Supplementary information: Forstner, Hofmann et al.
|
|
|
- Martin Lawson
- 5 years ago
- Views:
Transcription
1 Supplementary information: Forstner, Hofmann et al., Genome-wide analysis implicates micrornas and their target genes in the development of bipolar disorder Table of contents Supplementary Figure 1: Regional association plot of mir Supplementary Figure 2: Regional association plot of mir Supplementary Figure 3: Regional association plot of mir-644 and mir Supplementary Figure 4: Regional association plot of let-7g and mir-135a Legend: Supplementary Figures Supplementary Figure 5: Directed Acyclic Graphs (DAG) of the enriched GO categories...7 Supplementary Figure 6: Validation of mir-499 and mir-708 processing using a dual-luciferase reporter assay in rat hippocampal neurons...9 Supplementary Table 1: Subcategory enrichment for different p value thresholds Supplementary Table 2: Comparison of the results of the gene-based tests based on different LD structure Supplementary Table 3: Significant biological pathways in microrna target gene data sets Supplementary Box 1: List of the 107 brain-expressed microrna target genes associated with bipolar disorder at gene-based p <
2 Supplementary Figure 1: Regional association plot of mir-640 2
3 Supplementary Figure 2: Regional association plot of mir-581 3
4 Supplementary Figure 3: Regional association plot of mir-644 and mir-499 4
5 Supplementary Figure 4: Regional association plot of let-7g and mir-135a-1 5
6 Legend Supplementary Figures 1-4: Regional association results for all associated mirnas and their +/- 500 kb flanking regions were plotted using LocusZoom (Pruim et al., 2010). A signal was considered mirna-associated if the Top SNP of the region was located at the mirna locus or if it was in high or moderate LD (r 2 >0.6) with the mirna locus. 6
7 Supplementary Figure 5: Directed Acyclic Graphs (DAG) of the enriched GO categories A B 7
8 C Legend Supplementary Figure 5: Target gene sets of the three micrornas 499, 708 and 1908 were subjected to gene ontology (GO) analysis. For each microrna the results of the GO analysis are presented as directed acyclic graphs. Significant results (p < 0.05, Bonferroni corrected) for pathways which contained at least three target genes are shown in red. A mir-499 B mir-708 and C mir
9 Supplementary Figure 6: Validation of mir-499 and mir-708 processing using a dual-luciferase reporter assay in rat hippocampal neurons Legend Supplementary Figure 6: Relative Luciferase activity (RLA) of DIV 10 primary rat hippocampal neurons transfected with the pmirglo dual Luciferase reporter construct containing indicated primary-mirna genes in the Firefly 3 UTR. Processing of the primary-mirna genes leads to a loss of the protective poly-a tail, resulting in a fast degradation of the Firefly mrna and therefore to reduced luciferase activity. Data are presented as the mean of three independent experiments normalized to the empty reporter construct ± standard deviation; n.s. = non significant *p>0.05 **p>0.01 ***p<0.005 (t-test). 9
10 Supplementary Table 1: Subcategory enrichment for different p value thresholds p value threshold SNPs in mirna Intergenic SNPs in genes +/- 20 kb SNPs <1 x x 10-4 > x 10-6 & <1 x x 10-7 > x 10-4 & < x > > x Legend Supplementary Table 1: Subcategory testing for different BD-association p value thresholds for the categories SNPs in mirna loci (+/- 20 kb), SNPs in genes, intergenic SNPs. Enrichment was calculated using a one-sided fisher s exact test. 10
11 Supplementary Table 2: Comparison of the results of the gene-based tests for the nine micrornas that withstood Bonferroni correction based on the LD structure derived from HapMap phase 2 or the 1,000 Genomes Project MicroRNA Chr Top SNP p Top SNP nsnps HM2 p Gene HM2 nsnps 1000G p Gene 1000G mir rs x x x 10-6 mir rs x x x 10-6 mir rs x x x 10-6 mir rs x x x 10-5 mir rs x x x 10-6 mir-135a-1 3 rs x x x 10-5 let-7g 3 rs x x x 10-5 mir rs x x x 10-5 mir rs x x x 10-5 Legend Supplementary Table 2: Abbreviations: Chr = Chromosome; p Top SNP = p value of the Top SNP within gene; nsnps = number of investigated SNPs; HM2 = based on HapMap phase 2 data, 1000G = based on the 1,000 Genomes Project data, p Gene = nominal gene-based p value. 11
12 Supplementary Table 3: Significant biological pathways in microrna target gene data sets microrna Type Pathway no. genes in subcategory no. target genes p corr. gene symbol mir-499 KEGG Regulation of actin cytoskeleton ENAH, VAV3, PFN2 GO forebrain development LMX1A, BCL11B, EPHA5, CNTNAP2, SLC8A3, ETS1, HOOK3, ZEB2 GO limbic system development LMX1A, EPHA5, CNTNAP2, ETS1, ZEB2 GO telencephalon development LMX1A, BCL11B, EPHA5, CNTNAP2, SLC8A3, ZEB2 GO protein binding GO brain development GO neuronal cell body GO cell body AAK1, H2AFZ, ENAH, CNTNAP2, VPS13A, PFN2, ETS1, AP3S1, EEA1, RIMS1, CACNB2, QKI, PIM1, MARCKS, TBC1D15, CPSF6, TOP1, VAV3, CHD9, SLC8A3, DYNLT1, HOOK3, HNRNPC, ILF3, PTCH1, EFHC1, WDR82, UHRF1BP1, GPC6, KCNN3, PURB, FKBP5, SOX5, PTPN14, SPAST, PRKAR1A, ZEB2 BCL11B, SLC8A3, HOOK3, PTCH1, LMX1A, EPHA5, CNTNAP2, ETS1, ZEB2 EPHA5, EFHC1, CNTNAP2, TANC1, SLC8A3, TOP1 GO synaptic vesicle transport RIMS1, AP3S1, PFN2, EEA1 GO central nervous system development BCL11B, SLC8A3, HOOK3, PTCH1, LMX1A, EPHA5, CNTNAP2, ETS1, SOX5, ZEB2 GO multicellular organismal process GO single-multicellular organism process BCL11B, ENAH, CNTNAP2, VPS13A, PFN2, ETS1, KCNQ5, AP3S1, EEA1, RIMS1, CACNB2, QKI, PIM1, TOP1, VAV3, SLC8A3, TMEM2, DYNLT1, JPH1, HOOK3, PTCH1, EPHA5, TANC1, KCNN3, LMX1A, PURB, PTPN14, SPAST, SOX5, ZEB2, PRKAR1A, RNF114, ROD1/PTBP3 mir-708 GO protein C-terminus binding ATXN1, SHANK3, PFKM, FOXN3 mir-1908 GO neuron projection MINK1, SLC12A5, SLC17A7, NCDN, KLC2 GO nervous system development MINK1, PCDHA1, MDGA1, OTX1, GDF11, NRGN, NCDN GO cell projection MINK1, SLC12A5, SLC17A7, NCDN, KLC2 GO neuronal cell body GO cell body SLC12A5, NCDN, EEF1A2 12
13 Legend Supplementary Table 3: Significant pathways with a minimum of three genes included per set are depicted in Supplementary Table 3. Abbreviations: KEGG = Kyoto Encyclopedia of Genes and Genomes; GO = Gene Ontology; no. genes in subcategory = number of reference genes in KEGG/GO subcategory; no. target genes = number of microrna target genes contained in subcategory; p corr = Bonferroni corrected p value. Supplementary Box 1: List of the 107 brain-expressed microrna target genes associated with bipolar disorder at genebased p < 0.05 mir-499-5p: GPC6, C16orf72, WDR82, CACNB2, BCL11B, HNRNPC, EFHC1, ILF3, SLC8A3, QKI, ANKRD40, MARCKS, DYNLT1, AAK1, PTCH1, PRKAR1A, EPHA5, TANC1, VAV3, SOX5, MTX3, CPSF6, FKBP5, LHFPL2, RIMS1, RBMS3, TMEM2, RRP1B, RSBN1, UHRF1BP1, EEA1, HOOK3, AP3S1, CHD9, UTP18, KCNQ5, C20orf112, MAST4, ETS1, RNF114, TOP1, CNTNAP2, ENAH, ZEB2, ACBD5, MAMDC2, JPH1, PURB, VPS13A, SPAST, TBC1D15, PTPN14, PIM1, LMX1A, KCNN3, PFN2, H2AFZ, C1orf151/MINOS1, ROD1/PTBP3 mir-708-5p: NRAS, CREB1, HNRNPC, PSMF1, RPGRIP1L, ATXN1, ALG9, RFT1, PFKM, QKI, GRIP2, AAK1, FOXN3, SLC44A5, STK4, LIF, MDGA1, AS3MT, FAM135B, BSN, ETF1, CDC42SE1, SHANK3, C20orf112, INTS7, GLG1, RORA, SSH2, ITPKB, SLC39A14, KPNA4, RNF165, HS2ST1, KCNN3, GOLT1A, C1orf144/SZRD1 mir p: KLC2, NCDN, MDGA1, SLC12A5, HPD, NRGN, MINK1, PCDHA1, GDF11, OTX1, EEF1A2, SLC17A7 13
Supplementary Figure 1: Attenuation of association signals after conditioning for the lead SNP. a) attenuation of association signal at the 9p22.
