The mir-51 Family of MicroRNAs Functions in Diverse Regulatory Pathways in Caenorhbditis elegans
|
|
|
- Gary Lyons
- 5 years ago
- Views:
Transcription
1 Marquette University Biological Sciences Faculty Research and Publications Biological Sciences, Department of The mir-51 Family of MicroRNAs Functions in Diverse Regulatory Pathways in Caenorhbditis elegans Allison Abbott Marquette University, John L. Brenner Marquette University Benedict J. Kemp Marquette University Published version. PLoS, Vol. 7, No. 5 (2012): e DOI Brenner et al. Used with permission. Published under Creative Commons License 4.0.
2 The mir-51 Family of micrornas Functions in Diverse Regulatory Pathways in Caenorhabditiselegans John L. Brenner, Benedict J. Kemp, Allison L. Abbott* Department of Biological Sciences, Marquette University, Milwaukee, Wisconsin, United States of America Abstract The mir-51 family of micrornas (mirnas) in C. elegans are part of the deeply conserved mir-99/100 family. While loss of all six family members (mir-51-56) in C. elegans results in embryonic lethality, loss of individual mir-51 family members results in a suppression of retarded developmental timing defects associated with the loss of alg-1. The mechanism of this suppression of developmental timing defects is unknown. To address this, we characterized the function of the mir-51 family in the developmental timing pathway. We performed genetic analysis and determined that mir-51 family members regulate the developmental timing pathway in the L2 stage upstream of hbl-1. Loss of the mir-51 family member, mir-52, suppressed retarded developmental timing defects associated with the loss of let-7 family members and lin-46. Enhancement of precocious defects was observed for mutations in lin-14, hbl-1, and mir-48(ve33), but not later acting developmental timing genes. Interestingly, mir-51 family members showed genetic interactions with additional mirnaregulated pathways, which are regulated by the let-7 and mir-35 family mirnas, lsy-6, mir-240/786, and mir-1. Loss of mir- 52 likely does not suppress mirna-regulated pathways through an increase in mirna biogenesis or mirna activity. We found no increase in the levels of four mature mirnas, let-7, mir-58, mir-62 or mir-244, in mir-52 or mir-52/53/54/55/56 mutant worms. In addition, we observed no increase in the activity of ectopic lsy-6 in the repression of a downstream target in uterine cells in worms that lack mir-52. We propose that the mir-51 family functions broadly through the regulation of multiple targets, which have not yet been identified, in diverse regulatory pathways in C. elegans. Citation: Brenner JL, Kemp BJ, Abbott AL (2012) The mir-51 Family of micrornas Functions in Diverse Regulatory Pathways in Caenorhabditiselegans. PLoS ONE 7(5): e doi: /journal.pone Editor: Anne C. Hart, Brown University, United States of America Received March 1, 2012; Accepted April 17, 2012; Published May 16, 2012 Copyright: ß 2012 Brenner et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Funding: This work was funded by a National Institutes of Health grant, R15 GM awarded to ALA. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Competing Interests: The authors have declared that no competing interests exist. * allison.abbott@marquette.edu Introduction micrornas (mirnas) are,22 nucleotide, non-coding RNAs that post-transcriptionally regulate the expression of their downstream targets. mirnas bind to sites with imperfect complementarity in target mrnas [1], which, in most cases, results in lower target protein levels due to the inhibition of translation and the reduced stability of target mrnas [2,3]. The effects of mirna regulation on target protein levels can vary widely. In some cases, mirna binding to a target can function as a switch by directing the nearly complete suppression of target mrna translation. For example, the lin-4 and let-7 mirnas function as developmental switches to strongly down-regulate their respective targets, lin-14 and lin-41 [4-7]. In other cases, mirna binding to a target can function as a fine tuner to direct modest repression of target mrna translation. For example, in flies, mir-8 maintains the levels of atrophin in an optimal range [8]. However, in recent work, Mukherji et al. [9] demonstrate that the effect of mirna regulation upon target protein levels is not an inherent property of the mirna but rather depends on the stoichiometry and binding affinity of a mirna and its associated target mrnas. At low levels of target mrna, a mirna can act to strongly repress translation, whereas at high levels of the target mrna, a mirna can act to modestly repress translation [9]. While penetrant mutant phenotypes are observed in lin-4 and let-7 mutants, defects were not identified for most individual mirna mutants in C. elegans [10], though progress in identifying functions for mirnas is being made [11]. For some mirna mutants, like let-7 or mir-35 family mutants, the lack of observed defects is a result of functional redundancy among mirna family members, which share a six nucleotide 59 seed sequence [12,13]. For other mirna mutants, like lsy-6, the lack of obvious defects reflects highly specialized functions for individual mirnas that were not observed in broad-based phenotypic analyses [14]. Furthermore, since some mirnas function to modestly regulate, or fine tune, target gene expression, the loss of these mirnas may not result in obvious defects during normal growth conditions. Approaches that have examined mirna mutant worms under conditions of stress, such as altered environmental conditions [15,16] or genetic backgrounds [17], have been successful in identifying mutant phenotypes associated with the loss of individual mirnas. Using a sensitized genetic background, we characterized defects in 80% of the individual mirna mutants analyzed [17]. For that analysis, we used strains that lack one of two Argonaute proteins that function in the mirna pathway in C. elegans, ALG-1, as a sensitized background in which to identify mutant phenotypes. In alg-1 mutants, overall mirna levels are reduced, including the lin-4 and let-7 mirnas, which leads to observable defects in the developmental timing pathway [18]. This pathway controls the appropriate temporal execution of stage-specific developmental PLoS ONE 1 May 2012 Volume 7 Issue 5 e37185
3 programs through the four larval stages, L1 L4 [19]. Loss of alg-1 activity results in developmental timing defects including incomplete alae formation at the L4 to adult transition, an increased number of hypodermal seam cells, and a failure to exit the molting cycle [17,18,20 23]. Loss of mir-51 family members partially suppresses these developmental timing defects in alg-1 worms [17]. The mir-51 family is part of the larger mir-99/100 family, a mirna family that shows deep conservation from cnidarians through humans [24]. In C. elegans, the mir-51 family comprises six mirnas, mir-51 through mir-56. Loss of the entire mir-51 family in C. elegans results in embryonic lethality, due to a failure of pharyngeal attachment [25]. Loss of multiple members causes several mutant phenotypes including larval lethality and slow growth [13,25]. These pleiotropic phenotypes indicate that mir-51 family members likely function to regulate multiple downstream targets and pathways. The mechanism whereby loss of individual mir-51 family members suppresses alg-1 developmental timing defects is unclear. Unlike other genes that regulate developmental timing, mir-51 family members are expressed broadly and abundantly throughout the life of the worm [25 29]. We therefore wanted to determine the function of the mir-51 family members in the regulation of the developmental timing pathway. Here, we have defined the genetic interactions of mir-51 family members with components of the developmental timing pathway. Additionally, we report that the mir-51 family interacts with multiple, diverse, mirna regulated genetic pathways, including pathways regulated by the let-7 and mir-35 family mirnas, as well as lsy-6, mir-240/786, and mir-1. We provide evidence that is inconsistent with the model that the mir-51 family regulates mirna biogenesis or mirna activity. Instead, we propose that the mir-51 family functions to regulate multiple targets in diverse developmental pathways in C. elegans. Results Loss of mir-51 family members partially suppresses retarded developmental timing phenotypes The loss of mir-51 family members suppresses alg-1 developmental timing defects [17], suggesting a possible direct role in the regulation of the developmental timing pathway. However, mutants lacking individual mir-51 family members did not display developmental timing abnormalities such as defects in alae formation or defects in seam cell divisions (Table 1 and Table 2). Further, worms that are multiply mutant for 5 out of 6 members of the mir-51 family, mir-52/53/54/55/56, also do not display alae formation defects (Table 1 and Table 2), despite displaying other mutant phenotypes including slow growth and larval lethality [13,25]. Because the alg-1 developmental timing defects are similar to those associated with the loss of the let-7 family mirnas [18], we determined if loss of individual mir-51 family members was sufficient to suppress let-7 timing defects. To do this, we used a temperature sensitive let-7 allele, n2853. At 25uC, these let-7(ts) mutants display a repetition of a late larval program with failure to form complete alae and lethality due to bursting at the vulva at the L4 to adult transition ([5]; Table 1). Loss of mir-51 family members did not suppress either of these phenotypes in let-7 mutants (Table 1). The let-7 family members, mir-48, mir-84, and mir-241, function together to control the timing of the L3 stage program through down-regulation of their target, hbl-1 [12]. In the L2 stage, a subset of hypodermal seam cells undergo two rounds of cell division resulting in an increase in the number of seam cells from 10 to 16. In mutants lacking mir-48, mir-84 and mir-241 (hereafter referred to as mir-48/84/241), the L3 stage program is not executed properly and the L2 stage program is reiterated. This reiteration of the L2 stage program results in an increased number of seam cells [12]. mir-48/84/241 mutant worms often display defects in alae formation at the L4 to adult transition. In addition, many of these mutants burst at the L4 to adult transition or execute an extra adult-stage molt, which leads to the bag-of-worms phenotype [12]. mir-52; mir-48/84/241 had fewer seam cells than mir-48/84/ 241 worms, indicating a suppression of the L2 reiteration phenotype (Table 1). Additionally, loss of mir-52 suppressed the alae formation defects and bursting phenotypes of mir-48/84/241: 100% of mir-48/84/241 displayed incomplete alae formation and 56% of mir-48/84/241 worms burst at the L4 to adult transition compared to 51% and 3% of mir-52;mir-48/84/241 mutants, respectively (Table 1). However, 77% of mir-52; mir-48/84/241 worms showed the bag of worms phenotype, indicating an extra adult-stage molt. This likely reflects a partial suppression of mir- 48/84/241 phenotypes, rather than an inability to suppress molting since loss of mir-52 strongly suppresses the ectopic molting phenotype of alg-1 worms [17] as well as mir-48/84 double mutant worms (Table 1). Next, we examined the effect of elevated expression of mir-51 family members on the retarded development of mir-48 mir-241 (mir-48/241) mutant worms. To accomplish this, we used mjex160, an extrachromosomal array with the genomic fragment for mir- 54/55/56 that was previously shown to rescue the embryonic lethality of mir-51 family mutant worms [25] and the developmental timing phenotypes in mir-54/55/56 alg-1 mutant worms [17]. mjex160 enhanced developmental timing defects of mir-48/ 241 mutant worms (Table 1). mir-48/241 worms have 19.1 seam cells on average. This is increased to 22.1 in mir-48/241; mjex160 worms (Table 1). This indicates elevated expression of mir-51 family members enhances the L2 repetition phenotype. We determined if the loss of mir-51 family members can suppress the phenotypes of lin-46 and puf-9, mutants that display retarded developmental timing defects [30,31]. lin-46 functions in parallel to the let-7 family to control the timing of the L3 stage program [12,30]. lin-46 mutants fail to properly execute the L3 stage program and show reiteration of the L2 program at 15uC. lin-46 mutants display extra seam cells and incomplete alae formation [30]. Loss of mir-52 partially suppressed lin-46 developmental timing defects: mir-52; lin-46 double mutant worms had fewer seam cells and displayed weaker alae defects compared to lin-46 mutant worms (Table 1). Loss of the other mir-51 family members had no significant effect on lin-46 developmental timing defects (data not shown). puf-9 encodes a pumilio family protein that acts to negatively regulate hbl-1 through its 39UTR [31]. puf-9 mutant worms fail to form complete alae at the L4 to adult transition. Loss of mir-52 did not suppress the puf-9 alae defects (Table 1). This suggests that puf-9 may function downstream of the mir-51 family to regulate developmental timing. To determine if puf-9 is necessary for mir-52-mediated suppression of the let-7 family developmental timing defects, we examined worms multiply mutant for mir-52, puf-9, and let-7 family mirnas, mir-48 and mir-241 (mir-48/241). Loss of mir-52 suppressed the seam cell and alae formation defects in mir-48/ 241 mutants (Table 1). Loss of puf-9 did not affect the mir-52 mediated suppression of the extra seam cell phenotype of mir-48/ 241 mutant worms (Table 1). However, no suppression of alae formation defects was observed in mir-52; mir-48/241; puf-9 worms relative to mir-52; mir-48/241 (Table 1). This is consistent with a function for puf-9 later in development, after the L2 to L3 transition. Together, these data indicate that the mir-51 family functions to regulate the execution of the L3 stage program, acting either downstream or in parallel to the let-7 family mirnas and PLoS ONE 2 May 2012 Volume 7 Issue 5 e37185
