Automatic Myocardial Segmentation by Using A Deep Learning Network in Cardiac MRI
|
|
|
- Molly Owen
- 6 years ago
- Views:
Transcription
1 Automatic Myocardial Segmentation by Using A Deep Learning Network in Cardiac MRI arxiv: v1 [cs.cv] 24 Aug 2017 Ariel H. Curiale, Flavio D. Colavecchia, Pablo Kaluza, Roberto A. Isoardi and German Mato CONICET - FCEN, Universidad Nacional de Cuyo, Padre Jorge Contreras 1300, 5500 Mendoza, Argentina CNEA - Fundación Escuela Medicina Nuclear, Garibaldi 405, 5500, Mendoza, Argentina Comisión Nacional de Energía Atómica - CONICET Departamento de Física Médica, Centro Atómico Bariloche, Centro Integral de Medicina Nuclear y Radioterapia Bariloche, Avenida Bustillo 9500, 8400 S. C. de Bariloche, Río Negro, Argentina see Abstract Cardiac function is of paramount importance for both prognosis and treatment of different pathologies such as mitral regurgitation, ischemia, dyssynchrony and myocarditis. Cardiac behavior is determined by structural and functional features. In both cases, the analysis of medical imaging studies requires to detect and segment the myocardium. Nowadays, magnetic resonance imaging (MRI) is one of the most relevant and accurate non-invasive diagnostic tools for cardiac structure and function. In this work we propose to use a deep learning technique to assist the automatization of myocardial segmentation in cardiac MRI. We present several improvements to previous works in this paper: we propose to use the Jaccard distance as optimization objective function, we integrate a residual learning strategy into the code, and we introduce a batch normalization layer to train the fully convolutional neural network. Our results demonstrate that this architecture outperforms previous approaches based on a similar network architecture, and that provides a suitable approach for myocardial segmentation. Our benchmark shows that the automatic myocardial segmentation takes less than 22 seg. for a volume of 128 x 128 x 13 pixels in a 3.1 GHz intel core i7. Index Terms deep learning; convolutional neural networks; cardiac segmentation; batch normalization; residual learning; I. INTRODUCTION Quantitative characterization of cardiac function is highly desirable for both prognosis and treatment of different pathologies such as mitral regurgitation, ischemia, dyssynchrony and myocarditis [1], [2], [3], [4], [5]. In particular, cardiac function is characterized by structural and functional features. These features can be classified as global or regional, according to the type of information they provide. One of the most relevant global structural features are the left ventricular mass (LVM), the left ventricular volume (LV) and the ejection fraction (EF), which is directly derived from the LV at end-diastole (ED) and end-systole (ES). Besides, the most relevant functional features are those derived from myocardial deformation such as strain and strain rate. Functional information is commonly presented according to the 17-segment model proposed by the American Heart Association (AHA) [6]. In both cases, it is necessary to detect and segment the myocardium of the left ventricle. Magnetic resonance imaging (MRI) is one of the most important and accurate non-invasive diagnostic tools for imaging of cardiac structure and function [7]. It is usually considered as the gold standard for ventricular volume quantification [8]. Chamber segmentation from cardiac MRI datasets is an essential step for quantification of global features, which involves the estimation of ventricular volume, ejection fraction, mass and wall thickness, among others. Manual delineation by experts is currently the standard clinical practice for performing chamber segmentation. However, and despite the efforts of researchers and medical vendors, global quantification and volumetric analysis still remain time consuming tasks which heavily rely on user interaction. For example, the time required to accurately extract the most common volumetric indices for the left ventricle for a single patient is around 10 minutes [9]. Thus, there is still a significant need for tools allowing automatic 3D quantification. The main goal of this work is to provide a suitable and accurate automatic myocardial segmentation approach for cardiac MRI based on deep convolutional neural networks [10], [11]. Many different techniques have been applied in the past to treat the problem of automatic segmentation of the left ventricle. Most of these approaches depend on proper initialization step [12]. Many model-based left ventricle segmentation approaches have been studied[13]: active contours [14], levelset method [15], active shape models and active appearance models [16], [17]. Unfortunately, all of them have a strong dependence on the initial model placement. In general, improper initialization leads to unwanted local minima of the objective function. In contrast, non model-based approaches to the problem are expected to have a more robust behavior. Among these approaches are Convolutional Neural Networks (CNNs), which combine insights from the structure of receptive fields of the visual system with modern techniques of machine learning [18]. Several recent studies in computer vision and pattern recognition highlight the capabilities of these networks to solve challenging tasks such as classification, segmentation and object detection, achieving state-of-the-art performances [19]. Fully convolutional networks trained end-to-end have been recently used for medical image segmentation [20], [21]. These models, which serve as an inspiration for our
2 work, employ different types of network architectures and were trained to generate a segmentation mask that delineates the structures of interest in the image. One of the main disadvantages of deep neural networks, such as CNN, is that its training is complicated by the fact that the distribution of each layer s inputs changes along this process, as the parameters of the previous layers change. This phenomenon is called internal covariate shift [22]. As the networks start to converge, a degradation problem occurs: with increasing network depth, accuracy gets saturated. Unexpectedly, such degradation is not caused by overfitting, and adding more layers to a suitably deep model leads to higher training error, as reported [23], [24]. To overcome this problem the technique of batch normalization has been proposed [25]. This procedure involves the evaluation of the statistical properties of the neural activations that are present for a given batch of data in order to normalize the inputs to any layer to obtain some desired objective (such as zero mean and unit variance). At the same time, the architecture is modified in order to prevent loss of information that could arise from the normalization. This technique allows to use much higher learning rates and also acts as regularizer, in some cases eliminating the need for dropout [26]. Another recent performance improvement of deep neural networks has been achieved by reformulating the layers as learning residual functions with reference to the layer inputs, instead of learning unreferenced functions [27]. In other words, the parameters to be determined at a given stage generate only the difference (or residual) between the objective function to be learned and some fixed function such as the identity. It was empirically found that this approach gives rise to networks that are easier to optimize, which can also gain accuracy from considerably increased depth [27]. In this work, we analyze the use of these techniques in the U-net architecture to segment the myocardium of the left ventricle, similarly as proposed in [21] for studying the prostate. Unlike previous works, we propose to use the Jaccard distance [28] as optimization objective and a batch normalization layer for the training of the CNN. Results demonstrate that the proposed model outperforms previous approaches based on the U-net architecture and provides a suitable approach for myocardial segmentation. The paper is structured as follows: in Section 2 the U- net network with residual learning and batch normalization is introduced. Additionally, the training strategy and the objective function used for training the CNN is presented. In Section 3, the evaluation of the proposed method in cardiac MRI is presented. Finally, we present the conclusions in Section 4. Materials II. MATERIALS AND METHODS The present network for myocardial segmentation was trained and evaluated with the Sunnybrook Cardiac Dataset (SCD), also known as the 2009 Cardiac MR Left Ventricle Segmentation Challenge data [29]. The dataset consists of 45 cine-mri images from a mix of patients and pathologies: healthy, hypertrophy, heart failure with infarction and heart failure without infarction. A subset of this dataset was first used in the automated myocardium segmentation challenge from short-axis MRI, held by a MICCAI workshop in The whole complete dataset is now available in the Cardiac Atlas Project database with public domain license 1. The Sunnybrook Cardiac Dataset [29] contains 45 cardiac cine-mr datasets with expert contours, as part of the clinical routine. Cine steady state free precession (SSFP) MR short axis (SAX) images were obtained with a 1.5T GE Signa MRI. All the images were obtained during second breathholds with a temporal resolution of 20 cardiac phases over the heart cycle, and scanned from the ED phase. In these SAX MR acquisitions, endocardial and epicardial contours were drawn by an experienced cardiologist in all slices at ED and ES phases. All the contours were confirmed by another cardiologist. The manual segmentations will be used as the ground truth for evaluation purposes. Additionally, a ROI of 128 x 128 pixels with a spatial resolution of 1.36 x 1.36 mm in the short-axis around the heart was manually selected. The ROI size was defined by an expert to ensure that the entire left ventricle was included. Network architecture The modified U-net system that we use for myocardial segmentation, follows a typical encode-decode network architecture. In this architecture, depicted in Fig. 1, the network learns how to encode information about features presented in the training set (left side on Fig. 1). Then, in the decode path the network learns about the image reconstruction process from the encoded features learned previously. The specific feature of the U-net architecture lies on the concatenation between the output of the encode path, for each level, and the input of the decoding path (denoted as big gray arrows on Fig. 1). These concatenations provide the ability to localize high spatial resolution features to the fully convolutional network, thus, generating a more precise output based on this information. As mentioned in [20] this strategy allows the seamless segmentation of large images by an overlaptile strategy. In order to predict the pixels in the border region of the image, the missing context is extrapolated by the concatenation from the encoding path. The encoding path consists of the repeated application of two 3 x 3 convolutions, each followed by a rectified linear unit (ReLU) as it was originally proposed in [20]. After performing the convolution, a batch normalization is carried out to improve the accuracy and reduce learning time. Then, a residual learning is introduced just before performing the 2 x 2 max pooling operation with stride 2 for downsampling, in a similar way as it was proposed in [21]. At each downsampling step we double the number of feature channels, that is initially set to 64. Every step in the decoding path can be seen as the mirrored step of the encode path, i.e. each step in the decoding path 1
3 Input Image Myocardial Segmentation conv 3x3 ReLu + batch normalization copy max pool 2x2 up-conv 2x2 residual learning conv 1x1 Figure 1: Network Architecture proposed for myocardial segmentation in cardiac MRI. The number of channels is denoted on top of the box and the input layer dimension is provided at the lower left edge of the box according to the U-net diagram [20]. The arrows denote the different operations according to the legend. consists of an upsampling of the feature map followed by a 2 x 2 convolution ( up-convolution ) that halves the number of feature channels, a concatenation with the corresponding feature map from the encoding path, and two 3 x 3 convolutions, each followed by a ReLU and a batch normalization. Finally, a residual learning is introduced to get the input of the next level. At the final layer, a 1 x 1 convolution is carried out to map the 64 feature maps to the two classes used for the myocardial segmentation (myocardium and background). The output of the last layer, after soft-max non-linear function, represents the likelihood of a pixel belongs to the myocardium of the left ventricle. Indeed, only those voxels with higher likelihood (> 0.5) are considered as part of the left ventricle tissue. Training Half of the images and their corresponding myocardial segmentations were manually cropped into 128 x 28 pixels with a spatial resolution of 1.36 x 1.36 mm in the short-axis around the heart. This ROI size was defined by an expert to ensure that the entire left ventricle was included for all the images. Then, the cropped images and their corresponding myocardial segmentations were used to train the network with the stochastic gradient descent (Adaptive Moment Estimation) implementation of Keras [30]. In the proposed U-net we use the Jaccard distance as objective function instead of the Dice s coefficient commonly used in image processing. The Jaccard distance is defined as follows: Jd(myo pred, myo true ) = 1 i myopred i i myopred i myo true i + i myotrue i i myopred i, myo true i where myo pred and myo true are the myocardial segmentation prediction and the ground truth segmentation, and the i myo i refers to the pixel sum over the entire segmentation (prediction or ground truth). Annotated medical information like myocardial classification is not easy to obtain due to the fact that one or more experts are required to manually trace a reliable ground truth of the myocardial classification. So, in this work it was necessary to augment the original training dataset in order to increase the examples. Also, data augmentation is essential to teach the network the desired invariance and robustness properties. Heterogeneity in the cardiac MRI dataset is needed to teach the network some shift and rotation invariance, as well as robustness to deformations. With this intention, during the every training iteration, the input of the network was randomly deformed by means of a spatial shift in a range of 10% of the image size, a rotation in a range of 10 in the short axis, a zoom in a range of 2x or by using a gaussian deformation field (µ = 10 and σ = 20) and a B-spline interpolation. The data augmentation has been performed on-the-fly to reduce the data storage and achieve a total of 5500 new images by epoch. III. RESULTS Three sets of experiments are conducted to evaluate the proposed methodology on the Sunnybrooks dataset [29]. In the first two experiments, the papillary muscles (PM) were excluded on both, the automatic and manual segmentation. So, the proposed method will avoid to detect the PM as part of the myocardial tissue. In the first experiment, the proposed objective function loss, batch normalization and residual learning were evaluated on the classic U-net architecture with respect to the commonly used Dice s coefficient. Empirical results show that the Jaccard distances (0.6 ± 0.1 mean accuracy Dice s value) outperform the Dice s coefficient (0.58 ± 0.1 mean accuracy Dice s value) when it is used as the objective loss function (Fig. 2a). Moreover, Fig. 2a shows that this improvement (Jaccard distance vs. Dice s coefficient) is uncorrelated with the use of batch normalization and residual learning. As results show on Fig. 2a and Table I, the use of the batch normalization and residual learning strategies are of paramount importance to achieve an accurate myocardial segmentation.
