Nature Immunology: doi: /ni.3837
|
|
|
- Phyllis Townsend
- 5 years ago
- Views:
Transcription
1
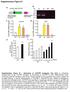
2 Supplementary Figure 1 T FR cell responses. (A) B6 mice were infected with PR8 and serial cryosections from the mln on day 3 were stained with anti- B22 (blue), anti-foxp3 (green) and anti-cd35 (red) or with anti-igd (green), anti-foxp3 (purple) and anti- CD35 (red). Stained cryosections were analyzed by fluorescence microscopy. White lines define the border of the B-cell follicle. Images are representative of three independent experiments. (B) B6 mice were infected with PR8 and serial cryosections from the mln on day 3 were stained with anti-b22 (blue), anti-foxp3 (red) and anti-cd4 (green). Stained cryosections were analyzed by fluorescence microscopy. Images are representative of two independent experiments. (C) B6 mice were infected with PR8 and cryosections of the mln were stained with anti-b22 (blue), anti-foxp3 (green) and anti-cd35 (red) on days 1 and 3. White lines define the border of the B cell follicle. Images are representative of three independent experiments. (D-E) B6 mice were immunized with 5 μg of hemaglutinin (HA) adsorbed to alum, and the frequency (D) and number (E) of Bcl- 6 hi CXCR5 hi T FR cells were calculated in the mln at the indicated time points. Representative plots were gated on FoxP3 + CD69 hi CD19 - CD4 + T cells. Data are representative of two independent experiments (mean SD of 4-5 mice per group). *P <.5, **P <.1, ***P <.1. P values were determined using a two-tailed Student s t-test. (F-G) B6 mice were infected with LCMV-Armstrong and the frequency (F) and number (G) of Bcl- 6 hi CXCR5 hi T FR cells were calculated in the mln at the indicated time-points. Representative plots were gated on FoxP3 + CD69 hi CD19 - CD4 + T cells. Data are representative of two independent experiments (mean SD of 5 mice per group). *P <.5, **P <.1, ***P <.1. P values were determined using a two-tailed Student s t- test.
3
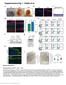
4 Supplementary Figure 2 CD25 lo Foxp3 + cells fail to up-regulate T FR cell markers at the peak of the infection. (A-C) B6 mice were infected with PR8 and CD69 hi FoxP3 + CD4 + T cells from the mln were analyzed for expression of CD25, Bcl-6, CXCR5 and PD-1 on day 1 (A) and day 3 (B) after infection by flow cytometry. Data are representative of five independent experiments. Data are shown as the mean ± SD (n=4-5 mice). (C) B6 mice were infected with PR8 and the frequency of Bcl-6 hi CXCR5 hi T FR cells within the CD25 lo CD69 hi FoxP3 + CD4 + T cell population were calculated by flow cytometry at the indicated timepoints. Data are shown as the mean ± SD (n=4-5 mice/time point). Data are representative of three independent experiments. (D-E). B6 mice were infected with LCMV (D) or immunized with HA adsorbed to alum (E) and the frequency of CD25 hi and CD25 lo FoxP3 + CD69 hi CD4 + T cells from the mlns with a Bcl-6 hi CXCR5 hi phenotype is shown. Data are representative of two independent experiments (mean SD of 5 mice per group). (F) GSEA showing enrichment in the IL-2 STAT5 Hallmark-signaling pathway in T reg cells vs T FR cells (adjusted p-value <.5, Log2fold change greater than or equal to 1). Three replicates for each cell type were obtained from three independent experiments.
5 CD44 A/PR8 HA-Alum Figure S % ± GFP/IL % ± Supplementary Figure 3 IL-2 production in PR8-infected and HA-alum immunized mice. Il21-mCherry-Il2-emGFP mice were infected with PR8 (left panel) or immunized with HA adsorbed to alum (right panel) and cells from the mln were analyzed on day 1. The frequency of GFP/IL-2 + cells within the CD4 + T-cell population is shown. Data are shown as the mean ± SD (n=3-5 mice per group). Data are representative of two independent experiments.
6 PD1 NPT FH cells (x1 3 ) FigureS4 a YFP/Cre B6 Bcl-6 f f / Foxp % 38.4% ±9.1 ± CXCR5 B6 Bcl-6 f f / b Foxp3 YFP/Cre Supplementary Figure 4 NP-specific T FH cell response in T FR cell-deficient mice (A-B) B6 and Bcl-6 fl/fl Foxp3 YFP/Cre mice were infected with PR8 and cells from the mln were analyzed by flow cytometry on day 3. The frequency (A) and number (B) of NP-specific CD4 + T cells with a CXCR5 hi PD-1 hi T FH cell phenotype were determined. Data are shown as the mean ± SD (n=3-5 mice). Data are representative of three independent experiments.
7 Bcl6 NP + T FH cells (x1 3 ) CD95 GC B cells (x1 4 ) ASC cells (x1 3 ) T FR cells (x1 3 ) Bcl6 T FH cells (x1 4 ) Bcl-6 T FH cells (x1 3 ) CD38 GC B cells (x1 3 ) Figure S5 a b Foxp3-WT Foxp3-Cxcr5 -/- 2.8% ± CXCR5 2.8% ± Foxp3 WT Foxp3 Cxcr5 -/- c Foxp3-WT Foxp3-Cxcr5 -/- 6.7% ± CD95 7.7% ± d Foxp3 WT Foxp3 Cxcr5 -/- e Foxp3 WT - Foxp3 Cxcr5 -/- + Foxp3 WT * + Foxp3 Cxcr5 -/- DTx f * Control ril-2 g Control %± %± CXCR5 +ril h Control +ril-2 i Control 33.7% ± CXCR5 +ril % ± j Control +ril-2 k Control %± %± GL-7 +ril-2 l Control +ril-2 Supplementary Figure 5 T FH cell and B cell responses in T FR cell-depleted mice (A-E) -/- mice were irradiated and reconstituted with a 5:5 mix of BM from CD FoxP3-DTR and CD B6 donors (FoxP3-B6) or from CD FoxP3-DTR and CD Cxcr5 -/- donors (FoxP3- Cxcr5 -/- ). Reconstituted FoxP3-B6 and FoxP3-Cxcr5 -/- chimeras were infected with PR8, treated with PBS or DT every four days starting 2 days after infection, and cells from the mln were analyzed on day 5. Frequency (A) and number (B) of Bcl6 hi CXCR5 hi T FH cells. Representative plots were gated on FoxP3 - CD19 - CD4 + T cells. Frequency (c) and number (D) of CD19 + CD138 - GL-7 + CD38 lo CD95 hi GC B cells. Number (E) of CD138 + ASCs. Data are shown as the mean ± SD (n=3-5 mice). Data are representative of three independent experiments. P values were determined using a two-tailed Student s t-test. (F-L) B6 mice were infected with
8 PR8 and treated daily with 15, U of ril-2 or PBS starting 2 days after infection. Cells from the mln were analyzed by flow cytometry on day 3. Number (F) of T FR cells in PBS and ril-2 treated mice. Frequency (G) and number (H) of Bcl-6 hi CXCR5 hi T FH cells. Representative plots were gated on FoxP3 - CD19 - CD4 + T cells. Frequency (I) and number (J) of NP-specific CD4 + T cells with a Bcl-6 hi CXCR5 hi T FH cell phenotype. Representative plots were gated on FoxP3 - CD19 - NP + CD4 + T cells. The frequency (K) and number (L) of CD19 + CD138 - GL-7 + CD95 hi GC B cells. Data are shown as the mean ± SD (n=4-5 mice). Data are representative of four independent experiments. P values were determined using a two-tailed Student s t-test.
9 Figure S6 B6 Control Infected naive 1 m Bcl-6 f f / Foxp3 YFP/Cre Infected naive Supplementary Figure 6 Influenza-infected Bcl-6 fl/fl Foxp3 YFP/Cre mice develop self-reactive ASC responses. The presence of ANAs in the serum from naïve and age-matched day 3 influenza-infected B6 (top panels) and Bcl6 fl/fl Foxp3 YFP/Cre mice (bottom panels) was determined by fluorescence microscopy using HEp-2 slides. Images are representative of two independent experiments (n=4-5 mice/group).
