10) phosphatidylinositol: 1,2-dipalmitoyl-sn-glycero-3-phosphoinositol (Avanti) Lipid fractionation Various solid-phase extraction chromatography procedures have been used to fractionate lipid classes. The standardized extraction procedure employed in our lab is illustrated in Fig. 1. After solvent extraction, the organic phase is divided into two parts. The first is directly analyzed by LC/MS n to measure high-abundance, positively charged lipids (e.g., phosphatidylcholines). The second part is fractionated by open-bed silica gel column chromatography (Silica gel mesh) prior to LC/MS n analyses. A serial elution with 9:1 and 1:1 chloroform/methanol (vol:vol) is used to separate lipid groups according to their relative polarities (Fig. 1) Step 2. Chromatographic separation Reversed-phase LC can be used to separate lipids based on to their fatty acyl chain length and degree of unsaturation. Lipids species containing longer acyl chains elute from the LC column later than do shorter-chain lipids (e.g. C18:0>C16:0>C14:0), while saturated acyl structures tend to elute later than do long polyunsaturated ones (e.g. C18:0>C18:1>C18:2). Generally, the first double bond on an acyl chain reduces the effective chain length by a little less than 2 carbon units, while additional double bonds have smaller effects on retention. The combinatorial nature of complex lipids makes possible only a partial separation of isomeric species. Therefore, to further characterize samples containing such lipids, LC separation is coupled with MS n, which allows each lipid species to be identified based on both retention time and MS n properties (Fig. 1) Analysis of small lipids Small lipids, which we define here as molecules containing a single fatty acyl group, can be separated using a reversed-phase C18 LC column packed with conventional porous silica particles of small diameter (Fig. 1). For small neutral lipids (e.g. esters and amides of fatty acids), a linear gradient of methanol (or acetonitrile) in water is used. More polar small lipids (e.g. fatty acids and their derivatives containing a free carboxylic group or lysophospholipids containing a phosphate group) can be separated using a linear gradient of methanol in water containing acetate buffer (5 mm ammonium acetate, 0.25% acetic acid, ph 3.8), which is included to improve peak shape. These reversed-phase LC conditions generally allow for the separation of small lipids that differ in double bond number and/or chain length Analysis of large apolar, anionic and cationic lipids Large lipids, defined here as molecules containing two or more fatty acyl groups, may be separated using a reversedphase C18 LC column packed with superficially porous particles (e.g., SB300 Poroshell column, Agilent-Technologies, CA, USA) (Fig. 1). A linear gradient of methanol (or acetonitrile) in water containing acetate buffer affords good peak shape at fast flow rates. This is probably due to the fact that large lipids rapidly penetrate the thin shell of the superficial coating material, reducing diffusion and peak tailing. A combination of high temperature and high flow velocities improves the separation speed, resulting in better peak shape Step 3. Intrasource ionization coupled to MS separation of lipid molecules Most lipids can be detected using either ESI or APCI techniques. Here, we illustrate an approach that uses ESI in either the positive or negative mode (Fig. 1).
6 Astarita and Piomelli Page Small lipids Small neutral lipids (e.g. esters or amides of fatty acids) are generally detected as protonated molecular ions or sodium and ammonium adducts in the positive ESI mode. In contrast, more polar small lipids (e.g. those containing free carboxylic or phosphate groups) are detected as deprotonated molecular ions in the negative ESI mode (Fig. 1) Large lipids Large apolar lipids (e.g., triacylglycerols, TAGs, and diacylglycerols, DAGs) are detected as sodium or ammonium adducts in the positive ESI mode. Large lipids containing a positively charged group (e.g. phosphatidylcholine), can be detected as positively charged molecular ions in the positive ESI mode. Alternatively, such lipids can be also detected as acetate adducts in the negative ESI mode. In contrast, large anionic lipids are detected as deprotonated molecular ions in the negative ESI mode (Fig. 1) LC/MS tools for metabolite discovery LC/MS imaging Traditional ways to represent LC/MS runs (peak intensity vs retention time) (Fig. 2A, top) provide limited information, owing to the coexistence in a single sample of multiple isobaric and isomeric molecular species. A practical solution to bypass this problem is to represent LC/MS chromatograms as 3D maps (LC elution time vs mass vs intensity) (Fig. 2A, bottom) [12,14]. This imaging tool allows for an intuitive visual inspection of the molecular content of a biological sample and can also be used for qualitative analysis [12,14] Gentle ESI/MS n fragmentation The combination of soft ionization techniques such as ESI and the gentle process of fragmentation produced by the ion trap instrument generates a series of fragment ions that may be found physiologically as neutral molecular species (Fig. 2B). For example, fragments generated by the ESI/MS n analysis of complex lipid species are also common products of enzymatic hydrolysis (e.g., lysophospholipids and fatty acids) or chemical rearrangement (e.g., products of intramolecular cyclization of phosphate groups) (Fig.2B) [12]. This suggests that a gentle fragmentation approach can be used as a discovery tool in biochemistry for the study of catabolic products of known biomolecules and the identification of novel biologically relevant metabolites. 3. Metabolism of anandamide Anandamide is biosynthesized through cleavage of a group of membrane glycerophospholipid precursors in response to extracellular signals (Fig. 3). In the next section, we illustrate the applications of a lipidomic approach (previously described in Section 2) to the analysis of anandamide formation and degradation in human brain samples (frontal cortex from healthy male subjects aging from 64 to 82; average post mortem interval 4.2 hours) obtained through the courtesy of Drs. Carl W. Cotman and Dr. Elizabeth Head (Institute for Brain Aging and Dementia and Alzheimer s Disease Research Center at University of California, Irvine) Lipid remodeling of anandamide precursors A likely physiological route for the formation of anandamide requires the generation of N- arachidonoyl-phosphatidylethanolamines (NAPEs), which are produced through the transfer of arachidonic acid from the sn-1 position of phospholipids (e.g., phosphatidylcholine, PC) to the primary amine of phosphatidylethanolamine (PE, Fig. 4; method details are in the figure legend) [12,28-30]. This reaction is catalyzed by an as-yet-uncharacterized N-acyltransferase (NAT), which may also be involved in the biosynthesis of other N-acyl PE [12,28-30] (Fig. 3) (see Section 3.5). Figure 5 shows the LC/MS n identification of a NAPE precursor, 1,2- diarachidonoyl-phosphatidylcholine, in frozen human brain sample (method details are in the figure legend).
7 Astarita and Piomelli Page Anandamide formation Starting from NAPEs (Fig. 6), three distinct biochemical pathways have been proposed for the formation of anandamide (Fig. 3): 1) the direct conversion of NAPE to anandamide by a NAPEspecific phospholipase D (PLD) [5,31]; 2) the hydrolysis of NAPE by a NAPE-specific phospholipase A1/A2 (e.g. α/β hydrolase-4, ABHD-4) to form the intermediates lyso-nape (Fig. 6) and glycerophospho-anandamide, which are then cleaved by PLD [32-34]; 3) the hydrolysis of NAPE catalyzed by a NAPE-specific phospholipase C to yield the intermediate phosphoanandamide, which can then be cleaved by a lipid phosphatase such as protein tyrosine phosphatase, non-receptor type 22[35,36] Anandamide degradation Anandamide degradation requires first its transport across the plasma membrane, which may occur by carrier-mediated facilitated diffusion [37-39]. Once inside the cell, anandamide is hydrolyzed to arachidonic acid and ethanolamine by intracellular, membrane-bound fatty acid amide hydrolases (FAAHs) [40,41]. FAAHs also hydrolyze other bioactive fatty acid amides (e.g. fatty acid primary amides and N-acyl amino acids, see Section 3.5) [42-44] Oxidative metabolism of anandamide Anandamide can be metabolized by cyclooxygenase-2 (COX-2), 12- and 15-lipoxygenase (LOX), and cytochrome P450 (CYP450) to generate prostaglandin ethanolamides (prostamides), hydroperoxides, hydroxy and epoxide metabolites (Fig. 3) [45-52]. Most of these molecules have low affinity for cannabinoid receptors and display moderate activity as FAAH inhibitors, which may result in a potentiation of anandamide signaling. However, because of their extremely low concentrations in tissues, their physiological functions remain unclear Fatty-acid ethanolamides (FAEs) Anandamide belongs to the fatty acid ethanolamide (FAEs) (or else N-acylethanolamides, NAEs) family of signaling lipids, which include both cannabimimetic and noncannabimimetic molecules that are present in most mammalian tissues. These molecules, which only differ in length and degree of unsaturation of their acyl chains, share several biosynthetic and degradative enzymes Cannabimimetic FAEs Two polyunsaturated FAEs, docosatetraenoylethanolamide (C22:4, Δ ) and dihomo-γ-linolenoylethanolamide (C20:3, Δ ) bind with high affinity to cannabinoid receptors [53,54] and produce pharmacological effects similar to those of anandamide. Notably, our analyses show that these two lipids are present in human brain tissue at concentrations comparable to those of anandamide (Fig. 7) Non-cannabimimetic FAEs Most endogenous FAEs do not productively bind cannabinoid receptors [55]. Yet, their levels are from 1-10 times higher than those of anandamide and they may serve important physiological functions. For example, palmitoylethanolamide (C16:0) and oleoylethanolamide (C18:1, Δ 9 ) (Fig. 7) are endogenous ligands for the nuclear receptor peroxisome-activated receptor-alpha (PPAR-alpha) and exert anorexic, analgesic and antiinflammatory effects [56-60]. Stearoylethanolamide (C18:0) (Fig. 7) also appears to produce antiinflammatory and anorexic effects [61-63]. The biological distribution of other FAEs such as linoleoylethanolamide (C18:2, Δ 9-12 ) and linolenoylethanolamide (C18:3, Δ ) in rat appears to be tissue-specific, with very high levels in peripheral tissues (e.g., intestine) [14,64], while almost undetectable levels in the brain.
