DEFINITIONS OF HISTOCOMPATIBILITY TYPING TERMS
|
|
|
- Anne Morrison
- 6 years ago
- Views:
Transcription
1 DEFINITIONS OF HISTOCOMPATIBILITY TYPING TERMS The definitions below are intended as general concepts. There will be exceptions to these general definitions. These definitions do not imply any specific requirements for HLA typing but are only meant to define useful terms. Definitions of typing resolution. HLA laboratories should have a written agreement with each entity requesting HLA typing regarding the specifications for the resolution of typing. The following terms might be used in the agreement. These definitions do not imply any specific requirements for typing; that decision is made by the HLA typing laboratory in compliance with testing standards and requirements of the test requesting entity. Allelic resolution: The DNA-based typing result is consistent with a single allele as defined in a given version of the WHO HLA Nomenclature Report as described on the reference web site, An allele is defined as a unique nucleotide sequence for a gene as defined by the use of all of the digits in a current allele name. Examples include A*01:01:01:01; A*02:07 (based on the IMGT/HLA Database version 3.1.0, July 2010). High resolution: A high resolution typing result is defined as a set of alleles that specify and encode the same protein sequence for the peptide binding region of an HLA molecule and that excludes alleles that are not expressed as cell-surface proteins. Examples are typings with the following characteristics: (1) includes only alleles within a P group designation eg., A*02:01P, DPB1*04:02P or (2) includes alleles within a G group designation (eg., A*02:01:01G, DRB1*12:01:01G) with the exception that it does not include non-expressed alleles with the same nucleotide sequence (e.g., A*02:01:01G without A*02:43N and A*02:83N). A list of P and G groups can be found at The appendix shows another example. Confirmed allele high resolution: A confirmed allele is one that has been identified in two or more unrelated individuals and is designated as confirmed in the IMGT/HLA database ( A confirmed allele high resolution typing result is defined as a set of alleles that 1. specify and encode the same protein sequence for the peptide binding region of an HLA molecule, and 2. exclude confirmed alleles that are not expressed as cell-surface proteins and 3. include alternative genotypes with two unconfirmed alleles that specify different protein sequences, when applicable. An example of confirmed allele high resolution typing is: A*02:01:01G in which the nonexpressed alleles, A*02:43N and A*02:83N, are not excluded because both are unconfirmed. The appendix shows another example. Page 1 of 9
2 Common and well-documented (CWD) allele high resolution: This resolution is based on a list of alleles designated as CWD and distinguished from rare alleles. An initial report defining CWD alleles has been published (Cano P, et.al., Common and welldocumented HLA alleles: report of the ad-hoc committee of the American Society for Histocompatibility and Immunogenetics. Hum Immunol 2007; 68(5): ); however, it is suggested that laboratories rely on the most current definition of CWD alleles. Common alleles appear with gene frequencies greater than in any reference population. Accurate frequency estimations can be made for alleles observed three or more times; thus, population studies with a sample size of 1,500 unrelated subjects (2n = 3,000 chromosomes) can detect with accuracy only allele frequencies of or greater. Well-documented alleles are a second category which includes those alleles that have been observed in at least three unrelated individuals. In certain cases, the gene frequency of well-documented alleles may not be estimated accurately. This is the case for alleles found only in isolated populations such that their frequencies are likely to be less than in any outbred population. Rare alleles have extremely low frequencies and are not likely to be found again in a significant number of unrelated subjects. Intermediate resolution: Intermediate resolution is defined as a DNA-based typing result that includes a subset of alleles sharing the digits in the first field of their allele name and that excludes some alleles sharing those digits. Examples include: A*02:01 or A*02:02 or A*02:07 or A*02:20 but not other A*02 alleles. There may be cases in which the subset of alleles includes one or more alleles with a name beginning with different digits but these alleles should be the exception i.e., the majority of the alleles should share the same first digits eg., A*01:01 or A*01:02 or A*01:14 or A*36:04. Low resolution: The DNA-based typing result is at the level of the digits comprising the first field in the DNA-based nomenclature. Examples include: A*01; A*02. The exception is B*15 in which low resolution is defined as a subset of alleles that might be considered as sharing a broad serologic type, either B15 or B70. Replacement of the term confirmatory typing. It was felt that the activities encompassed within the term confirmatory typing have become unclear. It is recommended that the following two terms be used in the place of the words confirmatory typing. Verification typing: HLA typing performed on an independent sample (or, for a cord blood unit, from an attached segment or from the unit itself) with the purpose of verifying concordance of that typing assignment with the initial HLA typing assignment. Concordance does not require identical levels of resolution for the two sets of typing but requires the two assignments to be consistent with one another. Page 2 of 9
3 Extended typing: HLA typing performed to add additional information to an existing HLA assignment. This additional HLA typing may: (1) extend the typing to include assignments at additional HLA loci (e.g., to type HLA-C for an HLA-A,-B,-DRB1 typed volunteer donor) and/or (2) include additional typing to increase the resolution at any previously typed HLA locus (e.g., to type an HLA-B serologically typed individual to identify the alleles potentially carried at the HLA-B locus at intermediate, at high or at allelic resolution. Format for the report of HLA assignments. HLA typing assignments must be clearly understood by the end user. HLA laboratories should have a written agreement with each entity requesting HLA typing regarding the specifications for the typing. This must include the loci to be tested, the level of resolution of the typing, criteria for selection of a donor for allogeneic transplant, and the format in which typing results will be reported. The impact of any uncertainty in HLA assignments of either the potential donor or patient on matching should be addressed in the report. Unresolved alternative assignments: It is strongly recommended that the report to the end user include all uncertainty in the typing assignment. This means that genotypes and/or alleles that have not been excluded should be listed in the final report. If it is not possible to provide a list of unresolved alternatives on the report, the laboratory should indicate that alternative assignments exist and might provide a rationale for the HLA assignment that is selected to be included in the report. Database used in interpretation of typing results: The version of the IMGT/HLA Database used to interpret the HLA results should be included in the report to the end user. Reporting a string of alleles: Slashes should be used to separate a string of alternative alleles e.g., A*02:01/02:02/02:07/02:20 to mean A*02:01 or A*02:02 or A*02:07 or A*02:20. When the laboratory wishes to shorten the length of the string, it is recommended that the laboratory use the following format: A*02:01/02/07/20; DRB1*01:01:01/02. The more complex assignment to represent A*02:01:01 or A*02:01:02 or A*02:05 would be A*02:01:01/02/A*02:05. Other notations may be utilized as long as the typing assignment is clearly understood by the end user. Matching: If the laboratory is providing HLA assignments for a potential donor and patient for allogeneic transplantation, the laboratory might include a description of the matching status of the pair. Matching denotes interpretation of the comparison of HLA types of patient (P) and a putative donor (D). The outcome of this interpretation is either a match (they share the type) or a mismatch (they do not). Directionality of match: The laboratory may wish to refer to directionality (or vector) of the mismatch in cases where the patient or a potential donor is homozygous at a locus Page 3 of 9