10 Supplementary Table 1. Differentially expressed genes in Tfr cells vs conventional Treg cells. Genes down-regulated in Tfr cells vs conventional Treg cells Socs2 Tph1 Aldh1a2 Nckap5 Stra6 Dkk3 Kcnj15 Gpr34 Col1a2 Il1rl1 Ighv9-4 Tcrg-C4 Il2ra Prkar1b Plekhd1 Ltb4r1 Fam19a3 Ptger3 Dnah7b Itgae Dnah7a Cish Ighv1-82 Gbp2b Rorc AI Dyrk4 Col23a1 Peli2 Ccl22 Drc1 Lad1 Gm4956 A6391E8Rik Genes up-regulated in Tfr cells vs conventional Treg cell H13Rik Chpt1 Fbln1 Mtmr11 Irak1bp1 Ube2n Tox Ska1 Cbx6 Dynlt1a Ube2v1 St3gal1 Slc24a3 Commd1 Nup62 Trav14-3 Spcs1 Eid2b Themis Orai3 Tor2a 63343K7Rik Trp53i13 Malt1 Zfp839 Trbv23 Gm5617 Rnls Nudt14 Zdhhc1 Trav14n-3 Zfp358 Lnx2 Tspan2
11 Selp Abhd12b Fgl2 Ptrf Fbln2 Lcn1 Gzmb Bend6 Slc22a2 Mmp9 Ryr2 Apol7c Tcrg-C1 Ccr2 Fsd1l Lars2 Cecr6 L1cam Col15a1 Gata1 Medag Gm4951 Src Ahr Tbc1d16 Ntn4 Rasip1 Gm15564 Pla2g4f Ighv1-26 Tmem154 Sorcs2 Gzmc Neb Itga2 Cpe Ncf1 Lrrc32 Gbp1 Ccr7 Gpld1 Tlr7 Gm26771 Depdc1b Cyfip1 1118F13Rik Pgk1 Fyn Ehd3 Cyp4f16 Dclre1c Fam11b Rhbdd3 Slc16a1 Hist1h2ap Arl2 Efnb2 Spc24 Cdca3 Gm1576 Lmna Stk39 Btla Praf2 Ctsb Bdh1 Otub2 Bik Stxbp1 Map2k3 Ift57 Snx1 Kif18b St6galnac6 Fut8 Aplp1 Fam221b Gapdh S1a6 Tnfsf14 Eea1 Ptger2 Cdk2ap2 Stard5 Dusp14 Pard6g
12 Maml1 Tcf7l1 St8sia6 Gm5431 Pros1 Treml2 Dennd5a L13Rik Scn5a Car12 Sytl4 Havcr2 Ppm1l Foxp3 Il7r Tdpoz4 Actn1 Gm47 Ahnak Gvin1 Specc1 Myo3b 11132F4Rik Gm626 Cd86 Gm E21Rik Gm2663 She Tgfbr1 Cx3cl L14Rik Ctla2a Scin Fggy Arhgap2 Rasgef1b Irak3 Fscn1 Fam12b Coro2a Tnfsf15 Pde8a Zfyve21 Sestd1 Gucy2g Slc16a5 Pstk Ggt7 Kif22 Myo1h Tmem18 Ccdc74a Sqrdl Gabarapl1 Hsf2bp Syce2 1717B5Rik Churc1 Cstb Cmc2 Ptpn5 Alad Alyref2 Srfbp1 1747I17Rik2 Hist1h1b Ppp1r14b Trav16 Itfg3 Maf Fgfr1op 11165P2Rik Arnt2 Gpsm2 Ccnb2 Rab31 Tom1 Bcl7c Slco3a1 Car5b Kras Trav4-4-dv1 Mthfs Synj2bp Amigo1
13 Spef2 Ecm1 Gm14446 Pkd1l3 159L16Rik Rxra Sapcd1 Acsbg1 Rragd Entpd1 Abcg3 Adamts6 Lfng Cd226 Adam12 Cpd Dpy19l1 Ankrd11 Gm26583 Gpr83 Gm113 Gfpt2 Slco5a1 Gm17641 Gm26561 Klrg1 Tle1 Chst2 C1qtnf6 Rara Pik3r5 Ccr6 Il17rb Tmem64 Atp6vd2 Dgkh Nod1 Dpt Maoa Smad7 Mgat4a Slc3a2 Dennd2c 24166E13Rik Egr2 Klk1b27 Tmem176b Nab1 Csnk1e Rras Lime1 Krt222 Epdr1 St3gal5 Dynlt1f Bspry Trav8-2 Pfkp Ift43 Pfn2 2117G23Rik Cdk1 Pex11a Aurka Ptpn11 Higd1a Pkm Ypel1 Asf1a Ccnb1 Oaz2 1118P14Rik D17H6S56E-5 Stom Tmem O7Rik Luzp1 Ccdc22 Pbk P2rx4 Gm194 Hes6 Pif1 Fam11a Elmo3 H2-Aa
14 Aqp3 Palm3 Ift8 Tcam1 Prkce Dclk2 Lama5 Larp1 Gpr56 Ptger B19Rik Zbtb18 Col2a1 Hcn3 Dmd Capn3 Gm4759 Dscam Dusp5 Klf2 Mical3_2 Itpka Adam19 Rras2 Itga5 P2rx5 Wdr66 Vprbp Pparg Notch2 Bank1 Gm17354 Nox1 Fnip2 Egr3 Tnfsf1 A6333H2Rik Camk2n1 Scml4 Ager Gpr132 Itga6 Qser1 Rab11a 23145N1Rik Tm7sf2 Ube2e3 Pptc7 Rpp25l Trav19 Tspan17 Myo1f Tpmt Efemp2 Adat3 Stat4 RP23-45G16.5 St3gal3 Lpp Gpd2 Tnfsf9 Errfi1 Pfkfb4 Cdc2 Syne2 Mapre2 Numbl A631G21Rik Zfyve28 Syde1 Trerf1 Txlnb Mafg_1 Fam19a Trp53inp2 Il23r BC3544 Tmem29_1 Tmem256 Slc25a1 Uap1l1 Grn Ttc39c Susd3 Ccdc57 Cdca8
15 Akt3 Wnt3 Gm17231 Ppp3ca Cysltr1 Mical3_1 Utp2 Tatdn2 Satb1 Nacad Ky Kbtbd11 Arhgef18 Serpina3h Gbp7 Gbp6 Il18r1 Sytl2 Cib2 Fam46a Mpst F8316B8Rik Mettl21d Bach2 Adnp Dennd4a Kynu Znrf3 Swap7 Furin Cd8a Grin2d Gm1225 Gm9875 Stk24 Sdc4 Scg5 Fam129a Dapk1 Frmd5 Hbegf Gm9938 Nckap1 Cisd1 Thtpa Fam89b Lamp1 Sapcd2 Tstd3 Trib3 Pfkl H2afx Trav16n Cdkn3 Lgalsl Crip2 Echdc2 Map2 Tgm2 Kif3a Gm5918 Trav7d-4 Hsd11b1 Ctsz Rgs11 Dennd3 Cd16 Camk2b Gm16894 Map1lc3a Dleu2 Peli3 Tmem16a Timp2 Susd4 Pgam1 Rab39b Edn3 Fibcd1 Serf1 Rab2 Il9r Plk1 Gas2l1 Ankrd9 Matk
16 Taf4b Atp13a3 Hopx Atp6va1 Ksr2 Sardh Abca1 Slc4a1 Magi1 Cyp4f39 Gpx3 Ust Vav2 Gbp J21Rik2 Sun2 1725G4Rik Npm3 Cpsf7 Jag2 Sytl1 Mcoln3 Il18rap Htt Kndc1 Rai14 Hs6st1 Zfyve9 Stk1 Rell2 Lclat1 Kctd12b Gm12185 Agpat4 Naip6 Atg4c Uhrf1bp1 Ncoa7 Gltscr1l Syp Plcd1 A4378G23Rik Sell Repin1 Crat Prss41 Oscp1 Grhpr Gsn Tmem3b Hmces Pkig Batf Tmem86a Ufsp1 Rhbdl3 Trim2 Ctsh D835J1Rik Plcxd1 Grcc1 Fuom 213C2Rik Slc35e4 Gss Plxnb2 Npdc1 Spock2 Hmgb3 Casp1 Tmem38a Arvcf Spsb1 Ms4a6d Trim8 Blk Gstt2 Eaf2 Dclk1 2119I3Rik Lgmn Trf Dus2l Cystm1 Fam188b Perp
17 Esyt1 Ighg3 Rbm38 Sorl1 Abca2 Gm13212 Ahcyl2 Mir17hg Rab21 Grap2 Sp6 Bcam Rgs1 Dip2c Armcx4 Rasgrp2 Hlf Atr Nr6a1 Smad4 Crls1 Sdc3 Rapgef6 Igf2r Slc17a9 Rasl11b Chrnb2 Plekhg1 Gm168 Gm8898 Tns1 Pygm Slc7a5 Slfn8 Thra B9341F14Rik_1 Sema6d Rims3 Vegfc Pik3r1 Vim Zfp652 Itpr3 Ccdc M7Rik Ube2s Sepn1 Bnip3 Cacna1s Trbv12-1 Dnajc12 Tspan5 Susd1 Mnd1 Ripk4 Morn2 Ccdc12a Ly86 Ikzf3 H1f App Acpl2 Zan Camk1 Trav6d-5 Snn Renbp Cass4 Cited2 Gm14167 Cpt1c Wdfy1 Mettl1 H2-Ab1 Jazf1 Lyz2 Rgcc Id1 Cc2d2a Emid1 Pank1 Ube2c Rnaset2b Ptges3l Myh J18Rik
18 Tfap A18Rik Camk2g Ifngr1 Gpr55 Prkd3 Ctla2b Zdhhc23 Ranbp2 Zscan2 Fnip1 Atp8a2 Hoxb4 Gm H17Rik Nfix Gm8995 Thy1 Atp11a Nin Ctsw Dync1h1 Stab1 Zc3h12c Svil Nfya Gbp3 Macf1 Gm26917 Dpysl2 Arl5a Dock9 Arhgap22 Rasgrf2 Abca5 Mfhas1 Pfas Sec31b Alpk1 Npas2 BC16423 Mdn1 Dyx1c1 Morn1 Tspan4 Rapgef5 Sap F4Rik Enpp6 Tnfsf13b Rnaset2a Tmem38b Akap5 Rbm4b Cxcr6 Gna13 Zc2hc1c Try1 RP24-289J12.1 Bace2 Ngly1 Gmfg Slc22a15 Tubg2 Aldoa Pla2g2d Reep2 Ccdc8 Fbxo44 Ier5l Tmem17 Csf1r Cdkn2c Tmie Desi1 Ly6c2 Gpr19 Mettl7a1 Dock4 Pim2 Psg16 Susd2 Man1c1 Hdhd3 Vamp5 Cd4lg
19 Kif1b Cyp2d22 Rhoq Rcc2 Rnf17 Lta Samd9l Mgst3 Vav3 Sirt1 Thrb Mga Dmxl1 Traf6 Selplg Klc2 Gucy2e Col11a2 Nlrc5 Crim1 Prickle1 Rcan3 Clstn1 Cd96 Gbp4 Trim16 Sbf2 Galnt1 Tmprss O19Rik Plcxd2 Foxk1 Ptch1 Atm Sgk3 Gm1355 Plxnd1 Kif21b Lphn1 Gm999 Ldlr Lipe Cd79b Tg Plxdc1 Ect2l Dennd2d Raf1 Ift81 Tmem25 Hspa2 Snx8 Nphp1 Fam132b Gm12728 Pbx3 Carhsp1 Csgalnact1 Gm15283 Vmn2r96 Gpm6b Arl6 Ly96 Acyp2 Trav21-dv12 Tceal3 Marcksl1 Dhrs1 Tmem141 Fer Farp2 Ggt5 Abhd4 Trav13n-3 Rasgef1a Nme4 Card1 Fgd1 Nmnat2 Rnase6 Ttc16 Cdh23 Ppfibp2 Ppfia4 Eif3j2 Lyn
20 Prrc2a Arl4c Pcsk4 Pdpr Heatr1 Ikzf4 Dgat2 Wdr78 Sacs Dock1 Tex15 Pglyrp2 Ung Nlrc3 Lilrb4 Zfp442 Atxn2 Gsto1 Prg4 Actr3b Il4ra Axin1 Slk Bcl9l Sfn Ifi23 Prdm1 Sav1 Ighg2b BC337 F13a1 Dst Golm1 Mprip Ccr5 1719G17Rik Pmepa1 Igfbp4 Clip2 Nedd4l Sprtn Plec Alpl Pop7 Tnfaip8_1 Lacc1 Tiam2 Opn3 Ramp3 Spo11 C538M17Rik B3gnt9 Smco4 Col5a3 Tsc22d1 Lzts3 Cox7a1 Hif1a Fam212a Cdkn1a C1qtnf4 Fam69c Cox6b2 Cetn4 Atp5l-ps1 Fah Hc Selm Nav2 Bub3 Ces2d-ps Sec16b Pdlim7 Fgf2 Ppic Kctd11 Pard3b Bcl2a1d Gm1617 Serp2 Rsph1 Tcf4 Hpcal1 Cyb561 Alpk2 Pxmp2
21 Cad Gan Akr1c12 Notch1 Trim13 Anxa6 Serpinc1 S1pr4 Adcy6 Fam11b Zfp658 Syne3 Ada Rbpj Itgam H2-Q4 Spin2c Fancf Adap1 Myc Pdp2 Fam129c Piezo1 Chdh Pcnx Huwe1 Pag1 Pds5a Hif3a Sipa1l3 Gm16976 Tacc2 Sppl3 Osm Ubr5 Tnrc6c Ly6g5b BC1857 Mink1 Xxylt1 Mettl2 Skil Hist2h4 Rap1gap Gstt3 Zfp3 Igj Morn4 Msl3l2 S1a11 Il1r2 Itga7 Celf3 Fndc7 Itgb5 Ago4 Rhbdf1 Ell3 Gm3839_1 Plekho1 Ddc Ctnnbip1 Dgki Cdk5r1 Ap3s1 Col1a1 Fam217b S1a1 Lmo4 Kdelr3 Cxcl3 Appl2 Elovl7 Sh2b2 Gm C22Rik Zscan1 Clu Nt5dc D16Rik Tm6sf1 Ccdc136 Cd29b Syde2 Rnf32 Enpp2
22 Mansc1 Crebl2 Klhl3 Iqgap2 Serpina3g Nlgn2 Ptprv Myh9 Ap1s3 Fam65b Txnrd3 Rnf28 Ptprs Rmnd5a Marf1 Lypd6b Slc35g1 Axin2 BC H11Rik Camkk1 Bcl2l1 Dvl1 Tnfrsf25 Rb1 Gbp8 Gm16157 Dnah8 Apol9a Naip5 Gm4955 Sema4b Cd97 Lrrc8c Slc12a6 Tdrkh Phc3 Vps54 Rnf213 Rnasel Trim24 Birc6 Pelp1 Ccdc141 Stfa3 Gm1612 Cyp39a1 Sh2d1a Naprt1 Gadd45a Pak6 Cttnbp2nl Epb4.1l5 Oaf Acta2 Liph Cenpv Fhl2 Fam213b Cdc25b Fam131a Msrb3 Tyrobp Tpi1 Rnf157 Wdr54 Il1rl2 Slc16a3 Pcbd1 C1ra Cstad Cacna1d Prx Gm11998 Chac2 Slc7a1 Ptprf A932I21Rik Slc9a9 Ltbp4 Clip3 Hhex Igf2bp3 Klrd1 Fam2c Nsun7
