Sulfur fertilization influences the sulphur species composition in Allium sativum: sulfomics using HPLC-ICP-MS/MS-ESI-MS/MS
|
|
|
- Iris Roberts
- 5 years ago
- Views:
Transcription
1 Electronic upplementary Material (EI) for Metallomics. This journal is The Royal ociety of Chemistry 2017 Electronic supplement for ulfur fertilization influences the sulphur species composition in Allium sativum: sulfomics using HPLC-ICP-M/M-EI-M/M Andrea Raab, Marilena Ronzan, Joerg Feldmann TELA (Trace Element peciation Laboratory), University of Aberdeen, Chemistry, Meston Walk, Aberdeen, AB243UE, cotland, UK Page number: Hoagland solution 2. Details of compounds (M spectra, EIC+ICP-M/M trace, fragmentation by M/M) Figures Cycloalliin, aliin & isoalliin 2.2. Methiin, propiin and methionine allyl-cysteine (AC, deoxyalliin) and derivatives 2.4. γ-glutamyl--allyl-cysteine (GA and γ-glutamyl--1-propenylcysteine (GP 2.5. γ-glutamyl--2-propenylcysteine sulfoxide GAC() and γ- glutamyl--1-propenylcysteine sulfoxide GPC() 2.6. γ-glutamyl--methyl-cysteine (GM and γ-glutamylhomocysteine (C264) 2.7. Glutathione (GH and GG) propenylmercaptoglutathione, -2- propenylmercaptoglutathione and -propylmercaptoglutathione (C379) 2.9. xidised forms of -1-propenylmercaptoglutathione and -2- propenylmercaptoglutathione (C395) Phytochelatin 2 (PC-2) and derivatives Precursor molecules for -alk(en)ylcysteine sulfoxides Amount of sulphur associated with non-resolved chromatographic peaks 3. Results from principal component analysis and Cluster analysis 3.1. Loading plot (PCA) 3.2. Dendrogram 4. Example of ICP-M/M traces for all plant parts fertilized with different amount of sulphur 5. Example of peak area integration 1 P age
2 1. Composition of Hoagland solution ulphur level 0.5 mm: 1.25 mm KN 3, 1.50 mm Ca(N 3 ) 2, 0.75 mm Mg 4, 0.50 mm KH 2 P 4, 50 µm KCl, 50 µm H 3 B 3, 10 µm Mn 4, 2.0 µm Zn 4, 1.5 µm Cu 4, 0.5 µm Na 2 Mo 4.2H 2 0, 0.1 mm Na 2 3 i, and 72 µm Fe- EDTA ph 6.0 ulphur level 0.1 mm: Plant group exposed to 0.1 M sulphur: Mg 4 was replaced by 0.75 mm MgCl 2 ulphur level 2 mm: Plant group exposed to 2 mm sulphur: Mg 4 2 mm Mg 4 was used instead of 0.75 mm 2 P age
3 2. Details of compounds (EI-M spectra, EIC+ICP-M/M trace, fragmentation by EI-M/M) Figures 1-26 The following tables summarize the molecular structure, molecular weight, theoretical and measured [M+H] + and retention time of the compounds and contain the average compound or sulphur concentrations Cycloalliin, aliin & isoalliin The identities of cyclo-alliin, alliin and isoalliin were deduced by comparison of their q-tf-ei-m/m spectra. E-M/M spectra of the q-tf instrument for alliin contained predominantly signals at m/z (C 3 H 7 N 3 ) and m/z (C 3 H 6 N 2 ) (Fig. 2). Isoalliin (Fig. 3) showed additionally a fragment at m/z identical to one of the major fragments of cyclo-alliin (C 5 H 8 N). The different position of the double bond compared to alliin allows stabilisation of isoalliinfragments by cyclisation. The other major fragments of cyclo-alliin were m/z (loss of water) and m/z (C 5 H 7 2 ) (Fig. 1). Table 1 Alliin and isoalliin content in root and bulb in mmol compound kg -1 d.w. (mean ± standard deviation, n = 9 per group), cylcoalliin; in brackets compound average in % of total sulphur, database ID: Chempider. H 3 C Cycloalliin* Alliin Isoalliin N H H C H 2 Molecular formula C 6 H 11 N 3 C 6 H 11 N 3 C 6 H 11 N 3 MW (g mol -1 ) [M+H] + (theoretical) [M + H] + (measured) RT (min) Database ID Root (0.1 mm ) 14 ± 15 a (7.1 %) 4.1 ± 3.4 a (2.0 %) Root (0.5 mm ) 19 ± 16 b (6.7%) 4.3 ± 4.3 b (1.5 %) Root (2 mm ) 79 ± 41 a,b (17%) 30 ± 27 a,b (6.4%) Bulb (0.1 2 mm ) 21 ± 8.1 (16 %) 3.7 ± 2.1 (2.8 %) *cycloalliin + methiin + methylcysteine, amount of associated sulphur see table 13 a,b : statistically significant difference between groups by ne Way ANVA p < H C H 3 2 H 3 P age
4 H 3 C A) N H H C 6 H 11 N 3 relative intensity ICP-M/M m/z 178 relative intensity EI-M CH 3 2 -CH 4 N -H 2 Fig. 1 Cycloalliin; panel A) structure; panel B: EIC of m/z + ICP-M/M trace of root extract exposed to 2 mm, panel q-tf EI-M-spectra, panel EI- M/M spectra with some annotated fragments, shown is the composition of the loss for the individual fragment calculated from the parent mass; for HPLC + ICP- M/M and EI-M conditions see main text under instrumentation. 4 P age
5 A) N H 2 C 6 H 11 N 3 CH 2 H relative intensity ICP-M/M m/z relative intensity EI-M -C 3 H 5 -C 3 H 6-2 Fig. 2 Alliin; panel A) structure; panel B: EIC of m/z + ICP-M/M trace of root extract exposed to 2 mm, panel q-tf EI-M-spectra, panel EI- M/M spectra with some annotated fragments, shown is the composition of the loss for the individual fragment calculated from the parent mass; for HPLC + ICP- M/M and EI-M conditions see main text under instrumentation. 5 P age
6 A) N H 2 CH 3 H relative intensity ICP-M/M m/z 178 relative intensity EI-M C 6 H 11 N CH 3 2 -C 3 H C 3 H 6-2 Fig. 3 Isoalliin; panel A) structure; panel B: EIC of m/z + ICP-M/M trace of root extract exposed to 2 mm, panel q-tf EI-M-spectra, panel EI- M/M spectra with some annotated fragments, shown is the composition of the loss for the individual fragment calculated from the parent mass; for HPLC + ICP- M/M and EI-M conditions see main text under instrumentation. 6 P age
7 2.2. Methiin, propiin and methionine It was not possible to confirm the identity of methiin other than from the accurate m/z ratio (Δppm = 0.54). The signal was not intense enough in any of the samples for fragmentation by either rbitrap EI-M or q-tf-ei-m (Fig. 4). The retention time is similar to those published by others. Cycloalliin (Table 1) and methylcysteine co-eluted with methiin. Propiin (Fig. 5) eluted together with glutathione (Fig. 17) and methionine (Fig. 6). In addition to the accurate [M+H] +, propiin and methionine showed the expected fragmentation patterns, with propiin fragmenting to m/z (loss of CH 2 2 ), m/z (loss of CH 3 3 ) and m/z (loss of 2 and C 3 H 7 ) (Fig. 5). Methionine fragments of m/z (loss of C 2 ) m/z (loss of 2 ) (Fig. 6) confirmed its presence together with its accurate mass. Table 2 methiin propiin and methionine, database ID: Chempider. C H 3 Methiin* Propiin + Methionine + 2 C H 3 2 C H 3 H H Molecular formula C 4 H 9 N 3 C 6 H 13 N 3 C 5 H 11 N 2 MW (g mol -1 ) [M+H] + (theoretical) [M+H] + (measured) RT (min) Database ID Root (0.1 mm ) Root (0.5 mm ) Root (2 mm ) Bulb (0.1 2 mm ) *cycloalliin + methiin + methylcysteine, amount of associated sulphur see table 13 + propiin + glutathione + methionine, amount of associated sulphur see table 13 H 2 7 P age
8 A) C H 3 2 H C 4 H 9 N 3 relative intensity ICP-M/M m/z 152 relative intensity EI-M Fig. 4 Methiin; panel A) structure; panel B: EIC of m/z + ICP-M/M trace of root extract exposed to 2 mm, panel q-tf EI-M-spectra, panel no fragmentation; for HPLC + ICP-M/M and EI-M conditions see main text under instrumentation. 8 P age
9 A) N H 2 C 6 H 13 N 3 CH 3 H relative intensity ICP-M/M m/z 180 relative intensity EI-M CH 2 2 -CH 3 3 -C 3 H Fig. 5 Propiin; panel A) structure; panel B: EIC of m/z + ICP-M/M trace of root extract exposed to 2 mm, panel q-tf EI-M-spectra, panel EI- M/M spectra with some annotated fragments, shown is the composition of the loss for the individual fragment calculated from the parent mass; for HPLC + ICP- M/M and EI-M conditions see main text under instrumentation. 9 P age
10 A) C 5 H 11 N 2 H H 2 N H 3 C relative intensity ICP-M/M m/z 150 relative intensity EI-M C 2-2 Fig. 6 Methionine; panel A) structure; panel B: EIC of m/z + ICP-M/M trace of root extract exposed to 2 mm, panel q-tf EI-M-spectra, panel EI-M/M spectra with some annotated fragments, shown is the composition of the loss for the individual fragment calculated from the parent mass; for HPLC + ICP- M/M and EI-M conditions see main text under instrumentation. 10 P age
11 2.3. -allyl-cysteine (AC, deoxyalliin) and derivatives Methyl-cysteine showed a fragment at m/z (loss of 2 ) (Fig. 7) and no other significant fragments due to settings of the mass range for both rbitrap EI- M and q-tf-ei-m. -allyl-cysteine showed fragments at m/z (amine loss), m/z (loss of C 2 ) and m/z (loss of 2 and C 2 ) (Fig. 8). AC was also identified in garlic by Yamazaki et al. 1 eluting after GMC identical to the retention behaviour under the conditions used here. -propyl-cysteine did not occur in either bulbs or roots. Table 3 -allyl-cysteine in bulb and root in µmol compound kg -1 d.w. (mean ± standard deviation, n = 9 per group), methyl cysteine, database ID: Chempider. -allyl-cysteine -propyl-cysteine Methylcysteine* C H 2 2 H Molecular formula C 6 H 12 N 2 C 6 H 14 N 2 C 4 H 9 N 2 MW (g mol -1 ) [M + H] + (theoretical) [M+H] + (measured) RT (min) Database ID Root (0.1 mm ) 325 ± Root (0.5 mm ) 256 ± Root (2 mm ) 414 ± Bulb (0.1 2 mm ) 143 ± *cycloalliin + methiin + methylcysteine, amount of associated sulphur see table 13 C H 3 2 H C H 3 2 H 11 P age
12 A) C H 3 2 H C 4 H 9 N 2 relative intensity ICP-M/M m/z 136 relative intensity EI-M Fig. 7 Methyl-cysteine; panel A) structure; panel B: EIC of m/z + ICP- M/M trace of root extract exposed to 2 mm, panel q-tf EI-M-spectra, panel EI-M/M spectra with some annotated fragments, shown is the composition of the loss for the individual fragment calculated from the parent mass; for HPLC + ICP-M/M and EI-M conditions see main text under instrumentation. 12 P age
13 A) N H 2 H relative intensity ICP-M/M m/z 162 relative intensity EI-M CH 2 C 6 H 12 N C 2 -C 2 Fig. 8 -allyl-cysteine; panel A) structure; panel B: EIC of m/z + ICP- M/M trace of root extract exposed to 2 mm, panel q-tf EI-M-spectra, panel EI-M/M spectra with some annotated fragments, shown is the composition of the loss for the individual fragment calculated from the parent mass; for HPLC + ICP-M/M and EI-M conditions see main text under instrumentation. 13 P age
