Table S2. Baseline characteristics of the Rotterdam Study for interaction analysis
|
|
|
- Bruce Marshall
- 5 years ago
- Views:
Transcription
1 Supplementary data Table S1. List of the primers for the cloning of precursor mir-4707 Table S2. Baseline characteristics of the Rotterdam Study for interaction analysis Table S3. List of the primers for the cloning of 3 UTR sequence of CARD10 Table S4. List of databases and web tools used for functional annotation of mirna-binding site SNPs associated with POAG endophenotypes Table S5. MiRNA genetic variants nominally associated with POAG endophenotypes Table S6. mir-612 and mir p putative target genes associated with and cup area Table S7. Expression of the putative mir-612 and mir p target genes associated with and cup area across different eye tissues Table S8. The mirna-target gene interaction analysis using the Rotterdam Study data Table S9. Forty-seven mirna-binding site variants within the 3 UTRs of 21 genes associated with POAG endophenotypes Table S10. Functional characteristics of 47 mirna-binding site variants associated with POAG endophenotypes Table 11. Expression of genes hosting the 47 mirna-binding site variants associated with POAG endophenotypes across eye tissues Table S12. Prioritization of 47 mirna-binding site variants associated with POAG endophenotypes Figure S1. The predicted hairpin structure of pre-mir-612 and pre-mir-4707 and the position of two identified variants associated with and cup area Figure S2. Manhattan plots showing the association of 72,052 mirna-binding site variants with POAG endophenotypes Figure S3. Regional plots showing the association of ten highlighted mirna-binding site variants with POAG endophenotypes
2 Table S1. List of the primers for the cloning of precursor mir-4707 Oligo name Sequence (5'-3') mir-4707_wt_f mir-4707_wt_r mir-4707_mut_f mir-4707_mut_r 5 TCGAGGGTTCCGGAGCCCCGGCGCGGGCGGGTTCTGGGGTGTAGACGCTGC TGGCCAGCCCGCCCCAGCCGAGGTTCTCGGCACCG 3 5 AATTCGGTGCCGAGAACCTCGGCTGGGGCGGGCTGGCCAGCAGCGTCTACA CCCCAGAACCCGCCCGCGCCGGGGCTCCGGAACCC 3 5 TCGAGGGTTCCGGAGCCCCGGCGCGGGCGGGTTCTGGGGTGTAGACGCTGC TGGCCAGCCCUCCCCAGCCGAGGTTCTCGGCACCG 3 5 AATTCGGTGCCGAGAACCTCGGCTGGGTAGGGCTGGCCAGCAGCGTCTACA CCCCAGAACCCGCCCGCGCCGGGGCTCCGGAACCC 3 WT, Wild type pre-mirna sequence containing allele G at the rs SNP site; Mut, Mutant premirna containing allele A at the rs SNP site; F, Forward; R, Reverse. The restriction enzymes XhoI and EcoRI (with sticky end) are shown in red and the position of SNP is underlined in the pre-mirna sequence.
3 Table S2. Baseline characteristics of the Rotterdam Study for interaction analysis Total Number of participants 10,404 Age (years) 64.4 (9.1) Sex (male) 4441 (42.7 %) Body mass index (kg/m 2 ) (9.14) Systolic blood pressure (mmhg) (25.66) Diastolic blood pressure (mmhg) (18.46) Data are mean (SD) or n (%)
4 Table S3. List of the primers for the cloning of 3 UTR of CARD10 gene Oligo name Sequence (5'-3') CARD10_F CARD10 _R 5 CTAGATGTAAATGTCACGCCCCACCCCTGTTTCATGTGGGCACTAACAC GTGTGCGTTCCTGGCGGGCACACTCAGGACGGGCC 3 5 CGTCCTGAGTGTGCCCGCCAGGAACGCACACGTGTTAGTGCCCACATGA AACAGGGGTGGGGCGTGACATTTACAT 3 F, Forward; R, Reverse. The restriction enzymes XbaI and ApaI (sticky end) are shown in red and the binding site of mir p within the 3 UTR of CARD10 is underlined.
5 Table S4. List of databases and web tools used for functional annotation of mirna-binding site SNPs associated with POAG endophenotypes Name Main feature URL mirnasnp v2 SNPs in mirna-related sequences PolymiRTS 3.0 SNPs in mirna binding sites Vienna RNA Prediction of SNP effect on mirna secondary structure TargetScan 7.0 mirna target prediction mirdb mirna target prediction / IPA A comprehensive software on biological and pathway analysis OTDB Gene expression data across different human eye tissues from the Ocular Tissue Database HMDD The human mirna expression database SNAP Proxy SNPs based on linkage disequilibrium and physical distance HaploReg v4.1 For exploring annotations of variants on haplotype blocks Genenetwork Whole blood eqtl data GTEx eqtl data in other tissue mirbase mirna information GeneCards LocusZoom A database of human genes Generating regional association plots
6 Table S5. MiRNA genetic variants nominally associated with POAG endophenotypes mirna ID Location SNP ID A1 A2 Freq A1 Chr Effect P-value Pheno in mir mir-1178 seed rs t c DA mir-1178 premir rs a g mir-1208 premir rs t g mir-1208 premir rs c g IOP mir-1258 premir rs t c mir-1269b seed rs c g CA mir-1269b seed rs c g mir premir rs a g CA mir-1343 premir rs t c DA mir-199b premir rs t c CA mir-300 premir rs t c DA mir-3144 mature rs a g CA mir-3144 premir rs a c CA mir-3152 mature rs a g CA mir-3152 mature rs a g mir-3166 premir rs a t IOP mir-320e premir rs a g CA mir-339 mature rs t c DA mir-339 mature rs t c IOP mir-3609 premir rs a t CA mir-3612 premir rs a g DA mir-3659 premir rs t c DA mir-3683 premir rs a t DA mir-3686 premir rs a g IOP mir-3909 premir rs a g DA mir-3928 premir rs a g mir-3936 premir rs t c DA mir-411 premir rs t c mir-4268 premir rs t c DA mir-4274 premir rs a g DA mir seed rs c g mir-4480 premir rs t g CA mir-4480 premir rs t g IOP mir-4482 premir rs t g IOP mir-4513 seed rs a g IOP mir-4532 seed rs a g DA mir-4532 seed rs a g mir-4634 premir rs a g mir-4655 mature rs t c IOP mir-4707 seed rs a c E-05 CA mir-4707 seed rs a c E-05
7 mir-4743 premir rs a g CA mir-4743 premir rs a g mir-4749 mature rs t c DA mir-4781 seed rs a g CA mir-4781 seed rs a g DA mir-4791 premir rs a g IOP mir-4792 premir rs t g IOP mir-492 premir rs c g IOP mir-499b mature rs a g CA mir-499b mature rs a g DA mir-499b mature rs a g IOP mir-520f premir rs a g CA mir-548ae-2 premir rs a g DA mir-548al seed rs a g CA mir-548al seed rs a g IOP mir-548al seed rs a g mir-548ap mature rs a g DA mir-548ap premir rs a t DA mir-548h-4 premir rs a g CA mir-548h-4 premir rs a g mir-550a-3 premir rs t c mir-550a-3 premir rs a g mir-564 premir rs c g CA mir-5680 premir rs t c CA mir-5682 premir rs a g IOP mir-5689 mature rs t c CA mir-5689 mature rs t c mir-5700 mature rs t c IOP mir-596 mature rs t c CA mir-597 premir rs a g DA mir-612 premir rs a g E-06 CA mir-612 premir rs a g E-09 mir-6130 premir rs a t DA mir-656 premir rs t c DA mir-656 premir rs t c mir-6719 premir rs a g mir p Mature rs a g DA mir p Mature rs a g DA mir premir rs t c Shown are 63 genetic variants located in 59 unique mirnas that are nominally (p < 0.05) associated with POAG endophenotypes in the publicly available IGGC GWAS data.
8 Table S6. The putative mir-612 and mir p target genes associated with and cup area mirna ID Phenotype All target genes (number of SNPs) mir (118,327) -cup areaassociated putative targets USH2A FAM101A JRK SIX4 Top SNP rs rs rs rs GWAS p-value 1.06E E E E-08 mir-612 Cup area 717 SIX4 rs E-17 (118,197) FAM101A rs E-13 mir p 1159 MYPN rs E-31 (131,887) CARD10 rs E-12 LTBP3 rs E-09 H1F0 rs E-08 FLNB rs E-08 SLC25A16 rs E-08 FRMD8 rs E-07 mir p Cup area 1159 FLNB rs E-10 (131,758) MYPN rs E-09 PFAS rs E-07 Shown are putative target genes of mir-612 and mir p associated with and cup area (using the publicly available GWAS data of POAG endophenotypes). TargetScan v7.0 (total context++ score > 0.01) and mirdb databases were used to extract the predicted target genes of the mirnas. The Bonferroni correction was used (based on the number of SNPs in the studied target genes of each mirna) to set the significance level, which is p-value < 4.2E-07 for mir-612 target genes and p-value < 3.8E-07 for mir-4707 target genes. The SNPs shown in the table are top SNPs in the mentioned target genes associated with and cup area. Bold target genes have been shown previously to be differentially expressed in the eye tissues (trabecular meshwork, aqueous humor or optic nerve) in the previous gene expression profiling studies of glaucoma patients.
9 Table S7. Expression of the putative mir-612 and mir p target genes associated with and cup area across different eye tissues Gene Probe ID USH2A FAM101A JRK SIX MYPN CARD LTBP H1F FLNB PFAS SLC25A FRMD Choroid RPE Ciliary Body Cornea Optic Nerve Retina Sclera Trabecular Meshwork Shown are expression of a number of mir-612 and mir p predicted target genes, which found to be significantly associated with and cup area, on eye tissues using the online Ocular Tissue Database. The gene expression is indicated as Affymetrix Probe Logarithmic Intensity Error (PLIER) number. The PLIER number were calculated by GC-background correction, PLIER normalization, log transformation and z-score calculation.