 Supplementary Figure 1: Attenuation of association signals after conditioning for the lead SNP. a) attenuation of association signal at the 9p22.32 PCOS locus after conditioning for the lead SNP rs10993397;
Supplementary Figure 1: Attenuation of association signals after conditioning for the lead SNP. a) attenuation of association signal at the 9p22.32 PCOS locus after conditioning for the lead SNP rs10993397;
Quantitative Real Time PCR (RT-qPCR) Differentiation of human preadipocytes to adipocytes
 Quantitative Real Time PCR (RT-qPCR) First strand cdna was synthesized and RT-qPCR was performed using RT2 first strand kits (Cat#330401) and RT2 qpcr Master Mix (SABioscience, Frederick, MD). Experiments
Quantitative Real Time PCR (RT-qPCR) First strand cdna was synthesized and RT-qPCR was performed using RT2 first strand kits (Cat#330401) and RT2 qpcr Master Mix (SABioscience, Frederick, MD). Experiments
mir-7a regulation of Pax6 in neural stem cells controls the spatial origin of forebrain dopaminergic neurons
 Supplemental Material mir-7a regulation of Pax6 in neural stem cells controls the spatial origin of forebrain dopaminergic neurons Antoine de Chevigny, Nathalie Coré, Philipp Follert, Marion Gaudin, Pascal
Supplemental Material mir-7a regulation of Pax6 in neural stem cells controls the spatial origin of forebrain dopaminergic neurons Antoine de Chevigny, Nathalie Coré, Philipp Follert, Marion Gaudin, Pascal
Table S2. Baseline characteristics of the Rotterdam Study for interaction analysis
 Supplementary data Table S1. List of the primers for the cloning of precursor mir-4707 Table S2. Baseline characteristics of the Rotterdam Study for interaction analysis Table S3. List of the primers for
Supplementary data Table S1. List of the primers for the cloning of precursor mir-4707 Table S2. Baseline characteristics of the Rotterdam Study for interaction analysis Table S3. List of the primers for
Supplementary Figure 1. mir124 does not change neuron morphology and synaptic
 Supplementary Figure 1. mir124 does not change neuron morphology and synaptic density. Hippocampal neurons were transfected with mir124 (containing DsRed) or DsRed as a control. 2 d after transfection,
Supplementary Figure 1. mir124 does not change neuron morphology and synaptic density. Hippocampal neurons were transfected with mir124 (containing DsRed) or DsRed as a control. 2 d after transfection,
Supplementary Figure S1. Gene expression analysis of epidermal marker genes and TP63.
 Supplementary Figure Legends Supplementary Figure S1. Gene expression analysis of epidermal marker genes and TP63. A. Screenshot of the UCSC genome browser from normalized RNAPII and RNA-seq ChIP-seq data
Supplementary Figure Legends Supplementary Figure S1. Gene expression analysis of epidermal marker genes and TP63. A. Screenshot of the UCSC genome browser from normalized RNAPII and RNA-seq ChIP-seq data
Supplementary Figure 1. Quantile-quantile (Q-Q) plots. (Panel A) Q-Q plot graphical
 Supplementary Figure 1. Quantile-quantile (Q-Q) plots. (Panel A) Q-Q plot graphical representation using all SNPs (n= 13,515,798) including the region on chromosome 1 including SORT1 which was previously
Supplementary Figure 1. Quantile-quantile (Q-Q) plots. (Panel A) Q-Q plot graphical representation using all SNPs (n= 13,515,798) including the region on chromosome 1 including SORT1 which was previously
Contribution of the intronic mir-338-3p and its Hosting Gene AATK to Compensatory β-cell Mass Expansion
 Contribution of the intronic mir-338-3p and its Hosting Gene AATK to Compensatory β-cell Mass Expansion Cécile Jacovetti 1, Veronica Jimenez 2, Eduard Ayuso 2#, Ross Laybutt 3, Marie-Line Peyot 4, Marc
Contribution of the intronic mir-338-3p and its Hosting Gene AATK to Compensatory β-cell Mass Expansion Cécile Jacovetti 1, Veronica Jimenez 2, Eduard Ayuso 2#, Ross Laybutt 3, Marie-Line Peyot 4, Marc
Gene-microRNA network module analysis for ovarian cancer
 Gene-microRNA network module analysis for ovarian cancer Shuqin Zhang School of Mathematical Sciences Fudan University Oct. 4, 2016 Outline Introduction Materials and Methods Results Conclusions Introduction
Gene-microRNA network module analysis for ovarian cancer Shuqin Zhang School of Mathematical Sciences Fudan University Oct. 4, 2016 Outline Introduction Materials and Methods Results Conclusions Introduction
Supplementary Figure S1A
 Supplementary Figure S1A-G. LocusZoom regional association plots for the seven new cross-cancer loci that were > 1 Mb from known index SNPs. Genes up to 500 kb on either side of each new index SNP are
Supplementary Figure S1A-G. LocusZoom regional association plots for the seven new cross-cancer loci that were > 1 Mb from known index SNPs. Genes up to 500 kb on either side of each new index SNP are
a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation,
 Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
Huntington s Disease and its therapeutic target genes: A global functional profile based on the HD Research Crossroads database
 Supplementary Analyses and Figures Huntington s Disease and its therapeutic target genes: A global functional profile based on the HD Research Crossroads database Ravi Kiran Reddy Kalathur, Miguel A. Hernández-Prieto
Supplementary Analyses and Figures Huntington s Disease and its therapeutic target genes: A global functional profile based on the HD Research Crossroads database Ravi Kiran Reddy Kalathur, Miguel A. Hernández-Prieto
SUPPLEMENTARY FIGURE LEGENDS
 SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1 Negative correlation between mir-375 and its predicted target genes, as demonstrated by gene set enrichment analysis (GSEA). 1 The correlation between
SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1 Negative correlation between mir-375 and its predicted target genes, as demonstrated by gene set enrichment analysis (GSEA). 1 The correlation between
SUPPLEMENTARY INFORMATION. Supp. Fig. 1. Autoimmunity. Tolerance APC APC. T cell. T cell. doi: /nature06253 ICOS ICOS TCR CD28 TCR CD28
 Supp. Fig. 1 a APC b APC ICOS ICOS TCR CD28 mir P TCR CD28 P T cell Tolerance Roquin WT SG Icos mrna T cell Autoimmunity Roquin M199R SG Icos mrna www.nature.com/nature 1 Supp. Fig. 2 CD4 + CD44 low CD4
Supp. Fig. 1 a APC b APC ICOS ICOS TCR CD28 mir P TCR CD28 P T cell Tolerance Roquin WT SG Icos mrna T cell Autoimmunity Roquin M199R SG Icos mrna www.nature.com/nature 1 Supp. Fig. 2 CD4 + CD44 low CD4
A genome-wide association study identifies vitiligo
 A genome-wide association study identifies vitiligo susceptibility loci at MHC and 6q27 Supplementary Materials Index Supplementary Figure 1 The principal components analysis (PCA) of 2,546 GWAS samples
A genome-wide association study identifies vitiligo susceptibility loci at MHC and 6q27 Supplementary Materials Index Supplementary Figure 1 The principal components analysis (PCA) of 2,546 GWAS samples
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1. Differential expression of mirnas from the pri-mir-17-92a locus.