4 0.9 Accuracy on Training performance 0.9 Accuracy on Training performance Dice's accuracy Jaccard dist. - BN Jaccard dist. Jaccard dist. - BN - RL Dice's coeff. - BN Dice's coeff. Dice's coeff. - BN - RL Number of epochs Dice's accuracy Jaccard dist. - BN Jaccard dist. - RL Jaccard dist. - BN - RL Number of epochs (a) Figure 2: Accuracy on training performance for 50 training iteration over the entire test dataset (50 epochs) with a shrink factor of 2 (i.e. the original input size was reduced by a factor of 2). In the picture is depicted the accuracy performance between Dice s coefficient and Jaccard distance (JD) objective loss function, batch normalization (BN) and residual learning (RL) on the U-net architecture. For clarity in (b) it is showed the effect of BN and RL only for the Jaccard distance loss function. (b) Furthermore, the combination of both strategies shows the higher myocardial segmentation accuracy. However, if the training accuracy with the strategies of batch normalization and residual learning is analyzed, we can conclude that the contribution of the batch normalization is higher than contribution of the residual learning (Fig. 2b). Also, we can conclude that the training accuracy of the residual learning is comparable to the basic U-net for our architecture (i.e. 5 levels, 2 conv. operation by level and the same number of features by level). Nonetheless, we strongly recommend to use both strategies together due to the accuracy achieved by them working together is remarkable higher than the individual contribution of each one as it can be seen in Fig. 2b. SF Architecture Dice s acc. MSE MAE U-net - Dice s U-net - JD U-net - BN - JD U-net - BN - RL - JD U-net - BN - RL - JD Table I: Myocardial segmentation accuracy for each convolutional neural network studied on the Sunnybrooks dataset for 50 training iteration over the entire test dataset (50 epochs). SF: shrink factor with respect to the original image (i.e. SF=2 means that the original input size was reduced by a factor of 2). MSE: mean squeared error. MAE: mean absolute error. JD: Jaccard distance. BN: Batch normalization. RL: Residual learning. The second experiment shows (Fig. 3) that our CNN for myocardial segmentation in cardiac MRI reaches suitable accuracy in a few training iterations. i.e. in 30 epochs. In this case, the present CCN reaches a value of 0.9 and for the Dice s coefficient and a mean squared error, respectively, which is comparable to manual segmentation. Dice's accuracy Accuracy on Training performance (63, 0.91) Number of epochs Figure 3: Accuracy on training performance of the proposed U-net architecture for myocardial segmentation in cardiac MRI. Finally, as it was expected, the third experiment revealed that the accuracy tend to decrease when the PM were taking into account as part of the myocardial tissue, since it makes the myocardial tissue segmentation harder. However, the proposed approach seems to be not affected and reaches a similar accuracy to the previous experiment. We obtained 0.89 for the Dice s coefficient and a mean squared error of 0.01.
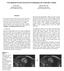
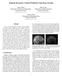
5 (a) Figure 4: Example of myocardial segmentation for the proposed convolutional neural network. (a) The proposed myocardial segmentation is presented in three orthogonal views. (b) 3D view of the proposed myocardial segmentation. (b) Figure 5: Qualitative results for a patient on Sunnybrook dataset. Different short axis slices are plotted from apex (upper-left) to base (down-right) of the left ventricle. In brown it is depicts those voxels where the manual myocardial segmentation and the proposed automatic segmentation overlaps, otherwise, the voxels are depicted in light-green. An example of the automatic 3D myocardial segmentation can be seen in Fig. 4. In particular, Fig. 4a shows the myocardial segmentation in three orthogonal views and Fig. 4b shows the segmentation in two 3D views. Qualitative details of the this Automatic 3D myocardial segmentation (Fig. 4) can be seen in several slices in Fig. 5. They show that the proposed myocardial segmentation provides a suitable approach for myocardial segmentation in cardiac MRI. In yellow it is depicts those voxels where the manual myocardial segmentation and the proposed automatic segmentation overlaps, otherwise, the voxels are depicted in magenta. As it can be seen only a reduced number of voxels correspond to a non-overlapping classification. IV. CONCLUSION In this paper, we have proposed an automatic 3D myocardial segmentation approach for cardiac MRI by using a deep learning network. Unlike previous approaches, our method makes use of the Jaccard distance as objective loss function, a batch normalization and residual learning strategies to provide a suitable approach for myocardial segmentation. Quantitative and qualitative results show that the proposed approach presents a high potential for being used to estimate
6 different structural and functional features for both prognosis and treatment of different pathologies. Thanks to real time data augmentation with free form deformations, it only need very few annotated images (22 cardiac MRI) and has a very reasonable training time of only 8.5 hours on a NVidia Tesla C2070 (6 GB) for reaching a suitable accuracy of 0.9 Dice s coefficient and mean squared error in 30 epochs. The segmentation and tracking of different cardiac structures, such as the left ventricle, have an important role in the treatment of different pathologies. However, accurate and reliable measurements for volumetric analysis and functional assessment heavily rely on user interaction. The results obtained in this work can be efficiently used for estimating different structural (left ventricle mass, left ventricle volume and ejection fraction among others) features. Additionally, this work leads to extensions for automatic detection and tracking of the right and left ventricle. Myocardial motion is useful in the evaluation of regional cardiac functions such as the strain and strain rate. ACKNOWLEDGMENTS This work was partially supported by Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) and by grants M SECTyP and L SECTyP, Universidad Nacional de Cuyo, Argentina; PICT , Agencia Nacional de Promoción Científica y Tecnológica, Argentina. German Mato and Pablo Kaluza acknowledge CONICET for the grant PIP and PIP respectively. REFERENCES [1] B. D. Lowes, E. A. Gill, W. T. Abraham, J.-R. Larrain, A. D. Robertson, M. R. Bristow, and E. M. Gilbert, Effects of carvedilol on left ventricular mass, chamber geometry, and mitral regurgitation in chronic heart failure, The American journal of cardiology, vol. 83, no. 8, pp , [2] T. M. Koelling, K. D. Aaronson, R. J. Cody, D. S. Bach, and W. F. Armstrong, Prognostic significance of mitral regurgitation and tricuspid regurgitation in patients with left ventricular systolic dysfunction, American heart journal, vol. 144, no. 3, pp , [3] M. G. Friedrich, U. Sechtem, J. Schulz-Menger, G. Holmvang, P. Alakija, L. T. Cooper, J. A. White, H. Abdel-Aty, M. Gutberlet, S. Prasad et al., Cardiovascular magnetic resonance in myocarditis: A jacc white paper, Journal of the American College of Cardiology, vol. 53, no. 17, pp , [4] T. Edvardsen, S. Urheim, H. Skulstad, K. Steine, H. Ihlen, and O. A. Smiseth, Quantification of left ventricular systolic function by tissue doppler echocardiography, Circulation, vol. 105, no. 17, pp , [5] M. S. Suffoletto, K. Dohi, M. Cannesson, S. Saba, and J. Gorcsan, Novel speckle-tracking radial strain from routine black-and-white echocardiographic images to quantify dyssynchrony and predict response to cardiac resynchronization therapy, Circulation, vol. 113, no. 7, pp , [6] M. D. Cerqueira, N. J. Weissman, V. Dilsizian, A. K. Jacobs, S. Kaul, W. K. Laskey, D. J. Pennell, J. A. Rumberger, T. Ryan, and M. S. Verani, Standardized myocardial segmentation and nomenclature for tomographic imaging of the heart, Circulation, vol. 105, no. 4, pp , Jan [7] J. W. Weinsaft, I. Klem, and R. M. Judd, Mri for the assessment of myocardial viability, Cardiology Clinics, vol. 25, no. 1, pp , [8] A. L. Gerche, G. Claessen, A. Van de Bruaene, N. Pattyn, J. Van Cleemput, M. Gewillig, J. Bogaert, S. Dymarkowski, P. Claus, and H. Heidbuchel, Cardiac mriclinical perspective, Circulation: Cardiovascular Imaging, vol. 6, no. 2, pp , [9] E. Heijman, J.