Automated identification of analyzable metaphase chromosomes depicted on microscopic digital images
|
|
|
- Thomasine Phelps
- 5 years ago
- Views:
Transcription
1 Available online at Journal of Biomedical Informatics 41 (2008) Automated identification of analyzable metaphase chromosomes depicted on microscopic digital images Xingwei Wang a, Shibo Li b, Hong Liu a, Marc Wood a, Wei R. Chen c, Bin Zheng d, * a Center for Bioengineering and School of Electrical and Computer Engineering, University of Oklahoma, Norman, OK 73019, USA b Department of Pediatrics, University of Oklahoma Health Science Center, Oklahoma City, OK 73104, USA c Department of Physics and Engineering, University of Central Oklahoma, Edmond, OK 73034, USA d Department of Radiology, University of Pittsburgh, 300 Halket Street, Suite 4200, Pittsburgh, PA , USA Received 27 February 2007 Available online 10 July 2007 Abstract Visual search and identification of analyzable metaphase chromosomes using optical microscopes is a very tedious and time-consuming task that is routinely performed in genetic laboratories to detect and diagnose cancers and genetic diseases. The purpose of this study is to develop and test a computerized scheme that can automatically identify chromosomes in metaphase stage and classify them into analyzable and un-analyzable groups. Two independent datasets involving 170 images are used to train and test the scheme. The scheme uses image filtering, threshold, and labeling algorithms to detect chromosomes, followed by computing a set of features for each individual chromosome as well as for each identified metaphase cell. Two machine learning classifiers including a decision tree (DT) based on the features of individual chromosomes and an artificial neural network (ANN) using the features of the metaphase cells are optimized and tested to classify between analyzable and un-analyzable cells. Using the DT based classifier the Kappa coefficients for agreement between the cytogeneticist and the scheme are 0.83 and 0.89 for the training and testing datasets, respectively. We apply an independent testing and a 2-fold cross-validation method to assess the performance of the ANN-based classifier. The area under and receiver operating characteristic (ROC) curve is 0.93 for the complete dataset. This preliminary study demonstrates the feasibility of developing a computerized scheme to automatically identify and classify metaphase chromosomes. Ó 2007 Elsevier Inc. All rights reserved. Keywords: Artificial neural network; Decision tree; Metaphase chromosomes; Microscopic digital images 1. Introduction Since Tjio and Levan discovered that the chromosome number of human being was 46 in 1956 [1], the knowledge about chromosomal abnormalities, as a cause of diseases, increased enormously. For example, in 1960 Nowell and Hungerford discovered a small chromosome marker, the Philadelphia chromosome, in patients with chronic myeloid leukemia (CML) [2]. This is proved to be the first consistent chromosomal abnormality in human cancer and it greatly stimulated interest in cancer cytogenetics. Currently, identification and classification of chromosomes * Corresponding author. Fax: address: zhengb@upmc.edu (B. Zheng). using optical microscopic images is an important laboratory and clinical procedure to screen and diagnose genetic disorders [3], cancers [4,5] and other diseases [6]. Specifically, chromosome abnormalities and mutations are the results of changes in chromosome structure. Studies have found that consistent chromosomal changes led to isolation of the genes involved in the cancer pathogenesis [7]. Detection of these consistent, recurrent chromosomal changes has allowed the division of patients into clinical groups which define their duration of remission and mean survival time [8]. Hence, better understanding the mechanism of abnormal chromosome can help oncologists evaluate cancer prognosis and select more effective treatment procedures. For this purpose, karyotyping of metaphase cells is the most common procedure to analyze and classify /$ - see front matter Ó 2007 Elsevier Inc. All rights reserved. doi: /j.jbi
2 X. Wang et al. / Journal of Biomedical Informatics 41 (2008) chromosomes [9]. This procedure requires generating a layout of chromosomes organized by decreasing size in pairs for each testing cell by the comparison between the chromosomes identified in the cell and the chromosomes stored in a pre-established standard database. Chromosomes are assigned to each of the 24 classes [10]. Then, variety of diseases, genetic disorders, and cancers, can be diagnosed based on the possible distortion of the banded patterns of different chromosome pairs [11]. Before performing karyotyping and other diagnostic procedures, a cytogenetic technologist uses an optical microscope to visually examine each glass slide prepared from samples (i.e., amniotic fluid, blood sample, skin or bone marrow) acquired from a patient to search for and identify the cells with analyzable chromosomes. Because the cells are very small (in the order of a few 100 lm), the technologist must move the slide under the microscope many times to thoroughly search the entire slide and frequently switch between two microscopic objectives of low (e.g., 10 ) and high magnification power (e.g., 100 ). It is desirable to find at least analyzable metaphase chromosome cells for each patient. Since, not all cells are engaged in cell division and not all dividing cells are in the analyzable metaphase stage, the technologist must look at a large number of cells before a cell with clearly imaged chromosomes can be found. Fig. 1 demonstrates two metaphase chromosome cells, one is considered analyzable and one is un-analyzable by an experienced cytogeneticist. In the clinical practice, examining as many as 5 10 microscopic sample slides is typically needed for one patient. Therefore, it takes tremendous effort and time for the technologist to obtain a sufficient number of analyzable metaphase cells before making an accurate laboratory diagnosis of abnormal human chromosomes that have been altered by disease or cancer mechanisms. This lengthy and inefficient process can also cause delay to the treatment of patients. Therefore, developing a computerized scheme could potentially significantly speed up the process of searching for and identifying analyzable chromosomes. It may also help cytogeneticists improve the diagnostic performance and minimize inter- and intra-reader variability. Fig. 1. Digital images of two metaphase chromosome cells in which (a) is considered un-analyzable cells that will be deleted and (b) is an analyzable cell that will be selected to perform karyotyping. Since 1980s a number of research groups has developed and tested different computerized schemes to analyze chromosome images including identifying analyzable metaphase cells, separating overlapped chromosomes, classifying different chromosomes (karyotyping), and detecting abnormal patterns depicted on the chromosomes. The detailed discussion of the previous studies (including the advantages and limitations of these studies) has been reported in a previously published review article [12]. For examples, one study developed an Athena system to semi-automatically identify analyzable metaphase chromosomes [13]. The second study reported an automated karyotyping system (AKS) using a novel recursive searching algorithm [14]. The third study developed a scheme to recognize metaphase spreads from nuclei and artifacts in microscopic images acquired at 10 magnification power with approximately 91% recognition rate [15]. The fourth study developed an automatic metaphase finder and reported a true positive rate of 80% with a false positive rate of 20% [16]. The fifth group used texture features to classify manually segmented objects into metaphase spreads with approximately true positive rate of 85% [17]. The sixth group applied a wavelet-based algorithm to improve the salient features of chromosome images for the better diagnosis [18]. This group also developed and reported a subspace-based model to classify chromosomes into 24 types after applying the semi-automated methods to pre-process all individual chromosomes (including straightening the bending chromosomes and identifying the orientation of p-arm of each chromosome) [19]. In addition, different statistical models and machine learning classifiers (i.e., artificial neural network and probabilistic Markov network) based on an optimal feature vector or pixel value distribution have been trained and implemented in computerized schemes to detect (or identify) abnormal patterns of chromosomes [20 23]. The success of the schemes for detecting chromosomal aberrations depends on several pre-conditions including that the analyzable metaphase chromosomes have been identified and the individual chromosomes have been separated (without overlapping) and correctly oriented (i.e., straightening the bending chromosomes and knowing p-arms). As a result, when applying these schemes to real clinical images, human intervention is often required to select analyzable chromosomes [24]. For examples, since early 1990s, several research groups have tested the feasibility of applying the semi-automated computer systems to detect aberrant chromosomes in different clinical images [25,26]. Despite the considerable research effort in developing computerized schemes to analyze chromosome images, one of the most importantly clinical issues of how to automatically and robustly detecting and identifying analyzable metaphase chromosomes depicted on the original microscopic sample slides remains un-solved. As a result, the technologists in genetic laboratories still visually search for and identify analyzable metaphase chromosomes in the routine clinical practice to date. In this study, we
3 266 X. Wang et al. / Journal of Biomedical Informatics 41 (2008) focused on developing and testing a new automated scheme to identify metaphase chromosome cells depicted on microscopic digital images and to classify them into analyzable and un-analyzable cells. The purpose of this study is to develop a simple and robust scheme that has potential to replace the time-consumed visual searching process. This scheme can also be the first and important step in developing other fully automated schemes for chromosome image analysis and diagnosis. 2. Materials and methods From a clinical database established at a genetic laboratory of University of Oklahoma Health Science Center, a cytogeneticist randomly selected 100 metaphase cells as one dataset (which was later used as a training dataset) and then selected another dataset involving 70 metaphase cells (a testing dataset). An optical microscope equipped with an objective of 100 magnification power and a digital camera was used to acquire digital images of these selected metaphase chromosome cells. These images have average size of pixels and the size of each pixel on the surface of the slides is approximately 0.2 lm 0.2 lm. In the training dataset, 35 cells are considered (visually recognized as) analyzable and 65 are un-analyzable by the cytogeneticist. In the test dataset, 37 cells are analyzable and 33 are un-analyzable, respectively. These visual classification results were saved in a truth file that is used as a standard to train our computerized scheme and test its performance. Our computerized scheme includes five image processing and feature classification stages to segment chromosome areas and to identify analyzable metaphase cells (Fig. 2). First, the scheme uses an image filter to pre-process the images that aims to enhance the image quality (i.e., increase signal-to-noise ratio). Because a median filter is a simple and effective filter to reduce random image noise while preserving image sharpness (minimizing image blurring) [27], we implement a median filter (with window size of 5 5 pixels) in the scheme to reduce the noise and artifact background in the originally digital chromosome images. Second, an adjustable threshold is applied to remove all pixels with gray (digital) values smaller than the threshold (because chromosomes in metaphase stages usually have greater gray value than majority of artifacts in the background). Third, a 4-connectivity component labeling algorithm and a raster scanning method are applied to label and group the detected pixels into connected areas and delete the isolated pixels. The rationale of selecting the 4-connectivity component labeling algorithm is because it is less sensitive to the image noise (or broken pixels) comparing to other labeling algorithm (e.g., 8-connectivity component labeling algorithm) [27]. Fourth, the scheme computes following five image features from the labeled regions in each microscopic digital image. Fig. 2. A flow diagram of a computerized scheme to segment chromosomes and classify metaphase cells into analyzable and un-analyzable cells. 1. The number of labeled regions: the scheme counts the total number of labeled regions (N M ) in one image slide. 2. The size of each labeled region: it is defined by counting the number of pixels involved in the region (S i = N M ). 3. The circularity of each labeled region: based on the size of the region (N M ), the scheme defines an equivalent circle originating at the gravity center of the region. For a circle with the same size as the labeled region, the scheme computes the number of pixels that are located inside the region contour and the circle (N C ). The circularity is defined as C i ¼ N C N M, the ratio of the region pixels covered by the circle and the total pixels inside the labeled region [28]. 4. Average gray value of each labeled region: it is computed as P an average digital value of pixels I ave ¼ 1 n n i¼1 I i. 5. The radial length of each region to the cell center: it is defined as the distance between the gravity center (x c,y c ) of total labeled regions in the image (center of a cell) and the center of one individual region (x k,y k ). It is computed as L k ¼ ðx c x k Þ 2 þðy c y k Þ 2 qffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffi. In these five features, feature #1 is global feature of the metaphase cell and the rest of four features are computed for each individual chromosome. In the last stage of the scheme, a machine learning classifier is applied to identify analyzable metaphase chromosomes and delete un-analyzable ones. Two classifiers, a decision tree (DT) and an artificial neural network
