A comparative study of fat storage quantitation in nematode Caenorhabditis elegans using label and label-free methods
|
|
|
- Francine Sanders
- 5 years ago
- Views:
Transcription
1 University of Massachusetts Medical School Program in Gene Function and Expression Publications and Presentations Molecular, Cell and Cancer Biology A comparative study of fat storage quantitation in nematode Caenorhabditis elegans using label and label-free methods Kelvin Yen University of Massachusetts Medical School Thuc T. Le Purdue University Ankita Bansal Univesity of Massachusetts Medical School See next page for additional authors Follow this and additional works at: Part of the Genetics and Genomics Commons Repository Citation Yen, Kelvin; Le, Thuc T.; Bansal, Ankita; Narasimhan, Sri Devi; Cheng, Ji-Xin; and Tissenbaum, Heidi A., "A comparative study of fat storage quantitation in nematode Caenorhabditis elegans using label and label-free methods" (2010). Program in Gene Function and Expression Publications and Presentations This material is brought to you by escholarship@umms. It has been accepted for inclusion in Program in Gene Function and Expression Publications and Presentations by an authorized administrator of escholarship@umms. For more information, please contact Lisa.Palmer@umassmed.edu.
2 A comparative study of fat storage quantitation in nematode Caenorhabditis elegans using label and label-free methods Authors Kelvin Yen, Thuc T. Le, Ankita Bansal, Sri Devi Narasimhan, Ji-Xin Cheng, and Heidi A. Tissenbaum This article is available at
3 A Comparative Study of Fat Storage Quantitation in Nematode Caenorhabditis elegans Using Label and Label-Free Methods Kelvin Yen 1., Thuc T. Le 2., Ankita Bansal 1, Sri Devi Narasimhan 1, Ji-Xin Cheng 2,3,4 *, Heidi A. Tissenbaum 1,5 * 1 Program in Gene Function and Expression, University of Massachusetts Medical School, Worcester, Massachusetts, United States of America, 2 Weldon School of Biomedical Engineering, Purdue University, West Lafayette, Indiana, United States of America, 3 Purdue Cancer Center, Purdue University, West Lafayette, Indiana, United States of America, 4 Department of Chemistry, Purdue University, West Lafayette, Indiana, United States of America, 5 Program in Molecular Medicine, University of Massachusetts Medical School, Worcester, Massachusetts, United States of America Abstract The nematode Caenorhabditis elegans has been employed as a model organism to study human obesity due to the conservation of the pathways that regulate energy metabolism. To assay for fat storage in C. elegans, a number of fatsoluble dyes have been employed including BODIPY, Nile Red, Oil Red O, and Sudan Black. However, dye-labeled assays produce results that often do not correlate with fat stores in C. elegans. An alternative label-free approach to analyze fat storage in C. elegans has recently been described with coherent anti-stokes Raman scattering (CARS) microscopy. Here, we compare the performance of CARS microscopy with standard dye-labeled techniques and biochemical quantification to analyze fat storage in wild type C. elegans and with genetic mutations in the insulin/igf-1 signaling pathway including the genes daf-2 (insulin/igf-1 receptor), rict-1 (rictor) and sgk-1 (serum glucocorticoid kinase). CARS imaging provides a direct measure of fat storage with unprecedented details including total fat stores as well as the size, number, and lipid-chain unsaturation of individual lipid droplets. In addition, CARS/TPEF imaging reveals a neutral lipid species that resides in both the hypodermis and the intestinal cells and an autofluorescent organelle that resides exclusively in the intestinal cells. Importantly, coherent addition of the CARS fields from the C-H abundant neutral lipid permits selective CARS imaging of the fat store, and further coupling of spontaneous Raman analysis provides unprecedented details including lipid-chain unsaturation of individual lipid droplets. We observe that although daf-2, rict-1, and sgk-1 mutants affect insulin/igf-1 signaling, they exhibit vastly different phenotypes in terms of neutral lipid and autofluorescent species. We find that CARS imaging gives quantification similar to standard biochemical triglyceride quantification. Further, we independently confirm that feeding worms with vital dyes does not lead to the staining of fat stores, but rather the sequestration of dyes in lysosome-related organelles. In contrast, fixative staining methods provide reproducible data but are prone to errors due to the interference of autofluorescent species and the non-specific staining of cellular structures other than fat stores. Importantly, both growth conditions and developmental stage should be considered when comparing methods of C. elegans lipid storage. Taken together, we confirm that CARS microscopy provides a direct, non-invasive, and label-free means to quantitatively analyze fat storage in living C. elegans. Citation: Yen K, Le TT, Bansal A, Narasimhan SD, Cheng J-X, et al. (2010) A Comparative Study of Fat Storage Quantitation in Nematode Caenorhabditis elegans Using Label and Label-Free Methods. PLoS ONE 5(9): e doi: /journal.pone Editor: Simon Melov, Buck Institute for Age Research, United States of America Received April 29, 2010; Accepted August 11, 2010; Published September 16, 2010 Copyright: ß 2010 Yen et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Funding: The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The following funders were used for this grant: Ellison Medical Foundation, Glenn Foundation, William Randolph Hearst Foundation, and grants F32HL089074, AG025891, and R01EB7243 from the National Institutes of Health. Competing Interests: The authors have declared that no competing interests exist. * jcheng@purdue.edu (J-XC); Heidi.Tissenbaum@umassmed.edu (HAT). These authors contributed equally to this work. Introduction One of the benefits of using C. elegans as a model organism is the ability to use powerful, high-throughput, forward genetic screens to discover important genes and pathways [1,2]. Several papers have used these methods to determine genes that are involved in the accumulation of fat stores with possible applications to human obesity [3 5]. The regulatory roles of many of these genes are assigned based on dye-labeled assays for fat stores. However, dyelabeled assays for fat stores produce highly inconsistent results depending on the type of dyes or whether the staining is performed in live or fixed worms [6,7]. Many dye-labeled assays for fat stores do not correlate with standard biochemical assays [6,7]. Consequently, the functional assignments of many genes related to fat storage based on a single method may not be accurate and potentially hinders the functional studies of genes that control fat accumulation in C. elegans. In recent years, an alternative to dye-labeled assay to quantify fat stores in C. elegans has been described with coherent anti-stokes Raman scattering (CARS) microscopy [8,9]. CARS microscopy is a label-free chemical imaging technique that relies on intrinsic molecular vibration as a contrast mechanism [10,11]. CARS PLoS ONE 1 September 2010 Volume 5 Issue 9 e12810
4 microscopy allows direct visualization of lipid-rich organelles due to the abundance of the CH 2 group that has the symmetric stretch vibration frequency of 2840 cm 21. In addition, CARS microscopy is highly multifunctional. It is capable of simultaneous chemical and fluorescent imaging, which allows label-free visualization of both neutral lipid droplets and autofluorescent granules [9]. Furthermore, CARS is capable of Raman spectral analysis, which allows assaying of lipid-chain unsaturation and lipid packing order of individual lipid droplets [8,9,12 14]. With such versatility, CARS microscopy has the potential to become a robust and reliable method to screen for the function of genes that control lipid metabolism in living C. elegans. However, technical challenges are hindering the widespread adoption of CARS microscopy for C. elegans research. First, the very expensive price tag for a CARS microscope places it beyond the affordability of most researchers. Secondly, setting up a CARS microscope system requires expertise in nonlinear optics, which may be a challenge to many biologists. Thirdly, dye-labeled assays are inexpensive and readily available which render them attractive choices. And lastly, many biologists are unfamiliar with the capability of CARS and other nonlinear optical imaging modalities. In this paper, we explore the capability of CARS microscopy for quantitative analysis of fat storage in wild type worms and three strains with mutations in genes involved in the insulin/igf-1 signaling pathway: daf-2(e1370), rict-1(ft7), and sgk-1(ok538) (Figure 1). We directly compare the abilities of CARS with several dyes commonly used in C. elegans, as well as triglyceride quantification to study fat storage. Each of these three mutants has been reported to have abnormal fat stores compared to wild type using a variety of techniques [6,7,15]. daf-2(e1370) mutants bear a mutation in the insulin/igf receptor [15], while rict-1 mutants have a mutation in the RICTOR (Rapamycin Insensitive Companion of mtor) gene [16,17]. sgk-1 mutants have a mutation in the serum/glucocorticoid regulated kinase (SGK) gene [18]. Previous studies using vital stains to assay for fat stores Figure 1. Simplified diagram of insulin/igf signaling. Bolded proteins are proteins that have been investigated in this study. doi: /journal.pone g001 has produced variable results that are inconsistent with standard biochemical assays [6,7]. We compare the performance of labelfree CARS imaging to both dye-labeled imaging and triglyceride quantification to further investigate quantitation of fat stores in C. elegans. We aim to introduce CARS microscopy to researchers who are unfamiliar with its capability for lipid studies in C. elegans. Using CARS, we are able to identify the sources of errors in dyelabeled assays. We also stress the importance of similar growth conditions as well as similar developmental stages when trying to compare methods. Therefore, these studies aim to help researchers improve the accuracy for fat store measurements by standard dye assays should CARS microscopy remain inaccessible. Results CARS/TPEF imaging reveals co-existing neutral lipid stores and autofluorescent granules in C. elegans Fat stores in C. elegans can be visualized by dye-labeled imaging or by label-free imaging (Figure 2, Figure S1, S2). Dye-labeled imaging of fat stores relies on the use of fat-soluble dyes such as Sudan Black, Oil Red O, Nile Red, or BODIPY (Figure 2A, Figure S1, S2). Sudan Black and Oil Red O are fixative-based dyes, whereas Nile Red and BODIPY are vital dyes, which are fed to live worms to assay fat stores [3,7,15,19]. In contrast, label-free imaging of fat stores with CARS microscopy relies on the intrinsic molecular vibration of CH 2 groups for the contrast mechanism (Figure 2B). Simultaneous CARS and two-photon excited fluorescent (TPEF) imaging allows label-free visualization of both neutral lipid droplets and autofluorescent gut granules [9]. Autofluorescent gut granules have been classified as lysosomerelated organelles (LROs) whose expression increases with aging and oxidative stress [20,21]. The partial overlap of the TPEF and CARS signals (Figure 2B) is possibly due to the insufficient depth resolution of our technique. We have observed that all autofluorescent granules contain some CARS signal although the intensity varies from granule to granule. However the converse is not true and all CARS signal is not associated with autofluorescent granules. Thus, multimodal CARS/TPEF imaging suggests the co-existence of 1) neutral lipid stores that resides in both the hypodermis and the intestinal cells, and 2) autofluorescent organelles that reside exclusively in the intestinal cells. Quantitative measurements of fat stores in living C. elegans CARS microscopy allows quantitative measurements of fat stores in living C. elegans. The CARS signal typically arises from both the cytoplasmic lipid droplets or fat stores and the phospholipid membranes (Figure 2B). Importantly, the coherent addition of CARS fields produces a signal from the C-H abundant lipid droplets much larger than that from other cellular organelles. To quantify the fat stores, a threshold can be set such that the CARS signal from the phospholipid membrane and other cellular structures is removed. Integration of the remaining CARS signal intensity yields a numerical value for total fat stores of the probed volume (Figure 3A). In addition, a CARS image can be converted to a binary image for measurements of number and size distribution of lipid droplets using the particle analysis function of the ImageJ software. This approach provides a robust means for lipid droplet analysis. However, when the lipid droplets are too close to one another, errors can be introduced. In such a case, limits can be set for additional optimization of the quantitation (Figure 3A). Furthermore, spontaneous Raman spectral analysis can provide critical information on the lipid chain of the lipid droplets (Figure 3B). The ratio of C = C over C-C or I 1660 /I 1445 PLoS ONE 2 September 2010 Volume 5 Issue 9 e12810
