AutoLigand: A Tool for Finding Ligand Binding Sites. CUHK workshop Dec, 2011
|
|
|
- Adam Sullivan
- 5 years ago
- Views:
Transcription
1 AutoLigand: A Tool for Finding Ligand Binding Sites CUHK workshop Dec, 2011 Stefano Forli, Ph.D., TSRI Rodney M. Harris, Ph.D., 1060 Discovery Engineering
2 Overview Description and uses of AutoLigand Algorithms (how it works) Flood fill Optimization Local minimum check Statistics and scoring Results
3 AutoLigand Uses Find unknown sites scan complete receptor molecule report top ten hits Optimize known site use energy per volume of fill identify atom types 1ac8 CYTOCHROME C PEROXIDASE
4 Plot of energy/volume vs volume
5 Finding the best binding site
6 Results HIV Protease Amprenavir (black) HIV Protease (CPK colors) Image generated with PMV 1.4
7 Fill Example HIV Protease 50 points from AutoLigand Image generated with PMV 1.4
8 Fill Example HIV Protease 100 points from AutoLigand Image generated with PMV 1.4
9 Fill Example HIV Protease 150 points from AutoLigand Image generated with PMV 1.4
10 Fill Example HIV Protease 200 points from AutoLigand Image generated with PMV 1.4
11 Algorithm Generate 1 Å grid using AutoGrid Pick starting points scan whole protein for 10 best or pick single point from GUI Fill loop: Flood fill (initial volume fill) Affinotaxis migration (optimization) Pseudopod extensions (extended search) Output results Reference: Harris, R. M., Olson, A. J., and Goodsell, D. S. Automated prediction of ligand-binding sites in proteins. Proteins 2008; 70:
12 AutoGrid Affinity Calculation Empirical force field free energy function with the following terms: Van der Waal Electrostatic Hydrogen bond Desolvation E repulsive r attractive
13 AutoGrid Carbon Affinity Map HIV Protease backbone Zones of high carbon atom affinity (gray shapes)
14 Fill Loop part I Fill initial volume (flood fill): Add starting point to Fill list (red point) Generate list of orthogonal Neighbor points (6 blue points) Find best Neighbor point (sort on affinity energy) and add to Fill list (change from blue to red) Remove that point from Neighbor list and generate new Neighbors (now have 10 blue points) This produces a shell of Neighbor points around the Fill list Continue adding points until user specified number is reached
15 Fill Loop part II Affinotaxis (volume migration to areas of higher affinity) Compare best Neighbor point to worst Fill point, if better add Neighbor point and remove Fill point But! Need to check if point to be removed is safe to remove if not, remove 2nd worst Fill point, etc. Assign State to point in two passes 1st pass: assign removable/non removable state 2nd pass: re evaluate non removable state point to see if in loop structure and thus removable
16 Second pass non-removables Only 11 points need to be re-evaluated for loops in 2nd pass Non-removable One of these may be removed from loop Still non-removable
17 Fill Loop part III The code must avoid local minima
18 Fill Loop part III Pseudopod extension searching (extend search over the hill for better affinity) Select best point in 6 Å shell about each point in turn Extend a set of points from Fill point to the selected point and sum total affinity energy Compare total energy of pseudopod to total energy of same number of removable worst points If extended ray is better, add to Fill and remove worst points then loop back to flow migration and pseudopod projection again
19 Testing the code Screen AutoDock calibration set of 187 receptor/ligand systems count the % of the ligand overlapped by the volume fill Ligand site is found in 95% of the systems 29% of systems overlap 100% of ligand 64% of systems overlap between 20% and 99% 2% of systems overlap < 20% of ligand 5% of systems have no overlap of ligand (a miss) Sounds Great?
20 Better Scoring of the Tests How do you tell how well you have overlapped the ligand with the fill? Extra fill Missed fill Answer: Use an Intersection over Union Low I/U High I/U AutoLigand finds binding sites with I/U >= 0.2 only 85% of the time.
21 Results for HIV (complex) Optimal ligand binding site and size
22 HIV Results 1hps with active site and exosites filled
23 HIV exo site
24 More Results p38a High Throughput Screen found second site AutoLigand result for second site
25 Rhodopsin Binding Site
26 Swine Flu N1 Binding Site Relenza Tamiflu
27 Conclusions AutoLigand is effective in finding ligand binding sites without a priori knowledge of ligand AutoLigand can find the size, shape, and atom type best fit for ligand sites AutoLigand can be used as a drug development tool by indicating regions to modify existing drugs or suggesting new drug structures altogether
28 Acknowledgments Art Olson David Goodsell Albert Beuscher Mike Pique Michel Sanner
News. Volume 8: November 2, 2009
 FightAIDS@Home News Volume 8: November 2, 2009 The FightAIDS@Home Project uses the volunteered computing power of the World Community Grid to test candidate compounds against the variations (or mutants
FightAIDS@Home News Volume 8: November 2, 2009 The FightAIDS@Home Project uses the volunteered computing power of the World Community Grid to test candidate compounds against the variations (or mutants
News. Volume 5: June 12th, The Research Team We re all members of Prof. Olson s Molecular Graphics Lab
 FightAIDS@Home News Volume 5: June 12th, 2008 Alex Perry man Ga rret t Mo r ri s Stefano Art Forl i Ol son Alex Gi l let The FightAIDS@Home Research Team We re all members of Prof. Olson s Molecular Graphics
FightAIDS@Home News Volume 5: June 12th, 2008 Alex Perry man Ga rret t Mo r ri s Stefano Art Forl i Ol son Alex Gi l let The FightAIDS@Home Research Team We re all members of Prof. Olson s Molecular Graphics
News. Volume 7: April 14, 2009
 FightAIDS@Home News Volume 7: April 14, 2009 The FightAIDS@Home Project uses the volunteered computer power of the World Community Grid to test candidate drug compounds against the variations (or mutants
FightAIDS@Home News Volume 7: April 14, 2009 The FightAIDS@Home Project uses the volunteered computer power of the World Community Grid to test candidate drug compounds against the variations (or mutants
Investigation of the binding of spermine and derivative polyamines on the 1U6M active site in the presence of Acetyl coenzyme-a
 Investigation of the binding of spermine and derivative polyamines on the 1U6M active site in the presence of Acetyl coenzyme-a Lindsay Mitchell lm04575n@pace.edu Pace University NYC Department of Chemistry
Investigation of the binding of spermine and derivative polyamines on the 1U6M active site in the presence of Acetyl coenzyme-a Lindsay Mitchell lm04575n@pace.edu Pace University NYC Department of Chemistry
List of Figures. List of Tables
 Supporting Information for: Signaling Domain of Sonic Hedgehog as Cannibalistic Calcium-Regulated Zinc-Peptidase Rocio Rebollido-Rios 1, Shyam Bandari 3, Christoph Wilms 1, Stanislav Jakuschev 1, Andrea
Supporting Information for: Signaling Domain of Sonic Hedgehog as Cannibalistic Calcium-Regulated Zinc-Peptidase Rocio Rebollido-Rios 1, Shyam Bandari 3, Christoph Wilms 1, Stanislav Jakuschev 1, Andrea
a) The statement is true for X = 400, but false for X = 300; b) The statement is true for X = 300, but false for X = 200;
 1. Consider the following statement. To produce one molecule of each possible kind of polypeptide chain, X amino acids in length, would require more atoms than exist in the universe. Given the size of
1. Consider the following statement. To produce one molecule of each possible kind of polypeptide chain, X amino acids in length, would require more atoms than exist in the universe. Given the size of
Guided Inquiry Skills Lab. Additional Lab 1 Making Models of Macromolecules. Problem. Introduction. Skills Focus. Materials.
