doi: /nature14508 Rappsilber et al.
|
|
|
- Audra Kelly
- 5 years ago
- Views:
Transcription
1 SUPPLEMENTARY INFORMATION doi:1.138/nature1458 Grosso et al. Barbosa et al Rappsilber et al. Supplementary Figure 1 a, Venn-Diagram of identified splice factors in the work of Barbossa et al., Grosso et al. and Rappsilber et al. A total number of 329 genes were indentified A C Supplementary Figure 2 Schematic representation of KHK pre-mrna. Alternative spliced exons 3A and 3C leading to expression of either KHK-A or KHK-C protein are highlighted in blue (exon 3A) and red (exon 3C). 1
2 RESEARCH SUPPLEMENTARY INFORMATION a Rel. mrna levels Rel. mrna levels sham 1K1C ** b Nppa Nppb αmhc βmhc sk. αactin sham 2 ** TAC c Rel. mrna levels d Rel. mrna levels sk. αactin Nppb αmhc βmhc NaCl ISO ** ** ** sk. αactin Nppb αmhc βmhc NPPA Ctrl HCM NPPB Rel. mrna levels NPPA Ctrl sten. * * NPPB Supplementary Figure 3 Gene expression of hypertrophic markers in mouse models of cardiac hypertrophy or heart disease patients and their corresponding controls, presented in Extended Data Fig. 1. a, b, Probes from left ventricles of 1K1C, TAC or ISO-treated mice and their corresponding controls were analyzed for mrna hypertrophic markers. All values were presented in relation to sham-operated controls set as 1. (n = 6; shown is mean ± SEM; *p<.5; **p<.1; p <.1; two-tailed unpaired t-test). c, d, Leftventricular biopsies of hypertrophic cardiomyopathy (HCM) patients and healthy controls were analysed for mrna expression hypertrophic markers (t). All values are presented in relation to healthy controls set as 1. (n = 6 for controls and n = 16 for patient samples; shown is mean ± SEM; *p<.5; p<.1; two-tailed unpaired t-test). 2
3 SUPPLEMENTARY INFORMATION RESEARCH Rel. mrna levels Sf3a3 Sf3b2 Sf3b3 Sf3b1 ** ** * ** ** shsf3a3 shsf3b2 shsf3b3 shsf3a3 shsf3b2 shsf3b3 ** HIF1αΔODD Supplementary Figure 4 Depletion of U2-snRNP components in NMC in the presence or absence of HIF1αΔODD. NMC were transduced with either scrambled shrna or shsf3a3, shsf3b2 or shsf3b3 were co-transduced with vector or HIF1αΔODD. Gene expression was evaluated by qrt-pcr. Values were normalised to / infected NMC (n = 3; shown is mean ± SD; *p <.5; **p <.1; p <.1; one-way ANOVA followed by Dunnett s multiple comparison post-test). 3
4 RESEARCH SUPPLEMENTARY INFORMATION OCR/ECAR [pmol/mph] ECAR [mph/min] OA shsf3b shns shsf3b1 Time (min) OCR [pmol/min] FCCP OA FCCP Supplementary Figure 5 Assessment of extracellular acidification rate (ECAR), and oleic acid and FCCP-induced oxygen consumption rate (OCR) in NMC transduced with and shsf3b1 lentiviruses. Highlighted ECAR measurements were done at baseline. All data are compared to -transduced NMC. (n = 9 per condition) 4
5 SUPPLEMENTARY INFORMATION RESEARCH mm Glc 2.5 mm Glc 1 mm Glc µm Frc µm Frc 5 µm Frc a KHK-A KHK-C KHK-A KHK-C KHK-A KHK-C KHK-A KHK-C KHK-A KHK-C KHK-A KHK-C Khk-A/C Khk-C α-actinin Rel. lipid accumulation e Rel. [ 3 H] count in lipid fraction / /HIF1αΔODD ** shsf3b1 -A -C shglut shsf3b1 21% O 2 3% O 2 b c d g OCR/ECAR [pmol/mph/min] ECAR [mph/min] A -C OA OCR [pmol/min] FCCP -A Time (min) -A OA FCCP Rel. lipid accumulation -A -C HIF1αΔODD h OCR/ECAR [pmol/mph/min] ECAR [mph/min] A -C shglut5 B d, NMC transduced and treated as indicated were incubated overnight with OA and stained with Nile Red. Stainings were quantified using a plate reader with 485/5 nm excitation/emission filters. Values were compared to control set as 1.. (n = 3; shown is mean ± SD; p >.1; one-way ANOVA followed by Dunnett s multiple comparison post-test).e, NMC infected as denoted were incubated with oleic acid (OA) and [ 3 H]fructose. A lipid extraction was done to assess incorporation of [ 3 H] into lipids (n = 4; shown is mean ± SD; p >.1; one-way ANOVA followed by Dunnett s multiple comparison post-test). f, shrna against Glut5 was evaluated by qpcr for its potential to inhibit Glut5 expression. Data are shown in relation to NMC (set to 1.), (n = 3; shown is mean ± SD; p >.1; two-tailed unpaired t-test). g, h, Assessment of ECAR and OA and FCCP-induced OCR in NMC infected with or -A lentiviruses (g) and or -C lentiviruses (h), Highlighted ECAR measurements were done at baseline. All data are compared to transduced NMC (n = 9; shown is mean ± SEM; p >.1; two-tailed unpaired t-test). KHK-A KHK-C OA Rel. lipid accumulation f Glut5 (Rel. mrna levels) C Time (min) OCR [pmol/min] FCCP -C OA FCCP mock PE shglut5 shsf3b1 -A -C Supplementary Figure 6 Downstream metabolic effects of fructose metabolism in NMC. a, Lysates of NMC transduced with or and coinfected with vector or KHK-A or KHK-C overexpression lentiviruses, respectively and cultured in media with different fructose and glucose concentrations as indicated were probed for protein expression of KHK-A, KHK-C and sarcomeric α-actinin by immunoblotting. 5
6 RESEARCH SUPPLEMENTARY INFORMATION a b Ad lacz Ad Cre Ad-lacZ / AAV9-fl/fl- GFP 1 x 1 x Ad-lacZ / AAV9-fl/fl-shSf3b1#1 GFP 1 x 1 x Ad-lacZ / AAV9-fl/fl-shSf3b1#2 1 x GFP 1 x shsf3b1#1 shsf3b1#2 shsf3b1#1 shsf3b1#2 GFP Ad-Cre / AAV9-fl/fl- Ad-Cre/ AAV9-fl/fl-shSf3b1#1 Ad-Cre/ AAV9-fl/fl-shSf3b1#2 Cre Sf3b1 Sf3b1 (Rel. mrna levels) Sf3b1 (Rel. mrna Levels) c d Mlc2v-cre Mlc2v-cre+ LV Liver Spleen Pancreas sk. Muscle * LV Liver Spleen Pancreas sk. Muscle * AAV9-fl/fl-shSf3b1#1 α-actinin WAT Intestine Gut right Kidney left Kidney Brain BAT Lungs AAV9-fl/fl-shSf3b1#1 WAT Intestine Gut right Kidney left Kidney Brain BAT Lungs Supplementary Figure 7 In vitro and in vivo validation of modified AAV9. a, b, NMC transduced with AAV9-fl/fl, AAV9-fl/fl-shSf3b1#1 and AAV9-fl/fl-shSf3b1#2 viruses in NMC were investigated for removal of the CMV-GFP/ stop reporter cassette by Adeno-Cre-mediated recombination using immunofluorescent microscopy (a) and immunoblot detection for indicated proteins (b) As a control served transductions with Adeno-lacZ viruses, where the GFP/stop reporter cassette is retained. c, d, Sf3b1 mrna expression was assessed using qpcr in different organs of Mlc2v-cre and Mlc2v-cre+ mice injected as in m. Data represent mean ± SEM; (n = 4 for Mlc2v-cre AAV9-fl/fl-shSf3b1#1 or Mlc2vcre+ AAV9-fl/fl-shSf3b1#1 mice and n = 3 for for Mlc2v-cre AAV9-fl/fl-shSf3b1#2 or Mlc2v-cre+ AAV9-fl/fl-shSf3b1#2 mice; shown is mean ± SEM; p <.1; two-tailed unpaired t-test). 6