8 Astarita and Piomelli Page Fatty-acid amides Besides FAEs, fatty acyl amide analogs of anandamide include two additional classes of endogenous molecules: 1) primary fatty acid amides (e.g. oleamide) [65]; 2) N-acyl amino acids (also called elmiric acids), which derive from the conjugation of fatty acids with amino acids or amino-acid derivatives (e.g. N-acyl glycine, N-acyl taurine, N-acyl serotonin and N- acyl dopamine) [55,66,67]. Although some of these amides have clear pharmacological effects, such as the activation of the TRPV-1 receptor channels, the elucidation of their physiological roles awaits further investigation. It is important to point out that several such amides (e.g., oleamide) are commonly used in the production of plasticware, and may thus leach into the biological samples during work-up [68] Assays for the metabolism of anandamide Several enzymatic assays have been developed to study anandamide metabolism in vitro [12, 14]. Here, we review some selected examples of assays relying on fast and ultra-fast LC/MS analysis NAPE biosynthesis [12] The N-acylation of PE (e.g. by NAT activity) can be measured by incubating (at 37 C in 50mM Tris-HCl, ph 8.0 and 0.1% Triton X-100) a protein preparation and synthetic 1,2-diheptadecanoyl-sn-glycero-3-phosphoethanolamine, which is used as a substrate. The reaction product, 1,2-diheptadecanoyl-sn-glycero-3- phosphoethanolamine-n-arachidonoyl, can be measured by LC/MS using 1,2-dipalmitoyl-snglycero-3-phosphoethanolamine-N-heptadecanoyl as an internal standard NAPE hydrolysis [14] The rate of NAPE hydrolysis (e.g. by NAPE-PLD activity) can be determined by incubating (at 37 C in 50 mm Tris-HCl, ph 7.4 and 0.1% Triton X-100, 1 mm phenylmethylsulphonylfluoride) a protein preparation in the presence of the synthetic 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-heptadecanoyl, which is used as substrate and can be synthesized as previously reported [12]. The final product, heptadecanoylethanolamide, can be monitored by LC/MS using d 4 - heptadecanoylethanolamide as an internal standard Anandamide degradation Anandamide hydrolysis (e.g. by FAAH activity) can be measured by incubating (at 37 C in 50mM Tris-HCl, ph 8.0 and 0.05% fatty acid-free bovine serum albumin) a protein preparation in the presence of synthetic heptadecanoylethanolamide, which is used as substrate and can be synthesized as previously reported [14]. The final product, heptadecanoic acid (C17:0), can be monitored by LC/MS using heptadecenoic acid (C17:1, Δ 10 ) as internal standard. 4. Metabolism of 2-AG 2-AG biosynthesis in neural cells is driven by a rise in intracellular calcium levels and/or activation of G q/11 -coupled receptors [2,15,69], and proceeds through a series of pathways that are distinct from those involved in anandamide production (Fig. 8). In the following section, we review current applications for the lipidomic analysis of 2-AG metabolism in human brain samples (Section 3) Lipid remodeling of 2-AG precursors 2-AG formation requires the generation of arachidonic acid-containing DAGs and lysoglycerophospholipid species (Fig. 8) DAGs DAGs can derive from three main enzymatic routes: 1) PLC-mediated hydrolysis of membrane phospholipids, particularly phosphoinositides; 2) phosphatase-
9 Astarita and Piomelli Page AG formation AG degradation mediated hydrolysis of phosphatidic acid (PA); 3) lipase-mediated hydrolysis of TAGs (Fig. 8). The fatty acid compositions of DAGs formed by these various routes reflect the composition of the parent lipid. In particular, those derived from inositol phospholipids are highly enriched in molecular species containing stearic acid in position sn-1 and arachidonic acid in position sn-2 (Fig. 9A) [15]. However, also phosphatidylcholine (Fig. 9B) and PA may serve as source of 1-acyl,2-arachidonoyl-DAG [70-72]. Furthermore, our analysis reveal that several arachidonoyl-containing TAG species are present in human brain, which might contribute to DAG and 2-AG formation (Fig.10). The biological significance of neuronal TAGs is unclear at present. Finally, recent evidence suggests the existence of preformed DAG pools in neurons, which may be directly hydrolyzed to 2-AG in response to stimuli [15,73] Lysophospholipids Arachidonoyl-containing lysophospholipids may contribute to 2-AG formation [74]. Such lysophospholipids may derive from the PLA 1 -mediated hydrolysis of phospholipids (Fig. 8). Three biosynthetic pathways have been proposed for 2-AG formation (Fig. 8) starting either from glycerolipids or glycerophospholipids: 1) diacylglycerol lipase (DGL)-mediated hydrolysis of 1-acyl,2-arachidonoyl-DAGs (Fig. 11) [75]; 2) lyso-plc-mediated hydrolysis of arachidonoyl-containing lysophospholipids (e.g. 2-arachidonoyl phosphatidylcholine) [74]; and 3) phosphatase-mediated hydrolysis of 2-arachidonoyl-lysophosphatidic acid (LPA) [71]. LPA may derive either from the PLA 1 -mediated hydrolysis of PA or the direct hydrolysis of lysophospholipids by a lyso-pld. The relative importance of each of these routes to the biosynthesis of 2-AG (Fig. 11) is currently under investigation. 2-AG is rapidly internalized by neuronal cells, and, after uptake, is cleaved by serine hydrolases to generate free arachidonic acid and glycerol. Approximately 85% of the 2-AG-hydrolyzing activity in the murine brain is accounted for by monoacylglycerol lipase (MGL) [76-78]. The remaining activity has been ascribed to α/β hydrolase-6 (ABHD-6) and α/β hydrolase-12 (ABHD-12) [76]. Notably, because 2-AG levels are in the order of nmoles per gram of tissue in most biological tissues, its hydrolysis can provide a significant source of arachidonic acid and eicosanoids (Fig. 8) [15,73] Oxidative metabolism As for anandamide, 2-AG can be metabolized by oxidative enzymes as effectively as arachidonic acid. LOXs-, COX-2-, and CYP450-mediated 2-AG oxygenation provides hydroxyperoxy, hydroxy, prostaglandin and epoxide metabolites (Fig. 8) [47,79-81]. Notably, because of the high physiological concentrations of 2-AG, the oxidative metabolism of this endocannabinoid may be physiologically relevant, and may give rise to a novel class of signal mediators [82,83]. For example, 2-(14,15-epoxyeicosatrienoyl)-glycerol, which is a 2-AG metabolite produced through action of CYP450, has been shown to bind with high affinity to cannabinoid receptors and exerts mitogenic activity [84]. Moreover, the hydrolysis of oxygenated derivatives of 2-AG could contribute to the mobilization of other eicosanoids (i.e. prostaglandin, hydroxy and epoxide derivatives of arachidonic acid) [45,46,82] Anabolic metabolism 2-AG can be converted into complex lipids by anabolic enzymes such as MAG kinases or acyltransferases, which generate 2-arachidonoyl-LPA and DAG, respectively. These lipids can be further converted into glycerophospholipids (through the cytidine diphosphate-dag pathway) or TAG (Fig. 8).
10 Astarita and Piomelli Page Ether and esters of glycerol 2-AG analogs include both cannabimimetic and non-cannabimimetic molecules Cannabimimetic analogs 2-Arachidonoyl glyceryl-ether or noladin ether is a metabolically stable analog of 2-AG that binds to cannabinoid receptors with high affinity [85]. This lipid was found in brain extracts, but this finding was later called into question [86,87] Non-cannabimimetic analogs 2-AG biosynthesis is accompanied by formation of various 2-acylglycerol esters such as 2-oleoyl-sn-glycerol and 2-linoleyl-sn-glycerol [15]. Although these molecules show no distinct activity at cannabinoid receptors, they might potentiate the activity of 2-AG by reducing its lipase-mediated hydrolysis [88] Assays for the metabolism of 2-AG 5. Conclusions Acknowledgments References Enzymatic assays of 2-AG metabolism that use fast and ultra-fast LC/MS analysis are reviewed below [15,89] DAG hydrolysis [15] DAG hydrolysis (e.g., by DGL activity) can be measured by incubating (at 37 C in 50 mm Tris-HCl, ph 7.0, containing 0.1% Triton X-100) a protein preparation and synthetic diheptadecanoin as substrate. The final product, monoheptanoyl glycerol, can be monitored by LC/MS using d 8-2AG as internal standard AG hydrolysis [89] 2-AG hydrolysis (e.g., by MGL activity) can be measured by incubating (at 37 C in 50mM Tris-HCl buffer, ph 8.0 containing 0.05% bovine serum albumin) a protein preparation and 2-oleoyl-sn-glycerol as substrate. The final product, oleic acid, can be monitored by LC/MS using heptadecanoic acid as internal standard. The use of LC/MS-based lipidomic approaches is leading to a better understanding of the interconnected lipid pathways that regulate endocannabinoid metabolism. Many questions still remain on the regulation of the biosynthesis and degradation of endocannabinoids. It can be assumed that the endocannabinoid field will greatly benefit from the development of new technologies and applications for the analysis of lipids. The contribution of the Agilent Technologies/University of California Irvine Analytical Discovery Facility, Center for Drug Discovery, the Agilent Technologies Foundation, Advanced Chemistry Development, Inc., the Institute for Brain Aging & Dementia and the UC Irvine Alzheimer s Disease Research Center is gratefully acknowledged. The authors would like also to thank Dr Faizy Ahmed for the numerous discussions and invaluable advices on chromatography. This work was supported by grants from the National Institute of Health (R21DA , R01DK , R01 DA , R01DA , RR / , RR / , 1RL1AA to D.P. and UCI ADRC P50 AG16573). 1. Katona I, Freund TF. Nat Med 2008;14:923. [PubMed: ] 2. Di Marzo V. Nat Rev Drug Discov 2008;7:438. [PubMed: ] 3. Piomelli D, Astarita G, Rapaka R. Nat Rev Neurosci 2007;8:743. [PubMed: ] 4. Devane WA, Hanus L, Breuer A, Pertwee RG, Stevenson LA, Griffin G, Gibson D, Mandelbaum A, Etinger A, Mechoulam R. Science 1992;258:1946. [PubMed: ]
