Concurrent Practical Session ACMG Classification
|
|
|
- Dayna Potter
- 5 years ago
- Views:
Transcription
1 Variant Effect Prediction Training Course 6-8 November 2017 Prague, Czech Republic Concurrent Practical Session ACMG Classification Andreas Laner / Anna Benet-Pagès 1
2 Content 1. Background Aim of the Workshop Outline... 4 Part A: Practical Variant Classification (75 min)... 4 Part B: Demonstration of inter-laboratory concordance in variant classification (15 min) Cases... 5 Case Notes:... 5 Case 2:... 6 Notes:... 6 Case 3:... 7 Notes:... 7 Case 4:... 8 Notes:... 8 Case 5:... 9 Notes: ACMG Tables Criteria for classifying pathogenic variants (ACMG Standards and Guidelines) Criteria for classifying benign variants (ACMG Standards and Guidelines) Rules for combining criteria to classify sequence variants (ACMG Standards and Guidelines) Recommended Literature Databases
3 1. Background The dramatic progress in sequence technology, lab automatization, and bio-it data processing in the last decade have made high-throughput sequencing applications the standard method in molecular diagnostics. Especially since the development of benchtop NGS machines, almost every lab is able to create vast amounts of high-quality sequence data. However, there are still some important hurdles to overcome, especially the interpretation of sequence variants with a view to providing accurate clinical recommendations, a process that is considered a major bottleneck. Evaluating the pathogenicity of a variant is challenging given the plethora of types of genetic evidence that laboratories need to consider. Deciding how to weigh each type of evidence is difficult, and standards have been set. In 2015, the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP) published guidelines for the assessment of variants in genes associated with Mendelian diseases (1) (hereafter: ACMG-AMP guidelines). The goal of these guidelines is to establish standardized classification, annotation, interpretation, and reporting of sequence variants. 2. Aim of the Workshop In this workshop, participants will be familiarized with the basic application of ACMG-AMP classification guidelines as well as with the limitations and pitfalls inherent in working with these guidelines on a daily basis. The following points will be addressed during the presentation: Familiarization with ACMG-AMP guidelines and their basic application Identification of classes of variants not covered by ACMG-AMP guidelines (e.g. somatic variants, pharmacogenomics, multigenic/complex disorders) or which must be considered cautiously (e.g. variants with low/moderate penetrance) Identification of top error-prone sources of information (e.g. ClinVar OMIM entries, old data sources, research submissions, disease areas, etc.) (5) Awareness of various possible errors in variant interpretation Awareness of the fact that a considerable number of inter-laboratory discrepancies in variant classification are the result of a lack of published internal data, special biology, and old or invalid data sources. (2, 3, 4, 5) 3
4 3. Outline The workshop is divided in a practical part and in a short demonstration part. Part A: Practical Variant Classification (75 min) In the first part of the workshop, variants from real cases will be discussed collectively. The variants have been selected to represent ACMG-AMP categories that are known to be challenging in the classification process (2, 3, 4, 5). These variants were sent to workshop participants in advance of the VEPTC (see cases 1 to 5 on the following pages). In order to focus discussion at the variant level, only one or two variants per case will be considered for the classification process. To facilitate classification an overview of the ACMG Criteria are provided in this document in table format (see Chaper 5). The tables describe the criteria to classify pathogenic, benign and unclassified variants. The first table lists the criteria for pathogenic variants and the second table lists criteria for benign variants. The third table describes the rules for the combination of the criteria to classify the variant. To make the workshop more interactive, participants were asked to collect evidence for and against pathogenicity for each case and to prepare possible questions and remarks in advance. During this practical section, differences, difficulties, and discrepancies in variant classification will be discussed for all five cases. Furthermore, we encourage participants to bring their own cases which can be discussed during or after the workshop, depending on the time. Part B: Demonstration of inter-laboratory concordance in variant classification (15 min) In the second part of the workshop recent publications reporting conflicting results regarding consistent variant classification using the ACMG-AMP guidelines will be discussed. Furthermore, the problem of discordant inter-laboratory variant classification will also be addressed. 4
5 4. Cases Case 1 PATIENT: PHENOTYPE: HPO-TERMS: Male, 46 years of age. Mental retardation, hypophosphatemia and spinal deformity (osteopenia). Family history negative regarding mental retardation or skeletal abnormalities. Hypophosphatemia, Osteopenia ANALYSED GENES: Clinical Exome-Kit (Illumina; genes) DETECTED VARIANT(S): Gene (GRCh37/hg19) Transcript Variant (HGVS) Zygosity Ref Alt Chr. Position dbsnp ID SLC9A3R 1 NM_ c.328c>g (p.leu110val) heterozygous C G chr17: rs Notes: 5
6 Case 2: PATIENT: PHENOTYPE: ANALYSED GENES: 12 Female, 38 years of age. Breast-cancer, mother and maternal aunt also affected by breast-cancer at age 45 and 59, respectively. Clinical suspicion of hereditary breast-cancer. No family segregation analysis performed. DETECTED VARIANT(S): Gene (GRCh37/hg19) Transcript Variant (HGVS) Zygosity Ref Alt Chr. Position dbsnp ID CHEK2 NM_ c.470t>c (p.ile157thr) heterozygous A G chr22: rs Notes: 6
7 Case 3: PATIENT: PHENOTYPE: ANALYSED GENES: 23 Male, 2 years of age. Childhood cardiomyopathy, poor feeding, and muscle hypotonia. DETECTED VARIANT(S): Gene (GRCh37/hg19) Transcript Variant (HGVS) Zygosity Ref Alt Chr. Position dbsnp ID SLC22A5 NM_ c.1463g>a (p.arg488his) homozygous G A chr5: rs Notes: 7
8 Case 4: PATIENT: PHENOTYPE: ANALYSED GENES: 22 Male, 30 years of age Intracerebral hemorrhage at age 27. Positive family history of porencephaly, father deceased of haemorrhagic stroke. DETECTED VARIANT(S): Gene (GRCh37/hg19) Transcript Variant (HGVS) Zygosity Ref Alt Chr. Position dbsnp ID COL4A1 NM_ c.3067g>a (p.gly1023arg) COL4A1 NM_ c.3484g>a (p.ala1162thr) heterozygou s heterozygou s C T chr13: C T chr13: rs Notes: 8
9 Case 5: PATIENT: PHENOTYPE: ANALYSED GENES: 94 Female, 46 years of age. Astrocytoma at 10 years of age, breast cancer at 29 years of age, colorectal cancer at 39 years of age, kidney cancer at 45 years of age. Microsatellite stable with expression of all mismatch repair genes. DETECTED VARIANT(S): Gene (GRCh37/hg19) Transcript Variant (HGVS) Zygosity Ref Alt Chr. Position dbsnp ID TP53 NM_ c.722c>t (p.ser241phe) heterozygous G A chr17: rs FH NM_ c.1431_1433dup (p.lys477dup) heterozygous - TTT chr1: rs Notes: 9
10 5. ACMG Tables Criteria for classifying pathogenic variants (ACMG Standards and Guidelines) Evidence of pathogenicit y Category Very strong PVS1 Null variant (nonsense, frameshift, canonical ±1 or 2 splice sites, initiation codon, single or multiexon deletion) in a gene where LOF is a known mechanism of disease. Caveats: Beware of genes where LOF is not a known disease mechanism (e.g., GFAP, MYH7) Use caution interpreting LOF variants at the extreme 3 end of a gene Use caution with splice variants that are predicted to lead to exon skipping but leave the remainder of the protein intact Use caution in the presence of multiple transcripts Strong PS1 PS2 PS3 PS4 Same amino acid change as a previously established pathogenic variant regardless of nucleotide change Example: Val Leu caused by either G>C or G>T in the same codon Caveat: Beware of changes that impact splicing rather than at the amino acid/protein level De novo (both maternity and paternity confirmed) in a patient with the disease and no family history Note: Confirmation of paternity only is insufficient. Egg donation, surrogate motherhood, errors in embryo transfer, and so on, can contribute to non maternity. Well-established in vitro or in vivo functional studies supportive of a damaging effect on the gene or gene product Note: Functional studies that have been validated and shown to be reproducible and robust in a clinical diagnostic laboratory setting are considered the most well established. The prevalence of the variant in affected individuals is significantly increased compared with the prevalence in controls Note 1: Relative risk or OR, as obtained from case control studies, is >5.0, and the confidence interval around the estimate of relative risk or OR does not include 1.0. See the article for detailed guidance. Note 2: In instances of very rare variants where case control studies may not reach statistical significance, the prior observation of the variant in multiple unrelated patients with the same phenotype, and its absence in controls, may be used as moderate level of evidence. PM1 PM2 Located in a mutational hot spot and/or critical and well-established functional domain (e.g., active site of an enzyme) without benign variation. Absent from controls (or at extremely low frequency if recessive) (Table 6) in Exome Sequencing Project, 1000 Genomes Project, or Exome Aggregation Consortium Caveat: Population data for insertions/deletions may be poorly called by nextgeneration sequencing. 10
