Live cell imaging and electron microscopy reveal dynamic processes of BAF-directed nuclear envelope assembly
|
|
|
- Derick Spencer
- 6 years ago
- Views:
Transcription
1 2540 Research Article Live cell imaging and electron microscopy reveal dynamic processes of BAF-directed nuclear envelope assembly Tokuko Haraguchi 1,2, *, Tomoko Kojidani 1, Takako Koujin 1, Takeshi Shimi 1, Hiroko Osakada 1, Chie Mori 1, Akitsugu Yamamoto 3 and Yasushi Hiraoka 1,2,4 1 CREST Research Project, Kobe Advanced ICT Research Center, National Institute of Information and Communications Technology, Iwaoka, Iwaoka-cho, Nishi-ku, Kobe , Japan 2 Graduate School of Science, Osaka University, 1-1 Machikaneyama, Toyonaka , Japan 3 Nagahama Institute of Bio-Science and Technology, 1266 Tamura-cho, Nagahama , Japan 4 Graduate School of Frontier Biosciences, Osaka University 1-3 Suita , Japan *Author for correspondence ( tokuko@nict.go.jp) Accepted19 May , Published by The Company of Biologists 2008 doi: /jcs Summary Assembly of the nuclear envelope (NE) in telophase is essential for higher eukaryotic cells to re-establish a functional nucleus. Time-lapse, FRAP and FRET analyses in human cells showed that barrier-to-autointegration factor (BAF), a DNA-binding protein, assembled first at the distinct core region of the telophase chromosome and formed an immobile complex by directly binding with other core-localizing NE proteins, such as lamin A and emerin. Correlative light and electron microscopy after live cell imaging, further showed that BAF formed an electron-dense structure on the chromosome surface of the core, close to spindle microtubules (MTs) prior to the attachment of precursor NE membranes, suggesting that MTs may mediate core assembly of BAF. Disruption of the spindle MTs consistently abolished BAF accumulation at the core. In addition, RNAi of BAF eliminated the core assembly of lamin A and emerin, caused abnormal cytoplasmic accumulation of precursor nuclear membranes and resulted in a significant delay of NE assembly. These results suggest that the MT-mediated BAF accumulation at the core facilitates NE assembly at the end of mitosis. Supplementary material available online at Key words: Barrier-to-autointegration factor, Chromatin, Emerin, Lamin A, Nuclear envelope, Microtubule Introduction The nuclear envelope (NE) is a cellular structure that encloses chromosomes and provides a physicochemical framework for genetic activities such as gene expression and DNA replication. The NE is a dynamic structure in eukaryotes, disassembling at prophase and reassembling at telophase in each mitosis. The reassembly of the NE is crucial to re-establish a functional nucleus for the next interphase and to ensure proper progression of the cell cycle. Several lines of evidence have shown that defects in NE architecture can cause diseases in animals. Recently, a variety of human disorders known as nuclear envelopathy or nuclear laminopathy have been reported (reviewed by Nagano and Arahata, 2000; Wilson et al., 2001; Worman and Courvalin, 2005). These disorders involve mutations in NE components. Mutations in the gene encoding lamin A (official protein symbol, LMNA) cause 12 human diseases (reviewed by Mattout et al., 2006) including Emery-Dreifuss muscular dystrophy (Bonne et al., 1999; Clements et al., 2000) and Hutchinson-Gilford progeria syndrome (Eriksson et al., 2003; Mounkes et al., 2003). Emery-Dreifuss muscular dystrophy can also be caused by mutations of the gene encoding emerin (official protein symbol, EMD) (Bione et al., 1994). The fact that defects in the NE can cause many different diseases indicates that the NE is involved in a wide variety of cellular functions. Thus, understanding the dynamic architecture of the NE will provide insight into the regulatory mechanisms for these cellular functions. Molecular mechanisms of NE formation have been well described in in vitro experimental systems (Hetzer et al., 2000; Harel et al., 2003; Hachet et al., 2004); however, compared with NE formation in vitro, NE reformation in vivo is poorly understood. It is believed that it begins with attachment of precursor membranes to telophase chromosomes, followed by fusion of the membranes and reassembly of integral membrane proteins, the NPC, and nuclear lamina into the NE. Chromatin-binding integral membrane proteins, such as lamin B receptor (LBR) and LAP2β (official symbol, LAP2B) are proposed to posses a role in early assembly of the NE (Gant and Wilson, 1997; Ulbert et al., 2006). Assembly of LEM-domain NE proteins such as emerin and LAP2β depends on barrier-toautointegration factor (BAF), a chromatin-binding protein, in human cells (Haraguchi et al., 2001). BAF forms dimers that bind nonspecifically to double-stranded DNA in vitro (Lee and Cragie, 1998) and can be phosphorylated in its N-terminal region by the vaccinia-related kinases (Nichols et al., 2006; Gorjánácz et al., 2007). BAF was first discovered as a cellular protein that prevents retroviral DNA from suicidal autointegration and ensures its integration into host DNA (Lee and Cragie, 1998: Jacque and Stevenson, 2006). It was later described as a binding partner of LEM-domain NE proteins (Furukawa, 1999; Lee et al., 2001; Shumaker et al., 2001). BAF is proposed to have a function in NE assembly in human cells (Haraguchi et al., 2000) and C. elegans (Margalit et al., 2005; Gorjánácz et al.,
2 BAF directs nuclear envelope assembly ) as well as in Xenopus oocyte extracts (Segura-Totten et al., 2002). Recently, it has also been reported to be important for the normal progression of S phase in human cells (Haraguchi et al., 2007). In human cells, BAF accumulates with emerin, at the central regions of the assembling nuclear rim close to the spindle microtubules (MTs) during telophase when the NE is reforming. This region is named the core region (Haraguchi et al., 2001). In our previous study, we found that emerin, LAP2β and lamin A accumulated in the core region during telophase in a BAF-dependent manner (Haraguchi et al., 2001). LAP2α (official symbol, LAP2A) also accumulates at the core (Dechat et al., 2004). Although the core structure seems important for NE assembly of the LEM-domain proteins and lamin A, its structural basis and dynamics in living cells are largely unknown. To understand the structural basis and biological significance of the core region, we have examined physical and chemical properties of the core region by time-lapse microscopy of living cells, FRAP and FRET analyses, and EM analysis combined with live-cell imaging. In addition, we used RNAi to selectively remove individual components of the core structure, BAF, lamin A and emerin, to examine their functions in living cells. Here we report that BAF forms an electron-dense stable structure at the core region of the telophase chromosome in a spindle-mt-dependent manner, and propose that this MT-dependent BAF accumulation at the core coordinates efficient rendezvous of nuclear membranes and chromosomes. Results BAF assembles first to the ʻcoreʼ region of the reforming telophase nuclear envelope Assembly of NE proteins at the end of mitosis is an essential step in the reformation of a functional nucleus in higher eukaryotic cells. In order to elucidate underlying molecular mechanisms of NE reformation in human cells, we examined spatial and temporal sequences of NE assembly by monitoring the behavior of NE- GFP fusion proteins in living cells. The NE proteins tested were BAF (which binds both chromatin and LEM-domain proteins); LAP2β, emerin and MAN1 (membrane-bound LEM-domain NE proteins); LAP2α (LEM-domain protein localized in the nucleus); lamin A and lamin B1 (major components of the nuclear lamina); LBR (classical integral nuclear membrane protein); and nucleoporin Nup35 (official protein symbol, NUP53). Time-lapse observation of HeLa cells expressing each one of these NE fusion proteins showed that BAF, LAP2α, LAP2β, emerin, MAN1 and lamin A accumulated at a specialized region of the telophase chromosome mass close to the spindle (the core region; see Fig. 1A and images indicated by arrows in Fig. 1B). This accumulation occurred during a limited period of telophase, as described previously (Haraguchi et al., 2000; Haraguchi et al., 2001; Dechat et al., 2004). In contrast with these core-localizing proteins, lamin B1, LBR and Nup35 did not show clear accumulation at the core region (Fig. 1B). The fluorescence intensity of NE proteins at the core region is plotted in Fig. 1C; the timing of core localization was determined as the time, in seconds after the metaphase-anaphase transition, at which 80% of maximum accumulation had occurred (Fig. 1D). Among the core-localizing proteins, BAF accumulated earliest at the core region, approximately 434 seconds after the metaphaseanaphase transition. LAP2α, lamin A and emerin accumulated approximately 30 seconds later, and LAP2β and MAN1 accumulated after approximately another 30 seconds (Fig. 1C,D). That BAF accumulated first was confirmed by observation of HeLa cells expressing mrfp-baf transiently co-transfected with GFP-emerin or GFP-lamin A. In these cells, mrfp-baf accumulated in the core region earlier than GFP-emerin or GFPlamin A (data not shown). These results indicate that BAF appears at the core region prior to the accumulation of other corelocalizing NE proteins. Since BAF binds both emerin and lamin A in vitro (Lee et al., 2001; Holaska et al., 2003), the early accumulation of BAF at the core region suggests the possibility that BAF provides a structural foothold to allow assembly of nuclear lamins and membrane-bound NE proteins. The time course of assembly to the reforming NE at telophase is schematically summarized in Fig. 1E. BAF forms an immobile structure at the core region From our time-lapse analysis, we hypothesized that BAF might form a foothold on the chromosome surface to allow assembly of other NE proteins. To test this hypothesis, we examined the mobility of BAF at the core region during telophase by FRAP analysis of HeLa cells stably expressing GFP-BAF. We previously reported that GFP-BAF during interphase was highly mobile with a half time recovery of 270 mseconds and an immobile fraction (IM) of approximately 3% (Shimi et al., 2004). GFP-BAF was also highly mobile during metaphase (data not shown). By contrast, GFP-BAF became highly immobile during telophase with an IM of 72±16% (n=7) (Fig. 2A,B), and remained immobile until the core region disappeared at the end of M phase; GFP-BAF regained its high mobility in early G1 phase (data not shown). This result indicates that physical attributes of BAF are regulated during the cell cycle, with BAF forming an immobile structure at the core region specifically in telophase, a characteristic also exhibited by LAP2α (Dechat et al., 2004). To further examine physical properties of the core structure, we determined the mobility of other core-localizing NE proteins, such as lamin A and emerin, at the core region. These NE components showed a similar mobility to that of BAF with an IM of 74±14% (n=5) for lamin A and 46±7% for emerin (n=5) (Fig. 2C,D). These results suggest that BAF may form a stable higher-order complex together with these NE proteins at the core. BAF directly interacts with BAF, lamin A and emerin at the core region Because BAF, lamin A and LEM-domain NE proteins all become immobile at the core region, the core structure may consist of a higher-order complex of BAF with lamin A and the LEM domain NE proteins. To test whether BAF directly interacts with these NE proteins at the core region, we used fluorescence Förster resonance energy transfer (FRET) analysis. In HeLa cells transiently coexpressing mvenus-baf and mcfp fusions of the second protein, FRET signals were detected by acceptor photobleaching (Fig. 3). The results showed that mvenus-baf produced FRET signals with mcfp-baf (Fig. 3A,B), mcfplamin A (Fig. 3C,D) and mcfp-emerin (Fig. 3E,F). In addition, time-lapse ratio imaging between mcfp-baf and YFP-BAF showed that FRET signals between BAF molecules dramatically increased in the core region during telophase (Fig. 3I). As BAF forms a dimer or dodecamer in vitro (Zheng et al., 2000), this high FRET signal may represent a higher-order complex structure. LAP2α fused with mcfp also produced FRET signals with mvenus-baf in the core (data not shown), consistent with the previous finding that LAP2α is colocalized with BAF at the core
3 (15) Fig. 1. See next page for legend.
