Magnetic Resonance Contrast Prediction Using Deep Learning
|
|
|
- Madison Holt
- 6 years ago
- Views:
Transcription
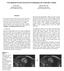
1 Magnetic Resonance Contrast Prediction Using Deep Learning Cagan Alkan Department of Electrical Engineering Stanford University Andrew Weitz Department of Bioengineering Stanford University John Cocjin Department of Bioengineering Stanford University Abstract A key strength of magnetic resonance imaging (MRI) is its ability to measure different tissue contrasts. However, each contrast is typically collected in series, making clinical MRI a slow and expensive procedure. In this paper, we present a novel approach for learning transformations from one MRI contrast to another. We train a fully convolutional neural network that takes T1-weighted MRI images as input, and generates the corresponding T2-weighted images as output. To evaluate our results, we compare different network depths, input features, and numbers of training subjects. We find that a nine-layer 2D convolutional neural network (CNN) using T1-intensity with eight tissue masks as input generates the best output images with lowest L2 error. Furthermore, the results can continue to improve with even larger training datasets. These results confirm the feasibility of using deep learning to predict one MRI contrast from another and accelerate clinical MRI acquisition. 1. Introduction Magnetic resonance images can represent many different tissue contrasts depending on the specific acquisition paradigm that is used. For example, two of the most common MRI contrasts are T1-relaxation and T2-relaxation. T1-weighted images are acquired using short echo and repetition times, while T2-weighted images are acquired using long echo and repetition times. Each contrast conveys specific information about the local tissue and its physical properties. Because T1- and T2-relaxation values differ across various tissue types, radiologists and scientists must use the information conveyed in both contrasts to make informed decisions. For example, in T2-weighted images, areas of the brain filled with water appear bright and tissues with high fat content appear dark. This can aid in localizing pathology since many brain lesions are associated with an increase in water content. Similarly, in T1-weighted images, tissues with high fat content appear bright and compartments filled with water appear dark. T1-weighted images are therefore useful for looking at detailed anatomical images. Examples of these two contrasts are provided in Figure 1. Figure 1. Example pair of T1-weighted (left) and corresponding T2-weighted (right) images. The proposed CNN predicts T2- weighted images from T1-weighted images. It is important to note that despite the tissue-specific changes in brightness associated with T1- and T2-weighted images described above, these two contrasts are not simply inverted versions of one another. Indeed, they each correspond to distinct tissue properties, and a low T1 value will not necessarily correspond to a high T2 value (or vice versa). Because the information provided by both T1- and T2- weighted images is important, clinical MRI protocols will often collect both. However, MRI acquisition is a slow and costly procedure. Therefore, a significant aim of modern MRI research is to accelerate the image acquisition process. Current approaches focus on both software (e.g. compressed sensing) and hardware (e.g. stronger gradient magnets). Here, we propose the use of convolutional neural networks and deep learning to predict one contrast from an- 1
2 other. If successful, this approach would eliminate the need for one acquisition altogether and dramatically cut down the time and costs of clinical MRI. We formulate the problem of T1-to-T2 mapping as a standard regression problem. The input to our algorithm is a two-dimensional T1-weighted image concatenated with eight additional channels that serve as binary masks to indicate different tissue types. We then use a CNN to output a predicted T2-weighted image. We choose to learn the mapping of T1-to-T2 transformations rather than vice versa, since the eight tissue mask channels can be easily computed from T1-weighted images. 2. Related Work In the last year, deep learning and CNNs have rapidly taken over the field of medical imaging and MRI. Researchers in the field have focused on two main problems: noiseless reconstruction of undersampled data [15, 19, 17, 7, 12], and automated segmentation of different tissue types [18, 14, 8, 2]. Regarding the former, various deep learning methods have been developed to reconstruct MRI images that are undersampled in the frequency (i.e. acquisition) domain. For example, Schlemper et al. [15] used a cascade of CNNs with data consistency layers to outperform stateof-the-art compressed sensing methods for reconstructing six-fold undersampled cardiac MRI images. In another case [19], researchers developed a generalized reconstruction algorithm composed of three fully-connected layers followed by a three-layer convolutional autoencoder. The researchers show that this approach can be used to noiselessly reconstruct undersampled and/or corrupted MRI images, independent of the sampling trajectory. Deep learning and CNNs have also been used for automated segmentation and detection of various pathologies or tissue types in MRI. For example, CNNs were used to segment brain tissue into white matter, gray matter, and cerebrospinal fluid in infant MRIs [18]. The authors used three modalities of imaging as input - T1, T2, and fractional anisotropy - and patched each image into 13x13 subimages. Their architecture included three convolutional layers and one fully connected layer with a soft-max loss function on the three different tissue classes. Using this approach, they were able to achieve results that significantly outperform previous methods of infant brain tissue segmentation. Despite the many applications of deep learning to medical imaging, there have been very few applications for performing image transformations - the problem we address in this paper. In one application, CNNs were used to predict computed tomography (CT) images from corresponding MRI images [13]. In another, CNNs were trained to predict high resolution MRI images acquired with a 7 Tesla scanner from low resolution images acquired with a 3 Tesla scanner [3]. Both approaches used relatively straightforward architectures three repeated convolutional layers followed by a loss function, with no pooling or fully connected layers. Both approaches also used patches to divide the original images into smaller subimages. Finally, both used the L2 loss function to learn their respective transformations. The latter network [3] used anatomical tissue masks as input to improve performance. We believe each of these features worked well for their respective problems, and adapt them here. Specifically, we use fully convolutional networks, the L2 loss function, small input patches, and eight tissue masks to serve as additional input channels. These methods are described in detail in the following sections. 3. Dataset and Features We utilized the Human Connectome Project (HCP) data release of March 2017, which consists of both raw and processed 3T MRI imaging data from 1206 healthy young adults [16]. Each subject includes T1- and T2- weighted image sets collected within the same scanning session. In a minority of subjects, more than one scanning session was conducted. For these, the image sets were preprocessed, registered, and averaged within contrast. All image sets were screened to exclude major neuroanatomical anomalies. Minor, benign variation such as a cyst is allowed, however, and is reported in ConnectomeDB, the data management platform for accessing HCP data. We do not distinguish between subjects with or without such variations. Below, we briefly describe preprocessing of the data that is relevant to the interpretation of our experiments. Further details can be found in [6]. All files specified below are available via the ConnectomeDB database. Separately, the raw T1- and T2- weighted image sets were corrected for nonlinearities induced by B0 field image distortions. The T1-weighted images underwent anterior commissure-posterior commissue (ACPC) alignment, and the T2-weighted images were then aligned to these using rigid body transformation. These steps produced the images stored in T1w acpc dc.nii.gz and T2w acpc dc.nii.gz that we use for T1-to-T2 contrast mapping. Images in both datasets are of size voxels with 0.7 mm isotropic resolution. The T1 volume was skull stripped and segmented into different tissue types using HCP-specific Freesurfer scripts [5]. The segmentation is provided in the output file wmparc.nii.gz and corresponds to a combination of white matter/grey matter cortical parcellation and subcortical segmentation. The brain mask derived from the skull-stripping step is provided in the file brainmask fs.nii.gz. We aggregate the brain mask and segmentation into eight label sets: brain, grey matter, white matter, cerebral spinal fluid, subcortical structure, vessel, cerebellum, and unknown. Binary masks were 2
