DeepLung: 3D Deep Convolutional Nets for Automated Pulmonary Nodule Detection and Classification
|
|
|
- Brenda Ramsey
- 6 years ago
- Views:
Transcription
1 DeepLung: 3D Deep Convolutional Nets for Automated Pulmonary Nodule Detection and Classification Wentao Zhu, Chaochun Liu, Wei Fan, Xiaohui Xie University of California, Irvine and Baidu Research and The emergence of large-scale dataset, LUNA16 (Setio et al. 2017), accelerates the nodule detection related research. Typically, nodule detection consists of two stages, region proposal generation and false positive reduction. Traditional approaches generally require manually designed features such as morphological features, voxel clustering and pixel thresholding (Murphy et al. 2009; Jacobs et al. 2014). Recently, deep convolutional networks, such as Faster R-CNN (Ren et al. 2015) and fully convolutional networks (Long, Shelhamer, and Darrell 2015), are employed to generate candidate bounding boxes (Ding et al. 2017; Dou et al. 2017). In the second stage, more advanced methods or complex features, such as more carefully designed texture features, are designed to remove false positive nodules. Due to the 3D nature of CT data and the effectiveness of Faster R-CNN for object detection in 2D natural images (Huang et al. 2016b), we design a 3D Faster R-CNN for nodule detection with U- net-like encoder-decoder structure to effectively learn latent features (Ronneberger, Fischer, and Brox 2015). Before the era of deep learning, feature engineering followed by classifiers is a general pipeline for nodule classification (Han et al. 2013). After the public large-scale dataset, LIDC-IDRI (Armato et al. 2011), becomes available, deep learning based methods have become dominant for nodule classification research (Shen et al. 2016). Multi-scale deep convolutional network with shared weights on different scales has been proposed for the nodule classification (Shen et al. 2015). The weight sharing scheme reduces the number of parameters and forces the multi-scale deep convolutional network to learn scale-invariant features. Inspired by the recent success of dual path network (DPN) on ImageNet (Chen et al. 2017; Deng et al. 2009), we propose a novel framework for CT nodule classification. First, we design a 3D deep dual path network to extract features. Then we use gradient boosting machines (GBM) with deep 3D dual path features, nodule size and cropped raw nodule CT pixels for the nodule classification (Friedman 2001). At last, we build a fully automated lung CT cancer diagnosis system, DeepLung, by combining the nodule detection network and nodule classification network together, as illustrated in Fig. 1. For a CT image, we first use the detection subnetwork to detect candidate nodules. Next, we employ the classification subnetwork to classify the detected nodules into either malignant or benign. Finally, the patient-level diarxiv: v1 [cs.cv] 16 Sep 2017 Abstract In this work, we present a fully automated lung CT cancer diagnosis system, DeepLung. DeepLung contains two parts, nodule detection and classification. Considering the 3D nature of lung CT data, two 3D networks are designed for the nodule detection and classification respectively. Specifically, a 3D Faster R-CNN is designed for nodule detection with a U-net-like encoder-decoder structure to effectively learn nodule features. For nodule classification, gradient boosting machine (GBM) with 3D dual path network (DPN) features is proposed. The nodule classification subnetwork is validated on a public dataset from LIDC-IDRI, on which it achieves better performance than state-of-the-art approaches, and surpasses the average performance of four experienced doctors. For the DeepLung system, candidate nodules are detected first by the nodule detection subnetwork, and nodule diagnosis is conducted by the classification subnetwork. Extensive experimental results demonstrate the DeepLung is comparable to the experienced doctors both for the nodule-level and patient-level diagnosis on the LIDC-IDRI dataset. Introduction Lung cancer is the most common cause of cancer-related death in men. Low-dose lung CT screening provides an effective way for early diagnosis, which can sharply reduce the lung cancer mortality rate. Advanced computer-aided diagnosis systems (CADs) are expected to have high sensitivities while at the same time maintaining low false positive rates. Recent advances in deep learning enable us to rethink the ways of clinician lung cancer diagnosis. Current lung CT analysis research mainly includes nodule detection (Dou et al. 2017; Ding et al. 2017), and nodule classification (Shen et al. 2015; 2016; Hussein et al. 2017; Yan et al. 2016). There is few work on building a complete lung CT cancer diagnosis system for fully automated lung CT cancer diagnosis, integrating both nodule detection and nodule classification. It is worth exploring a whole lung CT cancer diagnosis system and understanding how far the performance of current technology differs from that of experienced doctors. To our best knowledge, this is the first work for a fully automated and complete lung CT cancer diagnosis system. Copyright c 2018, Association for the Advancement of Artificial Intelligence ( All rights reserved.
2 Figure 1: The framework of DeepLung. DeepLung first employs 3D Faster R-CNN to generate candidate nodules. Then it uses 3D deep dual path network (DPN) to extract deep features from the detected and cropped nodules. Lastly, gradient boosting machine (GBM) with deep features, detected nodule size, and raw nodule pixels are employed for classification. Patient-level diagnosis can be achieved by fusing the classification results of detected nodules in the CT. agnosis result can be achieved for the whole CT by fusing the diagnosis result of each nodule. Our main contributions are as follows: 1) To our best knowledge, DeepLung is the first work for a complete automated lung CT cancer diagnosis system. 2) Two 3D deep convolutional networks are designed for nodule detection and classification. Specifically, inspired by the effective of Faster R-CNN for object detection and deep dual path network s success on ImageNet (Huang et al. 2016b; Chen et al. 2017), we propose 3D Faster R-CNN for nodule detection, and 3D deep dual path network for nodule classification. 3) Our classification framework achieves superior performance compared with state-of-the-art approaches, and the performance surpasses the average performance of experienced doctors on the largest public dataset, LIDC-IDRI dataset. 4) The DeepLung system is comparable to the average performance of experienced doctors both on nodulelevel and patient-level diagnosis. Related Work Traditional nodule detection requires manually designed features or descriptors (Lopez Torres et al. 2015). Recently, several works have been proposed to use deep convolutional networks for nodule detection to automatically learn features, which is proven to be much more effective than hand-crafted features. Setio et al. proposes multi-view convolutional network for false positive nodule reduction (Setio et al. 2016). Due to the 3D nature of CT scans, some work propose 3D convolutional networks to handle the challenge. The 3D fully convolutional network (FCN) is proposed to generate region candidates, and deep convolution network with weighted sampling is used in the false positive candidates reduction stage (Dou et al. 2017). Ding et al. uses the Faster R-CNN to generate candidate nodules, followed by 3D convolutional networks to remove false positive nodules (Ding et al. 2017). Due to the effective performance of Faster R-CNN (Huang et al. 2016b; Ren et al. 2015), we design a novel network, 3D Faster R- CNN, for the nodule detection. Further, U-net-like encoderdecoder scheme is employed for 3D Faster R-CNN to effectively learn the features (Ronneberger, Fischer, and Brox 2015). Nodule classification has traditionally been based on segmentation (El-Baz et al. 2011; Zhu and Xie 2016) and manual feature design (Aerts et al. 2014). Several works designed 3D contour feature, shape feature and texture feature for CT nodule diagnosis (Way et al. 2006; El-Baz et al. 2011; Han et al. 2013). Recently, deep networks have been shown to be effective for medical images. Artificial neural network was implemented for CT nodule diagnosis (Suzuki et al. 2005). More computationally effective network, multi-scale convolutional neural network with shared weights for different scales to learn scale-invariant features, is proposed for nodule classification (Shen et al. 2015). Deep transfer learning and multi-instance learning is used for patientlevel lung CT diagnosis (Shen et al. 2016). A comparison on 2D and 3D convolutional networks is conducted and shown that 3D convolutional network is superior than 2D convolutional network for 3D CT data (Yan et al. 2016). Further, a multi-task learning and transfer learning framework is proposed for nodule diagnosis (Hussein et al. 2017; Zhu et al. 2017). Different from their approaches, we propose a novel classification framework for CT nodule diagnosis. Inspired by the recent success of deep dual path network (DPN) on ImageNet (Chen et al. 2017), we design a novel totally 3D deep dual path network to extract features from raw CT nodules. Then, we employ gradient boosting machine (GBM) with the deep DPN features, nodule size, and raw nodule CT pixels for the nodule diagnosis. Patientlevel diagnosis can be achieved by fusing the nodule-level diagnosis. DeepLung Framework The fully automated lung CT cancer diagnosis system, DeepLung, consists of two parts, nodule detection and nodule classification. We design a 3D Faster R-CNN for nodule detection, and propose gradient boosting machine with deep 3D dual path network features, raw nodule CT pixels and nodule size for nodule classification.
