Supporting Information. Controlled Nucleation and Growth of DNA Tile Arrays within Prescribed DNA Origami Frames and Their Dynamics
|
|
|
- Marvin Daniels
- 5 years ago
- Views:
Transcription
1 Supporting Information Controlled Nucleation and Growth of DNA Tile Arrays within Prescribed DNA Origami Frames and Their Dynamics Wei Li, Yang Yang, Shuoxing Jiang, Hao Yan, Yan Liu Department of Chemistry and Biochemistry and The Biodesign Institute Arizona State University, Tempe, AZ S1
2 Experimental Materials and Methods Materials: All DNA helper strands used in the origami frame were purchased in 96-well plates from Integrated DNA Technologies, Inc. ( desalted, with concentrations normalized to 200 µm. Single stranded M13mp18 viral DNA and phi X 174 DNA were purchased from New England Biolabs, Inc. (NEB, catalog number: N4040S and N3023S). All DNA strands in the DNA origami frame were used without further purification. All DNA strands used in the DX tiles were purchased from Integrated DNA Technologies, Inc. ( in the format of desalted dry powder. The tile strands were all purified using denaturing polyacrylamide gel electrophoresis (10% 19:1 acrylamide/bisacrylamide, containing 50% urea) in 1 TBE buffer (ph 8.0, 89 mm tris base, 89 mm boric acid, 2 mm EDTA). The bands corresponding to the full length strands were individually excised from the gel, chopped into small pieces, soaked in 500 µl elution buffer (500 mm NH 4 OAc, 10 mm Mg(OAc) 2, and 2 mm EDTA) and then shaken overnight to allow the DNA strands to elute from the gel blocks into the solution. After filtering out the gel blocks, the solutions were then mixed with butanol to extract any organic residue. After removing the butanol layer, 1 ml of ethanol was mixed with each solution to precipitate the DNA molecules. The mixtures were kept at -20 o C to ensure rapid and complete DNA precipitation. Then the purified DNA strands were spun down using a centrifuge, and then dried under vacuum. The DNA strands were then reconstituted in pure water and their concentrations were measured by absorbance at 260 nm. Assembly Procedure: The DNA origami frame structure was assembled by mixing M13mp18 DNA (10 nm) and phi X 174 DNA (10 nm) with the helper strands in a 1:1:30 molar ratio in 1 TAE/Mg 2+ buffer (ph 8.0, 20 mm Tris base, 20 mm acetic acid, 2 mm EDTA, 12.5 mm Mg(OAc) 2 ). The final volume of the reaction was 100 µl. The solution was annealed in a PCR thermocycler with the temperature decreased from 90 o C to 70 o C at a rate of 1 o C every 5 minutes, from 70 o C to 40 o C at a rate of 1 o C every 15 minutes, then from 40 o C to 25 o C at a rate of 1 o C every 10 minutes, and finally kept at 4 o C. Following annealing, the origami frame was washed with 1 TAE/Mg 2+ buffer three times and passed through a 100 kd MWCO Microcon centrifugal filter device (Amicon, catalog number: UFC510096) to remove the excess helper strands. Each DNA DX tile was assembled by mixing all the strands in the tile in an equal molar ratio (1 mm) in 100 µl 1 TAE/Mg 2+ buffer. The solution was annealed in a PCR thermocycler with the temperature decreased from 90 o C to 25 o C at a rate of 4 o C every 5 minutes, and then kept at 25 o C. The DNA origami frame DX tile 2D array hybrid was assembled by mixing 1 pmol of purified DNA origami frame (100 µl, 10 nm) with the solutions of the four DX tiles. The amount of each tile was 100 pmol (100 µl, 1 mm). The final 500 µl solution was incubated at 25 o C S2
3 overnight. Then the mixture was concentrated to 100 µl using a 100 kd MWCO Amicon centrifugal filter device. Agarose Gel Electrophoresis Purification: The assembled frame-array hybrid was loaded onto an agarose gel (0.3% agarose containing 0.5 µg/ml ethidium bromide, 1 TAE/Mg 2+ buffer) and subjected to gel electrophoresis at 80 volts for one hour on an ice-water bath. The product band was excised from the gel and shredded. The shredded gel blocks were transferred into a Freeze 'N Squeeze DNA Gel Extraction Spin Column (Bio-Rad, catalog number: ) and centrifuged to recover the buffer containing the purified product. The product was then stored at 4 o C and characterized by AFM. Monomeric Avidin Resin Purification: 100 µl Monomeric Avidin Resin (Thermo Scientific, catalog number: 53146) suspension was transferred into a SigmaPrep TM spin column (Sigma, catalog number: SC1000). The resin was washed with 1 PBS buffer once (Sigma, catalog number: P4417), then washed with 2 mm biotin solution to block the non-reversible binding sites, and finally regenerated with glycine solution. The resin and biotin modified DNA origami frame 2D array hybrid were mixed and incubated for 30 minutes. The resin bound with the frame-array hybrid was then washed with 1 PBS buffer to remove the free 2D array and DX tiles. The purified frame-array hybrid was then displaced from the resin with 100 µl biotin (2 mm) solution. The solution containing the purified product was then stored at 4 o C and subjected to AFM characterization. AFM Imaging: The AFM imaging was performed using a Dimension FastScan AFM (Bruker). The samples (2 µl to 5 µl) were deposited onto freshly cleaved mica (Ted Pella, Inc.) and left to adsorb for 2 min. Buffer (1 TAE/Mg 2+, 100 µl) was added on top of the sample and the sample was imaged in ScanAsyst in Fluid mode, using ScanAssyst Fluid+ probes (Bruker). Fluorescence Kinetics: The fluorescence kinetics experiments were performed using a Nanolog fluorometer (Horiba Jobin Yvon). The origami frame was purified with 100 kd MWCO Microcon centrifugal filter devices (Amicon, catalog number: UFC510096) to remove excess helper strands. The concentration of the origami stock solution was 10 nm. The concentration of each tile stock solution was 1 μm. The sample chamber of the fluorometer was preset at 21 o C. 2.4 μl of Tile C solution (labeled with Fluorescein), and 2.4 μl of Tile D solution were added to a 120 μl quartz fluorescence cuvette. 1 TAE/Mg 2+ buffer was added to make the final volume 120 μl. To the reaction with tile/origami at a molar ratio of 100:1, 2.4 μl the purified origami solution was added. To the reaction with tile/origami at a molar ratio of 100:2 or 100:3, the volume of the origami stock solution added was doubled or tripled. The sample was placed in the fluorometer and the time dependence of the intensity was monitored. Then 2.4 μl of Tile A solution (labeled with a black quencher) and 2.4 μl of Tile B solution were added to the cuvette and mixed well. The fluorescence intensity was measured once every 30 seconds, with an integration time of 10 seconds. The fluorescence intensities were first corrected for the volume difference, to a total volume of μl after the addition of Tile A and B and then the data S3
4 were corrected for photo bleaching using a control with the same concentration of Tile C and Tile A. Fluorescence Data: For each reaction, the first trace is the original data collected by the fluorometer. The second trace is the data after correcting for the volume change. The third trace is the data after correcting for photo bleaching. The fourth trace is the data after normalization, which was used to generate the plots shown in Figure 4C and Figure S11B. Design of the DX Tiles Figure S1. The design of the four DX tiles. (A) Schematic design of the four tiles. The four tiles share the same sequences of Strands 2, 3, and 5. Each tile has a specific Strand 1 and 4. The sticky end pairing e.g. a, a are marked for each tile. (B) The detailed design of the four tiles. Each tile is four helical turns long. Strand 3 is 42 nts long. Strands 2 and 5 are both 37 nts long. Strands 1 and 4 are both 26 nts long. S4
5 PAGE Characterization of DX Tiles Figure S2. Native polyacrylamide gel electrophoresis characterization of the formation of the four tiles. Lanes 1 & 15: 10 bp DNA marker. Lane 2: the core structure of the four tiles: Strand A2 + Strand A3 + Strand A5. (For Tile B, C, and D, the core structures all have the same sequences as Tile A). Lane 3: core + Strand A1. Lane 4: core + Strand A4. Lane 5: full Tile A (core + Strand A1 + Strand A4). Lane 6-8: the same combinations as Lanes 3-5 for Tile B. Lane 9-11: the same combinations as Lanes 3-5 for Tile C. Lane 12-14: the same combinations as Lanes 3-5 for Tile D. S5
6 Design of the DNA Origami Frame Figure S3. Detailed design of the DNA origami frame. The origami frame is 210 nm wide, 60 nm and 95 nm tall (the two sides). The blue strand represents the phi X 174 scaffold and the red strand corresponds to the M13mp18 scaffold. The interior is decorated with sticky ends complementary to the sticky ends on Tiles A and B. At the outer ends of each helix, two extra thymine bases are added to prevent π-π stacking between origami. S6
