Joint co-clustering: Co-clustering of genomic and clinical bioimaging data
|
|
|
- Amy Phillips
- 5 years ago
- Views:
Transcription
1 Computers and Mathematics with Applications 55 (2008) Joint co-clustering: Co-clustering of genomic and clinical bioimaging data Elisa Ficarra a,, Giovanni De Micheli a, Sungroh Yoon b, Luca Benini c, Enrico Macii d a Ecole Polythecnique Federale de Lausanne (EPFL), LSI, Station 14, 1015 Lausanne, Switzerland b Computer Systems Laboratory, Stanford University, Stanford, CA 94305, USA c University of Bologna, DEIS, Viale Risorgimento, 2 Bologna, Italy d Politecnico di Torino, DAUIN, Corso Duca degli Abruzzi, 24 Torino, Italy Abstract For better understanding the genetic mechanisms underlying clinical observations, and better defining a group of potential candidates for protein-family-inhibiting therapy, it is interesting to determine the correlations between genomic, clinical data and data coming from high resolution and fluorescent microscopy. We introduce a computational method, called joint co-clustering, that can find co-clusters or groups of genes, bioimaging parameters and clinical traits that are believed to be closely related to each other based on the given empirical information. As bioimaging parameters, we quantify the expression of growth factor receptor EGFR/erb-B family in non-small cell lung carcinoma (NSCLC) through a fully-automated computer-aided analysis approach. This immunohistochemical analysis is usually performed by pathologists via visual inspection of tissue samples images. Our fully-automated techniques streamlines this error-prone and time-consuming process, thereby facilitating analysis and diagnosis. Experimental results for several real-life datasets demonstrate the high quantitative precision of our approach. The joint coclustering method was tested with the receptor EGFR/erb-B family data on non-small cell lung carcinoma (NSCLC) tissue and identified statistically significant co-clusters of genes, receptor protein expression and clinical traits. The validation of our results with the literature suggest that the proposed method can provide biologically meaningful co-clusters of genes and traits and that it is a very promising approach to analyse large-scale biological data and to study multi-factorial genetic pathologies through their genetic alterations. c 2007 Elsevier Ltd. All rights reserved. Keywords: Image processing; Gene clustering; Protein activities; Clinics 1. Introduction For better understanding the genetic mechanisms underlying clinical observations, it is interesting to determine which genes and clinical traits are interrelated. In the last few years a considerable amount of research in genomics has been done concerning correlation of gene expression to multi-factorial genetic pathologies. Microarray data Corresponding author. addresses: elisa.ficarra@polito.it (E. Ficarra), giovanni.demicheli@epfl.ch (G. De Micheli), sryoon@stanford.edu (S. Yoon), lbenini@deis.unibo.it (L. Benini), enrico.macii@polito.it (E. Macii) /$ - see front matter c 2007 Elsevier Ltd. All rights reserved. doi: /j.camwa
2 E. Ficarra et al. / Computers and Mathematics with Applications 55 (2008) analysis, as well as real-time PCR, are useful techniques exploited so far to this purpose [1,2]. Despite this effort, results obtained are strongly limited by the poor informative content provided by clustering techniques applied to gene expression data [3]. At the same time, in the field of biomedical and molecular imaging, new techniques have been shown to be effective in extracting clinical and functional biological information from images of molecules and tissues [4,5]. By observing processes as they happen within the cell, these techniques add an important extra dimension to the understanding of cell behavior and functioning for early disease detection and drug response. In clinics, new applications of conventional imaging technologies are likely to play increasingly important roles, particularly in oncology. These two independent sources of information, namely gene expression mining techniques and fully-automated bioimaging, can be correlated to enhance gene expression analysis and to increase the amount of confidence in the hypothesized gene expression paths. For this purpose, we developed a joint co-clustering technique able to extract clinical bioimaging parameters through a fully-automated computer-aided approach and to perform co-clustering technique between clinical bioimaging parameters and gene expression data. Our proposed method consists of two steps. As a first step, we had earlier developed a computational method that can deterministically find all the co-clusters, between clinical traits and gene expression data, satisfying specific input parameters in an efficient manner [6]. To measure the correlation between a gene and a clinical trait, existing approaches obtain a vector of the expression level of the gene over a number of samples and another vector of the value of the clinical trait over the same samples and then calculate the statistical correlation between the two vectors. By applying this procedure to many genes, we can identify some genes correlated to the clinical trait of interest. Proceeding one step further from prior methods that can reveal one-to-many relationships between a single trait and multiple genes (or vice versa), we developed a method that can find many-to-many relationships between genes and traits using a clustering technique called co-clustering. This method possesses clear advantages over heuristic methods that can provide only partial solutions and other exact algorithms that are not scalable to large-scale problems [7,8]. As a second step, we developed a fully-automated tissue image processing method, namely computer-aided protein quantification tool, able to extract a set of clinical parameters that give a characterization of the pathology dynamics. This tool was successfully tested on non-small cell lung carcinoma (NSCLC) tissue images in order to characterize and quantify, in a standardized way, the activation of the EGFR/erb-B protein receptor family that plays an important role in non-small cell lung carcinoma growing [9]. This type of analysis aims at characterizing each pathological cell, and on an average the whole tissue, by performing a standardized quantitative and qualitative measurement of protein activations. This information can be treated as a clinical parameter, and can be finally correlated with the genetic expression data on same lung carcinoma tissue in order to better define a group of potential candidates for protein-family-inhibiting therapy. For this purpose, we developed the proposed fully-automated joint co-clustering approach to find correlations between genetic data and clinical and bioimaging parameters. The tool was tested with the epidermal growth factor receptor EGFR/erb-B family dataset in the non-small cell lung carcinoma (NSCLC) tissue. The EGFR/erb-B family of receptors plays an important role for NSCLC development. Quantifying and classifying the EGFR expression and activity in NSCLC with special regard to the assessment of the prevalence of somatic EGFR mutations, as well as to ligand receptor interactions, could lead to new insights into the modulation of EGFR/erb-B in individual lung carcinomas. Thus, it is important to extract these information by using methodologies that give quantifiable, standardized and precise measurements [9]. We quantified the activity of the EGFR/erb-B receptors in NSCLC immunohistochemical images of 70 patients. Subsequently, we correlated these bioimaging parameters with the expression of genes that regulate the transcription of the EGFR/erb-B protein family, measured on same tissues and on the same datasets of 70 patients and other clinical traits (e.g. tumor classification, survival, follow-up etc.). Results show that there is a strong correlation between bioimaging parameters quantifying EGFR/erb-B protein family activations and their gene regulative expression. To justify our analyses, we present some supporting evidence for our results in the literature. Our experimental studies suggest that joint co-clustering is a very promising approach to analyse large-scale biological data and to study multi-factorial genetic pathologies through their genetic alterations. Moreover, this approach enables new opportunities for early diagnosis and provides information in future strategies for therapy. Section 2 explains the computer-aided protein quantification tool. Section 3 explains our method to find co-clusters. Experimental results and discussions are presented in Section 4, followed by concluding remarks in Section 5.
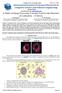
3 940 E. Ficarra et al. / Computers and Mathematics with Applications 55 (2008) Fig. 1. (a) Example for lung cancer tissue immunohistochemical image; (b) example for membranes detection, see big cell in the bottom-right part of the image. 2. Computer-aided protein quantification tool: Membrane detection and parameter extraction Direct monitoring of the activity of proteins involved in the genesis and development multi-factorial genetic pathologies is a very useful diagnostic tool. It leads to the classification of the pathology in a more accurate way, through its particular genetic alterations, and to create new opportunities for early diagnosis as well as to provide information in future strategies for therapy. An approach for monitoring and quantifying the activity of proteins is to analyse their localization and the intensity of their activity in pathological tissues by using, for example, images of the tissue where the localization of proteins, as well as their ligands, is highlighted by fluorescent-marked antibodies that can detect and link the target proteins. The antibodies are marked with a particular stain. The protein activity intensity is related to the intensity of the stains. This procedure is called immunohistochemistry (IHC). Fig. 1(a) shows an example for immunohistochemical image of lung cancer tissue. What is interesting to extract from these images is not a specific coloured area, that is almost the standard procedure with this kind of images [10,11]. Rather, the focus is cell-by-cell localization of the coloured areas in particular the cellular regions (i.e. membranes or cytoplasm or nuclei). Similarly, the quantification of the percentages of coloured areas at the location of interest is important because it relates to the activity of specific receptors. In other words, it is important to quantify if the proteins have a membrane activity or not (or cytoplasm or nucleus one), how much of that membrane is positive for the specific protein activity and, vice versa, if it is not active. This type of analysis aims at characterizing each pathological cell, and on an average the whole tissue, by performing a standardized quantitative and qualitative measurement of protein activations. In this section we describe a fully-automated procedure that provides standardized measures of protein activities, and related ligands, involved in the development of a pathology. This goal is reached (i) by identifying different cellular regions, (ii) quantifying the percentage of active areas with respect to each whole region, and (iii) quantifying the intensity of the protein activity. These analyses have traditionally been performed directly by pathologists in a very subjective and time-consuming way. The major contribution of this research is to provide an automated, fast and precise means for performing this kind of immunohistochemical image analysis. To the best of our knowledge the methodology presented in this paper is the first completely automated approach to this purpose. Much previous work in biomedical image processing focused on automated methods for segmentation of nuclei and cells [12 15]. Classical approaches, such as active contours or watersheds, are not effective when the objects to be identified lack specific geometrical features or gradient variations. Unfortunately, these critical conditions are very common in the images targeted by our work. Cancer tissue cells are characterized by non-predictable variations in shape that lead to a non-trivial determination of an effective approach based on shape-based segmentation. Moreover, in immunohistochemical cancer tissue images cells are not well separated and, in addition, they are usually not characterized by variations gradient magnitude. To address these issues, we developed a novel deterministic fully-automated approach for the quantification of protein activities and localization of molecular activities in tissue images.
4 E. Ficarra et al. / Computers and Mathematics with Applications 55 (2008) Immunohistochemical lung cancer tissue images are characterized by a blue stain as background colour and a brown stain where a receptor of the EGFR family is detected. We focus here on quantification of membrane receptor activity. Cell membrane segmentation is a hard problem because those membranes that are negative to the EGFR family of receptors, are generally not visible. In other words, they are not characterized by gradient magnitude variation. It is also possible that a cell has only some parts of its membrane positive to receptor activity. The automated procedure is composed of several sequential steps, as outlined in the following subsections. In this work, our description concentrates on the steps we customized Virtual cell membrane detection To reconstruct the cell membrane locations we first detected nucleus membranes using standard morphological segmentation approaches. For each nucleus, we detected seeds, applying noise filtering, colour filtering to detect nucleus regions, artifacts removal, filling of connected components and boundaries detection. These first steps lead to an approximate detection of nucleus boundaries. We used these nucleus boundaries as initial curves for the final detection of nucleus membranes. We completed the detection of nucleus membranes by applying the active contour algorithm presented in [16]. This algorithm was found very useful for nucleus membranes detection. Further details on seed detection and active contours are beyond the scope of this paper because they are obtained and implemented using standard approaches. The interested readers are directed to [16,14]. After detecting nucleus membranes, we implemented a procedure for virtual cell membrane detection. This is an important step in our approach. In fact, to perform membrane cell segmentation, we use virtual membranes as part of the final-detected cell membranes in those regions that are negative to the EGFR family of receptors and that are as a consequence not characterized by gradient magnitude variation. Virtual cell membranes are computed as set of connected points equidistant from closest nucleus membranes. Since our analysis concerns cells in tissues, the assumption that cellular membranes are equidistant from closest nucleus boundaries is reasonable as first-order approximation. Note that, we implemented a customized procedure for virtual cell membranes design because alternative traditional methods, such as Voronoi tessellation [17], build curves equidistant from points. Since these alternative methods fix as center of tessellation a point instead of complex shape membranes (i.e. nucleus membranes), they obtain virtual curves that have few edges and sharp angles between edges. These curves are thus a poor approximation of real cell membranes Colour filtering To select the region that are positive to receptor activations, we filtered the image on Hue Saturation Intensity (HSI) colour space. We chose the HSI space because the stains we used are well defined in (HSI) space. In particular, looking at several Hue histograms of the tissue images, we noticed well-separated bi-modal value distributions. To separate the two distributions there are several standard thresholding algorithms that can be successfully employed, such as [18 20]. As expression of receptor activity we chose brown pixels with hue components minor than a threshold automatically computed by using Ridler thresholding as detailed in [21] Cellular membrane detection The detection of cellular membranes is done in two steps. Beforehand, we perform membrane segmentation in the brown areas one cell at a time and we connect them with the virtual cell membrane in those regions that are not characterized by receptor reaction. To this purpose, we developed an ad hoc procedure, as described later in this section. The second step consists of a customized fitting procedure of the detected membrane points to complete the cellular membrane segmentation. The first step of cellular membrane detection is the Scanning procedure: to connect brown areas with the virtual membrane in those regions where there was no receptor reaction, the area across the virtual membrane is dilated in order to be able to reach, if they exist, brown regions of the cell. The level of dilation is an input parameter and it depends on image resolution. We set this value to 18 (pixels) for images with a resolution of about 3 nm. Then, we scan the dilated area with a scan line having one end on the center of the nucleus and the other one on the external border of dilated area.