 Supplementary Figure 1 Differential expression of mirnas from the pri-mir-17-92a locus. (a) The mir-17-92a expression unit in the third intron of the host mir-17hg transcript. (b,c) Impact of knockdown
Supplementary Figure 1 Differential expression of mirnas from the pri-mir-17-92a locus. (a) The mir-17-92a expression unit in the third intron of the host mir-17hg transcript. (b,c) Impact of knockdown
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells
 MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
Chapter 2. Investigation into mir-346 Regulation of the nachr α5 Subunit
 15 Chapter 2 Investigation into mir-346 Regulation of the nachr α5 Subunit MicroRNA s (mirnas) are small (< 25 base pairs), single stranded, non-coding RNAs that regulate gene expression at the post transcriptional
15 Chapter 2 Investigation into mir-346 Regulation of the nachr α5 Subunit MicroRNA s (mirnas) are small (< 25 base pairs), single stranded, non-coding RNAs that regulate gene expression at the post transcriptional
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10860 Supplementary Discussion It remains unclear why H3K9 demethylation appeared to be more sensitive to suppression than at least some other histone methylation marks as a result of
doi:10.1038/nature10860 Supplementary Discussion It remains unclear why H3K9 demethylation appeared to be more sensitive to suppression than at least some other histone methylation marks as a result of
SUPPLEMENTARY INFORMATION. Meta-analyses of genome-wide association studies identify multiple novel loci related to pulmonary function
 SUPPLEMENTARY INFORMATION Meta-analyses of genome-wide association studies identify multiple novel loci related to pulmonary function Dana B. Hancock, 1,29 Mark Eijgelsheim, 2,29 Jemma B. Wilk, 3,29 Sina
SUPPLEMENTARY INFORMATION Meta-analyses of genome-wide association studies identify multiple novel loci related to pulmonary function Dana B. Hancock, 1,29 Mark Eijgelsheim, 2,29 Jemma B. Wilk, 3,29 Sina
Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v)
 SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
Supplementary note: Comparison of deletion variants identified in this study and four earlier studies
 Supplementary note: Comparison of deletion variants identified in this study and four earlier studies Here we compare the results of this study to potentially overlapping results from four earlier studies
Supplementary note: Comparison of deletion variants identified in this study and four earlier studies Here we compare the results of this study to potentially overlapping results from four earlier studies
Supplementary Figure 1:
 Supplementary Figure 1: (A) Whole aortic cross-sections stained with Hematoxylin and Eosin (H&E), 7 days after porcine-pancreatic-elastase (PPE)-induced AAA compared to untreated, healthy control aortas
Supplementary Figure 1: (A) Whole aortic cross-sections stained with Hematoxylin and Eosin (H&E), 7 days after porcine-pancreatic-elastase (PPE)-induced AAA compared to untreated, healthy control aortas
Appendix table of content: Appendix Table S1. Appendix Table S2. Appendix Figure S1. Appendix Figure S2. Appendix Methods.
 Appendix table of content: Appendix Table S Appendix Table S Appendix Figure S Appendix Figure S Appendix Methods Appendix Legends Appendix Methods mir--3p putative targets in silico analysis: Predictions
Appendix table of content: Appendix Table S Appendix Table S Appendix Figure S Appendix Figure S Appendix Methods Appendix Legends Appendix Methods mir--3p putative targets in silico analysis: Predictions
Large-Scale Discovery of Enhancers from Human Heart Tissue
 Large-Scale Discovery of Enhancers from Human Heart Tissue Dalit May, Matthew J. Blow, Tommy Kaplan, David J. McCulley, Brian C. Jensen, Jennifer A. Akiyama, Amy Holt, Ingrid Plajzer-Frick, Malak Shoukry,
Large-Scale Discovery of Enhancers from Human Heart Tissue Dalit May, Matthew J. Blow, Tommy Kaplan, David J. McCulley, Brian C. Jensen, Jennifer A. Akiyama, Amy Holt, Ingrid Plajzer-Frick, Malak Shoukry,
Supporting Information
 Supporting Information Retinal expression of small non-coding RNAs in a murine model of proliferative retinopathy Chi-Hsiu Liu 1, Zhongxiao Wang 1, Ye Sun 1, John Paul SanGiovanni 2, Jing Chen 1, * 1 Department
Supporting Information Retinal expression of small non-coding RNAs in a murine model of proliferative retinopathy Chi-Hsiu Liu 1, Zhongxiao Wang 1, Ye Sun 1, John Paul SanGiovanni 2, Jing Chen 1, * 1 Department
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES 1 Supplementary Figure 1, Adult hippocampal QNPs and TAPs uniformly express REST a-b) Confocal images of adult hippocampal mouse sections showing GFAP (green), Sox2 (red), and REST
SUPPLEMENTARY FIGURES 1 Supplementary Figure 1, Adult hippocampal QNPs and TAPs uniformly express REST a-b) Confocal images of adult hippocampal mouse sections showing GFAP (green), Sox2 (red), and REST
Comprehensive Replication of the Relationship Between Myopia-Related Genes and Refractive Errors in a Large Japanese Cohort
 Genetics Comprehensive Replication of the Relationship Between Myopia-Related Genes and Refractive Errors in a Large Japanese Cohort Munemitsu Yoshikawa, 1 Kenji Yamashiro, 1 Masahiro Miyake, 1,2 Maho
Genetics Comprehensive Replication of the Relationship Between Myopia-Related Genes and Refractive Errors in a Large Japanese Cohort Munemitsu Yoshikawa, 1 Kenji Yamashiro, 1 Masahiro Miyake, 1,2 Maho
Selective filtering defect at the axon initial segment in Alzheimer s disease mouse models. Yu Wu
 Selective filtering defect at the axon initial segment in Alzheimer s disease mouse models Yu Wu Alzheimer s Disease (AD) Mouse models: APP/PS1, PS1δE9, APPswe, hps1 Wirths, O. et al, Acta neuropathologica
Selective filtering defect at the axon initial segment in Alzheimer s disease mouse models Yu Wu Alzheimer s Disease (AD) Mouse models: APP/PS1, PS1δE9, APPswe, hps1 Wirths, O. et al, Acta neuropathologica
Genetic variants in the bipolar disorder risk locus SYNE1 that affect CPG2 expression and protein function
 Molecular Psychiatry https://doi.org/1.138/s4138-18-314-z ARTICLE Genetic variants in the bipolar disorder risk locus SYNE1 that affect CPG2 expression and protein function Mette Rathje 1 Hannah Waxman
Molecular Psychiatry https://doi.org/1.138/s4138-18-314-z ARTICLE Genetic variants in the bipolar disorder risk locus SYNE1 that affect CPG2 expression and protein function Mette Rathje 1 Hannah Waxman
Figure S1. Reduction in glomerular mir-146a levels correlate with progression to higher albuminuria in diabetic patients.
