Structural Characterization of Prion-like Conformational Changes of the Neuronal Isoform of Aplysia CPEB
|
|
|
- Allison Harmon
- 5 years ago
- Views:
Transcription
1 Structural Characterization of Prion-like Conformational Changes of the Neuronal Isoform of Aplysia CPEB Bindu L. Raveendra, 1,5 Ansgar B. Siemer, 2,6 Sathyanarayanan V. Puthanveettil, 1,3,7 Wayne A. Hendrickson, 3,4 Eric R. Kandel, 1,3,4 Ann E. McDermott. 2 Supplementary Figures a b c d Supplementary Figure 1. Soluble form of Aplysia CPEB exists as oligomers in the native state. (a) Calibration curve for the superdex 200 column using 4 standards, (High Molecular weight Calibration Kit, GE, Cat No ). The X-axis shows the log (molecular weight) and Y-axis is Kav (gel phase distribution coefficient). Kav = (Ve-V0)/(Vc-V0). Where Ve: elution volume, V0: void volume (43.08 ml) and Vc: total column volume (120 ml). (b) Estimation of apparent molecular weight of the
2 fractions (number 22, 31 and 32 from Fig. 2b and Supplementary Fig. 1c) as interpreted based on the molecular weight calibration curve. Two preparations of Aplysia CPEB (from Aplysia central nervous system and E.coli) have similar elution times and therefore should have a similar apparent molar mass. Monomer mass of endogenous Aplysia CPEB is 78.2 kda and recombinant protein is 83 kda. (c) Western analysis of native E.coli extract (native lysis) after fractionation by SEC. The different fractions obtained after SEC are numbered sequentially from Aplysia CPEB in the soluble form in E.coli extract has similar pattern of molar mass distribution as of the Aplysia neuronal extract. Both of them showed mainly two populations: a small percentage of total population (< 20%) with a molecular weight corresponding to a dimer to tetramer ( kda) and a larger percentage (>80%) as aggregated high molecular weight population (> 10-mer, >720 kda at fraction 22). Purified recombinant Aplysia CPEB in the soluble form also showed similar behavior in SEC and Western analysis. (d) Control experiment using kinesin heavy chain antibody. Western analysis of native CNS extract of Aplysia after fractionation by SEC. The protein kinesin heavy chain is analyzed in a similar way as for CPEB. The data for kinesin heavy chain showed that in the soluble form it exists mainly in two populations. The first fraction has an apparent molar mass corresponding to a dimer and the second fraction has very high apparent molecular weight indicating that kinesin might also exists as a homo oligomer or a high molecular weight hetero oligomeric protein complex. Kinesin heavy chain known to exists as dimer and high molecular weight protein complexes, which is in agreement with the data we obtained from this experiment.
3 Supplementary Figure 2: Analysis of purified soluble Aplysia CPEB by size exclusion chromatography light scattering (SEC-LS). (a) Purified recombinant full length Aplysia CPEB in PBS with 2 M urea was analyzed by size exclusion chromatography followed by light scattering. SEC is used as a fractionation column. The solid line indicates the trace from the UV detector and dotted line is the signal from LS detector at 90º. The dots are the weight average molecular weight for each slice measured every second. The molecular weight is calculated from light scattering signal and is shown on the right side Y-axis. The oligomeric state has an average radius of gyration (Rg) around 25 nm. (b) The hydrodynamic radius (Rh) distribution of the purified Aplysia CPEB full length protein in PBS in the presence of 2 M urea. The dotted line shows the Rh distribution derived from the light scattering signal. The right side of the Y-axis is the molecular weight of the oligomer calculated from light scattering experiment. The Rh value for the oligomeric fraction of Aplysia CPEB is found similar to the observed radius of gyration (Rg), around 25 nm, suggesting a hollow spherical shape for these oligomers. (c) Comparison of SEC-LS analysis of purified recombinant full length Aplysia CPEB in PBS buffer with 2 M urea (blue) and 4 M urea (red). The solid line indicates the trace from the UV detector and dotted line from LS detector at 90 degree. Purified full length Aplysia CPEB in PBS with 4 M urea eluted at a lower elution volume (5 6.5 ml) in the SEC indicating a high apparent molecular weight. But from the light scattering data, this fraction has an average molecular weight of ~ kda equal to the mass of the monomer to the trimer. Despite the fraction showed a very low elution volume in SEC (5 6.5 ml) compared to the protein in PBS with 2 M
4 urea buffer, it is having a lower molecular weight by light scattering data. The larger size observed for this molecules might be due to the extended conformation resulting from the partial denaturing of the protein under this condition.
5 Supplementary Figure 3. Solid-state NMR of fiber form of full length Aplysia CPEB and isolated prion domain. (a) 13 C- 13 C DARR spectrum of full-length Aplysia CPEB recorded at a 1 H frequency of 900 MHz, 20 khz MAS, and mixing time of 50 ms. The assignment of some of the amino acid types is indicated. The average Cα-Cβ cross peak positions corresponding to an α-helical (red), extended β-sheet (orange), and random coil (blue) conformation for the amino acids Thr, Ser, Gln, and Ala, as well as the corresponding Cα-CO cross peaks for Thr and Gln are marked with ellipsoids. The average chemical shifts as well as the standard deviation of these shifts (corresponding to the width of the ellipsoids) were taken from Wang & Jardezky. 48 The full 13 C connectivity of Gln in α-helical and β-sheet conformation is shown with red and green bars, respectively. As illustrated in Fig. 7, this spectrum is very similar to the 2D DARR spectrum of the isolated prion domain of Aplysia CPEB. (b) Assignment of Gln 13 C shifts. A detail of the 13 C- 13 C DARR spectrum of Figure 7a is shown in red (CP DARR). The equivalent spectrum recorded with a direct 13 C excitation instead of 1 H- 13 C CP to create the initial polarization is shown in blue (DE DARR). The contours were adjusted in a way that only the most intense cross peaks (i.e. the cross peaks of Gln, the most abundant amino acid of the PRD) are visible. Both spectra show the entire 13 C spin system of Gln. However, the CP DARR (red) gave a higher intensity for the Gln in a β-sheet conformation (solid-lines) and the DE DARR (blue) for Gln in a random-coil and α-helical conformation (dashed lines). The figure illustrates that the high abundance of Gln (~40%) in the PRD and the very intense cross peaks resulting from that make it is possible to unambiguously assign the Gln 13 C shifts in our 2D DARR spectra.
6
7 d e 28 KDa! PRD 10min 30min! 7 KDa! 15 f Supplementary Figure 4: Proteolysis experiments on Aplysia CPEB ( a) Mass spectrometric data of the trypsin digested full length Aplysia CPEB fibers. The red color shows the sequence of the fragment obtained by mass spec analysis after digestion of full length Aplysia CPEB fibers by trypsin. (b) Mass spectrometric analysis of full length Aplysia CPEB in the soluble form. The red segments show the sequence of the fragments obtained by mass spec analysis after trypsin digestion. (c) Theoretical prediction of trypsin digestion sites for full length Aplysia CPEB using the program Expassy Peptide cutter. The trypsin cleavable sites are shown in red colored letters. (d) Coomassie stained PAGE gel showing a 20 kda trypsin resistant fragment obtained from full length Aplysia CPEB fibers after digestion. The presence high molecular weight bands (>20 KDa) in the gel indicates that the fragment might be very prone to aggregation resulting in high molecular weight oligomers. (e) Proteolysis (proteinase K) of the isolated prion domain (PRD) and full length Aplysia CPEB fibers prepared from purified recombinant protein. A 7 kda protease resistant fragment is present in both samples after the proteolysis. The presence of the same band from both full length Aplysia CPEB and PRD indicates that the fragment comes from the PRD of the protein. The identification of band by Mass spectrometry and N-terminal sequencing was unsuccessful. (f) Amino acid sequence of a protease- fragment cleaved from the full length Aplysia CPEB fibers. This sequence has four potential resistant trypsin cleavage sites (red) that are not cut by the enzyme when Aplysia CPEB is in the fibrillar form. This fragment is part of the PRD of the neuronal isoform of Aplysia CPEB and, therefore, not present in
8 the developmental isoform. In contrast, the soluble form of full length Aplysia CPEB does not exhibit such pronounced protease resistance.