4 Table 1. Genetic interactions of the mir-51 family with retarded developmental timing mutants. Alae at L4 to Adult transition Lethality Strain a seam cells b complete gapped none n % burst % bag of worms n RG733 wild type RF481 wild type RF491 mir RF499 mir RF483 mir RF399 mir-54/55/ RF692 mir-52/53/54/55/56 c MT d RF447 mir-51; 0 80 d RF448 mir-52; 7 73 d RF449 mir-53; 0 53 d RF442 mir-54/55/56; 7 21 d RF554 mir-48/84/ RF556 mir-52; mir-48/84/ e 49 f f RF553 mir-48/84/ RF555 mir-51; mir-48/84/ g RF557 mir-53; mir-48/84/ g RF558 mir-54/55/56; mir-48/84/ h 21 g g VT1064 mir-48/ RF451 mir-51; mir-48/ i 101 RF469 mir-52; mir-48/ i 148 RF454 mir-53; mir-48/ RF415 mir-54/55/56; mir- 48/ i 93 RF619 mir-48/ RF730 mir-48/241; mjex160[mir-54/55/56] 22.1 j k 24 k 136 k RF RF569 mir-52; 17.8 l 23 m VC894 puf RF578 mir-52; puf RF620 mir-52; mir-48/ n RF625 mir-48/241; puf RF626 mir-52; mir-48/241; puf o a Full genotype information can be found in Table S1. b seam cells counted in L4-stage worms using wis78 or wis79[scm::gfp], n$18 (range 19 30). c indicates results not determined. d alae scored categorized as partially or faintly visible rather than gapped as elsewhere. e indicates significant difference compared to RF554 mir-48/84/241 (student s t-test, p,0.05), which contained wis79. f indicates significant difference compared to RF554 mir-48/84/241 (x2, p,0.05) which contained wis79. g indicates significant difference compared to RF553 mir-48/84/241 (x2, p,0.05) which contained wis78. h indicates significant difference compared to RF553 mir-48/84/241 (student s t-test, p,0.05), which contained wis78. i indicates significant difference compared to VT1064 mir-48/84 (x2, p,0.05). j indicates significant difference comparing worms from the same strain, but +/2 for mjex160[mir-54/55/56] (student s t-test, p,0.05). k population scored for lethality is a mix of worms +/2 for mjex160[mir-54/55/56]. l indicates significant difference compared to RF568 lin-46 (student s t-test, p,0.05). m indicates significant difference compared to RF568 lin-46 (x2, p,0.05). n indicates significant difference compared to RF619 mir-48/241 (student s t-test, p,0.05). o indicates significant difference compared to RF625 mir-48/241; puf-9 (student s t-test, p,0.05). doi: /journal.pone t001 lin-46 and may have additional activity in the control of alae formation in late larval development. Loss of mir-52 enhances precocious developmental timing phenotypes We next characterized genetic interactions between mir-52 and a set of precocious developmental timing genes. Loss of mir-52 PLoS ONE 3 May 2012 Volume 7 Issue 5 e37185
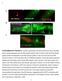
5 Table 2. Genetic interactions of mir-51 family with precocious developmental timing mutants. Precocious Alae b Strain a complete gapped none n RG733 wild type RF481 wild type RF491 mir RF499 mir RF483 mir RF399 mir-54/55/ RF692 mir /53/54/55/56 RG490 mir-48(ve33) RF583 mir-52; 0 88 d mir-48(ve33) RF534 hbl RF535 mir-52; 2 d hbl-1 RF RF588 mir-52; 76 d RF536 lin RF537 mir-52; lin-41 RF538 lin RF541 mir-52; lin-42 VT517 lin-28 c RF573 mir-52; lin-28 c a full genotype information can be found in Table S1. b alae were scored in L3 molt or early L4-stage worms, except where otherwise noted. c alae were scored in the L2 molt. d indicates significant difference between strains of same genotype +/2 mir-52 (x2, p,0.05). doi: /journal.pone t002 enhanced the developmental timing defects observed in three precocious mutants: mir-48(ve33), hbl-1(ve18), and lin-14(n179). (Table 2). First, mir-48(ve33) worms display early accumulation of mir-48 and precocious formation of adult-specific alae one stage early at the L3 to L4 transition [32]. Loss of mir-52 significantly enhanced this precocious alae formation in the mir-48(ve33) background (Table 2). We found that 55% of mir-48(ve33) mutants displayed precocious alae formation compared to 88% of mir- 52;mir-48(ve33) worms. Next, hbl-1 is a central regulator of the L2 versus L3 cell fate decision [33,34]. Loss of mir-52 enhanced the precocious alae phenotype of hbl-1(ve18) mutants: 76% of hbl- 1(ve18) worms displayed either complete or gapped precocious alae in the L4 stage compared to 97% of mir-52; hbl-1 double mutant worms (Table 2). Enhancement of the precocious phenotype of hbl-1(ve18) worms may reflect reduced activity of hbl-1 itself, since ve18 is a reduced function, not a null, allele [33]. Finally, lin-14 functions to regulate the timing of both L1 versus L2 and L2 versus L3 cell fate decisions [35]. To analyze genetic interactions with lin-14, we used the temperature sensitive allele, n179. At 25uC, 34% of lin-14(ts) worms form complete precocious alae at the L3 to L4 transition compared to 76% of mir-52; lin- 14(n179) worms (Table 2). Enhancement was not observed for the lin-41, lin-42, or lin-28 phenotypes (Table 2). The enhancement of the precocious developmental timing defects observed in mir- 48(ve33), hbl-1(ve18), and lin-14(n179ts) mutant worms is consistent with a role for the mir-51 family in the regulation of L2 versus L3 cell fate decisions. mir-51 family members function upstream of hbl-1, but not lin-28, to suppress developmental timing defects in let-7 family mutants Genetic interactions between mir-52 and let-7 family members as well as hbl-1(ve18) suggest that mir-52 may act upstream of hbl-1 to promote its activity. hbl-1 is robustly expressed in the hypodermis during embryonic and early larval development and then is subsequently down-regulated through its 39 UTR by the early L3 stage [33,34,36]. The down-regulation of hbl-1 in the hypodermis requires the let-7 family members, mir-48, mir-84, and mir-241 [12]. We therefore determined whether the observed suppression of developmental timing defects in mir-52; mir-48/84/241 reflects a suppression of hbl-1 misregulation. Indeed, loss of mir-52 partially suppressed the hbl-1 misexpression phenotype of mir-48/84/241 mutant worms: in 91% of mir-48/84/241 worms hbl-1::gfp::hbl-1 transgene expression remained high in L3, whereas only 62% of mir-52; mir-48/84/241 displayed high hbl-1::gfp::hbl-1 expression (Figure 1). This indicates that mir-52 acts upstream of hbl-1 expression in opposition to let-7 family activity. Like hbl-1, lin-28 is also a critical regulator of L2 versus L3 cell fate decisions. We used a lin-28::gfp::lin-28 transgene to determine whether mir-52 suppression is the result of a misregulation of lin- 28. However, no difference was observed in lin-28::gfp::lin-28 expression between mir-48/84/241 and mir-52; mir-48/84/241 in L2 molt stage worms (Figure 2). Thus, misregulation of lin-28 does not account for the observed suppression of developmental timing defects in mir-52; mir-48/84/241 worms. Together, these data are consistent with the mir-51 family functioning downstream or in parallel to lin-28, lin-46 and the let-7 family, but upstream of hbl-1 to regulate the L2 versus L3 cell fate decisions. Loss of mir-51 family members suppresses additional mirna-dependent regulatory pathways in C. elegans Genetic interactions with the developmental timing pathway may reflect a specific function for the mir-51 family mirnas in the regulation of targets in this pathway. Alternatively, these interactions may reflect a broader function for the mir-51 family in the regulation of mirna biogenesis or activity. For example, the developmental timing defects observed in alg-1 or ain-1 mutants [18,20] are due to lower overall mirna activity, including the lin-4 and let-7 family mirnas, rather than a specific function in the developmental timing pathway. Therefore, we tested whether the mir-51 family interacted with additional mirna-regulated pathways by determining if loss of mir-51 family members could suppress other mirna mutant phenotypes that are distinct from developmental timing, including lsy-6 regulation of neuronal asymmetry, let-7 family regulation of vulva cell fate specification, mir-240/786 regulation of defecation, mir-35 family regulation of embryonic development and mir-1 regulation of neuromuscular function. lsy-6. The lsy-6 mirna specifies the ASEL cell fate through the down-regulation of its target, cog-1 [14]. lsy-6 repression of cog- 1 is necessary for lim-6::gfp expression in the ASEL [14]. To achieve a genetic background with optimally compromised lsy-6 activity, we used heterozygous worms that carry a loss of function allele, ot149, and a reduced function allele, ot % of these lsy- PLoS ONE 4 May 2012 Volume 7 Issue 5 e37185
6 The mir-51 Family Regulates Diverse Pathways Figure 1. Loss of mir-52 suppresses hbl-1 misregulation in mir-48/84/241 mutants. Representative fluorescent micrographs of hbl-1::gfp::hbl-1 transgene expression in (A) mir-48/84/241 and (B) mir-52; mir-48/84/241 mutant worms in the L3 stage with corresponding DIC images (C and D, respectively). White arrow in A indicates a hyp7 nucleus. (E) Percentage of worms with hbl-1::gfp::hbl-1 expression in hypodermis of L3 stage worms, n$33 (range 33 37). * indicates significant difference (x2, p,0.01). doi: /journal.pone g001 Figure 2. Loss of mir-52 does not result in increased expression of lin-28::gfp::lin-28. (A, B) Representative fluorescent micrographs of lin28::gfp::lin-28 transgene expression at in (A) mir-48/84/241 and (B) mir-52; mir-48/84/241 worms in the L2 molt stage with corresponding DIC images, (C and D, respectively). Strains were scored for expression of lin-28::gfp::lin-28 at the L2 molt (n = 17). No significant difference was observed between strains (x2, p.0.05). doi: /journal.pone g002 PLoS ONE 5 May 2012 Volume 7 Issue 5 e37185
7 6(ot149lf)/lsy-6(ot150rf) heterozygous worms fail to express lim- 6::gfp in the ASEL neuron compared to 100% of lsy-6(ot149lf) and 14% of lsy-6(ot150rf) worms (Figure 3B; [14]). Loss of mir-52 partially suppressed mutant lim-6::gfp expression in lsy-6(ot149lf)/ lsy-6(ot150rf): 85% of lsy-6rf/lsy-6lf worms displayed mutant lim- 6::gfp expression compared to 61% of mir-52; lsy-6rf/lsy-6lf (Figure 3B). let-7 family regulation of vulva development. The let-7 family mirnas repress let-60/ras in the regulation of vulva development [37]. Worms with a gain-of-function mutation in let- 60 display defects in cell fate specification, which often results in a Muv phenotype with multiple vulva structures produced [38]. Overexpression of let-7 family members partially suppresses the let- 60gf Muv phenotype [37]. If the mir-51 family opposes let-7 activity in vulva development, as it did in the developmental timing pathway, then it would be expected that loss of mir-51 family members should suppress the let-60gf Muv phenotype. This is observed in mir-52;let-60gf worms (Figure 3E). Interestingly, loss of Figure 3. The mir-51 family members, mir-52 and mir-54/55/56, function in multiple mirna-dependent developmental pathways. (A, B) mir-52 suppresses ASEL specification defects of lsy-6(rf)/lsy-6(lf) worms. (A) Cartoon of lim-6::gfp expression in wild-type and lsy-6(lf) worms. A, anterior; P, posterior; L, left; R, right. (B) Worms of indicated genotypes were scored for lim-6::gfp expression in late larval and young adult stages, n$169. * indicates significant difference (x 2,p,0.01). (C E) Loss of mir-52 partially suppresses, while loss of mir-54/55/56 enhances, the multivulva (Muv) phenotype of let-60gf worms. (C) A wild type worm with one normal vulva, white arrow. (D) A let-60gf worm with one normal vulva, white arrow, and one ectopic vulva, black arrow. Bars represent 100 mm. (E) Synchronized L1 worms of the indicated genotype were allowed to develop at 25uC for 2 3 days and then scored as young adults for the Muv phenotype. n$100. * indicates significant difference (x 2,p,0.01). (F) Loss of mir-52 reduces the average defecation cycle time of mir-240/786 mutant worms. Average time between consecutive pboc contractions for n$5 worms. * indicates significant difference (student s t-test, p,0.01). Error bars indicate SEM values. (G) Loss of mir-54/55/56 enhances the embryonic lethality of mir-35 through 41 mutant worms. L4 worms of the indicated genotypes were shifted to 25u and the next day embryos from these worms were collected. After 24 hours, unhatched embryos were counted to determine the percentage of embryonic lethality (n$148). * indicates significant difference (x 2,p,0.01). (H) Loss of mir-52 modestly suppresses the resistance to levamisole of mir-1 worms. mir-52 mutants show weakly enhanced sensitivity to levamisole. * indicates significant difference compared to wild type at the indicated time point (x 2,p,0.05). ** indicates significant difference compared to mir-1 at the indicated time point (x 2,p,0.05). doi: /journal.pone g003 PLoS ONE 6 May 2012 Volume 7 Issue 5 e37185
