2005 LANDES BIOSCIENCE. DO NOT DISTRIBUTE.
|
|
|
- Julian Owen
- 5 years ago
- Views:
Transcription
1 [Autophagy 1:2, 84-91; July/August/September 2005]; 2005 Landes Bioscience Research Paper Lysosomal Turnover, but Not a Cellular Level, of Endogenous LC3 is a Marker for Autophagy Isei Tanida Naoko Minematsu-Ikeguchi Takashi Ueno Eiki Kominami* Department of Biochemistry; Juntendo University School of Medicine; Tokyo, Japan *Correspondence to: Eiki Kominami; Department of Biochemistry; Juntendo University School of Medicine; Hongo, Bunkyo-ku; Tokyo Japan; Tel.: ; Fax: ; kominami@med.juntendo.ac.jp Received 03/30/05; Accepted 03/31/05 Previously published online as an Autophagy E-publication: KEY WORDS autophagy, Atg8/Apg8/Aut7, autophagosome, LC3, lysosome, lysosomal protease, cathepsin, E64d, pepstatin A ABBREVIATIONS CBB DMEM GFP LC3 LC3-I LC3-II PAGE PVDF S6RP SDS ACKNOWLEDGEMENTS Coomassie brilliant blue Dulbecco s modified Eagle medium green fluorescent protein microtubule-associated protein 1 light chain 3 soluble unlipidated form of LC3 membrane-bound LC3-phospholipid conjugate modified by endogenous Atg7 and Atg3 polyacrylamide gel electrophoresis polyvinylidene fluoride S6 ribosomal protein sodium dodecyl sulfate This work was supported, in part, by grants-inaid (to I.T.), (to T.U.) and (to E.K.) for Scientific Research, and grant-in-aid (to E.K.) for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan, and The Science Research Promotion Fund from the Japan Private School Promotion Foundation (to E.K.). ABSTRACT During starvation-induced autophagy in mammals, autophagosomes form and fuse with lysosomes, leading to the degradation of the intra-autophagosomal contents by lysosomal proteases. During the formation of autophagosomes, LC3 is lipidated, and this LC3-phospholipid conjugate (LC3-II) is localized on autophagosomes and autolysosomes. While intra-autophagosomal LC3-II may be degraded by lysosomal hydrolases, recent studies have regarded LC3-II accumulation as marker of autophagy. The effect of lysosomal turnover of endogenous LC3-II in this process, however, has not been considered. We therefore investigated the lysosomal turnover of endogenous LC3-II during starvationinduced autophagy using E64d and pepstatin A, which inhibit lysosomal proteases, including cathepsins B, D and L. We found that endogenous LC3-II significantly accumulated in the presence of E64d and pepstatin A under starvation conditions, increasing about 3.5 fold in HEK293 cells and about 6.7 fold in HeLa cells compared with that in their absence, whereas the amount of LC3-II in their absence is cell-line dependent. Morphological analyses indicated that endogenous LC3-positive puncta and autolysosomes increased in HeLa cells under starvation conditions in the presence of these inhibitors. These results indicate that endogenous LC3-II is considerably degraded by lysosomal hydrolases after formation of autolysosomes, and suggest that lysosomal turnover, not a transient amount, of this protein reflects starvation-induced autophagic activity. INTRODUCTION Autophagy, the bulk degradation of intracellular components including organelles, is induced in cells under starvation conditions. 1-4 In mammals, autophagy shows significant associations with neurodegenerative diseases, cancer, cardiomyopathies, aging, type II programmed cell death, bacterial invasion, MHC class II antigen-presentation, and early neonatal stage, as well as cellular maintenance. 3,5-13 During autophagy, cup-shaped preautophagosomes (or phagophores) enwrap cytoplasmic components, including organelles, and become enclosed, forming autophagosomes, also known as autophagic vesicles containing undegraded cytoplasm (Avi) (Fig. 1, Step 1) Autophagosomes fuse with lysosomes to form autolysosomes, the degradative form of autophagic vacuoles (Avd) (Fig. 1, Step 2), in which intra-lumenal components are degraded by lysosomal hydrolases (Fig. 1, Step 3). Under starvation conditions in perfused rat liver, the populations of Avi and Avd attain a steady state within about 20 min. The autophagy-related genes, ATG/APG/AUT/CVT genes, have been isolated and characterized in yeast and mammals. 2,20,21 In mammalian cells, LC3, one of the homologues of yeast Atg8/Apg8/Aut7, is modified via a ubiquitylation-like system. 22,23 The carboxyl terminal region of LC3 is cleaved, generating a soluble form known as LC3-I and exposing a carboxyl terminal glycine essential for further reactions. 22,24,25 LC3-I, in turn, is modified to a membrane-bound form, LC3-II (a LC3-phospholipid conjugate), by mammalian Atg7/Apg7/Cvt2/Gsa7 and Atg3/Apg3/Aut1 homologues, which are E1- and E2-like enzymes, respectively 22,25-27 to localize to autophagosomes and autolysosomes. 22 Thus, the amount of LC3-II in mammalian cells is a good early marker for the formation of autophagosomes (Fig. 1, Step 1). 22 Thereafter, autophagosomes fuse with lysosomes to degrade enwrapped cytosolic components, forming autolysosomes (Fig. 1, Step 2). Therefore, it is probable that the degradation of intra-lumenal LC3-II will occur (Fig. 1, Step 3). However, there is little information available regarding the degradation of endogenous LC3-II in the autolysosomes during starvation-induced autophagy LANDES BIOSCIENCE. DO NOT DISTRIBUTE. 84 Autophagy 2005; Vol. 1 Issue 2
2 Several lines of evidence have indicated that intra-autophagosomal components are rapidly degraded by the lysosomal proteolytic system after formation of autophagosomes during autophagy. Morphological studies have indicated that, in primary cultured hepatocytes, only a few autophagosomes and autolysosomes are observed under starvation-conditions without inhibition of lysosomal hydrolases, 18,19,28,29 probably because the lysosomal fusion and degradation of autophagosomes occurs rapidly under starvation conditions. Therefore, autophagy, including pexophagy (the degradation of peroxisomes by the autophagic machinery), has been studied morphologically and biochemically by inhibiting lysosomal function using agents that inhibit cathepsins or lysosomal acidification. 18,19,28,30 Autolysosomes are easily isolated from leupeptin-treated starved rat liver, but not from untreated starved rat liver. These results have indicated that lysosomal degradation of intraautophagosomal components occurs rapidly after fusion of autophagosomes with lysosomes to form autolysosomes. Furthermore, in the yeast Sachharomyces cerevisiae, the vacuolar proteolytic system contributes considerably to the degradation of engulfed compartments during autophagy. 20,31-34 Accumulation of autophagic bodies in the vacuoles (yeast vacuoles containing autophagic bodies correspond to mammalian autolysosomes) is observed under starvation conditions in a pep4 or prb1 mutant, or following treatment with the serine protease inhibitor phenylmethylsulfonyl fluoride, but not in wild type yeast. 31 Pep4 (proteinase A) is a vacuolar aspartic protease required for the posttranslational precursor-maturation of vacuolar proteinases, whereas Prb1 (proteinase B) is a vacuolar serine protease involved in protein degradation in the vacuoles. 38,39 Therefore, these results indicate that the degradation of intraautophagosomal components in the vacuoles occurs very rapidly during starvation-induced autophagy in yeast. Atg8-PE, a yeast counterpart of LC3-II, is an autophagosomal marker localized on autophagosomes during both the cytosolic and lumenal faces. 32 Atg8-positive autophagic bodies accumulate in the vacuoles of pep4 mutant cells under starvation conditions, while little accumulates in the vacuoles of wild type yeast. 31,32,40 Taken together, these results indicate that, in wild type yeast, intra-autophagosomal Atg8-PE is dramatically degraded in the vacuoles during autophagy. Recently, the cellular level of LC3-II has been regarded as an indicator that reflects the activation of autophagy, without considering the lysosomal degradation of LC3-II. If the lysosomal degradation of LC3-II will be much slower than the formation of autophagosomes during autophagy, the cellular level of LC3-II reflects an activation of autophagy. However, considering the contribution of the lysosomal/vacuolar proteolytic system during autophagy in mammals and yeast as described above, we hypothesized that intra-autophagosomal Figure 1. Schematic representation of the relationship between formation of autophagosomes and autolysosomes and the amount of LC3-II during mammalian autophagy. Under starvation conditions, a cup-shaped preautophagosome (or isolation membrane) is formed and engulfs cytosolic compartments including organelles (Step 1). The preautophagosome finally becomes enclosed, forming an autophagosome as a double membrane structure. During this step, cytosolic LC3 (closed dot) is lipidated, becoming localized on autophagosomes as an LC3-phospholipid conjugate (LC3-II) on both the cytosolic and intra-lumenal faces. After the formation of the autophagosome, it fuses with a lysosome to form a autolysosome (Step 2). This fusion is mediated by a small GTPase, rab7. Following this fusion, cytosolically oriented LC3-II on the autophagosome is delipidated by human Atg4B and released into the cytosol. After the formation of the autolysosome, intra-autophagosomal components are rapidly degraded by lysosomal hydrolases, including cathepsins B, D and L (Step 3). Intra-autophagosomal LC3-II is also degraded at the same time. When E64d and pepstatin A are added to the medium, the proteolytic activities of these cathepsins are inhibited, and autolysosomes containing cytosolic components accumulate. LC3-II will be rapidly degraded by lysosomal proteases during autophagy following formation of autolysosomes. At present, little work is known about the lysosomal turnover of endogenous LC3-II under starvation conditions. We therefore examined the contribution of lysosomal turnover of endogenous LC3-II during starvationinduced autophagy using the protease inhibitors E64d and pepstatin A, and found that its lysosomal turnover dramatically occurs after autophagosome-formation during starvation-induced autophagy. MATERIALS AND METHODS Cell culture. HeLa and HEK293 cell lines, purchased from the ATCC (Manassas, VA), were cultured in 60-mm dishes in DMEM (Invitrogen Corp., Carlsbad, CA, USA) containing 10% fetal calf serum (FCS; Invitrogen) in a humidified 5% CO 2 atmosphere at 37 C. For starvation, cells were incubated in Krebs-Ringer medium at 37 C for the indicated times. 29,41 Materials, biochemical and molecular biological techniques. E64d and pepstatin A were purchased from the Peptide Institute (Osaka, Japan). Molecular biological and biochemical techniques were performed as described previously. 42 Coomassie brilliant blue staining of proteins on a polyvinylidene fluoride (PVDF) membrane after immunoblotting was performed using a Quick CBB kit (Wako, Tokyo, Japan). Total lysates of human cell lines cultured in DMEM containing 10% FCS were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies. Antiserum against thioredoxin-human LC3 was raised in rabbits, and anti-human LC3 antibody was purified by affinity chromatography on a glutathione-s-transferase-immobilized human LC3-Sepharose column Autophagy 85