7 SUPPLEMENTARY INFORMATION RESEARCH Sample ID LV 1J LV 2J LV 3J LV 26 LV 29 LV 3 LV 34 LV 39 LV 41 LV 43 LV 48 LV 33 LV 61 LV 62 LV 9 LV 92 LV11 Sample ID Sex Type age AVA (cm 2 ) male male male male male male Sex Type Age PCWP (mmhg) MPG (mmhg) PWD (mm) IVSD (mm) Antihyp. Drugs EF (%) Type 2 Diabetes yes yes yes 75 no yes 74 yes yes 6 yes yes 75 no yes 55 no 58 n.a. yes 65 yes yes 7 yes yes 7 yes yes yes 6 no yes yes yes yes yes yes 6 CI (l/min/ m 3 ) pmean (mmhg) IVSD (mm) Hyperte nsion EF (%) LVEDD (mm) LV 77 HCM yes 65 LV 91 HCM 5 yes 3 LV115 HCM yes 6 LV496 male HCM yes 6 76 LV5 male HCM yes LV552 HCM yes LV624 HCM yes 65 LV679 HCM yes 39 4 LV73 male HCM yes 3 LV712 HCM LV7 male HCM yes 4 53 yes 2 Supplementary Table 4 Demographic and clinical data of patients with and hypertrophic cardiomyopathy. LV = left ventricle; AVA = aortic valve area; MPG = mean pressure gradient; PWD = posterior wall diameter; IVSD = interventricular septum diameter; EF = ejection fraction; PCWP = pulmonary capillary wedge pressure; CI = cardiac index; pmean = mean pressure; LVEDD = left ventricular end-diastolic diameter. 7
8 RESEARCH SUPPLEMENTARY INFORMATION The protein coding sequence of (mouse) codon optimised Sf3b1 atggcgaagatcgccaagactcacgaagatatcgaagcacagattcgagaaattcaaggcaagaaggcagctcttgatga! agcccaaggagtgggccttgattccacaggttattatgaccaagaaatttatggtggaagtgatagcagatttgctggat! atgtgacatcaattgctgcaactgaacttgaagatgatgacgatgactactcatcatccacgagcttgctcggtcagaag! aagcctggatatcatgcccccgtggcgttgcttaatgatataccacagtcaacagagcagtatgatccatttgctgagca! tcgtcctccaaagattgccgatcgggaagatgaatacaaaaagcataggcggaccatgataatttccccagagcgtcttg! atccttttgcagatggagggaagactcccgatcctaaaatgaatgctagaacctacatggatgttatgcgagaacaacat! ttgactaaggaagagagagaaattaggcaacaactagcagaaaaagctaaagctggagaactaaaagtcgtcaatggagc! agcagcatcccagcctccctcaaaacgaaaacggcgctgggatcagaccgctgaccagactcctggtgccactcccaaaa! agctatcgagttgggatcaggcagagacccctgggcataccccatctttaagatgggatgagacaccgggtcgtgcaaaa! ggaagtgaaacacctggcgcaaccccaggctcaaaaatatgggaccctacaccgagtcatacacctgcgggagctgctac! tcctgggcgaggcgacacaccaggccatgcaaccccgggccatggtggtgcaacttccagtgctcgtaaaaacagatggg! atgagacccccaaaacagaaagagatactcctgggcatggaagtggatgggctgagactcctcgaacagatcgaggtgga! gactctattggtgaaacaccaactcctggagcaagtaaaagaaagtctcgttgggatgaaacgccggcgagtcaaatggg! tggaagcactcctgttctgactccaggaaaaacaccaattggcacaccagccatgaacatggctaccccgactccaggtc! acataatgagcatgacacctgaacagcttcaggcgtggcggtgggagagagaaattgatgaacgaaaccgcccactttct! gatgaagaattagatgctatgttcccagaaggatataaggtacttccccctccagctggttatgttcctattcgaactcc! agctcgaaagctgacagcaactcctacacctttgggtggtatgactggtttccacatgcaaactgaagacagaactatga! aaagtgtcaatgaccagccatctgggaatcttccatttttaaaacctgatgacattcagtactttgataaactattggtt! gatgtagatgaatcaacacttagtccagaagaacaaaaagaaagaaaaataatgaagttgcttttaaaaattaagaatgg! tacacctccaatgagaaaggctgccttacgtcagattactgataaagctcgagaatttggagccggaccactgttcaacc! agattctgccactgctgatgtcacctacactggaagaccaggagagacacctgctggtgaaggtcatcgaccggattctg! tataaactggacgatctggtgagaccatacgtccataaaatcctggtggtcattgagcccctgctgatcgacgaagatta! ctatgccagggtggagggcagagaaatcatttcaaacctggctaaggccgctggactggcaacaatgatcagcactatgc! ggcccgacattgataacatggatgagtatgtgagaaataccacagcacgggccttcgctgtggtcgcaagtgccctgggc! atcccctcactgctgccttttctgaaggccgtgtgcaaatctaagaaaagttggcaggcacggcacactgggatcaagat! tgtgcagcagatcgccattctgatgggctgtgctatcctgcctcacctgcgcagtctggtggagatcattgaacatggcc! tggtggacgagcagcagaaggtcaggactatctcagctctggcaattgcagccctggctgaggctgcaaccccttatgga! atcgaatcattcgatagcgtgctgaagccactgtggaaaggaattcgacagcataggggcaagggactggccgcttttct! gaaagccatcggctatctgattccactgatggacgccgagtacgctaactactatacacgggaagtgatgctgatcctga! ttcgcgagttccagagccccgacgaggaaatgaagaaaatcgtgctgaaggtggtcaaacagtgctgtggcacagatgga! gtggaggccaattatatcaaaactgaaattctgccccctttctttaagcacttttggcagcatcgcatggccctggacag! gagaaactaccgacagctggtggatactaccgtcgaactggctaataaggtcggagcagccgagatcattagcagaatcg! tcgacgatctgaaggatgaggccgaacagtaccggaaaatggtcatggagaccatcgaaaagattatggggaacctgggc! gctgcagacatcgatcacaagctggaggaacagctgatcgacgggattctgtatgccttccaggagcagacaactgaaga! ttccgtgatgctgaacgggtttggcaccgtggtcaatgctctgggaaagcgcgtgaaaccttacctgccacagatctgcg! ggacagtcctgtggcggctgaacaataagtctgcaaaagtgcgccagcaggccgctgacctgatcagtaggactgctgtg! gtcatgaagacctgtcaggaggaaaaactgatgggacacctgggggtggtcctgtacgaatatctgggggaggaataccc! agaggtgctggggtctatcctgggcgcgttaaaggccatcgtgaacgtcattggcatgcacaagatgaccccacccatca! aagatctgctgccacgactgacacccattctgaagaacaggcatgagaaagtgcaggaaaattgcattgacctggtggga! agaattgctgatcgcggagctgagtatgtgagtgcccgagaatggatgaggatctgtttcgagctgctggaactgctgaa! ggcacacaagaaagccattcggcgcgctactgtgaatacctttgggtacatcgccaaagctattggacctcatgacgtgc! tggccaccctgctgaacaatctgaaggtccaggagagacagaaccgggtgtgcaccacagtcgcaatcgccattgtggcc! gagacctgttcccctttcacagtcctgccagctctgatgaatgagtatagagtgcctgaactgaacgtgcagaatggcgt! cctgaaaagcctgtccttcctgtttgagtacatcggcgaaatgggaaaggactacatctacgcagtgacaccactgctgg! aggatgccctgatggaccgcgatctggtgcaccgacagactgctagcgcagtggtccagcatatgtccctgggagtgtat! ggattcgggtgcgaggactccctgaaccacctgctgaattacgtgtggcccaacgtctttgaaacctctcctcatgtgat! ccaggcagtcatgggagccctggagggactgagggtggccattggaccctgcagaatgctgcagtattgtctgcaggggc! tgttccaccctgctcgcaaggtgcgagacgtgtactggaagatctataactccatctacatcggctctcaggatgctctg! attgcacattaccctcgaatctacaatgacgacaagaacacttacatccgctatgaactggactacatcctgtga! Translation of the protein coding sequence of (mouse) codon optimised Sf3b1 MAKIAKTHEDIEAQIREIQGKKAALDEAQGVGLDSTGYYDQEIYGGSDSRFAGYVTSIAATELEDDDDDYSSSTSLLGQKKPGYHAPVAL LNDIPQSTEQYDPFAEHRPPKIADREDEYKKHRRTMIISPERLDPFADGGKTPDPKMNARTYMDVMREQHLTKEEREIRQQLAEKAKAGE LKVVNGAAASQPPSKRKRRWDQTADQTPGATPKKLSSWDQAETPGHTPSLRWDETPGRAKGSETPGATPGSKIWDPTPSHTPAGAATPGR GDTPGHATPGHGGATSSARKNRWDETPKTERDTPGHGSGWAETPRTDRGGDSIGETPTPGASKRKSRWDETPASQMGGSTPVLTPGKTPI GTPAMNMATPTPGHIMSMTPEQLQAWRWEREIDERNRPLSDEELDAMFPEGYKVLPPPAGYVPIRTPARKLTATPTPLGGMTGFHMQTED RTMKSVNDQPSGNLPFLKPDDIQYFDKLLVDVDESTLSPEEQKERKIMKLLLKIKNGTPPMRKAALRQITDKAREFGAGPLFNQILPLLM SPTLEDQERHLLVKVIDRILYKLDDLVRPYVHKILVVIEPLLIDEDYYARVEGREIISNLAKAAGLATMISTMRPDIDNMDEYVRNTTAR AFAVVASALGIPSLLPFLKAVCKSKKSWQARHTGIKIVQQIAILMGCAILPHLRSLVEIIEHGLVDEQQKVRTISALAIAALAEAATPYG IESFDSVLKPLWKGIRQHRGKGLAAFLKAIGYLIPLMDAEYANYYTREVMLILIREFQSPDEEMKKIVLKVVKQCCGTDGVEANYIKTEI LPPFFKHFWQHRMALDRRNYRQLVDTTVELANKVGAAEIISRIVDDLKDEAEQYRKMVMETIEKIMGNLGAADIDHKLEEQLIDGILYAF QEQTTEDSVMLNGFGTVVNALGKRVKPYLPQICGTVLWRLNNKSAKVRQQAADLISRTAVVMKTCQEEKLMGHLGVVLYEYLGEEYPEVL GSILGALKAIVNVIGMHKMTPPIKDLLPRLTPILKNRHEKVQENCIDLVGRIADRGAEYVSAREWMRICFELLELLKAHKKAIRRATVNT FGYIAKAIGPHDVLATLLNNLKVQERQNRVCTTVAIAIVAETCSPFTVLPALMNEYRVPELNVQNGVLKSLSFLFEYIGEMGKDYIYAVT PLLEDALMDRDLVHRQTASAVVQHMSLGVYGFGCEDSLNHLLNYVWPNVFETSPHVIQAVMGALEGLRVAIGPCRMLQYCLQGLFHPARK VRDVYWKIYNSIYIGSQDALIAHYPRIYNDDKNTYIRYELDYIL*! Supplementary Data 1 Coding and amino acid sequence of (mouse) codon optimised Sf3b1. 8