14 2.4. γ-glutamyl--allyl-cysteine (GA and γ-glutamyl--1- propenyl-cysteine (GP GAC and GPC were identified by their accurate m/z ratio and their fragmentation pattern. The assignment of the allyl respectively the propenyl-group to either compound was done based on comparison with published retention times, which all showed that compounds containing the allyl-group elute before the related compound containing a propenyl-group. The main fragment for both compounds was m/z (C 6 H 9 2 ) followed by m/z (C 6 H 12 N 2, loss of γ-glu) and m/z (C 5 H 8 N 3, loss of propenyl/allyl-cysteine) (Fig. 9 and 10). Identification and quantification of both compounds in bulbs was uncomplicated. Both compounds showed clear signals in the ICP-M/M trace and the EI-M extracted ion chromatogram (Fig. 11b). GAC in root was similarly clear to identify. GPC identification/quantification was more complicated, since the extracted ion chromatogram from roots showed two clearly separated signals at m/z (with the same fragmentation pattern) at RT 17.0 and 17.6 min overlapping one broad sulphur signal from the ICP-M/M (Fig. 10 and 11a). The extracted ion chromatogram of bulbs showed one dominant signal at 17.6 min associated with one sulphur signal from the ICP-M/M. It was not possible to identify any other coeluting sulphur compounds in root extracts and the existence of this double peak in the EIC remained unexplained (Fig. 11 comparison of bulb and root). Table 4 GAC and GPC in µmol compound kg -1 d.w. (mean ± standard deviation, n = 9 per group), database ID: Chempider. GAC GPC H 2 C H H 3 C H Molecular formula C 11 H 19 N 2 5 C 11 H 19 N 2 5 MW (g mol -1 ) [M+H] + (theoretical) [M+H] + (measured) RT (min) Database ID Root (0.1 mm ) 70 ± ± 50 a Root (0.5 mm ) 61 ± ± 59 b Root (2 mm ) 78 ± ± 97 a,b Bulb (0.1-2 mm ) 40 ± ± 660 a,b : statistically significant difference between groups by ne Way ANVA p < H 2 H 14 P age
15 A) H 2 N H H relative intensity ICP-M/M m/z 291 relative intensity EI-M C 11 H 19 N CH 2 -C 5 H 9 N 2 3 -allyl/propenyl-cys -γ-glu Fig. 9 GAC; panel A) structure; panel B: EIC of m/z + ICP-M/M trace of root extract exposed to 2 mm, panel q-tf EI-M-spectra, panel EI- M/M spectra with some annotated fragments, shown is the composition of the loss for the individual fragment calculated from the parent mass; for HPLC + ICP- M/M and EI-M conditions see main text under instrumentation. 15 P age
16 A) H H 2 N H relative intensity ICP-M/M m/z 291 relative intensity EI-M C 11 H 19 N 2 5 CH C 5 H 9 N 2 3 -allyl/propenyl-cys -γ-glu CH 2-2 Fig. 10 GPC; panel A) structure; panel B: EIC of m/z + ICP-M/M trace of root extract exposed to 2 mm, panel q-tf EI-M-spectra, panel EI- M/M spectra with some annotated fragments, shown is the composition of the loss for the individual fragment calculated from the parent mass; for HPLC + ICP- M/M and EI-M conditions see main text under instrumentation. 16 P age
17 A) relative intensity ICP-M/M m/z 291 relative intensity EI-M m/z 291 relative intensity ICP-M/M relative intensity EI-M Fig. 11 Comparison of EIC (GAC and GP and ICP-M/M trace for root (panel A) and bulb (panel b). 17 P age
18 2.5. γ-glutamyl--2-propenylcysteine sulfoxide GAC() and γ- glutamyl--1-propenylcysteine sulfoxide GPC() The main fragments of oxidised GAC and GPC were m/z (γ-glu) and m/z (Fig. 12 and 13). Also the loss of γ-glu at m/z was present in both M spectra, careful comparison of the fragment spectra showed that m/z (loss of C 8 H 12 N 3 ), m/z (C 3 H 6 N 2 ) and m/z (loss of C 4 H 7 ) were more intense in GAC() than GPC() (Fig. 14). Table 5 GAC() and GPC() in root and bulb in µmol compound kg -1 d.w. (mean ± standard deviation, n = 9 per group), database ID: Chempider. H 2 C GAC() H 3 C GPC() H H H H Molecular formula C 11 H 19 N 2 6 C 11 H 19 N 2 6 MW (g mol -1 ) [M+H] + (theoretical) [M+H] + (measured) RT (min) Database ID Root (0.1 mm ) 703 ± 398 a 836 ± 635 b Root (0.5 mm ) 1236 ± ± 1317 Root (2 mm ) 3065 ± 2471 a 1900 ± 856 b Bulb (0.1 2 mm ) 400 ± ± 241 a : statistically significant difference between groups by ne Way ANVA p < b : statistically significant difference between groups by ne Way ANVA p < P age
19 A) H 2 N H H relative intensity ICP-M/M m/z 307 relative intensity EI-M C 11 H 19 N CH 2 -C 6 H 10 N 3 -CH C 3 H 5 -C 3 H 5 -C 4 H 7 + H -C 3 H 5 Fig. 12 GAC(); panel A) structure; panel B: EIC of m/z + ICP-M/M trace of root extract exposed to 2 mm, panel q-tf EI-M-spectra, panel EI- M/M spectra with some annotated fragments, shown is the composition of the loss for the individual fragment calculated from the parent mass; for HPLC + ICP- M/M and EI-M conditions see main text under instrumentation. 19 P age
20 A) H 2 N H C 11 H 19 N 2 6 H 32- m/z 307 relative intensity ICP-M/M relative intensity EI-M CH 3 -C 6 H 10 N 3 -CH C 3 H 5 -C 3 H 5 -C 3 H 5 Fig. 13 GPC(); panel A) structure; panel B: EIC of m/z + ICP-M/M trace of root extract exposed to 2 mm, panel q-tf EI-M-spectra, panel EI- M/M spectra with some annotated fragments, shown is the composition of the loss for the individual fragment calculated from the parent mass; for HPLC + ICP- M/M and EI-M conditions see main text under instrumentation. 20 P age
21 A) Fig. 14 Details of q-tf EI-M/M spectra of GAC() and GPC(), main differences in occurring fragments marked by cycles. 21 P age
22 2.6. γ-glutamyl--methyl-cysteine (GM and γ-glutamylhomocysteine (C264) Three chromatographic peaks with the same molecular composition at m/z (GM were clearly detectable in the extracted ion chromatogram. The detailed study of the EI-M/M fragmentation patterns showed that one of the minor signals at 12.5 min was an in-source fragment of C393, whereas the signal at ca. 8.4 min was a genuine compound with the same m/z but a different fragmentation pattern not yet described in the literature (C264). GMC showed a main fragment at m/z (loss of C 5 H 9 N 2 3 ) (Fig. 15) and C264 s main fragment was m/z (loss C 5 H 8 N 3 ) (Fig. 16). To fragment this way C264 is likely to be γ-glu- HCy (2-amino-5-[(1-carboxy-3-sulfanylpropyl)amino]-5-oxopentanoic acid). Table 6 GMC and γ-glu-hcy in root and bulb in µmol compound kg -1 d.w. (mean ± standard deviation, n = 9 per group), database ID: Chempider. GMC C264 (γ-glu-hcy) C H 3 H H H Molecular formula C 9 H 17 N 2 5 C 9 H 17 N 2 5 MW (g mol -1 ) [M+H] + (theoretical) [M+H] + (measured) RT (min) Database ID Root (0.1 mm ) 699 ± 750 a 288 ± 207 Root (0.5 mm ) 1863 ± 1758 a 278 ± 222 Root (2 mm ) 1695 ± ± 69 Bulb (0.1 2 mm ) 79 ± ± 117 a : statistically significant difference between groups by ne Way ANVA p < H 2 H 22 P age
23 A) H H 2 N H C 9 H 17 N m/z 265 relative intensity ICP-M/M relative intensity EI-M CH C 5 H 9 N 2 3 -C 4 H 8 N 2 -CH 3 + CH CH 2 Fig. 15 GMC; panel A) structure; panel B: EIC of m/z + ICP-M/M trace of root extract exposed to 2 mm, panel q-tf EI-M-spectra, panel EI- M/M spectra with some annotated fragments, shown is the composition of the loss for the individual fragment calculated from the parent mass; for HPLC + ICP- M/M and EI-M conditions see main text under instrumentation. 23 P age
24 A) H H 2 N H C 9 H 17 N 2 5 H 32- m/z 265 relative intensity ICP-M/M relative intensity EI-M C 5 H 8 N 3 -C 5 H 9 N 2 3 -C 4 H 5 N 2 -CH H Fig. 16 C264 (γ-glu-hcy); panel A) structure; panel B: EIC of m/z + ICP- M/M trace of root extract exposed to 2 mm, panel q-tf EI-M-spectra, panel EI-M/M spectra with some annotated fragments, shown is the composition of the loss for the individual fragment calculated from the parent mass; for HPLC + ICP-M/M and EI-M conditions see main text under instrumentation. 24 P age
25 2.7. Glutathione (GH and GG) The molecular masses of reduced and oxidised glutathione were detectable by both EI-M instruments, but EI-M/M fragments of the single charge molecular ions were only found in the data of the rbitrap (shown in Fig. 17 and 18). The main fragments for GH were m/z (loss of γ-glu), m/z (C 5 H 8 N 3 ) and m/z (loss of Gly) (Fig. 17), whereas GG showed main fragments at m/z (loss of γ-glu) and m/z (loss of 2 γ-glu) (Fig. 18). Table 7 GG in µmol compound kg -1 d.w. (mean ± standard deviation, n = 9 per group), GH, database ID: Chempider. H GH + H H H H 2 GG 2 H H Molecular formula C 10 H 18 N 3 6 C 20 H 33 N MW (g mol -1 ) [M+H] + (theoretical) [M+H] + (measured) RT (min) Database ID Root (0.1 mm ) 174 ± 178 Root (0.5 mm ) 253 ± 137 Root (2 mm ) 275 ± 104 Bulb (0.1 2 mm ) 93 ± 88 + propiin + glutathione + methionine, amount of associated sulphur see table P age
26 H A) H H relative intensity ICP-M/M m/z 308 relative intensity EI-M C 10 H 18 N Glu 5 0 -C 6 H 9 N Gly m/z m/z Fig. 17 GH; panel A) structure; panel B: EIC of m/z + ICP-M/M trace of root extract exposed to 2 mm, panel EI-M-spectra, panel ) rbitrap- M/M spectra with some annotated fragments, shown is the composition of the loss for the individual fragment calculated from the parent mass; for HPLC + ICP- M/M and EI-M conditions see main text under instrumentation. 26 P age
27 A) H 2 N H H relative intensity ICP-M/M m/z 613 relative intensity EI-M H 2 N H H C 20 H 33 N * Glu -Glu m/z Fig. 18 GG; panel A) structure; panel B: EIC of m/z + ICP-M/M trace of root extract exposed to 2 mm, panel M-spectra, panel rbitrap-m/m spectra with some annotated fragments, shown is the composition of the loss for the individual fragment calculated from the parent mass; for HPLC + ICP-M/M and EI-M conditions see main text under instrumentation m/z -Glu + Gly -Gly P age
28 propenylmercaptoglutathione, -2- propenylmercaptoglutathione and -propylmercaptoglutathione Both compounds were described before by Nakabayashi et al. 2 Their main EI- M/M fragments were m/z (loss of C 5 H 7 N 3 ), m/z (loss of C 8 H 13 N 3 ), m/z (C 5 H 10 N 2 ), m/z (C 5 H 8 N 3 ) and m/z (C 3 H 5 2 ). We identified the same M fragments using q-tf-ei-m and rbitrap EI-M as identified by Nakabayashi and co-workers using an FT-ICR-M. The fragmentation spectra for both compounds also showed the by Nakabayashi et al identified characteristic ions for the allyl respectively propenyl compound (m/z (C 5 H 9 N ), Fig. 19, respectively m/z (C 8 H 13 N 2 3, Fig. 20). -propylmercaptoglutathione (C 13 H 23 N ), [M+H] + m/z , measured m/z ) not yet mentioned in the literature was identified by EI-M, eluting shortly after -2-propenylmercaptoglutathione. Its main EI-M/M fragments were m/z (loss of C 5 H 7 N 3 ), m/z (loss of C 8 H 13 N 3 ), m/z (loss of Gly) and m/z (C 5 H 8 N 3 ) (Fig. 21). Table 8-1-propenylmercaptglutathione and -2-propenylmercaptoglutathione in µmol compound kg -1 d.w. (mean ± standard deviation, n = 9 per group), - propylmercaptoglutathione, database ID: Chempider propylmercaptoglutathione* propenylmercaptoglutathione propenylmercaptoglutathione H H 2 H H 2 H H 2 CH 2 Molecular formula C 13 H 21 N C 13 H 21 N C 13 H 23 N MW (g mol -1 ) [M+H] + (theoretical) [M+H] + (measured) RT (min) Database ID Root (0.1 mm ) 245 ± ± 720 Root ( ± ± 724 CH 3 CH 3 28 P age