4 X. Wang et al. / Journal of Biomedical Informatics 41 (2008) (ANN), are optimized and tested in this study. DT is one of the most widely used and practical methods for inductive inference, which approximates discrete-valued functions that is robust to noisy data and is capable of learning disjunctive expressions. ANN is loosely motivated by biological neural systems to learn the real-valued and discretevalued functions from noisy or incomplete samples. In particular, the training algorithm of back-propagation using gradient descent can turn ANN parameters to best fit a training set of input output pairs. The limitations of these two classifiers include potentially inductive bias for DT and over-fitting for ANN. The detailed mathematic foundations and characteristics (including advantages and limitations) of DT and ANN can be found elsewhere [29]. Despite the potential limitations, DT and ANN are two of the most popular machine learning classifiers implemented in the computerized schemes for biomedical images including chromosome images [12]. DT implemented in this study is constructed based on the five computed features and used to classify which digital image depicts an analyzable metaphase chromosome cell or not. The structure of the DT is demonstrated in Fig. 3. This DT uses a simple classification criterion: analyzable metaphase chromosome cells contain more recognizable individual chromosomes than un-analyzable cells. The DT includes four (horizontal) layers (rows). The first layer includes a start node. The five computed images features are connected to the five nodes listed in the second row. The first node in this row is the number of labeled regions. If the number of regions is less than a threshold value (that is determined by the training dataset), this chromosome image is defined as not analyzable. Only the images depicted the labeled chromosomes (regions) within the range between N min = 19 and N max = 46 are further analyzed by the following nodes of the DT. Node 2 is the size of each labeled (individual) region and it aims to delete some very large connected areas (e.g., S > S max = 5000) and very small regions (e.g., S < S min = 90) in each image. Node 3 is the circularity of the region and it is designed to delete the circled regions (e.g., a nucleus) if the circularities are larger than a pre-determined threshold (e.g., C > C T = 0.90). Node 4 is average gray level of the region and it is used to delete the regions dominated with dirty substances or cells in each image. In general, the gray value of chromosomes is larger than those dirty substances and cells. Hence, if the average gray level of a region is smaller than the threshold (e.g., I ave < I T = 75), the region is deleted. Node 5 is the radial length of the region and it deletes the labeled regions that are far away from the center of the cell or cluster (e.g., L P L k = (maximum radial length standard deviation of radial length of all labeled regions)). Since in each of nodes 2 5, the DT may delete a number of labeled regions and result in the reduction of the total number of regions remained in one image, this image (or cell) is analyzed again based on the number of remaining regions (similar to the node 1) in the third layer Fig. 3. A five feature based DT for recognizing analyzable and un-analyzable metaphase chromosome cells. Note: F1, average size of each region; F2, circularity of each region; F3, average gray value of each region; F4, radial length of each region; Th1, number of regions is between N min = 19 and N max = 46; Th2, average size of each region is between S min = 90 and S max = 5000; Th3, circularity of each region is <C T = 0.9; Th4, average gray value of each region is PI T = 75; Th5, number of regions is either <N min =19 or >N max = 46; Th6, average size of each region is either <S min =90 or >S max = 5000; Th7, circularity of each region is PC T = 0.9; Th8, average gray value of each region is <I T = 75; Th9, radial length of each region is PL k (the maximum radial length standard deviation of radial length of all labeled regions); and Th10, radial length of each region is <L k.
5 268 X. Wang et al. / Journal of Biomedical Informatics 41 (2008) (row) of the tree. The last (fourth) layer contains a number of decision nodes. The second classifier tested in this study is an ANN. Unlike the DT (Fig. 3) that uses the features computed from the individual chromosomes, the ANN only uses the features computed from all labeled regions in one acquired image region of interest (ROI). The ANN has a simple three-layer feed-forward topology. The input layer includes six neurons that are represented by six features, which are (1) the number of labeled regions; (2) the average size of all labeled regions; (3) the standard deviation of region size; (4) the average pixel value of all regions; (5) the standard deviation of pixel values; and (6) the average radial length of all regions. The hidden layer of the ANN involves three neurons and the output layer contains one decision neuron. A standard back-propagation training algorithm is used to train the ANN. Due to the limited size of our training dataset and in order to minimize over-fitting and keep the robustness of the ANN performance when applied to new testing cases, a limited number of training iterations as well as a large ratio between the momentum and learning rate is used [30]. Hence, we limit the training iterations to 400; while the momentum and learning rate are set at 0.9 and 0.01, respectively. For each training or testing sample (chromosome cell), the ANN generates a classification score in the range from 0 to 1, where 0 means definitely un-analyzable and 1 indicates easily analyzable. Different methods are applied to assess the performance of two classifiers. When using DT, we tabulated the experimental data and computed the Kappa coefficients for agreement of the classification results between the cytogeneticist and the computerized scheme. The performance of using ANN to classify metaphase chromosome cells is evaluated using receiver operating characteristics (ROC), a standard method widely used to evaluate the performance of observers and computerized schemes in diagnosis and analysis of biomedical images [31]. A computer program converts the ANN-generated classification scores of all analyzable ( positive ) and un-analyzable ( negative ) samples in one dataset (either training or testing) into two histograms with 11 bins. Based on these two histograms (one for positive cases and one for negative cases), an un-smoothed ROC-type performance curve can be plotted. ROCFIT program [32] that uses maximum likelihood estimation method [33] is then used to fit ROC data (curve) and compute an area under the ROC curve (A Z value), an index to measure the scheme performance. Because two datasets were independently selected and size of each dataset is relatively small, two datasets may have substantially different distribution of image feature characteristics. To better estimate the performance of our scheme and minimize the potential bias when using an ANN as a classifier, we also applied a 2-fold cross-validation method to assess scheme performance. We trained and tested the ANN twice by switching between training and testing dataset, which means that each of two datasets is used once for training and once for testing. The testing classification score of each cell region is used to generate the final ROC curve for the complete dataset using ROCFIT program. 3. Results The difference of classification results between the truth (provided by the cytogeneticist) and the DT based scheme for both training and testing datasets is summarized and compared (as shown in Tables 1 and 2). The results indicate that 92.0% (92 out of 100) and 94.3% (66 out of 70) of queried cell regions in the training and testing datasets are assigned to the same group (either analyzable or un-analyzable ) by both the cytogeneticist in our genetic laboratory and DT based classifier. The corresponding Kappa coefficients for agreement are 0.83 and 0.89 for the training and testing datasets, respectively. Specifically, the DT based scheme achieves 94.3% (33 out of 35) of detection sensitivity ( true-positive classification rate) and 90.8% (59 out of 65) of specificity ( true-negative classification rate) for the training dataset. For the testing dataset, the sensitivity and specificity are 91.9% (34 out of 37) and 96.9% (32 out of 33), respectively. To verify the feature distribution difference between two datasets, we plotted a series of scatter diagrams between different pairs of features. For example, the scatter diagrams between the features of average pixel values and the number of labeled regions are shown in Figs. 4 and 5 for the original training and testing dataset. For these two features the analyzable cells are mostly located in the up-right corner of the diagram (which indicated the larger number of labeled regions and higher pixel value). Comparing between Figs. 4 and 5, we find that in the initial training dataset used for DT based classifier; several unanalyzable cells also involve larger number of labeled regions and higher average pixel values. These difficult un-analyzable cells reduce the classification performance Table 1 Comparison of classification results between a cytogeneticist and the DT based scheme for training dataset Data classified DT based scheme DT accuracy by a cytogeneticist Correct Wrong rate (%) Analyzable cells Un-analyzable cells Total cells Table 2 Comparison of classification results between a cytogeneticist and the DT based scheme for testing dataset Data classified DT based scheme DT accuracy by a cytogeneticist Correct Wrong rate (%) Analyzable cells Un-analyzable cells Total cells
