Noninvasive Determination of Gene Mutations in Clear Cell Renal Cell Carcinoma using Multiple Instance Decisions Aggregated CNN
|
|
|
- Amber Willis
- 5 years ago
- Views:
Transcription
1 Noninvasive Determination of Gene Mutations in Clear Cell Renal Cell Carcinoma using Multiple Instance Decisions Aggregated CNN Mohammad Arafat Hussain 1, Ghassan Hamarneh 2, and Rafeef Garbi 1 1 BiSICL, University of British Columbia, Vancouver, BC, Canada 2 Medical Image Analysis Lab, Simon Fraser University, Burnaby, BC, Canada {arafat,rafeef}@ece.ubc.ca, hamarneh@sfu.ca Abstract. Kidney clear cell renal cell carcinoma (ccrcc) is the major sub-type of RCC, constituting one the most common cancers worldwide accounting for a steadily increasing mortality rate with 350,000 new cases recorded in Understanding the underlying genetic mutations in ccrcc provides crucial information enabling malignancy staging and patient survival estimation thus plays a vital role in accurate ccrcc diagnosis, prognosis, treatment planning, and response assessment. Although the underlying gene mutations can be identified by whole genome sequencing of the ccrcc following invasive nephrectomy or kidney biopsy procedures, recent studies have suggested that such mutations may be noninvasively identified by studying image features of the ccrcc from Computed Tomography (CT) data. Such image feature identification currently relies on laborious manual processes based on visual inspection of 2D image slices that are time-consuming and subjective. In this paper, we propose a convolutional neural network approach for automatic detection of underlying ccrcc gene mutations from 3D CT volumes. We aggregate the mutation-presence/absence decisions for all the ccrcc slices in a kidney into a robust singular decision that determines whether the interrogated kidney bears a specific mutation or not. When validated on clinical CT datasets of 267 patients from the TCIA database, our method detected gene mutations with 94% accuracy. 1 Introduction Kidney cancer, or renal cell carcinomas (RCC) are a common group of chemotherapy resistant diseases that accounted for an estimated 62,000 new patients and 14,000 deaths in the United States in 2015 alone [1]. North America and Europe have recently reported the highest numbers of new cases of RCC in the world [2]. The most common histologic sub-type of RCC is clear cell RCC (ccrcc) [3], which is known to be a genetically heterogeneous disease [4]. Recent studies [5,6] have identified several mutations in genes associated with ccrcc. For example, the von Hippel-Lindau (VHL) tumor suppressor gene, BRCA1-associated protein 1 (BAP1) gene, polybromo 1 (PBRM1) gene, and SET domain containing 2 (SETD2) gene have been identified as the most commonly mutated genes in ccrcc [5].
2 2 M. A. Hussain et al. Traditionally, ccrcc underlying gene mutations are identified by genome sequencing of the ccrcc of the kidney samples after invasive nephrectomy or kidney biopsy [5]. This identification of genetic mutations is clinically important because advanced stages of ccrcc and poor patient survival have been found to be associated with the VHL, PBRM1, BAP1, SETD2, and lysine (K)-specific demethylase 5C (KDM5C) gene mutations [5,7]. Therefore, knowledge of the genetic make-up of a patient s kidney ccrcc has great prognostic value that is helpful for treatment planning [5,7]. Correlations between mutations in genes and different ccrcc features seen in patient CT images has been shown in recent work [4,8]. For example, an association between well-defined tumor margin, nodular enhancement and intratumoral vascularity with the VHL mutation has been reported [8]. Ill-defined tumor margin and renal vein invasion were also reported to be associated with the BAP1 mutation [4] whereas PBRM1 and SETD2 mutations are mostly seen in solid (non-cystic) ccrcc cases [8]. Such use of radiological imaging data as a noninvasive determinant of the mutational status and a complement to genomic analysis in characterizing disease biology is refereed to as Radiogenomics [4,8]. Radiogenomics requires robust image feature identification, which is typically performed by expert radiologists. However, relying on human visual inspection is laborious, time consuming, and suffers from high intra/inter-observer variability. A number of machine learning (ML) tools have been used to facilitate the processes of high-throughput quantitative feature extraction from volumetric medical images, with some subsequently used in treatment decision support [9]. This practice is generally known as Radiomics. Radiomics uses higher order statistics (on the medical images) combined with clinical and radiogenomic data to develop models that may potentially improve cancer diagnostic, prognostic, and predictive accuracy [9,10]. The typical tumor radiomic analysis workflow has 4 distinct steps (1) 3D imaging, (2) manual or automatic tumor detection and segmentation, (3) tumor phenotype quantification, and (4) data integration (i.e. phenotype+genotype+clinical+proteomic) and analysis [10]. Discriminant features needed for proper mutation detection are often not seen in the marginal ccrcc cross-sections (e.g. axial slices in the top and bottom regions of a ccrcc). This scenario makes single-instance ML approaches, especially convolutional neural network (CNN) very difficult to train, as some of the input slices do not contain discriminating features, thus do not correspond to the assigned mutation label. Another solution is to use the full 3D volume as a single instance. However, 3D CNNs are considerably more difficult to train as they contain significantly more parameters and consequently require many more training samples, while the use of the 3D volume itself severely reduces the available number of training samples than its 2D counterpart. An alternative approach is multiple-instance learning (MIL) [11], where the learner receives a set of labeled bags (e.g. mutation present/absent), each containing multiple instances (e.g. all the ccrcc slices in a kidney). A MIL model labels a bag with a class even if some or most of the instances within it are not members of that class. We propose a deep CNN approach that addresses the challenge of automatic mutation detection in kidney ccrcc. Our method is a variant of the conventional
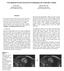
3 Gene Mutations Detection in Kidney ccrcc 3 MIL approach, where we use multiple instances for robust binary classification, while using single instances for training the CNN to facilitate higher number and variation of training data. The CNN automatically learns the ccrcc image features and the binary decisions (i.e. presence/absence of a mutation) for all the ccrcc slices in a particular kidney sample are aggregated into a robust singular decision that ultimately determines whether an interrogated kidney sample has undergone a certain mutation or not. Our method can be incorporated in the Radiomics step-3 given that the tumor boundary is already known in step-2. The estimated mutation data can subsequently be integrated in the step-4. The frequency of occurrence of various mutations in ccrcc varies significantly, e.g. VHL, PBRM1, BAP1, SETD2 and KDM5C were found in 76%, 43%, 14%, 14% and 8% of kidney samples of our dataset, respectively. In this study we consider the four most prevalent gene mutations (i.e. VHL, PBRM1, BAP1 and SETD2). We achieve this via four multiple instance decisions aggregation CNNs, however, our approach is directly extendable to more mutation types depending on the availability of sufficient training data. 2 Materials and Methods 2.1 Data We obtained access to 267 patients CT scans from The Cancer Imaging Archive (TCIA) database [12]. In this dataset, 138 scans contained at least one mutated gene because of ccrcc. For example, 105 patients had VHL, 60 patients had PBRM1, 60 patients had SETD2, and 20 patients had BAP1 mutations. In addition, some of the patients had multiple types of mutations. However, 9 patients had CT scans acquired after nephrectomy and, therefore, those patients data were not usable for this study. The images in our database included variations in CT scanner models, contrast administration, field of view, and spatial resolution. The in-plane pixel size ranged from 0.29 to 1.87 mm and the slice thickness ranged from 1.5 to 7.5 mm. Ground truth mutation labels were collected from the cbioportal for Cancer Genomics [7]. Table 1. Number of kidney samples used in training, validation and testing per mutation case. Acronym used M mutation. Genes # Training Samples # Validation Samples # Test Samples M-Present M-Absent M-Present M-Absent M-Present M-Absent VHL PBRM SETD BAP We show the number of kidney samples used in the training, validation and testing stages in Table 1. During training, validation and testing, we use only those slices of the kidney that contain ccrcc as our CNNs aim to learn ccrcc features. We form a 3-channel image from each scalar-valued CT slice by generating channel intensities [I, I-50, I-100] HU, where I represents the original intensities in a CT image slice tightly encompassing a kidney+ccrcc crosssection (Fig. 1), whereas I-50 and I-100 represent two variants of I with different