4 BAF directs nuclear envelope assembly 2543 Fig. 1. Localization of NE proteins on the telophase chromosome. (A) Schematic diagram of the core region of NE proteins on telophase chromosomes. The region marked in red represents the core region where BAF is initially accumulated. The region marked in green represents the non-core region outside the core region. (B) Living HeLa cells fluorescently stained with Hoechst for chromosomes and with GFP fusions of NE proteins (as indicated on the left) were observed every minute. The time-lapse images obtained are shown from left to right, with time 0 indicating the metaphaseanaphase transition. BAF, lamin A and all LEM domain proteins are localized at the core region. Arrows indicate the time of core localization obtained from quantification of core localization shown in C. Scale bar: 10 μm. (C) Fluorescence intensity at the core region is plotted as a function of time in minutes after the metaphase-anaphase transition. Fluorescence intensity is presented as % maximum intensity after subtracting the intensity at 4 minutes. (D) The time of core localization was estimated from the plots shown in C. The time of core localization is defined by the time required, in seconds after the metaphase-anaphase transition, for intensity at the core to reach 80% of the maximum. The s.d. and the number of cells tested (in parentheses) is also shown. (E) Schematic diagram of NE formation. BAF (red) localizes first to the core region, prior to other LEM domain proteins. structure (Dechat et al., 2004). A mutant emerin that does not bind to BAF (emerin-m24) either in vivo (Haraguchi et al., 2001) or in vitro (Lee et al., 2001) was used as a negative control. No FRET signals were detected between mcfp-emerin-m24 and mvenus- BAF (Fig. 3G,H), suggesting that the detected FRET signals are the result of specific interactions. Estimated FRET efficiencies are summarized in Table 1. These results suggest that BAF Table 1. FRET efficiency FRET efficiency (%)* Interphase NE Telophase core region mcfp-baf/mvenus-baf 12.3±8.7 (n=7) 26.7±14.2 (n=15) mcfp-baf/mvenus-emerin 15.2±10.6 (n=11) 14.6±13.2 (n=29) mcfp-baf/mvenus-emerin m24 ND 2.7±1.35 (n=10) mcfp-baf/mvenus-lamin A 3.1±8.2 (n=10) 23.0±12.0 (n=31) *FRET efficiency (%) = [1 (donor intensity before acceptor photobleaching/donor intensity after acceptor photobleaching)] 100. probably forms a polymer or a higher-order complex structure with BAF itself in the core region and also directly binds emerin, lamin A and LAP2α, assembling them into the core complex. BAF forms an electron-dense complex on the surface of the telophase chromosome mass To examine the core structure at high resolution, we used electron microscopy (EM). Because the core structure forms transiently, for only a few minutes, during telophase, we selected specimens by observing living HeLa cells expressing GFP-BAF. During live-cell observation, the cell was fixed when the core structure formed and embedded in epoxy resin to prepare EM specimens (Fig. 4A; see Materials and Methods). Three-dimensional fluorescence microscopy images of the fixed cell were also obtained (Fig. 4B). Thin EM sections were compared with fluorescence microscopy images to identify the cell observed during live-cell observation (Fig. 4C,D) and details of the core structure were investigated at high resolution (Fig. 4E,F). Images shown in a, b, and c in Fig. 4F correspond to the regions indicated in Fig. 4E. This method provides an opportunity to combine the temporal information and molecular selectivity of fluorescence live-cell imaging with the high-resolution imaging of EM: this method is designated live correlative light electron microscopy (live CLEM). Examples fixed at later stages are shown in Figs 5 and 6. The reliability of this method was confirmed in an immunoelectron micrograph for GFP- BAF localization using anti-gfp antibody (supplementary material Fig. S1): GFP-BAF was localized in the core region of telophase cells as seen in the live CLEM imaging technique. We applied live CLEM to cells Fig. 2. FRAP analysis of core-localizing NE proteins. (A) HeLa cells stably expressing core-localizing NE proteins were examined by FRAP analysis. Images of GFP-BAF are shown as an example. The bleached region is the 2 μm region indicated by the white square. (B-D) FRAP profiles of GFP-BAF (B), GFPlamin A (C) and GFP-emerin (D). Closed circles, fluorescence intensity in the bleached region; open circles, fluorescence intensity in the unbleached region. Bars represent s.d. Number of cells examined was 7 for GFP-BAF, 5 for GFP-lamin A and 5 for GFP-emerin. fixed at approximately 4 minutes (n=4), 7 minutes (n=17) and 10 minutes (n=3) after the metaphaseanaphase transition, (Figs 4, 5 and 6, respectively). The cell shown in Fig. 4 was fixed at 4 minutes 15 seconds after the metaphase-anaphase transition, before BAF became concentrated at the core. Comparison of EM images with those from fluorescence microscopy shows that
5 (15) Fig. 3. FRET analyses of core-localizing NE proteins. (A-H) HeLa cells transiently expressing two fusion proteins, as indicated, were analyzed by the acceptor photobleaching method: FRET signals were detected by an increase of donor fluorescence after photobleaching the acceptor chomophore. (A,C,E,G) Typical examples of images at 480 nm for mcfp and 520 nm for mvenus. The bleached area is indicated by the white square; the inset in the upper right corner is an enlarged image of the bleached area. (B,D,F,H) Fluorescence spectra measured in the bleached area before and after acceptor photobleaching. (I) HeLa cells transiently expressing CFP-BAF and YFP-BAF were analyzed by the ratio-imaging method. Images were obtained every minute. Time 0 represents timing of the metaphase-anaphase transition. Ratio images are represented by the color code shown on the right. Scale bars:10 μm (A,C,E,G).
6 BAF directs nuclear envelope assembly 2545 BAF was localized to only a limited region of the chromosome mass (superimposed with the green region in panel b of Fig. 4F); this region represents a trailing edge of the separating chromosomes, which might correspond to the telomeres, as suggested by Foisner and colleagues (Dechat et al., 2004). In the central region of the chromosomes where BAF localizes, spindle MTs approached and penetrated into the chromosome mass (superimposed with the orange lines in regions b and c of Fig. 4F). At this time, the NE (superimposed with the red hatches) was not being formed in the central region of the chromosome mass where the mitotic spindle was present; rather, the most peripheral regions ( non-core region in Fig. 1A) of the chromosome mass were being enveloped by the NE (see panel a of Fig. 4F; also see supplementary material Fig. S2A,B). The purple arrowheads indicate vesicular membranes. These vesicular membranes are presumably part of the ER tubules, the precursors of nuclear membranes (Anderson et al., 2007). It should be emphasized that almost no NE was formed in the central region (b and c in Fig. 4F) where spindle MTs are present, whereas the NE was well defined in the peripheral regions where spindle MTs are few or not present at all. The cell shown in Fig. 5 was fixed at 7 minutes 10 seconds after the metaphase-anaphase transition, when distinct core localization of BAF was observed. Strikingly, an electron-dense area, with a width of nm, overlapping with the localization of BAF (superimposed with green) was clearly seen on the surface of the chromosome mass (Fig. 5G). At this time, well-defined NE could be observed in this region of the chromosome mass, but the NE was not yet fully assembled at the sites of the electrondense areas (panel a in Fig. 5G). Vesicular membranes could be seen near these electron-dense areas (indicated by purple Fig. 4. Live-correlated EM analyses. (A) Time-lapse images of living HeLa cells expressing GFP-BAF were obtained every minute. Magenta represents chromosomes (Hoechst 33342) and green represents GFP-BAF. Cells were fixed after live-cell fluorescence imaging, and the same cell was subjected to EM analysis. (B) A series of deconvolved 3D images of the fixed cell. (C) A single section image from the indicated panel in B. (D) An EM image corresponding to the image in C. (E) Magnified view of the region indicated in D. (F) Highmagnification EM images of the regions a-c indicated in E. In the lower panels, drawings are superimposed on the EM images indicating GFP-BAF (green), the NE (red), MTs (orange) and vesicles (purple arrowheads). Scale bars: 10 μm (A,C), 2 μm (D) and 200 nm (F).
7 (15) arrowheads in Fig. 5G); some vesicles with a diameter of approximately 50 nm appeared to attach to the BAF-containing electron dense areas (see panel c in Fig. 5G and panel b in Fig. 6G). In addition, vesicular membranes at the site of the reforming NE at the core region appeared to be attached to MTs (see arrowheads in panel c in Fig. 5G and panel b in Fig. 6G). It could also be seen that some MTs had penetrated into the chromosome mass in the gap between two BAF-containing electron-dense areas (panel b in Fig. 5G), and others attached to the surface of the electron-dense area (panel d in Fig. 5G). At the same time, most Fig. 5. Live-correlated EM analyses. (A) Time-lapse images of living HeLa cells expressing GFP-BAF were obtained every minute. Magenta represents chromosomes (Hoechst 33342) and green represents GFP-BAF. Cells were fixed after live-cell fluorescence imaging, and the same cell was subjected to EM analysis. (B) A series of deconvolved 3D images of the fixed cell. (C) A single section image from the indicated panel in B. (D) An EM image corresponding to the image in C. (E,F) Magnified views of the regions indicated in D. (G) High-magnification EM images of the regions a-d indicated in E and F. In the lower panels, drawings are superimposed on the EM images indicating GFP-BAF (green), the NE (red), MTs (orange) and vesicles (purple arrowheads). Scale bars: 10 μm (A,C), 2 μm (D) and 200 nm (G). (H) Time-lapse images of living HeLa cells obtained every minute. Upper panels represent chromosomes stained with Hoechst and lower panels fluorescently conjugated NLS-BSA microinjected as described by Haraguchi et al. (Haraguchi et al., 2000). Time 0 represents the metaphase-anaphase transition. Arrows in the image taken at 7 minutes indicate the first detectable accumulation of fluorescent NLS signals.
8 BAF directs nuclear envelope assembly 2547 Fig. 6. Live-correlated EM analyses. (A) Time-lapse images of living HeLa cells expressing GFP-BAF were obtained every minute. Magenta represents chromosomes (Hoechst 33342) and green represents GFP-BAF. Cells were fixed after live-cell fluorescence imaging, and the same cell was subjected to EM analysis. (B) A series of deconvolved 3D images of the fixed cell. (C) A single section image from the indicated panel in B. (D) An EM image corresponding to the image in C. (E,F) Magnified views of the regions indicated in D. (G) High-magnification EM images of the regions a-c indicated in E and F. In the lower panels, drawings are superimposed on the EM images indicating GFP-BAF (green), the NE (red), MTs (orange), and vesicles (purple arrowheads). (H) Schematic diagram of NE reformation during telophase. Upper panel: cartoon representation of NE formation at the core region. BAF (green) forms a stable complex on the surface of the telophase chromosome mass in the presence of MTs. Then, LEM-domain proteins containing NE precursor vesicles attach to the BAF complex, fuse with each other, and finally enclose the chromosomes at the core region as MTs disassemble. Lower panel: EM images of the stages depicted by the cartoon representations. Scale bars: 10 μm (A,C), 2 μm (D), 200 nm (G) and 100 nm (H).
9 (15) Fig. 7. See next page for legend.