3 Figure 2: The 3D Faster R-CNN framework contains residual blocks and U-net-like encoder-decoder structure. The model employs 3 anchors and multitask learning loss, including coordinates (x, y, z) and diameter d regression, and candidate box classification. The numbers in boxes are feature map sizes in the format (#slices*#rows*#cols*#maps). The numbers above the connections are in the format (#filters #slices*#rows*#cols). 3D Faster R-CNN for Nodule Detection The 3D Faster R-CNN with U-net-like encoder-decoder structure is illustrated in Fig. 2. Due to the GPU memory limitation, the input of 3D Faster R-CNN is cropped from 3D reconstructed CT images with pixel size The encoder network is derived from ResNet-18 (He et al. 2016). Before the first max-pooling, two convolutional layers are used to generate features. After that, four residual blocks are employed in the encoder subnetwork. We integrate the U- net-like encoder-decoder design concept in the detection to learn the deep networks efficiently (Ronneberger, Fischer, and Brox 2015). In fact, for the region proposal generation, the 3D Faster R-CNN conducts pixel-wise multi-scale learning and the U-net is validated as an effective way for pixelwise labeling. This integration makes candidate nodule generation more effective. In the decoder network, the feature maps are processed by deconvolution layers and residual blocks, and are subsequently concatenated with the corresponding layers in the encoder network (Zeiler et al. 2010). Then a convolutional layer with dropout (dropout probability 0.5) is used for the second last layer. In the last layer, we design 3 anchors, 5, 10, 20, for scale references which are designed based on the distribution of nodule sizes. For each anchor, there are 5 parts in the loss function, classification loss L cls for whether the current box is a nodule or not, regression loss L reg for nodule coordinates x, y, z and nodule size d. If an anchor overlaps a ground truth bounding box with the intersection over union (IoU) higher than 0.5, we consider it as a positive anchor (p = 1). On the other hand, if an anchor has IoU with all ground truth boxes less than 0.02, we consider it as a negative anchor (p = 0). The multi-task loss function for the anchor i is defined as L(p i, t i ) = λl cls (p i, p i ) + p i L reg (t i, t i ), (1) where p i is the predicted probability for current anchor i being a nodule, t i is the regression predicted relative coordinates for nodule position, which is defined as t i = ( x x a d a, y y a, z z a, log( d )), (2) d a d a d a where (x, y, z, d) are the predicted nodule coordinates and diameter in the original space, (x a, y a, z a, d a ) are the coordinates and scale for the anchor i. For ground truth nodule position, it is defined as t i = ( x x a, y y a, z z a, log( d )), (3) d a d a d a d a where (x, y, z, d ) are nodule ground truth coordinates and diameter. The λ is set as 0.5. For L cls, we used binary cross entropy loss function. For L reg, we used smooth l 1 regression loss function (Girshick 2015). Gradient Boosting Machine with 3D Dual Path Net Feature for Nodule Classification Inspired by the success of dual path network on the ImageNet (Chen et al. 2017; Deng et al. 2009), we design a 3D deep dual path network framework for lung CT nodule classification in Fig. 4. Dual path connection benefits both from the advantage of residual learning and that of dense connection (He et al. 2016; Huang et al. 2016a). The shortcut connection in residual learning is an effective way to eliminate gradient vanishing phenomenon in very deep networks. From a learned feature weights sharing perspective, residual learning enables feature reuse, while dense connection has an advantage of keeping exploiting new features (Chen et al. 2017). And densely connected network has fewer parameters than residual learning because there is no need to relearn redundant feature maps. The assumption of dual path connection is that there might exist some redundancy in the exploited features. And dual path connection uses part of feature maps for dense connection and part of them for residual learning. In implementation, the dual path connection splits its feature maps into two parts. One part, F(x)[d :], is used for residual learning, the other part, F(x)[: d], is used for dense connection as shown in Fig. 3. Here d is the hyper-parameter for deciding how many new features to be exploited. The dual path connection can be formulated as y = G([F(x)[: d], F(x)[d :] + x]), (4) where y is the feature map for dual path connection, G is used as ReLU activation function, F is convolutional layer functions, and x is the input of dual path connection block. Dual path connection integrates the advantages of the two advanced frameworks, residual learning for feature reuse and dense connection for keeping exploiting new features, into a unified structure, which obtains success on the ImageNet dataset. For CT data, advanced method should be effective to extract 3D volume feature (Yan et al. 2016). We design a 3D deep dual path network for the 3D CT lung nodule classification in Fig. 4. We firstly crop CT data centered at predicted nodule locations with size After that, a convolutional layer is used to extract features. Then 4 dual path blocks are employed to learn higher level features. Lastly, the 3D average pooling and binary logistic regression layer are used for benign or malignant diagnosis. The 3D deep dual path network can be used as a classifier for nodule diagnosis directly. And it can also be employed to learn effective features. We construct feature by
4 Figure 3: Illustration of dual path connection (Chen et al. 2017), which benefits both from the advantage of residual learning (He et al. 2016) and that of dense connection (Huang et al. 2016a) from network structure design intrinsically. Figure 4: The 3D deep dual path network framework in the nodule classification subnetwork, which contains 4 dual path connection blocks. After the training, the deep 3D dual path network feature is extracted for gradient boosting machine to do nodule diagnosis. The numbers in the figure are of the same formats as Fig. 2. concatenating the learned 3D deep dual path network features (the second last layer, 2,560 dimension), nodule size, and raw 3D cropped nodule pixels. Given complete and effective features, gradient boosting machine (GBM) is a superior method to build an advanced classifier from these features (Friedman 2001). We validate the feature combining nodule size with raw 3D cropped nodule pixels, employ GBM as a classifier, and obtain 86.12% test accuracy averagely. Lastly, we employ GBM with the constructed feature and achieve the best diagnosis performance. DeepLung System: Fully Automated Lung CT Cancer Diagnosis The DeepLung system includes the nodule detection using the 3D Faster R-CNN, and nodule classification using gradient boosting machine (GBM) with constructed feature (deep dual path features, nodule size and raw nodule CT pixels) in Fig. 1. Due to the GPU memory limitation, we first split the whole CT into several patches, process them through the detector, and combine the detected results together. We only keep the detected boxes of detection probabilities larger than 0.12 (threshold as -2 before sigmoid function). After that, non-maximum suppression (NMS) is adopted based on detection probability with the intersection over union (IoU) threshold as 0.1. Here we expect to not miss too many ground truth nodules. After we get the detected nodules, we crop the nodule with the center as the detected center and size as The detected nodule size is kept for the classification model as a part of features. The 3D deep dual path network is employed to extract features. We use the GBM and construct features to conduct diagnosis for the detected nodules. For CT pixel feature, we use the cropped size as and center as the detected nodule center in the experiments. For patient-level diagnosis, if one of the detected nodules is positive (cancer), the patient is a cancer patient, and if all the detected nodules are negative, the patient is a negative (non-cancer) patient. Experiments We conduct extensive experiments to validate the DeepLung system. We perform 10-fold cross validation using the detector on LUNA16 dataset. For nodule classification, we use the LIDC-IDRI annotation, and employ the LUNA16 s patientlevel dataset split. Finally, we also validate the whole system based on the detected nodules both on patient-level diagnosis and nodule-level diagnosis. In the training, for each model, we use 150 epochs in total with stochastic gradient descent optimization and momentum as 0.9. The used batch size is relied on the GPU memory. We use weight decay as The initial learning rate is 0.01, at the half of training, and after the epoch 120. Datasets LUNA16 dataset is a subset of the largest public dataset for pulmonary nodules, the LIDC-IDRI dataset (Armato et al. 2011). LUNA16 dataset only has the detection annotations, while LIDC-IDRI contains almost all the related information for low-dose lung CTs including several doctors annotations on nodule sizes, locations, diagnosis results, nodule texture, nodule margin and other informations. LUNA16 dataset removes CTs with slice thickness greater than 3mm, slice spacing inconsistent or missing slices from LIDC-IDRI dataset, and explicitly gives the patient-level 10-fold cross validation split of the dataset. LUNA16 dataset contains 888 low-dose lung CTs, and LIDC-IDRI contains 1,018 lowdose lung CTs. Note that LUNA16 dataset removes the annotated nodules of size smaller than 3mm. For nodule classification, we extract nodule annotations from LIDC-IDRI dataset, find the mapping of different doctors nodule annotations with the LUNA16 s nodule annotations, and get the ground truth of nodule diagnosis by taking different doctors diagnosis averagely (Do not count the 0 score for diagnosis, which means N/A.). If the final average score is equal to 3 (uncertain about malignant or benign), we remove the nodule. For the nodules with score greater than 3, we label them as positive. Otherwise, we label them as negative. For doctors annotations, we only keep those of four doctors who label most of the nodules for comparison. We only keep the CTs within LUNA16 dataset, and use the same cross validation split as LUNA16 for classification. Preprocessing We firstly clip the raw data into [ 1200, 600]. Secondly, we transform the range linearly into [0, 1]. Thirdly, we use the
5 Table 1: Nodule classification comparisons on LIDC-IDRI dataset. Nodule Size All feat. Models 3D DPN +Pixel+GBM +GBM m-cnn Acc. (%) Table 2: Nodule-level diagnosis accuracy (%) between nodule classification subnetwork in DeepLung and experienced doctors on doctor s individually confident nodules. Dr 1 Dr 2 Dr 3 Dr 4 Average Doctors DeepLung Figure 5: Sensitivity (Recall) rate with respect to false positives per scan. The FROC (average recall rate at the false positives as 0.125, 0.25, 0.5, 1, 2, 4, 8) is 83.4%. The 3D Faster R-CNN has a recall rate 94.6% for all the nodules. The dash lines are lower bound and upper bound FROC for 95% confidence interval using bootstrapping with 1,000 bootstraps (Setio et al. 2017). The solid line is the interpolated FROC based on prediction. LUNA16 s given segmentation ground truth and remove the useless background. It is worth visualizing the processed data during processing because some segmentations are imperfect, and need to be removed. After that, the data can be processed by the deep networks. DeepLung for Nodule Detection We train and evaluate the detector on LUNA16 dataset following 10-fold cross validation with given patient-level split. In the training, we use the flipping, randomly scale from 0.75 to 1.25 for the cropped patches to augment the data. The evaluation metric, FROC, is the average recall rate at the average number of false positives as 0.125, 0.25, 0.5, 1, 2, 4, 8 per scan, which is the official evaluation metric for LUNA16 dataset. The FROC performance on LUNA16 is visualized in Fig. 5. The solid line is interpolated FROC based on true prediction, and the dash lines are upper bound and lower bound for the bootstrapped FROC performance. The 3D Faster R-CNN achieves 83.4% FROC without any false positive nodule reduction stage, which is comparable with 83.9% for the best method, which uses two-stage training (Dou et al. 2017). DeepLung for Nodule Classification We validate the nodule classification performance of the DeepLung system on the LIDC-IDRI dataset with the LUNA16 s split principle, 10-fold patient-level cross validation. There are 1,004 nodules left and 450 nodules are positive. In the training, we firstly pad the nodules of size into , randomly crop from the padded data, horizontal flip, vertical flip, z-axis flip the data for augmentation, randomly set patch as 0, and normalize the data with the mean and std. obtained from training data. The total number of epochs is 1,050. The learning rate is 0.01 at first, then became after epoch 525, and turned into after epoch 840. Due to time and resource limitation for training, we use the fold 1, 2, 3, 4, 5 for test, and the final performance is the average performance on the five test folds. The nodule classification performance is concluded in Table 1. From the table 1, our 3D deep dual path network (DPN) achieves better performance than that of multi-scale CNN (Shen et al. 2015) because of the power of 3D structure and deep dual path network. Because of the power of gradient boosting machine (GBM), GBM with nodule size and raw nodule pixels with crop size as achieves comparable performance as multi-scale CNN (Shen et al. 2015). Finally, we construct feature with 3D deep dual path network features, nodule size and raw nodule pixels, and obtain 90.44% accuracy. Compared with Experienced Doctors on Their Individually Confident Nodules We compare our predictions with those of four experienced doctors on their individually confident nodules (with individual score not as 3). Note that about 1/3 nodules are labeled as 3 for each doctor. Comparison results are concluded in Table 2. From Table 2, these doctors confident nodules are easy to be diagnosed nodules from the performance comparison between our model s performances in Table 1 and Table 2. To our surprise, the average performance of our model is 1.5% better than that of experienced doctors. In fact, our model s performance is better than 3 out of 4 doctors (doctor 1, 3, 4) on the nodule diagnosis task. The result validates deep network surpasses human-level performance for image classification (He et al. 2016), and the DeepLung is better suited for nodule diagnosis than experienced doctors. DeepLung for Fully Automated Lung CT Cancer Diagnosis We also validate the DeepLung for fully automated lung CT cancer diagnosis on the LIDC-IDRI dataset with the same protocol as LUNA16 s patient-level split. Firstly, we employ the 3D Faster R-CNN to detect suspicious nodules. Then we retrain the model from nodule classification model on the detected nodules dataset. If the center of detected nodule is
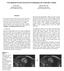
6 Table 3: Comparison between DeepLung s nodule classification on all detected nodules and doctors on all nodules. Method TP Set FP Set Doctors Acc. (%) Table 4: Patient-level diagnosis accuracy(%) between DeepLung and experienced doctors on doctor s individually confident CTs. Dr 1 Dr 2 Dr 3 Dr 4 Average Doctors DeepLung within the ground truth positive nodule, it is a positive nodule. Otherwise, it is a negative nodule. Through this mapping from the detected nodule and ground truth nodule, we can evaluate the performance and compare it with the performance of experienced doctors. We adopt the test fold 1, 2, 3, 4, 5 to validate the performance the same as that for nodule classification. Different from pure nodule classification, the fully automated lung CT nodule diagnosis relies on nodule detection. We evaluate the performance of DeepLung on the detection true positive (TP) set and detection false positive (FP) set individually in Table 3. If the detected nodule of center within one of ground truth nodule regions, it is in the TP set. If the detected nodule of center out of any ground truth nodule regions, it is in FP set. From Table 3, the DeepLung system using detected nodule region obtains 81.42% accuracy for all the detected TP nodules. Note that the experienced doctors obtain 78.36% accuracy for all the nodule diagnosis on average. The DeepLung system with fully automated lung CT nodule diagnosis still achieves above average performance of experienced doctors. On the FP set, our nodule classification subnetwork in the DeepLung can reduce 97.02% FP detected nodules, which guarantees that our fully automated system is effective for the lung CT cancer diagnosis. Compared with Experienced Doctors on Their Individually Confident CTs We employ the DeepLung for patientlevel diagnosis further. If the current CT has one nodule that is classified as positive, the diagnosis of the CT is positive. If all the nodules are classified as negative for the CT, the diagnosis of the CT is negative. We evaluate the DeepLung on the doctors individually confident CTs for benchmark comparison in Table 4. From Table 4, DeepLung achieves 81.41% patient-level diagnosis accuracy. The performance is 99% of the average performance of four experienced doctors, and the performance of DeepLung is better than that of doctor 4. Thus DeepLung can be used to help improve some doctors performance, like that of doctor 4, which is the goal for computer aided diagnosis system. We also employ Kappa coefficient, which is a common approach to evaluate the agreement between two raters, to test the agreement between DeepLung and the ground truth patient-level labels (Landis and Koch 1977). For comparison, we valid those of 4 individual doctors on their individual confident CTs. The Kappa coefficient of DeepLung is 63.02%, while the average Kappa coefficient of doctors is 64.46%. It shows the predictions of DeepLung are of good agreement with ground truths for patient-level diagnosis, and are comparable with those of experienced doctors. Discussion In this section, we are trying to explain the DeepLung by visualizing the nodule detection and classification results. Nodule Detection We randomly pick nodules from test fold 1 and visualize them in the first row and third row in Fig. 6. Detected nodules are visualized in the second, fourth and fifth row. The numbers below detected nodules are with the format (ground truth slice number-detected slice number-detection probability). The fifth row with red detection probabilies are the false positive detections. Here we only visualize the slice of center z. The yellow circles are the ground truth nodules or detected nodules respectively. The center of the circle is the nodule center and the diameter of the circle is relative to the nodule size. From the first four rows in Fig. 6, we observe the 3D Faster R-CNN works well for the nodules from test fold 1. The detection probability is also very high for these nodules. From the detected false positive nodules in the last row, we can find these false positives are suspicious to nodules visually if we observe them carefully. And the sizes of these false positive nodules are very small. Note that the nodules less than 3mm are removed in the ground truth annotations. Nodule Classification We also visualize the nodule classification results from test fold 1 in Fig. 7. The nodules in the first four rows are those the DeepLung predicted right, but some doctors predicted wrong. The first number is the DeepLung predicted malignant probability. The second number is which doctor predicted wrong. If the probability is large than 0.5, it is a malignant nodule. Otherwise, it is a benign nodule. The rest nodules are those the DeepLung predicted wrong with red numbers indicating the DeepLung predicted malignant probability. For an experienced doctor, if a nodule is big and has irregular shape, it has a high probability to be a malignant nodule. From the first 4 rows in Fig. 7, we can observe that doctors mis-diagnose some nodules. The reason is that, humans are not good at processing 3D CT data, which is of low signal to noise ratio. In fact, even for high quality 2D natural image, the performance of deep network surpasses that of humans (He et al. 2016). From the doctors individual experience point of view, some doctors intend to be optimistic to some nodules, and some doctors intend to be pessimistic to them. And they can just observe one slice each time. Some irregular boundaries are vague. The machine learning based methods can learn these complicated rules and high dimensional features from these doctors annotations, and avoid such bias.