6 X. Wang et al. / Journal of Biomedical Informatics 41 (2008) Fig. 4. A scatter diagram between two features of 100 training samples including 35 analyzable ( positive ) and 65 un-analyzable ( negative ) cells. Fig. 6. The distribution of the computed performance points for two data subsets ( training and test subsets) and the complete dataset. A ROCtype performance curve was generated based on fitting of the complete dataset using ROCFIT program. Fig. 5. A scatter diagram between two features of 70 testing samples including 37 analyzable ( positive ) and 33 un-analyzable ( negative ) cells. of the ANN when applying to this dataset comparing to the classification performance on the initial testing dataset (as shown in Fig. 6). A ROC curve generated based on complete dataset using 2-fold cross-validation method is also plotted in Fig. 6. The computed A Z values (the areas under ROC curve) are ± 0.015, ± 0.013, and ± 0.008, for the original training, testing, and complete dataset, respectively. 4. Discussion Because of its advantages and effectiveness over traditionally anatomical imaging modalities and techniques in detecting cancers and monitoring cancer treatment efficacy, molecular and chromosome imaging have been attracted extensive research interests. Chromosomal disorder is a powerful indicator in diagnosis of cancers (i.e., leukemia, skin and breast cancers) and other genetic diseases. Although identification of chromosomal aberrations (disorders) is routinely performed in the clinical laboratories to provide physicians the diagnostic results and help them decide and manage optimal therapeutic treatment plans for patients, this is a very tedious and time-consuming task. Currently, the laboratory technologists spend more valuable time to identify analyzable metaphase chromosome cells, rather than spend time to identify and analyze the chromosomal abnormal patterns. Our previous study has demonstrated that using computerized scheme could potentially help clinicians more efficiently and accurately diagnose (classify) different types of leukemia and predict the cancer prognosis [34]. However, the precondition of accurate diagnosis of cancers and genetic diseases using chromosome images is to identify a set of analyzable (or diagnosable) metaphase chromosome cells. In addition, developing a fully automated scheme to detect and identify analyzable metaphase chromosome cells is the first and probably the most important step to replace tediously manual searching process. Therefore, the motivation and purpose of this study is to develop a computerized scheme that has potential to replace the manual searching process and meet the requirement of other semi-automated schemes in chromosome classification by automatically identifying analyzable metaphase chromosomes depicted on microscopic digital images. For this purpose, we developed and tested a simple and unique computerized scheme. The scheme was directly optimized and applied to the originally cultured chromosome images used for the standard (or banded) chromo-
7 270 X. Wang et al. / Journal of Biomedical Informatics 41 (2008) some analysis routinely performed in our genetic laboratory for the diagnosis of cancers and genetic diseases. This approach of using real clinical images without pre-processing increases the application potential and robustness of our scheme in the clinical environment. We are aware that when different cultured or sample preparation methods are used in different laboratories for different diagnostic purpose, the banded patterns of the chromosomes could be different. However, this does not affect our scheme to detect and identify analyzable metaphase chromosomes because no specific band features and absolute size features of individual chromosomes are used in our scheme. Our scheme was also applied to the high-resolution (or high magnification) microscopic digital images. Comparing with previously reported studies using low-resolution images, which could only alert the laboratory technologists the location of potentially analyzable chromosome cells and visual examination is needed by switching to another microscopic objective with high magnification power to determine whether the cell is analyzable or not [15], our approach has two advantages. First, the analyzable metaphase chromosome cells detected and prompted by the scheme can be directly examined and analyzed by technologists (or cytogeneticists) for the diagnosis purpose without using the microscope. Second, the identified analyzable metaphase chromosomes can be used by other computeraided diagnosis scheme to potentially perform more comprehensive tasks (i.e., the detection of the distortion in chromosomes banded patterns). Hence, our scheme can be integrated with other available semi-automated schemes to develop fully automated computer schemes in the future studies. During the development and evaluation of a computer scheme involving a machine learning classifier, the researchers often face a number of biases, in particular the bias of case (learning sample) selection and validation method. We took several measures (procedures) in an attempt to avoid or minimize the bias of the classification results. First, the training and testing datasets used in this study were independently selected in our genetic laboratory by cytogeneticists. The researchers who developed and tested the computerized scheme did not involve in the case selection, which helps eliminate the bias in case selection. Second, due to the limitation of size of the dataset, avoiding or minimizing the bias in classification results (e.g., due to the potential over-training) is always a difficult challenge. Several optimization (training and testing) methods including jackknifing (leave-one-out), N-fold cross-validation, and use of independent testing dataset have been widely used in development and optimization of the computerized schemes. Previous studies have suggested that both leave-one-out and cross-validation methods were more likely to generate considerable bias and variance [35,36]. Although using independent testing dataset is the best way to reduce the classification bias, dividing limited dataset into two independent datasets reduces the size and diversity of training dataset and may also reduce the performance of testing result [35]. As a result, to minimize the classification bias and assess the optimal performance of the scheme, we used both independent testing and 2-fold cross-validation methods in this study. The results of this preliminary study are encouraging. The scheme achieved high performance on an independent testing dataset (i.e., Kappa = 0.89 for using the DT based classifier and A Z > 0.93 for using the ANN-based classifier). The results indicated that the scheme could correctly identify more than 90% of analyzable chromosome cells while eliminating majority of un-analyzable cells (e.g., >85%). However, there are several limitations in this study. First, the size of datasets was relatively limited (small). Second, the truth was determined by one cytogeneticist in our genetic laboratory. The issue of potential inter-observer variability has not been investigated. Third, all microscopic digital images of chromosomes were pre-selected and each image depicts one metaphase cell (either analyzable or un-analyzable one). In our future studies, we will further optimize and test our scheme using a much large and diverse image database. The truth file will be verified by a panel of cytogeneticists. We will also test and apply this scheme to the sequential images automatically acquired by a high-speed microscopic image scanner (that is currently under development in our laboratory). The new computerized scheme should first detect whether an image depicts a metaphase cell or not, since in the clinical environment most of images acquired by a high-speed microscopic image scanner contain no metaphase cells. After the metaphase cells are detected, the scheme then identifies analyzable metaphase cells and discards un-analyzable ones. Acknowledgments This research is supported in part by Grant CA to the University of Oklahoma from the National Cancer Institute, National Institutes of Health. The authors would like to acknowledge the support of Charles and Jean Smith Chair endowment fund as well.7 References [1] Tjio JH, Levan A. The chromosome number in man. Hereditas 1956;42:1 6. [2] Nowell PC, Hungerford DA. A minute chromosome in human chronic granulocytic leukemia. Science 1960;132:1497. [3] Piper J, Granum E, Rutovitz D, Ruttledge H. Automation of chromosome analysis. Signal Process 1980;2: [4] Truong K, Gibaud A, Zalcman G, Guilly M. Quantitative fish determination of chromosome 3 arm imbalances in lung tumors by automated image cytometry. Med Sci Monit 2004;10: [5] Shih LM, Wang TL. Apply innovative technologies to explore cancer genome. Curr Opin Oncol 2005;17:33 8. [6] Boehm D, Herold S, Kuechler A, Liehr T. Rapid detection of subtelomeric deletion/duplication by novel real-time quantitative PCR using SYBR-Green dye. Hum Mutat 2004;23: [7] Hoffbrand VA, Pettit JE. Color atlas of clinical hematology. 3rd ed. San Francisco, CA: Mosby Press; 2000.
8 X. Wang et al. / Journal of Biomedical Informatics 41 (2008) [8] LeBeau MM, Rowley JD. Recurring chromosomal abnormalities in leukemia and lymphoma. In: Rowly J, editor. Cancer survey, 3. Oxford, UK: Oxford University Press; p [9] Kyan MJ, Guan L, Amison MR, Cogswell CJ. Feature extraction of chromosomes from 3D confocal microscope images. Proc Int Conf Image Process 1999;2:24 8. [10] Harnden DG, Klinger HP, Jensen JT, Kaelbling M. An international system for human cytogenetic nomenclature (ISCN1985): report of the standing committee on human cytogenetic nomenclature. Basel, Switzerland: KAEGER; [11] Zimmerman SO, Johnston DA, Arrighi FE, Rupp ME. Automated homologue matching of human G-banded chromosomes. Comput Biol Med 1986;16: [12] Wang X, Zheng B, Wood M, Li S, Chen W, Liu H. Development and evaluation of automated systems for detection and classification of banded chromosomes: current status and future perspectives. J Phys D Appl Phys 2005;38: [13] Vliet LJ, Young IT, Mayall BH. The Athena semi-automated karyotyping system. Cytometry 1990;11:51 8. [14] Popescu M, Gader P, Keller J, Klein C. Automatic karyotyping of metaphase cells with overlapping chromosomes. Comput Biol Med 1999;29: [15] Cosio F, Vega L, Becerra A, Melendez C. Automatic identification of metaphase spreads and nuclei using neural networks. Med Biol Eng Comput 2001;39: [16] Castleman KR. The PSI automatic metaphase finder. J Radiat Res 1992;33: [17] Corkidi G, Vega L, Márquez J, Rojas E, Ostrosky P. Roughness feature of metaphase chromosome spreads and nuclei for automated cell proliferation analysis. Med Biol Eng Cornput 1998;36: [18] Wang Y, Wu Q, Castleman KR, Xiong Z. Chromosome image enhancement using multiscale differential operators. IEEE Trans Med Imaging 2003;22: [19] Wu Q, Liu Z, Chen T, Xiong Z, Castleman KR. Subspace-based prototyping and classification of chromosme images. IEEE Trans Image Processing 2005;14: [20] Piper J, Granum E. On fully automatic feature measurement for banded chromosome classification. Cytometry 1989;10: [21] Errington P, Graham J. Application of artificial neural networks to chromosome classification. Cytometry 1993;14: [22] Guthrie C, Gregor J, Thomason MG. Constrained Markov networks for automated analysis of G-banded chromosomes. Comput Biol Med 1993;23: [23] Stanley RJ, Keller JM, Gader P, Caldwell CW. Data-driven homologue matching for chromosome identification. IEEE Trans Med Imaging 1998;17: [24] Stanley R, Keller J, Gader P, Caldwell CW. Automated chromosome classification limitations due to image processing. Biomed Sci Instrum 1995;31: [25] Boschman GA, Manders EM, Rens W, Slater R, Aten JA. Semiautomated detection of aberrant chromosomes in bivariate flow karyotypes. Cytometry 1992;13: [26] Malet P, Benkhalifa M, Perissel B, Geneix A, Le Corvaisier B. Chromosome analysis by image processing in a computerized environment: clinical application. J Radiat Res 1992;33: [27] Gonzalez RC, Woods RE. Digital image processing. Reading, MA: Addison-Wesley Publishing Company; [28] Zheng B, Lu A, Hardesty LA, Sumkin JH, Gur D. A method to improve visual similarity of breast masses for an interactive computer-aided diagnosis environment. Med Phys 2006;33: [29] Mitchell TM. Machine learning. Boston, MA: WCB McGraw-Hill; [30] Hertz J, Krogh A, Palmer RG. Introduction to the theory of neural computation. Redwood City, CA: Addison-Wesley Publishing Company; [31] Obuchowski NA. ROC analysis. Am J Roentgenol 2005;184: [32] Metz CE. ROCFIT 0.9B Beta version. Chicago, IL: University of Chicago, [33] Metz CE, Herman BA, Shen JH. Maximum-likelihood estimation of receiver operating characteristic (ROC) curves from continuouslydistributed data. Stat Med 1998;17: [34] Wang XW, Li S, Liu H, Mulvihill JJ, Chen W, Zheng B. A computeraided method to expedite the evaluation of prognosis for childhood acute lymphoblastic leukemia. Technol Cancer Res Treat 2006;5: [35] Zheng B, Chang YH, Good WF, Gur D. Adequacy testing of training set sample sizes in the development of a computer-assisted diagnosis scheme. Acad Radiol 1997;4: [36] Li Q, Doi K. Reduction of bias and variance for evaluation of computer-aided diagnostic schemes. Med Phys 2006;33:
A Hierarchical Artificial Neural Network Model for Giemsa-Stained Human Chromosome Classification