4 or has a non-expressed allele. If the patient is homozygous and one of the HLA assignments is identical to an assignment of the heterozygous donor (e.g., patient A*01:01:01:01, potential donor A*01:01:01:01, A*23:01), the mismatch may be referred to as a mismatch in the host versus graft vector direction. If the potential donor is homozygous and one of the HLA assignments is identical to an assignment of the heterozygous patient, the mismatch may be referred to as a mismatch in the graft versus host vector direction. Matching within a family: HLA haplotypes identical by descent: This phrase may be used when (1) parental HLA assignments are available, (2) all four haplotypes are unequivocally defined in the family, (3) the HLA assignments of the parents are clearly distinguishable from one another, and (4) the assignments include HLA-A, -B, -C, DRB1, DQB1 and DPB1. Other loci (DRA, DQA1, DPA1, DRB3/4/5) may be included. The patient and potential donor who share both haplotypes may be described as HLA identical by descent. HLA identical for all loci tested: This phrase may be used to refer to matching of related donors who appear to share the HLA loci tested with the patient based on segregation within the family. This phrase would refer to matching in which not all HLA loci (that is, HLA-A, -B, -C, DRB1, DQB1 and DPB1) are tested (e.g. not DPB1) so the possibility of recombination is not excluded. Families where segregation to confirm identity by descent is not possible: For example, if there are two common haplotypes present within a family, and it has not been possible to exclude the possibility that a phenotypically identical sibling donor is actually only haploidentical, the phrases used to describe matching should be those used for an unrelated donor (see below). Matching of patient to unrelated donor or matching within a family where identity by descent can not be ascertained: Matched for {insert} at {insert} resolution: A phrase like this might be used to refer to the number of loci tested (i.e., two assignments at three loci yielding 6 assignments to include HLA-A,-B,-DRB1 or four loci yielding 8 assignments with the addition of HLA-C or 10 assignments with the addition of HLA-DQB1 or 12 assignments with DPB1), the potential identity of the assignments (e.g., 8/8 or 7/8 or 9/10), and the level of resolution used to determine the potential identity (e.g., confirmed allele high resolution or intermediate resolution). The number of loci to be included in this grading of match should be agreed locally with the service users. For example, an umbilical cord blood unit may be described as matched for 6/6 at low resolution for HLA-A and HLA-B and at allelic level resolution for HLA-DRB1 with the Page 4 of 9
5 patient. For example, an adult volunteer donor may be described as matched for 9/10 at high resolution with the patient. Reference web site: Information on the current status of this document can be found at Approval: These definitions have been reviewed by representatives from the following organizations. The extent to which each organization is able to incorporate the definitions above can be found on the web site of each organization. AABB American Society for Histocompatibility and Immunogenetics (ASHI) College of American Pathologists (CAP) European Federation for Immunogenetics (EFI) Foundation for the Accreditation of Cellular Therapy (FACT) National Marrow Donor Program (NMDP) World Marrow Donor Association (WMDA) American Society for Blood and Marrow Transplantation (ASBMT) Page 5 of 9
6 Appendix I: Examples of Resolution Typing required to reach resolution when the following alternative genotypes are present: A*01:01:01G+A*02:01:01G or A*01:01:13+A*02:01:02 or A*01:14+A*02:101 or A*02:36+A*36:04 The high resolution typing result would be: A*01:01:01:01/A*01:32/A*01:37/A*01:45 + A*02:01:01:01/A*02:01:01:02L/A*02:01:01:03/A*02:01:08/A*02:01:11/A*02:01:14/ A*02:01:15/A*02:01:21/A*02:09/A*02:66/A*02:75/A*02:89/A*02:97:01/A*02:97:02/A *02:132/A*02:134/A*02:140 or A*01:01:13+A*02:01:02 Alternative Genotypes Present A*01:01:01G+A*02:01:01G A*01:01:01G= A*01:01:01:01/ A*01:01:01:02N/ A*01:04N/ A*01:22N/ A*01:32/ A*01:34N/ A*01:37/ A*01:45 A*02:01:01G= A*02:01:01:01/ A*02:01:01:02L/ A*02:01:01:03/ A*02:01:08/ A*02:01:11/ A*02:01:14/ A*02:01:15/ A*02:01:21/ A*02:09/ A*02:43N/ A*02:66/ A*02:75/ A*02:83N/ A*02:89/ A*02:97:01/ A*02:97:02/ A*02:132/ A*02:134/ A*02:140 A*01:01:13+A*02:01:02 A*01:14+A*02:101 A*02:36+A*36:04 To Achieve a High Resolution Typing Result, The Typing Result Must Be Narrowed Down to the Following Alleles: Non-expressed alleles are excluded to give: A*01:01:01:01/ A*01:32/ A*01:37/ A*01:45 A*02:01:01:01/ A*02:01:01:02L/ A*02:01:01:03/ A*02:01:08/ A*02:01:11/ A*02:01:14/ A*02:01:15/ A*02:01:21/ A*02:09/ A*02:66/ A*02:75/ A*02:89/ A*02:97:01/ A*02:97:02/ A*02:132/ A*02:134/ A*02:140 Same peptide binding region so further resolution is not required A*01:01:13+A*02:01:02 Different peptide binding regions; Testing must exclude this genotype Different peptide binding regions; Testing must exclude this genotype Page 6 of 9
7 The confirmed allele high resolution typing would be: A*01:01:01:01/A*01:01:01:02N/A*01:22N/A*01:32/A*01:34N/A*01:37/A*01:45 + A*02:01:01G or A*01:01:13+A*02:01:02 or A*01:14+A*02:101 or A*02:36+A*36:04 Alternative Genotypes A*01:01:01G+A*02:01:01G A*01:01:01G= A*01:01:01:01/ A*01:01:01:02N/ A*01:04N/ A*01:22N/ A*01:32/ A*01:34N/ A*01:37/ A*01:45 A*02:01:01G= A*02:01:01:01/ A*02:01:01:02L/ A*02:01:01:03/ A*02:01:08/ A*02:01:11/ A*02:01:14/ A*02:01:15/ A*02:01:21/ A*02:09/ A*02:43N/ A*02:66/ A*02:75/ A*02:83N/ A*02:89/ A*02:97:01/ A*02:97:02/ A*02:132/ A*02:134/ A*02:140 A*01:01:13+A*02:01:02 A*01:14+A*02:101 A*02:36+A*36:04 To Achieve Confirmed Allele High Resolution Typing Result, The Typing Result Must Be Narrowed Down to the Following Alleles: Confirmed non-expressed alleles are excluded to give: A*01:01:01:01/ A*01:01:01:02N/ A*01:22N/ A*01:32/ A*01:34N/ A*01:37/ A*01:45 [Note: A*01:34N has been found in only one individual according to IMGT/HLA but has been sequenced by two labs] A*02:01:01:01/ A*02:01:01:02L/ A*02:01:01:03/ A*02:01:08/ A*02:01:11/ A*02:01:14/ A*02:01:15/ A*02:01:21/ A*02:09/ A*02:43N/ A*02:66/ A*02:75/ A*02:83N/ A*02:89/ A*02:97:01/ A*02:97:02/ A*02:132/ A*02:134/ A*02:140 Same peptide binding region so not required to resolve A*01:01:13+A*02:01:02 Both alleles unconfirmed so further typing to exclude this genotype is not required A*01:14+A*02:101 Both alleles unconfirmed so further typing to exclude this genotype not required A*02:36+A*36:04 Typing required to reach resolution when the following alternative genotypes are present: A*02:01:01G + A*68:02:01G or A*02:34 + A*68:31 or A*02:35:01 + A*68:15 The high resolution typing result would be: A*02:01:01:01/A*02:01:01:02L/A*02:01:01:03/A*02:01:08/A*02:01:11/A*02:01:14/A* Page 7 of 9
8 02:01:15/A*02:01:21/A*02:09/A*02:66/A*02:75/A*02:89/A*02:97:01/A*02:97:02/A*0 2:132/A*02:134/A*02:140 + A*68:02:01G The confirmed allele high resolution typing would be: A*02:01:01G + A*68:02:01G Alternative Genotypes A*02:01:01G + A*68:02:01G A*02:01:01G= A*02:01:01:01/ A*02:01:01:02L/ A*02:01:01:03/ A*02:01:08/ A*02:01:11/ A*02:01:14/ A*02:01:15/ A*02:01:21/ A*02:09/ A*02:43N/ A*02:66/ A*02:75/ A*02:83N/ A*02:89/ A*02:97:01/ A*02:97:02/ A*02:132/ A*02:134/ A*02:140 A*68:02:01G= A*68:02:01/ A*68:02:01:02/ A*68:01:02:03 A*02:34 + A*68:31 A*02:35:01 + A*68:15 To Achieve Confirmed Allele High Resolution Typing Result, The Typing Result Must Be Narrowed Down to the Following Alleles: Confirmed non-expressed alleles are excluded: A*02:01:01G (A*02:01:01:01/ A*02:01:01:02L/ A*02:01:01:03/ A*02:01:08/ A*02:01:11/ A*02:01:14/ A*02:01:15/ A*02:01:21/ A*02:09/ A*02:43N/ A*02:66/ A*02:75/ A*02:83N/ A*02:89/ A*02:97:01/ A*02:97:02/ A*02:132/ A*02:134/ A*02:140) A*68:02:01G One allele (A*68:31) is confirmed so further typing to exclude this genotype is required One allele (A*68:15) is confirmed so further typing to exclude this genotype is required Page 8 of 9