11 Astarita and Piomelli Page Okamoto Y, Morishita J, Wang J, Schmid PC, Krebsbach RJ, Schmid HH, Ueda N. Biochem J 2005;389:241. [PubMed: ] 6. Giuffrida A, Piomelli D. FEBS Lett 1998;422:373. [PubMed: ] 7. Giuffrida, A.; Piomelli, D. Lipid Second Messengers. Laychock, SG.; Rubin, RP., editors. CRC Press LLC; Boca Raton, FL: p Hansen HH, Hansen SH, Bjornsdottir I, Hansen HS. J Mass Spectrom 1999;34:761. [PubMed: ] 9. Schreiber D, Harlfinger S, Nolden BM, Gerth CW, Jaehde U, Schomig E, Klosterkotter J, Giuffrida A, Astarita G, Piomelli D, Markus Leweke F. Anal Biochem 2007;361:162. [PubMed: ] 10. Williams J, Wood J, Pandarinathan L, Karanian DA, Bahr BA, Vouros P, Makriyannis A. Anal Chem 2007;79:5582. [PubMed: ] 11. Lam PMW, Marczylo TH, El-Talatini M, Finney M, Nallendran V, Taylor AH, Konje JC. Analytical Biochemistry Astarita G, Ahmed F, Piomelli D. J Lipid Res 2008;49:48. [PubMed: ] 13. Weber A, Ni J, Ling KHJ, Acheampong A, Tang-Liu DDS, Burk R, Cravatt BF, Woodward D. Journal of Lipid Research 2004;45:757. [PubMed: ] 14. Fu J, Astarita G, Gaetani S, Kim J, Cravatt BF, Mackie K, Piomelli D. J Biol Chem 2007;282:1518. [PubMed: ] 15. Jung KM, Astarita G, Zhu C, Wallace M, Mackie K, Piomelli D. Mol Pharmacol 2007;72:612. [PubMed: ] 16. Marczylo TH, Lam PM, Nallendran V, Taylor AH, Konje JC. Anal Biochem 2009;384:106. [PubMed: ] 17. Kingsley PJ, Marnett LJ. Lipidomics and Bioactive Lipids: Specialized Analytical Methods and Lipids in Disease Wakelam MJ, Pettitt TR, Postle AD. Methods Enzymol 2007;432:233. [PubMed: ] 19. Christie, WW. Advances in lipid methodology Two. Dundee: Oily Press; p Schmid P, Calvert J, Steiner R. Physiol Chem Phys 1973;5: Bjerve KS, Daae LNW, Bremer J. Analytical Biochemistry 1974;58:238. [PubMed: ] 22. Radin NS. Methods Enzymol 1981;72:5. [PubMed: ] 23. Hara A, Radin NS. Analytical Biochemistry 1978;90:420. [PubMed: ] 24. Carlson LA. Clinica chimica acta 1985;149: Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D. The Journal of Lipid Research 2008;49: Folch J, Lees M, Stanley GHS. Journal of Biological Chemistry 1957;226:497. [PubMed: ] 27. Wenk MR, Lucast L, Di Paolo G, Romanelli AJ, Suchy SF, Nussbaum RL, Cline GW, Shulman GI, McMurray W, De Camilli P. Nat Biotechnol 2003;21:813. [PubMed: ] 28. Cadas H, di Tomaso E, Piomelli D. J Neurosci 1997;17:1226. [PubMed: ] 29. Cadas H, Gaillet S, Beltramo M, Venance L, Piomelli D. J Neurosci 1996;16:3934. [PubMed: ] 30. Cadas H, Schinelli S, Piomelli D. J Lipid Mediat Cell Signal 1996;14:63. [PubMed: ] 31. Wang J, Okamoto Y, Morishita J, Tsuboi K, Miyatake A, Ueda N. J Biol Chem 2006;281: [PubMed: ] 32. Simon GM, Cravatt BF. J Biol Chem 2006;281: [PubMed: ] 33. Leung D, Saghatelian A, Simon GM, Cravatt BF. Biochemistry 2006;45:4720. [PubMed: ] 34. Sun YX, Tsuboi K, Okamoto Y, Tonai T, Murakami M, Kudo I, Ueda N. Biochem J 2004;Pt 35. Liu J, Wang L, Harvey-White J, Huang BX, Kim HY, Luquet S, Palmiter RD, Krystal G, Rai R, Mahadevan A, Razdan RK, Kunos G. Neuropharmacology Liu J, Wang L, Harvey-White J, Osei-Hyiaman D, Razdan R, Gong Q, Chan AC, Zhou Z, Huang BX, Kim HY, Kunos G. Proc Natl Acad Sci U S A 2006;103: [PubMed: ] 37. Piomelli D. Nat Rev Neurosci 2003;4:873. [PubMed: ]
12 Astarita and Piomelli Page Piomelli D, Beltramo M, Glasnapp S, Lin SY, Goutopoulos A, Xie XQ, Makriyannis A. Proc Natl Acad Sci U S A 1999;96:5802. [PubMed: ] 39. Beltramo M, Stella N, Calignano A, Lin SY, Makriyannis A, Piomelli D. Science 1997;277:1094. [PubMed: ] 40. Cravatt BF, Giang DK, Mayfield SP, Boger DL, Lerner RA, Gilula NB. Nature 1996;384:83. [PubMed: ] 41. Wei BQ, Mikkelsen TS, McKinney MK, Lander ES, Cravatt BF. J Biol Chem 2006;281: [PubMed: ] 42. McKinney MK, Cravatt BF. Annu Rev Biochem 2005;74:411. [PubMed: ] 43. Want EJ, Cravatt BF, Siuzdak G. Chembiochem 2005;6:1941. [PubMed: ] 44. Saghatelian A, Trauger SA, Want EJ, Hawkins EG, Siuzdak G, Cravatt BF. Biochemistry 2004;43: [PubMed: ] 45. Kozak KR, Crews BC, Morrow JD, Wang LH, Ma YH, Weinander R, Jakobsson PJ, Marnett LJ. Journal of Biological Chemistry 2002;277: [PubMed: ] 46. Kozak KR, Crews BC, Ray JL, Tai HH, Morrow JD, Marnett LJ. Journal of Biological Chemistry 2001;276: [PubMed: ] 47. Kozak KR, Prusakiewicz JJ, Marnett LJ. Current Pharmaceutical Design 2004;10:659. [PubMed: ] 48. Snider NT, Sikora MJ, Sridar C, Feuerstein TJ, Rae JM, Hollenberg PF. Journal of Pharmacology and Experimental Therapeutics Hampson AJ, Hill WA, Zan-Phillips M, Makriyannis A, Leung E, Eglen RM, Bornheim LM. Biochim Biophys Acta 1995;1259:173. [PubMed: ] 50. Fowler CJ. British Journal of Pharmacology 2007;152:594. [PubMed: ] 51. Snider NT, Kornilov AM, Kent UM, Hollenberg PF. Journal of Pharmacology and Experimental Therapeutics 2007;321:590. [PubMed: ] 52. Ueda N, Yamamoto K, Yamamoto S, Tokunaga T, Shirakawa E, Shinkai H, Ogawa M, Sato T, Kudo I, Inoue K, et al. Biochim Biophys Acta 1995;1254:127. [PubMed: ] 53. Hanus L, Gopher A, Almog S, Mechoulam R. J Med Chem 1993;36:3032. [PubMed: ] 54. Felder CC, Joyce KE, Briley EM, Mansouri J, Mackie K, Blond O, Lai Y, Ma AL, Mitchell RL. Molecular Pharmacology 1995;48:443. [PubMed: ] 55. Farrell EK, Merkler DJ. Drug Discovery Today 2008;13:558. [PubMed: ] 56. Schwartz GJ, Fu J, Astarita G, Li X, Gaetani S, Campolongo P, Cuomo V, Piomelli D. Cell Metab 2008;8:281. [PubMed: ] 57. Astarita G, Di Giacomo B, Gaetani S, Oveisi F, Compton TR, Rivara S, Tarzia G, Mor M, Piomelli D. J Pharmacol Exp Ther 2006;318:563. [PubMed: ] 58. LoVerme J, Russo R, La Rana G, Fu J, Farthing J, Mattace-Raso G, Meli R, Hohmann A, Calignano A, Piomelli D. J Pharmacol Exp Ther 2006;319:1051. [PubMed: ] 59. Lo Verme J, Fu J, Astarita G, La Rana G, Russo R, Calignano A, Piomelli D. Mol Pharmacol 2005;67:15. [PubMed: ] 60. Fu J, Gaetani S, Oveisi F, Lo Verme J, Serrano A, Rodriguez de Fonseca F, Rosengarth A, Luecke H, Di Giacomo B, Tarzia G, Piomelli D. Nature 2003;425:90. [PubMed: ] 61. Maccarrone M, Cartoni A, Parolaro D, Margonelli A, Massi P, Bari M, Battista N, Finazzi-Agrò A. Molecular and Cellular Neuroscience 2002;21:126. [PubMed: ] 62. Terrazzino S, Berto F, Carbonare MD, Fabris M, Guiotto A, Bernardini D, Leon A. FASEB J 2004:1580. [PubMed: ] 63. Dalle Carbonare M, Del Giudice E, Stecca A, Colavito D, Fabris M, D Arrigo A, Bernardini D, Dam M, Leon A. Journal of Neuroendocrinology 2008;20:26. [PubMed: ] 64. Astarita G, Rourke BC, Andersen JB, Fu J, Kim JH, Bennett AF, Hicks JW, Piomelli D. Am J Physiol Regul Integr Comp Physiol 2006;290:R1407. [PubMed: ] 65. Cravatt BF, Próspero-García O, Siuzdak G, Gilula NB, Heriksen SJ, Boger DL, Lerner RA. Science 1995;268:1506. [PubMed: ] 66. Vuong LAQ, Mitchell VA, Vaughan CW. Neuropharmacology 2008;54:189. [PubMed: ]
13 Astarita and Piomelli Page Burstein S. Neuropharmacology McDonald GR, Hudson AL, Dunn SM, You H, Baker GB, Whittal RM, Martin JW, Jha A, Edmondson DE, Holt A. Science 2008;322:917. [PubMed: ] 69. Jung KM, Mangieri R, Stapleton C, Kim J, Fegley D, Wallace M, Mackie K, Piomelli D. Mol Pharmacol 2005;68:1196. [PubMed: ] 70. Bisogno T, Melck D, De Petrocellis L, Di Marzo V. Journal of Neurochemistry 1999;72:2113. [PubMed: ] 71. Nakane S, Oka S, Arai S, Waku K, Ishima Y, Tokumura A, Sugiura T. Arch Biochem Biophys 2002;402:51. [PubMed: ] 72. Oka S, Yanagimoto S, Ikeda S, Gokoh M, Kishimoto S, Waku K, Ishima Y, Sugiura T. Journal of Biological Chemistry 2005;280: [PubMed: ] 73. Nomura DK, Hudak CSS, Ward AM, Burston JJ, Issa RS, Fisher KJ, Abood ME, Wiley JL, Lichtman AH, Casida JE. Bioorganic & Medicinal Chemistry Letters Sugiura T, Kishimoto S, Oka S, Gokoh M. Progress in Lipid Research 2006;45:405. [PubMed: ] 75. Bisogno T, Howell F, Williams G, Minassi A, Cascio MG, Ligresti A, Matias I, Schiano-Moriello A, Paul P, Williams EJ, Gangadharan U, Hobbs C, Di Marzo V, Doherty P. J Cell Biol 2003;163:463. [PubMed: ] 76. Blankman JL, Simon GM, Cravatt BF. Chemistry & Biology 2007;14:1347. [PubMed: ] 77. Dinh TP, Kathuria S, Piomelli D. Molecular Pharmacology 2004;66:1260. [PubMed: ] 78. Dinh TP, Carpenter D, Leslie FM, Freund TF, Katona I, Sensi SL, Kathuria S, Piomelli D. Proc Natl Acad Sci U S A 2002;99: [PubMed: ] 79. Kozak KR, Gupta RA, Moody JS, Ji C, Boeglin WE, DuBois RN, Brash AR, Marnett LJ. J Biol Chem 2002;15: Moody JS, Kozak KR, Ji C, Marnett LJ. Biochemistry 2001;40:861. [PubMed: ] 81. Kozak KR, Rowlinson SW, Marnett LJ. J Biol Chem 2000;275: [PubMed: ] 82. Kozak KR, Marnett LJ. Prostaglandins Leukot Essent Fatty Acids 2002;66:211. [PubMed: ] 83. Woodward DF, Carling RWC, Cornell CL, Fliri HG, Martos JL, Pettit SN, Liang Y, Wang JW. Pharmacology and Therapeutics Chen JK, Chen J, Imig JD, Wei S, Hachey DL, Guthi JS, Falck JR, Capdevila JH, Harris RC. J Biol Chem Hanus L, Abu-Lafi S, Fride E, Breuer A, Vogel Z, Shalev DE, Kustanovich I, Mechoulam R. Proc Natl Acad Sci U S A 2001;98:3662. [PubMed: ] 86. Fezza F, Bisogno T, Minassi A, Appendino G, Mechoulam R, Di Marzo V. FEBS Lett 2002;513:294. [PubMed: ] 87. Oka S, Tsuchie A, Tokumura A, Muramatsu M, Suhara Y, Takayama H, Waku K, Sugiura T. J Neurochem 2003;85:1374. [PubMed: ] 88. Ben-Shabat S, Fride E, Sheskin T, Tamiri T, Rhee MH, Vogel Z, Bisogno T, De Petrocellis L, Di Marzo V, Mechoulam R. European Journal of Pharmacology 1998;353:23. [PubMed: ] 89. King AR, Duranti A, Tontini A, Rivara S, Rosengarth A, Clapper JR, Astarita G, Geaga JA, Luecke H, Mor M, Tarzia G, Piomelli D. Chem Biol 2007;14:1357. [PubMed: ] 90. Larsen A, Uran S, Jacobsen PB, Skotland T. Rapid Commun Mass Spectrom 2001;15:2393. [PubMed: ] 91. Stella N, Schweitzer P, Piomelli D. Nature 1997;388:773. [PubMed: ]
14 Astarita and Piomelli Page 13 Figure 1. Lipidomic analysis of the endocannabinoid metabolism Flow chart of the strategy used to profile endocannabinoid lipids from biological samples. Abbreviations: DAG, diacylglycerol; FA, fatty acid; FAE, fatty acid ethanolamide; MAG, monoacylglycerol; NAPE, N-acyl-phosphatidylethanolamine; LNAPE, lyso-nape; oxfa, oxygenated fatty acids; oxfae, oxygenated FAE; oxmag, oxygenated MAG; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PI, phosphatidylinositol; TAG, triacylglycerol.
15 Astarita and Piomelli Page 14 Figure 2. LC/MS metabolite discovery tools A, Representative total ion LC/MS chromatogram of a mixture of isobaric and isomeric N-acyl phosphatidylethanolamine (NAPE) species (top). A 3D LC/MS contour mapping (bottom) visualizes individual molecular species present in the mixture. The first dimension is elution time, the second is mass-to-charge ratio (m/z) and the third relative intensity of the signal, which is symbolized with a pseudocolor scale. LC/MS n conditions are those previously reported [12]. Three-dimensional maps were generated using MS Processor from Advanced Chemistry Development, Inc. (Toronto, Canada). B, Representative ESI/MS n analysis of N- arachidonoyl phosphoethanolamide species in the rat brain. MS 2 and MS 3 lead to fragment ions that are found physiologically as neutral molecular species: lyso-nape (LNAPE), phosphoanandamide (PAEA), oleic acid (OA), stearic aldehyde (SAd), cycliclysophosphatidic acid (clpa) and a cyclic glycerophospho-anandamide (cgpaea, which has not yet been identified in mammals).