9 SUPPLEMENTARY INFORMATION RESEARCH Agarose gel referring to Fig. 1c Agarose gel referring to Fig. 1d HIF1αΔODD shns shsf3b1 shns shsf3b1 sham 1K1C 1 bp 5 bp 4 bp 3 bp 2 bp 1 bp 1.5 % agarose gel, Gene Ruler 1 bp DNA Ladder 1 bp 5 bp 4 bp 3 bp 2 bp 1 bp 1.5 % agarose gel, Gene Ruler 1 bp DNA Ladder Agarose gels referring to Fig. 1g Primer set 1 Primer set 1 1 bp 5 bp 3 bp 2 bp 1 bp 5 bp 3 bp 2 bp 1.5 % agarose gel, Gene Ruler 1 bp DNA Ladder Primer set % agarose gel, Gene Ruler 1 bp DNA Ladder Primer set 2 1 bp 5 bp 3 bp 2 bp 5 bp 3 bp 2 bp 1.5 % agarose gel, Gene Ruler 1 bp DNA Ladder 1.5 % agarose gel, Gene Ruler 1 bp DNA Ladder 9
10 RESEARCH SUPPLEMENTARY INFORMATION Immunoblots Figure 1e Anti-HIF1α Anti-Sf3b1 Anti-Khk-A/C (l. E.) 13 1 Anti-Khk-A/C 75 Anti-αActinin 1
11 SUPPLEMENTARY INFORMATION RESEARCH Immunoblots Figure 1h Anti-SF3B1 Anti-HIF1α Anti-KHK-A/C 13 Anti-KHK-C 1 7 Anti-cardiac αactinin 11
12 RESEARCH SUPPLEMENTARY INFORMATION ChIP Fig. 2b 2 bp 1 bp 21% O 2 HIF1αΔODD 3% O 2 21% O 2 HIF1αΔODD shhif1α 3% O 2 shhif1α 5 bp 4 bp 3 bp 2 bp 1 bp 12
13 SUPPLEMENTARY INFORMATION RESEARCH Immunoblots Figure 4e Anti-Hif1α Anti-Sf3b1 15 Anti-GFP Anti-Khk-A/C Anti-Khk-C 13 1 Anti-αActinin 13
14 RESEARCH SUPPLEMENTARY INFORMATION Immunoblots Figure 4h Anti-Hif1α Anti-Sf3b1 5 Anti-GFP Anti-Khk-A/C 13 Anti-Khk-C 1 Anti-αActinin 14
15 SUPPLEMENTARY INFORMATION RESEARCH Immunoblots Figure 5d Anti-HIF1α Anti-Sf3b1 Anti-Khk-A/C Anti-Khk-C Anti-αActinin 15
16 RESEARCH SUPPLEMENTARY INFORMATION Immunoblots Figure 5h 13 1 Anti-HIF1α 15 Anti-Sf3b1 Anti-Khk-A/C Anti-Khk-C Anti-αActinin 16
Tcf21 MCM ; R26 mtmg Sham GFP Col 1/3 TAC 8W TAC 2W. Postn MCM ; R26 mtmg Sham GFP Col 1/3 TAC 8W TAC 2W
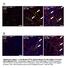 A Tcf21 MCM ; R26 mtmg Sham GFP Col 1/3 Tcf21 MCM ; R26 mtmg TAC 2W Tcf21 MCM ; R26 mtmg TAC 8W B Postn MCM ; R26 mtmg Sham GFP Col 1/3 Postn MCM ; R26 mtmg TAC 2W Postn MCM ; R26 mtmg TAC 8W Supplementary
A Tcf21 MCM ; R26 mtmg Sham GFP Col 1/3 Tcf21 MCM ; R26 mtmg TAC 2W Tcf21 MCM ; R26 mtmg TAC 8W B Postn MCM ; R26 mtmg Sham GFP Col 1/3 Postn MCM ; R26 mtmg TAC 2W Postn MCM ; R26 mtmg TAC 8W Supplementary
SUPPLEMENTARY INFORMATION
 a c e doi:10.1038/nature10407 b d f Supplementary Figure 1. SERCA2a complex analysis. (a) Two-dimensional SDS-PAGE gels of SERCA2a complexes. A silver-stained SDSPAGE gel is shown, which reveals a 12 kda
a c e doi:10.1038/nature10407 b d f Supplementary Figure 1. SERCA2a complex analysis. (a) Two-dimensional SDS-PAGE gels of SERCA2a complexes. A silver-stained SDSPAGE gel is shown, which reveals a 12 kda
E10.5 E18.5 P2 10w 83w NF1 HF1. Sham ISO. Bmi1. H3K9me3. Lung weight (g)
 Myociyte cross-sectional Relative mrna levels Relative levels Relative mrna levels Supplementary Figures and Legends a 8 6 4 2 Ezh2 E1.5 E18.5 P2 1w 83w b Ezh2 p16 amhc b-actin P2 43w kd 37 86 16 wt mouse
Myociyte cross-sectional Relative mrna levels Relative levels Relative mrna levels Supplementary Figures and Legends a 8 6 4 2 Ezh2 E1.5 E18.5 P2 1w 83w b Ezh2 p16 amhc b-actin P2 43w kd 37 86 16 wt mouse
BNP mrna expression in DR and DS rat left ventricles (n = 5). (C) Plasma norepinephrine
 Kanazawa, et al. Supplementary figure legends Supplementary Figure 1 DS rats had congestive heart failure. (A) DR and DS rat hearts. (B) QRT-PCR analysis of BNP mrna expression in DR and DS rat left ventricles
Kanazawa, et al. Supplementary figure legends Supplementary Figure 1 DS rats had congestive heart failure. (A) DR and DS rat hearts. (B) QRT-PCR analysis of BNP mrna expression in DR and DS rat left ventricles
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
Author Manuscript Faculty of Biology and Medicine Publication
 Serveur Académique Lausannois SERVAL serval.unil.ch Author Manuscript Faculty of Biology and Medicine Publication This paper has been peer-reviewed but dos not include the final publisher proof-corrections
Serveur Académique Lausannois SERVAL serval.unil.ch Author Manuscript Faculty of Biology and Medicine Publication This paper has been peer-reviewed but dos not include the final publisher proof-corrections
hemodynamic stress. A. Echocardiographic quantification of cardiac dimensions and function in
 SUPPLEMENTAL FIGURE LEGENDS Supplemental Figure 1. Fbn1 C1039G/+ hearts display normal cardiac function in the absence of hemodynamic stress. A. Echocardiographic quantification of cardiac dimensions and
SUPPLEMENTAL FIGURE LEGENDS Supplemental Figure 1. Fbn1 C1039G/+ hearts display normal cardiac function in the absence of hemodynamic stress. A. Echocardiographic quantification of cardiac dimensions and
Control. csarnt -/- Cre, f/f
 ody weight (g) A re,f/f re x f/f f/+ re, f/+ re,f/+ f/f x f/f f/+ cs -/- re, f/f re f/f re, f/f Normal chow Tamoxifen Tamoxifen Tamoxifen W 4W re f/f re, re/ff f/f re f/f re, re/ff f/f Normal chow Tamoxifen
ody weight (g) A re,f/f re x f/f f/+ re, f/+ re,f/+ f/f x f/f f/+ cs -/- re, f/f re f/f re, f/f Normal chow Tamoxifen Tamoxifen Tamoxifen W 4W re f/f re, re/ff f/f re f/f re, re/ff f/f Normal chow Tamoxifen
Gallic acid prevents isoproterenol-induced cardiac hypertrophy and fibrosis through regulation of JNK2 signaling and Smad3 binding activity
 Gallic acid prevents isoproterenol-induced cardiac hypertrophy and fibrosis through regulation of JNK2 signaling and Smad3 binding activity Yuhee Ryu 1,+, Li Jin 1,2+, Hae Jin Kee 1,, Zhe Hao Piao 3, Jae
Gallic acid prevents isoproterenol-induced cardiac hypertrophy and fibrosis through regulation of JNK2 signaling and Smad3 binding activity Yuhee Ryu 1,+, Li Jin 1,2+, Hae Jin Kee 1,, Zhe Hao Piao 3, Jae
Supplementary Figure S1. PTPN2 levels are not altered in proliferating CD8+ T cells. Lymph node (LN) CD8+ T cells from C57BL/6 mice were stained with
 Supplementary Figure S1. PTPN2 levels are not altered in proliferating CD8+ T cells. Lymph node (LN) CD8+ T cells from C57BL/6 mice were stained with CFSE and stimulated with plate-bound α-cd3ε (10µg/ml)
Supplementary Figure S1. PTPN2 levels are not altered in proliferating CD8+ T cells. Lymph node (LN) CD8+ T cells from C57BL/6 mice were stained with CFSE and stimulated with plate-bound α-cd3ε (10µg/ml)
Nature Immunology: doi: /ni Supplementary Figure 1. Id2 and Id3 define polyclonal T H 1 and T FH cell subsets.
