Spectrophotometric Determination of the Kinetic Parameters of β-fructofuranosidase and the Mechanism of Inhibition by Copper (II) Sulfate
|
|
|
- Lorin Walters
- 5 years ago
- Views:
Transcription
1 Spectrophotometric Determination of the Kinetic Parameters of β-fructofuranosidase and the Mechanism of Inhibition by Copper (II) Sulfate Allen Zhang Tyson Miao Science One Program The University of British Columbia Vancouver, Canada April 20
2 Abstract Excessive dosages of heavy metals can be hazardous to organisms and the environment. This is due to the ability of heavy metal ions to inhibit several kinds of biological enzymes. In this experiment, the kinetic parameters of β-fructofuranosidase, a fungal enzyme that catalyzes the breakdown of sucrose, were quantified through dinitrosalicylic acid assay and nonlinear regression. Furthermore, the activity of β-fructofuranosidase was measured in 6 different concentrations of copper (II) sulfate, an inhibitor, to determine the nature of inhibition. From the experiment, the results show that the nature of inhibition is noncompetitive at [CuSO 4 ] < M and competitive at [CuSO 4 ] > M, V max is 0.44 ± mmol, K min m is 7.6 ± 2.6mM, and K i is ± Introduction 2. Background Sucrose is a disaccharide composed of α-glucose and β-fructose joined by an α-,4-glycosidic linkage (Freeman et al., 20). This bond can be hydrolyzed by β-fructofuranosidase, an enzyme commonly known as invertase, to yield glucose and fructose in equal proportions (Greenwood Health Systems, 2009). The presence of heavy metal ions such as Cu 2+ may inhibit the activity of invertase. This is due to the high affinity of the sulfhydryl group on cysteine and methionine residues in a protein for the Cu 2+ ion (Greenwood Health Systems, 2009). Soft bases, including sulfhydryl groups, have a high affinity for soft acids, such as the Cu 2+ ion (Cotton et al., 976). When copper ions form complexes with sulfhydryl groups in amino acid residues, disulfide bridges are disrupted, modifying the enzyme s tertiary structure. Since the functions of an enzyme are highly correlated with its form, a change in the tertiary structure of invertase would likely inhibit its function (Cotton et al., 976). As a result of the close proximity of methionine residues to the glycosylation sites of invertase, CuSO 4 is suspected to be a noncompetitive inhibitor one that slows product formation, while not affecting substrate binding ( In order to test this hypothesis, the rates of sucrose hydrolysis by invertase at various concentrations of copper sulfate were experimentally determined, and a Lineweaver-Burk plot was used to determine the mechanism of inhibition. 2.2 Michaelis-Menten Kinetics Michaelis-Menten kinetics were used throughout the experiment to model the reaction rate as a function of the inhibitor concentration, and to test whether the mechanism of inhibition was noncompetitive, competitive, or mixed. Noncompetitive inhibition refers to the process by which an inhibitor binds to a non-active site in an enzyme, and slows down the rate of product formation. However, noncompetitive inhibitors do not change the affinity, as measured by the Michaelis-Menten constant, K m, of an enzyme for its substrate. On the other hand, competitive inhibition refers to the type of inhibition in which an inhibitor binds to an enzyme s active site and lowers the affinity of the enzyme for its substrate. Competitive inhibition does not change the maximal rate of reaction, measured as V max, due to the fact that the presence of a large
3 amount of substrate can saturate the active sites, preventing the inhibitor from binding. Mixed inhibition is a combination of competitive and noncompetitive inhibition; both V max and K m are changed. The general kinetic scheme for noncompetitive inhibition is as follows ( The reaction velocity, v, is: v = V max [S] (K m + [S])( + [I] K i ) where K i is the kinetic constant of inhibition, which measures the ratio between the concentration of the enzyme-inhibitor complex and free inhibitor ( Rearranging, v = ( + [I] K i ) K m V max [S] + ( + [I] K i ) V max is a linear equation in terms of [S] and v. This equation serves as the basis for the Lineweaver- Burk plot (/v vs. /[S]), for noncompetitive inhibition. Similar equations for competitive and mixed inhibition can be found in literature ( 2.3 Applications The study of enzyme kinetics is essential in the fields of medicine, biology, and chemistry. Baker s yeast (scientific name: Saccharomyces cerevisiae), a species of fungi that produces invertase, can be used to detect trace amounts of heavy metal ions, like cadmium and mercury, by analyzing the yeast s enzymatic activity over a fixed period of time, in solution (Narayana & Kumar, 200). Detecting heavy metal concentrations is vital in testing water quality. Enzyme inhibitors can also be used in antibiotics. Most antibiotics are administered with β- lactamase inhibitors, such as tazobactam and clavulanic acid, to prevent the bacterial enzyme β-lactamase from degrading antibiotics (Kuck et al., 989). By using an approach similar to the one carried out in this experiment, the enzyme kinetics of certain types of β-lactamases can be studied in order to elucidate the amount of tazobactam or clavulanic acid that should be administered to patients so as to minimize the probability of bacterial survival while maintaining reasonable production costs. 2
4 Figure : Diagram of the experimental setup for the treatment with no inhibitor. DNS reagent was added to each of the test tubes, and the test tubes were immersed in the 95 C water bath. 3 Methods 3. Beer s Law Calibration Various amounts of deionized water and glucose-fructose solution were added to 7 labelled test tubes to generate the glucose-fructose concentrations specified in Table..5mL of DNS reagent was then added to each test tube, and all of the test tubes were transferred to a C water bath. The test tubes were collected after 0 minutes and transferred to a cold water bath. After the test tubes returned to room temperature, the contents in each of the test tubes were diluted 00 times by mixing 0. ml of test tube solution with 9.9mL of deionized water. An aliquot of the diluted content for each test tube was transferred into individual borosilicate test tubes in order to measure the absorbance of the diluted contents at 540nm with a spectrophotometer. The blank used was the same as that for Measuring Enzyme Activity without Inhibitor (see below). All empty borosilicate test tubes were chosen within a range of A absorbances at a wavelength of 540nm for consistency. Table. Summary of the concentrations of glucose-fructose solution in each of the test tubes. Test tube Test tube 2 Test tube 3 Test tube 4 Test tube 5 Test tube 6 Test tube 7 [Glucose- Fructose] (M)
5 3.2 Measuring Enzyme Activity without Inhibitor Various amounts of deionized water and 50mg/mL sucrose solution were added to 6 labelled test tubes, exact quantities are specified in Table 2..5 ml of 0.04 mg/ml invertase solution (from Sigma-Aldrich I4504 Grade VII invertase) was then added to each test tube before the test tubes were transferred to a 55 C water bath. The sudden rise in temperature facilitated the reaction between invertase and sucrose. After 5 minutes, the test tubes were taken out of the 55 C water bath and.5 ml of the DNS Reagent was added to each test tube. Upon the addition of the DNS Reagent, the test tubes were immediately transferred to a C water bath; the second rise in temperature was intended to denature invertase and facilitate the bonding of DNS Reagent to glucose and fructose. The test tubes were collected after 0 minutes and transferred to a cold water bath. When the test tubes returned to room temperature, the content of each test tube was diluted 00 times by mixing 0. ml of the content with 9.9 ml of deionized water. An aliquot of the diluted content of each test tube was transferred into individual borosilicate test tubes in order to measure the absorbance of the diluted contents at 540nm, using a spectrophotometer; Test tube was used as the blank for calibration. Furthermore, all empty borosilicate test tubes were chosen within a range of 0.073A to 0.077A absorbances at 540nm for consistency. Table 2. Summary of the amounts of reagents added to each of the test tubes (see Measuring Enzyme Activity without Inhibitor). Test tube Test tube 2 Test tube 3 Test tube 4 Test tube 5 Test tube 6 Deionized H 2 O (ml) mg/ml Sucrose (ml) Measuring Enzyme Activity with Varying Concentrations of Inhibitor (CuSO 4 ) Similar to the setup of the previous section, various amounts of deionized water and 50mg/mL sucrose solution were added to labelled test tubes with new additions of 0. M CuSO 4 at different treatments levels. During the experiment, this section was conducted in sets of 6 test tubes; all sets contained the amounts of sucrose specified in Table 2 and the amount of CuSO 4 specified for each treatment level in Table 3. Although the amount of sucrose in each test tube remained constant from the last section, as illustrated by Table 2, the amount of deionized water required by each test tube varies between different treatments levels of CuSO 4. After sucrose and CuSO 4 were added, the volume of each test tube was adjusted up to.5 ml with deionized water. For each set, an additional.5 ml of 0.04 mg/ml invertase solution was added before the test tubes were transferred to a 55 C water bath. The sudden rise in temperature facilitated the reaction between invertase and sucrose. After 5 minutes, the test tubes were taken out of the 55 C water bath and.5 ml of the DNS Reagent was added to each test tube. Upon the addition of the DNS Reagent, the test tubes were immediately transferred to a C water bath; the second rise in temperature was intended to denature invertase and facilitate the bonding of DNS Reagent to glucose and fructose. The test tubes were collected after 0 minutes and transferred into a cooling water bath. When the test tubes returned to room temperature, the 4
