Infection, Genetics and Evolution
|
|
|
- Lee Warren
- 6 years ago
- Views:
Transcription
1 Infection, Genetics and Evolution 12 (2012) Contents lists available at SciVerse ScienceDirect Infection, Genetics and Evolution journal homepage: Review The revised Trypanosoma cruzi subspecific nomenclature: Rationale, epidemiological relevance and research applications Bianca Zingales a,, Michael A. Miles b, David A. Campbell c, Michel Tibayrenc d, Andrea M. Macedo e, Marta M.G. Teixeira f, Alejandro G. Schijman g, Martin S. Llewellyn b, Eliane Lages-Silva h, Carlos R. Machado e, Sonia G. Andrade i, Nancy R. Sturm c a Departamento de Bioquímica, Instituto de Química, Universidade de São Paulo, Avenida Professor Lineu Prestes 748, São Paulo, SP, Brazil b The London School of Hygiene and Tropical Medicine, Keppel Street, London WC1E 7HT, UK c Department of Microbiology, Immunology & Molecular Genetics, David Geffen School of Medicine, University of California at Los Angeles, Le Conte Ave, Los Angeles, CA , USA d Maladies Infectieuses et Vecteurs Ecologie, Génétique, Evolution etcontrôle, MIVEGEC/IDVEGEC, UM1-CNRS 5290-IRD 224, IRD Center, BP 64501, Montpellier Cedex 5, France e Departamento de Bioquímica e Imunologia, Instituto de Ciências Biológicas, Universidade Federal de Minas Gerais, CP 486, Belo Horizonte, MG, Brazil f Departamento de Parasitologia, Instituto de Biociências, Universidade de São Paulo, Avenida Professor Lineu Prestes 1374, São Paulo, SP, Brazil g Laboratorio de Biología Molecular de la Enfermedad de Chagas, INGEBI-CONICET, Vuelto Obligado 2490, Buenos Aires 1428, Argentina h Departamento de Ciências Biológicas, Universidade Federal do Triângulo Mineiro, Rua Frei Paulino 30, Uberaba, MG, Brazil i Centro de Pesquisas Gonçalo Moniz, Fundação Oswaldo Cruz, Rua Waldemar Falcão 121, Salvador, Brazil article info abstract Article history: Received 31 October 2011 Accepted 16 December 2011 Available online 27 December 2011 Keywords: Trypanosoma cruzi strains Discrete typing unit Genotyping Phylogeography Hybridization Pathology The protozoan Trypanosoma cruzi, its mammalian reservoirs, and vectors have existed in nature for millions of years. The human infection, named Chagas disease, is a major public health problem for Latin America. T. cruzi is genetically highly diverse and the understanding of the population structure of this parasite is critical because of the links to transmission cycles and disease. At present, T. cruzi is partitioned into six discrete typing units (DTUs), TcI TcVI. Here we focus on the current status of taxonomy-related areas such as population structure, phylogeographical and eco-epidemiological features, and the correlation of DTU with natural and experimental infection. We also summarize methods for DTU genotyping, available for widespread use in endemic areas. For the immediate future multilocus sequence typing is likely to be the gold standard for population studies. We conclude that greater advances in our knowledge on pathogenic and epidemiological features of these parasites are expected in the coming decade through the comparative analysis of the genomes from isolates of various DTUs. Ó 2012 Elsevier B.V. All rights reserved. Contents 1. Introduction The concept of discrete typing unit The clonal model of evolution in T. cruzi Discrete typing units and clonets Two major models for the origin of hybrid DTUs Phylogeography of the DTUs TcI and its extensive genetic diversity TcIII and TcIV TcII, TcV and TcVI An enigmatic T. cruzi genotype from bats (Tcbat) Abbreviations: ITS1 rdna, internal transcribed spacer 1 of rdna; MLEE, multilocus enzyme electrophoresis; MLST, multilocus sequence typing; NTS, non-transcribed spacer; RAPD, randomly amplified polymorphic DNA; RFLP, restriction fragment length polymorphism; SNPs, single-nucleotide polymorphisms; SL, spliced leader; SL-IR, spliced leader intergenic sequence. Corresponding author. Tel.: ; fax: address: zingales@iq.usp.br (B. Zingales) /$ - see front matter Ó 2012 Elsevier B.V. All rights reserved. doi: /j.meegid
2 B. Zingales et al. / Infection, Genetics and Evolution 12 (2012) Standardizing genotyping for identification of the six T. cruzi DTUs Comparative experimental pathology of the DTUs T. cruzi DTUs and human Chagas disease Clinical presentations Acute Chagas disease Chronic Chagas disease Chagas disease reactivation due to immunosuppression T. cruzi genomics Concluding remarks and perspectives Acknowledgements References Introduction Infection with Trypanosoma cruzi is a complex zoonosis, transmitted by many hematophagous triatomine species and sustained by over 70 genera of mammalian reservoir hosts. T. cruzi has a broad endemic range that extends from the Southern United States to Argentinean Patagonia. The human infection, named Chagas disease in recognition of Carlos Chagas who first discovered American trypanosomiasis in 1909, is found mostly in South and Central America, primarily affects poor rural populations, and is considered to be the most important parasitic infection in Latin America with serious consequences for public health and national economies. The spectrum of pathological outcomes associated with acute and chronic Chagas disease ranges from subclinical infection through the cardiac and digestive syndromes to death. Specific outcomes may be determined by a variety of non-exclusive factors including parasite genetics, host genetics, mixed infections, and cultural and geographical factors (Macedo et al., 2002, 2004; Buscaglia and Di Noia, 2003; Campbell et al., 2004). The diversity of the T. cruzi genome and multiplicity of its genotypes and phenotypes is well recognized (Dvorak et al., 1982; Barnabé et al., 2000; Brisse et al., 2000; Devera et al., 2003; Lewis et al., 2009a). Designation of ecologically and epidemiologically relevant groups for T. cruzi has oscillated between a few discrete groups (Miles and Cibulskis, 1986; Souto and Zingales, 1993; Souto et al., 1996; Zingales et al., 1999) and many (Tibayrenc and Ayala, 1988). Currently, six discrete typing units (DTUs) are assigned (Brisse et al., 2000). In 2009, these DTUs were renamed by consensus as TcI TcVI (Zingales et al., 2009). Several reviews already describe how these DTUs correspond with former nomenclatures and with prospective biological and host associations (Campbell et al., 2004; Miles et al., 2009; Sturm and Campbell, 2010; Zingales et al., 2009). The aim of this review is to explain further the rationale for naming TcI TcVI, with reference to their known molecular genetics, eco-epidemiological features and pathogenicity. We also summarize methods for DTU genotyping, and discuss a possible seventh T. cruzi branch, provisionally named Tcbat. An understanding of the T. cruzi DTUs and their epidemiological implications will provide new insights to guide research and future interventions against this devastating infectious disease. 2. The concept of discrete typing unit Since the late 1970s, T. cruzi has become one of the models for molecular epidemiologists and population geneticists, and consequently this protozoan parasite is a pathogenic agent for which evolution and population structure are among the best studied, although not necessarily the best understood. The emerging picture is that of a typical pattern of reticulate evolution, similar to that of many plant species (Avise, 2004). The concepts of DTUs and clonal evolution have been designed within the framework of evolutionary research on T. cruzi. Tibayrenc and co-workers devised descriptive concepts and terminology to make such research and its implications accessible to non-specialists, including medical professionals and epidemiologists, and to bypass certain demands of classical evolutionary biology definitions. Classical cladistic and population genetics approaches imperfectly depict the biological realities of the evolution of pathogenic microorganisms The clonal model of evolution in T. cruzi In the framework of this model, a clonal species refers to all cases where descendant multilocus genotypes are virtually identical to the founding genotype. The main parameter focused on in this scenario is the inhibition of genetic recombination. The term clone in this context refers to the population structure of the species under study, not to its precise mating system. Different methods of propagation can generate genetic clones, including classical cell division, several cases of parthenogenesis and gynogenesis. Following this definition (Tibayrenc et al., 1990), selfing and extreme inbreeding are not alternative hypotheses to clonality (Rougeron et al., 2009), but rather a particular case of it. Selfing refers to mating between identical genotypes, which can be issued from the same clone (Tibayrenc et al., 2010). Extreme inbreeding refers to mating between extremely similar genotypes. The result is a lack or extreme limitation of genetic recombination, hence genetic clonality. Stating that T. cruzi is a basically clonal species means neither that recombination is totally absent in the parasite s natural populations, nor that it does not have an impact on the evolutionary scale, but rather that it is too rare to break the prevalent pattern of clonality. The potential for genetic exchange is still present (Gaunt et al., 2003). Moreover, some localized transmission cycles suggest that genetic recombination does occur within DTUs of T. cruzi (Carranza et al., 2009; Ocaña-Mayorga et al., 2010). The possibility of limited genetic exchange between DTUs is also under debate (Lewis et al., 2011). However, the species T. cruzi considered as a whole shows all the signs for a typical clonal population structure: departures from panmictic expectations, strong linkage disequilibrium (non-random association of genotypes at different loci) within and especially between DTUs, and division into discrete genetic clusters (see DTUs, below) Discrete typing units and clonets Often the genetic subdivisions identified by evolutionary studies in pathogen species do not fulfill the criteria demanded by rigorous cladistic analysis. The main reason is that even in predominantly clonal species such as T. cruzi there is a certain amount of genetic recombination that clouds the distinction of
3 242 B. Zingales et al. / Infection, Genetics and Evolution 12 (2012) phylogenetic subdivisions. By definition, a clade represents an evolutionary unit that is strictly isolated from other evolutionary units. Moreover, some pathogen lineages may have a hybrid origin: in other words, two ancestors. This is the case of several T. cruzi genetic subdivisions (Sturm and Campbell, 2010). Thus the clade concept is not applicable in the case of T. cruzi. However, even when some genetic exchange occurs, discrete and stable subdivisions can be identified reliably in many instances. Thus the term discrete typing unit (Tibayrenc, 1998) was proposed to describe sets of stocks that are genetically more similar to each other than to any other stock, and are identifiable by common molecular markers sometimes referred to as tags. The DTUs constitute reliable units for analysis for molecular epidemiology and experimental studies of evolution. Genetic clusters within T. cruzi perfectly fit this definition, and thus the DTUs TcI TcVI have been assigned (Brisse et al., 2000; Zingales et al., 2009). Within these DTUs T. cruzi stocks that share profiles for a given panel of molecular markers are not necessarily genetically identical, and can often be distinguished with additional markers. Thus strains within DTUs should be considered as families of closely related clones, not as a single clone. Tibayrenc and Ayala (1991) coined the term clonet to refer to sets of stocks that appear to be indistinguishable with a given set of genetic markers in a basically clonal species, such as T. cruzi. The clonets are relevant units of analysis for molecular epidemiology, for example, with multilocus sequence types (MLSTs) or karoytypes derived from pulsedfield gel electrophoresis. However, it is crucial to keep in mind that the most recent common ancestor of a given clonet can be either a few weeks or hundreds of years old, depending on the marker s power of resolution and rate of evolutionary change (molecular clock). The latter parameter cannot be known a priori and may have considerable epidemiological relevance. 3. Two major models for the origin of hybrid DTUs T. cruzi is predominantly diploid (El-Sayed et al., 2005) and the known cell replication method is binary fission, i.e. it is an asexual process. Under the clonal model (above) new DTUs evolve with the accumulation of discrete mutations, unaffected by rare events of genetic exchange. However, consistent with the caveat that some genetic exchange events may occur, evidence for T. cruzi heterozygosity in nature emerged through the study of individual genes (Chapman et al., 1984; Bogliolo et al., 1996; Carrasco et al., 1996; Souto et al., 1996; Brisse et al., 1998). The pronounced heterozygosity observed in natural isolates of TcV and TcVI suggested that these DTUs are hybrids and derived from TcII and TcIII (Sturm et al., 2003; Sturm and Campbell, 2010). The remaining DTUs, TcI, TcII, TcIII, TcIV (and Tcbat, see below) show substantial allelic homozygosity. While the tenet of the clonal theory may still explain the common mode of T. cruzi population expansion, newer models incorporate hybridization events to explain the extant population structure that includes hybrid DTUs (Westenberger et al., 2005; Freitas et al., 2006). The Two-Hybridization model (Westenberger et al., 2005) and the Three Ancestor model (Freitas et al., 2006) both incorporate two hybridization events (Fig. 1). In the Three Ancestor model the two recent genetic exchange events between TcII and TcIII yield TcV and TcVI. The Two-Hybridization model invokes one ancient genetic exchange event between TcI and TcII, with loss of heterozygosity among progeny to produce TcIII and TcIV, followed by a second more recent hybridization event between TcII and TcIII to yield both TcV and TcVI. Analysis of single-nucleotide polymorphisms (SNPs) among the six DTUs by multilocus sequence typing (MLST) revealed four rather than six distinct DNA sequence classes, termed haplogroups (Machado and Ayala, 2001; Sturm et al., 2003; Broutin et al., 2006). Two of the four haplogroups were always present in TcV and TcVI, confirming the predominantly heterozygous nature of their alleles. Despite being similar by most standards TcV and TcVI are distinguishable by isoenzyme electrophoresis (Chapman et al., 1984; Barnabé et al., 2000), ribosomal RNA markers (Souto et al., 1996), restriction fragment length polymorphism (RFLP) assays (Rozas et al., 2008), some MLST markers (Yeo et al., 2011), and microsatellite analysis (Lewis et al., 2011). The nucleotide patterns in each TcV/TcVI haplogroup closely resemble TcII and TcIII alleles (Machado and Ayala, 2001; Brisse et al., 2003; Westenberger et al., 2005; Freitas et al., 2006; Yeo et al., 2011; Lewis et al., 2011), confirming that TcII and TcIII as the most likely parental types of TcV and TcVI. However, different MLST data indicated that the TcIII parental cells included characters derived from TcI and TcII (Sturm et al., 2003; Elias et al., 2005; Tomazi et al., 2009). Thus, TcIII, as well as TcV and TcVI, could be the product of a hybridization event (Westenberger et al., 2005; Ienne et al., 2010). The major difference between the Two Hybridization and the Three Ancestor models is therefore whether TcV and TcVI are progeny from a single hybridization event incorporating TcI alleles acquired via TcIII (Westenberger et al., 2005) or progeny of two Fig. 1. Comparison of (A) the Two-Hybridization and (B) the Three Ancestor models for the roles of genetic exchange during the clonal evolution of T. cruzi. Rectangles indicate the distinct DTUs. Fusion of two cells and genetic exchange is indicated by the ovals, with parental contribution indicated by the red arrows. The mitochondrial clades are shown by fill colors: Blue = clade A; Green = clade B; orange = clade C.
