APOLs with low ph dependence can kill all African trypanosomes
|
|
|
- Francis Welch
- 6 years ago
- Views:
Transcription
1 SUPPLEMENTARY INFORMATION Letters DOI: 1.138/s In the format provided by the authors and unedited. APOLs with low ph dependence can kill all African trypanosomes Frédéric Fontaine 1, Laurence Lecordier 1, Gilles Vanwalleghem 1,5, Pierrick Uzureau 1,6, Nick Van Reet 2, Martina Fontaine 3, Patricia Tebabi 1, Benoit Vanhollebeke 1, Philippe Büscher 2, David Pérez-Morga 1,4 and Etienne Pays 1 * 1 Laboratory of Molecular Parasitology, IBMM, Université Libre de Bruxelles, 12, rue des Profs Jeener et Brachet, B-641 Gosselies, Belgium. 2 Unit of Parasite Diagnostics, Institute of Tropical Medicine, 155, Nationalestraat, B-2 Antwerpen, Belgium. 3 Laboratory of Immunobiology, IBMM, Université Libre de Bruxelles, 12, rue des Profs Jeener et Brachet, B-641 Gosselies, Belgium. 4 Center for Microscopy and Molecular Imaging (CMMI), Université Libre de Bruxelles, 12, rue des Profs Jeener et Brachet, B-641 Gosselies, Belgium. Present addresses: 5 School of Biomedical Sciences, The University of Queensland, St Lucia, QLD 472, Australia. 6 Laboratoire de Médecine Expérimentale (ULB222), Hôpital André Vésale, Université Libre de Bruxelles, 76, route de Gozée, B-611 Montigny le Tilleul, Belgium. Frédéric Fontaine and Laurence Lecordier contributed equally to this work. * epays@ulb.ac.be Nature Microbiology Macmillan Publishers Limited, part of Springer Nature. All rights reserved.
2 Parasite density (x1 4 ml -1 ) kda Acetic acid + rapol1 1 + rapol2 + rapol3 + rapol Time (h) Acetic acid + rapol1 + rapol2 + rapol3 + rapol rppapol1 rapol1 rapol2 rapol3 rapol6 Suppl. Fig. 1 Supplementary Figure 1. In vitro trypanolytic potential of various human rapols. The effect of different rapols (1 µg ml -1 rapol1, 3 µg ml -1 rapol3, 5 µg ml -1 rapol2 and rapol6) on T.b. brucei growth kinetics and 24 h survival was evaluated (error bars: s.e.m.; 3 technical replicates; n=1). The panel at the right shows the SDS-PAGE pattern of the rapols, as assessed by Coomassie blue staining.
3 APOL3 APOL1 PpAPOL1 APOL3 APOL1 PpAPOL1 APOL3 APOL1 PpAPOL1 APOL3 APOL1 PpAPOL1 MGLGQGWGWEASCFACLIRSCCQVVTFTFPFGFQGISQSLENVSGYYADARLEVGSTQLRTAGSCSHSFKRS MEGAALLRVSVLCIWMSAL-----FLGVGVRAEE-----AGARVQQNVPSGTDTGDPQSKPLGD MEGAALLRLFVLCIWTSGL-----FLGVGVRAEE-----AAARVQQNVPSGTDTGDPQ *.:.: : *. : * *:. *: * **::... :*.. Helix 2 Helix 3 FL------EKKRFTEEATKYFRERVSPVHLQILLTNNEAWKRFVTAAELPRDEADALYEALKKLRTYAAIED WAAGTMDPESSIFIEDAIKYFKEKVSTQNLLLLLTDNEAWNGFVAAAELPRNEADELRKALDNLARQMIMKD ESNVSLEDYLNYFQKNVSPQEMLLLLSDHKAWERVVATAELPRDEADELYKALNKLIRHMVMKD *.. *: :**.:.**. : :**::::**:.*::*****:*** * :**.:* ::* Helix 4 Helix 5+6 Helix 7+8 EYVQQKDEQFREWFLKEFPQVKRKIQESIEKLRALANGIEEVHRGCTISNVVSSSTGAASGIMSLAGLVLAP KNWHDKGQQYRNWFLKEFPRLKSELEDNIRRLRALADGVQKVHKGTTIANVVSGSLSISSGILTLVGMGLAP KNWLEEVQQHRKRFLEEFPRLERELQDKIRRLCDLAGQVQKVHKGATIANAFSSTLGVASGVLTFLGLGLAP : :: :*.*:.**:***.:: ::::.*.* **. :::**.* **:*..*.:. :**:::: *: *** BH3 Helix 9 FTAGTSLALTAAGVGLGAASAVTGITTSIVEHSYTSSAEAEASRLTATSIDRLKVFKEVMRDITPNLLSLLN FTEGGSLVLLEPGMELGITAALTGITSSTMDYGKKWWTQAQAHDLVIKSLDKLKEVREFLGENISNFLSLAG FTAGSSLVLLEPVTGLGITAALTGITSGSVEYAKKRWAQAEAHELVNKSLDTVEEMNEFLYHNIPNFISLRV ** * **.*. ** ::*:****:. :::.. ::*:* *..*:* ::..*.:.*::** APOL3 APOL1 PpAPOL1 APOL3 APOL1 PpAPOL1 NYYEATQTIGSEIRAIRQARAR ARLPVTTWRISAGSGG------QAERTIAGTTRAVSRGARIL NTYQLTRGIGKDIRALRRARANLQSVPHASASRPRVTEPISAESGE------QVERVNEPSILEMSRGVKLT NLVKFTEDTGKAIRAIRQARANPHSVSHVPASLHRVTEPVSATSVEERARVVEMERVAESRTTEVIRGAKIV * : * *. ***:*.***. *.* :** * : **. : **..: C-terminal helix SATTSGIFLALDVVNLVYESKHLHEGAKSASAEELRRQAQELEENLMELTQIYQRLNPCHTH DVAPVSFFLVLDVVYLVYESKHLHEGAKSETAEELKKVAQELEEKLNILNNNYKILQADQEL DKVFEGALFVLDVVGLVCQLKHLHEGAKSKTAEELKKVAQELEKKLNILNKKYETLRQEP--... ::.**** ** : ********* :****.. *****::* *.: *: *. Supplementary Figure 2. Sequence comparison between APOLs. Asterisks and dots respectively refer to identical and similar amino acids. The helices have been defined in 6. BH3 = Bcl2 homology domain 3.
4 a Tb gambiense b k - FMK + FMK rapol3 rat anti-apol3 TgsGP KO Tb gambiense n rapol1 25 µg ml -1 rapol1 5 µg ml -1 rapol3 25 µg ml -1 Supplementary Figure 3. Uptake of rapol3 in trypanosomes. (a) Immunodetection of rapol3 in T.b. gambiense LiTat 1.3 parasites incubated or not in the presence of the cathepsin inhibitor FMK-24. (scale bar=5 µm; n=nucleus and k=kinetoplast, both stained with DAPI). (b) Effect of temperature on trypanolysis of T.b. gambiense TgsGP KO LiTat 1.3 parasites by rapol1 or rapol3. Results are expressed as % control growth at each temperature (error bars: s.e.m.; 3 technical replicates; n=2 independent experiments).
5 Supplementary Figure 4. Doxycycline-mediated induction of TbKIFC1 RNAi. Western blot of induced (+Dox) and non-induced (-Dox) parasite extracts. See 7 for details.
6 Nucleus Mitochondrion + rapol3 M FP K FP M K M K FP K M M Scale = 5 nm Supplementary Figure 5. Mitochondrial and nuclear phenotype of rapol3-mediated trypanolysis. TEM micrographs of T.b. brucei after 2 h incubation with 3 µg ml -1 rapol3 (FP; flagellar pocket; K=kinetoplast; M=mitochondrion; N=nucleus; arrows: heterochromatin patches).