10 Table S8. The mirna-target gene interaction analysis using the Rotterdam Study data mirna SNP ID Target gene Top SNP Cup area (p-value) (p-value) mir-612 rs SIX4 rs USH2A rs FAM101A rs JRK rs mir-4707 rs MYPN rs CARD10 rs LTBP3 rs H1F0 rs FRMD8 rs FLNB rs SLC25A16 rs PFAS rs This table shows the interaction analysis between two identified SNPs in mir-612 and mir-4707 with the top SNP in their target genes associated with glaucoma using the Rotterdam Study (n=10,404)., the mean of vertical cup to disc ratio which is between 0 and 1 (Mean= 0.44, SD= 0.18); Cup area is between 0 and 2 (Mean= 0.54, SD= 0.34). This analysis indicates a significant interaction between mir-4707 (rs ) and CARD10 (rs ) in relation to cup area, shown in bold.
11 Table S9. Forty-seven mirna-binding site variants within the 3 UTRs of 21 genes associated with POAG endophenotypes SNP ID Gene ID Chr. Position Asso. pheno MA effect p-value GWAS A1 mirna binding sites in presence of A1 A2 mirna binding sites in presence of A2 rs CDC DA C (-) 4.00E-08 t mir p c mir-4729, mir p mir-5696, mir-579-3p mir-664b-3p rs TMCO IOP C (+) 4.23E-11 c mir-296-3p, mir-323b-5p mir-410-5p, mir-494-5p mir-511-5p t mir p, mir p mir-4675, mir-4741 mir p, mir-626 mir p rs TMCO IOP T (-) 4.35E-11 c mir-4454, mir-598-5p t mir-4299, mir-4456 mir-548q, mir-7978 rs TRIB CA T (+) 1.13E-09 t mir-545-3p, mir-583 a mir-3125, mir-3916 mir-4310, mir p mir p, mir-877-5p rs7344 SRSF CA C (+) 3.95E-07 t hsa-let-7f-2-3p, mir p mir p, mir p rs CAV IOP T(+) 1.22E-10 c mir p, mir-4435 mir p, mir-588 mir p, mir-548s c t hsa-let-7e-3p, mir p mir-548al mir-4662a-5p, mir-575 mir p, mir-6858 rs CAV IOP G (+) 1.54E-10 a mir-216b-3p g mir p rs CAV IOP G (+) 1.90E-09 g mir-224-5p, mir-377-3p t - rs PSCA A (-) 2.82E-08 a mir-1273h-3p, mir-3166 mir-342-5p, mir p mir-4771, mir p g mir p rs PSCA G (+) 2.67E-08 t mir p g mir-597-5p, mir-668-3p rs PSCA A (-) 2.50E-08 g mir p a mir-296-5p, mir p mir p rs PSCA G (-) 2.66E-08 c mir p, mir-3188 mir-3975, mir p mir-4663, mir-485-5p mir p, mir-8077, mir-7160 rs PSCA G (-) 2.38E-08 c mir-3650, mir-4455 g mir p, mir-4679 mir p, mir p mir p, mir p rs PSCA T (-) 2.53E-08 c mir-4455, mir p t mir p, mir p mir p mir p, mir p rs PSCA A (-) 2.58E-08 g mir-4537 a mir p g mir-3155a, mir-3155b mir p, mir-484
12 rs PSCA A (-) 2.49E-08 g mir-4421, mir p mir p rs ABO IOP T (+) 1.05E-08 t mir p, mir p mir-6762, mir p, mir-885-3p rs CDKN2B , CA a mir p,mir-3655 mir-4431, mir-676-3p c - G (-) 5.31E-33 a mir-382-5p g mir p, mir-323b-5p mir-410-5p mir-494-5p rs CDKN2B CA, T (+) 4.07E-18 c mir p, mir-205-3p t mir-374c-5p, mir-655-3p rs HNRNPH DA T (-) 5.77E-08 a mir-489-3p t - rs HNRNPH DA, A (-) 1.35E-24 a mir p, mir-361-5p c mir-383-5p, mir p rs MYPN DA G (+) 2.52E-15 c mir p mir-636, mir-96-5p g - rs MYPN DA C (+) 8.41E-09 c mir p, mir-624-3p g mir-133a-5p,mir-320e mir-4490 rs MYPN DA A (+) 1.20E-08 a mir-339-5p, hsa-let-7a-2-3p, g mir p hsa-let-7g-3p, mir-486-5p mir p rs MYPN DA T (+) 2.67E-15 c mir p, mir-548g-3p t mir p mir p, mir p rs PBLD DA, A (-) 3.13E-84 a mir-133a-5p, mir p g mir-138-5p, mir-7978 rs PBLD DA, A (-) 1.29E-83 a mir p, mir-1265 c mir-4493, mir p mir-605-3p, mir-6126 rs PBLD DA, T (-) 4.14E-84 t mir-3143, mir-543 c mir-141-3p, mir-200a-3p rs RUFY DA, A (-) 8.54E-20 g mir-651-3p a mir p rs RUFY DA A (-) 1.18E-07 a - g mir-32-3p, mir p mir p, mir-921 rs RUFY DA, A (-) 4.26E-25 a mir-4719 g - rs RUFY DA, G (-) 4.64E-25 a mir-6074 g mir-548ap/h, mir-559 mir-8054 rs RUFY DA, C (-) 6.82E-25 t mir p c mir-132-5p rs RUFY DA, T (-) 9.99E-25 t mir-219b-3p, mir-3123 mir-6511b-5p, mir p c mir-1205, mir-146a-5p mir-146b-5p, mir p
13 mir-4418, mir p mir-509-5p, mir p rs RUFY DA, A (-) 1.66E-24 a mir p, mir p c mir p mir-326, mir-330-5p mir-518c-5p, mir p mir p, mir p mir p, mir-767-3p rs SLC25A DA, C (-) 5.37E-19 c mir-3622b-5p, mir p t mir-564 mir p, mir p rs SLC25A DA A (-) 1.35E-15 g mir-518 a mir-4422, mir p rs CELF IOP G (+) 2.32E-07 a mir-4721 g mir-1273c rs FOLH IOP A (+) 1.29E-08 g mir-186-5p, mir-3133 mir-4509, mir-4744 a mir p, mir-548c-3p rs MAP3K C (-) 2.47E-07 c mir-124-3p, mir-1295b t mir-3910, mir-520a-5p mir-1912, mir-500b-3p, mir-506-3p mir-525-5p rs PAX DA T (+) 5.53E-08 t mir p, mir-3135b c mir-1203 mir-6081 rs1506 PAX DA T (-) 6.34E-09 a mir p, mir p t mir-500b-3p mir-370-3p, mir-4718 mir p, mir p rs PAX DA C (-) 4.21E-09 t mir-5681b, mir-5696 mir-579-3p, mir-664a-3p mir-664b-3p c mir p rs RBM A (+) 6.53E-07 g mir-3924 a mir-190a-3p, mir p mir-6083 rs AAGAB IOP C (-) 2.89E-07 t mir-329-5p, mir-4659a-3p mir-4659b-3p, mir p c mir p, mir p mir p rs PFAS CA T (-) 5.53E-07 c mir-149-3p, mir p t mir-3175, mir-3975 mir-423-5p, mir-4489 mir p, mir-8085 mir p, mir-6085 mir p, mir p mir p, mir p rs TRIOBP A (+) 3.16E-07 g mir p a mir-30a-3p, mir-30d-3p mir-30e-3p, mir p Shown are 47 glaucoma-associated variants that are predicted to affect mirna-binding sites using PolymiRTS (v3.0). DA, disc area; CA, cup area; Chr, chromosome; MA, minor allele; A1, ancestral allele; A2, alternative allele. Bold mirnas have been shown previously to be differentially expressed in the eye tissues (trabecular meshwork, aqueous humor or optic nerve) in the previous mirna expression profiling studies of glaucoma patients.
14 Table S10. Functional characteristics of 47 mirna-binding site variants associated with POAG endophenotypes SNP ID Host gene A1 A2 Chr. Position MAF Proxy SNPs Non-syn. cis-eqtl Effect allele (Europe) (R 2 > 0.8) Proxies (tissue) in eqtl (β) rs CDC7 t c Brain, Nerve, Blood, other tissues C (-) rs TMCO1 c t Nerve, Blood, Artery, other tissues C (-) rs TMCO1 c t Nerve, Blood, Artery, other tissues T (-) rs TRIB2 t a Nerve, Whole blood T (-) rs7344 SRSF3 t c Nerve, Whole blood, Skin, Thyroid C (+) rs CAV2 c t Pancreases, Testis T (-) rs CAV2 a g Pancreases, Blood G (-) rs CAV2 g t Pancreases, Blood, Lung, Testis G (-) rs PSCA a g Brain, Nerve, Blood, other tissues A (+) rs PSCA t g Brain, Nerve, Blood, other tissues G (+) rs PSCA g a Brain, Nerve, Blood, other tissues A (+) rs PSCA c g Brain, Nerve, Blood, other tissues G (+) rs PSCA c g Brain, Nerve, Blood, other tissues G (+) rs PSCA c t Brain, Nerve, Blood, other tissues T (+) rs PSCA g a Brain, Nerve, Blood, other tissues A (+) rs PSCA g a Brain, Nerve, Blood, other tissues A (+) rs ABO t c Nerve, Adipose, Heart, Lung, Skin T (+) rs CDKN2B a g Temporal cortex G (-) rs CDKN2B c t Whole blood T (+) rs MYPN c g Muscle skeletal G (+) rs MYPN c t Muscle skeletal T (+) rs MYPN c g C (+) rs MYPN a g A (+)
15 rs PBLD a g Brain, Nerve, Liver, other tissues A (+) rs PBLD a c Brain, Nerve, Liver, other tissues A (+) rs PBLD t c Brain, Nerve, Liver, other tissues T (+) rs HNRNPH3 a t T (-) rs RUFY2 a g A (-) rs RUFY2 a g Brain, Nerve, Blood, other tissues A (+) rs RUFY2 a g Brain, Nerve, Blood, other tissues G (+) rs RUFY2 t c Brain, Nerve, Blood, other tissues C (+) rs RUFY2 t c Brain, Nerve, Blood, other tissues T (+) rs RUFY2 a c Brain, Nerve, Blood, other tissues A (+) rs RUFY2 g a Brain, Nerve, Blood, other tissues A (+) rs HNRNPH3 a c Brain, Nerve, Blood, other tissues A (-) rs SLC25A16 c t Brain, Nerve, Blood, other tissues C (-) rs SLC25A16 g a Brain, Nerve, Blood, other tissues A (-) rs CELF1 a g Nerve, Muscle, Blood, other tissues G (+) rs FOLH1 g a Skin, Testis A (+) rs MAP3K11 c t Artery, Adipose, Blood, Skin C (-) rs1506 PAX6 a t T (-) rs PAX6 t c C (-) rs PAX6 t c Lymphoblastoid T (-) rs RBM23 g a Brain, Nerve, Blood, other tissues A (-) rs AAGAB t c Adipose, Lung, Skin, Thyroid C (-) rs PFAS c t Nerve, Blood, Adipose, Colon T (-) rs TRIOBP g a Nerve, Blood, Adipose, other tissues A (-) The high LD proxies and eqtl of 47 mirna-binding site SNPs associated with POAG endophenotypes using HaploReg v4.1. In total, 723 intronic and 10 missense SNPs are in LD (R 2 > 0.8) with the binding site SNPs. Bold are the most significant SNPs among the correlated SNPs in one gene.