4 B. Zingales et al. / Infection, Genetics and Evolution 12 (2012) Fig. 2. Approximate geographical distribution of T. cruzi DTUs in domestic and silvatic transmission cycles. Table 1 Summary of ecotope, host, vector and disease associations of T. cruzi DTUs. a Genotype Ecotope/niche Silvatic hosts Silvatic vectors Geography b Chagas disease TcI TcII Primary: arboreal; palms (e.g Attalea), tree holes Secondary: arid, rocky; terrestrial in Amazonia Incompletely known; rare in silvatic cycles Primary: arboreal, semi-arboreal; especially Didelphis, other didelphids, arboreal rodents, primates, Tamandua Secondary: terrestrial rodents Incompletely known: Atlantic forest primates, didephids, Euphractus (Paraguay) TcIII Terrestrial, fossorial Armadillos, especially Dasypus, Chaetophractus, Euphractus; Didephids, Monodelphis TcIV Arboreal, and some Primates, Nasua nasua, terrestrial hosts in North America. TcV Rare in silvatic cycles Incompletely known: Dasypus, Euphractus, Octodon a Primary: Rhodnius species Secondary: Panstrongylus Triatoma Eratyrus Incompletely known: (Triatomini) South, Central and North America Southern Cone, sporadic further North North of the Amazon, sporadic in Southern Cone Cardiomyopathy Atlantic and Central Brazil. Cardiomyopathy, megasyndromes P. geniculatus South America Rare in humans (also domestic dogs). Acute cases in Amazonian Brazil. Clinical presentation poorly known Rhodnius, Panstrongylus, Triatoma Incompletely known TcVI Rare in silvatic cycles Incompletely known Incompletely known Ecotope host and vector associations are not exclusive. b See map in Fig. 2. North and South America Southern Cone, greater Gran Chaco, extreme South of Brazil Southern Cone, greater Gran Chaco Secondary cause of Chagas disease in Venezuela, sporadic elsewhere in South America Southern cone Cardiomyopathy, megasyndromes. Vector borne Southern cone Cardiomyopathy, megasyndromes hybridization events excluding TcI (Freitas et al., 2006) (Fig. 1). The participation of TcI in the generation of the extant heterozygous lines is supported by maxicircle sequences (Westenberger et al., 2006a; Ruvalcaba-Trejo and Sturm, 2011), MLST data (Tomazi et al., 2009), RFLP data (Rozas et al., 2008), and 195-bp satellite DNA sequences and distribution (Elias et al., 2005; Ienne et al., 2010). Conversely, maxicircle-encoded genes cytochrome oxidase subunit II and NADH dehydrogenase subunit 1 (Machado and Ayala, 2001; Freitas et al., 2006), microsatellite analyses (Freitas et al., 2006) and some other nuclear markers (Machado and Ayala, 2002; Rozas et al., 2007) did not detect participation of TcI. The comparison of the new genome sequence of the Sylvio X10/1 strain (Franzen et al., 2011), representative of TcI, with CL Brener, a TcVI hybrid that encompasses both TcII and TcIII genomes (El-Sayed et al., 2005), has confirmed that TcIII has an average higher genetic similarity at the genome sequence level with TcI than TcII. This observation is not inconsistent with the previous conclusion that TcIII may be the product of an ancient hybridization between TcI and TcII, as depicted in the Two Hybridization model of Fig. 1 (Elias et al., 2005; Westenberger et al., 2005). However, the possibility
5 244 B. Zingales et al. / Infection, Genetics and Evolution 12 (2012) that characteristics shared by TcI and TcIII are ancestral states that have diverged in TcII may need further consideration. 4. Phylogeography of the DTUs Setting aside the theoretical origins of the DTUs, divergent geographical and biological characteristics are apparent, as is their relevance to understanding of the eco-epidemiology of Chagas disease (Fig. 2). It is not surprising that, given the current level of sampling, the ecological history for all T. cruzi DTUs cannot yet be fully discerned. Relationships have been obscured by massive changes, from mammal migrations between the Americas, to climate induced retraction and expansion of habitats, and dramatic recent habitat destruction and urbanization by humans. A summary of ecotope, host, and vector associations of T. cruzi DTUs is given in Table 1, as has been extensively reviewed elsewhere (Miles et al., 2009). Here we will provide primarily more information on the phylogeography and the extensive genetic diversity of TcI, for which there has been recent rapid knowledge progress. In terms of propensity to cause severe Chagas disease, all six DTUs are known to be infective to humans, and clinical aspects are described in more detail in Section TcI and its extensive genetic diversity TcI is the most abundant and widely dispersed of all the T. cruzi DTUs in the Americas. It is found throughout the range of triatomine vector distribution, and can be associated with silvatic and domestic cycles. Human infection with TcI is concentrated in the north of South and Central America, and is associated with chagasic cardiomyopathy. There are only disparate reports of infection and disease south of the Amazon basin. Wild TcI isolates exist from Alabama in the United States (Roellig et al., 2008) at 32 North, to Limari, Chile (Apt et al., 1987)at30 South. Review of the literature reveals 52 mammalian genera naturally infected with this DTU, with representatives from Marsupialia, Rodentia, Primata, Chiroptera, Xenartha, Carnivora, and Artiodactyla in order of abundance, as well as all major genera of triatomine bugs (Llewellyn, unpublished records, updated 05/01/2010). The divergence date between TcI and TcII is ill defined, estimated between 88 and 37 million years ago, based on small subunit rdna (Briones et al., 1999; Kawashita et al., 2001) and between 16 and 3 million years ago, based on dihydrofolate reductase-thymidylate synthase and trypanothione reductase genes (Machado and Ayala, 2001). Unsurprisingly for a parasite so ancient and dispersed, significant genetic diversity has accumulated within TcI. The earliest recognition of TcI heterogeneity is manifest in the isoenzyme clonet typing scheme proposed by Tibayrenc and co-workers whereby 25 genotypes are assigned to TcI, with a much lower number assigned to any other of the other five DTUs (Tibayrenc et al., 1986). Sample size may have represented an early confounder, and we now know that substantial diversity has also accumulated within other lineages (Machado and Ayala, 2001; Westenberger et al., 2006b; Llewellyn et al., 2009a; Marcili et al., 2009a). Access to new, high-resolution genotyping techniques has seen a resurgence of interest in the delineation of TcI intra-dtu diversity. Saravia et al. (1987) examined genetic diversity among 54 Colombian T. cruzi isolates collected from silvatic and domestic localities at several foci in Meta, Casanare and Cundinamarca provinces using 13 isoenzyme markers. Of those isolates examined, most were TcI ( Z1-like ). Furthermore, among TcI isolates, marked genetic subdivision was observed between strains from domestic and silvatic transmission cycles, largely independent of geographic origin. This biological observation is now supported by spliced leader (SL, also known as mini-exon) intergenic region (SL-IR) sequence data from western Colombia (Herrera et al., 2007, 2009; Falla et al., 2009). Several other molecular methods have been applied to study TcI heterogeneity in Colombia, including molecular karyotypic analysis (Triana et al., 2006), SL probe hybridization (Triana et al., 2006) and minicircle random RFLP (Jaramillo et al., 1999). More recently, low-stringency single primer PCR (Rodriguez et al., 2009) provided important insight into the dynamics of TcI transmission among several communities in the Sierra Nevada de Santa Marta. Outside Colombia, multilocus enzyme electrophoresis (MLEE) and randomly amplified polymorphic DNA (RAPD) were used to reveal putative hybrid and parental strains at a focus of silvatic TcI transmission in Carajás, Pará State, Brazil (Carrasco et al., 1996). Using the same parental strains, an extant capacity for genetic exchange was demonstrated in vitro (Gaunt et al., 2003). Polymorphic microsatellites can resolve T. cruzi inter-specific variability (Oliveira et al., 1998), and Llewellyn et al. (2009a) demonstrated their efficiency in revealing TcI intra-dtu diversity at a continental scale. Crucially, the use of a multilocus typing system permits inference of linkage disequilibrium between markers within parasite populations and the extent of clonal vs. sexual reproduction. Thus, when the same markers were employed to analyze the molecular epidemiology of TcI transmission at restricted geographic foci in Ecuador (Ocaña-Mayorga et al., 2010), the first population genetic evidence for genetic exchange in TcI was uncovered among isolates not subdivided in space or time. The central observation of almost all analyses of TcI diversity in northern South America is the apparent subdivision between domestic and silvatic cycles of transmission (Saravia et al., 1987; Herrera et al., 2007; Falla et al., 2009; Llewellyn et al., 2009a,b; Ocaña-Mayorga et al., 2010). However, compiling data from these studies is frustrated by constraints of the different genotyping techniques employed. Certain types of population genetic data, especially those derived from microsatellites and isoenzyme loci, are difficult to standardize between studies. Sequence data, on the other hand, are a more versatile population genetic currency. Cura et al. (2010) demonstrated this in an ambitious multi-centric study using the SL-IR, and corrected the erroneous use of the term haplotype in the context of these data (Cura et al., 2010). On the basis of this dataset, the authors delineated several discrete TcI groups, some widely dispersed, as well as instances of mixed infections of genotypes in humans and vectors. Whilst the SL-IR represents an accessible marker as it is diverse, easy to amplify directly from biological samples, and straightforward to sequence with no internal primers required, there are several limitations associated with its use. First, it is a multicopy gene. Non-identical copies are tandemly repeated hundreds of times throughout the T. cruzi genome, and orthology between samples is impossible to ascertain. Second, polymorphic microsatellites located at the 5 0 end introduce numerous ambiguous alignments, with an adverse effect on phylogenetic stability (Tomasini et al., 2011). Third, significant insertions and/or deletions (indels) in this region with respect to other T. cruzi DTUs (Souto et al., 1996) prohibit the identification of a suitable outgroup. Perhaps the most important criticism, however, is not intrinsic to the SL-IR per se. Gene trees are not genome trees. The use of a single genetic locus to describe genetic diversity in an organism limits conclusions that can be drawn, especially in the context of genetic recombination, which may occur not only in TcI (Ocaña-Mayorga et al., 2010) but also other DTUs. Typing strategies must be improved and standardized if further progress is to be made. Sequence data are the ideal genotypic format for swift comparison. However, new, low copy number, highly discriminatory markers must be identified, aided by the publication of the Sylvio X10/1 genome (Franzen et al., 2011). Furthermore, the T. cruzi mitochondrial genome, used for bar-coding so many other