7 rmut4 probability rmut3 probability rapol3 WT probability TMHMM ph 5.5 ph 7.5 T. b. brucei T. b. gambiense transmembrane inside outside % Glc + 75 M IPTG mm NH 4 Cl mm NH4 Cl RGCTISNVVSSSTGAASGIMSLAGLVLAPFTAGTSLALTAAGVGLGAASAVTGITTSIVEHSYTSSA rapol3 (µg ml -1 ) rapol3 (µg ml -1 ) % Glc + 75 M IPTG mm NH4 Cl + 2 mm NH4 Cl RGCTISNVVSSSTGAASGIMSLAGLVLAPFTEGTSLALTAAGVGLGAASAVTGITTSIVEHSYTSSA rmut3 (µg ml -1 ) rmut3 (µg ml -1 ) % Glc + 75 M IPTG mm NH4 1 Cl + 2 mm NH4 Cl RGCTISNVVSSSTGAASGIMSLAGLVLAPFTAGTSLALTEAGVELGAASAVTGITTSIVEHSYTSSA rmut4 (µg ml -1 ) rmut4 (µg ml -1 ) Supplementary Figure 6. Relative activity of glutamic acid mutants in the helix 9 region of rapol3. In each row, the left panels show the predicted patterns of transmembrane spans (TMHMM program, Expasy) in the APOL sequences defined underneath (numbers refer to key helices in the pore-forming domain 6 ), followed by measurements of the colicin-like activity of the different rapols in E. coli as determined by scoring the plating efficiency after overnight incubation at 37 C, comparing induction of protein expression following IPTG addition with control following glucose (Glc) addition 16, and comparing the effect of ph. The right panels show the trypanolytic potential of the different rapols, as determined on T.b. brucei and T.b. gambiense LiTat 1.3 after 24 h incubation in vitro with or without 2 mm NH 4 Cl (error bars: s.e.m.; 3 technical replicates; n=3 independent experiments).
8 Supplementary Figure 7. Trypanolytic activity of 1 µg ml -1 rapol1/apol3 chimaeras (a) or 1 µg ml -1 rapol1 mutants (b) on T.b. brucei and T.b. gambiense LiTat 1.3 after 24 h incubation in vitro. The restriction endonuclease sites used to generate the constructs are indicated (italics: generated by site-directed mutagenesis without changing the amino acid code; underlined: inserted in the PCR primers used to generate the gene fragments). Panel (c) illustrates the activity measurements reported as positive in table a. C=chimaeras; M=mutants. (error bars: s.e.m.; 3 technical replicates; n=3 independent experiments).
9 Intensity value (A.U) Intensity value (A.U) kda Non injected h 24h 48h rapol1 (1µg) 72h 96h kda Non injected h 2h 6h rapol3 (1µg) 24h R 2 = R 2 = Time (h) Time (h) rapol1 half-life = ~5h rapol3 half-life = ~13h Supplementary Figure 8. Measurement of the rapol1 and rapol3 decay in mouse blood. Western blot detection in the blood of 6 weeks-old female BALB/c mice, of different rapols (1 µg) intravenously injected at t=. We used ImageJ to quantify the signal strength from Fig. 4a western blots images (middle panel). Using these values, we applied a linear regression to evaluate the recombinant proteins half-life in mice blood.
10 rmut-null probability rapol3 WT probability TMHMM ph 5.5 ph 7.5 T. b. brucei T. b. gambiense transmembrane inside outside % Glc + 75 M IPTG mm NH 4 Cl mm NH 4 Cl RGCTISNVVSSSTGAASGIMSLAGLVLAPFTAGTSLALTAAGVGLGAASAVTGITTSIVEHSYTSSA RGCTISNVVSSSTGAASGIMSLAGLVQSSFTASTSLALTAAGVGLGAASAVTGITTSIVEHSYTSSA % Glc + 75 M IPTG mm NH 4 Cl rapol3 (µg ml -1 ) rapol3 (µg ml -1 ) rmut-null (µg ml -1 ) rmut-null (µg ml -1 ) mm NH 4 Cl Supplementary Figure 9. Lack of trypanolytic activity of rapol3 137 QQSFTAS 143 (rmut-null). In each row, the left panels show the predicted patterns of transmembrane spans (TMHMM program, Expasy) in the APOL sequences defined underneath (numbers refer to key helices in the pore-forming domain 6 ), followed by measurements of the colicinlike activity of the different rapols in E. coli as determined by scoring the plating efficiency after overnight incubation at 37 C, comparing induction of protein expression following IPTG addition with control following glucose (Glc) addition 16, and comparing the effect of ph. The right panels show the trypanolytic potential of the different rapols, as determined on T.b. brucei and T.b. gambiense LiTat 1.3 after 24 h incubation in vitro with or without 2 mm NH 4 Cl (error bars: s.e.m.; 3 technical replicates; n=3 independent experiments).
11 Supplementary Figure 1. Trypanolysis differences between different T.b. gambiense populations: relative adaptation to in vitro growth conditions, influence of the type of VSG and TgsGP level. (a) Trypanolysis and growth curves of different cloned T.b. gambiense populations: in vitro adapted LiTat 1.3 clone from the ELIANE strain, LiTat 1.3 parasites freshly isolated from blood and bloodstream parasites from the MBA strain expressing the LiTat 1.3 isotype AnTat 11.8, or bloodstream parasites from the ELIANE strain expressing LiTat 1.2 (error bars: s.e.m.; 3 technical replicates; n=3 independent experiments). (b) Western blots of protein extracts from the different T.b. gambiense populations, incubated with anti-litat 1.3 VSG antibodies. (c) Expression levels of TgsGP in the different T.b. gambiense populations and in T.b. brucei negative control parasites, as measured by qrt- PCR.
12 Supplementary Figure 11. Selective precipitation of rapol3 in the trypanosome incubation medium. rapol1 and rapol3 (1 µg ml -1 each) were incubated for different periods at 37 C in the incubation medium, then the centrifugation supernatant was analyzed by Western blotting with anti-apol antibodies. =no protein.
13 From Fig. 4a From Suppl. Fig. 4 Supplementary Figure 12. Raw version of Western blots.
14 Supplementary Table Effect of intravenous injection of different rapols on mice health (3 mice per group; 1=no effect; 2=mobility reduced; 3=mobility reduced + fur bristling; 4=death).
Antigenic variation as adaptive process: the case of Trypanosoma brucei
 Antigenic variation as adaptive process: the case of Trypanosoma brucei African trypanosomes infect a wide spectrum of mammalian hosts, including humans Mechanisms of adaptation: I. Antigenic variation
Antigenic variation as adaptive process: the case of Trypanosoma brucei African trypanosomes infect a wide spectrum of mammalian hosts, including humans Mechanisms of adaptation: I. Antigenic variation
T rypanosoma evansi is a widely distributed hemoflagellate parasite
 The new england journal of medicine brief report Human Trypanosoma evansi Infection Linked to a Lack of Apolipoprotein L-I Benoit Vanhollebeke, Eng., Philippe Truc, Ph.D., Philippe Poelvoorde, M.Sc., Annette
The new england journal of medicine brief report Human Trypanosoma evansi Infection Linked to a Lack of Apolipoprotein L-I Benoit Vanhollebeke, Eng., Philippe Truc, Ph.D., Philippe Poelvoorde, M.Sc., Annette
Relative SOD1 activity. Relative SOD2 activity. Relative SOD activity (Infected:Mock) + CP + DDC
 Supplementary Figure 1. SOD1 activity is significantly increased relative to SOD1 levels. SOD1 and SOD2 activities in the infected mork13 cells are shown normalised to their corresponding levels and relative
Supplementary Figure 1. SOD1 activity is significantly increased relative to SOD1 levels. SOD1 and SOD2 activities in the infected mork13 cells are shown normalised to their corresponding levels and relative
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1. Generation and validation of mtef4-knockout mice.
 Supplementary Figure 1 Generation and validation of mtef4-knockout mice. (a) Alignment of EF4 (E. coli) with mouse, yeast and human EF4. (b) Domain structures of mouse mtef4 compared to those of EF4 (E.
Supplementary Figure 1 Generation and validation of mtef4-knockout mice. (a) Alignment of EF4 (E. coli) with mouse, yeast and human EF4. (b) Domain structures of mouse mtef4 compared to those of EF4 (E.