5 Figure 2. Label and label-free imaging methods to assay fat storage in C. elegans. (A) Labeled imaging of fat stores using Sudan Black, Oil Red O, and Nile Red staining of fixed worms and Nile Red and BODIPY-labeled fatty acids fed to live worms. (B) Label-free visualization of neutral lipid species and autofluorescent gut granules using simultaneous CARS and TPEF imaging, respectively. Note the co-localization of TPEF signal with CARS signal in the intestinal cells. Images are presented as 3-D stacks of 30 frames taken at 1 mm increment along the vertical axis. Rightmost panel is an enlargement of the overlaid image with the xy dimensions of 20 mm620 mm. It should be noted that the association of fluorescent signal with lipid signal can be found surrounding, at one end, above, or below the lipid signal. In addition, the lipid contents of the fluorescent particles vary from one to another. For the particular image presented, the fluorescent puncta do exhibit lipid signal when examined at a higher magnification. doi: /journal.pone g002 Figure 3. Quantitative analysis of the total fat stores and the size, number, and composition of lipid droplets with CARS microscopy. (A) Total fat stores are defined as the integrated CARS signal intensity over the probed volume minus the background signal arising from worm bodies (leftmost panel). The ImageJ software is used for binary image conversion of CARS images and for particle analysis to determine the size and number of lipid droplets. Upper limits (2 mm 2 ) can be manually set to minimize sizing errors due to lipid droplets being too close to each other. (B) The compositions of six lipid droplets (crosshairs) are analyzed using spontaneous Raman spectroscopy. Statistically insignificant variability in lipid composition is observed for lipid droplets within a single worm. Leftmost panel is an enlargement of Figure 2A with the xy dimensions of 20 mm620 mm. doi: /journal.pone g003 PLoS ONE 3 September 2010 Volume 5 Issue 9 e12810
6 is a reliable measure of lipid-chain unsaturation and the ratio of asymmetric to symmetric CH 2 stretch vibration or I 2880 /I 2845 can be used to measure lipid-chain packing order [12]. Taken together, CARS microscopy provides quantitative measurements of single lipid droplets that have not been described with standard dye-labeled assays. Comparison of CARS imaging with dyes in insulin/igf-1 signaling mutants To evaluate the performance of CARS microscopy, we compare CARS imaging assays with standard dye-labeled assays to quantitate fat stores. We chose three mutants that appeared to have different lipid profiles by CARS that also happen to affect insulin/igf signaling (Figure 1). We assessed lipid stores in all of the three mutants in comparison to wild type. As shown in Figure 3, we find that staining fat stores in fixed worms with fixative dyes Oil Red O and Sudan Black produce results that partially correlate with CARS imaging of living worms. Qualitatively, we observe that daf-2 mutants exhibit a significant increase in staining level, rict-1 worms exhibit a moderate increase in staining level, and sgk-1 worms exhibit a reduction in staining level as compared to wild type worms (Figure 4A, Figure S1, S2 and quantified in Figure S3). Notably, both fixative dyes give similar results. Similarly, label-free CARS imaging also shows increased fat stores in daf-2 mutants and reduced fat stores in sgk-1 worms as compared to wild type worms (Figure 4B). Thus, the data from the two fixative stains, Oil Red O and Sudan Black, agree with CARS imaging data on daf-2 and sgk-1 worms. Figure 4. Label and label-free imaging of fat storage in wild type and mutant C. elegans. (A) Visualization of fat stores in fixed worms using fixative dyes Sudan Black, Oil Red O, and Nile Red*. Arrows indicate the pharynx (B) Visualization of fat stores (upper panels) and autofluorescent gut granules (lower panels) using simultaneous CARS and TPEF imaging of living worms, respectively. Images are presented as 3-D stacks of 30 frames taken at 1 mm increment along the vertical axis. *Nile Red pictures were taken with a more sensitive black and white camera. doi: /journal.pone g004 PLoS ONE 4 September 2010 Volume 5 Issue 9 e12810
7 However, CARS imaging shows that rict-1 mutants have similar lipid stores when compared to wild type, which is in contrast to the dye staining. To further investigate these differences, we used additional analysis with CARS imaging. When we used simultaneous TPEF imaging, we observed that the level of autofluorescent granules is lower in daf-2 mutants, higher in rict-1 mutants, and similar in sgk-1 mutants when compared with wild type worms (Figure 4B). However, the autofluorescent phenotypes of these mutants have largely been omitted in the literature. It is possible that the dramatic increase in the level of autofluorescent species in rict-1 worms could interfere with the evaluation of neutral lipid species using fixative dyes. Fed Nile Red and Fed BODIPY do not reveal fat stores We next examined staining fat stores by feeding worms with vital dyes Nile Red and BODIPY. As shown in Figure 5, the results from feeding worms dyes to stain lipids produce data that do not correlate with CARS imaging of live worms. Quantifying the total fluorescence from the worms using ImageJ from vital labeling show reduced fat stores in daf-2 worms, increased fat stores in rict-1 worms, and similar fat stores in sgk-1 worms as compared to wild type worms (Figure 5A). We find that the Nile Red and BIODIPY vital labeling data do not agree with fixative labeling data using Oil Red O, Sudan Black, and Nile Red, or with label-free CARS imaging data. Our results show that there is an intrinsic difference between the capacities of dyes to stain fat stores in live and fixed worms. Previously, Nile Red staining of fixed worms has been shown to be a better proxy for fat stores than Nile Red staining of live worms [6]. Indeed, our assays for fat stores using Nile Red staining of fixed daf-2 and sgk-1 worms produce data that in general tend to agree with those obtained with fixative dyes and CARS imaging (Figure 5A). However, fixed Nile Red staining also shows an increase in fat stores in rict-1 worms, whereas, CARS imaging show comparable level of fat stores relative to wild type worms. Further verification of fat stores using biochemical methods corroborates the CARS findings (see Text S1 and Figure S4). Quantitation of lipids with CARS Next, we further extended our analysis of lipids in C. elegans using CARS imaging. In addition to assaying total fat stores, CARS imaging allows quantitative analysis of autofluorescent granules, size and number of lipid droplets, and the degree of lipidchain unsaturation (Figure 5B). Compared to wild type worms, daf-2 mutants exhibit a two-fold increase in the CARS signal and a three-fold reduction in the autofluorescent signal. rict-1 mutants when compared to wild type, exhibit similar levels of the CARS signal and a two-fold increase in the autofluorescent signal. sgk-1 mutants exhibit a three-fold decrease in the CARS signal and a twenty percent reduction in the autofluorescent signal when compared to wild type. When we examined the size of lipid droplets, we found that the lipid droplet size is approximately 1.4 mm 2 in wild type worms as well as daf-2 and rict-1 mutants, while sgk-1 mutants were approximately 1 mm 2. Further analysis showed that compared to wild type worms, daf-2 mutants have a two-fold increase in the number of lipid droplets and sgk-1 mutants have one-third reduction in lipid droplets (Figure 5B). The number of lipid droplets in rict-1 mutants is statistically comparable to that in wild type worms. In general, all three mutants have a lower degree of lipid-chain unsaturation compared to wild type worms. One possibility for this lower lipid-chain unsaturation is that mutation in either daf-2, rict-1, or sgk-1 might have a negative effect on the expression or activity of lipid desaturation enzymes, or the ability to uptake unsaturated dietary lipids. Figure 5. Quantitative analysis of fat storage in wild type and mutant C. elegans. (A) Total fat stores in mutant worms relative to wild type worms assayed by feeding worms with vital dyes Nile Red and BODIPY-labeled fatty acids, with CARS imaging, and with fixative staining with Nile Red. Data represent the average of 3 independent trials with an average of 18 worms quantified per trial for fed Nile Red, 20 worms quantified per trial for fed BODIPY, 9 worms quantified per trial for CARS imaging, and 23 worms quantified per trial for Nile Red on fixed worms. Data are normalized to 100 for wild type worms and comparatively for mutant worms. Error bars represent the standard error. (B) Quantitative analysis of autofluorescent granules using TPEF imaging and size, number, and lipid unsaturation of lipid droplets using CARS imaging. TPEF data are normalized to 100 for wild type worms and comparatively for mutant worms. Lipid unsaturation represents the ratio of C = C peak intensity over C-C peak intensity, or I 1660 /I Data represent the average of 3 independent trials with lipid droplets of 9 worms quantified per trial, or 27 worms total. Error bars represent the standard error. doi: /journal.pone g005 PLoS ONE 5 September 2010 Volume 5 Issue 9 e12810
8 Examining sources of errors with fat storage analysis in C. elegans CARS microscopy measures fat stores directly, whereas fixative or vital dyes serve as proxies for fat stores. Therefore, the inconsistencies between dye-labeled assays are likely due to labeling efficiency or errors during quantitation of the dye signal. To try to investigate the source of errors in dye-labeled assays, we first examined the spectral properties of two fluorescent dyes, Nile Red and BODIPY as well as the autofluorescent granules. We find that the emission spectra of autofluorescent granules and BODIPY peak at 530 nm and 520 nm, respectively, when excited with a 457 nm continuous-wave laser (Figure 6A). The broad emission spectrum of the autofluorescent granules therefore suggests that when fat stores are assayed with BODIPY, the readout inevitably includes the autofluorescent signal. Furthermore, a recent study shows that BODIPY does not stain fat stores when fed to live worms [7]. Taken together, BODIPY should not be used as a proxy for fat stores in live or fixed worms. On the other hand, the conditions for Nile Red signal detection vary among researchers. Some researchers used the Rhodamine emission filters that centers at 575 nm to detect the Nile Red signal [3]. At this wavelength, significant autofluorescent signal can still be detected according to the spectral data (Figure 6A). Thus, some errors in the Nile Red assays for fat stores can be attributed to the signal bleed-through from the autofluorescent granules. Ideally, the emission filter for Nile Red should center around 650 nm. Nonetheless, we observe minimal bleed-through of the autofluorescent signal using a 600 nm emission filter and twophoton excitation at 885 nm (Figure 6B). In fact, most researchers use the emission filters that center around 600 nm, yet vital staining of fat stores using Nile Red still does not correlate with biochemical assays [5 7]. Figure 6. Possible errors in dye-labeled assays to analyze fat stores in living C. elegans. (A) Emission spectra of autofluorescent granules, BODIPY, and Nile Red obtained with microspectrometry. The autofluorescent granules and BODIPY are excited at 457 nm using an Argon-Ion continuous-wave laser. The Nile Red is excited with a 543 nm Helium-Neon continuous-wave laser. (B) Insignificant bleed-through of autofluorescent granule signal is observed using a 600 nm bandpass filter and two-photon excitation at 885 nm. (C) CARS and TPEF imaging of two rict-1 worms fed with Nile Red. Autofluorescent granules (blue) are detected with a 520 nm bandpass filter and Nile Red (red) is detected with a 600 nm bandpass filter. Pink color is the result of overlapping blue and red colors which indicates co-localization of autofluorescent granules and Nile Red. Note the complete co-localization of autofluorescent granules and Nile Red in worm 1 (upper panels) and partial co-localization of autofluorescent granules and Nile Red in worm 2 (lower panels). Rightmost panels are enlargements of the CARS/Nile Red overlaid images with the xy dimensions of 20 mm620 mm. Note the poor co-localization of lipid droplets (grey) and Nile Red (red). Images are presented as 3-D stacks of 30 frames taken at 1 mm increment along the vertical axis. doi: /journal.pone g006 PLoS ONE 6 September 2010 Volume 5 Issue 9 e12810