6 B. Zingales et al. / Infection, Genetics and Evolution 12 (2012) species, should not be ignored (Machado and Ayala, 2001; Freitas et al., 2006; Spotorno et al., 2008; Carranza et al., 2009), although some incongruence between nuclear and mitochondrial markers is likely due to introgression events. Accordingly, and if intra-dtu genetic recombination is common, the delineation of fixed genetic groups within TcI represents at best a distraction. Studies targeted at TcI diversity must be designed with a specific biological or epidemiological hypothesis in mind, not undertaken merely to categorize diversity for its own sake. Irrespective of the genotyping system involved, studies like those of Saravia et al. (1987), Falla et al. (2009), Rodriguez et al. (2009) and Ocaña-Mayorga et al. (2010) all provide significant insight into the epidemiology of local transmission. Thus their conclusions inform future interventions to the benefit of the communities involved TcIII and TcIV TcIII is mostly associated with the silvatic cycle in Brazil and adjacent countries, and documented human infections are rare. Silvatic TcIII is associated with the terrestrial niche and with Dasypus novemcinctus, over a vast range from western Venezuela to the Argentine Chaco (Llewellyn et al., 2009a; Marcili et al., 2009a). TcIII is also isolated occasionally from domestic dogs (Chapman et al., 1984; Cardinal et al., 2008). TcIV, shows a similar pattern of distribution in South America to TcIII, with the exception of the Chaco, where it appears to be absent. Unlike TcIII, TcIV occurs fairly frequently in humans and is a secondary cause of Chagas disease in Venezuela (Miles et al., 1981). Five new isolates of TcIV from primates and eight from Rhodnius brethesi in the Amazon basin were recovered (Marcili et al., 2009b), confirming earlier indications (Yeo et al., 2005) that TcIV can have an arboreal ecotope. Evidence is accumulating that TcIV is split into distinct South and North American lineages (Lewis et al., 2009b; Marcili et al., 2009b). Further research is required to understand the history of TcIV and these complex ecological associations TcII, TcV and TcVI TcII is found predominantly in the southern and central regions of South America, but its true extent is not yet clear. Within its main geographic distribution TcII is associated with cardiac manifestations, and concomitant megaesophagus and megacolon may be present. It has been isolated mostly from domestic transmission cycles. The natural hosts and vectors of TcII have proven elusive and most of the reported isolations have been made in remaining fragments of the Atlantic forest of Brazil, from primates and sporadically from other mammal species (Fernandes et al., 1999; Zingales et al., 1999; Lisboa et al., 2007). TcV and TcVI are two similar hybrid DTUs associated with Chagas disease in southern and central South America. Even more so than TcII, TcV and VI are virtually unknown as silvatic isolates. Comparative molecular genetics have proven that TcV and TcVI are hybrids of TcII and TcIII (see above). Until recently genetic markers have not been of sufficient resolution to determine firstly, whether TcV and TcVI are the products of independent hybridization events (Freitas et al., 2006) or a single hybridization event followed by clonal divergence, and secondly, whether these hybridization(s) were evolutionarily ancient (Tibayrenc and Ayala, 2002; Brisse et al., 2003) or recent events (Machado and Ayala, 2001; Westenberger et al., 2005). Two recent studies have attempted to address these issues: Flores-López and Machado (2011) analyzed the evolution of 31 nuclear genes to show the hybridization events occurred less than 1 million years ago, concluding a single event prior to arrival of humans in the Americas. Lewis et al. (2011) analyzed higher resolution maxicircle sequences and multiple microsatellite loci and found evidence for two independent events dated to within the last 100,000 years, concluding that hybridization may conceivably have occurred as a result of human activities. TcII and TcIII may have met and hybridized as co-infections emerged in humans, peridomestic mammals, or domestic Triatoma infestans. Nevertheless, understanding of the ecology of TcII, V and VI is as yet vulnerable to the limited sampling of silvatic hosts and vectors. The paradigms may change. It has been suggested that TcII, V and VI are more widespread geographically than currently understood and might be found much further North (Zafra et al., 2008). If the known TcV and TcVI hybrids are also found in Central and North America it will most likely imply recent migration with humans or other carriers; if genetically distinct TcII/TcIII hybrids are observed, hybridization may be an ongoing phenomenon where such mixed infections occur. Indeed in some endemic areas, notably parts of Bolivia, mixed DTU infections are common. Microsatellite analysis reveals that even within a single mammal, there may be a remarkable range of mixed genotypes (Llewellyn et al., 2011) An enigmatic T. cruzi genotype from bats (Tcbat) Silvatic cycles of T. cruzi transmission are numerous and complex. DTUs circulate in relatively independent cycles with particular ecological niches and preferentially or opportunistically determined mammals and vectors. However, members of the same DTU can infect mammals of distinct species and orders, indicating that host-switching may be common among sympatric hosts (Gaunt and Miles, 2000; Yeo et al., 2005; Marcili et al., 2009a,b; Miles et al., 2009). Several species of the genus Trypanosoma occur in species of Chiroptera throughout the world, with more than 30 trypanosome species recorded from more than 100 species of bats. Insectivorous bat species are infected more frequently and can harbor stercorarian (subgenera Herpetosoma, Schizotrypanum and Megatrypanum) and salivarian (Trypanosoma evansi of the subgenus Trypanozoon) trypanosomes. An extensive summary of the prevalence of bat trypanosomes worldwide is available (Cavazzana et al., 2010). The strong association between bats and all Schizotrypanum spp. except T. cruzi suggests a long shared evolutionary history. However, the evolutionary processes that have led to the current phylogenetic structure of Schizotrypanum trypanosomes are understood poorly. Phylogenetic studies of chiropteran stercorarian species can enhance understanding of host-parasite interactions and reconstruction of T. cruzi evolutionary history (Stevens et al., 2001; Barnabé et al., 2003; Cavazzana et al., 2010). Brazilian bats infected with T. cruzi are reported from the Amazonian rainforest to urban areas of Central, Northeast and Southeast Brazil (references cited in Marcili et al., 2009c). To date, most in vitro adapted isolates from bats belong to the subgenus Schizotrypanum. Identification of T. cruzi from bats requires careful analysis; Schizotrypanum species are morphologically indistinguishable and generically named as T. cruzi-like. However, T. cruzi can be confirmed by the ability to infect mice. Since T. cruzi isolates from wild mammals may induce very low parasitemias in mice, as is the case for bat isolates, infections must be evaluated using immunocompromized mice and sensitive parasitological methods such as PCR and multiple haemocultures. Analysis of SSU rdna, ggapdh and cytochrome b sequences allows separation of T. cruzi from other trypanosomes infecting Brazilian bats, including Trypanosoma cruzi-marinkellei, Trypanosoma dionisii-like and Trypanosoma rangeli (Maia da Silva et al., 2009; Marcili et al., 2009c; Cavazzana et al., 2010). Traditional genotyping methods based on the SL (Fernandes et al., 2001) and LSU rdna (Souto et al., 1996) markers placed four bat isolates from Amazonia within TcI. However, 11 bat isolates from other Brazilian regions
7 246 B. Zingales et al. / Infection, Genetics and Evolution 12 (2012) yielded a new combination of genotypes, with a TcII-SL pattern and a novel LSU rdna product (Marcili et al., 2009c). This group of isolates earned the provisional title of Tcbat and awaits further characterization for definitive DTU assignment (Marcili et al., 2009c), potentially as a seventh DTU: TcVII. Tcbat is distinguished from the six DTUs by PCR-RFLP analysis of the internal transcribed spacer 1 of rdna (ITS1 rdna) (Marcili et al., 2009c). The method of fluorescent fragment length barcoding, developed to identify species of trypanosomes on the basis of polymorphisms of regions of the rdna locus, when applied to T. cruzi DTUs showed a unique barcoding pattern for Tcbat (Hamilton et al., 2011). Karyotype (Marcili et al., 2009c) and sequence analyses of SL gene repeats (D.A.C. and N.R.S., unpublished results) corroborated that Tcbat diverges from the known DTUs. All phylogenetic analyses using sets of molecular markers point to the placement of Tcbat in a distinct cluster, closer to TcI, but clearly separated from clusters comprising all the other DTUs (Marcili et al., 2009c; Cavazzana et al., 2010). MLST analysis corroborates some affinity of Tcbat to TcI (Teixeira and Yeo, unpublished observations). Tcbat develops within mammalian cells in vitro, similar to other T. cruzi DTUs (Marcili et al., 2009c). Unlike isolates of the six DTUs, Tcbat does not develop in the commonly available triatomine species reared in laboratory colonies: T. infestans, Rhodnius prolixus and Panstrongylus megistus. The Tcbat insect vector is unknown. Possible vectors are triatomine species encountered in bat refuges, or cimicids, vectors of T. dionisii in Europe, or bat ectoparasites (Cavazzana et al., 2010). Some other T. cruzi isolates also display unusually complex combinations of molecular markers (Lewis et al., 2009b; Marcili et al., 2009a,b,c). Thus, Tcbat is but one indicator that the complexity of T. cruzi is higher than currently defined, and will require revision of DTU relationships as more silvatic isolates are genotyped. Ideally, criteria for establishing new DTUs could be considered, for example using MLST data and a specified degree of divergence from existing DTUs. 5. Standardizing genotyping for identification of the six T. cruzi DTUs The standardized nomenclature for the six T. cruzi DTUs will improve scientific communication and guide future research on comparative epidemiology and pathology. To achieve this aim a straightforward and reproducible genotyping strategy is required for DTU identification, manageable in any laboratory and adopted by the T. cruzi research community. Over the years, numerous approaches have been used to characterize the biochemical and genetic diversity of T. cruzi isolates. No single genetic target allows complete DTU resolution, and reliance on a single target is also inadvisable because of the potential influence of genetic exchange. A PCR assay system based on the amplification of particular regions of the SL gene and 24Sa rdna (Souto and Zingales, 1993; Table 2 Size of PCR product (in bp) of T. cruzi DTUs. a DTU 24Sa rdna SL 18S rdna TcI TcII TcIII 110 None b 165 TcIV 120 None b 155 TcV TcVI None a Brisse et al. (2001). b TcIII and TcIV DTUs can be detected by multiplex PCR of the SL gene (Fernandes et al., 1998; 2001; Burgos et al., 2007). Souto et al., 1996) and 18S rdna (Clark and Pung, 1994) was proposed (Brisse et al., 2001) in which the size polymorphisms of the amplification products were suitable for T. cruzi assignment into each of the six DTUs (Table 2). However, assignments based on the absence rather than the presence of PCR products are problematic, thus an alternative set of criteria is preferable for a gold standard typing method. A multilocus PCR-RFLP analysis of genetic polymorphism of 12 loci was proposed for DTU genotyping (Rozas et al., 2007), several of which demonstrated inter-dtu differences, and a combination of one, two or three of these assays allowed identification of the complete DTU set. The major limitation of this strategy is the complexity of the analysis. A three-marker sequential typing strategy (Fig. 3A) was proposed (Lewis et al., 2009b) consisting of PCR amplification of the 24Sa rdna (Souto and Zingales, 1993; Souto et al., 1996) and PCR-RFLP of the heat shock protein 60 (HSP60) and glucose-6- phosphate isomerase (GPI) loci (Westenberger et al., 2005). The combined application of the three PCR-RFLP markers was sufficient to discriminate the six DTUs in 45 out of 48 analyzed strains (Lewis et al., 2009b). Another three-step assay (D Avila et al., 2009) is outlined in Fig. 3B. PCR-RFLP analysis of the COII gene (Freitas et al., 2006) allows discrimination of TcI and TcII from the other DTUs; amplification of the non-transcribed spacer (NTS) of SL genes (Burgos et al., 2007) of the unclassified strains defines two distinct clusters, one formed by TcIII and TcIV and another by TcV and TcVI; amplification of 24Sa rdna (Souto et al., 1996) then resolves the four DTUs. A third scheme using nested-hot-start PCR assays is technically more demanding but has the potential advantage of allowing direct DTU typing in biological (Cardinal et al., 2008; Marcet et al., 2006) and clinical (Burgos et al., 2007, 2010) samples (Fig. 3C), and has been improved by the use of four sequential multiplex Real Time PCR assays using TaqMan probes (Duffy et al., 2010). The first NTS-SL based PCR employs primers recognizing regions flanking a 50-bp insertion characteristic of TcIII and TcIV intergenic regions. The second NTS-SL based PCR is a hemi-nested reaction using TCC-TC1 and TCC-TC2 primers (Souto et al., 1996). The A10 PCR uses primers (Burgos et al., 2007) that recognize a dimorphic region within the A10 nuclear fragment (Brisse et al., 2000). At present the revised assay is being tested on biological samples. An approach based on fluorescent-labeled fragment barcoding that detects PCR products from four rdna domains has also been devised (Hamilton et al., 2011). This technique was able to differentiate many trypanosome species from South American mammals. Some T. cruzi DTUs (including Tcbat) could be clearly identified, however, TcV and TcVI could not be distinguished from TcIII and TcII, respectively. 