6 content of each test tube was diluted 00 times by mixing 0. ml of the content with 9.9 ml of deionized water. An aliquot of the diluted content of each test tube was transferred into individual borosilicate test tubes in order to measure the absorbance of the diluted contents at 540nm, using a spectrophotometer; Test tube was used as the blank for calibration. Furthermore, all empty borosilicate test tubes were chosen within a range of 0.073A to 0.077A absorbances at 540nm for consistency. Table 3. Summary of the volumes of 0.M CuSO 4 in each of the treatment levels with inhibitor. Treatment level (mls of CuSO 4 ) Results & Discussion 4. Measurements Table 4. Beer s Law Analysis. The following data was taken with a spectrophotometer by pouring the invertase solutions into borosilicate test tubes, and recording the absorbances at 540nm. The solutions in the test tubes were diluted 00X from the values listed below. The borosilicate test tubes had absorbances that ranged from 0.073A to 0.077A. The uncertainties were determined by taking half of this range (0.002A). [Glucose] (M ) Absorbance Uncertainty [Glucose] Absorbance [Glucose] 2 (M) (Model(Abs.)-Actual) 2 (M 2 ) The linear regression line has to go through the origin (the blank), so slope (m) and the uncertainty in slope (σ m ) were determined by minimizing χ 2 with: m = N i= x iy i N i= (x i) = M M = 48.0M ; σ 2 m = N N i= (y i mx i ) 2 N i= (x i) 2 =.2M 5
7 Table 5. Rates of sucrose hydrolysis by invertase under different concentrations of CuSO 4. The sucrose solutions were diluted 00X from the values listed. The rate of hydrolysis were determined using the absorbances and Beer s Law calibration techniques (see below). All rates listed below are in units of mm/min. Absorbances listed below are the means of three replicates for each treatment. The uncertainties (δ) were determined by taking the standard error for each of the sets of three replicates. No inhibitor 25µl CuSO 4 50µl CuSO 4 00µl CuSO 4 [Sucrose] (M ) Abs. δ Rate Abs. δ Rate Abs. δ Rate Abs. δ Rate µl CuSO 4 300µl CuSO 4 [Sucrose] (M ) Abs. δ Rate Abs. δ Rate The rates of hydrolysis were calculated from the absorbances with the slope from the Beer s Law plot (Table 4), as follows: R = 4.2 Uncertainty Calculations Abs (48.0M ) 5min 000mM M Table 6. Summary of the uncertainty calculations for the rate of sucrose hydrolysis under different concentrations of copper (II) sulfate. These uncertainties were obtained from propagating the uncertainties listed in Table 5 with the results from the Beer s Law calibration in Table 4. Uncertainty in Rate of Hydrolysis (mmol/min) [Sucrose] (M ) No inhibitor 25µl CuSO 4 50µl CuSO 4 00µl CuSO 4 200µl CuSO 4 300µl CuSO The rate uncertainties (δr) were calculated in the same way as the rates of hydrolysis R were calculated, with a few changes. A sample calculation for the no inhibitor, 0.004M sucrose 6
8 treatment, using the method of partial derivatives, is shown below: r δabs 2 (Abs)(δm) 2 000mM δr = ( ) +( ) m 5min M r m (0.064)(.2M ) 2 000mM = ( ) + ( ) 48.0M 48.0M 5min M mmol = min Figure 2: Graph of the changes in the hydrolysis rate of sucrose by invertase at 6 different concentrations of sucrose and 6 different concentrations of CuSO4 (n = 3). This graph was plotted with the data in Table 5, and the uncertainties in Table Lineweaver-Burk Plot A Lineweaver-Burk plot is a method for linearizing the Rate vs. [Substrate] plots in Figure 2. If ), then copper (II) sulfate is a noncompetitive inhibitor. This the lines intersect at the x-axis ( [S] is due to the fact that the x-intercept will be Km ( Likewise, if the lines intersect at the y-axis ( Rate ), then copper (II) sulfate is a competitive inhibitor, as the y-intercept will be Vmax ( If the lines intersect somewhere in the ( x, +y) quadrant, then the inhibition method is mixed. Table 7. Summary of the calculations used to construct the Lineweaver-Burk plots (Figure 2). The uncertainties were calculated through propagation; smaller uncertainties indicate higher precision. All min values for V (/rate) are in units of mmol. S (M ) No inhibitor 95% C.I V µl CuSO4 95% C.I. V µl CuSO4 95% C.I. V µl CuSO4 95% C.I. V µl CuSO4 95% C.I. V µl CuSO4 95% C.I. V
9 Figure 3: Linearized plots of /Hydrolysis Rate vs. /[Sucrose], commonly referred to as Lineweaver-Burk plots. Plots were constructed from the data in Table 7. 2a Lineweaver- Burk plot for the first four concentrations of CuSO 4, with the point of intersection near the -axis. 2b Lineweaver-Burk plot for the two highest concentrations of CuSO [Sucrose] 4; the point of intersection is at the -axis. V 4.4 Nonlinear Regression Results Due to the fact that using the Lineweaver-Burk method of linearizing to determine K m and V max magnifies experimental errors, GraphPad s Prism 5 R nonlinear regression software was used to determine the kinetic parameters of invertase. Table 8. Summary of the values of the kinetic parameters for invertase (with sucrose as a substrate and copper (II) sulfate as the inhibitor) as determined by nonlinear regression analysis with GraphPad s Prism 5 R software. V max (mmol/min) K m (mm) K i Best Fit Values Standard Error % Confidence Intervals Overall R-squared The R-squared value of is close to.00, which provides confidence in our regression lines. 4.5 Data Analysis 4.5. Kinetic Parameters From nonlinear regression analysis, it was found that K m = 7.6 ± 2.6mM (Table 8). This is comparable to the value of 8.3mM reported by Ribeiro and Vitolo (2006) for soluble invertase at ph 4.5. On the other hand, the value of V max obtained in this study, 0.44 ± mmol min (Table 8), was not in agreement with the results of the same study (V max = Units) ml (Ribeiro & Vitolo 2006). This can be explained by the different experimental conditions in which the two studies were conducted. K i, the kinetic constant of inhibition by copper (II) sulfate, was determined to be ± (Table 8). 8
10 4.5.2 Mechanism of Inhibition When interpreting the data in Figure 2, it is interesting to note that the shape of the ratesubstrate curve differs between concentrations of inhibitor. As inhibitor concentration is increased, the curve gradually becomes more sigmoidally (S) shaped, as opposed to the hyperbolic shape for the noninhibited reaction curve. This indicates that there may have been cooperative or noncooperative binding when the substrate of the reaction serves to facilitate or inhibit the binding of other substrate molecules or feedback inhibition (Combes & Monsan, 983). In fact, both the substrate, sucrose, and the products of the reaction, glucose and fructose, inhibit invertase (Combes & Monsan, 983). Despite this, inhibition due to glucose and fructose likely plays a minor role in the kinetics of the enzyme, as the affinity of sucrose for invertase (K m = 7.6 ± 2.6mM) is greater than that for fructose (K m = 28mM) and glucose (K m = 270mM) (Combes & Monsan, 983). Furthermore, from the Lineweaver-Burk plot in Figure 3a, it is apparent that the best fit lines intersect close to the x-axis, which is indicative of the noncompetitive nature of copper (II) sulfate inhibition, as K m stays nearly constant while the inhibitor concentrations are varied; this is in agreement with the hypothesis. However, the Lineweaver-Burk plot in Figure 3b shows that the best fit lines for the 200µl copper (II) sulfate and 300µl copper (II) sulfate treatments actually intersect at the y-axis. This suggests that, at high concentrations, copper (II) sulfate