7 Figure 6: Visualization of central slices for nodule ground truths and detection results. We randomly choose nodules (the first row and third row) from test fold 1. Detection results are shown in the second row and fourth row. The numbers below detected nodules are with the format (ground truth slice number-detected slice number-detection probability). The last row shows the false positive detected nodules and detection probabilities. Note that the ground truth nodules are of diameter greater than 3mm. The DeepLung performs well for detection. The rest nodules with red numbers are the nodules the DeepLung predicted wrong for test fold 1 in Fig. 7. If the predicted malignant probability is large than 0.5, the ground truth label is benign. Otherwise, the ground truth label is malignant. From the central slices, we can hardly decide the DeepLung s predictions are wrong. Maybe the DeepLung cannot find some weak irregular boundaries as for the bottom right 4 nodules, which is the possible reason why the DeepLung predicts them as benign. From the above analysis, the DeepLung can be considered as a tool to assist the diagnosis for doctors. Combing the DeepLung and doctor s own diagnosis is an effective way to improve diagnosis accuracy. Conclusion In this work, we propose a fully automated lung CT cancer diagnosis system, DeepLung. DeepLung consists two parts, nodule detection and classification. For nodule detec- Figure 7: Visualization of central slices for nodule classification results on test fold 1. We choose nodules that is predicted right, but annotated wrong by some doctors in the first 4 rows. The numbers below the nodules are (model predicted malignant probability-which doctor is wrong). The last 2 rows show the nodules our model predicted wrong. The red number is the wrongly predicted malignant probabilities (> 0.5, predict malignant; < 0.5, predict benign). The DeepLung avoids doctors individual bias and achieves better performance than the average performance of doctors. tion, we design a 3D Faster R-CNN with U-net-like encoderdecoder structure to detect suspicious nodules. Then we input the detected nodules into the nodule classification network. The 3D deep dual path network is designed to extract features. Further, gradient boosting machine with different features combined together to achieve the state-of-the-art performance, which surpasses that of experienced doctors. Extensive experimental results on public available largescale datasets, LUNA16 and LIDC-IDRI datasets, demonstrate the superior performance of the DeepLung. In the future, we will develop more advanced nodule detection method to further boost the performance of DeepLung. Further, a unified framework for automated lung CT cancer diagnosis is expected for nodule detection and classification. Integrating multi-modality data, such as electronic medical report history, into the DeepLung is another
8 direction to enhance the diagnosis performance. References Aerts, H. J.; Velazquez, E. R.; Leijenaar, R. T.; Parmar, C.; Grossmann, P.; Cavalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nature communications 5. Armato, S. G.; McLennan, G.; Bidaut, L.; McNitt-Gray, M. F.; Meyer, C. R.; Reeves, A. P.; Zhao, B.; Aberle, D. R.; Henschke, C. I.; Hoffman, E. A.; et al The lung image database consortium (lidc) and image database resource initiative (idri): a completed reference database of lung nodules on ct scans. Medical physics 38(2): Chen, Y.; Li, J.; Xiao, H.; Jin, X.; Yan, S.; and Feng, J Dual path networks. arxiv preprint arxiv: Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; and Fei-Fei, L Imagenet: A large-scale hierarchical image database. In CVPR, IEEE. Ding, J.; Li, A.; Hu, Z.; and Wang, L Accurate pulmonary nodule detection in computed tomography images using deep convolutional neural networks. arxiv preprint arxiv: Dou, Q.; Chen, H.; Jin, Y.; Lin, H.; Qin, J.; and Heng, P.-A Automated pulmonary nodule detection via 3d convnets with online sample filtering and hybrid-loss residual learning. arxiv preprint arxiv: El-Baz, A.; Nitzken, M.; Khalifa, F.; Elnakib, A.; Gimelfarb, G.; Falk, R.; and El-Ghar, M. A d shape analysis for early diagnosis of malignant lung nodules. In Biennial International Conference on Information Processing in Medical Imaging, Springer. Friedman, J. H Greedy function approximation: a gradient boosting machine. Annals of statistics Girshick, R Fast r-cnn. In ICCV, Han, F.; Zhang, G.; Wang, H.; Song, B.; Lu, H.; Zhao, D.; Zhao, H.; and Liang, Z A texture feature analysis for diagnosis of pulmonary nodules using lidc-idri database. In Medical Imaging Physics and Engineering (ICMIPE), 2013 IEEE International Conference on, IEEE. He, K.; Zhang, X.; Ren, S.; and Sun, J Deep residual learning for image recognition. In CVPR, Huang, G.; Liu, Z.; Weinberger, K. Q.; and van der Maaten, L. 2016a. Densely connected convolutional networks. arxiv preprint arxiv: Huang, J.; Rathod, V.; Sun, C.; Zhu, M.; Korattikara, A.; Fathi, A.; Fischer, I.; Wojna, Z.; Song, Y.; Guadarrama, S.; et al. 2016b. Speed/accuracy trade-offs for modern convolutional object detectors. arxiv preprint arxiv: Hussein, S.; Cao, K.; Song, Q.; and Bagci, U Risk stratification of lung nodules using 3d cnn-based multi-task learning. In IPMI, Springer. Jacobs, C.; van Rikxoort, E. M.; Twellmann, T.; Scholten, E. T.; de Jong, P. A.; Kuhnigk, J.-M.; Oudkerk, M.; de Koning, H. J.; Prokop, M.; Schaefer-Prokop, C.; et al Automatic detection of subsolid pulmonary nodules in thoracic computed tomography images. Medical image analysis 18(2): Landis, J. R., and Koch, G. G The measurement of observer agreement for categorical data. biometrics Long, J.; Shelhamer, E.; and Darrell, T Fully convolutional networks for semantic segmentation. In CVPR, Lopez Torres, E.; Fiorina, E.; Pennazio, F.; Peroni, C.; Saletta, M.; Camarlinghi, N.; Fantacci, M.; and Cerello, P Large scale validation of the m5l lung cad on heterogeneous ct datasets. Medical physics 42(4): Murphy, K.; van Ginneken, B.; Schilham, A. M.; De Hoop, B.; Gietema, H.; and Prokop, M A large-scale evaluation of automatic pulmonary nodule detection in chest ct using local image features and k-nearest-neighbour classification. Medical image analysis 13(5): Ren, S.; He, K.; Girshick, R.; and Sun, J Faster r-cnn: Towards real-time object detection with region proposal networks. In NIPS, Ronneberger, O.; Fischer, P.; and Brox, T U-net: Convolutional networks for biomedical image segmentation. In MICCAI, Springer. Setio, A. A. A.; Ciompi, F.; Litjens, G.; Gerke, P.; Jacobs, C.; van Riel, S. J.; Wille, M. M. W.; Naqibullah, M.; Sánchez, C. I.; and van Ginneken, B Pulmonary nodule detection in ct images: false positive reduction using multi-view convolutional networks. IEEE transactions on medical imaging 35(5): Setio, A. A. A.; Traverso, A.; de Bel, T.; Berens, M. S.; van den Bogaard, C.; Cerello, P.; Chen, H.; Dou, Q.; Fantacci, M. E.; Geurts, B.; et al Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: the luna16 challenge. Medical Image Analysis 42:1 13. Shen, W.; Zhou, M.; Yang, F.; Yang, C.; and Tian, J Multiscale convolutional neural networks for lung nodule classification. In IPMI, Springer. Shen, W.; Zhou, M.; Yang, F.; Dong, D.; Yang, C.; Zang, Y.; and Tian, J Learning from experts: Developing transferable deep features for patient-level lung cancer prediction. In MICCAI, Springer. Suzuki, K.; Li, F.; Sone, S.; and Doi, K Computer-aided diagnostic scheme for distinction between benign and malignant nodules in thoracic low-dose ct by use of massive training artificial neural network. IEEE Transactions on Medical Imaging 24(9): Way, T. W.; Hadjiiski, L. M.; Sahiner, B.; Chan, H.-P.; Cascade, P. N.; Kazerooni, E. A.; Bogot, N.; and Zhou, C Computeraided diagnosis of pulmonary nodules on ct scans: Segmentation and classification using 3d active contours. Medical Physics 33(7): Yan, X.; Pang, J.; Qi, H.; Zhu, Y.; Bai, C.; Geng, X.; Liu, M.; Terzopoulos, D.; and Ding, X Classification of lung nodule malignancy risk on computed tomography images using convolutional neural network: A comparison between 2d and 3d strategies. In ACCV, Springer. Zeiler, M. D.; Krishnan, D.; Taylor, G. W.; and Fergus, R Deconvolutional networks. In CVPR, IEEE. Zhu, W., and Xie, X Adversarial deep structural networks for mammographic mass segmentation. arxiv preprint arxiv: Zhu, W.; Lou, Q.; Vang, Y. S.; and Xie, X Deep multiinstance networks with sparse label assignment for whole mammogram classification. In MICCAI, Springer.
arxiv: v3 [cs.cv] 26 May 2018
![arxiv: v3 [cs.cv] 26 May 2018 arxiv: v3 [cs.cv] 26 May 2018](/thumbs/86/94630916.jpg) DeepEM: Deep 3D ConvNets With EM For Weakly Supervised Pulmonary Nodule Detection Wentao Zhu, Yeeleng S. Vang, Yufang Huang, and Xiaohui Xie University of California, Irvine Lenovo AI Lab {wentaoz1,ysvang,xhx}@uci.edu,
DeepEM: Deep 3D ConvNets With EM For Weakly Supervised Pulmonary Nodule Detection Wentao Zhu, Yeeleng S. Vang, Yufang Huang, and Xiaohui Xie University of California, Irvine Lenovo AI Lab {wentaoz1,ysvang,xhx}@uci.edu,
arxiv: v3 [cs.cv] 28 Mar 2017
![arxiv: v3 [cs.cv] 28 Mar 2017 arxiv: v3 [cs.cv] 28 Mar 2017](/thumbs/73/69121210.jpg) Discovery Radiomics for Pathologically-Proven Computed Tomography Lung Cancer Prediction Devinder Kumar 1*, Audrey G. Chung 1, Mohammad J. Shaifee 1, Farzad Khalvati 2,3, Masoom A. Haider 2,3, and Alexander
Discovery Radiomics for Pathologically-Proven Computed Tomography Lung Cancer Prediction Devinder Kumar 1*, Audrey G. Chung 1, Mohammad J. Shaifee 1, Farzad Khalvati 2,3, Masoom A. Haider 2,3, and Alexander
arxiv: v2 [cs.cv] 3 Jun 2018
![arxiv: v2 [cs.cv] 3 Jun 2018 arxiv: v2 [cs.cv] 3 Jun 2018](/thumbs/89/97775247.jpg) S4ND: Single-Shot Single-Scale Lung Nodule Detection Naji Khosravan and Ulas Bagci Center for Research in Computer Vision (CRCV), School of Computer Science, University of Central Florida, Orlando, FL.