 Additional Lab 1 Making Models of Macromolecules Guided Inquiry Skills Lab Problem How do monomers join together to form polymers? Introduction A small number of elements make up most of the mass of your
Additional Lab 1 Making Models of Macromolecules Guided Inquiry Skills Lab Problem How do monomers join together to form polymers? Introduction A small number of elements make up most of the mass of your
In Silico docking studies of lipoxygenase inhibitory activity of commercially available flavonoids
 Available online at www.scholarsresearchlibrary.com J. Comput. Method. Mol. Design, 2011, 1 (4):65-72 (http://scholarsresearchlibrary.com/archive.html) ISSN : 2231-3176 CODEN (USA): JCMMDA In Silico docking
Available online at www.scholarsresearchlibrary.com J. Comput. Method. Mol. Design, 2011, 1 (4):65-72 (http://scholarsresearchlibrary.com/archive.html) ISSN : 2231-3176 CODEN (USA): JCMMDA In Silico docking
Biology 2E- Zimmer Protein structure- amino acid kit
 Biology 2E- Zimmer Protein structure- amino acid kit Name: This activity will use a physical model to investigate protein shape and develop key concepts that govern how proteins fold into their final three-dimensional
Biology 2E- Zimmer Protein structure- amino acid kit Name: This activity will use a physical model to investigate protein shape and develop key concepts that govern how proteins fold into their final three-dimensional
Bioinformatics for molecular biology
 Bioinformatics for molecular biology Structural bioinformatics tools, predictors, and 3D modeling Structural Biology Review Dr Research Scientist Department of Microbiology, Oslo University Hospital -
Bioinformatics for molecular biology Structural bioinformatics tools, predictors, and 3D modeling Structural Biology Review Dr Research Scientist Department of Microbiology, Oslo University Hospital -
Exploring Energy and Fitness Landscapes of Proteins
 Exploring Energy and Fitness Landscapes of Proteins! Computer Simulations in Chemistry and Biophysics! Protein-Ligand Binding: structure based drug design! HIV Family and Kinase Family Protein Targets!
Exploring Energy and Fitness Landscapes of Proteins! Computer Simulations in Chemistry and Biophysics! Protein-Ligand Binding: structure based drug design! HIV Family and Kinase Family Protein Targets!
MOLECULAR DOCKING STUDIES OF NOVEL HERBAL DRUG DEVA CHOORNAM ON HUMAN IMMUNE DEFICIENCY VIRUS (HIV) PDB 1HNV REVERSE TRANSCRIPTASE ENZYME
 MOLECULAR DOCKING STUDIES OF NOVEL HERBAL DRUG DEVA CHOORNAM ON HUMAN IMMUNE DEFICIENCY VIRUS (HIV) PDB 1HNV REVERSE TRANSCRIPTASE ENZYME Dr. Thangadurai. K *1, Dr.Vinayak.S 2, Dr. Gayatri.R 3, Dr B.R
MOLECULAR DOCKING STUDIES OF NOVEL HERBAL DRUG DEVA CHOORNAM ON HUMAN IMMUNE DEFICIENCY VIRUS (HIV) PDB 1HNV REVERSE TRANSCRIPTASE ENZYME Dr. Thangadurai. K *1, Dr.Vinayak.S 2, Dr. Gayatri.R 3, Dr B.R
Secondary Structure North 72nd Street, Wauwatosa, WI Phone: (414) Fax: (414) dmoleculardesigns.com
 Secondary Structure In the previous protein folding activity, you created a generic or hypothetical 15-amino acid protein and learned that basic principles of chemistry determine how each protein spontaneously
Secondary Structure In the previous protein folding activity, you created a generic or hypothetical 15-amino acid protein and learned that basic principles of chemistry determine how each protein spontaneously
Growing knowledge of cell biology at the fundamental molecular level opened the door to new ways of inventing drugs
 E-tutorial 5 Rational drug design - The way we find new medicines was modernised as the 20 th and 21 st centuries progressed - Modernisation was driven by many factors, but above all by an improved understanding
E-tutorial 5 Rational drug design - The way we find new medicines was modernised as the 20 th and 21 st centuries progressed - Modernisation was driven by many factors, but above all by an improved understanding
Discovery of Potent, Highly Selective, Broad Spectrum Antivirals
 Discovery of Potent, Highly Selective, Broad Spectrum Antivirals Fred Hutch/Merck Infectious Disease Summit May 10, 2018 Sam Lee, Ph.D. 1 Forward Looking Statements This presentation contains forward-looking
Discovery of Potent, Highly Selective, Broad Spectrum Antivirals Fred Hutch/Merck Infectious Disease Summit May 10, 2018 Sam Lee, Ph.D. 1 Forward Looking Statements This presentation contains forward-looking
Supplementary materials
 Supplementary materials Chemical library from ChemBridge 50,240 structurally diverse small molecule compounds dissolved in DMSO Hits Controls: No virus added μ Primary screening at 20 g/ml of compounds
Supplementary materials Chemical library from ChemBridge 50,240 structurally diverse small molecule compounds dissolved in DMSO Hits Controls: No virus added μ Primary screening at 20 g/ml of compounds
SUPPLEMENTARY INFORMATION. Computational Assay of H7N9 Influenza Neuraminidase Reveals R292K Mutation Reduces Drug Binding Affinity
 SUPPLEMENTARY INFORMATION Computational Assay of H7N9 Influenza Neuraminidase Reveals R292K Mutation Reduces Drug Binding Affinity Christopher Woods 1, Maturos Malaisree 1, Ben Long 2, Simon McIntosh-Smith
SUPPLEMENTARY INFORMATION Computational Assay of H7N9 Influenza Neuraminidase Reveals R292K Mutation Reduces Drug Binding Affinity Christopher Woods 1, Maturos Malaisree 1, Ben Long 2, Simon McIntosh-Smith
MBB 694:407, 115:511. Please use BLOCK CAPITAL letters like this --- A, B, C, D, E. Not lowercase!