23 Frmd6 Pgpep1l Plekhn1 Ctdsp1 Zfp526 Lamb3 Arrdc4 Igf1r Itga4 Foxk2 Them6 Pikfyve Serpini1 Stx1a Tet2 Acot6 Tcf2 AY36118 Vps13b Usp18 Pla2g4b Marveld1 Pan3 Slamf1 Dfna5 Gdf11 Nek6 Nr1d1 Pdlim1 Arrb1 Dbp Ston1 Cd4 Plagl2 Usp9x Klf16 Ifi44 Pik3c2a Paqr8 C1336L24Rik Pip5k1b Naf1 Slc35d1 Npnt Slc7a7 Sgip1 Pld4 Nr2f6 Fmn1 Cd3lf Dync2li1 Rnf144a Trav6n-5 Prf1 Maged1 Cd9 Srcrb4d Dap Gpr62 Marco Fam135a Plekhs1 Asph Trpm6 Dnm3 Fam83g Lancl E2Rik Srgap3 B23217C12Rik_2 Gstt1 Cela1 Rab33a Bmp1 Cd29a Optn Pnpla3 Spsb2 Smox Etv4 Tmem15a Nhsl1 Epas1 Scel Slc43a1 Lrrn2
24 Tsc22d3 Marcks Kif13a Ifngr2 Anks1 Gm2897 S1pr1 Nup21 Atic Hgf Hivep1 KCTD12 Rtn4rl1 Nrarp LRRC8A Pa2g4 Fam117a Usp24 Cep192 Il1 Atxn1 Itpripl1 Slc39a8 Usp32 N4bp2 Smg1 Dyrk1a Adcy3 F7343M19Rik Fam22a_2 Ms4a4c Calcrl Zdhhc18 Nbeal2 Zbtb8a Otud4 Ago1 Atp2b4 Ncoa J21Rik1 Ssh1 Setx Ski Tmod F21Rik Ces2c Hes5 Ier3 Intu Kcnq4 Gm1975 Nrbp2 Fam171b Mef2c Ccdc28b Nanos1 1819J6Rik Ly6d Gm21987 S1pr2 Nfe2 Gna14 Bcl6 Fam43a Pla2g7 1719L3Rik Myl12b Napsa Pnck Cables1 Ltbr Stx11 Galnt9 Nfia Tmem26 Hap1 Evi2a Aldoc Lamb1 Myl12a Egln3 Slpi Dixdc1 Metrnl Atp6ve2 Srxn1
25 Wdtc1 Npc1 B4336N3Rik Hemk1 Ccdc88c Foxp4 Psme4 Padi2 Tub Pus7 Xcr1 Rab35 Sptbn1 Inpp4b Rreb1 Mmp11 Was Zfp28 Sik1 Psd Rarg Dnase1 1614C1Rik Wdr82 Bcr Lrp12 Tnfrsf1b Pydc4 Ppp2cb F2Rik Parp14 Fyco1 Usp31 Kif13b 26157B11Rik Itih5 Zfp69 Abca7 Rasgrp1 Zcchc11 Mib1 Ifrd2 Fam126a Mboat2 Cxcr5 Slc26a11 Tshz2 Palld Cap2 Rnf128 Ltk Fam2a Gm11454 Rnd3 Ehf Dmwd Arc St3gal6 Abhd15 Adamts3 Acy3 Nudt17 Tmem82 Arl4d Baiap3 Adra2a Itgax Arhgef25 Disc1 B2328H11Rik Asb2 Tmcc3 Slc16a11 Mnda Coro2b Muc D3Rik Dpep2 Pdcd1 Klk1 Ptrh1 Clec12a Dll1 Gm2663 Coprs Mcf2l
26 Smurf2 Zcchc24 Dennd1b Irf6 Tet1 A235M16Rik Bcor Herc2 Pstpip2 Akt1 Aak1 Airn Igsf9 Kpnb1 Acvrl1 Gm3411 Cytip Atrn Sos1 Ankrd5 Sox12 Ncoa1 Tdgf1 Fam78a Heca Arhgap24 Iigp1 Camk1d Plcb2 Ahctf1 Inf2 Tlk1 Proz Zfyve26 Wdr6 Fbrsl1 Alg1b Snx9 Epg5 Ppp1r13b Ccbl2 Taok1 Mapkbp1 Col16a1 Vat1l Atp2b2 H2-Q3 Gm5559 Sccpdh Ldlrad3 Dram1 Gm14718 Gab2 Ifi24 Efna4 Fam184a Ccl3 Cend1 171O22Rik Syt5 Anxa2 Vipr2 Gzma Ecel1 Nat14 Ccdc162 Rsph9 Olfml3 Spi1 BC22687 Plek H2-Q2 Slc37a2 Zeb2 Cd68 Atp6v1g3 Cyp1a1 Cebpa Vegfa Neurl2 Mzb1 Ddah2 Slamf7 Tubb6 Pltp Cdo1
27 Xpo4 Sec24a Slc24a5 Pprc1 Simc1 Mlkl Slfn1 Mybbp1a Slc3a1 Dnmt3b Gabbr1 Prdm11 Slc2a3 Amigo2 Phf11c Plp1 Gphn Ncoa2 C1galt1 Nbas Ppp1r12a Prkdc Slc2a4rg-ps Trio Acaca Zfp B9Rik Etohi1 Ttc7 Pelo Mettl13 Amer1 Pde1b A831M2Rik Rfwd2 Ephx4 Abcb1a Acvr2a Kntc1 P2ry1 Mfsd2a Rdx Vcl Ctxn1 Zfp768 Gm26512 Rasl11a Zcchc12 Gfra1 Iglc2 Ndrg4 Tubb2a Sox4 Faim3 Pipox Ablim2 Il1rn Serpinf1 Krt83 Hes1 Igkc Glis3_1 Gstm5 2315B5Rik Ntf5 Ispd Nr4a2 Ak4 Spry4 Pde3b Adck3 Car2 Klk1b9 Sox5 Jdp2 Phactr3 Cxcl13 Gdpd1 Tuba8 Siglech Atp1b1 Lypd1 Smtn Abcc3 Sort1 Coch
28 Vcam1 Man2b2 Asxl1 Nup188 Mycbp2 Hist1h2bc Wash Lrrk2 Etnk1 Erdr1 Styx Akap1 Agap2 Vps13d_1 Kmt2d Hipk2 Ints2 Tbx21 Pofut1 Helz2 Atg9b Zfp72 Fry Tgfb1 Slc45a4 Trim45 Gigyf1 Ccl1 Cdv3 Rrad Zfp68 Crmp1 Lmtk2 Pitpnm2 Armcx1 Akap13 Zcchc3 Urb1 Tbc1d3 Txnip Igtp BC51226 Mettl1 Ddx25 Hmox1 C334L19Rik Gm12498 Jup Ntrk3 Tnfrsf21 Tmc3 Samd3 Serpina9 Adam33 Cnga1 Lgals3 Ttyh1 Gm16754 Ces2g Pla1a Lamc3 Vdr N7Rik Ppp1r3d Cd24a Rassf6 Nedd4 Plekhg3 Vit D18Rik Dok M11Rik Dbh S1pr3 Chi3l1 Ltbp A3Rik Fam84a Tmem121 Tnfsf4 Serpine2 E5311L22Rik Gm1375 Cd19 Cpne7 Haao
29 Mllt6 Bcorl1 Zfml Arhgap31 Kmt2a Adam8 Rc3h2 Ifit2 Nt5dc3 Chd8 Ptger1 Tep1 Myo19 Gcn1l1 Brwd3 Slc22a5 Relt Mndal Rttn Ipo7 Arid3a Hsf2 Aldh2 Il12rb1 Rprd2 Cul9 Ankrd44 Dopey1 Prr E14Rik Bag6 Ccdc6 Ifit3 Ubr4 Btg3 Zfp493 Zfp955a Zhx2 Epn1 Ehbp1 Aoc2 Spg11 Mxd1 A931A2Rik Spint1 A9312O16Rik Adamtsl2 Art2a-ps Cyp4f18 Micall2 Fbn2 Cldn1 Blnk Fsd1 Prss3 Ryr1 Lpar3 Fam11a Gpr153 Popdc2 Gm684 Scimp Sstr3 Krt17 Ccdc67 Dio2 Akr1b8 Pawr Il13ra1 Dusp18 Mcoln2 Fads2 Tns4 Tubb2b Gm26686 Rtn4r Slc2a6 Cd22 Klhl P16Rik Ovol1 Nodal Rhov Slc1a4 Sox8 Slc6a9
30 Pip4k2b Ttll3 Ddit4 Znf512b Socs3 Asb7 Gm158 Cdk12 Snrnp2 Trim25 Hdgfrp3 Fasn Polr1b E19Rik Trim33 Nxpe3 Rnf169 B4galt5 Emp3 Gpr183 Slc35e2 Ralgapb Fam22a_1 Blm Itpkb Ttc39b Rrp12 Rad54l2 Hipk1 St8sia4 Sidt1 Ly6c1 Cd11 Snx25 Espn Map4k5 Braf Mdfic G7Rik Mknk2 Tbrg4 Tnpo1 Adss Pgam2 Myo1b Zfp651 Akap6 Tox2 Sytl3 Klrb1c Qrfp N21Rik Tgfb3 Gm1173 Spp1 Smim3 Ica1l Ccdc12 Reep1 Adssl1 Fgf18 Mpzl2 Dapk2 Grtp1 Maats1 Eps8l2 Smim1 Eomes Slc17a7 Ptprg Chn1 Tnc Kirrel3 Cebpd Cilp2 Syt6 Pou2af1 Klrb1a Kctd15 Plxnb3 Cxxc5 Rgs8 Ctsl Cpeb1 Prss5 Gpr35
31 Dhcr24 Zfp646 Slc29a2 Gtf2a1 Ppp1r37 Mex3c Ppp1r9b Taf15 Il4i1 Pole Rai1 Lats1 Nfrkb Ints1 Slfn3 Zfp275 Tmbim6 Ass1 Nr1d2 Zfp316 Oas2 Cnot1 Dqx1 Pdzk1ip1 Olfr56 Irgm2 Btaf1 Ly6k Dtx1 Setd1a Ppp1r12b Ddx6 Tiam1 Aida Irf4 BC3336 Kcnip2 Myo9b Samhd1 Fam6a Atxn7l1 Prpf8 Atxn1l Dnase1l3 Mtus1 Ighg2c Efna3 Ptprk Xcl1 Heyl Ptpn14 Nrgn 217H6Rik Cp Crtam Apoe Cxcl17 Proser2 Igf2bp2 Gstm1 Cbfa2t3 Bcar1 Vash2 Mgat3 Fst Col4a6 Calcb Mt3 Il4 Lrp1 Rnf39 Slc3a3 Sdc1 Il1r1 Tas1r1 Slc2a1 Mgarp Ppp1r3g Fam71b Lrrc52 Stx1b Lox Gm1374 Apoc L23Rik Klra17
32 Itgb7 Ar Nup25 Begain Il2rb Car11 Pi4ka Flna Nle1 D355H7Rik Cluh Chd3 Sptan1 Zfp22 Cds2 Magi3 Sec16a Apol1b Rfx7 Slc5a6 Med12 Rrp1b Fam12c Bcat1 Agap3 Xpo1 Inpp5f Kcnab2 Fktn Zfp827 Mfsd4 Socs1 Zfp937 Scrib Atp7a Zbtb37 Fchsd2 Fli1 Fam3c Bmp2k E339J7Rik Zfp445 Arfgef2 Mgst1 Ghrh Slc44a5 Ppp2r2c Spink4 Stc2 Kcnq1 P4ha2 Sbk2 Mycl Gzmk Padi4 Gm26975 Rgs6 Cacna1g Il21 Rtn4rl2 C3 Dpysl5 Atp1a3 Vangl1 Ascl2 Lipm Sostdc1 Vwde Klhl3 Fabp7 Gm9885 Gfra4 Gm1253 Scnn1g Alox12 Fam124a Mei1 Irg1 Apoc4 Gm12968 Olr1 Igkv3-2 BC18473 Gal Scnn1b
33 BC3734_1 Acss1 Atad2 Ifit1 Gm26733 Ercc2 Gm2458 Zfp292 Lemd3 Slc18a2 Ppp3cb Slfn5 Itgb3 Trip12 Daam1 Kpna4 Sesn3 Dicer1 Ttc37 Gucy1b3 BC94916 Ppp2r5a Chsy1 C336D17Rik Mdc1 A838D1Rik RP23-32A8.1 Apol9b Nacc1 Smad3 Usp3 Kdm3b Trib2 Zfx Klf11 Zfp449 Ttll4 Utf1 Fbxl17 Mgat5 Dopey2 Ube2cbp D1Ertd622e
34 Supplemental Table 2. Genes up-regulated by STAT5 in response to IL2 stimulation. ABCB1 ADAM19 AGER AHCY AHNAK AHR AKAP2 ALCAM AMACR ANXA4 APLP1 ARL4A BATF BATF3 BCL2 BCL2L1 BHLHE4 BMP2 BMPR2 CA2 CAPG CAPN3 CASP3 CCDC164 CCND2 CCND3 CCNE1 CCR4 CD44 CD48 CD79B CD81 CD83 CD86 CDC42SE2 CDC6 CDCP1 CDKN1C
35 CISH CKAP4 COCH COL6A1 CSF1 CSF2 CST7 CTLA4 CTSZ CXCL1 CYFIP1 DCPS DENND5A DHRS3 ECM1 EMP1 ENO3 ENPP1 EOMES ETV4 F2RL2 FAH FAM126B FGL2 FLT3LG FURIN GABARAPL1 GADD45B GALM GATA1 GBP4 GLIPR2 GPR65 GPR83 GPX4 GSTO1 GUCY1B3 HIPK2 HK2 HOPX HUWE1 ICOS IFITM3
36 IFNGR1 IGF1R IGF2R IKZF2 IKZF4 IL1 IL1RA IL13 IL18R1 IL1R2 IL1RL1 IL2RA IL2RB IL3RA IL4R IRF4 IRF6 IRF8 ITGA6 ITGAE ITGAV ITIH5 KLF6 LCLAT1 LIF LRIG1 LRRC8C LTB MAFF MAP3K8 MAP6 MAPKAPK2 METTL2 MUC1 MXD1 MYC MYO1C MYO1E N6AMT2 NCOA3 NCS1 NDRG1 NFIL3
37 NFKBIZ NOP2 NRP1 NT5E ODC1 P2RX4 P4HA1 PDCD2L PENK PHLDA1 PHTF2 PIM1 PLAGL1 PLEC PLIN2 PLSCR1 PNP POU2F1 PPAP2A PRAF2 PRKCH PRNP PTCH1 PTGER2 PTH1R PTRH2 PUS1 RABGAP1L RGS16 RHOB RHOH RNH1 RORA RRAGD S1A1 SCN9A SELL SELP SERPINB6 SERPINC1 SH3BGRL2 SHE SLC1A5