3 A B Figure 2. Lysosomal degradation of endogenous LC3 during autophagy. Endogenous LC3 in (A). HEK293 or (B) HeLa cells during autophagy. For Nutrient-rich conditions, cells were cultured in DMEM medium containing 10% FCS. Where indicated, cells were treated with the protease inhibitors, E64d (10 µg/ml) and pepstatin A (10 µg/ml) (Inhibitors +) for 4 h, or, as a negative control (Inhibitors -), with the solvent, dimethylsulfoxide. For Starvation conditions, cells were incubated for 4 hrs in Krebs-Ringer medium (KRB) in the presence (+) or absence (-) of protease inhibitors. The cells were lysed, total proteins (10 µg per lane) were separated by SDS-PAGE, and endogenous LC3 in the lysates was recognized by immunoblotting with an anti-lc3 antibody (WB: anti-lc3). LC3-I, soluble form of LC3; LC3-II, membrane-bound form of LC3. Antisera to phosphorylated and dephosphorylated forms of Tor, S6K and S6RP were used to monitor Tor activity. (C) Densitometric analyses of endogenous LC3-I and LC3-II in HEK293 (A) and HeLa (B) cells. The relative amounts of LC3-I (open bars) and LC3-II (closed bars) are shown. Bars indicate standard errors. D. Time course of the amount of endogenous LC3-II under starvation conditions in the absence (open box) or presence (closed box) of E64d (10 µg/ml) and pepstatin A (10 µg/ml) in HEK293 cells. C D The antibody did not cross-react with GATE-16 or GABARAP, the other mammalian Atg8 homologues. Affinity purified polyclonal rabbit antibodies against human Atg7 and Atg3 have been described. 26,27 Antiserum against glutathione-s-transferase-human beclin 1 43 was raised in rabbits, and the antibodies were purified by affinity chromatography on a thioredoxin-beclin 1- immobilized Sepharose column. Monoclonal mouse anti-green fluorescent protein (GFP) antibody was purchased from BD Biosciences (Palo Alto, CA). Polyclonal rabbit antibodies against mtor, phospho-mtor (Ser2448), p70 S6 kinase, phospho-p70 S6 kinase (Thr389), phospho-p70 S6 kinase (Thr421/Ser424), S6 ribosomal protein, and phospho-s6 ribosomal protein (Ser235/236) were purchased from Cell Signaling (Beverly, MA). Immunoblot analyses. Protein concentrations were determined using the BCA protein assay reagent (Pierce, Rockford, IL). Immunoblot analyses were carried out according to standard protocols using a chemiluminescent method with SuperSignal West Dura 86 Autophagy 2005; Vol. 1 Issue 2
4 Figure 3. Accumulation of endogenous LC3-positive puncta under starvation conditions in the presence of E64d and pepstatin A in HeLa cells. HeLa cells were cultured in DMEM containing 10% FCS (Nutrient-rich, A and B) or transferred to KRB medium for 4 h (Starvation, C, D and G J). E64d & pepstatin A (B, D, F, H and J) indicates that cells were incubated for 4 h with these protease inhibitors, whereas Non-treated (A, C and E) indicates addition of a solvent instead of these inhibitors. The cells were fixed and permeabilized with digitonin, and endogenous LC3 was recognized using Alexa Flour594-conjugated anti-human LC3 antibody. Intracellular fluorescence of endogenous LC3 in HeLa cells was observed. As a negative control, rabbit IgG was employed instead of anti-human LC3 antibody. A series of Z-scanned fluorescent images was deconvoluted by a program for 2D blind deconvolution with Aqua C-imaging software (Hamamatsu Photonics) (G J). Images (I and J) are 4 fold magnifications of the images shown in boxes with dotted lines in (G and H), respectively. An arrowhead indicates a cup-shaped preautophagosome, and an arrow indicates an autophagosome/autolysosome. A C B D Extended Duration Substrate or SuperSignal West Pico Chemiluminescent Substrate (Pierce). Immunofluorescence analysis. Endogenous LC3 in HeLa cells was stained using Alexa Flour594-conjugated anti-lc3 antibody as previously described, 44 and fluorescence was monitored with a Zeiss Axioplan2 fluorescence microscope (Carl Zeiss, Thornwood, NY) and ORCA-ER CCD camera (Hamamatsu Photonics, Tokyo, Japan). For 2D blind deconvolution of Z-scanned images, a series of fluorescent images were obtained with a Zeiss Axioplan2 fluorescence microscope (Carl Zeiss) and Aqua C-imaging system (Hamamatsu Photonics). Electron microscopy analysis. HeLa cells cultured on cover glass were prefixed in 4% glutaraldehyde in PBS at 4 C, treated with 1% OsO 4 for 2 h at 4 C, dehydrated in a graded series of ethanol, and flat embedded in epon. Ultra-thin sections were doubly stained with uranyl acetate and observed using a JEOL JEM2000EX electron microscope at 100kV (Peabody, MA). Other techniques. Quantification of the images was performed using Aqua C-imaging (Hamamatsu Photonics) and NIH image (Dr. W. Raeband, National Institute of Health, Bethesda, MD, USA) programs on Endeavor (EPSON direct, Tokyo, Japan) and Macintosh G4 (Apple Computer, NY) computers, respectively. RESULTS Endogenous LC3-II markedly accumulates in the presence of E64d and pepstatin A during starvation-induced autophagy in HEK293 and HeLa cells. During autophagy, intra-autophagosomal components are degraded by lysosomal hydrolases following the formation of autophagosomes and their fusion with lysosomes to form autolysosomes. Therefore, a significant amount of endogenous LC3-II on autophagosomes may be rapidly degraded by lysosomal hydrolases. To investigate the lysosomal turnover of intra-autophagosomal LC3-II, we employed two protease inhibitors, E64d, a membrane-permeable inhibitor of cathepsins B, H, and L, and pepstatin A, an inhibitor of cathepsins D and E. 29,47,48 Preliminary experiments with HEK293, HeLa, P19EC, C2C12, MPC, and B16-F1 cells and rat primary cultured hepatocytes treated with 10 µg/ml of E64d and 10 µg/ml of pepstatin A indicated that these inhibitors have little effect on cell viability, morphology and growth under nutrient-rich conditions for at least 24 h. Using E64d (10 µg/ml) and pepstatin A (10 µg/ml), we sought to determine the contribution of lysosomal degradation of LC3-II under starvation conditions. Under nutrient-rich conditions in HEK293 cells, endogenous LC3 is mainly present as the cytosolic form (LC3-I) (Fig. 2A, lane 1). Following the incubation of HEK293 cells in starvation medium, the amount of endogenous LC3-I decreased while that of LC3-II increased (Fig. 2A lane 3 vs. lane 1), indicating that LC3-lipidation E G I F H J Autophagy 87
5 A B D F Figure 4. Accumulation of autolysosomes under starvation conditions in the presence of protease inhibitors in HeLa cells. Representative electron microscopic images of the cytoplasmic regions of cells are shown. HeLa cells were cultured in DMEM containing 10% FCS (Nutrient-rich, A), or incubated in KRB medium in the absence (B and C) or presence (D G) of E64d and pepstatin A. Bar indicates 500 nm. An arrowhead indicates an autolysosome, and an arrow indicates an autophagosome. Double-membrane-structures, representative of autophagosomes, were observed in (B and C). Electron-dense structures, representative of autolysosomes, were accumulated in (D G). Autophagosomes and autolysosomes under these conditions were quantified (H). is facilitated under these conditions. LC3-II also markedly accumulated in the presence of E64d and pepstatin A under the same conditions (Fig. 2A, lane 3 vs. lane 4). Densitometric analysis indicated that, under starvation conditions, the amount of LC3-II in the presence of these inhibitors was H C E G about 3.5 fold higher than that in their absence in HEK293 cells (Fig. 2C, left panel). Time-course experiments showed that, in the absence of E64d and pepstatin A, the amount of endogenous LC3-II increased 2 hrs after the induction of starvation, but slightly decreased at 4 hrs (Fig. 2D, KRB). In the presence of E64d and pepstatin A, endogenous LC3-II dramatically increased in a time-dependent manner (Fig. 2D, KRB + inhibitors). While the Tor kinasesignaling pathway is inactivated under starvation conditions, these inhibitors have no effect on the Tor kinase-signaling pathway related to autophagy (Fig. 2A). These results suggested that endogenous LC3-II is rapidly degraded in the lysosomes after lipidation during starvation-induced autophagy in HEK293 cells. In HeLa cells, endogenous LC3 is mainly present as the membrane-bound form (LC3-II) (Fig. 2B, lane 1). Under starvation conditions, LC3-II markedly accumulated in the presence of E64d and pepstatin A, whereas it decreased in the absence of these inhibitors (Fig. 2B, lane 4 vs. lane 3), suggesting that LC3-II is degraded by lysosomal hydrolases soon after lipidation. Under starvation conditions, the amount of LC3-II in the presence of these inhibitors was about 6.7 fold higher than that in their absence in HeLa cells (Fig. 2C, right panel). These results suggested that LC3-II is dramatically degraded by lysosomal hydrolases under starvation conditions in both HeLa and HEK293 cells. LC3-positive puncta, indicative of autolysosomes, accumulate in the presence of E64d and pepstatin a under starvation conditions in HeLa cells. When expressed under the control of a constitutive CMV (or CAG) promoter in HeLa cells, GFP-rat LC3 has been reported to localize to punctatestructures (autophagosomes) under starvation conditions Although the expression of endogenous human LC3-II increases under starvation conditions, 22 little is known of its intracellular localization in the presence of these inhibitors under starvation conditions. Using immunofluorescence microscopy and an anti-human LC3 antibody, we assayed the intracellular localization of endogenous LC3 in HeLa cells during starvationinduced autophagy in the presence or absence of E64d and pepstatin A. We hypothesized that, if these inhibitors inhibit the lysosomal degradation of intra-autophagosomal components during autophagy, LC3-positive puncta would increase in their presence under starvation conditions. Even under nutrient-rich conditions, we observed immunofluorescence of puncta of endogenous LC3 in either the presence or absence of E64d and pepstatin A (Fig. 3A and B). When HeLa cells were transferred to starvation conditions for 4 h, a decrease in LC3-puncta fluorescence was observed (Fig. 3C). This decrease coincides with the decrease in endogenous LC3-II observed by 88 Autophagy 2005; Vol. 1 Issue 2