11 Moder ate PM3 PM4 PM5 PM6 For recessive disorders, detected in trans with a pathogenic variant Note: This requires testing of parents (or offspring) to determine phase. Protein length changes as a result of in-frame deletions/insertions in a nonrepeat region or stop-loss variants Novel missense change at an amino acid residue where a different missense change determined to be pathogenic has been seen before Example: Arg156His is pathogenic; now you observe Arg156Cys Caveat: Beware of changes that impact splicing rather than at the amino acid/protein level. Assumed de novo, but without confirmation of paternity and maternity Suppo rting PP1 PP2 PP3 PP4 PP5 Co segregation with disease in multiple affected family members in a gene definitively known to cause the disease Note: May be used as stronger evidence with increasing segregation data Missense variant in a gene that has a low rate of benign missense variation and in which missense variants are a common mechanism of disease Multiple lines of computational evidence support a deleterious effect on the gene or gene product (conservation, evolutionary, splicing impact, etc.) Caveat: Because many in-silico algorithms use the same or very similar input for their predictions, each algorithm should not be counted as an independent criterion. PP3 can be used only once in any evaluation of a variant. Patient s phenotype or family history is highly specific for a disease with a single genetic etiology Reputable source recently reports variant as pathogenic, but the evidence is not available to the laboratory to perform an independent evaluation Criteria for classifying benign variants (ACMG Standards and Guidelines) Evidence of benign impact Category Stand alone BA1 Allele frequency is >5% in Exome Sequencing Project, 1000 Genomes Project, or Exome Aggregation Consortium BS1 Allele frequency is greater than expected for disorder (see Table 6) Strong BS2 BS3 Observed in a healthy adult individual for a recessive (homozygous), dominant (heterozygous), or X-linked (hemizygous) disorder, with full penetrance expected at an early age Well-established in vitro or in vivo functional studies show no damaging effect on protein function or splicing Lack of segregation in affected members of a family 11
12 BS4 Caveat: The presence of phenocopies for common phenotypes (i.e., cancer, epilepsy) can mimic lack of segregation among affected individuals. Also, families may have more than one pathogenic variant contributing to an autosomal dominant disorder, further confounding an apparent lack of segregation. Suppo rting BP1 BP2 BP3 BP4 BP5 BP6 BP7 Missense variant in a gene for which primarily truncating variants are known to cause disease Observed in trans with a pathogenic variant for a fully penetrant dominant gene/disorder or observed in cis with a pathogenic variant in any inheritance pattern In-frame deletions/insertions in a repetitive region without a known function Multiple lines of computational evidence suggest no impact on gene or gene product (conservation, evolutionary, splicing impact, etc.) Caveat: Because many in silico algorithms use the same or very similar input for their predictions, each algorithm cannot be counted as an independent criterion. BP4 can be used only once in any evaluation of a variant. Variant found in a case with an alternate molecular basis for disease Reputable source recently reports variant as benign, but the evidence is not available to the laboratory to perform an independent evaluation A synonymous (silent) variant for which splicing prediction algorithms predict no impact to the splice consensus sequence nor the creation of a new splice site AND the nucleotide is not highly conserved Rules for combining criteria to classify sequence variants (ACMG Standards and Guidelines) (i) 1 Very strong (PVS1) AND a) 1 Strong (PS1 PS4) OR b) 2 Moderate (PM1 PM6) OR c) 1 Moderate (PM1 PM6) and 1 supporting (PP1 PP5) OR Pathogenic d) 2 Supporting (PP1 PP5) (ii) 2 Strong (PS1 PS4) OR (iii) 1 Strong (PS1 PS4) AND a) 3 Moderate (PM1 PM6) OR b) 2 Moderate (PM1 PM6) AND 2 Supporting (PP1 PP5) OR c) 1 Moderate (PM1 PM6) AND 4 supporting (PP1 PP5) (i) 1 Very strong (PVS1) AND 1 moderate (PM1 PM6) OR (ii) 1 Strong (PS1 PS4) AND 1 2 moderate (PM1 PM6) OR Likely (iii) 1 Strong (PS1 PS4) AND 2 supporting (PP1 PP5) OR 12
13 Likely pathogenic (iv) 3 Moderate (PM1 PM6) OR (v) 2 Moderate (PM1 PM6) AND 2 supporting (PP1 PP5) OR (vi) 1 Moderate (PM1 PM6) AND 4 supporting (PP1 PP5) Benign (i) 1 Stand-alone (BA1) OR (ii) 2 Strong (BS1 BS4) (i) 1 Strong (BS1 BS4) and 1 supporting (BP1 BP7) OR Likely benign (ii) 2 Supporting (BP1 BP7) Uncertain significance (i) Other criteria shown above are not met OR (ii) the criteria for benign and pathogenic are contradictory 6. Recommended Literature (1) Richards et al.; Genet. Med. 17, , 2015 (2) Amendola et al.; Am J Hum Genet 98, , June 2, 2016 (3) Harrison et al.; Genet. Med. Mar 16 (PMID: ) (4) Pepin et al.; Genet Med. Jan; 18(1) 20-4 (PMID: ) (5) Yang et al.; Genet. Med. Jun (PMID: ) 7. Databases Population databases Exome Aggregation Consortium Exome Variant Server Genomes Project dbsnp 13
14 dbvar GnomAD Disease databases ClinVar OMIM Human Gene Mutation Database Human Genome Variation Society Leiden Open Variation Database DECIPHER Sequence databases NCBI Genome RefSeqGene Locus Reference Genomic (LRG) MitoMap HumanMitoSeq 14
Variant Classification: ACMG recommendations. Andreas Laner MGZ München
 Variant Classification: ACMG recommendations Andreas Laner MGZ München laner@mgz-muenchen.de Overview Introduction ACMG-AMP Classification System Evaluation of inter-laboratory concordance in variant classification
Variant Classification: ACMG recommendations Andreas Laner MGZ München laner@mgz-muenchen.de Overview Introduction ACMG-AMP Classification System Evaluation of inter-laboratory concordance in variant classification
Variant Classification: ACMG recommendations. Andreas Laner MGZ München
 Variant Classification: ACMG recommendations Andreas Laner MGZ München laner@mgz-muenchen.de OVERVIEW Introduction ACMG-AMP Classification System Evaluation of inter-laboratory concordance in variant classification
Variant Classification: ACMG recommendations Andreas Laner MGZ München laner@mgz-muenchen.de OVERVIEW Introduction ACMG-AMP Classification System Evaluation of inter-laboratory concordance in variant classification
Merging single gene-level CNV with sequence variant interpretation following the ACMGG/AMP sequence variant guidelines
 Merging single gene-level CNV with sequence variant interpretation following the ACMGG/AMP sequence variant guidelines Tracy Brandt, Ph.D., FACMG Disclosure I am an employee of GeneDx, Inc., a wholly-owned
Merging single gene-level CNV with sequence variant interpretation following the ACMGG/AMP sequence variant guidelines Tracy Brandt, Ph.D., FACMG Disclosure I am an employee of GeneDx, Inc., a wholly-owned
6/12/2018. Disclosures. Clinical Genomics The CLIA Lab Perspective. Outline. COH HopeSeq Heme Panels
 Clinical Genomics The CLIA Lab Perspective Disclosures Raju K. Pillai, M.D. Hematopathologist / Molecular Pathologist Director, Pathology Bioinformatics City of Hope National Medical Center, Duarte, CA
Clinical Genomics The CLIA Lab Perspective Disclosures Raju K. Pillai, M.D. Hematopathologist / Molecular Pathologist Director, Pathology Bioinformatics City of Hope National Medical Center, Duarte, CA
Variant interpretation using the ACMG-AMP guidelines
 Variant interpretation using the ACMG-AMP guidelines 20 TH A P R I L 2 0 1 7 S T A C E Y D A N I E L S W E S S E X R E G I O N A L G E N E T I C S L A B O R A T O R Y Talk outline Overview of the ACMG-AMP
Variant interpretation using the ACMG-AMP guidelines 20 TH A P R I L 2 0 1 7 S T A C E Y D A N I E L S W E S S E X R E G I O N A L G E N E T I C S L A B O R A T O R Y Talk outline Overview of the ACMG-AMP
Variant interpretation exercise. ACGS Somatic Variant Interpretation Workshop Joanne Mason 21/09/18
 Variant interpretation exercise ACGS Somatic Variant Interpretation Workshop Joanne Mason 21/09/18 Format of exercise Compile a list of tricky variants across solid cancer and haematological malignancy.
