Investigation of the binding of spermine and derivative polyamines on the 1U6M active site in the presence of Acetyl coenzyme-a
|
|
|
- Timothy McBride
- 5 years ago
- Views:
Transcription
1 Investigation of the binding of spermine and derivative polyamines on the 1U6M active site in the presence of Acetyl coenzyme-a Lindsay Mitchell Pace University NYC Department of Chemistry and Physical Sciences Advisor: Dr. Demosthenes Athanasopoulos
2 TABLE OF CONTENTS LIST OF FIGURES AND TABLES 3 ABSTRACT...4 INTRODUCTION.5 MATERIALS & METHODS.13 RESULTS. 16 DISCUSSION...26 REFERENCES
3 LIST OF FIGURES AND TABLES Figure 1. PyRx User Interface Figure 2. Polyamine biosynthetic pathway in C. parvum Figure 3. Polyamine Structures Figure 4. Docking Quadrants.14 Table 1. Polyamine Binding Energies Table 2. GNAT Binding Energies...17 Table 3. Binding Positions Relative to Spermine..18 Table 4. Binding Site Locations Compared With and Without Coenzyme...20 Figure 4. Spermine (Quadrant A) without Coenzyme.22 Figure 5. Spermine (Quadrant A) with Coenzyme...22 Figure 6. Spermine (Quadrant B) without Coenzyme..23 Figure 7. Spermine (Quadrant B) with Coenzyme...23 Figure 8. Spermine (Quadrant C) without Coenzyme.24 Figure 9. Spermine (Quadrant C) with Coenzyme..24 Figure 10. Diaminobutane (Quadrant C) without Coenzyme.25 Figure 11. Diaminobutane (Quadrant C) with Coenzyme.. 25
4 ABSTRACT Cryptosporidiosis is a disease caused by Cryptosporidium parvum that is not curable for immune-compromised individuals. Spermidine/spermine N 1 -acetyltransferase (SSAT) is an enzyme in C. parvum that retro converts spermine to spermidine and spermidine to putrescine. Since a treatment for the disease does not exist, studies are ongoing to establish a difference between SSAT in C. parvum and SSAT in the human host cell to determine whether CpSSAT could be a good drug target. This study investigates binding sites of the 1U6M active site of the CpSSAT enzyme using various computer programs including PythonPrescription, AutoDockVina, and AutoDockTools. These programs analyze the binding affinity for spermine and derivative polyamines to specific binding sites within the 1U6M active site to investigate how the different ligands will bind, especially in the presence of acetyl-coenzyme A (CoA). It was found that the presence of acetyl-coa did not affect the binding energies of the polyamines. A direct trend between polyamine size and binding energies was also found. As polyamine size increased (closer to size or spermine) the binding energies decreased. Polyamines with similar sizes to spermine were found to have similar binding energies as well. Additionally, it was found that the enzyme was more specific for spermine and spermidine than lysine, Dapsone, and para-aminobenzoic acid indicating that the enzyme belongs to the SSAT protein family, not the general N-acetyltransferase (GNAT) group. 4
5 INTRODUCTION Molecular Docking Virtual docking, a computational method for determining possible binding sites of ligands on enzymes, is employed in this experiment to determine where spermine and it s derivative polyamines will bind to the spermidine/spermine N 1 -acetyltransferase (SSAT) enzyme in the presence of acetyl Co-enzyme A (acetyl-coa). There are many different virtual docking programs that exist but these computational packages all utilize two key functions, search algorithms and a scoring function, in order to determine the best possible docking positions for a ligand on the enzymatic protein (Onodera 2007). The docking methodology consists of scanning of the surface of the protein by the potentially bound ligand. In the beginning, we define a grid of points on the surface of the protein. This step requires a balanced approach: the resolution of the grid points must be enough for a thorough scanning of the surface of the protein, but not excessive since this would require long computing time. During the docking calculation, the ligand molecule is placed sequentially in each one of these points and the interactions between the ligand and the protein at this particular relative position is allowed to reach a minimum of the potential energy. At the end of the calculation, the program will select these grid points and conformations of the ligand that correspond to the lowest potential energy and therefore constitute probably binding sites. 5
6 The search algorithm in the AutoDock program, one of the programs used in this experiment, is comprised of a combination of a genetic algorithm and the Lamarkian genetic algorithm, which is a local search algorithm (Onodera 2007). In the case of molecular modeling, the algorithm searches all possible conformations of the ligand and scans it against the enzyme (Trott 2009). The user designates a search space, which is the space on the enzymatic protein that the computer will test within, and all the possible conformations are scanned against all possible locations within that search space. Smaller search spaces are better because it is easier for the algorithm to explore it (Trott 2009). The next part of the virtual screening process is assigning a scoring function, which is the evaluation of the local minima of the ligand-protein interaction. After scanning, the results are given a fitness value and the results with the highest values are reported. These fitness values correlate to the binding energy, in kcal/mol, required to bind a specific conformation of a ligand to the enzyme in any given location (Onodera 2007). AutoDock s scoring functions produces two results, which are the final docking energy (FDE) and the estimated final energy of binding (EFEB). The FDE is the first result, while the EFEB includes other factors such as van der Waals interactions and hydrogen binding (Onodera 2007). Python Prescription, PyRx, is a graphical interface for the preparation of the input data and the performance of the calculation in an integrated user friendly environment. It uses open source software including AutoDock4, AutoDock Vina, and AutoDockTools. 6
7 AutoDock 4 and AutoDock Vina are the two docking software programs that utilize the algorithm and scoring function to find the docking results. AutoDockTools is used to generate the input files,.pdb or.pdbqt files, which are used in the docking program (PyRx, Introduction 2009). AutoDockVina is the newer version of AutoDock4 with improved accuracy in regards to binding predictions (Trott 2009). PyRx uses both programs so the user can choose which one to work with. In this experiment, AutoDockVina was used. The user interface of PyRx is shown in Figure 1 below. Figure 1. This is the interface for the PyRx program. Along the bottom Controls panel, the user can select whether to use the Vina Wizard (AutoDockVina) of the AutoDock Wizard (AutoDock4). Also, in the Controls panel, the ligands and macromolecules to be docked are selected using the Select Molecules window. When selected they will appear in the Navigator panel under Molecules. AutoDockTools is a software program from The Scripps Research Institute in their Molecular Graphics Laboratory. It is comprised of three main applications, AutoDockTools (ADT), Python Molecular Viewer (PMV), and Vision. The first two 7
8 were used in this experiment. ADT is used for making the molecules run in AutoDock. PMV is a more advanced molecular viewer that can show molecules displayed in multiple ways, such ball and stick, ribbon, and molecular surface configurations (MGL Tools 2010). Enzyme Background An enzyme is a protein that speeds up the rate of a reaction by decreasing the activation energy (Dolphin 2008). The amino acid groups that make up the protein, as well as the secondary and tertiary structures, help determine the shape of the enzyme. It has an active site, where the substrate binds, which is specific to certain ligands present on the substrate (Dolphin 2008). When the substrate enters the active site of the enzyme it binds with the enzyme and sometimes the binding is facilitated by the presence of a coenzyme, which are usually metallic ions that help the substrate bind to the enzyme. After the ligand is bound to the active site, the enzyme changes its shape so that the substrate fits better in the enzyme. When bound to the active site of the enzyme an enzyme-substrate complex forms weak, non-covalent bonds. The covalent bonds in the substrate are stressed, breaking the bonds in the substrate and converting it to the products (Dolphin 2008). Enzymatic activity is dependent on multiple factors including temperature, ph, concentration of enzyme or substrate, and presence of inhibitors. Competitive inhibitors are compounds that have structures similar to the ligand, so the inhibitor can bind to the active site. When a competitive inhibitor binds to an active site, the substrate ligand 8
9 cannot bind to that same active site, resulting in reversible inhibition (Campbell and Reece 2008). Irreversible inhibitors covalently bind to the active site or they damage a functional group so that the intended ligand can no longer bind. Reversible inhibitors, however, are non-covalently bonded and do not change the active site so they can be removed (Nelson 2008). Cryptosporidium parvum Background Cryptosporidium parvum is a parasite that causes cryptosporidiosis, a diarrheal disease with symptoms such as dehydration, nausea/vomiting, stomach pains, and weight loss that affects approximately 748,000 people per year in the United States alone (CDC, Disease 2010 & CDC, Epidemiology & Risk Factors 2011). The different species of C. parvum, which can infect both humans and animals, are spread many different ways, the most common being through water (CDC, Parasite-Cryptosporidium). While treatment is usually not necessary for people with healthy immune systems, infection can last longer and be more severe for people with weaker immune systems. For example, for individuals with AIDS, cryptosporidiosis is not curable. Even if the symptoms disappear, they could return with the weakening of the immune system over time (CDC, Treatment 2012). Spermidine/spermine N 1 -acetyltransferase (SSAT) is an important enzyme for the metabolism of polyamines by C. parvum (Yarlett 2007). It was determined that this enzyme, along with polyamine oxidase, converts spermine to lower polyamines. SSAT in C. parvum (CpSSAT) has been compared to SSAT in the host cell (HsSSAT) to 9