10 BAF directs nuclear envelope assembly 2549 Fig. 7. RNAi knockdown of BAF and lamin A. (A) Western blotting analysis of HeLa cells after sirna treatment for BAF or lamin A. (B) Number of cells showing core localization after knockdown of BAF or lamin A. Loss of BAF abolished core localization of both emerin and lamin A. (C) Representative image data which was analyzed to generate the results shown in B. Scale bars: 10 μm. (D) Western blotting analysis of HeLa cells stably expressing GFP- BAF after sirna treatment for BAF, lamin A or emerin. (E) Electron microscopy images of HeLa cells subjected to sirna treatment for control luciferase (a) and BAF (c). The respective insets (b,d) are fluorescence images of the same specimen: magenta represents chromosomes and green represents BAF. The arrows show the BAF core (a,b), and the arrowheads indicate no accumulation of BAF (c,d). Scale bars: 2 μm (a,c), 10 μm (b,d). (F) Timelapse images of living cells stably expressing GFP-BAF are shown for control RNAi (luciferase RNAi), lamin A RNAi, and emerin RNAi. Numbers on the top represent the time in minutes after the metaphase-anaphase transition. Numbers on the left or the bottom represent the lifetime of the core region. Scale bars: 10 μm. (G) HeLa cells were subjected to sirna treatment for control luciferase, BAF, lamin A and emerin, as indicated on the left. After sirna treatment, cells expressing GFP-BAF were fixed with formaldehyde and observed (GFP-BAF, labeled at the top); cells without GFP-BAF were fixed with methanol and endogenous proteins (emerin, lamin A and lamin B as indicated) were detected by indirect immunofluorescence with specific antibodies. Scale bar: 10 μm. of the non-core region of telophase chromosomes was overlaid with NE (supplementary material Fig. S2C,D), suggesting that NE reformation in the core and non-core regions are regulated by distinct mechanisms. It should be pointed out that the core region is not fully covered by NE at 7 minutes; complete reassembly of the NE does not occur until approximately 10 minutes (Fig. 6). This is surprising because nuclear transport started at 7-8 minutes (Fig. 5H, arrows; also see Haraguchi et al., 2000). At this time, the NPCs that have been targeted to the NE were localized exclusively outside the core region (Fig. 1B; also see supplementary material Fig. S2C,D). Collectively, these results suggest that the NPCs on the reforming NE at 7-8 minutes after the metaphase-anaphase transition are functional and that nuclear transport through the NPCs may begin in the absence of a fully reassembled NE. The cell shown in Fig. 6 was fixed at 10 minutes 11 seconds after the metaphase-anaphase transition. At this time, most of the chromosome mass was surrounded by the NE; the BAF-containing electron-dense surface (green) was also enveloped within the NE (red). This is consistent with previous findings that emerincontaining and LBR-containing membranes fuse at this time (Haraguchi et al., 2000). Most spindle MTs have disappeared (panels a and b Fig. 6G), but some MTs were still present close to the chromosomes. In some cases, MT cables proximate to the electrondense chromosome areas were disrupted by the presence of the NE (panel c Fig. 6G). Importantly, electron-dense structures formed on the surface of the telophase chromosome mass in HeLa cells not expressing exogenous GFP-BAF (supplementary material Fig. S3), indicating that this structure is a native one, and not an artefact of GFP-BAF. A proposed order of NE formation at the core region is summarized in Fig. 6H. The NE forms directly on the chromosomes outside the core region, but forms on the BAF-containing dense structures at the core region. It appears that accumulation of the electron-dense material eliminates MT attachment to the chromosome mass as the electron-dense core grows. The BAFcontaining electron-dense structure then disappears after the NE covers the entire chromosome mass. BAF has a direct role on NE formation at the core region, and lamin A and emerin affect the lifetime of the core structure To determine the contribution of BAF to core formation and subsequent NE formation, we examined the effects of BAF knockdown by RNAi in HeLa cells. BAF was selectively removed in cells treated with BAF sirna (Fig. 7A,D). In those cells, core localization of emerin and lamin A was effectively inhibited (Fig. 7B); examples of microscopic images are shown in Fig. 7C. In addition, we applied live CLEM analysis to BAF sirna-treated HeLa cells expressing GFP-BAF, and observed cell structures especially focusing on the electron-dense structure, the NE, chromosomes, the MTs and membranes (Fig. 7E, Figs 8 and 9; also see supplementary material Fig. S4). In BAF sirna-treated cells (see Fig. 7D), the electron-dense structure at the periphery of the central region of the chromosome mass was lost (n=4; Fig. 7E, compare arrowheads in cells at 7 minutes after the metaphaseanaphase transition with arrows in the control cells), and chromosomes remained condensed (Fig. 8, compare G or M with B), indicating that BAF is required for formation of the core structure and for chromosome decondensation. In addition, the NE at the core region had not formed in BAF sirna-treated cells even at 12 minutes after the metaphase-anaphase transition (n=10; see Fig. 8I,J) whereas it was almost fully reassembled in luciferase sirna-treated control cells (n=4; Fig. 8D). In the BAF-depleted cells it was also frequently observed that relatively long MTs remained in the chromosomal region at this late period (compare Fig. 8J with 8D; also compare Fig. 9F with 9C). Finally, abnormal inclusion of cytoplasm inside the nucleus was also often seen these cells (Fig. 8P,Q; pale regions in nucleus). Interestingly, in addition to these phenotypes, BAF depletion caused a significant reduction of tubular membranes near the core region where the spindle MTs persisted, in contrast to an abundance of tubular membranes in the presence of BAF (compare Fig. 9E,F with 9B,C). Instead, in BAF RNAi cells, abnormal piles of threedimensionally extended ER membranes in the cytoplasm were generated (Fig. 9G,H; also see Fig. 8K,L, compare these with Fig. 8E): in many cases these abnormal membranes seemed to be NElike membranes assembling away from the chromosomes (Fig. 9G,H). These findings are schematically summarized in Fig. 9I. Taken together, these results indicate that BAF has a direct effect on NE formation at the core region, as suggested previously (Haraguchi et al., 2000), and suggest that the role of BAF in NE formation is to coordinate the recruitment of NE precursor membranes to the chromosome surface. RNAi knockdown of lamin A did not eliminate core localization of BAF or emerin, but did reduce the frequency of core localization (Fig. 7B). To test whether this reduced frequency reflected a shortened life of the core structure, we determined the duration of the core structure (Fig. 7F) in cells with specific RNAi-mediated knockdown of BAF, lamin A or emerin, as shown in Fig. 7D. Its duration was significantly shortened from 8 minutes 9 seconds (n=10) in control luciferase sirna cells to 1 minute 42 seconds (n=7) in lamin A sirna cells, suggesting that lamin A stabilizes the core structure (Fig. 7F). By contrast, in emerin sirna cells, the core structure remained intact significantly longer (more than 20 minutes) (n=7) compared with control RNAi cells (n=7) (Fig. 7F), often persisting up to early G1 phase (data not shown), suggesting that emerin destabilizes the core structure. Furthermore, knockdown of these proteins also affected localization to the NE in the subsequent interphase. Knockdown of BAF disrupted NE localization of emerin and lamin A,
11 (15) knockdown of lamin A disrupted NE localization of BAF and emerin, and knockdown of emerin disrupted NE localization of BAF and lamin A (Fig. 7G). These effects are specific because the same treatment did not affect lamin B localization (Fig. 7G). Taken together, these results suggest that BAF has a direct role on core formation and that lamin A and emerin affect the stability of the core structure. The spindle MTs mediate BAF assembly to the core region BAF forms an electron-dense core structure close to the MTattaching regions (Fig. 5). Therefore, the MTs could be involved in BAF assembly to the core region. To test this idea, we first examined whether BAF colocalizes with the spindle MTs during mitosis. Indirect immunofluorescence staining of cells fixed with trichloroacetic acid (TCA) showed that in mitotic cells BAF Fig. 8. Live-correlated EM analyses of HeLa cells after sirna treatment for BAF. (A) Time-lapse images for chromosomes and GFP-BAF were taken for control cells. Cells were fixed at 12 minutes after the metaphase-anaphase transition. Only images for GFP-BAF are shown. See supplementary material Fig. S4 for more details. (B) An EM image of the same cells shown in A. (C) A fluorescence micrograph of the same cells shown in A: selected focus from a series of 3D images. Magenta represents chromosomes (Hoechst 33342) and green represents GFPBAF. (D) Magnified view of the region indicated in the larger square in B. Drawings are superimposed on the EM images indicating the NE (red) and MTs (orange). (E) Highmagnification EM images of the small box indicated in B. (F) Timelapse images for chromosomes and GFP-BAF were taken for BAF RNAi cells. Cells were fixed at 12 minutes after the metaphase-anaphase transition. Only images for GFP-BAF are shown. See supplementary material Fig. S4 for more details. (G) EM images of the same cells shown in F. (H) A fluorescence micrograph of the same cells in F: a selected focus from a series of 3D images. Magenta represents chromosomes (Hoechst 33342) and green represents GFP-BAF. (I) Magnified view of the region indicated in the largest square in G. (J) Drawings are superimposed on the EM images of I, indicating the NE (red) and MTs (orange). (K,L) High-magnification EM images of the small boxes indicated in G: K, left box; L, right box. (M) An EM image of another example of BAF sirna-treated cells. (N) A fluorescence micrograph of the cells shown in M. Magenta represents chromosomes (Hoechst 33342) and green represents GFP-BAF. (O) Enlarged image of the boxed region in N and corresponding to the boxed region in M. (P) Magnified view of the boxed region in M and corresponding to the image shown in O. (Q) Drawings are superimposed on the EM images of P, indicating the NE (red) and chromosomes (magenta). Scale bars: 10 μm (A,F), 2 μm (B,C,G,H,M,N), 500 nm (D,J,P,Q) and 200 nm (L).
12 BAF directs nuclear envelope assembly 2551 localizes to the spindle MTs from metaphase to telophase (Fig. 10A). Since this result shows colocalization of BAF with the spindle MTs for the first time, we used BAF knockdown by BAF RNAi to confirm our initial observation; BAF RNAi treatment effectively removed the signal, indicating that BAF, at least in part, colocalizes with the spindle MTs (supplementary material Fig. S5). To further test a direct role of the spindle MTs in core formation, we examined the effect of nocodazole, a MT-depolymerizing reagent, on BAF assembly to the core region. In cells treated with nocodazole during anaphase, the spindle MTs disappeared and BAF core assembly was lost (Fig. 10B), indicating that the spindle MTs are required for BAF assembly to the core. Taken together, these results suggest that BAF is associated with the spindle MTs during metaphase through anaphase and assembles to the core region in early telophase, in a spindle-mt-dependent manner, where it forms a stable complex with other core-localizing proteins such as lamin A and emerin. Based on these findings, we propose a model, schematically illustrated in Fig. 10C, in which spindle MT function organizes BAF spatially, thereby engendering BAF-directed NE assembly. Discussion In this report, we have demonstrated that the BAF-dependent core structure is an immobile complex that appears as an electron-dense structure directly on chromosomes with a width of nm. This study shows for the first time that the spindle MTs are involved in NE reformation at the end of mitosis: spindle MTs mediate BAF assembly to the core region where BAF recruits selective NE components such as lamin A and emerin. The core structure is important in establishing a normal interphase NE and, consequently, normal nuclear function. Live CLEM observation revealed that the timing of NE reformation is different in the core region from that in the non-core region: NE reformation is significantly later in the core region (compare Figs 4-6 with supplementary material Fig. S2). This suggests that there are at least two different mechanisms for NE assembly; one that acts at the core region, and a second that acts at the non-core region. RanGTP would appear to be involved in NE reformation at the non-core region because it is known to be involved in the assembly of the NPCs in in vitro Xenopus oocyte extracts Fig. 9. Live-correlated EM analyses of HeLa cells expressing GFP-BAF after sirna treatment (D-H) or luciferase sirna treatment as a control (A-C). During timelapse imaging of living HeLa cells expressing GFP-BAF, cells were fixed 7 minutes after the metaphase-anaphase transition and subjected to EM analysis. A magnified view of the region indicated in A is shown in B and C. Magnified views of the regions indicated in D are shown in E and F (left square region), and G and H (right square region). In C, F and H, drawings are superimposed on the EM images indicating GFP-BAF (green), the NE (red), and MTs (orange). Scale bars: 2 μm (A,D), 500 nm (B,C,E-H). (I) Model of NE formation in the control sirna-treated (left) and BAF sirna-treated cell (right). For details, see text.
