Genome-editing via Oviductal Nucleic Acids Delivery (GONAD) system: a novel microinjection-independent genome engineering method in mice
|
|
|
- Griffin Osborne
- 5 years ago
- Views:
Transcription
1 Supplementary Information Genome-editing via Oviductal Nucleic Acids Delivery (GONAD) system: a novel microinjection-independent genome engineering method in mice Gou Takahashi, Channabasavaiah B Gurumurthy, Kenta Wada, Hiromi Miura, Masahiro Sato, Masato Ohtsuka
2 Supplementary Tables Supplementary Table S1 Supplementary Table S1 Delivery of egfp mrna to pre-implantation embryos in oviducts in vitro 1 Day 1.5 : 2-cell stages Day 2.5 : 8-cell to16-cell stages Day 3.5 : morula to blastocyst stages No. of embryos recoverd Total Normal Abnormal 2 No. of embryos normally developed (No. of embryos showing fluorescence) No. of embryos normally developed (No. of embryos showing fluorescence) (5) 16 (4) 1 egfp mrna (500 ng/!l) was electroporated into a dissected oviduct isolated from C57BL/6N mice (A-#01) as described in Methods. 2-cell embryos were flushed from the treated oviducts and cultured for two days up to morula or blastocyst stages. 2 Abnomal embryos were defined as those showing fragmentation, developmental arrest or degeneration.
3 Supplementary Table S2 Supplementary Table S2 Delivery of egfp mrna to pre-implantation embryos in oviducts in situ 1 Group Experimental group Control group Mice Fluorescence Day 2.5 : 8-cell to 16-cell stages No. of embryos recoverd (No. of embryos showing fluorescence) Day 3.5 : morula to blastocyst stages No. of embryos developed (No. of embryos showing fluorescence) of oviducts 2 Total Normal Abnormal 3 Total Normal Abnormal 3 B-# (1) 6 (0) 5 (1) 11 (1) 5 (0) 6 (1) B-# (6) 3 (3) 5 (3) 8 (4) 2 (2) 6 (2) B-# (1) 2 (1) 7 (0) 9 (1) 2 (1) 7 (0) B-# (0) 0 (0) 4 (0) 4 (0) 0 (0) 4 (0) B-# (5) 10 (4) 3 (1) 13 (4) 10 (3) 3 (1) B-# (0) 4 (0) 3 (0) 7 (0) 4 (0) 3 (0) Total 52 (13) 25 (8) 27 (5) 52 (10) 23 (6) 29 (4) B-# B-# B-# Total egfp mrna (500 ng/!l) was electroporated into an intact oviduct of ICR mice as described in Methods. One day after electroporation, embryos were flushed from treated oviducts and then cultured up to morula to blastocyst stages. No electroporation was performed for the control group. 2 At least one oviduct of the two oviducts per mouse exhibiting fluorescence was defined as +. 3 Abnormal embryos were defined those showing fragmentation, developmental arrest or degeneration.
4 Supplementary Table S3 Supplementary Table S3 Delivery of CRISPR/Cas9 RNAs to pre-implantation embryos in oviducts in vitro 1 Group Experimental group Mice RNAs injected (concentration) Day 1.5 : 2-cell stage No. of embryos recoverd Total Normal Abnormal 2 Day 2.5 : 8-cell to 16-cell stages No. of embryos normally developed (No. of embryos showing fluorescence) Day 3.5 : morula to blastocyst stages No. of embryos normally developed (No. of embryos showing fluorescence) C-# (6) 12 (6) 1 C-# (1) 10 (1) 0 egfp mrna (150 ng/!l) C-#03 Cas9 mrna (500 ng/!l) (2) 11 (2) 0 Hprt_Cr1_sgRNA (50 ng/!l) C-# (5) 22 (5) 0 C-# (0) 9 (0) 0 No. of embryos showing indel C-#06 egfp mrna (200 ng/!l) (1) 16 (1) 0 Cas9 mrna (400 ng/!l) C-#07 Hprt_Cr1_sgRNA (100 ng/!l) (2) 5 (2) 1 C-#08 Cas9_mRNA (923 ng/!l) (NA) 6 (NA) 0 C-#09 eef2_cr1_sgrna (243 ng/!l) (NA) 10 (NA) 2 C-#10 Cas9_mRNA (500 ng/!l) (NA) 24 (NA) 0 C-#11 eef2_cr2_sgrna (409 ng/!l) (NA) 9 (NA) 0 C-#12 Cas9 mrna (444 ng/!l) (NA) 5 (NA) 0 C-#13 egfp_sgrna (133 ng/!l) (NA) 7 (NA) 1 mutation 3 C-# N.D. Control group - C-# N.D. 1 RNAs were electroporated into a dissected oviduct isolated from C57BL/6N mice as described in Methods. 2-cell embryos were flushed from treated oviducts and then cultured for two days up to morula to blastocyst stages. No electroporation was performed for the control group. 2 Abnomal embryos were defined those showing fragmentation, developmental arrest or degeneration. 3 Indel mutation analysis was performed using Surveyor- or T7E1-assay from genomic DNA of separate embryos. Sequencing was performed to confirm the mutations in some cases. N.D.= not done.
5 Supplementary Table S4 Supplementary Table S4 Primer sets for sgrna synthesis, Surveyor- and T7E1-assay Primer sets for sgrna synthesis Primer sets for Surveyor- and T7E1-assay *Primers used for direct sequencing. Target Primer name Primer sequence (5' to 3') Product size TAATACGACTCACTATAGGGACC egfp F M940 AGGATGGGCACCACCCGTTTTA GAGCTAGAAATAGCAAG 122-bp R M939 AAAAAAAGCACCGACTCGG Hprt_Cr1 eef2_cr1 TAATACGACTCACTATAGGGCTA F PP105 CCGGAAAGGGCCCGGAGTTTTA F M938 TAATACGACTCACTATAGG GAGCTAGAAATAGCAAG R M939 AAAAAAAGCACCGACTCGG 122-bp 122-bp R M939 AAAAAAAGCACCGACTCGG eef2_cr2 egfp_1st PCR egfp_2nd PCR Hprt_Cr1_1st PCR Hprt_Cr1_2nd PCR eef2_cr1_1st PCR eef2_cr1_2nd PCR eef2_cr2_1st PCR eef2_cr2_2nd PCR F PP106 TAATACGACTCACTATAGGGGGA GCGACTTCCCAAGGCTGTTTTA GAGCTAGAAATAGCAAG R M939 AAAAAAAGCACCGACTCGG 1F M212 CTCCTGGGCAACGTGCTGGT 1R M495 AAGAAGATGGTGCGCTCCTG 2F M389 TCGCCACCATGGTGAGCAAGG GCGAG 2R M026* GGTGGTGCAGATGAACTTCAG 1F M925 TGATGCAGGACGATTTCAAG 1R M926 GGATGGTTGTGAGCCATGAT 2F M949* GCCAGCCAGATCCTATCTAAA 2R M950 TGACAAGAAGATGGGAATGG 1F PP113 ATATCTGCGCGTCCCTGA 1R PP114 TGAGGCTCAAGTCACATAACC 2F PP115* TTGTGCTTGGTGATGTGG 2R PP116 CACACACCTCTTCCCCAAA 1F PP117 TGGGGAGACCGGTGAGTA 1R PP118 GAGGCCCCTCGTATAGCA 2F PP119 GCCAATGGCAAGTTCAGTA 2R PP120 GCCTTGAGCAATGGCTTG 122-bp 376-bp 162-bp 609-bp 445-bp 525-bp 301-bp 527-bp 301-bp
6 Supplementary Figures Supplementary Figure S1 Supplementary Figure S1 egfp expression in 8- to 16-cell stage embryos recovered from the dissected oviducts that were in vitro electroporated with egfp mrna. The embryos were derived from A-#01 mouse (C57BL/6N strain) shown in Supplementary Table S1. The embryos exhibiting fluorescence throughout are shown with red arrows. Scale bar = 100 µm.