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk
 Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
Figure S1. PMVs from THP-1 cells expose phosphatidylserine and carry actin. A) Flow
 SUPPLEMENTARY DATA Supplementary Figure Legends Figure S1. PMVs from THP-1 cells expose phosphatidylserine and carry actin. A) Flow cytometry analysis of PMVs labelled with annexin-v-pe (Guava technologies)
SUPPLEMENTARY DATA Supplementary Figure Legends Figure S1. PMVs from THP-1 cells expose phosphatidylserine and carry actin. A) Flow cytometry analysis of PMVs labelled with annexin-v-pe (Guava technologies)
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells
 Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3461 In the format provided by the authors and unedited. Supplementary Figure 1 (associated to Figure 1). Cpeb4 gene-targeted mice develop liver steatosis. a, Immunoblot displaying CPEB4
DOI: 10.1038/ncb3461 In the format provided by the authors and unedited. Supplementary Figure 1 (associated to Figure 1). Cpeb4 gene-targeted mice develop liver steatosis. a, Immunoblot displaying CPEB4
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 Characterization of stable expression of GlucB and sshbira in the CT26 cell line (a) Live cell imaging of stable CT26 cells expressing green fluorescent protein
Supplementary Figures Supplementary Figure 1 Characterization of stable expression of GlucB and sshbira in the CT26 cell line (a) Live cell imaging of stable CT26 cells expressing green fluorescent protein
Nature Medicine: doi: /nm.4322
 1 2 3 4 5 6 7 8 9 10 11 Supplementary Figure 1. Predicted RNA structure of 3 UTR and sequence alignment of deleted nucleotides. (a) Predicted RNA secondary structure of ZIKV 3 UTR. The stem-loop structure
1 2 3 4 5 6 7 8 9 10 11 Supplementary Figure 1. Predicted RNA structure of 3 UTR and sequence alignment of deleted nucleotides. (a) Predicted RNA secondary structure of ZIKV 3 UTR. The stem-loop structure
SD-1 SD-1: Cathepsin B levels in TNF treated hch
 SD-1 SD-1: Cathepsin B levels in TNF treated hch. A. RNA and B. protein extracts from TNF treated and untreated human chondrocytes (hch) were analyzed via qpcr (left) and immunoblot analyses (right) for
SD-1 SD-1: Cathepsin B levels in TNF treated hch. A. RNA and B. protein extracts from TNF treated and untreated human chondrocytes (hch) were analyzed via qpcr (left) and immunoblot analyses (right) for
SUPPLEMENTARY INFORMATION
 Figure S1 Induction of non-apoptotic death of SV40-transformed and primary DKO MEFs, and DKO thymocytes. (A-F) STS-induced non-apoptotic death of DKO MEF. (A, B) Reduced viability of DKO MEFs after exposure
Figure S1 Induction of non-apoptotic death of SV40-transformed and primary DKO MEFs, and DKO thymocytes. (A-F) STS-induced non-apoptotic death of DKO MEF. (A, B) Reduced viability of DKO MEFs after exposure
Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein
 Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein content relative to GAPDH in two independent experiments.
Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein content relative to GAPDH in two independent experiments.
a b G75 G60 Sw-2 Sw-1 Supplementary Figure 1. Structure predictions by I-TASSER Server.
 a b G75 2 2 G60 Sw-2 Sw-1 Supplementary Figure 1. Structure predictions by I-TASSER Server. a. Overlay of top 10 models generated by I-TASSER illustrates the potential effect of 7 amino acid insertion
a b G75 2 2 G60 Sw-2 Sw-1 Supplementary Figure 1. Structure predictions by I-TASSER Server. a. Overlay of top 10 models generated by I-TASSER illustrates the potential effect of 7 amino acid insertion
Figure S1. (A) SDS-PAGE separation of GST-fusion proteins purified from E.coli BL21 strain is shown. An equal amount of GST-tag control, LRRK2 LRR
 Figure S1. (A) SDS-PAGE separation of GST-fusion proteins purified from E.coli BL21 strain is shown. An equal amount of GST-tag control, LRRK2 LRR and LRRK2 WD40 GST fusion proteins (5 µg) were loaded
Figure S1. (A) SDS-PAGE separation of GST-fusion proteins purified from E.coli BL21 strain is shown. An equal amount of GST-tag control, LRRK2 LRR and LRRK2 WD40 GST fusion proteins (5 µg) were loaded
Prolonged mitotic arrest induces a caspase-dependent DNA damage
 SUPPLEMENTARY INFORMATION Prolonged mitotic arrest induces a caspase-dependent DNA damage response at telomeres that determines cell survival Karolina O. Hain, Didier J. Colin, Shubhra Rastogi, Lindsey
SUPPLEMENTARY INFORMATION Prolonged mitotic arrest induces a caspase-dependent DNA damage response at telomeres that determines cell survival Karolina O. Hain, Didier J. Colin, Shubhra Rastogi, Lindsey
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature11429 S1a 6 7 8 9 Nlrc4 allele S1b Nlrc4 +/+ Nlrc4 +/F Nlrc4 F/F 9 Targeting construct 422 bp 273 bp FRT-neo-gb-PGK-FRT 3x.STOP S1c Nlrc4 +/+ Nlrc4 F/F casp1
SUPPLEMENTARY INFORMATION doi:10.1038/nature11429 S1a 6 7 8 9 Nlrc4 allele S1b Nlrc4 +/+ Nlrc4 +/F Nlrc4 F/F 9 Targeting construct 422 bp 273 bp FRT-neo-gb-PGK-FRT 3x.STOP S1c Nlrc4 +/+ Nlrc4 F/F casp1
Suppl Video: Tumor cells (green) and monocytes (white) are seeded on a confluent endothelial
 Supplementary Information Häuselmann et al. Monocyte induction of E-selectin-mediated endothelial activation releases VE-cadherin junctions to promote tumor cell extravasation in the metastasis cascade
Supplementary Information Häuselmann et al. Monocyte induction of E-selectin-mediated endothelial activation releases VE-cadherin junctions to promote tumor cell extravasation in the metastasis cascade
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
Lack of cadherins Celsr2 and Celsr3 impairs ependymal ciliogenesis, leading to fatal
 Lack of cadherins Celsr2 and Celsr3 impairs ependymal ciliogenesis, leading to fatal hydrocephalus Fadel TISSIR, Yibo QU, Mireille MONTCOUQUIOL, Libing ZHOU, Kouji KOMATSU, Dongbo SHI, Toshihiko FUJIMORI,
Lack of cadherins Celsr2 and Celsr3 impairs ependymal ciliogenesis, leading to fatal hydrocephalus Fadel TISSIR, Yibo QU, Mireille MONTCOUQUIOL, Libing ZHOU, Kouji KOMATSU, Dongbo SHI, Toshihiko FUJIMORI,
ERK1/2/MAPK pathway-dependent regulation of the telomeric factor TRF2
 ERK1/2/MAPK pathway-dependent regulation of the telomeric factor TRF2 SUPPLEMENTARY FIGURES AND TABLE Supplementary Figure S1: Conservation of the D domain throughout evolution. Alignment of TRF2 sequences
ERK1/2/MAPK pathway-dependent regulation of the telomeric factor TRF2 SUPPLEMENTARY FIGURES AND TABLE Supplementary Figure S1: Conservation of the D domain throughout evolution. Alignment of TRF2 sequences
Supplementary Figure 1 Expression of Crb3 in mouse sciatic nerve: biochemical analysis (a) Schematic of Crb3 isoforms, ERLI and CLPI, indicating the
 Supplementary Figure 1 Expression of Crb3 in mouse sciatic nerve: biochemical analysis (a) Schematic of Crb3 isoforms, ERLI and CLPI, indicating the location of the transmembrane (TM), FRM binding (FB)
Supplementary Figure 1 Expression of Crb3 in mouse sciatic nerve: biochemical analysis (a) Schematic of Crb3 isoforms, ERLI and CLPI, indicating the location of the transmembrane (TM), FRM binding (FB)
Supplementary Information. Tissue-wide immunity against Leishmania. through collective production of nitric oxide
 Supplementary Information Tissue-wide immunity against Leishmania through collective production of nitric oxide Romain Olekhnovitch, Bernhard Ryffel, Andreas J. Müller and Philippe Bousso Supplementary
Supplementary Information Tissue-wide immunity against Leishmania through collective production of nitric oxide Romain Olekhnovitch, Bernhard Ryffel, Andreas J. Müller and Philippe Bousso Supplementary
Suppl. Figure 1. T 3 induces autophagic flux in hepatic cells. (A) RFP-GFP-LC3 transfected HepG2/TRα cells were visualized and cells were quantified
 Suppl. Figure 1. T 3 induces autophagic flux in hepatic cells. (A) RFP-GFP-LC3 transfected HepG2/TRα cells were visualized and cells were quantified for RFP-LC3 puncta (red dots) representing both autolysosomes
Suppl. Figure 1. T 3 induces autophagic flux in hepatic cells. (A) RFP-GFP-LC3 transfected HepG2/TRα cells were visualized and cells were quantified for RFP-LC3 puncta (red dots) representing both autolysosomes
a surface permeabilized
 a surface permeabilized RAW 64.7 P388D1 J774 b CD11b + Ly-6G - Blood Monocytes WT Supplementary Figure 1. Cell surface expression on macrophages and DCs. (a) RAW64.7, P388D1, and J774 cells were subjected
a surface permeabilized RAW 64.7 P388D1 J774 b CD11b + Ly-6G - Blood Monocytes WT Supplementary Figure 1. Cell surface expression on macrophages and DCs. (a) RAW64.7, P388D1, and J774 cells were subjected
(a) Significant biological processes (upper panel) and disease biomarkers (lower panel)
 Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid.