13 (15) Fig. 10. Microtubule-dependent accumulation of BAF at the core. (A) Localization of BAF on the spindle MTs in metaphase and at the core in telophase. (B) Localization of BAF in HeLa cells treated with nocodazole during anaphase (bottom) and in untreated control cells (top). (C) Model of spindle MT-mediated, BAF-dependent NE formation. See text for details. Scale bars: 10 μm (A,B). (Zhang et al., 2002; Walther et al., 2003) and our observations indicate that NPCs assemble exclusively in the non-core region. We propose that BAF is a factor involved in NE assembly at the core region. Live cell imaging of various ER- or NE-associated integral membrane proteins have shown that most membranes are excluded from the area of the spindle MTs until early telophase (Ellenberg et al., 1997; Yang et al., 1997; Haraguchi et al., 2000; Anderson and Hetzer, 2007; Anderson and Hetzer, 2008). From these observations, we speculate that the mitotic spindle, or their associated proteins, may have an inhibitory effect on NE reformation, presumably by somehow repelling precursor membranes. As a consequence of this, fewer precursor membranes would be available for NE reformation in the core region. Here, we report that BAF associates with the spindle MTs, and that its targeting to the core region requires spindle MTs (Fig. 10B). These findings strongly support the idea that BAF acts to direct core NE reformation by recruiting selective NE protein-bearing membranes in the presence of the inhibitory spindle MTs. This hypothesis is further supported by the fact that loss of BAF by RNAi resulted in removal of most of the membranes from the area near the core region during telophase (Fig. 9E,F), and abnormal assembly of NE-like membranes in the cytoplasm far from the chromosomal region (Fig. 9G,H), and also caused lamin A and emerin to disperse into the cytoplasm (Fig. 7C,G). In addition, abnormal inclusion of cytoplasm inside the nucleus was frequently observed in cells lacking BAF (Fig. 8P,Q). As BAF is a key molecule for core formation, BAF targeting to the core region must be an important step for core formation. Recently, Gorjánácz and colleagues have reported that BAF-1 (C. elegans BAF homolog) is localized in a chromosomal region similar to the core during NE formation in dividing cells of early C. elegans embryos, and its core -like localization is abolished by sirna depletion of VRK-1 (C. elegans vaccinia-related kinase homolog) (Gorjánácz et al., 2007). Their results suggest that phosphorylation of BAF is essential for its core -like localization in C. elegans embryos. Human BAF can be phosphorylated in vitro on its N- terminal conserved amino acid residues, Ser4 (Bengtsson and Wilson, 2006) or all three amino acid residues Thr2-Thr3-Ser4, by vaccinia-related kinases (Nichols et al., 2006). Paradoxically, however, initial studies have indicated that phosphorylation of human BAF abolishes its interaction with DNA in vitro (Nichols et al., 2006), reduces its binding to the LEM proteins in vitro (Nichols et al., 2006), and disrupts emerin localization to the NE in vivo (Bengtsson and Wilson, 2006), suggesting that the N-terminal phosphorylated form of human BAF cannot attach to the chromosomal region at the core. One possibility to reconcile these results is that BAF phosphorylated by VRK1 binds spindle MTs, and then is able to localize to the core region once it is dephosphorylated at the end of mitosis. This hypothesis is supported by the following findings: (1) BAF associates with the spindle MTs (Fig. 10A); (2) BAF core accumulation is abolished by destruction of the spindle MTs (Fig. 10B); and (3) phosphorylated BAF is localized on the spindle MTs but not in the core region (T.H., unpublished result). However, it remains to be elucidated whether phosphorylation by Vrk1 is required for association with the spindle MTs. Cell-cycle dependent phosphorylation of emerin also regulates the interaction of emerin with BAF (Hirano et al., 2005; Ellis et al., 1998), suggesting that modification of emerin or other corelocalizing proteins may also contribute to the core formation and structural features of the core. Since mitotic progression is regulated by the spatial and temporal coordination of several cellcycle-regulating kinases and phosphatases, the combination of several protein modifications may cooperatively enforce core formation in telophase. Cooperative actions of BAF and core-localizing proteins in NE organization are supported by several observations. BAF, lamin A and emerin form a three-way complex in vitro (Holaska et al., 2003) and in vivo in C. elegans (Liu et al., 2003; Margalit et al., 2005; Gorjánácz et al., 2007). Retention of lamin A and emerin in the reforming NE requires their initial localization at the core region, and this localization is BAF dependent (see Fig. 7G). Core localization is also required for BAF to establish its own proper nuclear localization (Zheng et al., 2000; Margalit et al., 2005; Gorjánácz et al., 2007). Furthermore, the lifetime of the core structure seems to depend on specific core-localizing proteins such as emerin, lamin A and LAP2α. Loss of emerin by RNAi prolonged core lifetime (see Fig. 7F). A persistent core structure is also
14 BAF directs nuclear envelope assembly 2553 observed in HeLaS3 cells expressing high levels of lamin A, and this persistent core structure is enriched in emerin and lamin A (Maeshima et al., 2006). We also observed a prolonged core lifetime in cells overexpressing lamin A or LAP2α (data not shown). By contrast, loss of lamin A by RNAi significantly reduced the lifetime of the core structure (see Fig. 7F). These results suggest that emerin destabilizes the core, whereas lamin A and LAP2α stabilize the core structure. The balance between their competing activities may be important for regulating the formation of the transient core structure, the formation of the mature NE and the eventual formation of a functional nucleus. In summary, we conclude that BAF assembles on the surface of the telophase chromosome mass in regions where the chromosomes are interacting with spindle MTs, and that this assembly is MT dependent and requires coordinated BAF phosphorylation followed by BAF dephosphorylation. Chromosome-associated BAF makes a higher-order complex with itself, lamin A and emerin, resulting in formation of an electron-dense structure at the core which is required for proper reformation of the NE in this region and for the re-establishment of a functional nucleus. Furthermore, BAFdependent NE reformation at the core, which involves LEM-domain proteins containing NE precursor vesicles attaching to the BAF complex and then fusing with each other, is mechanistically distinct from NE reformation at the non-core region of the chromosome mass. Understanding the dynamics and the structural details of NE reformation at the end of mitosis will provide new insights into the development of lamin-a-dependent disease and other NE-associated human diseases. Materials and Methods Cells and reagents HeLa cells were obtained from the Riken Cell Bank (Tsukuba, Japan) and maintained in DME medium containing 10% calf serum. Double-stranded sirna for BAF, lamin A and emerin were purchased from Qiagen (Hilden, Germany). Mouse monoclonal anti-lamin A/C antibodies TiM92 (IgM) were generated as described previously (Haraguchi et al., 2001). Mouse monoclonal anti-lamin A/C antibodies MMS-107P (IgM) were purchased from Covance (Berkeley, CA). Mouse monoclonal anti-lamin B (101-B7) antibodies were obtained from MatriTech (Cambridge, MA). Rabbit polyclonal anti-emerin antibodies were a gift of the Arahata lab (Tokyo, Japan). Anti- BAF antibodies (PU38143) were generated against recombinant BAF in a rabbit and affinity purified. Plasmids encoding mcfp and mvenus (monomeric versions of ECFP and Venus), which were made by substitution of Ala for Lys (Zacharias et al., 2002), were gifts of Nagai, Sawano and Miyawaki (RIKEN, Wako, Japan). LAP2α-YFP and GFP-lamin B1 were gifts of R. Foisner (Medical University Vienna) and R. Goldman (Northwestern University), respectively. Plasmid construction Methods for constructing DNA plasmids encoding LBR-GFP, GFP-emerin, GFP- BAF, GFP-LAP2β and GFP-MAN1 were described previously (Haraguchi et al., 2000; Haraguchi et al., 2001, Shimi et al., 2004). To construct GFP-lamin A, the coding region was PCR-amplified from the RT-PCR product of lamin A using primers lam- 1 and lam-2 and inserted into the pegfp-c1 vector (Clontech Laboratories, Palo Alto, CA). To construct mcfp and mvenus fusion proteins, the coding region of mcfp or mvenus was PCR-amplified from ECFP-A206K/pRSETB or Venus- A206K/pRSETB, respectively, with primers cfp-1 and cfp-2 and inserted into the pegfp-c1 vector digested with NheI and BglII restriction enzymes to replace EGFP. The resulting mcfp and mvenus vectors were used for construction of mcfp-baf, mcfp-lamin A, mcfp-emerin, mcfp-emerin-m24 and mvenus-baf. To construct mcfp-baf or mvenus-baf, the coding region of BAF was PCR-amplified from pet15b-baf (Lee and Craigie, 1998) using primers baf-1 and baf-2 and inserted into the above described mcfp or mvenus vectors, respectively. To construct mvenusemerin or mvenus-emerin-24m, the coding region of emerin was PCR-amplified from a GFP-emerin construct (Haraguchi et al., 2000) using primers eme-1 and eme-2 and inserted into the mvenus vector. To construct mvenus-lamin A, the coding region of lamin A was PCR-amplified from the GFP-lamin A construct using primers lam-3 and lam-4 and inserted into the mvenus vectors. The coding sequence of human Nup35 was obtained from mrna in HeLa cells by RT-PCR using primers nup-1 and nup-2, cloned into the TA cloning vector pcr2.1 (Invitrogen), and re-cloned to the pegfp-c3 vector at the EcoRI sites. The DNA sequences of all fusion plasmids were confirmed using an ABI 310 or ABI 3100 Genetic Analyser (Applied Biosystems, Norwalk, CT). Primer sequences are listed in supplementary material Table S1. Fluorescence microscopy Cells were grown in a glass-bottom culture dish (MatTech, Ashland, MA). GFP fusion plasmids ( μg) were transfected into cells with LipofectaminePlus (Gibco BRL) according to the manufacturer s methods except that the incubation time with DNA was reduced to 1.5 hours. Time-lapse, multicolor images in living cells, as well as indirect immunofluorescence images in fixed cells, were obtained using the DeltaVision microscope system based on an Olympus wide-field fluorescence microscope IX70 (Applied Precision, Seattle, WA). For temperature control during live observation, the microscope was kept at 37 C in a temperature-controlled room as described previously (Haraguchi et al., 1999). A series of three-dimensional image data were obtained, and deconvolved with the SoftWorx software equipped with the DeltaVision as described previously (Haraguchi et al., 2001; Haraguchi et al., 2004). FRAP and FRET analyses FRAP and FRET studies were done using a laser-scanning confocal microscope, Zeiss LSM510 META (Carl Zeiss, Yena) equipped with a Kr laser (Coherent, Santa Clara, CA) (Haraguchi et al., 2002; Hiraoka et al., 2002). FRAP experiments were carried out as described previously (Shimi et al., 2004). Briefly, HeLa cells stably expressing GFP fusion proteins were imaged using a water-immersion objective lens (C- Apochromat 40 /NA1.2) on a Zeiss LSM510 META. Photobleaching was performed with full power of 488 nm light from the argon laser 10 iterations for a defined region of each cell. Image data were collected through a pinhole of 2.37 Airy units (corresponding to an optical section 2 μm thick) with 488 nm argon laser at 0.5% full power. For FRET experiments, living HeLa cells transiently expressing pairs of NE proteins fused with mcfp and mvenus were imaged using a water-immersion objective lens (C-Apochromat 40 /NA1.2) on a Zeiss LSM510 META. FRET was detected by acceptor photobleaching (Wouters et al., 2001). mvenus was photobleached by 514 nm light from the argon laser, with full power, 50 iterations for a defined area (2.1 μm square) in each cell. After photobleaching, time-lapse images were collected on the META detector from 470 nm to 545 nm through a pinhole of 2.18 Airy units (corresponding to an optical section 1.8 μm thick) by exciting with 413 nm light from the Kr laser at % full power. FRET efficiency was determined for fluorescence intensity of mcfp and mvenus after linear unmixing as described previously (Shimi et al., 2004). For time-lapse FRET ratio imaging, a fluorescence microscope IX70 (Olympus) equipped with a Nipkow disc confocal unit, CSU21 (Yokogawa, Japan) was used. Cells were illuminated with a 430 nm diode laser to excite CFP, and image data for CFP and YFP were simultaneously obtained on a CoolSnapHQ CCD camera (Photometrics, Roper, Tucson, Texas) through a dual viewer (Optical Insights), which separates emission light for wavelengths of 480 nm (corresponding to CFP) and 535 nm (corresponding to YFP). Ratio images between CFP and YFP were obtained by MetaMorph software (Universal Imaging Corporation, Downingtown, PA). Correlative light electron microscope analysis after live observation (live CLEM) Cells stably expressing GFP-BAF were cultured in a special glass-bottomed dish with an addressing grid (grid size, 175 μm) on the coverslip (Iwaki, Japan). After staining with Hoechst 33342, live cells were monitored on DeltaVision; at the desired time point after the metaphase-anaphase transition, cells were fixed with glutaraldehyde at a final concentration of 2.5% for 1 hour. Three-dimensional images (50-60 focal planes at 0.2 μm intervals) were obtained using an Olympus oilimmersion objective lens (PlanApo 60/NA1.4) and computationally processed by three-dimensional deconvolution (Agard et al., 1989). Electron microscope (EM) observation of the same cell was carried out as follows. Cells were post-fixed with 1% OsO 4 (Nisshin EM, Tokyo, Japan) in phosphate buffer, ph 7.4, for 1 hour, washed briefly with distilled water, and sequentially dehydrated with 50 and 70% ethanol. After staining with 2% uranyl acetate in 70% ethanol for 1 hour, cells were again sequentially dehydrated with 90% and 100% ethanol, and then embedded in epoxy resin by incubating with 50% (v/v) Epon812 (TAAB, Berkshire, UK) in ethanol for 30 minutes and 100% Epon812 for 1 hour. The epoxy block containing the same cell observed by a fluorescence microscope was trimmed according to the grid reference on the coverslip, and sliced to ultra-thin sections with a thickness of 80 nm using a microtome (Leica microsystems, Germany). Thin sections were stained with 2% uranyl acetate (Merck) for 1 hour and a commercial ready-to-use solution of lead citrate (Sigma) for 1 minute. Image data were collected using an electron microscope Hitachi H7600 (80 kv, Hitachi, Japan). Knockdown by RNAi Two to five micrograms of oligomeric double-stranded RNA for BAF, lamin A or emerin were transfected into HeLa cells cultured in a 35 mm culture dish using Oligofectamine (Invitrogen, Carlsbad, CA), Lipofectamine PLUS (Invitrogen), or RNAiFect (Qiagen) transfection reagents according to the manufacturer s instructions. The cells were incubated for 3 days in DMEM culture medium containing 10% calf serum.
