Analysis of Online Sleep Apnea Data Streams for Mobile Platforms
|
|
|
- Rodney Watson
- 5 years ago
- Views:
Transcription
1 Analysis of Online Sleep Apnea Data Streams for Mobile Platforms Steffen Lien Thesis submitted for the degree of Master in Informatics: Programming and Networks 60 credits Department of Informatics Faculty of mathematics and natural sciences UNIVERSITY OF OSLO Autumn 2016
2
3 Analysis of Online Sleep Apnea Data Streams for Mobile Platforms Steffen Lien 31st October 2016
4
5 Abstract Sleep apnea is a sleeping disorder characterized by disruption in the natural breathing cycles during sleep. The disorder is common and is linked together with other disabilities and disorders that have serious ramifications to a person s health. Diagnostic tools such as the polysomnography and portable monitoring devices can record physiological signals to determine a diagnose and severity rating of a patient. Before a diagnose can be determined, the recorded data needs to be manually analyzed by physicians which is a costly and time consuming process. With data mining, machine learning and event analysis the process can be redefined to be more efficient, automatic and require a fraction of the resources. Smart phone devices we have today are very capable of running advanced and complex software, and in some terms can be comparable to laptops. Data mining is a concept of classifying and predicting large amounts of data based on patterns and data analysis built using models. Esper is an open source Complex Event Processing engine and library component that use event series analysis to extract information and patterns in various types of data streams. In this thesis we design and implement four commonly used data mining methods: K-Nearest Neighbor, Support Vector Machine, Artificial Neural Network and Decision Tree. Along the data mining methods, we also design and implement two different detection methods in combination with statistical methods such as a Moving Average and Standard Deviation using the Event Processing Language utilized by Esper. We test both sets of classification methods on input data from three different database sources from PhysioNet. In addition, we analyze the performance utilization of the implementation with comparison with smart phone hardware to gather knowledge that can help develop a future automatic diagnosis application on smart phones. By streaming the data sets from a client to a server running Esper we achieve an accuracy of 90.89% using Decision Tree in a combination of four non-invasive signals. Overall accuracy results from all the data mining methods above 85% accuracy and close to 90% except for three signal combinations. Results from the two apnea detection methods score an accuracy of 93.26% and 93.13% for the Moving Average and Standard Deviation respectively. From the performance measurements we conclude that smart phones that is labeled as budget in terms of hardware and price is suitable to run data mining methods. The results overall show that both data mining methods and our designed detection method based on Esper combined with the performance metrics are suitable to implement as a standalone automatic diagnosis application on budget smart phones.
6 Acknowledgment I would like to thank my mom and dad for beign born, without them this thesis would have probably been done by someone else. I also want to thank Professor Dr. Vera Goebel for her great guidance and counselling regarding the work on this thesis. I would also like to thank "Assa-gjengen" for the five incredible years. I also want to thank Mari Hugaas Sønsteby for her amazing work in her thesis "Data Mining for the Detection of Disrupted Breathing Caused by Sleep Apnea - A Comparison of Methods". i
7 Contents 1 Introduction Background & Motivation Problem Statement Outline Sleep Apnea Characteristics Obstructive Sleep Apnea Symptoms Risk Factors Diagnosis Severity Rating Polysomnography Portable Monitoring Systems Questionnaires Treatments Challenges Data Mining Introduction Data Types Data Mining Tasks & Models Clustering Classification Regression Anomaly Detection Association Rules Summarization K-Nearest Neighbor Support Vector Machine Hard-margin Soft-margin Non-linear Decision Tree Artificial Neural Network Single-layer Perceptron Multi-layer Perceptron ii
8 4 Esper Complex Event Processing Rule-Based Classification Event Processing Language Performance Requirement Analysis Input Data Streaming Suitability Data Mining Sensor Simulation Apnea Detection Related Work Design System Overview Initial Design Revised Design PhysioNet Databases Apnea-ECG Database MIT-BIH Polysomnography Database St. Vincent s University Hospital Database Input Data Data Mining K-Nearest Neighbor Support Vector Machine Decision Tree Artificial Neural Network Apnea Detection Moving Average Standard Deviation Output Data Performance Evaluation Hardware Profiling Implementation System Environment Input Data WFDB Toolkit Apnea-ECG Database MIT-BIH Polysomnography & St. Vincent s Databases Client CLIParser ConnectionHandler EventEmitter Server ConfigurationLoader Classifier EventHandler QueryGenerator iii
9 7.4.5 SQLiteJDBC Data Mining K-Nearest Neighbor Support Vector Machine Decision Tree Artificial Neural Network Apnea Detection Moving Average Standard Deviation Storage Evaluation & Performance Analysis Performance Metrics & Configurations Data Mining Evaluation K-Nearest Neighbor Support Vector Machine Decision Tree Artificial Neural Network Apnea Detection Evaluation Moving Average Standard Deviation AHI Comparison Metric Comparison Offline Results Comparison Resource Performance & Utilization Virtual Machine Environment CPU Utilization Memory Utilization I/O Operations Conclusion Contributions Data Mining Apnea Detection Resource Profiling Discussion Future Work Sensor Data Detection Optimization Appendices 128 A Configuration & Runtime Setup 129 A.1 Dependencies A.1.1 Python A.1.2 Java A.2 Matlab A.2.1 Paths A.2.2 Runtime A.3 PhysioNet Databases iv
10 A.4 Pre-processing Scripts A.5 Validation Scripts A.6 Server & Client A.7 JProfiler B Results Overview 136 B.1 Data Mining Results B.2 Additional Graphs B.2.1 K-Nearest Neighbor B.2.2 Support Vector Machine B.2.3 Decision Tree B.2.4 Moving Average B.2.5 Standard Deviation v
11 Chapter 1 Introduction 1.1 Background & Motivation Sleep apnea is classified as a sleeping disorder with involuntary cessation of breathing that occurs while a person is asleep. The word "apnea" stems from the Latin language and literally translates to "temporary suspension of breathing" [56]. The disorder is said to be affecting over a 100 million people worldwide and the severity of the disorder ranges from mild to severe [38]. An apnea can last from ten seconds up to as much as two minutes depending on the severity of the subject. Pauses in respiration cause the oxygen levels in the blood to drop and initiate a buildup of carbon dioxide. When the oxygen deprivation in the blood stream reaches critical levels, the body signals the brain to wake up, forcing it to resume breathing. The abrupt awakening makes the subject regular gasp for air for a short period of time before falling back to sleep. The whole incident often happens while the subject is still half asleep and dazed resulting in no recollection of the episode. These apneas can occur several times during an hour and continue throughout the night which prevents the body to go into a deep sleep state. According to Helse Bergen, about one in six people in Norway suffer from Obstructive Sleep Apnea (OSA) in some sort of severity group [68]. In North America the estimated number of people suffering from a sleeping disorder is said to be reaching 18 to 22 million people. These estimations might just skim the surface of the problem as the disorder characteristics and symptoms combined with a time consuming and costly diagnostics process ensues a large number of undiagnosed and untreated subjects. Slow-wave sleep, often referred to as deep sleep or Rapid Eye Movement sleep (REM), is considered to be a natural phenomenon where the brain traverses different sleep stages in order to recover from everyday activities. Studies have shown that deep sleep is needed for the body and brain to fully recover and to consolidate new memories. Sleep apnea is heavily linked together with other illnesses and disorders, such as diabetes, stroke and depression. The lack of required sleep impacts a person s motor skills and reaction times which in terms can make daily activities such as driving a car or operating machinery accident prone [6]. In the US, the yearly estimated health care cost of subjects with moderate to severe OSA is around 65 to 165 billion dollars alone. This estimation concludes root causes, treatments, diagnosis and the impacts on general productivity in society [65]. Diagnosing sleep apnea can be both time consuming and costly. Today, the standard way of diagnosing sleep apnea subjects is by performing a polysomnography in a sleep laboratory at a hospital. A polysomnography is a comprehensive sleep study and consists of several measurements including electroencephalography (EEG), electrooculography (EOG), elec- 1
12 tromyography (EMG), electrocardiography (ECG), oxygen saturation and respiratory airflow. Oxygen saturation measurements are collected by using a pulse oximetry. Respiratory airflow is measured using a mask or strain gauge band. All of these signals and measurements show clear indications of apneas in subjects, with some measurements being more profound than others. Most of these signals are measured using sensors or electrodes that require wire attachments. Wires restrict movement during sleep and can make the subject feel uncomfortable. This can induce the sense of an unnatural sleeping environment impacting both the data and the diagnosis. The gathered data needs to be manually processed by a trained physician to get the correct diagnosis and treatment. The yearly cost of a single polysomnography is estimated to vary from four to six thousand dollars per patient [65]. Home measurement kits have been used at increasing rates as of late because of their low cost and time saving benefits, but they still lack automated data analysis. The data collected with these still needs to be manually checked when the equipment is returned. Merging the cost and time of the diagnostics process the demand for a quicker, cheaper and more automated screening and diagnostics solution is clearly becoming evident. Data mining is about analyzing and extracting patterns and relations in data. This field is a subfield in computer science that has several cross sections in machine learning and artificial intelligence to perform predictive analytical processing. Data mining is a big field and varies in terms of complexity, size and possible tasks such as automation, classification, prediction and pattern recognition. Its field of use is huge and spans multiple domains from business and finance to health and transportation. Data mining have become such a necessity because of the increasing volumes of data we produce every day. The amount of information in this data is impossible for a human to manually process. Computers on the other hand can process the data orders of magnitude faster and can run continuously for 24 hours seven days a week. The abilities and benefits of using data mining techniques makes this area relevant for detecting an abnormal breathing pattern in sleep apnea subjects. This thesis builds upon the results and conclusion of another thesis, "Data Mining for the Detection of Disrupted Breathing Caused by Sleep Apnea - A Comparison of Methods", by Mari Sønsteby Hugaas which we will reference throughout the thesis as "the offline analysis" [42]. In the thesis the focus was to analyze and compare four popular data mining methods, K-Nearest Neighbor (KNN), Support Vector Machine (SVM), Artificial Neural Network (ANN) and Decision Tree on non-invasive signals that is easy to use and monitor at home. Both oxygen saturation and various respiratory signals gathered from the chest, nose and abdomen were considered non-invasive. The goal was to measure how well the four methods performed in classifying epochs of abnormal breathing. Input data consisted of three separate databases from PhysioNet, a reliable source for medical data used in other related work. Evaluation of the data mining methods exerting k-fold cross-validation resulted in an accuracy of 96.6% using respiratory data from the chest and nose. An accuracy of 90% was achieved using a combination of all the signals. The K-Nearest Neighbor method scored the highest accuracy overall while Decision Tree performed the worst. From the results a conclusion was made that accuracy over 90% can be produced by using a single non-invasive signal and that data mining in general is very efficient at classifying epochs of abnormal breathing using data from sleep apnea subjects. 2
13 1.2 Problem Statement The goal for this thesis is to implement and incorporate the four data mining methods used in the offline analysis together with Esper to simulate real-time data streams using databases from PhysioNet. We also design and implement two separate methods for apnea detection using queries over data streams in Esper to detect periods of abnormal breathing in real-time. We compare the results from the four data mining methods and the apnea detection methods based on how well they can find and classify epochs of abnormal breathing. A performance evaluation of the implementation is performed as the last stage to measure resource usage with direct comparison to smart phone hardware. The thesis will be separated into two main tasks with a set of sub-tasks for each: Implement online analysis using Esper. Implement a design that conforms to data stream simulation using Esper using databases from PhysioNet. Integrate the four data mining methods used in the offline analysis. Find suitable methods in related work and incorporate apnea detection methods using EPL and custom functions in Esper. Run performance analysis to determine resource demands. Evaluate the performance of the four data mining methods and the apnea detection methods. Run performance analysis of the implementation to determine resource demands of CPU and memory usage, I/O operations and storage space requirements. The first main task focus on assessing if the four data mining methods and databases from PhysioNet used in the offline analysis is suitable for real-time data streaming using Esper. The databases from PhysioNet have proven to be a good fit for data mining classification in both the offline analysis and other related work using non-invasive signal types as a requirement. As all of the non-invasive signals clearly display epochs of abnormal breathing, one of the sub-tasks consist of designing and implementing apnea detection methods using Esper s Event Processing Language (EPL) to evaluate how well they can detect periods of abnormal breathing. EPL is an expressive query language used to extract, manipulate and aggregate data and events over streams and is used to find patterns and anomalies in various other domains. The second main task will consist of evaluating the results generated form the apnea detection methods and compare them to the four data mining methods implemented in the offline analysis. As a sub-task, we also profile our implementation by measuring performance in the form of resource usage and allocation to estimate if such a future application can indeed be made available for smart phones. The results from the performance evaluation will be a rough estimate, as the implementation will not be a direct representation of a smart phone application, but rather a skeleton concept implementing the main components. 1.3 Outline In Chapter 2 we present a summary of sleep apnea in regards to the different types, symptoms and treatments, as well as issues surrounding diagnosis and tools used today. Chapter 3 is an overview of data mining in general and a description of the four data mining methods used 3
14 in the offline analysis. Further, a brief outline of Esper, its area of use and the expressive EPL is presented in Chapter 4. In Chapter 5 we discuss and summarize the requirements established in the offline analysis and take a look at requirements needed for this thesis. Chapter 6 consist of design choices for our input data, implementation and performance measurements and environments. Details about the implementation is covered in Chapter 7, followed by performance and evaluation coverage in Chapter??. The last Chapter 9 presents the final a summary and discussion of the obtained results as well as future work beyond this thesis. Configuration details and a quick tutorial on how to run the implementation resides in Appendix A. In Appendix B an overview of all the results obtained from the tests is presented as we will summarize and focus on the most important findings in Chapter??. 4
15 Chapter 2 Sleep Apnea In this chapter we present an overview of sleep apnea with a general focus on the type Obstructive Sleep Apnea (OSA). In Section 2.1 we describe what characterizes the disorder, followed by a detailed overview of OSA. Relevant symptoms and risk factors of sleep apnea is presented in Section 2.3 and 2.4. A complete section of various diagnostic tools and procedures is listed in 2.5. Treatments and the surrounding challenges regarding sleep apnea is presented in Section 2.6 and Characteristics Sleep apnea is a type of sleeping disorder and is identified by abnormal breathing or complete cessation of airflow over longer periods while sleeping. As a consequence, the oxygen level retained in the blood stream begin to drop. If the oxygen level reaches a critical point the brain will force the person to wake up in order to restore normal oxygen levels. When the awakening occurs the person is often dazed and still half asleep, resulting in no recollection of it ever occurring. Each epoch of abnormal breathing can last up to as much as two minutes and occur several times a minute throughout the night. As a result, the sleeping pattern is disrupted and often the person never goes into a Rapid Eye Movement (REM) sleep stage. Each person spends approximately 20% of their sleep in a REM sleep stage. Studies show that the different sleep stages are a crucial part of body restoration and for the brain to consolidate memories [63]. The nightly ordeal can cause the person to become sleep deprived and it is estimated that around 70-80% of people affected by sleep apnea are undiagnosed and remain untreated [62]. If the disorder goes untreated it can have serious implications and be the lead cause of developing diabetes, heart disease and depression. Over 100 million people is said to suffer from a form of sleeping disorder worldwide [38]. Taken into the account that a large margin of people goes undiagnosed, the number is even higher. People affected of sleep apnea does not only increase the risk of developing other disabilities and disorders, but have an increased risk of harmful and fatal injuries as a result. Without proper restorative sleep the person is prone to decreased concentration, lack of awareness and fatigue. A recent study published results concluding that people suffering from sleep apnea is over two times likely to be the impacting cause of a car accident than healthy people [50]. The ramifications are huge and the amount of undiagnosed and untreated people is costly to society. 5
16 2.2 Obstructive Sleep Apnea OSA is regarded as the most common type of sleep apnea as it constitutes of about 84% of all patients diagnosed [72]. The type is characterized by complete or partial obstruction of the airways reducing or halting the passage of oxygen to the lungs. When the upper airways are blocked the lungs will try to force air through the air canals and the diaphragm, which can in terms lead to hypopneas and loud snoring. A blockage can be caused by different physiological traits, some that is genetically and some that develop over time. When a person is asleep, the body naturally relaxes its muscles and this can cause the tissue to block off the back of the throat and upper airways. This way of blockage is very prevalent in people which are overweight or have a larger neck with more surrounding fat and tissue. Figure 2.1: Illustration showing the diaphragm blocking the airflow [31]. As seen in Figure 2.1 the diaphragm connects with the back of the throat, effectively blocking off airflow passing through the mouth and nostrils. The amount of surrounding tissue will increase the pressure around the throat and airways which can lead to a partial or total blockage of air. Other physical traits that can cause OSA is abnormalities in growth of the jaw and mouth, larger tongue or tonsils and narrower air canals. Once the oxygen level in the blood drops to a critical level the brain forces the person to wake up. This causes the person to cough and gasp for air until the oxygen levels are back to normal. 2.3 Symptoms Sleep apnea can be hard to detect as the signs and symptoms are often generalized and misinterpreted as less serious causes. People suffering from OSA are often not aware of having the disorder but it is rather identified by a partner or spouse because of the loud snoring and repeated gasping while sleeping. If a person sleeps alone the risk of OSA going unnoticed is higher as it is hard to pin point the cause if the person is not aware of the medical related signs. Below is a general list of the symptoms of OSA [57]: Snoring Fatigue Lack of concentration Memory loss 6
17 High blood pressure Mood swings Snoring is the most common symptom related to OSA. Snoring is caused by hindered or obstructed airflow in the respiratory tract. Often people who snores will not experience this themselves but usually have a partner noticing the person snoring. The snoring can range from just normal rattling with the lips creating minimal noise to heavy and loud snoring. Snoring is regularly the first sign of OSA as the tongue and back of the throat blocks the airways. Heavy snoring can lead to lack of sleep and result in sleep deprivation and tiredness. Even though snoring is prevalent in OSA sufferers, not every person that snores have the disorder. Other symptoms not listed that can relate to OSA are headaches, night sweats and decreased libido [2]. Most of these symptoms overlap between the other type of sleep apnea, Central Sleep Apnea (CSA), which can make the diagnosis based purely on the signs alone even more difficult. 2.4 Risk Factors Sleep apnea is not distinct in a certain age or sex group but can occur in every person. Some groups are at a higher risk of developing sleep apnea mainly because of their life style. If a regular healthy person has sleep apnea it is often considered to be a genetic type that correlates to CSA. Men are more than twice as likely of developing sleep apnea than women. Beyond genetics there are multiple factors that come into play. Weight is the biggest contributor to develop OSA. The increase in body mass generates extra pressure and tension around the neck and chest areas. Other big factors are alcohol consumption and smoking. Excessive alcohol consumption combined with being overweight can increase the chance of the diaphragm blocking the airways as the muscles are relaxing while asleep. Smoking can contribute to less oxygen consumption in the lungs, making the gasping session even more acute. All these factors combined creates a dangerous setting as the person might not be able to recover their oxygen levels and have a chance of choking while asleep. People that suffers from OSA are also more likely to develop diabetes, heart diseases and depression along with other disorders. 2.5 Diagnosis There are mainly two types of diagnostic tools available today. The most common one is the polysomnography as it is seen as the golden standard tool for diagnosing sleeping disorders. Portable monitoring systems have seen an increased use in the recent years as production costs of hardware have decreased following technological advancements, but they are still costly and are not commercially available to regular consumers. There are some discussions surrounding the effectiveness between polysomnography and portable devices as the former tool is used to cover a wider scope of sleeping disorders, not only sleep apnea. But a study concluded that portable devices have a great effect on societies where health coverage is lacking [45]. Combined with the tools are questionnaires that are used as a screening tool to determine if a person is in the risk of developing sleep apnea or might already be affected by it. Frequently the questionnaires are used by physicians to set the required treatments and estimate a severity group of a patient. Some questionnaires can take time to perform and is thus not common practice among physicians and doctors. 7
18 In this section we describe the standard sleep study and the various sensors and signals recorded. Further we will discuss a few portable devices and home monitoring systems. At last we present the various questionnaires that are used today Severity Rating There are two indices that helps physician rate the disorder severity of a patient. The most common one is the Apnea-Hypopnea Index (AHI) and the other one is the Respiratory Disturbance Index (RDI) [1]. The AHI index have a total of four severity groups: normal, mild, moderate and severe. Each group have a range which determines which severity a person is part of. The index is generated by calculating the sum of the total number of apneas and hypopneas per hour. The normal group ranges from an index between 0-4, the mild between 5-14, the moderate between and severe is everything above an index of 30. The RDI index is similar to the AHI index, but is more extensive as it includes body movement and arousals when presenting a severity score. Both rating systems have similar criteria on how to characterize an apnea or hypopnea such as the amount of decrease in oxygen saturation and apnea duration [54] Polysomnography A polysomnography is a sleep study used to diagnose various sleeping disorders, including sleep apnea. The test is performed in a sleep laboratory where the person spends the night in a bed with multiple sensors attached to the body. The sensors record various signals such as electrocardiogram (ECG), electrooculography (EOG), electromyogram (EMG), electroencephalogram (EEG), respiratory airflow and oxygen saturation. Some sleep studies also include blood pressure and heart rate measurements. A list of the physiological signals recorded in the study is presented below [58]: ECG Electrocardiogram shows the heart s electrical activity by using electrodes attached around the chest area. EOG Electrooculography is used to record eye movement during sleep. The electrodes are placed in opposite directions around the eye to capture the eye movements. EMG Electromyogram measures the electrical impulses in the muscles sent by neurons. It is used to detect involuntary body movement that may be caused by a sleeping disorder. EEG Electroencephalogram is a test to measure the electrical activity of the brain. measurements track and record the wave patterns generated by brain activity. The Respiratory airflow Respiratory measurements are recorded from the nose, chest and abdomen. Both chest and abdomen share similar methods as they use an elastic band strapped around the chest or abdominal area. The elastic band expands on inhalation and contracts on exhalation. Measurements from the nose are either recorded using a mask or a slim tube that is inserted in the nostrils. These three ways of recording respiratory data is all seen as non-invasive. 8
19 Oxygen saturation Oxygen saturation is done by using a small clip that is attached to one finger. The clip measures the oxygen levels in the blood stream by using infrared light. The saturation is presented in percentage and a normal level for the human body is considered to be around %. Below 90% is considered low saturation and may result in hypoxemia and below 80% is critical as it can result in damaged organs [14]. Together with respiratory measurements the finger clip is also considered a very non-invasive method. The polysomnography requires trained personnel to attach the sensors and equipment before the session can being. The amount of wires and attached sensors is illustrated in Figure 2.2. With the amount of wires required, a person will likely feel uncomfortable and restrict sleeping positions and body movement. After the session is done the data recorded needs to be processed and analyzed by a physician to determine the diagnose and the resulting severity group of the disorder. A polysomnography requires resource in the form of time from both the patient and the personnel as well as the cost running the sleep laboratory and the equipment. Figure 2.2: Polysomnography with wires and sensors attached [32] Portable Monitoring Systems A person in need of a diagnose can see a polysomnography as uncomfortable as it requires the person to sleep a night in a sleeping laboratory with various sensors strapped to the body. People tend to be discouraged to get their condition checked due to the invasive process of a polysomnography, leaving the disorder untreated. Portable monitoring systems have been in increased use in the recent years because the device can be easily managed at home, relieving the time and resources needed by performing a polysomnography. The portable devices are usually distributed to people in the mild and moderate severity group. People with severe OSA are advised to do a polysomnography to get a more detailed examination because the disorder might have neurological effects as well [59]. Although the portable devices can be used at home they are still widely seen as cumbersome and impractical. These systems are not available to consumers and are sold to private and governmental health sectors for redistribution. Below is a list of three popular portable monitoring systems that are used today: WatchPAT WatchPAT is a non-invasive portable pulse oximetry monitor a person can use at home. It consists of a small device that can be strapped around the wrist with three sensors [76]. 9
20 There are two sensors that connects to the fingers that measure the blood oxygen levels and a sensor that sit on the chest measuring body movements and heart rate. The device is easy and can track real time changes of sleep apnea and is bundled with a software so the user can see the data collected and a suggested diagnose. ApneaLink Plus Another monitor is the ApneaLink Plus which is also non-invasive but has more sensors and wires to attach than the WatchPAT. Not only does it measure the blood oxygen levels but have also both chest and nasal respiratory sensors as well. The ApneaLink Plus have no visual information feedback for the user and the device needs to be sent back to an expert to get the data analyzed [3]. NOX-T3 NOX-T3 share similar sensors as the others above but have also a built in microphone to capture audio during sleep. The audio is to detect scenarios where a person might snore, gasp or stop breathing. A software is included to export the data and get an in detail view of sleeping sessions, but it lacks the ability to monitor it in real time [13] Questionnaires There are four questionnaires used today in a varying degree, the Berlin, STOP-BANG, G.A.S.P and Epworth Sleepiness Scale (EES). The questionnaires are used to assess the risk of a person having sleep apnea or developing the disorder. Questions range from everyday activities, health issues and personal body metrics such as age, weight and height. The questionnaires alone are not used to diagnose a person or to determine a person s severity group, but it is a tool to help determine further examination. A study has shown that although questionnaires are a good way to assess the likelihood of sleep apnea, they can be inaccurate based on the general population [61]. The questionnaires can be combined to give a more accurate score and are often performed with a full physical assessment. A more detailed description of each questionnaire is as follow: G.A.S.P G.A.S.P contains five questions and is the smallest questionnaire of them all. The questions are very general and determines subjects such as sleepiness, snoring, witnessed apneas, high blood pressure and overall weight. The survey is not very indepth and have three risk groups based on a total score of five which could make this prone to misclassifications if used by its own. Berlin The Berlin questionnaire is by far the most excessive questionnaire by having the highest number of questions and options. There are ten questions in total divided into three categories and each question have a number of options that range from two to five. Although the questionnaire at hand is the most complex and in-depth, there have been physicians unwilling to implement this questionnaire due to needing to manual score the answers with a key. The answers are not listed together with the survey as the other questionnaires [67]. STOP-BANG The STOP-BANG is less excessive than the Berlin questionnaire but is more in-depth than the G.A.S.P questionnaire. The questionnaire consists of eight questions and covers almost the same areas as the G.A.S.P questionnaire but have additional questions regarding age, BMI and neck size. 10
21 Epworth Sleepiness Scale ESS is survey to determine daily sleepiness of a person. Questions that needs to be answered is related to if a person is likely to get sleepy or doze off by doing everyday activities such as reading a book, watching television or driving. This survey is often combined with another questionnaire that is more specific to sleep apnea in this case. 2.6 Treatments There are a number of treatments available for patients suffering from OSA, including Continuous Positive Airway Pressure (CPAP) machines and life style advices. Treatments are derived together with a doctor or physician and is often tailored to the patient in terms of their severity group. Patients that are in the mild to moderate categories can improve symptoms and benefit mainly from life style changes. The large majority of patients presented with OSA are either overweight, heavy smokers or a combination of both. For patients that are overweight and are heavy smokers, losing weight and quit smoking will dramatically improve OSA symptoms [64]. Patients that are in the severe group having an AHI index over 30 might need more extensive treatments. CPAP is a device that generates positive air pressure through a mask that hinders the soft tissue in the neck to collapse during sleep. Despite that the machine is often the best treatment for severe OSA patients, people find the CPAP mask to be uncomfortable and refers it to sleeping in scuba gear [51]. Surgery is also a treatment option available, but is rather used as a last resort because it is invasive. The procedure is done by either removing the surrounding tissue around the neck and jaw, or cutting off the tonsils and the uvula to create more space in the back of the throat [66]. 2.7 Challenges Sleep apnea is a sleeping disorder that is hard to detect and often goes unnoticed because the symptoms can be misinterpreted as multiple other causes. People often trivialize problems that relate to sleep apnea because they have little to no knowledge about the disorder and how to acknowledge its symptoms. In societies where both education and health coverage is lacking, the awareness of the risks and symptoms is low or almost non-existent [45]. In situations where a person knows they are likely to have the disorder, some refuse to go to a physician for diagnosis as the process can be long and tiresome. There is a huge demand for a quicker and more wide spread solution to diagnose people with sleep apnea. The golden standard diagnostics tool today is the polysomnography which is time intensive and costly. The sleep study requires a lot of human resources to be able to collect the data and analyze it in order to give the right diagnose and severity group. Portable monitoring systems that exists offer a better experience but have disadvantages in the form availability restrictions and not being available for consumers. Smart phones and electronic devices have seen a rapid increase in adoption rate not only in western countries but in third world countries as well. The decreasing production costs for sensor chips and hardware drives the adoption rate even higher for each year. A proposed solution is to develop a cheaper off the shelf application and monitoring system that utilizes the hardware in smart phones that people already own. The smart phone application combined with an easy to handle noninvasive sensor can classify the sleeping pattern of a OSA patient in real-time. The solution 11
22 offers a cheap and more time efficient home monitoring system that can decrease the resources needed to diagnose patients in the future. 12
23 Chapter 3 Data Mining In this chapter we present an introductory overview of the data mining concept and a basic description of the four data mining methods used in the offline analysis. Most of the material used in this chapter is found in various books that intersect multiple domains such as data mining and machine learning, as well as material from the offline analysis [42] [78] [52]. In Section 3.1 we present an introduction to data mining and the concept behind data analysis, followed by an overview of the various data types, models and tasks in Sections 3.2 and 3.3. We proceed by presenting a description of the data mining methods used in the offline analysis. The K-Nearest Neighbor is presented in Section 3.4 followed by the Support Vector Machine in Section 3.5. A description of decision trees is listed in Section 3.6 while the last Section 3.7 includes the concepts of the Artificial Neural Network. 3.1 Introduction "Data Mining" as a term is the computational process of finding relevant, interesting and novel patterns. The patterns are often derived into descriptive and predictive models based on largescale data. There are many definitions that is valid for data mining, but the underlying process of finding patterns, derive models and analysis is broadly the same [52]. The term is relatively new, while the technology and methods used to analyze and summarize data into understandable information is not. The increase in computational power and the decrease in size and power consumption on devices in today s society means that the amount of data produced every day is also steadily increasing. Data mining plays a role in that the amount of data produced is too big for humans to analyze and comprehend by hand. The notion "Big Data" is used for large data sets with a complex structure that can be analyzed to find patterns, predictive models and analytical behavior that otherwise would be impossible with traditional processing methods [10]. The term is often used interchangeably with data mining. Data mining is part of a larger processes commonly referred to as "Knowledge Discovery in Databases" (KDD). Data mining is a step in the process which can consist of an arbitrary number of steps depending on its presentation, but the overall goal of the process is the same for each definition. A nine-step variant of the KDD process is listed below [52]: 1. Plan and develop an understanding of the application domain. 2. Selection and creation of a data set which discovery will be performed. 13
24 3. Pre-processing and cleaning of the data set to remove noise and artifacts. 4. Data transformation and complexity reduction. 5. Choosing a Data Mining task that suits the problem outline. 6. Choosing a relevant Data Mining algorithm for the application domain. 7. Employing and implementing the Data Mining algorithm. 8. Evaluating the performance of the mined patterns and extracted information. 9. Integrating and incorporating the discovered knowledge in other parts of a system. Each step in the process is iterative, meaning that even though the steps are dependent on each other they can be changed or altered at any time based on changes made in other steps. There are two different main types of Data Mining, verification-oriented and discovery-oriented. The former is for verification of a hypothesis while the latter is used to find new patterns and models autonomously by using Data Mining methods [52]. 3.2 Data Types There are two main types of data attributes that is considered. Both have different aspects and properties that need to be acknowledge to be able to incorporate a data mining task or algorithm as some data mining methods perform better with one than the other. The two types can further be divided into two sub-types: Numerical attributes Numeric attributes consist of any possible natural number. It can either be in a form of a real-valued or integer-valued domain. Examples of a numeric attribute can be age or weight of a person. There are two types of numeric attributes, discrete and continuous. The former is a numeric set of any values that is either finite or infinite in length, while the latter can be any real value. Further the attribute can be classified as either interval-scaled or ration-scaled, which determine how they are comparable to each other. Categorical attributes Categorical attributes can be seen as a set of values that not necessarily are numbers, but can contain any set of values. An example could be the sex of a person that would either be male or female. A categorical attribute can be split into two different types, either nominal or ordinal. Nominal attributes are ordered which means they cannot be compared to each other, while Ordinal attributes can be compared for equality or inequality such as military ranks for example. Some categorical attributes can also be transformed and represented as numerical attributes, but still hold the categorical properties. For example, the sex of a person can either be male or female, but can also be represented in binary as either 0 or 1. Most data sets containing either numerical or categorical attributes can be represented in a matrix. Even data sets that consist of image or video data, which is in a sequence of bytes can be represented in a matrix by using the correct feature extraction tools or methods. 14
25 3.3 Data Mining Tasks & Models In general, there are a total of six common data mining tasks [74]. Data mining models is commonly divided into two types, a predictive and a descriptive model. These models depend on the goal of the data mining task and have different characteristics. The outcome of the models has different learning properties and can further be divided into two more learning methods, supervised and unsupervised. A more complete description is listed below: Predictive models Predictive models are models that can predict an outcome based on statistics and data attributes. These models can utilize more than one classifiers to predict an outcome. Often the outcome is either a real number value, a class definition or a binary statement. Spam filters as an example use predictive models to determine and predict if an incoming is defined as real or junk. A training set of other pre-classified contexts are used to predict the outcome of new s. If the share similar context and properties to other s that are classified as spam, the outcome would be that the particular would also be spam. Any regression based model can be used to predict outcomes. Naive Bayes, Support Vector Machine, K-Nearest Neighbor and Artificial Neural Network are all popular data mining methods that can produce a predictive model. Descriptive models Models of descriptive type use the data processed to generate a model based on patterns and relationships. A descriptive model summarizes similarities and differences in the data. For example, in marketing a descriptive model can tell the business which predetermined age group orders products from a specific category. For example, old people living in houses are more likely to buy gardening tools than younger people living in urban cities. Marketing and business strategies are more relying on descriptive models to measure performance and profitable sectors. Clustering methods are good at producing descriptive models as they can group objects that have no natural connection. Supervised learning Methods that use supervised learning typically use a data set for training. Training a method relies on a training set as input with valued attributes that are paired up with corresponding target classes. The supervised learning method then produces a model that can be used to classify and map new instances as input. The quality of the model produced is often a direct product of the quality of the training data used. A high quality training set often leads to better results. Problems surrounding supervised learning methods is overfitting the model with too much training data or that the training data has a complex balance issue. This can make the model shift more away from generalization and towards a specific outcome. Having more attributes than observations in a training set will create a model with bad predictive properties. For example, in face recognition a model based on supervised learning need labeled training data in order to comprehend the notion of a face, or what the form of a face looks like. Unsupervised learning While supervised learning bases the model on relationships between the labeled training data and the test data used, unsupervised learning algorithm use unlabeled data. Clustering is a technique that conforms to unsupervised learning algorithms as instead of trying to use labeled examples which tells the algorithm what is a correct object or not, the algorithm tries to cluster different objects based on attributes so that it can clearly 15
26 see where the line is drawn between objects. For example, properties and attributes in various images can be distinguished by clustering such that a dog is not correctly classified together with a whale. Neural networks and K-means algorithms are popular unsupervised learning algorithms Clustering Clustering or cluster analysis is the task of grouping sets of data objects in a way that objects that have similar properties are grouped together in a feature space. While clustering is not synonymous with unsupervised learning, the supervised learning methods are called statistical classification and the unsupervised methods are called cluster analysis. Cluster analysis is not specified to one single algorithm but describes a group of algorithms that use clustering techniques. The main goal for clustering is to group subsets together to form categorical descriptive patterns that share resemblance. Figure 3.1: Plotted values with three distinct clusters [25]. In figure 3.1 we can see that there are three distinct clusters that forms. The color indicates that the objects are part of a different cluster and share similarities. What the dissimilar clusters represent is based on the properties of the objects and the domain. To determine if two objects are similar there are commonly two types that are used: distance measurement and similarity measurement. Distance measurement calculates the dimensional distance between the objects while measuring similarity compares the vector objects symmetrically [52] Classification Classification is a data mining method that determine items in a data set to categories or classes based on their similar traits. The potential goal for classification is to classify and arrange each test object to a specific class, depending on the structure and configurations of the model. It uses a supervised learning algorithm that models relationships based on their labeled context. Classification are used in multiple domains to determine a class or category of an unknown or unseen object. For example, new advancements in medical treatments can correctly diagnose a patient based on their symptoms. In this example the symptoms are the properties or attributes of the person, which is the object. The diagnose is the class or category. 16
27 The training process results in building a model that can generalize unknown objects. The model is built by a classification algorithm, often called a classifier, that maps relationships between the labeled training data. The summarized model is then used to predict the outcome of other objects. With data streams a traditional model needs to adapt to the changes in the data stream. If the data in a stream can be rationalized so that the upper and lower bounds of the data is already known, the model can be kept intact. Otherwise there is a need for an iterative process of building new models based on the data or transform the incoming data Regression Another form of classification is regression-based methods. The definition of regression analysis is broad but is in the lines of "to understand as far as possible with the available data how the conditional distribution of some response y varies across subpopulations determined by the possible values of the predictor or predictors" [16]. While classifiers are good at determine an object, regression predicts or estimates an outcome based on statistical probability among relationships between the data. Because the outcome typically is predicted or estimated at a certain point in time, a traditional model of sorts is not generated. There is a myriad of different regression-based techniques out there where some of them are linear, nonlinear or polynomial Anomaly Detection Finding objects that conform outside of the patterns might as well be as important as finding the patterns. Anomaly detection, also called outlier detection, is a technique by analyzing objects that is forming outside of the regular patterns [52]. Objects that lie outside of the normal patterns is assumed to be errors or noisy values that are cause of the displacement. Errors could be classified as several different things depending on the domain and situation. While anomaly detection is a good way to find deviations, it is often used as a pre-processing method to filter out noisy objects. Methods that utilize anomaly detection can be found in numerous systems such as bank fraud detection, network intrusion and weather satellites. For example, with credit card fraud the bank can analyze the transaction history of the victim to see if there are any irregularities in the pattern. In case a credit card was stolen, the usual anomaly would translate to high activity of online transactions that often goes outside the victim s regular history. To find the deviation three types of methods are suggested: distancemeasured methods, clustering methods and spatial methods [52] Association Rules Association rules mining is commonly used to mine information in marketing and trends. Rules are defined as "one out of three females buy x when ordering y". Maximizing profits based on trends and likelihood is something every retail business wants to achieve. A good example of using association rules to increase profits is called Market Basket Analysis [4]. Product placements in stores are heavily impacted by this analysis. For example, in stores that sells electronics, commonly the most popular products and categories are located in the back of the store rather than in the front. Smaller and cheaper products are placed around the cash register and the front entrance of the store. Businesses use this mining technique to see relationships between a pair of products or products that share similarities. A simple, yet powerful association rules mining algorithm is the apriori algorithm [52]. 17