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
Supplementary Table 1. Metabolic parameters in GFP and OGT-treated mice
 Supplementary Table 1. Metabolic parameters in GFP and OGT-treated mice Fasted Refed GFP OGT GFP OGT Liver G6P (mmol/g) 0.03±0.01 0.04±0.02 0.60±0.04 0.42±0.10 A TGs (mg/g of liver) 20.08±5.17 16.29±0.8
Supplementary Table 1. Metabolic parameters in GFP and OGT-treated mice Fasted Refed GFP OGT GFP OGT Liver G6P (mmol/g) 0.03±0.01 0.04±0.02 0.60±0.04 0.42±0.10 A TGs (mg/g of liver) 20.08±5.17 16.29±0.8
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Supplementary Figure 1. (A) Left, western blot analysis of ISGylated proteins in Jurkat T cells treated with 1000U ml -1 IFN for 16h (IFN) or left untreated (CONT); right, western
SUPPLEMENTARY FIGURES Supplementary Figure 1. (A) Left, western blot analysis of ISGylated proteins in Jurkat T cells treated with 1000U ml -1 IFN for 16h (IFN) or left untreated (CONT); right, western
Cellular and Molecular Remodeling of the Endocytic Pathway during Differentiation of Trypanosoma brucei Bloodstream Forms
 EUKARYOTIC CELL, Aug. 2010, p. 1272 1282 Vol. 9, No. 8 1535-9778/10/$12.00 doi:10.1128/ec.00076-10 Copyright 2010, American Society for Microbiology. All Rights Reserved. Cellular and Molecular Remodeling
EUKARYOTIC CELL, Aug. 2010, p. 1272 1282 Vol. 9, No. 8 1535-9778/10/$12.00 doi:10.1128/ec.00076-10 Copyright 2010, American Society for Microbiology. All Rights Reserved. Cellular and Molecular Remodeling
Supplementary Information Titles Journal: Nature Medicine
 Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
SUPPLEMENTARY INFORMATION. Supplementary Figures S1-S9. Supplementary Methods
 SUPPLEMENTARY INFORMATION SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane Jian Huang 1,2#, Jie Yan 1,2#, Jian Zhang 3#, Shiguo Zhu 1, Yanli Wang
SUPPLEMENTARY INFORMATION SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane Jian Huang 1,2#, Jie Yan 1,2#, Jian Zhang 3#, Shiguo Zhu 1, Yanli Wang
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Krenn et al., http://www.jcb.org/cgi/content/full/jcb.201110013/dc1 Figure S1. Levels of expressed proteins and demonstration that C-terminal
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Krenn et al., http://www.jcb.org/cgi/content/full/jcb.201110013/dc1 Figure S1. Levels of expressed proteins and demonstration that C-terminal
TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer
 Supplementary Information TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer Yabing Mu, Reshma Sundar, Noopur Thakur, Maria Ekman, Shyam Kumar Gudey, Mariya
Supplementary Information TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer Yabing Mu, Reshma Sundar, Noopur Thakur, Maria Ekman, Shyam Kumar Gudey, Mariya
Supplemental Figure 1. Western blot analysis indicated that MIF was detected in the fractions of
 Supplemental Figure Legends Supplemental Figure 1. Western blot analysis indicated that was detected in the fractions of plasma membrane and cytosol but not in nuclear fraction isolated from Pkd1 null
Supplemental Figure Legends Supplemental Figure 1. Western blot analysis indicated that was detected in the fractions of plasma membrane and cytosol but not in nuclear fraction isolated from Pkd1 null
Supplemental Figure 1. (A) The localization of Cre DNA recombinase in the testis of Cyp19a1-Cre mice was detected by immunohistchemical analyses
 Supplemental Figure 1. (A) The localization of Cre DNA recombinase in the testis of Cyp19a1-Cre mice was detected by immunohistchemical analyses using an anti-cre antibody; testes at 1 week (left panel),
Supplemental Figure 1. (A) The localization of Cre DNA recombinase in the testis of Cyp19a1-Cre mice was detected by immunohistchemical analyses using an anti-cre antibody; testes at 1 week (left panel),
Supplementary Figure 1. Ent inhibits LPO activity in a dose- and time-dependent fashion.
 Supplementary Information Supplementary Figure 1. Ent inhibits LPO activity in a dose- and time-dependent fashion. Various concentrations of Ent, DHBA or ABAH were pre-incubated for 10 min with LPO (50
Supplementary Information Supplementary Figure 1. Ent inhibits LPO activity in a dose- and time-dependent fashion. Various concentrations of Ent, DHBA or ABAH were pre-incubated for 10 min with LPO (50
Supplementary Fig. 1 eif6 +/- mice show a reduction in white adipose tissue, blood lipids and normal glycogen synthesis. The cohort of the original
 Supplementary Fig. 1 eif6 +/- mice show a reduction in white adipose tissue, blood lipids and normal glycogen synthesis. The cohort of the original phenotypic screening was n=40. For specific tests, the
Supplementary Fig. 1 eif6 +/- mice show a reduction in white adipose tissue, blood lipids and normal glycogen synthesis. The cohort of the original phenotypic screening was n=40. For specific tests, the
Mutual self-defence: the trypanolytic factor story
 Microbes and Infection 10 (2008) 985e989 www.elsevier.com/locate/micinf Original article Mutual self-defence: the trypanolytic factor story Etienne Pays*, Benoit Vanhollebeke Laboratory of Molecular Parasitology,
Microbes and Infection 10 (2008) 985e989 www.elsevier.com/locate/micinf Original article Mutual self-defence: the trypanolytic factor story Etienne Pays*, Benoit Vanhollebeke Laboratory of Molecular Parasitology,
GPR120 *** * * Liver BAT iwat ewat mwat Ileum Colon. UCP1 mrna ***
 a GPR120 GPR120 mrna/ppia mrna Arbitrary Units 150 100 50 Liver BAT iwat ewat mwat Ileum Colon b UCP1 mrna Fold induction 20 15 10 5 - camp camp SB202190 - - - H89 - - - - - GW7647 Supplementary Figure
a GPR120 GPR120 mrna/ppia mrna Arbitrary Units 150 100 50 Liver BAT iwat ewat mwat Ileum Colon b UCP1 mrna Fold induction 20 15 10 5 - camp camp SB202190 - - - H89 - - - - - GW7647 Supplementary Figure
Supplemental information
 Supplemental information PI(3)K p11δ controls the sucellular compartmentalization of TLR4 signaling and protects from endotoxic shock Ezra Aksoy, Salma Taoui, David Torres, Sandrine Delauve, Aderrahman
Supplemental information PI(3)K p11δ controls the sucellular compartmentalization of TLR4 signaling and protects from endotoxic shock Ezra Aksoy, Salma Taoui, David Torres, Sandrine Delauve, Aderrahman
Supplementary Materials for
 www.sciencetranslationalmedicine.org/cgi/content/full/4/117/117ra8/dc1 Supplementary Materials for Notch4 Normalization Reduces Blood Vessel Size in Arteriovenous Malformations Patrick A. Murphy, Tyson
www.sciencetranslationalmedicine.org/cgi/content/full/4/117/117ra8/dc1 Supplementary Materials for Notch4 Normalization Reduces Blood Vessel Size in Arteriovenous Malformations Patrick A. Murphy, Tyson
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2211 a! mir-143! b! mir-103/107! let-7a! mir-144! mir-122a! mir-126-3p! mir-194! mir-27a! mir-30c! Figure S1 Northern blot analysis of mir-143 expression dependent on feeding conditions.
