SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer.
|
|
|
- Noreen Hubbard
- 6 years ago
- Views:
Transcription
1 Supplementary Figure 1 SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer. Scatter plots comparing expression profiles of matched pretreatment tumor biopsies (x axis) and residual scars after complete response to radiotherapy (y axis) in two patients with renal cancer (RCa0003 and RCa0016, as indicated on top). Blue dots highlight signature genes corresponding to the SSM subtype, respectively, in the CRCA classifier (a), CCS classifier (b) and CCMS classifier (c).
2 Supplementary Figure 2 GSEA testing of CRC classifier signatures for differential expression in stromal versus neoplastic cells. The signature gene sets for the CCMS4, CRCA5, CCS3, CCMS2 and CRCA1 subtypes, as indicated in each panel, were tested for upregulation in stromal cell populations (FAP +, CD31 +, CD45 + ) versus epithelial cells (EpCAM + ), as indicated. NES, normalized enrichment score.
3 Supplementary Figure 3 Scatter plots for signal comparisons in Affymetrix HG-U133 Plus 2 arrays hybridized with human, mouse and mixed RNA. (a) Standard probe set analysis; x axis: average signal across 27 human tumors (GSE35144); y axis, maximum signal for 3 Mouse universal RNA samples (GSE49353). (b) Single-probe analysis: x axis, average signal for 27 human tumors; y axis, maximum signal for Mouse universal RNA. The horizontal red line indicates the signal threshold above which probes are considered to cross-hybridize with mouse sequences. (c) Signals of new probe sets obtained using H-spec CDF: x axis, average signal across 27 human tumors; y axis, maximum signal for Mouse universal RNA. (d) Standard probe set analysis: x axis, average signal for 27 human tumors; y axis, average signal for the 27 matched PDX samples (GSE35144).
4 Supplementary Figure 4 Scatter plot comparing probe signals of Illumina human gene expression arrays upon hybridization with crna derived from human and mouse samples. x axis, average probe signal of four human primary CRCs; y axis, maximum probe signal from a mouse CRC and a mouse endothelial cell sample.
5 Supplementary Figure 5 Human versus mouse expression of CRC subtype signature genes in PDX RNA-seq data. Scatter plots comparing, for each gene, human ortholog (x axis) versus mouse ortholog (y axis) RPM values from PDX RNA-seq data. Colored dots identify the various subtype signatures of the three CRC classifiers.
6 Supplementary Figure 6 Waterfall plots of various CRC signature gene lists, ranked, from left to right, by the fraction of their expression levels contributed by the stroma. Because of the varying size of the gene lists, the x axis reports the percentile and the y axis reports the stromal contribution to gene expression, calculated for each gene as the percentage of mouse reads over the total (mouse + human) reads. The color code for each gene list is reported at the bottom of the panel. Up and Down are relative to the differential expression as mentioned in the respective referenced works. In plots with a single gene list, the blue color indicates association with the phenotype described in the respective reference works. GSEA statistics for enrichment in the mouse or human fraction are reported in Supplementary Table 11.
7 References for the analyzed Signatures are reported in Supplementary Note c.
8 Supplementary Figure 7 Stromal expression of representative SSM genes. (a) Micrographs of IHC staining of ZEB1, MAP1B and TAGLN in rectal cancer preoperative biopsies classified as SSM. In all micrographs, specific staining of all antibodies is confined to the stromal components. ZEB1 is localized to the nuclei of fibroblasts,
9 leukocytes and other mesenchymal cells; MAP1B preferentially stains endothelial cells and nerve structures; and TAGLN is mainly expressed by smooth muscle cells. Scale bar, 20 µm. (b) RNA in situ hybridization for ZEB1 mrna (RNAscope 2.0 assay, Advanced Cell Diagnostics) in the CRC315 sample, classified as SSM and displaying weak cytoplasmic IHC positivity for ZEB1, as shown in Figure 4b. Scale bar, 20 µm.
10 Supplementary Figure 8 CRC proteomic subtype C signature expression is contributed by CAFs. In all panels except e, blue and pink dots/lines indicate transcripts coding for proteins upregulated and downregulated in subtype C, respectively. (a) Box plots reporting, for each proteomic subgroup, tumor purity estimated by Absolute analysis on TCGA CRC samples. (b) Scatter plots comparing, for each gene ortholog pair, mouse (x axis) and human (y axis) RPM values from PDX RNA-seq data. (c) Scatter plots comparing, for each gene tested on human arrays, average signals from human CRC samples (x axis) and the corresponding PDX derivatives (y axis). Left, Affymetrix human arrays on 27 sample pairs; right, Illumina human arrays on 4 sample pairs. (d) Waterfall plots of proteomic subtype C gene lists, ranked, from left to right, by the fraction of their expression levels contributed by the stroma. Because of the varying size of the gene lists, the x axis reports the percentile and the y axis reports the stromal contribution to gene expression, calculated for each gene as the percentage of mouse reads over the total (mouse + human) reads. (e) GSEA testing for upregulation of the C type gene set in stromal cell populations (FAP +, CD31 +, CD45 + ) versus epithelial cells (EpCAM + ). NES, normalized enrichment score.
11 Supplementary Figure 9 Definition and characterization of three stromal signatures. (a) Expression heat map of the three stromal signatures in sorted CRC cell subpopulations. Genes for the three stromal cell signatures (columns) distinguish the various sorted cell populations (rows): EpCAM +, epithelial cells; CD31 +, endothelial cells; CD45 +, leukocytes; FAP +, CAFs. (b) Dot plot reporting the 3 stromal scores (yellow, CAF; blue, leukocyte; red, endothelial; each as the percentile of its own distribution) in 450 TCGA samples, sorted by descending CAF score. (c) Venn diagrams showing the fractions of cases concordantly or discordantly falling in the top or bottom quartile of each stromal score.
12 Supplementary Figure 10 Reduced expression of endothelial genes after radiotherapy of rectal cancer. (a) GSEA testing for expression of endothelial score genes comparing pretreatment biopsies of SSM cases with surgical samples classified as SSM after radiotherapy. NES, normalized enrichment score. (b) Heat map displaying the GSEA leading edge ( core ) endothelial genes that lost expression in samples after radiotherapy.
13 Supplementary Figure 11 Prognosis of CRC samples stratified by the Estimate score. Kaplan-Meier analysis of disease-free survival on a data set of 226 CRC samples classified as having a high Estimate score (top quartile; blue line) or a low Estimate score (first to third quartiles; green line); the analysis was run on all 226 cases (a), on 138 cases that did not undergo any adjuvant therapy (b) and on 66 samples that underwent adjuvant chemotherapy after surgery (c).