3 genereated for each anatomical tissue label, and treated as a separate input channel in addition to the T1-weighted intensity values. Binary values of -1 and +1 were chosen to maintain zero-mean input. In this work, we operate over 2D images and treat each slice as an independent sample. This results in only 260 samples per subject. Therefore, to increase the size of our dataset, we split each slice into smaller subimages, or patches. We use a patch size of ( ) and a stride of 24, giving 12,480 samples per subject effectively increasing the dataset by a factor of 50. Prior to patching, the average overall intensity value was calculated across all training images and subtracted from the complete dataset. As described below, we ran experiments to explore the effect of the training set size on model output. We first developed the model using only a single subject. The 12,480 image patches were split into training, validation, and test sets, using an % breakdown, respectively. Thus, n=9,984 samples were used for training, and n=2,371 samples were used for testing. To determine how well our results generalized across subjects, we next trained the model using all samples from either one or fifty subjects, and used all images from five different subjects as the test set. In these cases, the total number of training samples was n=12,480 and n=624,000, respectively, while the total number of test samples was n=62, Methods 4.1. Network Architecture The network architecture implemented here relies on three main operations: two dimensional convolution layers, spatial batch normalization [9], and the rectified linear unit (ReLU) activation function. The 2D convolution layers compute the output using the relation shown in Equation 1 y[b, i, j, k] = X[b, s i i + d i, s j j + d j, q] f[d i, d j, q, k] d i,d j,q where X and y are the input and output tensors stored in NHWC format, respectively, f is the 4D tensor that has the same 2D filter along N and C dimensions, and s i and s j are the strides along H and W dimensions, respectively. Batch normalization is a recently developed technique that makes deep neural networks more robust to poor initialization [9]. Each batch normalization layer serves to normalize the previous layers activations into a distribution with learnable mean β and variance γ. Typically these values are initialized to 0 and 1, respectively, making the previous layer s output take on a zero-mean, unit-variance distribution. A momentum variable also allows these parameters to be updated as a running average. The batch normalization operation has the effect of implicit regularization, (1) and allows higher learning rates to be used with less concern over initialization parameters [9]. In the model implemented here, batch normalization parameters were chosen to be the same as those initially described in [9]. There are: momentum = 0.99, ɛ = 0.001, β and its moving average initialized to 0, and γ and its moving average initialized to 1. The ɛ parameter is a small float value that is added to the variance to avoid division by zero, and has negligible impact on results. The ReLU function takes the form f(x) = max(0, x). This activation function was favored over other options, such as the sigmoid and tanh functions, for its computational simplicity and accelerated convergence properties in stochastic gradient descent [11]. In the model used here, we pass input patches of size ( depth) through a nine-layer CNN. The input depth can either take on a value of 1 or 9, depending on whether tissue labels are used as additional input features. The first eight convolutional layers conform to the pattern [Conv2D]-[BatchNormalization]-[ReLU]. Each Conv2D operation includes 32 (3 3) filters. To collapse the hidden layer data into the desired output dimensions (i.e. to force a depth of one), the final convolutional layer has only one (3 3) filter. We note that no explicit regularization techniques were implemented in our model, since overfitting did not manifest in preliminary tests. A summary of the architecture is provided in Figure 2. Figure 2. Proposed model architecture for T1-to-T2 mapping. Note that input depth can be either 1 or 9, depending on whether tissue labels are used as additional features Training Training is implemented using the mean-square-error loss function shown in Equation 2 d 2 (I 1, I 2 ) = 1 N N (I 1 [n] I 2 [n]) 2 (2) n=1 where I 1 and I 2 are flattened images and N is the number of pixels. The Adam algorithm was used as the optimizer to minimize this loss [10]. This is a method for performing 3
4 stochastic gradient descent over a high-dimensional, nonconvex landscape. It computes individual adaptive learning rates for different parameters from estimates of first and second moments of the gradients. The full Adam algorithm is described in Equations 3a through 3c below, where m is a moving average of the gradient, v is a moving average of the gradient squared, β 1 is the momentum parameter of the gradients, β 2 is the momentum parameter for the gradients squared, λ is the learning rate, and ɛ is a small float added to avoid division by zero. A higher momentum value means that the current gradient will contribute less to the moving average. We implemented the Adam algorithm with hyperparameters suggested in its original formulation [10]. These are: β 1 = 0.9, β 2 = 0.999, and ɛ = 1e-09. m t+1 = β 1 m t + (1 β 1 ) dx v t+1 = β 2 v t + (1 β 2 ) dx 2 x t+1 = x t λ m t+1 /( v t+1 + ɛ) (3a) (3b) (3c) In our implementation, we anneal the learning rate λ in equation 3c following a step decay schedule. We reduce λ by a half every 4 epochs, starting with an initial learning rate of 0.1. Decaying the learning rate helps improve training performance by allowing the optimizer to settle down into the narrower parts of the loss function. As described in the experiments section below, we train our models using either data from a single subject or multiple subjects. The models that use a single subject for training are trained for 20 epochs. Per epoch training time for the models that use 50 subjects is considerably higher. Therefore, these models are trained only for 8 epochs. We use a batch size of 32 for the models trained on a single subject. In order to take advantage of the parallelism between CPU based patch extraction and GPU based model training, we select a batch size of 128 for training the models using training data from multiple subjects Implementation To implement our model, we use the Keras [4] API with a TensorFlow [1] backend. All computations are executed on Google Cloud using a CPU or NVIDIA Tesla K80 GPU. Due to the large size of the dataset, it is not possible to load the patches for all subjects into computer memory at once. In order to remedy this, Python generators are used to extract and generate patches for each batch separately. In addition, the NumPy memmap module is utilized to partially load data from disk on the fly and reduce memory usage. 5. Experiments, Results, and Discussion 5.1. Experiments Multiple experiments were run to evaluate the effect of network depth, training set size, and the addition of input features providing voxelwise tissue-type labeling. We evaluate model performance both by visual inspection and by using the mean-square-error loss values between the output T2-weighted images and the ground truth T2-weighted images. To facilitate comparison between models, we show the same images from the test set for the across-subject experiment results Results and Discussion Model Learning To confirm that the proposed model could learn, we first trained and tested the model using data from one subject. This model included nine convolutional layers and nine input channels (i.e. T1-intensity plus eight tissue masks). The loss curves shown in Figure 3 exhibit a steady decline followed by a plateau, indicating that the network could learn and that network parameters are updating in a proper and stable manner. The small gap between training and validation loss also suggest that the models are properly regularized. Having confirmed that the model successfully learned, we moved on to test the role that input features, network depth, and training set size have in the accuracy of our results. Figure 3. Learning curves for within-subject testing and training Effect of input features We first investigated the effect of using only the T1- weighted image intensity as input versus using the T1- weighted image combined with eight additional tissue mask channels. Visually, the resulting T2-weighted images looked much more similar to the ground-truth images when the tissue labels were included. This was true in the case of both deep (Figure 4c vs. 4d) and shallow (Figure 4e vs. 4f) networks when using one subject for training and using five different subjects for testing. It was also true in the case of a deep network when using 50 subjects for training (Figure 5c vs. 5d). Furthermore, as shown in Tables 1-3, providing 4
5 3 Layer Network 9 Layer Network No Tissue Masks 12,795 5,177 Tissue Masks 10,664 4,091 3 Layer Network 9 Layer Network Table 1. Within-subject loss values. Corresponding output images are not shown for simplicity. No Tissue Masks 20,385 12,643 Tissue Masks 16,627 9,783 Table 2. Across-subject loss values, training size = 1 subject. 9 Layer Network these tissue labels as input reduced the L2 error across all model variants tested - typically by 20%. We thus conclude that the addition of binary tissue label masks significantly enhances the prediction of T2-weighted images from T1weighted images. No Tissue Masks 9,093 Tissue Masks 7,728 Table 3. Across-subject loss values, training size = 50 subjects Effect of network size We next compared our results with those of a shallower network that uses only three convolutional layers instead of nine. Visually, we found that the resulting T2-weighted images looked much more similar to the ground-truth images when a deeper network was used (Figure 4c,d vs. 4e,f). In this case, we see that adding additional convolutional layers significantly enhances the prediction performance. As shown in Tables 1-2, increasing the number of convolutional layers reduced the L2 error across all model variants tested - typically by 40-60%. We thus conclude that the addition of more convolutional layers significantly enhances the prediction of T2-weighted images from T1-weighted images Effect of training set size Finally, we compared two scenarios to determine the improvement in performance achieved by training over larger datasets: (1) a single subject was used as the training set, while five other subjects were used as the test set, and (2) images from 50 subjects were used as the training set, while five other subjects were used as the test set. Comparing the loss values in Tables 2 and 3 indicates that the loss decreased when using the larger training set size. Furthermore, a comparison of the images shown in figures 4d and 5d shows that certain features were better preserved in the inclusion of multiple subjects, such as the apparent separation of adjacent gyri. In addition, subcortical blurring was significantly reduced on the images obtained using the model trained with a larger training set. Figure 4. Across-subject T2 prediction results using n=1 subject training set for four representative patches. (a) T1-weighted image. (b) Ground-truth T2-weighted image. (c) Predicted T2 image using nine-layer network without tissue masks. (d) Predicted T2 image using nine-layer network with tissue masks. (e) Predicted T2 image using three-layer network without tissue masks. (f) Predicted T2 image using three-layer network with tissue masks Tissue specificity As can be seen, the trained networks could preserve tissue traits in the mapping of T1-weighted to T2-weighted images. As a remarkable example, the CNN recognized inversion. For example, while the predicted T2 images show 5