Variant interpretation exercise ACGS Somatic Variant Interpretation Workshop Joanne Mason 21/09/18 Format of exercise Compile a list of tricky variants across solid cancer and haematological malignancy.
A guide to understanding variant classification
 White paper A guide to understanding variant classification In a diagnostic setting, variant classification forms the basis for clinical judgment, making proper classification of variants critical to your
White paper A guide to understanding variant classification In a diagnostic setting, variant classification forms the basis for clinical judgment, making proper classification of variants critical to your
Introduction to genetic variation. He Zhang Bioinformatics Core Facility 6/22/2016
 Introduction to genetic variation He Zhang Bioinformatics Core Facility 6/22/2016 Outline Basic concepts of genetic variation Genetic variation in human populations Variation and genetic disorders Databases
Introduction to genetic variation He Zhang Bioinformatics Core Facility 6/22/2016 Outline Basic concepts of genetic variation Genetic variation in human populations Variation and genetic disorders Databases
PALB2 c g>c is. VARIANT OF UNCERTAIN SIGNIFICANCE (VUS) CGI s summary of the available evidence is in Appendices A-C.
 Consultation sponsor (may not be the patient): First LastName [Patient identity withheld] Date received by CGI: 2 Sept 2017 Variant Fact Checker Report ID: 0000001.5 Date Variant Fact Checker issued: 12
Consultation sponsor (may not be the patient): First LastName [Patient identity withheld] Date received by CGI: 2 Sept 2017 Variant Fact Checker Report ID: 0000001.5 Date Variant Fact Checker issued: 12
Reporting TP53 gene analysis results in CLL
 Reporting TP53 gene analysis results in CLL Mutations in TP53 - From discovery to clinical practice in CLL Discovery Validation Clinical practice Variant diversity *Leroy at al, Cancer Research Review
Reporting TP53 gene analysis results in CLL Mutations in TP53 - From discovery to clinical practice in CLL Discovery Validation Clinical practice Variant diversity *Leroy at al, Cancer Research Review
Variant classification according to the ACMG Guidelines
 Variant classification according to the ACMG Guidelines The results from the variant classification in different Norwegian laboratories. Mari Ann Kulseth AMG - OUS 5 variants identified in 3 patients (UK-NEQAS)
Variant classification according to the ACMG Guidelines The results from the variant classification in different Norwegian laboratories. Mari Ann Kulseth AMG - OUS 5 variants identified in 3 patients (UK-NEQAS)
CHAPTER IV RESULTS Microcephaly General description
 47 CHAPTER IV RESULTS 4.1. Microcephaly 4.1.1. General description This study found that from a previous study of 527 individuals with MR, 48 (23 female and 25 male) unrelated individuals were identified
47 CHAPTER IV RESULTS 4.1. Microcephaly 4.1.1. General description This study found that from a previous study of 527 individuals with MR, 48 (23 female and 25 male) unrelated individuals were identified
Interpretation can t happen in isolation. Jonathan S. Berg, MD/PhD Assistant Professor Department of Genetics UNC Chapel Hill
 Interpretation can t happen in isolation Jonathan S. Berg, MD/PhD Assistant Professor Department of Genetics UNC Chapel Hill With the advent of genome-scale sequencing, variant interpretation is increasingly
Interpretation can t happen in isolation Jonathan S. Berg, MD/PhD Assistant Professor Department of Genetics UNC Chapel Hill With the advent of genome-scale sequencing, variant interpretation is increasingly
Welcome to the Genetic Code: An Overview of Basic Genetics. October 24, :00pm 3:00pm
 Welcome to the Genetic Code: An Overview of Basic Genetics October 24, 2016 12:00pm 3:00pm Course Schedule 12:00 pm 2:00 pm Principles of Mendelian Genetics Introduction to Genetics of Complex Disease
Welcome to the Genetic Code: An Overview of Basic Genetics October 24, 2016 12:00pm 3:00pm Course Schedule 12:00 pm 2:00 pm Principles of Mendelian Genetics Introduction to Genetics of Complex Disease
Key determinants of pathogenicity
 Key determinants of pathogenicity Session 6: Determining pathogenicity and genotype-phenotype correlation J. Peter van Tintelen MD PhD Clinical geneticist Academic Medical Center Amsterdam, the Netherlands
Key determinants of pathogenicity Session 6: Determining pathogenicity and genotype-phenotype correlation J. Peter van Tintelen MD PhD Clinical geneticist Academic Medical Center Amsterdam, the Netherlands
Identifying Mutations Responsible for Rare Disorders Using New Technologies
 Identifying Mutations Responsible for Rare Disorders Using New Technologies Jacek Majewski, Department of Human Genetics, McGill University, Montreal, QC Canada Mendelian Diseases Clear mode of inheritance
Identifying Mutations Responsible for Rare Disorders Using New Technologies Jacek Majewski, Department of Human Genetics, McGill University, Montreal, QC Canada Mendelian Diseases Clear mode of inheritance
How many disease-causing variants in a normal person? Matthew Hurles
 How many disease-causing variants in a normal person? Matthew Hurles Summary What is in a genome? What is normal? Depends on age What is a disease-causing variant? Different classes of variation Final
How many disease-causing variants in a normal person? Matthew Hurles Summary What is in a genome? What is normal? Depends on age What is a disease-causing variant? Different classes of variation Final
CITATION FILE CONTENT/FORMAT
 CITATION For any resultant publications using please cite: Matthew A. Field, Vicky Cho, T. Daniel Andrews, and Chris C. Goodnow (2015). "Reliably detecting clinically important variants requires both combined
CITATION For any resultant publications using please cite: Matthew A. Field, Vicky Cho, T. Daniel Andrews, and Chris C. Goodnow (2015). "Reliably detecting clinically important variants requires both combined
Genomic and Genetic Medicine in the Clinical Setting
 Genomic and Genetic Medicine in the Clinical Setting Objectives Define genomic and genetic medicine Introduce molecular genetic testing Outline clinical applications including those for: Mendelian disease
Genomic and Genetic Medicine in the Clinical Setting Objectives Define genomic and genetic medicine Introduce molecular genetic testing Outline clinical applications including those for: Mendelian disease
Using large-scale human genetic variation to inform variant prioritization in neuropsychiatric disorders
 Using large-scale human genetic variation to inform variant prioritization in neuropsychiatric disorders Kaitlin E. Samocha Hurles lab, Wellcome Trust Sanger Institute ACGS Summer Scientific Meeting 27
Using large-scale human genetic variation to inform variant prioritization in neuropsychiatric disorders Kaitlin E. Samocha Hurles lab, Wellcome Trust Sanger Institute ACGS Summer Scientific Meeting 27
CANCER GENETICS PROVIDER SURVEY
 Dear Participant, Previously you agreed to participate in an evaluation of an education program we developed for primary care providers on the topic of cancer genetics. This is an IRB-approved, CDCfunded
Dear Participant, Previously you agreed to participate in an evaluation of an education program we developed for primary care providers on the topic of cancer genetics. This is an IRB-approved, CDCfunded
