Kidney. Heart. Lung. Sirt1. Gapdh. Mouse IgG DAPI. Rabbit IgG DAPI
|
|
|
- Claribel Caldwell
- 6 years ago
- Views:
Transcription
1 a e Na V 1.5 Ad-LacZ Ad- 110KD b Scn5a/ (relative to Ad-LacZ) f p = 0.65 Ad-LacZ Ad- c Heart Lung Kidney Spleen 110KD d fl/fl c -/- DAPI 20 µm Na v KD fl/fl Rabbit IgG DAPI fl/fl Mouse IgG DAPI 40KD fl/fl c -/- fl/fl c -/- 20 µm Supplementary Fig. 1. Effect of overexpression or deletion on Na v 1.5 expression. (a & b) Effect of adenoviral overexpression of on expression of Na v 1.5 at protein (a) or mrna (b) level in rat neonatal cardiac myocytes. n = 4 for Ad-LacZ and Ad-. (c) expression in hearts of c -/- mouse with conditional deletion of cardiomyocyte. (d) Representative photomicrographs showing immunostaining for in the cardiomyocytes isolated from fl/fl and c -/- mouse. (e) Immunoblots showing Na v 1.5 expression in fl/fl and c -/- mouse hearts. (f) Negative controls for immunofluorescence using non-immune mouse (red) and rabbit (green) IgGs. Ad-: adenovirus expressing ; Ad-LacZ: control adenovirus expressing E. Coli LacZ. Uncropped blots are shown in supplementary figure 8. Independent sample t-test was used. For box-and-whiskers plots, center line shows median end of box shows IQR and whiskers show minima and maxima within 1.5 IQR. 1
2 a b Relative currents Na v 1.5 Na v 1.5+S irt1 Na v 1.5+ (H363Y) Relative conductance Relative currents Ex-242 Ex Relative conductance c d Relative currents Na v 1.5 Na v 1.5K 1479A Na v 1.5K 1479A + S irt1 Na v 1.5K 1479A +(H363Y) Relative conductance Relative currents Na v 1.5-W T Na v 1.5-K 1479R Na v 1.5-K 1479Q Relative conductance Membrane potential (mv) Membrane potential (mv) Supplementary Fig. 2. Activation and inactivation plots for wild type, mutant, and native Na v 1.5 with overexpression or inhibition. (a) Effect of and dominant negative (H363Y) expression on steady state activation and inactivation of Na v 1.5 expressed in HEK 293 cells. n =10 cells for Na v 1.5 and Na v 1.5+ (H363Y), n= 11 cells for Na v (b) Effect of inhibitor Ex-243 on steady state activation and inactivation of native Na v 1.5 in rat neonatal cardiac myocytes. n =6 cells for Ex-242, n= 5 cells for Ex-243. (c) Effect of and (H363Y) expression on steady state activation and inactivation currents in HEK 293 cells expressing K1479A-Na v 1.5. n= 8 cells for Na v 1.5, n = 6 cells for Na v 1.5+, and n=7 cells for Na v 1.5-K1479A and Na v 1.5-K1479A+ (H363Y). (d) Effect of K1479R and K1479Q mutations on steady state activation and inactivation currents in HEK 293 cells expressing Na v 1.5-WT, Na v 1.5-K 1479R or Na v 1.5-K 1479Q. n= 8 cells for Na v 1.5-WT, n = 9 cells for Na v 1.5- K 1479R, and n=10 cells for Na v 1.5-K 1479Q. Independent sample t-test was used. In the XY scatter plot data shown as mean and error bar represents s.e.m. Ex-242: inactive isomer of Ex-243. For XY scatter plots data is shown as mean and error bar represents s.e.m. 2
3 a 150 ms b Normalized current 50 ms 50 ms 0.2 mv 0.05 mv fl/fl c -/- 150 ms Late current (I Na, t150ms / I Na, peak ) p = 0.89 fl/fl c -/- c V max (V/s) * APD (ms) * fl/fl c -/- 0 d fl/fl * * c -/- APD 25 APD 50 APD 75 APD 90 * e Resting membrane potential (mv) p = 0.19 fl/fl c -/- Supplementary Fig. 3. Effect of cardiomyocyte deletion on late I Na, V max, cardiac action potential duration, and resting membrane potential. Late sodium current was measured in response to a 200 ms voltage steps to 20 mv from a holding potential of 120 mv. (a) Representative whole-cell I Na in ventricular cardiomyocytes isolated from fl/fl and c -/- mice (Normalized to the peak I Na ). Lower panel shows voltage-expanded view of the I Na. (b) Mean I Na at 150 ms. n = 7 cells for fl/fl and n = 5 cells for c -/-. (c & d) Maximum velocity (V max ) of upstroke of the action potential (AP) (c) and action potential duration (APD) (d) in ventricular myocytes isolated from c -/- compared to control fl/fl mice. P < 0.05, * vs. fl/fl. n = 7 cells for fl/fl and n = 6 cells for c -/-. (e) Resting membrane potential in cardiomyocytes isolated from c -/- and fl/fl mice. n = 11 cells for fl/fl and n = 8 cells for c -/-. Independent sample t-test was used. Data shown as mean and error represents s.e.m. For box-and-whiskers plots, center line shows median, end of box shows IQR, and whiskers show minima and maxima within 1.5 IQR. 3
4 a IP: Na V 1.5 Ace-K Na V 1.5 Myc- Ad-Lac-Z Ad-SIRT1 (H363Y) 220KD 220KD 110KD b Ace-K Na V 1.5(III-IV) NAM Ace-K Na V 1.5 Supplementary Fig. 4. Sirtuin1 and non-sirtuin HDACs regulate acetylation of Na v 1.5. (a) Effect of inhibition of by adenoviral overexpression of dominant negative (H363Y) on acetylation of endogenous Na v 1.5 in rat neonatal cardiac myocytes. (b) Effect of inhibition of with NAM, overexpression of, and activation of with resveratrol (Resv.) on acetylation of - Na v 1.5 (III-IV) peptide expressed in HEK 293 cells (c) Effect of Sirtuin inhibition with nicotinamide (NAM 10mM) and HDAC inhibition with trichostatin A (TSA 100nM) on K1479 acetylation of Na v 1.5 in HEK 293 cells expressing Na v 1.5. Uncropped blots are shown in supplementary figure 8. Resveratrol c Control NAM TSA 4
5 a Ace-K p Na V (III-IV) + + b Intensity (counts) c Na V 1.5(III-IV) p Relative peak intensity + d Fold specificity Antibody concentration (ng/ml) e Ace.-K Na V 1.5 Na V Na V (acetyl K 1479 ) 0.02 µg 0.20 µg 2.00 µg Antibody concentration (2.5 ng/ml) Amount of peptide Ace.-K Na V 1.5 IB: Na V 1.5(III/IV) pcdna p300/cbp + + Supplementary Fig. 5. Mass spectrometry identifying Lysine 1479 in Na v 1.5 as targeted for deacetylation by ; generation of acetyl-k1479 antibody (a) Effect of p300/cbp and on lysine acetylation of recombinant -Na v 1.5 (III-IV) peptide in vitro. (b) Representative annotated MS/MS fragmentation spectra of -Na v 1.5 (III-IV) peptide treated with p300/cbp. The acetylated K1479-containing peptide in Na v 1.5 identified with LC-MS/MS analysis was not observed in cells not treated with p300/cbp, or in cells which were sequentially treated with after being incubated with p300/cbp. (c) Relative quantities of ionized acetylated peptide in -Na v 1.5 (III-IV) peptide treated with p300 and p (d-f) Specificity and sensitivity of acetyl- K 1479 antibody for acetylated K1479 of Na v 1.5. (d) Fold specificity of acetyl-k1479 antibody by a quantitative immuno-adsorption assay of antibody binding to a membrane pre-coated with nonacetylated or acetylated peptide. Fold specificity = ratio of affinity of antibody to acetylated vs. non-acetylated peptide. Orange-bar represents the concentration range in which antibody was used for western blot and immunohistochemistry experiments. Dotted line indicates background reading. (e) Dot blot of the non-acetylated or K1479 acetylated peptide (a.a ) of Na v 1.5 with the acetyl-k 1479 antibody. (f) Western blot using acetyl-k1479 antibody on recombinant -Na v 1.5 (III-IV) peptide treated with p300/cbp in vitro. Uncropped blots are shown in supplementary figure 8. f + IP: + 5
