Supplementary Information
|
|
|
- Dale Conley
- 6 years ago
- Views:
Transcription
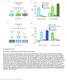
1 Supplementary Information Supplementary s Supplementary 1 All three types of foods suppress subsequent feeding in both sexes when the same food is used in the pre-feeding test feeding. (a) Adjusted pre-feeding paradigm. In each group, the same food was used in the pre-feeding test feeding. (b) The amount of food consumed decreased with feeding time. After 30 min feeding of normal food, tryptone, or sucrose, both females males were satiated stopped eating. n=5-7. 1
2 Supplementary 2 Protein intake up-regulates fit expression level. (a) The expression levels of Rp49 are comparable among starvation group (Agar) three feeding groups. Dp Actin 5C were used as the internal controls, respectively. n=3. 2
3 (b) Fit expression levels in WT flies greatly increased after 30 min feeding of tryptone, when analyzed with the Rp49, Dp, or Actin 5C as the internal control. n = 3. (c) Fit expression levels significantly increased after the consumption of normal food or tryptone, but not after sucrose or lipid intake. n = 3. (d) Fit expression levels increased after 30 min feeding of tryptone in a concentration-dependent manner. n = 3. (e) BCAAs promote Fit expression, while non-bcaas have little effect. Sucrose served as the sweetener, showing no effect on Fit expression on its own. n = 3. (f) Food consumption was decreased when DMSO was added into the food, while it did not further decrease when Rapamycin was added. Sucrose-tryptone-mixed food was used. n = 4. Data were analyzed by unpaired Student's t test in b by one-way ANOVA, LSD s post hoc test in other panels. *, p < 0.05; **, p < 0.01; ***, p < n.s. indicates no statistical significance. 3
4 Supplementary 3 Fit KO flies develop normally. (a) Fit KO flies were generated by homologue recombination. Two genomic fragments adjacent to the Fit gene were cloned as left (3.4 Kb) right (5.4 Kb) arms into the vector pendsout-riko flanking the white gene (upper panel). KO cidates were selected based on red-eye phenotype, screened by PCR (lower panel) using the primers shown by arrows in the upper panel. (b) FIT antibody detected FIT expression in head section of WT but not Fit 81 mutant flies. The FB region on the head section is circled by dashed lines. Scale bar, 100 μm. (c) Representive images of Fit 81 mutant WT control flies. Scale bar, 1 mm. (d, e f) Body weight (d, n = 15), protein content (e, n = 9), lipid content (f, n = 6) are comparable between Fit 81 mutant WT flies in both sexes. Data were analyzed by unpaired Student's t test in d, e, f. *, p < 0.05; n.s. indicates no statistical significance. 4
5 Supplementary 4 Feeding behavior in Fit KO flies. (a,,b) Compared to WT control flies, Fit KO flies showed normal basal feeding (a, n = 20-28), but exhibited increased food intake after starvation (b, n = 9-19). (c) In the modified two-choice feeding assay, Fit KO flies showed similar preference for the tryptone-sucrose mix versus sucrose only. Left panel shows the Choice Ratio (CR) for two nutrients. Right panel shows the Preference Index calculated as CR S+T CR S. n = 6. Data were analyzed by unpaired Student's t test in a, c by one-way ANOVA, LSD s post hoc test in b. *, p < 0.05; ***, p < n.s. indicates no statistical significance. 5
6 Supplementary 5 Feeding behavior of two WT fly strains in the pre-feeding assay. In pre-feeding assay (a), WT flies of wcs (b, c) CS (d, e) displayed significant differences between sexes in tryptone pre-feeding groups, but little sexual difference in normal food or sucrose pre-feeding groups. n = Data were analyzed by One-Way ANOVA in b, d by two-way ANOVA, Bonferroni test in c, e. P(Food*Sex) = 4.01E-4 (two-way ANOVA, Bonferroni test) in c. P(Food*Sex) = 4.43E-10 (two-way ANOVA, Bonferroni test) in e. *, p < 0.05; **, p < 0.01; ***, p < n.s. indicates no statistical significance. 6
7 Supplementary 6 WT flies show sexual differences in modified pre-feeding assay. 7
8 (a) Pre-feeding of AA mix significantly suppressed subsequent feeding in female flies. n = 6. Data were analyzed by unpaired Student's t test. (b) When tryptone-sucrose mixed food (T+S mixed food) was used instead of normal food in test feeding phase, WT flies also exhibited significant sexual differences in the tryptone pre-feeding groups. n = 8. Data were analyzed by one-way ANOVA, LSD s post hoc test in Food Consumption experiments. For Suppression Index analysis, P (Food*Sex) =1.31E-9 (two-way ANOVA, Bonferroni test). (c) After pre-feeding of tryptone at different concentration, flies exhibited gradually decreased tryptone intake in the test feeding. n = 6. One-way ANOVA, LSD s post hoc test. (d) Fit expression levels were comparable between mated virgin females. n = 4. P (Food*Mated) = (two-way ANOVA, Bonferroni test). (e) Mated virgin females exhibited similar PIFI effect, the SI are comparable between them. n = 7-9. P (Food*Mated) = (two-way ANOVA, Bonferroni test). For Suppression Index analysis, data were analyzed by unpaired Student's t test. *, p < 0.05; **, p < 0.01; ***, p < n.s. indicates no statistical significance. 8
9 Supplementary 7 Fit mutant flies are deficient in protein feeding. (a) The suppression indexes of normal food sucrose were comparable between Fit 81 mutant WT flies. n = 8-9. Data were analyzed by unpaired Student's t test. (b, c) The suppressive effect of tryptone pre-feeding was significantly reduced in Fit 52 9
10 mutant flies (b) flies with FB-knock down of Fit (c). n = Data were analyzed by unpaired Student's t test in b two-way ANOVA, Bonferroni test in c. P (Food*Genotype) = in female in male (c). (d, e) Knock down of Fit in the FB (with Fit-Gal4), but not in the nervous system (with Elav-Gal4 or Dilp2-Gal4), resulted in a significant reduction in the PIFI effect. n = 8-9. P (Food*Genotype) = (two-way ANOVA, Bonferroni test) in d. Data were analyzed by one-way ANOVA, LSD s post hoc test in e. (f) The reduction in feeding suppression observed in Fit 81 female flies was rescued by FB-expression of FIT. n = P (Food*Genotype)= in female in male (two-way ANOVA, Bonferroni test). *, p < 0.05; **, p < 0.01; ***, p < n.s. indicates no statistical significance. 10
11 Supplementary 8 The feeding suppression effect of tryptone in WT female flies is diminished following blocking of the AA sensing pathway (a, b) Upon Rapamycin treatment, the suppressive effect of tryptone was significantly reduced. n = Sucrose served as the sweetener. P (Drug*Sex) = in female in male (two-way ANOVA, Bonferroni test) in a. Data were analyzed by one-way ANOVA, LSD s post hoc test in b. (c, d) The suppressive effect of tryptone pre-feeding was abolished in female flies when AA transporter Slif was knocked down (with Slif-anti), or when the negative regulator of 11