9 Appendix II: Further justification and information HLA typing assigns genotypes or pairs of alleles. The results of DNA based typing are genotypes, that is, combinations of two HLA alleles that might be carried by an individual. For example, the typing result may be consistent with the following 3 genotypes: B*08:01:01G+B*15:18:01 B*08:21+B*15:93 B*08:35+B*15:10:01 This result, which includes alternative genotypes, is called ambiguous because it is not known which of the 3 genotypes the individual actually carries. Depending on the typing method, the list of genotypes can be very long. If the laboratory decides not to resolve the ambiguity, it has three alternatives for reporting the HLA typing of this individual. (1) The list of 3 genotypes can be reported. (2) The result might be collapsed into B*08:01/21/35, B*15:10/18/93 or B*08:MDY (01/21/35), B*15:DZBP (10/18/93). Furthermore, if the result included alleles that might be represented by a G or P code, this designation might be used. (3) Based on allele and haplotype frequencies, the lab might predict the most likely genotype and indicate this on the report. For example, since B*08:21, B*08:35, and B*15:93 are on the NMDP s rare allele list, the lab might indicate that B*08:01:01:01+B*15:18:01 is the genotype expected to be present. In this case, the report should note alternative genotypes that were not excluded by testing. If the actual genotype carried by a donor and/or a patient is not known, the transplant center will not know whether or not the pair is an allele match, and this uncertainty should be clearly stated. A list of ambiguous alleles and ambiguous genotypes can be found on the HLA nomenclature web site ( and the IMGT/HLA website ( Page 9 of 9
Documentation of Changes to EFI Standards: v 5.6.1
 Modified Standard B - PERSONNEL QUALIFICATIONS B1.000 The laboratory must employ one or more individuals who meet the qualifications and fulfil the responsibilities of the Director/Co-Director, Technical
Modified Standard B - PERSONNEL QUALIFICATIONS B1.000 The laboratory must employ one or more individuals who meet the qualifications and fulfil the responsibilities of the Director/Co-Director, Technical
Histocompatibility Evaluations for HSCT at JHMI. M. Sue Leffell, PhD. Professor of Medicine Laboratory Director
 Histocompatibility Evaluations for HSCT at JHMI M. Sue Leffell, PhD Professor of Medicine Laboratory Director JHMI Patient Population Adults Peds NMDP data >20,000 HSCT JHMI HSCT Protocols Bone marrow
Histocompatibility Evaluations for HSCT at JHMI M. Sue Leffell, PhD Professor of Medicine Laboratory Director JHMI Patient Population Adults Peds NMDP data >20,000 HSCT JHMI HSCT Protocols Bone marrow
Human Leukocyte Antigens and donor selection
 Human Leukocyte Antigens and donor selection Duangtawan Thammanichanond, MD. PhD. Histocompatibility and Immunogenetics Laboratory, Faculty of Medicine Ramathibodi Hospital, Mahidol University Outline
Human Leukocyte Antigens and donor selection Duangtawan Thammanichanond, MD. PhD. Histocompatibility and Immunogenetics Laboratory, Faculty of Medicine Ramathibodi Hospital, Mahidol University Outline
HLA-A * L
 What is Nomenclature? HLA Nomenclature 3.0 Signifies DNA Subtype Differences outside the coding region (introns) HLA-A * 4 0 01 0 L Steve Spellman Sr. Manager, NMDP Asst. Scientific Director, CIBMTR Locus
What is Nomenclature? HLA Nomenclature 3.0 Signifies DNA Subtype Differences outside the coding region (introns) HLA-A * 4 0 01 0 L Steve Spellman Sr. Manager, NMDP Asst. Scientific Director, CIBMTR Locus
Completing the CIBMTR Confirmation of HLA Typing Form (Form 2005)
 Completing the CIBMTR Confirmation of HLA Typing Form (Form 2005) Stephen Spellman Research Manager NMDP Scientific Services Maria Brown Scientific Services Specialist Data Management Conference 2007 1
Completing the CIBMTR Confirmation of HLA Typing Form (Form 2005) Stephen Spellman Research Manager NMDP Scientific Services Maria Brown Scientific Services Specialist Data Management Conference 2007 1
HLA Mismatches. Professor Steven GE Marsh. Anthony Nolan Research Institute EBMT Anthony Nolan Research Institute
 HLA Mismatches Professor Steven GE Marsh HLA Mismatches HLA Genes, Structure, Polymorphism HLA Nomenclature HLA Mismatches in HSCT Defining a mismatch HLA Mismatches HLA Genes, Structure, Polymorphism
HLA Mismatches Professor Steven GE Marsh HLA Mismatches HLA Genes, Structure, Polymorphism HLA Nomenclature HLA Mismatches in HSCT Defining a mismatch HLA Mismatches HLA Genes, Structure, Polymorphism
The Human Major Histocompatibility Complex
 The Human Major Histocompatibility Complex 1 Location and Organization of the HLA Complex on Chromosome 6 NEJM 343(10):702-9 2 Inheritance of the HLA Complex Haplotype Inheritance (Family Study) 3 Structure
The Human Major Histocompatibility Complex 1 Location and Organization of the HLA Complex on Chromosome 6 NEJM 343(10):702-9 2 Inheritance of the HLA Complex Haplotype Inheritance (Family Study) 3 Structure
ASHI Proficiency Testing Program Summary Report. Survey 2013-HT1 / HLA Typing
 ASHI Proficiency Testing Program Summary Report Survey 2013-HT1 / HLA Typing Shipping Date: February 26,2013 / Results Due Date:April 5,2013 / Report Date: May 17,2013 The ASHI HT proficiency testing survey
ASHI Proficiency Testing Program Summary Report Survey 2013-HT1 / HLA Typing Shipping Date: February 26,2013 / Results Due Date:April 5,2013 / Report Date: May 17,2013 The ASHI HT proficiency testing survey
Minimal Requirements for Histocompatibility & Immunogenetics Laboratory
 Minimal Requirements for Histocompatibility & Immunogenetics Laboratory The 4 th WBMT Congress and Workshop Riyadh, KSA - January 15-17, 2017 HLA Discovery, 1958 The Nobel Prize in Physiology or Medicine
Minimal Requirements for Histocompatibility & Immunogenetics Laboratory The 4 th WBMT Congress and Workshop Riyadh, KSA - January 15-17, 2017 HLA Discovery, 1958 The Nobel Prize in Physiology or Medicine
OPTN/UNOS Policy Notice Review of HLA Tables (2016)
 Review of HLA Tables (2016) Sponsoring Committee: Policy/Bylaws Affected: Histocompatibility Policy 4.10 (Reference Tables of HLA Antigen Values and Split Equivalences) Public Comment: July 31, 2017 October
Review of HLA Tables (2016) Sponsoring Committee: Policy/Bylaws Affected: Histocompatibility Policy 4.10 (Reference Tables of HLA Antigen Values and Split Equivalences) Public Comment: July 31, 2017 October
Significance of the MHC
 CHAPTER 7 Major Histocompatibility Complex (MHC) What is is MHC? HLA H-2 Minor histocompatibility antigens Peter Gorer & George Sneell (1940) Significance of the MHC role in immune response role in organ
CHAPTER 7 Major Histocompatibility Complex (MHC) What is is MHC? HLA H-2 Minor histocompatibility antigens Peter Gorer & George Sneell (1940) Significance of the MHC role in immune response role in organ
MATCHMAKER, MATCHMAKER, MAKE ME A MATCH, FIND ME A MISMATCHED TRANSPLANT TO CATCH
 MATCHMAKER, MATCHMAKER, MAKE ME A MATCH, FIND ME A MISMATCHED TRANSPLANT TO CATCH TEJASWINI M. DHAWALE, M.D. HEME FELLOWS CONFERENCE NOVEMBER 08, 2013 CASE PRESENTATION 51 yo M with history of MDS (unilinear
MATCHMAKER, MATCHMAKER, MAKE ME A MATCH, FIND ME A MISMATCHED TRANSPLANT TO CATCH TEJASWINI M. DHAWALE, M.D. HEME FELLOWS CONFERENCE NOVEMBER 08, 2013 CASE PRESENTATION 51 yo M with history of MDS (unilinear
The MHC and Transplantation Brendan Clark. Transplant Immunology, St James s University Hospital, Leeds, UK
 The MHC and Transplantation Brendan Clark Transplant Immunology, St James s University Hospital, Leeds, UK Blood Groups Immunofluorescent staining has revealed blood group substance in the cell membranes
The MHC and Transplantation Brendan Clark Transplant Immunology, St James s University Hospital, Leeds, UK Blood Groups Immunofluorescent staining has revealed blood group substance in the cell membranes
Handling Immunogenetic Data Managing and Validating HLA Data
 Handling Immunogenetic Data Managing and Validating HLA Data Steven J. Mack PhD Children s Hospital Oakland Research Institute 16 th IHIW & Joint Conference Sunday 3 June, 2012 Overview 1. Master Analytical
Handling Immunogenetic Data Managing and Validating HLA Data Steven J. Mack PhD Children s Hospital Oakland Research Institute 16 th IHIW & Joint Conference Sunday 3 June, 2012 Overview 1. Master Analytical
Role of NMDP Repository in the Evolution of HLA Matching and Typing for Unrelated Donor HCT
 Role of NMDP Repository in the Evolution of HLA Matching and Typing for Unrelated Donor HCT Stephen Spellman, MBS Director, Immunobiology and Observational Research Assistant Scientific Director CIBMTR,
Role of NMDP Repository in the Evolution of HLA Matching and Typing for Unrelated Donor HCT Stephen Spellman, MBS Director, Immunobiology and Observational Research Assistant Scientific Director CIBMTR,
Title: SECTION 7.0 NZBMDR STANDARDS PROCESS FOR DONOR IDENTIFICATION
 Title: SECTION 7.0 NZBMDR STANDARDS PROCESS FOR DONOR IDENTIFICATION Authorised by: Executive Officer Contributing Authors: ABMDR Scientific Expert Advisory Committee Sally Gordon Dr Hilary Blacklock Raewyn
Title: SECTION 7.0 NZBMDR STANDARDS PROCESS FOR DONOR IDENTIFICATION Authorised by: Executive Officer Contributing Authors: ABMDR Scientific Expert Advisory Committee Sally Gordon Dr Hilary Blacklock Raewyn
EBMT2008_1_21:EBMT :06 Pagina 46 * CHAPTER 3. Immunogenetics of allogeneic HSCT * 3.1. The role of HLA in HSCT. J.M.