14 Isella et al., Supplementary Note (a) Information sheets for three selected genes of the SSM signature for which public IHC data are available through the Human Protein Atlas portal. ZEB1 Features: Present in CRCA5 UP signature, IHC used also for CCS3 UP Not expressed by cancer cells in PDX Supportive antibody in the Human Protein Atlas (CAB058686): AMAb90510 Normal tissue expression: Colon: Rectum: Colorectal Ca expression:
15 TAGLN Features: Present in the CRCA5 UP and CCMS4 UP gene signatures Supportive antibody in the Human Protein Atlas (HPA019467): lang=it®ion=it Normal tissue expression: Colon: Rectum: Colorectal Ca expression:
16 MAP1B Features: Present in CRCA5 UP, CCMS4 UP and CCS3 UP signatures Supportive antibody in the Human Protein Atlas (HPA022275): lang=it®ion=it Normal tissue expression: Colon: Rectum: Colorectal Ca expression:
17 Isella et al., Supplementary Note (b) Database entries to the TCGA Digital Slide Archive for representative CRC cases with high or low stromal scores. The TCGA provides H&E micrographs of most samples in the Digital Slide Archive (DSA: In this document we include the sample IDs and DSA search indications (COAD or READ) for five samples in the following categories: (i) 5 cases with triple high score; (ii) 5 cases with triple low score; (iii) 5 cases with CAF-only high score; (iv) 5 cases with Endo-only high score (v) 5 cases with Leuco-only high score. Indeed, a clear correlation emerges between the scores and the corresponding morphology of the tissue section, particularly evident for cases in which only one score is high. Specifically, while samples with high CAF score alone showed prominent desmoplasia, samples with high Leuco score alone displayed marked inflammatory infiltration, and samples with high Endo score alone displayed well-evident vascularization. When all three scores were concordantly high or low the overall stroma was, respectively, overabundant or scarce. Representative micrographs are also reported here for selected cases. 1. Five samples with triple HIGH score COAD TCGA-F4-6703à Micrograph reported above READ TCGA-AF-2690 COAD TCGA-AY-6196 COAD TCGA-D COAD TCGA-A6-6651
18 2. Five samples with triple LOW score COAD TCGA-AA-A01F à Micrograph reported above COAD TCGA-AD-6888 COAD TCGA-AZ-6608 COAD TCGA-AA-A02J READ TCGA-AG-A Five samples with CAF-only HIGH score READ TCGA-AG-3893 à Micrograph reported above COAD TCGA-CM-6170 COAD TCGA-AA-3968 COAD TCGA-A COAD TCGA-CM-5349
19 4. Five samples with Endo-only HIGH score COAD TCGA-G à Micrograph reported above READ TCGA-AG-3885 COAD TCGA-AA-A00O READ TCGA-AG-3726 COAD TCGA-CK Five samples with Leuco-only HIGH score READ TCGA-AG-3892 à Micrograph reported above COAD TCGA-CK-6746 COAD TCGA-AA-3710 COAD TCGA-D COAD TCGA-AA-3856
20 Isella et al., Supplementary Note (c) Reference list for supplementary figure Salazar, R. et al. Gene expression signature to improve prognosis prediction of stage II and III colorectal cancer. J Clin Oncol 29, (2011) 2. Popovici, V. et al. Identification of a poor-prognosis BRAF-mutant-like population of patients with colon cancer. J Clin Oncol 30, (2012) 3. Loboda, A. et al. EMT is the dominant program in human colon cancer. BMC Med Genomics 4, 9 (2011). 4. de Sousa E Melo, F. et al. Methylation of cancer-stem-cell-associated Wnt target genes predicts poor prognosis in colorectal cancer patients. Cell Stem Cell 9, (2011). 5. Oh, S.C. et al. Prognostic gene expression signature associated with two molecularly distinct subtypes of colorectal cancer. Gut 61, (2012). 6. Jorissen, R.N. et al. Metastasis-Associated Gene Expression Changes Predict Poor Outcomes in Patients with Dukes Stage B and C Colorectal Cancer. Clin Cancer Res 15, (2009). 7. Smith, J.J. et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology 138, (2010). 8. Merlos-Suarez, A. et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell 8, (2011).
Nature Genetics: doi: /ng Supplementary Figure 1. Phenotypic characterization of MES- and ADRN-type cells.
 Supplementary Figure 1 Phenotypic characterization of MES- and ADRN-type cells. (a) Bright-field images showing cellular morphology of MES-type (691-MES, 700-MES, 717-MES) and ADRN-type (691-ADRN, 700-
Supplementary Figure 1 Phenotypic characterization of MES- and ADRN-type cells. (a) Bright-field images showing cellular morphology of MES-type (691-MES, 700-MES, 717-MES) and ADRN-type (691-ADRN, 700-
Supplementary Table S1. List of PTPRK-RSPO3 gene fusions in TCGA's colon cancer cohort. Chr. # of Gene 2. Chr. # of Gene 1
 Supplementary Tale S1. List of PTPRK-RSPO3 gene fusions in TCGA's colon cancer cohort TCGA Case ID Gene-1 Gene-2 Chr. # of Gene 1 Chr. # of Gene 2 Genomic coordiante of Gene 1 at fusion junction Genomic
Supplementary Tale S1. List of PTPRK-RSPO3 gene fusions in TCGA's colon cancer cohort TCGA Case ID Gene-1 Gene-2 Chr. # of Gene 1 Chr. # of Gene 2 Genomic coordiante of Gene 1 at fusion junction Genomic
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10866 a b 1 2 3 4 5 6 7 Match No Match 1 2 3 4 5 6 7 Turcan et al. Supplementary Fig.1 Concepts mapping H3K27 targets in EF CBX8 targets in EF H3K27 targets in ES SUZ12 targets in ES
doi:10.1038/nature10866 a b 1 2 3 4 5 6 7 Match No Match 1 2 3 4 5 6 7 Turcan et al. Supplementary Fig.1 Concepts mapping H3K27 targets in EF CBX8 targets in EF H3K27 targets in ES SUZ12 targets in ES
Title: Human breast cancer associated fibroblasts exhibit subtype specific gene expression profiles
 Author's response to reviews Title: Human breast cancer associated fibroblasts exhibit subtype specific gene expression profiles Authors: Julia Tchou (julia.tchou@uphs.upenn.edu) Andrew V Kossenkov (akossenkov@wistar.org)
Author's response to reviews Title: Human breast cancer associated fibroblasts exhibit subtype specific gene expression profiles Authors: Julia Tchou (julia.tchou@uphs.upenn.edu) Andrew V Kossenkov (akossenkov@wistar.org)
Supplementary Figure 1. Metabolic landscape of cancer discovery pipeline. RNAseq raw counts data of cancer and healthy tissue samples were downloaded
 Supplementary Figure 1. Metabolic landscape of cancer discovery pipeline. RNAseq raw counts data of cancer and healthy tissue samples were downloaded from TCGA and differentially expressed metabolic genes
Supplementary Figure 1. Metabolic landscape of cancer discovery pipeline. RNAseq raw counts data of cancer and healthy tissue samples were downloaded from TCGA and differentially expressed metabolic genes
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb3399 a b c d FSP DAPI 5mm mm 5mm 5mm e Correspond to melanoma in-situ Figure a DCT FSP- f MITF mm mm MlanaA melanoma in-situ DCT 5mm FSP- mm mm mm mm mm g melanoma in-situ MITF MlanaA mm mm
DOI:.38/ncb3399 a b c d FSP DAPI 5mm mm 5mm 5mm e Correspond to melanoma in-situ Figure a DCT FSP- f MITF mm mm MlanaA melanoma in-situ DCT 5mm FSP- mm mm mm mm mm g melanoma in-situ MITF MlanaA mm mm
Nature Immunology: doi: /ni Supplementary Figure 1. RNA-Seq analysis of CD8 + TILs and N-TILs.