5 942 E. Ficarra et al. / Computers and Mathematics with Applications 55 (2008) At each step, the points of the membrane are computed as weighted barycentre B of brown pixels among the scan line, as shown in Eq. (1) c j I j j B = j, c j I j j where j is the coordinate on the scan line. This coordinate is 0 on the virtual membrane, negative in the inner part of the dilated area and positive in the outer part. I j is the value of the jth pixel and c j is a coefficient for barycentre computation. The coefficient c j is 1 for pixels on scan line negative coordinate while for positive coordinates the coefficient has a negative parabolic trend as function of coordinate j. In this way, when a brown region branches off, the scanning procedure is forced to choose as points belonging to the membrane those pixels that lie on the path closest to the nucleus. Moreover, we assigned to pixels of the scan line the value of 1 if they belong or precede to the virtual membrane pixels. This has been done when there are no brown pixels in the scan line, to choose as points belonging to membrane those pixels that are close to the virtual membrane. Finally, we set to 0 the pixels that are neither brown nor virtual membrane ones. The second step in the detection of cellular membranes is the Fitting and complete membranes detection: to complete the detection of cellular membranes, we implemented an iterative fitting procedure in which outlier pixels are deleted at each step. We defined outlier pixels as the pixels located far away from the fitting line more than three times the standard deviation. An example for membrane detection is shown in Fig. 1(b) Clinical parameter computation We quantify the activity of membrane EGFR/erb-B receptors through the computation of percentage of active areas with respect to each whole membrane region. Then, the final parameter is the average value of all single-cell parameters on the image. 3. Co-clustering method The co-clustering approach finds groups of genes and clinical parameters that are believed to be closely related to each other based on the given empirical information. In particular, it can find many-to-many relationships between genes and traits using a clustering technique called co-clustering. Here the term co-clustering refers to an unsupervised learning technique that performs simultaneous clustering of rows and columns in a matrix to find (possibly) overlapping submatrices covering the matrix. More specifically, given gene expression data and clinical parameter values, we first create a matrix called correlation matrix that can collectively represent the degree of correlation between genes and clinical traits. Each row and column of this matrix corresponds to a gene and a clinical trait, respectively. Then, our method searches co-clusters or submatrices (with some semantics to be defined) covering the correlation matrix Definitions Let S represent a set of clinical samples. For each sample in S, gene expression levels are measured by the DNA microarray technology of choice. Let G be the set of genes in the measurement. Clinical traits are recorded for each sample. Let T be the set of the recorded traits. The input of the proposed approach is composed of two data matrices. One is a gene expression data matrix denoted by pair A = (G, S), where, A R G S, and the element a ik of the matrix A represents the expression level of gene i for sample k. The other matrix is denoted by pair B = (T, S), and the element b jk of the matrix B is the value of trait j for sample k. The columns of A and B are arranged in the same order. Depending upon the type of trait j, b jk may be quantitative, categorical, or others. The output is a set of co-clusters. A co-cluster is composed of a gene set I G and a trait set J T and represents a group of genes and traits closely related to each other, given the input matrices A and B. A co-cluster can formally be defined by the following series of definitions. (1)
6 E. Ficarra et al. / Computers and Mathematics with Applications 55 (2008) Fig. 2. Construction of the correlation matrix. A co-cluster appears as a submatrix of the correlation matrix C. Definition 1. For V, a vector on R, the range of V, denoted by RANGE(V ), is the absolute difference between the largest and the smallest elements of V. Definition 2. Given V and W, two real vectors of the same dimension, the linear deviation of V and W, denoted by LIN-DEV(V, W ), is defined as min{range(v W ), RANGE(V + W )}. (2) Definition 3. Given the input matrices A and B, a correlation matrix, denoted by C, is a matrix where the row set and the column set of C are G and T, respectively, and the element c i j is the statistic indicating the degree of correlation between gene i and trait j and is defined in significance analysis of microarrays (SAM) [8], namely, c i j = r i j s i j + s 0, where r i j is a score to measure the degree of correlation between the expression level of gene i and the value of clinical trait j, s i j is the gene-specific scatter or the standard deviation of repeated expression measurements, and s 0 is a fudge factor to prevent the computed statistic from becoming too large when s i j is close to zero [23]. Fig. 2 shows the correlation matrix construction scheme. Definition 4. Given the correlation matrix C = (G, T ) and thresholds τ 0 and π > 0, a co-cluster is a matrix, denoted by D = (I, J), satisfying the following conditions: (1) I G and J T ; (2) for any two column vectors V and W of size I in D, LIN-DEV(V, W ) τ. Condition (1) indicates that D is a submatrix of the correlation matrix C. Condition (2) is to require that every pair of I -dimensional column vectors from D exhibit correlation with respect to the metric LIN-DEV Algorithm overview As detailed in [6], the proposed co-clustering algorithm consists of three steps: First, an intermediate data matrix called correlation matrix is constructed from the input matrices. Then, special co-clusters called pairwise co-clusters are found in the correlation matrix. Finally, co-clusters are derived from the pairwise co-clusters. An overview of the algorithm is presented below, and more details can be found in [6]. In the first step, each element c i j of the correlation matrix is calculated using the SAM procedure. When calculating c i j, we must follow a procedure for multiple comparisons, thus ensuring that too many falsely significant ones are not declared [22,23]. To this end, the false discovery rate (FDR) is estimated for each c i j by random permutation of the data for gene expression among the different experimental arms. The SAM procedure can be outlined as follows [8]: (1) For given j, compute statistic c i j for i = 1, 2,..., G, where G is the number of genes in the gene expression matrix. (2) Compute order statistics c (1) c (2) c ( G ). (3)
7 944 E. Ficarra et al. / Computers and Mathematics with Applications 55 (2008) (3) Take M sets of permutations of the vector associated with trait j. For each permutation m, compute statistics ci m j and corresponding order statistics. (4) From the set of M permutations, estimate the expected order statistics by c (i) = (1/M) m c m (i) for i = 1, 2,..., G. (5) Plot the values of c (i) versus the values of c (i). (6) For, a fixed threshold, find the first i = i 1 such that c (i) c (i) >, starting at the origin and moving up to the right. All genes past i 1 are called significant positive. Similarly, find significant negative genes. For each, define the upper cut-point cut up ( ) as the smallest c i j among the significant positive genes, and similarly define the lower cut-point cut low ( ). (7) For a grid of values, compute the total number of significant genes (from the previous step), and the median number of falsely called genes, by computing the median number of values among each of the M sets of c m (i) (for i = 1, 2,..., G ) that fall above cut up ( ) or below cut low ( ). (8) Estimate P 0, the proportion of true null (unaffected) genes in the dataset (see [8] for details). (9) The median of the number of falsely called genes from Step 6 is scaled appropriately, according to the value of P 0 (see [8] for details). (10) A value of can be specified by the user and the significant genes are listed. (11) The FDR is computed as the median of the number of falsely called genes divided by the number of genes called significant. After having computed the correlation matrix, the next step is to find a special type of co-cluster called pairwise cocluster. A pairwise co-cluster is a co-cluster with only two traits and can therefore be represented by a submatrix (of the correlation matrix) with two columns. Pairwise co-clusters are used later as seeds to find (non-pairwise) co-clusters. To find a pairwise co-cluster in the correlation matrix C = (G, T ), we first select two distinct columns v, w T and construct from them two G -dimensional column vectors V = (c 1v, c 2v,..., c G v ) and W = (c 1w, c 2w,..., c G w ). Then, we compare V and W to identify I, a set of dimensions over which V and W are correlated (I G). Finally, we remove all i I such that the p-value of c iv or c iw is greater than a given threshold. By definition, the matrix denoted by pair (I, {v, w}) represents a co-cluster, and this co-cluster with only a pair of traits is called pairwise co-cluster. In the last step of our method, co-clusters are derived from pairwise co-clusters. Recall that T is the set of clinical traits or the set of column indices in the correlation matrix C = (G, T ). Our co-clustering method examines elements J 2 T in such an order that efficient enumeration is possible to find a co-cluster (I, J). To this end, a data structure called prefix tree or trie [24] is employed to systematically represent the elements of the power set 2 T. Each node in the trie represents candidates for co-clusters, and using an efficient traversal method nodes are gradually merged and pruned, resulting in co-clusters in their final form Remarks To assess the degree of correlation, in Definition 2 we introduced a metric called linear deviation. This is not to deny the effectiveness of a conventional statistic such as the Pearson correlation coefficient [22] but to transform it to a computation-efficient form, minimizing loss in the detection power. It is possible to see the relationship between LIN-DEV and the Pearson correlation coefficient, as shown in [6]: a lower value of LIN-DEV typically corresponds to a higher level of either positive or negative correlation. The specific definition of r i j in Definition 3 varies depending upon the type of clinical trait j. For example, if clinical trait j has quantitative values then r i j is defined in terms of the Pearson correlation coefficient [22] between the ith row vector of the matrix A and the jth row vector of the matrix B Joint co-clustering method The joint co-clustering approach is a fully-automated framework that aims to extract receptor and protein expressions from tissue images and correlate these bioimaging parameters with other clinical traits and the gene regulative expression of same receptors and proteins evaluated on same tissues. Thus, the joint co-clustering framework consists on the co-clustering algorithm where clinical traits are obtained through the fully-automated protein quantification tool.