16 Table 11. Expression of genes hosting the 47 mirna-binding site variants associated with POAG endophenotypes in eye tissues Gene ID Probe ID Trabecular Optic Choroid Ciliary Cornea Lens Retina Sclera Meshwork Nerve RPE Body AAGAB ABO CAV CDC CDKN2B CELF FOLH HNRNPH MAP3K MYPN PAX PBLD PFAS PSCA RBM RUFY SRSF SLC25A TMCO TRIB TRIOBP Expression of 21 genes hosting the 47 mirna-binding sites associated with glaucoma endophenotypes across human eye tissues (using The Ocular Tissue Database). The gene expression is indicated as Affymetrix Probe Logarithmic Intensity Error (PLIER) number. The PLIER number were calculated by GC-background correction, PLIER normalization, log transformation and z-score calculation.
17 Table S12. Prioritization of 47 mirna-binding site variants associated with glaucoma endophenotypes SNP ID Top in GWAS eqtl MA (effect) Proxy SNPs (# non-syn) Gene ID Gene exp in the eye Gene asso with dis MiRNA-binding site (disruption/creation) mir exp in the eye mir asso with dis rs T (+) 4 (0) CDKN2B mir p 9 rs G (-) 29 (0) CDKN2B mir-323-5p 8 rs G (-) 11 (0) CAV2 mir-224-5p 8 rs C (-) 74 (0) AAGAB mir-329-5p - 8 rs A (+) 48 (0) PSCA mir-342-5p, mir p - 8 rs T (-) 5 (0) PFAS mir-149-3p, mir-423-5p - 8 rs T (-) 10 (0) TRIB2 mir-877-5p - 8 rs A (-) 1 (0) RBM23 mir-190a-3p - 8 rs T (+) 7 (1) ABO mir-885-3p - 8 rs7344 C (+) 64 (0) SFRS3 mir-548al, Let-7f-2-3p - 8 rs G (+) 48 (0) PSCA mir-597-5p - 8 rs G (+) 81 (0) RUFY2 mir-548ap/h, mir rs A (+) 81 (0) RUFY2 mir p - 7 rs C (+) 81 (0) RUFY2 mir-132-5p - 7 rs A (+) 81 (0) RUFY2 mir p - 7 rs T (+) 81 (0) RUFY2 mir-219b-3p - 7 rs T (-) 10 (1) CAV2 mir p, mir rs C (-) 33 (0) TMCO1 mir-323-5p, mir-410-5p - 7 rs G (+) 48 (0) PSCA mir-485-5p - 7 rs G (+) 11 (0) MYPN mir p, mir-96-5p - 6 rs A (+) 48 (0) PSCA mir-676-3p - 6 rs A (+) 30 (1) PBLD mir p rs T (+) 11 (0) MYPN mir-548g-3p - 6 rs A (+) 48 (0) PSCA mir-296-5p - 6 rs G (-) 10 (1) CAV2 mir p - 6 rs C (-) 15 (0) CDC7 mir-579-3p rs C (+) 22 (0) MYPN mir-320e, mir-133a - 6 Score
18 rs G (+) 30 (0) CELF1 mir-1273c rs C (-) 4 (0) PAX6 mir-664a-3p - 5 rs T (-) 4 (0) PAX6 mir-370-3p - 5 rs G (+) 48 (0) PSCA mir p rs C (-) 20 (0) MAP3K11 mir-520a, mir rs T (-) 33 (0) TMCO1 mir-548q - 5 rs T (+) 30 (1) PBLD mir rs A (+) 60 (2) FOLH1 mir-186-5p - 5 rs A (-) 61 (2) TRIOBP mir-30a-3p - 5 rs A (+) 30 (1) PBLD mir-133a-5p - 5 rs A (+) 81 (0) RUFY2 mir rs T (-) 3 (0) PAX6 mir-3135b rs C (-) 26 (0) SLC25A16 mir rs A (+) 22 (0) MYPN mir-339-5p, Let-7a-2-3p - 5 rs A (-) 81 (0) HNRNPH3 mir-361-5p - 4 rs A (-) 60 (0) SLC25A16 mir rs A (+) 48 (0) PSCA mir p rs T (+) 48 (0) PSCA mir p rs A (-) 30 (1) RUFY2 mir-32-3p - 3 rs T (-) 30 (1) HNRNPH3 mir-489-3p - 3 Prioritization of the 47 mirna-binding site SNPs associated with POAG endophenotypes. A functional score (between 1 and 10) was calculated for each SNP by combining the results of our predefined criteria including the strength of association, LD pattern, eqtl data, expression and importance of the related mirna and target gene in the eye. 1. The strength of the association in GWAS results: Lead SNP (2), One of the top 10% SNPs in the locus (1), Only passed the threshold (0). 2. The eqtl data: Correlation between SNP and expression of the host gene (2), Correlation of SNPs with expression of the nearby genes in the locus (1), No correlation (0). 3. The LD pattern: No or just a few synonymous proxies (2), Several synonymous proxies, but no non-synonymous (1), Several proxies including nonsynonymous proxy SNPs (0). 4. Host gene: Expressed in the eye and previously reported to be involved in POAG (2), Expressed in the eye or involved in POAG (1), Not expressed in the eye and no evidence to be involved in POAG (0). 5. mirna: Expressed in the eye and previously reported to be involved in the eye disease (2), Expressed in the eye or involved in the eye disease (1), Not expressed in the eye and no evidence to be involved in the eye disease (0). One extra point for the mirna-target interactions that are validated.
19 Figure S1. The predicted hairpin structure of pre-mir-612 and pre-mir-4707 and the position of two identified variants associated with and cup area pre-mir ca --a c cu c - -a a 5' ucc ucugg cc ug ggg agggcuucug agcuccuu gc c 3' agg ggacc gg ac ccc ucccggggac ucggggag cg u acucuc aca gac u cu - c ga a rs (G/A) pre-mir-4707 u - c gc -- ug - u 5' ggu ccg gagcc cggc gggcggguu c g ggug a 3' cca ggc cuugg gccg cccgcccga g c ucgc g c u a ac cc gu g a rs (T/G)
20 Figure S2. Manhattan plots showing the association of 72,052 mirna-binding site variants with POAG endophenotypes
21
22 Figure S3. Regional plots showing the association of ten highlighted mirna-binding site variants with POAG endophenotypes
23
Supplementary Figure 1: Attenuation of association signals after conditioning for the lead SNP. a) attenuation of association signal at the 9p22.