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Lu et al., http://www.jcb.org/cgi/content/full/jcb.201012063/dc1 Figure S1. Kinetics of nuclear envelope assembly, recruitment of Nup133
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Lu et al., http://www.jcb.org/cgi/content/full/jcb.201012063/dc1 Figure S1. Kinetics of nuclear envelope assembly, recruitment of Nup133
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Edens and Levy, http://www.jcb.org/cgi/content/full/jcb.201406004/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Nuclear shrinking does not depend on the cytoskeleton
Supplemental material Edens and Levy, http://www.jcb.org/cgi/content/full/jcb.201406004/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Nuclear shrinking does not depend on the cytoskeleton
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Krenn et al., http://www.jcb.org/cgi/content/full/jcb.201110013/dc1 Figure S1. Levels of expressed proteins and demonstration that C-terminal
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Krenn et al., http://www.jcb.org/cgi/content/full/jcb.201110013/dc1 Figure S1. Levels of expressed proteins and demonstration that C-terminal
Supplemental Information. 3D-CLEM Reveals that a Major Portion. of Mitotic Chromosomes Is Not Chromatin
 Molecular Cell, Volume 64 Supplemental Information 3D-CLEM Reveals that a Major Portion of Mitotic Chromosomes Is Not Chromatin Daniel G. Booth, Alison J. Beckett, Oscar Molina, Itaru Samejima, Hiroshi
Molecular Cell, Volume 64 Supplemental Information 3D-CLEM Reveals that a Major Portion of Mitotic Chromosomes Is Not Chromatin Daniel G. Booth, Alison J. Beckett, Oscar Molina, Itaru Samejima, Hiroshi
Imaging of Chromosomes at Nanometer-Scale Resolution, Using Soft X-Ray Microscope
 Imaging of Chromosomes at Nanometer-Scale Resolution, Using Soft X-Ray Microscope K. Takemoto, A. Yamamoto 1, I. Komura 2, K. Nakanishi 3, H. Namba 2 and H. Kihara Abstract In order to clarify the process
Imaging of Chromosomes at Nanometer-Scale Resolution, Using Soft X-Ray Microscope K. Takemoto, A. Yamamoto 1, I. Komura 2, K. Nakanishi 3, H. Namba 2 and H. Kihara Abstract In order to clarify the process
Regulators of Cell Cycle Progression
 Regulators of Cell Cycle Progression Studies of Cdk s and cyclins in genetically modified mice reveal a high level of plasticity, allowing different cyclins and Cdk s to compensate for the loss of one
Regulators of Cell Cycle Progression Studies of Cdk s and cyclins in genetically modified mice reveal a high level of plasticity, allowing different cyclins and Cdk s to compensate for the loss of one
Supplementary Figure 1. Properties of various IZUMO1 monoclonal antibodies and behavior of SPACA6. (a) (b) (c) (d) (e) (f) (g) .
 Supplementary Figure 1. Properties of various IZUMO1 monoclonal antibodies and behavior of SPACA6. (a) The inhibitory effects of new antibodies (Mab17 and Mab18). They were investigated in in vitro fertilization
Supplementary Figure 1. Properties of various IZUMO1 monoclonal antibodies and behavior of SPACA6. (a) The inhibitory effects of new antibodies (Mab17 and Mab18). They were investigated in in vitro fertilization
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb222 / b. WB anti- WB anti- ulin Mitotic index (%) 14 1 6 2 T (h) 32 48-1 1 2 3 4 6-1 4 16 22 28 3 33 e. 6 4 2 Time (min) 1-6- 11-1 > 1 % cells Figure S1 depletion leads to mitotic defects
DOI: 1.138/ncb222 / b. WB anti- WB anti- ulin Mitotic index (%) 14 1 6 2 T (h) 32 48-1 1 2 3 4 6-1 4 16 22 28 3 33 e. 6 4 2 Time (min) 1-6- 11-1 > 1 % cells Figure S1 depletion leads to mitotic defects
Cell Cycle, Mitosis, and Microtubules. LS1A Final Exam Review Friday 1/12/07. Processes occurring during cell cycle
 Cell Cycle, Mitosis, and Microtubules LS1A Final Exam Review Friday 1/12/07 Processes occurring during cell cycle Replicate chromosomes Segregate chromosomes Cell divides Cell grows Cell Growth 1 The standard
Cell Cycle, Mitosis, and Microtubules LS1A Final Exam Review Friday 1/12/07 Processes occurring during cell cycle Replicate chromosomes Segregate chromosomes Cell divides Cell grows Cell Growth 1 The standard
Molecular Cell Biology - Problem Drill 22: The Mechanics of Cell Division
 Molecular Cell Biology - Problem Drill 22: The Mechanics of Cell Division Question No. 1 of 10 1. Which of the following statements about mitosis is correct? Question #1 (A) Mitosis involves the dividing
Molecular Cell Biology - Problem Drill 22: The Mechanics of Cell Division Question No. 1 of 10 1. Which of the following statements about mitosis is correct? Question #1 (A) Mitosis involves the dividing
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells
 Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
TFEB-mediated increase in peripheral lysosomes regulates. Store Operated Calcium Entry
 TFEB-mediated increase in peripheral lysosomes regulates Store Operated Calcium Entry Luigi Sbano, Massimo Bonora, Saverio Marchi, Federica Baldassari, Diego L. Medina, Andrea Ballabio, Carlotta Giorgi
TFEB-mediated increase in peripheral lysosomes regulates Store Operated Calcium Entry Luigi Sbano, Massimo Bonora, Saverio Marchi, Federica Baldassari, Diego L. Medina, Andrea Ballabio, Carlotta Giorgi
Supplementary Material and Methods
 Online Supplement Kockx et al, Secretion of Apolipoprotein E from Macrophages 1 Supplementary Material and Methods Cloning of ApoE-GFP Full-length human apoe3 cdna (pcdna3.1/zeo + -apoe) was kindly provided
Online Supplement Kockx et al, Secretion of Apolipoprotein E from Macrophages 1 Supplementary Material and Methods Cloning of ApoE-GFP Full-length human apoe3 cdna (pcdna3.1/zeo + -apoe) was kindly provided
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Beck et al., http://www.jcb.org/cgi/content/full/jcb.201011027/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Membrane binding of His-tagged proteins to Ni-liposomes.
Supplemental material Beck et al., http://www.jcb.org/cgi/content/full/jcb.201011027/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Membrane binding of His-tagged proteins to Ni-liposomes.
The subcortical maternal complex controls symmetric division of mouse zygotes by
 The subcortical maternal complex controls symmetric division of mouse zygotes by regulating F-actin dynamics Xing-Jiang Yu 1,2, Zhaohong Yi 1, Zheng Gao 1,2, Dan-dan Qin 1,2, Yanhua Zhai 1, Xue Chen 1,
The subcortical maternal complex controls symmetric division of mouse zygotes by regulating F-actin dynamics Xing-Jiang Yu 1,2, Zhaohong Yi 1, Zheng Gao 1,2, Dan-dan Qin 1,2, Yanhua Zhai 1, Xue Chen 1,
Supplementary Figure S1
 Supplementary Figure S1 Supplementary Figure S1. PARP localization patterns using GFP-PARP and PARP-specific antibody libraries GFP-PARP localization in non-fixed (A) and formaldehyde fixed (B) GFP-PARPx
Supplementary Figure S1 Supplementary Figure S1. PARP localization patterns using GFP-PARP and PARP-specific antibody libraries GFP-PARP localization in non-fixed (A) and formaldehyde fixed (B) GFP-PARPx
10-2 Cell Division. Chromosomes
 Cell Division In eukaryotes, cell division occurs in two major stages. The first stage, division of the cell nucleus, is called mitosis. The second stage, division of the cell cytoplasm, is called cytokinesis.
Cell Division In eukaryotes, cell division occurs in two major stages. The first stage, division of the cell nucleus, is called mitosis. The second stage, division of the cell cytoplasm, is called cytokinesis.
Chapter 10 Cell Growth and Division
 Chapter 10 Cell Growth and Division 10 1 Cell Growth 2 Limits to Cell Growth The larger a cell becomes, the more demands the cell places on its DNA. In addition, the cell has more trouble moving enough
Chapter 10 Cell Growth and Division 10 1 Cell Growth 2 Limits to Cell Growth The larger a cell becomes, the more demands the cell places on its DNA. In addition, the cell has more trouble moving enough
klp-18 (RNAi) Control. supplementary information. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach
![klp-18 (RNAi) Control. supplementary information. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach klp-18 (RNAi) Control. supplementary information. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach](/thumbs/91/104639484.jpg) DOI: 10.1038/ncb1891 A. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach embryos let hatch overnight transfer to RNAi plates; incubate 5 days at 15 C RNAi food L1 worms adult worms
DOI: 10.1038/ncb1891 A. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach embryos let hatch overnight transfer to RNAi plates; incubate 5 days at 15 C RNAi food L1 worms adult worms
(a) Significant biological processes (upper panel) and disease biomarkers (lower panel)
 Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1.
