Context aware decision support in neurosurgical oncology based on an efficient classification of endomicroscopic data
|
|
|
- Erin Eaton
- 5 years ago
- Views:
Transcription
1 International Journal of Computer Assisted Radiology and Surgery (2018) 13: ORIGINAL ARTICLE Context aware decision support in neurosurgical oncology based on an efficient classification of endomicroscopic data Yachun Li 2 Patra Charalampaki 3,4 Yong Liu 2 Guang-Zhong Yang 1 Stamatia Giannarou 1 Received: 27 January 2018 / Accepted: 4 June 2018 / Published online: 13 June 2018 The Author(s) 2018 Abstract Purpose Probe-based confocal laser endomicroscopy (pcle) enables in vivo, in situ tissue characterisation without changes in the surgical setting and simplifies the oncological surgical workflow. The potential of this technique in identifying residual cancer tissue and improving resection rates of brain tumours has been recently verified in pilot studies. The interpretation of endomicroscopic information is challenging, particularly for surgeons who do not themselves routinely review histopathology. Also, the diagnosis can be examiner-dependent, leading to considerable inter-observer variability. Therefore, automatic tissue characterisation with pcle would support the surgeon in establishing diagnosis as well as guide robot-assisted intervention procedures. Methods The aim of this work is to propose a deep learning-based framework for brain tissue characterisation for context aware diagnosis support in neurosurgical oncology. An efficient representation of the context information of pcle data is presented by exploring state-of-the-art CNN models with different tuning configurations. A novel video classification framework based on the combination of convolutional layers with long-range temporal recursion has been proposed to estimate the probability of each tumour class. The video classification accuracy is compared for different network architectures and data representation and video segmentation methods. Results We demonstrate the application of the proposed deep learning framework to classify Glioblastoma and Meningioma brain tumours based on endomicroscopic data. Results show significant improvement of our proposed image classification framework over state-of-the-art feature-based methods. The use of video data further improves the classification performance, achieving accuracy equal to 99.49%. Conclusions This work demonstrates that deep learning can provide an efficient representation of pcle data and accurately classify Glioblastoma and Meningioma tumours. The performance evaluation analysis shows the potential clinical value of the technique. Keywords Brain tissue characterisation Image and video classification Deep learning Introduction B Stamatia Giannarou stamatia.giannarou@imperial.ac.uk 1 The Hamlyn Centre for Robotic Surgery, Imperial College London, London, UK 2 Institute of Cyber-Systems and Control, Zhejiang University, Hangzhou, China 3 Department of Neurosurgery, Cologne Medical Center, University Witten, Cologne, Germany 4 Department of Neurosurgery, Heinrich Heine University Duesseldorf, Duesseldorf, Germany Biophotonics techniques such as probe-based confocal laser endomicroscopy (pcle) have enabled direct visualisation of tissue at a microscopic level, with recent pilot studies suggesting it has a clear role in identifying residual cancer tissue and improving resection rates of brain tumours [16]. However, the interpretation of endomicroscopic information remains challenging, particularly for surgeons who do not themselves routinely review histopathology. Also, even among experts, the diagnosis can be examiner-dependent, leading to considerable inter-observer variability. Automatic tissue characterisation with pcle, based on a database of previously annotated data by expert physicians with diagnosis
2 1188 International Journal of Computer Assisted Radiology and Surgery (2018) 13: confirmed by histology, would support the surgeon in establishing diagnosis and could also guide autonomous robotic tissue scanning to focus locally on pathological areas. State-of-the-art computer-aided diagnosis (CAD) systems designed for the analysis of pcle data have mainly focused on the classification of colonic cancers. A content-based image and video retrieval framework based on the Bag-ofvisual Words (BoW) approach has been proposed in [2] for the differentiation of neoplastic and benign colorectal polyps. A local dense multiscale image descriptor has been introduced to extract a set of features which are clustered into a number of visual words, whose frequencies build the image signature. The similarity between two images can be estimated by calculating the distance between their signatures. This image retrieval method has been extended for video retrieval by relying on the coarse registration of images taken at different times. Clinical evaluation of this framework presented in [1] shows that automated classification of pcle videos of colonic polyps achieves high performance, comparable to off-line diagnosis of pcle videos established by expert endoscopists. The above video retrieval framework has been extended in [3] to include high-level knowledge for the pathological interpretation of pcle images. For that purpose, binary semantic concepts commonly used by expert endoscopists to diagnose pcle videos of colonic polyps have been extracted and used as additional information that complements the visual outputs of the retrieval framework. Recently, approaches to content-based image retrieval of pcle data have focused on learning discriminative visual features to improve the retrieval accuracy. In [10], a Multi- View Multi-Modal Embedding (MVMME) framework has been proposed to learn discriminative features of pcle videos by exploiting both mosaics and histology images. Visual features were extracted from the pcle mosaics using multiple operators such as SIFT and HOG and a mapping from these features to a latent space was learned in a supervised way to generate robust latent features which are more discriminative than unimodal features from mosaics alone. This work has been extended in [9] where an unsupervised multimodal graph mining (UMGM) approach has been proposed to learn the discriminative features for pcle mosaics of breast tissue. A multiscale multimodal graph is built based on pcle mosaics and histology patches, and a latent feature space is created to learn discriminative features without supervision. Wan et al. [22] extended the content-based image retrieval framework proposed in [2] using an efficient feature encoding scheme based on codeword proximity. A set of keypoints has been uniformly sampled from the pcle images, and different state-of-the-art descriptors such as SIFT and FREAK have been used to describe the keypoints. The keypoint descriptors are encoded with the proposed linear locality constraint (LLC) scheme which exploits spatial locality of the visual features instead of the hard quantization employed in BoW-based approaches. The above framework was applied for the characterisation of brain tissue. A support vector machine (SVM) classifier was used to classify from pcle images two types of commonly diagnosed brain tumours, namely Glioblastomas and Meningiomas. This work has been extended in [12] by exploring more encoding schemes for data description and using a majority voting-based classification scheme for video classification. The above CAD systems are composed of three main steps, namely visual feature extraction, feature encoding, and supervised classification. The performance of these systems relies on the design of handcrafted morphological or textural descriptors, capable of extracting discriminative image features which could facilitate the classification task. However, the selection of efficient features is problem-dependent and it is not easy to find a generic feature extraction method which can perform equally well in different applications. Hence, the above CAD systems can have significant performance variations depending on the data and the surgical context. Recently, significant progress has been made in image feature extraction and representation, mainly due to the revival of deep learning models such as convolutional neural networks (CNN). These architectures enable the learning of data-driven and highly representative image features from a large set of training data. This alleviates the need to design explicit handcrafted image features and also allows the feature extraction, encoding and classification tasks to be done within the optimisation of the same deep architecture. Hence, the network performance can be easily tuned, in a systematic fashion. The success of deep learning models in general computer vision tasks has motivated their application to medical image analysis [20]. More specifically, they have been used to detect and segment organs from multiple imaging modalities [4,6], semantic segmentation in histological images [23], multimodal registration, image super-resolution, image classification [15] and workflow analysis [21]. Deep learning has also gained a lot of attention for the design of CAD systems, focusing on abnormality detection and tissue state classification [5,17]. CNN models have been extensively employed for this purpose and extract image features either by training the models from scratch or using transfer learning when a limited number of training data is available. Transfer learning has shown promising performance and extracts image features using off-the-shelf CNNs pre-trained on different data (such as natural images) or performing pre-training of the models on big datasets and then supervised fine-tuning on medical data. The aim of this paper is to propose a brain tissue characterisation framework for context aware decision support in neurosurgical oncology. This will be part of a platform which can extract, interpret and use context information to
3 International Journal of Computer Assisted Radiology and Surgery (2018) 13: provide the surgeon with a set of clinical actions that can be made in the current situation and guide intraoperative resection. The context in our case refers to the discriminative information which is extracted from the pcle data and is used to characterise the state of the tissue. In particular, we focus on the classification of brain tumours into Glioblastoma and Meningioma which are the most common malignant and benign tumours, respectively, in neurosurgery. Our classification is based on the analysis of endomicroscopic data, and we propose two deep learning-based frameworks for the classification of image and video pcle data. To the best of our knowledge, this is the first work on brain tissue characterisation based on deep learning. The novel aspects of our work include: An efficient representation of the context information of pcle images by exploring state-of-the-art CNN models and proposing different tuning configurations. The best configurations have been selected and used to classify image and video data into the above tumour types. A novel video classification framework based on the combination of convolutional layers with long-range temporal recursion to estimate the probability of each tumour class. A set of network architectures and video segmentation methods which has been combined with different pcle data representations to increase the video classification accuracy. The proposed framework has been validated on ex vivo pcle data and has been compared to state-of-the-art brain tumour classification approaches. The performance evaluation analysis shows the potential clinical value of the proposed framework. Methods In this work, we propose two deep learning-based frameworks for brain tissue characterisation based on the classification of image and video pcle data into two tumour types, namely Glioblastoma and Meningioma. Image classification Our image-based tissue characterisation approach employs convolutional neural networks for context representation and classification of pcle images. A CNN for classification can be thought of as the composition of a number of convolutional functions f (x) = f L (... f 2 ( f 1 (x; w 1 ); w 2 )...; w L ), where L is the number of layers, x is an input image and w =[w 1,...,w L ] are the network parameters learned during training. The convolutional operations are applied using a sliding window over the image to detect generic features. The convolutions are followed by a nonlinear function, the Rectified Linear Unit (ReLU), applied to each element of the convolution output. The feature map extracted at each layer becomes input for the next layer. In order to reduce the dimensionality of the extracted features, the maps from each convolution are downsampled through a max-pooling layer which keeps the maximum value of the features in the pool. After the convolution and pooling layers, fully connected layers are introduced where each pixel is considered as a separate neuron just like a regular neural network. The final fully connected layer has the same number of outputs as the number of labels in the classification task and those values represent the likelihood of each label, estimated using the softmax function. In this work we employ three state-of-the-art CNN architectures namely, the AlexNet [14], the VGG16 [18] and the Inception-v3 [19] to classify two types of brain tumour. The AlexNet is an eight-layer CNN which is composed of 5 convolutional layers followed by 3 fully connected layers. The VGG16 network is much deeper, consisting of 13 convolutional layers and 3 fully connected layers. We cluster the convolutional layers into 5 groups which are separated by max-pooling layers as shown in Fig. 1. In the rest of our analysis, we denote the convolutional layer groups as conv1, conv2,..., conv5 and the fully connected layers as fc6, fc7, fc8. The Inception is a fully convolutional network including various Inception modules with parallel structure. Different to AlexNet and VGG16, which have 3 fully connected layers at the end, the Inception network has only one fully connected layer combined with a softmax layer. As we only need to differentiate two types of tumour, we change the output of the last layer in the above networks from 1000 to 2. Also, since these networks require colour images as input and the endomicroscopic data in our dataset is greyscale, we simply copy the grey values to the second and third channel to produce 3-channel input images. Upon initialization, a CNN is simply a function which through numerous operations transforms highly dimensional data into several numerical values which at this point are random. It is through training that the model function z = f (x; w) learns to achieve a desired goal. More specifically, during training the parameters w are learned by evaluating the performance of the network using a loss function which in the case of classification is directly related to the probabilities allocated to the correct classes. In this work, in order to efficiently represent the endomicroscopic data, we choose not to train our CNNs from scratch or fine-tune all the layers. Instead, we propose two approaches to tune the above CNN architectures. In the first approach, called feature extractor, we randomly initialise the weights and biases of several of the 8 layer groups and then train those variables as usual. More specifi-
4 1190 International Journal of Computer Assisted Radiology and Surgery (2018) 13: Fig. 1 Layer clustering in the employed CNN architectures. a AlexNet, b VGG16 cally, we tune the first (8 n), (n = 1, 2, 3,..., 8) layers using parameters from pre-trained models and do not update them during training. The rest n layers are initialised randomly, with the weights sampled from Gaussian distributions and the biases set to a constant value. Another approach is the fine-tuning, where we initialise the weights and biases of some trainable layers from pre-trained models and then finetune those values using a smaller learning rate. Similar to the above tuning approach, we copy to the first (8 n) layers the weights and biases of the employed CNN models pre-trained on the ImageNet dataset and do not update them during training. For the later n layers, we transfer the pre-trained values and update them at a learning rate equal to the 1/10th of the pre-training rate [8] as pre-training has already provided good initialisation for those layers. The parameter n is here referred as the tuning depth. Since the Inception network has a different architecture, we retrain only the final fully connected layer which corresponds to tuning depth n = 1. For the last layer of both architectures, we cannot use the values from pre-trained models since fc8 has been modified to fit our binary classification task. So, the last layer is always randomly initialised in both approaches. Training the CNN from scratch is a special case of the feature extractor method where we optimise all the layers. The case of fine-tuning all the layers is covered when we use the fine-tuning approach with tuning depth n = 8. Video classification For the classification of video pcle data, we use a deep learning architecture which combines convolutional layers and long-range temporal recursion. Specifically, we extract visual features from a set of consecutive endomicroscopic images using a CNN network and then feed the ordered feature sequences to a recurrent neural network (RNN) to perform classification. At the end of the network, a fully connected layer with softmax is included to generate a prediction about the tumour class. An outline of the proposed architecture is shown in Fig. 2.