 EBMT2008_1_21:EBMT2008 6-11-2008 9:06 Pagina 46 * CHAPTER 3 Immunogenetics of allogeneic HSCT * 3.1 The role of HLA in HSCT J.M. Tiercy EBMT2008_1_21:EBMT2008 6-11-2008 9:06 Pagina 47 CHAPTER 3.1 HLA and
EBMT2008_1_21:EBMT2008 6-11-2008 9:06 Pagina 46 * CHAPTER 3 Immunogenetics of allogeneic HSCT * 3.1 The role of HLA in HSCT J.M. Tiercy EBMT2008_1_21:EBMT2008 6-11-2008 9:06 Pagina 47 CHAPTER 3.1 HLA and
25/10/2017. Clinical Relevance of the HLA System in Blood Transfusion. Outline of talk. Major Histocompatibility Complex
 Clinical Relevance of the HLA System in Blood Transfusion Dr Colin J Brown PhD FRCPath. October 2017 Outline of talk HLA genes, structure and function HLA and immune complications of transfusion TA-GVHD
Clinical Relevance of the HLA System in Blood Transfusion Dr Colin J Brown PhD FRCPath. October 2017 Outline of talk HLA genes, structure and function HLA and immune complications of transfusion TA-GVHD
National Marrow Donor Program HLA-Matching Guidelines for Unrelated Marrow Transplants
 Biology of Blood and Marrow Transplantation 9:610-615 (2003) 2003 American Society for Blood and Marrow Transplantation 1083-8791/03/0910-0003$30.00/0 doi:10.1016/s1083-8791(03)00329-x National Marrow
Biology of Blood and Marrow Transplantation 9:610-615 (2003) 2003 American Society for Blood and Marrow Transplantation 1083-8791/03/0910-0003$30.00/0 doi:10.1016/s1083-8791(03)00329-x National Marrow
How to Find an Unrelated Donor Theory & Technology
 How to Find an Unrelated Donor Theory & Technology Carlheinz R. Müller Zentrales Knochenmarkspender-Register für die Bundesrepublik Deutschland (ZKRD) Ulm, Germany How to Find an Unrelated Donor HLA-Basics
How to Find an Unrelated Donor Theory & Technology Carlheinz R. Müller Zentrales Knochenmarkspender-Register für die Bundesrepublik Deutschland (ZKRD) Ulm, Germany How to Find an Unrelated Donor HLA-Basics
21/05/2018. Continuing Education. Presentation Recording. learn.immucor.com
 Transplant Webinar Series: Ep. 6 Donor Selection for Haematopoietic Stem Cell Transplantation Future Webinars The Role of NGS in the Transplant Setting Featuring Dr Sujatha Krishnakumar Sirona Genomics,
Transplant Webinar Series: Ep. 6 Donor Selection for Haematopoietic Stem Cell Transplantation Future Webinars The Role of NGS in the Transplant Setting Featuring Dr Sujatha Krishnakumar Sirona Genomics,
Factors Influencing Haematopoietic Progenitor cell transplant outcome Optimising donor selection
 Factors Influencing Haematopoietic Progenitor cell transplant outcome Optimising donor selection Alison Logan Transplantation Laboratory Manchester Royal Infirmary Haematopoietic progenitor cell transplants
Factors Influencing Haematopoietic Progenitor cell transplant outcome Optimising donor selection Alison Logan Transplantation Laboratory Manchester Royal Infirmary Haematopoietic progenitor cell transplants
CHAPTER 3 LABORATORY PROCEDURES
 CHAPTER 3 LABORATORY PROCEDURES CHAPTER 3 LABORATORY PROCEDURES 3.1 HLA TYPING Molecular HLA typing will be performed for all donor cord blood units and patients in the three reference laboratories identified
CHAPTER 3 LABORATORY PROCEDURES CHAPTER 3 LABORATORY PROCEDURES 3.1 HLA TYPING Molecular HLA typing will be performed for all donor cord blood units and patients in the three reference laboratories identified
Outcomes of pediatric bone marrow transplantation for leukemia and myelodysplasia using matched. unrelated donors
 Outcomes of pediatric bone marrow transplantation for leukemia and myelodysplasia using matched sibling, mismatched related or matched unrelated donors Immunobiology Working Committee PIs: Peter Shaw and
Outcomes of pediatric bone marrow transplantation for leukemia and myelodysplasia using matched sibling, mismatched related or matched unrelated donors Immunobiology Working Committee PIs: Peter Shaw and
Clinical Relevance of the HLA System in Blood Transfusion. Dr Colin J Brown PhD FRCPath. October 2017
 Clinical Relevance of the HLA System in Blood Transfusion Dr Colin J Brown PhD FRCPath. October 2017 Outline of talk HLA genes, structure and function HLA and immune complications of transfusion TA-GVHD
Clinical Relevance of the HLA System in Blood Transfusion Dr Colin J Brown PhD FRCPath. October 2017 Outline of talk HLA genes, structure and function HLA and immune complications of transfusion TA-GVHD
Evaluation of MIA FORA NGS HLA test and software. Lisa Creary, PhD Department of Pathology Stanford Blood Center Research & Development Group
 Evaluation of MIA FORA NGS HLA test and software Lisa Creary, PhD Department of Pathology Stanford Blood Center Research & Development Group Disclosure Alpha and Beta Studies Sirona Genomics Reagents,
Evaluation of MIA FORA NGS HLA test and software Lisa Creary, PhD Department of Pathology Stanford Blood Center Research & Development Group Disclosure Alpha and Beta Studies Sirona Genomics Reagents,
2/10/2016. Evaluation of MIA FORA NGS HLA test and software. Disclosure. NGS-HLA typing requirements for the Stanford Blood Center
 Evaluation of MIA FORA NGS HLA test and software Lisa Creary, PhD Department of Pathology Stanford Blood Center Research & Development Group Disclosure Alpha and Beta Studies Sirona Genomics Reagents,
Evaluation of MIA FORA NGS HLA test and software Lisa Creary, PhD Department of Pathology Stanford Blood Center Research & Development Group Disclosure Alpha and Beta Studies Sirona Genomics Reagents,
CHAPTER 2 PROTOCOL DESIGN
 CHAPTER 2 PROTOCOL DESIGN CHAPTER 2 PROTOCOL DESIGN 2.1 ELIGIBILITY CRITERIA Participants fulfilling the following criteria will be eligible for enrollment in the protocol: 1. Participant is diagnosed
CHAPTER 2 PROTOCOL DESIGN CHAPTER 2 PROTOCOL DESIGN 2.1 ELIGIBILITY CRITERIA Participants fulfilling the following criteria will be eligible for enrollment in the protocol: 1. Participant is diagnosed
HLA and new technologies. Vicky Van Sandt
 HLA and new technologies. Vicky Van Sandt Life-threatning malignant and non malignant blood disorders can be cured by hematopoetic stem cell transplantation (HSCT). GVHD is the 2nd most prevalent cause
HLA and new technologies. Vicky Van Sandt Life-threatning malignant and non malignant blood disorders can be cured by hematopoetic stem cell transplantation (HSCT). GVHD is the 2nd most prevalent cause
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Gragert L, Eapen M, Williams E, et al. HLA match likelihoods
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Gragert L, Eapen M, Williams E, et al. HLA match likelihoods
OPERATIONS AND PATIENT SERVICES USER GUIDE. Procedures for transplant centres and international establishments
 OPERATIONS AND PATIENT SERVICES USER GUIDE Procedures for transplant centres and international establishments 1. INTRODUCTION 9 1.1. Introduction 9 1.2. User Guide 9 1.3. Services Provided by Anthony Nolan
OPERATIONS AND PATIENT SERVICES USER GUIDE Procedures for transplant centres and international establishments 1. INTRODUCTION 9 1.1. Introduction 9 1.2. User Guide 9 1.3. Services Provided by Anthony Nolan
EZER MIZION PROCEDURES FOR TRANSPLANT CENTERS AND COOPERATIVE REGISTRIES
 EZER MIZION PROCEDURES FOR TRANSPLANT CENTERS AND COOPERATIVE REGISTRIES Table of content 1. Purpose and Scope... 2 2. Introduction... 2 3. Procedure for Requesting a Search... 3 4. The Search Process...
EZER MIZION PROCEDURES FOR TRANSPLANT CENTERS AND COOPERATIVE REGISTRIES Table of content 1. Purpose and Scope... 2 2. Introduction... 2 3. Procedure for Requesting a Search... 3 4. The Search Process...
6/19/2012. Who is in the room today? What is your level of understanding of Donor Antigens and Candidate Unacceptables in KPD?
 6/19/212 KPD Webinar Series: Part 3 Demystifying the OPTN Kidney Paired Donation Pilot Program Unacceptable HLA antigens: The key to finding a compatible donor for your patient M. Sue Leffell, Ph.D. J.