10 determine whether SSAT would be a good target for drug design, and the two SSAT s were in fact found to have different properties making it a good target (Yarlett 2007). Biochemical Mechanism of Spermine Interaction with SSAT As was mentioned before, CpSSAT metabolizes polyamines in C. parvum. More specifically, CpSSAT retro-converts spermine to spermidine and spermidine to putrescence (diaminobutane) in the presence of acetyl-coa (Yarlett 2007). This pathway is shown below in Figure 2. While some information about pathway is known, it is not known if the complex is ternary or substituted. It is also unknown whether the reaction is ordered or random. Using the 1U6M active site of the CpSSAT, docking can be performed on the enzyme to help determine whether or not it is ternary or substituted as well as random or ordered. Figure 2. Polyamine biosynthetic pathway in C. parvum. spermine to spermidine then to putrescine. SSAT retro-converts The 1U6M active site is chosen for two main reasons. First, since it is represents the active site, it contains the substrate binding domain of CpSSAT. Second, the entire SSAT is so large that the computer cannot determine the fold of the entire molecule. With this particular part however, the computer can use HsSSAT as a guide for folding 10
11 because the two SSAT s share a high degree of amino acid similarity in the active site. The HsSSAT contains five arginine residues, while the CpSSAT contains eight arginine residues (five of which are in the same location as the human enzyme). Investigation of Activity of Other Polyamines Figure 3. The structures of the three main polyamines used in this experiment. From top to bottom: spermine, spermidine and 1,4-diaminobutane. These are the polyamines that are used in the SSAT pathway shown in Figure 2. Spermine and Spermidine (shown above in Figure 3) are both cationic molecules involved in cellular metabolism. Spermine is the preferred polyamine in C. parvum SSAT, while spermidine is preferred in the human enzyme meaning that CpSSAT exhibited higher enzyme activity for spermine, while HsSSAT exhibited higher enzyme activity for spermidine (Yarlett 2007). Since it is known that spermine is acetylated to N- acetyl spermine, which can be further metabolized to spermidine, N-acetylspermidine or putrescine, it is of interest to note whether or not the other polyamines of different sizes can also bind to the 1U6M active site. The additional polyamines scanned are 1,3- diaminopropane, 1,5-diaminopentane, 1,6-diaminohexane, 1,7-diaminoheptane, 1,8- diaminooctane, 1,9-diaminononane, 1,10-diaminodecane, and 1,11-diaminendecane. It 11
12 was already found that a molecule larger than spermine was able to dock because it made boomerang curve allowing the amine groups on the end of the molecule to interact with the active site. By using a series of larger polyamines that only contain amino groups on the end, it is possible to determine whether the size affects binding with the enzyme, and if so, at what size is there a change in ability to bind. Activity of Non-Polyamine Ligands Another use for the molecular docking is to determine whether or not the enzyme in question is actually CpSSAT or a Gcn5-related N-acetyltransferases (GNAT). GNAT s are a group of enzymes that function similarly to SSAT in that they also transfer the acetyl group to the ligand (Vetting 2005). There are three molecules, Histone II A, Dapsone, and p-aminobenzoic acid that typically bind with GNAT s. Lysine will be docked in place of Histone II A because it is the active residue of Histone II A that binds to the enzyme active site. If these ligands selectively bind to the 1U6M active site, it infers the enzyme belongs to the GNAT class. If these ligands do not selectively bind however, it would support previous enzymatic data that the enzyme is a true SSAT. Our Computational Approach Our calculations aim to investigate, at the molecular level, the interactions of all the above mentioned ligands with the SSAT. In particular, to identify the possible binding sites on the protein surface, the binding affinities for each ligand and each binding position. Furthermore, we address the effect of the pre-bonded coenzyme in the ligand/protein interactions. 12
13 MATERIALS & METHODS 1. PyRx/Vina Computational Package Polyamines The Vina Computational Package is used to run a virtual screening of the possible binding sites of spermine, spermidine, diaminodecane, diaminoendecane, diaminopentane, diaminoheptane, diaminohexane, diaminononane, diaminooctane, diaminopentane, and diaminopropane, and acetyl-coa on the enzyme. The enzyme is separated into four quadrants, A, B, C, and D (shown on the next page in Figure 4), and the 9 binding sites with the highest binding energies are found in each part totaling 32 binding sites for each polyamine. First, the enzyme was screened in the presence of spermine in quadrant A and the 9 binding sites were obtained. Then it was screened in quadrant B, followed by quadrant C and D. This same process was repeated for spermidine, the rest of the polyamines, and acetyl-coa. Next, the polyamines were all screened again in quadrant A in the presence of both the polyamine and acetyl-coa. This is used to determine whether or not there is a difference between the binding sites and energies with or without the presence of acetyl-coa. GNAT Experiment The same procedures will also be followed using lysine, Dapsone, and paraaminobenzoic acid. 13
14 Figure 4. This figure shows the four docking quadrants in respect to the enzyme as well as the coordinates of each quadrant. Quadrant A is shown in the top left, B in the top right, C in the bottom left and D in the bottom right. 14
15 2. AutoDockTools The runs from the Vina computational program are then analyzed using AutoDockTools1.5.4 by AutoDockTools. This program visually shows the binding energies and locations of the polyamines in regard to the enzyme. The highest binding energies are recorded for each polyamine/ligand in quadrants A, B, C, and D without the coenzyme. Then they are recorded for quadrant A in the presence of the coenzyme. In the case that all nine locations are similar, only the lowest binding energies are recorded. If the locations are scattered within the quadrant, the lowest binding energy at each location is recorded. 15
16 RESULTS Table 1. Polyamine Binding Energies Polyamine Binding Energy w/o Coenzyme Binding Energy w/ Coenzyme Spermine A Spermine B -4.4, , -4.4, -4.2 Spermine C -4.4, -4.1, , -4.1 Spermine D -4.3, -4.1, -4.5, -4.2 Spermidine A -3.9, -3.9, , -4.0, -3.8 Spermidine B -4.0, , -3.8, -3.8, -3.7 Spermidine C -3.9, , -3.7 Spermidine D -4.0, -3.6, , -3.7 Diaminopropane A -3.1, -3.1, , -3.1, -3.0, -2.9, -2.9 Diaminopropane B -3.1, -3.0, -2.8, , -3.1, -2.9, -2.9, -2.8 Diaminopropane C -3.0, -2.8, , -3.0, -2.8, -2.7 Diaminopropane D -3.1, -2.9, , -3.0, -2.8, Diaminobutane A -3.4, -3.3, , -3.4, -3.3 Diaminobutane B -3.3, -3.3, , -3.4, -3.4 Diaminobutane C -3.3, -3.2, -3.1, -3.3, -3.1, -2.9, -2.9 Diaminobutane D -3.4, -3.2, , -3.4, -3.2 Diaminopentane A -3.6, -3.5, -3.4, , -3.5, -3.4, -3.3 Diaminopentane B -3.5, -3.3, , -3.5, -3.5, -3.4 Diaminopentane C -3.5, -3.5, -3.3, , -3.6, -3.3 Diaminopentane D -3.6, -3.5, -3.3, , -3.5, -3.3 Diaminohexane A -3.8, , -3.6 Diaminohexane B -3.7, -3.5, , -3.5, -3.5 Diaminohexane C -3.7, -3.6, -3.6, , -3.7, -3.7 Diaminohexane D -3.7, -3.7, , -3.6, -3.5, -3.4, -3.4 Diaminoheptane A -4.0, , -3.7, -3.7 Diaminoheptane B -4.0, -3.6, , -3.7, -3.7, -3.7 Diaminoheptane C -4.0, -3.8, -3.8, , -3.7, -3.5 Diaminoheptane D -3.9, -3.8, , -3.7, -3.6, -3.5 Diaminooctane A -4.3, Diaminooctane B -3.9, -3.7, , -3.9, -3.7 Diaminooctane C -4.4, -4.1, -3.9, , -3.7 Diaminooctane D -3.7, , -3.9, Diaminononane A
17 Diaminononane B -4.0, , -4.0, -3.8, -3.8 Diaminononane C -4.0, -3.9, , -4.0, -3.8 Diaminononane D -4.2, , -3.8, -3.7 Diaminodecane A -4.9, Diaminodecane B -4.2, , -4.3 Diaminodecane C -4.2, -4.2, -4.0, , -3.9, Diaminodecane D -4.2, , -3.7, Diaminoendecane A Diaminoendecane B -4.7, , -3.9, -3.9, -3.8 Diaminoendecane -4.6, -4.5, , -4.2, -4.0, -4.0 Diaminoendecane D -4.2, , -4.0 This table shows the binding energies obtained without the presence of the coenzyme (first column) as well as in the presence of the coenzyme (column 2). A, B, C, and D refers to the quadrant of the macromolecule in which it was tested. Multiple binding energies per polyamine correlate to multiple possible binding sites. The green highlighting indicates that the first binding posiiton of the polyamine was the same with or without the coenzyme. 17
18 Table 2. GNAT Experiment binding energies Polyamine Binding Energy w/o Coenzyme Binding Energy w/ Coenzyme Spermine A Spermine B -4.4, , -4.4, -4.2 Spermine C -4.4, -4.1, , -4.1 Spermine D -4.3, , -4.2 Spermidine A -3.9, -3.9, , -4.0, -3.8 Spermidine B -4.0, , -3.8, -3.8, -3.7 Spermidine C -3.9, , -3.7 Spermidine D -4.0, -3.6, , -3.7 Histone A -4.7, -4.3, , -4.1, -4.0, -4.0 Histone B -4.3, , -4.3, -4.3, -4.3, -4.0, -4.0 Histone C -4.3, -4.0, -3.8, , -4.5, -4.5 Histone D -4.7, -4.1, -4.3, -4.1, -4.0, -4.0 Dapsone A , -6.7 Dapsone B -6.5, -5.7, , -6.4, -6.1, -6.1 Dapsone C -6.9, , -6.0, -5.9, -5.9 Dapsone D -7.0, -5.8, -5.6, , -5.7, -5.6, -5.6 p-aminobenzoic acid A -5.5, -5.3, -5.1, , -5.3, -4.8 p-aminobenzoic acid B -5.3, -5.1, -5.0, -4.7, , -5.3, -5.3, -5.0, -4.9 p-aminobenzoic acid C -5.1, -5.0, -4.8, , -5.0, -4.9, -4.6 p-aminobenzoic acid D -5.4, -5.0, , -5.0 These are the binging energies obtained with and without the coenzyme for the following ligands: Histone IIa, Dapsone, and p-aminobenzoic acid. Multiple binding energies per polyamine correlate to multiple possible binding sites. The ligands highlighted in green had a similar first binding position with and without the coenzyme present. 18
