Introduction Session 1 and 2 (Ubiquitin, proteasome and human disease)
|
|
|
- Lewis Young
- 6 years ago
- Views:
Transcription
1 Introduction Session 1 and 2 (Ubiquitin, proteasome and human disease) Courtesy of Sam Griffiths-Jones. Used with permission. Source: "Peptide models for protein beta-sheets." PhD thesis, University of Nottingham, Figure by MIT OCW. After Goodsell, D.S. The Oncologist 8, Appendage of ubiquitin monomers to a protein substrate. Ubiquitin is a small, 76 aa protein which gets appended to another proteins, as a label. The protein substrate has amino groups in the side chains of its Lys aa residues. Ubiquitin has a C-terminal Gly. The carboxyl group of this Gly forms an isopeptide bond with the amino group of the Lys in the protein substrate (see figure). Ubiquitin also has several Lys that can act as internal acceptors for binding to the C-t Gly of new ubiquitin molecules, allowing the formation of a chain.
2 Courtesy of Sam Griffiths-Jones. Used with permission. Source: "Peptide models for protein beta-sheets." PhD thesis, University of Nottingham, DISCOVERY OF THE ROLE OF UBIQUITINN IN PROTEIN DEGRADATION.. HISTORICAL FACTS.. (1975) Ubiquitin was first isolated by Gideon Goldstein and colleagues from the thymus (reason why it was originally thought to be a thymic hormone). But because it was later found in all tissues and eukaryotic organisms it received the name of UBIQUITIN (for ubiquitous protein). (1977) Harris Goldknopf and Ira Busch found a DNA-associated protein that had one C-t but two N-t! The short arm of this Y- shaped unusual protein was joined through its C-terminal to the ε- amino group of an internal Lys of the histone H2A. Margaret Dayhoff soon identified it as Ubiquitin (a protein initially described as free by Goldstein). ( ) Avram Hershko studies regulation of tyrosine aminotransferase by its degradation Æ he found that degradation of the enzyme was arrested by inhibitors of cellular energy production (fluoride, azide) That was the first indication that an as-yet-unknown energydependent proteolytic system must exist. ( s) Hershko decided to identify this energy-dependent system responsible for the degradation of proteins, by means of classi cal biochemistry. His aims were: - to reproduce the ATP-dependent protein breakdown in a cell-free system. - to fractionate such system to find the mode of action of its components.
3 (1977) Etlinger and Goldberg discovered an ATP-dependent proteolytic system occurring in reticulocytes (red blood cells). Therefore, Hershko, helped by his grad student Aron ciechanover and Irwin Rose (Fox Chase cancer Center, Philadelphia) decided to isolate the ATPdependent proteolytic system from these cells. Reticulocyte lysates were fractionated on DEAE-cellulose (anionic exchange chromatography) into two crude fractions: Æ Fxn1(not adsorbed), with most of the hemoglobin. Æ Fxn2, proteins adsorbed to the resin and eluted with high salt. Fxn2 had lost most of the ATP-dependent proteolytic activity and only was restored when combining back Fxn1 with 2. The active component in Fxn1 was isolated in basis of its high stability upon heat treatment: APF-1, for ATP-dependent proteolysis factor 1. possible activator or regulatory subunit of other component/s of the system?? Radiolabeling APF-1 they saw substantial ATP-dependent binding to high-molecular weight proteins (gel filtration chromatography). Interaction extremely stable in the presence of conditions that disrupt non- covalent interactions: interaction should be covalent (peptidic or amidic linkage). APF-1 interacting not with an active component of the proteolytic system but with protein substrates??
4 By using a good substrate for ATP-dependent proteolysis, lysozyme, they found: 1) that similar high-molecular weight derivatives were for med when 125 I-labeled APF-1 was incubated with unlabeled lysozyme than when 125 I-labeled lysozyme was incubated with unlabeled APF-1. 2) analysis of the ratio of radiactivity in APF-1 and lysozyme indicated that the various derivatives consisted of increasing numbers of APF-1 molecules linked to one molecule of lysozyme. (Hershko et al., 1980) Courtesy of A. Hershko. Used with permission of the author. Source: Figure 1 in Hershko et al. "Proposed role of ATP in protein breakdown: conjugation of protein with multiple chains of the polypeptide of ATP-dependent proteolysis." PNAS 1980 April; 77(4):
5 (Model of action proposed byy Hershkoet al., 1980)) Courtesy of A. Hershko. Used with permission of the author. Source: Figure 6 in Hershko et al. "Proposed role of ATP in protein breakdown: conjugation of protein with multiple chains of the polypeptide of ATP-dependent proteolysis." PNAS 1980 April; 77(4): Several molecules of APF-1 linked to ε-amino groups of the protein substrate by an APF-1-protein amide synthetase (step1). Proteins ligated to several APF-1 are broken down by a specific protease that recognizes such conjugates (step 3). The protein is broken down to free amino acids and to APF-1 still linked by isopeptide linkage to a lysine or a small peptide, APF-1-X. Finally, free APF-1 would be released for re-use by specific amidase-isopeptidase (step 4). A hypothetical correcting isopeptidase would release free APF-1 and substrate protein from erroneous ligations (editing activity; step 2). Short after Hershko s model proposal, Keith Wilkinson and Arthur Haas (pot-doctoral fellows in Irwin Rose s lab) indicated that APF-1 was indeed UBIQUITIN.
6 Ubiquitin proteolytic pathwayy Courtesy of Annual Reviews. Used with permission. Source: Figure 1A in Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67: A: Conjugation of ubiquitin to the target substrate Image removed due to copyright considerations. See Figure 1 in Ciechanover A, Orian A, Schwartz AL. "Ubiquitin-mediated proteolysis: biological regulation via destruction." Bioessays May; 22 (5): E1: Ub-activating enzyme E2: Ub-conjugating enzyme; Ubc s E3: Ub-ligase B: Degradation of the polyubiquitinated substrate by the 26S proteasome complex and Ub recycling by deubiquitinating enzymes (Ub C-terminal hydrolases, DUBs or UCH s)
7 (1980 s) Hershko and Ciechanover purified the components of the first part of the reaction (APF-1-protein amide synthetase): E1, E2 and E3 enzymes, by covalent affinity chromatography over immobilized Ub. The protease step was further clarified with the characterization and purification of the 26 proteasome complex, by Martin Rechsteiner and colleagues (1986). It was first thought that multiple ubiquitin attached to different Lys sites in the substrate protein but Hershko (1985) and then Alexander Varshavsky and colleagues (1989) demonstrated that a single poly ubiquitin chain could be form through attachment to a single internal Lys residue. Attachment of subsequent Ub to the first one occurs through linkages of the Gly C-t of the next Ub to specific lysines present in the precedent Ub. Poly-ubiquitin chains might have different roles depending on the Lys utilized: K48 Æ recognition by proteasome K63 Æ signal for traffic and degradation in the lysosome, vacuole; subunit selectivity K29, K6, etc.. First evidence showing that ubiquitin constitutes a signal for protein degradation in vivo (Hershko lab; 1982): immunochemical analysis of Ub adducts in cells Æ by using anti-ub Abs they saw that, after incubation of cells in the presence of aa analogs the resulting abnormal proteins were short-lived and their rapid degradation was accompanied by a transient increase in Ub adducts. But the strongest confirmation came with Alexander Varshavsky s work.
8 UBIQUITIN CONJUGATION REQUIRED FOR PROTEIN DEGRADATION IN VIVO (first evidence through the studies carried out by Alexander Varshavsky, Dan Finley and Aaron Ciechanover) Varshavsky studied chromosome structure and regulation of gene expression and came to Boston in the fall of 1977 where, a month later, became a faculty member in the Biology Department here at MIT. He became interested in the role of protein degradation in gene regulation and expression at the light of Hershko s and Wilkinson s experiments (showing that the ATP-dependent proteolytic factor 1, was ubiquitin). Considering that he already knew that Ub had been found attached to histones, he became very excited when in 1980 he read a paper, by Yamada and coll. that described a conditionally lethal, temperature sensitive mouse cell line called ts85 (they had seen that in this cell line at restrictive temperature a nuclear protein disappeared and Varshavsky confirmed it was UbH2A). ÆDan Finley, from his lab, and A. Ciechanover helped him to study ts85 and saw that cells were arrested in S/G2 phase of cell division cycle and had induced synthesis of heat shock proteins at non permi ssive temperature. They found that ts85 mouse cells had a temperature-sensitive Ub-activating enzyme (E1) and that these cells stopped degrading the bulk of their normally short-lived proteins at the nonpermissive temperature. Later studies (by Hunt and coll.), with rapidly dividing fertilized clam eggs, led to the discovery of CYCLINS and in 1984 Varshavsky proposed that cyclins were degraded at the exit from mitosis by the ubiquitin system.