7 Supplementary Figure S2 Supplementary Figure S2 CRISPR/Cas9-mediated disruption of Hprt locus achieved through in vitro electroporation of oviducts. (a) egfp fluorescence in the blastocysts, one day after culturing of embryos recovered from an in vitro electroporated mouse with egfp mrna (used as an indicator of electroporation), Cas9 mrna and Hprt_Cr1 sgrna (C-#01 mouse: Supplementary Table S3). Scale bar = 100 µm. (b) Agarose gel electrophoresis of a few Surveyor-treated PCR products derived from egfp fluorescence-positive embryos (C-#01_No.36, 42, 43 and 44). The red arrows indicate the cleavage products generated in Surveyor assay and the wild-type sized band is indicated by a black arrow M: 100-bp DNA ladder markers. N.C: a Negative Control embryo derived from a non-instilled and non-electroporated mouse. (c) Direct sequencing of the PCR products from wild-type mouse or a mutated blastocyst (C-#01_No.42, shown in (b)). The black arrow below the electropherogram shows overlapping peaks indicative of indel mutations (see text for details).
8 Supplementary Figure S3 Supplementary Figure S3 CRISPR/Cas9-mediated disruption of eef2 locus achieved through in vitro electroporation of oviducts. (a) Bright field micrograph of embryos collected and cultured for two-days after in vitro electroporation of CRISPR/Cas9 RNAs against the eef2 gene (egfp mrna, as an indicator of electroporation, was not included in this experiment). The embryos from two different mice (C57BL/6N strain) are shown: the upper panel embryos were recovered from the C-#08 mouse (Supplementary Table S3) and the bottom panel embryos were recovered from the C-#09 mouse. Scale bars = 100 µm. (b) Agarose gel electrophoresis of T7E1-treated PCR products derived from the embryos shown in (a). The red arrows indicate the cleavage products generated in T7E1 assay and the wild-type sized band is indicated by a black arrow. D.W: distilled water used as negative control. M: 100-bp DNA Ladder. (c) Direct sequencing of the PCR products of the target region amplified from samples C-#09_No.14 and _No.16. The black arrow below the electropherogram shows overlapping peaks indicative of indel mutations (see text for details).
9 Supplementary Figure S4 Supplementary Figure S4 CRISPR/Cas9-mediated disruption of egfp transgene achieved through in vitro electroporation of oviducts. (a) egfp fluorescence in the blastocysts, two-days after culturing of embryos recovered from an in vitro electroporated mouse (C57BL/6N strain mated with hemizygote egfp Tg males) with Cas9 mrna, and egfp sgrna. One embryos each from two different mice are shown: the upper panel embryo showing normal egfp fluorescence recovered from the C-#12 mouse (Supplementary Table S3) and the bottom panel embryo showing greatly reduced fluorescence was recovered from the C-#13 mouse. Scale bars: 100 µm. (b) Direct sequencing of the PCR product amplified from the target region from bottom panel s embryo in (a). The black arrow below the electropherogram shows overlapping peaks indicative of indel mutations (see text for details).
10 Supplementary Figure S5 Supplementary Figure S5 CRISPR/Cas9 mediated disruption of Hprt locus using the GONAD system. (a) egfp fluorescence at the 8-cell to 16-cell embryos (left side panels) and blastocyst embryos (right side panels) that were collected from GONAD procedure performed using egfp mrna (used as an indicator of electroporation), Cas9 mrna and Hprt_Cr1 sgrna (Table 1). Scale bars = 100 µm. (b) Agarose gel electrophoresis of T7E1-treated PCR products derived from 7 selected blastocysts. The red arrows indicate the cleavage products generated in T7E1 assay and the wild-type sized band is indicated by a black arrow. M: 100-bp DNA ladder marker. (c) Direct sequencing of PCR products amplified from the target region from wild-type, D-#05_No.5, _No.6, and _No.7 samples. The black arrow below the electropherogram shows overlapping peaks indicative of indel mutations (see text for details).
11 Supplementary Figure S6 Figure S6 Mutated egfp alleles possessed in founder fetuses. The changes in the nucleotide sequence are shown in red and the type of changes (insertions: + or deletions: Δ) are indicated on the right-side of the sequences. All fetuses had only one type of mutation except the fetus 7 of Figure 4 which was a mosaic with two mutant alleles (shown in a box).
Supplementary methods:
 Supplementary methods: Primers sequences used in real-time PCR analyses: β-actin F: GACCTCTATGCCAACACAGT β-actin [11] R: AGTACTTGCGCTCAGGAGGA MMP13 F: TTCTGGTCTTCTGGCACACGCTTT MMP13 R: CCAAGCTCATGGGCAGCAACAATA
Supplementary methods: Primers sequences used in real-time PCR analyses: β-actin F: GACCTCTATGCCAACACAGT β-actin [11] R: AGTACTTGCGCTCAGGAGGA MMP13 F: TTCTGGTCTTCTGGCACACGCTTT MMP13 R: CCAAGCTCATGGGCAGCAACAATA
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Analysis of hair bundle morphology in Ush1c c.216g>a mice at P18 by SEM.