 Supplementary Figure 1. Visualization of endoplasmic reticulum-mitochondria interaction by in situ proximity ligation assay. A) Illustration of targeted proteins in mitochondria (M), endoplasmic reticulum
Supplementary Figure 1. Visualization of endoplasmic reticulum-mitochondria interaction by in situ proximity ligation assay. A) Illustration of targeted proteins in mitochondria (M), endoplasmic reticulum
Supplemental information contains 7 movies and 4 supplemental Figures
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplemental information contains 7 movies and 4 supplemental Figures Movies: Movie 1. Single virus tracking of A4-mCherry-WR MV
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplemental information contains 7 movies and 4 supplemental Figures Movies: Movie 1. Single virus tracking of A4-mCherry-WR MV
Supplementary Figure 1. Mother centrioles can reduplicate while in the close association
 C1-GFP distance (nm) C1-GFP distance (nm) a arrested HeLa cell expressing C1-GFP and Plk1TD-RFP -3 s 1 2 3 4 5 6 7 8 9 11 12 13 14 16 17 18 19 2 21 22 23 24 26 27 28 29 3 b 9 8 7 6 5 4 3 2 arrested HeLa
C1-GFP distance (nm) C1-GFP distance (nm) a arrested HeLa cell expressing C1-GFP and Plk1TD-RFP -3 s 1 2 3 4 5 6 7 8 9 11 12 13 14 16 17 18 19 2 21 22 23 24 26 27 28 29 3 b 9 8 7 6 5 4 3 2 arrested HeLa
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
Nature Biotechnology: doi: /nbt.3828
 Supplementary Figure 1 Development of a FRET-based MCS. (a) Linker and MA2 modification are indicated by single letter amino acid code. indicates deletion of amino acids and N or C indicate the terminus
Supplementary Figure 1 Development of a FRET-based MCS. (a) Linker and MA2 modification are indicated by single letter amino acid code. indicates deletion of amino acids and N or C indicate the terminus
Bio 401 Sp2014. Multiple Choice (Circle the letter corresponding to the correct answer)
 MIDTERM EXAM KEY (60 pts) You should have 5 pages. Please put your name on every page. You will have 70 minutes to complete the exam. You may begin working as soon as you receive the exam. NOTE: the RED
MIDTERM EXAM KEY (60 pts) You should have 5 pages. Please put your name on every page. You will have 70 minutes to complete the exam. You may begin working as soon as you receive the exam. NOTE: the RED
10-2 Cell Division mitosis. cytokinesis. Chromosomes chromosomes Slide 1 of 38
 In eukaryotes, cell division occurs in two major stages. The first stage, division of the cell nucleus, is called mitosis. The second stage, division of the cell cytoplasm, is called cytokinesis. Chromosomes
In eukaryotes, cell division occurs in two major stages. The first stage, division of the cell nucleus, is called mitosis. The second stage, division of the cell cytoplasm, is called cytokinesis. Chromosomes
1. The diagram shows four stages in mitosis. Only one pair of homologous chromosomes is shown. A B C D ... (1) ... (1)
 1. The diagram shows four stages in mitosis. Only one pair of homologous chromosomes is shown. X A B C D (a) Place stages A, B, C and D in the correct order.... (b) Name the structures labelled X.... Describe
1. The diagram shows four stages in mitosis. Only one pair of homologous chromosomes is shown. X A B C D (a) Place stages A, B, C and D in the correct order.... (b) Name the structures labelled X.... Describe
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3076 Supplementary Figure 1 btrcp targets Cep68 for degradation during mitosis. a) Cep68 immunofluorescence in interphase and metaphase. U-2OS cells were transfected with control sirna
DOI: 10.1038/ncb3076 Supplementary Figure 1 btrcp targets Cep68 for degradation during mitosis. a) Cep68 immunofluorescence in interphase and metaphase. U-2OS cells were transfected with control sirna
10-2 Cell Division. Slide 1 of 38. End Show. Copyright Pearson Prentice Hall
 1 of 38 Cell Division In eukaryotes, cell division occurs in two major stages. The first stage, division of the cell nucleus, is called mitosis. The second stage, division of the cell cytoplasm, is called
1 of 38 Cell Division In eukaryotes, cell division occurs in two major stages. The first stage, division of the cell nucleus, is called mitosis. The second stage, division of the cell cytoplasm, is called
Nature Immunology: doi: /ni.3631
 Supplementary Figure 1 SPT analyses of Zap70 at the T cell plasma membrane. (a) Total internal reflection fluorescent (TIRF) excitation at 64-68 degrees limits single molecule detection to 100-150 nm above
Supplementary Figure 1 SPT analyses of Zap70 at the T cell plasma membrane. (a) Total internal reflection fluorescent (TIRF) excitation at 64-68 degrees limits single molecule detection to 100-150 nm above
Fig. S1. Subcellular localization of overexpressed LPP3wt-GFP in COS-7 and HeLa cells. Cos7 (top) and HeLa (bottom) cells expressing for 24 h human
 Fig. S1. Subcellular localization of overexpressed LPP3wt-GFP in COS-7 and HeLa cells. Cos7 (top) and HeLa (bottom) cells expressing for 24 h human LPP3wt-GFP, fixed and stained for GM130 (A) or Golgi97
Fig. S1. Subcellular localization of overexpressed LPP3wt-GFP in COS-7 and HeLa cells. Cos7 (top) and HeLa (bottom) cells expressing for 24 h human LPP3wt-GFP, fixed and stained for GM130 (A) or Golgi97
Prentice Hall Biology Slide 1 of 38
 Prentice Hall Biology 1 of 38 2 of 38 In eukaryotes, cell division occurs in two major stages. The first stage, division of the cell nucleus, is called mitosis. The second stage, division of the cell cytoplasm,
Prentice Hall Biology 1 of 38 2 of 38 In eukaryotes, cell division occurs in two major stages. The first stage, division of the cell nucleus, is called mitosis. The second stage, division of the cell cytoplasm,
Mitosis. AND Cell DiVISION
 Mitosis AND Cell DiVISION Cell Division Characteristic of living things: ability to reproduce their own kind. Cell division purpose: When unicellular organisms such as amoeba divide to form offspring reproduction
Mitosis AND Cell DiVISION Cell Division Characteristic of living things: ability to reproduce their own kind. Cell division purpose: When unicellular organisms such as amoeba divide to form offspring reproduction
Figure S1. HP1α localizes to centromeres in mitosis and interacts with INCENP. (A&B) HeLa
 SUPPLEMENTARY FIGURES Figure S1. HP1α localizes to centromeres in mitosis and interacts with INCENP. (A&B) HeLa tet-on cells that stably express HP1α-CFP, HP1β-CFP, or HP1γ-CFP were monitored with livecell
SUPPLEMENTARY FIGURES Figure S1. HP1α localizes to centromeres in mitosis and interacts with INCENP. (A&B) HeLa tet-on cells that stably express HP1α-CFP, HP1β-CFP, or HP1γ-CFP were monitored with livecell
U3.2.3: Eukaryotic chromosomes are linear DNA molecules associated with histone proteins. (Oxford Biology Course Companion page 151).
 Cell Division Study Guide U3.2.3: Eukaryotic chromosomes are linear DNA molecules associated with histone proteins. (Oxford Biology Course Companion page 151). 1. Describe the structure of eukaryotic DNA
Cell Division Study Guide U3.2.3: Eukaryotic chromosomes are linear DNA molecules associated with histone proteins. (Oxford Biology Course Companion page 151). 1. Describe the structure of eukaryotic DNA
Bacterial cell. Origin of replication. Septum
 Bacterial cell Bacterial chromosome: Double-stranded DNA Origin of replication Septum 1 2 3 Chromosome Rosettes of Chromatin Loops Scaffold protein Chromatin Loop Solenoid Scaffold protein Chromatin loop
Bacterial cell Bacterial chromosome: Double-stranded DNA Origin of replication Septum 1 2 3 Chromosome Rosettes of Chromatin Loops Scaffold protein Chromatin Loop Solenoid Scaffold protein Chromatin loop
Origin of replication. Septum
 Bacterial cell Bacterial chromosome: Double-stranded DNA Origin of replication Septum 1 2 3 Chromosome Rosettes of Chromatin Loops Chromatin Loop Solenoid Scaffold protein Scaffold protein Chromatin loop
Bacterial cell Bacterial chromosome: Double-stranded DNA Origin of replication Septum 1 2 3 Chromosome Rosettes of Chromatin Loops Chromatin Loop Solenoid Scaffold protein Scaffold protein Chromatin loop
Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v)
 SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2566 Figure S1 CDKL5 protein expression pattern and localization in mouse brain. (a) Multiple-tissue western blot from a postnatal day (P) 21 mouse probed with an antibody against CDKL5.
DOI: 10.1038/ncb2566 Figure S1 CDKL5 protein expression pattern and localization in mouse brain. (a) Multiple-tissue western blot from a postnatal day (P) 21 mouse probed with an antibody against CDKL5.
Lab title: Cell Division author: Dr. Ruth Dahlquist-Willard (modified by D. Bell)
 Corresponding Readings: Lab title: Cell Division author: Dr. Ruth Dahlquist-Willard (modified by D. Bell) Campbell Ch. 8 BIOL-100L Safety Information: We will be using laboratory glassware such as microscope
Corresponding Readings: Lab title: Cell Division author: Dr. Ruth Dahlquist-Willard (modified by D. Bell) Campbell Ch. 8 BIOL-100L Safety Information: We will be using laboratory glassware such as microscope
Unduplicated. Chromosomes. Telophase
 10-2 Cell Division The Cell Cycle Interphase Mitosis Prophase Cytokinesis G 1 S G 2 Chromatin in Parent Nucleus & Daughter Cells Chromatin Daughter Nuclei Telophase Mitotic Anaphase Metaphase Use what
10-2 Cell Division The Cell Cycle Interphase Mitosis Prophase Cytokinesis G 1 S G 2 Chromatin in Parent Nucleus & Daughter Cells Chromatin Daughter Nuclei Telophase Mitotic Anaphase Metaphase Use what
Nature Structural and Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Mutational analysis of the SA2-Scc1 interaction in vitro and in human cells. (a) Autoradiograph (top) and Coomassie stained gel (bottom) of 35 S-labeled Myc-SA2 proteins (input)
Supplementary Figure 1 Mutational analysis of the SA2-Scc1 interaction in vitro and in human cells. (a) Autoradiograph (top) and Coomassie stained gel (bottom) of 35 S-labeled Myc-SA2 proteins (input)
Biology is the only subject in which multiplication is the same thing as division
 Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division Ch. 10 Where it all began You started as a cell smaller than a period
Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division Ch. 10 Where it all began You started as a cell smaller than a period
General Biology. Overview: The Key Roles of Cell Division The continuity of life is based upon the reproduction of cells, or cell division
 General Biology Course No: BNG2003" Credits: 3.00 " " " 8. The Cell Cycle Prof. Dr. Klaus Heese Overview: The Key Roles of Cell Division The continuity of life is based upon the reproduction of cells,
General Biology Course No: BNG2003" Credits: 3.00 " " " 8. The Cell Cycle Prof. Dr. Klaus Heese Overview: The Key Roles of Cell Division The continuity of life is based upon the reproduction of cells,
CELL CYCLE INTRODUCTION PART I ANIMAL CELL CYCLE INTERPHASE
 CELL CYCLE INTRODUCTION The nuclei in cells of eukaryotic organisms contain chromosomes with clusters of genes, discrete units of hereditary information consisting of double-stranded DNA. Structural proteins
CELL CYCLE INTRODUCTION The nuclei in cells of eukaryotic organisms contain chromosomes with clusters of genes, discrete units of hereditary information consisting of double-stranded DNA. Structural proteins
General Biology. Overview: The Key Roles of Cell Division. Unicellular organisms
 General Biology Course No: BNG2003 Credits: 3.00 8. The Cell Cycle Prof. Dr. Klaus Heese Overview: The Key Roles of Cell Division The continuity of life is based upon the reproduction of cells, or cell
General Biology Course No: BNG2003 Credits: 3.00 8. The Cell Cycle Prof. Dr. Klaus Heese Overview: The Key Roles of Cell Division The continuity of life is based upon the reproduction of cells, or cell
Biology 4A Laboratory MITOSIS Asexual Reproduction OBJECTIVE