6 incorporate pathological images with tissue abnormalities. If the model cannot successfully predict these abnormalities, the training set can be revised to include this type of data. There are a number of hyperparameter options left to test. One other item to test in future work is any potential improvement by using 3D convolutions instead of 2D convolutions. 3D convolutions were initially attempted here, but found to be infeasible due to memory constraints. Running the model with higher specification computers and employing the NumPy memory map module used here are likely to make this approach more feasible. Another possibility is to sweep across patch or 1st layer kernel sizes to vary the amount of spatial information that the network is provided at the input level. If network optimization were to reach satisfactory mapping, as validated by radiologists or imaging scientists, it would be of interest to transfer the network to other contrast problems. Figure 5. Across-subject T2 prediction results using n=50 subject training set for four representative patches. (a) T1-weighted image. (b) Ground-truth T2-weighted image. (c) Predicted T2 image using nine-layer network without tissue masks. (d) Predicted T2 image using nine-layer network with tissue masks. dark white matter and bright gray matter (the opposite from the input T1 images), the skull and ventricle intensities generally do not change. All networks are able to produce these inversions. That the network without tissue labels could learn these mappings suggests that tissue specific mappings are latent in intensive patterning of the patches, that the patches provided enough view to sufficiently distinguish large scale structure, or both. 6. Conclusions and Future Work Our experiments demonstrate the feasibility of applying deep convolutional neural networks towards the problem of predicting one MRI contrast from another. Our results indicate that deep CNNs perform better than shallow CNNs, and that including tissue type feature labels as additional inputs improves performance. Finally, we find that larger training datasets have potential in improving the performance of the network but require significant training time, even on a GPU. A critical question moving forward with this approach is whether the algorithm can accurately predict T2-weighted images in the presence of abnormalities. Because MRI images are acquired for diagnostic purposes, this ability is neccessary if it were to ever be deployed. Although the dataset used here included healthy subjects only, future work will References [1] M. Abadi, A. Agarwal, P. Barham, E. Brevdo, Z. Chen, C. Citro, G. S. Corrado, A. Davis, J. Dean, M. Devin, et al. Tensorflow: Large-scale machine learning on heterogeneous distributed systems. arxiv preprint arxiv: , [2] M. Avendi, A. Kheradvar, and H. Jafarkhani. A combined deep-learning and deformable-model approach to fully automatic segmentation of the left ventricle in cardiac mri. Medical image analysis, 30: , [3] K. Bahrami, F. Shi, I. Rekik, and D. Shen. Convolutional Neural Network for Reconstruction of 7T-like Images from 3T MRI Using Appearance and Anatomical Features, pages Springer International Publishing, Cham, [4] F. Chollet et al. Keras. fchollet/keras, [5] B. Fischl. Freesurfer. Neuroimage, 62(2): , [6] M. F. Glasser, S. N. Sotiropoulos, J. A. Wilson, T. S. Coalson, B. Fischl, J. L. Andersson, J. Xu, S. Jbabdi, M. Webster, J. R. Polimeni, et al. The minimal preprocessing pipelines for the human connectome project. Neuroimage, 80: , [7] Y. S. Han, J. Yoo, and J. C. Ye. Deep learning with domain adaptation for accelerated projection reconstruction mr. arxiv preprint arxiv: , [8] M. Havaei, A. Davy, D. Warde-Farley, A. Biard, A. Courville, Y. Bengio, C. Pal, P.-M. Jodoin, and H. Larochelle. Brain tumor segmentation with deep neural networks. Medical image analysis, 35:18 31, [9] S. Ioffe and C. Szegedy. Batch normalization: Accelerating deep network training by reducing internal covariate shift. arxiv preprint arxiv: , [10] D. Kingma and J. Ba. Adam: A method for stochastic optimization. arxiv preprint arxiv: , [11] A. Krizhevsky, I. Sutskever, and G. E. Hinton. Imagenet classification with deep convolutional neural networks. In 6
7 Advances in neural information processing systems, pages , [12] D. Lee, J. Yoo, and J. C. Ye. Deep artifact learning for compressed sensing and parallel mri. arxiv preprint arxiv: , [13] D. Nie, X. Cao, Y. Gao, L. Wang, and D. Shen. Estimating CT Image from MRI Data Using 3D Fully Convolutional Networks, pages Springer International Publishing, Cham, [14] A. Prasoon, K. Petersen, C. Igel, F. Lauze, E. Dam, and M. Nielsen. Deep feature learning for knee cartilage segmentation using a triplanar convolutional neural network. In International conference on medical image computing and computer-assisted intervention, pages Springer, [15] J. Schlemper, J. Caballero, J. V. Hajnal, A. Price, and D. Rueckert. A deep cascade of convolutional neural networks for mr image reconstruction. arxiv preprint arxiv: , [16] D. C. Van Essen, S. M. Smith, D. M. Barch, T. E. Behrens, E. Yacoub, K. Ugurbil, W.-M. H. Consortium, et al. The wuminn human connectome project: an overview. Neuroimage, 80:62 79, [17] S. Wang, Z. Su, L. Ying, X. Peng, S. Zhu, F. Liang, D. Feng, and D. Liang. Accelerating magnetic resonance imaging via deep learning. In Biomedical Imaging (ISBI), 2016 IEEE 13th International Symposium on, pages IEEE, [18] W. Zhang, R. Li, H. Deng, L. Wang, W. Lin, S. Ji, and D. Shen. Deep convolutional neural networks for multimodality isointense infant brain image segmentation. NeuroImage, 108: , [19] B. Zhu, J. Z. Liu, B. R. Rosen, and M. S. Rosen. Image reconstruction by domain transform manifold learning. arxiv preprint arxiv: ,
CSE Introduction to High-Perfomance Deep Learning ImageNet & VGG. Jihyung Kil
 CSE 5194.01 - Introduction to High-Perfomance Deep Learning ImageNet & VGG Jihyung Kil ImageNet Classification with Deep Convolutional Neural Networks Alex Krizhevsky, Ilya Sutskever, Geoffrey E. Hinton,
CSE 5194.01 - Introduction to High-Perfomance Deep Learning ImageNet & VGG Jihyung Kil ImageNet Classification with Deep Convolutional Neural Networks Alex Krizhevsky, Ilya Sutskever, Geoffrey E. Hinton,
arxiv: v1 [cs.cv] 9 Sep 2017
![arxiv: v1 [cs.cv] 9 Sep 2017 arxiv: v1 [cs.cv] 9 Sep 2017](/thumbs/79/79077141.jpg) Sequential 3D U-Nets for Biologically-Informed Brain Tumor Segmentation Andrew Beers 1, Ken Chang 1, James Brown 1, Emmett Sartor 2, CP Mammen 3, Elizabeth Gerstner 1,4, Bruce Rosen 1, and Jayashree Kalpathy-Cramer
Sequential 3D U-Nets for Biologically-Informed Brain Tumor Segmentation Andrew Beers 1, Ken Chang 1, James Brown 1, Emmett Sartor 2, CP Mammen 3, Elizabeth Gerstner 1,4, Bruce Rosen 1, and Jayashree Kalpathy-Cramer
Differentiating Tumor and Edema in Brain Magnetic Resonance Images Using a Convolutional Neural Network
 Original Article Differentiating Tumor and Edema in Brain Magnetic Resonance Images Using a Convolutional Neural Network Aida Allahverdi 1, Siavash Akbarzadeh 1, Alireza Khorrami Moghaddam 2, Armin Allahverdy
Original Article Differentiating Tumor and Edema in Brain Magnetic Resonance Images Using a Convolutional Neural Network Aida Allahverdi 1, Siavash Akbarzadeh 1, Alireza Khorrami Moghaddam 2, Armin Allahverdy
Lung Nodule Segmentation Using 3D Convolutional Neural Networks
 Lung Nodule Segmentation Using 3D Convolutional Neural Networks Research paper Business Analytics Bernard Bronmans Master Business Analytics VU University, Amsterdam Evert Haasdijk Supervisor VU University,
Lung Nodule Segmentation Using 3D Convolutional Neural Networks Research paper Business Analytics Bernard Bronmans Master Business Analytics VU University, Amsterdam Evert Haasdijk Supervisor VU University,
POC Brain Tumor Segmentation. vlife Use Case
 Brain Tumor Segmentation vlife Use Case 1 Automatic Brain Tumor Segmentation using CNN Background Brain tumor segmentation seeks to separate healthy tissue from tumorous regions such as the advancing tumor,
Brain Tumor Segmentation vlife Use Case 1 Automatic Brain Tumor Segmentation using CNN Background Brain tumor segmentation seeks to separate healthy tissue from tumorous regions such as the advancing tumor,
arxiv: v2 [cs.cv] 19 Dec 2017
![arxiv: v2 [cs.cv] 19 Dec 2017 arxiv: v2 [cs.cv] 19 Dec 2017](/thumbs/89/97557782.jpg) An Ensemble of Deep Convolutional Neural Networks for Alzheimer s Disease Detection and Classification arxiv:1712.01675v2 [cs.cv] 19 Dec 2017 Jyoti Islam Department of Computer Science Georgia State University
An Ensemble of Deep Convolutional Neural Networks for Alzheimer s Disease Detection and Classification arxiv:1712.01675v2 [cs.cv] 19 Dec 2017 Jyoti Islam Department of Computer Science Georgia State University
A CONVOLUTION NEURAL NETWORK ALGORITHM FOR BRAIN TUMOR IMAGE SEGMENTATION
 A CONVOLUTION NEURAL NETWORK ALGORITHM FOR BRAIN TUMOR IMAGE SEGMENTATION 1 Priya K, 2 Dr.O. Saraniya 1 PG Scholar, 2 Assistant Professor Department Of ECE Government College of Technology, Coimbatore,
A CONVOLUTION NEURAL NETWORK ALGORITHM FOR BRAIN TUMOR IMAGE SEGMENTATION 1 Priya K, 2 Dr.O. Saraniya 1 PG Scholar, 2 Assistant Professor Department Of ECE Government College of Technology, Coimbatore,
Convolutional Neural Networks for Estimating Left Ventricular Volume
 Convolutional Neural Networks for Estimating Left Ventricular Volume Ryan Silva Stanford University rdsilva@stanford.edu Maksim Korolev Stanford University mkorolev@stanford.edu Abstract End-systolic and
Convolutional Neural Networks for Estimating Left Ventricular Volume Ryan Silva Stanford University rdsilva@stanford.edu Maksim Korolev Stanford University mkorolev@stanford.edu Abstract End-systolic and
Early Diagnosis of Autism Disease by Multi-channel CNNs
 Early Diagnosis of Autism Disease by Multi-channel CNNs Guannan Li 1,2, Mingxia Liu 2, Quansen Sun 1(&), Dinggang Shen 2(&), and Li Wang 2(&) 1 School of Computer Science and Engineering, Nanjing University
Early Diagnosis of Autism Disease by Multi-channel CNNs Guannan Li 1,2, Mingxia Liu 2, Quansen Sun 1(&), Dinggang Shen 2(&), and Li Wang 2(&) 1 School of Computer Science and Engineering, Nanjing University
HHS Public Access Author manuscript Med Image Comput Comput Assist Interv. Author manuscript; available in PMC 2018 January 04.