9 Supplementary Figure 5: Comparison of combined MS scans from soluble and insoluble forms of Aplysia CPEB after Trypsin digestion. Scans across the identical time range for both analyses were combined using the combine function of the MassLynx 4.1 software and compared. (a,b) Combined scans from soluble and insoluble forms, respectively, showing the 3+ ion of peptide (EQLQQQQLQLQQQLQQQLQHIQK) present in the soluble form only. (c,d) Combined scans from the soluble and insoluble forms, respectively, showing the 3+ion of peptide (QQLQQQQQQEQLQQQQLQLQQQLQQQLQHIQK) present in the soluble form only. (e,f) combined scans from the soluble and insoluble forms, respectively, showing the 2+ion of peptide (EPSSHTYTPGSPELQSVLNYANVPLSK) present in the soluble form only.
10 Supplementary Table 1 (NMR): Comparison of the chemical shifts extracted from the 1 H- 13 C HETCOR spectrum shown in Fig. 5c to the average chemical shifts of α-helical, β-sheet, and random coil secondary structure elements reported by Wang & Jardetzky 1 (the standard deviation is given in parenthesis). This analysis shows that the chemical shifts of the dynamic parts detected in Aplysia CPEB fibers are in agreement with random-coil values or, in some cases, α-helical values. 1. Wang, Y. & Jardetzky, O. Probability-based protein secondary structure identification using combined NMR chemical-shift data. Protein science : a publication of the Protein Society 11, , (2002).
Supplementary Figure 1 Preparation, crystallization and structure determination of EpEX. (a), Purified EpEX and EpEX analyzed on homogenous 12.
 Supplementary Figure 1 Preparation, crystallization and structure determination of EpEX. (a), Purified EpEX and EpEX analyzed on homogenous 12.5 % SDS-PAGE gel under reducing and non-reducing conditions.
Supplementary Figure 1 Preparation, crystallization and structure determination of EpEX. (a), Purified EpEX and EpEX analyzed on homogenous 12.5 % SDS-PAGE gel under reducing and non-reducing conditions.
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/2/4/e1500980/dc1 Supplementary Materials for The crystal structure of human dopamine -hydroxylase at 2.9 Å resolution Trine V. Vendelboe, Pernille Harris, Yuguang
advances.sciencemag.org/cgi/content/full/2/4/e1500980/dc1 Supplementary Materials for The crystal structure of human dopamine -hydroxylase at 2.9 Å resolution Trine V. Vendelboe, Pernille Harris, Yuguang
Double charge of 33kD peak A1 A2 B1 B2 M2+ M/z. ABRF Proteomics Research Group - Qualitative Proteomics Study Identifier Number 14146
 Abstract The 2008 ABRF Proteomics Research Group Study offers participants the chance to participate in an anonymous study to identify qualitative differences between two protein preparations. We used
Abstract The 2008 ABRF Proteomics Research Group Study offers participants the chance to participate in an anonymous study to identify qualitative differences between two protein preparations. We used
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Design of isolated protein and RNC constructs, and homogeneity of purified RNCs. (a) Schematic depicting the design and nomenclature used for all the isolated proteins and RNCs used
Supplementary Figure 1 Design of isolated protein and RNC constructs, and homogeneity of purified RNCs. (a) Schematic depicting the design and nomenclature used for all the isolated proteins and RNCs used
Multiple-Choice Questions Answer ALL 20 multiple-choice questions on the Scantron Card in PENCIL
 Multiple-Choice Questions Answer ALL 20 multiple-choice questions on the Scantron Card in PENCIL For Questions 1-10 choose ONE INCORRECT answer. 1. Which ONE of the following statements concerning the
Multiple-Choice Questions Answer ALL 20 multiple-choice questions on the Scantron Card in PENCIL For Questions 1-10 choose ONE INCORRECT answer. 1. Which ONE of the following statements concerning the
This exam consists of two parts. Part I is multiple choice. Each of these 25 questions is worth 2 points.
 MBB 407/511 Molecular Biology and Biochemistry First Examination - October 1, 2002 Name Social Security Number This exam consists of two parts. Part I is multiple choice. Each of these 25 questions is
MBB 407/511 Molecular Biology and Biochemistry First Examination - October 1, 2002 Name Social Security Number This exam consists of two parts. Part I is multiple choice. Each of these 25 questions is
Lane: 1. Spectra BR protein ladder 2. PFD 3. TERM 4. 3-way connector 5. 2-way connector
 kda 1 2 3 4 5 26 14 1 7 Lane: 1. Spectra BR protein ladder 2. PFD 3. TERM 4. 3-way connector 5. 2-way connector 5 4 35 25 15 1 Supplementary Figure 1. SDS-PAGE of acterially expressed and purified proteins.
kda 1 2 3 4 5 26 14 1 7 Lane: 1. Spectra BR protein ladder 2. PFD 3. TERM 4. 3-way connector 5. 2-way connector 5 4 35 25 15 1 Supplementary Figure 1. SDS-PAGE of acterially expressed and purified proteins.
Supplementary Figure 1 (previous page). EM analysis of full-length GCGR. (a) Exemplary tilt pair images of the GCGR mab23 complex acquired for Random
 S1 Supplementary Figure 1 (previous page). EM analysis of full-length GCGR. (a) Exemplary tilt pair images of the GCGR mab23 complex acquired for Random Conical Tilt (RCT) reconstruction (left: -50,right:
S1 Supplementary Figure 1 (previous page). EM analysis of full-length GCGR. (a) Exemplary tilt pair images of the GCGR mab23 complex acquired for Random Conical Tilt (RCT) reconstruction (left: -50,right:
Nafith Abu Tarboush DDS, MSc, PhD
 Nafith Abu Tarboush DDS, MSc, PhD natarboush@ju.edu.jo www.facebook.com/natarboush Protein conformation Many conformations are possible for proteins due to flexibility of amino acids linked by peptide
Nafith Abu Tarboush DDS, MSc, PhD natarboush@ju.edu.jo www.facebook.com/natarboush Protein conformation Many conformations are possible for proteins due to flexibility of amino acids linked by peptide
Nature Methods: doi: /nmeth Supplementary Figure 1. Salipro lipid particles.
 Supplementary Figure 1 Salipro lipid particles. (a) Gel filtration analysis of Saposin A after incubation with the indicated detergent solubilised lipid solutions. The generation of Saposin A-lipid complexes
Supplementary Figure 1 Salipro lipid particles. (a) Gel filtration analysis of Saposin A after incubation with the indicated detergent solubilised lipid solutions. The generation of Saposin A-lipid complexes
Sheet #5 Dr. Mamoun Ahram 8/7/2014
 P a g e 1 Protein Structure Quick revision - Levels of protein structure: primary, secondary, tertiary & quaternary. - Primary structure is the sequence of amino acids residues. It determines the other
P a g e 1 Protein Structure Quick revision - Levels of protein structure: primary, secondary, tertiary & quaternary. - Primary structure is the sequence of amino acids residues. It determines the other
Unveiling transient protein-protein interactions that modulate inhibition of alpha-synuclein aggregation
 Supplementary information Unveiling transient protein-protein interactions that modulate inhibition of alpha-synuclein aggregation by beta-synuclein, a pre-synaptic protein that co-localizes with alpha-synuclein.