5 International Journal of Computer Assisted Radiology and Surgery (2018) 13: Fig. 2 Overview of proposed approach for video classification. The LSTM in the green box may contain more than one LSTM layers To extract visual features from the pcle images, we employed the same CNN models that were used for image classification. For temporal recursion, we use the long shortterm memory (LSTM) network [11]. We explore 3 different architectures with different width (number of hidden units) and depth (number of LSTM layers), namely the Standard LSTM with 2 LSTM layers, the Wide LSTM with only one but wider layer and the Deep LSTM of 3 layers as shown in Fig. 3. In this work, we test two different configurations to combine the CNN and the LSTM networks, namely the Before-Final-fc (BFF) and After-Final-fc) (AFF).Inthe former configuration, we pass to the LSTM network the input of the final fully connected layer. This is a feature vector of length 2048, 4096, 4096 for the Inception network, the AlexNet and the VGGNet, respectively. In the latter configuration, the input of the LSTM network is the output of the last fully connected with softmax layer of the CNN network which is a vector of size equal to the number of classes in the classification task and represents the predictions of the classes. The above architecture is different to the long-term recurrent convolutional networks proposed in [7] as our visual feature extraction is not based on the hybrid CaffeNet and ZFNet model. Also, the LSTM configuration in [7] includes only one layer with different number of hidden units. In our framework, we train the CNN and LSTM networks separately rather than training the architecture end-to-end which provides a more efficient training and allows us to train the LSTM with various features. Finally, our final prediction is based on the probability at time step T while [7]is using the average of all label probabilities from time step 1 to T. Experiments In this section, we first describe the preprocessing steps that we apply on our endomicroscopic data. Then we evaluate the performance of the proposed image and video classification methods. Data preprocessing The validation of the proposed classification framework is based on ex vivo pcle data of Meningioma and Glioblastoma brain tumours, collected during brain tumour resection procedures at the Merheim Hospital in Germany. The examined tissue samples were scanned in the surgical theatre next to the patient, immediately after removing the tumour. As contrast agent, acriflavine hydrochloride AF from Sigma Pharmaceuticals, Victoria, Australia, was used. The data were collected using the Cellvizio system (Mauna Kea Technologies, Paris, France) and were classified into the above two types by expert histopathologists. Each pcle video represents one tumour type and corresponds to a different patient. Our dataset is a subset 1 of the dataset used in [22] and includes 16 videos for Glioblastoma and 17 for Menin- 1 Due to data ownership reasons, only a subset of the data used in [22] was made available to us and was used for our experimental results.
6 1192 International Journal of Computer Assisted Radiology and Surgery (2018) 13: Fig. 3 LSTM architectures with different width and depth gioma, with 25,000 frames in total (13,862 for Glioblastoma and 11,616 for Meningioma). Sample frames of our data are shown in Fig. 4. The frames of our dataset are rectangular images of size As the employed CNNs require square images as input, we first centrally crop the pcle images into images of size by removing the black area on their left and right site and then resize them into the appropriate size for each network ( for AlexNet, for VGGNet, and for Inception). For the video classification approach, to expand the dataset for both training and validation, we segment the videos into short video clips. The proposed video classification framework is tested using overlapping and non-overlapping video clips. In the non-overlapping case, no frame appears in two video clips at the same time as shown in Fig. 5a. Overlapping video clips are generated as shown in Fig. 5bbyshiftingthe frame selection block by one frame at a time. In both types of clipping, we ensure that no video clip contains frames from two different videos. To evaluate the performance of the different models and configurations proposed in this work, we split the dataset into 3 parts, namely the training set (66%), the validation set (17%) and the test set (17%). The training set is used to fit the parameters of each model while the validation set to compare their performance and decide which model to select. The performance of the selected models is then assessed on the test set. To ensure independence of each set, we perform patient-level (i.e. video-level) splitting and no frame from one set ends up on the other sets. To eliminate data splitting bias we employ sixfold cross-validation, where the test and validation sets are selected such that they are different for each run, while the rest of the dataset is used for training. The size of the test set for each fold during image and video classification is shown in Table 1. The test set size for each fold changes slightly because of our video-level splitting. All experiments report average classification performance on the sixfold. Image classification The hyperparameters we apply when training our models are those used for AlexNet in [14]. The AlexNet and VGG16 models are trained for 1000 iterations (approximately 10 epochs depending on the size of the training set), using gradient descent with mini-batch size 128. The learning rate is fixed to 0.01 for the feature extraction method, while for the fine-tuning it is 10 times smaller, i.e Our network minimises the cross-entropy loss function during the training optimisation process. For the Inception network, we set the number of training iterations to The last layer is
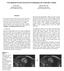
7 International Journal of Computer Assisted Radiology and Surgery (2018) 13: Fig. 4 Sample images of our pcle dataset. a Glioblastoma, b Meningioma Fig. 5 Video dataset expansion. In a m = L/k and in b n = L (k 1), wherek is the length of the video clip and L the length of the video sequence. a Video segmentation with non-overlapping video clips, b video segmentation with overlapping video clips Table 1 Test set size, in number of frames and clips, for each cross-validation fold during image and video classification, respectively Method Fold 1 Fold 2 Fold 3 Fold 4 Fold 5 Fold 6 Total Image classification ,478 Video classification Overlapping ,927 Non-overlapping retrained using the gradient descent optimiser with learning rate 0.01, batch size 100 and cross-entropy as the loss function. We use the TensorFlow framework to train our models. To constrain the set of classification models for validation, we first assess the performance of all possible combinations of tuning methods and tuning depths proposed in Image classification section without cross-validation, to select the best classifiers. We refer to tuning depths n = 1, 2 as shallow tuning, n = 3, 4, 5, 6 as medium tuning and n = 7, 8 as deep tuning. The models with the best performance were the (a) Alex extract n = 5, (b) Alex tune n = 5, (c) Alex tune n = 3, (d) VGG tune n = 5, (e) VGG tune n = 3. These models together with the Inception tune n = 1, were selected for further performance evaluation with sixfold cross-validation. The performance of the selected models on the validation and test datasets is shown in Table 2, and their classification accuracy is compared in Fig. 6. From the above results, it can be seen that the best configuration for the AlexNet is the feature extractor with tuning from conv4 to fc8, achieving 97.97% classification accuracy on the test set. For the
8 1194 International Journal of Computer Assisted Radiology and Surgery (2018) 13: Table 2 Image classification cross-validation results on the validation and test sets Dataset Configuration Accuracy TPR FPR Precision F 1 measure Validation Alex extract n = Alex tune n = Alex tune n = VGG tune n = VGG tune n = Inception tune n = Test Alex extract n = VGG tune n = Inception tune n = Bold value indicates the best models with the highest accuracy are very different from the dataset used for pre-training, random initialisation can generate efficient data representation when used with simple networks such as the AlexNet. Video classification Fig. 6 Image classification accuracy of sixfold cross-validation on the validation set. Error bars represent the 95% confidence interval of mean accuracy VGGNet, the fine-tuning with tuning from fc6 to fc8 is the best with 98.51% classification accuracy on the test set. The Inception network has the lowest accuracy, equal to 96.61%. It is also clear from Fig. 6 that the VGG tune n = 3, with the highest accuracy and lowest variance, outperforms the other models. In general, as we increase the depth of tuning n, the features tend to be more low-level. The above results indicate that model tuning of medium depth can generate discriminative features and provide an efficient representation of our pcle data. This enables the selected classifiers to recognise the delicate difference between the two tumour types and generate accurate predictions. In our classification task, deep tuning results in lower classification accuracy possibly due to the relatively small size of our training dataset. In the above analysis, we observe that the AlexNet works better with the feature extractor tuning method while the VGGNet achieves more accurate results using the fine-tuning method. This shows that loading pre-trained weights helps the convergence of deeper and complex networks. Since pcle data To extract visual features for video classification, we selected the CNN models that performed the best in the image classification task, namely the Alex extract n = 5, the VGG tune n = 3 and the Inception tune n = 1. To train the LSTM model, we set the batch size to 32 and use the Adam method [13] for optimisation with the default values proposed in [13]. The training epoch is 4 for the overlapping video clip case and 80 for the non-overlapping case. The cross-entropy is still used as our loss function. For evaluation, we use fixed-length video clips, 48 frames long and report the accuracy of classifying video clips rather than entire videos. The different CNN-LSTM combination approaches (BFF and AFF), video segmentation methods (overlapping and non-overlapping video clips) and LSTM configurations (Standard LSTM, Wide LSTM, Deep LSTM) result in 36 models which are evaluated on the validation dataset and compared in Fig. 7. Detailed performance evaluation results are presented in Table 3. As it can be observed, all the models are able to achieve accurate predictions with classification rate higher than 95%. The configuration AFF overlapping seems to perform the best and for the VGGNet model with deep LSTM achieves 99.49% accuracy on the test dataset. The superior performance of our video classification framework verifies that temporal information assists the classification of pcle data. From the performance evaluation results in Table 3, it can be deduced that the AFF configuration outperforms the BFF. This is because in the BFF configuration the RNN has to learn what the fully connected layers at the end of the CNN are already doing, namely mapping the feature vector to a prediction. On the other hand, the AFF directly inputs to the LSTM of the probabilities of two classes and then
9 International Journal of Computer Assisted Radiology and Surgery (2018) 13: Fig. 7 Video classification accuracy of sixfold cross-validation on the validation set. Error bars represent the 95% confidence interval of mean accuracy Table 3 Video classification cross-validation results on the validation and test set CNN Dataset Configurations Video classification Standard Wide Deep AlexNet Validation BFF overlapping BFF non-overlapping AFF overlapping AFF non-overlapping Test AFF non-overlapping VGGNet Validation BFF overlapping BFF non-overlapping AFF overlapping AFF non-overlapping Test AFF overlapping Inception Validation BFF overlapping BFF non-overlapping AFF overlapping AFF non-overlapping Test AFF overlapping Bold value indicates the best models with the highest accuracy the LSTM is trained to learn the connections between the different frames. Since the CNN already obtains very accurate results through prior training, the LSTM based on AFF further leverages the predictions from the neural network and achieves very good classification accuracy. The different video-clip generation types achieve very similar performance which indicates that extending the size of the training dataset with overlapping clips does not improve the classification accuracy significantly. As for the architecture of the LSTM, wide architectures are able to achieve slightly greater performance in most situations verifying that they are better at sequence learning. The qualitative performance evaluation results in Fig. 8 for VGG tune n = 3, the best video classification configuration, show that the proposed framework can correctly classify challenging data such as the frames in Fig. 8a, c which are not very similar to the typical tumour class data. However, when the appearance of the data differs significantly from the typical tumour class appearance, such as in Fig. 8b or when the data quality is impaired as in Fig. 8d, the proposed video classification fails. The performance of our proposed video classification framework has also been compared to score averaging and majority voting of the image classification results on the same temporal window for the non-overlapping and overlapping dataset segmentation methods. Score averaging is done by averaging the output of the softmax layer for each frame on the temporal window and using this average for class pre-