 Discriminative Localization in CNNs for Weakly-Supervised Segmentation of Pulmonary Nodules Xinyang Feng 1, Jie Yang 1, Andrew F. Laine 1, and Elsa D. Angelini 1,2 1 Department of Biomedical Engineering,
Discriminative Localization in CNNs for Weakly-Supervised Segmentation of Pulmonary Nodules Xinyang Feng 1, Jie Yang 1, Andrew F. Laine 1, and Elsa D. Angelini 1,2 1 Department of Biomedical Engineering,
IEEE TRANSACTIONS ON BIOMEDICAL ENGINEERING 1
 IEEE TRANSACTIONS ON BIOMEDICAL ENGINEERING Joint Classification and Regression via Deep Multi-Task Multi-Channel Learning for Alzheimer s Disease Diagnosis Mingxia Liu, Jun Zhang, Ehsan Adeli, Dinggang
IEEE TRANSACTIONS ON BIOMEDICAL ENGINEERING Joint Classification and Regression via Deep Multi-Task Multi-Channel Learning for Alzheimer s Disease Diagnosis Mingxia Liu, Jun Zhang, Ehsan Adeli, Dinggang
Classification and Statistical Analysis of Auditory FMRI Data Using Linear Discriminative Analysis and Quadratic Discriminative Analysis
 International Journal of Innovative Research in Computer Science & Technology (IJIRCST) ISSN: 2347-5552, Volume-2, Issue-6, November-2014 Classification and Statistical Analysis of Auditory FMRI Data Using
International Journal of Innovative Research in Computer Science & Technology (IJIRCST) ISSN: 2347-5552, Volume-2, Issue-6, November-2014 Classification and Statistical Analysis of Auditory FMRI Data Using
Highly Accurate Brain Stroke Diagnostic System and Generative Lesion Model. Junghwan Cho, Ph.D. CAIDE Systems, Inc. Deep Learning R&D Team
 Highly Accurate Brain Stroke Diagnostic System and Generative Lesion Model Junghwan Cho, Ph.D. CAIDE Systems, Inc. Deep Learning R&D Team Established in September, 2016 at 110 Canal st. Lowell, MA 01852,
Highly Accurate Brain Stroke Diagnostic System and Generative Lesion Model Junghwan Cho, Ph.D. CAIDE Systems, Inc. Deep Learning R&D Team Established in September, 2016 at 110 Canal st. Lowell, MA 01852,
Deep Learning of Brain Lesion Patterns for Predicting Future Disease Activity in Patients with Early Symptoms of Multiple Sclerosis
 Deep Learning of Brain Lesion Patterns for Predicting Future Disease Activity in Patients with Early Symptoms of Multiple Sclerosis Youngjin Yoo 1,2,5(B),LisaW.Tang 2,3,5, Tom Brosch 1,2,5,DavidK.B.Li
Deep Learning of Brain Lesion Patterns for Predicting Future Disease Activity in Patients with Early Symptoms of Multiple Sclerosis Youngjin Yoo 1,2,5(B),LisaW.Tang 2,3,5, Tom Brosch 1,2,5,DavidK.B.Li
Synthesizing Missing PET from MRI with Cycle-consistent Generative Adversarial Networks for Alzheimer s Disease Diagnosis
 Synthesizing Missing PET from MRI with Cycle-consistent Generative Adversarial Networks for Alzheimer s Disease Diagnosis Yongsheng Pan 1,2, Mingxia Liu 2, Chunfeng Lian 2, Tao Zhou 2,YongXia 1(B), and
Synthesizing Missing PET from MRI with Cycle-consistent Generative Adversarial Networks for Alzheimer s Disease Diagnosis Yongsheng Pan 1,2, Mingxia Liu 2, Chunfeng Lian 2, Tao Zhou 2,YongXia 1(B), and
A Novel Method using Convolutional Neural Network for Segmenting Brain Tumor in MRI Images
 A Novel Method using Convolutional Neural Network for Segmenting Brain Tumor in MRI Images K. Meena*, S. Pavitra**, N. Nishanthi*** & M. Nivetha**** *UG Student, Department of Electronics and Communication
A Novel Method using Convolutional Neural Network for Segmenting Brain Tumor in MRI Images K. Meena*, S. Pavitra**, N. Nishanthi*** & M. Nivetha**** *UG Student, Department of Electronics and Communication
Deep CNNs for Diabetic Retinopathy Detection
 Deep CNNs for Diabetic Retinopathy Detection Alex Tamkin Stanford University atamkin@stanford.edu Iain Usiri Stanford University iusiri@stanford.edu Chala Fufa Stanford University cfufa@stanford.edu 1
Deep CNNs for Diabetic Retinopathy Detection Alex Tamkin Stanford University atamkin@stanford.edu Iain Usiri Stanford University iusiri@stanford.edu Chala Fufa Stanford University cfufa@stanford.edu 1
Recurrent Fully Convolutional Neural Networks for Multi-slice MRI Cardiac Segmentation
 Recurrent Fully Convolutional Neural Networks for Multi-slice MRI Cardiac Segmentation Rudra P K Poudel, Pablo Lamata and Giovanni Montana Department of Biomedical Engineering, King s College London, SE1
Recurrent Fully Convolutional Neural Networks for Multi-slice MRI Cardiac Segmentation Rudra P K Poudel, Pablo Lamata and Giovanni Montana Department of Biomedical Engineering, King s College London, SE1
DIAGNOSTIC CLASSIFICATION OF LUNG NODULES USING 3D NEURAL NETWORKS
 DIAGNOSTIC CLASSIFICATION OF LUNG NODULES USING 3D NEURAL NETWORKS Raunak Dey Zhongjie Lu Yi Hong Department of Computer Science, University of Georgia, Athens, GA, USA First Affiliated Hospital, School
DIAGNOSTIC CLASSIFICATION OF LUNG NODULES USING 3D NEURAL NETWORKS Raunak Dey Zhongjie Lu Yi Hong Department of Computer Science, University of Georgia, Athens, GA, USA First Affiliated Hospital, School
arxiv: v2 [cs.cv] 3 Jun 2018
![arxiv: v2 [cs.cv] 3 Jun 2018 arxiv: v2 [cs.cv] 3 Jun 2018](/thumbs/89/97775247.jpg) S4ND: Single-Shot Single-Scale Lung Nodule Detection Naji Khosravan and Ulas Bagci Center for Research in Computer Vision (CRCV), School of Computer Science, University of Central Florida, Orlando, FL.
S4ND: Single-Shot Single-Scale Lung Nodule Detection Naji Khosravan and Ulas Bagci Center for Research in Computer Vision (CRCV), School of Computer Science, University of Central Florida, Orlando, FL.
Final Report: Automated Semantic Segmentation of Volumetric Cardiovascular Features and Disease Assessment
 Final Report: Automated Semantic Segmentation of Volumetric Cardiovascular Features and Disease Assessment Tony Lindsey 1,3, Xiao Lu 1 and Mojtaba Tefagh 2 1 Department of Biomedical Informatics, Stanford
Final Report: Automated Semantic Segmentation of Volumetric Cardiovascular Features and Disease Assessment Tony Lindsey 1,3, Xiao Lu 1 and Mojtaba Tefagh 2 1 Department of Biomedical Informatics, Stanford
arxiv: v1 [cs.cv] 13 Jul 2018
![arxiv: v1 [cs.cv] 13 Jul 2018 arxiv: v1 [cs.cv] 13 Jul 2018](/thumbs/84/89699669.jpg) Multi-Scale Convolutional-Stack Aggregation for Robust White Matter Hyperintensities Segmentation Hongwei Li 1, Jianguo Zhang 3, Mark Muehlau 2, Jan Kirschke 2, and Bjoern Menze 1 arxiv:1807.05153v1 [cs.cv]
Multi-Scale Convolutional-Stack Aggregation for Robust White Matter Hyperintensities Segmentation Hongwei Li 1, Jianguo Zhang 3, Mark Muehlau 2, Jan Kirschke 2, and Bjoern Menze 1 arxiv:1807.05153v1 [cs.cv]
arxiv: v1 [stat.ml] 23 Jan 2017
![arxiv: v1 [stat.ml] 23 Jan 2017 arxiv: v1 [stat.ml] 23 Jan 2017](/thumbs/81/83414183.jpg) Learning what to look in chest X-rays with a recurrent visual attention model arxiv:1701.06452v1 [stat.ml] 23 Jan 2017 Petros-Pavlos Ypsilantis Department of Biomedical Engineering King s College London
Learning what to look in chest X-rays with a recurrent visual attention model arxiv:1701.06452v1 [stat.ml] 23 Jan 2017 Petros-Pavlos Ypsilantis Department of Biomedical Engineering King s College London
A new Method on Brain MRI Image Preprocessing for Tumor Detection
 2015 IJSRSET Volume 1 Issue 1 Print ISSN : 2395-1990 Online ISSN : 2394-4099 Themed Section: Engineering and Technology A new Method on Brain MRI Preprocessing for Tumor Detection ABSTRACT D. Arun Kumar
2015 IJSRSET Volume 1 Issue 1 Print ISSN : 2395-1990 Online ISSN : 2394-4099 Themed Section: Engineering and Technology A new Method on Brain MRI Preprocessing for Tumor Detection ABSTRACT D. Arun Kumar
Automated diagnosis of pneumothorax using an ensemble of convolutional neural networks with multi-sized chest radiography images
 Automated diagnosis of pneumothorax using an ensemble of convolutional neural networks with multi-sized chest radiography images Tae Joon Jun, Dohyeun Kim, and Daeyoung Kim School of Computing, KAIST,
Automated diagnosis of pneumothorax using an ensemble of convolutional neural networks with multi-sized chest radiography images Tae Joon Jun, Dohyeun Kim, and Daeyoung Kim School of Computing, KAIST,
arxiv: v3 [stat.ml] 27 Mar 2018
![arxiv: v3 [stat.ml] 27 Mar 2018 arxiv: v3 [stat.ml] 27 Mar 2018](/thumbs/82/87026346.jpg) ATTACKING THE MADRY DEFENSE MODEL WITH L 1 -BASED ADVERSARIAL EXAMPLES Yash Sharma 1 and Pin-Yu Chen 2 1 The Cooper Union, New York, NY 10003, USA 2 IBM Research, Yorktown Heights, NY 10598, USA sharma2@cooper.edu,
ATTACKING THE MADRY DEFENSE MODEL WITH L 1 -BASED ADVERSARIAL EXAMPLES Yash Sharma 1 and Pin-Yu Chen 2 1 The Cooper Union, New York, NY 10003, USA 2 IBM Research, Yorktown Heights, NY 10598, USA sharma2@cooper.edu,
On Training of Deep Neural Network. Lornechen
 On Training of Deep Neural Network Lornechen 2016.04.20 1 Outline Introduction Layer-wise Pre-training & Fine-tuning Activation Function Initialization Method Advanced Layers and Nets 2 Neural Network
On Training of Deep Neural Network Lornechen 2016.04.20 1 Outline Introduction Layer-wise Pre-training & Fine-tuning Activation Function Initialization Method Advanced Layers and Nets 2 Neural Network
arxiv: v2 [cs.cv] 8 Mar 2018
![arxiv: v2 [cs.cv] 8 Mar 2018 arxiv: v2 [cs.cv] 8 Mar 2018](/thumbs/87/97094636.jpg) Automated soft tissue lesion detection and segmentation in digital mammography using a u-net deep learning network Timothy de Moor a, Alejandro Rodriguez-Ruiz a, Albert Gubern Mérida a, Ritse Mann a, and