 Supplementary Materials Supplementary Figures Figure S1. Reduction in glomerular mir-146a levels correlate with progression to higher albuminuria in diabetic patients. Figure S2. Expression level of podocyte
Supplementary Materials Supplementary Figures Figure S1. Reduction in glomerular mir-146a levels correlate with progression to higher albuminuria in diabetic patients. Figure S2. Expression level of podocyte
Nature Medicine: doi: /nm.4439
 Figure S1. Overview of the variant calling and verification process. This figure expands on Fig. 1c with details of verified variants identification in 547 additional validation samples. Somatic variants
Figure S1. Overview of the variant calling and verification process. This figure expands on Fig. 1c with details of verified variants identification in 547 additional validation samples. Somatic variants
Supplementary information
 Supplementary information Human Cytomegalovirus MicroRNA mir-us4-1 Inhibits CD8 + T Cell Response by Targeting ERAP1 Sungchul Kim, Sanghyun Lee, Jinwook Shin, Youngkyun Kim, Irini Evnouchidou, Donghyun
Supplementary information Human Cytomegalovirus MicroRNA mir-us4-1 Inhibits CD8 + T Cell Response by Targeting ERAP1 Sungchul Kim, Sanghyun Lee, Jinwook Shin, Youngkyun Kim, Irini Evnouchidou, Donghyun
Nature Genetics: doi: /ng.3731
 Supplementary Figure 1 Circadian profiles of Adarb1 transcript and ADARB1 protein in mouse tissues. (a) Overlap of rhythmic transcripts identified in the previous transcriptome analyses. The mouse liver
Supplementary Figure 1 Circadian profiles of Adarb1 transcript and ADARB1 protein in mouse tissues. (a) Overlap of rhythmic transcripts identified in the previous transcriptome analyses. The mouse liver
Nature Immunology: doi: /ni Supplementary Figure 1. Huwe1 has high expression in HSCs and is necessary for quiescence.
 Supplementary Figure 1 Huwe1 has high expression in HSCs and is necessary for quiescence. (a) Heat map visualizing expression of genes with a known function in ubiquitin-mediated proteolysis (KEGG: Ubiquitin
Supplementary Figure 1 Huwe1 has high expression in HSCs and is necessary for quiescence. (a) Heat map visualizing expression of genes with a known function in ubiquitin-mediated proteolysis (KEGG: Ubiquitin
Supplementary Figure 1. The mir-182 binding site of SMAD7 3 UTR and the. mutated sequence.
 Supplementary Figure 1. The mir-182 binding site of SMAD7 3 UTR and the mutated sequence. 1 Supplementary Figure 2. Expression of mir-182 and SMAD7 in various cell lines. (A) Basal levels of mir-182 expression
Supplementary Figure 1. The mir-182 binding site of SMAD7 3 UTR and the mutated sequence. 1 Supplementary Figure 2. Expression of mir-182 and SMAD7 in various cell lines. (A) Basal levels of mir-182 expression
Additional file: Cell cycle, oncogenic and tumor suppressor pathways regulate numerous long and macro non-protein coding RNAs
 Additional file: Cell cycle, oncogenic and tumor suppressor pathways regulate numerous long and macro non-protein coding RNAs February 6, 2014 Jö rg Hackermü ller 1,2,,, Kristin Reiche 1,2,,, Christian
Additional file: Cell cycle, oncogenic and tumor suppressor pathways regulate numerous long and macro non-protein coding RNAs February 6, 2014 Jö rg Hackermü ller 1,2,,, Kristin Reiche 1,2,,, Christian
Supplementary Information
 Supplementary Information Index of Supplementary Materials Supplementary Figure. Distribution of methylation levels, characteristics of cis-meqtls and association plots for meqtls in methylation-related
Supplementary Information Index of Supplementary Materials Supplementary Figure. Distribution of methylation levels, characteristics of cis-meqtls and association plots for meqtls in methylation-related
Circular RNAs (circrnas) act a stable mirna sponges
 Circular RNAs (circrnas) act a stable mirna sponges cernas compete for mirnas Ancestal mrna (+3 UTR) Pseudogene RNA (+3 UTR homolgy region) The model holds true for all RNAs that share a mirna binding
Circular RNAs (circrnas) act a stable mirna sponges cernas compete for mirnas Ancestal mrna (+3 UTR) Pseudogene RNA (+3 UTR homolgy region) The model holds true for all RNAs that share a mirna binding
injected subcutaneously into flanks of 6-8 week old athymic male nude mice (LNCaP SQ) and body
 SUPPLEMENTAL FIGURE LEGENDS Figure S1: Generation of ENZR Xenografts and Cell Lines: (A) 1x10 6 LNCaP cells in matrigel were injected subcutaneously into flanks of 6-8 week old athymic male nude mice (LNCaP
SUPPLEMENTAL FIGURE LEGENDS Figure S1: Generation of ENZR Xenografts and Cell Lines: (A) 1x10 6 LNCaP cells in matrigel were injected subcutaneously into flanks of 6-8 week old athymic male nude mice (LNCaP
Supplementary Material
 Supplementary Material Summary: The supplementary information includes 1 table (Table S1) and 4 figures (Figure S1 to S4). Supplementary Figure Legends Figure S1 RTL-bearing nude mouse model. (A) Tumor
Supplementary Material Summary: The supplementary information includes 1 table (Table S1) and 4 figures (Figure S1 to S4). Supplementary Figure Legends Figure S1 RTL-bearing nude mouse model. (A) Tumor
Integrated network analysis reveals distinct regulatory roles of transcription factors and micrornas
 Integrated network analysis reveals distinct regulatory roles of transcription factors and micrornas Yu Guo 1,2,4, Katherine Alexander 1, Andrew G Clark 1, Andrew Grimson 1 and Haiyuan Yu 2,3* 1 Department
Integrated network analysis reveals distinct regulatory roles of transcription factors and micrornas Yu Guo 1,2,4, Katherine Alexander 1, Andrew G Clark 1, Andrew Grimson 1 and Haiyuan Yu 2,3* 1 Department
Global variation in copy number in the human genome
 Global variation in copy number in the human genome Redon et. al. Nature 444:444-454 (2006) 12.03.2007 Tarmo Puurand Study 270 individuals (HapMap collection) Affymetrix 500K Whole Genome TilePath (WGTP)
Global variation in copy number in the human genome Redon et. al. Nature 444:444-454 (2006) 12.03.2007 Tarmo Puurand Study 270 individuals (HapMap collection) Affymetrix 500K Whole Genome TilePath (WGTP)
Supplementary Figure (OH) 22 nanoparticles did not affect cell viability and apoposis. MDA-MB-231, MCF-7, MCF-10A and BT549 cells were
 Supplementary Figure 1. Gd@C 82 (OH) 22 nanoparticles did not affect cell viability and apoposis. MDA-MB-231, MCF-7, MCF-10A and BT549 cells were treated with PBS, Gd@C 82 (OH) 22, C 60 (OH) 22 or GdCl
Supplementary Figure 1. Gd@C 82 (OH) 22 nanoparticles did not affect cell viability and apoposis. MDA-MB-231, MCF-7, MCF-10A and BT549 cells were treated with PBS, Gd@C 82 (OH) 22, C 60 (OH) 22 or GdCl