7 AFM Image of Empty Origami Frame Figure S4. AFM image of the empty origami frame. (A) Zoom-out AFM image of the empty origami frame. Most of the origami frames are well formed. There are several aggregated structures in the image that may be caused by crosslinking of multiple scaffold strands. (B) Zoom-in AFM image of selected well-formed empty origami frame. The scale bar is 100 nm. S7
8 Examination of the spontaneous formation of the DX tile arrays Figure S5. Unregulated growth of 2D arrays of DX tiles. The four DX tiles were mixed together to a final concentration of 250 nm each. The mixture was incubated at 25 o C overnight and characterized by AFM. The four tiles form 2D arrays as designed. S8
9 Agarose Gel Image of the Purification of the DNA Origami Frame 2D Array Hybrid Figure S6. Image of agarose gel electrophoresis showing the purification of the origami-2d array hybrid. Lane 1: 1kb DNA ladder. Lane 2: Empty origami frame without purification. The fastest intense band corresponds to the extra helper strands. The second fastest band corresponds to the empty origami frame. Upper faint bands are aggregated structures (see Figure S4). Lane 3: Origami frame and the four tiles incubated overnight at r.t. The faster band and the smear after it correspond to uncontrolled 2D tile-array of various sizes. The slower band corresponds to the origami-array hybrid, which runs faster than the empty origami frame in Lane 2, because once the frame is fully filled, the structure gets more solid. Lane 4: The four tiles incubated overnight at r.t. without the origami frame. The band and smear correspond to uncontrolled 2D tile-array of various sizes. S9
10 AFM Image of DNA Origami Frame 2D Array Hybrid Purified by Agarose Gel Electrophoresis Figure S7. AFM image of Frame-array hybrid purified by agarose gel electrophoresis. (A) Zoom-out AFM image of Frame-array hybrid purified by agarose gel electrophoresis. There were quite a few pieces of free 2D array of DX tiles that were not cleanly removed. Note that these 2D arrays had similar sizes as the frame-array hybrid, which mostly showed a filled interior. (B) Zoom-in AFM image of selected Frame-array hybrid purified by agarose gel electrophoresis. The scale bar is 100 nm. S10
11 Boitin Modified DNA Origami Frame 2D Array Hybrid Purified with Monomeric Avidin Resin Figure S8. AFM images of Boitin modified frame-array hybrid after purification with monomeric avidin resin. The origami frame was modified with biotin. When purifying with monomeric avidin resin, unmodified tiles and 2D arrays were washed away while the boitin modified frame-array hybrids were bound to the resin. The purified product was then washed off with excess biotin solution. (A) & (B) The AFM images show that using this purification method, fewer free 2D array residues remained. (C) Zoom-in AFM image of selected Frame-array hybrid purified with monomeric avidin resin. The scale bar is 100 nm. S11
12 DNA Origami Frame 2D Array Hybrid Before Purification Figure S9. AFM image of unpurified frame-array hybrid. Several, but not all of, distinguishable frame-array hybrid structures are marked in the image. S12
13 Defects of DNA Origami Frame 2D Array Hybrid Figure S10. Three major classes of defects in the frame-array hybrids. (A) The shrunken framearray hybrid caused by sticky ends on tiles hybridizing with another row of non-neighboring tiles. (B) The widened frame-array hybrid caused by inserting one or two rows of tiles between neighboring rows. (C) The bent frame-array hybrid caused by association of sticky ends between non-neighboring columns of tiles. Each image in the figure is 610 nm 610 nm. S13
14 Dynamics of the Nucleation of DX Tiles in the Origami Frame Figure S11. FS-AFM images showing the dynamics of nucleation and growth of DX tiles into the DNA origami frame. (A) This is another example of the experiment shown in Figure 3. Each frame was collected over 87 seconds. Each frame is 287 nm 287 nm. (B) The full set of images in Figure 3. Each frame was collected over 87 seconds. The scale bar is 100 nm. S14
15 Kinetics of the Nucleation Process of the Four Tiles Figure S12. Characterization of the kinetics of the nucleation process. (A) The modification of the tiles with a fluorophore and dark quencher. The 5 end of Strand A1 was modified with an Iowa Black Dark Quencher. The 3 end of Strand C2 was modified with 6-FAM. Upon sticky end association in the tile array formation, the fluorophore and the quencher are brought into close proximity and fluorescence quenching is expected. (B) Normalized fluorescence decrease. The concentration of each of the tiles was 20 nm in all experiments. The legend indicates the molar ratio between the tiles and the origami frame. Each experiment was conducted in duplicate, the data of which coincided with each other. All curves shown are after correction for photo-bleaching. (C) Logarithm of the data in Panel B to the base e. The average of the curves of the reactions without origami seed in Panel B are subtracted from all other curves. Then ln(i/i ini ) is plotted against time. The data are then fit by Equation 5 in the main text. S15
16 DNA Sequences Sequences of tile strands: A1: AGGAACCATGAACCCTGCAGCATGTC A2: GCTGCAGGCGGAATCCGACCCTGTGGCGTTGCACCAT A3: GTCGGATTCCGCTGGCTTGCCTAGAGTCACCAACGCCACAGG A4: ACTCAATGGTGCACTAAACCTCTAAG A5: AGGTTTAGTGGTGACTCTAGGCAAGCCAGGTTCATGG B1: GTGATCCATGAACCCTGCAGCAGAAC B2=A2 B3=A3 B4: TAACGATGGTGCACTAAACCTAAGCT B5=A5 C1: TGAGTCCATGAACCCTGCAGCAGCTT C2=A2 C3=A3 C4: TTCCTATGGTGCACTAAACCTGTTCT C5=A5 D1: CGTTACCATGAACCCTGCAGCCTTAG D2=A2 D3=A3 D4: ATCACATGGTGCACTAAACCTGACAT D5=A5 S16
17 Sequences of the helper strands and sticky end strands in the DNA origami frame: Helper 1 GTATTAACTCACTTGCCTGAGTAGACCGTTGTAGCAATACTTCTTTGATTTT Helper 2 AGAGTCTGTCCATCACGCAAATTAAAGAACTC Helper 3 CAGCAGAAGGCCTTGCTGGTAATACGAGTAAA Helper 4 AAACCGTCTATCAGTGAGGCCACTCCAGAA Helper 5 ACATCGCCCCGCCAGCCATTGCAAAGGGCGAA Helper 6 AAAGAACGTGGACTCCAACGTCAACAGGAAAA Helper 7 TAGTCTTTGGAAATACCTACATTTCCACTATT Helper 8 TTGTTCCAGTTTGGAACAAGAGTTGACGCT Helper 9 CGTGGCACTGAAATGGATTATTTAGTTGAGTG Helper 10 ATCAAAAGAATAGCCCGAGATAGGCATTGGCA Helper 11 TAGAACCCAGTCACACGACCAGTACCTTATAA Helper 12 CCTGTTTGATGGTGGTTCCGAAATCGGCAAAATCATAAAAGGGAAAAATTTT Helper 13 GTCAACCCCGGCGTTATAACCTCAGCGAAAAT Helper 14 TCCACGCTGGTTTGCCCCAGCAGCACTCAA Helper 15 CCTAAGCACACGAAGTCATGATTGGCAAGCGG Helper 16 CCGCCTGGCCCTGAGAGAGTTGCAAATCGCGA Helper 17 CGAGAAATCAGATTGCGATAAACGGCCCTTCA Helper 18 AGTGAGACGGGCAACAGCTGATTGTCACAT Helper 19 CAGCTTATACCTGACTATTCCACTTTTTCACC Helper 20 GCGGTTTGCGTATTGGGCGCCAGGGTGGTTTTTCGCAACAACTGAACGGACT Helper 21 TAAAACAGTGGTCATAATCATGGTGGGGAGAG Helper 22 GCATTAATGAATCGGCCAACGCGCGGCGAATA Helper 23 TTAGTAATAACAACCGCCTGCATT Helper 24 AAACTATCGATAAAACAGAGGTGAAAATGAAA Helper 25 CAATATTAATTAAAAATACCGAACCTCAAA Helper 26 ACGCTCATAATGCGCGAACTGATAGTCAGTTG Helper 27 CAATCGTCAGACAATATTTTTGAGAGGAAG Helper 28 GATTCACCTTCTGACCTGAAAGCGACTAACAA Helper 29 TACCGCTTCTCAGCGGCAAAAATTCATTCTGGCCATAATACATTTGAGGATT Helper 30 TCTTTTATGAAAACCTACCGCGCATTCGAC Helper 31 GTGGTCGGAAAAGTCTGAAACATGAACGTTAT Helper 32 TAAATTTACAGAAAAAAAGTTTGTATCATT Helper 33 GGAAACACGTGCCGAAGAAGCTGGAGTAACAGAATGCAATGAAGAAAACCAC Helper 34 AGTACGCGTGACGATGTAGCTTTATATCAAAA Helper 35 AAGATGATGCTGAGAGCCAGCAGCGGCGGTCA Helper 36 CGAATTATGCATCACCTTGCTGAACGAACCAC Helper 37 TTGAATACCCTCAATCAATATCTGGCCCTAAA Helper 38 GGGAGAAAACAGTTGAAAGGAATTATGGCTAT Helper 39 GTTAACCATTTTACGGAACGTCAGATGAATATAAATATCTTTAGGAGCTAAGAATA Helper 40 GAAATTGCTTAGAGCCGTCAATAGAACAGAGA Helper 41 ACCTACCATTAGACTTTACAAACATTCGCTTG Helper 42 TGGCAATTAAAGTTTGAGTAACATAATTATGG Helper 43 GCCAGAGTGCGTATCAAGGAGCGGAATTATCACAAAGAAACCACCAGAGTGAGAAC Helper 44 ATAGCCAGGCATTAACCGTCAAACGGTGTCTG Helper 45 TTACAGTGCCACGAAACAAACATT S17
18 Helper 46 AATCTAAATCATTTCAATTACCTGTTAAGTGG Helper 47 TATCAAACCAAGTTACAAAATCGAACCTGA Helper 48 GCAAATCACAATAACGGATTCGCCTTAGTAGC Helper 49 GTTATCTAACAGTAACAGTACCTACCAACA Helper 50 CTAATAGAGTAGATTTTCAGGTTTGGAAGGACGTCAATAGTCGGACAAGC Helper 51 TAGAAGTATATCAAAATTATTTGCACGTAAAACAGGTATAATAACCACCATC Helper 52 TAATTTTACATCAATATAATCCTGGAAGAAGA Helper 53 TTGCGGAATCATATTCCTGATTAAAATTTA Helper 54 CATTACCAGGCGTTGACAGATGTATCCATCTGAAGCACCAACAGAAACAACCTAGAGGAC Helper 55 TATAACGTCGTTTGGTCAGTTCCAGCGCATGA Helper 56 CATTTGAAAAAATTAATTACATTTAGCAAAAG Helper 57 AGCACCAAAAATAATCTCTTTAATCGCAGAGG Helper 58 GGTAAAGTTAGACCAAACCATGAATTTACATC Helper 59 ATGGCGACCATTCAAAGGATAAACGGGTTAGA Helper 60 CTCAAAGCGAACCAAACAGGCAAATCAGATGA Helper 61 TTTCAAGAAAACTTACCTTTTTTT Helper 62 CTGGAGACACATAAATCACCTCACTATGTGAG Helper 63 TTCAGCGAGCAGAAGCAATACCGGCCTCCA Helper 64 AGATGGCGTTGAGGCAGTCGGGAGGGTAGTCGGGATCGGAGG Helper 65 CAAGTAAAGGACGGTTGTCAGCGTAAAACTGG Helper 66 TAGCGATAAGTACATAAATCAATAAACAATTT Helper 67 TAATTAATCTTGCTTCTGTAAATCCCAGCAAT Helper 68 TTTAATGGAAACGCTTAGATTATT Helper 69 TGAATAACTTTCCCTTAGAATCCTAATACCAG Helper 70 AACAATTTGGCGGCTTTTTGACCTATCGGT Helper 71 AATCATAGAAGAGTCAATAGTGAATGAAAACA Helper 72 ATTAGAGCATGCCTACAGTATTGTGTCGCTAT Helper 73 TTAGACGCTGAGGTCTGAGAGATT Helper 74 CATCACCCCTTGAATGGCAGATTTTGGGTTAT Helper 75 AGCAAGCAGCGGCCTCATCAGGGACCAGCT Helper 76 AAATATATAACCTCCGGCTTAGGTTTTATCAA Helper 77 TCGCAAGAATGTAAATGCTGATGCTTAGGAAC Helper 78 TTCTACCTTTTTTTTAGTTAATTT Helper 79 ATAACTATCAAAGAACGCGAGAAACTTGCCAC Helper 80 TTAGCCATTTCAAGAAGTCCTTTTATCAGA Helper 81 ACCGACCGGACCTAAATTTAATGGACTTTTTC Helper 82 ATCCTTTCACCAAATCAAGCAACTAAATCCAA Helper 83 TTTTCATCTTCTTGTGATAAATTT Helper 84 CAAGTCCACTTTATCAGCGGCAGAGAATCATA Helper 85 AACGGCAGGCAGCAGCAAGATAAAGCACCA Helper 86 CGCTCAACATAAGAATAAACACCGTTTGAAAT Helper 87 CGTTATACAAAAAGCCTGTTTAGTTCACGAGT Helper 88 TTAAGGCGTTAAAGTAGGGCTTTT Helper 89 ATTACTAGAAATTCTTACCAGTATCTCTTTCT Helper 90 GCACGCTCAGCAGAGGAAGCATCGCTCTTT Helper 91 GTAATTTACGCCATATTTAACAACAAAGCCAA Helper 92 AGTCTCATAGTTGCATTTTAGTAAATCATATG S18