 A Hierarchical Artificial Neural Network Model for Giemsa-Stained Human Chromosome Classification JONGMAN CHO 1 1 Department of Biomedical Engineering, Inje University, Gimhae, 621-749, KOREA minerva@ieeeorg
A Hierarchical Artificial Neural Network Model for Giemsa-Stained Human Chromosome Classification JONGMAN CHO 1 1 Department of Biomedical Engineering, Inje University, Gimhae, 621-749, KOREA minerva@ieeeorg
Classification of benign and malignant masses in breast mammograms
 Classification of benign and malignant masses in breast mammograms A. Šerifović-Trbalić*, A. Trbalić**, D. Demirović*, N. Prljača* and P.C. Cattin*** * Faculty of Electrical Engineering, University of
Classification of benign and malignant masses in breast mammograms A. Šerifović-Trbalić*, A. Trbalić**, D. Demirović*, N. Prljača* and P.C. Cattin*** * Faculty of Electrical Engineering, University of
Investigating the performance of a CAD x scheme for mammography in specific BIRADS categories
 Investigating the performance of a CAD x scheme for mammography in specific BIRADS categories Andreadis I., Nikita K. Department of Electrical and Computer Engineering National Technical University of
Investigating the performance of a CAD x scheme for mammography in specific BIRADS categories Andreadis I., Nikita K. Department of Electrical and Computer Engineering National Technical University of
Application of Artificial Neural Network-Based Survival Analysis on Two Breast Cancer Datasets
 Application of Artificial Neural Network-Based Survival Analysis on Two Breast Cancer Datasets Chih-Lin Chi a, W. Nick Street b, William H. Wolberg c a Health Informatics Program, University of Iowa b
Application of Artificial Neural Network-Based Survival Analysis on Two Breast Cancer Datasets Chih-Lin Chi a, W. Nick Street b, William H. Wolberg c a Health Informatics Program, University of Iowa b
Neural Network based Heart Arrhythmia Detection and Classification from ECG Signal
 Neural Network based Heart Arrhythmia Detection and Classification from ECG Signal 1 M. S. Aware, 2 V. V. Shete *Dept. of Electronics and Telecommunication, *MIT College Of Engineering, Pune Email: 1 mrunal_swapnil@yahoo.com,
Neural Network based Heart Arrhythmia Detection and Classification from ECG Signal 1 M. S. Aware, 2 V. V. Shete *Dept. of Electronics and Telecommunication, *MIT College Of Engineering, Pune Email: 1 mrunal_swapnil@yahoo.com,
COMMUNICATION ENGINEERING & TECHNOLOGY (IJECET) DETECTION OF ACUTE LEUKEMIA USING WHITE BLOOD CELLS SEGMENTATION BASED ON BLOOD SAMPLES
 International INTERNATIONAL Journal of Electronics JOURNAL and Communication OF ELECTRONICS Engineering & Technology AND (IJECET), COMMUNICATION ENGINEERING & TECHNOLOGY (IJECET) ISSN 0976 6464(Print)
International INTERNATIONAL Journal of Electronics JOURNAL and Communication OF ELECTRONICS Engineering & Technology AND (IJECET), COMMUNICATION ENGINEERING & TECHNOLOGY (IJECET) ISSN 0976 6464(Print)
Cancer Cells Detection using OTSU Threshold Algorithm
 Cancer Cells Detection using OTSU Threshold Algorithm Nalluri Sunny 1 Velagapudi Ramakrishna Siddhartha Engineering College Mithinti Srikanth 2 Velagapudi Ramakrishna Siddhartha Engineering College Kodali
Cancer Cells Detection using OTSU Threshold Algorithm Nalluri Sunny 1 Velagapudi Ramakrishna Siddhartha Engineering College Mithinti Srikanth 2 Velagapudi Ramakrishna Siddhartha Engineering College Kodali
Early Detection of Lung Cancer
 Early Detection of Lung Cancer Aswathy N Iyer Dept Of Electronics And Communication Engineering Lymie Jose Dept Of Electronics And Communication Engineering Anumol Thomas Dept Of Electronics And Communication
Early Detection of Lung Cancer Aswathy N Iyer Dept Of Electronics And Communication Engineering Lymie Jose Dept Of Electronics And Communication Engineering Anumol Thomas Dept Of Electronics And Communication
Hidden Markov Mqdels.for Chromosome Identification
 Hidden Markov Mqdels.for Chromosome Identification John M. Conroy Center for Computzizg Scneiices Instztute for Defense Analyses Bowae, Maryland Robert L. Becker, Jr. William Lefkowitz Kewi L. Christopher
Hidden Markov Mqdels.for Chromosome Identification John M. Conroy Center for Computzizg Scneiices Instztute for Defense Analyses Bowae, Maryland Robert L. Becker, Jr. William Lefkowitz Kewi L. Christopher
Mammogram Analysis: Tumor Classification
 Mammogram Analysis: Tumor Classification Term Project Report Geethapriya Raghavan geeragh@mail.utexas.edu EE 381K - Multidimensional Digital Signal Processing Spring 2005 Abstract Breast cancer is the
Mammogram Analysis: Tumor Classification Term Project Report Geethapriya Raghavan geeragh@mail.utexas.edu EE 381K - Multidimensional Digital Signal Processing Spring 2005 Abstract Breast cancer is the
Classification of mammogram masses using selected texture, shape and margin features with multilayer perceptron classifier.
 Biomedical Research 2016; Special Issue: S310-S313 ISSN 0970-938X www.biomedres.info Classification of mammogram masses using selected texture, shape and margin features with multilayer perceptron classifier.
Biomedical Research 2016; Special Issue: S310-S313 ISSN 0970-938X www.biomedres.info Classification of mammogram masses using selected texture, shape and margin features with multilayer perceptron classifier.
A framework for the Recognition of Human Emotion using Soft Computing models
 A framework for the Recognition of Human Emotion using Soft Computing models Md. Iqbal Quraishi Dept. of Information Technology Kalyani Govt Engg. College J Pal Choudhury Dept. of Information Technology
A framework for the Recognition of Human Emotion using Soft Computing models Md. Iqbal Quraishi Dept. of Information Technology Kalyani Govt Engg. College J Pal Choudhury Dept. of Information Technology
Australian Journal of Basic and Applied Sciences
 ISSN:1991-8178 Australian Journal of Basic and Applied Sciences Journal home page: www.ajbasweb.com Improved Accuracy of Breast Cancer Detection in Digital Mammograms using Wavelet Analysis and Artificial
ISSN:1991-8178 Australian Journal of Basic and Applied Sciences Journal home page: www.ajbasweb.com Improved Accuracy of Breast Cancer Detection in Digital Mammograms using Wavelet Analysis and Artificial
Assessment of Reliability of Hamilton-Tompkins Algorithm to ECG Parameter Detection
 Proceedings of the 2012 International Conference on Industrial Engineering and Operations Management Istanbul, Turkey, July 3 6, 2012 Assessment of Reliability of Hamilton-Tompkins Algorithm to ECG Parameter
Proceedings of the 2012 International Conference on Industrial Engineering and Operations Management Istanbul, Turkey, July 3 6, 2012 Assessment of Reliability of Hamilton-Tompkins Algorithm to ECG Parameter
Improved Intelligent Classification Technique Based On Support Vector Machines
 Improved Intelligent Classification Technique Based On Support Vector Machines V.Vani Asst.Professor,Department of Computer Science,JJ College of Arts and Science,Pudukkottai. Abstract:An abnormal growth
Improved Intelligent Classification Technique Based On Support Vector Machines V.Vani Asst.Professor,Department of Computer Science,JJ College of Arts and Science,Pudukkottai. Abstract:An abnormal growth
TWO HANDED SIGN LANGUAGE RECOGNITION SYSTEM USING IMAGE PROCESSING
 134 TWO HANDED SIGN LANGUAGE RECOGNITION SYSTEM USING IMAGE PROCESSING H.F.S.M.Fonseka 1, J.T.Jonathan 2, P.Sabeshan 3 and M.B.Dissanayaka 4 1 Department of Electrical And Electronic Engineering, Faculty
134 TWO HANDED SIGN LANGUAGE RECOGNITION SYSTEM USING IMAGE PROCESSING H.F.S.M.Fonseka 1, J.T.Jonathan 2, P.Sabeshan 3 and M.B.Dissanayaka 4 1 Department of Electrical And Electronic Engineering, Faculty
Sparse Coding in Sparse Winner Networks
 Sparse Coding in Sparse Winner Networks Janusz A. Starzyk 1, Yinyin Liu 1, David Vogel 2 1 School of Electrical Engineering & Computer Science Ohio University, Athens, OH 45701 {starzyk, yliu}@bobcat.ent.ohiou.edu
Sparse Coding in Sparse Winner Networks Janusz A. Starzyk 1, Yinyin Liu 1, David Vogel 2 1 School of Electrical Engineering & Computer Science Ohio University, Athens, OH 45701 {starzyk, yliu}@bobcat.ent.ohiou.edu
COMPUTER AIDED DIAGNOSTIC SYSTEM FOR BRAIN TUMOR DETECTION USING K-MEANS CLUSTERING
 COMPUTER AIDED DIAGNOSTIC SYSTEM FOR BRAIN TUMOR DETECTION USING K-MEANS CLUSTERING Urmila Ravindra Patil Tatyasaheb Kore Institute of Engineering and Technology, Warananagar Prof. R. T. Patil Tatyasaheb
COMPUTER AIDED DIAGNOSTIC SYSTEM FOR BRAIN TUMOR DETECTION USING K-MEANS CLUSTERING Urmila Ravindra Patil Tatyasaheb Kore Institute of Engineering and Technology, Warananagar Prof. R. T. Patil Tatyasaheb
Extraction of Blood Vessels and Recognition of Bifurcation Points in Retinal Fundus Image
 International Journal of Research Studies in Science, Engineering and Technology Volume 1, Issue 5, August 2014, PP 1-7 ISSN 2349-4751 (Print) & ISSN 2349-476X (Online) Extraction of Blood Vessels and
International Journal of Research Studies in Science, Engineering and Technology Volume 1, Issue 5, August 2014, PP 1-7 ISSN 2349-4751 (Print) & ISSN 2349-476X (Online) Extraction of Blood Vessels and
Predicting Breast Cancer Survivability Rates
 Predicting Breast Cancer Survivability Rates For data collected from Saudi Arabia Registries Ghofran Othoum 1 and Wadee Al-Halabi 2 1 Computer Science, Effat University, Jeddah, Saudi Arabia 2 Computer
Predicting Breast Cancer Survivability Rates For data collected from Saudi Arabia Registries Ghofran Othoum 1 and Wadee Al-Halabi 2 1 Computer Science, Effat University, Jeddah, Saudi Arabia 2 Computer
AN ALGORITHM FOR EARLY BREAST CANCER DETECTION IN MAMMOGRAMS
 AN ALGORITHM FOR EARLY BREAST CANCER DETECTION IN MAMMOGRAMS Isaac N. Bankman', William A. Christens-Barryl, Irving N. Weinberg2, Dong W. Kim3, Ralph D. Semmell, and William R. Brody2 The Johns Hopkins
AN ALGORITHM FOR EARLY BREAST CANCER DETECTION IN MAMMOGRAMS Isaac N. Bankman', William A. Christens-Barryl, Irving N. Weinberg2, Dong W. Kim3, Ralph D. Semmell, and William R. Brody2 The Johns Hopkins
COMPUTERIZED SYSTEM DESIGN FOR THE DETECTION AND DIAGNOSIS OF LUNG NODULES IN CT IMAGES 1
 ISSN 258-8739 3 st August 28, Volume 3, Issue 2, JSEIS, CAOMEI Copyright 26-28 COMPUTERIZED SYSTEM DESIGN FOR THE DETECTION AND DIAGNOSIS OF LUNG NODULES IN CT IMAGES ALI ABDRHMAN UKASHA, 2 EMHMED SAAID
ISSN 258-8739 3 st August 28, Volume 3, Issue 2, JSEIS, CAOMEI Copyright 26-28 COMPUTERIZED SYSTEM DESIGN FOR THE DETECTION AND DIAGNOSIS OF LUNG NODULES IN CT IMAGES ALI ABDRHMAN UKASHA, 2 EMHMED SAAID
Segmentation of Tumor Region from Brain Mri Images Using Fuzzy C-Means Clustering And Seeded Region Growing
 IOSR Journal of Computer Engineering (IOSR-JCE) e-issn: 2278-0661,p-ISSN: 2278-8727, Volume 18, Issue 5, Ver. I (Sept - Oct. 2016), PP 20-24 www.iosrjournals.org Segmentation of Tumor Region from Brain
IOSR Journal of Computer Engineering (IOSR-JCE) e-issn: 2278-0661,p-ISSN: 2278-8727, Volume 18, Issue 5, Ver. I (Sept - Oct. 2016), PP 20-24 www.iosrjournals.org Segmentation of Tumor Region from Brain
Automated Brain Tumor Segmentation Using Region Growing Algorithm by Extracting Feature
 Automated Brain Tumor Segmentation Using Region Growing Algorithm by Extracting Feature Shraddha P. Dhumal 1, Ashwini S Gaikwad 2 1 Shraddha P. Dhumal 2 Ashwini S. Gaikwad ABSTRACT In this paper, we propose
Automated Brain Tumor Segmentation Using Region Growing Algorithm by Extracting Feature Shraddha P. Dhumal 1, Ashwini S Gaikwad 2 1 Shraddha P. Dhumal 2 Ashwini S. Gaikwad ABSTRACT In this paper, we propose
MRI Image Processing Operations for Brain Tumor Detection
 MRI Image Processing Operations for Brain Tumor Detection Prof. M.M. Bulhe 1, Shubhashini Pathak 2, Karan Parekh 3, Abhishek Jha 4 1Assistant Professor, Dept. of Electronics and Telecommunications Engineering,
MRI Image Processing Operations for Brain Tumor Detection Prof. M.M. Bulhe 1, Shubhashini Pathak 2, Karan Parekh 3, Abhishek Jha 4 1Assistant Professor, Dept. of Electronics and Telecommunications Engineering,
Proceedings of the UGC Sponsored National Conference on Advanced Networking and Applications, 27 th March 2015
 Brain Tumor Detection and Identification Using K-Means Clustering Technique Malathi R Department of Computer Science, SAAS College, Ramanathapuram, Email: malapraba@gmail.com Dr. Nadirabanu Kamal A R Department
Brain Tumor Detection and Identification Using K-Means Clustering Technique Malathi R Department of Computer Science, SAAS College, Ramanathapuram, Email: malapraba@gmail.com Dr. Nadirabanu Kamal A R Department
1 Introduction. Abstract: Accurate optic disc (OD) segmentation and fovea. Keywords: optic disc segmentation, fovea detection.