8 E. Ficarra et al. / Computers and Mathematics with Applications 55 (2008) Fig. 3. Results for the first dataset: the plot shows the automated procedure measurements versus the manual-trace ones and the regression line. 4. Experimental results and discussion Experimental results were separately obtained for the computer-aided protein quantification tool and the co-clustering to demonstrate their accuracy and robustness. Afterwards, we present experimental results for joint co-clustering method Computer-aided protein quantification results We tested the algorithm on four datasets. All of them are composed by real lung cancer tissue immunohistochemical images. For each dataset, the images show different portions of the same IHC tissue. The four datasets present positive reactions at the EGFR/erb-B receptor activation. These reactions are localized in the cellular membranes. The four datasets differ because of different levels of positivity intensity. For each dataset, we first localized each cellular membrane in the images, as described in Section 2. Afterwards, we computed for each cell the percentage of area characterized by positive activation of receptor EGFR/erb-B with respect to the whole cellular membrane surface. At the end, we computed the final parameter as an average value of all single-cell parameters on each image. This final parameter is the clinical parameter that characterizes the percentage of receptors that is active in the lung cancer tissue. In order to evaluate the performance of our approach, positive protein reaction parameters have also been computed on membranes drawn manually by pathologists for taking advantage of the knowledge and skills of experts in that field. Manual analysis has been performed on all the datasets. These manual measurements were thus compared with the positive protein reaction parameters computed through our fully-automated approach. We show in this paper the results for two datasets in order to demonstrate the accuracy and robustness of our approach. On the other datasets, we obtained similar results and performance. Details can be found in [25]. Results are reported as follow. For each dataset, we compute the average error and the root mean square error (RMSE) incurred by our automated approach with respect to manual-trace measurements. We then computed the coefficient of correlation between each set of automated results and the corresponding manual-trace measurements. Finally, we performed a linear regression between automated manual-trace results to evaluate the level of confidence of the regression coefficient through the Student t-test. We first evaluate the correlation between the automated and the manual-trace measurements on the first immunohistochemical lung cancer tissue image set. Our analysis shows that these two sets of measurements are highly correlated, with a coefficient of correlation We then computed a linear regression of automated measures on manual-trace ones. We performed the Student t-test under the null hypothesis on the regression coefficients in order to estimate the confidence level of this regression. As a result, we rejected this hypothesis at significance level less than 1% obtaining a coefficient of the regression line of 0.96 with a region of acceptance of the hypothesis of the range Thus, the two sets of measures are highly correlated with a confidence level greater than 99%. Fig. 3 shows the results obtained for EGF-R protein activation measurements on the first immunohistochemical lung cancer
9 946 E. Ficarra et al. / Computers and Mathematics with Applications 55 (2008) Table 1 Results for percentage computation of receptor EGFR family activation on the three tissue image experimental datasets: first column shows the clinical parameter while the other ones indicate the average error and the root mean square error incurred by the automated procedure Positive membrane reaction (%) Average error (%) RMSE (%) Fig. 4. Results for the second dataset: the plot shows the automated procedure measurements versus the manual-trace ones and the regression line. tissue image set. The figure shows the automated measurements versus the manual-trace ones as well as the regression line. Moreover, we computed the difference between automated and manual-trace measurements and we performed the same Student t-test. We found that the difference between the two typologies of measurements is not significant and the average of the differences between automatic and manual measurements is 0.773%. Finally, the RMSE of our automated measurements is 3.3% (with a confidence of 99%), as shown in the first row of Table 1. Table 1 shows, in the first column, the computed percentage of receptor activation in the lung cancer tissue. In this first dataset that percentage is 58.65%. We also performed the same analysis on the second dataset and dataset of immunohistochemical lung cancer tissue images. On the second dataset, our analysis showed that automated and manual-trace measurements were highly correlated with a coefficient of correlation Performing the Student t-test under the null hypothesis on the regression coefficients we finally rejected this hypothesis at significance level less than 1%. We obtained a coefficient of the regression line of 0.85 with a region of acceptance of the hypothesis of the range Fig. 4 shows these results for EGF-R protein activation measurements on the second immunohistochemical lung cancer tissue image set. By performing the Student t-test on the difference between automated and manual-trace measurements we found that the difference between these two typologies of measurement is not significant. Moreover, the average of difference between automatic and manual measurements is 0.25% and the RMSE of our automated measurements is about 1.6%, as shown in the second row of Table 1. The percentage of receptors active in this second lung cancer tissue set is 95.89%. In this dataset the EGF-R receptor is highly active in most of the cells on the tissue. Looking at Fig. 4, we notice that almost all the measurements are clustered around a very high value while only a few measures are slightly smaller. This leads to a very little dispersion of the measures. At the same time, since the significance is computed with respect to the dispersion, lower values on the dataset slightly affect the slope of the regression line thus increasing the level of the significance of the test. Nevertheless, in this case also, the automated and manual-trace measurements are correlated with a confidence level greater than 99%.
10 E. Ficarra et al. / Computers and Mathematics with Applications 55 (2008) Co-clustering and joint co-clustering results The co-clustering was first tested with the Acute Myelogenous Leukemia (AML) dataset [7]. The AML dataset used included two matrices. One was a gene expression data matrix with 6283 genes and 119 samples. The other was a matrix of 15 clinical parameters measured from the identical samples. We used the procedure described in Section 3 to produce the correlation matrix. We identified 43 co-clusters. To justify the grouping of certain genes and clinical traits by the co-clusters found from the AML data, we present some supporting evidence for co-clustered genes and traits from the literature. In addition, we show that certain Gene Ontology terms annotating genes in some co-clusters are significantly over-represented. Taken together, these experimental studies suggest that our method can find biologically meaningful co-clusters. Details on these results can be found in [6]. The joint co-clustering was tested with the epidermal growth factor receptor EGFR/erb-B family dataset in the nonsmall cell lung carcinoma (NSCLC) tissue. The EGFR/erb-B family of receptors plays an important role for NSCLC development. Quantifying and classifying the EGFR/erb-B expression and activity in NSCLC with special regard to the assessment of the prevalence of somatic EGFR/erb-B mutations, as well as to ligand receptor interactions, could lead to new insights into the modulation of EGFR/erb-B in individual lung carcinomas. Thus, it is important to extract these information by using methodologies that give quantifiable, standardized and precise measurements. We quantified the activity of the EGFR/erb-B receptors in NSCLC immunohistochemical images of 70 patients. Subsequently, we correlated these bioimaging parameters with the expression of genes that regulate the transcription of the EGFR/erb-B protein family, measured on the same tissues and on the same datasets of 70 patients and other clinical traits (such as tumor type classifications, namely diagnosis, T, N, stage, size, survival, etc). Furthermore, we found supporting evidence for our results in the literature. Note that results for EGFR/erb-B protein expression (i.e. quantification) have been already given in this section (see Section 4.1). As a result of joint co-clustering between EGFR/erb-B protein expression and the regulative expression of the EGFR/erb-B protein transcripts, we found out significant correlations in about 83% of the studied cases. Among this percentage, we found co-clusters characterized by up-regulation of the transcripts and over-expression of the proteins. Among tumors that did not exhibit over-expression, i.e., the tumors that showed low protein positivity or negative staining, no gene up-regulation was observed. Moreover, high-level regulation was significantly more frequent in tumors with highest staining than in tumors with medium staining. Similar results were reported in recent studies [26,27]. The remaining 17% of the studied cases presents activation of EGFR protein family (visible through image analysis) but no up-regulation of the expression of the protein transcripts. In these particular cases 70% of the tumor was squamous cell carcinoma (SqCa), while 30% was adenocarcinoma (AdCa). Similar findings have been reported not only in lung carcinomas but also in other tumors, such as renal, pancreatic, breast, and colon carcinomas. Although protein over-expression in these tumors probably is caused by transcriptional or post-transcriptional activation, various theories have been proposed to explain the underlying mechanisms [28,26]. Post-translational changes as well as changes in genetic enhancer elements [29,30] were shown to be associated with an increased EGFR expression. Recently, a polymorphic CA-repeat in intron 1 of EGFR has been shown to have an important impact on EGFR transcription and expression, and seems to be a major target of EGFR mutations [31,27]. In the literature it has been found that this mechanism can explain protein over-expression in about 18.7% of the cases [27], that is in accordance with our results. We also found out that trait diagnosis (e.g., SqCA, AdCa, LCa) is correlated with genes erb-1, erb-2 and TGFalpha with an FDR of 1%. Similarly, size trait is correlated with erb-1 gene and survival trait is correlated with the erb-2 gene, as confirmed in [32,33]. Finally, we identified co-clusters of erb-1 and erb-2 proteins. These co-clusters consisted of 54% of the studied cases and were characterized, in particular, by erb-1 and erb-2 protein expression, their genetic regulation and SqCA/AdCA type classification of tumor. As a result, it can be seen that the over-expression of erb-1 (EGFR) impacts the SqCa tumors more than AdCa ones (56% of cases vs. 32%) and vice versa for the expression of erb-2 protein and the AdCa tumors (63% vs. 26%). Evidence for our results was found in literature [34,35]. We also found out coclusters that presented over-expression and gene up-regulation for erb-1 protein and did not exhibit gene up-regulation nor over-expression for erb-2. These co-clusters were characterized by a percentage of SqCa tumors higher than those of AdCa ones (67% vs. 0%). The reverse was found for co-clusters that presented over-expression and gene upregulation for erb-2 protein and which did not exhibit gene up-regulation or over-expression for erb-1 (100% of AdCa
11 948 E. Ficarra et al. / Computers and Mathematics with Applications 55 (2008) tumors). SqCA and AdCA tumors were found also in co-clusters characterized by either gene up-regulation or overexpression for both erb-1 and erb-2 (67% of AdCa and 27% SqCa). We also found supporting evidence for these last analyses in the literature [27]. 5. Conclusions We presented a fully-automated framework for finding co-clusters of genes and clinical traits using microarray data and bioimaging and clinical parameter information. We first quantified the expression of receptors in carcinoma tissue images by using our fully-automated protein quantification tool. This immunohistochemical analysis (IHC) is usually performed by pathologists via visual inspection of tissue samples images. Our techniques streamlines this error-prone and time-consuming process, thereby facilitating analysis and diagnosis. In particular, our method leads to classify protein reactions according to a specific cell region and to quantify the percentage and the intensity of this protein activity. The effectiveness of the proposed method has been tested using immunohistochemical non-small cell lung carcinoma tissue images. Results of comparison with manual-trace method on several real-life datasets demonstrate the high quantitative precision of our approach. Data coming from IHC images can be treated as a clinical parameter, and can be finally correlated with the genetic expression data of same lung carcinoma tissue (and same set of patients) in order to better define a group of potential candidates for protein-family-inhibiting therapy. For this purpose, we developed the proposed fully-automated joint co-clustering approach. An intermediate data matrix called correlation matrix was computed from microarray data and bioimaging and clinical parameter information by means of a statistical method. We then modeled a co-cluster by a submatrix of the correlation matrix with some semantics and aimed at finding statistically significant co-clusters. In order to validate our approach, we found supporting evidence for our analysis in the literature. Results show that there is a strong correlation between bioimaging parameters quantifying EGFR/erb-B protein family activations and their gene regulative expression measured on same tissues. These preliminary results show that the joint co-clustering is a very promising approach to analyse large-scale biological data and to study multi-factorial genetic pathologies through their genetic alterations. Moreover, this approach enables new opportunities for early diagnosis and provides information in future strategies for therapy. Acknowledgments We wish to acknowledge the Swiss Institute for Experimental Cancer Research (ISREC) in Lausanne, Switzerland and the Department of Pathology at the S. Luigi Hospital, Turin, Italy for their helpful and valuable advices and discussions. References [1] S. Saviozzi, G. Iazzetti, E. Caserta, A. Guffanti, R.A. Calogero, Microarray data analysis and mining, Methods in Molecular Medicine 94 (2004) [2] S. Yoon, C. Nardini, L. Benini, G. De Micheli, Enhanced pclustering and its applications to gene expression data, Proceedings of IEEE Bioinformatics and Bioengineering (2004) [3] O.G. Troyanskaya, Putting microarrays in a context: Integrated analysis of diverse biological data, Briefings in Bioinformatics 6 (1) (2005) [4] A. Hengerer, A. Wunder, D.J. Wagenaar, A.H. Vija, M. Shah, J. Grimm, From genomics to clinical molecular imaging, Proceedings of the IEEE 93 (4) (2005) [5] W. Chen, M. Reiss, D.J. Foran, Unsupervised tissue microarray analysis for cancer research and diagnostics, IEEE Transactions on Information Technology in Biomedicine 8 (2) (2004) [6] S. Yoon, L. Benini, G. De Micheli, Finding co-clusters of genes and clinical parameters, in: Proc. of the 27th Annual International Conference of the IEEE EMBS, 2005, pp [7] L. Bullinger, et al., Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia, New England Journal Medicine 350 (16) (2004) [8] V.G. Tusher, R. Tibshirani, G. Chu, Significance analysis of microarrays applied to the ionizing radiation response, Proceedings of the National Academy of Sciences of the United States of America 98 (9) (2001) [9] T.K. Taneja, S.K. Sharma, Markers of small cell lung cancer, World Journal of Surgical Oncology 2 (10) (2004). [10] E.M. Brey, et al., Automated selection of DAB-labeled tissue for immunohistochemical quantification, The Journal of Histochemistry and Cytochemistry 51 (5) (2003)