19 Helper 93 Helper 94 Helper 95 Helper 96 Helper 97 Helper 98 Helper 99 Helper 100 Helper 101 Helper 102 Helper 103 Helper 104 Helper 105 Helper 106 Helper 107 Helper 108 Helper 109 Helper 110 Helper 111 Helper 112 Helper 113 Helper 114 Helper 115 Helper 116 Helper 117 Helper 118 Helper 119 Helper 120 Helper 121 Helper 122 Helper 123 Helper 124 Helper 125 Helper 126 Helper 127 Helper 128 Helper 129 Helper 130 Helper 131 Helper 132 Helper 133 Helper 134 Helper 135 Helper 136 Helper 137 Helper 138 Helper 139 Helper 140 TTAATTGAGAATGGCAGAGGCATT GATTGTCCTTTGCATCTCGGCAATAAAGTACC TTGATTCTTGAATGCCAGCAATCCAGACGA ACTGAACAAGTAATAAGAGAATATGCCAACAT AAAACAGGGTAAAGTAATTCTGTCTCTTTTTG AAATAGCAAACAACATGTTCAGCTGCGTGAAG ATATACCTGGTCTTTCGTATTCTGAATGCAGA AGAAACGAGTTTATCAACAATAGATTTTGTGC AACAGCCAAAAAATAATATCCCATAGACTCGGCGATGCT CGGATCTGAATACGCAACGCGAGCAGTCCTAATTT AATCTCGGAAACCTGCTGTTGCTTGGAAAGATTGAATCGGCTGTCTTTCCTT GCTACAATAAGAACGGGTATTAAATGGCGCAT TTCGCTCATCTCAGCCGTTTGAGCTTGAGTAACTCCGACGAC TTTGATTTGGTCATTGGTAAAATACCGTTTTT AACCTCCCCGTAGGAATCATTACCGTCATTTC CGGTATTCCAAATCAGATATAGAAAACTACCAGATGCAA GCATCCTTGGTTCTGCGTTTGCTGATGTATTTCCTAGACAAATTA AACATACAACCATCAGCTTTACCGAATATGAG AGAAATATCCTTTGCAGTAGCGCCTCTTTCCA TTTTTTCGAGCCCCCTGAACAATT GACAAAAGGAAGCGCATTAGACGGTCAGAGAG CGACAATAGCCTTTACAGAGAGACCCAATA ACGCGCCTTTTTTTGTTTAACGTCGCAATAGC TGAACAAGTATTATTTATCCCAAAAAAGTA ACGAGCATGCCTAATTTGCCAGTTAGAAGGAA ATCATTCCTTTATCCTGAATCTTACCAACGCTAAAATACCCAAACAAACTCA ATTTTCATGACTTGCGGGAGGTTTACTCAACG ATAGCAAGTAAGAACGCGAGGCGTCTTCCA GAGCCAATATTGGGAGGGTGTCAATCCTGACGGTGCTTATGGAAGCCAAGCA GAAATTGTGCCTCCAAGATTTGGATGCCACAA TCAACCGATAATTGAGCGCTAATAGAGAATTA GTTTACCACAAGAATTGAGTTAAGATAACATA ATTTTGTCAAGAAACAATGAAATAAAAAATGA AAAGAAACCGAAGCCCTTTTTAAGTCCAAATA GAAAATACGCCGAACAAAGTTACCACAAAATA ACTCCTTAAACGCAATAATAACGGCGAGCGTC CCATTAACGTCAGAAGCAGCCTTATGCACCCA GGGAGCACATATCACCATTATCGATGAAGCCT GGTGGTCTACGAAAAGACAGAATCTTTTAGCG TCTAAAAAATGCGGTTATCCATCTGGCTTATC GCAGCCAGTGAGAAAGAGTAGAAAGGCATGAA TTAGTCAGAGGGTTGAGGGAGGTT ATAACCCAGCGCCAAAGACAAAAGCATTAAAG ATAAGAGCACAATCAATAGAAAAGAGCCAT TATCTTACGCAAAGACACCACGGAACCAGTAG AGCAGATAATACATAAAGGTGGCAAACGTC ACCGAGGATTACGCAGTATGTTAGACCGTAAT TCACGAACTTCTCAGTAACAGATAAGAACTGGCACTTTAGCGTCAGACTGTA S19
20 Helper 141 Helper 142 Helper 143 Helper 144 Helper 145 Helper 146 Helper 147 Helper 148 Helper 149 Helper 150 Helper 151 Helper 152 Helper 153 Helper 154 Helper 155 Helper 156 Helper 157 Helper 158 Helper 159 Helper 160 Helper 161 Helper 162 Helper 163 Helper 164 Helper 165 Helper 166 Helper 167 Helper 168 Helper 169 Helper 170 Helper 171 Helper 172 Helper 173 Helper 174 Helper 175 Helper 176 Helper 177 Helper 178 Helper 179 Helper 180 Helper 181 Helper 182 Helper 183 Helper 184 Helper 185 Helper 186 Helper 187 Helper 188 CAACATACATTGTAGCATTGTGCTCATAGC CCCTGCATATAGTGTTATTAATATTTCATAAT AGAGCTTGCCATTTTTCGTCCCCCACCGGA TTGGGGATCTTGCGGCAAAACTGCGTAACCGTCTCTCAGAACCGCCACCCTC GCCTCAATCGAATATCCTTAAGAGCTGAATAG TTGAAGGTAAATATTGACGGAAATTATTGGCGACAT GTGAATTATCACCGTCACCGACTTTTCATATG TTGGGAATTAGAGCCAGCAAAATCATAAGTTT CACCATTACCATTAGCAAGGCCGGAACATATA ACCAATGAAACCATCGATAGCAGCCAAACGTA CAGTAGCGACAGAATCAAGTTTGCTGATTAAG GCGCGTTTTCATCGGCATTTTCGGCAATTCAT CCCCTTATTAGCGTTTGCCATCTTCAAGTTGG CAAAATCACCGGAACCAGAGCCACTTCGGGGC ACCGCCTCCCTCAGAGCCGCCACCTCTCGTTC CACCACCACACCCTCAGAGCCGCCGGCGTTCA AGAGCCACGAGCCGCCGCTT CAAAGCCTTTGCATTCATCAAACGTCAGACGA AATTTACCAGGAGGTTGAGGCAGGACCAGAAC TTCAGCATTGACGTTCCAGTAATT TTGGCCTTCCAGAATGGAAAGCGCCTTGCGAC ACTGGTAATGGCTTTTGATGATACAGTCTCTG TTGCGTCATACATAAGTTTTAATT CCTCGGCACGTGTGAATCATTAGCCCCGTATA TTCGGGGTCAGTCTCAAGAGAATT TGAGACTCGCCTTGAGTAACAGTGAGGAGTGT GCGGATAATAGCGGGGTTTTGCTCTAAGAGGC AACAGTTATGAAACATGAAAGTATGCTATTTA TTGGATTAGGATGTGCCGTCGATT ACTGGCGGGCCACGTATTTTGCAAATAGGTGT GCGTAACGATAAGTATAGCCCGGAAGTACCAG TTGAGGGTTGATATCTAAAGTTTT ATCACCGTTTCCACAGACAGCCCTTGAATTTT TAAAGGAATCCAGACGTTAGTAAACATAGTTA TTTTGTCGTCTTTTGCGAATAATT CTGTATGGGGAGTGAGAATAGAAAAAAAAAAG TTTAATTTTTTCACGTTGAAAATCTCCAGGAACAAC GCTCCAAAAGGAGCCTTTAATTGTTTTCAACA AACAAGCGTTCTTGCAAATCACCATGCCAGCT CTTTCCAGTCGGGAAACCTGTCGGAAGGCG GAATCTCTATGAATGGGAAGCCTTACTGCCCG ACTCACATTAATTGCGTTGCGCTCCAAGAAGG ATAAGTCAAGGAGAAACATACGAAGTGAGCTA GTAAAGCCTGGGGTGCCTAATGAGGCGCAT CATACAAACACTGACCCTCAGCAACATAAAGT TCCACACAACATACGAGCCGGAAGTCTTAAAC CTTCATAGCGAATCACCAGAACGGCTCACAAT TTCCTGTGTGAAATTGTTATCCGCGCCATT S20
21 Helper 189 CTGGTGCCAGGCTGCGCAACTGTTATAGCTGT Helper 190 CCCGGGTACCGAGCTCGAATTCGTAATCATGGTCGGGAAGGGCGATCGGTGC Helper 191 CCTCAGGATCGCTATTACGCCAGCAGAGGATC Helper 192 CTTGCATGCCTGCAGGTCGACTCTTGGCGAAA Helper 193 CTGCCAGTTGCTGCAAGGCGATTAGTGCCAAG Helper 194 TTGTCACGACGTTGTAAAACGACGGCCAAGTTGGG Helper 195 GTTCCTGATTAGTCGCAGTAGGCGCCATGC Helper 196 TGATAAGCAAGCACCTTTAGCGTTGATTGTAT Helper 197 AACGATACACAGGGTCGCCAGCATTAATAT Helper 198 TTCTTAGAAAATTTCACGCGGCGGTTGTTAAA Helper 199 CGCCATTCGGAAACCAGGCAAAGAACGCCA Helper 200 GGGCCTCTAGATCGCACTCCAGCCAGCTTTCCGGTCCTGTAGCCAGCTTTCA Helper 201 GGGGGATGTTGAGGGGACGACGACCAACCCGT Helper 202 TAACGCCAATGGGCGCATCGTAACGGATTG Helper 203 TTCGTTGGTGTAGGGGTTTTCCCATT Helper 204 CCTAACGACAAGAGTAAACATAGTGGAAAACG Helper 205 TCAGAAAAATTTAAATTGTAAACGATATCGGT Helper 206 TGACCGCTATATAAGCTAAAACTAGCATGTCAAATTCGCATTAAATTTCAAGTTGC Helper 207 AAGAGAATTTTTTTAACCAATAGGAAAACATC Helper 208 TCAGGTCAAATTCGCGTCTGGCCTCACCGCTT Helper 209 TGCCGGAGAAATGTGAGCGAGTAAAGTATCGG Helper 210 ATGATATTCCGTGGGAACAAACGGCCGTGCAT Helper 211 AAGCAAATGCCCCAAAAACAGGAAAACATCAT Helper 212 TTTGTTAAATCATATGTACCCCGGTTCTTG Helper 213 TCAGCTCACGATGAACGGTAATCGAGCTTGCA Helper 214 TCAAAAATTTGCCTGAGAGTCTGTAGAAGT Helper 215 TCAACATTAGGGTAGCTATTTTTGAGAGATCTACCTCAGGAG Helper 216 CGGATTCTCAACCGTTCTAGCTGAGCAACGGA Helper 217 ACCGTAATGAGACAGTCAAATCAATGTGTA Helper 218 TTGAGAAAGGCCGGGGATAGGTCATT Helper 219 GGTAACGCTGCATGAAGTAATCACGTTGATAA Helper 220 CGTCATTTGGCGAGAAAGCTCAGTAAAGGCTA Helper 221 CGGCGCTTTGTTTTTGAGATGGCATAAATTAA Helper 222 TTAGGGTTCGAGCATCATCTTGATCCATCAAT Helper 223 CCTACTGATCGGAGGTTTTACCTCCAAATGAATGGACAGCCA Helper 224 GACCCATAACCGTGCTCA Helper 225 AACCATAAAGCCTCGGTACGGTCATACTTTTG Helper 226 GGTAAAGATGCAATGCCTGAGTAAAGGATA Helper 227 TTTATATTTTAAATTCAAAAGGGTTT Helper 228 TTAGGGATTTCAAATAACCCTGAAGGCATCCA Helper 229 ACCAAAAAGCCTTTATTTCAACGCTAAGCTCA Helper 230 CGGGAGAACATTATGACCCTGTAAGGCATGGT Helper 231 AAAATTTTGAGCATAAAGCTAAAAGGCAAA Helper 232 TTATAAAGCCTCATAGAACCCTCATT Helper 233 AATCCACTTCGTGCCAAGAAAAGCACAAATGC Helper 234 GGAGTGGCCCAGTAGTGTTAACAGTCGGTTGT Helper 235 CAATATAAATTAACACCATCCTTCATTTTCAT S21
22 Helper 236 Helper 237 Helper 238 Helper 239 Helper 240 Helper 241 Helper 242 Helper 243 Helper 244 Helper 245 Helper 246 Helper 247 Helper 248 Helper 249 Helper 250 Helper 251 Helper 252 Helper 253 Helper 254 Helper 255 Helper 256 Helper 257 Helper 258 Helper 259 Helper 260 Helper 261 Helper 262 Helper 263 Helper 264 Helper 265 Helper 266 Helper 267 Helper 268 Helper 269 Helper 270 Helper 271 Helper 272 Helper 273 Helper 274 Helper 275 Helper 276 Helper 277 Helper 278 Helper 279 Helper 280 Helper 281 Helper 282 Helper 283 GAATTAGCTAAATCATACAGGCACATCAAT TTTTAACATCCAAAAAATTAAGCATT TCTACAGTTGAGGGACATAAAAAGATGAACTT ATGGTCAACGAGCTGAAAAGGTGGTCGGGAGA TTGGGGCGTAACCTGTTTAGCTATACGGAGAG TCTACTAATGACCATTAGATACAAGTTGAT TTTAGATTTAGTTTAGTAGTAGCATT ACGACCAAGACGCAATGGAGAAAGTAAAAATG GCGATAACGCGTCCATCTCGAAGGTTTCGCAA CGCCAACGCGGAGTAGTTGAAATGTAATTGCT TCCCAATTTTCATTCCATATAACGTTTTAA TTTGTCTGGAAGTCTGCGAACGAGTT CGCTCGGCAGATGGGAAAGGTCATGTAATAAG CATTTTTGTGCTGTAGCTCAACATAGTCGCCA GAATATAACGGATGGCTTAGAGCTAAGGGGCC ATATGCAAAATTGCTCCTTTTGAAGCAAAC TTAGAGTACCTTTCTAAAGTACGGTT TTTAGTACTAATTTATCCTCAAGTGCGGCATA ACATACCATGCAATTAAAATTGTTTAAGAGGT GAAGCCCCAAGACGAGCGCCTTTAGATTGCAT TCCAACAGGCGAACCAGACCGGAAAGACTT TTCGAGCTTCAAAGTCAGGATTAGTT ATAAAAAATCCAAGTATCGGCAACACGACATT CTGGCCAATCACAACCACACCAGAAGCAGCATAGCAATCATA TTACCTTTCCAGGGCGAGCGCCAGCGCTTGCC ACTATTATTTAAGAGGAAGCCCGAGACCACCT CAAAAAGAAGTCAGAAGCAAAGCGCGCACGTT CAAATATCTCAAAAATCAGGTCTTGCTTTA TTATGACCATAAAGCGTTTTAATTTT ACTCATCGAGCAGGTTTAAGAGCCAACGAACC TCAGCGGCCGCACGTAATTTTTGAAACGTTTT CTGCGCGTCGTCAGTAAGAACGTCTTACCCTG GCTCAAAGACCTTTCTTTTTGGGTGGAGGC TTCTTCTGACACGCAAGGTAAACGAGAGGGGG AACAGTTCTGAATCCCCCTCAAATAGCGTC TTTAAATATTCATAGAAAACGAGATT CTGTCGCACTACGCGATTTCATAGGTAATTAT CAGCGCCTCATTAATAATGTTTTCCGAACAAT GGCTTTTGAAATGTTTAGACTGGAAGTGTTTC CTCCAGCAATAAACCAACCATCATAATCGG TAATAGTACAAAAGAAGTTTTGCCTGAACATA CAATACTGCGATAAAAACCAAAAAAGAGCA TTTTACCAGACGACGGAATCGTCATT TGAGTTTCACCGCCACCCTCAGAAAGCGTCCT CCACAACCAACCAGAACGTGAAAACCGCCACC CAAGCCCACACCACCCTCATTTTCTCAACAGG CATAGAAAGCCACTTCTCCTCATCGTGCCGATCCGTCTG ACCAGAGTCGGCCAGTCCTTGACGAACCAACGCGT S22