9 To further investigate the source of errors by obtaining CARS and fluorescent images of live worms fed with Nile Red dye, we obtained fluorescent images at both 520 nm for autofluorescent granules and 600 nm for Nile Red dye (Figure 6C). We observed that in some worms Nile Red signal completely co-localizes with the autofluorescent signal (Figure 6C top row). This observation is consistent with many previous findings where autofluorescent granules were identified as LROs and Nile Red was reported to stain LROs [7,20]. The presence of more autofluorescent signal than the Nile Red signal could be due to insufficient Nile Red concentration or due to the degradation of the Nile Red dye. In some other worms, only some of the Nile Red signal co-localizes with the autofluorescent signal (Figure 6C bottom row). However, in all worms, the Nile Red signal localizes poorly with the CARS signal for fat stores. This observation is quite surprising given the proven ability of Nile Red to stain lipid droplets in mammalian cells and in fixed worms [6,22]. It is highly probable that Nile Red enters the intestinal cells of C. elegans via endocytosis, and then remains in the endosomes which eventually mature into lysosomes [20]. It is unlikely that Nile Red enters the cytoplasm where the lipid droplets reside because in this scenario Nile Red should stain lipid droplets given its nonspecific affinity for lipids in general. Our data clearly suggests that Nile Red should not be used as a proxy for fat stores in live worms. Discussion As C. elegans is increasingly used as a model system to study the basic mechanisms of fat storage and energy homeostasis, it is critical to ensure that the established assays accurately determine and reflect fat content. To that effect, here we study the performance of existing techniques including vital and fixative labeling and label-free assays to evaluate fat stores. We find that vital staining with the fat-soluble dyes Nile Red and BODIPY labeled fatty acids cannot serve as proxies for fat stores. Nile Red and BODIPY-labeled fatty acids do not stain fat stores but stain autofluorescent organelles previously identified as LROs in living worms [7]. It is possible that Nile red and BODIPY-labeled fatty acids are actively transported into intestinal cells. In fact, a specific transporter for Nile Red has been identified in yeast [23]. Similar transporters in worms may sequester Nile Red and BODIPYlabeled fatty acids, thus shielding them from the fat stores in living worms. Furthermore, Clokey and Jacobson reported over two decades ago that exogenous fluorescent probes are taken up by endocytosis and accumulate within the autofluorescent lipofuscin granules, which are the secondary lysosomes and active recipients of endocytosed fluorescent probes [20]. Our findings, together with other independent reports, strongly suggest that vital staining using fat-soluble dyes including both Nile Red and BODIPY, should not be used to assay fat stores in living C. elegans. We find that fixative staining using Sudan Black, Oil Red O, and Nile Red produce consistent and reproducible data on fat stores (Table 1). Qualitatively, fixative staining data agree with CARS imaging data on the level of fat stores in mutant daf-2 and sgk-1 relative to wild type worms. However, our fixative staining data differ with CARS imaging data and biochemical analysis on the level of fat stores in rict-1 mutants where there is a high level of autofluorescent gut granules/lipofuscin. These results are in contrast to those found by Soukas et al. [16] and are further discussed in Text S1. Since CARS microscopy provides a direct measure of fat stores instead of a proxy of fat storage, it is possible that this difference could be attributable to errors introduced during fixation or staining procedures. Additional error could also be due to the following reasons. First, lipofuscin is a hydrophobic Table 1. Qualitative comparison of fat stores in mutants relative to wild type worms. Strain Method daf-2 (e1370) rict-1 (ft7) sgk-1 (ok538) Fed Nile Red decrease increase no change Fed BODIPY fatty acids decrease no change no change Fixed Nile Red increase increase decrease Fixed Sudan Black increase increase decrease Fixed Oil-Red-O increase increase decrease CARS (neutral lipids) increase no change decrease CARS (autofluorescence) decrease increase no change doi: /journal.pone t001 accumulation of highly oxidized cellular proteins and lipids that have characteristic fluorescent properties [24,25]. Their hydrophobic properties pose a problem for lipophilic stains because both Sudan Black and Oil Red O stain for lipofuscin as well as triglycerides [26,27]. As Nile Red is also a hydrophilic stain, it is possible that it could also stain lipofuscin. Therefore, fixative staining cannot distinguish neutral fat stores from lipofuscin. Second, the autofluorescence of lipofuscin might contribute to errors in the quantitation of fluorescent stains such as Nile Red and BODIPY. Third, formaldehyde fixation of the worms introduces additional non-specific fluorescence due to the fluorescent properties of the Schiff bases produced [28]. Finally, non-specific staining of dyes, variable dye concentration, and thoroughness of washing procedures are among the other factors that might contribute to the inaccuracy in assaying for fat stores using labeling techniques [29 31]. Therefore, although fixative staining offers a simple and reproducible means to assay fat stores in C. elegans, we suggest that errors may arise due to interference from autofluorescence granules and non-specific staining of lipofuscin and other cellular organelles. Thus, fixative staining to assay for fat stores in C. elegans should also present data on autofluorescent granules. In contrast to labeling techniques, label-free CARS imaging provides a direct and reliable means to assay fat stores in living C. elegans. In addition, CARS imaging allows for detailed analysis of the number, size, and lipid-chain unsaturation of single lipid droplets. Multimodal CARS and TPEF imaging further allows simultaneous visualization of neutral lipids and autofluorescent lipofuscins. Together, CARS microscopy allows for the assaying of various aspects of fat storage in living C. elegans in a non-invasive and label-free manner. Fat storage in the form of lipid droplets has recently emerged as a complex process that is dependent on insulin signaling, phospholipid synthesis, fatty acids synthesis and desaturation, fatty acids uptake and transport, activation of nuclear hormone receptors, and upregulation of a wide range of lipid synthesis enzymes [32 39]. Genetic mutations of genes that control lipid metabolism in C. elegans and other organisms have been shown to impact the total fat stores, as well as the size, number, and composition of lipid droplets [36,40,41]. Furthermore, a strong correlation between fat storage, oxidative stress, and the lifespan of C. elegans has been revealed in recent years [15 17,42 44]. Therefore, the versatility of CARS microscopy is critical not only for forward genotype-phenotype screening of genes that control fat accumulation, but also for the studies of potential interactions of lipid metabolism genes. Most importantly, CARS microscopy is currently the only reliable method to assay for fat stores in living C. elegans. When combined with recent PLoS ONE 7 September 2010 Volume 5 Issue 9 e12810
10 advances in microfluidic devices [45 47], this capability should allow dynamic studies of the correlation between lipid metabolism, behavioral response, and aging over the lifetime of a single worm. Conclusions We have used several methods including fixative stains, live stains and live imaging to analyze C. elegans fat storage. Our data supports previous publications that vital dyes should be used with caution for lipid analysis in C. elegans. Using CARS microscopy we show Nile Red itself stains the autofluorescent granules and this is the confounding problem. Importantly, we show that similar stages and growth conditions should be used when comparing different methods (Figure S5). We suggest that CARS is the optimal method for C. elegans fat storage. However, when not available, we suggest using one of the fixative methods as well as checking autofluorescent levels. Materials and Methods Worm strains and growing conditions All C. elegans strains used in this work were obtained from the Caenorhabditis Genetics Center. Wild type N2, daf-2(e1370), sgk- 1(ok538) and rict-1(ft7) mutants were maintained at 15uC using standard C. elegans techniques [48]. All strains except for daf- 2(e1370) were shifted to room temperature at 22.5uC overnight, while daf-2(e1370) worms were shifted to 20uC overnight, prior to label or label-free assays. All assays were repeated for a total of three times using worms at the L3/L4 stage. Worm fixation Worms were washed twice with 1 x PBS and then suspended in 120 ml of PBS to which an equal volume of 2X MRWB buffer (160 mm KCl, 40 mm NaCl, 14 mm Na 2 EGTA, PIPES ph 7.4, 1 mm Spermidine, 0.4 mm Spermine, 30 mm, 2% Paraformaldehyde, 0.2% beta-mercaptoethanol) was added. The worms were taken through 3 freeze-thaw cycles between dry ice/ethanol and warm running tap water, followed by spinning at 14000g washing once in PBS to remove paraformaldehyde. Nile Red and BODIPY-labeled fatty acid feeding protocol Nile Red (Cat. No. N1142, Invitrogen, Carlsbad, CA, USA) diluted in water (100 ng/ml final concentration) or BODIPYlabeled fatty acids (Cat. No. D-3823, Invitrogen, Carlsbad, CA, USA) diluted in water (1 mm final concentration) was overlaid on an NGM media plate and allowed to dry overnight. Worms were then transferred to the plate for at least 24 hours before fixing the worms. Fixed Nile Red Staining A stock solution was made by dissolving Nile Red in acetone. It was then diluted in water (100 ng/ml final concentration) and fixed worms were incubated overnight in the working solution. Sudan Black Staining Sudan Black (Cat. No , Sigma-Aldrich, St. Louis, MO, USA) staining of stored fat was performed as previously described [15], except for the fixation step as described above. After fixation, worms were sequentially dehydrated by washes in 25%, 50% and 70% ethanol. Saturated Sudan Black solution was prepared fresh in 70% ethanol. The fixed worms were incubated overnight in 250 ml of Sudan Black solution, on a shaker at room temperature. Quantification of Sudan Black staining Using ImageJ, we measured the average pixel intensity for a 30- pixel radius immediately behind the pharynx of each animal. A minimum of 9 animals was measured for each strain and we repeated the experiments an additional 2 times. Significance was determined by Student s t-test. Oil Red O Staining Oil Red O staining was performed as previously described [16]. After fixation, worms were resuspended and dehydrated in 60% isopropanol. Approximately 250 ml of 60% Oil Red O stain (Cat. No. O9755, Sigma-Aldrich, St. Louis, MO, USA) was added to each sample, and samples were incubated overnight at room temperature. Oil Red O was prepared as follows: 0.5 g of Oil Red O powder was dissolved in 100 ml isopropanol solution and equilibrated for several days. The solution was then freshly diluted with 40% water to get a 60% stock and allowed to sit 10 minutes at room temperature and filtered using 0.2 to 0.4 mm filters. Quantification of Oil Red O