12 E. Ficarra et al. / Computers and Mathematics with Applications 55 (2008) [11] A. Ruifrok, R. Katz, D. Johnston, Comparison of quantification of histochemical staining by hue-saturation-intensity (HSI) transformation and color deconvolution, Applied Immunohistochemistry and Molecular Morphology 11 (1) (2004) [12] L. Yang, P. Meer, D.J. Foran, Unsupervised segmentation based on robust estimation and color active contour models, IEEE Transactions on Information Technology in Biomedicine 9 (3) (2005) [13] D.P. Mukherjee, N. Ray, S.T. Acton, Level set analysis for leukocyte detection and tracking, IEEE Transaction on Image Processing 13 (4) (2004) [14] A. Elmoataz, S. Schupp, R. Clouard, P. Herlin, D. Bloyet, Using active contours and mathematical morphology tools for quantification of immunohistochemical images, Signal Processing 71 (1998) [15] N. Malpica, et al., Applying watershed algorithms to the segmentation of clustered nuclei, Cytometry 28 (1997) [16] M. Jacob, T. Blu, M. Unser, Efficient energies and algorithms for parametric snakes, IEEE Transactions on Image Processing 13 (9) (2004) [17] F. Aurenhammer, R. Klein, in: J.-R. Sack, J. Urrutia (Eds.), Voronoi Diagrams, Handbook of Computational Geometry, 2000, pp (Chapter 5). [18] T.W. Ridler, S. Calvard, Picture thresholding using an iterative selection method, IEEE Transactions on Systems Man and Cybernetics 8 (8) (1978) [19] J. Kittler, J. Illingworth, C.Y. Suen, Minimum error thresholding, Pattern Recognition 19 (1986) [20] N. Otsu, A threshold selection method from gray level histograms, IEEE Transactions on Systems Man and Cybernetics 9 (1979) 62. [21] E. Ficarra, L. Benini, E. Macii, G. Zuccheri, Automated DNA fragments recognition and sizing through AFM image processing, IEEE Transactions on Information Technology in Biomedicine 9 (4) (2005) [22] B. Rosner, Fundamentals of Biostatistics, 5th edition, Duxbury Edit., [23] S. Draghici, Data Analysis Tools for DNA Microarrays, Chapman & Hall/CRC, [24] A.V. Aho, J.E. Hopcroft, J.D. Ullman, Data Structures and Algorithms, Addison-Wesley, Reading, MA, [25] E. Ficarra, E. Macii, L. Benini, G. De Micheli, Computer-aided evaluation of protein expression in pathological tissue images, in: Proc. of IEEE International Symposium on Computer-Based Medical Systems, CBMS 2006, 2006, pp [26] S. Suzuki, Y. Dobashi, H. Sakurai, K. Nishikawa, M. Hanawa, A. Ooi, Protein overexpression and gene amplification of epidermal growth factor receptor in nonsmall cell lung carcinomas, Cancer 103 (6) (2006) [27] C. Kersting, N. Tidow, H. Schmidt, et al., Gene dosage PCR and fluorescence in situ hybridization reveal low frequency of egfr amplifications despite protein overexpression in invasive breast carcinoma, Laboratory Investigation 84 (2004) [28] J. Amann, S. Kalyankrishna, P.P. Massion, et al., Aberrant epidermal growth factor receptor signaling and enhanced sensitivity to EGFR inhibitors in lung cancer, Cancer Research 65 (2005) [29] F. Gebhardt, H. Burger, B. Brandt, Modulation of EGFR gene transcription by secondary structures, a polymorphic repetitive sequence and mutationsa link between genetics and epigenetics, Histol Histopathol 15 (2000) [30] J.M. McInerney, M.A. Wilson, K.J. Strand, et al., A strong intronic enhancer element of the EGFR gene is preferentially active in high EGFR expressing breast cancer cells, Journal of Cellular Biochemistry 4 (2001) [31] N. Tidow, A. Boecker, H. Schmidt, et al., Distinct amplification of an untranslated regulatory sequence in the egfr gene contributes to early steps in breast cancer development, Cancer Research 6 (2003) [32] Z. Zheng, G. Bepler, A. Cantor, E.B. Haura, Small tumor size and limited smoking history predicts activated epidermal growth factor receptor in early-stage non-small cell lung cancer, Chest 128 (2005) [33] J. Brabender, K.D. Danenberg, R. Metzger, P.M. Schneider, J.M. Park, D. Salonga, A.H. Hölscher, P.V. Danenberg, Epidermal growth factor receptor and HER2-neu mrna expression in Non-Small Cell Lung Cancer is correlated with Survival, Clinical Cancer Research 7 (2001) [34] Y. Zhou, S. Li, Y.P. Hu, J. Wang, J. Hauser, A.N. Conway, M.A. Vinci, L. Humphrey, E. Zborowska, J.K.V. Willson, M.G. Brattain, Blockade of EGFR and ErbB2 by the novel dual EGFR and ErbB2 Tyrosine Kinase inhibitor GW sensitizes human colon carcinoma GEO cells to apoptosis, Cancer Research 66 (2006) [35] S. Dacic, M. Flanagan, K. Cieply, S. Ramalingam, J. Luketich, C. Belani, S.A. Yousem, Significance of EGFR protein expression and gene amplification in non-small cell lung carcinoma, American Journal of Clinical Pathology 125 (6) (2006)
Bioimaging and Functional Genomics
 Bioimaging and Functional Genomics Elisa Ficarra, EPF Lausanne Giovanni De Micheli, EPF Lausanne Sungroh Yoon, Stanford University Luca Benini, University of Bologna Enrico Macii,, Politecnico di Torino
Bioimaging and Functional Genomics Elisa Ficarra, EPF Lausanne Giovanni De Micheli, EPF Lausanne Sungroh Yoon, Stanford University Luca Benini, University of Bologna Enrico Macii,, Politecnico di Torino
T. R. Golub, D. K. Slonim & Others 1999
 T. R. Golub, D. K. Slonim & Others 1999 Big Picture in 1999 The Need for Cancer Classification Cancer classification very important for advances in cancer treatment. Cancers of Identical grade can have
T. R. Golub, D. K. Slonim & Others 1999 Big Picture in 1999 The Need for Cancer Classification Cancer classification very important for advances in cancer treatment. Cancers of Identical grade can have
Chapter 1. Introduction
 Chapter 1 Introduction 1.1 Motivation and Goals The increasing availability and decreasing cost of high-throughput (HT) technologies coupled with the availability of computational tools and data form a
Chapter 1 Introduction 1.1 Motivation and Goals The increasing availability and decreasing cost of high-throughput (HT) technologies coupled with the availability of computational tools and data form a
1 Introduction. Abstract: Accurate optic disc (OD) segmentation and fovea. Keywords: optic disc segmentation, fovea detection.
 Current Directions in Biomedical Engineering 2017; 3(2): 533 537 Caterina Rust*, Stephanie Häger, Nadine Traulsen and Jan Modersitzki A robust algorithm for optic disc segmentation and fovea detection
Current Directions in Biomedical Engineering 2017; 3(2): 533 537 Caterina Rust*, Stephanie Häger, Nadine Traulsen and Jan Modersitzki A robust algorithm for optic disc segmentation and fovea detection
Gene expression analysis. Roadmap. Microarray technology: how it work Applications: what can we do with it Preprocessing: Classification Clustering
 Gene expression analysis Roadmap Microarray technology: how it work Applications: what can we do with it Preprocessing: Image processing Data normalization Classification Clustering Biclustering 1 Gene
Gene expression analysis Roadmap Microarray technology: how it work Applications: what can we do with it Preprocessing: Image processing Data normalization Classification Clustering Biclustering 1 Gene
EARLY STAGE DIAGNOSIS OF LUNG CANCER USING CT-SCAN IMAGES BASED ON CELLULAR LEARNING AUTOMATE
 EARLY STAGE DIAGNOSIS OF LUNG CANCER USING CT-SCAN IMAGES BASED ON CELLULAR LEARNING AUTOMATE SAKTHI NEELA.P.K Department of M.E (Medical electronics) Sengunthar College of engineering Namakkal, Tamilnadu,
EARLY STAGE DIAGNOSIS OF LUNG CANCER USING CT-SCAN IMAGES BASED ON CELLULAR LEARNING AUTOMATE SAKTHI NEELA.P.K Department of M.E (Medical electronics) Sengunthar College of engineering Namakkal, Tamilnadu,
AUTOMATIC MEASUREMENT ON CT IMAGES FOR PATELLA DISLOCATION DIAGNOSIS
 AUTOMATIC MEASUREMENT ON CT IMAGES FOR PATELLA DISLOCATION DIAGNOSIS Qi Kong 1, Shaoshan Wang 2, Jiushan Yang 2,Ruiqi Zou 3, Yan Huang 1, Yilong Yin 1, Jingliang Peng 1 1 School of Computer Science and
AUTOMATIC MEASUREMENT ON CT IMAGES FOR PATELLA DISLOCATION DIAGNOSIS Qi Kong 1, Shaoshan Wang 2, Jiushan Yang 2,Ruiqi Zou 3, Yan Huang 1, Yilong Yin 1, Jingliang Peng 1 1 School of Computer Science and
Earlier Detection of Cervical Cancer from PAP Smear Images
 , pp.181-186 http://dx.doi.org/10.14257/astl.2017.147.26 Earlier Detection of Cervical Cancer from PAP Smear Images Asmita Ray 1, Indra Kanta Maitra 2 and Debnath Bhattacharyya 1 1 Assistant Professor
, pp.181-186 http://dx.doi.org/10.14257/astl.2017.147.26 Earlier Detection of Cervical Cancer from PAP Smear Images Asmita Ray 1, Indra Kanta Maitra 2 and Debnath Bhattacharyya 1 1 Assistant Professor
Detection and Classification of Lung Cancer Using Artificial Neural Network
 Detection and Classification of Lung Cancer Using Artificial Neural Network Almas Pathan 1, Bairu.K.saptalkar 2 1,2 Department of Electronics and Communication Engineering, SDMCET, Dharwad, India 1 almaseng@yahoo.co.in,
Detection and Classification of Lung Cancer Using Artificial Neural Network Almas Pathan 1, Bairu.K.saptalkar 2 1,2 Department of Electronics and Communication Engineering, SDMCET, Dharwad, India 1 almaseng@yahoo.co.in,
Identification of Tissue Independent Cancer Driver Genes
 Identification of Tissue Independent Cancer Driver Genes Alexandros Manolakos, Idoia Ochoa, Kartik Venkat Supervisor: Olivier Gevaert Abstract Identification of genomic patterns in tumors is an important
Identification of Tissue Independent Cancer Driver Genes Alexandros Manolakos, Idoia Ochoa, Kartik Venkat Supervisor: Olivier Gevaert Abstract Identification of genomic patterns in tumors is an important
Automated Nuclei Segmentation Approach based on Mathematical Morphology for Cancer Scoring in Breast Tissue Images
 Automated Nuclei Segmentation Approach based on Mathematical Morphology for Cancer 915 Automated Nuclei Segmentation Approach based on Mathematical Morphology for Cancer Scoring in Breast Tissue Images
Automated Nuclei Segmentation Approach based on Mathematical Morphology for Cancer 915 Automated Nuclei Segmentation Approach based on Mathematical Morphology for Cancer Scoring in Breast Tissue Images
Early Detection of Lung Cancer
 Early Detection of Lung Cancer Aswathy N Iyer Dept Of Electronics And Communication Engineering Lymie Jose Dept Of Electronics And Communication Engineering Anumol Thomas Dept Of Electronics And Communication
Early Detection of Lung Cancer Aswathy N Iyer Dept Of Electronics And Communication Engineering Lymie Jose Dept Of Electronics And Communication Engineering Anumol Thomas Dept Of Electronics And Communication
Final Project Report Sean Fischer CS229 Introduction
 Introduction The field of pathology is concerned with identifying and understanding the biological causes and effects of disease through the study of morphological, cellular, and molecular features in
Introduction The field of pathology is concerned with identifying and understanding the biological causes and effects of disease through the study of morphological, cellular, and molecular features in
Gene-microRNA network module analysis for ovarian cancer
 Gene-microRNA network module analysis for ovarian cancer Shuqin Zhang School of Mathematical Sciences Fudan University Oct. 4, 2016 Outline Introduction Materials and Methods Results Conclusions Introduction
Gene-microRNA network module analysis for ovarian cancer Shuqin Zhang School of Mathematical Sciences Fudan University Oct. 4, 2016 Outline Introduction Materials and Methods Results Conclusions Introduction
Automated Assessment of Diabetic Retinal Image Quality Based on Blood Vessel Detection
 Y.-H. Wen, A. Bainbridge-Smith, A. B. Morris, Automated Assessment of Diabetic Retinal Image Quality Based on Blood Vessel Detection, Proceedings of Image and Vision Computing New Zealand 2007, pp. 132
Y.-H. Wen, A. Bainbridge-Smith, A. B. Morris, Automated Assessment of Diabetic Retinal Image Quality Based on Blood Vessel Detection, Proceedings of Image and Vision Computing New Zealand 2007, pp. 132
Bayesian Bi-Cluster Change-Point Model for Exploring Functional Brain Dynamics
 Int'l Conf. Bioinformatics and Computational Biology BIOCOMP'18 85 Bayesian Bi-Cluster Change-Point Model for Exploring Functional Brain Dynamics Bing Liu 1*, Xuan Guo 2, and Jing Zhang 1** 1 Department
Int'l Conf. Bioinformatics and Computational Biology BIOCOMP'18 85 Bayesian Bi-Cluster Change-Point Model for Exploring Functional Brain Dynamics Bing Liu 1*, Xuan Guo 2, and Jing Zhang 1** 1 Department
MRI Image Processing Operations for Brain Tumor Detection
 MRI Image Processing Operations for Brain Tumor Detection Prof. M.M. Bulhe 1, Shubhashini Pathak 2, Karan Parekh 3, Abhishek Jha 4 1Assistant Professor, Dept. of Electronics and Telecommunications Engineering,
MRI Image Processing Operations for Brain Tumor Detection Prof. M.M. Bulhe 1, Shubhashini Pathak 2, Karan Parekh 3, Abhishek Jha 4 1Assistant Professor, Dept. of Electronics and Telecommunications Engineering,
Breast cancer. Risk factors you cannot change include: Treatment Plan Selection. Inferring Transcriptional Module from Breast Cancer Profile Data
 Breast cancer Inferring Transcriptional Module from Breast Cancer Profile Data Breast Cancer and Targeted Therapy Microarray Profile Data Inferring Transcriptional Module Methods CSC 177 Data Warehousing
Breast cancer Inferring Transcriptional Module from Breast Cancer Profile Data Breast Cancer and Targeted Therapy Microarray Profile Data Inferring Transcriptional Module Methods CSC 177 Data Warehousing
LUNG NODULE SEGMENTATION IN COMPUTED TOMOGRAPHY IMAGE. Hemahashiny, Ketheesan Department of Physical Science, Vavuniya Campus
 LUNG NODULE SEGMENTATION IN COMPUTED TOMOGRAPHY IMAGE Hemahashiny, Ketheesan Department of Physical Science, Vavuniya Campus tketheesan@vau.jfn.ac.lk ABSTRACT: The key process to detect the Lung cancer
LUNG NODULE SEGMENTATION IN COMPUTED TOMOGRAPHY IMAGE Hemahashiny, Ketheesan Department of Physical Science, Vavuniya Campus tketheesan@vau.jfn.ac.lk ABSTRACT: The key process to detect the Lung cancer
Keywords: Leukaemia, Image Segmentation, Clustering algorithms, White Blood Cells (WBC), Microscopic images.