6. Comparative experimental pathology of the DTUs Since the discovery of Chagas disease in 1909, heterogeneity of parasite strains has been considered one factor implicated in different clinical presentations of the disease. Andrade (1974) attempted to discriminate several distinct T. cruzi morphobiological and behavioral phenotypes in murine models using the criteria of virulence (capacity of multiplication in the host) and pathogenicity (ability to produce tissue lesions and immunological responses). These studies defined three main strain phenotypes (Andrade, 1974; Andrade et al., 1983), subsequently designated as biodemes I III (Andrade and Magalhães, 1997), as follows: Biodeme type I: strains with rapid multiplication rates, maximum parasitemia, and mortality from 7 to 11 days after infection; predominance of slender forms, and macrophagotropism during the early phase of infection. Neuronal alterations are more
8 B. Zingales et al. / Infection, Genetics and Evolution 12 (2012) Fig. 3. Typing approaches for DTU assignment. Panel A: triple-assay proposed by Lewis et al. (2009b). Exceptions pointed by the authors for two T. cruzi IV strains from North America: characteristic 130 bp 24Sa rdna PCR product; two bands instead of 3 for GPI-HhaI PCR-RFLP. Panel B: triple-assay proposed by D Avila et al. (2009). Panel C: Heminested-PCR assay proposed by Marcet et al. (2006), Burgos et al. (2007, 2010). See details in the text. frequent and intense in biodeme type I infections. Zymodeme patterns of this biodeme correspond to zymodeme Z2b strains, a variant of Z2. Biodeme type II: strains with slow multiplication rates and irregular parasitemia peaks days after infection, when mortality rates reach a maximum; predominance of broad forms, myotropism, with predominant myocardial involvement. Zymodeme patterns of this biodeme type correspond to zymodeme Z2 strains. According to the 1999 revised nomenclature, this biodeme should be classified into the major group T. cruzi II (Anonymous, 1999). Biodeme type III: slow multiplication strains with late and high parasitemia peaks days following infection and late mortality, usually from day 30 after infection; predominance of broad forms and myotropism, with myocardial and skeletal muscle involvement. Biodeme type III corresponds to zymodeme Z1 strains and to the major group T. cruzi I(Anonymous, 1999). Due to the intense chronic myocarditis, strains of biodeme type III are considered the most pathogenic in mice (Andrade, 1974). Nevertheless, the degree of strain virulence may vary within the same biodeme and between clones of the same strain (Andrade, 1974; Postan et al., 1987). Further studies in murine experimental models support and expand these observations (Andrade et al., 1985) and clearly indicate that both the parasite and host genotypes are important in determining the tissue distribution, physiopathology and eventual outcome of T. cruzi infection. For example, simultaneous infection of four mouse lineages with the Colombiana Col1.7G2 clone (TcI) and the JG clone (TcII) showed identical tissue distributions in chronic phase infections of BALB/c and DBA-2 mice, but very different distributions in C57BL/6 (H-2b) and outbred Swiss mice (Andrade et al., 1999, 2002). Inoculation of Sylvio X10/4 clone (TcI) into the C3H/HePAS mouse strain caused intense cardiac inflammatory lesions, whereas in A/J mice chronic inflammatory lesions were found in the liver and skeletal muscle without detectable cardiac pathology (Marinho et al., 2004). Huge differences in virulence in experimental infections were reported for TcI isolates from Chile (Andersson et al., 2003),
9 248 B. Zingales et al. / Infection, Genetics and Evolution 12 (2012) Mexico (Espinoza et al., 2010) and the United States (Roellig and Yabsley, 2010). Tibayrenc and co-workers undertook long-term comparative studies on the association between T. cruzi subspecific genetic diversity and the parasite s biological properties, including behavior in axenic and mammalian cell culture, drug sensitivity in vitro, transmissibility through the insect vector and pathogenicity in mice (for example, see Laurent et al., 1997; de Lana et al., 1998; Revollo et al., 1998). The general pattern is that different DTUs exhibit statistically different biological properties, but with some overlap between different DTUs. Interestingly, in several cases mixtures of two clones behaved differently from a simple summation of their behavior, suggesting interaction between the genotypes (Pinto et al., 1998). As one approach to the underlying molecular basis of biological differences between DTUs, Telleria et al. (2010) performed phylogenetic character mapping of gene expression (proteomic diversity) between the six T. cruzi DTUs. The authors found a correlation between genetic distances measured by various markers (MLEE, RAPDs, MLST) and proteomic differences between DTUs, showing a tight correlation between genetic evolution and proteic divergence, and they identified several proteins with DTU-specific expression. Overall these studies indicate extensive intra-dtu phenotypic diversity, complicating the identification of genetic determinants of pathogenesis and virulence and requiring higher resolution intra-dtu genetic markers or methods to cross experimentally strains with different virulence and to analyze the genotypes and phenotypes of resultant progeny. 7. T. cruzi DTUs and human Chagas disease 7.1. Clinical presentations T. cruzi is transmitted to humans mainly by triatomine insect vectors, blood transfusion, infected mothers during pregnancy, and oral infection by consumption of food contaminated with triatomines or their feces. Following infection, a short acute phase is recognized only in 1 2% of the infected individuals, characterized by an abundant parasitemia and mild symptoms that spontaneously decline after 4 8 weeks. The disease proceeds to a chronic phase with scarce parasitemia and an unpredictable clinical course. Most of the chronic individuals are asymptomatic and show no electrocardiographic or radiologic alterations in the heart, esophagus or colon. The individuals present positive serological tests for T. cruzi infection and in many the xenodiagnosis and PCR results may be repeatedly positive for many years. These persons with the indeterminate form will remain asymptomatic for decades, if not the rest of their lives. Each year approximately 3% will develop lesions in the heart or gastrointestinal tract (Dias, 2006). Chronic cardiomyopathy, or chronic Chagas heart disease is the most common and severe manifestation in humans, affecting approximately 30% of the patients. In endemic areas, it represents the main cause of disability and mortality. The basic lesions of chronic Chagas heart disease are focal or extensive myocardial fibrosis, which result from myocardial cell destruction due to direct parasite action, inflammatory response, and neuronal involvement. The gastrointestinal manifestations consist of progressive enlargement of the esophagus or colon caused by chronic inflammation and destruction of parasympathetic neurons. Great regional diversity of Chagas disease severity and the nature of the chronic infection has been reported, attributed to a set of complex interactions among the genetic make-up of the parasite, the host immunogenetic background, and environmental factors (reviewed by Campbell et al., 2004; Macedo et al., 2004). A goal of T. cruzi taxonomic studies is to identify links between the infecting DTUs and the clinical presentation of disease. No proven associations are evident at present. The search for these associations will drive the ultimate criterion for defining the clinically meaningful number of biological subdivisions within this species Acute Chagas disease The control of vector and blood transfusion transmission in several Latin American countries has promoted the steady reduction of acute infections. In recent years, most of these cases are linked to oral and congenital transmission, blood transfusion or laboratory accidents (Coura, 2006, 2007). Several acute cases were documented in the Amazon region, most caused by TcI and, less frequently, by TcIII and TcIV (Coura, 2007). In northern countries of South America TcI is also the major cause of human acute cases (Miles et al., 1981; Añez et al., 2004; Llewellyn et al., 2009b and cited references). Acute cases resulting from oral contamination have been documented for outbreaks in different localities, most frequently in the Amazon region. Most of these cases were due to TcI, with rare cases due to TcIII and TcIV (Coura, 2007; Marcili et al., 2009a,b; Valente et al., 2009), with TcI in Venezuela and French Guiana (Alarcón de Noya et al., 2010; Cura et al., 2010) and TcII in southern Brazil (Steindel et al., 2008). The morbidity and mortality may vary depending on the parasite burden and parasite genotype ingested. The incidence of congenital transmission is estimated at more than 15,000 cases annually in the Americas and is one of the main modes of transmission in non-endemic countries. The risk factors determining transmission of the parasite to the fetus are largely unknown. Cases of congenital infection with all DTUs except TcIV were reported in Argentina, Bolivia, Chile, Colombia, and Paraguay (for examples, see García et al., 2001; Svoboda et al., 2005; Virreira et al., 2006; Burgos et al., 2007; Corrales et al., 2009; Del Puerto et al., 2010). The prevalence of specific DTUs among congenital cases appears to be in accordance with their presence in the infected population. However, there seems to be a disparate prevalence of congenital cases in endemic regions, with few cases reported, for example from Venezuela and Brazil, with the exception of southern Brazil (Carlier and Truyens, 2010) Chronic Chagas disease TcI is implicated with human disease in Amazonia, the Andean region, Central America, and Mexico (Bosseno et al., 2002; Montilla et al., 2002; Añez et al., 2004; Higo et al., 2004; Sánchez-Guillén et al., 2006). Clinical presentations of TcI include chagasic cardiomyopathy and in immunocopromized hosts severe cases of meningoencephalitis. In the Southern Cone region, where T. infestans is the main vector, TcII, TcV and TcVI are the main causes of Chagas disease. TcII predominates in eastern and central Brazil, TcV in Argentina, Bolivia, and Paraguay, and TcVI in the Gran Chaco (Chapman et al., 1984; Zingales et al., 1999; Brenière et al., 2002; Diosque et al., 2003; Higo et al., 2004; Coronado et al., 2006; Burgos et al., 2007; Cardinal et al., 2008; Carranza et al., 2009; Del Puerto et al., 2010). Throughout the Southern Cone region chagasic cardiomyopathy can be severe, and a proportion of cases may develop megaesophagus and megacolon (Luquetti et al., 1986; Freitas et al., 2005; Lages-Silva et al., 2006). The disparate geographical distribution of the megasyndromes may reflect the divergent phylogeographies of T. cruzi DTUs (Miles et al., 1981), a hypothesis supported by circumstantial evidence. Chagasic megaesophagus and megacolon are considered rare in northern South America and Central America (Miles et al., 2009).