Supplementary information Unveiling transient protein-protein interactions that modulate inhibition of alpha-synuclein aggregation by beta-synuclein, a pre-synaptic protein that co-localizes with alpha-synuclein.
Light Scattering Coupled to Refractive Index and UV. Homo- and Hetero Associations
 Light Scattering Coupled to Refractive Index and UV Absorption Measurements in Studies of Membrane Proteins Homo- and Hetero Associations Ewa Folta-Stogniew Biophysics Resource; Keck Laboratory Yale School
Light Scattering Coupled to Refractive Index and UV Absorption Measurements in Studies of Membrane Proteins Homo- and Hetero Associations Ewa Folta-Stogniew Biophysics Resource; Keck Laboratory Yale School
Structural analysis of fungus-derived FAD glucose dehydrogenase
 Structural analysis of fungus-derived FAD glucose dehydrogenase Hiromi Yoshida 1, Genki Sakai 2, Kazushige Mori 3, Katsuhiro Kojima 3, Shigehiro Kamitori 1, and Koji Sode 2,3,* 1 Life Science Research
Structural analysis of fungus-derived FAD glucose dehydrogenase Hiromi Yoshida 1, Genki Sakai 2, Kazushige Mori 3, Katsuhiro Kojima 3, Shigehiro Kamitori 1, and Koji Sode 2,3,* 1 Life Science Research
Supporting Information. Lysine Propionylation to Boost Proteome Sequence. Coverage and Enable a Silent SILAC Strategy for
 Supporting Information Lysine Propionylation to Boost Proteome Sequence Coverage and Enable a Silent SILAC Strategy for Relative Protein Quantification Christoph U. Schräder 1, Shaun Moore 1,2, Aaron A.
Supporting Information Lysine Propionylation to Boost Proteome Sequence Coverage and Enable a Silent SILAC Strategy for Relative Protein Quantification Christoph U. Schräder 1, Shaun Moore 1,2, Aaron A.
SUPPLEMENTAL INFORMATION
 SUPPLEMENTAL INFORMATION EXPERIMENTAL PROCEDURES Tryptic digestion protection experiments - PCSK9 with Ab-3D5 (1:1 molar ratio) in 50 mm Tris, ph 8.0, 150 mm NaCl was incubated overnight at 4 o C. The
SUPPLEMENTAL INFORMATION EXPERIMENTAL PROCEDURES Tryptic digestion protection experiments - PCSK9 with Ab-3D5 (1:1 molar ratio) in 50 mm Tris, ph 8.0, 150 mm NaCl was incubated overnight at 4 o C. The
Protein structure. Dr. Mamoun Ahram Summer semester,
 Protein structure Dr. Mamoun Ahram Summer semester, 2017-2018 Overview of proteins Proteins have different structures and some have repeating inner structures, other do not. A protein may have gazillion
Protein structure Dr. Mamoun Ahram Summer semester, 2017-2018 Overview of proteins Proteins have different structures and some have repeating inner structures, other do not. A protein may have gazillion
SDS-Assisted Protein Transport Through Solid-State Nanopores
 Supplementary Information for: SDS-Assisted Protein Transport Through Solid-State Nanopores Laura Restrepo-Pérez 1, Shalini John 2, Aleksei Aksimentiev 2 *, Chirlmin Joo 1 *, Cees Dekker 1 * 1 Department
Supplementary Information for: SDS-Assisted Protein Transport Through Solid-State Nanopores Laura Restrepo-Pérez 1, Shalini John 2, Aleksei Aksimentiev 2 *, Chirlmin Joo 1 *, Cees Dekker 1 * 1 Department
Mass Spectrometry. Mass spectrometer MALDI-TOF ESI/MS/MS. Basic components. Ionization source Mass analyzer Detector
 Mass Spectrometry MALDI-TOF ESI/MS/MS Mass spectrometer Basic components Ionization source Mass analyzer Detector 1 Principles of Mass Spectrometry Proteins are separated by mass to charge ratio (limit
Mass Spectrometry MALDI-TOF ESI/MS/MS Mass spectrometer Basic components Ionization source Mass analyzer Detector 1 Principles of Mass Spectrometry Proteins are separated by mass to charge ratio (limit
Detergent solubilised 5 TMD binds pregnanolone at the Q245 neurosteroid potentiation site.
 Supplementary Figure 1 Detergent solubilised 5 TMD binds pregnanolone at the Q245 neurosteroid potentiation site. (a) Gel filtration profiles of purified 5 TMD samples at 100 nm, heated beforehand for
Supplementary Figure 1 Detergent solubilised 5 TMD binds pregnanolone at the Q245 neurosteroid potentiation site. (a) Gel filtration profiles of purified 5 TMD samples at 100 nm, heated beforehand for
Biological Mass Spectrometry. April 30, 2014
 Biological Mass Spectrometry April 30, 2014 Mass Spectrometry Has become the method of choice for precise protein and nucleic acid mass determination in a very wide mass range peptide and nucleotide sequencing
Biological Mass Spectrometry April 30, 2014 Mass Spectrometry Has become the method of choice for precise protein and nucleic acid mass determination in a very wide mass range peptide and nucleotide sequencing
Characterization of Disulfide Linkages in Proteins by 193 nm Ultraviolet Photodissociation (UVPD) Mass Spectrometry. Supporting Information
 Characterization of Disulfide Linkages in Proteins by 193 nm Ultraviolet Photodissociation (UVPD) Mass Spectrometry M. Montana Quick, Christopher M. Crittenden, Jake A. Rosenberg, and Jennifer S. Brodbelt
Characterization of Disulfide Linkages in Proteins by 193 nm Ultraviolet Photodissociation (UVPD) Mass Spectrometry M. Montana Quick, Christopher M. Crittenden, Jake A. Rosenberg, and Jennifer S. Brodbelt
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/398/rs12/dc1 Supplementary Materials for Quantitative phosphoproteomics reveals new roles for the protein phosphatase PP6 in mitotic cells Scott F. Rusin, Kate
www.sciencesignaling.org/cgi/content/full/8/398/rs12/dc1 Supplementary Materials for Quantitative phosphoproteomics reveals new roles for the protein phosphatase PP6 in mitotic cells Scott F. Rusin, Kate
TECHNICAL BULLETIN. R 2 GlcNAcβ1 4GlcNAcβ1 Asn
 GlycoProfile II Enzymatic In-Solution N-Deglycosylation Kit Product Code PP0201 Storage Temperature 2 8 C TECHNICAL BULLETIN Product Description Glycosylation is one of the most common posttranslational
GlycoProfile II Enzymatic In-Solution N-Deglycosylation Kit Product Code PP0201 Storage Temperature 2 8 C TECHNICAL BULLETIN Product Description Glycosylation is one of the most common posttranslational
Protein Structure and Function
 Protein Structure and Function Protein Structure Classification of Proteins Based on Components Simple proteins - Proteins containing only polypeptides Conjugated proteins - Proteins containing nonpolypeptide
Protein Structure and Function Protein Structure Classification of Proteins Based on Components Simple proteins - Proteins containing only polypeptides Conjugated proteins - Proteins containing nonpolypeptide
Supplementary Figure 1. Structure models of the c4 variant proteins based on the structures of maltose binding protein (MBP) in red and TEM-1
 Supplementary Figure 1. Structure models of the c4 variant proteins based on the structures of maltose binding protein (MBP) in red and TEM-1 β-lactamase (BLA) in blue. (a) c4 model and the close-up view
Supplementary Figure 1. Structure models of the c4 variant proteins based on the structures of maltose binding protein (MBP) in red and TEM-1 β-lactamase (BLA) in blue. (a) c4 model and the close-up view
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/4/3/eaaq0762/dc1 Supplementary Materials for Structures of monomeric and oligomeric forms of the Toxoplasma gondii perforin-like protein 1 Tao Ni, Sophie I. Williams,
advances.sciencemag.org/cgi/content/full/4/3/eaaq0762/dc1 Supplementary Materials for Structures of monomeric and oligomeric forms of the Toxoplasma gondii perforin-like protein 1 Tao Ni, Sophie I. Williams,