8 mir-54/55/56 enhanced the Muv phenotype of let-60gf (Figure 3E). This may reflect distinct activities of individual mir-51 family members in the control of vulva development. Identification of mir-51 family targets in the vulva specification pathway is required to elucidate the functions of individual mir-51 family members. mir-240/786. mir-240/786 is necessary for the normal rhythmicity of the defecation motor program [10]. In wild type worms, a defecation motor program occurs every,50 seconds [39]. In mir-240/786 mutant worms, the average defecation cycle time is increased [10]. We found that loss of mir-52 significantly reduced the average defecation cycle time of mir-240/786 worms (Figure 3F). Loss of mir-54/55/56 had no effect on the mean defecation cycle time of mir-240/786 worms (data not shown). mir-35 family. The mir-35 family comprises eight mirnas, mir-35 through mir-42. These family members are redundantly required for embryonic development and mutants lacking mir-35 through mir-41 exhibit temperature sensitive embryonic lethality [13]. We found that loss of mir-54/55/56 did not suppress the embryonic lethal phenotype of mir-35/41 mutants, but rather significantly enhances this phenotype (Figure 3G). mir-1. mir-1 is necessary for normal neuromuscular function [40]. mir-1 mutants display a resistance to levamisole-induced paralysis due to an increase in levels of its targets, UNC-29 and UNC-63 [40]. We found that loss of mir-52 weakly suppressed the levamisole resistance phenotype of mir-1 worms (Figure 3H). We found that after 140 minutes on 200 mm levamisole mir-52; mir-1 worms were less resistant to levamisole compared to mir-1 (Figure 3H). We also found that mir-52 worms appeared to be slightly more sensitive to levamisole compared to wild type worms. Loss of mir-54/55/56 had no effect on levamisole sensitivity of mir- 1 or wild-type worms (data not shown). Loss of mir-51 family members does not broadly enhance mirna biogenesis or activity To account for the observation that the loss of mir-52 suppressed multiple mirna-dependent phenotypes, we proposed that mir-52 may act to broadly regulate mirna biogenesis or activity. To examine if the mir-51 family regulates the mirna pathway, we measured mature mirna levels for a set of mirnas that display various expression and biogenesis characteristics. We analyzed levels of the let-7 mirna, a developmentally-regulated mirna that functions in the developmental timing pathway in the hypodermis [5], mir-58, a highly abundant mirna [26], mir- 62, a mirtron that displays Drosha independent biogenesis [41], and mir-244, a mirna that is expressed at lower levels primarily in hypodermal seam cells [29]. We found that the levels of these mirnas are unchanged in mir-52 mutants as well as in mir-52/53/ 54/55/56 mutants (Figure 4). mir-52/53/54/55/56 mutant worms display impenetrant embryonic lethality, slow growth, and mating defects [13,25] indicating that mir-51 family targets are sufficiently misregulated to result in severe, penetrant mutant phenotypes. However, no change in mirna levels were detected for the four mirnas analyzed. These results indicate that the observed suppression of developmental timing defects is not likely due to an increase in overall mirna levels and that mir-51 family mirnas likely do not function broadly to regulate mirna biogenesis. In order to determine if loss of mir-52 can act to enhance mirna activity, we analyzed the activity of ectopically expressed lsy-6 in the repression of a cog-1::gfp reporter [14]. Ectopic expression of lsy-6 under the control of the cog-1 promoter allowed us to examine the activity of the lsy-6 mirna in cells where it is normally not found, including uterine and vulva cells [14]. We found that in 60% of worms examined, ectopic expression of the Figure 4. Levels of mature let-7, mir-58, mir-62, and mir-244 are unchanged in the absence of mir-51 family members. Levels of mature mirnas in wild type, mir-52, and mir-52/53/54/55/56 mutant worms were measured and normalized to the average of two control RNAs, U18 and sn2343. The graph represents the level of mature mirnas relative to wild type. Error bars represent the standard deviation (SD) between biological replicates. No differences in mature mirna expression was observed (student9s t-test, p.0.24). doi: /journal.pone g004 lsy-6 mirna resulted in the down-regulation of cog-1::gfp in uterine cells (Figure 5). We found that loss of mir-52 had no effect on the activity of ectopic lsy-6 repression of cog-1 (Figure 5). These data indicate that lsy-6 activity is not enhanced in the absence of mir-52, thereby suggesting that the mir-51 family does not function broadly to regulate the activity of mirnas. Discussion The goal of this study was to define the mechanism whereby loss of mir-51 family members can suppress the developmental timing defects of alg-1 mutant worms. Our genetic evidence indicates that mir-51 family members act early in the developmental timing pathway to regulate L2 versus L3 cell fate decisions. We observed that loss of the mir-51 family member, mir-52, strongly suppressed the L2 stage reiteration phenotype of mir-48/84/241 mutants and lin-46 mutants. No significant suppression was observed with later acting genes in the developmental timing pathway, such as let-7 and puf-9. Similarly, we observed genetic enhancement of precocious phenotypes due to mutations that result in omissions of early larval stage programs, like lin-14 and hbl-1, but not mutations that result in omission of later larval stage programs, like lin-41. This suggests that the developmental timing pathway is the most sensitive to the loss of mir-52 in the L2 stage. In many species, including humans and flies, mir-100, let-7, and lin-4 family members are located in a genomic cluster [42-44]. In flies, these three mirnas are polycistronic and function together to regulate adult behaviors [42]. Although this clustered organization in the genome is not observed in worms, evidence herein supports a functional relationship between the let-7 and mir-51 family of mirnas in the regulation of the developmental timing pathway. Although mir-51 family members interact with developmental timing genes, such as let-7 family members and lin-46, mir-51 family members are atypical developmental timing genes. First, unlike other developmental timing mirnas, such as lin-4 and let- 7, mir-51 family members do not display stage-specific expression PLoS ONE 7 May 2012 Volume 7 Issue 5 e37185
9 Figure 5. Loss of mir-52 does not enhance the ability of ectopically-expressed lsy-6 to regulate its target, cog-1. (A E) Effect of mir-52 on lsy-6 mediated regulation of cog-1::gfp::cog-1 expression. Representative fluorescent image of cog-1::gfp::cog-1 transgene expression in (A) wild type worms and (B) worms with cog-1::lsy-6 transgene with corresponding DIC images (C and D, respectively). White triangles point to uterine cells. Bars represent 10 mm. (E) Percentage of worms of given genotype with cog-1::gfp expression in either uterine cell, n$20 (range 20 68). Worms were scored in mid-to-late L4 stage. doi: /journal.pone g005 but rather display nearly ubiquitous expression throughout development. In addition, loss of nearly all mir-51 family members, which results in multiple defects including slow growth and embryonic lethality, did not result in developmental timing defects [13,25; Table 1 and Table 2]. We therefore propose that the mir- 51 family mirnas are not themselves regulators of developmental timing decisions but that they likely act downstream in the execution phase of developmental programs. Surprisingly, we found that mir-51 family members displayed genetic interactions with multiple mirna genes. These mirnas function in diverse developmental and physiological processes, which include developmental timing, vulva fate specification, neuronal fate specification, defecation, and neuromuscular function. Loss of the mir-51 family member, mir-52, partially suppressed the vulva cell fate defects of let-60gf mutants, the ASEL cell fate defects of lsy-6 mutants, the defecation defects of mir-240/786 mutants, and the levamisole resistance of mir-1 mutants. These activities for the mir-51 family may reflect the regulation of a single target that functions broadly in many pathways or the regulation of multiple targets that each function in distinct pathways. Our analysis of candidate targets failed to conclusively identify downstream mir-51 family targets (data not shown). One hypothesis to account for the observed suppression of multiple mirna-regulated pathways is that the loss of an abundant mirna such as mir-52 frees up mirna-induced silencing complex (mirisc) so that it is available for binding by other mirnas in a cell. Consistent with this model, the mir-51 family is both abundantly [26 28] and broadly expressed in tissues in which we observed a genetic interaction, including the hypodermis, the ASEL neuron, the vulva, the intestine, and muscle [25,29,45,46]. In this model, excess mirnas would compete for a limited pool of mirisc in wild-type worms in order to effectively repress their targets. In genetic backgrounds in which the activity of mirisc factors are reduced, such as in alg-1 mutants, mirisc becomes limiting as evidenced by an increased amount of stem-loop mirna precursors in both worms [18] and in human cells [47]. In human cells, overexpression of Argonauteencoding genes results in an elevation of ectopically-expressed mature mirnas [47]. However, it was not determined if endogenous mirnas were elevated following Argonaute overexpression. In wild-type worms, precursor mirnas are often detected in relatively low abundance [18,48,49]. These low levels of mirna precursors that are detected may indicate either the competition for limited mirisc or the normal, steady state level of mirna precursor in the biogenesis pathway. Although our genetic data are consistent with this limiting mirisc model, our PLoS ONE 8 May 2012 Volume 7 Issue 5 e37185
10 molecular and transgene expression data are inconsistent with this model. First, it is expected that overall levels of all mature mirnas would be elevated in mutants that lack the abundant mir-52 due to increased loading into mirisc. However, no such elevation in the mature mirna levels was observed for the four mirnas that were analyzed in the absence of mir-52 or mir-52/53/54/55/56 activities. In addition, loss of mir-52 did not result in an enhancement of ectopic lsy-6 activity in uterine cells as would be predicted by the limiting mirisc model. Future research will be directed at testing this model in order to determine if critical mirisc factors, such as Argonaute proteins, are limiting. Interestingly, in human cells and early Xenopus embryos, Argonaute protein levels are low and can be readily saturated by exogenous sirnas [50,51]. We found that the interactions with multiple mirna-dependent pathways likely does not reflect the regulation of the mirna pathway by the mir-51 family. As described above, neither increased mirna biogenesis nor increased mirna activity was observed in mir-51 family mutant backgrounds. Thus, mir-51 family members likely do not function to regulate the core pathway required for all mirna biogenesis or activity. However, it remains possible that mir-51 family members may function to modulate mirna activity in specific cells or for only a subset of mirnas not included in our analysis. We propose that the broad activity of mir-51 family members reflects the repression of a target or multiple target mrnas that act in distinct genetic pathways, possibly acting to fine tune, or buffer target protein levels. This broad activity described for the mir-51 family is unlike that of previously described mirnas in C. elegans. A direct target or targets of the mir-51 family in the diverse development pathways described herein remain unknown. Of the 293 conserved targets predicted by Targetscan [52,53], only 6 contain more than one binding site for the mir-51 family and none of these 6 have more than two binding sites for the mir-51 family. Because the mir-51 family is broadly and abundantly expressed throughout development in C. elegans [25 28], multiple binding sites for the mir-51 family within the 39UTR of a gene would be expected to cause robust repression of that gene throughout development. Multiple sites or sites with high binding affinity may therefore be selected against during evolution. We speculate that the function of mir-51 family members may be to weakly repress or fine-tune the protein levels for a large set of diverse downstream targets. Materials and Methods Nematode Methods C. elegans were maintained using standard conditions. Strains used in this study are listed in Table S1. Worms were kept at 20uC unless otherwise indicated. All strains were constructed using standard genetic techniques [54]. PCR was used to confirm the genotype of strains that contained mirna deletion alleles [17]. Fluorescence and differential interference contrast (DIC) microscopy were performed with a Nikon Eclipse 80i compound microscope equipped with a Photometrics CoolSNAP HQ2 monochrome digital camera and RS Image software (Roper Scientific) or with NIS Elements software (Nikon). References 1. Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136: doi: /j.cell Defecation assay Worms were transferred to NGM plates at room temperature and allowed to equilibrate on plates for at least 5 minutes prior to scoring the time between consecutive pboc contractions. 10 consecutive contractions was scored per worm. Levamisole sensitivity assay Worms were transferred to NGM plates supplemented with 200 mm levamisole at room temperature, as done elsewhere [40]. Paralysis was assessed every 20 minutes over the course of 140 minutes by prodding worms with a wire. 20 worms were scored per strain in three independent assays. GFP reporter transgenes The syis63 transgene was used to monitor cog-1 repression in the presence of ectopic lsy-6 expression driven from the otex1450 transgene array expressing cog-1 prom ::lsy-6 hairpin as described in Johnston and Hobert (2003) [14]. This array was chromosomally integrated to generate otis193 (kindly provided by L. Cochella and O. Hobert). RNA Preparation A mix of 1000 late L4 and L4 molt worms were collected for wild type, mir-52, and mir-52/53/54/55/56 mutant worms. Total RNA was prepared using Trizol (Invitrogen) followed by isopropanol precipitation. RNA samples were DNase treated (DNA-free Kit, Ambion). qrt- PCR 10 ng of total RNA was used to analyze the levels of mature mirnas with Applied Biosystems Taqman mirna assays following the manufacturers protocol. Data was analyzed using 2 2DDCt analysis [55] with the mean of U18 and sn2343 as reference. Two technical replicates were performed using two independently isolated total RNA samples for each strain. Each qpcr reaction was performed in triplicate. Student s t-tests were used to statistically compare the fold change of mirna expression relative to wild type. No significant difference was identified between strains, p RNA isolated from N2 worms was used to determine the % efficiency for each PCR assay, which was found to be.95% in each case. Supporting Information Table S1 (PDF) Strains used in this study. Acknowledgments Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR). We thank Luisa Cochella and Oliver Hobert for providing a strain carrying the otis193 chromosomallyintegrated transgene array. Author Contributions Conceived and designed the experiments: JLB BJK ALA. Performed the experiments: JLB BJK. Analyzed the data: JLB BJK ALA. Contributed reagents/materials/analysis tools: JLB BJK. Wrote the paper: JLB ALA. 2. Filipowicz W, Bhattacharyya SN, Sonenberg N (2008) Mechanisms of posttranscriptional regulation by micrornas: are the answers in sight? Nat Rev Genet 9: doi: /nrg2290. PLoS ONE 9 May 2012 Volume 7 Issue 5 e37185
11 3. Djuranovic S, Nahvi A, Green R (2011) A parsimonious model for gene regulation by mirnas. Science 331: doi: /science Wightman B, Ha I, Ruvkun G (1993) Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75: Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, et al. (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditiselegans. Nature 403: doi: / Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, et al. (2005) Regulation by let- 7 and lin-4 mirnas Results in Target mrna Degradation. Cell 122: doi: /j.cell Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: Karres JS, Hilgers V, Carrera I, Treisman J, Cohen SM (2007) The conserved microrna mir-8 tunes atrophin levels to prevent neurodegeneration in Drosophila. Cell 131: doi: /j.cell Mukherji S, Ebert MS, Zheng GXY, Tsang JS, Sharp PA, et al. (2011) MicroRNAs can generate thresholds in target gene expression. Nat Genet 43: doi: /ng Miska EA, Alvarez-Saavedra E, Abbott AL, Lau NC, Hellman AB, et al. (2007) Most Caenorhabditiselegans micrornas are individually not essential for development or viability. PLoS Genet 3: e215. doi: /journal.pgen Abbott AL (2011) Uncovering New Functions for MicroRNAs in Caenorhabditiselegans. Curr Biol 21: R668 R671. doi: /j.cub Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, et al. (2005) The let-7 MicroRNA Family Members mir-48, mir-84, and mir-241 Function Together to Regulate Developmental Timing in Caenorhabditiselegans. Dev Cell 9: doi: /j.devcel Alvarez-Saavedra E, Horvitz HR (2010) Many families of C. elegans micrornas are not essential for development or viability. Curr Biol 20: doi: /j.cub Johnston RJ, Hobert O (2003) A microrna controlling left/right neuronal asymmetry in Caenorhabditiselegans. Nature 426: doi:doi / nature de Lencastre A, Pincus Z, Zhou K, Kato M, Lee SS, et al. (2010) MicroRNAs both promote and antagonize longevity in C. elegans. Curr Biol 20: doi: /j.cub Kato M, Paranjape T, Müller RU, Ullrich R, Nallur S, et al. (2009) The mir-34 microrna is required for the DNA damage response in vivo in C. elegans and in vitro in human breast cancer cells. Oncogene 28: doi: / onc Brenner JL, Jasiewicz KL, Fahley AF, Kemp BJ, Abbott AL (2010) Loss of individual micrornas causes mutant phenotypes in sensitized genetic backgrounds in C. elegans. Curr Biol 20: doi: / j.cub Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, et al. (2001) Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106: Rougvie AE (2001) Control of developmental timing in animals. Nat Rev Genet 2: doi: / Ding L, Spencer A, Morita K, Han M (2005) The developmental timing regulator AIN-1 interacts with miriscs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol Cell 19: doi: /j.molcel Morita K, Han M (2006) Multiple mechanisms are involved in regulating the expression of the developmental timing regulator lin-28 in Caenorhabditiselegans. EMBO J 25: doi: /sj.emboj Hammell CM, Lubin I, Boag PR, Blackwell TK, Ambros V (2009) nhl-2 Modulates microrna activity in Caenorhabditiselegans. Cell 136: doi: /j.cell Ren H, Zhang H (2010) Wnt signaling controls temporal identities of seam cells in Caenorhabditiselegans. Developmental Biology doi: /j.ydbio Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, et al. (2008) Early origins and evolution of micrornas and Piwi-interacting RNAs in animals. Nature 455: doi: /nature Shaw WR, Armisen J, Lehrbach NJ, Miska EA (2010) The conserved mir-51 microrna family is redundantly required for embryonic development and pharynx attachment in Caenorhabditiselegans. Genetics 185: doi: /genetics Kato M, de Lencastre A, Pincus Z, Slack FJ (2009) Dynamic expression of small non-coding RNAs, including novel micrornas and pirnas/21u-rnas, during Caenorhabditiselegans development. Genome Biol 10: R54. doi: /gb r Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, et al. (2003) The micrornas of Caenorhabditiselegans. Genes Dev 17: doi: / gad Ruby JG, Jan C, Player C, Axtell MJ, Lee W, et al. (2006) Large-scale sequencing reveals 21U-RNAs and additional micrornas and endogenous sirnas in C. elegans. Cell 127: doi: /j.cell Martinez NJ, Ow MC, Reece-Hoyes JS, Barrasa MI, Ambros VR, et al. (2008) Genome-scale spatiotemporal analysis of Caenorhabditiselegans microrna promoter activity. Genome Research 18: doi: / gr Pepper AS-R, McCane JE, Kemper K, Yeung DA, Lee RC, et al. (2004) The C. elegans heterochronic gene lin-46 affects developmental timing at two larval stages and encodes a relative of the scaffolding protein gephyrin. Development 131: doi: /dev Nolde MJ, Saka N, Reinert KL, Slack FJ (2007) The Caenorhabditiselegans pumilio homolog, puf-9, is required for the 39UTR-mediated repression of the let-7 microrna target gene, hbl-1. Developmental Biology 305: doi: /j.ydbio Li M, Jones-Rhoades MW, Lau NC, Bartel DP, Rougvie AE (2005) Regulatory mutations of mir-48, a C. elegans let-7 family MicroRNA, cause developmental timing defects. Dev Cell 9: doi: /j.devcel Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, et al. (2003) The Caenorhabditiselegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by micrornas. Dev Cell 4: Lin S-Y, Johnson SM, Abraham M, Vella MC, Pasquinelli A, et al. (2003) The C elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microrna target. Dev Cell 4: Ambros V, Horvitz HR (1987) The lin-14 locus of Caenorhabditiselegans controls the time of expression of specific postembryonic developmental events. Genes Dev 1: Fay DS, Stanley HM, Han M, Wood WB (1999) A Caenorhabditiselegans homologue of hunchback is required for late stages of development but not early embryonic patterning. Developmental Biology 205: doi: / dbio Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, et al. (2005) RAS is regulated by the let-7 microrna family. Cell 120: doi: / j.cell Eisenmann DM, Kim SK (1997) Mechanism of activation of the Caenorhabditiselegans ras homologue let-60 by a novel, temperature-sensitive, gain-offunction mutation. Genetics 146: Thomas JH (1990) Genetic analysis of defecation in Caenorhabditiselegans. Genetics 124: Simon DJ, Madison JM, Conery AL, Thompson-Peer KL, Soskis M, et al. (2008) The MicroRNA mir-1 Regulates a MEF-2-Dependent Retrograde Signal at Neuromuscular Junctions. Cell 133: doi: / j.cell Ruby JG, Jan CH, Bartel DP (2007) Intronic microrna precursors that bypass Drosha processing. Nature 448: doi: /nature Sokol NS, Xu P, Jan YN, Ambros V (2008) Drosophila let-7 microrna is required for remodeling of the neuromusculature during metamorphosis. Genes Dev 22: doi: /gad Sempere LF, Sokol NS, Dubrovsky EB, Berger EM, Ambros V (2003) Temporal regulation of microrna expression in Drosophila melanogaster mediated by hormonal signals and broad-complex gene activity. Developmental Biology 259: Prochnik SE, Rokhsar DS, Aboobaker AA (2007) Evidence for a microrna expansion in the bilaterian ancestor. Dev Genes Evol 217: doi: / s Didiano D, Cochella L, Tursun B, Hobert O (2010) Neuron-type specific regulation of a 39UTR through redundant and combinatorially acting cisregulatory elements. RNA 16: doi: /rna Zhang H, Emmons SW (2009) Regulation of the Caenorhabditiselegans posterior Hox gene egl-5 by microrna and the polycomb-like gene sop-2. Dev Dyn 238: doi: /dvdy Diederichs S, Haber DA (2007) Dual role for argonautes in microrna processing and posttranscriptional regulation of microrna expression. Cell 131: doi: /j.cell Lee RC, Ambros V (2001) An extensive class of small RNAs in Caenorhabditiselegans. Science 294: doi: /science Lau NC, Lim LP, Weinstein EG, Bartel DP (2001) An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditiselegans. Science 294: doi: /science Lund E, Sheets MD, Imboden SB, Dahlberg JE (2011) Limiting Ago protein restricts RNAi and microrna biogenesis during early development in Xenopus laevis. Genes Dev 25: doi: /gad Khan AA, Betel D, Miller ML, Sander C, Leslie CS, et al. (2009) Transfection of small RNAs globally perturbs gene regulation by endogenous micrornas. Nat Biotechnol 27: doi: /nbt Jan CH, Friedman RC, Ruby JG, Bartel DP (2011) Formation, regulation and evolution of Caenorhabditiselegans 39UTRs. Nature 469: doi: / nature Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microrna targets. Cell 120: doi: /j.cell Brenner S (1974) The genetics of Caenorhabditiselegans. Genetics 77: Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: doi: /meth PLoS ONE 10 May 2012 Volume 7 Issue 5 e37185
Report. Loss of Individual MicroRNAs Causes Mutant Phenotypes in Sensitized Genetic Backgrounds in C. elegans
 Current Biology 20, 1321 1325, July 27, 2010 ª2010 Elsevier Ltd All rights reserved DOI 10.1016/j.cub.2010.05.062 Loss of Individual MicroRNAs Causes Mutant Phenotypes in Sensitized Genetic Backgrounds
Current Biology 20, 1321 1325, July 27, 2010 ª2010 Elsevier Ltd All rights reserved DOI 10.1016/j.cub.2010.05.062 Loss of Individual MicroRNAs Causes Mutant Phenotypes in Sensitized Genetic Backgrounds
Noncoding or non-messenger RNAs are
 The functions of animal micrornas Victor Ambros Dartmouth Medical School, Department of Genetics, Hanover, New Hampshire 03755, USA (e-mail: vra@dartmouth.edu) MicroRNAs (mirnas) are small RNAs that regulate