This article appeared in a journal published by Elsevier. The attached copy is furnished to the author for internal non-commercial research and
 This article appeared in a journal published by Elsevier. The attached copy is furnished to the author for internal non-commercial research and education use, including for instruction at the authors institution
This article appeared in a journal published by Elsevier. The attached copy is furnished to the author for internal non-commercial research and education use, including for instruction at the authors institution
Infection, Genetics and Evolution
 Infection, Genetics and Evolution 11 (2011) 44 51 Contents lists available at ScienceDirect Infection, Genetics and Evolution journal homepage: www.elsevier.com/locate/meegid Identification and lineage
Infection, Genetics and Evolution 11 (2011) 44 51 Contents lists available at ScienceDirect Infection, Genetics and Evolution journal homepage: www.elsevier.com/locate/meegid Identification and lineage
Neglected Diseases (NDs) Landscape in Brazil and South America
 Frontiers in Science on Neglected Diseases 13 th November 2014 Neglected Diseases (NDs) Landscape in Brazil and South America Jeffrey Shaw São Paulo University Biomedical Sciences Institute jeffreyj@usp.br
Frontiers in Science on Neglected Diseases 13 th November 2014 Neglected Diseases (NDs) Landscape in Brazil and South America Jeffrey Shaw São Paulo University Biomedical Sciences Institute jeffreyj@usp.br
Epidemiology, Biochemistry and Evolution of Trypanosoma cruzi Lineages Based on Ribosomal RNA Sequences
 Mem Inst Oswaldo Cruz, Rio de Janeiro, Vol. 94, Suppl. I: 59-64, 999 59 Epidemiology, Biochemistry and Evolution of Trypanosoma cruzi Lineages Based on Ribosomal RNA Sequences Bianca Zingales/ +, Beatriz
Mem Inst Oswaldo Cruz, Rio de Janeiro, Vol. 94, Suppl. I: 59-64, 999 59 Epidemiology, Biochemistry and Evolution of Trypanosoma cruzi Lineages Based on Ribosomal RNA Sequences Bianca Zingales/ +, Beatriz
Chagas Cardiomyopathy Manifestations and Trypanosoma cruzi Genotypes Circulating in Chronic Chagasic Patients
 Chagas Cardiomyopathy Manifestations and Trypanosoma cruzi Genotypes Circulating in Chronic Chagasic Patients Juan David Ramírez 1, Felipe Guhl 1 *, Lina María Rendón 1, Fernando Rosas 2, Jose A. Marin-Neto
Chagas Cardiomyopathy Manifestations and Trypanosoma cruzi Genotypes Circulating in Chronic Chagasic Patients Juan David Ramírez 1, Felipe Guhl 1 *, Lina María Rendón 1, Fernando Rosas 2, Jose A. Marin-Neto
Trypanosomiasis WRAIR- GEIS 'Operational Clinical Infectious Disease' Course
 Trypanosomiasis WRAIR- GEIS 'Operational Clinical Infectious Disease' Course UNCLASSIFIED Disclaimer The views expressed in this presentation are those of the speaker and do not reflect the official policy
Trypanosomiasis WRAIR- GEIS 'Operational Clinical Infectious Disease' Course UNCLASSIFIED Disclaimer The views expressed in this presentation are those of the speaker and do not reflect the official policy
Chagas disease reactivation in a heart transplant patient infected by domestic Trypanosoma cruzi discrete typing unit I (TcI DOM )
 Costales et al. Parasites & Vectors (2015) 8:435 DOI 10.1186/s13071-015-1039-3 SHORT REPORT Open Access Chagas disease reactivation in a heart transplant patient infected by domestic Trypanosoma cruzi
Costales et al. Parasites & Vectors (2015) 8:435 DOI 10.1186/s13071-015-1039-3 SHORT REPORT Open Access Chagas disease reactivation in a heart transplant patient infected by domestic Trypanosoma cruzi
Variability of Kinetoplast DNA Gene Signatures of Trypanosoma cruzi II Strains from Patients with Different Clinical Forms of Chagas Disease in Brazil
 JOURNAL OF CLINICAL MICROBIOLOGY, June 2006, p. 2167 2171 Vol. 44, No. 6 0095-1137/06/$08.00 0 doi:10.1128/jcm.02124-05 Copyright 2006, American Society for Microbiology. All Rights Reserved. Variability
JOURNAL OF CLINICAL MICROBIOLOGY, June 2006, p. 2167 2171 Vol. 44, No. 6 0095-1137/06/$08.00 0 doi:10.1128/jcm.02124-05 Copyright 2006, American Society for Microbiology. All Rights Reserved. Variability
Parasitic Protozoa, Helminths, and Arthropod Vectors
 PowerPoint Lecture Slides for MICROBIOLOGY ROBERT W. BAUMAN Chapter 23 Parasitic Protozoa, Helminths, and Arthropod Vectors Parasitic Diseases Protozoan and helminthic parasites are emerging as serious
PowerPoint Lecture Slides for MICROBIOLOGY ROBERT W. BAUMAN Chapter 23 Parasitic Protozoa, Helminths, and Arthropod Vectors Parasitic Diseases Protozoan and helminthic parasites are emerging as serious
WRAIR- GEIS 'Operational Clinical Infectious Disease' Course
 Trypanosomiasis WRAIR- GEIS 'Operational Clinical Infectious Disease' Course The opinions or assertions contained herein are the private views of the author, and are not to be construed as official, or
Trypanosomiasis WRAIR- GEIS 'Operational Clinical Infectious Disease' Course The opinions or assertions contained herein are the private views of the author, and are not to be construed as official, or
Departamento de Microbiologia, Imunologia e Parasitologia, Universidade Federal de São Paulo, São Paulo, SP, Brasil
 Haplotype distribution of five nuclear genes based on network genealogies and Bayesian inference indicates that Trypanosoma cruzi hybrid strains are polyphyletic L. Tomazi 1 *, S.Y. Kawashita 1 *, P.M.
Haplotype distribution of five nuclear genes based on network genealogies and Bayesian inference indicates that Trypanosoma cruzi hybrid strains are polyphyletic L. Tomazi 1 *, S.Y. Kawashita 1 *, P.M.
CURRENT GENETIC TESTING TOOLS IN NEONATAL MEDICINE. Dr. Bahar Naghavi
 2 CURRENT GENETIC TESTING TOOLS IN NEONATAL MEDICINE Dr. Bahar Naghavi Assistant professor of Basic Science Department, Shahid Beheshti University of Medical Sciences, Tehran,Iran 3 Introduction Over 4000
2 CURRENT GENETIC TESTING TOOLS IN NEONATAL MEDICINE Dr. Bahar Naghavi Assistant professor of Basic Science Department, Shahid Beheshti University of Medical Sciences, Tehran,Iran 3 Introduction Over 4000
University of Warwick institutional repository:
 University of Warwick institutional repository: http://go.warwick.ac.uk/wrap This paper is made available online in accordance with publisher policies. Please scroll down to view the document itself. Please
University of Warwick institutional repository: http://go.warwick.ac.uk/wrap This paper is made available online in accordance with publisher policies. Please scroll down to view the document itself. Please
PopGen4: Assortative mating
 opgen4: Assortative mating Introduction Although random mating is the most important system of mating in many natural populations, non-random mating can also be an important mating system in some populations.
opgen4: Assortative mating Introduction Although random mating is the most important system of mating in many natural populations, non-random mating can also be an important mating system in some populations.
Genetics and Genomics in Medicine Chapter 8 Questions
 Genetics and Genomics in Medicine Chapter 8 Questions Linkage Analysis Question Question 8.1 Affected members of the pedigree above have an autosomal dominant disorder, and cytogenetic analyses using conventional
Genetics and Genomics in Medicine Chapter 8 Questions Linkage Analysis Question Question 8.1 Affected members of the pedigree above have an autosomal dominant disorder, and cytogenetic analyses using conventional
Chagas disease. (American trypanosomiasis)
 Chagas disease (American trypanosomiasis) Epidemiology 7 to 8 million people worldwide are infected Endemic in Latin America and South America USA, Canada, Europe http://www.who.int/mediacentre/factsheets/fs340/en/index.html
Chagas disease (American trypanosomiasis) Epidemiology 7 to 8 million people worldwide are infected Endemic in Latin America and South America USA, Canada, Europe http://www.who.int/mediacentre/factsheets/fs340/en/index.html
SALSA MLPA KIT P060-B2 SMA
 SALSA MLPA KIT P6-B2 SMA Lot 111, 511: As compared to the previous version B1 (lot 11), the 88 and 96 nt DNA Denaturation control fragments have been replaced (QDX2). Please note that, in contrast to the
SALSA MLPA KIT P6-B2 SMA Lot 111, 511: As compared to the previous version B1 (lot 11), the 88 and 96 nt DNA Denaturation control fragments have been replaced (QDX2). Please note that, in contrast to the
MODULE NO.14: Y-Chromosome Testing
 SUBJECT Paper No. and Title Module No. and Title Module Tag FORENSIC SIENCE PAPER No.13: DNA Forensics MODULE No.21: Y-Chromosome Testing FSC_P13_M21 TABLE OF CONTENTS 1. Learning Outcome 2. Introduction:
SUBJECT Paper No. and Title Module No. and Title Module Tag FORENSIC SIENCE PAPER No.13: DNA Forensics MODULE No.21: Y-Chromosome Testing FSC_P13_M21 TABLE OF CONTENTS 1. Learning Outcome 2. Introduction:
Trypanosoma cruzi-infected Panstrongylus geniculatus and Rhodnius robustus adults invade households in the Tropics of Cochabamba region of Bolivia
 Rojas-Cortez et al. Parasites & Vectors (2016) 9:158 DOI 10.1186/s13071-016-1445-1 SHORT REPORT Open Access Trypanosoma cruzi-infected Panstrongylus geniculatus and Rhodnius robustus adults invade households
Rojas-Cortez et al. Parasites & Vectors (2016) 9:158 DOI 10.1186/s13071-016-1445-1 SHORT REPORT Open Access Trypanosoma cruzi-infected Panstrongylus geniculatus and Rhodnius robustus adults invade households
SEX. Genetic Variation: The genetic substrate for natural selection. Sex: Sources of Genotypic Variation. Genetic Variation
 Genetic Variation: The genetic substrate for natural selection Sex: Sources of Genotypic Variation Dr. Carol E. Lee, University of Wisconsin Genetic Variation If there is no genetic variation, neither
Genetic Variation: The genetic substrate for natural selection Sex: Sources of Genotypic Variation Dr. Carol E. Lee, University of Wisconsin Genetic Variation If there is no genetic variation, neither
HOST-PARASITE INTERPLAY
 HOST-PARASITE INTERPLAY Adriano Casulli EURLP, ISS (Rome, Italy) HOST-PARASITE INTERPLAY WP3 (parasite virulence vs human immunity) (Parasite) Task 3.1: Genotypic characterization Task 3.6: Transcriptome
HOST-PARASITE INTERPLAY Adriano Casulli EURLP, ISS (Rome, Italy) HOST-PARASITE INTERPLAY WP3 (parasite virulence vs human immunity) (Parasite) Task 3.1: Genotypic characterization Task 3.6: Transcriptome
Trypanosoma cruzi, the etiologic agent of Chagas
 ORIGINAL RESEARCH Frequency of Trypanosoma cruzi parasitemia among infected blood donors with a potential association between parasite lineage and transfusion transmission David A. Leiby, 1 * Megan L.
ORIGINAL RESEARCH Frequency of Trypanosoma cruzi parasitemia among infected blood donors with a potential association between parasite lineage and transfusion transmission David A. Leiby, 1 * Megan L.
MRC-Holland MLPA. Description version 19;
 SALSA MLPA probemix P6-B2 SMA Lot B2-712, B2-312, B2-111, B2-511: As compared to the previous version B1 (lot B1-11), the 88 and 96 nt DNA Denaturation control fragments have been replaced (QDX2). SPINAL
SALSA MLPA probemix P6-B2 SMA Lot B2-712, B2-312, B2-111, B2-511: As compared to the previous version B1 (lot B1-11), the 88 and 96 nt DNA Denaturation control fragments have been replaced (QDX2). SPINAL
Evolution of influenza
 Evolution of influenza Today: 1. Global health impact of flu - why should we care? 2. - what are the components of the virus and how do they change? 3. Where does influenza come from? - are there animal
Evolution of influenza Today: 1. Global health impact of flu - why should we care? 2. - what are the components of the virus and how do they change? 3. Where does influenza come from? - are there animal
Everything you ever wanted to know about Zika Virus Disease
 Everything you ever wanted to know about Zika Virus Disease (in 14 slides) Jon Temte, MD/PhD University of Wisconsin School of Medicine and Public Health 28 January 2016 Zika Virus mosquito-borne flavivirus
Everything you ever wanted to know about Zika Virus Disease (in 14 slides) Jon Temte, MD/PhD University of Wisconsin School of Medicine and Public Health 28 January 2016 Zika Virus mosquito-borne flavivirus
Charlotte L. Kerr 1, Tapan Bhattacharyya 1*, Samanta C. C. Xavier 2, Juliana H. Barros 2, Valdirene S. Lima 2, Ana M. Jansen 2 and Michael A.
 Kerr et al. Parasites & Vectors (2016) 9:584 DOI 10.1186/s13071-016-1873-y RESEARCH Open Access Lineage-specific serology confirms Brazilian Atlantic forest lion tamarins, Leontopithecus chrysomelas and
Kerr et al. Parasites & Vectors (2016) 9:584 DOI 10.1186/s13071-016-1873-y RESEARCH Open Access Lineage-specific serology confirms Brazilian Atlantic forest lion tamarins, Leontopithecus chrysomelas and
Criteria of Chagas Disease Cure
 Mem Inst Oswaldo Cruz, Rio de Janeiro, Vol. 94, Suppl. I: 331-335, 1999 331 Criteria of Chagas Disease Cure J Romeu Cançado Cadeira de Terapêutica Clínica, Universidade Federal de Minas Gerais, Caixa Postal
Mem Inst Oswaldo Cruz, Rio de Janeiro, Vol. 94, Suppl. I: 331-335, 1999 331 Criteria of Chagas Disease Cure J Romeu Cançado Cadeira de Terapêutica Clínica, Universidade Federal de Minas Gerais, Caixa Postal
Chagas Disease in non-endemic Countries
 Chagas Disease in non-endemic Countries Tsutomu Takeuchi Institute of Tropical Medicine, Nagasaki University, Nagasaki Apparent distribution of Triatoma infestans 1983 2003 Migration flow of immigrants
Chagas Disease in non-endemic Countries Tsutomu Takeuchi Institute of Tropical Medicine, Nagasaki University, Nagasaki Apparent distribution of Triatoma infestans 1983 2003 Migration flow of immigrants
Mechanisms of Evolution. Macroevolution. Speciation. MICROEVOLUTION - A change in the frequency of alleles. Review population genetics Ch. 23.