Supplementary Figure-1. SDS PAGE analysis of purified designed carbonic anhydrase enzymes. M1-M4 shown in lanes 1-4, respectively, with molecular
 Supplementary Figure-1. SDS PAGE analysis of purified designed carbonic anhydrase enzymes. M1-M4 shown in lanes 1-4, respectively, with molecular weight markers (M). Supplementary Figure-2. Overlay of
Supplementary Figure-1. SDS PAGE analysis of purified designed carbonic anhydrase enzymes. M1-M4 shown in lanes 1-4, respectively, with molecular weight markers (M). Supplementary Figure-2. Overlay of
Purification of Glucagon3 Interleukin-2 Fusion Protein Derived from E. coli
 Purification of Glucagon3 Interleukin-2 Fusion Protein Derived from E. coli Hye Soon Won Dept. of Chem. Eng. Chungnam National University INTRODUCTION Human interleukin-2(hil-2) - known as T Cell Growth
Purification of Glucagon3 Interleukin-2 Fusion Protein Derived from E. coli Hye Soon Won Dept. of Chem. Eng. Chungnam National University INTRODUCTION Human interleukin-2(hil-2) - known as T Cell Growth
Improve Protein Analysis with the New, Mass Spectrometry- Compatible ProteasMAX Surfactant
 Improve Protein Analysis with the New, Mass Spectrometry- Compatible Surfactant ABSTRACT Incomplete solubilization and digestion and poor peptide recovery are frequent limitations in protein sample preparation
Improve Protein Analysis with the New, Mass Spectrometry- Compatible Surfactant ABSTRACT Incomplete solubilization and digestion and poor peptide recovery are frequent limitations in protein sample preparation
CRY2 binding to CIB1N w/ MTHF
 Supplemental Figures: CRY2 binding to CIB1N w/ MTHF.36 Polarization.34.32.3.28 Blue.26 5 1 15 [Cry2] in nm Figure S1: Addition of MTHF does not significantly change CRY2- CIB1N binding. Direct fluorescence
Supplemental Figures: CRY2 binding to CIB1N w/ MTHF.36 Polarization.34.32.3.28 Blue.26 5 1 15 [Cry2] in nm Figure S1: Addition of MTHF does not significantly change CRY2- CIB1N binding. Direct fluorescence
Chapter 3. Structure of Enzymes. Enzyme Engineering
 Chapter 3. Structure of Enzymes Enzyme Engineering 3.1 Introduction With purified protein, Determining M r of the protein Determining composition of amino acids and the primary structure Determining the
Chapter 3. Structure of Enzymes Enzyme Engineering 3.1 Introduction With purified protein, Determining M r of the protein Determining composition of amino acids and the primary structure Determining the
Project Title: Value-added Utilization of GEM Normal and High-amylose Line Starch
 Project Title: Value-added Utilization of GEM Normal and High-amylose Line Starch Prepared by Jay-lin Jane, Hongxin Jiang, and Li Li, Department of Food Science and Human Nutrition, Iowa State University,
Project Title: Value-added Utilization of GEM Normal and High-amylose Line Starch Prepared by Jay-lin Jane, Hongxin Jiang, and Li Li, Department of Food Science and Human Nutrition, Iowa State University,
Systematic analysis of protein-detergent complexes applying dynamic light scattering to optimize solutions for crystallization trials
 Supporting information 1 2 3 Volume 71 (2015) Supporting information for article: 4 5 6 7 8 Systematic analysis of protein-detergent complexes applying dynamic light scattering to optimize solutions for
Supporting information 1 2 3 Volume 71 (2015) Supporting information for article: 4 5 6 7 8 Systematic analysis of protein-detergent complexes applying dynamic light scattering to optimize solutions for
Lecture 3. Tandem MS & Protein Sequencing
 Lecture 3 Tandem MS & Protein Sequencing Nancy Allbritton, M.D., Ph.D. Department of Physiology & Biophysics 824-9137 (office) nlallbri@uci.edu Office- Rm D349 Medical Science D Bldg. Tandem MS Steps:
Lecture 3 Tandem MS & Protein Sequencing Nancy Allbritton, M.D., Ph.D. Department of Physiology & Biophysics 824-9137 (office) nlallbri@uci.edu Office- Rm D349 Medical Science D Bldg. Tandem MS Steps:
Supplementary Figure 1
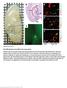 Supplementary Figure 1 AAV-GFP injection in the MEC of the mouse brain C57Bl/6 mice at 4 months of age were injected with AAV-GFP into the MEC and sacrificed at 7 days post injection (dpi). (a) Brains
Supplementary Figure 1 AAV-GFP injection in the MEC of the mouse brain C57Bl/6 mice at 4 months of age were injected with AAV-GFP into the MEC and sacrificed at 7 days post injection (dpi). (a) Brains
BIOCHEMISTRY I HOMEWORK III DUE 10/15/03 66 points total + 2 bonus points = 68 points possible Swiss-PDB Viewer Exercise Attached
 BIOCHEMISTRY I HOMEWORK III DUE 10/15/03 66 points total + 2 bonus points = 68 points possible Swiss-PDB Viewer Exercise Attached 1). 20 points total T or F (2 points each; if false, briefly state why
BIOCHEMISTRY I HOMEWORK III DUE 10/15/03 66 points total + 2 bonus points = 68 points possible Swiss-PDB Viewer Exercise Attached 1). 20 points total T or F (2 points each; if false, briefly state why
PTM Discovery Method for Automated Identification and Sequencing of Phosphopeptides Using the Q TRAP LC/MS/MS System
 Application Note LC/MS PTM Discovery Method for Automated Identification and Sequencing of Phosphopeptides Using the Q TRAP LC/MS/MS System Purpose This application note describes an automated workflow
Application Note LC/MS PTM Discovery Method for Automated Identification and Sequencing of Phosphopeptides Using the Q TRAP LC/MS/MS System Purpose This application note describes an automated workflow
Biology Open (2014) 000, 1 10 doi: /bio
 (2014) 000, 1 10 doi:10.1242/bio.201410041 Supplementary Material Michael Brauchle et al. doi: 10.1242/bio.201410041 Fig. S1. Alignment of GFP, sfgfp, egfp, eyfp, mcherry and mruby2. Sequence-based alignment
(2014) 000, 1 10 doi:10.1242/bio.201410041 Supplementary Material Michael Brauchle et al. doi: 10.1242/bio.201410041 Fig. S1. Alignment of GFP, sfgfp, egfp, eyfp, mcherry and mruby2. Sequence-based alignment
Supplementary Figure 1. Comparative analysis of thermal denaturation of enzymatically
 Supplementary Figures Supplementary Figure 1. Comparative analysis of thermal denaturation of enzymatically synthesized polyubiquitin chains of different length. a, Differential scanning calorimetry traces
Supplementary Figures Supplementary Figure 1. Comparative analysis of thermal denaturation of enzymatically synthesized polyubiquitin chains of different length. a, Differential scanning calorimetry traces
Heparin Sodium ヘパリンナトリウム
 Heparin Sodium ヘパリンナトリウム Add the following next to Description: Identification Dissolve 1 mg each of Heparin Sodium and Heparin Sodium Reference Standard for physicochemical test in 1 ml of water, and
Heparin Sodium ヘパリンナトリウム Add the following next to Description: Identification Dissolve 1 mg each of Heparin Sodium and Heparin Sodium Reference Standard for physicochemical test in 1 ml of water, and
Proteins. Amino acids, structure and function. The Nobel Prize in Chemistry 2012 Robert J. Lefkowitz Brian K. Kobilka
 Proteins Amino acids, structure and function The Nobel Prize in Chemistry 2012 Robert J. Lefkowitz Brian K. Kobilka O O HO N N HN OH Ser65-Tyr66-Gly67 The Nobel prize in chemistry 2008 Osamu Shimomura,
Proteins Amino acids, structure and function The Nobel Prize in Chemistry 2012 Robert J. Lefkowitz Brian K. Kobilka O O HO N N HN OH Ser65-Tyr66-Gly67 The Nobel prize in chemistry 2008 Osamu Shimomura,
bio-mof-1 DMASM Wavenumber (cm -1 ) Supplementary Figure S1 FTIR spectra of bio-mof-1, DMASMI, and bio-mof-1 DMASM.