 MBB 694:407, 115:511 First Test Severinov/Deis Tue. Sep. 30, 2003 Name Index number (not SSN) Row Letter Seat Number This exam consists of two parts. Part I is multiple choice. Each of these 25 questions
MBB 694:407, 115:511 First Test Severinov/Deis Tue. Sep. 30, 2003 Name Index number (not SSN) Row Letter Seat Number This exam consists of two parts. Part I is multiple choice. Each of these 25 questions
Activities for the α-helix / β-sheet Construction Kit
 Activities for the α-helix / β-sheet Construction Kit The primary sequence of a protein, composed of amino acids, determines the organization of the sequence into the secondary structure. There are two
Activities for the α-helix / β-sheet Construction Kit The primary sequence of a protein, composed of amino acids, determines the organization of the sequence into the secondary structure. There are two
A Random Forest Model for the Analysis of Chemical Descriptors for the Elucidation of HIV 1 Protease Protein Ligand Interactions
 A Random Forest Model for the Analysis of Chemical Descriptors for the Elucidation of HIV 1 Protease Protein Ligand Interactions Gene M. Ko, A. Srinivas Reddy, Sunil Kumar, Barbara A. Bailey, and Rajni
A Random Forest Model for the Analysis of Chemical Descriptors for the Elucidation of HIV 1 Protease Protein Ligand Interactions Gene M. Ko, A. Srinivas Reddy, Sunil Kumar, Barbara A. Bailey, and Rajni
Integrative Studies: Translational Biomedical Research
 Integrative Studies: Translational Biomedical Research Wilfred W. Li, Ph.D. wilfred@ucsd.edu National Biomedical Computation Resource Center for Research in Biological Systems San Diego Supercomputer Center
Integrative Studies: Translational Biomedical Research Wilfred W. Li, Ph.D. wilfred@ucsd.edu National Biomedical Computation Resource Center for Research in Biological Systems San Diego Supercomputer Center
Workshop on Analysis and prediction of contacts in proteins
 Workshop on Analysis and prediction of contacts in proteins 1.09.09 Eran Eyal 1, Vladimir Potapov 2, Ronen Levy 3, Vladimir Sobolev 3 and Marvin Edelman 3 1 Sheba Medical Center, Ramat Gan, Israel; Departments
Workshop on Analysis and prediction of contacts in proteins 1.09.09 Eran Eyal 1, Vladimir Potapov 2, Ronen Levy 3, Vladimir Sobolev 3 and Marvin Edelman 3 1 Sheba Medical Center, Ramat Gan, Israel; Departments
Bjoern Peters La Jolla Institute for Allergy and Immunology Buenos Aires, Oct 31, 2012
 www.iedb.org Bjoern Peters bpeters@liai.org La Jolla Institute for Allergy and Immunology Buenos Aires, Oct 31, 2012 Overview 1. Introduction to the IEDB 2. Application: 2009 Swine-origin influenza virus
www.iedb.org Bjoern Peters bpeters@liai.org La Jolla Institute for Allergy and Immunology Buenos Aires, Oct 31, 2012 Overview 1. Introduction to the IEDB 2. Application: 2009 Swine-origin influenza virus
A Study on Docking Mode of HIV Protease and Their Inhibitors
 J. Chem. Software, Vol. 7, No. 3, p. 103 114 (2001) A Study on Docking Mode of HIV Protease and Their Inhibitors Eiichi AKAHO a *, Garret MORRIS b, David GOODSELL b, David WONG b and Arthur OLSON b a Faculty
J. Chem. Software, Vol. 7, No. 3, p. 103 114 (2001) A Study on Docking Mode of HIV Protease and Their Inhibitors Eiichi AKAHO a *, Garret MORRIS b, David GOODSELL b, David WONG b and Arthur OLSON b a Faculty
BCH Graduate Survey of Biochemistry
 BCH 5045 Graduate Survey of Biochemistry Instructor: Charles Guy Producer: Ron Thomas Director: Glen Graham Lecture 10 Slide sets available at: http://hort.ifas.ufl.edu/teach/guyweb/bch5045/index.html
BCH 5045 Graduate Survey of Biochemistry Instructor: Charles Guy Producer: Ron Thomas Director: Glen Graham Lecture 10 Slide sets available at: http://hort.ifas.ufl.edu/teach/guyweb/bch5045/index.html
MITOCW watch?v=xms9dyhqhi0
 MITOCW watch?v=xms9dyhqhi0 The following content is provided under a Creative Commons license. Your support will help MIT OpenCourseWare continue to offer high-quality, educational resources for free.
MITOCW watch?v=xms9dyhqhi0 The following content is provided under a Creative Commons license. Your support will help MIT OpenCourseWare continue to offer high-quality, educational resources for free.
Simulated Docking of Darunavir with the HIV-1 L76V Mutant Protease Active Site
 Simulated Docking of Darunavir with the HIV-1 L76V Mutant Protease Active Site Jack K. Horner P.O. Box 266 Los Alamos NM 87544 USA Abstract Darunavir is a second-generation HIV-1 protease inhibitor. Here
Simulated Docking of Darunavir with the HIV-1 L76V Mutant Protease Active Site Jack K. Horner P.O. Box 266 Los Alamos NM 87544 USA Abstract Darunavir is a second-generation HIV-1 protease inhibitor. Here
Docking Analysis of Darunavir as HIV Protease Inhibitors
 Journal of Computational Methods in Molecular Design, 2012, 2 (1):39-43 Scholars Research Library (http://scholarsresearchlibrary.com/archive.html) ISSN : 2231-3176 CODEN (USA): JCMMDA Docking Analysis
Journal of Computational Methods in Molecular Design, 2012, 2 (1):39-43 Scholars Research Library (http://scholarsresearchlibrary.com/archive.html) ISSN : 2231-3176 CODEN (USA): JCMMDA Docking Analysis
BIOCHEMISTRY I HOMEWORK III DUE 10/15/03 66 points total + 2 bonus points = 68 points possible Swiss-PDB Viewer Exercise Attached
 BIOCHEMISTRY I HOMEWORK III DUE 10/15/03 66 points total + 2 bonus points = 68 points possible Swiss-PDB Viewer Exercise Attached 1). 20 points total T or F (2 points each; if false, briefly state why
BIOCHEMISTRY I HOMEWORK III DUE 10/15/03 66 points total + 2 bonus points = 68 points possible Swiss-PDB Viewer Exercise Attached 1). 20 points total T or F (2 points each; if false, briefly state why
Secondary Structure. by hydrogen bonds
 Secondary Structure In the previous protein folding activity, you created a hypothetical 15-amino acid protein and learned that basic principles of chemistry determine how each protein spontaneously folds
Secondary Structure In the previous protein folding activity, you created a hypothetical 15-amino acid protein and learned that basic principles of chemistry determine how each protein spontaneously folds
Prerequisite Knowledge: Students should have already been introduced to the inputs and outputs of photosynthesis.
 www.ngsslifescience.com. Topic: Metabolism Chemistry Model Summary: Students will learn act out polymerization by performing dehydration synthesis and hydrolysis using chemistry models. Students will also
www.ngsslifescience.com. Topic: Metabolism Chemistry Model Summary: Students will learn act out polymerization by performing dehydration synthesis and hydrolysis using chemistry models. Students will also
INTERACTION DRUG BODY
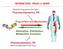 INTERACTION DRUG BODY What the drug does to the body What the body does to the drug Receptors - intracellular receptors - membrane receptors - Channel receptors - G protein-coupled receptors - Tyrosine-kinase
INTERACTION DRUG BODY What the drug does to the body What the body does to the drug Receptors - intracellular receptors - membrane receptors - Channel receptors - G protein-coupled receptors - Tyrosine-kinase
Life Sciences 1a. Practice Problems 4
 Life Sciences 1a Practice Problems 4 1. KcsA, a channel that allows K + ions to pass through the membrane, is a protein with four identical subunits that form a channel through the center of the tetramer.