 Supplementary Figure 1 Id2 and Id3 define polyclonal T H 1 and T FH cell subsets. Id2 YFP/+ (a) or Id3 GFP/+ (b) mice were analyzed 7 days after LCMV infection. T H 1 (SLAM + CXCR5 or CXCR5 PD-1 ), T FH
Supplementary Figure 1 Id2 and Id3 define polyclonal T H 1 and T FH cell subsets. Id2 YFP/+ (a) or Id3 GFP/+ (b) mice were analyzed 7 days after LCMV infection. T H 1 (SLAM + CXCR5 or CXCR5 PD-1 ), T FH
SUPPLEMENTARY INFORMATION
 Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Supplementary Figure 1. Confocal immunofluorescence showing mitochondrial translocation of Drp1. Cardiomyocytes treated with H 2 O 2 were prestained
 Supplementary Figure 1. Confocal immunofluorescence showing mitochondrial translocation of Drp1. Cardiomyocytes treated with H 2 O 2 were prestained with MitoTracker (red), then were immunostained with
Supplementary Figure 1. Confocal immunofluorescence showing mitochondrial translocation of Drp1. Cardiomyocytes treated with H 2 O 2 were prestained with MitoTracker (red), then were immunostained with
Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous
 Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous LRP5 in intact adult mouse ventricular myocytes (AMVMs)
Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous LRP5 in intact adult mouse ventricular myocytes (AMVMs)
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11095 Supplementary Table 1. Summary of the binding between Angptls and various Igdomain containing receptors as determined by flow cytometry analysis. The results were summarized from
doi:10.1038/nature11095 Supplementary Table 1. Summary of the binding between Angptls and various Igdomain containing receptors as determined by flow cytometry analysis. The results were summarized from
In vivo bromodeoxyuridine (BrdU) incorporation was performed to analyze cell
 Supplementary Methods BrdU incorporation in vivo In vivo bromodeoxyuridine (BrdU) incorporation was performed to analyze cell proliferation in the heart. Mice were subjected to LI-TAC, and 5 days later
Supplementary Methods BrdU incorporation in vivo In vivo bromodeoxyuridine (BrdU) incorporation was performed to analyze cell proliferation in the heart. Mice were subjected to LI-TAC, and 5 days later
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12215 Supplementary Figure 1. The effects of full and dissociated GR agonists in supporting BFU-E self-renewal divisions. BFU-Es were cultured in self-renewal medium with indicated GR
doi:10.1038/nature12215 Supplementary Figure 1. The effects of full and dissociated GR agonists in supporting BFU-E self-renewal divisions. BFU-Es were cultured in self-renewal medium with indicated GR
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10188 Supplementary Figure 1. Embryonic epicardial genes are down-regulated from midgestation stages and barely detectable post-natally. Real time qrt-pcr revealed a significant down-regulation
doi:10.1038/nature10188 Supplementary Figure 1. Embryonic epicardial genes are down-regulated from midgestation stages and barely detectable post-natally. Real time qrt-pcr revealed a significant down-regulation
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature11429 S1a 6 7 8 9 Nlrc4 allele S1b Nlrc4 +/+ Nlrc4 +/F Nlrc4 F/F 9 Targeting construct 422 bp 273 bp FRT-neo-gb-PGK-FRT 3x.STOP S1c Nlrc4 +/+ Nlrc4 F/F casp1
SUPPLEMENTARY INFORMATION doi:10.1038/nature11429 S1a 6 7 8 9 Nlrc4 allele S1b Nlrc4 +/+ Nlrc4 +/F Nlrc4 F/F 9 Targeting construct 422 bp 273 bp FRT-neo-gb-PGK-FRT 3x.STOP S1c Nlrc4 +/+ Nlrc4 F/F casp1
Supporting Information
 Supporting Information Palmisano et al. 10.1073/pnas.1202174109 Fig. S1. Expression of different transgenes, driven by either viral or human promoters, is up-regulated by amino acid starvation. (A) Quantification
Supporting Information Palmisano et al. 10.1073/pnas.1202174109 Fig. S1. Expression of different transgenes, driven by either viral or human promoters, is up-regulated by amino acid starvation. (A) Quantification
Kidney. Heart. Lung. Sirt1. Gapdh. Mouse IgG DAPI. Rabbit IgG DAPI
 a e Na V 1.5 Ad-LacZ Ad- 110KD b Scn5a/ (relative to Ad-LacZ) f 150 100 50 0 p = 0.65 Ad-LacZ Ad- c Heart Lung Kidney Spleen 110KD d fl/fl c -/- DAPI 20 µm Na v 1.5 250KD fl/fl Rabbit IgG DAPI fl/fl Mouse
a e Na V 1.5 Ad-LacZ Ad- 110KD b Scn5a/ (relative to Ad-LacZ) f 150 100 50 0 p = 0.65 Ad-LacZ Ad- c Heart Lung Kidney Spleen 110KD d fl/fl c -/- DAPI 20 µm Na v 1.5 250KD fl/fl Rabbit IgG DAPI fl/fl Mouse
Supplemental Figure 1. Intracranial transduction of a modified ptomo lentiviral vector in the mouse
 Supplemental figure legends Supplemental Figure 1. Intracranial transduction of a modified ptomo lentiviral vector in the mouse hippocampus targets GFAP-positive but not NeuN-positive cells. (A) Stereotaxic
Supplemental figure legends Supplemental Figure 1. Intracranial transduction of a modified ptomo lentiviral vector in the mouse hippocampus targets GFAP-positive but not NeuN-positive cells. (A) Stereotaxic
Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2)
 Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
Supplementary. presence of the. (c) mrna expression. Error. in naive or
 Figure 1. (a) Naive CD4 + T cells were activated in the presence of the indicated cytokines for 3 days. Enpp2 mrna expression was measured by qrt-pcrhr, infected with (b, c) Naive CD4 + T cells were activated
Figure 1. (a) Naive CD4 + T cells were activated in the presence of the indicated cytokines for 3 days. Enpp2 mrna expression was measured by qrt-pcrhr, infected with (b, c) Naive CD4 + T cells were activated
EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH
 EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH Supplementary Figure 1. Supplementary Figure 1. Characterization of KP and KPH2 autochthonous UPS tumors. a) Genotyping of KPH2
EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH Supplementary Figure 1. Supplementary Figure 1. Characterization of KP and KPH2 autochthonous UPS tumors. a) Genotyping of KPH2
Supplementary Figure 1
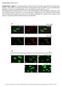 Supplementary Figure 1. (A) Representative confocal analysis of cardiac mesenchymal cells from nondiabetic (ND-CMSC) and diabetic (D-CMSC) immunostained with anti-h3k9ac and anti-h3k27me3 antibodies. Analysis
Supplementary Figure 1. (A) Representative confocal analysis of cardiac mesenchymal cells from nondiabetic (ND-CMSC) and diabetic (D-CMSC) immunostained with anti-h3k9ac and anti-h3k27me3 antibodies. Analysis
c Ischemia (30 min) Reperfusion (8 w) Supplementary Figure bp 300 bp Ischemia (30 min) Reperfusion (4 h) Dox 20 mg/kg i.p.
 a Marker Ripk3 +/ 5 bp 3 bp b Ischemia (3 min) Reperfusion (4 h) d 2 mg/kg i.p. 1 w 5 w Sacrifice for IF size A subset for echocardiography and morphological analysis c Ischemia (3 min) Reperfusion (8
a Marker Ripk3 +/ 5 bp 3 bp b Ischemia (3 min) Reperfusion (4 h) d 2 mg/kg i.p. 1 w 5 w Sacrifice for IF size A subset for echocardiography and morphological analysis c Ischemia (3 min) Reperfusion (8
Supplemental Table 1. Plasma NEFA and liver triglyceride levels in ap2-hif1ako and ap2-hif2ako mice under control and high fat diets.
 Supplemental Table 1. Plasma NEFA and liver triglyceride levels in Hif1aKO and Hif2aKO mice under control and high fat diets. Hif1a (n=6) Hif1aK O (n=6) Hif2a Hif2aK O Hif1a (n=5) Hif1aKO (n=5) Hif2a Hif2aK
Supplemental Table 1. Plasma NEFA and liver triglyceride levels in Hif1aKO and Hif2aKO mice under control and high fat diets. Hif1a (n=6) Hif1aK O (n=6) Hif2a Hif2aK O Hif1a (n=5) Hif1aKO (n=5) Hif2a Hif2aK
FUNDAMENTALS OF HEMODYNAMICS, VASOACTIVE DRUGS AND IABP IN THE FAILING HEART
 FUNDAMENTALS OF HEMODYNAMICS, VASOACTIVE DRUGS AND IABP IN THE FAILING HEART CINDY BITHER, MSN, ANP, ANP, AACC, CHFN CHIEF NP, ADV HF PROGRAM MEDSTAR WASHINGTON HOSPITAL CENTER CONFLICTS OF INTEREST NONE
FUNDAMENTALS OF HEMODYNAMICS, VASOACTIVE DRUGS AND IABP IN THE FAILING HEART CINDY BITHER, MSN, ANP, ANP, AACC, CHFN CHIEF NP, ADV HF PROGRAM MEDSTAR WASHINGTON HOSPITAL CENTER CONFLICTS OF INTEREST NONE
Figure S1. Reduction in glomerular mir-146a levels correlate with progression to higher albuminuria in diabetic patients.