38 SLC29A2 SLC2A3 SLC39A8 SMPDL3A SNX14 SNX9 SOCS1 SOCS2 SPP1 SPRED2 SPRY4 ST3GAL4 SWAP7 SYNGR2 SYT11 TGM2 TIAM1 TLR7 TNFRSF18 TNFRSF1B TNFRSF21 TNFRSF4 TNFRSF8 TNFRSF9 TNFSF1 TNFSF11 TRAF1 TTC39B TWSG1 UCK2 UMPS WLS XBP1
39 Supplementary Table 3. Genes of the Hallmark IL2-STAT5 signaling pathway down-regulated in Tfr cells vs conventional Treg cells. IL4R IL2RB TNFSF1 IL1 MYC MXD1 TNFRSF1B IL18R1 SELL AHR SOCS1 CD86 IRF4 GBP4 FGL2 SELP NCOA3 IFNGR1 IL2RA SLC2A3 DENND5A TIAM1 IGF1R CISH FURIN HIPK2 ECM1 ABCB1 PTCH1 IGF2R BCL2L1 SLC29A2 ITIH5 SERPINC1 RRAGD GATA1 GSTO1 SHE
40 HUWE1 GPR83 TTC39B AGER SWAP7 LCLAT1 METTL2 GUCY1B3 ITGA6 IRF6 ITGAE TLR7 SNX9 PLEC SLC39A8 IKZF4 AHNAK CAPN3 LRRC8C HOPX ADAM19 SOCS2 IL1RL1 CD79B
HTG EdgeSeq Immuno-Oncology Assay Gene List
 A2M ABCB1 ABCB11 ABCC2 ABCG2 ABL1 ABL2 ACTB ADA ADAM17 ADGRE5 ADORA2A AICDA AKT3 ALCAM ALO5 ANA1 APAF1 APP ATF1 ATF2 ATG12 ATG16L1 ATG5 ATG7 ATM ATP5F1 AL BATF BA BCL10 BCL2 BCL2L1 BCL6 BID BIRC5 BLNK
A2M ABCB1 ABCB11 ABCC2 ABCG2 ABL1 ABL2 ACTB ADA ADAM17 ADGRE5 ADORA2A AICDA AKT3 ALCAM ALO5 ANA1 APAF1 APP ATF1 ATF2 ATG12 ATG16L1 ATG5 ATG7 ATM ATP5F1 AL BATF BA BCL10 BCL2 BCL2L1 BCL6 BID BIRC5 BLNK
Supplementary Table 1: List of the 242 hypoxia/reoxygenation marker genes collected from literature and databases
 Supplementary Tables: Supplementary Table 1: List of the 242 hypoxia/reoxygenation marker genes collected from literature and databases Supplementary Table 2: Contingency tables used in the fisher exact
Supplementary Tables: Supplementary Table 1: List of the 242 hypoxia/reoxygenation marker genes collected from literature and databases Supplementary Table 2: Contingency tables used in the fisher exact
** *** population. (A) Representative FACS plots showing the gating strategy for T N
 Sca-1 (MFI) CD122 (MFI) CD44 A CD8 + gated: resting spleen CD8 + gated: T Mem -derived CD8 + gated: T N -derived, expanded alone B CD62L CD44 low CD62L +, resting CD44 high, T Mem -derived CD44 low CD62L
Sca-1 (MFI) CD122 (MFI) CD44 A CD8 + gated: resting spleen CD8 + gated: T Mem -derived CD8 + gated: T N -derived, expanded alone B CD62L CD44 low CD62L +, resting CD44 high, T Mem -derived CD44 low CD62L
Supplemental Table 1: Enriched GO categories per cluster
 Supplemental Table 1: Enriched GO categories per cluster Cluster Enriched with #genes Raw p-value Corrected p-value Frequency in set (%) Cluster 1 immune response - GO:0006955 32 2.36E-14 0.001 13.5 Cluster
Supplemental Table 1: Enriched GO categories per cluster Cluster Enriched with #genes Raw p-value Corrected p-value Frequency in set (%) Cluster 1 immune response - GO:0006955 32 2.36E-14 0.001 13.5 Cluster
Supplementary Figures and Tables. An inflammatory gene signature distinguishes neurofibroma Schwann cells and
 Supplementary Figures and Tables An inflammatory gene signature distinguishes neurofibroma Schwann cells and macrophages from cells in the normal peripheral nervous system Kwangmin Choi 1, Kakajan Komurov
Supplementary Figures and Tables An inflammatory gene signature distinguishes neurofibroma Schwann cells and macrophages from cells in the normal peripheral nervous system Kwangmin Choi 1, Kakajan Komurov
Supplementary Table 1. Information on the 174 single nucleotide variants identified by whole-exome sequencing
 Supplementary Table 1. Information on the 174 single nucleotide s identified by whole-exome sequencing Chr Position Type AF exomes Database 1 10163148 A/G UBE4B missense 0.000326 1534 2 98 1 4.44 Damaging
Supplementary Table 1. Information on the 174 single nucleotide s identified by whole-exome sequencing Chr Position Type AF exomes Database 1 10163148 A/G UBE4B missense 0.000326 1534 2 98 1 4.44 Damaging
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nature11986 relative IL-6 expression Viable intracellular Bp per well 18 16 1 1 5 5 3 1 6 +DG 8 8 8 Control DG Time (h) hours B. pertussis IL-6 (pg/ml) 15 Control DG
SUPPLEMENTARY INFORMATION doi:1.138/nature11986 relative IL-6 expression Viable intracellular Bp per well 18 16 1 1 5 5 3 1 6 +DG 8 8 8 Control DG Time (h) hours B. pertussis IL-6 (pg/ml) 15 Control DG
Eosinophils! 40! 30! 20! 10! 0! NS!
 A Macrophages Lymphocytes Eosinophils Neutrophils Percentage (%) 1 ** 4 * 1 1 MMA SA B C Baseline FEV1, % predicted 15 p = 1.11 X 10-9 5 CD4:CD8 ratio 1 Supplemental Figure 1. Cellular infiltrate in the
A Macrophages Lymphocytes Eosinophils Neutrophils Percentage (%) 1 ** 4 * 1 1 MMA SA B C Baseline FEV1, % predicted 15 p = 1.11 X 10-9 5 CD4:CD8 ratio 1 Supplemental Figure 1. Cellular infiltrate in the
Supplementary Materials
 Supplementary Materials Supplemental figure 1. Absolute cell count of naïve/memory compartment of CD4 and CD8 T cells in early years Absolute cell number of naïve (CD45RA+CCR7+), Stem cell memory T cell
Supplementary Materials Supplemental figure 1. Absolute cell count of naïve/memory compartment of CD4 and CD8 T cells in early years Absolute cell number of naïve (CD45RA+CCR7+), Stem cell memory T cell
Gene expression of muscular and neuronal pathways is cooperatively dysregulated in patients
 Gene expression of muscular and neuronal pathways is cooperatively dysregulated in patients with idiopathic achalasia Orazio Palmieri 1, Tommaso Mazza 2, Antonio Merla 1, Caterina Fusilli 2, Antonello
Gene expression of muscular and neuronal pathways is cooperatively dysregulated in patients with idiopathic achalasia Orazio Palmieri 1, Tommaso Mazza 2, Antonio Merla 1, Caterina Fusilli 2, Antonello
Supplemental Data for Toischer et al., Cardiomyocyte proliferation prevents failure in
 Supplemental Data, Page 1 Supplemental Data for Toischer et al., Cardiomyocyte proliferation prevents failure in pressure but not volume overload. Contents: Page 3: Supplemental Figure 1 - Echocardiographic
Supplemental Data, Page 1 Supplemental Data for Toischer et al., Cardiomyocyte proliferation prevents failure in pressure but not volume overload. Contents: Page 3: Supplemental Figure 1 - Echocardiographic
Supplementary Table 1. Candidate loci that were associated with diabetes- and/or obesity-related traits.
 Supplementary Table 1. Candidate loci that were associated with diabetes- and/or obesity-related traits. All With Fst in the With long With SNP(s) at the 5' top 5% bracket haplotypes regulatory region
Supplementary Table 1. Candidate loci that were associated with diabetes- and/or obesity-related traits. All With Fst in the With long With SNP(s) at the 5' top 5% bracket haplotypes regulatory region
Supplementary Information
 1 Supplementary Information Human TSC2 Null Fibroblast-Like Cells Induce Hair Follicle Neogenesis and Hamartoma Morphogenesis Shaowei Li 1, Rajesh L. Thangapazham 1, Ji-an Wang 1, Sangeetha Rajesh 1, Tzu-Cheg
1 Supplementary Information Human TSC2 Null Fibroblast-Like Cells Induce Hair Follicle Neogenesis and Hamartoma Morphogenesis Shaowei Li 1, Rajesh L. Thangapazham 1, Ji-an Wang 1, Sangeetha Rajesh 1, Tzu-Cheg
Table SІ. List of genes upregulated by IL-4/IL-13-treatment of NHEK
 Supplemental legends Table SІ. List of genes upregulated by IL-4/IL-13-treatment of NHEK Table SП. List of genes in cluster 1 identified using two-dimensional hierarchical clustering analysis Table SШ.