 bio-mof-1 Transmittance bio-mof-1 DMASM DMASMI 2000 1500 1000 500 Wavenumber (cm -1 ) Supplementary Figure S1 FTIR spectra of bio-mof-1, DMASMI, and bio-mof-1 DMASM. Intensity (a.u.) bio-mof-1 DMASM as
bio-mof-1 Transmittance bio-mof-1 DMASM DMASMI 2000 1500 1000 500 Wavenumber (cm -1 ) Supplementary Figure S1 FTIR spectra of bio-mof-1, DMASMI, and bio-mof-1 DMASM. Intensity (a.u.) bio-mof-1 DMASM as
Q: How do I get the protein concentration in mg/ml from the standard curve if the X-axis is in units of µg.
 Photometry Frequently Asked Questions Q: How do I get the protein concentration in mg/ml from the standard curve if the X-axis is in units of µg. Protein standard curves are traditionally presented as
Photometry Frequently Asked Questions Q: How do I get the protein concentration in mg/ml from the standard curve if the X-axis is in units of µg. Protein standard curves are traditionally presented as
Nature Methods: doi: /nmeth Supplementary Figure 1
 Supplementary Figure 1 Subtiligase-catalyzed ligations with ubiquitin thioesters and 10-mer biotinylated peptides. (a) General scheme for ligations between ubiquitin thioesters and 10-mer, biotinylated
Supplementary Figure 1 Subtiligase-catalyzed ligations with ubiquitin thioesters and 10-mer biotinylated peptides. (a) General scheme for ligations between ubiquitin thioesters and 10-mer, biotinylated
Supplementary Materials. Wild-type and mutant SOD1 share an aberrant conformation and
 Supplementary Materials Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS Daryl A. Bosco, Gerardo Morfini, Murat Karabacak, Yuyu Song, Francois Gros-Louis,
Supplementary Materials Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS Daryl A. Bosco, Gerardo Morfini, Murat Karabacak, Yuyu Song, Francois Gros-Louis,
Tivadar Orban, Beata Jastrzebska, Sayan Gupta, Benlian Wang, Masaru Miyagi, Mark R. Chance, and Krzysztof Palczewski
 Structure, Volume Supplemental Information Conformational Dynamics of Activation for the Pentameric Complex of Dimeric G Protein-Coupled Receptor and Heterotrimeric G Protein Tivadar Orban, Beata Jastrzebska,
Structure, Volume Supplemental Information Conformational Dynamics of Activation for the Pentameric Complex of Dimeric G Protein-Coupled Receptor and Heterotrimeric G Protein Tivadar Orban, Beata Jastrzebska,
Supplementary Fig. 1. Identification of acetylation of K68 of SOD2
 Supplementary Fig. 1. Identification of acetylation of K68 of SOD2 A B H. sapiens 54 KHHAAYVNNLNVTEEKYQEALAK 75 M. musculus 54 KHHAAYVNNLNATEEKYHEALAK 75 X. laevis 55 KHHATYVNNLNITEEKYAEALAK 77 D. rerio
Supplementary Fig. 1. Identification of acetylation of K68 of SOD2 A B H. sapiens 54 KHHAAYVNNLNVTEEKYQEALAK 75 M. musculus 54 KHHAAYVNNLNATEEKYHEALAK 75 X. laevis 55 KHHATYVNNLNITEEKYAEALAK 77 D. rerio
Development of a Bioanalytical Method for Quantification of Amyloid Beta Peptides in Cerebrospinal Fluid
 Development of a Bioanalytical Method for Quantification of Amyloid Beta Peptides in Cerebrospinal Fluid Joanne ( 乔安妮 ) Mather Senior Scientist Waters Corporation Data courtesy of Erin Chambers and Mary
Development of a Bioanalytical Method for Quantification of Amyloid Beta Peptides in Cerebrospinal Fluid Joanne ( 乔安妮 ) Mather Senior Scientist Waters Corporation Data courtesy of Erin Chambers and Mary
MS/MS as an LC Detector for the Screening of Drugs and Their Metabolites in Race Horse Urine
 Application Note: 346 MS/MS as an LC Detector for the Screening of Drugs and Their Metabolites in Race Horse Urine Gargi Choudhary and Diane Cho, Thermo Fisher Scientific, San Jose, CA Wayne Skinner and
Application Note: 346 MS/MS as an LC Detector for the Screening of Drugs and Their Metabolites in Race Horse Urine Gargi Choudhary and Diane Cho, Thermo Fisher Scientific, San Jose, CA Wayne Skinner and
Characterization of Starch Polysaccharides in Aqueous Systems: de facto molar masses vs supermolecular structures
 Characterization of Starch Polysaccharides in Aqueous Systems: de facto molar masses vs supermolecular structures Werner Praznik and Anton uber Department of Chemistry BKU - University of Natural Resourses
Characterization of Starch Polysaccharides in Aqueous Systems: de facto molar masses vs supermolecular structures Werner Praznik and Anton uber Department of Chemistry BKU - University of Natural Resourses
Separation of Main Proteins in Plasma and Serum
 BCH 471 Experiment (2) Separation of Main Proteins in Plasma and Serum PLASMA PROTEINS Mw The main plasma proteins are: þ Albumin (36-50 g/l), Mw 66.241kDa. þ Globulins (18-32 g/l), Mw of globulins Cover
BCH 471 Experiment (2) Separation of Main Proteins in Plasma and Serum PLASMA PROTEINS Mw The main plasma proteins are: þ Albumin (36-50 g/l), Mw 66.241kDa. þ Globulins (18-32 g/l), Mw of globulins Cover
The Amyloid Precursor Protein Has a Flexible Transmembrane Domain and Binds Cholesterol
 The Amyloid Precursor Protein Has a Flexible Transmembrane Domain and Binds Cholesterol Science 336, 1171 (2013) Coach Prof. : Dr. Chung-I Chang Sit-in Prof.: Dr. Wei Yuan Yang Presenter: Han-Ying Wu Date:
The Amyloid Precursor Protein Has a Flexible Transmembrane Domain and Binds Cholesterol Science 336, 1171 (2013) Coach Prof. : Dr. Chung-I Chang Sit-in Prof.: Dr. Wei Yuan Yang Presenter: Han-Ying Wu Date:
ATP-independent reversal of a membrane protein aggregate by a chloroplast SRP
 ATP-independent reversal of a membrane protein aggregate by a chloroplast SRP Peera Jaru-Ampornpan 1, Kuang Shen 1,3, Vinh Q. Lam 1,3, Mona Ali 2, Sebastian Doniach 2, Tony Z. Jia 1, Shu-ou Shan 1 Supplementary
ATP-independent reversal of a membrane protein aggregate by a chloroplast SRP Peera Jaru-Ampornpan 1, Kuang Shen 1,3, Vinh Q. Lam 1,3, Mona Ali 2, Sebastian Doniach 2, Tony Z. Jia 1, Shu-ou Shan 1 Supplementary
HOMEWORK II and Swiss-PDB Viewer Tutorial DUE 9/26/03 62 points total. The ph at which a peptide has no net charge is its isoelectric point.