6 a Connexin 43 b Connexin 43 fl/fl c -/- Vehicle Ex-527 c Ace-K Na V 1.5 d 80 32KD 18KD 45KD Bip Chop Vehicle Ex-527 Vehicle Ex-527 Supplementary Fig. 6. Effect of deletion or inhibition of cardiomyocyte on connexin 43, Na v 1.5 acetylation, and unfolded protein response. (a & b) Immunohistochemistry and immunoblotting for cardiac connexin 43 in hearts from -/- and fl/fl mice (a) or mice treated with the inhibitor Ex-527 (2mg/kg/day, 14 days) (b). (c) K1479 acetylation of Na v 1.5 in the hearts of mice with treated with Ex-527. (d) Bip and Chop expression in the hearts of mice treated with Ex-527. Uncropped blots are shown in supplementary figure 8. 6
7 Fig. 1a Fig. 1c Fig. 1d Fig. 2a IP: N-IgG IP: Na V 1.5 IP: N-IgG IP: Na V 1.5 cmyc IP: Na V 1.5 IB: Ace-K IP: Na V 1.5 IB: Ace-K Na V 1.5 Rat neonatal CM Na V 1.5 Mouse heart Na v 1.5 Na v 1.5+ Na v 1.5+ (H363Y) Rat neonatal CM IP: Na V 1.5 IB: Na V 1.5 Ex-242 Ex-243 Na V 1.5 (GFP) Control Resv. Fig. 2b Fig. 2c Fig. 2d Fig. 2e -Na V 1.5 (III-IV)+p300 Exp-2 -Na V 1.5(III-IV) -Na V 1.5 (III-IV)+p300 Exp-1 -Na V 1.5(III-IV) Ace-K Ace-K 1479 Na v 1.5 Ace-K 1479 Na v KD 110KD Na v KD Fig. 2f Ace-K 1479 Na v Na V 1.5(III-IV) Ace-CoA p SIRT1 NAD KD 40KD pcdna (H363Y) Fig. 2h Fig. 4f Ace-K 1479 Na v ETN Ace-K 1479 Na v 1.5 GAPDH 100KD 75KD 42KD 34KD SIRT1 Na v 1.5 (N-term) 250KD GAPDH c -/- WT ID No ID No GAPDH ID No Control samples Supplementary Fig. 7. Uncropped immunoblots of indicated figures. 7
8 Suppl. Fig. 1c 110KD Suppl. Fig. 1e Na v 1.5 (N-term) 250KD fl/fl Heart Lung Kidney Spleen c -/- fl/fl Suppl. Fig. 6b c -/- Connexin 43 Vehicle Suppl. Fig. 6c Ace-K 1479 Na v 1.5 Ex-527 Vehicle Ex-527 Suppl. Fig. 4a IP: Na V 1.5 IB: Ace-K IP: Na V 1.5 IB: Na V 1.5 Myc- Suppl. Fig. 4b IP: IB: Ace-K IP: IB: Suppl. Fig. 6d Bip Untreat. NAM Ad-Lac-Z Resveratrol Ad-SIRT1 (H363Y) 220KD 220KD 110KD Suppl. Fig. 5e Ace.-K Na V 1.5 Na V Na V (acetyl K 1479 ) Suppl. Fig. 4c IB: Ace.-K Na V µg 0.20 µg Suppl. Fig. 5a IP: IB: Ace-K IP: IB: -Na V 1.5 (III-IV) -Na V 1.5 (III-IV) Amount of peptide µg p Suppl. Fig. 5f IP: IB: Ace.-K Na V 1.5 IB: Chop 2.00 µg - Na V 1.5(III/IV) pcdna p300/cbp Vehicle Ex-527 Supplementary Fig. 8. Uncropped immunoblots of indicated figures. 8
9 Supplementary Table 1. Kinetics of I Na conducted by wild type or K1479A Na v 1.5 in HEK 293 cells expressing wild type or dominant negative (H363Y), and of I Na conducted by native Na v 1.5 in rat neonatal cardiac myocytes treated with Ex-242 or Ex-243. HEK 293 cells Steady State Activation (V 50, mv) Steady State Inactivation (V 50, mv) Development of Slow Inactivation (ms) Na v ± ± ± 35.2 Recovery from Slow & Fast Inactivation (ms) Slow;174.6 ± 18.6 Fast;10.4 ± 15.0 Mean cell Capacitance (pf) 9.36±0.58 Na v ± ± ± 41.0 Slow;208.9 ± 40.4 Fast;6.4 ± ±0.51 Na v (H363Y) ± ± ± 47.4 K1479A-Na v ± ± ± 41.7 K1479A-Na v K1479A-Na v (H363Y) ± ± ± ± ± ± 25.9 Slow; ± 23.5 Fast;13.9 ± 2.6 Slow;166.8 ± 28.3 Fast;12.8 ± 3.6 Slow;193.8 ± 15.4 Fast;18.3 ± 1.6 Slow;158.6 ± 32.7 Fast; 14.3 ± ± ± ± ±0.88 Rat neonatal cardiac myocytes Ex ± ± ±0.78 Ex ± ± ±0.91 Rat neonatal cardiac myocytes were treated with inhibitor Ex-243 or its inactive control isomer Ex-242. Data shown as mean and error bar represents s.e.m. 9
10 Supplementary Table 2. Voltage-gating of I Na in cardiomyocytes isolated from c -/- and fl/fl mice. Mouse ventricular cardiomyocytes Steady State Inactivation (V 50, mv) Time Constants of the Decay Phase of I Na (ms) fl/fl ± 1.3 (n=7) 1.7 ± 0.1 (n=7) Recovery from Slow & Fast Inactivation (ms) Slow;137.6 ± 31.9 (n=4) Fast;5.6 ± 0.3 c -/ ± 1.8, (n=5) 1.3 ± 0.3 (n=4) Slow;165.1 ± 60.3 (n=4) Fast;4.6 ± 1.6 p ns ns ns ns; p > 0.05 vs. fl/fl. Independent sample t-test was used. Data shown as mean and error bar represents s.e.m. 10
11 Supplementary Table 3. Kinetic properties of wild type, K1479A, and K1479Q Na v 1.5 expressed in HEK 293 cells. Steady State Activation (V 50, mv) Steady State Inactivation (V 50, mv) Na v 1.5-WT (n) Na v 1.5-K 1479R (n) p value Na v 1.5-K 1479Q (n) p value ± 2.7 (8) ± 1.7 (10) ± 2.1(10) ± 2.0 (9) ± 2.0 (9) ± 1.5 (11)
12 Supplementary Table 4. EKG features of c -/- mice and fl/fl mice. Genotype Age (months) N Sex (% Male) HR (min -1 ) PR (msec) QRS (msec) QT (msec) QTc (msec) fl/fl 4.2 ± ± ± ± ± 3 59 ± 3 c -/- 4.4 ± ± ± ± ± 5 54 ± 3 fl/fl 8.3 ± ± ± ± ± 5 49 ± 3 c -/- 9.2 ± ± ± ± 1.0* 74 ± 4* 63 ± 3* * p < 0.05 vs. fl/fl. Independent sample t-test was used. Data shown as mean and error bar represents s.e.m. 12
13 Supplementary Table 5. Effect of inhibition with Ex-527 on EKG features in Scn5a +/+ and Scn5a +/- mice. Scn5a +/+ Mice Basal Ex-527 N 5 4 Age (months) 10.6 ± 0.4 p value RR interval (ms) PR interval (ms) QRS interval (ms) 121 ± ± ± ± ± ± QTc duration (ms) 50.9 ± ± Scn5a +/- Mice Basal Ex-527 N 8 8 Age (months) 11.1 ± 0.2 RR interval (ms) PR interval (ms) QRS interval (ms) 125 ± ± ± ± ± ± QTc duration (ms) 66.5 ± ±
14 Supplementary Table 6. Ambulatory telemetry of c -/- and fl/fl mice. Parameters fl/fl c -/- p value N Age (Months) 7.2 ± ± Sex (male/female) 3/7 8/ Maximum heart rate (beats/min) 751 ± ± Minimum heart rate (beats/min) 360 ± ± Mice with nonsustained VT Longest ventricular pause (msec) 331 ± ± Frequent sinus pauses (>5/hr) o Degree heart block (in 12 hrs) 1.3 ± ± Nonsustained VT, 3 consecutive ventricular beats; Sinus pause, cycle length increase of >50% to >150 msec. Independent sample t-test was used. Data shown as mean and error bar represents s.e.m. 14
15 Supplementary Table 7. General, gross, and echocardiographic features of male fl/fl and c -/- mice. General Parameters fl/fl c -/- p value n 5 6 Age (Months) 5.6 ± ± Heart weight (mg) 157 ± ± Lung weight (mg) 180 ± ± Body weight (grams) 32.7 ± ± Heart weight/body weight ± ± Lung weight/body weight ± ± Echocardiogram Parameters fl/fl c -/- p value n 5 7 Age (Months) 5.6 ± ± Heart rate (beats/minute) 612 ± ± End diastolic Volume (µl) 40 ± 4 51 ± End systolic Volume (µl) 8 ± 2 18 ± LV ejection fraction (%) 80 ± 4 63 ± Cardiac output (µl/minute) 19.4 ± ± Independent sample t-test was used. Data shown as mean and error bar represents s.e.m. 15
16 Supplementary Table 8. Demographic, gross, EKG, and echocardiographic features of hearts from normal donors and patients with idiopathic dilated cardiomyopathy. Normal Parameters Cardiomyopathy Donors Diagnosis Normal Idiopathic Dilated N 4 7 Conduction abnormality - 5 Heart rate ± 8.2 p value PR interval (ms) ± 15.9 Normal <200 QRS interval (ms) ± 13.8 Normal <120 QTc interval (ms) ± 16.3 Normal <450 Ethnicity (% Caucasian) Patient Weight (kg) 70.8 ± ± Patient Height (cm) ± ± Age (Y) 51.0 ± ± Heart Weight (g) ± ± LVEF % 66.3 ± ± 2.5 < CMP; Cardiomyopathy, LVEF%; left ventricular ejection fraction. Independent sample t-test was used. Data shown as mean and error represents s.e.m. 16
SUPPLEMENTARY INFORMATION
 a c e doi:10.1038/nature10407 b d f Supplementary Figure 1. SERCA2a complex analysis. (a) Two-dimensional SDS-PAGE gels of SERCA2a complexes. A silver-stained SDSPAGE gel is shown, which reveals a 12 kda
a c e doi:10.1038/nature10407 b d f Supplementary Figure 1. SERCA2a complex analysis. (a) Two-dimensional SDS-PAGE gels of SERCA2a complexes. A silver-stained SDSPAGE gel is shown, which reveals a 12 kda
Supplementary Figure 1. MAT IIα is Acetylated at Lysine 81.