10 1196 International Journal of Computer Assisted Radiology and Surgery (2018) 13: Fig. 8 Qualitative results from our video classification using VGG tune n = 3 for feature extraction, AFF combination, overlapping segmentation and deep LSTM. a True negative. True label: Glioblastoma, prediction: Glioblastoma. b False positive. True label: Glioblastoma, prediction: Meningioma. c True positive. True label: Meningioma, prediction: Meningioma. d False negative. True label: Meningioma, prediction: Glioblastoma Table 4 Classification accuracy of score averaging and majority voting of the image classification results on the original dataset Dataset Configurations Score averaging Majority voting Inception AlexNet VGGNet Inception AlexNet VGGNet Validation Overlapping Non-overlapping Test Overlapping Non-overlapping diction. Majority voting refers to averaging over the class predictions for each frame on the temporal window. As it is shown in Table 4, for video sequences which represent a single tumour class, these approaches can achieve high classification accuracy, comparable to our proposed video classification. However, in practice, during tissue scanning the pcle video data might contain frames from multiple tissue classes (for instance, healthy tissue and tumour). Since our dataset does not include healthy tissue pcle data, to simulate the above scenario, we generated using our original dataset, video sequences containing frames from both tumour classes and evaluated the classification accuracy of the above three approaches. The mixed video sequences were created following two different frame mixing rules, namely regular replacement and group replacement. In the former case, a mixed video sequence is generated by replacing, in the original sequence in regular intervals, a number of frames with random frames
11 International Journal of Computer Assisted Radiology and Surgery (2018) 13: Table 5 Classification accuracy of score averaging and majority voting of the image classification results on the mixed video sequences Dataset Percent Configurations Regular replace Group replace LSTM Majority voting Score averaging LSTM Majority voting Score averaging Standard Wide Deep Standard Wide Deep Validation 2/12 BFF overlapping AFF overlapping BFF non-overlapping AFF non-overlapping /12 BFF overlapping AFF overlapping BFF non-overlapping AFF non-overlapping /12 BFF overlapping AFF overlapping BFF non-overlapping AFF non-overlapping Test 2/12 AFF overlapping BFF non-overlapping /12 AFF overlapping BFF non-overlapping AFF non-overlapping /12 AFF overlapping BFF non-overlapping Bold value indicates the best models with the highest accuracy
12 1198 International Journal of Computer Assisted Radiology and Surgery (2018) 13: of the opposite tumour class. In the latter case, a group of consecutive frames is replaced considering temporal windows of the same size as our video clips. In addition, different percentage of replaced frames were considered. For example, a video clip with regular replacement of (2/12) 100% Meningioma (MEN) frames on a Glioblastoma (GBM) sequence would have the following structure [10GBM, 2MEN, 10GBM, 2MEN, 10GBM, 2MEN, 10GBM, 2MEN], while the group replacement would result in [40GBM, 8MEN]. By considering overlapping video clips in our performance evaluation, we introduce randomness in the distribution of the replaced frames in the examined temporal window. The superiority of our proposed video classification approach compared to score averaging and majority voting of the image classification results is shown in Table 5. The improvement in the classification accuracy provided by our framework is more significant when the percentage of replaced frames is high. Also, from the above performance evaluation study it can be deduced that the frame mixing rule does not affect the classification performance significantly. The performance of our proposed video classification is higher when non-overlapping video clips are used for video segmentation. This is because in that case the LSTM learns the temporal pattern of the replaced frames which is consistent among the datasets. This also explains the good performance of the BFF configuration in the non-overlapping case which is comparable to the AFF configuration. From the above performance evaluation study, we can deduce that our deep learning-based frameworks can provide an efficient representation of the pcle data and accurately classify Glioblastoma and Meningioma tumours. The proposed image classification approach with 98.51% classification accuracy outperforms the feature-based brain tumour classification approach presented in [22] with close to 90% accuracy and in [12] with 84.32%. The combination of the VGGNet and LSTM networks for video classification achieves improved performance with classification accuracy equal to 99.49%. These results verify the potential of deep learning-based methods in classifying pcle data, with significant gain in classification accuracy. The average running time of our image classification on a GTX 1080 Ti GPU for the AlexNet, the VGG16 and the Inception network is 3.5, 9 and 25 msec, respectively. Our video classification based on pre-computed features (e.g. AFF with VGG) runs at about 1 s. Conclusions In this work, we have proposed a deep learning-based brain tissue characterisation framework which can classify brain tumours into Glioblastoma and Meningioma based on the analysis of pcle image and video data. For image classification, an efficient representation of the context information of pcle images has been proposed by exploring different CNN models and tuning configurations. Our video classification is based on a novel combination of convolutional layers with long-range temporal recursion to estimate the probability of each tumour class. The proposed deep learning-based image classification approach with 98.51% classification accuracy outperforms state-of-the-art feature-based brain tumour classification methods. The use of video data further improves the classification performance with accuracy equal to 99.49%. These results verify the potential of deep learning-based methods in classifying pcle data, with significant gain in classification accuracy. The focus of our future work is to build up a decision support platform which can characterise different states of brain tissue. For this purpose, more brain tissue classes will be examined with pcle data collected during intraoperative tissue scanning using a contrast agent which has been approved for use in humans. Acknowledgements The authors are grateful for support from the Royal Society, the Hamlyn Centre for Robotic Surgery, Imperial College London and the Institute of Global Health Innovations (IGHI), Imperial College London. Funding Dr. Stamatia Giannarou is supported by the Royal Society (UF140290). Compliance with ethical standards Conflict of interest The authors declare that they have no conflict of interest. Ethical approval All studies on human subjects were performed according to the requirements of the local ethic committee and in agreement with the Declaration of Helsinki. Informed consent Informed consent was obtained from all individual participants included in the study. Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License ( ons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. References 1. André B, Vercauteren T, Buchner AM, Krishna M, Ayache N, Wallace MB (2012) Software for automated classification of probebased confocal laser endomicroscopy videos of colorectal polyps. World J Gastroenterol 18(39): André B, Vercauteren T, Buchner AM, Wallace MB, Ayache N (2011) A smart atlas for endomicroscopy using automated video retrieval. Med Image Anal 15(4):
13 International Journal of Computer Assisted Radiology and Surgery (2018) 13: André B, Vercauteren T, Buchner AM, Wallace MB, Ayache N (2012) Learning semantic and visual similarity for endomicroscopy video retrieval. IEEE Trans Med Imaging 31(6): Cai J, Lu L, Zhang Z, Xing F, Yang L, Yin Q (2016) Pancreas segmentation in MRI using graph-based decision fusion on convolutional neural networks. In: Medical image computing and computer-assisted intervention (MICCAI), pp Cheng JZ, Ni D, Chou YH, Qin J, Tiu CM, Chang YC, Huang CS, Shen D, Chen CM (2016) Computer-aided diagnosis with deep learning architecture: applications to breast lesions in US images and pulmonary nodules in CT scans. Sci Rep 6: Christ PF, Elshaer MEA, Ettlinger F, Tatavarty S, Bickel M, Bilic P, Rempfler M, Armbruster M, Hofmann F, D Anastasi M, Sommer WH, Ahmadi S, Menze BH (2016) Automatic liver and lesion segmentation in CT using cascaded fully convolutional neural networks and 3D conditional random fields. In: Medical image computing and computer-assisted intervention (MICCAI), pp Donahue J, Hendricks LA, Rohrbach M, Venugopalan S, Guadarrama S, Saenko K, Darrell T (2017) Long-term recurrent convolutional networks for visual recognition and description. IEEE Trans Pattern Anal Mach Intell 39(4): Girshick R, Donahue J, Darrell T, Malik J (2016) Region-based convolutional networks for accurate object detection and segmentation. IEEE Trans Pattern Anal Mach Intell 38(1): Gu Y, Vyas K, Yang J, Yang GZ (2017) Unsupervised feature learning for endomicroscopy image retrieval. In: Medical image computing and computer-assisted intervention (MICCAI), pp Gu Y, Yang J, Yang GZ (2016) Multi-view multi-modal feature embedding for endomicroscopy mosaic classification. In: 2016 IEEE conference on computer vision and pattern recognition workshops (CVPRW), pp Hochreiter S, Schmidhuber J (1997) Long short-term memory. Neural Comput 9(8): Kamen A, Sun S, Wan S, Kluckner S, Chen T, Gigler AM, Simon E, Fleischer M, Javed M, Daali S, Igressa A, Charalampaki P (2016) Automatic tissue differentiation based on confocal endomicroscopic images for intraoperative guidance in neurosurgery. BioMed Res Int 2016: Kingma D, Ba J (2014) Adam: a method for stochastic optimization. arxiv preprint arxiv: Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks. In: Annual conference on neural information processing systems (NIPS). Neural Information Processing System Foundation, pp Li Q, Cai W, Wang X, Zhou Y, Feng DD, Chen M (2014) Medical image classification with convolutional neural network. In: International conference on control, automation, robotics & vision (ICARCV), pp Pavlov V, Meyronet D, Meyer-Bisch V, Armoiry X, Pikul B, Dumot C, Beuriat PA, Signorelli F, Guyotat J (2016) Intraoperative probebased confocal laser endomicroscopy in surgery and stereotactic biopsy of low-grade and high-grade gliomas: a feasibility study in humans. Neurosurgery 79(4): Pereira S, Pinto A, Alves V, Silva CA (2016) Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans Med Imaging 35(5): Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arxiv preprint arxiv: Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2016) Rethinking the inception architecture for computer vision. In: IEEE conference on computer vision and pattern recognition (CVPR), pp Tajbakhsh N, Shin JY, Gurudu SR, Hurst RT, Kendall CB, Gotway MB, Liang J (2016) Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Trans Med Imaging 35(5): Twinanda AP, Shehata S, Mutter D, Marescaux J, de Mathelin M, Padoy N (2017) Endonet: a deep architecture for recognition tasks on laparoscopic videos. IEEE Trans Med Imaging 36(1): Wan S, Sun S, Bhattacharya S, Kluckner S, Gigler A, Simon E, Fleischer M, Charalampaki P, Chen T, Kamen A (2015) Towards an efficient computational framework for guiding surgical resection through intra-operative endo-microscopic pathology. In: Medical image computing and computer-assisted intervention (MICCAI), pp Wang S, Yao J, Xu Z, Huang J (2016) Subtype cell detection with an accelerated deep convolution neural network. In: Medical image computing and computer-assisted intervention (MICCAI), pp
CSE Introduction to High-Perfomance Deep Learning ImageNet & VGG. Jihyung Kil
 CSE 5194.01 - Introduction to High-Perfomance Deep Learning ImageNet & VGG Jihyung Kil ImageNet Classification with Deep Convolutional Neural Networks Alex Krizhevsky, Ilya Sutskever, Geoffrey E. Hinton,
CSE 5194.01 - Introduction to High-Perfomance Deep Learning ImageNet & VGG Jihyung Kil ImageNet Classification with Deep Convolutional Neural Networks Alex Krizhevsky, Ilya Sutskever, Geoffrey E. Hinton,
arxiv: v1 [stat.ml] 23 Jan 2017
![arxiv: v1 [stat.ml] 23 Jan 2017 arxiv: v1 [stat.ml] 23 Jan 2017](/thumbs/81/83414183.jpg) Learning what to look in chest X-rays with a recurrent visual attention model arxiv:1701.06452v1 [stat.ml] 23 Jan 2017 Petros-Pavlos Ypsilantis Department of Biomedical Engineering King s College London
Learning what to look in chest X-rays with a recurrent visual attention model arxiv:1701.06452v1 [stat.ml] 23 Jan 2017 Petros-Pavlos Ypsilantis Department of Biomedical Engineering King s College London
HHS Public Access Author manuscript Med Image Comput Comput Assist Interv. Author manuscript; available in PMC 2018 January 04.