 Supplementary Figure 1 Analysis of hair bundle morphology in Ush1c c.216g>a mice at P18 by SEM. (a-c) Heterozygous c.216ga mice displayed normal hair bundle morphology at P18. (d-i) Disorganized hair bundles
Supplementary Figure 1 Analysis of hair bundle morphology in Ush1c c.216g>a mice at P18 by SEM. (a-c) Heterozygous c.216ga mice displayed normal hair bundle morphology at P18. (d-i) Disorganized hair bundles
Supplemental Information. Fluorescence-based visualization of autophagic activity predicts mouse embryo
 Supplemental Information Fluorescence-based visualization of autophagic activity predicts mouse embryo viability Satoshi Tsukamoto*, Taichi Hara, Atsushi Yamamoto, Seiji Kito, Naojiro Minami, Toshiro Kubota,
Supplemental Information Fluorescence-based visualization of autophagic activity predicts mouse embryo viability Satoshi Tsukamoto*, Taichi Hara, Atsushi Yamamoto, Seiji Kito, Naojiro Minami, Toshiro Kubota,
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1171320/dc1 Supporting Online Material for A Frazzled/DCC-Dependent Transcriptional Switch Regulates Midline Axon Guidance Long Yang, David S. Garbe, Greg J. Bashaw*
www.sciencemag.org/cgi/content/full/1171320/dc1 Supporting Online Material for A Frazzled/DCC-Dependent Transcriptional Switch Regulates Midline Axon Guidance Long Yang, David S. Garbe, Greg J. Bashaw*
Nature Medicine: doi: /nm.4322
 1 2 3 4 5 6 7 8 9 10 11 Supplementary Figure 1. Predicted RNA structure of 3 UTR and sequence alignment of deleted nucleotides. (a) Predicted RNA secondary structure of ZIKV 3 UTR. The stem-loop structure
1 2 3 4 5 6 7 8 9 10 11 Supplementary Figure 1. Predicted RNA structure of 3 UTR and sequence alignment of deleted nucleotides. (a) Predicted RNA secondary structure of ZIKV 3 UTR. The stem-loop structure
The subcortical maternal complex controls symmetric division of mouse zygotes by
 The subcortical maternal complex controls symmetric division of mouse zygotes by regulating F-actin dynamics Xing-Jiang Yu 1,2, Zhaohong Yi 1, Zheng Gao 1,2, Dan-dan Qin 1,2, Yanhua Zhai 1, Xue Chen 1,
The subcortical maternal complex controls symmetric division of mouse zygotes by regulating F-actin dynamics Xing-Jiang Yu 1,2, Zhaohong Yi 1, Zheng Gao 1,2, Dan-dan Qin 1,2, Yanhua Zhai 1, Xue Chen 1,
Supplementary Materials and Methods
 Supplementary Materials and Methods Whole Mount X-Gal Staining Whole tissues were collected, rinsed with PBS and fixed with 4% PFA. Tissues were then rinsed in rinse buffer (100 mm Sodium Phosphate ph
Supplementary Materials and Methods Whole Mount X-Gal Staining Whole tissues were collected, rinsed with PBS and fixed with 4% PFA. Tissues were then rinsed in rinse buffer (100 mm Sodium Phosphate ph
Conditional and reversible disruption of essential herpesvirus protein functions
 nature methods Conditional and reversible disruption of essential herpesvirus protein functions Mandy Glaß, Andreas Busche, Karen Wagner, Martin Messerle & Eva Maria Borst Supplementary figures and text:
nature methods Conditional and reversible disruption of essential herpesvirus protein functions Mandy Glaß, Andreas Busche, Karen Wagner, Martin Messerle & Eva Maria Borst Supplementary figures and text:
Supplementary Information. Supplementary Figure 1
 Supplementary Information Supplementary Figure 1 1 Supplementary Figure 1. Functional assay of the hcas9-2a-mcherry construct (a) Gene correction of a mutant EGFP reporter cell line mediated by hcas9 or
Supplementary Information Supplementary Figure 1 1 Supplementary Figure 1. Functional assay of the hcas9-2a-mcherry construct (a) Gene correction of a mutant EGFP reporter cell line mediated by hcas9 or
Joint Department of Biomedical Engineering
 Electronic Supplementary Material (ESI) for Biomaterials Science. This journal is The Royal Society of Chemistry 2018 Supplementary Data Systemic Delivery of CRISPR/Cas9 with PEG-PLGA Nanoparticles for
Electronic Supplementary Material (ESI) for Biomaterials Science. This journal is The Royal Society of Chemistry 2018 Supplementary Data Systemic Delivery of CRISPR/Cas9 with PEG-PLGA Nanoparticles for
Inhibition of Cdk5 Promotes β-cell Differentiation from Ductal Progenitors
 Inhibition of Cdk5 Promotes β-cell Differentiation from Ductal Progenitors Ka-Cheuk Liu, Gunter Leuckx, Daisuke Sakano, Philip A. Seymour, Charlotte L. Mattsson, Linn Rautio, Willem Staels, Yannick Verdonck,
Inhibition of Cdk5 Promotes β-cell Differentiation from Ductal Progenitors Ka-Cheuk Liu, Gunter Leuckx, Daisuke Sakano, Philip A. Seymour, Charlotte L. Mattsson, Linn Rautio, Willem Staels, Yannick Verdonck,
Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid.
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
Host Double Strand Break Repair Generates HIV-1 Strains Resistant to CRISPR/Cas9
 Host Double Strand Break Repair Generates HIV-1 Strains Resistant to CRISPR/Cas9 Kristine E. Yoder, a * and Ralf Bundschuh b a Department of Molecular Virology, Immunology and Medical Genetics, Center
Host Double Strand Break Repair Generates HIV-1 Strains Resistant to CRISPR/Cas9 Kristine E. Yoder, a * and Ralf Bundschuh b a Department of Molecular Virology, Immunology and Medical Genetics, Center
Nature Biotechnology: doi: /nbt Supplementary Figure 1. PL gene expression in tomato fruit.
 Supplementary Figure 1 PL gene expression in tomato fruit. Relative expression of five PL-coding genes measured in at least three fruit of each genotype (cv. Alisa Craig) at four stages of development,
Supplementary Figure 1 PL gene expression in tomato fruit. Relative expression of five PL-coding genes measured in at least three fruit of each genotype (cv. Alisa Craig) at four stages of development,
Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated
 Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C genes. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated from 7 day-old seedlings treated with or without
Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C genes. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated from 7 day-old seedlings treated with or without
Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein
 Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein content relative to GAPDH in two independent experiments.
Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein content relative to GAPDH in two independent experiments.
reads observed in trnas from the analysis of RNAs carrying a 5 -OH ends isolated from cells induced to express
 Supplementary Figure 1. VapC-mt4 cleaves trna Ala2 in E. coli. Histograms representing the fold change in reads observed in trnas from the analysis of RNAs carrying a 5 -OH ends isolated from cells induced
Supplementary Figure 1. VapC-mt4 cleaves trna Ala2 in E. coli. Histograms representing the fold change in reads observed in trnas from the analysis of RNAs carrying a 5 -OH ends isolated from cells induced
Supplementary Figure 1. Schematic diagram of o2n-seq. Double-stranded DNA was sheared, end-repaired, and underwent A-tailing by standard protocols.
 Supplementary Figure 1. Schematic diagram of o2n-seq. Double-stranded DNA was sheared, end-repaired, and underwent A-tailing by standard protocols. A-tailed DNA was ligated to T-tailed dutp adapters, circularized
Supplementary Figure 1. Schematic diagram of o2n-seq. Double-stranded DNA was sheared, end-repaired, and underwent A-tailing by standard protocols. A-tailed DNA was ligated to T-tailed dutp adapters, circularized
Supplementary Material
 Supplementary Material Summary: The supplementary information includes 1 table (Table S1) and 4 figures (Figure S1 to S4). Supplementary Figure Legends Figure S1 RTL-bearing nude mouse model. (A) Tumor
Supplementary Material Summary: The supplementary information includes 1 table (Table S1) and 4 figures (Figure S1 to S4). Supplementary Figure Legends Figure S1 RTL-bearing nude mouse model. (A) Tumor
Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A.
 Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A. Upper part, three-primer PCR strategy at the Mcm3 locus yielding
Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A. Upper part, three-primer PCR strategy at the Mcm3 locus yielding
Generating Mouse Models of Pancreatic Cancer
 Generating Mouse Models of Pancreatic Cancer Aom Isbell http://www2.massgeneral.org/cancerresourceroom/types/gi/index.asp Spring/Summer 1, 2012 Alexandros Tzatsos, MD PhD Bardeesy Lab: Goals and Objectives
Generating Mouse Models of Pancreatic Cancer Aom Isbell http://www2.massgeneral.org/cancerresourceroom/types/gi/index.asp Spring/Summer 1, 2012 Alexandros Tzatsos, MD PhD Bardeesy Lab: Goals and Objectives
EGFR shrna A: CCGGCGCAAGTGTAAGAAGTGCGAACTCGAGTTCGCACTTCTTACACTTGCG TTTTTG. EGFR shrna B: CCGGAGAATGTGGAATACCTAAGGCTCGAGCCTTAGGTATTCCACATTCTCTT TTTG
 Supplementary Methods Sequence of oligonucleotides used for shrna targeting EGFR EGFR shrna were obtained from the Harvard RNAi consortium. The following oligonucleotides (forward primer) were used to
Supplementary Methods Sequence of oligonucleotides used for shrna targeting EGFR EGFR shrna were obtained from the Harvard RNAi consortium. The following oligonucleotides (forward primer) were used to
Abstract. Introduction. RBMOnline - Vol 8. No Reproductive BioMedicine Online; on web 10 December 2003
 RBMOnline - Vol 8. No 2. 224-228 Reproductive BioMedicine Online; www.rbmonline.com/article/1133 on web 10 December 2003 Article Preimplantation genetic diagnosis for early-onset torsion dystonia Dr Svetlana
RBMOnline - Vol 8. No 2. 224-228 Reproductive BioMedicine Online; www.rbmonline.com/article/1133 on web 10 December 2003 Article Preimplantation genetic diagnosis for early-onset torsion dystonia Dr Svetlana
Supplemental Figure 1. Genes showing ectopic H3K9 dimethylation in this study are DNA hypermethylated in Lister et al. study.