 IP: Flag a Mascot PTM Modified Mass Error Position Gene Names Score Score Sequence m/z [ppm] 81 MAT2A;AMS2;MATA2 35.6 137.28 _AAVDYQK(ac)VVR_ 595.83-2.28 b Pre-immu After-immu Flag- WT K81R WT K81R / Flag
IP: Flag a Mascot PTM Modified Mass Error Position Gene Names Score Score Sequence m/z [ppm] 81 MAT2A;AMS2;MATA2 35.6 137.28 _AAVDYQK(ac)VVR_ 595.83-2.28 b Pre-immu After-immu Flag- WT K81R WT K81R / Flag
Supplementary Figure 1. Confocal immunofluorescence showing mitochondrial translocation of Drp1. Cardiomyocytes treated with H 2 O 2 were prestained
 Supplementary Figure 1. Confocal immunofluorescence showing mitochondrial translocation of Drp1. Cardiomyocytes treated with H 2 O 2 were prestained with MitoTracker (red), then were immunostained with
Supplementary Figure 1. Confocal immunofluorescence showing mitochondrial translocation of Drp1. Cardiomyocytes treated with H 2 O 2 were prestained with MitoTracker (red), then were immunostained with
c Ischemia (30 min) Reperfusion (8 w) Supplementary Figure bp 300 bp Ischemia (30 min) Reperfusion (4 h) Dox 20 mg/kg i.p.
 a Marker Ripk3 +/ 5 bp 3 bp b Ischemia (3 min) Reperfusion (4 h) d 2 mg/kg i.p. 1 w 5 w Sacrifice for IF size A subset for echocardiography and morphological analysis c Ischemia (3 min) Reperfusion (8
a Marker Ripk3 +/ 5 bp 3 bp b Ischemia (3 min) Reperfusion (4 h) d 2 mg/kg i.p. 1 w 5 w Sacrifice for IF size A subset for echocardiography and morphological analysis c Ischemia (3 min) Reperfusion (8
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells
 Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous
 Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous LRP5 in intact adult mouse ventricular myocytes (AMVMs)
Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous LRP5 in intact adult mouse ventricular myocytes (AMVMs)
Supplementary Figure 1
 Supplementary Figure 1 how HFD how HFD Epi WT p p Hypothalamus p p Inguinal WT T Liver Lean mouse adipocytes p p p p p p Obese mouse adipocytes Kidney Muscle Spleen Heart p p p p p p p p Extracellular
Supplementary Figure 1 how HFD how HFD Epi WT p p Hypothalamus p p Inguinal WT T Liver Lean mouse adipocytes p p p p p p Obese mouse adipocytes Kidney Muscle Spleen Heart p p p p p p p p Extracellular
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb2822 a MTC02 FAO cells EEA1 b +/+ MEFs /DAPI -/- MEFs /DAPI -/- MEFs //DAPI c HEK 293 cells WCE N M C P AKT TBC1D7 Lamin A/C EEA1 VDAC d HeLa cells WCE N M C P AKT Lamin A/C EEA1 VDAC Figure
DOI:.38/ncb2822 a MTC02 FAO cells EEA1 b +/+ MEFs /DAPI -/- MEFs /DAPI -/- MEFs //DAPI c HEK 293 cells WCE N M C P AKT TBC1D7 Lamin A/C EEA1 VDAC d HeLa cells WCE N M C P AKT Lamin A/C EEA1 VDAC Figure
Control. csarnt -/- Cre, f/f
 ody weight (g) A re,f/f re x f/f f/+ re, f/+ re,f/+ f/f x f/f f/+ cs -/- re, f/f re f/f re, f/f Normal chow Tamoxifen Tamoxifen Tamoxifen W 4W re f/f re, re/ff f/f re f/f re, re/ff f/f Normal chow Tamoxifen
ody weight (g) A re,f/f re x f/f f/+ re, f/+ re,f/+ f/f x f/f f/+ cs -/- re, f/f re f/f re, f/f Normal chow Tamoxifen Tamoxifen Tamoxifen W 4W re f/f re, re/ff f/f re f/f re, re/ff f/f Normal chow Tamoxifen
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Supplementary Figure 1. (A) Left, western blot analysis of ISGylated proteins in Jurkat T cells treated with 1000U ml -1 IFN for 16h (IFN) or left untreated (CONT); right, western
SUPPLEMENTARY FIGURES Supplementary Figure 1. (A) Left, western blot analysis of ISGylated proteins in Jurkat T cells treated with 1000U ml -1 IFN for 16h (IFN) or left untreated (CONT); right, western
Supplementary Figure 1. A. Bar graph representing the expression levels of the 19 indicated genes in the microarrays analyses comparing human lung
 Supplementary Figure 1. A. Bar graph representing the expression levels of the 19 indicated genes in the microarrays analyses comparing human lung immortalized broncho-epithelial cells (AALE cells) expressing
Supplementary Figure 1. A. Bar graph representing the expression levels of the 19 indicated genes in the microarrays analyses comparing human lung immortalized broncho-epithelial cells (AALE cells) expressing
Postn MCM Smad2 fl/fl Postn MCM Smad3 fl/fl Postn MCM Smad2/3 fl/fl. Postn MCM. Tgfbr1/2 fl/fl TAC
 A Smad2 fl/fl Smad3 fl/fl Smad2/3 fl/fl Tgfbr1/2 fl/fl 1. mm B Tcf21 MCM Tcf21 MCM Smad3 fl/fl Tcf21 MCM Smad2/3 fl/fl Tcf21 MCM Tgfbr1/2 fl/fl αmhc MCM C 1. mm 1. mm D Smad2 fl/fl Smad3 fl/fl Smad2/3
A Smad2 fl/fl Smad3 fl/fl Smad2/3 fl/fl Tgfbr1/2 fl/fl 1. mm B Tcf21 MCM Tcf21 MCM Smad3 fl/fl Tcf21 MCM Smad2/3 fl/fl Tcf21 MCM Tgfbr1/2 fl/fl αmhc MCM C 1. mm 1. mm D Smad2 fl/fl Smad3 fl/fl Smad2/3
Predictive PP1Ca binding region in BIG3 : 1,228 1,232aa (-KAVSF-) HEK293T cells *** *** *** KPL-3C cells - E E2 treatment time (h)
 Relative expression ERE-luciferase activity activity (pmole/min) activity (pmole/min) activity (pmole/min) activity (pmole/min) MCF-7 KPL-3C ZR--1 BT-474 T47D HCC15 KPL-1 HBC4 activity (pmole/min) a d
Relative expression ERE-luciferase activity activity (pmole/min) activity (pmole/min) activity (pmole/min) activity (pmole/min) MCF-7 KPL-3C ZR--1 BT-474 T47D HCC15 KPL-1 HBC4 activity (pmole/min) a d
(a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable
 Supplementary Figure 1. Frameshift (FS) mutation in UVRAG. (a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable A 10 DNA repeat, generating a premature stop codon
Supplementary Figure 1. Frameshift (FS) mutation in UVRAG. (a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable A 10 DNA repeat, generating a premature stop codon
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation.
 Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
Supplementary Information
 Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
hemodynamic stress. A. Echocardiographic quantification of cardiac dimensions and function in
 SUPPLEMENTAL FIGURE LEGENDS Supplemental Figure 1. Fbn1 C1039G/+ hearts display normal cardiac function in the absence of hemodynamic stress. A. Echocardiographic quantification of cardiac dimensions and
SUPPLEMENTAL FIGURE LEGENDS Supplemental Figure 1. Fbn1 C1039G/+ hearts display normal cardiac function in the absence of hemodynamic stress. A. Echocardiographic quantification of cardiac dimensions and
Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was
 Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was painted on the shaved back skin of CBL/J and BALB/c mice for consecutive days. (a, b) Phenotypic presentation of mouse back skin
Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was painted on the shaved back skin of CBL/J and BALB/c mice for consecutive days. (a, b) Phenotypic presentation of mouse back skin
Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus
 Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus changes in corresponding proteins between wild type and Gprc5a-/-
Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus changes in corresponding proteins between wild type and Gprc5a-/-
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature06994 A phosphatase cascade by which rewarding stimuli control nucleosomal response A. Stipanovich*, E. Valjent*, M. Matamales*, A. Nishi, J.H. Ahn, M. Maroteaux, J. Bertran-Gonzalez,
doi: 10.1038/nature06994 A phosphatase cascade by which rewarding stimuli control nucleosomal response A. Stipanovich*, E. Valjent*, M. Matamales*, A. Nishi, J.H. Ahn, M. Maroteaux, J. Bertran-Gonzalez,
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Krenn et al., http://www.jcb.org/cgi/content/full/jcb.201110013/dc1 Figure S1. Levels of expressed proteins and demonstration that C-terminal
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Krenn et al., http://www.jcb.org/cgi/content/full/jcb.201110013/dc1 Figure S1. Levels of expressed proteins and demonstration that C-terminal
Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures
 Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures Type of file: MOV Title of file for HTML: Supplementary Movie 1 Description: NLRP3 is moving along
Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures Type of file: MOV Title of file for HTML: Supplementary Movie 1 Description: NLRP3 is moving along
Supplementary Figure 1: Expression of NFAT proteins in Nfat2-deleted B cells (a+b) Protein expression of NFAT2 (a) and NFAT1 (b) in isolated splenic
 Supplementary Figure 1: Expression of NFAT proteins in Nfat2-deleted B cells (a+b) Protein expression of NFAT2 (a) and NFAT1 (b) in isolated splenic B cells from WT Nfat2 +/+, TCL1 Nfat2 +/+ and TCL1 Nfat2
Supplementary Figure 1: Expression of NFAT proteins in Nfat2-deleted B cells (a+b) Protein expression of NFAT2 (a) and NFAT1 (b) in isolated splenic B cells from WT Nfat2 +/+, TCL1 Nfat2 +/+ and TCL1 Nfat2
E10.5 E18.5 P2 10w 83w NF1 HF1. Sham ISO. Bmi1. H3K9me3. Lung weight (g)
 Myociyte cross-sectional Relative mrna levels Relative levels Relative mrna levels Supplementary Figures and Legends a 8 6 4 2 Ezh2 E1.5 E18.5 P2 1w 83w b Ezh2 p16 amhc b-actin P2 43w kd 37 86 16 wt mouse
Myociyte cross-sectional Relative mrna levels Relative levels Relative mrna levels Supplementary Figures and Legends a 8 6 4 2 Ezh2 E1.5 E18.5 P2 1w 83w b Ezh2 p16 amhc b-actin P2 43w kd 37 86 16 wt mouse
Supplementary Figures Supplementary Figure 1. Development of the camp biosensor targeted to the SERCA2a microdomain.
 Supplementary Figures Supplementary Figure 1. Development of the camp biosensor targeted to the SERCA2a microdomain. A B C (A) Schematic representation of the new constructs designed for local camp imaging.
Supplementary Figures Supplementary Figure 1. Development of the camp biosensor targeted to the SERCA2a microdomain. A B C (A) Schematic representation of the new constructs designed for local camp imaging.
Supplementary Figures and Figure Legends
 1 Supplementary Figures and Figure Legends 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 Supplementary Figure 1: camp levels in myocytes confirm studies in perfused hearts. (a) Time-resolved camp dynamics (presented
1 Supplementary Figures and Figure Legends 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 Supplementary Figure 1: camp levels in myocytes confirm studies in perfused hearts. (a) Time-resolved camp dynamics (presented
Supplementary Figure 1. c Human
 Supplementary Figure 1 a b c Human Mouse d Gapdh Amino acid sequence and baseline expression of MYDGF N-terminal signal peptides (S-scores) and signal peptide cleavage sites (C-scores) of (a) human MYDGF
Supplementary Figure 1 a b c Human Mouse d Gapdh Amino acid sequence and baseline expression of MYDGF N-terminal signal peptides (S-scores) and signal peptide cleavage sites (C-scores) of (a) human MYDGF
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
Fetal gene upregulation by 1-wk TAC is significantly increased in mice lacking RGS2.