19 Table 3. Binding Positions Relative to Spermine QUAD A w/o POLYAMINES coenzyme Spermidine Diaminopropane Diaminobutane Diaminopentane Diaminohexane Diaminoheptane Diaminooctane Diaminononane Diaminodecane Diaminoendecane LIGANDS Histone Dapsone p-aminobenzoic acid QUAD C w/o POLYAMINES coenzyme Spermidine Diaminopropane Diaminobutane Diaminopentane Diaminohexane Diaminoheptane Diaminooctane Diaminononane Diaminodecane Diaminoendecane LIGANDS Histone Dapsone p-aminobenzoic acid w/ coenzyme w/ coenzyme QUAD B w/o POLYAMINES coenzyme Spermidine Diaminopropane Diaminobutane Diaminopentane Diaminohexane Diaminoheptane Diaminooctane Diaminononane Diaminodecane Diaminoendecane LIGANDS Histone Dapsone p-aminobenzoic acid QUAD D w/o POLYAMINES coenzyme Spermidine Diaminopropane Diaminobutane Diaminopentane Diaminohexane Diaminoheptane Diaminooctane Diaminononane Diaminodecane Diaminoendecane LIGANDS Histone Dapsone p-aminobenzoic acid w/ coenzyme w/ coenzyme All outputs 5 or more outputs 4 outputs less than 4 This table shows the binding positions of the polyamines/ligands relative to spermine. Light blue indicates that all ligand output positions were the same as 19
20 spermine positions. Purple indicated 5 or more outputs were the same, purple indicates 4, and orange indicated less than 4 outputs were the same as spermine. Table 4. Binding Site Locations Compared With and Without Coenzyme Ligand Site Locations Spermine A same site Spermine B introduces 1 new binding site Spermine C one binding site missing Spermine D same sites Spermidine A same sites Spermidine B introduces 2 new binding sites Spermidine C 2nd binding site moved Spermidine D one site missing Diaminopropane A introduces 2 new binding sites in relative same area Diaminopropane B one site missing; one introduced Diaminopropane C introduces 1 new binding site Diaminopropane D same sites Diaminobutane A same sites Diaminobutane B roughly same sites Diaminobutane C 3rd binding site moved, introduces 4th site Diaminobutane D 1 binding site moved Diaminopentane A same sites Diaminopentane B 1 binding site moved; 1 binding site splits into two Diaminopentane C one site missing; one introduced Diaminopentane D introduces 1 new binding site Diaminohexane A same sites Diaminohexane B one site moved Diaminohexane C one site missing Diaminohexane D introduces 2 new binding sites Diaminoheptane A introduces 1 new binding site Diaminoheptane B 1 binding site moved; 1 introduced Diaminoheptane C 2 binding sites condense into 1 Diaminoheptane D introduces 1 new binding site Diaminooctane A one site missing Diaminooctane B 2 sites missing; 2 introduced Diaminooctane C 2 binding sites missing Diaminooctane D same sites Diaminononane A same site Diaminononane B introduces 2 new binding sites 20
21 Diaminononane C Diaminononane D Diaminodecane A Diaminodecane B Diaminodecane C Diaminodecane D Diaminoendecane A Diaminoendecane B Diaminoendecane C Diaminoendecane D Histone A Histone B Histone C Histone D Dapsone A Dapsone B Dapsone C Dapsone D p-aminobenzoic acid A p-aminobenzoic acid B p-aminobenzoic acid C p-aminobenzoic acid D one site moved slightly, one site introduced binding sites become 1, introduced 2 new sites 2nd binding site disappears one binding site moves 2 binding sites disappear same sites same site 2 sites introduced introduces 1 new binding site same sites one site moved, introduces new site introduces 4 new sites 1 binding site disappears introduces 2 new sites introduces 1 new site one site remains the same one site remains the same one site moves one site moves, 1 introduced 3 binding sites are the same same sites one site missing This table indicates how the presence of the coenzyme affected binding positions. 21
22 Figure 4. Spermine (Quadrant A) without Coenzyme Figure 5. Spermine (Quadrant A) with Coenzyme These figures show that the same binding site exist with or without the coenzyme. 22
23 Figure 6. Spermine (Quadrant B) without Coenzyme Figure 7. Spermine (Quadrant B) with Coenzyme These figures show the addition of a new binding site in the presence of the coenzyme. 23
24 Figure 8. Spermine (Quadrant C) without Coenzyme Figure 9. Spermine (Quadrant C) with Coenzyme These two figures indicate that the presence of the coenzyme takes away a binding position. 24
25 Figure 10. Diaminobutane (Quadrant C) without Coenzyme Figure 11. Diaminobutane (Quadrant C) with Coenzyme These figures show the removal of one site and addition of two more sites in the presence of the coenzyme. 25
26 DISCUSSION Polyamine Binding Energies Overall, it was found that the presence of the acetyl-coa did not significantly affect the binding energies of the polyamines. Additionally, all except 15 polyamines had the same first binding position regardless of the presence of acetyl-coa. In the cases where the polyamine did not bind in the same position first, the second or third binding positions were similar. Compared to spermine, the positions of the ligands were relatively similar. In only 13 out of the 104 cases, did less than half of the outputs not match up with the spermine outputs. This indicates that the reaction is most likely a ternary reaction since the introduction of a coenzyme does not significantly affect the binding energy or positions. A direct trend between polyamine size and binding energies was also found. As the size of the polyamine increases, the binding energies become more negative and therefore stronger. The binding energies of the polyamines are consistent with that of spermine. Spermine has a 10-Carbon backbone (with 2 amine groups in the carbon chain and 2 more amine groups on the ends) and its energies are consistent with 1,9-diaminononane and 1,10-diaminodecane, which are molecules with 9-carbon and 10-carbon backbones respectively. GNAT Experiment Three ligands, lysine, Dapsone, and para-aminobenzoic acid were docked in order to determine whether or not the enzyme being worked with is in fact SSAT or if it is a 26
27 GNAT, an enzyme that functions similarly to SSAT. These three ligands were tested because they are known to bind well with GNAT s. The binding energies for the three ligands were more negative than spermine and spermidine, which could possibly indicate that they bind stronger. However, all the binding energies are so similar, it is not possibly to say that one definitely binds stronger than another. It is possible to determine, though, that spermine and spermidine are more specific for this enzyme than the lysine, Dapsone, and p-aminobenzoic acid. This would indicate that the enzyme is in fact SSAT, and not a GNAT. 27
28 REFERENCES 1. Onodera K, Satou K, Hirota H. Evaluations of Molecular Docking Programs for Virtual Screening. J Chem Inf Model 2007;47: Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. Journal of Computational Chemistry 2010;31: Introduction. Python Prescription Virtual Screening Tool. PyRx, 22 September Web 4. Dallakyan Sargis. MGL Tools Website-Welcome. MGL Tools. The Scripps Research Institute, 19 April Web. 5. Dolphin, W.D. Biological Investigations: Form, Function, Diversity and Process. 2008; Campbell, N.A., Reece, J.B. Biology. 2008; 8 7. Nelson DL, Cox MM. Principles of Biochemistry. 2008; Disease. Centers for Disease Control and Prevention. CDC, 2 November Web. 9. Epidemiology & Risk Factors. Centers for Disease Control and Prevention. CDC, 9 March Web. 10. Parasite-Cryptosporidium. Centers for Disease Control and Prevention. CDC, 16 January Web. 11. Treatments. Centers for Disease Control and Prevention. CDC, 1 March Web. 12. Yarlett N, Wu G, Waters WR, Harp JA, Wannemuehler MJ, Morada M, Athanasopoulos D, Martinez MP, Upton SJ, Marton LJ, Frydman BJ. 28
29 Cryptosporidium parvum spermidine/spermine N 1 -acetyltransferase exhibits different characteristics from the host enzyme. Mol Biochem Parisitol. 2007;152: Vetting MW. Structure and functions of the GNAT superfamily of acetyltransferases. Archives of Biochemistry and Biophysics. 2005;433:
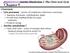
Project Manual Bio3055. Cholesterol Homeostasis: HMG-CoA Reductase
 Project Manual Bio3055 Cholesterol Homeostasis: HMG-CoA Reductase Bednarski 2003 Funded by HHMI Cholesterol Homeostasis: HMG-CoA Reductase Introduction: HMG-CoA Reductase is an enzyme in the cholesterol
Project Manual Bio3055 Cholesterol Homeostasis: HMG-CoA Reductase Bednarski 2003 Funded by HHMI Cholesterol Homeostasis: HMG-CoA Reductase Introduction: HMG-CoA Reductase is an enzyme in the cholesterol
Final Report. Identification of Residue Mutations that Increase the Binding Affinity of LSTc to HA RBD. Wendy Fong ( 方美珍 ) CNIC; Beijing, China
 Final Report Identification of Residue Mutations that Increase the Binding Affinity of LSTc to HA RBD Wendy Fong ( 方美珍 ) CNIC; Beijing, China Influenza s Viral Life Cycle 1. HA on virus binds to sialic
Final Report Identification of Residue Mutations that Increase the Binding Affinity of LSTc to HA RBD Wendy Fong ( 方美珍 ) CNIC; Beijing, China Influenza s Viral Life Cycle 1. HA on virus binds to sialic
Energy and catalysts. Enzymes. Contents. 1 Energy and catalysts 2 Enzymes
 Contents 1 Energy and catalysts 2 Enzymes Energy and catalysts In Biological systems, energy is roughly defined as the capacity to do work. Molecules are held together by electrons. Breaking and building
Contents 1 Energy and catalysts 2 Enzymes Energy and catalysts In Biological systems, energy is roughly defined as the capacity to do work. Molecules are held together by electrons. Breaking and building
AutoLigand: A Tool for Finding Ligand Binding Sites. CUHK workshop Dec, 2011
 AutoLigand: A Tool for Finding Ligand Binding Sites CUHK workshop Dec, 2011 Stefano Forli, Ph.D., TSRI Rodney M. Harris, Ph.D., 1060 Discovery Engineering Overview Description and uses of AutoLigand Algorithms
AutoLigand: A Tool for Finding Ligand Binding Sites CUHK workshop Dec, 2011 Stefano Forli, Ph.D., TSRI Rodney M. Harris, Ph.D., 1060 Discovery Engineering Overview Description and uses of AutoLigand Algorithms
Review of Energetics Intro
 Review of Energetics Intro Learning Check The First Law of Thermodynamics states that energy can be Created Destroyed Converted All of the above Learning Check The second law of thermodynamics essentially
Review of Energetics Intro Learning Check The First Law of Thermodynamics states that energy can be Created Destroyed Converted All of the above Learning Check The second law of thermodynamics essentially
The building blocks of life.