 mc mc mc mc SUP mc mc Supplemental Figure. Genes showing ectopic HK9 dimethylation in this study are DNA hypermethylated in Lister et al. study. Representative views of genes that gain HK9m marks in their
mc mc mc mc SUP mc mc Supplemental Figure. Genes showing ectopic HK9 dimethylation in this study are DNA hypermethylated in Lister et al. study. Representative views of genes that gain HK9m marks in their
CRISPR-mediated Editing of Hematopoietic Stem Cells for the Treatment of β-hemoglobinopathies
 CRISPR-mediated Editing of Hematopoietic Stem Cells for the Treatment of β-hemoglobinopathies Jennifer Gori American Society of Gene & Cell Therapy May 11, 2017 editasmedicine.com 1 Highlights Developed
CRISPR-mediated Editing of Hematopoietic Stem Cells for the Treatment of β-hemoglobinopathies Jennifer Gori American Society of Gene & Cell Therapy May 11, 2017 editasmedicine.com 1 Highlights Developed
mir-7a regulation of Pax6 in neural stem cells controls the spatial origin of forebrain dopaminergic neurons
 Supplemental Material mir-7a regulation of Pax6 in neural stem cells controls the spatial origin of forebrain dopaminergic neurons Antoine de Chevigny, Nathalie Coré, Philipp Follert, Marion Gaudin, Pascal
Supplemental Material mir-7a regulation of Pax6 in neural stem cells controls the spatial origin of forebrain dopaminergic neurons Antoine de Chevigny, Nathalie Coré, Philipp Follert, Marion Gaudin, Pascal
Supplementary Information
 Supplementary Information Overexpression of Fto leads to increased food intake and results in obesity Chris Church, Lee Moir, Fiona McMurray, Christophe Girard, Gareth T Banks, Lydia Teboul, Sara Wells,
Supplementary Information Overexpression of Fto leads to increased food intake and results in obesity Chris Church, Lee Moir, Fiona McMurray, Christophe Girard, Gareth T Banks, Lydia Teboul, Sara Wells,
Fig. S1. Weight of embryos during development. Embryos are collected at different time points (E12.5, E14.5, E16.5 and E18.5) from matings between
 Fig. S1. Weight of embryos during development. Embryos are collected at different time points (E12.5, E14.5, E16.5 and E18.5) from matings between Myod +/ or Myod / females and Myod +/ ;Igf2 +/ males and
Fig. S1. Weight of embryos during development. Embryos are collected at different time points (E12.5, E14.5, E16.5 and E18.5) from matings between Myod +/ or Myod / females and Myod +/ ;Igf2 +/ males and
Soluble ADAM33 initiates airway remodeling to promote susceptibility for. Elizabeth R. Davies, Joanne F.C. Kelly, Peter H. Howarth, David I Wilson,
 Revised Suppl. Data: Soluble ADAM33 1 Soluble ADAM33 initiates airway remodeling to promote susceptibility for allergic asthma in early life Elizabeth R. Davies, Joanne F.C. Kelly, Peter H. Howarth, David
Revised Suppl. Data: Soluble ADAM33 1 Soluble ADAM33 initiates airway remodeling to promote susceptibility for allergic asthma in early life Elizabeth R. Davies, Joanne F.C. Kelly, Peter H. Howarth, David
100 mm Sucrose. +Berberine +Quinine
 8 mm Sucrose Probability (%) 7 6 5 4 3 Wild-type Gr32a / 2 +Caffeine +Berberine +Quinine +Denatonium Supplementary Figure 1: Detection of sucrose and bitter compounds is not affected in Gr32a / flies.
8 mm Sucrose Probability (%) 7 6 5 4 3 Wild-type Gr32a / 2 +Caffeine +Berberine +Quinine +Denatonium Supplementary Figure 1: Detection of sucrose and bitter compounds is not affected in Gr32a / flies.
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Breeding scheme, transgenes, histological analysis and site distribution of SB-mutagenized osteosarcoma.
 Supplementary Figure 1 Breeding scheme, transgenes, histological analysis and site distribution of SB-mutagenized osteosarcoma. (a) Breeding scheme. R26-LSL-SB11 homozygous mice were bred to Trp53 LSL-R270H/+
Supplementary Figure 1 Breeding scheme, transgenes, histological analysis and site distribution of SB-mutagenized osteosarcoma. (a) Breeding scheme. R26-LSL-SB11 homozygous mice were bred to Trp53 LSL-R270H/+
Supplementary Information
 Supplementary Information 1 Supplementary information, Figure S1 Establishment of PG-haESCs. (A) Summary of derivation of PG-haESCs. (B) Upper, Flow analysis of DNA content of established PG-haES cell
Supplementary Information 1 Supplementary information, Figure S1 Establishment of PG-haESCs. (A) Summary of derivation of PG-haESCs. (B) Upper, Flow analysis of DNA content of established PG-haES cell
Doctor of Philosophy
 Regulation of Gene Expression of the 25-Hydroxyvitamin D la-hydroxylase (CYP27BI) Promoter: Study of A Transgenic Mouse Model Ivanka Hendrix School of Molecular and Biomedical Science The University of
Regulation of Gene Expression of the 25-Hydroxyvitamin D la-hydroxylase (CYP27BI) Promoter: Study of A Transgenic Mouse Model Ivanka Hendrix School of Molecular and Biomedical Science The University of
Supplemental Information For: The genetics of splicing in neuroblastoma
 Supplemental Information For: The genetics of splicing in neuroblastoma Justin Chen, Christopher S. Hackett, Shile Zhang, Young K. Song, Robert J.A. Bell, Annette M. Molinaro, David A. Quigley, Allan Balmain,
Supplemental Information For: The genetics of splicing in neuroblastoma Justin Chen, Christopher S. Hackett, Shile Zhang, Young K. Song, Robert J.A. Bell, Annette M. Molinaro, David A. Quigley, Allan Balmain,
SUPPLEMENTARY INFORMATION
 Supplementary text Collectively, we were able to detect ~14,000 expressed genes with RPKM (reads per kilobase per million) > 1 or ~16,000 with RPKM > 0.1 in at least one cell type from oocyte to the morula
Supplementary text Collectively, we were able to detect ~14,000 expressed genes with RPKM (reads per kilobase per million) > 1 or ~16,000 with RPKM > 0.1 in at least one cell type from oocyte to the morula
Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was
 Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was painted on the shaved back skin of CBL/J and BALB/c mice for consecutive days. (a, b) Phenotypic presentation of mouse back skin
Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was painted on the shaved back skin of CBL/J and BALB/c mice for consecutive days. (a, b) Phenotypic presentation of mouse back skin
Figure S1. Molecular confirmation of the precise insertion of the AsMCRkh2 cargo into the kh w locus.
 Supporting Information Appendix Table S1. Larval and adult phenotypes of G 2 progeny of lines 10.1 and 10.2 G 1 outcrosses to wild-type mosquitoes. Table S2. List of oligonucleotide primers. Table S3.