 3562-RG-1 Supplementary Figure 1 Fetal gene upregulation by 1-wk is significantly increased in mice lacking RGS2. ANP(Nppa) /BNP(Nppb) A-type and B-type natriuretic peptide; β-mhc (Myh7) beta myosin heavy
3562-RG-1 Supplementary Figure 1 Fetal gene upregulation by 1-wk is significantly increased in mice lacking RGS2. ANP(Nppa) /BNP(Nppb) A-type and B-type natriuretic peptide; β-mhc (Myh7) beta myosin heavy
Supplemental Figure 1
 Supplemental Figure 1 1a 1c PD-1 MFI fold change 6 5 4 3 2 1 IL-1α IL-2 IL-4 IL-6 IL-1 IL-12 IL-13 IL-15 IL-17 IL-18 IL-21 IL-23 IFN-α Mut Human PD-1 promoter SBE-D 5 -GTCTG- -1.2kb SBE-P -CAGAC- -1.kb
Supplemental Figure 1 1a 1c PD-1 MFI fold change 6 5 4 3 2 1 IL-1α IL-2 IL-4 IL-6 IL-1 IL-12 IL-13 IL-15 IL-17 IL-18 IL-21 IL-23 IFN-α Mut Human PD-1 promoter SBE-D 5 -GTCTG- -1.2kb SBE-P -CAGAC- -1.kb
p = formed with HCI-001 p = Relative # of blood vessels that formed with HCI-002 Control Bevacizumab + 17AAG Bevacizumab 17AAG
 A.. Relative # of ECs associated with HCI-001 1.4 1.2 1.0 0.8 0.6 0.4 0.2 0.0 ol b p < 0.001 Relative # of blood vessels that formed with HCI-001 1.4 1.2 1.0 0.8 0.6 0.4 0.2 0.0 l b p = 0.002 Control IHC:
A.. Relative # of ECs associated with HCI-001 1.4 1.2 1.0 0.8 0.6 0.4 0.2 0.0 ol b p < 0.001 Relative # of blood vessels that formed with HCI-001 1.4 1.2 1.0 0.8 0.6 0.4 0.2 0.0 l b p = 0.002 Control IHC:
EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH
 EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH Supplementary Figure 1. Supplementary Figure 1. Characterization of KP and KPH2 autochthonous UPS tumors. a) Genotyping of KPH2
EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH Supplementary Figure 1. Supplementary Figure 1. Characterization of KP and KPH2 autochthonous UPS tumors. a) Genotyping of KPH2
SUPPLEMENTAL FIGURE LEGENDS
 SUPPLEMENTAL FIGURE LEGENDS Supplemental Figure S1: Endogenous interaction between RNF2 and H2AX: Whole cell extracts from 293T were subjected to immunoprecipitation with anti-rnf2 or anti-γ-h2ax antibodies
SUPPLEMENTAL FIGURE LEGENDS Supplemental Figure S1: Endogenous interaction between RNF2 and H2AX: Whole cell extracts from 293T were subjected to immunoprecipitation with anti-rnf2 or anti-γ-h2ax antibodies
Supplemental Figure 1. Western blot analysis indicated that MIF was detected in the fractions of
 Supplemental Figure Legends Supplemental Figure 1. Western blot analysis indicated that was detected in the fractions of plasma membrane and cytosol but not in nuclear fraction isolated from Pkd1 null
Supplemental Figure Legends Supplemental Figure 1. Western blot analysis indicated that was detected in the fractions of plasma membrane and cytosol but not in nuclear fraction isolated from Pkd1 null
This PDF file includes: Supplementary Figures 1 to 6 Supplementary Tables 1 to 2 Supplementary Methods Supplementary References
 Structure of the catalytic core of p300 and implications for chromatin targeting and HAT regulation Manuela Delvecchio, Jonathan Gaucher, Carmen Aguilar-Gurrieri, Esther Ortega, Daniel Panne This PDF file
Structure of the catalytic core of p300 and implications for chromatin targeting and HAT regulation Manuela Delvecchio, Jonathan Gaucher, Carmen Aguilar-Gurrieri, Esther Ortega, Daniel Panne This PDF file
(A) SW480, DLD1, RKO and HCT116 cells were treated with DMSO or XAV939 (5 µm)
 Supplementary Figure Legends Figure S1. Tankyrase inhibition suppresses cell proliferation in an axin/β-catenin independent manner. (A) SW480, DLD1, RKO and HCT116 cells were treated with DMSO or XAV939
Supplementary Figure Legends Figure S1. Tankyrase inhibition suppresses cell proliferation in an axin/β-catenin independent manner. (A) SW480, DLD1, RKO and HCT116 cells were treated with DMSO or XAV939
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11700 Figure 1: RIP3 as a potential Sirt2 interacting protein. Transfected Flag-tagged Sirt2 was immunoprecipitated from cells and eluted from the Sepharose beads using Flag peptide.
doi:10.1038/nature11700 Figure 1: RIP3 as a potential Sirt2 interacting protein. Transfected Flag-tagged Sirt2 was immunoprecipitated from cells and eluted from the Sepharose beads using Flag peptide.
Nature Structural and Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Mutational analysis of the SA2-Scc1 interaction in vitro and in human cells. (a) Autoradiograph (top) and Coomassie stained gel (bottom) of 35 S-labeled Myc-SA2 proteins (input)
Supplementary Figure 1 Mutational analysis of the SA2-Scc1 interaction in vitro and in human cells. (a) Autoradiograph (top) and Coomassie stained gel (bottom) of 35 S-labeled Myc-SA2 proteins (input)
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supplementary Figure 1. Long-term protection studies. 45 minutes of ischemia was induced in wild type (S1pr2 +/+ ) and S1pr2 -/- by MCAO. A) 5 days later brains were harvested
SUPPLEMENTARY INFORMATION Supplementary Figure 1. Long-term protection studies. 45 minutes of ischemia was induced in wild type (S1pr2 +/+ ) and S1pr2 -/- by MCAO. A) 5 days later brains were harvested
Incorporation of photo-caged lysine (pc-lys) at K273 of human LCK allows specific control of the enzyme activity.
 Supplementary Figure 1 Incorporation of photo-caged lysine (pc-lys) at K273 of human LCK allows specific control of the enzyme activity. (a) Modeling of the kinase domain of LCK with ATP (left) or pc-lys
Supplementary Figure 1 Incorporation of photo-caged lysine (pc-lys) at K273 of human LCK allows specific control of the enzyme activity. (a) Modeling of the kinase domain of LCK with ATP (left) or pc-lys
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature11429 S1a 6 7 8 9 Nlrc4 allele S1b Nlrc4 +/+ Nlrc4 +/F Nlrc4 F/F 9 Targeting construct 422 bp 273 bp FRT-neo-gb-PGK-FRT 3x.STOP S1c Nlrc4 +/+ Nlrc4 F/F casp1
SUPPLEMENTARY INFORMATION doi:10.1038/nature11429 S1a 6 7 8 9 Nlrc4 allele S1b Nlrc4 +/+ Nlrc4 +/F Nlrc4 F/F 9 Targeting construct 422 bp 273 bp FRT-neo-gb-PGK-FRT 3x.STOP S1c Nlrc4 +/+ Nlrc4 F/F casp1
SUPPLEMENTARY INFORMATION
 Supplementary Figure 1. Normal AMPAR-mediated fepsp input-output curve in CA3-Psen cdko mice. Input-output curves, which are plotted initial slopes of the evoked fepsp as function of the amplitude of the
Supplementary Figure 1. Normal AMPAR-mediated fepsp input-output curve in CA3-Psen cdko mice. Input-output curves, which are plotted initial slopes of the evoked fepsp as function of the amplitude of the
LPS LPS P6 - + Supplementary Fig. 1.
 P6 LPS - - - + + + - LPS + + - - P6 + Supplementary Fig. 1. Pharmacological inhibition of the JAK/STAT blocks LPS-induced HMGB1 nuclear translocation. RAW 267.4 cells were stimulated with LPS in the absence
P6 LPS - - - + + + - LPS + + - - P6 + Supplementary Fig. 1. Pharmacological inhibition of the JAK/STAT blocks LPS-induced HMGB1 nuclear translocation. RAW 267.4 cells were stimulated with LPS in the absence
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Analysis of hair bundle morphology in Ush1c c.216g>a mice at P18 by SEM.