-P. Aben, C. Penners, P. Niessen, R. Guillaume, G. van Eys, K. Nicolay, and G. J. Strijkers, Evaluation of manual and automatic segmentation of the mouse heart from cine mr images, Journal of Magnetic Resonance Imaging, vol. 27, no. 1, pp , [10] A. Krizhevsky, I. Sutskever, and G. E. Hinton, Imagenet classification with deep convolutional neural networks, in Advances in neural information processing systems, 2012, pp [11] K. Simonyan and A. Zisserman, Very deep convolutional networks for large-scale image recognition, arxiv preprint arxiv: , [12] A. K. Jain, Y. Zhong, and M.-P. Dubuisson-Jolly, Deformable template models: A review, Signal Proccessing, vol. 71, no. 2, pp , Dec [13] C. Petitjean and J.-N. Dacher, A review of segmentation methods in short axis cardiac MR images, Medical Image Analysis, vol. 15, no. 2, pp , Apr [14] M. Kass, A. Witkin, and D. Terzopoulos, Snakes: Active contour models, Int. J. Comput. Vision, vol. 1, no. 4, pp , Jan [15] S. Osher and J. A. Sethian, Fronts propagating with curvaturedependent speed: Algorithms based on hamilton-jacobi formulations, J. Comput. Phys., vol. 79, no. 1, pp , Nov [16] T. Cootes and C. Taylor, Active shape models: Smart snakes, in Proc. British Machine Vision Conference, vol Citeseer, [17] T. Cootes, G. Edwards, and C. Taylor, Active appearance models, Computer Vision and Image Understanding, vol. 61, no. 1, pp , [18] Y. Lecun, L. Bottou, Y. Bengio, and P. Haffner, Gradient-based learning applied to document recognition, Proceedings of the IEEE, vol. 86, no. 11, pp , Nov [19] G. Litjens, T. Kooi, B. E. Bejnordi, A. A. A. Setio, F. Ciompi, M. Ghafoorian, J. A. van der Laak, B. van Ginneken, and C. I. Sánchez, A survey on deep learning in medical image analysis, arxiv preprint arxiv: , [20] O. Ronneberger, P. Fischer, and T. Brox, U-net: Convolutional networks for biomedical image segmentation, in Medical Image Computing and Computer-Assisted Intervention MICCAI 2015: 18th International Conference, Munich, Germany, October 5-9, 2015, Proceedings, Part III, N. Navab, J. Hornegger, W. M. Wells, and A. F. Frangi, Eds. Cham: Springer International Publishing, 2015, pp [21] F. Milletari, N. Navab, and S. A. Ahmadi, V-net: Fully convolutional neural networks for volumetric medical image segmentation, in 2016 Fourth International Conference on 3D Vision (3DV), Oct 2016, pp [22] H. Shimodaira, Improving predictive inference under covariate shift by weighting the log-likelihood function, Journal of statistical planning and inference, vol. 90, no. 2, pp , [23] K. He and J. Sun, Convolutional neural networks at constrained time cost, in The IEEE Conference on Computer Vision and Pattern Recognition (CVPR), June [24] R. K. Srivastava, K. Greff, and J. Schmidhuber, Highway networks, arxiv preprint arxiv: , [25] S. Ioffe and C. Szegedy, Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift, ArXiv e-prints, Feb [26] N. Srivastava, G. E. Hinton, A. Krizhevsky, I. Sutskever, and R. Salakhutdinov, Dropout: a simple way to prevent neural networks from overfitting. Journal of Machine Learning Research, vol. 15, no. 1, pp , [27] K. He, X. Zhang, S. Ren, and J. Sun, Deep residual learning for image recognition, in The IEEE Conference on Computer Vision and Pattern Recognition (CVPR), June [28] R. Toldo and A. Fusiello, Robust multiple structures estimation with j-linkage, Computer Vision ECCV 2008, pp , [29] P. Radau, Y. Lu, K. Connelly, G. Paul, A. J. Dick, and G. A. W. GA., Evaluation framework for algorithms segmenting short axis cardiac mri, The MIDAS Journal -Cardiac MR Left Ventricle Segmentation Challenge, [30] F. Chollet, Keras,
Recurrent Fully Convolutional Neural Networks for Multi-slice MRI Cardiac Segmentation
 Recurrent Fully Convolutional Neural Networks for Multi-slice MRI Cardiac Segmentation Rudra P K Poudel, Pablo Lamata and Giovanni Montana Department of Biomedical Engineering, King s College London, SE1
Recurrent Fully Convolutional Neural Networks for Multi-slice MRI Cardiac Segmentation Rudra P K Poudel, Pablo Lamata and Giovanni Montana Department of Biomedical Engineering, King s College London, SE1
Convolutional Neural Networks for Estimating Left Ventricular Volume
 Convolutional Neural Networks for Estimating Left Ventricular Volume Ryan Silva Stanford University rdsilva@stanford.edu Maksim Korolev Stanford University mkorolev@stanford.edu Abstract End-systolic and
Convolutional Neural Networks for Estimating Left Ventricular Volume Ryan Silva Stanford University rdsilva@stanford.edu Maksim Korolev Stanford University mkorolev@stanford.edu Abstract End-systolic and
CSE Introduction to High-Perfomance Deep Learning ImageNet & VGG. Jihyung Kil
 CSE 5194.01 - Introduction to High-Perfomance Deep Learning ImageNet & VGG Jihyung Kil ImageNet Classification with Deep Convolutional Neural Networks Alex Krizhevsky, Ilya Sutskever, Geoffrey E. Hinton,
CSE 5194.01 - Introduction to High-Perfomance Deep Learning ImageNet & VGG Jihyung Kil ImageNet Classification with Deep Convolutional Neural Networks Alex Krizhevsky, Ilya Sutskever, Geoffrey E. Hinton,
Automated Volumetric Cardiac Ultrasound Analysis
 Whitepaper Automated Volumetric Cardiac Ultrasound Analysis ACUSON SC2000 Volume Imaging Ultrasound System Bogdan Georgescu, Ph.D. Siemens Corporate Research Princeton, New Jersey USA Answers for life.
Whitepaper Automated Volumetric Cardiac Ultrasound Analysis ACUSON SC2000 Volume Imaging Ultrasound System Bogdan Georgescu, Ph.D. Siemens Corporate Research Princeton, New Jersey USA Answers for life.
arxiv: v1 [cs.cv] 13 Jul 2018
![arxiv: v1 [cs.cv] 13 Jul 2018 arxiv: v1 [cs.cv] 13 Jul 2018](/thumbs/84/89699669.jpg) Multi-Scale Convolutional-Stack Aggregation for Robust White Matter Hyperintensities Segmentation Hongwei Li 1, Jianguo Zhang 3, Mark Muehlau 2, Jan Kirschke 2, and Bjoern Menze 1 arxiv:1807.05153v1 [cs.cv]
Multi-Scale Convolutional-Stack Aggregation for Robust White Matter Hyperintensities Segmentation Hongwei Li 1, Jianguo Zhang 3, Mark Muehlau 2, Jan Kirschke 2, and Bjoern Menze 1 arxiv:1807.05153v1 [cs.cv]
Final Report: Automated Semantic Segmentation of Volumetric Cardiovascular Features and Disease Assessment
 Final Report: Automated Semantic Segmentation of Volumetric Cardiovascular Features and Disease Assessment Tony Lindsey 1,3, Xiao Lu 1 and Mojtaba Tefagh 2 1 Department of Biomedical Informatics, Stanford
Final Report: Automated Semantic Segmentation of Volumetric Cardiovascular Features and Disease Assessment Tony Lindsey 1,3, Xiao Lu 1 and Mojtaba Tefagh 2 1 Department of Biomedical Informatics, Stanford
arxiv: v1 [cs.cv] 9 Oct 2018
![arxiv: v1 [cs.cv] 9 Oct 2018 arxiv: v1 [cs.cv] 9 Oct 2018](/thumbs/95/126101195.jpg) Automatic Segmentation of Thoracic Aorta Segments in Low-Dose Chest CT Julia M. H. Noothout a, Bob D. de Vos a, Jelmer M. Wolterink a, Ivana Išgum a a Image Sciences Institute, University Medical Center
Automatic Segmentation of Thoracic Aorta Segments in Low-Dose Chest CT Julia M. H. Noothout a, Bob D. de Vos a, Jelmer M. Wolterink a, Ivana Išgum a a Image Sciences Institute, University Medical Center
HHS Public Access Author manuscript Med Image Comput Comput Assist Interv. Author manuscript; available in PMC 2018 January 04.