Supporting Information Appendix Table S1. Larval and adult phenotypes of G 2 progeny of lines 10.1 and 10.2 G 1 outcrosses to wild-type mosquitoes. Table S2. List of oligonucleotide primers. Table S3.
CRISPR/Cas9 cleavage of viral DNA efficiently suppresses hepatitis B virus
 0 0 CRISPR/Cas cleavage of viral DNA efficiently suppresses hepatitis B virus Authors: Vyas Ramanan,, Amir Shlomai,, David B.T. Cox,,,, Robert E. Schwartz,,, Eleftherios Michailidis, Ankit Bhatta, David
0 0 CRISPR/Cas cleavage of viral DNA efficiently suppresses hepatitis B virus Authors: Vyas Ramanan,, Amir Shlomai,, David B.T. Cox,,,, Robert E. Schwartz,,, Eleftherios Michailidis, Ankit Bhatta, David
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Amelio et al., http://www.jcb.org/cgi/content/full/jcb.201203134/dc1 Figure S1. mir-24 regulates proliferation and by itself induces
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Amelio et al., http://www.jcb.org/cgi/content/full/jcb.201203134/dc1 Figure S1. mir-24 regulates proliferation and by itself induces
Lack of cadherins Celsr2 and Celsr3 impairs ependymal ciliogenesis, leading to fatal
 Lack of cadherins Celsr2 and Celsr3 impairs ependymal ciliogenesis, leading to fatal hydrocephalus Fadel TISSIR, Yibo QU, Mireille MONTCOUQUIOL, Libing ZHOU, Kouji KOMATSU, Dongbo SHI, Toshihiko FUJIMORI,
Lack of cadherins Celsr2 and Celsr3 impairs ependymal ciliogenesis, leading to fatal hydrocephalus Fadel TISSIR, Yibo QU, Mireille MONTCOUQUIOL, Libing ZHOU, Kouji KOMATSU, Dongbo SHI, Toshihiko FUJIMORI,
Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v)
 SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
Postn MCM Smad2 fl/fl Postn MCM Smad3 fl/fl Postn MCM Smad2/3 fl/fl. Postn MCM. Tgfbr1/2 fl/fl TAC
 A Smad2 fl/fl Smad3 fl/fl Smad2/3 fl/fl Tgfbr1/2 fl/fl 1. mm B Tcf21 MCM Tcf21 MCM Smad3 fl/fl Tcf21 MCM Smad2/3 fl/fl Tcf21 MCM Tgfbr1/2 fl/fl αmhc MCM C 1. mm 1. mm D Smad2 fl/fl Smad3 fl/fl Smad2/3
A Smad2 fl/fl Smad3 fl/fl Smad2/3 fl/fl Tgfbr1/2 fl/fl 1. mm B Tcf21 MCM Tcf21 MCM Smad3 fl/fl Tcf21 MCM Smad2/3 fl/fl Tcf21 MCM Tgfbr1/2 fl/fl αmhc MCM C 1. mm 1. mm D Smad2 fl/fl Smad3 fl/fl Smad2/3
The Human Major Histocompatibility Complex
 The Human Major Histocompatibility Complex 1 Location and Organization of the HLA Complex on Chromosome 6 NEJM 343(10):702-9 2 Inheritance of the HLA Complex Haplotype Inheritance (Family Study) 3 Structure
The Human Major Histocompatibility Complex 1 Location and Organization of the HLA Complex on Chromosome 6 NEJM 343(10):702-9 2 Inheritance of the HLA Complex Haplotype Inheritance (Family Study) 3 Structure
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supplementary Figure 1. Generation of a conditional allele of the Kindlin-2 gene. (A) A restriction map of the relevant genomic region of Kindlin-2 (top), the targeting construct
SUPPLEMENTARY INFORMATION Supplementary Figure 1. Generation of a conditional allele of the Kindlin-2 gene. (A) A restriction map of the relevant genomic region of Kindlin-2 (top), the targeting construct
The coiled-coil domain containing protein CCDC40 is essential for motile cilia function and left-right axis formation
 The coiled-coil domain containing protein CCDC40 is essential for motile cilia function and left-right axis formation Anita Becker-Heck#, Irene Zohn#, Noriko Okabe#, Andrew Pollock#, Kari Baker Lenhart,
The coiled-coil domain containing protein CCDC40 is essential for motile cilia function and left-right axis formation Anita Becker-Heck#, Irene Zohn#, Noriko Okabe#, Andrew Pollock#, Kari Baker Lenhart,
Appendix. Table of Contents
 Appendix Table of Contents Appendix Figures Figure S1: Gp78 is not required for the degradation of mcherry-cl1 in Hela Cells. Figure S2: Indel formation in the MARCH6 sgrna targeted HeLa clones. Figure
Appendix Table of Contents Appendix Figures Figure S1: Gp78 is not required for the degradation of mcherry-cl1 in Hela Cells. Figure S2: Indel formation in the MARCH6 sgrna targeted HeLa clones. Figure
The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep
 SUPPLEMENTARY INFORMATION The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep degradation associated with lymphocyte and dendritic cell hyperresponsiveness Jinyi Zhang, Naima
SUPPLEMENTARY INFORMATION The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep degradation associated with lymphocyte and dendritic cell hyperresponsiveness Jinyi Zhang, Naima
(A) PCR primers (arrows) designed to distinguish wild type (P1+P2), targeted (P1+P2) and excised (P1+P3)14-
 1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
Supplementary Figure 1: Hsp60 / IEC mice are embryonically lethal (A) Light microscopic pictures show mouse embryos at developmental stage E12.
 Supplementary Figure 1: Hsp60 / IEC mice are embryonically lethal (A) Light microscopic pictures show mouse embryos at developmental stage E12.5 and E13.5 prepared from uteri of dams and subsequently genotyped.
Supplementary Figure 1: Hsp60 / IEC mice are embryonically lethal (A) Light microscopic pictures show mouse embryos at developmental stage E12.5 and E13.5 prepared from uteri of dams and subsequently genotyped.
GEMouse. Added Service Guide
 Contact mouse : mouse@macrogen.com Payment Inquiry: payment@macrogen.com Technical Support Macrogen Korea: support@macrogen.com Macrogen-Europe: support-europe@macrogen.com Update 68 GEMouse Added Service
Contact mouse : mouse@macrogen.com Payment Inquiry: payment@macrogen.com Technical Support Macrogen Korea: support@macrogen.com Macrogen-Europe: support-europe@macrogen.com Update 68 GEMouse Added Service
Problem Set 5 KEY
 2006 7.012 Problem Set 5 KEY ** Due before 5 PM on THURSDAY, November 9, 2006. ** Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. You are studying the development
2006 7.012 Problem Set 5 KEY ** Due before 5 PM on THURSDAY, November 9, 2006. ** Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. You are studying the development
pplementary Figur Supplementary Figure 1. a.
 pplementary Figur Supplementary Figure 1. a. Quantification by RT-qPCR of YFV-17D and YFV-17D pol- (+) RNA in the supernatant of cultured Huh7.5 cells following viral RNA electroporation of respective
pplementary Figur Supplementary Figure 1. a. Quantification by RT-qPCR of YFV-17D and YFV-17D pol- (+) RNA in the supernatant of cultured Huh7.5 cells following viral RNA electroporation of respective
Prevalence of HPV-16 genomic variant carrying a 63-bp duplicated sequence within the E1 gene in Slovenian women
 Prevalence of HPV-16 genomic variant carrying a 63-bp duplicated sequence within the E1 gene in Slovenian women K E Y WORDS HPV-16, cervical cancer, E6-T350G variant A BSTRACT High-risk HPV, particularly
Prevalence of HPV-16 genomic variant carrying a 63-bp duplicated sequence within the E1 gene in Slovenian women K E Y WORDS HPV-16, cervical cancer, E6-T350G variant A BSTRACT High-risk HPV, particularly
Balancing intestinal and systemic inflammation through cell type-specific expression of
 Supplementary Information Balancing intestinal and systemic inflammation through cell type-specific expression of the aryl hydrocarbon receptor repressor Olga Brandstätter 1,2,6, Oliver Schanz 1,6, Julia
Supplementary Information Balancing intestinal and systemic inflammation through cell type-specific expression of the aryl hydrocarbon receptor repressor Olga Brandstätter 1,2,6, Oliver Schanz 1,6, Julia
Supplementary Fig. S1. Schematic diagram of minigenome segments.