 Volume 6, Issue 10, October 2016 ISSN: 2277 128X International Journal of Advanced Research in Computer Science and Software Engineering Research Paper Available online at: www.ijarcsse.com A Study on
Volume 6, Issue 10, October 2016 ISSN: 2277 128X International Journal of Advanced Research in Computer Science and Software Engineering Research Paper Available online at: www.ijarcsse.com A Study on
ANALYSIS AND DETECTION OF BRAIN TUMOUR USING IMAGE PROCESSING TECHNIQUES
 ANALYSIS AND DETECTION OF BRAIN TUMOUR USING IMAGE PROCESSING TECHNIQUES P.V.Rohini 1, Dr.M.Pushparani 2 1 M.Phil Scholar, Department of Computer Science, Mother Teresa women s university, (India) 2 Professor
ANALYSIS AND DETECTION OF BRAIN TUMOUR USING IMAGE PROCESSING TECHNIQUES P.V.Rohini 1, Dr.M.Pushparani 2 1 M.Phil Scholar, Department of Computer Science, Mother Teresa women s university, (India) 2 Professor
Diagnosis of Liver Tumor Using 3D Segmentation Method for Selective Internal Radiation Therapy
 Diagnosis of Liver Tumor Using 3D Segmentation Method for Selective Internal Radiation Therapy K. Geetha & S. Poonguzhali Department of Electronics and Communication Engineering, Campus of CEG Anna University,
Diagnosis of Liver Tumor Using 3D Segmentation Method for Selective Internal Radiation Therapy K. Geetha & S. Poonguzhali Department of Electronics and Communication Engineering, Campus of CEG Anna University,
Lung Region Segmentation using Artificial Neural Network Hopfield Model for Cancer Diagnosis in Thorax CT Images
 Automation, Control and Intelligent Systems 2015; 3(2): 19-25 Published online March 20, 2015 (http://www.sciencepublishinggroup.com/j/acis) doi: 10.11648/j.acis.20150302.12 ISSN: 2328-5583 (Print); ISSN:
Automation, Control and Intelligent Systems 2015; 3(2): 19-25 Published online March 20, 2015 (http://www.sciencepublishinggroup.com/j/acis) doi: 10.11648/j.acis.20150302.12 ISSN: 2328-5583 (Print); ISSN:
MULTIPLE LINEAR REGRESSION 24.1 INTRODUCTION AND OBJECTIVES OBJECTIVES
 24 MULTIPLE LINEAR REGRESSION 24.1 INTRODUCTION AND OBJECTIVES In the previous chapter, simple linear regression was used when you have one independent variable and one dependent variable. This chapter
24 MULTIPLE LINEAR REGRESSION 24.1 INTRODUCTION AND OBJECTIVES In the previous chapter, simple linear regression was used when you have one independent variable and one dependent variable. This chapter
AN ALGORITHM FOR EARLY BREAST CANCER DETECTION IN MAMMOGRAMS
 AN ALGORITHM FOR EARLY BREAST CANCER DETECTION IN MAMMOGRAMS Isaac N. Bankman', William A. Christens-Barryl, Irving N. Weinberg2, Dong W. Kim3, Ralph D. Semmell, and William R. Brody2 The Johns Hopkins
AN ALGORITHM FOR EARLY BREAST CANCER DETECTION IN MAMMOGRAMS Isaac N. Bankman', William A. Christens-Barryl, Irving N. Weinberg2, Dong W. Kim3, Ralph D. Semmell, and William R. Brody2 The Johns Hopkins
COMMUNICATION ENGINEERING & TECHNOLOGY (IJECET) DETECTION OF ACUTE LEUKEMIA USING WHITE BLOOD CELLS SEGMENTATION BASED ON BLOOD SAMPLES
 International INTERNATIONAL Journal of Electronics JOURNAL and Communication OF ELECTRONICS Engineering & Technology AND (IJECET), COMMUNICATION ENGINEERING & TECHNOLOGY (IJECET) ISSN 0976 6464(Print)
International INTERNATIONAL Journal of Electronics JOURNAL and Communication OF ELECTRONICS Engineering & Technology AND (IJECET), COMMUNICATION ENGINEERING & TECHNOLOGY (IJECET) ISSN 0976 6464(Print)
Efficacy of the Extended Principal Orthogonal Decomposition Method on DNA Microarray Data in Cancer Detection
 202 4th International onference on Bioinformatics and Biomedical Technology IPBEE vol.29 (202) (202) IASIT Press, Singapore Efficacy of the Extended Principal Orthogonal Decomposition on DA Microarray
202 4th International onference on Bioinformatics and Biomedical Technology IPBEE vol.29 (202) (202) IASIT Press, Singapore Efficacy of the Extended Principal Orthogonal Decomposition on DA Microarray
Evaluating Classifiers for Disease Gene Discovery
 Evaluating Classifiers for Disease Gene Discovery Kino Coursey Lon Turnbull khc0021@unt.edu lt0013@unt.edu Abstract Identification of genes involved in human hereditary disease is an important bioinfomatics
Evaluating Classifiers for Disease Gene Discovery Kino Coursey Lon Turnbull khc0021@unt.edu lt0013@unt.edu Abstract Identification of genes involved in human hereditary disease is an important bioinfomatics
Segmentation of Tumor Region from Brain Mri Images Using Fuzzy C-Means Clustering And Seeded Region Growing
 IOSR Journal of Computer Engineering (IOSR-JCE) e-issn: 2278-0661,p-ISSN: 2278-8727, Volume 18, Issue 5, Ver. I (Sept - Oct. 2016), PP 20-24 www.iosrjournals.org Segmentation of Tumor Region from Brain
IOSR Journal of Computer Engineering (IOSR-JCE) e-issn: 2278-0661,p-ISSN: 2278-8727, Volume 18, Issue 5, Ver. I (Sept - Oct. 2016), PP 20-24 www.iosrjournals.org Segmentation of Tumor Region from Brain
Implementation of Automatic Retina Exudates Segmentation Algorithm for Early Detection with Low Computational Time
 www.ijecs.in International Journal Of Engineering And Computer Science ISSN: 2319-7242 Volume 5 Issue 10 Oct. 2016, Page No. 18584-18588 Implementation of Automatic Retina Exudates Segmentation Algorithm
www.ijecs.in International Journal Of Engineering And Computer Science ISSN: 2319-7242 Volume 5 Issue 10 Oct. 2016, Page No. 18584-18588 Implementation of Automatic Retina Exudates Segmentation Algorithm
Extraction of Blood Vessels and Recognition of Bifurcation Points in Retinal Fundus Image
 International Journal of Research Studies in Science, Engineering and Technology Volume 1, Issue 5, August 2014, PP 1-7 ISSN 2349-4751 (Print) & ISSN 2349-476X (Online) Extraction of Blood Vessels and
International Journal of Research Studies in Science, Engineering and Technology Volume 1, Issue 5, August 2014, PP 1-7 ISSN 2349-4751 (Print) & ISSN 2349-476X (Online) Extraction of Blood Vessels and
Abstract. Optimization strategy of Copy Number Variant calling using Multiplicom solutions APPLICATION NOTE. Introduction
 Optimization strategy of Copy Number Variant calling using Multiplicom solutions Michael Vyverman, PhD; Laura Standaert, PhD and Wouter Bossuyt, PhD Abstract Copy number variations (CNVs) represent a significant
Optimization strategy of Copy Number Variant calling using Multiplicom solutions Michael Vyverman, PhD; Laura Standaert, PhD and Wouter Bossuyt, PhD Abstract Copy number variations (CNVs) represent a significant
Segmentation of Color Fundus Images of the Human Retina: Detection of the Optic Disc and the Vascular Tree Using Morphological Techniques
 Segmentation of Color Fundus Images of the Human Retina: Detection of the Optic Disc and the Vascular Tree Using Morphological Techniques Thomas Walter and Jean-Claude Klein Centre de Morphologie Mathématique,
Segmentation of Color Fundus Images of the Human Retina: Detection of the Optic Disc and the Vascular Tree Using Morphological Techniques Thomas Walter and Jean-Claude Klein Centre de Morphologie Mathématique,
Noise-Robust Speech Recognition Technologies in Mobile Environments
 Noise-Robust Speech Recognition echnologies in Mobile Environments Mobile environments are highly influenced by ambient noise, which may cause a significant deterioration of speech recognition performance.
Noise-Robust Speech Recognition echnologies in Mobile Environments Mobile environments are highly influenced by ambient noise, which may cause a significant deterioration of speech recognition performance.