S4ND: Single-Shot Single-Scale Lung Nodule Detection Naji Khosravan and Ulas Bagci Center for Research in Computer Vision (CRCV), School of Computer Science, University of Central Florida, Orlando, FL.
DIAGNOSTIC CLASSIFICATION OF LUNG NODULES USING 3D NEURAL NETWORKS
 DIAGNOSTIC CLASSIFICATION OF LUNG NODULES USING 3D NEURAL NETWORKS Raunak Dey Zhongjie Lu Yi Hong Department of Computer Science, University of Georgia, Athens, GA, USA First Affiliated Hospital, School
DIAGNOSTIC CLASSIFICATION OF LUNG NODULES USING 3D NEURAL NETWORKS Raunak Dey Zhongjie Lu Yi Hong Department of Computer Science, University of Georgia, Athens, GA, USA First Affiliated Hospital, School
HHS Public Access Author manuscript Med Image Comput Comput Assist Interv. Author manuscript; available in PMC 2018 January 04.
 Discriminative Localization in CNNs for Weakly-Supervised Segmentation of Pulmonary Nodules Xinyang Feng 1, Jie Yang 1, Andrew F. Laine 1, and Elsa D. Angelini 1,2 1 Department of Biomedical Engineering,
Discriminative Localization in CNNs for Weakly-Supervised Segmentation of Pulmonary Nodules Xinyang Feng 1, Jie Yang 1, Andrew F. Laine 1, and Elsa D. Angelini 1,2 1 Department of Biomedical Engineering,
Copyright 2008 Society of Photo Optical Instrumentation Engineers. This paper was published in Proceedings of SPIE, vol. 6915, Medical Imaging 2008:
 Copyright 2008 Society of Photo Optical Instrumentation Engineers. This paper was published in Proceedings of SPIE, vol. 6915, Medical Imaging 2008: Computer Aided Diagnosis and is made available as an
Copyright 2008 Society of Photo Optical Instrumentation Engineers. This paper was published in Proceedings of SPIE, vol. 6915, Medical Imaging 2008: Computer Aided Diagnosis and is made available as an
arxiv: v2 [cs.cv] 8 Mar 2018
![arxiv: v2 [cs.cv] 8 Mar 2018 arxiv: v2 [cs.cv] 8 Mar 2018](/thumbs/87/97094636.jpg) Automated soft tissue lesion detection and segmentation in digital mammography using a u-net deep learning network Timothy de Moor a, Alejandro Rodriguez-Ruiz a, Albert Gubern Mérida a, Ritse Mann a, and
Automated soft tissue lesion detection and segmentation in digital mammography using a u-net deep learning network Timothy de Moor a, Alejandro Rodriguez-Ruiz a, Albert Gubern Mérida a, Ritse Mann a, and
Lung Nodule Segmentation Using 3D Convolutional Neural Networks
 Lung Nodule Segmentation Using 3D Convolutional Neural Networks Research paper Business Analytics Bernard Bronmans Master Business Analytics VU University, Amsterdam Evert Haasdijk Supervisor VU University,
Lung Nodule Segmentation Using 3D Convolutional Neural Networks Research paper Business Analytics Bernard Bronmans Master Business Analytics VU University, Amsterdam Evert Haasdijk Supervisor VU University,
arxiv: v1 [cs.cv] 2 Jul 2018
![arxiv: v1 [cs.cv] 2 Jul 2018 arxiv: v1 [cs.cv] 2 Jul 2018](/thumbs/87/96052165.jpg) A Pulmonary Nodule Detection Model Based on Progressive Resolution and Hierarchical Saliency Junjie Zhang 1, Yong Xia 1,2, and Yanning Zhang 1 1 Shaanxi Key Lab of Speech & Image Information Processing
A Pulmonary Nodule Detection Model Based on Progressive Resolution and Hierarchical Saliency Junjie Zhang 1, Yong Xia 1,2, and Yanning Zhang 1 1 Shaanxi Key Lab of Speech & Image Information Processing
arxiv: v2 [cs.cv] 7 Jun 2018
![arxiv: v2 [cs.cv] 7 Jun 2018 arxiv: v2 [cs.cv] 7 Jun 2018](/thumbs/79/79761281.jpg) Deep supervision with additional labels for retinal vessel segmentation task Yishuo Zhang and Albert C.S. Chung Lo Kwee-Seong Medical Image Analysis Laboratory, Department of Computer Science and Engineering,
Deep supervision with additional labels for retinal vessel segmentation task Yishuo Zhang and Albert C.S. Chung Lo Kwee-Seong Medical Image Analysis Laboratory, Department of Computer Science and Engineering,
arxiv: v1 [cs.cv] 17 Aug 2017
![arxiv: v1 [cs.cv] 17 Aug 2017 arxiv: v1 [cs.cv] 17 Aug 2017](/thumbs/78/78278805.jpg) Deep Learning for Medical Image Analysis Mina Rezaei, Haojin Yang, Christoph Meinel Hasso Plattner Institute, Prof.Dr.Helmert-Strae 2-3, 14482 Potsdam, Germany {mina.rezaei,haojin.yang,christoph.meinel}@hpi.de
Deep Learning for Medical Image Analysis Mina Rezaei, Haojin Yang, Christoph Meinel Hasso Plattner Institute, Prof.Dr.Helmert-Strae 2-3, 14482 Potsdam, Germany {mina.rezaei,haojin.yang,christoph.meinel}@hpi.de
Comparison of Two Approaches for Direct Food Calorie Estimation
 Comparison of Two Approaches for Direct Food Calorie Estimation Takumi Ege and Keiji Yanai Department of Informatics, The University of Electro-Communications, Tokyo 1-5-1 Chofugaoka, Chofu-shi, Tokyo
Comparison of Two Approaches for Direct Food Calorie Estimation Takumi Ege and Keiji Yanai Department of Informatics, The University of Electro-Communications, Tokyo 1-5-1 Chofugaoka, Chofu-shi, Tokyo
Copyright 2007 IEEE. Reprinted from 4th IEEE International Symposium on Biomedical Imaging: From Nano to Macro, April 2007.
 Copyright 27 IEEE. Reprinted from 4th IEEE International Symposium on Biomedical Imaging: From Nano to Macro, April 27. This material is posted here with permission of the IEEE. Such permission of the
Copyright 27 IEEE. Reprinted from 4th IEEE International Symposium on Biomedical Imaging: From Nano to Macro, April 27. This material is posted here with permission of the IEEE. Such permission of the
CSE Introduction to High-Perfomance Deep Learning ImageNet & VGG. Jihyung Kil
 CSE 5194.01 - Introduction to High-Perfomance Deep Learning ImageNet & VGG Jihyung Kil ImageNet Classification with Deep Convolutional Neural Networks Alex Krizhevsky, Ilya Sutskever, Geoffrey E. Hinton,
CSE 5194.01 - Introduction to High-Perfomance Deep Learning ImageNet & VGG Jihyung Kil ImageNet Classification with Deep Convolutional Neural Networks Alex Krizhevsky, Ilya Sutskever, Geoffrey E. Hinton,
Differentiating Tumor and Edema in Brain Magnetic Resonance Images Using a Convolutional Neural Network
 Original Article Differentiating Tumor and Edema in Brain Magnetic Resonance Images Using a Convolutional Neural Network Aida Allahverdi 1, Siavash Akbarzadeh 1, Alireza Khorrami Moghaddam 2, Armin Allahverdy
Original Article Differentiating Tumor and Edema in Brain Magnetic Resonance Images Using a Convolutional Neural Network Aida Allahverdi 1, Siavash Akbarzadeh 1, Alireza Khorrami Moghaddam 2, Armin Allahverdy
arxiv: v1 [cs.cv] 3 Apr 2018
![arxiv: v1 [cs.cv] 3 Apr 2018 arxiv: v1 [cs.cv] 3 Apr 2018](/thumbs/90/103707026.jpg) Towards whole-body CT Bone Segmentation André Klein 1,2, Jan Warszawski 2, Jens Hillengaß 3, Klaus H. Maier-Hein 1 arxiv:1804.00908v1 [cs.cv] 3 Apr 2018 1 Division of Medical Image Computing, Deutsches
Towards whole-body CT Bone Segmentation André Klein 1,2, Jan Warszawski 2, Jens Hillengaß 3, Klaus H. Maier-Hein 1 arxiv:1804.00908v1 [cs.cv] 3 Apr 2018 1 Division of Medical Image Computing, Deutsches
arxiv: v2 [cs.cv] 19 Dec 2017
![arxiv: v2 [cs.cv] 19 Dec 2017 arxiv: v2 [cs.cv] 19 Dec 2017](/thumbs/89/97557782.jpg) An Ensemble of Deep Convolutional Neural Networks for Alzheimer s Disease Detection and Classification arxiv:1712.01675v2 [cs.cv] 19 Dec 2017 Jyoti Islam Department of Computer Science Georgia State University
An Ensemble of Deep Convolutional Neural Networks for Alzheimer s Disease Detection and Classification arxiv:1712.01675v2 [cs.cv] 19 Dec 2017 Jyoti Islam Department of Computer Science Georgia State University
Y-Net: Joint Segmentation and Classification for Diagnosis of Breast Biopsy Images
 Y-Net: Joint Segmentation and Classification for Diagnosis of Breast Biopsy Images Sachin Mehta 1, Ezgi Mercan 1, Jamen Bartlett 2, Donald Weaver 2, Joann G. Elmore 1, and Linda Shapiro 1 1 University
Y-Net: Joint Segmentation and Classification for Diagnosis of Breast Biopsy Images Sachin Mehta 1, Ezgi Mercan 1, Jamen Bartlett 2, Donald Weaver 2, Joann G. Elmore 1, and Linda Shapiro 1 1 University
arxiv: v1 [cs.cv] 13 Jul 2018
![arxiv: v1 [cs.cv] 13 Jul 2018 arxiv: v1 [cs.cv] 13 Jul 2018](/thumbs/84/89699669.jpg) Multi-Scale Convolutional-Stack Aggregation for Robust White Matter Hyperintensities Segmentation Hongwei Li 1, Jianguo Zhang 3, Mark Muehlau 2, Jan Kirschke 2, and Bjoern Menze 1 arxiv:1807.05153v1 [cs.cv]
Multi-Scale Convolutional-Stack Aggregation for Robust White Matter Hyperintensities Segmentation Hongwei Li 1, Jianguo Zhang 3, Mark Muehlau 2, Jan Kirschke 2, and Bjoern Menze 1 arxiv:1807.05153v1 [cs.cv]
Early Detection of Lung Cancer
 Early Detection of Lung Cancer Aswathy N Iyer Dept Of Electronics And Communication Engineering Lymie Jose Dept Of Electronics And Communication Engineering Anumol Thomas Dept Of Electronics And Communication
Early Detection of Lung Cancer Aswathy N Iyer Dept Of Electronics And Communication Engineering Lymie Jose Dept Of Electronics And Communication Engineering Anumol Thomas Dept Of Electronics And Communication
Highly Accurate Brain Stroke Diagnostic System and Generative Lesion Model. Junghwan Cho, Ph.D. CAIDE Systems, Inc. Deep Learning R&D Team
 Highly Accurate Brain Stroke Diagnostic System and Generative Lesion Model Junghwan Cho, Ph.D. CAIDE Systems, Inc. Deep Learning R&D Team Established in September, 2016 at 110 Canal st. Lowell, MA 01852,
Highly Accurate Brain Stroke Diagnostic System and Generative Lesion Model Junghwan Cho, Ph.D. CAIDE Systems, Inc. Deep Learning R&D Team Established in September, 2016 at 110 Canal st. Lowell, MA 01852,