4 4 M. A. Hussain et al. a b c d e f g h Fig. 1. Illustration of CT features of ccrcc seen in the data of this study. (a) Cystic tumor architecture, (b) calcification, (c) exophytic tumor, (d) endophytic tumor, (e) necrosis, (f) ill-defined tumor margin, (g) nodular enhancement, and (h) renal vein invasion. Arrow indicates feature of interest in each image. HU values. We add these variations in channel intensity values as similar ccrcc features may have different X-ray attenuation properties across patients [4]. We resized all the image data by resampling into a size of pixels. We augmented the number of training samples by a factor of 24 by flipping and rotating the 3-ch image slices as well as by re-ordering the 3 channels in each image. We normalized the training and validation data before training by subtracting the image mean and dividing by the image standard deviation. 2.2 Multiple Instance Decision Aggregation for Mutation Detection Typically, ccrcc grows in different regions of the kidney and is clinically scored on the basis of their CT slice-based image features, such as size, margin (well- or ill-defined), composition (solid or cystic), necrosis, growth pattern (endophytic or exophytic), calcification etc. [4]. Some of these features seen in our dataset are shown in Fig. 1. We propose to learn corresponding features from the CT images using four different CNNs VHL-CNN, PBRM1-CNN, SETD2-CNN and BAP1- CNN, each for one of the four mutations (VHL, PBRM1, SETD2 and BAP1). Using a separate CNN per mutation alleviates the problem of data imbalance among mutation types, given that the mutations are not mutually exclusive. CNN Architecture All the CNNs in this study (i.e. VHL-CNN, PBRM1- CNN, SETD2-CNN and BAP1-CNN) have similar configuration but are trained separately (Fig. 2). Each CNN has twelve layers excluding the input five convolutional (Conv) layers; three fully connected (FC) layers; one softmax layer; one average pooling layer; and two thresholding layers. All but the last three layers contain trainable weights. The input is the pixel image slice containing the kidney+ccrcc. We train these CNNs (layers 1 9) using a balanced dataset for each mutation case separately (i.e. a particular mutation-present and absent). During training, images are fed to the CNNs in a randomly shuffled single instance fashion. Typically, Conv layers are known for sequentially learning the high-level non-linear spatial image features (e.g. ccrcc size, orientation, edge variation, etc). We used five Conv layers as the 5th Conv layer typically grabs an entire object (e.g. ccrcc shape) in an image even if there is a significant pose variation [13]. Subsequent FC layers prepare those features for optimal classification of an interrogated image. In our case, three FC layers are deployed to make the decision on the learned features from the 3-ch images to decide if a particular gene mutation is probable or not. The number of FC layers plays a vital role as the overall depth of the model is important for obtaining good performance [13], and we achieve optimal performance with three FC layers. s
5 Gene Mutations Detection in Kidney ccrcc 5 55X55X96 27X27X256 13X13X384 13X13X384 13X13X X1 4096X1 2X1 227X227X3 Image C1 R1 P1 N1 C2 R2 P2 N2 C3 R3 C4 R4 C5 R5 P5 F6 R6D6 F7 R7D7 F8 11X11X3 (4) 3X3 (2) 5X5X96 3X3 (2) 3X3X256 3X3X384 3X3X256 3X3 (2) Filter Size (stride) Cx Convolutional Rx ReLU Px Max Pooling Nx Local Response Normalization Fx Fully Connected Dx Dropout 1-channel slices 3-channel slices (RGB) VHL-CNN/ PBRM1-CNN/ SETD2-CNN/ BAP1-CNN Softmax 0.5 Average Pooling N X 1 E avg 0.5 >= 0.5 < 0.5 Mutation present Mutation absent Fig. 2. Multiple instance decisions aggregated CNN for gene mutation detection. 10, 11 and 12 (i.e. two thresholding and one average pooling layers) of the CNNs are used during the testing phase and do not contain any trainable weights. Solver These networks were trained by minimizing the softmax loss between the expected and detected labels (1 mutation present and 0 mutation absent). We used the Adam optimization method [14]. All the parameters for this solver were set to the suggested (by [14]) default values, i.e. β 1 = 0.9, β 2 = and ɛ = We also employed a Dropout unit (Dx) that drops 50% of units in both F6 and F7 layers (Fig. 2) and used a weight decay of The base learning rate was set to 0.01 and was decreased by a factor of 0.1 to over 250,000 iterations with a batch of 256 images processed at each iteration. Training was performed on a workstation with Intel 4.0 GHz Core-i7 processor, an Nvidia GeForce Titan Xp GPU with 12 GB of VRAM, and 32 GB of RAM. Mutation Detection After all the CNNs are trained (from layer 1 to 9), we use the full configuration (from layer 1 to 12) in the testing phase. Although we use only ccrcc containing kidney slices during training and validation, often not all the ccrcc cross-sections contains the discriminating features for proper mutation detection. Therefore, our trained CNN (from layer 1 to 9) often missclassifies the interrogated image slice based on the probability estimated at the layer 9 (i.e. softmax layer). In order to address this misclassification by our CNNs, we adopt a multiple instance decision aggregation procedure. In this procedure, we feed all the candidate image slices of a particular kidney to the trained CNN and accumulate the slice-wise binary classification labels (0 or 1) at layer 10 (the thresholding layer). These labels are then fed into a N 1 average pooling layer, where N is the total number of 3-channel axial slices of an interrogated kidney. Finally, the estimated average (E avg ) from layer 11 is fed to the second thresholding layer (layer 12), where E avg 0.5 indicates the presence of the mutation in that kidney, and no-mutation otherwise (see Fig. 2).
6 6 M. A. Hussain et al. Table 2. Automatic gene mutation detection performance of different methods. We use acronyms as M mutation, x one of VHL/PBRM1/SETD2/BAP1, Aug augmentation, SI single instance, MI multiple instance, 3ch 3-channel data with augmentation by channel re-ordering, F augmentation by flipping, and R augmentation by rotation. Methods Genes # Test Samples # Correct Detection Overall Mean x M-present M-absent M-present M-absent Error (%) Error (%) Random VHL Forest PBRM (SI+1ch) SETD No Aug BAP x-cnn VHL (SI+1ch) PBRM No Aug SETD BAP VHL x-cnn PBRM (SI+3ch) SETD BAP x-cnn VHL (SI+1ch PBRM F+R) SETD BAP x-cnn VHL (SI+3ch PBRM F+R) SETD BAP Proposed VHL (MI+3ch PBRM F+R SETD Average) BAP Results We compare the mutation detection performance by a wide range of methods. At first, we tested the performance using a single instance (SI)-based random forest (RF) approach, where hand-engineered image features were used. In a typical SI-based classification approach, the class-label is decided from the maximum among the predicted class-probabilities [15]. Similarly in our SI-based approaches, presence or absence of a certain mutation is decided from the maximum among the estimated probabilities associated with all the ccrcc image slices in a particular kidney. Then we demonstrate the effectiveness of automatic feature learning compared to the hand-engineered features generation using the CNN approach. Afterwards, we show the effect of incorporating augmented data in the training dataset and compared the mutation detection performance for three different types of augmentation (i.e. image flipping+rotation, 3-ch re-ordering and those combined). Finally, we demonstrated the effectiveness of using multiple instance decisions aggregation in our proposed method. In row 1 of the comparison Table 2, we show results of a traditional RF approach with hand-engineered image features shown to be effective in anatomy classification task [15] histogram of oriented gradient, Haar features, and local binary patterns. Here, we did not augment any manually transformed data to the
7 Gene Mutations Detection in Kidney ccrcc 7 training samples. We trained four RFs for the four different mutation cases and as we see in Table 2, the resulting mean detection error was the highest ( 54%) among all contrasted methods. Row 2 shows the results of a deep CNN (namely, x-cnn, where x VHL/PBRM1/SETD2/BAP1 (see Fig. 2)) approach with no data augmentation. Since the CNN learns the image features automatically, it may have helped this CNN method perform better (mean error 42%) than that of the hand-engineered features-based RF approach. Row 3 shows results for x- CNN, where we used data augmentation by deploying 3-ch data and re-ordering of channels (see Sect. 2.1). These data were fed to x-cnn and it can be seen how the SI-based mutation detection performance by this approach (mean error 29%) outperformed that with no data augmentation. Thus, including channels with different intensity ranges, mimicking the tumor intensity variation across patients, have shown positive impact on the mutation detection task. Row 4 shows results for x-cnn with a different augmentation process, which deploys the flipping and rotating of the 1-ch training samples. This approach (mean error 22%) outperformed that with 3-ch augmentation. So it is clear that the flipping+rotation-based augmentation introduced more variation in the training data than that by the 3-ch augmentation, resulting in better generalization of the model. In the method shown in row 5, we combined the flipping+rotation augmentation with the 3-ch re-ordering augmentation. The performance of the x- CNN with these data was better in mutation detection (mean error 14%) than that of flipping+rotation or 3-ch augmentation alone (see Table 2). Finally, row 6 demonstrates results of our proposed method, where flipping, rotation and 3-ch re-ordering augmentations were used. In addition, binary classification was performed based on the multiple instance decisions aggregation. We see in the Table 2 that the mean mutation detection error by our method is 6%, which is the lowest tested. In addition, detection errors for individual mutation cases were also low and in the range of 10%. Thus, our multiple instance decisions aggregation procedure made our CNN models more robust on SI-based missclassification. 4 Conclusions In this paper, we proposed a multiple instance decision aggregation-based deep CNN approach for automatic mutation detection in kidney ccrcc. We have shown how our approach automatically learned discriminating ccrcc features from CT images and aggregated the binary decisions on the mutation-presence/ absence for all the ccrcc slices in a particular kidney sample. This aggregation produced a robust decision on the presence of a certain mutation in an interrogated kidney sample. In addition, our multiple instance decision aggregation approach achieved better accuracy in mutation detection than that of a typical single instance-based approach. On the other hand, better performance by conventional MIL approaches is subject to the availability of sufficient number of data, while in applications such as ours, there are usually very few data samples for some of the mutation cases. Therefore, an end-to-end MIL approach will most likely fail for those mutation cases with few data samples. However, this paper