 Supplementary Figure 1 Analysis of hair bundle morphology in Ush1c c.216g>a mice at P18 by SEM. (a-c) Heterozygous c.216ga mice displayed normal hair bundle morphology at P18. (d-i) Disorganized hair bundles
Supplementary Figure 1 Analysis of hair bundle morphology in Ush1c c.216g>a mice at P18 by SEM. (a-c) Heterozygous c.216ga mice displayed normal hair bundle morphology at P18. (d-i) Disorganized hair bundles
Supporting Information. FADD regulates NF-кB activation and promotes ubiquitination of cflip L to induce. apoptosis
 1 2 Supporting Information 3 4 5 FADD regulates NF-кB activation and promotes ubiquitination of cflip L to induce apoptosis 6 7 Kishu Ranjan and Chandramani Pathak* 8 9 Department of Cell Biology, School
1 2 Supporting Information 3 4 5 FADD regulates NF-кB activation and promotes ubiquitination of cflip L to induce apoptosis 6 7 Kishu Ranjan and Chandramani Pathak* 8 9 Department of Cell Biology, School
(a) Significant biological processes (upper panel) and disease biomarkers (lower panel)
 Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1
 Supplementary Figure 1 a γ-h2ax MDC1 RNF8 FK2 BRCA1 U2OS Cells sgrna-1 ** 60 sgrna 40 20 0 % positive Cells (>5 foci per cell) b ** 80 sgrna sgrna γ-h2ax MDC1 γ-h2ax RNF8 FK2 MDC1 BRCA1 RNF8 FK2 BRCA1
Supplementary Figure 1 a γ-h2ax MDC1 RNF8 FK2 BRCA1 U2OS Cells sgrna-1 ** 60 sgrna 40 20 0 % positive Cells (>5 foci per cell) b ** 80 sgrna sgrna γ-h2ax MDC1 γ-h2ax RNF8 FK2 MDC1 BRCA1 RNF8 FK2 BRCA1
a surface permeabilized
 a surface permeabilized RAW 64.7 P388D1 J774 b CD11b + Ly-6G - Blood Monocytes WT Supplementary Figure 1. Cell surface expression on macrophages and DCs. (a) RAW64.7, P388D1, and J774 cells were subjected
a surface permeabilized RAW 64.7 P388D1 J774 b CD11b + Ly-6G - Blood Monocytes WT Supplementary Figure 1. Cell surface expression on macrophages and DCs. (a) RAW64.7, P388D1, and J774 cells were subjected
Supplementary Information Titles Journal: Nature Medicine
 Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
Supplementary fig. 1. Crystals induce necroptosis does not involve caspases, TNF receptor or NLRP3. A. Mouse tubular epithelial cells were pretreated
 Supplementary fig. 1. Crystals induce necroptosis does not involve caspases, TNF receptor or NLRP3. A. Mouse tubular epithelial cells were pretreated with zvad-fmk (10µM) and exposed to calcium oxalate
Supplementary fig. 1. Crystals induce necroptosis does not involve caspases, TNF receptor or NLRP3. A. Mouse tubular epithelial cells were pretreated with zvad-fmk (10µM) and exposed to calcium oxalate
Tcf21 MCM ; R26 mtmg Sham GFP Col 1/3 TAC 8W TAC 2W. Postn MCM ; R26 mtmg Sham GFP Col 1/3 TAC 8W TAC 2W
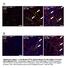 A Tcf21 MCM ; R26 mtmg Sham GFP Col 1/3 Tcf21 MCM ; R26 mtmg TAC 2W Tcf21 MCM ; R26 mtmg TAC 8W B Postn MCM ; R26 mtmg Sham GFP Col 1/3 Postn MCM ; R26 mtmg TAC 2W Postn MCM ; R26 mtmg TAC 8W Supplementary
A Tcf21 MCM ; R26 mtmg Sham GFP Col 1/3 Tcf21 MCM ; R26 mtmg TAC 2W Tcf21 MCM ; R26 mtmg TAC 8W B Postn MCM ; R26 mtmg Sham GFP Col 1/3 Postn MCM ; R26 mtmg TAC 2W Postn MCM ; R26 mtmg TAC 8W Supplementary
Supplementary Figure S1 Supplementary Figure S2
 Supplementary Figure S A) The blots shown in Figure B were qualified by using Gel-Pro analyzer software (Rockville, MD, USA). The ratio of LC3II/LC3I to actin was then calculated. The data are represented
Supplementary Figure S A) The blots shown in Figure B were qualified by using Gel-Pro analyzer software (Rockville, MD, USA). The ratio of LC3II/LC3I to actin was then calculated. The data are represented
Supplementary Figure 1: Co-localization of reconstituted L-PTC and dendritic cells
 a CD11c Na + K + ATPase Na + K + ATPase CD11c x-y CD11c Na + K + ATPase Na + K + ATPase CD11c x-z c b x-y view BoNT NAPs CD11c BoNT CD11c NAPs BoNT NAPs CD11c 90 x-z view Apical Basolateral Supplementary
a CD11c Na + K + ATPase Na + K + ATPase CD11c x-y CD11c Na + K + ATPase Na + K + ATPase CD11c x-z c b x-y view BoNT NAPs CD11c BoNT CD11c NAPs BoNT NAPs CD11c 90 x-z view Apical Basolateral Supplementary
condition. Left panel, the HCT-116 cells were lysed with RIPA buffer containing 0.1%
 FIGURE LEGENDS Supplementary Fig 1 (A) sumoylation pattern detected under denaturing condition. Left panel, the HCT-116 cells were lysed with RIPA buffer containing 0.1% SDS in the presence and absence
FIGURE LEGENDS Supplementary Fig 1 (A) sumoylation pattern detected under denaturing condition. Left panel, the HCT-116 cells were lysed with RIPA buffer containing 0.1% SDS in the presence and absence
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/278/rs11/dc1 Supplementary Materials for In Vivo Phosphoproteomics Analysis Reveals the Cardiac Targets of β-adrenergic Receptor Signaling Alicia Lundby,* Martin
www.sciencesignaling.org/cgi/content/full/6/278/rs11/dc1 Supplementary Materials for In Vivo Phosphoproteomics Analysis Reveals the Cardiac Targets of β-adrenergic Receptor Signaling Alicia Lundby,* Martin
Figure S1. Sorting nexin 9 (SNX9) specifically binds psmad3 and not psmad 1/5/8. Lysates from AKR-2B cells untreated (-) or stimulated (+) for 45 min
 Figure S1. Sorting nexin 9 (SNX9) specifically binds psmad3 and not psmad 1/5/8. Lysates from AKR2B cells untreated () or stimulated () for 45 min with 5 ng/ml TGFβ or 10 ng/ml BMP4 were incubated with
Figure S1. Sorting nexin 9 (SNX9) specifically binds psmad3 and not psmad 1/5/8. Lysates from AKR2B cells untreated () or stimulated () for 45 min with 5 ng/ml TGFβ or 10 ng/ml BMP4 were incubated with
Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis
 Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis of PD-L1 in ovarian cancer cells. (c) Western blot analysis
Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis of PD-L1 in ovarian cancer cells. (c) Western blot analysis
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
Supplementary Figure 1 CD4 + T cells from PKC-θ null mice are defective in NF-κB activation during T cell receptor signaling. CD4 + T cells were
 Supplementary Figure 1 CD4 + T cells from PKC-θ null mice are defective in NF-κB activation during T cell receptor signaling. CD4 + T cells were isolated from wild type (PKC-θ- WT) or PKC-θ null (PKC-θ-KO)
Supplementary Figure 1 CD4 + T cells from PKC-θ null mice are defective in NF-κB activation during T cell receptor signaling. CD4 + T cells were isolated from wild type (PKC-θ- WT) or PKC-θ null (PKC-θ-KO)
Supplemental Table I
 Supplemental Table I Cortex Hippocampus Age (weeks) Genotype STE 61 pste 61 STE 61 pste 61 8 100.0 ± 4.4 96.2 ± 6.0 99.8 ± 8.7 167.5 ± 7.8* 100.0 ± 7.0 90.5 ± 13.8 100.0 ± 12.8 260.4 ± 33.9** 12 100.0
Supplemental Table I Cortex Hippocampus Age (weeks) Genotype STE 61 pste 61 STE 61 pste 61 8 100.0 ± 4.4 96.2 ± 6.0 99.8 ± 8.7 167.5 ± 7.8* 100.0 ± 7.0 90.5 ± 13.8 100.0 ± 12.8 260.4 ± 33.9** 12 100.0
Probe. Hind III Q,!?R'!! /0!!!!D1"?R'! vector. Homologous recombination
 Supple-Zhang Page 1 Wild-type locus Targeting construct Targeted allele Exon Exon3 Exon Probe P1 P P3 FRT FRT loxp loxp neo vector amh I Homologous recombination neo P1 P P3 FLPe recombination Q,!?R'!!
Supple-Zhang Page 1 Wild-type locus Targeting construct Targeted allele Exon Exon3 Exon Probe P1 P P3 FRT FRT loxp loxp neo vector amh I Homologous recombination neo P1 P P3 FLPe recombination Q,!?R'!!
Supplementary Figure 1. ETBF activate Stat3 in B6 and Min mice colons
 Supplementary Figure 1 ETBF activate Stat3 in B6 and Min mice colons a pstat3 controls Pos Neg ETBF 1 2 3 4 b pstat1 pstat2 pstat3 pstat4 pstat5 pstat6 Actin Figure Legend: (a) ETBF induce predominantly
Supplementary Figure 1 ETBF activate Stat3 in B6 and Min mice colons a pstat3 controls Pos Neg ETBF 1 2 3 4 b pstat1 pstat2 pstat3 pstat4 pstat5 pstat6 Actin Figure Legend: (a) ETBF induce predominantly
Supplementary Table I Blood pressure and heart rate measurements pre- and post-stroke
 SUPPLEMENTARY INFORMATION doi:10.1038/nature09511 Supplementary Table I Blood pressure and heart rate measurements pre- and post-stroke Pre Post 7-days Systolic Diastolic BPM Systolic Diastolic BPM Systolic
SUPPLEMENTARY INFORMATION doi:10.1038/nature09511 Supplementary Table I Blood pressure and heart rate measurements pre- and post-stroke Pre Post 7-days Systolic Diastolic BPM Systolic Diastolic BPM Systolic
Supplementary Figure 1. Repression of hepcidin expression in the liver of mice treated with
 Supplementary Figure 1. Repression of hepcidin expression in the liver of mice treated with DMN Immunohistochemistry for hepcidin and H&E staining (left). qrt-pcr assays for hepcidin in the liver (right).