28 3.3.6 Summarization Summarization is defined as a way to generate a sub-context, a portion of the context from a document or file to be able to get the full context. A sub-set is extracted from the original context together with meta data such as length, number of words and other relevant attributes. Summarization is the part of data mining that is regularly used in search engines to reduce the amount of stored information. One of the two ways of dealing with summarization is extraction of paragraphs, phrases, words and meta data to build a summary. The other way is called abstraction and generates a semantic representation of the context as a summary [52]. 3.4 K-Nearest Neighbor The Nearest Neighbor classifier is considered a very simple classification model, but performance wise can be comparable to other more advanced and sophisticated classifiers. The classifier predicts a class based on the surrounding neighbors in a data set. Nearest Neighbors is considered a "lazy learner" as it has no form of training phase and only do the computation based on a local approximation. While other classifiers generate a model based on training data to generalize and find patterns among the data, this classifier only stores the training data and configuration to compare and classify the test data during run-time. The K-Nearest Neighbor is a variant of the Nearest Neighbor classifier which employs a variable k that determines the number of neighbors to be considered in the calculation. It is used for either classification or regression purposes. With classification an actually class object is the output result rather than a property value derived from an average of the nearest objects. A k is selected to represent the number of neighbors to be considered in the computation out of a training set D. All the objects in D represent a class and a set of properties. For each test object z the method computes the distance between all the objects in D and test object z. The output class for the test object is the class majority of the closest k objects. An example of this computational process is illustrated in Figure 3.2. The green circle in the figure represents test Figure 3.2: Illustration of the computational decision of K-Nearest Neighbor [29]. object z while the blue squares and the red triangles represents a distinct class and form the training set D. The solid circle illustrates when k = 3 and the dashed circle when k = 5. Picking the solid circle as the decision boundary results in the triangle class being the majority class chosen, while picking the dashed line results in the square class being the majority. Picking the right value for k is crucial to the classifiers performance, but it all depends on the training data applied and the test data used. Depending on the number of classes in the data set a problem with picking k as either an odd or even number can result in a tie in choosing the 18
29 right class. In this case a tie breaker is purposed to force a resulting class. There some common ways to apply tie breakers that eliminates the problem with class calculations resulting in ties. Weighting and distance measurement methods are used to determine the correct class in cases of ties. There are three common methods of determine a class if there is a tie. Picking a k value that is odd when the number of class labels are even, and vice versa will never result in a tie situation. Another tie breaker is to calculate and pick the class that has the closets object to the test object. In situations where the distance also becomes a tie, as in both the closest objects have the same distance and different classes, the class is determined by a random factor. The value of k also impacts the classification accuracy if the data set contains noisy artifacts. Advantages of using K-Nearest Neighbor is that it is easy to implement and understand, while still being regarded as an effective classification model. The classification model has no training phase and stores the data for run-time classification, this means that the method consumes more resources than "eager learners" and have an impact on classification times and memory consumption. 3.5 Support Vector Machine The Support Vector Machine is an advanced and mathematically complex classifier that conforms to both classification and regression problems. The classifier was introduced by Vladimir Naumovich Vapnik over two decades ago and is one of the most commonly used classifiers in practice. Vapnik proved that a data set that can be separated linearly by a hyperplane can be bounded in regards to a margin. SVM was introduced as a binary classifier, but has and can be used to classify objects and patterns concerning multiple classes [42]. Training data that can be separated linearly can produce hyperplanes like in Figure 3.3. There Figure 3.3: Hyperplanes separating clusters of two data object classes. The red line is considered the maximum margin hyperplane [34]. are two separated classes that is represented by the white and black dots. The hyperplanes H 1, H 2 and H 3 are represented by different colored lines. The green hyperplane is not able to clearly divide the classes while both the blue and the red hyperplane is able to separate the two classes. Even though the blue hyperplane is able to separate the classes, it is only doing so with a minimal margin. The red hyperplane on the other hand is known as a maximum margin hyperplane because it is able to find and separate the closest two data objects with the largest distance in between them. The margin is defined as the minimal distance of an example to the distance surface [78]. 19
30 3.5.1 Hard-margin The maximum margin hyperplane or in a more general term, the hard-margin, is illustrated in Figure 3.4. The dashed lines in the figure is the boundaries for the maximum margin for each support vector, which is the two closest data objects to the margin calculation. The line in the middle is the maximum margin hyperplane, which is represented in Equation 3.1 together with the boundaries in Equation 3.2 and 3.3. The w represents the weight vector and is relative to the distance of the class vector x i, while the b is the bias added to computation. w x + b = 0 (3.1) w x + b = 1 (3.2) w x + b = 1 (3.3) Figure 3.4: The maximum margin hyperplane [34]. When the boundary is found between the class objects, the objects should be either above or below 1 or -1, like in Figure 3.4. In a binary classification problem, the 1 will always indicate one class and -1 will always indicate the other class. The distance between two support vectors x 1 and x 2 from separate classes are given in Equation 3.4 which results in the Equation 3.5. w w (x 1 x 2 ) (3.4) 2 w (3.5) To maximize the margin between all the possible hyperplanes we need to minimize w which results in a convex optimization problem trying to construct the optimal hyperplane. The optimization problem can be solved introducing the Lagrange multipliers and the Lagrange function [78] Soft-margin In cases where the input space of the data set is not fully linearly separable we introduce a Softmargin Support Vector Machine. Hard-margin separates two support vectors by a maximum possible distance to each other, for the Soft-margin we allow some of the data objects to overlap the decision boundary. A slack variable is introduced so that some of the data objects may be able to stray over. Adjusting for the slack variable we get the following soft-margin Equations 20
31 3.6 and 3.7. The problem with adding a slack variable is that the overall error rate will increase, even though the classifier is able to separate two classes while not being fully linear. w x i + b 1 ε i i f y i = 1 (3.6) w x i + b 1 + ε i i f y i = 1 (3.7) Figure 3.5: Example of a Soft-margin hyperplane [36]. The optimization problem is that the soft-margin reaches a tradeoff between how many points that are allowed to cross the boundaries and how many points needs to be moved around in order to get a better hyperplane. In some cases, we can accept the tradeoff for a data set that is not linearly separable because it can help to produce a more generalizable model Non-linear Problems that cannot be separate linearly can be solved by dividing the input space into a higher dimensional space. Input space coming from the data set in use can be transformed into a n-dimension feature space by using feature extraction methods and techniques. Figure 3.6: Input transformation of the data to a 3-dimensional feature space [35]. In Figure 3.6 the input space to the left is transformed to the 3-dimensional feature space on the right. The margin is represented by the red line cross-sectioning the black and white data objects. The margin is calculated along the closest data objects for both classes. The problem here is that finding the perfect amount of slack so that the margin can separate the classes and the complexity of working with a higher dimensional model is a balancing act. Here the 21
32 performance impact is not only the accuracy of the classification model, but also the amount of computational power needed to perform the dot product calculations and function requirements to find the optimal decision boundaries. The transformation is performed by using a kernel trick that is often referred to as a kernel function or method. The raw input space is not actually transformed in the fact that the data attributes are manipulated, but we create an abstraction of the input space which is often cheaper to compute than to transform the input space directly. Kernel functions are commonly used to transform data sequences, graphs, images and other data that is hard to process and separate linearly [78]. 3.6 Decision Tree A decision tree is produced by an algorithm that map and identifies possible ways to split the data into branching segments forming a tree like structure. The tree is inverted in that it the entry point is always represented as a root node on top of the tree, splitting into branches moving downwards. The objects of analysis are evaluated at each node to specific decision terms based on their properties with all objects starting at the root node. The name of the rule and the expression of the decision rule is commonly displayed in the tree along with the logical conditions leading to each branch. Each resulting leaf node, also called the target node, holds a conclusion or a target class that is the result of the object analysis progressing through the tree. A visual representation of a decision tree is illustrated in Figure 3.7. The example tree is from a savings plan that bases the decision on a person s income to evaluate if the person is likely to upgrade to one or the other savings plan. The expression at the root node analyze the object which consists of a numerical attribute, the income. Each node is split binary, meaning we have two possible paths for each expression. If the income is over a certain value we go to left node, if the income is less we go to the right node. These branches have another set of decision rules which can result into four different outcomes, even though the target nodes only hold two different classes. Expression in the tree can evaluate both categorical and numerical attributes depending on the use case and the size of the data set. Figure 3.7: Representation of a decision tree for a savings plan [26]. The process of generating the decision rules comes from methods that extract relationships between objects of relevancy and the properties of the splitting branches or segments. When 22
33 the extraction of relationships is done, decision rules can be derived that describes the correlating objects of analysis and the target branches. The result of this process generates what we call a model. The model can be used to analyze objects that not necessarily have any coherent relationship with the decisions or branches, resulting in classification of unknown objects. There are many factors that play a role in the tree s performance and complexity that needs to be addressed when generating a decision tree. First, finding good split conditions can be a challenge if the data set have higher entropy. This means that the relationships between objects have a high mixture of possible classes at the target nodes, resulting in leaf nodes with no majority class. There are various metric methods that calculate the best possible splits to counter higher entropy, the term purity is used to represent child nodes that are split based on the best possible case of having the same target class. Given an example of a tree with three different classes, a node that ultimately ends up having none of the two classes, but contains just samples of one class is identified as a pure node and needs to further processing [78]. Overfitting can be a problem when generating a decision tree if the resulting model is too complex and struggles to generalize well to data set used for training. This problem can be solved by introducing various pruning techniques. Pruning in decision trees is a technique by removing unwanted and unnecessary parts of the tree that is not contributing to the overall classification accuracy. For example, further branches below a node that is considered pure can be removed as it will not have any effective impact on the accuracy, thus simplifying the tree and decreasing its complexity. Pruning can be done during the building phase of the tree, called pre-pruning, or after the tree is generated which is called post-pruning. A combination of the two can also be used. Together with pruning, using the right stopping criteria is also a technique that can reduce the overall complexity without impacting the classification accuracy. Decision trees have some advantages because of its simplicity. A tree structure is easy to visualize and interpret that make it easy to see where the results derive from. The tree can contain both numerical and categorical data which makes them useable in various business and system domains, including easy integration of rule sets that conforms to business processes. While having advantages, a big disadvantage of using decision tree is based on the data used, if the majority of the data set is based on numerical data the tree can become overall complex if the splitting is done binary, which is often the case for tree-like structures. 3.7 Artificial Neural Network Artificial Neural Networks are inspired by the concept and inner works of the human brain. Our biological brain consists of billions of neurons that communicate with each other by sending electrical signals between junctions. The signals pass through the neurons and is eventually sent through an axon [11]. The Artificial Neural Network tries to resemble similar conceptual mechanics by a computational abstraction of nodes and layers. While the neural networks are similar in concept to a biological brain, they are not comparable in size nor complexity. All though they are not comparable, they share similar traits as they have parallel processing of information and can learn and generalize from events. There are numerous variants and different models of neural networks that each serve a purpose and is suited for specific tasks. The most common variants are the Single-layer Perceptron and the Multi-layer Perceptron [42]. Both are classified as a feed-forward network where the connections between the nodes and layers do not form a cycle. 23
34 3.7.1 Single-layer Perceptron The Single-layer Perceptron is one of the simplest variant of a neural network. The network model consists of either one or more input nodes that is interconnected with one or more output nodes. In classification problems with binary classes, a single output node can fire or activate for one class and not fire for the other class. An illustration of the perceptron is presented in Figure 3.8. The perceptron consists of three input nodes u 1, u 2 and u 3 that are connected to the output node g. Figure 3.8: Illustration of the Single-layer Perceptron [33]. Each connection between input nodes and the output node have weights attached to them. A single bias weight is also included with the output node which is added to the overall calculation score. The weights control the relevancy of the attribute to a given class. For example, classifying animals in regards to if the animal is a dog or not will sway more towards animals that have four legs rather than two legs. The O represents the class, or the output information. These small networks can be combined together to form a sophisticated network where each output node work individual or isolation to a section of the classification problem. The learning process is iterative, meaning that initializing the weights is difficult without known the whole relationship of the training data. Instead the weights are initialized to either 0 or a random number and updated in steps to improve the performance and accuracy of the input nodes. For each iteration, the overall sum is calculated and compared to the class of the training data used. This is done multiple times until the weights are optimal and can no longer be improved in regards to the training data. If the output node fires incorrectly, the weights either get adjusted with a smaller or larger number depending on the overall score difference between the class in the training data and the attributes in the training data. This way the perceptron contains optimal weight values. As the perceptron is a linear classifier, problems with learning can occur in that it cannot separate the positive instances from the negative ones and thus the learning will never be able to learn. This means that the weights never settles and are updated based on training data that cannot be separated by a hyperplane. There are two algorithm variants that solve this problem and are called the pocket and the Maxover algorithm Multi-layer Perceptron The Multi-layer Perceptron is most common neural network variant used across various application domains. Multi-layer Perceptron is a feed-forward neural network that is very suit- 24
35 able for solving problems surrounding relationships between input nodes that corresponds to a set of output variables. Feed-forward neural networks are also appealing for data mining tasks as they are very good at classification and prediction. A Multi-layered Perceptron network consists of a set of connected nodes that form a directed acyclic graph from multiple sources to one or more sinks. It is similar to the Single-layer Perceptron, but the connected nodes are organized in layers, and a network always consist of three or more layers depending on the complexity and specification of the network. Each node works individually and commits a simple task of processing the incoming information to an output variable. An example of a feed-forward Multi-layered Perceptron is illustrated in Figure 3.9. The network consists of three layers: input layer, a hidden layer and an output layer. Usually a network of this type always consist of at least an input and output layer, while the amount of hidden layers between these can vary depending on the specifications and needs. The input layer in Figure 3.9 consist of four input nodes that are interconnected to all the five hidden nodes in the hidden layer. Input nodes are passive, meaning they do not process the information they receive but simply send it along to the hidden nodes. The five hidden nodes are connected to both the input and output nodes. They receive complex data patterns from the input nodes and process the information that is then sent to the output node. The output node in this case is a binary output node, meaning when it reaches a certain threshold based on the input value it might fire or not fire based on the configuration and training data applied. It is able to represent two possible outcomes or classes, but a network can also consist of more than one output node. Figure 3.9: Illustration of the Multi-layer Perceptron [30]. All the information from the input nodes to the output nodes are one-directional, hence the name feed-forward. Activation functions, or more commonly called transfer functions are methods that process the information gathered and produces an output value. The most commonly used transfer functions is the Sigmoid function and the Hyperbolic Tangent function. In problems where the desired output is of either a categorical or binary type the Sigmoid function is the most popular choice as it is the simplest one [78]. A key factor to learning of the information they receive is using and continuously adjust weights of the incoming and outgoing connections like with the Single-layer Perceptron. In the training phase of the network training data is fed through the network. For each training 25
36 pattern that is processed a sum of the weights are derived at the hidden nodes and is sent to the output nodes. At the output nodes the weights are calculated and compared to the correct values in the terms of annotations or other variables. This process can iterate many times before an optimal network adjustment is found. One of the most popular training algorithms to solve error rates is backpropagation. Backpropagation works by calculating the partial derivatives of the cost function with respect to any of the weights in the network. This process if often used together with optimization functions to find the smallest possible error rate. While Multi-layer Perceptron is commonly used to solve classification problems there are a number of issues that is a factor in generating an effective and accurate neural network. One issues are learning and generalization of the network model in regards to bias and variance, both impacts the model s usefulness. Another issue is the requirement for data quality in training and data sets. Noisy artifacts and errors in the data will affect the adaption to the error rate and will overall decrease the accuracy of the network model. The last issue is the splitting of the training set for the iterative phases of updating weights. All of these factors contribute to finding best configuration and optimizing the neural network. 26
37 Chapter 4 Esper Esper is an event series analysis and Complex Event Processing (CEP) engine providing scalable and memory efficient processing of historical and real-time data streams. Esper enables detection of event situations in data streams and the creation of custom actions to trigger on occurring event conditions. Esper is one of the few open source CEP engines available and implements both event-driven programming and architecture [22]. A simplified overview of components and workflow in Esper is presented in Figure 4.1. Figure 4.1: Component overview of Esper [27]. Esper handles data streams with support of multiple input adapters and can process a wide variety of popular data formats such as JavaScript Object Notation (JSON) and Extensible Markup Language (XML). The engine is made to easily be supported and incorporated with existing services and applications. Query statements are stored and initialized in the Esper engine instance and the incoming data from the input adapters are mapped to Plain Old Java Objects (POJOs), which in terms is represented as events. Different actions can be triggered based on the queries stored with the engine such as storing data values from events in a relational database for later comparison, or enable handlers for more complex tasks. Output adapters can be attached or integrated into other services and applications. A list of the following input and output adapters for Esper is presented below [20]: File adapter The file adapter offers both read and write support for various file formats and includes a dedicated adapter for reading Comma Separated Value (CSV) files. The CSV adapter provides many features such as content playback and pausing the input reader. Spring framework Spring is a popular open source framework for modern enterprise applications built on 27
38 the Java platform. The adapter makes use of Java Message Service (JMS) combined with context loaders to send and receive data. Advanced Message Queue Protocol Advanced Message Queue Protocol (AMQP) is a standardized protocol for messageoriented middleware. The protocol can solve problems such as interoperability across platforms and environments, security and open standards. The adapter uses the same principles of publishers and subscribers in the form of sources and sinks and can be integrated into services using AMPQ. Socket adapter In a two-way communication a network socket is one of the end-points. The socket adapter can be used to send events into the Esper engine instance which makes it possible to integrate various platforms and devices. Hypertext Transfer Protocol Hypertext Transfer Protocol (HTTP) is a foundation protocol of the World Wide Web (WWW). The HTTP adapter that Esper provides make it easy for developers to integrate CEP concepts into web applications. The adapter can handle incoming and outgoing HTTP requests directly from an Esper engine instance. Relational database adapter With the relational database adapter, events processed by the Esper engine instance can be stored to compatible relational databases. This means that historical events can be combined and compared with new incoming events from a data stream. Support relational databases offers possibilities to incorporate Esper into different organizational layers in a business. In Section 4.1 we present a short description and overview of the concept of CEP, followed by a summary of steps for rule-based classification. A short description of the Event Processing Language (EPL) is presented in Section 4.3 together with a small example using the EPL. The last Section 4.4 contains performance and benchmark results for Esper. 4.1 Complex Event Processing CEP is a concept of processing events to find or detect complex occurrences in data from multiple different sources [19]. Events can be a broad range of representations such as customer or sales orders, stock trading or filters. Other data that can be represented as something that changes state based on conditions. Temperature changes from multiple sensor sent to a central hub for processing is a good example of events that changes state and comes from multiple sources. With these changes, solutions using the CEP concept can extract complex weather patterns from new correlating events. Continuous Queries (CQs) are a big part of the CEP concept. CQs are processed in a repeated fashion over new incoming data from data streams to evaluate new results or events. They tend to be running for longer periods of time to extract results continuously from new arriving events. CQs commonly extends a core query languages and offer the same type of relational operators we see in relational databases [7]. Although CQ share similar traits as standard relation query languages, they work a bit different. Operators in a relational query languages are termed to be blocking operators because they need a complete data set to be able to output a result. This is not feasible when processing data streams as the stream have no defined end and 28
39 can in terms be infinite. To overcome this problem CQs introduces the concept of windows. In situations where a data stream is infinite a window is defined as a subset or segmented finite section of the data stream. Most Database Management Systems (DBMSs) and standalone Figure 4.2: Event processing in Esper [27]. solutions incorporating the CEP concept usually consist of two different types. Aggregationoriented systems focus on having continuous queries trigger execution of algorithms in order to derive a result of incoming data events. A stock trading system often use algorithms to continuously calculate graphs of trading events. Detection-oriented systems on the other hand focus on detection of patterns or anomalies in a data stream sequence. Security measurements in a network can utilize this to detect abnormal activity or burst of activity in order to counter the attacking threat [15]. 4.2 Rule-Based Classification Rule-based classification can be expressed as any method or scheme that use a hierarchy or a collection of "IF-THEN" rules. A typical rule set consist of two main concepts, the antecedent represents the cause or the conditional state of the rule, and the consequent represents the consequence or the result of a conditional state. Rule-based classification is a popular concept and is often used in machine learning and artificial intelligence. A rule-based classification scheme usually consists of the following steps [73]: The rules need to be extracted from the data in order to be suitable for classification. Different sequential covering algorithms or generation of decision trees are often used to extract the rule set. When the rules have been established there is a need to determine the usefulness of the rules and to remove unnecessary rules that can impact classification accuracy. This step also focuses on giving rules weights in order to improve ranking, which can impact decisions. Data with unknown class objects is used to test the rules extracted by the first step. Multiple rules can be aggregated and optimized by rankings from step two to output the correct classified object. 4.3 Event Processing Language Esper provides the EPL to create continuous query statements that are used to process and analyze events. The EPL is based on the core Standard Query Language (SQL) and converges 29
40 event stream processing such as filtering, joining, aggregation and causality. Esper also supports the creation of custom functions that can be expressed directly in EPL queries. The EPL also have additional features for data flow control, pattern matching and window sequences. Code 4.1: Example query in written in EPL. SELECT count (*) FROM SensorEvent ( temp = 10). win : time (3 min ) HAVING count (*) >= 5 OUTPUT first every 3 minutes In Code 4.1 we detect n number of events within three minutes where each event has a temperature of ten. With the output operator we suppress the output of events every third minute for the first occurring match. The query is very simple and more advanced examples utilizing more of the custom functions and operators available in the EPL is listed in our implementation in Chapter 7. Additional examples are also available on Esper s web page [22]. 4.4 Performance Esper is able to process over events per second on a machine running dual Intel Xeon processors with a marginal latency of below three microseconds on average. Esper also have great linear scalability on the same configuration from events per second to events per second [21]. For some of the tests the more advanced queries are limited by the network throughput rather than the CPU utilization. Unofficial attempts of benchmarking Esper have been performed using the included benchmarking kit and the YourKit profiler tool [53]. The hardware configurations used in the benchmark test is presented in Table 4.1. Both the client and the server are virtual machines running on a physical machine with dual Intel Xeon E5-2620V2 2.1GHz processors and 32GB of memory. The experiment measures throughput, latency, memory utilization and CPU usage for each query statement at increasing input rates. CPU Client QEMU Virtual CPU version 1.0 2GHz Server Memory 1GB 4GB OS Network JVM Ubuntu LTS Server Realtek RTL-8139/8139C/8139C+ 100Mbit/s IcedTea bit JVM Options : -Xms1024m -Xmx1024m Table 4.1: Hardware configurations for the Virtual Machines. With the hardware and environment configuration presented in Table 4.1 the results matched over events per second on average for EPL queries using operations such as selection, projection, disjunction aggregation. The conjunction, sequence and negation operations averaged above events per second. The average heap memory usage ranged between 30
41 MB and all the SQL operations except sequence maxed out below 250MB. In contrast to these benchmark results EsperTech has published results form a similar experiment on a laptop [23]. The laptop hardware consisted of dual core Intel Centrino T GHz and 4GB of memory. Although the results show that the laptop performed five times worse than a high performance server the amount of throughput is still above events per second and having comparable latency results. The overall benchmark results indicates that Esper is very suitable to be run on smaller or thinner hardware clients such as smart phones or even smart devices like clocks and wristbands. 31
42 Chapter 5 Requirement Analysis This thesis focuses on classification and analysis of sleep apnea data towards a future goal of having a fully self-sustained solution for use at home. The main goal is to use existing classification techniques and methods in combination with event analysis using Esper. We want to design and implement a skeleton concept that can detect sequences of apneas in subjects in real-time, classify the sequences and store the results and related information in a database for future use. We conduct performance profiling of the implementation to directly compare the resource utilization with hardware that is comparable in thin and portable devices, such as smart phones. As this thesis builds upon the data mining results and conclusion of the offline analysis we share a lot of similar requirements. In Section 5.1 we present a summary of the input data requirements from the offline analysis and discuss these with input data requirements for online analysis. We will analysis and determine if the requirements established are suitable for this thesis. In Section 5.2 we first summarize the four data mining methods suitable for offline analysis and determine if they can be used in this thesis. Further in Section 5.3 we set requirements and discuss the use of Esper to simulate a sensor and data streams. Lastly in Section 5.4 we present an analysis and discussion of suitable apnea detection methods using Esper. A complementary list of related work is presented in Section Input Data Polysomnography is the golden standard diagnosis tool for sleeping disorders performed today. The sleep study is performed in a laboratory where multiple electrodes and devices are attached to the subject to measure a wide range of signals. The list of measurements is broad as the sleep study tries to identify more than one disorder. Although the signal measurements are painless, some of them can be classified as being invasive to the patient by creating an unnatural sleeping environment and constricting movement. In the offline analysis the focus is on using non-invasive signal types as the goal was to give patients the ability to monitor and diagnose sleep apnea at home. Recording at home sets requirements to what kind of measurements can be performed at home by untrained personnel. The non-invasive signals are seen as being less constricting and prompt a more natural sleeping environment. ECG, EOG, EMG and EEG are all considered invasive as they require the use of electrodes and a heavy use of wires. Respiratory signals from the chest, abdomen and nose, in addition to oxygen saturation is used in the offline analysis as they require minimal 32
43 amounts of attachments and cables. Respiratory measurements from the chest and abdomen make use of an elastic band that measure the expansion and contraction when patient breaths. Oxygen saturation measures the oxygen levels in the blood stream by using infrared light in a clip attached to a finger. All of these non-invasive signals show clear epochs of abnormal breathing according to the results from the offline analysis. PhysioNet is a large online medical database that offers a large collection of recordings of various physiological signals. The web page is hosted by Harvard-MIT Division of Health Sciences and Technology and is a reliable source of data for analysis. The databases are well maintained and most of them are suitable for machine learning. PhysioToolkit is a set of Command-Line Interface (CLI) functions that can freely be used to extract the information from the different databases. In the offline analysis, out of the four sleep apnea databases available three of them were chosen. Annotations are included in two of the databases and can be used for supervised learning. The following PhysioNet databases are used [42]: Apnea-ECG Database The database consists of 70 records from 35 subjects with varying degrees of sleep apnea. The database is divided into 35 records included annotations which can be used for data mining and 35 records for testing. The sampling rate is noted as 100 Hz and apnea events are annotated every minute. Included is a file with additional information of the subjects used in the recordings, they all vary in terms of sex, age, height and weight. In addition, duration, number of apnea minutes and the AHI index is included. Eight of the 35 records contain respiratory signals from he chest, abdomen and nose as well as oxygen saturation. MIT-BIH Polysomnography Database The MIT-BIH Polysomnography database holds a total of 18 records from 16 different subjects, where two of the records is from the same subject. Signal sampling rate is just above the minimum requirements set by American Sleep Apnea Association (ASAA) at 250 Hz [5]. Included signals are ECG, EOG, EMG, EEG, respiratory from the chest, abdomen and nose, oxygen saturation, as well as blood pressure and cardiac stroke volume. While all the records in the Apnea-ECG contain the same number of signals, the MIT-BIH Polysomnography database lacks the consistency in signal combinations. The annotations included is more complex than the ones in Apnea-ECG. Annotations is every 30 seconds and have a slight more complex representation than the Apnea-ECG. Instead of annotating an epoch either as apnea or non-apnea the MIT-BIH Polysomnography database have multiple classes to determine if the epoch is either a hypopnea or apnea, either obstructive or a type of central apnea, and if the occurrence contain an arousal or not. St. Vincent s University Hospital Database The last database consists of 25 records and includes signals such as ECG, EOG, EEG, EMG, respiratory from the chest, abdomen and nose, oxygen saturation, snoring and the current body position. Similar to the Apnea-ECG database, all the records contain the same signal combinations. In the offline analysis a sampling rate of 128 Hz is mentioned as the correct one after inspecting the time frame. The sampling rate is a bit confusing, as none of the other databases contain multiple sampling rates in their records. A related work also presents the sampling rate to be around 256 Hz. None of these makes sense to us as the only value we get from the records are 8 Hz, the timestamps match 8 samples per second, not per second as stated in the offline analysis. After further inspection, 33
44 we can determine the signals are sampled at different rates. All respiratory and oxygen saturation signals are sampled at 8 Hz while the EMG signals are sampled at 64 Hz and the ECG sampled at 128 Hz. The annotations included are not the same as in the other two databases. Annotations are not segmented into time slices, but rather marked at where an apnea have occurred giving the timestamp Streaming Suitability In this thesis the focus is on simulating a physical sensor attached to a patient. In order to get the desired results, we need to determine if the signals and databases chosen in the offline analysis is suitable for streaming. Because we share a similar goal of home recording of sleep apnea data as with the offline analysis we want to set the same requirements for noninvasive signals. The records from the databases have already been streamed from the sensors or devices to a computer in a polysomnography, so we have already determined that the signals are fit for streaming. Data streams are just a sequence of data packets, the contents and origin of the data is irrelevant to its representation. The three databases used in the offline analysis have certain aspects that differs. Out of all the three, the Apnea-ECG seem to be the best fit in terms of records, diversity in subjects and consistency in signals. The database also achieved the best performance results for all the data mining methods evaluated in the offline analysis. The only issues are the low number of records that is suitable. The thought was that the database had been cleaned for noisy objects and artifacts, but that is not the case [18]. In terms of subject classification and cross-database classification the results from the offline analysis show poor results. Subject classification using one record for training and another for testing is very susceptible for class imbalance when one of the records is over represented with one of either class. Cross-database classification can be influenced in a number of ways as the databases might differ slightly, but causing the evaluation to be totally different. As we already know the databases are sampled at different rates, we can assume that also much of the sensor equipment differ from each other. There are many factors that can cause cross-database evaluation to produce poor results. Based on these factors we will only use the Apnea-ECG for evaluating the offline data mining methods using data streams as the database have shown best results and have overall the best data quality. As all the database contain non-invasive signals that can be used for apnea detection we will use all the databases to evaluate the two apnea detection methods. 5.2 Data Mining Picking the correct data mining techniques is critical to be able to classify apneas and generate an automatic diagnose based on the data gathered from the sensors. Similar to the input data we use, the ground work done in the offline analysis makes our choice an easier task to accomplish. As stated in the offline analysis, related work that have been using classification and sleep apnea have focused towards the medical aspect of the study rather than the technological aspect. The two most common methods used in related work are Support Vector Machine and Artificial Neural Network. The Decision Tree method has been of limited use in similar studies, but is less complex than the Support Vector Machine and Artificial Neural Network and is easier to implement and visualize. As both the Support Vector Machine and Artificial Neural Network are black-box methods, Decision Tree is included despite drawbacks when it comes to splitting trees on numerical data because it is a white-box method. K-Nearest Neighbor was 34
45 added as a fourth data mining method in the offline analysis. While Support Vector Machine, Artificial Neural Network and Decision Tree are all "eager learners", K-Nearest Neighbor is a "lazy learner". The offline analysis stated that the K-Nearest Neighbor method might not be a good fit for online analysis because of the increased training data size required for the method to classify. The required storage space might not be suitable for smaller devices, such as a smart phone. Based on the requirements and results from the offline analysis we want to test the same four data mining methods using data streams as input. We want to determine the performance impact by simulating online data streams using the offline data and data mining methods. As a secondary goal we want to measure how much resources are used in terms of CPU time, memory and storage. 5.3 Sensor Simulation We have established both the input and data mining requirements for this thesis. Our focus is to do online analysis of the sleep apnea data in real-time. Using a physical sensor, recording signals from patients is a time consuming effort as we need to collect a large amount of data to be able to correctly evaluate the methods we use. Even if we were able to collect the data we need, we would need to verify the quality and have a physician process the data to mark apnea events. Not only that, but developing or implementing a sensor that can produce these types of data is part of a total different field that this thesis is currently focused on. Therefore, we want to use the same data as in the offline analysis, but the data transmission will be different as we want to stream the data from the databases. Data streams is a broad term and covers everything from wireless data packets to simply reading a file from disk into memory. Instead of implementing a system or approach form the ground we want to use a system that is already available and proven to be suitable. Esper is an open source component library and event analysis engine developed by Esper- Tech. It is one of the few free open source data stream management systems out there and have been used to detect myocardial ischemia in related work and intrusion detection in wireless sensor networks in less relevant work [17] [71]. With EPL we can create complex queries and incorporate these into functions to handle and manipulate the data stream. Esper have multiple built-in functions to handle windows and control the flow of data streams. Incoming data streams can be branched off to create new temporary data streams that conforms to other sets of queries. Adding this to the fact that it is fully supported on the Java platform is less constricting and can contribute to the overall goal of automatic diagnosis. Esper is also well documented and supports a wide range of adapters to handle input data, which fits our task, even though the learning curve is a bit rough to understand all the concepts and options available for the EPL. 5.4 Apnea Detection For a monitoring system used at home we can see the benefit of implementing various triggers and alarms that can evoke at different stages of sleep, or when abnormal breathing patterns reach critical levels. We have established the input data and the use of offline data mining methods so our next focus is to find detection methods that we can incorporate and adapt into a rule-based classification approach using EPL. 35
46 In the offline analysis they use a set of proven and tested data mining methods to classify apnea epochs. The case in our thesis is a bit different as we do not nearly have the same amount of related work that have both used Esper for classification and using any form of data from sleeping disorders. The vast majority of research papers that use Esper for detection is focused on network intrusion detection. Esper have been used to detect myocardial ischemia using ECG and QRS detection over sliding windows. Results showed that Esper and more or less simple EPL queries are able to separate heart beats and cardiac anomalies [71]. This concept can further be developed into a system where personnel without programming skills can create detection schemas through a simple interface. Based on these results and that event processing have been used to detect anomalies in sensor networks make us believe that we can detect epochs of abnormal breathing by using simple EPL queries over sleep apnea data streams without the need for advanced classifiers and pre-generated models with training data. A common way to detect anomalies and patterns is to use statistical methods. Statistics is the analysis and presentation of unstructured data. Anomaly detection is to identify events or objects that does not conform to the rest of the data set. In our case the anomalies are the epochs of abnormal breathing, the apneas that causes the signal measurements to shift, often with a high degree of random spikes. Moving Average is a statistical method that calculates the mean average over a set of data points in a sub set of a larger population. Moving average is a popular method often used in economics and stock markets to even out the fluctuations in the data. The method is capable of eliminating short-term spikes and bring out long-term peaks and lows in a data series. Standard deviation is another method taken from statistics which focus on calculating the amount of variance or disruption in a sub set of values. A higher variance or deviation means that the data values in the sub set is further in-between than a lower variance or deviation. Standard deviation can be used to calculate the incline or decline in a sub set of values represented in a graph. Both these statistical methods can be incorporated into EPL queries and made into custom functions using Esper. Standard deviation is already supported as a built-in function in Esper, but it lacks some properties that we otherwise can develop with a custom function. Figure 5.1 presents a small plotted segment of the four non-invasive signals suitable for this thesis. The data the graphs represent is taken from the Apnea-ECG database using the record from subject "a02r". Even though all the signals suit offline data mining, for a rule-based approach using EPL we have to look at how well we can extract a pattern based on the attributes that forms from each signal. All of the four signals are non-invasive and can without trouble be handled by a person lacking experience and training. Respiratory from the chest and abdomen are recorded using a band that can constrict the patient s movement and can also become trapped in certain positions that make the patient uncomfortable and give inaccurate readings. Respiratory measurements taken from the nose is either recorded using a mask that covers the mouth and nose area or a small tube that is partially inserted into the nostrils. Respiratory measurements from the nose are less prone to inaccurate readings cause of body positions but can still be considered uncomfortable as the mask is susceptible for condensation. Oxygen saturation is recorded using a small sensor that is attached to one of the fingers with a clip. Even though all the signals mentioned are non-invasive, oxygen saturation is the least invasive out of them. The clip is easy to attach and does not require long cords as the sensor equipment can reside on top of a glove or around the wrist. As for signal attributes all of the signals show epochs of abnormal breathing. In Figure 5.1 we see that the respiratory signals indicate lack of breathing as segments of little to no fluctu- 36
