Identification of Domains in Gag Important for Prototypic Foamy Virus Egress
|
|
|
- Tobias Gallagher
- 6 years ago
- Views:
Transcription
1 JOURNAL OF VIROLOGY, May 2005, p Vol. 79, No X/05/$ doi: /jvi Copyright 2005, American Society for Microbiology. All Rights Reserved. Identification of Domains in Gag Important for Prototypic Foamy Virus Egress Gillian S. Patton, 1 Stephen A. Morris, 1 Wayne Chung, 2 Paul D. Bieniasz, 2 and Myra O. McClure 1 * Jefferiss Research Trust Laboratories, Wright-Fleming Institute, Faculty of Medicine, Imperial College London, St. Mary s Campus, Norfolk Place, London W2 1PG, United Kingdom, 1 and Aaron Diamond AIDS Research Center and The Rockefeller University, New York, New York 2 Received 24 August 2004/Accepted 8 December 2004 Sequence motifs (L domains) have been described in viral structural proteins. Mutations in these lead to a defect at a late stage in virus assembly and budding. For several viruses, recruitment of an endosomal sorting complexes required for transport 1 subunit (Tsg101), a component of the class E vacuolar protein sorting (EVPS) machinery, is a prerequisite for virion budding. To effect this, Tsg101 interacts with the PT/SAP L domain. We have identified candidate L-domain motifs, PSAP, PPPI, and YEIL, in the prototypic foamy virus (PFV) Gag protein, based on their homology to known viral L domains. Mutation of the PSAP and PPPI motifs individually reduced PFV egress, and their combined mutation had an additive effect. When PSAP was mutated, residual infectious PFV release was unaffected by dominant negative Vps4 (an ATPase involved in the final stages of budding), and sensitivity to dominant negative Tsg101 was dramatically reduced, suggesting that the PSAP motif functions as a conventional class E VPS-dependent L domain. Consistent with this notion, yeast two-hybrid analysis showed a PSAP motif-dependent interaction between PFV Gag and Tsg101. Surprisingly, PFV release which is dependent on the PPPI motif was Vps4-independent and was partially inhibited by dominant negative Tsg101, suggesting that PPPI functions by an unconventional mechanism to facilitate PFV egress. Mutation of the YEIL sequence completely abolished particle formation and also reduced the rate of Gag processing by the viral protease, suggesting that the integrity of YEIL is required at an assembly step prior to budding and YEIL is not acting as an L domain. Viruses rely on using normal cellular processes for their replication, assembly, and egress from the infected cell. An essential stage in this process for enveloped viruses is budding through a cellular membrane to acquire their envelope, and, in the case of budding through the plasma membrane, to exit the cell. The major structural proteins of some enveloped viruses (e.g., retrovirus Gag proteins, filovirus matrix proteins, and rhabdovirus M proteins) are both necessary and sufficient for virus-like particle assembly and egress. These proteins contain late-budding domains (L domains), which play an essential role in virus particle release. Mutations in these sequences lead to a late budding defect in which virus particles assemble but are unable to complete the final stages in budding from cellular membranes and, hence, remain tethered to the cell (11). To date, three classes of L domain have been identified in a number of retrovirus Gag proteins, including those of human immunodeficiency virus type 1 (14, 19), murine leukemia virus (52), and equine infectious anemia virus (39), as well as in the structural proteins of Ebola virus (16, 30), rabies virus (17), and arenaviruses (35). These are characterized by the sequence motifs PT/SAP, PPXY, and YPDL/LXXLF, which can apparently occur individually or in close proximity within viral structural proteins (5, 15, 19, 29, 37, 39, 41, 46). A number of recent * Corresponding author. Mailing address: Jefferiss Research Trust Laboratories, Wright-Fleming Institute, Division of Medicine, Imperial College London, St Mary s Campus, Norfolk Place, London W2 1PG, United Kingdom. Phone: Fax: m.mcclure@imperial.ac.uk. G.S.P and S.A.M contributed equally to this work. studies have shown that at least two of these motifs bind to components of the class E vacuolar protein sorting (VPS) pathway which functions during vesicular budding into multivesicular bodies (MVB) (5, 7, 12, 30 32, 38, 41, 42). This process likely involves the sequential recruitment of a number of multiprotein complexes, termed endosomal sorting complexes required for transport (ESCRT), to endosomal membranes. Initially, an ubiquitin-based sorting signal on cargo proteins is recognized by the hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs) (2, 22, 28, 38). This recruits ESCRT-I, which further recruits the ESCRT-II/III complexes either directly or via an interaction with AIP-1/ALIX (a mammalian orthologue of the yeast VPS factor Bro1) (31, 41, 44). A final step prior to membrane fusion requires the disassembly of ESCRT complexes catalyzed by the homomultimeric ATPase Vps4 (1). Several studies have established that the PT/SAP motif functions during viral budding by recruiting the class E VPS pathway via a direct interaction with a component of the ESCRT-I complex, Tsg101 (tumor-susceptibility gene product 101) (7, 12, 30, 42). Conversely, the binding partner for the YPDL domain was initially suggested to be the AP-2 adaptor complex involved in clathrin-mediated endocytosis from the plasma membrane (40). However, more recent studies have suggested that the functional interaction is more likely to be with a class E VPS factor, namely, AIP-1/ALIX (32, 41). Additionally, expression of dominant negative forms of Vps4 inhibits the budding of all retroviruses tested thus far (12, 32, 41). It thus seems likely that the mechanism of particle budding in viruses containing L domains is making use of the cellular pathways 6392
2 VOL. 79, 2005 FUNCTIONAL L DOMAINS IN PFV Gag 6393 normally used to sort cargo into MVBs. This model is particularly attractive since MVB formation and virus budding are the only known examples of sorting/budding of cellular membranes away from the cytoplasm. However, while the PPXY motif is thought to interact with the WW domains of the Nedd4-like E3 ubiquitin ligases (16, 23, 48), precisely how this leads to a dependence on class E VPS factors for budding in most, but apparently not all, cases (20) is unknown. The prototypic foamy virus is unusual among retroviruses in that it has a number of unique features to its replication cycle (25). The provirus genome organization is similar to that of other complex retroviruses, but there are important differences in the way the Pol protein is expressed (3, 9, 27, 51), the nature of the nucleic acid in the virion (25, 51), the regulation of gene expression (26), and the structure and function of the Env protein (24). Some of these features would appear to resemble the hepadnavirus replication strategy more closely than that of classical retroviruses (51). The mechanism of foamy virus particle assembly and budding further shows important deviations from that of other retroviruses. These include differences in the minimal structural protein requirement for particle assembly and budding, particularly a requirement for Env in addition to Gag, and the site of budding which occurs primarily in an intracellular compartment, thought to be related to the endoplasmic reticulum (ER) (10, 36). The prototypic foamy virus (PFV) Env protein contains an ER retrieval signal within its cytoplasmic C terminus (13), and the N-terminal 15 amino acids are essential for virus particle formation and budding (24). The majority of virus particles remain cell associated in vitro. Standard procedures, such as cell lysis by sonication, do nothing to facilitate virion egress. This is relevant to the fact that foamy viruses offer an as yet untapped alternative vector system for gene therapy, given their lack of disease association, broad host range, larger than average retroviral packaging capacity, and ability to integrate into cells. Currently, their major disadvantage is the modest titer that foamy virus-derived vectors have compared to other viral vectors. An understanding of why this virus tends to remain cell associated and the mechanisms that might be employed to enhance their release would undoubtedly strengthen the case for their exploitation as vectors. In this study, we identified three sequences with homology to known L domains, PSAP, PPPI and YEIL, in the PFV Gag protein and show that at least one of these acts as an interface with the class E VPS pathway to facilitate PFV egress. Specifically, the PSAP motif functions as a conventional Tsg101 and Vps4-dependent L domain and the PPPI motif appears to facilitate PFV egress by an unconventional mechanism that may share some features with that of classical L domains. However, the YEIL sequence does not function as an L domain, but its integrity is nevertheless important for PFV assembly and/or Gag processing. Cell lines, plasmids, and mutagenesis. Human fibrosarcoma (HT1080), dog fibroblast (D17), human embryonic kidney (293T) cells, and baby hamster kidney cells stably transfected with the lacz gene under the transcriptional control of the PFV long terminal repeat (BHLL cells) (4) were cultivated in Dulbecco s modified Eagle minimal essential medium supplemented with 10% fetal calf serum, penicillin, streptomycinm and 2 mm L-glutamine. The PFV vector was produced using the vector plasmid pmh71 (18) and pczhfvenv encoding the HFV envelope protein (36). The wild-type PFV was produced using the construct pczhsrv2 (33). Amino acid mutations were introduced into the Gag proteins of MH71 and pczhsrv2 by site-directed mutagenesis using a QuikChange XL kit (Stratagene). Following mutagenesis, the integrity of all constructs was confirmed by sequencing. Plasmids expressing dominant negative forms of Vps4, Vps4B/SKD1, Tsg101, and AIP-1/ALIX in mammalian cells, namely, pcr3.1/gfp-vps4 223 M, pd- SRED-VPS4B E228Q (a gift from Wes Sundquist, University of Utah), pcr3.1/ YFP-Tsg101 (1-157), and pcr3.1/yfp-alix(d1-176), have been described previously (12, 31, 32). Plasmids based on pgbkt7 and expressing a GAL-4 DNA binding domain fused to Tsg101 or AIP-1/ALIX have been previously described (32), as have pvp16-based plasmids expressing VP16 activation domains fused to Tsg101, AIP-1/ALIX, and EIAV-p9. Additional plasmids expressing GAL4 or VP16 fused to wild-type and mutant forms of PFV Gag were generated by PCR amplification of the latter from the pmh71-based constructs and insertion of the amplicons into pgbkt7 and pvp16, respectively. PFV vector and virus production. The PFV vector and virus, and their mutated forms, were produced by transient transfection of 293T cells using polyethylenimine (34). Briefly, 293T cells were seeded into 12-well plates at cells per well and incubated overnight at 37 C. For comparison of virion particle production by wild-type and L-domain mutants, the cells were transfected with 1.5 g MH71 and 0.5 g pczhfvenv or 3 g pczhsrv2. To analyze the effect of dominant negative Tsg101, Vps4, Vps4B/SKD1, or AIP-1/ALIX proteins on PFV egress, 293T cells were cotransfected with 20 ng pcr3.1/yfp-tsg101 (1-157), 20 ng wild-type or mutant pcr3.1/gfp-vps4, 20 ng pdsred-c1/dsredvps4b- E228Q, or 100 ng pcr3.1/yfp-alix(d1-176). The supernatants were harvested 24, 48, or 72 h after transfection and clarified through a m-pore-size filter. For Western blot analysis, vector or viral particles were further purified by centrifugation (twice for 10 min at 2,000 g followed by centrifugation through a 20% sucrose cushion at 100,000 g for 25 min). All centrifugation steps were performed at 4 C and transfected cells or viral pellets resuspended in Laemmli polyacrylamide gel electrophoresis-sodium dodecyl sulfate (PAGE-SDS) lysis buffer (30). Vector and virus infectivity assays. D17 cells were used as target cells to measure egfp-expressing PFV vector titers, and BHLL cells were used to titer replication-competent virus. The cells were seeded into 96-well plates at cells per well and incubated at 37 C overnight. All retroviral supernatant stocks were serially diluted 10-fold (to 10 5 ) in growth medium, and then 75 l of each dilution was added to triplicate wells. Transduced cells were incubated for 3 days at 37 C and fixed in 4% formaldehyde and the egfp-expressing D17 cells detected using fluorescence microscopy. The PFV-infected BHLL cells expressing -galactosidase were detected by the insoluble X-Gal (5-bromo-4-chloro-3- indolyl- -D-galactopyranoside assay, as described previously (4). Western blot analysis. Virion and cell lysates were separated on 10% acrylamide gels and proteins transferred to nitrocellulose membranes (Amersham). The blots were incubated with human anti-pfv serum (diluted 1:500) overnight at 4 C, probed with peroxidase-conjugated anti-human secondary antibody (Sigma), and developed with metal-enhanced diaminobenzidine (Pierce). Electron microscopy (EM) analysis. 