The functions of animal micrornas Victor Ambros Dartmouth Medical School, Department of Genetics, Hanover, New Hampshire 03755, USA (e-mail: vra@dartmouth.edu) MicroRNAs (mirnas) are small RNAs that regulate
he micrornas of Caenorhabditis elegans (Lim et al. Genes & Development 2003)
 MicroRNAs: Genomics, Biogenesis, Mechanism, and Function (D. Bartel Cell 2004) he micrornas of Caenorhabditis elegans (Lim et al. Genes & Development 2003) Vertebrate MicroRNA Genes (Lim et al. Science
MicroRNAs: Genomics, Biogenesis, Mechanism, and Function (D. Bartel Cell 2004) he micrornas of Caenorhabditis elegans (Lim et al. Genes & Development 2003) Vertebrate MicroRNA Genes (Lim et al. Science
Improved annotation of C. elegans micrornas by deep sequencing reveals structures associated with processing by Drosha and Dicer
 BIOINFORMATICS Improved annotation of C. elegans micrornas by deep sequencing reveals structures associated with processing by Drosha and Dicer M. BRYAN WARF, 1 W. EVAN JOHNSON, 2 and BRENDA L. BASS 1
BIOINFORMATICS Improved annotation of C. elegans micrornas by deep sequencing reveals structures associated with processing by Drosha and Dicer M. BRYAN WARF, 1 W. EVAN JOHNSON, 2 and BRENDA L. BASS 1
MicroRNA expression profiling and functional analysis in prostate cancer. Marco Folini s.c. Ricerca Traslazionale DOSL
 MicroRNA expression profiling and functional analysis in prostate cancer Marco Folini s.c. Ricerca Traslazionale DOSL What are micrornas? For almost three decades, the alteration of protein-coding genes
MicroRNA expression profiling and functional analysis in prostate cancer Marco Folini s.c. Ricerca Traslazionale DOSL What are micrornas? For almost three decades, the alteration of protein-coding genes
MicroRNA and Male Infertility: A Potential for Diagnosis
 Review Article MicroRNA and Male Infertility: A Potential for Diagnosis * Abstract MicroRNAs (mirnas) are small non-coding single stranded RNA molecules that are physiologically produced in eukaryotic
Review Article MicroRNA and Male Infertility: A Potential for Diagnosis * Abstract MicroRNAs (mirnas) are small non-coding single stranded RNA molecules that are physiologically produced in eukaryotic
Circular RNAs (circrnas) act a stable mirna sponges
 Circular RNAs (circrnas) act a stable mirna sponges cernas compete for mirnas Ancestal mrna (+3 UTR) Pseudogene RNA (+3 UTR homolgy region) The model holds true for all RNAs that share a mirna binding
Circular RNAs (circrnas) act a stable mirna sponges cernas compete for mirnas Ancestal mrna (+3 UTR) Pseudogene RNA (+3 UTR homolgy region) The model holds true for all RNAs that share a mirna binding
The Biology and Genetics of Cells and Organisms The Biology of Cancer
 The Biology and Genetics of Cells and Organisms The Biology of Cancer Mendel and Genetics How many distinct genes are present in the genomes of mammals? - 21,000 for human. - Genetic information is carried
The Biology and Genetics of Cells and Organisms The Biology of Cancer Mendel and Genetics How many distinct genes are present in the genomes of mammals? - 21,000 for human. - Genetic information is carried
Bi 8 Lecture 17. interference. Ellen Rothenberg 1 March 2016
 Bi 8 Lecture 17 REGulation by RNA interference Ellen Rothenberg 1 March 2016 Protein is not the only regulatory molecule affecting gene expression: RNA itself can be negative regulator RNA does not need
Bi 8 Lecture 17 REGulation by RNA interference Ellen Rothenberg 1 March 2016 Protein is not the only regulatory molecule affecting gene expression: RNA itself can be negative regulator RNA does not need
MicroRNAs, RNA Modifications, RNA Editing. Bora E. Baysal MD, PhD Oncology for Scientists Lecture Tue, Oct 17, 2017, 3:30 PM - 5:00 PM
 MicroRNAs, RNA Modifications, RNA Editing Bora E. Baysal MD, PhD Oncology for Scientists Lecture Tue, Oct 17, 2017, 3:30 PM - 5:00 PM Expanding world of RNAs mrna, messenger RNA (~20,000) trna, transfer
MicroRNAs, RNA Modifications, RNA Editing Bora E. Baysal MD, PhD Oncology for Scientists Lecture Tue, Oct 17, 2017, 3:30 PM - 5:00 PM Expanding world of RNAs mrna, messenger RNA (~20,000) trna, transfer
mirna Dr. S Hosseini-Asl
 mirna Dr. S Hosseini-Asl 1 2 MicroRNAs (mirnas) are small noncoding RNAs which enhance the cleavage or translational repression of specific mrna with recognition site(s) in the 3 - untranslated region
mirna Dr. S Hosseini-Asl 1 2 MicroRNAs (mirnas) are small noncoding RNAs which enhance the cleavage or translational repression of specific mrna with recognition site(s) in the 3 - untranslated region
Cross species analysis of genomics data. Computational Prediction of mirnas and their targets
 02-716 Cross species analysis of genomics data Computational Prediction of mirnas and their targets Outline Introduction Brief history mirna Biogenesis Why Computational Methods? Computational Methods
02-716 Cross species analysis of genomics data Computational Prediction of mirnas and their targets Outline Introduction Brief history mirna Biogenesis Why Computational Methods? Computational Methods
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10643 Supplementary Table 1. Identification of hecw-1 coding polymorphisms at amino acid positions 322 and 325 in 162 strains of C. elegans. WWW.NATURE.COM/NATURE 1 Supplementary Figure
doi:10.1038/nature10643 Supplementary Table 1. Identification of hecw-1 coding polymorphisms at amino acid positions 322 and 325 in 162 strains of C. elegans. WWW.NATURE.COM/NATURE 1 Supplementary Figure
Chapter 2. Investigation into mir-346 Regulation of the nachr α5 Subunit
 15 Chapter 2 Investigation into mir-346 Regulation of the nachr α5 Subunit MicroRNA s (mirnas) are small (< 25 base pairs), single stranded, non-coding RNAs that regulate gene expression at the post transcriptional
15 Chapter 2 Investigation into mir-346 Regulation of the nachr α5 Subunit MicroRNA s (mirnas) are small (< 25 base pairs), single stranded, non-coding RNAs that regulate gene expression at the post transcriptional
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1171320/dc1 Supporting Online Material for A Frazzled/DCC-Dependent Transcriptional Switch Regulates Midline Axon Guidance Long Yang, David S. Garbe, Greg J. Bashaw*
www.sciencemag.org/cgi/content/full/1171320/dc1 Supporting Online Material for A Frazzled/DCC-Dependent Transcriptional Switch Regulates Midline Axon Guidance Long Yang, David S. Garbe, Greg J. Bashaw*
MicroRNA function in animal development
 FEBS 29869 FEBS Letters xxx (2005) xxx xxx Minireview MicroRNA function in animal development Erno Wienholds, Ronald H.A. Plasterk * Hubrecht Laboratory, Uppsalalaan 8, 3584 CT Utrecht, The Netherlands
FEBS 29869 FEBS Letters xxx (2005) xxx xxx Minireview MicroRNA function in animal development Erno Wienholds, Ronald H.A. Plasterk * Hubrecht Laboratory, Uppsalalaan 8, 3584 CT Utrecht, The Netherlands
Programmed Cell Death (apoptosis)
 Programmed Cell Death (apoptosis) Stereotypic death process includes: membrane blebbing nuclear fragmentation chromatin condensation and DNA framentation loss of mitochondrial integrity and release of
Programmed Cell Death (apoptosis) Stereotypic death process includes: membrane blebbing nuclear fragmentation chromatin condensation and DNA framentation loss of mitochondrial integrity and release of
CRS4 Seminar series. Inferring the functional role of micrornas from gene expression data CRS4. Biomedicine. Bioinformatics. Paolo Uva July 11, 2012
 CRS4 Seminar series Inferring the functional role of micrornas from gene expression data CRS4 Biomedicine Bioinformatics Paolo Uva July 11, 2012 Partners Pharmaceutical company Fondazione San Raffaele,
CRS4 Seminar series Inferring the functional role of micrornas from gene expression data CRS4 Biomedicine Bioinformatics Paolo Uva July 11, 2012 Partners Pharmaceutical company Fondazione San Raffaele,
Phenomena first observed in petunia
 Vectors for RNAi Phenomena first observed in petunia Attempted to overexpress chalone synthase (anthrocyanin pigment gene) in petunia. (trying to darken flower color) Caused the loss of pigment. Bill Douherty
Vectors for RNAi Phenomena first observed in petunia Attempted to overexpress chalone synthase (anthrocyanin pigment gene) in petunia. (trying to darken flower color) Caused the loss of pigment. Bill Douherty
Most Caenorhabditis elegans micrornas Are Individually Not Essential for Development or Viability
 Most Caenorhabditis elegans micrornas Are Individually Not Essential for Development or Viability Eric A. Miska 1,2 a[, Ezequiel Alvarez-Saavedra 1,2[, Allison L. Abbott 3[ b, Nelson C. Lau 4 c, Andrew
Most Caenorhabditis elegans micrornas Are Individually Not Essential for Development or Viability Eric A. Miska 1,2 a[, Ezequiel Alvarez-Saavedra 1,2[, Allison L. Abbott 3[ b, Nelson C. Lau 4 c, Andrew
MicroRNA-mediated incoherent feedforward motifs are robust
 The Second International Symposium on Optimization and Systems Biology (OSB 8) Lijiang, China, October 31 November 3, 8 Copyright 8 ORSC & APORC, pp. 62 67 MicroRNA-mediated incoherent feedforward motifs
The Second International Symposium on Optimization and Systems Biology (OSB 8) Lijiang, China, October 31 November 3, 8 Copyright 8 ORSC & APORC, pp. 62 67 MicroRNA-mediated incoherent feedforward motifs
(,, ) microrna(mirna) 19~25 nt RNA, RNA mirna mirna,, ;, mirna mirna. : microrna (mirna); ; ; ; : R321.1 : A : X(2015)
 35 1 Vol.35 No.1 2015 1 Jan. 2015 Reproduction & Contraception doi: 10.7669/j.issn.0253-357X.2015.01.0037 E-mail: randc_journal@163.com microrna ( 450052) microrna() 19~25 nt RNA RNA ; : microrna (); ;
35 1 Vol.35 No.1 2015 1 Jan. 2015 Reproduction & Contraception doi: 10.7669/j.issn.0253-357X.2015.01.0037 E-mail: randc_journal@163.com microrna ( 450052) microrna() 19~25 nt RNA RNA ; : microrna (); ;
SUPPLEMENTARY FIGURE S1: nlp-22 is expressed in the RIA interneurons and is secreted. (a) An animal expressing both the RIA specific reporter
 1 SUPPLEMENTARY FIGURE S1: nlp-22 is expressed in the RIA interneurons and is secreted. (a) An animal expressing both the RIA specific reporter Pglr-3:mCherry (red) and Pnlp-22:gfp (green) shows co-localization
1 SUPPLEMENTARY FIGURE S1: nlp-22 is expressed in the RIA interneurons and is secreted. (a) An animal expressing both the RIA specific reporter Pglr-3:mCherry (red) and Pnlp-22:gfp (green) shows co-localization
micrornas (mirna) and Biomarkers
 micrornas (mirna) and Biomarkers Small RNAs Make Big Splash mirnas & Genome Function Biomarkers in Cancer Future Prospects Javed Khan M.D. National Cancer Institute EORTC-NCI-ASCO November 2007 The Human
micrornas (mirna) and Biomarkers Small RNAs Make Big Splash mirnas & Genome Function Biomarkers in Cancer Future Prospects Javed Khan M.D. National Cancer Institute EORTC-NCI-ASCO November 2007 The Human
Overview: Conducting the Genetic Orchestra Prokaryotes and eukaryotes alter gene expression in response to their changing environment
 Overview: Conducting the Genetic Orchestra Prokaryotes and eukaryotes alter gene expression in response to their changing environment In multicellular eukaryotes, gene expression regulates development
Overview: Conducting the Genetic Orchestra Prokaryotes and eukaryotes alter gene expression in response to their changing environment In multicellular eukaryotes, gene expression regulates development
micrornas Under the Microscope
 DEVELOPMENTAL DYNAMICS 235:846 853, 2006 WHAT S COOL IN DEV BIO micrornas Under the Microscope Julie C. Kiefer* Once regarded as a biological anomaly, micrornas (mirnas) have since been recognized as a
DEVELOPMENTAL DYNAMICS 235:846 853, 2006 WHAT S COOL IN DEV BIO micrornas Under the Microscope Julie C. Kiefer* Once regarded as a biological anomaly, micrornas (mirnas) have since been recognized as a
Impact of mirna Sequence on mirna Expression and Correlation between mirna Expression and Cell Cycle Regulation in Breast Cancer Cells
 Impact of mirna Sequence on mirna Expression and Correlation between mirna Expression and Cell Cycle Regulation in Breast Cancer Cells Zijun Luo 1 *, Yi Zhao 1, Robert Azencott 2 1 Harbin Institute of
Impact of mirna Sequence on mirna Expression and Correlation between mirna Expression and Cell Cycle Regulation in Breast Cancer Cells Zijun Luo 1 *, Yi Zhao 1, Robert Azencott 2 1 Harbin Institute of
Mutations in the Caenorhabditis elegans U2AF Large Subunit UAF-1 Al= of a 3' Splice Site In Vivo
 Mutations in the Caenorhabditis elegans U2AF Large Subunit UAF-1 Al= of a 3' Splice Site In Vivo The MIT Faculty has made this article openly available. Please share how this access benefits you. Your
Mutations in the Caenorhabditis elegans U2AF Large Subunit UAF-1 Al= of a 3' Splice Site In Vivo The MIT Faculty has made this article openly available. Please share how this access benefits you. Your
Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells.
 SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression.
 Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
mir-355 Functions as An Important Link between p38 MAPK Signaling and Insulin Signaling in the Regulation of Innate Immunity
 www.nature.com/scientificreports Received: 7 July 2017 Accepted: 24 October 2017 Published: xx xx xxxx OPEN mir-355 Functions as An Important Link between p38 MAPK Signaling and Insulin Signaling in the
www.nature.com/scientificreports Received: 7 July 2017 Accepted: 24 October 2017 Published: xx xx xxxx OPEN mir-355 Functions as An Important Link between p38 MAPK Signaling and Insulin Signaling in the
klp-18 (RNAi) Control. supplementary information. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach
![klp-18 (RNAi) Control. supplementary information. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach klp-18 (RNAi) Control. supplementary information. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach](/thumbs/91/104639484.jpg) DOI: 10.1038/ncb1891 A. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach embryos let hatch overnight transfer to RNAi plates; incubate 5 days at 15 C RNAi food L1 worms adult worms
DOI: 10.1038/ncb1891 A. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach embryos let hatch overnight transfer to RNAi plates; incubate 5 days at 15 C RNAi food L1 worms adult worms
MicroRNAs Modulate Hematopoietic Lineage Differentiation
 MicroRNAs Modulate Hematopoietic Lineage Differentiation Harvey F. Lodish, Chang-Zheng Chen, David P. Bartel Whitehead Institute for Biomedical Research, Department of Biology, Massachusetts Institute
MicroRNAs Modulate Hematopoietic Lineage Differentiation Harvey F. Lodish, Chang-Zheng Chen, David P. Bartel Whitehead Institute for Biomedical Research, Department of Biology, Massachusetts Institute
Research Article Base Composition Characteristics of Mammalian mirnas
 Journal of Nucleic Acids Volume 2013, Article ID 951570, 6 pages http://dx.doi.org/10.1155/2013/951570 Research Article Base Composition Characteristics of Mammalian mirnas Bin Wang Department of Chemistry,
Journal of Nucleic Acids Volume 2013, Article ID 951570, 6 pages http://dx.doi.org/10.1155/2013/951570 Research Article Base Composition Characteristics of Mammalian mirnas Bin Wang Department of Chemistry,
Supplementary Figure 1
 Supplementary Figure 1 Asymmetrical function of 5p and 3p arms of mir-181 and mir-30 families and mir-142 and mir-154. (a) Control experiments using mirna sensor vector and empty pri-mirna overexpression
Supplementary Figure 1 Asymmetrical function of 5p and 3p arms of mir-181 and mir-30 families and mir-142 and mir-154. (a) Control experiments using mirna sensor vector and empty pri-mirna overexpression
Detection of let-7a MicroRNA by Real-time PCR in Colorectal Cancer: a Single-centre Experience from China
 The Journal of International Medical Research 2007; 35: 716 723 Detection of let-7a MicroRNA by Real-time PCR in Colorectal Cancer: a Single-centre Experience from China W-J FANG 1, C-Z LIN 2, H-H ZHANG
The Journal of International Medical Research 2007; 35: 716 723 Detection of let-7a MicroRNA by Real-time PCR in Colorectal Cancer: a Single-centre Experience from China W-J FANG 1, C-Z LIN 2, H-H ZHANG
Src-INACTIVE / Src-INACTIVE
 Biology 169 -- Exam 1 February 2003 Answer each question, noting carefully the instructions for each. Repeat- Read the instructions for each question before answering!!! Be as specific as possible in each
Biology 169 -- Exam 1 February 2003 Answer each question, noting carefully the instructions for each. Repeat- Read the instructions for each question before answering!!! Be as specific as possible in each
Correspondence: mirna regulation of Sdf1 chemokine signaling provides genetic robustness to germ cell migration
 Correspondence: mirna regulation of Sdf1 chemokine signaling provides genetic robustness to germ cell migration Alison A. Staton, Holger Knaut, and Antonio J. Giraldez Supplementary Note Materials and
Correspondence: mirna regulation of Sdf1 chemokine signaling provides genetic robustness to germ cell migration Alison A. Staton, Holger Knaut, and Antonio J. Giraldez Supplementary Note Materials and
SUPPLEMENTARY FIGURE LEGENDS
 SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1 Negative correlation between mir-375 and its predicted target genes, as demonstrated by gene set enrichment analysis (GSEA). 1 The correlation between
SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1 Negative correlation between mir-375 and its predicted target genes, as demonstrated by gene set enrichment analysis (GSEA). 1 The correlation between
MicroRNAs and Cancer
 MicroRNAs and Cancer MicroRNAs are an abundant class of small (20-25 nucleotides) non-protein coding RNAs that function as negative gene regulators. The human genome contains up to 1000 micrornas which
MicroRNAs and Cancer MicroRNAs are an abundant class of small (20-25 nucleotides) non-protein coding RNAs that function as negative gene regulators. The human genome contains up to 1000 micrornas which
Molecular Mechanism of MicroRNA-203 Mediated Epidermal Stem Cell Differentiation
 University of Colorado, Boulder CU Scholar Undergraduate Honors Theses Honors Program Spring 2011 Molecular Mechanism of MicroRNA-203 Mediated Epidermal Stem Cell Differentiation Sarah Jackson University
University of Colorado, Boulder CU Scholar Undergraduate Honors Theses Honors Program Spring 2011 Molecular Mechanism of MicroRNA-203 Mediated Epidermal Stem Cell Differentiation Sarah Jackson University
Implications of microrna-197 downregulated expression in esophageal cancer with poor prognosis
 Implications of microrna-197 downregulated expression in esophageal cancer with poor prognosis T.-Y. Wang 1, S.-G. Liu 2, B.-S. Zhao 2, B. Qi 2, X.-G. Qin 2 and W.-J. Yao 2 1 Department of Biochemistry
Implications of microrna-197 downregulated expression in esophageal cancer with poor prognosis T.-Y. Wang 1, S.-G. Liu 2, B.-S. Zhao 2, B. Qi 2, X.-G. Qin 2 and W.-J. Yao 2 1 Department of Biochemistry
Cellular MicroRNA and P Bodies Modulate Host-HIV-1 Interactions. 指導教授 : 張麗冠博士 演講者 : 黃柄翰 Date: 2009/10/19
 Cellular MicroRNA and P Bodies Modulate Host-HIV-1 Interactions 指導教授 : 張麗冠博士 演講者 : 黃柄翰 Date: 2009/10/19 1 MicroRNA biogenesis 1. Pri mirna: primary mirna 2. Drosha: RNaseIII 3. DCR 1: Dicer 1 RNaseIII
Cellular MicroRNA and P Bodies Modulate Host-HIV-1 Interactions 指導教授 : 張麗冠博士 演講者 : 黃柄翰 Date: 2009/10/19 1 MicroRNA biogenesis 1. Pri mirna: primary mirna 2. Drosha: RNaseIII 3. DCR 1: Dicer 1 RNaseIII
RNA interference induced hepatotoxicity results from loss of the first synthesized isoform of microrna-122 in mice
 SUPPLEMENTARY INFORMATION RNA interference induced hepatotoxicity results from loss of the first synthesized isoform of microrna-122 in mice Paul N Valdmanis, Shuo Gu, Kirk Chu, Lan Jin, Feijie Zhang,
SUPPLEMENTARY INFORMATION RNA interference induced hepatotoxicity results from loss of the first synthesized isoform of microrna-122 in mice Paul N Valdmanis, Shuo Gu, Kirk Chu, Lan Jin, Feijie Zhang,
Novel RNAs along the Pathway of Gene Expression. (or, The Expanding Universe of Small RNAs)
 Novel RNAs along the Pathway of Gene Expression (or, The Expanding Universe of Small RNAs) Central Dogma DNA RNA Protein replication transcription translation Central Dogma DNA RNA Spliced RNA Protein
Novel RNAs along the Pathway of Gene Expression (or, The Expanding Universe of Small RNAs) Central Dogma DNA RNA Protein replication transcription translation Central Dogma DNA RNA Spliced RNA Protein
Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v)
 SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
High AU content: a signature of upregulated mirna in cardiac diseases
 https://helda.helsinki.fi High AU content: a signature of upregulated mirna in cardiac diseases Gupta, Richa 2010-09-20 Gupta, R, Soni, N, Patnaik, P, Sood, I, Singh, R, Rawal, K & Rani, V 2010, ' High
https://helda.helsinki.fi High AU content: a signature of upregulated mirna in cardiac diseases Gupta, Richa 2010-09-20 Gupta, R, Soni, N, Patnaik, P, Sood, I, Singh, R, Rawal, K & Rani, V 2010, ' High
Post-transcriptional regulation of an intronic microrna
 Post-transcriptional regulation of an intronic microrna Carl Novina Dana-Farber Cancer Institute Harvard Medical School Broad Institute of Harvard and MIT Qiagen Webinar 05-17-11 Outline 1. The biology
Post-transcriptional regulation of an intronic microrna Carl Novina Dana-Farber Cancer Institute Harvard Medical School Broad Institute of Harvard and MIT Qiagen Webinar 05-17-11 Outline 1. The biology
Small RNAs and how to analyze them using sequencing
 Small RNAs and how to analyze them using sequencing Jakub Orzechowski Westholm (1) Long- term bioinforma=cs support, Science For Life Laboratory Stockholm (2) Department of Biophysics and Biochemistry,
Small RNAs and how to analyze them using sequencing Jakub Orzechowski Westholm (1) Long- term bioinforma=cs support, Science For Life Laboratory Stockholm (2) Department of Biophysics and Biochemistry,
Problem Set 5 KEY
 2006 7.012 Problem Set 5 KEY ** Due before 5 PM on THURSDAY, November 9, 2006. ** Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. You are studying the development
2006 7.012 Problem Set 5 KEY ** Due before 5 PM on THURSDAY, November 9, 2006. ** Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. You are studying the development
C. elegans Embryonic Development
 Autonomous Specification in Tunicate development Autonomous & Conditional Specification in C. elegans Embryonic Development Figure 8.36 Bilateral Symmetry in the Egg of the Tunicate Styela partita Fig.