8 8 M. A. Hussain et al. included a number of meaningful comparisons to highlight the effects of different augmentation, pooling schemes etc within the context of insufficient data, which we believe provide more interesting findings and appears to be suitable for ccrcc Radiomics, where the learned mutations would aid in better ccrcc diagnosis, prognosis and treatment response assessment. Our experimental results demonstrated an approximately 94% accuracy in kidney-wise mutation detection. References 1. Siegel, R.L., Miller, K.D., Jemal, A. Cancer statistics, CA a cancer journal for clinicians 65(1) (2015) Ridge, C.A., Pua, B.B., Madoff, D.C. Epidemiology and staging of renal cell carcinoma. In Seminars in interventional radiology. Volume 31., Thieme Medical Publishers (2014) Lam, J.S., Shvarts, O., Leppert, J.T., Figlin, R.A., Belldegrun, A.S. Renal cell carcinoma 2005 New frontiers in staging, prognostication and targeted molecular therapy. The Journal of urology 173(6) (2005) Shinagare, A.B., Vikram, R., Jaffe, C., Akin, O., Kirby, J., et al. Radiogenomics of clear cell renal cell carcinoma preliminary findings of The Cancer Genome Atlas Renal Cell Carcinoma (TCGA RCC) Imaging Research Group. Abdominal imaging 40(6) (2015) Network, C.G.A.R., et al. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 499(7456) (2013) Guo, G., Gui, Y., Gao, S., Tang, A., Hu, X., Huang, Y., Jia, W., et al. Frequent mutations of genes encoding ubiquitin-mediated proteolysis pathway components in clear cell renal cell carcinoma. Nature genetics 44(1) (2012) Gao, J., Aksoy, B.A., Dogrusoz, U., Dresdner, G., Gross, B., Sumer, S.O., Sun, Y., Jacobsen, A., et al. Integrative analysis of complex cancer genomics and clinical profiles using the cbioportal. Sci. Signal. 6(269) (2013) pl1 pl1 8. Karlo, C.A., Di Paolo, P.L., Chaim, J., Hakimi, A.A., et al. Radiogenomics of clear cell renal cell carcinoma associations between ct imaging features and mutations. Radiology 270(2) (2014) Gillies, R.J., Kinahan, P.E., Hricak, H. Radiomics images are more than pictures, they are data. Radiology 278(2) (2015) Aerts, H.J. The potential of radiomic-based phenotyping in precision medicine a review. JAMA oncology 2(12) (2016) Dietterich, T.G., Lathrop, R.H., Lozano-Pérez, T. Solving the multiple instance problem with axis-parallel rectangles. Artificial intelligence 89(1-2) (1997) Clark, K., Vendt, B., Smith, K., Freymann, J., Kirby, J., Koppel, P., Moore, S.,, et al. The Cancer Imaging Archive (TCIA) maintaining and operating a public information repository. Journal of digital imaging 26(6) (2013) Zeiler, M.D., Fergus, R. Visualizing and understanding convolutional networks. In European conference on computer vision, Springer (2014) Kingma, D.P., Ba, J. Adam A method for stochastic optimization. arxiv preprint arxiv (2014) 15. Wang, H., Moradi, M., Gur, Y., Prasanna, P., Syeda-Mahmood, T. A multiatlas approach to region of interest detection for medical image classification. In International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer (2017)
CSE Introduction to High-Perfomance Deep Learning ImageNet & VGG. Jihyung Kil
 CSE 5194.01 - Introduction to High-Perfomance Deep Learning ImageNet & VGG Jihyung Kil ImageNet Classification with Deep Convolutional Neural Networks Alex Krizhevsky, Ilya Sutskever, Geoffrey E. Hinton,
CSE 5194.01 - Introduction to High-Perfomance Deep Learning ImageNet & VGG Jihyung Kil ImageNet Classification with Deep Convolutional Neural Networks Alex Krizhevsky, Ilya Sutskever, Geoffrey E. Hinton,
Lung Nodule Segmentation Using 3D Convolutional Neural Networks
 Lung Nodule Segmentation Using 3D Convolutional Neural Networks Research paper Business Analytics Bernard Bronmans Master Business Analytics VU University, Amsterdam Evert Haasdijk Supervisor VU University,
Lung Nodule Segmentation Using 3D Convolutional Neural Networks Research paper Business Analytics Bernard Bronmans Master Business Analytics VU University, Amsterdam Evert Haasdijk Supervisor VU University,
Mutation stratification of desmoid-type fibromatosis using a radiomics approach preliminary results
 Mutation stratification of desmoid-type fibromatosis using a radiomics approach preliminary results DTRF 2018 Milea J.M. Timbergen 1,2 *, MD, PhD candidate Department of Surgical Oncology, Department of
Mutation stratification of desmoid-type fibromatosis using a radiomics approach preliminary results DTRF 2018 Milea J.M. Timbergen 1,2 *, MD, PhD candidate Department of Surgical Oncology, Department of
EARLY STAGE DIAGNOSIS OF LUNG CANCER USING CT-SCAN IMAGES BASED ON CELLULAR LEARNING AUTOMATE
 EARLY STAGE DIAGNOSIS OF LUNG CANCER USING CT-SCAN IMAGES BASED ON CELLULAR LEARNING AUTOMATE SAKTHI NEELA.P.K Department of M.E (Medical electronics) Sengunthar College of engineering Namakkal, Tamilnadu,
EARLY STAGE DIAGNOSIS OF LUNG CANCER USING CT-SCAN IMAGES BASED ON CELLULAR LEARNING AUTOMATE SAKTHI NEELA.P.K Department of M.E (Medical electronics) Sengunthar College of engineering Namakkal, Tamilnadu,
The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters.
 Radiogenomics of clear cell renal cell carcinoma: preliminary findings of The Cancer Genome Atlas Renal Cell Carcinoma (TCGA RCC) Imaging Research Group The Harvard community has made this article openly
Radiogenomics of clear cell renal cell carcinoma: preliminary findings of The Cancer Genome Atlas Renal Cell Carcinoma (TCGA RCC) Imaging Research Group The Harvard community has made this article openly
Computer based delineation and follow-up multisite abdominal tumors in longitudinal CT studies
 Research plan submitted for approval as a PhD thesis Submitted by: Refael Vivanti Supervisor: Professor Leo Joskowicz School of Engineering and Computer Science, The Hebrew University of Jerusalem Computer
Research plan submitted for approval as a PhD thesis Submitted by: Refael Vivanti Supervisor: Professor Leo Joskowicz School of Engineering and Computer Science, The Hebrew University of Jerusalem Computer
arxiv: v3 [cs.cv] 28 Mar 2017
![arxiv: v3 [cs.cv] 28 Mar 2017 arxiv: v3 [cs.cv] 28 Mar 2017](/thumbs/73/69121210.jpg) Discovery Radiomics for Pathologically-Proven Computed Tomography Lung Cancer Prediction Devinder Kumar 1*, Audrey G. Chung 1, Mohammad J. Shaifee 1, Farzad Khalvati 2,3, Masoom A. Haider 2,3, and Alexander
Discovery Radiomics for Pathologically-Proven Computed Tomography Lung Cancer Prediction Devinder Kumar 1*, Audrey G. Chung 1, Mohammad J. Shaifee 1, Farzad Khalvati 2,3, Masoom A. Haider 2,3, and Alexander
HHS Public Access Author manuscript Med Image Comput Comput Assist Interv. Author manuscript; available in PMC 2018 January 04.
 Discriminative Localization in CNNs for Weakly-Supervised Segmentation of Pulmonary Nodules Xinyang Feng 1, Jie Yang 1, Andrew F. Laine 1, and Elsa D. Angelini 1,2 1 Department of Biomedical Engineering,
Discriminative Localization in CNNs for Weakly-Supervised Segmentation of Pulmonary Nodules Xinyang Feng 1, Jie Yang 1, Andrew F. Laine 1, and Elsa D. Angelini 1,2 1 Department of Biomedical Engineering,
Oncology - Evolution of imaging From helpful to essential
 Oncology - Evolution of imaging From helpful to essential 1990s 2000 1980s 18 FDG PET/CT MRI/MRSI 2010 1970s MRI 1960s CT 2015 HP13C-MRSI Ultrasound Nuc Med X-Ray - IVU MRI/PET Imaging 2016: Essential
Oncology - Evolution of imaging From helpful to essential 1990s 2000 1980s 18 FDG PET/CT MRI/MRSI 2010 1970s MRI 1960s CT 2015 HP13C-MRSI Ultrasound Nuc Med X-Ray - IVU MRI/PET Imaging 2016: Essential
POC Brain Tumor Segmentation. vlife Use Case
 Brain Tumor Segmentation vlife Use Case 1 Automatic Brain Tumor Segmentation using CNN Background Brain tumor segmentation seeks to separate healthy tissue from tumorous regions such as the advancing tumor,
Brain Tumor Segmentation vlife Use Case 1 Automatic Brain Tumor Segmentation using CNN Background Brain tumor segmentation seeks to separate healthy tissue from tumorous regions such as the advancing tumor,
arxiv: v2 [cs.cv] 3 Jun 2018
![arxiv: v2 [cs.cv] 3 Jun 2018 arxiv: v2 [cs.cv] 3 Jun 2018](/thumbs/89/97775247.jpg) S4ND: Single-Shot Single-Scale Lung Nodule Detection Naji Khosravan and Ulas Bagci Center for Research in Computer Vision (CRCV), School of Computer Science, University of Central Florida, Orlando, FL.
S4ND: Single-Shot Single-Scale Lung Nodule Detection Naji Khosravan and Ulas Bagci Center for Research in Computer Vision (CRCV), School of Computer Science, University of Central Florida, Orlando, FL.