Supplemental legends Table SІ. List of genes upregulated by IL-4/IL-13-treatment of NHEK Table SП. List of genes in cluster 1 identified using two-dimensional hierarchical clustering analysis Table SШ.
Supplementary Figure 1. Microarray data mining and validation
 Supplementary Figure 1. Microarray data mining and validation Microarray (>47,000 probes) Raw data Background subtraction NanoString (124 genes) Set cut-off and threshold values Raw data Normalized to
Supplementary Figure 1. Microarray data mining and validation Microarray (>47,000 probes) Raw data Background subtraction NanoString (124 genes) Set cut-off and threshold values Raw data Normalized to
Deconvolution of Heterogeneous Wound Tissue Samples into Relative Macrophage Phenotype Composition via Models based on Gene Expression
 Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2017 Supplementary Information for: Deconvolution of Heterogeneous Wound Tissue Samples into
Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2017 Supplementary Information for: Deconvolution of Heterogeneous Wound Tissue Samples into
Quantitative Real Time PCR (RT-qPCR) Differentiation of human preadipocytes to adipocytes
 Quantitative Real Time PCR (RT-qPCR) First strand cdna was synthesized and RT-qPCR was performed using RT2 first strand kits (Cat#330401) and RT2 qpcr Master Mix (SABioscience, Frederick, MD). Experiments
Quantitative Real Time PCR (RT-qPCR) First strand cdna was synthesized and RT-qPCR was performed using RT2 first strand kits (Cat#330401) and RT2 qpcr Master Mix (SABioscience, Frederick, MD). Experiments
A genome-wide association study identifies vitiligo
 A genome-wide association study identifies vitiligo susceptibility loci at MHC and 6q27 Supplementary Materials Index Supplementary Figure 1 The principal components analysis (PCA) of 2,546 GWAS samples
A genome-wide association study identifies vitiligo susceptibility loci at MHC and 6q27 Supplementary Materials Index Supplementary Figure 1 The principal components analysis (PCA) of 2,546 GWAS samples
devseek (Sequence(Analysis(Panel(for(Neurodevelopmental(Disorders
 ACSL4 Mental/retardation,/XBlinked/63 XLBR ADSL Adenylosuccinase/Deficiency AR AFF2 Mental/retardation,/XBlinked,/FRAXE/type XLBR ALG6 Congenital/disorder/of/glycosylation/type/Ic AR ANK3 Mental/retardation,/autosomal/recessive/37
ACSL4 Mental/retardation,/XBlinked/63 XLBR ADSL Adenylosuccinase/Deficiency AR AFF2 Mental/retardation,/XBlinked,/FRAXE/type XLBR ALG6 Congenital/disorder/of/glycosylation/type/Ic AR ANK3 Mental/retardation,/autosomal/recessive/37
Lions and Tigers and Bears.Oh My!
 Lions and Tigers and Bears.Oh My! TAKING THE FEAR OUT OF GENETIC TESTING Lisa Butterfield, MS, CGC Genetic Counselor Clinical Assistant Professor University of Kansas Health System Why is Genetic Testing
Lions and Tigers and Bears.Oh My! TAKING THE FEAR OUT OF GENETIC TESTING Lisa Butterfield, MS, CGC Genetic Counselor Clinical Assistant Professor University of Kansas Health System Why is Genetic Testing
Moyamoya disease susceptibility gene RNF213 links inflammatory and angiogenic
 Supplementary Information for Moyamoya disease susceptibility gene RNF213 links inflammatory and angiogenic signals in endothelial cells Kazuhiro Ohkubo 1,, Yasunari Sakai 1,*,, Hirosuke Inoue 1, Satoshi
Supplementary Information for Moyamoya disease susceptibility gene RNF213 links inflammatory and angiogenic signals in endothelial cells Kazuhiro Ohkubo 1,, Yasunari Sakai 1,*,, Hirosuke Inoue 1, Satoshi
Table S1. CDE Sequences and Globin Reporter mrna Half-Lives in NIH3T3 Cells, Related to Figures 1 and 2 Construct Sequence t1/2 ± SD (hrs)
 Table S1. CDE Sequences and Globin Reporter mrna Half-Lives in NIH3T3 Cells, Related to Figures 1 and 2 Construct Sequence t 1/2 ± SD (hrs) n TNF -CDE 37 -V2 AGATCCTTCAGACAGACATGTTTTCTGTGAAAACGGAGCTGAGCTAGATCT
Table S1. CDE Sequences and Globin Reporter mrna Half-Lives in NIH3T3 Cells, Related to Figures 1 and 2 Construct Sequence t 1/2 ± SD (hrs) n TNF -CDE 37 -V2 AGATCCTTCAGACAGACATGTTTTCTGTGAAAACGGAGCTGAGCTAGATCT
Supplementary Information for nature05543: Foxp3-dependent programme of regulatory T-cell differentiation
 Supplementary Information for nature5543: Foxp3-dependent programme of regulatory T-cell differentiation Supplementary Methods Mice. The Foxp3 gfpko targeting vector was generated by inserting the stop
Supplementary Information for nature5543: Foxp3-dependent programme of regulatory T-cell differentiation Supplementary Methods Mice. The Foxp3 gfpko targeting vector was generated by inserting the stop
Editorial Type 2 Diabetes and More Gene Panel: A Predictive Genomics Approach for a Polygenic Disease
 Cronicon OPEN ACCESS EC DIABETES AND METABOLIC RESEARCH Editorial Type 2 Diabetes and More Gene Panel: A Predictive Genomics Approach for a Polygenic Disease Amr TM Saeb* University Diabetes Center, College
Cronicon OPEN ACCESS EC DIABETES AND METABOLIC RESEARCH Editorial Type 2 Diabetes and More Gene Panel: A Predictive Genomics Approach for a Polygenic Disease Amr TM Saeb* University Diabetes Center, College
An Integrative Pharmacogenomic Approach Identifies Two-drug. Combination Therapies for Personalized Cancer Medicine
 An Integrative Pharmacogemic Approach Identifies Two-drug Combination Therapies for Personalized Cancer Medicine Yin Liu 1,2,3, Teng Fei 4,5, Xiaoqi Zheng 6, Myles Brown 4,5, Peng Zhang 7*, X. Shirley
An Integrative Pharmacogemic Approach Identifies Two-drug Combination Therapies for Personalized Cancer Medicine Yin Liu 1,2,3, Teng Fei 4,5, Xiaoqi Zheng 6, Myles Brown 4,5, Peng Zhang 7*, X. Shirley
CD Marker Antibodies. atlasantibodies.com
 CD Marker Antibodies atlasantibodies.com CD Marker Antibodies Product Name Product Number Validated Applications Anti-ACE HPA029298 IHC,WB Anti-ACKR1 HPA016421 IHC Anti-ACKR1 HPA017672 IHC Anti-ADAM17
CD Marker Antibodies atlasantibodies.com CD Marker Antibodies Product Name Product Number Validated Applications Anti-ACE HPA029298 IHC,WB Anti-ACKR1 HPA016421 IHC Anti-ACKR1 HPA017672 IHC Anti-ADAM17
Nature Immunology: doi: /ni.3745
 Supplementary Figure 1 Experimental settings. (a) Experimental setup and bioinformatic data analysis of transcriptomic changes induced in neonatal and adult monocytes by stimulation with 10 ng/ml LPS for
Supplementary Figure 1 Experimental settings. (a) Experimental setup and bioinformatic data analysis of transcriptomic changes induced in neonatal and adult monocytes by stimulation with 10 ng/ml LPS for
CPM (x 10-3 ) Tregs +Teffs. Tregs alone ICOS CLTA-4
 A 2,5 B 4 Number of cells (x 1-6 ) 2, 1,5 1, 5 CPM (x 1-3 ) 3 2 1 5 1 15 2 25 3 Days of culture 1/1 1/2 1/4 1/8 1/16 1/32 Treg/Teff ratio C alone alone alone alone CD25 FoxP3 GITR CD44 ICOS CLTA-4 CD127
A 2,5 B 4 Number of cells (x 1-6 ) 2, 1,5 1, 5 CPM (x 1-3 ) 3 2 1 5 1 15 2 25 3 Days of culture 1/1 1/2 1/4 1/8 1/16 1/32 Treg/Teff ratio C alone alone alone alone CD25 FoxP3 GITR CD44 ICOS CLTA-4 CD127
Supplementary Material. Table S1. Summary of mapping results
 Supplementary Material Table S1. Summary of mapping results Sample Total reads Mapped (%) HNE0-1 99687516 80786083 (81.0%) HNE0-2 94318720 77047792 (81.7%) HNE0-3 104033900 84348102 (81.1%) HNE15-1 94426598
Supplementary Material Table S1. Summary of mapping results Sample Total reads Mapped (%) HNE0-1 99687516 80786083 (81.0%) HNE0-2 94318720 77047792 (81.7%) HNE0-3 104033900 84348102 (81.1%) HNE15-1 94426598
BD Rhapsody Immune Response Panel Hs
 Doc ID: 55665 Rev. 2.0 PRODUCT INSERT BD Rhapsody Immune Response Panel Hs BD Biosciences Kit Part Number: 633750 Purpose The BD Rhapsody Immune Response Panel Hs is a set of ready-to-use primer pools
Doc ID: 55665 Rev. 2.0 PRODUCT INSERT BD Rhapsody Immune Response Panel Hs BD Biosciences Kit Part Number: 633750 Purpose The BD Rhapsody Immune Response Panel Hs is a set of ready-to-use primer pools
Supplemental Figure 1. Immune phenotype of mcd19 targeted CAR T and dose titration of in vivo
 Supplemental Materials Supplemental Methods Supplemental Figure 1. Immune phenotype of mcd19 targeted CAR T and dose titration of in vivo efficacy. Supplemental Figure 2. Gene expression of fluorescent-protein
Supplemental Materials Supplemental Methods Supplemental Figure 1. Immune phenotype of mcd19 targeted CAR T and dose titration of in vivo efficacy. Supplemental Figure 2. Gene expression of fluorescent-protein
Regulation of eif4 and p70s6k signaling. Chondroitin and dermatan biosynthesis Interferon signaling
 Supplemental Information Table S1: Differentially regulated pathways in cortical versus cerebellar human astrocytes following treatment with PBS or IFN-β Pathway Regulation of eif4 and p70s6k signaling
Supplemental Information Table S1: Differentially regulated pathways in cortical versus cerebellar human astrocytes following treatment with PBS or IFN-β Pathway Regulation of eif4 and p70s6k signaling
Supplementary Table S1: Gene expression targets
 Supplementary Table S1: Gene expression targets Probe ID Gene ID Gene Descriptions Cell type Antigen-dependent activation MmugDNA.32422.1.S1_at CD180 CD180 molecule B 1 MmugDNA.18254.1.S1_at CD19 CD19
Supplementary Table S1: Gene expression targets Probe ID Gene ID Gene Descriptions Cell type Antigen-dependent activation MmugDNA.32422.1.S1_at CD180 CD180 molecule B 1 MmugDNA.18254.1.S1_at CD19 CD19
Cardiovascular Disease Products
 Cardiovascular Disease Products For more information, visit: www.bosterbio.com Cardiovascular Disease Research Cardiovascular disease is the leading cause of death in developed nations. Boster Bio aims
Cardiovascular Disease Products For more information, visit: www.bosterbio.com Cardiovascular Disease Research Cardiovascular disease is the leading cause of death in developed nations. Boster Bio aims
Supplementary information. Dual targeting of ANGPT1 and TGFBR2 genes by mir-204 controls
 Supplementary information Dual targeting of ANGPT1 and TGFBR2 genes by mir-204 controls angiogenesis in breast cancer Ali Flores-Pérez, Laurence A. Marchat, Sergio Rodríguez-Cuevas, Verónica Bautista-Piña,
Supplementary information Dual targeting of ANGPT1 and TGFBR2 genes by mir-204 controls angiogenesis in breast cancer Ali Flores-Pérez, Laurence A. Marchat, Sergio Rodríguez-Cuevas, Verónica Bautista-Piña,
Targeted Genes and Methodology Details for Epilepsy/Seizure Genetic Panels
 Targeted s and Methodology Details for Epilepsy/Seizure tic Panels Reference transcripts based on build GRCh37 (hg19) interrogated by Epilepsy/Seizure tic Panels Epilepsy Expanded Panel Epilepsy Expanded
Targeted s and Methodology Details for Epilepsy/Seizure tic Panels Reference transcripts based on build GRCh37 (hg19) interrogated by Epilepsy/Seizure tic Panels Epilepsy Expanded Panel Epilepsy Expanded
103.5 Membrane metalloendopeptidase TGA GGG GTC ACG ATT TTA GG ATG ATG GTG AGG AGC AGG AC
 Supplementary Table SA Primers used for real-time RT-PCR gene regulation and their confirmation Gene Name Accession no Size Region Forward Primer Reverse Primer Efficiency% AKT v-akt oncogene homolog NM_00563
Supplementary Table SA Primers used for real-time RT-PCR gene regulation and their confirmation Gene Name Accession no Size Region Forward Primer Reverse Primer Efficiency% AKT v-akt oncogene homolog NM_00563
Supplementary information: Forstner, Hofmann et al.