 Current Directions in Biomedical Engineering 2017; 3(2): 533 537 Caterina Rust*, Stephanie Häger, Nadine Traulsen and Jan Modersitzki A robust algorithm for optic disc segmentation and fovea detection
Current Directions in Biomedical Engineering 2017; 3(2): 533 537 Caterina Rust*, Stephanie Häger, Nadine Traulsen and Jan Modersitzki A robust algorithm for optic disc segmentation and fovea detection
EXTRACT THE BREAST CANCER IN MAMMOGRAM IMAGES
 International Journal of Civil Engineering and Technology (IJCIET) Volume 10, Issue 02, February 2019, pp. 96-105, Article ID: IJCIET_10_02_012 Available online at http://www.iaeme.com/ijciet/issues.asp?jtype=ijciet&vtype=10&itype=02
International Journal of Civil Engineering and Technology (IJCIET) Volume 10, Issue 02, February 2019, pp. 96-105, Article ID: IJCIET_10_02_012 Available online at http://www.iaeme.com/ijciet/issues.asp?jtype=ijciet&vtype=10&itype=02
ECG Beat Recognition using Principal Components Analysis and Artificial Neural Network
 International Journal of Electronics Engineering, 3 (1), 2011, pp. 55 58 ECG Beat Recognition using Principal Components Analysis and Artificial Neural Network Amitabh Sharma 1, and Tanushree Sharma 2
International Journal of Electronics Engineering, 3 (1), 2011, pp. 55 58 ECG Beat Recognition using Principal Components Analysis and Artificial Neural Network Amitabh Sharma 1, and Tanushree Sharma 2
Mammogram Analysis: Tumor Classification
 Mammogram Analysis: Tumor Classification Literature Survey Report Geethapriya Raghavan geeragh@mail.utexas.edu EE 381K - Multidimensional Digital Signal Processing Spring 2005 Abstract Breast cancer is
Mammogram Analysis: Tumor Classification Literature Survey Report Geethapriya Raghavan geeragh@mail.utexas.edu EE 381K - Multidimensional Digital Signal Processing Spring 2005 Abstract Breast cancer is
Mammographic Cancer Detection and Classification Using Bi Clustering and Supervised Classifier
 Mammographic Cancer Detection and Classification Using Bi Clustering and Supervised Classifier R.Pavitha 1, Ms T.Joyce Selva Hephzibah M.Tech. 2 PG Scholar, Department of ECE, Indus College of Engineering,
Mammographic Cancer Detection and Classification Using Bi Clustering and Supervised Classifier R.Pavitha 1, Ms T.Joyce Selva Hephzibah M.Tech. 2 PG Scholar, Department of ECE, Indus College of Engineering,
PET in Radiation Therapy. Outline. Tumor Segmentation in PET and in Multimodality Images for Radiation Therapy. 1. Tumor segmentation in PET
 Tumor Segmentation in PET and in Multimodality Images for Radiation Therapy Wei Lu, Ph.D. Department of Radiation Oncology Mallinckrodt Institute of Radiology Washington University in St. Louis Outline
Tumor Segmentation in PET and in Multimodality Images for Radiation Therapy Wei Lu, Ph.D. Department of Radiation Oncology Mallinckrodt Institute of Radiology Washington University in St. Louis Outline
arxiv:physics/ v1 [physics.med-ph] 4 Jan 2007
![arxiv:physics/ v1 [physics.med-ph] 4 Jan 2007 arxiv:physics/ v1 [physics.med-ph] 4 Jan 2007](/thumbs/74/70454308.jpg) arxiv:physics/0701053v1 [physics.med-ph] 4 Jan 2007 An Automatic System to Discriminate Malignant from Benign Massive Lesions on Mammograms A. Retico a, P. Delogu ab, M.E. Fantacci ab, P. Kasae c a Istituto
arxiv:physics/0701053v1 [physics.med-ph] 4 Jan 2007 An Automatic System to Discriminate Malignant from Benign Massive Lesions on Mammograms A. Retico a, P. Delogu ab, M.E. Fantacci ab, P. Kasae c a Istituto
Computerized image analysis: Estimation of breast density on mammograms
 Computerized image analysis: Estimation of breast density on mammograms Chuan Zhou, Heang-Ping Chan, a) Nicholas Petrick, Mark A. Helvie, Mitchell M. Goodsitt, Berkman Sahiner, and Lubomir M. Hadjiiski
Computerized image analysis: Estimation of breast density on mammograms Chuan Zhou, Heang-Ping Chan, a) Nicholas Petrick, Mark A. Helvie, Mitchell M. Goodsitt, Berkman Sahiner, and Lubomir M. Hadjiiski
Automatic Hemorrhage Classification System Based On Svm Classifier
 Automatic Hemorrhage Classification System Based On Svm Classifier Abstract - Brain hemorrhage is a bleeding in or around the brain which are caused by head trauma, high blood pressure and intracranial
Automatic Hemorrhage Classification System Based On Svm Classifier Abstract - Brain hemorrhage is a bleeding in or around the brain which are caused by head trauma, high blood pressure and intracranial
Detection of Glaucoma and Diabetic Retinopathy from Fundus Images by Bloodvessel Segmentation
 International Journal of Engineering and Advanced Technology (IJEAT) ISSN: 2249 8958, Volume-5, Issue-5, June 2016 Detection of Glaucoma and Diabetic Retinopathy from Fundus Images by Bloodvessel Segmentation
International Journal of Engineering and Advanced Technology (IJEAT) ISSN: 2249 8958, Volume-5, Issue-5, June 2016 Detection of Glaucoma and Diabetic Retinopathy from Fundus Images by Bloodvessel Segmentation
Deep Learning Analytics for Predicting Prognosis of Acute Myeloid Leukemia with Cytogenetics, Age, and Mutations
 Deep Learning Analytics for Predicting Prognosis of Acute Myeloid Leukemia with Cytogenetics, Age, and Mutations Andy Nguyen, M.D., M.S. Medical Director, Hematopathology, Hematology and Coagulation Laboratory,
Deep Learning Analytics for Predicting Prognosis of Acute Myeloid Leukemia with Cytogenetics, Age, and Mutations Andy Nguyen, M.D., M.S. Medical Director, Hematopathology, Hematology and Coagulation Laboratory,
COMPUTER-AIDED HER-2/neu EVALUATION IN EXTERNAL QUALITY ASSURANCE (EQA) OF BREAST CANCER SCREENING PROGRAMME
 COMPUTER-AIDED HER-2/neu EVALUATION IN EXTERNAL QUALITY ASSURANCE (EQA) OF BREAST CANCER SCREENING PROGRAMME Maria Lunardi MD Anatomic Pathology Fracastoro Hospital San Bonifacio, Verona -Italy HER2-neu
COMPUTER-AIDED HER-2/neu EVALUATION IN EXTERNAL QUALITY ASSURANCE (EQA) OF BREAST CANCER SCREENING PROGRAMME Maria Lunardi MD Anatomic Pathology Fracastoro Hospital San Bonifacio, Verona -Italy HER2-neu
LUNG CANCER continues to rank as the leading cause
 1138 IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 24, NO. 9, SEPTEMBER 2005 Computer-Aided Diagnostic Scheme for Distinction Between Benign and Malignant Nodules in Thoracic Low-Dose CT by Use of Massive
1138 IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 24, NO. 9, SEPTEMBER 2005 Computer-Aided Diagnostic Scheme for Distinction Between Benign and Malignant Nodules in Thoracic Low-Dose CT by Use of Massive
Development of novel algorithm by combining Wavelet based Enhanced Canny edge Detection and Adaptive Filtering Method for Human Emotion Recognition
 International Journal of Engineering Research and Development e-issn: 2278-067X, p-issn: 2278-800X, www.ijerd.com Volume 12, Issue 9 (September 2016), PP.67-72 Development of novel algorithm by combining
International Journal of Engineering Research and Development e-issn: 2278-067X, p-issn: 2278-800X, www.ijerd.com Volume 12, Issue 9 (September 2016), PP.67-72 Development of novel algorithm by combining
Detection of microcalcifications in digital mammogram using wavelet analysis
 American Journal of Engineering Research (AJER) e-issn : 2320-0847 p-issn : 2320-0936 Volume-02, Issue-11, pp-80-85 www.ajer.org Research Paper Open Access Detection of microcalcifications in digital mammogram
American Journal of Engineering Research (AJER) e-issn : 2320-0847 p-issn : 2320-0936 Volume-02, Issue-11, pp-80-85 www.ajer.org Research Paper Open Access Detection of microcalcifications in digital mammogram
A Survey on Brain Tumor Detection Technique
 (International Journal of Computer Science & Management Studies) Vol. 15, Issue 06 A Survey on Brain Tumor Detection Technique Manju Kadian 1 and Tamanna 2 1 M.Tech. Scholar, CSE Department, SPGOI, Rohtak
(International Journal of Computer Science & Management Studies) Vol. 15, Issue 06 A Survey on Brain Tumor Detection Technique Manju Kadian 1 and Tamanna 2 1 M.Tech. Scholar, CSE Department, SPGOI, Rohtak
arxiv: v2 [cs.cv] 8 Mar 2018
![arxiv: v2 [cs.cv] 8 Mar 2018 arxiv: v2 [cs.cv] 8 Mar 2018](/thumbs/87/97094636.jpg) Automated soft tissue lesion detection and segmentation in digital mammography using a u-net deep learning network Timothy de Moor a, Alejandro Rodriguez-Ruiz a, Albert Gubern Mérida a, Ritse Mann a, and
Automated soft tissue lesion detection and segmentation in digital mammography using a u-net deep learning network Timothy de Moor a, Alejandro Rodriguez-Ruiz a, Albert Gubern Mérida a, Ritse Mann a, and
arxiv: v1 [stat.ml] 23 Jan 2017