47 Figure 5.1: The various signals plotted in a graph using data from Apnea-ECG. ations in the data stream followed by a smaller segment of spikes, indicating breathing. The latter segment has huge spikes as the body gasps for air to regain stability. Oxygen saturation presents more stable properties as the person stops breathing we see a steady decline in oxygen saturation. When the person regains stable breathing, the oxygen levels in the blood steadily increases to normal. While the upper and lower boundaries of respiratory signals can in theory scale indefinitely and vary based on the data presentation, oxygen saturation on the other hand have pre-defined boundaries as it is measured in percentage. A normal breathing pattern would have the graph linger between 92-96%, clearly having less fluctuations. With the two statistical methods above we have two separate approaches to detecting epochs of abnormal breathing. Based on time constraints and the related work we want to use oxygen saturation as the signal for apnea detection as this is the least invasive and best physiological signal to monitor in a home setting. A combination is also possible but would require more time to incorporate them into the overall design, this will not be done in this thesis. 5.5 Related Work Related works from the offline analysis focus more towards the medical aspects of the research and analysis rather than the technological. The vast majority of the results are based on ECG signals as input which is something we have regarded as intrusive and discarded from our list of suitable signals. It is hard to find related work that touches upon all the different aspects in our thesis: the online analysis of sleep apnea subjects, a purposed concept design of an application and performance measurements with low-end devices in mind, and utilizing complex event processing with Esper. We had to put focus on related works that purposed ideas and analysis using online or real-time analysis of sleep apnea data. A lot of the related work regarding using CEP and classifying data using rule-based classification techniques and EPL is not related to using sleep apnea data, but the methods and approaches are still valid 37
48 to our thesis. Finding public articles and information regarding usage of Esper or concepts using Esper explicitly is hard to find. In addition to most of the related work in the offline analysis, we have also collected a list of some related work that contributes to the design and implementation of this thesis in Table 5.1. Reference [42] Data Mining for the Detection of Disrupted Breathing Caused by Sleep Apnea - A Comparison of Methods [12] On-Line Detection of Apnea/Hypopnea Events Using SpO2 Signal: A Rule-Based Approach Employing Binary Classifier Models [17] Online Analysis of Myocardial Ischemia From Medical Sensor Data Streams with Esper [69] Deviation Detection in Automated Home Care using CommonSens Focus Data Mining & Classification Anomaly Detection Anomaly Detection & CEP Detection, CEP & Esper [55] Online Association Rule Mining over Fast Data Data Mining & CEP [48] Adaptive Classification System for Real-Time Detection of Apnea and Hypopnea Events [40] An Online Sleep Apnea Detection Method Based on Recurrence Quantification Analysis Detection & Data Mining Detection & Data Mining [44] Unobtrusive Online Monitoring of Sleep at Home Sleep Monitoring [24] A Generic Intrusion Detection and Diagnoser System Based on Complex Event Processing [75] Asynchronous Standard Deviation Method for Fault Detection [71] Complex Event Processing for Object Tracking and Intrusion Detection in Wireless Sensor Networks Anomaly Detection & CEP Standard Deviation & Detection Detection, Esper & CEP Table 5.1: List of used related works to determine our requirements for the design in this thesis. 38
49 Chapter 6 Design In Chapter 5 we summarized the requirements used in the offline analysis and expanded on them by establishing requirements for our thesis. First we reviewed the physiological signals and the need for non-invasive sensor equipment. We determined that the four signals, respiratory from the nose, chest and abdomen, together with oxygen saturation is suitable for online analysis. Further we analyzed the data sources used as input for the offline analysis. All of the databases considered is hosted by PhysioNet which is a reliable source of medical data. The three databases in question is Apnea-ECG, MIT-BIH Polysomnography and St. Vincent s University Hospital. These databases have been used as input data to evaluate the performance of the data mining methods which achieved good results. We concluded that in order to evaluate the performance of the data mining methods using data streams as input, we want to use the same input data so that the we get a more accurate comparison. The input data can be used to evaluate the apnea detection methods as well. K-Nearest Neighbor, Support Vector Machine, Artificial Neural Network and Decision Tree are the four data mining methods chosen in the offline analysis. Both Support Vector Machine and Artificial Neural Network have been extensively used in related work to classify apneas using various signals as input. All of them besides Decision Tree achieved good results when classifying epochs of abnormal breathing. We want to incorporate the same data mining methods into our design and implementation to evaluate their performance on data streams and to assess the resource utilization. Further we concluded that we want to use EPL to create two separate detection approaches using statistical methods. Moving Average and Standard Deviation are extensively used in economics and statistical analysis and can be integrated into custom aggregate functions and queries using Esper. In Section 6.1 we discuss designs for our implementation with focus on simulating streaming using Esper in the best possible way to create a realistic scenario. We go through the components needed and the architecture we plan to adopt. Section 6.2 and 6.3 presents a detailed overview of the three databases chosen in Chapter 5, how they are structured and a design for pre-processing of the data. In Section 6.4 we summarize the design choices of the data mining methods discussed in the offline analysis and determine how we incorporate these into our implementation and use with EPL queries. The design for our apnea detection methods is explained in detail in Section 6.5. As a complementary task, we discuss output storage solutions in Section 6.6 and the last Section 6.7 we present an outline of our evaluation process for the data mining methods, the apnea detection methods and the resource profiling. 39
50 6.1 System Overview In this section we first present an initial design that we developed. This design had some flaws and disadvantages under early testing which led to the revised design which is detailed later in the section. The two design alternatives both work as is, but the drawbacks of the initial design led to the revised design. Before we go into detail about the two designs we need to discuss some restriction and requirements that comes with using Esper and our approach to simulate a sensor under a realistic scenario: Esper is only available for the.net and Java platforms. We want a future application to transmit the data from the sensor to the smart phone over a wireless connection. We want to evaluate the resource utilization of the main components of the implementation. Based on our experience with the Java platform and our general inexperience with the.net platform we chose to go with an implementation using the Java programming language. Although Esper only supports specific platforms, they have a wide range of input adapters that can send and receive different data types based on the integration. This means that Esper can be integrated into an already active system or be designed as a stand-alone system communicating via HTTP requests or sockets. The different input adapters are: File and CSV Spring JMS AMQP HTTP Socket Relational Database Along with great input support Esper also have output support for most of the popular data formats such as JSON and XML. How our data is presented is not so relevant for our case because we want to store the data in a database to measure the performance when it comes to I/O operations. The data stored in the database can later on be used to any other scenario if needed Initial Design Our first design is presented in Figure 6.1. This design incorporates all the main components we need to get a functioning implementation. Records from the three different PhysioNet databases is read by the input handler component. The component reads the records by requesting a system call to the WFDB Toolkit functions and processes the records directly. We implement the pre-processing of the data and align the data with all the annotations and record details in an input handler. The input handler creates event objects that is sent to the event handler component and then sent to the Esper engine. Before the input handler starts 40
51 to parse the records the system loads the configuration file. The file contains various options such as which data mining or apnea detection method to use and information about the record properties. Once the input handler starts to parse the records the event handler sends the specific event segments to a chosen data mining or apnea detection method. The method chosen classifies the segment and sends the result to the storage component which inserts the information into a database table. Figure 6.1: Architectural overview of the initial design. This design is compact and is able to stream the records in the form of events to the Esper engine, classifying the annotation segments and store the results and other related data to a database. But the design purposes some drawbacks and does not fulfill two of our requirements mentioned in Section 6.1. As data streaming is a broad term, the records are read by using file descriptors that streams the data to the input handler which again creates events that are sent to the Esper engine for processing. This solution gets the job done and in theory simulates data streams because it is processed by Esper, but the connection-less solution does not fulfill the requirement of a transmission over a wireless communication channel. The second drawback is the compact design. It is less complex than to split the design into standalone services, but it comes at a cost. When profiling the implementation using a profiling tool we immediately see that the system generates a lot of overhead when processing the records. In terms of a real sensor, the data is either in the form of raw data or it can be transformed 41
52 into a more suitable format so that the application is able process the data directly from the communication channel. Most likely, when a real sensor will be used to transmit the data to an application, the data objects and handling of the data will be tailored and optimized to each other. The extra resources spent transforming and reading the data from the PhysioNet records can be done by the sensor and transmitted to the application for classification. These drawbacks are the reason for the revised design that is a bit more complex but eliminates the drawbacks and is an overall better solution to evaluate the resource utilization Revised Design The revised design is based on the initial design but we use a client-server architecture to improve the resource utilization and to illustration and test a more realistic simulation. The system overview for this design is illustrated in Figure 6.2. The overall server design remains very similar to the initial design. the main difference is that we have separated the input handling to a separate process. When the server starts the configuration loader component loads the configuration file. The file contains the same information as in the initial design, but some of the less static options that does not control the behavior of the classification output is done by the CLI parser. Once the specified options are loaded the server waits for an incoming connection. The client reads the CLI options that specifies connection information and the output rate to send. We want to ensure that we can control the output from the client so that we can test the implementation at different output rates. Which output rate is specified impacts the resource usage when profiling the implementation. Figure 6.2: Architectural overview of the revised design. The client establishes a connection to the server and start to process the records. The records are read per line and the values are wrapped in Java objects that forms the events. The event class is registered with the engine and is processed through Esper and sent over the connection once it triggers. The objects are sent over the socketed connection to the server and is received by the event handler. Upon loading the configuration file, the server generates the queries and registers them and the event object with the Esper engine. The events that are received 42
53 is directly sent to the classification method specified on server initialization. By classification method, we mean either one of the four data mining methods in the offline analysis, or one of the two apnea detection methods derived from EPL queries. Once the classification method processes the events and produces a result, the result is then sent to the SQLite component and stored in a database table. Now with this design we divide the workload onto a separate process to keep the resource footprint from the implementation as tiny as possible while improving the simulation. The design mimics a client acting as a sensor streaming signal data to a server which is acting as the smart phone application. Responsibilities are almost the same as with the initial design but with the segmented input processing we eliminate the overhead generated when we start to measure resource usage with a profiling tool. This results in a more accurate evaluate of the resource usage, even though the implementation is just a concept and does not represent a finished application. 6.2 PhysioNet Databases The three databases used in this thesis is explained in detail in Sections 6.2.1, and We list all the records we use in to evaluate the data mining and apnea detection methods in tables. All of the databases have some similarities and some discrepancies that we elaborate and handle in Section Apnea-ECG Database The Apnea-ECG database consists of 70 records in total. Only eight of the records contain the signals we have set as a requirement, the 62 other records contain just ECG signals or a combination of ECG and the respiratory signals, excluding oxygen saturation. The files with just the ECG measurements have no postfix and the records containing ECG and respiratory have the "er" postfix. Sampling rate for the database is set at a 100 Hz, which is a bit low considering newer guidelines set by the ASAA [5], which suggest a minimum sampling rate of 250 Hz. In Table 6.1 we have listed the records we use in this thesis, including their name, total length in minutes, how many non-apneic and apneic segments are included and the AHI index. Record File Total Length Non-apnea Annotations Apnea Annotations AHI a01r.dat a02r.dat a03r.dat a04r.dat b01r.dat c01r.dat c02r.dat c03r.dat Table 6.1: Records from the Apnea-ECG database. 43
54 All the records contain the same signals and have the same composition. The first column is the sample index followed by respiratory from the chest, abdomen and nose on the second, third and fourth column. The last column contains the oxygen saturation signal. Annotations is set to every 60 seconds, meaning we have a total of 6000 data rows per annotation segment. The annotation files correspond to each record with the same name with ".apn" as the file extension. The annotation files have seven columns where the second and third column contain the start index for the annotation segment and the apnea class respectively. The classes are stored as either N for segments containing no apneas and A for segments containing one or more apneas. The AHI index for the records span from very severe to perfectly healthy subjects which might impact the performance evaluation as we have no subjects that are in between these two groups MIT-BIH Polysomnography Database The MIT-BIH Polysomnography Database contains 18 records from 16 unique subjects. Two of the records are from the same subject and is have the same name but differentiated by an "a" and a "b" postfix. Out of the three, the MIT-BIH Polysomnography database is the smallest one. The records are sampled at a rate of 250 Hz which is the minimum requirement according to guidelines. Despite having all the required signals, there is no record containing all of them. The records have an arbitrary number of signal combinations. As we want to use the data to test the apnea detection methods we will use the records that contain oxygen saturation, they are listed in Table 6.2. Record File Total Length Non-apnea Annotations Apnea Annotations AHI slp59.dat slp60.dat slp61.dat slp66.dat slp67x.dat Table 6.2: Records from the MIT-BIH Polysomnography database. Which data is contained in which column is controlled by the signal combination of the given record. Annotation segments are marked every 30 seconds which corresponds to 7500 data rows per segment. The annotation files use the ".st" extension and the annotation classes are presented a bit different than in the Apnea-ECG database. The structure of the annotation file is almost the same as with the Apnea-ECG database, but some of the columns have shifted. The third column is still the start index for the first annotation segment, the ninth column without a header contain the class annotations. Instead of having two classes the classes are divided into types of sleep apnea. The different class annotations and what they represent is listed in Table
55 Annotation CA CAA H HA OA X Representation Central Apnea Central Apnea with arousal Hypopnea Hypopnea with arousal Obstructive apnea Obstructive apnea with arousal Table 6.3: List of annotation types MIT-BIH Polysomnography database. Both this thesis and the offline analysis will not distinguish between the different types of sleep apneas. Our general focus is to separate epochs of normal and abnormal breathing. This means that we will mark the different annotations listed in Table 6.3 as all being an apnea, or a period of abnormal breathing. We will not distinguish the differences between the types St. Vincent s University Hospital Database St. Vincent s Database is the largest database we will use. The database holds 25 records and share similar structure as the Apnea-ECG containing all the required non-invasive signals. Each record contains the same amount of signals, with an addition of other signals that are not relevant for this thesis. In the offline analysis there was some confusion surrounding the sampling rate. The records contain different sampling rates for the various signals. The sampling rate in the header file is set at 8 Hz, while each row in the file lists 64 Hz for body movement and EMG and 128 Hz for the EEG. For all the signals we use the sampling rate is left unchanged at 8 Hz. A list of all the records is in Table 6.4. This database has a good variation in subjects from different severity groups. 45
56 Record File Total Length Apnea Annotations AHI ucddb002.rec ucddb003.rec ucddb005.rec ucddb006.rec ucddb007.rec ucddb008.rec ucddb009.rec ucddb010.rec ucddb011.rec ucddb012.rec ucddb013.rec ucddb014.rec ucddb015.rec ucddb017.rec ucddb018.rec ucddb019.rec ucddb020.rec ucddb021.rec ucddb022.rec ucddb023.rec ucddb024.rec ucddb025.rec ucddb026.rec ucddb027.rec ucddb028.rec Table 6.4: Records from the St. Vincent s database. In column one we have the sample index, the seventh column contains the oxygen saturation, the ninth column contain the airflow from the nose, and respiratory from the chest and abdomen are presented in twelfth and thirteenth column. The annotations are denoted a bit different than the Apnea-ECG and MIT-BIH Polysomnography database. Instead of slicing the data into annotation segments of a given length, the included annotations just mark the timestamp of where the apneas occur. A list of annotation types is presented in table 6.5. These are taken from the annotation files which is not the same as the ones listed on the PhysioNet web page. Like with the MIT-BIH Polysomnography database the different types will be combined to represent one class. 46
57 Annotation HYP-O HYP-C HYP-M APNEA-O APNEA-C APNEA-M POSSIBLE Representation Obstructive hypopnea Central hypopnea Mixed hypopnea Obstructive apnea Central apnea Mixed apnea Either hypopnea or apnea, unclear type Table 6.5: List of annotation types for St Vincent s database. 6.3 Input Data In order to evaluate the data mining and the apnea detection methods we need to prepare the input data. The events represented in Esper is Java objects containing local variables, get methods and set methods. So how we process the records can be done in multiple ways. To make it simple we have chosen to create test and training files in the CSV format. Esper have an input adapter that supports CSV files, but we can also create a custom input adapter if we need to. Since we have chosen to use just the Apnea-ECG database to evaluate the data mining methods, we only need to create training files from this particular database. To evaluate the apnea detection methods, we create test files from all the databases. The offline analysis state that since the signals show change over seconds and that an apnea can last for over a minute, the sampling rate can be scaled down to 1 Hz and still be fit for classification and clearly show epochs of abnormal breathing. We will create three different Python scripts to process the records using the PhysioNet toolkit into ready to read CSV files. The three different scripts will be tailored to each specific database to handle the differences between them. As the pre-processing phase is not so important to our performance evaluation, we see no point in integrating this process in the client, but rather have ready to go records that can be sent to the server. The script that processes the Apnea-ECG database will read all the eight records into memory. Since we want to test the same data mining methods using data streams we also want to use the same evaluation method used in the offline analysis. Ten-fold cross-validation is a validation method to assess the performance of how a data mining technique will generalize to an independent data set. This means that we need to divide the eight records combined into ten testing sets with ten corresponding training sets. Most of the records in the Apnea-ECG database contain more data than there are annotations. As all the annotation indices in this database is set at the start of the records, the solve the issue by slicing of the last part of the data set that corresponds to the number of annotation segments in the annotation file. So each annotation is 6000 rows, we multiply the number with the number of annotation segments and get the end index for the data sets. Once we have read and aligned the eight records and corresponding annotations we divide the data into ten equal parts. For each part we assign as a testing set, we select the nine other parts as a training set. The corresponding training set is first processed by averaging 6000 rows. Once the training set have been aggregated we append the classes read from the annotation files. The training set is then written to a CSV file having 47
58 the same index in the file name as the testing set that correlate. To create the test files for the apnea detection evaluation we create a separate CSV file for each record read. As the methods are based on detection of the fluctuations in the oxygen saturation, a combination of the records will generate unnatural inclines or declines once the data from one record goes over to another record. Once a record is read we align the data based on the annotations and write the column containing the oxygen saturation data to a CSV file. There is no need to average the test files as this is done by Esper on arrival of new sensor events. To test if normalization have an impact on the data we also create the same test and training files for the data mining evaluation using minimum-maximum normalization. The eight records chosen from the Apnea-ECG database have an almost equal number of classes. Class imbalance can cause the data mining methods to sway in favor of one or the other class if the training data is over-saturated with one specific class. If we use K-Nearest Neighbor and the class balance is equal to one out of ten, using a k higher than one will always result in a classification with the highest majority. We eliminate this problem by combining the records into a big data set and use cross-validation which tests the whole data set using training data with an almost equal balance of classes. A balanced distribution is key to evaluate a data mining method as the results can be good, but in theory the data mining methods can perform average at best if using an unbalanced testing or training set. For example, training data that contain almost no apneas will create a model that is over fitted with one specific class. This results in a model that can have almost a 100% accuracy when classifying records that contain no apneas, or of patients that is not diagnosed with sleep apnea. Classy fining records with a moderate to a high number of apneas will result in a high misclassification rate. The pre-processing scripts for the MIT-BIH Polysomnography and the St. Vincent s databases are very similar to each other. They both read each record and create a corresponding CSV file that can be streamed by the client. For the MIT-BIH Polysomnography database the annotation segment starts at different indices and needs to be aligned so that we have the correct class corresponding to the correct segment of data. The records slp59 and slp61 starts at index and respectively. Alignment is done by cutting of the first parts corresponding to the start index. With the St. Vincent s database, this is not an issue as the annotations for the records are not divided into timed segments like with the other two databases. Instead the apneas are denoted when they occur. 6.4 Data Mining We want to evaluate the data mining performance over data streams using the same data mining methods used in the offline analysis. Finding a suitable library for Java that have all these four methods, as well as coming from a reliable source is a difficult task. We have evaluated some different libraries, such as WEKA and Neuroph, but found the best solution to be using the Matlab toolkit. Matlab supports compilation of custom functions including most of the machine learning toolbox to external libraries. This makes it possible to compile the data mining functions to a JAR file which can be imported and used as regular functions in Java. In the four sections below we present a short summary of the design for each method used in the offline analysis and their available options. Our focus is not to test these extensively as this is already done for us, so we will use the best available settings that produced the best results according to the evaluation in the offline analysis. All the options and settings for each data mining method from the offline analysis is determined by what is available in the Matlab 48
59 toolbox K-Nearest Neighbor The K-Nearest Neighbor method is regarded as the simplest out of all the data mining methods used. It has no form of training phase and only do the computation using a local approximation. It works by measuring the distance between k set of objects in correlation to the test object. The method is used for both classification and regression purposes. There are some things to consider about this method. When selecting a k value that is an even number there is a chance that the majority inside the circle may be equal. This is not a problem when select k as an odd number as it is not possible to produce this scenario. There are three tie breakers that can decide the class output. We can select the class that has the smallest index, select the class of the nearest object or select the class at random. The second thing to consider is a weighting option for objects that is further away from the test object. In situations where a tie breaker is not enough a weight to the objects can be considered. No weighting, inverse and squared inverse is three common weighting schemes. Inverse weighting is the weight of 1/d, where the distance to the neighbor. Squared inverse is the weight of 1/d 2. The third consideration is the distance function to apply. Matlab supports in total eleven different distance functions. The difference in these functions is the way they compute the distance between objects in a data set. One of the more popular function used in related work is the Euclidean function. The offline analysis used one, five and ten as the values for k. More neighbors than ten did not improve the overall performance. As for the tie breaker, the smallest index was used as this had a slight advantage in initial tests than the others, but the results from all the tie breakers were pretty similar with tiny variations. No weighting of the objects was used because the data set objects had clear clusters and little to no overlapping objects and the results from all the weighting options were similar. Even though the Euclidean function is the most commonly used, the offline analysis used the Cityblock function as it produced slight better results in initial testing and most of the other functions produced similar results. The setup for this thesis is identical to the one used in the offline analysis with us just using a k value of one. Results from the 15 signal combinations from the Apnea-ECG database were in favor of k being one. For the MIT-BIH Polysomnography database having k set to five had a slight favor in the results, but we will only be using the Apnea-ECG database as it is the most consistent one and produced the best overall results. A summary of the options used for this method is listed in Table 6.6. Option k Value 1 Tie Breaker Weight Function Distance Function Value Smallest Index No weight Cityblock Table 6.6: List of used options for K-Nearest Neighbor. 49
60 6.4.2 Support Vector Machine Support Vector Machine is the one most advanced and common data mining method used in the related works. This data mining method is complex and mathematically heavy which requires a thorough knowledge of mathematics to understand all the aspects of this algorithm. The method works well with problems that are binary and as we have only two classes to consider, the method is suitable for sleep apnea classification. SVM calculates and finds a hyperplane between the classes represented in the training and data set. The hyperplane is the maximum distance between the closes objects from each class in the vector space. Once the hyperplane is derived, a test object that falls on one either of the sides of the maximum margin hyperplane is determined to belong to the closest class. In the offline analysis they state that an optimal Support Vector Machine configuration is not possible to know in advance. This means that most of the configuration options were derived from initial test experiments. In order to solve the problem of convex optimization they chose to use the Dual Lagrangian multiplier, which is the most common method and the default one in the Matlab toolbox. The convex optimization compares the similarity between objects by utilizing the dot product. They picked a soft-margin instead of a hard-margin so that some of the objects overlap and can be misclassified. Because of this we need to introduce a penalty to the misclassifications to keep the error rate as low as possible. A higher penalty leads to worse training times. From the three available kernel functions in Matlab the Radial Basis/Gaussian function performed overall the best. Because of this all the options and settings remain the same as in the offline analysis except the kernel functions, we will only use the Radial Basis/Gaussian function. Option Margin Kernel Function Misclassification Penalty 1 Value Soft-margin Radial Basis/Gaussian Table 6.7: List of used options for Support Vector Machine Decision Tree For the Decision Tree method, we need to look at which aspects we need to consider in order to generate an optimal tree structure. A tree structure consists of a root node and multiple levels with parent and children nodes, with leaf nodes at the bottom of the tree. As the tree grows because of splits in the numerical values it increases the trees complexity. In order to stop the tree from becoming more complex and larger than it needs to be we need to look at how we split the values, the stopping criteria to halt the tree growth and pruning. There are two ways of splitting values either by using a heuristic search method or an exhaustive search method. The exhaustive search method derives all the possibilities from each attribute object, while the heuristic search method evaluates the splits based on the available information. There are three splitting criterion available in Matlab, Gini s Diversity Index, Deviance and Twoing. The Gini s Diversity Index calculates an index for each node in the tree. 50
61 When the node s index is set to zero the node is a pure node, and all children nodes result in the same class. The Deviance method evaluates how observations deviate from being a pure node. The last method, the Twoing, evaluates each class leaf nodes and splits the observations in fractions. Then it traverses up the tree to check deviations in purity of parent and children nodes. The second consideration is tree growth and to set the optimal stopping criterion. As with the splitting criterion, we have three available options in Matlab. The first criterion is if a node is pure, only containing child nodes with the same class. The second criterion is controlled by maximum observations from the parent node. And the third is controlled by a minimum requirement of observations for a leaf node on split value. Pruning of the tree, alongside the stopping criterion, is a method to minimize tree complexity and growth. Pruning is the operation of removing a sub tree from the tree structure that has no benefit. We can either pre-prune, post-prune or do a combination of the two. The two different running approaches is if the prune process is done while the tree is generated or after the tree is complete. The offline analysis used all the default values that is set by Matlab. They used an exhaustive search combined with Gini s Diversity Index to find and split values. Exhaustive search is sufficient as we only have two classes to think about and all the splitting criterions had similar performance in initial testing. The stopping criterion is to check when a node is a pure node. Because we have two classes we want to limit the tree growth to a pure node as this will generate an optimal tree and remove unnecessary levels of complexity. As for pruning, the offline analysis chose to pre-prune the tree. Which pruning method was chosen had no benefits in performance. Because the offline analysis used all the default option values in their testing and adjusting the values had no increase or decrease in performance we use the same settings for our Decision Tree implementation. A list of the options used for our Decision Tree design is in Table 6.8. Option Tree Structure Split Finding Split Evaluation Stopping Criteria 1 Stopping Criteria 2 Stopping Criteria 3 Value Binary Exhaustive Gini s Diversity Index Node is pure Node contains less observations than parent (10 observations) Potential child nodes would contain less observations (1 observation) Table 6.8: List of used options for Decision Tree Artificial Neural Network One of the two most popular data mining methods used in related work is Artificial Neural Network. A network can either be a Single-Layer Perceptron model or a Multi-Layer Perceptron model. The difference between the two is that the Multi-Layer model can generate additional layers of abstraction and decision boundaries to the network based on the number 51
62 of hidden layers in between the input and output layer. A Single-Layer model can only separate the data linearly. The most common variant of the Multi-Layer model is the feed-forward variant. The other variant is the recurrent neural network where the nodes in the layers form a direct cycle, which means the variant is better at solving other tasks that is not the focus of this thesis. How many input nodes corresponds to the number of attributes in the database chosen and the signal combinations. For the Apnea-ECG database we have 60 attributes per signal which equals to 60, 120, 180 or 240 total input nodes respectively. The number of output nodes is related to the number of classes in the given data set. For two classes an option of either using one output node or two output nodes that represent true or false can be used. A single output node will only trigger for either of the classes. The number of hidden layers in a network impacts the complexity of the network. A more complex network does not necessarily mean better results as there are claims that a network with more than two hidden layers is not needed. The amount of nodes in the hidden layer can vary depending on the data set and output goal. Additional key factors of the network are the initialization of the weight and biases on the layer nodes. The Artificial Neural Network function in the Matlab toolbox has Nguyen-Widrow algorithm as a default function for initialization of weights. The weights combined with a reference threshold and an activation function is used to produce the output outcome. The weight initialization is done in a random fashion and does not result in the same weighting scheme. Another key aspect is the learning algorithm, also known as backpropagation. Backpropagation networks are the most prevalent neural networks used and is the key to make the network learn from errors. The idea behind backpropagation is to calculate gradient descent from all the connected weights in the hidden layer. The gradient is then fed to an optimization function to update the weights to minimize the potential error rate when classifying. The following general steps are performed in backpropagation [8]: 1. (Propagation Phase) Forward propagation of a training pattern s input through the neural network in order to generate the propagation s output activations. 2. (Propagation Phase) backward propagation of the propagation s output activations through the neural network using the training pattern target in order to generate the deltas (the difference between the targeted and actual output values) of all output and hidden neurons. 3. (Update Phase) Multiply its output delta and input activation to get the gradient of the weight. 4. (Update Phase) Subtract a ratio (percentage) from the gradient of the weight. The subtracted ratio from the gradient is also called the learning rate. This propagation and update process is repeated in an effort to find the optimal training speed while still producing a good accuracy. The toolbox function in Matlab allows for twelve different backpropagation functions. The final aspect to consider is the activation function, also called the transfer function, for the hidden and the output layer. The data fed to the input nodes are seen as activation values 52
63 and are sent throughout the network. Each node has an activation function which derives an output value based on the activation value. The activation functions sum the received activation values and produces an output value. Activation ranges that conform to either a firing or not firing is up to the activation function used as some have a range from 0 to 1 and others have a range from -1 to 1. In Matlab we have a total of 16 different transfer functions to choose from. The offline analysis chose to use a Multi-Layered network using the feed-forward variant and a single output node in the output layer. Because we only have two classes, normal and abnormal breathing, we can evaluate the class based on if the output node fires or not. Triggering the output node is a positive indication of an abnormal breathing period, if there is no trigger the result is negative. The number of input nodes correlates to the database and signal combination used for training. As for the initialization of weights they used the default function, the Nguyen-Widrow function. There was no reason to try any other functions as it required more experience and knowledge about the relationship among the data set and the attributes. Increasing the learning rate and epoch updates when training the network had no performance gain and remained at the default values. After trying different combinations of learning algorithms to find the best suitable one, they landed on the Scaled Conjugate Gradient algorithm. All the conjugate algorithms performed seemingly similar to each other. Even though Matlab has 16 available transfer functions. The Sigmoid and Softmax functions were chosen as the combination from the results of the initial experiments. Using the Sigmoid Hyperbolic Tangent function at the hidden layer and the Softmax function at the output layer results in the best performance in the offline analysis. Results from the offline analysis concluded that 20 hidden nodes produced the overall best results, but not by much. Because the results were so similar we will use the same number of hidden nodes in our design and performance evaluation. Besides the hidden nodes, there was not used any other variable settings in testing of the data mining method. All initial experiments performed in the offline analysis resulted in exclusion of bad performing options and combinations. As the focus for this thesis is to only test and evaluate the data mining functions, our design results in the same optimal settings. A summary of the options in the offline analysis with the addition of our options is listed in Table
64 Option Value Network Type Multi-Layer Network Network Model Feed-Forward Input Nodes 60, 120, 180 and 240 Output Nodes 1 Hidden Nodes 20 Hidden Layers 1 Backpropagation Scaled Conjugate Gradient Learning Rate Adjusted by SCG Weight/Bias Nguyen-Widrow Function Max Update Epochs 1000 Activation of Hidden Neurons Hyperbolic Tangent Function Activation of Output Node Softmax Function Table 6.9: List of used options for Artificial Neural Network. 6.5 Apnea Detection Besides integrating the four data mining methods in the offline analysis we want to utilize Esper to implement online apnea detection. In this section we present the design for two different approaches using statistical methods such as Moving Average and Standard Deviation to detect periods of abnormal breathing over sleep apnea data streams. The two methods in question are presented in detail in section and Other related work has used sliding window segments in combination with data mining classification methods as an approach to classify apneas. We purpose an alternative solution by creating EPL queries and use the functions available in Esper Moving Average A moving average is a statistical method to calculate the mean or average value over a subset of values from a full data series. The idea is simple and is wildly used in economics and trade marketing domains to smooth data in a time series to eliminate small spikes and to highlight bigger tendencies and trends. The time series in our case is the incoming sleep apnea data from the sensor. There are multiple variations of moving average, where simple, cumulative and weighted are the ones most commonly used: Simple Cumulative Weighted Exponential Both Cumulative Moving Average (CMA) and Weighted Moving Average (WMA) have slight different properties to how they work compared to the simple variant. CMA adds up and 54
65 derives a new moving average for each incoming new value from a fixed point in the time series. This means that there is no set window length and that the old values are kept in the subset rather than being discarded. WMA adds weight to each number in the subset where the most recent events get weighted more than old events. In our case, all the numbers in the subset is equally important and using the weighted variant is not a good option. The cumulative variant over time results in the same moving average as the simple variant with a large enough window length, so there is no benefit of using cumulative in our tests. So our design is based on Simple Moving Average (SMA) seen in Equation 6.1 and 6.2, with slight adjustments to fit our data. SMA = p M + p M p M (n 1) n = 1 n n 1 p M i (6.1) i=0 SMA today = SMA yesterday + p M n p M n (6.2) n One thing we need to consider when adapting the moving average to our data stream is the window length of the subset of incoming numbers. The length determines the factor of smoothing on the subset data. With a smaller window length, the averaged total is more influenced by new incoming values which cause the moving average to be closer to the original data series. A larger window length has little to no fluctuations depending on how large the window is. Considering how oxygen saturation operates, where the drop is often very significant and each drop is separated with a range of normal top values, we can determine that the effect of a very large window length has no impact. In initial testing a window length smaller than 20 caused the moving average to skew the cross sections which impacted the number of apneas found. Since both the Apnea-ECG and MIT-BIH Polysomnography databases have annotations for every minute and every 30 seconds respectively, we choose to use these same numbers as our window lengths. Another thing to consider is where the cross sections occur and how large the apnea range becomes. In figure 6.3, the cross sections between the data series and the moving average is approximately half way down the curvature on both sides. This becomes a problem as parts of the apnea range are missed when triggering the start and end points of the drop. To counter this, we add a fixed weight on the moving average, moving the cross sections closer to when the actual oxygen saturation starts to drop and when it has gone back to a normal range. Each yellow segment indicated in figure 6.3 is identified as an apnea. To correctly classify each segment, we need to look at what criteria determines an apnea event. According to ASAA an apnea is defined as a pause in breathing of more than ten consecutive seconds. For the SpO2 signal, the overall drop in this period of time needs to be more than three or four percent. When the SpO2 value of incoming events drops below the moving average, we trigger the starting point for a possible apnea. All the values from new incoming events are then accumulated into a new stream up to a point where the SpO2 value is higher than the moving average, triggering the end point. With the gathered sensor events in the new stream we use aggregation functions in Esper to check if the period of abnormal breathing is meeting the criteria to be classified as an apnea Standard Deviation Standard deviation is another simple method used in statistics to calculate the amount of dispersion in a subset of values. Unlike a moving average which focus on finding the mean across a subset, standard deviation focus on finding the amount of variation in a subset. The 55