Y-Net: Joint Segmentation and Classification for Diagnosis of Breast Biopsy Images
 Y-Net: Joint Segmentation and Classification for Diagnosis of Breast Biopsy Images Sachin Mehta 1, Ezgi Mercan 1, Jamen Bartlett 2, Donald Weaver 2, Joann G. Elmore 1, and Linda Shapiro 1 1 University
Y-Net: Joint Segmentation and Classification for Diagnosis of Breast Biopsy Images Sachin Mehta 1, Ezgi Mercan 1, Jamen Bartlett 2, Donald Weaver 2, Joann G. Elmore 1, and Linda Shapiro 1 1 University
Object Detectors Emerge in Deep Scene CNNs
 Object Detectors Emerge in Deep Scene CNNs Bolei Zhou, Aditya Khosla, Agata Lapedriza, Aude Oliva, Antonio Torralba Presented By: Collin McCarthy Goal: Understand how objects are represented in CNNs Are
Object Detectors Emerge in Deep Scene CNNs Bolei Zhou, Aditya Khosla, Agata Lapedriza, Aude Oliva, Antonio Torralba Presented By: Collin McCarthy Goal: Understand how objects are represented in CNNs Are
Improving 3D Ultrasound Scan Adequacy Classification Using a Three-Slice Convolutional Neural Network Architecture
 EPiC Series in Health Sciences Volume 2, 2018, Pages 152 156 CAOS 2018. The 18th Annual Meeting of the International Society for Computer Assisted Orthopaedic Surgery Health Sciences Improving 3D Ultrasound
EPiC Series in Health Sciences Volume 2, 2018, Pages 152 156 CAOS 2018. The 18th Annual Meeting of the International Society for Computer Assisted Orthopaedic Surgery Health Sciences Improving 3D Ultrasound
Highly Accurate Brain Stroke Diagnostic System and Generative Lesion Model. Junghwan Cho, Ph.D. CAIDE Systems, Inc. Deep Learning R&D Team
 Highly Accurate Brain Stroke Diagnostic System and Generative Lesion Model Junghwan Cho, Ph.D. CAIDE Systems, Inc. Deep Learning R&D Team Established in September, 2016 at 110 Canal st. Lowell, MA 01852,
Highly Accurate Brain Stroke Diagnostic System and Generative Lesion Model Junghwan Cho, Ph.D. CAIDE Systems, Inc. Deep Learning R&D Team Established in September, 2016 at 110 Canal st. Lowell, MA 01852,
Skin cancer reorganization and classification with deep neural network
 Skin cancer reorganization and classification with deep neural network Hao Chang 1 1. Department of Genetics, Yale University School of Medicine 2. Email: changhao86@gmail.com Abstract As one kind of skin
Skin cancer reorganization and classification with deep neural network Hao Chang 1 1. Department of Genetics, Yale University School of Medicine 2. Email: changhao86@gmail.com Abstract As one kind of skin
arxiv: v2 [cs.cv] 8 Mar 2018
![arxiv: v2 [cs.cv] 8 Mar 2018 arxiv: v2 [cs.cv] 8 Mar 2018](/thumbs/87/97094636.jpg) Automated soft tissue lesion detection and segmentation in digital mammography using a u-net deep learning network Timothy de Moor a, Alejandro Rodriguez-Ruiz a, Albert Gubern Mérida a, Ritse Mann a, and
Automated soft tissue lesion detection and segmentation in digital mammography using a u-net deep learning network Timothy de Moor a, Alejandro Rodriguez-Ruiz a, Albert Gubern Mérida a, Ritse Mann a, and
Differentiating Tumor and Edema in Brain Magnetic Resonance Images Using a Convolutional Neural Network
 Original Article Differentiating Tumor and Edema in Brain Magnetic Resonance Images Using a Convolutional Neural Network Aida Allahverdi 1, Siavash Akbarzadeh 1, Alireza Khorrami Moghaddam 2, Armin Allahverdy
Original Article Differentiating Tumor and Edema in Brain Magnetic Resonance Images Using a Convolutional Neural Network Aida Allahverdi 1, Siavash Akbarzadeh 1, Alireza Khorrami Moghaddam 2, Armin Allahverdy
Leukemia Blood Cell Image Classification Using Convolutional Neural Network
 Leukemia Blood Cell Image Classification Using Convolutional Neural Network T. T. P. Thanh, Caleb Vununu, Sukhrob Atoev, Suk-Hwan Lee, and Ki-Ryong Kwon Abstract Acute myeloid leukemia is a type of malignant
Leukemia Blood Cell Image Classification Using Convolutional Neural Network T. T. P. Thanh, Caleb Vununu, Sukhrob Atoev, Suk-Hwan Lee, and Ki-Ryong Kwon Abstract Acute myeloid leukemia is a type of malignant
arxiv: v1 [cs.cv] 9 Oct 2018
![arxiv: v1 [cs.cv] 9 Oct 2018 arxiv: v1 [cs.cv] 9 Oct 2018](/thumbs/95/126101195.jpg) Automatic Segmentation of Thoracic Aorta Segments in Low-Dose Chest CT Julia M. H. Noothout a, Bob D. de Vos a, Jelmer M. Wolterink a, Ivana Išgum a a Image Sciences Institute, University Medical Center
Automatic Segmentation of Thoracic Aorta Segments in Low-Dose Chest CT Julia M. H. Noothout a, Bob D. de Vos a, Jelmer M. Wolterink a, Ivana Išgum a a Image Sciences Institute, University Medical Center
arxiv: v1 [stat.ml] 23 Jan 2017
![arxiv: v1 [stat.ml] 23 Jan 2017 arxiv: v1 [stat.ml] 23 Jan 2017](/thumbs/81/83414183.jpg) Learning what to look in chest X-rays with a recurrent visual attention model arxiv:1701.06452v1 [stat.ml] 23 Jan 2017 Petros-Pavlos Ypsilantis Department of Biomedical Engineering King s College London
Learning what to look in chest X-rays with a recurrent visual attention model arxiv:1701.06452v1 [stat.ml] 23 Jan 2017 Petros-Pavlos Ypsilantis Department of Biomedical Engineering King s College London
Supplementary Online Content
 Supplementary Online Content Ting DS, Cheung CY-L, Lim G, et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic
Supplementary Online Content Ting DS, Cheung CY-L, Lim G, et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic
Final Report: Automated Semantic Segmentation of Volumetric Cardiovascular Features and Disease Assessment
 Final Report: Automated Semantic Segmentation of Volumetric Cardiovascular Features and Disease Assessment Tony Lindsey 1,3, Xiao Lu 1 and Mojtaba Tefagh 2 1 Department of Biomedical Informatics, Stanford
Final Report: Automated Semantic Segmentation of Volumetric Cardiovascular Features and Disease Assessment Tony Lindsey 1,3, Xiao Lu 1 and Mojtaba Tefagh 2 1 Department of Biomedical Informatics, Stanford
Automatic Quality Assessment of Cardiac MRI
 Automatic Quality Assessment of Cardiac MRI Ilkay Oksuz 02.05.2018 Contact: ilkay.oksuz@kcl.ac.uk http://kclmmag.org 1 Cardiac MRI Quality Issues Need for high quality images Wide range of artefacts Manual
Automatic Quality Assessment of Cardiac MRI Ilkay Oksuz 02.05.2018 Contact: ilkay.oksuz@kcl.ac.uk http://kclmmag.org 1 Cardiac MRI Quality Issues Need for high quality images Wide range of artefacts Manual
for the TCGA Breast Phenotype Research Group
 Decoding Breast Cancer with Quantitative Radiomics & Radiogenomics: Imaging Phenotypes in Breast Cancer Risk Assessment, Diagnosis, Prognosis, and Response to Therapy Maryellen Giger & Yuan Ji The University
Decoding Breast Cancer with Quantitative Radiomics & Radiogenomics: Imaging Phenotypes in Breast Cancer Risk Assessment, Diagnosis, Prognosis, and Response to Therapy Maryellen Giger & Yuan Ji The University
A deep learning model integrating FCNNs and CRFs for brain tumor segmentation
 A deep learning model integrating FCNNs and CRFs for brain tumor segmentation Xiaomei Zhao 1,2, Yihong Wu 1, Guidong Song 3, Zhenye Li 4, Yazhuo Zhang,3,4,5,6, and Yong Fan 7 1 National Laboratory of Pattern
A deep learning model integrating FCNNs and CRFs for brain tumor segmentation Xiaomei Zhao 1,2, Yihong Wu 1, Guidong Song 3, Zhenye Li 4, Yazhuo Zhang,3,4,5,6, and Yong Fan 7 1 National Laboratory of Pattern
ARTIFICIAL INTELLIGENCE FOR DIGITAL PATHOLOGY. Kyunghyun Paeng, Co-founder and Research Scientist, Lunit Inc.
 ARTIFICIAL INTELLIGENCE FOR DIGITAL PATHOLOGY Kyunghyun Paeng, Co-founder and Research Scientist, Lunit Inc. 1. BACKGROUND: DIGITAL PATHOLOGY 2. APPLICATIONS AGENDA BREAST CANCER PROSTATE CANCER 3. DEMONSTRATIONS
ARTIFICIAL INTELLIGENCE FOR DIGITAL PATHOLOGY Kyunghyun Paeng, Co-founder and Research Scientist, Lunit Inc. 1. BACKGROUND: DIGITAL PATHOLOGY 2. APPLICATIONS AGENDA BREAST CANCER PROSTATE CANCER 3. DEMONSTRATIONS
arxiv: v2 [cs.cv] 7 Jun 2018
![arxiv: v2 [cs.cv] 7 Jun 2018 arxiv: v2 [cs.cv] 7 Jun 2018](/thumbs/79/79761281.jpg) Deep supervision with additional labels for retinal vessel segmentation task Yishuo Zhang and Albert C.S. Chung Lo Kwee-Seong Medical Image Analysis Laboratory, Department of Computer Science and Engineering,
Deep supervision with additional labels for retinal vessel segmentation task Yishuo Zhang and Albert C.S. Chung Lo Kwee-Seong Medical Image Analysis Laboratory, Department of Computer Science and Engineering,
Classification of breast cancer histology images using transfer learning
 Classification of breast cancer histology images using transfer learning Sulaiman Vesal 1 ( ), Nishant Ravikumar 1, AmirAbbas Davari 1, Stephan Ellmann 2, Andreas Maier 1 1 Pattern Recognition Lab, Friedrich-Alexander-Universität
Classification of breast cancer histology images using transfer learning Sulaiman Vesal 1 ( ), Nishant Ravikumar 1, AmirAbbas Davari 1, Stephan Ellmann 2, Andreas Maier 1 1 Pattern Recognition Lab, Friedrich-Alexander-Universität
arxiv: v1 [cs.cv] 3 Apr 2018
![arxiv: v1 [cs.cv] 3 Apr 2018 arxiv: v1 [cs.cv] 3 Apr 2018](/thumbs/90/103707026.jpg) Towards whole-body CT Bone Segmentation André Klein 1,2, Jan Warszawski 2, Jens Hillengaß 3, Klaus H. Maier-Hein 1 arxiv:1804.00908v1 [cs.cv] 3 Apr 2018 1 Division of Medical Image Computing, Deutsches