![arxiv: v1 [stat.ml] 23 Jan 2017 arxiv: v1 [stat.ml] 23 Jan 2017](/thumbs/81/83414183.jpg) Learning what to look in chest X-rays with a recurrent visual attention model arxiv:1701.06452v1 [stat.ml] 23 Jan 2017 Petros-Pavlos Ypsilantis Department of Biomedical Engineering King s College London
Learning what to look in chest X-rays with a recurrent visual attention model arxiv:1701.06452v1 [stat.ml] 23 Jan 2017 Petros-Pavlos Ypsilantis Department of Biomedical Engineering King s College London
[Kiran, 2(1): January, 2015] ISSN:
![[Kiran, 2(1): January, 2015] ISSN: [Kiran, 2(1): January, 2015] ISSN:](/thumbs/72/66395926.jpg) AN EFFICIENT LUNG CANCER DETECTION BASED ON ARTIFICIAL NEURAL NETWORK Shashi Kiran.S * Assistant Professor, JNN College of Engineering, Shimoga, Karnataka, India Keywords: Artificial Neural Network (ANN),
AN EFFICIENT LUNG CANCER DETECTION BASED ON ARTIFICIAL NEURAL NETWORK Shashi Kiran.S * Assistant Professor, JNN College of Engineering, Shimoga, Karnataka, India Keywords: Artificial Neural Network (ANN),
Efficacy of the Extended Principal Orthogonal Decomposition Method on DNA Microarray Data in Cancer Detection
 202 4th International onference on Bioinformatics and Biomedical Technology IPBEE vol.29 (202) (202) IASIT Press, Singapore Efficacy of the Extended Principal Orthogonal Decomposition on DA Microarray
202 4th International onference on Bioinformatics and Biomedical Technology IPBEE vol.29 (202) (202) IASIT Press, Singapore Efficacy of the Extended Principal Orthogonal Decomposition on DA Microarray
Lung Cancer Diagnosis from CT Images Using Fuzzy Inference System
 Lung Cancer Diagnosis from CT Images Using Fuzzy Inference System T.Manikandan 1, Dr. N. Bharathi 2 1 Associate Professor, Rajalakshmi Engineering College, Chennai-602 105 2 Professor, Velammal Engineering
Lung Cancer Diagnosis from CT Images Using Fuzzy Inference System T.Manikandan 1, Dr. N. Bharathi 2 1 Associate Professor, Rajalakshmi Engineering College, Chennai-602 105 2 Professor, Velammal Engineering
Primary Level Classification of Brain Tumor using PCA and PNN
 Primary Level Classification of Brain Tumor using PCA and PNN Dr. Mrs. K.V.Kulhalli Department of Information Technology, D.Y.Patil Coll. of Engg. And Tech. Kolhapur,Maharashtra,India kvkulhalli@gmail.com
Primary Level Classification of Brain Tumor using PCA and PNN Dr. Mrs. K.V.Kulhalli Department of Information Technology, D.Y.Patil Coll. of Engg. And Tech. Kolhapur,Maharashtra,India kvkulhalli@gmail.com
Comparison of volume estimation methods for pancreatic islet cells
 Comparison of volume estimation methods for pancreatic islet cells Jiří Dvořák a,b, Jan Švihlíkb,c, David Habart d, and Jan Kybic b a Department of Probability and Mathematical Statistics, Faculty of Mathematics
Comparison of volume estimation methods for pancreatic islet cells Jiří Dvořák a,b, Jan Švihlíkb,c, David Habart d, and Jan Kybic b a Department of Probability and Mathematical Statistics, Faculty of Mathematics
Improved Detection of Lung Nodule Overlapping with Ribs by using Virtual Dual Energy Radiology
 Improved Detection of Lung Nodule Overlapping with Ribs by using Virtual Dual Energy Radiology G.Maria Dhayana Latha Associate Professor, Department of Electronics and Communication Engineering, Excel
Improved Detection of Lung Nodule Overlapping with Ribs by using Virtual Dual Energy Radiology G.Maria Dhayana Latha Associate Professor, Department of Electronics and Communication Engineering, Excel
Nature Neuroscience: doi: /nn Supplementary Figure 1. Behavioral training.
 Supplementary Figure 1 Behavioral training. a, Mazes used for behavioral training. Asterisks indicate reward location. Only some example mazes are shown (for example, right choice and not left choice maze
Supplementary Figure 1 Behavioral training. a, Mazes used for behavioral training. Asterisks indicate reward location. Only some example mazes are shown (for example, right choice and not left choice maze
J2.6 Imputation of missing data with nonlinear relationships
 Sixth Conference on Artificial Intelligence Applications to Environmental Science 88th AMS Annual Meeting, New Orleans, LA 20-24 January 2008 J2.6 Imputation of missing with nonlinear relationships Michael
Sixth Conference on Artificial Intelligence Applications to Environmental Science 88th AMS Annual Meeting, New Orleans, LA 20-24 January 2008 J2.6 Imputation of missing with nonlinear relationships Michael
Brain Tumour Diagnostic Support Based on Medical Image Segmentation
 Brain Tumour Diagnostic Support Based on Medical Image Segmentation Z. Měřínský, E. Hošťálková, A. Procházka Institute of Chemical Technology, Prague Department of Computing and Control Engineering Abstract
Brain Tumour Diagnostic Support Based on Medical Image Segmentation Z. Měřínský, E. Hošťálková, A. Procházka Institute of Chemical Technology, Prague Department of Computing and Control Engineering Abstract
EARLY STAGE DIAGNOSIS OF LUNG CANCER USING CT-SCAN IMAGES BASED ON CELLULAR LEARNING AUTOMATE
 EARLY STAGE DIAGNOSIS OF LUNG CANCER USING CT-SCAN IMAGES BASED ON CELLULAR LEARNING AUTOMATE SAKTHI NEELA.P.K Department of M.E (Medical electronics) Sengunthar College of engineering Namakkal, Tamilnadu,
EARLY STAGE DIAGNOSIS OF LUNG CANCER USING CT-SCAN IMAGES BASED ON CELLULAR LEARNING AUTOMATE SAKTHI NEELA.P.K Department of M.E (Medical electronics) Sengunthar College of engineering Namakkal, Tamilnadu,
Computer aided detection of clusters of microcalcifications on full field digital mammograms
 Computer aided detection of clusters of microcalcifications on full field digital mammograms Jun Ge, a Berkman Sahiner, Lubomir M. Hadjiiski, Heang-Ping Chan, Jun Wei, Mark A. Helvie, and Chuan Zhou Department
Computer aided detection of clusters of microcalcifications on full field digital mammograms Jun Ge, a Berkman Sahiner, Lubomir M. Hadjiiski, Heang-Ping Chan, Jun Wei, Mark A. Helvie, and Chuan Zhou Department
Detection of suspicious lesion based on Multiresolution Analysis using windowing and adaptive thresholding method.
 Detection of suspicious lesion based on Multiresolution Analysis using windowing and adaptive thresholding method. Ms. N. S. Pande Assistant Professor, Department of Computer Science and Engineering,MGM
Detection of suspicious lesion based on Multiresolution Analysis using windowing and adaptive thresholding method. Ms. N. S. Pande Assistant Professor, Department of Computer Science and Engineering,MGM
Applications of Machine learning in Prediction of Breast Cancer Incidence and Mortality
 Applications of Machine learning in Prediction of Breast Cancer Incidence and Mortality Nadia Helal and Eman Sarwat Radiation Safety Dep. NCNSRC., Atomic Energy Authority, 3, Ahmed El Zomor St., P.Code
Applications of Machine learning in Prediction of Breast Cancer Incidence and Mortality Nadia Helal and Eman Sarwat Radiation Safety Dep. NCNSRC., Atomic Energy Authority, 3, Ahmed El Zomor St., P.Code
I. INTRODUCTION III. OVERALL DESIGN
 Inherent Selection Of Tuberculosis Using Graph Cut Segmentation V.N.Ilakkiya 1, Dr.P.Raviraj 2 1 PG Scholar, Department of computer science, Kalaignar Karunanidhi Institute of Technology, Coimbatore, Tamil
Inherent Selection Of Tuberculosis Using Graph Cut Segmentation V.N.Ilakkiya 1, Dr.P.Raviraj 2 1 PG Scholar, Department of computer science, Kalaignar Karunanidhi Institute of Technology, Coimbatore, Tamil
LUNG NODULE SEGMENTATION IN COMPUTED TOMOGRAPHY IMAGE. Hemahashiny, Ketheesan Department of Physical Science, Vavuniya Campus
 LUNG NODULE SEGMENTATION IN COMPUTED TOMOGRAPHY IMAGE Hemahashiny, Ketheesan Department of Physical Science, Vavuniya Campus tketheesan@vau.jfn.ac.lk ABSTRACT: The key process to detect the Lung cancer
LUNG NODULE SEGMENTATION IN COMPUTED TOMOGRAPHY IMAGE Hemahashiny, Ketheesan Department of Physical Science, Vavuniya Campus tketheesan@vau.jfn.ac.lk ABSTRACT: The key process to detect the Lung cancer
Automatic Classification of Breast Masses for Diagnosis of Breast Cancer in Digital Mammograms using Neural Network
 IJSTE - International Journal of Science Technology & Engineering Volume 1 Issue 11 May 2015 ISSN (online): 2349-784X Automatic Classification of Breast Masses for Diagnosis of Breast Cancer in Digital
IJSTE - International Journal of Science Technology & Engineering Volume 1 Issue 11 May 2015 ISSN (online): 2349-784X Automatic Classification of Breast Masses for Diagnosis of Breast Cancer in Digital
A REVIEW ON CLASSIFICATION OF BREAST CANCER DETECTION USING COMBINATION OF THE FEATURE EXTRACTION MODELS. Aeronautical Engineering. Hyderabad. India.