 Supplementary Materials Supplementary Figures Figure S1. Reduction in glomerular mir-146a levels correlate with progression to higher albuminuria in diabetic patients. Figure S2. Expression level of podocyte
Supplementary Materials Supplementary Figures Figure S1. Reduction in glomerular mir-146a levels correlate with progression to higher albuminuria in diabetic patients. Figure S2. Expression level of podocyte
Supplementary Figure 1. A. Bar graph representing the expression levels of the 19 indicated genes in the microarrays analyses comparing human lung
 Supplementary Figure 1. A. Bar graph representing the expression levels of the 19 indicated genes in the microarrays analyses comparing human lung immortalized broncho-epithelial cells (AALE cells) expressing
Supplementary Figure 1. A. Bar graph representing the expression levels of the 19 indicated genes in the microarrays analyses comparing human lung immortalized broncho-epithelial cells (AALE cells) expressing
Supplementary Figure 1: Additional metabolic parameters of obesity mouse models and controls. (a) Body weight, (b) blood glucose and (c) insulin
 Supplementary Figure 1: Additional metabolic parameters of obesity mouse models and controls. (a) Body weight, (b) blood glucose and (c) insulin resistance index of homeostatic model assessment (HOMA IR)
Supplementary Figure 1: Additional metabolic parameters of obesity mouse models and controls. (a) Body weight, (b) blood glucose and (c) insulin resistance index of homeostatic model assessment (HOMA IR)
Supplementary methods:
 Supplementary methods: Primers sequences used in real-time PCR analyses: β-actin F: GACCTCTATGCCAACACAGT β-actin [11] R: AGTACTTGCGCTCAGGAGGA MMP13 F: TTCTGGTCTTCTGGCACACGCTTT MMP13 R: CCAAGCTCATGGGCAGCAACAATA
Supplementary methods: Primers sequences used in real-time PCR analyses: β-actin F: GACCTCTATGCCAACACAGT β-actin [11] R: AGTACTTGCGCTCAGGAGGA MMP13 F: TTCTGGTCTTCTGGCACACGCTTT MMP13 R: CCAAGCTCATGGGCAGCAACAATA
Supplementary Figure 1. Blood glucose and insulin levels in mice during 4-day infusion.
 Supplementary Figure 1. Blood glucose and insulin levels in mice during 4-day infusion. (A-B) WT and HT mice infused with saline or glucose had overlapping achieved blood glucose and insulin levels, necessitating
Supplementary Figure 1. Blood glucose and insulin levels in mice during 4-day infusion. (A-B) WT and HT mice infused with saline or glucose had overlapping achieved blood glucose and insulin levels, necessitating
Postn MCM Smad2 fl/fl Postn MCM Smad3 fl/fl Postn MCM Smad2/3 fl/fl. Postn MCM. Tgfbr1/2 fl/fl TAC
 A Smad2 fl/fl Smad3 fl/fl Smad2/3 fl/fl Tgfbr1/2 fl/fl 1. mm B Tcf21 MCM Tcf21 MCM Smad3 fl/fl Tcf21 MCM Smad2/3 fl/fl Tcf21 MCM Tgfbr1/2 fl/fl αmhc MCM C 1. mm 1. mm D Smad2 fl/fl Smad3 fl/fl Smad2/3
A Smad2 fl/fl Smad3 fl/fl Smad2/3 fl/fl Tgfbr1/2 fl/fl 1. mm B Tcf21 MCM Tcf21 MCM Smad3 fl/fl Tcf21 MCM Smad2/3 fl/fl Tcf21 MCM Tgfbr1/2 fl/fl αmhc MCM C 1. mm 1. mm D Smad2 fl/fl Smad3 fl/fl Smad2/3
Supplementary Figure 1. DJ-1 modulates ROS concentration in mouse skeletal muscle.
 Supplementary Figure 1. DJ-1 modulates ROS concentration in mouse skeletal muscle. (a) mrna levels of Dj1 measured by quantitative RT-PCR in soleus, gastrocnemius (Gastroc.) and extensor digitorum longus
Supplementary Figure 1. DJ-1 modulates ROS concentration in mouse skeletal muscle. (a) mrna levels of Dj1 measured by quantitative RT-PCR in soleus, gastrocnemius (Gastroc.) and extensor digitorum longus
SUPPLEMENTAL MATERIAL. Supplementary Methods
 SUPPLEMENTAL MATERIAL Supplementary Methods Culture of cardiomyocytes, fibroblasts and cardiac microvascular endothelial cells The isolation and culturing of neonatal rat ventricular cardiomyocytes was
SUPPLEMENTAL MATERIAL Supplementary Methods Culture of cardiomyocytes, fibroblasts and cardiac microvascular endothelial cells The isolation and culturing of neonatal rat ventricular cardiomyocytes was
1.5 ASK1KO fed. fasted 16 hrs w/o water. Fed. 4th. 4th WT ASK1KO N=29, 11(WT), ,5(ASK1KO) ASK1KO ASK1KO **** Time [h]
![1.5 ASK1KO fed. fasted 16 hrs w/o water. Fed. 4th. 4th WT ASK1KO N=29, 11(WT), ,5(ASK1KO) ASK1KO ASK1KO **** Time [h] 1.5 ASK1KO fed. fasted 16 hrs w/o water. Fed. 4th. 4th WT ASK1KO N=29, 11(WT), ,5(ASK1KO) ASK1KO ASK1KO **** Time [h]](/thumbs/87/97258189.jpg) 7: 13: 19: 1: 7: 151117 a 151117 4th 4th b c RQ.95 KO.9.85.8.75.7 light dark light dark.65 7: 19: 7: 19: 7: Means ± SEM, N=6 RQ 1..9.8.7.6.6 KO CL (-) CL (+) ibat weight ratio (/body weight) [%].5.4.3.2.1
7: 13: 19: 1: 7: 151117 a 151117 4th 4th b c RQ.95 KO.9.85.8.75.7 light dark light dark.65 7: 19: 7: 19: 7: Means ± SEM, N=6 RQ 1..9.8.7.6.6 KO CL (-) CL (+) ibat weight ratio (/body weight) [%].5.4.3.2.1
and follicular helper T cells is Egr2-dependent. (a) Diagrammatic representation of the
 Supplementary Figure 1. LAG3 + Treg-mediated regulation of germinal center B cells and follicular helper T cells is Egr2-dependent. (a) Diagrammatic representation of the experimental protocol for the
Supplementary Figure 1. LAG3 + Treg-mediated regulation of germinal center B cells and follicular helper T cells is Egr2-dependent. (a) Diagrammatic representation of the experimental protocol for the
Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A.
 Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A. Upper part, three-primer PCR strategy at the Mcm3 locus yielding
Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A. Upper part, three-primer PCR strategy at the Mcm3 locus yielding
doi: /nature10632
 SUPPLEMENTARY INFORMATION doi:10.1038/nature10632 Supplementary Figure 1 Lyn mediates neutrophil wound responses as a redox sensor. a, A schematic model. Wounded epithelial cells release H 2 O 2 by an
SUPPLEMENTARY INFORMATION doi:10.1038/nature10632 Supplementary Figure 1 Lyn mediates neutrophil wound responses as a redox sensor. a, A schematic model. Wounded epithelial cells release H 2 O 2 by an
well for 2 h at rt. Each dot represents an individual mouse and bar is the mean ±
 Supplementary data: Control DC Blimp-1 ko DC 8 6 4 2-2 IL-1β p=.5 medium 8 6 4 2 IL-2 Medium p=.16 8 6 4 2 IL-6 medium p=.3 5 4 3 2 1-1 medium IL-1 n.s. 25 2 15 1 5 IL-12(p7) p=.15 5 IFNγ p=.65 4 3 2 1
Supplementary data: Control DC Blimp-1 ko DC 8 6 4 2-2 IL-1β p=.5 medium 8 6 4 2 IL-2 Medium p=.16 8 6 4 2 IL-6 medium p=.3 5 4 3 2 1-1 medium IL-1 n.s. 25 2 15 1 5 IL-12(p7) p=.15 5 IFNγ p=.65 4 3 2 1
Supplemental Fig. 1. Relative mrna Expression. Relative mrna Expression WT KO WT KO RT 4 0 C
 Supplemental Fig. 1 A 1.5 1..5 Hdac11 (ibat) n=4 n=4 n=4 n=4 n=4 n=4 n=4 n=4 WT KO WT KO WT KO WT KO RT 4 C RT 4 C Supplemental Figure 1. Hdac11 mrna is undetectable in KO adipose tissue. Quantitative
Supplemental Fig. 1 A 1.5 1..5 Hdac11 (ibat) n=4 n=4 n=4 n=4 n=4 n=4 n=4 n=4 WT KO WT KO WT KO WT KO RT 4 C RT 4 C Supplemental Figure 1. Hdac11 mrna is undetectable in KO adipose tissue. Quantitative
Supplementary Figure 1 IL-27 IL
 Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
18s AAACGGCTACCACATCCAAG CCTCCAATGGATCCTCGTTA. 36b4 GTTCTTGCCCATCAGCACC AGATGCAGCAGATCCGCAT. Acc1 AGCAGATCCGCAGCTTG ACCTCTGCTCGCTGAGTGC
 Supplementary Table 1. Quantitative PCR primer sequences Gene symbol Sequences (5 to 3 ) Forward Reverse 18s AAACGGCTACCACATCCAAG CCTCCAATGGATCCTCGTTA 36b4 GTTCTTGCCCATCAGCACC AGATGCAGCAGATCCGCAT Acc1
Supplementary Table 1. Quantitative PCR primer sequences Gene symbol Sequences (5 to 3 ) Forward Reverse 18s AAACGGCTACCACATCCAAG CCTCCAATGGATCCTCGTTA 36b4 GTTCTTGCCCATCAGCACC AGATGCAGCAGATCCGCAT Acc1
Figure S1. Generation of inducible PTEN deficient mice and the BMMCs (A) B6.129 Pten loxp/loxp mice were mated with B6.