 The building blocks of life. The 4 Major Organic Biomolecules The large molecules (biomolecules OR polymers) are formed when smaller building blocks (monomers) bond covalently. via anabolism Small molecules
The building blocks of life. The 4 Major Organic Biomolecules The large molecules (biomolecules OR polymers) are formed when smaller building blocks (monomers) bond covalently. via anabolism Small molecules
News. Volume 8: November 2, 2009
 FightAIDS@Home News Volume 8: November 2, 2009 The FightAIDS@Home Project uses the volunteered computing power of the World Community Grid to test candidate compounds against the variations (or mutants
FightAIDS@Home News Volume 8: November 2, 2009 The FightAIDS@Home Project uses the volunteered computing power of the World Community Grid to test candidate compounds against the variations (or mutants
Terminology-Amino Acids
 Enzymes 1 2 Terminology-Amino Acids Primary Structure: is a polypeptide (large number of aminoacid residues bonded together in a chain) chain of amino acids linked with peptide bonds. Secondary Structure-
Enzymes 1 2 Terminology-Amino Acids Primary Structure: is a polypeptide (large number of aminoacid residues bonded together in a chain) chain of amino acids linked with peptide bonds. Secondary Structure-
Hemoglobin & Sickle Cell Anemia Exercise
 Name StarBiochem Hemoglobin & Sickle Cell Anemia Exercise Learning Objectives In this exercise, you will use StarBiochem, a protein 3D viewer, to explore the structure of the normal hemoglobin protein
Name StarBiochem Hemoglobin & Sickle Cell Anemia Exercise Learning Objectives In this exercise, you will use StarBiochem, a protein 3D viewer, to explore the structure of the normal hemoglobin protein
Simulate enzymatic actions. Explain enzymatic specificity. Investigate two types of enzyme inhibitors used in regulating enzymatic activity.
 Name: Enzymes in Action Objectives: You will use the model pieces in the kit to: Simulate enzymatic actions. Explain enzymatic specificity. Investigate two types of enzyme inhibitors used in regulating
Name: Enzymes in Action Objectives: You will use the model pieces in the kit to: Simulate enzymatic actions. Explain enzymatic specificity. Investigate two types of enzyme inhibitors used in regulating
Hemoglobin & Sickle Cell Anemia Exercise
 Name StarBiochem Hemoglobin & Sickle Cell Anemia Exercise Background Hemoglobin is the protein in red blood cells responsible for carrying oxygen from the lungs to the rest of the body and for returning
Name StarBiochem Hemoglobin & Sickle Cell Anemia Exercise Background Hemoglobin is the protein in red blood cells responsible for carrying oxygen from the lungs to the rest of the body and for returning
6.5 Enzymes. Enzyme Active Site and Substrate Specificity
 180 Chapter 6 Metabolism 6.5 Enzymes By the end of this section, you will be able to: Describe the role of enzymes in metabolic pathways Explain how enzymes function as molecular catalysts Discuss enzyme
180 Chapter 6 Metabolism 6.5 Enzymes By the end of this section, you will be able to: Describe the role of enzymes in metabolic pathways Explain how enzymes function as molecular catalysts Discuss enzyme
The building blocks of life.
 The building blocks of life. All the functions of the cell are based on chemical reactions. the building blocks of organisms BIOMOLECULE MONOMER POLYMER carbohydrate monosaccharide polysaccharide lipid
The building blocks of life. All the functions of the cell are based on chemical reactions. the building blocks of organisms BIOMOLECULE MONOMER POLYMER carbohydrate monosaccharide polysaccharide lipid
List of Figures. List of Tables
 Supporting Information for: Signaling Domain of Sonic Hedgehog as Cannibalistic Calcium-Regulated Zinc-Peptidase Rocio Rebollido-Rios 1, Shyam Bandari 3, Christoph Wilms 1, Stanislav Jakuschev 1, Andrea
Supporting Information for: Signaling Domain of Sonic Hedgehog as Cannibalistic Calcium-Regulated Zinc-Peptidase Rocio Rebollido-Rios 1, Shyam Bandari 3, Christoph Wilms 1, Stanislav Jakuschev 1, Andrea
Transient β-hairpin Formation in α-synuclein Monomer Revealed by Coarse-grained Molecular Dynamics Simulation
 Transient β-hairpin Formation in α-synuclein Monomer Revealed by Coarse-grained Molecular Dynamics Simulation Hang Yu, 1, 2, a) Wei Han, 1, 3, b) Wen Ma, 1, 2 1, 2, 3, c) and Klaus Schulten 1) Beckman
Transient β-hairpin Formation in α-synuclein Monomer Revealed by Coarse-grained Molecular Dynamics Simulation Hang Yu, 1, 2, a) Wei Han, 1, 3, b) Wen Ma, 1, 2 1, 2, 3, c) and Klaus Schulten 1) Beckman
Chapter 5 Microbial Metabolism: The Chemical Crossroads of Life
 Chapter 5 Microbial Metabolism: The Chemical Crossroads of Life Copyright The McGraw-Hill Companies, Inc. Permission required for reproduction or display. The Metabolism of Microbes metabolism all chemical
Chapter 5 Microbial Metabolism: The Chemical Crossroads of Life Copyright The McGraw-Hill Companies, Inc. Permission required for reproduction or display. The Metabolism of Microbes metabolism all chemical
Human Biochemistry. Enzymes
 Human Biochemistry Enzymes Characteristics of Enzymes Enzymes are proteins which catalyze biological chemical reactions In enzymatic reactions, the molecules at the beginning of the process are called
Human Biochemistry Enzymes Characteristics of Enzymes Enzymes are proteins which catalyze biological chemical reactions In enzymatic reactions, the molecules at the beginning of the process are called
Term Definition Example Amino Acids
 Name 1. What are some of the functions that proteins have in a living organism. 2. Define the following and list two amino acids that fit each description. Term Definition Example Amino Acids Hydrophobic
Name 1. What are some of the functions that proteins have in a living organism. 2. Define the following and list two amino acids that fit each description. Term Definition Example Amino Acids Hydrophobic
Enzymes: The Catalysts of Life
 Chapter 6 Enzymes: The Catalysts of Life Lectures by Kathleen Fitzpatrick Simon Fraser University Activation Energy and the Metastable State Many thermodynamically feasible reactions in a cell that could
Chapter 6 Enzymes: The Catalysts of Life Lectures by Kathleen Fitzpatrick Simon Fraser University Activation Energy and the Metastable State Many thermodynamically feasible reactions in a cell that could
BIOCHEMISTRY I HOMEWORK III DUE 10/15/03 66 points total + 2 bonus points = 68 points possible Swiss-PDB Viewer Exercise Attached
 BIOCHEMISTRY I HOMEWORK III DUE 10/15/03 66 points total + 2 bonus points = 68 points possible Swiss-PDB Viewer Exercise Attached 1). 20 points total T or F (2 points each; if false, briefly state why
BIOCHEMISTRY I HOMEWORK III DUE 10/15/03 66 points total + 2 bonus points = 68 points possible Swiss-PDB Viewer Exercise Attached 1). 20 points total T or F (2 points each; if false, briefly state why
7 Cellular Respiration and Fermentation
 CAMPBELL BIOLOGY IN FOCUS URRY CAIN WASSERMAN MINORSKY REECE 7 Cellular Respiration and Fermentation Lecture Presentations by Kathleen Fitzpatrick and Nicole Tunbridge, Simon Fraser University SECOND EDITION
CAMPBELL BIOLOGY IN FOCUS URRY CAIN WASSERMAN MINORSKY REECE 7 Cellular Respiration and Fermentation Lecture Presentations by Kathleen Fitzpatrick and Nicole Tunbridge, Simon Fraser University SECOND EDITION
Introduction to proteins and protein structure
 Introduction to proteins and protein structure The questions and answers below constitute an introduction to the fundamental principles of protein structure. They are all available at [link]. What are
Introduction to proteins and protein structure The questions and answers below constitute an introduction to the fundamental principles of protein structure. They are all available at [link]. What are
Guided Inquiry Skills Lab. Additional Lab 1 Making Models of Macromolecules. Problem. Introduction. Skills Focus. Materials.