 Discriminative Localization in CNNs for Weakly-Supervised Segmentation of Pulmonary Nodules Xinyang Feng 1, Jie Yang 1, Andrew F. Laine 1, and Elsa D. Angelini 1,2 1 Department of Biomedical Engineering,
Discriminative Localization in CNNs for Weakly-Supervised Segmentation of Pulmonary Nodules Xinyang Feng 1, Jie Yang 1, Andrew F. Laine 1, and Elsa D. Angelini 1,2 1 Department of Biomedical Engineering,
Differentiating Tumor and Edema in Brain Magnetic Resonance Images Using a Convolutional Neural Network
 Original Article Differentiating Tumor and Edema in Brain Magnetic Resonance Images Using a Convolutional Neural Network Aida Allahverdi 1, Siavash Akbarzadeh 1, Alireza Khorrami Moghaddam 2, Armin Allahverdy
Original Article Differentiating Tumor and Edema in Brain Magnetic Resonance Images Using a Convolutional Neural Network Aida Allahverdi 1, Siavash Akbarzadeh 1, Alireza Khorrami Moghaddam 2, Armin Allahverdy
3D Deep Learning for Multi-modal Imaging-Guided Survival Time Prediction of Brain Tumor Patients
 3D Deep Learning for Multi-modal Imaging-Guided Survival Time Prediction of Brain Tumor Patients Dong Nie 1,2, Han Zhang 1, Ehsan Adeli 1, Luyan Liu 1, and Dinggang Shen 1(B) 1 Department of Radiology
3D Deep Learning for Multi-modal Imaging-Guided Survival Time Prediction of Brain Tumor Patients Dong Nie 1,2, Han Zhang 1, Ehsan Adeli 1, Luyan Liu 1, and Dinggang Shen 1(B) 1 Department of Radiology
Efficient Deep Model Selection
 Efficient Deep Model Selection Jose Alvarez Researcher Data61, CSIRO, Australia GTC, May 9 th 2017 www.josemalvarez.net conv1 conv2 conv3 conv4 conv5 conv6 conv7 conv8 softmax prediction???????? Num Classes
Efficient Deep Model Selection Jose Alvarez Researcher Data61, CSIRO, Australia GTC, May 9 th 2017 www.josemalvarez.net conv1 conv2 conv3 conv4 conv5 conv6 conv7 conv8 softmax prediction???????? Num Classes
arxiv: v2 [cs.cv] 19 Dec 2017
![arxiv: v2 [cs.cv] 19 Dec 2017 arxiv: v2 [cs.cv] 19 Dec 2017](/thumbs/89/97557782.jpg) An Ensemble of Deep Convolutional Neural Networks for Alzheimer s Disease Detection and Classification arxiv:1712.01675v2 [cs.cv] 19 Dec 2017 Jyoti Islam Department of Computer Science Georgia State University
An Ensemble of Deep Convolutional Neural Networks for Alzheimer s Disease Detection and Classification arxiv:1712.01675v2 [cs.cv] 19 Dec 2017 Jyoti Islam Department of Computer Science Georgia State University
Hierarchical Convolutional Features for Visual Tracking
 Hierarchical Convolutional Features for Visual Tracking Chao Ma Jia-Bin Huang Xiaokang Yang Ming-Husan Yang SJTU UIUC SJTU UC Merced ICCV 2015 Background Given the initial state (position and scale), estimate
Hierarchical Convolutional Features for Visual Tracking Chao Ma Jia-Bin Huang Xiaokang Yang Ming-Husan Yang SJTU UIUC SJTU UC Merced ICCV 2015 Background Given the initial state (position and scale), estimate
Multi-attention Guided Activation Propagation in CNNs
 Multi-attention Guided Activation Propagation in CNNs Xiangteng He and Yuxin Peng (B) Institute of Computer Science and Technology, Peking University, Beijing, China pengyuxin@pku.edu.cn Abstract. CNNs
Multi-attention Guided Activation Propagation in CNNs Xiangteng He and Yuxin Peng (B) Institute of Computer Science and Technology, Peking University, Beijing, China pengyuxin@pku.edu.cn Abstract. CNNs
Classification of breast cancer histology images using transfer learning
 Classification of breast cancer histology images using transfer learning Sulaiman Vesal 1 ( ), Nishant Ravikumar 1, AmirAbbas Davari 1, Stephan Ellmann 2, Andreas Maier 1 1 Pattern Recognition Lab, Friedrich-Alexander-Universität
Classification of breast cancer histology images using transfer learning Sulaiman Vesal 1 ( ), Nishant Ravikumar 1, AmirAbbas Davari 1, Stephan Ellmann 2, Andreas Maier 1 1 Pattern Recognition Lab, Friedrich-Alexander-Universität
Rich feature hierarchies for accurate object detection and semantic segmentation
 Rich feature hierarchies for accurate object detection and semantic segmentation Ross Girshick, Jeff Donahue, Trevor Darrell, Jitendra Malik UC Berkeley Tech Report @ http://arxiv.org/abs/1311.2524! Detection
Rich feature hierarchies for accurate object detection and semantic segmentation Ross Girshick, Jeff Donahue, Trevor Darrell, Jitendra Malik UC Berkeley Tech Report @ http://arxiv.org/abs/1311.2524! Detection
Object Detectors Emerge in Deep Scene CNNs
 Object Detectors Emerge in Deep Scene CNNs Bolei Zhou, Aditya Khosla, Agata Lapedriza, Aude Oliva, Antonio Torralba Presented By: Collin McCarthy Goal: Understand how objects are represented in CNNs Are
Object Detectors Emerge in Deep Scene CNNs Bolei Zhou, Aditya Khosla, Agata Lapedriza, Aude Oliva, Antonio Torralba Presented By: Collin McCarthy Goal: Understand how objects are represented in CNNs Are
Automated diagnosis of pneumothorax using an ensemble of convolutional neural networks with multi-sized chest radiography images
 Automated diagnosis of pneumothorax using an ensemble of convolutional neural networks with multi-sized chest radiography images Tae Joon Jun, Dohyeun Kim, and Daeyoung Kim School of Computing, KAIST,
Automated diagnosis of pneumothorax using an ensemble of convolutional neural networks with multi-sized chest radiography images Tae Joon Jun, Dohyeun Kim, and Daeyoung Kim School of Computing, KAIST,
Convolutional Neural Networks (CNN)
 Convolutional Neural Networks (CNN) Algorithm and Some Applications in Computer Vision Luo Hengliang Institute of Automation June 10, 2014 Luo Hengliang (Institute of Automation) Convolutional Neural Networks
Convolutional Neural Networks (CNN) Algorithm and Some Applications in Computer Vision Luo Hengliang Institute of Automation June 10, 2014 Luo Hengliang (Institute of Automation) Convolutional Neural Networks
Facial Expression Classification Using Convolutional Neural Network and Support Vector Machine
 Facial Expression Classification Using Convolutional Neural Network and Support Vector Machine Valfredo Pilla Jr, André Zanellato, Cristian Bortolini, Humberto R. Gamba and Gustavo Benvenutti Borba Graduate
Facial Expression Classification Using Convolutional Neural Network and Support Vector Machine Valfredo Pilla Jr, André Zanellato, Cristian Bortolini, Humberto R. Gamba and Gustavo Benvenutti Borba Graduate
Computational modeling of visual attention and saliency in the Smart Playroom
 Computational modeling of visual attention and saliency in the Smart Playroom Andrew Jones Department of Computer Science, Brown University Abstract The two canonical modes of human visual attention bottomup
Computational modeling of visual attention and saliency in the Smart Playroom Andrew Jones Department of Computer Science, Brown University Abstract The two canonical modes of human visual attention bottomup
arxiv: v1 [cs.cv] 17 Aug 2017
![arxiv: v1 [cs.cv] 17 Aug 2017 arxiv: v1 [cs.cv] 17 Aug 2017](/thumbs/78/78278805.jpg) Deep Learning for Medical Image Analysis Mina Rezaei, Haojin Yang, Christoph Meinel Hasso Plattner Institute, Prof.Dr.Helmert-Strae 2-3, 14482 Potsdam, Germany {mina.rezaei,haojin.yang,christoph.meinel}@hpi.de
Deep Learning for Medical Image Analysis Mina Rezaei, Haojin Yang, Christoph Meinel Hasso Plattner Institute, Prof.Dr.Helmert-Strae 2-3, 14482 Potsdam, Germany {mina.rezaei,haojin.yang,christoph.meinel}@hpi.de
Shu Kong. Department of Computer Science, UC Irvine
 Ubiquitous Fine-Grained Computer Vision Shu Kong Department of Computer Science, UC Irvine Outline 1. Problem definition 2. Instantiation 3. Challenge 4. Fine-grained classification with holistic representation
Ubiquitous Fine-Grained Computer Vision Shu Kong Department of Computer Science, UC Irvine Outline 1. Problem definition 2. Instantiation 3. Challenge 4. Fine-grained classification with holistic representation
A HMM-based Pre-training Approach for Sequential Data
 A HMM-based Pre-training Approach for Sequential Data Luca Pasa 1, Alberto Testolin 2, Alessandro Sperduti 1 1- Department of Mathematics 2- Department of Developmental Psychology and Socialisation University
A HMM-based Pre-training Approach for Sequential Data Luca Pasa 1, Alberto Testolin 2, Alessandro Sperduti 1 1- Department of Mathematics 2- Department of Developmental Psychology and Socialisation University
Quantifying Radiographic Knee Osteoarthritis Severity using Deep Convolutional Neural Networks
 Quantifying Radiographic Knee Osteoarthritis Severity using Deep Convolutional Neural Networks Joseph Antony, Kevin McGuinness, Noel E O Connor, Kieran Moran Insight Centre for Data Analytics, Dublin City
Quantifying Radiographic Knee Osteoarthritis Severity using Deep Convolutional Neural Networks Joseph Antony, Kevin McGuinness, Noel E O Connor, Kieran Moran Insight Centre for Data Analytics, Dublin City
A CONVOLUTION NEURAL NETWORK ALGORITHM FOR BRAIN TUMOR IMAGE SEGMENTATION
 A CONVOLUTION NEURAL NETWORK ALGORITHM FOR BRAIN TUMOR IMAGE SEGMENTATION 1 Priya K, 2 Dr.O. Saraniya 1 PG Scholar, 2 Assistant Professor Department Of ECE Government College of Technology, Coimbatore,
A CONVOLUTION NEURAL NETWORK ALGORITHM FOR BRAIN TUMOR IMAGE SEGMENTATION 1 Priya K, 2 Dr.O. Saraniya 1 PG Scholar, 2 Assistant Professor Department Of ECE Government College of Technology, Coimbatore,
Shu Kong. Department of Computer Science, UC Irvine
 Ubiquitous Fine-Grained Computer Vision Shu Kong Department of Computer Science, UC Irvine Outline 1. Problem definition 2. Instantiation 3. Challenge and philosophy 4. Fine-grained classification with
Ubiquitous Fine-Grained Computer Vision Shu Kong Department of Computer Science, UC Irvine Outline 1. Problem definition 2. Instantiation 3. Challenge and philosophy 4. Fine-grained classification with