Clinical Spectrum and Genetic Mechanism of GLUT1-DS. Yasushi ITO (Tokyo Women s Medical University, Japan)
 Clinical Spectrum and Genetic Mechanism of GLUT1-DS Yasushi ITO (Tokyo Women s Medical University, Japan) Glucose transporter type 1 (GLUT1) deficiency syndrome Mutation in the SLC2A1 / GLUT1 gene Deficiency
Clinical Spectrum and Genetic Mechanism of GLUT1-DS Yasushi ITO (Tokyo Women s Medical University, Japan) Glucose transporter type 1 (GLUT1) deficiency syndrome Mutation in the SLC2A1 / GLUT1 gene Deficiency
Deciphering Developmental Disorders (DDD) DDG2P
 Deciphering Developmental Disorders (DDD) DDG2P David FitzPatrick MRC Human Genetics Unit, University of Edinburgh Deciphering Developmental Disorders objectives research understand genetics of DD translation
Deciphering Developmental Disorders (DDD) DDG2P David FitzPatrick MRC Human Genetics Unit, University of Edinburgh Deciphering Developmental Disorders objectives research understand genetics of DD translation
Molecular Diagnostic Laboratory 18 Sequencing St, Gene Town, ZY Tel: Fax:
 Molecular Diagnostic Laboratory 18 Sequencing St, Gene Town, ZY 01234 Tel: 555-920-3333 Fax: 555-920-3334 www.moldxlaboratory.com Patient Name: Jane Doe Specimen type: Blood, peripheral DOB: 04/05/1990
Molecular Diagnostic Laboratory 18 Sequencing St, Gene Town, ZY 01234 Tel: 555-920-3333 Fax: 555-920-3334 www.moldxlaboratory.com Patient Name: Jane Doe Specimen type: Blood, peripheral DOB: 04/05/1990
NGS panels in clinical diagnostics: Utrecht experience. Van Gijn ME PhD Genome Diagnostics UMCUtrecht
 NGS panels in clinical diagnostics: Utrecht experience Van Gijn ME PhD Genome Diagnostics UMCUtrecht 93 Gene panels UMC Utrecht Cardiovascular disease (CAR) (5 panels) Epilepsy (EPI) (11 panels) Hereditary
NGS panels in clinical diagnostics: Utrecht experience Van Gijn ME PhD Genome Diagnostics UMCUtrecht 93 Gene panels UMC Utrecht Cardiovascular disease (CAR) (5 panels) Epilepsy (EPI) (11 panels) Hereditary
Clinical Genetics. Functional validation in a diagnostic context. Robert Hofstra. Leading the way in genetic issues
 Clinical Genetics Leading the way in genetic issues Functional validation in a diagnostic context Robert Hofstra Clinical Genetics Leading the way in genetic issues Future application of exome sequencing
Clinical Genetics Leading the way in genetic issues Functional validation in a diagnostic context Robert Hofstra Clinical Genetics Leading the way in genetic issues Future application of exome sequencing
Genetic Testing and Analysis. (858) MRN: Specimen: Saliva Received: 07/26/2016 GENETIC ANALYSIS REPORT
 GBinsight Sample Name: GB4411 Race: Gender: Female Reason for Testing: Type 2 diabetes, early onset MRN: 0123456789 Specimen: Saliva Received: 07/26/2016 Test ID: 113-1487118782-4 Test: Type 2 Diabetes
GBinsight Sample Name: GB4411 Race: Gender: Female Reason for Testing: Type 2 diabetes, early onset MRN: 0123456789 Specimen: Saliva Received: 07/26/2016 Test ID: 113-1487118782-4 Test: Type 2 Diabetes
JULY 21, Genetics 101: SCN1A. Katie Angione, MS CGC Certified Genetic Counselor CHCO Neurology
 JULY 21, 2018 Genetics 101: SCN1A Katie Angione, MS CGC Certified Genetic Counselor CHCO Neurology Disclosures: I have no financial interests or relationships to disclose. Objectives 1. Review genetic
JULY 21, 2018 Genetics 101: SCN1A Katie Angione, MS CGC Certified Genetic Counselor CHCO Neurology Disclosures: I have no financial interests or relationships to disclose. Objectives 1. Review genetic
Computational Systems Biology: Biology X
 Bud Mishra Room 1002, 715 Broadway, Courant Institute, NYU, New York, USA L#4:(October-0-4-2010) Cancer and Signals 1 2 1 2 Evidence in Favor Somatic mutations, Aneuploidy, Copy-number changes and LOH
Bud Mishra Room 1002, 715 Broadway, Courant Institute, NYU, New York, USA L#4:(October-0-4-2010) Cancer and Signals 1 2 1 2 Evidence in Favor Somatic mutations, Aneuploidy, Copy-number changes and LOH
Proposal form for the evaluation of a genetic test for NHS Service Gene Dossier/Additional Provider
 Proposal form for the evaluation of a genetic test for NHS Service Gene Dossier/Additional Provider TEST DISORDER/CONDITION POPULATION TRIAD Submitting laboratory: Exeter RGC Approved: Sept 2013 1. Disorder/condition
Proposal form for the evaluation of a genetic test for NHS Service Gene Dossier/Additional Provider TEST DISORDER/CONDITION POPULATION TRIAD Submitting laboratory: Exeter RGC Approved: Sept 2013 1. Disorder/condition
CHR POS REF OBS ALLELE BUILD CLINICAL_SIGNIFICANCE
 CHR POS REF OBS ALLELE BUILD CLINICAL_SIGNIFICANCE is_clinical dbsnp MITO GENE chr1 13273 G C heterozygous - - -. - DDX11L1 chr1 949654 A G Homozygous 52 - - rs8997 - ISG15 chr1 1021346 A G heterozygous
CHR POS REF OBS ALLELE BUILD CLINICAL_SIGNIFICANCE is_clinical dbsnp MITO GENE chr1 13273 G C heterozygous - - -. - DDX11L1 chr1 949654 A G Homozygous 52 - - rs8997 - ISG15 chr1 1021346 A G heterozygous
What we know about Li-Fraumeni syndrome
 What we know about Li-Fraumeni syndrome Dr Helen Hanson Consultant in Cancer Genetics St Georges Hospital, South-West Thames Regional Genetics Service History of LFS 1969 Li and Fraumeni describe four
What we know about Li-Fraumeni syndrome Dr Helen Hanson Consultant in Cancer Genetics St Georges Hospital, South-West Thames Regional Genetics Service History of LFS 1969 Li and Fraumeni describe four
p.arg119gly p.arg119his p.ala179thr c.540+1g>a c.617_633+6del Prediction basis structure
 a Missense ATG p.arg119gly p.arg119his p.ala179thr p.ala189val p.gly206trp p.gly206arg p.arg251his p.ala257thr TGA 5 UTR 1 2 3 4 5 6 7 3 UTR Splice Site, Frameshift b Mutation p.gly206trp p.gly4fsx50 c.138+1g>a
a Missense ATG p.arg119gly p.arg119his p.ala179thr p.ala189val p.gly206trp p.gly206arg p.arg251his p.ala257thr TGA 5 UTR 1 2 3 4 5 6 7 3 UTR Splice Site, Frameshift b Mutation p.gly206trp p.gly4fsx50 c.138+1g>a
Chapter 4 PEDIGREE ANALYSIS IN HUMAN GENETICS
 Chapter 4 PEDIGREE ANALYSIS IN HUMAN GENETICS Chapter Summary In order to study the transmission of human genetic traits to the next generation, a different method of operation had to be adopted. Instead
Chapter 4 PEDIGREE ANALYSIS IN HUMAN GENETICS Chapter Summary In order to study the transmission of human genetic traits to the next generation, a different method of operation had to be adopted. Instead
Issues arising from UKNEQAS schemes. Ottie O Brien, Northern Genetics Service, Newcastle, UK 15 th May 2014
 Issues arising from UKNEQAS schemes Ottie O Brien, Northern Genetics Service, Newcastle, UK 15 th May 2014 2013 schemes There was great variation in the way HGVS nomenclature was applied Scheme would like
Issues arising from UKNEQAS schemes Ottie O Brien, Northern Genetics Service, Newcastle, UK 15 th May 2014 2013 schemes There was great variation in the way HGVS nomenclature was applied Scheme would like
Multiple Copy Number Variations in a Patient with Developmental Delay ASCLS- March 31, 2016
 Multiple Copy Number Variations in a Patient with Developmental Delay ASCLS- March 31, 2016 Marwan Tayeh, PhD, FACMG Director, MMGL Molecular Genetics Assistant Professor of Pediatrics Department of Pediatrics
Multiple Copy Number Variations in a Patient with Developmental Delay ASCLS- March 31, 2016 Marwan Tayeh, PhD, FACMG Director, MMGL Molecular Genetics Assistant Professor of Pediatrics Department of Pediatrics
TP53 mutational profile in CLL : A retrospective study of the FILO group.