 Mechanisms of Evolution Macroevolution Speciation MICROEVOLUTION - A change in the frequency of alleles. Review population genetics Ch. 23. MACROEVOLUTION - Speciation (or emergence of higher taxonomic
Mechanisms of Evolution Macroevolution Speciation MICROEVOLUTION - A change in the frequency of alleles. Review population genetics Ch. 23. MACROEVOLUTION - Speciation (or emergence of higher taxonomic
 . /////////////////// /////////////////// / Berenice? . BLOOD HEPATITIS B HEPATITIS C HIV T. pallidum T. cruzi TRANSFUSION HOST The Southern Cone Initiative, 1991 The Ministers of Health of Argentina,
. /////////////////// /////////////////// / Berenice? . BLOOD HEPATITIS B HEPATITIS C HIV T. pallidum T. cruzi TRANSFUSION HOST The Southern Cone Initiative, 1991 The Ministers of Health of Argentina,
Infestation of peridomestic Attalea phalerata palms by Rhodnius stali, a vector of Trypanosoma cruzi in the Alto Beni, Bolivia
 Tropical Medicine and International Health doi:10.1111/j.1365-3156.2010.02527.x volume 15 no 6 pp 727 732 june 2010 Infestation of peridomestic Attalea phalerata palms by Rhodnius stali, a vector of Trypanosoma
Tropical Medicine and International Health doi:10.1111/j.1365-3156.2010.02527.x volume 15 no 6 pp 727 732 june 2010 Infestation of peridomestic Attalea phalerata palms by Rhodnius stali, a vector of Trypanosoma
SURVEILLANCE TECHNICAL
 CHAPTER 5 SURVEILLANCE TECHNICAL ASPECTS 55 Protect - detect - protect Polio eradication strategies can be summed up as protect and detect protect children against polio by vaccinating them, and detect
CHAPTER 5 SURVEILLANCE TECHNICAL ASPECTS 55 Protect - detect - protect Polio eradication strategies can be summed up as protect and detect protect children against polio by vaccinating them, and detect
Micropathology Ltd. University of Warwick Science Park, Venture Centre, Sir William Lyons Road, Coventry CV4 7EZ
 www.micropathology.com info@micropathology.com Micropathology Ltd Tel 24hrs: +44 (0) 24-76 323222 Fax / Ans: +44 (0) 24-76 - 323333 University of Warwick Science Park, Venture Centre, Sir William Lyons
www.micropathology.com info@micropathology.com Micropathology Ltd Tel 24hrs: +44 (0) 24-76 323222 Fax / Ans: +44 (0) 24-76 - 323333 University of Warwick Science Park, Venture Centre, Sir William Lyons
Genome - Wide Linkage Mapping
 Biological Sciences Initiative HHMI Genome - Wide Linkage Mapping Introduction This activity is based on the work of Dr. Christine Seidman et al that was published in Circulation, 1998, vol 97, pgs 2043-2048.
Biological Sciences Initiative HHMI Genome - Wide Linkage Mapping Introduction This activity is based on the work of Dr. Christine Seidman et al that was published in Circulation, 1998, vol 97, pgs 2043-2048.
Acute chagas outbreaks: molecular and biological features of Trypanosoma cruzi isolates, and clinical aspects of acute cases in Santander, Colombia
 Díaz et al. Parasites & Vectors (2015) 8:608 DOI 10.1186/s13071-015-1218-2 RESEARCH Open Access Acute chagas outbreaks: molecular and biological features of Trypanosoma cruzi isolates, and clinical aspects
Díaz et al. Parasites & Vectors (2015) 8:608 DOI 10.1186/s13071-015-1218-2 RESEARCH Open Access Acute chagas outbreaks: molecular and biological features of Trypanosoma cruzi isolates, and clinical aspects
Diagnostic Methods of HBV and HDV infections
 Diagnostic Methods of HBV and HDV infections Zohreh Sharifi,ph.D Blood Transfusion Research Center, High Institute for Research and Education in Transfusion Medicine Hepatitis B-laboratory diagnosis Detection
Diagnostic Methods of HBV and HDV infections Zohreh Sharifi,ph.D Blood Transfusion Research Center, High Institute for Research and Education in Transfusion Medicine Hepatitis B-laboratory diagnosis Detection
Ch. 24 Speciation BIOL 221
 Ch. 24 Speciation BIOL 221 Speciation Speciation Origin of new, is at the focal point of evolutionary theory Microevolution consists of adaptations that evolve within a population confined to one gene
Ch. 24 Speciation BIOL 221 Speciation Speciation Origin of new, is at the focal point of evolutionary theory Microevolution consists of adaptations that evolve within a population confined to one gene
KINETOPLASTIDS. Kinetoplast. Nucleus
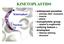 KINETOPLASTIDS Kinetoplast Nucleus widespread parasites animals (fish humans) insects plants monophyletic group related to euglenoids unifying feature = kinetoplast Giemsa staining structure KINETOPLAST
KINETOPLASTIDS Kinetoplast Nucleus widespread parasites animals (fish humans) insects plants monophyletic group related to euglenoids unifying feature = kinetoplast Giemsa staining structure KINETOPLAST
Drug discovery for Chagas disease should consider Trypanosoma cruzi strain diversity
 828 Mem Inst Oswaldo Cruz, Rio de Janeiro, Vol. 109(6): 828-833, September 2014 READER S OPINION Drug discovery for Chagas disease should consider Trypanosoma cruzi strain diversity Bianca Zingales 1 /
828 Mem Inst Oswaldo Cruz, Rio de Janeiro, Vol. 109(6): 828-833, September 2014 READER S OPINION Drug discovery for Chagas disease should consider Trypanosoma cruzi strain diversity Bianca Zingales 1 /
George R. Honig Junius G. Adams III. Human Hemoglobin. Genetics. Springer-Verlag Wien New York
 George R. Honig Junius G. Adams III Human Hemoglobin Genetics Springer-Verlag Wien New York George R. Honig, M.D., Ph.D. Professor and Head Department of Pediatrics, College of Medicine University of Illinois
George R. Honig Junius G. Adams III Human Hemoglobin Genetics Springer-Verlag Wien New York George R. Honig, M.D., Ph.D. Professor and Head Department of Pediatrics, College of Medicine University of Illinois
Within-host temporal fluctuations of Trypanosoma cruzi discrete typing units: the case of the wild reservoir rodent Octodon degus
 Rojo et al. Parasites & Vectors (2017) 10:380 DOI 10.1186/s13071-017-2314-2 RESEARCH Open Access Within-host temporal fluctuations of Trypanosoma cruzi discrete typing units: the case of the wild reservoir
Rojo et al. Parasites & Vectors (2017) 10:380 DOI 10.1186/s13071-017-2314-2 RESEARCH Open Access Within-host temporal fluctuations of Trypanosoma cruzi discrete typing units: the case of the wild reservoir
Proposal form for the evaluation of a genetic test for NHS Service Gene Dossier
 Proposal form for the evaluation of a genetic test for NHS Service Gene Dossier Test Disease Population Triad Disease name Choroideremia OMIM number for disease 303100 Disease alternative names please
Proposal form for the evaluation of a genetic test for NHS Service Gene Dossier Test Disease Population Triad Disease name Choroideremia OMIM number for disease 303100 Disease alternative names please
Kinetoplastids Handout
 Kinetoplastids Handout 1 Kinetoplastids widespread group of flagellated protozoa parasitize virtually all animal groups as well as plants and insects 3 distinct kinetoplastid species cause human disease
Kinetoplastids Handout 1 Kinetoplastids widespread group of flagellated protozoa parasitize virtually all animal groups as well as plants and insects 3 distinct kinetoplastid species cause human disease
Trypanosomiasis. Introduction. Epidemiology. Global Epidemiology. Trypanosomiasis Risk in UK Travellers
 Trypanosomiasis Introduction Epidemiology Risk for travellers Transmission Signs and symptoms Treatment Prevention References Reading list Links Introduction Trypanosomiasis is caused by parasitic protozoa
Trypanosomiasis Introduction Epidemiology Risk for travellers Transmission Signs and symptoms Treatment Prevention References Reading list Links Introduction Trypanosomiasis is caused by parasitic protozoa
The plant of the day Pinus longaeva Pinus aristata
 The plant of the day Pinus longaeva Pinus aristata Today s Topics Non-random mating Genetic drift Population structure Big Questions What are the causes and evolutionary consequences of non-random mating?
The plant of the day Pinus longaeva Pinus aristata Today s Topics Non-random mating Genetic drift Population structure Big Questions What are the causes and evolutionary consequences of non-random mating?
SALSA MLPA KIT P050-B2 CAH
 SALSA MLPA KIT P050-B2 CAH Lot 0510, 0909, 0408: Compared to lot 0107, extra control fragments have been added at 88, 96, 100 and 105 nt. The 274 nt probe gives a higher signal in lot 0510 compared to
SALSA MLPA KIT P050-B2 CAH Lot 0510, 0909, 0408: Compared to lot 0107, extra control fragments have been added at 88, 96, 100 and 105 nt. The 274 nt probe gives a higher signal in lot 0510 compared to
OVERVIEW OF CURRENT IDENTIFICATION SYSTEMS AND DATABASES
 OVERVIEW OF CURRENT IDENTIFICATION SYSTEMS AND DATABASES EVERY STEP OF THE WAY 1 EVERY STEP OF THE WAY MICROBIAL IDENTIFICATION METHODS DNA RNA Genotypic Sequencing of ribosomal RNA regions of bacteria
OVERVIEW OF CURRENT IDENTIFICATION SYSTEMS AND DATABASES EVERY STEP OF THE WAY 1 EVERY STEP OF THE WAY MICROBIAL IDENTIFICATION METHODS DNA RNA Genotypic Sequencing of ribosomal RNA regions of bacteria
Quantification of early stage lesions for loss of p53 should be shown in the main figures.