staining Using ImageJ, we separated out each color image into its RGB channel components. As it has been reported that Oil Red O absorbs light at 510 nm, we used the green channel for further analysis [49]. We measured the average pixel intensity for a 40 pixel radius immediately behind the pharynx of each animal. In addition, we measured a 40 pixel radius of the background, which was later subtracted from the values obtained from the staining. A minimum of 9 animals was measured for each strain and we repeated the experiments an additional 2 times. Significance was determined by Student s t-test. Triglyceride Assay Biovision s Triglyceride Quantification Kit (Mountain View, CA) was used to assay for triglyceride content. Worms were synchronized by hypochlorite treatment and then grown at 15uC. They were then upshifted to 20uC for 8 hours until they were at the L3/L4 stage. Worms were then collected and washed with S basal solution. A 5% Triton X-100 solution with 1x protease inhibitors (Roche complete mini, EDTA-free Indianapolis, IN) was added 1:1 to a 50 ul worm pellet. The worms were sonicated with a Bioruptor (Diagenode, Sparta, NJ) using power output 4 for 10 seconds. Protein content was estimated by BCA method. Lipids were dissolved by heating the lysate to 90uC for 5 minutes followed by vortexing. This was preformed twice before the lysate was cleared by centrifugation. The supernatant was then used for the triglyceride assay per the manufacturers instructions. At least 2 biological replicates were used for each strain and three technical replicates were used for each biological replicate. Imaging conditions for dye-labeled assays Fixed worms were mounted on slides and visualized using a Zeiss Axioscope 2+ microscope. ImageJ software was used to quantify total fluorescence from pictures obtained from the microscope. Exposure times were kept constant within each trial. NR and SB images were obtained using an ORCA ER CCD camera (Hamamatsu Photonics, Japan). ORO images were obtained using an Axiocam HRc CCD camera (Carl Zeiss, Thornwood, NY). Fluorescent dyes were excited using a 100W HBO mercury lamp in conjunction with a FITC (480ex/535em) or TRITC (540ex/605em) filter set (Chroma Technology, Bellows Falls, VT) for BODIPY or NR respectively. A multifunctional CARS microscope Two mode-locked 5-ps Ti:sapphire lasers (Tsunami, Spectra- Physics, Mountain View, CA) were synchronized to each other through an electronic module controller (Lok-to-Clock, Spectra- PLoS ONE 8 September 2010 Volume 5 Issue 9 e12810
11 Physics). The two parallel-polarized laser beams, pump and Stokes, were collinearly combined and sent into a laser scanning confocal microscope (FV300/IX71, Olympus America, Center Valley, PA). Pump and Stokes lasers were tuned to cm 21 (or 707 nm) and cm 21 (or 885 nm), respectively, to be in resonance with the CH 2 symmetric stretch vibration at 2840 cm 21. The combined beams were focused into the sample through a 60x water immersion microscope objective with a 1.2 numerical aperture. Forward-detected CARS signal was collected by an air condenser with a 0.55 numerical aperture, transmitted through a 600/65 nm bandpass filter (Cat. No , Ealing Catalog, Rocklin, CA), and detected by a photomultiplier tube (PMT, H , Hamamatsu, Japan). Simultaneously, backreflected TPEF signal was collected by the same illuminating objective, spectrally separated from the excitation source, transmitted through a 520/40 nm bandpass filter (Cat. No , Chroma Technology, Bellows Falls, VT), and detected by a photomultiplier tube (PMT, H , Hamamatsu, Japan) mounted at the back port of the microscope. To detect Nile Red fluorescence, the pump beam was blocked and the Stokes beam at 885 nm was used for excitation. To obtain Raman spectra of lipid droplets, the Stokes beam was blocked and the pump-laser induced Raman scattering signal was directed toward a spectrometer to permit spectral analysis from 830 cm 21 to 3100 cm 21, which covers both the fingerprint and the CH-stretch vibration regions. A spectrometer with a 300 g/mm grating and a TE cooled back-illuminated EMCCD (Newton 920-BRD, Andor Technology, Belfast, Ireland) was mounted to the side port of the microscope to allow spontaneous Raman spectral analysis on the same microscope platform. Average acquisition time for a pixels CARS image was 1.12 second and a full-spectral Raman analysis was 4 seconds. The combined Stokes and pump laser power was kept constantly at 40 mw. For all Raman spectral measurements, pump laser power was reduced to 10 mw. CARS imaging conditions and data analysis All C. elegans were anesthetized in a droplet of 100 mm sodium azide and mounted on fresh 2% agarose slides prior to imaging. To evaluate the expression level of neutral and autofluorescent bodies, a probe volume with xyz dimensions of 125 mm6125 mm629 mm was defined at the mid-section of wild type or mutant worms. Simultaneous depth imaging with CARS and TPEF along the vertical (z) axis was performed at 1 mm step size to obtain 30 frames. Total CARS and TPEF signal arising from worms were integrated over 30 frames and divided by the worm volumes to obtain average pixel intensity values. Thus, the average pixel intensity values were not affected by size variability among worms. Background CARS and TPEF signal were defined as those arising from the worm bodies devoid of lipid droplets or autofluorescent granules. Background CARS and TPEF pixel intensity were subtracted from the average pixel intensity of CARS and TPEF signal to obtain CARS signal for fat stores and TPEF signal for autofluorescent granules, respectively. Fluoview software (Olympus America, Center Valley, PA) was used for image acquisition and display. ImageJ software was used for particle analysis. Supporting Information Text S1 Found at: doi: /journal.pone s001 (0.03 MB DOC) Figure S1 Additional Sudan Black stained worms. Arrow indicates the pharynx of the worm. Found at: doi: /journal.pone s002 (2.28 MB TIF) Figure S2 Additional Oil Red O stained worms. Arrow indicates the pharynx of the worm. Found at: doi: /journal.pone s003 (3.65 MB TIF) Figure S3 Quantification of Sudan Black and Oil Red O staining. Both daf-2 and rict-1 have increased staining compared to wild type. sgk-1 mutants have decreased staining. * indicates a significant difference compared to wild type. + in 2 out of 3 trials there was a significant difference from wild type. Found at: doi: /journal.pone s004 (1.44 MB TIF) Figure S4 Quantification of triglyceride stores. Corroborating the data from the CARS quantification, there is a significant increase in triglycerides in daf-2 worms, a significant decrease in triglycerides in sgk-1 worms, and no change in triglycerides in rict- 1 worms. Triglycerides has been normalized to total protein levels and the units for the Y-axis is nmoles of triglycerides/gram of protein. Found at: doi: /journal.pone s005 (0.49 MB TIF) Figure S5 Differential fat stores in larval stage 3 (L3) worms compared to larval stage 4 (L4) worms. Typical Oil Red O staining of an L3 sgk-1 worm compared to an L4 sgk-1 worm. The top panel is an L3 worm and the bottom is an L4 worm. Found at: doi: /journal.pone s006 (7.78 MB TIF) Acknowledgments We thank Nina Bhabhalia and John Davis for assistance. We thank members of the Tissenbaum lab for advice. Strains were kindly provided by the Caenorhabditis Genetics Center. Author Contributions Conceived and designed the experiments: KY TTL JXC HAT. Performed the experiments: KY TTL AB SDN. Analyzed the data: KY TTL AB SDN JXC HAT. Contributed reagents/materials/analysis tools: KY TTL JXC. Wrote the paper: KY TTL JXC HAT. References 1. Kaletta T, Hengartner MO (2006) Finding function in novel targets: C. elegans as a model organism. Nat Rev Drug Discov 5: McKay RM, McKay JP, Avery L, Graff JM (2003) C-elegans: A model for exploring the genetics of fat storage. Dev Cell 4: Ashrafi K, Chang FY, Watts JL, Fraser AG, Kamath RS, et al. (2003) Genomewide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 421: Mak HY, Nelson LS, Basson M, Johnson CD, Ruvkun G (2006) Polygenic control of Caenorhabditis elegans fat storage. Nat Genet 38: Srinivasan S, Sadegh L, Elle IC, Christensen AGL, Faergeman NJ, et al. (2008) Serotonin regulates C. elegans fat and feeding through independent molecular mechanisms. Cell Metab 7: Brooks KK, Liang B, Watts JL (2009) The Influence of Bacterial Diet on Fat Storage in C. elegans. PLoS One 4(10): e O Rourke EJ, Soukas AA, Carr CE, Ruvkun G (2009) C. elegans Major Fats Are Stored in Vesicles Distinct from Lysosome-Related Organelles. Cell Metab 10: Hellerer T, Axang C, Brackmann C, Hillertz P, Pilon M, et al. (2007) Monitoring of lipid storage in Caenorhabditis elegans using coherent anti-stokes Raman scattering (CARS) microscopy. Proc Natl Acad Sci U S A 104: Le TT, Duren HM, Slipchenko MN, Hu CD, Cheng JX (2010) Label-free quantitative analysis of lipid metabolism in living Caenorhabditis elegans. J Lipid Res 51: Cheng JX, Xie XS (2004) Coherent anti-stokes Raman scattering microscopy: Instrumentation, theory, and applications. J Phys Chem B 108: Evans CL, Xie XS (2008) Coherent Anti-Stokes Raman Scattering Microscopy: Chemically Selective Imaging for Biology and Medicine. Annu Rev Anal Chem 1: PLoS ONE 9 September 2010 Volume 5 Issue 9 e12810
12 12. Rinia HA, Burger KNJ, Bonn M, Muller M (2008) Quantitative Label-Free Imaging of Lipid Composition and Packing of Individual Cellular Lipid Droplets Using Multiplex CARS Microscopy. Biophys J 95: Slipchenko MN, Le TT, Chen H, Cheng JX (2009) High-speed vibrational imaging and spectral analysis of lipid bodies by compound Raman microscopy. J Phys Chem B 113: Freudiger CW, Min W, Saar BG, Lu S, Holtom GR, et al. (2008) Label-Free Biomedical Imaging with High Sensitivity by Stimulated Raman Scattering Microscopy. Science 322: Kimura KD, Tissenbaum HA, Liu YX, Ruvkun G (1997) daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277: Soukas AA, Kane EA, Carr CE, Melo JA, Ruvkun G (2009) Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes Dev 23: Jones KT, Greer ER, Pearce D, Ashrafi K (2009) Rictor/TORC2 Regulates Caenorhabditis elegans Fat Storage, Body Size, and Development through sgk-1. PLoS Bio 7: Hertweck M, Gobel C, Baumeister R (2004) C. elegans SGK-1 Is the Critical Component in the Akt/PKB Kinase Complex to Control Stress Response and Life Span. Dev Cell 6(4): Padmanabhan S, Mukhopadhyay A, Narasimhan SD, Tesz G, Czech MP, et al. (2009) A PP2A Regulatory Subunit Regulates C. elegans Insulin/IGF-1 Signaling by Modulating AKT-1 Phosphorylation. Cell 136: Clokey GV, Jacobson LA (1986) The Autofluorescent Lipofuscin Granules in the Intestinal-Cells of Caenorhabditis-Elegans Are Secondary Lysosomes. Mech Ageing Dev 35: Hosokawa H, Ishii N, Ishida H, Ichimori K, Nakazawa H, et al. (1994) Rapid Accumulation of Fluorescent Material with Aging in an Oxygen-Sensitive Mutant Mev-1 of Caenorhabditis-Elegans. Mech Ageing Dev 74: Greenspan P, Mayer EP, Fowler SD (1985) Nile Red - a Selective Fluorescent Stain for Intracellular Lipid Droplets. J Cell Bio 100: Ivnitski-Steele I, Holmes AR, Lamping E, Monk BC, Cannon RD, et al. (2009) Identification of Nile red as a fluorescent substrate of the Candida albicans ATPbinding cassette transporters Cdr1p and Cdr2p and the major facilitator superfamily transporter Mdr1p. Anal Biochem 394: Jung T, Bader N, Grune T (2007) Lipofuscin: Formation, distribution, and metabolic consequences. Ann N Y Acad Sci 1119: Brunk UT, Terman A (2002) Lipofuscin: Mechanisms of age-related accumulation and influence on cell function. Free Radic Biol Med 33: Jung T, Hohn A, Grune T (2010) Lipofuscin: Detection and Quantification by Microscopic Techniques Armstrong D, editor. New York: Humana Press. pp Bancroft JD, Gamble M (2007) Theory and Practice of Histological Techniques. New York: Churchill Livingstone. 