 TP53 mutational profile in CLL : A retrospective study of the FILO group. Fanny Baran-Marszak Hopital Avicenne Bobigny France 2nd ERIC workshop on TP53 analysis in CLL, Stresa 2017 TP53 abnormalities :
TP53 mutational profile in CLL : A retrospective study of the FILO group. Fanny Baran-Marszak Hopital Avicenne Bobigny France 2nd ERIC workshop on TP53 analysis in CLL, Stresa 2017 TP53 abnormalities :
Variant Annotation and Functional Prediction
 Variant Annotation and Functional Prediction Copyrighted 2018 Isabelle Schrauwen and Suzanne M. Leal This exercise touches on several functionalities of the program ANNOVAR to annotate and interpret candidate
Variant Annotation and Functional Prediction Copyrighted 2018 Isabelle Schrauwen and Suzanne M. Leal This exercise touches on several functionalities of the program ANNOVAR to annotate and interpret candidate
Proposal form for the evaluation of a genetic test for NHS Service Gene Dossier
 Proposal form for the evaluation of a genetic test for NHS Service Gene Dossier Test Disease Population Triad Disease name Epileptic encephalopathy, early infantile 4. OMIM number for disease 612164 Disease
Proposal form for the evaluation of a genetic test for NHS Service Gene Dossier Test Disease Population Triad Disease name Epileptic encephalopathy, early infantile 4. OMIM number for disease 612164 Disease
Variant Detection & Interpretation in a diagnostic context. Christian Gilissen
 Variant Detection & Interpretation in a diagnostic context Christian Gilissen c.gilissen@gen.umcn.nl 28-05-2013 So far Sequencing Johan den Dunnen Marja Jakobs Ewart de Bruijn Mapping Victor Guryev Variant
Variant Detection & Interpretation in a diagnostic context Christian Gilissen c.gilissen@gen.umcn.nl 28-05-2013 So far Sequencing Johan den Dunnen Marja Jakobs Ewart de Bruijn Mapping Victor Guryev Variant
Assessing Laboratory Performance for Next Generation Sequencing Based Detection of Germline Variants through Proficiency Testing
 Assessing Laboratory Performance for Next Generation Sequencing Based Detection of Germline Variants through Proficiency Testing Karl V. Voelkerding, MD Professor of Pathology University of Utah Medical
Assessing Laboratory Performance for Next Generation Sequencing Based Detection of Germline Variants through Proficiency Testing Karl V. Voelkerding, MD Professor of Pathology University of Utah Medical
Variant Classification. Author: Mike Thiesen, Golden Helix, Inc.
 Variant Classification Author: Mike Thiesen, Golden Helix, Inc. Overview Sequencing pipelines are able to identify rare variants not found in catalogs such as dbsnp. As a result, variants in these datasets
Variant Classification Author: Mike Thiesen, Golden Helix, Inc. Overview Sequencing pipelines are able to identify rare variants not found in catalogs such as dbsnp. As a result, variants in these datasets
Functional analysis of DNA variants
 Functional analysis of DNA variants GS011143, Introduction to Bioinformatics The University of Texas GSBS program, Fall 2012 Ken Chen, Ph.D. Department of Bioinformatics and Computational Biology UT MD
Functional analysis of DNA variants GS011143, Introduction to Bioinformatics The University of Texas GSBS program, Fall 2012 Ken Chen, Ph.D. Department of Bioinformatics and Computational Biology UT MD
Pedigree Construction Notes
 Name Date Pedigree Construction Notes GO TO à Mendelian Inheritance (http://www.uic.edu/classes/bms/bms655/lesson3.html) When human geneticists first began to publish family studies, they used a variety
Name Date Pedigree Construction Notes GO TO à Mendelian Inheritance (http://www.uic.edu/classes/bms/bms655/lesson3.html) When human geneticists first began to publish family studies, they used a variety
Cardiology Genetics Report Long QT Syndrome (LQTS) Panel
 Patient Name: Test(s) Requested: The ordered test and the genes analyzed are Genes Evaluated (Disease): Result: Only mutations detected appear here KCNQ1 (LQT1 and Jervell and LangeNielsen syndrome), KCNH2
Patient Name: Test(s) Requested: The ordered test and the genes analyzed are Genes Evaluated (Disease): Result: Only mutations detected appear here KCNQ1 (LQT1 and Jervell and LangeNielsen syndrome), KCNH2
NGS for Cancer Predisposition
 NGS for Cancer Predisposition Colin Pritchard MD, PhD University of Washington Dept. of Lab Medicine AMP Companion Society Meeting USCAP Boston March 22, 2015 Disclosures I am an employee of the University
NGS for Cancer Predisposition Colin Pritchard MD, PhD University of Washington Dept. of Lab Medicine AMP Companion Society Meeting USCAP Boston March 22, 2015 Disclosures I am an employee of the University
Proposal form for the evaluation of a genetic test for NHS Service Gene Dossier
 Proposal form for the evaluation of a genetic test for NHS Service Gene Dossier Test Disease Population Triad Disease name Leber congenital amaurosis OMIM number for disease 204000 Disease alternative
Proposal form for the evaluation of a genetic test for NHS Service Gene Dossier Test Disease Population Triad Disease name Leber congenital amaurosis OMIM number for disease 204000 Disease alternative
Basic Definitions. Dr. Mohammed Hussein Assi MBChB MSc DCH (UK) MRCPCH
 Basic Definitions Chromosomes There are two types of chromosomes: autosomes (1-22) and sex chromosomes (X & Y). Humans are composed of two groups of cells: Gametes. Ova and sperm cells, which are haploid,
Basic Definitions Chromosomes There are two types of chromosomes: autosomes (1-22) and sex chromosomes (X & Y). Humans are composed of two groups of cells: Gametes. Ova and sperm cells, which are haploid,
Names for ABO (ISBT 001) Blood Group Alleles
 Names for ABO (ISBT 001) blood group alleles v1.1 1102 Names for ABO (ISBT 001) Blood Group Alleles General description: The ABO system was discovered as in 1900 and is considered the first and clinically
Names for ABO (ISBT 001) blood group alleles v1.1 1102 Names for ABO (ISBT 001) Blood Group Alleles General description: The ABO system was discovered as in 1900 and is considered the first and clinically
Germline Testing for Hereditary Cancer with Multigene Panel
 Germline Testing for Hereditary Cancer with Multigene Panel Po-Han Lin, MD Department of Medical Genetics National Taiwan University Hospital 2017-04-20 Disclosure No relevant financial relationships with
Germline Testing for Hereditary Cancer with Multigene Panel Po-Han Lin, MD Department of Medical Genetics National Taiwan University Hospital 2017-04-20 Disclosure No relevant financial relationships with
Lecture 17: Human Genetics. I. Types of Genetic Disorders. A. Single gene disorders
 Lecture 17: Human Genetics I. Types of Genetic Disorders A. Single gene disorders B. Multifactorial traits 1. Mutant alleles at several loci acting in concert C. Chromosomal abnormalities 1. Physical changes
Lecture 17: Human Genetics I. Types of Genetic Disorders A. Single gene disorders B. Multifactorial traits 1. Mutant alleles at several loci acting in concert C. Chromosomal abnormalities 1. Physical changes
Genome Summary. Sequencing Coverage: Variation Counts: Known Phenotype Summary: 131 disease or trait variations are found in this genome
 Report Date: August 2, 215 Software Annotation Version: 8 Genome Summary Name: Greg Mendel Genome ID: genome_greg_mendel_dad Sequencing Provider: 23andMe Sequencing Type: Genotyping SNP Array Sequencing
Report Date: August 2, 215 Software Annotation Version: 8 Genome Summary Name: Greg Mendel Genome ID: genome_greg_mendel_dad Sequencing Provider: 23andMe Sequencing Type: Genotyping SNP Array Sequencing
PERSONALIZED GENETIC REPORT CLIENT-REPORTED DATA PURPOSE OF THE X-SCREEN TEST
 INCLUDED IN THIS REPORT: REVIEW OF YOUR GENETIC INFORMATION RELEVANT TO ENDOMETRIOSIS PERSONAL EDUCATIONAL INFORMATION RELEVANT TO YOUR GENES INFORMATION FOR OBTAINING YOUR ENTIRE X-SCREEN DATA FILE PERSONALIZED
INCLUDED IN THIS REPORT: REVIEW OF YOUR GENETIC INFORMATION RELEVANT TO ENDOMETRIOSIS PERSONAL EDUCATIONAL INFORMATION RELEVANT TO YOUR GENES INFORMATION FOR OBTAINING YOUR ENTIRE X-SCREEN DATA FILE PERSONALIZED
Dr Rick Tearle Senior Applications Specialist, EMEA Complete Genomics Complete Genomics, Inc.