29 mm ) Root (2 mm ) Bulb (0.1-2 mm ) 344 ± ± ± ± 93 *co-eluting with one isomer of -allyl/propenyl-pc-2, amount of associated sulphur see table P age
30 H H H 2 N A) C 13 H 21 N relative intensity ICP-M/M m/z relative intensity EI-M H 2 C Glu + C 3 H Glu + C 3 H 4 N 3 -C 8 H 13 N Glu +C 3 H 5 -Glu Fig propenylmercaptoglutathione; panel A) structure; panel B: EIC of m/z + ICP-M/M trace of root extract exposed to 2 mm, panel q-tf EI- M-spectra, panel EI-M/M spectra with some annotated fragments, shown is the composition of the loss for the individual fragment calculated from the parent mass; for HPLC + ICP-M/M and EI-M conditions see main text under instrumentation; cycle marks specific fragment for this isomer. 30 P age
31 A) H H H 2 N C 13 H 21 N relative intensity ICP-M/M m/z relative intensity EI-M H 3 C -C 8 H 13 N Glu + C 3 H 4 N Gly + C 3 H 5 + -Glu Glu + C 3 H 5 Fig propenylmercaptoglutathione; panel A) structure; panel B: EIC of m/z + ICP-M/M trace of root extract exposed to 2 mm, panel q-tf EI- M-spectra, panel EI-M/M spectra with some annotated fragments, shown is the composition of the loss for the individual fragment calculated from the parent mass; for HPLC + ICP-M/M and EI-M conditions see main text under instrumentation; cycle marks specific fragment for this isomer. 31 P age
32 A) H H 2 N C 13 H 23 N H relative intensity ICP-M/M m/z relative intensity EI-M H 3 C -C 8 H 15 N Glu + C 3 H 4 N 3 -Glu + C3 H Glu Fig. 21 -propylmercaptoglutathione; panel A) structure; panel B: EIC of m/z + ICP-M/M trace of root extract exposed to 2 mm, panel q-tf EI-Mspectra, panel EI-M/M spectra with some annotated fragments, shown is the composition of the loss for the individual fragment calculated from the parent mass; for HPLC + ICP-M/M and EI-M conditions see main text under instrumentation. 32 P age
33 2.9. xidised forms of -1-propenylmercaptoglutathione and -2- propenylmercaptoglutathione Just as the oxidised relatives of GAC and GPC are present oxidised forms of - 1 and -2-propenylmercaptoglutathion were present in the extracts of root and bulb. The fragmentation patterns for 3 of the compounds by qtf-ei-m were very similar. Compounds 1a and b showed more pronounced fragments at m/z (C 4 H 7 N 2 3 ) and m/z (C 8 H 11 N ) (Fig. 22 and 23). Compound 2a showed a more pronounced fragment at m/z (C 3 H 6 N 2 ) (Fig. 23). The last of the mentioned fragments may indicate that for this compound the H-group of GH was oxidised. No further identification with regard to the influence of double bond position and oxygen position on retention time and therefore compound characterisation was possible. Table 9 xidised forms of -1-propenylmercaptglutathione and -2- propenylmercaptoglutathione; 4 isomers (forms 1a, 1b, 2a and 2b) were found, double bond and position of -group can vary, compound 1a co-eluted with PC 2 oxidised. um of compounds (1b, 2a and 2b) in µmol compound kg -1 d.w. (mean ± standard deviation, n = 9 per group); quantification of individual compounds in Table 10, database ID: Chempider. -1- propenylmercaptoglutathione oxidised CH 2-2- propenylmercaptoglutathione oxidised CH 3 H H H H 2 2 Molecular formula C 13 H 21 N C 13 H 21 N MW (g mol -1 ) [M+H] + (theoretical) [M+H] + (measured) RT (min) 11.1, 11.7, 13.6, 14.3 Database ID - - Root (0.1 mm ) 265 ± 125* Root (0.5 mm ) 187 ± 96* Root (2 mm ) 395 ± 219* Bulb (0.1 2 mm ) 85 34* * -1-propenylmercaptoglutathione and -2-propenylmercaptoglutathione compounds 1b + 2a + 2b 33 P age
34 Table 10 xidised forms of -1-propenylmercaptoglutathione and -2- propenylmercaptoglutathione compounds 1b, 2a and 2b in µmol compound kg -1 d.w. (mean ± standard deviation, n = 9 per group) (mean ± standard deviation, n = 9 per group). compound 1 a (11.3 min)* compound 1b (11.7 min) compound 2a (13.6 min) compound 2b (14.3 min) Root (0.1 mm ) 104 ± ± 3 24 ± 2 Root (0.5 mm ) 63 ± ± 5 26 ± 10 Root (2 mm ) 153 ± ± 6 43 ± 12 Bulb (0.1 2 mm ) 54 ± ± 8 16 ± 5 *PC-2 + oxidised form of -1-propenylmercaptoglutathione or -2-propenylmercaptoglutathione compound 1a, amount of associated sulphur see table P age
35 A) H relative intensity ICP-M/M m/z 396 relative intensity EI-M H 2 N H CH 2 C 13 H 21 N Glu + C 3 H C 8 H 13 N Glu + C 3 H 4 N 3 -Glu + C 4 H 7 2 -C 3 H 5 2 Fig. 22 oxidised forms of -1-propenylmercaptoglutathione and -2- propenylmercaptoglutathione; panel A) structure; panel B: EIC of m/z + ICP- M/M trace of root extract exposed to 2 mm, panel q-tf EI-M-spectra, panel EI-M/M spectra with some annotated fragments, shown is the composition of the loss for the individual fragment calculated from the parent mass; for HPLC + ICP-M/M and EI-M conditions see main text under instrumentation. 35 P age
36 A) Fig. 23 Details of q-tf EI-M/M spectra of compounds 1a (panel A), 1b (panel and 2a (panel of oxidised forms of -1-propenylmercaptoglutathione and -2-propenylmercaptoglutathione; main differences in occurring fragments marked by cycles. 36 P age
37 2.10. Phytochelatin 2 (PC-2) and derivatives The main EI-M/M fragments of oxidised PC-2 were m/z (C 8 H 11 N 2 4 ), m/z (C 10 H 15 N 3 6 ), m/z (loss of γ-glu) and m/z (loss of γ-glu + Gly) (Fig. 24). The yet undescribed, -allyl/propenyl-containing PC-2 (C611) was present in both root and bulb in at least four different isomeric forms (Fig. 24), indicating that the -allyl/propenyl-group can be bound to either H-group of PC2. The main EI-M/M fragments were m/z (loss of γ-glu + Gly), m/z (loss of γ-glu), m/z (loss of γ-glu-cys) and m/z (C 10 H 15 N 3 6 ) (Fig. 25). Table 11 PC-2 oxidised and -allyl/propenyl-pc-2 (mean ± standard deviation, n = 9 per group), database ID: Chempider. H H 2 N PC-2 oxidised* HN H -allyl/propenyl-containing PC-2 + H 2 C N H H H N H H N H 2 H Molecular formula C 18 H 27 N C 21 H 33 N MW (g mol -1 ) [M+H] + (theoretical) [M+H] + (measured) RT (min) Database ID - - Root (0.1 mm ) Root (0.5 mm ) Root (2 mm ) Bulb (0.1-2 mm ) * PC propenylmercaptoglutathione compound1a, amount of associated sulphur see table allyl/propenyl-containing PC-2 + -propymercaptoglutathione, amount of associated sulphur see table P age
38 A) H HN H 2 N H relative intensity ICP-M/M m/z 538 relative intensity EI-M C 18 H 27 N H -Gly + γ-glu-cys -Gly + C 6 H 7 N 4 -Gly + Glu -Glu Fig. 24 oxidised PC-2; panel A) structure; panel B: EIC of m/z + ICP-M/M trace of root extract exposed to 2 mm, panel q-tf EI-M-spectra, panel EI-M/M spectra with some annotated fragments, shown is the composition of the loss for the individual fragment calculated from the parent mass; for HPLC + ICP- M/M and EI-M conditions see main text under instrumentation. 38 P age
39 A) H HN CH 2 relative intensity ICP-M/M m/z 612 relative intensity EI-M H HN C 21 H 33 N H 2 N H Cys-γ-Glu-Cys(-allyl)-Gly γ-glucys + -ally -Glu + Gly -Glu Fig. 25 -allyl/propenyl-pc-2; panel A) structure; panel B: EIC of m/z + ICP- M/M trace of root extract exposed to 2 mm, panel q-tf EI-M-spectra, panel EI-M/M spectra with some annotated fragments, shown is the composition of the loss for the individual fragment calculated from the parent mass; for HPLC + ICP-M/M and EI-M conditions see main text under instrumentation. 39 P age
40 2.11. Precursor molecules for -alk(en)ylcysteine sulfoxides The main fragments of C207 were m/z (C 4 H 7 2 ) and m/z (loss of 3 ) (Fig. 26). For C393 the main fragments were m/z (C 4 H 7 2 ), m/z (γ-glu), m/z (C 6 H 12 N 2 ), m/z (C 9 H 12 N 4 ) and m/z (loss of Gly) (Fig. 27). Table 12 C207 and C393 µmol compound kg -1 d.w. (mean ± standard deviation, n = 9 per group), database ID: Chempider. C207 (2-amino-3-[(2- carboxypropyl)sulfany l]-propanoic acid) C336 (2-amino-5-({1- carboxy-2-[(2- carboxypropyl)sulfany l]ethyl}amino)-5- oxopentanoic acid) C393 (2-amino-5- ({1-[(carboxy- methyl)amino]-3- [(2- carboxypropyl)sulf anyl]-1-oxopropan- 2-yl}amino)-5- oxopentanoic acid) C H 3 H 2 H H 3 C H H H H 3 C H H H Molecular formula C 7 H 13 N 4 C 12 H 20 N 2 7 C 14 H 23 N 3 8 MW (g mol -1 ) [M+H] + (theoretical) [M+H] + (measured) RT (min) Database ID Root (0.1 mm ) 669 ± 305 a - 83 ± 36 c Root (0.5 mm ) 1272 ± 859 b ± 104 d Root (2 mm ) 3214 ± 1827 a,b ± 375 c,d Bulb (0.1 2 mm ) 158 ± ± 216 a,b : statistically significant difference between groups by ne-way ANVA p < 0.05 c,d : statistically significant difference between groups by ne-way ANVA p < P age
41 A) 32- m/z 208 C H 3 H 2 H C 7 H 13 N 4 relative intensity ICP-M/M relative intensity EI-M C 3 H 6 N 2 -CH 2-2 Fig. 26 C207; panel A) structure; panel B: EIC of m/z + ICP-M/M trace of root extract exposed to 2 mm, panel q-tf EI-M-spectra, panel EI- M/M spectra with some annotated fragments, shown is the composition of the loss for the individual fragment calculated from the parent mass; for HPLC + ICP- M/M and EI-M conditions see main text under instrumentation. 41 P age
42 A) H 2 N H H H CH 3 C 14 H 23 N 3 8 relative intensity ICP-M/M m/z relative intensity EI-M -Glu + C 3 H 4 N 3 - CH C 3 H 4 N Glu + Gly + C C 10 H 16 N Gly Fig. 27 C393; panel A) structure; panel B: EIC of m/z + ICP-M/M trace of root extract exposed to 2 mm, panel q-tf EI-M-spectra, panel EI- M/M spectra with some annotated fragments, shown is the composition of the loss for the individual fragment calculated from the parent mass; for HPLC + ICP- M/M and EI-M conditions see main text under instrumentation. 42 P age
43 Table 13 Amount of sulphur associated with co-eluting compounds in mmol kg - 1 d.w. at the given retention times Root (0.1 mm ) Root (0.5 mm ) Root (2 mm ) Bulb (0.1 2 mm ) RT 2.2 min RT 3.7 min RT 11.1 min RT 27.3 min PC2 ox. & γ- propylmercaptoglutathione cycloalliin propiin, glutamyl--2- methiin & methinone & propenylcysteine & -allyl/propenylcontaining PC-2 methylcysteine GH sulfoxide 1a 3.5 ± 3.6 (1.7 %) 1.2 ± 0.53 a 0.19 ± ± ± 4.5 (2.2 %) 1.5 ± ± ± ± 1.5 (2.2%) 3.2 ± 2.2 a 0.32 ± ± ± 0.66 (0.67 %) 0.95 ± ± ± P age
44 3. Results from PCA and cluster analysis 3.1. Loading plot Loading Plot allyl/propenyl- GAC Methiin + cyclo + methylcystein -2-PMG GPC() -1-PMG GG 0.2 -allycysteine GPC PC2 0.1 C PC2 GMC Alliin C207-1/2 Propiin + GH + Met C393 GAC() Isoalliin PC Fig. 28 Loading plot for the PCA in which all plants from the three different - fertilization stage (0.1, 0.5 and 2 mm ) by utilisation of all identified low molecular weight -containing metabolites. 44 P age