23 Helper 284 Helper 285 Helper 286 Helper 287 Helper 288 Helper 289 Helper 290 Helper 291 Helper 292 Helper 293 Helper 294 Helper 295 Helper 296 Helper 297 Helper 298 Helper 299 Helper 300 Helper 301 Helper 302 Helper 303 Helper 304 Helper 305 Helper 306 Helper 307 Helper 308 Helper 309 Helper 310 Helper 311 Helper 312 Helper 313 Helper 314 Helper 315 Helper 316 Helper 317 Helper 318 Helper 319 Helper 320 Helper 321 Helper 322 Helper 323 Helper 324 Helper 325 Helper 326 Helper 327 Helper 328 Helper 329 Helper 330 Helper 331 AGAATCTCTACCATGAACAAAATGATGGCG GCAAGGATCAAAGTAAGAGCTTCTTCAACAAG CTCAGAGCATAGGAACCCATGTACGGAAGTAG CTTTAAGCCCAACAGCCATATAAGTTCCAT CAGTTTTTACTTTTTGTTAACGTAGCAAGGTC AAAGGTCGAGGTCGAATTTTCTCCGTAAAC TAAGGGAACCGAACAAGATAATTTTTCGACT GTGAGCATTCTGAACAGCTTCTTGCGTAACAC GGATTAAGTGGTTTTTAGTGAGTTAGGGATAG GGCGTCGCTCCTAGACCTTTAGCATTTAGCCA TTTTTGCGCCACTTCGATTTAATTATTTTCCG GTAACTTTGTAATTCCTGCTTTATCGAGCTGC CGACAGCTCACTCCGTGGACAGATTTCTTAAA TCTTTAGCGTCGTAACCCAGCTTGACAATG CATATCTGTTCTGCTTCAATATCTCCGATATA AAGCAGTATCCCAGCCTCAATCTGTTAAAG CATCAGAAAGCGATAAAACTCGCCGCCAAAACGTTCAGCAGCGAAAGACAGC TATCAGCTTGCTTTCGAGGTGAATTTGTCATT CAGCTTGATACCGATAGTTGCGCCGGTAAGTT ACAACAACCATCGCCCACGCATAAGGTTGAAC TTCGGTCGCTGAGGCTTGCAGGGACATCTCTC GCCGCTTTTGCGGGATCGTCACCCCGGCTACA ATCGGAACGAGGGTAGCAACGGCTACTTCTGC CATTAAAGGATATTCACAAACAAAGCATGAGC TCGTCAGCATCATAAAACGCCTCCAATATC GCAGTCGGGCAAGAACCATACGACTAAATCCT ACGAAAATTCAGGCACACAAAAACGCATGG TATTATTCATGCCCCCTGCCTATTTACTGATA ACCATAAGCGATTGCGTACCCGACTCGGAACC AAATGAAGCCGCATAAAGTGCACGACCAAA ATTAGGGTCGAACTGCGATGGGCACCGCCA CTGTAGCAACTCAGGAGGTTTAGTATACTGTA CCCTCAGAGTCACCAGTACAAACGCGGCTC GTTTCAGCGATTTTGCTAAACAACTACAACGC ATTCTGATTTTCATCCCGAAGTTATCGGTT AGCGTACCTTGAATGTTGACGGGACGTAAATT GAGCAGGATGACGGCAGCAATAAATAGCGAGA ATAAGCAAAAGCGAGGGTATCCCAAGAAAGAT ACACTATCATTACGAGGCATAGTCACATTC TTCGCCAAAAGGAATAACCCTCGTTT TAGCAATCAGCGACGAGCACGAGACAAAGTCC TAACGGAATGAGATTTAGGAATACCTCAACAG TCATCAGTCAACATTATTACAGGTTCGGTTAA AACTAATGAAATCTACGTTAATAAACTGGC TTGTTGGGAAGAACAGATACATAATT TGTTCAGTAAAATCGAAATCATCTGCGGTCAG GGACTCAGACCTATTAGTGGTTGAGTACGGAT ATCCAAAAAAAGCGGTCTGGAAACAAACACCA S23
24 Helper 332 Helper 333 Helper 334 Helper 335 Helper 336 Helper 337 Helper 338 Helper 339 Helper 340 Helper 341 Helper 342 Helper 343 Helper 344 Helper 345 Helper 346 Helper 347 Helper 348 Helper 349 Helper 350 TAGAGGCCCGGCAGAAGCCTGAATAAACGAAC GTGAATAAAGTAAATTGGGCTTGAGAGCTTAA TCATTATATTATGCGATTTTAAGGATGGTT TTTGTGAATTACCCCAGTCAGGACTT GAACGAGTGGCTTGCCCTGACGAGAGGCGCAT TAATTTCAAACGTAACAAAGCTGTAATCTT TTTTACCCAAATCACTTTAATCATTT CGAGGCGCGAACGGTGTACAGACCACAGCATC TCCGCGACTGACCTTCATCAAGAGCTCATTCA AGGCTGGCCTGCTCCATGTTACTTAAAACACT GACAAGAATCGCCTGATAAATTGCCAAGCG TTGATTTGTATCACCGGATATTCATT GTCATGGAATATCCGAAAGTGTTAAGCCGGAA ACGAAAGAACCCCCAGCGATTATATGTCGAAA CATCTTTGGGCAAAAGAATACACTACAGAGGC CGAAACAACACTACGAAGGCACCGAGGAAGTTTCCATTAAACGGGTAAATT TTATACGTAATGCAGTACAACGGATT TTTGAGGACTAAAGACTTTTTCATAACCTAAA CTTTGAAAGAGGACAGATAGACGGTCAATCA Sticky End Left 1 Sticky End Left 2 Sticky End Left 3 Sticky End Left 4 Sticky End Left 5 Sticky End Left 6 Sticky End Left 7 Sticky End Left 8 Sticky End Left 9 Sticky End Left 10 Sticky End Left 11 Sticky End Left 12 Sticky End Left 13 Sticky End Left 14 Sticky End Left 15 Sticky End Right 1 Sticky End Right 2 Sticky End Right 3 Sticky End Right 4 Sticky End Right 5 Sticky End Right 6 Sticky End Right 7 Sticky End Right 8 Sticky End Right 9 Sticky End Right 10 Sticky End Right 11 Sticky End Right 12 ATCACGGCCGCTGCACCAGCAAGAAACCAATCCGCGGCATTGATTGCT TTCCTACTGACGGATGCCACCGGAAGACATGGCGCCTGTATGGGTTCT ATCACTGGTACCTCAAAACTAGGGCATCACCTTGAAGTCACTGGACAT TTCCTCCATGAACTGCAACGTACCAGCACCAGAAACGTATCGCGTTCT ATCACGGTGGTCAGCTCAGGAAATAAGTGCCAGCCGCCGTCCAGACAT TTCCTTCCTTGACTCAACCATACCCCAAGCATTAAAGCACGACGTTCT TCGCCGACCAAATCCGGCGCAGGCCAGGACAT ATCACAGGTACACGAATCCGGGACAT TTCCTCGCTCCGTAGCGTGACATTATGAAAAATATACTTATACGTTCT ATCACCACACGCTTCATCCTTAATTCAAAATAATCGCCGTCCAGACAT TTCCTTCTAGATCTGTCAAAAACGATCTTGAACACTCTCTTAAGTTCT ATCACGGTTCGCAGCATTGGGATTCAACGTGAGAGCGGAAGTCGACAT TTCCTACAAACGTCTGTACCATACAGTCACGCAAACTTCCTTCGTTCT ATCACCAGCGTATGTAGGAAGTGTACGGCCATTAGAAGCTTCAGACAT GCGTGTAGCAACGCTACCTTGCGCCTAGTTCT GAAGCGGAGCAGTCCAAATAAAATAGTTCCAGGAGCCTTAG TGAGTTCGCTGATGTTATAGATATTTATTGGTATATGCCGCAGCTT CGTTATCGGCCAATCAGGGTTAAGTTCACCATATGTTATGTCTTAG TGAGTCTTGCGGCAACGTGACGAAGAGTCAATATGTCAAGCAGCTT CGTTAATCTACGTGCAAGGCCACTCTGACCAGCAGCCGAGACTTAG TGAGTCCACCTATAAGGAAGCCAGCCAGTTTGATGAGCACTAGCTT CGTTAGTTCGGATATATTAGACACTCGCAACGGCTAATGGCCTTAG TGAGTGGCCGAGACTGCGGACGAAGACATTACAGGTAGTCCAGCTT CGTTAGGACAGCGTCACTCCTTCTTTAACCGGAGGTGGCCGCTTAG TGAGTCTGAACACCGCTCGACGCTCCATGATGACAGGAACAAGCTT CGTTACACGCGGAGACAGGCCGTATAAACGCAATTATAGGCCTTAG TGAGTGTGCTCGCGCCTCAACGCCAAACTTTGTCAGTCCTCAGCTT S24
25 Sticky End Right 13 CGTTACACTCTTCTACTCGTCAGAACTTGACTCATCGCCGACTTAG Sticky End Right 14 TGAGTATAGACGCATGATTTCTTATAGTAATCCACGCTCTTTTAAAATGCTGACCAA Sticky End Up 1 TAAACGTTATTGCCCGGCGCCAGGTCCAGCTT Sticky End Up 2 TTCCTCCGAAGAGTCACACAGTCCTTGACGAAATAAA Sticky End Up 3 AACTCGTATTCTGAATAATGGAAATCATGGAGCTGGCTTAG Sticky End Up 4 ATCACCCAGTGCCGAACCATTGTTTGGATTATACTTAAATCCTTTGCCCGATTAAACT Sticky End Up 5 GGGTCGGCATCAAAAGCAATCGGCCGCAGCTT Sticky End Up 6 TTCCTCGAGCCAGCCTGATTAGCATGCCCAGAGATTAGATCAACATC Sticky End Up 7 TCAGGAACGTTGAACACGACCAGCATAAAGCCTCTTCTTAG Sticky End Up 8 ATCACTGCTACAGGAAATGAATGTTTATAGGTCTAAAGAAACGCGGCACAAAGGTACT Sticky End Up 9 GTCAGTATGCAAATTAGCAACCAGTGGAGCTT Sticky End Up 10 TTCCTAGAGCTCCATGTCAATAGATGTGGGAGCAAAC Sticky End Down 1 TGAGTGCGGACGCCCTTCTGTTGATAAGCAAGCATCTCATAAGTCC Sticky End Down 2 TTTCCAGAGTAGAAACCAATCAATGTGTTTTCCATAATAGAATTAGGCGTTCT Sticky End Down 3 CGTTAAGTAAGGTGCATTCCAAGTACCGCACTCGATTAGTTGCTATTTTGGCCGT Sticky End Down 4 TAAATCAAATCGAGAACAAGCAAGCTGACGGAAATGCGACAT Sticky End Down 5 TGAGTGCCGGCCTAGTCAACCTCAGCACTAACCTTGCGAGCGCCCA Sticky End Down 6 AAGAGCCATACCGCTGATCAAGAACTGTTCT Sticky End Down 7 CGTTACGTGTTGGCAGTGAGCTTTATCAATACCCAGAAGGGTAATAAGTCGATAC Sticky End Down 8 CGTTATTCAGTCGAAGCATATTAAGGCTCACCTTTAGCACTGGTAG Sticky End Down 9 CCAGCAGTGACAGAATCGTTAGTTGTGACTCATATCTAAATGGCCAGGGACAT Sticky End-Scaffold Linker Left 1 Sticky End-Scaffold Linker Left 2 Sticky End-Scaffold Linker Left 3 Sticky End-Scaffold Linker Left 4 Sticky End-Scaffold Linker Left 5 Sticky End-Scaffold Linker Left 6 Sticky End-Scaffold Linker Left 7 Sticky End-Scaffold Linker Left 8 Sticky End-Scaffold Linker Left 9 Sticky End-Scaffold Linker Left 10 Sticky End-Scaffold Linker Left 11 Sticky End-Scaffold Linker Left 12 Sticky End-Scaffold Linker Left 13 Sticky End-Scaffold Linker Up 1 Sticky End-Scaffold Linker Up 2 Sticky End-Scaffold Linker Up 3 CCATACAGCAGCGGCC CAGTGACTCCGTCAGT GCGATACGAGGTACCA TGGACGGCGTTCATGG GTCGTGCTTGACCACC CTGGCCTGGTCAAGGA GTATAAGTGTGTACCT TGGACGGCACGGAGCG TTAAGAGAAGCGTGTG GACTTCCGGATCTAGA GAAGGAAGTGCGAACC TGAAGCTTACGTTTGT TAGGCGCAATACGCTG GGACCTGGAAATGGTTAACGCTTGTCCGACTCTTCGG CCAGCTCCCAACGCCATCTCCTCCGATCCGGCACTGG GCGGCCGACGCACTCTGGCGTCCTCTAGGCTGGCTCG S25
26 Sticky End-Scaffold Linker Up 4 Sticky End-Scaffold Linker Up 5 AAGAGGCTCGATCAGTAGGTGGCTGTCCACTGTAGCA CCACTGGTTATAGCGGTCATGAGCACGGTGGAGCTCT Sticky End-Scaffold Linker Right 1 Sticky End-Scaffold Linker Right 2 Sticky End-Scaffold Linker Right 3 Sticky End-Scaffold Linker Right 4 Sticky End-Scaffold Linker Right 5 Sticky End-Scaffold Linker Right 6 Sticky End-Scaffold Linker Right 7 Sticky End-Scaffold Linker Right 8 Sticky End-Scaffold Linker Right 9 Sticky End-Scaffold Linker Right 10 Sticky End-Scaffold Linker Right 11 Sticky End-Scaffold Linker Right 12 Sticky End-Scaffold Linker Right 13 Sticky End-Scaffold Linker Down 1 Sticky End-Scaffold Linker Down 2 Sticky End-Scaffold Linker Down 3 Sticky End-Scaffold Linker Down 4 Sticky End-Scaffold Linker Down 5 GCTCCTGGATCAGCGA GCGGCATATTGGCCGA ACATAACAGCCGCAAG GCTTGACAACGTAGAT TCTCGGCTATAGGTGG AGTGCTCAATCCGAAC GCCATTAGTCTCGGCC GGACTACCCGCTGTCC CGGCCACCGTGTTCAG TGTTCCTGTCCGCGTG GCCTATAAGCGAGCAC GAGGACTGGAAGAGTG TCGGCGATGCGTCTAT GCCTAATTATTCAGATCCGAGCATCGCCGGCGTCCGC GCATTTCCAGATGAGCGAAGTCGTCGGAGACCTTACT AGTTCTTGACCAAGGATGCTTGCATCTGGAGGCCGGC CCGGATTCTGATTGGCCAGTATGATTGCTCCAACACG CCTGGCCACCGACTCTGGTCAGACGGATCCGACTGAA Helpers modified with biotin: Biotin Helper 158 Biotin Helper 159 Biotin Helper 161 Biotin Helper 162 CAAAGCCTTTGCATTCATCAAACGTCAGACGATTTTTTTTTTTTTTTTTTTT AATTTACCAGGAGGTTGAGGCAGGACCAGAACTTTTTTTTTTTTTTTTTTTT TTGGCCTTCCAGAATGGAAAGCGCCTTGCGACTTTTTTTTTTTTTTTTTTTT ACTGGTAATGGCTTTTGATGATACAGTCTCTGTTTTTTTTTTTTTTTTTTTT Biotin 20A [5 biotin]aaaaaaaaaaaaaaaaaaaa S26
Fig.S1 ESI-MS spectrum of reaction of ApA and THPTb after 16 h.