 Additional Lab 1 Making Models of Macromolecules Guided Inquiry Skills Lab Problem How do monomers join together to form polymers? Introduction A small number of elements make up most of the mass of your
Additional Lab 1 Making Models of Macromolecules Guided Inquiry Skills Lab Problem How do monomers join together to form polymers? Introduction A small number of elements make up most of the mass of your
Molecular Graphics Perspective of Protein Structure and Function
 Molecular Graphics Perspective of Protein Structure and Function VMD Highlights > 20,000 registered Users Platforms: Unix (16 builds) Windows MacOS X Display of large biomolecules and simulation trajectories
Molecular Graphics Perspective of Protein Structure and Function VMD Highlights > 20,000 registered Users Platforms: Unix (16 builds) Windows MacOS X Display of large biomolecules and simulation trajectories
Workshop on Analysis and prediction of contacts in proteins
 Workshop on Analysis and prediction of contacts in proteins 1.09.09 Eran Eyal 1, Vladimir Potapov 2, Ronen Levy 3, Vladimir Sobolev 3 and Marvin Edelman 3 1 Sheba Medical Center, Ramat Gan, Israel; Departments
Workshop on Analysis and prediction of contacts in proteins 1.09.09 Eran Eyal 1, Vladimir Potapov 2, Ronen Levy 3, Vladimir Sobolev 3 and Marvin Edelman 3 1 Sheba Medical Center, Ramat Gan, Israel; Departments
Protein. An Introduction to Protein Structure and Denaturation by Acidification and Temperature Acid. By Noel Ways
 Protein An Introduction to Protein Structure and Denaturation by Acidification and Temperature Acid By oel Ways Amino Acids and Protein Structure A protein is a polymer of amino acids that obtains a unique
Protein An Introduction to Protein Structure and Denaturation by Acidification and Temperature Acid By oel Ways Amino Acids and Protein Structure A protein is a polymer of amino acids that obtains a unique
7.014 Problem Set 2 Solutions
 7.014 Problem Set 2 Solutions Please print out this problem set and record your answers on the printed copy. Answers to this problem set are to be turned in at the box outside 68-120 by 11:45 Friday, February
7.014 Problem Set 2 Solutions Please print out this problem set and record your answers on the printed copy. Answers to this problem set are to be turned in at the box outside 68-120 by 11:45 Friday, February
Ras and Cell Signaling Exercise
 Ras and Cell Signaling Exercise Learning Objectives In this exercise, you will use, a protein 3D- viewer, to explore: the structure of the Ras protein the active and inactive state of Ras and the amino
Ras and Cell Signaling Exercise Learning Objectives In this exercise, you will use, a protein 3D- viewer, to explore: the structure of the Ras protein the active and inactive state of Ras and the amino
Metabolism Energy Pathways Biosynthesis. Catabolism Anabolism Enzymes
 Topics Microbial Metabolism Metabolism Energy Pathways Biosynthesis 2 Metabolism Catabolism Catabolism Anabolism Enzymes Breakdown of complex organic molecules in order to extract energy and dform simpler
Topics Microbial Metabolism Metabolism Energy Pathways Biosynthesis 2 Metabolism Catabolism Catabolism Anabolism Enzymes Breakdown of complex organic molecules in order to extract energy and dform simpler
Supplementary Figure 1 (previous page). EM analysis of full-length GCGR. (a) Exemplary tilt pair images of the GCGR mab23 complex acquired for Random
 S1 Supplementary Figure 1 (previous page). EM analysis of full-length GCGR. (a) Exemplary tilt pair images of the GCGR mab23 complex acquired for Random Conical Tilt (RCT) reconstruction (left: -50,right:
S1 Supplementary Figure 1 (previous page). EM analysis of full-length GCGR. (a) Exemplary tilt pair images of the GCGR mab23 complex acquired for Random Conical Tilt (RCT) reconstruction (left: -50,right:
III. 6. Test. Respiració cel lular
 III. 6. Test. Respiració cel lular Chapter Questions 1) What is the term for metabolic pathways that release stored energy by breaking down complex molecules? A) anabolic pathways B) catabolic pathways
III. 6. Test. Respiració cel lular Chapter Questions 1) What is the term for metabolic pathways that release stored energy by breaking down complex molecules? A) anabolic pathways B) catabolic pathways
AP Biology Summer Assignment Chapter 3 Quiz
 AP Biology Summer Assignment Chapter 3 Quiz 2016-17 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. All of the following are found in a DNA nucleotide
AP Biology Summer Assignment Chapter 3 Quiz 2016-17 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. All of the following are found in a DNA nucleotide
Excerpt from J. Mol. Biol. (2002) 320, :
 Excerpt from J. Mol. Biol. (2002) 320, 1095 1108: Crystal Structure of the Ternary Complex of the Catalytic Domain of Human Phenylalanine Hydroxylase with Tetrahydrobiopterin and 3-(2-Thienyl)-L-alanine,
Excerpt from J. Mol. Biol. (2002) 320, 1095 1108: Crystal Structure of the Ternary Complex of the Catalytic Domain of Human Phenylalanine Hydroxylase with Tetrahydrobiopterin and 3-(2-Thienyl)-L-alanine,
Good Afternoon! 11/30/18
 Good Afternoon! 11/30/18 1. The term polar refers to a molecule that. A. Is cold B. Has two of the same charges C. Has two opposing charges D. Contains a hydrogen bond 2. Electrons on a water molecule
Good Afternoon! 11/30/18 1. The term polar refers to a molecule that. A. Is cold B. Has two of the same charges C. Has two opposing charges D. Contains a hydrogen bond 2. Electrons on a water molecule
Foundations in Microbiology Seventh Edition
 Lecture PowerPoint to accompany Foundations in Microbiology Seventh Edition Talaro Chapter 8 An Introduction to Microbial Metabolism Copyright The McGraw-Hill Companies, Inc. Permission required for reproduction
Lecture PowerPoint to accompany Foundations in Microbiology Seventh Edition Talaro Chapter 8 An Introduction to Microbial Metabolism Copyright The McGraw-Hill Companies, Inc. Permission required for reproduction
AP Biology. Metabolism & Enzymes
 Metabolism & Enzymes From food webs to the life of a cell energy energy energy Flow of energy through life Life is built on chemical reactions transforming energy from one form to another organic molecules
Metabolism & Enzymes From food webs to the life of a cell energy energy energy Flow of energy through life Life is built on chemical reactions transforming energy from one form to another organic molecules
CHAPTER 2- ENZYMES PROTEINS B. AMINO ACID- 10/4/2016
 CHAPTER 2- ENZYMES BIOL. 1 AB KENNEDY PROTEINS A. DEFINITION- LARGE MACROMOLECULES MADE OF CARBON, HYDROGEN, NITROGEN, OXYGEN, AND SULFUR THEIR PRIMARY BUILDING BLOCK IS THE AMINO ACID THEY FUNCTION AS
CHAPTER 2- ENZYMES BIOL. 1 AB KENNEDY PROTEINS A. DEFINITION- LARGE MACROMOLECULES MADE OF CARBON, HYDROGEN, NITROGEN, OXYGEN, AND SULFUR THEIR PRIMARY BUILDING BLOCK IS THE AMINO ACID THEY FUNCTION AS
Supporting Information Identification of Amino Acids with Sensitive Nanoporous MoS 2 : Towards Machine Learning-Based Prediction
 Supporting Information Identification of Amino Acids with Sensitive Nanoporous MoS 2 : Towards Machine Learning-Based Prediction Amir Barati Farimani, Mohammad Heiranian, Narayana R. Aluru 1 Department
Supporting Information Identification of Amino Acids with Sensitive Nanoporous MoS 2 : Towards Machine Learning-Based Prediction Amir Barati Farimani, Mohammad Heiranian, Narayana R. Aluru 1 Department
Biological Science 101 General Biology
 Lecture Seven: Cellular Respiration Ch. 9, Pgs. 163-181 Figs. 9.2-9.20 Biological Science 101 General Biology Cellular Respiration: - A series of processes that is involved in converting food to energy
Lecture Seven: Cellular Respiration Ch. 9, Pgs. 163-181 Figs. 9.2-9.20 Biological Science 101 General Biology Cellular Respiration: - A series of processes that is involved in converting food to energy
Biological Molecules B Lipids, Proteins and Enzymes. Triglycerides. Glycerol
 Glycerol www.biologymicro.wordpress.com Biological Molecules B Lipids, Proteins and Enzymes Lipids - Lipids are fats/oils and are present in all cells- they have different properties for different functions
Glycerol www.biologymicro.wordpress.com Biological Molecules B Lipids, Proteins and Enzymes Lipids - Lipids are fats/oils and are present in all cells- they have different properties for different functions
Enzymes Topic 3.6 & 7.6 SPEED UP CHEMICAL REACTIONS!!!!!!!
 Enzymes Topic 3.6 & 7.6 SPEED UP CHEMICAL REACTIONS!!!!!!! Key Words Enzyme Substrate Product Active Site Catalyst Activation Energy Denature Enzyme-Substrate Complex Lock & Key model Induced fit model
Enzymes Topic 3.6 & 7.6 SPEED UP CHEMICAL REACTIONS!!!!!!! Key Words Enzyme Substrate Product Active Site Catalyst Activation Energy Denature Enzyme-Substrate Complex Lock & Key model Induced fit model
Org/Biochem Final Lec Form, Spring 2012 Page 1 of 6
 Page 1 of 6 Missing Complete Protein and Question #45 Key Terms: Fill in the blank in the following 25 statements with one of the key terms in the table. Each key term may only be used once. Print legibly.