28. Viegas MS, Martins TC, Seco F, do Carmo A (2007) An improved and costeffective methodology for the reduction of autofluorescence in direct immunofluorescence studies on formalin-fixed paraffin-embedded tissues. Eur J Histochem 51(1): Thumser AE, Storch J (2007) Characterization of a BODIPY-labeled fluorescent fatty acid analogue. Binding to fatty acid-binding proteins, intracellular localization, and metabolism. Mol Cell Biochem 299: Le TT, Huff TB, Cheng JX (2009) Coherent anti-stokes Raman scattering imaging of lipids in cancer metastasis. BMC Cancer 9: Mukherjee S, Raghuraman H, Chattopadhyay A (2007) Membrane localization and dynamics of Nile Red: Effect of cholesterol. Biochim Biophys Acta Biomembranes 1768: Saltiel AR, Kahn CR (2001) Insulin signalling and the regulation of glucose and lipid metabolism. Nature 414: Le TT, Cheng JX (2009) Single-cell profiling reveals the origin of phenotypic variability in adipogenesis. PLoS ONE 4(4): e Rosen ED, MacDougald OA (2006) Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol 7: Guo Y, Cordes KR, Farese RV, Walther TC (2009) Lipid droplets at a glance. J Cell Sci 122: Guo Y, Walther TC, Rao M, Stuurman N, Goshima G, et al. (2008) Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature 453: Martin S, Parton RG (2006) Lipid droplets: a unified view of a dynamic organelle. Nat Rev Mol Cell Biol 7: Farese RV, Walther TC (2009) Lipid Droplets Finally Get a Little R-E-S-P-E-C- T. Cell 139: Walther TC, Farese RV (2009) The life of lipid droplets. Biochim Biophys Acta - Molecular and Cell Biology of Lipids 1791: Yang FJ, Vought BW, Satterlee JS, Walker AK, Sun ZYJ, et al. (2006) An ARC/ Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature 442: Brock TJ, Browse J, Watts JL (2007) Fatty acid desaturation and the regulation of adiposity in Caenorhabditis elegans. Genetics 176: Schulz TJ, Zarse K, Voigt A, Urban N, Birringer M, et al. (2007) Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab 6: Narasimhan SD, Yen K, Tissenbaum HA (2009) Converging Pathways in Lifespan Regulation. Curr Biol 19: R657 R Oh SW, Mukhopadhyay A, Dixit BL, Raha T, Green MR, et al. (2006) Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat Genet 38: Chung KH, Crane MM, Lu H (2008) Automated on-chip rapid microscopy, phenotyping and sorting of C. elegans. Nat Methods 5: Chronis N, Zimmer M, Bargmann CI (2007) Microfluidics for in vivo imaging of neuronal and behavioral activity in Caenorhabditis elegans. Nat Methods 4: Hulme SE, Shevkoplyas SS, McGuigan AP, Apfeld J, Fontana W, et al. (2010) Lifespan-on-a-chip: microfluidic chambers for performing lifelong observation of C. elegans. Lab Chip 10: Stiernagle T (2006) Maintenance of C. elegans. In: Community TCeR, ed. WormBook. 49. Ramírez-Zacarías JL, Castro-Muñozledo F, Kuri-Harcuch W (1992) Quantitation of adipose conversion and triglycerides by staining intracytoplasmic lipids with Oil red O. Histochemistry 97(6): PLoS ONE 10 September 2010 Volume 5 Issue 9 e12810
Specific polyunsaturated fatty acids modulate lipid delivery and oocyte development in C. elegans revealed by molecular-selective label-free imaging
 Specific polyunsaturated fatty acids modulate lipid delivery and oocyte development in C. elegans revealed by molecular-selective label-free imaging Wei-Wen Chen 1,2,3,+, Yung-Hsiang Yi 4,5,+, Cheng-Hao
Specific polyunsaturated fatty acids modulate lipid delivery and oocyte development in C. elegans revealed by molecular-selective label-free imaging Wei-Wen Chen 1,2,3,+, Yung-Hsiang Yi 4,5,+, Cheng-Hao
Label-free Quantitative Analysis of Lipid Metabolism in Living Caenorhabditis elegans
 Label-free Quantitative Analysis of Lipid Metabolism in Living Caenorhabditis elegans Thuc T. Le 1, Holli M. Duren 2, Mikhail N. Slipchenko 1, Chang-Deng Hu 2,3*, Ji-Xin Cheng 1,3,4* 1 Weldon School of
Label-free Quantitative Analysis of Lipid Metabolism in Living Caenorhabditis elegans Thuc T. Le 1, Holli M. Duren 2, Mikhail N. Slipchenko 1, Chang-Deng Hu 2,3*, Ji-Xin Cheng 1,3,4* 1 Weldon School of
ab CytoPainter Golgi/ER Staining Kit
 ab139485 CytoPainter Golgi/ER Staining Kit Instructions for Use Designed to detect Golgi bodies and endoplasmic reticulum by microscopy This product is for research use only and is not intended for diagnostic
ab139485 CytoPainter Golgi/ER Staining Kit Instructions for Use Designed to detect Golgi bodies and endoplasmic reticulum by microscopy This product is for research use only and is not intended for diagnostic
ab Adipogenesis Assay Kit (Cell-Based)
 ab133102 Adipogenesis Assay Kit (Cell-Based) Instructions for Use For the study of induction and inhibition of adipogenesis in adherent cells. This product is for research use only and is not intended
ab133102 Adipogenesis Assay Kit (Cell-Based) Instructions for Use For the study of induction and inhibition of adipogenesis in adherent cells. This product is for research use only and is not intended
Free Fatty Acid Assay Kit
 Free Fatty Acid Assay Kit Catalog Number KA1667 100 assays Version: 05 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 General Information...
Free Fatty Acid Assay Kit Catalog Number KA1667 100 assays Version: 05 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 General Information...
ROS Activity Assay Kit
 ROS Activity Assay Kit Catalog Number KA3841 200 assays Version: 03 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General Information... 4 Materials
ROS Activity Assay Kit Catalog Number KA3841 200 assays Version: 03 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General Information... 4 Materials
Mammalian Membrane Protein Extraction Kit
 Mammalian Membrane Protein Extraction Kit Catalog number: AR0155 Boster s Mammalian Membrane Protein Extraction Kit is a simple, rapid and reproducible method to prepare cellular protein fractions highly
Mammalian Membrane Protein Extraction Kit Catalog number: AR0155 Boster s Mammalian Membrane Protein Extraction Kit is a simple, rapid and reproducible method to prepare cellular protein fractions highly
A ph-dependent Charge Reversal Peptide for Cancer Targeting
 Supporting Information A ph-dependent Charge Reversal Peptide for Cancer Targeting Naoko Wakabayashi 1, Yoshiaki Yano 1, Kenichi Kawano 1, and Katsumi Matsuzaki 1 1 Graduate School of Pharmaceutical Sciences,
Supporting Information A ph-dependent Charge Reversal Peptide for Cancer Targeting Naoko Wakabayashi 1, Yoshiaki Yano 1, Kenichi Kawano 1, and Katsumi Matsuzaki 1 1 Graduate School of Pharmaceutical Sciences,
Instructions. Fuse-It-mRNA easy. Shipping and Storage. Overview. Kit Contents. Specifications. Note: Important Guidelines
 Membrane fusion is a highly efficient method for transfecting various molecules and particles into mammalian cells, even into sensitive and primary cells. The Fuse-It reagents are cargo-specific liposomal
Membrane fusion is a highly efficient method for transfecting various molecules and particles into mammalian cells, even into sensitive and primary cells. The Fuse-It reagents are cargo-specific liposomal
Free Fatty Acid Uptake Assay Kit (Fluorometric)
 ab176768 Free Fatty Acid Uptake Assay Kit (Fluorometric) Instructions for Use For measurement of fatty acid uptake in cells containing fatty acid transporters. This product is for research use only and
ab176768 Free Fatty Acid Uptake Assay Kit (Fluorometric) Instructions for Use For measurement of fatty acid uptake in cells containing fatty acid transporters. This product is for research use only and
Human Carbamylated LDL ELISA Kit (CBL-LDL Quantitation)
 Product Manual Human Carbamylated LDL ELISA Kit (CBL-LDL Quantitation) Catalog Number MET-5032 96 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Lipoproteins are submicroscopic
Product Manual Human Carbamylated LDL ELISA Kit (CBL-LDL Quantitation) Catalog Number MET-5032 96 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Lipoproteins are submicroscopic
Supplemental Information. Octopamine Neurons Mediate Flight-Induced Modulation of Visual Processing in Drosophila. Supplemental Inventory
 1 Current Biology, Volume 22 Supplemental Information Octopamine Neurons Mediate Flight-Induced Modulation of Visual Processing in Drosophila Marie P. Suver, Akira Mamiya, and Michael H. Dickinson Supplemental
1 Current Biology, Volume 22 Supplemental Information Octopamine Neurons Mediate Flight-Induced Modulation of Visual Processing in Drosophila Marie P. Suver, Akira Mamiya, and Michael H. Dickinson Supplemental
Glucose Uptake Colorimetric Assay Kit
 ab136955 Glucose Uptake Colorimetric Assay Kit Instructions for Use For the sensitive and accurate measurement of Glucose uptake in various samples This product is for research use only and is not intended
ab136955 Glucose Uptake Colorimetric Assay Kit Instructions for Use For the sensitive and accurate measurement of Glucose uptake in various samples This product is for research use only and is not intended
ab Hepatic Lipid Accumulation/ Steatosis Assay Kit
 ab133131 Hepatic Lipid Accumulation/ Steatosis Assay Kit Instructions for Use For evaluating steatosis risk of drug candidates using Oil Red O to stain neutral lipids in hepatocytes. This product is for
ab133131 Hepatic Lipid Accumulation/ Steatosis Assay Kit Instructions for Use For evaluating steatosis risk of drug candidates using Oil Red O to stain neutral lipids in hepatocytes. This product is for
Total Phosphatidic Acid Assay Kit
 Product Manual Total Phosphatidic Acid Assay Kit Catalog Number MET- 5019 100 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Phosphatidic Acid (PA) is a critical precursor
Product Manual Total Phosphatidic Acid Assay Kit Catalog Number MET- 5019 100 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Phosphatidic Acid (PA) is a critical precursor
LDL Uptake Cell-Based Assay Kit
 LDL Uptake Cell-Based Assay Kit Catalog Number KA1327 100 assays Version: 07 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 Principle of the Assay...
LDL Uptake Cell-Based Assay Kit Catalog Number KA1327 100 assays Version: 07 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 Principle of the Assay...
High-Speed Vibrational Imaging and Spectral Analysis of Lipid Bodies by Compound Raman Microscopy
 J. Phys. Chem. B 2009, 113, 7681 7686 7681 High-Speed Vibrational Imaging and Spectral Analysis of Lipid Bodies by Compound Raman Microscopy Mikhail N. Slipchenko, Thuc T. Le, Hongtao Chen, and Ji-Xin
J. Phys. Chem. B 2009, 113, 7681 7686 7681 High-Speed Vibrational Imaging and Spectral Analysis of Lipid Bodies by Compound Raman Microscopy Mikhail N. Slipchenko, Thuc T. Le, Hongtao Chen, and Ji-Xin
Superior Fluorescent Labeling Dyes Spanning the Full Visible Spectrum...1. Trademarks: HiLyte Fluor (AnaSpec, Inc.)
 Table of Contents Fluor TM Labeling Dyes Superior Fluorescent Labeling Dyes Spanning the Full Visible Spectrum....1 Fluor TM 405 Dye, an Excellent Alternative to Alexa Fluor 405 & DyLight 405....2 Fluor
Table of Contents Fluor TM Labeling Dyes Superior Fluorescent Labeling Dyes Spanning the Full Visible Spectrum....1 Fluor TM 405 Dye, an Excellent Alternative to Alexa Fluor 405 & DyLight 405....2 Fluor
1. Stowers Institute for Medical Research, Kansas City, MO 64110
 Lipid droplets as fat storage organelles in Caenorhabditis elegans Ho Yi Mak 1, 2 1. Stowers Institute for Medical Research, Kansas City, MO 64110 2. Department of Molecular and Integrative Physiology,
Lipid droplets as fat storage organelles in Caenorhabditis elegans Ho Yi Mak 1, 2 1. Stowers Institute for Medical Research, Kansas City, MO 64110 2. Department of Molecular and Integrative Physiology,
Human Leptin ELISA Kit
 Product Manual Human Leptin ELISA Kit Catalog Numbers MET-5057 MET-5057-5 96 assays 5 x 96 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Leptin is a polypeptide hormone
Product Manual Human Leptin ELISA Kit Catalog Numbers MET-5057 MET-5057-5 96 assays 5 x 96 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Leptin is a polypeptide hormone
ab Membrane fluidity kit Instructions for Use For the detection of membrane fluidity in cells
 ab189819 Membrane fluidity kit Instructions for Use For the detection of membrane fluidity in cells This product is for research use only and is not intended for diagnostic use. Version 1 Last Updated
ab189819 Membrane fluidity kit Instructions for Use For the detection of membrane fluidity in cells This product is for research use only and is not intended for diagnostic use. Version 1 Last Updated
For the rapid, sensitive and accurate measurement of Triglyceride in various samples.
 ab65336 Triglyceride Quantification Kit Instructions for Use For the rapid, sensitive and accurate measurement of Triglyceride in various samples. This product is for research use only and is not intended
ab65336 Triglyceride Quantification Kit Instructions for Use For the rapid, sensitive and accurate measurement of Triglyceride in various samples. This product is for research use only and is not intended
Nature Protocols: doi: /nprot Supplementary Figure 1. Fluorescent titration of probe CPDSA.