Automated soft tissue lesion detection and segmentation in digital mammography using a u-net deep learning network Timothy de Moor a, Alejandro Rodriguez-Ruiz a, Albert Gubern Mérida a, Ritse Mann a, and
Convolutional Neural Networks (CNN)
 Convolutional Neural Networks (CNN) Algorithm and Some Applications in Computer Vision Luo Hengliang Institute of Automation June 10, 2014 Luo Hengliang (Institute of Automation) Convolutional Neural Networks
Convolutional Neural Networks (CNN) Algorithm and Some Applications in Computer Vision Luo Hengliang Institute of Automation June 10, 2014 Luo Hengliang (Institute of Automation) Convolutional Neural Networks
Synthesis of Gadolinium-enhanced MRI for Multiple Sclerosis patients using Generative Adversarial Network
 Medical Application of GAN Synthesis of Gadolinium-enhanced MRI for Multiple Sclerosis patients using Generative Adversarial Network Sumana Basu School of Computer Science McGill University 260727568 sumana.basu@mail.mcgill.ca
Medical Application of GAN Synthesis of Gadolinium-enhanced MRI for Multiple Sclerosis patients using Generative Adversarial Network Sumana Basu School of Computer Science McGill University 260727568 sumana.basu@mail.mcgill.ca
End-To-End Alzheimer s Disease Diagnosis and Biomarker Identification
 End-To-End Alzheimer s Disease Diagnosis and Biomarker Identification Soheil Esmaeilzadeh 1, Dimitrios Ioannis Belivanis 1, Kilian M. Pohl 2, and Ehsan Adeli 1 1 Stanford University 2 SRI International
End-To-End Alzheimer s Disease Diagnosis and Biomarker Identification Soheil Esmaeilzadeh 1, Dimitrios Ioannis Belivanis 1, Kilian M. Pohl 2, and Ehsan Adeli 1 1 Stanford University 2 SRI International
Automatic Prostate Cancer Classification using Deep Learning. Ida Arvidsson Centre for Mathematical Sciences, Lund University, Sweden
 Automatic Prostate Cancer Classification using Deep Learning Ida Arvidsson Centre for Mathematical Sciences, Lund University, Sweden Outline Autoencoders, theory Motivation, background and goal for prostate
Automatic Prostate Cancer Classification using Deep Learning Ida Arvidsson Centre for Mathematical Sciences, Lund University, Sweden Outline Autoencoders, theory Motivation, background and goal for prostate
International Journal of Research (IJR) Vol-1, Issue-6, July 2014 ISSN
 Developing an Approach to Brain MRI Image Preprocessing for Tumor Detection Mr. B.Venkateswara Reddy 1, Dr. P. Bhaskara Reddy 2, Dr P. Satish Kumar 3, Dr. S. Siva Reddy 4 1. Associate Professor, ECE Dept,
Developing an Approach to Brain MRI Image Preprocessing for Tumor Detection Mr. B.Venkateswara Reddy 1, Dr. P. Bhaskara Reddy 2, Dr P. Satish Kumar 3, Dr. S. Siva Reddy 4 1. Associate Professor, ECE Dept,
arxiv: v1 [cs.cv] 9 Oct 2018
![arxiv: v1 [cs.cv] 9 Oct 2018 arxiv: v1 [cs.cv] 9 Oct 2018](/thumbs/95/126101195.jpg) Automatic Segmentation of Thoracic Aorta Segments in Low-Dose Chest CT Julia M. H. Noothout a, Bob D. de Vos a, Jelmer M. Wolterink a, Ivana Išgum a a Image Sciences Institute, University Medical Center
Automatic Segmentation of Thoracic Aorta Segments in Low-Dose Chest CT Julia M. H. Noothout a, Bob D. de Vos a, Jelmer M. Wolterink a, Ivana Išgum a a Image Sciences Institute, University Medical Center
A HMM-based Pre-training Approach for Sequential Data
 A HMM-based Pre-training Approach for Sequential Data Luca Pasa 1, Alberto Testolin 2, Alessandro Sperduti 1 1- Department of Mathematics 2- Department of Developmental Psychology and Socialisation University
A HMM-based Pre-training Approach for Sequential Data Luca Pasa 1, Alberto Testolin 2, Alessandro Sperduti 1 1- Department of Mathematics 2- Department of Developmental Psychology and Socialisation University
Automated detection of abnormal changes in cortical thickness: A tool to help diagnosis in neocortical focal epilepsy
 Automated detection of abnormal changes in cortical thickness: A tool to help diagnosis in neocortical focal epilepsy 1. Introduction Epilepsy is a common neurological disorder, which affects about 1 %
Automated detection of abnormal changes in cortical thickness: A tool to help diagnosis in neocortical focal epilepsy 1. Introduction Epilepsy is a common neurological disorder, which affects about 1 %
2D-Sigmoid Enhancement Prior to Segment MRI Glioma Tumour
 2D-Sigmoid Enhancement Prior to Segment MRI Glioma Tumour Pre Image-Processing Setyawan Widyarto, Siti Rafidah Binti Kassim 2,2 Department of Computing, Faculty of Communication, Visual Art and Computing,
2D-Sigmoid Enhancement Prior to Segment MRI Glioma Tumour Pre Image-Processing Setyawan Widyarto, Siti Rafidah Binti Kassim 2,2 Department of Computing, Faculty of Communication, Visual Art and Computing,
Group-Wise FMRI Activation Detection on Corresponding Cortical Landmarks
 Group-Wise FMRI Activation Detection on Corresponding Cortical Landmarks Jinglei Lv 1,2, Dajiang Zhu 2, Xintao Hu 1, Xin Zhang 1,2, Tuo Zhang 1,2, Junwei Han 1, Lei Guo 1,2, and Tianming Liu 2 1 School
Group-Wise FMRI Activation Detection on Corresponding Cortical Landmarks Jinglei Lv 1,2, Dajiang Zhu 2, Xintao Hu 1, Xin Zhang 1,2, Tuo Zhang 1,2, Junwei Han 1, Lei Guo 1,2, and Tianming Liu 2 1 School
arxiv: v1 [cs.cv] 17 Aug 2017
![arxiv: v1 [cs.cv] 17 Aug 2017 arxiv: v1 [cs.cv] 17 Aug 2017](/thumbs/78/78278805.jpg) Deep Learning for Medical Image Analysis Mina Rezaei, Haojin Yang, Christoph Meinel Hasso Plattner Institute, Prof.Dr.Helmert-Strae 2-3, 14482 Potsdam, Germany {mina.rezaei,haojin.yang,christoph.meinel}@hpi.de
Deep Learning for Medical Image Analysis Mina Rezaei, Haojin Yang, Christoph Meinel Hasso Plattner Institute, Prof.Dr.Helmert-Strae 2-3, 14482 Potsdam, Germany {mina.rezaei,haojin.yang,christoph.meinel}@hpi.de
Reduction of Overfitting in Diabetes Prediction Using Deep Learning Neural Network
 Reduction of Overfitting in Diabetes Prediction Using Deep Learning Neural Network Akm Ashiquzzaman *, Abdul Kawsar Tushar *, Md. Rashedul Islam *, 1, and Jong-Myon Kim **, 2 * Department of CSE, University
Reduction of Overfitting in Diabetes Prediction Using Deep Learning Neural Network Akm Ashiquzzaman *, Abdul Kawsar Tushar *, Md. Rashedul Islam *, 1, and Jong-Myon Kim **, 2 * Department of CSE, University
Segmentation of Cell Membrane and Nucleus by Improving Pix2pix
 Segmentation of Membrane and Nucleus by Improving Pix2pix Masaya Sato 1, Kazuhiro Hotta 1, Ayako Imanishi 2, Michiyuki Matsuda 2 and Kenta Terai 2 1 Meijo University, Siogamaguchi, Nagoya, Aichi, Japan
Segmentation of Membrane and Nucleus by Improving Pix2pix Masaya Sato 1, Kazuhiro Hotta 1, Ayako Imanishi 2, Michiyuki Matsuda 2 and Kenta Terai 2 1 Meijo University, Siogamaguchi, Nagoya, Aichi, Japan
Skin cancer reorganization and classification with deep neural network
 Skin cancer reorganization and classification with deep neural network Hao Chang 1 1. Department of Genetics, Yale University School of Medicine 2. Email: changhao86@gmail.com Abstract As one kind of skin
Skin cancer reorganization and classification with deep neural network Hao Chang 1 1. Department of Genetics, Yale University School of Medicine 2. Email: changhao86@gmail.com Abstract As one kind of skin
A deep learning model integrating FCNNs and CRFs for brain tumor segmentation
 A deep learning model integrating FCNNs and CRFs for brain tumor segmentation Xiaomei Zhao 1,2, Yihong Wu 1, Guidong Song 3, Zhenye Li 4, Yazhuo Zhang,3,4,5,6, and Yong Fan 7 1 National Laboratory of Pattern
A deep learning model integrating FCNNs and CRFs for brain tumor segmentation Xiaomei Zhao 1,2, Yihong Wu 1, Guidong Song 3, Zhenye Li 4, Yazhuo Zhang,3,4,5,6, and Yong Fan 7 1 National Laboratory of Pattern
COMP9444 Neural Networks and Deep Learning 5. Convolutional Networks
 COMP9444 Neural Networks and Deep Learning 5. Convolutional Networks Textbook, Sections 6.2.2, 6.3, 7.9, 7.11-7.13, 9.1-9.5 COMP9444 17s2 Convolutional Networks 1 Outline Geometry of Hidden Unit Activations
COMP9444 Neural Networks and Deep Learning 5. Convolutional Networks Textbook, Sections 6.2.2, 6.3, 7.9, 7.11-7.13, 9.1-9.5 COMP9444 17s2 Convolutional Networks 1 Outline Geometry of Hidden Unit Activations
arxiv: v2 [cs.cv] 7 Jun 2018
![arxiv: v2 [cs.cv] 7 Jun 2018 arxiv: v2 [cs.cv] 7 Jun 2018](/thumbs/79/79761281.jpg) Deep supervision with additional labels for retinal vessel segmentation task Yishuo Zhang and Albert C.S. Chung Lo Kwee-Seong Medical Image Analysis Laboratory, Department of Computer Science and Engineering,
Deep supervision with additional labels for retinal vessel segmentation task Yishuo Zhang and Albert C.S. Chung Lo Kwee-Seong Medical Image Analysis Laboratory, Department of Computer Science and Engineering,
The Impact of Visual Saliency Prediction in Image Classification
 Dublin City University Insight Centre for Data Analytics Universitat Politecnica de Catalunya Escola Tècnica Superior d Enginyeria de Telecomunicacions de Barcelona Eric Arazo Sánchez The Impact of Visual
Dublin City University Insight Centre for Data Analytics Universitat Politecnica de Catalunya Escola Tècnica Superior d Enginyeria de Telecomunicacions de Barcelona Eric Arazo Sánchez The Impact of Visual