Supplementary. properties of. network types. randomly sampled. subsets (75%
 Supplementary Information Gene co-expression network analysis reveals common system-level prognostic genes across cancer types properties of Supplementary Figure 1 The robustness and overlap of prognostic
Supplementary Information Gene co-expression network analysis reveals common system-level prognostic genes across cancer types properties of Supplementary Figure 1 The robustness and overlap of prognostic
Palindromic amplification of the ERBB2 oncogene in human primary breast tumors. A common pattern of copy number transition of chromosome 17 in HER2-
 Supplemental Figures Table of Contents Palindromic amplification of the ERBB2 oncogene in human primary breast tumors Michael Marotta 1,5, Taku Onodera 3,5, Jeffrey Johnson 4, Thomas Budd 2, Takaaki Watanabe
Supplemental Figures Table of Contents Palindromic amplification of the ERBB2 oncogene in human primary breast tumors Michael Marotta 1,5, Taku Onodera 3,5, Jeffrey Johnson 4, Thomas Budd 2, Takaaki Watanabe
c Tuj1(-) apoptotic live 1 DIV 2 DIV 1 DIV 2 DIV Tuj1(+) Tuj1/GFP/DAPI Tuj1 DAPI GFP
 Supplementary Figure 1 Establishment of the gain- and loss-of-function experiments and cell survival assays. a Relative expression of mature mir-484 30 20 10 0 **** **** NCP mir- 484P NCP mir- 484P b Relative
Supplementary Figure 1 Establishment of the gain- and loss-of-function experiments and cell survival assays. a Relative expression of mature mir-484 30 20 10 0 **** **** NCP mir- 484P NCP mir- 484P b Relative
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2566 Figure S1 CDKL5 protein expression pattern and localization in mouse brain. (a) Multiple-tissue western blot from a postnatal day (P) 21 mouse probed with an antibody against CDKL5.
DOI: 10.1038/ncb2566 Figure S1 CDKL5 protein expression pattern and localization in mouse brain. (a) Multiple-tissue western blot from a postnatal day (P) 21 mouse probed with an antibody against CDKL5.
(A) Cells grown in monolayer were fixed and stained for surfactant protein-c (SPC,
 Supplemental Figure Legends Figure S1. Cell line characterization (A) Cells grown in monolayer were fixed and stained for surfactant protein-c (SPC, green) and co-stained with DAPI to visualize the nuclei.
Supplemental Figure Legends Figure S1. Cell line characterization (A) Cells grown in monolayer were fixed and stained for surfactant protein-c (SPC, green) and co-stained with DAPI to visualize the nuclei.
mir-509-5p and mir-1243 increase the sensitivity to gemcitabine by inhibiting
 mir-509-5p and mir-1243 increase the sensitivity to gemcitabine by inhibiting epithelial-mesenchymal transition in pancreatic cancer Hidekazu Hiramoto, M.D. 1,3, Tomoki Muramatsu, Ph.D. 1, Daisuke Ichikawa,
mir-509-5p and mir-1243 increase the sensitivity to gemcitabine by inhibiting epithelial-mesenchymal transition in pancreatic cancer Hidekazu Hiramoto, M.D. 1,3, Tomoki Muramatsu, Ph.D. 1, Daisuke Ichikawa,
Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor suppressor genes
 Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor suppressor genes Kaifu Chen 1,2,3,4,5,10, Zhong Chen 6,10, Dayong Wu 6, Lili Zhang 7, Xueqiu Lin 1,2,8,
Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor suppressor genes Kaifu Chen 1,2,3,4,5,10, Zhong Chen 6,10, Dayong Wu 6, Lili Zhang 7, Xueqiu Lin 1,2,8,
Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression.
 Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Supplementary Table 1. Candidate loci that were associated with diabetes- and/or obesity-related traits.
 Supplementary Table 1. Candidate loci that were associated with diabetes- and/or obesity-related traits. All With Fst in the With long With SNP(s) at the 5' top 5% bracket haplotypes regulatory region
Supplementary Table 1. Candidate loci that were associated with diabetes- and/or obesity-related traits. All With Fst in the With long With SNP(s) at the 5' top 5% bracket haplotypes regulatory region
MicroRNA in Cancer Karen Dybkær 2013
 MicroRNA in Cancer Karen Dybkær RNA Ribonucleic acid Types -Coding: messenger RNA (mrna) coding for proteins -Non-coding regulating protein formation Ribosomal RNA (rrna) Transfer RNA (trna) Small nuclear
MicroRNA in Cancer Karen Dybkær RNA Ribonucleic acid Types -Coding: messenger RNA (mrna) coding for proteins -Non-coding regulating protein formation Ribosomal RNA (rrna) Transfer RNA (trna) Small nuclear
A genome-wide association study identifies two new cervical cancer susceptibility loci at 4q12 and 17q12. Supplementary Materials
 A genome-wide association study identifies two new cervical cancer susceptibility loci at 4q12 and 17q12 Supplementary Materials Yongyong Shi 1-4,42, Li Li 5,42, Zhibin Hu 6,7,42, Shuang Li 1,42, Shixuan
A genome-wide association study identifies two new cervical cancer susceptibility loci at 4q12 and 17q12 Supplementary Materials Yongyong Shi 1-4,42, Li Li 5,42, Zhibin Hu 6,7,42, Shuang Li 1,42, Shixuan
Somatic intronic microsatellite loci differentiate glioblastoma from lower-grade gliomas
 1 Somatic intronic microsatellite loci differentiate glioblastoma from lower-grade gliomas 2 3 FIGURE S1: Sensitivity & Specificity receiver operator characteristics (ROC) of Signature Cancer-Associated
1 Somatic intronic microsatellite loci differentiate glioblastoma from lower-grade gliomas 2 3 FIGURE S1: Sensitivity & Specificity receiver operator characteristics (ROC) of Signature Cancer-Associated
Nature Genetics: doi: /ng Supplementary Figure 1. Study design.
 Supplementary Figure 1 Study design. Leukopenia was classified as early when it occurred within the first 8 weeks of thiopurine therapy and as late when it occurred more than 8 weeks after the start of
Supplementary Figure 1 Study design. Leukopenia was classified as early when it occurred within the first 8 weeks of thiopurine therapy and as late when it occurred more than 8 weeks after the start of
Supplementary Figure 1. Quantile-quantile (Q-Q) plot of the log 10 p-value association results from logistic regression models for prostate cancer
 Supplementary Figure 1. Quantile-quantile (Q-Q) plot of the log 10 p-value association results from logistic regression models for prostate cancer risk in stage 1 (red) and after removing any SNPs within
Supplementary Figure 1. Quantile-quantile (Q-Q) plot of the log 10 p-value association results from logistic regression models for prostate cancer risk in stage 1 (red) and after removing any SNPs within
Supplemental Table 1. List of genes and mature shrna sequences identified from the shrna screen in BRCA2-mutant PEO1 cells.