45 3.2. Dendrogram Cluster analysis: Dendrogram 0.00 imilarity Fig. 29 Cluster analysis in which all plants from the three different - fertilization stage (0.1, 0.5 and 2 mm ) by utilisation of all identified low molecular weight -containing metabolites plants with the different sulphur fertilization (mm ) P age
46 4. Example of HPLC-ICP-M/M traces for all plant parts fertilized with different amount of sulphur root 0.1 mm root 2 mm bulb 0.5 mm root 0.5 mm bulb 0.1 mm bulb 2 mm relative intensity ICP-M/M Fig. 30 comparison of 32 -trace of root and bulb extracts (one each at different fertilizer concentrations); for HPLC + ICP-M/M and EI-M conditions see main text under instrumentation. relative intensity ICP-M/M root 0.1 mm root 0.5 mm root 2 mm bulb 0.1 mm bulb 0.5 mm bulb 2 mm Fig. 31 comparison of 32 -trace of root and bulb extracts (one each at different fertilizer concentrations); for HPLC + ICP-M/M and EI-M conditions see main text under instrumentation. 46 P age
47 5. Example of peak area integration Fig. 28 Example of peak area integration using PeakFit. Top panel: measured signal (line) and calculated line (dots) using the described model for calculation in logarithmic y-scale, bottom panel: showing individual peaks (linear y-scale); R 2 = for peak area covered 1 Y. Yamazaki, T. Tokunaga, T. kuno, Quantitative determination of eleven flavor precursors (-alk(en)yl cysteine derivatives) in garlic with HPLC method, Nippon hokuhin Kagaku Kogaku Kaishi, 2005, 52, ; in E. Block, Garlic and other Alliums The Lore and the cience, Royal ociety of Chemistry: Cambridge, U.K., R. Nakabayashi, Y. awada, M. Aoyagi, Y. Yamada, M. Y. Hirai, T. akurai, T. Kamoi, D. D. Rowan, K. aito, Chemical Assignment of tructural Isomers of ulfur-containing Metabolites in Garlic by Liquid Chromatography-Fourier Transform Ion Cyclotron Resonance-Mass pectrometry, J. Nutr., 2016, 146, P age
Time (min) Supplementary Figure 1: Gas decomposition products of irradiated DMC.
 200000 C 2 CH 3 CH 3 DMC 180000 160000 140000 Intensity 120000 100000 80000 60000 40000 C 2 H 6 CH 3 CH 2 CH 3 CH 3 CCH 3 EMC DEC 20000 C 3 H 8 HCCH 3 5 10 15 20 25 Time (min) Supplementary Figure 1: Gas
200000 C 2 CH 3 CH 3 DMC 180000 160000 140000 Intensity 120000 100000 80000 60000 40000 C 2 H 6 CH 3 CH 2 CH 3 CH 3 CCH 3 EMC DEC 20000 C 3 H 8 HCCH 3 5 10 15 20 25 Time (min) Supplementary Figure 1: Gas
Accurate determination of protein methionine oxidation by stable isotope labeling and LC-MS analysis
 Accurate determination of protein methionine oxidation by stable isotope labeling and LC-MS analysis Hongcheng Liu*, Gomathinayagam Ponniah, Alyssa Neil, Rekha Patel, Bruce Andrien Protein Characterization,
Accurate determination of protein methionine oxidation by stable isotope labeling and LC-MS analysis Hongcheng Liu*, Gomathinayagam Ponniah, Alyssa Neil, Rekha Patel, Bruce Andrien Protein Characterization,
Organic Chemistry Laboratory Fall Lecture 3 Gas Chromatography and Mass Spectrometry June
 344 Organic Chemistry Laboratory Fall 2013 Lecture 3 Gas Chromatography and Mass Spectrometry June 19 2013 Chromatography Chromatography separation of a mixture into individual components Paper, Column,
344 Organic Chemistry Laboratory Fall 2013 Lecture 3 Gas Chromatography and Mass Spectrometry June 19 2013 Chromatography Chromatography separation of a mixture into individual components Paper, Column,
2. Ionization Sources 3. Mass Analyzers 4. Tandem Mass Spectrometry
 Dr. Sanjeeva Srivastava 1. Fundamental of Mass Spectrometry Role of MS and basic concepts 2. Ionization Sources 3. Mass Analyzers 4. Tandem Mass Spectrometry 2 1 MS basic concepts Mass spectrometry - technique
Dr. Sanjeeva Srivastava 1. Fundamental of Mass Spectrometry Role of MS and basic concepts 2. Ionization Sources 3. Mass Analyzers 4. Tandem Mass Spectrometry 2 1 MS basic concepts Mass spectrometry - technique
SUPPORTING INFORMATION. Lysine Carbonylation is a Previously Unrecognized Contributor. to Peroxidase Activation of Cytochrome c by Chloramine-T
 Electronic Supplementary Material (ESI) for Chemical Science. This journal is The Royal Society of Chemistry 2019 SUPPORTING INFORMATION Lysine Carbonylation is a Previously Unrecognized Contributor to
Electronic Supplementary Material (ESI) for Chemical Science. This journal is The Royal Society of Chemistry 2019 SUPPORTING INFORMATION Lysine Carbonylation is a Previously Unrecognized Contributor to
Accumulation and transformation of inorganic and organic arsenic in rice and role of
 Supplementary Information: Accumulation and transformation of inorganic and organic arsenic in rice and role of thiol-complexation to restrict their translocation to shoot Seema Mishra 1,2 *, Jürgen Mattusch
Supplementary Information: Accumulation and transformation of inorganic and organic arsenic in rice and role of thiol-complexation to restrict their translocation to shoot Seema Mishra 1,2 *, Jürgen Mattusch
Supplemental Data. Deinlein et al. Plant Cell. (2012) /tpc
 µm Zn 2+ 15 µm Zn 2+ Growth (% of control) empty vector NS1 NS2 NS3 NS4 S. pombe zhfδ Supplemental Figure 1. Functional characterization of. halleri NS genes in Zn 2+ hypersensitive S. pombe Δzhf mutant
µm Zn 2+ 15 µm Zn 2+ Growth (% of control) empty vector NS1 NS2 NS3 NS4 S. pombe zhfδ Supplemental Figure 1. Functional characterization of. halleri NS genes in Zn 2+ hypersensitive S. pombe Δzhf mutant
Analysis of FAMEs Using Cold EI GC/MS for Enhanced Molecular Ion Selectivity
 APPLICATION NOTE Gas Chromatography/ Mass Spectrometry Author: Adam Patkin PerkinElmer, Inc. Shelton, CT Analysis of FAMEs Using GC/MS for Enhanced Molecular Ion Selectivity Introduction Characterization
APPLICATION NOTE Gas Chromatography/ Mass Spectrometry Author: Adam Patkin PerkinElmer, Inc. Shelton, CT Analysis of FAMEs Using GC/MS for Enhanced Molecular Ion Selectivity Introduction Characterization
Preparation and Characterization of Cysteine Adducts of Deoxynivalenol
 Preparation and Characterization of Cysteine Adducts of Deoxynivalenol Ana Stanic, Silvio Uhlig, Anita Solhaug, Frode Rise, Alistair L. Wilkins, Christopher O. Miles S1 Figure S1. 1 H spectrum of 1 (DON)
Preparation and Characterization of Cysteine Adducts of Deoxynivalenol Ana Stanic, Silvio Uhlig, Anita Solhaug, Frode Rise, Alistair L. Wilkins, Christopher O. Miles S1 Figure S1. 1 H spectrum of 1 (DON)
Ultra High Definition Optimizing all Analytical Dimensions
 Ultra High Definition Optimizing all Analytical Dimensions Sensitivity Dynamic Range Signal Response Linearity Separation Speed Peak Capacity Chromatogram Mass Spectrum Mass Accuracy Resolving Power Acquisition
Ultra High Definition Optimizing all Analytical Dimensions Sensitivity Dynamic Range Signal Response Linearity Separation Speed Peak Capacity Chromatogram Mass Spectrum Mass Accuracy Resolving Power Acquisition
Comparison of Relative Quantification of Monoclonal Antibody N-glycans Using Fluorescence and MS Detection
 Comparison of Relative Quantification of Monoclonal ntibody N-glycans Using Fluorescence and MS Detection pplication Note iotherapeutics & iologics uthors scar Potter and Gregory Staples gilent Technologies,
Comparison of Relative Quantification of Monoclonal ntibody N-glycans Using Fluorescence and MS Detection pplication Note iotherapeutics & iologics uthors scar Potter and Gregory Staples gilent Technologies,
The use of mass spectrometry in lipidomics. Outlines
 The use of mass spectrometry in lipidomics Jeevan Prasain jprasain@uab.edu 6-2612 utlines Brief introduction to lipidomics Analytical methodology: MS/MS structure elucidation of phospholipids Phospholipid
The use of mass spectrometry in lipidomics Jeevan Prasain jprasain@uab.edu 6-2612 utlines Brief introduction to lipidomics Analytical methodology: MS/MS structure elucidation of phospholipids Phospholipid
Supporting Information
 Supporting Information Campholenic aldehyde ozonolysis: A possible mechanism for the formation of specific biogenic secondary organic aerosol constituents Ariane Kahnt 1,2, Yoshiteru Iinuma 1, Anke Mutzel
Supporting Information Campholenic aldehyde ozonolysis: A possible mechanism for the formation of specific biogenic secondary organic aerosol constituents Ariane Kahnt 1,2, Yoshiteru Iinuma 1, Anke Mutzel
Benefits and Characteristic Applications of High Resolution GC/MS and LC/MS. Frank David RIC and Ghent University
 Benefits and Characteristic Applications of High Resolution GC/MS and LC/MS. Frank David RIC and Ghent University Mass Spectrometry Structure Elucidation Selective and Sensitive Detection Identification
Benefits and Characteristic Applications of High Resolution GC/MS and LC/MS. Frank David RIC and Ghent University Mass Spectrometry Structure Elucidation Selective and Sensitive Detection Identification
MASS SPECTROMETRY IN METABOLOMICS
 For personal use only. Please do not reuse or reproduce without the author s permission MASS SPECTRMETRY IN METABLMICS Pavel Aronov Stanford Mass Spectrometry Users Meeting August 21, 2008 rigin of Metabolomics
For personal use only. Please do not reuse or reproduce without the author s permission MASS SPECTRMETRY IN METABLMICS Pavel Aronov Stanford Mass Spectrometry Users Meeting August 21, 2008 rigin of Metabolomics
Lecture 3. Tandem MS & Protein Sequencing
 Lecture 3 Tandem MS & Protein Sequencing Nancy Allbritton, M.D., Ph.D. Department of Physiology & Biophysics 824-9137 (office) nlallbri@uci.edu Office- Rm D349 Medical Science D Bldg. Tandem MS Steps:
Lecture 3 Tandem MS & Protein Sequencing Nancy Allbritton, M.D., Ph.D. Department of Physiology & Biophysics 824-9137 (office) nlallbri@uci.edu Office- Rm D349 Medical Science D Bldg. Tandem MS Steps:
Electronic Supplementary Information NOVEL NON-TARGET ANALYSIS FOR FLUORINE COMPOUNDS USING ICPMS/MS AND HPLC-ICPMS/MS
 Electronic Supplementary Material (ESI) for Journal of Analytical Atomic Spectrometry. This journal is The Royal Society of Chemistry 2017 Electronic Supplementary Information NOVEL NON-TARGET ANALYSIS
Electronic Supplementary Material (ESI) for Journal of Analytical Atomic Spectrometry. This journal is The Royal Society of Chemistry 2017 Electronic Supplementary Information NOVEL NON-TARGET ANALYSIS
 Studies on the Sulfur-containing Am TitleCompounds in Garlic. (I) : Assimila Garlic Author(s) Sugii, Michiyasu; Suzuki, Tomoji; K Jyoji Citation Bulletin of the Institute for Chemi University (1964), 42(4):
Studies on the Sulfur-containing Am TitleCompounds in Garlic. (I) : Assimila Garlic Author(s) Sugii, Michiyasu; Suzuki, Tomoji; K Jyoji Citation Bulletin of the Institute for Chemi University (1964), 42(4):