Image Classification with TensorFlow: Radiomics 1p/19q Chromosome Status Classification Using Deep Learning
 Image Classification with TensorFlow: Radiomics 1p/19q Chromosome Status Classification Using Deep Learning Charles Killam, LP.D. Certified Instructor, NVIDIA Deep Learning Institute NVIDIA Corporation
Image Classification with TensorFlow: Radiomics 1p/19q Chromosome Status Classification Using Deep Learning Charles Killam, LP.D. Certified Instructor, NVIDIA Deep Learning Institute NVIDIA Corporation
Efficient Deep Model Selection
 Efficient Deep Model Selection Jose Alvarez Researcher Data61, CSIRO, Australia GTC, May 9 th 2017 www.josemalvarez.net conv1 conv2 conv3 conv4 conv5 conv6 conv7 conv8 softmax prediction???????? Num Classes
Efficient Deep Model Selection Jose Alvarez Researcher Data61, CSIRO, Australia GTC, May 9 th 2017 www.josemalvarez.net conv1 conv2 conv3 conv4 conv5 conv6 conv7 conv8 softmax prediction???????? Num Classes
MAMMOGRAM AND TOMOSYNTHESIS CLASSIFICATION USING CONVOLUTIONAL NEURAL NETWORKS
 University of Kentucky UKnowledge Theses and Dissertations--Computer Science Computer Science 2017 MAMMOGRAM AND TOMOSYNTHESIS CLASSIFICATION USING CONVOLUTIONAL NEURAL NETWORKS Xiaofei Zhang University
University of Kentucky UKnowledge Theses and Dissertations--Computer Science Computer Science 2017 MAMMOGRAM AND TOMOSYNTHESIS CLASSIFICATION USING CONVOLUTIONAL NEURAL NETWORKS Xiaofei Zhang University
An Artificial Neural Network Architecture Based on Context Transformations in Cortical Minicolumns
 An Artificial Neural Network Architecture Based on Context Transformations in Cortical Minicolumns 1. Introduction Vasily Morzhakov, Alexey Redozubov morzhakovva@gmail.com, galdrd@gmail.com Abstract Cortical
An Artificial Neural Network Architecture Based on Context Transformations in Cortical Minicolumns 1. Introduction Vasily Morzhakov, Alexey Redozubov morzhakovva@gmail.com, galdrd@gmail.com Abstract Cortical
3D Deep Learning for Multi-modal Imaging-Guided Survival Time Prediction of Brain Tumor Patients
 3D Deep Learning for Multi-modal Imaging-Guided Survival Time Prediction of Brain Tumor Patients Dong Nie 1,2, Han Zhang 1, Ehsan Adeli 1, Luyan Liu 1, and Dinggang Shen 1(B) 1 Department of Radiology
3D Deep Learning for Multi-modal Imaging-Guided Survival Time Prediction of Brain Tumor Patients Dong Nie 1,2, Han Zhang 1, Ehsan Adeli 1, Luyan Liu 1, and Dinggang Shen 1(B) 1 Department of Radiology
Elad Hoffer*, Itay Hubara*, Daniel Soudry
 Train longer, generalize better: closing the generalization gap in large batch training of neural networks Elad Hoffer*, Itay Hubara*, Daniel Soudry *Equal contribution Better models - parallelization
Train longer, generalize better: closing the generalization gap in large batch training of neural networks Elad Hoffer*, Itay Hubara*, Daniel Soudry *Equal contribution Better models - parallelization
Y-Net: Joint Segmentation and Classification for Diagnosis of Breast Biopsy Images
 Y-Net: Joint Segmentation and Classification for Diagnosis of Breast Biopsy Images Sachin Mehta 1, Ezgi Mercan 1, Jamen Bartlett 2, Donald Weaver 2, Joann G. Elmore 1, and Linda Shapiro 1 1 University
Y-Net: Joint Segmentation and Classification for Diagnosis of Breast Biopsy Images Sachin Mehta 1, Ezgi Mercan 1, Jamen Bartlett 2, Donald Weaver 2, Joann G. Elmore 1, and Linda Shapiro 1 1 University
Classification of breast cancer histology images using transfer learning
 Classification of breast cancer histology images using transfer learning Sulaiman Vesal 1 ( ), Nishant Ravikumar 1, AmirAbbas Davari 1, Stephan Ellmann 2, Andreas Maier 1 1 Pattern Recognition Lab, Friedrich-Alexander-Universität
Classification of breast cancer histology images using transfer learning Sulaiman Vesal 1 ( ), Nishant Ravikumar 1, AmirAbbas Davari 1, Stephan Ellmann 2, Andreas Maier 1 1 Pattern Recognition Lab, Friedrich-Alexander-Universität
arxiv: v1 [cs.cv] 28 Feb 2018
![arxiv: v1 [cs.cv] 28 Feb 2018 arxiv: v1 [cs.cv] 28 Feb 2018](/thumbs/93/111675272.jpg) Brain Tumor Segmentation and Radiomics Survival Prediction: Contribution to the BRATS 2017 Challenge arxiv:1802.10508v1 [cs.cv] 28 Feb 2018 Fabian Isensee 1, Philipp Kickingereder 2, Wolfgang Wick 3, Martin
Brain Tumor Segmentation and Radiomics Survival Prediction: Contribution to the BRATS 2017 Challenge arxiv:1802.10508v1 [cs.cv] 28 Feb 2018 Fabian Isensee 1, Philipp Kickingereder 2, Wolfgang Wick 3, Martin
ACCELERATING EMPHYSEMA DIAGNOSIS ON LUNG CT IMAGES USING EMPHYSEMA PRE-DETECTION METHOD
 ACCELERATING EMPHYSEMA DIAGNOSIS ON LUNG CT IMAGES USING EMPHYSEMA PRE-DETECTION METHOD 1 KHAIRUL MUZZAMMIL BIN SAIPULLAH, 2 DEOK-HWAN KIM, 3 NURUL ATIQAH ISMAIL 1 Lecturer, 3 Student, Faculty of Electronic
ACCELERATING EMPHYSEMA DIAGNOSIS ON LUNG CT IMAGES USING EMPHYSEMA PRE-DETECTION METHOD 1 KHAIRUL MUZZAMMIL BIN SAIPULLAH, 2 DEOK-HWAN KIM, 3 NURUL ATIQAH ISMAIL 1 Lecturer, 3 Student, Faculty of Electronic
Deep Supervision for Pancreatic Cyst Segmentation in Abdominal CT Scans
 Deep Supervision for Pancreatic Cyst Segmentation in Abdominal CT Scans Yuyin Zhou 1, Lingxi Xie 2( ), Elliot K. Fishman 3, Alan L. Yuille 4 1,2,4 The Johns Hopkins University, Baltimore, MD 21218, USA
Deep Supervision for Pancreatic Cyst Segmentation in Abdominal CT Scans Yuyin Zhou 1, Lingxi Xie 2( ), Elliot K. Fishman 3, Alan L. Yuille 4 1,2,4 The Johns Hopkins University, Baltimore, MD 21218, USA
Accurate Automated Volumetry of Cartilage of the Knee using Convolutional Neural Networks: Data from the Osteoarthritis Initiative 1
 Zuse Institute Berlin Takustr. 7 14195 Berlin Germany ALEXANDER TACK, STEFAN ZACHOW Accurate Automated Volumetry of Cartilage of the Knee using Convolutional Neural Networks: Data from the Osteoarthritis
Zuse Institute Berlin Takustr. 7 14195 Berlin Germany ALEXANDER TACK, STEFAN ZACHOW Accurate Automated Volumetry of Cartilage of the Knee using Convolutional Neural Networks: Data from the Osteoarthritis
Image-Based Estimation of Real Food Size for Accurate Food Calorie Estimation
 Image-Based Estimation of Real Food Size for Accurate Food Calorie Estimation Takumi Ege, Yoshikazu Ando, Ryosuke Tanno, Wataru Shimoda and Keiji Yanai Department of Informatics, The University of Electro-Communications,
Image-Based Estimation of Real Food Size for Accurate Food Calorie Estimation Takumi Ege, Yoshikazu Ando, Ryosuke Tanno, Wataru Shimoda and Keiji Yanai Department of Informatics, The University of Electro-Communications,
arxiv: v1 [cs.cv] 12 Jun 2018
![arxiv: v1 [cs.cv] 12 Jun 2018 arxiv: v1 [cs.cv] 12 Jun 2018](/thumbs/90/102210908.jpg) Multiview Two-Task Recursive Attention Model for Left Atrium and Atrial Scars Segmentation Jun Chen* 1, Guang Yang* 2, Zhifan Gao 3, Hao Ni 4, Elsa Angelini 5, Raad Mohiaddin 2, Tom Wong 2,Yanping Zhang
Multiview Two-Task Recursive Attention Model for Left Atrium and Atrial Scars Segmentation Jun Chen* 1, Guang Yang* 2, Zhifan Gao 3, Hao Ni 4, Elsa Angelini 5, Raad Mohiaddin 2, Tom Wong 2,Yanping Zhang
Retinopathy Net. Alberto Benavides Robert Dadashi Neel Vadoothker
 Retinopathy Net Alberto Benavides Robert Dadashi Neel Vadoothker Motivation We were interested in applying deep learning techniques to the field of medical imaging Field holds a lot of promise and can
Retinopathy Net Alberto Benavides Robert Dadashi Neel Vadoothker Motivation We were interested in applying deep learning techniques to the field of medical imaging Field holds a lot of promise and can
arxiv: v1 [cs.cv] 21 Jul 2017
![arxiv: v1 [cs.cv] 21 Jul 2017 arxiv: v1 [cs.cv] 21 Jul 2017](/thumbs/89/100163556.jpg) A Multi-Scale CNN and Curriculum Learning Strategy for Mammogram Classification William Lotter 1,2, Greg Sorensen 2, and David Cox 1,2 1 Harvard University, Cambridge MA, USA 2 DeepHealth Inc., Cambridge
A Multi-Scale CNN and Curriculum Learning Strategy for Mammogram Classification William Lotter 1,2, Greg Sorensen 2, and David Cox 1,2 1 Harvard University, Cambridge MA, USA 2 DeepHealth Inc., Cambridge
A Survey on Brain Tumor Detection Technique
 (International Journal of Computer Science & Management Studies) Vol. 15, Issue 06 A Survey on Brain Tumor Detection Technique Manju Kadian 1 and Tamanna 2 1 M.Tech. Scholar, CSE Department, SPGOI, Rohtak
(International Journal of Computer Science & Management Studies) Vol. 15, Issue 06 A Survey on Brain Tumor Detection Technique Manju Kadian 1 and Tamanna 2 1 M.Tech. Scholar, CSE Department, SPGOI, Rohtak
A Reliable Method for Brain Tumor Detection Using Cnn Technique
 IOSR Journal of Electrical and Electronics Engineering (IOSR-JEEE) e-issn: 2278-1676,p-ISSN: 2320-3331, PP 64-68 www.iosrjournals.org A Reliable Method for Brain Tumor Detection Using Cnn Technique Neethu
IOSR Journal of Electrical and Electronics Engineering (IOSR-JEEE) e-issn: 2278-1676,p-ISSN: 2320-3331, PP 64-68 www.iosrjournals.org A Reliable Method for Brain Tumor Detection Using Cnn Technique Neethu
Research Article Multiscale CNNs for Brain Tumor Segmentation and Diagnosis
 Computational and Mathematical Methods in Medicine Volume 2016, Article ID 8356294, 7 pages http://dx.doi.org/10.1155/2016/8356294 Research Article Multiscale CNNs for Brain Tumor Segmentation and Diagnosis