6 immunoblotting with anti-human LC3 antibody. When E64d and pepstatin A were added to the medium under starvation conditions, endogenous LC3-positive puncta accumulated (Fig. 3D). We further analyzed Z-scanned fluorescent images of LC3-positive puncta in HeLa cells using a program for 2D blind deconvolution (Fig. 3G J). Under starvation conditions, and in the absence of E64d and pepstatin A, cup-shaped structures, representative of preautophagosomes, were observed (Fig. 3G and I). In the presence of E64d and pepstatin A under starvation conditions, both ring-shaped structures, representative of autophagosomes/autolysosomes, and cup-shaped structures were observed (Fig. 3H and J). Together with our finding that the amount of endogenous LC3-II increased under starvation conditions in the presence of E64d and pepstatin A (Fig. 3B), these results indicate that this increase in LC3-II correlates with an increase in LC3-positive puncta. Autolysosomes are dramatically increased under starvation conditions by inhibiting lysosomal hydrolases. We previously showed that endogenous LC3-II accumulates in autolysosomes purified from leupeptintreated rat liver. 22 Previous morphological studies of autophagosomes and autolysosomes suggest that the increase of LC3-positive puncta observed here are reflective of an increase of autophagosomes or autolysosomes. We therefore investigated whether autophagosomes or autolysosomes accumulated in HeLa cells in the presence of E64d and pepstatin A under starvation conditions by electron microscopy (Fig. 4). In the absence of E64d and pepstatin A, a few autolysosomes were observed in HeLa cell under both nutrient-rich (Fig. 4A and H) and starvation (Fig. 4B, C, and H) conditions. In the presence of these inhibitors, however, electron-dense structures corresponding to autolysosomes accumulated in these cells under starvation conditions (Fig. 4D G and H). Higher magnification of the images showed that these electron-dense structures contain cytosolic components, including mitochondria and vesicular like structures (Fig. 4F and G). Measurement of the number of autolysosomes per 100 µm 2 of cytoplasmic area indicated that, under starvation conditions, the number of autolysosomes in the presence of these inhibitors is about 7.3 fold higher than that in their absence (Fig. 4H, autolysosomes). Taken together with the increase in LC3-II (Fig. 2B) and the accumulation of LC3-positive puncta (Fig. 3D) under the same conditions in HeLa cells, these results suggest that the increase of LC3-II is a marker for an increase in the number of autolysosomes under starvation conditions in the presence of protease inhibitors. Endogenous LC3 is present as LC3-II under nutrient-rich conditions independent of inactivation of the Tor kinase-pathway in human cell-lines. In HeLa cells, endogenous LC3 is present as LC3-II even under nutrient-rich conditions. While it has been reported that LC3-I is rapidly lipidated to LC3-II under starvation conditions, it is possible that, in other human cell lines in addition to HeLa cells, endogenous LC3-II is present even under nutrient-rich conditions. We then investigated whether the cytosolic form of LC3, LC3-I, is the major form of endogenous LC3 under nutrient-rich conditions in human cell lines (Fig. 5). Endogenous LC3-II accumulates in several human cell lines, especially, A423, MDA-MB-231, Figure 5. Endogenous LC3-I and LC3-II in human cell lines under nutrient-rich conditions. Expression of endogenous Atg proteins in human cell lines. Total proteins (10 µg per lane) of each cell lysate from cells grown in DMEM containing 10% FCS were separated by SDS-PAGE, and endogenous human LC3 (LC3-I and LC3-II), Atg7, Atg3, and beclin 1 were recognized by immunoblotting using anti-human LC3, Atg7, Atg3, and beclin 1 antibodies, respectively. Tor kinase-signaling pathway inactivation was determined by assaying S6 ribosomal protein (S6RP) and phosphorylated S6 ribosomal protein (phosphos6rp) using anti-s6rp and anti-phosphos6rp antibodies. As an internal control, tubulin was assayed using an anti-tubulin antibody. and SK-N-SH cells, even under nutrient-rich conditions. The amount of endogenous LC3-II is not dependent on the expression of endogenous human Atg7 (E1-like enzyme for LC3-lipidation), 26 human Atg3 (E2-like enzyme for LC3-lipidation) 27 or beclin 1 (mammalian Vps30/Apg6/Atg6 homolog). 43 One explanation for these results is that Tor kinase may be inactivated in cells in which endogenous LC3-II accumulates under nutrient-rich conditions. We therefore assayed S6RP phosphorylation as a downstream marker of the Tor kinase pathway. We observed considerable S6RP phosphorylation in A431, MDA-MB-231, and SK-N-SH cells, all cell lines in which LC3-II accumulates (Fig. 5, S6RP vs. phospho-s6rp), indicating that, in these cell lines, endogenous LC3-II is present even under nutrient-rich conditions and also suggested that the cellular level of LC3-II is not a simple marker for autophagic activity. DISCUSSION While the lysosomal contribution during autophagy has been well studied, there have been few studies of the lysosomal turnover of endogenous LC3-II during autophagy. We have shown here that a significant amount of LC3-II is inevitably degraded by lysosomal hydrolases during starvation-induced autophagy. Using E64d and pepstatin A, we next showed that a significant amount of Autophagy 89
7 intra-autophagosomal LC3-II is degraded by lysosomal hydrolases. During autophagy in HEK293 and HeLa cells, the amount of endogenous LC3-II in the presence of E64d and pepstatin A is about 3.5 and 6.7 fold higher, respectively, than in their absence, indicating that a significant amount of LC3-II is degraded by lysosomal hydrolases during starvation-induced autophagy. Immunofluorescent and electron microscopy analyses suggested that LC3-positive puncta and autolysosomes accumulate in the presence of E64d and pepstatin A under starvation conditions. We further attempted to investigate the relationship between LC3 and autolysosomes in the presence of these inhibitors by immunoelectron microscopy using an anti-human LC3 antibody, but unfortunately, this antibody was not suitable for immunoelectron microscopic analysis. However, several lines of evidence suggest that LC3-positive puncta, i.e., autolysosomes, highly accumulate in the presence of E64d and pepstatin A under starvation conditions, indicating that LC3-II is dramatically degraded in autolysosomes/lysosomes during starvation-induced autophagy. We next showed that, in some cell lines, including A431, MDA-MB-231, and SK-N-SH cells, a significant amount of LC3-II is present under nutrient-rich conditions, independent of the inactivation of Tor kinase, which occurs under starvation conditions. These results are in agreement with a series of previous morphological and biochemical studies of autophagy. Furthermore, these results indicated that the amount of LC3-II does not accurately reflect the activation of starvation-induced autophagy: however, monitoring LC3-II levels in the presence and absence of inhibitors can be used to estimate lysosomal degradation during autophagy. Even without considering lysosomal turnover of LC3-II during autophagy, an increase in the amount of LC3-II has been regarded as marker for autophagy. In some cell lines, however, LC3-II remains present under nutrient-rich conditions without inactivation of Tor kinase. Furthermore, in HeLa cells, LC3-II decreases under starvation conditions in the absence of E64d and pepstatin A, while it increases under the same conditions in HEK293 cells. In the presence of these inhibitors, LC3-II dramatically accumulates under starvation conditions in both HeLa and HEK293 cells, as do LC3-positive puncta and autolysosomal structures. These results indicate that the lysosomal degradation of intra-autophagosomal components (Fig. 1, Step 3) significantly affects the amount of endogenous LC3-II after the formation of autophagosomes. Furthermore, inhibiting the fusion of autophagosomes with lysosomes using rab7 RNAi led to the accumulation of LC3-II, indicating that during the impairment of autophagosome-lysosome fusion LC3 is lipidated and LC3-II accumulates. 52 Therefore, without assaying the lysosomal turnover of LC3-II, the cellular level of LC3-II itself is far from conclusive regarding the activation of autophagy. Using GFP-tagged LC3, it has been reported that an increase of GFP-positive puncta and LC3-II may be a marker of autophagy. However, an increase in the amount of endogenous LC3-II can occur under three conditions: (1) lipidation of LC3-I to form LC3-II during autophagosome-formation, (2) inhibition of the fusion of autophagosomes with lysosomes, and (3) inhibition of lysosomal degradation. Under the first condition, an increase in endogenous LC3-II may be regarded as a marker for autophagy. Under the latter two conditions, however, an increase in endogenous LC3-II is indicative of the inhibition of autophagic processes (Fig. 1, Steps 2 and 3). Thus, to distinguish the first condition from the other two, it is necessary to assay lysosomal turnover of endogenous LC3-II using E64d and pepstatin A. When we compared the lysosomal degradation of endogenous LC3 with that of GFP-human LC3, we found that GFP-human LC3 tended to accumulate less than endogenous LC3 in the presence of E64d and pepstatin A under starvation conditions (Tanida I, Ueno T, Kominami E, unpublished results Therefore, measurement of endogenous LC3 is better for estimating lysosomal degradation. In some cell lines, including HeLa, A431, MDA-MB-231, and SK-N-SH cells, endogenous LC3-II is present even under nutrientrich conditions without inactivation of Tor kinase. LC3-II may have two possible functions under nutrient-rich conditions in these cells. The first is that basal autophagic activity in these cells may be higher than in other cells. It has been reported that, even under nutrientrich conditions, a basal level of autophagic activity is present in rat primary cultured hepatocytes. 53 If the basal level of autophagic activity is high under nutrient-rich conditions in these cells, LC3 would likely be present as LC3-II. Another possibility is that a pathway analogous to the yeast Cvt (Cytoplasm-to-vacuole targeting)- like pathway is activated in these cells. 54 In yeast, aminopeptidase I and α-mannosidase are synthesized in the cytosol and are directly transported to the vacuoles, the yeast counterparts of mammalian lysosomes. 55,56 This Cvt pathway requires most of the yeast Atg proteins, 21 indicating that autophagy and the Cvt-pathway use a common molecular machinery to some extent. At present, there is no evidence for a Cvt-like pathway in mammalian cells, but, if present, this pathway may be activated in these cell lines. References 1. Seglen PO, Berg TO, Blankson H, Fengsrud M, Holen I, Stromhaug PE. Structural aspects of autophagy. Adv Exp Med Biol 1996; 389: Ohsumi Y. Molecular dissection of autophagy: Two ubiquitin-like systems. Nat Rev Mol Cell Biol 2001; 2: Levine B, Klionsky DJ. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev Cell 2004; 6: Meijer AJ, Codogno P. Regulation and role of autophagy in mammalian cells. Int J Biochem Cell Biol 2004; 36: Nixon RA, Cataldo AM, Mathews PM. The endosomal-lysosomal system of neurons in Alzheimer s disease pathogenesis: A review. Neurochem Res 2000; 25: Bursch W. The autophagosomal-lysosomal compartment in programmed cell death. Cell Death Differ 2001; 8: Lemasters JJ, Qian T, He L, Kim JS, Elmore SP, Cascio WE, Brenner DA. Role of mitochondrial inner membrane permeabilization in necrotic cell death, apoptosis, and autophagy. Antioxid Redox Signal 2002; 4: Ogier-Denis E, Codogno P. Autophagy: A barrier or an adaptive response to cancer. Biochim Biophys Acta 2003; 1603: Bergamini E, Cavallini G, Donati A, Gori Z. The role of macroautophagy in the ageing process, anti-ageing intervention and age-associated diseases. Int J Biochem Cell Biol 2004; 36: Keller JN, Dimayuga E, Chen Q, Thorpe J, Gee J, Ding Q. Autophagy, proteasomes, lipofuscin, and oxidative stress in the aging brain. Int J Biochem Cell Biol 2004; 36: Klionsky DJ. Cell biology: Regulated self-cannibalism. Nature 2004; 431: Liberski PP, Sikorska B, Bratosiewicz-Wasik J, Gajdusek DC, Brown P. Neuronal cell death in transmissible spongiform encephalopathies (prion diseases) revisited: From apoptosis to autophagy. Int J Biochem Cell Biol 2004; 36: Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature 2004; 432: De Duve C, Wattiaux R. Functions of lysosomes. Annu Rev Physiol 1966; 28: Arstila AU, Jauregui HO, Chang J, Trump BF. Studies on cellular autophagocytosis. Relationship between heterophagy and autophagy in HeLa cells. Lab Invest 1971; 24: Arstila AU, Nuuja IJ, Trump BF. Studies on cellular autophagocytosis. Vinblastine-induced autophagy in the rat liver. Exp Cell Res 1974; 87: Pfeifer U, Bertling J. A morphometric study of the inhibition of autophagic degradation during restorative growth of liver cells in rats refed after starvation. Virchows Arch B Cell Pathol 1977; 24: Pfeifer U. Functional morphology of the lysosomal apparatus. In: Glaumann H, Ballard FJ, eds. Lysosomes: Their role in protein breakdown. London: Academic Press, 1987: Seglen PO. Regulation of autophagic protein degradation in isolated liver cells. In: Glaumann H, Ballard FJ, eds. Lysosomes: Their role in protein breakdown. London: Academic Press, 1987: Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett 1993; 333: Autophagy 2005; Vol. 1 Issue 2