Quantification of labile and stable non-polar arsenolipids in commercial fish meals and edible seaweed samples
 Electronic Supplementary Material (ESI) for Journal of Analytical Atomic Spectrometry. This journal is The Royal Society of Chemistry 7 Quantification of labile and stable non-polar arsenolipids in commercial
Electronic Supplementary Material (ESI) for Journal of Analytical Atomic Spectrometry. This journal is The Royal Society of Chemistry 7 Quantification of labile and stable non-polar arsenolipids in commercial
An Investigative Study of Reactions Involving Glucosinolates and Isothiocyanates
 An Investigative Study of Reactions Involving Glucosinolates and Isothiocyanates Alzea Chrisel H. Alea 1, Diane Elaine T. Co 2 and Marissa G Noel 3* 1,2,3Chemistry Department, De La Salle University, 2401
An Investigative Study of Reactions Involving Glucosinolates and Isothiocyanates Alzea Chrisel H. Alea 1, Diane Elaine T. Co 2 and Marissa G Noel 3* 1,2,3Chemistry Department, De La Salle University, 2401
Identification & Confirmation of Structurally Related Degradation Products of Simvastatin
 Identification & Confirmation of Structurally Related Degradation Products of Simvastatin Power of QTRAP Systems for Identification and Confirmation of Degradation Products Dilip Reddy 1, Chandra Sekar
Identification & Confirmation of Structurally Related Degradation Products of Simvastatin Power of QTRAP Systems for Identification and Confirmation of Degradation Products Dilip Reddy 1, Chandra Sekar
Metabolomics: quantifying the phenotype
 Metabolomics: quantifying the phenotype Metabolomics Promises Quantitative Phenotyping What can happen GENOME What appears to be happening Bioinformatics TRANSCRIPTOME What makes it happen PROTEOME Systems
Metabolomics: quantifying the phenotype Metabolomics Promises Quantitative Phenotyping What can happen GENOME What appears to be happening Bioinformatics TRANSCRIPTOME What makes it happen PROTEOME Systems
Supporting Information
 Supporting Information Mass Spectrometry Imaging Shows Cocaine and Methylphenidate have Opposite Effects on Major Lipids in Drosophila Brain Mai H. Philipsen *, Nhu T. N. Phan *, John S. Fletcher *, Per
Supporting Information Mass Spectrometry Imaging Shows Cocaine and Methylphenidate have Opposite Effects on Major Lipids in Drosophila Brain Mai H. Philipsen *, Nhu T. N. Phan *, John S. Fletcher *, Per
CHM 424L Organic Laboratory, Dr. Laurie S. Starkey Introduction to Mass Spectrometry
 CM 424L rganic Laboratory, Dr. Laurie S. Starkey Introduction to Mass Spectrometry Mass spectrometry is used to determine a sample's molecular mass and molecular formula. Some structural information can
CM 424L rganic Laboratory, Dr. Laurie S. Starkey Introduction to Mass Spectrometry Mass spectrometry is used to determine a sample's molecular mass and molecular formula. Some structural information can
Engineering of Ministry of Education, Institute of Molecular Science, Shanxi University, Taiyuan , China.
 Indicator approach to develop a chemosensor for the colorimetric sensing of thiol-containing in water and its application for the thiol detection in plasma Fang-Jun Huo, a Yu-Tao Yang, b Jing Su, b Yuan-Qiang
Indicator approach to develop a chemosensor for the colorimetric sensing of thiol-containing in water and its application for the thiol detection in plasma Fang-Jun Huo, a Yu-Tao Yang, b Jing Su, b Yuan-Qiang
Amadeo R. Fernández-Alba
 % of compounds % of compounds % of compounds % of compounds Amadeo R. Fernández-Alba LC-Orbitrap QExactive Focus Instrumental LOQ 1% 9% 8% 7% 6% 5% 4% 3% 2% 1% %.1 mg/g.2 mg/g Tomato.5 mg/g ddms2 Target
% of compounds % of compounds % of compounds % of compounds Amadeo R. Fernández-Alba LC-Orbitrap QExactive Focus Instrumental LOQ 1% 9% 8% 7% 6% 5% 4% 3% 2% 1% %.1 mg/g.2 mg/g Tomato.5 mg/g ddms2 Target
Characterization of Disulfide Linkages in Proteins by 193 nm Ultraviolet Photodissociation (UVPD) Mass Spectrometry. Supporting Information
 Characterization of Disulfide Linkages in Proteins by 193 nm Ultraviolet Photodissociation (UVPD) Mass Spectrometry M. Montana Quick, Christopher M. Crittenden, Jake A. Rosenberg, and Jennifer S. Brodbelt
Characterization of Disulfide Linkages in Proteins by 193 nm Ultraviolet Photodissociation (UVPD) Mass Spectrometry M. Montana Quick, Christopher M. Crittenden, Jake A. Rosenberg, and Jennifer S. Brodbelt
CONTENT. i iv ix. SVKM s NMIMS, School of Pharmacy and Technology Management
 CONTENT Chapter No. Title Page No. Abbreviations List of Figures List of Tables 1 Introduction 1 1.1 Practical aspects 4 1.2 Stability-indicating assay method (SIAM) 5 1.3 Regulatory aspects 6 1.4 Techniques
CONTENT Chapter No. Title Page No. Abbreviations List of Figures List of Tables 1 Introduction 1 1.1 Practical aspects 4 1.2 Stability-indicating assay method (SIAM) 5 1.3 Regulatory aspects 6 1.4 Techniques
Thermo Scientific LipidSearch Software for Lipidomics Workflows. Automated Identification and Relative. Quantitation of Lipids by LC/MS
 Thermo Scientific LipidSearch Software for Lipidomics Workflows Automated Identification and Relative of Lipids by LC/MS The promise of lipidomics Lipidomics is a new field of study crucial for understanding
Thermo Scientific LipidSearch Software for Lipidomics Workflows Automated Identification and Relative of Lipids by LC/MS The promise of lipidomics Lipidomics is a new field of study crucial for understanding
LC/QTOF Discovery of Previously Unreported Microcystins in Alberta Lake Waters
 LC/QTOF Discovery of Previously Unreported Microcystins in Alberta Lake Waters Ralph Hindle Vogon Laboratory Services Ltd. Cochrane, Alberta, Canada Xu Zhang David W. Kinniburgh Alberta Centre for Toxicology
LC/QTOF Discovery of Previously Unreported Microcystins in Alberta Lake Waters Ralph Hindle Vogon Laboratory Services Ltd. Cochrane, Alberta, Canada Xu Zhang David W. Kinniburgh Alberta Centre for Toxicology
Metabolomic and Proteomics Solutions for Integrated Biology. Christine Miller Omics Market Manager ASMS 2015
 Metabolomic and Proteomics Solutions for Integrated Biology Christine Miller Omics Market Manager ASMS 2015 Integrating Biological Analysis Using Pathways Protein A R HO R Protein B Protein X Identifies
Metabolomic and Proteomics Solutions for Integrated Biology Christine Miller Omics Market Manager ASMS 2015 Integrating Biological Analysis Using Pathways Protein A R HO R Protein B Protein X Identifies
An Introduction to the Use of itraq Reagents for Amino Acid Analysis
 An Introduction to the Use of itraq Reagents for Amino Acid Analysis Lisa Sapp Clinical Research and Forensic Toxicology Product Manager Mass Spectrometry Systems Amino Acid Analysis Using itraq Reagents
An Introduction to the Use of itraq Reagents for Amino Acid Analysis Lisa Sapp Clinical Research and Forensic Toxicology Product Manager Mass Spectrometry Systems Amino Acid Analysis Using itraq Reagents
Supplementary Figure 1 (previous page). EM analysis of full-length GCGR. (a) Exemplary tilt pair images of the GCGR mab23 complex acquired for Random
 S1 Supplementary Figure 1 (previous page). EM analysis of full-length GCGR. (a) Exemplary tilt pair images of the GCGR mab23 complex acquired for Random Conical Tilt (RCT) reconstruction (left: -50,right:
S1 Supplementary Figure 1 (previous page). EM analysis of full-length GCGR. (a) Exemplary tilt pair images of the GCGR mab23 complex acquired for Random Conical Tilt (RCT) reconstruction (left: -50,right:
Sue D Antonio Application Chemist Cedar Creek, TX
 Sue D Antonio Application Chemist Cedar Creek, TX What is Hemp Oil? CBD hemp oil is a natural botanical extract of the common hemp plant. CBD hemp oil is derived from the seeds and stem of the Cannabis
Sue D Antonio Application Chemist Cedar Creek, TX What is Hemp Oil? CBD hemp oil is a natural botanical extract of the common hemp plant. CBD hemp oil is derived from the seeds and stem of the Cannabis
[ APPLICATION NOTE ] High Sensitivity Intact Monoclonal Antibody (mab) HRMS Quantification APPLICATION BENEFITS INTRODUCTION WATERS SOLUTIONS KEYWORDS
![[ APPLICATION NOTE ] High Sensitivity Intact Monoclonal Antibody (mab) HRMS Quantification APPLICATION BENEFITS INTRODUCTION WATERS SOLUTIONS KEYWORDS [ APPLICATION NOTE ] High Sensitivity Intact Monoclonal Antibody (mab) HRMS Quantification APPLICATION BENEFITS INTRODUCTION WATERS SOLUTIONS KEYWORDS](/thumbs/79/80328542.jpg) Yun Wang Alelyunas, Henry Shion, Mark Wrona Waters Corporation, Milford, MA, USA APPLICATION BENEFITS mab LC-MS method which enables users to achieve highly sensitive bioanalysis of intact trastuzumab
Yun Wang Alelyunas, Henry Shion, Mark Wrona Waters Corporation, Milford, MA, USA APPLICATION BENEFITS mab LC-MS method which enables users to achieve highly sensitive bioanalysis of intact trastuzumab
Supporting information
 Supporting information Figure legends Supplementary Table 1. Specific product ions obtained from fragmentation of lithium adducts in the positive ion mode comparing the different positional isomers of
Supporting information Figure legends Supplementary Table 1. Specific product ions obtained from fragmentation of lithium adducts in the positive ion mode comparing the different positional isomers of
Characterization of an Unknown Compound Using the LTQ Orbitrap
 Characterization of an Unknown Compound Using the LTQ rbitrap Donald Daley, Russell Scammell, Argenta Discovery Limited, 8/9 Spire Green Centre, Flex Meadow, Harlow, Essex, CM19 5TR, UK bjectives unknown
Characterization of an Unknown Compound Using the LTQ rbitrap Donald Daley, Russell Scammell, Argenta Discovery Limited, 8/9 Spire Green Centre, Flex Meadow, Harlow, Essex, CM19 5TR, UK bjectives unknown
Identification of Imidacloprid Metabolites in Onions Using High Resolution Accurate Mass Spectrometry (LC/Q-TOF MS) and Accurate Mass Tools
 Identification of Imidacloprid Metabolites in Onions Using High Resolution Accurate Mass Spectrometry (LC/Q-TOF MS) and Accurate Mass Tools Application ote Food Authors E. Michael Thurman and Imma Ferrer
Identification of Imidacloprid Metabolites in Onions Using High Resolution Accurate Mass Spectrometry (LC/Q-TOF MS) and Accurate Mass Tools Application ote Food Authors E. Michael Thurman and Imma Ferrer
SimGlycan. A high-throughput glycan and glycopeptide data analysis tool for LC-, MALDI-, ESI- Mass Spectrometry workflows.