competitively inhibits invertase. While this apparent discrepancy can be attributed to experimental error as well as the non-michaelis-menten (sigmoidal) nature of inhibition, it is also possible that Cu 2+ ions interact with the active sites of invertase when the allosteric sites are saturated. Research shows that one methionine residue (residue number 40) is in close proximity to one of the active sites of invertase (residue number 42); furthermore, residue number 224, cysteine, is located next to another substrate binding site in invertase (residue number 223) (Taussig & Carlson, 983). Therefore, it can be conjectured that Cu 2+ ions preferentially bind to the allosteric sites in invertase (noncompetitive inhibition), but at high concentrations of Cu 2+, the ions may also bind to the active sites in invertase (competitive inhibition) Biological and Chemical Applications From the results of this study, CuSO 4 has been classified as a noncompetitive inhibitor of invertase, even when [CuSO 4 ] is low. While there are inherent differences among invertase and other biological enzymes, other sources have reported that heavy metal ions are noncompetitive inhibitors for most enzymes (Narayana & Kumar, 200). Therefore, it is likely that the ingestion of a substantial amount of Cu 2+, and possibly also other heavy metal ions may be life-threatening for organisms (heavy metal poisoning), as the essential enzymes that support life may be denatured in the presence of metal ions. With the kinetic parameters determined in this experiment (see Kinetic Parameters), a precise value for the decrease in the activity of invertase in the presence of a certain amount of CuSO 4 can be elucidated. Consequently, [Cu 2+ ] can be determined by adding known amounts of invertase and sucrose into a solution, assuming it does not contain any other inhibitors. A similar experiment and analysis to the ones used in this study can be carried out, for other heavy metal ions. Through this process, the qualities and metal ion contents of water samples can be ascertained. Furthermore, due to the noncompetitive nature of invertase inhibition by CuSO 4, the Michaelis- Menten model of enzyme inhibition can be extended to an enzyme-inhibitor system involv- 9
11 ing certain types of β-lactamases and tazobactam, a noncompetitive inhibitor of particular β- lactamases (Kuck et al., 989). After determining the various kinetic constants K m, V max, and K i for the β-lactamase-tazobactam system, the degree of inhibition at various concentrations of tazobactam can be related to the reaction rates between β-lactamases and their substrates. 5 Conclusion Throughout the experiment, the kinetic parameters of invertase with respect to its substrate, sucrose, were determined as K m = 7.6 ± 2.6mM and V max = 0.44 ± mmol. Furthermore, it was found that CuSO 4 is a noncompetitive inhibitor of invertase at low [CuSO 4 ] min (below M), and CuSO 4 is a competitive inhibitor of invertase at higher [CuSO 4 ] (above M). The inhibition constant, K i, was found to be ± However, further studies should be conducted, with more precise equipment, to determine the kinetic constants to a greater degree of accuracy, and to confirm or refute our conjecture (see Data Analysis). 6 Acknowledgements We would like to thank Dr. Carl Douglas for his guidance and assistance throughout the conception and presentation of this project; Mrs. Xueqin Huang and Mrs. Mingxin Guo for the use of various equipment in the Biology Lab; ; Dr. Eric Cytrynbaum for his help in deriving the appropriate mathematical equations; and Meiying Zhuang and Christie Chan for their assistance in editing this report. 7 References Combes, D. & Monsan, P. (983). Sucrose hydrolysis by invertase. Characterization of products and substrate inhibition. Carbohydrate Research, 7, doi:0.06/ (83) Cotton, F.A., Wilkinson, G. & Gaus, P.L. (976). Basic Inorganic Chemistry. New Baskerville: John Wiley & Sons. Freeman, S., Harrington, M. & Sharp, J.C. (20). Biological Science. Toronto: Pearson Canada. Greenwood Health Systems. (2009). Invertase. Retrieved March 0, 20, from odhealth.net/np/invertase.htm. Kuck, N.A., Jacobus, N.V., Petersen, P.J., Weiss, W.J. & Testa, R.T. (989). Comparative In Vitro and In Vivo Activities of Piperacillin Combined with the -Lactamase Inhibitors Tazobactam, Clavulanic Acid, and Sulbactam. Antimicrobial Agents and Chemotherapy, 33, Narayana, N.N.H. & Kumar, N.V.N. (200). Detection of Compounds of Mercury, Cadmium and Copper by Baker s Dry Yeast Enzyme Inhibition. Middle-East Journal of Scientific Education, 6, Ribeiro, R.R. & Vitolo, M. (2006). Anion exchange resin support for invertase immobilization. Journal of Basic and Applied Pharmaceutical Sciences, 26, Taussig, R. & Carlson, M. (983). Nucleotide sequence of the yeast SUC2 gene for invertase. Nucleic Acids Research,, doi: 0.093/nar/
12 University of Waterloo. (200). Enzyme kinetics. Retrieved March 0, 20, from civil.uwaterloo.ca/enve375 /Background.htm#Enzyme.
4-The effect of sucrose concentration on the rate of reaction catalyzed by β-fructofuranosidase enzyme.
 Kinetics analysis of β-fructofuranosidase enzyme 4-The effect of sucrose concentration on the rate of reaction catalyzed by β-fructofuranosidase enzyme. One of the important parameter affecting the rate
Kinetics analysis of β-fructofuranosidase enzyme 4-The effect of sucrose concentration on the rate of reaction catalyzed by β-fructofuranosidase enzyme. One of the important parameter affecting the rate
Β-FRUCTOFURANOSIDASE ENZYME
 KINETICS ANALYSIS OF Β-FRUCTOFURANOSIDASE ENZYME 2-The effects of enzyme concentration on the rate of an enzyme catalyzed reaction. Systematic names and numbers β-fructofuranosidase (EC 3.2.1.26) Reactions
KINETICS ANALYSIS OF Β-FRUCTOFURANOSIDASE ENZYME 2-The effects of enzyme concentration on the rate of an enzyme catalyzed reaction. Systematic names and numbers β-fructofuranosidase (EC 3.2.1.26) Reactions
Kinetics analysis of β-fructofuranosidase enzyme. 1-Effect of Time Incubation On The Rate Of An Enzymatic Reaction
 Kinetics analysis of β-fructofuranosidase enzyme 1-Effect of Time Incubation On The Rate Of An Enzymatic Reaction Enzyme kinetics It is the study of the chemical reactions that are catalyzed by enzymes.
Kinetics analysis of β-fructofuranosidase enzyme 1-Effect of Time Incubation On The Rate Of An Enzymatic Reaction Enzyme kinetics It is the study of the chemical reactions that are catalyzed by enzymes.
The MOLECULES of LIFE
 The MOLECULES of LIFE Physical and Chemical Principles Solutions Manual Prepared by James Fraser and Samuel Leachman Chapter 16 Principles of Enzyme Catalysis Problems True/False and Multiple Choice 1.
The MOLECULES of LIFE Physical and Chemical Principles Solutions Manual Prepared by James Fraser and Samuel Leachman Chapter 16 Principles of Enzyme Catalysis Problems True/False and Multiple Choice 1.
Past Years Questions Chpater 6
 Past Years Questions Chpater 6 **************************************** 1) Which of the following about enzymes is Incorrect? A) Most enzymes are proteins. B) Enzymes are biological catalysts. C) Enzymes
Past Years Questions Chpater 6 **************************************** 1) Which of the following about enzymes is Incorrect? A) Most enzymes are proteins. B) Enzymes are biological catalysts. C) Enzymes
Biochemistry Department. Level 1 Lecture No : 3 Date : 1 / 10 / Enzymes kinetics
 Biochemistry Department Level 1 Lecture No : 3 Date : 1 / 10 / 2017 Enzymes kinetics 1 Intended Learning Outcomes By the end of this lecture, the student will be able to: 1.Understand what is meant by
Biochemistry Department Level 1 Lecture No : 3 Date : 1 / 10 / 2017 Enzymes kinetics 1 Intended Learning Outcomes By the end of this lecture, the student will be able to: 1.Understand what is meant by
The effects of ph on Type VII-NA Bovine Intestinal Mucosal Alkaline Phosphatase Activity
 The effects of ph on Type VII-NA Bovine Intestinal Mucosal Alkaline Phosphatase Activity ANDREW FLYNN, DYLAN JONES, ERIC MAN, STEPHEN SHIPMAN, AND SHERMAN TUNG Department of Microbiology and Immunology,
The effects of ph on Type VII-NA Bovine Intestinal Mucosal Alkaline Phosphatase Activity ANDREW FLYNN, DYLAN JONES, ERIC MAN, STEPHEN SHIPMAN, AND SHERMAN TUNG Department of Microbiology and Immunology,
Biology 2180 Laboratory #3. Enzyme Kinetics and Quantitative Analysis
 Biology 2180 Laboratory #3 Name Introduction Enzyme Kinetics and Quantitative Analysis Catalysts are agents that speed up chemical processes and the catalysts produced by living cells are called enzymes.
Biology 2180 Laboratory #3 Name Introduction Enzyme Kinetics and Quantitative Analysis Catalysts are agents that speed up chemical processes and the catalysts produced by living cells are called enzymes.