 Supplementary Figure 1 RNA-Seq analysis of CD8 + TILs and N-TILs. (a) Schematic representation of the tumor and cell types used for the study. HNSCC, head and neck squamous cell cancer; NSCLC, non-small
Supplementary Figure 1 RNA-Seq analysis of CD8 + TILs and N-TILs. (a) Schematic representation of the tumor and cell types used for the study. HNSCC, head and neck squamous cell cancer; NSCLC, non-small
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
Nature Medicine: doi: /nm.3967
 Supplementary Figure 1. Network clustering. (a) Clustering performance as a function of inflation factor. The grey curve shows the median weighted Silhouette widths for varying inflation factors (f [1.6,
Supplementary Figure 1. Network clustering. (a) Clustering performance as a function of inflation factor. The grey curve shows the median weighted Silhouette widths for varying inflation factors (f [1.6,
a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation,
 Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH
 EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH Supplementary Figure 1. Supplementary Figure 1. Characterization of KP and KPH2 autochthonous UPS tumors. a) Genotyping of KPH2
EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH Supplementary Figure 1. Supplementary Figure 1. Characterization of KP and KPH2 autochthonous UPS tumors. a) Genotyping of KPH2
10/15/2012. Biologic Subtypes of TNBC. Topics. Topics. Histopathology Molecular pathology Clinical relevance
 Biologic Subtypes of TNBC Andrea L. Richardson M.D. Ph.D. Brigham and Women s Hospital Dana-Farber Cancer Institute Harvard Medical School Boston, MA Topics Histopathology Molecular pathology Clinical
Biologic Subtypes of TNBC Andrea L. Richardson M.D. Ph.D. Brigham and Women s Hospital Dana-Farber Cancer Institute Harvard Medical School Boston, MA Topics Histopathology Molecular pathology Clinical
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
TEB. Id4 p63 DAPI Merge. Id4 CK8 DAPI Merge
 a Duct TEB b Id4 p63 DAPI Merge Id4 CK8 DAPI Merge c d e Supplementary Figure 1. Identification of Id4-positive MECs and characterization of the Comma-D model. (a) IHC analysis of ID4 expression in the
a Duct TEB b Id4 p63 DAPI Merge Id4 CK8 DAPI Merge c d e Supplementary Figure 1. Identification of Id4-positive MECs and characterization of the Comma-D model. (a) IHC analysis of ID4 expression in the
SUPPLEMENTARY FIGURE LEGENDS
 SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1 Negative correlation between mir-375 and its predicted target genes, as demonstrated by gene set enrichment analysis (GSEA). 1 The correlation between
SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1 Negative correlation between mir-375 and its predicted target genes, as demonstrated by gene set enrichment analysis (GSEA). 1 The correlation between
Molecular subtyping: how useful is it?
 Molecular subtyping: how useful is it? Daniela E. Aust, Institute for Pathology, University Hospital Dresden, Germany Center for Molecular Tumor Diagnostics at the NCT-Partner Site Dresden CMTD Disclosure
Molecular subtyping: how useful is it? Daniela E. Aust, Institute for Pathology, University Hospital Dresden, Germany Center for Molecular Tumor Diagnostics at the NCT-Partner Site Dresden CMTD Disclosure
Cancer Biology Course. Invasion and Metastasis
 Cancer Biology Course Invasion and Metastasis 2016 Lu-Hai Wang NHRI Cancer metastasis Major problem: main reason for killing cancer patients, without it cancer can be cured or controlled. Challenging questions:
Cancer Biology Course Invasion and Metastasis 2016 Lu-Hai Wang NHRI Cancer metastasis Major problem: main reason for killing cancer patients, without it cancer can be cured or controlled. Challenging questions:
Supplementary Figure 1. A. Bar graph representing the expression levels of the 19 indicated genes in the microarrays analyses comparing human lung
 Supplementary Figure 1. A. Bar graph representing the expression levels of the 19 indicated genes in the microarrays analyses comparing human lung immortalized broncho-epithelial cells (AALE cells) expressing
Supplementary Figure 1. A. Bar graph representing the expression levels of the 19 indicated genes in the microarrays analyses comparing human lung immortalized broncho-epithelial cells (AALE cells) expressing
Supplementary Figure 1: STAT3 suppresses Kras-induced lung tumorigenesis
 Supplementary Figure 1: STAT3 suppresses Kras-induced lung tumorigenesis (a) Immunohistochemical (IHC) analysis of tyrosine 705 phosphorylation status of STAT3 (P- STAT3) in tumors and stroma (all-time
Supplementary Figure 1: STAT3 suppresses Kras-induced lung tumorigenesis (a) Immunohistochemical (IHC) analysis of tyrosine 705 phosphorylation status of STAT3 (P- STAT3) in tumors and stroma (all-time
Nature Genetics: doi: /ng Supplementary Figure 1. SEER data for male and female cancer incidence from
 Supplementary Figure 1 SEER data for male and female cancer incidence from 1975 2013. (a,b) Incidence rates of oral cavity and pharynx cancer (a) and leukemia (b) are plotted, grouped by males (blue),
Supplementary Figure 1 SEER data for male and female cancer incidence from 1975 2013. (a,b) Incidence rates of oral cavity and pharynx cancer (a) and leukemia (b) are plotted, grouped by males (blue),
SUPPLEMENTARY APPENDIX
 SUPPLEMENTARY APPENDIX 1) Supplemental Figure 1. Histopathologic Characteristics of the Tumors in the Discovery Cohort 2) Supplemental Figure 2. Incorporation of Normal Epidermal Melanocytic Signature
SUPPLEMENTARY APPENDIX 1) Supplemental Figure 1. Histopathologic Characteristics of the Tumors in the Discovery Cohort 2) Supplemental Figure 2. Incorporation of Normal Epidermal Melanocytic Signature
Supplemental Figure S1. RANK expression on human lung cancer cells.
 Supplemental Figure S1. RANK expression on human lung cancer cells. (A) Incidence and H-Scores of RANK expression determined from IHC in the indicated primary lung cancer subgroups. The overall expression
Supplemental Figure S1. RANK expression on human lung cancer cells. (A) Incidence and H-Scores of RANK expression determined from IHC in the indicated primary lung cancer subgroups. The overall expression
The 16th KJC Bioinformatics Symposium Integrative analysis identifies potential DNA methylation biomarkers for pan-cancer diagnosis and prognosis
 The 16th KJC Bioinformatics Symposium Integrative analysis identifies potential DNA methylation biomarkers for pan-cancer diagnosis and prognosis Tieliu Shi tlshi@bio.ecnu.edu.cn The Center for bioinformatics
The 16th KJC Bioinformatics Symposium Integrative analysis identifies potential DNA methylation biomarkers for pan-cancer diagnosis and prognosis Tieliu Shi tlshi@bio.ecnu.edu.cn The Center for bioinformatics
Supplementary Figure S1 Expression of mir-181b in EOC (A) Kaplan-Meier
 Supplementary Figure S1 Expression of mir-181b in EOC (A) Kaplan-Meier curves for progression-free survival (PFS) and overall survival (OS) in a cohort of patients (N=52) with stage III primary ovarian
Supplementary Figure S1 Expression of mir-181b in EOC (A) Kaplan-Meier curves for progression-free survival (PFS) and overall survival (OS) in a cohort of patients (N=52) with stage III primary ovarian
7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans.
 Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
Supplementary Table 1 Clinicopathological characteristics of 35 patients with CRCs
 Supplementary Table Clinicopathological characteristics of 35 patients with CRCs Characteristics Type-A CRC Type-B CRC P value Sex Male / Female 9 / / 8.5 Age (years) Median (range) 6. (9 86) 6.5 (9 76).95
Supplementary Table Clinicopathological characteristics of 35 patients with CRCs Characteristics Type-A CRC Type-B CRC P value Sex Male / Female 9 / / 8.5 Age (years) Median (range) 6. (9 86) 6.5 (9 76).95
Colorectal cancer Chapelle, J Clin Oncol, 2010
 Colorectal cancer Chapelle, J Clin Oncol, 2010 Early-Stage Colorectal cancer: Microsatellite instability, multigene assay & emerging molecular strategy Asit Paul, MD, PhD 11/24/15 Mr. X: A 50 yo asymptomatic
Colorectal cancer Chapelle, J Clin Oncol, 2010 Early-Stage Colorectal cancer: Microsatellite instability, multigene assay & emerging molecular strategy Asit Paul, MD, PhD 11/24/15 Mr. X: A 50 yo asymptomatic
WHAT SHOULD WE DO WITH TUMOUR BUDDING IN EARLY COLORECTAL CANCER?
 CANCER STAGING TNM and prognosis in CRC WHAT SHOULD WE DO WITH TUMOUR BUDDING IN EARLY COLORECTAL CANCER? Alessandro Lugli, MD Institute of Pathology University of Bern Switzerland Maastricht, June 19
CANCER STAGING TNM and prognosis in CRC WHAT SHOULD WE DO WITH TUMOUR BUDDING IN EARLY COLORECTAL CANCER? Alessandro Lugli, MD Institute of Pathology University of Bern Switzerland Maastricht, June 19
Supplemental Information
 Supplemental Information Prediction of Prostate Cancer Recurrence using Quantitative Phase Imaging Shamira Sridharan 1, Virgilia Macias 2, Krishnarao Tangella 3, André Kajdacsy-Balla 2 and Gabriel Popescu
Supplemental Information Prediction of Prostate Cancer Recurrence using Quantitative Phase Imaging Shamira Sridharan 1, Virgilia Macias 2, Krishnarao Tangella 3, André Kajdacsy-Balla 2 and Gabriel Popescu
Prosigna BREAST CANCER PROGNOSTIC GENE SIGNATURE ASSAY
 Prosigna BREAST CANCER PROGNOSTIC GENE SIGNATURE ASSAY Methodology The test is based on the reported 50-gene classifier algorithm originally named PAM50 and is performed on the ncounter Dx Analysis System
Prosigna BREAST CANCER PROGNOSTIC GENE SIGNATURE ASSAY Methodology The test is based on the reported 50-gene classifier algorithm originally named PAM50 and is performed on the ncounter Dx Analysis System
Prosigna BREAST CANCER PROGNOSTIC GENE SIGNATURE ASSAY
 Prosigna BREAST CANCER PROGNOSTIC GENE SIGNATURE ASSAY GENE EXPRESSION PROFILING WITH PROSIGNA What is Prosigna? Prosigna Breast Cancer Prognostic Gene Signature Assay is an FDA-approved assay which provides
Prosigna BREAST CANCER PROGNOSTIC GENE SIGNATURE ASSAY GENE EXPRESSION PROFILING WITH PROSIGNA What is Prosigna? Prosigna Breast Cancer Prognostic Gene Signature Assay is an FDA-approved assay which provides
Nature Genetics: doi: /ng Supplementary Figure 1. HOX fusions enhance self-renewal capacity.
 Supplementary Figure 1 HOX fusions enhance self-renewal capacity. Mouse bone marrow was transduced with a retrovirus carrying one of three HOX fusion genes or the empty mcherry reporter construct as described
Supplementary Figure 1 HOX fusions enhance self-renewal capacity. Mouse bone marrow was transduced with a retrovirus carrying one of three HOX fusion genes or the empty mcherry reporter construct as described
Nature Methods: doi: /nmeth.3115
 Supplementary Figure 1 Analysis of DNA methylation in a cancer cohort based on Infinium 450K data. RnBeads was used to rediscover a clinically distinct subgroup of glioblastoma patients characterized by
Supplementary Figure 1 Analysis of DNA methylation in a cancer cohort based on Infinium 450K data. RnBeads was used to rediscover a clinically distinct subgroup of glioblastoma patients characterized by
CTC in clinical studies: Latest reports on GI cancers
 CTC in clinical studies: Latest reports on GI cancers François-Clément Bidard, MD PhD GI cancers are characterized by Multimodal treatment strategies Treatments are adapted to tumor burden & prognosis
CTC in clinical studies: Latest reports on GI cancers François-Clément Bidard, MD PhD GI cancers are characterized by Multimodal treatment strategies Treatments are adapted to tumor burden & prognosis
Nature Genetics: doi: /ng Supplementary Figure 1. Somatic coding mutations identified by WES/WGS for 83 ATL cases.
 Supplementary Figure 1 Somatic coding mutations identified by WES/WGS for 83 ATL cases. (a) The percentage of targeted bases covered by at least 2, 10, 20 and 30 sequencing reads (top) and average read
Supplementary Figure 1 Somatic coding mutations identified by WES/WGS for 83 ATL cases. (a) The percentage of targeted bases covered by at least 2, 10, 20 and 30 sequencing reads (top) and average read
Supplementary Note. Nature Genetics: doi: /ng.2928
 Supplementary Note Loss of heterozygosity analysis (LOH). We used VCFtools v0.1.11 to extract only singlenucleotide variants with minimum depth of 15X and minimum mapping quality of 20 to create a ped
Supplementary Note Loss of heterozygosity analysis (LOH). We used VCFtools v0.1.11 to extract only singlenucleotide variants with minimum depth of 15X and minimum mapping quality of 20 to create a ped
Colorectal cancer: pathology
 UK NEQAS for Molecular Pathology Colorectal cancer: pathology Nick West Pathology & Tumour Biology May 2013 Colorectal cancer (CRC) 40,695 new cases in 2010 15,708 deaths Management of CRC Surgery Main
UK NEQAS for Molecular Pathology Colorectal cancer: pathology Nick West Pathology & Tumour Biology May 2013 Colorectal cancer (CRC) 40,695 new cases in 2010 15,708 deaths Management of CRC Surgery Main
Supplementary Figure 1. IDH1 and IDH2 mutation site sequences on WHO grade III
 Supplementary Materials: Supplementary Figure 1. IDH1 and IDH2 mutation site sequences on WHO grade III patient samples. Genomic DNA samples extracted from punch biopsies from either FFPE or frozen tumor
Supplementary Materials: Supplementary Figure 1. IDH1 and IDH2 mutation site sequences on WHO grade III patient samples. Genomic DNA samples extracted from punch biopsies from either FFPE or frozen tumor
Supplementary Figure 1
 Supplementary Figure 1 5 microns C7 B6 unclassified H19 C7 signal H19 guide signal H19 B6 signal C7 SNP spots H19 RNA spots B6 SNP spots colocalization H19 RNA classification Supplementary Figure 1. Allele-specific
Supplementary Figure 1 5 microns C7 B6 unclassified H19 C7 signal H19 guide signal H19 B6 signal C7 SNP spots H19 RNA spots B6 SNP spots colocalization H19 RNA classification Supplementary Figure 1. Allele-specific
Nature Genetics: doi: /ng Supplementary Figure 1. Assessment of sample purity and quality.