 open reading frame 1565 (segment 5) 47 (-) 3 5 (+) 76 101 125 149 173 197 221 246 287 open reading frame 890 (segment 8) 60 (-) 3 5 (+) 172 Supplementary Fig. S1. Schematic diagram of minigenome segments.
open reading frame 1565 (segment 5) 47 (-) 3 5 (+) 76 101 125 149 173 197 221 246 287 open reading frame 890 (segment 8) 60 (-) 3 5 (+) 172 Supplementary Fig. S1. Schematic diagram of minigenome segments.
Supplementary Figure 1
 Supplementary Figure 1 Isolation of mt-trnas and RNA-MS analysis of mt-trna Asn from M. nudus (a)m. nudus mt-trnas were isolated by RCC and resolved by 10% denaturing PAGE. The gel was stained with SYBR
Supplementary Figure 1 Isolation of mt-trnas and RNA-MS analysis of mt-trna Asn from M. nudus (a)m. nudus mt-trnas were isolated by RCC and resolved by 10% denaturing PAGE. The gel was stained with SYBR
Supplementary Table 1. List of primers used in this study
 Supplementary Table 1. List of primers used in this study Gene Forward primer Reverse primer Rat Met 5 -aggtcgcttcatgcaggt-3 5 -tccggagacacaggatgg-3 Rat Runx1 5 -cctccttgaaccactccact-3 5 -ctggatctgcctggcatc-3
Supplementary Table 1. List of primers used in this study Gene Forward primer Reverse primer Rat Met 5 -aggtcgcttcatgcaggt-3 5 -tccggagacacaggatgg-3 Rat Runx1 5 -cctccttgaaccactccact-3 5 -ctggatctgcctggcatc-3
BIOL2005 WORKSHEET 2008
 BIOL2005 WORKSHEET 2008 Answer all 6 questions in the space provided using additional sheets where necessary. Hand your completed answers in to the Biology office by 3 p.m. Friday 8th February. 1. Your
BIOL2005 WORKSHEET 2008 Answer all 6 questions in the space provided using additional sheets where necessary. Hand your completed answers in to the Biology office by 3 p.m. Friday 8th February. 1. Your
Supplemental Information. Myocardial Polyploidization Creates a Barrier. to Heart Regeneration in Zebrafish
 Developmental Cell, Volume 44 Supplemental Information Myocardial Polyploidization Creates a Barrier to Heart Regeneration in Zebrafish Juan Manuel González-Rosa, Michka Sharpe, Dorothy Field, Mark H.
Developmental Cell, Volume 44 Supplemental Information Myocardial Polyploidization Creates a Barrier to Heart Regeneration in Zebrafish Juan Manuel González-Rosa, Michka Sharpe, Dorothy Field, Mark H.
Supplementary Figure 1. Generation of knockin mice expressing L-selectinN138G. (a) Schematics of the Sellg allele (top), the targeting vector, the
 Supplementary Figure 1. Generation of knockin mice expressing L-selectinN138G. (a) Schematics of the Sellg allele (top), the targeting vector, the targeted allele in ES cells, and the mutant allele in
Supplementary Figure 1. Generation of knockin mice expressing L-selectinN138G. (a) Schematics of the Sellg allele (top), the targeting vector, the targeted allele in ES cells, and the mutant allele in
Antiviral restriction factor transgenesis in the domestic cat
 Nature Methods Antiviral restriction factor transgenesis in the domestic cat Pimprapar Wongsrikeao, Dyana Saenz, Tommy Rinkoski, Takeshige Otoi & Eric Poeschla Supplementary Figure 1 Supplementary Figure
Nature Methods Antiviral restriction factor transgenesis in the domestic cat Pimprapar Wongsrikeao, Dyana Saenz, Tommy Rinkoski, Takeshige Otoi & Eric Poeschla Supplementary Figure 1 Supplementary Figure
M.Sc. (Reproductive Biology & Clinical Embryology)
 M.Sc. (Reproductive Biology & Clinical Embryology) ACADEMIC SCHEDULE Theory (Didactic) Lectures + Seminars + Journal Clubs Semester I & II (2 hours / day) Practicals (Hands-on + Demo) Semester I & II (first
M.Sc. (Reproductive Biology & Clinical Embryology) ACADEMIC SCHEDULE Theory (Didactic) Lectures + Seminars + Journal Clubs Semester I & II (2 hours / day) Practicals (Hands-on + Demo) Semester I & II (first
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Fig. 1: Quality assessment of formalin-fixed paraffin-embedded (FFPE)-derived DNA and nuclei. (a) Multiplex PCR analysis of unrepaired and repaired bulk FFPE gdna from
Supplementary Figure 1 Supplementary Fig. 1: Quality assessment of formalin-fixed paraffin-embedded (FFPE)-derived DNA and nuclei. (a) Multiplex PCR analysis of unrepaired and repaired bulk FFPE gdna from
Supplemental Figure 1. (A) The localization of Cre DNA recombinase in the testis of Cyp19a1-Cre mice was detected by immunohistchemical analyses
 Supplemental Figure 1. (A) The localization of Cre DNA recombinase in the testis of Cyp19a1-Cre mice was detected by immunohistchemical analyses using an anti-cre antibody; testes at 1 week (left panel),
Supplemental Figure 1. (A) The localization of Cre DNA recombinase in the testis of Cyp19a1-Cre mice was detected by immunohistchemical analyses using an anti-cre antibody; testes at 1 week (left panel),
Supplementary Figure 1. Quantile-quantile (Q-Q) plots. (Panel A) Q-Q plot graphical
 Supplementary Figure 1. Quantile-quantile (Q-Q) plots. (Panel A) Q-Q plot graphical representation using all SNPs (n= 13,515,798) including the region on chromosome 1 including SORT1 which was previously
Supplementary Figure 1. Quantile-quantile (Q-Q) plots. (Panel A) Q-Q plot graphical representation using all SNPs (n= 13,515,798) including the region on chromosome 1 including SORT1 which was previously
Influenza A virus hemagglutinin and neuraminidase act as novel motile machinery. Tatsuya Sakai, Shin I. Nishimura, Tadasuke Naito, and Mineki Saito
 Supplementary Information for: Influenza A virus hemagglutinin and neuraminidase act as novel motile machinery Tatsuya Sakai, Shin I. Nishimura, Tadasuke Naito, and Mineki Saito Supplementary Methods Supplementary
Supplementary Information for: Influenza A virus hemagglutinin and neuraminidase act as novel motile machinery Tatsuya Sakai, Shin I. Nishimura, Tadasuke Naito, and Mineki Saito Supplementary Methods Supplementary
Supplementary Figure 1. Expression of phospho-sik3 in normal and osteoarthritic articular cartilage in the knee. (a) Semiserial histological sections
 Supplementary Figure 1. Expression of phospho-sik3 in normal and osteoarthritic articular cartilage in the knee. (a) Semiserial histological sections of normal cartilage were stained with safranin O-fast
Supplementary Figure 1. Expression of phospho-sik3 in normal and osteoarthritic articular cartilage in the knee. (a) Semiserial histological sections of normal cartilage were stained with safranin O-fast
Nature Immunology: doi: /ni Supplementary Figure 1. Gene expression profile of CD4 + T cells and CTL responses in Bcl6-deficient mice.