 Reviewer #1 (Remarks to the Author): Expert in prostate cancer The manuscript "Clonal dynamics following p53 loss of heterozygosity in Kras-driven cancers" uses a number of novel genetically engineered
Reviewer #1 (Remarks to the Author): Expert in prostate cancer The manuscript "Clonal dynamics following p53 loss of heterozygosity in Kras-driven cancers" uses a number of novel genetically engineered
New genomic typing method MLST
 New genomic typing method MLST Bon KIMURA fingerprinting PFGE DNA multilocus sequence typingmlst alleles PFGE MLST 1990 PCR 1 PCR DNA PFGE 1 PFGE RAPDrandomly amplified polymorphic DNA 3 AFLPAmplified
New genomic typing method MLST Bon KIMURA fingerprinting PFGE DNA multilocus sequence typingmlst alleles PFGE MLST 1990 PCR 1 PCR DNA PFGE 1 PFGE RAPDrandomly amplified polymorphic DNA 3 AFLPAmplified
PALEOPATHOLOGY IN CHAGAS DISEASE
 PALEOPATHOLOGY IN CHAGAS DISEASE MARVIN J. ALLISON AND ENRIQUE GERSZTEN PROFESSORS OF PATHOLOGY VIRGINIA COMMONWEALTH UNIVERSITY MEDICAL COLLEGE OF VIRGINIA CAMPUS RICHMOND, VA, USA REDUVIID (KISSING BUG-
PALEOPATHOLOGY IN CHAGAS DISEASE MARVIN J. ALLISON AND ENRIQUE GERSZTEN PROFESSORS OF PATHOLOGY VIRGINIA COMMONWEALTH UNIVERSITY MEDICAL COLLEGE OF VIRGINIA CAMPUS RICHMOND, VA, USA REDUVIID (KISSING BUG-
The Improbable Transmission of Trypanosoma cruzi to Human: The Missing Link in the Dynamics and Control of Chagas Disease
 The Improbable Transmission of Trypanosoma cruzi to Human: The Missing Link in the Dynamics and Control of Chagas Disease Pierre Nouvellet 1,2,3, Eric Dumonteil 1,4,Sébastien Gourbière 2,5 * 1 Laboratorio
The Improbable Transmission of Trypanosoma cruzi to Human: The Missing Link in the Dynamics and Control of Chagas Disease Pierre Nouvellet 1,2,3, Eric Dumonteil 1,4,Sébastien Gourbière 2,5 * 1 Laboratorio
outbred colonies are stocks inbred colonies are strains 3/22/2012 Mouse strains 2.500
 Nomenclature for rodents Stock vs strain outbred colonies are stocks Kai Õkva inbred colonies are strains Outbred nomenclature First three letters reveal place where stock is maintained (Kuo) Followed
Nomenclature for rodents Stock vs strain outbred colonies are stocks Kai Õkva inbred colonies are strains Outbred nomenclature First three letters reveal place where stock is maintained (Kuo) Followed
The selfish gene. mitochondrium
 The selfish gene selection acts mostly for the benefit of the individual sometimes selection may act for the benefit of relatives rarely, selection acts for the benefit of the group mitochondrium in asexual
The selfish gene selection acts mostly for the benefit of the individual sometimes selection may act for the benefit of relatives rarely, selection acts for the benefit of the group mitochondrium in asexual
Generating Spontaneous Copy Number Variants (CNVs) Jennifer Freeman Assistant Professor of Toxicology School of Health Sciences Purdue University
 Role of Chemical lexposure in Generating Spontaneous Copy Number Variants (CNVs) Jennifer Freeman Assistant Professor of Toxicology School of Health Sciences Purdue University CNV Discovery Reference Genetic
Role of Chemical lexposure in Generating Spontaneous Copy Number Variants (CNVs) Jennifer Freeman Assistant Professor of Toxicology School of Health Sciences Purdue University CNV Discovery Reference Genetic
AIDS - Knowledge and Dogma. Conditions for the Emergence and Decline of Scientific Theories Congress, July 16/ , Vienna, Austria
 AIDS - Knowledge and Dogma Conditions for the Emergence and Decline of Scientific Theories Congress, July 16/17 2010, Vienna, Austria Reliability of PCR to detect genetic sequences from HIV Juan Manuel
AIDS - Knowledge and Dogma Conditions for the Emergence and Decline of Scientific Theories Congress, July 16/17 2010, Vienna, Austria Reliability of PCR to detect genetic sequences from HIV Juan Manuel
Whole genome sequencing & new strain typing methods in IPC. Lyn Gilbert ACIPC conference Hobart, November 2015
 Whole genome sequencing & new strain typing methods in IPC Lyn Gilbert ACIPC conference Hobart, November 2015 Why do strain typing? Evolution, population genetics, geographic distribution 2 Why strain
Whole genome sequencing & new strain typing methods in IPC Lyn Gilbert ACIPC conference Hobart, November 2015 Why do strain typing? Evolution, population genetics, geographic distribution 2 Why strain
Systems of Mating: Systems of Mating:
 8/29/2 Systems of Mating: the rules by which pairs of gametes are chosen from the local gene pool to be united in a zygote with respect to a particular locus or genetic system. Systems of Mating: A deme
8/29/2 Systems of Mating: the rules by which pairs of gametes are chosen from the local gene pool to be united in a zygote with respect to a particular locus or genetic system. Systems of Mating: A deme
Ebola Virus. Emerging Diseases. Biosciences in the 21 st Century Dr. Amber Rice December 4, 2017
 Ebola Virus Emerging Diseases Biosciences in the 21 st Century Dr. Amber Rice December 4, 2017 Outline Disease emergence: a case study How do pathogens shift hosts? Evolution within hosts: The evolution
Ebola Virus Emerging Diseases Biosciences in the 21 st Century Dr. Amber Rice December 4, 2017 Outline Disease emergence: a case study How do pathogens shift hosts? Evolution within hosts: The evolution
Trypanosoma cruzi strain TcI is associated with chronic Chagas disease in the Brazilian Amazon
 Santana et al. Parasites & Vectors 2014, 7:267 RESEARCH Open Access Trypanosoma cruzi strain TcI is associated with chronic Chagas disease in the Brazilian Amazon Rosa Amélia Gonçalves Santana 1, Laylah
Santana et al. Parasites & Vectors 2014, 7:267 RESEARCH Open Access Trypanosoma cruzi strain TcI is associated with chronic Chagas disease in the Brazilian Amazon Rosa Amélia Gonçalves Santana 1, Laylah
EPIDEMIOLOGICAL HETEROGENEITY OF INFECTIONS WITH HEPATITIS B AND D VIRUSES IN BRAZIL
 PREVENTION AND CONTROL OF VIRAL HEPATITIS IN BRASILAND OTHER LATIN AMERICAN COUNTRIES BRASÍLIA, BRAZIL 21 DE MARCH 2014 EPIDEMIOLOGICAL HETEROGENEITY OF INFECTIONS WITH HEPATITIS B AND D VIRUSES IN BRAZIL
PREVENTION AND CONTROL OF VIRAL HEPATITIS IN BRASILAND OTHER LATIN AMERICAN COUNTRIES BRASÍLIA, BRAZIL 21 DE MARCH 2014 EPIDEMIOLOGICAL HETEROGENEITY OF INFECTIONS WITH HEPATITIS B AND D VIRUSES IN BRAZIL
More than 98 countries affected by Leishmaniasis
 More than 98 countries affected by Leishmaniasis ACL In many urban & sub urban areas (8 provinces) Tehran, Mashhad, Neishabur, Shiraz, Kerman, Bam (Sharifi et al. 2011) ZCL In 2012, totally ( ZCL & ACL)
More than 98 countries affected by Leishmaniasis ACL In many urban & sub urban areas (8 provinces) Tehran, Mashhad, Neishabur, Shiraz, Kerman, Bam (Sharifi et al. 2011) ZCL In 2012, totally ( ZCL & ACL)
West Nile Virus. By Frank Riusech
 West Nile Virus By Frank Riusech Disease Etiology: West Nile virus(wnv), genus, flavivirus is positive- stranded RNA arbovirus (arthropod- borne), belonging to the Flaviviridae family. Included in this
West Nile Virus By Frank Riusech Disease Etiology: West Nile virus(wnv), genus, flavivirus is positive- stranded RNA arbovirus (arthropod- borne), belonging to the Flaviviridae family. Included in this
Computational Systems Biology: Biology X
 Bud Mishra Room 1002, 715 Broadway, Courant Institute, NYU, New York, USA L#4:(October-0-4-2010) Cancer and Signals 1 2 1 2 Evidence in Favor Somatic mutations, Aneuploidy, Copy-number changes and LOH
Bud Mishra Room 1002, 715 Broadway, Courant Institute, NYU, New York, USA L#4:(October-0-4-2010) Cancer and Signals 1 2 1 2 Evidence in Favor Somatic mutations, Aneuploidy, Copy-number changes and LOH
Chronic HIV-1 Infection Frequently Fails to Protect against Superinfection
 Chronic HIV-1 Infection Frequently Fails to Protect against Superinfection Anne Piantadosi 1,2[, Bhavna Chohan 1,2[, Vrasha Chohan 3, R. Scott McClelland 3,4,5, Julie Overbaugh 1,2* 1 Division of Human
Chronic HIV-1 Infection Frequently Fails to Protect against Superinfection Anne Piantadosi 1,2[, Bhavna Chohan 1,2[, Vrasha Chohan 3, R. Scott McClelland 3,4,5, Julie Overbaugh 1,2* 1 Division of Human
Chagas Disease in the Americas: Impacts and Opportunities for Control
 Chagas Disease in the Americas: Impacts and Opportunities for Control Rick Tarleton Center for Tropical and Emerging Global Diseases University of Georgia The Chagas Disease Foundation Trypanosoma cruzi
Chagas Disease in the Americas: Impacts and Opportunities for Control Rick Tarleton Center for Tropical and Emerging Global Diseases University of Georgia The Chagas Disease Foundation Trypanosoma cruzi
Ultrastructural Studies on Plasmodium vivax
 Characterization of Human Malaria Parasites Ultrastructural Studies on Plasmodium vivax For the first time a detailed ultrastructural study was carried out on P. vivax. Fine structural analysis of growth
Characterization of Human Malaria Parasites Ultrastructural Studies on Plasmodium vivax For the first time a detailed ultrastructural study was carried out on P. vivax. Fine structural analysis of growth
By Mir Mohammed Abbas II PCMB 'A' CHAPTER CONCEPT NOTES
 Chapter Notes- Genetics By Mir Mohammed Abbas II PCMB 'A' 1 CHAPTER CONCEPT NOTES Relationship between genes and chromosome of diploid organism and the terms used to describe them Know the terms Terms
Chapter Notes- Genetics By Mir Mohammed Abbas II PCMB 'A' 1 CHAPTER CONCEPT NOTES Relationship between genes and chromosome of diploid organism and the terms used to describe them Know the terms Terms
Answering your daily challenges. in the ELISA technology I N FECTI OUS DISEASES. bioelisa
 Answering your daily challenges in the ELISA technology I N FECTI OUS DISEASES bioelisa RETROVIRUS HIV is the cause of the most important global pandemic. Due to the lack of vaccination, early diagnosis
Answering your daily challenges in the ELISA technology I N FECTI OUS DISEASES bioelisa RETROVIRUS HIV is the cause of the most important global pandemic. Due to the lack of vaccination, early diagnosis
Colorful patterns of evolution mark butterflies and bumblebees 15 May 2014, by David Pacchioli
 Colorful patterns of evolution mark butterflies and bumblebees 15 May 2014, by David Pacchioli thought she was looking at many members of a single species. Peering closer, however, she noted slight differences
Colorful patterns of evolution mark butterflies and bumblebees 15 May 2014, by David Pacchioli thought she was looking at many members of a single species. Peering closer, however, she noted slight differences
WORLD HEALTH ORGANIZATION. Smallpox eradication: destruction of Variola virus stocks
 WORLD HEALTH ORGANIZATION EXECUTIVE BOARD EB111/5 111th Session 23 December 2002 Provisional agenda item 5.3 Smallpox eradication: destruction of Variola virus stocks Report by the Secretariat 1. The WHO
WORLD HEALTH ORGANIZATION EXECUTIVE BOARD EB111/5 111th Session 23 December 2002 Provisional agenda item 5.3 Smallpox eradication: destruction of Variola virus stocks Report by the Secretariat 1. The WHO
Variations of the External Male Genitalia in Three Populations of Triatoma infestans Klug, 1834
 Mem Inst Oswaldo Cruz, Rio de Janeiro, Vol. 93(4): 479-483, Jul./Aug. 1998 Variations of the External Male Genitalia in Three Populations of Triatoma infestans Klug, 1834 Herton Helder Rocha Pires/ +,
Mem Inst Oswaldo Cruz, Rio de Janeiro, Vol. 93(4): 479-483, Jul./Aug. 1998 Variations of the External Male Genitalia in Three Populations of Triatoma infestans Klug, 1834 Herton Helder Rocha Pires/ +,
Jay M. Baraban MD, PhD January 2007 GENES AND BEHAVIOR
 Jay M. Baraban MD, PhD jay.baraban@gmail.com January 2007 GENES AND BEHAVIOR Overview One of the most fascinating topics in neuroscience is the role that inheritance plays in determining one s behavior.
Jay M. Baraban MD, PhD jay.baraban@gmail.com January 2007 GENES AND BEHAVIOR Overview One of the most fascinating topics in neuroscience is the role that inheritance plays in determining one s behavior.