Segmentation of Cell Membrane and Nucleus by Improving Pix2pix
 Segmentation of Membrane and Nucleus by Improving Pix2pix Masaya Sato 1, Kazuhiro Hotta 1, Ayako Imanishi 2, Michiyuki Matsuda 2 and Kenta Terai 2 1 Meijo University, Siogamaguchi, Nagoya, Aichi, Japan
Segmentation of Membrane and Nucleus by Improving Pix2pix Masaya Sato 1, Kazuhiro Hotta 1, Ayako Imanishi 2, Michiyuki Matsuda 2 and Kenta Terai 2 1 Meijo University, Siogamaguchi, Nagoya, Aichi, Japan
Multi-attention Guided Activation Propagation in CNNs
 Multi-attention Guided Activation Propagation in CNNs Xiangteng He and Yuxin Peng (B) Institute of Computer Science and Technology, Peking University, Beijing, China pengyuxin@pku.edu.cn Abstract. CNNs
Multi-attention Guided Activation Propagation in CNNs Xiangteng He and Yuxin Peng (B) Institute of Computer Science and Technology, Peking University, Beijing, China pengyuxin@pku.edu.cn Abstract. CNNs
Predictive Data Mining for Lung Nodule Interpretation
 Predictive Data Mining for Lung Nodule Interpretation William Horsthemke, Ekarin Varutbangkul, Daniela Raicu, Jacob Furst DePaul University, Chicago, IL USA {whorsthe,evarutba}@students.depaul.edu, {draicu,
Predictive Data Mining for Lung Nodule Interpretation William Horsthemke, Ekarin Varutbangkul, Daniela Raicu, Jacob Furst DePaul University, Chicago, IL USA {whorsthe,evarutba}@students.depaul.edu, {draicu,
Lung Cancer Diagnosis from CT Images Using Fuzzy Inference System
 Lung Cancer Diagnosis from CT Images Using Fuzzy Inference System T.Manikandan 1, Dr. N. Bharathi 2 1 Associate Professor, Rajalakshmi Engineering College, Chennai-602 105 2 Professor, Velammal Engineering
Lung Cancer Diagnosis from CT Images Using Fuzzy Inference System T.Manikandan 1, Dr. N. Bharathi 2 1 Associate Professor, Rajalakshmi Engineering College, Chennai-602 105 2 Professor, Velammal Engineering
Dual Path Network and Its Applications
 Learning and Vision Group (NUS), ILSVRC 2017 - CLS-LOC & DET tasks Dual Path Network and Its Applications National University of Singapore: Yunpeng Chen, Jianan Li, Huaxin Xiao, Jianshu Li, Xuecheng Nie,
Learning and Vision Group (NUS), ILSVRC 2017 - CLS-LOC & DET tasks Dual Path Network and Its Applications National University of Singapore: Yunpeng Chen, Jianan Li, Huaxin Xiao, Jianshu Li, Xuecheng Nie,
arxiv: v1 [cs.cv] 21 Jul 2017
![arxiv: v1 [cs.cv] 21 Jul 2017 arxiv: v1 [cs.cv] 21 Jul 2017](/thumbs/89/100163556.jpg) A Multi-Scale CNN and Curriculum Learning Strategy for Mammogram Classification William Lotter 1,2, Greg Sorensen 2, and David Cox 1,2 1 Harvard University, Cambridge MA, USA 2 DeepHealth Inc., Cambridge
A Multi-Scale CNN and Curriculum Learning Strategy for Mammogram Classification William Lotter 1,2, Greg Sorensen 2, and David Cox 1,2 1 Harvard University, Cambridge MA, USA 2 DeepHealth Inc., Cambridge
Using Deep Convolutional Neural Networks to Predict Semantic Features of Lesions in Mammograms
 Using Deep Convolutional Neural Networks to Predict Semantic Features of Lesions in Mammograms Vibhu Agarwal Stanford University vibhua@stanford.edu Clayton Carson Stanford University carsoncz@stanford.edu
Using Deep Convolutional Neural Networks to Predict Semantic Features of Lesions in Mammograms Vibhu Agarwal Stanford University vibhua@stanford.edu Clayton Carson Stanford University carsoncz@stanford.edu
Automatic Prostate Cancer Classification using Deep Learning. Ida Arvidsson Centre for Mathematical Sciences, Lund University, Sweden
 Automatic Prostate Cancer Classification using Deep Learning Ida Arvidsson Centre for Mathematical Sciences, Lund University, Sweden Outline Autoencoders, theory Motivation, background and goal for prostate
Automatic Prostate Cancer Classification using Deep Learning Ida Arvidsson Centre for Mathematical Sciences, Lund University, Sweden Outline Autoencoders, theory Motivation, background and goal for prostate
Convolutional Neural Networks (CNN)
 Convolutional Neural Networks (CNN) Algorithm and Some Applications in Computer Vision Luo Hengliang Institute of Automation June 10, 2014 Luo Hengliang (Institute of Automation) Convolutional Neural Networks
Convolutional Neural Networks (CNN) Algorithm and Some Applications in Computer Vision Luo Hengliang Institute of Automation June 10, 2014 Luo Hengliang (Institute of Automation) Convolutional Neural Networks
Cervical cytology intelligent diagnosis based on object detection technology
 Cervical cytology intelligent diagnosis based on object detection technology Meiquan Xu xumeiquan@126.com Weixiu Zeng Semptian Co., Ltd. Machine Learning Lab. zengweixiu@gmail.com Hunhui Wu 736886978@qq.com
Cervical cytology intelligent diagnosis based on object detection technology Meiquan Xu xumeiquan@126.com Weixiu Zeng Semptian Co., Ltd. Machine Learning Lab. zengweixiu@gmail.com Hunhui Wu 736886978@qq.com
arxiv: v1 [cs.cv] 2 Jun 2017
![arxiv: v1 [cs.cv] 2 Jun 2017 arxiv: v1 [cs.cv] 2 Jun 2017](/thumbs/74/71415505.jpg) INTEGRATED DEEP AND SHALLOW NETWORKS FOR SALIENT OBJECT DETECTION Jing Zhang 1,2, Bo Li 1, Yuchao Dai 2, Fatih Porikli 2 and Mingyi He 1 1 School of Electronics and Information, Northwestern Polytechnical
INTEGRATED DEEP AND SHALLOW NETWORKS FOR SALIENT OBJECT DETECTION Jing Zhang 1,2, Bo Li 1, Yuchao Dai 2, Fatih Porikli 2 and Mingyi He 1 1 School of Electronics and Information, Northwestern Polytechnical
Age Estimation based on Multi-Region Convolutional Neural Network
 Age Estimation based on Multi-Region Convolutional Neural Network Ting Liu, Jun Wan, Tingzhao Yu, Zhen Lei, and Stan Z. Li 1 Center for Biometrics and Security Research & National Laboratory of Pattern
Age Estimation based on Multi-Region Convolutional Neural Network Ting Liu, Jun Wan, Tingzhao Yu, Zhen Lei, and Stan Z. Li 1 Center for Biometrics and Security Research & National Laboratory of Pattern
doi: /
 Yiting Xie ; Matthew D. Cham ; Claudia Henschke ; David Yankelevitz ; Anthony P. Reeves; Automated coronary artery calcification detection on low-dose chest CT images. Proc. SPIE 9035, Medical Imaging
Yiting Xie ; Matthew D. Cham ; Claudia Henschke ; David Yankelevitz ; Anthony P. Reeves; Automated coronary artery calcification detection on low-dose chest CT images. Proc. SPIE 9035, Medical Imaging
POC Brain Tumor Segmentation. vlife Use Case
 Brain Tumor Segmentation vlife Use Case 1 Automatic Brain Tumor Segmentation using CNN Background Brain tumor segmentation seeks to separate healthy tissue from tumorous regions such as the advancing tumor,
Brain Tumor Segmentation vlife Use Case 1 Automatic Brain Tumor Segmentation using CNN Background Brain tumor segmentation seeks to separate healthy tissue from tumorous regions such as the advancing tumor,
Computer-aided detection of pulmonary nodules: a comparative study using the public LIDC/IDRI database
 Eur Radiol (2016) 26:2139 2147 DOI 10.1007/s00330-015-4030-7 COMPUTER APPLICATIONS Computer-aided detection of pulmonary nodules: a comparative study using the public LIDC/IDRI database Colin Jacobs 1
Eur Radiol (2016) 26:2139 2147 DOI 10.1007/s00330-015-4030-7 COMPUTER APPLICATIONS Computer-aided detection of pulmonary nodules: a comparative study using the public LIDC/IDRI database Colin Jacobs 1
arxiv: v1 [cs.cv] 30 May 2018
![arxiv: v1 [cs.cv] 30 May 2018 arxiv: v1 [cs.cv] 30 May 2018](/thumbs/85/92577534.jpg) A Robust and Effective Approach Towards Accurate Metastasis Detection and pn-stage Classification in Breast Cancer Byungjae Lee and Kyunghyun Paeng Lunit inc., Seoul, South Korea {jaylee,khpaeng}@lunit.io
A Robust and Effective Approach Towards Accurate Metastasis Detection and pn-stage Classification in Breast Cancer Byungjae Lee and Kyunghyun Paeng Lunit inc., Seoul, South Korea {jaylee,khpaeng}@lunit.io
Computational modeling of visual attention and saliency in the Smart Playroom
 Computational modeling of visual attention and saliency in the Smart Playroom Andrew Jones Department of Computer Science, Brown University Abstract The two canonical modes of human visual attention bottomup
Computational modeling of visual attention and saliency in the Smart Playroom Andrew Jones Department of Computer Science, Brown University Abstract The two canonical modes of human visual attention bottomup
Improved Intelligent Classification Technique Based On Support Vector Machines
 Improved Intelligent Classification Technique Based On Support Vector Machines V.Vani Asst.Professor,Department of Computer Science,JJ College of Arts and Science,Pudukkottai. Abstract:An abnormal growth
Improved Intelligent Classification Technique Based On Support Vector Machines V.Vani Asst.Professor,Department of Computer Science,JJ College of Arts and Science,Pudukkottai. Abstract:An abnormal growth
Lung Cancer Detection using CT Scan Images
 Available online at www.sciencedirect.com ScienceDirect Procedia Computer Science 125 (2018) 107 114 6th International Conference on Smart Computing and Communications, ICSCC 2017, 7-8 December 2017, Kurukshetra,
Available online at www.sciencedirect.com ScienceDirect Procedia Computer Science 125 (2018) 107 114 6th International Conference on Smart Computing and Communications, ICSCC 2017, 7-8 December 2017, Kurukshetra,
Efficient Deep Model Selection