66 Figure 6.3: Illustration of the moving average design. SpO2 values are represented in blue while the moving average is represented in green. The yellow segments are periods of abnormal breathing. higher the variation, the further the numbers are from the mean in the subset. As seen in figure 6.4, as the slopes in the graph increases, the higher the deviation becomes. There are two types Figure 6.4: A plot illustration of deviations in a bell-curve [28]. of standard deviation we need to review: Sample Standard Deviation and Population Standard Deviation. Both are very similar to each other with a slight adjustment to how calculations are made. Population Standard Deviation as seen in Equation 6.3, calculates the standard deviation as if the whole subset n is part of the parent population. Sample Standard Deviation in Equation 6.4 on the other hand, is valid for a subset n that is part of a larger population. If the subset is a sample of a parent population that is larger than the subset, we subtract n with one. This is called the Bessel s correction [9]. σ = s = 1 n n i=1 1 n 1 (x i µ) 2 (6.3) n i=1 (x i x) 2 (6.4) The idea behind this is to counter bias towards population variance. The impact this has in our thesis is minimal for either one we decide to go for. We are not interested to store or use the standard deviation on anything other than trigger start and end points of a slope. Our subset consists of SpO2 values that are part of a larger SpO2 population, the population is growing continuous as new incoming sensor events arrive. Bessel s correction is only used if the mean 56
67 across a population is unknown, which it is in our case as we simply calculate the mean from the sliding subset. We decided to go with Sample Standard Deviation in our design based on these facts. Like with a moving average, we need to decide a sliding window length for the standard deviation subset. The factor in window length for standard deviation is the opposite of a moving average in that a smaller window length has larger variance. We are interested in the calculated variance between each new sliding window to generate the triggers we need. Early testing of four, six and eight as our window lengths were all viable at finding the start and end points. In a related work [49], they use a smaller window length of eight with a two second window jump to classify segments. These segments are labeled desaturation, resaturation and steady using three differently trained SVM classifiers. A larger window segment is overlapping the smaller ones to determine if four of the smaller segments are periods of normal or abnormal breathing using another trained SVM classifier. We use the same approach to standard deviation as with moving average, thus dividing two separate streams into segments is unnecessary. The window lengths we consider to test are four, six and eight, where eight is the one used in the related work. We will also be testing a solution with sliding window, but during inital testing the adding jump to the window had almost no impact, the jump option will still be available as an option in the configuration file. Besides the window length, we need to set some trigger thresholds to indicate the start and end points of an apnea. In early tests we used minus one as a threshold for a descending slope, and one as a threshold for an ascending slope. Deviation values in between these two thresholds are classified as steady periods of little to no variance. This was tested using a window length of eight and a jumping window of two lengths. A combination of window lengths and sliding options needs to be tested accordingly to determine the thresholds. We need to create EPL queries to trigger the start and end of an apnea, therefore we design three separate rules to check if the slope either descends, ascends or flattens out. These three rules combined detect the apnea event. Figure?? illustrate how the rules are triggered based on the deviation: The rules are built on top of each other so that the first rule needs to be followed by the second and the second followed by the third. Once the deviation drops below the positive threshold the second rule is in effect and triggers an end of the apnea event. 1. If the standard deviation is lower than the negative threshold, the slope descends. Trigger a start of an apnea. 2. If the standard deviation is higher than the negative threshold but lower than the positive threshold, the slope flattens out. 3. If the standard deviation is higher than the positive threshold, the slope ascends 57
68 Figure 6.5: The red line represents the first rule, yellow line represents the second rule and the green line represents the third rule. Because of the way the standard deviation is calculated we have no way of telling if the slope is dropping or going back up because we only have positive deviations to work with. Esper has numerous built in functions to aggregate event data in the data streams. The included function to calculate standard deviation in Esper has the same problem. To solve this issue, we construct a custom aggregate function that calculates the sliding window and checks if the previous event to the last arriving event in the sliding window is higher. If that is the case, the returned result from the custom function is multiplied by minus one to indicate a dropping trend. The start and end points mark the apnea range where we insert all the incoming sensor events into a new data stream. Like with the Moving Average method, we use the same criteria to correctly classify an apnea using aggregate functions available in Esper. We want to count the number of ticks in the apnea range to determine the duration. Measure of the SpO2 drop is done by calculating the difference between the SpO2 value at the start of the apnea and the minimum value over the entire apnea event. 6.6 Output Data Considering a storage solution is based on some key factors. As the focus of this thesis is towards using smart phones as hardware we need to see the available storage options for the three major platforms, ios, Android and Windows Phone. Storage solutions on mobile platforms needs to be light weight, efficient and easy to maintain and incorporate. Although there are multiple data storage solutions that have support libraries for the mobile platforms in question, we decided to go for SQLite. SQLite is a light weight and efficient software library database engine that is widely used in all three platforms. The database engine implements a number of features including zero configuration, ACID properties and single cross-platform disk file storage [70]. The engine is written in C and has good cross-platform support. Both Android and ios have great support for SQLite as the database engine is implemented into the operating system and is also used 58
69 as the storage solution for various OS related information such as contacts. Recommended use of SQLite is for applications, embedded platforms and IoT-devices because of its low overhead and storage footprint. Windows Phone support for SQLite is fully supported for their Windows Universal Platform, while on Windows Phone 8 it requires some more work and extensions to function. The tiny amount of data we want to store and the low throughput of read and write operations make SQLite suitable for our needs. For the storage we create three separate relational database tables. The session table will store all the basic information about a newly started session such as date, timestamp, AHI index, number of apneas, as well as id s to the two other tables. The tick table will consist of incoming sensor events. As we want to down scale the sample rate to 1 Hz, each row in this table consist of data from the aggregated sensor event object as a result from Esper. The data from the tick table can be used to display graphs and generate data for remote diagnosis. The data consist of all the respiratory signals, the oxygen saturation and the index. To store the relevant classification information, we need an apnea table to store the class and the index of where the apnea starts and ends. For the data mining methods, the start and end values is the start and end indices of the current apnea segment. For the apnea detection methods, we can more precise and register the start and end indices of where the actual apnea starts and ends. But because we need a reference to the annotations from the record in order to evaluate the accuracy we need to store both. An overview of the planned tables and attributes is illustrated in Figure 6.6. Figure 6.6: Schema design for the application storage. What and how we store the data is not so relevant for this thesis as long as we get a performance evaluation on how much data is generated and stored with input and output throughput analysis. The data stored in the apnea table is used in the validation scripts to evaluate the performance of the data mining and apnea detection methods. We have covered the basic information needed for diagnosis and analysis. 59
70 6.7 Performance Evaluation In the offline analysis they use an evaluation method called ten-fold cross-validation and the hold out method to evaluate the data mining methods. The ten-fold cross-validation method is a standard method to evaluate classification techniques and how well they perform. The hold out method is used as a supplementary validation method to evaluate cross-database and cross-record classification performance. The results from the hold-out evaluation was poor and we see no point in doing the same for this thesis. As we already have the evaluation results from the offline analysis we do not need to test the data mining methods so extensively. We have decided to use the database with the best results as input and to use the best settings and options of each specific data mining method to evaluate them. The Apnea-ECG database had the best results out of all the databases. It has all the signals we have as a requirement, is not pre-processed and has overall the best data quality. The only negative thing is the number of records we have and the sampling rate, which is a bit low looking at newer guidelines. Using the Apnea-ECG database we divide the data from the eight records into ten almost equal partitions. One of the partitions are used to test the data mining method and the nine others are used as training data to generate a model. This process is repeated ten times shifting the testing and training set for each step so that we cover the whole eight records. We use the same metrics as in the offline analysis, accuracy, specificity and sensitivity. Normalization over data streams is either not used or it is used per a set interval. Measuring minimum and maximum values over a data stream is useless as we do not know when the stream ends. But for the sake of the evaluation we will test both normalized and not normalized data sets to compare the differences in performance. For the apnea detection methods there is no training process involved as we simply evaluate the data in real-time using the incoming sensor data. Because of this there is no reason to use the cross-validation method as the oxygen saturation data needs to be intact, fluctuations in the SpO2 data can impact the results. We use two different approaches to evaluate the detection methods. The first approach is to measure the accuracy, specificity and sensitivity of each method and the options available. We run each record listed in Chapter 5 once for each option combination with the two apnea detection methods. We use the same query that generate annotation segments for the specific database used and store the latest index where an apnea has occurred. Instead of sending the aggregated data segment to a classifier like with the data mining methods, each apnea detection method stores the index of where the last apnea occurred. This means that when Esper triggers the annotation segment query we check if the index stored is the same as the segment index. If it is, the segment is classified and stored in the database table as true, if not the segment contains no apneas and is set to false. As a supplementary evaluation we want to measure the AHI index and how many apneas the methods can detect. We want to select two records form each database to manually process these and annotate the apneas. We will follow the same criteria when manually annotate the records and the six records in total will be plotted in a graph. The criteria for picking the data sets for manual processing is based on a number of factors, but mostly they were picked at random with a slight favor in ones that had moderate to severe sleep apnea. As a disclaimer, we do not have the knowledge or experience of annotating data from a polysomnography. To get accurate results the plotting must be done by a trained physician, which will not be done in this thesis. 60
71 6.7.1 Hardware Profiling In order to evaluate the resource usage and hardware utilization of the implementation we need a profiler tool. Profiling and testing the implementation without a tool would result in tedious work and a need for knowledge of the inner works of the JVM. There are three profiling contenders where one is a free tool included in the Java SDK and the two others are licensed tools. We were able to get license for JProfiler as an academic and open source project. The other licensed option was the YourKit profiling tool. Both seem to be equal in features and performance so either one would suffice for this thesis. Using JProfiler we can measure the resource usage and hardware utilization of our implementation in greater detail. The resources in a laptop or a PC is not comparable to the chipset and hardware in a smart phone. The difference in architecture makes it impossible to compare the devices directly. A modern laptop outperforms a smart phone in benchmarks by a good margin. We did some early testing of three approaches. The first was using a Raspberry Pi 3 (Model B) as this have the same architecture and ARM chipset used in most smart phones to date. The comparison would be ideal in this approach, but neither the Matlab Runtime Compiler (MCR) or Esper have support for the ARM architecture. The second approach was using Docker as a virtual environment to encapsulate the implementation and restrict the resource pool to the container. Unfortunately, the JProfiler is not able to scale the resource usage of the implementation inside the container itself, but rather it is profiling the resource usage of the container. Based on the initial tests we do not know if this is caused by the JProfiler or the Docker container, or if this is even a feature that is possible. This is a problem when we set the number of cores and CPU time of the container to mimic a smart phone as we would get incorrect results and performance measurements that are not relevant for our thesis. The approach we chose to go for is create virtual machines using Oracle s VirtualBox. We create virtual machines and scale them down by configuring the number of cores used and the CPU time of the VM process. As we want to compare the results based on smart phone hardware we use a cross-platform benchmarking tool to generate a benchmark score that corresponds to the benchmark score from various smart phones. A more detailed overview of the benchmark scores is presented in Chapter??. 61
72 Chapter 7 Implementation In Chapter 5 we established a few requirements for our implementation and design based on the goal for this thesis and the requirements set for the offline analysis. The majority of the requirements we established are similar to the ones in the offline analysis. We found out that the input data from the offline analysis is suitable for data stream evaluation and the purpose of this thesis. Three databases from PhysioNet were chosen, the Apnea-ECG database, the MIT-BIH Polysomnography database and the St. Vincent s University Hospital database. Four classification methods were chosen: K-Nearest Neighbor, Support Vector Machine, Artificial Neural Network and Decision Tree. All except Decision Tree produced overall good results in the evaluation performance in the offline analysis using test and training data from the databases above. We want to evaluate their performance based on online data streams in contrast to offline data. Moving Average and Standard Deviation are two popular statistical methods which we chose to use in our apnea detection methods. We established that we want to test all the signal combinations when evaluating the data mining methods and use only oxygen saturation as our input signal when evaluating the apnea detection methods. Chapter 6 presents a detailed explanation of our design choices for the implementation based on the established requirements in Chapter 5. This chapter consist of an overview of the implementation following our design. First, in Section 7.1 we present a short description of our system environment as well as the environment configuration of our virtual machine setup. The implementation of our pre-processing scripts and an overview of the input data structure is listed in Section 7.2. The two main parts of our implementation is presented in Section 7.3 and 7.4 and describes the implementation and integration of Esper and the Matlab data mining methods for the client and server respectively. The code and implementation of the four data mining methods are presented in Section 7.5, followed by code and a detailed description of our apnea detection methods and the EPL queries in Section 7.6. Finally, we give a short description of our storage implementation using SQLite in Section 7.7. Setup configuration and examples on how to execute the implementation is presented in Appendix A. As the implementation is over 5000 lines of code we chose to only add the most relevant parts in our component explanations for the client and server sections. The code base is available as a Github repository which is explained in Appendix A. 62
73 7.1 System Environment Finding good and trustworthy data mining libraries is a challenge as with the Java platform there is no single library that come from a universal place containing all the needed data mining methods. WEKA is a data mining library for Java that come close to what we need but is lacking proper Artificial Neural Network support. A good library that supports Artificial Neural Network is called Neuroph, but dealing with libraries from two different sources is not ideal. To implement Java-based libraries we would have to combine multiple libraries from different sources to achieve our goal which is not an ideal solution. Thankfully, Matlab supports all the data mining methods we need and has support for external library compilation of the custom functions and most of the toolbox functions available. The methods have already been used and tested in the offline analysis so we chose to go for the same methods from the Matlab toolbox. We used the free academic license for Matlab (2016a) provided by the University of Oslo. As for the MCR the corresponding version to Matlab (2016a) is the version We use Java as our platform of choice based on two factors. Java is one of our main programming language when it comes to experience. The second factors are Esper which is only available as library component for the Java platform and for the.net platform, renamed NEsper. Java is also the main application platform for the Android operating system which use a lot of the same optimization techniques. By using Java as our platform we can integrate the required Matlab toolbox functions as JAR-files together with the Esper library component. For Java we use the version as Matlab still does not support CPU Memory HDD OS Hardware Configuration AMD FX GHz (6 cores) 16GB DDR3 1600MHz (667MHz) HP 250GB 3G 7.2K 3.5 SATA HDD Ubuntu Server LTS Table 7.1: Base hardware configuration for the VM. JProfiler is a profiling tool developed by EJ Technologies for the Java platform. The tool features extensive profiling for applications and services running on the Java platform to help developers optimize and debug their application in regards to resource usage and bottlenecks. The performance tests use version of JProfiler. In order to performance evaluate our implementation and compare it to the average hardware in a smart phone we need to scale and adapt our environment to meet the same threshold. We cannot compare the hardware in a laptop or stationary PC directly to the hardware in a smart phone. The hardware configurations and architecture between a smart phone and PC is not the same and cannot be compared. Instead we need to find a solution to test the implementation on hardware that is similar in performance. In the design we mentioned that we tried using the Raspberry Pi 3 (Model B), but came up short as both Matlab and Esper do not support the architecture. Instead we use Oracle s VirtualBox which is a full virtualizer for x86 hardware and features performance management scaling by adjusting the VM s CPU time and the number of cores available. We use the VirtualBox version to run our performance tests. The virtual machine environment is based on the hardware configuration in Table 7.1. The hardware we use to run the VM on is not that relevant as we use benchmark scores from 63
74 a multi-platform benchmarking tool to determine the scale and performance of the VM. The different VM configurations and benchmark results are presented in detail in Chapter??. 7.2 Input Data In this section we first describe the WFDB toolkit functions we use in our pre-processing scripts. Later we go into detail on the implementation of our preprocessing scripts for the three databases we plan to use. Because we only want to use the Apnea-ECG database to evaluate the data mining method, the implementation and script is a bit more complex than the two other implementations for the MIT-BIH Polysomnography and the St. Vincent s databases. The two databases share very similar implementation and code as the scripts basically do the same task, but on different formatted data WFDB Toolkit Each record in the PhysioNet databases we use is stored in a compressed bytecode format. Without knowing the actual schema of the format we need to use the toolkit that is available on their web page. The toolkit provides basic functions to extract, configure and transform the record data and annotation files for the databases. Below is a list of the three toolkit functions that we implement [77]: rdsamp The rdsamp function is used to read signal files for a specified record by using the -r option. The function supports many types of options to adjust the data output. The default output reads the file from the beginning till the end of the record and separate the signals in tabbed columns. In our implementation scripts we use the -c option to use commas as a delimiter rather than tab for easier processing. For the MIT-BIH Polysomnography and the St. Vincent s databases we also use the -pe option to convert the oxygen saturation to percent rather than an obscure number we are unsure of what represents. The -pe option is only to convert the timestamp precision format, but we do not know why it also changes the data format of the columns. rdann Rdann is the function to read the annotations form the corresponding annotation files for each record. The function needs the same -r option as rdsamp to read the record as well as the -a option to specify the file extension of the annotation file. The Apnea-ECG and the MIT-BIH Polysomnography databases have apn and st as file extensions for the annotation files respectively. The annotation files for the St. Vincent s database is stored in plain text files and we process them using regular read methods in Python. edf2mit The records from the St. Vincent s are all stored in the EDF format. We use the edf2mit function to convert the records to a WFDB-compatible format so that we are able to process them using rdsamp Apnea-ECG Database From the Apnea-ECG database we use eight records, these records contain all the signals we use in this thesis. The original sampling rate is set to 100 Hz which in terms mean that each 64
75 annotation interval at 60 seconds corresponds to 6000 rows. As with the offline analysis we aggregate each 6000 rows to align the data and get a reference for each annotation segment in the annotation file. This is need to be able to generate a model out of the four data mining methods. The preprocessing script for the Apnea-ECG database executes the following preprocessing steps: Reads each annotation file from the record list. Reads and stores the record data from the list into an array. Reshapes the data array and the annotations to a suitable dimension. Segments the arrays into ten equally folds. Iterates over the folds writing a CSV file containing one test fold segment while writing a corresponding CSV file with nine training fold segments. Writes a record file containing the SpO2 signal data for each record processed to be used for apnea detection testing. Code 7.1: The code for the Apnea-ECG preprocessing. import numpy as np import subprocess import sklearn. preprocessing as pp import sys import csv import math RECORDS = [ a01r, a02r, a03r, a04r, b01r, c01r, c02r, c03r ] # files to be processed SIGNALS = [ respchest, respabdomen, respnasal, spo2, class ] # signals in each record plus the class / ann SAMPLE_ RATE = 100 # original 100 hz sampling rate ANN_ INTERVAL = 60 # 60 seconds annotation interval K_FOLD = 10 # uses rdann to return a list of annotations from record r def get_ record_ annotations ( r): cmd = [ rdann, -r, r, -a, apn ] out, err = subprocess. Popen (cmd, stdout = subprocess. PIPE ). communicate () annotations = [] for row in out. splitlines (): [ annotations. append (1) if A in row. decode ( encoding = UTF -8 ) else annotations. append (0) ] return np. array ( annotations ). reshape (-1, 1) # calculates the mean for every 6000 row and appends the annotation column def create_ training_ set ( data, annotations ): data = data. transpose (). reshape ( -1,( SAMPLE_RATE * ANN_INTERVAL )). mean (1). reshape (4, -1). transpose () return np. concatenate (( data, annotations ), 1) # generates test folds def write_test_files ( data ): step = int ( data. shape [0]/ K_FOLD ) f_idx = 0 65
76 for i in range (0, data. shape [0], step ): print ( generating test fold : + str ( f_idx )) with open ( test_fold_ + str ( f_idx )+. csv, w ) as f: writer = csv. writer (f) writer. writerow ( SIGNALS [:4]) writer. writerows ( data [i:i+ step ]) f_idx += 1 # generates training folds def write_training_files ( data ): data = data [0:( data. shape [0] -7) ] step = int ( data. shape [0]/ K_FOLD ) f_idx = 0 for i in range (0, data. shape [0], step ): print ( generating training fold : + str ( f_idx )) with open ( training_fold_ + str ( f_idx )+. csv, w ) as f: writer = csv. writer (f) writer. writerow ( SIGNALS ) writer. writerows (np. delete (data, list ( range (i, i+ step )), axis =0) ) f_idx += 1 # generates apnea detection records def write_test_ap_full (data, r): data = data [:,3:] with open ( test_ +r+ _ap. csv, w ) as f: print ( generating apnea detection record : +r) writer = csv. writer (f) writer. writerow ([ spo2 ]) writer. writerows ( data ) # reads records from RECORDS one by one def read_ records (): test_container = np. empty ( shape =(0,4) ) training_container = np. empty ( shape =(0,5) ) for r in RECORDS : print ( record : %s % r) annotations = get_ record_ annotations ( r) cmd = [ rdsamp, -c, -r, r] p = subprocess. Popen (cmd, stdout = subprocess. PIPE ) data = np. genfromtxt (p. stdout, delimiter =, ) data = data [: annotations. shape [0]*( SAMPLE_ RATE * ANN_ INTERVAL ),1:] write_test_ap_full (data, r) if len ( sys. argv ) > 1 and sys. argv [1] is n : data = pp. normalize (data, axis =0, norm = max ) test_ container = np. concatenate (( test_container, data )) training_ container = np. concatenate (( training_ container, create_training_set (data, annotations ))) write_ test_ files ( test_ container ) write_ training_ files ( training_ container ) read_ records () The code for the script is presented in 7.1. The entry point for the code is the function read_records which iterates and reads all files in the RECORDS list by using a process call to rdsamp. Annotations are also read for each record and the record data is trimmed to align with the number of annotation segments in the annotation array. The annotations are represented as N for normal breathing and A for abnormal breathing. We replace these with numerical values instead which gives us N = 0 and A = 1. In this database there is more data than there are annotation segments, which means we need to calculate the amount of data to be 66
77 kept in regards to the number of annotation segments for the record. For each record read the data is passed to the write_test_ap_full function that selects the array column that contains the SpO2 signal and writes the data to a CSV-file which will later be used for apnea detection testing. The annotations for each record is added to the training array using the numpy function np.concatenate. The training array make use the function create_training_set which reshapes and calculates the average of all the signal columns per 6000 rows. Once all the records and annotations are read, the arrays are sent to the functions write_test_files and write_training_files. The former function divides the entire data array into ten equal segments and writes each one to a file containing an index. The write_training_files function operates in a similar way, but writes the remaining nine data segments left out from the test segment to a corresponding training file with the same index. The script takes one argument on execution. If given the n argument the data that is read for each record is normalized using min-max normalization. If not argument is given it will process the data using the raw output from rdsamp MIT-BIH Polysomnography & St. Vincent s Databases We use only five records from the MIT-BIH Polysomnography database and all the available records from the St. Vincent s database that contains SpO2 signal data. The records for the former database are sampled at 250 Hz and have annotation intervals at every 30 seconds. This means that for each 7500 row we have a corresponding annotation segment. The St. Vincent s database contains different sampling rates depending on the signals recorded. The included header files list the sampling rate at 8 Hz, but the sampling rate for EEG is set to 128 Hz while the body movements and EMG signals are set to 64 Hz. The annotation files are also a bit different in that they are not segmented into time slices but rather denotes the timestamp of an occurring apnea. This means that we do not have any annotations to conform to. The pre-processing scripts for these two databases are almost identical except for minor adjustments to fix the structure of the data. The records from the St. Vincent s database needs to be converted from the EDF format to a MIT-compatible format. The pre-processing script for the two databases executes the following preprocessing steps: Reads each annotation file from the record list. Reshapes the data array and the annotations to alignment. Writes the SpO2 signal data to a CSV-file. For the records from the St. Vincent s database, conversion from EDF to MIT format. The code is similar to the code presented in Code 7.1, but is less complex as it only does one task. First each record from the RECORDS list is processed and read using rdsamp. The process call using rdsamp use the -pe option to get the SpO2 signal data to represent percentage rather than an unknown metric. This was not a problem with the Apnea-ECG database as it already presented the SpO2 signal as percentage. Once a record is read and processed the data is sent to the function write_test_files that writes the entire data array to a CSV-file matching the record name. Specific adjustments to records for the MIT-BIH Polysomnography database is needed to set 67
78 the correct start index for the data. The third column in the annotation files represents the starting index of the first annotation segment, except for the record slp67x where the start index is in the second column. The records for the St. Vincent s database is stored in the EDF format which means we need to use the edf2mit function provided with the WFDB toolkit to convert the contents to be MITcompatible. The edf2mit function in the script makes an external process call to convert to the different formats. As all the records start at the same index, no index extraction is needed. 7.3 Client The client implementation consists of three components, one entry point class and an event object class which is used by Esper. Most of the tasks performed by the client is done in the EventEmitter component which is responsible for the reading the CSV-file containing the record data, process it by sending to the Esper engine instance and send it over the socket connection to the server. It serves as an input handler that feeds the server running the Esper engine instance. The client is responsible for the following tasks: Parse the CLI arguments on execution. Connect to the specified server instance. Read the CSV-file and process it through the Esper engine instance. Send the event objects over the socket to the server. The entry point for the client implementation is in the class Client. First the client makes a call to the CLIParser component to read and parse the arguments using the parseargs() function. Then it tries to establish a connection to the server with the specified arguments, if it is a success the client proceeds by initializing the EventEmitter and starts the sensor simulation by calling the startsensor() function. If the connection fails, the client terminates. The SensorEvent class consist of five variables that holds all the information we need from the record. We store the signal data in the variables respchest, respnasal, respabdomen and spo2. All of them have corresponding get and set functions which is needed for the Esper engine instance to retrieve the data from the variables. The eventindex variable is used to store the event object index and represents time. It is easier to work with indices as integers that correlates to the other components and with the database rather than using timestamps which would make us have to convert starting indices for some records to timestamps and vice versa. Each object that is sent holds data for all the signals and the server is responsible for using the correct specified signal combinations. The constructor takes three parameters, an event index, a string containing the record data and another string that contains the actual name of the signals we want to extract. These are stored in the correct variable by name definitions. In order to send the object over the socket connection with ObjectOutputStream, the SensorEvent class needs to implement Serializable. Serializable is able to convert the Java objects to a series of bytes and the other way around CLIParser The CLIParser performs just a single task and parses the arguments given at execution time. We have five parameters in total that are used to specify the configuration of the components. The 68
79 file argument is the path or filename of the CSV-file that holds the record data. The hostname and port arguments are needed for the ConnectionHandler to establish a connection to the server. Both output-rate and esper-rate is used to control the flow of objects that is emitted over the server connection. A more detailed explanation of their use is presented in Section ConnectionHandler Like with the CLIParser component, the ConnectionHandler serves a single task. It tries to establish a socket connection using the hostname and port number specified at execution time from the CLIParser. Further it creates a new ObjectOuputStream class that is used in EventEmitter to send the event objects across the socket connection EventEmitter EventEmitter is the largest component and performs the majority of the tasks of the client. The component has three main tasks, initializing the Esper engine instance, reading data from the CSV-file and creation of SensorEvent objects and sending the aggregated objects over the socket connection to the server. The component includes four main functions and two helper functions. The init() function is responsible for initializing the CSVReader component, the Esper engine instance and the RateLimiter. First the function initializes the CSVReader using the specified filename or path. Then it proceeds to read the first line from the CSV-file to get the column names that is later used to generate the EPL query we use to aggregate the event objects. Code 7.2: EPL query to aggregate event objects in the Client. INSERT INTO SensorStream SELECT avg ([ signal ]) AS [ signal ] FROM SensorEvent. win : length_batch ([ esper - rate ]); The value from output-rate is used to initialize the RateLimiter. The class is from an external library called Google Guava and functions as a way to block further execution of code at certain intervals. It takes one argument which is the number of pass-through or unblocks per second. In order to initialize the Esper engine we need to pass a Configuration object to the instance. The Configuration object contains a great amount of changeable variables to alter the properties of the engine instance, Btu we will run the engine with the default settings, only register the necessary event object class. An EPL query is also registered with the engine to aggregate the event objects for data flow control. The createsensorstreamquery() function generates a query string based on properties of the record and the arguments given by CLIParser, the generated query is presented in Code 7.2. We average the signal data over a length batch corresponding to the esper-rate given at execution. This means we can down sample the raw sampling rate down to 1 Hz, simulating a sensor giving of data once per second. Together with the query we register a corresponding listener to the event. Code for the UpdateListener is presented in 7.4. Code 7.3: The startsensor() function from EventEmitter. public static void startsensor () { try { String [] line ; // Process each line as an event while (( line = csvreader. readnext ())!= null ) { esperengine. sendevent ( new SensorEvent (0, line, getcolumnnames ())); 69
80 } } } catch ( Exception e) { e. printstacktrace (); } The startsensor() function is called from the Client class at start up once the other components are initialized. In Code 7.3 we have a while-loop that continuously reads new lines from the CSV-file and sends new SensorEvent objects to the Esper engine instance. Each new event object is processed and aggregated by Esper based on the EPL query we have registered. When the amount of aggregated batch reaches the value of esper-rate the execution jumps to the registered UpdateListener which we call the EventReadyListener. First we if the limit of outgoing objects has not been reached by making a call to the acquire() function. If the limit is reached the execution is blocked further until we get a permit to proceed by RateLimiter. Once we proceed we first extract the aggregated values from the EventBean object generated by the engine instance. Every event that is generated by the engine is mapped to an EventBean object. The object contains the selected signal values from the registered query string. The extracteventvalues() function takes one EventBean object as parameter and returns a new SensorEvent object containing the new averaged values and sends it over the connection using the output stream. At times we need to reset the stream buffer in order to delete old objects that are accumulated. This is to limit the memory impact when we evaluate the performance with JProfiler. Code 7.4: The UpdateListener implementation from EventEmitter. public static class EventReadyListener implements UpdateListener { int resetlimit = public void update ( EventBean [] eventbeans, EventBean [] eventbeans1 ) { if( CLIParser. getoutputrate () > 0) ratelimiter. acquire (); try { ConnectionHandler. getoutputstream (). writeobject ( extracteventvalues ( eventbeans [0]) ); if( resetlimit >= CLIParser. getesperrate ()) { ConnectionHandler. getoutputstream (). reset (); resetlimit = 0; } resetlimit ++; } catch ( Exception e) { e. printstacktrace (); ConnectionHandler. disconnect (); System. exit (1) ; } } } 7.4 Server The server implementation is far more extensive than the client. It consists of six main components with six sub-components that either extends a super class or have unique specified properties but share similarities with a main component. The largest component is the EventHandler which is the contrast to the EventEmitter component in the client. There are 70
81 three classes that handles query generation and four unique classes that generates a model for each of the data mining methods. The server is responsible for the following tasks: Load and initialize the configuration options. Read training file and generate a data mining model. Connect and generate necessary database tables. Initialize and register detection queries with the Esper engine. Process incoming event objects and classify batches. Add sensor data and classification results to specified database tables. Similar to the client we have a Server class that is our entry point. The class first reads and parses the command line arguments on execution and reads the configuration options in the config.properties file. Once the options and properties are loaded the class checks the data mining methods specified. We have three options to choose from that determines the data mining method. The value determines if the program is to generate a model based on the Matlab JAR library that is either K-Nearest Neighbor, Support Vector Machine, Artificial Neural Network or Decision Tree or proceed with the two other options that are the apnea detection methods that do not generate models, but rather EPL queries that detect the anomalies in incoming objects. The last steps are initializing the database and the tables if it does not already exist before the server starts to handle incoming objects with a call to receiveevents(). The server shares similar CLIParser and ConnectionHandler components. The only difference is the change in available parameters in the CLIParser. There are additional options to select data mining or apnea detection methods, as well as options for which signal combination to be used for classification. The CustomEsperFunction, Segment and StandardDeviationEvent are helper classes that we explain in detail in the sections below ConfigurationLoader The ConfigurationLoader component loads the config.properties file and stores the values in a Properties class. This makes it easier to retrieve and use properties within other components by using the get functions from this utility class. Below is a list of the properties that can be changed: signals The name representation of the different signals. Is set to default values respchest, respnasal, respabdomen and spo2 across the server, the client and the records. ann_interval The annotation interval that is set by the annotation files for a specific PhysioNet database. It is either 100, 250 or 8 for the Apnea-ECG, MIT-BIH Polysomnography and St. Vincent s databases respectively. It is used by the EventHandler to align incoming events into segments corresponding with the included annotations. spo2_drop The drop in oxygen saturation is an option for both the apnea detection methods. It can be set to any value, but the criteria to record an apnea is around 3-4%. 71
82 ma_win_length The window length of the moving average and can be any positive value. ma_weight The weight of the moving average. It can help to shift the start and end cross-sections for each apnea segment. ma_win_jump The jumping length of the moving average window. sd_win_length he window length of the moving average and can be any positive value. Very high values make it impossible to detect deviations in the oxygen saturation. sd_threshold The threshold is a value to determine which segment the standard deviation calculation belongs to, if it is to be labeled a steady, desaturation or resaturation period. sd_win_jump The jumping length of the standard deviation window Classifier Classifier is a superclass that is inherited by one of the four data mining methods from the Matlab JAR library. We implement four almost identical data mining classes, KNearestNeighbor, ArtificialNeuralNetwork, SupportVectorMachine and DecisionTree. Each of these classes extends Classifier and the only difference is what Matlab function they use and belong to. When the classifier object is created the constructor first calls the constructor of the superclass with the super() function. The Classifier has a function called generateset() which reads the specified CSV-file used as training data. All columns besides the last one is processed and stored in MWNumericArray data representation. This data representation is used as the default array type for Matlab and makes it possible to send and receive data when using the compiled Matlab functions. The last column in the CSV-file is the annotations which is put in a separate array. Both the training data and the annotations can be fetched by calling the get functions from the superclass. Code 7.5: The implementation code for the K-Nearest Neighbor classifier. public class KNearestNeighbor extends Classifier { private KNN knn ; private ArrayList < Double > classified ; public KNearestNeighbor () { super (); System. out. println (" Building model (" + this. getclass (). getname () + " )"); long starttime = System. nanotime (); try { knn = new KNN (); buildmodel (); classified = new ArrayList < >(); } catch ( Exception e) { e. printstacktrace (); } 72
83 } long endtime = System. nanotime (); System. out. println (" Model built (" + ( endtime - starttime ) / " public double classifyevent ( double [] eventvalues ) { Object [] results = null ; MWNumericArray dataset ; try { dataset = new MWNumericArray ( eventvalues, MWClassID. DOUBLE ); results = knn. matlab_knn (1, dataset ); } catch ( Exception e) { e. printstacktrace (); } Double r = Double. parsedouble ( results [0]. tostring ()); classified. add (r); return r; } private void buildmodel () throws MWException { knn. matlab_ knn (1, gettrainingset (), getannotations ()); } } The code in 7.5 references the implementation of K-Nearest Neighbor. Once the superclass has generated the two arrays containing training and annotation data, the constructor initializes the KNN class which is a direct reference to the Matlab function. Then we proceed by calling the buildmodel() function that sends three arguments to the Matlab function. The first argument is a numeric value that represents how many results we want in return. The second and third parameters is arrays for the training data and annotations. The model resides in the KNN object once it is called. All the other data mining methods have identical implementation, except for Decision Tree that need a list of the column names to generate the tree structure. The classifyevent() function operates by converting the test data that is included in the event- Values array to a Matlab-compatible MWNumericArray type. The converted array is sent as an argument when calling the matlab_knn function and that returns an object array with the results from the classification process. The result is parsed to a double and returned. The implementation for the specific data mining functions in Matlab is given in more detail in Section EventHandler This is the biggest component and does most of the work on incoming events and classification of the test data. The class contains four functions, two of them which are helper functions and three different inner class listeners implementing Esper s UpdateListener class. The first task of the EventHandler component is to initialize the configuration of the Esper engine and register the needed EPL queries and listeners to the engine instance. The second task is to handle incoming events by sending them to the engine while keeping track of the event index. The third task is to store the classification result in the database table. In the first task, the Server class calls EventHandler s init() function that initializes a new Configuration object while registering the SensorEvent class. Like with the client, the event class 73
84 holds all the sensor data sent from the client. The event class needs to be identical to each other because of serializability. Esper have function support for calculating the standard deviation of values, but as we pointed out in our design we also want to be able to determine if the deviation is in a decline or incline state. For us to be able to do that we need to implement a custom function and register it with the engine instance. The line configuration.addpluginsinglerowfunction("mystddev", "CustomEsperFunction", "computestandarddeviation") registers an EPL operator called mystddev using the code computestandarddeviation() function from the CustomEsperFunction class. The implementation of the custom function is presented in Code 7.6. Code 7.6: Implementation of our custom standard deviation calculation. public static double computestandarddeviation ( Double [] window ) { double dev = new StandardDeviation (). evaluate ( ArrayUtils. toprimitive ( window ), 0, window. length ); double incline = 0; if ( window. length > 1) { for ( int i = window. length - 1; i > 0; i - -) { incline += ( window [i] - window [i - 1]) ; } } if ( incline < 0) { return dev * -1; } return dev ; } The function takes one parameter, a double array containing the SpO2 values for the current window. We then use a math library to compute the standard deviation over using the window values. We then proceed by checking the first incoming event value in the window to the previous to see if it is lower. This is done to determine if the window is either is either leaving a declining state or entering an inclining state. If the window still has inclining values, the returned deviation value is multiplied by -1 so that a negative result presents a desaturation segment. The usage of this custom function is explained in Section 7.6. When the engine and the configuration is initialized the Server make a call to the receiveevents() function that first registers the two general EPL queries to the engine instance. The first is to aggregate all incoming events to down sample the values to 1 Hz. The second query is responsible for aggregating a set number of events that corresponds to the annotation segments set by the database records. If either of the two apnea detection methods are chosen, specific EPL queries from their respective query generator classes is registered to the engine as well. The execution ends up in a while loop that continuously receive and process the incoming events sent by the client by resending them to the engine instance via the sendevent() function. Code 7.7: The listener attached to the annotation segment query. public static class AnnotationStreamListener implements UpdateListener public void update ( EventBean [] eventbeans, EventBean [] eventbeans1 ) { // Triggered when using apnea detection // If false the data from the event bean is sent to the offline data mining method if ( CLIParser. getapmethod () == 1 CLIParser. getapmethod () == 2) { 74