293T cells in 35-mm-diameter dishes were transfected with plasmids to produce wild-type vector and incubated at 37 C for 48 h. The cells were fixed for 30 min in 200 mm cacodylate 0.5% glutaraldehyde and then washed in 200 mm cacodylate and postfixed in 1% osmium tetroxide and 1.5% potassium ferrocyanide for 1 h. After extensive washing in water, the cells were incubated in 0.5% magnesium uranyl acetate overnight at 4 C, dehydrated in ethanol and propylene oxide, and embedded in epon resin. Ultrathin sections were cut and collected on transmission electron microscopy grids, and lead citrate was added as a contrast agent. Sections were analyzed using an FEI Technai G2 transmission electron microscope, and digital images were captured using Soft Imaging Software and processed in Adobe Photoshop 7. Yeast two-hybrid assays. Yeast Y190 cells were transformed with pgbkt7 and pvp16 derivatives and transformants selected on medium lacking tryptophan and leucine. Protein-protein interactions were measured by induction of -galactosidase reporter activity, using assays described previously (30). MATERIALS AND METHODS RESULTS Prototypic foamy virus Gag protein contains functional L domains. Three classes of L-domain sequence, based on the peptide sequences PT/SAP, PPPY, and YPDL/LXXLF, have
3 6394 PATTON ET AL. J. VIROL. FIG. 1. Schematic representation of the PFV Gag protein. Possible positions of the matrix (MA), capsid (CA), and nucleocapsid (NC) regions are indicated in brackets. The positions of putative L domains PSAP, PPPI, and YEIL are underlined along with the mutations made. The dotted line indicates the major proteolytic cleavage site for the viral protease. been described in retroviruses, two of which are known to bind class E VPS factors. An analysis of the PFV Gag protein sequence identified candidate L-domain motifs based on sequence similarity (Fig. 1). To test whether these motifs were required for virion budding, each candidate L domain was mutated separately or in combination (Fig. 1). Mutations were introduced into both the intact proviral plasmid pczhsrv2 and the PFV vector construct pmh71. These mutant plasmids were then transfected into 293T cells, either alone for pczhsrv2- based constructs or with pczhfvenv, which expresses PFV Env, for the production of pmh71-based vectors. The viral or vector titer in the corresponding culture supernatants was determined (Fig. 2). Mutation of the L1 domain, from PSAP to AARA, resulted in titer reductions of 10.1-fold for the intact virus and 13.4-fold for the PFV vector. Mutation of the L2 domain, from FIG. 2. The PSAP and PPPI motifs are required for optimal release of PFV vector and wild-type virus. The solid columns show titers of vector released from 293T cells transfected with either the wild-type (MH71), PSAP-mutated (MH71 L1), PPPI-mutated (MH71 L2), PSAP/PPPI dual-mutated (MH71 L1/L2), or YEIL-mutated (MH71 L3) vector constructs, all cotransfected with the pczhfvenv construct. Vector was titrated on D17 cells. The hashed columns show titers of virus released from 293T cells transfected with either the wild-type (pczhsrv2), PSAP-mutated (pczhsrv2 L1), PPPI-mutated (pczhsrv2 L2), PSAP/PPPI dual-mutated (pczhsrv2 L1/L2), or YEIL-mutated (pczhsrv2 L3) virus constructs. Virus is titrated on BHLL cells. Results shown are the means and standard errors of three independent experiments. PPPI to AAAA, resulted in a reduction in titer by 2.5-fold for the virus and 10-fold for the vector. Mutating both L1 and L2 motifs reduced the titer by 15.9-fold for virus and 24.1-fold for vector and mutation of the L3 domain from YEIL to AAAA reduced the titer to below the detection threshold ( 10 infectious particles/ ml) in both cases. Mutation of PFV potential L domains results in defects of virus particle budding but not in Gag synthesis or particle infectivity. Reductions in titer could reflect a reduced level of Gag expression, a failure of virus particles to bud from the cell, or budding of normal quantities of less infectious particles. In order to differentiate between these possibilities, lysates and culture supernatants from cells expressing wild-type and mutant PFV vectors were analyzed by SDS-PAGE and Western blotting for the presence of PFV proteins. This analysis showed that in all cases, the mutant forms of Gag were expressed as full-length proteins at levels comparable to those of wild-type Gag (Fig. 3A). Normally, a significant proportion of the fulllength 72-kDa PFV Gag undergoes a posttranslational maturation, with a 4-kDa fragment being removed by the viralencoded protease to yield a 68-kDa form. The degree and rate of this cleavage appears normal for Gag with mutated L1, L2, and L1 and L2 domains (Fig. 3A). However, Gag processing was significantly reduced when the L3 domain was mutated (Fig. 3B). Having shown that the reduction in titer was not simply a reflection of reduced expression of mutant Gag, we analyzed the level of Gag released to the culture supernatant that could be pelleted through a sucrose cushion. In each case, mutation of the candidate L domain resulted in a reduction in the fraction of Gag that was present in the virion pellets (Fig. 3C). Although the Western blot analysis was not quantitative, these data suggest that the reduced PFV titers obtained upon L1 and L2 mutation predominantly reflect a reduced level of particle egress, rather than release of noninfectious particles. In the case of the L3 mutant, where no infectious particles could be detected, some Gag was detectable in the pellet. This was all in the unprocessed form, in agreement with a previous study which showed that PFV particles containing unprocessed Gag are noninfectious (8). No particles were detected in supernatants from cells transfected with wild-type Gag in the absence of expression of Env (data not shown). PFV budding is inhibited by a blockage of the VPS pathway. One of the final steps in budding via the class E VPS pathway requires the assembled ESCRT protein complexes to be actively removed by the action of the ATPase Vps4. Thus, expression of a dominant negative form of this enzyme results in inhibition of class E VPS-dependent viral budding. For a number of retroviruses, this leads to the accumulation of virions that appear, in electron micrographs, to be blocked at a late stage in budding. We, therefore, coexpressed dominant negative forms of Vps4 and the closely related Vps4B/SKD1 (50) as GFP or DS-RED fusion proteins in 293T cells transfected with PFV virus and vector constructs. Unfused GFP or DS-RED was used as a control for nonspecific effects. Dominant negative Vps4 reduced the level of PFV virus and vector production by 11.5-fold and 34.8-fold, respectively (Fig. 4A), and dominant negative Vps4B/SKD1 reduced vector production by 8.7- fold (Fig. 4B). EM analysis of 293T cells cotransfected with PFV vector and dominant negative Vps4 revealed a phenotype
4 VOL. 79, 2005 FUNCTIONAL L DOMAINS IN PFV Gag 6395 FIG. 3. Western blots showing the PFV Gag proteins for vector and L-domain mutants. (A) Gag proteins from lysates of 293T cells transfected with the pczhfvenv construct and either the wild-type (MH71), PSAP/PPPI dual-mutated (MH71 L1/L2), PPPI-mutated (MH71 L2), or PSAP-mutated (MH71 L1) vector constructs and harvested at day 3 posttransfection. Normal Gag processing is seen for the L1, L2, and L1/L2 double mutants. (B) Gag in lysates from cells 3 days posttransfection with the pczhfvenv construct and either the wildtype (MH71) or YEIL mutation (MH71 L3) where little of the 68-kDa cleavage product is observed. In this case, the 293T cells were transfected with half the normal amount of MH71 or MH71 L3 construct. For panels A and B, identical results were obtained when lysates were prepared on days 1 and 2 posttransfection. (C) Gag proteins of sucrose-cushion purified vector particles from the wild-type and L-domain mutant vector preparations. A reduced amount of Gag is seen in the mutant vector preparations consistent with the reduced titer. In the case of the L3 mutant, only unprocessed Gag is detected in the culture supernatant. similar to that seen for other retroviruses known to require an intact VPS pathway during budding, namely, an accumulation of assembled virus particles arrested at a late stage of budding (Fig. 5B). In addition, we frequently observed fully assembled, but not yet enveloped, capsids in close proximity to cell membranes, as well as apparently partially enveloped forms. Taken together, these results show that inhibition of the VPS pathway inhibits PFV budding and perhaps envelopment. We then asked if the budding of particles with mutated L1 and L2 domains was also sensitive to expression of dominant negative Vps4. Only PFV with an intact L1 domain was sensitive to dominant negative Vps4, with infectious virus or vector production being inhibited by 4.6-fold and by 7.5-fold, respectively (Fig. 4A). An identical result was obtained with dominant negative Vps4B/SKD1 (Fig. 4B). L domains of the PT/SAP (L1) type interact directly with the Tsg101 component of the ESCRT-I complex (30). We therefore examined the effect of a dominant negative, PT/SAPbinding form of Tsg101 consisting of the N-terminal 157 amino acids on budding of PFV vector (Fig. 4C). Consistent with the aforementioned inhibition by dominant negative Vps4, wildtype PFV vector, as well as mutants in which Gag had an intact L1 domain, were inhibited by the overexpression of dominant negative Tsg101. Constructs in which both L1 and L2 domains were mutated were completely insensitive to dominant negative Tsg101. Surprisingly, when Gag had a mutated L1 (PSAP) domain but an intact L2 (PPPI), residual infectious virus release was inhibited to a significant degree (approximately 10- fold) by dominant negative Tsg101. However, the degree of inhibition was 10- to 100-fold less than that for Gag with an intact L1 domain. Analysis