Supplementary Figure 1. Repression of hepcidin expression in the liver of mice treated with DMN Immunohistochemistry for hepcidin and H&E staining (left). qrt-pcr assays for hepcidin in the liver (right).
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
B. SDS-PAGE. Western blot MW (kda) Supplementary Information. Supplementary Figure 1. MW (kda) Glutelin. Prolamin. 3 E.coli. 3 E.
 Supplementary Information Supplementary Figure 1. A. MW (kda) B. SDS-PAGE Western blot MW (kda) 50 40 100 75 50 37 30 Glutelin 25 20 15 20 Prolamin 10 ARP1 1 2 3 E.coli MucoRice ARP1 (RNAi +) 4 5 ARP1
Supplementary Information Supplementary Figure 1. A. MW (kda) B. SDS-PAGE Western blot MW (kda) 50 40 100 75 50 37 30 Glutelin 25 20 15 20 Prolamin 10 ARP1 1 2 3 E.coli MucoRice ARP1 (RNAi +) 4 5 ARP1
SUPPLEMENTARY INFORMATION. Supplementary Figures S1-S9. Supplementary Methods
 SUPPLEMENTARY INFORMATION SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane Jian Huang 1,2#, Jie Yan 1,2#, Jian Zhang 3#, Shiguo Zhu 1, Yanli Wang
SUPPLEMENTARY INFORMATION SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane Jian Huang 1,2#, Jie Yan 1,2#, Jian Zhang 3#, Shiguo Zhu 1, Yanli Wang
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk
 Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 Relative expression of K IR2.1 transcript to enos was reduced 29-fold in capillaries from knockout animals. Relative expression of K IR2.1 transcript to enos was reduced 29-fold
Supplementary Figure 1 Relative expression of K IR2.1 transcript to enos was reduced 29-fold in capillaries from knockout animals. Relative expression of K IR2.1 transcript to enos was reduced 29-fold
Supplementary Figure 1 Validation of Per2 deletion in neuronal cells in N Per2 -/- mice. (a) Western blot from liver extracts of mice held under ad
 Supplementary Figure 1 Validation of Per2 deletion in neuronal cells in N Per2 -/- mice. (a) Western blot from liver extracts of mice held under ad libitum conditions detecting PER2 protein in brain and
Supplementary Figure 1 Validation of Per2 deletion in neuronal cells in N Per2 -/- mice. (a) Western blot from liver extracts of mice held under ad libitum conditions detecting PER2 protein in brain and
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 Increased ABHD5 expression in human colon cancer associated macrophages. (a) Murine peritoneal macrophages were treated with regular culture medium (Ctrl) or
Supplementary Figures Supplementary Figure 1 Increased ABHD5 expression in human colon cancer associated macrophages. (a) Murine peritoneal macrophages were treated with regular culture medium (Ctrl) or
Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein
 Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein content relative to GAPDH in two independent experiments.
Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein content relative to GAPDH in two independent experiments.
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 Characterization of stable expression of GlucB and sshbira in the CT26 cell line (a) Live cell imaging of stable CT26 cells expressing green fluorescent protein
Supplementary Figures Supplementary Figure 1 Characterization of stable expression of GlucB and sshbira in the CT26 cell line (a) Live cell imaging of stable CT26 cells expressing green fluorescent protein
Expanded View Figures
 PEX13 functions in selective autophagy Ming Y Lee et al Expanded View Figures Figure EV1. PEX13 is required for Sindbis virophagy. A, B Quantification of mcherry-capsid puncta per cell (A) and GFP-LC3
PEX13 functions in selective autophagy Ming Y Lee et al Expanded View Figures Figure EV1. PEX13 is required for Sindbis virophagy. A, B Quantification of mcherry-capsid puncta per cell (A) and GFP-LC3
293T cells were transfected with indicated expression vectors and the whole-cell extracts were subjected
 SUPPLEMENTARY INFORMATION Supplementary Figure 1. Formation of a complex between Slo1 and CRL4A CRBN E3 ligase. (a) HEK 293T cells were transfected with indicated expression vectors and the whole-cell
SUPPLEMENTARY INFORMATION Supplementary Figure 1. Formation of a complex between Slo1 and CRL4A CRBN E3 ligase. (a) HEK 293T cells were transfected with indicated expression vectors and the whole-cell
Supplementary Figure 1
 VO (ml kg - min - ) VCO (ml kg - min - ) Respiratory exchange ratio Energy expenditure (cal kg - min - ) Locomotor activity (x count) Body temperature ( C) Relative mrna expression TA Sol EDL PT Heart
VO (ml kg - min - ) VCO (ml kg - min - ) Respiratory exchange ratio Energy expenditure (cal kg - min - ) Locomotor activity (x count) Body temperature ( C) Relative mrna expression TA Sol EDL PT Heart
Supplemental Figures
 Supplemental Figures Supplemental Figure 1. Fasting-dependent regulation of the SREBP ortholog SBP-1 and lipid homeostasis mediated by the SIRT1 ortholog SIR-2.1 in C. elegans. (A) Wild-type or sir-2.1(lof)
Supplemental Figures Supplemental Figure 1. Fasting-dependent regulation of the SREBP ortholog SBP-1 and lipid homeostasis mediated by the SIRT1 ortholog SIR-2.1 in C. elegans. (A) Wild-type or sir-2.1(lof)
GPR120 *** * * Liver BAT iwat ewat mwat Ileum Colon. UCP1 mrna ***
 a GPR120 GPR120 mrna/ppia mrna Arbitrary Units 150 100 50 Liver BAT iwat ewat mwat Ileum Colon b UCP1 mrna Fold induction 20 15 10 5 - camp camp SB202190 - - - H89 - - - - - GW7647 Supplementary Figure
a GPR120 GPR120 mrna/ppia mrna Arbitrary Units 150 100 50 Liver BAT iwat ewat mwat Ileum Colon b UCP1 mrna Fold induction 20 15 10 5 - camp camp SB202190 - - - H89 - - - - - GW7647 Supplementary Figure
doi: /nature14508 Rappsilber et al.
 SUPPLEMENTARY INFORMATION doi:1.138/nature1458 Grosso et al. Barbosa et al. 74 72 45 33 47 7 51 Rappsilber et al. Supplementary Figure 1 a, Venn-Diagram of identified splice factors in the work of Barbossa
SUPPLEMENTARY INFORMATION doi:1.138/nature1458 Grosso et al. Barbosa et al. 74 72 45 33 47 7 51 Rappsilber et al. Supplementary Figure 1 a, Venn-Diagram of identified splice factors in the work of Barbossa
Tbk1-TKO! DN cells (%)! 15! 10!
 a! T Cells! TKO! B Cells! TKO! b! CD4! 8.9 85.2 3.4 2.88 CD8! Tbk1-TKO! 1.1 84.8 2.51 2.54 c! DN cells (%)! 4 3 2 1 DP cells (%)! 9 8 7 6 CD4 + SP cells (%)! 5 4 3 2 1 5 TKO! TKO! TKO! TKO! 15 1 5 CD8
a! T Cells! TKO! B Cells! TKO! b! CD4! 8.9 85.2 3.4 2.88 CD8! Tbk1-TKO! 1.1 84.8 2.51 2.54 c! DN cells (%)! 4 3 2 1 DP cells (%)! 9 8 7 6 CD4 + SP cells (%)! 5 4 3 2 1 5 TKO! TKO! TKO! TKO! 15 1 5 CD8
SUPPLEMENTARY FIGURES AND TABLE
 SUPPLEMENTARY FIGURES AND TABLE Supplementary Figure S1: Characterization of IRE1α mutants. A. U87-LUC cells were transduced with the lentiviral vector containing the GFP sequence (U87-LUC Tet-ON GFP).
SUPPLEMENTARY FIGURES AND TABLE Supplementary Figure S1: Characterization of IRE1α mutants. A. U87-LUC cells were transduced with the lentiviral vector containing the GFP sequence (U87-LUC Tet-ON GFP).
Supplementary Figure 1. mir124 does not change neuron morphology and synaptic
 Supplementary Figure 1. mir124 does not change neuron morphology and synaptic density. Hippocampal neurons were transfected with mir124 (containing DsRed) or DsRed as a control. 2 d after transfection,
Supplementary Figure 1. mir124 does not change neuron morphology and synaptic density. Hippocampal neurons were transfected with mir124 (containing DsRed) or DsRed as a control. 2 d after transfection,
Figure S1. Reduction in glomerular mir-146a levels correlate with progression to higher albuminuria in diabetic patients.