 Supplementary Figure 1 Fluorescent titration of probe CPDSA. Fluorescent titration of probe CPDSA (10 um) upon addition of GSH in HEPES (10 mm, ph = 7.4) containing 10% DMSO. Each spectrum was recorded
Supplementary Figure 1 Fluorescent titration of probe CPDSA. Fluorescent titration of probe CPDSA (10 um) upon addition of GSH in HEPES (10 mm, ph = 7.4) containing 10% DMSO. Each spectrum was recorded
Fluorescence Microscopy
 Fluorescence Microscopy Imaging Organelles Mitochondria Lysosomes Nuclei Endoplasmic Reticulum Plasma Membrane F-Actin AAT Bioquest Introduction: Organelle-Selective Stains Organelles are tiny, specialized
Fluorescence Microscopy Imaging Organelles Mitochondria Lysosomes Nuclei Endoplasmic Reticulum Plasma Membrane F-Actin AAT Bioquest Introduction: Organelle-Selective Stains Organelles are tiny, specialized
ab65336 Triglyceride Quantification Assay Kit (Colorimetric/ Fluorometric)
 Version 10 Last updated 19 December 2017 ab65336 Triglyceride Quantification Assay Kit (Colorimetric/ Fluorometric) For the measurement of triglycerides in various samples. This product is for research
Version 10 Last updated 19 December 2017 ab65336 Triglyceride Quantification Assay Kit (Colorimetric/ Fluorometric) For the measurement of triglycerides in various samples. This product is for research
TFEB-mediated increase in peripheral lysosomes regulates. Store Operated Calcium Entry
 TFEB-mediated increase in peripheral lysosomes regulates Store Operated Calcium Entry Luigi Sbano, Massimo Bonora, Saverio Marchi, Federica Baldassari, Diego L. Medina, Andrea Ballabio, Carlotta Giorgi
TFEB-mediated increase in peripheral lysosomes regulates Store Operated Calcium Entry Luigi Sbano, Massimo Bonora, Saverio Marchi, Federica Baldassari, Diego L. Medina, Andrea Ballabio, Carlotta Giorgi
ab Lipid Peroxidation (MDA) Assay kit (Colorimetric/ Fluorometric)
 Version 10b Last updated 19 December 2018 ab118970 Lipid Peroxidation (MDA) Assay kit (Colorimetric/ Fluorometric) For the measurement of Lipid Peroxidation in plasma, cell culture and tissue extracts.
Version 10b Last updated 19 December 2018 ab118970 Lipid Peroxidation (MDA) Assay kit (Colorimetric/ Fluorometric) For the measurement of Lipid Peroxidation in plasma, cell culture and tissue extracts.
Biol110L-Cell Biology Lab Spring Quarter 2012 Module 1-4 Friday April 13, 2012 (Start promptly; work fast; the protocols take ~4 h)
 Biol110L-Cell Biology Lab Spring Quarter 2012 Module 1-4 Friday April 13, 2012 (Start promptly; work fast; the protocols take ~4 h) A. Microscopic Examination of the Plasma Membrane and Its Properties
Biol110L-Cell Biology Lab Spring Quarter 2012 Module 1-4 Friday April 13, 2012 (Start promptly; work fast; the protocols take ~4 h) A. Microscopic Examination of the Plasma Membrane and Its Properties
OxiSelect Human Oxidized LDL ELISA Kit (OxPL-LDL Quantitation)
 Product Manual OxiSelect Human Oxidized LDL ELISA Kit (OxPL-LDL Quantitation) Catalog Number STA-358 96 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Lipoproteins are submicroscopic
Product Manual OxiSelect Human Oxidized LDL ELISA Kit (OxPL-LDL Quantitation) Catalog Number STA-358 96 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Lipoproteins are submicroscopic
SensoLyte 520 HIV-1 Protease Assay Kit *Fluorimetric*
 SensoLyte 520 HIV-1 Protease Assay Kit *Fluorimetric* Catalog # 71147 Kit Size 100 assays (96-well) or 500 assays (384-well) Convenient Format: Complete kit including all the assay components. Optimized
SensoLyte 520 HIV-1 Protease Assay Kit *Fluorimetric* Catalog # 71147 Kit Size 100 assays (96-well) or 500 assays (384-well) Convenient Format: Complete kit including all the assay components. Optimized
ab Glucose Uptake Assay Kit (colorimetric) 1
 Version 16 Last updated 10 January 2018 ab136955 Glucose Uptake Assay Kit (Colorimetric) For the measurement of Glucose uptake in a variety of cells. This product is for research use only and is not intended
Version 16 Last updated 10 January 2018 ab136955 Glucose Uptake Assay Kit (Colorimetric) For the measurement of Glucose uptake in a variety of cells. This product is for research use only and is not intended
Free Fatty Acid Assay Kit (Fluorometric)
 Product Manual Free Fatty Acid Assay Kit (Fluorometric) Catalog Number STA-619 100 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Triglycerides (TAG) are a type of lipid
Product Manual Free Fatty Acid Assay Kit (Fluorometric) Catalog Number STA-619 100 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Triglycerides (TAG) are a type of lipid
For the rapid, sensitive and accurate measurement of Glycerol in cell cultures.
 ab185433 Lipolysis Assay Kit (Colorimetric) Instructions for Use For the rapid, sensitive and accurate measurement of Glycerol in cell cultures. This product is for research use only and is not intended
ab185433 Lipolysis Assay Kit (Colorimetric) Instructions for Use For the rapid, sensitive and accurate measurement of Glycerol in cell cultures. This product is for research use only and is not intended
Phospholipid Assay Kit
 Phospholipid Assay Kit Catalog Number KA1635 100 assays Version: 05 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 General Information...
Phospholipid Assay Kit Catalog Number KA1635 100 assays Version: 05 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 General Information...
Cholesterol/Cholesteryl Ester Detection Kit
 ab102515 Cholesterol/Cholesteryl Ester Detection Kit Instructions for Use For the rapid, sensitive and accurate measurement of free cholesterol, cholesteryl esters, or both in various samples. This product
ab102515 Cholesterol/Cholesteryl Ester Detection Kit Instructions for Use For the rapid, sensitive and accurate measurement of free cholesterol, cholesteryl esters, or both in various samples. This product
For the rapid, sensitive and accurate detection of long-chain Free Fatty Acids in various samples.
 ab65341 Free Fatty Acid Quantification Kit Instructions for Use For the rapid, sensitive and accurate detection of long-chain Free Fatty Acids in various samples. This product is for research use only
ab65341 Free Fatty Acid Quantification Kit Instructions for Use For the rapid, sensitive and accurate detection of long-chain Free Fatty Acids in various samples. This product is for research use only
Lipid (Oil Red O) staining Kit
 Lipid (Oil Red O) staining Kit Catalog Number KA4541 1 Kit Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General Information... 4 Materials
Lipid (Oil Red O) staining Kit Catalog Number KA4541 1 Kit Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General Information... 4 Materials
Kit for assay of thioredoxin
 FkTRX-02-V2 Kit for assay of thioredoxin The thioredoxin system is the major protein disulfide reductase in cells and comprises thioredoxin, thioredoxin reductase and NADPH (1). Thioredoxin systems are
FkTRX-02-V2 Kit for assay of thioredoxin The thioredoxin system is the major protein disulfide reductase in cells and comprises thioredoxin, thioredoxin reductase and NADPH (1). Thioredoxin systems are
LDL Uptake Cell-Based Assay Kit
 LDL Uptake Cell-Based Assay Kit Item No. 10011125 www.caymanchem.com Customer Service 800.364.9897 Technical Support 888.526.5351 1180 E. Ellsworth Rd Ann Arbor, MI USA TABLE OF CONTENTS GENERAL INFORMATION
LDL Uptake Cell-Based Assay Kit Item No. 10011125 www.caymanchem.com Customer Service 800.364.9897 Technical Support 888.526.5351 1180 E. Ellsworth Rd Ann Arbor, MI USA TABLE OF CONTENTS GENERAL INFORMATION
Product Manual. Human LDLR ELISA Kit. Catalog Number. FOR RESEARCH USE ONLY Not for use in diagnostic procedures
 Product Manual Human LDLR ELISA Kit Catalog Number STA-386 96 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Cholesterol is an essential component of cellular membranes,
Product Manual Human LDLR ELISA Kit Catalog Number STA-386 96 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Cholesterol is an essential component of cellular membranes,
4 Development of an ESR online-method for the monitoring of in vitro fat digestion
 4 Development of an ESR online-method for the monitoring of in vitro fat digestion 4.1 Introduction When regarding the oral administration of lipid-based nanocapsules, gastrointestinal digestion will play
4 Development of an ESR online-method for the monitoring of in vitro fat digestion 4.1 Introduction When regarding the oral administration of lipid-based nanocapsules, gastrointestinal digestion will play
Qualifying Fat Storage Phenotypes Using Gas Chromatography in Caenorhabditis elegans
 University of Colorado, Boulder CU Scholar Undergraduate Honors Theses Honors Program Spring 2014 Qualifying Fat Storage Phenotypes Using Gas Chromatography in Caenorhabditis elegans Sabrina Fechtner University
University of Colorado, Boulder CU Scholar Undergraduate Honors Theses Honors Program Spring 2014 Qualifying Fat Storage Phenotypes Using Gas Chromatography in Caenorhabditis elegans Sabrina Fechtner University
Multi-Parameter Apoptosis Assay Kit
 Multi-Parameter Apoptosis Assay Kit Catalog Number KA1335 5 x 96 assays Version: 05 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 Principle of the Assay...
Multi-Parameter Apoptosis Assay Kit Catalog Number KA1335 5 x 96 assays Version: 05 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 Principle of the Assay...
Chromatin IP (Isw2) Fix soln: 11% formaldehyde, 0.1 M NaCl, 1 mm EDTA, 50 mm Hepes-KOH ph 7.6. Freshly prepared. Do not store in glass bottles.
 Chromatin IP (Isw2) 7/01 Toshi last update: 06/15 Reagents Fix soln: 11% formaldehyde, 0.1 M NaCl, 1 mm EDTA, 50 mm Hepes-KOH ph 7.6. Freshly prepared. Do not store in glass bottles. 2.5 M glycine. TBS:
Chromatin IP (Isw2) 7/01 Toshi last update: 06/15 Reagents Fix soln: 11% formaldehyde, 0.1 M NaCl, 1 mm EDTA, 50 mm Hepes-KOH ph 7.6. Freshly prepared. Do not store in glass bottles. 2.5 M glycine. TBS:
Dental Research Institute, Faculty of Dentistry, University of Toronto, Toronto, Canada *For correspondence:
 Zymogram Assay for the Detection of Peptidoglycan Hydrolases in Streptococcus mutans Delphine Dufour and Céline M. Lévesque * Dental Research Institute, Faculty of Dentistry, University of Toronto, Toronto,
Zymogram Assay for the Detection of Peptidoglycan Hydrolases in Streptococcus mutans Delphine Dufour and Céline M. Lévesque * Dental Research Institute, Faculty of Dentistry, University of Toronto, Toronto,
klp-18 (RNAi) Control. supplementary information. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach
![klp-18 (RNAi) Control. supplementary information. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach klp-18 (RNAi) Control. supplementary information. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach](/thumbs/91/104639484.jpg) DOI: 10.1038/ncb1891 A. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach embryos let hatch overnight transfer to RNAi plates; incubate 5 days at 15 C RNAi food L1 worms adult worms
DOI: 10.1038/ncb1891 A. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach embryos let hatch overnight transfer to RNAi plates; incubate 5 days at 15 C RNAi food L1 worms adult worms
Investigating Lipids
 Investigating Lipids In today s culture, there is a stigma associated with the word fat. While it is true that too much fat can lead to health problems, fats (or lipids) play very important roles in biology.