 Guillemette_Supplemental Table 1 Gene Mature Sequence Gene Mature Sequence ATTCATAGGATGTCAGCAG LOC157193 TATGCGTAAGTAATAAATTGCT TTAGTTCTGTCTTGGAGG LOC152225 CGCATATATGTTCTGACTT ALDH2 CACTTCAGTGTATGCCT
Guillemette_Supplemental Table 1 Gene Mature Sequence Gene Mature Sequence ATTCATAGGATGTCAGCAG LOC157193 TATGCGTAAGTAATAAATTGCT TTAGTTCTGTCTTGGAGG LOC152225 CGCATATATGTTCTGACTT ALDH2 CACTTCAGTGTATGCCT
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/2/1/ra81/dc1 Supplementary Materials for Delivery of MicroRNA-126 by Apoptotic Bodies Induces CXCL12- Dependent Vascular Protection Alma Zernecke,* Kiril Bidzhekov,
www.sciencesignaling.org/cgi/content/full/2/1/ra81/dc1 Supplementary Materials for Delivery of MicroRNA-126 by Apoptotic Bodies Induces CXCL12- Dependent Vascular Protection Alma Zernecke,* Kiril Bidzhekov,
Gene Ontology 2 Function/Pathway Enrichment. Biol4559 Thurs, April 12, 2018 Bill Pearson Pinn 6-057
 Gene Ontology 2 Function/Pathway Enrichment Biol4559 Thurs, April 12, 2018 Bill Pearson wrp@virginia.edu 4-2818 Pinn 6-057 Function/Pathway enrichment analysis do sets (subsets) of differentially expressed
Gene Ontology 2 Function/Pathway Enrichment Biol4559 Thurs, April 12, 2018 Bill Pearson wrp@virginia.edu 4-2818 Pinn 6-057 Function/Pathway enrichment analysis do sets (subsets) of differentially expressed
Supplementary Figure S1: Pial and periventricular vascular networks and the dual GABA neuron streams of the early embryonic mouse telencephalon
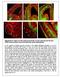 Supplementary Figure S1: Pial and periventricular vascular networks and the dual GABA neuron streams of the early embryonic mouse telencephalon a) The profile of GABA neurons arriving at the pallial-subpallial
Supplementary Figure S1: Pial and periventricular vascular networks and the dual GABA neuron streams of the early embryonic mouse telencephalon a) The profile of GABA neurons arriving at the pallial-subpallial
Research Article Identifying Liver Cancer-Related Enhancer SNPs by Integrating GWAS and Histone Modification ChIP-seq Data
 BioMed Volume 2016, Article ID 2395341, 6 pages http://dx.doi.org/10.1155/2016/2395341 Research Article Identifying Liver Cancer-Related Enhancer SNPs by Integrating GWAS and Histone Modification ChIP-seq
BioMed Volume 2016, Article ID 2395341, 6 pages http://dx.doi.org/10.1155/2016/2395341 Research Article Identifying Liver Cancer-Related Enhancer SNPs by Integrating GWAS and Histone Modification ChIP-seq
sirna count per 50 kb small RNAs matching the direct strand Repeat length (bp) per 50 kb repeats in the chromosome
 Qi et al. 26-3-2564C Qi et al., Figure S1 sirna count per 5 kb small RNAs matching the direct strand sirna count per 5 kb small RNAs matching the complementary strand Repeat length (bp) per 5 kb repeats
Qi et al. 26-3-2564C Qi et al., Figure S1 sirna count per 5 kb small RNAs matching the direct strand sirna count per 5 kb small RNAs matching the complementary strand Repeat length (bp) per 5 kb repeats
Novel Insights into Breast Cancer Genetic Variance through RNA Sequencing
 Insights into Breast Cancer Genetic Variance through RNA Sequencing SUPPLEMENTARY DATA Anelia Horvath, 1,2* Suresh Babu Pakala, 2* Prakriti Mudvari, 1* Sirigiri Divijendra Natha Reddy, 2 Kazufumi Ohshiro,
Insights into Breast Cancer Genetic Variance through RNA Sequencing SUPPLEMENTARY DATA Anelia Horvath, 1,2* Suresh Babu Pakala, 2* Prakriti Mudvari, 1* Sirigiri Divijendra Natha Reddy, 2 Kazufumi Ohshiro,
SUPPLEMENTARY INFORMATION
 doi: 1.138/nature89 IFN- (ng ml ) 5 4 3 1 Splenocytes NS IFN- (ng ml ) 6 4 Lymph node cells NS Nfkbiz / Nfkbiz / Nfkbiz / Nfkbiz / IL- (ng ml ) 3 1 Splenocytes IL- (ng ml ) 1 8 6 4 *** ** Lymph node cells
doi: 1.138/nature89 IFN- (ng ml ) 5 4 3 1 Splenocytes NS IFN- (ng ml ) 6 4 Lymph node cells NS Nfkbiz / Nfkbiz / Nfkbiz / Nfkbiz / IL- (ng ml ) 3 1 Splenocytes IL- (ng ml ) 1 8 6 4 *** ** Lymph node cells
QTL Studies- Past, Present and Future. David Evans
 QTL Studies Past, Present and Future David Evans Genetic studies of complex diseases have not met anticipated success Glazier et al, Science (2002) 298:23452349 Korstanje & Pagan (2002) Nat Genet Korstanje
QTL Studies Past, Present and Future David Evans Genetic studies of complex diseases have not met anticipated success Glazier et al, Science (2002) 298:23452349 Korstanje & Pagan (2002) Nat Genet Korstanje
Lions and Tigers and Bears.Oh My!
 Lions and Tigers and Bears.Oh My! TAKING THE FEAR OUT OF GENETIC TESTING Lisa Butterfield, MS, CGC Genetic Counselor Clinical Assistant Professor University of Kansas Health System Why is Genetic Testing
Lions and Tigers and Bears.Oh My! TAKING THE FEAR OUT OF GENETIC TESTING Lisa Butterfield, MS, CGC Genetic Counselor Clinical Assistant Professor University of Kansas Health System Why is Genetic Testing
Supplementary Figure 1. Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Nature Immunology: doi: /ni.
 Supplementary Figure 1 Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Expression of Mll4 floxed alleles (16-19) in naive CD4 + T cells isolated from lymph nodes and
Supplementary Figure 1 Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Expression of Mll4 floxed alleles (16-19) in naive CD4 + T cells isolated from lymph nodes and
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Effect of HSP90 inhibition on expression of endogenous retroviruses. (a) Inducible shrna-mediated Hsp90 silencing in mouse ESCs. Immunoblots of total cell extract expressing the
Supplementary Figure 1 Effect of HSP90 inhibition on expression of endogenous retroviruses. (a) Inducible shrna-mediated Hsp90 silencing in mouse ESCs. Immunoblots of total cell extract expressing the
Supplementary Figures. Supplementary Figure 1. Dinucleotide variant proportions. These are described and quantitated. for each lesion type.