 Dr Rick Tearle Senior Applications Specialist, EMEA Complete Genomics Topics Overview of Data Processing Pipeline Overview of Data Files 2 DNA Nano-Ball (DNB) Read Structure Genome : acgtacatgcattcacacatgcttagctatctctcgccag
Dr Rick Tearle Senior Applications Specialist, EMEA Complete Genomics Topics Overview of Data Processing Pipeline Overview of Data Files 2 DNA Nano-Ball (DNB) Read Structure Genome : acgtacatgcattcacacatgcttagctatctctcgccag
TumorNext-HRD with OvaNext: Paired Germline and Tumor Analyses of Genes Involved in
 SAMPLE REPORT Ordered By Contact ID:1251298 Example, Doctor, MD MOCKORG44 (10829) 123 Somewhere LaneSuite 4 Heaven NV 78872 US Ph:123-123-1234 Fx:123-123-1223 Org ID:8141 Normal Specimen Accession #: 00-086947
SAMPLE REPORT Ordered By Contact ID:1251298 Example, Doctor, MD MOCKORG44 (10829) 123 Somewhere LaneSuite 4 Heaven NV 78872 US Ph:123-123-1234 Fx:123-123-1223 Org ID:8141 Normal Specimen Accession #: 00-086947
Mutational and phenotypical spectrum of phenylalanine hydroxylase deficiency in Denmark
 Clin Genet 2016: 90: 247 251 Printed in Singapore. All rights reserved Short Report 2015 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd CLINICAL GENETICS doi: 10.1111/cge.12692 Mutational and
Clin Genet 2016: 90: 247 251 Printed in Singapore. All rights reserved Short Report 2015 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd CLINICAL GENETICS doi: 10.1111/cge.12692 Mutational and
variant led to a premature stop codon p.k316* which resulted in nonsense-mediated mrna decay. Although the exact function of the C19L1 is still
 157 Neurological disorders primarily affect and impair the functioning of the brain and/or neurological system. Structural, electrical or metabolic abnormalities in the brain or neurological system can
157 Neurological disorders primarily affect and impair the functioning of the brain and/or neurological system. Structural, electrical or metabolic abnormalities in the brain or neurological system can
CURRENT GENETIC TESTING TOOLS IN NEONATAL MEDICINE. Dr. Bahar Naghavi
 2 CURRENT GENETIC TESTING TOOLS IN NEONATAL MEDICINE Dr. Bahar Naghavi Assistant professor of Basic Science Department, Shahid Beheshti University of Medical Sciences, Tehran,Iran 3 Introduction Over 4000
2 CURRENT GENETIC TESTING TOOLS IN NEONATAL MEDICINE Dr. Bahar Naghavi Assistant professor of Basic Science Department, Shahid Beheshti University of Medical Sciences, Tehran,Iran 3 Introduction Over 4000
Genetic Testing for Li-Fraumeni Syndrome
 Protocol Genetic Testing for Li-Fraumeni Syndrome (204101) Medical Benefit Effective Date: 10/01/18 Next Review Date: 07/19 Preauthorization Yes Review Dates: 07/14, 07/15, 07/16, 07/17, 07/18 Preauthorization
Protocol Genetic Testing for Li-Fraumeni Syndrome (204101) Medical Benefit Effective Date: 10/01/18 Next Review Date: 07/19 Preauthorization Yes Review Dates: 07/14, 07/15, 07/16, 07/17, 07/18 Preauthorization
Protein Domain-Centric Approach to Study Cancer Somatic Mutations from High-throughput Sequencing Studies
 Protein Domain-Centric Approach to Study Cancer Somatic Mutations from High-throughput Sequencing Studies Dr. Maricel G. Kann Assistant Professor Dept of Biological Sciences UMBC 2 The term protein domain
Protein Domain-Centric Approach to Study Cancer Somatic Mutations from High-throughput Sequencing Studies Dr. Maricel G. Kann Assistant Professor Dept of Biological Sciences UMBC 2 The term protein domain
MEDICAL GENOMICS LABORATORY. Non-NF1 RASopathy panel by Next-Gen Sequencing and Deletion/Duplication Analysis of SPRED1 (NNP-NG)
 Non-NF1 RASopathy panel by Next-Gen Sequencing and Deletion/Duplication Analysis of SPRED1 (NNP-NG) Ordering Information Acceptable specimen types: Blood (3-6ml EDTA; no time limitations associated with
Non-NF1 RASopathy panel by Next-Gen Sequencing and Deletion/Duplication Analysis of SPRED1 (NNP-NG) Ordering Information Acceptable specimen types: Blood (3-6ml EDTA; no time limitations associated with
Finding subtle mutations with the Shannon human mrna splicing pipeline
 Finding subtle mutations with the Shannon human mrna splicing pipeline Presentation at the CLC bio Medical Genomics Workshop American Society of Human Genetics Annual Meeting November 9, 2012 Peter K Rogan
Finding subtle mutations with the Shannon human mrna splicing pipeline Presentation at the CLC bio Medical Genomics Workshop American Society of Human Genetics Annual Meeting November 9, 2012 Peter K Rogan
Investigating rare diseases with Agilent NGS solutions
 Investigating rare diseases with Agilent NGS solutions Chitra Kotwaliwale, Ph.D. 1 Rare diseases affect 350 million people worldwide 7,000 rare diseases 80% are genetic 60 million affected in the US, Europe
Investigating rare diseases with Agilent NGS solutions Chitra Kotwaliwale, Ph.D. 1 Rare diseases affect 350 million people worldwide 7,000 rare diseases 80% are genetic 60 million affected in the US, Europe
Putative low penetrance or susceptibility variants: sodium channel genes in painful neuropathy as an example
 Putative low penetrance or susceptibility variants: sodium channel genes in painful neuropathy as an example Carl Fratter 1 Kate Sergeant 1, Julie C Evans 1, Anneke Seller 1, David Bennett 2 1 Oxford Medical
Putative low penetrance or susceptibility variants: sodium channel genes in painful neuropathy as an example Carl Fratter 1 Kate Sergeant 1, Julie C Evans 1, Anneke Seller 1, David Bennett 2 1 Oxford Medical
Molecular Diagnosis of Genetic Diseases: From 1 Gene to 1000s
 ROMA, 23 GIUGNO 2011 Molecular Diagnosis of Genetic Diseases: From 1 Gene to 1000s USSD Lab Genetica Medica Ospedali Riuniti Bergamo The problem What does the clinician/patient want to know?? Diagnosis,
ROMA, 23 GIUGNO 2011 Molecular Diagnosis of Genetic Diseases: From 1 Gene to 1000s USSD Lab Genetica Medica Ospedali Riuniti Bergamo The problem What does the clinician/patient want to know?? Diagnosis,
Hands-On Ten The BRCA1 Gene and Protein
 Hands-On Ten The BRCA1 Gene and Protein Objective: To review transcription, translation, reading frames, mutations, and reading files from GenBank, and to review some of the bioinformatics tools, such
Hands-On Ten The BRCA1 Gene and Protein Objective: To review transcription, translation, reading frames, mutations, and reading files from GenBank, and to review some of the bioinformatics tools, such
Single Gene (Monogenic) Disorders. Mendelian Inheritance: Definitions. Mendelian Inheritance: Definitions
 Single Gene (Monogenic) Disorders Mendelian Inheritance: Definitions A genetic locus is a specific position or location on a chromosome. Frequently, locus is used to refer to a specific gene. Alleles are
Single Gene (Monogenic) Disorders Mendelian Inheritance: Definitions A genetic locus is a specific position or location on a chromosome. Frequently, locus is used to refer to a specific gene. Alleles are
The coiled-coil domain containing protein CCDC40 is essential for motile cilia function and left-right axis formation
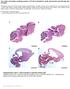 The coiled-coil domain containing protein CCDC40 is essential for motile cilia function and left-right axis formation Anita Becker-Heck#, Irene Zohn#, Noriko Okabe#, Andrew Pollock#, Kari Baker Lenhart,
The coiled-coil domain containing protein CCDC40 is essential for motile cilia function and left-right axis formation Anita Becker-Heck#, Irene Zohn#, Noriko Okabe#, Andrew Pollock#, Kari Baker Lenhart,
Spectrum of mutations in monogenic diabetes genes identified from high-throughput DNA sequencing of 6888 individuals
 Bansal et al. BMC Medicine (2017) 15:213 DOI 10.1186/s12916-017-0977-3 RESEARCH ARTICLE Spectrum of mutations in monogenic diabetes genes identified from high-throughput DNA sequencing of 6888 individuals
Bansal et al. BMC Medicine (2017) 15:213 DOI 10.1186/s12916-017-0977-3 RESEARCH ARTICLE Spectrum of mutations in monogenic diabetes genes identified from high-throughput DNA sequencing of 6888 individuals
Clinical Genetics in Cardiomyopathies
 Clinical Genetics in Cardiomyopathies Γεώργιος Κ Ευθυμιάδης Αναπληρωτής Καθηγητής Καρδιολογίας ΑΠΘ No conflict of interest Genetic terms Proband: The first individual diagnosed in a family Mutation: A
Clinical Genetics in Cardiomyopathies Γεώργιος Κ Ευθυμιάδης Αναπληρωτής Καθηγητής Καρδιολογίας ΑΠΘ No conflict of interest Genetic terms Proband: The first individual diagnosed in a family Mutation: A
Pedigree Analysis Why do Pedigrees? Goals of Pedigree Analysis Basic Symbols More Symbols Y-Linked Inheritance
 Pedigree Analysis Why do Pedigrees? Punnett squares and chi-square tests work well for organisms that have large numbers of offspring and controlled mating, but humans are quite different: Small families.