Page 1 of 6 Missing Complete Protein and Question #45 Key Terms: Fill in the blank in the following 25 statements with one of the key terms in the table. Each key term may only be used once. Print legibly.
GRU3L1 Metabolism & Enzymes. AP Biology
 GRU3L1 Metabolism & Enzymes From food webs to the life of a cell energy energy energy Flow of energy through life Life is built on chemical reactions u transforming energy from one form to organic molecules
GRU3L1 Metabolism & Enzymes From food webs to the life of a cell energy energy energy Flow of energy through life Life is built on chemical reactions u transforming energy from one form to organic molecules
Lecture Sixteen: METABOLIC ENERGY: [Based on GENERATION Chapter 15
 Lecture Sixteen: METABOLIC ENERGY: [Based on GENERATION Chapter 15 AND STORAGE Berg, (Figures in red are for the 7th Edition) Tymoczko (Figures in Blue are for the 8th Edition) & Stryer] Two major questions
Lecture Sixteen: METABOLIC ENERGY: [Based on GENERATION Chapter 15 AND STORAGE Berg, (Figures in red are for the 7th Edition) Tymoczko (Figures in Blue are for the 8th Edition) & Stryer] Two major questions
Lecture 10 More about proteins
 Lecture 10 More about proteins Today we're going to extend our discussion of protein structure. This may seem far-removed from gene cloning, but it is the path to understanding the genes that we are cloning.
Lecture 10 More about proteins Today we're going to extend our discussion of protein structure. This may seem far-removed from gene cloning, but it is the path to understanding the genes that we are cloning.
1) DNA unzips - hydrogen bonds between base pairs are broken by special enzymes.
 Biology 12 Cell Cycle To divide, a cell must complete several important tasks: it must grow, during which it performs protein synthesis (G1 phase) replicate its genetic material /DNA (S phase), and physically
Biology 12 Cell Cycle To divide, a cell must complete several important tasks: it must grow, during which it performs protein synthesis (G1 phase) replicate its genetic material /DNA (S phase), and physically
Enzymes and Metabolism
 PowerPoint Lecture Slides prepared by Vince Austin, University of Kentucky Enzymes and Metabolism Human Anatomy & Physiology, Sixth Edition Elaine N. Marieb 1 Protein Macromolecules composed of combinations
PowerPoint Lecture Slides prepared by Vince Austin, University of Kentucky Enzymes and Metabolism Human Anatomy & Physiology, Sixth Edition Elaine N. Marieb 1 Protein Macromolecules composed of combinations
Prerequisite Knowledge: Students should have already been introduced to the inputs and outputs of photosynthesis.
 www.ngsslifescience.com. Topic: Metabolism Chemistry Model Summary: Students will learn act out polymerization by performing dehydration synthesis and hydrolysis using chemistry models. Students will also
www.ngsslifescience.com. Topic: Metabolism Chemistry Model Summary: Students will learn act out polymerization by performing dehydration synthesis and hydrolysis using chemistry models. Students will also
FIRST BIOCHEMISTRY EXAM Tuesday 25/10/ MCQs. Location : 102, 105, 106, 301, 302
 FIRST BIOCHEMISTRY EXAM Tuesday 25/10/2016 10-11 40 MCQs. Location : 102, 105, 106, 301, 302 The Behavior of Proteins: Enzymes, Mechanisms, and Control General theory of enzyme action, by Leonor Michaelis
FIRST BIOCHEMISTRY EXAM Tuesday 25/10/2016 10-11 40 MCQs. Location : 102, 105, 106, 301, 302 The Behavior of Proteins: Enzymes, Mechanisms, and Control General theory of enzyme action, by Leonor Michaelis
Chapter 6. Flow of energy through life. Chemical reactions of life. Examples. Examples. Chemical reactions & energy 9/7/2012. Enzymes & Metabolism
 Flow of energy through life Chapter 6 Life is built on chemical reactions Enzymes & Metabolism Chemical reactions of life Examples Metabolism Forming bonds between molecules Dehydration synthesis Anabolic
Flow of energy through life Chapter 6 Life is built on chemical reactions Enzymes & Metabolism Chemical reactions of life Examples Metabolism Forming bonds between molecules Dehydration synthesis Anabolic
Putrescine Lyndsay Grover Biotechnology Technician - Health CHEM April 4 th 2011
 Putrescine Lyndsay Grover Biotechnology Technician - Health CHEM 10002 April 4 th 2011 Introduction Putrescine is found in living and dead organisms due to the breakdown of amino acids and is the main
Putrescine Lyndsay Grover Biotechnology Technician - Health CHEM 10002 April 4 th 2011 Introduction Putrescine is found in living and dead organisms due to the breakdown of amino acids and is the main
Large Biological Molecules Multiple Choice Review
 New Jersey enter for Teaching and Learning Slide 1 / 43 Progressive Science Initiative This material is made freely available at www.njctl.org and is intended for the non-commercial use of students and
New Jersey enter for Teaching and Learning Slide 1 / 43 Progressive Science Initiative This material is made freely available at www.njctl.org and is intended for the non-commercial use of students and
Enzymes. Ch 3: Macromolecules
 Enzymes Ch 3: Macromolecules Living things use different chemical reactions to get the energy needed for life Chemical Reactions Reactants = substance that is changed Products = new substance that forms
Enzymes Ch 3: Macromolecules Living things use different chemical reactions to get the energy needed for life Chemical Reactions Reactants = substance that is changed Products = new substance that forms
Biological Molecules Ch 2: Chemistry Comes to Life
 Outline Biological Molecules Ch 2: Chemistry Comes to Life Biol 105 Lecture 3 Reading Chapter 2 (pages 31 39) Biological Molecules Carbohydrates Lipids Amino acids and Proteins Nucleotides and Nucleic
Outline Biological Molecules Ch 2: Chemistry Comes to Life Biol 105 Lecture 3 Reading Chapter 2 (pages 31 39) Biological Molecules Carbohydrates Lipids Amino acids and Proteins Nucleotides and Nucleic
Chapter 6. Metabolism & Enzymes. AP Biology
 Chapter 6. Metabolism & Enzymes Flow of energy through life Life is built on chemical reactions Chemical reactions of life Metabolism forming bonds between molecules dehydration synthesis anabolic reactions
Chapter 6. Metabolism & Enzymes Flow of energy through life Life is built on chemical reactions Chemical reactions of life Metabolism forming bonds between molecules dehydration synthesis anabolic reactions
MITOCW watch?v=lcih8faydgk
 MITOCW watch?v=lcih8faydgk The following content is provided under a Creative Commons license. Your support will help MIT OpenCourseWare continue to offer high-quality educational resources for free. To
MITOCW watch?v=lcih8faydgk The following content is provided under a Creative Commons license. Your support will help MIT OpenCourseWare continue to offer high-quality educational resources for free. To
Trilateral Project WM4
 ANNEX 2: Comments of the JPO Trilateral Project WM4 Comparative studies in new technologies Theme: Comparative study on protein 3-dimensional (3-D) structure related claims 1. Introduction As more 3-D
ANNEX 2: Comments of the JPO Trilateral Project WM4 Comparative studies in new technologies Theme: Comparative study on protein 3-dimensional (3-D) structure related claims 1. Introduction As more 3-D
WHY IS THIS IMPORTANT?
 CHAPTER 3 ESSENTIALS OF METABOLISM WHY IS THIS IMPORTANT? It is important to have a basic understanding of metabolism because it governs the survival and growth of microorganisms The growth of microorganisms
CHAPTER 3 ESSENTIALS OF METABOLISM WHY IS THIS IMPORTANT? It is important to have a basic understanding of metabolism because it governs the survival and growth of microorganisms The growth of microorganisms
Metabolism. Chapter 8 Microbial Metabolism. Metabolic balancing act. Catabolism Anabolism Enzymes. Topics. Metabolism Energy Pathways Biosynthesis
 Chapter 8 Microbial Metabolism Topics Metabolism Energy Pathways Biosynthesis Catabolism Anabolism Enzymes Metabolism 1 2 Metabolic balancing act Catabolism and anabolism simple model Catabolism Enzymes
Chapter 8 Microbial Metabolism Topics Metabolism Energy Pathways Biosynthesis Catabolism Anabolism Enzymes Metabolism 1 2 Metabolic balancing act Catabolism and anabolism simple model Catabolism Enzymes
! Proteins are involved functionally in almost everything: " Receptor Proteins - Respond to external stimuli. " Storage Proteins - Storing amino acids
 Proteins Most structurally & functionally diverse group! Proteins are involved functionally in almost everything: Proteins Multi-purpose molecules 2007-2008 Enzymatic proteins - Speed up chemical reactions!
Proteins Most structurally & functionally diverse group! Proteins are involved functionally in almost everything: Proteins Multi-purpose molecules 2007-2008 Enzymatic proteins - Speed up chemical reactions!