 Supplementary Figures Supplementary Figure 1. Dinucleotide variant proportions. These are described and quantitated for each lesion type. a b Supplementary Figure 2. Non-negative matrix factorization-derived
Supplementary Figures Supplementary Figure 1. Dinucleotide variant proportions. These are described and quantitated for each lesion type. a b Supplementary Figure 2. Non-negative matrix factorization-derived
Supplementary Files. Novel blood-based microrna biomarker panel for early diagnosis of chronic pancreatitis
 Supplementary Files Novel blood-based microrna biomarker panel for early diagnosis of chronic pancreatitis Lei Xin 1 *, M.D., Jun Gao 1 *, Ph.D., Dan Wang 1 *, M.D., Jin-Huan Lin 1, M.D., Zhuan Liao 1,
Supplementary Files Novel blood-based microrna biomarker panel for early diagnosis of chronic pancreatitis Lei Xin 1 *, M.D., Jun Gao 1 *, Ph.D., Dan Wang 1 *, M.D., Jin-Huan Lin 1, M.D., Zhuan Liao 1,
York criteria, 6 RA patients and 10 age- and gender-matched healthy controls (HCs).
 MATERIALS AND METHODS Study population Blood samples were obtained from 15 patients with AS fulfilling the modified New York criteria, 6 RA patients and 10 age- and gender-matched healthy controls (HCs).
MATERIALS AND METHODS Study population Blood samples were obtained from 15 patients with AS fulfilling the modified New York criteria, 6 RA patients and 10 age- and gender-matched healthy controls (HCs).
FH- FH+ DM. 52 Volunteers. Oral & IV Glucose Tolerance Test Hyperinsulinemic Euglycemic Clamp in Non-DM Subjects ACADSB MYSM1. Mouse Skeletal Muscle
 A 52 Volunteers B 6 5 4 3 2 FH- FH+ DM 1 Oral & IV Glucose Tolerance Test Hyperinsulinemic Euglycemic Clamp in Non-DM Subjects ZYX EGR2 NR4A1 SRF target TPM1 ACADSB MYSM1 Non SRF target FH- FH+ DM2 C SRF
A 52 Volunteers B 6 5 4 3 2 FH- FH+ DM 1 Oral & IV Glucose Tolerance Test Hyperinsulinemic Euglycemic Clamp in Non-DM Subjects ZYX EGR2 NR4A1 SRF target TPM1 ACADSB MYSM1 Non SRF target FH- FH+ DM2 C SRF
HLA-Cw*16:02. rs Supplementary Figure 1: Genetic association of imputed markers and HLA types in the MHC region.
 -log10 P HLA-B*51 rs1050502 rs116799036 HLA-Cw*16:02 rs12525170 rs114854070 Uncondition Condition on HLA-B*51 HLA-F HLA-A HLA-B HLA-G HLA-C MICA Supplementary Figure 1: Genetic association of imputed markers
-log10 P HLA-B*51 rs1050502 rs116799036 HLA-Cw*16:02 rs12525170 rs114854070 Uncondition Condition on HLA-B*51 HLA-F HLA-A HLA-B HLA-G HLA-C MICA Supplementary Figure 1: Genetic association of imputed markers
Supplementary Figures
 Supplementary Figures Supplementary Fig 1. Comparison of sub-samples on the first two principal components of genetic variation. TheBritishsampleisplottedwithredpoints.The sub-samples of the diverse sample
Supplementary Figures Supplementary Fig 1. Comparison of sub-samples on the first two principal components of genetic variation. TheBritishsampleisplottedwithredpoints.The sub-samples of the diverse sample
NMDAR-dependent Argonaute 2 phosphorylation regulates mirna activity and dendritic spine plasticity
 Article NMDAR-dependent Argonaute 2 phosphorylation regulates mirna activity and dendritic spine plasticity Dipen Rajgor 1, Thomas M Sanderson 2, Mascia Amici 2, Graham L Collingridge 2,3,4 & Jonathan
Article NMDAR-dependent Argonaute 2 phosphorylation regulates mirna activity and dendritic spine plasticity Dipen Rajgor 1, Thomas M Sanderson 2, Mascia Amici 2, Graham L Collingridge 2,3,4 & Jonathan
How To Use SubpathwayGMir
 How To Use SubpathwayGMir Li Feng, Chunquan Li and Xia Li May 20, 2015 Contents 1 Overview 1 2 The experimentally verified mirna-target interactions 2 3 Reconstruct KEGG metabolic pathways 2 3.1 Embed
How To Use SubpathwayGMir Li Feng, Chunquan Li and Xia Li May 20, 2015 Contents 1 Overview 1 2 The experimentally verified mirna-target interactions 2 3 Reconstruct KEGG metabolic pathways 2 3.1 Embed
Scaffold function of long noncoding RNA HOTAIR in protein ubiquitination
 Yoon et al, page Scaffold function of long noncoding RNA HOTAIR in protein ubiquitination Je-Hyun Yoon,, Kotb Abdelmohsen, Jiyoung Kim, Xiaoling Yang, Jennifer L. Martindale, Kumiko Tominaga-Yamanaka,
Yoon et al, page Scaffold function of long noncoding RNA HOTAIR in protein ubiquitination Je-Hyun Yoon,, Kotb Abdelmohsen, Jiyoung Kim, Xiaoling Yang, Jennifer L. Martindale, Kumiko Tominaga-Yamanaka,
Supplemental Figure 1. Small RNA size distribution from different soybean tissues.
 Supplemental Figure 1. Small RNA size distribution from different soybean tissues. The size of small RNAs was plotted versus frequency (percentage) among total sequences (A, C, E and G) or distinct sequences
Supplemental Figure 1. Small RNA size distribution from different soybean tissues. The size of small RNAs was plotted versus frequency (percentage) among total sequences (A, C, E and G) or distinct sequences
SALSA MLPA Probemix P014-B1 Chromosome 8 Lot B and B
 SALSA MLPA Probemix P014-B1 Chromosome 8 Lot B1-0916 and B1-0713. Copy number changes of the human chromosome 8 are common in many types of tumours. In most cases, losses of 8p sequences and gains of 8q
SALSA MLPA Probemix P014-B1 Chromosome 8 Lot B1-0916 and B1-0713. Copy number changes of the human chromosome 8 are common in many types of tumours. In most cases, losses of 8p sequences and gains of 8q
Editorial Type 2 Diabetes and More Gene Panel: A Predictive Genomics Approach for a Polygenic Disease
 Cronicon OPEN ACCESS EC DIABETES AND METABOLIC RESEARCH Editorial Type 2 Diabetes and More Gene Panel: A Predictive Genomics Approach for a Polygenic Disease Amr TM Saeb* University Diabetes Center, College
Cronicon OPEN ACCESS EC DIABETES AND METABOLIC RESEARCH Editorial Type 2 Diabetes and More Gene Panel: A Predictive Genomics Approach for a Polygenic Disease Amr TM Saeb* University Diabetes Center, College
Supplementary Information
 Supplementary Information Supplementary Figure 1. Effect of mir mimics and anti-mirs on DTPs a, Representative fluorescence microscopy images of GFP vector control or mir mimicexpressing parental and DTP
Supplementary Information Supplementary Figure 1. Effect of mir mimics and anti-mirs on DTPs a, Representative fluorescence microscopy images of GFP vector control or mir mimicexpressing parental and DTP
SUPPLEMENTARY INFORMATION
 doi: 1.138/nature8645 Physical coverage (x haploid genomes) 11 6.4 4.9 6.9 6.7 4.4 5.9 9.1 7.6 125 Neither end mapped One end mapped Chimaeras Correct Reads (million ns) 1 75 5 25 HCC1187 HCC1395 HCC1599
doi: 1.138/nature8645 Physical coverage (x haploid genomes) 11 6.4 4.9 6.9 6.7 4.4 5.9 9.1 7.6 125 Neither end mapped One end mapped Chimaeras Correct Reads (million ns) 1 75 5 25 HCC1187 HCC1395 HCC1599
(a) Significant biological processes (upper panel) and disease biomarkers (lower panel)
 Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. Nature Genetics: doi: /ng.3736
 Supplementary Figure 1 Genetic correlations of five personality traits between 23andMe discovery and GPC samples. (a) The values in the colored squares are genetic correlations (r g ); (b) P values of
Supplementary Figure 1 Genetic correlations of five personality traits between 23andMe discovery and GPC samples. (a) The values in the colored squares are genetic correlations (r g ); (b) P values of
Supplemental figure 1. Genome-wide plots of P-values for all 17 traits.