Enzymes: The Catalysts of Life
 Chapter 6 Enzymes: The Catalysts of Life Lectures by Kathleen Fitzpatrick Simon Fraser University Activation Energy and the Metastable State Many thermodynamically feasible reactions in a cell that could
Chapter 6 Enzymes: The Catalysts of Life Lectures by Kathleen Fitzpatrick Simon Fraser University Activation Energy and the Metastable State Many thermodynamically feasible reactions in a cell that could
Experiment 2 Introduction
 Characterization of Invertase from Saccharomyces cerevisiae Experiment 2 Introduction The method we used in A Manual for Biochemistry I Laboratory: Experiment 7 worked well to detect any created reducing
Characterization of Invertase from Saccharomyces cerevisiae Experiment 2 Introduction The method we used in A Manual for Biochemistry I Laboratory: Experiment 7 worked well to detect any created reducing
Human Biochemistry. Enzymes
 Human Biochemistry Enzymes Characteristics of Enzymes Enzymes are proteins which catalyze biological chemical reactions In enzymatic reactions, the molecules at the beginning of the process are called
Human Biochemistry Enzymes Characteristics of Enzymes Enzymes are proteins which catalyze biological chemical reactions In enzymatic reactions, the molecules at the beginning of the process are called
FIRST BIOCHEMISTRY EXAM Tuesday 25/10/ MCQs. Location : 102, 105, 106, 301, 302
 FIRST BIOCHEMISTRY EXAM Tuesday 25/10/2016 10-11 40 MCQs. Location : 102, 105, 106, 301, 302 The Behavior of Proteins: Enzymes, Mechanisms, and Control General theory of enzyme action, by Leonor Michaelis
FIRST BIOCHEMISTRY EXAM Tuesday 25/10/2016 10-11 40 MCQs. Location : 102, 105, 106, 301, 302 The Behavior of Proteins: Enzymes, Mechanisms, and Control General theory of enzyme action, by Leonor Michaelis
Biochem sheet (5) done by: razan krishan corrected by: Shatha Khtoum DATE :4/10/2016
 Biochem sheet (5) done by: razan krishan corrected by: Shatha Khtoum DATE :4/10/2016 Note about the last lecture: you must know the classification of enzyme Sequentially. * We know that a substrate binds
Biochem sheet (5) done by: razan krishan corrected by: Shatha Khtoum DATE :4/10/2016 Note about the last lecture: you must know the classification of enzyme Sequentially. * We know that a substrate binds
Six Types of Enzyme Catalysts
 Six Types of Enzyme Catalysts Although a huge number of reactions occur in living systems, these reactions fall into only half a dozen types. The reactions are: 1. Oxidation and reduction. Enzymes that
Six Types of Enzyme Catalysts Although a huge number of reactions occur in living systems, these reactions fall into only half a dozen types. The reactions are: 1. Oxidation and reduction. Enzymes that
Nafith Abu Tarboush DDS, MSc, PhD
 Nafith Abu Tarboush DDS, MSc, PhD natarboush@ju.edu.jo www.facebook.com/natarboush Biochemical Kinetics: the science that studies rates of chemical reactions An example is the reaction (A P), The velocity,
Nafith Abu Tarboush DDS, MSc, PhD natarboush@ju.edu.jo www.facebook.com/natarboush Biochemical Kinetics: the science that studies rates of chemical reactions An example is the reaction (A P), The velocity,
SAFETY & DISPOSAL onpg is a potential irritant. Be sure to wash your hands after the lab.
 OVERVIEW In this lab we will explore the reaction between the enzyme lactase and its substrate lactose (i.e. its target molecule). Lactase hydrolyzes lactose to form the monosaccharides glucose and galactose.
OVERVIEW In this lab we will explore the reaction between the enzyme lactase and its substrate lactose (i.e. its target molecule). Lactase hydrolyzes lactose to form the monosaccharides glucose and galactose.
Exam 3 Fall 2015 Dr. Stone 8:00. V max = k cat x E t. ΔG = -RT lnk eq K m + [S]
![Exam 3 Fall 2015 Dr. Stone 8:00. V max = k cat x E t. ΔG = -RT lnk eq K m + [S] Exam 3 Fall 2015 Dr. Stone 8:00. V max = k cat x E t. ΔG = -RT lnk eq K m + [S]](/thumbs/75/71985573.jpg) Exam 3 Fall 2015 Dr. Stone 8:00 Name There are 106 possible points (6 bonus points) on this exam. There are 8 pages. v o = V max x [S] k cat = kt e - ΔG /RT V max = k cat x E t ΔG = -RT lnk eq K m + [S]
Exam 3 Fall 2015 Dr. Stone 8:00 Name There are 106 possible points (6 bonus points) on this exam. There are 8 pages. v o = V max x [S] k cat = kt e - ΔG /RT V max = k cat x E t ΔG = -RT lnk eq K m + [S]
Experiment 9. NATURE OF α-amylase ACTIVITY ON STARCH
 Experiment 9 NATURE OF α-amylase ACTIVITY ON STARC In Experiment 1 we described the action of α-amylase on starch as that of catalyzing the hydrolysis of α-1,4-glucosidic bonds at random in the interior
Experiment 9 NATURE OF α-amylase ACTIVITY ON STARC In Experiment 1 we described the action of α-amylase on starch as that of catalyzing the hydrolysis of α-1,4-glucosidic bonds at random in the interior
Biology 104: Human Biology Lab-Like Activity Four
 1 Biology 104: Human Biology Lab-Like Activity Four A scientific study: How to collect data, analysis, graph, and interpret the significance of the study This is to be handed in, in class, on the last
1 Biology 104: Human Biology Lab-Like Activity Four A scientific study: How to collect data, analysis, graph, and interpret the significance of the study This is to be handed in, in class, on the last
Can you explain that monomers are smaller units from which larger molecules are made?
 Biological molecules Can you explain that all living things have a similar biochemical basis? Can you explain that monomers are smaller units from which larger molecules are made? Can you describe polymers
Biological molecules Can you explain that all living things have a similar biochemical basis? Can you explain that monomers are smaller units from which larger molecules are made? Can you describe polymers
Qualitative test of protein-lab2
 1- Qualitative chemical reactions of amino acid protein functional groups: Certain functional groups in proteins can react to produce characteristically colored products. The color intensity of the product
1- Qualitative chemical reactions of amino acid protein functional groups: Certain functional groups in proteins can react to produce characteristically colored products. The color intensity of the product
Exploring enzyme kinetics Monitoring invertase activity using DNSA reagent
 Dean Madden National Centre for Biotechnology Education, University of Reading 2 Earley Gate, Reading RG6 6AU D.R.Madden@reading.ac.uk Exploring enzyme kinetics Monitoring invertase activity using DNSA
Dean Madden National Centre for Biotechnology Education, University of Reading 2 Earley Gate, Reading RG6 6AU D.R.Madden@reading.ac.uk Exploring enzyme kinetics Monitoring invertase activity using DNSA
Biological Sciences 4087 Exam I 9/20/11
 Name: Biological Sciences 4087 Exam I 9/20/11 Total: 100 points Be sure to include units where appropriate. Show all calculations. There are 5 pages and 11 questions. 1.(20pts)A. If ph = 4.6, [H + ] =
Name: Biological Sciences 4087 Exam I 9/20/11 Total: 100 points Be sure to include units where appropriate. Show all calculations. There are 5 pages and 11 questions. 1.(20pts)A. If ph = 4.6, [H + ] =
1. Measurement of the rate constants for simple enzymatic reaction obeying Michaelis- Menten kinetics gave the following results: =3x10-5 = 30μM
 1. Measurement of the rate constants for simple enzymatic reaction obeying Michaelis- Menten kinetics gave the following results: k 1 = 2 x 10 8 M -1 s -1, k 2 = 1 x 10 3 s -1, k 3 = 5 x 10 3 s -1 a) What
1. Measurement of the rate constants for simple enzymatic reaction obeying Michaelis- Menten kinetics gave the following results: k 1 = 2 x 10 8 M -1 s -1, k 2 = 1 x 10 3 s -1, k 3 = 5 x 10 3 s -1 a) What
Chapter 11: Enzyme Catalysis
 Chapter 11: Enzyme Catalysis Matching A) high B) deprotonated C) protonated D) least resistance E) motion F) rate-determining G) leaving group H) short peptides I) amino acid J) low K) coenzymes L) concerted
Chapter 11: Enzyme Catalysis Matching A) high B) deprotonated C) protonated D) least resistance E) motion F) rate-determining G) leaving group H) short peptides I) amino acid J) low K) coenzymes L) concerted
Enzyme Analysis using Tyrosinase. Evaluation copy
 Enzyme Analysis using Tyrosinase Computer 15 Enzymes are molecules that regulate the chemical reactions that occur in all living organisms. Almost all enzymes are globular proteins that act as catalysts,
Enzyme Analysis using Tyrosinase Computer 15 Enzymes are molecules that regulate the chemical reactions that occur in all living organisms. Almost all enzymes are globular proteins that act as catalysts,
CHEM 527 SECOND EXAM FALL 2006
 CEM 527 SECD EXAM FALL 2006 YUR AME: TES: 1. Where appropriate please show work if in doubt show it anyway. 2. Pace yourself you may want to do the easier questions first. 3. Please note the point value
CEM 527 SECD EXAM FALL 2006 YUR AME: TES: 1. Where appropriate please show work if in doubt show it anyway. 2. Pace yourself you may want to do the easier questions first. 3. Please note the point value
االمتحان النهائي لعام 1122
 االمتحان النهائي لعام 1122 Amino Acids : 1- which of the following amino acid is unlikely to be found in an alpha-helix due to its cyclic structure : -phenylalanine -tryptophan -proline -lysine 2- : assuming
االمتحان النهائي لعام 1122 Amino Acids : 1- which of the following amino acid is unlikely to be found in an alpha-helix due to its cyclic structure : -phenylalanine -tryptophan -proline -lysine 2- : assuming
Experiment 1. Isolation of Glycogen from rat Liver
 Experiment 1 Isolation of Glycogen from rat Liver Figure 35: FIG-2, Liver, PAS, 100x. Note the presence of a few scattered glycogen granules (GG). Objective To illustrate the method for isolating glycogen.