 Supplementary Figure 1: Attenuation of association signals after conditioning for the lead SNP. a) attenuation of association signal at the 9p22.32 PCOS locus after conditioning for the lead SNP rs10993397;
Supplementary Figure 1: Attenuation of association signals after conditioning for the lead SNP. a) attenuation of association signal at the 9p22.32 PCOS locus after conditioning for the lead SNP rs10993397;
Supplementary Figure 1. Quantile-quantile (Q-Q) plot of the log 10 p-value association results from logistic regression models for prostate cancer
 Supplementary Figure 1. Quantile-quantile (Q-Q) plot of the log 10 p-value association results from logistic regression models for prostate cancer risk in stage 1 (red) and after removing any SNPs within
Supplementary Figure 1. Quantile-quantile (Q-Q) plot of the log 10 p-value association results from logistic regression models for prostate cancer risk in stage 1 (red) and after removing any SNPs within
Supplementary Figure 1. Quantile-quantile (Q-Q) plots. (Panel A) Q-Q plot graphical
 Supplementary Figure 1. Quantile-quantile (Q-Q) plots. (Panel A) Q-Q plot graphical representation using all SNPs (n= 13,515,798) including the region on chromosome 1 including SORT1 which was previously
Supplementary Figure 1. Quantile-quantile (Q-Q) plots. (Panel A) Q-Q plot graphical representation using all SNPs (n= 13,515,798) including the region on chromosome 1 including SORT1 which was previously
a) Primary cultures derived from the pancreas of an 11-week-old Pdx1-Cre; K-MADM-p53
 1 2 3 4 5 6 7 8 9 10 Supplementary Figure 1. Induction of p53 LOH by MADM. a) Primary cultures derived from the pancreas of an 11-week-old Pdx1-Cre; K-MADM-p53 mouse revealed increased p53 KO/KO (green,
1 2 3 4 5 6 7 8 9 10 Supplementary Figure 1. Induction of p53 LOH by MADM. a) Primary cultures derived from the pancreas of an 11-week-old Pdx1-Cre; K-MADM-p53 mouse revealed increased p53 KO/KO (green,
SUPPLEMENTARY INFORMATION. Meta-analyses of genome-wide association studies identify multiple novel loci related to pulmonary function
 SUPPLEMENTARY INFORMATION Meta-analyses of genome-wide association studies identify multiple novel loci related to pulmonary function Dana B. Hancock, 1,29 Mark Eijgelsheim, 2,29 Jemma B. Wilk, 3,29 Sina
SUPPLEMENTARY INFORMATION Meta-analyses of genome-wide association studies identify multiple novel loci related to pulmonary function Dana B. Hancock, 1,29 Mark Eijgelsheim, 2,29 Jemma B. Wilk, 3,29 Sina
Global variation in copy number in the human genome
 Global variation in copy number in the human genome Redon et. al. Nature 444:444-454 (2006) 12.03.2007 Tarmo Puurand Study 270 individuals (HapMap collection) Affymetrix 500K Whole Genome TilePath (WGTP)
Global variation in copy number in the human genome Redon et. al. Nature 444:444-454 (2006) 12.03.2007 Tarmo Puurand Study 270 individuals (HapMap collection) Affymetrix 500K Whole Genome TilePath (WGTP)
Supplementary Figure 1
 Supplementary Figure 1 Asymmetrical function of 5p and 3p arms of mir-181 and mir-30 families and mir-142 and mir-154. (a) Control experiments using mirna sensor vector and empty pri-mirna overexpression
Supplementary Figure 1 Asymmetrical function of 5p and 3p arms of mir-181 and mir-30 families and mir-142 and mir-154. (a) Control experiments using mirna sensor vector and empty pri-mirna overexpression
c Tuj1(-) apoptotic live 1 DIV 2 DIV 1 DIV 2 DIV Tuj1(+) Tuj1/GFP/DAPI Tuj1 DAPI GFP
 Supplementary Figure 1 Establishment of the gain- and loss-of-function experiments and cell survival assays. a Relative expression of mature mir-484 30 20 10 0 **** **** NCP mir- 484P NCP mir- 484P b Relative
Supplementary Figure 1 Establishment of the gain- and loss-of-function experiments and cell survival assays. a Relative expression of mature mir-484 30 20 10 0 **** **** NCP mir- 484P NCP mir- 484P b Relative
Inventory of Supplemental Data: Supplemental Figures 1-7. Supplemental Tables 1-8. Supplemental References. Supplemental Video:
 Inventory of Supplemental Data: Supplemental Figures -7 Supplemental Tables -8 Supplemental References Supplemental Video: Note that the video shows JHU-9 cells transfected with mir-93a antagomir or control
Inventory of Supplemental Data: Supplemental Figures -7 Supplemental Tables -8 Supplemental References Supplemental Video: Note that the video shows JHU-9 cells transfected with mir-93a antagomir or control
Supplementary Table 3. 3 UTR primer sequences. Primer sequences used to amplify and clone the 3 UTR of each indicated gene are listed.
 Supplemental Figure 1. DLKI-DIO3 mirna/mrna complementarity. Complementarity between the indicated DLK1-DIO3 cluster mirnas and the UTR of SOX2, SOX9, HIF1A, ZEB1, ZEB2, STAT3 and CDH1with mirsvr and PhastCons
Supplemental Figure 1. DLKI-DIO3 mirna/mrna complementarity. Complementarity between the indicated DLK1-DIO3 cluster mirnas and the UTR of SOX2, SOX9, HIF1A, ZEB1, ZEB2, STAT3 and CDH1with mirsvr and PhastCons
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Heatmap of GO terms for differentially expressed genes. The terms were hierarchically clustered using the GO term enrichment beta. Darker red, higher positive
Supplementary Figures Supplementary Figure 1. Heatmap of GO terms for differentially expressed genes. The terms were hierarchically clustered using the GO term enrichment beta. Darker red, higher positive
3/16/2018. Ultrasound Biomicroscopy in Glaucoma By Ahmed Salah Abdel Rehim. Prof. of Ophthalmology Al-Azhar University
 Ultrasound Biomicroscopy in Glaucoma By Ahmed Salah Abdel Rehim Prof. of Ophthalmology Al-Azhar University 1 Ultrasound biomicroscopy (UBM) is a recent technique to visualize anterior segment with the
Ultrasound Biomicroscopy in Glaucoma By Ahmed Salah Abdel Rehim Prof. of Ophthalmology Al-Azhar University 1 Ultrasound biomicroscopy (UBM) is a recent technique to visualize anterior segment with the
ab FirePlex mirna Panel Kidney Toxicity
 Version 1 Last updated 30 May 2017 ab219508 FirePlex mirna Panel Kidney Toxicity This product is for research use only and is not intended for diagnostic use. Copyright 2017 Abcam. All rights reserved.
Version 1 Last updated 30 May 2017 ab219508 FirePlex mirna Panel Kidney Toxicity This product is for research use only and is not intended for diagnostic use. Copyright 2017 Abcam. All rights reserved.
Investigating causality in the association between 25(OH)D and schizophrenia
 Investigating causality in the association between 25(OH)D and schizophrenia Amy E. Taylor PhD 1,2,3, Stephen Burgess PhD 1,4, Jennifer J. Ware PhD 1,2,5, Suzanne H. Gage PhD 1,2,3, SUNLIGHT consortium,
Investigating causality in the association between 25(OH)D and schizophrenia Amy E. Taylor PhD 1,2,3, Stephen Burgess PhD 1,4, Jennifer J. Ware PhD 1,2,5, Suzanne H. Gage PhD 1,2,3, SUNLIGHT consortium,
Ct=28.4 WAT 92.6% Hepatic CE (mg/g) P=3.6x10-08 Plasma Cholesterol (mg/dl)
 a Control AAV mtm6sf-shrna8 Ct=4.3 Ct=8.4 Ct=8.8 Ct=8.9 Ct=.8 Ct=.5 Relative TM6SF mrna Level P=.5 X -5 b.5 Liver WAT Small intestine Relative TM6SF mrna Level..5 9.6% Control AAV mtm6sf-shrna mtm6sf-shrna6
a Control AAV mtm6sf-shrna8 Ct=4.3 Ct=8.4 Ct=8.8 Ct=8.9 Ct=.8 Ct=.5 Relative TM6SF mrna Level P=.5 X -5 b.5 Liver WAT Small intestine Relative TM6SF mrna Level..5 9.6% Control AAV mtm6sf-shrna mtm6sf-shrna6
Supplementary Document
 Supplementary Document 1. Supplementary Table legends 2. Supplementary Figure legends 3. Supplementary Tables 4. Supplementary Figures 5. Supplementary References 1. Supplementary Table legends Suppl.
Supplementary Document 1. Supplementary Table legends 2. Supplementary Figure legends 3. Supplementary Tables 4. Supplementary Figures 5. Supplementary References 1. Supplementary Table legends Suppl.
Nature Genetics: doi: /ng Supplementary Figure 1
 Supplementary Figure 1 LD (r 2 ) between the A3AB deletion and all markers in a 400-kb APOBEC3 region in 1000 Genomes Project populations. Populations: CEU, individuals of European ancestry from Utah,
Supplementary Figure 1 LD (r 2 ) between the A3AB deletion and all markers in a 400-kb APOBEC3 region in 1000 Genomes Project populations. Populations: CEU, individuals of European ancestry from Utah,
P. Mathijs Voorhoeve, Carlos le Sage, Mariette Schrier, Ad J.M. Gillis, Hans Stoop,
 Supplemental Data A Genetic Screen Implicates mirna-372 and mirna-373 As Oncogenes in Testicular Germ Cell Tumors P. Mathijs Voorhoeve, Carlos le Sage, Mariette Schrier, Ad J.M. Gillis, Hans Stoop, Remco
Supplemental Data A Genetic Screen Implicates mirna-372 and mirna-373 As Oncogenes in Testicular Germ Cell Tumors P. Mathijs Voorhoeve, Carlos le Sage, Mariette Schrier, Ad J.M. Gillis, Hans Stoop, Remco
Supplemental Figure 1. Small RNA size distribution from different soybean tissues.
 Supplemental Figure 1. Small RNA size distribution from different soybean tissues. The size of small RNAs was plotted versus frequency (percentage) among total sequences (A, C, E and G) or distinct sequences
Supplemental Figure 1. Small RNA size distribution from different soybean tissues. The size of small RNAs was plotted versus frequency (percentage) among total sequences (A, C, E and G) or distinct sequences
SUPPLEMENTARY INFORMATION
 SUPPLEMENTRY INFORMTION Supplementary Figure 1. Q-Q plot of RE/NIMH autism family TDT results. The quantile-quantile plot of the expected and observed P-values is shown. The blue circles represent the
SUPPLEMENTRY INFORMTION Supplementary Figure 1. Q-Q plot of RE/NIMH autism family TDT results. The quantile-quantile plot of the expected and observed P-values is shown. The blue circles represent the
Genome-wide association study identifies variants in TMPRSS6 associated with hemoglobin levels.
 Supplementary Online Material Genome-wide association study identifies variants in TMPRSS6 associated with hemoglobin levels. John C Chambers, Weihua Zhang, Yun Li, Joban Sehmi, Mark N Wass, Delilah Zabaneh,
Supplementary Online Material Genome-wide association study identifies variants in TMPRSS6 associated with hemoglobin levels. John C Chambers, Weihua Zhang, Yun Li, Joban Sehmi, Mark N Wass, Delilah Zabaneh,
Association-heterogeneity mapping identifies an Asian-specific association of the GTF2I locus with rheumatoid arthritis
 Supplementary Material Association-heterogeneity mapping identifies an Asian-specific association of the GTF2I locus with rheumatoid arthritis Kwangwoo Kim 1,, So-Young Bang 1,, Katsunori Ikari 2,3, Dae
Supplementary Material Association-heterogeneity mapping identifies an Asian-specific association of the GTF2I locus with rheumatoid arthritis Kwangwoo Kim 1,, So-Young Bang 1,, Katsunori Ikari 2,3, Dae
Retinal Nerve Fiber Analysis: the Role It Plays in Assessing Glaucoma Grace Martin, CPOT
 Retinal Nerve Fiber Analysis: the Role It Plays in Assessing Glaucoma Grace Martin, CPOT Every day, patients experiencing visual difficulties are seen in the optometric practice. This can range from a
Retinal Nerve Fiber Analysis: the Role It Plays in Assessing Glaucoma Grace Martin, CPOT Every day, patients experiencing visual difficulties are seen in the optometric practice. This can range from a
THE CHRONIC GLAUCOMAS
 THE CHRONIC GLAUCOMAS WHAT IS GLAUCOMA? People with glaucoma have lost some of their field of all round vision. It is often the edge or periphery that is lost. That is why the condition can be missed until
THE CHRONIC GLAUCOMAS WHAT IS GLAUCOMA? People with glaucoma have lost some of their field of all round vision. It is often the edge or periphery that is lost. That is why the condition can be missed until
Supplemental Table 1 Age and gender-specific cut-points used for MHO.