9 Stress Death Survival Glucocorticoids (Radiation) Receptor Receptor Ubiquitination Mdm2 Receptor Ubiquitination Ubiquitination Bcl-2 cflip Ubiquitination p53 Mitochondrion NFxB IxB Ubiquitination Image removed for copyright considerations. Source: Figure 5 in Muratani M, W. P. Tansey. "How the Ubiquitin-proteasome System Controls Transcription." Nat Rev Mol Cell Biol. 4, no. 3 (Mar 2003): Smac/Diablo Cytochrome c Omi NFxB IxB ub ub Ubiquitination IAPs ub ubub Ubiquitination Caspase-Dependent Apoptosis Pathway Degradation Figure by MIT OCW. After Yang and Xu, "Regulation of apoptosis: the ubiquitous way." FASEB J 17 (2003) Figure by MIT OCW.
10 How are the substrates recognized by the E3s? Image removed due to copyright considerations. See Figure 2 in Ciechanover A. et al. "Ubiquitin-mediated proteolysis: biological regulation via destruction." Bioessays May; 22(5):
11 N-End Rule Discoveryy Hershko and Ciechanover saw a discrepancy while attempting to purify APF-1 (Ub): the preparation had a high dry-weight to low proteincontent ratio. Was APF-1 a ribonucleoprotein?? When they treated the preparation with RNase A this abrogated the degradation of BSA me diated by APF-1, although not that of lysozyme. They later found the weight to protein content discrepancy was in fact caused by the protein detection method (Lowry assay) which is based in the detection of aromatic aa s (Ub had a low content on those). Then what caused the RNase effect on BSA Ub-dependent degradation? Some proteins in order to be degraded must be recognized by specific destabilizing aa residues located in their N-t ( N-End rule ). In some cases these destabilizing residues are added by post-translational modification, carried out by specific amidases and transferases. In the specific case of BSA, the RNase was destroying the trna Arg that was necessary along with arginyl-trna protein transferase to convert the N-t acidic (Asp) residue of BSA to Arg. Only this modi fied form of BSA can bind to the E3α. (The lysozyme has a Lys in the N-t so doesn t need to undergo this post translational modification to be recognized by the E3). Varshavsky, Finley and others discovered further aspects of the N- End rule and established a family of N-t degradation signals called N- Degrons, N-t destabilizing residue internal Lys (site of Ub attachment)
12 The ubiquitin system of S. cerevisiaee Image removed due to copyright considerations. See Figure 5 in Hershko, A. et al. "Basic Medical Research Award. The ubiquitin system." Nature Medicine 2000 Oct; 6 (10):
13 Ubiquitin is not the only protein used as a tag in cells: Figure by MIT OCW. After Table 1 in Schwartz and Hochstrasser, TIBS 28 (2003)
14 Two main kinds of E3 ubiquitin ligasess 1) HECT- domain proteins (homologous to E6-AP carboxyl terminus): coordinate the transfer of Ub to the substrate by forming a thiol-ester bond with Ub prior to transferring it to a substrate. Example: Rsp5p Figure by MIT OCW. 2) RING finger proteins: coordinate the transfer of Ub to the substrate by bringing the E2-ubiquitin complex to the substrate. Consensus sequence: 8 conserved Cys and His that coordinate 2 Zn ions in a cross-braced fashion. Some act singly, such as Ubr1/E3α; others exist in complex, such as the SCF complexes (Skp1, yeast Cdc53, or mammalian cullin, and F-box protein)
15 F-box protein: substrate binding. Fbws bind substrate through WD - 40 domains; Fbls, through Leu-rich repeats; Fbxs, use different motifs. Skp1: scaffold that keeps together the cullin and the F-box. Cullin: mediates attachment of Nedd8, Skp1, and Roc1. Roc1/Rbx1 (Hrt1 in yeast): C-t RING finger domain; stabilizing interaction between the E2 and the rest of the complex. Figure by MIT OCW. Nedd8 (Rub1 in yeast): Ubi-like protein that enhances the ubiquitin-ligating activity of the complex.
16 Ubiquitination involving degradation of proteins targeted to the secretory pathway: ERAD or Endoplasmicc Reticulumm Associated Degradationn Figure by MIT OCW. After Fang et al. "RING finger ubiquitin protein ligases: implications for tumorigenesis, metastasis and for molecular targets in cancer." Semin Cancer Biol Feb; 13 (1): Vacuolar/ Lysosomal Degradation Figure by MIT OCW. After Fig. 5 in Horák, J. (2003). The role of ubiquitin in down-regulation and intracellular sorting of membrane proteins: insights from yeast. Biochim. Biophys. Acta 1614,
17 Infectious agents hijack cell machineryy involved inn ubiquitination and protein degradationn Figure by MIT OCW. After Fig. 2 in Amara, A and Littman, DR After Hrs with HIV. J. Cell Biol., 162: Example: HIV virus budding off cells by using host machinery involved in traffic of membranes
18 What can go wrong in the UPS (Ubiquitinn Proteasome System) to cause disorderr and/or disease?? Disease Normal Function Lower Steady State Level Protein Substrate Protein Substrate Normal Steady State Level Accelerated Degradation Normal Degradation Ancillary Viral Protein The Ubiquitin System Mutation in a System Enzyme or a Substrate Decreased Degradation Normal Degradation Higher Steady State Level Protein Substrate Protein Substrate Normal Function Normal Steady State Level Disease Figure by MIT OCW. After Fig 3 in Hershko, A. et al. "Basic Medical Research Award. The ubiquitin system." Nature Medicine 2000 Oct; 6 (10):
19 Example: The UPS and the pathogenesis of neurodegeneration Image removed due to copyright considerations. See Figure 2 in Ciechanover, A. and Brundin, P The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron 40:
The Nobel Prize in Chemistry 2004
 The Nobel Prize in Chemistry 2004 Ubiquitous Quality Control of Life C S Karigar and K R Siddalinga Murthy The Nobel Prize in Chemistry for 2004 is shared by Aaron Ciechanover, Avram Hershko and Irwin
The Nobel Prize in Chemistry 2004 Ubiquitous Quality Control of Life C S Karigar and K R Siddalinga Murthy The Nobel Prize in Chemistry for 2004 is shared by Aaron Ciechanover, Avram Hershko and Irwin
The ubiquitin system
 The ubiquitin system AVRAM HERSHKO, AARON CIECHANOVER & ALEXANDER VARSHAVSKY iquitin-mediated protein degradation: The early days It has been often stated that until recently the ubiquitin system was thought
The ubiquitin system AVRAM HERSHKO, AARON CIECHANOVER & ALEXANDER VARSHAVSKY iquitin-mediated protein degradation: The early days It has been often stated that until recently the ubiquitin system was thought
Study of different types of ubiquitination
 Study of different types of ubiquitination Rudi Beyaert (rudi.beyaert@irc.vib-ugent.be) VIB UGent Center for Inflammation Research Ghent, Belgium VIB Training Novel Proteomics Tools: Identifying PTMs October
Study of different types of ubiquitination Rudi Beyaert (rudi.beyaert@irc.vib-ugent.be) VIB UGent Center for Inflammation Research Ghent, Belgium VIB Training Novel Proteomics Tools: Identifying PTMs October
Protein methylation CH 3
 Protein methylation CH 3 methionine S-adenosylmethionine (SAM or adomet) Methyl group of the methionine is activated by the + charge of the adjacent sulfur atom. SAM S-adenosylhomocysteine homocysteine
Protein methylation CH 3 methionine S-adenosylmethionine (SAM or adomet) Methyl group of the methionine is activated by the + charge of the adjacent sulfur atom. SAM S-adenosylhomocysteine homocysteine
Molecular Graphics Perspective of Protein Structure and Function
 Molecular Graphics Perspective of Protein Structure and Function VMD Highlights > 20,000 registered Users Platforms: Unix (16 builds) Windows MacOS X Display of large biomolecules and simulation trajectories