Experiment 1 Isolation of Glycogen from rat Liver Figure 35: FIG-2, Liver, PAS, 100x. Note the presence of a few scattered glycogen granules (GG). Objective To illustrate the method for isolating glycogen.
Enzymes - Exercise 3 - Rockville
 Enzymes - Exercise 3 - Rockville Objectives -Understand the function of an enzyme. -Know what the substrate, enzyme, and the product of the reaction for this lab. -Understand how at various environments
Enzymes - Exercise 3 - Rockville Objectives -Understand the function of an enzyme. -Know what the substrate, enzyme, and the product of the reaction for this lab. -Understand how at various environments
بسم هللا الرحمن الرحيم
 بسم هللا الرحمن الرحيم Q1: the overall folding of a single protein subunit is called : -tertiary structure -primary structure -secondary structure -quaternary structure -all of the above Q2 : disulfide
بسم هللا الرحمن الرحيم Q1: the overall folding of a single protein subunit is called : -tertiary structure -primary structure -secondary structure -quaternary structure -all of the above Q2 : disulfide
Qualitative chemical reaction of functional group in protein
 Qualitative chemical reaction of functional group in protein Certain functional groups in proteins can react to produce characteristically colored products. The color intensity of the product formed by
Qualitative chemical reaction of functional group in protein Certain functional groups in proteins can react to produce characteristically colored products. The color intensity of the product formed by
Lecture 6: Allosteric regulation of enzymes
 Chem*3560 Lecture 6: Allosteric regulation of enzymes Metabolic pathways do not run on a continuous basis, but are regulated according to need Catabolic pathways run if there is demand for ATP; for example
Chem*3560 Lecture 6: Allosteric regulation of enzymes Metabolic pathways do not run on a continuous basis, but are regulated according to need Catabolic pathways run if there is demand for ATP; for example
v o = V max [S] rate = kt[s] e V max = k cat E t ΔG = -RT lnk eq K m + [S]
![v o = V max [S] rate = kt[s] e V max = k cat E t ΔG = -RT lnk eq K m + [S] v o = V max [S] rate = kt[s] e V max = k cat E t ΔG = -RT lnk eq K m + [S]](/thumbs/81/84404059.jpg) Exam 3 Spring 2017 Dr. Stone 8:00 Name There are 100 possible points on this exam. -ΔG / RT v o = V max [S] rate = kt[s] e V max = k cat E t ΔG = -RT lnk eq K m + [S] h rate forward = k forward [reactants]
Exam 3 Spring 2017 Dr. Stone 8:00 Name There are 100 possible points on this exam. -ΔG / RT v o = V max [S] rate = kt[s] e V max = k cat E t ΔG = -RT lnk eq K m + [S] h rate forward = k forward [reactants]
Enzymes Topic 3.6 & 7.6 SPEED UP CHEMICAL REACTIONS!!!!!!!
 Enzymes Topic 3.6 & 7.6 SPEED UP CHEMICAL REACTIONS!!!!!!! Key Words Enzyme Substrate Product Active Site Catalyst Activation Energy Denature Enzyme-Substrate Complex Lock & Key model Induced fit model
Enzymes Topic 3.6 & 7.6 SPEED UP CHEMICAL REACTIONS!!!!!!! Key Words Enzyme Substrate Product Active Site Catalyst Activation Energy Denature Enzyme-Substrate Complex Lock & Key model Induced fit model
Bridging task for 2016 entry. AS/A Level Biology. Why do I need to complete a bridging task?
 Bridging task for 2016 entry AS/A Level Biology Why do I need to complete a bridging task? The task serves two purposes. Firstly, it allows you to carry out a little bit of preparation before starting
Bridging task for 2016 entry AS/A Level Biology Why do I need to complete a bridging task? The task serves two purposes. Firstly, it allows you to carry out a little bit of preparation before starting
Review of Biochemistry
 Review of Biochemistry Chemical bond Functional Groups Amino Acid Protein Structure and Function Proteins are polymers of amino acids. Each amino acids in a protein contains a amino group, - NH 2,
Review of Biochemistry Chemical bond Functional Groups Amino Acid Protein Structure and Function Proteins are polymers of amino acids. Each amino acids in a protein contains a amino group, - NH 2,
pharmaceutical formulations. Anagliptin shows absorption maximum at 246 nm and obeys beer s law in the
 ISSN: 0975-766X CODEN: IJPTFI Available Online through Research Article www.ijptonline.com DEVELOPMENT AND VALIDATION OF ANALYTICAL METHOD FOR ESTIMATION OF ANAGLIPTIN IN TABLET DOSAGE FORM BY U.V. SPECTROPHOTOMETRIC
ISSN: 0975-766X CODEN: IJPTFI Available Online through Research Article www.ijptonline.com DEVELOPMENT AND VALIDATION OF ANALYTICAL METHOD FOR ESTIMATION OF ANAGLIPTIN IN TABLET DOSAGE FORM BY U.V. SPECTROPHOTOMETRIC
Written Answers. (i) No inhibitor. (ii) Noncompetitive inhibitor. (iii) Competitive inhibitor. (iv) Mixed inhibitor
 Written Answers 1. (a) If the K M of an enzyme for its substrate remains constant as the concentration of the inhibitor increases, what can be said about the mode of inhibition? (b) The kinetic data for
Written Answers 1. (a) If the K M of an enzyme for its substrate remains constant as the concentration of the inhibitor increases, what can be said about the mode of inhibition? (b) The kinetic data for
MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question.
 Practice Quiz 1 AP Bio Sept 2016 Name MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question. 1) The element present in all organic molecules is A) hydrogen.
Practice Quiz 1 AP Bio Sept 2016 Name MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question. 1) The element present in all organic molecules is A) hydrogen.
HiPer Carbohydrates Estimation Teaching Kit (Quantitative)
 HiPer Carbohydrates Estimation Teaching Kit (Quantitative) Product Code: HTBC003 Number of experiments that can be performed: 10 Duration of Experiment Protocol DNSA Method :1 hour Phenol Sulphuric Acid
HiPer Carbohydrates Estimation Teaching Kit (Quantitative) Product Code: HTBC003 Number of experiments that can be performed: 10 Duration of Experiment Protocol DNSA Method :1 hour Phenol Sulphuric Acid
ENZYME ACTIVITY. Readings: Review pp , and in your text (POHS, 5 th ed.).
 ENZYME ACTIVITY Readings: Review pp. 51-58, and 128-139 in your text (POHS, 5 th ed.). Introduction Enzymes are biological catalysts; that is, enzymes are able to mediate the conversion of substrate into
ENZYME ACTIVITY Readings: Review pp. 51-58, and 128-139 in your text (POHS, 5 th ed.). Introduction Enzymes are biological catalysts; that is, enzymes are able to mediate the conversion of substrate into
Tala Saleh. Ahmad Attari. Mamoun Ahram
 23 Tala Saleh Ahmad Attari Minna Mushtaha Mamoun Ahram In the previous lecture, we discussed the mechanisms of regulating enzymes through inhibitors. Now, we will start this lecture by discussing regulation
23 Tala Saleh Ahmad Attari Minna Mushtaha Mamoun Ahram In the previous lecture, we discussed the mechanisms of regulating enzymes through inhibitors. Now, we will start this lecture by discussing regulation
Name. The following exam contains 44 questions, valued at 2.6 points/question. 2. Which of the following is not a principal use of proteins?