Towards whole-body CT Bone Segmentation André Klein 1,2, Jan Warszawski 2, Jens Hillengaß 3, Klaus H. Maier-Hein 1 arxiv:1804.00908v1 [cs.cv] 3 Apr 2018 1 Division of Medical Image Computing, Deutsches
Tumor Cellularity Assessment. Rene Bidart
 Tumor Cellularity Assessment Rene Bidart 1 Goals Find location and class of all cell nuclei: Lymphocyte Cells Normal Epithelial Cells Malignant Epithelial Cells Why: Test effectiveness of pre-surgery treatment
Tumor Cellularity Assessment Rene Bidart 1 Goals Find location and class of all cell nuclei: Lymphocyte Cells Normal Epithelial Cells Malignant Epithelial Cells Why: Test effectiveness of pre-surgery treatment
ImageCLEF2018: Transfer Learning for Deep Learning with CNN for Tuberculosis Classification
 ImageCLEF2018: Transfer Learning for Deep Learning with CNN for Tuberculosis Classification Amilcare Gentili 1-2[0000-0002-5623-7512] 1 San Diego VA Health Care System, San Diego, CA USA 2 University of
ImageCLEF2018: Transfer Learning for Deep Learning with CNN for Tuberculosis Classification Amilcare Gentili 1-2[0000-0002-5623-7512] 1 San Diego VA Health Care System, San Diego, CA USA 2 University of
Automatic Prostate Cancer Classification using Deep Learning. Ida Arvidsson Centre for Mathematical Sciences, Lund University, Sweden
 Automatic Prostate Cancer Classification using Deep Learning Ida Arvidsson Centre for Mathematical Sciences, Lund University, Sweden Outline Autoencoders, theory Motivation, background and goal for prostate
Automatic Prostate Cancer Classification using Deep Learning Ida Arvidsson Centre for Mathematical Sciences, Lund University, Sweden Outline Autoencoders, theory Motivation, background and goal for prostate
Background Information
 Background Information Erlangen, November 26, 2017 RSNA 2017 in Chicago: South Building, Hall A, Booth 1937 Artificial intelligence: Transforming data into knowledge for better care Inspired by neural
Background Information Erlangen, November 26, 2017 RSNA 2017 in Chicago: South Building, Hall A, Booth 1937 Artificial intelligence: Transforming data into knowledge for better care Inspired by neural
ANALYSIS AND DETECTION OF BRAIN TUMOUR USING IMAGE PROCESSING TECHNIQUES
 ANALYSIS AND DETECTION OF BRAIN TUMOUR USING IMAGE PROCESSING TECHNIQUES P.V.Rohini 1, Dr.M.Pushparani 2 1 M.Phil Scholar, Department of Computer Science, Mother Teresa women s university, (India) 2 Professor
ANALYSIS AND DETECTION OF BRAIN TUMOUR USING IMAGE PROCESSING TECHNIQUES P.V.Rohini 1, Dr.M.Pushparani 2 1 M.Phil Scholar, Department of Computer Science, Mother Teresa women s university, (India) 2 Professor
Sleep Staging with Deep Learning: A convolutional model
 Sleep Staging with Deep Learning: A convolutional model Isaac Ferna ndez-varela1, Dimitrios Athanasakis2, Samuel Parsons3 Elena Herna ndez-pereira1, and Vicente Moret-Bonillo1 1- Universidade da Corun
Sleep Staging with Deep Learning: A convolutional model Isaac Ferna ndez-varela1, Dimitrios Athanasakis2, Samuel Parsons3 Elena Herna ndez-pereira1, and Vicente Moret-Bonillo1 1- Universidade da Corun
MRI Image Processing Operations for Brain Tumor Detection
 MRI Image Processing Operations for Brain Tumor Detection Prof. M.M. Bulhe 1, Shubhashini Pathak 2, Karan Parekh 3, Abhishek Jha 4 1Assistant Professor, Dept. of Electronics and Telecommunications Engineering,
MRI Image Processing Operations for Brain Tumor Detection Prof. M.M. Bulhe 1, Shubhashini Pathak 2, Karan Parekh 3, Abhishek Jha 4 1Assistant Professor, Dept. of Electronics and Telecommunications Engineering,
Extraction of Blood Vessels and Recognition of Bifurcation Points in Retinal Fundus Image
 International Journal of Research Studies in Science, Engineering and Technology Volume 1, Issue 5, August 2014, PP 1-7 ISSN 2349-4751 (Print) & ISSN 2349-476X (Online) Extraction of Blood Vessels and
International Journal of Research Studies in Science, Engineering and Technology Volume 1, Issue 5, August 2014, PP 1-7 ISSN 2349-4751 (Print) & ISSN 2349-476X (Online) Extraction of Blood Vessels and
ARTERY/VEIN CLASSIFICATION IN FUNDUS IMAGES USING CNN AND LIKELIHOOD SCORE PROPAGATION
 ARTERY/VEIN CLASSIFICATION IN FUNDUS IMAGES USING CNN AND LIKELIHOOD SCORE PROPAGATION Fantin Girard and Farida Cheriet Liv4D, Polytechnique Montreal fantin.girard@polymtl.ca MOTIVATION Changes in blood
ARTERY/VEIN CLASSIFICATION IN FUNDUS IMAGES USING CNN AND LIKELIHOOD SCORE PROPAGATION Fantin Girard and Farida Cheriet Liv4D, Polytechnique Montreal fantin.girard@polymtl.ca MOTIVATION Changes in blood
Early Detection of Lung Cancer
 Early Detection of Lung Cancer Aswathy N Iyer Dept Of Electronics And Communication Engineering Lymie Jose Dept Of Electronics And Communication Engineering Anumol Thomas Dept Of Electronics And Communication
Early Detection of Lung Cancer Aswathy N Iyer Dept Of Electronics And Communication Engineering Lymie Jose Dept Of Electronics And Communication Engineering Anumol Thomas Dept Of Electronics And Communication
A Multi-Resolution Deep Learning Framework for Lung Adenocarcinoma Growth Pattern Classification
 MIUA2018, 053, v4 (final): Multi-Resolution CNN for Lung Growth Pattern Classification 1 A Multi-Resolution Deep Learning Framework for Lung Adenocarcinoma Growth Pattern Classification Najah Alsubaie
MIUA2018, 053, v4 (final): Multi-Resolution CNN for Lung Growth Pattern Classification 1 A Multi-Resolution Deep Learning Framework for Lung Adenocarcinoma Growth Pattern Classification Najah Alsubaie
arxiv: v2 [cs.cv] 19 Dec 2017
![arxiv: v2 [cs.cv] 19 Dec 2017 arxiv: v2 [cs.cv] 19 Dec 2017](/thumbs/89/97557782.jpg) An Ensemble of Deep Convolutional Neural Networks for Alzheimer s Disease Detection and Classification arxiv:1712.01675v2 [cs.cv] 19 Dec 2017 Jyoti Islam Department of Computer Science Georgia State University
An Ensemble of Deep Convolutional Neural Networks for Alzheimer s Disease Detection and Classification arxiv:1712.01675v2 [cs.cv] 19 Dec 2017 Jyoti Islam Department of Computer Science Georgia State University
arxiv: v1 [cs.cv] 25 Jan 2018
![arxiv: v1 [cs.cv] 25 Jan 2018 arxiv: v1 [cs.cv] 25 Jan 2018](/thumbs/74/70727868.jpg) 1 Convolutional Invasion and Expansion Networks for Tumor Growth Prediction Ling Zhang, Le Lu, Senior Member, IEEE, Ronald M. Summers, Electron Kebebew, and Jianhua Yao arxiv:1801.08468v1 [cs.cv] 25 Jan
1 Convolutional Invasion and Expansion Networks for Tumor Growth Prediction Ling Zhang, Le Lu, Senior Member, IEEE, Ronald M. Summers, Electron Kebebew, and Jianhua Yao arxiv:1801.08468v1 [cs.cv] 25 Jan
Large-scale Histopathology Image Analysis for Colon Cancer on Azure
 Large-scale Histopathology Image Analysis for Colon Cancer on Azure Yan Xu 1, 2 Tao Mo 2 Teng Gao 2 Maode Lai 4 Zhuowen Tu 2,3 Eric I-Chao Chang 2 1 Beihang University; 2 Microsoft Research Asia; 3 UCSD;
Large-scale Histopathology Image Analysis for Colon Cancer on Azure Yan Xu 1, 2 Tao Mo 2 Teng Gao 2 Maode Lai 4 Zhuowen Tu 2,3 Eric I-Chao Chang 2 1 Beihang University; 2 Microsoft Research Asia; 3 UCSD;
COMP9444 Neural Networks and Deep Learning 5. Convolutional Networks
 COMP9444 Neural Networks and Deep Learning 5. Convolutional Networks Textbook, Sections 6.2.2, 6.3, 7.9, 7.11-7.13, 9.1-9.5 COMP9444 17s2 Convolutional Networks 1 Outline Geometry of Hidden Unit Activations
COMP9444 Neural Networks and Deep Learning 5. Convolutional Networks Textbook, Sections 6.2.2, 6.3, 7.9, 7.11-7.13, 9.1-9.5 COMP9444 17s2 Convolutional Networks 1 Outline Geometry of Hidden Unit Activations
arxiv: v1 [cs.cv] 24 Jul 2018
![arxiv: v1 [cs.cv] 24 Jul 2018 arxiv: v1 [cs.cv] 24 Jul 2018](/thumbs/82/86822918.jpg) Multi-Class Lesion Diagnosis with Pixel-wise Classification Network Manu Goyal 1, Jiahua Ng 2, and Moi Hoon Yap 1 1 Visual Computing Lab, Manchester Metropolitan University, M1 5GD, UK 2 University of
Multi-Class Lesion Diagnosis with Pixel-wise Classification Network Manu Goyal 1, Jiahua Ng 2, and Moi Hoon Yap 1 1 Visual Computing Lab, Manchester Metropolitan University, M1 5GD, UK 2 University of
Convolutional Neural Networks (CNN)
 Convolutional Neural Networks (CNN) Algorithm and Some Applications in Computer Vision Luo Hengliang Institute of Automation June 10, 2014 Luo Hengliang (Institute of Automation) Convolutional Neural Networks
Convolutional Neural Networks (CNN) Algorithm and Some Applications in Computer Vision Luo Hengliang Institute of Automation June 10, 2014 Luo Hengliang (Institute of Automation) Convolutional Neural Networks
arxiv: v3 [stat.ml] 27 Mar 2018
![arxiv: v3 [stat.ml] 27 Mar 2018 arxiv: v3 [stat.ml] 27 Mar 2018](/thumbs/82/87026346.jpg) ATTACKING THE MADRY DEFENSE MODEL WITH L 1 -BASED ADVERSARIAL EXAMPLES Yash Sharma 1 and Pin-Yu Chen 2 1 The Cooper Union, New York, NY 10003, USA 2 IBM Research, Yorktown Heights, NY 10598, USA sharma2@cooper.edu,
ATTACKING THE MADRY DEFENSE MODEL WITH L 1 -BASED ADVERSARIAL EXAMPLES Yash Sharma 1 and Pin-Yu Chen 2 1 The Cooper Union, New York, NY 10003, USA 2 IBM Research, Yorktown Heights, NY 10598, USA sharma2@cooper.edu,
A fully automated framework for lung tumour detection, segmentation and analysis
 A fully automated framework for lung tumour detection, segmentation and analysis Devesh Walawalkar dwalawal@andrew.cmu.in Abstract Early and correct diagnosis is a very important aspect of cancer treatment.