 Supplementary Figure 1 Assessment of sample purity and quality. (a) Hematoxylin and eosin staining of formaldehyde-fixed, paraffin-embedded sections from a human testis biopsy collected concurrently with
Supplementary Figure 1 Assessment of sample purity and quality. (a) Hematoxylin and eosin staining of formaldehyde-fixed, paraffin-embedded sections from a human testis biopsy collected concurrently with
* * * * Supplementary Figure 1. DS Lv CK HSA CK HSA. CK Col-3. CK Col-3. See overleaf for figure legend. Cancer cells
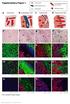 Supplementary Figure 1 Cancer cells Desmoplastic stroma Hepatocytes Pre-existing sinusoidal blood vessel New blood vessel a Normal liver b Desmoplastic HGP c Pushing HGP d Replacement HGP e f g h i DS
Supplementary Figure 1 Cancer cells Desmoplastic stroma Hepatocytes Pre-existing sinusoidal blood vessel New blood vessel a Normal liver b Desmoplastic HGP c Pushing HGP d Replacement HGP e f g h i DS
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Pan-cancer analysis of global and local DNA methylation variation a) Variations in global DNA methylation are shown as measured by averaging the genome-wide
Supplementary Figures Supplementary Figure 1. Pan-cancer analysis of global and local DNA methylation variation a) Variations in global DNA methylation are shown as measured by averaging the genome-wide
microrna Presented for: Presented by: Date:
 microrna Presented for: Presented by: Date: 2 micrornas Non protein coding, endogenous RNAs of 21-22nt length Evolutionarily conserved Regulate gene expression by binding complementary regions at 3 regions
microrna Presented for: Presented by: Date: 2 micrornas Non protein coding, endogenous RNAs of 21-22nt length Evolutionarily conserved Regulate gene expression by binding complementary regions at 3 regions
Nature Genetics: doi: /ng Supplementary Figure 1. Workflow of CDR3 sequence assembly from RNA-seq data.
 Supplementary Figure 1 Workflow of CDR3 sequence assembly from RNA-seq data. Paired-end short-read RNA-seq data were mapped to human reference genome hg19, and unmapped reads in the TCR regions were extracted
Supplementary Figure 1 Workflow of CDR3 sequence assembly from RNA-seq data. Paired-end short-read RNA-seq data were mapped to human reference genome hg19, and unmapped reads in the TCR regions were extracted
Type of file: PDF Size of file: 0 KB Title of file for HTML: Supplementary Information Description: Supplementary Figures
 Type of file: PDF Size of file: 0 KB Title of file for HTML: Supplementary Information Description: Supplementary Figures Supplementary Figure 1 mir-128-3p is highly expressed in chemoresistant, metastatic
Type of file: PDF Size of file: 0 KB Title of file for HTML: Supplementary Information Description: Supplementary Figures Supplementary Figure 1 mir-128-3p is highly expressed in chemoresistant, metastatic
fl/+ KRas;Atg5 fl/+ KRas;Atg5 fl/fl KRas;Atg5 fl/fl KRas;Atg5 Supplementary Figure 1. Gene set enrichment analyses. (a) (b)
 KRas;At KRas;At KRas;At KRas;At a b Supplementary Figure 1. Gene set enrichment analyses. (a) GO gene sets (MSigDB v3. c5) enriched in KRas;Atg5 fl/+ as compared to KRas;Atg5 fl/fl tumors using gene set
KRas;At KRas;At KRas;At KRas;At a b Supplementary Figure 1. Gene set enrichment analyses. (a) GO gene sets (MSigDB v3. c5) enriched in KRas;Atg5 fl/+ as compared to KRas;Atg5 fl/fl tumors using gene set
Nature Genetics: doi: /ng Supplementary Figure 1. Clinical timeline for the discovery WES cases.
 Supplementary Figure 1 Clinical timeline for the discovery WES cases. This illustrates the timeline of the disease events during the clinical course of each patient s disease, further indicating the available
Supplementary Figure 1 Clinical timeline for the discovery WES cases. This illustrates the timeline of the disease events during the clinical course of each patient s disease, further indicating the available
Cancer Informatics Lecture
 Cancer Informatics Lecture Mayo-UIUC Computational Genomics Course June 22, 2018 Krishna Rani Kalari Ph.D. Associate Professor 2017 MFMER 3702274-1 Outline The Cancer Genome Atlas (TCGA) Genomic Data Commons
Cancer Informatics Lecture Mayo-UIUC Computational Genomics Course June 22, 2018 Krishna Rani Kalari Ph.D. Associate Professor 2017 MFMER 3702274-1 Outline The Cancer Genome Atlas (TCGA) Genomic Data Commons
Nature Immunology: doi: /ni Supplementary Figure 1. Characteristics of SEs in T reg and T conv cells.
 Supplementary Figure 1 Characteristics of SEs in T reg and T conv cells. (a) Patterns of indicated transcription factor-binding at SEs and surrounding regions in T reg and T conv cells. Average normalized
Supplementary Figure 1 Characteristics of SEs in T reg and T conv cells. (a) Patterns of indicated transcription factor-binding at SEs and surrounding regions in T reg and T conv cells. Average normalized
SUPPLEMENTAL INFORMATIONS
 1 SUPPLEMENTAL INFORMATIONS Figure S1 Cumulative ZIKV production by testis explants over a 9 day-culture period. Viral titer values presented in Figure 1B (viral release over a 3 day-culture period measured
1 SUPPLEMENTAL INFORMATIONS Figure S1 Cumulative ZIKV production by testis explants over a 9 day-culture period. Viral titer values presented in Figure 1B (viral release over a 3 day-culture period measured
Basement membrane in lobule.
 Bahram Memar, MD Basement membrane in lobule. Normal lobule-luteal phase Normal lobule-follicular phase Lactating breast Greater than 95% are adenocarcinomas in situ carcinomas and invasive carcinomas.
Bahram Memar, MD Basement membrane in lobule. Normal lobule-luteal phase Normal lobule-follicular phase Lactating breast Greater than 95% are adenocarcinomas in situ carcinomas and invasive carcinomas.
SUPPLEMENTARY FIGURE LEGENDS
 SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1. Hippocampal sections from new-born Pten+/+ and PtenFV/FV pups were stained with haematoxylin and eosin (H&E) and were imaged at (a) low and (b) high
SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1. Hippocampal sections from new-born Pten+/+ and PtenFV/FV pups were stained with haematoxylin and eosin (H&E) and were imaged at (a) low and (b) high
mir-509-5p and mir-1243 increase the sensitivity to gemcitabine by inhibiting
 mir-509-5p and mir-1243 increase the sensitivity to gemcitabine by inhibiting epithelial-mesenchymal transition in pancreatic cancer Hidekazu Hiramoto, M.D. 1,3, Tomoki Muramatsu, Ph.D. 1, Daisuke Ichikawa,
mir-509-5p and mir-1243 increase the sensitivity to gemcitabine by inhibiting epithelial-mesenchymal transition in pancreatic cancer Hidekazu Hiramoto, M.D. 1,3, Tomoki Muramatsu, Ph.D. 1, Daisuke Ichikawa,
Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types.