of interactions between PFV Gag and Tsg101 using yeast 2-hybrid assays showed a clear interaction between wild-type Gag and Tsg101. This interaction was dependent on the L1 (PSAP) motif, and no interaction between L2 (PPPI) and Tsg101 was observed (Table 1). Using yeast two hybrid assays, we tested whether PFV Gag containing an intact or mutated PPPI motif could bind to the WW domains from an variety of HECT ubiqutin ligases, including Nedd4, Nedd4-L, Bul-1, WWP1, and Itch. These results were universally negative (data not shown). The L3 (YPXL/LXXLF) L domain type apparently recruits components of the class E VPS pathway at a stage downstream of Tsg101, via an interaction with the protein AIP-1/ALIX (23, 32, 41), which bridges the ESCRT-I and ESCRT-III complexes. We, therefore, determined whether the expression of a dominant negative form of AIP-1/ALIX (lacking amino acids 1 through 176) inhibited PFV vector budding. While this protein has previously been shown to inhibit potently YPDL-dependent EIAV budding (32), PFV release was not significantly affected, irrespective of whether the other candidate L domains were intact or mutated (Fig. 4D). Additionally, no interaction between PFV Gag and AIP-1/ALIX was observed using the yeast two-hybrid assay (Table 1). DISCUSSION Three classes of L-domain sequences based on the peptide motifs PT/SAP, PPPY, and YPDL/LXXLF within the structural proteins of retroviruses and several other enveloped viruses interact with cellular machinery involved in vesicle budding (reviewed in reference 11). Our examination of the PFV Gag sequence revealed three tetrapeptide motifs with significant homology to each of the three known classes of viral L domains, specifically, PSAP, PPPI, and YEIL. While the PSAP and PPPI motifs are present in only PFV and the other primate foamy viruses, the YXXL domain is conserved in all known foamy virus Gag proteins. In the context of both intact PFV and in a PFV-based vector system, mutation of each individual candidate L domain had no effect on levels of Gag protein synthesis, but resulted in a significant reduction in the yield of virus particles. Mutation of either the PSAP or PPPI domain reduced viral titer by 2.5- to 10-fold, an effect similar in magnitude to that observed upon mutation of the L domains of other retroviruses (5, 15, 16, 45, 47). L domains frequently occur in combination and bind separate cofactors. We, therefore, constructed PFV containing a Gag protein with muta-
5 6396 PATTON ET AL. J. VIROL. FIG. 4. Effect of the dominant negative Vps4, Tsg101 or AIP-1/ALIX proteins on release of PFV virus or vector. Hatched bars indicate titers of virus produced from 293T cells transfected with pczhsrv2 (wild type or mutant), solid bars show titers of vector from cells transfected with MH71 (wild type or mutant). All vector constructs were cotransfected with pczhfvenv. In all cases, negative controls were also cotransfected with pcr3.1gfp or pdsred-c1. (A) Effect of expression of dominant negative Vps4 (VPS4-223 M-GFP). (B) Expression of dominant negative Vps4B/SKD1 E228Q. (C) Expression of dominant negative truncated Tsg101 (pcr3.1-yfp-tsg101 [1-157]). (D) Expression of dominant negative truncated AIP-1/ALIX [pcr3.1-yfp-alix(d1-176)]. tions in both PSAP and PPPI domains. This had a greater effect on viral particle release than did each single mutation, confirming that these domains act in a nonredundant fashion to promote the formation of PFV virions. The known L domains of enveloped viruses hijack the cellular machinery normally used in budding of vesicles into MVBs. This involves the sequential recruitment of ESCRT complexes, with the AAA-ATPase Vps4 subsequently being required for their dissociation from the membrane and recycling. PT/SAP based L domains bind directly to the Tsg101 component of the ESCRT-I complex (12, 42), YPXL motifs bind AIP-1/ALIX (32, 41), and PPPY motifs the WW domains of Nedd4-like ubiquitin ligases (48, 49), respectively. We found that expression of dominant negative versions of the Tsg101 component of ESCRT -I and Vps4 significantly reduced the budding of PFV. This suggests that, like other retroviruses, PFV budding is dependent, at least in part, on the class E VPS machinery. However, expression of a dominant negative form of AIP-1/ALIX had no effect on PFV budding, suggesting that it is not required in the budding of PFV. Consistent with these observations, PFV Gag bound to Tsg101 but not to AIP-1/ ALIX. Of note, ESCRT complex assembly does not require AIP-1/ALIX in the yeast Saccharomyces cerevisiae (6) or human cells (44). An analysis of the action of dominant negative Tsg101 and Vps4 proteins on mutant PFV budding revealed discrepancies between the current models of L-domain exploitation of ESCRT machinery and that apparently used by PFV. Current models predict that if both PSAP and PPPI domains act by recruiting components of the class E VPS pathway, then a dominant negative form of Vps4 should be inhibitory for both. However, for both replication-competent PFV and the PFVbased vector, dominant negative Vps4 and Vps4B/SKD1 reduced virion yield only in the presence of an intact PSAP motif. Dominant negative Tsg101 inhibited both PSAP and PPPI domain function. This might suggest that the PPPI motif is not a true L domain and facilitates PFV egress by some mechanism completely independent of the class E VPS pathway. However, this interpretation is difficult to reconcile with the observation that the presence of the PPPI motif both stimulates PFV egress and confers at least some degree of sensitivity to dominant negative Tsg101 in the absence of PSAP. Conversely, absence of the PPPI motif apparently renders PSAP-dependent PFV egress hypersensitive to dominant negative Tsg101. This find-
6 VOL. 79, 2005 FUNCTIONAL L DOMAINS IN PFV Gag 6397 TABLE 1. Direct interaction of PFV Gag with Tsg101 shown by yeast two-hybrid assay a GAL4 (DNA binding) fusion VP16 (activation domain) fusion -Galactosidase activity (OD 540 ) b None None 0.01 PFV Gag (WT) None 0.01 PFV Gag (L1) None 0.01 PFV Gag (L2) None 0.01 PFV Gag (L1 L2) None 0.01 PFV Gag (WT) Tsg PFV Gag (L1) Tsg PFV Gag (L2) Tsg PFV Gag (L1 L2) Tsg FIG. 5. EM analysis of 293T cells transfected with PFV vector constructs. (A) Wild-type PFV vector constructs. (B) Wild-type PFV vector constructs cotransfected with dominant negative Vps4. ing contrasts with previous studies which showed that PPXYdependent MuLV budding was unaffected by dominant negative Tsg101 fragments (7, 31) and insensitive to Tsg101 depletion using sirna (12). These observations hint at a functional relationship between the PPPI motif and Tsg101, but the PFV PPPI clearly does not bind directly to Tsg101 in the yeast two-hybrid assays. It is possible that either the presence of an isoleucine in the PFV PPPI motif (as opposed to a tyrosine in conventional PPXY L domains), and/or that the cellular location of PFV budding (ER rather than plasma membrane or endosomal membranes) changes the nature of the factors recruited by the PFV PPPI domain and confers its partial dependence on Tsg101. It will be interesting to determine why PPPI-dependent PFV budding appears Tsg101 dependent but Vps4 independent and whether PFV budding shares features with that of Rhabdoviruses which has been reported to be Vps4 independent (20). PFV Gag also contains the sequence YEIL, which is homologous to the YPDL-based L domain found in EIAV. Mutation of this sequence completely abolished the ability of the PFV Gag protein to support infectious particle formation. While the YPDL motif from EIAV binds to AIP-1/ALIX (12, 32, 41), the PFV YEIL motif lacks this activity, since we find no direct interaction of PFV Gag and AIP-1/ALIX in yeast two-hybrid assays. Predictably therefore, a dominant negative form of AIP-1/ALIX failed to inhibit PFV budding. While this class of L domain was originally described as YXXL rather than Tsg101 None 0.01 Tsg101 PFV Gag (WT) Tsg101 PFV Gag (L1) 0.01 Tsg101 PFV Gag (L2) Tsg101 PFV Gag (L1 L2) 0.01 PFV Gag AIP1/ALIX 0.01 AIP1/ALIX PFV Gag 0.01 AIP1/ALIX EIAV p a Yeast (Y190) cells were transformed with the indicated GAL4 DNA binding domain and VP16 activation domain fusion protein expression plasmids. Transformants were selected on media lacking tryptophan and leucine, and -galactosidase levels in cell lysates were determined. WT, wild type. b OD 540, optical density at 540 nm. YPXL (39) it has been shown that the proline plays an important role in EIAV budding (39) and binding of AIP-1/ALIX (43). Thus we conclude that PFV budding probably proceeds independently of AIP-1/ALIX. At present, it is therefore unclear what role the YEIL motif plays in PFV assembly. It may simply be required for the structural integrity of Gag monomers or multimers at an earlier stage in the assembly process. Indeed, the virally encoded protease cleaves 4 kda from the C terminus of PFV Gag (8, 10). This occurs prior to budding and is thought to be required for the viral particles to be infectious (8). Mutation of the PSAP and PPPI domains had no effect on this processing. However, mutation of YEIL significantly reduced the kinetics of processing. The YEIL domain lies in a region of sequence previously shown to contain a splice acceptor sight for the production of the Pol mrna transcript (21, 51). These studies found that minor nucleotide changes in this region, which conserve the Gag coding sequence, abolish the expression of Pol and prevent the processing of Gag. In our hands, mutation of the YEIL domain to AAAA does not completely abolish processing of Gag but slows its kinetics, possibly reflecting a reduced expression of Pol. In conclusion, we have identified three motifs within PFV Gag with significant homology to known L domains. One of these, PSAP, functions as a conventional L domain by binding to Tsg101. A second, PPPI, which does not bind directly Tsg101, also plays a role in virus assembly or budding and confers sensitivity to dominant negative Tsg101. The third candidate L-domain motif, YEIL, is required for virus assembly but appears to act by a mechanism distinct from that of known L domains. Differences in the mechanism by which these domains function in PFV compared to other retroviruses may reflect differences in budding mechanisms across specific cel-