 Chemistry 131 Exam 3 Practice Proteins, Enzymes, and Carbohydrates Spring 2018 Name The following exam contains 44 questions, valued at 2.6 points/question 1. Which of the following is a protein? a. Amylase
Chemistry 131 Exam 3 Practice Proteins, Enzymes, and Carbohydrates Spring 2018 Name The following exam contains 44 questions, valued at 2.6 points/question 1. Which of the following is a protein? a. Amylase
Unit 3: Chemistry of Life Mr. Nagel Meade High School
 Unit 3: Chemistry of Life Mr. Nagel Meade High School IB Syllabus Statements 3.2.1 Distinguish between organic and inorganic compounds. 3.2.2 Identify amino acids, glucose, ribose and fatty acids from
Unit 3: Chemistry of Life Mr. Nagel Meade High School IB Syllabus Statements 3.2.1 Distinguish between organic and inorganic compounds. 3.2.2 Identify amino acids, glucose, ribose and fatty acids from
Chapter 5 Structure and Function Of Large Biomolecules
 Formation of Macromolecules Monomers Polymers Macromolecules Smaller larger Chapter 5 Structure and Function Of Large Biomolecules monomer: single unit dimer: two monomers polymer: three or more monomers
Formation of Macromolecules Monomers Polymers Macromolecules Smaller larger Chapter 5 Structure and Function Of Large Biomolecules monomer: single unit dimer: two monomers polymer: three or more monomers
GLYCOGEN BEFORE THE LAB YOU HAVE TO READ ABOUT:
 GLYCGEN BEFRE THE LAB YU HAVE T READ ABUT:. Glycogen structure. 2. Glycogen synthesis and degradation (reactions with structural formulas and enzymes). 3. The role of glycogen in liver and muscles. INTRDUCTIN
GLYCGEN BEFRE THE LAB YU HAVE T READ ABUT:. Glycogen structure. 2. Glycogen synthesis and degradation (reactions with structural formulas and enzymes). 3. The role of glycogen in liver and muscles. INTRDUCTIN
MCB 102 Discussion, Spring 2012
 MB Discussion, Spring 2012 Practice Problems 1. Effect of enzymes on reactions Which of the listed effects would be brought about by any enzyme catalyzing the following simple reaction? k 1 S P where K
MB Discussion, Spring 2012 Practice Problems 1. Effect of enzymes on reactions Which of the listed effects would be brought about by any enzyme catalyzing the following simple reaction? k 1 S P where K
Student Biochemistry I Homework III Due 10/13/04 64 points total (48 points based on text; 16 points for Swiss-PDB viewer exercise)
 Biochemistry I Homework III Due 10/13/04 64 points total (48 points based on text; 16 points for Swiss-PDB viewer exercise) 1). 20 points total T or F; if false, provide a brief rationale as to why. Only
Biochemistry I Homework III Due 10/13/04 64 points total (48 points based on text; 16 points for Swiss-PDB viewer exercise) 1). 20 points total T or F; if false, provide a brief rationale as to why. Only
Analysis of Polyphenoloxidase Enzyme Activity from Potato Extract Biochemistry Lab I (CHEM 4401)
 Analysis of Polyphenoloxidase Enzyme Activity from Potato Extract Biochemistry Lab I (CHEM 4401) Background Enzymes are protein molecules (primarily) that serve as biological catalysts. They are responsible
Analysis of Polyphenoloxidase Enzyme Activity from Potato Extract Biochemistry Lab I (CHEM 4401) Background Enzymes are protein molecules (primarily) that serve as biological catalysts. They are responsible
Development and validation of UV-visible spectrophotometric method for estimation of rifapentine in bulk and dosage form
 Available online at www.derpharmachemica.com Scholars Research Library Der Pharma Chemica, 2013, 5(2):251-255 (http://derpharmachemica.com/archive.html) ISSN 0975-413X CODEN (USA): PCHHAX Development and
Available online at www.derpharmachemica.com Scholars Research Library Der Pharma Chemica, 2013, 5(2):251-255 (http://derpharmachemica.com/archive.html) ISSN 0975-413X CODEN (USA): PCHHAX Development and
Terminology-Amino Acids
 Enzymes 1 2 Terminology-Amino Acids Primary Structure: is a polypeptide (large number of aminoacid residues bonded together in a chain) chain of amino acids linked with peptide bonds. Secondary Structure-
Enzymes 1 2 Terminology-Amino Acids Primary Structure: is a polypeptide (large number of aminoacid residues bonded together in a chain) chain of amino acids linked with peptide bonds. Secondary Structure-
Enzymes. Gibbs Free Energy of Reaction. Parameters affecting Enzyme Catalysis. Enzyme Commission Number
 SCBC203 Enzymes Jirundon Yuvaniyama, Ph.D. Department of Biochemistry Faculty of Science Mahidol University Gibbs Free Energy of Reaction Free Energy A B + H 2 O A OH + B H Activation Energy Amount of
SCBC203 Enzymes Jirundon Yuvaniyama, Ph.D. Department of Biochemistry Faculty of Science Mahidol University Gibbs Free Energy of Reaction Free Energy A B + H 2 O A OH + B H Activation Energy Amount of
Dr. Nafeth Abu-Tarbou sh Introduction to Biochemist ry 15/08/2014 Sec 1,2, 3 Sheet #21 P a g e 1 Written by Baha Aldeen Alshraideh
 P a g e 1 Enzyme Kinetics Vmax: The Maximal rate - The rate of reaction when the enzyme is saturated with substrate. -You can calculate it by the following equation: Vmax = k2 [E] T [E]T :Total enzyme
P a g e 1 Enzyme Kinetics Vmax: The Maximal rate - The rate of reaction when the enzyme is saturated with substrate. -You can calculate it by the following equation: Vmax = k2 [E] T [E]T :Total enzyme
Name: Student Number
 UNIVERSITY OF GUELPH CHEM 454 ENZYMOLOGY Winter 2003 Quiz #1: February 13, 2003, 11:30 13:00 Instructor: Prof R. Merrill Instructions: Time allowed = 80 minutes. Total marks = 34. This quiz represents
UNIVERSITY OF GUELPH CHEM 454 ENZYMOLOGY Winter 2003 Quiz #1: February 13, 2003, 11:30 13:00 Instructor: Prof R. Merrill Instructions: Time allowed = 80 minutes. Total marks = 34. This quiz represents
LECTURE 4: REACTION MECHANISM & INHIBITORS
 LECTURE 4: REACTION MECHANISM & INHIBITORS Chymotrypsin 1 LECTURE OUTCOMES After mastering the present lecture materials, students will be able to 1. to explain reaction mechanisms of between enzyme and
LECTURE 4: REACTION MECHANISM & INHIBITORS Chymotrypsin 1 LECTURE OUTCOMES After mastering the present lecture materials, students will be able to 1. to explain reaction mechanisms of between enzyme and
EXPERIMENT 3 ENZYMATIC QUANTITATION OF GLUCOSE
 EXPERIMENT 3 ENZYMATIC QUANTITATION OF GLUCOSE This is a team experiment. Each team will prepare one set of reagents; each person will do an individual unknown and each team will submit a single report.
EXPERIMENT 3 ENZYMATIC QUANTITATION OF GLUCOSE This is a team experiment. Each team will prepare one set of reagents; each person will do an individual unknown and each team will submit a single report.
Chapter 6. Metabolism & Enzymes. AP Biology
 Chapter 6. Metabolism & Enzymes Flow of energy through life Life is built on chemical reactions Chemical reactions of life Metabolism forming bonds between molecules dehydration synthesis anabolic reactions
Chapter 6. Metabolism & Enzymes Flow of energy through life Life is built on chemical reactions Chemical reactions of life Metabolism forming bonds between molecules dehydration synthesis anabolic reactions
Examples. Chapter 8. Metabolism & Enzymes. Flow of energy through life. Examples. Chemical reactions of life. Chemical reactions & energy
 WH Examples dehydration synthesis Chapter 8 Metabolism & Enzymes + H 2 O hydrolysis + H 2 O Flow of energy through life Life is built on chemical reactions Examples dehydration synthesis hydrolysis 2005-2006
WH Examples dehydration synthesis Chapter 8 Metabolism & Enzymes + H 2 O hydrolysis + H 2 O Flow of energy through life Life is built on chemical reactions Examples dehydration synthesis hydrolysis 2005-2006
Research Article Derivative Spectrophotometric Method for Estimation of Metformin Hydrochloride in Bulk Drug and Dosage Form
 Research Article Derivative Spectrophotometric Method for Estimation of Metformin Hydrochloride in Bulk Drug and Dosage Form Gowekar NM, Lawande YS*, Jadhav DP, Hase RS and Savita N. Gowekar Department
Research Article Derivative Spectrophotometric Method for Estimation of Metformin Hydrochloride in Bulk Drug and Dosage Form Gowekar NM, Lawande YS*, Jadhav DP, Hase RS and Savita N. Gowekar Department
Figure 1 Original Advantages of biological reactions being catalyzed by enzymes:
 Enzyme basic concepts, Enzyme Regulation I III Carmen Sato Bigbee, Ph.D. Objectives: 1) To understand the bases of enzyme catalysis and the mechanisms of enzyme regulation. 2) To understand the role of
Enzyme basic concepts, Enzyme Regulation I III Carmen Sato Bigbee, Ph.D. Objectives: 1) To understand the bases of enzyme catalysis and the mechanisms of enzyme regulation. 2) To understand the role of
CLASS SET. Modeling Life s Important Compounds. AP Biology
 Modeling Life s Important Compounds AP Biology CLASS SET OBJECTIVES: Upon completion of this activity, you will be able to: Explain the connection between the sequence and the subcomponents of a biological
Modeling Life s Important Compounds AP Biology CLASS SET OBJECTIVES: Upon completion of this activity, you will be able to: Explain the connection between the sequence and the subcomponents of a biological
Fall 2005: CH395G - Exam 2 - Multiple Choice (2 pts each)
 Fall 2005: CH395G - Exam 2 - Multiple Choice (2 pts each) These constants may be helpful in some of your calculations: Avogadro s number = 6.02 x 10 23 molecules/mole; Gas constant (R) = 8.3145 x 10-3
Fall 2005: CH395G - Exam 2 - Multiple Choice (2 pts each) These constants may be helpful in some of your calculations: Avogadro s number = 6.02 x 10 23 molecules/mole; Gas constant (R) = 8.3145 x 10-3
Yeast Extracts containing Mannoproteins (Tentative)
 0 out of 6 Residue Monograph prepared by the meeting of the Joint FAO/WHO Expert Committee on Food Additives (JECFA), 84th meeting 2017 Yeast Extracts containing Mannoproteins (Tentative) This monograph
0 out of 6 Residue Monograph prepared by the meeting of the Joint FAO/WHO Expert Committee on Food Additives (JECFA), 84th meeting 2017 Yeast Extracts containing Mannoproteins (Tentative) This monograph
THE MALATE DEHYDROGENASE LABORATORIES
 THE MALATE DEHYDROGENASE LABORATORIES Laboratory Page Overview of the Enzyme Kinetics Block of Laboratories 1 Introduction to the Study of Enzyme Kinetics and Enzyme Mechanisms 2 Review of the Roles of
THE MALATE DEHYDROGENASE LABORATORIES Laboratory Page Overview of the Enzyme Kinetics Block of Laboratories 1 Introduction to the Study of Enzyme Kinetics and Enzyme Mechanisms 2 Review of the Roles of
2.1. thebiotutor. Unit F212: Molecules, Biodiversity, Food and Health. 1.1 Biological molecules. Answers
 thebiotutor Unit F212: Molecules, Biodiversity, Food and Health 1.1 Biological molecules Answers 1 1. δ + H hydrogen bond δ + H O δ - O δ - H H δ + δ+ 1 hydrogen bond represented as, horizontal / vertical,
thebiotutor Unit F212: Molecules, Biodiversity, Food and Health 1.1 Biological molecules Answers 1 1. δ + H hydrogen bond δ + H O δ - O δ - H H δ + δ+ 1 hydrogen bond represented as, horizontal / vertical,
Question Expected Answers Mark Additional Guidance 1 (a) (i) peptide (bond / link) ; 1 DO NOT CREDIT dipeptide 1 (a) (ii) hydrolysis ;
 Question Expected Answers Mark Additional Guidance 1 (a) (i) peptide (bond / link) ; 1 DO NOT CREDIT dipeptide 1 (a) (ii) hydrolysis ; IGNORE name of bond 1 (b) 1 water / H O, is, added / used / needed
Question Expected Answers Mark Additional Guidance 1 (a) (i) peptide (bond / link) ; 1 DO NOT CREDIT dipeptide 1 (a) (ii) hydrolysis ; IGNORE name of bond 1 (b) 1 water / H O, is, added / used / needed
Enzymes. Enzymes : are protein catalysts that increase the rate of reactions without being changed in the overall process.