 Supplementary Figure 1 Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types. (a) Pearson correlation heatmap among open chromatin profiles of different
Supplementary Figure 1 Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types. (a) Pearson correlation heatmap among open chromatin profiles of different
Circulating Stromal Cells in Immunotherapy Utilizing a Total Blood Based Biopsy
 Circulating Stromal Cells in Immunotherapy Utilizing a Total Blood Based Biopsy Cancer associated macrophage CTCs Daniel Adams Senior Research Scientist/Head of Clinical Core Laboratory Creatv MicroTech,
Circulating Stromal Cells in Immunotherapy Utilizing a Total Blood Based Biopsy Cancer associated macrophage CTCs Daniel Adams Senior Research Scientist/Head of Clinical Core Laboratory Creatv MicroTech,
Nature Immunology: doi: /ni Supplementary Figure 1. Transcriptional program of the TE and MP CD8 + T cell subsets.
 Supplementary Figure 1 Transcriptional program of the TE and MP CD8 + T cell subsets. (a) Comparison of gene expression of TE and MP CD8 + T cell subsets by microarray. Genes that are 1.5-fold upregulated
Supplementary Figure 1 Transcriptional program of the TE and MP CD8 + T cell subsets. (a) Comparison of gene expression of TE and MP CD8 + T cell subsets by microarray. Genes that are 1.5-fold upregulated
Expression of VEGF and Semaphorin Genes Define Subgroups of Triple Negative Breast Cancer
 Expression of VEGF and Semaphorin Genes Define Subgroups of Triple Negative Breast Cancer R. Joseph Bender, Feilim Mac Gabhann* Institute for Computational Medicine and Department of Biomedical Engineering,
Expression of VEGF and Semaphorin Genes Define Subgroups of Triple Negative Breast Cancer R. Joseph Bender, Feilim Mac Gabhann* Institute for Computational Medicine and Department of Biomedical Engineering,
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Experimental design and workflow utilized to generate the WMG Protein Atlas.
 Supplementary Figure 1 Experimental design and workflow utilized to generate the WMG Protein Atlas. (a) Illustration of the plant organs and nodule infection time points analyzed. (b) Proteomic workflow
Supplementary Figure 1 Experimental design and workflow utilized to generate the WMG Protein Atlas. (a) Illustration of the plant organs and nodule infection time points analyzed. (b) Proteomic workflow
Supplemental Information. Aryl Hydrocarbon Receptor Controls. Monocyte Differentiation. into Dendritic Cells versus Macrophages
 Immunity, Volume 47 Supplemental Information Aryl Hydrocarbon Receptor Controls Monocyte Differentiation into Dendritic Cells versus Macrophages Christel Goudot, Alice Coillard, Alexandra-Chloé Villani,
Immunity, Volume 47 Supplemental Information Aryl Hydrocarbon Receptor Controls Monocyte Differentiation into Dendritic Cells versus Macrophages Christel Goudot, Alice Coillard, Alexandra-Chloé Villani,
Pathologic Stage. Lymph node Stage
 ASC ASC a c Patient ID BMI Age Gleason score Non-obese PBMC 1 22.1 81 6 (3+3) PBMC 2 21.9 6 6 (3+3) PBMC 3 22 84 8 (4+4) PBMC 4 24.6 68 7 (3+4) PBMC 24. 6 (3+3) PBMC 6 24.7 73 7 (3+4) PBMC 7 23. 67 7 (3+4)
ASC ASC a c Patient ID BMI Age Gleason score Non-obese PBMC 1 22.1 81 6 (3+3) PBMC 2 21.9 6 6 (3+3) PBMC 3 22 84 8 (4+4) PBMC 4 24.6 68 7 (3+4) PBMC 24. 6 (3+3) PBMC 6 24.7 73 7 (3+4) PBMC 7 23. 67 7 (3+4)
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/375/ra41/dc1 Supplementary Materials for Actin cytoskeletal remodeling with protrusion formation is essential for heart regeneration in Hippo-deficient mice
www.sciencesignaling.org/cgi/content/full/8/375/ra41/dc1 Supplementary Materials for Actin cytoskeletal remodeling with protrusion formation is essential for heart regeneration in Hippo-deficient mice
Applying Tissue Phenomics to Colorectal Clinical Questions
 Applying Tissue Phenomics to Colorectal Clinical Questions International Symposium for Tissue Phenomics San Francisco October 2014 Peter Caie Senior Research Fellow University of St Andrews Systems Pathology
Applying Tissue Phenomics to Colorectal Clinical Questions International Symposium for Tissue Phenomics San Francisco October 2014 Peter Caie Senior Research Fellow University of St Andrews Systems Pathology
SUPPLEMENTARY INFORMATION
 doi:.38/nature8975 SUPPLEMENTAL TEXT Unique association of HOTAIR with patient outcome To determine whether the expression of other HOX lincrnas in addition to HOTAIR can predict patient outcome, we measured
doi:.38/nature8975 SUPPLEMENTAL TEXT Unique association of HOTAIR with patient outcome To determine whether the expression of other HOX lincrnas in addition to HOTAIR can predict patient outcome, we measured
Multi-omics data integration colon cancer using proteogenomics approach
 Dept. of Medical Oncology Multi-omics data integration colon cancer using proteogenomics approach DTL Focus meeting, 29 August 2016 Thang Pham OncoProteomics Laboratory, Dept. of Medical Oncology VU University
Dept. of Medical Oncology Multi-omics data integration colon cancer using proteogenomics approach DTL Focus meeting, 29 August 2016 Thang Pham OncoProteomics Laboratory, Dept. of Medical Oncology VU University
NGS in tissue and liquid biopsy
 NGS in tissue and liquid biopsy Ana Vivancos, PhD Referencias So, why NGS in the clinics? 2000 Sanger Sequencing (1977-) 2016 NGS (2006-) ABIPrism (Applied Biosystems) Up to 2304 per day (96 sequences
NGS in tissue and liquid biopsy Ana Vivancos, PhD Referencias So, why NGS in the clinics? 2000 Sanger Sequencing (1977-) 2016 NGS (2006-) ABIPrism (Applied Biosystems) Up to 2304 per day (96 sequences
gliomas. Fetal brain expected who each low-
 Supplementary Figure S1. Grade-specificity aberrant expression of HOXA genes in gliomas. (A) Representative RT-PCR analyses of HOXA gene expression in human astrocytomas. Exemplified glioma samples include
Supplementary Figure S1. Grade-specificity aberrant expression of HOXA genes in gliomas. (A) Representative RT-PCR analyses of HOXA gene expression in human astrocytomas. Exemplified glioma samples include
Agilent GeneSpring/MPP Metadata Analysis Framework
 Agilent GeneSpring/MPP Metadata Analysis Framework Technical Overview Authors Srikanthi R., Pritha Aggarwal, Durairaj R., Maria Kammerer, and Pramila Tata Strand Life Sciences Bangalore, India Michael
Agilent GeneSpring/MPP Metadata Analysis Framework Technical Overview Authors Srikanthi R., Pritha Aggarwal, Durairaj R., Maria Kammerer, and Pramila Tata Strand Life Sciences Bangalore, India Michael
Supplementary Tables. Supplementary Figures
 Supplementary Files for Zehir, Benayed et al. Mutational Landscape of Metastatic Cancer Revealed from Prospective Clinical Sequencing of 10,000 Patients Supplementary Tables Supplementary Table 1: Sample
Supplementary Files for Zehir, Benayed et al. Mutational Landscape of Metastatic Cancer Revealed from Prospective Clinical Sequencing of 10,000 Patients Supplementary Tables Supplementary Table 1: Sample
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2638 Figure S1 Morphological characteristics of fetal testes and ovaries from 6.5-20 developmental weeks. Representative images of Hematoxylin and Eosin staining of testes and ovaries over
DOI: 10.1038/ncb2638 Figure S1 Morphological characteristics of fetal testes and ovaries from 6.5-20 developmental weeks. Representative images of Hematoxylin and Eosin staining of testes and ovaries over
SUPPLEMENTAL INFORMATION
 SUPPLEMENTAL INFORMATION GO term analysis of differentially methylated SUMIs. GO term analysis of the 458 SUMIs with the largest differential methylation between human and chimp shows that they are more
SUPPLEMENTAL INFORMATION GO term analysis of differentially methylated SUMIs. GO term analysis of the 458 SUMIs with the largest differential methylation between human and chimp shows that they are more
Clasificación Molecular del Cáncer de Próstata. JM Piulats
 Clasificación Molecular del Cáncer de Próstata JM Piulats Introduction The Gleason score is the major method for prostate cancer tissue grading and the most important prognostic factor in this disease.