 PREMIER Biosoft SimGlycan A high-throughput glycan and glycopeptide data analysis tool for LC-, MALDI-, ESI- Mass Spectrometry workflows SimGlycan software processes and interprets the MS/MS and higher
PREMIER Biosoft SimGlycan A high-throughput glycan and glycopeptide data analysis tool for LC-, MALDI-, ESI- Mass Spectrometry workflows SimGlycan software processes and interprets the MS/MS and higher
Metabolite identification in metabolomics: Database and interpretation of MSMS spectra
 Metabolite identification in metabolomics: Database and interpretation of MSMS spectra Jeevan K. Prasain, PhD Department of Pharmacology and Toxicology, UAB jprasain@uab.edu utline Introduction Putative
Metabolite identification in metabolomics: Database and interpretation of MSMS spectra Jeevan K. Prasain, PhD Department of Pharmacology and Toxicology, UAB jprasain@uab.edu utline Introduction Putative
DETECTION OF BEESWAX ADULTERATION WITH HYDROCARBONS USING GAS CHROMATOGRAPHY WITH MASS DETECTOR (GC-MS)
 DETECTION OF BEESWAX ADULTERATION WITH HYDROCARBONS USING GAS CHROMATOGRAPHY WITH MASS DETECTOR (GC-MS) Ewa Waś,, Helena Rybak-Chmielewska, Teresa Szczęsna e-mail: ewa.was@man.pulawy.pl Research Institute
DETECTION OF BEESWAX ADULTERATION WITH HYDROCARBONS USING GAS CHROMATOGRAPHY WITH MASS DETECTOR (GC-MS) Ewa Waś,, Helena Rybak-Chmielewska, Teresa Szczęsna e-mail: ewa.was@man.pulawy.pl Research Institute
PHOTOCATALYTIC DECONTAMINATION OF CHLORANTRANILIPROLE RESIDUES IN WATER USING ZnO NANOPARTICLES. DR. A. RAMESH, Ph.D, D.Sc.,
 PHOTOCATALYTIC DECONTAMINATION OF CHLORANTRANILIPROLE RESIDUES IN WATER USING ZnO NANOPARTICLES DR. A. RAMESH, Ph.D, D.Sc., raamesh_a@yahoo.co.in 1 OBJECTIVES Determination of persistence and photolysis
PHOTOCATALYTIC DECONTAMINATION OF CHLORANTRANILIPROLE RESIDUES IN WATER USING ZnO NANOPARTICLES DR. A. RAMESH, Ph.D, D.Sc., raamesh_a@yahoo.co.in 1 OBJECTIVES Determination of persistence and photolysis
Supporting Information. Lysine Propionylation to Boost Proteome Sequence. Coverage and Enable a Silent SILAC Strategy for
 Supporting Information Lysine Propionylation to Boost Proteome Sequence Coverage and Enable a Silent SILAC Strategy for Relative Protein Quantification Christoph U. Schräder 1, Shaun Moore 1,2, Aaron A.
Supporting Information Lysine Propionylation to Boost Proteome Sequence Coverage and Enable a Silent SILAC Strategy for Relative Protein Quantification Christoph U. Schräder 1, Shaun Moore 1,2, Aaron A.
Application Note # FTMS-46 solarix XR: Analysis of Complex Mixtures
 Application Note # FTMS-46 solarix XR: Analysis of Complex Mixtures Introduction Natural organic matter (NOM) as well as crude oil and crude oil fractions are very complex mixtures of organic compounds
Application Note # FTMS-46 solarix XR: Analysis of Complex Mixtures Introduction Natural organic matter (NOM) as well as crude oil and crude oil fractions are very complex mixtures of organic compounds
Introduction to Peptide Sequencing
 Introduction to Peptide equencing Quadrupole Ion Traps tructural Biophysics Course December 3, 2014 12/8/14 Introduction to Peptide equencing - athan Yates 1 Why are ion traps used to sequence peptides?
Introduction to Peptide equencing Quadrupole Ion Traps tructural Biophysics Course December 3, 2014 12/8/14 Introduction to Peptide equencing - athan Yates 1 Why are ion traps used to sequence peptides?
Development of a Bioanalytical Method for Quantification of Amyloid Beta Peptides in Cerebrospinal Fluid
 Development of a Bioanalytical Method for Quantification of Amyloid Beta Peptides in Cerebrospinal Fluid Joanne ( 乔安妮 ) Mather Senior Scientist Waters Corporation Data courtesy of Erin Chambers and Mary
Development of a Bioanalytical Method for Quantification of Amyloid Beta Peptides in Cerebrospinal Fluid Joanne ( 乔安妮 ) Mather Senior Scientist Waters Corporation Data courtesy of Erin Chambers and Mary
Iron depletion enhances production of antimicrobials by Pseudomonas
 Iron depletion enhances production of antimicrobials by Pseudomonas aeruginosa. Angela T. Nguyen 1, Jace W. Jones 1, Max A. Ruge 1, Maureen A. Kane 1, and Amanda G. Oglesby-Sherrouse 1,2 * University of
Iron depletion enhances production of antimicrobials by Pseudomonas aeruginosa. Angela T. Nguyen 1, Jace W. Jones 1, Max A. Ruge 1, Maureen A. Kane 1, and Amanda G. Oglesby-Sherrouse 1,2 * University of
Analysis of Uncomplexed and Copper-complexed Methanobactin with UV/Visible Spectrophotometry, Mass Spectrometry and NMR Spectrometry
 Analysis of Uncomplexed and Copper-complexed Methanobactin with UV/Visible Spectrophotometry, Mass Spectrometry and NMR Spectrometry Lee Behling, Alan DiSpirito, Scott Hartsel, Larry Masterson, Gianluigi
Analysis of Uncomplexed and Copper-complexed Methanobactin with UV/Visible Spectrophotometry, Mass Spectrometry and NMR Spectrometry Lee Behling, Alan DiSpirito, Scott Hartsel, Larry Masterson, Gianluigi
Identification of Unknown Pesticides in Food Using Both LC/MSD TOF and Ion Trap MS n Application
 Identification of Unknown Pesticides in Food Using Both LC/MD TF and Ion Trap M n Application Food afety Author E. Michael Thurman and Imma Ferrer Pesticide Residue Research Group University of Almería
Identification of Unknown Pesticides in Food Using Both LC/MD TF and Ion Trap M n Application Food afety Author E. Michael Thurman and Imma Ferrer Pesticide Residue Research Group University of Almería
Mass Spectrometry based metabolomics
 Mass Spectrometry based metabolomics Metabolomics- A realm of small molecules (
Mass Spectrometry based metabolomics Metabolomics- A realm of small molecules (
Kazusa DNA Res Inst. Daisuke Shibata
 Workshop Argentina-Japan Bioscience and Biotechnology for the promotion of Agricutulure and Food Production August 3 rd to 7 th 2009- Metabolite Metabolomics Databases approaches for Plant for Agro-Biotechnology
Workshop Argentina-Japan Bioscience and Biotechnology for the promotion of Agricutulure and Food Production August 3 rd to 7 th 2009- Metabolite Metabolomics Databases approaches for Plant for Agro-Biotechnology
John P. McCauley and Rui Chen Waters Corporation, Milford, MA, USA INTRODUCTION APPLICATION BENEFITS WATERS SOLUTIONS KEY WORDS
 Enantiomeric and diastereomeric separations of fragrance and essential oil components using the ACQUITY UPC 2 System with ACQUITY UPC 2 Trefoil Columns John P. McCauley and Rui Chen Waters Corporation,
Enantiomeric and diastereomeric separations of fragrance and essential oil components using the ACQUITY UPC 2 System with ACQUITY UPC 2 Trefoil Columns John P. McCauley and Rui Chen Waters Corporation,
Advanced oxidation processes for the removal of [bmim][sal] third generation ionic liquid: Effect of water matrices and intermediates identification
![Advanced oxidation processes for the removal of [bmim][sal] third generation ionic liquid: Effect of water matrices and intermediates identification Advanced oxidation processes for the removal of [bmim][sal] third generation ionic liquid: Effect of water matrices and intermediates identification](/thumbs/96/128474283.jpg) Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2016 Electronic Supplementary Information Advanced oxidation processes for the removal of [bmim][sal]
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2016 Electronic Supplementary Information Advanced oxidation processes for the removal of [bmim][sal]
Progenesis QI: complex data processing in plant metabolomics. Introducing an extra gear for. Geert Goeminne
 Progenesis QI: Introducing an extra gear for complex data processing in plant metabolomics. Geert Goeminne gegoe@psb.vib-ugent.be Why Plants? EM Rubin Nature 454, 841-845 (2008) doi:10.1038/nature07190
Progenesis QI: Introducing an extra gear for complex data processing in plant metabolomics. Geert Goeminne gegoe@psb.vib-ugent.be Why Plants? EM Rubin Nature 454, 841-845 (2008) doi:10.1038/nature07190
SUPPLEMENTARY DATA. Materials and Methods
 SUPPLEMENTARY DATA Materials and Methods HPLC-UV of phospholipid classes and HETE isomer determination. Fractionation of platelet lipid classes was undertaken on a Spherisorb S5W 150 x 4.6 mm column (Waters
SUPPLEMENTARY DATA Materials and Methods HPLC-UV of phospholipid classes and HETE isomer determination. Fractionation of platelet lipid classes was undertaken on a Spherisorb S5W 150 x 4.6 mm column (Waters
Biological Mass Spectrometry. April 30, 2014
 Biological Mass Spectrometry April 30, 2014 Mass Spectrometry Has become the method of choice for precise protein and nucleic acid mass determination in a very wide mass range peptide and nucleotide sequencing
Biological Mass Spectrometry April 30, 2014 Mass Spectrometry Has become the method of choice for precise protein and nucleic acid mass determination in a very wide mass range peptide and nucleotide sequencing
Determination of red blood cell fatty acid profiles in clinical research
 Application Note Clinical Research Determination of red blood cell fatty acid profiles in clinical research Chemical ionization gas chromatography tandem mass spectrometry Authors Yvonne Schober 1, Hans
Application Note Clinical Research Determination of red blood cell fatty acid profiles in clinical research Chemical ionization gas chromatography tandem mass spectrometry Authors Yvonne Schober 1, Hans
Profiling Analysis of Polysulfide Silane Coupling Agent
 C146-E154 Profiling Analysis of Polysulfide lane Coupling Agent Technical Report vol.42 1. Introduction Recently, in line with the oil-conservation movement, silica, as a non-petroleum resource, is increasingly
C146-E154 Profiling Analysis of Polysulfide lane Coupling Agent Technical Report vol.42 1. Introduction Recently, in line with the oil-conservation movement, silica, as a non-petroleum resource, is increasingly
Library identifications for Lemnaceae
 Library identifications for Lemnaceae NP-001984 ESI+ NP-001984 present in, enriched in (15.47min) NP-001984 -MS^E CH 3 Moupinamide H NH NP-005512 + NP-016675 ESI- s NP-005512 NP-016675 NP-005512 present
Library identifications for Lemnaceae NP-001984 ESI+ NP-001984 present in, enriched in (15.47min) NP-001984 -MS^E CH 3 Moupinamide H NH NP-005512 + NP-016675 ESI- s NP-005512 NP-016675 NP-005512 present
Dr. Erin E. Chambers Waters Corporation. Presented by Dr. Diego Rodriguez Cabaleiro Waters Europe Waters Corporation 1
 Development of an SPE-LC/MS/MS Assay for the Simultaneous Quantification of Amyloid Beta Peptides in Cerebrospinal Fluid in Support of Alzheimer s Research Dr. Erin E. Chambers Waters Corporation Presented
Development of an SPE-LC/MS/MS Assay for the Simultaneous Quantification of Amyloid Beta Peptides in Cerebrospinal Fluid in Support of Alzheimer s Research Dr. Erin E. Chambers Waters Corporation Presented
Improved method for the quantification of lysophospholipids including enol ether
 Supplemental Material Improved method for the quantification of lysophospholipids including enol ether species by liquid chromatography-tandem mass spectrometry James G. Bollinger *, Hiromi Ii*, Martin
Supplemental Material Improved method for the quantification of lysophospholipids including enol ether species by liquid chromatography-tandem mass spectrometry James G. Bollinger *, Hiromi Ii*, Martin
Essential Lipidomics Experiments using the LTQ Orbitrap Hybrid Mass Spectrometer
 Application Note: 367 Essential Lipidomics Experiments using the LTQ rbitrap Hybrid Mass Spectrometer Thomas Moehring 1, Michaela Scigelova 2, Christer S. Ejsing 3, Dominik Schwudke 3, Andrej Shevchenko
Application Note: 367 Essential Lipidomics Experiments using the LTQ rbitrap Hybrid Mass Spectrometer Thomas Moehring 1, Michaela Scigelova 2, Christer S. Ejsing 3, Dominik Schwudke 3, Andrej Shevchenko
Development and Validation of a Polysorbate 20 Assay in a Therapeutic Antibody Formulation by RP-HPLC and Charged Aerosol Detector (CAD)
 LIFE SCIENCES AR ENOUGH? HOW DO YOU KNOW RAW MATERIALS ARE PURE? HOW DO YOU EVALUATE PRODUCT PACKAGING? HOW DO YOU DO YOU KNOW WHAT ANALYTICAL TECHNIQUE TO USE? HOW DO YOU SIMULTANEOUSLY TEST FOR TWO BYPRODUCTS?