85 } } int eventindex = transformeventindex (( int ) eventbeans [0]. get ( " eventindex ")); if ( eventindex == currentapneastopindex ) { SQLiteJDBC. insertclassifiedevent ( currentapneastopindex, 1); } else { SQLiteJDBC. insertclassifiedevent ( eventindex, 0); } } else { double [] values = extracteventvalues ( eventbeans [0]) ; SQLiteJDBC. insertclassifiedevent (( int ) eventbeans [0]. get (" eventindex "), classifier. classifyevent ( values )); } The query responsible for creating the annotation segments is attached to the AnnotationStream- Listener presented in Code 7.7. Just after the engine triggers the query a resulted EventBean object is sent to the listener. The listener is used for all the classification methods presented in this thesis, but we handle the apnea detection methods a bit different than the data mining methods. We first check to see if either of the detection methods are selected on startup, if not the values from the EventBean object is extracted and stored in the SQLite database. Before the execution enters the insertclassifiedevent() function we make a call to the classifier to classify the values using the compiled Matlab function we provide. The result from the classifyevent() is stored in the database table alongside current event index. If one of the apnea detection methods are specified, we then extract the event index and check if the index is the same as the value of currentapneastopindex. The currentapneastopindex is set by another listener which is explained in Section 7.6, it basically means that the detection method has triggered in an annotation segment and that the segment contains apneas QueryGenerator The QueryGenerator is a helper class that generates the needed EPL queries we use for the data mining and the apnea detection methods. There are three similar classes, named Query- Generator, MovingAverageQueryGenerator and StandardDeviationQueryGenerator. They consist of functions that returns String data types that we register with the Esper engine instance on initialization. The queries returned from QueryGenerator are used in all the data mining and detection methods while the other two are specific for either of the detection methods. The queries for Moving Average and Standard Deviation is presented in Section 7.6. QueryGenerator has two functions that resolves to two EPL queries. Code 7.8 represents the generated EPL query by calling function createsensorstreamquery() and Code 7.9 represents the EPL query generated by calling function createannotationstreamquery(). First we need to generate an aggregation query to down sample the incoming events to 1 Hz. We select the eventindex and the signal combination that is specified on execution time. Once we aggregate enough events set by the input-rate value we insert the aggregated event object and values into a new stream called SensorStream which can be used later on by other EPL queries. Code 7.8: EPL query for aggregation of incoming events. INSERT INTO SensorStream SELECT eventindex, avg ([ signal ]) AS [ signal ] FROM SensorEvent. win : length_batch ([ input - rate ]); 75
86 The second EPL query works by creating the annotation segments that correlates to the incoming data and the annotations from the records of the database in use. We select the event index and the signals specified from a larger length batch. The line SensorEvent.win:length_batch([input-rate]*[ann-interval]) aggregates all the events by multiplying the input rate of incoming events and the annotation segment interval value. For the Apnea-ECG database for example we have a raw sampling rate of 100 Hz and an annotation interval of per 60 seconds that in terms is equivalent to 6000 events per annotation segment as a length batch in the EPL query. Code 7.9: EPL query for generating annotation segments of incoming events. SELECT eventindex, avg ([ signal ]) AS [ signal ] FROM SensorEvent. win : length_batch ([ input - rate ]*[ ann - interval ]); SQLiteJDBC The SQLiteJDBC is a utility class to handle all SQLite database related operations. We need to include an external library to be able to connection and do operations on a SQLite database. The class has three main functions and a constructor that first checks to see if the database already exists, if that is not the case it proceeds by creating the three tables needed for storage which is presented in our design in Section?? and a further detailed overview of the SQL queries for the insert operations is presented in Section Data Mining In this section we list a detail description and overview of the four implemented data mining methods from the offline analysis. Our implementation of the data mining methods is very similar, and to some extent more simplified to the ones in the offline analysis. The implementations are very similar to each other except for Decision Tree where we need to handle the column names to generate the tree and for Artificial Neural Network which have a bit more steps to generate the network. We start by introducing K-Nearest Neighbor in Section followed by Support Vector Machine in Section Decision Tree and Artificial Neural Network is presented in Section and All the implementations are built up the same way. First we declare a function that returns a results object containing either a one or a zero, indicating either a normal or abnormal breathing segment. We use the same function to both train a model based on training data and to evaluate once the training phase is done. To be able to do this we use the varargin list which enables us to handle an any number of input arguments. If varargin contains more arguments than 1, we presume that it is in the initialization phase because we only give test data as argument once we want to evaluate a segment. The model is stored as a global variable after the initialization phase is done and is used to evaluate the test data K-Nearest Neighbor In Code 7.10 we present the implementation of K-Nearest Neighbor. We already established the design parameters to be used for the method based on the results and evaluation form the offline analysis. We use the Cityblock distance function and a total of 5 neighbors in our tests, even though the offline analysis tested neighbors of 1, 5 and 10. The K-Nearest Neighbor has 76
87 no training phase but simply store the training data to be evaluated once test data evaluated. The fitcknn() function is used to create a model, but not in the sense of a trained model but more like an object containing the training data and the settings to be used once evaluating. The NumNeighbors argument followed by a number sets the number of neighbors and to set the distance function we use the Distance argument followed by the name of the pre-determined function. Code 7.10: The code for the K-Nearest Neighbor implementation in Matlab. function [ results ] = matlab_ knn ( varargin ) end global mdl ; results = ; if nargin > 1 mdl = fitcknn ( varargin {1}, varargin {2}, NumNeighbors, 5, Distance, cityblock ); else results = predict ( mdl, varargin {1}) ; end The varargin{1} and varargin{2} arguments represent the training data and the annotations. In the offline analysis they enabled ten-fold cross-validation by using the CrossVal argument, but the training data we use is already divided into ten folds by the pre-processing scripts Support Vector Machine The Support Vector Machine is identical to the K-Nearest Neighbor implementation; the only difference is the settings in the training phase. The implementation is presented in Code We use the fitcsvm() function to generate a Support Vector Machine model using the Gaussian kernel function. As with the implementation of K-Nearest Neighbor, varargin{1} and varargin{2} represents the training data and annotations. Code 7.11: The code for the Support Vector Machine implementation in Matlab. function [ results ] = matlab_ svm ( varargin ) end global mdl ; results = ; if nargin > 1 mdl = fitcsvm ( varargin {1}, varargin {2}, KernelFunction, gaussian ) ; else results = predict ( mdl, varargin {1}) ; end Decision Tree For Decision Tree we only use the default settings available for the Matlab function. Generating the tree is done by using the fitctree() function. The function takes exactly the same input 77
88 arguments as with K-Nearest Neighbor and Support Vector Machine. The PredictorNames argument is strictly not necessary but if we want to generate an overview of the tree generated we need to give the function the names of the different signals for each column. In order to do so the data structure for the signals need to be altered as we had issues extracting the String data types from the list argument. Code 7.12: The code for the Decision Tree implementation in Matlab. function [ results ] = matlab_ dt ( varargin ) end global mdl ; results = ; if nargin > 1 % Transforms and reshapes the input data to extract the predictor % names form the Java implementation. data = cellstr ( varargin {3}) ; data = reshape (data,1,[]) ; data = data (1: length ( data )); mdl = fitctree ( varargin {1}, varargin {2}, PredictorNames, data ); else results = predict ( mdl, varargin {1}) ; end Artificial Neural Network The implementation for the Artificial Neural Network is presented in Code 7.13 and has the same structure as the other data mining implementations, but a quite few more settings. In Section we list quite a few settings for our design. Matlab offers a number of different neural networks but we decided to go with the same network topology used in the offline analysis. First we initialize the network by calling the patternnet() function. The function s argument is 20, indicating we want the network to consist of 20 hidden nodes for the hidden layer. Further we divide the training data into two portions, one that is for training and one that is for validation. This is not the same as cross-validation, as these ratios is used to iterate and improve performance of the network. The showwindow option is to enable or disable the pop-up window in the Matlab editor, despite us having trouble disabling it. Code 7.13: The code for the Artificial Neural Network implementation in Matlab. function [ results ] = matlab_ ann ( varargin ) global mdl ; results = ; if nargin > 1 % Inits patternnet with a hidden layer size of 20. net = patternnet (20) ; % Divides the set into training and validation ratios. net. divideparam. trainratio = 70/100; net. divideparam. valratio = 30/100; 78
89 % Turns of the GUI popup ( doesn t really seem to work ). net. trainparam. showwindow = 0; % Set the backpropagation and activation functions. net. trainfcn = trainscg ; net. layers {1}. transferfcn = tansig ; net. layers {2}. transferfcn = softmax ; [mdl,~] = train (net, varargin {1}, varargin {2} ) ; else % Classifies input data, everything above 0.5 is seen as a fire. results = mdl ( varargin {1} ) > 0.5; end end In the line net.trainfcn = trainscg we set the backpropagation to be the Scaled Conjugate Gradient, while the two activation functions are set below, the tansig representing the Hyper Tangent Function, and the softmax representing the Softmax function. These are the default values for the feed-forward network in Matlab, but for the sake of the thesis we want to show how they can be set. As a last step we generate the network by calling the train() function with the network, training data and annotations as input arguments. 7.6 Apnea Detection In this section we will explain the two different detection methods we have implemented using Esper and the EPL. In Section we present a detailed overview of our detection method using Moving Average, followed by a similar overview of the other detection method using Standard Deviation in Section Both the data mining methods and the detection methods make use of the queries generated by the QueryGenerator class. To be able to check for criteria that fit an apnea and store the right segment with the corresponding class we attach the ApneaValidationListener class. The listener attached is shared between both the detection methods and the EventBean object contains the same data values in both. For the Moving Average method, the listener is registered with the query in Code 7.21 and for the Standard Deviation method it is registered with the query in Code The listener extracts the maxspo2, minspo2 and duration values from the EventBean object. Then it checks if the duration is over ten seconds and that the difference between the maximum and minimum SpO2 value is higher than the threshold set in the configuration file. The event index for the last event in the apnea segment is stored in the variable currentapneastopindex and is used in the AnnotationStreamListener to annotate the current annotation segment as either abnormal or normal breathing Moving Average All the queries for this detection method are generated and resides in the MovingAverageQuery- Generator class. There are a total of nine functions in the class which all have the same structure and returns a String data object containing the query itself. The initialization of the detection method is in the receiveevents() function in the EventHandler. The order the queries are registered with the engine instance is important as the queries have references in between them to other queries. If the queries are not registered in order or a reference name is misspelt, an 79
90 exception is raised. In our design we want the moving average to be generated across the incoming data events over a sliding window to be able to trigger starting and ending apneas over the oxygen saturation. In order to do so, we need to create some context variables and triggers using the context variables. In Code 7.14 we declare a variable of the Boolean type with the default value as "false". This variable is important as it is used to check if an apnea have already been triggered. Once we have the variable in place, we also need to create a custom window reference with the signal values we need. The custom window is used to collect the incoming data between a start and end trigger point of the moving average. In Code 7.15 we establish a new window called ApneaStream which always aggregates the incoming events and keep a record of the last one. All the properties we need to check for criteria of an apnea is present, such as total oxygen saturation drop and the duration. Code 7.14: EPL custom variable query. CREATE VARIABLE boolean apneadetected = false Code 7.15: EPL custom window query. CREATE WINDOW ApneaStream. std : lastevent () AS ( eventindex integer, maxspo2 double, minspo2 double, duration long ) The triggers are presented in Code 7.16 and The ON-operator indicates that once the ApneaStartTrigger or ApneaEndTrigger event occurs, the variable we declared at the start is to be set to either true or false, depending on the trigger outcome. Code 7.16: Start trigger using the custom variable. ON ApneaStartTrigger SET apneadetected = true Code 7.17: End trigger using the custom variable. ON ApneaEndTrigger SET apneadetected = false The context in Code 7.18 is needed to make the custom window start and stop and the right time. If we do not create a context the aggregation continues beyond each apnea window which impacts the oxygen saturation drop calculation since it will never reset. So when the ApneaEndTrigger event happens we want the aggregations for the custom window to reset and start over once a new ApneaStartTrigger event happens. Code 7.18: Context declaration for the ApneaRange. CREATE CONTEXT ApneaRange start ApneaStartTrigger end ApneaEndTrigger To generate the moving average over the incoming events we need to aggregate the oxygen saturation signal over a window length. In Code 7.19 we insert the aggregated SpO2 values together with the corresponding event index for the first arriving event. The length of the sliding window is determining by the [length] variable, which resides in the properties file for the implementation. For every aggregated event inserted into MovingAverageStream we can use the stream in other queries. Both Code 7.20 and 7.21 use the moving average stream to trigger 80
91 start and end events. The trigger query for the start of an apnea first selects the event index and the SpO2 value from the moving average stream once a new incoming event have a SpO2 value lower than the value in the moving average window and if the apneadetected is false. These conditions indicate that an apnea has started and the query in Code 7.16 sets the value of apneadetected to true. The custom window we declared above will continue to accumulate and aggregated events as long as the value of the incoming events is lower than the value of the moving average. In Code 7.21 we insert events into ApneaEndTrigger to set an end point to the apnea. The query works similar to the start trigger query; the difference is that we extract the aggregated values from the custom window we declared as the ApneaStream. To trigger the end of an apnea we check if a new incoming event has a higher SpO2 value than the moving average and if apneadetected is true. Code 7.19: The Moving Average stream query. INSERT INTO MovingAverageStream SELECT eventindex, avg ( spo2 ) +[ weight ] AS mavg FROM SensorStream. win : length ([ length ]) Code 7.20: The start trigger for an apnea range. INSERT INTO ApneaStartTrigger SELECT b. eventindex AS eventindex, b. spo2 AS spo2 FROM MovingAverageStream. std : lastevent () AS a, SensorStream. std : lastevent () AS b WHERE a. eventindex = b. eventindex HAVING a. mavg > b. spo2 AND apneadetected = false Code 7.21: The end trigger for an apnea range. INSERT INTO ApneaEndTrigger SELECT c. eventindex AS endindex, maxspo2, minspo2, duration FROM MovingAverageStream. win : length (1) AS a, SensorStream. win : length (1) AS b, ApneaStream AS c WHERE a. eventindex = b. eventindex HAVING a. mavg < b. spo2 AND apneadetected = true Code 7.22 is the query that implements the custom window by using the context we declared at the start. This query will only run if the context is triggered based on the start and end of an apnea sequence. Code 7.22: The ApneaRange context query. CONTEXT ApneaRange INSERT INTO ApneaStream SELECT eventindex AS eventindex, max ( spo2 ) AS maxspo2, min ( spo2 ) AS minspo2, count (*) AS duration FROM SensorStream To summarize, this detection method uses a variable to detect if an apnea segment has started in the incoming event stream. We have two triggers, one indicating the start of the apnea and one for the end of the apnea. A custom window and a query is created to collect the data in between these triggers so that the information is available once the end trigger activates. 81
92 7.6.2 Standard Deviation The Standard Deviation method works a bit different than the Moving Average method. Instead of using triggers to specify the start and end points of the apnea we use a standard deviation calculation to determine inclines and declines of the incoming event stream based on a rule set and pattern operators available in the EPL. For this detection method we also use an enum class called Segment and a new temporary event class named StandardDeviationEvent. The class contains four variables, the event index, standard deviation value, SpO2 value and the Segment enum. There are three enums that we use, DESATURATION, RESATURATION and STEADY. As with the Moving Average method we create a context for the apnea range to determine where the starting and end segment is. The code is identical to the one in the other detection method as seen in Code 7.14 and The standard deviation calculation is done in the same way as with the moving average calculation, we have a query with a sliding window of a specific length that recalculates the standard deviation once new incoming events arrive. The difference is the custom function we implemented in Code 7.6. In Code 7.23 we select the event index and the SpO2 signal value from the last arriving event from the sensor stream. In the line mystddev(window(spo2)) we use the built-in window() function and the SpO2 value as input argument to our custom aggregation function. The length of the window is determined by the length of the FROM clause. The segment for each StandardDeviationEvent is set upon the assignment of the standard deviation value. In the class we have a set method for each declared variable and in the set function for the standard deviation we also set the segment based on the value. If the value is above the assigned threshold in the configuration file, the segment is set to RESATURATION, if the value is below on the negative scale of the threshold it is assigned as DESATURATION. Anything in between the thresholds is assigned as STEADY. After the creation of the new event we insert them into a new stream with the query in Code Code 7.23: The Standard Deviation calculation event stream. INSERT INTO StandardDeviationEvent SELECT eventindex, mystddev ( window ( spo2 )) AS sd, spo2 FROM SensorStream. win : length ([ length ]) Code 7.24: Temporary stream to handle segment assignment. INSERT INTO StandardDeviationSegmentEventStream SELECT eventindex, sd, spo2, seg FROM StandardDeviationEvent The queries in Code 7.25 and 7.26 is for finding the start and end of the apnea range. The first query inserts a new event into a new event stream called ApneaStart. The pattern subexpression is used together with the FROM clause. Instead of querying a specific reference table we tell Esper to look for a pattern based on references combined with pattern operators. The every sub-expression controls repetition of events and conditions and matches on every conditions that holds true for the following expression. The -> operator, also called the followed-by operator, conforms to true if a match of the parent condition is followed by another event or condition that also holds true. So for our query, we are looking for every incoming StandardDeviationEvent that has a STEADY segment that is followed-by another StandardDeviationEvent with the DESATURATION segment. We also add the AND NOT operators to the followed-by operator to decline conditions where events go from STEADY segments to 82
93 either another STEADY or RESATURATION segment. We do this because of fluctuations in the SpO2 signal stream as the breathing patterns for a human will always have random elements from person to person. We use annotation that instructs Esper to discard all but the first matching pattern condition for multiple overlapping matches because of performance and memory issues. We are only interested in the pattern form one point in time to another point in time, so every other match in between or before this is irrelevant to us. The query to find the end point of the apnea is similar to the start query. The query inserts an event into ApneaEnd once the pattern expression condition matches. First we want to find every ApneaStart event that is followed-by a StandardDeviationEvent that has a RESATURA- TION segment. If this condition matches, we want this to be followed-by another StandardDeviationEvent with the STEADY segment. The two queries combined will first find an event with a STEADY segment followed-by a series of events containing the DESATURATION segment, that shift onto a series of events having the RESATURATION segment that is followed-by another STEADY segment. Any fluctuations in between these two queries are ignored as we only want to find the pattern that conforms to a person that stops breathing and entering a desaturation state, and a state of resaturation when the person starts breathing again. Code 7.25: The start pattern expression for the apnea range. INSERT INTO ApneaStart SELECT a. eventindex AS eventindex FROM SuppressOverlappingMatches [ every a= StandardDeviationSegmentEventStream ( seg = Segment. STEADY ) -> ( b= StandardDeviationSegmentEventStream ( seg = Segment. DESATURATION ) AND NOT ( StandardDeviationSegmentEventStream ( seg = Segment. STEADY ) OR StandardDeviationSegmentEventStream ( seg = Segment. RESATURATION )))] Code 7.26: The end pattern expression for the apnea range. INSERT INTO ApneaEnd SELECT a. eventindex AS startindex, c. eventindex AS endindex FROM SuppressOverlappingMatches [ every a= ApneaStart -> b= StandardDeviationSegmentEventStream ( seg = Segment. RESATURATION ) -> c= StandardDeviationSegmentEventStream ( seg = Segment. STEADY )] Code 7.27: Extraction of values from the apnea range. SELECT b. endindex as endindex, a. minspo2 AS minspo2, a. maxspo2 AS maxspo2, a. duration AS duration FROM ApneaStream. std : lastevent () AS a, ApneaEnd. std : lastevent () AS b WHERE a. eventindex = b. endindex The last query in Code 7.27 selects the four variables needed by the attached listener to check if the apnea range is valid or not. The values are extracted from ApneaStream and is trigger by the last incoming event inserted into the ApneaEnd stream. 83
94 7.7 Storage All the storage operations are done by using the class SQLiteJDBC. In the class we have a constructor that handles the creation of the database tables if they do not exist already in a present database, and three main functions that add information to the database. Based on our design we chose to go with three tables, even though there are numerous schema designs that are viable for this sort of solution based on the complexity of the application. We use an external SQLite adapter that we include in our implementation to be able to connect and operate the SQLite database. The three tables generated by the constructor is presented in Code 7.28, 7.29 and Code 7.28: The SQL syntax for the Session table. CREATE TABLE IF NOT EXISTS session ( rowid INTEGER PRIMARY KEY AUTOINCREMENT, ticktableid INTEGER FOREIGN KEY ( ticktable ) REFERENCES tick ( rowid ), apneatableid INTEGER FOREIGN KEY ( apneatable ) REFERENCES apnea ( rowid ), sessionstart TIMESTAMP DEFAULT CURRENT_ TIMESTAMP NOT NULL, sessionstart TIMESTAMP, sessionahi DOUBLE, sessionapneas INTEGER ); CREATE TABLE IF NOT EXISTS tick ( rowid INTEGER PRIMARY KEY, sessionid INTEGER NOT NULL, tickindex INTEGER NOT NULL, tickrespchest DOUBLE, tickrespnasal DOUBLE, tickrespabdomen DOUBLE, tickspo2 DOUBLE ); Code 7.29: The SQL syntax for the Tick table. CREATE TABLE IF NOT EXISTS apnea ( rowid INTEGER PRIMARY KEY, sessionid INTEGER NOT NULL, apneastartindex INTEGER, apneaendindex INTERGER, apneastartsegment INTEGER NOT NULL, apneaendsegment INTEGER NOT NULL, class DOUBLE NOT NULL ); Code 7.30: The SQL syntax for the Apnea table. We implement three functions named insertsession(), inserttickevent() and insertapneaevent(). A session is created once the server is executed and for each run the session gets a new auto incremented id. As for this implementation, the same id is used in the tick and apnea tables. The inserttickevent() function inserts data values from the aggregated EventBean objects that is generated by the first EPL query. As for the apnea table, the start and end indices are only used by the apnea detection methods because they are able to not only classify a segment if it is either a period of normal or abnormal breathing, but also detecting when the apnea started and ended. The value for the start and end segments indicates which segment that is currently stored, while the last value is the actual classification results for the given segment. When the 84
95 AnnotationStreamListener triggers we call the insertapneaevent() to store the results from either the apnea detection or data mining methods, along with the indices and segment numbers. 85
96 Chapter 8 Evaluation & Performance Analysis In Chapter 6 we present an overview of our design in regards to four data mining methods used in the offline analysis, the approach to streaming simulation and the two apnea detection methods using Esper. Chapter 7 describe in detail the implementation of our client-server design together with the four data mining methods and the two apnea detection methods. In this chapter we present an overview of the performance evaluation of our methods. We also perform a profiling evaluation of the implementation, with a comparison to hardware related in smart phones. In Section 8.1 we present the metrics for our evaluation methods and a description on how we perform the evaluations. The section also includes benchmark results from a set of smart phones, as well as configuration combinations for our VirtualBox VMs. Section 8.2 lists an evaluation overview of three data mining methods: K-Nearest Neighbor, Support Vector Machine, Decision Tree. Evaluation results from Artificial Neural Network were unobtainable as we came across serious issue in running the library function in our implementation. More details surrounding the issue is presented in Section Our evaluation of the apnea detection methods is included in Section 8.3, followed by a comparison of the AHI results from the manual processing and in general for the records we used in the test sessions. A comparison evaluation of the metrics across the four data mining methods and our apnea detection methods is presented in Section 8.5. Section 8.6 consist of a summary of the comparison between our results and the results from the offline analysis. Lastly, in Section 8.7 we present a resource usage evaluation of our server implementation. 8.1 Performance Metrics & Configurations The offline analysis performs a broad and extensive evaluation of the four data mining methods, as well as a thoroughly comparison between the different databases from PhysioNet. Ten-fold cross-validation is used as the primary evaluation method while the second evaluation method is the hold-out method. N-fold cross-validation is one of the most common evaluation methods to measure the performance of classifiers. The hold-out method is also a very popular method that is similar to the cross-validation. Three metrics are used to evaluate the classifiers: accuracy, sensitivity and specificity. Accuracy measures the number of correctly classified annotation segments to the total number of segments. Sensitivity, also called true positive rate, measures the number of positive classified segments out of the total number of positive segments. Specificity, also called the true negative rate, measures the number of negative classified segments out of the number of total negative segments. They 86
97 are represented in Equations 8.1, 8.2 and 8.3. Accuracy = (TP + FN)/(TP + FN + FN + TN) (8.1) Sensitivity = TP/(TP + FN) (8.2) Speci f icity = TN/(TN + FP) (8.3) The positive class represents the class that contains epochs of abnormal breathing and the negative epochs of normal breathing. The offline analysis evaluates two different settings where the major one is epoch classification and a supplementary subject classification. The epoch classification used ten-fold cross-validation to test the generalization of the data mining methods on the three different databases from PhysioNet. The supplementary subject classification consists of the hold-out method to focus on calculating the AHI index for each subject to see if the numbers match up with the included AHI indices. They in addition evaluated crossdatabase analysis, which have poor performance overall. There is also performed a database and signal evaluation to determine the overall quality of the non-invasive signals and databases. This thesis will focus on evaluating the same data mining methods using the same data sources from PhysioNet, but using the best resulting settings for each method from the offline analysis. Based on this factor we see no point in changing the evaluation scheme. Using other metrics and evaluation settings will make the results even more difficult to compare. We already established that we want to use ten-fold cross-validation to evaluate the data mining methods. We will not be doing a cross-database or subject classification as it results in poor performance. There are so many factors that can impact the quality and properties of the data coming from separate databases that it is nearly impossible to directly compare. We will be using the same three metrics used in the offline analysis for our data mining and apnea detection evaluation. For the apnea detection evaluation, we will not be using cross-validation nor the hold-out method. The way we evaluate them are based on running each record, from each database with a series of different apnea detection settings to determine the overall best performing settings. For each test session we use the validation scripts to calculate the accuracy, sensitivity and specificity by using the stored results in the SQLite database. As a supplementary evaluation, we also want to know how many apneas our detection methods is able to find because we already know from initial testing that some of the records contain multiple apneas in a minute, at least for the Apnea-ECG database. In addition, we also want to see how accurate the detection methods can evaluate the AHI index based on the recorded apneas. A problem is that none of the databases include the number of apneas, so we do not have anything to compare our results with. Fortunately, the AHI index is present for all the records. In order to determine the number of apneas we will have to manually process and annotate records. This process is usually done by trained personnel with experience in knowing how to look for apneas in graphical signal variations. Due to time constraints, we do not have the time to get these annotated by a trained physician. Instead, we will pick two records from each database and manually process a plotted graph of the oxygen saturation levels based on the criteria established for an apnea. The criteria are that it should be a continuous drop and remain below optimal levels for more than ten seconds and a total drop of 3-4% in oxygen saturation in the blood stream. When it comes to resource evaluation and profiling of the implementation we need to analyze the best possible way to compare hardware. As already stated in previous chapters, the 87
98 ideal situation would be to design and implement a concept solution directly on a smart phone running either Android or ios. Not only does this require a very good understanding of the different APIs of the mobile platforms, but because both Matlab and Esper have no support for the chipset architecture we would have find similar supported classification libraries and streaming components. There is no official port of Esper and Matlab, and there might not ever be as Matlab is mostly used by the research and scientific community. The architectural difference in the chipset across mobile platforms compared to laptops or desktop machines is significant, and it is impossible to compare the systems directly. A processor having the same amount of cores and clock frequency can perform vastly different based on a number of factors such as architecture, power consumption, size and memory cache levels. So in order to actually evaluate and profile a conceptual diagnosis application to answer if it could be developed on the different mobile platforms, we use benchmarking scores to compare the results. Geekbench is a popular multi-platform benchmarking tool that performs varied calculation tasks on the CPU to determine how powerful a system is. The benchmarking result is a score that is given based on how fast the CPU can complete the different tasks. A baseline score for Geekbench is calibrated at around 2500 using an Intel Core i5-2520m. The tasks are divided into three different categories [43]: Integer performance Floating point performance Memory performance There are also three separate score types, but we will only be using the regular Geekbench score as it uses weighted arithmetic between the various score types so that an overall performance measurement can be compared to completely different systems. The higher the benchmark score, the better the performance, while double the score indicates double the performance of the CPU. We collect a list of benchmark results from individual smart phones from the Geekbench results page [37]. The results for various systems are freely available to everyone and contains both single and multi-core scores, as well as individual scores for each task performed. As there is a wide variety of smart phone brands, we have picked out the smart phones based on theses criteria in order of priority: 1. Benchmark score available 2. Price range 3. Consumer popularity 4. Technical reviews Price range is probably the biggest factor in picking a phone for consumers. Great technological advancements in devices the recent years have pushed the price of smart phones to match, and in some areas, surpass laptops. Which phone is popular also plays a factor as people tend to consistently buy phones from the same brand or product series. We have divided the smart phones into three tiers: budget, consumer and top. These are based on the criteria we set for the smart phones. A list of the individual smart phones and their year of release, as well as their benchmark score and tier is presented in Table 8.1. The top tier is based on phones that have been marketed as the most powerful products in recent years. The 88
99 Smart Phone Score (Single-core) Score (Multi-core) Tier Apple iphone 6s (2015) Top Samsung Galaxy S7 (2016) Top HTC 10 (2016) Top Apple iphone 5c (2013) Consumer LG Nexus 5X (2015) Consumer Sony Xperia Z3 Compact (2014) Consumer Motorola Moto G4 (2016) Budget Samsung Galaxy J5 (2015) Budget Sony Xperia M (2013) Budget Table 8.1: List of smart phones used as comparison to our VM setup. consumer tier are phones that has a price range below the top models, but is still considered as powerful in terms of hardware to price ratio. Looking from a price standpoint, budget phones can also include top tier phones that is now considered old and has dropped in price. We have discarded any phones running the Windows Phone operating system as these phones have such a low market share in comparison. 8.2 Data Mining Evaluation In this section we first discuss and present an overview of the evaluation results with a spread across all the three different data mining methods. In Section we present a more detailed results overview of the K-Nearest Neighbor method, followed by a similar overview for the Support Vector Machine in Section Section presents the results from the Decision Tree method. At last, in Section 8.6 we focus on comparing our achieved results to the results in the offline analysis. In Table 8.2 we present the best result for each signal combination from our evaluation analysis. The table consist of the signal combination, the data mining method and the accuracy, sensitivity and specificity. The data mining in the table represents the method that produced the best results overall, sorted by the signal combination. This is the result form just the Apnea-ECG database. The results are good, but we can already see that some signal combinations produce poor results than compared to others. Our highest accuracy is achieved using Decision Tree that produce an accuracy of 90.89% using all the signals in combination. Two methods stand out in our evaluation, the Support Vector Machine and the Decision Tree. Out of the 15 signal combinations, the Support Vector Machine produce the best result in eight of the results listed while the Decision Tree is the clear winner in six. Because we performed two separate test runs, one with pre-normalized data and one with normalized data, the results in Table 8.2 is not surprisingly all from the normalized results. The average accuracy across the normalized results are 80.50% in contrast to the pre-normalized data which is 74.01%. That is a difference of 6.57% between the pre-normalized and the normalized data. The difference might seem big, but looking at all the results from the runs we see that the low average accuracy for the pre-normalized results are caused by the Support Vector Machine not doing well with values above 1 or below -1, we elaborate this in more detail in
100 Signal Combination(s) Method Accuracy Sensitivity Specificity Resp. C DT 60.89% 64.86% 55.13% Resp. A DT 63.73% 69.03% 56.06% Resp. N SVM 86.29% 81.25% 93.60% SpO2 SVM 87.23% 95.84% 74.77% Resp. C, Resp. A DT 67.59% 68.30% 66.56% Resp. C, Resp. N SVM 86.68% 81.34% 94.41% Resp. C, SpO2 SVM 87.82% 96.18% 75.70% Resp. A, Resp. N SVM 86.40% 81.30% 93.78% Resp. A, SpO2 SVM 87.41% 95.84% 75.20% Resp. N, SpO2 KNN 87.28% 89.19% 84.52% Resp. C, Resp. A, Resp. N SVM 86.80% 81.30% 94.78% Resp. C, Resp. A, SpO2 SVM 87.79% 96.05% 75.82% Resp. C, Resp. N, SpO2 DT 89.44% 91.38% 86.64% Resp. A, Resp. N, SpO2 DT 89.97% 91.38% 87.94% Resp. C, Resp. A, Resp. N, SpO2 DT 90.89% 91.38% 90.18% Table 8.2: Best results sorted by signal combination. Respiratory from the chest and abdomen clearly produce worse results than the other signal combinations. The combination of using both of these signals resulted in an accuracy increase of 3.86%. While all the other combinations had an accuracy of over 85%, these two signals measured alone had both an accuracy of below 65%. As we already know, the Apnea-ECG database is not pre-processed, the results should present an overall feeling of the data quality for the records in the database [18]. A theory as to why the results for these signals are worse might be because of the way they are measured. When we did initial testing we plotted all the signals for some of the records and we could clearly see that both respiratory from the chest and abdomen had a more general fluctuation than respiratory from the nose and the oxygen saturation. We already know that measuring respiratory signals with an elastic band are prone to noisy artifacts as the band can either get trapped, or the sudden body movements can interfere with the results. For us, the results indicate that this is indeed the case. Figure 8.1 presents a graph with the average accuracy obtained for each signal over all the data mining methods. The green bars are represented as the normalized session results, while the blue ones are for the pre-normalized session. In general, we can see that the normalized results are higher than the pre-normalized ones, except for the signal combinations that produce poor results. Respiratory from the chest alone and in combination with respiratory from the abdomen produce better accuracy results with the pre-normalized data. Respiratory from the abdomen produced better with the normalized data, but only marginally. This only strengthen our belief that the data quality from these two signals are subpar compared to respiratory from the nose and oxygen saturation. Looking at our worst results, our lowest accuracy score come from respiratory from the chest and in combination with respiratory from the abdomen, both 90
101 Figure 8.1: Graph showing the difference between pre-normalized and normalized data. produce an accuracy of 39.21%. Our ten lowest results are dominated with bad accuracy performance for the Support Vector Machine with pre-normalized data. All the results from these test sessions have roughly an accuracy of 59%. In Section 8.6 we will compare our results from the data mining and apnea detection methods to correlating results in the offline analysis to see what were the main differences between our evaluation K-Nearest Neighbor The K-Nearest Neighbor produce overall good results for both the pre-normalized and normalized data. We only ran the method with one set of options as in contrast to the offline analysis which could optimize the results using different numbers of neighbors. The five best signal combinations are given for the pre-normalized data in Table 8.3 and for the normalized data in Table 8.4. Our best performing signal combination for the pre-normalized data use all the available signals which produce an accuracy score of 84.52%. For the normalized data we achieve an even higher accuracy score of 88.86%. The difference is around 3-4% between the two test sessions. Looking at the sensitivity and specificity we see that the method is better at classifying epochs of abnormal breathing rather than normal breathing, but the metrics are very similar by having the specificity a bit lower with around 5%. Our worst results are not surprisingly a use of signal combinations and standalone measurements of respiratory from the chest and abdomen. The worst we can achieve is 60.05% using respiratory from the chest. The average accuracy for the normalized data is 81.03% and for our pre-normalized data it is 77.83%. The average accuracy could have been higher, but were impacted by the bad results from respiratory from the chest and abdomen. 91
102 Signal Combination(s) Accuracy Sensitivity Specificity Resp. C, Resp. A, Resp. N, SpO % 86.19% 82.10% Resp. C, Resp. A, Resp. N 84.47% 86.01% 82.22% Resp. C, Resp. N, SpO % 86.87% 80.80% Resp. C, Resp. N 84.24% 86.19% 81.42% Resp. N, SpO % 85.29% 81.73% Table 8.3: Top five results for the K-Nearest Neighbor using pre-normalized data. Signal Combination(s) Accuracy Sensitivity Specificity Resp. C, Resp. N, SpO % 89.92% 87.32% Resp. A, Resp. N, SpO % 90.95% 85.46% Resp. C, Resp. A, Resp. N, SpO % 89.96% 86.45% Resp. N, SpO % 89.19% 84.52% Resp. C, Resp. A, SpO % 83.53% 83.53% Table 8.4: Top five results for the K-Nearest Neighbor using normalized data Support Vector Machine The Support Vector Machine produce very good results in our tests for normalized data, but using pre-normalized data the situation is quite the opposite. All though the Support Vector machine have eight out of the 15 total best results, the method also contribute to some of the worst accuracy scores we have. Our best accuracy score for this method is 87.82% using a combination of respiratory from the chest and oxygen saturation on normalized data. For prenormalized, a score of 85.84% is achieved using just oxygen saturation alone. From the results already presented in Table 8.2, we see that the Support Vector Machine have problems with values that are not normalized. This we also see in Table 8.5 as there are just two results that achieve an accuracy score of over 80%. The fourth and fifth best result achieves very poor results and have trouble classifying both epochs of normal and abnormal breathing. Taking a look at the results in Table 8.6 the results are much better. All the results have an accuracy over 85% and also a sensitivity of over 95%. Only two signal combinations are present in both the tables below. The pre-normalized results have two signal combinations using only one signal as a measurement. Signal Combination(s) Accuracy Sensitivity Specificity SpO % 88.07% 82.60% Resp. N, SpO % 72.80% 94.97% Resp. N 79.64% 77.22% % Resp. A, SpO % 64.18% % Resp. C, SpO % 61.82% 56.68% Table 8.5: Top five results for the Support Vector Machine using pre-normalized data. 92
103 Signal Combination(s) Accuracy Sensitivity Specificity Resp. C, SpO % 96.18% 75.70% Resp. C, Resp. A, SpO % 96.05% 75.82% Resp. C, Resp. N, SpO % 95.90% 81.94% Resp. C, Resp. A, Resp. N, SpO % 95.46% 82.02% Resp. A, SpO % 95.84% 75.20% Table 8.6: Top five results for the Support Vector Machine using normalized data. Mentioning the worst performing signal combinations using Support Vector Machine is a bit meaningless as all the results from the normalized data is heavily affected by the distance function not being very good at handling values greater or smaller than 1 and -1. This indicates that if a solution using the Support Vector Machine is being viable, there is a need to normalize the value scales of the data either by the sensor or the application before using the data for classification or use a different kernel function. Even though the normalized test runs are very good compared to the pre-normalized, we get the lowest accuracy score by using a combination of respiratory form the chest and abdomen, having an accuracy of 39.21%. In contrast, the same signal combination using normalized data achieves an accuracy of 59.16%. But the pre-normalized data have an average accuracy of 60.70%, while the normalized averages 78.91% in accuracy and is very affected by the two results that have the abysmal accuracy Decision Tree The Decision Tree achieve our best result out of all the data mining methods with an accuracy of 90.89%. The difference between the pre-normalized and normalized session is also not that huge as the pre-normalized data achieves an accuracy of 89.14%. The Decision Tree is on average our best performing data mining method reaching an average accuracy of 81.80% with K-Nearest Neighbor being a close second with 81.03%. Indications from the offline analysis and our design should suggest that the Decision Tree method would perform worse because it can have issues by separating the numerical values from our training set in binary. Based on our results it did not affect the results as much as one would expect. The highest accuracy using all the signal in combination with no standalone signals, reach our top five for each test session. The closest results using just one signal we get from with oxygen saturation having an accuracy of 85.25%. The overall sensitivity and specificity is good for both the pre-normalized and normalized test runs. The top five results for each run is presented in Table 8.7 and 8.8. Signal Combination(s) Accuracy Sensitivity Specificity Resp. C, Resp. A, Resp. N, SpO % 87.76% 90.09% Resp. A, Resp. N, SpO % 87.32% 89.66% Resp. C, Resp. N, SpO % 86.08% 90.35% Resp. N, SpO % 85.33% 85.97% Resp. C, Resp. A, Resp. N 85.46% 84.65% 86.01% Table 8.7: Top five results for the Decision Tree using pre-normalized data. 93