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2014 Experiment Cleavage of dinucleotides Dinucleotides (ApA, CpC, GpG, UpU) were purchased from
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2014 Experiment Cleavage of dinucleotides Dinucleotides (ApA, CpC, GpG, UpU) were purchased from
SUPPLEMENTARY MATERIAL
 SUPPLEMENTARY MATERIAL Purification and biochemical properties of SDS-stable low molecular weight alkaline serine protease from Citrullus Colocynthis Muhammad Bashir Khan, 1,3 Hidayatullah khan, 2 Muhammad
SUPPLEMENTARY MATERIAL Purification and biochemical properties of SDS-stable low molecular weight alkaline serine protease from Citrullus Colocynthis Muhammad Bashir Khan, 1,3 Hidayatullah khan, 2 Muhammad
Supporting Information
 Supporting Information The Effects of Spacer Length and Composition on Aptamer-Mediated Cell-Specific Targeting with Nanoscale PEGylated Liposomal Doxorubicin Hang Xing +, [a] Ji Li +, [a] Weidong Xu,
Supporting Information The Effects of Spacer Length and Composition on Aptamer-Mediated Cell-Specific Targeting with Nanoscale PEGylated Liposomal Doxorubicin Hang Xing +, [a] Ji Li +, [a] Weidong Xu,
INSTRUCTION MANUAL. RNA Clean & Concentrator -5 Catalog Nos. R1015 & R1016. Highlights. Contents
 INSTRUCTION MANUAL Catalog Nos. R1015 & R1016 Highlights Quick (5 minute) method for cleaning and concentrating RNA. Ideal for purification of RNA from aqueous phase following an acid phenol extraction.
INSTRUCTION MANUAL Catalog Nos. R1015 & R1016 Highlights Quick (5 minute) method for cleaning and concentrating RNA. Ideal for purification of RNA from aqueous phase following an acid phenol extraction.
Pinpoint Slide RNA Isolation System II Catalog No. R1007
 INSTRUCTION MANUAL Pinpoint Slide RNA Isolation System II Catalog No. R1007 Highlights Allows for the isolation of total RNA from paraffin-embedded tissue sections on glass slides Simple procedure combines
INSTRUCTION MANUAL Pinpoint Slide RNA Isolation System II Catalog No. R1007 Highlights Allows for the isolation of total RNA from paraffin-embedded tissue sections on glass slides Simple procedure combines
Europium Labeling Kit
 Europium Labeling Kit Catalog Number KA2096 100ug *1 Version: 03 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 Principle of the Assay...
Europium Labeling Kit Catalog Number KA2096 100ug *1 Version: 03 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 Principle of the Assay...
Product Manual. Omni-Array Sense Strand mrna Amplification Kit, 2 ng to 100 ng Version Catalog No.: Reactions
 Genetic Tools and Reagents Universal mrna amplification, sense strand amplification, antisense amplification, cdna synthesis, micro arrays, gene expression, human, mouse, rat, guinea pig, cloning Omni-Array
Genetic Tools and Reagents Universal mrna amplification, sense strand amplification, antisense amplification, cdna synthesis, micro arrays, gene expression, human, mouse, rat, guinea pig, cloning Omni-Array
Nature Methods: doi: /nmeth Supplementary Figure 1
 Supplementary Figure 1 Subtiligase-catalyzed ligations with ubiquitin thioesters and 10-mer biotinylated peptides. (a) General scheme for ligations between ubiquitin thioesters and 10-mer, biotinylated
Supplementary Figure 1 Subtiligase-catalyzed ligations with ubiquitin thioesters and 10-mer biotinylated peptides. (a) General scheme for ligations between ubiquitin thioesters and 10-mer, biotinylated
Plasmonic blood glucose monitor based on enzymatic. etching of gold nanorods
 Plasmonic blood glucose monitor based on enzymatic etching of gold nanorods Xin Liu, Shuya Zhang, Penglong Tan, Jiang Zhou, Yan Huang, Zhou Nie* and Shouzhuo Yao State Key Laboratory of Chemo/Biosensing
Plasmonic blood glucose monitor based on enzymatic etching of gold nanorods Xin Liu, Shuya Zhang, Penglong Tan, Jiang Zhou, Yan Huang, Zhou Nie* and Shouzhuo Yao State Key Laboratory of Chemo/Biosensing
EXPERIMENT 26: Detection of DNA-binding Proteins using an Electrophoretic Mobility Shift Assay Gel shift
 EXPERIMENT 26: Detection of DNA-binding Proteins using an Electrophoretic Mobility Shift Assay Gel shift Remember to use sterile conditions (tips, tubes, etc.) throughout this experiment Day 1: Biotinylation
EXPERIMENT 26: Detection of DNA-binding Proteins using an Electrophoretic Mobility Shift Assay Gel shift Remember to use sterile conditions (tips, tubes, etc.) throughout this experiment Day 1: Biotinylation
Protocol for Gene Transfection & Western Blotting
 The schedule and the manual of basic techniques for cell culture Advanced Protocol for Gene Transfection & Western Blotting Schedule Day 1 26/07/2008 Transfection Day 3 28/07/2008 Cell lysis Immunoprecipitation
The schedule and the manual of basic techniques for cell culture Advanced Protocol for Gene Transfection & Western Blotting Schedule Day 1 26/07/2008 Transfection Day 3 28/07/2008 Cell lysis Immunoprecipitation
Chromatin Immunoprecipitation (ChIPs) Protocol (Mirmira Lab)
 Chromatin Immunoprecipitation (ChIPs) Protocol (Mirmira Lab) Updated 12/3/02 Reagents: ChIP sonication Buffer (1% Triton X-100, 0.1% Deoxycholate, 50 mm Tris 8.1, 150 mm NaCl, 5 mm EDTA): 10 ml 10 % Triton
Chromatin Immunoprecipitation (ChIPs) Protocol (Mirmira Lab) Updated 12/3/02 Reagents: ChIP sonication Buffer (1% Triton X-100, 0.1% Deoxycholate, 50 mm Tris 8.1, 150 mm NaCl, 5 mm EDTA): 10 ml 10 % Triton
Product Contents. 1 Specifications 1 Product Description. 2 Buffer Preparation... 3 Protocol. 3 Ordering Information 4 Related Products..
 INSTRUCTION MANUAL Quick-RNA MidiPrep Catalog No. R1056 Highlights 10 minute method for isolating RNA (up to 1 mg) from a wide range of cell types and tissue samples. Clean-Spin column technology allows
INSTRUCTION MANUAL Quick-RNA MidiPrep Catalog No. R1056 Highlights 10 minute method for isolating RNA (up to 1 mg) from a wide range of cell types and tissue samples. Clean-Spin column technology allows
For Research Use Only Ver
 INSTRUCTION MANUAL Quick-cfDNA/cfRNA Serum & Plasma Kit Catalog No. R1072 Highlights High-quality cell-free DNA and RNA are easily and robustly purified from up to 3 ml of plasma, serum, or other biological
INSTRUCTION MANUAL Quick-cfDNA/cfRNA Serum & Plasma Kit Catalog No. R1072 Highlights High-quality cell-free DNA and RNA are easily and robustly purified from up to 3 ml of plasma, serum, or other biological
SUPPLEMENTAL INFORMATION
 SUPPLEMENTAL INFORMATION EXPERIMENTAL PROCEDURES Tryptic digestion protection experiments - PCSK9 with Ab-3D5 (1:1 molar ratio) in 50 mm Tris, ph 8.0, 150 mm NaCl was incubated overnight at 4 o C. The
SUPPLEMENTAL INFORMATION EXPERIMENTAL PROCEDURES Tryptic digestion protection experiments - PCSK9 with Ab-3D5 (1:1 molar ratio) in 50 mm Tris, ph 8.0, 150 mm NaCl was incubated overnight at 4 o C. The
Product Contents. 1 Specifications 1 Product Description. 2 Buffer Preparation... 3 Protocol. 3 Ordering Information 4
 INSTRUCTION MANUAL Quick-RNA Midiprep Kit Catalog No. R1056 Highlights 10 minute method for isolating RNA (up to 1 mg) from a wide range of cell types and tissue samples. Clean-Spin column technology allows
INSTRUCTION MANUAL Quick-RNA Midiprep Kit Catalog No. R1056 Highlights 10 minute method for isolating RNA (up to 1 mg) from a wide range of cell types and tissue samples. Clean-Spin column technology allows
Hepatitis B Virus Genemer
 Product Manual Hepatitis B Virus Genemer Primer Pair for amplification of HBV Viral Specific Fragment Catalog No.: 60-2007-10 Store at 20 o C For research use only. Not for use in diagnostic procedures
Product Manual Hepatitis B Virus Genemer Primer Pair for amplification of HBV Viral Specific Fragment Catalog No.: 60-2007-10 Store at 20 o C For research use only. Not for use in diagnostic procedures
Caution: For Laboratory Use. A product for research purposes only. Eu-W1284 Iodoacetamido Chelate & Europium Standard. Product Number: AD0014
 TECHNICAL DATA SHEET Lance Caution: For Laboratory Use. A product for research purposes only. Eu-W1284 Iodoacetamido Chelate & Europium Standard Product Number: AD0014 INTRODUCTION: Iodoacetamido-activated
TECHNICAL DATA SHEET Lance Caution: For Laboratory Use. A product for research purposes only. Eu-W1284 Iodoacetamido Chelate & Europium Standard Product Number: AD0014 INTRODUCTION: Iodoacetamido-activated
Supplementary material: Materials and suppliers
 Supplementary material: Materials and suppliers Electrophoresis consumables including tris-glycine, acrylamide, SDS buffer and Coomassie Brilliant Blue G-2 dye (CBB) were purchased from Ameresco (Solon,
Supplementary material: Materials and suppliers Electrophoresis consumables including tris-glycine, acrylamide, SDS buffer and Coomassie Brilliant Blue G-2 dye (CBB) were purchased from Ameresco (Solon,
Dental Research Institute, Faculty of Dentistry, University of Toronto, Toronto, Canada *For correspondence:
 Zymogram Assay for the Detection of Peptidoglycan Hydrolases in Streptococcus mutans Delphine Dufour and Céline M. Lévesque * Dental Research Institute, Faculty of Dentistry, University of Toronto, Toronto,
Zymogram Assay for the Detection of Peptidoglycan Hydrolases in Streptococcus mutans Delphine Dufour and Céline M. Lévesque * Dental Research Institute, Faculty of Dentistry, University of Toronto, Toronto,
Use of double- stranded DNA mini- circles to characterize the covalent topoisomerase- DNA complex
 SUPPLEMENTARY DATA Use of double- stranded DNA mini- circles to characterize the covalent topoisomerase- DNA complex Armêl Millet 1, François Strauss 1 and Emmanuelle Delagoutte 1 1 Structure et Instabilité
SUPPLEMENTARY DATA Use of double- stranded DNA mini- circles to characterize the covalent topoisomerase- DNA complex Armêl Millet 1, François Strauss 1 and Emmanuelle Delagoutte 1 1 Structure et Instabilité
Tunable Hydrophobicity in DNA Micelles Anaya, Milena; Kwak, Minseok; Musser, Andrew J.; Muellen, Klaus; Herrmann, Andreas; Müllen, Klaus
 University of Groningen Tunable Hydrophobicity in DNA Micelles Anaya, Milena; Kwak, Minseok; Musser, Andrew J.; Muellen, Klaus; Herrmann, Andreas; Müllen, Klaus Published in: Chemistry DOI: 10.1002/chem.201001816
University of Groningen Tunable Hydrophobicity in DNA Micelles Anaya, Milena; Kwak, Minseok; Musser, Andrew J.; Muellen, Klaus; Herrmann, Andreas; Müllen, Klaus Published in: Chemistry DOI: 10.1002/chem.201001816
Supplementary Information
 Supplementary Information Supplementary Figure 1. CD4 + T cell activation and lack of apoptosis after crosslinking with anti-cd3 + anti-cd28 + anti-cd160. (a) Flow cytometry of anti-cd160 (5D.10A11) binding
Supplementary Information Supplementary Figure 1. CD4 + T cell activation and lack of apoptosis after crosslinking with anti-cd3 + anti-cd28 + anti-cd160. (a) Flow cytometry of anti-cd160 (5D.10A11) binding
For Research Use Only Ver
 INSTRUCTION MANUAL Quick-RNA Miniprep Kit Catalog Nos. R1054 & R1055 Highlights High-quality total RNA (including small RNAs) from a wide range of samples. You can opt to isolate small and large RNAs in
INSTRUCTION MANUAL Quick-RNA Miniprep Kit Catalog Nos. R1054 & R1055 Highlights High-quality total RNA (including small RNAs) from a wide range of samples. You can opt to isolate small and large RNAs in
ph Switchable and Fluorescent Ratiometric Squarylium Indocyanine Dyes as Extremely Alkaline Sensors
 ph Switchable and Fluorescent Ratiometric Squarylium Indocyanine Dyes as Extremely Alkaline Sensors Jie Li, Chendong Ji, Wantai Yang, Meizhen Yin* State Key Laboratory of Chemical Resource Engineering,
ph Switchable and Fluorescent Ratiometric Squarylium Indocyanine Dyes as Extremely Alkaline Sensors Jie Li, Chendong Ji, Wantai Yang, Meizhen Yin* State Key Laboratory of Chemical Resource Engineering,
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 Minicircle topoisomer generation. a, Generation of supercoiled topoisomers. Minicircles were nicked with the sequence-specific nicking endonuclease, Nb.BbvCI.