12 TOR pathway TSC1/2 was overexpressed in the FB. n = P (Food*Genotype) = in female in male (two-way ANOVA, Bonferroni test) in c. Data were analyzed by one-way ANOVA, LSD s post hoc test in d. *, p < 0.05; **, p < 0.01; ***, p < n.s. indicates no statistical significance. 12
13 Supplementary 9 Temporary expression of FIT in the nervous system suppresses feeding behavior. (a, b) The full size pictures of Western gels. HA signals were detected in medium (a) or in fly hemolymph (b) when FIT-HA was expressed, not however when FITΔSP-HA was expressed. (c) Ectopic expressing FIT in the nervous system suppressed feeding, whereas expression of FITΔSP showed no such effect. n = Data were analyzed by unpaired Student's t 13
14 test. (d) RU486-induced neuronal expression of FIT suppressed food consumption. n = 8. P (Drug*Genotype) = (two-way ANOVA, Bonferroni test). (e) The difference between overexpression groups their parental controls showing as ΔCR for Tryptone = (CR Overexpression CR control )/CR control. Overexpression of FIT resulted in a significant decease in the CR for tryptone, while overexpression of InR DN or FIT together with InR DN did not affect this choice ratio. One-way ANOVA, LSD s post hoc test. *, p < 0.05; ***, p < n.s. indicates no statistical significance. 14
15 Supplementary 10 Quantification of DILP2 immunostaining signals. (a) Compared to that in agar (A) group, the overall mean of DILP2 signals were reduced in female flies after feeding of normal food (N) or tryptone (T), but not after sucrose (S) feeding, while it was not reduced in male flies after tryptone feeding. The total volume remained unchanged in all groups. n = P (Food*Sex) = in Overall Mean Intensity of DILP in Total Volume of DILP2 Signal (two-way ANOVA, 15
16 Bonferroni test). (b) The overall mean of DILP2 signals did not reduce after tryptone feeding in female flies with the secretion of IPCs blocked. The total volume remained unchanged. n = P (Food*Genotype) = 3.49E-5 in Overall Mean Intensity of DILP in Total Volume of DILP2 Signal (two-way ANOVA, Bonferroni test). (c) The overall mean of DILP2 signals did not reduce after tryptone feeding in Fit 81 mutant female male flies. The total volume remained unchanged. n = P (Food*Genotype) = in female in male in Overall Mean Intensity of DILP2 (two-way ANOVA, Bonferroni test). P (Food*Genotype) = in female in male in Total Volume of DILP2 Signal (two-way ANOVA, Bonferroni test). (d) The overall mean of DILP2 signals significantly reduced in brains incubated with FITbut not FITΔSP- conditioned medium. The total volume remained unchanged. n = One-way ANOVA, LSD s post hoc test. (e, f) Expressing Shi ts1 in IPCs abolishes the reduction of DILP2 signal induced by FIT medium incubation. n = Scale bar, 20 μm. P (Medium*Genotype) = in Total Fluorescence Intensity, 1.80E-5 in Overall Mean Intensity of DILP in Total Volume of DILP2 Signal (two-way ANOVA, Bonferroni test). **, p < 0.01; ***, p < n.s. indicates no statistical significance. 16
17 Supplementary Table 1 The composition of amino acid mixture Amino Acids Ala Arg Asp Cys Glu Gly IIe Leu Lys g/l Amino Acids Met Phe Pro Ser Thr Trp Tyr Val SUM g/l Supplemental Table 2 Primers for gene cloning or qpcr analysis Description Cloning the coding sequences of FIT for UAS-FIT-HA flies Cloning the coding sequences of FITΔSP for UAS-FITΔSP-HA flies Cloning the genome sequence of Fit promoter for Fit-GAL4 flies Cloning the genome sequences of Fit left arm for Fit knock-out flies Cloning the genome sequences of Fit right arm for Fit knock-out flies Screen primers for Fit knock-out flies Primers Fit-cDNA-s-EcoRI GGAATTCCATGAACTCAACTCTGGTGATTCTAC TTC Fit-cDNA-as-XbaI GCTCTAGAGCTTACATTCCATTCGCCTGGCTCA TTTG Fit-cDNA-Δ SP-EcoRI-B-F CGGAATTCATTCGTTGGTCTGAGGAGGA Fit-cDNA-as-XbaI GCTCTAGAGCTTACATTCCATTCGCCTGGCTCA TTTG Fit-pro-s-StuI AAGGCCTTCGGACCTGTTTCGTTCATAAAAAAG ATC Fit-pro-as EcoRI GGAATTCCTTCAACGATCGCTGTTAAACTGTGC Fit-KO-left-s-SacII TCCCCGCGGGACCTTGATTACTGAGCACAGATG G Fit-KO-left-as-KpnI GGGGTACCCCGTAAGCTCACGACTTGGTGATC AG Fit-KO-right-s-SpeI GACTAGTCAACGATCGCTGTTAAACTGTGCC Fit-KO-right-as-AscI AGGCGCGCCACGAGCAGGAGTGTGGTTTGAAC Fit-KO-exleft-F ATCGATTCAAGGAGCAGCTC Fit-KO-exright-R 17
18 Cloning FIT-HA into pcdna3.1 vector for cell culture Cloning FITΔSP-HA into pcdna3.1 vector for cell culture qpcr primers for Fit qpcr primers for Dilp2 qpcr primers for Rp49 qpcr primers for CG8147 qpcr primers for CG14688 qpcr primers for CG14867 qpcr primers for CG30431 qpcr primers for Sug CTGAGTAGCAACATTGTGAGG Fit-pcDNA3.1-EcoRI-B-F CGGAATTCATGAACTCAACTCTGGTGATTCTA Fit-pcDNA3.1-HA-XhoI-B-R CCGCTCGAGCTATGCGTAATCTGGAACATCG Fit- pcdna3.1-δsp-ecori-b-f CGGAATTCATTCGTTGGTCTGAGGAGGA Fit-pcDNA3.1-HA-XhoI-B-R CCGCTCGAGCTATGCGTAATCTGGAACATCG Fit-qPCR-F TTGGTGCAGGCCAGGAATAT Fit-qPCR-R CACAGGGCCAGTTGACAGAGT dilp2-qpcr-f GTATGGTGTGCGAGGAGTAT dilp2-qpcr-r TGAGTACACCCCCAAGATAG rp49-qpcr-f AGGGTATCGACAACAGAGTG rp49-qpcr-r CACCAGGAACTTCTTGAATC CG8147-qPCR-F CAAAGGCGTGACTGATTCCA CG8147-qPCR-R GATTGCGGCCATCCAAAC CG14688-qPCR-F ACGGAAATGTGGTGCACAAG CG14688-qPCR-R GCGTGGCGACTCTTTTGC CG14867-qPCR-F TCAGCGACAGCGTCCAAA CG14867-qPCR-R GGATTTCCCGTATTGTTCTGGTT CG30431-qPCR-F CCCGAGTGCGAAAAGAAGTT CG30431-qPCR-R GGCTGTCATGTGCAGCTTCA Sug-qPCR-F TCGGTGCGCTGCGTTATA Sug-qPCR-R 18
19 qpcr primers for CG5773 qpcr primers for CG6129 qpcr primers for CG7227 qpcr primers for CG9451 qpcr primers for CG13607 qpcr primers for CG15282 qpcr primers for Lsp2 qpcr primers for CG12116 CGCCGTTCCAAGTCCAAT CG5773-qPCR-F TGTTTTGCTCGCCGGATT CG5773-qPCR-R ACGGGTGCCGCATTCA CG6129-qPCR-F AGCTGCGGAGACTATGACAACA CG6129-qPCR-R TCCGGGTCCACATCAATTG CG7227-qPCR-F GCCAGTCCCAGGTGCAAGT CG7227-qPCR-R CAGCGGTATTGGATGGAATTC CG9451-qPCR-F TCAGATGCCCCGCTTCTC CG9451-qPCR-R TTTGCGGGTTTTGATGCA CG13607-qPCR-F CGTGGCTAGTGGTAAGGGATCA CG13607-qPCR-R ACGGTGGTGCAGGTCACAT CG15282-qPCR-F TTCTTGGTGATCGTTTTTGTGGC CG15282-qPCR-R CACCGAATCC TCCGAATCCA Lsp2-qPCR-F TCCGGCTATGCCAGTAACCT Lsp2-qPCR-R GATTCTCCCGGTGCCATTG CG12116-qPCR-F CGGTGGCGCTCAATCAG CG12116-qPCR-R TCCACTCGCTCCAAGTGCTT 19
20 Supplementary Table 3 Genotype Sex Assays Antibodies Statistic Methods 1a 1b 1c 1d 1e 2a 2b 2c 2d Pre-feeding (Tryptone) Test (Tryptone or Sucrose) qpcr Two-Way ANOVA 10min vs 0min: p = 1.01E-14, 20min vs 0min: p = 1.40E-21, 30min vs 0min: p = 3.39E-24. qpcr - Two-Way ANOVA 30min vs 0min in Tryptone: p = 2.03E-5, 60min vs 0min in Tryptone: p = 1.56E-4. qpcr - One-Way ANOVA Trptone vs Agar: p = 2.13E-4, Agar with L-Arabinose vs Agar: p = 0.62, AA Mix vs Agar: p = 0.002, BCAAs vs Agar: p = 0.012, Trptone vs AA Mixr: p = qpcr - Two-Way ANOVA Tryptone vs Agar in DMSO: p = 4.34E-4, Tryptone vs Agar in Rapamycin: p = Fit-Gal4 /UAS-GFP Fit expression pattern - - Fit-Gal4 / UAS-GFP Immunohistochemical stain ing Fit-Gal4 / UAS-GFP Fit-Gal4 / UAS-GFP Immunohistochemical stain ing Qualification of staining signal Nile Red - anti-fit, Nile Red anti-fit, Nile Red - One-Way ANOVA female fed with Protein Food vs male fed with 20