6/19/212 KPD Webinar Series: Part 3 Demystifying the OPTN Kidney Paired Donation Pilot Program Unacceptable HLA antigens: The key to finding a compatible donor for your patient M. Sue Leffell, Ph.D. J.
New trends in donor selection in Europe: "best match" versus haploidentical. Prof Jakob R Passweg
 New trends in donor selection in Europe: "best match" versus haploidentical Prof Jakob R Passweg HSCT change in donor type: 1990-2015 9000 H S C T 8000 7000 6000 5000 4000 HLA identical sibling/twin Haplo-identical
New trends in donor selection in Europe: "best match" versus haploidentical Prof Jakob R Passweg HSCT change in donor type: 1990-2015 9000 H S C T 8000 7000 6000 5000 4000 HLA identical sibling/twin Haplo-identical
Basel - 6 September J.-M. Tiercy National Reference Laboratory for Histocompatibility (LNRH) University Hospital Geneva
 Basel - 6 eptember 2012 J.-M. Tiercy National Reference Laboratory for Histocompatibility (LNRH) University Hospital Geneva Outline the HLA system is (a) complex anti-hla immunisation and alloreactivity
Basel - 6 eptember 2012 J.-M. Tiercy National Reference Laboratory for Histocompatibility (LNRH) University Hospital Geneva Outline the HLA system is (a) complex anti-hla immunisation and alloreactivity
Update to the Human Leukocyte Antigens (HLA) Equivalency Tables
 Update to the Human Leukocyte s (HLA) Equivalency Tables Sponsoring Committee: Histocompatibility Policy/Bylaws Affected: Policy 2.11.A: Required Information for Deceased Kidney Donors, Policy 2.11.B:
Update to the Human Leukocyte s (HLA) Equivalency Tables Sponsoring Committee: Histocompatibility Policy/Bylaws Affected: Policy 2.11.A: Required Information for Deceased Kidney Donors, Policy 2.11.B:
HISTOCOMPATIBILITY LABORATORIES
 HISTOCOMPATIBILITY LABORATORIES SERVICE PROVISION USER GUIDE DOC45 Version 004 September 2015 Page 1 of 23 CONTENTS CONTENTS... 1 SERVICE PROVISION INTRODUCTION... 3 SUMMARY... 4 PROCEDURES UNDERTAKEN
HISTOCOMPATIBILITY LABORATORIES SERVICE PROVISION USER GUIDE DOC45 Version 004 September 2015 Page 1 of 23 CONTENTS CONTENTS... 1 SERVICE PROVISION INTRODUCTION... 3 SUMMARY... 4 PROCEDURES UNDERTAKEN
Haplo vs Cord vs URD Debate
 3rd Annual ASBMT Regional Conference for NPs, PAs and Fellows Haplo vs Cord vs URD Debate Claudio G. Brunstein Associate Professor University of Minnesota Medical School Take home message Finding a donor
3rd Annual ASBMT Regional Conference for NPs, PAs and Fellows Haplo vs Cord vs URD Debate Claudio G. Brunstein Associate Professor University of Minnesota Medical School Take home message Finding a donor
Cover Page. The handle holds various files of this Leiden University dissertation.
 Cover Page The handle http://hdl.handle.net/1887/20898 holds various files of this Leiden University dissertation. Author: Jöris, Monique Maria Title: Challenges in unrelated hematopoietic stem cell transplantation.
Cover Page The handle http://hdl.handle.net/1887/20898 holds various files of this Leiden University dissertation. Author: Jöris, Monique Maria Title: Challenges in unrelated hematopoietic stem cell transplantation.
HISTOCOMPATIBILITY LABORATORIES
 HISTOCOMPATIBILITY LABORATORIES SERVICE PROVISION USER GUIDE Page 1 of 22 CONTENTS SERVICE PROVISION INTRODUCTION... 3 SUMMARY... 4 PROCEDURES UNDERTAKEN FOR HAEMATOPOIETIC STEM CELL TRANSPLATATION...
HISTOCOMPATIBILITY LABORATORIES SERVICE PROVISION USER GUIDE Page 1 of 22 CONTENTS SERVICE PROVISION INTRODUCTION... 3 SUMMARY... 4 PROCEDURES UNDERTAKEN FOR HAEMATOPOIETIC STEM CELL TRANSPLATATION...
The National Marrow Donor Program. Graft Sources for Hematopoietic Cell Transplantation. Simon Bostic, URD Transplant Recipient
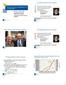 1988 199 1992 1994 1996 1998 2 22 24 26 28 21 212 214 216 218 Adult Donors Cord Blood Units The National Donor Program Graft Sources for Hematopoietic Cell Transplantation Dennis L. Confer, MD Chief Medical
1988 199 1992 1994 1996 1998 2 22 24 26 28 21 212 214 216 218 Adult Donors Cord Blood Units The National Donor Program Graft Sources for Hematopoietic Cell Transplantation Dennis L. Confer, MD Chief Medical
Indian Journal of Nephrology Indian J Nephrol 2001;11: 88-97
 88 Indian Journal of Nephrology Indian J Nephrol 2001;11: 88-97 ARTICLE HLA gene and haplotype frequency in renal transplant recipients and donors of Uttar Pradesh (North India) S Agrawal, AK Singh, RK
88 Indian Journal of Nephrology Indian J Nephrol 2001;11: 88-97 ARTICLE HLA gene and haplotype frequency in renal transplant recipients and donors of Uttar Pradesh (North India) S Agrawal, AK Singh, RK
STEP-programme S-candiaT-ransplant kidney E-xchange P-rogramme (STEP) Version 1.8
 STEP-programme S-candiaT-ransplant kidney E-xchange P-rogramme (STEP) Version 1.8 June 12, 2017 General guidelines Purpose To increase the likelihood of offering a suitable kidney graft to immunized (HI)
STEP-programme S-candiaT-ransplant kidney E-xchange P-rogramme (STEP) Version 1.8 June 12, 2017 General guidelines Purpose To increase the likelihood of offering a suitable kidney graft to immunized (HI)
Chapter 10. Histocompatibility Testing
 Chapter 10 Histocompatibility Testing Chapter 10 Histocompatibility Testing Table of Contents 10.1 General... 3 10.1.1 Registration of renal transplant patients...3 10.1.2 Material for histocompatibility
Chapter 10 Histocompatibility Testing Chapter 10 Histocompatibility Testing Table of Contents 10.1 General... 3 10.1.1 Registration of renal transplant patients...3 10.1.2 Material for histocompatibility
Virtual Crossmatch in Kidney Transplantation
 Virtual Crossmatch in Kidney Transplantation Shiva Samavat Associate Professor of Nephrology Labbafinejad Hospital SBMU 2018.11.21 All transplant candidates are screened to determine the degree of humoral
Virtual Crossmatch in Kidney Transplantation Shiva Samavat Associate Professor of Nephrology Labbafinejad Hospital SBMU 2018.11.21 All transplant candidates are screened to determine the degree of humoral
Significance of the MHC
 CHAPTER 8 Major Histocompatibility Complex (MHC) What is is MHC? HLA H-2 Minor histocompatibility antigens Peter Gorer & George Sneell (1940) Significance of the MHC role in immune response role in organ
CHAPTER 8 Major Histocompatibility Complex (MHC) What is is MHC? HLA H-2 Minor histocompatibility antigens Peter Gorer & George Sneell (1940) Significance of the MHC role in immune response role in organ
Allele and Haplotype Frequencies of Human Leukocyte Antigen-A, -B, -C, -DRB1, and -DQB1 From Sequence- Based DNA Typing Data in Koreans
 Original Article Diagnostic Immunology Ann Lab Med 2015;35:429-435 http://dx.doi.org/10.3343/alm.2015.35.4.429 ISSN 2234-3806 eissn 2234-3814 Allele and Haplotype Frequencies of Human Leukocyte Antigen-A,
Original Article Diagnostic Immunology Ann Lab Med 2015;35:429-435 http://dx.doi.org/10.3343/alm.2015.35.4.429 ISSN 2234-3806 eissn 2234-3814 Allele and Haplotype Frequencies of Human Leukocyte Antigen-A,
Objectives. The need for national registry Beginning of SSCDR SSCDR collaborations Achievements so far Our future goals How can we help
 Objectives The need for national registry Beginning of SSCDR SSCDR collaborations Achievements so far Our future goals How can we help The start of unrelated donor registries 1971 Anthony Nolan born with
Objectives The need for national registry Beginning of SSCDR SSCDR collaborations Achievements so far Our future goals How can we help The start of unrelated donor registries 1971 Anthony Nolan born with
IMMUNOLOGY. Elementary Knowledge of Major Histocompatibility Complex and HLA Typing
 IMMUNOLOGY Elementary Knowledge of Major Histocompatibility Complex and HLA Typing Tapasya Srivastava and Subrata Sinha Department of Biochemistry All India Institute of Medical Sciences New Delhi - 110029
IMMUNOLOGY Elementary Knowledge of Major Histocompatibility Complex and HLA Typing Tapasya Srivastava and Subrata Sinha Department of Biochemistry All India Institute of Medical Sciences New Delhi - 110029
Expanding Candidate and Deceased Donor HLA Typing Requirements to Provide Greater Consistency Across Organ Types
 At-a-Glance Expanding Candidate and Deceased Donor HLA Typing Requirements to Provide Greater Consistency Across Organ Types Affected/Proposed Policy: 2.8.A (Required Information for Deceased Kidney Donors);
At-a-Glance Expanding Candidate and Deceased Donor HLA Typing Requirements to Provide Greater Consistency Across Organ Types Affected/Proposed Policy: 2.8.A (Required Information for Deceased Kidney Donors);
Specimen Collection Requirements. Test Name Specimen Type Storage Time Storage Conditions
 1 Specimen Collection Requirements PURPOSE/PRINCIPLE: To outline proper specimen collection for molecular HLA testing, including tube type, minimum specimen amount, proper tube and requisition labeling,