Life Sciences 1a Practice Problems 4 1. KcsA, a channel that allows K + ions to pass through the membrane, is a protein with four identical subunits that form a channel through the center of the tetramer.
Probability-Based Protein Identification for Post-Translational Modifications and Amino Acid Variants Using Peptide Mass Fingerprint Data
 Probability-Based Protein Identification for Post-Translational Modifications and Amino Acid Variants Using Peptide Mass Fingerprint Data Tong WW, McComb ME, Perlman DH, Huang H, O Connor PB, Costello
Probability-Based Protein Identification for Post-Translational Modifications and Amino Acid Variants Using Peptide Mass Fingerprint Data Tong WW, McComb ME, Perlman DH, Huang H, O Connor PB, Costello
Practice Problems 3. a. What is the name of the bond formed between two amino acids? Are these bonds free to rotate?
 Life Sciences 1a Practice Problems 3 1. Draw the oligopeptide for Ala-Phe-Gly-Thr-Asp. You do not need to indicate the stereochemistry of the sidechains. Denote with arrows the bonds formed between the
Life Sciences 1a Practice Problems 3 1. Draw the oligopeptide for Ala-Phe-Gly-Thr-Asp. You do not need to indicate the stereochemistry of the sidechains. Denote with arrows the bonds formed between the
Crystal Structure of the Subtilisin Carlsberg: OMTKY3 Complex
 John Clizer & Greg Ralph Crystal Structure of the Subtilisin Carlsberg: OMTKY3 Complex The turkey ovomucoid third domain (OMTKY3) is considered to be one of the most studied protein inhibitors. 1 Ovomucin
John Clizer & Greg Ralph Crystal Structure of the Subtilisin Carlsberg: OMTKY3 Complex The turkey ovomucoid third domain (OMTKY3) is considered to be one of the most studied protein inhibitors. 1 Ovomucin
Design and Modeling Studies on Liriodenine derivatives as novel topoisomerase II inhibitors
 International Journal of ChemTech Research CODEN( USA): IJCRGG ISSN : 0974-4290 Vol. 3, No.3,pp 1622-1627, July-Sept 2011 Design and Modeling Studies on Liriodenine derivatives as novel topoisomerase II
International Journal of ChemTech Research CODEN( USA): IJCRGG ISSN : 0974-4290 Vol. 3, No.3,pp 1622-1627, July-Sept 2011 Design and Modeling Studies on Liriodenine derivatives as novel topoisomerase II
Introduction to proteins and protein structure
 Introduction to proteins and protein structure The questions and answers below constitute an introduction to the fundamental principles of protein structure. They are all available at [link]. What are
Introduction to proteins and protein structure The questions and answers below constitute an introduction to the fundamental principles of protein structure. They are all available at [link]. What are
Draw how two amino acids form the peptide bond. Draw in the space below:
 Name Date Period Modeling Protein Folding Draw how two amino acids form the peptide bond. Draw in the space below: What we are doing today: The core idea in life sciences is that there is a fundamental
Name Date Period Modeling Protein Folding Draw how two amino acids form the peptide bond. Draw in the space below: What we are doing today: The core idea in life sciences is that there is a fundamental
Interactions of Polyethylenimines with Zwitterionic and. Anionic Lipid Membranes
 Interactions of Polyethylenimines with Zwitterionic and Anionic Lipid Membranes Urszula Kwolek, Dorota Jamróz, Małgorzata Janiczek, Maria Nowakowska, Paweł Wydro, Mariusz Kepczynski Faculty of Chemistry,
Interactions of Polyethylenimines with Zwitterionic and Anionic Lipid Membranes Urszula Kwolek, Dorota Jamróz, Małgorzata Janiczek, Maria Nowakowska, Paweł Wydro, Mariusz Kepczynski Faculty of Chemistry,
Ras and Cell Signaling Exercise
 Ras and Cell Signaling Exercise Learning Objectives In this exercise, you will use, a protein 3D- viewer, to explore: the structure of the Ras protein the active and inactive state of Ras and the amino
Ras and Cell Signaling Exercise Learning Objectives In this exercise, you will use, a protein 3D- viewer, to explore: the structure of the Ras protein the active and inactive state of Ras and the amino
SDS-Assisted Protein Transport Through Solid-State Nanopores
 Supplementary Information for: SDS-Assisted Protein Transport Through Solid-State Nanopores Laura Restrepo-Pérez 1, Shalini John 2, Aleksei Aksimentiev 2 *, Chirlmin Joo 1 *, Cees Dekker 1 * 1 Department
Supplementary Information for: SDS-Assisted Protein Transport Through Solid-State Nanopores Laura Restrepo-Pérez 1, Shalini John 2, Aleksei Aksimentiev 2 *, Chirlmin Joo 1 *, Cees Dekker 1 * 1 Department
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/2/9/e1600292/dc1 Supplementary Materials for Native phasing of x-ray free-electron laser data for a G protein coupled receptor Alexander Batyuk, Lorenzo Galli,
advances.sciencemag.org/cgi/content/full/2/9/e1600292/dc1 Supplementary Materials for Native phasing of x-ray free-electron laser data for a G protein coupled receptor Alexander Batyuk, Lorenzo Galli,
7.014 Problem Set 2 Solutions
 7.014 Problem Set 2 Solutions Please print out this problem set and record your answers on the printed copy. Answers to this problem set are to be turned in at the box outside 68-120 by 11:45 Friday, February
7.014 Problem Set 2 Solutions Please print out this problem set and record your answers on the printed copy. Answers to this problem set are to be turned in at the box outside 68-120 by 11:45 Friday, February
Trilateral Project WM4
 ANNEX 2: Comments of the JPO Trilateral Project WM4 Comparative studies in new technologies Theme: Comparative study on protein 3-dimensional (3-D) structure related claims 1. Introduction As more 3-D
ANNEX 2: Comments of the JPO Trilateral Project WM4 Comparative studies in new technologies Theme: Comparative study on protein 3-dimensional (3-D) structure related claims 1. Introduction As more 3-D
EARLY STAGE DIAGNOSIS OF LUNG CANCER USING CT-SCAN IMAGES BASED ON CELLULAR LEARNING AUTOMATE