7 6398 PATTON ET AL. J. VIROL. lular membranes. Elucidation of the reasons for these differences provides scope for future study. ACKNOWLEDGMENTS We thank Wes Sundquist for providing dominant negative Vps4B/ SKD1 constructs and comments on the manuscript and also Mike Hollinshead for assistance with electron microscopy. This work was supported by grants from the Wellcome Trust and the Jefferiss Research Trust to M.O.M. and the NIH (R01A and R01A150111) to P.D.B. REFERENCES 1. Babst, M., B. Wendland, E. J. Estepa, and S. D. Emr The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17: Bache, K. G., A. Brech, A. Mehlum, and H. Stenmark Hrs regulates multivesicular body formation via ESCRT recruitment to endosomes. J. Cell Biol. 162: Baldwin, D. N., and M. L. Linial The roles of Pol and Env in the assembly pathway of human foamy virus. J. Virol. 72: Bieniasz, P. D., A. Rethwilm, R. Pitman, M. D. Daniel, I. Chrystie, and M. O. McClure A comparative study of higher primate foamy viruses, including a new virus from a gorilla. Virology 207: Bouamr, F., J. A. Melillo, M. Q. Wang, K. Nagashima, M. D. Los Santos, A. Rein, and S. P. Goff PPPYEPTAP motif is the late domain of human T-cell leukemia virus type 1 GaG and mediates its functional interaction with cellular proteins Nedd4 and Tsg101. J. Virol. 77: Bowers, K., J. Lottridge, S. B. Helliwell, L. M. Goldthwaite, J. P. Luzio, and T. H. Stevens Protein-protein interactions of ESCRT complexes in the yeast Saccharomyces cerevisiae. Traffic 5: Demirov, D. G., A. Ono, J. M. Orenstein, and E. O. Freed Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. USA 99: Eastman, S. W., and M. L. Linial Identification of a conserved residue of foamy virus Gag required for intracellular capsid assembly. J. Virol. 75: Enssle, J., I. Jordan, B. Mauer, and A. Rethwilm Foamy virus reverse transcriptase is expressed independently from the Gag protein. Proc. Natl. Acad. Sci. USA 93: Fischer, N., M. Heinkelein, D. Lindemann, J. Enssle, C. Baum, E. Werder, H. Zentgraf, J. G. Muller, and A. Rethwilm Foamy virus particle formation. J. Virol. 72: Freed, E. O Viral late domains. J. Virol. 76: Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107: Goepfert, P. A., K. Shaw, G. Wang, A. Bansal, B. H. Edwards, and M. J. Mulligan An endoplasmic reticulum retrieval signal partitions human foamy virus maturation to intracytoplasmic membranes. J. Virol. 73: Gottlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 88: Gottwein, E., J. Bodem, B. Muller, A. Schmechel, H. Zentgraf, and H. G. Krausslich The Mason-Pfizer monkey virus PPPY and PSAP motifs both contribute to virus release. J. Virol. 77: Harty, R. N., M. E. Brown, G. Wang, J. Huibregtse, and F. P. Hayes A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. USA 97: Harty, R. N., J. Paragas, M. Sudol, and P. Palese A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J. Virol. 73: Heinkelein, M., M. Rammling, T. Juretzek, D. Lindemann, and A. Rethwilm Retrotransposition and cell-to-cell transfer of foamy viruses. J. Virol. 77: Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed p6gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69: Irie, T., J. M. Licata, J. P. McGettigan, M. J. Schnell, and R. N. Harty Budding of PPxY-containing rhabdoviruses is not dependent on host proteins TGS101 and VPS4A. J. Virol. 78: Jordan, I., J. Enssle, E. Guttler, B. Mauer, and A. Rethwilm Expression of human foamy virus reverse transcriptase involves a spliced pol mrna. Virology 224: Katzmann, D. J., C. J. Stefan, M. Babst, and S. D. Emr Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J. Cell Biol. 162: Kikonyogo, A., F. Bouamr, M. L. Vana, Y. Xiang, A. Aiyar, C. Carter, and J. Leis Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc. Natl. Acad. Sci. USA 98: Lindemann, D., T. Pietschmann, M. Picard-Maureau, A. Berg, M. Heinkelein, J. Thurow, P. Knaus, H. Zentgraf, and A. Rethwilm A particleassociated glycoprotein signal peptide essential for virus maturation and infectivity. J. Virol. 75: Linial, M. L Foamy viruses are unconventional retroviruses. J. Virol. 73: Lochelt, M Foamy virus transactivation and gene expression, p In A. Rethwilm (ed.), Foamy viruses. Springer-Verlag, Berlin, Germany. 27. Lochelt, M., and R. M. Flugel The human foamy virus pol gene is expressed as a Pro-Pol polyprotein and not as a Gag-Pol fusion protein. J. Virol. 70: Lu, Q., L. W. Q. Hope, M. Brasch, C. Reinhard, and S. N. Cohen TSG101 interaction with HRS mediates endosomal trafficking and receptor down-regulation. Proc. Natl. Acad. Sci. USA 100: Martin-Serrano, J., D. Perez-Caballero, and P. D. Bieniasz Contextdependent effects of L domains and ubiquitination on viral budding. J. Virol. 78: Martin-Serrano, J., T. Zang, and P. D. Bieniasz HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7: Martin-Serrano, J., T. Zang, and P. D. Bieniasz Role of ESCRT-I in retroviral budding. J. Virol. 77: Martin-Serrano, J., A. Yaravoy, D. Perez-Caballero, and P. D. Bieniasz Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc. Natl. Acad. Sci. USA 100: Moebes, A., J. Enssle, P. D. Bieniasz, M. Heinkelein, D. Lindemann, M. Bock, M. O. McClure, and A. Rethwilm Human foamy virus reverse transcription that occurs late in the viral replication cycle. J. Virol. 71: Patton, G. S., O. Erlwein, and M. O. McClure Cell cycle dependence of foamy virus vectors. J. Gen. Virol. 85: Perez, M., R. C. Craven, and J. C. de la Torre The small RING finger protein Z drives arenavirus budding: implications for antiviral strategies. Proc. Natl. Acad. Sci. USA 100: Pietschmann, T., M. Heinkelein, M. Heldmann, H. Zentgraf, A. Rethwilm, and D. Lindemann Foamy virus capsids require the cognate envelope protein for particle export. J. Virol. 73: Pornillos, O., J. E. Garrus, and W. I. Sundquist Mechanisms of enveloped RNA virus budding. Trends Cell Biol. 12: Pornillos, O., D. S. Higginson, K. M. Stray, R. D. Fisher, J. E. Garrus, M. Payne, G. P. He, H. E. Wang, S. G. Morham, and W. I. Sundquist HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein. J. Cell Biol. 162: Puffer, B. A., L. J. Parent, J. W. Wills, and R. C. Montelaro Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 71: Puffer, B. A., S. C. Watkins, and R. C. Montelaro Equine infectious anemia virus Gag polyprotein late domain specifically recruits cellular AP-2 adapter protein complexes during virion assembly. J. Virol. 72: Strack, B., A. Calistri, S. Craig, E. Popova, and H. G. Gottlinger AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114: VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc. Natl. Acad. Sci. USA. 98: Vincent, O., L. Rainbow, J. Tilburn, H. N. Arst, Jr., and M. A. Peñalva YPXL/I is a protein interaction motif recognized by Aspergillus PalA and its human homologue, AIP1/Alix. Mol. Cell. Biol. 23: von Schwedler, U. K., M. Stuchell, B. Muller, D. M. Ward, H. Y. Chung, E. Morita, H. E. Wang, T. Davis, G. P. He, D. M. Cimbora, A. Scott, H. G. Krausslich, J. Kaplan, S. G. Morham, and W. I. Sundquist The protein network of HIV budding. Cell 114: Wang, H. T., N. J. Machesky, and L. M. Mansky Both the PPPY and PTAP motifs are involved in human T-cell leukemia virus type 1 particle release. J. Virol. 78: Xiang, Y., C. E. Cameron, J. W. Wills, and J. Leis Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J. Virol. 70: Yasuda, J., and E. Hunter A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J. Virol. 72: Yasuda, J., E. Hunter, M. Nakao, and H. Shida Functional involvement of a novel Nedd4-like ubiquitin ligase on retrovirus budding. EMBO Rep. 3:
8 VOL. 79, 2005 FUNCTIONAL L DOMAINS IN PFV Gag Yasuda, J., M. Nakao, Y. Kawaoka, and H. Shida Nedd4 regulates egress of Ebola virus-like particles from host cells. J. Virol. 77: Yoshimori, T., F. Yamagata, A. Yamamoto, N. Mizushima, Y. Kabeya, A. Nara, I. Miwako, M. Ohashi, M. Ohsumi, and Y. Ohsumi The mouse SKD1, a homologue of yeast Vps4p, is required for normal endosomal trafficking and morphology in mammalian cells. Mol. Biol. Cell 11: Yu, S. F., D. N. Baldwin, S. R. Gwynn, S. Yendapalli, and M. L. Linial Human foamy virus replication: a pathway distinct from that of retroviruses and hepadnaviruses. Science 271: Yuan, B., X. Li, and S. P. Goff Mutations altering the moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 18:
Cellular Factors Required for Lassa Virus Budding
 JOURNAL OF VIROLOGY, Apr. 2006, p. 4191 4195 Vol. 80, No. 8 0022-538X/06/$08.00 0 doi:10.1128/jvi.80.8.4191 4195.2006 Copyright 2006, American Society for Microbiology. All Rights Reserved. Cellular Factors
JOURNAL OF VIROLOGY, Apr. 2006, p. 4191 4195 Vol. 80, No. 8 0022-538X/06/$08.00 0 doi:10.1128/jvi.80.8.4191 4195.2006 Copyright 2006, American Society for Microbiology. All Rights Reserved. Cellular Factors
Role of ESCRT-I in Retroviral Budding
 JOURNAL OF VIROLOGY, Apr. 2003, p. 4794 4804 Vol. 77, No. 8 0022-538X/03/$08.00 0 DOI: 10.1128/JVI.77.8.4794 4804.2003 Copyright 2003, American Society for Microbiology. All Rights Reserved. Role of ESCRT-I
JOURNAL OF VIROLOGY, Apr. 2003, p. 4794 4804 Vol. 77, No. 8 0022-538X/03/$08.00 0 DOI: 10.1128/JVI.77.8.4794 4804.2003 Copyright 2003, American Society for Microbiology. All Rights Reserved. Role of ESCRT-I
Retroviral Gag proteins are necessary and sufficient for the
 Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function Dimiter G. Demirov*, Akira Ono*, Jan M. Orenstein, and Eric O. Freed* *Laboratory of Molecular
Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function Dimiter G. Demirov*, Akira Ono*, Jan M. Orenstein, and Eric O. Freed* *Laboratory of Molecular
Late Domain-Independent Rescue of a Release-Deficient Moloney Murine Leukemia Virus by the Ubiquitin Ligase Itch
 JOURNAL OF VIROLOGY, Jan. 2010, p. 704 715 Vol. 84, No. 2 0022-538X/10/$12.00 doi:10.1128/jvi.01319-09 Late Domain-Independent Rescue of a Release-Deficient Moloney Murine Leukemia Virus by the Ubiquitin
JOURNAL OF VIROLOGY, Jan. 2010, p. 704 715 Vol. 84, No. 2 0022-538X/10/$12.00 doi:10.1128/jvi.01319-09 Late Domain-Independent Rescue of a Release-Deficient Moloney Murine Leukemia Virus by the Ubiquitin
Late budding domains and host proteins in enveloped virus release
 Virology 344 (2006) 55 63 www.elsevier.com/locate/yviro Late budding domains and host proteins in enveloped virus release Paul D. Bieniasz Aaron Diamond AIDS Research Center and Laboratory of Retrovirology,
Virology 344 (2006) 55 63 www.elsevier.com/locate/yviro Late budding domains and host proteins in enveloped virus release Paul D. Bieniasz Aaron Diamond AIDS Research Center and Laboratory of Retrovirology,
Received 15 August 2002/Accepted 1 November 2002
 JOURNAL OF VIROLOGY, Feb. 2003, p. 1812 1819 Vol. 77, No. 3 0022-538X/03/$08.00 0 DOI: 10.1128/JVI.77.3.1812 1819.2003 Copyright 2003, American Society for Microbiology. All Rights Reserved. Overlapping
JOURNAL OF VIROLOGY, Feb. 2003, p. 1812 1819 Vol. 77, No. 3 0022-538X/03/$08.00 0 DOI: 10.1128/JVI.77.3.1812 1819.2003 Copyright 2003, American Society for Microbiology. All Rights Reserved. Overlapping
Ebola Virus VP40 Late Domains Are Not Essential for Viral Replication in Cell Culture
 JOURNAL OF VIROLOGY, Aug. 2005, p. 10300 10307 Vol. 79, No. 16 0022-538X/05/$08.00 0 doi:10.1128/jvi.79.16.10300 10307.2005 Copyright 2005, American Society for Microbiology. All Rights Reserved. Ebola
JOURNAL OF VIROLOGY, Aug. 2005, p. 10300 10307 Vol. 79, No. 16 0022-538X/05/$08.00 0 doi:10.1128/jvi.79.16.10300 10307.2005 Copyright 2005, American Society for Microbiology. All Rights Reserved. Ebola
JCB. After Hrs with HIV. The Journal of Cell Biology. Comment. Ali Amara 1 and Dan R. Littman 1,2
 JCB Comment After Hrs with HIV Ali Amara 1 and Dan R. Littman 1,2 1 Molecular Pathogenesis Program and 2 Howard Hughes Medical Institute, Skirball Institute of Biomolecular Medicine, New York University
JCB Comment After Hrs with HIV Ali Amara 1 and Dan R. Littman 1,2 1 Molecular Pathogenesis Program and 2 Howard Hughes Medical Institute, Skirball Institute of Biomolecular Medicine, New York University
Deletion of a Cys His motif from the Alpharetrovirus nucleocapsid domain reveals late domain mutant-like budding defects
 Virology 347 (2006) 226 233 www.elsevier.com/locate/yviro Deletion of a Cys His motif from the Alpharetrovirus nucleocapsid domain reveals late domain mutant-like budding defects Eun-Gyung Lee, Maxine
Virology 347 (2006) 226 233 www.elsevier.com/locate/yviro Deletion of a Cys His motif from the Alpharetrovirus nucleocapsid domain reveals late domain mutant-like budding defects Eun-Gyung Lee, Maxine
Mechanisms for enveloped virus budding: Can some viruses do without an ESCRT?