 Discriminative Localization in CNNs for Weakly-Supervised Segmentation of Pulmonary Nodules Xinyang Feng 1, Jie Yang 1, Andrew F. Laine 1, and Elsa D. Angelini 1,2 1 Department of Biomedical Engineering,
Discriminative Localization in CNNs for Weakly-Supervised Segmentation of Pulmonary Nodules Xinyang Feng 1, Jie Yang 1, Andrew F. Laine 1, and Elsa D. Angelini 1,2 1 Department of Biomedical Engineering,
Multi-attention Guided Activation Propagation in CNNs
 Multi-attention Guided Activation Propagation in CNNs Xiangteng He and Yuxin Peng (B) Institute of Computer Science and Technology, Peking University, Beijing, China pengyuxin@pku.edu.cn Abstract. CNNs
Multi-attention Guided Activation Propagation in CNNs Xiangteng He and Yuxin Peng (B) Institute of Computer Science and Technology, Peking University, Beijing, China pengyuxin@pku.edu.cn Abstract. CNNs
arxiv: v1 [cs.cv] 17 Aug 2017
![arxiv: v1 [cs.cv] 17 Aug 2017 arxiv: v1 [cs.cv] 17 Aug 2017](/thumbs/78/78278805.jpg) Deep Learning for Medical Image Analysis Mina Rezaei, Haojin Yang, Christoph Meinel Hasso Plattner Institute, Prof.Dr.Helmert-Strae 2-3, 14482 Potsdam, Germany {mina.rezaei,haojin.yang,christoph.meinel}@hpi.de
Deep Learning for Medical Image Analysis Mina Rezaei, Haojin Yang, Christoph Meinel Hasso Plattner Institute, Prof.Dr.Helmert-Strae 2-3, 14482 Potsdam, Germany {mina.rezaei,haojin.yang,christoph.meinel}@hpi.de
Impaired Regional Myocardial Function Detection Using the Standard Inter-Segmental Integration SINE Wave Curve On Magnetic Resonance Imaging
 Original Article Impaired Regional Myocardial Function Detection Using the Standard Inter-Segmental Integration Ngam-Maung B, RT email : chaothawee@yahoo.com Busakol Ngam-Maung, RT 1 Lertlak Chaothawee,
Original Article Impaired Regional Myocardial Function Detection Using the Standard Inter-Segmental Integration Ngam-Maung B, RT email : chaothawee@yahoo.com Busakol Ngam-Maung, RT 1 Lertlak Chaothawee,
Automatic cardiac contour propagation in short axis cardiac MR images
 International Congress Series 1281 (2005) 351 356 www.ics-elsevier.com Automatic cardiac contour propagation in short axis cardiac MR images G.L.T.F. Hautvast a,b, T, M. Breeuwer a, S. Lobregt a, A. Vilanova
International Congress Series 1281 (2005) 351 356 www.ics-elsevier.com Automatic cardiac contour propagation in short axis cardiac MR images G.L.T.F. Hautvast a,b, T, M. Breeuwer a, S. Lobregt a, A. Vilanova
arxiv: v2 [cs.cv] 19 Dec 2017
![arxiv: v2 [cs.cv] 19 Dec 2017 arxiv: v2 [cs.cv] 19 Dec 2017](/thumbs/89/97557782.jpg) An Ensemble of Deep Convolutional Neural Networks for Alzheimer s Disease Detection and Classification arxiv:1712.01675v2 [cs.cv] 19 Dec 2017 Jyoti Islam Department of Computer Science Georgia State University
An Ensemble of Deep Convolutional Neural Networks for Alzheimer s Disease Detection and Classification arxiv:1712.01675v2 [cs.cv] 19 Dec 2017 Jyoti Islam Department of Computer Science Georgia State University
Differentiating Tumor and Edema in Brain Magnetic Resonance Images Using a Convolutional Neural Network
 Original Article Differentiating Tumor and Edema in Brain Magnetic Resonance Images Using a Convolutional Neural Network Aida Allahverdi 1, Siavash Akbarzadeh 1, Alireza Khorrami Moghaddam 2, Armin Allahverdy
Original Article Differentiating Tumor and Edema in Brain Magnetic Resonance Images Using a Convolutional Neural Network Aida Allahverdi 1, Siavash Akbarzadeh 1, Alireza Khorrami Moghaddam 2, Armin Allahverdy
arxiv: v2 [cs.cv] 7 Jun 2018
![arxiv: v2 [cs.cv] 7 Jun 2018 arxiv: v2 [cs.cv] 7 Jun 2018](/thumbs/79/79761281.jpg) Deep supervision with additional labels for retinal vessel segmentation task Yishuo Zhang and Albert C.S. Chung Lo Kwee-Seong Medical Image Analysis Laboratory, Department of Computer Science and Engineering,
Deep supervision with additional labels for retinal vessel segmentation task Yishuo Zhang and Albert C.S. Chung Lo Kwee-Seong Medical Image Analysis Laboratory, Department of Computer Science and Engineering,
Fully Convolutional Network-based Multi-Task Learning for Rectum and Rectal Cancer Segmentation
 Fully Convolutional Network-based Multi-Task Learning for Rectum and Rectal Cancer Segmentation Joohyung Lee 1, Ji Eun Oh 1, Min Ju Kim 2, Bo Yun Hur 2, Sun Ah Cho 1 1, 2*, and Dae Kyung Sohn 1 Innovative
Fully Convolutional Network-based Multi-Task Learning for Rectum and Rectal Cancer Segmentation Joohyung Lee 1, Ji Eun Oh 1, Min Ju Kim 2, Bo Yun Hur 2, Sun Ah Cho 1 1, 2*, and Dae Kyung Sohn 1 Innovative
arxiv: v1 [cs.cv] 3 Apr 2018
![arxiv: v1 [cs.cv] 3 Apr 2018 arxiv: v1 [cs.cv] 3 Apr 2018](/thumbs/90/103707026.jpg) Towards whole-body CT Bone Segmentation André Klein 1,2, Jan Warszawski 2, Jens Hillengaß 3, Klaus H. Maier-Hein 1 arxiv:1804.00908v1 [cs.cv] 3 Apr 2018 1 Division of Medical Image Computing, Deutsches
Towards whole-body CT Bone Segmentation André Klein 1,2, Jan Warszawski 2, Jens Hillengaß 3, Klaus H. Maier-Hein 1 arxiv:1804.00908v1 [cs.cv] 3 Apr 2018 1 Division of Medical Image Computing, Deutsches
Comparative Study of K-means, Gaussian Mixture Model, Fuzzy C-means algorithms for Brain Tumor Segmentation
 Comparative Study of K-means, Gaussian Mixture Model, Fuzzy C-means algorithms for Brain Tumor Segmentation U. Baid 1, S. Talbar 2 and S. Talbar 1 1 Department of E&TC Engineering, Shri Guru Gobind Singhji
Comparative Study of K-means, Gaussian Mixture Model, Fuzzy C-means algorithms for Brain Tumor Segmentation U. Baid 1, S. Talbar 2 and S. Talbar 1 1 Department of E&TC Engineering, Shri Guru Gobind Singhji
THE first objective of this thesis was to explore possible shape parameterizations
 8 SUMMARY Columbus is not the only person who has discovered a new continent. So too have I. Anak Semua Bangsa (Child of All Nations) PRAMOEDYA ANANTA TOER 8.1 Myocardial wall motion modeling THE first
8 SUMMARY Columbus is not the only person who has discovered a new continent. So too have I. Anak Semua Bangsa (Child of All Nations) PRAMOEDYA ANANTA TOER 8.1 Myocardial wall motion modeling THE first
arxiv: v1 [stat.ml] 23 Jan 2017
![arxiv: v1 [stat.ml] 23 Jan 2017 arxiv: v1 [stat.ml] 23 Jan 2017](/thumbs/81/83414183.jpg) Learning what to look in chest X-rays with a recurrent visual attention model arxiv:1701.06452v1 [stat.ml] 23 Jan 2017 Petros-Pavlos Ypsilantis Department of Biomedical Engineering King s College London
Learning what to look in chest X-rays with a recurrent visual attention model arxiv:1701.06452v1 [stat.ml] 23 Jan 2017 Petros-Pavlos Ypsilantis Department of Biomedical Engineering King s College London
MEDVISO WHITE PAPER ON STRAIN ANALYSIS IN TAGGED MR IMAGES
 Purpose of document The purpose of this document is to document validation of the Strain analysis module in Segment software packages. Intended audience The intended audiences of this document are: Engineering
Purpose of document The purpose of this document is to document validation of the Strain analysis module in Segment software packages. Intended audience The intended audiences of this document are: Engineering
MICCAI 2018 VISDMML Tutorial Image Analysis Meets Deep Learning and Visualization in Cardiology and Cardiac Surgery
 MICCAI 2018 VISDMML Tutorial Image Analysis Meets Deep Learning and Visualization in Cardiology and Cardiac Surgery Dr. Sandy Engelhardt Faculty of Computer Science, Mannheim University of Applied Sciences,
MICCAI 2018 VISDMML Tutorial Image Analysis Meets Deep Learning and Visualization in Cardiology and Cardiac Surgery Dr. Sandy Engelhardt Faculty of Computer Science, Mannheim University of Applied Sciences,
arxiv: v1 [cs.cv] 12 Jun 2018
![arxiv: v1 [cs.cv] 12 Jun 2018 arxiv: v1 [cs.cv] 12 Jun 2018](/thumbs/90/102210908.jpg) Multiview Two-Task Recursive Attention Model for Left Atrium and Atrial Scars Segmentation Jun Chen* 1, Guang Yang* 2, Zhifan Gao 3, Hao Ni 4, Elsa Angelini 5, Raad Mohiaddin 2, Tom Wong 2,Yanping Zhang
Multiview Two-Task Recursive Attention Model for Left Atrium and Atrial Scars Segmentation Jun Chen* 1, Guang Yang* 2, Zhifan Gao 3, Hao Ni 4, Elsa Angelini 5, Raad Mohiaddin 2, Tom Wong 2,Yanping Zhang
Automatic Quality Assessment of Cardiac MRI
 Automatic Quality Assessment of Cardiac MRI Ilkay Oksuz 02.05.2018 Contact: ilkay.oksuz@kcl.ac.uk http://kclmmag.org 1 Cardiac MRI Quality Issues Need for high quality images Wide range of artefacts Manual
Automatic Quality Assessment of Cardiac MRI Ilkay Oksuz 02.05.2018 Contact: ilkay.oksuz@kcl.ac.uk http://kclmmag.org 1 Cardiac MRI Quality Issues Need for high quality images Wide range of artefacts Manual
Fully Automatic Segmentation of Papillary Muscles in 3-D LGE-MRI
 Fully Automatic Segmentation of Papillary Muscles in 3-D LGE-MRI Tanja Kurzendorfer 1, Alexander Brost 2, Christoph Forman 3, Michaela Schmidt 3, Christoph Tillmanns 4, Andreas Maier 1 1 Pattern Recognition
Fully Automatic Segmentation of Papillary Muscles in 3-D LGE-MRI Tanja Kurzendorfer 1, Alexander Brost 2, Christoph Forman 3, Michaela Schmidt 3, Christoph Tillmanns 4, Andreas Maier 1 1 Pattern Recognition
POC Brain Tumor Segmentation. vlife Use Case
 Brain Tumor Segmentation vlife Use Case 1 Automatic Brain Tumor Segmentation using CNN Background Brain tumor segmentation seeks to separate healthy tissue from tumorous regions such as the advancing tumor,
Brain Tumor Segmentation vlife Use Case 1 Automatic Brain Tumor Segmentation using CNN Background Brain tumor segmentation seeks to separate healthy tissue from tumorous regions such as the advancing tumor,
Efficient Deep Model Selection
 Efficient Deep Model Selection Jose Alvarez Researcher Data61, CSIRO, Australia GTC, May 9 th 2017 www.josemalvarez.net conv1 conv2 conv3 conv4 conv5 conv6 conv7 conv8 softmax prediction???????? Num Classes