Pedigree Analysis Why do Pedigrees? Punnett squares and chi-square tests work well for organisms that have large numbers of offspring and controlled mating, but humans are quite different: Small families.
Lab Activity Report: Mendelian Genetics - Genetic Disorders
 Name Date Period Lab Activity Report: Mendelian Genetics - Genetic Disorders Background: Sometimes genetic disorders are caused by mutations to normal genes. When the mutation has been in the population
Name Date Period Lab Activity Report: Mendelian Genetics - Genetic Disorders Background: Sometimes genetic disorders are caused by mutations to normal genes. When the mutation has been in the population
Re-classification of variants: Implications from the cardiac perspective
 Re-classification of variants: Implications from the cardiac perspective Karen McGuire Oxford Medical Genetics Laboratories, Churchill Hospital, Old Road, Oxford, OX3 7LE. Re-classification New evidence
Re-classification of variants: Implications from the cardiac perspective Karen McGuire Oxford Medical Genetics Laboratories, Churchill Hospital, Old Road, Oxford, OX3 7LE. Re-classification New evidence
Introduction. Introduction
 Introduction We are leveraging genome sequencing data from The Cancer Genome Atlas (TCGA) to more accurately define mutated and stable genes and dysregulated metabolic pathways in solid tumors. These efforts
Introduction We are leveraging genome sequencing data from The Cancer Genome Atlas (TCGA) to more accurately define mutated and stable genes and dysregulated metabolic pathways in solid tumors. These efforts
Psych 3102 Lecture 5
 Psych 3102 Lecture 5 Extensions of Mendel - continued Multiple alleles where more than two alleles are present for the trait in the population Example: ABO blood group system in humans antigen antibody
Psych 3102 Lecture 5 Extensions of Mendel - continued Multiple alleles where more than two alleles are present for the trait in the population Example: ABO blood group system in humans antigen antibody
Analysis with SureCall 2.1
 Analysis with SureCall 2.1 Danielle Fletcher Field Application Scientist July 2014 1 Stages of NGS Analysis Primary analysis, base calling Control Software FASTQ file reads + quality 2 Stages of NGS Analysis
Analysis with SureCall 2.1 Danielle Fletcher Field Application Scientist July 2014 1 Stages of NGS Analysis Primary analysis, base calling Control Software FASTQ file reads + quality 2 Stages of NGS Analysis
Bioinformatics Laboratory Exercise
 Bioinformatics Laboratory Exercise Biology is in the midst of the genomics revolution, the application of robotic technology to generate huge amounts of molecular biology data. Genomics has led to an explosion
Bioinformatics Laboratory Exercise Biology is in the midst of the genomics revolution, the application of robotic technology to generate huge amounts of molecular biology data. Genomics has led to an explosion
HHS Public Access Author manuscript Hum Mutat. Author manuscript; available in PMC 2016 April 16.
 GeneMatcher: A Matching Tool for Connecting Investigators with an Interest in the Same Gene Nara Sobreira 1,*, François Schiettecatte 2, David Valle 1, and Ada Hamosh 1 1 Institute of Genetic Medicine,
GeneMatcher: A Matching Tool for Connecting Investigators with an Interest in the Same Gene Nara Sobreira 1,*, François Schiettecatte 2, David Valle 1, and Ada Hamosh 1 1 Institute of Genetic Medicine,
Compound heterozygous mutations in UBA5 causing early-onset epileptic encephalopathy in two sisters
 Arnadottir et al. BMC Medical Genetics (2017) 18:103 DOI 10.1186/s12881-017-0466-8 CASE REPORT Open Access Compound heterozygous mutations in UBA5 causing early-onset epileptic encephalopathy in two sisters
Arnadottir et al. BMC Medical Genetics (2017) 18:103 DOI 10.1186/s12881-017-0466-8 CASE REPORT Open Access Compound heterozygous mutations in UBA5 causing early-onset epileptic encephalopathy in two sisters
De novo mutational profile in RB1 clarified using a mutation rate modeling algorithm
 Aggarwala et al. BMC Genomics (2017) 18:155 DOI 10.1186/s12864-017-3522-z RESEARCH ARTICLE Open Access De novo mutational profile in RB1 clarified using a mutation rate modeling algorithm Varun Aggarwala
Aggarwala et al. BMC Genomics (2017) 18:155 DOI 10.1186/s12864-017-3522-z RESEARCH ARTICLE Open Access De novo mutational profile in RB1 clarified using a mutation rate modeling algorithm Varun Aggarwala
Genetic Counselling in relation to genetic testing
 Genetic Counselling in relation to genetic testing Dr Julie Vogt Consultant Geneticist West Midlands Regional Genetics Service September 2016 Disclosures for Research Support/P.I. Employee Consultant Major
Genetic Counselling in relation to genetic testing Dr Julie Vogt Consultant Geneticist West Midlands Regional Genetics Service September 2016 Disclosures for Research Support/P.I. Employee Consultant Major
Home Brewed Personalized Genomics
 Home Brewed Personalized Genomics The Quest for Meaningful Analysis Results of a 23andMe Exome Pilot Trio of Myself, Wife, and Son February 22, 2013 Gabe Rudy, Vice President of Product Development Exome
Home Brewed Personalized Genomics The Quest for Meaningful Analysis Results of a 23andMe Exome Pilot Trio of Myself, Wife, and Son February 22, 2013 Gabe Rudy, Vice President of Product Development Exome
Genome 371, Autumn 2018 Quiz Section 9: Genetics of Cancer Worksheet
 Genome 371, Autumn 2018 Quiz Section 9: Genetics of Cancer Worksheet All cancer is due to genetic mutations. However, in cancer that clusters in families (familial cancer) at least one of these mutations
Genome 371, Autumn 2018 Quiz Section 9: Genetics of Cancer Worksheet All cancer is due to genetic mutations. However, in cancer that clusters in families (familial cancer) at least one of these mutations
CASE STUDY. Germline Cancer Testing in A Proband: Extension of Benefits to Unaffected Family Members. Introduction. Patient Profile.