LIFE SCIENCES AR ENOUGH? HOW DO YOU KNOW RAW MATERIALS ARE PURE? HOW DO YOU EVALUATE PRODUCT PACKAGING? HOW DO YOU DO YOU KNOW WHAT ANALYTICAL TECHNIQUE TO USE? HOW DO YOU SIMULTANEOUSLY TEST FOR TWO BYPRODUCTS?
GlycanPac AXR-1 Columns
 CHRMATGRAPHY GlycanPac AXR- Columns For High Resolution Glycan Analysis Product Specifications The Thermo Scientific GlycanPac AXR- columns are highperformance, silica-based HPLC columns for simultaneous
CHRMATGRAPHY GlycanPac AXR- Columns For High Resolution Glycan Analysis Product Specifications The Thermo Scientific GlycanPac AXR- columns are highperformance, silica-based HPLC columns for simultaneous
Waters ASMS Users Meeting May 30 th, Overview
 Application of accurate mass spectrometry to mechanistic studies of metabolismdependent inhibition of cytochrome P45 enzymes Joanna Barbara, Ph.D. XenoTech Waters AM 29 Users Meeting May 3, 29 verview
Application of accurate mass spectrometry to mechanistic studies of metabolismdependent inhibition of cytochrome P45 enzymes Joanna Barbara, Ph.D. XenoTech Waters AM 29 Users Meeting May 3, 29 verview
VaTx1 VaTx2 VaTx3. VaTx min Retention Time (min) Retention Time (min)
 a Absorbance (mau) 5 2 5 3 4 5 6 7 8 9 6 2 3 4 5 6 VaTx2 High Ca 2+ Low Ca 2+ b 38.2 min Absorbance (mau) 3 2 3 4 5 3 2 VaTx2 39.3 min 3 4 5 3 2 4. min 3 4 5 Supplementary Figure. Toxin Purification For
a Absorbance (mau) 5 2 5 3 4 5 6 7 8 9 6 2 3 4 5 6 VaTx2 High Ca 2+ Low Ca 2+ b 38.2 min Absorbance (mau) 3 2 3 4 5 3 2 VaTx2 39.3 min 3 4 5 3 2 4. min 3 4 5 Supplementary Figure. Toxin Purification For
Identification of Aromatic Fatty Acid Ethyl Esters
 Chapter 3.2 Identification of Aromatic Fatty Acid Ethyl Esters The only use of gas chromatography is not sufficient to determine which compounds are eluting from the catalytic bed. At the beginning of
Chapter 3.2 Identification of Aromatic Fatty Acid Ethyl Esters The only use of gas chromatography is not sufficient to determine which compounds are eluting from the catalytic bed. At the beginning of
SWATH Acquisition Enables the Ultra-Fast and Accurate Determination of Novel Synthetic Opioids
 SWATH Acquisition Enables the Ultra-Fast and Accurate Determination of Novel Synthetic Opioids Data Independent Acquisition on TripleTOF and X-Series QTOF Systems for Seized Drug Analysis Oscar G. Cabrices
SWATH Acquisition Enables the Ultra-Fast and Accurate Determination of Novel Synthetic Opioids Data Independent Acquisition on TripleTOF and X-Series QTOF Systems for Seized Drug Analysis Oscar G. Cabrices
Improving Selectivity in Quantitative Analysis Using MS 3 on a Hybrid Quadrupole-Linear Ion Trap Mass Spectrometer
 Improving Selectivity in Quantitative Analysis Using MS 3 on a Hybrid Quadrupole-Linear Ion Trap Mass Spectrometer Overview Increased selectivity is achieved in quantitative analysis by using MS 3. The
Improving Selectivity in Quantitative Analysis Using MS 3 on a Hybrid Quadrupole-Linear Ion Trap Mass Spectrometer Overview Increased selectivity is achieved in quantitative analysis by using MS 3. The
Mass Spectrometry Infrastructure
 Mass Spectrometry Infrastructure Todd Williams, Ph.D. Director KU Mass Spectrometry and Analytical Proteomics Laboratory Mass Spectrometry Lab B025 Malott Hall Mission The Mass Spectrometry and analytical
Mass Spectrometry Infrastructure Todd Williams, Ph.D. Director KU Mass Spectrometry and Analytical Proteomics Laboratory Mass Spectrometry Lab B025 Malott Hall Mission The Mass Spectrometry and analytical
Terminology. Metabolite: substance produced or used during metabolism such as lipids, sugars and amino acids
 Terminology Metabolite: substance produced or used during metabolism such as lipids, sugars and amino acids A metabolite may be described as a compound which is internalized, chemically converted or secreted
Terminology Metabolite: substance produced or used during metabolism such as lipids, sugars and amino acids A metabolite may be described as a compound which is internalized, chemically converted or secreted
Supplementary Information. Effects of Perfluorooctanoic Acid on Metabolic Profiles in Brain and Liver of Mouse by a
 Supplementary Information Effects of Perfluorooctanoic Acid on Metabolic Profiles in Brain and Liver of Mouse by a High-throughput Targeted Metabolomics Approach Nanyang Yu, Si Wei, *, Meiying Li, Jingping
Supplementary Information Effects of Perfluorooctanoic Acid on Metabolic Profiles in Brain and Liver of Mouse by a High-throughput Targeted Metabolomics Approach Nanyang Yu, Si Wei, *, Meiying Li, Jingping
First identification of PLTX-like molecules in the Atlantic coral species. Palythoa canariensis
 Supporting information First identification of PLTX-like molecules in the Atlantic coral species Palythoa canariensis M Fraga 1, N Vilariño 1, MC Louzao 1, L Molina 2, Y López 2, M Poli 3 and LM Botana
Supporting information First identification of PLTX-like molecules in the Atlantic coral species Palythoa canariensis M Fraga 1, N Vilariño 1, MC Louzao 1, L Molina 2, Y López 2, M Poli 3 and LM Botana
Separation of Macrocyclic Lactones (Avermectins) on FLARE C18 MM & FLARE C18+ Columns
 Separation of Macrocyclic Lactones (Avermectins) on FLARE C8 MM & FLARE C8+ Columns Introduction Diamond Analytics Technical Note: T05- Avermectins are a series of 6-membered macrocyclic lactone derivatives
Separation of Macrocyclic Lactones (Avermectins) on FLARE C8 MM & FLARE C8+ Columns Introduction Diamond Analytics Technical Note: T05- Avermectins are a series of 6-membered macrocyclic lactone derivatives
Databehandling. 3. Mark e.g. the first fraction (1: 0-45 min, 2: min, 3; min, 4: min, 5: min, 6: min).