 Available online at www.sciencedirect.com Virology 372 (2008) 221 232 www.elsevier.com/locate/yviro Minireview Mechanisms for enveloped virus budding: Can some viruses do without an ESCRT? Benjamin J.
Available online at www.sciencedirect.com Virology 372 (2008) 221 232 www.elsevier.com/locate/yviro Minireview Mechanisms for enveloped virus budding: Can some viruses do without an ESCRT? Benjamin J.
Human Immunodeficiency Virus Type 1 Gag Engages the Bro1 Domain of ALIX/AIP1 through the Nucleocapsid
 JOURNAL OF VIROLOGY, Feb. 2008, p. 1389 1398 Vol. 82, No. 3 0022-538X/08/$08.00 0 doi:10.1128/jvi.01912-07 Copyright 2008, American Society for Microbiology. All Rights Reserved. Human Immunodeficiency
JOURNAL OF VIROLOGY, Feb. 2008, p. 1389 1398 Vol. 82, No. 3 0022-538X/08/$08.00 0 doi:10.1128/jvi.01912-07 Copyright 2008, American Society for Microbiology. All Rights Reserved. Human Immunodeficiency
Fayth K. Yoshimura, Ph.D. September 7, of 7 RETROVIRUSES. 2. HTLV-II causes hairy T-cell leukemia
 1 of 7 I. Diseases Caused by Retroviruses RETROVIRUSES A. Human retroviruses that cause cancers 1. HTLV-I causes adult T-cell leukemia and tropical spastic paraparesis 2. HTLV-II causes hairy T-cell leukemia
1 of 7 I. Diseases Caused by Retroviruses RETROVIRUSES A. Human retroviruses that cause cancers 1. HTLV-I causes adult T-cell leukemia and tropical spastic paraparesis 2. HTLV-II causes hairy T-cell leukemia
Pregenomic RNA Is Required for Efficient Incorporation of Pol Polyprotein into Foamy Virus Capsids
 JOURNAL OF VIROLOGY, Oct. 2002, p. 10069 10073 Vol. 76, No. 19 0022-538X/02/$04.00 0 DOI: 10.1128/JVI.76.19.10069 10073.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Pregenomic
JOURNAL OF VIROLOGY, Oct. 2002, p. 10069 10073 Vol. 76, No. 19 0022-538X/02/$04.00 0 DOI: 10.1128/JVI.76.19.10069 10073.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Pregenomic
Regulation of Marburg virus (MARV) budding by Nedd4.1: a different WW domain of Nedd4.1 is critical for binding to MARV and Ebola virus VP40
 Journal of General Virology (2010), 91, 228 234 DOI 10.1099/vir.0.015495-0 Regulation of Marburg virus (MARV) budding by Nedd4.1: a different WW domain of Nedd4.1 is critical for binding to MARV and Ebola
Journal of General Virology (2010), 91, 228 234 DOI 10.1099/vir.0.015495-0 Regulation of Marburg virus (MARV) budding by Nedd4.1: a different WW domain of Nedd4.1 is critical for binding to MARV and Ebola
Journal of Biology. BioMed Central
 Journal of Biology BioMed Central Open Access Research article Endophilins interact with Moloney murine leukemia virus Gag and modulate virion production Margaret Q Wang*, Wankee Kim, Guangxia Gao, Ted
Journal of Biology BioMed Central Open Access Research article Endophilins interact with Moloney murine leukemia virus Gag and modulate virion production Margaret Q Wang*, Wankee Kim, Guangxia Gao, Ted
Differential Effects of Actin Cytoskeleton Dynamics on Equine Infectious Anemia Virus Particle Production
 JOURNAL OF VIROLOGY, Jan. 2004, p. 882 891 Vol. 78, No. 2 0022-538X/04/$08.00 0 DOI: 10.1128/JVI.78.2.882 891.2004 Copyright 2004, American Society for Microbiology. All Rights Reserved. Differential Effects
JOURNAL OF VIROLOGY, Jan. 2004, p. 882 891 Vol. 78, No. 2 0022-538X/04/$08.00 0 DOI: 10.1128/JVI.78.2.882 891.2004 Copyright 2004, American Society for Microbiology. All Rights Reserved. Differential Effects
VIROLOGY. Engineering Viral Genomes: Retrovirus Vectors
 VIROLOGY Engineering Viral Genomes: Retrovirus Vectors Viral vectors Retrovirus replicative cycle Most mammalian retroviruses use trna PRO, trna Lys3, trna Lys1,2 The partially unfolded trna is annealed
VIROLOGY Engineering Viral Genomes: Retrovirus Vectors Viral vectors Retrovirus replicative cycle Most mammalian retroviruses use trna PRO, trna Lys3, trna Lys1,2 The partially unfolded trna is annealed
Vacuolar Protein Sorting Pathway Contributes to the Release of Marburg Virus
 JOURNAL OF VIROLOGY, Mar. 2009, p. 2327 2337 Vol. 83, No. 5 0022-538X/09/$08.00 0 doi:10.1128/jvi.02184-08 Copyright 2009, American Society for Microbiology. All Rights Reserved. Vacuolar Protein Sorting
JOURNAL OF VIROLOGY, Mar. 2009, p. 2327 2337 Vol. 83, No. 5 0022-538X/09/$08.00 0 doi:10.1128/jvi.02184-08 Copyright 2009, American Society for Microbiology. All Rights Reserved. Vacuolar Protein Sorting
Retroviruses. ---The name retrovirus comes from the enzyme, reverse transcriptase.
 Retroviruses ---The name retrovirus comes from the enzyme, reverse transcriptase. ---Reverse transcriptase (RT) converts the RNA genome present in the virus particle into DNA. ---RT discovered in 1970.
Retroviruses ---The name retrovirus comes from the enzyme, reverse transcriptase. ---Reverse transcriptase (RT) converts the RNA genome present in the virus particle into DNA. ---RT discovered in 1970.
October 26, Lecture Readings. Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell
 October 26, 2006 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
October 26, 2006 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
Julianne Edwards. Retroviruses. Spring 2010
 Retroviruses Spring 2010 A retrovirus can simply be referred to as an infectious particle which replicates backwards even though there are many different types of retroviruses. More specifically, a retrovirus
Retroviruses Spring 2010 A retrovirus can simply be referred to as an infectious particle which replicates backwards even though there are many different types of retroviruses. More specifically, a retrovirus
Role of the C terminus Gag protein in human immunodefieieney virus type 1 virion assembly and maturation
 Journal of General Virology (1995), 76, 3171-3179. Printed in Great Britain 3171 Role of the C terminus Gag protein in human immunodefieieney virus type 1 virion assembly and maturation X.-F. Yu, ~ Z.
Journal of General Virology (1995), 76, 3171-3179. Printed in Great Britain 3171 Role of the C terminus Gag protein in human immunodefieieney virus type 1 virion assembly and maturation X.-F. Yu, ~ Z.
Mechanisms of enveloped RNA virus budding
 569 44 Brown, S.A. et al. (1998) Holoprosencephaly due to mutations in ZIC2, a homologue of Drosophila odd-paired. Nat. Genet. 20, 180 183 45 Aruga, J. et al. (2002) Zic2 controls cerebellar development
569 44 Brown, S.A. et al. (1998) Holoprosencephaly due to mutations in ZIC2, a homologue of Drosophila odd-paired. Nat. Genet. 20, 180 183 45 Aruga, J. et al. (2002) Zic2 controls cerebellar development
HIV-1 Virus-like Particle Budding Assay Nathan H Vande Burgt, Luis J Cocka * and Paul Bates
 HIV-1 Virus-like Particle Budding Assay Nathan H Vande Burgt, Luis J Cocka * and Paul Bates Department of Microbiology, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, USA
HIV-1 Virus-like Particle Budding Assay Nathan H Vande Burgt, Luis J Cocka * and Paul Bates Department of Microbiology, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, USA
HIV-1 genome organization
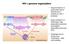 HIV-1 genome organization Gag encodes for a polyprotein that is cleaved into 4 proteins by HIV-1 protease. Gag is present in 2 transcripts: Gag polyprotein precursor and GagPol polyprotein precursor (5%).
HIV-1 genome organization Gag encodes for a polyprotein that is cleaved into 4 proteins by HIV-1 protease. Gag is present in 2 transcripts: Gag polyprotein precursor and GagPol polyprotein precursor (5%).