Computational and Mathematical Methods in Medicine Volume 2016, Article ID 8356294, 7 pages http://dx.doi.org/10.1155/2016/8356294 Research Article Multiscale CNNs for Brain Tumor Segmentation and Diagnosis
arxiv: v1 [cs.cv] 24 Jul 2018
![arxiv: v1 [cs.cv] 24 Jul 2018 arxiv: v1 [cs.cv] 24 Jul 2018](/thumbs/82/86822918.jpg) Multi-Class Lesion Diagnosis with Pixel-wise Classification Network Manu Goyal 1, Jiahua Ng 2, and Moi Hoon Yap 1 1 Visual Computing Lab, Manchester Metropolitan University, M1 5GD, UK 2 University of
Multi-Class Lesion Diagnosis with Pixel-wise Classification Network Manu Goyal 1, Jiahua Ng 2, and Moi Hoon Yap 1 1 Visual Computing Lab, Manchester Metropolitan University, M1 5GD, UK 2 University of
arxiv: v2 [cs.lg] 1 Jun 2018
![arxiv: v2 [cs.lg] 1 Jun 2018 arxiv: v2 [cs.lg] 1 Jun 2018](/thumbs/89/101161632.jpg) Shagun Sodhani 1 * Vardaan Pahuja 1 * arxiv:1805.11016v2 [cs.lg] 1 Jun 2018 Abstract Self-play (Sukhbaatar et al., 2017) is an unsupervised training procedure which enables the reinforcement learning agents
Shagun Sodhani 1 * Vardaan Pahuja 1 * arxiv:1805.11016v2 [cs.lg] 1 Jun 2018 Abstract Self-play (Sukhbaatar et al., 2017) is an unsupervised training procedure which enables the reinforcement learning agents
arxiv: v1 [cs.cv] 3 Apr 2018
![arxiv: v1 [cs.cv] 3 Apr 2018 arxiv: v1 [cs.cv] 3 Apr 2018](/thumbs/90/103707026.jpg) Towards whole-body CT Bone Segmentation André Klein 1,2, Jan Warszawski 2, Jens Hillengaß 3, Klaus H. Maier-Hein 1 arxiv:1804.00908v1 [cs.cv] 3 Apr 2018 1 Division of Medical Image Computing, Deutsches
Towards whole-body CT Bone Segmentation André Klein 1,2, Jan Warszawski 2, Jens Hillengaß 3, Klaus H. Maier-Hein 1 arxiv:1804.00908v1 [cs.cv] 3 Apr 2018 1 Division of Medical Image Computing, Deutsches
Satoru Hiwa, 1 Kenya Hanawa, 2 Ryota Tamura, 2 Keisuke Hachisuka, 3 and Tomoyuki Hiroyasu Introduction
 Computational Intelligence and Neuroscience Volume 216, Article ID 1841945, 9 pages http://dx.doi.org/1.1155/216/1841945 Research Article Analyzing Brain Functions by Subject Classification of Functional
Computational Intelligence and Neuroscience Volume 216, Article ID 1841945, 9 pages http://dx.doi.org/1.1155/216/1841945 Research Article Analyzing Brain Functions by Subject Classification of Functional
AUTOMATING NEUROLOGICAL DISEASE DIAGNOSIS USING STRUCTURAL MR BRAIN SCAN FEATURES
 AUTOMATING NEUROLOGICAL DISEASE DIAGNOSIS USING STRUCTURAL MR BRAIN SCAN FEATURES ALLAN RAVENTÓS AND MOOSA ZAIDI Stanford University I. INTRODUCTION Nine percent of those aged 65 or older and about one
AUTOMATING NEUROLOGICAL DISEASE DIAGNOSIS USING STRUCTURAL MR BRAIN SCAN FEATURES ALLAN RAVENTÓS AND MOOSA ZAIDI Stanford University I. INTRODUCTION Nine percent of those aged 65 or older and about one
LUNG CANCER continues to rank as the leading cause
 1138 IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 24, NO. 9, SEPTEMBER 2005 Computer-Aided Diagnostic Scheme for Distinction Between Benign and Malignant Nodules in Thoracic Low-Dose CT by Use of Massive
1138 IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 24, NO. 9, SEPTEMBER 2005 Computer-Aided Diagnostic Scheme for Distinction Between Benign and Malignant Nodules in Thoracic Low-Dose CT by Use of Massive
arxiv: v3 [stat.ml] 28 Oct 2017
![arxiv: v3 [stat.ml] 28 Oct 2017 arxiv: v3 [stat.ml] 28 Oct 2017](/thumbs/81/83820195.jpg) Interpretable Deep Learning applied to Plant Stress Phenotyping arxiv:1710.08619v3 [stat.ml] 28 Oct 2017 Sambuddha Ghosal sghosal@iastate.edu Asheesh K. Singh singhak@iastate.edu Arti Singh arti@iastate.edu
Interpretable Deep Learning applied to Plant Stress Phenotyping arxiv:1710.08619v3 [stat.ml] 28 Oct 2017 Sambuddha Ghosal sghosal@iastate.edu Asheesh K. Singh singhak@iastate.edu Arti Singh arti@iastate.edu
Training deep Autoencoders for collaborative filtering Oleksii Kuchaiev & Boris Ginsburg
 Training deep Autoencoders for collaborative filtering Oleksii Kuchaiev & Boris Ginsburg Motivation Personalized recommendations 2 Key points (spoiler alert) 1. Deep autoencoder for collaborative filtering
Training deep Autoencoders for collaborative filtering Oleksii Kuchaiev & Boris Ginsburg Motivation Personalized recommendations 2 Key points (spoiler alert) 1. Deep autoencoder for collaborative filtering
Sign Language Recognition using Convolutional Neural Networks
 Sign Language Recognition using Convolutional Neural Networks Lionel Pigou, Sander Dieleman, Pieter-Jan Kindermans, Benjamin Schrauwen Ghent University, ELIS, Belgium Abstract. There is an undeniable communication
Sign Language Recognition using Convolutional Neural Networks Lionel Pigou, Sander Dieleman, Pieter-Jan Kindermans, Benjamin Schrauwen Ghent University, ELIS, Belgium Abstract. There is an undeniable communication
Smaller, faster, deeper: University of Edinburgh MT submittion to WMT 2017
 Smaller, faster, deeper: University of Edinburgh MT submittion to WMT 2017 Rico Sennrich, Alexandra Birch, Anna Currey, Ulrich Germann, Barry Haddow, Kenneth Heafield, Antonio Valerio Miceli Barone, Philip
Smaller, faster, deeper: University of Edinburgh MT submittion to WMT 2017 Rico Sennrich, Alexandra Birch, Anna Currey, Ulrich Germann, Barry Haddow, Kenneth Heafield, Antonio Valerio Miceli Barone, Philip
arxiv: v1 [cs.cv] 2 Jul 2018
![arxiv: v1 [cs.cv] 2 Jul 2018 arxiv: v1 [cs.cv] 2 Jul 2018](/thumbs/87/96052165.jpg) A Pulmonary Nodule Detection Model Based on Progressive Resolution and Hierarchical Saliency Junjie Zhang 1, Yong Xia 1,2, and Yanning Zhang 1 1 Shaanxi Key Lab of Speech & Image Information Processing
A Pulmonary Nodule Detection Model Based on Progressive Resolution and Hierarchical Saliency Junjie Zhang 1, Yong Xia 1,2, and Yanning Zhang 1 1 Shaanxi Key Lab of Speech & Image Information Processing
Learning Convolutional Neural Networks for Graphs
 GA-65449 Learning Convolutional Neural Networks for Graphs Mathias Niepert Mohamed Ahmed Konstantin Kutzkov NEC Laboratories Europe Representation Learning for Graphs Telecom Safety Transportation Industry
GA-65449 Learning Convolutional Neural Networks for Graphs Mathias Niepert Mohamed Ahmed Konstantin Kutzkov NEC Laboratories Europe Representation Learning for Graphs Telecom Safety Transportation Industry
Automatic Detection of Knee Joints and Quantification of Knee Osteoarthritis Severity using Convolutional Neural Networks
 Automatic Detection of Knee Joints and Quantification of Knee Osteoarthritis Severity using Convolutional Neural Networks Joseph Antony 1, Kevin McGuinness 1, Kieran Moran 1,2 and Noel E O Connor 1 Insight
Automatic Detection of Knee Joints and Quantification of Knee Osteoarthritis Severity using Convolutional Neural Networks Joseph Antony 1, Kevin McGuinness 1, Kieran Moran 1,2 and Noel E O Connor 1 Insight
arxiv: v1 [cs.ai] 28 Nov 2017
![arxiv: v1 [cs.ai] 28 Nov 2017 arxiv: v1 [cs.ai] 28 Nov 2017](/thumbs/78/77416856.jpg) : a better way of the parameters of a Deep Neural Network arxiv:1711.10177v1 [cs.ai] 28 Nov 2017 Guglielmo Montone Laboratoire Psychologie de la Perception Université Paris Descartes, Paris montone.guglielmo@gmail.com
: a better way of the parameters of a Deep Neural Network arxiv:1711.10177v1 [cs.ai] 28 Nov 2017 Guglielmo Montone Laboratoire Psychologie de la Perception Université Paris Descartes, Paris montone.guglielmo@gmail.com
Handling Unbalanced Data in Deep Image Segmentation
 Handling Unbalanced Data in Deep Image Segmentation Harriet Small Brown University harriet small@brown.edu Jonathan Ventura University of Colorado, Colorado Springs jventura@uccs.edu Abstract We approach
Handling Unbalanced Data in Deep Image Segmentation Harriet Small Brown University harriet small@brown.edu Jonathan Ventura University of Colorado, Colorado Springs jventura@uccs.edu Abstract We approach
Two-Stage Convolutional Neural Network for Breast Cancer Histology Image Classification
 Two-Stage Convolutional Neural Network for Breast Cancer Histology Image Classification Kamyar Nazeri, Azad Aminpour, and Mehran Ebrahimi Faculty of Science, University of Ontario Institute of Technology
Two-Stage Convolutional Neural Network for Breast Cancer Histology Image Classification Kamyar Nazeri, Azad Aminpour, and Mehran Ebrahimi Faculty of Science, University of Ontario Institute of Technology
CS-E Deep Learning Session 4: Convolutional Networks
 CS-E4050 - Deep Learning Session 4: Convolutional Networks Jyri Kivinen Aalto University 23 September 2015 Credits: Thanks to Tapani Raiko for slides material. CS-E4050 - Deep Learning Session 4: Convolutional
CS-E4050 - Deep Learning Session 4: Convolutional Networks Jyri Kivinen Aalto University 23 September 2015 Credits: Thanks to Tapani Raiko for slides material. CS-E4050 - Deep Learning Session 4: Convolutional
Multi-attention Guided Activation Propagation in CNNs
 Multi-attention Guided Activation Propagation in CNNs Xiangteng He and Yuxin Peng (B) Institute of Computer Science and Technology, Peking University, Beijing, China pengyuxin@pku.edu.cn Abstract. CNNs
Multi-attention Guided Activation Propagation in CNNs Xiangteng He and Yuxin Peng (B) Institute of Computer Science and Technology, Peking University, Beijing, China pengyuxin@pku.edu.cn Abstract. CNNs
Copyright 2007 IEEE. Reprinted from 4th IEEE International Symposium on Biomedical Imaging: From Nano to Macro, April 2007.