Molecular Graphics Perspective of Protein Structure and Function VMD Highlights > 20,000 registered Users Platforms: Unix (16 builds) Windows MacOS X Display of large biomolecules and simulation trajectories
Proteasomes. When Death Comes a Knock n. Warren Gallagher Chem412, Spring 2001
 Proteasomes When Death Comes a Knock n Warren Gallagher Chem412, Spring 2001 I. Introduction Introduction The central dogma Genetic information is used to make proteins. DNA RNA Proteins Proteins are the
Proteasomes When Death Comes a Knock n Warren Gallagher Chem412, Spring 2001 I. Introduction Introduction The central dogma Genetic information is used to make proteins. DNA RNA Proteins Proteins are the
UBIQUITIN AT FOX CHASE
 UBIQUITIN AT FOX CHASE Nobel Lecture, December 8, 2004 by Irwin Rose University of California, Department of Physiology and Biophysics, College of Medicine, Irvine, CA 92697 4560, USA. My interest in protein
UBIQUITIN AT FOX CHASE Nobel Lecture, December 8, 2004 by Irwin Rose University of California, Department of Physiology and Biophysics, College of Medicine, Irvine, CA 92697 4560, USA. My interest in protein
Ubiquitin-aldehyde: A general inhibitor of ubiquitinrecycling
 Proc. Nati. Acad. Sci. USA Vol. 84, pp. 1829-1833, April 1987 Biochemistry Ubiquitin-aldehyde: A general inhibitor of ubiquitinrecycling processes (protein breakdown/isopeptidase/c-terminal hydrolase)
Proc. Nati. Acad. Sci. USA Vol. 84, pp. 1829-1833, April 1987 Biochemistry Ubiquitin-aldehyde: A general inhibitor of ubiquitinrecycling processes (protein breakdown/isopeptidase/c-terminal hydrolase)
Cellular functions of protein degradation
 Protein Degradation Cellular functions of protein degradation 1. Elimination of misfolded and damaged proteins: Environmental toxins, translation errors and genetic mutations can damage proteins. Misfolded
Protein Degradation Cellular functions of protein degradation 1. Elimination of misfolded and damaged proteins: Environmental toxins, translation errors and genetic mutations can damage proteins. Misfolded
Biochemistry 2 Recita0on Amino Acid Metabolism
 Biochemistry 2 Recita0on Amino Acid Metabolism 04-20- 2015 Glutamine and Glutamate as key entry points for NH 4 + Amino acid catabolism Glutamine synthetase enables toxic NH 4 + to combine with glutamate
Biochemistry 2 Recita0on Amino Acid Metabolism 04-20- 2015 Glutamine and Glutamate as key entry points for NH 4 + Amino acid catabolism Glutamine synthetase enables toxic NH 4 + to combine with glutamate
1. to understand how proteins find their destination in prokaryotic and eukaryotic cells 2. to know how proteins are bio-recycled
 Protein Targeting Objectives 1. to understand how proteins find their destination in prokaryotic and eukaryotic cells 2. to know how proteins are bio-recycled As a protein is being synthesized, decisions
Protein Targeting Objectives 1. to understand how proteins find their destination in prokaryotic and eukaryotic cells 2. to know how proteins are bio-recycled As a protein is being synthesized, decisions
Degradation needs no energy or does it?
 English Swedish The Nobel Prize in Chemistry 2004 Information for the Public 6 October 2004 A human cell contains some hundred thousand different proteins. These have numerous important functions: as accelerators
English Swedish The Nobel Prize in Chemistry 2004 Information for the Public 6 October 2004 A human cell contains some hundred thousand different proteins. These have numerous important functions: as accelerators
ubiquitin, protein degradation, proteasome, proteolysis SIGNALS IN PROTEINS FOR UBIQUITINYLATION AND DEGRADATION...438
 Annu. Rev. Biochem. 1998. 67:425 79 Copyright c 1998 by Annual Reviews. All rights reserved THE UBIQUITIN SYSTEM Avram Hershko and Aaron Ciechanover Unit of Biochemistry, Faculty of Medicine and the Rappaport
Annu. Rev. Biochem. 1998. 67:425 79 Copyright c 1998 by Annual Reviews. All rights reserved THE UBIQUITIN SYSTEM Avram Hershko and Aaron Ciechanover Unit of Biochemistry, Faculty of Medicine and the Rappaport
Ub-CHOP-Reporter Deubiquitination Assay Kit Catalog Number PR1001
 - 1 - MANUAL Ub-CHOP-Reporter Deubiquitination Assay Kit Catalog Number PR1001 all products are for research use only not intended for human or animal diagnostic or therapeutic uses LifeSensors, Inc.,
- 1 - MANUAL Ub-CHOP-Reporter Deubiquitination Assay Kit Catalog Number PR1001 all products are for research use only not intended for human or animal diagnostic or therapeutic uses LifeSensors, Inc.,
Amino acids. Side chain. -Carbon atom. Carboxyl group. Amino group
 PROTEINS Amino acids Side chain -Carbon atom Amino group Carboxyl group Amino acids Primary structure Amino acid monomers Peptide bond Peptide bond Amino group Carboxyl group Peptide bond N-terminal (
PROTEINS Amino acids Side chain -Carbon atom Amino group Carboxyl group Amino acids Primary structure Amino acid monomers Peptide bond Peptide bond Amino group Carboxyl group Peptide bond N-terminal (
BIOL 4374/BCHS 4313 Cell Biology Exam #1 February 13, 2001
 BIOL 4374/BCHS 4313 Cell Biology Exam #1 February 13, 2001 SS# Name This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses. Good luck! 1. (2) The
BIOL 4374/BCHS 4313 Cell Biology Exam #1 February 13, 2001 SS# Name This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses. Good luck! 1. (2) The
SHORT LINEAR MOTIFS. Holger Dinkel EMBO Practical Course Computational analysis of protein-protein interactions From sequences to networks
 Holger Dinkel EMBO Practical Course Computational analysis of protein-protein interactions From sequences to networks IMPORTANCE OF 2 / 19 IMPORTANCE OF 2 / 19 IMPORTANCE OF 2 / 19 IMPORTANCE OF 2 / 19
Holger Dinkel EMBO Practical Course Computational analysis of protein-protein interactions From sequences to networks IMPORTANCE OF 2 / 19 IMPORTANCE OF 2 / 19 IMPORTANCE OF 2 / 19 IMPORTANCE OF 2 / 19
Cell Quality Control. Peter Takizawa Department of Cell Biology
 Cell Quality Control Peter Takizawa Department of Cell Biology Cellular quality control reduces production of defective proteins. Cells have many quality control systems to ensure that cell does not build
Cell Quality Control Peter Takizawa Department of Cell Biology Cellular quality control reduces production of defective proteins. Cells have many quality control systems to ensure that cell does not build
There are approximately 30,000 proteasomes in a typical human cell Each proteasome is approximately 700 kda in size The proteasome is made up of 3
 Proteasomes Proteasomes Proteasomes are responsible for degrading proteins that have been damaged, assembled improperly, or that are of no profitable use to the cell. The unwanted protein is literally
Proteasomes Proteasomes Proteasomes are responsible for degrading proteins that have been damaged, assembled improperly, or that are of no profitable use to the cell. The unwanted protein is literally
Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy
 (Courtesy of Magda Stumpfova. Used with permission.) Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy M.D. Gomes, S.H. Lecker, R.T. Jagoe, A. Navon, and A.L. Goldberg PNAS,
(Courtesy of Magda Stumpfova. Used with permission.) Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy M.D. Gomes, S.H. Lecker, R.T. Jagoe, A. Navon, and A.L. Goldberg PNAS,
Molecular Cell Biology (Bio 5068) Cell Cycle I. Ron Bose, MD PhD November 14, 2017
 Molecular Cell Biology (Bio 5068) Cell Cycle I Ron Bose, MD PhD November 14, 2017 CELL DIVISION CYCLE M G2 S G1 DISCOVERY AND NAMING OF CYCLINS A protein (called cyclin ) was observed to increase as cells