Ch. 23 The Evolution of Populations
 Ch. 23 The Evolution of Populations 1 Essential question: Do populations evolve? 2 Mutation and Sexual reproduction produce genetic variation that makes evolution possible What is the smallest unit of
Ch. 23 The Evolution of Populations 1 Essential question: Do populations evolve? 2 Mutation and Sexual reproduction produce genetic variation that makes evolution possible What is the smallest unit of
Single Gene (Monogenic) Disorders. Mendelian Inheritance: Definitions. Mendelian Inheritance: Definitions
 Single Gene (Monogenic) Disorders Mendelian Inheritance: Definitions A genetic locus is a specific position or location on a chromosome. Frequently, locus is used to refer to a specific gene. Alleles are
Single Gene (Monogenic) Disorders Mendelian Inheritance: Definitions A genetic locus is a specific position or location on a chromosome. Frequently, locus is used to refer to a specific gene. Alleles are
Coevolution. Coevolution
 Coevolution Fitness is a genotype-by-environment interaction. The environment for one species includes other species For species that interact, they form part of each other s environment As one species
Coevolution Fitness is a genotype-by-environment interaction. The environment for one species includes other species For species that interact, they form part of each other s environment As one species
Lecture-7- Hazem Al-Khafaji 2016
 TOXOPLASMOSIS Lecture-7- Hazem Al-Khafaji 2016 TOXOPLASMOSIS It is a disease caused by Toxoplasma gondii which is a protozoan parasite that is infects a variety of mammals and birds throughout the world.
TOXOPLASMOSIS Lecture-7- Hazem Al-Khafaji 2016 TOXOPLASMOSIS It is a disease caused by Toxoplasma gondii which is a protozoan parasite that is infects a variety of mammals and birds throughout the world.
MRC-Holland MLPA. Description version 08; 30 March 2015
 SALSA MLPA probemix P351-C1 / P352-D1 PKD1-PKD2 P351-C1 lot C1-0914: as compared to the previous version B2 lot B2-0511 one target probe has been removed and three reference probes have been replaced.
SALSA MLPA probemix P351-C1 / P352-D1 PKD1-PKD2 P351-C1 lot C1-0914: as compared to the previous version B2 lot B2-0511 one target probe has been removed and three reference probes have been replaced.
GENETICS - NOTES-
 GENETICS - NOTES- Warm Up Exercise Using your previous knowledge of genetics, determine what maternal genotype would most likely yield offspring with such characteristics. Use the genotype that you came
GENETICS - NOTES- Warm Up Exercise Using your previous knowledge of genetics, determine what maternal genotype would most likely yield offspring with such characteristics. Use the genotype that you came
MRC-Holland MLPA. Description version 14; 28 September 2016
 SALSA MLPA probemix P279-B3 CACNA1A Lot B3-0816. As compared to version B2 (lot B2-1012), one reference probe has been replaced and the length of several probes has been adjusted. Voltage-dependent calcium
SALSA MLPA probemix P279-B3 CACNA1A Lot B3-0816. As compared to version B2 (lot B2-1012), one reference probe has been replaced and the length of several probes has been adjusted. Voltage-dependent calcium
On the origin of species, Really
 On the origin of species, Really Mohamed Noor Duke University Life on our planet is highly diverse but that life seems to exist in discrete clusters at multiple levels We readily recognize these clusters
On the origin of species, Really Mohamed Noor Duke University Life on our planet is highly diverse but that life seems to exist in discrete clusters at multiple levels We readily recognize these clusters
Rama Nada. - Malik
 - 2 - Rama Nada - - Malik 1 P a g e We talked about HAV in the previous lecture, now we ll continue the remaining types.. Hepatitis E It s similar to virus that infect swine, so its most likely infect
- 2 - Rama Nada - - Malik 1 P a g e We talked about HAV in the previous lecture, now we ll continue the remaining types.. Hepatitis E It s similar to virus that infect swine, so its most likely infect
Trypanosoma cruzi: ancestral genomes and population structure
 108 Mem Inst Oswaldo Cruz, Rio de Janeiro, Vol. 104(Suppl. I): 108-114, 2009 Trypanosoma cruzi: ancestral genomes and population structure Sérgio DJ Pena/ +, Carlos Renato Machado, Andréa Mara Macedo Departamento
108 Mem Inst Oswaldo Cruz, Rio de Janeiro, Vol. 104(Suppl. I): 108-114, 2009 Trypanosoma cruzi: ancestral genomes and population structure Sérgio DJ Pena/ +, Carlos Renato Machado, Andréa Mara Macedo Departamento
BA, BSc, and MSc Degree Examinations
 Examination Candidate Number: Desk Number: BA, BSc, and MSc Degree Examinations 2017-8 Department : BIOLOGY Title of Exam: Evolutionary ecology Time Allowed: 2 hours Marking Scheme: Total marks available
Examination Candidate Number: Desk Number: BA, BSc, and MSc Degree Examinations 2017-8 Department : BIOLOGY Title of Exam: Evolutionary ecology Time Allowed: 2 hours Marking Scheme: Total marks available
Usefulness of PCR-based assays to assess drug efficacy in Chagas disease chemotherapy: value and limitations
 122 Mem Inst Oswaldo Cruz, Rio de Janeiro, Vol. 104(Suppl. I): 122-135, 2009 Usefulness of PCR-based assays to assess drug efficacy in Chagas disease chemotherapy: value and limitations Constança Carvalho
122 Mem Inst Oswaldo Cruz, Rio de Janeiro, Vol. 104(Suppl. I): 122-135, 2009 Usefulness of PCR-based assays to assess drug efficacy in Chagas disease chemotherapy: value and limitations Constança Carvalho
Mammalogy Lecture 16 Conservation Genetics (with a side emphasis on Marine Mammals)
 Mammalogy Lecture 16 Conservation Genetics (with a side emphasis on Marine Mammals) Initial definition: Allele An allele is a viable DNA coding that occupies a given locus (position) on a chromosome (plural
Mammalogy Lecture 16 Conservation Genetics (with a side emphasis on Marine Mammals) Initial definition: Allele An allele is a viable DNA coding that occupies a given locus (position) on a chromosome (plural
5/2/18. After this class students should be able to: Stephanie Moon, Ph.D. - GWAS. How do we distinguish Mendelian from non-mendelian traits?
 corebio II - genetics: WED 25 April 2018. 2018 Stephanie Moon, Ph.D. - GWAS After this class students should be able to: 1. Compare and contrast methods used to discover the genetic basis of traits or
corebio II - genetics: WED 25 April 2018. 2018 Stephanie Moon, Ph.D. - GWAS After this class students should be able to: 1. Compare and contrast methods used to discover the genetic basis of traits or
Genomic structural variation
 Genomic structural variation Mario Cáceres The new genomic variation DNA sequence differs across individuals much more than researchers had suspected through structural changes A huge amount of structural
Genomic structural variation Mario Cáceres The new genomic variation DNA sequence differs across individuals much more than researchers had suspected through structural changes A huge amount of structural
The Application of Latent Class Analysis for Diagnostic Test Validation of Chronic Trypanosoma cruzi Infection in Blood Donors
 BJID 2002; 6 (August) 181 The Application of Latent Class Analysis for Diagnostic Test Validation of Chronic Trypanosoma cruzi Infection in Blood Donors Dante M. Langhi Junior, José O. Bordin, Adauto Castelo,
BJID 2002; 6 (August) 181 The Application of Latent Class Analysis for Diagnostic Test Validation of Chronic Trypanosoma cruzi Infection in Blood Donors Dante M. Langhi Junior, José O. Bordin, Adauto Castelo,
Epidemiology and Laboratory Diagnosis of Fungal Diseases
 Medical Mycology (BIOL 4849) Summer 2007 Dr. Cooper Epidemiology of Mycoses Epidemiology and Laboratory Diagnosis of Fungal Diseases Mycosis (pl., mycoses) - an infection caused by a fungus Two broad categories
Medical Mycology (BIOL 4849) Summer 2007 Dr. Cooper Epidemiology of Mycoses Epidemiology and Laboratory Diagnosis of Fungal Diseases Mycosis (pl., mycoses) - an infection caused by a fungus Two broad categories
M I C R O B I O L O G Y WITH DISEASES BY TAXONOMY, THIRD EDITION
 M I C R O B I O L O G Y WITH DISEASES BY TAXONOMY, THIRD EDITION Chapter 14 Infection, Infectious Diseases, and Epidemiology Lecture prepared by Mindy Miller-Kittrell, University of Tennessee, Knoxville
M I C R O B I O L O G Y WITH DISEASES BY TAXONOMY, THIRD EDITION Chapter 14 Infection, Infectious Diseases, and Epidemiology Lecture prepared by Mindy Miller-Kittrell, University of Tennessee, Knoxville
SpringerBriefs in Food, Health, and Nutrition
 SpringerBriefs in Food, Health, and Nutrition Editor-in-Chief Richard W. Hartel, University of Wisconsin Madison, USA Associate Editors John W. Finley, Louisiana State University, Louisiana, USA David
SpringerBriefs in Food, Health, and Nutrition Editor-in-Chief Richard W. Hartel, University of Wisconsin Madison, USA Associate Editors John W. Finley, Louisiana State University, Louisiana, USA David
Ménard D, Barnadas C et al, Proc Natl Acad Sci U S A Mar 30;107(13):
 Plasmodium vivax malaria in Madagascar Pvivaxinfection P. infection & clinical malaria in Duffy-negative Malagasy people Ménard D, Barnadas C et al, Proc Natl Acad Sci U S A. 2010 Mar 30;107(13):5967-71
Plasmodium vivax malaria in Madagascar Pvivaxinfection P. infection & clinical malaria in Duffy-negative Malagasy people Ménard D, Barnadas C et al, Proc Natl Acad Sci U S A. 2010 Mar 30;107(13):5967-71
Roadmap. Inbreeding How inbred is a population? What are the consequences of inbreeding?
 1 Roadmap Quantitative traits What kinds of variation can selection work on? How much will a population respond to selection? Heritability How can response be restored? Inbreeding How inbred is a population?
1 Roadmap Quantitative traits What kinds of variation can selection work on? How much will a population respond to selection? Heritability How can response be restored? Inbreeding How inbred is a population?
Genetic Disorders and Congenital Defects in Latin America and the Caribbean
 Genetic Disorders and Congenital Defects in Latin America and the Caribbean Victor B. Penchaszadeh, MD, MSPH Columbia University, New York and Pan American Health Organization CAPABILITY Workshop Lund,
Genetic Disorders and Congenital Defects in Latin America and the Caribbean Victor B. Penchaszadeh, MD, MSPH Columbia University, New York and Pan American Health Organization CAPABILITY Workshop Lund,
OUT-TB Web. Ontario Universal Typing of Tuberculosis: Surveillance and Communication System
 OUT-TB Web Ontario Universal Typing of Tuberculosis: Surveillance and Communication System Dr. Frances Jamieson, Ontario Public Health Laboratories November 30 th, 2009 Tuberculosis : A Global Problem
OUT-TB Web Ontario Universal Typing of Tuberculosis: Surveillance and Communication System Dr. Frances Jamieson, Ontario Public Health Laboratories November 30 th, 2009 Tuberculosis : A Global Problem
The Biology and Genetics of Cells and Organisms The Biology of Cancer
 The Biology and Genetics of Cells and Organisms The Biology of Cancer Mendel and Genetics How many distinct genes are present in the genomes of mammals? - 21,000 for human. - Genetic information is carried
The Biology and Genetics of Cells and Organisms The Biology of Cancer Mendel and Genetics How many distinct genes are present in the genomes of mammals? - 21,000 for human. - Genetic information is carried
Progress Toward Rubella and Congenital Rubella Syndrome Elimination in the Western Hemisphere,
 1 Introduction: Progress Toward Rubella and Congenital Rubella Syndrome Elimination in the Western Hemisphere, 2003-2008 1 Enhanced measles elimination activities in the Region of the Americas during the
1 Introduction: Progress Toward Rubella and Congenital Rubella Syndrome Elimination in the Western Hemisphere, 2003-2008 1 Enhanced measles elimination activities in the Region of the Americas during the
Q: Do platypus have more functional genes on their Y s than other mammals?
 One minute responses Q: Do platypus have more functional genes on their Y s than other mammals? A: I wasn t able to find out I did learn that the platypus X s and Y s are not homologous to normal mammal
One minute responses Q: Do platypus have more functional genes on their Y s than other mammals? A: I wasn t able to find out I did learn that the platypus X s and Y s are not homologous to normal mammal
Human Genetics 542 Winter 2018 Syllabus
 Human Genetics 542 Winter 2018 Syllabus Monday, Wednesday, and Friday 9 10 a.m. 5915 Buhl Course Director: Tony Antonellis Jan 3 rd Wed Mapping disease genes I: inheritance patterns and linkage analysis
Human Genetics 542 Winter 2018 Syllabus Monday, Wednesday, and Friday 9 10 a.m. 5915 Buhl Course Director: Tony Antonellis Jan 3 rd Wed Mapping disease genes I: inheritance patterns and linkage analysis