Efficient Deep Model Selection Jose Alvarez Researcher Data61, CSIRO, Australia GTC, May 9 th 2017 www.josemalvarez.net conv1 conv2 conv3 conv4 conv5 conv6 conv7 conv8 softmax prediction???????? Num Classes
arxiv: v2 [cs.cv] 8 Mar 2018
![arxiv: v2 [cs.cv] 8 Mar 2018 arxiv: v2 [cs.cv] 8 Mar 2018](/thumbs/87/97094636.jpg) Automated soft tissue lesion detection and segmentation in digital mammography using a u-net deep learning network Timothy de Moor a, Alejandro Rodriguez-Ruiz a, Albert Gubern Mérida a, Ritse Mann a, and
Automated soft tissue lesion detection and segmentation in digital mammography using a u-net deep learning network Timothy de Moor a, Alejandro Rodriguez-Ruiz a, Albert Gubern Mérida a, Ritse Mann a, and
Global left ventricular circumferential strain is a marker for both systolic and diastolic myocardial function
 Global left ventricular circumferential strain is a marker for both systolic and diastolic myocardial function Toshinari Onishi 1, Samir K. Saha 2, Daniel Ludwig 1, Erik B. Schelbert 1, David Schwartzman
Global left ventricular circumferential strain is a marker for both systolic and diastolic myocardial function Toshinari Onishi 1, Samir K. Saha 2, Daniel Ludwig 1, Erik B. Schelbert 1, David Schwartzman
Convolutional Neural Networks (CNN)
 Convolutional Neural Networks (CNN) Algorithm and Some Applications in Computer Vision Luo Hengliang Institute of Automation June 10, 2014 Luo Hengliang (Institute of Automation) Convolutional Neural Networks
Convolutional Neural Networks (CNN) Algorithm and Some Applications in Computer Vision Luo Hengliang Institute of Automation June 10, 2014 Luo Hengliang (Institute of Automation) Convolutional Neural Networks
arxiv: v1 [cs.cv] 16 Jan 2018
![arxiv: v1 [cs.cv] 16 Jan 2018 arxiv: v1 [cs.cv] 16 Jan 2018](/thumbs/94/118494581.jpg) Fully Convolutional Multi-scale Residual DenseNets for Cardiac Segmentation and Automated Cardiac Diagnosis using Ensemble of Classifiers Mahendra Khened a, Varghese Alex a, Ganapathy Krishnamurthi a,
Fully Convolutional Multi-scale Residual DenseNets for Cardiac Segmentation and Automated Cardiac Diagnosis using Ensemble of Classifiers Mahendra Khened a, Varghese Alex a, Ganapathy Krishnamurthi a,
An Artificial Neural Network Architecture Based on Context Transformations in Cortical Minicolumns
 An Artificial Neural Network Architecture Based on Context Transformations in Cortical Minicolumns 1. Introduction Vasily Morzhakov, Alexey Redozubov morzhakovva@gmail.com, galdrd@gmail.com Abstract Cortical
An Artificial Neural Network Architecture Based on Context Transformations in Cortical Minicolumns 1. Introduction Vasily Morzhakov, Alexey Redozubov morzhakovva@gmail.com, galdrd@gmail.com Abstract Cortical
Segmentation of Cell Membrane and Nucleus by Improving Pix2pix
 Segmentation of Membrane and Nucleus by Improving Pix2pix Masaya Sato 1, Kazuhiro Hotta 1, Ayako Imanishi 2, Michiyuki Matsuda 2 and Kenta Terai 2 1 Meijo University, Siogamaguchi, Nagoya, Aichi, Japan
Segmentation of Membrane and Nucleus by Improving Pix2pix Masaya Sato 1, Kazuhiro Hotta 1, Ayako Imanishi 2, Michiyuki Matsuda 2 and Kenta Terai 2 1 Meijo University, Siogamaguchi, Nagoya, Aichi, Japan
Velocity Vector Imaging as a new approach for cardiac magnetic resonance: Comparison with echocardiography
 Velocity Vector Imaging as a new approach for cardiac magnetic resonance: Comparison with echocardiography Toshinari Onishi 1, Samir K. Saha 2, Daniel Ludwig 1, Erik B. Schelbert 1, David Schwartzman 1,
Velocity Vector Imaging as a new approach for cardiac magnetic resonance: Comparison with echocardiography Toshinari Onishi 1, Samir K. Saha 2, Daniel Ludwig 1, Erik B. Schelbert 1, David Schwartzman 1,
Highly Accurate Brain Stroke Diagnostic System and Generative Lesion Model. Junghwan Cho, Ph.D. CAIDE Systems, Inc. Deep Learning R&D Team
 Highly Accurate Brain Stroke Diagnostic System and Generative Lesion Model Junghwan Cho, Ph.D. CAIDE Systems, Inc. Deep Learning R&D Team Established in September, 2016 at 110 Canal st. Lowell, MA 01852,
Highly Accurate Brain Stroke Diagnostic System and Generative Lesion Model Junghwan Cho, Ph.D. CAIDE Systems, Inc. Deep Learning R&D Team Established in September, 2016 at 110 Canal st. Lowell, MA 01852,
Object Detectors Emerge in Deep Scene CNNs
 Object Detectors Emerge in Deep Scene CNNs Bolei Zhou, Aditya Khosla, Agata Lapedriza, Aude Oliva, Antonio Torralba Presented By: Collin McCarthy Goal: Understand how objects are represented in CNNs Are
Object Detectors Emerge in Deep Scene CNNs Bolei Zhou, Aditya Khosla, Agata Lapedriza, Aude Oliva, Antonio Torralba Presented By: Collin McCarthy Goal: Understand how objects are represented in CNNs Are
A convolutional neural network to classify American Sign Language fingerspelling from depth and colour images
 A convolutional neural network to classify American Sign Language fingerspelling from depth and colour images Ameen, SA and Vadera, S http://dx.doi.org/10.1111/exsy.12197 Title Authors Type URL A convolutional
A convolutional neural network to classify American Sign Language fingerspelling from depth and colour images Ameen, SA and Vadera, S http://dx.doi.org/10.1111/exsy.12197 Title Authors Type URL A convolutional
Skin cancer reorganization and classification with deep neural network
 Skin cancer reorganization and classification with deep neural network Hao Chang 1 1. Department of Genetics, Yale University School of Medicine 2. Email: changhao86@gmail.com Abstract As one kind of skin
Skin cancer reorganization and classification with deep neural network Hao Chang 1 1. Department of Genetics, Yale University School of Medicine 2. Email: changhao86@gmail.com Abstract As one kind of skin
Y-Net: Joint Segmentation and Classification for Diagnosis of Breast Biopsy Images
 Y-Net: Joint Segmentation and Classification for Diagnosis of Breast Biopsy Images Sachin Mehta 1, Ezgi Mercan 1, Jamen Bartlett 2, Donald Weaver 2, Joann G. Elmore 1, and Linda Shapiro 1 1 University
Y-Net: Joint Segmentation and Classification for Diagnosis of Breast Biopsy Images Sachin Mehta 1, Ezgi Mercan 1, Jamen Bartlett 2, Donald Weaver 2, Joann G. Elmore 1, and Linda Shapiro 1 1 University
B657: Final Project Report Holistically-Nested Edge Detection
 B657: Final roject Report Holistically-Nested Edge Detection Mingze Xu & Hanfei Mei May 4, 2016 Abstract Holistically-Nested Edge Detection (HED), which is a novel edge detection method based on fully
B657: Final roject Report Holistically-Nested Edge Detection Mingze Xu & Hanfei Mei May 4, 2016 Abstract Holistically-Nested Edge Detection (HED), which is a novel edge detection method based on fully
CHAPTER. Quantification in cardiac MRI. This chapter was adapted from:
 CHAPTER Quantification in cardiac MRI This chapter was adapted from: Quantification in cardiac MRI Rob J. van der Geest, Johan H.C. Reiber Journal of Magnetic Resonance Imaging 1999, Volume 10, Pages 602-608.
CHAPTER Quantification in cardiac MRI This chapter was adapted from: Quantification in cardiac MRI Rob J. van der Geest, Johan H.C. Reiber Journal of Magnetic Resonance Imaging 1999, Volume 10, Pages 602-608.
arxiv: v1 [cs.cv] 9 Sep 2017
![arxiv: v1 [cs.cv] 9 Sep 2017 arxiv: v1 [cs.cv] 9 Sep 2017](/thumbs/79/79077141.jpg) Sequential 3D U-Nets for Biologically-Informed Brain Tumor Segmentation Andrew Beers 1, Ken Chang 1, James Brown 1, Emmett Sartor 2, CP Mammen 3, Elizabeth Gerstner 1,4, Bruce Rosen 1, and Jayashree Kalpathy-Cramer
Sequential 3D U-Nets for Biologically-Informed Brain Tumor Segmentation Andrew Beers 1, Ken Chang 1, James Brown 1, Emmett Sartor 2, CP Mammen 3, Elizabeth Gerstner 1,4, Bruce Rosen 1, and Jayashree Kalpathy-Cramer
Research Article Multiscale CNNs for Brain Tumor Segmentation and Diagnosis
 Computational and Mathematical Methods in Medicine Volume 2016, Article ID 8356294, 7 pages http://dx.doi.org/10.1155/2016/8356294 Research Article Multiscale CNNs for Brain Tumor Segmentation and Diagnosis
Computational and Mathematical Methods in Medicine Volume 2016, Article ID 8356294, 7 pages http://dx.doi.org/10.1155/2016/8356294 Research Article Multiscale CNNs for Brain Tumor Segmentation and Diagnosis
Sign Language Recognition using Convolutional Neural Networks
 Sign Language Recognition using Convolutional Neural Networks Lionel Pigou, Sander Dieleman, Pieter-Jan Kindermans, Benjamin Schrauwen Ghent University, ELIS, Belgium Abstract. There is an undeniable communication
Sign Language Recognition using Convolutional Neural Networks Lionel Pigou, Sander Dieleman, Pieter-Jan Kindermans, Benjamin Schrauwen Ghent University, ELIS, Belgium Abstract. There is an undeniable communication
A CONVOLUTION NEURAL NETWORK ALGORITHM FOR BRAIN TUMOR IMAGE SEGMENTATION
 A CONVOLUTION NEURAL NETWORK ALGORITHM FOR BRAIN TUMOR IMAGE SEGMENTATION 1 Priya K, 2 Dr.O. Saraniya 1 PG Scholar, 2 Assistant Professor Department Of ECE Government College of Technology, Coimbatore,
A CONVOLUTION NEURAL NETWORK ALGORITHM FOR BRAIN TUMOR IMAGE SEGMENTATION 1 Priya K, 2 Dr.O. Saraniya 1 PG Scholar, 2 Assistant Professor Department Of ECE Government College of Technology, Coimbatore,
Automated Image Biometrics Speeds Ultrasound Workflow
 Whitepaper Automated Image Biometrics Speeds Ultrasound Workflow ACUSON SC2000 Volume Imaging Ultrasound System S. Kevin Zhou, Ph.D. Siemens Corporate Research Princeton, New Jersey USA Answers for life.
Whitepaper Automated Image Biometrics Speeds Ultrasound Workflow ACUSON SC2000 Volume Imaging Ultrasound System S. Kevin Zhou, Ph.D. Siemens Corporate Research Princeton, New Jersey USA Answers for life.