Autonomous Specification in Tunicate development Autonomous & Conditional Specification in C. elegans Embryonic Development Figure 8.36 Bilateral Symmetry in the Egg of the Tunicate Styela partita Fig.
A Genetic Program for Embryonic Development
 Concept 18.4: A program of differential gene expression leads to the different cell types in a multicellular organism During embryonic development, a fertilized egg gives rise to many different cell types
Concept 18.4: A program of differential gene expression leads to the different cell types in a multicellular organism During embryonic development, a fertilized egg gives rise to many different cell types
Repressive Transcription
 Repressive Transcription The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation As Published Publisher Guenther, M. G., and R. A.
Repressive Transcription The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation As Published Publisher Guenther, M. G., and R. A.
MicroRNA dysregulation in cancer. Systems Plant Microbiology Hyun-Hee Lee
 MicroRNA dysregulation in cancer Systems Plant Microbiology Hyun-Hee Lee Contents 1 What is MicroRNA? 2 mirna dysregulation in cancer 3 Summary What is MicroRNA? What is MicroRNA? MicroRNAs (mirnas) -
MicroRNA dysregulation in cancer Systems Plant Microbiology Hyun-Hee Lee Contents 1 What is MicroRNA? 2 mirna dysregulation in cancer 3 Summary What is MicroRNA? What is MicroRNA? MicroRNAs (mirnas) -
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS)
 Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Computational Analysis of mirna and Target mrna Interactions: Combined Effects of The Quantity and Quality of Their Binding Sites *
 www.pibb.ac.cn!"#$%!""&'( Progress in Biochemistry and Biophysics 2009, 36(5): 608~615 Computational Analysis of mirna and Target mrna Interactions: Combined Effects of The Quantity and Quality of Their
www.pibb.ac.cn!"#$%!""&'( Progress in Biochemistry and Biophysics 2009, 36(5): 608~615 Computational Analysis of mirna and Target mrna Interactions: Combined Effects of The Quantity and Quality of Their
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10495 WWW.NATURE.COM/NATURE 1 2 WWW.NATURE.COM/NATURE WWW.NATURE.COM/NATURE 3 4 WWW.NATURE.COM/NATURE WWW.NATURE.COM/NATURE 5 6 WWW.NATURE.COM/NATURE WWW.NATURE.COM/NATURE 7 8 WWW.NATURE.COM/NATURE
doi:10.1038/nature10495 WWW.NATURE.COM/NATURE 1 2 WWW.NATURE.COM/NATURE WWW.NATURE.COM/NATURE 3 4 WWW.NATURE.COM/NATURE WWW.NATURE.COM/NATURE 5 6 WWW.NATURE.COM/NATURE WWW.NATURE.COM/NATURE 7 8 WWW.NATURE.COM/NATURE
MicroRNA in Cancer Karen Dybkær 2013
 MicroRNA in Cancer Karen Dybkær RNA Ribonucleic acid Types -Coding: messenger RNA (mrna) coding for proteins -Non-coding regulating protein formation Ribosomal RNA (rrna) Transfer RNA (trna) Small nuclear
MicroRNA in Cancer Karen Dybkær RNA Ribonucleic acid Types -Coding: messenger RNA (mrna) coding for proteins -Non-coding regulating protein formation Ribosomal RNA (rrna) Transfer RNA (trna) Small nuclear
Lesson Flowchart. Why Gene Expression Regulation?; microrna in Gene Regulation: an overview; microrna Genomics and Biogenesis;
 micrornas Lesson Flowchart Why Gene Expression Regulation?; microrna in Gene Regulation: an overview; microrna Genomics and Biogenesis; How do micrornas function? micrornas and complex cellular circuits;
micrornas Lesson Flowchart Why Gene Expression Regulation?; microrna in Gene Regulation: an overview; microrna Genomics and Biogenesis; How do micrornas function? micrornas and complex cellular circuits;
Identification of mirnas in Eucalyptus globulus Plant by Computational Methods
 International Journal of Pharmaceutical Science Invention ISSN (Online): 2319 6718, ISSN (Print): 2319 670X Volume 2 Issue 5 May 2013 PP.70-74 Identification of mirnas in Eucalyptus globulus Plant by Computational
International Journal of Pharmaceutical Science Invention ISSN (Online): 2319 6718, ISSN (Print): 2319 670X Volume 2 Issue 5 May 2013 PP.70-74 Identification of mirnas in Eucalyptus globulus Plant by Computational
Nature Genetics: doi: /ng Supplementary Figure 1. Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms.
 Supplementary Figure 1 Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms. IF images of wild-type (wt) and met-2 set-25 worms showing the loss of H3K9me2/me3 at the indicated developmental
Supplementary Figure 1 Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms. IF images of wild-type (wt) and met-2 set-25 worms showing the loss of H3K9me2/me3 at the indicated developmental
Strategies to determine the biological function of micrornas
 Strategies to determine the biological function of micrornas Jan Krützfeldt, Matthew N Poy & Markus Stoffel MicroRNAs (mirnas) are regulators of gene expression that control many biological processes in
Strategies to determine the biological function of micrornas Jan Krützfeldt, Matthew N Poy & Markus Stoffel MicroRNAs (mirnas) are regulators of gene expression that control many biological processes in
Early Embryonic Development
 Early Embryonic Development Maternal effect gene products set the stage by controlling the expression of the first embryonic genes. 1. Transcription factors 2. Receptors 3. Regulatory proteins Maternal
Early Embryonic Development Maternal effect gene products set the stage by controlling the expression of the first embryonic genes. 1. Transcription factors 2. Receptors 3. Regulatory proteins Maternal
PREPARED FOR: U.S. Army Medical Research and Materiel Command Fort Detrick, Maryland
 AD Award Number: W81XWH-10-1-1029 TITLE: PRINCIPAL INVESTIGATOR: Mu-Shui Dai, M.D., Ph.D. CONTRACTING ORGANIZATION: Oregon Health Science niversity, Portland, Oregon 97239 REPORT DATE: October 2013 TYPE
AD Award Number: W81XWH-10-1-1029 TITLE: PRINCIPAL INVESTIGATOR: Mu-Shui Dai, M.D., Ph.D. CONTRACTING ORGANIZATION: Oregon Health Science niversity, Portland, Oregon 97239 REPORT DATE: October 2013 TYPE
Genetic suppressors and enhancers provide clues to gene regulation and genetic pathways
 Genetic suppressors and enhancers provide clues to gene regulation and genetic pathways Suppressor mutation: a second mutation results in a less severe phenotype than the original mutation Suppressor mutations
Genetic suppressors and enhancers provide clues to gene regulation and genetic pathways Suppressor mutation: a second mutation results in a less severe phenotype than the original mutation Suppressor mutations
Ch. 18 Regulation of Gene Expression
 Ch. 18 Regulation of Gene Expression 1 Human genome has around 23,688 genes (Scientific American 2/2006) Essential Questions: How is transcription regulated? How are genes expressed? 2 Bacteria regulate
Ch. 18 Regulation of Gene Expression 1 Human genome has around 23,688 genes (Scientific American 2/2006) Essential Questions: How is transcription regulated? How are genes expressed? 2 Bacteria regulate
Vertebrate Limb Patterning
 Vertebrate Limb Patterning What makes limb patterning an interesting/useful developmental system How limbs develop Key events in limb development positioning and specification initiation of outgrowth establishment
Vertebrate Limb Patterning What makes limb patterning an interesting/useful developmental system How limbs develop Key events in limb development positioning and specification initiation of outgrowth establishment
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells
 MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
SUPPLEMENTARY INFORMATION
 Supplementary Figure 1. Formation of the AA5x. a, Camera lucida drawing of embryo at 48 hours post fertilization (hpf, modified from Kimmel et al. Dev Dyn. 1995 203:253-310). b, Confocal microangiogram
Supplementary Figure 1. Formation of the AA5x. a, Camera lucida drawing of embryo at 48 hours post fertilization (hpf, modified from Kimmel et al. Dev Dyn. 1995 203:253-310). b, Confocal microangiogram
MicroRNAs: novel regulators in skin research
 MicroRNAs: novel regulators in skin research Eniko Sonkoly, Andor Pivarcsi KI, Department of Medicine, Unit of Dermatology and Venerology What are micrornas? Small, ~21-mer RNAs 1993: The first mirna discovered,
MicroRNAs: novel regulators in skin research Eniko Sonkoly, Andor Pivarcsi KI, Department of Medicine, Unit of Dermatology and Venerology What are micrornas? Small, ~21-mer RNAs 1993: The first mirna discovered,
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/3/8/e1701143/dc1 Supplementary Materials for Impaired DNA replication derepresses chromatin and generates a transgenerationally inherited epigenetic memory Adam
advances.sciencemag.org/cgi/content/full/3/8/e1701143/dc1 Supplementary Materials for Impaired DNA replication derepresses chromatin and generates a transgenerationally inherited epigenetic memory Adam
RAS Is Regulated by the let-7 MicroRNA Family
 Cell, Vol. 120, 635 647, March 11, 2005, Copyright 2005 by Elsevier Inc. DOI 10.1016/j.cell.2005.01.014 RAS Is Regulated by the let-7 MicroRNA Family Steven M. Johnson, 1 Helge Grosshans, 1 expression
Cell, Vol. 120, 635 647, March 11, 2005, Copyright 2005 by Elsevier Inc. DOI 10.1016/j.cell.2005.01.014 RAS Is Regulated by the let-7 MicroRNA Family Steven M. Johnson, 1 Helge Grosshans, 1 expression
10/31/2017. micrornas and cancer. From the one gene-one enzyme hypothesis to. microrna DNA RNA. Transcription factors.
 micrornas and cancer Cellular and Molecular Biology of Cancer (PATH G4500-001) November 1 st, 2017 -Katia Basso- Columbia University Katia Basso, PhD Office: ICRC RM506 E-mail: kb451@cumc.columbia.edu
micrornas and cancer Cellular and Molecular Biology of Cancer (PATH G4500-001) November 1 st, 2017 -Katia Basso- Columbia University Katia Basso, PhD Office: ICRC RM506 E-mail: kb451@cumc.columbia.edu
Alternative Splicing and Genomic Stability
 Alternative Splicing and Genomic Stability Kevin Cahill cahill@unm.edu http://dna.phys.unm.edu/ Abstract In a cell that uses alternative splicing, the total length of all the exons is far less than in
Alternative Splicing and Genomic Stability Kevin Cahill cahill@unm.edu http://dna.phys.unm.edu/ Abstract In a cell that uses alternative splicing, the total length of all the exons is far less than in
UC San Diego UC San Diego Electronic Theses and Dissertations
 UC San Diego UC San Diego Electronic Theses and Dissertations Title MicroRNAs in the Making: Post-transcriptional Regulation of MicroRNA Biogenesis in Caenorhabditis elegans Permalink https://escholarship.org/uc/item/3f93318b
UC San Diego UC San Diego Electronic Theses and Dissertations Title MicroRNAs in the Making: Post-transcriptional Regulation of MicroRNA Biogenesis in Caenorhabditis elegans Permalink https://escholarship.org/uc/item/3f93318b
Study of mirna based gene regulation involved in Squamous Lung Carcinoma by assistance of Argonaute Protein
 January - February 2018; 7(1): 2913-2917 International Journal of Research and Development in Pharmacy & Life Science An International open access peer reviewed journal ISSN (P): 2393-932X, ISSN (E): 2278-0238
January - February 2018; 7(1): 2913-2917 International Journal of Research and Development in Pharmacy & Life Science An International open access peer reviewed journal ISSN (P): 2393-932X, ISSN (E): 2278-0238
Concentration memory-dependent synaptic plasticity of a taste circuit regulates salt chemotaxis in Caenorhabditis elegans
 Supplementary Information for: Concentration memory-dependent synaptic plasticity of a taste circuit regulates salt chemotaxis in Caenorhabditis elegans Hirofumi Kunitomo, Hirofumi Sato, Ryo Iwata, Yohsuke
Supplementary Information for: Concentration memory-dependent synaptic plasticity of a taste circuit regulates salt chemotaxis in Caenorhabditis elegans Hirofumi Kunitomo, Hirofumi Sato, Ryo Iwata, Yohsuke
Chapter 11. How Genes Are Controlled. Lectures by Edward J. Zalisko
 Chapter 11 How Genes Are Controlled PowerPoint Lectures for Campbell Essential Biology, Fifth Edition, and Campbell Essential Biology with Physiology, Fourth Edition Eric J. Simon, Jean L. Dickey, and
Chapter 11 How Genes Are Controlled PowerPoint Lectures for Campbell Essential Biology, Fifth Edition, and Campbell Essential Biology with Physiology, Fourth Edition Eric J. Simon, Jean L. Dickey, and
Regulation of Gene Expression in Eukaryotes
 Ch. 19 Regulation of Gene Expression in Eukaryotes BIOL 222 Differential Gene Expression in Eukaryotes Signal Cells in a multicellular eukaryotic organism genetically identical differential gene expression
Ch. 19 Regulation of Gene Expression in Eukaryotes BIOL 222 Differential Gene Expression in Eukaryotes Signal Cells in a multicellular eukaryotic organism genetically identical differential gene expression
CONTRACTING ORGANIZATION: Baylor College of Medicine Houston, TX 77030
 AD Award Number: W81XWH-05-1-0428 TITLE: MicroRNA and Breast Cancer Progression PRINCIPAL INVESTIGATOR: Konstantin Galaktionov, Ph.D. CONTRACTING ORGANIZATION: Baylor College of Medicine Houston, TX 77030
AD Award Number: W81XWH-05-1-0428 TITLE: MicroRNA and Breast Cancer Progression PRINCIPAL INVESTIGATOR: Konstantin Galaktionov, Ph.D. CONTRACTING ORGANIZATION: Baylor College of Medicine Houston, TX 77030
Prediction of micrornas and their targets
 Prediction of micrornas and their targets Introduction Brief history mirna Biogenesis Computational Methods Mature and precursor mirna prediction mirna target gene prediction Summary micrornas? RNA can
Prediction of micrornas and their targets Introduction Brief history mirna Biogenesis Computational Methods Mature and precursor mirna prediction mirna target gene prediction Summary micrornas? RNA can
Chapter 5 A Dose Dependent Screen for Modifiers of Kek5
 Chapter 5 A Dose Dependent Screen for Modifiers of Kek5 "#$ ABSTRACT Modifier screens in Drosophila have proven to be a powerful tool for uncovering gene interaction and elucidating molecular pathways.