16 Astarita and Piomelli Page 15 Figure 3. Anandamide metabolism: an overview Postulated pathways of anandamide metabolism. Abbreviations: PC, phosphatidylcholine; PE, phosphatidylethanolamine; NAT, N-acyl transferase; LPA, lysophosphatidic acid; PA, phosphatidic acid; NAPE, N-acyl-phosphatidylethanolamine; LNAPE, 1-lyso,2-acyl-snglycero-3-phosphoethanolamine-N-acyl; GP-anadamide, glycerophospho-anandamide; PAEA, phosphoanandamide; PLA, phospholipase A; ABHD, α/β hydrolase; NAPE-PLD, NAPE phospholipase D; PLC, phospholipase C; FAAH, fatty acid amide hydrolase; P, phosphatase; PG, prostaglandin; EET, epoxyeicosatrienoic acid; HETE, hydroxyeicosatetraenoic acid; COX, cyclooxygenase; PGS, prostaglandin synthase; LOX, lipoxygenase; CYP450; cytochrome P450, PDE, phosphodiesterase.
17 Astarita and Piomelli Page 16 Figure 4. Lipid remodeling and anandamide metabolism: analysis of phosphatidylethanolamine (PE) species Representative LC/MS analysis of PE species in a human brain. LC/MS chromatogram (top) and MS 2 fragmentation pattern using an ion trap mass spectrometer (bottom). PE species were detected as deprotonated molecular ions in the negative mode. In MS 2, the most prominent fragments are the sn-2 lyso-pe in combination with the sn-1 and sn-2 carboxylate anions [90]. Sample characteristics are described in the text. Lipids were extracted from approximately 50 mg of brain tissue and resuspended in 0.1 ml of methanol. Injection volume was 10 μl. Separation was performed on a SB300 Poroshell column (Agilent-Technologies coating layer of 0.25 μm on total particle diameter of 5 μm) using a linear gradient of methanol in water containing 0.25% acetic acid and 5 mm ammonium acetate (from 85% to 100% of methanol in 5 min) at a flow rate of 1.0 ml/min with column temperature set at 50 C. Capillary voltage was 4.5kV, skim1-40v, and capillary exit -151V. N 2 was used as drying gas at a flow rate of 12 liters/min, temperature of 350 C and nebulizer pressure of 80 PSI. Helium was used as collision gas. Abbreviations: R 1 =sn-1 aliphatic chain; R 2 =sn-2 aliphatic chain.
18 Astarita and Piomelli Page 17 Figure 5. Lipid remodeling and anandamide metabolism: analysis of phosphatidylcholine (PC) species Identification of 1,2-diarachidonoyl PC in human brain samples. LC/MS chromatogram (top) and fragmentation pattern in MS 2 and MS 3 using an ion trap instrument (bottom). Sample characteristics are described in the text and LC/MS conditions in Fig. 4 legend. Lipids were extracted from approximately 50 mg of brain tissue and resuspended in 0.1 ml of methanol. Injection volume was 10 μl. PC species were detected as acetate adducts of the molecular ions using ESI set in the negative mode. The MS 2 fragmentation pattern is characterized by neutral loss of the acetate adduct of the N-methyl group. MS 3 of the ion with m/z yields the lysophospholipid with neutral loss of ketene in combination with the sn-1 and sn-2 carboxylate anions. Abbreviations: R 1 =sn-1 aliphatic chain; R 2 =sn-2 aliphatic chain.
19 Astarita and Piomelli Page 18 Figure 6. Anandamide precursors: analysis of NAPE and lyso-nape species Identification of endogenous NAPE (A) and lyso-nape species (B) in a human brain sample using LC coupled to an ion trap instrument. Sample characteristics are described in the text and LC/MS conditions in Fig. 4 legend. Lipids were extracted from approximately 50 mg of brain tissue and resuspended in 0.1 ml of methanol. Injection volume was 10 μl. A 3D LC/MS contour mapping (A, top) of lipid extracts visualized the individual NAPE species, which were detected as deprotonated molecular ion in the negative ion mode. The first dimension is the elution time, the second is m/z ratio and the third the relative intensity of the signal, which is represented by a pseudocolor scale. In MS 2, the product ions are mainly sn-2 lysophospholipid with neutral loss of ketene, the sn-2 carboxylate anion and a series of characteristic minor fragments (A, bottom) [12]. Lyso-NAPE species were separated by PE and NAPE species using the chromatographic conditions described in Fig. 4 legend (B, top). In MS 2, the lyso- NAPE yielded the putative cyclic glycerophospho-anandamide and sn-1 carboxylate anions (B, bottom) [90].
20 Astarita and Piomelli Page 19 Figure 7. Anandamide analogs: fatty acid ethanolamides (FAEs) LC/MS n analysis of endogenous cannabimimetic and non-cannabimimetic fatty acid ethanolamides (FAEs) in human brain. Sample characteristics are described in the text. Lipids were extracted from approximately 50 mg of brain tissue and resuspended in 0.1 ml of methanol. Injection volume was 10 μl. FAEs were detected as sodium adducts of the molecular ions using ESI set in the positive mode. LC/MS conditions are as described [64].
Mass Spectrometry based metabolomics
 Mass Spectrometry based metabolomics Metabolomics- A realm of small molecules (
Mass Spectrometry based metabolomics Metabolomics- A realm of small molecules (
LC/MS Method for Comprehensive Analysis of Plasma Lipids
 Application Note omics LC/MS Method for Comprehensive Analysis of Plasma s Authors Tomas Cajka and Oliver Fiehn West Coast Metabolomics Center, University of California Davis, 451 Health Sciences Drive,
Application Note omics LC/MS Method for Comprehensive Analysis of Plasma s Authors Tomas Cajka and Oliver Fiehn West Coast Metabolomics Center, University of California Davis, 451 Health Sciences Drive,
The use of mass spectrometry in lipidomics. Outlines
 The use of mass spectrometry in lipidomics Jeevan Prasain jprasain@uab.edu 6-2612 utlines Brief introduction to lipidomics Analytical methodology: MS/MS structure elucidation of phospholipids Phospholipid
The use of mass spectrometry in lipidomics Jeevan Prasain jprasain@uab.edu 6-2612 utlines Brief introduction to lipidomics Analytical methodology: MS/MS structure elucidation of phospholipids Phospholipid
Mass-Spectrometric Analysis of Lipids (Lipidomics)
 Mass-Spectrometric Analysis of Lipids (Lipidomics) 1. Identification 2. Quantification 3. Metabolism Why to do lipidomics? Biology: Functions of different lipids? Medicine: Diagnostics and Therapy Industry:
Mass-Spectrometric Analysis of Lipids (Lipidomics) 1. Identification 2. Quantification 3. Metabolism Why to do lipidomics? Biology: Functions of different lipids? Medicine: Diagnostics and Therapy Industry:
Glycerolipid Analysis. LC/MS/MS Analytical Services
 Glycerolipid Analysis LC/MS/MS Analytical Services Molecular Characterization and Quantitation of Glycerophospholipids in Commercial Lecithins by High Performance Liquid Chromatography with Mass Spectrometric
Glycerolipid Analysis LC/MS/MS Analytical Services Molecular Characterization and Quantitation of Glycerophospholipids in Commercial Lecithins by High Performance Liquid Chromatography with Mass Spectrometric
Relative Quantitation of Human Polymorphonuclear Leukocyte Cell Membrane GPEtn Lipids
 Relative Quantitation of Human Polymorphonuclear Leukocyte Cell Membrane GPEtn Lipids Using the QTRAP System with mtraq Reagents Karin A. Zemski-Berry 1, John M. Hevko 2, and Robert C. Murphy 1 1 Department
Relative Quantitation of Human Polymorphonuclear Leukocyte Cell Membrane GPEtn Lipids Using the QTRAP System with mtraq Reagents Karin A. Zemski-Berry 1, John M. Hevko 2, and Robert C. Murphy 1 1 Department
SUPPLEMENTARY DATA. Materials and Methods
 SUPPLEMENTARY DATA Materials and Methods HPLC-UV of phospholipid classes and HETE isomer determination. Fractionation of platelet lipid classes was undertaken on a Spherisorb S5W 150 x 4.6 mm column (Waters
SUPPLEMENTARY DATA Materials and Methods HPLC-UV of phospholipid classes and HETE isomer determination. Fractionation of platelet lipid classes was undertaken on a Spherisorb S5W 150 x 4.6 mm column (Waters
Impact of Chromatography on Lipid Profiling of Liver Tissue Extracts
 Impact of Chromatography on Lipid Profiling of Liver Tissue Extracts Application Note Clinical Research Authors Mark Sartain and Theodore Sana Agilent Technologies, Inc. Santa Clara, California, USA Introduction
Impact of Chromatography on Lipid Profiling of Liver Tissue Extracts Application Note Clinical Research Authors Mark Sartain and Theodore Sana Agilent Technologies, Inc. Santa Clara, California, USA Introduction
Enrichment of Phospholipids from Biological Matrices with Zirconium Oxide-Modified Silica Sorbents
 Enrichment of Phospholipids from Biological Matrices with Zirconium Oxide-Modified Silica Sorbents Xiaoning Lu, Jennifer E. Claus, and David S. Bell Supelco, Div. of Sigma-Aldrich Bellefonte, PA 16823
Enrichment of Phospholipids from Biological Matrices with Zirconium Oxide-Modified Silica Sorbents Xiaoning Lu, Jennifer E. Claus, and David S. Bell Supelco, Div. of Sigma-Aldrich Bellefonte, PA 16823
New Frontiers for MS in Metabolipidomics
 New Frontiers for MS in Metabolipidomics Giuseppe Astarita, PhD Principal Scientist Discovery & Life Sciences Milford, MA USA 2011 Waters Corporation 1 Why Metabolipidomics? Biomedical Sciences o Biomarker
New Frontiers for MS in Metabolipidomics Giuseppe Astarita, PhD Principal Scientist Discovery & Life Sciences Milford, MA USA 2011 Waters Corporation 1 Why Metabolipidomics? Biomedical Sciences o Biomarker
Phospholipids Metabolism
 Chapter VI: Phospholipids Metabolism Dr. Sameh Sarray Hlaoui Phospholipids Features: Amphipatic: - Hydrophobic head: fatty acids - Hydropholic head: P group+ alcohol Composed of alcohol attached by a phosphodiester
Chapter VI: Phospholipids Metabolism Dr. Sameh Sarray Hlaoui Phospholipids Features: Amphipatic: - Hydrophobic head: fatty acids - Hydropholic head: P group+ alcohol Composed of alcohol attached by a phosphodiester
MEMBRANE LIPIDS I and II: GLYCEROPHOSPHOLIPIDS AND SPHINGOLIPIDS
 December 6, 2011 Lecturer: Eileen M. Lafer MEMBRANE LIPIDS I and II: GLYCEROPHOSPHOLIPIDS AND SPHINGOLIPIDS Reading: Stryer Edition 6: Chapter 26 Images: All images in these notes were taken from Lehninger,
December 6, 2011 Lecturer: Eileen M. Lafer MEMBRANE LIPIDS I and II: GLYCEROPHOSPHOLIPIDS AND SPHINGOLIPIDS Reading: Stryer Edition 6: Chapter 26 Images: All images in these notes were taken from Lehninger,
UC Irvine UC Irvine Previously Published Works
 UC Irvine UC Irvine Previously Published Works Title Carrier-mediated transport and enzymatic hydrolysis of the endogenous cannabinoid 2- arachidonylglycerol Permalink https://escholarship.org/uc/item/92w2j8v2