DOI: 10.1038/ncb2211 a! mir-143! b! mir-103/107! let-7a! mir-144! mir-122a! mir-126-3p! mir-194! mir-27a! mir-30c! Figure S1 Northern blot analysis of mir-143 expression dependent on feeding conditions.
African Americans suffer from kidney failure
 Association of Trypanolytic ApoL Variants with Kidney Disease in African Americans Giulio Genovese,, * David J. Friedman,, * Michael D. Ross, Laurence Lecordier, Pierrick Uzureau, Barry I. Freedman, Donald
Association of Trypanolytic ApoL Variants with Kidney Disease in African Americans Giulio Genovese,, * David J. Friedman,, * Michael D. Ross, Laurence Lecordier, Pierrick Uzureau, Barry I. Freedman, Donald
Figure S1. Sorting nexin 9 (SNX9) specifically binds psmad3 and not psmad 1/5/8. Lysates from AKR-2B cells untreated (-) or stimulated (+) for 45 min
 Figure S1. Sorting nexin 9 (SNX9) specifically binds psmad3 and not psmad 1/5/8. Lysates from AKR2B cells untreated () or stimulated () for 45 min with 5 ng/ml TGFβ or 10 ng/ml BMP4 were incubated with
Figure S1. Sorting nexin 9 (SNX9) specifically binds psmad3 and not psmad 1/5/8. Lysates from AKR2B cells untreated () or stimulated () for 45 min with 5 ng/ml TGFβ or 10 ng/ml BMP4 were incubated with
Supplemental Table 1. Primers used for RT-PCR analysis of inflammatory cytokines Gene Primer Sequence
 Supplemental Table 1. Primers used for RT-PCR analysis of inflammatory cytokines Gene Primer Sequence IL-1α Forward primer 5 -CAAGATGGCCAAAGTTCGTGAC-3' Reverse primer 5 -GTCTCATGAAGTGAGCCATAGC-3 IL-1β
Supplemental Table 1. Primers used for RT-PCR analysis of inflammatory cytokines Gene Primer Sequence IL-1α Forward primer 5 -CAAGATGGCCAAAGTTCGTGAC-3' Reverse primer 5 -GTCTCATGAAGTGAGCCATAGC-3 IL-1β
Supplemental information Acid-sensing ion channel 1a contributes to hippocampal LTP inducibility through multiple mechanisms
 Supplemental information Acid-sensing ion channel 1a contributes to hippocampal LTP inducibility through multiple mechanisms Ming-Gang Liu, Hu-Song Li, Wei-Guang Li, Yan-Jiao Wu, Shi-Ning Deng, Chen Huang,
Supplemental information Acid-sensing ion channel 1a contributes to hippocampal LTP inducibility through multiple mechanisms Ming-Gang Liu, Hu-Song Li, Wei-Guang Li, Yan-Jiao Wu, Shi-Ning Deng, Chen Huang,
Supplementary Figure 1 Chemokine and chemokine receptor expression during muscle regeneration (a) Analysis of CR3CR1 mrna expression by real time-pcr
 Supplementary Figure 1 Chemokine and chemokine receptor expression during muscle regeneration (a) Analysis of CR3CR1 mrna expression by real time-pcr at day 0, 1, 4, 10 and 21 post- muscle injury. (b)
Supplementary Figure 1 Chemokine and chemokine receptor expression during muscle regeneration (a) Analysis of CR3CR1 mrna expression by real time-pcr at day 0, 1, 4, 10 and 21 post- muscle injury. (b)
The development of new diagnostic tools for sleeping sickness
 The development of new diagnostic tools for sleeping sickness P. Büscher Institute of Tropical Medicine, Antwerp WHO CC for Research and Training on Human African Trypanosomiasis Diagnosis 1 1 Distribution
The development of new diagnostic tools for sleeping sickness P. Büscher Institute of Tropical Medicine, Antwerp WHO CC for Research and Training on Human African Trypanosomiasis Diagnosis 1 1 Distribution
Supplementary Figure 1.
 Supplementary Figure 1. Increased β cell mass and islet diameter in βtsc2 -/- mice up to 35 weeks A: Reconstruction of multiple anti-insulin immunofluorescence images showing differences in β cell mass
Supplementary Figure 1. Increased β cell mass and islet diameter in βtsc2 -/- mice up to 35 weeks A: Reconstruction of multiple anti-insulin immunofluorescence images showing differences in β cell mass
SUPPLEMENTARY FIGURES AND TABLE
 SUPPLEMENTARY FIGURES AND TABLE Supplementary Figure S1: Characterization of IRE1α mutants. A. U87-LUC cells were transduced with the lentiviral vector containing the GFP sequence (U87-LUC Tet-ON GFP).
SUPPLEMENTARY FIGURES AND TABLE Supplementary Figure S1: Characterization of IRE1α mutants. A. U87-LUC cells were transduced with the lentiviral vector containing the GFP sequence (U87-LUC Tet-ON GFP).
Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS
 Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS nucleotide sequences (a, b) or amino acid sequences (c) from
Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS nucleotide sequences (a, b) or amino acid sequences (c) from
Luminescent Multiplex Viability Assay for T.b. gambiense
 Luminescent Multiplex Viability Assay for T.b. gambiense Van Reet N., Pyana P., Rogé S., Claes F. and Büscher, P. Institute of Tropical Medicine Antwerp 32nd ISCTRC conference 8th 12th September 2013 Introduction
Luminescent Multiplex Viability Assay for T.b. gambiense Van Reet N., Pyana P., Rogé S., Claes F. and Büscher, P. Institute of Tropical Medicine Antwerp 32nd ISCTRC conference 8th 12th September 2013 Introduction
Nature Immunology: doi: /ni eee Supplementary Figure 1
 eee Supplementary Figure 1 Hyphae induce NET release, but yeast do not. (a) NET release by human peripheral neutrophils stimulated with a hgc1 yeast-locked C. albicans mutant (yeast) or pre-formed WT C.
eee Supplementary Figure 1 Hyphae induce NET release, but yeast do not. (a) NET release by human peripheral neutrophils stimulated with a hgc1 yeast-locked C. albicans mutant (yeast) or pre-formed WT C.
Supplementary figure legends
 Supplementary figure legends Supplementary Figure 1. Exposure of CRT occurs independently from the apoptosisassociated loss of the mitochondrial membrane potential (MMP). (A) HeLa cells treated with MTX
Supplementary figure legends Supplementary Figure 1. Exposure of CRT occurs independently from the apoptosisassociated loss of the mitochondrial membrane potential (MMP). (A) HeLa cells treated with MTX
p = formed with HCI-001 p = Relative # of blood vessels that formed with HCI-002 Control Bevacizumab + 17AAG Bevacizumab 17AAG
 A.. Relative # of ECs associated with HCI-001 1.4 1.2 1.0 0.8 0.6 0.4 0.2 0.0 ol b p < 0.001 Relative # of blood vessels that formed with HCI-001 1.4 1.2 1.0 0.8 0.6 0.4 0.2 0.0 l b p = 0.002 Control IHC:
A.. Relative # of ECs associated with HCI-001 1.4 1.2 1.0 0.8 0.6 0.4 0.2 0.0 ol b p < 0.001 Relative # of blood vessels that formed with HCI-001 1.4 1.2 1.0 0.8 0.6 0.4 0.2 0.0 l b p = 0.002 Control IHC:
F-actin VWF Vinculin. F-actin. Vinculin VWF
 a F-actin VWF Vinculin b F-actin VWF Vinculin Supplementary Fig. 1. WPBs in HUVECs are located along stress fibers and at focal adhesions. (a) Immunofluorescence images of f-actin (cyan), VWF (yellow),
a F-actin VWF Vinculin b F-actin VWF Vinculin Supplementary Fig. 1. WPBs in HUVECs are located along stress fibers and at focal adhesions. (a) Immunofluorescence images of f-actin (cyan), VWF (yellow),
B. SDS-PAGE. Western blot MW (kda) Supplementary Information. Supplementary Figure 1. MW (kda) Glutelin. Prolamin. 3 E.coli. 3 E.