21 2e 3a 3b 3c 3d - - Pre-feeding (Agar, Normal Food, Tryptone, Sucrose) Test (Tryptone or Sucrose) Fit 81 Protein Food: p = 7.86E-5, female fed with Protein Food vs female fed without Protein Food: p = 2.06E-6. qpcr - Two-Way ANOVA femle: head vs body: p = 1.06E-4, fat body vs body: p = 1.56E-4. male: head vs body: p = 0.03, fat body vs body: p = 7.23E-3. Pre-feeding Assay, as 3a Pre-feeding Assay, as 3a Pre-feeding Assay, as 3a One-Way ANOVA female: Normal Food vs Agar: p = 1.17E-12, Tryptone vs Agar: p = 2.43E-10, Sucrose vs Agar: p = 9.30E-10, Normal Food vs Tryptone: p = 0.053, Tryptone vs Sucrose: p = male: Normal Food vs Agar: p = 4.15E-11, Tryptone vs Agar: p = 0.094, Sucrose vs Agar: p = 1.96E-11, Normal Food vs Tryptone: p = 2.37E-8, Tryptone vs Sucrose: p = 1.08E-7. - Two-Way ANOVA - One-Way ANOVA female: Normal Food vs Agar: p = 1.56E-12, Tryptone vs Agar: p = , Sucrose vs Agar: p =3.13E-11. male: Normal Food vs Agar: p =8.02E-15, Tryptone vs Agar: p = 0.059, Sucrose vs Agar: p = 3.32E
22 3e 3f 3g 4a Fit 81 Fit 52 Fit-Gal4/+ Fit-Gal4/+;Fit-RNAi/+ Fit-Gal4/+; Fit 81 / Fit 81 UAS-FIT/+; Fit 81 / Fit 81 Fit-Gal4/UAS-FIT ;Fit 81 / Fit 81 Pre-feeding Assay, as 3a Pre-feeding Assay, as 3a Pre-feeding Assay, as 3a - One-Way ANOVA female vs Fit 81 female: p = , female vs Fit 81 female: p = , female vs male: p = , female vs Fit 81 male: p = , female vs Fit 52 male: p = Unpaired Student t test female: Fit-Gal4/+ vs Fit-Gal4>UAS-Fit-RNAi: p = 9.07E-5. male: Fit-Gal4/+ vs Fit-Gal4>UAS-Fit-RNAi : p = One-Way ANOVA female: Fit-Gal4;Fit 81 /Fit 81 vs Fit-Gal4/UAS-FIT;Fit 8 /Fit 811 : p = 4.93E-6, UAS-FIT;Fit 81 /Fit 81 vs Fit-Gal4/UAS-FIT;Fit 8 /Fit 811 : p = 3.37E-7, Fit-Gal4;Fit 81 /Fit 81 vs UAS-FIT;Fit 8 /Fit 811 : p = male: Fit-Gal4;Fit 8 /Fit 811 vs Fit-Gal4/UAS-FIT;Fit 8 /Fit 811 :p = 0.78, UAS-FIT;Fit 81 /Fit 81 vs Fit-Gal4/UAS-FIT;Fit 8 /Fit 811 :p = 0.92, Fit-Gal4;Fit 81 /Fit 81 vs UAS-FIT;Fit 8 /Fit 811 : p = Fit 81 Two-choice feeding assay - Unpaired Student t test Sucrose pre-feeding condition: Sucrose selection in vs Fit 81 : p = 0.57, Tryptone selection in vs Fit 81 : p = No pre-feeding condition: Sucrose selection in vs Fit 81 : p = 0.012, Tryptone selection in vs Fit 81 : p = Tryptone pre-feeding condition: Sucrose selection in vs Fit 81 : p = 7.09E-4, Tryptone selection in vs Fit 81 : p = 3.70E-4. 22
23 4b 4c Fit 81 Two-choice feeding assay - Unpaired Student t test Sucrose pre-feeding condition: Sucrose selection in vs Fit 81 : p = 0.90, Tryptone selection in vs Fit 81 : p = No pre-feeding condition: Sucrose selection in vs Fit 81 : p = 0.72, Tryptone selection in vs Fit 81 : p = Tryptone pre-feeding condition: Sucrose selection in vs Fit 81 : p = 0.57, Tryptone selection in vs Fit 81 : p = Fit-Gal4/+ +/UAS-FIT Fit-Gal4/UAS-FIT Two-choice feeding assay - One-Way ANOVA Sucrose pre-feeding condition: Sucrose selection in Fit-Gal4/+ vs +/UAS-FIT: p = 0.82, Sucrose selection in Fit-Gal4/+ vs Fit-Gal4/UAS-FIT: p = 0.39, Sucrose selection in +/UAS-FIT vs Fit-Gal4/UAS-FIT: p = 0.28; Tryptone selection in Fit-Gal4/+ vs +/UAS-FIT: p = 0.31, Tryptone selection in Fit-Gal4/+ vs Fit-Gal4/UAS-FIT: p = 3.11E-9, Tryptone selection in +/UAS-FIT vs Fit-Gal4/UAS-FIT: p = 1.05E-8. No pre-feeding condition: Sucrose selection in Fit-Gal4/+ vs +/UAS-FIT: p = 0.76, Sucrose selection in Fit-Gal4/+ vs Fit-Gal4>UAS-FIT: p = , Sucrose selection in +/UAS-FIT vs Fit-Gal4/UAS-FIT: p = ; Tryptone selection in Fit-Gal4/+ vs +/UAS-FIT: p = 0.58, Tryptone selection in Fit-Gal4/+ vs Fit-Gal4/UAS-FIT: p = , Tryptone selection in +/UAS-FIT vs Fit-Gal4/UAS-FIT: p = Tryptone pre-feeding condition: Sucrose selection in Fit-Gal4/+ vs +/UAS-FIT: p = 0.56, Sucrose selection in Fit-Gal4/+ vs Fit-Gal4/UAS-FIT: p = 0.73, Sucrose 23
24 4d Fit-Gal4/+ +/UAS-FIT Fit-Gal4/UAS-FIT 24 selection in +/UAS-FIT vs Fit-Gal4/UAS-FIT: p = 0.81; Tryptone selection in Fit-Gal4/+ vs +/UAS-FIT: p = 0.36, Tryptone selection in Fit-Gal4/+ vs Fit-Gal4/UAS-FIT: p = 0.72, Tryptone selection in + /UAS-FIT vs Fit-Gal4/UAS-FIT: p = Two-choice feeding assay - One-Way ANOVA Sucrose pre-feeding condition: Sucrose selection in Fit-Gal4/+ vs +/UAS-FIT: p=0.67, Sucrose selection in Fit-Gal4/+ vs Fit-Gal4/UAS-FIT: p = 0.24, Sucrose selection in +/UAS-FIT vs Fit-Gal4/UAS-FIT: p = 0.43; Tryptone selection in Fit-Gal4/+ vs +/UAS-FIT: p = 0.28, Tryptone selection in Fit-Gal4/+ vs Fit-Gal4/UAS-FIT: p = 2.27E-5, Tryptone selection in +/UAS-FIT vs Fit-Gal4/UAS-FIT: p = 1.84E-4. No pre-feeding condition: Sucrose selection in Fit-Gal4/+ vs +/UAS-FIT: p = 0.78, Sucrose selection in Fit-Gal4/+ vs Fit-Gal4/UAS-FIT: p = 0.23, Sucrose selection in +/UAS-FIT vs Fit-Gal4/UAS-FIT: p = 0.35; Tryptone selection in Fit-Gal4/+ vs +/UAS-FIT: p = 0.84, Tryptone selection in Fit-Gal4/+ vs Fit-Gal4/UAS-FIT: p = 0.13, Tryptone selection in + /UAS-FIT vs Fit-Gal4/UAS-FIT: p = Tryptone pre-feeding condition: Sucrose selection in Fit-Gal4/+ vs +/UAS-FIT: p=0.78, Sucrose selection in Fit-Gal4/+ vs Fit-Gal4/UAS-FIT: p = 0.56, Sucrose selection in + /UAS-FIT vs Fit-Gal4/UAS-FIT: p = 0.76; Tryptone selection in Fit-Gal4/+ vs +/UAS-FIT: p = 0.39, Tryptone selection in Fit-Gal4/+ vs Fit-Gal4/UAS-FIT: p = 0.28, Tryptone selection in + /UAS-FIT vs Fit-Gal4/UAS-FIT: p = Signal peptide prediction in - -
25 5a SignalP 4.1 5b 5c 5d 5e 5f 6a 6b - - Western Blot HA, GAPDH - Fit-Gal4/UAS-FIT-HA Fit-Gal4/+;+/UAS-FITΔSP-HA UAS-FIT /+ Fit-Gal4/UAS-FIT UAS-FIT /+; +/Lsp2-Gal4 UAS-FIT /Ppl-Gal4 UAS-FITΔSP /+ Ppl-Gal4/+;+/UAS-FITΔSP-HA Elav-Gal4/+ UAS-FIT /+ Elav-Gal4/+;+ /UAS-FIT UAS-InR DN /+ Elav-Gal4/+;+ / UAS-InR DN UAS-FIT/ UAS-InR DN Elav-Gal4/+; UAS-FIT/ UAS-InR DN Elav-Gal4/+ UAS-FIT /+ Elav-Gal4/+;+ /UAS-FIT UAS-InR DN /+ Elav-Gal4/+;+ / UAS-InR DN UAS-Fit/ UAS-InR DN Elav-Gal4/+; UAS-FIT/ UAS-InR DN - Western Blot HA, GAPDH - - Feeding Assay: - Feeding Assay: - Two Choice Assay: Pre-feed (Sucrose) Test (Tryptone or Sucrose) Immunohistochemical stain ing Signal intensity statistics of staining 25 - Unpaired Student t test +/UAS-FIT vs Fit-Gal4>UAS-FIT: p = , +/UAS-FIT vs Lsp2-Gal4>UAS-FIT: p = 2.26E-4, +/UAS-FIT vs Ppl-Gal4>UAS-FIT: p = 2.01E-8, +/UAS-FIT SP vs Fit-Gal4>UAS-FIT SP: p = Unpaired Student t test +/UAS-FIT vs Elav-Gal4>UAS-FIT: p = 4.70E-5, +/UAS-InR DN vs Elav-Gal4>UAS-InR DN : p = 0.015, UAS-FIT ;UAS- InR DN vs Elav-Gal4/+; UAS-FIT/UAS-InR DN : p = , Elav-Gal4>UAS- InR DN vs Elav-Gal4/+; UAS-FIT/UAS-InR DN : p = Unpaired Student t test +/UAS-FIT vs Elav-Gal4>UAS-FIT: p = 1.23E-6, +/UAS-InR DN vs Elav-Gal4>UAS-InR DN : p = 0.65, UAS-FIT; UAS-InR DN vs Elav-Gal4/+;UAS-FIT/ UAS-InR DN : p = 0.26, Elav-Gal4>UAS-InR DN vs Elav-Gal4/+;UAS-FIT/UAS- InR DN : p = DILP2 - DILP2 Two-Way ANOVA female: pn-a = 9.77E-6, pt-a = 1.57E-6, ps-a = male: pt-a = 0.06.