1 Specimen Collection Requirements PURPOSE/PRINCIPLE: To outline proper specimen collection for molecular HLA testing, including tube type, minimum specimen amount, proper tube and requisition labeling,
Calculation Tables. Olerup SSP Kits without Taq Polymeras
 Calculation Tables lerup SSP Kits without Taq Polymeras Table 1: Volumes of the components needed per test for different numbers of when using Master Mix without. The recommended volumes listed below include
Calculation Tables lerup SSP Kits without Taq Polymeras Table 1: Volumes of the components needed per test for different numbers of when using Master Mix without. The recommended volumes listed below include
Diversity and Frequencies of HLA Class I and Class II Genes of an East African Population
 Open Journal of Genetics, 2014, 4, 99-124 Published Online April 2014 in SciRes. http://www.scirp.org/journal/ojgen http://dx.doi.org/10.4236/ojgen.2014.42013 Diversity and Frequencies of HLA Class I and
Open Journal of Genetics, 2014, 4, 99-124 Published Online April 2014 in SciRes. http://www.scirp.org/journal/ojgen http://dx.doi.org/10.4236/ojgen.2014.42013 Diversity and Frequencies of HLA Class I and
Calculation Tables. Olerup SSP Kits without Taq Polymeras
 Calculation Tables lerup SSP Kits without Polymeras Table 1: Volumes of the components needed per test for different numbers of when using Master Mix without. The recommended volumes listed below include
Calculation Tables lerup SSP Kits without Polymeras Table 1: Volumes of the components needed per test for different numbers of when using Master Mix without. The recommended volumes listed below include
Validation of the MIA FORA NGS FLEX Assay Using Buccal Swabs as the Sample Source
 S. Krishnakumar, M. Li, C. Wang, M, Osada, R. Kuehn, M. Fukushima and Y. Thorstenson Immucor, Inc. S. Krishnakumar, M. Li, C. Wang, M, Osada, R. Kuehn, M. Fukushima and Y. Thorstenson Immucor, Inc. Introduction
S. Krishnakumar, M. Li, C. Wang, M, Osada, R. Kuehn, M. Fukushima and Y. Thorstenson Immucor, Inc. S. Krishnakumar, M. Li, C. Wang, M, Osada, R. Kuehn, M. Fukushima and Y. Thorstenson Immucor, Inc. Introduction
Sylwia Mizia, 1 Dorota Dera-Joachimiak, 1 Malgorzata Polak, 1 Katarzyna Koscinska, 1 Mariola Sedzimirska, 1 and Andrzej Lange 1, 2. 1.
 Bone Marrow Research Volume 2012, Article ID 873695, 5 pages doi:10.1155/2012/873695 Clinical Study Both Optimal Matching and Procedure Duration Influence Survival of Patients after Unrelated Donor Hematopoietic
Bone Marrow Research Volume 2012, Article ID 873695, 5 pages doi:10.1155/2012/873695 Clinical Study Both Optimal Matching and Procedure Duration Influence Survival of Patients after Unrelated Donor Hematopoietic
BE THE MATCH. The Role of HLA in Finding a Match for Bone Marrow or Peripheral Blood Stem Cell Transplantation
 BE THE MATCH The Role of HLA in Finding a Match for Bone Marrow or Peripheral Blood Stem Cell Transplantation Leukemia Leukemia is a type of cancer of the blood or bone marrow. It is characterized by an
BE THE MATCH The Role of HLA in Finding a Match for Bone Marrow or Peripheral Blood Stem Cell Transplantation Leukemia Leukemia is a type of cancer of the blood or bone marrow. It is characterized by an
The Use of HLA /HPA Selected Platelets
 The Use of HLA /HPA Selected Platelets Dr Colin Brown PhD FRCPath. Consultant Clinical Scientist Histocompatibility and Immunogenetics HLA and Transfusion Human Leucocyte Antigens HLA Human Platelet Antigens
The Use of HLA /HPA Selected Platelets Dr Colin Brown PhD FRCPath. Consultant Clinical Scientist Histocompatibility and Immunogenetics HLA and Transfusion Human Leucocyte Antigens HLA Human Platelet Antigens
Significance of the MHC
 CHAPTER 8 Major Histocompatibility Complex (MHC) What is MHC? HLA H-2 Minor histocompatibility antigens Peter Gorer & George Sneell (1940) - MHC molecules were initially discovered during studies aimed
CHAPTER 8 Major Histocompatibility Complex (MHC) What is MHC? HLA H-2 Minor histocompatibility antigens Peter Gorer & George Sneell (1940) - MHC molecules were initially discovered during studies aimed
The Anthony Nolan Trust Histocompatibility Laboratories & Operations Department Service Provision
 The Anthony Nolan Trust Histocompatibility Laboratories & Operations Department Registered Charity 803716/SC038827 Contents 1. Introduction... 2. Summary... 3. Procedures Undertaken for Haematopoietic
The Anthony Nolan Trust Histocompatibility Laboratories & Operations Department Registered Charity 803716/SC038827 Contents 1. Introduction... 2. Summary... 3. Procedures Undertaken for Haematopoietic
Robert B. Colvin, M.D. Department of Pathology Massachusetts General Hospital Harvard Medical School
 Harvard-MIT Division of Health Sciences and Technology HST.035: Principle and Practice of Human Pathology Dr. Robert B. Colvin Transplantation: Friendly organs in a hostile environment Robert B. Colvin,
Harvard-MIT Division of Health Sciences and Technology HST.035: Principle and Practice of Human Pathology Dr. Robert B. Colvin Transplantation: Friendly organs in a hostile environment Robert B. Colvin,
Dan Koller, Ph.D. Medical and Molecular Genetics
 Design of Genetic Studies Dan Koller, Ph.D. Research Assistant Professor Medical and Molecular Genetics Genetics and Medicine Over the past decade, advances from genetics have permeated medicine Identification
Design of Genetic Studies Dan Koller, Ph.D. Research Assistant Professor Medical and Molecular Genetics Genetics and Medicine Over the past decade, advances from genetics have permeated medicine Identification
HLA Amino Acid Polymorphisms and Kidney Allograft Survival. Supplemental Digital Content
 HLA Amino Acid Polymorphisms and Kidney Allograft Survival Supplemental Digital Content Malek Kamoun, MD, PhD a, Keith P. McCullough, MS b, Martin Maiers, MS c, Marcelo A. Fernandez Vina, MD, PhD d, Hongzhe
HLA Amino Acid Polymorphisms and Kidney Allograft Survival Supplemental Digital Content Malek Kamoun, MD, PhD a, Keith P. McCullough, MS b, Martin Maiers, MS c, Marcelo A. Fernandez Vina, MD, PhD d, Hongzhe
Effects of age-at-diagnosis and duration of diabetes on GADA and IA-2A positivity
 Effects of age-at-diagnosis and duration of diabetes on GADA and IA-2A positivity Duration of diabetes was inversely correlated with age-at-diagnosis (ρ=-0.13). However, as backward stepwise regression
Effects of age-at-diagnosis and duration of diabetes on GADA and IA-2A positivity Duration of diabetes was inversely correlated with age-at-diagnosis (ρ=-0.13). However, as backward stepwise regression
Antigen Presentation to T lymphocytes
 Antigen Presentation to T lymphocytes Immunology 441 Lectures 6 & 7 Chapter 6 October 10 & 12, 2016 Jessica Hamerman jhamerman@benaroyaresearch.org Office hours by arrangement Antigen processing: How are
Antigen Presentation to T lymphocytes Immunology 441 Lectures 6 & 7 Chapter 6 October 10 & 12, 2016 Jessica Hamerman jhamerman@benaroyaresearch.org Office hours by arrangement Antigen processing: How are
Summary of Changes BMT CTN 1101 Version 7.0 to 8.0 Dated: January 18, Original text: Changed to: Rationale
 BMT CTN 1101 RIC ducb vs. Haplo Page 1 of 10 Date: January 20, 2017 Summary of Changes BMT CTN 1101 Version 7.0 to 8.0 Dated: January 18, 2017 A Multi-Center, Phase III, Randomized Trial of Reduced Intensity(RIC)
BMT CTN 1101 RIC ducb vs. Haplo Page 1 of 10 Date: January 20, 2017 Summary of Changes BMT CTN 1101 Version 7.0 to 8.0 Dated: January 18, 2017 A Multi-Center, Phase III, Randomized Trial of Reduced Intensity(RIC)
Available online at , 2(1):27-37
 Available online at www.apjbg.com Asia-Pacific Journal of Blood Types and Genes 2018, 2(1):27-37 APJBG Allele and haplotype frequencies of human leukocyte antigen-a, -B, -C, -DRB1, and -DQB1 in Chinese
Available online at www.apjbg.com Asia-Pacific Journal of Blood Types and Genes 2018, 2(1):27-37 APJBG Allele and haplotype frequencies of human leukocyte antigen-a, -B, -C, -DRB1, and -DQB1 in Chinese
Transplantation and Cancer
 Transplantation and Cancer Immunology Bởi: OpenStaxCollege The immune responses to transplanted organs and to cancer cells are both important medical issues. With the use of tissue typing and anti-rejection
Transplantation and Cancer Immunology Bởi: OpenStaxCollege The immune responses to transplanted organs and to cancer cells are both important medical issues. With the use of tissue typing and anti-rejection
Seventy percent of people who are candidates for allogeneic hematopoietic
 274 7th Biennial Symposium on Minorities, the Medically Underserved and Cancer Supplement to Cancer The National Marrow Donor Program Meeting the Needs of the Medically Underserved Dennis L. Confer, M.D.