8 21. Klionsky DJ, Cregg JM, Dunn Jr WA, Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M, Ohsumi Y. A unified nomenclature for yeast autophagy-related genes. Dev Cell 2003; 5: Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 2000; 19: Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol 2004; 36: Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-ii formation. J Cell Sci 2004; 117: Tanida I, Sou YS, Ezaki J, Minematsu-Ikeguchi N, Ueno T, Kominami E. HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phospholipid conjugates. J Biol Chem 2004; 279: Tanida I, Tanida-Miyake E, Ueno T, Kominami E. The human homolog of Saccharomyces cerevisiae Apg7p is a Protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3. J Biol Chem 2001; 276: Tanida I, Tanida-Miyake E, Komatsu M, Ueno T, Kominami E. Human Apg3p/Aut1p homologue is an authentic E2 enzyme for multiple substrates, GATE-16, GABARAP, and MAP-LC3, and facilitates the conjugation of hapg12p to hapg5p. J Biol Chem 2002; 277: Andersson G, Glaumann H. Current Methods for the isolation of coated vesicles, endocytic vesicles, the Golgi complex, lysosomes and autophagic vacuoles. In: Glaumann H, Ballard FJ, eds. Lysosomes: Their role in protein breakdown. London: Academic Press, 1987: Ueno T, Kominami E. Mechanism and regulation of lysosomal sequestration and proteolysis. Biomed Biochim Acta 1991; 50: Yokota S, Himeno M, Kato K. Formation of autophagosomes during degradation of excess peroxisomes induced by di-(2-ethylhexyl)-phthalate treatment. III. Fusion of early autophagosomes with lysosomal compartments. Eur J Cell Biol 1995; 66: Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol 1992; 119: Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol 1999; 147: Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, Ohsumi Y. A ubiquitin-like system mediates protein lipidation. Nature 2000; 408: Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The preautophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J 2001; 20: Jones EW, Zubenko GS, Parker RR. PEP4 gene function is required for expression of several vacuolar hydrolases in Saccharomyces cerevisiae. Genetics 1982; 102: Zubenko GS, Park FJ, Jones EW. Mutations in PEP4 locus of Saccharomyces cerevisiae block final step in maturation of two vacuolar hydrolases. Proc Natl Acad Sci USA 1983; 80: Ammerer G, Hunter CP, Rothman JH, Saari GC, Valls LA, Stevens TH. PEP4gene of Saccharomyces cerevisiae encodes proteinase A, a vacuolar enzyme required for processing of vacuolar precursors. Mol Cell Biol 1986; 6: Zubenko GS, Jones EW. Catabolite inactivation of gluconeogenic enzymes in mutants of yeast deficient in proteinase B. Proc Natl Acad Sci USA 1979; 76: Zubenko GS, Mitchell AP, Jones EW. Septum formation, cell division, and sporulation in mutants of yeast deficient in proteinase B. Proc Natl Acad Sci USA 1979; 76: Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol 2000; 151: Ueno T, Ishidoh K, Mineki R, Tanida I, Murayama K, Kadowaki M, Kominami E. Autolysosomal membrane-associated betaine homocysteine methyltransferase. Limited degradation fragment of a sequestered cytosolic enzyme monitoring autophagy. J Biol Chem 1999; 274: Tanida I, Ueno T, Kominami E. Human light chain 3/MAP1LC3B is cleaved at its carboxyl-terminal Met 121 to expose Gly 120 for lipidation and targeting to autophagosomal membranes. J Biol Chem 2004; 279: Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 1999; 402: Asanuma K, Tanida I, Shirato I, Ueno T, Takahara H, Nishitani T, Kominami E, Tomino Y. MAP-LC3, a promising autophagosomal marker, is processed during the differentiation and recovery of podocytes from PAN nephrosis. FASEB J 2003; 17: Tamai M, Matsumoto K, Omura S, Koyama I, Ozawa Y, Hanada K. In vitro and in vivo inhibition of cysteine proteinases by EST, a new analog of E-64. J Pharmacobiodyn 1986; 9: Tamai M, Yokoo C, Murata M, Oguma K, Sota K, Sato E, Kanaoka Y. Efficient synthetic method for ethyl (+)-(2S,3S)-3-[(S)-3-methyl- 1-(3-methylbutylcarbamoyl)butylcarbamoyl]-2-oxiranecarb oxylate (EST), a new inhibitor of cysteine proteinases. Chem Pharm Bull (Tokyo) 1987; 35: Kirschke H, Barrett AJ. Chemistry of lysosomal proteases. In: Glaumann H, Ballard FJ, eds. Lysosomes: Their role in protein breakdown. London: Academic Press, 1987: Umezawa H, Aoyagi T, Morishima H, Matsuzaki M, Hamada M. Pepstatin, a new pepsin inhibitor produced by Actinomycetes. J Antibiot (Tokyo) 1970; 23: Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol 2001; 152: Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell 2004; 15: Mizushima N. Methods for monitoring autophagy. Int J Biochem Cell Biol 2004; 36: Jager S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, Eskelinen EL. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci 2004; 117: Schworer CM, Shiffer KA, Mortimore GE. Quantitative relationship between autophagy and proteolysis during graded amino acid deprivation in perfused rat liver. J Biol Chem 1981; 256: Teter SA, Klionsky DJ. How to get a folded protein across a membrane. Trends Cell Biol 1999; 9: Harding TM, Morano KA, Scott SV, Klionsky DJ. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol 1995; 131: Hutchins MU, Klionsky DJ. Vacuolar localization of oligomeric α -mannosidase requires the cytoplasm to vacuole targeting and autophagy pathway components in Saccharomyces cerevisiae. J Biol Chem 2001; 276: Autophagy 91
Chapter 2 History of Autophagy After 1963
 Chapter 2 History of Autophagy After 1963 Abstract This chapter gives a historical perspective of the landmark studies that have allowed the autophagy field to progress to its current status. It begins
Chapter 2 History of Autophagy After 1963 Abstract This chapter gives a historical perspective of the landmark studies that have allowed the autophagy field to progress to its current status. It begins
Critical Review. Protein Turnover. Yoshinori Ohsumi National Institute for Basic Biology, Okazaki, Japan INTRODUCTION
 IUBMB Life, 58(5 6): 363 369, May June 2006 Critical Review Protein Turnover Yoshinori Ohsumi National Institute for Basic Biology, Okazaki, Japan Summary The mechanisms and physiological meanings of protein
IUBMB Life, 58(5 6): 363 369, May June 2006 Critical Review Protein Turnover Yoshinori Ohsumi National Institute for Basic Biology, Okazaki, Japan Summary The mechanisms and physiological meanings of protein
SUPPLEMENTARY INFORMATION In format provided by JAATTELA (NOVEMBER 2005)
 Box S1: Methods for studying lysosomal function and integrity Volume and distribution of the acidic compartment. Acridine orange is a metachromatic fluorochrome and a weak base that accumulates in the
Box S1: Methods for studying lysosomal function and integrity Volume and distribution of the acidic compartment. Acridine orange is a metachromatic fluorochrome and a weak base that accumulates in the
Chapter 2 The Core Molecular Machinery of Autophagosome Formation
 Chapter 2 The Core Molecular Machinery of Autophagosome Formation Meiyan Jin and Daniel J. Klionsky Abstract Autophagy is a conserved cytoplasmic process from yeast to mammals, by which cells degrade and
Chapter 2 The Core Molecular Machinery of Autophagosome Formation Meiyan Jin and Daniel J. Klionsky Abstract Autophagy is a conserved cytoplasmic process from yeast to mammals, by which cells degrade and
A novel assay to study autophagy: regulation of autophagosome vacuole size by amino acid deprivation
 RESEARCH ARTICLE 3619 A novel assay to study autophagy: regulation of autophagosome vacuole size by amino acid deprivation Daniela B. Munafó and María I. Colombo* Laboratorio de Biología Celular y Molecular-Instituto
RESEARCH ARTICLE 3619 A novel assay to study autophagy: regulation of autophagosome vacuole size by amino acid deprivation Daniela B. Munafó and María I. Colombo* Laboratorio de Biología Celular y Molecular-Instituto
Summary of Endomembrane-system
 Summary of Endomembrane-system 1. Endomembrane System: The structural and functional relationship organelles including ER,Golgi complex, lysosome, endosomes, secretory vesicles. 2. Membrane-bound structures
Summary of Endomembrane-system 1. Endomembrane System: The structural and functional relationship organelles including ER,Golgi complex, lysosome, endosomes, secretory vesicles. 2. Membrane-bound structures
2007 LANDES BIOSCIENCE. DO NOT DISTRIBUTE.
 [Autophagy 3:5, 452-460; September/October 2007]; 2007 Landes Bioscience Research Paper Dissection of the Autophagosome Maturation Process by a Novel Reporter Protein, Tandem Fluorescent-Tagged LC3 Shunsuke
[Autophagy 3:5, 452-460; September/October 2007]; 2007 Landes Bioscience Research Paper Dissection of the Autophagosome Maturation Process by a Novel Reporter Protein, Tandem Fluorescent-Tagged LC3 Shunsuke
6. TNF-α regulates oxidative stress, mitochondrial function and autophagy in neuronal cells
 6. TNF-α regulates oxidative stress, mitochondrial function and autophagy in neuronal cells 6.1 TNF-α induces mitochondrial oxidative stress in SH-SY5Y cells. The dysregulation of mitochondria and oxidative
6. TNF-α regulates oxidative stress, mitochondrial function and autophagy in neuronal cells 6.1 TNF-α induces mitochondrial oxidative stress in SH-SY5Y cells. The dysregulation of mitochondria and oxidative
THE ROLE OF ALTERED CALCIUM AND mtor SIGNALING IN THE PATHOGENESIS OF CYSTINOSIS
 Research Foundation, 18 month progress report THE ROLE OF ALTERED CALCIUM AND mtor SIGNALING IN THE PATHOGENESIS OF CYSTINOSIS Ekaterina Ivanova, doctoral student Elena Levtchenko, MD, PhD, PI Antonella
Research Foundation, 18 month progress report THE ROLE OF ALTERED CALCIUM AND mtor SIGNALING IN THE PATHOGENESIS OF CYSTINOSIS Ekaterina Ivanova, doctoral student Elena Levtchenko, MD, PhD, PI Antonella
Supplementary Fig. 1 V-ATPase depletion induces unique and robust phenotype in Drosophila fat body cells.