 Figure S1. Generation of inducible PTEN deficient mice and the BMMCs (A) B6.129 Pten loxp/loxp mice were mated with B6.129-Gt(ROSA)26Sor tm1(cre/ert2)tyj /J mice. To induce deletion of the Pten locus,
Figure S1. Generation of inducible PTEN deficient mice and the BMMCs (A) B6.129 Pten loxp/loxp mice were mated with B6.129-Gt(ROSA)26Sor tm1(cre/ert2)tyj /J mice. To induce deletion of the Pten locus,
Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was
 Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was painted on the shaved back skin of CBL/J and BALB/c mice for consecutive days. (a, b) Phenotypic presentation of mouse back skin
Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was painted on the shaved back skin of CBL/J and BALB/c mice for consecutive days. (a, b) Phenotypic presentation of mouse back skin
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/1/8/e1500296/dc1 Supplementary Materials for Transcriptional regulation of APOBEC3 antiviral immunity through the CBF- /RUNX axis This PDF file includes: Brett
advances.sciencemag.org/cgi/content/full/1/8/e1500296/dc1 Supplementary Materials for Transcriptional regulation of APOBEC3 antiviral immunity through the CBF- /RUNX axis This PDF file includes: Brett
Nature Genetics: doi: /ng Supplementary Figure 1
 Supplementary Figure 1 MSI2 interactors are associated with the riboproteome and are functionally relevant. (a) Coomassie blue staining of FLAG-MSI2 immunoprecipitated complexes. (b) GO analysis of MSI2-interacting
Supplementary Figure 1 MSI2 interactors are associated with the riboproteome and are functionally relevant. (a) Coomassie blue staining of FLAG-MSI2 immunoprecipitated complexes. (b) GO analysis of MSI2-interacting
GFP/Iba1/GFAP. Brain. Liver. Kidney. Lung. Hoechst/Iba1/TLR9!
 Supplementary information a +KA Relative expression d! Tlr9 5!! 5! NSC Neuron Astrocyte Microglia! 5! Tlr7!!!! NSC Neuron Astrocyte! GFP/Sβ/! Iba/Hoechst Microglia e Hoechst/Iba/TLR9! GFP/Iba/GFAP f Brain
Supplementary information a +KA Relative expression d! Tlr9 5!! 5! NSC Neuron Astrocyte Microglia! 5! Tlr7!!!! NSC Neuron Astrocyte! GFP/Sβ/! Iba/Hoechst Microglia e Hoechst/Iba/TLR9! GFP/Iba/GFAP f Brain
Serum cytokine levels in control and tumor-bearing male and female mice at day 15.
 Supplementary Table 1. Serum cytokine levels in control and tumor-bearing male and female mice at day 15. Male Female Cytokine Control C-26 Control C-26 IL-1β 2.0 ± 0.8 9.6 ± 1.5* 1.8 ± 0.2 6.8 ± 1.4*
Supplementary Table 1. Serum cytokine levels in control and tumor-bearing male and female mice at day 15. Male Female Cytokine Control C-26 Control C-26 IL-1β 2.0 ± 0.8 9.6 ± 1.5* 1.8 ± 0.2 6.8 ± 1.4*
Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus
 Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus changes in corresponding proteins between wild type and Gprc5a-/-
Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus changes in corresponding proteins between wild type and Gprc5a-/-
Relative Rates. SUM159 CB- 839-Resistant *** n.s Intracellular % Labeled by U- 13 C-Asn 0.
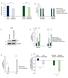 A Relative Growth Rates 1.2 1.8.6.4.2 B Relative Rates 1.6 1.4 1.2 1.8.6.4.2 LPS2 Parental LPS2 Q-Independent SUM159 Parental SUM159 CB-839-Resistant LPS2 Parental LPS2 Q- Independent SUM159 Parental SUM159
A Relative Growth Rates 1.2 1.8.6.4.2 B Relative Rates 1.6 1.4 1.2 1.8.6.4.2 LPS2 Parental LPS2 Q-Independent SUM159 Parental SUM159 CB-839-Resistant LPS2 Parental LPS2 Q- Independent SUM159 Parental SUM159
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES 1 Supplementary Figure 1, Adult hippocampal QNPs and TAPs uniformly express REST a-b) Confocal images of adult hippocampal mouse sections showing GFAP (green), Sox2 (red), and REST
SUPPLEMENTARY FIGURES 1 Supplementary Figure 1, Adult hippocampal QNPs and TAPs uniformly express REST a-b) Confocal images of adult hippocampal mouse sections showing GFAP (green), Sox2 (red), and REST
Supplementary Figure 1
 Supplementary Figure 1 YAP negatively regulates IFN- signaling. (a) Immunoblot analysis of Yap knockdown efficiency with sh-yap (#1 to #4 independent constructs) in Raw264.7 cells. (b) IFN- -Luc and PRDs
Supplementary Figure 1 YAP negatively regulates IFN- signaling. (a) Immunoblot analysis of Yap knockdown efficiency with sh-yap (#1 to #4 independent constructs) in Raw264.7 cells. (b) IFN- -Luc and PRDs
SUPPLEMENTARY FIGURE LEGENDS. atypical adenomatous hyperplasias (AAH); Grade II: adenomas; Grade III: adenocarcinomas;
 SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure S1: Tumor grades in Ras G12D ; p53 / lung tumors. Representative histology (H&E) of K-Ras G12D ; p53 / lung tumors 13 weeks after tumor initiation. Grade
SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure S1: Tumor grades in Ras G12D ; p53 / lung tumors. Representative histology (H&E) of K-Ras G12D ; p53 / lung tumors 13 weeks after tumor initiation. Grade
Supplementary Figure 1. c Human
 Supplementary Figure 1 a b c Human Mouse d Gapdh Amino acid sequence and baseline expression of MYDGF N-terminal signal peptides (S-scores) and signal peptide cleavage sites (C-scores) of (a) human MYDGF
Supplementary Figure 1 a b c Human Mouse d Gapdh Amino acid sequence and baseline expression of MYDGF N-terminal signal peptides (S-scores) and signal peptide cleavage sites (C-scores) of (a) human MYDGF
Supplementary Figure 1. Characterization of human carotid plaques. (a) Flash-frozen human plaques were separated into vulnerable (V) and stable (S),
 Supplementary Figure 1. Characterization of human carotid plaques. (a) Flash-frozen human plaques were separated into vulnerable (V) and stable (S), regions which were then quantified for mean fluorescence
Supplementary Figure 1. Characterization of human carotid plaques. (a) Flash-frozen human plaques were separated into vulnerable (V) and stable (S), regions which were then quantified for mean fluorescence
Supplementary Information. Glycogen shortage during fasting triggers liver-brain-adipose. neurocircuitry to facilitate fat utilization
 Supplementary Information Glycogen shortage during fasting triggers liver-brain-adipose neurocircuitry to facilitate fat utilization Supplementary Figure S1. Liver-Brain-Adipose neurocircuitry Starvation
Supplementary Information Glycogen shortage during fasting triggers liver-brain-adipose neurocircuitry to facilitate fat utilization Supplementary Figure S1. Liver-Brain-Adipose neurocircuitry Starvation
AAV-TBGp-Cre treatment resulted in hepatocyte-specific GH receptor gene recombination
 AAV-TBGp-Cre treatment resulted in hepatocyte-specific GH receptor gene recombination Supplementary Figure 1. Generation of the adult-onset, liver-specific GH receptor knock-down (alivghrkd, Kd) mouse
AAV-TBGp-Cre treatment resulted in hepatocyte-specific GH receptor gene recombination Supplementary Figure 1. Generation of the adult-onset, liver-specific GH receptor knock-down (alivghrkd, Kd) mouse
SUPPLEMENTARY FIGURES AND TABLES
 SUPPLEMENTARY FIGURES AND TABLES Supplementary Figure S1: CaSR expression in neuroblastoma models. A. Proteins were isolated from three neuroblastoma cell lines and from the liver metastasis of a MYCN-non
SUPPLEMENTARY FIGURES AND TABLES Supplementary Figure S1: CaSR expression in neuroblastoma models. A. Proteins were isolated from three neuroblastoma cell lines and from the liver metastasis of a MYCN-non
7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans.
 Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
Supplementary Figure 1. EC-specific Deletion of Snail1 Does Not Affect EC Apoptosis. (a,b) Cryo-sections of WT (a) and Snail1 LOF (b) embryos at
 Supplementary Figure 1. EC-specific Deletion of Snail1 Does Not Affect EC Apoptosis. (a,b) Cryo-sections of WT (a) and Snail1 LOF (b) embryos at E10.5 were double-stained for TUNEL (red) and PECAM-1 (green).
Supplementary Figure 1. EC-specific Deletion of Snail1 Does Not Affect EC Apoptosis. (a,b) Cryo-sections of WT (a) and Snail1 LOF (b) embryos at E10.5 were double-stained for TUNEL (red) and PECAM-1 (green).
SUPPLEMENTARY LEGENDS...
 TABLE OF CONTENTS SUPPLEMENTARY LEGENDS... 2 11 MOVIE S1... 2 FIGURE S1 LEGEND... 3 FIGURE S2 LEGEND... 4 FIGURE S3 LEGEND... 5 FIGURE S4 LEGEND... 6 FIGURE S5 LEGEND... 7 FIGURE S6 LEGEND... 8 FIGURE
TABLE OF CONTENTS SUPPLEMENTARY LEGENDS... 2 11 MOVIE S1... 2 FIGURE S1 LEGEND... 3 FIGURE S2 LEGEND... 4 FIGURE S3 LEGEND... 5 FIGURE S4 LEGEND... 6 FIGURE S5 LEGEND... 7 FIGURE S6 LEGEND... 8 FIGURE
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Jewell et al., http://www.jcb.org/cgi/content/full/jcb.201007176/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. IR Munc18c association is independent of IRS-1. (A and
Supplemental material Jewell et al., http://www.jcb.org/cgi/content/full/jcb.201007176/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. IR Munc18c association is independent of IRS-1. (A and
Expanded View Figures
 PEX13 functions in selective autophagy Ming Y Lee et al Expanded View Figures Figure EV1. PEX13 is required for Sindbis virophagy. A, B Quantification of mcherry-capsid puncta per cell (A) and GFP-LC3
PEX13 functions in selective autophagy Ming Y Lee et al Expanded View Figures Figure EV1. PEX13 is required for Sindbis virophagy. A, B Quantification of mcherry-capsid puncta per cell (A) and GFP-LC3
IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia
 Supplementary Figures IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia Yaming Wang, Kristy J. Szretter, William Vermi, Susan Gilfillan, Cristina
Supplementary Figures IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia Yaming Wang, Kristy J. Szretter, William Vermi, Susan Gilfillan, Cristina
Lack of GPR88 enhances medium spiny neuron activity and alters. motor- and cue- dependent behaviors
 Lack of GPR88 enhances medium spiny neuron activity and alters motor- and cue- dependent behaviors Albert Quintana, Elisenda Sanz, Wengang Wang, Granville P. Storey, Ali D. Güler Matthew J. Wanat, Bryan
Lack of GPR88 enhances medium spiny neuron activity and alters motor- and cue- dependent behaviors Albert Quintana, Elisenda Sanz, Wengang Wang, Granville P. Storey, Ali D. Güler Matthew J. Wanat, Bryan
Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein
 Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein content relative to GAPDH in two independent experiments.
Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein content relative to GAPDH in two independent experiments.
ECHOCARDIOGRAPHY DATA REPORT FORM
 Patient ID Patient Study ID AVM - - Date of form completion / / 20 Initials of person completing the form mm dd yyyy Study period Preoperative Postoperative Operative 6-month f/u 1-year f/u 2-year f/u
Patient ID Patient Study ID AVM - - Date of form completion / / 20 Initials of person completing the form mm dd yyyy Study period Preoperative Postoperative Operative 6-month f/u 1-year f/u 2-year f/u
Tbk1-TKO! DN cells (%)! 15! 10!
 a! T Cells! TKO! B Cells! TKO! b! CD4! 8.9 85.2 3.4 2.88 CD8! Tbk1-TKO! 1.1 84.8 2.51 2.54 c! DN cells (%)! 4 3 2 1 DP cells (%)! 9 8 7 6 CD4 + SP cells (%)! 5 4 3 2 1 5 TKO! TKO! TKO! TKO! 15 1 5 CD8
a! T Cells! TKO! B Cells! TKO! b! CD4! 8.9 85.2 3.4 2.88 CD8! Tbk1-TKO! 1.1 84.8 2.51 2.54 c! DN cells (%)! 4 3 2 1 DP cells (%)! 9 8 7 6 CD4 + SP cells (%)! 5 4 3 2 1 5 TKO! TKO! TKO! TKO! 15 1 5 CD8
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells
 MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
Supplementary Figure 1: Hsp60 / IEC mice are embryonically lethal (A) Light microscopic pictures show mouse embryos at developmental stage E12.
 Supplementary Figure 1: Hsp60 / IEC mice are embryonically lethal (A) Light microscopic pictures show mouse embryos at developmental stage E12.5 and E13.5 prepared from uteri of dams and subsequently genotyped.
Supplementary Figure 1: Hsp60 / IEC mice are embryonically lethal (A) Light microscopic pictures show mouse embryos at developmental stage E12.5 and E13.5 prepared from uteri of dams and subsequently genotyped.
Nature Genetics: doi: /ng Supplementary Figure 1. Parameters and consequences of mononuclear cardiomyocyte frequency.
 Supplementary Figure 1 Parameters and consequences of mononuclear cardiomyocyte frequency. (a) Correlation of the frequency of mononuclear cardiomyocytes to the frequency of cardiomyocytes with three or
Supplementary Figure 1 Parameters and consequences of mononuclear cardiomyocyte frequency. (a) Correlation of the frequency of mononuclear cardiomyocytes to the frequency of cardiomyocytes with three or
GPR120 *** * * Liver BAT iwat ewat mwat Ileum Colon. UCP1 mrna ***
 a GPR120 GPR120 mrna/ppia mrna Arbitrary Units 150 100 50 Liver BAT iwat ewat mwat Ileum Colon b UCP1 mrna Fold induction 20 15 10 5 - camp camp SB202190 - - - H89 - - - - - GW7647 Supplementary Figure
a GPR120 GPR120 mrna/ppia mrna Arbitrary Units 150 100 50 Liver BAT iwat ewat mwat Ileum Colon b UCP1 mrna Fold induction 20 15 10 5 - camp camp SB202190 - - - H89 - - - - - GW7647 Supplementary Figure
X P. Supplementary Figure 1. Nature Medicine: doi: /nm Nilotinib LSK LT-HSC. Cytoplasm. Cytoplasm. Nucleus. Nucleus
 a b c Supplementary Figure 1 c-kit-apc-eflu780 Lin-FITC Flt3-Linc-Kit-APC-eflu780 LSK Sca-1-PE-Cy7 d e f CD48-APC LT-HSC CD150-PerCP-cy5.5 g h i j Cytoplasm RCC1 X Exp 5 mir 126 SPRED1 SPRED1 RAN P SPRED1
a b c Supplementary Figure 1 c-kit-apc-eflu780 Lin-FITC Flt3-Linc-Kit-APC-eflu780 LSK Sca-1-PE-Cy7 d e f CD48-APC LT-HSC CD150-PerCP-cy5.5 g h i j Cytoplasm RCC1 X Exp 5 mir 126 SPRED1 SPRED1 RAN P SPRED1
pplementary Figur Supplementary Figure 1. a.