 Supplemental figure 1. Genome-wide plots of P-values for all 17 traits. Supplemental figure 2. Regional plots for (A) the 10p11.21 NDSS drive/priority locus, (B) the 15q22.2 FTND time to first cigarette
Supplemental figure 1. Genome-wide plots of P-values for all 17 traits. Supplemental figure 2. Regional plots for (A) the 10p11.21 NDSS drive/priority locus, (B) the 15q22.2 FTND time to first cigarette
A Normal Exencephaly Craniora- Spina bifida Microcephaly chischisis. Midbrain Forebrain/ Forebrain/ Hindbrain Spinal cord Hindbrain Hindbrain
 A Normal Exencephaly Craniora- Spina bifida Microcephaly chischisis NTD Number of embryos % among NTD Embryos Exencephaly 52 74.3% Craniorachischisis 6 8.6% Spina bifida 5 7.1% Microcephaly 7 1% B Normal
A Normal Exencephaly Craniora- Spina bifida Microcephaly chischisis NTD Number of embryos % among NTD Embryos Exencephaly 52 74.3% Craniorachischisis 6 8.6% Spina bifida 5 7.1% Microcephaly 7 1% B Normal
Supplementary Figure 1 IL-27 IL
 Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
Whole-genome detection of disease-associated deletions or excess homozygosity in a case control study of rheumatoid arthritis
 HMG Advance Access published December 21, 2012 Human Molecular Genetics, 2012 1 13 doi:10.1093/hmg/dds512 Whole-genome detection of disease-associated deletions or excess homozygosity in a case control
HMG Advance Access published December 21, 2012 Human Molecular Genetics, 2012 1 13 doi:10.1093/hmg/dds512 Whole-genome detection of disease-associated deletions or excess homozygosity in a case control
Supplementary Information
 Supplementary Information SUPPLEMENTARY FIGURES: MCF-7 G/C Hs578 G/C T47D G/G 4T1 C/C Supplementary Figure 1. rs1071738 SNP genotype of breast cancer cell lines. Sanger sequencing of regions flanking the
Supplementary Information SUPPLEMENTARY FIGURES: MCF-7 G/C Hs578 G/C T47D G/G 4T1 C/C Supplementary Figure 1. rs1071738 SNP genotype of breast cancer cell lines. Sanger sequencing of regions flanking the
Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells.
 SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
Imaging Genetics: Heritability, Linkage & Association
 Imaging Genetics: Heritability, Linkage & Association David C. Glahn, PhD Olin Neuropsychiatry Research Center & Department of Psychiatry, Yale University July 17, 2011 Memory Activation & APOE ε4 Risk
Imaging Genetics: Heritability, Linkage & Association David C. Glahn, PhD Olin Neuropsychiatry Research Center & Department of Psychiatry, Yale University July 17, 2011 Memory Activation & APOE ε4 Risk
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/393/rs9/dc1 Supplementary Materials for Identification of potential drug targets for tuberous sclerosis complex by synthetic screens combining CRISPR-based knockouts
www.sciencesignaling.org/cgi/content/full/8/393/rs9/dc1 Supplementary Materials for Identification of potential drug targets for tuberous sclerosis complex by synthetic screens combining CRISPR-based knockouts
Big Data Training for Translational Omics Research. Session 1, Day 3, Liu. Case Study #2. PLOS Genetics DOI: /journal.pgen.
 Session 1, Day 3, Liu Case Study #2 PLOS Genetics DOI:10.1371/journal.pgen.1005910 Enantiomer Mirror image Methadone Methadone Kreek, 1973, 1976 Methadone Maintenance Therapy Long-term use of Methadone
Session 1, Day 3, Liu Case Study #2 PLOS Genetics DOI:10.1371/journal.pgen.1005910 Enantiomer Mirror image Methadone Methadone Kreek, 1973, 1976 Methadone Maintenance Therapy Long-term use of Methadone
Supplementary Materials for VAMP4 directs synaptic vesicles to a pool that selectively maintains asynchronous neurotransmission
 Supplementary Materials for VAMP4 directs synaptic vesicles to a pool that selectively maintains asynchronous neurotransmission Jesica Raingo, Mikhail Khvotchev, Pei Liu, Frederic Darios, Ying C. Li, Denise
Supplementary Materials for VAMP4 directs synaptic vesicles to a pool that selectively maintains asynchronous neurotransmission Jesica Raingo, Mikhail Khvotchev, Pei Liu, Frederic Darios, Ying C. Li, Denise
Supplementary figure legends
 Supplementary figure legends SUPPLEMENTRY FIGURE S1. Lentiviral construct. Schematic representation of the PCR fragment encompassing the genomic locus of mir-33a that was introduced in the lentiviral construct.
Supplementary figure legends SUPPLEMENTRY FIGURE S1. Lentiviral construct. Schematic representation of the PCR fragment encompassing the genomic locus of mir-33a that was introduced in the lentiviral construct.
MicroRNA and Male Infertility: A Potential for Diagnosis
 Review Article MicroRNA and Male Infertility: A Potential for Diagnosis * Abstract MicroRNAs (mirnas) are small non-coding single stranded RNA molecules that are physiologically produced in eukaryotic
Review Article MicroRNA and Male Infertility: A Potential for Diagnosis * Abstract MicroRNAs (mirnas) are small non-coding single stranded RNA molecules that are physiologically produced in eukaryotic
SUPPLEMENTAL DATA AGING, July 2014, Vol. 6 No. 7
 SUPPLEMENTAL DATA Figure S1. Muscle mass changes in different anatomical regions with age. (A) The TA and gastrocnemius muscle showed a significant loss of weight in aged mice (24 month old) compared to
SUPPLEMENTAL DATA Figure S1. Muscle mass changes in different anatomical regions with age. (A) The TA and gastrocnemius muscle showed a significant loss of weight in aged mice (24 month old) compared to
CONTRACTING ORGANIZATION: Texas A&M Health Science Center Temple, TX 76504
 Award Number: W81XWH-11-2-0166 TITLE: The Root Cause of Post-traumatic and Developmental Stress Disorder PRINCIPAL INVESTIGATOR: Keith A. Young, Ph.D. CONTRACTING ORGANIZATION: Texas A&M Health Science
Award Number: W81XWH-11-2-0166 TITLE: The Root Cause of Post-traumatic and Developmental Stress Disorder PRINCIPAL INVESTIGATOR: Keith A. Young, Ph.D. CONTRACTING ORGANIZATION: Texas A&M Health Science