Beyond R-CNN detection: Learning to Merge Contextual Attribute
 Brain Unleashing Series - Beyond R-CNN detection: Learning to Merge Contextual Attribute Shu Kong CS, ICS, UCI 2015-1-29 Outline 1. RCNN is essentially doing classification, without considering contextual
Brain Unleashing Series - Beyond R-CNN detection: Learning to Merge Contextual Attribute Shu Kong CS, ICS, UCI 2015-1-29 Outline 1. RCNN is essentially doing classification, without considering contextual
Chair for Computer Aided Medical Procedures (CAMP) Seminar on Deep Learning for Medical Applications. Shadi Albarqouni Christoph Baur
 Chair for (CAMP) Seminar on Deep Learning for Medical Applications Shadi Albarqouni Christoph Baur Results of matching system obtained via matching.in.tum.de 108 Applicants 9 % 10 % 9 % 14 % 30 % Rank
Chair for (CAMP) Seminar on Deep Learning for Medical Applications Shadi Albarqouni Christoph Baur Results of matching system obtained via matching.in.tum.de 108 Applicants 9 % 10 % 9 % 14 % 30 % Rank
Y-Net: Joint Segmentation and Classification for Diagnosis of Breast Biopsy Images
 Y-Net: Joint Segmentation and Classification for Diagnosis of Breast Biopsy Images Sachin Mehta 1, Ezgi Mercan 1, Jamen Bartlett 2, Donald Weaver 2, Joann G. Elmore 1, and Linda Shapiro 1 1 University
Y-Net: Joint Segmentation and Classification for Diagnosis of Breast Biopsy Images Sachin Mehta 1, Ezgi Mercan 1, Jamen Bartlett 2, Donald Weaver 2, Joann G. Elmore 1, and Linda Shapiro 1 1 University
Highly Accurate Brain Stroke Diagnostic System and Generative Lesion Model. Junghwan Cho, Ph.D. CAIDE Systems, Inc. Deep Learning R&D Team
 Highly Accurate Brain Stroke Diagnostic System and Generative Lesion Model Junghwan Cho, Ph.D. CAIDE Systems, Inc. Deep Learning R&D Team Established in September, 2016 at 110 Canal st. Lowell, MA 01852,
Highly Accurate Brain Stroke Diagnostic System and Generative Lesion Model Junghwan Cho, Ph.D. CAIDE Systems, Inc. Deep Learning R&D Team Established in September, 2016 at 110 Canal st. Lowell, MA 01852,
DEEP CONVOLUTIONAL ACTIVATION FEATURES FOR LARGE SCALE BRAIN TUMOR HISTOPATHOLOGY IMAGE CLASSIFICATION AND SEGMENTATION
 DEEP CONVOLUTIONAL ACTIVATION FEATURES FOR LARGE SCALE BRAIN TUMOR HISTOPATHOLOGY IMAGE CLASSIFICATION AND SEGMENTATION Yan Xu1,2, Zhipeng Jia2,, Yuqing Ai2,, Fang Zhang2,, Maode Lai4, Eric I-Chao Chang2
DEEP CONVOLUTIONAL ACTIVATION FEATURES FOR LARGE SCALE BRAIN TUMOR HISTOPATHOLOGY IMAGE CLASSIFICATION AND SEGMENTATION Yan Xu1,2, Zhipeng Jia2,, Yuqing Ai2,, Fang Zhang2,, Maode Lai4, Eric I-Chao Chang2
Deep CNNs for Diabetic Retinopathy Detection
 Deep CNNs for Diabetic Retinopathy Detection Alex Tamkin Stanford University atamkin@stanford.edu Iain Usiri Stanford University iusiri@stanford.edu Chala Fufa Stanford University cfufa@stanford.edu 1
Deep CNNs for Diabetic Retinopathy Detection Alex Tamkin Stanford University atamkin@stanford.edu Iain Usiri Stanford University iusiri@stanford.edu Chala Fufa Stanford University cfufa@stanford.edu 1
Comparison of Two Approaches for Direct Food Calorie Estimation
 Comparison of Two Approaches for Direct Food Calorie Estimation Takumi Ege and Keiji Yanai Department of Informatics, The University of Electro-Communications, Tokyo 1-5-1 Chofugaoka, Chofu-shi, Tokyo
Comparison of Two Approaches for Direct Food Calorie Estimation Takumi Ege and Keiji Yanai Department of Informatics, The University of Electro-Communications, Tokyo 1-5-1 Chofugaoka, Chofu-shi, Tokyo
Automatic Detection of Knee Joints and Quantification of Knee Osteoarthritis Severity using Convolutional Neural Networks
 Automatic Detection of Knee Joints and Quantification of Knee Osteoarthritis Severity using Convolutional Neural Networks Joseph Antony 1, Kevin McGuinness 1, Kieran Moran 1,2 and Noel E O Connor 1 Insight
Automatic Detection of Knee Joints and Quantification of Knee Osteoarthritis Severity using Convolutional Neural Networks Joseph Antony 1, Kevin McGuinness 1, Kieran Moran 1,2 and Noel E O Connor 1 Insight
arxiv: v2 [cs.cv] 29 Jan 2019
![arxiv: v2 [cs.cv] 29 Jan 2019 arxiv: v2 [cs.cv] 29 Jan 2019](/thumbs/88/117548796.jpg) Comparison of Deep Learning Approaches for Multi-Label Chest X-Ray Classification Ivo M. Baltruschat 1,2,, Hannes Nickisch 3, Michael Grass 3, Tobias Knopp 1,2, and Axel Saalbach 3 arxiv:1803.02315v2 [cs.cv]
Comparison of Deep Learning Approaches for Multi-Label Chest X-Ray Classification Ivo M. Baltruschat 1,2,, Hannes Nickisch 3, Michael Grass 3, Tobias Knopp 1,2, and Axel Saalbach 3 arxiv:1803.02315v2 [cs.cv]
arxiv: v1 [cs.cv] 13 Jul 2018
![arxiv: v1 [cs.cv] 13 Jul 2018 arxiv: v1 [cs.cv] 13 Jul 2018](/thumbs/84/89699669.jpg) Multi-Scale Convolutional-Stack Aggregation for Robust White Matter Hyperintensities Segmentation Hongwei Li 1, Jianguo Zhang 3, Mark Muehlau 2, Jan Kirschke 2, and Bjoern Menze 1 arxiv:1807.05153v1 [cs.cv]
Multi-Scale Convolutional-Stack Aggregation for Robust White Matter Hyperintensities Segmentation Hongwei Li 1, Jianguo Zhang 3, Mark Muehlau 2, Jan Kirschke 2, and Bjoern Menze 1 arxiv:1807.05153v1 [cs.cv]
Motivation: Attention: Focusing on specific parts of the input. Inspired by neuroscience.
 Outline: Motivation. What s the attention mechanism? Soft attention vs. Hard attention. Attention in Machine translation. Attention in Image captioning. State-of-the-art. 1 Motivation: Attention: Focusing
Outline: Motivation. What s the attention mechanism? Soft attention vs. Hard attention. Attention in Machine translation. Attention in Image captioning. State-of-the-art. 1 Motivation: Attention: Focusing
arxiv: v2 [cs.cv] 22 Mar 2018
![arxiv: v2 [cs.cv] 22 Mar 2018 arxiv: v2 [cs.cv] 22 Mar 2018](/thumbs/94/122062874.jpg) Deep saliency: What is learnt by a deep network about saliency? Sen He 1 Nicolas Pugeault 1 arxiv:1801.04261v2 [cs.cv] 22 Mar 2018 Abstract Deep convolutional neural networks have achieved impressive performance
Deep saliency: What is learnt by a deep network about saliency? Sen He 1 Nicolas Pugeault 1 arxiv:1801.04261v2 [cs.cv] 22 Mar 2018 Abstract Deep convolutional neural networks have achieved impressive performance
A Reliable Method for Brain Tumor Detection Using Cnn Technique
 IOSR Journal of Electrical and Electronics Engineering (IOSR-JEEE) e-issn: 2278-1676,p-ISSN: 2320-3331, PP 64-68 www.iosrjournals.org A Reliable Method for Brain Tumor Detection Using Cnn Technique Neethu
IOSR Journal of Electrical and Electronics Engineering (IOSR-JEEE) e-issn: 2278-1676,p-ISSN: 2320-3331, PP 64-68 www.iosrjournals.org A Reliable Method for Brain Tumor Detection Using Cnn Technique Neethu
Automatic Diagnosis of Ovarian Carcinomas via Sparse Multiresolution Tissue Representation
 Automatic Diagnosis of Ovarian Carcinomas via Sparse Multiresolution Tissue Representation Aïcha BenTaieb, Hector Li-Chang, David Huntsman, Ghassan Hamarneh Medical Image Analysis Lab, Simon Fraser University,
Automatic Diagnosis of Ovarian Carcinomas via Sparse Multiresolution Tissue Representation Aïcha BenTaieb, Hector Li-Chang, David Huntsman, Ghassan Hamarneh Medical Image Analysis Lab, Simon Fraser University,
COMP9444 Neural Networks and Deep Learning 5. Convolutional Networks
 COMP9444 Neural Networks and Deep Learning 5. Convolutional Networks Textbook, Sections 6.2.2, 6.3, 7.9, 7.11-7.13, 9.1-9.5 COMP9444 17s2 Convolutional Networks 1 Outline Geometry of Hidden Unit Activations
COMP9444 Neural Networks and Deep Learning 5. Convolutional Networks Textbook, Sections 6.2.2, 6.3, 7.9, 7.11-7.13, 9.1-9.5 COMP9444 17s2 Convolutional Networks 1 Outline Geometry of Hidden Unit Activations
Medical Image Analysis
 Medical Image Analysis 1 Co-trained convolutional neural networks for automated detection of prostate cancer in multiparametric MRI, 2017, Medical Image Analysis 2 Graph-based prostate extraction in t2-weighted
Medical Image Analysis 1 Co-trained convolutional neural networks for automated detection of prostate cancer in multiparametric MRI, 2017, Medical Image Analysis 2 Graph-based prostate extraction in t2-weighted
arxiv: v1 [cs.cv] 2 Jun 2017
![arxiv: v1 [cs.cv] 2 Jun 2017 arxiv: v1 [cs.cv] 2 Jun 2017](/thumbs/74/71415505.jpg) INTEGRATED DEEP AND SHALLOW NETWORKS FOR SALIENT OBJECT DETECTION Jing Zhang 1,2, Bo Li 1, Yuchao Dai 2, Fatih Porikli 2 and Mingyi He 1 1 School of Electronics and Information, Northwestern Polytechnical
INTEGRATED DEEP AND SHALLOW NETWORKS FOR SALIENT OBJECT DETECTION Jing Zhang 1,2, Bo Li 1, Yuchao Dai 2, Fatih Porikli 2 and Mingyi He 1 1 School of Electronics and Information, Northwestern Polytechnical
arxiv: v2 [cs.cv] 8 Mar 2018
![arxiv: v2 [cs.cv] 8 Mar 2018 arxiv: v2 [cs.cv] 8 Mar 2018](/thumbs/87/97094636.jpg) Automated soft tissue lesion detection and segmentation in digital mammography using a u-net deep learning network Timothy de Moor a, Alejandro Rodriguez-Ruiz a, Albert Gubern Mérida a, Ritse Mann a, and
Automated soft tissue lesion detection and segmentation in digital mammography using a u-net deep learning network Timothy de Moor a, Alejandro Rodriguez-Ruiz a, Albert Gubern Mérida a, Ritse Mann a, and
arxiv: v1 [cs.cv] 21 Jul 2017
![arxiv: v1 [cs.cv] 21 Jul 2017 arxiv: v1 [cs.cv] 21 Jul 2017](/thumbs/89/100163556.jpg) A Multi-Scale CNN and Curriculum Learning Strategy for Mammogram Classification William Lotter 1,2, Greg Sorensen 2, and David Cox 1,2 1 Harvard University, Cambridge MA, USA 2 DeepHealth Inc., Cambridge
A Multi-Scale CNN and Curriculum Learning Strategy for Mammogram Classification William Lotter 1,2, Greg Sorensen 2, and David Cox 1,2 1 Harvard University, Cambridge MA, USA 2 DeepHealth Inc., Cambridge
arxiv: v3 [cs.cv] 28 Mar 2017