Comparative Study of K-means, Gaussian Mixture Model, Fuzzy C-means algorithms for Brain Tumor Segmentation
 Comparative Study of K-means, Gaussian Mixture Model, Fuzzy C-means algorithms for Brain Tumor Segmentation U. Baid 1, S. Talbar 2 and S. Talbar 1 1 Department of E&TC Engineering, Shri Guru Gobind Singhji
Comparative Study of K-means, Gaussian Mixture Model, Fuzzy C-means algorithms for Brain Tumor Segmentation U. Baid 1, S. Talbar 2 and S. Talbar 1 1 Department of E&TC Engineering, Shri Guru Gobind Singhji
Copyright 2008 Society of Photo Optical Instrumentation Engineers. This paper was published in Proceedings of SPIE, vol. 6915, Medical Imaging 2008:
 Copyright 2008 Society of Photo Optical Instrumentation Engineers. This paper was published in Proceedings of SPIE, vol. 6915, Medical Imaging 2008: Computer Aided Diagnosis and is made available as an
Copyright 2008 Society of Photo Optical Instrumentation Engineers. This paper was published in Proceedings of SPIE, vol. 6915, Medical Imaging 2008: Computer Aided Diagnosis and is made available as an
Comparison of volume estimation methods for pancreatic islet cells
 Comparison of volume estimation methods for pancreatic islet cells Jiří Dvořák a,b, Jan Švihlíkb,c, David Habart d, and Jan Kybic b a Department of Probability and Mathematical Statistics, Faculty of Mathematics
Comparison of volume estimation methods for pancreatic islet cells Jiří Dvořák a,b, Jan Švihlíkb,c, David Habart d, and Jan Kybic b a Department of Probability and Mathematical Statistics, Faculty of Mathematics
A Comparison of Collaborative Filtering Methods for Medication Reconciliation
 A Comparison of Collaborative Filtering Methods for Medication Reconciliation Huanian Zheng, Rema Padman, Daniel B. Neill The H. John Heinz III College, Carnegie Mellon University, Pittsburgh, PA, 15213,
A Comparison of Collaborative Filtering Methods for Medication Reconciliation Huanian Zheng, Rema Padman, Daniel B. Neill The H. John Heinz III College, Carnegie Mellon University, Pittsburgh, PA, 15213,
Clustering mass spectrometry data using order statistics
 Proteomics 2003, 3, 1687 1691 DOI 10.1002/pmic.200300517 1687 Douglas J. Slotta 1 Lenwood S. Heath 1 Naren Ramakrishnan 1 Rich Helm 2 Malcolm Potts 3 1 Department of Computer Science 2 Department of Wood
Proteomics 2003, 3, 1687 1691 DOI 10.1002/pmic.200300517 1687 Douglas J. Slotta 1 Lenwood S. Heath 1 Naren Ramakrishnan 1 Rich Helm 2 Malcolm Potts 3 1 Department of Computer Science 2 Department of Wood
Proceedings of the UGC Sponsored National Conference on Advanced Networking and Applications, 27 th March 2015
 Brain Tumor Detection and Identification Using K-Means Clustering Technique Malathi R Department of Computer Science, SAAS College, Ramanathapuram, Email: malapraba@gmail.com Dr. Nadirabanu Kamal A R Department
Brain Tumor Detection and Identification Using K-Means Clustering Technique Malathi R Department of Computer Science, SAAS College, Ramanathapuram, Email: malapraba@gmail.com Dr. Nadirabanu Kamal A R Department
Cancer Cells Detection using OTSU Threshold Algorithm
 Cancer Cells Detection using OTSU Threshold Algorithm Nalluri Sunny 1 Velagapudi Ramakrishna Siddhartha Engineering College Mithinti Srikanth 2 Velagapudi Ramakrishna Siddhartha Engineering College Kodali
Cancer Cells Detection using OTSU Threshold Algorithm Nalluri Sunny 1 Velagapudi Ramakrishna Siddhartha Engineering College Mithinti Srikanth 2 Velagapudi Ramakrishna Siddhartha Engineering College Kodali
Comments on Significance of candidate cancer genes as assessed by the CaMP score by Parmigiani et al.
 Comments on Significance of candidate cancer genes as assessed by the CaMP score by Parmigiani et al. Holger Höfling Gad Getz Robert Tibshirani June 26, 2007 1 Introduction Identifying genes that are involved
Comments on Significance of candidate cancer genes as assessed by the CaMP score by Parmigiani et al. Holger Höfling Gad Getz Robert Tibshirani June 26, 2007 1 Introduction Identifying genes that are involved
Outlier Analysis. Lijun Zhang
 Outlier Analysis Lijun Zhang zlj@nju.edu.cn http://cs.nju.edu.cn/zlj Outline Introduction Extreme Value Analysis Probabilistic Models Clustering for Outlier Detection Distance-Based Outlier Detection Density-Based
Outlier Analysis Lijun Zhang zlj@nju.edu.cn http://cs.nju.edu.cn/zlj Outline Introduction Extreme Value Analysis Probabilistic Models Clustering for Outlier Detection Distance-Based Outlier Detection Density-Based
A ROBUST MORPHOLOGICAL ANALYSIS OF NORMAL AND ABNORMAL LEUKEMIC CELLS POPULATIONS IN ACUTE LYMPHOBLASTIC LEUKEMIA
 A ROBUST MORPHOLOGICAL ANALYSIS OF NORMAL AND ABNORMAL LEUKEMIC CELLS POPULATIONS IN ACUTE LYMPHOBLASTIC LEUKEMIA 1 TRUPTI A. KULKARNI, 2 DILIP S. BHOSALE 1 E & TC Department JSPM s, Bhivarabai Sawant
A ROBUST MORPHOLOGICAL ANALYSIS OF NORMAL AND ABNORMAL LEUKEMIC CELLS POPULATIONS IN ACUTE LYMPHOBLASTIC LEUKEMIA 1 TRUPTI A. KULKARNI, 2 DILIP S. BHOSALE 1 E & TC Department JSPM s, Bhivarabai Sawant
Unsupervised MRI Brain Tumor Detection Techniques with Morphological Operations
 Unsupervised MRI Brain Tumor Detection Techniques with Morphological Operations Ritu Verma, Sujeet Tiwari, Naazish Rahim Abstract Tumor is a deformity in human body cells which, if not detected and treated,
Unsupervised MRI Brain Tumor Detection Techniques with Morphological Operations Ritu Verma, Sujeet Tiwari, Naazish Rahim Abstract Tumor is a deformity in human body cells which, if not detected and treated,
A Biclustering Based Classification Framework for Cancer Diagnosis and Prognosis
 A Biclustering Based Classification Framework for Cancer Diagnosis and Prognosis Baljeet Malhotra and Guohui Lin Department of Computing Science, University of Alberta, Edmonton, Alberta, Canada T6G 2E8
A Biclustering Based Classification Framework for Cancer Diagnosis and Prognosis Baljeet Malhotra and Guohui Lin Department of Computing Science, University of Alberta, Edmonton, Alberta, Canada T6G 2E8
Measuring Focused Attention Using Fixation Inner-Density
 Measuring Focused Attention Using Fixation Inner-Density Wen Liu, Mina Shojaeizadeh, Soussan Djamasbi, Andrew C. Trapp User Experience & Decision Making Research Laboratory, Worcester Polytechnic Institute
Measuring Focused Attention Using Fixation Inner-Density Wen Liu, Mina Shojaeizadeh, Soussan Djamasbi, Andrew C. Trapp User Experience & Decision Making Research Laboratory, Worcester Polytechnic Institute
EXTRACTION OF RETINAL BLOOD VESSELS USING IMAGE PROCESSING TECHNIQUES
 EXTRACTION OF RETINAL BLOOD VESSELS USING IMAGE PROCESSING TECHNIQUES T.HARI BABU 1, Y.RATNA KUMAR 2 1 (PG Scholar, Dept. of Electronics and Communication Engineering, College of Engineering(A), Andhra
EXTRACTION OF RETINAL BLOOD VESSELS USING IMAGE PROCESSING TECHNIQUES T.HARI BABU 1, Y.RATNA KUMAR 2 1 (PG Scholar, Dept. of Electronics and Communication Engineering, College of Engineering(A), Andhra
Using Perceptual Grouping for Object Group Selection
 Using Perceptual Grouping for Object Group Selection Hoda Dehmeshki Department of Computer Science and Engineering, York University, 4700 Keele Street Toronto, Ontario, M3J 1P3 Canada hoda@cs.yorku.ca
Using Perceptual Grouping for Object Group Selection Hoda Dehmeshki Department of Computer Science and Engineering, York University, 4700 Keele Street Toronto, Ontario, M3J 1P3 Canada hoda@cs.yorku.ca
LUNG NODULE DETECTION SYSTEM
 LUNG NODULE DETECTION SYSTEM Kalim Bhandare and Rupali Nikhare Department of Computer Engineering Pillai Institute of Technology, New Panvel, Navi Mumbai, India ABSTRACT: The Existing approach consist
LUNG NODULE DETECTION SYSTEM Kalim Bhandare and Rupali Nikhare Department of Computer Engineering Pillai Institute of Technology, New Panvel, Navi Mumbai, India ABSTRACT: The Existing approach consist
Mammogram Analysis: Tumor Classification
 Mammogram Analysis: Tumor Classification Term Project Report Geethapriya Raghavan geeragh@mail.utexas.edu EE 381K - Multidimensional Digital Signal Processing Spring 2005 Abstract Breast cancer is the
Mammogram Analysis: Tumor Classification Term Project Report Geethapriya Raghavan geeragh@mail.utexas.edu EE 381K - Multidimensional Digital Signal Processing Spring 2005 Abstract Breast cancer is the
International Journal of Digital Application & Contemporary research Website: (Volume 1, Issue 1, August 2012) IJDACR.