Supplementary Figures Supplementary Figure 1 Minicircle topoisomer generation. a, Generation of supercoiled topoisomers. Minicircles were nicked with the sequence-specific nicking endonuclease, Nb.BbvCI.
For Research Use Only Ver
 INSTRUCTION MANUAL Quick-cfDNA Serum & Plasma Kit Catalog No. D4076 Highlights High-quality DNA, including cell-free, is easily and robustly purified from up to 10 ml of serum/plasma, up to 1 ml amniotic
INSTRUCTION MANUAL Quick-cfDNA Serum & Plasma Kit Catalog No. D4076 Highlights High-quality DNA, including cell-free, is easily and robustly purified from up to 10 ml of serum/plasma, up to 1 ml amniotic
UV Tracer TM Maleimide NHS ester
 UV Tracer TM Maleimide HS ester Product o.: 1020 Product ame: UV-Tracer TM Maleimide-HS ester Chemical Structure: Chemical Composition: C 41 H 67 5 18 Molecular Weight: 1014.08 Appearance: Storage: Yellow
UV Tracer TM Maleimide HS ester Product o.: 1020 Product ame: UV-Tracer TM Maleimide-HS ester Chemical Structure: Chemical Composition: C 41 H 67 5 18 Molecular Weight: 1014.08 Appearance: Storage: Yellow
HiPer Western Blotting Teaching Kit
 HiPer Western Blotting Teaching Kit Product Code: HTI009 Number of experiments that can be performed: 5/20 Duration of Experiment: ~ 2 days Day 1: 6-8 hours (SDS- PAGE and Electroblotting) Day 2: 3 hours
HiPer Western Blotting Teaching Kit Product Code: HTI009 Number of experiments that can be performed: 5/20 Duration of Experiment: ~ 2 days Day 1: 6-8 hours (SDS- PAGE and Electroblotting) Day 2: 3 hours
Annexure III SOLUTIONS AND REAGENTS
 Annexure III SOLUTIONS AND REAGENTS A. STOCK SOLUTIONS FOR DNA ISOLATION 0.5M Ethylene-diamine tetra acetic acid (EDTA) (ph=8.0) 1M Tris-Cl (ph=8.0) 5M NaCl solution Red cell lysis buffer (10X) White cell
Annexure III SOLUTIONS AND REAGENTS A. STOCK SOLUTIONS FOR DNA ISOLATION 0.5M Ethylene-diamine tetra acetic acid (EDTA) (ph=8.0) 1M Tris-Cl (ph=8.0) 5M NaCl solution Red cell lysis buffer (10X) White cell
Tumor Targeting of Functionalized Quantum Dot- Liposome Hybrids by Intravenous Administration
 Tumor Targeting of Functionalized Quantum Dot- Liposome Hybrids by Intravenous Administration Wafa T. Al-Jamal 1, Khuloud T. Al-Jamal 1, Bowen Tian 1, Andrew Cakebread 2, John M. Halket 2 and Kostas Kostarelos
Tumor Targeting of Functionalized Quantum Dot- Liposome Hybrids by Intravenous Administration Wafa T. Al-Jamal 1, Khuloud T. Al-Jamal 1, Bowen Tian 1, Andrew Cakebread 2, John M. Halket 2 and Kostas Kostarelos
For the 5 GATC-overhang two-oligo adaptors set up the following reactions in 96-well plate format:
 Supplementary Protocol 1. Adaptor preparation: For the 5 GATC-overhang two-oligo adaptors set up the following reactions in 96-well plate format: Per reaction X96 10X NEBuffer 2 10 µl 10 µl x 96 5 -GATC
Supplementary Protocol 1. Adaptor preparation: For the 5 GATC-overhang two-oligo adaptors set up the following reactions in 96-well plate format: Per reaction X96 10X NEBuffer 2 10 µl 10 µl x 96 5 -GATC
EXO-DNA Circulating and EV-associated DNA extraction kit
 Datasheet EXO-DNA Circulating and EV-associated DNA extraction kit This product is for research use only. It is highly recommended to read this users guide in its entirety prior to using this product.
Datasheet EXO-DNA Circulating and EV-associated DNA extraction kit This product is for research use only. It is highly recommended to read this users guide in its entirety prior to using this product.
Data Sheet. PCSK9[Biotinylated]-LDLR Binding Assay Kit Catalog # 72002
![Data Sheet. PCSK9[Biotinylated]-LDLR Binding Assay Kit Catalog # 72002 Data Sheet. PCSK9[Biotinylated]-LDLR Binding Assay Kit Catalog # 72002](/thumbs/79/80181612.jpg) Data Sheet PCSK9[Biotinylated]-LDLR Binding Assay Kit Catalog # 72002 DESCRIPTION: The PCSK9[Biotinylated]-LDLR Binding Assay Kit is designed for screening and profiling purposes. PCSK9 is known to function
Data Sheet PCSK9[Biotinylated]-LDLR Binding Assay Kit Catalog # 72002 DESCRIPTION: The PCSK9[Biotinylated]-LDLR Binding Assay Kit is designed for screening and profiling purposes. PCSK9 is known to function
Supporting Information for. Boronic Acid Functionalized Aza-Bodipy (azabdpba) based Fluorescence Optodes for the. analysis of Glucose in Whole Blood
 Supporting Information for Boronic Acid Functionalized Aza-Bodipy (azabdpba) based Fluorescence Optodes for the analysis of Glucose in Whole Blood Yueling Liu, Jingwei Zhu, Yanmei Xu, Yu Qin*, Dechen Jiang*
Supporting Information for Boronic Acid Functionalized Aza-Bodipy (azabdpba) based Fluorescence Optodes for the analysis of Glucose in Whole Blood Yueling Liu, Jingwei Zhu, Yanmei Xu, Yu Qin*, Dechen Jiang*
The Schedule and the Manual of Basic Techniques for Cell Culture
 The Schedule and the Manual of Basic Techniques for Cell Culture 1 Materials Calcium Phosphate Transfection Kit: Invitrogen Cat.No.K2780-01 Falcon tube (Cat No.35-2054:12 x 75 mm, 5 ml tube) Cell: 293
The Schedule and the Manual of Basic Techniques for Cell Culture 1 Materials Calcium Phosphate Transfection Kit: Invitrogen Cat.No.K2780-01 Falcon tube (Cat No.35-2054:12 x 75 mm, 5 ml tube) Cell: 293
Caution: For Laboratory Use. A product for research purposes only. Eu-W1024 ITC Chelate & Europium Standard. Product Number: AD0013
 TECHNICAL DATA SHEET Lance Caution: For Laboratory Use. A product for research purposes only. Eu-W1024 ITC Chelate & Europium Standard Product Number: AD0013 INTRODUCTION: Fluorescent isothiocyanato-activated
TECHNICAL DATA SHEET Lance Caution: For Laboratory Use. A product for research purposes only. Eu-W1024 ITC Chelate & Europium Standard Product Number: AD0013 INTRODUCTION: Fluorescent isothiocyanato-activated
For Research Use Only Ver
 INSTRUCTION MANUAL Quick-RNA Miniprep Kit Catalog Nos. R1054 & R1055 Highlights High-quality total RNA (including small RNAs) from a wide range of samples. You can opt to isolate small and large RNAs in
INSTRUCTION MANUAL Quick-RNA Miniprep Kit Catalog Nos. R1054 & R1055 Highlights High-quality total RNA (including small RNAs) from a wide range of samples. You can opt to isolate small and large RNAs in
Data are contained in multiple tabs in Excel spreadsheets and in CSV files.
 Contents Overview Curves Methods Measuring enzymatic activity (figure 2) Enzyme characterisation (Figure S1, S2) Enzyme kinetics (Table 3) Effect of ph on activity (figure 3B) Effect of metals and inhibitors
Contents Overview Curves Methods Measuring enzymatic activity (figure 2) Enzyme characterisation (Figure S1, S2) Enzyme kinetics (Table 3) Effect of ph on activity (figure 3B) Effect of metals and inhibitors
Protein MultiColor Stable, Low Range
 Product Name: DynaMarker Protein MultiColor Stable, Low Range Code No: DM670L Lot No: ******* Size: 200 μl x 3 (DM670 x 3) (120 mini-gel lanes) Storage: 4 C Stability: 12 months at 4 C Storage Buffer:
Product Name: DynaMarker Protein MultiColor Stable, Low Range Code No: DM670L Lot No: ******* Size: 200 μl x 3 (DM670 x 3) (120 mini-gel lanes) Storage: 4 C Stability: 12 months at 4 C Storage Buffer:
EXO-DNAc Circulating and EV-associated DNA extraction kit
 Datasheet EXO-DNAc Circulating and EV-associated DNA extraction kit This product is for research use only. It is highly recommended to read this users guide in its entirety prior to using this product.
Datasheet EXO-DNAc Circulating and EV-associated DNA extraction kit This product is for research use only. It is highly recommended to read this users guide in its entirety prior to using this product.
Exo-spin Exosome Purification Kit For cell culture media/urine/saliva and other low-protein biological fluids
 User Guide Exo-spin Exosome Purification Kit For cell culture media/urine/saliva and other low-protein biological fluids Cat EX01 Protocol Version 5.6 Contents 1. Storage 3 2. Product Components 3 3. Product
User Guide Exo-spin Exosome Purification Kit For cell culture media/urine/saliva and other low-protein biological fluids Cat EX01 Protocol Version 5.6 Contents 1. Storage 3 2. Product Components 3 3. Product
Protocol for protein SDS PAGE and Transfer
 Protocol for protein SDS PAGE and Transfer According to Laemmli, (1970) Alaa El -Din Hamid Sayed, Alaa_h254@yahoo.com Serum Selection of a protein source cell cultures (bacteria, yeast, mammalian, etc.)
Protocol for protein SDS PAGE and Transfer According to Laemmli, (1970) Alaa El -Din Hamid Sayed, Alaa_h254@yahoo.com Serum Selection of a protein source cell cultures (bacteria, yeast, mammalian, etc.)