26 6c 6d 6e 7a 7b 7c 7d 7e 7f 1b Dilp2-Gal4/+ Immunohistochemical stain Dilp2-Gal4/UAS-shi ts1 ing Dilp2-Gal4/+ Signal intensity statistics of Dilp2-Gal4/UAS-shi ts1 staining Fit 81 Immunohistochemical stain ing Signal intensity statistics of Fit 81 staining DILP2 - DILP2 DILP2 - DILP2 qpcr DILP2 Unpaired Student t test Fit 81 Immunohistochemical stain ing Signal intensity statistics of staining DILP2 - DILP Starvation (Agar) Pre-feeding (Normal Food, Tryptone, Sucrose) Test (Normal Food, Tryptone, Sucrose). - - Two-Way ANOVA Tryptone vs Agar in Dilp2-Gal4: p = 3.92E-5, Tryptone vs Agar in Dilp2-Gal4>UAS-shits1: p = One-Way ANOVA, Two-Way ANOVA female: Tryptone vs Agar in : p = 0.045, Tryptone vs Agar in Fit 81 : p = 0.15, Agar in Fit 81 vs Agar in : p = 2.78E-5. males: Tryptone vs Agar in p = 0.87, Tryptone vs Agar in Fit 81 p = One-Way ANOVA qpcr - One-Way ANOVA 26
27 2a,c, d, e 2b 2f 3a 3b 3d-f 4a 4b 4c qpcr - Unpaired Student t test Starvation (Agar) Pre-feeding (Agar, Agar+DMSO, Agar+DMSO+Rapamycin) Fit 81 Fit 52 PCR - - Immunohistochemical stain Fit 81 ing Body Weight per Fly (d) Fit 81 Protein/ Body Weight (e) TAG/Body Weight (f) Fit 81 Fit 81 Fit 52 Fit 81 wcs anti-fit Nile-Red One-Way ANOVA - - Unpaired Student t test CAFE - Unpaired Student t test - One-Way ANOVA Two-choice feeding assay - Unpaired Student t test Starvation (Agar) - One-Way ANOVA 27
28 5b Pre-feeding (Agar, Normal Food, Tryptone, Sucrose) 5c 5d 5e 6a 6b 6c wcs CS CS Starvation (Agar) Pre-feeding (Agar, Normal Food, Tryptone, Sucrose) Starvation (Agar) Pre-feeding (Agar, Normal Food, Tryptone, Sucrose) Starvation (Agar) Pre-feeding (Agar, Normal Food, Tryptone, Sucrose) Starvation (Agar) Pre-feeding (Agar, AA Mix) Starvation (Agar) Pre-feeding (Agar, T+S Mixed Food, Tryptone, Sucrose) Test (T+S Mixed Food) Starvation (Agar) Pre-feeding (Tryptone at - Two-Way ANOVA - One-Way ANOVA - Two-Way ANOVA - Unpaired Student t test - Unpaired Student t test Two-Way ANOVA - One-Way ANOVA 28
29 0%, 0.5%, 1.1%, 1.7%, or 2.3% ) Test (Tryptone at 1.7%) 6d : Mated Virgin qpcr - Two-Way ANOVA 6e-f : Mated Virgin Pre-feeding Tryptone) (Agar, - Unpaired Student t test 7a Fit 81 Pre-feeding (Agar, Normal Food, Sucrose) - Unpaired Student t test 7b Fit 52 Pre-feeding Assay : Pre-feeding (Agar, - Unpaired Student t test Tryptone) 7c Fit-Gal4/+ Fit-Gal4/+;Fit-RNAi/+ Pre-feeding (Agar, - Two-Way ANOVA Tryptone) 7d Fit-RNAi/+ Fit-Gal4/+;Fit-RNAi/+ Elav-Gal4/+;Fit-RNAi/+ Dilp2-Gal4/+;Fit-RNAi/+ Pre-feeding Tryptone) (Agar, - d: Two-Way ANOVA e: One-Way ANOVA 29
30 7e Fit-RNAi/+ Fit-Gal4/+;Fit-RNAi/+ Elav-Gal4/+;Fit-RNAi/+ Dilp2-Gal4/+;Fit-RNAi/+ Pre-feeding Tryptone) (Agar, - One-Way ANOVA 7f Fit-Gal4/+;Fit 81 UAS-FIT /+;Fit 81 Fit-Gal4/ UAS-FIT ;Fit 81 Pre-feeding (Agar, - Two-Way ANOVA Tryptone) 8a Pre-feeding (Agar, Tryptone) - Two-Way ANOVA 8b Pre-feeding (Agar, Tryptone) - One-Way ANOVA 8c Fit-Gal4/ slif-anti Fit-Gal4/TSC1/2 Pre-feeding (Agar, - Two-Way ANOVA Tryptone) 8d Fit-Gal4/ slif-anti Fit-Gal4/TSC1/2 Pre-feeding (Agar, - One-Way ANOVA Tryptone) 30
31 9a 9b 9c 9d 9e 10a 10b 10c Fit-Gal4/UAS-FIT-HA Fit-Gal4/+;+/UAS-FITΔSP-HA - Western Blot HA, GAPDH Western Blot HA, GAPDH - UAS-FIT /+ Dilp2-Gal4/UAS-FIT/+ Elav-Gal4/+; UAS-FIT /+ +/UAS-FITΔSP-HA Elav-Gal4/+; +/UAS-FITΔSP-HA Elav-GSG/ UAS-FIT Elav-Gal4/+ Elav-Gal4/+;+ /UAS-FIT Elav-Gal4/+;+ / UAS-InR DN Elav-Gal4/+; UAS-FIT/ UAS-InR DN - Unpaired Student t test - Two-Way ANOVA - Two Choice Assay - One-Way ANOVA Overall Mean Intensity of DILP2, Total Volume of DILP2 Signal Dilp2-Gal4/+ Dilp2-Gal4/UAS-Shi ts1 Overall Mean Intensity of DILP2, Total Volume of DILP2 Signal Overall Mean Intensity of Fit 81 DILP2, Total Volume of DILP2 Signal DILP2 DILP2 DILP2 Two-Way ANOVA Two-Way ANOVA Two-Way ANOVA 31
32 10d 10e 10f Overall Mean Intensity of DILP2, Total Volume of DILP2 Signal UAS-FIT /+;+/Dilp2-Gal4 Dilp2-Gal4/+;UAS-FITΔSP-HA /+ Dilp2-Gal4/UAS-shi ts1 ;+/UAS-FITΔSP-HA Dilp2-Gal4/UAS-shi ts1 ; UAS-FIT /+ UAS-FIT /+;+/Dilp2-Gal4 Dilp2-Gal4/+;UAS-FITΔSP-HA /+ Dilp2-Gal4/UAS-shi ts1 ;+/UAS-FITΔSP-HA Dilp2-Gal4/UAS-shi ts1 ; UAS-FIT /+ Immunohistochemical stain ing Total Fluorescence Intensity, Overall Mean Intensity of DILP2, Total Volume of DILP2 Signal. DILP2 One-Way ANOVA DILP2 - DILP2 Two-Way ANOVA 32
effects on organ development. a-f, Eye and wing discs with clones of ε j2b10 show no
 Supplementary Figure 1. Loss of function clones of 14-3-3 or 14-3-3 show no significant effects on organ development. a-f, Eye and wing discs with clones of 14-3-3ε j2b10 show no obvious defects in Elav
Supplementary Figure 1. Loss of function clones of 14-3-3 or 14-3-3 show no significant effects on organ development. a-f, Eye and wing discs with clones of 14-3-3ε j2b10 show no obvious defects in Elav
marker. DAPI labels nuclei. Flies were 20 days old. Scale bar is 5 µm. Ctrl is
 Supplementary Figure 1. (a) Nos is detected in glial cells in both control and GFAP R79H transgenic flies (arrows), but not in deletion mutant Nos Δ15 animals. Repo is a glial cell marker. DAPI labels
Supplementary Figure 1. (a) Nos is detected in glial cells in both control and GFAP R79H transgenic flies (arrows), but not in deletion mutant Nos Δ15 animals. Repo is a glial cell marker. DAPI labels
Supplementary Figure 1. Procedures to independently control fly hunger and thirst states.
 Supplementary Figure 1 Procedures to independently control fly hunger and thirst states. (a) Protocol to produce exclusively hungry or thirsty flies for 6 h water memory retrieval. (b) Consumption assays
Supplementary Figure 1 Procedures to independently control fly hunger and thirst states. (a) Protocol to produce exclusively hungry or thirsty flies for 6 h water memory retrieval. (b) Consumption assays
50mM D-Glucose. Percentage PI. L-Glucose
 Monica Dus, Minrong Ai, Greg S.B Suh. Taste-independent nutrient selection is mediated by a brain-specific Na+/solute cotransporter in Drosophila. Control + Phlorizin mm D-Glucose 1 2mM 1 L-Glucose Gr5a;Gr64a
Monica Dus, Minrong Ai, Greg S.B Suh. Taste-independent nutrient selection is mediated by a brain-specific Na+/solute cotransporter in Drosophila. Control + Phlorizin mm D-Glucose 1 2mM 1 L-Glucose Gr5a;Gr64a
Supplementary Figure 1 hlrrk2 promotes CAP dependent protein translation.
 ` Supplementary Figure 1 hlrrk2 promotes CAP dependent protein translation. (a) Overexpression of hlrrk2 in HeLa cells enhances total protein synthesis in [35S] methionine/cysteine incorporation assays.
` Supplementary Figure 1 hlrrk2 promotes CAP dependent protein translation. (a) Overexpression of hlrrk2 in HeLa cells enhances total protein synthesis in [35S] methionine/cysteine incorporation assays.
Supplementary Figures
 J. Cell Sci. 128: doi:10.1242/jcs.173807: Supplementary Material Supplementary Figures Fig. S1 Fig. S1. Description and/or validation of reagents used. All panels show Drosophila tissues oriented with
J. Cell Sci. 128: doi:10.1242/jcs.173807: Supplementary Material Supplementary Figures Fig. S1 Fig. S1. Description and/or validation of reagents used. All panels show Drosophila tissues oriented with
Transgenic Mice and Genetargeting
 Transgenic Mice and Genetargeting mice In Biomedical Science Techniques of transgenic and gene-targeting mice are indispensable for analyses of in vivo functions of particular genes and roles of their
Transgenic Mice and Genetargeting mice In Biomedical Science Techniques of transgenic and gene-targeting mice are indispensable for analyses of in vivo functions of particular genes and roles of their
SUPPLEMENTARY INFORMATION
 Supplementary Figure 1. Behavioural effects of ketamine in non-stressed and stressed mice. Naive C57BL/6 adult male mice (n=10/group) were given a single dose of saline vehicle or ketamine (3.0 mg/kg,
Supplementary Figure 1. Behavioural effects of ketamine in non-stressed and stressed mice. Naive C57BL/6 adult male mice (n=10/group) were given a single dose of saline vehicle or ketamine (3.0 mg/kg,
SUPPLEMENTARY INFORMATION Glucosylceramide synthase (GlcT-1) in the fat body controls energy metabolism in Drosophila
 SUPPLEMENTARY INFORMATION Glucosylceramide synthase (GlcT-1) in the fat body controls energy metabolism in Drosophila Ayako Kohyama-Koganeya, 1,2 Takuji Nabetani, 1 Masayuki Miura, 2,3 Yoshio Hirabayashi
SUPPLEMENTARY INFORMATION Glucosylceramide synthase (GlcT-1) in the fat body controls energy metabolism in Drosophila Ayako Kohyama-Koganeya, 1,2 Takuji Nabetani, 1 Masayuki Miura, 2,3 Yoshio Hirabayashi
BIOCHEMISTRY REVIEW. Overview of Biomolecules. Chapter 4 Protein Sequence
 BIOCHEMISTRY REVIEW Overview of Biomolecules Chapter 4 Protein Sequence 2 3 4 Are You Getting It?? A molecule of hemoglobin is compared with a molecule of lysozyme. Which characteristics do they share?
BIOCHEMISTRY REVIEW Overview of Biomolecules Chapter 4 Protein Sequence 2 3 4 Are You Getting It?? A molecule of hemoglobin is compared with a molecule of lysozyme. Which characteristics do they share?