2. Which of the following amino acids is most likely to be found on the outer surface of a properly folded protein?
 Name: WHITE Student Number: Answer the following questions on the computer scoring sheet. 1 mark each 1. Which of the following amino acids would have the highest relative mobility R f in normal thin layer
Name: WHITE Student Number: Answer the following questions on the computer scoring sheet. 1 mark each 1. Which of the following amino acids would have the highest relative mobility R f in normal thin layer
Previous Class. Today. Term test I discussions. Detection of enzymatic intermediates: chymotrypsin mechanism
 Term test I discussions Previous Class Today Detection of enzymatic intermediates: chymotrypsin mechanism Mechanistic Understanding of Enzymemediated Reactions Ultimate goals: Identification of the intermediates,
Term test I discussions Previous Class Today Detection of enzymatic intermediates: chymotrypsin mechanism Mechanistic Understanding of Enzymemediated Reactions Ultimate goals: Identification of the intermediates,
Proteins. (b) Protein Structure and Conformational Change
 Proteins (b) Protein Structure and Conformational Change Protein Structure and Conformational Change Proteins contain the elements carbon (C), hydrogen (H), oxygen (O2) and nitrogen (N2) Some may also
Proteins (b) Protein Structure and Conformational Change Protein Structure and Conformational Change Proteins contain the elements carbon (C), hydrogen (H), oxygen (O2) and nitrogen (N2) Some may also
Enzymes. Ms. Paxson. From food webs to the life of a cell. Enzymes. Metabolism. Flow of energy through life. Examples. Examples
 From food webs to the life of a cell energy energy energy Flow of energy through life Life is built on chemical reactions sun transforming energy from one form to another solar energy ATP & organic molecules
From food webs to the life of a cell energy energy energy Flow of energy through life Life is built on chemical reactions sun transforming energy from one form to another solar energy ATP & organic molecules
Chapter 20. Proteins & Enzymes. Proteins & Enzymes - page 1
 Chapter 20 Proteins & Enzymes Proteins & Enzymes - page 1 Proteins & Enzymes Part 1: Amino Acids The building blocks of proteins are -amino acids, small molecules that contain a carboxylic acid and an
Chapter 20 Proteins & Enzymes Proteins & Enzymes - page 1 Proteins & Enzymes Part 1: Amino Acids The building blocks of proteins are -amino acids, small molecules that contain a carboxylic acid and an
Life History of A Drug
 DRUG ACTION & PHARMACODYNAMIC M. Imad Damaj, Ph.D. Associate Professor Pharmacology and Toxicology Smith 652B, 828-1676, mdamaj@hsc.vcu.edu Life History of A Drug Non-Specific Mechanims Drug-Receptor Interaction
DRUG ACTION & PHARMACODYNAMIC M. Imad Damaj, Ph.D. Associate Professor Pharmacology and Toxicology Smith 652B, 828-1676, mdamaj@hsc.vcu.edu Life History of A Drug Non-Specific Mechanims Drug-Receptor Interaction
How Did Energy-Releasing Pathways Evolve? (cont d.)
 How Did Energy-Releasing Pathways Evolve? (cont d.) 7.1 How Do Cells Access the Chemical Energy in Sugars? In order to use the energy stored in sugars, cells must first transfer it to ATP The energy transfer
How Did Energy-Releasing Pathways Evolve? (cont d.) 7.1 How Do Cells Access the Chemical Energy in Sugars? In order to use the energy stored in sugars, cells must first transfer it to ATP The energy transfer
Student Guide. Concluding module. Visualizing proteins
 Student Guide Concluding module Visualizing proteins Developed by bioinformaticsatschool.eu (part of NBIC) Text Hienke Sminia Illustrations Bioinformaticsatschool.eu Yasara.org All the included material
Student Guide Concluding module Visualizing proteins Developed by bioinformaticsatschool.eu (part of NBIC) Text Hienke Sminia Illustrations Bioinformaticsatschool.eu Yasara.org All the included material
Human Biochemistry Option B
 Human Biochemistry Option B A look ahead... Your body has many functions to perform every day: Structural support, genetic information, communication, energy supply, metabolism Right now, thousands of
Human Biochemistry Option B A look ahead... Your body has many functions to perform every day: Structural support, genetic information, communication, energy supply, metabolism Right now, thousands of
Review of Biochemistry
 Review of Biochemistry Chemical bond Functional Groups Amino Acid Protein Structure and Function Proteins are polymers of amino acids. Each amino acids in a protein contains a amino group, - NH 2,
Review of Biochemistry Chemical bond Functional Groups Amino Acid Protein Structure and Function Proteins are polymers of amino acids. Each amino acids in a protein contains a amino group, - NH 2,
BY: RASAQ NURUDEEN OLAJIDE
 BY: RASAQ NURUDEEN OLAJIDE LECTURE CONTENT INTRODUCTION CITRIC ACID CYCLE (T.C.A) PRODUCTION OF ACETYL CoA REACTIONS OF THE CITIRC ACID CYCLE THE AMPHIBOLIC NATURE OF THE T.C.A CYCLE THE GLYOXYLATE CYCLE
BY: RASAQ NURUDEEN OLAJIDE LECTURE CONTENT INTRODUCTION CITRIC ACID CYCLE (T.C.A) PRODUCTION OF ACETYL CoA REACTIONS OF THE CITIRC ACID CYCLE THE AMPHIBOLIC NATURE OF THE T.C.A CYCLE THE GLYOXYLATE CYCLE
1- Which of the following statements is TRUE in regards to eukaryotic and prokaryotic cells?
 Name: NetID: Exam 3 - Version 1 October 23, 2017 Dr. A. Pimentel Each question has a value of 4 points and there are a total of 160 points in the exam. However, the maximum score of this exam will be capped
Name: NetID: Exam 3 - Version 1 October 23, 2017 Dr. A. Pimentel Each question has a value of 4 points and there are a total of 160 points in the exam. However, the maximum score of this exam will be capped
Chapter 8. An Introduction to Microbial Metabolism
 Chapter 8 An Introduction to Microbial Metabolism The metabolism of microbes Metabolism sum of all chemical reactions that help cells function Two types of chemical reactions: Catabolism -degradative;
Chapter 8 An Introduction to Microbial Metabolism The metabolism of microbes Metabolism sum of all chemical reactions that help cells function Two types of chemical reactions: Catabolism -degradative;
Design and Modeling Studies on Liriodenine derivatives as novel topoisomerase II inhibitors
 International Journal of ChemTech Research CODEN( USA): IJCRGG ISSN : 0974-4290 Vol. 3, No.3,pp 1622-1627, July-Sept 2011 Design and Modeling Studies on Liriodenine derivatives as novel topoisomerase II
International Journal of ChemTech Research CODEN( USA): IJCRGG ISSN : 0974-4290 Vol. 3, No.3,pp 1622-1627, July-Sept 2011 Design and Modeling Studies on Liriodenine derivatives as novel topoisomerase II
Studying the Effect of Hydrogen Peroxide Substrate Concentration on Catalase Induced Reaction
 Studying the Effect of Hydrogen Peroxide Substrate Concentration on Catalase Induced Reaction Submitted by: [Student Name] [Course Name] [University Name] Table of Contents 1.0 Aim... 3 2.0 Background
Studying the Effect of Hydrogen Peroxide Substrate Concentration on Catalase Induced Reaction Submitted by: [Student Name] [Course Name] [University Name] Table of Contents 1.0 Aim... 3 2.0 Background
Arginine side chain interactions and the role of arginine as a mobile charge carrier in voltage sensitive ion channels. Supplementary Information
 Arginine side chain interactions and the role of arginine as a mobile charge carrier in voltage sensitive ion channels Craig T. Armstrong, Philip E. Mason, J. L. Ross Anderson and Christopher E. Dempsey
Arginine side chain interactions and the role of arginine as a mobile charge carrier in voltage sensitive ion channels Craig T. Armstrong, Philip E. Mason, J. L. Ross Anderson and Christopher E. Dempsey
Proteins and their structure
 Proteins and their structure Proteins are the most abundant biological macromolecules, occurring in all cells and all parts of cells. Proteins also occur in great variety; thousands of different kinds,
Proteins and their structure Proteins are the most abundant biological macromolecules, occurring in all cells and all parts of cells. Proteins also occur in great variety; thousands of different kinds,
Foundations in Microbiology Seventh Edition
 Lecture PowerPoint to accompany Foundations in Microbiology Seventh Edition Talaro Chapter 8 To run the animations you must be in Slideshow View. Use the buttons on the animation to play, pause, and turn
Lecture PowerPoint to accompany Foundations in Microbiology Seventh Edition Talaro Chapter 8 To run the animations you must be in Slideshow View. Use the buttons on the animation to play, pause, and turn
Metabolism & Enzymes. From food webs to the life of a cell. Flow of energy through life. Life is built on chemical reactions
 Metabolism & Enzymes 2007-2008 From food webs to the life of a cell energy energy energy Flow of energy through life Life is built on chemical reactions transforming energy from one form to another organic
Metabolism & Enzymes 2007-2008 From food webs to the life of a cell energy energy energy Flow of energy through life Life is built on chemical reactions transforming energy from one form to another organic
Section B: The Process of Cellular Respiration
 CHAPTER 9 CELLULAR RESPIRATION: HARVESTING CHEMICAL ENERGY Section B: The Process of Cellular Respiration 1. Respiration involves glycolysis, the Krebs cycle, and electron transport: an overview 2. Glycolysis
CHAPTER 9 CELLULAR RESPIRATION: HARVESTING CHEMICAL ENERGY Section B: The Process of Cellular Respiration 1. Respiration involves glycolysis, the Krebs cycle, and electron transport: an overview 2. Glycolysis
IB Biology BIOCHEMISTRY. Biological Macromolecules SBI3U7. Topic 3. Thursday, October 4, 2012
 + IB Biology SBI3U7 BIOCHEMISTRY Topic 3 Biological Macromolecules Essential Questions: 1.What are the 4 main types of biological macromolecules and what is their function within cells? 2.How does the
+ IB Biology SBI3U7 BIOCHEMISTRY Topic 3 Biological Macromolecules Essential Questions: 1.What are the 4 main types of biological macromolecules and what is their function within cells? 2.How does the
Biological Chemistry. Is biochemistry fun? - Find it out!