UC Irvine UC Irvine Previously Published Works Title Carrier-mediated transport and enzymatic hydrolysis of the endogenous cannabinoid 2- arachidonylglycerol Permalink https://escholarship.org/uc/item/92w2j8v2
Supplementary Information. Effects of Perfluorooctanoic Acid on Metabolic Profiles in Brain and Liver of Mouse by a
 Supplementary Information Effects of Perfluorooctanoic Acid on Metabolic Profiles in Brain and Liver of Mouse by a High-throughput Targeted Metabolomics Approach Nanyang Yu, Si Wei, *, Meiying Li, Jingping
Supplementary Information Effects of Perfluorooctanoic Acid on Metabolic Profiles in Brain and Liver of Mouse by a High-throughput Targeted Metabolomics Approach Nanyang Yu, Si Wei, *, Meiying Li, Jingping
Lipid Analysis. Andréina Laffargue, IRD CRYMCEPT Montpellier workshop, October 17th Introduction to lipid structures
 Lipid Analysis Andréina Laffargue, IRD CRYMCEPT Montpellier workshop, October 17th 2005 Introduction to lipid structures Fatty acids Acylglycerols Glycerophospholipids Sterols Strategies involved in lipid
Lipid Analysis Andréina Laffargue, IRD CRYMCEPT Montpellier workshop, October 17th 2005 Introduction to lipid structures Fatty acids Acylglycerols Glycerophospholipids Sterols Strategies involved in lipid
Lipid Class Separation Using UPC 2 /MS
 Michael D. Jones, 1,3 Giorgis Isaac, 1 Giuseppe Astarita, 1 Andrew Aubin, 1 John Shockcor, 1 Vladimir Shulaev, 2 Cristina Legido-Quigley, 3 and Norman Smith 3 1 Waters Corporation, Milford, MA, USA 2 Department
Michael D. Jones, 1,3 Giorgis Isaac, 1 Giuseppe Astarita, 1 Andrew Aubin, 1 John Shockcor, 1 Vladimir Shulaev, 2 Cristina Legido-Quigley, 3 and Norman Smith 3 1 Waters Corporation, Milford, MA, USA 2 Department
Principles of Shotgun Lipidomics
 Principles of Shotgun Lipidomics Xianlin Han Diabetes and Obesity Research Center Sanford-Burnham Medical Research Institute Lake Nona Orlando, FL 32827 What is shotgun lipidomics? Original definition
Principles of Shotgun Lipidomics Xianlin Han Diabetes and Obesity Research Center Sanford-Burnham Medical Research Institute Lake Nona Orlando, FL 32827 What is shotgun lipidomics? Original definition
A Definitive Lipidomics Workflow for Human Plasma Utilizing Off-line Enrichment and Class Specific Separation of Phospholipids
 A Definitive Lipidomics Workflow for Human Plasma Utilizing Off-line Enrichment and Class Specific Separation of Phospholipids Jeremy Netto, 1 Stephen Wong, 1 Federico Torta, 2 Pradeep Narayanaswamy, 2
A Definitive Lipidomics Workflow for Human Plasma Utilizing Off-line Enrichment and Class Specific Separation of Phospholipids Jeremy Netto, 1 Stephen Wong, 1 Federico Torta, 2 Pradeep Narayanaswamy, 2
Rapid Lipid Profiling of Serum by Reverse Phase UPLC-Tandem Quadrupole MS
 Rapid Lipid Profiling of Serum by Reverse Phase UPLC-Tandem Quadrupole MS Mark Ritchie and Evelyn Goh Waters Pacific Pte Ltd., Singapore A P P L I C AT ION B E N E F I T S Delivers a rapid 10-min MRM method
Rapid Lipid Profiling of Serum by Reverse Phase UPLC-Tandem Quadrupole MS Mark Ritchie and Evelyn Goh Waters Pacific Pte Ltd., Singapore A P P L I C AT ION B E N E F I T S Delivers a rapid 10-min MRM method
Lipid Analysis ISOLATION, SEPARATION, IDENTIFICATION AND. Bridgwater, England LIPIDOMIC ANALYSIS. Fourth Edition. Invergowrie, Dundee, Scotland
 Lipid Analysis ISOLATION, SEPARATION, IDENTIFICATION AND LIPIDOMIC ANALYSIS Fourth Edition WILLIAM W.CHRISTIE MRS Lipid Analysis Unit, Scottish Crop Research Institute, Dundee, Scotland Invergowrie, and
Lipid Analysis ISOLATION, SEPARATION, IDENTIFICATION AND LIPIDOMIC ANALYSIS Fourth Edition WILLIAM W.CHRISTIE MRS Lipid Analysis Unit, Scottish Crop Research Institute, Dundee, Scotland Invergowrie, and
Inactivation of N-Acyl Phosphatidylethanolamine Phospholipase D Reveals Multiple Mechanisms for the Biosynthesis of Endocannabinoids
 4720 Biochemistry 2006, 45, 4720-4726 Articles Inactivation of N-Acyl Phosphatidylethanolamine Phospholipase D Reveals Multiple Mechanisms for the Biosynthesis of Endocannabinoids Donmienne Leung, Alan
4720 Biochemistry 2006, 45, 4720-4726 Articles Inactivation of N-Acyl Phosphatidylethanolamine Phospholipase D Reveals Multiple Mechanisms for the Biosynthesis of Endocannabinoids Donmienne Leung, Alan
Essential Lipidomics Experiments using the LTQ Orbitrap Hybrid Mass Spectrometer
 Application Note: 367 Essential Lipidomics Experiments using the LTQ rbitrap Hybrid Mass Spectrometer Thomas Moehring 1, Michaela Scigelova 2, Christer S. Ejsing 3, Dominik Schwudke 3, Andrej Shevchenko
Application Note: 367 Essential Lipidomics Experiments using the LTQ rbitrap Hybrid Mass Spectrometer Thomas Moehring 1, Michaela Scigelova 2, Christer S. Ejsing 3, Dominik Schwudke 3, Andrej Shevchenko
Comprehensive Lipid Profiling of Human Liver Tissue Extracts of Non-Alcoholic Fatty Liver Disease
 Comprehensive Lipid Profiling of Human Liver Tissue Extracts of Non-Alcoholic Fatty Liver Disease Multiplexed Precursor Ion Scanning and LipidView Software Brigitte Simons 1 and Bianca Arendt 2 1 AB SCIEX,
Comprehensive Lipid Profiling of Human Liver Tissue Extracts of Non-Alcoholic Fatty Liver Disease Multiplexed Precursor Ion Scanning and LipidView Software Brigitte Simons 1 and Bianca Arendt 2 1 AB SCIEX,
High-throughput lipidomic analysis of fatty acid derived eicosanoids and N-acylethanolamines
 High-throughput lipidomic analysis of fatty acid derived eicosanoids and N-acylethanolamines Darren S. Dumlao, Matthew W. Buczynski, Paul C. Norris, Richard Harkewicz and Edward A. Dennis. Biochimica et
High-throughput lipidomic analysis of fatty acid derived eicosanoids and N-acylethanolamines Darren S. Dumlao, Matthew W. Buczynski, Paul C. Norris, Richard Harkewicz and Edward A. Dennis. Biochimica et
LC/MS/MS Separation of Cholesterol and Related Sterols in Plasma on an Agilent InfinityLab Poroshell 120 EC C18 Column
 Application Note Clinical Research LC/MS/MS Separation of Cholesterol and Related Sterols in Plasma on an Agilent InfinityLab Poroshell EC C8 Column Authors Rongjie Fu and Zhiming Zhang Agilent Technologies
Application Note Clinical Research LC/MS/MS Separation of Cholesterol and Related Sterols in Plasma on an Agilent InfinityLab Poroshell EC C8 Column Authors Rongjie Fu and Zhiming Zhang Agilent Technologies
Core E Analysis of Neutral Lipids from Human Plasma June 4, 2010 Thomas J. Leiker and Robert M. Barkley
 Core E Analysis of Neutral Lipids from Human Plasma June 4, 2010 Thomas J. Leiker and Robert M. Barkley This protocol describes the extraction and direct measurement of cholesterol esters (CEs) and triacylglycerols
Core E Analysis of Neutral Lipids from Human Plasma June 4, 2010 Thomas J. Leiker and Robert M. Barkley This protocol describes the extraction and direct measurement of cholesterol esters (CEs) and triacylglycerols
Supporting information
 Supporting information Figure legends Supplementary Table 1. Specific product ions obtained from fragmentation of lithium adducts in the positive ion mode comparing the different positional isomers of
Supporting information Figure legends Supplementary Table 1. Specific product ions obtained from fragmentation of lithium adducts in the positive ion mode comparing the different positional isomers of
O O H. Robert S. Plumb and Paul D. Rainville Waters Corporation, Milford, MA, U.S. INTRODUCTION EXPERIMENTAL. LC /MS conditions
 Simplifying Qual/Quan Analysis in Discovery DMPK using UPLC and Xevo TQ MS Robert S. Plumb and Paul D. Rainville Waters Corporation, Milford, MA, U.S. INTRODUCTION The determination of the drug metabolism
Simplifying Qual/Quan Analysis in Discovery DMPK using UPLC and Xevo TQ MS Robert S. Plumb and Paul D. Rainville Waters Corporation, Milford, MA, U.S. INTRODUCTION The determination of the drug metabolism
The detergent-solubilized and gel filtration purified rhodopsin was partitioned against
 Supplement Jastrzebska et al. Materials and Methods The detergent-solubilized and gel filtration purified rhodopsin was partitioned against H 2 O/MeOH/CHCl 3, and the bottom layer was removed, dried down,
Supplement Jastrzebska et al. Materials and Methods The detergent-solubilized and gel filtration purified rhodopsin was partitioned against H 2 O/MeOH/CHCl 3, and the bottom layer was removed, dried down,
Fast Separation of Triacylglycerols in Oils using UltraPerformance Convergence Chromatography (UPC 2 )
 Fast Separation of Triacylglycerols in Oils using UltraPerformance Convergence Chromatography (UPC 2 ) Mehdi Ashraf-Khorassani, 1 Larry T. Taylor, 1 Jinchuan Yang, 2 and Giorgis Isaac 2 1 Department of
Fast Separation of Triacylglycerols in Oils using UltraPerformance Convergence Chromatography (UPC 2 ) Mehdi Ashraf-Khorassani, 1 Larry T. Taylor, 1 Jinchuan Yang, 2 and Giorgis Isaac 2 1 Department of
The Raptor HILIC-Si Column
 The Raptor HILIC-Si Column With Raptor LC columns, Restek chemists became the first to combine the speed of superficially porous particles (also known as SPP or core-shell particles) with the resolution
The Raptor HILIC-Si Column With Raptor LC columns, Restek chemists became the first to combine the speed of superficially porous particles (also known as SPP or core-shell particles) with the resolution
CELLULAR METABOLISM. Metabolic pathways can be linear, branched, cyclic or spiral
 CHM333 LECTURE 24 & 25: 3/27 29/13 SPRING 2013 Professor Christine Hrycyna CELLULAR METABOLISM What is metabolism? - How cells acquire, transform, store and use energy - Study reactions in a cell and how
CHM333 LECTURE 24 & 25: 3/27 29/13 SPRING 2013 Professor Christine Hrycyna CELLULAR METABOLISM What is metabolism? - How cells acquire, transform, store and use energy - Study reactions in a cell and how
ARTICLE IN PRESS. Prostaglandins & other Lipid Mediators xxx (2014) xxx xxx. Contents lists available at ScienceDirect
 Prostaglandins & other Lipid Mediators xxx (2014) xxx xxx Contents lists available at ScienceDirect Prostaglandins and Other Lipid Mediators 1 2 3 Q1 4 5 6 7 Review Endocannabinoid metabolism by cytochrome
Prostaglandins & other Lipid Mediators xxx (2014) xxx xxx Contents lists available at ScienceDirect Prostaglandins and Other Lipid Mediators 1 2 3 Q1 4 5 6 7 Review Endocannabinoid metabolism by cytochrome
rapid communication Misidentification of prostamides as prostaglandins Michelle Glass, 1, * Jiwon Hong,* Timothy A. Sato, and Murray D.