 pplementary Figur Supplementary Figure 1. a. Quantification by RT-qPCR of YFV-17D and YFV-17D pol- (+) RNA in the supernatant of cultured Huh7.5 cells following viral RNA electroporation of respective
pplementary Figur Supplementary Figure 1. a. Quantification by RT-qPCR of YFV-17D and YFV-17D pol- (+) RNA in the supernatant of cultured Huh7.5 cells following viral RNA electroporation of respective
Supplementary Information Epigenetic modulation of inflammation and synaptic plasticity promotes resilience against stress in mice
 Supplementary Information Epigenetic modulation of inflammation and synaptic plasticity promotes resilience against stress in mice Wang et. al. IL-6 in plasma (pg/ml) Rac1/HPRT (% of control) PSD9/HPRT
Supplementary Information Epigenetic modulation of inflammation and synaptic plasticity promotes resilience against stress in mice Wang et. al. IL-6 in plasma (pg/ml) Rac1/HPRT (% of control) PSD9/HPRT
Supplementary Figure 1: STAT3 suppresses Kras-induced lung tumorigenesis
 Supplementary Figure 1: STAT3 suppresses Kras-induced lung tumorigenesis (a) Immunohistochemical (IHC) analysis of tyrosine 705 phosphorylation status of STAT3 (P- STAT3) in tumors and stroma (all-time
Supplementary Figure 1: STAT3 suppresses Kras-induced lung tumorigenesis (a) Immunohistochemical (IHC) analysis of tyrosine 705 phosphorylation status of STAT3 (P- STAT3) in tumors and stroma (all-time
Supplementary Table 1 Gene clone ID for ShRNA-mediated gene silencing TNFα downstream signals in in vitro Symbol Gene ID RefSeqID Clone ID
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 Supplementary Table 1 Gene clone ID for ShRNA-mediated gene silencing TNFα downstream
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 Supplementary Table 1 Gene clone ID for ShRNA-mediated gene silencing TNFα downstream
Supplemental Information. Increased 4E-BP1 Expression Protects. against Diet-Induced Obesity and Insulin. Resistance in Male Mice
 Cell Reports, Volume 16 Supplemental Information Increased 4E-BP1 Expression Protects against Diet-Induced Obesity and Insulin Resistance in Male Mice Shih-Yin Tsai, Ariana A. Rodriguez, Somasish G. Dastidar,
Cell Reports, Volume 16 Supplemental Information Increased 4E-BP1 Expression Protects against Diet-Induced Obesity and Insulin Resistance in Male Mice Shih-Yin Tsai, Ariana A. Rodriguez, Somasish G. Dastidar,
Supporting Information
 Supporting Information van der Windt et al. 10.1073/pnas.1221740110 SI Materials and Methods Mice and Reagents. C57BL/6 and major histocompatibility complex class I-restricted OVA-specific T-cell receptor
Supporting Information van der Windt et al. 10.1073/pnas.1221740110 SI Materials and Methods Mice and Reagents. C57BL/6 and major histocompatibility complex class I-restricted OVA-specific T-cell receptor
Supplementary Figures Supplementary Figure 1. Development of the camp biosensor targeted to the SERCA2a microdomain.
 Supplementary Figures Supplementary Figure 1. Development of the camp biosensor targeted to the SERCA2a microdomain. A B C (A) Schematic representation of the new constructs designed for local camp imaging.
Supplementary Figures Supplementary Figure 1. Development of the camp biosensor targeted to the SERCA2a microdomain. A B C (A) Schematic representation of the new constructs designed for local camp imaging.
Supplementary Figure 1. Generation of knockin mice expressing L-selectinN138G. (a) Schematics of the Sellg allele (top), the targeting vector, the
 Supplementary Figure 1. Generation of knockin mice expressing L-selectinN138G. (a) Schematics of the Sellg allele (top), the targeting vector, the targeted allele in ES cells, and the mutant allele in
Supplementary Figure 1. Generation of knockin mice expressing L-selectinN138G. (a) Schematics of the Sellg allele (top), the targeting vector, the targeted allele in ES cells, and the mutant allele in
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 DOT1L regulates the expression of epithelial and mesenchymal markers. (a) The expression levels and cellular localizations of EMT markers were confirmed by
Supplementary Figures Supplementary Figure 1 DOT1L regulates the expression of epithelial and mesenchymal markers. (a) The expression levels and cellular localizations of EMT markers were confirmed by
Supplementary Figure S1. Generation of LSL-EZH2 conditional transgenic mice.
 Downstream Col1A locus S P P P EP Genotyping with P1, P2 frt PGKneopA + frt hygro-pa Targeting vector Genotyping with P3, P4 P1 pcag-flpe P2 P3 P4 frt SApA CAG LSL PGKATG frt hygro-pa C. D. E. ormal KRAS
Downstream Col1A locus S P P P EP Genotyping with P1, P2 frt PGKneopA + frt hygro-pa Targeting vector Genotyping with P3, P4 P1 pcag-flpe P2 P3 P4 frt SApA CAG LSL PGKATG frt hygro-pa C. D. E. ormal KRAS
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 Characterization of stable expression of GlucB and sshbira in the CT26 cell line (a) Live cell imaging of stable CT26 cells expressing green fluorescent protein
Supplementary Figures Supplementary Figure 1 Characterization of stable expression of GlucB and sshbira in the CT26 cell line (a) Live cell imaging of stable CT26 cells expressing green fluorescent protein
SUPPLEMENTARY INFORMATION
 doi: 1.138/nature7221 Brown fat selective genes 12 1 Control Q-RT-PCR (% of Control) 8 6 4 2 Ntrk3 Cox7a1 Cox8b Cox5b ATPase b2 ATPase f1a1 Sirt3 ERRα Elovl3/Cig3 PPARα Zic1 Supplementary Figure S1. stimulates
doi: 1.138/nature7221 Brown fat selective genes 12 1 Control Q-RT-PCR (% of Control) 8 6 4 2 Ntrk3 Cox7a1 Cox8b Cox5b ATPase b2 ATPase f1a1 Sirt3 ERRα Elovl3/Cig3 PPARα Zic1 Supplementary Figure S1. stimulates
Supplementary Figure 1. DNA methylation of the adiponectin promoter R1, Pparg2, and Tnfa promoter in adipocytes is not affected by obesity.
 Supplementary Figure 1. DNA methylation of the adiponectin promoter R1, Pparg2, and Tnfa promoter in adipocytes is not affected by obesity. (a) Relative amounts of adiponectin, Ppar 2, C/ebp, and Tnf mrna
Supplementary Figure 1. DNA methylation of the adiponectin promoter R1, Pparg2, and Tnfa promoter in adipocytes is not affected by obesity. (a) Relative amounts of adiponectin, Ppar 2, C/ebp, and Tnf mrna
Supplementary Information
 Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
SUPPLEMENTARY INFORMATION
 Haematopoietic stem cell release is regulated by circadian oscillations Simón Méndez-Ferrer *, Daniel Lucas *, Michela Battista * and Paul S. Frenette * Mount Sinai School of Medicine, *Departments of
Haematopoietic stem cell release is regulated by circadian oscillations Simón Méndez-Ferrer *, Daniel Lucas *, Michela Battista * and Paul S. Frenette * Mount Sinai School of Medicine, *Departments of
(a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable
 Supplementary Figure 1. Frameshift (FS) mutation in UVRAG. (a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable A 10 DNA repeat, generating a premature stop codon
Supplementary Figure 1. Frameshift (FS) mutation in UVRAG. (a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable A 10 DNA repeat, generating a premature stop codon
MII. Supplement Figure 1. CapZ β2. Merge. 250ng. 500ng DIC. Merge. Journal of Cell Science Supplementary Material. GFP-CapZ β2 DNA
 A GV GVBD MI DNA CapZ β2 CapZ β2 Merge B DIC GFP-CapZ β2 Merge CapZ β2-gfp 250ng 500ng Supplement Figure 1. MII A early MI late MI Control RNAi CapZαβ DNA Actin Tubulin B Phalloidin Intensity(A.U.) n=10
A GV GVBD MI DNA CapZ β2 CapZ β2 Merge B DIC GFP-CapZ β2 Merge CapZ β2-gfp 250ng 500ng Supplement Figure 1. MII A early MI late MI Control RNAi CapZαβ DNA Actin Tubulin B Phalloidin Intensity(A.U.) n=10
Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS
 Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS nucleotide sequences (a, b) or amino acid sequences (c) from
Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS nucleotide sequences (a, b) or amino acid sequences (c) from
S1a S1b S1c. S1d. S1f S1g S1h SUPPLEMENTARY FIGURE 1. - si sc Il17rd Il17ra bp. rig/s IL-17RD (ng) -100 IL-17RD
 SUPPLEMENTARY FIGURE 1 0 20 50 80 100 IL-17RD (ng) S1a S1b S1c IL-17RD β-actin kda S1d - si sc Il17rd Il17ra rig/s15-574 - 458-361 bp S1f S1g S1h S1i S1j Supplementary Figure 1. Knockdown of IL-17RD enhances
SUPPLEMENTARY FIGURE 1 0 20 50 80 100 IL-17RD (ng) S1a S1b S1c IL-17RD β-actin kda S1d - si sc Il17rd Il17ra rig/s15-574 - 458-361 bp S1f S1g S1h S1i S1j Supplementary Figure 1. Knockdown of IL-17RD enhances
Supplementary Fig. 1 eif6 +/- mice show a reduction in white adipose tissue, blood lipids and normal glycogen synthesis. The cohort of the original
 Supplementary Fig. 1 eif6 +/- mice show a reduction in white adipose tissue, blood lipids and normal glycogen synthesis. The cohort of the original phenotypic screening was n=40. For specific tests, the
Supplementary Fig. 1 eif6 +/- mice show a reduction in white adipose tissue, blood lipids and normal glycogen synthesis. The cohort of the original phenotypic screening was n=40. For specific tests, the
Supplementary Figure 1.
 Supplementary Figure 1. Transduction of adipocytes after intra-ewat administration of AAV vectors. A: Immunostaining against GFP (green) in sections of ewat two weeks after the intra-ewat administration
Supplementary Figure 1. Transduction of adipocytes after intra-ewat administration of AAV vectors. A: Immunostaining against GFP (green) in sections of ewat two weeks after the intra-ewat administration
SUPPLEMENTARY MATERIAL
 SUPPLEMENTARY MATERIAL IL-1 signaling modulates activation of STAT transcription factors to antagonize retinoic acid signaling and control the T H 17 cell it reg cell balance Rajatava Basu 1,5, Sarah K.
SUPPLEMENTARY MATERIAL IL-1 signaling modulates activation of STAT transcription factors to antagonize retinoic acid signaling and control the T H 17 cell it reg cell balance Rajatava Basu 1,5, Sarah K.
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells
 Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