![arxiv: v3 [cs.cv] 28 Mar 2017 arxiv: v3 [cs.cv] 28 Mar 2017](/thumbs/73/69121210.jpg) Discovery Radiomics for Pathologically-Proven Computed Tomography Lung Cancer Prediction Devinder Kumar 1*, Audrey G. Chung 1, Mohammad J. Shaifee 1, Farzad Khalvati 2,3, Masoom A. Haider 2,3, and Alexander
Discovery Radiomics for Pathologically-Proven Computed Tomography Lung Cancer Prediction Devinder Kumar 1*, Audrey G. Chung 1, Mohammad J. Shaifee 1, Farzad Khalvati 2,3, Masoom A. Haider 2,3, and Alexander
Hierarchical Approach for Breast Cancer Histopathology Images Classification
 Hierarchical Approach for Breast Cancer Histopathology Images Classification Nidhi Ranjan, Pranav Vinod Machingal, Sunil Sri Datta Jammalmadka, Veena Thenkanidiyoor Department of Computer Science and Engineering
Hierarchical Approach for Breast Cancer Histopathology Images Classification Nidhi Ranjan, Pranav Vinod Machingal, Sunil Sri Datta Jammalmadka, Veena Thenkanidiyoor Department of Computer Science and Engineering
DIAGNOSTIC CLASSIFICATION OF LUNG NODULES USING 3D NEURAL NETWORKS
 DIAGNOSTIC CLASSIFICATION OF LUNG NODULES USING 3D NEURAL NETWORKS Raunak Dey Zhongjie Lu Yi Hong Department of Computer Science, University of Georgia, Athens, GA, USA First Affiliated Hospital, School
DIAGNOSTIC CLASSIFICATION OF LUNG NODULES USING 3D NEURAL NETWORKS Raunak Dey Zhongjie Lu Yi Hong Department of Computer Science, University of Georgia, Athens, GA, USA First Affiliated Hospital, School
Research Article Multiscale CNNs for Brain Tumor Segmentation and Diagnosis
 Computational and Mathematical Methods in Medicine Volume 2016, Article ID 8356294, 7 pages http://dx.doi.org/10.1155/2016/8356294 Research Article Multiscale CNNs for Brain Tumor Segmentation and Diagnosis
Computational and Mathematical Methods in Medicine Volume 2016, Article ID 8356294, 7 pages http://dx.doi.org/10.1155/2016/8356294 Research Article Multiscale CNNs for Brain Tumor Segmentation and Diagnosis
arxiv: v1 [cs.cv] 9 Oct 2018
![arxiv: v1 [cs.cv] 9 Oct 2018 arxiv: v1 [cs.cv] 9 Oct 2018](/thumbs/95/126101195.jpg) Automatic Segmentation of Thoracic Aorta Segments in Low-Dose Chest CT Julia M. H. Noothout a, Bob D. de Vos a, Jelmer M. Wolterink a, Ivana Išgum a a Image Sciences Institute, University Medical Center
Automatic Segmentation of Thoracic Aorta Segments in Low-Dose Chest CT Julia M. H. Noothout a, Bob D. de Vos a, Jelmer M. Wolterink a, Ivana Išgum a a Image Sciences Institute, University Medical Center
arxiv: v1 [cs.cv] 26 Feb 2018
![arxiv: v1 [cs.cv] 26 Feb 2018 arxiv: v1 [cs.cv] 26 Feb 2018](/thumbs/95/125817543.jpg) Classification of breast cancer histology images using transfer learning Sulaiman Vesal 1, Nishant Ravikumar 1, AmirAbbas Davari 1, Stephan Ellmann 2, Andreas Maier 1 arxiv:1802.09424v1 [cs.cv] 26 Feb
Classification of breast cancer histology images using transfer learning Sulaiman Vesal 1, Nishant Ravikumar 1, AmirAbbas Davari 1, Stephan Ellmann 2, Andreas Maier 1 arxiv:1802.09424v1 [cs.cv] 26 Feb
arxiv: v1 [cs.cv] 15 Aug 2018
![arxiv: v1 [cs.cv] 15 Aug 2018 arxiv: v1 [cs.cv] 15 Aug 2018](/thumbs/83/87651811.jpg) Ensemble of Convolutional Neural Networks for Dermoscopic Images Classification Tomáš Majtner 1, Buda Bajić 2, Sule Yildirim 3, Jon Yngve Hardeberg 3, Joakim Lindblad 4,5 and Nataša Sladoje 4,5 arxiv:1808.05071v1
Ensemble of Convolutional Neural Networks for Dermoscopic Images Classification Tomáš Majtner 1, Buda Bajić 2, Sule Yildirim 3, Jon Yngve Hardeberg 3, Joakim Lindblad 4,5 and Nataša Sladoje 4,5 arxiv:1808.05071v1
Lung Nodule Segmentation Using 3D Convolutional Neural Networks
 Lung Nodule Segmentation Using 3D Convolutional Neural Networks Research paper Business Analytics Bernard Bronmans Master Business Analytics VU University, Amsterdam Evert Haasdijk Supervisor VU University,
Lung Nodule Segmentation Using 3D Convolutional Neural Networks Research paper Business Analytics Bernard Bronmans Master Business Analytics VU University, Amsterdam Evert Haasdijk Supervisor VU University,
Early Diagnosis of Autism Disease by Multi-channel CNNs
 Early Diagnosis of Autism Disease by Multi-channel CNNs Guannan Li 1,2, Mingxia Liu 2, Quansen Sun 1(&), Dinggang Shen 2(&), and Li Wang 2(&) 1 School of Computer Science and Engineering, Nanjing University
Early Diagnosis of Autism Disease by Multi-channel CNNs Guannan Li 1,2, Mingxia Liu 2, Quansen Sun 1(&), Dinggang Shen 2(&), and Li Wang 2(&) 1 School of Computer Science and Engineering, Nanjing University
End-To-End Alzheimer s Disease Diagnosis and Biomarker Identification
 End-To-End Alzheimer s Disease Diagnosis and Biomarker Identification Soheil Esmaeilzadeh 1, Dimitrios Ioannis Belivanis 1, Kilian M. Pohl 2, and Ehsan Adeli 1 1 Stanford University 2 SRI International
End-To-End Alzheimer s Disease Diagnosis and Biomarker Identification Soheil Esmaeilzadeh 1, Dimitrios Ioannis Belivanis 1, Kilian M. Pohl 2, and Ehsan Adeli 1 1 Stanford University 2 SRI International
Visual interpretation in pathology
 13 Visual interpretation in pathology Tissue architecture (alteration) evaluation e.g., for grading prostate cancer Immunohistochemistry (IHC) staining scoring e.g., HER2 in breast cancer (companion diagnostic
13 Visual interpretation in pathology Tissue architecture (alteration) evaluation e.g., for grading prostate cancer Immunohistochemistry (IHC) staining scoring e.g., HER2 in breast cancer (companion diagnostic
CS231n Project: Prediction of Head and Neck Cancer Submolecular Types from Patholoy Images
 CSn Project: Prediction of Head and Neck Cancer Submolecular Types from Patholoy Images Kuy Hun Koh Yoo Energy Resources Engineering Stanford University Stanford, CA 90 kohykh@stanford.edu Markus Zechner
CSn Project: Prediction of Head and Neck Cancer Submolecular Types from Patholoy Images Kuy Hun Koh Yoo Energy Resources Engineering Stanford University Stanford, CA 90 kohykh@stanford.edu Markus Zechner
Network Dissection: Quantifying Interpretability of Deep Visual Representation
 Name: Pingchuan Ma Student number: 3526400 Date: August 19, 2018 Seminar: Explainable Machine Learning Lecturer: PD Dr. Ullrich Köthe SS 2018 Quantifying Interpretability of Deep Visual Representation
Name: Pingchuan Ma Student number: 3526400 Date: August 19, 2018 Seminar: Explainable Machine Learning Lecturer: PD Dr. Ullrich Köthe SS 2018 Quantifying Interpretability of Deep Visual Representation
Convolutional Neural Networks for Estimating Left Ventricular Volume
 Convolutional Neural Networks for Estimating Left Ventricular Volume Ryan Silva Stanford University rdsilva@stanford.edu Maksim Korolev Stanford University mkorolev@stanford.edu Abstract End-systolic and
Convolutional Neural Networks for Estimating Left Ventricular Volume Ryan Silva Stanford University rdsilva@stanford.edu Maksim Korolev Stanford University mkorolev@stanford.edu Abstract End-systolic and
Discovery of Rare Phenotypes in Cellular Images Using Weakly Supervised Deep Learning
 Discovery of Rare Phenotypes in Cellular Images Using Weakly Supervised Deep Learning Heba Sailem *1, Mar Arias Garcia 2, Chris Bakal 2, Andrew Zisserman 1, and Jens Rittscher 1 1 Department of Engineering
Discovery of Rare Phenotypes in Cellular Images Using Weakly Supervised Deep Learning Heba Sailem *1, Mar Arias Garcia 2, Chris Bakal 2, Andrew Zisserman 1, and Jens Rittscher 1 1 Department of Engineering
Comparative Study of K-means, Gaussian Mixture Model, Fuzzy C-means algorithms for Brain Tumor Segmentation
 Comparative Study of K-means, Gaussian Mixture Model, Fuzzy C-means algorithms for Brain Tumor Segmentation U. Baid 1, S. Talbar 2 and S. Talbar 1 1 Department of E&TC Engineering, Shri Guru Gobind Singhji
Comparative Study of K-means, Gaussian Mixture Model, Fuzzy C-means algorithms for Brain Tumor Segmentation U. Baid 1, S. Talbar 2 and S. Talbar 1 1 Department of E&TC Engineering, Shri Guru Gobind Singhji
A deep learning model integrating FCNNs and CRFs for brain tumor segmentation
 A deep learning model integrating FCNNs and CRFs for brain tumor segmentation Xiaomei Zhao 1,2, Yihong Wu 1, Guidong Song 3, Zhenye Li 4, Yazhuo Zhang,3,4,5,6, and Yong Fan 7 1 National Laboratory of Pattern
A deep learning model integrating FCNNs and CRFs for brain tumor segmentation Xiaomei Zhao 1,2, Yihong Wu 1, Guidong Song 3, Zhenye Li 4, Yazhuo Zhang,3,4,5,6, and Yong Fan 7 1 National Laboratory of Pattern
Deep Supervision for Pancreatic Cyst Segmentation in Abdominal CT Scans
 Deep Supervision for Pancreatic Cyst Segmentation in Abdominal CT Scans Yuyin Zhou 1, Lingxi Xie 2( ), Elliot K. Fishman 3, Alan L. Yuille 4 1,2,4 The Johns Hopkins University, Baltimore, MD 21218, USA
Deep Supervision for Pancreatic Cyst Segmentation in Abdominal CT Scans Yuyin Zhou 1, Lingxi Xie 2( ), Elliot K. Fishman 3, Alan L. Yuille 4 1,2,4 The Johns Hopkins University, Baltimore, MD 21218, USA
Recurrent Fully Convolutional Neural Networks for Multi-slice MRI Cardiac Segmentation
 Recurrent Fully Convolutional Neural Networks for Multi-slice MRI Cardiac Segmentation Rudra P K Poudel, Pablo Lamata and Giovanni Montana Department of Biomedical Engineering, King s College London, SE1
Recurrent Fully Convolutional Neural Networks for Multi-slice MRI Cardiac Segmentation Rudra P K Poudel, Pablo Lamata and Giovanni Montana Department of Biomedical Engineering, King s College London, SE1
arxiv: v2 [cs.cv] 3 Jun 2018
![arxiv: v2 [cs.cv] 3 Jun 2018 arxiv: v2 [cs.cv] 3 Jun 2018](/thumbs/89/97775247.jpg) S4ND: Single-Shot Single-Scale Lung Nodule Detection Naji Khosravan and Ulas Bagci Center for Research in Computer Vision (CRCV), School of Computer Science, University of Central Florida, Orlando, FL.