 CASE STUDY Germline Cancer Testing in A Proband: Extension of Benefits to Unaffected Family Members Introduction Most cancers are caused by genetic damage resulting from random mutations and exposure to
CASE STUDY Germline Cancer Testing in A Proband: Extension of Benefits to Unaffected Family Members Introduction Most cancers are caused by genetic damage resulting from random mutations and exposure to
Screen Positive Follow-up In The Clinic
 Screen Positive Follow-up In The Clinic NBS Molecular Training Workshop April 2016 Sherly Pardo-Reoyo, M.D. Medical Geneticist Assistant Professor Dept. Biochemistry & Pediatrics UPR-School of Medicine
Screen Positive Follow-up In The Clinic NBS Molecular Training Workshop April 2016 Sherly Pardo-Reoyo, M.D. Medical Geneticist Assistant Professor Dept. Biochemistry & Pediatrics UPR-School of Medicine
Guidelines for Variant Classification
 Guidelines for Variant Classification By the BeSHG BELMOLGEN variant classification working group: Wim Wuyts, Anniek Corveleyn, Valérie Benoit, Rydlewski Catherine, Emilie Castermans, Xavier Pepermans,
Guidelines for Variant Classification By the BeSHG BELMOLGEN variant classification working group: Wim Wuyts, Anniek Corveleyn, Valérie Benoit, Rydlewski Catherine, Emilie Castermans, Xavier Pepermans,
Case presentations from diagnostic exome sequencing results in ion channels M. Koko, U. Hedrich, H. Lerche DASNE meeting 2017, Eisenach
 Case presentations from diagnostic exome sequencing results in ion channels M. Koko, U. Hedrich, H. Lerche DASNE meeting 2017, Eisenach CASE 1: Clinical picture Patient s presentation: 38 year old jewish
Case presentations from diagnostic exome sequencing results in ion channels M. Koko, U. Hedrich, H. Lerche DASNE meeting 2017, Eisenach CASE 1: Clinical picture Patient s presentation: 38 year old jewish
The Promise of Epilepsy Genetics A Personal & Scientific Perspective December 3, 2012
 The Promise of Epilepsy Genetics A Personal & Scientific Perspective December 3, 2012 Tracy Dixon-Salazar, Ph.D. University of California, San Diego American Epilepsy Society Annual Meeting 1 Disclosure
The Promise of Epilepsy Genetics A Personal & Scientific Perspective December 3, 2012 Tracy Dixon-Salazar, Ph.D. University of California, San Diego American Epilepsy Society Annual Meeting 1 Disclosure
Whole Exome Sequencing (WES): Questions and Answers for Providers
 Whole Exome Sequencing (WES): Questions and Answers for Providers 1. What is Whole Exome Sequencing?... 2 2. What is the difference between Whole Exome Sequencing (WES) and Whole Exome Sequencing Plus
Whole Exome Sequencing (WES): Questions and Answers for Providers 1. What is Whole Exome Sequencing?... 2 2. What is the difference between Whole Exome Sequencing (WES) and Whole Exome Sequencing Plus
Advances in genetic diagnosis of neurological disorders
 Acta Neurol Scand 2014: 129 (Suppl. 198): 20 25 DOI: 10.1111/ane.12232 2014 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd ACTA NEUROLOGICA SCANDINAVICA Review Article Advances in genetic diagnosis
Acta Neurol Scand 2014: 129 (Suppl. 198): 20 25 DOI: 10.1111/ane.12232 2014 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd ACTA NEUROLOGICA SCANDINAVICA Review Article Advances in genetic diagnosis
Evaluation of Next-Generation Sequencing as a clinical and research modality in the diagnosis of hereditary breast cancer
 Boston University OpenBU Theses & Dissertations http://open.bu.edu Boston University Theses & Dissertations 2015 Evaluation of Next-Generation Sequencing as a clinical and research modality in the diagnosis
Boston University OpenBU Theses & Dissertations http://open.bu.edu Boston University Theses & Dissertations 2015 Evaluation of Next-Generation Sequencing as a clinical and research modality in the diagnosis
Lecture 20. Disease Genetics
 Lecture 20. Disease Genetics Michael Schatz April 12 2018 JHU 600.749: Applied Comparative Genomics Part 1: Pre-genome Era Sickle Cell Anaemia Sickle-cell anaemia (SCA) is an abnormality in the oxygen-carrying
Lecture 20. Disease Genetics Michael Schatz April 12 2018 JHU 600.749: Applied Comparative Genomics Part 1: Pre-genome Era Sickle Cell Anaemia Sickle-cell anaemia (SCA) is an abnormality in the oxygen-carrying
Corporate Medical Policy
 Corporate Medical Policy Moderate Penetrance Variants Associated with Breast Cancer in File Name: Origination: Last CAP Review: Next CAP Review: Last Review: moderate_penetrance_variants_associated_with_breast_cancer_
Corporate Medical Policy Moderate Penetrance Variants Associated with Breast Cancer in File Name: Origination: Last CAP Review: Next CAP Review: Last Review: moderate_penetrance_variants_associated_with_breast_cancer_
Name: PS#: Biol 3301 Midterm 1 Spring 2012
 Name: PS#: Biol 3301 Midterm 1 Spring 2012 Multiple Choice. Circle the single best answer. (4 pts each) 1. Which of the following changes in the DNA sequence of a gene will produce a new allele? a) base
Name: PS#: Biol 3301 Midterm 1 Spring 2012 Multiple Choice. Circle the single best answer. (4 pts each) 1. Which of the following changes in the DNA sequence of a gene will produce a new allele? a) base
Novel Heterozygous Mutation in YAP1 in A Family with Isolated Ocular Colobomas
 Novel Heterozygous Mutation in YAP1 in A Family with Isolated Ocular Colobomas Julius T. Oatts 1, Sarah Hull 2, Michel Michaelides 2, Gavin Arno 2, Andrew R. Webster 2*, Anthony T. Moore 1,2* 1. Department
Novel Heterozygous Mutation in YAP1 in A Family with Isolated Ocular Colobomas Julius T. Oatts 1, Sarah Hull 2, Michel Michaelides 2, Gavin Arno 2, Andrew R. Webster 2*, Anthony T. Moore 1,2* 1. Department
Personalis ACE Clinical Exome The First Test to Combine an Enhanced Clinical Exome with Genome- Scale Structural Variant Detection
 Personalis ACE Clinical Exome The First Test to Combine an Enhanced Clinical Exome with Genome- Scale Structural Variant Detection Personalis, Inc. 1350 Willow Road, Suite 202, Menlo Park, California 94025
Personalis ACE Clinical Exome The First Test to Combine an Enhanced Clinical Exome with Genome- Scale Structural Variant Detection Personalis, Inc. 1350 Willow Road, Suite 202, Menlo Park, California 94025
Beta Thalassemia Case Study Introduction to Bioinformatics
 Beta Thalassemia Case Study Sami Khuri Department of Computer Science San José State University San José, California, USA sami.khuri@sjsu.edu www.cs.sjsu.edu/faculty/khuri Outline v Hemoglobin v Alpha
Beta Thalassemia Case Study Sami Khuri Department of Computer Science San José State University San José, California, USA sami.khuri@sjsu.edu www.cs.sjsu.edu/faculty/khuri Outline v Hemoglobin v Alpha
MutationTaster & RegulationSpotter
 MutationTaster & RegulationSpotter Pathogenicity Prediction of Sequence Variants: Past, Present and Future Dr. rer. nat. Jana Marie Schwarz Klinik für Pädiatrie m. S. Neurologie Exzellenzcluster NeuroCure
MutationTaster & RegulationSpotter Pathogenicity Prediction of Sequence Variants: Past, Present and Future Dr. rer. nat. Jana Marie Schwarz Klinik für Pädiatrie m. S. Neurologie Exzellenzcluster NeuroCure
Non-Mendelian inheritance
 Non-Mendelian inheritance Focus on Human Disorders Peter K. Rogan, Ph.D. Laboratory of Human Molecular Genetics Children s Mercy Hospital Schools of Medicine & Computer Science and Engineering University
Non-Mendelian inheritance Focus on Human Disorders Peter K. Rogan, Ph.D. Laboratory of Human Molecular Genetics Children s Mercy Hospital Schools of Medicine & Computer Science and Engineering University
Dr. Ayman Mohsen Mashi, MBBS Consultant Hematology & Blood Transfusion Department Head, Laboratory & Blood Bank King Fahad Central Hospital, Gazan,
 Dr. Ayman Mohsen Mashi, MBBS Consultant Hematology & Blood Transfusion Department Head, Laboratory & Blood Bank King Fahad Central Hospital, Gazan, KSA amashi@moh.gov.sa 24/02/2018 β-thalassemia syndromes
Dr. Ayman Mohsen Mashi, MBBS Consultant Hematology & Blood Transfusion Department Head, Laboratory & Blood Bank King Fahad Central Hospital, Gazan, KSA amashi@moh.gov.sa 24/02/2018 β-thalassemia syndromes