 Supplementary Fig. 1 V-ATPase depletion induces unique and robust phenotype in Drosophila fat body cells. a. Schematic of the V-ATPase proton pump macro-complex structure. The V1 complex is composed of
Supplementary Fig. 1 V-ATPase depletion induces unique and robust phenotype in Drosophila fat body cells. a. Schematic of the V-ATPase proton pump macro-complex structure. The V1 complex is composed of
A. Generation and characterization of Ras-expressing autophagycompetent
 Supplemental Material Supplemental Figure Legends Fig. S1 A. Generation and characterization of Ras-expressing autophagycompetent and -deficient cell lines. HA-tagged H-ras V12 was stably expressed in
Supplemental Material Supplemental Figure Legends Fig. S1 A. Generation and characterization of Ras-expressing autophagycompetent and -deficient cell lines. HA-tagged H-ras V12 was stably expressed in
Sestrin2 and BNIP3 (Bcl-2/adenovirus E1B 19kDa-interacting. protein3) regulate autophagy and mitophagy in renal tubular cells in. acute kidney injury
 Sestrin2 and BNIP3 (Bcl-2/adenovirus E1B 19kDa-interacting protein3) regulate autophagy and mitophagy in renal tubular cells in acute kidney injury by Masayuki Ishihara 1, Madoka Urushido 2, Kazu Hamada
Sestrin2 and BNIP3 (Bcl-2/adenovirus E1B 19kDa-interacting protein3) regulate autophagy and mitophagy in renal tubular cells in acute kidney injury by Masayuki Ishihara 1, Madoka Urushido 2, Kazu Hamada
Protein Trafficking in the Secretory and Endocytic Pathways
 Protein Trafficking in the Secretory and Endocytic Pathways The compartmentalization of eukaryotic cells has considerable functional advantages for the cell, but requires elaborate mechanisms to ensure
Protein Trafficking in the Secretory and Endocytic Pathways The compartmentalization of eukaryotic cells has considerable functional advantages for the cell, but requires elaborate mechanisms to ensure
endomembrane system internal membranes origins transport of proteins chapter 15 endomembrane system
 endo system chapter 15 internal s endo system functions as a coordinated unit divide cytoplasm into distinct compartments controls exocytosis and endocytosis movement of molecules which cannot pass through
endo system chapter 15 internal s endo system functions as a coordinated unit divide cytoplasm into distinct compartments controls exocytosis and endocytosis movement of molecules which cannot pass through
SUPPLEMENT. Materials and methods
 SUPPLEMENT Materials and methods Cell culture and reagents Cell media and reagents were from Invitrogen unless otherwise indicated. Antibiotics and Tet-certified serum were from Clontech. In experiments
SUPPLEMENT Materials and methods Cell culture and reagents Cell media and reagents were from Invitrogen unless otherwise indicated. Antibiotics and Tet-certified serum were from Clontech. In experiments
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Krenn et al., http://www.jcb.org/cgi/content/full/jcb.201110013/dc1 Figure S1. Levels of expressed proteins and demonstration that C-terminal
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Krenn et al., http://www.jcb.org/cgi/content/full/jcb.201110013/dc1 Figure S1. Levels of expressed proteins and demonstration that C-terminal
Effect of starvation-induced autophagy on cell cycle of tumor cells
 [Chinese Journal of Cancer 27:8, 102-108; August 2008]; 2008 Sun Yat-Sen University Cancer Center Basic Research Paper Effect of starvation-induced autophagy on cell cycle of tumor cells Jun-Na Ge, 1 Dan
[Chinese Journal of Cancer 27:8, 102-108; August 2008]; 2008 Sun Yat-Sen University Cancer Center Basic Research Paper Effect of starvation-induced autophagy on cell cycle of tumor cells Jun-Na Ge, 1 Dan
1. to understand how proteins find their destination in prokaryotic and eukaryotic cells 2. to know how proteins are bio-recycled
 Protein Targeting Objectives 1. to understand how proteins find their destination in prokaryotic and eukaryotic cells 2. to know how proteins are bio-recycled As a protein is being synthesized, decisions
Protein Targeting Objectives 1. to understand how proteins find their destination in prokaryotic and eukaryotic cells 2. to know how proteins are bio-recycled As a protein is being synthesized, decisions
Cell Quality Control. Peter Takizawa Department of Cell Biology
 Cell Quality Control Peter Takizawa Department of Cell Biology Cellular quality control reduces production of defective proteins. Cells have many quality control systems to ensure that cell does not build
Cell Quality Control Peter Takizawa Department of Cell Biology Cellular quality control reduces production of defective proteins. Cells have many quality control systems to ensure that cell does not build
Beclin phosphatidylinositol 3-kinase complex functions at the trans-golgi network. Akio Kihara, Yukiko Kabeya, Yoshinori Ohsumi & Tamotsu Yoshimori +
 EMBO reports Beclin phosphatidylinositol 3-kinase complex functions at the trans-golgi network Akio Kihara, Yukiko Kabeya, Yoshinori Ohsumi & Tamotsu Yoshimori + Department of Cell Biology, National Institute
EMBO reports Beclin phosphatidylinositol 3-kinase complex functions at the trans-golgi network Akio Kihara, Yukiko Kabeya, Yoshinori Ohsumi & Tamotsu Yoshimori + Department of Cell Biology, National Institute
Peroxisome turnover by micropexophagy: an autophagy-related process
 Review TRENDS in Cell Biology Vol.14 No.9 September 2004 Peroxisome turnover by micropexophagy: an autophagy-related process Jean-Claude Farré and Suresh Subramani Section of Molecular Biology, Division
Review TRENDS in Cell Biology Vol.14 No.9 September 2004 Peroxisome turnover by micropexophagy: an autophagy-related process Jean-Claude Farré and Suresh Subramani Section of Molecular Biology, Division
Methods for Monitoring Autophagy from Yeast to Human
 [Autophagy 3:3, 181-206; May/June 2007]; 2007 Landes Bioscience Review Methods for Monitoring Autophagy from Yeast to Human Daniel J. Klionsky 1, * Ana Maria Cuervo 2 Per O. Seglen 3 1 Life Sciences Institute
[Autophagy 3:3, 181-206; May/June 2007]; 2007 Landes Bioscience Review Methods for Monitoring Autophagy from Yeast to Human Daniel J. Klionsky 1, * Ana Maria Cuervo 2 Per O. Seglen 3 1 Life Sciences Institute
SUPPLEMENTARY INFORMATION
 Figure S1 Induction of non-apoptotic death of SV40-transformed and primary DKO MEFs, and DKO thymocytes. (A-F) STS-induced non-apoptotic death of DKO MEF. (A, B) Reduced viability of DKO MEFs after exposure
Figure S1 Induction of non-apoptotic death of SV40-transformed and primary DKO MEFs, and DKO thymocytes. (A-F) STS-induced non-apoptotic death of DKO MEF. (A, B) Reduced viability of DKO MEFs after exposure
Practice Exam 2 MCBII
 1. Which feature is true for signal sequences and for stop transfer transmembrane domains (4 pts)? A. They are both 20 hydrophobic amino acids long. B. They are both found at the N-terminus of the protein.
1. Which feature is true for signal sequences and for stop transfer transmembrane domains (4 pts)? A. They are both 20 hydrophobic amino acids long. B. They are both found at the N-terminus of the protein.
Suppl. Figure 1. T 3 induces autophagic flux in hepatic cells. (A) RFP-GFP-LC3 transfected HepG2/TRα cells were visualized and cells were quantified
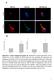 Suppl. Figure 1. T 3 induces autophagic flux in hepatic cells. (A) RFP-GFP-LC3 transfected HepG2/TRα cells were visualized and cells were quantified for RFP-LC3 puncta (red dots) representing both autolysosomes
Suppl. Figure 1. T 3 induces autophagic flux in hepatic cells. (A) RFP-GFP-LC3 transfected HepG2/TRα cells were visualized and cells were quantified for RFP-LC3 puncta (red dots) representing both autolysosomes
Name Animal source Vendor Cat # Dilutions
 Supplementary data Table S1. Primary and Secondary antibody sources Devi et al, TXNIP in mitophagy A. Primary Antibodies Name Animal source Vendor Cat # Dilutions 1. TXNIP mouse MBL KO205-2 1:2000 (WB)
Supplementary data Table S1. Primary and Secondary antibody sources Devi et al, TXNIP in mitophagy A. Primary Antibodies Name Animal source Vendor Cat # Dilutions 1. TXNIP mouse MBL KO205-2 1:2000 (WB)
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
Lysosomes, Peroxisomes and Centrioles. Hüseyin Çağsın
 Lysosomes, Peroxisomes and Centrioles Hüseyin Çağsın Lysosomes Outline Endosomes Molecule transport to the lysosomes Endocytosis Exocytosis Autophagy Vacuoles Peroxisomes Centrioles Lysosomes Lysosomes
Lysosomes, Peroxisomes and Centrioles Hüseyin Çağsın Lysosomes Outline Endosomes Molecule transport to the lysosomes Endocytosis Exocytosis Autophagy Vacuoles Peroxisomes Centrioles Lysosomes Lysosomes
Trehalose, sucrose and raffinose are novel activators of autophagy in human. keratinocytes through an mtor-independent pathway
 Title page Trehalose, sucrose and raffinose are novel activators of autophagy in human keratinocytes through an mtor-independent pathway Xu Chen 1*, Min Li 1*, Li Li 1, Song Xu 1, Dan Huang 1, Mei Ju 1,
Title page Trehalose, sucrose and raffinose are novel activators of autophagy in human keratinocytes through an mtor-independent pathway Xu Chen 1*, Min Li 1*, Li Li 1, Song Xu 1, Dan Huang 1, Mei Ju 1,
PpAtg30 Tags Peroxisomes for Turnover by Selective Autophagy
 Article PpAtg30 Tags Peroxisomes for Turnover by Selective Autophagy Jean-Claude Farré, 1 Ravi Manjithaya, 1 Richard D. Mathewson, 1 and Suresh Subramani 1, * 1 Section of Molecular Biology, Division of
Article PpAtg30 Tags Peroxisomes for Turnover by Selective Autophagy Jean-Claude Farré, 1 Ravi Manjithaya, 1 Richard D. Mathewson, 1 and Suresh Subramani 1, * 1 Section of Molecular Biology, Division of
Assessing Autophagy with the guava easycyte Benchtop Flow Cytometer
 Application Note Assessing Autophagy with the guava easycyte Benchtop Flow Cytometer Mark Santos, Kevin Su, Luke Armstrong, Angelica Olcott, Jason Whalley, and Matthew Hsu EMD Millipore Introduction Autophagy
Application Note Assessing Autophagy with the guava easycyte Benchtop Flow Cytometer Mark Santos, Kevin Su, Luke Armstrong, Angelica Olcott, Jason Whalley, and Matthew Hsu EMD Millipore Introduction Autophagy
Selective Mitophagy in budding yeast, a mitochondrial self-eating quality control
 European Journal of Biophysics 2014; 2(5): 49-60 Published online November 10, 2014 (http://www.sciencepublishinggroup.com/j/ejb) doi: 10.11648/j.ejb.20140205.11 ISSN: 2329-1745 (Print); ISSN: 2329-1737
European Journal of Biophysics 2014; 2(5): 49-60 Published online November 10, 2014 (http://www.sciencepublishinggroup.com/j/ejb) doi: 10.11648/j.ejb.20140205.11 ISSN: 2329-1745 (Print); ISSN: 2329-1737
TFEB-mediated increase in peripheral lysosomes regulates. Store Operated Calcium Entry
 TFEB-mediated increase in peripheral lysosomes regulates Store Operated Calcium Entry Luigi Sbano, Massimo Bonora, Saverio Marchi, Federica Baldassari, Diego L. Medina, Andrea Ballabio, Carlotta Giorgi
TFEB-mediated increase in peripheral lysosomes regulates Store Operated Calcium Entry Luigi Sbano, Massimo Bonora, Saverio Marchi, Federica Baldassari, Diego L. Medina, Andrea Ballabio, Carlotta Giorgi
genome edited transient transfection, CMV promoter
 Supplementary Figure 1. In the absence of new protein translation, overexpressed caveolin-1-gfp is degraded faster than caveolin-1-gfp expressed from the endogenous caveolin 1 locus % loss of total caveolin-1-gfp
Supplementary Figure 1. In the absence of new protein translation, overexpressed caveolin-1-gfp is degraded faster than caveolin-1-gfp expressed from the endogenous caveolin 1 locus % loss of total caveolin-1-gfp
MCB130 Midterm. GSI s Name:
 1. Peroxisomes are small, membrane-enclosed organelles that function in the degradation of fatty acids and in the degradation of H 2 O 2. Peroxisomes are not part of the secretory pathway and peroxisomal
1. Peroxisomes are small, membrane-enclosed organelles that function in the degradation of fatty acids and in the degradation of H 2 O 2. Peroxisomes are not part of the secretory pathway and peroxisomal
2013 John Wiley & Sons, Inc. All rights reserved. PROTEIN SORTING. Lecture 10 BIOL 266/ Biology Department Concordia University. Dr. S.