 Biological Chemistry Is biochemistry fun? - Find it out! 1. Key concepts Outline 2. Condensation and Hydrolysis Reactions 3. Carbohydrates 4. Lipids 5. Proteins 6. Nucleic Acids Key Concepts: 1. Organic
Biological Chemistry Is biochemistry fun? - Find it out! 1. Key concepts Outline 2. Condensation and Hydrolysis Reactions 3. Carbohydrates 4. Lipids 5. Proteins 6. Nucleic Acids Key Concepts: 1. Organic
Objectives. Carbon Bonding. Carbon Bonding, continued. Carbon Bonding
 Biochemistry Table of Contents Objectives Distinguish between organic and inorganic compounds. Explain the importance of carbon bonding in biological molecules. Identify functional groups in biological
Biochemistry Table of Contents Objectives Distinguish between organic and inorganic compounds. Explain the importance of carbon bonding in biological molecules. Identify functional groups in biological
Enzymes. Enzyme Structure. How do enzymes work?
 Page 1 of 6 Enzymes Enzymes are biological catalysts. There are about 40,000 different enzymes in human cells, each controlling a different chemical reaction. They increase the rate of reactions by a factor
Page 1 of 6 Enzymes Enzymes are biological catalysts. There are about 40,000 different enzymes in human cells, each controlling a different chemical reaction. They increase the rate of reactions by a factor
Chapter 9 Overview. Aerobic Metabolism I: The Citric Acid Cycle. Live processes - series of oxidation-reduction reactions. Aerobic metabolism I
 n n Chapter 9 Overview Aerobic Metabolism I: The Citric Acid Cycle Live processes - series of oxidation-reduction reactions Ingestion of proteins, carbohydrates, lipids Provide basic building blocks for
n n Chapter 9 Overview Aerobic Metabolism I: The Citric Acid Cycle Live processes - series of oxidation-reduction reactions Ingestion of proteins, carbohydrates, lipids Provide basic building blocks for
General Biology 1004 Chapter 6 Lecture Handout, Summer 2005 Dr. Frisby
 Slide 1 CHAPTER 6 Cellular Respiration: Harvesting Chemical Energy PowerPoint Lecture Slides for Essential Biology, Second Edition & Essential Biology with Physiology Presentation prepared by Chris C.
Slide 1 CHAPTER 6 Cellular Respiration: Harvesting Chemical Energy PowerPoint Lecture Slides for Essential Biology, Second Edition & Essential Biology with Physiology Presentation prepared by Chris C.
Energy and life. Generation of Biochemical Energy Chapter 21. Energy. Energy and biochemical reactions: 4/5/09
 Energy and life Generation of Biochemical Energy Chapter 21 1 Biological systems are powered by oxidation of biomolecules made mainly of C, H and O. The food biomolecules are mainly Lipids (fats) Carbohydrates
Energy and life Generation of Biochemical Energy Chapter 21 1 Biological systems are powered by oxidation of biomolecules made mainly of C, H and O. The food biomolecules are mainly Lipids (fats) Carbohydrates
Lecture Series 2 Macromolecules: Their Structure and Function
 Lecture Series 2 Macromolecules: Their Structure and Function Reading Assignments Read Chapter 4 (Protein structure & Function) Biological Substances found in Living Tissues The big four in terms of macromolecules
Lecture Series 2 Macromolecules: Their Structure and Function Reading Assignments Read Chapter 4 (Protein structure & Function) Biological Substances found in Living Tissues The big four in terms of macromolecules
7.014 Problem Set 2 Answers to this problem set are to be turned in. Problem sets will not be accepted late. Solutions will be posted on the web.
 MIT Department of Biology 7.014 Introductory Biology, Spring 2005 Name: Section : 7.014 Problem Set 2 Answers to this problem set are to be turned in. Problem sets will not be accepted late. Solutions
MIT Department of Biology 7.014 Introductory Biology, Spring 2005 Name: Section : 7.014 Problem Set 2 Answers to this problem set are to be turned in. Problem sets will not be accepted late. Solutions
a) The statement is true for X = 400, but false for X = 300; b) The statement is true for X = 300, but false for X = 200;
 1. Consider the following statement. To produce one molecule of each possible kind of polypeptide chain, X amino acids in length, would require more atoms than exist in the universe. Given the size of
1. Consider the following statement. To produce one molecule of each possible kind of polypeptide chain, X amino acids in length, would require more atoms than exist in the universe. Given the size of
Chapter 23 Enzymes 1
 Chapter 23 Enzymes 1 Enzymes Ribbon diagram of cytochrome c oxidase, the enzyme that directly uses oxygen during respiration. 2 Enzyme Catalysis Enzyme: A biological catalyst. With the exception of some
Chapter 23 Enzymes 1 Enzymes Ribbon diagram of cytochrome c oxidase, the enzyme that directly uses oxygen during respiration. 2 Enzyme Catalysis Enzyme: A biological catalyst. With the exception of some
Lecture Series 2 Macromolecules: Their Structure and Function
 Lecture Series 2 Macromolecules: Their Structure and Function Reading Assignments Read Chapter 4 (Protein structure & Function) Biological Substances found in Living Tissues The big four in terms of macromolecules
Lecture Series 2 Macromolecules: Their Structure and Function Reading Assignments Read Chapter 4 (Protein structure & Function) Biological Substances found in Living Tissues The big four in terms of macromolecules
6/9/2015. Unit 15: Organic Chemistry Lesson 15.2: Substituted Hydrocarbons & Functional Groups
 1-chloropropane 2-methylpropane 1-iodobutane Ethanoic Acid Unit 15: Organic Chemistry Lesson 15.2: Substituted Hydrocarbons & Functional Groups 43 It Ain t Just Hydrocarbons There are all sorts of organic
1-chloropropane 2-methylpropane 1-iodobutane Ethanoic Acid Unit 15: Organic Chemistry Lesson 15.2: Substituted Hydrocarbons & Functional Groups 43 It Ain t Just Hydrocarbons There are all sorts of organic
CHEM-643 Biochemistry Mid-term Examination 8:00 10:00, Monday, 24 October 2005
 CHEM-643 Biochemistry Mid-term Examination 8:00 10:00, Monday, 24 October 2005 Name Dr. H. White - Instructor There are 8 pages to this examination including this page. In addition, you will get a metabolic
CHEM-643 Biochemistry Mid-term Examination 8:00 10:00, Monday, 24 October 2005 Name Dr. H. White - Instructor There are 8 pages to this examination including this page. In addition, you will get a metabolic
Microbial Metabolism
 PowerPoint Lecture Slides for MICROBIOLOGY ROBERT W. BAUMAN Chapter 5 Microbial Metabolism Microbial Metabolism The sum total of chemical reactions that take place within cells (of an organism) Metabolic
PowerPoint Lecture Slides for MICROBIOLOGY ROBERT W. BAUMAN Chapter 5 Microbial Metabolism Microbial Metabolism The sum total of chemical reactions that take place within cells (of an organism) Metabolic
Biology 2E- Zimmer Protein structure- amino acid kit
 Biology 2E- Zimmer Protein structure- amino acid kit Name: This activity will use a physical model to investigate protein shape and develop key concepts that govern how proteins fold into their final three-dimensional
Biology 2E- Zimmer Protein structure- amino acid kit Name: This activity will use a physical model to investigate protein shape and develop key concepts that govern how proteins fold into their final three-dimensional
ENZYMES: CLASSIFICATION, STRUCTURE
 ENZYMES: CLASSIFICATION, STRUCTURE Enzymes - catalysts of biological reactions Accelerate reactions by a millions fold Common features for enzymes and inorganic catalysts: 1. Catalyze only thermodynamically
ENZYMES: CLASSIFICATION, STRUCTURE Enzymes - catalysts of biological reactions Accelerate reactions by a millions fold Common features for enzymes and inorganic catalysts: 1. Catalyze only thermodynamically
MBB 694:407, 115:511. Please use BLOCK CAPITAL letters like this --- A, B, C, D, E. Not lowercase!
 MBB 694:407, 115:511 First Test Severinov/Deis Tue. Sep. 30, 2003 Name Index number (not SSN) Row Letter Seat Number This exam consists of two parts. Part I is multiple choice. Each of these 25 questions
MBB 694:407, 115:511 First Test Severinov/Deis Tue. Sep. 30, 2003 Name Index number (not SSN) Row Letter Seat Number This exam consists of two parts. Part I is multiple choice. Each of these 25 questions
Table S1: Kinetic parameters of drug and substrate binding to wild type and HIV-1 protease variants. Data adapted from Ref. 6 in main text.
 Dynamical Network of HIV-1 Protease Mutants Reveals the Mechanism of Drug Resistance and Unhindered Activity Rajeswari Appadurai and Sanjib Senapati* BJM School of Biosciences and Department of Biotechnology,
Dynamical Network of HIV-1 Protease Mutants Reveals the Mechanism of Drug Resistance and Unhindered Activity Rajeswari Appadurai and Sanjib Senapati* BJM School of Biosciences and Department of Biotechnology,
Enzymes what are they?
 Topic 11 (ch8) Microbial Metabolism Topics Metabolism Energy Pathways Biosynthesis 1 Catabolism Anabolism Enzymes Metabolism 2 Metabolic balancing act Catabolism Enzymes involved in breakdown of complex
Topic 11 (ch8) Microbial Metabolism Topics Metabolism Energy Pathways Biosynthesis 1 Catabolism Anabolism Enzymes Metabolism 2 Metabolic balancing act Catabolism Enzymes involved in breakdown of complex