 Supplementary information: Forstner, Hofmann et al., Genome-wide analysis implicates micrornas and their target genes in the development of bipolar disorder Table of contents Supplementary Figure 1: Regional
Supplementary information: Forstner, Hofmann et al., Genome-wide analysis implicates micrornas and their target genes in the development of bipolar disorder Table of contents Supplementary Figure 1: Regional
NGS data processing and alignment: Raw reads generated from the Illumina HiSeq2500
 Supplemental aterials ethods NGS data processing and alignment: Raw reads generated from the Illumina HiSeq25 sequencer were demultiplexed using configurebcl2fastq.pl version 1.8.4. Quality filtering and
Supplemental aterials ethods NGS data processing and alignment: Raw reads generated from the Illumina HiSeq25 sequencer were demultiplexed using configurebcl2fastq.pl version 1.8.4. Quality filtering and
Supplementary Figures. Supplementary Figure 1. Dinucleotide variant proportions. These are described and quantitated. for each lesion type.
 Supplementary Figures Supplementary Figure 1. Dinucleotide variant proportions. These are described and quantitated for each lesion type. a b Supplementary Figure 2. Non-negative matrix factorization-derived
Supplementary Figures Supplementary Figure 1. Dinucleotide variant proportions. These are described and quantitated for each lesion type. a b Supplementary Figure 2. Non-negative matrix factorization-derived
Comprehensive genetic testing for hearing and vision loss
 Comprehensive genetic testing for hearing and vision loss Hearing and vision loss can result from both genetic and non-genetic etiologies In general, there is a genetic basis for up to 50% of prelingual
Comprehensive genetic testing for hearing and vision loss Hearing and vision loss can result from both genetic and non-genetic etiologies In general, there is a genetic basis for up to 50% of prelingual
WT-TP53 DLBCL WT-TP53 GCB-DLBCL
 SUPPLEMENTAL TABLES AND FIGURES Supplemental Table S1. Summary of gene expression profiling analysis results listing counts of significant differentially expressed transcripts between defined two groups
SUPPLEMENTAL TABLES AND FIGURES Supplemental Table S1. Summary of gene expression profiling analysis results listing counts of significant differentially expressed transcripts between defined two groups
Delineation of Diverse Macrophage Activation Programs in Response to Intracellular Parasites and Cytokines
 Delineation of Diverse Macrophage Activation Programs in Response to Intracellular Parasites and Cytokines Shuyi Zhang 1,2, Charles C. Kim 3, Sajeev Batra 3, James H. McKerrow 1,2, P ng Loke 4 * 1 Department
Delineation of Diverse Macrophage Activation Programs in Response to Intracellular Parasites and Cytokines Shuyi Zhang 1,2, Charles C. Kim 3, Sajeev Batra 3, James H. McKerrow 1,2, P ng Loke 4 * 1 Department
Supplemental Figure 1: Anti-GDF15 antibody blocks binding of GDF15 to hgfral.
 Supplemental Figure 1: Anti-GDF15 antibody blocks binding of GDF15 to hgfral. Anti-GDF15 antibody (mab26, a neutralizing monoclonal antibody to wtgdf15) blocks binding of GDF15 to human GFRAL in a dose-dependent
Supplemental Figure 1: Anti-GDF15 antibody blocks binding of GDF15 to hgfral. Anti-GDF15 antibody (mab26, a neutralizing monoclonal antibody to wtgdf15) blocks binding of GDF15 to human GFRAL in a dose-dependent
Table S1. Differentially expressed transcripts between hepatic inkt cells. sorted form WT and plck-hcd1d tg mice. Differentially expressed
 Supplementary Information Table S1. Differentially expressed transcripts between hepatic inkt cells sorted form and plck-hcd1d tg mice. Differentially expressed transcripts identified by gene expression
Supplementary Information Table S1. Differentially expressed transcripts between hepatic inkt cells sorted form and plck-hcd1d tg mice. Differentially expressed transcripts identified by gene expression
CD80 and PD-L2 define functionally distinct memory B cell subsets that are. Griselda V Zuccarino-Catania, Saheli Sadanand, Florian J Weisel, Mary M
 Supplementary Figures CD8 and PD-L define functionally distinct memory B cell subsets that are independent of antibody isotype Running title: Memory B Cell Subset Function Griselda V Zuccarino-Catania,
Supplementary Figures CD8 and PD-L define functionally distinct memory B cell subsets that are independent of antibody isotype Running title: Memory B Cell Subset Function Griselda V Zuccarino-Catania,
What Do We Know About Individual Variability and Its
 What Do We Know About Individual Variability and Its Contribution to Disease? Nathaniel Rothman, MD, MPH, MHS Occupational and Environmental Epidemiology Branch, Division of Cancer Epidemiology and Genetics,
What Do We Know About Individual Variability and Its Contribution to Disease? Nathaniel Rothman, MD, MPH, MHS Occupational and Environmental Epidemiology Branch, Division of Cancer Epidemiology and Genetics,
Jocelyn Chapman, MD Division of Gynecologic Oncology. Julie S. Mak, MS, MSc Genetic Counselor Hereditary Cancer Clinic
 Jocelyn Chapman, MD Division of Gynecologic Oncology Julie S. Mak, MS, MSc Genetic Counselor Hereditary Cancer Clinic Genetics underlies all cancers Somatic or tumor genetics Germline or inherited genetics
Jocelyn Chapman, MD Division of Gynecologic Oncology Julie S. Mak, MS, MSc Genetic Counselor Hereditary Cancer Clinic Genetics underlies all cancers Somatic or tumor genetics Germline or inherited genetics
The expression of genes contributing to pancreatic adenocarcinoma progression is influenced by the respective environment Sagini et al
 The expression of genes contributing to pancreatic adenocarcinoma progression is influenced by the respective environment Sagini et al Supplementary Figure 1: Target genes regulated by TGM2. Figure represents
The expression of genes contributing to pancreatic adenocarcinoma progression is influenced by the respective environment Sagini et al Supplementary Figure 1: Target genes regulated by TGM2. Figure represents
Supplement Results, Figures and Tables
 Supplement Results, Figures and Tables Results PET imaging The SUV was significantly higher in tumours of the parental HSC-3 cell line than tumours of the EV and the MMP-8 overexpressing cell lines (Figure
Supplement Results, Figures and Tables Results PET imaging The SUV was significantly higher in tumours of the parental HSC-3 cell line than tumours of the EV and the MMP-8 overexpressing cell lines (Figure
b kb
 Supplementary Figure S1 COX7RP mrna/gapdh COX activity (OD 45 nm/mg protein) Relative DNA amount COX7RP mrna/gapdh a c pcxn2-flag-cox7rp CAG Flag CMV enhancer Chiken b-actin promoter 1..8.6 COX7RP Muscle
Supplementary Figure S1 COX7RP mrna/gapdh COX activity (OD 45 nm/mg protein) Relative DNA amount COX7RP mrna/gapdh a c pcxn2-flag-cox7rp CAG Flag CMV enhancer Chiken b-actin promoter 1..8.6 COX7RP Muscle
Supplemental Table 1. List of genes and mature shrna sequences identified from the shrna screen in BRCA2-mutant PEO1 cells.
 Guillemette_Supplemental Table 1 Gene Mature Sequence Gene Mature Sequence ATTCATAGGATGTCAGCAG LOC157193 TATGCGTAAGTAATAAATTGCT TTAGTTCTGTCTTGGAGG LOC152225 CGCATATATGTTCTGACTT ALDH2 CACTTCAGTGTATGCCT
Guillemette_Supplemental Table 1 Gene Mature Sequence Gene Mature Sequence ATTCATAGGATGTCAGCAG LOC157193 TATGCGTAAGTAATAAATTGCT TTAGTTCTGTCTTGGAGG LOC152225 CGCATATATGTTCTGACTT ALDH2 CACTTCAGTGTATGCCT
Figure S1. Cos-7. Cos-7 GFP. pjnk C2C12 C2C12 GFP * 60. pjnk. WT lamin A. H222P lamin A
 Figure S1 A Cos-7 C NT WT lamin A Cos-7 H222P lamin A 100 % JNK nucleus / positives GFP GFP pjnk 80 60 ** 40 20 0 NT D C2C12 NT WT lamin A GFP pjnk H222P lamin A C2C12 H222P lamin A 100 % JNK nucleus /
Figure S1 A Cos-7 C NT WT lamin A Cos-7 H222P lamin A 100 % JNK nucleus / positives GFP GFP pjnk 80 60 ** 40 20 0 NT D C2C12 NT WT lamin A GFP pjnk H222P lamin A C2C12 H222P lamin A 100 % JNK nucleus /
SUPPLEMENTAL DATA. Table S1. Genes in each pathway selected for analysis.
 SUPPLEMENTAL DATA Table S1. Genes in each pathway selected for analysis. Gene Chr Start End bp Pathway 1 AKR1C1 10 5000044 5022159 22115 COX 2 AKR1C2 10 5029967 5060207 30240 COX 3 AKR1C3 10 5077549 5149878
SUPPLEMENTAL DATA Table S1. Genes in each pathway selected for analysis. Gene Chr Start End bp Pathway 1 AKR1C1 10 5000044 5022159 22115 COX 2 AKR1C2 10 5029967 5060207 30240 COX 3 AKR1C3 10 5077549 5149878
Jennifer Hauenstein Oncology Cytogenetics Emory University Hospital Atlanta, GA
 Comparison of Genomic Coverage using Affymetrix OncoScan Array and Illumina TruSight Tumor 170 NGS Panel for Detection of Copy Number Abnormalities in Clinical GBM Specimens Jennifer Hauenstein Oncology
Comparison of Genomic Coverage using Affymetrix OncoScan Array and Illumina TruSight Tumor 170 NGS Panel for Detection of Copy Number Abnormalities in Clinical GBM Specimens Jennifer Hauenstein Oncology
Expression Profiling of KLF4
 Expression Profiling of KLF4 AJCR0000006 Supplemental Data Figure S1. Snapshot of enriched gene sets identified by GSEA in Klf4-null MEFs. Figure S2. Snapshot of enriched gene sets identified by GSEA in
Expression Profiling of KLF4 AJCR0000006 Supplemental Data Figure S1. Snapshot of enriched gene sets identified by GSEA in Klf4-null MEFs. Figure S2. Snapshot of enriched gene sets identified by GSEA in
Cover Page. The handle holds various files of this Leiden University dissertation.