Protein-Mediated Anti-Adhesion Surface against Oral Bacteria
 Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 2018 Supporting Information Protein-Mediated Anti-Adhesion Surface against Oral Bacteria Xi Liu a,b,
Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 2018 Supporting Information Protein-Mediated Anti-Adhesion Surface against Oral Bacteria Xi Liu a,b,
Data Sheet. PCSK9[Biotinylated]-LDLR Binding Assay Kit Catalog # 72002
![Data Sheet. PCSK9[Biotinylated]-LDLR Binding Assay Kit Catalog # 72002 Data Sheet. PCSK9[Biotinylated]-LDLR Binding Assay Kit Catalog # 72002](/thumbs/79/80181511.jpg) Data Sheet PCSK9[Biotinylated]-LDLR Binding Assay Kit Catalog # 72002 DESCRIPTION: The PCSK9[Biotinylated]-LDLR Binding Assay Kit is designed for screening and profiling purposes. PCSK9 is known to function
Data Sheet PCSK9[Biotinylated]-LDLR Binding Assay Kit Catalog # 72002 DESCRIPTION: The PCSK9[Biotinylated]-LDLR Binding Assay Kit is designed for screening and profiling purposes. PCSK9 is known to function
LANCE Eu-W1024 ITC Chelate & Europium Standard AD0013 Development grade
 AD0017P-4 (en) 1 LANCE Eu-W1024 ITC Chelate & Europium Standard AD0013 Development grade INTRODUCTION Fluorescent isothiocyanato-activated (ITC-activated) Eu-W1024 chelate is optimized for labelling proteins
AD0017P-4 (en) 1 LANCE Eu-W1024 ITC Chelate & Europium Standard AD0013 Development grade INTRODUCTION Fluorescent isothiocyanato-activated (ITC-activated) Eu-W1024 chelate is optimized for labelling proteins
SensoLyte 490 HIV-1 Protease Assay Kit *Fluorimetric*
 SensoLyte 490 HIV-1 Protease Assay Kit *Fluorimetric* Catalog # 71127 Unit Size Kit Size 1 kit 500 assays (96-well) or 1250 assays (384-well) This kit is optimized to detect the activity of human immunodeficiency
SensoLyte 490 HIV-1 Protease Assay Kit *Fluorimetric* Catalog # 71127 Unit Size Kit Size 1 kit 500 assays (96-well) or 1250 assays (384-well) This kit is optimized to detect the activity of human immunodeficiency
Fluorescent probes for detecting monoamine oxidase activity and cell imaging
 Fluorescent probes for detecting monoamine oxidase activity and cell imaging Xuefeng Li, Huatang Zhang, Yusheng Xie, Yi Hu, Hongyan Sun *, Qing Zhu * Supporting Information Table of Contents 1. General
Fluorescent probes for detecting monoamine oxidase activity and cell imaging Xuefeng Li, Huatang Zhang, Yusheng Xie, Yi Hu, Hongyan Sun *, Qing Zhu * Supporting Information Table of Contents 1. General
Mouse Cathepsin B ELISA Kit
 GenWay Biotech, Inc. 6777 Nancy Ridge Drive San Diego, CA 92121 Phone: 858.458.0866 Fax: 858.458.0833 Email: techline@genwaybio.com http://www.genwaybio.com Mouse Cathepsin B ELISA Kit Catalog No. GWB-ZZD154
GenWay Biotech, Inc. 6777 Nancy Ridge Drive San Diego, CA 92121 Phone: 858.458.0866 Fax: 858.458.0833 Email: techline@genwaybio.com http://www.genwaybio.com Mouse Cathepsin B ELISA Kit Catalog No. GWB-ZZD154
Chromatin IP (Isw2) Fix soln: 11% formaldehyde, 0.1 M NaCl, 1 mm EDTA, 50 mm Hepes-KOH ph 7.6. Freshly prepared. Do not store in glass bottles.
 Chromatin IP (Isw2) 7/01 Toshi last update: 06/15 Reagents Fix soln: 11% formaldehyde, 0.1 M NaCl, 1 mm EDTA, 50 mm Hepes-KOH ph 7.6. Freshly prepared. Do not store in glass bottles. 2.5 M glycine. TBS:
Chromatin IP (Isw2) 7/01 Toshi last update: 06/15 Reagents Fix soln: 11% formaldehyde, 0.1 M NaCl, 1 mm EDTA, 50 mm Hepes-KOH ph 7.6. Freshly prepared. Do not store in glass bottles. 2.5 M glycine. TBS:
ab Lipid Peroxidation (MDA) Assay kit (Colorimetric/ Fluorometric)
 Version 10b Last updated 19 December 2018 ab118970 Lipid Peroxidation (MDA) Assay kit (Colorimetric/ Fluorometric) For the measurement of Lipid Peroxidation in plasma, cell culture and tissue extracts.
Version 10b Last updated 19 December 2018 ab118970 Lipid Peroxidation (MDA) Assay kit (Colorimetric/ Fluorometric) For the measurement of Lipid Peroxidation in plasma, cell culture and tissue extracts.
Supplementary Figures
 Supplementary Figures Absorption 4 3 2 1 Intensity Energy U(R) relaxation ~~~ ~~~~~~ 2 3 4 1 S S 1 2 3 4 1 Fluoescence 4 3 2 1 Intensity H-aggregation ~~~~ J-aggregation Absorption Emission Vibrational
Supplementary Figures Absorption 4 3 2 1 Intensity Energy U(R) relaxation ~~~ ~~~~~~ 2 3 4 1 S S 1 2 3 4 1 Fluoescence 4 3 2 1 Intensity H-aggregation ~~~~ J-aggregation Absorption Emission Vibrational
DELFIA Eu-DTPA ITC Chelate & Europium Standard
 AD0026P-3 (en) 1 DELFIA Eu-DTPA ITC Chelate & AD0021 Europium Standard For Research Use Only INTRODUCTION DELFIA Eu-DTPA ITC Chelate is optimized for the europium labelling of proteins and peptides for
AD0026P-3 (en) 1 DELFIA Eu-DTPA ITC Chelate & AD0021 Europium Standard For Research Use Only INTRODUCTION DELFIA Eu-DTPA ITC Chelate is optimized for the europium labelling of proteins and peptides for
A ph-dependent Charge Reversal Peptide for Cancer Targeting
 Supporting Information A ph-dependent Charge Reversal Peptide for Cancer Targeting Naoko Wakabayashi 1, Yoshiaki Yano 1, Kenichi Kawano 1, and Katsumi Matsuzaki 1 1 Graduate School of Pharmaceutical Sciences,
Supporting Information A ph-dependent Charge Reversal Peptide for Cancer Targeting Naoko Wakabayashi 1, Yoshiaki Yano 1, Kenichi Kawano 1, and Katsumi Matsuzaki 1 1 Graduate School of Pharmaceutical Sciences,
Mouse TrkB ELISA Kit
 Mouse TrkB ELISA Kit CATALOG NO: IRKTAH5472 LOT NO: SAMPLE INTENDED USE For quantitative detection of mouse TrkB in cell culture supernates, cell lysates and tissue homogenates. BACKGROUND TrkB receptor
Mouse TrkB ELISA Kit CATALOG NO: IRKTAH5472 LOT NO: SAMPLE INTENDED USE For quantitative detection of mouse TrkB in cell culture supernates, cell lysates and tissue homogenates. BACKGROUND TrkB receptor
VaTx1 VaTx2 VaTx3. VaTx min Retention Time (min) Retention Time (min)
 a Absorbance (mau) 5 2 5 3 4 5 6 7 8 9 6 2 3 4 5 6 VaTx2 High Ca 2+ Low Ca 2+ b 38.2 min Absorbance (mau) 3 2 3 4 5 3 2 VaTx2 39.3 min 3 4 5 3 2 4. min 3 4 5 Supplementary Figure. Toxin Purification For
a Absorbance (mau) 5 2 5 3 4 5 6 7 8 9 6 2 3 4 5 6 VaTx2 High Ca 2+ Low Ca 2+ b 38.2 min Absorbance (mau) 3 2 3 4 5 3 2 VaTx2 39.3 min 3 4 5 3 2 4. min 3 4 5 Supplementary Figure. Toxin Purification For
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION DOI: 10.1038/NNANO.2012.80 Protein-Inorganic Hybrid Nanoflowers Jun Ge, Jiandu Lei, and Richard N. Zare Supporting Online Material Materials Proteins including albumin from bovine
SUPPLEMENTARY INFORMATION DOI: 10.1038/NNANO.2012.80 Protein-Inorganic Hybrid Nanoflowers Jun Ge, Jiandu Lei, and Richard N. Zare Supporting Online Material Materials Proteins including albumin from bovine
XCF TM COMPLETE Exosome and cfdna Isolation Kit (for Serum & Plasma)
 XCF TM COMPLETE Exosome and cfdna Isolation Kit (for Serum & Plasma) Cat# XCF100A-1 User Manual Store kit components at +4ºC and +25ºC Version 1 2/2/2017 A limited-use label license covers this product.
XCF TM COMPLETE Exosome and cfdna Isolation Kit (for Serum & Plasma) Cat# XCF100A-1 User Manual Store kit components at +4ºC and +25ºC Version 1 2/2/2017 A limited-use label license covers this product.
Instructions for Use. RealStar Influenza Screen & Type RT-PCR Kit /2017 EN
 Instructions for Use RealStar Influenza Screen & Type RT-PCR Kit 4.0 05/2017 EN RealStar Influenza Screen & Type RT-PCR Kit 4.0 For research use only! (RUO) 164003 INS-164000-EN-S01 96 05 2017 altona
Instructions for Use RealStar Influenza Screen & Type RT-PCR Kit 4.0 05/2017 EN RealStar Influenza Screen & Type RT-PCR Kit 4.0 For research use only! (RUO) 164003 INS-164000-EN-S01 96 05 2017 altona
In situ drug-receptor binding kinetics in single cells: a quantitative label-free study of anti-tumor drug resistance
 Supplementary Information for In situ drug-receptor binding kinetics in single cells: a quantitative label-free study of anti-tumor drug resistance Wei Wang 1, Linliang Yin 2,3, Laura Gonzalez-Malerva
Supplementary Information for In situ drug-receptor binding kinetics in single cells: a quantitative label-free study of anti-tumor drug resistance Wei Wang 1, Linliang Yin 2,3, Laura Gonzalez-Malerva
Triptycene-Based Small Molecules Modulate (CAG) (CTG) Repeat Junctions
 Electronic Supplementary Material (ESI) for Chemical Science. This journal is The Royal Society of Chemistry 2015 Triptycene-Based Small Molecules Modulate (CAG) (CTG) Repeat Junctions Stephanie A. Barros
Electronic Supplementary Material (ESI) for Chemical Science. This journal is The Royal Society of Chemistry 2015 Triptycene-Based Small Molecules Modulate (CAG) (CTG) Repeat Junctions Stephanie A. Barros
Cholesterol determination using protein-templated fluorescent gold nanocluster probes
 Electronic Supplementary Information for Cholesterol determination using protein-templated fluorescent gold nanocluster probes Xi Chen and Gary A. Baker* Department of Chemistry, University of Missouri-Columbia,
Electronic Supplementary Information for Cholesterol determination using protein-templated fluorescent gold nanocluster probes Xi Chen and Gary A. Baker* Department of Chemistry, University of Missouri-Columbia,
Trypsin Mass Spectrometry Grade
 058PR-03 G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name Trypsin Mass Spectrometry Grade A Chemically Modified, TPCK treated, Affinity Purified
058PR-03 G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name Trypsin Mass Spectrometry Grade A Chemically Modified, TPCK treated, Affinity Purified
HIV-1 p24 ELISA Pair Set Cat#: orb54951 (ELISA Manual)
 HIV-1 p24 ELISA Pair Set Cat#: orb54951 (ELISA Manual) BACKGROUND Human Immunodeficiency Virus ( HIV ) can be divided into two major types, HIV type 1 (HIV-1) and HIV type 2 (HIV-2). HIV-1 is related to
HIV-1 p24 ELISA Pair Set Cat#: orb54951 (ELISA Manual) BACKGROUND Human Immunodeficiency Virus ( HIV ) can be divided into two major types, HIV type 1 (HIV-1) and HIV type 2 (HIV-2). HIV-1 is related to
PicoProbe Acetyl CoA Assay Kit
 ab87546 PicoProbe Acetyl CoA Assay Kit Instructions for Use For the rapid, sensitive and accurate measurement of Acetyl CoA levels in various samples. This product is for research use only and is not intended
ab87546 PicoProbe Acetyl CoA Assay Kit Instructions for Use For the rapid, sensitive and accurate measurement of Acetyl CoA levels in various samples. This product is for research use only and is not intended
Trypsin Digestion Mix
 G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name 239PR Trypsin Digestion Mix Provides optimal buffered conditions for in gel trypsin digestion
G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name 239PR Trypsin Digestion Mix Provides optimal buffered conditions for in gel trypsin digestion
Characterization of the DNA-mediated Oxidation of Dps, a Bacterial Ferritin
 SUPPORTING INFORMATION Characterization of the DNA-mediated Oxidation of Dps, a Bacterial Ferritin Anna R. Arnold, Andy Zhou, and Jacqueline K. Barton Division of Chemistry and Chemical Engineering, California
SUPPORTING INFORMATION Characterization of the DNA-mediated Oxidation of Dps, a Bacterial Ferritin Anna R. Arnold, Andy Zhou, and Jacqueline K. Barton Division of Chemistry and Chemical Engineering, California
Lipoprotein Lipase Activity Assay Kit (Fluorometric)
 Lipoprotein Lipase Activity Assay Kit (Fluorometric) Catalog Number KA4538 100 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General
Lipoprotein Lipase Activity Assay Kit (Fluorometric) Catalog Number KA4538 100 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General
Human Cathepsin D ELISA Kit
 GenWay Biotech, Inc. 6777 Nancy Ridge Drive San Diego, CA 92121 Phone: 858.458.0866 Fax: 858.458.0833 Email: techline@genwaybio.com http://www.genwaybio.com Human Cathepsin D ELISA Kit Catalog No. GWB-J4JVV9
GenWay Biotech, Inc. 6777 Nancy Ridge Drive San Diego, CA 92121 Phone: 858.458.0866 Fax: 858.458.0833 Email: techline@genwaybio.com http://www.genwaybio.com Human Cathepsin D ELISA Kit Catalog No. GWB-J4JVV9
PRODUCT: RNAzol BD for Blood May 2014 Catalog No: RB 192 Storage: Store at room temperature
 PRODUCT: RNAzol BD for Blood May 2014 Catalog No: RB 192 Storage: Store at room temperature PRODUCT DESCRIPTION. RNAzol BD is a reagent for isolation of total RNA from whole blood, plasma or serum of human
PRODUCT: RNAzol BD for Blood May 2014 Catalog No: RB 192 Storage: Store at room temperature PRODUCT DESCRIPTION. RNAzol BD is a reagent for isolation of total RNA from whole blood, plasma or serum of human
Supplemental Data Figure S1 Effect of TS2/4 and R6.5 antibodies on the kinetics of CD16.NK-92-mediated specific lysis of SKBR-3 target cells.