 EARLY STAGE DIAGNOSIS OF LUNG CANCER USING CT-SCAN IMAGES BASED ON CELLULAR LEARNING AUTOMATE SAKTHI NEELA.P.K Department of M.E (Medical electronics) Sengunthar College of engineering Namakkal, Tamilnadu,
EARLY STAGE DIAGNOSIS OF LUNG CANCER USING CT-SCAN IMAGES BASED ON CELLULAR LEARNING AUTOMATE SAKTHI NEELA.P.K Department of M.E (Medical electronics) Sengunthar College of engineering Namakkal, Tamilnadu,
Chapter 6. X-ray structure analysis of D30N tethered HIV-1 protease. dimer/saquinavir complex
 Chapter 6 X-ray structure analysis of D30N tethered HIV-1 protease dimer/saquinavir complex 6.1 Introduction: The arrival of HIV protease inhibitors (PIs) in late 1995 marked the beginning of an important
Chapter 6 X-ray structure analysis of D30N tethered HIV-1 protease dimer/saquinavir complex 6.1 Introduction: The arrival of HIV protease inhibitors (PIs) in late 1995 marked the beginning of an important
OUTLIER SUBJECTS PROTOCOL (art_groupoutlier)
 OUTLIER SUBJECTS PROTOCOL (art_groupoutlier) Paul K. Mazaika 2/23/2009 Outlier subjects are a problem in fmri data sets for clinical populations. This protocol and program are a method to identify outlier
OUTLIER SUBJECTS PROTOCOL (art_groupoutlier) Paul K. Mazaika 2/23/2009 Outlier subjects are a problem in fmri data sets for clinical populations. This protocol and program are a method to identify outlier
Hemoglobin & Sickle Cell Anemia Exercise
 Name StarBiochem Hemoglobin & Sickle Cell Anemia Exercise Learning Objectives In this exercise, you will use StarBiochem, a protein 3D viewer, to explore the structure of the normal hemoglobin protein
Name StarBiochem Hemoglobin & Sickle Cell Anemia Exercise Learning Objectives In this exercise, you will use StarBiochem, a protein 3D viewer, to explore the structure of the normal hemoglobin protein
Citric Acid Cycle and Oxidative Phosphorylation
 Citric Acid Cycle and Oxidative Phosphorylation Page by: OpenStax Summary The Citric Acid Cycle In eukaryotic cells, the pyruvate molecules produced at the end of glycolysis are transported into mitochondria,
Citric Acid Cycle and Oxidative Phosphorylation Page by: OpenStax Summary The Citric Acid Cycle In eukaryotic cells, the pyruvate molecules produced at the end of glycolysis are transported into mitochondria,
Introduction to Protein Structure Collection
 Introduction to Protein Structure Collection Teaching Points This collection is designed to introduce students to the concepts of protein structure and biochemistry. Different activities guide students
Introduction to Protein Structure Collection Teaching Points This collection is designed to introduce students to the concepts of protein structure and biochemistry. Different activities guide students
2. Which of the following amino acids is most likely to be found on the outer surface of a properly folded protein?
 Name: WHITE Student Number: Answer the following questions on the computer scoring sheet. 1 mark each 1. Which of the following amino acids would have the highest relative mobility R f in normal thin layer
Name: WHITE Student Number: Answer the following questions on the computer scoring sheet. 1 mark each 1. Which of the following amino acids would have the highest relative mobility R f in normal thin layer
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10913 Supplementary Figure 1 2F o -F c electron density maps of cognate and near-cognate trna Leu 2 in the A site of the 70S ribosome. The maps are contoured at 1.2 sigma and some of
doi:10.1038/nature10913 Supplementary Figure 1 2F o -F c electron density maps of cognate and near-cognate trna Leu 2 in the A site of the 70S ribosome. The maps are contoured at 1.2 sigma and some of
Ultrasonic Inspection of Adhesive Joints of Composite Pipelines
 Ultrasonic Inspection of Adhesive Joints of Composite Pipelines Priscila Duarte de Almeida a, João Marcos Alcoforado Rebello b, Gabriela Ribeiro Pereira b, Sérgio Damasceno Soares c, and Roman Fernandez
Ultrasonic Inspection of Adhesive Joints of Composite Pipelines Priscila Duarte de Almeida a, João Marcos Alcoforado Rebello b, Gabriela Ribeiro Pereira b, Sérgio Damasceno Soares c, and Roman Fernandez
Empowering Teachers Through Modeling
 Empowering Teachers Through Modeling Many Models of Membranes! WORKSHOP LEARNING OBJECTIVES Educators attending this workshop will: o Utilize tools that can support the NGSS student learning outcomes of
Empowering Teachers Through Modeling Many Models of Membranes! WORKSHOP LEARNING OBJECTIVES Educators attending this workshop will: o Utilize tools that can support the NGSS student learning outcomes of
Lecture 10 More about proteins
 Lecture 10 More about proteins Today we're going to extend our discussion of protein structure. This may seem far-removed from gene cloning, but it is the path to understanding the genes that we are cloning.
Lecture 10 More about proteins Today we're going to extend our discussion of protein structure. This may seem far-removed from gene cloning, but it is the path to understanding the genes that we are cloning.
NDT PERFORMANCE DEMONSTRATION USING SIMULATION
 NDT PERFORMANCE DEMONSTRATION USING SIMULATION N. Dominguez, F. Jenson, CEA P. Dubois, F. Foucher, EXTENDE nicolas.dominguez@cea.fr CEA 10 AVRIL 2012 PAGE 1 NDT PERFORMANCE DEMO. USING SIMULATION OVERVIEW
NDT PERFORMANCE DEMONSTRATION USING SIMULATION N. Dominguez, F. Jenson, CEA P. Dubois, F. Foucher, EXTENDE nicolas.dominguez@cea.fr CEA 10 AVRIL 2012 PAGE 1 NDT PERFORMANCE DEMO. USING SIMULATION OVERVIEW
Life History of A Drug
 DRUG ACTION & PHARMACODYNAMIC M. Imad Damaj, Ph.D. Associate Professor Pharmacology and Toxicology Smith 652B, 828-1676, mdamaj@hsc.vcu.edu Life History of A Drug Non-Specific Mechanims Drug-Receptor Interaction
DRUG ACTION & PHARMACODYNAMIC M. Imad Damaj, Ph.D. Associate Professor Pharmacology and Toxicology Smith 652B, 828-1676, mdamaj@hsc.vcu.edu Life History of A Drug Non-Specific Mechanims Drug-Receptor Interaction
Allosteric Inhibition of SHP2: Identification of a Potent, Selective, and Orally Efficacious Phosphatase Inhibitor!