Molecular Cell Biology (Bio 5068) Cell Cycle I Ron Bose, MD PhD November 14, 2017 CELL DIVISION CYCLE M G2 S G1 DISCOVERY AND NAMING OF CYCLINS A protein (called cyclin ) was observed to increase as cells
Ubiquitylation: its role and medical significance. Dr. Zoltán Lipinszki Institute of Biochemistry, HAS-BRC 24 October, 2018
 Ubiquitylation: its role and medical significance Dr. Zoltán Lipinszki Institute of Biochemistry, HAS-BRC 24 October, 2018 Cellular homeostasis The proteins of the body are in a dynamic state of synthesis
Ubiquitylation: its role and medical significance Dr. Zoltán Lipinszki Institute of Biochemistry, HAS-BRC 24 October, 2018 Cellular homeostasis The proteins of the body are in a dynamic state of synthesis
The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein
 THESIS BOOK The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein Orsolya Buzás-Bereczki Supervisors: Dr. Éva Bálint Dr. Imre Miklós Boros University of
THESIS BOOK The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein Orsolya Buzás-Bereczki Supervisors: Dr. Éva Bálint Dr. Imre Miklós Boros University of
VII SYNOPSIS. Cancer is a disease, causes due to failure of control of growth and. cell proliferation mechanisms. There are genetic control systems,
 VII SYPSIS Cancer is a disease, causes due to failure of control of growth and cell proliferation mechanisms. There are genetic control systems, regulate the balance between cell birth and death in response
VII SYPSIS Cancer is a disease, causes due to failure of control of growth and cell proliferation mechanisms. There are genetic control systems, regulate the balance between cell birth and death in response
Lecture 10. G1/S Regulation and Cell Cycle Checkpoints. G1/S regulation and growth control G2 repair checkpoint Spindle assembly or mitotic checkpoint
 Lecture 10 G1/S Regulation and Cell Cycle Checkpoints Outline: G1/S regulation and growth control G2 repair checkpoint Spindle assembly or mitotic checkpoint Paper: The roles of Fzy/Cdc20 and Fzr/Cdh1
Lecture 10 G1/S Regulation and Cell Cycle Checkpoints Outline: G1/S regulation and growth control G2 repair checkpoint Spindle assembly or mitotic checkpoint Paper: The roles of Fzy/Cdc20 and Fzr/Cdh1
Chapter 31. Completing the Protein Life Cycle: Folding, Processing and Degradation. Biochemistry by Reginald Garrett and Charles Grisham
 Chapter 31 Completing the Protein Life Cycle: Folding, Processing and Degradation Biochemistry by Reginald Garrett and Charles Grisham Essential Question How are newly synthesized polypeptide chains transformed
Chapter 31 Completing the Protein Life Cycle: Folding, Processing and Degradation Biochemistry by Reginald Garrett and Charles Grisham Essential Question How are newly synthesized polypeptide chains transformed
The ubiquitin system for protein degradation and some of its roles in the control of the cell division cycle*
 (2005) 12, 1191 1197 & 2005 Nature Publishing Group All rights reserved 1350-9047/05 $30.00 www.nature.com/cdd Review: Nobel Lecture The ubiquitin system for protein degradation and some of its roles in
(2005) 12, 1191 1197 & 2005 Nature Publishing Group All rights reserved 1350-9047/05 $30.00 www.nature.com/cdd Review: Nobel Lecture The ubiquitin system for protein degradation and some of its roles in
This exam consists of two parts. Part I is multiple choice. Each of these 25 questions is worth 2 points.
 MBB 407/511 Molecular Biology and Biochemistry First Examination - October 1, 2002 Name Social Security Number This exam consists of two parts. Part I is multiple choice. Each of these 25 questions is
MBB 407/511 Molecular Biology and Biochemistry First Examination - October 1, 2002 Name Social Security Number This exam consists of two parts. Part I is multiple choice. Each of these 25 questions is
The ubiquitin-proteasome system. in bone biology. University of Nottingham. ECTS PhD Training July 5 th Dr Rob Layfield
 The ubiquitin-proteasome system in bone biology Dr Rob Layfield University of Nottingham ECTS PhD Training July 5 th 2009 Images reproduced from: http://www.bostonbiochem.com/overview.php?prod=ubchains
The ubiquitin-proteasome system in bone biology Dr Rob Layfield University of Nottingham ECTS PhD Training July 5 th 2009 Images reproduced from: http://www.bostonbiochem.com/overview.php?prod=ubchains
Previous Class. Today. Detection of enzymatic intermediates: Protein tyrosine phosphatase mechanism. Protein Kinase Catalytic Properties
 Previous Class Detection of enzymatic intermediates: Protein tyrosine phosphatase mechanism Today Protein Kinase Catalytic Properties Protein Phosphorylation Phosphorylation: key protein modification
Previous Class Detection of enzymatic intermediates: Protein tyrosine phosphatase mechanism Today Protein Kinase Catalytic Properties Protein Phosphorylation Phosphorylation: key protein modification
Problem-solving Test: The Mechanism of Protein Synthesis
 Q 2009 by The International Union of Biochemistry and Molecular Biology BIOCHEMISTRY AND MOLECULAR BIOLOGY EDUCATION Vol. 37, No. 1, pp. 58 62, 2009 Problem-based Learning Problem-solving Test: The Mechanism
Q 2009 by The International Union of Biochemistry and Molecular Biology BIOCHEMISTRY AND MOLECULAR BIOLOGY EDUCATION Vol. 37, No. 1, pp. 58 62, 2009 Problem-based Learning Problem-solving Test: The Mechanism
INTRACELLULAR PROTEIN DEGRADATION: FROM A VAGUE IDEA THRU THE LYSOSOME AND THE UBIQUITIN-PROTEASOME SYSTEM AND ONTO HUMAN DISEASES AND DRUG TARGETING
 INTRACELLULAR PROTEIN DEGRADATION: FROM A VAGUE IDEA THRU THE LYSOSOME AND THE UBIQUITIN-PROTEASOME SYSTEM AND ONTO HUMAN DISEASES AND DRUG TARGETING Nobel Lecture, December 8, 2004 by Aaron Ciechanover
INTRACELLULAR PROTEIN DEGRADATION: FROM A VAGUE IDEA THRU THE LYSOSOME AND THE UBIQUITIN-PROTEASOME SYSTEM AND ONTO HUMAN DISEASES AND DRUG TARGETING Nobel Lecture, December 8, 2004 by Aaron Ciechanover
Degrading the Barriers to Drug Discovery in Ubiquitin E3 Ligase Pathways
 Degrading the Barriers to Drug Discovery in iquitin E3 Ligase Pathways Blaine N. Armbruster, PhD. Sr. Manager - Discovery & Development Solutions EMD Millipore 1 Oct, 2012 Outline Introduction ti to EMD
Degrading the Barriers to Drug Discovery in iquitin E3 Ligase Pathways Blaine N. Armbruster, PhD. Sr. Manager - Discovery & Development Solutions EMD Millipore 1 Oct, 2012 Outline Introduction ti to EMD
CELLS. Cells. Basic unit of life (except virus)
 Basic unit of life (except virus) CELLS Prokaryotic, w/o nucleus, bacteria Eukaryotic, w/ nucleus Various cell types specialized for particular function. Differentiation. Over 200 human cell types 56%
Basic unit of life (except virus) CELLS Prokaryotic, w/o nucleus, bacteria Eukaryotic, w/ nucleus Various cell types specialized for particular function. Differentiation. Over 200 human cell types 56%
The Ubiquitin-Proteasome Proteolytic Pathway: Destruction for the Sake of Construction
 Physiol Rev 82: 373 428, 2002; 10.1152/physrev.00027.2001. The Ubiquitin-Proteasome Proteolytic Pathway: Destruction for the Sake of Construction MICHAEL H. GLICKMAN AND AARON CIECHANOVER Faculty of Biology
Physiol Rev 82: 373 428, 2002; 10.1152/physrev.00027.2001. The Ubiquitin-Proteasome Proteolytic Pathway: Destruction for the Sake of Construction MICHAEL H. GLICKMAN AND AARON CIECHANOVER Faculty of Biology
Introduction to Biochemistry Midterm exam )ومن أحياها(
 Introduction to Biochemistry Midterm exam 2016-2017 )ومن أحياها( 1. Which of the following amino (in a peptide chain) would probably be found at a beta bend or turn? a. lysine * b. Gly c. arg d. asn 2.