 Supplementary Figure 1 Gene expression profile of CD4 + T cells and CTL responses in Bcl6-deficient mice. (a) Gene expression profile in the resting CD4 + T cells were analyzed by an Affymetrix microarray
Supplementary Figure 1 Gene expression profile of CD4 + T cells and CTL responses in Bcl6-deficient mice. (a) Gene expression profile in the resting CD4 + T cells were analyzed by an Affymetrix microarray
Supplementary Figure 1
 Supplementary Figure 1 5 microns C7 B6 unclassified H19 C7 signal H19 guide signal H19 B6 signal C7 SNP spots H19 RNA spots B6 SNP spots colocalization H19 RNA classification Supplementary Figure 1. Allele-specific
Supplementary Figure 1 5 microns C7 B6 unclassified H19 C7 signal H19 guide signal H19 B6 signal C7 SNP spots H19 RNA spots B6 SNP spots colocalization H19 RNA classification Supplementary Figure 1. Allele-specific
marker. DAPI labels nuclei. Flies were 20 days old. Scale bar is 5 µm. Ctrl is
 Supplementary Figure 1. (a) Nos is detected in glial cells in both control and GFAP R79H transgenic flies (arrows), but not in deletion mutant Nos Δ15 animals. Repo is a glial cell marker. DAPI labels
Supplementary Figure 1. (a) Nos is detected in glial cells in both control and GFAP R79H transgenic flies (arrows), but not in deletion mutant Nos Δ15 animals. Repo is a glial cell marker. DAPI labels
Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers
 Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers Gordon Blackshields Senior Bioinformatician Source BioScience 1 To Cancer Genetics Studies
Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers Gordon Blackshields Senior Bioinformatician Source BioScience 1 To Cancer Genetics Studies
(A) Cells grown in monolayer were fixed and stained for surfactant protein-c (SPC,
 Supplemental Figure Legends Figure S1. Cell line characterization (A) Cells grown in monolayer were fixed and stained for surfactant protein-c (SPC, green) and co-stained with DAPI to visualize the nuclei.
Supplemental Figure Legends Figure S1. Cell line characterization (A) Cells grown in monolayer were fixed and stained for surfactant protein-c (SPC, green) and co-stained with DAPI to visualize the nuclei.
Supplemental Information. Figures. Figure S1
 Supplemental Information Figures Figure S1 Identification of JAGGER T-DNA insertions. A. Positions of T-DNA and Ds insertions in JAGGER are indicated by inverted triangles, the grey box represents the
Supplemental Information Figures Figure S1 Identification of JAGGER T-DNA insertions. A. Positions of T-DNA and Ds insertions in JAGGER are indicated by inverted triangles, the grey box represents the
EMMA Cryopreservation Workshop Novel concepts in mouse production and preservation
 EMMA Cryopreservation Workshop Novel concepts in mouse production and preservation Consejo Superior de Investigaciones Cientificas Madrid, Espagna May 7-8, 2012 K. C. Kent Lloyd, DVM, PhD University of
EMMA Cryopreservation Workshop Novel concepts in mouse production and preservation Consejo Superior de Investigaciones Cientificas Madrid, Espagna May 7-8, 2012 K. C. Kent Lloyd, DVM, PhD University of
Nature Biotechnology: doi: /nbt Supplementary Figure 1
 Supplementary Figure 1 Overview of the transplant procedure and supplementary data to Figure 1. a. Under isofluorane anesthesia, the lumen of the colon is washed by a gentle PBS enema. b. Using a p200
Supplementary Figure 1 Overview of the transplant procedure and supplementary data to Figure 1. a. Under isofluorane anesthesia, the lumen of the colon is washed by a gentle PBS enema. b. Using a p200
Nature Genetics: doi: /ng Supplementary Figure 1. Brain magnetic resonance imaging in patient 9 at age 3.6 years.
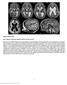 Supplementary Figure 1 Brain magnetic resonance imaging in patient 9 at age 3.6 years. (a d) Axial T2-weighted images show a marked degree of ventriculomegaly. Note the diencephalic mesencephalic junction
Supplementary Figure 1 Brain magnetic resonance imaging in patient 9 at age 3.6 years. (a d) Axial T2-weighted images show a marked degree of ventriculomegaly. Note the diencephalic mesencephalic junction
sirna count per 50 kb small RNAs matching the direct strand Repeat length (bp) per 50 kb repeats in the chromosome
 Qi et al. 26-3-2564C Qi et al., Figure S1 sirna count per 5 kb small RNAs matching the direct strand sirna count per 5 kb small RNAs matching the complementary strand Repeat length (bp) per 5 kb repeats
Qi et al. 26-3-2564C Qi et al., Figure S1 sirna count per 5 kb small RNAs matching the direct strand sirna count per 5 kb small RNAs matching the complementary strand Repeat length (bp) per 5 kb repeats
SALSA MLPA KIT P050-B2 CAH
 SALSA MLPA KIT P050-B2 CAH Lot 0510, 0909, 0408: Compared to lot 0107, extra control fragments have been added at 88, 96, 100 and 105 nt. The 274 nt probe gives a higher signal in lot 0510 compared to
SALSA MLPA KIT P050-B2 CAH Lot 0510, 0909, 0408: Compared to lot 0107, extra control fragments have been added at 88, 96, 100 and 105 nt. The 274 nt probe gives a higher signal in lot 0510 compared to
Probe. Hind III Q,!?R'!! /0!!!!D1"?R'! vector. Homologous recombination
 Supple-Zhang Page 1 Wild-type locus Targeting construct Targeted allele Exon Exon3 Exon Probe P1 P P3 FRT FRT loxp loxp neo vector amh I Homologous recombination neo P1 P P3 FLPe recombination Q,!?R'!!
Supple-Zhang Page 1 Wild-type locus Targeting construct Targeted allele Exon Exon3 Exon Probe P1 P P3 FRT FRT loxp loxp neo vector amh I Homologous recombination neo P1 P P3 FLPe recombination Q,!?R'!!
Nature Genetics: doi: /ng Supplementary Figure 1. Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms.
 Supplementary Figure 1 Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms. IF images of wild-type (wt) and met-2 set-25 worms showing the loss of H3K9me2/me3 at the indicated developmental
Supplementary Figure 1 Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms. IF images of wild-type (wt) and met-2 set-25 worms showing the loss of H3K9me2/me3 at the indicated developmental
Name Animal source Vendor Cat # Dilutions
 Supplementary data Table S1. Primary and Secondary antibody sources Devi et al, TXNIP in mitophagy A. Primary Antibodies Name Animal source Vendor Cat # Dilutions 1. TXNIP mouse MBL KO205-2 1:2000 (WB)
Supplementary data Table S1. Primary and Secondary antibody sources Devi et al, TXNIP in mitophagy A. Primary Antibodies Name Animal source Vendor Cat # Dilutions 1. TXNIP mouse MBL KO205-2 1:2000 (WB)
Zhu et al, page 1. Supplementary Figures
 Zhu et al, page 1 Supplementary Figures Supplementary Figure 1: Visual behavior and avoidance behavioral response in EPM trials. (a) Measures of visual behavior that performed the light avoidance behavior
Zhu et al, page 1 Supplementary Figures Supplementary Figure 1: Visual behavior and avoidance behavioral response in EPM trials. (a) Measures of visual behavior that performed the light avoidance behavior
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES 1 Supplementary Figure 1, Adult hippocampal QNPs and TAPs uniformly express REST a-b) Confocal images of adult hippocampal mouse sections showing GFAP (green), Sox2 (red), and REST
SUPPLEMENTARY FIGURES 1 Supplementary Figure 1, Adult hippocampal QNPs and TAPs uniformly express REST a-b) Confocal images of adult hippocampal mouse sections showing GFAP (green), Sox2 (red), and REST
Supplementary Figure 1. Properties of various IZUMO1 monoclonal antibodies and behavior of SPACA6. (a) (b) (c) (d) (e) (f) (g) .