 Biology 4A Laboratory MITOSIS Asexual Reproduction OBJECTIVE To study the cell cycle and understand how, when and why cells divide. To study and identify the major stages of cell division. To relate the
Biology 4A Laboratory MITOSIS Asexual Reproduction OBJECTIVE To study the cell cycle and understand how, when and why cells divide. To study and identify the major stages of cell division. To relate the
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Choi YL, Soda M, Yamashita Y, et al. EML4-ALK mutations in
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Choi YL, Soda M, Yamashita Y, et al. EML4-ALK mutations in
Supplemental Materials. STK16 regulates actin dynamics to control Golgi organization and cell cycle
 Supplemental Materials STK16 regulates actin dynamics to control Golgi organization and cell cycle Juanjuan Liu 1,2,3, Xingxing Yang 1,3, Binhua Li 1, Junjun Wang 1,2, Wenchao Wang 1, Jing Liu 1, Qingsong
Supplemental Materials STK16 regulates actin dynamics to control Golgi organization and cell cycle Juanjuan Liu 1,2,3, Xingxing Yang 1,3, Binhua Li 1, Junjun Wang 1,2, Wenchao Wang 1, Jing Liu 1, Qingsong
Cell Division Questions. Mitosis and Meiosis
 Cell Division Questions Mitosis and Meiosis 1 10 Do not write outside the box 5 Figure 3 shows a pair of chromosomes at the start of meiosis. The letters represent alleles. Figure 3 E E e e F F f f 5 (a)
Cell Division Questions Mitosis and Meiosis 1 10 Do not write outside the box 5 Figure 3 shows a pair of chromosomes at the start of meiosis. The letters represent alleles. Figure 3 E E e e F F f f 5 (a)
The Cell Cycle and How Cells Divide
 The Cell Cycle and How Cells Divide 1 Phases of the Cell Cycle The cell cycle consists of Interphase normal cell activity The mitotic phase cell divsion INTERPHASE Growth G 1 (DNA synthesis) Growth G 2
The Cell Cycle and How Cells Divide 1 Phases of the Cell Cycle The cell cycle consists of Interphase normal cell activity The mitotic phase cell divsion INTERPHASE Growth G 1 (DNA synthesis) Growth G 2
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS)
 Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary information. The Light Intermediate Chain 2 Subpopulation of Dynein Regulates Mitotic. Spindle Orientation
 Supplementary information The Light Intermediate Chain 2 Subpopulation of Dynein Regulates Mitotic Spindle Orientation Running title: Dynein LICs distribute mitotic functions. Sagar Mahale a, d, *, Megha
Supplementary information The Light Intermediate Chain 2 Subpopulation of Dynein Regulates Mitotic Spindle Orientation Running title: Dynein LICs distribute mitotic functions. Sagar Mahale a, d, *, Megha
SUPPLEMENTARY INFORMATION
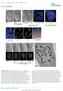 DOI: 0.038/ncb33 a b c 0 min 6 min 7 min (fixed) DIC -GFP, CenpF 3 µm Nocodazole Single optical plane -GFP, CenpF Max. intensity projection d µm -GFP, CenpF, -GFP CenpF 3-D rendering e f 0 min 4 min 0
DOI: 0.038/ncb33 a b c 0 min 6 min 7 min (fixed) DIC -GFP, CenpF 3 µm Nocodazole Single optical plane -GFP, CenpF Max. intensity projection d µm -GFP, CenpF, -GFP CenpF 3-D rendering e f 0 min 4 min 0
Cell Division (Mitosis)
 Cell Division (Mitosis) Chromosomes The essential part of a chromosome is a single very long strand of DNA. This DNA contains all the genetic information for creating and running the organism. Each chromosome
Cell Division (Mitosis) Chromosomes The essential part of a chromosome is a single very long strand of DNA. This DNA contains all the genetic information for creating and running the organism. Each chromosome
Name. A.P. Biology Chapter 12 The Cell Cycle
 A.P. Biology Chapter 12 The Cell Cycle Name Living species MUST possess the ability to r if they are to flourish. The Cell Cycle follows the life of a cell from its o until its d. The Key Roles Of Cell
A.P. Biology Chapter 12 The Cell Cycle Name Living species MUST possess the ability to r if they are to flourish. The Cell Cycle follows the life of a cell from its o until its d. The Key Roles Of Cell
Cell Cycle and Cell Division
 122 Cell Cycle and Cell Division 1. Meiosis I is reductional division. Meiosis II is equational division due to [1988] (a) pairing of homologous chromosomes (b) crossing over (c) separation of chromatids
122 Cell Cycle and Cell Division 1. Meiosis I is reductional division. Meiosis II is equational division due to [1988] (a) pairing of homologous chromosomes (b) crossing over (c) separation of chromatids
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Chen et al., http://www.jcb.org/cgi/content/full/jcb.201210119/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Lack of fast reversibility of UVR8 dissociation. (A) HEK293T
Supplemental material Chen et al., http://www.jcb.org/cgi/content/full/jcb.201210119/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Lack of fast reversibility of UVR8 dissociation. (A) HEK293T
Effects of UBL5 knockdown on cell cycle distribution and sister chromatid cohesion
 Supplementary Figure S1. Effects of UBL5 knockdown on cell cycle distribution and sister chromatid cohesion A. Representative examples of flow cytometry profiles of HeLa cells transfected with indicated
Supplementary Figure S1. Effects of UBL5 knockdown on cell cycle distribution and sister chromatid cohesion A. Representative examples of flow cytometry profiles of HeLa cells transfected with indicated
Name: Date: Block: 10-2 Cell Division Worksheet
 10-2 Cell Division Worksheet W hat do you think would happen if a cell were simple to split into two, without any advance preparation? Would each daughter cell have everything it needed to survive? Because
10-2 Cell Division Worksheet W hat do you think would happen if a cell were simple to split into two, without any advance preparation? Would each daughter cell have everything it needed to survive? Because
Chapter 3. Expression of α5-megfp in Mouse Cortical Neurons. on the β subunit. Signal sequences in the M3-M4 loop of β nachrs bind protein factors to
 22 Chapter 3 Expression of α5-megfp in Mouse Cortical Neurons Subcellular localization of the neuronal nachr subtypes α4β2 and α4β4 depends on the β subunit. Signal sequences in the M3-M4 loop of β nachrs
22 Chapter 3 Expression of α5-megfp in Mouse Cortical Neurons Subcellular localization of the neuronal nachr subtypes α4β2 and α4β4 depends on the β subunit. Signal sequences in the M3-M4 loop of β nachrs
Stages of Mitosis. Introduction
 Name: Due: Stages of Mitosis Introduction Mitosis, also called karyokinesis, is division of the nucleus and its chromosomes. It is followed by division of the cytoplasm known as cytokinesis. Both mitosis
Name: Due: Stages of Mitosis Introduction Mitosis, also called karyokinesis, is division of the nucleus and its chromosomes. It is followed by division of the cytoplasm known as cytokinesis. Both mitosis
The Cell Cycle. Dr. SARRAY Sameh, Ph.D
 The Cell Cycle Dr. SARRAY Sameh, Ph.D Overview When an organism requires additional cells (either for growth or replacement of lost cells), new cells are produced by cell division (mitosis) Somatic cells
The Cell Cycle Dr. SARRAY Sameh, Ph.D Overview When an organism requires additional cells (either for growth or replacement of lost cells), new cells are produced by cell division (mitosis) Somatic cells
The Process of Cell Division
 Lesson Overview 10.2 The Process of Cell Division THINK ABOUT IT What role does cell division play in your life? Does cell division stop when you are finished growing? Chromosomes What is the role of chromosomes
Lesson Overview 10.2 The Process of Cell Division THINK ABOUT IT What role does cell division play in your life? Does cell division stop when you are finished growing? Chromosomes What is the role of chromosomes
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells
 MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Posch et al., http://www.jcb.org/cgi/content/full/jcb.200912046/dc1 Figure S1. Biochemical characterization of the interaction between
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Posch et al., http://www.jcb.org/cgi/content/full/jcb.200912046/dc1 Figure S1. Biochemical characterization of the interaction between
Fig. 1. Simple columnar epithelial cells lining the small intestine.
 Mitosis Prelab Reading Fig. 1. Simple columnar epithelial cells lining the small intestine. The tall cells pictured in Fig. 1 form the lining of the small intestine in humans and other animals. These cells
Mitosis Prelab Reading Fig. 1. Simple columnar epithelial cells lining the small intestine. The tall cells pictured in Fig. 1 form the lining of the small intestine in humans and other animals. These cells
10-2 Cell Division. Copyright Pearson Prentice Hall
 10-2 Cell Division Copyright Pearson Prentice Hall Cell Growth and Division In multicellular organisms, cell division makes new cells To replace old or damaged ones So organisms can grow In single-celled
10-2 Cell Division Copyright Pearson Prentice Hall Cell Growth and Division In multicellular organisms, cell division makes new cells To replace old or damaged ones So organisms can grow In single-celled
CELL CYCLE INTRODUCTION PART I ANIMAL CELL CYCLE INTERPHASE EVOLUTION/HEREDITY UNIT. Activity #3
 AP BIOLOGY EVOLUTION/HEREDITY UNIT Unit 1 Part 3 Chapter 12 Activity #3 INTRODUCTION CELL CYCLE NAME DATE PERIOD The nuclei in cells of eukaryotic organisms contain chromosomes with clusters of genes,
AP BIOLOGY EVOLUTION/HEREDITY UNIT Unit 1 Part 3 Chapter 12 Activity #3 INTRODUCTION CELL CYCLE NAME DATE PERIOD The nuclei in cells of eukaryotic organisms contain chromosomes with clusters of genes,
EPI TIR-FM min
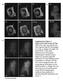 a 15 b EPI 45 75 min TIR-FM c min Supplementary figure 1. Fluorescence microscopy of Gag- GFP. HeLa cells were transfected with Gag and Gag-GFP and imaged at 5-6 hpt. a, Images of a single cell observed
a 15 b EPI 45 75 min TIR-FM c min Supplementary figure 1. Fluorescence microscopy of Gag- GFP. HeLa cells were transfected with Gag and Gag-GFP and imaged at 5-6 hpt. a, Images of a single cell observed
Cell Growth and Division
 Name Class Date 10 Cell Growth and Division Big idea Growth, Development, and Reproduction Q: How does a cell produce a new cell? WHAT I KNOW WHAT I LEARNED 10.1 Why do cells divide? 10.2 How do cells
Name Class Date 10 Cell Growth and Division Big idea Growth, Development, and Reproduction Q: How does a cell produce a new cell? WHAT I KNOW WHAT I LEARNED 10.1 Why do cells divide? 10.2 How do cells
Chapter 8: Cellular Reproduction
 Chapter 8: Cellular Reproduction 1. The Cell Cycle 2. Mitosis 3. Meiosis 2 Types of Cell Division 2n 1n Mitosis: occurs in somatic cells (almost all cells of the body) generates cells identical to original
Chapter 8: Cellular Reproduction 1. The Cell Cycle 2. Mitosis 3. Meiosis 2 Types of Cell Division 2n 1n Mitosis: occurs in somatic cells (almost all cells of the body) generates cells identical to original
(i) List these events in the correct order, starting with D.... (1)... (1)... (1)
 Q1. (a) Boxes A to E show some of the events of the cell cycle. A Chromatids seperate B Nuclear envelopes disappears C Cytoplasm divides D Chromosomes condense and become visible E Chromosomes on the equator
Q1. (a) Boxes A to E show some of the events of the cell cycle. A Chromatids seperate B Nuclear envelopes disappears C Cytoplasm divides D Chromosomes condense and become visible E Chromosomes on the equator
Cell Cycle and Mitosis
 Cell Cycle and Mitosis Name Period A# THE CELL CYCLE The cell cycle, or cell-division cycle, is the series of events that take place in a eukaryotic cell between its formation and the moment it replicates
Cell Cycle and Mitosis Name Period A# THE CELL CYCLE The cell cycle, or cell-division cycle, is the series of events that take place in a eukaryotic cell between its formation and the moment it replicates
Supplementary Information for. Shi and King, Chromosome Nondisjunction Yields Tetraploid Rather than Aneuploid Cells in Human Cell Lines.