 Segmentation of Brain MRI Images for Tumor extraction by combining C-means clustering and Watershed algorithm with Genetic Algorithm Kailash Sinha 1 1 Department of Electronics & Telecommunication Engineering,
Segmentation of Brain MRI Images for Tumor extraction by combining C-means clustering and Watershed algorithm with Genetic Algorithm Kailash Sinha 1 1 Department of Electronics & Telecommunication Engineering,
Predicting Breast Cancer Survivability Rates
 Predicting Breast Cancer Survivability Rates For data collected from Saudi Arabia Registries Ghofran Othoum 1 and Wadee Al-Halabi 2 1 Computer Science, Effat University, Jeddah, Saudi Arabia 2 Computer
Predicting Breast Cancer Survivability Rates For data collected from Saudi Arabia Registries Ghofran Othoum 1 and Wadee Al-Halabi 2 1 Computer Science, Effat University, Jeddah, Saudi Arabia 2 Computer
Data Mining in Bioinformatics Day 7: Clustering in Bioinformatics
 Data Mining in Bioinformatics Day 7: Clustering in Bioinformatics Karsten Borgwardt February 21 to March 4, 2011 Machine Learning & Computational Biology Research Group MPIs Tübingen Karsten Borgwardt:
Data Mining in Bioinformatics Day 7: Clustering in Bioinformatics Karsten Borgwardt February 21 to March 4, 2011 Machine Learning & Computational Biology Research Group MPIs Tübingen Karsten Borgwardt:
Automatic Definition of Planning Target Volume in Computer-Assisted Radiotherapy
 Automatic Definition of Planning Target Volume in Computer-Assisted Radiotherapy Angelo Zizzari Department of Cybernetics, School of Systems Engineering The University of Reading, Whiteknights, PO Box
Automatic Definition of Planning Target Volume in Computer-Assisted Radiotherapy Angelo Zizzari Department of Cybernetics, School of Systems Engineering The University of Reading, Whiteknights, PO Box
A Hierarchical Artificial Neural Network Model for Giemsa-Stained Human Chromosome Classification
 A Hierarchical Artificial Neural Network Model for Giemsa-Stained Human Chromosome Classification JONGMAN CHO 1 1 Department of Biomedical Engineering, Inje University, Gimhae, 621-749, KOREA minerva@ieeeorg
A Hierarchical Artificial Neural Network Model for Giemsa-Stained Human Chromosome Classification JONGMAN CHO 1 1 Department of Biomedical Engineering, Inje University, Gimhae, 621-749, KOREA minerva@ieeeorg
Machine Learning to Inform Breast Cancer Post-Recovery Surveillance
 Machine Learning to Inform Breast Cancer Post-Recovery Surveillance Final Project Report CS 229 Autumn 2017 Category: Life Sciences Maxwell Allman (mallman) Lin Fan (linfan) Jamie Kang (kangjh) 1 Introduction
Machine Learning to Inform Breast Cancer Post-Recovery Surveillance Final Project Report CS 229 Autumn 2017 Category: Life Sciences Maxwell Allman (mallman) Lin Fan (linfan) Jamie Kang (kangjh) 1 Introduction
Research Methods in Forest Sciences: Learning Diary. Yoko Lu December Research process
 Research Methods in Forest Sciences: Learning Diary Yoko Lu 285122 9 December 2016 1. Research process It is important to pursue and apply knowledge and understand the world under both natural and social
Research Methods in Forest Sciences: Learning Diary Yoko Lu 285122 9 December 2016 1. Research process It is important to pursue and apply knowledge and understand the world under both natural and social
Detection of aneuploidy in a single cell using the Ion ReproSeq PGS View Kit
 APPLICATION NOTE Ion PGM System Detection of aneuploidy in a single cell using the Ion ReproSeq PGS View Kit Key findings The Ion PGM System, in concert with the Ion ReproSeq PGS View Kit and Ion Reporter
APPLICATION NOTE Ion PGM System Detection of aneuploidy in a single cell using the Ion ReproSeq PGS View Kit Key findings The Ion PGM System, in concert with the Ion ReproSeq PGS View Kit and Ion Reporter
Bootstrapped Integrative Hypothesis Test, COPD-Lung Cancer Differentiation, and Joint mirnas Biomarkers
 Bootstrapped Integrative Hypothesis Test, COPD-Lung Cancer Differentiation, and Joint mirnas Biomarkers Kai-Ming Jiang 1,2, Bao-Liang Lu 1,2, and Lei Xu 1,2,3(&) 1 Department of Computer Science and Engineering,
Bootstrapped Integrative Hypothesis Test, COPD-Lung Cancer Differentiation, and Joint mirnas Biomarkers Kai-Ming Jiang 1,2, Bao-Liang Lu 1,2, and Lei Xu 1,2,3(&) 1 Department of Computer Science and Engineering,
Automated Brain Tumor Segmentation Using Region Growing Algorithm by Extracting Feature
 Automated Brain Tumor Segmentation Using Region Growing Algorithm by Extracting Feature Shraddha P. Dhumal 1, Ashwini S Gaikwad 2 1 Shraddha P. Dhumal 2 Ashwini S. Gaikwad ABSTRACT In this paper, we propose
Automated Brain Tumor Segmentation Using Region Growing Algorithm by Extracting Feature Shraddha P. Dhumal 1, Ashwini S Gaikwad 2 1 Shraddha P. Dhumal 2 Ashwini S. Gaikwad ABSTRACT In this paper, we propose
Tumor cut segmentation for Blemish Cells Detection in Human Brain Based on Cellular Automata
 Tumor cut segmentation for Blemish Cells Detection in Human Brain Based on Cellular Automata D.Mohanapriya 1 Department of Electronics and Communication Engineering, EBET Group of Institutions, Kangayam,
Tumor cut segmentation for Blemish Cells Detection in Human Brain Based on Cellular Automata D.Mohanapriya 1 Department of Electronics and Communication Engineering, EBET Group of Institutions, Kangayam,
Introduction to Statistical Data Analysis I
 Introduction to Statistical Data Analysis I JULY 2011 Afsaneh Yazdani Preface What is Statistics? Preface What is Statistics? Science of: designing studies or experiments, collecting data Summarizing/modeling/analyzing
Introduction to Statistical Data Analysis I JULY 2011 Afsaneh Yazdani Preface What is Statistics? Preface What is Statistics? Science of: designing studies or experiments, collecting data Summarizing/modeling/analyzing
Computer-based 3d Puzzle Solving For Pre-operative Planning Of Articular Fracture Reductions In The Ankle, Knee, And Hip
 Computer-based 3d Puzzle Solving For Pre-operative Planning Of Articular Fracture Reductions In The Ankle, Knee, And Hip Andrew M. Kern, MS, Donald Anderson. University of Iowa, Iowa City, IA, USA. Disclosures:
Computer-based 3d Puzzle Solving For Pre-operative Planning Of Articular Fracture Reductions In The Ankle, Knee, And Hip Andrew M. Kern, MS, Donald Anderson. University of Iowa, Iowa City, IA, USA. Disclosures:
Brain Tumor Detection using Watershed Algorithm
 Brain Tumor Detection using Watershed Algorithm Dawood Dilber 1, Jasleen 2 P.G. Student, Department of Electronics and Communication Engineering, Amity University, Noida, U.P, India 1 P.G. Student, Department
Brain Tumor Detection using Watershed Algorithm Dawood Dilber 1, Jasleen 2 P.G. Student, Department of Electronics and Communication Engineering, Amity University, Noida, U.P, India 1 P.G. Student, Department
Blood Microscopic Image Segmentation & Acute Leukemia Detection Tejashri G. Patil *, V. B. Raskar E & TC Department & S.P. Pune, Maharashtra, India
 International Journal of Emerging Research in Management &Technology Research Article September 215 Blood Microscopic Image Segmentation & Acute Leukemia Detection Tejashri G. Patil *, V. B. Raskar E &
International Journal of Emerging Research in Management &Technology Research Article September 215 Blood Microscopic Image Segmentation & Acute Leukemia Detection Tejashri G. Patil *, V. B. Raskar E &
Educator Navigation Guide
 Decoding Breast Cancer Virtual Lab Educator Navigation Guide Decoding Cancer Nav Guide 2 Introduction In this virtual lab, students test tissue samples from different patients with breast cancer in order
Decoding Breast Cancer Virtual Lab Educator Navigation Guide Decoding Cancer Nav Guide 2 Introduction In this virtual lab, students test tissue samples from different patients with breast cancer in order
Utilizing machine learning techniques to rapidly identify. MUC2 expression in colon cancer tissues
 1 Utilizing machine learning techniques to rapidly identify MUC2 expression in colon cancer tissues Thesis by Preethi Periyakoil In partial fulfillment of the requirements for the degree of Bachelor of
1 Utilizing machine learning techniques to rapidly identify MUC2 expression in colon cancer tissues Thesis by Preethi Periyakoil In partial fulfillment of the requirements for the degree of Bachelor of
Type II Fuzzy Possibilistic C-Mean Clustering
 IFSA-EUSFLAT Type II Fuzzy Possibilistic C-Mean Clustering M.H. Fazel Zarandi, M. Zarinbal, I.B. Turksen, Department of Industrial Engineering, Amirkabir University of Technology, P.O. Box -, Tehran, Iran
IFSA-EUSFLAT Type II Fuzzy Possibilistic C-Mean Clustering M.H. Fazel Zarandi, M. Zarinbal, I.B. Turksen, Department of Industrial Engineering, Amirkabir University of Technology, P.O. Box -, Tehran, Iran
Automated Volumetric Cardiac Ultrasound Analysis
 Whitepaper Automated Volumetric Cardiac Ultrasound Analysis ACUSON SC2000 Volume Imaging Ultrasound System Bogdan Georgescu, Ph.D. Siemens Corporate Research Princeton, New Jersey USA Answers for life.
Whitepaper Automated Volumetric Cardiac Ultrasound Analysis ACUSON SC2000 Volume Imaging Ultrasound System Bogdan Georgescu, Ph.D. Siemens Corporate Research Princeton, New Jersey USA Answers for life.
Reliability of Ordination Analyses
 Reliability of Ordination Analyses Objectives: Discuss Reliability Define Consistency and Accuracy Discuss Validation Methods Opening Thoughts Inference Space: What is it? Inference space can be defined
Reliability of Ordination Analyses Objectives: Discuss Reliability Define Consistency and Accuracy Discuss Validation Methods Opening Thoughts Inference Space: What is it? Inference space can be defined
Primary Level Classification of Brain Tumor using PCA and PNN
 Primary Level Classification of Brain Tumor using PCA and PNN Dr. Mrs. K.V.Kulhalli Department of Information Technology, D.Y.Patil Coll. of Engg. And Tech. Kolhapur,Maharashtra,India kvkulhalli@gmail.com
Primary Level Classification of Brain Tumor using PCA and PNN Dr. Mrs. K.V.Kulhalli Department of Information Technology, D.Y.Patil Coll. of Engg. And Tech. Kolhapur,Maharashtra,India kvkulhalli@gmail.com
Classification of cancer profiles. ABDBM Ron Shamir
 Classification of cancer profiles 1 Background: Cancer Classification Cancer classification is central to cancer treatment; Traditional cancer classification methods: location; morphology, cytogenesis;
Classification of cancer profiles 1 Background: Cancer Classification Cancer classification is central to cancer treatment; Traditional cancer classification methods: location; morphology, cytogenesis;
Lung Tumour Detection by Applying Watershed Method
 International Journal of Computational Intelligence Research ISSN 0973-1873 Volume 13, Number 5 (2017), pp. 955-964 Research India Publications http://www.ripublication.com Lung Tumour Detection by Applying
International Journal of Computational Intelligence Research ISSN 0973-1873 Volume 13, Number 5 (2017), pp. 955-964 Research India Publications http://www.ripublication.com Lung Tumour Detection by Applying
EXTRACT THE BREAST CANCER IN MAMMOGRAM IMAGES
 International Journal of Civil Engineering and Technology (IJCIET) Volume 10, Issue 02, February 2019, pp. 96-105, Article ID: IJCIET_10_02_012 Available online at http://www.iaeme.com/ijciet/issues.asp?jtype=ijciet&vtype=10&itype=02
International Journal of Civil Engineering and Technology (IJCIET) Volume 10, Issue 02, February 2019, pp. 96-105, Article ID: IJCIET_10_02_012 Available online at http://www.iaeme.com/ijciet/issues.asp?jtype=ijciet&vtype=10&itype=02
Unit 1 Exploring and Understanding Data
 Unit 1 Exploring and Understanding Data Area Principle Bar Chart Boxplot Conditional Distribution Dotplot Empirical Rule Five Number Summary Frequency Distribution Frequency Polygon Histogram Interquartile
Unit 1 Exploring and Understanding Data Area Principle Bar Chart Boxplot Conditional Distribution Dotplot Empirical Rule Five Number Summary Frequency Distribution Frequency Polygon Histogram Interquartile