Investigating Lipids In today s culture, there is a stigma associated with the word fat. While it is true that too much fat can lead to health problems, fats (or lipids) play very important roles in biology.
Fluorescence based Fixative and Vital Staining of Lipid Droplets in C. elegans. reveal Fat Stores using Microscopic and Flow Cytometry Approaches
 1 Fluorescence based Fixative and Vital Staining of Lipid Droplets in C. elegans reveal Fat Stores using Microscopic and Flow Cytometry Approaches Maja Klapper 1, Madeleine Ehmke 1, Daniela Palgunow 1,
1 Fluorescence based Fixative and Vital Staining of Lipid Droplets in C. elegans reveal Fat Stores using Microscopic and Flow Cytometry Approaches Maja Klapper 1, Madeleine Ehmke 1, Daniela Palgunow 1,
Glucose Uptake Assay Kit (Fluorometric)
 ab136956 Glucose Uptake Assay Kit (Fluorometric) Instructions for Use For the sensitive and accurate measurement of Glucose uptake in various samples This product is for research use only and is not intended
ab136956 Glucose Uptake Assay Kit (Fluorometric) Instructions for Use For the sensitive and accurate measurement of Glucose uptake in various samples This product is for research use only and is not intended
Lipid Droplets Fluorescence Assay Kit
 Lipid Droplets Fluorescence Assay Kit Item No. 500001 www.caymanchem.com Customer Service 800.364.9897 Technical Support 888.526.5351 1180 E. Ellsworth Rd Ann Arbor, MI USA TABLE OF CONTENTS GENERAL INFORMATION
Lipid Droplets Fluorescence Assay Kit Item No. 500001 www.caymanchem.com Customer Service 800.364.9897 Technical Support 888.526.5351 1180 E. Ellsworth Rd Ann Arbor, MI USA TABLE OF CONTENTS GENERAL INFORMATION
Human Oxidized LDL ELISA Kit (MDA-LDL Quantitation), General
 Human Oxidized LDL ELISA Kit (MDA-LDL Quantitation), General For the detection and quantitation of human OxLDL in plasma, serum or other biological fluid samples Cat. No. KT-959 For Research Use Only.
Human Oxidized LDL ELISA Kit (MDA-LDL Quantitation), General For the detection and quantitation of human OxLDL in plasma, serum or other biological fluid samples Cat. No. KT-959 For Research Use Only.
Coenzyme A Assay Kit. Catalog Number KA assays Version: 02. Intended for research use only.
 Coenzyme A Assay Kit Catalog Number KA0809 100 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General Information... 4 Materials Supplied...
Coenzyme A Assay Kit Catalog Number KA0809 100 assays Version: 02 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General Information... 4 Materials Supplied...
ab LDL Uptake Assay Kit (Cell-Based)
 ab133127 LDL Uptake Assay Kit (Cell-Based) Instructions for Use For the detection of LDL uptake into cultured cells. This product is for research use only and is not intended for diagnostic use. Version
ab133127 LDL Uptake Assay Kit (Cell-Based) Instructions for Use For the detection of LDL uptake into cultured cells. This product is for research use only and is not intended for diagnostic use. Version
RayBio Phenylalanine Fluorometric Assay Kit
 RayBio Phenylalanine Fluorometric Assay Kit User Manual Version 1.0 June 25, 2014 RayBio Phenylalanine Fluorometric (Cat#: 68-Phenyl-S100) RayBiotech, Inc. We Provide You With Excellent Support And Service
RayBio Phenylalanine Fluorometric Assay Kit User Manual Version 1.0 June 25, 2014 RayBio Phenylalanine Fluorometric (Cat#: 68-Phenyl-S100) RayBiotech, Inc. We Provide You With Excellent Support And Service
doi: /j.bbrc
 doi: 10.1016/j.bbrc.2009.11.006 Fatty Acid Metabolism is Involved in Stress Resistance Mechanisms of Caenorhabditis elegans Makoto Horikawa a, b, Kazuichi Sakamoto a, 1 a Graduate School of Life and Environmental
doi: 10.1016/j.bbrc.2009.11.006 Fatty Acid Metabolism is Involved in Stress Resistance Mechanisms of Caenorhabditis elegans Makoto Horikawa a, b, Kazuichi Sakamoto a, 1 a Graduate School of Life and Environmental
Chromatin Immunoprecipitation (ChIPs) Protocol (Mirmira Lab)
 Chromatin Immunoprecipitation (ChIPs) Protocol (Mirmira Lab) Updated 12/3/02 Reagents: ChIP sonication Buffer (1% Triton X-100, 0.1% Deoxycholate, 50 mm Tris 8.1, 150 mm NaCl, 5 mm EDTA): 10 ml 10 % Triton
Chromatin Immunoprecipitation (ChIPs) Protocol (Mirmira Lab) Updated 12/3/02 Reagents: ChIP sonication Buffer (1% Triton X-100, 0.1% Deoxycholate, 50 mm Tris 8.1, 150 mm NaCl, 5 mm EDTA): 10 ml 10 % Triton
Lipase Detection Kit II (Colorimetric)
 ab102525 Lipase Detection Kit II (Colorimetric) Instructions for Use For the rapid, sensitive and accurate measurement of Lipase activity in various samples. This product is for research use only and is
ab102525 Lipase Detection Kit II (Colorimetric) Instructions for Use For the rapid, sensitive and accurate measurement of Lipase activity in various samples. This product is for research use only and is
DAG (Diacylglycerol) Assay Kit
 Product Manual DAG (Diacylglycerol) Assay Kit Catalog Number MET-5028 100 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Diacylglycerols (DAG) are key intermediates in the
Product Manual DAG (Diacylglycerol) Assay Kit Catalog Number MET-5028 100 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Diacylglycerols (DAG) are key intermediates in the
SensoLyte Rh110 Cathepsin K Assay Kit *Fluorimetric* Revision#1.2 Last Updated: May 2017 Catalog # Kit Size
 SensoLyte Rh110 Cathepsin K Assay Kit *Fluorimetric* Revision#1.2 Last Updated: May 2017 Catalog # 72152 Kit Size 100 Assays (96-well plate) Optimized Performance: This kit detects Cathepsin K activity.
SensoLyte Rh110 Cathepsin K Assay Kit *Fluorimetric* Revision#1.2 Last Updated: May 2017 Catalog # 72152 Kit Size 100 Assays (96-well plate) Optimized Performance: This kit detects Cathepsin K activity.
Supplementary material: Materials and suppliers
 Supplementary material: Materials and suppliers Electrophoresis consumables including tris-glycine, acrylamide, SDS buffer and Coomassie Brilliant Blue G-2 dye (CBB) were purchased from Ameresco (Solon,
Supplementary material: Materials and suppliers Electrophoresis consumables including tris-glycine, acrylamide, SDS buffer and Coomassie Brilliant Blue G-2 dye (CBB) were purchased from Ameresco (Solon,
Serum Triglyceride Quantification Kit (Colorimetric)
 Product Manual Serum Triglyceride Quantification Kit (Colorimetric) Catalog Number STA-396 100 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Triglycerides (TAG) are a type
Product Manual Serum Triglyceride Quantification Kit (Colorimetric) Catalog Number STA-396 100 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Triglycerides (TAG) are a type
Human LDL Receptor / LDLR ELISA Pair Set
 Human LDL Receptor / LDLR ELISA Pair Set Catalog Number : SEK10231 To achieve the best assay results, this manual must be read carefully before using this product and the assay is run as summarized in
Human LDL Receptor / LDLR ELISA Pair Set Catalog Number : SEK10231 To achieve the best assay results, this manual must be read carefully before using this product and the assay is run as summarized in
2-Deoxyglucose (2DG) Uptake Measurement kit
 Document#:K2DG13516E For research use only. Not for clinical diagnosis. Catalog No. CSR-OKP-PMG-K1E 2-Deoxyglucose (2DG) Uptake Measurement kit Introduction Measurement of 2-deoxyglucose (2DG) uptake in
Document#:K2DG13516E For research use only. Not for clinical diagnosis. Catalog No. CSR-OKP-PMG-K1E 2-Deoxyglucose (2DG) Uptake Measurement kit Introduction Measurement of 2-deoxyglucose (2DG) uptake in
2-Deoxyglucose Assay Kit (Colorimetric)
 2-Deoxyglucose Assay Kit (Colorimetric) Catalog Number KA3753 100 assays Version: 01 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General Information...
2-Deoxyglucose Assay Kit (Colorimetric) Catalog Number KA3753 100 assays Version: 01 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General Information...
Phospholipid Assay Kit
 Phospholipid Assay Kit Catalog Number KA1635 100 assays Version: 06 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 General Information...
Phospholipid Assay Kit Catalog Number KA1635 100 assays Version: 06 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 General Information...
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION FOR Liver X Receptor α mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 (G0S2) expression I: SUPPLEMENTARY METHODS II: SUPPLEMENTARY FIGURES
SUPPLEMENTARY INFORMATION FOR Liver X Receptor α mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 (G0S2) expression I: SUPPLEMENTARY METHODS II: SUPPLEMENTARY FIGURES
ab SREBP-2 Translocation Assay Kit (Cell-Based)
 ab133114 SREBP-2 Translocation Assay Kit (Cell-Based) Instructions for Use For analysis of translocation of SREBP-2 into nuclei. This product is for research use only and is not intended for diagnostic
ab133114 SREBP-2 Translocation Assay Kit (Cell-Based) Instructions for Use For analysis of translocation of SREBP-2 into nuclei. This product is for research use only and is not intended for diagnostic
Glycerol- 3- Phosphate (G3P) Assay Kit (Colorimetric)
 Product Manual Glycerol- 3- Phosphate (G3P) Assay Kit (Colorimetric) Catalog Number MET- 5075 100 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Glycerol-3-phosphate (G3P)
Product Manual Glycerol- 3- Phosphate (G3P) Assay Kit (Colorimetric) Catalog Number MET- 5075 100 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Glycerol-3-phosphate (G3P)
LDL Uptake Flow Cytometry Assay Kit
 LDL Uptake Flow Cytometry Assay Kit Item No. 601470 www.caymanchem.com Customer Service 800.364.9897 Technical Support 888.526.5351 1180 E. Ellsworth Rd Ann Arbor, MI USA TABLE OF CONTENTS GENERAL INFORMATION
LDL Uptake Flow Cytometry Assay Kit Item No. 601470 www.caymanchem.com Customer Service 800.364.9897 Technical Support 888.526.5351 1180 E. Ellsworth Rd Ann Arbor, MI USA TABLE OF CONTENTS GENERAL INFORMATION
Mammalian Tissue Protein Extraction Reagent
 Mammalian Tissue Protein Extraction Reagent Catalog number: AR0101 Boster s Mammalian Tissue Protein Extraction Reagent is a ready-to-use Western blot related reagent solution used for efficient extraction
Mammalian Tissue Protein Extraction Reagent Catalog number: AR0101 Boster s Mammalian Tissue Protein Extraction Reagent is a ready-to-use Western blot related reagent solution used for efficient extraction
Intracellular (Total) ROS Activity Assay Kit (Red)
 Intracellular (Total) ROS Activity Assay Kit (Red) Catalog Number KA2525 200 assays Version: 04 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General
Intracellular (Total) ROS Activity Assay Kit (Red) Catalog Number KA2525 200 assays Version: 04 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General
CoQ10(Coenzyme Q10) ELISA Kit
 CoQ10(Coenzyme Q10) ELISA Kit Catalogue No.: EU0196 Size: 48T/96T Reactivity: Universal Detection Range: 0.781-50ng/ml Sensitivity:
CoQ10(Coenzyme Q10) ELISA Kit Catalogue No.: EU0196 Size: 48T/96T Reactivity: Universal Detection Range: 0.781-50ng/ml Sensitivity:
CytoPainter Lysosomal Staining Kit - Blue Fluorescence
 ab112135 CytoPainter Lysosomal Staining Kit - Blue Fluorescence Instructions for Use For staining Lysosomes in suspension and adherent cells by using our proprietary blue fluorescence probe This product
ab112135 CytoPainter Lysosomal Staining Kit - Blue Fluorescence Instructions for Use For staining Lysosomes in suspension and adherent cells by using our proprietary blue fluorescence probe This product
Lipid Accumulation in HepG2 Cells Exposed to Free Fatty Acids Image-Based Assay to Model Non-Alcoholic Steatohepatitis (NASH)
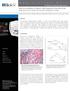 A p p l i c a t i o n N o t e Lipid Accumulation in HepG2 Cells Exposed to Free Fatty Acids Image-Based Assay to Model Non-Alcoholic Steatohepatitis (NASH) Paul Held, Ph.D., Laboratory Manager, Applications
A p p l i c a t i o n N o t e Lipid Accumulation in HepG2 Cells Exposed to Free Fatty Acids Image-Based Assay to Model Non-Alcoholic Steatohepatitis (NASH) Paul Held, Ph.D., Laboratory Manager, Applications
Quantifying Lipid Contents in Enveloped Virus Particles with Plasmonic Nanoparticles
 Quantifying Lipid Contents in Enveloped Virus Particles with Plasmonic Nanoparticles Amin Feizpour Reinhard Lab Department of Chemistry and the Photonics Center, Boston University, Boston, MA May 2014
Quantifying Lipid Contents in Enveloped Virus Particles with Plasmonic Nanoparticles Amin Feizpour Reinhard Lab Department of Chemistry and the Photonics Center, Boston University, Boston, MA May 2014
Biology 12. Biochemistry. Water - a polar molecule Water (H 2 O) is held together by covalent bonds.