 rapid communication Misidentification of prostamides as prostaglandins Michelle Glass, 1, * Jiwon Hong,* Timothy A. Sato, and Murray D. Mitchell*,, Department of Pharmacology* and Liggins Institute, University
rapid communication Misidentification of prostamides as prostaglandins Michelle Glass, 1, * Jiwon Hong,* Timothy A. Sato, and Murray D. Mitchell*,, Department of Pharmacology* and Liggins Institute, University
JNJ : selective and slowly reversible FAAH inhibitor. Central and Peripheral PK/PD
 JNJ-42165279: selective and slowly reversible FAAH inhibitor Central and Peripheral PK/PD The Endocannabinoid System Research initiated by efforts to elucidate the active substance of Cannabis (THC in
JNJ-42165279: selective and slowly reversible FAAH inhibitor Central and Peripheral PK/PD The Endocannabinoid System Research initiated by efforts to elucidate the active substance of Cannabis (THC in
Tandem mass spectrometry analysis of prostaglandins and isoprostanes
 Tandem mass spectrometry analysis of prostaglandins and isoprostanes Jeevan Prasain jprasain@uab.edu 6-2612 verview Introduction to PGs and their synthesis Mass spectrometry characterization of PGs and
Tandem mass spectrometry analysis of prostaglandins and isoprostanes Jeevan Prasain jprasain@uab.edu 6-2612 verview Introduction to PGs and their synthesis Mass spectrometry characterization of PGs and
A Novel Solution for Vitamin K₁ and K₂ Analysis in Human Plasma by LC-MS/MS
 A Novel Solution for Vitamin K₁ and K₂ Analysis in Human Plasma by LC-MS/MS By Shun-Hsin Liang and Frances Carroll Abstract Vitamin K₁ and K₂ analysis is typically complex and time-consuming because these
A Novel Solution for Vitamin K₁ and K₂ Analysis in Human Plasma by LC-MS/MS By Shun-Hsin Liang and Frances Carroll Abstract Vitamin K₁ and K₂ analysis is typically complex and time-consuming because these
Detection of Cannabinoids in Oral Fluid with the Agilent 7010 GC-MS/MS System
 Application Note Forensics, Workplace Drug Testing Detection of Cannabinoids in Oral Fluid with the Agilent 7010 GC-MS/MS System Authors Fred Feyerherm and Anthony Macherone Agilent Technologies, Inc.
Application Note Forensics, Workplace Drug Testing Detection of Cannabinoids in Oral Fluid with the Agilent 7010 GC-MS/MS System Authors Fred Feyerherm and Anthony Macherone Agilent Technologies, Inc.
Improved method for the quantification of lysophospholipids including enol ether
 Supplemental Material Improved method for the quantification of lysophospholipids including enol ether species by liquid chromatography-tandem mass spectrometry James G. Bollinger *, Hiromi Ii*, Martin
Supplemental Material Improved method for the quantification of lysophospholipids including enol ether species by liquid chromatography-tandem mass spectrometry James G. Bollinger *, Hiromi Ii*, Martin
Supplementary Material (ESI) for Chemical Communications This journal is (c) The Royal Society of Chemistry 2008
 Experimental Details Unless otherwise noted, all chemicals were purchased from Sigma-Aldrich Chemical Company and were used as received. 2-DOS and neamine were kindly provided by Dr. F. Huang. Paromamine
Experimental Details Unless otherwise noted, all chemicals were purchased from Sigma-Aldrich Chemical Company and were used as received. 2-DOS and neamine were kindly provided by Dr. F. Huang. Paromamine
Hydrolysis of Acylglycerols and Phospholipids of Milled Rice Surface Lipids During Storage 1
 RICE QUALITY AND PROCESSING Hydrolysis of Acylglycerols and Phospholipids of Milled Rice Surface Lipids During Storage 1 H.S. Lam and A. Proctor ABSTRACT The relative contribution of acylglycerols and
RICE QUALITY AND PROCESSING Hydrolysis of Acylglycerols and Phospholipids of Milled Rice Surface Lipids During Storage 1 H.S. Lam and A. Proctor ABSTRACT The relative contribution of acylglycerols and
Life Science and Chemical Analysis Solutions. Baljit Ubhi, Jeff Patrick, David Alonso, and Joe Shambaugh 1 2 3
 Complementary LC- and GC- Mass Spectrometry Techniques Provide Broader Coverage of the Metabolome Cross-Platform Metabolomics Data Analysis Combining AB SCIEX LC/MS, LECO GC/MS, and Genedata Software 1
Complementary LC- and GC- Mass Spectrometry Techniques Provide Broader Coverage of the Metabolome Cross-Platform Metabolomics Data Analysis Combining AB SCIEX LC/MS, LECO GC/MS, and Genedata Software 1
Ammonia chemical ionization mass spectrometry of intact diacyl phosphatidylcholine
 Ammonia chemical ionization mass spectrometry of intact diacyl phosphatidylcholine C. G. Crawford and R. D. Plattner Northern Regional Research Center, Agricultural Research Service, United States Department
Ammonia chemical ionization mass spectrometry of intact diacyl phosphatidylcholine C. G. Crawford and R. D. Plattner Northern Regional Research Center, Agricultural Research Service, United States Department
Application Note. Abstract. Authors. Pharmaceutical
 Analysis of xycodone and Its Metabolites-oroxycodone, xymorphone, and oroxymorphone in Plasma by LC/MS with an Agilent ZRBAX StableBond SB-C18 LC Column Application ote Pharmaceutical Authors Linda L.
Analysis of xycodone and Its Metabolites-oroxycodone, xymorphone, and oroxymorphone in Plasma by LC/MS with an Agilent ZRBAX StableBond SB-C18 LC Column Application ote Pharmaceutical Authors Linda L.
LC-Based Lipidomics Analysis on QTRAP Instruments
 LC-Based Lipidomics Analysis on QTRAP Instruments Junhua Wang and Paul RS Baker SCIEX LC-Based Lipidomics Analysis Topics Covered Lipid extraction techniques Hydrophilic Interaction Chromatography (HILIC)
LC-Based Lipidomics Analysis on QTRAP Instruments Junhua Wang and Paul RS Baker SCIEX LC-Based Lipidomics Analysis Topics Covered Lipid extraction techniques Hydrophilic Interaction Chromatography (HILIC)
Phospholipid characterization by a TQ-MS data based identification scheme
 P-CN1716E Phospholipid characterization by a TQ-MS data based identification scheme ASMS 2017 MP-406 Tsuyoshi Nakanishi 1, Masaki Yamada 1, Ningombam Sanjib Meitei 2, 3 1 Shimadzu Corporation, Kyoto, Japan,
P-CN1716E Phospholipid characterization by a TQ-MS data based identification scheme ASMS 2017 MP-406 Tsuyoshi Nakanishi 1, Masaki Yamada 1, Ningombam Sanjib Meitei 2, 3 1 Shimadzu Corporation, Kyoto, Japan,
Standardization of a LC/MS/MS Method for the Determination of Acyl Glucuronides and Their Isomers
 1 1 2 2 3 3 2 1 S.Bolze,.Lacombe, G.Durand, P.Chaimbault, F.Massière, C.Gay-Feutry,.Bromet * and T.Hulot 1. LIPHA, 115, avenue Lacassagne, 69003 Lyon, France 2. BITEC CETRE, 10 avenue aude Guillemin, 45071
1 1 2 2 3 3 2 1 S.Bolze,.Lacombe, G.Durand, P.Chaimbault, F.Massière, C.Gay-Feutry,.Bromet * and T.Hulot 1. LIPHA, 115, avenue Lacassagne, 69003 Lyon, France 2. BITEC CETRE, 10 avenue aude Guillemin, 45071
Lipids Analysis. Lipids
 Lipids Analysis Stephen Barnes 3 5 15 Lipids Lipids are mostly very hydrophobic Most are conjugates of fatty acids of a variety of chain lengths, which have different degrees of unsaturation, cis trans
Lipids Analysis Stephen Barnes 3 5 15 Lipids Lipids are mostly very hydrophobic Most are conjugates of fatty acids of a variety of chain lengths, which have different degrees of unsaturation, cis trans
Factors to Consider in the Study of Biomolecules
 Factors to Consider in the Study of Biomolecules What are the features of the basic building blocks? (ex: monosaccharides, alcohols, fatty acids, amino acids) 1) General structure and functional groups
Factors to Consider in the Study of Biomolecules What are the features of the basic building blocks? (ex: monosaccharides, alcohols, fatty acids, amino acids) 1) General structure and functional groups
Identification & Confirmation of Structurally Related Degradation Products of Simvastatin
 Identification & Confirmation of Structurally Related Degradation Products of Simvastatin Power of QTRAP Systems for Identification and Confirmation of Degradation Products Dilip Reddy 1, Chandra Sekar
Identification & Confirmation of Structurally Related Degradation Products of Simvastatin Power of QTRAP Systems for Identification and Confirmation of Degradation Products Dilip Reddy 1, Chandra Sekar
SUPPLEMENTARY INFORMATION FOR. (R)-Profens Are Substrate-Selective Inhibitors of Endocannabinoid Oxygenation. by COX-2
 SUPPLEMENTARY INFORMATION FOR (R)-Profens Are Substrate-Selective Inhibitors of Endocannabinoid Oxygenation by COX-2 Kelsey C. Duggan, Daniel J. Hermanson, Joel Musee, Jeffery J. Prusakiewicz, Jami L.
SUPPLEMENTARY INFORMATION FOR (R)-Profens Are Substrate-Selective Inhibitors of Endocannabinoid Oxygenation by COX-2 Kelsey C. Duggan, Daniel J. Hermanson, Joel Musee, Jeffery J. Prusakiewicz, Jami L.
Ozonolysis of phospholipid double bonds during electrospray. ionization: a new tool for structure determination
 Ozonolysis of phospholipid double bonds during electrospray ionization: a new tool for structure determination Michael C. Thomas, Todd W. Mitchell, Stephen J. Blanksby Departments of Chemistry and Biomedical
Ozonolysis of phospholipid double bonds during electrospray ionization: a new tool for structure determination Michael C. Thomas, Todd W. Mitchell, Stephen J. Blanksby Departments of Chemistry and Biomedical
SEPARATION OF BRANCHED PFOS ISOMERS BY UPLC WITH MS/MS DETECTION
 SEPARATION OF BRANCHED PFOS ISOMERS BY UPLC WITH MS/MS DETECTION Marian Twohig 1, Nicholas Ellor 1, Keith Worrall 2, Tim Jenkins 2, Gordon Kearney 2 1 Waters Corporation, 1 Cummings Center, Beverly, MA
SEPARATION OF BRANCHED PFOS ISOMERS BY UPLC WITH MS/MS DETECTION Marian Twohig 1, Nicholas Ellor 1, Keith Worrall 2, Tim Jenkins 2, Gordon Kearney 2 1 Waters Corporation, 1 Cummings Center, Beverly, MA
Metabolomic fingerprinting of serum samples by direct infusion mass spectrometry
 Metabolomic fingerprinting of serum samples by direct infusion mass spectrometry Raúl González-Domínguez * Department of Chemistry, Faculty of Experimental Sciences. University of Huelva, Spain. * Corresponding
Metabolomic fingerprinting of serum samples by direct infusion mass spectrometry Raúl González-Domínguez * Department of Chemistry, Faculty of Experimental Sciences. University of Huelva, Spain. * Corresponding
Test Bank for Lehninger Principles of Biochemistry 5th Edition by Nelson
 Test Bank for Lehninger Principles of Biochemistry 5th Edition by Nelson Link download full: http://testbankair.com/download/test-bank-forlehninger-principles-of-biochemistry-5th-edition-by-nelson/ Chapter
Test Bank for Lehninger Principles of Biochemistry 5th Edition by Nelson Link download full: http://testbankair.com/download/test-bank-forlehninger-principles-of-biochemistry-5th-edition-by-nelson/ Chapter
Simple and Robust Measurement of Blood Plasma Lysophospholipids Using Liquid Chromatography Mass Spectrometry
 LETTER Vol. 8, No. 4, 2017 ISSN 2233-4203/ e-issn 2093-8950 www.msletters.org Mass Spectrometry Letters Simple and Robust Measurement of Blood Plasma Lysophospholipids Using Liquid Chromatography Mass
LETTER Vol. 8, No. 4, 2017 ISSN 2233-4203/ e-issn 2093-8950 www.msletters.org Mass Spectrometry Letters Simple and Robust Measurement of Blood Plasma Lysophospholipids Using Liquid Chromatography Mass
BCM 221 LECTURES OJEMEKELE O.