 Allosteric Inhibition of SHP2: Identification of a Potent, Selective, and Orally Efficacious Phosphatase Inhibitor Allosteric pocket SHP2 Phosphatase ovel allosteric Phosphatase inhibitor Evan Carder Wipf
Allosteric Inhibition of SHP2: Identification of a Potent, Selective, and Orally Efficacious Phosphatase Inhibitor Allosteric pocket SHP2 Phosphatase ovel allosteric Phosphatase inhibitor Evan Carder Wipf
FCC2 5CY7 FCC1 5CY8. Actinonin 5CVQ
 Table S1. Data collection and refinement statistics Data collection Apo 5E5D MA 5CPD MAS 5CP0 Actinonin 5CVQ FCC1 5CY8 FCC2 5CY7 FCC 5CWY FCC4 5CVK FCC5 5CWX FCC6 5CVP X-ray source 17A-KEK PAL4A PAL4A
Table S1. Data collection and refinement statistics Data collection Apo 5E5D MA 5CPD MAS 5CP0 Actinonin 5CVQ FCC1 5CY8 FCC2 5CY7 FCC 5CWY FCC4 5CVK FCC5 5CWX FCC6 5CVP X-ray source 17A-KEK PAL4A PAL4A
Lecture 15. Membrane Proteins I
 Lecture 15 Membrane Proteins I Introduction What are membrane proteins and where do they exist? Proteins consist of three main classes which are classified as globular, fibrous and membrane proteins. A
Lecture 15 Membrane Proteins I Introduction What are membrane proteins and where do they exist? Proteins consist of three main classes which are classified as globular, fibrous and membrane proteins. A
How Do Antacids Work?
 How Do Antacids Work? Materials Needed: Bottle of antacid (the tablets should be broken into smaller pieces) A Mortar and pestle Dropper bottle with fake stomach juice = (0.01M HCl, or use vinegar) Round
How Do Antacids Work? Materials Needed: Bottle of antacid (the tablets should be broken into smaller pieces) A Mortar and pestle Dropper bottle with fake stomach juice = (0.01M HCl, or use vinegar) Round
SIEVE 2.1 Proteomics Example
 SIEVE 2.1 Proteomics Example Software Overview What is SIEVE? SIEVE is Thermo Scientific s differential software solution. SIEVE will continue to enhance our current product for label-free differential
SIEVE 2.1 Proteomics Example Software Overview What is SIEVE? SIEVE is Thermo Scientific s differential software solution. SIEVE will continue to enhance our current product for label-free differential
AP Biology Summer Assignment Chapter 3 Quiz
 AP Biology Summer Assignment Chapter 3 Quiz 2016-17 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. All of the following are found in a DNA nucleotide
AP Biology Summer Assignment Chapter 3 Quiz 2016-17 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. All of the following are found in a DNA nucleotide
1. Describe the difference between covalent and ionic bonds. What are the electrons doing?
 Exam 1 Review Bio 212: 1. Describe the difference between covalent and ionic bonds. What are the electrons doing? 2. Label each picture either a Carbohydrate, Protein, Nucleic Acid, or Fats(Lipid). a.
Exam 1 Review Bio 212: 1. Describe the difference between covalent and ionic bonds. What are the electrons doing? 2. Label each picture either a Carbohydrate, Protein, Nucleic Acid, or Fats(Lipid). a.
Background Information. Instructions. Problem Statement. HOMEWORK INSTRUCTIONS Homework #2 HIV Statistics Problem
 Background Information HOMEWORK INSTRUCTIONS The scourge of HIV/AIDS has had an extraordinary impact on the entire world. The spread of the disease has been closely tracked since the discovery of the HIV
Background Information HOMEWORK INSTRUCTIONS The scourge of HIV/AIDS has had an extraordinary impact on the entire world. The spread of the disease has been closely tracked since the discovery of the HIV
Protein Primer, Lumry I, Chapter 10, Enzyme structure, middle
 Protein Primer, Lumry I, Chapter 10, Enzyme structure, 5-15-03 10-1 Chapter 10. Enzyme structure A major feature of the trypsin family of proteases is the palindromic pattern taken by the B factors of
Protein Primer, Lumry I, Chapter 10, Enzyme structure, 5-15-03 10-1 Chapter 10. Enzyme structure A major feature of the trypsin family of proteases is the palindromic pattern taken by the B factors of
Supporting Information Identification of Amino Acids with Sensitive Nanoporous MoS 2 : Towards Machine Learning-Based Prediction
 Supporting Information Identification of Amino Acids with Sensitive Nanoporous MoS 2 : Towards Machine Learning-Based Prediction Amir Barati Farimani, Mohammad Heiranian, Narayana R. Aluru 1 Department
Supporting Information Identification of Amino Acids with Sensitive Nanoporous MoS 2 : Towards Machine Learning-Based Prediction Amir Barati Farimani, Mohammad Heiranian, Narayana R. Aluru 1 Department
Jmol and Crystal. Bob Hanson St. Olaf College, Northfield, MN
 Jmol and Crystal Bob Hanson St. Olaf College, Northfield, MN http://www.stolaf.edu/people/hansonr Crystal Workshop MSSC2018 l'università di Torino, Italia 4 Sep 2018 Bob Hanson St. Olaf College Jmol Topics
Jmol and Crystal Bob Hanson St. Olaf College, Northfield, MN http://www.stolaf.edu/people/hansonr Crystal Workshop MSSC2018 l'università di Torino, Italia 4 Sep 2018 Bob Hanson St. Olaf College Jmol Topics
INTERACTIONS OF GANODERIOL-F WITH ASPARTIC PROTEASES OF HIV AND PLASMEPSIN FOR ANTI-HIV AND ANTI-MALARIA DISCOVERY
 Innovare Academic Sciences International Journal of Pharmacy and Pharmaceutical Sciences ISSN- 0975-1491 Vol 6, Issue 5, 2014 Original Article INTERACTIONS OF GANODERIOL-F WITH ASPARTIC PROTEASES OF HIV
Innovare Academic Sciences International Journal of Pharmacy and Pharmaceutical Sciences ISSN- 0975-1491 Vol 6, Issue 5, 2014 Original Article INTERACTIONS OF GANODERIOL-F WITH ASPARTIC PROTEASES OF HIV
Supporting material. Membrane permeation induced by aggregates of human islet amyloid polypeptides
 Supporting material Membrane permeation induced by aggregates of human islet amyloid polypeptides Chetan Poojari Forschungszentrum Jülich GmbH, Institute of Complex Systems: Structural Biochemistry (ICS-6),
Supporting material Membrane permeation induced by aggregates of human islet amyloid polypeptides Chetan Poojari Forschungszentrum Jülich GmbH, Institute of Complex Systems: Structural Biochemistry (ICS-6),
Summer Assignment 2016
 Welcome to IB Biology 11 th graders. Please complete the following three components over the summer. 1) Chemistry of life review - on a separate piece of paper, please answer the following by hand: Lewis
Welcome to IB Biology 11 th graders. Please complete the following three components over the summer. 1) Chemistry of life review - on a separate piece of paper, please answer the following by hand: Lewis
Some Thoughts on Calibrating LaModel
 Some Thoughts on Calibrating LaModel Dr. Keith A. Heasley Professor Department of Mining Engineering West Virginia University Introduction Recent mine collapses and pillar failures have highlighted hli
Some Thoughts on Calibrating LaModel Dr. Keith A. Heasley Professor Department of Mining Engineering West Virginia University Introduction Recent mine collapses and pillar failures have highlighted hli
Visualizing Biopolymers and Their Building Blocks
 Visualizing Biopolymers and Their Building Blocks By Sharlene Denos (UIUC) & Kathryn Hafner (Danville High) Living things are primarily composed of carbon-based (organic) polymers. These are made up many
Visualizing Biopolymers and Their Building Blocks By Sharlene Denos (UIUC) & Kathryn Hafner (Danville High) Living things are primarily composed of carbon-based (organic) polymers. These are made up many
Proteins and their structure
 Proteins and their structure Proteins are the most abundant biological macromolecules, occurring in all cells and all parts of cells. Proteins also occur in great variety; thousands of different kinds,
Proteins and their structure Proteins are the most abundant biological macromolecules, occurring in all cells and all parts of cells. Proteins also occur in great variety; thousands of different kinds,
PHARMACOPHORE MODELING
 CHAPTER 9 PHARMACOPHORE MODELING 9.1 PHARMACOPHORES AS VIRTUAL SCREENING TOOLS The pharmacophore concept has been widely used over the past decades for rational drug design approaches and has now been
CHAPTER 9 PHARMACOPHORE MODELING 9.1 PHARMACOPHORES AS VIRTUAL SCREENING TOOLS The pharmacophore concept has been widely used over the past decades for rational drug design approaches and has now been
Pharmacologic Principles. Dr. Alia Shatanawi
 Pharmacologic Principles Dr. Alia Shatanawi Definitions Drug: It is any chemical that affect living processes. It modifies an already existing function, and does not create a new function. 2 What is Pharmacology?