7/6/2015. Cancer Related Deaths: United States. Management of NSCLC TODAY. Emerging mutations as predictive biomarkers in lung cancer: Overview
 Emerging mutations as predictive biomarkers in lung cancer: Overview Kirtee Raparia, MD Assistant Professor of Pathology Cancer Related Deaths: United States Men Lung and bronchus 28% Prostate 10% Colon
Emerging mutations as predictive biomarkers in lung cancer: Overview Kirtee Raparia, MD Assistant Professor of Pathology Cancer Related Deaths: United States Men Lung and bronchus 28% Prostate 10% Colon
Cancer Informatics Lecture
 Cancer Informatics Lecture Mayo-UIUC Computational Genomics Course June 22, 2018 Krishna Rani Kalari Ph.D. Associate Professor 2017 MFMER 3702274-1 Outline The Cancer Genome Atlas (TCGA) Genomic Data Commons
Cancer Informatics Lecture Mayo-UIUC Computational Genomics Course June 22, 2018 Krishna Rani Kalari Ph.D. Associate Professor 2017 MFMER 3702274-1 Outline The Cancer Genome Atlas (TCGA) Genomic Data Commons
HER2 CISH pharmdx TM Kit Interpretation Guide Breast Cancer
 P A T H O L O G Y HER2 CISH pharmdx TM Kit Interpretation Guide Breast Cancer FROM CERTAINTY COMES TRUST For in vitro diagnostic use HER2 CISH pharmdx Kit HER2 CISH pharmdx Kit is intended for dual-color
P A T H O L O G Y HER2 CISH pharmdx TM Kit Interpretation Guide Breast Cancer FROM CERTAINTY COMES TRUST For in vitro diagnostic use HER2 CISH pharmdx Kit HER2 CISH pharmdx Kit is intended for dual-color
Machine Learning! Robert Stengel! Robotics and Intelligent Systems MAE 345,! Princeton University, 2017
 Machine Learning! Robert Stengel! Robotics and Intelligent Systems MAE 345,! Princeton University, 2017 A.K.A. Artificial Intelligence Unsupervised learning! Cluster analysis Patterns, Clumps, and Joining
Machine Learning! Robert Stengel! Robotics and Intelligent Systems MAE 345,! Princeton University, 2017 A.K.A. Artificial Intelligence Unsupervised learning! Cluster analysis Patterns, Clumps, and Joining
Predicting Breast Cancer Survival Using Treatment and Patient Factors
 Predicting Breast Cancer Survival Using Treatment and Patient Factors William Chen wchen808@stanford.edu Henry Wang hwang9@stanford.edu 1. Introduction Breast cancer is the leading type of cancer in women
Predicting Breast Cancer Survival Using Treatment and Patient Factors William Chen wchen808@stanford.edu Henry Wang hwang9@stanford.edu 1. Introduction Breast cancer is the leading type of cancer in women
Enhanced Detection of Lung Cancer using Hybrid Method of Image Segmentation
 Enhanced Detection of Lung Cancer using Hybrid Method of Image Segmentation L Uma Maheshwari Department of ECE, Stanley College of Engineering and Technology for Women, Hyderabad - 500001, India. Udayini
Enhanced Detection of Lung Cancer using Hybrid Method of Image Segmentation L Uma Maheshwari Department of ECE, Stanley College of Engineering and Technology for Women, Hyderabad - 500001, India. Udayini
Mammogram Analysis: Tumor Classification
 Mammogram Analysis: Tumor Classification Literature Survey Report Geethapriya Raghavan geeragh@mail.utexas.edu EE 381K - Multidimensional Digital Signal Processing Spring 2005 Abstract Breast cancer is
Mammogram Analysis: Tumor Classification Literature Survey Report Geethapriya Raghavan geeragh@mail.utexas.edu EE 381K - Multidimensional Digital Signal Processing Spring 2005 Abstract Breast cancer is
Computerized Detection System for Acute Myelogenous Leukemia in Blood Microscopic Images
 Computerized Detection System for Acute Myelogenous Leukemia in Blood Microscopic Images Yogesh Ambadas Gajul 1, Rupali Shelke 2 P.G. Student, Department of Electronics Engineering, Walchand Institute
Computerized Detection System for Acute Myelogenous Leukemia in Blood Microscopic Images Yogesh Ambadas Gajul 1, Rupali Shelke 2 P.G. Student, Department of Electronics Engineering, Walchand Institute
Grading of Vertebral Rotation
 Chapter 5 Grading of Vertebral Rotation The measurement of vertebral rotation has become increasingly prominent in the study of scoliosis. Apical vertebral deformity demonstrates significance in both preoperative
Chapter 5 Grading of Vertebral Rotation The measurement of vertebral rotation has become increasingly prominent in the study of scoliosis. Apical vertebral deformity demonstrates significance in both preoperative
Automatic Ascending Aorta Detection in CTA Datasets
 Automatic Ascending Aorta Detection in CTA Datasets Stefan C. Saur 1, Caroline Kühnel 2, Tobias Boskamp 2, Gábor Székely 1, Philippe Cattin 1,3 1 Computer Vision Laboratory, ETH Zurich, 8092 Zurich, Switzerland
Automatic Ascending Aorta Detection in CTA Datasets Stefan C. Saur 1, Caroline Kühnel 2, Tobias Boskamp 2, Gábor Székely 1, Philippe Cattin 1,3 1 Computer Vision Laboratory, ETH Zurich, 8092 Zurich, Switzerland
Brain Tumor Segmentation: A Review Dharna*, Priyanshu Tripathi** *M.tech Scholar, HCE, Sonipat ** Assistant Professor, HCE, Sonipat
 International Journal of scientific research and management (IJSRM) Volume 4 Issue 09 Pages 4467-4471 2016 Website: www.ijsrm.in ISSN (e): 2321-3418 Brain Tumor Segmentation: A Review Dharna*, Priyanshu
International Journal of scientific research and management (IJSRM) Volume 4 Issue 09 Pages 4467-4471 2016 Website: www.ijsrm.in ISSN (e): 2321-3418 Brain Tumor Segmentation: A Review Dharna*, Priyanshu
Computer Assisted Detection and Analysis of Tall Cell Variant Papillary Thyroid Carcinoma in Histological Images
 Computer Assisted Detection and Analysis of Tall Cell Variant Papillary Thyroid Carcinoma in Histological Images Edward Kim 1, Zubair Baloch 2, Caroline Kim 3 1 Department of Computing Sciences, Villanova
Computer Assisted Detection and Analysis of Tall Cell Variant Papillary Thyroid Carcinoma in Histological Images Edward Kim 1, Zubair Baloch 2, Caroline Kim 3 1 Department of Computing Sciences, Villanova
WDHS Curriculum Map Probability and Statistics. What is Statistics and how does it relate to you?
 WDHS Curriculum Map Probability and Statistics Time Interval/ Unit 1: Introduction to Statistics 1.1-1.3 2 weeks S-IC-1: Understand statistics as a process for making inferences about population parameters
WDHS Curriculum Map Probability and Statistics Time Interval/ Unit 1: Introduction to Statistics 1.1-1.3 2 weeks S-IC-1: Understand statistics as a process for making inferences about population parameters
Reveal Relationships in Categorical Data
 SPSS Categories 15.0 Specifications Reveal Relationships in Categorical Data Unleash the full potential of your data through perceptual mapping, optimal scaling, preference scaling, and dimension reduction
SPSS Categories 15.0 Specifications Reveal Relationships in Categorical Data Unleash the full potential of your data through perceptual mapping, optimal scaling, preference scaling, and dimension reduction
Application of Local Control Strategy in analyses of the effects of Radon on Lung Cancer Mortality for 2,881 US Counties
 Application of Local Control Strategy in analyses of the effects of Radon on Lung Cancer Mortality for 2,881 US Counties Bob Obenchain, Risk Benefit Statistics, August 2015 Our motivation for using a Cut-Point
Application of Local Control Strategy in analyses of the effects of Radon on Lung Cancer Mortality for 2,881 US Counties Bob Obenchain, Risk Benefit Statistics, August 2015 Our motivation for using a Cut-Point
Biostatistical modelling in genomics for clinical cancer studies
 This work was supported by Entente Cordiale Cancer Research Bursaries Biostatistical modelling in genomics for clinical cancer studies Philippe Broët JE 2492 Faculté de Médecine Paris-Sud In collaboration
This work was supported by Entente Cordiale Cancer Research Bursaries Biostatistical modelling in genomics for clinical cancer studies Philippe Broët JE 2492 Faculté de Médecine Paris-Sud In collaboration
Automatic Hemorrhage Classification System Based On Svm Classifier
 Automatic Hemorrhage Classification System Based On Svm Classifier Abstract - Brain hemorrhage is a bleeding in or around the brain which are caused by head trauma, high blood pressure and intracranial
Automatic Hemorrhage Classification System Based On Svm Classifier Abstract - Brain hemorrhage is a bleeding in or around the brain which are caused by head trauma, high blood pressure and intracranial
International Journal of Computer Science Trends and Technology (IJCST) Volume 5 Issue 1, Jan Feb 2017
 RESEARCH ARTICLE Classification of Cancer Dataset in Data Mining Algorithms Using R Tool P.Dhivyapriya [1], Dr.S.Sivakumar [2] Research Scholar [1], Assistant professor [2] Department of Computer Science
RESEARCH ARTICLE Classification of Cancer Dataset in Data Mining Algorithms Using R Tool P.Dhivyapriya [1], Dr.S.Sivakumar [2] Research Scholar [1], Assistant professor [2] Department of Computer Science
Frequency Tracking: LMS and RLS Applied to Speech Formant Estimation
 Aldebaro Klautau - http://speech.ucsd.edu/aldebaro - 2/3/. Page. Frequency Tracking: LMS and RLS Applied to Speech Formant Estimation ) Introduction Several speech processing algorithms assume the signal
Aldebaro Klautau - http://speech.ucsd.edu/aldebaro - 2/3/. Page. Frequency Tracking: LMS and RLS Applied to Speech Formant Estimation ) Introduction Several speech processing algorithms assume the signal
Threshold Based Segmentation Technique for Mass Detection in Mammography
 Threshold Based Segmentation Technique for Mass Detection in Mammography Aziz Makandar *, Bhagirathi Halalli Department of Computer Science, Karnataka State Women s University, Vijayapura, Karnataka, India.
Threshold Based Segmentation Technique for Mass Detection in Mammography Aziz Makandar *, Bhagirathi Halalli Department of Computer Science, Karnataka State Women s University, Vijayapura, Karnataka, India.
AUTOMATIC DIABETIC RETINOPATHY DETECTION USING GABOR FILTER WITH LOCAL ENTROPY THRESHOLDING
 AUTOMATIC DIABETIC RETINOPATHY DETECTION USING GABOR FILTER WITH LOCAL ENTROPY THRESHOLDING MAHABOOB.SHAIK, Research scholar, Dept of ECE, JJT University, Jhunjhunu, Rajasthan, India Abstract: The major
AUTOMATIC DIABETIC RETINOPATHY DETECTION USING GABOR FILTER WITH LOCAL ENTROPY THRESHOLDING MAHABOOB.SHAIK, Research scholar, Dept of ECE, JJT University, Jhunjhunu, Rajasthan, India Abstract: The major
A novel and automatic pectoral muscle identification algorithm for mediolateral oblique (MLO) view mammograms using ImageJ
 A novel and automatic pectoral muscle identification algorithm for mediolateral oblique (MLO) view mammograms using ImageJ Chao Wang Wolfson Institute of Preventive Medicine Queen Mary University of London
A novel and automatic pectoral muscle identification algorithm for mediolateral oblique (MLO) view mammograms using ImageJ Chao Wang Wolfson Institute of Preventive Medicine Queen Mary University of London
DYNAMIC SEGMENTATION OF BREAST TISSUE IN DIGITIZED MAMMOGRAMS
 DYNAMIC SEGMENTATION OF BREAST TISSUE IN DIGITIZED MAMMOGRAMS J. T. Neyhart 1, M. D. Ciocco 1, R. Polikar 1, S. Mandayam 1 and M. Tseng 2 1 Department of Electrical & Computer Engineering, Rowan University,
DYNAMIC SEGMENTATION OF BREAST TISSUE IN DIGITIZED MAMMOGRAMS J. T. Neyhart 1, M. D. Ciocco 1, R. Polikar 1, S. Mandayam 1 and M. Tseng 2 1 Department of Electrical & Computer Engineering, Rowan University,
MBios 478: Systems Biology and Bayesian Networks, 27 [Dr. Wyrick] Slide #1. Lecture 27: Systems Biology and Bayesian Networks
![MBios 478: Systems Biology and Bayesian Networks, 27 [Dr. Wyrick] Slide #1. Lecture 27: Systems Biology and Bayesian Networks MBios 478: Systems Biology and Bayesian Networks, 27 [Dr. Wyrick] Slide #1. Lecture 27: Systems Biology and Bayesian Networks](/thumbs/80/82116384.jpg) MBios 478: Systems Biology and Bayesian Networks, 27 [Dr. Wyrick] Slide #1 Lecture 27: Systems Biology and Bayesian Networks Systems Biology and Regulatory Networks o Definitions o Network motifs o Examples
MBios 478: Systems Biology and Bayesian Networks, 27 [Dr. Wyrick] Slide #1 Lecture 27: Systems Biology and Bayesian Networks Systems Biology and Regulatory Networks o Definitions o Network motifs o Examples