Myocardial Strain Imaging in Cardiac Diseases and Cardiomyopathies.
 Myocardial Strain Imaging in Cardiac Diseases and Cardiomyopathies. Session: Cardiomyopathy Tarun Pandey MD, FRCR. Associate Professor University of Arkansas for Medical Sciences Disclosures No relevant
Myocardial Strain Imaging in Cardiac Diseases and Cardiomyopathies. Session: Cardiomyopathy Tarun Pandey MD, FRCR. Associate Professor University of Arkansas for Medical Sciences Disclosures No relevant
Lung Nodule Segmentation Using 3D Convolutional Neural Networks
 Lung Nodule Segmentation Using 3D Convolutional Neural Networks Research paper Business Analytics Bernard Bronmans Master Business Analytics VU University, Amsterdam Evert Haasdijk Supervisor VU University,
Lung Nodule Segmentation Using 3D Convolutional Neural Networks Research paper Business Analytics Bernard Bronmans Master Business Analytics VU University, Amsterdam Evert Haasdijk Supervisor VU University,
arxiv: v2 [cs.cv] 3 Jun 2018
![arxiv: v2 [cs.cv] 3 Jun 2018 arxiv: v2 [cs.cv] 3 Jun 2018](/thumbs/89/97775247.jpg) S4ND: Single-Shot Single-Scale Lung Nodule Detection Naji Khosravan and Ulas Bagci Center for Research in Computer Vision (CRCV), School of Computer Science, University of Central Florida, Orlando, FL.
S4ND: Single-Shot Single-Scale Lung Nodule Detection Naji Khosravan and Ulas Bagci Center for Research in Computer Vision (CRCV), School of Computer Science, University of Central Florida, Orlando, FL.
Myocardial Delineation via Registration in a Polar Coordinate System
 Myocardial Delineation via Registration in a Polar Coordinate System Nicholas M.I. Noble, Derek L.G. Hill, Marcel Breeuwer 2, JuliaA. Schnabel, David J. Hawkes, FransA. Gerritsen 2, and Reza Razavi Computer
Myocardial Delineation via Registration in a Polar Coordinate System Nicholas M.I. Noble, Derek L.G. Hill, Marcel Breeuwer 2, JuliaA. Schnabel, David J. Hawkes, FransA. Gerritsen 2, and Reza Razavi Computer
Hierarchical Convolutional Features for Visual Tracking
 Hierarchical Convolutional Features for Visual Tracking Chao Ma Jia-Bin Huang Xiaokang Yang Ming-Husan Yang SJTU UIUC SJTU UC Merced ICCV 2015 Background Given the initial state (position and scale), estimate
Hierarchical Convolutional Features for Visual Tracking Chao Ma Jia-Bin Huang Xiaokang Yang Ming-Husan Yang SJTU UIUC SJTU UC Merced ICCV 2015 Background Given the initial state (position and scale), estimate
CS-E Deep Learning Session 4: Convolutional Networks
 CS-E4050 - Deep Learning Session 4: Convolutional Networks Jyri Kivinen Aalto University 23 September 2015 Credits: Thanks to Tapani Raiko for slides material. CS-E4050 - Deep Learning Session 4: Convolutional
CS-E4050 - Deep Learning Session 4: Convolutional Networks Jyri Kivinen Aalto University 23 September 2015 Credits: Thanks to Tapani Raiko for slides material. CS-E4050 - Deep Learning Session 4: Convolutional
Cronfa - Swansea University Open Access Repository
 Cronfa - Swansea University Open Access Repository This is an author produced version of a paper published in: Medical Image Understanding and Analysis Cronfa URL for this paper: http://cronfa.swan.ac.uk/record/cronfa40803
Cronfa - Swansea University Open Access Repository This is an author produced version of a paper published in: Medical Image Understanding and Analysis Cronfa URL for this paper: http://cronfa.swan.ac.uk/record/cronfa40803
LV FUNCTION ASSESSMENT: WHAT IS BEYOND EJECTION FRACTION
 LV FUNCTION ASSESSMENT: WHAT IS BEYOND EJECTION FRACTION Jamilah S AlRahimi Assistant Professor, KSU-HS Consultant Noninvasive Cardiology KFCC, MNGHA-WR Introduction LV function assessment in Heart Failure:
LV FUNCTION ASSESSMENT: WHAT IS BEYOND EJECTION FRACTION Jamilah S AlRahimi Assistant Professor, KSU-HS Consultant Noninvasive Cardiology KFCC, MNGHA-WR Introduction LV function assessment in Heart Failure:
Image-Based Estimation of Real Food Size for Accurate Food Calorie Estimation
 Image-Based Estimation of Real Food Size for Accurate Food Calorie Estimation Takumi Ege, Yoshikazu Ando, Ryosuke Tanno, Wataru Shimoda and Keiji Yanai Department of Informatics, The University of Electro-Communications,
Image-Based Estimation of Real Food Size for Accurate Food Calorie Estimation Takumi Ege, Yoshikazu Ando, Ryosuke Tanno, Wataru Shimoda and Keiji Yanai Department of Informatics, The University of Electro-Communications,
Deep Learning of Brain Lesion Patterns for Predicting Future Disease Activity in Patients with Early Symptoms of Multiple Sclerosis
 Deep Learning of Brain Lesion Patterns for Predicting Future Disease Activity in Patients with Early Symptoms of Multiple Sclerosis Youngjin Yoo 1,2,5(B),LisaW.Tang 2,3,5, Tom Brosch 1,2,5,DavidK.B.Li
Deep Learning of Brain Lesion Patterns for Predicting Future Disease Activity in Patients with Early Symptoms of Multiple Sclerosis Youngjin Yoo 1,2,5(B),LisaW.Tang 2,3,5, Tom Brosch 1,2,5,DavidK.B.Li
Automated diagnosis of pneumothorax using an ensemble of convolutional neural networks with multi-sized chest radiography images
 Automated diagnosis of pneumothorax using an ensemble of convolutional neural networks with multi-sized chest radiography images Tae Joon Jun, Dohyeun Kim, and Daeyoung Kim School of Computing, KAIST,
Automated diagnosis of pneumothorax using an ensemble of convolutional neural networks with multi-sized chest radiography images Tae Joon Jun, Dohyeun Kim, and Daeyoung Kim School of Computing, KAIST,
Automatic Layer Generation for Scar Transmurality Visualization
 Automatic Layer Generation for Scar Transmurality Visualization S. Reiml 1,4, T. Kurzendorfer 1,4, D. Toth 2,5, P. Mountney 6, M. Panayiotou 2, J. M. Behar 2,3, C. A. Rinaldi 2,3, K. Rhode 2, A. Maier
Automatic Layer Generation for Scar Transmurality Visualization S. Reiml 1,4, T. Kurzendorfer 1,4, D. Toth 2,5, P. Mountney 6, M. Panayiotou 2, J. M. Behar 2,3, C. A. Rinaldi 2,3, K. Rhode 2, A. Maier
arxiv: v2 [cs.cv] 22 Mar 2018
![arxiv: v2 [cs.cv] 22 Mar 2018 arxiv: v2 [cs.cv] 22 Mar 2018](/thumbs/94/122062874.jpg) Deep saliency: What is learnt by a deep network about saliency? Sen He 1 Nicolas Pugeault 1 arxiv:1801.04261v2 [cs.cv] 22 Mar 2018 Abstract Deep convolutional neural networks have achieved impressive performance
Deep saliency: What is learnt by a deep network about saliency? Sen He 1 Nicolas Pugeault 1 arxiv:1801.04261v2 [cs.cv] 22 Mar 2018 Abstract Deep convolutional neural networks have achieved impressive performance
Fast left ventricle tracking using localized anatomical affine optical flow SUMMARY
 Fast left ventricle tracking using localized anatomical affine optical flow Sandro Queirós 1,2,3, João L. Vilaça 1,4, Pedro Morais 1,2,5, Jaime C. Fonseca 3, Jan D hooge 2, Daniel Barbosa 1 1 ICVS/3B s
Fast left ventricle tracking using localized anatomical affine optical flow Sandro Queirós 1,2,3, João L. Vilaça 1,4, Pedro Morais 1,2,5, Jaime C. Fonseca 3, Jan D hooge 2, Daniel Barbosa 1 1 ICVS/3B s
Reduction of Overfitting in Diabetes Prediction Using Deep Learning Neural Network
 Reduction of Overfitting in Diabetes Prediction Using Deep Learning Neural Network Akm Ashiquzzaman *, Abdul Kawsar Tushar *, Md. Rashedul Islam *, 1, and Jong-Myon Kim **, 2 * Department of CSE, University
Reduction of Overfitting in Diabetes Prediction Using Deep Learning Neural Network Akm Ashiquzzaman *, Abdul Kawsar Tushar *, Md. Rashedul Islam *, 1, and Jong-Myon Kim **, 2 * Department of CSE, University
Magnetic Resonance Contrast Prediction Using Deep Learning
 Magnetic Resonance Contrast Prediction Using Deep Learning Cagan Alkan Department of Electrical Engineering Stanford University calkan@stanford.edu Andrew Weitz Department of Bioengineering Stanford University
Magnetic Resonance Contrast Prediction Using Deep Learning Cagan Alkan Department of Electrical Engineering Stanford University calkan@stanford.edu Andrew Weitz Department of Bioengineering Stanford University
Detection of Cardiac Events in Echocardiography using 3D Convolutional Recurrent Neural Networks
 Detection of Cardiac Events in Echocardiography using 3D Convolutional Recurrent Neural Networks Adrian Meidell Fiorito Dept. of Engineering Cybernetics adrianf@stud.ntnu.no Andreas Østvik andreas.ostvik@ntnu.no
Detection of Cardiac Events in Echocardiography using 3D Convolutional Recurrent Neural Networks Adrian Meidell Fiorito Dept. of Engineering Cybernetics adrianf@stud.ntnu.no Andreas Østvik andreas.ostvik@ntnu.no
arxiv: v1 [cs.cv] 30 Dec 2018