 PROTEIN SORTING Lecture 10 BIOL 266/4 2014-15 Dr. S. Azam Biology Department Concordia University Introduction Membranes divide the cytoplasm of eukaryotic cells into distinct compartments. The endomembrane
PROTEIN SORTING Lecture 10 BIOL 266/4 2014-15 Dr. S. Azam Biology Department Concordia University Introduction Membranes divide the cytoplasm of eukaryotic cells into distinct compartments. The endomembrane
by Freeze-Replica Method
 CELL STRUCTURE AND FUNCTION 20: 465-471 (1995) 1995 by Japan Society for Cell Biology Anaysis of the MembraneStructures by Freeze-Replica Method Involved in Autophagy in Yeast Misuzu Baba, Masako Osumi*,
CELL STRUCTURE AND FUNCTION 20: 465-471 (1995) 1995 by Japan Society for Cell Biology Anaysis of the MembraneStructures by Freeze-Replica Method Involved in Autophagy in Yeast Misuzu Baba, Masako Osumi*,
Intracellular vesicular traffic. B. Balen
 Intracellular vesicular traffic B. Balen Three types of transport in eukaryotic cells Figure 12-6 Molecular Biology of the Cell ( Garland Science 2008) Endoplasmic reticulum in all eucaryotic cells Endoplasmic
Intracellular vesicular traffic B. Balen Three types of transport in eukaryotic cells Figure 12-6 Molecular Biology of the Cell ( Garland Science 2008) Endoplasmic reticulum in all eucaryotic cells Endoplasmic
Autophagy-related Products Catalogue
 Research Use Only Autophagy-related Products Catalogue Antigen presentation Virus infection Cancer Bacterial infection Cell death Starvation-adaptation Neurodegenerative disease Autophagy was first discovered
Research Use Only Autophagy-related Products Catalogue Antigen presentation Virus infection Cancer Bacterial infection Cell death Starvation-adaptation Neurodegenerative disease Autophagy was first discovered
Intracellular Compartments and Protein Sorting
 Intracellular Compartments and Protein Sorting Intracellular Compartments A eukaryotic cell is elaborately subdivided into functionally distinct, membrane-enclosed compartments. Each compartment, or organelle,
Intracellular Compartments and Protein Sorting Intracellular Compartments A eukaryotic cell is elaborately subdivided into functionally distinct, membrane-enclosed compartments. Each compartment, or organelle,
SUPPLEMENTARY INFORMATION
 Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
supplementary information
 Figure S1 Nucleotide binding status of RagA mutants. Wild type and mutant forms of MycRagA was transfected into HEK293 cells and the transfected cells were labeled with 32 Pphosphate. MycRagA was immunoprecipitated
Figure S1 Nucleotide binding status of RagA mutants. Wild type and mutant forms of MycRagA was transfected into HEK293 cells and the transfected cells were labeled with 32 Pphosphate. MycRagA was immunoprecipitated
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk
 Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Vesicle Transport. Vesicle pathway: many compartments, interconnected by trafficking routes 3/17/14
 Vesicle Transport Vesicle Formation Curvature (Self Assembly of Coat complex) Sorting (Sorting Complex formation) Regulation (Sar1/Arf1 GTPases) Fission () Membrane Fusion SNARE combinations Tethers Regulation
Vesicle Transport Vesicle Formation Curvature (Self Assembly of Coat complex) Sorting (Sorting Complex formation) Regulation (Sar1/Arf1 GTPases) Fission () Membrane Fusion SNARE combinations Tethers Regulation
Antibodies for Unfolded Protein Response
 Novus-lu-2945 Antibodies for Unfolded rotein Response Unfolded roteins ER lumen GR78 IRE-1 GR78 ERK Cytosol GR78 TRAF2 ASK1 JNK Activator Intron RIDD elf2α Degraded mrna XB1 mrna Translation XB1-S (p50)
Novus-lu-2945 Antibodies for Unfolded rotein Response Unfolded roteins ER lumen GR78 IRE-1 GR78 ERK Cytosol GR78 TRAF2 ASK1 JNK Activator Intron RIDD elf2α Degraded mrna XB1 mrna Translation XB1-S (p50)
PROTEIN TRAFFICKING. Dr. SARRAY Sameh, Ph.D
 PROTEIN TRAFFICKING Dr. SARRAY Sameh, Ph.D Overview Proteins are synthesized either on free ribosomes or on ribosomes bound to endoplasmic reticulum (RER). The synthesis of nuclear, mitochondrial and peroxisomal
PROTEIN TRAFFICKING Dr. SARRAY Sameh, Ph.D Overview Proteins are synthesized either on free ribosomes or on ribosomes bound to endoplasmic reticulum (RER). The synthesis of nuclear, mitochondrial and peroxisomal
BIOL 4374/BCHS 4313 Cell Biology Exam #1 February 13, 2001
 BIOL 4374/BCHS 4313 Cell Biology Exam #1 February 13, 2001 SS# Name This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses. Good luck! 1. (2) The
BIOL 4374/BCHS 4313 Cell Biology Exam #1 February 13, 2001 SS# Name This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses. Good luck! 1. (2) The
Supplementary Figure 1 CD4 + T cells from PKC-θ null mice are defective in NF-κB activation during T cell receptor signaling. CD4 + T cells were
 Supplementary Figure 1 CD4 + T cells from PKC-θ null mice are defective in NF-κB activation during T cell receptor signaling. CD4 + T cells were isolated from wild type (PKC-θ- WT) or PKC-θ null (PKC-θ-KO)
Supplementary Figure 1 CD4 + T cells from PKC-θ null mice are defective in NF-κB activation during T cell receptor signaling. CD4 + T cells were isolated from wild type (PKC-θ- WT) or PKC-θ null (PKC-θ-KO)
General information. Cell mediated immunity. 455 LSA, Tuesday 11 to noon. Anytime after class.
 General information Cell mediated immunity 455 LSA, Tuesday 11 to noon Anytime after class T-cell precursors Thymus Naive T-cells (CD8 or CD4) email: lcoscoy@berkeley.edu edu Use MCB150 as subject line
General information Cell mediated immunity 455 LSA, Tuesday 11 to noon Anytime after class T-cell precursors Thymus Naive T-cells (CD8 or CD4) email: lcoscoy@berkeley.edu edu Use MCB150 as subject line
supplementary information
 DOI: 1.138/ncb1 Control Atg7 / NAC 1 1 1 1 (mm) Control Atg7 / NAC 1 1 1 1 (mm) Lamin B Gstm1 Figure S1 Neither the translocation of into the nucleus nor the induction of antioxidant proteins in autophagydeficient
DOI: 1.138/ncb1 Control Atg7 / NAC 1 1 1 1 (mm) Control Atg7 / NAC 1 1 1 1 (mm) Lamin B Gstm1 Figure S1 Neither the translocation of into the nucleus nor the induction of antioxidant proteins in autophagydeficient
Supplemental Data. Wang et al. (2013). Plant Cell /tpc
 Supplemental Data. Wang et al. (2013). Plant Cell 10.1105/tpc.112.108993 Supplemental Figure 1. 3-MA Treatment Reduces the Growth of Seedlings. Two-week-old Nicotiana benthamiana seedlings germinated on
Supplemental Data. Wang et al. (2013). Plant Cell 10.1105/tpc.112.108993 Supplemental Figure 1. 3-MA Treatment Reduces the Growth of Seedlings. Two-week-old Nicotiana benthamiana seedlings germinated on
Growth and Differentiation Phosphorylation Sampler Kit
 Growth and Differentiation Phosphorylation Sampler Kit E 0 5 1 0 1 4 Kits Includes Cat. Quantity Application Reactivity Source Akt (Phospho-Ser473) E011054-1 50μg/50μl IHC, WB Human, Mouse, Rat Rabbit
Growth and Differentiation Phosphorylation Sampler Kit E 0 5 1 0 1 4 Kits Includes Cat. Quantity Application Reactivity Source Akt (Phospho-Ser473) E011054-1 50μg/50μl IHC, WB Human, Mouse, Rat Rabbit
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb2822 a MTC02 FAO cells EEA1 b +/+ MEFs /DAPI -/- MEFs /DAPI -/- MEFs //DAPI c HEK 293 cells WCE N M C P AKT TBC1D7 Lamin A/C EEA1 VDAC d HeLa cells WCE N M C P AKT Lamin A/C EEA1 VDAC Figure
DOI:.38/ncb2822 a MTC02 FAO cells EEA1 b +/+ MEFs /DAPI -/- MEFs /DAPI -/- MEFs //DAPI c HEK 293 cells WCE N M C P AKT TBC1D7 Lamin A/C EEA1 VDAC d HeLa cells WCE N M C P AKT Lamin A/C EEA1 VDAC Figure
Edinburgh Research Explorer
 Edinburgh Research Explorer Interaction between FIP200 and ATG16L1 distinguishes ULK1 complex-dependent and -independent autophagy Citation for published version: Noor, G, Florey, O, Overholtzer, M & Jiang,
Edinburgh Research Explorer Interaction between FIP200 and ATG16L1 distinguishes ULK1 complex-dependent and -independent autophagy Citation for published version: Noor, G, Florey, O, Overholtzer, M & Jiang,
RayBio KinaseSTAR TM Akt Activity Assay Kit
 Activity Assay Kit User Manual Version 1.0 March 13, 2015 RayBio KinaseSTAR TM Akt Activity Kit Protocol (Cat#: 68AT-Akt-S40) RayBiotech, Inc. We Provide You With Excellent Support And Service Tel:(Toll
Activity Assay Kit User Manual Version 1.0 March 13, 2015 RayBio KinaseSTAR TM Akt Activity Kit Protocol (Cat#: 68AT-Akt-S40) RayBiotech, Inc. We Provide You With Excellent Support And Service Tel:(Toll
Posttranslational Modification and Targeting of Proteins
 Posttranslational Modification and Targeting of Proteins Graduate Biochemistry Term 2/2016 Assist. Prof. Dr. Panida Khunkaewla School of Chemistry, Institute of Science Suranaree University of Technology
Posttranslational Modification and Targeting of Proteins Graduate Biochemistry Term 2/2016 Assist. Prof. Dr. Panida Khunkaewla School of Chemistry, Institute of Science Suranaree University of Technology
Supplemental Information. Autophagy in Oncogenic K-Ras. Promotes Basal Extrusion. of Epithelial Cells by Degrading S1P. Current Biology, Volume 24
 Current Biology, Volume 24 Supplemental Information Autophagy in Oncogenic K-Ras Promotes Basal Extrusion of Epithelial Cells by Degrading S1P Gloria Slattum, Yapeng Gu, Roger Sabbadini, and Jody Rosenblatt
Current Biology, Volume 24 Supplemental Information Autophagy in Oncogenic K-Ras Promotes Basal Extrusion of Epithelial Cells by Degrading S1P Gloria Slattum, Yapeng Gu, Roger Sabbadini, and Jody Rosenblatt
Supplementary Materials and Methods