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
Nature Methods: doi: /nmeth Supplementary Figure 1
 Supplementary Figure 1 Subtiligase-catalyzed ligations with ubiquitin thioesters and 10-mer biotinylated peptides. (a) General scheme for ligations between ubiquitin thioesters and 10-mer, biotinylated
Supplementary Figure 1 Subtiligase-catalyzed ligations with ubiquitin thioesters and 10-mer biotinylated peptides. (a) General scheme for ligations between ubiquitin thioesters and 10-mer, biotinylated
Algal Biofuels Research: Using basic science to maximize fuel output. Jacob Dums, PhD candidate, Heike Sederoff Lab March 9, 2015
 Algal Biofuels Research: Using basic science to maximize fuel output Jacob Dums, PhD candidate, jtdums@ncsu.edu Heike Sederoff Lab March 9, 2015 Outline Research Approach Dunaliella Increase Oil Content
Algal Biofuels Research: Using basic science to maximize fuel output Jacob Dums, PhD candidate, jtdums@ncsu.edu Heike Sederoff Lab March 9, 2015 Outline Research Approach Dunaliella Increase Oil Content
Supporting Information
 Supporting Information Fig. S1. Overexpression of Rpr causes progenitor cell death. (A) TUNEL assay of control intestines. No progenitor cell death could be observed, except that some ECs are undergoing
Supporting Information Fig. S1. Overexpression of Rpr causes progenitor cell death. (A) TUNEL assay of control intestines. No progenitor cell death could be observed, except that some ECs are undergoing
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature06994 A phosphatase cascade by which rewarding stimuli control nucleosomal response A. Stipanovich*, E. Valjent*, M. Matamales*, A. Nishi, J.H. Ahn, M. Maroteaux, J. Bertran-Gonzalez,
doi: 10.1038/nature06994 A phosphatase cascade by which rewarding stimuli control nucleosomal response A. Stipanovich*, E. Valjent*, M. Matamales*, A. Nishi, J.H. Ahn, M. Maroteaux, J. Bertran-Gonzalez,
100 mm Sucrose. +Berberine +Quinine
 8 mm Sucrose Probability (%) 7 6 5 4 3 Wild-type Gr32a / 2 +Caffeine +Berberine +Quinine +Denatonium Supplementary Figure 1: Detection of sucrose and bitter compounds is not affected in Gr32a / flies.
8 mm Sucrose Probability (%) 7 6 5 4 3 Wild-type Gr32a / 2 +Caffeine +Berberine +Quinine +Denatonium Supplementary Figure 1: Detection of sucrose and bitter compounds is not affected in Gr32a / flies.
Figure S1. Effect of bafilomycin on EGF-induced Akt and Erk signaling. Effect of chloroquine on EGF-stimulated mtorc1, Akt and Erk
 EGF induced VATPase assembly and mtorc1 activation Supplemental Information Supplemental Figure Legends Figure S1. Effect of bafilomycin on EGFinduced Akt and Erk signaling. A. Hepatocytes were treated
EGF induced VATPase assembly and mtorc1 activation Supplemental Information Supplemental Figure Legends Figure S1. Effect of bafilomycin on EGFinduced Akt and Erk signaling. A. Hepatocytes were treated
Supplementary Figure-1. SDS PAGE analysis of purified designed carbonic anhydrase enzymes. M1-M4 shown in lanes 1-4, respectively, with molecular
 Supplementary Figure-1. SDS PAGE analysis of purified designed carbonic anhydrase enzymes. M1-M4 shown in lanes 1-4, respectively, with molecular weight markers (M). Supplementary Figure-2. Overlay of
Supplementary Figure-1. SDS PAGE analysis of purified designed carbonic anhydrase enzymes. M1-M4 shown in lanes 1-4, respectively, with molecular weight markers (M). Supplementary Figure-2. Overlay of
Supplementary Fig. 1 V-ATPase depletion induces unique and robust phenotype in Drosophila fat body cells.
 Supplementary Fig. 1 V-ATPase depletion induces unique and robust phenotype in Drosophila fat body cells. a. Schematic of the V-ATPase proton pump macro-complex structure. The V1 complex is composed of
Supplementary Fig. 1 V-ATPase depletion induces unique and robust phenotype in Drosophila fat body cells. a. Schematic of the V-ATPase proton pump macro-complex structure. The V1 complex is composed of
Nature Neuroscience: doi: /nn Supplementary Figure 1. Neuron class-specific arrangements of Khc::nod::lacZ label in dendrites.
 Supplementary Figure 1 Neuron class-specific arrangements of Khc::nod::lacZ label in dendrites. Staining with fluorescence antibodies to detect GFP (Green), β-galactosidase (magenta/white). (a, b) Class
Supplementary Figure 1 Neuron class-specific arrangements of Khc::nod::lacZ label in dendrites. Staining with fluorescence antibodies to detect GFP (Green), β-galactosidase (magenta/white). (a, b) Class
Supplementary Figure 1. Flies form water-reward memory only in the thirsty state
 1 2 3 4 5 6 7 Supplementary Figure 1. Flies form water-reward memory only in the thirsty state Thirsty but not sated wild-type flies form robust 3 min memory. For the thirsty group, the flies were water-deprived
1 2 3 4 5 6 7 Supplementary Figure 1. Flies form water-reward memory only in the thirsty state Thirsty but not sated wild-type flies form robust 3 min memory. For the thirsty group, the flies were water-deprived
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
Supplementary Table 1. The primers used for quantitative RT-PCR. Gene name Forward (5 > 3 ) Reverse (5 > 3 )
 770 771 Supplementary Table 1. The primers used for quantitative RT-PCR. Gene name Forward (5 > 3 ) Reverse (5 > 3 ) Human CXCL1 GCGCCCAAACCGAAGTCATA ATGGGGGATGCAGGATTGAG PF4 CCCCACTGCCCAACTGATAG TTCTTGTACAGCGGGGCTTG
770 771 Supplementary Table 1. The primers used for quantitative RT-PCR. Gene name Forward (5 > 3 ) Reverse (5 > 3 ) Human CXCL1 GCGCCCAAACCGAAGTCATA ATGGGGGATGCAGGATTGAG PF4 CCCCACTGCCCAACTGATAG TTCTTGTACAGCGGGGCTTG
LAB#23: Biochemical Evidence of Evolution Name: Period Date :
 LAB#23: Biochemical Evidence of Name: Period Date : Laboratory Experience #23 Bridge Worth 80 Lab Minutes If two organisms have similar portions of DNA (genes), these organisms will probably make similar
LAB#23: Biochemical Evidence of Name: Period Date : Laboratory Experience #23 Bridge Worth 80 Lab Minutes If two organisms have similar portions of DNA (genes), these organisms will probably make similar
Supplementary Table 1. List of primers used in this study
 Supplementary Table 1. List of primers used in this study Gene Forward primer Reverse primer Rat Met 5 -aggtcgcttcatgcaggt-3 5 -tccggagacacaggatgg-3 Rat Runx1 5 -cctccttgaaccactccact-3 5 -ctggatctgcctggcatc-3
Supplementary Table 1. List of primers used in this study Gene Forward primer Reverse primer Rat Met 5 -aggtcgcttcatgcaggt-3 5 -tccggagacacaggatgg-3 Rat Runx1 5 -cctccttgaaccactccact-3 5 -ctggatctgcctggcatc-3
A post-ingestive amino acid sensor promotes food consumption in Drosophila
 www.nature.com/cr www.cell-research.com ARTICLE OPEN A post-ingestive amino acid sensor promotes food consumption in Drosophila Zhe Yang 1,2, Rui Huang 3,4, Xin Fu 5,6,7, Gaohang Wang 1,2, Wei Qi 1,2,
www.nature.com/cr www.cell-research.com ARTICLE OPEN A post-ingestive amino acid sensor promotes food consumption in Drosophila Zhe Yang 1,2, Rui Huang 3,4, Xin Fu 5,6,7, Gaohang Wang 1,2, Wei Qi 1,2,
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2566 Figure S1 CDKL5 protein expression pattern and localization in mouse brain. (a) Multiple-tissue western blot from a postnatal day (P) 21 mouse probed with an antibody against CDKL5.
DOI: 10.1038/ncb2566 Figure S1 CDKL5 protein expression pattern and localization in mouse brain. (a) Multiple-tissue western blot from a postnatal day (P) 21 mouse probed with an antibody against CDKL5.
Supplementary Figure 1 Madm is not required in GSCs and hub cells. (a,b) Act-Gal4-UAS-GFP (a), Act-Gal4-UAS- GFP.nls (b,c) is ubiquitously expressed
 Supplementary Figure 1 Madm is not required in GSCs and hub cells. (a,b) Act-Gal4-UAS-GFP (a), Act-Gal4-UAS- GFP.nls (b,c) is ubiquitously expressed in the testes. The testes were immunostained with GFP
Supplementary Figure 1 Madm is not required in GSCs and hub cells. (a,b) Act-Gal4-UAS-GFP (a), Act-Gal4-UAS- GFP.nls (b,c) is ubiquitously expressed in the testes. The testes were immunostained with GFP
Arginine side chain interactions and the role of arginine as a mobile charge carrier in voltage sensitive ion channels. Supplementary Information
 Arginine side chain interactions and the role of arginine as a mobile charge carrier in voltage sensitive ion channels Craig T. Armstrong, Philip E. Mason, J. L. Ross Anderson and Christopher E. Dempsey
Arginine side chain interactions and the role of arginine as a mobile charge carrier in voltage sensitive ion channels Craig T. Armstrong, Philip E. Mason, J. L. Ross Anderson and Christopher E. Dempsey
SUPPLEMENTARY FIGURES AND TABLE
 SUPPLEMENTARY FIGURES AND TABLE Supplementary Figure S1: Characterization of IRE1α mutants. A. U87-LUC cells were transduced with the lentiviral vector containing the GFP sequence (U87-LUC Tet-ON GFP).
SUPPLEMENTARY FIGURES AND TABLE Supplementary Figure S1: Characterization of IRE1α mutants. A. U87-LUC cells were transduced with the lentiviral vector containing the GFP sequence (U87-LUC Tet-ON GFP).
1. Describe the relationship of dietary protein and the health of major body systems.
 Food Explorations Lab I: The Building Blocks STUDENT LAB INVESTIGATIONS Name: Lab Overview In this investigation, you will be constructing animal and plant proteins using beads to represent the amino acids.