 Cover Page The handle http://hdl.handle.net/1887/43190 holds various files of this Leiden University dissertation. Author: Raeven, R.H.M. Title: Systems vaccinology : molecular signatures of immunity to
Cover Page The handle http://hdl.handle.net/1887/43190 holds various files of this Leiden University dissertation. Author: Raeven, R.H.M. Title: Systems vaccinology : molecular signatures of immunity to
The circulating T helper subsets and regulatory T cells in patients with common variable immunodeficiency with no monogenic disease
 1 Supplementary data The circulating T helper subsets and regulatory T cells in patients with common variable immunodeficiency with no monogenic disease Azizi et al. 2 Table S1. Excluded 373 genes variants
1 Supplementary data The circulating T helper subsets and regulatory T cells in patients with common variable immunodeficiency with no monogenic disease Azizi et al. 2 Table S1. Excluded 373 genes variants
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nature12912 Figure S1. Distribution of mutation rates and spectra across 4,742 tumor-normal (TN) pairs from 21 tumor types, as in Figure 1 from Lawrence et al. Nature
SUPPLEMENTARY INFORMATION doi:1.138/nature12912 Figure S1. Distribution of mutation rates and spectra across 4,742 tumor-normal (TN) pairs from 21 tumor types, as in Figure 1 from Lawrence et al. Nature
COMPLETE A FORM FOR EACH SAMPLE SUBMITTED ETHNIC BACKGROUND REPORTING INFORMATION
 DATE SAMPLE DRAWN: SAMPLE TYPE: BLOOD OTHER (SPECIFY): COMPLETE A FORM FOR EACH SAMPLE SUBMITTED PATIENT FIRST NAME: LAST NAME: DOB: SEX: M F UNKNOWN ADDRESS: HOME PHONE: ( ) WORK: ( ) EUROPEAN CAUCASIAN
DATE SAMPLE DRAWN: SAMPLE TYPE: BLOOD OTHER (SPECIFY): COMPLETE A FORM FOR EACH SAMPLE SUBMITTED PATIENT FIRST NAME: LAST NAME: DOB: SEX: M F UNKNOWN ADDRESS: HOME PHONE: ( ) WORK: ( ) EUROPEAN CAUCASIAN
Plasma-Seq conducted with blood from male individuals without cancer.
 Supplementary Figures Supplementary Figure 1 Plasma-Seq conducted with blood from male individuals without cancer. Copy number patterns established from plasma samples of male individuals without cancer
Supplementary Figures Supplementary Figure 1 Plasma-Seq conducted with blood from male individuals without cancer. Copy number patterns established from plasma samples of male individuals without cancer
Category GOTERM_CC_FAT GOTERM_CC_FAT GOTERM_CC_FAT GOTERM_BP_FAT GOTERM_MF_FAT GOTERM_CC_FAT GOTERM_MF_FAT GOTERM_CC_FAT GOTERM_BP_FAT GOTERM_BP_FAT
 Category KEGG_PATHWAY KEGG_PATHWAY Term GO:0005764~lysosome GO:0000323~lytic vacuole GO:0005773~vacuole GO:0006898~receptor-mediated endocytosis GO:0019842~vitamin binding GO:0005667~transcription factor
Category KEGG_PATHWAY KEGG_PATHWAY Term GO:0005764~lysosome GO:0000323~lytic vacuole GO:0005773~vacuole GO:0006898~receptor-mediated endocytosis GO:0019842~vitamin binding GO:0005667~transcription factor
Differentially expressed genes in ATZ treated testes, cut-off value 2
 Table S1 Differentially expressed genes in ATZ treated testes, cut-off value 2 Gene Name FC Gene full name/ function Entpd4 0.4 Ectonucleoside triphosphate diphosphohydrolase 4, role in salvaging of nucleotides,
Table S1 Differentially expressed genes in ATZ treated testes, cut-off value 2 Gene Name FC Gene full name/ function Entpd4 0.4 Ectonucleoside triphosphate diphosphohydrolase 4, role in salvaging of nucleotides,
Supplementary Table 1. mirna modulated in D HF and ND HF patients. Table shows in a linear scale the same values of Fig. 2.
 Supplementary Table 1. mirna modulated in D HF and ND HF patients. Table shows in a linear scale the same values of Fig. 2. mir D HF VS CTR ND HF VS CTR D HF VS ND HF FOLD CHANGE p FOLD CHANGE p FOLD CHANGE
Supplementary Table 1. mirna modulated in D HF and ND HF patients. Table shows in a linear scale the same values of Fig. 2. mir D HF VS CTR ND HF VS CTR D HF VS ND HF FOLD CHANGE p FOLD CHANGE p FOLD CHANGE
Supplementary Information
 Supplementary Information Index of Supplementary Materials Supplementary Figure. Distribution of methylation levels, characteristics of cis-meqtls and association plots for meqtls in methylation-related
Supplementary Information Index of Supplementary Materials Supplementary Figure. Distribution of methylation levels, characteristics of cis-meqtls and association plots for meqtls in methylation-related
Subject ID: Date Test Started: Specimen Type: Peripheral Blood Date of Report: Date Specimen Obtained:
 TEST PERFORMED Whole exome sequencing (WES) with Focused Diagnostic Panel(s): Neuropathy, Leukodystrophy, Myopathy See below for a complete list of genes analyzed. RESULTS 1 sequence variant with potential
TEST PERFORMED Whole exome sequencing (WES) with Focused Diagnostic Panel(s): Neuropathy, Leukodystrophy, Myopathy See below for a complete list of genes analyzed. RESULTS 1 sequence variant with potential
Integrative Radiation Biology
 Dr. Kristian Unger Integrative Biology Group Research Unit of Radiation Cytogenetics Department of Radiation Sciences Helmholtz-Zentrum München 1 Key Questions Molecular mechanisms of radiation-induced
Dr. Kristian Unger Integrative Biology Group Research Unit of Radiation Cytogenetics Department of Radiation Sciences Helmholtz-Zentrum München 1 Key Questions Molecular mechanisms of radiation-induced
Precision Oncology: Experience at UW
 Precision Oncology: Experience at UW Colin Pritchard MD, PhD University of Washington, Department of Lab Medicine WSMOS Meeting November 1, 2013 Conflict of Interest Disclosures I declare the following,
Precision Oncology: Experience at UW Colin Pritchard MD, PhD University of Washington, Department of Lab Medicine WSMOS Meeting November 1, 2013 Conflict of Interest Disclosures I declare the following,
Bezzi et al., Supplementary Figure 1 *** Nature Medicine: doi: /nm Pten pc-/- ;Zbtb7a pc-/- Pten pc-/- ;Pml pc-/- Pten pc-/- ;Trp53 pc-/-
 Gr-1 Gr-1 Gr-1 Bezzi et al., Supplementary Figure 1 a Gr1-CD11b 3 months Spleen T cells 3 months Spleen B cells 3 months Spleen Macrophages 3 months Spleen 15 4 8 6 c CD11b+/Gr1+ cells [%] 1 5 b T cells
Gr-1 Gr-1 Gr-1 Bezzi et al., Supplementary Figure 1 a Gr1-CD11b 3 months Spleen T cells 3 months Spleen B cells 3 months Spleen Macrophages 3 months Spleen 15 4 8 6 c CD11b+/Gr1+ cells [%] 1 5 b T cells
Nature Neuroscience: doi: /nn.4295
 Supplementary Figure 1 Gene expression profiling of EGFRvIII-expressing human brain tumor stem cells (BTSCs) and mouse astrocytes (a) The protein lysates of human BTSCs were analyzed by immunoblotting
Supplementary Figure 1 Gene expression profiling of EGFRvIII-expressing human brain tumor stem cells (BTSCs) and mouse astrocytes (a) The protein lysates of human BTSCs were analyzed by immunoblotting
SUPPLEMENTAL MATERIAL. Characterization and discovery of novel mirnas and mornas in JAK2V617F. mutated SET2 cells
 SUPPLEMENTAL MATERIAL Characterization and discovery of novel mirnas and mornas in JAK2V617F mutated SET2 cells Stefania Bortoluzzi 1*, Andrea Bisognin 1*, et al. 1 Supplemental methods Short RNA library
SUPPLEMENTAL MATERIAL Characterization and discovery of novel mirnas and mornas in JAK2V617F mutated SET2 cells Stefania Bortoluzzi 1*, Andrea Bisognin 1*, et al. 1 Supplemental methods Short RNA library
The Genetics of Alcoholism. Robert Hitzemann, Ph.D. Departments of Behavioral Neuroscience & Psychiatry Oregon Health & Science University
 The Genetics of Alcoholism Robert Hitzemann, Ph.D. Departments of Behavioral Neuroscience & Psychiatry Oregon Health & Science University Genetics, Genomics and Alcoholism Robert Hitzemann, Ph.D. Departments
The Genetics of Alcoholism Robert Hitzemann, Ph.D. Departments of Behavioral Neuroscience & Psychiatry Oregon Health & Science University Genetics, Genomics and Alcoholism Robert Hitzemann, Ph.D. Departments
Äijö et al. BMC Genomics 2012, 13:572
 Äijö et al. BMC Genomics 2012, 13:572 RESEARCH ARTICLE Open Access An integrative computational systems biology approach identifies differentially regulated dynamic transcriptome signatures which drive
Äijö et al. BMC Genomics 2012, 13:572 RESEARCH ARTICLE Open Access An integrative computational systems biology approach identifies differentially regulated dynamic transcriptome signatures which drive
Copyright Warning & Restrictions
 Copyright Warning & Restrictions The copyright law of the United States (Title 17, United States Code) governs the making of photocopies or other reproductions of copyrighted material. Under certain conditions
Copyright Warning & Restrictions The copyright law of the United States (Title 17, United States Code) governs the making of photocopies or other reproductions of copyrighted material. Under certain conditions
HO-1 inhibits preadipocyte proliferation and differentiation at the onset of obesity via ROS dependent activation of Akt2
 SUPPLEMENTAL INFORMATION referring to: HO-1 inhibits preadipocyte proliferation and differentiation at the onset of obesity via ROS dependent activation of Akt2 Gabriel Wagner 1, Josefine Lindroos-Christensen
SUPPLEMENTAL INFORMATION referring to: HO-1 inhibits preadipocyte proliferation and differentiation at the onset of obesity via ROS dependent activation of Akt2 Gabriel Wagner 1, Josefine Lindroos-Christensen
EMT and ECM. Jesse Liang, Ph.D. Sample & Assay Technologies
 EMT and ECM Jesse Liang, Ph.D. 1 Epithelial Cells.1. Closely adjoined.2. Polarized.Epithelial Markers: E-Cadherin (adherens junctions) Claudins (tight junctions) Occludin (tight junctions) Desmoplakin
EMT and ECM Jesse Liang, Ph.D. 1 Epithelial Cells.1. Closely adjoined.2. Polarized.Epithelial Markers: E-Cadherin (adherens junctions) Claudins (tight junctions) Occludin (tight junctions) Desmoplakin
Supplemental Figure 1. Characterization of CNS lysosomal pathology in 2-month-old IDS-deficient mice. (A) Measurement of IDS activity in brain
 Supplemental Figure 1. Characterization of CNS lysosomal pathology in 2-month-old IDS-deficient mice. (A) Measurement of IDS activity in brain extracts from wild-type (WT) and IDS-deficient males. IDS
Supplemental Figure 1. Characterization of CNS lysosomal pathology in 2-month-old IDS-deficient mice. (A) Measurement of IDS activity in brain extracts from wild-type (WT) and IDS-deficient males. IDS
Nature Genetics: doi: /ng Supplementary Figure 1
 Supplementary Figure 1 Characterization of HPV Tg/+ ; Krt17 / mice. (a) Macroscopic images of HPV16 Tg/+ mouse ears at P40, P70 and P120. (b) Immunostaining of HPV16 Tg/+ ear tissue sections for K17 (red)
Supplementary Figure 1 Characterization of HPV Tg/+ ; Krt17 / mice. (a) Macroscopic images of HPV16 Tg/+ mouse ears at P40, P70 and P120. (b) Immunostaining of HPV16 Tg/+ ear tissue sections for K17 (red)
p5+mir30: AATGATACGGCGACCACCGACTAAAGTAGCCCCTTGAATTC