![arxiv: v1 [cs.cv] 30 Dec 2018 arxiv: v1 [cs.cv] 30 Dec 2018](/thumbs/92/111028397.jpg) Cascaded V-Net using ROI masks for brain tumor segmentation Adrià Casamitjana, Marcel Catà, Irina Sánchez, Marc Combalia and Verónica Vilaplana Signal Theory and Communications Department, Universitat
Cascaded V-Net using ROI masks for brain tumor segmentation Adrià Casamitjana, Marcel Catà, Irina Sánchez, Marc Combalia and Verónica Vilaplana Signal Theory and Communications Department, Universitat
A HMM-based Pre-training Approach for Sequential Data
 A HMM-based Pre-training Approach for Sequential Data Luca Pasa 1, Alberto Testolin 2, Alessandro Sperduti 1 1- Department of Mathematics 2- Department of Developmental Psychology and Socialisation University
A HMM-based Pre-training Approach for Sequential Data Luca Pasa 1, Alberto Testolin 2, Alessandro Sperduti 1 1- Department of Mathematics 2- Department of Developmental Psychology and Socialisation University
Tissue Doppler Imaging in Congenital Heart Disease
 Tissue Doppler Imaging in Congenital Heart Disease L. Youngmin Eun, M.D. Department of Pediatrics, Division of Pediatric Cardiology, Kwandong University College of Medicine The potential advantage of ultrasound
Tissue Doppler Imaging in Congenital Heart Disease L. Youngmin Eun, M.D. Department of Pediatrics, Division of Pediatric Cardiology, Kwandong University College of Medicine The potential advantage of ultrasound
Advanced Multi-Layer Speckle Strain Permits Transmural Myocardial Function Analysis in Health and Disease:
 Advanced Multi-Layer Speckle Strain Permits Transmural Myocardial Function Analysis in Health and Disease: Clinical Case Examples Jeffrey C. Hill, BS, RDCS Echocardiography Laboratory, University of Massachusetts
Advanced Multi-Layer Speckle Strain Permits Transmural Myocardial Function Analysis in Health and Disease: Clinical Case Examples Jeffrey C. Hill, BS, RDCS Echocardiography Laboratory, University of Massachusetts
arxiv: v1 [cs.cv] 2 Aug 2017
![arxiv: v1 [cs.cv] 2 Aug 2017 arxiv: v1 [cs.cv] 2 Aug 2017](/thumbs/76/73977344.jpg) Automatic 3D ardiovascular MR Segmentation with Densely-onnected Volumetric onvnets Lequan Yu 1, Jie-Zhi heng 2, Qi Dou 1, Xin Yang 1, Hao hen 1, Jing Qin 3, and Pheng-Ann Heng 1,4 arxiv:1708.00573v1 [cs.v]
Automatic 3D ardiovascular MR Segmentation with Densely-onnected Volumetric onvnets Lequan Yu 1, Jie-Zhi heng 2, Qi Dou 1, Xin Yang 1, Hao hen 1, Jing Qin 3, and Pheng-Ann Heng 1,4 arxiv:1708.00573v1 [cs.v]
Supplementary Online Content
 Supplementary Online Content Ting DS, Cheung CY-L, Lim G, et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic
Supplementary Online Content Ting DS, Cheung CY-L, Lim G, et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic
arxiv: v1 [cs.cv] 21 Jul 2017
![arxiv: v1 [cs.cv] 21 Jul 2017 arxiv: v1 [cs.cv] 21 Jul 2017](/thumbs/89/100163556.jpg) A Multi-Scale CNN and Curriculum Learning Strategy for Mammogram Classification William Lotter 1,2, Greg Sorensen 2, and David Cox 1,2 1 Harvard University, Cambridge MA, USA 2 DeepHealth Inc., Cambridge
A Multi-Scale CNN and Curriculum Learning Strategy for Mammogram Classification William Lotter 1,2, Greg Sorensen 2, and David Cox 1,2 1 Harvard University, Cambridge MA, USA 2 DeepHealth Inc., Cambridge
Normal LV Ejection Fraction Limits Using 4D-MSPECT: Comparisons of Gated Perfusion and Gated Blood Pool SPECT Data with Planar Blood Pool
 Normal LV Ejection Fraction Limits Using 4D-MSPECT: Comparisons of Gated Perfusion and Gated Blood Pool SPECT Data with Planar Blood Pool EP Ficaro, JN Kritzman, JR Corbett University of Michigan Health
Normal LV Ejection Fraction Limits Using 4D-MSPECT: Comparisons of Gated Perfusion and Gated Blood Pool SPECT Data with Planar Blood Pool EP Ficaro, JN Kritzman, JR Corbett University of Michigan Health
ANALYSIS AND DETECTION OF BRAIN TUMOUR USING IMAGE PROCESSING TECHNIQUES
 ANALYSIS AND DETECTION OF BRAIN TUMOUR USING IMAGE PROCESSING TECHNIQUES P.V.Rohini 1, Dr.M.Pushparani 2 1 M.Phil Scholar, Department of Computer Science, Mother Teresa women s university, (India) 2 Professor
ANALYSIS AND DETECTION OF BRAIN TUMOUR USING IMAGE PROCESSING TECHNIQUES P.V.Rohini 1, Dr.M.Pushparani 2 1 M.Phil Scholar, Department of Computer Science, Mother Teresa women s university, (India) 2 Professor
EARLY STAGE DIAGNOSIS OF LUNG CANCER USING CT-SCAN IMAGES BASED ON CELLULAR LEARNING AUTOMATE
 EARLY STAGE DIAGNOSIS OF LUNG CANCER USING CT-SCAN IMAGES BASED ON CELLULAR LEARNING AUTOMATE SAKTHI NEELA.P.K Department of M.E (Medical electronics) Sengunthar College of engineering Namakkal, Tamilnadu,
EARLY STAGE DIAGNOSIS OF LUNG CANCER USING CT-SCAN IMAGES BASED ON CELLULAR LEARNING AUTOMATE SAKTHI NEELA.P.K Department of M.E (Medical electronics) Sengunthar College of engineering Namakkal, Tamilnadu,
International Journal of Engineering Trends and Applications (IJETA) Volume 4 Issue 2, Mar-Apr 2017
 RESEARCH ARTICLE OPEN ACCESS Knowledge Based Brain Tumor Segmentation using Local Maxima and Local Minima T. Kalaiselvi [1], P. Sriramakrishnan [2] Department of Computer Science and Applications The Gandhigram
RESEARCH ARTICLE OPEN ACCESS Knowledge Based Brain Tumor Segmentation using Local Maxima and Local Minima T. Kalaiselvi [1], P. Sriramakrishnan [2] Department of Computer Science and Applications The Gandhigram
arxiv: v1 [cs.cv] 2 Jul 2018
![arxiv: v1 [cs.cv] 2 Jul 2018 arxiv: v1 [cs.cv] 2 Jul 2018](/thumbs/87/96052165.jpg) A Pulmonary Nodule Detection Model Based on Progressive Resolution and Hierarchical Saliency Junjie Zhang 1, Yong Xia 1,2, and Yanning Zhang 1 1 Shaanxi Key Lab of Speech & Image Information Processing
A Pulmonary Nodule Detection Model Based on Progressive Resolution and Hierarchical Saliency Junjie Zhang 1, Yong Xia 1,2, and Yanning Zhang 1 1 Shaanxi Key Lab of Speech & Image Information Processing
arxiv: v1 [cs.cv] 28 Feb 2018
![arxiv: v1 [cs.cv] 28 Feb 2018 arxiv: v1 [cs.cv] 28 Feb 2018](/thumbs/93/111675272.jpg) Brain Tumor Segmentation and Radiomics Survival Prediction: Contribution to the BRATS 2017 Challenge arxiv:1802.10508v1 [cs.cv] 28 Feb 2018 Fabian Isensee 1, Philipp Kickingereder 2, Wolfgang Wick 3, Martin
Brain Tumor Segmentation and Radiomics Survival Prediction: Contribution to the BRATS 2017 Challenge arxiv:1802.10508v1 [cs.cv] 28 Feb 2018 Fabian Isensee 1, Philipp Kickingereder 2, Wolfgang Wick 3, Martin
Echocardiographic Assessment of the Left Ventricle
 Echocardiographic Assessment of the Left Ventricle Theodora Zaglavara, MD, PhD, BSCI/BSCCT Department of Cardiovascular Imaging INTERBALKAN EUROPEAN MEDICAL CENTER 2015 The quantification of cardiac chamber
Echocardiographic Assessment of the Left Ventricle Theodora Zaglavara, MD, PhD, BSCI/BSCCT Department of Cardiovascular Imaging INTERBALKAN EUROPEAN MEDICAL CENTER 2015 The quantification of cardiac chamber
Combining Deep Learning and Level Set for the Automated Segmentation of the Left Ventricle of the Heart from Cardiac Cine Magnetic Resonance
 Combining Deep Learning and Level Set for the Automated Segmentation of the Left Ventricle of the Heart from Cardiac Cine Magnetic Resonance Tuan Anh Ngo,1, Zhi Lu,1 and Gustavo Carneiro,2 Vietnam National
Combining Deep Learning and Level Set for the Automated Segmentation of the Left Ventricle of the Heart from Cardiac Cine Magnetic Resonance Tuan Anh Ngo,1, Zhi Lu,1 and Gustavo Carneiro,2 Vietnam National
Comparison of Two Approaches for Direct Food Calorie Estimation
 Comparison of Two Approaches for Direct Food Calorie Estimation Takumi Ege and Keiji Yanai Department of Informatics, The University of Electro-Communications, Tokyo 1-5-1 Chofugaoka, Chofu-shi, Tokyo
Comparison of Two Approaches for Direct Food Calorie Estimation Takumi Ege and Keiji Yanai Department of Informatics, The University of Electro-Communications, Tokyo 1-5-1 Chofugaoka, Chofu-shi, Tokyo
Enhanced Endocardial Boundary Detection in Echocardiography Images using B-Spline and Statistical Method
 Copyright 2014 American Scientific Publishers Advanced Science Letters All rights reserved Vol. 20, 1876 1880, 2014 Printed in the United States of America Enhanced Endocardial Boundary Detection in Echocardiography
Copyright 2014 American Scientific Publishers Advanced Science Letters All rights reserved Vol. 20, 1876 1880, 2014 Printed in the United States of America Enhanced Endocardial Boundary Detection in Echocardiography
10/7/2013. Systolic Function How to Measure, How Accurate is Echo, Role of Contrast. Thanks to our Course Director: Neil J.