Food Explorations Lab I: The Building Blocks STUDENT LAB INVESTIGATIONS Name: Lab Overview In this investigation, you will be constructing animal and plant proteins using beads to represent the amino acids.
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1 Schematic depiction of the tandem Fc GDF15. Supplementary Figure 2 Supplementary Figure 2 Gfral mrna levels in the brains of both wild-type and knockout Gfral
Supplementary Figure 1 Supplementary Figure 1 Schematic depiction of the tandem Fc GDF15. Supplementary Figure 2 Supplementary Figure 2 Gfral mrna levels in the brains of both wild-type and knockout Gfral
Supplementary Figure S1 Supplementary Figure S2
 Supplementary Figure S A) The blots shown in Figure B were qualified by using Gel-Pro analyzer software (Rockville, MD, USA). The ratio of LC3II/LC3I to actin was then calculated. The data are represented
Supplementary Figure S A) The blots shown in Figure B were qualified by using Gel-Pro analyzer software (Rockville, MD, USA). The ratio of LC3II/LC3I to actin was then calculated. The data are represented
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
EGFR shrna A: CCGGCGCAAGTGTAAGAAGTGCGAACTCGAGTTCGCACTTCTTACACTTGCG TTTTTG. EGFR shrna B: CCGGAGAATGTGGAATACCTAAGGCTCGAGCCTTAGGTATTCCACATTCTCTT TTTG
 Supplementary Methods Sequence of oligonucleotides used for shrna targeting EGFR EGFR shrna were obtained from the Harvard RNAi consortium. The following oligonucleotides (forward primer) were used to
Supplementary Methods Sequence of oligonucleotides used for shrna targeting EGFR EGFR shrna were obtained from the Harvard RNAi consortium. The following oligonucleotides (forward primer) were used to
TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer
 Supplementary Information TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer Yabing Mu, Reshma Sundar, Noopur Thakur, Maria Ekman, Shyam Kumar Gudey, Mariya
Supplementary Information TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer Yabing Mu, Reshma Sundar, Noopur Thakur, Maria Ekman, Shyam Kumar Gudey, Mariya
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 Characterization of stable expression of GlucB and sshbira in the CT26 cell line (a) Live cell imaging of stable CT26 cells expressing green fluorescent protein
Supplementary Figures Supplementary Figure 1 Characterization of stable expression of GlucB and sshbira in the CT26 cell line (a) Live cell imaging of stable CT26 cells expressing green fluorescent protein
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1. Generation and validation of mtef4-knockout mice.
 Supplementary Figure 1 Generation and validation of mtef4-knockout mice. (a) Alignment of EF4 (E. coli) with mouse, yeast and human EF4. (b) Domain structures of mouse mtef4 compared to those of EF4 (E.
Supplementary Figure 1 Generation and validation of mtef4-knockout mice. (a) Alignment of EF4 (E. coli) with mouse, yeast and human EF4. (b) Domain structures of mouse mtef4 compared to those of EF4 (E.
Supplementary Materials and Methods
 Supplementary Materials and Methods Reagents and antibodies was purchased from iaffin GmbH & Co KG. Cisplatin (ristol-myers Squibb Co.) and etoposide (Sandoz Pharma Ltd.) were used. Antibodies recognizing
Supplementary Materials and Methods Reagents and antibodies was purchased from iaffin GmbH & Co KG. Cisplatin (ristol-myers Squibb Co.) and etoposide (Sandoz Pharma Ltd.) were used. Antibodies recognizing
Zhu et al, page 1. Supplementary Figures
 Zhu et al, page 1 Supplementary Figures Supplementary Figure 1: Visual behavior and avoidance behavioral response in EPM trials. (a) Measures of visual behavior that performed the light avoidance behavior
Zhu et al, page 1 Supplementary Figures Supplementary Figure 1: Visual behavior and avoidance behavioral response in EPM trials. (a) Measures of visual behavior that performed the light avoidance behavior
Supplementary Fig. 1: TBR2+ cells in different brain regions.
 Hip SVZ OB Cere Hypo Supplementary Fig. 1: TBR2 + cells in different brain regions. Three weeks after the last tamoxifen injection, TBR2 immunostaining images reveal a large reduction of TBR2 + cells in
Hip SVZ OB Cere Hypo Supplementary Fig. 1: TBR2 + cells in different brain regions. Three weeks after the last tamoxifen injection, TBR2 immunostaining images reveal a large reduction of TBR2 + cells in
Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis
 Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis of PD-L1 in ovarian cancer cells. (c) Western blot analysis
Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis of PD-L1 in ovarian cancer cells. (c) Western blot analysis
GFP/Iba1/GFAP. Brain. Liver. Kidney. Lung. Hoechst/Iba1/TLR9!
 Supplementary information a +KA Relative expression d! Tlr9 5!! 5! NSC Neuron Astrocyte Microglia! 5! Tlr7!!!! NSC Neuron Astrocyte! GFP/Sβ/! Iba/Hoechst Microglia e Hoechst/Iba/TLR9! GFP/Iba/GFAP f Brain
Supplementary information a +KA Relative expression d! Tlr9 5!! 5! NSC Neuron Astrocyte Microglia! 5! Tlr7!!!! NSC Neuron Astrocyte! GFP/Sβ/! Iba/Hoechst Microglia e Hoechst/Iba/TLR9! GFP/Iba/GFAP f Brain
Properties of amino acids in proteins
 Properties of amino acids in proteins one of the primary roles of DNA (but far from the only one!!!) is to code for proteins A typical bacterium builds thousands types of proteins, all from ~20 amino acids
Properties of amino acids in proteins one of the primary roles of DNA (but far from the only one!!!) is to code for proteins A typical bacterium builds thousands types of proteins, all from ~20 amino acids
Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated
 Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C genes. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated from 7 day-old seedlings treated with or without
Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C genes. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated from 7 day-old seedlings treated with or without
Amino Acids. Amino Acids. Fundamentals. While their name implies that amino acids are compounds that contain an NH. 3 and CO NH 3
 Fundamentals While their name implies that amino acids are compounds that contain an 2 group and a 2 group, these groups are actually present as 3 and 2 respectively. They are classified as α, β, γ, etc..
Fundamentals While their name implies that amino acids are compounds that contain an 2 group and a 2 group, these groups are actually present as 3 and 2 respectively. They are classified as α, β, γ, etc..
SUPPLEMENTARY INFORMATION. Supplementary Figures S1-S9. Supplementary Methods
 SUPPLEMENTARY INFORMATION SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane Jian Huang 1,2#, Jie Yan 1,2#, Jian Zhang 3#, Shiguo Zhu 1, Yanli Wang
SUPPLEMENTARY INFORMATION SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane Jian Huang 1,2#, Jie Yan 1,2#, Jian Zhang 3#, Shiguo Zhu 1, Yanli Wang
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells
 Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Table S1. Gal4 Driver Lines in This Study, Related to Figures 1, 2, 3, 4, 5, 6, S1, S2, S3, S4, S5, and S6
 Table S1. Gal4 Driver Lines in This Study, Related to s 1, 2, 3, 4, 5, 6, S1, S2, S3, S4, S5, and S6 Genotype Abbreviation Expression patterns Ubi-Gal4 Ubi Ubiquitous dome-gal4;elav-gal80 dome MZ Progenitor-specific
Table S1. Gal4 Driver Lines in This Study, Related to s 1, 2, 3, 4, 5, 6, S1, S2, S3, S4, S5, and S6 Genotype Abbreviation Expression patterns Ubi-Gal4 Ubi Ubiquitous dome-gal4;elav-gal80 dome MZ Progenitor-specific
Four melanocyte-stimulating hormones have the following amino acid sequences:
 Assignment 14: Melanocyte-stimulating hormone belongs to a group called the melanocortins. This group includes ACTH, alpha-msh, beta-msh and gamma-msh; these peptides are all cleavage products of a large
Assignment 14: Melanocyte-stimulating hormone belongs to a group called the melanocortins. This group includes ACTH, alpha-msh, beta-msh and gamma-msh; these peptides are all cleavage products of a large
Amino Acids : Towards Precise Nutrition in Monogastric Animals
 [AFMA FORUM 2016 SESSION 4] Amino Acids : Towards Precise Nutrition in Monogastric Animals AFMA 2016 CORRENT AMINO ACIDS 2016 All rights reserved AJINOMOTO EUROLYSINE S.A.S. 1 Dietary crude protein reduction
[AFMA FORUM 2016 SESSION 4] Amino Acids : Towards Precise Nutrition in Monogastric Animals AFMA 2016 CORRENT AMINO ACIDS 2016 All rights reserved AJINOMOTO EUROLYSINE S.A.S. 1 Dietary crude protein reduction
Title: Smooth muscle cell-specific Tgfbr1 deficiency promotes aortic aneurysm formation by stimulating multiple signaling events
 Title: Smooth muscle cell-specific Tgfbr1 deficiency promotes aortic aneurysm formation by stimulating multiple signaling events Pu Yang 1, 3, radley M. Schmit 1, Chunhua Fu 1, Kenneth DeSart 1, S. Paul
Title: Smooth muscle cell-specific Tgfbr1 deficiency promotes aortic aneurysm formation by stimulating multiple signaling events Pu Yang 1, 3, radley M. Schmit 1, Chunhua Fu 1, Kenneth DeSart 1, S. Paul
Histone acetyltransferase CBP-related H3K23 acetylation contributes to courtship learning in Drosophila
 Li et al. BMC Developmental Biology (2018) 18:20 https://doi.org/10.1186/s12861-018-0179-z RESEARCH ARTICLE Open Access Histone acetyltransferase CBP-related H3K23 acetylation contributes to courtship
Li et al. BMC Developmental Biology (2018) 18:20 https://doi.org/10.1186/s12861-018-0179-z RESEARCH ARTICLE Open Access Histone acetyltransferase CBP-related H3K23 acetylation contributes to courtship
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Supplementary Figure 1. (A) Left, western blot analysis of ISGylated proteins in Jurkat T cells treated with 1000U ml -1 IFN for 16h (IFN) or left untreated (CONT); right, western
SUPPLEMENTARY FIGURES Supplementary Figure 1. (A) Left, western blot analysis of ISGylated proteins in Jurkat T cells treated with 1000U ml -1 IFN for 16h (IFN) or left untreated (CONT); right, western
TGF-β Signaling Regulates Neuronal C1q Expression and Developmental Synaptic Refinement