 Supplemental Table 1 Age and gender-specific cut-points used for MHO. Age SBP (mmhg) DBP (mmhg) HDL-C (mmol/l) TG (mmol/l) FG (mmol/l) Boys 6-11 90th * 90th * 1.03 1.24 5.6 12 121 76 1.13 1.44 5.6 13 123
Supplemental Table 1 Age and gender-specific cut-points used for MHO. Age SBP (mmhg) DBP (mmhg) HDL-C (mmol/l) TG (mmol/l) FG (mmol/l) Boys 6-11 90th * 90th * 1.03 1.24 5.6 12 121 76 1.13 1.44 5.6 13 123
To test the possible source of the HBV infection outside the study family, we searched the Genbank
 Supplementary Discussion The source of hepatitis B virus infection To test the possible source of the HBV infection outside the study family, we searched the Genbank and HBV Database (http://hbvdb.ibcp.fr),
Supplementary Discussion The source of hepatitis B virus infection To test the possible source of the HBV infection outside the study family, we searched the Genbank and HBV Database (http://hbvdb.ibcp.fr),
Supplementary information for: A functional variation in BRAP confers risk of myocardial infarction in Asian populations
 Supplementary information for: A functional variation in BRAP confers risk of myocardial infarction in Asian populations Kouichi Ozaki 1, Hiroshi Sato 2, Katsumi Inoue 3, Tatsuhiko Tsunoda 4, Yasuhiko
Supplementary information for: A functional variation in BRAP confers risk of myocardial infarction in Asian populations Kouichi Ozaki 1, Hiroshi Sato 2, Katsumi Inoue 3, Tatsuhiko Tsunoda 4, Yasuhiko
Supplementary Figure 1. Nature Genetics: doi: /ng.3736
 Supplementary Figure 1 Genetic correlations of five personality traits between 23andMe discovery and GPC samples. (a) The values in the colored squares are genetic correlations (r g ); (b) P values of
Supplementary Figure 1 Genetic correlations of five personality traits between 23andMe discovery and GPC samples. (a) The values in the colored squares are genetic correlations (r g ); (b) P values of
Management of Angle Closure Glaucoma Hospital Authority Convention 18 May 2015
 Management of Angle Closure Glaucoma Hospital Authority Convention 18 May 2015 Jimmy Lai Clinical Professor Department of Ophthalmology The University of Hong Kong 1 Primary Angle Closure Glaucoma PACG
Management of Angle Closure Glaucoma Hospital Authority Convention 18 May 2015 Jimmy Lai Clinical Professor Department of Ophthalmology The University of Hong Kong 1 Primary Angle Closure Glaucoma PACG
Supplementary Figure S1A
 Supplementary Figure S1A-G. LocusZoom regional association plots for the seven new cross-cancer loci that were > 1 Mb from known index SNPs. Genes up to 500 kb on either side of each new index SNP are
Supplementary Figure S1A-G. LocusZoom regional association plots for the seven new cross-cancer loci that were > 1 Mb from known index SNPs. Genes up to 500 kb on either side of each new index SNP are
THE CHRONIC GLAUCOMAS
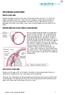 THE CHRONIC GLAUCOMAS WHAT IS GLAUCOMA People with glaucoma have lost some of their field of all round vision. It is often the edge or periphery that is lost. That is why the condition can be missed until
THE CHRONIC GLAUCOMAS WHAT IS GLAUCOMA People with glaucoma have lost some of their field of all round vision. It is often the edge or periphery that is lost. That is why the condition can be missed until
Patrocles: a database of polymorphic mirna-mediated gene regulation
 Patrocles: a database of polymorphic mirna-mediated gene regulation Satellite Eadgene Course "A primer in mirna biology" Liège, 3 march 2008 S. Hiard, D. Baurain, W. Coppieters, X. Tordoir, C. Charlier,
Patrocles: a database of polymorphic mirna-mediated gene regulation Satellite Eadgene Course "A primer in mirna biology" Liège, 3 march 2008 S. Hiard, D. Baurain, W. Coppieters, X. Tordoir, C. Charlier,
A genome-wide association study identifies two new cervical cancer susceptibility loci at 4q12 and 17q12. Supplementary Materials
 A genome-wide association study identifies two new cervical cancer susceptibility loci at 4q12 and 17q12 Supplementary Materials Yongyong Shi 1-4,42, Li Li 5,42, Zhibin Hu 6,7,42, Shuang Li 1,42, Shixuan
A genome-wide association study identifies two new cervical cancer susceptibility loci at 4q12 and 17q12 Supplementary Materials Yongyong Shi 1-4,42, Li Li 5,42, Zhibin Hu 6,7,42, Shuang Li 1,42, Shixuan
Supplementary Materials
 Supplementary Materials 1 Supplementary Table 1. List of primers used for quantitative PCR analysis. Gene name Gene symbol Accession IDs Sequence range Product Primer sequences size (bp) β-actin Actb gi
Supplementary Materials 1 Supplementary Table 1. List of primers used for quantitative PCR analysis. Gene name Gene symbol Accession IDs Sequence range Product Primer sequences size (bp) β-actin Actb gi
A total of 2,822 Mexican dyslipidemic cases and controls were recruited at INCMNSZ in
 Supplemental Material The N342S MYLIP polymorphism is associated with high total cholesterol and increased LDL-receptor degradation in humans by Daphna Weissglas-Volkov et al. Supplementary Methods Mexican
Supplemental Material The N342S MYLIP polymorphism is associated with high total cholesterol and increased LDL-receptor degradation in humans by Daphna Weissglas-Volkov et al. Supplementary Methods Mexican
Genetic Variants Associated With Different Risks for High Tension Glaucoma and Normal Tension Glaucoma in a Chinese Population
 Genetics Genetic Variants Associated With Different Risks for High Tension Glaucoma and Normal Tension Glaucoma in a Chinese Population Yuhong Chen, 1 Guy Hughes, 2 Xueli Chen, 1 Shaohong Qian, 1 Wenjun
Genetics Genetic Variants Associated With Different Risks for High Tension Glaucoma and Normal Tension Glaucoma in a Chinese Population Yuhong Chen, 1 Guy Hughes, 2 Xueli Chen, 1 Shaohong Qian, 1 Wenjun
ASSOCIATION OF KCNJ1 VARIATION WITH CHANGE IN FASTING GLUCOSE AND NEW ONSET DIABETES DURING HCTZ TREATMENT
 ONLINE SUPPLEMENT ASSOCIATION OF KCNJ1 VARIATION WITH CHANGE IN FASTING GLUCOSE AND NEW ONSET DIABETES DURING HCTZ TREATMENT Jason H Karnes, PharmD 1, Caitrin W McDonough, PhD 1, Yan Gong, PhD 1, Teresa
ONLINE SUPPLEMENT ASSOCIATION OF KCNJ1 VARIATION WITH CHANGE IN FASTING GLUCOSE AND NEW ONSET DIABETES DURING HCTZ TREATMENT Jason H Karnes, PharmD 1, Caitrin W McDonough, PhD 1, Yan Gong, PhD 1, Teresa
Supplementary Information. Common variants associated with general and MMR vaccine-related febrile seizures
 Supplementary Information Common variants associated with general and MMR vaccine-related febrile seizures Bjarke Feenstra, Björn Pasternak, Frank Geller, Lisbeth Carstensen, Tongfei Wang, Fen Huang, Jennifer
Supplementary Information Common variants associated with general and MMR vaccine-related febrile seizures Bjarke Feenstra, Björn Pasternak, Frank Geller, Lisbeth Carstensen, Tongfei Wang, Fen Huang, Jennifer
Supplementary information: Forstner, Hofmann et al.
 Supplementary information: Forstner, Hofmann et al., Genome-wide analysis implicates micrornas and their target genes in the development of bipolar disorder Table of contents Supplementary Figure 1: Regional
Supplementary information: Forstner, Hofmann et al., Genome-wide analysis implicates micrornas and their target genes in the development of bipolar disorder Table of contents Supplementary Figure 1: Regional
A rare variant in MYH6 confers high risk of sick sinus syndrome. Hilma Hólm ESC Congress 2011 Paris, France
 A rare variant in MYH6 confers high risk of sick sinus syndrome Hilma Hólm ESC Congress 2011 Paris, France Disclosures I am an employee of decode genetics, Reykjavik, Iceland. Sick sinus syndrome SSS is
A rare variant in MYH6 confers high risk of sick sinus syndrome Hilma Hólm ESC Congress 2011 Paris, France Disclosures I am an employee of decode genetics, Reykjavik, Iceland. Sick sinus syndrome SSS is
Genome-wide Association Analysis Applied to Asthma-Susceptibility Gene. McCaw, Z., Wu, W., Hsiao, S., McKhann, A., Tracy, S.