Chapter 5 A Dose Dependent Screen for Modifiers of Kek5 "#$ ABSTRACT Modifier screens in Drosophila have proven to be a powerful tool for uncovering gene interaction and elucidating molecular pathways.
Small RNAs and how to analyze them using sequencing
 Small RNAs and how to analyze them using sequencing RNA-seq Course November 8th 2017 Marc Friedländer ComputaAonal RNA Biology Group SciLifeLab / Stockholm University Special thanks to Jakub Westholm for
Small RNAs and how to analyze them using sequencing RNA-seq Course November 8th 2017 Marc Friedländer ComputaAonal RNA Biology Group SciLifeLab / Stockholm University Special thanks to Jakub Westholm for
MicroRNA target interactions: new insights from genome-wide approaches
 Ann. N.Y. Acad. Sci. ISSN 0077-8923 ANNALS OF THE NEW YORK ACADEMY OF SCIENCES Issue: Nutrition and Physical Activity in Aging, Obesity, and Cancer MicroRNA target interactions: new insights from genome-wide
Ann. N.Y. Acad. Sci. ISSN 0077-8923 ANNALS OF THE NEW YORK ACADEMY OF SCIENCES Issue: Nutrition and Physical Activity in Aging, Obesity, and Cancer MicroRNA target interactions: new insights from genome-wide
Chapter 11 How Genes Are Controlled
 Chapter 11 How Genes Are Controlled PowerPoint Lectures for Biology: Concepts & Connections, Sixth Edition Campbell, Reece, Taylor, Simon, and Dickey Copyright 2009 Pearson Education, Inc. Lecture by Mary
Chapter 11 How Genes Are Controlled PowerPoint Lectures for Biology: Concepts & Connections, Sixth Edition Campbell, Reece, Taylor, Simon, and Dickey Copyright 2009 Pearson Education, Inc. Lecture by Mary
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. nrg1 bns101/bns101 embryos develop a functional heart and survive to adulthood (a-b) Cartoon of Talen-induced nrg1 mutation with a 14-base-pair deletion in
Supplementary Figures Supplementary Figure 1. nrg1 bns101/bns101 embryos develop a functional heart and survive to adulthood (a-b) Cartoon of Talen-induced nrg1 mutation with a 14-base-pair deletion in
mirna-guided regulation at the molecular level
 molecular level Hervé Seitz IGH du CNRS, Montpellier, France March 3, 2016 microrna target prediction . microrna target prediction mirna: target: 2 7 5 N NNNNNNNNNNNNNN 3 NNNNNN the seed microrna target
molecular level Hervé Seitz IGH du CNRS, Montpellier, France March 3, 2016 microrna target prediction . microrna target prediction mirna: target: 2 7 5 N NNNNNNNNNNNNNN 3 NNNNNN the seed microrna target
Supplementary Figures
 J. Cell Sci. 128: doi:10.1242/jcs.173807: Supplementary Material Supplementary Figures Fig. S1 Fig. S1. Description and/or validation of reagents used. All panels show Drosophila tissues oriented with
J. Cell Sci. 128: doi:10.1242/jcs.173807: Supplementary Material Supplementary Figures Fig. S1 Fig. S1. Description and/or validation of reagents used. All panels show Drosophila tissues oriented with
MicroRNAs in Development
 Review Article TheScientificWorldJOURNAL (2006) 6, 1828-1840 TSW Development & Embryology ISSN 1537-744X; DOI 10.1100/tsw.2006.313 MicroRNAs in Development Danielle Maatouk and Brian D. Harfe* University
Review Article TheScientificWorldJOURNAL (2006) 6, 1828-1840 TSW Development & Embryology ISSN 1537-744X; DOI 10.1100/tsw.2006.313 MicroRNAs in Development Danielle Maatouk and Brian D. Harfe* University
ns ns hp761(lf); daf-28(lf) daf-28(gf) Figure S1
 A ns ns B 100 100 80 80 60 60 40 40 20 20 0 0% 0 0% hp761(lf); daf-28(lf) daf-28(gf) Figure S1 A 100 80 15 o C 22 o C 25 o C % Dauers 60 40 20 0 0% 0% * 0% 0% 0% Class I alleles Class II alleles daf-2(lf;ts)
A ns ns B 100 100 80 80 60 60 40 40 20 20 0 0% 0 0% hp761(lf); daf-28(lf) daf-28(gf) Figure S1 A 100 80 15 o C 22 o C 25 o C % Dauers 60 40 20 0 0% 0% * 0% 0% 0% Class I alleles Class II alleles daf-2(lf;ts)
V16: involvement of micrornas in GRNs
 What are micrornas? V16: involvement of micrornas in GRNs How can one identify micrornas? What is the function of micrornas? Elisa Izaurralde, MPI Tübingen Huntzinger, Izaurralde, Nat. Rev. Genet. 12,
What are micrornas? V16: involvement of micrornas in GRNs How can one identify micrornas? What is the function of micrornas? Elisa Izaurralde, MPI Tübingen Huntzinger, Izaurralde, Nat. Rev. Genet. 12,
Elisabet Pujadas PCBio Summer High-throughput screening of enhancer elements and mirna-target interactions
 Elisabet Pujadas PCBio Summer 2008 High-throughput screening of enhancer elements and mirna-target interactions Differences in gene regulation are known to underlie phenotypic disparities between cell
Elisabet Pujadas PCBio Summer 2008 High-throughput screening of enhancer elements and mirna-target interactions Differences in gene regulation are known to underlie phenotypic disparities between cell
GW182 proteins cause PABP dissociation from silenced mirna targets in the absence of deadenylation
 Manuscript EMBO-2012-83275 GW182 proteins cause PABP dissociation from silenced mirna targets in the absence of deadenylation Latifa Zekri, Duygu Kuzuoğlu-Öztürk and Elisa Izaurralde Corresponding author:
Manuscript EMBO-2012-83275 GW182 proteins cause PABP dissociation from silenced mirna targets in the absence of deadenylation Latifa Zekri, Duygu Kuzuoğlu-Öztürk and Elisa Izaurralde Corresponding author:
microrna as a Potential Vector for the Propagation of Robustness in Protein Expression and Oscillatory Dynamics within a cerna Network
 microrna as a Potential Vector for the Propagation of Robustness in Protein Expression and Oscillatory Dynamics within a cerna Network Claude Gérard*,Béla Novák Oxford Centre for Integrative Systems Biology,
microrna as a Potential Vector for the Propagation of Robustness in Protein Expression and Oscillatory Dynamics within a cerna Network Claude Gérard*,Béla Novák Oxford Centre for Integrative Systems Biology,
BIOL2005 WORKSHEET 2008
 BIOL2005 WORKSHEET 2008 Answer all 6 questions in the space provided using additional sheets where necessary. Hand your completed answers in to the Biology office by 3 p.m. Friday 8th February. 1. Your
BIOL2005 WORKSHEET 2008 Answer all 6 questions in the space provided using additional sheets where necessary. Hand your completed answers in to the Biology office by 3 p.m. Friday 8th February. 1. Your
Report. Many Families of C. elegans MicroRNAs Are Not Essential for Development or Viability. Ezequiel Alvarez-Saavedra 1 and H. Robert Horvitz 1, * 1
 Current Biology 20, 367 373, Feruary 23, 2010 ª2010 Elsevier Ltd All rights reserved DOI 10.1016/j.cu.2009.12.051 Many Families of C. elegans MicroRNAs Are Not Essential for Development or Viaility Report
Current Biology 20, 367 373, Feruary 23, 2010 ª2010 Elsevier Ltd All rights reserved DOI 10.1016/j.cu.2009.12.051 Many Families of C. elegans MicroRNAs Are Not Essential for Development or Viaility Report
Identification of target genes of micrornas in retinoic acid-induced neuronal differentiation*
 Pure Appl. Chem., Vol. 77, No. 1, pp. 313 318, 2005. DOI: 10.1351/pac200577010313 2005 IUPAC Identification of target genes of micrornas in retinoic acid-induced neuronal differentiation* Hiroaki Kawasaki
Pure Appl. Chem., Vol. 77, No. 1, pp. 313 318, 2005. DOI: 10.1351/pac200577010313 2005 IUPAC Identification of target genes of micrornas in retinoic acid-induced neuronal differentiation* Hiroaki Kawasaki
An Unexpected Function of the Prader-Willi Syndrome Imprinting Center in Maternal Imprinting in Mice
 An Unexpected Function of the Prader-Willi Syndrome Imprinting Center in Maternal Imprinting in Mice Mei-Yi Wu 1 *, Ming Jiang 1, Xiaodong Zhai 2, Arthur L. Beaudet 2, Ray-Chang Wu 1 * 1 Department of
An Unexpected Function of the Prader-Willi Syndrome Imprinting Center in Maternal Imprinting in Mice Mei-Yi Wu 1 *, Ming Jiang 1, Xiaodong Zhai 2, Arthur L. Beaudet 2, Ray-Chang Wu 1 * 1 Department of
RNA interference (RNAi)
 RN interference (RNi) Natasha aplen ene Silencing Section Office of Science and Technology Partnerships Office of the Director enter for ancer Research National ancer Institute ncaplen@mail.nih.gov Plants
RN interference (RNi) Natasha aplen ene Silencing Section Office of Science and Technology Partnerships Office of the Director enter for ancer Research National ancer Institute ncaplen@mail.nih.gov Plants
Supplementary information
 Supplementary information Human Cytomegalovirus MicroRNA mir-us4-1 Inhibits CD8 + T Cell Response by Targeting ERAP1 Sungchul Kim, Sanghyun Lee, Jinwook Shin, Youngkyun Kim, Irini Evnouchidou, Donghyun
Supplementary information Human Cytomegalovirus MicroRNA mir-us4-1 Inhibits CD8 + T Cell Response by Targeting ERAP1 Sungchul Kim, Sanghyun Lee, Jinwook Shin, Youngkyun Kim, Irini Evnouchidou, Donghyun