104 Signal Combination(s) Accuracy Sensitivity Specificity Resp. C, Resp. A, Resp. N, SpO2 90,89% 90,18% 91,38% Resp. A, Resp. N, SpO2 89,97% 87,94% 91,38% Resp. C, Resp. N, SpO2 89,44% 86,64% 91,38% Resp. N, SpO2 86,78% 84,28% 88,50% Resp. C, Resp. A, SpO2 86,50% 85,39% 87,26% Table 8.8: Top five results for the Decision Tree using normalized data. The average accuracy difference between the two test runs is a merely 1.15%, which is far better than both Support Vector Machine and K-Nearest Neighbor having a difference of 6.91% and 3.2% respectively. The worst accuracy we achieve is by using just the respiratory signal from the chest with a score of 60% using pre-normalized data and 60.89% using normalized data Artificial Neural Network The implementation for Artificial Neural Network is based on the same implementation from the offline analysis. We have no issues running the Matlab function using pre-loaded data included in Matlab, so this is not the cause. We ran into issues that are not linked to our implementation nor the data sources we use. According to the Matlab web page, the Matlab Compiler should support compilation of the Neural Network toolbox functions. The problem is related to the patternnet function in some form. The error message is: Default value is not a member of type "nntype.training_fcn", which is directly linked to the patternnet function that we use to generate the neural network in Matlab. Finding out what is causing the problem is hard, as a lot of the search results in an effort to troubleshoot the problem results in Chinese forums, meaning the problem is not that broad to begin with. There are two theories from two different sources that can be the cause of our Artificial Neural Network implementation not working. The toolbox support for Artificial Neural Network only allows for pre-trained networks [39] [41]. Based on these facts we have also tried to build a JAR library, including a pre-processed and aligned training set. This do not work and gave us the same error message as earlier. It is difficult to know what they mean by pre-trained, as the three other toolbox functions work out of the box without any issues. But this is probably the best answer to the issues we are having with the toolbox function. From the Chinese source, they mention that there is an issue with licensing or copyright issues with Matlab and the toolbox function that hinders full support when the function is compiled to another language as a library [60]. This seem like an odd problem considering that Matlab is reliable and used across various scientific and research domains. Most likely, this is not the root of the problem at all. 8.3 Apnea Detection Evaluation This section contains an overview of the results from both the apnea detection methods. We go through the bigger differences between the databases in contrasts to the methods we use. In Section we present and discuss the results and the available options in clearer detail for our Moving Average method. Section is presenting the same detailed information, but 94
105 for the Standard Deviation method. The two Tables 8.9 and 8.10 showcase some metric ranges from the Moving Average and Standard Deviation methods. First we list the database that produce the results followed by the average accuracy across all options and records, the accuracy range, sensitivity range and specificity range. An additional graph is presented in Figure 8.2 where the average accuracy from all the test runs is displayed next to each other. Database Avg. Accuracy Accuracy Range Sensitivity Range Specificity Range Apnea-ECG 91.35% % % % MIT-BIH 67.06% % % % St. Vincent s 70.01% % % % Table 8.9: Metric ranges and average accuracy of the three databases using the Moving Average method. Database Avg. Accuracy Accuracy Range Sensitivity Range Specificity Range Apnea-ECG 88.03% % % % MIT-BIH 66.57% % % % St. Vincent s 71.93% % % % Table 8.10: Metric ranges and average accuracy of the three databases using the Standard Deviation method. What we can already see is that between the two methods the performance difference is minimal. We achieve an average accuracy of 91.35% using the Moving Average method on the Apnea-ECG database, which is our highest score for the detection methods. Between the two methods there is just a difference in accuracy of 0.63%, with the Moving Average method coming out ahead. The difference is minimal and might indicate that either of the methods can be suitable to detect apneas. When it comes to their accuracy range we see that the Moving Average is far better across the board as the difference between the smallest and highest session is 4% in contrast to the Standard Deviation which ranges a difference of 15%. When looking at the sensitivity and specificity ranges we also see that our methods are clearly better at finding epoch of abnormal breathing than normal breathing. Ideally we would like the ranges to be very similar, but our test runs can be affected by simply one record to another so that the numbers will not represent the overall performance. The score we get from the specificity ranges is very poor, but that might be because the way our methods are designed and implemented. We classify them as "apnea detection" methods, and they rely on finding apneas in segments rather than not finding apneas. Also, in initial tests we plotted a great deal of records from the various databases to see how the data would visually be represented. It is very clear that the data quality in Apnea-ECG far surpasses the data quality in MIT-BIH Polysomnography and St. Vincent s databases. A lot of the records in both the two databases have artifacts in terms of random spikes and drops that will impact our methods. Some periods the oxygen saturation will spike and have a value greater than 100%, and in other periods the oxygen saturation drastically drop to 0%. This 95
106 only strengthens our theory in that the data quality across the databases is a key factor in our results. Figure 8.2: Graph showing the accuracy difference between the detection methods and the three databases Moving Average The Moving Average method consist of three options that can be adjusted to optimize the window segments and the criteria for classifying a segment as an epoch of abnormal breathing. The three Tables 8.11, 8.12 and 8.13 represents the top ten option combinations with the best results from the three databases. The first column includes weight, which is added to the moving average calculation so that the cross-sections between the SpO2 value stream and the moving average is better aligned with the apneas. In the second column, the length controls the amount of events in the calculation of the moving average at any given time. The SpO2 column is the amount of oxygen saturation of the apnea segment. The results from the databases are very consistent as the options make a very small impact on the outcome. The accuracy varies from 1-2% depending on the options selected, but the sensitivity and specificity are more inconsistent. Apnea-ECG Database The results from the Apnea-ECG database are very good as we achieve an overall best accuracy of 93.26% from two options as seen in Table The best weight scheme we can achieve with this database is 4 and 3. We may obtain a higher accuracy if we try a higher weight, but it can also result in a lower accuracy. We chose the weight value to stop at 4, similar to the SpO2 drop maximum value. This means that the database responds well to higher weights than lower. 96
107 The window length of the moving average is shared equally between 20 and 30, with 20 coming out as the best in combination with a weight of 4. There are no window lengths of 60 at the upper scale of the results which might indicate that a higher window length than 20 or 30 is meaningless. The amount of SpO2 drop seem to not affect the outcome that much. The lowest accuracy we achieve using records form the Apnea-ECG is 89.10%. An issue with these numbers from this database is that they are impacted by the low record number and the division between records that are clear opposites in terms of severity. Weight Length SpO2 Accuracy Sensitivity Specificity % 93.26% 88.49% 63.23% % 93.26% 88.49% 63.23% % 93.03% 89.15% 61.65% % 92.98% 89.15% 61.57% % 92.76% 85.22% 64.99% % 92.73% 85.06% 65.03% % 92.49% 81.06% 66.30% % 92.49% 81.22% 66.23% % 92.11% 86.26% 63.45% % 92.06% 86.14% 63.49% Table 8.11: Top ten results based on Moving Average options for the Apnea-ECG database. MIT-BIH Polysomnography Database Results from the MIT-BIH Polysomnography database is listed in Table Like with the results from the Apnea-ECG database we see the overall impact the options have on the outcome is minimal. In comparison to the Apnea-ECG database, we get better results with a weight of 2 rather than 3 or 4. The window lengths seem to be inconsistent in terms of affecting the results, here we see all the window length options in the top ten results. The SpO2 drop also have no effect on improving the accuracy results. We see that the specificity is low and we cannot link the low numbers with any options we chose as they all sway between 10-35%, but on the other hand the sensitivity numbers are very good. If we improve on the specificity, we can get higher accuracy results. The lowest accuracy we achieve is 65.50%, and is 3% shy of the top result. Our results from the MIT-BIH Polysomnography database may share the similar problem with the Apnea-ECG database in that it has a small number of records. 97
108 Weight Length SpO2 Accuracy Sensitivity Specificity % 68.57% 85.06% 33.31% % 68.39% 88.86% 27.05% % 68.24% 89.47% 25.93% % 67.98% 89.07% 26.09% % 67.94% 88.49% 26.58% % 67.78% 86.01% 30.58% % 67.69% 93.36% 19.49% % 67.69% 93.36% 19.49% % 67.68% 86.50% 28.95% % 67.41% 94.35% 17.41% Table 8.12: Top ten results based on Moving Average options for the MIT-BIH Polysomnography database. St Vincent s Database With the St. Vincent s database, we achieve a highest accuracy of 73.43% across all the records. The top ten results are presented in 8.13 where we see that the weighting scheme for the top results either consist of the highest possible weight, or no weight at all. There seem to be no difference in the window length as all the results have varying window lengths. We see a clear favor of using 4% as the SpO2 drop decider in the top results. As with the MIT-BIH Polysomnography database the specificity is very low and is even lower for this database. The reason for this can be a number of factors. We already know that the data quality for the MIT-BIH Polysomnography and the St. Vincent s database is not as good as the Apnea-ECG database. These two databases contain artifacts and spikes that can impact a large portion of the results. Besides, if we look at the plotted SpO2 graph for the St. Vincent s database we see that a lot of the records contain almost as much apneas as there are hypopneas which can be misclassified as regular apneas. The lowest accuracy we achieve is 64.90%. There is a bigger difference between the best and the worst accuracy score for this database compared to the others. 98
109 Weight Length SpO2 Accuracy Sensitivity Specificity % 73.43% 90.28% 10.79% % 73.40% 90.63% 10.28% % 72.97% 90.36% 9.84% % 72.95% 89.37% 11.21% % 72.94% 87.62% 13.74% % 72.60% 89.82% 10.35% % 72.55% 89.19% 11.41% % 72.54% 89.84% 10.35% % 72.39% 89.13% 11.23% % 72.28% 87.77% 12.85% Table 8.13: Top ten results based on Moving Average options for the St. Vincent s database Standard Deviation Similar to the Moving Average, the Standard Deviation method have three options that can be adjusted to optimize the method properties. The threshold is a set value that is used as a decider when comparing the threshold and the calculated value of the standard deviation of the SpO2 value. The length option is similar to the window length in the Moving Average, but is a bit smaller. When the length is higher than 12, which is the maximum, the deviation when computing the standard deviation has less impact in finding if the SpO2 value is increasing or decreasing. The SpO2 value is also added as some decider criteria to find and correctly validate an apnea segment. Apnea-ECG Database The best results from the Apnea-ECG database is listed in Table We have two option combinations that achieve an accuracy of 93.13% and close to similar sensitivity and specificity. The options seem to not favor in any direction as the threshold, length and SpO2 drop values are very inconsistent, but achieves over a 90% accuracy with sensitivity following suit. The window lengths shift more in the direction of smaller windows rather than bigger which is no surprise when we think of the possible highest standard deviation computation we can achieve is by having a sliding window length of 2. 99
110 Threshold Length SpO2 Accuracy Sensitivity Specificity 1 4 3% 93.13% 88.02% 60.83% 1 4 4% 93.13% 88.46% 60.75% 1 2 3% 93.06% 87.20% 60.93% 1 2 4% 93.01% 87.69% 60.74% % 92.12% 83.40% 62.64% % 91.53% 78.88% 63.92% % 91.07% 83.48% 59.07% % 91.05% 82.67% 59.07% % 90.41% 92.76% 56.21% % 90.16% 93.08% 55.65% Table 8.14: Top ten results based on Standard Deviation options for the Apnea-ECG database. MIT-BIH Polysomnography Database For the MIT-BIH Polysomnography database we see similar results as with the Apnea-ECG database. Inconsistency in the option combinations is present and they do not impact the results all that much. We achieve an accuracy of 68.35% using a threshold of 1, window length of 8 and a SpO2 drop of 3. For these results we favor towards bigger window lengths in contrast to the Apnea-ECG results that produce better results with smaller window lengths. The sensitivity is better across the board but like with the Moving Average method the specificity is awful, meaning that the database might be the problem rather than our methods. The Standard Deviation method produce at worst an accuracy of 63.84%. Threshold Length SpO2 Accuracy Sensitivity Specificity 1 8 3% 68.35% 86.20% 31.45% % 67.85% 92.17% 21.42% % 67.71% 92.33% 20.91% 1 2 4% 67.39% 94.05% 16.97% 1 2 3% 67.39% 94% 17.03% % 67.35% 88.42% 26.01% 1 8 4% 67.31% 87.72% 27.25% % 67.28% 88.86% 25.22% % 67.03% 87.58% 26.38% % 66.99% 90.16% 22.38% Table 8.15: Top ten results based on Standard Deviation options for the MIT-BIH Polysomnography database. 100
111 St Vincent s Database Results from Table 8.16 share similar outcomes as with the two other databases. There are no option combinations that favor among the top results. Considering the data quality and artifacts for this database we assumed the accuracy would be terrible, but we are able to achieve an accuracy of 74.19%. The sensitivity and specificity is almost a mirror of the results from the MIT-BIH Polysomnography database as the method is very good at finding and classifying the apneas in the records, but lacks the properties to filter out the segments containing normal breathing, which is not a surprise. But with such a huge difference in specificity between the lacking databases and the Apnea-ECG database we are unsure if this has something to do with the data we use or how our methods work. Threshold Length SpO2 Accuracy Sensitivity Specificity 1 4 4% 74.19% 94.09% 5.87% % 73.94% 93.33% 7.27% 1 4 3% 73.85% 93.94% 5.99% % 73.64% 92.28% 8.26% % 73.28% 90.84% 9.67% 1 2 4% 73.14% 89.64% 11.25% % 73.03% 90.12% 10.68% % 73% 87.95% 13.49% % 72.97% 88.48% 13.02% 1 2 3% 72.39% 88.63% 12.67% Table 8.16: Top ten results based on Standard Deviation options for the St. Vincent s database. 8.4 AHI Comparison The AHI index is calculated by the number of apneas or hypopneas per hour. In the offline analysis the results subject evaluation calculate the AHI based on the amount of correctly classified segments per hour, which means that for the Apnea-ECG database a maximum AHI index of 60. The problem is that the results will never be a 100% correct as patients with a high severity can, and most likely, have multiple apneas in a minute. For the manual processing we pick six records, two from each database that we manually annotate. From the Apnea-ECG database we chose record "a01r" and "a03r". Record "a01r" have a high severity and AHI index and record "a03r" contain an almost equally number of segments containing normal and abnormal breathing. For the MIT-BIH Polysomnography database we chose the records "slp60" and "slp66". They are basically chosen at random as we have a small number of records to choose from considering that record "slp67x" have a short duration and contain almost no apneas. Records "ucddb008" and "ucddb023" are chosen from the St. Vincent s database. Record "ucddb008 is chosen because of the low AHI index and a long duration, and the record "ucddb023" is chosen at random on the scope of having a moderate to high AHI index. Besides the selection of records, we use the best options for each database when testing our apnea detection methods ability to detect the number of apneas. 101
112 The results for the manual processing is presented in Table The first column is the record we test, the second column is the manual annotated apneas done by us, and the rest of the columns represent the method abbreviated as either "MA" or "SD" and the number of apneas and the AHI index. The results show that the methods can very accurately represent the records for the Apnea-ECG database, while the other records from the other databases are very inconsistent. The manual annotated apneas for the St. Vincent s database compared to the apneas processed by the methods are far off. When going through the records and looking at some plotted graphs we clearly could tell that the data from St. Vincent s database lacks annotations. We compared the annotation files to the actually plotted graph and found that the graph shows far more clear apneas than what is listed as annotations. This combined with artifacts and spikes, as well as the low sample rate we can conclude that the records in the St. Vincent s database is not suitable for classification. The AHI comparison in the three Tables Record Manual Apneas MA Apneas MA AHI SD Apneas SD AHI a01r a03r slp slp ucddb ucddb Table 8.17: Apnea and AHI comparison using manual annotation processing on selected records. 8.18, 8.20 and 8.19 are processed and compared across our apnea detection methods used in this thesis. The AHI index we produce as a results is not a 100% accurate representation because we will calculate the AHI index based on the number of segments for the Apnea-ECG and the MIT-BIH Polysomnography databases. For the St. Vincent s database, we have the number of apneas in the annotation file, but as we have stated above, the number of apneas is not correlating with the annotation list. The AHI comparison results from all the databases are not very good. The Apnea-ECG database is the closest one overall to accurately represent the AHI index listed on PhysioNet. Both the MIT-BIH Polysomnography and the St. Vincent s database miss the AHI index by a big margin. This can because of the data quality which we have mentioned numerous times throughout the other sections. In order to conclude a more accurate evaluation of the AHI capabilities of the detection methods we suggest more data with a higher quality to be used with corresponding annotations, similar to the Apnea-ECG database. 102
113 Record PhysioNet AHI MA AHI SD AHI a01r a02r a03r a04r b01r c01r c02r c03r Table 8.18: AHI comparison of the Apnea-ECG database records. Record PhysioNet AHI MA AHI SD AHI ucddb ucddb ucddb ucddb ucddb ucddb ucddb ucddb ucddb ucddb ucddb ucddb ucddb ucddb ucddb ucddb ucddb ucddb ucddb ucddb ucddb ucddb ucddb ucddb ucddb Table 8.19: AHI comparison of the St. Vincent s database records. 103
114 Record PhysioNet AHI MA AHI SD AHI slp slp slp slp slp67x Table 8.20: AHI comparison of the MIT-BIH Polysomnography database records. 8.5 Metric Comparison For our metric comparison we have three graphs that are presented in the Figures 8.3, 8.4 and 8.5. We took the best performing signal combinations for each data mining method for their respective metric. The graphs bring up some interesting patterns. The most interesting is the metrics from the results of the Support Vector Machine. For the accuracy it sorts of follows a similar pattern to the K-Nearest Neighbor and the Decision Tree. Meanwhile, looking at the sensitivity and specificity metrics there is clear discrepancy in the patterns. In order to see if this was a mistake we ran two sets of test session that produced the same results regardless. We do not know if this is a runtime error or a problem with the toolbox functions when they are compiled to JAR files. Even though the sensitivity and specificity generate an abnormal pattern, the accuracy remains relatively similar to the other data mining methods. Except for the pattern generated by the Support Vector Machine, both the K-Nearest Neighbor and the Decision Tree have an almost identical pattern for all the metrics. Figure 8.3: methods. Accuracy comparison for signal combinations across the three data mining Respiratory signals from the chest and abdomen sticks out as it did with the results from the data mining evaluation. The data mining methods seem to struggle when using these alone or 104
115 Figure 8.4: methods. Sensitivity comparison for signal combinations across the three data mining in a combination. When combined with either respiratory from the nose and oxygen saturation the increase in accuracy, sensitivity and specificity are huge. This means that the classification score for these two signals overrides the lacking signals. Figure 8.5: methods. Specificity comparison for signal combinations across the three data mining 8.6 Offline Results Comparison One of the main parts of the data mining evaluation is to compare our results with the results generated in the offline analysis. In this section we will focus on comparing the results from the data mining methods from our thesis and the offline analysis. We will also compare our 105
116 apnea detection methods with the data mining methods, even though they cannot be compared directly because they are not tested with the same evaluation method. Due to the difference in our approach to design, implementation, environment and test configurations we already know that we cannot achieve the same level of accuracy as the offline analysis. In the offline analysis they test the data mining method on both the Apnea-ECG and the MIT-BIH databases. In our thesis we only use the Apnea-ECG database because of the lack of data quality of the other databases. The offline analysis achieves an accuracy of 96.6% as the highest with the K-Nearest Neighbor method using a signal combination consisting of respiratory from the chest and nose. In our thesis, the highest accuracy we achieve is 90.89% using the Decision Tree on all the signals in combination. There are some interesting patterns we need to acknowledge. Looking at the results from the offline analysis we see that K-Nearest Neighbor performed the best overall, followed close by the Support Vector Machine and the Artificial Neural Network. The Decision Tree performed the worst, even though not by much, meaning there is no data mining method that performs bad. In contrast to our results, we see that the Support Vector Machine performs the best out of all, considering the data is normalized else the selected distance function would cause the classification to be very poor. The second best method is the Decision Tree, a direct opposite result from the results in the offline analysis. On average our accuracy compared to the offline accuracy is between 5-10% worse. It is hard to find a root cause of these results as there are many factors that can impact our results. Firstly, we only use the data mining configurations from the best results, meaning we have less leeway to optimize the method to the specific signal combination. Secondly, although we use the same toolbox functions from Matlab, there can be underlying factors that such as code optimization that can affect the float precision. The data from the initial records from PhysioNet is processed, transferred and classified through four different layers of programming languages and environment. This can impact the outcome of the data mining methods. In theory, as long as the pre-processing methods generate the same data and we use the same data mining library, the outcome should be exactly the same. Streaming the data should have no impact as it is the same data, it just arrives in sequences. For now, it is uncertain why the accuracy is worse, but the goal of the comparison was to see if online analysis is viable, which we think it is with and average accuracy of over 85%. If we compare the database results from the offline analysis with our apnea detection methods, we see a clear pattern emerge. The offline analysis concludes that the data quality for the Apnea-ECG is better by a good margin. That is also our conclusion taken from our results. On average we achieve roughly the same accuracy results for the MIT-BIH Polysomnography database with our apnea detection methods. The offline analysis achieves higher specificity than our methods, but lower overall sensitivity. This is not strange as our methods are more designed to find the apneas rather than distinguish the different segments. The offline analysis lacks result for the St. Vincent s database. We achieve an average accuracy of 71.93% using the St. Vincent s database. When it comes to metric comparison and the detailed results for the different data mining methods we can see that the pattern our results generate in Figures 8.3, 8.4 and 8.5 share similarities with the graphs included in the offline analysis. respiratory from the chest and abdomen, as standalone or in combination performs far worse than in combination with respiratory from the nose and oxygen saturation. 106
117 8.7 Resource Performance & Utilization In this section we present the performance measurements of our server implementation recorded by JProfiler. A list of all the virtual machine configurations is described in Section Evaluation results of the CPU and memory utilization is presented in Sections and 8.7.3, followed by a performance summary of the I/O operations of the storage capabilities in Section JProfiler use an agent service that attaches to a running JVM process to measure the implementation. This can either be attached by using the GUI interface or with the included offline mode. The offline mode is simply CLI commands that are included so that we can incorporate profiling with scripts or other automation methods such as cron jobs. This means that we do not need to have a running process of the GUI interface to attach the agent to the JVM. While we would like to have a profiling tool that does not impact the implementation performance in any way, this is surely impossible as there will always be some overhead to take into account when analyzing the structure of an implementation. The amount of overhead is impossible to estimate as JProfiler have no idea about the characteristics of our implementation. There are no absolute values for us to make a judgement call upon, we merely have to guess the amount of overhead. In the JProfiler application there is an indicator that can help us estimate the increase or decrease in overhead due to enabling certain features, a figure of this is presented in Appendix A. In initial tests we found the overhead indicator to be lacking when running tests using one CPU core and adding a low execution cap to our VM. This might be because the amount of CPU resources is so low that any JProfiler feature is causing a great deal of overhead. So even if the indicator was at normal levels the overhead clearly impacted the results. Because of the uncertainties we decided to keep the bar at a threshold that is below the middle of the bar indicator. When doing initial testing experiments we sometimes had irregularities in the data collected from JProfiler. At some point we had the CPU load go above 100%, event with just one core. In addition, there are no periods where the CPU load would be stable and suddenly spike up and down from 0% CPU load to an average higher CPU load than the rest. I could replicate this on virtual machines running on two different machines, although it was a random occurring event Virtual Machine Environment A list of benchmark scores for the select smart phones in this thesis is presented in Table 8.1. This list will set the baseline for our VM configurations so that the performance of each VM is correlating to the performance of the various smart phones. We used the Geekbench 4 CLI tool to benchmark each VM. The scores for each configuration is listed in Table CPU Cores Execution Cap Score (Single-core) Score (Multi-core) Tier 2 90% Top 3 35% Consumer 2 25% Budget Table 8.21: The different VM configurations and their benchmarking scores. 107
118 We need to adjust both the number of CPU cores and the CPU execution cap in the VM configuration before starting the VM. The combination of these two parameters gives is the score we have to work with when comparing the profiling results to the list of smart phones. In addition to these two settings, we will also set the amount of memory available. The memory limit will be set to 4GB, which should be more than sufficient considering that the VM is only running the operating system, and some necessary services and our implementation. Most of the smart phones today come with at least 1GB of memory and higher. Besides these, we use the default configuration that comes with VirtualBox. The hardware the VMs will be running on is presented in Table 7.1. The benchmark scores in the table is taken directly from a single run of Geekbench. There is no point in testing configurations that by far exceeds the top tier smart phones, but for the sake of a baseline comparison we have run Geekbench on a VM running 6 CPU cores and no execution cap. We were able to achieve a single-core score of 2409 and a multi-core score of We have decided to base our VM configurations based on the average score for both single-core and multi-core for each tier of smart phones from the Table in 8.1. We are not able to match the exact benchmark score, but as close as possible. Each JProfiler agent and test session will run for approximately 70 minutes. We ran some initial tests with sessions having a duration of over eight hours, simulating a whole night s worth of sleep. That resulted in the CPU load slowly declining until it came to a steady point around the 1-hour mark, meaning that running the test for a longer period will not result in any significant data than what we already can collect in an hour. The reason for the steady decline in CPU load is probably a cause of Just-In-Time (JIT) optimization of the Java implementation. Code that is often used during runtime might get highly optimized and over time require less resources than during execution. To achieve our results, we run the performance tests with a combination of conditions. We want to test how much impact the sampling rate from a real sensor has on a smart phone CPU. This means we chose to go for three options of 1 Hz, 100 Hz and a maximum of 250 Hz. We will use one record from the Apnea-ECG database to test both 1 Hz and 100 Hz sampling rates and one record from the MIT-BIH Polysomnography database to test a sampling rate of 250 Hz. All the data mining and apnea detection methods will be tested with each of the three sampling rates for an hour. All the combinations of methods and sampling rates will also be tested on the separate VMs so we collect data for the different tiers of smart phones CPU Utilization In this section we present the results from the combined profiling session for each individual VM configuration. The CPU load sampling rate is set at 5 ms and runs the default profiling options in JProfiler. The hardware the VM scales off is included in Table 7.1. It is running on a server, with mostly other processes stopped to keep the interference at a minimum. An overview of the results is listed in Tables 8.22, 8.23 and The top most row lists the abbreviation of the data mining and apnea detection methods, followed by a row of their correlating average, minimum and maximum CPU load, these values are presented in percentage. On the far left we mark each row with the sampling rate used for the test runs. From the results we see that the average CPU load from all the VM configuration regardless 108
119 of data mining method and sampling rate remains almost identical. The minimum CPU load is inconclusive as all the sessions had periods where the CPU load will dip to zero. Sessions running 250 Hz sampling rate had issues of spiking that produces some odd numbers in the maximum column. For the maximum CPU load, if we ignore the odd values, shows clearly that the first few segments prone to classification using the data mining methods from Matlab causes a higher CPU load than the detection methods. If we compare the values with all the VM configurations, the consumer tier VM is overall better in terms of CPU load comparing all the values. Despite the fact that the overall bench mark score for both single and multi-core is better on the top tier, the consumer tier VM has three running cores in contrast to the top tier which have two running cores. This is the reason for the better results for the consumer tier as the operating system schedules the work load on more cores, giving us a lower overall CPU load. There are cases of some anomalies that for us is has an unknown cause as we cannot pinpoint where the cause is because we are not able to reproduce the issue. For some sessions that ran 250 Hz sampling rate the CPU load had a strange pattern over a period of minutes where the CPU load would spike higher than a 100% followed by a period of no CPU load at all. This would repeat itself for some minutes throughout the profiling session. The problem is very random and we had trouble in trying to reproduce the issue as it occurs in some profiling session and might not occur in others. The link to the issue seem to be with the sessions running 250 Hz sampling rate and is not linked to neither the VM configuration, data mining method nor the data source used. Even though there is no clear indicator to what is causing the spikes, we have a theory that it is either JProfiler that queues the profiling data and measure it in one sudden operation or that something is blocking the implementation, filling up the network buffer with events and we see a sudden spike in processing of the events that is queued. The problem persisted even on profiling session that ran on two different machines, but the problem still had random occurrences. The results might indicate that a smart phone may struggle with a transfer rate of 250 events per second from the sensor, but we cannot conclude that this is the case as there are a number of factors that can be the root cause of this problem. It could either be the implementation, the profiling tool, the hardware or the number of different libraries and layers. It is impossible to determine without further testing and research. In Figures 8.6, 8.7 and 8.8 VM Configuration (Budget) KNN SVM DT MA SD Avg Min Max Avg Min Max Avg Min Max Avg Min Max Avg Min Max 1 Hz Hz Hz Table 8.22: Cross-table overview of the average, minimum and maximum CPU load from the profiling sessions running the all the methods on the budget tier VM. we plot the average CPU load pattern in combination between all the methods for each VM configuration. The results are not that all surprising except for the pattern created by sessions running 250 Hz sampling rate. The fluctuations in the pattern indicates that there are some problems processing the queued events in the network buffer. Once the client connects to the server we initialize the rest of the class objects that handles the incoming events. While this 109
120 VM Configuration (Consumer) KNN SVM DT MA SD Avg Min Max Avg Min Max Avg Min Max Avg Min Max Avg Min Max 1 Hz Hz Hz Table 8.23: Cross-table overview of the average, minimum and maximum CPU load from the profiling sessions running the all the methods on the consumer tier VM. VM Configuration (Top) KNN SVM DT MA SD Avg Min Max Avg Min Max Avg Min Max Avg Min Max Avg Min Max 1 Hz Hz Hz Table 8.24: Cross-table overview of the average, minimum and maximum CPU load from the profiling sessions running the all the methods on the top tier VM. happens, the client keeps sending events. The server then spends a lot of time processing the events that are queued in the buffer trying to catch up. Besides this, the issue surrounding the spikes can be the cause of the pattern if they occur at approximately the same point in the timeline. The peak at the start in timeline and the diminishing CPU load as the timeline goes on is caused by the optimization enforced JIT and the operating system. The JProfiler agent will attach itself to the JVM after our server implementation to has initialized the data mining methods. We tried to get measurements of the CPU load when the implementation starts the initialization of the methods, but ran into problems as the implementation and JProfiler would crash if we ran the default settings in our offline mode. Only the CPU profiling worked with Matlab libraries meaning we have run some small tests to measure the CPU load on the library initialization. The results are presented in Table 8.25 with an average and maximum column from the time the server is executed to the time the building of the models and initializing is done. It seems like the K-Nearest Neighbor do not benefit that much by more cores, but rather the execution cap that is set to 90%, meaning we only restrict the CPU by 10%. Considering that a future application on a smart phone will most likely integrate the initialization phase components needed to classify apneas into a loading interface, these results are not that relevant. KNN SVM DT Avg Max Avg Max Avg Max Budget Consumer Top Table 8.25: The CPU load of the initialization of the data mining methods from the Matlab toolbox. 110
121 Figure 8.6: CPU load plotted in a graph showing the pattern over time. This is for the budget tier VM configuration. Figure 8.7: CPU load plotted in a graph showing the pattern over time. This is for the consumer tier VM configuration. Figure 8.8: CPU load plotted in a graph showing the pattern over time. This is for the top tier VM configuration. 111
122 8.7.3 Memory Utilization In this section we discuss and present the details about the memory utilization of our server implementation and how it is affected by the sampling rate and other factors. We will only present the details about the heap memory and note that some of our components in the implementation is implemented as static classes meaning they impact the overall total memory pool, but not by much. The average, minimum and maximum used memory is presented similar to the CPU utilization and resides in the Tables 8.26, 8.27 and We see the same strange arbitrary pattern in some of these results like with the CPU load. The results in Table 8.26 the average, minimum and maximum is affected by something that causes them to use a lot more memory than the other test session in comparison. The test session running the Decision Tree method having a sample rate of a 100 Hz on the budget tier VM has an average use of memory by 123MB and a maximum of 284MB, far beyond normal values. A detailed look in the snapshot the memory pattern for an unknown reason keeps increasing until around 30 minutes where it then starts to decrease. We notice similar patterns for other test session, which is clear by the results in the table. Both the results from the CPU and memory utilization brings us to believe that the problem of random high values and spikes in the load stems from the JProfiler agent rather than our implementation or the hardware configuration we use. Besides the occasionally high numbers there is no other interesting patterns to take from this. The implementation across all the methods use more memory for each VM configuration but this is because the garbage collector is activated more frequent in the budget tier VM than the top tier VM. The activation of the garbage collector is also one of the causes to the strange patterns in memory consumption. A detailed look at the snapshot of the example above in question it is clear that once the garbage collector is done the amount of used memory is back down to around 20MB. So with a more frequent garbage collector the memory pattern would remain smaller than what the results tell us. VM Configuration (Budget) KNN SVM DT MA SD Avg Min Max Avg Min Max Avg Min Max Avg Min Max Avg Min Max 1 Hz Hz Hz Table 8.26: Cross-table overview of the average, minimum and maximum memory used from the profiling sessions running the all the methods on the budget tier VM. VM Configuration (Consumer) KNN SVM DT MA SD Avg Min Max Avg Min Max Avg Min Max Avg Min Max Avg Min Max 1 Hz Hz Hz Table 8.27: Cross-table overview of the average, minimum and maximum memory used from the profiling sessions running the all the methods on the consumer tier VM. 112
123 VM Configuration (Top) KNN SVM DT MA SD Avg Min Max Avg Min Max Avg Min Max Avg Min Max Avg Min Max 1 Hz Hz Hz Table 8.28: Cross-table overview of the average, minimum and maximum memory used from the profiling sessions running the all the methods on the top tier VM. Below we present graph plots of the used memory pattern during the session on a timeline. We select only a few examples in this section, but the rest of the plotted memory graphs are listed in Appendix B for reference. In Figure 8.9 we present the memory pattern for the K-Nearest Neighbor method on the budget tier VM. The green area in the graph represents the total allocated memory and the blue outline is the memory currently in use. A strange scenario is in the test session running a sampling rate of 1 Hz. The JVM allocates over 120MB memory for the implementation but it only uses below 30MB of this at any given time during the session. The garbage collector is never once activated as the results just grows steadily in an almost straight line. In Figure 8.10 and 8.11 the pattern is more normalized in terms of allocated memory. 113
124 Figure 8.9: Memory pattern for the K-Nearest Neighbor and the three sampling rates. The graph is plotted with the results from the budget tier VM. If we compare the graphs over sampling rates in-between the various VM configurations, the test session running a sampling rate of 1 Hz produce a very similar memory pattern. The only impact is the activation of the garbage collector as shown as sudden dips in the blue outline. The JVM adapts more of the allocated memory in the test session running 250 Hz sampling rate. For some graphs with the sampling rate set to 250 Hz the memory pattern seems to be erratic and do not follow the "wave"-like pattern that is generated by the majority. This erratic pattern correlates with the same abnormalities in the CPU load pattern generated in Section There we see sudden drops and spikes in the load which can translate to the same conclusion in the memory patterns for the 250 Hz sampling rate. Because the JProfiler and the server implementation crashes when we try to run the agent during data mining initialization with more than the minimal options enabled, we are uncertain if the used memory includes the memory used by the MCR. There is no indication of that as the heap walker feature in JProfiler only show the size of the pointers to the data mining class created by the implementation and not the actual size of the model created. Using the memory function in Matlab to display the memory information is not an option as it is only supported on the Windows operating system. 114
125 Figure 8.10: Memory pattern for the K-Nearest Neighbor and the three sampling rates. The graph is plotted with the results from the budget tier VM. The consumer and top tier VM configuration use more memory than the budget tier. The increase is minimal and may be induced by the operating system because of the low performance of the budget tier VM. The operating system may reduce the allowed memory event though the total amount of physical memory is set to the same for each VM, at 4GB in total. In theory they memory footprint should be the same, but can be affected by underlying factors in the JVM and operating system that make us of more memory when the performance cap is lifted. 2 For classes and objects, looking at the heap walker in JProfiler tells us that most of our memory consisted of SensorEvent objects and Double data types, which is no surprise. 115
126 Figure 8.11: Memory pattern for the K-Nearest Neighbor and the three sampling rates. The graph is plotted with the results from the budget tier VM I/O Operations In our implementation the database write operations will only write once per second to the tick table and another write operation for the classification either every 30 or 60 seconds. This has no impact on performance on a smart phone, yet alone our VM. Ideally we would like to measure the write operations when the there is no sample rate limit set in place. We were not able to get the database probe to work with JProfiler on our implementation meaning we have no measurements of the actual I/O from the database. The JProfiler supports the JDBC adapter which we used to connect to the SQLite database, but we find the documentation on this part a bit lacking as it does not mention if it only supports remote database connections. Also there is no examples to go for on how to connect to a database so we can specify the option ourselves. But considering that small amount of data we work with and the few operations that are used, the performance of the database is not that relevant. Besides, a future application with this main goal in mind will have an optimized database schema and even compression, making our results redundant. Our database schema when initialized containing three simple tables is around 5KB in size. We ran some tests with the full records from the Apnea-ECG database. These records contain data that roughly illustrates a whole night of sleep as all the records contain at least six hour s worth of data. From all the tests we get a size increase of 600KB to the database. That includes the data aggregated from a 100 Hz down to 1 Hz stored in the tick table and the apnea classification segments in the apnea table. A future application on a smart phone will use more or less storage data depending on how the application is implemented. Storage data can be optimized by using a better schema and the data can be compressed as there is no unknown factors to what we want to store. On the other hand, the Matlab JAR file combined is 69MB in size, this is without the MCR that is needed to run these libraries. So the database storage capabilities are not in any sort a problem for this type of application. We were able to get the network through put from the test session, although our measurements 116
127 are performed on a cable network. Making a performance comparison with these results are pointless because a smart phone application would either be built around using a Bluetooth or Wi-Fi connection to transfer the raw sensor data to the application. Our comparison will largely be impacted based on the cable network and the buffer size of the network card that is installed in the hardware configuration in our tests. The network buffer in a smart phone might be considerably smaller than the ones used in a desktop motherboard. 117
128 Chapter 9 Conclusion This thesis builds upon the work of another thesis that had a focus and goal of analyzing the possibilities of using popular data mining methods to classify epochs of abnormal breathing on data from PhysioNet. The future goal of this is to create an automatic diagnostics tool to help physicians in evaluating sleep apnea patients. This thesis had a goal of implementing the work in the related thesis and to integrate it with Esper. We implemented the data mining methods and two apnea detection methods based on the EPL that is offered by Esper. As a supplementary evaluation we also profiled hardware resources and usage of the concept implementation. The performance evaluation used smart phones as a base benchmark, as the future goal is to create smart applications that automatically diagnose and measure the sleep and breathing patterns of sleep apnea patients. Our main contributions are discussed in Section 9.1. In Section 9.2 we discuss the conclusion derived out from our work and results done in this thesis. Section 9.3 contains discussions and suggestions on future work and progress towards the main goal for the thesis. 9.1 Contributions We verified, summarized and analyzed the results and built upon the goal and focus from the work done in the offline analysis. In addition, we designed and implemented detection methods using Esper processing sleep apnea data from PhysioNet, performed profiling of resource usage of the implementation with emphasis on smart phone hardware performance. Most of the related work that have tried to use data mining techniques to classify sleep apnea data have used mostly regression based methods. We went with a different approach and wanted to see if EPL queries and rule-based decision making can generate similar results. The analysis of the offline data mining methods is presented and discussed in Section In Section we discuss our contribution towards automatic real-time detection of apneas using Esper, followed by another contribution discussion where we go through our resource profiling Data Mining From the offline analysis the four data mining methods were implemented: the K-Nearest Neighbor, The Support Vector Machine, the Artificial Neural Network and the Decision Tree. For the data mining options for each method we used the best performing options for each. In the offline analysis the best performing data mining method was the K-Nearest Neighbor with an accuracy of 96.6% and their worst scoring method was the Decision Tree. Our results had 118