 Supplementary Materials and Methods Immunoblotting Immunoblot analysis was performed as described previously (1). Due to high-molecular weight of MUC4 (~ 950 kda) and MUC1 (~ 250 kda) proteins, electrophoresis
Supplementary Materials and Methods Immunoblotting Immunoblot analysis was performed as described previously (1). Due to high-molecular weight of MUC4 (~ 950 kda) and MUC1 (~ 250 kda) proteins, electrophoresis
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1. Neither the activation nor suppression of the MAPK pathway affects the ASK1/Vif interaction. (a, b) HEK293 cells were cotransfected with plasmids encoding
Supplementary Figure 1 Supplementary Figure 1. Neither the activation nor suppression of the MAPK pathway affects the ASK1/Vif interaction. (a, b) HEK293 cells were cotransfected with plasmids encoding
Molecular switches between apoptosis and autophagy. 15_bmca_2011
 Molecular switches between apoptosis and autophagy Beclin 1: a BH3-only protein that fails to induce apoptosis Model for recruitment of Atg proteins and DFCP1 to the ER erived membrane and formation of
Molecular switches between apoptosis and autophagy Beclin 1: a BH3-only protein that fails to induce apoptosis Model for recruitment of Atg proteins and DFCP1 to the ER erived membrane and formation of
Antigen Presentation to T lymphocytes
 Antigen Presentation to T lymphocytes Immunology 441 Lectures 6 & 7 Chapter 6 October 10 & 12, 2016 Jessica Hamerman jhamerman@benaroyaresearch.org Office hours by arrangement Antibodies and T cell receptors
Antigen Presentation to T lymphocytes Immunology 441 Lectures 6 & 7 Chapter 6 October 10 & 12, 2016 Jessica Hamerman jhamerman@benaroyaresearch.org Office hours by arrangement Antibodies and T cell receptors
Fluorescence Microscopy
 Fluorescence Microscopy Imaging Organelles Mitochondria Lysosomes Nuclei Endoplasmic Reticulum Plasma Membrane F-Actin AAT Bioquest Introduction: Organelle-Selective Stains Organelles are tiny, specialized
Fluorescence Microscopy Imaging Organelles Mitochondria Lysosomes Nuclei Endoplasmic Reticulum Plasma Membrane F-Actin AAT Bioquest Introduction: Organelle-Selective Stains Organelles are tiny, specialized
1. endoplasmic reticulum This is the location where N-linked oligosaccharide is initially synthesized and attached to glycoproteins.
 Biology 4410 Name Spring 2006 Exam 2 A. Multiple Choice, 2 pt each Pick the best choice from the list of choices, and write it in the space provided. Some choices may be used more than once, and other
Biology 4410 Name Spring 2006 Exam 2 A. Multiple Choice, 2 pt each Pick the best choice from the list of choices, and write it in the space provided. Some choices may be used more than once, and other
Tumor suppressor Spred2 interaction with LC3 promotes autophagosome maturation and induces autophagy-dependent cell death
 www.impactjournals.com/oncotarget/ Oncotarget, Supplementary Materials 2016 Tumor suppressor Spred2 interaction with LC3 promotes autophagosome maturation and induces autophagy-dependent cell death Supplementary
www.impactjournals.com/oncotarget/ Oncotarget, Supplementary Materials 2016 Tumor suppressor Spred2 interaction with LC3 promotes autophagosome maturation and induces autophagy-dependent cell death Supplementary
Proteomic profiling of small-molecule inhibitors reveals dispensability of MTH1 for cancer cell survival
 Supplementary Information for Proteomic profiling of small-molecule inhibitors reveals dispensability of MTH1 for cancer cell survival Tatsuro Kawamura 1, Makoto Kawatani 1, Makoto Muroi, Yasumitsu Kondoh,
Supplementary Information for Proteomic profiling of small-molecule inhibitors reveals dispensability of MTH1 for cancer cell survival Tatsuro Kawamura 1, Makoto Kawatani 1, Makoto Muroi, Yasumitsu Kondoh,
Supporting Information
 Supporting Information Fig. S1. Overexpression of Rpr causes progenitor cell death. (A) TUNEL assay of control intestines. No progenitor cell death could be observed, except that some ECs are undergoing
Supporting Information Fig. S1. Overexpression of Rpr causes progenitor cell death. (A) TUNEL assay of control intestines. No progenitor cell death could be observed, except that some ECs are undergoing
Nature Structural & Molecular Biology: doi: /nsmb.3218
 Supplementary Figure 1 Endogenous EGFR trafficking and responses depend on biased ligands. (a) Lysates from HeLa cells stimulated for 2 min. with increasing concentration of ligands were immunoblotted
Supplementary Figure 1 Endogenous EGFR trafficking and responses depend on biased ligands. (a) Lysates from HeLa cells stimulated for 2 min. with increasing concentration of ligands were immunoblotted
Peroxisome Size Provides Insights into the Function of Autophagy-related Proteins
 Molecular Biology of the Cell Vol. 20, 3828 3839, September 1, 2009 Peroxisome Size Provides Insights into the Function of Autophagy-related Proteins Taras Y. Nazarko, Jean-Claude Farré, and Suresh Subramani
Molecular Biology of the Cell Vol. 20, 3828 3839, September 1, 2009 Peroxisome Size Provides Insights into the Function of Autophagy-related Proteins Taras Y. Nazarko, Jean-Claude Farré, and Suresh Subramani
Divergent modes of autophagy in the methylotrophic yeast Pichia pastoris
 Journal of Cell Science 108, 25-35 (1995) Printed in Great Britain The Company of Biologists Limited 1995 25 Divergent modes of autophagy in the methylotrophic yeast Pichia pastoris Daniel L. Tuttle and
Journal of Cell Science 108, 25-35 (1995) Printed in Great Britain The Company of Biologists Limited 1995 25 Divergent modes of autophagy in the methylotrophic yeast Pichia pastoris Daniel L. Tuttle and
Bortezomib blocks the catabolic process of. autophagy via a cathepsin-dependent mechanism, affects endoplasmic reticulum stress, and
 Autophagy ISSN: 1554-8627 (Print) 1554-8635 (Online) Journal homepage: http://www.tandfonline.com/loi/kaup20 Bortezomib blocks the catabolic process of autophagy via a cathepsin-dependent mechanism, affects
Autophagy ISSN: 1554-8627 (Print) 1554-8635 (Online) Journal homepage: http://www.tandfonline.com/loi/kaup20 Bortezomib blocks the catabolic process of autophagy via a cathepsin-dependent mechanism, affects
Lysosomes and endocytic pathways 9/27/2012 Phyllis Hanson
 Lysosomes and endocytic pathways 9/27/2012 Phyllis Hanson General principles Properties of lysosomes Delivery of enzymes to lysosomes Endocytic uptake clathrin, others Endocytic pathways recycling vs.
Lysosomes and endocytic pathways 9/27/2012 Phyllis Hanson General principles Properties of lysosomes Delivery of enzymes to lysosomes Endocytic uptake clathrin, others Endocytic pathways recycling vs.
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells
 Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Human SH-SY5Y neuroblastoma cells (A.T.C.C., Manassas, VA) were cultured in DMEM, F-12
 SUPPLEMENTARY METHODS Cell cultures Human SH-SY5Y neuroblastoma cells (A.T.C.C., Manassas, VA) were cultured in DMEM, F-12 Ham with 25 mm HEPES and NaHCO 3 (1:1) and supplemented with 10% (v/v) FBS, 1.0
SUPPLEMENTARY METHODS Cell cultures Human SH-SY5Y neuroblastoma cells (A.T.C.C., Manassas, VA) were cultured in DMEM, F-12 Ham with 25 mm HEPES and NaHCO 3 (1:1) and supplemented with 10% (v/v) FBS, 1.0
1. This is the location where N-linked oligosaccharide is initially synthesized and attached to glycoproteins.
 Biology 4410 Name Spring 2006 Exam 2 A. Multiple Choice, 2 pt each Pick the best choice from the list of choices, and write it in the space provided. Some choices may be used more than once, and other
Biology 4410 Name Spring 2006 Exam 2 A. Multiple Choice, 2 pt each Pick the best choice from the list of choices, and write it in the space provided. Some choices may be used more than once, and other
-Cyclodextrin-threaded Biocleavable Polyrotaxanes Ameliorate Impaired Autophagic Flux in Niemann-Pick Type C Disease
 -Cyclodextrin-threaded Biocleavable Polyrotaxanes Ameliorate Impaired Autophagic Flux in Niemann-Pick Type C Disease Atsushi Tamura and Nobuhiko Yui Department of Organic Biomaterials, Institute of Biomaterials
-Cyclodextrin-threaded Biocleavable Polyrotaxanes Ameliorate Impaired Autophagic Flux in Niemann-Pick Type C Disease Atsushi Tamura and Nobuhiko Yui Department of Organic Biomaterials, Institute of Biomaterials
Critical Review. Trafficking and Signaling in Mammalian Autophagy
 IUBMB Life, 62(7): 503 508, July 2010 Critical Review Trafficking and Signaling in Mammalian Autophagy Sharon A. Tooze*, Harold B. J. Jefferies, Eyal Kalie, Andrea Longatti, Fiona E. Mcalpine, Nicole C.
IUBMB Life, 62(7): 503 508, July 2010 Critical Review Trafficking and Signaling in Mammalian Autophagy Sharon A. Tooze*, Harold B. J. Jefferies, Eyal Kalie, Andrea Longatti, Fiona E. Mcalpine, Nicole C.
Homework Hanson section MCB Course, Fall 2014
 Homework Hanson section MCB Course, Fall 2014 (1) Antitrypsin, which inhibits certain proteases, is normally secreted into the bloodstream by liver cells. Antitrypsin is absent from the bloodstream of
Homework Hanson section MCB Course, Fall 2014 (1) Antitrypsin, which inhibits certain proteases, is normally secreted into the bloodstream by liver cells. Antitrypsin is absent from the bloodstream of
Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2)
 Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
Problem Set #5 4/3/ Spring 02
 Question 1 Chloroplasts contain six compartments outer membrane, intermembrane space, inner membrane, stroma, thylakoid membrane, and thylakoid lumen each of which is populated by specific sets of proteins.