Clasificación Molecular del Cáncer de Próstata JM Piulats Introduction The Gleason score is the major method for prostate cancer tissue grading and the most important prognostic factor in this disease.
Molecular in vitro diagnostic test for the quantitative detection of the mrna expression of ERBB2, ESR1, PGR and MKI67 in breast cancer tissue.
 Innovation for your breast cancer diagnostics PGR G ATA G C G A C G AT C G A A A G A A G T TA G ATA G C G A C G AT C G A A A G A A G T TA G ATA G C G A C G AT C G A A A G A A G T TA G ATA G C G A C ERBB2
Innovation for your breast cancer diagnostics PGR G ATA G C G A C G AT C G A A A G A A G T TA G ATA G C G A C G AT C G A A A G A A G T TA G ATA G C G A C G AT C G A A A G A A G T TA G ATA G C G A C ERBB2
SUPPLEMENTARY FIGURES: Supplementary Figure 1
 SUPPLEMENTARY FIGURES: Supplementary Figure 1 Supplementary Figure 1. Glioblastoma 5hmC quantified by paired BS and oxbs treated DNA hybridized to Infinium DNA methylation arrays. Workflow depicts analytic
SUPPLEMENTARY FIGURES: Supplementary Figure 1 Supplementary Figure 1. Glioblastoma 5hmC quantified by paired BS and oxbs treated DNA hybridized to Infinium DNA methylation arrays. Workflow depicts analytic
Supplementary Figure 1: LUMP Leukocytes unmethylabon to infer tumor purity
 Supplementary Figure 1: LUMP Leukocytes unmethylabon to infer tumor purity A Consistently unmethylated sites (30%) in 21 cancer types 174,696
Supplementary Figure 1: LUMP Leukocytes unmethylabon to infer tumor purity A Consistently unmethylated sites (30%) in 21 cancer types 174,696
Supplementary Figure 1: Comparison of acgh-based and expression-based CNA analysis of tumors from breast cancer GEMMs.
 Supplementary Figure 1: Comparison of acgh-based and expression-based CNA analysis of tumors from breast cancer GEMMs. (a) CNA analysis of expression microarray data obtained from 15 tumors in the SV40Tag
Supplementary Figure 1: Comparison of acgh-based and expression-based CNA analysis of tumors from breast cancer GEMMs. (a) CNA analysis of expression microarray data obtained from 15 tumors in the SV40Tag
Triple Negative Breast Cancer
 Triple Negative Breast Cancer Prof. Dr. Pornchai O-charoenrat Division of Head-Neck & Breast Surgery Department of Surgery Faculty of Medicine Siriraj Hospital Breast Cancer Classification Traditional
Triple Negative Breast Cancer Prof. Dr. Pornchai O-charoenrat Division of Head-Neck & Breast Surgery Department of Surgery Faculty of Medicine Siriraj Hospital Breast Cancer Classification Traditional
Layered-IHC (L-IHC): A novel and robust approach to multiplexed immunohistochemistry So many markers and so little tissue
 Page 1 The need for multiplex detection of tissue biomarkers. There is a constant and growing demand for increased biomarker analysis in human tissue specimens. Analysis of tissue biomarkers is key to
Page 1 The need for multiplex detection of tissue biomarkers. There is a constant and growing demand for increased biomarker analysis in human tissue specimens. Analysis of tissue biomarkers is key to
Supplementary Figure 1: Tissue of Origin analysis on 152 cell lines. (a) Heatmap representation of the 30 Tissue scores for the 152 cell lines.
 Supplementary Figure 1: Tissue of Origin analysis on 152 cell lines. (a) Heatmap representation of the 30 Tissue scores for the 152 cell lines. The scores summarize the global expression of the tissue
Supplementary Figure 1: Tissue of Origin analysis on 152 cell lines. (a) Heatmap representation of the 30 Tissue scores for the 152 cell lines. The scores summarize the global expression of the tissue
The Avatar System TM Yields Biologically Relevant Results
 Application Note The Avatar System TM Yields Biologically Relevant Results Liquid biopsies stand to revolutionize the cancer field, enabling early detection and noninvasive monitoring of tumors. In the
Application Note The Avatar System TM Yields Biologically Relevant Results Liquid biopsies stand to revolutionize the cancer field, enabling early detection and noninvasive monitoring of tumors. In the
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Effect of HSP90 inhibition on expression of endogenous retroviruses. (a) Inducible shrna-mediated Hsp90 silencing in mouse ESCs. Immunoblots of total cell extract expressing the
Supplementary Figure 1 Effect of HSP90 inhibition on expression of endogenous retroviruses. (a) Inducible shrna-mediated Hsp90 silencing in mouse ESCs. Immunoblots of total cell extract expressing the
RNA preparation from extracted paraffin cores:
 Supplementary methods, Nielsen et al., A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor positive breast cancer.
Supplementary methods, Nielsen et al., A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor positive breast cancer.
Reviewing Immunotherapy for Bladder Carcinoma In Situ
 Reviewing Immunotherapy for Bladder Carcinoma In Situ Samir Bidnur Dept of Urologic Sciences, Grand Rounds March 1 st, 2017 Checkpoint Inhibition and Bladder Cancer, an evolving story with immunotherapy
Reviewing Immunotherapy for Bladder Carcinoma In Situ Samir Bidnur Dept of Urologic Sciences, Grand Rounds March 1 st, 2017 Checkpoint Inhibition and Bladder Cancer, an evolving story with immunotherapy
Digitizing the Proteomes From Big Tissue Biobanks
 Digitizing the Proteomes From Big Tissue Biobanks Analyzing 24 Proteomes Per Day by Microflow SWATH Acquisition and Spectronaut Pulsar Analysis Jan Muntel 1, Nick Morrice 2, Roland M. Bruderer 1, Lukas
Digitizing the Proteomes From Big Tissue Biobanks Analyzing 24 Proteomes Per Day by Microflow SWATH Acquisition and Spectronaut Pulsar Analysis Jan Muntel 1, Nick Morrice 2, Roland M. Bruderer 1, Lukas
Supplementary. properties of. network types. randomly sampled. subsets (75%
 Supplementary Information Gene co-expression network analysis reveals common system-level prognostic genes across cancer types properties of Supplementary Figure 1 The robustness and overlap of prognostic
Supplementary Information Gene co-expression network analysis reveals common system-level prognostic genes across cancer types properties of Supplementary Figure 1 The robustness and overlap of prognostic
High expression of fibroblast activation protein is an adverse prognosticator in gastric cancer.