 Supplementary Materials Supplementary Figures Figure S1. Reduction in glomerular mir-146a levels correlate with progression to higher albuminuria in diabetic patients. Figure S2. Expression level of podocyte
Supplementary Materials Supplementary Figures Figure S1. Reduction in glomerular mir-146a levels correlate with progression to higher albuminuria in diabetic patients. Figure S2. Expression level of podocyte
Nature Genetics: doi: /ng Supplementary Figure 1. Parameters and consequences of mononuclear cardiomyocyte frequency.
 Supplementary Figure 1 Parameters and consequences of mononuclear cardiomyocyte frequency. (a) Correlation of the frequency of mononuclear cardiomyocytes to the frequency of cardiomyocytes with three or
Supplementary Figure 1 Parameters and consequences of mononuclear cardiomyocyte frequency. (a) Correlation of the frequency of mononuclear cardiomyocytes to the frequency of cardiomyocytes with three or
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1. Differential expression of mirnas from the pri-mir-17-92a locus.
 Supplementary Figure 1 Differential expression of mirnas from the pri-mir-17-92a locus. (a) The mir-17-92a expression unit in the third intron of the host mir-17hg transcript. (b,c) Impact of knockdown
Supplementary Figure 1 Differential expression of mirnas from the pri-mir-17-92a locus. (a) The mir-17-92a expression unit in the third intron of the host mir-17hg transcript. (b,c) Impact of knockdown
Supplemental Fig. 1. Relative mrna Expression. Relative mrna Expression WT KO WT KO RT 4 0 C
 Supplemental Fig. 1 A 1.5 1..5 Hdac11 (ibat) n=4 n=4 n=4 n=4 n=4 n=4 n=4 n=4 WT KO WT KO WT KO WT KO RT 4 C RT 4 C Supplemental Figure 1. Hdac11 mrna is undetectable in KO adipose tissue. Quantitative
Supplemental Fig. 1 A 1.5 1..5 Hdac11 (ibat) n=4 n=4 n=4 n=4 n=4 n=4 n=4 n=4 WT KO WT KO WT KO WT KO RT 4 C RT 4 C Supplemental Figure 1. Hdac11 mrna is undetectable in KO adipose tissue. Quantitative
Social deficits in Shank3-deficient mouse models of autism are rescued by histone deacetylase (HDAC) inhibition
 SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41593-018-0110-8 In the format provided by the authors and unedited. Social deficits in Shank3-deficient mouse models of autism are rescued by
SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41593-018-0110-8 In the format provided by the authors and unedited. Social deficits in Shank3-deficient mouse models of autism are rescued by
Supplementary Table 1. List of primers used in this study
 Supplementary Table 1. List of primers used in this study Gene Forward primer Reverse primer Rat Met 5 -aggtcgcttcatgcaggt-3 5 -tccggagacacaggatgg-3 Rat Runx1 5 -cctccttgaaccactccact-3 5 -ctggatctgcctggcatc-3
Supplementary Table 1. List of primers used in this study Gene Forward primer Reverse primer Rat Met 5 -aggtcgcttcatgcaggt-3 5 -tccggagacacaggatgg-3 Rat Runx1 5 -cctccttgaaccactccact-3 5 -ctggatctgcctggcatc-3
Supplementary Figure 1. Deletion of Smad3 prevents B16F10 melanoma invasion and metastasis in a mouse s.c. tumor model.
 A B16F1 s.c. Lung LN Distant lymph nodes Colon B B16F1 s.c. Supplementary Figure 1. Deletion of Smad3 prevents B16F1 melanoma invasion and metastasis in a mouse s.c. tumor model. Highly invasive growth
A B16F1 s.c. Lung LN Distant lymph nodes Colon B B16F1 s.c. Supplementary Figure 1. Deletion of Smad3 prevents B16F1 melanoma invasion and metastasis in a mouse s.c. tumor model. Highly invasive growth
supplementary information
 DOI: 1.138/ncb1 Control Atg7 / NAC 1 1 1 1 (mm) Control Atg7 / NAC 1 1 1 1 (mm) Lamin B Gstm1 Figure S1 Neither the translocation of into the nucleus nor the induction of antioxidant proteins in autophagydeficient
DOI: 1.138/ncb1 Control Atg7 / NAC 1 1 1 1 (mm) Control Atg7 / NAC 1 1 1 1 (mm) Lamin B Gstm1 Figure S1 Neither the translocation of into the nucleus nor the induction of antioxidant proteins in autophagydeficient
Supplementary material page 1/10
 Supplementary Figure 1. Metoprolol administration during ongoing AMI reduces MVO in STEMI patients (a, b) Complete representative CMR exams (short-axis covering the entire left ventricle (LV) from base
Supplementary Figure 1. Metoprolol administration during ongoing AMI reduces MVO in STEMI patients (a, b) Complete representative CMR exams (short-axis covering the entire left ventricle (LV) from base
J Jpn Coll Angiol, 2009, 49:
 Online publication October 6, 2009 48 2 20 J Jpn Coll Angiol, 2009, 49: 293 297 atrial natriuretic peptide, brain natriuretic peptide, guanylyl cyclase-a receptor, cardiac remodeling, cardiac hypertrophy
Online publication October 6, 2009 48 2 20 J Jpn Coll Angiol, 2009, 49: 293 297 atrial natriuretic peptide, brain natriuretic peptide, guanylyl cyclase-a receptor, cardiac remodeling, cardiac hypertrophy
Nature Immunology doi: /ni.3268
 Supplementary Figure 1 Loss of Mst1 and Mst2 increases susceptibility to bacterial sepsis. (a) H&E staining of colon and kidney sections from wild type and Mst1 -/- Mst2 fl/fl Vav-Cre mice. Scale bar,
Supplementary Figure 1 Loss of Mst1 and Mst2 increases susceptibility to bacterial sepsis. (a) H&E staining of colon and kidney sections from wild type and Mst1 -/- Mst2 fl/fl Vav-Cre mice. Scale bar,
Plasma exposure levels from individual mice 4 hours post IP administration at the
 Supplemental Figure Legends Figure S1. Plasma exposure levels of MKC-3946 in mice. Plasma exposure levels from individual mice 4 hours post IP administration at the indicated dose mg/kg. Data represent
Supplemental Figure Legends Figure S1. Plasma exposure levels of MKC-3946 in mice. Plasma exposure levels from individual mice 4 hours post IP administration at the indicated dose mg/kg. Data represent
TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer
 Supplementary Information TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer Yabing Mu, Reshma Sundar, Noopur Thakur, Maria Ekman, Shyam Kumar Gudey, Mariya
Supplementary Information TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer Yabing Mu, Reshma Sundar, Noopur Thakur, Maria Ekman, Shyam Kumar Gudey, Mariya
Nature Medicine: doi: /nm.4322
 1 2 3 4 5 6 7 8 9 10 11 Supplementary Figure 1. Predicted RNA structure of 3 UTR and sequence alignment of deleted nucleotides. (a) Predicted RNA secondary structure of ZIKV 3 UTR. The stem-loop structure
1 2 3 4 5 6 7 8 9 10 11 Supplementary Figure 1. Predicted RNA structure of 3 UTR and sequence alignment of deleted nucleotides. (a) Predicted RNA secondary structure of ZIKV 3 UTR. The stem-loop structure
Supplementary Figure 1
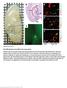 Supplementary Figure 1 AAV-GFP injection in the MEC of the mouse brain C57Bl/6 mice at 4 months of age were injected with AAV-GFP into the MEC and sacrificed at 7 days post injection (dpi). (a) Brains
Supplementary Figure 1 AAV-GFP injection in the MEC of the mouse brain C57Bl/6 mice at 4 months of age were injected with AAV-GFP into the MEC and sacrificed at 7 days post injection (dpi). (a) Brains
A Central Role of MG53 in Metabolic Syndrome. and Type-2 Diabetes
 A Central Role of MG53 in Metabolic Syndrome and Type-2 Diabetes Yan Zhang, Chunmei Cao, Rui-Ping Xiao Institute of Molecular Medicine (IMM) Peking University, Beijing, China Accelerated Aging in China
A Central Role of MG53 in Metabolic Syndrome and Type-2 Diabetes Yan Zhang, Chunmei Cao, Rui-Ping Xiao Institute of Molecular Medicine (IMM) Peking University, Beijing, China Accelerated Aging in China
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