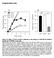 Supplemental Data Figure S1. Effect of TS2/4 and R6.5 antibodies on the kinetics of CD16.NK-92-mediated specific lysis of SKBR-3 target cells. (A) Specific lysis of IFN-γ-treated SKBR-3 cells in the absence
Supplemental Data Figure S1. Effect of TS2/4 and R6.5 antibodies on the kinetics of CD16.NK-92-mediated specific lysis of SKBR-3 target cells. (A) Specific lysis of IFN-γ-treated SKBR-3 cells in the absence
RayBio Human Granzyme B ELISA Kit
 RayBio Human Granzyme B ELISA Kit Catalog #: ELH-GZMB User Manual Last revised April 15, 2016 Caution: Extraordinarily useful information enclosed ISO 13485 Certified 3607 Parkway Lane, Suite 100 Norcross,
RayBio Human Granzyme B ELISA Kit Catalog #: ELH-GZMB User Manual Last revised April 15, 2016 Caution: Extraordinarily useful information enclosed ISO 13485 Certified 3607 Parkway Lane, Suite 100 Norcross,
ExoQuick Exosome Isolation and RNA Purification Kits
 ExoQuick Exosome Isolation and RNA Purification Kits Cat # EQ806A-1, EQ806TC-1, EQ808A-1 User Manual Storage: Please see individual components Version 2 8/14/2018 A limited-use label license covers this
ExoQuick Exosome Isolation and RNA Purification Kits Cat # EQ806A-1, EQ806TC-1, EQ808A-1 User Manual Storage: Please see individual components Version 2 8/14/2018 A limited-use label license covers this
Supporting Information
 Translation of DNA into Synthetic N-Acyloxazolidines Xiaoyu Li, Zev. J. Gartner, Brian N. Tse and David R. Liu* Department of Chemistry and Chemical Biology, Harvard University, Cambridge, Massachusetts
Translation of DNA into Synthetic N-Acyloxazolidines Xiaoyu Li, Zev. J. Gartner, Brian N. Tse and David R. Liu* Department of Chemistry and Chemical Biology, Harvard University, Cambridge, Massachusetts
Supplementary Data. Different volumes of ethanol or calcium solution were slowly added through one of four
 Supplementary Data METHODS Liposome preparation Different volumes of ethanol or calcium solution were slowly added through one of four methods: Method I, no ethanol or calcium solution; Method II, exactly
Supplementary Data METHODS Liposome preparation Different volumes of ethanol or calcium solution were slowly added through one of four methods: Method I, no ethanol or calcium solution; Method II, exactly
Data Sheet. Fluorogenic HDAC 8 Assay Kit Catalog #: 50068
 Data Sheet Fluorogenic HDAC 8 Assay Kit Catalog #: 50068 DESCRIPTION: The Fluorogenic HDAC 8 Assay Kit is a complete assay system designed to measure histone deacetylase (HDAC) 8 activity for screening
Data Sheet Fluorogenic HDAC 8 Assay Kit Catalog #: 50068 DESCRIPTION: The Fluorogenic HDAC 8 Assay Kit is a complete assay system designed to measure histone deacetylase (HDAC) 8 activity for screening
DELFIA Tb-N1 DTA Chelate & Terbium Standard
 AD0029P-1 (en) 1 DELFIA Tb-N1 DTA Chelate & AD0012 Terbium Standard For Research Use Only INTRODUCTION DELFIA Tb-N1 DTA Chelate is optimized for the terbium labeling of proteins and peptides for use in
AD0029P-1 (en) 1 DELFIA Tb-N1 DTA Chelate & AD0012 Terbium Standard For Research Use Only INTRODUCTION DELFIA Tb-N1 DTA Chelate is optimized for the terbium labeling of proteins and peptides for use in
Instructions for Use. APO-AB Annexin V-Biotin Apoptosis Detection Kit 100 tests
 3URGXFW,QIRUPDWLRQ Sigma TACS Annexin V Apoptosis Detection Kits Instructions for Use APO-AB Annexin V-Biotin Apoptosis Detection Kit 100 tests For Research Use Only. Not for use in diagnostic procedures.
3URGXFW,QIRUPDWLRQ Sigma TACS Annexin V Apoptosis Detection Kits Instructions for Use APO-AB Annexin V-Biotin Apoptosis Detection Kit 100 tests For Research Use Only. Not for use in diagnostic procedures.
ab Homocysteine Assay Kit (Fluorometric) 1
 Version 1 Last updated 25 May 2018 ab208559 Homocysteine Assay Kit (Fluorometric) For the measurement of homocysteine in mammalian plasma and serum. This product is for research use only and is not intended
Version 1 Last updated 25 May 2018 ab208559 Homocysteine Assay Kit (Fluorometric) For the measurement of homocysteine in mammalian plasma and serum. This product is for research use only and is not intended
Supplementary Figure 1. Method development.
 Supplementary Figure 1 Method development. Titration experiments to determine standard antibody:lysate concentration. Lysates (~2 mg of total proteins) were prepared from cells expressing FLAG- tagged
Supplementary Figure 1 Method development. Titration experiments to determine standard antibody:lysate concentration. Lysates (~2 mg of total proteins) were prepared from cells expressing FLAG- tagged
Human Carbamylated LDL ELISA Kit (CBL-LDL Quantitation)
 Product Manual Human Carbamylated LDL ELISA Kit (CBL-LDL Quantitation) Catalog Number MET-5032 96 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Lipoproteins are submicroscopic
Product Manual Human Carbamylated LDL ELISA Kit (CBL-LDL Quantitation) Catalog Number MET-5032 96 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Lipoproteins are submicroscopic
Work-flow: protein sample preparation Precipitation methods Removal of interfering substances Specific examples:
 Dr. Sanjeeva Srivastava IIT Bombay Work-flow: protein sample preparation Precipitation methods Removal of interfering substances Specific examples: Sample preparation for serum proteome analysis Sample
Dr. Sanjeeva Srivastava IIT Bombay Work-flow: protein sample preparation Precipitation methods Removal of interfering substances Specific examples: Sample preparation for serum proteome analysis Sample
Human Immunodeficiency Virus type 1 (HIV-1) gp120 / Glycoprotein 120 ELISA Pair Set
 Human Immunodeficiency Virus type 1 (HIV-1) gp120 / Glycoprotein 120 ELISA Pair Set Catalog Number : SEK11233 To achieve the best assay results, this manual must be read carefully before using this product
Human Immunodeficiency Virus type 1 (HIV-1) gp120 / Glycoprotein 120 ELISA Pair Set Catalog Number : SEK11233 To achieve the best assay results, this manual must be read carefully before using this product
2-Deoxyglucose Assay Kit (Colorimetric)
 2-Deoxyglucose Assay Kit (Colorimetric) Catalog Number KA3753 100 assays Version: 01 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General Information...
2-Deoxyglucose Assay Kit (Colorimetric) Catalog Number KA3753 100 assays Version: 01 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General Information...
PFK Activity Assay Kit (Colorimetric)
 PFK Activity Assay Kit (Colorimetric) Catalog Number KA3761 100 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General Information...
PFK Activity Assay Kit (Colorimetric) Catalog Number KA3761 100 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General Information...
Alkaline Phosphatase Assay Kit (Fluorometric)
 Alkaline Phosphatase Assay Kit (Fluorometric) Catalog Number KA0820 500 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General Information...
Alkaline Phosphatase Assay Kit (Fluorometric) Catalog Number KA0820 500 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General Information...
Analysis of small RNAs from Drosophila Schneider cells using the Small RNA assay on the Agilent 2100 bioanalyzer. Application Note
 Analysis of small RNAs from Drosophila Schneider cells using the Small RNA assay on the Agilent 2100 bioanalyzer Application Note Odile Sismeiro, Jean-Yves Coppée, Christophe Antoniewski, and Hélène Thomassin
Analysis of small RNAs from Drosophila Schneider cells using the Small RNA assay on the Agilent 2100 bioanalyzer Application Note Odile Sismeiro, Jean-Yves Coppée, Christophe Antoniewski, and Hélène Thomassin
For Research Use Only Ver
 INSTRUCTION MANUAL Quick-DNA/RNA Pathogen Miniprep Catalog Nos. R1042 & R1043 Highlights Spin-column purification of pathogen (virus, bacteria, protozoa) DNA/RNA from a wide variety of vectors (mosquitoes,
INSTRUCTION MANUAL Quick-DNA/RNA Pathogen Miniprep Catalog Nos. R1042 & R1043 Highlights Spin-column purification of pathogen (virus, bacteria, protozoa) DNA/RNA from a wide variety of vectors (mosquitoes,
Determination of Tetracyclines in Chicken by Solid-Phase Extraction and High-Performance Liquid Chromatography
 Determination of Tetracyclines in Chicken by Solid-Phase Extraction and High-Performance Liquid Chromatography Application ote Food Safety Authors Chen-Hao Zhai and Yun Zou Agilent Technologies Co. Ltd.
Determination of Tetracyclines in Chicken by Solid-Phase Extraction and High-Performance Liquid Chromatography Application ote Food Safety Authors Chen-Hao Zhai and Yun Zou Agilent Technologies Co. Ltd.
Human IL-2. Pre-Coated ELISA Kit
 Human IL-2 (Interleukin 2) Pre-Coated ELISA Kit Catalog No: 90-2083 1 96 well Format (96 tests) Detection Range: 31.2 2000 pg/ml Sensitivity: < 18.75 pg/ml This immunoassay kit allows for the in vitro
Human IL-2 (Interleukin 2) Pre-Coated ELISA Kit Catalog No: 90-2083 1 96 well Format (96 tests) Detection Range: 31.2 2000 pg/ml Sensitivity: < 18.75 pg/ml This immunoassay kit allows for the in vitro
Supplementary Figure 1. Chemical structures of activity-based probes (ABPs) and of click reagents used in this study.
 Supplementary Figure 1. Chemical structures of activity-based probes (ABPs) and of click reagents used in this study. In this study, one fluorophosphonate (FP, 1), three nitrophenol phosphonate probes
Supplementary Figure 1. Chemical structures of activity-based probes (ABPs) and of click reagents used in this study. In this study, one fluorophosphonate (FP, 1), three nitrophenol phosphonate probes
ABIOpure TM Viral (version 2.0)
 ABIOpure TM Viral (version 2.0) DNA/RNA Extraction Handbook Cat No: M561VT50 FOR RESEARCH USE ONLY Table of Contents Contents Page Kit Components 3 Precautions 3 Stability & Storage 4 General Description
ABIOpure TM Viral (version 2.0) DNA/RNA Extraction Handbook Cat No: M561VT50 FOR RESEARCH USE ONLY Table of Contents Contents Page Kit Components 3 Precautions 3 Stability & Storage 4 General Description
For in vitro Veterinary Diagnostics only. Kylt Rotavirus A. Real-Time RT-PCR Detection.
 For in vitro Veterinary Diagnostics only. Kylt Rotavirus A Real-Time RT-PCR Detection www.kylt.eu DIRECTION FOR USE Kylt Rotavirus A Real-Time RT-PCR Detection A. General Kylt Rotavirus A products are
For in vitro Veterinary Diagnostics only. Kylt Rotavirus A Real-Time RT-PCR Detection www.kylt.eu DIRECTION FOR USE Kylt Rotavirus A Real-Time RT-PCR Detection A. General Kylt Rotavirus A products are