A fully automated framework for lung tumour detection, segmentation and analysis Devesh Walawalkar dwalawal@andrew.cmu.in Abstract Early and correct diagnosis is a very important aspect of cancer treatment.
ITERATIVELY TRAINING CLASSIFIERS FOR CIRCULATING TUMOR CELL DETECTION
 ITERATIVELY TRAINING CLASSIFIERS FOR CIRCULATING TUMOR CELL DETECTION Yunxiang Mao 1, Zhaozheng Yin 1, Joseph M. Schober 2 1 Missouri University of Science and Technology 2 Southern Illinois University
ITERATIVELY TRAINING CLASSIFIERS FOR CIRCULATING TUMOR CELL DETECTION Yunxiang Mao 1, Zhaozheng Yin 1, Joseph M. Schober 2 1 Missouri University of Science and Technology 2 Southern Illinois University
Automated Volumetric Cardiac Ultrasound Analysis
 Whitepaper Automated Volumetric Cardiac Ultrasound Analysis ACUSON SC2000 Volume Imaging Ultrasound System Bogdan Georgescu, Ph.D. Siemens Corporate Research Princeton, New Jersey USA Answers for life.
Whitepaper Automated Volumetric Cardiac Ultrasound Analysis ACUSON SC2000 Volume Imaging Ultrasound System Bogdan Georgescu, Ph.D. Siemens Corporate Research Princeton, New Jersey USA Answers for life.
arxiv: v2 [cs.cv] 29 Jan 2019
![arxiv: v2 [cs.cv] 29 Jan 2019 arxiv: v2 [cs.cv] 29 Jan 2019](/thumbs/88/117548796.jpg) Comparison of Deep Learning Approaches for Multi-Label Chest X-Ray Classification Ivo M. Baltruschat 1,2,, Hannes Nickisch 3, Michael Grass 3, Tobias Knopp 1,2, and Axel Saalbach 3 arxiv:1803.02315v2 [cs.cv]
Comparison of Deep Learning Approaches for Multi-Label Chest X-Ray Classification Ivo M. Baltruschat 1,2,, Hannes Nickisch 3, Michael Grass 3, Tobias Knopp 1,2, and Axel Saalbach 3 arxiv:1803.02315v2 [cs.cv]
Image Classification with TensorFlow: Radiomics 1p/19q Chromosome Status Classification Using Deep Learning
 Image Classification with TensorFlow: Radiomics 1p/19q Chromosome Status Classification Using Deep Learning Charles Killam, LP.D. Certified Instructor, NVIDIA Deep Learning Institute NVIDIA Corporation
Image Classification with TensorFlow: Radiomics 1p/19q Chromosome Status Classification Using Deep Learning Charles Killam, LP.D. Certified Instructor, NVIDIA Deep Learning Institute NVIDIA Corporation
Clear Cell Renal Cell Carcinoma: Associations Between CT Features and Patient Survival
 Genitourinary Imaging Original Research Hötker et al. Associations Between CT Features and Survival of Patients With ccrcc Genitourinary Imaging Original Research Andreas M. Hötker 1,2 Christoph A. Karlo
Genitourinary Imaging Original Research Hötker et al. Associations Between CT Features and Survival of Patients With ccrcc Genitourinary Imaging Original Research Andreas M. Hötker 1,2 Christoph A. Karlo
A CONVOLUTION NEURAL NETWORK ALGORITHM FOR BRAIN TUMOR IMAGE SEGMENTATION
 A CONVOLUTION NEURAL NETWORK ALGORITHM FOR BRAIN TUMOR IMAGE SEGMENTATION 1 Priya K, 2 Dr.O. Saraniya 1 PG Scholar, 2 Assistant Professor Department Of ECE Government College of Technology, Coimbatore,
A CONVOLUTION NEURAL NETWORK ALGORITHM FOR BRAIN TUMOR IMAGE SEGMENTATION 1 Priya K, 2 Dr.O. Saraniya 1 PG Scholar, 2 Assistant Professor Department Of ECE Government College of Technology, Coimbatore,
arxiv: v3 [stat.ml] 28 Oct 2017
![arxiv: v3 [stat.ml] 28 Oct 2017 arxiv: v3 [stat.ml] 28 Oct 2017](/thumbs/81/83820195.jpg) Interpretable Deep Learning applied to Plant Stress Phenotyping arxiv:1710.08619v3 [stat.ml] 28 Oct 2017 Sambuddha Ghosal sghosal@iastate.edu Asheesh K. Singh singhak@iastate.edu Arti Singh arti@iastate.edu
Interpretable Deep Learning applied to Plant Stress Phenotyping arxiv:1710.08619v3 [stat.ml] 28 Oct 2017 Sambuddha Ghosal sghosal@iastate.edu Asheesh K. Singh singhak@iastate.edu Arti Singh arti@iastate.edu
BLADDERSCAN PRIME PLUS TM DEEP LEARNING
 BLADDERSCAN PRIME PLUS TM DEEP LEARNING BladderScan Prime Plus from Verathon Takes Accuracy and Ease of Use to a New Level Powered by ImageSense TM deep learning technology, an advanced implementation
BLADDERSCAN PRIME PLUS TM DEEP LEARNING BladderScan Prime Plus from Verathon Takes Accuracy and Ease of Use to a New Level Powered by ImageSense TM deep learning technology, an advanced implementation
Automatic Diagnosis of Ovarian Carcinomas via Sparse Multiresolution Tissue Representation
 Automatic Diagnosis of Ovarian Carcinomas via Sparse Multiresolution Tissue Representation Aïcha BenTaieb, Hector Li-Chang, David Huntsman, Ghassan Hamarneh Medical Image Analysis Lab, Simon Fraser University,
Automatic Diagnosis of Ovarian Carcinomas via Sparse Multiresolution Tissue Representation Aïcha BenTaieb, Hector Li-Chang, David Huntsman, Ghassan Hamarneh Medical Image Analysis Lab, Simon Fraser University,
Automated diagnosis of pneumothorax using an ensemble of convolutional neural networks with multi-sized chest radiography images
 Automated diagnosis of pneumothorax using an ensemble of convolutional neural networks with multi-sized chest radiography images Tae Joon Jun, Dohyeun Kim, and Daeyoung Kim School of Computing, KAIST,
Automated diagnosis of pneumothorax using an ensemble of convolutional neural networks with multi-sized chest radiography images Tae Joon Jun, Dohyeun Kim, and Daeyoung Kim School of Computing, KAIST,
GTC 2018, San Jose, USA
 Automated Segmentation of Suspicious Breast Masses from Ultrasound Images Viksit Kumar, Jeremy Webb, Adriana Gregory, Mostafa Fatemi, Azra Alizad GTC 2018, San Jose, USA SIGNIFICANCE Breast cancer is most
Automated Segmentation of Suspicious Breast Masses from Ultrasound Images Viksit Kumar, Jeremy Webb, Adriana Gregory, Mostafa Fatemi, Azra Alizad GTC 2018, San Jose, USA SIGNIFICANCE Breast cancer is most
NMF-Density: NMF-Based Breast Density Classifier
 NMF-Density: NMF-Based Breast Density Classifier Lahouari Ghouti and Abdullah H. Owaidh King Fahd University of Petroleum and Minerals - Department of Information and Computer Science. KFUPM Box 1128.
NMF-Density: NMF-Based Breast Density Classifier Lahouari Ghouti and Abdullah H. Owaidh King Fahd University of Petroleum and Minerals - Department of Information and Computer Science. KFUPM Box 1128.