REGULATION OF TSG101 EXPRESSION BY THE STEADINESS BOX: A ROLE OF TSG101-ASSOCIATED LIGASE (TAL)
 REGULATION OF TSG101 EXPRESSION BY THE STEADINESS BOX: A ROLE OF TSG101-ASSOCIATED LIGASE (TAL) Bethan McDonald and Juan Martin-Serrano Department of Infectious Diseases; King's College London School of
REGULATION OF TSG101 EXPRESSION BY THE STEADINESS BOX: A ROLE OF TSG101-ASSOCIATED LIGASE (TAL) Bethan McDonald and Juan Martin-Serrano Department of Infectious Diseases; King's College London School of
Expression of Human Immunodeficiency Virus Type 1 Gag Modulates Ligand-Induced Downregulation of EGF Receptor
 JOURNAL OF VIROLOGY, Nov. 2004, p. 12386 12394 Vol. 78, No. 22 0022-538X/04/$08.00 0 DOI: 10.1128/JVI.78.22.12386 12394.2004 Copyright 2004, American Society for Microbiology. All Rights Reserved. Expression
JOURNAL OF VIROLOGY, Nov. 2004, p. 12386 12394 Vol. 78, No. 22 0022-538X/04/$08.00 0 DOI: 10.1128/JVI.78.22.12386 12394.2004 Copyright 2004, American Society for Microbiology. All Rights Reserved. Expression
Materials and Methods , The two-hybrid principle.
 The enzymatic activity of an unknown protein which cleaves the phosphodiester bond between the tyrosine residue of a viral protein and the 5 terminus of the picornavirus RNA Introduction Every day there
The enzymatic activity of an unknown protein which cleaves the phosphodiester bond between the tyrosine residue of a viral protein and the 5 terminus of the picornavirus RNA Introduction Every day there
SUPPLEMENTARY INFORMATION
 sirna pool: Control Tetherin -HA-GFP HA-Tetherin -Tubulin Supplementary Figure S1. Knockdown of HA-tagged tetherin expression by tetherin specific sirnas. HeLa cells were cotransfected with plasmids expressing
sirna pool: Control Tetherin -HA-GFP HA-Tetherin -Tubulin Supplementary Figure S1. Knockdown of HA-tagged tetherin expression by tetherin specific sirnas. HeLa cells were cotransfected with plasmids expressing
In Vivo Interference of Rous Sarcoma Virus Budding by cis Expression of a WW Domain
 JOURNAL OF VIROLOGY, Mar. 2002, p. 2789 2795 Vol. 76, No. 6 0022-538X/02/$04.00 0 DOI: 10.1128/JVI.76.6.2789 2795.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. In Vivo Interference
JOURNAL OF VIROLOGY, Mar. 2002, p. 2789 2795 Vol. 76, No. 6 0022-538X/02/$04.00 0 DOI: 10.1128/JVI.76.6.2789 2795.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. In Vivo Interference
Supplementary information. MARCH8 inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins
 Supplementary information inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins Takuya Tada, Yanzhao Zhang, Takayoshi Koyama, Minoru Tobiume, Yasuko Tsunetsugu-Yokota, Shoji
Supplementary information inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins Takuya Tada, Yanzhao Zhang, Takayoshi Koyama, Minoru Tobiume, Yasuko Tsunetsugu-Yokota, Shoji
Packaging and Abnormal Particle Morphology
 JOURNAL OF VIROLOGY, OCt. 1990, p. 5230-5234 0022-538X/90/105230-05$02.00/0 Copyright 1990, American Society for Microbiology Vol. 64, No. 10 A Mutant of Human Immunodeficiency Virus with Reduced RNA Packaging
JOURNAL OF VIROLOGY, OCt. 1990, p. 5230-5234 0022-538X/90/105230-05$02.00/0 Copyright 1990, American Society for Microbiology Vol. 64, No. 10 A Mutant of Human Immunodeficiency Virus with Reduced RNA Packaging
Recombinant Protein Expression Retroviral system
 Recombinant Protein Expression Retroviral system Viruses Contains genome DNA or RNA Genome encased in a protein coat or capsid. Some viruses have membrane covering protein coat enveloped virus Ø Essential
Recombinant Protein Expression Retroviral system Viruses Contains genome DNA or RNA Genome encased in a protein coat or capsid. Some viruses have membrane covering protein coat enveloped virus Ø Essential
The Interferon-Induced Gene ISG15 Blocks Retrovirus Release from Cells Late in the Budding Process
 JOURNAL OF VIROLOGY, May 2010, p. 4725 4736 Vol. 84, No. 9 0022-538X/10/$12.00 doi:10.1128/jvi.02478-09 Copyright 2010, American Society for Microbiology. All Rights Reserved. The Interferon-Induced Gene
JOURNAL OF VIROLOGY, May 2010, p. 4725 4736 Vol. 84, No. 9 0022-538X/10/$12.00 doi:10.1128/jvi.02478-09 Copyright 2010, American Society for Microbiology. All Rights Reserved. The Interferon-Induced Gene
Fayth K. Yoshimura, Ph.D. September 7, of 7 HIV - BASIC PROPERTIES
 1 of 7 I. Viral Origin. A. Retrovirus - animal lentiviruses. HIV - BASIC PROPERTIES 1. HIV is a member of the Retrovirus family and more specifically it is a member of the Lentivirus genus of this family.
1 of 7 I. Viral Origin. A. Retrovirus - animal lentiviruses. HIV - BASIC PROPERTIES 1. HIV is a member of the Retrovirus family and more specifically it is a member of the Lentivirus genus of this family.
Gag. Gag-Gag. TSG101 Nedd4. Two-Hybrid HIV Gag. Two-Hybrid. Two-Hybrid
 CytoTrap HIV Gag AnnexinA2 AUP1 AUP1 HIV AUP1 N Gag CytoTrap Gag-Gag TSG101 Nedd4 Two-Hybrid Two-Hybrid HIVGag Gag Two-Hybrid HIV Gag Gag HIV Two-Hybrid CytoTrap cdna Gag Gag CytoTrap Gag CytoTrap Gag-Gag
CytoTrap HIV Gag AnnexinA2 AUP1 AUP1 HIV AUP1 N Gag CytoTrap Gag-Gag TSG101 Nedd4 Two-Hybrid Two-Hybrid HIVGag Gag Two-Hybrid HIV Gag Gag HIV Two-Hybrid CytoTrap cdna Gag Gag CytoTrap Gag CytoTrap Gag-Gag
Identification of late assembly domains of the human endogenous retrovirus-k(hml-2)
 Chudak et al. Retrovirology 2013, 10:140 RESEARCH Identification of late assembly domains of the human endogenous retrovirus-k(hml-2) Open Access Claudia Chudak 1, Nadine Beimforde 1, Maja George 2, Anja
Chudak et al. Retrovirology 2013, 10:140 RESEARCH Identification of late assembly domains of the human endogenous retrovirus-k(hml-2) Open Access Claudia Chudak 1, Nadine Beimforde 1, Maja George 2, Anja
7.012 Quiz 3 Answers
 MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel Friday 11/12/04 7.012 Quiz 3 Answers A > 85 B 72-84
MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel Friday 11/12/04 7.012 Quiz 3 Answers A > 85 B 72-84
Tsg101 and the Vacuolar Protein Sorting Pathway Are Essential for HIV-1 Budding
 Cell, Vol. 107, 55 65, October 5, 2001, Copyright 2001 by Cell Press Tsg101 and the Vacuolar Protein Sorting Pathway Are Essential for HIV-1 Budding Jennifer E. Garrus, 1,5 Uta K. von Schwedler, 1,5 Owen
Cell, Vol. 107, 55 65, October 5, 2001, Copyright 2001 by Cell Press Tsg101 and the Vacuolar Protein Sorting Pathway Are Essential for HIV-1 Budding Jennifer E. Garrus, 1,5 Uta K. von Schwedler, 1,5 Owen
~Lentivirus production~
 ~Lentivirus production~ May 30, 2008 RNAi core R&D group member Lentivirus Production Session Lentivirus!!! Is it health threatening to lab technician? What s so good about this RNAi library? How to produce
~Lentivirus production~ May 30, 2008 RNAi core R&D group member Lentivirus Production Session Lentivirus!!! Is it health threatening to lab technician? What s so good about this RNAi library? How to produce
Mutations of the Human Immunodeficiency Virus Type 1 p6 Gag Domain Result in Reduced Retention of Pol Proteins during Virus Assembly
 JOURNAL OF VIROLOGY, Apr. 1998, p. 3412 3417 Vol. 72, No. 4 0022-538X/98/$04.00 0 Copyright 1998, American Society for Microbiology Mutations of the Human Immunodeficiency Virus Type 1 p6 Gag Domain Result
JOURNAL OF VIROLOGY, Apr. 1998, p. 3412 3417 Vol. 72, No. 4 0022-538X/98/$04.00 0 Copyright 1998, American Society for Microbiology Mutations of the Human Immunodeficiency Virus Type 1 p6 Gag Domain Result
Introduction retroposon
 17.1 - Introduction A retrovirus is an RNA virus able to convert its sequence into DNA by reverse transcription A retroposon (retrotransposon) is a transposon that mobilizes via an RNA form; the DNA element
17.1 - Introduction A retrovirus is an RNA virus able to convert its sequence into DNA by reverse transcription A retroposon (retrotransposon) is a transposon that mobilizes via an RNA form; the DNA element
Involvement of host cellular multivesicular body functions in hepatitis B virus budding
 Involvement of host cellular multivesicular body functions in hepatitis B virus budding Tokiko Watanabe*, Ericka M. Sorensen*, Akira Naito*, Meghan Schott*, Seungtaek Kim*, and Paul Ahlquist* *Institute
Involvement of host cellular multivesicular body functions in hepatitis B virus budding Tokiko Watanabe*, Ericka M. Sorensen*, Akira Naito*, Meghan Schott*, Seungtaek Kim*, and Paul Ahlquist* *Institute
Helper virus-free transfer of human immunodeficiency virus type 1 vectors
 Journal of General Virology (1995), 76, 691 696. Printed in Great Britabz 691 Helper virus-free transfer of human immunodeficiency virus type 1 vectors Jennifer H. Riehardson,~ Jane F. Kaye, Lisa A. Child
Journal of General Virology (1995), 76, 691 696. Printed in Great Britabz 691 Helper virus-free transfer of human immunodeficiency virus type 1 vectors Jennifer H. Riehardson,~ Jane F. Kaye, Lisa A. Child
JCB Article. HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein. The Journal of Cell Biology
 JCB Article HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein Owen Pornillos, 1 Daniel S. Higginson, 1 Kirsten M. Stray, 1 Robert D. Fisher, 1 Jennifer E. Garrus, 1 Marielle Payne,
JCB Article HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein Owen Pornillos, 1 Daniel S. Higginson, 1 Kirsten M. Stray, 1 Robert D. Fisher, 1 Jennifer E. Garrus, 1 Marielle Payne,
The Clathrin Adaptor Complex AP-1 Binds HIV-1 and MLV Gag and Facilitates Their Budding D
 Vol. 18, 3193 3203, August 2007 The Clathrin Adaptor Complex AP-1 Binds HIV-1 and MLV Gag and Facilitates Their Budding D Grégory Camus,* Carolina Segura-Morales, Dorothee Molle, Sandra Lopez-Vergès,*
Vol. 18, 3193 3203, August 2007 The Clathrin Adaptor Complex AP-1 Binds HIV-1 and MLV Gag and Facilitates Their Budding D Grégory Camus,* Carolina Segura-Morales, Dorothee Molle, Sandra Lopez-Vergès,*
Structural vs. nonstructural proteins
 Why would you want to study proteins associated with viruses or virus infection? Receptors Mechanism of uncoating How is gene expression carried out, exclusively by viral enzymes? Gene expression phases?