Introduction to Biochemistry Midterm exam 2016-2017 )ومن أحياها( 1. Which of the following amino (in a peptide chain) would probably be found at a beta bend or turn? a. lysine * b. Gly c. arg d. asn 2.
Copyright 2008 Pearson Education, Inc., publishing as Pearson Benjamin Cummings
 Concept 5.4: Proteins have many structures, resulting in a wide range of functions Proteins account for more than 50% of the dry mass of most cells Protein functions include structural support, storage,
Concept 5.4: Proteins have many structures, resulting in a wide range of functions Proteins account for more than 50% of the dry mass of most cells Protein functions include structural support, storage,
PAPER No. : 16, Bioorganic and biophysical chemistry MODULE No. : 22, Mechanism of enzyme catalyst reaction (I) Chymotrypsin
 Subject Paper No and Title 16 Bio-organic and Biophysical Module No and Title 22 Mechanism of Enzyme Catalyzed reactions I Module Tag CHE_P16_M22 Chymotrypsin TABLE OF CONTENTS 1. Learning outcomes 2.
Subject Paper No and Title 16 Bio-organic and Biophysical Module No and Title 22 Mechanism of Enzyme Catalyzed reactions I Module Tag CHE_P16_M22 Chymotrypsin TABLE OF CONTENTS 1. Learning outcomes 2.
Lecture 14 - The cell cycle and cell death
 02.17.10 Lecture 14 - The cell cycle and cell death The cell cycle: cells duplicate their contents and divide The cell cycle may be divided into 4 phases The cell cycle triggers essential processes (DNA
02.17.10 Lecture 14 - The cell cycle and cell death The cell cycle: cells duplicate their contents and divide The cell cycle may be divided into 4 phases The cell cycle triggers essential processes (DNA
E2Select Ubiquitin Conjugation Kit
 E2Select Ubiquitin Conjugation Kit Catalog Number K-982 This kit contains reagents sufficient to perform 2 Ubiquitination reactions for each E2 included in the kit. This package insert must be read in
E2Select Ubiquitin Conjugation Kit Catalog Number K-982 This kit contains reagents sufficient to perform 2 Ubiquitination reactions for each E2 included in the kit. This package insert must be read in
Point total. Page # Exam Total (out of 90) The number next to each intermediate represents the total # of C-C and C-H bonds in that molecule.
 This exam is worth 90 points. Pages 2- have questions. Page 1 is for your reference only. Honor Code Agreement - Signature: Date: (You agree to not accept or provide assistance to anyone else during this
This exam is worth 90 points. Pages 2- have questions. Page 1 is for your reference only. Honor Code Agreement - Signature: Date: (You agree to not accept or provide assistance to anyone else during this
Molecular Biology. general transfer: occurs normally in cells. special transfer: occurs only in the laboratory in specific conditions.
 Chapter 9: Proteins Molecular Biology replication general transfer: occurs normally in cells transcription special transfer: occurs only in the laboratory in specific conditions translation unknown transfer:
Chapter 9: Proteins Molecular Biology replication general transfer: occurs normally in cells transcription special transfer: occurs only in the laboratory in specific conditions translation unknown transfer:
Chemical Mechanism of Enzymes
 Chemical Mechanism of Enzymes Enzyme Engineering 5.2 Definition of the mechanism 1. The sequence from substrate(s) to product(s) : Reaction steps 2. The rates at which the complex are interconverted 3.
Chemical Mechanism of Enzymes Enzyme Engineering 5.2 Definition of the mechanism 1. The sequence from substrate(s) to product(s) : Reaction steps 2. The rates at which the complex are interconverted 3.
Ionization of amino acids
 Amino Acids 20 common amino acids there are others found naturally but much less frequently Common structure for amino acid COOH, -NH 2, H and R functional groups all attached to the a carbon Ionization
Amino Acids 20 common amino acids there are others found naturally but much less frequently Common structure for amino acid COOH, -NH 2, H and R functional groups all attached to the a carbon Ionization
October 26, Lecture Readings. Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell
 October 26, 2006 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
October 26, 2006 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
BIOB111 - Tutorial activity for Session 14
 BIOB111 - Tutorial activity for Session 14 General topics for week 7 Session 14 Amino acids and proteins Students review the concepts learnt and answer the selected questions from the textbook. General
BIOB111 - Tutorial activity for Session 14 General topics for week 7 Session 14 Amino acids and proteins Students review the concepts learnt and answer the selected questions from the textbook. General
Cell Cycle. Trends in Cell Biology
 Cell Cycle Trends in Cell Biology Cell Cycle The orderly sequence of events by which a cell duplicates its contents and divides into two Daughter Cells Activities of a cell from one cell division to the
Cell Cycle Trends in Cell Biology Cell Cycle The orderly sequence of events by which a cell duplicates its contents and divides into two Daughter Cells Activities of a cell from one cell division to the
Effects of Second Messengers
 Effects of Second Messengers Inositol trisphosphate Diacylglycerol Opens Calcium Channels Binding to IP 3 -gated Channel Cooperative binding Activates Protein Kinase C is required Phosphorylation of many
Effects of Second Messengers Inositol trisphosphate Diacylglycerol Opens Calcium Channels Binding to IP 3 -gated Channel Cooperative binding Activates Protein Kinase C is required Phosphorylation of many
Lipids: diverse group of hydrophobic molecules
 Lipids: diverse group of hydrophobic molecules Lipids only macromolecules that do not form polymers li3le or no affinity for water hydrophobic consist mostly of hydrocarbons nonpolar covalent bonds fats
Lipids: diverse group of hydrophobic molecules Lipids only macromolecules that do not form polymers li3le or no affinity for water hydrophobic consist mostly of hydrocarbons nonpolar covalent bonds fats
Objective: You will be able to explain how the subcomponents of
 Objective: You will be able to explain how the subcomponents of nucleic acids determine the properties of that polymer. Do Now: Read the first two paragraphs from enduring understanding 4.A Essential knowledge:
Objective: You will be able to explain how the subcomponents of nucleic acids determine the properties of that polymer. Do Now: Read the first two paragraphs from enduring understanding 4.A Essential knowledge:
Cell cycle and Apoptosis. Chalermchai Mitrpant
 Cell cycle and Apoptosis 2556 Chalermchai Mitrpant Overview of the cell cycle Outline Regulatory mechanisms controlling cell cycle Progression of the cell cycle Checkpoint of the cell cycle Phases of the
Cell cycle and Apoptosis 2556 Chalermchai Mitrpant Overview of the cell cycle Outline Regulatory mechanisms controlling cell cycle Progression of the cell cycle Checkpoint of the cell cycle Phases of the
ab E3 Ligase Auto- Ubiquitilylation Assay Kit
 ab139469 E3 Ligase Auto- Ubiquitilylation Assay Kit Instructions for Use For testing ubiquitin E3 ligase activity through assessment of their ability to undergo auto-ubiquitinylation This product is for
ab139469 E3 Ligase Auto- Ubiquitilylation Assay Kit Instructions for Use For testing ubiquitin E3 ligase activity through assessment of their ability to undergo auto-ubiquitinylation This product is for
Cell Cycle, Mitosis, and Microtubules. LS1A Final Exam Review Friday 1/12/07. Processes occurring during cell cycle
 Cell Cycle, Mitosis, and Microtubules LS1A Final Exam Review Friday 1/12/07 Processes occurring during cell cycle Replicate chromosomes Segregate chromosomes Cell divides Cell grows Cell Growth 1 The standard
Cell Cycle, Mitosis, and Microtubules LS1A Final Exam Review Friday 1/12/07 Processes occurring during cell cycle Replicate chromosomes Segregate chromosomes Cell divides Cell grows Cell Growth 1 The standard
UbcH (E2) Enzyme Kit. Catalog Number K-980B. This kit contains reagents for the qualitative analysis of UbcH Enzyme usage.