 Supplementary Information Title: TGF-β Signaling Regulates Neuronal C1q Expression and Developmental Synaptic Refinement Authors: Allison R. Bialas and Beth Stevens Supplemental Figure 1. In vitro characterization
Supplementary Information Title: TGF-β Signaling Regulates Neuronal C1q Expression and Developmental Synaptic Refinement Authors: Allison R. Bialas and Beth Stevens Supplemental Figure 1. In vitro characterization
Page 8/6: The cell. Where to start: Proteins (control a cell) (start/end products)
 Page 8/6: The cell Where to start: Proteins (control a cell) (start/end products) Page 11/10: Structural hierarchy Proteins Phenotype of organism 3 Dimensional structure Function by interaction THE PROTEIN
Page 8/6: The cell Where to start: Proteins (control a cell) (start/end products) Page 11/10: Structural hierarchy Proteins Phenotype of organism 3 Dimensional structure Function by interaction THE PROTEIN
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 Expression of escargot (esg) and genetic approach for achieving IPC-specific knockdown. (a) esg MH766 -Gal4 UAS-cd8GFP (green) and esg-lacz B7-2-22 (red) show similar expression
Supplementary Figure 1 Expression of escargot (esg) and genetic approach for achieving IPC-specific knockdown. (a) esg MH766 -Gal4 UAS-cd8GFP (green) and esg-lacz B7-2-22 (red) show similar expression
reads observed in trnas from the analysis of RNAs carrying a 5 -OH ends isolated from cells induced to express
 Supplementary Figure 1. VapC-mt4 cleaves trna Ala2 in E. coli. Histograms representing the fold change in reads observed in trnas from the analysis of RNAs carrying a 5 -OH ends isolated from cells induced
Supplementary Figure 1. VapC-mt4 cleaves trna Ala2 in E. coli. Histograms representing the fold change in reads observed in trnas from the analysis of RNAs carrying a 5 -OH ends isolated from cells induced
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature07173 SUPPLEMENTARY INFORMATION Supplementary Figure Legends: Supplementary Figure 1: Model of SSC and CPC divisions a, Somatic stem cells (SSC) reside adjacent to the hub (red), self-renew
doi: 10.1038/nature07173 SUPPLEMENTARY INFORMATION Supplementary Figure Legends: Supplementary Figure 1: Model of SSC and CPC divisions a, Somatic stem cells (SSC) reside adjacent to the hub (red), self-renew
SIMPLE BASIC METABOLISM
 SIMPLE BASIC METABOLISM When we eat food such as a tuna fish sandwich, the polysaccharides, lipids, and proteins are digested to smaller molecules that are absorbed into the cells of our body. As these
SIMPLE BASIC METABOLISM When we eat food such as a tuna fish sandwich, the polysaccharides, lipids, and proteins are digested to smaller molecules that are absorbed into the cells of our body. As these
Supplementary Figure 1. Establishment of prostacyclin-secreting hmscs. (a) PCR showed the integration of the COX-1-10aa-PGIS transgene into the
 Supplementary Figure 1. Establishment of prostacyclin-secreting hmscs. (a) PCR showed the integration of the COX-1-10aa-PGIS transgene into the genomic DNA of hmscs (PGI2- hmscs). Native hmscs and plasmid
Supplementary Figure 1. Establishment of prostacyclin-secreting hmscs. (a) PCR showed the integration of the COX-1-10aa-PGIS transgene into the genomic DNA of hmscs (PGI2- hmscs). Native hmscs and plasmid
SUPPLEMENTARY LEGENDS...
 TABLE OF CONTENTS SUPPLEMENTARY LEGENDS... 2 11 MOVIE S1... 2 FIGURE S1 LEGEND... 3 FIGURE S2 LEGEND... 4 FIGURE S3 LEGEND... 5 FIGURE S4 LEGEND... 6 FIGURE S5 LEGEND... 7 FIGURE S6 LEGEND... 8 FIGURE
TABLE OF CONTENTS SUPPLEMENTARY LEGENDS... 2 11 MOVIE S1... 2 FIGURE S1 LEGEND... 3 FIGURE S2 LEGEND... 4 FIGURE S3 LEGEND... 5 FIGURE S4 LEGEND... 6 FIGURE S5 LEGEND... 7 FIGURE S6 LEGEND... 8 FIGURE
(A) RT-PCR for components of the Shh/Gli pathway in normal fetus cell (MRC-5) and a
 Supplementary figure legends Supplementary Figure 1. Expression of Shh signaling components in a panel of gastric cancer. (A) RT-PCR for components of the Shh/Gli pathway in normal fetus cell (MRC-5) and
Supplementary figure legends Supplementary Figure 1. Expression of Shh signaling components in a panel of gastric cancer. (A) RT-PCR for components of the Shh/Gli pathway in normal fetus cell (MRC-5) and
The Basics: A general review of molecular biology:
 The Basics: A general review of molecular biology: DNA Transcription RNA Translation Proteins DNA (deoxy-ribonucleic acid) is the genetic material It is an informational super polymer -think of it as the
The Basics: A general review of molecular biology: DNA Transcription RNA Translation Proteins DNA (deoxy-ribonucleic acid) is the genetic material It is an informational super polymer -think of it as the
Figure S1. Reduction in glomerular mir-146a levels correlate with progression to higher albuminuria in diabetic patients.
 Supplementary Materials Supplementary Figures Figure S1. Reduction in glomerular mir-146a levels correlate with progression to higher albuminuria in diabetic patients. Figure S2. Expression level of podocyte
Supplementary Materials Supplementary Figures Figure S1. Reduction in glomerular mir-146a levels correlate with progression to higher albuminuria in diabetic patients. Figure S2. Expression level of podocyte
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk
 Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Supplementary Information
 Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
Santulli G. et al. A microrna-based strategy to suppress restenosis while preserving endothelial function
 ONLINE DATA SUPPLEMENTS Santulli G. et al. A microrna-based strategy to suppress restenosis while preserving endothelial function Supplementary Figures Figure S1 Effect of Ad-p27-126TS on the expression
ONLINE DATA SUPPLEMENTS Santulli G. et al. A microrna-based strategy to suppress restenosis while preserving endothelial function Supplementary Figures Figure S1 Effect of Ad-p27-126TS on the expression
genome edited transient transfection, CMV promoter
 Supplementary Figure 1. In the absence of new protein translation, overexpressed caveolin-1-gfp is degraded faster than caveolin-1-gfp expressed from the endogenous caveolin 1 locus % loss of total caveolin-1-gfp
Supplementary Figure 1. In the absence of new protein translation, overexpressed caveolin-1-gfp is degraded faster than caveolin-1-gfp expressed from the endogenous caveolin 1 locus % loss of total caveolin-1-gfp
Supplementary Figure 1) GABAergic enhancement by leptin hyperpolarizes POMC neurons A) Representative recording samples showing the membrane
 Supplementary Figure 1) GABAergic enhancement by leptin hyperpolarizes POMC neurons A) Representative recording samples showing the membrane potential recorded from POMC neurons following treatment with
Supplementary Figure 1) GABAergic enhancement by leptin hyperpolarizes POMC neurons A) Representative recording samples showing the membrane potential recorded from POMC neurons following treatment with
What is most limiting?
 The Amino Acid Content of Rumen Microbes, Feed, Milk and Tissue after Multiple Hydrolysis Times and Implications for the CNCPS M. E. Van Amburgh, A. F. Ortega, S. W. Fessenden, D. A. Ross, and P. A. LaPierre
The Amino Acid Content of Rumen Microbes, Feed, Milk and Tissue after Multiple Hydrolysis Times and Implications for the CNCPS M. E. Van Amburgh, A. F. Ortega, S. W. Fessenden, D. A. Ross, and P. A. LaPierre
Optimizing Protein in a Carbohydrate World
 Optimizing Protein in a Carbohydrate World Donald K. Layman, Ph.D. Professor Emeritus Department of Food Science & Human Nutrition University of Illinois at Urbana Champaign The confused consumer 1 Myth:
Optimizing Protein in a Carbohydrate World Donald K. Layman, Ph.D. Professor Emeritus Department of Food Science & Human Nutrition University of Illinois at Urbana Champaign The confused consumer 1 Myth:
Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A.
 Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A. Upper part, three-primer PCR strategy at the Mcm3 locus yielding
Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A. Upper part, three-primer PCR strategy at the Mcm3 locus yielding
Free Amino Acid Changes in Serum throughout Rat Gestation and Lactation. Evolution of the Plasma/Serum Relationships
 J. Clin. Biochem. Nutr., 3, 73-77, 1987 Free Amino Acid Changes in Serum throughout Rat Gestation and Lactation. Evolution of the Plasma/Serum Relationships M. PASTOR-ANGLADA, D. LOPEZ-TEJERO, and X. REMESAR*
J. Clin. Biochem. Nutr., 3, 73-77, 1987 Free Amino Acid Changes in Serum throughout Rat Gestation and Lactation. Evolution of the Plasma/Serum Relationships M. PASTOR-ANGLADA, D. LOPEZ-TEJERO, and X. REMESAR*
Supplemental Figure 1. Western blot analysis indicated that MIF was detected in the fractions of
 Supplemental Figure Legends Supplemental Figure 1. Western blot analysis indicated that was detected in the fractions of plasma membrane and cytosol but not in nuclear fraction isolated from Pkd1 null
Supplemental Figure Legends Supplemental Figure 1. Western blot analysis indicated that was detected in the fractions of plasma membrane and cytosol but not in nuclear fraction isolated from Pkd1 null
Amino acids-incorporated nanoflowers with an
 Amino acids-incorporated nanoflowers with an intrinsic peroxidase-like activity Zhuo-Fu Wu 1,2,+, Zhi Wang 1,+, Ye Zhang 3, Ya-Li Ma 3, Cheng-Yan He 4, Heng Li 1, Lei Chen 1, Qi-Sheng Huo 3, Lei Wang 1,*
Amino acids-incorporated nanoflowers with an intrinsic peroxidase-like activity Zhuo-Fu Wu 1,2,+, Zhi Wang 1,+, Ye Zhang 3, Ya-Li Ma 3, Cheng-Yan He 4, Heng Li 1, Lei Chen 1, Qi-Sheng Huo 3, Lei Wang 1,*
Supplementary Figure 1: Additional metabolic parameters of obesity mouse models and controls. (a) Body weight, (b) blood glucose and (c) insulin
 Supplementary Figure 1: Additional metabolic parameters of obesity mouse models and controls. (a) Body weight, (b) blood glucose and (c) insulin resistance index of homeostatic model assessment (HOMA IR)
Supplementary Figure 1: Additional metabolic parameters of obesity mouse models and controls. (a) Body weight, (b) blood glucose and (c) insulin resistance index of homeostatic model assessment (HOMA IR)
Supplementary Figure S I: Effects of D4F on body weight and serum lipids in apoe -/- mice.