 BCM 221 LECTURES BY OJEMEKELE O. OUTLINE INTRODUCTION TO LIPID CHEMISTRY STORAGE OF ENERGY IN ADIPOCYTES MOBILIZATION OF ENERGY STORES IN ADIPOCYTES KETONE BODIES AND KETOSIS PYRUVATE DEHYDROGENASE COMPLEX
BCM 221 LECTURES BY OJEMEKELE O. OUTLINE INTRODUCTION TO LIPID CHEMISTRY STORAGE OF ENERGY IN ADIPOCYTES MOBILIZATION OF ENERGY STORES IN ADIPOCYTES KETONE BODIES AND KETOSIS PYRUVATE DEHYDROGENASE COMPLEX
Metabolomics: quantifying the phenotype
 Metabolomics: quantifying the phenotype Metabolomics Promises Quantitative Phenotyping What can happen GENOME What appears to be happening Bioinformatics TRANSCRIPTOME What makes it happen PROTEOME Systems
Metabolomics: quantifying the phenotype Metabolomics Promises Quantitative Phenotyping What can happen GENOME What appears to be happening Bioinformatics TRANSCRIPTOME What makes it happen PROTEOME Systems
Chapter 8. Functions of Lipids. Structural Nature of Lipids. BCH 4053 Spring 2001 Chapter 8 Lecture Notes. Slide 1. Slide 2.
 BCH 4053 Spring 2001 Chapter 8 Lecture Notes 1 Chapter 8 Lipids 2 Functions of Lipids Energy Storage Thermal Insulation Structural Components of Membranes Protective Coatings of Plants and Insects Hormonal
BCH 4053 Spring 2001 Chapter 8 Lecture Notes 1 Chapter 8 Lipids 2 Functions of Lipids Energy Storage Thermal Insulation Structural Components of Membranes Protective Coatings of Plants and Insects Hormonal
Characterization of the fatty-acid amide hydrolase inhibitor URB597: Effects on
 JPET Fast This Forward. article has not Published been copyedited on and December formatted. The 3, final 2004 version as DOI:10.1124/jpet.104.078980 may differ from this version. JPET # 78980 Characterization
JPET Fast This Forward. article has not Published been copyedited on and December formatted. The 3, final 2004 version as DOI:10.1124/jpet.104.078980 may differ from this version. JPET # 78980 Characterization
2D-LC as an Automated Desalting Tool for MSD Analysis
 2D-LC as an Automated Desalting Tool for MSD Analysis Direct Mass Selective Detection of a Pharmaceutical Peptide from an MS-Incompatible USP Method Application Note Biologics and Biosimilars Author Sonja
2D-LC as an Automated Desalting Tool for MSD Analysis Direct Mass Selective Detection of a Pharmaceutical Peptide from an MS-Incompatible USP Method Application Note Biologics and Biosimilars Author Sonja
What You Can t See Can Hurt You. How MS/MS Specificity Can Bite Your Backside
 What You Can t See Can Hurt You How MS/MS Specificity Can Bite Your Backside Johan van den Heever, Tom Thompson, and Don Noot Agri-Food Laboratories Branch Advances in Trace rganic Residue Analysis early
What You Can t See Can Hurt You How MS/MS Specificity Can Bite Your Backside Johan van den Heever, Tom Thompson, and Don Noot Agri-Food Laboratories Branch Advances in Trace rganic Residue Analysis early
Detection of Lipid Peroxidation Products From Free Radical and Enzymatic Processes. Jason D. Morrow M.D. Vanderbilt University School of Medicine
 Detection of Lipid Peroxidation Products From Free Radical and Enzymatic Processes Jason D. Morrow M.D. Vanderbilt University School of Medicine Question? Which one of the following is the most accurate
Detection of Lipid Peroxidation Products From Free Radical and Enzymatic Processes Jason D. Morrow M.D. Vanderbilt University School of Medicine Question? Which one of the following is the most accurate
Determination of red blood cell fatty acid profiles in clinical research
 Application Note Clinical Research Determination of red blood cell fatty acid profiles in clinical research Chemical ionization gas chromatography tandem mass spectrometry Authors Yvonne Schober 1, Hans
Application Note Clinical Research Determination of red blood cell fatty acid profiles in clinical research Chemical ionization gas chromatography tandem mass spectrometry Authors Yvonne Schober 1, Hans
Automated Sample Preparation for Profiling Fatty Acids in Blood and Plasma using the Agilent 7693 ALS
 Automated Sample Preparation for Profiling Fatty Acids in Blood and Plasma using the Agilent 7693 ALS Application Note Clinical Research Authors Frank David and Bart Tienpont, Research Institute for Chromatography,
Automated Sample Preparation for Profiling Fatty Acids in Blood and Plasma using the Agilent 7693 ALS Application Note Clinical Research Authors Frank David and Bart Tienpont, Research Institute for Chromatography,
Biosynthesis of anandamide and related acylethanolamides in mouse J774 macrophages and N 18 neuroblastoma cells
 Biochem. J. (1996) 316, 977 984 (Printed in Great Britain) 977 Biosynthesis of anandamide and related acylethanolamides in mouse J774 macrophages and N 18 neuroblastoma cells Vincenzo DI MARZO* Luciano
Biochem. J. (1996) 316, 977 984 (Printed in Great Britain) 977 Biosynthesis of anandamide and related acylethanolamides in mouse J774 macrophages and N 18 neuroblastoma cells Vincenzo DI MARZO* Luciano
Fatty Acid Mass Spectrometry Protocol Updated 10/11/2007 By Daren Stephens
 Fatty Acid Mass Spectrometry Protocol Updated 10/11/2007 By Daren Stephens Synopsis: This protocol describes the standard method for extracting and quantifying free fatty acids found in cells and media
Fatty Acid Mass Spectrometry Protocol Updated 10/11/2007 By Daren Stephens Synopsis: This protocol describes the standard method for extracting and quantifying free fatty acids found in cells and media
Supporting Information (SI)
 Electronic Supplementary Material (ESI) for Analyst. This journal is The Royal Society of Chemistry 2015 Supporting Information (SI) Title: Optimization of Metabolite Extraction of Human Vein Tissue for
Electronic Supplementary Material (ESI) for Analyst. This journal is The Royal Society of Chemistry 2015 Supporting Information (SI) Title: Optimization of Metabolite Extraction of Human Vein Tissue for
Comprehensive Two-Dimensional HPLC and Informative Data Processing for Pharmaceuticals and Lipids
 PO-CON1576E Comprehensive Two-Dimensional HPLC and Informative Data Processing for Pharmaceuticals and Lipids HPLC 2015 PSB-MULTI-06 Yoshiyuki WATABE, Tetsuo IIDA, Daisuke NAKAYAMA, Kanya TSUJII, Saki
PO-CON1576E Comprehensive Two-Dimensional HPLC and Informative Data Processing for Pharmaceuticals and Lipids HPLC 2015 PSB-MULTI-06 Yoshiyuki WATABE, Tetsuo IIDA, Daisuke NAKAYAMA, Kanya TSUJII, Saki
Integration Of Metabolism
 Integration Of Metabolism Metabolism Consist of Highly Interconnected Pathways The basic strategy of catabolic metabolism is to form ATP, NADPH, and building blocks for biosyntheses. 1. ATP is the universal
Integration Of Metabolism Metabolism Consist of Highly Interconnected Pathways The basic strategy of catabolic metabolism is to form ATP, NADPH, and building blocks for biosyntheses. 1. ATP is the universal
Nafith Abu Tarboush DDS, MSc, PhD
 Nafith Abu Tarboush DDS, MSc, PhD natarboush@ju.edu.jo www.facebook.com/natarboush Lipids (cholesterol, cholesterol esters, phospholipids & triacylglycerols) combined with proteins (apolipoprotein) in
Nafith Abu Tarboush DDS, MSc, PhD natarboush@ju.edu.jo www.facebook.com/natarboush Lipids (cholesterol, cholesterol esters, phospholipids & triacylglycerols) combined with proteins (apolipoprotein) in
Designer Cannabinoids
 Liquid Chromatography Mass Spectrometry SSI-LCMS-010 Designer Cannabinoids LCMS-8030 Summary A rapid LC-MS-MS method for determination of designer cannabinoids in smokeable herbs was developed. Background
Liquid Chromatography Mass Spectrometry SSI-LCMS-010 Designer Cannabinoids LCMS-8030 Summary A rapid LC-MS-MS method for determination of designer cannabinoids in smokeable herbs was developed. Background
Choosing the metabolomics platform
 Choosing the metabolomics platform Stephen Barnes, PhD Department of Pharmacology & Toxicology University of Alabama at Birmingham sbarnes@uab.edu Challenges Unlike DNA, RNA and proteins, the metabolome
Choosing the metabolomics platform Stephen Barnes, PhD Department of Pharmacology & Toxicology University of Alabama at Birmingham sbarnes@uab.edu Challenges Unlike DNA, RNA and proteins, the metabolome
Metabolite identification in metabolomics: Database and interpretation of MSMS spectra
 Metabolite identification in metabolomics: Database and interpretation of MSMS spectra Jeevan K. Prasain, PhD Department of Pharmacology and Toxicology, UAB jprasain@uab.edu utline Introduction Putative
Metabolite identification in metabolomics: Database and interpretation of MSMS spectra Jeevan K. Prasain, PhD Department of Pharmacology and Toxicology, UAB jprasain@uab.edu utline Introduction Putative
Analysis of FAMEs Using Cold EI GC/MS for Enhanced Molecular Ion Selectivity
 APPLICATION NOTE Gas Chromatography/ Mass Spectrometry Author: Adam Patkin PerkinElmer, Inc. Shelton, CT Analysis of FAMEs Using GC/MS for Enhanced Molecular Ion Selectivity Introduction Characterization
APPLICATION NOTE Gas Chromatography/ Mass Spectrometry Author: Adam Patkin PerkinElmer, Inc. Shelton, CT Analysis of FAMEs Using GC/MS for Enhanced Molecular Ion Selectivity Introduction Characterization
Quantitative Analysis of Underivatized Amino Acids in Plant Matrix by Hydrophilic Interaction Chromatography (HILIC) with LC/MS Detection
 Application Note Food Testing, Metabolomics, Agricultural Chemistry, Environmental Quantitative Analysis of Underivatized Amino Acids in Plant Matrix by Hydrophilic Interaction Chromatography (HILIC) with
Application Note Food Testing, Metabolomics, Agricultural Chemistry, Environmental Quantitative Analysis of Underivatized Amino Acids in Plant Matrix by Hydrophilic Interaction Chromatography (HILIC) with
Lipids. Lipids. Jiří Jonák and Lenka Fialová Institute of Medical Biochemistry, 1st Medical Faculty of the Charles University, Prague
 Lipids Jiří Jonák and Lenka Fialová Institute of Medical Biochemistry, 1st Medical Faculty of the Charles University, Prague Lipids 1. General introduction 2. Nomenclature of fatty acids 3. Degradation
Lipids Jiří Jonák and Lenka Fialová Institute of Medical Biochemistry, 1st Medical Faculty of the Charles University, Prague Lipids 1. General introduction 2. Nomenclature of fatty acids 3. Degradation
Molecular BioSystems
 PAPER www.rsc.org/molecularbiosystems Molecular BioSystems Characterization of mice lacking candidate N-acyl ethanolamine biosynthetic enzymes provides evidence for multiple pathways that contribute to
PAPER www.rsc.org/molecularbiosystems Molecular BioSystems Characterization of mice lacking candidate N-acyl ethanolamine biosynthetic enzymes provides evidence for multiple pathways that contribute to