 Databehandling Data analysis 1. Choose Open in the Data analysis window. 2. Press the Open folder and choose the desired analysis. Click the + button, so that the Chromatograms line appears. Click the
Databehandling Data analysis 1. Choose Open in the Data analysis window. 2. Press the Open folder and choose the desired analysis. Click the + button, so that the Chromatograms line appears. Click the
O O H. Robert S. Plumb and Paul D. Rainville Waters Corporation, Milford, MA, U.S. INTRODUCTION EXPERIMENTAL. LC /MS conditions
 Simplifying Qual/Quan Analysis in Discovery DMPK using UPLC and Xevo TQ MS Robert S. Plumb and Paul D. Rainville Waters Corporation, Milford, MA, U.S. INTRODUCTION The determination of the drug metabolism
Simplifying Qual/Quan Analysis in Discovery DMPK using UPLC and Xevo TQ MS Robert S. Plumb and Paul D. Rainville Waters Corporation, Milford, MA, U.S. INTRODUCTION The determination of the drug metabolism
Supporting Information (SI)
 Electronic Supplementary Material (ESI) for Analyst. This journal is The Royal Society of Chemistry 2015 Supporting Information (SI) Title: Optimization of Metabolite Extraction of Human Vein Tissue for
Electronic Supplementary Material (ESI) for Analyst. This journal is The Royal Society of Chemistry 2015 Supporting Information (SI) Title: Optimization of Metabolite Extraction of Human Vein Tissue for
Supporting information for. Determination of sub nm levels of low molecular mass (LMM) thiols in natural waters by liquid
 Supporting information for Determination of sub nm levels of low molecular mass (LMM) thiols in natural waters by liquid chromatography tandem mass spectrometry after derivatization with p-(hydroxymercuri)
Supporting information for Determination of sub nm levels of low molecular mass (LMM) thiols in natural waters by liquid chromatography tandem mass spectrometry after derivatization with p-(hydroxymercuri)
LOCALISATION, IDENTIFICATION AND SEPARATION OF MOLECULES. Gilles Frache Materials Characterization Day October 14 th 2016
 LOCALISATION, IDENTIFICATION AND SEPARATION OF MOLECULES Gilles Frache Materials Characterization Day October 14 th 2016 1 MOLECULAR ANALYSES Which focus? LOCALIZATION of molecules by Mass Spectrometry
LOCALISATION, IDENTIFICATION AND SEPARATION OF MOLECULES Gilles Frache Materials Characterization Day October 14 th 2016 1 MOLECULAR ANALYSES Which focus? LOCALIZATION of molecules by Mass Spectrometry
Analyzing Different Compositions of Polygala from Different Regions Using the X500R QTOF System
 Analyzing Different Compositions of Polygala from Different Regions Using the X500R QTF System Li Zhiyuan, Cheng Haiyan, Liu Ting, Li Lijun, Jin Wenhai SCIEX, Pacific Applications Support Center (Beijing),
Analyzing Different Compositions of Polygala from Different Regions Using the X500R QTF System Li Zhiyuan, Cheng Haiyan, Liu Ting, Li Lijun, Jin Wenhai SCIEX, Pacific Applications Support Center (Beijing),
Topic 6 Structure Determination Revision Notes
 1) Introduction Topic 6 Structure Determination Revision Notes Mass spectrometry, infrared spectroscopy and NMR spectroscopy can be used to determine the structure of unknown compounds 2) Mass spectrometry
1) Introduction Topic 6 Structure Determination Revision Notes Mass spectrometry, infrared spectroscopy and NMR spectroscopy can be used to determine the structure of unknown compounds 2) Mass spectrometry
Supplementary Information
 Supplementary Information The conifer biomarkers dehydroabietic and abietic acids are widespread in Cyanobacteria Maria Sofia Costa, Adriana Rego, Vitor Ramos, Tiago Afonso, Sara Freitas, Marco Preto,
Supplementary Information The conifer biomarkers dehydroabietic and abietic acids are widespread in Cyanobacteria Maria Sofia Costa, Adriana Rego, Vitor Ramos, Tiago Afonso, Sara Freitas, Marco Preto,
Supporting Information
 Supporting Information Wiley-VCH 2008 69451 Weinheim, Germany Chemoselective Peptide Cyclization via Induced Traceless Staudinger Ligation Rolf Kleineweischede, Christian P.R. Hackenberger* Institute for
Supporting Information Wiley-VCH 2008 69451 Weinheim, Germany Chemoselective Peptide Cyclization via Induced Traceless Staudinger Ligation Rolf Kleineweischede, Christian P.R. Hackenberger* Institute for
Development and Validation of a Simultaneous HPLC Method for Quantification of Amlodipine Besylate and Metoprolol Tartrate in Tablets
 Journal of PharmaSciTech 0; ():- Research Article Development and Validation of a Simultaneous HPLC Method for Quantification of Amlodipine Besylate and Metoprolol Tartrate in Tablets * Sayyed Hussain,
Journal of PharmaSciTech 0; ():- Research Article Development and Validation of a Simultaneous HPLC Method for Quantification of Amlodipine Besylate and Metoprolol Tartrate in Tablets * Sayyed Hussain,
Application of LC/Electrospray Ion Trap Mass Spectrometry for Identification and Quantification of Pesticides in Complex Matrices
 Application ote #LCMS-2 esquire series Application of LC/Electrospray Ion Trap Mass Spectrometry for Identification and Quantification of Pesticides in Complex Matrices Introduction The simple monitoring
Application ote #LCMS-2 esquire series Application of LC/Electrospray Ion Trap Mass Spectrometry for Identification and Quantification of Pesticides in Complex Matrices Introduction The simple monitoring
Michal Godula Thermo Fisher Scientific. The world leader in serving science
 Resolving the food authenticity challenges using advanced isotopic ratio and Thermo Scientific Orbitrap high resolution mass spectrometry tools in practice Michal Godula Thermo Fisher Scientific The world
Resolving the food authenticity challenges using advanced isotopic ratio and Thermo Scientific Orbitrap high resolution mass spectrometry tools in practice Michal Godula Thermo Fisher Scientific The world
Jose Castro-Perez, Henry Shion, Kate Yu, John Shockcor, Emma Marsden-Edwards, Jeff Goshawk Waters Corporation, Milford, MA, U.S. and Manchester, UK
 HIGH-THRUGHPUT REACTIVE METABLITE SCREEIG FR DICLFEAC BY UPLC AD XEV TQ MS WITH SCAWAVE Jose Castro-Perez, Henry Shion, Kate Yu, John Shockcor, Emma Marsden-Edwards, Jeff Goshawk Waters Corporation, Milford,
HIGH-THRUGHPUT REACTIVE METABLITE SCREEIG FR DICLFEAC BY UPLC AD XEV TQ MS WITH SCAWAVE Jose Castro-Perez, Henry Shion, Kate Yu, John Shockcor, Emma Marsden-Edwards, Jeff Goshawk Waters Corporation, Milford,
Supplementary Results
 upplementary Results upplementary Table 1. Primers used in this study. upplementary Figure 1. PCP3-TE assay in the absence of ul. upplementary Figure 2. Multiple alignment of the sulfazecin TE, highlighting
upplementary Results upplementary Table 1. Primers used in this study. upplementary Figure 1. PCP3-TE assay in the absence of ul. upplementary Figure 2. Multiple alignment of the sulfazecin TE, highlighting
3. AMINO ACID AND PEPTIDES
 3. AMINO ACID AND PEPTIDES 3.1 Amino Acids and Peptides General structure - Only 20 amino-acids are found in proteins - Amino group and carboxyl group - α-carbon and side chain group 3.1 Amino Acids and
3. AMINO ACID AND PEPTIDES 3.1 Amino Acids and Peptides General structure - Only 20 amino-acids are found in proteins - Amino group and carboxyl group - α-carbon and side chain group 3.1 Amino Acids and
Key Words: Brassica oleraceae, glucosinolate, liquid chromatography mass spectrometry, FNH-I-003
 IDENTIFICATION OF MAJOR GLUCOSINOLATES IN BROCCOLI (Brassica oleracea var. italica) BY LIQUID CHROMATOGRAPHY MASS SPECTROMETRY (LC-MS) AND DETERMINATION OF ANTICANCER PROPERTIES OF BROCCOLI EXTRACTS Carlos
IDENTIFICATION OF MAJOR GLUCOSINOLATES IN BROCCOLI (Brassica oleracea var. italica) BY LIQUID CHROMATOGRAPHY MASS SPECTROMETRY (LC-MS) AND DETERMINATION OF ANTICANCER PROPERTIES OF BROCCOLI EXTRACTS Carlos
In-stream attenuation of neuro-active pharmaceuticals and their
 In-stream attenuation of neuro-active pharmaceuticals and their metabolites Supporting Information Analytical Methods and Figures (10 pages) Jeffrey H. Writer, Ronald C. Antweiler, Imma Ferrer, Joseph
In-stream attenuation of neuro-active pharmaceuticals and their metabolites Supporting Information Analytical Methods and Figures (10 pages) Jeffrey H. Writer, Ronald C. Antweiler, Imma Ferrer, Joseph
4-Fluoroethamphetamine
 NMS Labs 2300 Stratford Ave Willow Grove, PA 19090 4-Fluoroethamphetamine Sample Type: Seized Material Latest Revision: January 3, 2019 Date Received: July 31, 2018 Date of Report: January 3, 2019 1. GENERAL
NMS Labs 2300 Stratford Ave Willow Grove, PA 19090 4-Fluoroethamphetamine Sample Type: Seized Material Latest Revision: January 3, 2019 Date Received: July 31, 2018 Date of Report: January 3, 2019 1. GENERAL
Supporting Information
 Supporting Information Wiley-VCH 2008 69451 Weinheim, Germany Supporting Information Enantioselective Cu-catalyzed 1,4-Addition of Various Grignard Reagents to Cyclohexenone using Taddol-derived Phosphine-Phosphite
Supporting Information Wiley-VCH 2008 69451 Weinheim, Germany Supporting Information Enantioselective Cu-catalyzed 1,4-Addition of Various Grignard Reagents to Cyclohexenone using Taddol-derived Phosphine-Phosphite
[ APPLICATION NOTE ] Profiling Mono and Disaccharides in Milk and Infant Formula Using the ACQUITY Arc System and ACQUITY QDa Detector
![[ APPLICATION NOTE ] Profiling Mono and Disaccharides in Milk and Infant Formula Using the ACQUITY Arc System and ACQUITY QDa Detector [ APPLICATION NOTE ] Profiling Mono and Disaccharides in Milk and Infant Formula Using the ACQUITY Arc System and ACQUITY QDa Detector](/thumbs/83/88757464.jpg) Profiling Mono and Disaccharides in Milk and Infant Formula Using the ACQUITY Arc System and ACQUITY QDa Detector Mark Benvenuti, Gareth Cleland, and Jennifer Burgess Waters Corporation, Milford, MA, USA
Profiling Mono and Disaccharides in Milk and Infant Formula Using the ACQUITY Arc System and ACQUITY QDa Detector Mark Benvenuti, Gareth Cleland, and Jennifer Burgess Waters Corporation, Milford, MA, USA
CEU MASS MEDIATOR USER'S MANUAL Version 2.0, 31 st July 2017
 CEU MASS MEDIATOR USER'S MANUAL Version 2.0, 31 st July 2017 1. Introduction... 2 1.1. System Requirements... 2 2. Peak search... 3 2.1. Simple Search... 3 2.2. Advanced Search... 5 2.3. Batch Search...
CEU MASS MEDIATOR USER'S MANUAL Version 2.0, 31 st July 2017 1. Introduction... 2 1.1. System Requirements... 2 2. Peak search... 3 2.1. Simple Search... 3 2.2. Advanced Search... 5 2.3. Batch Search...
Quantitative Analysis of -Hydroxybutyrate in Hair Using Target Analyte Finding Processing of Comprehensive GC-HRT Data
 Quantitative Analysis of -Hydroxybutyrate in Hair Using Target Analyte Finding Processing of Comprehensive GC-HRT Data LECO Corporation; Saint Joseph, Michigan USA Key Words: Pegasus GC-HRT, GHB, Hair,
Quantitative Analysis of -Hydroxybutyrate in Hair Using Target Analyte Finding Processing of Comprehensive GC-HRT Data LECO Corporation; Saint Joseph, Michigan USA Key Words: Pegasus GC-HRT, GHB, Hair,
AMINO ACIDS AND PEPTIDES Proteins/3 rd class of pharmacy
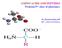 AMINO ACIDS AND PEPTIDES Proteins/3 rd class of pharmacy Dr. Basima Sadiq Jaff PhD. Clinical Biochemistry MEDICAL AND BIOLOGICAL IMPORTANCE 1. building blocks of proteins. Some amino acids are found in
AMINO ACIDS AND PEPTIDES Proteins/3 rd class of pharmacy Dr. Basima Sadiq Jaff PhD. Clinical Biochemistry MEDICAL AND BIOLOGICAL IMPORTANCE 1. building blocks of proteins. Some amino acids are found in
ANALYTICAL REPORT 1 3C-P (C14H23NO3) 1-(3,5-dimethoxy-4-propoxylphenyl)propan-2-amine.
 Vodovodna 95, 1000 LJUBLJANA, SLOVENIA T: +386 (0)1 428 44 93 E: nfl@policija.si www.policija.si Remark other NPS detected: none ANALYTICAL REPORT 1 3C-P (C14H23NO3) 1-(3,5-dimethoxy-4-propoxylphenyl)propan-2-amine
Vodovodna 95, 1000 LJUBLJANA, SLOVENIA T: +386 (0)1 428 44 93 E: nfl@policija.si www.policija.si Remark other NPS detected: none ANALYTICAL REPORT 1 3C-P (C14H23NO3) 1-(3,5-dimethoxy-4-propoxylphenyl)propan-2-amine
Development of non-aqueous single stage derivatisation method for the determination of putrescine and cadaverine using GC-MS
 Cent. Eur. J. Chem. 6(2) 2008 229 236 DOI: 10.2478/s11532-008-0007-6 Central European Journal of Chemistry Development of non-aqueous single stage derivatisation method for the determination of putrescine
Cent. Eur. J. Chem. 6(2) 2008 229 236 DOI: 10.2478/s11532-008-0007-6 Central European Journal of Chemistry Development of non-aqueous single stage derivatisation method for the determination of putrescine
Tivadar Orban, Beata Jastrzebska, Sayan Gupta, Benlian Wang, Masaru Miyagi, Mark R. Chance, and Krzysztof Palczewski
 Structure, Volume Supplemental Information Conformational Dynamics of Activation for the Pentameric Complex of Dimeric G Protein-Coupled Receptor and Heterotrimeric G Protein Tivadar Orban, Beata Jastrzebska,
Structure, Volume Supplemental Information Conformational Dynamics of Activation for the Pentameric Complex of Dimeric G Protein-Coupled Receptor and Heterotrimeric G Protein Tivadar Orban, Beata Jastrzebska,