 BIOCHEMISTRY I HOMEWORK II and Swiss-PDB Viewer Tutorial DUE 9/26/03 62 points total 1). 8 points total T or F (2 points each; if false, briefly state why it is false) The ph at which a peptide has no
BIOCHEMISTRY I HOMEWORK II and Swiss-PDB Viewer Tutorial DUE 9/26/03 62 points total 1). 8 points total T or F (2 points each; if false, briefly state why it is false) The ph at which a peptide has no
MBB 694:407, 115:511. Please use BLOCK CAPITAL letters like this --- A, B, C, D, E. Not lowercase!
 MBB 694:407, 115:511 First Test Severinov/Deis Tue. Sep. 30, 2003 Name Index number (not SSN) Row Letter Seat Number This exam consists of two parts. Part I is multiple choice. Each of these 25 questions
MBB 694:407, 115:511 First Test Severinov/Deis Tue. Sep. 30, 2003 Name Index number (not SSN) Row Letter Seat Number This exam consists of two parts. Part I is multiple choice. Each of these 25 questions
Manja Henze, Dorothee Merker and Lothar Elling. 1. Characteristics of the Recombinant β-glycosidase from Pyrococcus
 S1 of S17 Supplementary Materials: Microwave-Assisted Synthesis of Glycoconjugates by Transgalactosylation with Recombinant Thermostable β-glycosidase from Pyrococcus Manja Henze, Dorothee Merker and Lothar
S1 of S17 Supplementary Materials: Microwave-Assisted Synthesis of Glycoconjugates by Transgalactosylation with Recombinant Thermostable β-glycosidase from Pyrococcus Manja Henze, Dorothee Merker and Lothar
Sequence Identification And Spatial Distribution of Rat Brain Tryptic Peptides Using MALDI Mass Spectrometric Imaging
 Sequence Identification And Spatial Distribution of Rat Brain Tryptic Peptides Using MALDI Mass Spectrometric Imaging AB SCIEX MALDI TOF/TOF* Systems Patrick Pribil AB SCIEX, Canada MALDI mass spectrometric
Sequence Identification And Spatial Distribution of Rat Brain Tryptic Peptides Using MALDI Mass Spectrometric Imaging AB SCIEX MALDI TOF/TOF* Systems Patrick Pribil AB SCIEX, Canada MALDI mass spectrometric
2. Which of the following amino acids is most likely to be found on the outer surface of a properly folded protein?
 Name: WHITE Student Number: Answer the following questions on the computer scoring sheet. 1 mark each 1. Which of the following amino acids would have the highest relative mobility R f in normal thin layer
Name: WHITE Student Number: Answer the following questions on the computer scoring sheet. 1 mark each 1. Which of the following amino acids would have the highest relative mobility R f in normal thin layer
Defense Antibodies, interferons produced in response to infection Coordination and growth (signaling) Hormones (e.g. insulin, growth hormone) Communic
 Proteins Chapter 3 An Introduction to Organic Compounds Most varied of the biomolecules Also called polypeptides Make up more than half the dry weight of cells Categorized by function Lecture 3: Proteins
Proteins Chapter 3 An Introduction to Organic Compounds Most varied of the biomolecules Also called polypeptides Make up more than half the dry weight of cells Categorized by function Lecture 3: Proteins
2. Ionization Sources 3. Mass Analyzers 4. Tandem Mass Spectrometry
 Dr. Sanjeeva Srivastava 1. Fundamental of Mass Spectrometry Role of MS and basic concepts 2. Ionization Sources 3. Mass Analyzers 4. Tandem Mass Spectrometry 2 1 MS basic concepts Mass spectrometry - technique
Dr. Sanjeeva Srivastava 1. Fundamental of Mass Spectrometry Role of MS and basic concepts 2. Ionization Sources 3. Mass Analyzers 4. Tandem Mass Spectrometry 2 1 MS basic concepts Mass spectrometry - technique
130327SCH4U_biochem April 09, 2013
 Option B: B1.1 ENERGY Human Biochemistry If more energy is taken in from food than is used up, weight gain will follow. Similarly if more energy is used than we supply our body with, weight loss will occur.
Option B: B1.1 ENERGY Human Biochemistry If more energy is taken in from food than is used up, weight gain will follow. Similarly if more energy is used than we supply our body with, weight loss will occur.
(B D) Three views of the final refined 2Fo-Fc electron density map of the Vpr (red)-ung2 (green) interacting region, contoured at 1.4σ.
 Supplementary Figure 1 Overall structure of the DDB1 DCAF1 Vpr UNG2 complex. (A) The final refined 2Fo-Fc electron density map, contoured at 1.4σ of Vpr, illustrating well-defined side chains. (B D) Three
Supplementary Figure 1 Overall structure of the DDB1 DCAF1 Vpr UNG2 complex. (A) The final refined 2Fo-Fc electron density map, contoured at 1.4σ of Vpr, illustrating well-defined side chains. (B D) Three
L-Carnosine-Derived Fmoc-Tripeptides Forming ph- Sensitive and Proteolytically Stable Supramolecular
 Supporting Information: L-Carnosine-Derived Fmoc-Tripeptides Forming ph- Sensitive and Proteolytically Stable Supramolecular Hydrogels Rita Das Mahapatra, a Joykrishna Dey* a, and Richard G. Weiss b a
Supporting Information: L-Carnosine-Derived Fmoc-Tripeptides Forming ph- Sensitive and Proteolytically Stable Supramolecular Hydrogels Rita Das Mahapatra, a Joykrishna Dey* a, and Richard G. Weiss b a
Organic Chemistry Laboratory Fall Lecture 3 Gas Chromatography and Mass Spectrometry June
 344 Organic Chemistry Laboratory Fall 2013 Lecture 3 Gas Chromatography and Mass Spectrometry June 19 2013 Chromatography Chromatography separation of a mixture into individual components Paper, Column,
344 Organic Chemistry Laboratory Fall 2013 Lecture 3 Gas Chromatography and Mass Spectrometry June 19 2013 Chromatography Chromatography separation of a mixture into individual components Paper, Column,
Peanut Allergen: Arachis hypogaea 1. Monica Trejo. Copyright 2014 by Monica Trejo and Koni Stone
 Peanut Allergen: Arachis hypogaea 1 Monica Trejo Copyright 2014 by Monica Trejo and Koni Stone A food allergy is an unfavorable immune response to a substance in food being digested. According to the Food
Peanut Allergen: Arachis hypogaea 1 Monica Trejo Copyright 2014 by Monica Trejo and Koni Stone A food allergy is an unfavorable immune response to a substance in food being digested. According to the Food
SYNOPSIS STUDIES ON THE PREPARATION AND CHARACTERISATION OF PROTEIN HYDROLYSATES FROM GROUNDNUT AND SOYBEAN ISOLATES
 1 SYNOPSIS STUDIES ON THE PREPARATION AND CHARACTERISATION OF PROTEIN HYDROLYSATES FROM GROUNDNUT AND SOYBEAN ISOLATES Proteins are important in food processing and food product development, as they are
1 SYNOPSIS STUDIES ON THE PREPARATION AND CHARACTERISATION OF PROTEIN HYDROLYSATES FROM GROUNDNUT AND SOYBEAN ISOLATES Proteins are important in food processing and food product development, as they are
CS612 - Algorithms in Bioinformatics
 Spring 2016 Protein Structure February 7, 2016 Introduction to Protein Structure A protein is a linear chain of organic molecular building blocks called amino acids. Introduction to Protein Structure Amine
Spring 2016 Protein Structure February 7, 2016 Introduction to Protein Structure A protein is a linear chain of organic molecular building blocks called amino acids. Introduction to Protein Structure Amine
Superose 6 Increase columns