 Efficient Deep Model Selection Jose Alvarez Researcher Data61, CSIRO, Australia GTC, May 9 th 2017 www.josemalvarez.net conv1 conv2 conv3 conv4 conv5 conv6 conv7 conv8 softmax prediction???????? Num Classes
Efficient Deep Model Selection Jose Alvarez Researcher Data61, CSIRO, Australia GTC, May 9 th 2017 www.josemalvarez.net conv1 conv2 conv3 conv4 conv5 conv6 conv7 conv8 softmax prediction???????? Num Classes
arxiv: v1 [cs.cv] 9 Oct 2018
![arxiv: v1 [cs.cv] 9 Oct 2018 arxiv: v1 [cs.cv] 9 Oct 2018](/thumbs/95/126101195.jpg) Automatic Segmentation of Thoracic Aorta Segments in Low-Dose Chest CT Julia M. H. Noothout a, Bob D. de Vos a, Jelmer M. Wolterink a, Ivana Išgum a a Image Sciences Institute, University Medical Center
Automatic Segmentation of Thoracic Aorta Segments in Low-Dose Chest CT Julia M. H. Noothout a, Bob D. de Vos a, Jelmer M. Wolterink a, Ivana Išgum a a Image Sciences Institute, University Medical Center
Healthcare Research You
 DL for Healthcare Goals Healthcare Research You What are high impact problems in healthcare that deep learning can solve? What does research in AI applications to medical imaging look like? How can you
DL for Healthcare Goals Healthcare Research You What are high impact problems in healthcare that deep learning can solve? What does research in AI applications to medical imaging look like? How can you
Design of Palm Acupuncture Points Indicator
 Design of Palm Acupuncture Points Indicator Wen-Yuan Chen, Shih-Yen Huang and Jian-Shie Lin Abstract The acupuncture points are given acupuncture or acupressure so to stimulate the meridians on each corresponding
Design of Palm Acupuncture Points Indicator Wen-Yuan Chen, Shih-Yen Huang and Jian-Shie Lin Abstract The acupuncture points are given acupuncture or acupressure so to stimulate the meridians on each corresponding
3D Deep Learning for Multi-modal Imaging-Guided Survival Time Prediction of Brain Tumor Patients
 3D Deep Learning for Multi-modal Imaging-Guided Survival Time Prediction of Brain Tumor Patients Dong Nie 1,2, Han Zhang 1, Ehsan Adeli 1, Luyan Liu 1, and Dinggang Shen 1(B) 1 Department of Radiology
3D Deep Learning for Multi-modal Imaging-Guided Survival Time Prediction of Brain Tumor Patients Dong Nie 1,2, Han Zhang 1, Ehsan Adeli 1, Luyan Liu 1, and Dinggang Shen 1(B) 1 Department of Radiology
QUANTITATIVE LUNG NODULE ANALYSIS SYSTEM (NAS) FROM CHEST CT
 QUANTITATIVE LUNG NODULE ANALYSIS SYSTEM (NAS) FROM CHEST CT Amal Farag 1, PhD, Salwa Elshazly 1, Asem Ali 2, PhD, Islam Alkabbany 2, Albert Seow 2, MD and Aly Farag 2, PhD 1 Kentucky Imaging Technologies,
QUANTITATIVE LUNG NODULE ANALYSIS SYSTEM (NAS) FROM CHEST CT Amal Farag 1, PhD, Salwa Elshazly 1, Asem Ali 2, PhD, Islam Alkabbany 2, Albert Seow 2, MD and Aly Farag 2, PhD 1 Kentucky Imaging Technologies,
A Reliable Method for Brain Tumor Detection Using Cnn Technique
 IOSR Journal of Electrical and Electronics Engineering (IOSR-JEEE) e-issn: 2278-1676,p-ISSN: 2320-3331, PP 64-68 www.iosrjournals.org A Reliable Method for Brain Tumor Detection Using Cnn Technique Neethu
IOSR Journal of Electrical and Electronics Engineering (IOSR-JEEE) e-issn: 2278-1676,p-ISSN: 2320-3331, PP 64-68 www.iosrjournals.org A Reliable Method for Brain Tumor Detection Using Cnn Technique Neethu
Hierarchical Convolutional Features for Visual Tracking
 Hierarchical Convolutional Features for Visual Tracking Chao Ma Jia-Bin Huang Xiaokang Yang Ming-Husan Yang SJTU UIUC SJTU UC Merced ICCV 2015 Background Given the initial state (position and scale), estimate
Hierarchical Convolutional Features for Visual Tracking Chao Ma Jia-Bin Huang Xiaokang Yang Ming-Husan Yang SJTU UIUC SJTU UC Merced ICCV 2015 Background Given the initial state (position and scale), estimate
Computer aided detection of clusters of microcalcifications on full field digital mammograms
 Computer aided detection of clusters of microcalcifications on full field digital mammograms Jun Ge, a Berkman Sahiner, Lubomir M. Hadjiiski, Heang-Ping Chan, Jun Wei, Mark A. Helvie, and Chuan Zhou Department
Computer aided detection of clusters of microcalcifications on full field digital mammograms Jun Ge, a Berkman Sahiner, Lubomir M. Hadjiiski, Heang-Ping Chan, Jun Wei, Mark A. Helvie, and Chuan Zhou Department
LUNG CANCER continues to rank as the leading cause
 1138 IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 24, NO. 9, SEPTEMBER 2005 Computer-Aided Diagnostic Scheme for Distinction Between Benign and Malignant Nodules in Thoracic Low-Dose CT by Use of Massive
1138 IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 24, NO. 9, SEPTEMBER 2005 Computer-Aided Diagnostic Scheme for Distinction Between Benign and Malignant Nodules in Thoracic Low-Dose CT by Use of Massive
Automated detection of masses on whole breast volume ultrasound scanner: false positive reduction using deep convolutional neural network
 Automated detection of masses on whole breast volume ultrasound scanner: false positive reduction using deep convolutional neural network Yuya Hiramatsu a, Chisako Muramatsu* a, Hironobu Kobayashi b, Takeshi
Automated detection of masses on whole breast volume ultrasound scanner: false positive reduction using deep convolutional neural network Yuya Hiramatsu a, Chisako Muramatsu* a, Hironobu Kobayashi b, Takeshi
arxiv: v1 [stat.ml] 23 Jan 2017
![arxiv: v1 [stat.ml] 23 Jan 2017 arxiv: v1 [stat.ml] 23 Jan 2017](/thumbs/81/83414183.jpg) Learning what to look in chest X-rays with a recurrent visual attention model arxiv:1701.06452v1 [stat.ml] 23 Jan 2017 Petros-Pavlos Ypsilantis Department of Biomedical Engineering King s College London
Learning what to look in chest X-rays with a recurrent visual attention model arxiv:1701.06452v1 [stat.ml] 23 Jan 2017 Petros-Pavlos Ypsilantis Department of Biomedical Engineering King s College London
arxiv: v1 [cs.lg] 4 Feb 2019
![arxiv: v1 [cs.lg] 4 Feb 2019 arxiv: v1 [cs.lg] 4 Feb 2019](/thumbs/95/123488545.jpg) Machine Learning for Seizure Type Classification: Setting the benchmark Subhrajit Roy [000 0002 6072 5500], Umar Asif [0000 0001 5209 7084], Jianbin Tang [0000 0001 5440 0796], and Stefan Harrer [0000
Machine Learning for Seizure Type Classification: Setting the benchmark Subhrajit Roy [000 0002 6072 5500], Umar Asif [0000 0001 5209 7084], Jianbin Tang [0000 0001 5440 0796], and Stefan Harrer [0000
A CONVOLUTION NEURAL NETWORK ALGORITHM FOR BRAIN TUMOR IMAGE SEGMENTATION
 A CONVOLUTION NEURAL NETWORK ALGORITHM FOR BRAIN TUMOR IMAGE SEGMENTATION 1 Priya K, 2 Dr.O. Saraniya 1 PG Scholar, 2 Assistant Professor Department Of ECE Government College of Technology, Coimbatore,
A CONVOLUTION NEURAL NETWORK ALGORITHM FOR BRAIN TUMOR IMAGE SEGMENTATION 1 Priya K, 2 Dr.O. Saraniya 1 PG Scholar, 2 Assistant Professor Department Of ECE Government College of Technology, Coimbatore,
International Journal of Advances in Engineering Research. (IJAER) 2018, Vol. No. 15, Issue No. IV, April e-issn: , p-issn:
 SUPERVISED MACHINE LEARNING ALGORITHMS: DEVELOPING AN EFFECTIVE USABILITY OF COMPUTERIZED TOMOGRAPHY DATA IN THE EARLY DETECTION OF LUNG CANCER IN SMALL CELL Pushkar Garg Delhi Public School, R.K. Puram,
SUPERVISED MACHINE LEARNING ALGORITHMS: DEVELOPING AN EFFECTIVE USABILITY OF COMPUTERIZED TOMOGRAPHY DATA IN THE EARLY DETECTION OF LUNG CANCER IN SMALL CELL Pushkar Garg Delhi Public School, R.K. Puram,
arxiv: v1 [cs.cv] 2 Aug 2017
![arxiv: v1 [cs.cv] 2 Aug 2017 arxiv: v1 [cs.cv] 2 Aug 2017](/thumbs/76/73977344.jpg) Automatic 3D ardiovascular MR Segmentation with Densely-onnected Volumetric onvnets Lequan Yu 1, Jie-Zhi heng 2, Qi Dou 1, Xin Yang 1, Hao hen 1, Jing Qin 3, and Pheng-Ann Heng 1,4 arxiv:1708.00573v1 [cs.v]
Automatic 3D ardiovascular MR Segmentation with Densely-onnected Volumetric onvnets Lequan Yu 1, Jie-Zhi heng 2, Qi Dou 1, Xin Yang 1, Hao hen 1, Jing Qin 3, and Pheng-Ann Heng 1,4 arxiv:1708.00573v1 [cs.v]
Automatic Hemorrhage Classification System Based On Svm Classifier
 Automatic Hemorrhage Classification System Based On Svm Classifier Abstract - Brain hemorrhage is a bleeding in or around the brain which are caused by head trauma, high blood pressure and intracranial
Automatic Hemorrhage Classification System Based On Svm Classifier Abstract - Brain hemorrhage is a bleeding in or around the brain which are caused by head trauma, high blood pressure and intracranial
Firearm Detection in Social Media
 Erik Valldor and David Gustafsson Swedish Defence Research Agency (FOI) C4ISR, Sensor Informatics Linköping SWEDEN erik.valldor@foi.se david.gustafsson@foi.se ABSTRACT Manual analysis of large data sets
Erik Valldor and David Gustafsson Swedish Defence Research Agency (FOI) C4ISR, Sensor Informatics Linköping SWEDEN erik.valldor@foi.se david.gustafsson@foi.se ABSTRACT Manual analysis of large data sets
Automatic Detection of Knee Joints and Quantification of Knee Osteoarthritis Severity using Convolutional Neural Networks
 Automatic Detection of Knee Joints and Quantification of Knee Osteoarthritis Severity using Convolutional Neural Networks Joseph Antony 1, Kevin McGuinness 1, Kieran Moran 1,2 and Noel E O Connor 1 Insight
Automatic Detection of Knee Joints and Quantification of Knee Osteoarthritis Severity using Convolutional Neural Networks Joseph Antony 1, Kevin McGuinness 1, Kieran Moran 1,2 and Noel E O Connor 1 Insight
LUNG NODULE SEGMENTATION IN COMPUTED TOMOGRAPHY IMAGE. Hemahashiny, Ketheesan Department of Physical Science, Vavuniya Campus