 Volume 116 No. 21 2017, 203-208 ISSN: 1311-8080 (printed version); ISSN: 1314-3395 (on-line version) url: http://www.ijpam.eu A REVIEW ON CLASSIFICATION OF BREAST CANCER DETECTION USING COMBINATION OF
Volume 116 No. 21 2017, 203-208 ISSN: 1311-8080 (printed version); ISSN: 1314-3395 (on-line version) url: http://www.ijpam.eu A REVIEW ON CLASSIFICATION OF BREAST CANCER DETECTION USING COMBINATION OF
Gene Selection for Tumor Classification Using Microarray Gene Expression Data
 Gene Selection for Tumor Classification Using Microarray Gene Expression Data K. Yendrapalli, R. Basnet, S. Mukkamala, A. H. Sung Department of Computer Science New Mexico Institute of Mining and Technology
Gene Selection for Tumor Classification Using Microarray Gene Expression Data K. Yendrapalli, R. Basnet, S. Mukkamala, A. H. Sung Department of Computer Science New Mexico Institute of Mining and Technology
Threshold Based Segmentation Technique for Mass Detection in Mammography
 Threshold Based Segmentation Technique for Mass Detection in Mammography Aziz Makandar *, Bhagirathi Halalli Department of Computer Science, Karnataka State Women s University, Vijayapura, Karnataka, India.
Threshold Based Segmentation Technique for Mass Detection in Mammography Aziz Makandar *, Bhagirathi Halalli Department of Computer Science, Karnataka State Women s University, Vijayapura, Karnataka, India.
CHAPTER 5 WAVELET BASED DETECTION OF VENTRICULAR ARRHYTHMIAS WITH NEURAL NETWORK CLASSIFIER
 57 CHAPTER 5 WAVELET BASED DETECTION OF VENTRICULAR ARRHYTHMIAS WITH NEURAL NETWORK CLASSIFIER 5.1 INTRODUCTION The cardiac disorders which are life threatening are the ventricular arrhythmias such as
57 CHAPTER 5 WAVELET BASED DETECTION OF VENTRICULAR ARRHYTHMIAS WITH NEURAL NETWORK CLASSIFIER 5.1 INTRODUCTION The cardiac disorders which are life threatening are the ventricular arrhythmias such as
Diagnosis of Liver Tumor Using 3D Segmentation Method for Selective Internal Radiation Therapy
 Diagnosis of Liver Tumor Using 3D Segmentation Method for Selective Internal Radiation Therapy K. Geetha & S. Poonguzhali Department of Electronics and Communication Engineering, Campus of CEG Anna University,
Diagnosis of Liver Tumor Using 3D Segmentation Method for Selective Internal Radiation Therapy K. Geetha & S. Poonguzhali Department of Electronics and Communication Engineering, Campus of CEG Anna University,
DYNAMIC SEGMENTATION OF BREAST TISSUE IN DIGITIZED MAMMOGRAMS
 DYNAMIC SEGMENTATION OF BREAST TISSUE IN DIGITIZED MAMMOGRAMS J. T. Neyhart 1, M. D. Ciocco 1, R. Polikar 1, S. Mandayam 1 and M. Tseng 2 1 Department of Electrical & Computer Engineering, Rowan University,
DYNAMIC SEGMENTATION OF BREAST TISSUE IN DIGITIZED MAMMOGRAMS J. T. Neyhart 1, M. D. Ciocco 1, R. Polikar 1, S. Mandayam 1 and M. Tseng 2 1 Department of Electrical & Computer Engineering, Rowan University,
Automated Assessment of Diabetic Retinal Image Quality Based on Blood Vessel Detection
 Y.-H. Wen, A. Bainbridge-Smith, A. B. Morris, Automated Assessment of Diabetic Retinal Image Quality Based on Blood Vessel Detection, Proceedings of Image and Vision Computing New Zealand 2007, pp. 132
Y.-H. Wen, A. Bainbridge-Smith, A. B. Morris, Automated Assessment of Diabetic Retinal Image Quality Based on Blood Vessel Detection, Proceedings of Image and Vision Computing New Zealand 2007, pp. 132
A dynamic approach for optic disc localization in retinal images
 ISSN 2395-1621 A dynamic approach for optic disc localization in retinal images #1 Rutuja Deshmukh, #2 Karuna Jadhav, #3 Nikita Patwa 1 deshmukhrs777@gmail.com #123 UG Student, Electronics and Telecommunication
ISSN 2395-1621 A dynamic approach for optic disc localization in retinal images #1 Rutuja Deshmukh, #2 Karuna Jadhav, #3 Nikita Patwa 1 deshmukhrs777@gmail.com #123 UG Student, Electronics and Telecommunication
Research Article. Automated grading of diabetic retinopathy stages in fundus images using SVM classifer
 Available online www.jocpr.com Journal of Chemical and Pharmaceutical Research, 2016, 8(1):537-541 Research Article ISSN : 0975-7384 CODEN(USA) : JCPRC5 Automated grading of diabetic retinopathy stages
Available online www.jocpr.com Journal of Chemical and Pharmaceutical Research, 2016, 8(1):537-541 Research Article ISSN : 0975-7384 CODEN(USA) : JCPRC5 Automated grading of diabetic retinopathy stages
Quantitative Evaluation of Edge Detectors Using the Minimum Kernel Variance Criterion
 Quantitative Evaluation of Edge Detectors Using the Minimum Kernel Variance Criterion Qiang Ji Department of Computer Science University of Nevada Robert M. Haralick Department of Electrical Engineering
Quantitative Evaluation of Edge Detectors Using the Minimum Kernel Variance Criterion Qiang Ji Department of Computer Science University of Nevada Robert M. Haralick Department of Electrical Engineering
A comparative study of machine learning methods for lung diseases diagnosis by computerized digital imaging'"
 A comparative study of machine learning methods for lung diseases diagnosis by computerized digital imaging'" Suk Ho Kang**. Youngjoo Lee*** Aostract I\.' New Work to be 1 Introduction Presented U Mater~al
A comparative study of machine learning methods for lung diseases diagnosis by computerized digital imaging'" Suk Ho Kang**. Youngjoo Lee*** Aostract I\.' New Work to be 1 Introduction Presented U Mater~al
Epileptic seizure detection using EEG signals by means of stationary wavelet transforms
 I J C T A, 9(4), 2016, pp. 2065-2070 International Science Press Epileptic seizure detection using EEG signals by means of stationary wavelet transforms P. Grace Kanmani Prince 1, R. Rani Hemamalini 2,
I J C T A, 9(4), 2016, pp. 2065-2070 International Science Press Epileptic seizure detection using EEG signals by means of stationary wavelet transforms P. Grace Kanmani Prince 1, R. Rani Hemamalini 2,
DESIGN OF ULTRAFAST IMAGING SYSTEM FOR THYROID NODULE DETECTION
 DESIGN OF ULTRAFAST IMAGING SYSTEM FOR THYROID NODULE DETECTION Aarthipoornima Elangovan 1, Jeyaseelan.T 2 1 PG Student, Department of Electronics and Communication Engineering Kings College of Engineering,
DESIGN OF ULTRAFAST IMAGING SYSTEM FOR THYROID NODULE DETECTION Aarthipoornima Elangovan 1, Jeyaseelan.T 2 1 PG Student, Department of Electronics and Communication Engineering Kings College of Engineering,
Enhanced Detection of Lung Cancer using Hybrid Method of Image Segmentation
 Enhanced Detection of Lung Cancer using Hybrid Method of Image Segmentation L Uma Maheshwari Department of ECE, Stanley College of Engineering and Technology for Women, Hyderabad - 500001, India. Udayini
Enhanced Detection of Lung Cancer using Hybrid Method of Image Segmentation L Uma Maheshwari Department of ECE, Stanley College of Engineering and Technology for Women, Hyderabad - 500001, India. Udayini
A Review on Sleep Apnea Detection from ECG Signal
 A Review on Sleep Apnea Detection from ECG Signal Soumya Gopal 1, Aswathy Devi T. 2 1 M.Tech Signal Processing Student, Department of ECE, LBSITW, Kerala, India 2 Assistant Professor, Department of ECE,
A Review on Sleep Apnea Detection from ECG Signal Soumya Gopal 1, Aswathy Devi T. 2 1 M.Tech Signal Processing Student, Department of ECE, LBSITW, Kerala, India 2 Assistant Professor, Department of ECE,
Analysis of Microscopic Images of Blood Cells for Detection of Leukemia
 Analysis of Microscopic Images of Blood Cells for Detection of Leukemia Purnima.S 1, Manikandan.S 2, Mohan Raj. S 3, Nishanth. A 4, Selvakumar. A 5 1 Asst.Professor, 2,3,4,5 UG Scholar, Department of Biomedical
Analysis of Microscopic Images of Blood Cells for Detection of Leukemia Purnima.S 1, Manikandan.S 2, Mohan Raj. S 3, Nishanth. A 4, Selvakumar. A 5 1 Asst.Professor, 2,3,4,5 UG Scholar, Department of Biomedical
Time-to-Recur Measurements in Breast Cancer Microscopic Disease Instances
 Time-to-Recur Measurements in Breast Cancer Microscopic Disease Instances Ioannis Anagnostopoulos 1, Ilias Maglogiannis 1, Christos Anagnostopoulos 2, Konstantinos Makris 3, Eleftherios Kayafas 3 and Vassili
Time-to-Recur Measurements in Breast Cancer Microscopic Disease Instances Ioannis Anagnostopoulos 1, Ilias Maglogiannis 1, Christos Anagnostopoulos 2, Konstantinos Makris 3, Eleftherios Kayafas 3 and Vassili
Multiclass Classification of Cervical Cancer Tissues by Hidden Markov Model
 Multiclass Classification of Cervical Cancer Tissues by Hidden Markov Model Sabyasachi Mukhopadhyay*, Sanket Nandan*; Indrajit Kurmi ** *Indian Institute of Science Education and Research Kolkata **Indian
Multiclass Classification of Cervical Cancer Tissues by Hidden Markov Model Sabyasachi Mukhopadhyay*, Sanket Nandan*; Indrajit Kurmi ** *Indian Institute of Science Education and Research Kolkata **Indian
Algorithms in Nature. Pruning in neural networks
 Algorithms in Nature Pruning in neural networks Neural network development 1. Efficient signal propagation [e.g. information processing & integration] 2. Robust to noise and failures [e.g. cell or synapse
Algorithms in Nature Pruning in neural networks Neural network development 1. Efficient signal propagation [e.g. information processing & integration] 2. Robust to noise and failures [e.g. cell or synapse
arxiv: v1 [cs.lg] 4 Feb 2019
![arxiv: v1 [cs.lg] 4 Feb 2019 arxiv: v1 [cs.lg] 4 Feb 2019](/thumbs/95/123488545.jpg) Machine Learning for Seizure Type Classification: Setting the benchmark Subhrajit Roy [000 0002 6072 5500], Umar Asif [0000 0001 5209 7084], Jianbin Tang [0000 0001 5440 0796], and Stefan Harrer [0000
Machine Learning for Seizure Type Classification: Setting the benchmark Subhrajit Roy [000 0002 6072 5500], Umar Asif [0000 0001 5209 7084], Jianbin Tang [0000 0001 5440 0796], and Stefan Harrer [0000
Automatic Definition of Planning Target Volume in Computer-Assisted Radiotherapy
 Automatic Definition of Planning Target Volume in Computer-Assisted Radiotherapy Angelo Zizzari Department of Cybernetics, School of Systems Engineering The University of Reading, Whiteknights, PO Box
Automatic Definition of Planning Target Volume in Computer-Assisted Radiotherapy Angelo Zizzari Department of Cybernetics, School of Systems Engineering The University of Reading, Whiteknights, PO Box
REVIEW ON ARRHYTHMIA DETECTION USING SIGNAL PROCESSING
 REVIEW ON ARRHYTHMIA DETECTION USING SIGNAL PROCESSING Vishakha S. Naik Dessai Electronics and Telecommunication Engineering Department, Goa College of Engineering, (India) ABSTRACT An electrocardiogram
REVIEW ON ARRHYTHMIA DETECTION USING SIGNAL PROCESSING Vishakha S. Naik Dessai Electronics and Telecommunication Engineering Department, Goa College of Engineering, (India) ABSTRACT An electrocardiogram
Keywords: Leukaemia, Image Segmentation, Clustering algorithms, White Blood Cells (WBC), Microscopic images.