 Data file 9-99- AA Size-exclusion chromatography Superose Increase columns Superose Increase prepacked columns (Fig ) are designed for rapid separation and analysis of proteins and other biomolecules by
Data file 9-99- AA Size-exclusion chromatography Superose Increase columns Superose Increase prepacked columns (Fig ) are designed for rapid separation and analysis of proteins and other biomolecules by
Supplementary Figure 1. Using DNA barcode-labeled MHC multimers to generate TCR fingerprints
 Supplementary Figure 1 Using DNA barcode-labeled MHC multimers to generate TCR fingerprints (a) Schematic overview of the workflow behind a TCR fingerprint. Each peptide position of the original peptide
Supplementary Figure 1 Using DNA barcode-labeled MHC multimers to generate TCR fingerprints (a) Schematic overview of the workflow behind a TCR fingerprint. Each peptide position of the original peptide
SUPPLEMENTAL FIGURE LEGENDS
 SUPPLEMENTAL FIGURE LEGENDS Fig. S1. SDSPAGE of crosslinked Aβ42 oligomers after SEC. After crosslinking of Aβ42 with or without Myr or RA, APS and Ru(bpy) were removed by SEC. The resulting products were
SUPPLEMENTAL FIGURE LEGENDS Fig. S1. SDSPAGE of crosslinked Aβ42 oligomers after SEC. After crosslinking of Aβ42 with or without Myr or RA, APS and Ru(bpy) were removed by SEC. The resulting products were
Structure of proteins
 Structure of proteins Presented by Dr. Mohammad Saadeh The requirements for the Pharmaceutical Biochemistry I Philadelphia University Faculty of pharmacy Structure of proteins The 20 a.a commonly found
Structure of proteins Presented by Dr. Mohammad Saadeh The requirements for the Pharmaceutical Biochemistry I Philadelphia University Faculty of pharmacy Structure of proteins The 20 a.a commonly found
Amino Acids. Review I: Protein Structure. Amino Acids: Structures. Amino Acids (contd.) Rajan Munshi
 Review I: Protein Structure Rajan Munshi BBSI @ Pitt 2005 Department of Computational Biology University of Pittsburgh School of Medicine May 24, 2005 Amino Acids Building blocks of proteins 20 amino acids
Review I: Protein Structure Rajan Munshi BBSI @ Pitt 2005 Department of Computational Biology University of Pittsburgh School of Medicine May 24, 2005 Amino Acids Building blocks of proteins 20 amino acids
RAPID SAMPLE PREPARATION METHODS FOR THE ANALYSIS OF N-LINKED GLYCANS
 RAPID SAMPLE PREPARATION METHODS FOR THE ANALYSIS OF N-LINKED GLYCANS Zoltan Szabo, András Guttman, Tomas Rejtar and Barry L. Karger Barnett Institute, Boston, MA, USA PCT Workshop,Boston, 21 May, 2010.
RAPID SAMPLE PREPARATION METHODS FOR THE ANALYSIS OF N-LINKED GLYCANS Zoltan Szabo, András Guttman, Tomas Rejtar and Barry L. Karger Barnett Institute, Boston, MA, USA PCT Workshop,Boston, 21 May, 2010.
BIOCHEMISTRY REVIEW. Overview of Biomolecules. Chapter 4 Protein Sequence
 BIOCHEMISTRY REVIEW Overview of Biomolecules Chapter 4 Protein Sequence 2 3 4 Are You Getting It?? A molecule of hemoglobin is compared with a molecule of lysozyme. Which characteristics do they share?
BIOCHEMISTRY REVIEW Overview of Biomolecules Chapter 4 Protein Sequence 2 3 4 Are You Getting It?? A molecule of hemoglobin is compared with a molecule of lysozyme. Which characteristics do they share?
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
Quantitative LC-MS/MS Analysis of Glucagon. Veniamin Lapko, Ph.D June 21, 2011
 Quantitative LC-MS/MS Analysis of Glucagon Veniamin Lapko, Ph.D June 21, 2011 Contents Comparison with small molecule LC-MS/MS LC-MS/MS sensitivity of peptides detection Stability: neat vs. matrix solutions
Quantitative LC-MS/MS Analysis of Glucagon Veniamin Lapko, Ph.D June 21, 2011 Contents Comparison with small molecule LC-MS/MS LC-MS/MS sensitivity of peptides detection Stability: neat vs. matrix solutions
Supporting Information. Electrophoretic Deformation of Individual Transfer. RNA Molecules Reveals Their Identity
 Supporting Information Electrophoretic Deformation of Individual Transfer RNA Molecules Reveals Their Identity Robert Y. Henley a, Brian Alan Ashcroft f, Ian Farrell c, Barry S. Cooperman d, Stuart M.
Supporting Information Electrophoretic Deformation of Individual Transfer RNA Molecules Reveals Their Identity Robert Y. Henley a, Brian Alan Ashcroft f, Ian Farrell c, Barry S. Cooperman d, Stuart M.
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10962 Supplementary Figure 1. Expression of AvrAC-FLAG in protoplasts. Total protein extracted from protoplasts described in Fig. 1a was subjected to anti-flag immunoblot to detect AvrAC-FLAG
doi:10.1038/nature10962 Supplementary Figure 1. Expression of AvrAC-FLAG in protoplasts. Total protein extracted from protoplasts described in Fig. 1a was subjected to anti-flag immunoblot to detect AvrAC-FLAG
Supplementary Information: Liquid-liquid phase coexistence in lipid membranes observed by natural abundance 1 H 13 C solid-state NMR
 Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the wner Societies 28 Supplementary Information: Liquid-liquid phase coexistence in lipid membranes observed
Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the wner Societies 28 Supplementary Information: Liquid-liquid phase coexistence in lipid membranes observed
Supporting information
 S1 Supporting information Biodegradable Injectable Polymer Systems Exhibiting Temperature-Responsive Irreversible Sol-to-Gel Transition by Covalent Bond Formation Yasuyuki YOSHIDA 1,2, Keisuke KAWAHARA
S1 Supporting information Biodegradable Injectable Polymer Systems Exhibiting Temperature-Responsive Irreversible Sol-to-Gel Transition by Covalent Bond Formation Yasuyuki YOSHIDA 1,2, Keisuke KAWAHARA
Biosci., Vol. 5, Number 4, December 1983, pp
 Biosci., Vol. 5, Number 4, December 1983, pp. 311-320. Printed in India. Denaturation of the high molecular weight protein fraction of mustard (Brassica juncea) and rapeseed (Brassica campestris) by urea
Biosci., Vol. 5, Number 4, December 1983, pp. 311-320. Printed in India. Denaturation of the high molecular weight protein fraction of mustard (Brassica juncea) and rapeseed (Brassica campestris) by urea
Supplementary Table 1. Properties of lysates of E. coli strains expressing CcLpxI point mutants
 Supplementary Table 1. Properties of lysates of E. coli strains expressing CcLpxI point mutants Species UDP-2,3- diacylglucosamine hydrolase specific activity (nmol min -1 mg -1 ) Fold vectorcontrol specific
Supplementary Table 1. Properties of lysates of E. coli strains expressing CcLpxI point mutants Species UDP-2,3- diacylglucosamine hydrolase specific activity (nmol min -1 mg -1 ) Fold vectorcontrol specific
Supplementary Information Janssen et al.
 Supplementary Information Janssen et al. Insights into complement convertase formation based on the structure of the factor B CVF complex Bert J.C. Janssen 1, Lucio Gomes 1, Roman I. Koning 2, Dmitri I.
Supplementary Information Janssen et al. Insights into complement convertase formation based on the structure of the factor B CVF complex Bert J.C. Janssen 1, Lucio Gomes 1, Roman I. Koning 2, Dmitri I.