 UbcH (E2) Enzyme Kit Catalog Number K-980B This kit contains reagents for the qualitative analysis of UbcH Enzyme usage. This package insert must be read in its entirety before using this product. For
UbcH (E2) Enzyme Kit Catalog Number K-980B This kit contains reagents for the qualitative analysis of UbcH Enzyme usage. This package insert must be read in its entirety before using this product. For
Short polymer. Dehydration removes a water molecule, forming a new bond. Longer polymer (a) Dehydration reaction in the synthesis of a polymer
 HO 1 2 3 H HO H Short polymer Dehydration removes a water molecule, forming a new bond Unlinked monomer H 2 O HO 1 2 3 4 H Longer polymer (a) Dehydration reaction in the synthesis of a polymer HO 1 2 3
HO 1 2 3 H HO H Short polymer Dehydration removes a water molecule, forming a new bond Unlinked monomer H 2 O HO 1 2 3 4 H Longer polymer (a) Dehydration reaction in the synthesis of a polymer HO 1 2 3
Phenylketonuria (PKU) Structure of Phenylalanine Hydroxylase. Biol 405 Molecular Medicine
 Phenylketonuria (PKU) Structure of Phenylalanine Hydroxylase Biol 405 Molecular Medicine 1998 Crystal structure of phenylalanine hydroxylase solved. The polypeptide consists of three regions: Regulatory
Phenylketonuria (PKU) Structure of Phenylalanine Hydroxylase Biol 405 Molecular Medicine 1998 Crystal structure of phenylalanine hydroxylase solved. The polypeptide consists of three regions: Regulatory
Protein Modification Overview DEFINITION The modification of selected residues in a protein and not as a component of synthesis
 Lecture Four: Protein Modification & Cleavage [Based on Chapters 2, 9, 10 & 11 Berg, Tymoczko & Stryer] (Figures in red are for the 7th Edition) (Figures in Blue are for the 8th Edition) Protein Modification
Lecture Four: Protein Modification & Cleavage [Based on Chapters 2, 9, 10 & 11 Berg, Tymoczko & Stryer] (Figures in red are for the 7th Edition) (Figures in Blue are for the 8th Edition) Protein Modification
Regulating the Cell Cycle. Lesson Overview THINK ABOUT IT. How do cells know when to divide? Review: Why do cells divide?
 THINK ABOUT IT How do cells know when to divide? Review: Why do cells divide? Controls on Cell Division How is the cell cycle regulated? The cell cycle is controlled by regulatory proteins both inside
THINK ABOUT IT How do cells know when to divide? Review: Why do cells divide? Controls on Cell Division How is the cell cycle regulated? The cell cycle is controlled by regulatory proteins both inside
Summary of Endomembrane-system
 Summary of Endomembrane-system 1. Endomembrane System: The structural and functional relationship organelles including ER,Golgi complex, lysosome, endosomes, secretory vesicles. 2. Membrane-bound structures
Summary of Endomembrane-system 1. Endomembrane System: The structural and functional relationship organelles including ER,Golgi complex, lysosome, endosomes, secretory vesicles. 2. Membrane-bound structures
Methionine (Met or M)
 Fig. 5-17 Nonpolar Fig. 5-17a Nonpolar Glycine (Gly or G) Alanine (Ala or A) Valine (Val or V) Leucine (Leu or L) Isoleucine (Ile or I) Methionine (Met or M) Phenylalanine (Phe or F) Polar Trypotphan (Trp
Fig. 5-17 Nonpolar Fig. 5-17a Nonpolar Glycine (Gly or G) Alanine (Ala or A) Valine (Val or V) Leucine (Leu or L) Isoleucine (Ile or I) Methionine (Met or M) Phenylalanine (Phe or F) Polar Trypotphan (Trp
THE UBIQUITIN-PROTEASOME SYSTEM DURING PROTEOTOXIC STRESS
 Cell and Molecular Biology (CMB) Karolinska Institutet Stockholm, Sweden THE UBIQUITIN-PROTEASOME SYSTEM DURING PROTEOTOXIC STRESS Victoria Menéndez Benito Stockholm 2006 Front cover: Ub-R-YFP MelJuSo
Cell and Molecular Biology (CMB) Karolinska Institutet Stockholm, Sweden THE UBIQUITIN-PROTEASOME SYSTEM DURING PROTEOTOXIC STRESS Victoria Menéndez Benito Stockholm 2006 Front cover: Ub-R-YFP MelJuSo
Ch5: Macromolecules. Proteins
 Ch5: Macromolecules Proteins Essential Knowledge 4.A.1 The subcomponents of biological molecules and their sequence determine the properties of that molecule A. Structure and function of polymers are derived
Ch5: Macromolecules Proteins Essential Knowledge 4.A.1 The subcomponents of biological molecules and their sequence determine the properties of that molecule A. Structure and function of polymers are derived
Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin proteasome system and onto human diseases and drug targeting*
 (2005) 12, 1178 1190 & 2005 Nature Publishing Group All rights reserved 1350-9047/05 $30.00 Review: Nobel Lecture www.nature.com/cdd Intracellular protein degradation: from a vague idea thru the lysosome
(2005) 12, 1178 1190 & 2005 Nature Publishing Group All rights reserved 1350-9047/05 $30.00 Review: Nobel Lecture www.nature.com/cdd Intracellular protein degradation: from a vague idea thru the lysosome
The role of deubiquitinating enzymes in disease processes
 The role of deubiquitinating enzymes in disease processes Thesis submitted to Department of Pharmaceutical Biosciences, School of Pharmacy, Faculty of Mathematics and Natural Sciences, University of Oslo
The role of deubiquitinating enzymes in disease processes Thesis submitted to Department of Pharmaceutical Biosciences, School of Pharmacy, Faculty of Mathematics and Natural Sciences, University of Oslo
Ubiquitylation: its role and medical significance
 Practice-oriented, student-friendly modernization of the biomedical education for strengthening the international competitiveness of the rural Hungarian universities TÁMOP-4.1.1.C-13/1/KONV-2014-0001 Ubiquitylation:
Practice-oriented, student-friendly modernization of the biomedical education for strengthening the international competitiveness of the rural Hungarian universities TÁMOP-4.1.1.C-13/1/KONV-2014-0001 Ubiquitylation:
Saccharomyces cerevisiae*
 THE JOURNAL OF BIOLOGICAL CHEMISTRY 1988 by The American Society for Biochemistry and Molecular Biology, Inc. Vol. 263, No. 29, Issue of October 15, pp. 14948-14955, 1988 Printed in U.S.A. Purification
THE JOURNAL OF BIOLOGICAL CHEMISTRY 1988 by The American Society for Biochemistry and Molecular Biology, Inc. Vol. 263, No. 29, Issue of October 15, pp. 14948-14955, 1988 Printed in U.S.A. Purification
20S Proteasome Activity Assay Kit