 SUPPORTING INFORMATION METHODS Massively parallel sequencing and shrna screen data analysis pipeline Genomic DNA extraction and purification from surviving MCF7 cells in the genome wide screen was carried
SUPPORTING INFORMATION METHODS Massively parallel sequencing and shrna screen data analysis pipeline Genomic DNA extraction and purification from surviving MCF7 cells in the genome wide screen was carried
Transcriptome profiling of granulosa cells of bovine ovarian follicles during growth from small to large antral sizes
 Transcriptome profiling of granulosa cells of bovine ovarian follicles during growth from small to large antral sizes Author Hatzirodos, Nicholas, F. Irving-Rodgers, Helen, Hummitzsch, Katja, L. Harland,
Transcriptome profiling of granulosa cells of bovine ovarian follicles during growth from small to large antral sizes Author Hatzirodos, Nicholas, F. Irving-Rodgers, Helen, Hummitzsch, Katja, L. Harland,
Schrader et al. Cell Communication and Signaling 2012, 10:43
 Schrader et al. Cell Communication and Signaling 2012, 10:43 RESEARCH Open Access Global gene expression changes of in vitro stimulated human transformed germinal centre B cells as surrogate for oncogenic
Schrader et al. Cell Communication and Signaling 2012, 10:43 RESEARCH Open Access Global gene expression changes of in vitro stimulated human transformed germinal centre B cells as surrogate for oncogenic
Inferring the Mammalian Cardiogenic Gene Regulatory Network. Jason Bazil 10/18/13 Cardiac Physiome Workshop 2013
 Inferring the Mammalian Cardiogenic Gene Regulatory Network Jason Bazil 1/18/13 Cardiac Physiome Workshop 213 Reverse Engineering Gene Regulatory Networks Two main questions need addressed: 1) What s connected
Inferring the Mammalian Cardiogenic Gene Regulatory Network Jason Bazil 1/18/13 Cardiac Physiome Workshop 213 Reverse Engineering Gene Regulatory Networks Two main questions need addressed: 1) What s connected
Supplemental Information. High-Throughput Microfluidic Labyrinth for the. Label-free Isolation of Circulating Tumor Cells
 Cell Systems, Volume 5 Supplemental Information High-Throughput Microfluidic Labyrinth for the Label-free Isolation of Circulating Tumor Cells Eric Lin, Lianette Rivera-Báez, Shamileh Fouladdel, Hyeun
Cell Systems, Volume 5 Supplemental Information High-Throughput Microfluidic Labyrinth for the Label-free Isolation of Circulating Tumor Cells Eric Lin, Lianette Rivera-Báez, Shamileh Fouladdel, Hyeun
Supplemental Figures/Tables. Methylation forward GTCGGGGCGTATTTAGTTC. Methylation reverse AACGACGTAAACGAAAATATCG
 Supplemental Figures/Tables Supplementary Tables Table S1. Primer sequences MSP Primers SPARC2 Unmethylation forward TTTTTTAGATTGTTTGGAGAGTG Unmethylation reverse AACTAACAACATAAACAAAAATATC Methylation
Supplemental Figures/Tables Supplementary Tables Table S1. Primer sequences MSP Primers SPARC2 Unmethylation forward TTTTTTAGATTGTTTGGAGAGTG Unmethylation reverse AACTAACAACATAAACAAAAATATC Methylation
Human lung epithelial cells progressed to malignancy through specific oncogenic. manipulations
 Supplementary Material for: Human lung epithelial cells progressed to malignancy through specific oncogenic manipulations Mitsuo Sato* 1,10, Jill E. Larsen* 1, Woochang Lee 1,11, Han Sun 2, David S. Shames
Supplementary Material for: Human lung epithelial cells progressed to malignancy through specific oncogenic manipulations Mitsuo Sato* 1,10, Jill E. Larsen* 1, Woochang Lee 1,11, Han Sun 2, David S. Shames
Deciphering the transcriptional network of the dendritic cell lineage
 Deciphering the transcriptional network of the dendritic cell lineage Jennifer C Miller 1,2, Brian D Brown 1,3, Tal Shay 4, Emmanuel L Gautier 1,5,6, Vladimir Jojic 7,1, Ariella Cohain 3, Gaurav Pandey
Deciphering the transcriptional network of the dendritic cell lineage Jennifer C Miller 1,2, Brian D Brown 1,3, Tal Shay 4, Emmanuel L Gautier 1,5,6, Vladimir Jojic 7,1, Ariella Cohain 3, Gaurav Pandey
Glia in Alzheimer's disease and aging: Molecular mechanisms underlying astrocyte and microglia reactivity Orre, A.M.
 UvA-DARE (Digital Academic Repository) Glia in Alzheimer's disease and aging: Molecular mechanisms underlying astrocyte and microglia reactivity Orre, A.M. Link to publication Citation for published version
UvA-DARE (Digital Academic Repository) Glia in Alzheimer's disease and aging: Molecular mechanisms underlying astrocyte and microglia reactivity Orre, A.M. Link to publication Citation for published version
Supplemental materials Calcium/Calmodulin Dependent Kinase Kinase 2 (CaMKK2) regulates hematopoietic stem and progenitor cell regeneration
 Supplemental materials Calcium/Calmodulin Dependent Kinase Kinase 2 (CaMKK2) regulates hematopoietic stem and progenitor cell regeneration Luigi Racioppi 1,2^, William Lento^1, Wei Huang 1, Stephanie Arvai
Supplemental materials Calcium/Calmodulin Dependent Kinase Kinase 2 (CaMKK2) regulates hematopoietic stem and progenitor cell regeneration Luigi Racioppi 1,2^, William Lento^1, Wei Huang 1, Stephanie Arvai
Differential gene expression profiling of the sciatic nerve in type 1 and type 2 diabetic mice
 BIOMEDICAL REPORTS 9: 291-304, 2018 Differential gene expression profiling of the sciatic nerve in type 1 and type 2 diabetic mice YU GU *, ZHUO-LIN QIU *, DE-ZHAO LIU, GUO-LIANG SUN, YING-CHAO GUAN, ZI-QING
BIOMEDICAL REPORTS 9: 291-304, 2018 Differential gene expression profiling of the sciatic nerve in type 1 and type 2 diabetic mice YU GU *, ZHUO-LIN QIU *, DE-ZHAO LIU, GUO-LIANG SUN, YING-CHAO GUAN, ZI-QING
Investigation of genes in chronic and acute morphine-treated mice using microarray datasets
 Investigation of genes in chronic and acute morphine-treated mice using microarray datasets L. Ding 1, J.L. Zhang 2, S.H. Yu 1 and L.F. Sheng 1 1 Department of Anesthesiology, The People s Hospital of
Investigation of genes in chronic and acute morphine-treated mice using microarray datasets L. Ding 1, J.L. Zhang 2, S.H. Yu 1 and L.F. Sheng 1 1 Department of Anesthesiology, The People s Hospital of
Sample Metrics. Allele Frequency (%) Read Depth Ploidy. Gene CDS Effect Protein Effect. LN Metastasis Tumor Purity Computational Pathology 80% 60%
 Supplemental Table 1: Estimated tumor purity, allele frequency, and independent read depth for all gene mutations classified as either potentially pathogenic or VUS in the metatastic and primary tumor
Supplemental Table 1: Estimated tumor purity, allele frequency, and independent read depth for all gene mutations classified as either potentially pathogenic or VUS in the metatastic and primary tumor
Supplementary Materials
 Supplementary Materials Using Multi objective Optimization to Identify Dynamical Network Biomarkers as Early warning Signals of Complex Diseases Fatemeh Vafaee 1, 2 1 Charles Perkins Centre, University
Supplementary Materials Using Multi objective Optimization to Identify Dynamical Network Biomarkers as Early warning Signals of Complex Diseases Fatemeh Vafaee 1, 2 1 Charles Perkins Centre, University
Journal: Nature Medicine. ETS factors reprogram the androgen receptor cistrome and prime prostate tumorigenesis in response to PTEN loss
 Journal: Nature Medicine ETS factors reprogram the androgen receptor cistrome and prime prostate tumorigenesis in response to PTEN loss Yu Chen, Ping Chi, Shira Rockowitz, Phillip J. Iaquinta, Tambudzai
Journal: Nature Medicine ETS factors reprogram the androgen receptor cistrome and prime prostate tumorigenesis in response to PTEN loss Yu Chen, Ping Chi, Shira Rockowitz, Phillip J. Iaquinta, Tambudzai
Assessing the Economics of Genomic Medicine. Kenneth Offit, MD MPH Chief, Clinical Genetics Service Memorial Sloan-Kettering Cancer Center
 Assessing the Economics of Genomic Medicine Kenneth Offit, MD MPH Chief, Clinical Genetics Service Memorial Sloan-Kettering Cancer Center DISCLOSURES No conflicts Patents unenforced Consultancies unpaid
Assessing the Economics of Genomic Medicine Kenneth Offit, MD MPH Chief, Clinical Genetics Service Memorial Sloan-Kettering Cancer Center DISCLOSURES No conflicts Patents unenforced Consultancies unpaid
Supporting Information
 Supporting Information Materials and Methods Isolation of primary hepatocytes and adenovirus infection. Primary hepatocytes were isolated from male C57BL/6J mice at 8-12 weeks of age according to the procedure
Supporting Information Materials and Methods Isolation of primary hepatocytes and adenovirus infection. Primary hepatocytes were isolated from male C57BL/6J mice at 8-12 weeks of age according to the procedure
Supporting Information
 Supporting Information Lin et al. 10.1073/pnas.1408759111 Fig. S1. Safety and potential toxicity evaluation of M1 in immunocompetent mice. Immunocompetent BALB/c mice received two doses of i.v. infusion
Supporting Information Lin et al. 10.1073/pnas.1408759111 Fig. S1. Safety and potential toxicity evaluation of M1 in immunocompetent mice. Immunocompetent BALB/c mice received two doses of i.v. infusion
Nature Immunology: doi: /ni.3494
 Supplementary Figure 1 High-throughput screen for novel inducer of T FH cell differentiation. (a) Schematic of primary screen. Purified human naïve CD4 T cells were stimulated by anti-cd3/cd28 beads on
Supplementary Figure 1 High-throughput screen for novel inducer of T FH cell differentiation. (a) Schematic of primary screen. Purified human naïve CD4 T cells were stimulated by anti-cd3/cd28 beads on
Optimized between-group classification: a new jackknife-based gene selection procedure for genome-wide expression data Supplementary information
 Optimized between-group classification: a new jackknife-based gene selection procedure for genome-wide expression data Supplementary information Florent Baty 1 florent.baty@unibas.ch Michel P Bihl 1 michel.bihl@unibas.ch
Optimized between-group classification: a new jackknife-based gene selection procedure for genome-wide expression data Supplementary information Florent Baty 1 florent.baty@unibas.ch Michel P Bihl 1 michel.bihl@unibas.ch
 Supplementary Table 1. Annotated candidate SNV and indels in melanoma tumors. Patient C1 Patient C2 Patient C3 Patient C4 Patient C5 Patient C6 Patient C7 Patient C10 Chr Position Reference Allele Sample
Supplementary Table 1. Annotated candidate SNV and indels in melanoma tumors. Patient C1 Patient C2 Patient C3 Patient C4 Patient C5 Patient C6 Patient C7 Patient C10 Chr Position Reference Allele Sample
Strategy for Integrating Molecular Landscapes in Human Insulinomas
 Supplementary Figure 1 Strategy for Integrating Molecular Landscapes in Human Insulinomas 1 Supplementary Figure 2. Gene dendrogam and module colors 0.8 Height 0.9 1.00 module' The Complete Insulinoma
Supplementary Figure 1 Strategy for Integrating Molecular Landscapes in Human Insulinomas 1 Supplementary Figure 2. Gene dendrogam and module colors 0.8 Height 0.9 1.00 module' The Complete Insulinoma