Hierarchical Convolutional Features for Visual Tracking
 Hierarchical Convolutional Features for Visual Tracking Chao Ma Jia-Bin Huang Xiaokang Yang Ming-Husan Yang SJTU UIUC SJTU UC Merced ICCV 2015 Background Given the initial state (position and scale), estimate
Hierarchical Convolutional Features for Visual Tracking Chao Ma Jia-Bin Huang Xiaokang Yang Ming-Husan Yang SJTU UIUC SJTU UC Merced ICCV 2015 Background Given the initial state (position and scale), estimate
arxiv: v1 [cs.cv] 28 Feb 2018
![arxiv: v1 [cs.cv] 28 Feb 2018 arxiv: v1 [cs.cv] 28 Feb 2018](/thumbs/93/111675272.jpg) Brain Tumor Segmentation and Radiomics Survival Prediction: Contribution to the BRATS 2017 Challenge arxiv:1802.10508v1 [cs.cv] 28 Feb 2018 Fabian Isensee 1, Philipp Kickingereder 2, Wolfgang Wick 3, Martin
Brain Tumor Segmentation and Radiomics Survival Prediction: Contribution to the BRATS 2017 Challenge arxiv:1802.10508v1 [cs.cv] 28 Feb 2018 Fabian Isensee 1, Philipp Kickingereder 2, Wolfgang Wick 3, Martin
DIAGNOSTIC CLASSIFICATION OF LUNG NODULES USING 3D NEURAL NETWORKS
 DIAGNOSTIC CLASSIFICATION OF LUNG NODULES USING 3D NEURAL NETWORKS Raunak Dey Zhongjie Lu Yi Hong Department of Computer Science, University of Georgia, Athens, GA, USA First Affiliated Hospital, School
DIAGNOSTIC CLASSIFICATION OF LUNG NODULES USING 3D NEURAL NETWORKS Raunak Dey Zhongjie Lu Yi Hong Department of Computer Science, University of Georgia, Athens, GA, USA First Affiliated Hospital, School
Healthcare Research You
 DL for Healthcare Goals Healthcare Research You What are high impact problems in healthcare that deep learning can solve? What does research in AI applications to medical imaging look like? How can you
DL for Healthcare Goals Healthcare Research You What are high impact problems in healthcare that deep learning can solve? What does research in AI applications to medical imaging look like? How can you
Retinopathy Net. Alberto Benavides Robert Dadashi Neel Vadoothker
 Retinopathy Net Alberto Benavides Robert Dadashi Neel Vadoothker Motivation We were interested in applying deep learning techniques to the field of medical imaging Field holds a lot of promise and can
Retinopathy Net Alberto Benavides Robert Dadashi Neel Vadoothker Motivation We were interested in applying deep learning techniques to the field of medical imaging Field holds a lot of promise and can
London Medical Imaging & Artificial Intelligence Centre for Value-Based Healthcare. Professor Reza Razavi Centre Director
 London Medical Imaging & Artificial Intelligence Centre for Value-Based Healthcare Professor Reza Razavi Centre Director The traditional healthcare business model is changing Fee-for-service Fee-for-outcome
London Medical Imaging & Artificial Intelligence Centre for Value-Based Healthcare Professor Reza Razavi Centre Director The traditional healthcare business model is changing Fee-for-service Fee-for-outcome
Deep learning and non-negative matrix factorization in recognition of mammograms
 Deep learning and non-negative matrix factorization in recognition of mammograms Bartosz Swiderski Faculty of Applied Informatics and Mathematics Warsaw University of Life Sciences, Warsaw, Poland bartosz_swiderski@sggw.pl
Deep learning and non-negative matrix factorization in recognition of mammograms Bartosz Swiderski Faculty of Applied Informatics and Mathematics Warsaw University of Life Sciences, Warsaw, Poland bartosz_swiderski@sggw.pl
arxiv: v1 [cs.cv] 2 Jul 2018
![arxiv: v1 [cs.cv] 2 Jul 2018 arxiv: v1 [cs.cv] 2 Jul 2018](/thumbs/87/96052165.jpg) A Pulmonary Nodule Detection Model Based on Progressive Resolution and Hierarchical Saliency Junjie Zhang 1, Yong Xia 1,2, and Yanning Zhang 1 1 Shaanxi Key Lab of Speech & Image Information Processing
A Pulmonary Nodule Detection Model Based on Progressive Resolution and Hierarchical Saliency Junjie Zhang 1, Yong Xia 1,2, and Yanning Zhang 1 1 Shaanxi Key Lab of Speech & Image Information Processing
Early Diagnosis of Autism Disease by Multi-channel CNNs
 Early Diagnosis of Autism Disease by Multi-channel CNNs Guannan Li 1,2, Mingxia Liu 2, Quansen Sun 1(&), Dinggang Shen 2(&), and Li Wang 2(&) 1 School of Computer Science and Engineering, Nanjing University
Early Diagnosis of Autism Disease by Multi-channel CNNs Guannan Li 1,2, Mingxia Liu 2, Quansen Sun 1(&), Dinggang Shen 2(&), and Li Wang 2(&) 1 School of Computer Science and Engineering, Nanjing University
LUNG CANCER continues to rank as the leading cause
 1138 IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 24, NO. 9, SEPTEMBER 2005 Computer-Aided Diagnostic Scheme for Distinction Between Benign and Malignant Nodules in Thoracic Low-Dose CT by Use of Massive
1138 IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 24, NO. 9, SEPTEMBER 2005 Computer-Aided Diagnostic Scheme for Distinction Between Benign and Malignant Nodules in Thoracic Low-Dose CT by Use of Massive
arxiv: v1 [cs.cv] 17 Aug 2017
![arxiv: v1 [cs.cv] 17 Aug 2017 arxiv: v1 [cs.cv] 17 Aug 2017](/thumbs/78/78278805.jpg) Deep Learning for Medical Image Analysis Mina Rezaei, Haojin Yang, Christoph Meinel Hasso Plattner Institute, Prof.Dr.Helmert-Strae 2-3, 14482 Potsdam, Germany {mina.rezaei,haojin.yang,christoph.meinel}@hpi.de
Deep Learning for Medical Image Analysis Mina Rezaei, Haojin Yang, Christoph Meinel Hasso Plattner Institute, Prof.Dr.Helmert-Strae 2-3, 14482 Potsdam, Germany {mina.rezaei,haojin.yang,christoph.meinel}@hpi.de
A HMM-based Pre-training Approach for Sequential Data
 A HMM-based Pre-training Approach for Sequential Data Luca Pasa 1, Alberto Testolin 2, Alessandro Sperduti 1 1- Department of Mathematics 2- Department of Developmental Psychology and Socialisation University
A HMM-based Pre-training Approach for Sequential Data Luca Pasa 1, Alberto Testolin 2, Alessandro Sperduti 1 1- Department of Mathematics 2- Department of Developmental Psychology and Socialisation University
Patch-based Head and Neck Cancer Subtype Classification
 Patch-based Head and Neck Cancer Subtype Classification Wanyi Qian, Guoli Yin, Frances Liu, Advisor: Olivier Gevaert, Mu Zhou, Kevin Brennan Stanford University wqian2@stanford.edu, guoliy@stanford.edu,
Patch-based Head and Neck Cancer Subtype Classification Wanyi Qian, Guoli Yin, Frances Liu, Advisor: Olivier Gevaert, Mu Zhou, Kevin Brennan Stanford University wqian2@stanford.edu, guoliy@stanford.edu,
Convolutional Neural Networks for Estimating Left Ventricular Volume
 Convolutional Neural Networks for Estimating Left Ventricular Volume Ryan Silva Stanford University rdsilva@stanford.edu Maksim Korolev Stanford University mkorolev@stanford.edu Abstract End-systolic and
Convolutional Neural Networks for Estimating Left Ventricular Volume Ryan Silva Stanford University rdsilva@stanford.edu Maksim Korolev Stanford University mkorolev@stanford.edu Abstract End-systolic and
LUNG NODULE SEGMENTATION IN COMPUTED TOMOGRAPHY IMAGE. Hemahashiny, Ketheesan Department of Physical Science, Vavuniya Campus
 LUNG NODULE SEGMENTATION IN COMPUTED TOMOGRAPHY IMAGE Hemahashiny, Ketheesan Department of Physical Science, Vavuniya Campus tketheesan@vau.jfn.ac.lk ABSTRACT: The key process to detect the Lung cancer
LUNG NODULE SEGMENTATION IN COMPUTED TOMOGRAPHY IMAGE Hemahashiny, Ketheesan Department of Physical Science, Vavuniya Campus tketheesan@vau.jfn.ac.lk ABSTRACT: The key process to detect the Lung cancer
Using Deep Convolutional Networks for Gesture Recognition in American Sign Language
 Using Deep Convolutional Networks for Gesture Recognition in American Sign Language Abstract In the realm of multimodal communication, sign language is, and continues to be, one of the most understudied
Using Deep Convolutional Networks for Gesture Recognition in American Sign Language Abstract In the realm of multimodal communication, sign language is, and continues to be, one of the most understudied
Using Deep Convolutional Neural Networks to Predict Semantic Features of Lesions in Mammograms
 Using Deep Convolutional Neural Networks to Predict Semantic Features of Lesions in Mammograms Vibhu Agarwal Stanford University vibhua@stanford.edu Clayton Carson Stanford University carsoncz@stanford.edu
Using Deep Convolutional Neural Networks to Predict Semantic Features of Lesions in Mammograms Vibhu Agarwal Stanford University vibhua@stanford.edu Clayton Carson Stanford University carsoncz@stanford.edu
Efficient Deep Model Selection
 Efficient Deep Model Selection Jose Alvarez Researcher Data61, CSIRO, Australia GTC, May 9 th 2017 www.josemalvarez.net conv1 conv2 conv3 conv4 conv5 conv6 conv7 conv8 softmax prediction???????? Num Classes
Efficient Deep Model Selection Jose Alvarez Researcher Data61, CSIRO, Australia GTC, May 9 th 2017 www.josemalvarez.net conv1 conv2 conv3 conv4 conv5 conv6 conv7 conv8 softmax prediction???????? Num Classes
Lung Region Segmentation using Artificial Neural Network Hopfield Model for Cancer Diagnosis in Thorax CT Images
 Automation, Control and Intelligent Systems 2015; 3(2): 19-25 Published online March 20, 2015 (http://www.sciencepublishinggroup.com/j/acis) doi: 10.11648/j.acis.20150302.12 ISSN: 2328-5583 (Print); ISSN:
Automation, Control and Intelligent Systems 2015; 3(2): 19-25 Published online March 20, 2015 (http://www.sciencepublishinggroup.com/j/acis) doi: 10.11648/j.acis.20150302.12 ISSN: 2328-5583 (Print); ISSN:
Developing ML Models for semantic segmentation of medical images
 Developing ML Models for semantic segmentation of medical images Raj Jena University of Cambridge Microsoft Research Cambridge Disclosures I perform consultancy work for InnerEye team at Microsoft Research
Developing ML Models for semantic segmentation of medical images Raj Jena University of Cambridge Microsoft Research Cambridge Disclosures I perform consultancy work for InnerEye team at Microsoft Research
Automatic Classification of Perceived Gender from Facial Images
 Automatic Classification of Perceived Gender from Facial Images Joseph Lemley, Sami Abdul-Wahid, Dipayan Banik Advisor: Dr. Razvan Andonie SOURCE 2016 Outline 1 Introduction 2 Faces - Background 3 Faces
Automatic Classification of Perceived Gender from Facial Images Joseph Lemley, Sami Abdul-Wahid, Dipayan Banik Advisor: Dr. Razvan Andonie SOURCE 2016 Outline 1 Introduction 2 Faces - Background 3 Faces
arxiv: v1 [cs.ai] 28 Nov 2017
![arxiv: v1 [cs.ai] 28 Nov 2017 arxiv: v1 [cs.ai] 28 Nov 2017](/thumbs/78/77416856.jpg) : a better way of the parameters of a Deep Neural Network arxiv:1711.10177v1 [cs.ai] 28 Nov 2017 Guglielmo Montone Laboratoire Psychologie de la Perception Université Paris Descartes, Paris montone.guglielmo@gmail.com
: a better way of the parameters of a Deep Neural Network arxiv:1711.10177v1 [cs.ai] 28 Nov 2017 Guglielmo Montone Laboratoire Psychologie de la Perception Université Paris Descartes, Paris montone.guglielmo@gmail.com
arxiv: v2 [cs.lg] 1 Jun 2018
![arxiv: v2 [cs.lg] 1 Jun 2018 arxiv: v2 [cs.lg] 1 Jun 2018](/thumbs/89/101161632.jpg) Shagun Sodhani 1 * Vardaan Pahuja 1 * arxiv:1805.11016v2 [cs.lg] 1 Jun 2018 Abstract Self-play (Sukhbaatar et al., 2017) is an unsupervised training procedure which enables the reinforcement learning agents
Shagun Sodhani 1 * Vardaan Pahuja 1 * arxiv:1805.11016v2 [cs.lg] 1 Jun 2018 Abstract Self-play (Sukhbaatar et al., 2017) is an unsupervised training procedure which enables the reinforcement learning agents
Big Image-Omics Data Analytics for Clinical Outcome Prediction
 Big Image-Omics Data Analytics for Clinical Outcome Prediction Junzhou Huang, Ph.D. Associate Professor Dept. Computer Science & Engineering University of Texas at Arlington Dept. CSE, UT Arlington Scalable
Big Image-Omics Data Analytics for Clinical Outcome Prediction Junzhou Huang, Ph.D. Associate Professor Dept. Computer Science & Engineering University of Texas at Arlington Dept. CSE, UT Arlington Scalable
Attentional Masking for Pre-trained Deep Networks
 Attentional Masking for Pre-trained Deep Networks IROS 2017 Marcus Wallenberg and Per-Erik Forssén Computer Vision Laboratory Department of Electrical Engineering Linköping University 2014 2017 Per-Erik
Attentional Masking for Pre-trained Deep Networks IROS 2017 Marcus Wallenberg and Per-Erik Forssén Computer Vision Laboratory Department of Electrical Engineering Linköping University 2014 2017 Per-Erik
Visual interpretation in pathology
 13 Visual interpretation in pathology Tissue architecture (alteration) evaluation e.g., for grading prostate cancer Immunohistochemistry (IHC) staining scoring e.g., HER2 in breast cancer (companion diagnostic
13 Visual interpretation in pathology Tissue architecture (alteration) evaluation e.g., for grading prostate cancer Immunohistochemistry (IHC) staining scoring e.g., HER2 in breast cancer (companion diagnostic
White Paper Estimating Complex Phenotype Prevalence Using Predictive Models
 White Paper 23-12 Estimating Complex Phenotype Prevalence Using Predictive Models Authors: Nicholas A. Furlotte Aaron Kleinman Robin Smith David Hinds Created: September 25 th, 2015 September 25th, 2015
White Paper 23-12 Estimating Complex Phenotype Prevalence Using Predictive Models Authors: Nicholas A. Furlotte Aaron Kleinman Robin Smith David Hinds Created: September 25 th, 2015 September 25th, 2015
Segmentation of Cell Membrane and Nucleus by Improving Pix2pix
 Segmentation of Membrane and Nucleus by Improving Pix2pix Masaya Sato 1, Kazuhiro Hotta 1, Ayako Imanishi 2, Michiyuki Matsuda 2 and Kenta Terai 2 1 Meijo University, Siogamaguchi, Nagoya, Aichi, Japan
Segmentation of Membrane and Nucleus by Improving Pix2pix Masaya Sato 1, Kazuhiro Hotta 1, Ayako Imanishi 2, Michiyuki Matsuda 2 and Kenta Terai 2 1 Meijo University, Siogamaguchi, Nagoya, Aichi, Japan
Design of Palm Acupuncture Points Indicator
 Design of Palm Acupuncture Points Indicator Wen-Yuan Chen, Shih-Yen Huang and Jian-Shie Lin Abstract The acupuncture points are given acupuncture or acupressure so to stimulate the meridians on each corresponding
Design of Palm Acupuncture Points Indicator Wen-Yuan Chen, Shih-Yen Huang and Jian-Shie Lin Abstract The acupuncture points are given acupuncture or acupressure so to stimulate the meridians on each corresponding
Improving Network Accuracy with Augmented Imagery Training Data
 Improving Network Accuracy with Augmented Imagery Training Data Ted Hromadka 1, LCDR Niels Olson 2 MD 1, 2 US Navy NMCSD Presented at GTC 2017 Goals Determine effectiveness of various affine transforms
Improving Network Accuracy with Augmented Imagery Training Data Ted Hromadka 1, LCDR Niels Olson 2 MD 1, 2 US Navy NMCSD Presented at GTC 2017 Goals Determine effectiveness of various affine transforms
CS231n Project: Prediction of Head and Neck Cancer Submolecular Types from Patholoy Images
 CSn Project: Prediction of Head and Neck Cancer Submolecular Types from Patholoy Images Kuy Hun Koh Yoo Energy Resources Engineering Stanford University Stanford, CA 90 kohykh@stanford.edu Markus Zechner
CSn Project: Prediction of Head and Neck Cancer Submolecular Types from Patholoy Images Kuy Hun Koh Yoo Energy Resources Engineering Stanford University Stanford, CA 90 kohykh@stanford.edu Markus Zechner
Flexible, High Performance Convolutional Neural Networks for Image Classification
 Flexible, High Performance Convolutional Neural Networks for Image Classification Dan C. Cireşan, Ueli Meier, Jonathan Masci, Luca M. Gambardella, Jürgen Schmidhuber IDSIA, USI and SUPSI Manno-Lugano,
Flexible, High Performance Convolutional Neural Networks for Image Classification Dan C. Cireşan, Ueli Meier, Jonathan Masci, Luca M. Gambardella, Jürgen Schmidhuber IDSIA, USI and SUPSI Manno-Lugano,
arxiv: v1 [cs.cv] 30 May 2018
![arxiv: v1 [cs.cv] 30 May 2018 arxiv: v1 [cs.cv] 30 May 2018](/thumbs/85/92577534.jpg) A Robust and Effective Approach Towards Accurate Metastasis Detection and pn-stage Classification in Breast Cancer Byungjae Lee and Kyunghyun Paeng Lunit inc., Seoul, South Korea {jaylee,khpaeng}@lunit.io
A Robust and Effective Approach Towards Accurate Metastasis Detection and pn-stage Classification in Breast Cancer Byungjae Lee and Kyunghyun Paeng Lunit inc., Seoul, South Korea {jaylee,khpaeng}@lunit.io
Network Dissection: Quantifying Interpretability of Deep Visual Representation
 Name: Pingchuan Ma Student number: 3526400 Date: August 19, 2018 Seminar: Explainable Machine Learning Lecturer: PD Dr. Ullrich Köthe SS 2018 Quantifying Interpretability of Deep Visual Representation
Name: Pingchuan Ma Student number: 3526400 Date: August 19, 2018 Seminar: Explainable Machine Learning Lecturer: PD Dr. Ullrich Köthe SS 2018 Quantifying Interpretability of Deep Visual Representation
arxiv: v1 [cs.cv] 26 Feb 2018
![arxiv: v1 [cs.cv] 26 Feb 2018 arxiv: v1 [cs.cv] 26 Feb 2018](/thumbs/95/125817543.jpg) Classification of breast cancer histology images using transfer learning Sulaiman Vesal 1, Nishant Ravikumar 1, AmirAbbas Davari 1, Stephan Ellmann 2, Andreas Maier 1 arxiv:1802.09424v1 [cs.cv] 26 Feb
Classification of breast cancer histology images using transfer learning Sulaiman Vesal 1, Nishant Ravikumar 1, AmirAbbas Davari 1, Stephan Ellmann 2, Andreas Maier 1 arxiv:1802.09424v1 [cs.cv] 26 Feb
A comparative study of machine learning methods for lung diseases diagnosis by computerized digital imaging'"
 A comparative study of machine learning methods for lung diseases diagnosis by computerized digital imaging'" Suk Ho Kang**. Youngjoo Lee*** Aostract I\.' New Work to be 1 Introduction Presented U Mater~al
A comparative study of machine learning methods for lung diseases diagnosis by computerized digital imaging'" Suk Ho Kang**. Youngjoo Lee*** Aostract I\.' New Work to be 1 Introduction Presented U Mater~al
Computational modeling of visual attention and saliency in the Smart Playroom
 Computational modeling of visual attention and saliency in the Smart Playroom Andrew Jones Department of Computer Science, Brown University Abstract The two canonical modes of human visual attention bottomup
Computational modeling of visual attention and saliency in the Smart Playroom Andrew Jones Department of Computer Science, Brown University Abstract The two canonical modes of human visual attention bottomup
B657: Final Project Report Holistically-Nested Edge Detection
 B657: Final roject Report Holistically-Nested Edge Detection Mingze Xu & Hanfei Mei May 4, 2016 Abstract Holistically-Nested Edge Detection (HED), which is a novel edge detection method based on fully
B657: Final roject Report Holistically-Nested Edge Detection Mingze Xu & Hanfei Mei May 4, 2016 Abstract Holistically-Nested Edge Detection (HED), which is a novel edge detection method based on fully