Why would you want to study proteins associated with viruses or virus infection? Receptors Mechanism of uncoating How is gene expression carried out, exclusively by viral enzymes? Gene expression phases?
Ubiquitination of Human Immunodeficiency Virus Type 1 Gag Is Highly Dependent on Gag Membrane Association
 JOURNAL OF VIROLOGY, Sept. 2007, p. 9193 9201 Vol. 81, No. 17 0022-538X/07/$08.00 0 doi:10.1128/jvi.00044-07 Copyright 2007, American Society for Microbiology. All Rights Reserved. Ubiquitination of Human
JOURNAL OF VIROLOGY, Sept. 2007, p. 9193 9201 Vol. 81, No. 17 0022-538X/07/$08.00 0 doi:10.1128/jvi.00044-07 Copyright 2007, American Society for Microbiology. All Rights Reserved. Ubiquitination of Human
There are approximately 30,000 proteasomes in a typical human cell Each proteasome is approximately 700 kda in size The proteasome is made up of 3
 Proteasomes Proteasomes Proteasomes are responsible for degrading proteins that have been damaged, assembled improperly, or that are of no profitable use to the cell. The unwanted protein is literally
Proteasomes Proteasomes Proteasomes are responsible for degrading proteins that have been damaged, assembled improperly, or that are of no profitable use to the cell. The unwanted protein is literally
Supplementary Material
 Supplementary Material Nuclear import of purified HIV-1 Integrase. Integrase remains associated to the RTC throughout the infection process until provirus integration occurs and is therefore one likely
Supplementary Material Nuclear import of purified HIV-1 Integrase. Integrase remains associated to the RTC throughout the infection process until provirus integration occurs and is therefore one likely
MedChem 401~ Retroviridae. Retroviridae
 MedChem 401~ Retroviridae Retroviruses plus-sense RNA genome (!8-10 kb) protein capsid lipid envelop envelope glycoproteins reverse transcriptase enzyme integrase enzyme protease enzyme Retroviridae The
MedChem 401~ Retroviridae Retroviruses plus-sense RNA genome (!8-10 kb) protein capsid lipid envelop envelope glycoproteins reverse transcriptase enzyme integrase enzyme protease enzyme Retroviridae The
Supplementary Information. Supplementary Figure 1
 Supplementary Information Supplementary Figure 1 1 Supplementary Figure 1. Functional assay of the hcas9-2a-mcherry construct (a) Gene correction of a mutant EGFP reporter cell line mediated by hcas9 or
Supplementary Information Supplementary Figure 1 1 Supplementary Figure 1. Functional assay of the hcas9-2a-mcherry construct (a) Gene correction of a mutant EGFP reporter cell line mediated by hcas9 or
Peptide hydrolysis uncatalyzed half-life = ~450 years HIV protease-catalyzed half-life = ~3 seconds
 Uncatalyzed half-life Peptide hydrolysis uncatalyzed half-life = ~450 years IV protease-catalyzed half-life = ~3 seconds Life Sciences 1a Lecture Slides Set 9 Fall 2006-2007 Prof. David R. Liu In the absence
Uncatalyzed half-life Peptide hydrolysis uncatalyzed half-life = ~450 years IV protease-catalyzed half-life = ~3 seconds Life Sciences 1a Lecture Slides Set 9 Fall 2006-2007 Prof. David R. Liu In the absence
Lecture Readings. Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell
 October 26, 2006 1 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
October 26, 2006 1 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
Chapter 13 Viruses, Viroids, and Prions. Biology 1009 Microbiology Johnson-Summer 2003
 Chapter 13 Viruses, Viroids, and Prions Biology 1009 Microbiology Johnson-Summer 2003 Viruses Virology-study of viruses Characteristics: acellular obligate intracellular parasites no ribosomes or means
Chapter 13 Viruses, Viroids, and Prions Biology 1009 Microbiology Johnson-Summer 2003 Viruses Virology-study of viruses Characteristics: acellular obligate intracellular parasites no ribosomes or means
RNA and Protein Requirements for Incorporation of the Pol Protein into Foamy Virus Particles
 REFERENCES CONTENT ALERTS RNA and Protein Requirements for Incorporation of the Pol Protein into Foamy Virus Particles Katrin Peters, Tatiana Wiktorowicz, Martin Heinkelein and Axel Rethwilm J. Virol.
REFERENCES CONTENT ALERTS RNA and Protein Requirements for Incorporation of the Pol Protein into Foamy Virus Particles Katrin Peters, Tatiana Wiktorowicz, Martin Heinkelein and Axel Rethwilm J. Virol.
number Done by Corrected by Doctor Ashraf
 number 4 Done by Nedaa Bani Ata Corrected by Rama Nada Doctor Ashraf Genome replication and gene expression Remember the steps of viral replication from the last lecture: Attachment, Adsorption, Penetration,
number 4 Done by Nedaa Bani Ata Corrected by Rama Nada Doctor Ashraf Genome replication and gene expression Remember the steps of viral replication from the last lecture: Attachment, Adsorption, Penetration,
The clathrin adaptor Numb regulates intestinal cholesterol. absorption through dynamic interaction with NPC1L1
 The clathrin adaptor Numb regulates intestinal cholesterol absorption through dynamic interaction with NPC1L1 Pei-Shan Li 1, Zhen-Yan Fu 1,2, Ying-Yu Zhang 1, Jin-Hui Zhang 1, Chen-Qi Xu 1, Yi-Tong Ma
The clathrin adaptor Numb regulates intestinal cholesterol absorption through dynamic interaction with NPC1L1 Pei-Shan Li 1, Zhen-Yan Fu 1,2, Ying-Yu Zhang 1, Jin-Hui Zhang 1, Chen-Qi Xu 1, Yi-Tong Ma
Quantifying Lipid Contents in Enveloped Virus Particles with Plasmonic Nanoparticles
 Quantifying Lipid Contents in Enveloped Virus Particles with Plasmonic Nanoparticles Amin Feizpour Reinhard Lab Department of Chemistry and the Photonics Center, Boston University, Boston, MA May 2014
Quantifying Lipid Contents in Enveloped Virus Particles with Plasmonic Nanoparticles Amin Feizpour Reinhard Lab Department of Chemistry and the Photonics Center, Boston University, Boston, MA May 2014
Herpesviruses. Virion. Genome. Genes and proteins. Viruses and hosts. Diseases. Distinctive characteristics
 Herpesviruses Virion Genome Genes and proteins Viruses and hosts Diseases Distinctive characteristics Virion Enveloped icosahedral capsid (T=16), diameter 125 nm Diameter of enveloped virion 200 nm Capsid
Herpesviruses Virion Genome Genes and proteins Viruses and hosts Diseases Distinctive characteristics Virion Enveloped icosahedral capsid (T=16), diameter 125 nm Diameter of enveloped virion 200 nm Capsid
Hepatitis B Antiviral Drug Development Multi-Marker Screening Assay
 Hepatitis B Antiviral Drug Development Multi-Marker Screening Assay Background ImQuest BioSciences has developed and qualified a single-plate method to expedite the screening of antiviral agents against
Hepatitis B Antiviral Drug Development Multi-Marker Screening Assay Background ImQuest BioSciences has developed and qualified a single-plate method to expedite the screening of antiviral agents against
Virus Research 150 (2010) Contents lists available at ScienceDirect. Virus Research. journal homepage:
 Virus Research 150 (2010) 153 157 Contents lists available at ScienceDirect Virus Research journal homepage: www.elsevier.com/locate/virusres Short communication In vitro assembly of the feline immunodeficiency
Virus Research 150 (2010) 153 157 Contents lists available at ScienceDirect Virus Research journal homepage: www.elsevier.com/locate/virusres Short communication In vitro assembly of the feline immunodeficiency
CANCER. Inherited Cancer Syndromes. Affects 25% of US population. Kills 19% of US population (2nd largest killer after heart disease)
 CANCER Affects 25% of US population Kills 19% of US population (2nd largest killer after heart disease) NOT one disease but 200-300 different defects Etiologic Factors In Cancer: Relative contributions
CANCER Affects 25% of US population Kills 19% of US population (2nd largest killer after heart disease) NOT one disease but 200-300 different defects Etiologic Factors In Cancer: Relative contributions
Supplemental Materials and Methods Plasmids and viruses Quantitative Reverse Transcription PCR Generation of molecular standard for quantitative PCR
 Supplemental Materials and Methods Plasmids and viruses To generate pseudotyped viruses, the previously described recombinant plasmids pnl4-3-δnef-gfp or pnl4-3-δ6-drgfp and a vector expressing HIV-1 X4
Supplemental Materials and Methods Plasmids and viruses To generate pseudotyped viruses, the previously described recombinant plasmids pnl4-3-δnef-gfp or pnl4-3-δ6-drgfp and a vector expressing HIV-1 X4
Polyomaviridae. Spring
 Polyomaviridae Spring 2002 331 Antibody Prevalence for BK & JC Viruses Spring 2002 332 Polyoma Viruses General characteristics Papovaviridae: PA - papilloma; PO - polyoma; VA - vacuolating agent a. 45nm
Polyomaviridae Spring 2002 331 Antibody Prevalence for BK & JC Viruses Spring 2002 332 Polyoma Viruses General characteristics Papovaviridae: PA - papilloma; PO - polyoma; VA - vacuolating agent a. 45nm
Running Head: AN UNDERSTANDING OF HIV- 1, SYMPTOMS, AND TREATMENTS. An Understanding of HIV- 1, Symptoms, and Treatments.
 Running Head: AN UNDERSTANDING OF HIV- 1, SYMPTOMS, AND TREATMENTS An Understanding of HIV- 1, Symptoms, and Treatments Benjamin Mills Abstract HIV- 1 is a virus that has had major impacts worldwide. Numerous
Running Head: AN UNDERSTANDING OF HIV- 1, SYMPTOMS, AND TREATMENTS An Understanding of HIV- 1, Symptoms, and Treatments Benjamin Mills Abstract HIV- 1 is a virus that has had major impacts worldwide. Numerous
Section 6. Junaid Malek, M.D.