Pharmacologic Principles Dr. Alia Shatanawi Definitions Drug: It is any chemical that affect living processes. It modifies an already existing function, and does not create a new function. 2 What is Pharmacology?
Table S1. X-ray data collection and refinement statistics
 Table S1. X-ray data collection and refinement statistics Data collection H7.167 Fab-Sh2/H7 complex Beamline SSRL 12-2 Wavelength (Å) 0.97950 Space group I2 1 3 Unit cell parameters (Å, º) a = b = c=207.3,
Table S1. X-ray data collection and refinement statistics Data collection H7.167 Fab-Sh2/H7 complex Beamline SSRL 12-2 Wavelength (Å) 0.97950 Space group I2 1 3 Unit cell parameters (Å, º) a = b = c=207.3,
metal-organic compounds
 metal-organic compounds Acta Crystallographica Section E Structure Reports Online ISSN 1600-5368 Chlorido(2-chloronicotinato)triphenylantimony(V) Li Quan, Handong Yin* and Daqi Wang College of Chemistry
metal-organic compounds Acta Crystallographica Section E Structure Reports Online ISSN 1600-5368 Chlorido(2-chloronicotinato)triphenylantimony(V) Li Quan, Handong Yin* and Daqi Wang College of Chemistry
Citric Acid Cycle and Oxidative Phosphorylation
 Citric Acid Cycle and Oxidative Phosphorylation Bởi: OpenStaxCollege The Citric Acid Cycle In eukaryotic cells, the pyruvate molecules produced at the end of glycolysis are transported into mitochondria,
Citric Acid Cycle and Oxidative Phosphorylation Bởi: OpenStaxCollege The Citric Acid Cycle In eukaryotic cells, the pyruvate molecules produced at the end of glycolysis are transported into mitochondria,
Supplementary Figure 1 (previous page). EM analysis of full-length GCGR. (a) Exemplary tilt pair images of the GCGR mab23 complex acquired for Random
 S1 Supplementary Figure 1 (previous page). EM analysis of full-length GCGR. (a) Exemplary tilt pair images of the GCGR mab23 complex acquired for Random Conical Tilt (RCT) reconstruction (left: -50,right:
S1 Supplementary Figure 1 (previous page). EM analysis of full-length GCGR. (a) Exemplary tilt pair images of the GCGR mab23 complex acquired for Random Conical Tilt (RCT) reconstruction (left: -50,right:
Natural Flavonoids as CDK5 inhibitors: A Homology modeling and Molecular Docking Study
 International Journal of ChemTech Research CDEN (USA): IJCRGG, ISSN: 0974-4290, ISSN(nline):2455-9555 Vol.9, No.09 pp 210-215, 2016 Natural Flavonoids as CDK5 inhibitors: A Homology modeling and Molecular
International Journal of ChemTech Research CDEN (USA): IJCRGG, ISSN: 0974-4290, ISSN(nline):2455-9555 Vol.9, No.09 pp 210-215, 2016 Natural Flavonoids as CDK5 inhibitors: A Homology modeling and Molecular
Hemoglobin & Sickle Cell Anemia Exercise
 Name StarBiochem Hemoglobin & Sickle Cell Anemia Exercise Background Hemoglobin is the protein in red blood cells responsible for carrying oxygen from the lungs to the rest of the body and for returning
Name StarBiochem Hemoglobin & Sickle Cell Anemia Exercise Background Hemoglobin is the protein in red blood cells responsible for carrying oxygen from the lungs to the rest of the body and for returning
AEROBIC RESPIRATION. Chapter 8
 AEROBIC RESPIRATION Chapter 8 AEROBIC RESPIRATION Aerobic respiration is the next step after Glycolysis if the cell can obtain oxygen. We won t need it until the last step but we still need it. Remember
AEROBIC RESPIRATION Chapter 8 AEROBIC RESPIRATION Aerobic respiration is the next step after Glycolysis if the cell can obtain oxygen. We won t need it until the last step but we still need it. Remember
Using CART to Mine SELDI ProteinChip Data for Biomarkers and Disease Stratification
 Using CART to Mine SELDI ProteinChip Data for Biomarkers and Disease Stratification Kenna Mawk, D.V.M. Informatics Product Manager Ciphergen Biosystems, Inc. Outline Introduction to ProteinChip Technology
Using CART to Mine SELDI ProteinChip Data for Biomarkers and Disease Stratification Kenna Mawk, D.V.M. Informatics Product Manager Ciphergen Biosystems, Inc. Outline Introduction to ProteinChip Technology
Determination of Copper in Green Olives using ICP-OES
 Application Note Food and Agriculture Determination of Copper in Green Olives using ICP-OES Intelligent Rinse function reduced analysis time by 60%, saving 191.4 L of argon Authors Ryley Burgess, Agilent
Application Note Food and Agriculture Determination of Copper in Green Olives using ICP-OES Intelligent Rinse function reduced analysis time by 60%, saving 191.4 L of argon Authors Ryley Burgess, Agilent
Name Date Period. Go to:
 Name Date Period In this online investigation, you will examine foods to determine what type of predominant organic compounds can be found in each. Make sure to read the whole paper and answer all questions.
Name Date Period In this online investigation, you will examine foods to determine what type of predominant organic compounds can be found in each. Make sure to read the whole paper and answer all questions.