 20S Proteasome Activity Assay Kit For 100 Assays Cat. No. APT280 FOR RESEARCH USE ONLY NOT FOR USE IN DIAGNOSTIC PROCEDURES USA & Canada Phone: +1(800) 437-7500 Fax: +1 (951) 676-9209 Europe +44 (0) 23
20S Proteasome Activity Assay Kit For 100 Assays Cat. No. APT280 FOR RESEARCH USE ONLY NOT FOR USE IN DIAGNOSTIC PROCEDURES USA & Canada Phone: +1(800) 437-7500 Fax: +1 (951) 676-9209 Europe +44 (0) 23
Life Sciences 1A Midterm Exam 2. November 13, 2006
 Name: TF: Section Time Life Sciences 1A Midterm Exam 2 November 13, 2006 Please write legibly in the space provided below each question. You may not use calculators on this exam. We prefer that you use
Name: TF: Section Time Life Sciences 1A Midterm Exam 2 November 13, 2006 Please write legibly in the space provided below each question. You may not use calculators on this exam. We prefer that you use
The Basics: A general review of molecular biology:
 The Basics: A general review of molecular biology: DNA Transcription RNA Translation Proteins DNA (deoxy-ribonucleic acid) is the genetic material It is an informational super polymer -think of it as the
The Basics: A general review of molecular biology: DNA Transcription RNA Translation Proteins DNA (deoxy-ribonucleic acid) is the genetic material It is an informational super polymer -think of it as the
Biological systems interact, and these systems and their interactions possess complex properties. STOP at enduring understanding 4A
 Biological systems interact, and these systems and their interactions possess complex properties. STOP at enduring understanding 4A Homework Watch the Bozeman video called, Biological Molecules Objective:
Biological systems interact, and these systems and their interactions possess complex properties. STOP at enduring understanding 4A Homework Watch the Bozeman video called, Biological Molecules Objective:
MOLECULAR CELL BIOLOGY
 1 Lodish Berk Kaiser Krieger scott Bretscher Ploegh Matsudaira MOLECULAR CELL BIOLOGY SEVENTH EDITION CHAPTER 13 Moving Proteins into Membranes and Organelles Copyright 2013 by W. H. Freeman and Company
1 Lodish Berk Kaiser Krieger scott Bretscher Ploegh Matsudaira MOLECULAR CELL BIOLOGY SEVENTH EDITION CHAPTER 13 Moving Proteins into Membranes and Organelles Copyright 2013 by W. H. Freeman and Company
The Structure and Function of Macromolecules
 The Structure and Function of Macromolecules Macromolecules are polymers Polymer long molecule consisting of many similar building blocks. Monomer the small building block molecules. Carbohydrates, proteins
The Structure and Function of Macromolecules Macromolecules are polymers Polymer long molecule consisting of many similar building blocks. Monomer the small building block molecules. Carbohydrates, proteins
! Proteins are involved functionally in almost everything: " Receptor Proteins - Respond to external stimuli. " Storage Proteins - Storing amino acids
 Proteins Most structurally & functionally diverse group! Proteins are involved functionally in almost everything: Proteins Multi-purpose molecules 2007-2008 Enzymatic proteins - Speed up chemical reactions!
Proteins Most structurally & functionally diverse group! Proteins are involved functionally in almost everything: Proteins Multi-purpose molecules 2007-2008 Enzymatic proteins - Speed up chemical reactions!
Molecular Medicine: Gleevec and Chronic Myelogenous Leukemia. Dec 14 & 19, 2006 Prof. Erin O Shea Prof. Dan Kahne
 Molecular Medicine: Gleevec and Chronic Myelogenous Leukemia Dec 14 & 19, 2006 Prof. Erin Shea Prof. Dan Kahne 1 Cancer, Kinases and Gleevec: 1. What is CML? a. Blood cell maturation b. Philadelphia Chromosome
Molecular Medicine: Gleevec and Chronic Myelogenous Leukemia Dec 14 & 19, 2006 Prof. Erin Shea Prof. Dan Kahne 1 Cancer, Kinases and Gleevec: 1. What is CML? a. Blood cell maturation b. Philadelphia Chromosome
Genetic information flows from mrna to protein through the process of translation
 Genetic information flows from mrn to protein through the process of translation TYPES OF RN (RIBONUCLEIC CID) RN s job - protein synthesis (assembly of amino acids into proteins) Three main types: 1.
Genetic information flows from mrn to protein through the process of translation TYPES OF RN (RIBONUCLEIC CID) RN s job - protein synthesis (assembly of amino acids into proteins) Three main types: 1.
االمتحان النهائي لعام 1122
 االمتحان النهائي لعام 1122 Amino Acids : 1- which of the following amino acid is unlikely to be found in an alpha-helix due to its cyclic structure : -phenylalanine -tryptophan -proline -lysine 2- : assuming
االمتحان النهائي لعام 1122 Amino Acids : 1- which of the following amino acid is unlikely to be found in an alpha-helix due to its cyclic structure : -phenylalanine -tryptophan -proline -lysine 2- : assuming
A. B. C. D. E. F. G. H. I. J. K. Ser/Thr. Ser/Thr. Ser/Thr. Ser/Thr. Ser/Thr. Asn. Asn. Asn. Asn. Asn. Asn
 A. B. C. D. E. F. "3 "3!4!3 Ser/Thr "3!4!3!4 Asn Asn Ser/Thr Asn!3!6 Ser/Thr G. H. I. J. K.!3 Ser/Thr Ser/Thr 4 4 2 2 6 3 6 Asn Asn Asn Glycosidases and Glycosyltransferases Introduction to Inverting/Retaining
A. B. C. D. E. F. "3 "3!4!3 Ser/Thr "3!4!3!4 Asn Asn Ser/Thr Asn!3!6 Ser/Thr G. H. I. J. K.!3 Ser/Thr Ser/Thr 4 4 2 2 6 3 6 Asn Asn Asn Glycosidases and Glycosyltransferases Introduction to Inverting/Retaining
Chapter 23 Enzymes 1
 Chapter 23 Enzymes 1 Enzymes Ribbon diagram of cytochrome c oxidase, the enzyme that directly uses oxygen during respiration. 2 Enzyme Catalysis Enzyme: A biological catalyst. With the exception of some
Chapter 23 Enzymes 1 Enzymes Ribbon diagram of cytochrome c oxidase, the enzyme that directly uses oxygen during respiration. 2 Enzyme Catalysis Enzyme: A biological catalyst. With the exception of some
Biomolecules: amino acids
 Biomolecules: amino acids Amino acids Amino acids are the building blocks of proteins They are also part of hormones, neurotransmitters and metabolic intermediates There are 20 different amino acids in
Biomolecules: amino acids Amino acids Amino acids are the building blocks of proteins They are also part of hormones, neurotransmitters and metabolic intermediates There are 20 different amino acids in
Student name ID # 2. (4 pts) What is the terminal electron acceptor in respiration? In photosynthesis?