 Copyright 27 IEEE. Reprinted from 4th IEEE International Symposium on Biomedical Imaging: From Nano to Macro, April 27. This material is posted here with permission of the IEEE. Such permission of the
Copyright 27 IEEE. Reprinted from 4th IEEE International Symposium on Biomedical Imaging: From Nano to Macro, April 27. This material is posted here with permission of the IEEE. Such permission of the
MRI Image Processing Operations for Brain Tumor Detection
 MRI Image Processing Operations for Brain Tumor Detection Prof. M.M. Bulhe 1, Shubhashini Pathak 2, Karan Parekh 3, Abhishek Jha 4 1Assistant Professor, Dept. of Electronics and Telecommunications Engineering,
MRI Image Processing Operations for Brain Tumor Detection Prof. M.M. Bulhe 1, Shubhashini Pathak 2, Karan Parekh 3, Abhishek Jha 4 1Assistant Professor, Dept. of Electronics and Telecommunications Engineering,
Patch-based Head and Neck Cancer Subtype Classification
 Patch-based Head and Neck Cancer Subtype Classification Wanyi Qian, Guoli Yin, Frances Liu, Advisor: Olivier Gevaert, Mu Zhou, Kevin Brennan Stanford University wqian2@stanford.edu, guoliy@stanford.edu,
Patch-based Head and Neck Cancer Subtype Classification Wanyi Qian, Guoli Yin, Frances Liu, Advisor: Olivier Gevaert, Mu Zhou, Kevin Brennan Stanford University wqian2@stanford.edu, guoliy@stanford.edu,
OPTIMIZING CHANNEL SELECTION FOR SEIZURE DETECTION
 OPTIMIZING CHANNEL SELECTION FOR SEIZURE DETECTION V. Shah, M. Golmohammadi, S. Ziyabari, E. Von Weltin, I. Obeid and J. Picone Neural Engineering Data Consortium, Temple University, Philadelphia, Pennsylvania,
OPTIMIZING CHANNEL SELECTION FOR SEIZURE DETECTION V. Shah, M. Golmohammadi, S. Ziyabari, E. Von Weltin, I. Obeid and J. Picone Neural Engineering Data Consortium, Temple University, Philadelphia, Pennsylvania,
Development of Novel Approach for Classification and Detection of Brain Tumor
 International Journal of Latest Technology in Engineering & Management (IJLTEM) www.ijltem.com ISSN: 245677 Development of Novel Approach for Classification and Detection of Brain Tumor Abstract This paper
International Journal of Latest Technology in Engineering & Management (IJLTEM) www.ijltem.com ISSN: 245677 Development of Novel Approach for Classification and Detection of Brain Tumor Abstract This paper
Shared Response Model Tutorial! What works? How can it help you? Po-Hsuan (Cameron) Chen and Hejia Hasson Lab! Feb 15, 2017!
 Shared Response Model Tutorial! What works? How can it help you? Po-Hsuan (Cameron) Chen and Hejia Zhang!!! @ Hasson Lab! Feb 15, 2017! Outline! SRM Theory! SRM on fmri! Hands-on SRM with BrainIak! SRM
Shared Response Model Tutorial! What works? How can it help you? Po-Hsuan (Cameron) Chen and Hejia Zhang!!! @ Hasson Lab! Feb 15, 2017! Outline! SRM Theory! SRM on fmri! Hands-on SRM with BrainIak! SRM
Functional MRI Mapping Cognition
 Outline Functional MRI Mapping Cognition Michael A. Yassa, B.A. Division of Psychiatric Neuro-imaging Psychiatry and Behavioral Sciences Johns Hopkins School of Medicine Why fmri? fmri - How it works Research
Outline Functional MRI Mapping Cognition Michael A. Yassa, B.A. Division of Psychiatric Neuro-imaging Psychiatry and Behavioral Sciences Johns Hopkins School of Medicine Why fmri? fmri - How it works Research
Deep Learning based FACS Action Unit Occurrence and Intensity Estimation
 Deep Learning based FACS Action Unit Occurrence and Intensity Estimation Amogh Gudi, H. Emrah Tasli, Tim M. den Uyl, Andreas Maroulis Vicarious Perception Technologies, Amsterdam, The Netherlands Abstract
Deep Learning based FACS Action Unit Occurrence and Intensity Estimation Amogh Gudi, H. Emrah Tasli, Tim M. den Uyl, Andreas Maroulis Vicarious Perception Technologies, Amsterdam, The Netherlands Abstract
arxiv: v3 [cs.cv] 28 Mar 2017
![arxiv: v3 [cs.cv] 28 Mar 2017 arxiv: v3 [cs.cv] 28 Mar 2017](/thumbs/73/69121210.jpg) Discovery Radiomics for Pathologically-Proven Computed Tomography Lung Cancer Prediction Devinder Kumar 1*, Audrey G. Chung 1, Mohammad J. Shaifee 1, Farzad Khalvati 2,3, Masoom A. Haider 2,3, and Alexander
Discovery Radiomics for Pathologically-Proven Computed Tomography Lung Cancer Prediction Devinder Kumar 1*, Audrey G. Chung 1, Mohammad J. Shaifee 1, Farzad Khalvati 2,3, Masoom A. Haider 2,3, and Alexander
Automatic Definition of Planning Target Volume in Computer-Assisted Radiotherapy
 Automatic Definition of Planning Target Volume in Computer-Assisted Radiotherapy Angelo Zizzari Department of Cybernetics, School of Systems Engineering The University of Reading, Whiteknights, PO Box
Automatic Definition of Planning Target Volume in Computer-Assisted Radiotherapy Angelo Zizzari Department of Cybernetics, School of Systems Engineering The University of Reading, Whiteknights, PO Box
Neural Network for Detecting Head Impacts from Kinematic Data. Michael Fanton, Nicholas Gaudio, Alissa Ling CS 229 Project Report
 Neural Network for Detecting Head Impacts from Kinematic Data Michael Fanton, Nicholas Gaudio, Alissa Ling CS 229 Project Report 1. Abstract Mild Traumatic Brain Injury (mtbi) is a serious health concern,
Neural Network for Detecting Head Impacts from Kinematic Data Michael Fanton, Nicholas Gaudio, Alissa Ling CS 229 Project Report 1. Abstract Mild Traumatic Brain Injury (mtbi) is a serious health concern,
arxiv: v1 [cs.cv] 11 Jul 2017
![arxiv: v1 [cs.cv] 11 Jul 2017 arxiv: v1 [cs.cv] 11 Jul 2017](/thumbs/86/94145429.jpg) Adversarial training and dilated convolutions for brain MRI segmentation Pim Moeskops 1, Mitko Veta 1, Maxime W. Lafarge 1, Koen A.J. Eppenhof 1, and Josien P.W. Pluim 1 Medical Image Analysis Group, Department
Adversarial training and dilated convolutions for brain MRI segmentation Pim Moeskops 1, Mitko Veta 1, Maxime W. Lafarge 1, Koen A.J. Eppenhof 1, and Josien P.W. Pluim 1 Medical Image Analysis Group, Department
ANALYSIS AND DETECTION OF BRAIN TUMOUR USING IMAGE PROCESSING TECHNIQUES
 ANALYSIS AND DETECTION OF BRAIN TUMOUR USING IMAGE PROCESSING TECHNIQUES P.V.Rohini 1, Dr.M.Pushparani 2 1 M.Phil Scholar, Department of Computer Science, Mother Teresa women s university, (India) 2 Professor
ANALYSIS AND DETECTION OF BRAIN TUMOUR USING IMAGE PROCESSING TECHNIQUES P.V.Rohini 1, Dr.M.Pushparani 2 1 M.Phil Scholar, Department of Computer Science, Mother Teresa women s university, (India) 2 Professor
EARLY STAGE DIAGNOSIS OF LUNG CANCER USING CT-SCAN IMAGES BASED ON CELLULAR LEARNING AUTOMATE
 EARLY STAGE DIAGNOSIS OF LUNG CANCER USING CT-SCAN IMAGES BASED ON CELLULAR LEARNING AUTOMATE SAKTHI NEELA.P.K Department of M.E (Medical electronics) Sengunthar College of engineering Namakkal, Tamilnadu,
EARLY STAGE DIAGNOSIS OF LUNG CANCER USING CT-SCAN IMAGES BASED ON CELLULAR LEARNING AUTOMATE SAKTHI NEELA.P.K Department of M.E (Medical electronics) Sengunthar College of engineering Namakkal, Tamilnadu,
Copyright 2008 Society of Photo Optical Instrumentation Engineers. This paper was published in Proceedings of SPIE, vol. 6915, Medical Imaging 2008:
 Copyright 2008 Society of Photo Optical Instrumentation Engineers. This paper was published in Proceedings of SPIE, vol. 6915, Medical Imaging 2008: Computer Aided Diagnosis and is made available as an
Copyright 2008 Society of Photo Optical Instrumentation Engineers. This paper was published in Proceedings of SPIE, vol. 6915, Medical Imaging 2008: Computer Aided Diagnosis and is made available as an
Less is More: Culling the Training Set to Improve Robustness of Deep Neural Networks
 Less is More: Culling the Training Set to Improve Robustness of Deep Neural Networks Yongshuai Liu, Jiyu Chen, and Hao Chen University of California, Davis {yshliu, jiych, chen}@ucdavis.edu Abstract. Deep
Less is More: Culling the Training Set to Improve Robustness of Deep Neural Networks Yongshuai Liu, Jiyu Chen, and Hao Chen University of California, Davis {yshliu, jiych, chen}@ucdavis.edu Abstract. Deep