 Genome-wide Association Analysis Applied to Asthma-Susceptibility Gene McCaw, Z., Wu, W., Hsiao, S., McKhann, A., Tracy, S. December 17, 2014 1 Introduction Asthma is a chronic respiratory disease affecting
Genome-wide Association Analysis Applied to Asthma-Susceptibility Gene McCaw, Z., Wu, W., Hsiao, S., McKhann, A., Tracy, S. December 17, 2014 1 Introduction Asthma is a chronic respiratory disease affecting
Written by Administrator Wednesday, 13 January :27 - Last Updated Thursday, 21 January :34
 angle closure glaucoma A type of glaucoma caused by a sudden and severe rise in eye pressure. Occurs when the pupil enlarges too much or too quickly, and the outer edge of the iris blocks the eye s drainage
angle closure glaucoma A type of glaucoma caused by a sudden and severe rise in eye pressure. Occurs when the pupil enlarges too much or too quickly, and the outer edge of the iris blocks the eye s drainage
Nature Genetics: doi: /ng Supplementary Figure 1
 Supplementary Figure 1 Replicability of blood eqtl effects in ileal biopsies from the RISK study. eqtls detected in the vicinity of SNPs associated with IBD tend to show concordant effect size and direction
Supplementary Figure 1 Replicability of blood eqtl effects in ileal biopsies from the RISK study. eqtls detected in the vicinity of SNPs associated with IBD tend to show concordant effect size and direction
5/2/18. After this class students should be able to: Stephanie Moon, Ph.D. - GWAS. How do we distinguish Mendelian from non-mendelian traits?
 corebio II - genetics: WED 25 April 2018. 2018 Stephanie Moon, Ph.D. - GWAS After this class students should be able to: 1. Compare and contrast methods used to discover the genetic basis of traits or
corebio II - genetics: WED 25 April 2018. 2018 Stephanie Moon, Ph.D. - GWAS After this class students should be able to: 1. Compare and contrast methods used to discover the genetic basis of traits or
Comprehensive Evaluation of the Association of APOE Genetic Variation with Plasma Lipoprotein Traits in U.S. Whites and African Blacks
 RESEARCH ARTICLE Comprehensive Evaluation of the Association of APOE Genetic Variation with Plasma Lipoprotein Traits in U.S. Whites and African Blacks Zaheda H. Radwan 1, Xingbin Wang 1, Fahad Waqar 1,
RESEARCH ARTICLE Comprehensive Evaluation of the Association of APOE Genetic Variation with Plasma Lipoprotein Traits in U.S. Whites and African Blacks Zaheda H. Radwan 1, Xingbin Wang 1, Fahad Waqar 1,
Ovarian cancer variant rs is associated with HOXD1 and HOXD3 gene expression
 /, 2017, Vol. 8, (No. 61), pp: 103410-103414 Ovarian cancer variant rs2072590 is associated with HOXD1 and HOXD3 gene expression Liyuan Guo 1,*, Yan Peng 2,*, Lei Sun 3,*, Xia Han 4, Juan Xu 4 and Dongwei
/, 2017, Vol. 8, (No. 61), pp: 103410-103414 Ovarian cancer variant rs2072590 is associated with HOXD1 and HOXD3 gene expression Liyuan Guo 1,*, Yan Peng 2,*, Lei Sun 3,*, Xia Han 4, Juan Xu 4 and Dongwei
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Sherman SI, Wirth LJ, Droz J-P, et al. Motesanib diphosphate
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Sherman SI, Wirth LJ, Droz J-P, et al. Motesanib diphosphate
ARTICLE Genetic Risk Factors for Type 2 Diabetes: A Trans-Regulatory Genetic Architecture?
 ARTICLE Genetic Risk Factors for Type 2 Diabetes: A Trans-Regulatory Genetic Architecture? Steven C. Elbein, 1,7,8 Eric R. Gamazon, 2,7 Swapan K. Das, 1 Neda Rasouli, 3,4 Philip A. Kern, 5,6 and Nancy
ARTICLE Genetic Risk Factors for Type 2 Diabetes: A Trans-Regulatory Genetic Architecture? Steven C. Elbein, 1,7,8 Eric R. Gamazon, 2,7 Swapan K. Das, 1 Neda Rasouli, 3,4 Philip A. Kern, 5,6 and Nancy
Genome-wide association study dissects the genetic architecture of
 Supplementary Information for Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels Hui Li 1,6, Zhiyu Peng 2,6, Xiaohong Yang 1,6, Weidong Wang 1,6, Junjie
Supplementary Information for Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels Hui Li 1,6, Zhiyu Peng 2,6, Xiaohong Yang 1,6, Weidong Wang 1,6, Junjie
Genetic Testing and Analysis. (858) MRN: Specimen: Saliva Received: 07/26/2016 GENETIC ANALYSIS REPORT
 GBinsight Sample Name: GB4411 Race: Gender: Female Reason for Testing: Type 2 diabetes, early onset MRN: 0123456789 Specimen: Saliva Received: 07/26/2016 Test ID: 113-1487118782-4 Test: Type 2 Diabetes
GBinsight Sample Name: GB4411 Race: Gender: Female Reason for Testing: Type 2 diabetes, early onset MRN: 0123456789 Specimen: Saliva Received: 07/26/2016 Test ID: 113-1487118782-4 Test: Type 2 Diabetes
relative s privacy, do not identify your relative by full name in any assignment.
 Overview Do you or a family member have glaucoma? Do you wonder what this diagnosis means? Glaucoma affects tens of millions of people worldwide. Despite its prevalence, many people lack accurate information
Overview Do you or a family member have glaucoma? Do you wonder what this diagnosis means? Glaucoma affects tens of millions of people worldwide. Despite its prevalence, many people lack accurate information
Supplementary Figure 1 MicroRNA expression in human synovial fibroblasts from different locations. MicroRNA, which were identified by RNAseq as most
 Supplementary Figure 1 MicroRNA expression in human synovial fibroblasts from different locations. MicroRNA, which were identified by RNAseq as most differentially expressed between human synovial fibroblasts
Supplementary Figure 1 MicroRNA expression in human synovial fibroblasts from different locations. MicroRNA, which were identified by RNAseq as most differentially expressed between human synovial fibroblasts
Computational methods to model disease and genetic effects on optic nerve head structure
 University of Iowa Iowa Research Online Theses and Dissertations 2015 Computational methods to model disease and genetic effects on optic nerve head structure Mark Allen Christopher University of Iowa
University of Iowa Iowa Research Online Theses and Dissertations 2015 Computational methods to model disease and genetic effects on optic nerve head structure Mark Allen Christopher University of Iowa
Imaging Genetics: Heritability, Linkage & Association
 Imaging Genetics: Heritability, Linkage & Association David C. Glahn, PhD Olin Neuropsychiatry Research Center & Department of Psychiatry, Yale University July 17, 2011 Memory Activation & APOE ε4 Risk
Imaging Genetics: Heritability, Linkage & Association David C. Glahn, PhD Olin Neuropsychiatry Research Center & Department of Psychiatry, Yale University July 17, 2011 Memory Activation & APOE ε4 Risk
Understanding. Glaucoma. National Glaucoma Research
 Understanding National Research Understanding What is? is not just one disease, but a group of eye diseases that damage the optic nerve the bundle of nerve fibers that carries information from the eye
Understanding National Research Understanding What is? is not just one disease, but a group of eye diseases that damage the optic nerve the bundle of nerve fibers that carries information from the eye
University of Groningen. Metabolic risk in people with psychotic disorders Bruins, Jojanneke
 University of Groningen Metabolic risk in people with psychotic disorders Bruins, Jojanneke IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from
University of Groningen Metabolic risk in people with psychotic disorders Bruins, Jojanneke IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from
Supplementary Figure 1
 Supplementary Figure 1 a Location of GUUUCA motif relative to RNA position 4e+05 3e+05 Reads 2e+05 1e+05 0e+00 35 40 45 50 55 60 65 Distance upstream b 1: Egg 2: L3 3: Adult 4: HES 4e+05 3e+05 2e+05 start
Supplementary Figure 1 a Location of GUUUCA motif relative to RNA position 4e+05 3e+05 Reads 2e+05 1e+05 0e+00 35 40 45 50 55 60 65 Distance upstream b 1: Egg 2: L3 3: Adult 4: HES 4e+05 3e+05 2e+05 start
Genetics of COPD Prof. Ian P Hall
 Genetics of COPD 1 Prof. Ian P. Hall Dean, Faculty of Medicine and Health Sciences The University of Nottingham Medical School Ian.Hall@nottingham.ac.uk Chronic obstructive pulmonary disease (COPD) 900,000
Genetics of COPD 1 Prof. Ian P. Hall Dean, Faculty of Medicine and Health Sciences The University of Nottingham Medical School Ian.Hall@nottingham.ac.uk Chronic obstructive pulmonary disease (COPD) 900,000
Influence of Genetic Variants in Type I Interferon Genes on Melanoma Survival and Therapy
 Influence of Genetic Variants in Type I Interferon Genes on Melanoma Survival and Therapy Romina Elizabeth Lenci 1, Melanie Bevier 1, Andreas Brandt 1, Justo Lorenzo Bermejo 2, Antje Sucker 3, Iris Moll
Influence of Genetic Variants in Type I Interferon Genes on Melanoma Survival and Therapy Romina Elizabeth Lenci 1, Melanie Bevier 1, Andreas Brandt 1, Justo Lorenzo Bermejo 2, Antje Sucker 3, Iris Moll
Association Between F9 Malmö, Factor IX And Deep Vein Thrombosis
 G T G A G A T G A T A T T T C G A A G A A T A A A G A T G C C C T G G C T T T G G C T T G A T C T C T G G T A C C T T A T G T T T A A A G A A G G A T G G G A A Association Between F9 Malmö, Factor IX And
G T G A G A T G A T A T T T C G A A G A A T A A A G A T G C C C T G G C T T T G G C T T G A T C T C T G G T A C C T T A T G T T T A A A G A A G G A T G G G A A Association Between F9 Malmö, Factor IX And
Supplementary Methods
 Supplementary Methods Populations ascertainment and characterization Our genotyping strategy included 3 stages of SNP selection, with individuals from 3 populations (Europeans, Indian Asians and Mexicans).
Supplementary Methods Populations ascertainment and characterization Our genotyping strategy included 3 stages of SNP selection, with individuals from 3 populations (Europeans, Indian Asians and Mexicans).