 LUNG NODULE SEGMENTATION IN COMPUTED TOMOGRAPHY IMAGE Hemahashiny, Ketheesan Department of Physical Science, Vavuniya Campus tketheesan@vau.jfn.ac.lk ABSTRACT: The key process to detect the Lung cancer
LUNG NODULE SEGMENTATION IN COMPUTED TOMOGRAPHY IMAGE Hemahashiny, Ketheesan Department of Physical Science, Vavuniya Campus tketheesan@vau.jfn.ac.lk ABSTRACT: The key process to detect the Lung cancer
ITERATIVELY TRAINING CLASSIFIERS FOR CIRCULATING TUMOR CELL DETECTION
 ITERATIVELY TRAINING CLASSIFIERS FOR CIRCULATING TUMOR CELL DETECTION Yunxiang Mao 1, Zhaozheng Yin 1, Joseph M. Schober 2 1 Missouri University of Science and Technology 2 Southern Illinois University
ITERATIVELY TRAINING CLASSIFIERS FOR CIRCULATING TUMOR CELL DETECTION Yunxiang Mao 1, Zhaozheng Yin 1, Joseph M. Schober 2 1 Missouri University of Science and Technology 2 Southern Illinois University
Skin cancer reorganization and classification with deep neural network
 Skin cancer reorganization and classification with deep neural network Hao Chang 1 1. Department of Genetics, Yale University School of Medicine 2. Email: changhao86@gmail.com Abstract As one kind of skin
Skin cancer reorganization and classification with deep neural network Hao Chang 1 1. Department of Genetics, Yale University School of Medicine 2. Email: changhao86@gmail.com Abstract As one kind of skin
COMPUTERIZED SYSTEM DESIGN FOR THE DETECTION AND DIAGNOSIS OF LUNG NODULES IN CT IMAGES 1
 ISSN 258-8739 3 st August 28, Volume 3, Issue 2, JSEIS, CAOMEI Copyright 26-28 COMPUTERIZED SYSTEM DESIGN FOR THE DETECTION AND DIAGNOSIS OF LUNG NODULES IN CT IMAGES ALI ABDRHMAN UKASHA, 2 EMHMED SAAID
ISSN 258-8739 3 st August 28, Volume 3, Issue 2, JSEIS, CAOMEI Copyright 26-28 COMPUTERIZED SYSTEM DESIGN FOR THE DETECTION AND DIAGNOSIS OF LUNG NODULES IN CT IMAGES ALI ABDRHMAN UKASHA, 2 EMHMED SAAID
Automatic Classification of Breast Masses for Diagnosis of Breast Cancer in Digital Mammograms using Neural Network
 IJSTE - International Journal of Science Technology & Engineering Volume 1 Issue 11 May 2015 ISSN (online): 2349-784X Automatic Classification of Breast Masses for Diagnosis of Breast Cancer in Digital
IJSTE - International Journal of Science Technology & Engineering Volume 1 Issue 11 May 2015 ISSN (online): 2349-784X Automatic Classification of Breast Masses for Diagnosis of Breast Cancer in Digital
Mammogram Analysis: Tumor Classification
 Mammogram Analysis: Tumor Classification Term Project Report Geethapriya Raghavan geeragh@mail.utexas.edu EE 381K - Multidimensional Digital Signal Processing Spring 2005 Abstract Breast cancer is the
Mammogram Analysis: Tumor Classification Term Project Report Geethapriya Raghavan geeragh@mail.utexas.edu EE 381K - Multidimensional Digital Signal Processing Spring 2005 Abstract Breast cancer is the
arxiv: v1 [cs.cv] 24 Jul 2018
![arxiv: v1 [cs.cv] 24 Jul 2018 arxiv: v1 [cs.cv] 24 Jul 2018](/thumbs/82/86822918.jpg) Multi-Class Lesion Diagnosis with Pixel-wise Classification Network Manu Goyal 1, Jiahua Ng 2, and Moi Hoon Yap 1 1 Visual Computing Lab, Manchester Metropolitan University, M1 5GD, UK 2 University of
Multi-Class Lesion Diagnosis with Pixel-wise Classification Network Manu Goyal 1, Jiahua Ng 2, and Moi Hoon Yap 1 1 Visual Computing Lab, Manchester Metropolitan University, M1 5GD, UK 2 University of
Medical Image Analysis
 Medical Image Analysis 1 Co-trained convolutional neural networks for automated detection of prostate cancer in multiparametric MRI, 2017, Medical Image Analysis 2 Graph-based prostate extraction in t2-weighted
Medical Image Analysis 1 Co-trained convolutional neural networks for automated detection of prostate cancer in multiparametric MRI, 2017, Medical Image Analysis 2 Graph-based prostate extraction in t2-weighted
Big Image-Omics Data Analytics for Clinical Outcome Prediction
 Big Image-Omics Data Analytics for Clinical Outcome Prediction Junzhou Huang, Ph.D. Associate Professor Dept. Computer Science & Engineering University of Texas at Arlington Dept. CSE, UT Arlington Scalable
Big Image-Omics Data Analytics for Clinical Outcome Prediction Junzhou Huang, Ph.D. Associate Professor Dept. Computer Science & Engineering University of Texas at Arlington Dept. CSE, UT Arlington Scalable
Synthesizing Missing PET from MRI with Cycle-consistent Generative Adversarial Networks for Alzheimer s Disease Diagnosis
 Synthesizing Missing PET from MRI with Cycle-consistent Generative Adversarial Networks for Alzheimer s Disease Diagnosis Yongsheng Pan 1,2, Mingxia Liu 2, Chunfeng Lian 2, Tao Zhou 2,YongXia 1(B), and
Synthesizing Missing PET from MRI with Cycle-consistent Generative Adversarial Networks for Alzheimer s Disease Diagnosis Yongsheng Pan 1,2, Mingxia Liu 2, Chunfeng Lian 2, Tao Zhou 2,YongXia 1(B), and
Network Dissection: Quantifying Interpretability of Deep Visual Representation
 Name: Pingchuan Ma Student number: 3526400 Date: August 19, 2018 Seminar: Explainable Machine Learning Lecturer: PD Dr. Ullrich Köthe SS 2018 Quantifying Interpretability of Deep Visual Representation
Name: Pingchuan Ma Student number: 3526400 Date: August 19, 2018 Seminar: Explainable Machine Learning Lecturer: PD Dr. Ullrich Köthe SS 2018 Quantifying Interpretability of Deep Visual Representation
Improved Detection of Lung Nodule Overlapping with Ribs by using Virtual Dual Energy Radiology
 Improved Detection of Lung Nodule Overlapping with Ribs by using Virtual Dual Energy Radiology G.Maria Dhayana Latha Associate Professor, Department of Electronics and Communication Engineering, Excel
Improved Detection of Lung Nodule Overlapping with Ribs by using Virtual Dual Energy Radiology G.Maria Dhayana Latha Associate Professor, Department of Electronics and Communication Engineering, Excel
A comparative study of machine learning methods for lung diseases diagnosis by computerized digital imaging'"
 A comparative study of machine learning methods for lung diseases diagnosis by computerized digital imaging'" Suk Ho Kang**. Youngjoo Lee*** Aostract I\.' New Work to be 1 Introduction Presented U Mater~al
A comparative study of machine learning methods for lung diseases diagnosis by computerized digital imaging'" Suk Ho Kang**. Youngjoo Lee*** Aostract I\.' New Work to be 1 Introduction Presented U Mater~al
Convolutional Neural Networks for Estimating Left Ventricular Volume
 Convolutional Neural Networks for Estimating Left Ventricular Volume Ryan Silva Stanford University rdsilva@stanford.edu Maksim Korolev Stanford University mkorolev@stanford.edu Abstract End-systolic and
Convolutional Neural Networks for Estimating Left Ventricular Volume Ryan Silva Stanford University rdsilva@stanford.edu Maksim Korolev Stanford University mkorolev@stanford.edu Abstract End-systolic and
An improved U-Net architecture for simultaneous. arteriole and venule segmentation in fundus image
 MIUA2018, 042, v3 (final): An improved U-Net architecture for simultaneous arteriole... 1 An improved U-Net architecture for simultaneous arteriole and venule segmentation in fundus image Xiayu Xu 1,2*,
MIUA2018, 042, v3 (final): An improved U-Net architecture for simultaneous arteriole... 1 An improved U-Net architecture for simultaneous arteriole and venule segmentation in fundus image Xiayu Xu 1,2*,
Automated Mass Detection from Mammograms using Deep Learning and Random Forest
 Automated Mass Detection from Mammograms using Deep Learning and Random Forest Neeraj Dhungel 1 Gustavo Carneiro 1 Andrew P. Bradley 2 1 ACVT, University of Adelaide, Australia 2 University of Queensland,
Automated Mass Detection from Mammograms using Deep Learning and Random Forest Neeraj Dhungel 1 Gustavo Carneiro 1 Andrew P. Bradley 2 1 ACVT, University of Adelaide, Australia 2 University of Queensland,
Tumor Cellularity Assessment. Rene Bidart
 Tumor Cellularity Assessment Rene Bidart 1 Goals Find location and class of all cell nuclei: Lymphocyte Cells Normal Epithelial Cells Malignant Epithelial Cells Why: Test effectiveness of pre-surgery treatment
Tumor Cellularity Assessment Rene Bidart 1 Goals Find location and class of all cell nuclei: Lymphocyte Cells Normal Epithelial Cells Malignant Epithelial Cells Why: Test effectiveness of pre-surgery treatment
B657: Final Project Report Holistically-Nested Edge Detection
 B657: Final roject Report Holistically-Nested Edge Detection Mingze Xu & Hanfei Mei May 4, 2016 Abstract Holistically-Nested Edge Detection (HED), which is a novel edge detection method based on fully
B657: Final roject Report Holistically-Nested Edge Detection Mingze Xu & Hanfei Mei May 4, 2016 Abstract Holistically-Nested Edge Detection (HED), which is a novel edge detection method based on fully
Lung Region Segmentation using Artificial Neural Network Hopfield Model for Cancer Diagnosis in Thorax CT Images
 Automation, Control and Intelligent Systems 2015; 3(2): 19-25 Published online March 20, 2015 (http://www.sciencepublishinggroup.com/j/acis) doi: 10.11648/j.acis.20150302.12 ISSN: 2328-5583 (Print); ISSN:
Automation, Control and Intelligent Systems 2015; 3(2): 19-25 Published online March 20, 2015 (http://www.sciencepublishinggroup.com/j/acis) doi: 10.11648/j.acis.20150302.12 ISSN: 2328-5583 (Print); ISSN:
A REVIEW ON CLASSIFICATION OF BREAST CANCER DETECTION USING COMBINATION OF THE FEATURE EXTRACTION MODELS. Aeronautical Engineering. Hyderabad. India.