 Enzymes Enzymes Enzymes : are protein catalysts that increase the rate of reactions without being changed in the overall process. All reactions in the body are mediated by enzymes A + B E C A, B: substrate
Enzymes Enzymes Enzymes : are protein catalysts that increase the rate of reactions without being changed in the overall process. All reactions in the body are mediated by enzymes A + B E C A, B: substrate
Exercise 3. A Study of Enzyme Specificity. A Kinetic Analysis of Glucose Oxidase
 R e p r i n t e d f r o m G a l l i k S., C e l l B i o l o g y O L M P a g e 1 Exercise 3. A Study of Enzyme Specificity. A Kinetic Analysis of Glucose Oxidase A. Introduction Enzymes are one of the largest
R e p r i n t e d f r o m G a l l i k S., C e l l B i o l o g y O L M P a g e 1 Exercise 3. A Study of Enzyme Specificity. A Kinetic Analysis of Glucose Oxidase A. Introduction Enzymes are one of the largest
HEMICELLULASE from ASPERGILLUS NIGER, var.
 HEMICELLULASE from ASPERGILLUS NIGER, var. Prepared at the 55th JECFA (2000) and published in FNP 52 Add 8 (2000), superseding tentative specifications prepared at the 31st JECFA (1987) and published in
HEMICELLULASE from ASPERGILLUS NIGER, var. Prepared at the 55th JECFA (2000) and published in FNP 52 Add 8 (2000), superseding tentative specifications prepared at the 31st JECFA (1987) and published in
CELLULASE from PENICILLIUM FUNICULOSUM
 CELLULASE from PENICILLIUM FUNICULOSUM Prepared at the 55th JECFA (2000) and published in FNP 52 Add 8 (2000), superseding tentative specifications prepared at the 31st JECFA (1987) and published in FNP
CELLULASE from PENICILLIUM FUNICULOSUM Prepared at the 55th JECFA (2000) and published in FNP 52 Add 8 (2000), superseding tentative specifications prepared at the 31st JECFA (1987) and published in FNP
BACKGROUND INFORMATION:
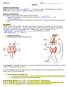 BIOLOGY 12 ENZYMES NAME: BACKGROUND INFORMATION: Energy: is defined as the ability to do or bring about change. A living organism must constantly perform work in order to maintain its organization, to
BIOLOGY 12 ENZYMES NAME: BACKGROUND INFORMATION: Energy: is defined as the ability to do or bring about change. A living organism must constantly perform work in order to maintain its organization, to
What Is the Relationship Between the Amount of Transmitted Light Through a Solution and Its Concentration?
 What Is the Relationship Between the Amount of Transmitted Light Through a and Its Concentration? Blue Food Dye Treats Spine Injury in Rats HTTP://WWW.WIRED.COM/WIREDSCIENCE/2009/07/BLUERATS/ BY HADLEY
What Is the Relationship Between the Amount of Transmitted Light Through a and Its Concentration? Blue Food Dye Treats Spine Injury in Rats HTTP://WWW.WIRED.COM/WIREDSCIENCE/2009/07/BLUERATS/ BY HADLEY
Student Manual. Background STUDENT MANUAL BACKGROUND. Enzymes
 Background Enzymes Enzymes are typically proteins (some nucleic acids have also been found to be enzymes) that act as catalysts, speeding up chemical reactions that would take far too long to occur on
Background Enzymes Enzymes are typically proteins (some nucleic acids have also been found to be enzymes) that act as catalysts, speeding up chemical reactions that would take far too long to occur on
The effect of incubation time on the rate of an enzyme catalyzed reaction
 The effect of incubation time on the rate of an enzyme catalyzed reaction Objectives To monitor the progress of an enzyme catalyzed reaction (Acid phosphatase). To determine the initial rate of the reaction
The effect of incubation time on the rate of an enzyme catalyzed reaction Objectives To monitor the progress of an enzyme catalyzed reaction (Acid phosphatase). To determine the initial rate of the reaction
DNA and Protein Synthesis Practice
 Biology 12 DNA and Protein Synthesis Practice Name: 1. DNA is often called the "code of life". Actually it contains the code for a) the sequence of amino acids in a protein b) the sequence of base pairs
Biology 12 DNA and Protein Synthesis Practice Name: 1. DNA is often called the "code of life". Actually it contains the code for a) the sequence of amino acids in a protein b) the sequence of base pairs
Chapter 4. Further Enzyme Studies
 Chapter 4 Further Enzyme Studies 4.1. Introduction Enzymes are a group of proteins possessing catalytic ability. The mechanism of an enzyme-catalysed reaction can be expressed as below: Figure 4-1 Single-substrate
Chapter 4 Further Enzyme Studies 4.1. Introduction Enzymes are a group of proteins possessing catalytic ability. The mechanism of an enzyme-catalysed reaction can be expressed as below: Figure 4-1 Single-substrate
Determining the Concentration of Iron in Vitamin Supplements
 Teacher Guide Determining the Concentration of Iron in Vitamin Supplements Background Information Colorimetry is the science of measuring color. Colorimetry is a useful technique for determining the concentration
Teacher Guide Determining the Concentration of Iron in Vitamin Supplements Background Information Colorimetry is the science of measuring color. Colorimetry is a useful technique for determining the concentration
USING CENTRAL COMPOSITE DESIGNS - RESPONSE SURFACE METHODOLOGY TO OPTIMIZE INVERATSE ACTIVITY CONDITIONS FOR FRUCTOSE PRODUCTION
 Conference proceedings of Biotechnology for Green Solutions and Sustainable Environment: 8-87, 010 USING CENTRAL COMPOSITE DESIGNS - RESPONSE SURFACE METHODOLOGY TO OPTIMIZE INVERATSE ACTIVITY CONDITIONS
Conference proceedings of Biotechnology for Green Solutions and Sustainable Environment: 8-87, 010 USING CENTRAL COMPOSITE DESIGNS - RESPONSE SURFACE METHODOLOGY TO OPTIMIZE INVERATSE ACTIVITY CONDITIONS
BIOL 347L Laboratory Three
 Introduction BIOL 347L Laboratory Three Osmosis in potato and carrot samples Osmosis is the movement of water molecules through a selectively permeable membrane into a region of higher solute concentration,
Introduction BIOL 347L Laboratory Three Osmosis in potato and carrot samples Osmosis is the movement of water molecules through a selectively permeable membrane into a region of higher solute concentration,
Kit for assay of thioredoxin
 FkTRX-02-V2 Kit for assay of thioredoxin The thioredoxin system is the major protein disulfide reductase in cells and comprises thioredoxin, thioredoxin reductase and NADPH (1). Thioredoxin systems are
FkTRX-02-V2 Kit for assay of thioredoxin The thioredoxin system is the major protein disulfide reductase in cells and comprises thioredoxin, thioredoxin reductase and NADPH (1). Thioredoxin systems are
24. What is the half-life of a compound (reactant) and does it depend on the concentration of this compound (reactant) in a first order process? 25. W
 Water 1. Why is water so different compared to methane although they have nearly the same molecular weight? 2. What are the main differences between water and methane which has nearly the same molecular
Water 1. Why is water so different compared to methane although they have nearly the same molecular weight? 2. What are the main differences between water and methane which has nearly the same molecular
PhosFree TM Phosphate Assay Biochem Kit
 PhosFree TM Phosphate Assay Biochem Kit (Cat. # BK050) ORDERING INFORMATION To order by phone: (303) - 322-2254 To order by Fax: (303) - 322-2257 To order by e-mail: cservice@cytoskeleton.com Technical
PhosFree TM Phosphate Assay Biochem Kit (Cat. # BK050) ORDERING INFORMATION To order by phone: (303) - 322-2254 To order by Fax: (303) - 322-2257 To order by e-mail: cservice@cytoskeleton.com Technical
DEVELOPMENT AND VALIDATION OF COLORIMETRIC METHODS FOR THE DETERMINATION OF RITONAVIR IN TABLETS
 Int. J. Chem. Sci.: 8(1), 2010, 711-715 DEVELOPMENT AND VALIDATION OF COLORIMETRIC METHODS FOR THE DETERMINATION OF RITONAVIR IN TABLETS R. PRIYADARSINI *, V. NIRAIMATHI, T. SARASWATHY, V. GOPI and R.