129 the Decision Tree scoring the best with an accuracy of 90.89% using a combination of all the signals. Most our results had an accuracy of above 85% except for three signal combinations that performed very poor. Respiratory from the chest and abdomen were contributing to the poor results as they barely were able to achieve an accuracy over 60%. Unfortunately, we had trouble getting the Artificial Neural Network functions to work with our implementation as it only supported pre-trained networks. We tried to pre-train our network with the data we used for the other methods, but still were not able to see it working. It is interesting that we were not able to achieve a higher accuracy from the methods and a score that is closer to the ones in the offline analysis. We are aware that they individual option combinations for each method helps to achieve the offline results that is, but then again we use same exact data sources and identical pre-processing scheme to transform the data. In theory the results should be closer as streaming should have no effect on the result because it is just a way of transferring the data. The amount of different layers and programming languages may affect the accuracy, but not by that much. The best results were also normalized and gave a higher accuracy and what is obtainable without normalization Apnea Detection Two detection methods have been implemented using the EPL in combination with Esper and sleep apnea data in three separate databases from PhysioNet. The goal of this was to find out if CEP can contribute to finding apneas in patients in real-time. The methods were designed with the use of statistical methods such as a Moving Average and Standard Deviation that were incorporated into the EPL queries either by using the built-in functions in Esper or creating custom standalone functions. The two methods operate on the basis of rule-based classification and have similar properties although they are different in their implementation. Our best method was the Moving Average which achieved an accuracy of 93.26% on average using the records from the Apnea-ECG database. The Standard Deviation method was not far behind, achieving an accuracy of 93.13% using the same records. The options we used in for the two methods across all the tests had minimal impact on the data. There were some instances where one option would be better than the other, but for the most part the options only skewed the accuracy results for the methods marginally. Our AHI evaluation performed good on the Apnea-ECG database but had serious trouble in identifying the correct AHI index for the records from the MIT-BIH Polysomnography and St. Vincent s database. This could be concerning in terms of using these methods further to evaluate sleep apnea epochs, but for the same reason given in the offline analysis the data quality among the records from these databases is subpar compared to the Apnea-ECG, and we come to the same conclusion with our results. Even though the databases are fit for data mining and contain sleep apnea data, the general quality makes it hard to justify their use in these evaluations. Besides the bad data quality, our two detection methods were able to achieve an average accuracy of 66.81% over all the records from the MIT-BIH Polysomnography database and an average accuracy of 70,97% for the records in the St. Vincent s database. We achieved great results using the methods, but they still lack optimization and seem overly complex as creating efficient EPL queries takes time to master. We believe that with optimization and addition of more methods using EPL queries combined with a greater pool of sleep apnea data with good data quality we can achieve even greater results. 119
130 9.1.3 Resource Profiling One of our goals was to measure the resource usage of our components and our implementation with a comparison towards smart phone hardware in order to get an indication on how well such a future solution would perform. In the offline analysis they mentioned that a sampling rate of 1 Hz is enough to be able to classify the sleep apnea epochs. As the apnea often occurs over several seconds up to over a minute, a very high sampling rate will not benefit the classification. We derived a list of suitable smart phones that we classified as either in a top, consumer or budget tier based on the hardware specifications, popularity and price. A collection of benchmark scores for the smart phones was collected in order to get a baseline for our VMs. We configured our VMs to be capped at a certain threshold that would produce similar benchmark results. To measure the resource usage of our implementation we used a licensed profiling tool called JProfiler. With JProfiler we attached an agent service to our server implementation in offline mode and store the measurements in snapshot which we later analyzed. The performance evaluation results show us that all the data mining and apnea detection methods are suitable within a certain sampling rate. At most the sampling rate should be at a maximum of 100 Hz. When testing sampling rates of 250 Hz either the server implementation or JProfiler started to generate unexplained fluctuations in both memory and CPU load. We are not sure what is the cause of this but it might be bound to the performance cap and the amount of data that is queued in the network buffer. 9.2 Discussion With the results from our three data mining methods we conclude that they are all suitable for online classification of sleep apnea data. We scored them based on three metrics: accuracy, sensitivity and specificity. Our test results are not as good as with the offline analysis, but that might be due to design decision in our thesis and that our implementation. Further, our implementation is not optimized seeing as much of the time spent was on doing research and understanding the basics regarding Matlab, Esper and JProfiler. Even though we did not achieve the same good results from the data mining methods we think that an accuracy of between 85-90% is very good. The offline analysis concluded that the data quality in the Apnea-ECG database was far better than the rest of the databases. Analyzing our results, both from the data mining and the apnea detection, we came to the same conclusion. The reason for this is still hard to explain but might be because of the difference in measurement equipment and especially for the St. Vincent s database, the sampling rate of 8 Hz. Three signal combinations from our analysis achieved bad results. The signals in question was respiratory from the chest and abdomen, and a combination of both. These results correlates with the results in the offline analysis as well. The signals are non-invasive but suffer from inaccurate reads when a person moves or when the band is not correctly attached. On the bright side, almost all of our results that achieved good results were with a signal combination that either consisted of respiratory from the nose or oxygen saturation. Meaning, either of these can potentially be used in a future application with good performance. With our two apnea detection methods we were able to achieve ad good accuracy score of mostly above 90%. The results can be a bit misleading as we only did the tests using subject classification. With these methods there is no need for training data as all the decision making is bound to rules that conforms to the criteria of the characteristics of an apnea event. It did not 120
131 make sense to use any cross-validation or hold-out method because of this. This might indicate that the results are very bound to the record that is tested and we wished we had more records of the same data quality like with the Apnea-ECG records. The results were best achieved using the records from the Apnea-ECG database, like with the other results we produced. We produce the results using just the SpO2 signal. With optimization and a multi-signal approach to the design and implementation we believe that a solution based on CEP can achieve even better results without training data and with more available options. Although both methods are a bit similar in how they are implemented we expected the difference in design to give us a more varied result overall, but they both generated around the same accuracy within a degree of 1-2%. The options we used also had minimal impact on the result. This is a cause of the small amount of variance in the values we chose and for future reference we could reduce the amount of options and instead increased the distance in values. We also added a secondary supplementary test to our apnea detection methods to tell how accurate they were to find the number of apneas and determine an AHI index for the records tested. A huge problem here was the time and effort it took to manually annotate the six records we used. We are by no means experience in this process and the results will not reflect an accurate measurement done by a trained physician. The methods were surprisingly accurate when determining the number of apneas and the AHI index for the Apnea-ECG database but had trouble with the other two databases. This is probably related to the issues we have mentioned earlier with the difference in data quality. As for the resource analysis we had questionable results because of a strange problem affecting our measurements. Even if the resource usage had no errors the general contribution from this analysis is uncertain. Our implementation is only to be considered a skeleton concept application and in no way can 100% accurately be compared to a more finished application running on smart phones. So how much these numbers and measurements means is up for discussion. However, we do think that the measurements we gathered indicates that a smart phone, even if it is classified as a budget one, can indeed run an automatic diagnosis application without any problems. This is of course based on the sampling rate as we presented with our results a sampling rate of 250 Hz will most likely not positively impact the results and can cause artifacts and performance issues. Running at a sampling rate of 250 Hz also requires a lot more memory to retain all the incoming events before it can be aggregated down to a more reasonable sampling size. To summarize, this thesis helped to prove the suitability of automatic sleep apnea classification using both regular regression methods and with a simpler rule-based approach using Esper on similar hardware that is comparable to smart phones. Some of our areas need further research and contributions in order to fully see the potential in developing a smart phone application with the features mentioned in this thesis. 9.3 Future Work In this section we will discuss possibilities for future work from the results we produced in this thesis as well as what we learned from them. The results from the data mining methods are good enough to justify them being used in a smart phone application, but it can be hard to implement them correctly. The methods cannot classify the data in real-time but need to batch the incoming data in order to classify. The problem is trying to figure out how much data needs to be batched together in order to get good results and also being effective as a 121
132 monitor application. An application that only relies on diagnosis of the nightly session can use regular regression methods such as the Support Vector Machine to batch a minute of incoming data and classify it as either a period of normal or abnormal breathing. If we want to have the incoming data to trigger alarms, find the specific periods in a minute where the apnea started and ended we cannot only use regular regression methods to handle this. We can either combine multiple data mining methods at a cost complexity and resource usage. With the results from our apnea detection methods using Esper we have proven the suitability of using EPL queries and CEP to classify epochs of abnormal breathing. The methods also have the ability to trigger many other actions depending on the design and choices, as well as giving the user a more accurate real-time analysis of their recorded data. A realization is that the data we use is not enough to correctly conclude that the methods are enough. We identify two main problems and improvements that can be used to further improve the research to achieve our main goal of creating a real-time diagnosis application running on smart phones Sensor Data How our data is produced and which tools have been used is still an unknown variable when it comes to the data from PhysioNet. We know that the data in the Apnea-ECG database have not been pre-processed and it still offers the best data quality out of the three databases we use. The problem with Apnea-ECG is not the data quality, but rather the low number of records to choose from. This is the same issue for the MIT-BIH Polysomnography database because of the inconsistency in signal combinations among the records. Records from this database also contains artifacts such as strange spikes and abnormal values from time to time. The St. Vincent s database contains a good selection of records, but lacks the overall data quality. This brings us to believe that with a proper collection of data either produced by a real sensor within our control or from a reliable local source such as a hospital can improve our methods further. The data quality and quantity is important and below is a list of criteria for the data and records: There must be two separate sets of the records: one that is pre-processed and one that is not pre-processed. For future work we want to test our methods and implementation in how it can handle data that contains noise and artifacts. If the sets contain artifacts such as values going beyond or below their respective measurement scales, we need the sections to be annotated so that we can improve and explain the problems rather than guessing. This means that if a sensor falls of while sleeping and is re-attached, there must be information about it happening. All records must contain the four non-invasive signals tested in this thesis. The overall variation of subjects must span across all the severity groups derived from the AHI index. The severity groups are: mild, moderate and severe, as well as subjects having very little to no apneas. With these criteria we hope to generate sleep apnea data that can be used to more accurately portray sleep apnea patients so that more in-depth and profound classification and testing can be achieved. 122
133 9.3.2 Detection Optimization Even though we were able to produce good results using EPL queries to distinguish normal breathing from abnormal breathing, these type of methods can be further improved and built upon. In this thesis we only had time to design, implement and test a solution using one signal. We decided to go with the most non-invasive signal which is the oxygen saturation. A more robust solution would be to incorporate the detection using multiple signals. From our results the respiratory measurements from the chest and abdomen were not so good and can most likely be discarded unless they can be improved. The two remaining signals are the respiratory from the nose and oxygen saturation which produced very good results from both the data mining and the detection methods. A combination of these two can be used to further improve the detection of apnea segments in a subject. One stream with the oxygen saturation can be simultaneously compared to another stream with respiratory data from the nose. Different methods can be tailored to the specific signal stream in order to optimize the classification of apnea segments. If the system detects an apnea in the stream containing respiratory airflow, it can also validate the segment by having a method detecting apneas in the stream containing oxygen saturation. The values and patterns generated by each signal is different and there is a need to design either different methods for each, or a method that dynamically can handle more signal patterns. Another improvement is to optimize and build better methods using EPL queries. Learning how to use Esper and EPL is one thing, how to use it efficiently is another. With such a large scope and an in-depth documentation, we have reason to believe that there is more than one way to detect anomalies such as apneas in a stream. Optimizing the queries or even redesign some of them to be less resource intensive and to filter out unwanted artifacts would be achievable. 123
134 Bibliography [1] AASM. Clinical Practice Guideline for the Treatment of Obstructive Sleep Apnea and Snoring with Oral Appliance Therapy. In: (2015). URL: http : / / www. aasmnet. org / Resources/clinicalguidelines/Oral_appliance-OSA.pdf. [2] An Unexpected Benefit of Treating OSA: Increased Libido. [Online; accessed 12-September- 2016]. URL: [3] ApneaLink Plus. [Online; accessed 09-January-2016]. URL: healthcare-professional/products/diagnostics/apnealink-plus.html. [4] Association Rules (Market Basket Analysis). [Online; accessed 16-September-2016]. URL: [5] American Sleep Apnea Association. Clinical Guideline for the Evaluation, Management and Long-term Care. In: (). [Online; accessed 18-September-2016]. URL: http : / / www. aasmnet.org/resources/clinicalguidelines/osa_adults.pdf. [6] Automobile Accidents in Patients with Sleep Apnea Syndrome. In: (26th Oct. 2015). URL: [7] Shivnath Babu and Jennifer Widom. Continuous Queries over Data Streams. In: (). [Online; accessed 18-September-2016]. URL: [8] Backpropagation. [Online; accessed 22-September-2016]. URL: en. wikipedia. org/ wiki/backpropagation. [9] Bessel s correction. [Online; accessed 22-September-2016]. URL: com/besselscorrection.html. [10] Big Data. [Online; accessed 16-September-2016]. URL: data. [11] Biological Neural Network. [Online; accessed 16-September-2016]. URL: org/wiki/biological_neural_network. [12] Koley BL and Dey D. On-Line Detection of Apnea/Hypopnea Events Using SpO2 Signal: A Rule-Based Approach Employing Binary Classifier Models. In: (2014). [13] CareFusion NOX-T3. [Online; accessed 09-January-2016]. URL: http : / / www. carefusion. com/our-products/respiratory-care/sleep-diagnostics-and-therapy/nox-t3-portable. [14] Mayo Clinic. Hypoxemia (low blood oxygen). [Online; accessed 11-September-2016]. URL: [15] Complex event processing. [Online; accessed 11-September-2016]. URL: org/wiki/complex_event_processing. 124
135 [16] Sandford Weisberg Cook D.R. Applied Regression Including Computing and Graphics. In: (1999). [17] India Department of IT - MIT - Anna University of Chennai. Complex Event Processing for Object Tracking and Intrusion Detection in Wireless Sensor Networks. In: (2010). [18] . Personal sent to Prof. T Penzel, responsible for the collection of data. [19] Esper Introduction. [Online; accessed 16-September-2016]. URL: http : / / www. espertech. com/products/esper.php. [20] EsperIO Documentation. [Online; accessed 16-September-2016]. URL: http : / / www. espertech.com/esper/release-5.2.0/esperio-reference/html_single/index.html. [21] EsperTech. EsperTech Documentation: Performance. [Online; accessed 25-Januar-2016]. URL: [22] EsperTech. EsperTech - Esper. [Online; accessed 21-January-2016]. URL: http : / / ww. espertech.com/esper. [23] EsperTech. EsperTech - Esper. [Online; accessed 21-January-2016]. URL: http : / / www. espertech.com/esper/performance.php. [24] Massimo Ficco and Luigi Romano. A Generic Intrusion Detection and Diagnoser System Based on Complex Event Processing. In: (2011). [25] Figure: Cluster Analysis. URL: [26] Figure: Decision Tree. URL: https : / / www. siggraph. org / education / materials / HyperVis / applicat/data_mining/data_mining.html. [27] Figure: Esper Component Overview. URL: [28] Figure: Esper Window Streaming. URL: [29] Figure: K-Nearest Neighbor. URL: nearest_neighbors_ algorithm. [30] Figure: Multi-layer Perceptron. URL: https : / / www. siggraph. org / education / materials / HyperVis/applicat/data_mining/data_mining.htm:// [31] Figure: Obstructive Sleep Apnea. URL: care/serviceareas/neurology/diseases-and-conditions/sleep-apnea. [32] Figure: Polysomnography. URL: [33] Figure: Single-layer Perceptron. URL: https : / / www. siggraph. org / education / materials / HyperVis/applicat/data_mining/data_mining.html. [34] Figure: Support Vector Machine. URL: algorith://en.wikipedia.org/wiki/support_vector_machine. [35] Figure: SVM Non-linear transformation. URL: htm. [36] Figure: SVM Soft-margin. URL: [37] Geekbench Browser. [Online; accessed 25-September-2016]. URL: https : / / browser. primatelabs.com/. [38] Global surveillance, prevention and control of chronic respiratory diseases. In: (2015). URL: 125
136 [39] GUi run properly in matlab file but when converted to exe file doesn t work. [Online; accessed 17-October-2016]. URL: [40] Nguyen HD et al. An Online Sleep Apnea Detection Method Based on Recurrence Quantification Analysis. In: (2014). [41] How to configure matlab toolbox when I use c-sharp both? [Online; accessed 17-October-2016]. URL: [42] Mari Sønsteby Hugaas. Data Mining for the Detection of Disrubted Breathing Caused by Sleep Apnea - A Comparison of Methods [43] Interpreting Geekbench 3 Scores. [Online; accessed 22-September-2016]. URL: http : / / support.primatelabs.com/kb/geekbench/interpreting-geekbench-3-scores. [44] Paalasmaa J1 et al. Unobtrusive Online Monitoring of Sleep at Home. In: (2012). [45] JABFM. Feasibility of Portable Sleep Monitors to Detect Obstructive Sleep Apnea (OSA) in a Vulnerable Urban Population. In: (Apr. 2015). [46] JProfiler Documentation. [Online; accessed 14-October-2016]. URL: http : / / resources. ej - technologies.com/jprofiler/help/doc/. [47] JProfiler Overview. [Online; accessed 14-October-2016]. URL: com/products/jprofiler/overview.html. [48] B. Koley and D. Dey. Adaptive Classification System for Real-Time Detection of Apnea and Hypopnea Events. In: (2013). [49] B. Koley and D. Dey. On-Line Detection of Apnea/Hypopnea Events Using SpO2 Signal: A Rule-Based Approach Employing Binary Classifier Models. In: (2014). [50] Karimi M et al. Sleep apnea-related risk of motor vehicle accidents is reduced by continuous positive airway pressure: Swedish Traffic Accident Registry data. In: (2015). URL: [51] Kris Maher. The New Face of Sleep. [Online; accessed 10-January-2016]. URL: wsj.com/articles/sb [52] Oded Maimon and Lior Rokach. Data Mining and Knowledge Discovery Handbook, pp. 1 19, , , [53] Arun Mathew. Benchmarking of Complex Event Processing Engine - Esper. In: (). [54] Walter T. McNicholas. Proceedings of the American Thoracic Society, pp [55] Erdi Ölmezoğulları and Ismail Ari. Online Association Rule Mining over Fast Data. In: (2013). [56] Online Etymology Dictionary. In: (26th Oct. 2015). URL: browse/apnea. [57] OSA Symptoms. [Online; accessed 26-July-2016]. URL: [58] Polysomnography. [Online; accessed 12-September-2016]. URL: tests-procedures/polysomnography/basics/what-you-can-expect/prc [59] Portalbe Monitors OK for Spotting Sleep Apnea: Doc. [Online; accessed 12-September-2016]. URL: http : / / www. webmd. com / sleep - disorders / sleep - apnea / news / / portable - monitors-ok-for-spotting-sleep-apnea-new-guidelines#1. 126
137 [60] Problem matlab compiler to compile neural network toolbox function. [Online; accessed 17- October-2016]. URL: [61] The Akershus Sleep Apnea Project. A Norwegian population-based study on the risk and prevalence of obstructive sleep apnea. In: (2011). [62] Punjabi. Proceedings of the American Thoracic Society. 2008, pp [63] Rasch and Born. About Sleep s Role in Memory. 2013, pp [64] Department of Cardia - Italy Respiratory Pathophysiology Division. Recent advances in the diagnosis and management of obstructive sleep apnoea. In: (). URL: ncbi.nlm.nih.gov/pubmed/ [65] Harvard Medical School. The Price of Fatigue: The surprising economic costs of unmanaged sleep apnea. In: (1st Dec. 2010). [66] Sleep Apnea - Treatment Overview. [Online; accessed 12-September-2016]. URL: http : / / [67] Sleep Questionnaires. [Online; accessed 07-January-2016]. URL: content.cfm?article=26. [68] Snorking og søvnapné. In: (26th Oct. 2015). URL: http : / / www. helse - bergen. no / no / OmOss/Avdelinger/sovno/sovn-og-sovnsykdommer/Sider/sovnapne.aspx. [69] Jarle Søberg, Vera Goebel and Thomas Plagemann. Deviation Detection in Automated Home Care using CommonSens. In: (). [70] SQLite. SQLite Page: About. [Online; accessed 10-March-2016]. URL: org/about.html. [71] Stig Støa, Morten Lindeberg and Vera Goebel. Online Analysis of Myocardial Ischemia From Medical Sensor Data Streams with Esper. In: (). [72] Viktor Hanak Timothy I. Morgenthaler Vadim Kagramanov and Paul A. Complex Sleep Apnea Syndrome: Is It a Unique Clinical Syndrome? In: (2009). URL: http : / / www. journalsleep.org/viewabstract.aspx?pid= [73] Anthony K. H. Tung. Rule-based Classification. In: (), pp [74] Padhraic Smyth Usama Fayyad Gregory Piatetsky-Shapiro. From Data Mining to Knowledge Discovery in Databases. In: (1996). [75] Kaijun Wang. Asynchronous Standard Deviation Method for Fault Detection. In: (). [76] WatchPAT. [Online; accessed 09-January-2016]. URL: medical.com/ WatchPAT%C3%A2%C2%84%C2%A2. [77] WFDB Application Documentation. [Online; accessed 22-September-2016]. URL: physionet.org/physiotools/wag/wag.htm. [78] MOHAMMED J. ZAKI and WAGNER MEIRA JR. DATA MINING AND ANALYSIS - Fundamental Concepts and Algorithms, pp. 1 25, 33, 63, , , , ,
138 Appendices 128
139 Appendix A Configuration & Runtime Setup This appendix is a general manual for configuration and setup of the environment. As the implementation relies on different environments and programming languages we try to list the dependencies that needs to be installed on the targeted computer. In Section A.1 we list libraries that are used for the Java implementation and the Python scripts. In Section A.2 we present a guide on how to compile the offline data mining methods into a JAR file, while in Section A.3 we present how to obtain the three databases used in this thesis. How to run the pre-processing and validation scripts is presented in Section A.4 and A.5. A guide on how to run the server and connect the client is presented in Section A.6. The Section A.7 describes a way to connect the JProfiler agent to the JVM and record various resource usages both with a script in offline mode and through the GUI. The entire implementation and the scripts are available in the Git repository: uio.no/steffeli/master-implementation. A.1 Dependencies We have included a list of the libraries that are used in the scripts and implementations. Older or newer versions of these libraries might work, but we have not tested anything else than the ones mentioned. A.1.1 Python For the preprocessing scripts and validation scripts we use Python 3.5.1, but every 3.x version of Python is sufficient. The following libraries are used in the scripts: A.1.2 numpy scipy Java The implementation uses multiple libraries to handle certain features such as streaming, arguments and database storage. The libraries are included in the lib folder in the implementation repository but can also be downloaded using Maven. The following libraries are used for the server and client implementations: antlr4-runtime
140 args4j 2.33 cglib-nodep commons-lang 3.4 commons-logging 1.2 commons-math esper log4j opencsv 3.7 abego-treelayout sqlite-jdbc guava 19.0 A.2 Matlab We use the Matlab version R2016a which was downloaded from the University of Oslo s own application repository. The version uses an academic license, but Matlab can also be bought and downloaded from MathWork s web page. We compiled and tested the data mining methods on both Mac OSX (El Capitan ) and Linux on the computers at Department of Informatics at University of Oslo. The included Matlab scripts in the scripts folder of the implementation repository need to be copied or moved to the current working directory assigned by Matlab. Under the "Apps" tab in the top there is a button to compile libraries. In the pop-up window enter the name "matlab_mining" and add the four Matlab scripts to the project clicking the plus icon. A picture of the library compilation window is presented in Figure A.1. In Figure A.1: Window of the Matlab compiler options screen. Section A.2.1 three environmental variables must be set for the compiler and runtime to work. The paths might also be set directly in the Matlab console if there are any problems compiling 130
141 the JAR files. If the compilation is successful, the compiled JAR file must be moved to the lib folder in the server implementation. The compiled JAR file is platform specific and will not work on any other platform other than the one it was compiled on. Compilation in Matlab without the GUI can be done by using the following command and have been tested on machines running at the Department of Informatics: mcc -v -W java:matlab_datamining classknn:matlab_knn.m classsvm:matlab_svm.m classann:matlab_ann.m classdt:matlab_dt.m A.2.1 Paths MCR_ROOT LD_LIBRARY_PATH MATLAB_JAVA [mcr-root]/[version]/runtime/glxna64/ [mcr-root]/[version]/runtime/glxna64/: [mcr-root]/[version]/bin/glxna64/: [mcr-root]/[version]/sys/os/glxna64/: [mcr-root]/[version]/sys/opengl/lib/glxna64/ test For the Mac OSX, the LD_LIBRARY_PATH equivalent is named DYLD_LIBRARY_PATH. The last part of the path also needs to be changed to maci64 as glxna64 is for Linux. A.2.2 Runtime For the implementation to be able to run the included data mining methods in the JAR file, Matlab Runtime needs to be installed on the machine. The runtime compiler is available for download at MathWork s web page for all platforms in 32-bit and 64-bit versions. A.3 PhysioNet Databases Each database used in this thesis can be downloaded on PhysioNet s web page. The records can be manually downloaded one by one or the entire record collection can be downloaded by using the wget command in the terminal. To download the Apnea-ECG database issue the command: wget -r -np The other databases can be downloaded by substituting "apnea-ecg" in the URL with "slpdb" or "ucddb". The records reside in a corresponding folder inside a parent folder named " The pre-processing and validation scripts assumes the records resides in the same folder. For the Apnea-ECG database the files with extensions.dat and.apn needs to be moved to the scripts folder in the implementation. For the MIT-BIH Polysomnography database the same goes for files having.dat and.st extensions. St. Vincent s have the.rec extension for the data files and a "_respevt.txt" postfix for the annotation files. The pre-processing and validation scripts make use of functions in the WFDB Toolbox from PhysioNet. This toolbox needs to be downloaded and installed for the specific platform from PhysioNet s web page. 131
142 A.4 Pre-processing Scripts The three pre-processing scripts are named preprocess_apnea_ecg.py, preprocess_mit_bih.py and preprocess_st_vincents.py. The scripts are coded in Python 3.5 and can be run by issuing python3 [script]. These scripts need to reside either in the folder where the PhysioNet records resides, or move the correct record files into the same scripts folder of the implementation. The preprocess_apnea_ecg.py script takes one argument. Given the n argument the script produces normalized folded CSV files instead of the regular folded CSV files. A.5 Validation Scripts Similar with the pre-processing scripts, the validation scripts automate the validation process of the data mining methods. The validate_dm_apnea_ecg.py script iterates over all the different combinations of data mining methods, records and signals. For each iteration, the method run_evaluations() starts one sub-process for the server and one for the client. Once a connection is established between the two, the test starts running. All the results will be stored in the database until the client disconnects. The method generate_results() then retrieves the results from the SQLite database and the annotations for the specific record tested. The two data arrays are transformed to be aligned correctly, and then use to calculate accuracy, sensitivity and specificity. All the calculations and other various information of the test is written to a CSV file using the method write_results(). Before a new iteration starts, the script deletes the database so that no other data is present once an iteration starts. The scripts: validate_ap_apnea_ecg.py, validate_ap_apnea_ecg.py and validate_ap_apnea_ecg.py do exactly the same as the validation script above, but these focus on validating the apnea detection methods for the individual databases instead. All the scripts can be executed in the scripts folder as is, as long as the pre-processing script have been executed and generated the needed record files in the same folder. They can be executed with the following command: python3 [script]. A.6 Server & Client The server implementation is Java based. Because of the MCR, compiling the server implementation with Java version 1.8 do not work. We have tried both version 1.6 and 1.7 which worked. The implementation can be compiled using the following command: javac - cp "lib/*:[javabuilder_path]" *.java. The classpath argument includes all the libraries in the lib folder, while the javabuilder is a JAR library that comes with the MCR. The javabuilder is located at [matlab_root]/toolbox/javabuilder/jar/javabuilder.jar. The root folder can reside at different locations depending on the operating system or if it is installed at a custom location. The server supports a total of six input arguments: -d ( data-mining), -p ( port), -t ( trainingset), -i ( input-rate), -s ( signals) and -a ( apneadetection). The port and input rate is required, the rest is optional to some degree of combinations. To execute the server implementation, the following command is used: java -cp ".:lib/*:[javabuilder_path]" Server [arguments...]. To make a bit easier a shell script is included with the name run.sh. The script determines the javabuilder path, compiles the implementation and runs the implementation with the given 132
143 input arguments. To use the script, simply run the script with the input arguments that is required for the implementation:./run.sh [arguments..]. The javabuilder paths may need to be changed in the script depending on the operating system and the location of the root folder. For the client, compilation and execution is exactly the same as with the server, except the differences in input arguments. The input arguments are: -p ( port), -h ( hostname), -f ( file), -o ( output-rate) and -e ( esper-rate). The port and the hostname is needed to be able to connect the client to the server, the port needs to be the same on the server obviously. The file is the record file, while the output-rate is the amount of objects sent per second to the server. The esper-rate is the amount of events that Esper aggregates before it is sent as one object to the server. For example, if we want to test sending records from the Apnea-ECG database and down sample the sampling rate to 1 Hz instead of the native 100 Hz, the value of output-rate needs to set to 1 and esper-rate set to 100. On the server side, the input-rate needs to be also set to 1, as we assume we will receive 1 event per second from the client. The client also has a helper script named run.sh Once the client and server are connected, the sessions starts automatically sending events and classifying the segments. A.7 JProfiler JProfiler is a profiling tool developed by EJ Technologies for the Java Platform. We use the version The tool has both a GUI interface and various smaller CLI tools that can be run in offline mode using a configuration file. The GUI interface is intuitive and easy to figure out. Once JProfiler is started, a starting prompt will pop up with a set of options to choose from. A picture of the start prompt is presented in Figure A.2. There are three general options to choose from. First, an existing profiling session can be opened or one can create a new session. A session consists of a huge number of options to choose from such as basic applications settings and JVM information, triggers, filters, and various probes and database adapters. Filters is a feature to filter out the information flow when the tool monitors objects, methods or variables in the code and their realtion to each other, combined with the load on CPU and memory. Triggers are actions one can set to be performed at certain events. For example, we can create a custom logging script that we want JProfiler to trigger if the CPU load of the JVM goes above a certain threshold. Another option is called "Quick attach", which basically attached the JProfiler agent to an already running JVM. A list is displayed with all of the running JVMs on the local machine. There is also an option to quick attach an agent to a JVM running on another machine, such as a server, using a hostname and port to connect. All the information that is analyzed by the tool can be stored in snapshots. These snapshots can be opened which will load all the analyzed data into graphs, lists and views in the GUI interface. Storing a snapshot of the analysis is great as we can tailor a script to run the profiling tool in offline mode, which can run for several hours without manually needing to monitor the analysis via a GUI interface. The snapshot can then be opened up at any time to evaluate the performance or to find bottlenecks in the implementation. Another option available is to compare several snapshots which can help developers find and correct bottlenecks and bugs in their applications using different versions. We will not explain how to set up the offline variant, but rather explain how to attach the agent via the GUI interface to an already running JVM. A step-by-step guide is listed below: 133
144 Figure A.2: Starting prompt upon running JProfiler. 1. Execute the server implementation by either running the command: java -cp ".:lib/*" Server [arguments...] or./run [arguments...]. Running the first command requires the server implementation to be compiled first using the command presented in Section A Once the server is up and running, start JProfiler and wait for it to load. 3. In the start center window, click the "Quick attach" tab in the top. A list of available JVMs on the local machine is displayed. A picture of the list is presented in A.4, here we see a running JVM without server implementation. 4. Pressing the start button attached the agent to the server JVM. After a short period, an overview display of different graphs will start to be active and accumulate data. 5. To get data form a running session, execute the client and establish a connection by using the command: java -cp ".:lib/*" Client [arguments...] or./run [arguments...]. Once both the implementation and JProfiler is up and running, a menu to the far right is available. Some options in the icons may not be available because it depends on the features selected at start up. Clicking through the icons displays different resource monitoring views such as graphs for the CPU, memory, garbage collection and system threads. There are also extensive lists and flow charts of the objects generated and methods used. In the offline mode, we use a script with a pre-defined configuration to attach the agent to the JVM. The configuration contains some triggers that activates at certain events. For us to run the session for 70 minutes, and then save the data to a snapshot we set up a trigger that saves a snapshot when 70 minutes have passed. This can also be set in the trigger tab referenced in Figure A.3. Some options impact the overhead when profiling. More information about the features and options can be found on the EJ Technologies web page [47]. The documentation for the profiling tool and also be found on their web page [46]. 134
145 Figure A.3: Profiling setup tab, showing the overhead bar indicator. Figure A.4: List of running JVMs in JProfiler. 135
Obstructive Sleep Apnea
 Obstructive Sleep Apnea Introduction Obstructive sleep apnea is an interruption in breathing during sleep. It is caused by throat and tongue muscles collapsing and relaxing. This blocks, or obstructs,
Obstructive Sleep Apnea Introduction Obstructive sleep apnea is an interruption in breathing during sleep. It is caused by throat and tongue muscles collapsing and relaxing. This blocks, or obstructs,
OSA - Obstructive sleep apnoea What you need to know if you think you might have OSA
 OSA - Obstructive sleep apnoea What you need to know if you think you might have OSA Obstructive sleep apnoea, or OSA, is a breathing problem that happens when you sleep. It can affect anyone men, women
OSA - Obstructive sleep apnoea What you need to know if you think you might have OSA Obstructive sleep apnoea, or OSA, is a breathing problem that happens when you sleep. It can affect anyone men, women
SNORING AND OBSTRUCTIVE SLEEP APNOEA WAYS TO DEAL WITH THESE PROBLEMS
 SNORING AND OBSTRUCTIVE SLEEP APNOEA WAYS TO DEAL WITH THESE PROBLEMS Laugh and the world laughs with you; snore and you sleep alone. These words by novelist Anthony Vergess ring true with all too many
SNORING AND OBSTRUCTIVE SLEEP APNOEA WAYS TO DEAL WITH THESE PROBLEMS Laugh and the world laughs with you; snore and you sleep alone. These words by novelist Anthony Vergess ring true with all too many
Data mining for Obstructive Sleep Apnea Detection. 18 October 2017 Konstantinos Nikolaidis
 Data mining for Obstructive Sleep Apnea Detection 18 October 2017 Konstantinos Nikolaidis Introduction: What is Obstructive Sleep Apnea? Obstructive Sleep Apnea (OSA) is a relatively common sleep disorder
Data mining for Obstructive Sleep Apnea Detection 18 October 2017 Konstantinos Nikolaidis Introduction: What is Obstructive Sleep Apnea? Obstructive Sleep Apnea (OSA) is a relatively common sleep disorder
Alaska Sleep Education Center
 Alaska Sleep Education Center The 3 Types of Sleep Apnea Explained: Obstructive, Central, & Mixed Posted by Kevin Phillips on Jan 28, 2015 6:53:00 PM Sleep apnea is a very common sleep disorder, affecting
Alaska Sleep Education Center The 3 Types of Sleep Apnea Explained: Obstructive, Central, & Mixed Posted by Kevin Phillips on Jan 28, 2015 6:53:00 PM Sleep apnea is a very common sleep disorder, affecting
Sleep Apnea. What is sleep apnea? How does it occur? What are the symptoms?
 What is sleep apnea? Sleep Apnea Sleep apnea is a serious sleep problem. If you have it, you stop breathing for more than 10 seconds at a time many times while you sleep. Another term for this problem
What is sleep apnea? Sleep Apnea Sleep apnea is a serious sleep problem. If you have it, you stop breathing for more than 10 seconds at a time many times while you sleep. Another term for this problem
Sleep Apnoea. Introduction Symptoms Causes Obtaining a Diagnosis Treatment Complications
 The most common forms of sleep apnoea are normally referred to as obstructive sleep apnoea (OSA) and relate to a condition which causes interruptions in breathing during sleep. People with obstructive
The most common forms of sleep apnoea are normally referred to as obstructive sleep apnoea (OSA) and relate to a condition which causes interruptions in breathing during sleep. People with obstructive
WHAT YOU NEED TO KNOW ABOUT SLEEP APNEA
 WHAT YOU NEED TO KNOW ABOUT SLEEP APNEA Wayne Driscoll Clinical Education Specialist 2 SLEEP APNEA IN THE NEWS Carrie Fisher died from sleep apnea, other factors, coroner says USA Today NJ Transit engineer
WHAT YOU NEED TO KNOW ABOUT SLEEP APNEA Wayne Driscoll Clinical Education Specialist 2 SLEEP APNEA IN THE NEWS Carrie Fisher died from sleep apnea, other factors, coroner says USA Today NJ Transit engineer
Obstructive sleep apnea
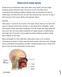 Page1 Obstructive sleep apnea People who have obstructive sleep apnea often snore heavily and have longer breathing pauses while they sleep. Snoring on its own is harmless. But in combination with breathing
Page1 Obstructive sleep apnea People who have obstructive sleep apnea often snore heavily and have longer breathing pauses while they sleep. Snoring on its own is harmless. But in combination with breathing
in China Shanghai Office Beijing Office (+86) (+86)
 SLEEP Apnea in China Guide 2018-2019 Shanghai Office (+86) 21 2426 6400 Beijing Office (+86) 010 6464 0611 www.pacificprime.cn Follow us on WeChat t A comprehensive overview of sleep apnea Perhaps you
SLEEP Apnea in China Guide 2018-2019 Shanghai Office (+86) 21 2426 6400 Beijing Office (+86) 010 6464 0611 www.pacificprime.cn Follow us on WeChat t A comprehensive overview of sleep apnea Perhaps you
A friend of mine, a 69-year-old pulmonologist in Port Arthur, Texas, who had OSA, died of a heart attack during sleep on July 13, 2015.
 SINCE U.S. Associate Justice Antonin Scalia, 79, a guest at the Cibolo Creek Ranch, a 30,000-acre luxury resort in West Texas, was found dead in his bedroom, speculation started that he could have died
SINCE U.S. Associate Justice Antonin Scalia, 79, a guest at the Cibolo Creek Ranch, a 30,000-acre luxury resort in West Texas, was found dead in his bedroom, speculation started that he could have died
Sleep apnea. What is sleep apnea? What are the symptoms of sleep apnea? Symptoms during the day. Symptoms during sleep
 Sleep apnea Your doctor thinks you may have sleep apnea. The purpose of this fact sheet is to inform you about this disease. It also gives you details about the tests done to diagnose it and possible treatments.
Sleep apnea Your doctor thinks you may have sleep apnea. The purpose of this fact sheet is to inform you about this disease. It also gives you details about the tests done to diagnose it and possible treatments.
Polysomnography (PSG) (Sleep Studies), Sleep Center
 Policy Number: 1036 Policy History Approve Date: 07/09/2015 Effective Date: 07/09/2015 Preauthorization All Plans Benefit plans vary in coverage and some plans may not provide coverage for certain service(s)
Policy Number: 1036 Policy History Approve Date: 07/09/2015 Effective Date: 07/09/2015 Preauthorization All Plans Benefit plans vary in coverage and some plans may not provide coverage for certain service(s)
Continuous Positive Airway Pressure (CPAP)
 Patient Information Continuous Positive Airway Pressure (CPAP) Author: Pulmonary physiology, sleep and ventilation Produced and designed by the Communications Team Issue date July 2017 Expiry date July
Patient Information Continuous Positive Airway Pressure (CPAP) Author: Pulmonary physiology, sleep and ventilation Produced and designed by the Communications Team Issue date July 2017 Expiry date July
OSA in children. About this information. What is obstructive sleep apnoea (OSA)?
 About this information This information explains all about sleep-related breathing problems in children, focusing on the condition obstructive sleep apnoea (OSA). It tells you what the risk factors are
About this information This information explains all about sleep-related breathing problems in children, focusing on the condition obstructive sleep apnoea (OSA). It tells you what the risk factors are
Development of OSA Event Detection Using Threshold Based Automatic Classification
 Development of OSA Event Detection Using Threshold Based Automatic Classification Laiali Almazaydeh, Khaled Elleithy, Varun Pande and Miad Faezipour Department of Computer Science and Engineering University
Development of OSA Event Detection Using Threshold Based Automatic Classification Laiali Almazaydeh, Khaled Elleithy, Varun Pande and Miad Faezipour Department of Computer Science and Engineering University
GOT SLEEP? YOUR COMPLETE GUIDE TO UNDERSTANDING SLEEP APNEA
 Count me. I dare you. GOT SLEEP? YOUR COMPLETE GUIDE TO UNDERSTANDING SLEEP APNEA PRESENTED BY INTRODUCTION So what s the point of this ebook anyway? At The Maxillofacial Surgery Center, we want to inform
Count me. I dare you. GOT SLEEP? YOUR COMPLETE GUIDE TO UNDERSTANDING SLEEP APNEA PRESENTED BY INTRODUCTION So what s the point of this ebook anyway? At The Maxillofacial Surgery Center, we want to inform
 Sleep Studies Sleep studies are tests that measure how well you sleep and how your body responds to sleep problems. These tests can help your doctor find out whether you have a sleep disorder and how severe
Sleep Studies Sleep studies are tests that measure how well you sleep and how your body responds to sleep problems. These tests can help your doctor find out whether you have a sleep disorder and how severe
Obstructive Sleep Apnea Syndrome. Common sleep disorder causes high blood pressure and heart attacks
 Obstructive Sleep Apnea Syndrome Common sleep disorder causes high blood pressure and heart attacks Message: Sleep apnea is very common. It is estimated that 158 million people worldwide suffer from sleep
Obstructive Sleep Apnea Syndrome Common sleep disorder causes high blood pressure and heart attacks Message: Sleep apnea is very common. It is estimated that 158 million people worldwide suffer from sleep
Helping You to Breathe Better, Sleep Easy & Live Well
 Helping You to Breathe Better, Sleep Easy & Live Well Your Guide to CPAP Therapy info@cansleep.ca Vancouver Island Lower Mainland Fraser Valley Sleep Apnea & Symptoms Obstructive Sleep Apnea (OSA) occurs
Helping You to Breathe Better, Sleep Easy & Live Well Your Guide to CPAP Therapy info@cansleep.ca Vancouver Island Lower Mainland Fraser Valley Sleep Apnea & Symptoms Obstructive Sleep Apnea (OSA) occurs
Anyone of any shape or size may snore, but there are certain features which significantly increase the chance of snoring.