 Supplementary Information for Shi and King, Chromosome Nondisjunction Yields Tetraploid Rather than Aneuploid Cells in Human Cell Lines Contains Supplementary Methods Supplementary Figures 1-7 Supplementary
Supplementary Information for Shi and King, Chromosome Nondisjunction Yields Tetraploid Rather than Aneuploid Cells in Human Cell Lines Contains Supplementary Methods Supplementary Figures 1-7 Supplementary
10.2 The Cell Cycle *
 OpenStax-CNX module: m52672 1 10.2 The Cell Cycle * Shannon McDermott Based on The Cell Cycle by OpenStax This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution License
OpenStax-CNX module: m52672 1 10.2 The Cell Cycle * Shannon McDermott Based on The Cell Cycle by OpenStax This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution License
Biology is the only subject in which multiplication is the same thing as division
 Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division 2007-2008 Where it all began You started as a cell smaller than a
Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division 2007-2008 Where it all began You started as a cell smaller than a
APGRU4L1 Chap 12 Extra Reading Cell Cycle and Mitosis
 APGRU4L1 Chap 12 Extra Reading Cell Cycle and Mitosis Dr. Ramesh Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division 2007-2008
APGRU4L1 Chap 12 Extra Reading Cell Cycle and Mitosis Dr. Ramesh Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division 2007-2008
3 The abbreviations used are: NE, nuclear envelope; LB, lamin B; LA, lamin A; Ig,
 crossmark THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 290, NO. 46, pp. 27557 27571, November 13, 2015 2015 by The American Society for Biochemistry and Molecular Biology, Inc. Published in the U.S.A. Concentration-dependent
crossmark THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 290, NO. 46, pp. 27557 27571, November 13, 2015 2015 by The American Society for Biochemistry and Molecular Biology, Inc. Published in the U.S.A. Concentration-dependent
Sestrin2 and BNIP3 (Bcl-2/adenovirus E1B 19kDa-interacting. protein3) regulate autophagy and mitophagy in renal tubular cells in. acute kidney injury
 Sestrin2 and BNIP3 (Bcl-2/adenovirus E1B 19kDa-interacting protein3) regulate autophagy and mitophagy in renal tubular cells in acute kidney injury by Masayuki Ishihara 1, Madoka Urushido 2, Kazu Hamada
Sestrin2 and BNIP3 (Bcl-2/adenovirus E1B 19kDa-interacting protein3) regulate autophagy and mitophagy in renal tubular cells in acute kidney injury by Masayuki Ishihara 1, Madoka Urushido 2, Kazu Hamada
SUPPLEMENTARY LEGENDS...
 TABLE OF CONTENTS SUPPLEMENTARY LEGENDS... 2 11 MOVIE S1... 2 FIGURE S1 LEGEND... 3 FIGURE S2 LEGEND... 4 FIGURE S3 LEGEND... 5 FIGURE S4 LEGEND... 6 FIGURE S5 LEGEND... 7 FIGURE S6 LEGEND... 8 FIGURE
TABLE OF CONTENTS SUPPLEMENTARY LEGENDS... 2 11 MOVIE S1... 2 FIGURE S1 LEGEND... 3 FIGURE S2 LEGEND... 4 FIGURE S3 LEGEND... 5 FIGURE S4 LEGEND... 6 FIGURE S5 LEGEND... 7 FIGURE S6 LEGEND... 8 FIGURE
The Cell Cycle. Packet #9. Thursday, August 20, 2015
 1 The Cell Cycle Packet #9 2 Introduction Cell Cycle An ordered sequence of events in the life of a dividing eukaryotic cell and is a cellular asexual reproduction. The contents of the parent s cell nucleus
1 The Cell Cycle Packet #9 2 Introduction Cell Cycle An ordered sequence of events in the life of a dividing eukaryotic cell and is a cellular asexual reproduction. The contents of the parent s cell nucleus
Mitosis Notes AP Biology Mrs. Laux
 I. Cell Cycle-includes interphase and mitosis (IPPMAT) A. Interphase 1. accounts for 90% of the cycle 2. cell grows and copies its chromosomes in preparation for cell division 3. produces proteins and
I. Cell Cycle-includes interphase and mitosis (IPPMAT) A. Interphase 1. accounts for 90% of the cycle 2. cell grows and copies its chromosomes in preparation for cell division 3. produces proteins and
The clathrin adaptor Numb regulates intestinal cholesterol. absorption through dynamic interaction with NPC1L1
 The clathrin adaptor Numb regulates intestinal cholesterol absorption through dynamic interaction with NPC1L1 Pei-Shan Li 1, Zhen-Yan Fu 1,2, Ying-Yu Zhang 1, Jin-Hui Zhang 1, Chen-Qi Xu 1, Yi-Tong Ma
The clathrin adaptor Numb regulates intestinal cholesterol absorption through dynamic interaction with NPC1L1 Pei-Shan Li 1, Zhen-Yan Fu 1,2, Ying-Yu Zhang 1, Jin-Hui Zhang 1, Chen-Qi Xu 1, Yi-Tong Ma
The Cell Cycle CHAPTER 12
 The Cell Cycle CHAPTER 12 The Key Roles of Cell Division cell division = reproduction of cells All cells come from pre-exisiting cells Omnis cellula e cellula Unicellular organisms division of 1 cell reproduces
The Cell Cycle CHAPTER 12 The Key Roles of Cell Division cell division = reproduction of cells All cells come from pre-exisiting cells Omnis cellula e cellula Unicellular organisms division of 1 cell reproduces
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/7/334/rs4/dc1 Supplementary Materials for Rapidly rendering cells phagocytic through a cell surface display technique and concurrent Rac activation Hiroki Onuma,
www.sciencesignaling.org/cgi/content/full/7/334/rs4/dc1 Supplementary Materials for Rapidly rendering cells phagocytic through a cell surface display technique and concurrent Rac activation Hiroki Onuma,
The Cell Cycle CAMPBELL BIOLOGY IN FOCUS SECOND EDITION URRY CAIN WASSERMAN MINORSKY REECE
 CAMPBELL BIOLOGY IN FOCUS URRY CAIN WASSERMAN MINORSKY REECE 9 The Cell Cycle Lecture Presentations by Kathleen Fitzpatrick and Nicole Tunbridge, Simon Fraser University SECOND EDITION Overview: The Key
CAMPBELL BIOLOGY IN FOCUS URRY CAIN WASSERMAN MINORSKY REECE 9 The Cell Cycle Lecture Presentations by Kathleen Fitzpatrick and Nicole Tunbridge, Simon Fraser University SECOND EDITION Overview: The Key
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Dunsch et al., http://www.jcb.org/cgi/content/full/jcb.201202112/dc1 Figure S1. Characterization of HMMR and CHICA antibodies. (A) HeLa
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Dunsch et al., http://www.jcb.org/cgi/content/full/jcb.201202112/dc1 Figure S1. Characterization of HMMR and CHICA antibodies. (A) HeLa
Supplementary table 1
 Supplementary table 1 S. pombe strain list Fig. 1A JX38 h + ade6-m216 nda3-km311 PX476 PW775 PX545 PX546 h- ade6-m216 sgo2::ura4 + nda3-km311 h 9 mad2::ura4 + nda3-km311 h + ade6-m21 nda3-km311 rad21 +
Supplementary table 1 S. pombe strain list Fig. 1A JX38 h + ade6-m216 nda3-km311 PX476 PW775 PX545 PX546 h- ade6-m216 sgo2::ura4 + nda3-km311 h 9 mad2::ura4 + nda3-km311 h + ade6-m21 nda3-km311 rad21 +
Supplementary Material
 Supplementary Material Nuclear import of purified HIV-1 Integrase. Integrase remains associated to the RTC throughout the infection process until provirus integration occurs and is therefore one likely
Supplementary Material Nuclear import of purified HIV-1 Integrase. Integrase remains associated to the RTC throughout the infection process until provirus integration occurs and is therefore one likely
Cell Cycle Phase. Interphase (G 1, S, G 2 ) Mitotic Phase (M phase) Prophase. Metaphase. Anaphase. Telophase
 Part I: The Cell Cycle Use your resources at hand and the Explore Student Guide to outline what occurs within the cell during each stage of the cell cycle. Record this information in Table 1 below. Cell
Part I: The Cell Cycle Use your resources at hand and the Explore Student Guide to outline what occurs within the cell during each stage of the cell cycle. Record this information in Table 1 below. Cell
BIOLOGY LTF DIAGNOSTIC TEST CELL CYCLE & MITOSIS
 Biology Multiple Choice 016044 BIOLOGY LTF DIAGNOSTIC TEST CELL CYCLE & MITOSIS TEST CODE: 016044 Directions: Each of the questions or incomplete statements below is followed by five suggested answers
Biology Multiple Choice 016044 BIOLOGY LTF DIAGNOSTIC TEST CELL CYCLE & MITOSIS TEST CODE: 016044 Directions: Each of the questions or incomplete statements below is followed by five suggested answers
SUPPLEMENTARY INFORMATION
 SUPPLEENTRY INFORTION DOI: 1.138/ncb2577 Early Telophase Late Telophase B icrotubules within the ICB (percent of total cells in telophase) D G ultinucleate cells (% total) 8 6 4 2 2 15 1 5 T without gaps
SUPPLEENTRY INFORTION DOI: 1.138/ncb2577 Early Telophase Late Telophase B icrotubules within the ICB (percent of total cells in telophase) D G ultinucleate cells (% total) 8 6 4 2 2 15 1 5 T without gaps
Biology is the only subject in which multiplication is the same thing as division
 Biology is the only subject in which multiplication is the same thing as division The Cell Cycle: Cell Growth, Cell Division 2007-2008 2007-2008 Getting from there to here Going from egg to baby. the original
Biology is the only subject in which multiplication is the same thing as division The Cell Cycle: Cell Growth, Cell Division 2007-2008 2007-2008 Getting from there to here Going from egg to baby. the original
Mitosis and Cytokinesis
 B-2.6 Summarize the characteristics of the cell cycle: interphase (called G1, S, G2); the phases of mitosis (called prophase, metaphase, anaphase, and telophase); and plant and animal cytokinesis. The
B-2.6 Summarize the characteristics of the cell cycle: interphase (called G1, S, G2); the phases of mitosis (called prophase, metaphase, anaphase, and telophase); and plant and animal cytokinesis. The
Supplementary Information for. A cancer-associated BRCA2 mutation reveals masked nuclear. export signals controlling localization
 Supplementary Information for A cancer-associated BRCA2 mutation reveals masked nuclear export signals controlling localization Anand D Jeyasekharan 1, Yang Liu 1, Hiroyoshi Hattori 1,3, Venkat Pisupati
Supplementary Information for A cancer-associated BRCA2 mutation reveals masked nuclear export signals controlling localization Anand D Jeyasekharan 1, Yang Liu 1, Hiroyoshi Hattori 1,3, Venkat Pisupati
The cell cycle dependent mislocalisation of emerin may contribute to the Emery-Dreifuss muscular dystrophy phenotype
 Research Article 341 The cell cycle dependent mislocalisation of emerin may contribute to the Emery-Dreifuss muscular dystrophy phenotype Elizabeth A. L. Fairley 1, *, Andrew Riddell 2, Juliet A. Ellis
Research Article 341 The cell cycle dependent mislocalisation of emerin may contribute to the Emery-Dreifuss muscular dystrophy phenotype Elizabeth A. L. Fairley 1, *, Andrew Riddell 2, Juliet A. Ellis
HEK293FT cells were transiently transfected with reporters, N3-ICD construct and
 Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
Ploidy and Human Cell Types. Cell Cycle and Mitosis. DNA and Chromosomes. Where It All Began 11/19/2014. Chapter 12 Pg
 Ploidy and Human Cell Types Cell Cycle and Mitosis Chapter 12 Pg. 228 245 Cell Types Somatic cells (body cells) have 46 chromosomes, which is the diploid chromosome number. A diploid cell is a cell with
Ploidy and Human Cell Types Cell Cycle and Mitosis Chapter 12 Pg. 228 245 Cell Types Somatic cells (body cells) have 46 chromosomes, which is the diploid chromosome number. A diploid cell is a cell with