DYSREGULATION OF WT1 (-KTS) IS ASSOCIATED WITH THE KIDNEY-SPECIFIC EFFECTS OF THE LMX1B R246Q MUTATION
 DYSREGULATION OF WT1 (-KTS) IS ASSOCIATED WITH THE KIDNEY-SPECIFIC EFFECTS OF THE LMX1B R246Q MUTATION Gentzon Hall 1,2#, Brandon Lane 1,3#, Megan Chryst-Ladd 1,3, Guanghong Wu 1,3, Jen-Jar Lin 4, XueJun
DYSREGULATION OF WT1 (-KTS) IS ASSOCIATED WITH THE KIDNEY-SPECIFIC EFFECTS OF THE LMX1B R246Q MUTATION Gentzon Hall 1,2#, Brandon Lane 1,3#, Megan Chryst-Ladd 1,3, Guanghong Wu 1,3, Jen-Jar Lin 4, XueJun
indicated in shaded lowercase letters (hg19, Chr2: 217,955, ,957,266).
 Legend for Supplementary Figures Figure S1: Sequence of 2q35 encnv. The DNA sequence of the 1,375bp 2q35 encnv is indicated in shaded lowercase letters (hg19, Chr2: 217,955,892-217,957,266). Figure S2:
Legend for Supplementary Figures Figure S1: Sequence of 2q35 encnv. The DNA sequence of the 1,375bp 2q35 encnv is indicated in shaded lowercase letters (hg19, Chr2: 217,955,892-217,957,266). Figure S2:
Radhakrishnan Sabarinathan 1., Anne Wenzel 1., Peter Novotny 2, Xiaojia Tang 3, Krishna R. Kalari 3,4, Jan Gorodkin 1 * Abstract.
 Transcriptome-Wide Analysis of UTRs in Non-Small Cell Lung Cancer Reveals Cancer-Related Genes with SNV- Induced Changes on RNA Secondary Structure and mirna Target Sites Radhakrishnan Sabarinathan 1.,
Transcriptome-Wide Analysis of UTRs in Non-Small Cell Lung Cancer Reveals Cancer-Related Genes with SNV- Induced Changes on RNA Secondary Structure and mirna Target Sites Radhakrishnan Sabarinathan 1.,
Clinical Genetics. Functional validation in a diagnostic context. Robert Hofstra. Leading the way in genetic issues
 Clinical Genetics Leading the way in genetic issues Functional validation in a diagnostic context Robert Hofstra Clinical Genetics Leading the way in genetic issues Future application of exome sequencing
Clinical Genetics Leading the way in genetic issues Functional validation in a diagnostic context Robert Hofstra Clinical Genetics Leading the way in genetic issues Future application of exome sequencing
Supplementary Materials
 1 Supplementary Materials Rotger et al. Table S1A: Demographic characteristics of study participants. VNP RP EC CP (n=6) (n=66) (n=9) (n=5) Male gender, n(%) 5 (83) 54 (82) 5 (56) 3 (60) White ethnicity,
1 Supplementary Materials Rotger et al. Table S1A: Demographic characteristics of study participants. VNP RP EC CP (n=6) (n=66) (n=9) (n=5) Male gender, n(%) 5 (83) 54 (82) 5 (56) 3 (60) White ethnicity,
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 U1 inhibition causes a shift of RNA-seq reads from exons to introns. (a) Evidence for the high purity of 4-shU-labeled RNAs used for RNA-seq. HeLa cells transfected with control
Supplementary Figure 1 U1 inhibition causes a shift of RNA-seq reads from exons to introns. (a) Evidence for the high purity of 4-shU-labeled RNAs used for RNA-seq. HeLa cells transfected with control
02/03/2014. Average Length: 23mm (Infant ~16mm) Approximately the size of a quarter Volume: ~5mL
 Identify the anatomy of the eye. Explain the basic physiology of the parts of the eye. Briefly discuss various surgeries related to different parts of the anatomy. Average Length: 23mm (Infant ~16mm) Approximately
Identify the anatomy of the eye. Explain the basic physiology of the parts of the eye. Briefly discuss various surgeries related to different parts of the anatomy. Average Length: 23mm (Infant ~16mm) Approximately
Association of genetic polymorphisms with chronic obstructive pulmonary disease in the Chinese Han population: a case control study
 Guo et al. BMC Medical Genomics 2012, 5:64 RESEARCH ARTICLE Open Access Association of genetic polymorphisms with chronic obstructive pulmonary disease in the Chinese Han population: a case control study
Guo et al. BMC Medical Genomics 2012, 5:64 RESEARCH ARTICLE Open Access Association of genetic polymorphisms with chronic obstructive pulmonary disease in the Chinese Han population: a case control study
Table S1. Primers used to quantitatively amplify the human mirnas precursors and indicated genes
 Table S1. Primers used to quantitatively amplify the human mirnas precursors and indicated genes Forward primer (5 3 ) Rervese primer (5 3 ) U6 CTCGCTTCGGCAGCACA AACGCTTCACGAATTTGCGT 5S TACGGCCATACCACCCTGAA
Table S1. Primers used to quantitatively amplify the human mirnas precursors and indicated genes Forward primer (5 3 ) Rervese primer (5 3 ) U6 CTCGCTTCGGCAGCACA AACGCTTCACGAATTTGCGT 5S TACGGCCATACCACCCTGAA
Heritability enrichment of differentially expressed genes. Hilary Finucane PGC Statistical Analysis Call January 26, 2016
 Heritability enrichment of differentially expressed genes Hilary Finucane PGC Statistical Analysis Call January 26, 2016 1 Functional genomics + GWAS gives insight into disease relevant tissues Trynka
Heritability enrichment of differentially expressed genes Hilary Finucane PGC Statistical Analysis Call January 26, 2016 1 Functional genomics + GWAS gives insight into disease relevant tissues Trynka
Nature Genetics: doi: /ng Supplementary Figure 1. Study design.
 Supplementary Figure 1 Study design. Leukopenia was classified as early when it occurred within the first 8 weeks of thiopurine therapy and as late when it occurred more than 8 weeks after the start of
Supplementary Figure 1 Study design. Leukopenia was classified as early when it occurred within the first 8 weeks of thiopurine therapy and as late when it occurred more than 8 weeks after the start of
Efficacy of latanoprost in management of chronic angle closure glaucoma. Kumar S 1, Malik A 2 Singh M 3, Sood S 4. Abstract
 Original article Efficacy of latanoprost in management of chronic angle closure glaucoma Kumar S 1, Malik A 2 Singh M 3, Sood S 4 1 Associate Professor, 2 Assistant Professor, 4 Professor, Department of
Original article Efficacy of latanoprost in management of chronic angle closure glaucoma Kumar S 1, Malik A 2 Singh M 3, Sood S 4 1 Associate Professor, 2 Assistant Professor, 4 Professor, Department of
SC-L-H shared(37) Specific (1)
 A. Brain (total 2) Tissue-specific (2) Brain-heart shared (8) Specific (2) CNS-heart shared(68) Specific () Heart (total 4) Tissue-specific () B-L-H shared (37) specific () B. Brain (total 2) Tissue-specific
A. Brain (total 2) Tissue-specific (2) Brain-heart shared (8) Specific (2) CNS-heart shared(68) Specific () Heart (total 4) Tissue-specific () B-L-H shared (37) specific () B. Brain (total 2) Tissue-specific
Variation in linkage disequilibrium patterns between populations of different production types VERONIKA KUKUČKOVÁ, NINA MORAVČÍKOVÁ, RADOVAN KASARDA
 Variation in linkage disequilibrium patterns between populations of different production types VERONIKA KUKUČKOVÁ, NINA MORAVČÍKOVÁ, RADOVAN KASARDA AIM OF THE STUDY THE COMPARISONS INCLUDED DIFFERENCES
Variation in linkage disequilibrium patterns between populations of different production types VERONIKA KUKUČKOVÁ, NINA MORAVČÍKOVÁ, RADOVAN KASARDA AIM OF THE STUDY THE COMPARISONS INCLUDED DIFFERENCES
Prediction of micrornas and their targets
 Prediction of micrornas and their targets Introduction Brief history mirna Biogenesis Computational Methods Mature and precursor mirna prediction mirna target gene prediction Summary micrornas? RNA can
Prediction of micrornas and their targets Introduction Brief history mirna Biogenesis Computational Methods Mature and precursor mirna prediction mirna target gene prediction Summary micrornas? RNA can
Genome Summary. Sequencing Coverage: Variation Counts: Known Phenotype Summary: 131 disease or trait variations are found in this genome
 Report Date: August 2, 215 Software Annotation Version: 8 Genome Summary Name: Greg Mendel Genome ID: genome_greg_mendel_dad Sequencing Provider: 23andMe Sequencing Type: Genotyping SNP Array Sequencing
Report Date: August 2, 215 Software Annotation Version: 8 Genome Summary Name: Greg Mendel Genome ID: genome_greg_mendel_dad Sequencing Provider: 23andMe Sequencing Type: Genotyping SNP Array Sequencing
Training Checking Vision Tonometry
 Training 101-2 Checking Vision Tonometry Checking Vision The classic example of an eye chart is the Snellen eye chart, in general they show 11 rows of capital letters. The top row contains one letter (usually
Training 101-2 Checking Vision Tonometry Checking Vision The classic example of an eye chart is the Snellen eye chart, in general they show 11 rows of capital letters. The top row contains one letter (usually
The genetic architecture of type 2 diabetes appears
 ORIGINAL ARTICLE A 100K Genome-Wide Association Scan for Diabetes and Related Traits in the Framingham Heart Study Replication and Integration With Other Genome-Wide Datasets Jose C. Florez, 1,2,3 Alisa
ORIGINAL ARTICLE A 100K Genome-Wide Association Scan for Diabetes and Related Traits in the Framingham Heart Study Replication and Integration With Other Genome-Wide Datasets Jose C. Florez, 1,2,3 Alisa
Supplemental Table 1. Components of MDS and AHEI
 Supplemental Table 1. Components of MDS and AHEI MDS AHEI Vegetable Fruit SSB & fruit juice Nut Legume Whole grain Fish Red meat MUFA/SAT ratio EPA & DHA PUFA Trans-fat Alcohol Sodium MDS: Mediterranean-style
Supplemental Table 1. Components of MDS and AHEI MDS AHEI Vegetable Fruit SSB & fruit juice Nut Legume Whole grain Fish Red meat MUFA/SAT ratio EPA & DHA PUFA Trans-fat Alcohol Sodium MDS: Mediterranean-style
OIS Presentation October 25, 2018