 Supplementary Information Supplementary Figure 1. A. MW (kda) B. SDS-PAGE Western blot MW (kda) 50 40 100 75 50 37 30 Glutelin 25 20 15 20 Prolamin 10 ARP1 1 2 3 E.coli MucoRice ARP1 (RNAi +) 4 5 ARP1
Supplementary Information Supplementary Figure 1. A. MW (kda) B. SDS-PAGE Western blot MW (kda) 50 40 100 75 50 37 30 Glutelin 25 20 15 20 Prolamin 10 ARP1 1 2 3 E.coli MucoRice ARP1 (RNAi +) 4 5 ARP1
Supplementary Figure S1. Distinct compartmentalization of proinsulin in obese db/db mouse islet ß- cells.
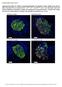 Supplementary Figure S1. Distinct compartmentalization of proinsulin in obese db/db mouse islet ß- cells. Pancreata from 16-weeks-old 6J +/+, 6J db/db, KS +/+ and KS db/db mice were harvested, fixed with
Supplementary Figure S1. Distinct compartmentalization of proinsulin in obese db/db mouse islet ß- cells. Pancreata from 16-weeks-old 6J +/+, 6J db/db, KS +/+ and KS db/db mice were harvested, fixed with
Supplementary Table 1. The primers used for quantitative RT-PCR. Gene name Forward (5 > 3 ) Reverse (5 > 3 )
 770 771 Supplementary Table 1. The primers used for quantitative RT-PCR. Gene name Forward (5 > 3 ) Reverse (5 > 3 ) Human CXCL1 GCGCCCAAACCGAAGTCATA ATGGGGGATGCAGGATTGAG PF4 CCCCACTGCCCAACTGATAG TTCTTGTACAGCGGGGCTTG
770 771 Supplementary Table 1. The primers used for quantitative RT-PCR. Gene name Forward (5 > 3 ) Reverse (5 > 3 ) Human CXCL1 GCGCCCAAACCGAAGTCATA ATGGGGGATGCAGGATTGAG PF4 CCCCACTGCCCAACTGATAG TTCTTGTACAGCGGGGCTTG
Evaluation of an EDTA version of CATT/Trypanosoma brucei gambiense. Prince Leopold Institute of Tropical Medicine, Department of Parasitology,
 Evaluation of an EDTA version of CATT/Trypanosoma brucei gambiense for serological screening of human blood samples Eddy Magnus 1 *, Veerle Lejon 1, Dominique Bayon 2, Dirk Buyse 3, Pere Simarro 4, Didier
Evaluation of an EDTA version of CATT/Trypanosoma brucei gambiense for serological screening of human blood samples Eddy Magnus 1 *, Veerle Lejon 1, Dominique Bayon 2, Dirk Buyse 3, Pere Simarro 4, Didier
Supplementary Information for. A cancer-associated BRCA2 mutation reveals masked nuclear. export signals controlling localization
 Supplementary Information for A cancer-associated BRCA2 mutation reveals masked nuclear export signals controlling localization Anand D Jeyasekharan 1, Yang Liu 1, Hiroyoshi Hattori 1,3, Venkat Pisupati
Supplementary Information for A cancer-associated BRCA2 mutation reveals masked nuclear export signals controlling localization Anand D Jeyasekharan 1, Yang Liu 1, Hiroyoshi Hattori 1,3, Venkat Pisupati
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1 Schematic depiction of the tandem Fc GDF15. Supplementary Figure 2 Supplementary Figure 2 Gfral mrna levels in the brains of both wild-type and knockout Gfral
Supplementary Figure 1 Supplementary Figure 1 Schematic depiction of the tandem Fc GDF15. Supplementary Figure 2 Supplementary Figure 2 Gfral mrna levels in the brains of both wild-type and knockout Gfral
Supplementary Figure S I: Effects of D4F on body weight and serum lipids in apoe -/- mice.
 Supplementary Figures: Supplementary Figure S I: Effects of D4F on body weight and serum lipids in apoe -/- mice. Male apoe -/- mice were fed a high-fat diet for 8 weeks, and given PBS (model group) or
Supplementary Figures: Supplementary Figure S I: Effects of D4F on body weight and serum lipids in apoe -/- mice. Male apoe -/- mice were fed a high-fat diet for 8 weeks, and given PBS (model group) or
SUPPLEMENTARY INFORMATION
 Supplemental Figure 1. Furin is efficiently deleted in CD4 + and CD8 + T cells. a, Western blot for furin and actin proteins in CD4cre-fur f/f and fur f/f Th1 cells. Wild-type and furin-deficient CD4 +
Supplemental Figure 1. Furin is efficiently deleted in CD4 + and CD8 + T cells. a, Western blot for furin and actin proteins in CD4cre-fur f/f and fur f/f Th1 cells. Wild-type and furin-deficient CD4 +
Supplementary Figure 1. Gene expression analysis of GSNOR1 and NIA2 in genotypes with altered NO signalling. Relative expression of GSNOR1 in leaves
 Supplementary Figure 1. Gene expression analysis of GSNOR1 and NIA2 in genotypes with altered NO signalling. Relative expression of GSNOR1 in leaves (a) and roots (b) and NIA2 in leaves (c) and roots (d)
Supplementary Figure 1. Gene expression analysis of GSNOR1 and NIA2 in genotypes with altered NO signalling. Relative expression of GSNOR1 in leaves (a) and roots (b) and NIA2 in leaves (c) and roots (d)
Supplementary Figure 1. Confocal immunofluorescence showing mitochondrial translocation of Drp1. Cardiomyocytes treated with H 2 O 2 were prestained
 Supplementary Figure 1. Confocal immunofluorescence showing mitochondrial translocation of Drp1. Cardiomyocytes treated with H 2 O 2 were prestained with MitoTracker (red), then were immunostained with
Supplementary Figure 1. Confocal immunofluorescence showing mitochondrial translocation of Drp1. Cardiomyocytes treated with H 2 O 2 were prestained with MitoTracker (red), then were immunostained with
Study on endocytosis and hemoglobin uptake in different developmental stages of Trypanosoma congolense IL3000 strain
 J. Protozool. Res. 23, 14-20 (2013) Copyright 2008, National Research Center for Protozoan Diseases Study on endocytosis and hemoglobin uptake in different developmental stages of Trypanosoma congolense
J. Protozool. Res. 23, 14-20 (2013) Copyright 2008, National Research Center for Protozoan Diseases Study on endocytosis and hemoglobin uptake in different developmental stages of Trypanosoma congolense
A. Generation and characterization of Ras-expressing autophagycompetent
 Supplemental Material Supplemental Figure Legends Fig. S1 A. Generation and characterization of Ras-expressing autophagycompetent and -deficient cell lines. HA-tagged H-ras V12 was stably expressed in
Supplemental Material Supplemental Figure Legends Fig. S1 A. Generation and characterization of Ras-expressing autophagycompetent and -deficient cell lines. HA-tagged H-ras V12 was stably expressed in
SUPPORTING INFORMATION
 SUPPORTING INFORMATION SUPPLEMENTARY FIGURE LEGENDS Fig. S1. Separation of non-dissolved nanoparticles. Tests were conducted on the separation of non-dissolved nanoparticles added in excess to BEGM (A)
SUPPORTING INFORMATION SUPPLEMENTARY FIGURE LEGENDS Fig. S1. Separation of non-dissolved nanoparticles. Tests were conducted on the separation of non-dissolved nanoparticles added in excess to BEGM (A)
Supplementary Figure 1: Digitoxin induces apoptosis in primary human melanoma cells but not in normal melanocytes, which express lower levels of the
 Supplementary Figure 1: Digitoxin induces apoptosis in primary human melanoma cells but not in normal melanocytes, which express lower levels of the cardiac glycoside target, ATP1A1. (a) The percentage
Supplementary Figure 1: Digitoxin induces apoptosis in primary human melanoma cells but not in normal melanocytes, which express lower levels of the cardiac glycoside target, ATP1A1. (a) The percentage