 1. Membrane transport. A. (4 pts) What ion couples primary and secondary active transport in animal cells? What ion serves the same function in plant cells? 2. (4 pts) What is the terminal electron acceptor
1. Membrane transport. A. (4 pts) What ion couples primary and secondary active transport in animal cells? What ion serves the same function in plant cells? 2. (4 pts) What is the terminal electron acceptor
Molecular Cell Biology. Prof. D. Karunagaran. Department of Biotechnology. Indian Institute of Technology Madras
 Molecular Cell Biology Prof. D. Karunagaran Department of Biotechnology Indian Institute of Technology Madras Module 6 Cell Division, Cell Cycle, Cell Growth and Differentiation, Programmed Cell Death
Molecular Cell Biology Prof. D. Karunagaran Department of Biotechnology Indian Institute of Technology Madras Module 6 Cell Division, Cell Cycle, Cell Growth and Differentiation, Programmed Cell Death
REGULATION OF ENZYME ACTIVITY. Medical Biochemistry, Lecture 25
 REGULATION OF ENZYME ACTIVITY Medical Biochemistry, Lecture 25 Lecture 25, Outline General properties of enzyme regulation Regulation of enzyme concentrations Allosteric enzymes and feedback inhibition
REGULATION OF ENZYME ACTIVITY Medical Biochemistry, Lecture 25 Lecture 25, Outline General properties of enzyme regulation Regulation of enzyme concentrations Allosteric enzymes and feedback inhibition
Translation. Host Cell Shutoff 1) Initiation of eukaryotic translation involves many initiation factors
 Translation Questions? 1) How does poliovirus shutoff eukaryotic translation? 2) If eukaryotic messages are not translated how can poliovirus get its message translated? Host Cell Shutoff 1) Initiation
Translation Questions? 1) How does poliovirus shutoff eukaryotic translation? 2) If eukaryotic messages are not translated how can poliovirus get its message translated? Host Cell Shutoff 1) Initiation
Eukaryotic Gene Regulation
 Eukaryotic Gene Regulation Chapter 19: Control of Eukaryotic Genome The BIG Questions How are genes turned on & off in eukaryotes? How do cells with the same genes differentiate to perform completely different,
Eukaryotic Gene Regulation Chapter 19: Control of Eukaryotic Genome The BIG Questions How are genes turned on & off in eukaryotes? How do cells with the same genes differentiate to perform completely different,
Glycosidic bond cleavage
 Glycosidases and Glycosyltransferases Introduction to Inverting/Retaining Mechanisms Inhibitor design Chemical Reaction Proposed catalytic mechanisms Multiple slides courtesy of Harry Gilbert with Wells
Glycosidases and Glycosyltransferases Introduction to Inverting/Retaining Mechanisms Inhibitor design Chemical Reaction Proposed catalytic mechanisms Multiple slides courtesy of Harry Gilbert with Wells
بسم هللا الرحمن الرحيم
 بسم هللا الرحمن الرحيم Q1: the overall folding of a single protein subunit is called : -tertiary structure -primary structure -secondary structure -quaternary structure -all of the above Q2 : disulfide
بسم هللا الرحمن الرحيم Q1: the overall folding of a single protein subunit is called : -tertiary structure -primary structure -secondary structure -quaternary structure -all of the above Q2 : disulfide
Regulation of Gene Expression in Eukaryotes
 Ch. 19 Regulation of Gene Expression in Eukaryotes BIOL 222 Differential Gene Expression in Eukaryotes Signal Cells in a multicellular eukaryotic organism genetically identical differential gene expression
Ch. 19 Regulation of Gene Expression in Eukaryotes BIOL 222 Differential Gene Expression in Eukaryotes Signal Cells in a multicellular eukaryotic organism genetically identical differential gene expression
Why do cellular proteins linked to K63-polyubiquitin chains not associate with proteasomes?
 The EMBO Journal (2013) 32, 552 565 www.embojournal.org Why do cellular proteins linked to K63-polyubiquitin chains not associate with proteasomes? THE EMBO JOURNAL James A Nathan 1,2, Hyoung Tae Kim 1,
The EMBO Journal (2013) 32, 552 565 www.embojournal.org Why do cellular proteins linked to K63-polyubiquitin chains not associate with proteasomes? THE EMBO JOURNAL James A Nathan 1,2, Hyoung Tae Kim 1,
CHAPTER 10: REGULATORY STRATEGIES. Traffic signals control the flow of traffic
 CHAPTER 10: REGULATORY STRATEGIES Traffic signals control the flow of traffic INTRODUCTION CHAPTER 10 The activity of enzymes must often be regulated so that they function at the proper time and place.
CHAPTER 10: REGULATORY STRATEGIES Traffic signals control the flow of traffic INTRODUCTION CHAPTER 10 The activity of enzymes must often be regulated so that they function at the proper time and place.
基醫所. The Cell Cycle. Chi-Wu Chiang, Ph.D. IMM, NCKU
 基醫所 The Cell Cycle Chi-Wu Chiang, Ph.D. IMM, NCKU 1 1 Introduction to cell cycle and cell cycle checkpoints 2 2 Cell cycle A cell reproduces by performing an orderly sequence of events in which it duplicates
基醫所 The Cell Cycle Chi-Wu Chiang, Ph.D. IMM, NCKU 1 1 Introduction to cell cycle and cell cycle checkpoints 2 2 Cell cycle A cell reproduces by performing an orderly sequence of events in which it duplicates
Bio 100 Serine Proteases 9/26/11
 Assigned Reading: 4th ed. 6.4.1 The Chymotrypsin Mechanism Involves Acylation And Deacylation Of A Ser Residue p. 213 BOX 20-1 Penicillin and β-lactamase p. 779 6.5.7 Some Enzymes Are Regulated By Proteolytic
Assigned Reading: 4th ed. 6.4.1 The Chymotrypsin Mechanism Involves Acylation And Deacylation Of A Ser Residue p. 213 BOX 20-1 Penicillin and β-lactamase p. 779 6.5.7 Some Enzymes Are Regulated By Proteolytic
Cleaning house is one way plants respond to inside and outside changes
 Cleaning house is one way plants respond to inside and outside changes Thank you! ADVANCE Nominators Colleagues Students Judy Callis, Department of MCB, UC-Davis Improving plant productivity impacts: Plants
Cleaning house is one way plants respond to inside and outside changes Thank you! ADVANCE Nominators Colleagues Students Judy Callis, Department of MCB, UC-Davis Improving plant productivity impacts: Plants
Molecular Biology (BIOL 4320) Exam #2 May 3, 2004
 Molecular Biology (BIOL 4320) Exam #2 May 3, 2004 Name SS# This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses after the question number. Good
Molecular Biology (BIOL 4320) Exam #2 May 3, 2004 Name SS# This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses after the question number. Good
HIV-1 genome organization
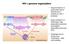 HIV-1 genome organization Gag encodes for a polyprotein that is cleaved into 4 proteins by HIV-1 protease. Gag is present in 2 transcripts: Gag polyprotein precursor and GagPol polyprotein precursor (5%).
HIV-1 genome organization Gag encodes for a polyprotein that is cleaved into 4 proteins by HIV-1 protease. Gag is present in 2 transcripts: Gag polyprotein precursor and GagPol polyprotein precursor (5%).
number Done by Corrected by Doctor Dr. Diala
 number 30 Done by Dergam Al-Tarawneh Corrected by Zaid Emad Doctor Dr. Diala 1 After we ve finished talking about lipids metabolism pathways, today we will start talking about another pathway that takes
number 30 Done by Dergam Al-Tarawneh Corrected by Zaid Emad Doctor Dr. Diala 1 After we ve finished talking about lipids metabolism pathways, today we will start talking about another pathway that takes
Chapter 10. Regulatory Strategy
 Chapter 10 Regulatory Strategy Regulation of enzymatic activity: 1. Allosteric Control. Allosteric proteins have a regulatory site(s) and multiple functional sites Activity of proteins is regulated by
Chapter 10 Regulatory Strategy Regulation of enzymatic activity: 1. Allosteric Control. Allosteric proteins have a regulatory site(s) and multiple functional sites Activity of proteins is regulated by
All intracellular proteins and many extracellular proteins
 Review Protein Degradation by the Ubiquitin Proteasome Pathway in Normal and Disease States Stewart H. Lecker,* Alfred L. Goldberg, and William E. Mitch *Nephrology Division, Beth Israel Deaconess, and
Review Protein Degradation by the Ubiquitin Proteasome Pathway in Normal and Disease States Stewart H. Lecker,* Alfred L. Goldberg, and William E. Mitch *Nephrology Division, Beth Israel Deaconess, and
The Structure and Function of Large Biological Molecules Part 4: Proteins Chapter 5
 Key Concepts: The Structure and Function of Large Biological Molecules Part 4: Proteins Chapter 5 Proteins include a diversity of structures, resulting in a wide range of functions Proteins Enzymatic s
Key Concepts: The Structure and Function of Large Biological Molecules Part 4: Proteins Chapter 5 Proteins include a diversity of structures, resulting in a wide range of functions Proteins Enzymatic s
PDF hosted at the Radboud Repository of the Radboud University Nijmegen
 PDF hosted at the Radboud Repository of the Radboud University Nijmegen The following full text is a publisher's version. For additional information about this publication click this link. http://hdl.handle.net/2066/142604
PDF hosted at the Radboud Repository of the Radboud University Nijmegen The following full text is a publisher's version. For additional information about this publication click this link. http://hdl.handle.net/2066/142604
Protein and membrane trafficking
 Protein and membrane trafficking Lodish et al, 2000 The secretory pathway cisternal progression Lodish et al, 2000 Cells can be specialized for the secretion of specific proteins Lodish et al, 2000 The
Protein and membrane trafficking Lodish et al, 2000 The secretory pathway cisternal progression Lodish et al, 2000 Cells can be specialized for the secretion of specific proteins Lodish et al, 2000 The