Protein Refinement in Discovery Studio 1.7. Presented by: Francisco Hernandez-Guzman, PhD
 Protein Refinement in Discovery Studio 1.7 Presented by: Francisco Hernandez-Guzman, PhD February 2007 Agenda Part I: Protein Refinement in Discovery Studio 1.7 (20mins) The problem Novel Algorithms at
Protein Refinement in Discovery Studio 1.7 Presented by: Francisco Hernandez-Guzman, PhD February 2007 Agenda Part I: Protein Refinement in Discovery Studio 1.7 (20mins) The problem Novel Algorithms at
DO NOW: What is a catalyst?
 AGENDA ABSENT Block Nov 5 th /6 th -Week 13 TOPIC: Enzymes OBJ : 1-4 DO NOW: What is a catalyst? Science of Life EXT: Enzyme Lab DUE DATE: 11-10 DW: -----------------------------------------------------------------------------------------------------------------
AGENDA ABSENT Block Nov 5 th /6 th -Week 13 TOPIC: Enzymes OBJ : 1-4 DO NOW: What is a catalyst? Science of Life EXT: Enzyme Lab DUE DATE: 11-10 DW: -----------------------------------------------------------------------------------------------------------------
Coarse grained simulations of Lipid Bilayer Membranes
 Coarse grained simulations of Lipid Bilayer Membranes P. B. Sunil Kumar Department of Physics IIT Madras, Chennai 600036 sunil@iitm.ac.in Atomistic MD: time scales ~ 10 ns length scales ~100 nm 2 To study
Coarse grained simulations of Lipid Bilayer Membranes P. B. Sunil Kumar Department of Physics IIT Madras, Chennai 600036 sunil@iitm.ac.in Atomistic MD: time scales ~ 10 ns length scales ~100 nm 2 To study
[ APPLICATION NOTE ] High Sensitivity Intact Monoclonal Antibody (mab) HRMS Quantification APPLICATION BENEFITS INTRODUCTION WATERS SOLUTIONS KEYWORDS
![[ APPLICATION NOTE ] High Sensitivity Intact Monoclonal Antibody (mab) HRMS Quantification APPLICATION BENEFITS INTRODUCTION WATERS SOLUTIONS KEYWORDS [ APPLICATION NOTE ] High Sensitivity Intact Monoclonal Antibody (mab) HRMS Quantification APPLICATION BENEFITS INTRODUCTION WATERS SOLUTIONS KEYWORDS](/thumbs/79/80328542.jpg) Yun Wang Alelyunas, Henry Shion, Mark Wrona Waters Corporation, Milford, MA, USA APPLICATION BENEFITS mab LC-MS method which enables users to achieve highly sensitive bioanalysis of intact trastuzumab
Yun Wang Alelyunas, Henry Shion, Mark Wrona Waters Corporation, Milford, MA, USA APPLICATION BENEFITS mab LC-MS method which enables users to achieve highly sensitive bioanalysis of intact trastuzumab
Protein structure. Dr. Mamoun Ahram Summer semester,
 Protein structure Dr. Mamoun Ahram Summer semester, 2017-2018 Overview of proteins Proteins have different structures and some have repeating inner structures, other do not. A protein may have gazillion
Protein structure Dr. Mamoun Ahram Summer semester, 2017-2018 Overview of proteins Proteins have different structures and some have repeating inner structures, other do not. A protein may have gazillion
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Binding capacity of DNA-barcoded MHC multimers and recovery of antigen specificity
 Supplementary Figure 1 Binding capacity of DNA-barcoded MHC multimers and recovery of antigen specificity (a, b) Fluorescent-based determination of the binding capacity of DNA-barcoded MHC multimers (+barcode)
Supplementary Figure 1 Binding capacity of DNA-barcoded MHC multimers and recovery of antigen specificity (a, b) Fluorescent-based determination of the binding capacity of DNA-barcoded MHC multimers (+barcode)
Molecular docking analysis of UniProtKB nitrate reductase enzyme with known natural flavonoids
 www.bioinformation.net Volume 12(12) Hypothesis Molecular docking analysis of UniProtKB nitrate reductase enzyme with known natural flavonoids Ayub Shaik 1, Vishnu Thumma 2, Aruna Kumari Kotha 2, Sandhya
www.bioinformation.net Volume 12(12) Hypothesis Molecular docking analysis of UniProtKB nitrate reductase enzyme with known natural flavonoids Ayub Shaik 1, Vishnu Thumma 2, Aruna Kumari Kotha 2, Sandhya
SAM Teachers Guide Lipids and Carbohydrates
 SAM Teachers Guide Lipids and Carbohydrates Overview Students will explore the structure and function of two of the four major macromolecules, lipids and carbohydrates. They will look specifically at the
SAM Teachers Guide Lipids and Carbohydrates Overview Students will explore the structure and function of two of the four major macromolecules, lipids and carbohydrates. They will look specifically at the
Enzyme Activity Lecture. Every reaction has energy requirement. The minimum amount of energy required is termed activation energy.
 Enzyme Activity Lecture Every reaction has energy requirement. The minimum amount of energy required is termed activation energy. Living organisms have optimum temperature requirement so elevating the
Enzyme Activity Lecture Every reaction has energy requirement. The minimum amount of energy required is termed activation energy. Living organisms have optimum temperature requirement so elevating the
Learning Target: Describe characteristics and functions of carbohydrates, lipids, and proteins. Compare and contrast the classes of organic
 Learning Target: Describe characteristics and functions of carbohydrates, lipids, and proteins. Compare and contrast the classes of organic compounds. What are inorganic molecules? Molecules that CANNOT
Learning Target: Describe characteristics and functions of carbohydrates, lipids, and proteins. Compare and contrast the classes of organic compounds. What are inorganic molecules? Molecules that CANNOT
Study on Different types of Structure based Properties in Human Membrane Proteins
 2017 IJSRST Volume 3 Issue 7 Print ISSN: 2395-6011 Online ISSN: 2395-602X Themed Section: Science and Technology Study on Different types of Structure based Properties in Human Membrane Proteins S. C.
2017 IJSRST Volume 3 Issue 7 Print ISSN: 2395-6011 Online ISSN: 2395-602X Themed Section: Science and Technology Study on Different types of Structure based Properties in Human Membrane Proteins S. C.
Energy Flow. Chapter 7. Cellular Respiration: Overview. Cellular Respiration. Cellular Respiration. Cellular Respiration occurs in three stages
 Energy Flow Chapter 7 Cellular Respiration hotosynthesis uses solar energy to produce glucose and O from CO and H O Cellular respiration makes and consumes O during the oxidation of glucose to CO and H
Energy Flow Chapter 7 Cellular Respiration hotosynthesis uses solar energy to produce glucose and O from CO and H O Cellular respiration makes and consumes O during the oxidation of glucose to CO and H
Supplementary Information
 Supplementary Information Two common structural motifs for TCR recognition by staphylococcal enterotoxins Karin Erica Johanna Rödström 1, Paulina Regenthal 1, Christopher Bahl 2, Alex Ford 2, David Baker
Supplementary Information Two common structural motifs for TCR recognition by staphylococcal enterotoxins Karin Erica Johanna Rödström 1, Paulina Regenthal 1, Christopher Bahl 2, Alex Ford 2, David Baker