nature methods Organelle-specific, rapid induction of molecular activities and membrane tethering
 nature methods Organelle-specific, rapid induction of molecular activities and membrane tethering Toru Komatsu, Igor Kukelyansky, J Michael McCaffery, Tasuku Ueno, Lidenys C Varela & Takanari Inoue Supplementary
nature methods Organelle-specific, rapid induction of molecular activities and membrane tethering Toru Komatsu, Igor Kukelyansky, J Michael McCaffery, Tasuku Ueno, Lidenys C Varela & Takanari Inoue Supplementary
Aggregated neutrophil extracellular traps limit inflammation by degrading cytokines and chemokines
 CORRECTION NOTICE Nat. Med. doi:10.1038/nm.3547; corrected online 25 August 2014 Aggregated neutrophil extracellular traps limit inflammation by degrading cytokines and chemokines Christine Schauer, Christina
CORRECTION NOTICE Nat. Med. doi:10.1038/nm.3547; corrected online 25 August 2014 Aggregated neutrophil extracellular traps limit inflammation by degrading cytokines and chemokines Christine Schauer, Christina
sequences of a styx mutant reveals a T to A transversion in the donor splice site of intron 5
 sfigure 1 Styx mutant mice recapitulate the phenotype of SHIP -/- mice. (A) Analysis of the genomic sequences of a styx mutant reveals a T to A transversion in the donor splice site of intron 5 (GTAAC
sfigure 1 Styx mutant mice recapitulate the phenotype of SHIP -/- mice. (A) Analysis of the genomic sequences of a styx mutant reveals a T to A transversion in the donor splice site of intron 5 (GTAAC
Supplementary Table 1. Primer sequences for conventional RT-PCR on mouse islets
 Supplementary Table 1. Primer sequences for conventional RT-PCR on mouse islets Gene 5 Forward 3 5 Reverse 3.T. Product (bp) ( C) mnox1 GTTCTTGGGCTGCCTTGG GCTGGGGCGGCGG 60 300 mnoxa1 GCTTTGCCGCGTGC GGTTCGGGTCCTTTGTGC
Supplementary Table 1. Primer sequences for conventional RT-PCR on mouse islets Gene 5 Forward 3 5 Reverse 3.T. Product (bp) ( C) mnox1 GTTCTTGGGCTGCCTTGG GCTGGGGCGGCGG 60 300 mnoxa1 GCTTTGCCGCGTGC GGTTCGGGTCCTTTGTGC
Supplemental Figure 1
 Supplemental Figure 1 A S100A4: SFLGKRTDEAAFQKLMSNLDSNRDNEVDFQEYCVFLSCIAMMCNEFFEGFPDK Overlap: SF G DE KLM LD N D VDFQEY VFL I M N FF G PD S100A2: SFVGEKVDEEGLKKLMGSLDENSDQQVDFQEYAVFLALITVMCNDFFQGCPDR
Supplemental Figure 1 A S100A4: SFLGKRTDEAAFQKLMSNLDSNRDNEVDFQEYCVFLSCIAMMCNEFFEGFPDK Overlap: SF G DE KLM LD N D VDFQEY VFL I M N FF G PD S100A2: SFVGEKVDEEGLKKLMGSLDENSDQQVDFQEYAVFLALITVMCNDFFQGCPDR
Supplementary Fig. S1. Schematic diagram of minigenome segments.
 open reading frame 1565 (segment 5) 47 (-) 3 5 (+) 76 101 125 149 173 197 221 246 287 open reading frame 890 (segment 8) 60 (-) 3 5 (+) 172 Supplementary Fig. S1. Schematic diagram of minigenome segments.
open reading frame 1565 (segment 5) 47 (-) 3 5 (+) 76 101 125 149 173 197 221 246 287 open reading frame 890 (segment 8) 60 (-) 3 5 (+) 172 Supplementary Fig. S1. Schematic diagram of minigenome segments.
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS)
 Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
on January 28, 2019 by guest
 INFECTION AND IMMUNITY, Feb. 2000, p. 848 860 Vol. 68, No. 2 0019-9567/00/$04.00 0 Copyright 2000, American Society for Microbiology. All Rights Reserved. Comparative Analysis of Antibody Responses against
INFECTION AND IMMUNITY, Feb. 2000, p. 848 860 Vol. 68, No. 2 0019-9567/00/$04.00 0 Copyright 2000, American Society for Microbiology. All Rights Reserved. Comparative Analysis of Antibody Responses against
Figure S1A. Blood glucose levels in mice after glucose injection
 ## Figure S1A. Blood glucose levels in mice after glucose injection Blood glucose (mm/l) 25 2 15 1 5 # 15 3 6 3+3 Time after glucose injection (min) # Figure S1B. α-kg levels in mouse livers after glucose
## Figure S1A. Blood glucose levels in mice after glucose injection Blood glucose (mm/l) 25 2 15 1 5 # 15 3 6 3+3 Time after glucose injection (min) # Figure S1B. α-kg levels in mouse livers after glucose
Procaspase-3. Cleaved caspase-3. actin. Cytochrome C (10 M) Z-VAD-fmk. Procaspase-3. Cleaved caspase-3. actin. Z-VAD-fmk
 A HeLa actin - + + - - + Cytochrome C (1 M) Z-VAD-fmk PMN - + + - - + actin Cytochrome C (1 M) Z-VAD-fmk Figure S1. (A) Pan-caspase inhibitor z-vad-fmk inhibits cytochrome c- mediated procaspase-3 cleavage.
A HeLa actin - + + - - + Cytochrome C (1 M) Z-VAD-fmk PMN - + + - - + actin Cytochrome C (1 M) Z-VAD-fmk Figure S1. (A) Pan-caspase inhibitor z-vad-fmk inhibits cytochrome c- mediated procaspase-3 cleavage.
Supplementary Figure 1. Supernatants electrophoresis from CD14+ and dendritic cells. Supernatants were resolved by SDS-PAGE and stained with
 Supplementary Figure 1. Supernatants electrophoresis from CD14+ and dendritic cells. Supernatants were resolved by SDS-PAGE and stained with Coomassie brilliant blue. One µg/ml recombinant human (rh) apo-e
Supplementary Figure 1. Supernatants electrophoresis from CD14+ and dendritic cells. Supernatants were resolved by SDS-PAGE and stained with Coomassie brilliant blue. One µg/ml recombinant human (rh) apo-e
Luminescent platforms for monitoring changes in the solubility of amylin and huntingtin in living cells
 Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2016 Contents Supporting Information Luminescent platforms for monitoring changes in the
Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2016 Contents Supporting Information Luminescent platforms for monitoring changes in the
LPS LPS P6 - + Supplementary Fig. 1.
 P6 LPS - - - + + + - LPS + + - - P6 + Supplementary Fig. 1. Pharmacological inhibition of the JAK/STAT blocks LPS-induced HMGB1 nuclear translocation. RAW 267.4 cells were stimulated with LPS in the absence
P6 LPS - - - + + + - LPS + + - - P6 + Supplementary Fig. 1. Pharmacological inhibition of the JAK/STAT blocks LPS-induced HMGB1 nuclear translocation. RAW 267.4 cells were stimulated with LPS in the absence
Soluble ADAM33 initiates airway remodeling to promote susceptibility for. Elizabeth R. Davies, Joanne F.C. Kelly, Peter H. Howarth, David I Wilson,
 Revised Suppl. Data: Soluble ADAM33 1 Soluble ADAM33 initiates airway remodeling to promote susceptibility for allergic asthma in early life Elizabeth R. Davies, Joanne F.C. Kelly, Peter H. Howarth, David
Revised Suppl. Data: Soluble ADAM33 1 Soluble ADAM33 initiates airway remodeling to promote susceptibility for allergic asthma in early life Elizabeth R. Davies, Joanne F.C. Kelly, Peter H. Howarth, David
Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated
 Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C genes. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated from 7 day-old seedlings treated with or without
Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C genes. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated from 7 day-old seedlings treated with or without
Supplementary material. Supplementary Figure legends
 Supplementary material Supplementary Figure legends Supplementary Figure 1: Senescence-associated proliferation stop in response to oncogenic N-RAS expression Proliferation of NHEM cells without (ctrl.)