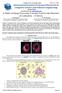 Volume 6, Issue 10, October 2016 ISSN: 2277 128X International Journal of Advanced Research in Computer Science and Software Engineering Research Paper Available online at: www.ijarcsse.com A Study on
Volume 6, Issue 10, October 2016 ISSN: 2277 128X International Journal of Advanced Research in Computer Science and Software Engineering Research Paper Available online at: www.ijarcsse.com A Study on
Local Image Structures and Optic Flow Estimation
 Local Image Structures and Optic Flow Estimation Sinan KALKAN 1, Dirk Calow 2, Florentin Wörgötter 1, Markus Lappe 2 and Norbert Krüger 3 1 Computational Neuroscience, Uni. of Stirling, Scotland; {sinan,worgott}@cn.stir.ac.uk
Local Image Structures and Optic Flow Estimation Sinan KALKAN 1, Dirk Calow 2, Florentin Wörgötter 1, Markus Lappe 2 and Norbert Krüger 3 1 Computational Neuroscience, Uni. of Stirling, Scotland; {sinan,worgott}@cn.stir.ac.uk
Brain Tumour Detection of MR Image Using Naïve Beyer classifier and Support Vector Machine
 International Journal of Scientific Research in Computer Science, Engineering and Information Technology 2018 IJSRCSEIT Volume 3 Issue 3 ISSN : 2456-3307 Brain Tumour Detection of MR Image Using Naïve
International Journal of Scientific Research in Computer Science, Engineering and Information Technology 2018 IJSRCSEIT Volume 3 Issue 3 ISSN : 2456-3307 Brain Tumour Detection of MR Image Using Naïve
QUANTIFICATION OF PROGRESSION OF RETINAL NERVE FIBER LAYER ATROPHY IN FUNDUS PHOTOGRAPH
 QUANTIFICATION OF PROGRESSION OF RETINAL NERVE FIBER LAYER ATROPHY IN FUNDUS PHOTOGRAPH Hyoun-Joong Kong *, Jong-Mo Seo **, Seung-Yeop Lee *, Hum Chung **, Dong Myung Kim **, Jeong Min Hwang **, Kwang
QUANTIFICATION OF PROGRESSION OF RETINAL NERVE FIBER LAYER ATROPHY IN FUNDUS PHOTOGRAPH Hyoun-Joong Kong *, Jong-Mo Seo **, Seung-Yeop Lee *, Hum Chung **, Dong Myung Kim **, Jeong Min Hwang **, Kwang
Computer based delineation and follow-up multisite abdominal tumors in longitudinal CT studies
 Research plan submitted for approval as a PhD thesis Submitted by: Refael Vivanti Supervisor: Professor Leo Joskowicz School of Engineering and Computer Science, The Hebrew University of Jerusalem Computer
Research plan submitted for approval as a PhD thesis Submitted by: Refael Vivanti Supervisor: Professor Leo Joskowicz School of Engineering and Computer Science, The Hebrew University of Jerusalem Computer
Brain Tumor Segmentation of Noisy MRI Images using Anisotropic Diffusion Filter
 Available Online at www.ijcsmc.com International Journal of Computer Science and Mobile Computing A Monthly Journal of Computer Science and Information Technology IJCSMC, Vol. 3, Issue. 7, July 2014, pg.744
Available Online at www.ijcsmc.com International Journal of Computer Science and Mobile Computing A Monthly Journal of Computer Science and Information Technology IJCSMC, Vol. 3, Issue. 7, July 2014, pg.744
Development of 2-Channel Eeg Device And Analysis Of Brain Wave For Depressed Persons
 Development of 2-Channel Eeg Device And Analysis Of Brain Wave For Depressed Persons P.Amsaleka*, Dr.S.Mythili ** * PG Scholar, Applied Electronics, Department of Electronics and Communication, PSNA College
Development of 2-Channel Eeg Device And Analysis Of Brain Wave For Depressed Persons P.Amsaleka*, Dr.S.Mythili ** * PG Scholar, Applied Electronics, Department of Electronics and Communication, PSNA College
LUNG NODULE DETECTION SYSTEM
 LUNG NODULE DETECTION SYSTEM Kalim Bhandare and Rupali Nikhare Department of Computer Engineering Pillai Institute of Technology, New Panvel, Navi Mumbai, India ABSTRACT: The Existing approach consist
LUNG NODULE DETECTION SYSTEM Kalim Bhandare and Rupali Nikhare Department of Computer Engineering Pillai Institute of Technology, New Panvel, Navi Mumbai, India ABSTRACT: The Existing approach consist
NAÏVE BAYESIAN CLASSIFIER FOR ACUTE LYMPHOCYTIC LEUKEMIA DETECTION
 NAÏVE BAYESIAN CLASSIFIER FOR ACUTE LYMPHOCYTIC LEUKEMIA DETECTION Sriram Selvaraj 1 and Bommannaraja Kanakaraj 2 1 Department of Biomedical Engineering, P.S.N.A College of Engineering and Technology,
NAÏVE BAYESIAN CLASSIFIER FOR ACUTE LYMPHOCYTIC LEUKEMIA DETECTION Sriram Selvaraj 1 and Bommannaraja Kanakaraj 2 1 Department of Biomedical Engineering, P.S.N.A College of Engineering and Technology,
A Pictorial Review and an Algorithm for the Determination of Sickle Cell Anemia
 International Journal of Engineering and Advanced Technology (IJEAT) ISSN: 2249 8958, Volume-5 Issue-2, December 2015 A Pictorial Review and an Algorithm for the Determination of Sickle Cell Anemia Hariharan.S,
International Journal of Engineering and Advanced Technology (IJEAT) ISSN: 2249 8958, Volume-5 Issue-2, December 2015 A Pictorial Review and an Algorithm for the Determination of Sickle Cell Anemia Hariharan.S,
NUCLEI SEGMENTATION OF MICROSCOPY IMAGES OF THYROID NODULES VIA ACTIVE CONTOURS AND K-MEANS CLUSTERING
 1 st International Conference on Experiments/Process/System Modelling/Simulation/Optimization 1 st IC-EpsMsO Athens, 6-9 July, 2005 IC-EpsMsO NUCLEI SEGMENTATION OF MICROSCOPY IMAGES OF THYROID NODULES
1 st International Conference on Experiments/Process/System Modelling/Simulation/Optimization 1 st IC-EpsMsO Athens, 6-9 July, 2005 IC-EpsMsO NUCLEI SEGMENTATION OF MICROSCOPY IMAGES OF THYROID NODULES
Neural Network Based Technique to Locate and Classify Microcalcifications in Digital Mammograms
 Neural Network Based Technique to Locate and Classify Microcalcifications in Digital Mammograms Author Verma, Brijesh Published 1998 Conference Title 1998 IEEE World Congress on Computational Intelligence
Neural Network Based Technique to Locate and Classify Microcalcifications in Digital Mammograms Author Verma, Brijesh Published 1998 Conference Title 1998 IEEE World Congress on Computational Intelligence
Implementation of Automatic Retina Exudates Segmentation Algorithm for Early Detection with Low Computational Time
 www.ijecs.in International Journal Of Engineering And Computer Science ISSN: 2319-7242 Volume 5 Issue 10 Oct. 2016, Page No. 18584-18588 Implementation of Automatic Retina Exudates Segmentation Algorithm
www.ijecs.in International Journal Of Engineering And Computer Science ISSN: 2319-7242 Volume 5 Issue 10 Oct. 2016, Page No. 18584-18588 Implementation of Automatic Retina Exudates Segmentation Algorithm
Automatic Detection of Heart Disease Using Discreet Wavelet Transform and Artificial Neural Network
 e-issn: 2349-9745 p-issn: 2393-8161 Scientific Journal Impact Factor (SJIF): 1.711 International Journal of Modern Trends in Engineering and Research www.ijmter.com Automatic Detection of Heart Disease
e-issn: 2349-9745 p-issn: 2393-8161 Scientific Journal Impact Factor (SJIF): 1.711 International Journal of Modern Trends in Engineering and Research www.ijmter.com Automatic Detection of Heart Disease
1 Introduction. Fig. 1 Examples of microaneurysms
 3rd International Conference on Multimedia Technology(ICMT 2013) Detection of Microaneurysms in Color Retinal Images Using Multi-orientation Sum of Matched Filter Qin Li, Ruixiang Lu, Shaoguang Miao, Jane
3rd International Conference on Multimedia Technology(ICMT 2013) Detection of Microaneurysms in Color Retinal Images Using Multi-orientation Sum of Matched Filter Qin Li, Ruixiang Lu, Shaoguang Miao, Jane
Copyright 2008 Society of Photo Optical Instrumentation Engineers. This paper was published in Proceedings of SPIE, vol. 6915, Medical Imaging 2008:
 Copyright 2008 Society of Photo Optical Instrumentation Engineers. This paper was published in Proceedings of SPIE, vol. 6915, Medical Imaging 2008: Computer Aided Diagnosis and is made available as an
Copyright 2008 Society of Photo Optical Instrumentation Engineers. This paper was published in Proceedings of SPIE, vol. 6915, Medical Imaging 2008: Computer Aided Diagnosis and is made available as an
Earlier Detection of Cervical Cancer from PAP Smear Images
 , pp.181-186 http://dx.doi.org/10.14257/astl.2017.147.26 Earlier Detection of Cervical Cancer from PAP Smear Images Asmita Ray 1, Indra Kanta Maitra 2 and Debnath Bhattacharyya 1 1 Assistant Professor
, pp.181-186 http://dx.doi.org/10.14257/astl.2017.147.26 Earlier Detection of Cervical Cancer from PAP Smear Images Asmita Ray 1, Indra Kanta Maitra 2 and Debnath Bhattacharyya 1 1 Assistant Professor