Levels of Protein Structure:
 Levels of Protein Structure: PRIMARY STRUCTURE (1 ) - Defined, non-random sequence of amino acids along the peptide backbone o Described in two ways: Amino acid composition Amino acid sequence M-L-D-G-C-G
Levels of Protein Structure: PRIMARY STRUCTURE (1 ) - Defined, non-random sequence of amino acids along the peptide backbone o Described in two ways: Amino acid composition Amino acid sequence M-L-D-G-C-G
SUPPLEMENTARY MATERIAL
 SUPPLEMENTARY MATERIAL Purification and biochemical properties of SDS-stable low molecular weight alkaline serine protease from Citrullus Colocynthis Muhammad Bashir Khan, 1,3 Hidayatullah khan, 2 Muhammad
SUPPLEMENTARY MATERIAL Purification and biochemical properties of SDS-stable low molecular weight alkaline serine protease from Citrullus Colocynthis Muhammad Bashir Khan, 1,3 Hidayatullah khan, 2 Muhammad
Chapter 3. Protein Structure and Function
 Chapter 3 Protein Structure and Function Broad functional classes So Proteins have structure and function... Fine! -Why do we care to know more???? Understanding functional architechture gives us POWER
Chapter 3 Protein Structure and Function Broad functional classes So Proteins have structure and function... Fine! -Why do we care to know more???? Understanding functional architechture gives us POWER
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/278/rs11/dc1 Supplementary Materials for In Vivo Phosphoproteomics Analysis Reveals the Cardiac Targets of β-adrenergic Receptor Signaling Alicia Lundby,* Martin
www.sciencesignaling.org/cgi/content/full/6/278/rs11/dc1 Supplementary Materials for In Vivo Phosphoproteomics Analysis Reveals the Cardiac Targets of β-adrenergic Receptor Signaling Alicia Lundby,* Martin
Figure S1. (A) SDS-PAGE separation of GST-fusion proteins purified from E.coli BL21 strain is shown. An equal amount of GST-tag control, LRRK2 LRR
 Figure S1. (A) SDS-PAGE separation of GST-fusion proteins purified from E.coli BL21 strain is shown. An equal amount of GST-tag control, LRRK2 LRR and LRRK2 WD40 GST fusion proteins (5 µg) were loaded
Figure S1. (A) SDS-PAGE separation of GST-fusion proteins purified from E.coli BL21 strain is shown. An equal amount of GST-tag control, LRRK2 LRR and LRRK2 WD40 GST fusion proteins (5 µg) were loaded
Catalysis & specificity: Proteins at work
 Catalysis & specificity: Proteins at work Introduction Having spent some time looking at the elements of structure of proteins and DNA, as well as their ability to form intermolecular interactions, it
Catalysis & specificity: Proteins at work Introduction Having spent some time looking at the elements of structure of proteins and DNA, as well as their ability to form intermolecular interactions, it
Proteins? Protein function. Protein folding. Protein folding diseases. Protein interactions. Macromolecular assemblies. The end product of Genes
 Proteins? Protein function Protein folding Protein folding diseases Protein interactions Macromolecular assemblies The end product of Genes Protein Unfolding DOD Acid Catalysis DOD HDOD + N H N D C N C
Proteins? Protein function Protein folding Protein folding diseases Protein interactions Macromolecular assemblies The end product of Genes Protein Unfolding DOD Acid Catalysis DOD HDOD + N H N D C N C
Supporting Information for. Interaction of Spin-Labeled HPMA-based Nanoparticles with Human Blood Plasma Proteins
 Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 2018 Supporting Information for Interaction of Spin-Labeled HPMA-based Nanoparticles with Human Blood
Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 2018 Supporting Information for Interaction of Spin-Labeled HPMA-based Nanoparticles with Human Blood
Supporting InformationCathepsin-Mediated Cleavage of Peptides from Peptide Amphiphiles Leads to Enhanced Intracellular Peptide Accumulation
 Supporting InformationCathepsin-Mediated Cleavage of Peptides from Peptide Amphiphiles Leads to Enhanced Intracellular Peptide Accumulation Handan Acar, PhD,, Ravand Samaeekia, MS,, Mathew R. Schnorenberg,,
Supporting InformationCathepsin-Mediated Cleavage of Peptides from Peptide Amphiphiles Leads to Enhanced Intracellular Peptide Accumulation Handan Acar, PhD,, Ravand Samaeekia, MS,, Mathew R. Schnorenberg,,
Chapter 9. Protein Folding, Dynamics, and Structural Evolution
 Chapter 9 Protein Folding, Dynamics, and Structural Evolution Proteins folding Self-assembly Primary sequence dictates three dimensional structure Folding accessory proteins Protein disulfide isomerases
Chapter 9 Protein Folding, Dynamics, and Structural Evolution Proteins folding Self-assembly Primary sequence dictates three dimensional structure Folding accessory proteins Protein disulfide isomerases
BIO 311C Spring Lecture 15 Friday 26 Feb. 1
 BIO 311C Spring 2010 Lecture 15 Friday 26 Feb. 1 Illustration of a Polypeptide amino acids peptide bonds Review Polypeptide (chain) See textbook, Fig 5.21, p. 82 for a more clear illustration Folding and
BIO 311C Spring 2010 Lecture 15 Friday 26 Feb. 1 Illustration of a Polypeptide amino acids peptide bonds Review Polypeptide (chain) See textbook, Fig 5.21, p. 82 for a more clear illustration Folding and
Overview of the Expressway Cell-Free Expression Systems. Expressway Mini Cell-Free Expression System
 Overview of the Expressway Cell-Free Expression Systems The Expressway Cell-Free Expression Systems use an efficient coupled transcription and translation reaction to produce up to milligram quantities
Overview of the Expressway Cell-Free Expression Systems The Expressway Cell-Free Expression Systems use an efficient coupled transcription and translation reaction to produce up to milligram quantities
Osmolytes Stabilize Ribonuclease S by Stabilizing Its Fragments S Protein and S Peptide to Compact Folding-competent States*
 THE JOURNAL OF BIOLOGICAL CHEMISTRY Vol. 276, No. 31, Issue of August 3, pp. 28789 28798, 2001 2001 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in U.S.A. Osmolytes Stabilize
THE JOURNAL OF BIOLOGICAL CHEMISTRY Vol. 276, No. 31, Issue of August 3, pp. 28789 28798, 2001 2001 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in U.S.A. Osmolytes Stabilize
Trypsin Mass Spectrometry Grade
 058PR-03 G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name Trypsin Mass Spectrometry Grade A Chemically Modified, TPCK treated, Affinity Purified
058PR-03 G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name Trypsin Mass Spectrometry Grade A Chemically Modified, TPCK treated, Affinity Purified
Supplementary Material
 Supplementary Material HLA-DM Captures Partially Empty HLA-DR Molecules for Catalyzed Peptide Removal Anne-Kathrin Anders, Melissa J. Call, Monika-Sarah E. D. Schulze, Kevin D. Fowler, David A. Schuert,
Supplementary Material HLA-DM Captures Partially Empty HLA-DR Molecules for Catalyzed Peptide Removal Anne-Kathrin Anders, Melissa J. Call, Monika-Sarah E. D. Schulze, Kevin D. Fowler, David A. Schuert,
Heterogeneity in Molecular Dimensions (V e ): mass molar fraction profiles
 molar fraction eterogeneity in Molecular Dimensions (V e ): mass molar fraction profiles R = e, i 3 V 4 π e, i 1 3 m _ R e D _ d n _ R e D _ d 0.08 0.08 sphere equivalent radii distribution (R e D) of
molar fraction eterogeneity in Molecular Dimensions (V e ): mass molar fraction profiles R = e, i 3 V 4 π e, i 1 3 m _ R e D _ d n _ R e D _ d 0.08 0.08 sphere equivalent radii distribution (R e D) of