 OIS Presentation October 25, 2018 Forward looking statements This presentation has been prepared by Aerpio Pharmaceuticals ( we, us or, the Company ) and includes forward-looking statements. All statements
OIS Presentation October 25, 2018 Forward looking statements This presentation has been prepared by Aerpio Pharmaceuticals ( we, us or, the Company ) and includes forward-looking statements. All statements
The Pathology and Pathogenesis of Acute Glaucoma in Dogs. Richard R Dubielzig
 The Pathology and Pathogenesis of Acute Glaucoma in Dogs Richard R Dubielzig Overview of Glaucoma Intraocular Pressure too High to Support a Healthy Optic Nerve Terminology used in the classification of
The Pathology and Pathogenesis of Acute Glaucoma in Dogs Richard R Dubielzig Overview of Glaucoma Intraocular Pressure too High to Support a Healthy Optic Nerve Terminology used in the classification of
Analysis Summary: Male Pattern Baldness
 Analysis Summary: Male Pattern Baldness Phenotype Description The male pattern baldness phenotype was defined for use in a recent meta analysis (Li et al., PLoS Genet 8: e1002746, 2012). All subjects are
Analysis Summary: Male Pattern Baldness Phenotype Description The male pattern baldness phenotype was defined for use in a recent meta analysis (Li et al., PLoS Genet 8: e1002746, 2012). All subjects are
SUPPLEMENTARY DATA. 1. Characteristics of individual studies
 1. Characteristics of individual studies 1.1. RISC (Relationship between Insulin Sensitivity and Cardiovascular disease) The RISC study is based on unrelated individuals of European descent, aged 30 60
1. Characteristics of individual studies 1.1. RISC (Relationship between Insulin Sensitivity and Cardiovascular disease) The RISC study is based on unrelated individuals of European descent, aged 30 60
Closed Angle Glaucoma Or Narrow Angle Glaucoma. What s is a closed angle type of glaucoma,
 Closed Angle Glaucoma Or Narrow Angle Glaucoma What s is a closed angle type of glaucoma, This is where the iris is found to be blocking the drainage of the eye through the trabecular meshwork. The eye
Closed Angle Glaucoma Or Narrow Angle Glaucoma What s is a closed angle type of glaucoma, This is where the iris is found to be blocking the drainage of the eye through the trabecular meshwork. The eye
Determination of genetic effects of ATF3 and CDKN1A genes on milk yield and compositions in Chinese Holstein population
 Han et al. BMC Genetics (2017) 18:47 DOI 10.1186/s12863-017-0516-4 RESEARCH ARTICLE Open Access Determination of genetic effects of ATF3 and CDKN1A genes on milk yield and compositions in Chinese Holstein
Han et al. BMC Genetics (2017) 18:47 DOI 10.1186/s12863-017-0516-4 RESEARCH ARTICLE Open Access Determination of genetic effects of ATF3 and CDKN1A genes on milk yield and compositions in Chinese Holstein
A LITTLE ANATOMY. three layers of eye: 1. outer: corneosclera. 2. middle - uvea. anterior - iris,ciliary body. posterior - choroid
 GLAUCOMA A LITTLE ANATOMY three layers of eye: 1. outer: corneosclera 2. middle - uvea anterior - iris,ciliary body posterior - choroid connection at the pars plana between post and ant uvea 3. retina
GLAUCOMA A LITTLE ANATOMY three layers of eye: 1. outer: corneosclera 2. middle - uvea anterior - iris,ciliary body posterior - choroid connection at the pars plana between post and ant uvea 3. retina
Supplementary Information IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy.
 Supplementary Information IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Vijayaprakash Suppiah, Max Moldovan, Golo Ahlenstiel, Thomas Berg, Martin Weltman,
Supplementary Information IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Vijayaprakash Suppiah, Max Moldovan, Golo Ahlenstiel, Thomas Berg, Martin Weltman,
Supplementary Information
 Supplementary Information Index of Supplementary Materials Supplementary Figure. Distribution of methylation levels, characteristics of cis-meqtls and association plots for meqtls in methylation-related
Supplementary Information Index of Supplementary Materials Supplementary Figure. Distribution of methylation levels, characteristics of cis-meqtls and association plots for meqtls in methylation-related
BST227 Introduction to Statistical Genetics. Lecture 4: Introduction to linkage and association analysis
 BST227 Introduction to Statistical Genetics Lecture 4: Introduction to linkage and association analysis 1 Housekeeping Homework #1 due today Homework #2 posted (due Monday) Lab at 5:30PM today (FXB G13)
BST227 Introduction to Statistical Genetics Lecture 4: Introduction to linkage and association analysis 1 Housekeeping Homework #1 due today Homework #2 posted (due Monday) Lab at 5:30PM today (FXB G13)
Supplementary Figure 1
 Supplementary Figure 1 5 microns C7 B6 unclassified H19 C7 signal H19 guide signal H19 B6 signal C7 SNP spots H19 RNA spots B6 SNP spots colocalization H19 RNA classification Supplementary Figure 1. Allele-specific
Supplementary Figure 1 5 microns C7 B6 unclassified H19 C7 signal H19 guide signal H19 B6 signal C7 SNP spots H19 RNA spots B6 SNP spots colocalization H19 RNA classification Supplementary Figure 1. Allele-specific
Supplementary Figure 1 a
 Supplementary Figure a Normalized expression/tbp (A.U.).6... Trip-br transcripts Trans Trans Trans b..5. Trip-br Ctrl LPS Normalized expression/tbp (A.U.) c Trip-br transcripts. adipocytes.... Trans Trans
Supplementary Figure a Normalized expression/tbp (A.U.).6... Trip-br transcripts Trans Trans Trans b..5. Trip-br Ctrl LPS Normalized expression/tbp (A.U.) c Trip-br transcripts. adipocytes.... Trans Trans
Supplementary information for: Human micrornas co-silence in well-separated groups and have different essentialities
 Supplementary information for: Human micrornas co-silence in well-separated groups and have different essentialities Gábor Boross,2, Katalin Orosz,2 and Illés J. Farkas 2, Department of Biological Physics,
Supplementary information for: Human micrornas co-silence in well-separated groups and have different essentialities Gábor Boross,2, Katalin Orosz,2 and Illés J. Farkas 2, Department of Biological Physics,
Deliverable 2.1 List of relevant genetic variants for pre-emptive PGx testing
 GA N 668353 H2020 Research and Innovation Deliverable 2.1 List of relevant genetic variants for pre-emptive PGx testing WP N and Title: WP2 - Towards shared European Guidelines for PGx Lead beneficiary:
GA N 668353 H2020 Research and Innovation Deliverable 2.1 List of relevant genetic variants for pre-emptive PGx testing WP N and Title: WP2 - Towards shared European Guidelines for PGx Lead beneficiary:
Association between vitamin D receptor gene polymorphism (TaqI) and obesity in Chinese population
 c Indian Academy of Sciences RESEARCH NOTE Association between vitamin D receptor gene polymorphism (TaqI) and obesity in Chinese population HUI-RU FAN, LI-QUN LIN, HAO MA, YING LI and CHANG-HAO SUN National
c Indian Academy of Sciences RESEARCH NOTE Association between vitamin D receptor gene polymorphism (TaqI) and obesity in Chinese population HUI-RU FAN, LI-QUN LIN, HAO MA, YING LI and CHANG-HAO SUN National
Nutritional Epidemiology in the Genomic Age. Saroja Voruganti, PhD Assistant Professor Department of Nutrition and UNC Nutrition Research Institute
 Nutritional Epidemiology in the Genomic Age Saroja Voruganti, PhD Assistant Professor Department of Nutrition and UNC Nutrition Research Institute Learning objectives Types of genetic association approaches
Nutritional Epidemiology in the Genomic Age Saroja Voruganti, PhD Assistant Professor Department of Nutrition and UNC Nutrition Research Institute Learning objectives Types of genetic association approaches
Figure S1: Heat map based on the relative expression of genes and mirnas in human
 Figure S1: Heat map based on the relative expression of genes and mirnas in human prostate tissue. Columns represent genes and mirnas; rows represent prostate tissue samples (red squares: Tu samples, green
Figure S1: Heat map based on the relative expression of genes and mirnas in human prostate tissue. Columns represent genes and mirnas; rows represent prostate tissue samples (red squares: Tu samples, green
Rare Variant Burden Tests. Biostatistics 666
 Rare Variant Burden Tests Biostatistics 666 Last Lecture Analysis of Short Read Sequence Data Low pass sequencing approaches Modeling haplotype sharing between individuals allows accurate variant calls
Rare Variant Burden Tests Biostatistics 666 Last Lecture Analysis of Short Read Sequence Data Low pass sequencing approaches Modeling haplotype sharing between individuals allows accurate variant calls
Tutorial on Genome-Wide Association Studies
 Tutorial on Genome-Wide Association Studies Assistant Professor Institute for Computational Biology Department of Epidemiology and Biostatistics Case Western Reserve University Acknowledgements Dana Crawford
Tutorial on Genome-Wide Association Studies Assistant Professor Institute for Computational Biology Department of Epidemiology and Biostatistics Case Western Reserve University Acknowledgements Dana Crawford
RmiR package vignette
 RmiR package vignette Francesco Favero * October 17, 2016 Contents 1 Introduction 1 2 Coupling mirna and gene expression data 2 2.1 Default parameters.............................. 3 2.2 Change the at.least
RmiR package vignette Francesco Favero * October 17, 2016 Contents 1 Introduction 1 2 Coupling mirna and gene expression data 2 2.1 Default parameters.............................. 3 2.2 Change the at.least