 Biomedical Research 2017; 28 (18): 7779-7783 ISSN 0970-938X www.biomedres.info High expression of fibroblast activation protein is an adverse prognosticator in gastric cancer. Hu Song 1, Qi-yu Liu 2, Zhi-wei
Biomedical Research 2017; 28 (18): 7779-7783 ISSN 0970-938X www.biomedres.info High expression of fibroblast activation protein is an adverse prognosticator in gastric cancer. Hu Song 1, Qi-yu Liu 2, Zhi-wei
Nature Methods: doi: /nmeth Supplementary Figure 1
 Supplementary Figure 1 Finite-element analysis of cell cluster dynamics in different cluster trap architectures. (a) Cluster-Chip (b) Filter (c) A structure identical to the Cluster-Chip except that one
Supplementary Figure 1 Finite-element analysis of cell cluster dynamics in different cluster trap architectures. (a) Cluster-Chip (b) Filter (c) A structure identical to the Cluster-Chip except that one
Supplementary Material. for. A cell-centered meta-analysis reveals baseline predictors of anti-tnf nonresponse. in biopsy and blood of IBD patients
 Supplementary Material for A cell-centered meta-analysis reveals baseline predictors of anti-tnf nonresponse in biopsy and blood of IBD patients This document includes Supplementary Methods, Figures and
Supplementary Material for A cell-centered meta-analysis reveals baseline predictors of anti-tnf nonresponse in biopsy and blood of IBD patients This document includes Supplementary Methods, Figures and
Adjuvan Chemotherapy in Breast Cancer
 Adjuvan Chemotherapy in Breast Cancer Prof Dr Adnan Aydıner Istanbul University, Oncology Institute aa1 Slide 1 aa1 adnan aydiner; 17.02.2008 15-Year Reductions in Recurrence and Disease-Specific Mortality
Adjuvan Chemotherapy in Breast Cancer Prof Dr Adnan Aydıner Istanbul University, Oncology Institute aa1 Slide 1 aa1 adnan aydiner; 17.02.2008 15-Year Reductions in Recurrence and Disease-Specific Mortality
RNA-Seq Preparation Comparision Summary: Lexogen, Standard, NEB
 RNA-Seq Preparation Comparision Summary: Lexogen, Standard, NEB CSF-NGS January 22, 214 Contents 1 Introduction 1 2 Experimental Details 1 3 Results And Discussion 1 3.1 ERCC spike ins............................................
RNA-Seq Preparation Comparision Summary: Lexogen, Standard, NEB CSF-NGS January 22, 214 Contents 1 Introduction 1 2 Experimental Details 1 3 Results And Discussion 1 3.1 ERCC spike ins............................................
fraction (soluble sugars, starch and total NSC). Conditional and marginal R 2 values are given for
 1 2 Martínez-Vilalta, Sala, Asensio, Galiano, Hoch, Palacio, Piper and Lloret. Dynamics of non- structural carbohydrates in terrestrial plants: a global synthesis. Ecological Monographs. 3 4 APPENDIX S2.
1 2 Martínez-Vilalta, Sala, Asensio, Galiano, Hoch, Palacio, Piper and Lloret. Dynamics of non- structural carbohydrates in terrestrial plants: a global synthesis. Ecological Monographs. 3 4 APPENDIX S2.
Next-Gen Analytics in Digital Pathology
 Next-Gen Analytics in Digital Pathology Cliff Hoyt, CTO Cambridge Research & Instrumentation April 29, 2010 Seeing life in a new light 1 Digital Pathology Today Acquisition, storage, dissemination, remote
Next-Gen Analytics in Digital Pathology Cliff Hoyt, CTO Cambridge Research & Instrumentation April 29, 2010 Seeing life in a new light 1 Digital Pathology Today Acquisition, storage, dissemination, remote
2.28E E AKT/CAT P-3-4:
 Supplemental Table 1: IPA network analysis of the microarray data presented in Figure 6 showing the most significant molecular and cellular function classifications along with the respective number of
Supplemental Table 1: IPA network analysis of the microarray data presented in Figure 6 showing the most significant molecular and cellular function classifications along with the respective number of
Supplementary Material
 Supplementary Material Fig. S1. Gpa33 -/- mice do not express Gpa33 mrna or GPA33 protein. (A) The Gpa33 null allele contains a premature translational stop codon (STOP) in exon 2, and almost all of the
Supplementary Material Fig. S1. Gpa33 -/- mice do not express Gpa33 mrna or GPA33 protein. (A) The Gpa33 null allele contains a premature translational stop codon (STOP) in exon 2, and almost all of the
Nature Neuroscience doi: /nn Supplementary Figure 1. Characterization of viral injections.
 Supplementary Figure 1 Characterization of viral injections. (a) Dorsal view of a mouse brain (dashed white outline) after receiving a large, unilateral thalamic injection (~100 nl); demonstrating that
Supplementary Figure 1 Characterization of viral injections. (a) Dorsal view of a mouse brain (dashed white outline) after receiving a large, unilateral thalamic injection (~100 nl); demonstrating that
ARTICLE RESEARCH. Macmillan Publishers Limited. All rights reserved
 Extended Data Figure 6 Annotation of drivers based on clinical characteristics and co-occurrence patterns. a, Putative drivers affecting greater than 10 patients were assessed for enrichment in IGHV mutated
Extended Data Figure 6 Annotation of drivers based on clinical characteristics and co-occurrence patterns. a, Putative drivers affecting greater than 10 patients were assessed for enrichment in IGHV mutated
Published in: Cancer Immunology Research. Document Version: Peer reviewed version
 Immune-derived PD-L1 gene expression defines a subgroup of stage II/III colorectal cancer patients with favorable prognosis that may be harmed by adjuvant chemotherapy Dunne, P. D., McArt, D. G., O'Reilly,
Immune-derived PD-L1 gene expression defines a subgroup of stage II/III colorectal cancer patients with favorable prognosis that may be harmed by adjuvant chemotherapy Dunne, P. D., McArt, D. G., O'Reilly,
Nature Structural & Molecular Biology: doi: /nsmb.2419
 Supplementary Figure 1 Mapped sequence reads and nucleosome occupancies. (a) Distribution of sequencing reads on the mouse reference genome for chromosome 14 as an example. The number of reads in a 1 Mb
Supplementary Figure 1 Mapped sequence reads and nucleosome occupancies. (a) Distribution of sequencing reads on the mouse reference genome for chromosome 14 as an example. The number of reads in a 1 Mb
Cellecta Overview. Started Operations in 2007 Headquarters: Mountain View, CA
 Cellecta Overview Started Operations in 2007 Headquarters: Mountain View, CA Focus: Development of flexible, scalable, and broadly parallel genetic screening assays to expedite the discovery and characterization
Cellecta Overview Started Operations in 2007 Headquarters: Mountain View, CA Focus: Development of flexible, scalable, and broadly parallel genetic screening assays to expedite the discovery and characterization
Case Studies on High Throughput Gene Expression Data Kun Huang, PhD Raghu Machiraju, PhD
 Case Studies on High Throughput Gene Expression Data Kun Huang, PhD Raghu Machiraju, PhD Department of Biomedical Informatics Department of Computer Science and Engineering The Ohio State University Review
Case Studies on High Throughput Gene Expression Data Kun Huang, PhD Raghu Machiraju, PhD Department of Biomedical Informatics Department of Computer Science and Engineering The Ohio State University Review