S4ND: Single-Shot Single-Scale Lung Nodule Detection Naji Khosravan and Ulas Bagci Center for Research in Computer Vision (CRCV), School of Computer Science, University of Central Florida, Orlando, FL.
Deep Learning of Brain Lesion Patterns for Predicting Future Disease Activity in Patients with Early Symptoms of Multiple Sclerosis
 Deep Learning of Brain Lesion Patterns for Predicting Future Disease Activity in Patients with Early Symptoms of Multiple Sclerosis Youngjin Yoo 1,2,5(B),LisaW.Tang 2,3,5, Tom Brosch 1,2,5,DavidK.B.Li
Deep Learning of Brain Lesion Patterns for Predicting Future Disease Activity in Patients with Early Symptoms of Multiple Sclerosis Youngjin Yoo 1,2,5(B),LisaW.Tang 2,3,5, Tom Brosch 1,2,5,DavidK.B.Li
The Impact of Visual Saliency Prediction in Image Classification
 Dublin City University Insight Centre for Data Analytics Universitat Politecnica de Catalunya Escola Tècnica Superior d Enginyeria de Telecomunicacions de Barcelona Eric Arazo Sánchez The Impact of Visual
Dublin City University Insight Centre for Data Analytics Universitat Politecnica de Catalunya Escola Tècnica Superior d Enginyeria de Telecomunicacions de Barcelona Eric Arazo Sánchez The Impact of Visual
Image Captioning using Reinforcement Learning. Presentation by: Samarth Gupta
 Image Captioning using Reinforcement Learning Presentation by: Samarth Gupta 1 Introduction Summary Supervised Models Image captioning as RL problem Actor Critic Architecture Policy Gradient architecture
Image Captioning using Reinforcement Learning Presentation by: Samarth Gupta 1 Introduction Summary Supervised Models Image captioning as RL problem Actor Critic Architecture Policy Gradient architecture
NMF-Density: NMF-Based Breast Density Classifier
 NMF-Density: NMF-Based Breast Density Classifier Lahouari Ghouti and Abdullah H. Owaidh King Fahd University of Petroleum and Minerals - Department of Information and Computer Science. KFUPM Box 1128.
NMF-Density: NMF-Based Breast Density Classifier Lahouari Ghouti and Abdullah H. Owaidh King Fahd University of Petroleum and Minerals - Department of Information and Computer Science. KFUPM Box 1128.
ANALYSIS AND DETECTION OF BRAIN TUMOUR USING IMAGE PROCESSING TECHNIQUES
 ANALYSIS AND DETECTION OF BRAIN TUMOUR USING IMAGE PROCESSING TECHNIQUES P.V.Rohini 1, Dr.M.Pushparani 2 1 M.Phil Scholar, Department of Computer Science, Mother Teresa women s university, (India) 2 Professor
ANALYSIS AND DETECTION OF BRAIN TUMOUR USING IMAGE PROCESSING TECHNIQUES P.V.Rohini 1, Dr.M.Pushparani 2 1 M.Phil Scholar, Department of Computer Science, Mother Teresa women s university, (India) 2 Professor
Flexible, High Performance Convolutional Neural Networks for Image Classification
 Flexible, High Performance Convolutional Neural Networks for Image Classification Dan C. Cireşan, Ueli Meier, Jonathan Masci, Luca M. Gambardella, Jürgen Schmidhuber IDSIA, USI and SUPSI Manno-Lugano,
Flexible, High Performance Convolutional Neural Networks for Image Classification Dan C. Cireşan, Ueli Meier, Jonathan Masci, Luca M. Gambardella, Jürgen Schmidhuber IDSIA, USI and SUPSI Manno-Lugano,
arxiv: v1 [cs.cv] 30 May 2018
![arxiv: v1 [cs.cv] 30 May 2018 arxiv: v1 [cs.cv] 30 May 2018](/thumbs/85/92577534.jpg) A Robust and Effective Approach Towards Accurate Metastasis Detection and pn-stage Classification in Breast Cancer Byungjae Lee and Kyunghyun Paeng Lunit inc., Seoul, South Korea {jaylee,khpaeng}@lunit.io
A Robust and Effective Approach Towards Accurate Metastasis Detection and pn-stage Classification in Breast Cancer Byungjae Lee and Kyunghyun Paeng Lunit inc., Seoul, South Korea {jaylee,khpaeng}@lunit.io
Patch-based Head and Neck Cancer Subtype Classification
 Patch-based Head and Neck Cancer Subtype Classification Wanyi Qian, Guoli Yin, Frances Liu, Advisor: Olivier Gevaert, Mu Zhou, Kevin Brennan Stanford University wqian2@stanford.edu, guoliy@stanford.edu,
Patch-based Head and Neck Cancer Subtype Classification Wanyi Qian, Guoli Yin, Frances Liu, Advisor: Olivier Gevaert, Mu Zhou, Kevin Brennan Stanford University wqian2@stanford.edu, guoliy@stanford.edu,
Improving 3D Ultrasound Scan Adequacy Classification Using a Three-Slice Convolutional Neural Network Architecture
 EPiC Series in Health Sciences Volume 2, 2018, Pages 152 156 CAOS 2018. The 18th Annual Meeting of the International Society for Computer Assisted Orthopaedic Surgery Health Sciences Improving 3D Ultrasound
EPiC Series in Health Sciences Volume 2, 2018, Pages 152 156 CAOS 2018. The 18th Annual Meeting of the International Society for Computer Assisted Orthopaedic Surgery Health Sciences Improving 3D Ultrasound
POC Brain Tumor Segmentation. vlife Use Case
 Brain Tumor Segmentation vlife Use Case 1 Automatic Brain Tumor Segmentation using CNN Background Brain tumor segmentation seeks to separate healthy tissue from tumorous regions such as the advancing tumor,
Brain Tumor Segmentation vlife Use Case 1 Automatic Brain Tumor Segmentation using CNN Background Brain tumor segmentation seeks to separate healthy tissue from tumorous regions such as the advancing tumor,
Image-Based Estimation of Real Food Size for Accurate Food Calorie Estimation
 Image-Based Estimation of Real Food Size for Accurate Food Calorie Estimation Takumi Ege, Yoshikazu Ando, Ryosuke Tanno, Wataru Shimoda and Keiji Yanai Department of Informatics, The University of Electro-Communications,
Image-Based Estimation of Real Food Size for Accurate Food Calorie Estimation Takumi Ege, Yoshikazu Ando, Ryosuke Tanno, Wataru Shimoda and Keiji Yanai Department of Informatics, The University of Electro-Communications,
Synthesizing Missing PET from MRI with Cycle-consistent Generative Adversarial Networks for Alzheimer s Disease Diagnosis
 Synthesizing Missing PET from MRI with Cycle-consistent Generative Adversarial Networks for Alzheimer s Disease Diagnosis Yongsheng Pan 1,2, Mingxia Liu 2, Chunfeng Lian 2, Tao Zhou 2,YongXia 1(B), and
Synthesizing Missing PET from MRI with Cycle-consistent Generative Adversarial Networks for Alzheimer s Disease Diagnosis Yongsheng Pan 1,2, Mingxia Liu 2, Chunfeng Lian 2, Tao Zhou 2,YongXia 1(B), and
ARTIFICIAL INTELLIGENCE FOR DIGITAL PATHOLOGY. Kyunghyun Paeng, Co-founder and Research Scientist, Lunit Inc.
 ARTIFICIAL INTELLIGENCE FOR DIGITAL PATHOLOGY Kyunghyun Paeng, Co-founder and Research Scientist, Lunit Inc. 1. BACKGROUND: DIGITAL PATHOLOGY 2. APPLICATIONS AGENDA BREAST CANCER PROSTATE CANCER 3. DEMONSTRATIONS
ARTIFICIAL INTELLIGENCE FOR DIGITAL PATHOLOGY Kyunghyun Paeng, Co-founder and Research Scientist, Lunit Inc. 1. BACKGROUND: DIGITAL PATHOLOGY 2. APPLICATIONS AGENDA BREAST CANCER PROSTATE CANCER 3. DEMONSTRATIONS
Large-scale Histopathology Image Analysis for Colon Cancer on Azure
 Large-scale Histopathology Image Analysis for Colon Cancer on Azure Yan Xu 1, 2 Tao Mo 2 Teng Gao 2 Maode Lai 4 Zhuowen Tu 2,3 Eric I-Chao Chang 2 1 Beihang University; 2 Microsoft Research Asia; 3 UCSD;
Large-scale Histopathology Image Analysis for Colon Cancer on Azure Yan Xu 1, 2 Tao Mo 2 Teng Gao 2 Maode Lai 4 Zhuowen Tu 2,3 Eric I-Chao Chang 2 1 Beihang University; 2 Microsoft Research Asia; 3 UCSD;
A comparative study of machine learning methods for lung diseases diagnosis by computerized digital imaging'"
 A comparative study of machine learning methods for lung diseases diagnosis by computerized digital imaging'" Suk Ho Kang**. Youngjoo Lee*** Aostract I\.' New Work to be 1 Introduction Presented U Mater~al
A comparative study of machine learning methods for lung diseases diagnosis by computerized digital imaging'" Suk Ho Kang**. Youngjoo Lee*** Aostract I\.' New Work to be 1 Introduction Presented U Mater~al
IEEE TRANSACTIONS ON BIOMEDICAL ENGINEERING 1
 IEEE TRANSACTIONS ON BIOMEDICAL ENGINEERING Joint Classification and Regression via Deep Multi-Task Multi-Channel Learning for Alzheimer s Disease Diagnosis Mingxia Liu, Jun Zhang, Ehsan Adeli, Dinggang
IEEE TRANSACTIONS ON BIOMEDICAL ENGINEERING Joint Classification and Regression via Deep Multi-Task Multi-Channel Learning for Alzheimer s Disease Diagnosis Mingxia Liu, Jun Zhang, Ehsan Adeli, Dinggang
arxiv: v3 [stat.ml] 27 Mar 2018
![arxiv: v3 [stat.ml] 27 Mar 2018 arxiv: v3 [stat.ml] 27 Mar 2018](/thumbs/82/87026346.jpg) ATTACKING THE MADRY DEFENSE MODEL WITH L 1 -BASED ADVERSARIAL EXAMPLES Yash Sharma 1 and Pin-Yu Chen 2 1 The Cooper Union, New York, NY 10003, USA 2 IBM Research, Yorktown Heights, NY 10598, USA sharma2@cooper.edu,
ATTACKING THE MADRY DEFENSE MODEL WITH L 1 -BASED ADVERSARIAL EXAMPLES Yash Sharma 1 and Pin-Yu Chen 2 1 The Cooper Union, New York, NY 10003, USA 2 IBM Research, Yorktown Heights, NY 10598, USA sharma2@cooper.edu,
Final Report: Automated Semantic Segmentation of Volumetric Cardiovascular Features and Disease Assessment
 Final Report: Automated Semantic Segmentation of Volumetric Cardiovascular Features and Disease Assessment Tony Lindsey 1,3, Xiao Lu 1 and Mojtaba Tefagh 2 1 Department of Biomedical Informatics, Stanford
Final Report: Automated Semantic Segmentation of Volumetric Cardiovascular Features and Disease Assessment Tony Lindsey 1,3, Xiao Lu 1 and Mojtaba Tefagh 2 1 Department of Biomedical Informatics, Stanford
Deep Compression and EIE: Efficient Inference Engine on Compressed Deep Neural Network
 Deep Compression and EIE: Efficient Inference Engine on Deep Neural Network Song Han*, Xingyu Liu, Huizi Mao, Jing Pu, Ardavan Pedram, Mark Horowitz, Bill Dally Stanford University Our Prior Work: Deep