 Supplementary Figure 1. Properties of various IZUMO1 monoclonal antibodies and behavior of SPACA6. (a) The inhibitory effects of new antibodies (Mab17 and Mab18). They were investigated in in vitro fertilization
Supplementary Figure 1. Properties of various IZUMO1 monoclonal antibodies and behavior of SPACA6. (a) The inhibitory effects of new antibodies (Mab17 and Mab18). They were investigated in in vitro fertilization
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2566 Figure S1 CDKL5 protein expression pattern and localization in mouse brain. (a) Multiple-tissue western blot from a postnatal day (P) 21 mouse probed with an antibody against CDKL5.
DOI: 10.1038/ncb2566 Figure S1 CDKL5 protein expression pattern and localization in mouse brain. (a) Multiple-tissue western blot from a postnatal day (P) 21 mouse probed with an antibody against CDKL5.
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Choi YL, Soda M, Yamashita Y, et al. EML4-ALK mutations in
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Choi YL, Soda M, Yamashita Y, et al. EML4-ALK mutations in
Genome 371, Autumn 2018 Quiz Section 9: Genetics of Cancer Worksheet
 Genome 371, Autumn 2018 Quiz Section 9: Genetics of Cancer Worksheet All cancer is due to genetic mutations. However, in cancer that clusters in families (familial cancer) at least one of these mutations
Genome 371, Autumn 2018 Quiz Section 9: Genetics of Cancer Worksheet All cancer is due to genetic mutations. However, in cancer that clusters in families (familial cancer) at least one of these mutations
Problem Set 8 Key 1 of 8
 7.06 2003 Problem Set 8 Key 1 of 8 7.06 2003 Problem Set 8 Key 1. As a bright MD/PhD, you are interested in questions about the control of cell number in the body. Recently, you've seen three patients
7.06 2003 Problem Set 8 Key 1 of 8 7.06 2003 Problem Set 8 Key 1. As a bright MD/PhD, you are interested in questions about the control of cell number in the body. Recently, you've seen three patients
SUPPORTING ONLINE MATERIAL
 SUPPORTING ONLINE MATERIAL SUPPORTING ONLINE TEXT Efficiency of SCNT Alive fetuses at mid-gestation The rate of viable (beating heart) embryos at day 12.5-14.5 dpc was assessed after sacrifice of foster
SUPPORTING ONLINE MATERIAL SUPPORTING ONLINE TEXT Efficiency of SCNT Alive fetuses at mid-gestation The rate of viable (beating heart) embryos at day 12.5-14.5 dpc was assessed after sacrifice of foster
Supplementary Information
 Supplementary Information Precursors of trnas are stabilized by methylguanosine cap structures Takayuki Ohira and Tsutomu Suzuki Department of Chemistry and Biotechnology, Graduate School of Engineering,
Supplementary Information Precursors of trnas are stabilized by methylguanosine cap structures Takayuki Ohira and Tsutomu Suzuki Department of Chemistry and Biotechnology, Graduate School of Engineering,
Supplementary Figure 1: Features of IGLL5 Mutations in CLL: a) Representative IGV screenshot of first
 Supplementary Figure 1: Features of IGLL5 Mutations in CLL: a) Representative IGV screenshot of first intron IGLL5 mutation depicting biallelic mutations. Red arrows highlight the presence of out of phase
Supplementary Figure 1: Features of IGLL5 Mutations in CLL: a) Representative IGV screenshot of first intron IGLL5 mutation depicting biallelic mutations. Red arrows highlight the presence of out of phase
Evaluating Variability of Allergens in Commodity Crops (Peanuts)
 Evaluating Variability of Allergens in Commodity Crops (Peanuts) Peggy Ozias-Akins Laura Ramos Paola Faustinelli Ye Chu University of Georgia Manel Jordana Katherine Arias McMaster University Soheila Maleki
Evaluating Variability of Allergens in Commodity Crops (Peanuts) Peggy Ozias-Akins Laura Ramos Paola Faustinelli Ye Chu University of Georgia Manel Jordana Katherine Arias McMaster University Soheila Maleki
Imagine Innovate Integrate. Using the NSET for embryo transfer and artificial insemination in mice and rats Kendra Steele, Ph.D.
 Imagine Innovate Integrate Using the NSET for embryo transfer and artificial insemination in mice and rats Kendra Steele, Ph.D. Outline- Here to make your life easier Non-surgical embryo transfer in mice
Imagine Innovate Integrate Using the NSET for embryo transfer and artificial insemination in mice and rats Kendra Steele, Ph.D. Outline- Here to make your life easier Non-surgical embryo transfer in mice
AVENIO family of NGS oncology assays ctdna and Tumor Tissue Analysis Kits
 AVENIO family of NGS oncology assays ctdna and Tumor Tissue Analysis Kits Accelerating clinical research Next-generation sequencing (NGS) has the ability to interrogate many different genes and detect
AVENIO family of NGS oncology assays ctdna and Tumor Tissue Analysis Kits Accelerating clinical research Next-generation sequencing (NGS) has the ability to interrogate many different genes and detect
Supporting Information
 Supporting Information Franco et al. 10.1073/pnas.1015557108 SI Materials and Methods Drug Administration. PD352901 was dissolved in 0.5% (wt/vol) hydroxyl-propyl-methylcellulose, 0.2% (vol/vol) Tween
Supporting Information Franco et al. 10.1073/pnas.1015557108 SI Materials and Methods Drug Administration. PD352901 was dissolved in 0.5% (wt/vol) hydroxyl-propyl-methylcellulose, 0.2% (vol/vol) Tween
a) SSR with core motif > 2 and repeats number >3. b) MNR with repeats number>5.
 1 2 APPENDIX Legends to figures 3 4 5 Figure A1: Distribution of perfect SSR along chromosome 1 of V. cholerae (El-Tor N191). a) SSR with core motif > 2 and repeats number >3. b) MNR with repeats number>5.
1 2 APPENDIX Legends to figures 3 4 5 Figure A1: Distribution of perfect SSR along chromosome 1 of V. cholerae (El-Tor N191). a) SSR with core motif > 2 and repeats number >3. b) MNR with repeats number>5.
Supporting Information Table of Contents
 Supporting Information Table of Contents Supporting Information Figure 1 Page 2 Supporting Information Figure 2 Page 4 Supporting Information Figure 3 Page 5 Supporting Information Figure 4 Page 6 Supporting
Supporting Information Table of Contents Supporting Information Figure 1 Page 2 Supporting Information Figure 2 Page 4 Supporting Information Figure 3 Page 5 Supporting Information Figure 4 Page 6 Supporting
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 Characterization of stable expression of GlucB and sshbira in the CT26 cell line (a) Live cell imaging of stable CT26 cells expressing green fluorescent protein
Supplementary Figures Supplementary Figure 1 Characterization of stable expression of GlucB and sshbira in the CT26 cell line (a) Live cell imaging of stable CT26 cells expressing green fluorescent protein