274 7th Biennial Symposium on Minorities, the Medically Underserved and Cancer Supplement to Cancer The National Marrow Donor Program Meeting the Needs of the Medically Underserved Dennis L. Confer, M.D.
HLA AND KIR GENE POLYMORPHISM IN HEMATOPOIETIC STEM CELL TRANSPLANTATION
 FROM THE DEPARTMENT OF LABORATORY MEDICINE, DIVISION OF CLINICAL IMMUNOLOGY KAROLINSKA INSTITUTET, STOCKHOLM, SWEDEN HLA AND KIR GENE POLYMORPHISM IN HEMATOPOIETIC STEM CELL TRANSPLANTATION Marie Schaffer
FROM THE DEPARTMENT OF LABORATORY MEDICINE, DIVISION OF CLINICAL IMMUNOLOGY KAROLINSKA INSTITUTET, STOCKHOLM, SWEDEN HLA AND KIR GENE POLYMORPHISM IN HEMATOPOIETIC STEM CELL TRANSPLANTATION Marie Schaffer
HLA and antigen presentation. Department of Immunology Charles University, 2nd Medical School University Hospital Motol
 HLA and antigen presentation Department of Immunology Charles University, 2nd Medical School University Hospital Motol MHC in adaptive immunity Characteristics Specificity Innate For structures shared
HLA and antigen presentation Department of Immunology Charles University, 2nd Medical School University Hospital Motol MHC in adaptive immunity Characteristics Specificity Innate For structures shared
White Paper Estimating Genotype-Specific Incidence for One or Several Loci
 White Paper 23-01 Estimating Genotype-Specific Incidence for One or Several Loci Authors: Mike Macpherson Brian Naughton Andro Hsu Joanna Mountain Created: September 5, 2007 Last Edited: November 18, 2007
White Paper 23-01 Estimating Genotype-Specific Incidence for One or Several Loci Authors: Mike Macpherson Brian Naughton Andro Hsu Joanna Mountain Created: September 5, 2007 Last Edited: November 18, 2007
General Terms: Appendix B. National Marrow Donor Program and The Medical College of Wisconsin
 Glossary of Terms This appendix is divided into two sections. The first section, General Terms, defines terms used throughout the CIBMTR data collection forms. The second section, FormsNet TM 2 Terms,
Glossary of Terms This appendix is divided into two sections. The first section, General Terms, defines terms used throughout the CIBMTR data collection forms. The second section, FormsNet TM 2 Terms,
BDC Keystone Genetics Type 1 Diabetes. Immunology of diabetes book with Teaching Slides
 BDC Keystone Genetics Type 1 Diabetes www.barbaradaviscenter.org Immunology of diabetes book with Teaching Slides PRACTICAL Trailnet screens relatives and new onset patients for autoantibodies and HLA
BDC Keystone Genetics Type 1 Diabetes www.barbaradaviscenter.org Immunology of diabetes book with Teaching Slides PRACTICAL Trailnet screens relatives and new onset patients for autoantibodies and HLA
Profiling HLA motifs by large scale peptide sequencing Agilent Innovators Tour David K. Crockett ARUP Laboratories February 10, 2009
 Profiling HLA motifs by large scale peptide sequencing 2009 Agilent Innovators Tour David K. Crockett ARUP Laboratories February 10, 2009 HLA Background The human leukocyte antigen system (HLA) is the
Profiling HLA motifs by large scale peptide sequencing 2009 Agilent Innovators Tour David K. Crockett ARUP Laboratories February 10, 2009 HLA Background The human leukocyte antigen system (HLA) is the
Samples Available for Recipient Only. Samples Available for Recipient and Donor
 Unrelated HCT Research Sample Inventory - Summary for First Allogeneic Transplants in CRF and TED with biospecimens available through the CIBMTR Repository stratified by availability of paired samples,
Unrelated HCT Research Sample Inventory - Summary for First Allogeneic Transplants in CRF and TED with biospecimens available through the CIBMTR Repository stratified by availability of paired samples,
The Relationship between Minor Histocompatibility Antigens and Graft Versus Host Disease in Unrelated Peripheral Blood Stem Cell Transplants
 mhags and GVHD Page 1 The Relationship between Minor Histocompatibility Antigens and Graft Versus Host Disease in Unrelated Peripheral Blood Stem Cell Transplants Running Title: mhags and GVHD Conflict
mhags and GVHD Page 1 The Relationship between Minor Histocompatibility Antigens and Graft Versus Host Disease in Unrelated Peripheral Blood Stem Cell Transplants Running Title: mhags and GVHD Conflict
Dr. Yi-chi M. Kong August 8, 2001 Benjamini. Ch. 19, Pgs Page 1 of 10 TRANSPLANTATION
 Benjamini. Ch. 19, Pgs 379-399 Page 1 of 10 TRANSPLANTATION I. KINDS OF GRAFTS II. RELATIONSHIPS BETWEEN DONOR AND RECIPIENT Benjamini. Ch. 19, Pgs 379-399 Page 2 of 10 II.GRAFT REJECTION IS IMMUNOLOGIC
Benjamini. Ch. 19, Pgs 379-399 Page 1 of 10 TRANSPLANTATION I. KINDS OF GRAFTS II. RELATIONSHIPS BETWEEN DONOR AND RECIPIENT Benjamini. Ch. 19, Pgs 379-399 Page 2 of 10 II.GRAFT REJECTION IS IMMUNOLOGIC
Support of Unrelated Stem Cell Donor Searches by Donor Center-Initiated HLA Typing of Potentially Matching Donors
 Support of Unrelated Stem Cell Donor Searches by Donor Center-Initiated HLA Typing of Potentially Matching Donors Alexander H. Schmidt 1 *, Ute V. Solloch 1, Daniel Baier 1, Alois Grathwohl 1, Jan Hofmann
Support of Unrelated Stem Cell Donor Searches by Donor Center-Initiated HLA Typing of Potentially Matching Donors Alexander H. Schmidt 1 *, Ute V. Solloch 1, Daniel Baier 1, Alois Grathwohl 1, Jan Hofmann
OPTN/UNOS Histocompatibility Committee Report to the Board of Directors November 12-13, 2014 St. Louis, MO
 OPTN/UNOS Histocompatibility Committee OPTN/UNOS Histocompatibility Committee Report to the Board of Directors November 12-13, 2014 St. Louis, MO Dolly Tyan, Ph.D., Chair Robert Bray, Ph.D., Vice Chair
OPTN/UNOS Histocompatibility Committee OPTN/UNOS Histocompatibility Committee Report to the Board of Directors November 12-13, 2014 St. Louis, MO Dolly Tyan, Ph.D., Chair Robert Bray, Ph.D., Vice Chair
Minutes Tissue Typers Meeting Aarhus, 21st September 2018
 Minutes Tissue Typers Meeting Aarhus, 21st September 2018 Location: Aarhus University Hospital, Palle Juul-Jensens Boulevard 99, DK-8200 Aarhus N, relocation to C104 patiently accepted by all participants.
Minutes Tissue Typers Meeting Aarhus, 21st September 2018 Location: Aarhus University Hospital, Palle Juul-Jensens Boulevard 99, DK-8200 Aarhus N, relocation to C104 patiently accepted by all participants.