 Volume 116 No. 21 2017, 203-208 ISSN: 1311-8080 (printed version); ISSN: 1314-3395 (on-line version) url: http://www.ijpam.eu A REVIEW ON CLASSIFICATION OF BREAST CANCER DETECTION USING COMBINATION OF
Volume 116 No. 21 2017, 203-208 ISSN: 1311-8080 (printed version); ISSN: 1314-3395 (on-line version) url: http://www.ijpam.eu A REVIEW ON CLASSIFICATION OF BREAST CANCER DETECTION USING COMBINATION OF
arxiv: v1 [cs.cv] 1 Oct 2018
![arxiv: v1 [cs.cv] 1 Oct 2018 arxiv: v1 [cs.cv] 1 Oct 2018](/thumbs/87/95687522.jpg) Augmented Mitotic Cell Count using Field Of Interest Proposal Marc Aubreville 1, Christof A. Bertram 2, Robert Klopfleisch 2, Andreas Maier 1 arxiv:1810.00850v1 [cs.cv] 1 Oct 2018 1 Pattern Recognition
Augmented Mitotic Cell Count using Field Of Interest Proposal Marc Aubreville 1, Christof A. Bertram 2, Robert Klopfleisch 2, Andreas Maier 1 arxiv:1810.00850v1 [cs.cv] 1 Oct 2018 1 Pattern Recognition
Fast lung nodule detection in chest CT images using cylindrical nodule-enhancement filter
 Int J CARS (2013) 8:193 205 DOI 10.1007/s11548-012-0767-5 ORIGINAL ARTICLE Fast lung nodule detection in chest CT images using cylindrical nodule-enhancement filter Atsushi Teramoto Hiroshi Fujita Received:
Int J CARS (2013) 8:193 205 DOI 10.1007/s11548-012-0767-5 ORIGINAL ARTICLE Fast lung nodule detection in chest CT images using cylindrical nodule-enhancement filter Atsushi Teramoto Hiroshi Fujita Received:
arxiv: v1 [cs.cv] 31 Jul 2017
![arxiv: v1 [cs.cv] 31 Jul 2017 arxiv: v1 [cs.cv] 31 Jul 2017](/thumbs/85/92902149.jpg) Synthesis of Positron Emission Tomography (PET) Images via Multi-channel Generative Adversarial Networks (GANs) Lei Bi 1, Jinman Kim 1, Ashnil Kumar 1, Dagan Feng 1,2, and Michael Fulham 1,3,4 1 School
Synthesis of Positron Emission Tomography (PET) Images via Multi-channel Generative Adversarial Networks (GANs) Lei Bi 1, Jinman Kim 1, Ashnil Kumar 1, Dagan Feng 1,2, and Michael Fulham 1,3,4 1 School
A Deep Learning Approach for Breast Cancer Mass Detection
 A Deep Learning Approach for Breast Cancer Mass Detection Wael E.Fathy 1, Amr S. Ghoneim 2 Teaching Assistant 1, Assistant Professor 2 Department of Computer Science, Faculty of Computers and Information
A Deep Learning Approach for Breast Cancer Mass Detection Wael E.Fathy 1, Amr S. Ghoneim 2 Teaching Assistant 1, Assistant Professor 2 Department of Computer Science, Faculty of Computers and Information
Deep Supervision for Pancreatic Cyst Segmentation in Abdominal CT Scans
 Deep Supervision for Pancreatic Cyst Segmentation in Abdominal CT Scans Yuyin Zhou 1, Lingxi Xie 2( ), Elliot K. Fishman 3, Alan L. Yuille 4 1,2,4 The Johns Hopkins University, Baltimore, MD 21218, USA
Deep Supervision for Pancreatic Cyst Segmentation in Abdominal CT Scans Yuyin Zhou 1, Lingxi Xie 2( ), Elliot K. Fishman 3, Alan L. Yuille 4 1,2,4 The Johns Hopkins University, Baltimore, MD 21218, USA
Deep-Learning Based Semantic Labeling for 2D Mammography & Comparison of Complexity for Machine Learning Tasks
 Deep-Learning Based Semantic Labeling for 2D Mammography & Comparison of Complexity for Machine Learning Tasks Paul H. Yi, MD, Abigail Lin, BSE, Jinchi Wei, BSE, Haris I. Sair, MD, Ferdinand K. Hui, MD,
Deep-Learning Based Semantic Labeling for 2D Mammography & Comparison of Complexity for Machine Learning Tasks Paul H. Yi, MD, Abigail Lin, BSE, Jinchi Wei, BSE, Haris I. Sair, MD, Ferdinand K. Hui, MD,
arxiv: v1 [cs.cv] 18 Dec 2016
![arxiv: v1 [cs.cv] 18 Dec 2016 arxiv: v1 [cs.cv] 18 Dec 2016](/thumbs/82/85669475.jpg) Deep Multi-instance Networks with Sparse Label Assignment for Whole Mammogram Classification Wentao Zhu, Qi Lou, Yeeleng Scott Vang, and Xiaohui Xie {wentaoz1, xhx}@ics.uci.edu, {qlou, ysvang}@uci.edu
Deep Multi-instance Networks with Sparse Label Assignment for Whole Mammogram Classification Wentao Zhu, Qi Lou, Yeeleng Scott Vang, and Xiaohui Xie {wentaoz1, xhx}@ics.uci.edu, {qlou, ysvang}@uci.edu
arxiv: v1 [cs.cv] 28 Dec 2018
![arxiv: v1 [cs.cv] 28 Dec 2018 arxiv: v1 [cs.cv] 28 Dec 2018](/thumbs/93/111330693.jpg) Salient Object Detection via High-to-Low Hierarchical Context Aggregation Yun Liu 1 Yu Qiu 1 Le Zhang 2 JiaWang Bian 1 Guang-Yu Nie 3 Ming-Ming Cheng 1 1 Nankai University 2 A*STAR 3 Beijing Institute
Salient Object Detection via High-to-Low Hierarchical Context Aggregation Yun Liu 1 Yu Qiu 1 Le Zhang 2 JiaWang Bian 1 Guang-Yu Nie 3 Ming-Ming Cheng 1 1 Nankai University 2 A*STAR 3 Beijing Institute
arxiv: v2 [cs.cv] 22 Mar 2018
![arxiv: v2 [cs.cv] 22 Mar 2018 arxiv: v2 [cs.cv] 22 Mar 2018](/thumbs/94/122062874.jpg) Deep saliency: What is learnt by a deep network about saliency? Sen He 1 Nicolas Pugeault 1 arxiv:1801.04261v2 [cs.cv] 22 Mar 2018 Abstract Deep convolutional neural networks have achieved impressive performance
Deep saliency: What is learnt by a deep network about saliency? Sen He 1 Nicolas Pugeault 1 arxiv:1801.04261v2 [cs.cv] 22 Mar 2018 Abstract Deep convolutional neural networks have achieved impressive performance
ACCELERATING EMPHYSEMA DIAGNOSIS ON LUNG CT IMAGES USING EMPHYSEMA PRE-DETECTION METHOD
 ACCELERATING EMPHYSEMA DIAGNOSIS ON LUNG CT IMAGES USING EMPHYSEMA PRE-DETECTION METHOD 1 KHAIRUL MUZZAMMIL BIN SAIPULLAH, 2 DEOK-HWAN KIM, 3 NURUL ATIQAH ISMAIL 1 Lecturer, 3 Student, Faculty of Electronic
ACCELERATING EMPHYSEMA DIAGNOSIS ON LUNG CT IMAGES USING EMPHYSEMA PRE-DETECTION METHOD 1 KHAIRUL MUZZAMMIL BIN SAIPULLAH, 2 DEOK-HWAN KIM, 3 NURUL ATIQAH ISMAIL 1 Lecturer, 3 Student, Faculty of Electronic
Copyright 2008 Society of Photo Optical Instrumentation Engineers. This paper was published in Proceedings of SPIE, vol. 6915, Medical Imaging 2008:
 Copyright 28 Society of Photo Optical Instrumentation Engineers. This paper was published in Proceedings of SPIE, vol. 695, Medical Imaging 28: Computer Aided Diagnosis and is made available as an electronic
Copyright 28 Society of Photo Optical Instrumentation Engineers. This paper was published in Proceedings of SPIE, vol. 695, Medical Imaging 28: Computer Aided Diagnosis and is made available as an electronic
Classification of breast cancer histology images using transfer learning
 Classification of breast cancer histology images using transfer learning Sulaiman Vesal 1 ( ), Nishant Ravikumar 1, AmirAbbas Davari 1, Stephan Ellmann 2, Andreas Maier 1 1 Pattern Recognition Lab, Friedrich-Alexander-Universität
Classification of breast cancer histology images using transfer learning Sulaiman Vesal 1 ( ), Nishant Ravikumar 1, AmirAbbas Davari 1, Stephan Ellmann 2, Andreas Maier 1 1 Pattern Recognition Lab, Friedrich-Alexander-Universität
NMF-Density: NMF-Based Breast Density Classifier
 NMF-Density: NMF-Based Breast Density Classifier Lahouari Ghouti and Abdullah H. Owaidh King Fahd University of Petroleum and Minerals - Department of Information and Computer Science. KFUPM Box 1128.
NMF-Density: NMF-Based Breast Density Classifier Lahouari Ghouti and Abdullah H. Owaidh King Fahd University of Petroleum and Minerals - Department of Information and Computer Science. KFUPM Box 1128.
Early Diagnosis of Autism Disease by Multi-channel CNNs
 Early Diagnosis of Autism Disease by Multi-channel CNNs Guannan Li 1,2, Mingxia Liu 2, Quansen Sun 1(&), Dinggang Shen 2(&), and Li Wang 2(&) 1 School of Computer Science and Engineering, Nanjing University
Early Diagnosis of Autism Disease by Multi-channel CNNs Guannan Li 1,2, Mingxia Liu 2, Quansen Sun 1(&), Dinggang Shen 2(&), and Li Wang 2(&) 1 School of Computer Science and Engineering, Nanjing University
Investigating the performance of a CAD x scheme for mammography in specific BIRADS categories
 Investigating the performance of a CAD x scheme for mammography in specific BIRADS categories Andreadis I., Nikita K. Department of Electrical and Computer Engineering National Technical University of
Investigating the performance of a CAD x scheme for mammography in specific BIRADS categories Andreadis I., Nikita K. Department of Electrical and Computer Engineering National Technical University of
Multi-Scale Salient Object Detection with Pyramid Spatial Pooling
 Multi-Scale Salient Object Detection with Pyramid Spatial Pooling Jing Zhang, Yuchao Dai, Fatih Porikli and Mingyi He School of Electronics and Information, Northwestern Polytechnical University, China.
Multi-Scale Salient Object Detection with Pyramid Spatial Pooling Jing Zhang, Yuchao Dai, Fatih Porikli and Mingyi He School of Electronics and Information, Northwestern Polytechnical University, China.
Deep Learning of Brain Lesion Patterns for Predicting Future Disease Activity in Patients with Early Symptoms of Multiple Sclerosis
 Deep Learning of Brain Lesion Patterns for Predicting Future Disease Activity in Patients with Early Symptoms of Multiple Sclerosis Youngjin Yoo 1,2,5(B),LisaW.Tang 2,3,5, Tom Brosch 1,2,5,DavidK.B.Li
Deep Learning of Brain Lesion Patterns for Predicting Future Disease Activity in Patients with Early Symptoms of Multiple Sclerosis Youngjin Yoo 1,2,5(B),LisaW.Tang 2,3,5, Tom Brosch 1,2,5,DavidK.B.Li
LUNG NODULE DETECTION SYSTEM
 LUNG NODULE DETECTION SYSTEM Kalim Bhandare and Rupali Nikhare Department of Computer Engineering Pillai Institute of Technology, New Panvel, Navi Mumbai, India ABSTRACT: The Existing approach consist
LUNG NODULE DETECTION SYSTEM Kalim Bhandare and Rupali Nikhare Department of Computer Engineering Pillai Institute of Technology, New Panvel, Navi Mumbai, India ABSTRACT: The Existing approach consist
Automated diagnosis of pneumothorax using an ensemble of convolutional neural networks with multi-sized chest radiography images
 Automated diagnosis of pneumothorax using an ensemble of convolutional neural networks with multi-sized chest radiography images Tae Joon Jun, Dohyeun Kim, and Daeyoung Kim School of Computing, KAIST,
Automated diagnosis of pneumothorax using an ensemble of convolutional neural networks with multi-sized chest radiography images Tae Joon Jun, Dohyeun Kim, and Daeyoung Kim School of Computing, KAIST,
DeepMiner: Discovering Interpretable Representations for Mammogram Classification and Explanation
 DeepMiner: Discovering Interpretable Representations for Mammogram Classification and Explanation Jimmy Wu 1, Bolei Zhou 1, Diondra Peck 2, Scott Hsieh 3, Vandana Dialani, MD 4 Lester Mackey 5, and Genevieve
DeepMiner: Discovering Interpretable Representations for Mammogram Classification and Explanation Jimmy Wu 1, Bolei Zhou 1, Diondra Peck 2, Scott Hsieh 3, Vandana Dialani, MD 4 Lester Mackey 5, and Genevieve