Supplementary material Supplementary Figure legends Supplementary Figure 1: Senescence-associated proliferation stop in response to oncogenic N-RAS expression Proliferation of NHEM cells without (ctrl.)
genome edited transient transfection, CMV promoter
 Supplementary Figure 1. In the absence of new protein translation, overexpressed caveolin-1-gfp is degraded faster than caveolin-1-gfp expressed from the endogenous caveolin 1 locus % loss of total caveolin-1-gfp
Supplementary Figure 1. In the absence of new protein translation, overexpressed caveolin-1-gfp is degraded faster than caveolin-1-gfp expressed from the endogenous caveolin 1 locus % loss of total caveolin-1-gfp
Supplementary Figure 1. Properties of various IZUMO1 monoclonal antibodies and behavior of SPACA6. (a) (b) (c) (d) (e) (f) (g) .
 Supplementary Figure 1. Properties of various IZUMO1 monoclonal antibodies and behavior of SPACA6. (a) The inhibitory effects of new antibodies (Mab17 and Mab18). They were investigated in in vitro fertilization
Supplementary Figure 1. Properties of various IZUMO1 monoclonal antibodies and behavior of SPACA6. (a) The inhibitory effects of new antibodies (Mab17 and Mab18). They were investigated in in vitro fertilization
Supplementary Figure 1. Characterization of basophils after reconstitution of SCID mice
 Supplementary figure legends Supplementary Figure 1. Characterization of after reconstitution of SCID mice with CD4 + CD62L + T cells. (A-C) SCID mice (n = 6 / group) were reconstituted with 2 x 1 6 CD4
Supplementary figure legends Supplementary Figure 1. Characterization of after reconstitution of SCID mice with CD4 + CD62L + T cells. (A-C) SCID mice (n = 6 / group) were reconstituted with 2 x 1 6 CD4
fl/+ KRas;Atg5 fl/+ KRas;Atg5 fl/fl KRas;Atg5 fl/fl KRas;Atg5 Supplementary Figure 1. Gene set enrichment analyses. (a) (b)
 KRas;At KRas;At KRas;At KRas;At a b Supplementary Figure 1. Gene set enrichment analyses. (a) GO gene sets (MSigDB v3. c5) enriched in KRas;Atg5 fl/+ as compared to KRas;Atg5 fl/fl tumors using gene set
KRas;At KRas;At KRas;At KRas;At a b Supplementary Figure 1. Gene set enrichment analyses. (a) GO gene sets (MSigDB v3. c5) enriched in KRas;Atg5 fl/+ as compared to KRas;Atg5 fl/fl tumors using gene set
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb3399 a b c d FSP DAPI 5mm mm 5mm 5mm e Correspond to melanoma in-situ Figure a DCT FSP- f MITF mm mm MlanaA melanoma in-situ DCT 5mm FSP- mm mm mm mm mm g melanoma in-situ MITF MlanaA mm mm
DOI:.38/ncb3399 a b c d FSP DAPI 5mm mm 5mm 5mm e Correspond to melanoma in-situ Figure a DCT FSP- f MITF mm mm MlanaA melanoma in-situ DCT 5mm FSP- mm mm mm mm mm g melanoma in-situ MITF MlanaA mm mm
marker. DAPI labels nuclei. Flies were 20 days old. Scale bar is 5 µm. Ctrl is
 Supplementary Figure 1. (a) Nos is detected in glial cells in both control and GFAP R79H transgenic flies (arrows), but not in deletion mutant Nos Δ15 animals. Repo is a glial cell marker. DAPI labels
Supplementary Figure 1. (a) Nos is detected in glial cells in both control and GFAP R79H transgenic flies (arrows), but not in deletion mutant Nos Δ15 animals. Repo is a glial cell marker. DAPI labels
GFP/Iba1/GFAP. Brain. Liver. Kidney. Lung. Hoechst/Iba1/TLR9!
 Supplementary information a +KA Relative expression d! Tlr9 5!! 5! NSC Neuron Astrocyte Microglia! 5! Tlr7!!!! NSC Neuron Astrocyte! GFP/Sβ/! Iba/Hoechst Microglia e Hoechst/Iba/TLR9! GFP/Iba/GFAP f Brain
Supplementary information a +KA Relative expression d! Tlr9 5!! 5! NSC Neuron Astrocyte Microglia! 5! Tlr7!!!! NSC Neuron Astrocyte! GFP/Sβ/! Iba/Hoechst Microglia e Hoechst/Iba/TLR9! GFP/Iba/GFAP f Brain
Supplementary Figure 1. Generation of knockin mice expressing L-selectinN138G. (a) Schematics of the Sellg allele (top), the targeting vector, the
 Supplementary Figure 1. Generation of knockin mice expressing L-selectinN138G. (a) Schematics of the Sellg allele (top), the targeting vector, the targeted allele in ES cells, and the mutant allele in
Supplementary Figure 1. Generation of knockin mice expressing L-selectinN138G. (a) Schematics of the Sellg allele (top), the targeting vector, the targeted allele in ES cells, and the mutant allele in
Programmed necrosis, not apoptosis, is a key mediator of cell loss and DAMP-mediated inflammation in dsrna-induced retinal degeneration
 Programmed necrosis, not apoptosis, is a key mediator of cell loss and DAMP-mediated inflammation in dsrna-induced retinal degeneration The Harvard community has made this article openly available. Please
Programmed necrosis, not apoptosis, is a key mediator of cell loss and DAMP-mediated inflammation in dsrna-induced retinal degeneration The Harvard community has made this article openly available. Please
Supplementary Figure 1: Co-localization of reconstituted L-PTC and dendritic cells
 a CD11c Na + K + ATPase Na + K + ATPase CD11c x-y CD11c Na + K + ATPase Na + K + ATPase CD11c x-z c b x-y view BoNT NAPs CD11c BoNT CD11c NAPs BoNT NAPs CD11c 90 x-z view Apical Basolateral Supplementary
a CD11c Na + K + ATPase Na + K + ATPase CD11c x-y CD11c Na + K + ATPase Na + K + ATPase CD11c x-z c b x-y view BoNT NAPs CD11c BoNT CD11c NAPs BoNT NAPs CD11c 90 x-z view Apical Basolateral Supplementary
Kidney. Heart. Lung. Sirt1. Gapdh. Mouse IgG DAPI. Rabbit IgG DAPI
 a e Na V 1.5 Ad-LacZ Ad- 110KD b Scn5a/ (relative to Ad-LacZ) f 150 100 50 0 p = 0.65 Ad-LacZ Ad- c Heart Lung Kidney Spleen 110KD d fl/fl c -/- DAPI 20 µm Na v 1.5 250KD fl/fl Rabbit IgG DAPI fl/fl Mouse
a e Na V 1.5 Ad-LacZ Ad- 110KD b Scn5a/ (relative to Ad-LacZ) f 150 100 50 0 p = 0.65 Ad-LacZ Ad- c Heart Lung Kidney Spleen 110KD d fl/fl c -/- DAPI 20 µm Na v 1.5 250KD fl/fl Rabbit IgG DAPI fl/fl Mouse
Zhu et al, page 1. Supplementary Figures
 Zhu et al, page 1 Supplementary Figures Supplementary Figure 1: Visual behavior and avoidance behavioral response in EPM trials. (a) Measures of visual behavior that performed the light avoidance behavior
Zhu et al, page 1 Supplementary Figures Supplementary Figure 1: Visual behavior and avoidance behavioral response in EPM trials. (a) Measures of visual behavior that performed the light avoidance behavior