Int. J. Chem. Sci.: 8(1), 2010, 711-715 DEVELOPMENT AND VALIDATION OF COLORIMETRIC METHODS FOR THE DETERMINATION OF RITONAVIR IN TABLETS R. PRIYADARSINI *, V. NIRAIMATHI, T. SARASWATHY, V. GOPI and R.
Name: Period: Date: Testing for Biological Macromolecules Lab
 Testing for Biological Macromolecules Lab Introduction: All living organisms are composed of various types of organic molecules, such as carbohydrates, starches, proteins, lipids and nucleic acids. These
Testing for Biological Macromolecules Lab Introduction: All living organisms are composed of various types of organic molecules, such as carbohydrates, starches, proteins, lipids and nucleic acids. These
Review of Energetics Intro
 Review of Energetics Intro Learning Check The First Law of Thermodynamics states that energy can be Created Destroyed Converted All of the above Learning Check The second law of thermodynamics essentially
Review of Energetics Intro Learning Check The First Law of Thermodynamics states that energy can be Created Destroyed Converted All of the above Learning Check The second law of thermodynamics essentially
Will s Pre-Test. (4) A collection of cells that work together to perform a function is termed a(n): a) Organelle b) Organ c) Cell d) Tissue e) Prison
 Will s Pre-Test This is a representative of Exam I that you will take Tuesday September 18, 2007. The actual exam will be 50 multiple choice questions. (1) The basic structural and functional unit of the
Will s Pre-Test This is a representative of Exam I that you will take Tuesday September 18, 2007. The actual exam will be 50 multiple choice questions. (1) The basic structural and functional unit of the
AP Biology Protein Structure and Enzymes
 AP Biology Protein Structure and Enzymes Connection to the Nitrogen-cycle Amino acids (protein) Nucleic acids (RNA and DNA) ATP 78% 1. Assimilation of nitrate by photosynthetic eukaryotes 2. Nitrogen fixation
AP Biology Protein Structure and Enzymes Connection to the Nitrogen-cycle Amino acids (protein) Nucleic acids (RNA and DNA) ATP 78% 1. Assimilation of nitrate by photosynthetic eukaryotes 2. Nitrogen fixation
Enzyme Activity Lecture. Every reaction has energy requirement. The minimum amount of energy required is termed activation energy.
 Enzyme Activity Lecture Every reaction has energy requirement. The minimum amount of energy required is termed activation energy. Living organisms have optimum temperature requirement so elevating the
Enzyme Activity Lecture Every reaction has energy requirement. The minimum amount of energy required is termed activation energy. Living organisms have optimum temperature requirement so elevating the
BIOLOGICAL MOLECULES REVIEW-UNIT 1 1. The factor being tested in an experiment is the A. data. B. variable. C. conclusion. D. observation. 2.
 BIOLOGICAL MOLECULES REVIEW-UNIT 1 1. The factor being tested in an experiment is the A. data. B. variable. C. conclusion. D. observation. 2. A possible explanation for an event that occurs in nature is
BIOLOGICAL MOLECULES REVIEW-UNIT 1 1. The factor being tested in an experiment is the A. data. B. variable. C. conclusion. D. observation. 2. A possible explanation for an event that occurs in nature is
Supplementary methods 1: Parameter estimation
 Supplementary methods : Parameter estimation A number of estimates of the parameters in model () of the main text can be obtained from the literature. However these estimates are not necessarily obtained
Supplementary methods : Parameter estimation A number of estimates of the parameters in model () of the main text can be obtained from the literature. However these estimates are not necessarily obtained
CHM 341 C: Biochemistry I. Test 2: October 24, 2014
 CHM 341 C: Biochemistry I Test 2: ctober 24, 2014 This test consists of 14 questions worth points. Make sure that you read the entire question and answer each question clearly and completely. To receive
CHM 341 C: Biochemistry I Test 2: ctober 24, 2014 This test consists of 14 questions worth points. Make sure that you read the entire question and answer each question clearly and completely. To receive
Chapter 6. Flow of energy through life. Chemical reactions of life. Examples. Examples. Chemical reactions & energy 9/7/2012. Enzymes & Metabolism
 Flow of energy through life Chapter 6 Life is built on chemical reactions Enzymes & Metabolism Chemical reactions of life Examples Metabolism Forming bonds between molecules Dehydration synthesis Anabolic
Flow of energy through life Chapter 6 Life is built on chemical reactions Enzymes & Metabolism Chemical reactions of life Examples Metabolism Forming bonds between molecules Dehydration synthesis Anabolic
Lecture 12 Enzymes: Inhibition
 Lecture 12 Enzymes: Inhibition Reading: Berg, Tymoczko & Stryer, 6th ed., Chapter 8, pp. 225-236 Problems: pp. 238-239, chapter 8, #1, 2, 4a,b, 5a,b, 7, 10 Jmol structure: cyclooxygenase/non-steroidal
Lecture 12 Enzymes: Inhibition Reading: Berg, Tymoczko & Stryer, 6th ed., Chapter 8, pp. 225-236 Problems: pp. 238-239, chapter 8, #1, 2, 4a,b, 5a,b, 7, 10 Jmol structure: cyclooxygenase/non-steroidal
Enzymes. Enzyme. Aim: understanding the basic concepts of enzyme catalysis and enzyme kinetics
 Enzymes Substrate Enzyme Product Aim: understanding the basic concepts of enzyme catalysis and enzyme kinetics Enzymes are efficient Enzyme Reaction Uncatalysed (k uncat s -1 ) Catalysed (k cat s -1 )
Enzymes Substrate Enzyme Product Aim: understanding the basic concepts of enzyme catalysis and enzyme kinetics Enzymes are efficient Enzyme Reaction Uncatalysed (k uncat s -1 ) Catalysed (k cat s -1 )
BASIC ENZYMOLOGY 1.1
 BASIC ENZYMOLOGY 1.1 1.2 BASIC ENZYMOLOGY INTRODUCTION Enzymes are synthesized by all living organisms including man. These life essential substances accelerate the numerous metabolic reactions upon which
BASIC ENZYMOLOGY 1.1 1.2 BASIC ENZYMOLOGY INTRODUCTION Enzymes are synthesized by all living organisms including man. These life essential substances accelerate the numerous metabolic reactions upon which
Extracellular Enzymes Lab
 Biochemistry Extracellular Enzymes Lab All organisms convert small organic compounds, such as glucose, into monomers required for the production of macromolecules; e.g., Building Blocks Monomers Macromolecules
Biochemistry Extracellular Enzymes Lab All organisms convert small organic compounds, such as glucose, into monomers required for the production of macromolecules; e.g., Building Blocks Monomers Macromolecules
HPLC Analysis of Sugars
 HPLC Analysis of Sugars Pre-Lab Exercise: 1) Read about HPLC, sugars and the experiment and its background. 2) Prepare a flowchart as appropriate for the lab exercise. 3) Note the various sugar concentrations
HPLC Analysis of Sugars Pre-Lab Exercise: 1) Read about HPLC, sugars and the experiment and its background. 2) Prepare a flowchart as appropriate for the lab exercise. 3) Note the various sugar concentrations
Nonlinear Pharmacokinetics
 Nonlinear Pharmacokinetics Non linear pharmacokinetics: In some cases, the kinetics of a pharmacokinetic process change from predominantly first order to predominantly zero order with increasing dose or
Nonlinear Pharmacokinetics Non linear pharmacokinetics: In some cases, the kinetics of a pharmacokinetic process change from predominantly first order to predominantly zero order with increasing dose or