 Biology 12 Biochemistry Water - a polar molecule Water (H 2 O) is held together by covalent bonds. Electrons in these bonds spend more time circulating around the larger Oxygen atom than the smaller Hydrogen
Biology 12 Biochemistry Water - a polar molecule Water (H 2 O) is held together by covalent bonds. Electrons in these bonds spend more time circulating around the larger Oxygen atom than the smaller Hydrogen
STAT1 (ps727) (Human/Mouse) ELISA Kit
 STAT1 (ps727) (Human/Mouse) ELISA Kit Catalog Number KA2171 96 assays Version: 01 Intended for research use only www.abnova.com I. INTRODUCTION STAT1 (ps727) (Human/Mouse) ELISA (Enzyme-Linked Immunosorbent
STAT1 (ps727) (Human/Mouse) ELISA Kit Catalog Number KA2171 96 assays Version: 01 Intended for research use only www.abnova.com I. INTRODUCTION STAT1 (ps727) (Human/Mouse) ELISA (Enzyme-Linked Immunosorbent
Glucose 6 Phosphate Assay Kit (Colorimetric)
 ab83426 Glucose 6 Phosphate Assay Kit (Colorimetric) Instructions for Use For the rapid, sensitive and accurate measurement of Glucose 6 Phosphate levels in various samples This product is for research
ab83426 Glucose 6 Phosphate Assay Kit (Colorimetric) Instructions for Use For the rapid, sensitive and accurate measurement of Glucose 6 Phosphate levels in various samples This product is for research
Data sheet. TBARS Assay kit. (Colorimetric/Fluorometric) Kit Contents. MDA-TBA Adduct. 2-Thiobarbituric Acid. Cat. No: CA995.
 Data sheet Cat. No: CA995 TBARS Assay kit (Colorimetric/Fluorometric) Introduction Oxidative stress in the cellular environment results in the formation of highly reactive and unstable lipid hydroperoxides.
Data sheet Cat. No: CA995 TBARS Assay kit (Colorimetric/Fluorometric) Introduction Oxidative stress in the cellular environment results in the formation of highly reactive and unstable lipid hydroperoxides.
Triglyceride Assay Kit-Bulk 5 Plate Kit Cat# TG-5-RB
 Triglyceride Assay Kit-Bulk 5 Plate Kit Cat# TG-5-RB INSTRUCTION MANUAL (ZBM-14) STORAGE CONDITIONS Reagents A & B, Buffers: Store at 2-8 C. Glycerol Standard -20 C For Research Use Only Not For Use In
Triglyceride Assay Kit-Bulk 5 Plate Kit Cat# TG-5-RB INSTRUCTION MANUAL (ZBM-14) STORAGE CONDITIONS Reagents A & B, Buffers: Store at 2-8 C. Glycerol Standard -20 C For Research Use Only Not For Use In
Cell Lysis Buffer. Catalog number: AR0103
 Cell Lysis Buffer Catalog number: AR0103 Boster s Cell Lysis Buffer is a ready-to-use Western blot related reagent solution used for efficient extraction of total soluble protein in nondenatured state
Cell Lysis Buffer Catalog number: AR0103 Boster s Cell Lysis Buffer is a ready-to-use Western blot related reagent solution used for efficient extraction of total soluble protein in nondenatured state
Cholesterol determination using protein-templated fluorescent gold nanocluster probes
 Electronic Supplementary Information for Cholesterol determination using protein-templated fluorescent gold nanocluster probes Xi Chen and Gary A. Baker* Department of Chemistry, University of Missouri-Columbia,
Electronic Supplementary Information for Cholesterol determination using protein-templated fluorescent gold nanocluster probes Xi Chen and Gary A. Baker* Department of Chemistry, University of Missouri-Columbia,
ab Lysosome/Cytotoxicity Dual Staining Kit
 ab133078 Lysosome/Cytotoxicity Dual Staining Kit Instructions for Use For studying lysosome function at the cellular level. This product is for research use only and is not intended for diagnostic use.
ab133078 Lysosome/Cytotoxicity Dual Staining Kit Instructions for Use For studying lysosome function at the cellular level. This product is for research use only and is not intended for diagnostic use.
Human LDL ELISA Kit. Innovative Research, Inc.
 Human LDL ELISA Kit Catalog No: IRKTAH2582 Lot No: SAMPLE INTRODUCTION Human low-density lipoprotein (LDL) transports cholesterol from the liver to tissues where it is incorporated into cell membranes.
Human LDL ELISA Kit Catalog No: IRKTAH2582 Lot No: SAMPLE INTRODUCTION Human low-density lipoprotein (LDL) transports cholesterol from the liver to tissues where it is incorporated into cell membranes.
Phenylalanine Assay Kit
 ab83376 Phenylalanine Assay Kit Instructions for Use For the rapid, sensitive and accurate measurement of Phenylalanine levels in various samples This product is for research use only and is not intended
ab83376 Phenylalanine Assay Kit Instructions for Use For the rapid, sensitive and accurate measurement of Phenylalanine levels in various samples This product is for research use only and is not intended
Exo-Glow TM Exosome Labeling Kits
 Exo-Glow TM Exosome Labeling Kits Cat# EXOR100A-1 Cat# EXOG200A-1 Cat# EXOC300A-1 User Manual Store kit at -20 o C on receipt Version 8 3/10/2017 A limited-use label license covers this product. By use
Exo-Glow TM Exosome Labeling Kits Cat# EXOR100A-1 Cat# EXOG200A-1 Cat# EXOC300A-1 User Manual Store kit at -20 o C on receipt Version 8 3/10/2017 A limited-use label license covers this product. By use
TRANSIL ASSAY KITS. Frequently Asked Questions (FAQs)
 TRANSIL ASSAY KITS Frequently Asked Questions (FAQs) 1. 2. 3. What is the principle of TRANSIL technology? What membrane is on the surface of the What are the key advantages of the TRANSIL technology?
TRANSIL ASSAY KITS Frequently Asked Questions (FAQs) 1. 2. 3. What is the principle of TRANSIL technology? What membrane is on the surface of the What are the key advantages of the TRANSIL technology?
SensoLyte 520 HDAC Activity Assay Kit *Fluorimetric*
 SensoLyte 520 HDAC Activity Assay Kit *Fluorimetric* Catalog # 72084 Kit Size 100 Assays (96-well plate) Optimized Performance: This kit is optimized to detect HDAC activity. Enhanced Value: It provides
SensoLyte 520 HDAC Activity Assay Kit *Fluorimetric* Catalog # 72084 Kit Size 100 Assays (96-well plate) Optimized Performance: This kit is optimized to detect HDAC activity. Enhanced Value: It provides
Supplementary Table 1. Primer sequences for conventional RT-PCR on mouse islets
 Supplementary Table 1. Primer sequences for conventional RT-PCR on mouse islets Gene 5 Forward 3 5 Reverse 3.T. Product (bp) ( C) mnox1 GTTCTTGGGCTGCCTTGG GCTGGGGCGGCGG 60 300 mnoxa1 GCTTTGCCGCGTGC GGTTCGGGTCCTTTGTGC
Supplementary Table 1. Primer sequences for conventional RT-PCR on mouse islets Gene 5 Forward 3 5 Reverse 3.T. Product (bp) ( C) mnox1 GTTCTTGGGCTGCCTTGG GCTGGGGCGGCGG 60 300 mnoxa1 GCTTTGCCGCGTGC GGTTCGGGTCCTTTGTGC
EpiQuik Circulating Acetyl Histone H3K18 ELISA Kit (Colorimetric)
 EpiQuik Circulating Acetyl Histone H3K18 ELISA Kit (Colorimetric) Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE Uses: The EpiQuik Circulating Acetyl Histone H3K18 ELISA Kit (Colorimetric)
EpiQuik Circulating Acetyl Histone H3K18 ELISA Kit (Colorimetric) Base Catalog # PLEASE READ THIS ENTIRE USER GUIDE BEFORE USE Uses: The EpiQuik Circulating Acetyl Histone H3K18 ELISA Kit (Colorimetric)
Cholesterol/Cholesteryl Ester Quantitation Assay kit (Colorimetric/Fluorometric)
 ab65359 Cholesterol/Cholesteryl Ester Quantitation Assay kit (Colorimetric/Fluorometric) Instructions for Use For the rapid, sensitive and accurate measurement of free cholesterol, cholesteryl esters,
ab65359 Cholesterol/Cholesteryl Ester Quantitation Assay kit (Colorimetric/Fluorometric) Instructions for Use For the rapid, sensitive and accurate measurement of free cholesterol, cholesteryl esters,
Choline/Acetylcholine Assay Kit
 ab65345 Choline/Acetylcholine Assay Kit Instructions for Use For the rapid, sensitive and accurate measurement of Choline/Acetylcholine levels in various samples. This product is for research use only
ab65345 Choline/Acetylcholine Assay Kit Instructions for Use For the rapid, sensitive and accurate measurement of Choline/Acetylcholine levels in various samples. This product is for research use only
Nature Biotechnology: doi: /nbt.3828
 Supplementary Figure 1 Development of a FRET-based MCS. (a) Linker and MA2 modification are indicated by single letter amino acid code. indicates deletion of amino acids and N or C indicate the terminus
Supplementary Figure 1 Development of a FRET-based MCS. (a) Linker and MA2 modification are indicated by single letter amino acid code. indicates deletion of amino acids and N or C indicate the terminus
A genetically targeted optical sensor to monitor calcium signals in astrocyte processes
 A genetically targeted optical sensor to monitor calcium signals in astrocyte processes 1 Eiji Shigetomi, 1 Sebastian Kracun, 2 Michael V. Sofroniew & 1,2 *Baljit S. Khakh Ψ 1 Departments of Physiology
A genetically targeted optical sensor to monitor calcium signals in astrocyte processes 1 Eiji Shigetomi, 1 Sebastian Kracun, 2 Michael V. Sofroniew & 1,2 *Baljit S. Khakh Ψ 1 Departments of Physiology