4th Multidimensional Chromatography Workshop Toronto (January, 2013) Herman C. Lam, Ph.D. Calibration & Validation Group
 4th Multidimensional Chromatography Workshop Toronto (January, 2013) Herman C. Lam, Ph.D. Calibration & Validation Group MDLC for Shotgun Proteomics Introduction General concepts Advantages Challenges
4th Multidimensional Chromatography Workshop Toronto (January, 2013) Herman C. Lam, Ph.D. Calibration & Validation Group MDLC for Shotgun Proteomics Introduction General concepts Advantages Challenges
Fatty Acid Amide Hydrolase Inhibitor Screening Assay Kit
 Fatty cid mide Hydrolase Inhibitor Screening ssay Kit Item No. 10005196 www.caymanchem.com Customer Service 800.364.9897 Technical Support 888.526.5351 1180 E. Ellsworth Rd nn rbor, MI US TBLE OF CONTENTS
Fatty cid mide Hydrolase Inhibitor Screening ssay Kit Item No. 10005196 www.caymanchem.com Customer Service 800.364.9897 Technical Support 888.526.5351 1180 E. Ellsworth Rd nn rbor, MI US TBLE OF CONTENTS
I. Structure and Properties of Lipids
 I. Structure and Properties of Lipids Lipids: A diverse group of compounds characterized by their low solubility in water and a high solubility in organic solvents such as chloroform and methanol. Nonpolar
I. Structure and Properties of Lipids Lipids: A diverse group of compounds characterized by their low solubility in water and a high solubility in organic solvents such as chloroform and methanol. Nonpolar
Direct determination of MCPD esters and glycidyl esters by LC-TOFMS
 1 AOCS Meeting May 16-19, Phoenix, AZ Direct determination of MCPD esters and glycidyl esters by LC-TOFMS Mark W. Collison, Ph. D., Director, Analytical Chemistry, Troy Haines, Analytical Laboratory Manager,
1 AOCS Meeting May 16-19, Phoenix, AZ Direct determination of MCPD esters and glycidyl esters by LC-TOFMS Mark W. Collison, Ph. D., Director, Analytical Chemistry, Troy Haines, Analytical Laboratory Manager,
PHOSPHOLIPIDS METABOLISM. BY Dr. Walid Said Zaki Dr. Marwa Ali LECTURER OF BIOCHEMISTRY AND MOLECULAR BIOLOGY
 PHOSPHOLIPIDS METABOLISM BY Dr. Walid Said Zaki Dr. Marwa Ali LECTURER OF BIOCHEMISTRY AND MOLECULAR BIOLOGY 1. State the definition and classification of Phospholipids. 2. Describe the general structure
PHOSPHOLIPIDS METABOLISM BY Dr. Walid Said Zaki Dr. Marwa Ali LECTURER OF BIOCHEMISTRY AND MOLECULAR BIOLOGY 1. State the definition and classification of Phospholipids. 2. Describe the general structure
The HPLC Preparative Scale-Up of Soybean Phospholipids Application
 The HPLC Preparative Scale-Up of Soybean Phospholipids Application Food and Flavors Authors Cliff Woodward and Ron Majors Agilent Technologies, Inc. 285 Centerville Road Wilmington, DE 1988-161 USA Abstract
The HPLC Preparative Scale-Up of Soybean Phospholipids Application Food and Flavors Authors Cliff Woodward and Ron Majors Agilent Technologies, Inc. 285 Centerville Road Wilmington, DE 1988-161 USA Abstract
Applying a Novel Glycan Tagging Reagent, RapiFluor-MS, and an Integrated UPLC-FLR/QTof MS System for Low Abundant N-Glycan Analysis
 Applying a Novel Glycan Tagging Reagent, RapiFluor-MS, and an Integrated UPLC-FLR/QTof MS System for Low Abundant N-Glycan Analysis Ying Qing Yu Waters Corporation, Milford, MA, USA APPLICATION BENEFITS
Applying a Novel Glycan Tagging Reagent, RapiFluor-MS, and an Integrated UPLC-FLR/QTof MS System for Low Abundant N-Glycan Analysis Ying Qing Yu Waters Corporation, Milford, MA, USA APPLICATION BENEFITS
Analysis of anti-epileptic drugs in human serum using an Agilent Ultivo LC/TQ
 Application Note Clinical Research Analysis of anti-epileptic drugs in human serum using an Agilent Ultivo LC/TQ Authors Jennifer Hitchcock 1, Lauren Frick 2, Peter Stone 1, and Vaughn Miller 2 1 Agilent
Application Note Clinical Research Analysis of anti-epileptic drugs in human serum using an Agilent Ultivo LC/TQ Authors Jennifer Hitchcock 1, Lauren Frick 2, Peter Stone 1, and Vaughn Miller 2 1 Agilent
Metabolite identification in metabolomics: Metlin Database and interpretation of MSMS spectra
 Metabolite identification in metabolomics: Metlin Database and interpretation of MSMS spectra Jeevan K. Prasain, PhD Department of Pharmacology and Toxicology, UAB jprasain@uab.edu Outline Introduction
Metabolite identification in metabolomics: Metlin Database and interpretation of MSMS spectra Jeevan K. Prasain, PhD Department of Pharmacology and Toxicology, UAB jprasain@uab.edu Outline Introduction
Introduction to LC/MS/MS
 Trends in 2006 Introduction to LC/MS/MS By Crystal Holt, LC/MS Product Specialist, Varian Inc. Toxicology laboratories Increased use of LC/MS Excellent LD Cheaper (still expensive) Much more robust Solves
Trends in 2006 Introduction to LC/MS/MS By Crystal Holt, LC/MS Product Specialist, Varian Inc. Toxicology laboratories Increased use of LC/MS Excellent LD Cheaper (still expensive) Much more robust Solves
Separation of Vitamin D and Vitamin D Metabolites on FLARE C18 MM (Mixed Mode) HPLC Column
 Diamond Analytics Technical Note: T131-1 Separation of Vitamin D and Vitamin D Metabolites on FLARE C1 MM (Mixed Mode) HPLC Column Introduction In this technical note, the versatility of the diamond-based
Diamond Analytics Technical Note: T131-1 Separation of Vitamin D and Vitamin D Metabolites on FLARE C1 MM (Mixed Mode) HPLC Column Introduction In this technical note, the versatility of the diamond-based
Organic Chemistry Laboratory Fall Lecture 3 Gas Chromatography and Mass Spectrometry June
 344 Organic Chemistry Laboratory Fall 2013 Lecture 3 Gas Chromatography and Mass Spectrometry June 19 2013 Chromatography Chromatography separation of a mixture into individual components Paper, Column,
344 Organic Chemistry Laboratory Fall 2013 Lecture 3 Gas Chromatography and Mass Spectrometry June 19 2013 Chromatography Chromatography separation of a mixture into individual components Paper, Column,
Ultra Performance Liquid Chromatography Coupled to Orthogonal Quadrupole TOF MS(MS) for Metabolite Identification
 22 SEPARATION SCIENCE REDEFINED MAY 2005 Ultra Performance Liquid Chromatography Coupled to Orthogonal Quadrupole TOF MS(MS) for Metabolite Identification In the drug discovery process the detection and
22 SEPARATION SCIENCE REDEFINED MAY 2005 Ultra Performance Liquid Chromatography Coupled to Orthogonal Quadrupole TOF MS(MS) for Metabolite Identification In the drug discovery process the detection and
Supporting Information
 Supporting Information Schlosburg et al. 10.1073/pnas.1219159110 SI Materials and Methods: Quantification of Heroin Metabolites Sample Collection. Trunk blood was collected in a 1:1 ratio with acetate
Supporting Information Schlosburg et al. 10.1073/pnas.1219159110 SI Materials and Methods: Quantification of Heroin Metabolites Sample Collection. Trunk blood was collected in a 1:1 ratio with acetate
Biosynthesis of Fatty Acids
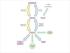 Biosynthesis of Fatty Acids Fatty acid biosynthesis takes place in the cytosol rather than the mitochondria and requires a different activation mechanism and different enzymes and coenzymes than fatty
Biosynthesis of Fatty Acids Fatty acid biosynthesis takes place in the cytosol rather than the mitochondria and requires a different activation mechanism and different enzymes and coenzymes than fatty
Lipids and Membranes
 Lipids and Membranes Presented by Dr. Mohammad Saadeh The requirements for the Pharmaceutical Biochemistry I Philadelphia University Faculty of pharmacy Lipids and Membranes I. overview Lipids are related
Lipids and Membranes Presented by Dr. Mohammad Saadeh The requirements for the Pharmaceutical Biochemistry I Philadelphia University Faculty of pharmacy Lipids and Membranes I. overview Lipids are related
PHOTOCATALYTIC DECONTAMINATION OF CHLORANTRANILIPROLE RESIDUES IN WATER USING ZnO NANOPARTICLES. DR. A. RAMESH, Ph.D, D.Sc.,
 PHOTOCATALYTIC DECONTAMINATION OF CHLORANTRANILIPROLE RESIDUES IN WATER USING ZnO NANOPARTICLES DR. A. RAMESH, Ph.D, D.Sc., raamesh_a@yahoo.co.in 1 OBJECTIVES Determination of persistence and photolysis
PHOTOCATALYTIC DECONTAMINATION OF CHLORANTRANILIPROLE RESIDUES IN WATER USING ZnO NANOPARTICLES DR. A. RAMESH, Ph.D, D.Sc., raamesh_a@yahoo.co.in 1 OBJECTIVES Determination of persistence and photolysis
Biological role of lipids
 Lipids Lipids Organic compounds present in living organisms, insoluble in water but able to be extracted by organic solvents such as: chloroform, acetone, benzene. Extraction = the action of taking out
Lipids Lipids Organic compounds present in living organisms, insoluble in water but able to be extracted by organic solvents such as: chloroform, acetone, benzene. Extraction = the action of taking out
Lipids and Classification:
 Lipids and Classification: Lipids: Biological lipids are a chemically diverse group of organic compounds which are insoluble or only poorly soluble in water. They are readily soluble in non-polar solvents
Lipids and Classification: Lipids: Biological lipids are a chemically diverse group of organic compounds which are insoluble or only poorly soluble in water. They are readily soluble in non-polar solvents
Endocannabinoids and related N-acylethanolamines: biological activities and metabolism
 Tsuboi et al. Inflammation and Regeneration (2018) 38:28 https://doi.org/10.1186/s41232-018-0086-5 Inflammation and Regeneration REVIEW Endocannabinoids and related N-acylethanolamines: biological activities
Tsuboi et al. Inflammation and Regeneration (2018) 38:28 https://doi.org/10.1186/s41232-018-0086-5 Inflammation and Regeneration REVIEW Endocannabinoids and related N-acylethanolamines: biological activities
Quantitative Analysis of Vit D Metabolites in Human Plasma using Exactive System
 Quantitative Analysis of Vit D Metabolites in Human Plasma using Exactive System Marta Kozak Clinical Research Applications Group Thermo Fisher Scientific San Jose CA Clinical Research use only, Not for
Quantitative Analysis of Vit D Metabolites in Human Plasma using Exactive System Marta Kozak Clinical Research Applications Group Thermo Fisher Scientific San Jose CA Clinical Research use only, Not for
Physiology Unit 1 CELL SIGNALING: CHEMICAL MESSENGERS AND SIGNAL TRANSDUCTION PATHWAYS
 Physiology Unit 1 CELL SIGNALING: CHEMICAL MESSENGERS AND SIGNAL TRANSDUCTION PATHWAYS In Physiology Today Cell Communication Homeostatic mechanisms maintain a normal balance of the body s internal environment
Physiology Unit 1 CELL SIGNALING: CHEMICAL MESSENGERS AND SIGNAL TRANSDUCTION PATHWAYS In Physiology Today Cell Communication Homeostatic mechanisms maintain a normal balance of the body s internal environment