 Systolic Function How to Measure, How Accurate is Echo, Role of Contrast Neil J. Weissman, MD MedStar Health Research Institute & Professor of Medicine Georgetown University Washington, D.C. No Disclosures
Systolic Function How to Measure, How Accurate is Echo, Role of Contrast Neil J. Weissman, MD MedStar Health Research Institute & Professor of Medicine Georgetown University Washington, D.C. No Disclosures
Deep CNNs for Diabetic Retinopathy Detection
 Deep CNNs for Diabetic Retinopathy Detection Alex Tamkin Stanford University atamkin@stanford.edu Iain Usiri Stanford University iusiri@stanford.edu Chala Fufa Stanford University cfufa@stanford.edu 1
Deep CNNs for Diabetic Retinopathy Detection Alex Tamkin Stanford University atamkin@stanford.edu Iain Usiri Stanford University iusiri@stanford.edu Chala Fufa Stanford University cfufa@stanford.edu 1
Automated quantification of myocardial infarction using graph cuts on contrast delayed enhanced magnetic resonance images
 Original Article Automated quantification of myocardial infarction using graph cuts on contrast delayed enhanced magnetic resonance images Yingli Lu 1, Yuesong Yang 1, Kim A. Connelly 2,3, Graham A. Wright
Original Article Automated quantification of myocardial infarction using graph cuts on contrast delayed enhanced magnetic resonance images Yingli Lu 1, Yuesong Yang 1, Kim A. Connelly 2,3, Graham A. Wright
Three-dimensional Wall Motion Tracking:
 Three-dimensional Wall Motion Tracking: A Novel Echocardiographic Method for the Assessment of Ventricular Volumes, Strain and Dyssynchrony Jeffrey C. Hill, BS, RDCS, FASE Jennifer L. Kane, RCS Gerard
Three-dimensional Wall Motion Tracking: A Novel Echocardiographic Method for the Assessment of Ventricular Volumes, Strain and Dyssynchrony Jeffrey C. Hill, BS, RDCS, FASE Jennifer L. Kane, RCS Gerard
Deep Supervision for Pancreatic Cyst Segmentation in Abdominal CT Scans
 Deep Supervision for Pancreatic Cyst Segmentation in Abdominal CT Scans Yuyin Zhou 1, Lingxi Xie 2( ), Elliot K. Fishman 3, Alan L. Yuille 4 1,2,4 The Johns Hopkins University, Baltimore, MD 21218, USA
Deep Supervision for Pancreatic Cyst Segmentation in Abdominal CT Scans Yuyin Zhou 1, Lingxi Xie 2( ), Elliot K. Fishman 3, Alan L. Yuille 4 1,2,4 The Johns Hopkins University, Baltimore, MD 21218, USA
Flexible, High Performance Convolutional Neural Networks for Image Classification
 Flexible, High Performance Convolutional Neural Networks for Image Classification Dan C. Cireşan, Ueli Meier, Jonathan Masci, Luca M. Gambardella, Jürgen Schmidhuber IDSIA, USI and SUPSI Manno-Lugano,
Flexible, High Performance Convolutional Neural Networks for Image Classification Dan C. Cireşan, Ueli Meier, Jonathan Masci, Luca M. Gambardella, Jürgen Schmidhuber IDSIA, USI and SUPSI Manno-Lugano,
Discovery of Rare Phenotypes in Cellular Images Using Weakly Supervised Deep Learning
 Discovery of Rare Phenotypes in Cellular Images Using Weakly Supervised Deep Learning Heba Sailem *1, Mar Arias Garcia 2, Chris Bakal 2, Andrew Zisserman 1, and Jens Rittscher 1 1 Department of Engineering
Discovery of Rare Phenotypes in Cellular Images Using Weakly Supervised Deep Learning Heba Sailem *1, Mar Arias Garcia 2, Chris Bakal 2, Andrew Zisserman 1, and Jens Rittscher 1 1 Department of Engineering
Right Ventricular Strain in Normal Healthy Adult Filipinos: A Retrospective, Cross- Sectional Pilot Study
 Right Ventricular Strain in Normal Healthy Adult Filipinos: A Retrospective, Cross- Sectional Pilot Study By Julius Caesar D. de Vera, MD Jonnah Fatima B. Pelat, MD Introduction Right ventricle contributes
Right Ventricular Strain in Normal Healthy Adult Filipinos: A Retrospective, Cross- Sectional Pilot Study By Julius Caesar D. de Vera, MD Jonnah Fatima B. Pelat, MD Introduction Right ventricle contributes
arxiv: v1 [cs.cv] 30 May 2018
![arxiv: v1 [cs.cv] 30 May 2018 arxiv: v1 [cs.cv] 30 May 2018](/thumbs/85/92577534.jpg) A Robust and Effective Approach Towards Accurate Metastasis Detection and pn-stage Classification in Breast Cancer Byungjae Lee and Kyunghyun Paeng Lunit inc., Seoul, South Korea {jaylee,khpaeng}@lunit.io
A Robust and Effective Approach Towards Accurate Metastasis Detection and pn-stage Classification in Breast Cancer Byungjae Lee and Kyunghyun Paeng Lunit inc., Seoul, South Korea {jaylee,khpaeng}@lunit.io
Image Captioning using Reinforcement Learning. Presentation by: Samarth Gupta
 Image Captioning using Reinforcement Learning Presentation by: Samarth Gupta 1 Introduction Summary Supervised Models Image captioning as RL problem Actor Critic Architecture Policy Gradient architecture
Image Captioning using Reinforcement Learning Presentation by: Samarth Gupta 1 Introduction Summary Supervised Models Image captioning as RL problem Actor Critic Architecture Policy Gradient architecture
arxiv: v3 [cs.cv] 26 May 2018
![arxiv: v3 [cs.cv] 26 May 2018 arxiv: v3 [cs.cv] 26 May 2018](/thumbs/86/94630916.jpg) DeepEM: Deep 3D ConvNets With EM For Weakly Supervised Pulmonary Nodule Detection Wentao Zhu, Yeeleng S. Vang, Yufang Huang, and Xiaohui Xie University of California, Irvine Lenovo AI Lab {wentaoz1,ysvang,xhx}@uci.edu,
DeepEM: Deep 3D ConvNets With EM For Weakly Supervised Pulmonary Nodule Detection Wentao Zhu, Yeeleng S. Vang, Yufang Huang, and Xiaohui Xie University of California, Irvine Lenovo AI Lab {wentaoz1,ysvang,xhx}@uci.edu,
Automatic Prostate Cancer Classification using Deep Learning. Ida Arvidsson Centre for Mathematical Sciences, Lund University, Sweden
 Automatic Prostate Cancer Classification using Deep Learning Ida Arvidsson Centre for Mathematical Sciences, Lund University, Sweden Outline Autoencoders, theory Motivation, background and goal for prostate
Automatic Prostate Cancer Classification using Deep Learning Ida Arvidsson Centre for Mathematical Sciences, Lund University, Sweden Outline Autoencoders, theory Motivation, background and goal for prostate
Deep Learning Quantitative Analysis of Cardiac Function
 Deep Learning Quantitative Analysis of Cardiac Function Validation and Implementation in Clinical Practice A L B E R T H S I A O, M D, P H D A S S I S TA N T P R O F E S S O R O F R A D I O LO G Y, I N
Deep Learning Quantitative Analysis of Cardiac Function Validation and Implementation in Clinical Practice A L B E R T H S I A O, M D, P H D A S S I S TA N T P R O F E S S O R O F R A D I O LO G Y, I N
Early Diagnosis of Autism Disease by Multi-channel CNNs
 Early Diagnosis of Autism Disease by Multi-channel CNNs Guannan Li 1,2, Mingxia Liu 2, Quansen Sun 1(&), Dinggang Shen 2(&), and Li Wang 2(&) 1 School of Computer Science and Engineering, Nanjing University
Early Diagnosis of Autism Disease by Multi-channel CNNs Guannan Li 1,2, Mingxia Liu 2, Quansen Sun 1(&), Dinggang Shen 2(&), and Li Wang 2(&) 1 School of Computer Science and Engineering, Nanjing University
End-To-End Alzheimer s Disease Diagnosis and Biomarker Identification
 End-To-End Alzheimer s Disease Diagnosis and Biomarker Identification Soheil Esmaeilzadeh 1, Dimitrios Ioannis Belivanis 1, Kilian M. Pohl 2, and Ehsan Adeli 1 1 Stanford University 2 SRI International
End-To-End Alzheimer s Disease Diagnosis and Biomarker Identification Soheil Esmaeilzadeh 1, Dimitrios Ioannis Belivanis 1, Kilian M. Pohl 2, and Ehsan Adeli 1 1 Stanford University 2 SRI International
Cardiac MRI in Small Rodents
 Cardiac MRI in Small Rodents Andreas Pohlmann, PhD Berlin Ultrahigh Field Facility, Max Delbrück Center for Molecular Medicine (MDC), Berlin, Germany Introduction The art of producing animal models has
Cardiac MRI in Small Rodents Andreas Pohlmann, PhD Berlin Ultrahigh Field Facility, Max Delbrück Center for Molecular Medicine (MDC), Berlin, Germany Introduction The art of producing animal models has
Photon Attenuation Correction in Misregistered Cardiac PET/CT
 Photon Attenuation Correction in Misregistered Cardiac PET/CT A. Martinez-Möller 1,2, N. Navab 2, M. Schwaiger 1, S. G. Nekolla 1 1 Nuklearmedizinische Klinik der TU München 2 Computer Assisted Medical
Photon Attenuation Correction in Misregistered Cardiac PET/CT A. Martinez-Möller 1,2, N. Navab 2, M. Schwaiger 1, S. G. Nekolla 1 1 Nuklearmedizinische Klinik der TU München 2 Computer Assisted Medical
A Framework Combining Multi-sequence MRI for Fully Automated Quantitative Analysis of Cardiac Global And Regional Functions
 A Framework Combining Multi-sequence MRI for Fully Automated Quantitative Analysis of Cardiac Global And Regional Functions Xiahai Zhuang 1,, Wenzhe Shi 2,SimonDuckett 3, Haiyan Wang 2, Reza Razavi 3,DavidHawkes
A Framework Combining Multi-sequence MRI for Fully Automated Quantitative Analysis of Cardiac Global And Regional Functions Xiahai Zhuang 1,, Wenzhe Shi 2,SimonDuckett 3, Haiyan Wang 2, Reza Razavi 3,DavidHawkes
Automatic functional analysis of left ventricle in cardiac cine MRI
 Original Article Automatic functional analysis of left ventricle in cardiac cine MRI Ying-Li Lu 1, Kim A. Connelly 2,3, Alexander J. Dick 4, Graham A. Wright 1,5, Perry E. Radau 1 1 Imaging Research, Sunnybrook
Original Article Automatic functional analysis of left ventricle in cardiac cine MRI Ying-Li Lu 1, Kim A. Connelly 2,3, Alexander J. Dick 4, Graham A. Wright 1,5, Perry E. Radau 1 1 Imaging Research, Sunnybrook