 Snoring Snoring is a common sleep related problem affecting more than 20% of the population at some stage in their lives. Snoring occurs when various parts of the throat, including the soft palate, tonsils
Snoring Snoring is a common sleep related problem affecting more than 20% of the population at some stage in their lives. Snoring occurs when various parts of the throat, including the soft palate, tonsils
Dental Sleep Medicine Basics
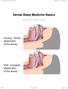 Dental Sleep Medicine Basics Written By: Patrick Tessier 2018 www.tap.wiki/ Page 1 of 8 INTRODUCTION Here are some basic aspect of Dental Sleep Medicine. This viewpoint is from an industry participant,
Dental Sleep Medicine Basics Written By: Patrick Tessier 2018 www.tap.wiki/ Page 1 of 8 INTRODUCTION Here are some basic aspect of Dental Sleep Medicine. This viewpoint is from an industry participant,
ROBERT C. PRITCHARD DIRECTOR MICHAEL O. FOSTER ASSISTANT DIR. SLEEP APNEA
 ROBERT C. PRITCHARD DIRECTOR MICHAEL O. FOSTER ASSISTANT DIR. SLEEP APNEA A Person is physically qualified to drive a motor vehicle if that person; -(5) has no established medical history or clinical diagnosis
ROBERT C. PRITCHARD DIRECTOR MICHAEL O. FOSTER ASSISTANT DIR. SLEEP APNEA A Person is physically qualified to drive a motor vehicle if that person; -(5) has no established medical history or clinical diagnosis
Web-Based Home Sleep Testing
 Editorial Web-Based Home Sleep Testing Authors: Matthew Tarler, Ph.D., Sarah Weimer, Craig Frederick, Michael Papsidero M.D., Hani Kayyali Abstract: Study Objective: To assess the feasibility and accuracy
Editorial Web-Based Home Sleep Testing Authors: Matthew Tarler, Ph.D., Sarah Weimer, Craig Frederick, Michael Papsidero M.D., Hani Kayyali Abstract: Study Objective: To assess the feasibility and accuracy
Sleep Disordered Breathing
 Sleep Disordered Breathing SDB SDB Is an Umbrella Term for Many Disorders characterized by a lack of drive to breathe Results n repetitive pauses in breathing with no effort Occurs for a minimum of 10
Sleep Disordered Breathing SDB SDB Is an Umbrella Term for Many Disorders characterized by a lack of drive to breathe Results n repetitive pauses in breathing with no effort Occurs for a minimum of 10
Balboa Island Dentistry (949)
 Do You Snore? Are you always tired? Snoring is no laughing matter! It may be more than an annoying habit. It may be a sign of. How well do you sleep? Just about everyone snores occasionally. Even a baby
Do You Snore? Are you always tired? Snoring is no laughing matter! It may be more than an annoying habit. It may be a sign of. How well do you sleep? Just about everyone snores occasionally. Even a baby
Snoring. Forty-five percent of normal adults snore at least occasionally and 25
 Snoring Insight into sleeping disorders and sleep apnea Forty-five percent of normal adults snore at least occasionally and 25 percent are habitual snorers. Problem snoring is more frequent in males and
Snoring Insight into sleeping disorders and sleep apnea Forty-five percent of normal adults snore at least occasionally and 25 percent are habitual snorers. Problem snoring is more frequent in males and
Frequently Asked Questions
 Q- What is Sleep Apnea? Frequently Asked Questions A- Sleep Apnea, sometimes known as the "silent killer" although there is usually nothing silent about it. It is associated with periodic loud snoring
Q- What is Sleep Apnea? Frequently Asked Questions A- Sleep Apnea, sometimes known as the "silent killer" although there is usually nothing silent about it. It is associated with periodic loud snoring
Helping You to Breathe Better, Sleep Easy & Live Well
 Helping You to Breathe Better, Sleep Easy & Live Well Your Guide to CPAP Therapy info@cansleep.ca Lower Mainland Vancouver Island Fraser Valley Sleep Apnea & Symptoms Obstructive Sleep Apnea (OSA) occurs
Helping You to Breathe Better, Sleep Easy & Live Well Your Guide to CPAP Therapy info@cansleep.ca Lower Mainland Vancouver Island Fraser Valley Sleep Apnea & Symptoms Obstructive Sleep Apnea (OSA) occurs
Obstructive Sleep Apnoea
 Obstructive Sleep Apnoea Feeling excessively tired during the day? Waking up tired? Snoring? Finding it difficult to stay awake?... or does your partner say you stop breathing while asleep? It could be
Obstructive Sleep Apnoea Feeling excessively tired during the day? Waking up tired? Snoring? Finding it difficult to stay awake?... or does your partner say you stop breathing while asleep? It could be
SLEEP APNOEA DR TAN KAH LEONG ALVIN CO-DIRECTOR SLEEP LABORATORY SITE CHIEF SDDC (SLEEP) DEPARTMENT OF OTORHINOLARYNGOLOGY, HEAD & NECK SURGERY
 SLEEP APNOEA DR TAN KAH LEONG ALVIN CO-DIRECTOR SLEEP LABORATORY SITE CHIEF SDDC (SLEEP) DEPARTMENT OF OTORHINOLARYNGOLOGY, HEAD & NECK SURGERY
SLEEP APNOEA DR TAN KAH LEONG ALVIN CO-DIRECTOR SLEEP LABORATORY SITE CHIEF SDDC (SLEEP) DEPARTMENT OF OTORHINOLARYNGOLOGY, HEAD & NECK SURGERY
SLEEP APNOEA AND DYSFUNCTIONAL BREATHING
 SLEEP APNOEA AND DYSFUNCTIONAL BREATHING The link missed by the Sleep Study industry. By Roger Price Respiratory Physiologist I would like to commence by stating that there is no doubt in my mind that
SLEEP APNOEA AND DYSFUNCTIONAL BREATHING The link missed by the Sleep Study industry. By Roger Price Respiratory Physiologist I would like to commence by stating that there is no doubt in my mind that
Sleep Screening Questionnaire
 Version: SLPQV1 Sleep Screening Questionnaire OFFICE USE Patient ID: NAME: CURRENT DATE: / / DATE OF BIRTH: / / MALE FEMALE Referring Physician: Contact ID: Number Number #1 = the most severe symptom #1
Version: SLPQV1 Sleep Screening Questionnaire OFFICE USE Patient ID: NAME: CURRENT DATE: / / DATE OF BIRTH: / / MALE FEMALE Referring Physician: Contact ID: Number Number #1 = the most severe symptom #1
Introducing the WatchPAT 200 # 1 Home Sleep Study Device
 Introducing the WatchPAT 200 # 1 Home Sleep Study Device Top 10 Medical Innovation for 2010 Cleveland Clinic Fidelis Diagnostics & Itamar Medical Fidelis Diagnostics founded in 2004, is a privately-held
Introducing the WatchPAT 200 # 1 Home Sleep Study Device Top 10 Medical Innovation for 2010 Cleveland Clinic Fidelis Diagnostics & Itamar Medical Fidelis Diagnostics founded in 2004, is a privately-held
Brian Palmer, D.D.S, Kansas City, Missouri, USA. April, 2001
 Brian Palmer, D.D.S, Kansas City, Missouri, USA A1 April, 2001 Disclaimer The information in this presentation is for basic information only and is not to be construed as a diagnosis or treatment for any
Brian Palmer, D.D.S, Kansas City, Missouri, USA A1 April, 2001 Disclaimer The information in this presentation is for basic information only and is not to be construed as a diagnosis or treatment for any
SLEEP DISORDERED BREATHING The Clinical Conditions
 SLEEP DISORDERED BREATHING The Clinical Conditions Robert G. Hooper, M.D. In the previous portion of this paper, the definitions of the respiratory events that are the hallmarks of problems with breathing
SLEEP DISORDERED BREATHING The Clinical Conditions Robert G. Hooper, M.D. In the previous portion of this paper, the definitions of the respiratory events that are the hallmarks of problems with breathing
How To Win Your War Against Snoring And Sleep Apnea
 Page 1 of 1 Contents What Is Sleep Apnea?... 9 Treatments For Central Sleep Apnea... 10 Learning About Sleep Apnea... 11 What Are The Symptoms Of Sleep Apnea?... 12 What Is Causing My Obstructive Sleep
Page 1 of 1 Contents What Is Sleep Apnea?... 9 Treatments For Central Sleep Apnea... 10 Learning About Sleep Apnea... 11 What Are The Symptoms Of Sleep Apnea?... 12 What Is Causing My Obstructive Sleep
Dr. Don McLaughlin N Main St. Rutland VT (802)
 Dr. Don McLaughlin www.snoresnomore.com dr.don@snoresnomore.com 206 N Main St. Rutland VT 05706 (802) 773-7000 Booklet for Better Sleep Working with Sleep Apnea Part 1 of 3 Sick and Tired of Always Being
Dr. Don McLaughlin www.snoresnomore.com dr.don@snoresnomore.com 206 N Main St. Rutland VT 05706 (802) 773-7000 Booklet for Better Sleep Working with Sleep Apnea Part 1 of 3 Sick and Tired of Always Being
Tired of being tired?
 Tired of being tired? Narval CC MRD ResMed.com/Narval Sleepiness and snoring are possible symptoms of sleep apnea. Did you know that one in every four adults has some form of sleep disordered-breathing
Tired of being tired? Narval CC MRD ResMed.com/Narval Sleepiness and snoring are possible symptoms of sleep apnea. Did you know that one in every four adults has some form of sleep disordered-breathing
WINDSOR DENTAL CARE 2224 WALKER ROAD SUITE 20 WINDSOR, ON N8W 5L7 PHONE FAX
 The quality of your sleep can impact you emotionally, physically and your overall general health. Poor sleep can cause chronic fatigue, daytime drowsiness, irritability and loss of focus. It affects your
The quality of your sleep can impact you emotionally, physically and your overall general health. Poor sleep can cause chronic fatigue, daytime drowsiness, irritability and loss of focus. It affects your
Assessment of Sleep Disorders DR HUGH SELSICK
 Assessment of Sleep Disorders DR HUGH SELSICK Goals Understand the importance of history taking Be able to take a basic sleep history Be aware the technology used to assess sleep disorders. Understand
Assessment of Sleep Disorders DR HUGH SELSICK Goals Understand the importance of history taking Be able to take a basic sleep history Be aware the technology used to assess sleep disorders. Understand
What You Need to Know About Sleep Apnea
 What You Need to Know About Sleep Apnea Statement of Rights You May NOT Change this Book or Sell it. But You Can Freely Share it Copyright Message Rights Acquired by White Dove Books 2006 What YOU Need
What You Need to Know About Sleep Apnea Statement of Rights You May NOT Change this Book or Sell it. But You Can Freely Share it Copyright Message Rights Acquired by White Dove Books 2006 What YOU Need
Positive Airway Pressure and Oral Devices for the Treatment of Obstructive Sleep Apnea
 Positive Airway Pressure and Oral Devices for the Treatment of Obstructive Sleep Apnea Policy Number: Original Effective Date: MM.01.009 11/01/2009 Line(s) of Business: Current Effective Date: HMO; PPO;
Positive Airway Pressure and Oral Devices for the Treatment of Obstructive Sleep Apnea Policy Number: Original Effective Date: MM.01.009 11/01/2009 Line(s) of Business: Current Effective Date: HMO; PPO;
Snoring and Sleep Apnea
 The Patient s Guide To Snoring and Sleep Apnea The Complications May Be More Severe Than You Think Unit #245, 520 3rd Ave SW Calgary, AB T2P 0R3 Tel: (587) 353-5060 www.centennialsmiles.ca CONTENTS Introduction
The Patient s Guide To Snoring and Sleep Apnea The Complications May Be More Severe Than You Think Unit #245, 520 3rd Ave SW Calgary, AB T2P 0R3 Tel: (587) 353-5060 www.centennialsmiles.ca CONTENTS Introduction
Sleep Apnea Exercises Cheat Sheet
 Sleep Apnea Exercises Cheat Sheet Thank you once again for taking my Sleep Apnea Exercises e-course! This cheat sheet* is an unadvertised bonus for subscribers who have taken the e-course, and is a companion
Sleep Apnea Exercises Cheat Sheet Thank you once again for taking my Sleep Apnea Exercises e-course! This cheat sheet* is an unadvertised bonus for subscribers who have taken the e-course, and is a companion
Sleep Apnoea. The Story of a Pause
 Sleep Apnoea The Story of a Pause There is almost zero awareness in India that many amongst us maybe living with Sleep Apnoea, which left untreated could be life threatening tomorrow. This largely undiagnosed
Sleep Apnoea The Story of a Pause There is almost zero awareness in India that many amongst us maybe living with Sleep Apnoea, which left untreated could be life threatening tomorrow. This largely undiagnosed
Patient Adult Information History
 Patient Adult Information History Patient name: Age: Date: What is the main reason for today s evaluation? Infant History Birth delivery: Normal C-section Delayed Epidural Premature: No Yes If yes, how
Patient Adult Information History Patient name: Age: Date: What is the main reason for today s evaluation? Infant History Birth delivery: Normal C-section Delayed Epidural Premature: No Yes If yes, how
Positive Airway Pressure and Oral Devices for the Treatment of Obstructive Sleep Apnea
 Positive Airway Pressure and Oral Devices for the Treatment of Obstructive Sleep Apnea Policy Number: Original Effective Date: MM.01.009 11/01/2009 Line(s) of Business: Current Effective Date: HMO; PPO
Positive Airway Pressure and Oral Devices for the Treatment of Obstructive Sleep Apnea Policy Number: Original Effective Date: MM.01.009 11/01/2009 Line(s) of Business: Current Effective Date: HMO; PPO
A New, Clinically Proven Sleep Apnea Therapy for people unable to use CPAP.
 A New, Clinically Proven Sleep Apnea Therapy for people unable to use CPAP. Take Heart. If You Have OSA, You re Not Alone. Like you, more than 18 million Americans are estimated to have Obstructive Sleep
A New, Clinically Proven Sleep Apnea Therapy for people unable to use CPAP. Take Heart. If You Have OSA, You re Not Alone. Like you, more than 18 million Americans are estimated to have Obstructive Sleep
SLEEP HISTORY QUESTIONNAIRE
 Date of birth: Today s date: Dear Patient: SLEEP HISTORY QUESTIONNAIRE Thank you for taking the time to fill out a sleep history questionnaire. This will help our healthcare team to provide the best possible
Date of birth: Today s date: Dear Patient: SLEEP HISTORY QUESTIONNAIRE Thank you for taking the time to fill out a sleep history questionnaire. This will help our healthcare team to provide the best possible
Denver, CO Welcome Packet
 Fax: (303) 957-5414 or 720-542-8699 For any after-hours questions, please call (303) 956-5145 Dear Mountain Sleep Patient, You have been scheduled for a sleep study at 1210 S Parker Road, Suite 101, Denver,
Fax: (303) 957-5414 or 720-542-8699 For any after-hours questions, please call (303) 956-5145 Dear Mountain Sleep Patient, You have been scheduled for a sleep study at 1210 S Parker Road, Suite 101, Denver,
Use of Technology in the Assessment of Type 2 Diabetes and Sleep Apnea
 Use of Technology in the Assessment of Type 2 Diabetes and Sleep Apnea Eileen R. Chasens, PhD Associate Professor University of Pittsburgh September 3, 2014 Disclosure I do not own a Smart Phone, I have
Use of Technology in the Assessment of Type 2 Diabetes and Sleep Apnea Eileen R. Chasens, PhD Associate Professor University of Pittsburgh September 3, 2014 Disclosure I do not own a Smart Phone, I have
Effective Treatment for Obstructive Sleep Apnoea
 Effective Treatment for Obstructive Sleep Apnoea The Series of Positive Airway Pressure devices from DeVilbiss Healthcare is designed to meet the varied needs of people suffering from Obstructive Sleep
Effective Treatment for Obstructive Sleep Apnoea The Series of Positive Airway Pressure devices from DeVilbiss Healthcare is designed to meet the varied needs of people suffering from Obstructive Sleep
Littleton, CO Welcome Packet 8151 Southpark Lane, Suite 200 Littleton, CO 80120
 Littleton, CO Welcome Packet For any after-hours questions, please call (303) 956-5145 Dear Mountain Sleep Patient, You have been scheduled for a sleep study at 8151 Southpark Lane, Suite 200, Littleton,
Littleton, CO Welcome Packet For any after-hours questions, please call (303) 956-5145 Dear Mountain Sleep Patient, You have been scheduled for a sleep study at 8151 Southpark Lane, Suite 200, Littleton,
Questions: What tests are available to diagnose sleep disordered breathing? How do you calculate overall AHI vs obstructive AHI?
 Pediatric Obstructive Sleep Apnea Case Study : Margaret-Ann Carno PhD, CPNP, D,ABSM for the Sleep Education for Pulmonary Fellows and Practitioners, SRN ATS Committee April 2014. Facilitator s guide Part
Pediatric Obstructive Sleep Apnea Case Study : Margaret-Ann Carno PhD, CPNP, D,ABSM for the Sleep Education for Pulmonary Fellows and Practitioners, SRN ATS Committee April 2014. Facilitator s guide Part
Non-contact Screening System with Two Microwave Radars in the Diagnosis of Sleep Apnea-Hypopnea Syndrome
 Medinfo2013 Decision Support Systems and Technologies - II Non-contact Screening System with Two Microwave Radars in the Diagnosis of Sleep Apnea-Hypopnea Syndrome 21 August 2013 M. Kagawa 1, K. Ueki 1,
Medinfo2013 Decision Support Systems and Technologies - II Non-contact Screening System with Two Microwave Radars in the Diagnosis of Sleep Apnea-Hypopnea Syndrome 21 August 2013 M. Kagawa 1, K. Ueki 1,
Home Sleep Testing Questionnaire
 Home Sleep Testing Questionnaire Patient Name: DOB: / / Gender: Male Female Study Date: / / Marital Status: Married Cohabitate Single Divorced Widow/Widower Email: Phone: Height: Weight: Neck Size: What
Home Sleep Testing Questionnaire Patient Name: DOB: / / Gender: Male Female Study Date: / / Marital Status: Married Cohabitate Single Divorced Widow/Widower Email: Phone: Height: Weight: Neck Size: What
EXPLORE NEW POSSIBILITIES
 EXPLORE NEW POSSIBILITIES TREATING SNORING AND SLEEP APNOEA HAS CHANGED FOREVER Introducing the Oventus O 2 Vent, a custom made, comfortable oral appliance with a unique airway design for the treatment
EXPLORE NEW POSSIBILITIES TREATING SNORING AND SLEEP APNOEA HAS CHANGED FOREVER Introducing the Oventus O 2 Vent, a custom made, comfortable oral appliance with a unique airway design for the treatment
WELCOME TO... Please read this brochure & the Provent Instructions For Use before starting Provent Sleep Apnea Therapy.
 WELCOME TO... Please read this brochure & the Provent Instructions For Use before starting Provent Sleep Apnea Therapy. Obstructive Sleep Apnea (OSA) is a serious medical condition characterized by pauses
WELCOME TO... Please read this brochure & the Provent Instructions For Use before starting Provent Sleep Apnea Therapy. Obstructive Sleep Apnea (OSA) is a serious medical condition characterized by pauses
Medicare CPAP/BIPAP Coverage Criteria
 Medicare CPAP/BIPAP Coverage Criteria For any item to be covered by Medicare, it must 1) be eligible for a defined Medicare benefit category, 2) be reasonable and necessary for the diagnosis or treatment
Medicare CPAP/BIPAP Coverage Criteria For any item to be covered by Medicare, it must 1) be eligible for a defined Medicare benefit category, 2) be reasonable and necessary for the diagnosis or treatment
QUESTIONS FOR DELIBERATION
 New England Comparative Effectiveness Public Advisory Council Public Meeting Hartford, Connecticut Diagnosis and Treatment of Obstructive Sleep Apnea in Adults December 6, 2012 UPDATED: November 28, 2012
New England Comparative Effectiveness Public Advisory Council Public Meeting Hartford, Connecticut Diagnosis and Treatment of Obstructive Sleep Apnea in Adults December 6, 2012 UPDATED: November 28, 2012
Diabetes & Obstructive Sleep Apnoea risk. Jaynie Pateraki MSc RGN
 Diabetes & Obstructive Sleep Apnoea risk Jaynie Pateraki MSc RGN Non-REM - REM - Both - Unrelated - Common disorders of Sleep Sleep Walking Night terrors Periodic leg movements Sleep automatism Nightmares
Diabetes & Obstructive Sleep Apnoea risk Jaynie Pateraki MSc RGN Non-REM - REM - Both - Unrelated - Common disorders of Sleep Sleep Walking Night terrors Periodic leg movements Sleep automatism Nightmares
Positive Pressure Therapy
 Positive Pressure Therapy Positive Pressure Therapy...2 What is Sleep Apnea?....2 Positive Pressure Machines..................................................... 4 Types..................................................................................
Positive Pressure Therapy Positive Pressure Therapy...2 What is Sleep Apnea?....2 Positive Pressure Machines..................................................... 4 Types..................................................................................
Obstructive sleep apnoea How to identify?
 Obstructive sleep apnoea How to identify? Walter McNicholas MD Newman Professor in Medicine, St. Vincent s University Hospital, University College Dublin, Ireland. Potential conflict of interest None Obstructive
Obstructive sleep apnoea How to identify? Walter McNicholas MD Newman Professor in Medicine, St. Vincent s University Hospital, University College Dublin, Ireland. Potential conflict of interest None Obstructive
Causes and Consequences of Respiratory Centre Depression and Hypoventilation
 Causes and Consequences of Respiratory Centre Depression and Hypoventilation Lou Irving Director Respiratory and Sleep Medicine, RMH louis.irving@mh.org.au Capacity of the Respiratory System At rest During
Causes and Consequences of Respiratory Centre Depression and Hypoventilation Lou Irving Director Respiratory and Sleep Medicine, RMH louis.irving@mh.org.au Capacity of the Respiratory System At rest During
THN. Sleep Therapy Study. ImThera. Information for Participants. Caution: Investigational device. Limited by United States law to investigational use.
 THN Sleep Therapy Study Information for Participants Caution: Investigational device. Limited by United States law to investigational use. ImThera Obstructive sleep apnea (OSA) is a very serious condition.
THN Sleep Therapy Study Information for Participants Caution: Investigational device. Limited by United States law to investigational use. ImThera Obstructive sleep apnea (OSA) is a very serious condition.
Commissioning Policy Individual Funding Request
 Commissioning Policy Individual Funding Request Continuous Positive Airway Pressure (CPAP) Treatment of Obstructive Sleep Apnoea/Hypopnoea Syndrome (OSAHS) Criteria Based Access Policy Date Adopted: 13
Commissioning Policy Individual Funding Request Continuous Positive Airway Pressure (CPAP) Treatment of Obstructive Sleep Apnoea/Hypopnoea Syndrome (OSAHS) Criteria Based Access Policy Date Adopted: 13
DECLARATION OF CONFLICT OF INTEREST
 DECLARATION OF CONFLICT OF INTEREST Obstructive sleep apnoea How to identify? Walter McNicholas MD Newman Professor in Medicine, St. Vincent s University Hospital, University College Dublin, Ireland. Potential
DECLARATION OF CONFLICT OF INTEREST Obstructive sleep apnoea How to identify? Walter McNicholas MD Newman Professor in Medicine, St. Vincent s University Hospital, University College Dublin, Ireland. Potential
Sleep Diordered Breathing (Part 1)
 Sleep Diordered Breathing (Part 1) History (for more topics & presentations, visit ) Obstructive sleep apnea - first described by Charles Dickens in 1836 in Papers of the Pickwick Club, Dickens depicted
Sleep Diordered Breathing (Part 1) History (for more topics & presentations, visit ) Obstructive sleep apnea - first described by Charles Dickens in 1836 in Papers of the Pickwick Club, Dickens depicted
Management of OSA in the Acute Care Environment. Robert S. Campbell, RRT FAARC HRC, Philips Healthcare May, 2018
 Management of OSA in the Acute Care Environment Robert S. Campbell, RRT FAARC HRC, Philips Healthcare May, 2018 1 Learning Objectives Upon completion, the participant should be able to: Understand pathology
Management of OSA in the Acute Care Environment Robert S. Campbell, RRT FAARC HRC, Philips Healthcare May, 2018 1 Learning Objectives Upon completion, the participant should be able to: Understand pathology
Types of Sleep Studies 8/28/2018. Ronald S. Prehn, ThM, DDS. Type 1 Attended in-lab polysomnography (PSG) 18 leads
 Ronald S. Prehn, ThM, DDS rprehn@tmjtexas.com Board Certified in Dental Sleep Medicine Board Certified in Orofacial Pain Types of Sleep Studies Type 1 Attended in-lab polysomnography (PSG) 18 leads Type
Ronald S. Prehn, ThM, DDS rprehn@tmjtexas.com Board Certified in Dental Sleep Medicine Board Certified in Orofacial Pain Types of Sleep Studies Type 1 Attended in-lab polysomnography (PSG) 18 leads Type
A Sleeping Monitor for Snoring Detection
 EECS 395/495 - mhealth McCormick School of Engineering A Sleeping Monitor for Snoring Detection By Hongwei Cheng, Qian Wang, Tae Hun Kim Abstract Several studies have shown that snoring is the first symptom
EECS 395/495 - mhealth McCormick School of Engineering A Sleeping Monitor for Snoring Detection By Hongwei Cheng, Qian Wang, Tae Hun Kim Abstract Several studies have shown that snoring is the first symptom
OSA and COPD: What happens when the two OVERLAP?
 2011 ISRC Seminar 1 COPD OSA OSA and COPD: What happens when the two OVERLAP? Overlap Syndrome 1 OSA and COPD: What happens when the two OVERLAP? ResMed 10 JAN Global leaders in sleep and respiratory medicine
2011 ISRC Seminar 1 COPD OSA OSA and COPD: What happens when the two OVERLAP? Overlap Syndrome 1 OSA and COPD: What happens when the two OVERLAP? ResMed 10 JAN Global leaders in sleep and respiratory medicine
Inspire Therapy for Sleep Apnea
 Inspire Therapy for Sleep Apnea Take Heart. If you have OSA, you re not alone. More than 18 million Americans are estimated to have Obstructive Sleep Apnea (OSA). OSA occurs when the tongue and other soft
Inspire Therapy for Sleep Apnea Take Heart. If you have OSA, you re not alone. More than 18 million Americans are estimated to have Obstructive Sleep Apnea (OSA). OSA occurs when the tongue and other soft
Procedures/Risks: pulmonology, sleep, critical care
 Procedures/Risks: pulmonology, sleep, critical care Bronchoscopy (and bronchoaveolar lavage) Purpose: The purpose of the bronchoscopy is to collect cells and fluid from the lung, so that information can
Procedures/Risks: pulmonology, sleep, critical care Bronchoscopy (and bronchoaveolar lavage) Purpose: The purpose of the bronchoscopy is to collect cells and fluid from the lung, so that information can
THE SLEEP DISORDERS CLINIC Medical Director: Dr Raymond Gottschalk PATIENT QUESTIONNAIRE
 THE SLEEP DISORDERS CLINIC Medical Director: Dr Raymond Gottschalk 55 Frid Street, Unit 7, Hamilton, Ontario L8P 4M3 Phone:905-529-2259 Fax: 905-529-2262 282 Linwell Road, Suite 118, St. Catharines, Ontario
THE SLEEP DISORDERS CLINIC Medical Director: Dr Raymond Gottschalk 55 Frid Street, Unit 7, Hamilton, Ontario L8P 4M3 Phone:905-529-2259 Fax: 905-529-2262 282 Linwell Road, Suite 118, St. Catharines, Ontario
Inspire Therapy for Sleep Apnea
 Inspire Therapy for Sleep Apnea Patient Guide Giving You the FREEDOM TO SLEEP Like Everyone Else Take Comfort. Inspire therapy can help. Inspire therapy is a breakthrough implantable treatment option for
Inspire Therapy for Sleep Apnea Patient Guide Giving You the FREEDOM TO SLEEP Like Everyone Else Take Comfort. Inspire therapy can help. Inspire therapy is a breakthrough implantable treatment option for
Baptist Health Floyd 1850 State Street New Albany, IN Sleep Disorders Center Lung & Sleep Specialists. Date of Birth: Age:
 Page 1 of 7 GENERAL INFORMATION Name: Date of Birth: Age: Social Security #: Sex: Height: Weight: Address: City: State: Zip: Home Phone: Cell Phone: Work Phone: Employer s Name: Marital Status: Married
Page 1 of 7 GENERAL INFORMATION Name: Date of Birth: Age: Social Security #: Sex: Height: Weight: Address: City: State: Zip: Home Phone: Cell Phone: Work Phone: Employer s Name: Marital Status: Married
Make a Smart IT & BT Devices for Future Warning for Heart Failure, Hypoglycemia, and Hyperglycemia Smart Watch. LP Lab LP Watch
 Index Technology Index Description Function of Hart attack Device Function of Hypoglycemia Device Abut Us Technology1 Technology2 Warning Sale Make a Smart IT & BT Devices for Future Warning for Heart
Index Technology Index Description Function of Hart attack Device Function of Hypoglycemia Device Abut Us Technology1 Technology2 Warning Sale Make a Smart IT & BT Devices for Future Warning for Heart
SLEEP SCREENING QUESTIONNAIRE
 Patient Information 433 W. University Dr. Rochester, MI 48307 www.rochesteradvanceddentistry.com +1 248 656-2020 SLEEP SCREENING QUESTIONNAIRE Name: DOB: Age: Address: Employer: SS# Home Phone: Work Phone:
Patient Information 433 W. University Dr. Rochester, MI 48307 www.rochesteradvanceddentistry.com +1 248 656-2020 SLEEP SCREENING QUESTIONNAIRE Name: DOB: Age: Address: Employer: SS# Home Phone: Work Phone:
Therapy options. snoring & obstructive sleep apnea. Mandibular advancement devices Nasal dilators Supine position preventers
 Therapy options for the treatment of snoring & obstructive sleep apnea Therapy options Mandibular advancement devices Nasal dilators Supine position preventers Snoring and obstructive sleep apnea A portion
Therapy options for the treatment of snoring & obstructive sleep apnea Therapy options Mandibular advancement devices Nasal dilators Supine position preventers Snoring and obstructive sleep apnea A portion
Upper airway resistance syndrome (UARS) Information for patients
 Upper airway resistance syndrome (UARS) Information for patients This leaflet answers common questions about upper airway resistance syndrome. If you would like further information, or have any concerns,
Upper airway resistance syndrome (UARS) Information for patients This leaflet answers common questions about upper airway resistance syndrome. If you would like further information, or have any concerns,
What is Oral Appliance Therapy?
 You may have been diagnosed by your physician as required treatment for sleep-disordered breathing (snoring and/or obstructive sleep apnea). This condition may pose serious health risks since it disrupts
You may have been diagnosed by your physician as required treatment for sleep-disordered breathing (snoring and/or obstructive sleep apnea). This condition may pose serious health risks since it disrupts
The most accurate predictors of arterial hypertension in patients with Obstructive Sleep Apnea Syndrome
 The most accurate predictors of arterial hypertension in patients with Obstructive Sleep Apnea Syndrome Natsios Georgios University Hospital of Larissa, Greece Definitions Obstructive Sleep Apnea (OSA)
The most accurate predictors of arterial hypertension in patients with Obstructive Sleep Apnea Syndrome Natsios Georgios University Hospital of Larissa, Greece Definitions Obstructive Sleep Apnea (OSA)
Drowsy Driving. Awareness and Prevention
 Drowsy Driving Awareness and Prevention 2 OUR THANKS! This educational program was funded by the National Highway Traffic Safety Administration with a grant from the New York Governor's Traffic Safety
Drowsy Driving Awareness and Prevention 2 OUR THANKS! This educational program was funded by the National Highway Traffic Safety Administration with a grant from the New York Governor's Traffic Safety
Healthy for Life Newsletter September/October 2010 Vol. 7 No. 5. Fibromyalgia a Sleep Disorder
 Healthy for Life Newsletter September/October 2010 Vol. 7 No. 5 Fibromyalgia a Sleep Disorder Introduction I have lived with fibromyalgia for over 30 years. No, I don t have fibromyalgia; however, my wife
Healthy for Life Newsletter September/October 2010 Vol. 7 No. 5 Fibromyalgia a Sleep Disorder Introduction I have lived with fibromyalgia for over 30 years. No, I don t have fibromyalgia; however, my wife
Sleep Disorders Diagnostic Center 9733 Healthway Drive, Berlin, MD , ext. 5118
 Sleep Questionnaire *Please complete the following as accurate as possible. Please bring your completed questionnaire, insurance card, photo ID, Pre-Authorization and/or Insurance referral form, and all
Sleep Questionnaire *Please complete the following as accurate as possible. Please bring your completed questionnaire, insurance card, photo ID, Pre-Authorization and/or Insurance referral form, and all
Rest Stop #101. Sleep & Fatigue-What s the difference and what to do about it.
 Rest Stop #101 This Photo by Unknown Author is licensed under CC BY-NC-ND Sleep & Fatigue-What s the difference and what to do about it. Presented by Mary Convey, Director of Key Accounts & Risk Mitigation
Rest Stop #101 This Photo by Unknown Author is licensed under CC BY-NC-ND Sleep & Fatigue-What s the difference and what to do about it. Presented by Mary Convey, Director of Key Accounts & Risk Mitigation
Diagnostic Accuracy of the Multivariable Apnea Prediction (MAP) Index as a Screening Tool for Obstructive Sleep Apnea
 Original Article Diagnostic Accuracy of the Multivariable Apnea Prediction (MAP) Index as a Screening Tool for Obstructive Sleep Apnea Ahmad Khajeh-Mehrizi 1,2 and Omid Aminian 1 1. Occupational Sleep
Original Article Diagnostic Accuracy of the Multivariable Apnea Prediction (MAP) Index as a Screening Tool for Obstructive Sleep Apnea Ahmad Khajeh-Mehrizi 1,2 and Omid Aminian 1 1. Occupational Sleep
About VirtuOx. Was marketed exclusively by Phillips Healthcare division, Respironics for 3 years
 About VirtuOx VirtuOx, Inc. assists physicians and Durable Medical Equipment (DME)( companies diagnose respiratory diseases and qualify patients for home respiratory equipment under the guidelines of CMS
About VirtuOx VirtuOx, Inc. assists physicians and Durable Medical Equipment (DME)( companies diagnose respiratory diseases and qualify patients for home respiratory equipment under the guidelines of CMS
Patient Instructions
 Home Sleep Testing... Patient Instructions VirtuOx Patient Support: 1-877-337-7111 www.virtuox.net Testing process is time sensitive. You must complete the testing and return the kit within 48 hours of
Home Sleep Testing... Patient Instructions VirtuOx Patient Support: 1-877-337-7111 www.virtuox.net Testing process is time sensitive. You must complete the testing and return the kit within 48 hours of
NATIONAL COMPETENCY SKILL STANDARDS FOR PERFORMING POLYSOMNOGRAPHY/SLEEP TECHNOLOGY
 NATIONAL COMPETENCY SKILL STANDARDS FOR PERFORMING POLYSOMNOGRAPHY/SLEEP TECHNOLOGY Polysomnography/Sleep Technology providers practice in accordance with the facility policy and procedure manual which
NATIONAL COMPETENCY SKILL STANDARDS FOR PERFORMING POLYSOMNOGRAPHY/SLEEP TECHNOLOGY Polysomnography/Sleep Technology providers practice in accordance with the facility policy and procedure manual which
Sleep and the Heart Reversing the Effects of Sleep Apnea to Better Manage Heart Disease
 1 Sleep and the Heart Reversing the Effects of Sleep Apnea to Better Manage Heart Disease Rami Khayat, MD Professor of Internal Medicine Director, OSU Sleep Heart Program Medical Director, Department of
1 Sleep and the Heart Reversing the Effects of Sleep Apnea to Better Manage Heart Disease Rami Khayat, MD Professor of Internal Medicine Director, OSU Sleep Heart Program Medical Director, Department of
Your physician has ordered a sleep study for you on. Your arrival time is scheduled for.
 Dear Patient: Your physician has ordered a sleep study for you on. Your arrival time is scheduled for. The Texas State Sleep Lab is located in the Health Professions Building on the Texas State University
Dear Patient: Your physician has ordered a sleep study for you on. Your arrival time is scheduled for. The Texas State Sleep Lab is located in the Health Professions Building on the Texas State University
Inspire. therapy for sleep apnea. Giving you the freedom to sleep like everyone else
 Inspire therapy for sleep apnea Giving you the freedom to sleep like everyone else Take Comfort. Take Action. Inspire therapy can help. Here are some reasons people like you have chosen Inspire therapy
Inspire therapy for sleep apnea Giving you the freedom to sleep like everyone else Take Comfort. Take Action. Inspire therapy can help. Here are some reasons people like you have chosen Inspire therapy
Hypoventilation? Obstructive Sleep Apnea? Different Tests, Different Treatment
 Hypoventilation? Obstructive Sleep Apnea? Different Tests, Different Treatment Judith R. Fischer, MSLS, Editor, Ventilator-Assisted Living (fischer.judith@sbcglobal.net) Thanks to Josh Benditt, MD, University
Hypoventilation? Obstructive Sleep Apnea? Different Tests, Different Treatment Judith R. Fischer, MSLS, Editor, Ventilator-Assisted Living (fischer.judith@sbcglobal.net) Thanks to Josh Benditt, MD, University
FAQ CODING & REIMBURSEMENT. WatchPAT TM Home Sleep Test
 FAQ CODING & REIMBURSEMENT WatchPAT TM Home Sleep Test TABLE OF CONTENTS PATIENT SELECTION CRITERIA 3 CODING & MODIFIERS 4-6 PLACE OF SERVICE 6 FREQUENCY 7 ACCREDITATION 7 SLEEP MEDICINE GLOSSARY AND ACRONYMS
FAQ CODING & REIMBURSEMENT WatchPAT TM Home Sleep Test TABLE OF CONTENTS PATIENT SELECTION CRITERIA 3 CODING & MODIFIERS 4-6 PLACE OF SERVICE 6 FREQUENCY 7 ACCREDITATION 7 SLEEP MEDICINE GLOSSARY AND ACRONYMS
Inspire Therapy for Sleep Apnea
 Inspire Therapy for Sleep Apnea Patient Guide Giving You the FREEDOM TO SLEEP Like Everyone Else Take Comfort. Inspire therapy can help. Inspire therapy is a breakthrough implantable treatment option for
Inspire Therapy for Sleep Apnea Patient Guide Giving You the FREEDOM TO SLEEP Like Everyone Else Take Comfort. Inspire therapy can help. Inspire therapy is a breakthrough implantable treatment option for
Jill D. Marshall. Professor Boye. MPH 510: Applied Epidemiology. Section 01 Summer A June 28, 2013
 1 Obstructive Sleep Apnea: Capstone Screening Project By Jill D. Marshall Professor Boye MPH 510: Applied Epidemiology Section 01 Summer A 2013 June 28, 2013 2 Sufficient sleep should be considered a vital
1 Obstructive Sleep Apnea: Capstone Screening Project By Jill D. Marshall Professor Boye MPH 510: Applied Epidemiology Section 01 Summer A 2013 June 28, 2013 2 Sufficient sleep should be considered a vital
Sleep Center. Have you had a previous sleep study? Yes No If so, when and where? Name of facility Address
 Patient Label For office use only Appt date: Clinician: Sleep Center Main Campus Highlands Ranch Location 1400 Jackson Street 8671 S. Quebec St., Ste 120 Denver, CO 80206 Highlands Ranch, CO 80130 Leading
Patient Label For office use only Appt date: Clinician: Sleep Center Main Campus Highlands Ranch Location 1400 Jackson Street 8671 S. Quebec St., Ste 120 Denver, CO 80206 Highlands Ranch, CO 80130 Leading