 Supplementary Figures: Supplementary Figure S I: Effects of D4F on body weight and serum lipids in apoe -/- mice. Male apoe -/- mice were fed a high-fat diet for 8 weeks, and given PBS (model group) or
Supplementary Figures: Supplementary Figure S I: Effects of D4F on body weight and serum lipids in apoe -/- mice. Male apoe -/- mice were fed a high-fat diet for 8 weeks, and given PBS (model group) or
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES 1 Supplementary Figure 1, Adult hippocampal QNPs and TAPs uniformly express REST a-b) Confocal images of adult hippocampal mouse sections showing GFAP (green), Sox2 (red), and REST
SUPPLEMENTARY FIGURES 1 Supplementary Figure 1, Adult hippocampal QNPs and TAPs uniformly express REST a-b) Confocal images of adult hippocampal mouse sections showing GFAP (green), Sox2 (red), and REST
Drosophila Cytokine Unpaired 2 Regulates Physiological Homeostasis by Remotely Controlling Insulin Secretion
 Drosophila Cytokine Unpaired 2 Regulates Physiological Homeostasis by Remotely Controlling Insulin Secretion Akhila Rajan 1, * and Norbert Perrimon 1,2, * 1 Department of Genetics 2 Howard Hughes Medical
Drosophila Cytokine Unpaired 2 Regulates Physiological Homeostasis by Remotely Controlling Insulin Secretion Akhila Rajan 1, * and Norbert Perrimon 1,2, * 1 Department of Genetics 2 Howard Hughes Medical
Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was
 Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was painted on the shaved back skin of CBL/J and BALB/c mice for consecutive days. (a, b) Phenotypic presentation of mouse back skin
Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was painted on the shaved back skin of CBL/J and BALB/c mice for consecutive days. (a, b) Phenotypic presentation of mouse back skin
c Ischemia (30 min) Reperfusion (8 w) Supplementary Figure bp 300 bp Ischemia (30 min) Reperfusion (4 h) Dox 20 mg/kg i.p.
 a Marker Ripk3 +/ 5 bp 3 bp b Ischemia (3 min) Reperfusion (4 h) d 2 mg/kg i.p. 1 w 5 w Sacrifice for IF size A subset for echocardiography and morphological analysis c Ischemia (3 min) Reperfusion (8
a Marker Ripk3 +/ 5 bp 3 bp b Ischemia (3 min) Reperfusion (4 h) d 2 mg/kg i.p. 1 w 5 w Sacrifice for IF size A subset for echocardiography and morphological analysis c Ischemia (3 min) Reperfusion (8
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS)
 Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid.
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
and follicular helper T cells is Egr2-dependent. (a) Diagrammatic representation of the
 Supplementary Figure 1. LAG3 + Treg-mediated regulation of germinal center B cells and follicular helper T cells is Egr2-dependent. (a) Diagrammatic representation of the experimental protocol for the
Supplementary Figure 1. LAG3 + Treg-mediated regulation of germinal center B cells and follicular helper T cells is Egr2-dependent. (a) Diagrammatic representation of the experimental protocol for the
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10643 Supplementary Table 1. Identification of hecw-1 coding polymorphisms at amino acid positions 322 and 325 in 162 strains of C. elegans. WWW.NATURE.COM/NATURE 1 Supplementary Figure
doi:10.1038/nature10643 Supplementary Table 1. Identification of hecw-1 coding polymorphisms at amino acid positions 322 and 325 in 162 strains of C. elegans. WWW.NATURE.COM/NATURE 1 Supplementary Figure
Supplementary Figure 1: Hsp60 / IEC mice are embryonically lethal (A) Light microscopic pictures show mouse embryos at developmental stage E12.
 Supplementary Figure 1: Hsp60 / IEC mice are embryonically lethal (A) Light microscopic pictures show mouse embryos at developmental stage E12.5 and E13.5 prepared from uteri of dams and subsequently genotyped.
Supplementary Figure 1: Hsp60 / IEC mice are embryonically lethal (A) Light microscopic pictures show mouse embryos at developmental stage E12.5 and E13.5 prepared from uteri of dams and subsequently genotyped.
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2211 a! mir-143! b! mir-103/107! let-7a! mir-144! mir-122a! mir-126-3p! mir-194! mir-27a! mir-30c! Figure S1 Northern blot analysis of mir-143 expression dependent on feeding conditions.
DOI: 10.1038/ncb2211 a! mir-143! b! mir-103/107! let-7a! mir-144! mir-122a! mir-126-3p! mir-194! mir-27a! mir-30c! Figure S1 Northern blot analysis of mir-143 expression dependent on feeding conditions.
Expanded View Figures
 PEX13 functions in selective autophagy Ming Y Lee et al Expanded View Figures Figure EV1. PEX13 is required for Sindbis virophagy. A, B Quantification of mcherry-capsid puncta per cell (A) and GFP-LC3
PEX13 functions in selective autophagy Ming Y Lee et al Expanded View Figures Figure EV1. PEX13 is required for Sindbis virophagy. A, B Quantification of mcherry-capsid puncta per cell (A) and GFP-LC3
Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus
 Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus changes in corresponding proteins between wild type and Gprc5a-/-
Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus changes in corresponding proteins between wild type and Gprc5a-/-
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Supplementary Figure 1.
 Supplementary Figure 1. Increased β cell mass and islet diameter in βtsc2 -/- mice up to 35 weeks A: Reconstruction of multiple anti-insulin immunofluorescence images showing differences in β cell mass
Supplementary Figure 1. Increased β cell mass and islet diameter in βtsc2 -/- mice up to 35 weeks A: Reconstruction of multiple anti-insulin immunofluorescence images showing differences in β cell mass
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature11429 S1a 6 7 8 9 Nlrc4 allele S1b Nlrc4 +/+ Nlrc4 +/F Nlrc4 F/F 9 Targeting construct 422 bp 273 bp FRT-neo-gb-PGK-FRT 3x.STOP S1c Nlrc4 +/+ Nlrc4 F/F casp1
SUPPLEMENTARY INFORMATION doi:10.1038/nature11429 S1a 6 7 8 9 Nlrc4 allele S1b Nlrc4 +/+ Nlrc4 +/F Nlrc4 F/F 9 Targeting construct 422 bp 273 bp FRT-neo-gb-PGK-FRT 3x.STOP S1c Nlrc4 +/+ Nlrc4 F/F casp1
(A) SW480, DLD1, RKO and HCT116 cells were treated with DMSO or XAV939 (5 µm)
 Supplementary Figure Legends Figure S1. Tankyrase inhibition suppresses cell proliferation in an axin/β-catenin independent manner. (A) SW480, DLD1, RKO and HCT116 cells were treated with DMSO or XAV939
Supplementary Figure Legends Figure S1. Tankyrase inhibition suppresses cell proliferation in an axin/β-catenin independent manner. (A) SW480, DLD1, RKO and HCT116 cells were treated with DMSO or XAV939
AP Bio. Protiens Chapter 5 1
 Concept.4: Proteins have many structures, resulting in a wide range of functions Proteins account for more than 0% of the dry mass of most cells Protein functions include structural support, storage, transport,
Concept.4: Proteins have many structures, resulting in a wide range of functions Proteins account for more than 0% of the dry mass of most cells Protein functions include structural support, storage, transport,
Objective: You will be able to explain how the subcomponents of
 Objective: You will be able to explain how the subcomponents of nucleic acids determine the properties of that polymer. Do Now: Read the first two paragraphs from enduring understanding 4.A Essential knowledge:
Objective: You will be able to explain how the subcomponents of nucleic acids determine the properties of that polymer. Do Now: Read the first two paragraphs from enduring understanding 4.A Essential knowledge:
Supplementary Figure 1
 Combination index (CI) Supplementary Figure 1 2. 1.5 1. Ishikawa AN3CA Nou-1 Hec-18.5...2.4.6.8 1. Fraction affected (Fa) Supplementary Figure 1. The synergistic effect of PARP inhibitor and PI3K inhibitor
Combination index (CI) Supplementary Figure 1 2. 1.5 1. Ishikawa AN3CA Nou-1 Hec-18.5...2.4.6.8 1. Fraction affected (Fa) Supplementary Figure 1. The synergistic effect of PARP inhibitor and PI3K inhibitor
Supplementary Figure 1. DNA methylation of the adiponectin promoter R1, Pparg2, and Tnfa promoter in adipocytes is not affected by obesity.
 Supplementary Figure 1. DNA methylation of the adiponectin promoter R1, Pparg2, and Tnfa promoter in adipocytes is not affected by obesity. (a) Relative amounts of adiponectin, Ppar 2, C/ebp, and Tnf mrna
Supplementary Figure 1. DNA methylation of the adiponectin promoter R1, Pparg2, and Tnfa promoter in adipocytes is not affected by obesity. (a) Relative amounts of adiponectin, Ppar 2, C/ebp, and Tnf mrna
Supplementary information
 Supplementary information 1 Supplementary Figure 1. CALM regulates autophagy. (a). Quantification of LC3 levels in the experiment described in Figure 1A. Data are mean +/- SD (n > 3 experiments for each
Supplementary information 1 Supplementary Figure 1. CALM regulates autophagy. (a). Quantification of LC3 levels in the experiment described in Figure 1A. Data are mean +/- SD (n > 3 experiments for each