Question 1 Chloroplasts contain six compartments outer membrane, intermembrane space, inner membrane, stroma, thylakoid membrane, and thylakoid lumen each of which is populated by specific sets of proteins.
Figure S1. Western blot analysis of clathrin RNA interference in human DCs Human immature DCs were transfected with 100 nm Clathrin SMARTpool or
 Figure S1. Western blot analysis of clathrin RNA interference in human DCs Human immature DCs were transfected with 100 nm Clathrin SMARTpool or control nontargeting sirnas. At 90 hr after transfection,
Figure S1. Western blot analysis of clathrin RNA interference in human DCs Human immature DCs were transfected with 100 nm Clathrin SMARTpool or control nontargeting sirnas. At 90 hr after transfection,
MOLECULAR CELL BIOLOGY
 1 Lodish Berk Kaiser Krieger scott Bretscher Ploegh Matsudaira MOLECULAR CELL BIOLOGY SEVENTH EDITION CHAPTER 13 Moving Proteins into Membranes and Organelles Copyright 2013 by W. H. Freeman and Company
1 Lodish Berk Kaiser Krieger scott Bretscher Ploegh Matsudaira MOLECULAR CELL BIOLOGY SEVENTH EDITION CHAPTER 13 Moving Proteins into Membranes and Organelles Copyright 2013 by W. H. Freeman and Company
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Supplementary Figure 1. (A) Left, western blot analysis of ISGylated proteins in Jurkat T cells treated with 1000U ml -1 IFN for 16h (IFN) or left untreated (CONT); right, western
SUPPLEMENTARY FIGURES Supplementary Figure 1. (A) Left, western blot analysis of ISGylated proteins in Jurkat T cells treated with 1000U ml -1 IFN for 16h (IFN) or left untreated (CONT); right, western
Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes
 3888 Research Article Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes Andrew R. J. Young 1, Edmond Y. W. Chan 1, Xiao Wen Hu 1, Robert Köchl 1, Samuel G. Crawshaw
3888 Research Article Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes Andrew R. J. Young 1, Edmond Y. W. Chan 1, Xiao Wen Hu 1, Robert Köchl 1, Samuel G. Crawshaw
Islet viability assay and Glucose Stimulated Insulin Secretion assay RT-PCR and Western Blot
 Islet viability assay and Glucose Stimulated Insulin Secretion assay Islet cell viability was determined by colorimetric (3-(4,5-dimethylthiazol-2-yl)-2,5- diphenyltetrazolium bromide assay using CellTiter
Islet viability assay and Glucose Stimulated Insulin Secretion assay Islet cell viability was determined by colorimetric (3-(4,5-dimethylthiazol-2-yl)-2,5- diphenyltetrazolium bromide assay using CellTiter
Cell morphology. Cell organelles structure and function. Chapter 1: UNIT 1. Dr. Charushila Rukadikar
 UNIT 1 Cell morphology Cell organelles structure and function Chapter 1: Dr. Charushila Rukadikar Assistant Professor Department Of Physiology ZMCH, Dahod Physiology The science that is concerned with
UNIT 1 Cell morphology Cell organelles structure and function Chapter 1: Dr. Charushila Rukadikar Assistant Professor Department Of Physiology ZMCH, Dahod Physiology The science that is concerned with
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
Phospho-AKT Sampler Kit
 Phospho-AKT Sampler Kit E 0 5 1 0 0 3 Kits Includes Cat. Quantity Application Reactivity Source Akt (Ab-473) Antibody E021054-1 50μg/50μl IHC, WB Human, Mouse, Rat Rabbit Akt (Phospho-Ser473) Antibody
Phospho-AKT Sampler Kit E 0 5 1 0 0 3 Kits Includes Cat. Quantity Application Reactivity Source Akt (Ab-473) Antibody E021054-1 50μg/50μl IHC, WB Human, Mouse, Rat Rabbit Akt (Phospho-Ser473) Antibody
In Vivo and in Vitro Reconstitution of Atg8 Conjugation Essential for Autophagy*
 THE JOURNAL OF BIOLOGICAL CHEMISTRY Vol. 279, No. 39, Issue of September 24, pp. 40584 40592, 2004 2004 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in U.S.A. In Vivo and
THE JOURNAL OF BIOLOGICAL CHEMISTRY Vol. 279, No. 39, Issue of September 24, pp. 40584 40592, 2004 2004 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in U.S.A. In Vivo and
Broad Spectrum Protease Inhibitor Cocktail
 Broad Spectrum Protease Inhibitor Cocktail Catalog number: AR1182 Boster s Broad Spectrum Protease Inhibitor Cocktail is a complex of various protease inhibitors, which has been tested for inhibiting proteases
Broad Spectrum Protease Inhibitor Cocktail Catalog number: AR1182 Boster s Broad Spectrum Protease Inhibitor Cocktail is a complex of various protease inhibitors, which has been tested for inhibiting proteases
Rabbit Polyclonal antibody to NFkB p65 (v-rel reticuloendotheliosis viral oncogene homolog A (avian))
 Datasheet GeneTex, Inc : Toll Free 1-877-GeneTex (1-877-436-3839) Fax:1-949-309-2888 info@genetex.com GeneTex International Corporation : Tel:886-3-6208988 Fax:886-3-6208989 infoasia@genetex.com Date :
Datasheet GeneTex, Inc : Toll Free 1-877-GeneTex (1-877-436-3839) Fax:1-949-309-2888 info@genetex.com GeneTex International Corporation : Tel:886-3-6208988 Fax:886-3-6208989 infoasia@genetex.com Date :
Kinase Inhibitor p21 WAF1/CIP1 in Apoptosis and Autophagy
 Pivotal Role of the Cyclin-dependent Kinase Inhibitor p21 WAF1/CIP1 in Apoptosis and Autophagy Keishi Fujiwara, Shigeru Daido, Akitsugu Yamamoto, Ryuji Kobayash, Tomohisa Yokoyama, Hiroshi Aok, Eiji Iwado,
Pivotal Role of the Cyclin-dependent Kinase Inhibitor p21 WAF1/CIP1 in Apoptosis and Autophagy Keishi Fujiwara, Shigeru Daido, Akitsugu Yamamoto, Ryuji Kobayash, Tomohisa Yokoyama, Hiroshi Aok, Eiji Iwado,
Bioluminescence Resonance Energy Transfer (BRET)-based studies of receptor dynamics in living cells with Berthold s Mithras
 Bioluminescence Resonance Energy Transfer (BRET)-based studies of receptor dynamics in living cells with Berthold s Mithras Tarik Issad, Ralf Jockers and Stefano Marullo 1 Because they play a pivotal role
Bioluminescence Resonance Energy Transfer (BRET)-based studies of receptor dynamics in living cells with Berthold s Mithras Tarik Issad, Ralf Jockers and Stefano Marullo 1 Because they play a pivotal role
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/7/334/rs4/dc1 Supplementary Materials for Rapidly rendering cells phagocytic through a cell surface display technique and concurrent Rac activation Hiroki Onuma,
www.sciencesignaling.org/cgi/content/full/7/334/rs4/dc1 Supplementary Materials for Rapidly rendering cells phagocytic through a cell surface display technique and concurrent Rac activation Hiroki Onuma,
The Role of Parkin and Mitophagy in Acetaminophen and Alcohol-induced Liver Injuries. Jessica A. Williams
 The Role of Parkin and Mitophagy in Acetaminophen and Alcohol-induced Liver Injuries By Jessica A. Williams Submitted to the graduate degree program in Pharmacology, Toxicology, and Therapeutics and the
The Role of Parkin and Mitophagy in Acetaminophen and Alcohol-induced Liver Injuries By Jessica A. Williams Submitted to the graduate degree program in Pharmacology, Toxicology, and Therapeutics and the
Protein sorting (endoplasmic reticulum) Dr. Diala Abu-Hsasan School of Medicine
 Protein sorting (endoplasmic reticulum) Dr. Diala Abu-Hsasan School of Medicine dr.abuhassand@gmail.com An overview of cellular components Endoplasmic reticulum (ER) It is a network of membrane-enclosed
Protein sorting (endoplasmic reticulum) Dr. Diala Abu-Hsasan School of Medicine dr.abuhassand@gmail.com An overview of cellular components Endoplasmic reticulum (ER) It is a network of membrane-enclosed
Autophagy and Human Disease
 Cell Cycle ISSN: 1538-4101 (Print) 1551-4005 (Online) Journal homepage: http://www.tandfonline.com/loi/kccy20 Autophagy and Human Disease Ju Huang & Daniel J. Klionsky To cite this article: Ju Huang &
Cell Cycle ISSN: 1538-4101 (Print) 1551-4005 (Online) Journal homepage: http://www.tandfonline.com/loi/kccy20 Autophagy and Human Disease Ju Huang & Daniel J. Klionsky To cite this article: Ju Huang &
2011LandesBioscience. Donotdistribute. Seeing is believing The impact of electron microscopy on autophagy research
 review Autophagy 7:9, 935-956; September 2011; 2011 Landes Bioscience REVIEW Seeing is believing The impact of electron microscopy on autophagy research Eeva-Liisa Eskelinen, 1, * Fulvio Reggiori, 2 Misuzu
review Autophagy 7:9, 935-956; September 2011; 2011 Landes Bioscience REVIEW Seeing is believing The impact of electron microscopy on autophagy research Eeva-Liisa Eskelinen, 1, * Fulvio Reggiori, 2 Misuzu
04_polarity. The formation of synaptic vesicles
 Brefeldin prevents assembly of the coats required for budding Nocodazole disrupts microtubules Constitutive: coatomer-coated Selected: clathrin-coated The formation of synaptic vesicles Nerve cells (and
Brefeldin prevents assembly of the coats required for budding Nocodazole disrupts microtubules Constitutive: coatomer-coated Selected: clathrin-coated The formation of synaptic vesicles Nerve cells (and
Binding of macrophages and phospholipid flip-flop in supported lipid bilayers
 Volume 192, number 1 FEBS 3073 November 1985 Binding of macrophages and phospholipid flip-flop in supported lipid bilayers Mamoru Nakanishi*, Kazuko Matsumoto and Seizo Takahashi ~Fac~~ty of Pharmaceutical
Volume 192, number 1 FEBS 3073 November 1985 Binding of macrophages and phospholipid flip-flop in supported lipid bilayers Mamoru Nakanishi*, Kazuko Matsumoto and Seizo Takahashi ~Fac~~ty of Pharmaceutical
Chapter 13: Vesicular Traffic
 Chapter 13: Vesicular Traffic Know the terminology: ER, Golgi, vesicle, clathrin, COP-I, COP-II, BiP, glycosylation, KDEL, microtubule, SNAREs, dynamin, mannose-6-phosphate, M6P receptor, endocytosis,
Chapter 13: Vesicular Traffic Know the terminology: ER, Golgi, vesicle, clathrin, COP-I, COP-II, BiP, glycosylation, KDEL, microtubule, SNAREs, dynamin, mannose-6-phosphate, M6P receptor, endocytosis,