Samples Available for Recipient and Donor
 Unrelated HCT Research Sample Inventory - Summary for First Allogeneic Transplants in CRF and TED with biospecimens available through the CIBMTR Repository stratified by availability of paired samples,
Unrelated HCT Research Sample Inventory - Summary for First Allogeneic Transplants in CRF and TED with biospecimens available through the CIBMTR Repository stratified by availability of paired samples,
Samples Available for Recipient Only. Samples Available for Recipient and Donor
 Unrelated HCT Research Sample Inventory - Summary for First Allogeneic Transplants in CRF and TED with biospecimens available through the CIBMTR Repository stratified by availability of paired samples,
Unrelated HCT Research Sample Inventory - Summary for First Allogeneic Transplants in CRF and TED with biospecimens available through the CIBMTR Repository stratified by availability of paired samples,
ASSESSMENT OF THE RISK FOR TYPE 1 DIABETES MELLITUS CONFERRED BY HLA CLASS II GENES. Irina Durbală
 ASSESSMENT OF THE RISK FOR TYPE 1 DIABETES MELLITUS CONFERRED BY HLA CLASS II GENES Summary Irina Durbală CELL AND MOLECULAR BIOLOGY DEPARTMENT FACULTY OF MEDICINE, OVIDIUS UNIVERSITY CONSTANŢA Class II
ASSESSMENT OF THE RISK FOR TYPE 1 DIABETES MELLITUS CONFERRED BY HLA CLASS II GENES Summary Irina Durbală CELL AND MOLECULAR BIOLOGY DEPARTMENT FACULTY OF MEDICINE, OVIDIUS UNIVERSITY CONSTANŢA Class II
MUD SCT. Pimjai Niparuck Division of Hematology, Department of Medicine Ramathibodi Hospital, Mahidol University
 MUD SCT Pimjai Niparuck Division of Hematology, Department of Medicine Ramathibodi Hospital, Mahidol University Outlines Optimal match criteria for unrelated adult donors Role of ATG in MUD-SCT Post-transplant
MUD SCT Pimjai Niparuck Division of Hematology, Department of Medicine Ramathibodi Hospital, Mahidol University Outlines Optimal match criteria for unrelated adult donors Role of ATG in MUD-SCT Post-transplant
DNA Analysis Techniques for Molecular Genealogy. Luke Hutchison Project Supervisor: Scott R. Woodward
 DNA Analysis Techniques for Molecular Genealogy Luke Hutchison (lukeh@email.byu.edu) Project Supervisor: Scott R. Woodward Mission: The BYU Center for Molecular Genealogy To establish the world s most
DNA Analysis Techniques for Molecular Genealogy Luke Hutchison (lukeh@email.byu.edu) Project Supervisor: Scott R. Woodward Mission: The BYU Center for Molecular Genealogy To establish the world s most
10/18/2012. A primer in HLA: The who, what, how and why. What?
 A primer in HLA: The who, what, how and why What? 1 First recognized in mice during 1930 s and 1940 s. Mouse (murine) experiments with tumors Independent observations were made in humans with leukoagglutinating
A primer in HLA: The who, what, how and why What? 1 First recognized in mice during 1930 s and 1940 s. Mouse (murine) experiments with tumors Independent observations were made in humans with leukoagglutinating
Related haploidentical donors versus matched unrelated donors
 Related haploidentical donors versus matched unrelated donors Bronwen Shaw, MD PhD Professor of Medicine, MCW Senior Scientific Director, CIBMTR Definition Matched Unrelated donor Refers to HLA matching
Related haploidentical donors versus matched unrelated donors Bronwen Shaw, MD PhD Professor of Medicine, MCW Senior Scientific Director, CIBMTR Definition Matched Unrelated donor Refers to HLA matching
HLA and more. Ilias I.N. Doxiadis. Geneva 03/04/2012.
 www.ebmt.org HLA and more Ilias I.N. Doxiadis Geneva 03/04/2012 HLA and more HLA and more / Doxiadis 2 Topic of the day Compatibility testing is a type of testing used to ensure compatibility of the system/application/website
www.ebmt.org HLA and more Ilias I.N. Doxiadis Geneva 03/04/2012 HLA and more HLA and more / Doxiadis 2 Topic of the day Compatibility testing is a type of testing used to ensure compatibility of the system/application/website
Transplantation. Immunology Unit College of Medicine King Saud University
 Transplantation Immunology Unit College of Medicine King Saud University Objectives To understand the diversity among human leukocyte antigens (HLA) or major histocompatibility complex (MHC) To know the
Transplantation Immunology Unit College of Medicine King Saud University Objectives To understand the diversity among human leukocyte antigens (HLA) or major histocompatibility complex (MHC) To know the
Structural Variation and Medical Genomics
 Structural Variation and Medical Genomics Andrew King Department of Biomedical Informatics July 8, 2014 You already know about small scale genetic mutations Single nucleotide polymorphism (SNPs) Deletions,
Structural Variation and Medical Genomics Andrew King Department of Biomedical Informatics July 8, 2014 You already know about small scale genetic mutations Single nucleotide polymorphism (SNPs) Deletions,
Historical definition of Antigen. An antigen is a foreign substance that elicits the production of antibodies that specifically binds to the antigen.
 Historical definition of Antigen An antigen is a foreign substance that elicits the production of antibodies that specifically binds to the antigen. Historical definition of Antigen An antigen is a foreign
Historical definition of Antigen An antigen is a foreign substance that elicits the production of antibodies that specifically binds to the antigen. Historical definition of Antigen An antigen is a foreign
Blood Types and Genetics
 Blood Types and Genetics Human blood type is determined by codominant alleles. An allele is one of several different forms of genetic information that is present in our DNA at a specific location on a
Blood Types and Genetics Human blood type is determined by codominant alleles. An allele is one of several different forms of genetic information that is present in our DNA at a specific location on a
Transplant Booklet D Page 1
 Booklet D Pretest Correct Answers 4. (A) is correct. Technically, performing a hematopoietic stem cell transplant is one of the simplest transplantation procedures. The hematopoietic stem cells are infused
Booklet D Pretest Correct Answers 4. (A) is correct. Technically, performing a hematopoietic stem cell transplant is one of the simplest transplantation procedures. The hematopoietic stem cells are infused
HLA Selected Platelets
 HLA Selected Platelets Dr PhD Clinical Scientist Department of Histocompatibility & Immunogenetics NHS Blood and Transplant Colindale Poor increment (
HLA Selected Platelets Dr PhD Clinical Scientist Department of Histocompatibility & Immunogenetics NHS Blood and Transplant Colindale Poor increment (
Management of platelet refractory patients, why does your patient keep on bleeding? Dr Colin Brown, H&I Dept, NHSBT Colindale
 Management of platelet refractory patients, why does your patient keep on bleeding? Dr Colin Brown, H&I Dept, NHSBT Colindale Pooled Platelets (Buffy Coat Derived) Adult dose from 4 ABO-identical donors
Management of platelet refractory patients, why does your patient keep on bleeding? Dr Colin Brown, H&I Dept, NHSBT Colindale Pooled Platelets (Buffy Coat Derived) Adult dose from 4 ABO-identical donors
UNIT 3 GENETICS LESSON #30: TRAITS, GENES, & ALLELES. Many things come in many forms. Give me an example of something that comes in many forms.
 UNIT 3 GENETICS LESSON #30: TRAITS, GENES, & ALLELES Many things come in many forms. Give me an example of something that comes in many forms. Genes, too, come in many forms. Main Idea #1 The same gene
UNIT 3 GENETICS LESSON #30: TRAITS, GENES, & ALLELES Many things come in many forms. Give me an example of something that comes in many forms. Genes, too, come in many forms. Main Idea #1 The same gene
Biology. Chapter 13. Observing Patterns in Inherited Traits. Concepts and Applications 9e Starr Evers Starr. Cengage Learning 2015
 Biology Concepts and Applications 9e Starr Evers Starr Chapter 13 Observing Patterns in Inherited Traits Cengage Learning 2015 Cengage Learning 2015 After completing today s activities, students should
Biology Concepts and Applications 9e Starr Evers Starr Chapter 13 Observing Patterns in Inherited Traits Cengage Learning 2015 Cengage Learning 2015 After completing today s activities, students should
(b) What is the allele frequency of the b allele in the new merged population on the island?
 2005 7.03 Problem Set 6 KEY Due before 5 PM on WEDNESDAY, November 23, 2005. Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. Two populations (Population One
2005 7.03 Problem Set 6 KEY Due before 5 PM on WEDNESDAY, November 23, 2005. Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. Two populations (Population One
The Major Histocompatibility Complex (MHC)
 Advanced molecular immunology-2011 The Major Histocompatibility Complex (MHC) Youmin Kang 2011.04.06 Activation IFN-g IL-4 IL-2 YYYYYY YYYYY B cells MHC class II pathway Discovery of MHC MHC Class I pathway
Advanced molecular immunology-2011 The Major Histocompatibility Complex (MHC) Youmin Kang 2011.04.06 Activation IFN-g IL-4 IL-2 YYYYYY YYYYY B cells MHC class II pathway Discovery of MHC MHC Class I pathway