Deep Compression and EIE: Efficient Inference Engine on Deep Neural Network Song Han*, Xingyu Liu, Huizi Mao, Jing Pu, Ardavan Pedram, Mark Horowitz, Bill Dally Stanford University Our Prior Work: Deep
CS-E Deep Learning Session 4: Convolutional Networks
 CS-E4050 - Deep Learning Session 4: Convolutional Networks Jyri Kivinen Aalto University 23 September 2015 Credits: Thanks to Tapani Raiko for slides material. CS-E4050 - Deep Learning Session 4: Convolutional
CS-E4050 - Deep Learning Session 4: Convolutional Networks Jyri Kivinen Aalto University 23 September 2015 Credits: Thanks to Tapani Raiko for slides material. CS-E4050 - Deep Learning Session 4: Convolutional
Age Estimation based on Multi-Region Convolutional Neural Network
 Age Estimation based on Multi-Region Convolutional Neural Network Ting Liu, Jun Wan, Tingzhao Yu, Zhen Lei, and Stan Z. Li 1 Center for Biometrics and Security Research & National Laboratory of Pattern
Age Estimation based on Multi-Region Convolutional Neural Network Ting Liu, Jun Wan, Tingzhao Yu, Zhen Lei, and Stan Z. Li 1 Center for Biometrics and Security Research & National Laboratory of Pattern
Design of Palm Acupuncture Points Indicator
 Design of Palm Acupuncture Points Indicator Wen-Yuan Chen, Shih-Yen Huang and Jian-Shie Lin Abstract The acupuncture points are given acupuncture or acupressure so to stimulate the meridians on each corresponding
Design of Palm Acupuncture Points Indicator Wen-Yuan Chen, Shih-Yen Huang and Jian-Shie Lin Abstract The acupuncture points are given acupuncture or acupressure so to stimulate the meridians on each corresponding
arxiv: v1 [cs.cv] 12 Jun 2018
![arxiv: v1 [cs.cv] 12 Jun 2018 arxiv: v1 [cs.cv] 12 Jun 2018](/thumbs/90/102210908.jpg) Multiview Two-Task Recursive Attention Model for Left Atrium and Atrial Scars Segmentation Jun Chen* 1, Guang Yang* 2, Zhifan Gao 3, Hao Ni 4, Elsa Angelini 5, Raad Mohiaddin 2, Tom Wong 2,Yanping Zhang
Multiview Two-Task Recursive Attention Model for Left Atrium and Atrial Scars Segmentation Jun Chen* 1, Guang Yang* 2, Zhifan Gao 3, Hao Ni 4, Elsa Angelini 5, Raad Mohiaddin 2, Tom Wong 2,Yanping Zhang
arxiv: v1 [cs.lg] 4 Feb 2019
![arxiv: v1 [cs.lg] 4 Feb 2019 arxiv: v1 [cs.lg] 4 Feb 2019](/thumbs/95/123488545.jpg) Machine Learning for Seizure Type Classification: Setting the benchmark Subhrajit Roy [000 0002 6072 5500], Umar Asif [0000 0001 5209 7084], Jianbin Tang [0000 0001 5440 0796], and Stefan Harrer [0000
Machine Learning for Seizure Type Classification: Setting the benchmark Subhrajit Roy [000 0002 6072 5500], Umar Asif [0000 0001 5209 7084], Jianbin Tang [0000 0001 5440 0796], and Stefan Harrer [0000
Firearm Detection in Social Media
 Erik Valldor and David Gustafsson Swedish Defence Research Agency (FOI) C4ISR, Sensor Informatics Linköping SWEDEN erik.valldor@foi.se david.gustafsson@foi.se ABSTRACT Manual analysis of large data sets
Erik Valldor and David Gustafsson Swedish Defence Research Agency (FOI) C4ISR, Sensor Informatics Linköping SWEDEN erik.valldor@foi.se david.gustafsson@foi.se ABSTRACT Manual analysis of large data sets
CPSC81 Final Paper: Facial Expression Recognition Using CNNs
 CPSC81 Final Paper: Facial Expression Recognition Using CNNs Luis Ceballos Swarthmore College, 500 College Ave., Swarthmore, PA 19081 USA Sarah Wallace Swarthmore College, 500 College Ave., Swarthmore,
CPSC81 Final Paper: Facial Expression Recognition Using CNNs Luis Ceballos Swarthmore College, 500 College Ave., Swarthmore, PA 19081 USA Sarah Wallace Swarthmore College, 500 College Ave., Swarthmore,
Action Recognition. Computer Vision Jia-Bin Huang, Virginia Tech. Many slides from D. Hoiem
 Action Recognition Computer Vision Jia-Bin Huang, Virginia Tech Many slides from D. Hoiem This section: advanced topics Convolutional neural networks in vision Action recognition Vision and Language 3D
Action Recognition Computer Vision Jia-Bin Huang, Virginia Tech Many slides from D. Hoiem This section: advanced topics Convolutional neural networks in vision Action recognition Vision and Language 3D
Automated Mass Detection from Mammograms using Deep Learning and Random Forest
 Automated Mass Detection from Mammograms using Deep Learning and Random Forest Neeraj Dhungel 1 Gustavo Carneiro 1 Andrew P. Bradley 2 1 ACVT, University of Adelaide, Australia 2 University of Queensland,
Automated Mass Detection from Mammograms using Deep Learning and Random Forest Neeraj Dhungel 1 Gustavo Carneiro 1 Andrew P. Bradley 2 1 ACVT, University of Adelaide, Australia 2 University of Queensland,
arxiv: v2 [cs.cv] 7 Jun 2018
![arxiv: v2 [cs.cv] 7 Jun 2018 arxiv: v2 [cs.cv] 7 Jun 2018](/thumbs/79/79761281.jpg) Deep supervision with additional labels for retinal vessel segmentation task Yishuo Zhang and Albert C.S. Chung Lo Kwee-Seong Medical Image Analysis Laboratory, Department of Computer Science and Engineering,
Deep supervision with additional labels for retinal vessel segmentation task Yishuo Zhang and Albert C.S. Chung Lo Kwee-Seong Medical Image Analysis Laboratory, Department of Computer Science and Engineering,
MAMMOGRAM AND TOMOSYNTHESIS CLASSIFICATION USING CONVOLUTIONAL NEURAL NETWORKS
 University of Kentucky UKnowledge Theses and Dissertations--Computer Science Computer Science 2017 MAMMOGRAM AND TOMOSYNTHESIS CLASSIFICATION USING CONVOLUTIONAL NEURAL NETWORKS Xiaofei Zhang University
University of Kentucky UKnowledge Theses and Dissertations--Computer Science Computer Science 2017 MAMMOGRAM AND TOMOSYNTHESIS CLASSIFICATION USING CONVOLUTIONAL NEURAL NETWORKS Xiaofei Zhang University
DeepMiner: Discovering Interpretable Representations for Mammogram Classification and Explanation
 DeepMiner: Discovering Interpretable Representations for Mammogram Classification and Explanation Jimmy Wu 1, Bolei Zhou 1, Diondra Peck 2, Scott Hsieh 3, Vandana Dialani, MD 4 Lester Mackey 5, and Genevieve
DeepMiner: Discovering Interpretable Representations for Mammogram Classification and Explanation Jimmy Wu 1, Bolei Zhou 1, Diondra Peck 2, Scott Hsieh 3, Vandana Dialani, MD 4 Lester Mackey 5, and Genevieve
Convolutional Neural Networks for Text Classification
 Convolutional Neural Networks for Text Classification Sebastian Sierra MindLab Research Group July 1, 2016 ebastian Sierra (MindLab Research Group) NLP Summer Class July 1, 2016 1 / 32 Outline 1 What is
Convolutional Neural Networks for Text Classification Sebastian Sierra MindLab Research Group July 1, 2016 ebastian Sierra (MindLab Research Group) NLP Summer Class July 1, 2016 1 / 32 Outline 1 What is
Deep Learning Human Mind for Automated Visual Classification
 Deep Learning Human Mind for Automated Visual Classification C. Spampinato, S. Palazzo, I. Kavasidis, D. Giordano Department of Electrical, Electronics and Computer Engineering - PeRCeiVe Lab Viale Andrea
Deep Learning Human Mind for Automated Visual Classification C. Spampinato, S. Palazzo, I. Kavasidis, D. Giordano Department of Electrical, Electronics and Computer Engineering - PeRCeiVe Lab Viale Andrea
Deep Learning Human Mind for Automated Visual Classification
 Deep Learning Human Mind for Automated Visual Classification C. Spampinato, S. Palazzo, I. Kavasidis, D. Giordano Department of Electrical, Electronics and Computer Engineering - PeRCeiVe Lab Viale Andrea
Deep Learning Human Mind for Automated Visual Classification C. Spampinato, S. Palazzo, I. Kavasidis, D. Giordano Department of Electrical, Electronics and Computer Engineering - PeRCeiVe Lab Viale Andrea
arxiv: v2 [cs.lg] 1 Jun 2018
![arxiv: v2 [cs.lg] 1 Jun 2018 arxiv: v2 [cs.lg] 1 Jun 2018](/thumbs/89/101161632.jpg) Shagun Sodhani 1 * Vardaan Pahuja 1 * arxiv:1805.11016v2 [cs.lg] 1 Jun 2018 Abstract Self-play (Sukhbaatar et al., 2017) is an unsupervised training procedure which enables the reinforcement learning agents
Shagun Sodhani 1 * Vardaan Pahuja 1 * arxiv:1805.11016v2 [cs.lg] 1 Jun 2018 Abstract Self-play (Sukhbaatar et al., 2017) is an unsupervised training procedure which enables the reinforcement learning agents
Deep Networks and Beyond. Alan Yuille Bloomberg Distinguished Professor Depts. Cognitive Science and Computer Science Johns Hopkins University
 Deep Networks and Beyond Alan Yuille Bloomberg Distinguished Professor Depts. Cognitive Science and Computer Science Johns Hopkins University Artificial Intelligence versus Human Intelligence Understanding
Deep Networks and Beyond Alan Yuille Bloomberg Distinguished Professor Depts. Cognitive Science and Computer Science Johns Hopkins University Artificial Intelligence versus Human Intelligence Understanding
Attentional Masking for Pre-trained Deep Networks
 Attentional Masking for Pre-trained Deep Networks IROS 2017 Marcus Wallenberg and Per-Erik Forssén Computer Vision Laboratory Department of Electrical Engineering Linköping University 2014 2017 Per-Erik
Attentional Masking for Pre-trained Deep Networks IROS 2017 Marcus Wallenberg and Per-Erik Forssén Computer Vision Laboratory Department of Electrical Engineering Linköping University 2014 2017 Per-Erik
Dilated Recurrent Neural Network for Short-Time Prediction of Glucose Concentration
 Dilated Recurrent Neural Network for Short-Time Prediction of Glucose Concentration Jianwei Chen, Kezhi Li, Pau Herrero, Taiyu Zhu, Pantelis Georgiou Department of Electronic and Electrical Engineering,
Dilated Recurrent Neural Network for Short-Time Prediction of Glucose Concentration Jianwei Chen, Kezhi Li, Pau Herrero, Taiyu Zhu, Pantelis Georgiou Department of Electronic and Electrical Engineering,