 Section 6 Junaid Malek, M.D. The Golgi and gp160 gp160 transported from ER to the Golgi in coated vesicles These coated vesicles fuse to the cis portion of the Golgi and deposit their cargo in the cisternae
Section 6 Junaid Malek, M.D. The Golgi and gp160 gp160 transported from ER to the Golgi in coated vesicles These coated vesicles fuse to the cis portion of the Golgi and deposit their cargo in the cisternae
Dr. Ahmed K. Ali Attachment and entry of viruses into cells
 Lec. 6 Dr. Ahmed K. Ali Attachment and entry of viruses into cells The aim of a virus is to replicate itself, and in order to achieve this aim it needs to enter a host cell, make copies of itself and
Lec. 6 Dr. Ahmed K. Ali Attachment and entry of viruses into cells The aim of a virus is to replicate itself, and in order to achieve this aim it needs to enter a host cell, make copies of itself and
Chapter 19: Viruses. 1. Viral Structure & Reproduction. 2. Bacteriophages. 3. Animal Viruses. 4. Viroids & Prions
 Chapter 19: Viruses 1. Viral Structure & Reproduction 2. Bacteriophages 3. Animal Viruses 4. Viroids & Prions 1. Viral Structure & Reproduction Chapter Reading pp. 393-396 What exactly is a Virus? Viruses
Chapter 19: Viruses 1. Viral Structure & Reproduction 2. Bacteriophages 3. Animal Viruses 4. Viroids & Prions 1. Viral Structure & Reproduction Chapter Reading pp. 393-396 What exactly is a Virus? Viruses
Sequences in the 5 and 3 R Elements of Human Immunodeficiency Virus Type 1 Critical for Efficient Reverse Transcription
 JOURNAL OF VIROLOGY, Sept. 2000, p. 8324 8334 Vol. 74, No. 18 0022-538X/00/$04.00 0 Copyright 2000, American Society for Microbiology. All Rights Reserved. Sequences in the 5 and 3 R Elements of Human
JOURNAL OF VIROLOGY, Sept. 2000, p. 8324 8334 Vol. 74, No. 18 0022-538X/00/$04.00 0 Copyright 2000, American Society for Microbiology. All Rights Reserved. Sequences in the 5 and 3 R Elements of Human
Chapter 19: Viruses. 1. Viral Structure & Reproduction. What exactly is a Virus? 11/7/ Viral Structure & Reproduction. 2.
 Chapter 19: Viruses 1. Viral Structure & Reproduction 2. Bacteriophages 3. Animal Viruses 4. Viroids & Prions 1. Viral Structure & Reproduction Chapter Reading pp. 393-396 What exactly is a Virus? Viruses
Chapter 19: Viruses 1. Viral Structure & Reproduction 2. Bacteriophages 3. Animal Viruses 4. Viroids & Prions 1. Viral Structure & Reproduction Chapter Reading pp. 393-396 What exactly is a Virus? Viruses
Human Genome Complexity, Viruses & Genetic Variability
 Human Genome Complexity, Viruses & Genetic Variability (Learning Objectives) Learn the types of DNA sequences present in the Human Genome other than genes coding for functional proteins. Review what you
Human Genome Complexity, Viruses & Genetic Variability (Learning Objectives) Learn the types of DNA sequences present in the Human Genome other than genes coding for functional proteins. Review what you
Chapter 6- An Introduction to Viruses*
 Chapter 6- An Introduction to Viruses* *Lecture notes are to be used as a study guide only and do not represent the comprehensive information you will need to know for the exams. 6.1 Overview of Viruses
Chapter 6- An Introduction to Viruses* *Lecture notes are to be used as a study guide only and do not represent the comprehensive information you will need to know for the exams. 6.1 Overview of Viruses
An Endoplasmic Reticulum Retrieval Signal Partitions Human Foamy Virus Maturation to Intracytoplasmic Membranes
 JOURNAL OF VIROLOGY, Sept. 1999, p. 7210 7217 Vol. 73, No. 9 0022-538X/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. An Endoplasmic Reticulum Retrieval Signal Partitions
JOURNAL OF VIROLOGY, Sept. 1999, p. 7210 7217 Vol. 73, No. 9 0022-538X/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. An Endoplasmic Reticulum Retrieval Signal Partitions
VIRUSES. 1. Describe the structure of a virus by completing the following chart.
 AP BIOLOGY MOLECULAR GENETICS ACTIVITY #3 NAME DATE HOUR VIRUSES 1. Describe the structure of a virus by completing the following chart. Viral Part Description of Part 2. Some viruses have an envelope
AP BIOLOGY MOLECULAR GENETICS ACTIVITY #3 NAME DATE HOUR VIRUSES 1. Describe the structure of a virus by completing the following chart. Viral Part Description of Part 2. Some viruses have an envelope
Chapter 13B: Animal Viruses
 Chapter 13B: Animal Viruses 1. Overview of Animal Viruses 2. DNA Viruses 3. RNA Viruses 4. Prions 1. Overview of Animal Viruses Life Cycle of Animal Viruses The basic life cycle stages of animal viruses
Chapter 13B: Animal Viruses 1. Overview of Animal Viruses 2. DNA Viruses 3. RNA Viruses 4. Prions 1. Overview of Animal Viruses Life Cycle of Animal Viruses The basic life cycle stages of animal viruses
Benchmarks Attenuated protein expression vectors for use in sirna rescue experiments
 Benchmarks Attenuated protein expression vectors for use in sirna rescue experiments Eiji Morita *, Jun Arii *, Devin Christensen, Jörg Votteler, and Wesley I. Sundquist Department of Biochemistry, University
Benchmarks Attenuated protein expression vectors for use in sirna rescue experiments Eiji Morita *, Jun Arii *, Devin Christensen, Jörg Votteler, and Wesley I. Sundquist Department of Biochemistry, University
AND JEREMY LUBAN 1,2 *
 JOURNAL OF VIROLOGY, Oct. 1999, p. 8527 8540 Vol. 73, No. 10 0022-538X/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. Human Immunodeficiency Virus Type 1 Gag Polyprotein
JOURNAL OF VIROLOGY, Oct. 1999, p. 8527 8540 Vol. 73, No. 10 0022-538X/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. Human Immunodeficiency Virus Type 1 Gag Polyprotein
HIV-1 p24 ELISA Pair Set Cat#: orb54951 (ELISA Manual)
 HIV-1 p24 ELISA Pair Set Cat#: orb54951 (ELISA Manual) BACKGROUND Human Immunodeficiency Virus ( HIV ) can be divided into two major types, HIV type 1 (HIV-1) and HIV type 2 (HIV-2). HIV-1 is related to
HIV-1 p24 ELISA Pair Set Cat#: orb54951 (ELISA Manual) BACKGROUND Human Immunodeficiency Virus ( HIV ) can be divided into two major types, HIV type 1 (HIV-1) and HIV type 2 (HIV-2). HIV-1 is related to
Human Immunodeficiency Virus
 Human Immunodeficiency Virus Virion Genome Genes and proteins Viruses and hosts Diseases Distinctive characteristics Viruses and hosts Lentivirus from Latin lentis (slow), for slow progression of disease
Human Immunodeficiency Virus Virion Genome Genes and proteins Viruses and hosts Diseases Distinctive characteristics Viruses and hosts Lentivirus from Latin lentis (slow), for slow progression of disease
Coronaviruses. Virion. Genome. Genes and proteins. Viruses and hosts. Diseases. Distinctive characteristics
 Coronaviruses Virion Genome Genes and proteins Viruses and hosts Diseases Distinctive characteristics Virion Spherical enveloped particles studded with clubbed spikes Diameter 120-160 nm Coiled helical
Coronaviruses Virion Genome Genes and proteins Viruses and hosts Diseases Distinctive characteristics Virion Spherical enveloped particles studded with clubbed spikes Diameter 120-160 nm Coiled helical
Epstein-Barr Virus: Stimulation By 5 '-Iododeoxy uridine or 5 '-Brom odeoxy uridine in Human Lymphoblastoid Cells F ro m a Rhabdom yosarcom a*
 A n n a ls o f C l i n i c a l L a b o r a t o r y S c i e n c e, Vol. 3, No. 6 Copyright 1973, Institute for Clinical Science Epstein-Barr Virus: Stimulation By 5 '-Iododeoxy uridine or 5 '-Brom odeoxy
A n n a ls o f C l i n i c a l L a b o r a t o r y S c i e n c e, Vol. 3, No. 6 Copyright 1973, Institute for Clinical Science Epstein-Barr Virus: Stimulation By 5 '-Iododeoxy uridine or 5 '-Brom odeoxy
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells
 MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
Specific Incorporation of Heat Shock Protein 70 Family Members into Primate Lentiviral Virions
 JOURNAL OF VIROLOGY, May 2002, p. 4666 4670 Vol. 76, No. 9 0022-538X/02/$04.00 0 DOI: 10.1128/JVI.76.9.4666 4670.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Specific Incorporation
JOURNAL OF VIROLOGY, May 2002, p. 4666 4670 Vol. 76, No. 9 0022-538X/02/$04.00 0 DOI: 10.1128/JVI.76.9.4666 4670.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Specific Incorporation
Supplementary Figure 1.
 Supplementary Figure 1. Visualization of endoplasmic reticulum-mitochondria interaction by in situ proximity ligation assay. A) Illustration of targeted proteins in mitochondria (M), endoplasmic reticulum
Supplementary Figure 1. Visualization of endoplasmic reticulum-mitochondria interaction by in situ proximity ligation assay. A) Illustration of targeted proteins in mitochondria (M), endoplasmic reticulum
Carboxy-Terminal Cleavage of the Human Foamy Virus Gag Precursor Molecule Is an Essential Step in the Viral Life Cycle
 JOURNAL OF VIROLOGY, Oct. 1997, p. 7312 7317 Vol. 71, No. 10 0022-538X/97/$04.00 0 Copyright 1997, American Society for Microbiology Carboxy-Terminal Cleavage of the Human Foamy Virus Gag Precursor Molecule
JOURNAL OF VIROLOGY, Oct. 1997, p. 7312 7317 Vol. 71, No. 10 0022-538X/97/$04.00 0 Copyright 1997, American Society for Microbiology Carboxy-Terminal Cleavage of the Human Foamy Virus Gag Precursor Molecule
An N-terminal domain helical motif of Prototype Foamy Virus Gag with dual functions essential for particle egress and viral infectivity
 Reh et al. Retrovirology 2013, 10:45 RESEARCH Open Access An N-terminal domain helical motif of Prototype Foamy Virus Gag with dual functions essential for particle egress and viral infectivity Juliane
Reh et al. Retrovirology 2013, 10:45 RESEARCH Open Access An N-terminal domain helical motif of Prototype Foamy Virus Gag with dual functions essential for particle egress and viral infectivity Juliane
LESSON 4.4 WORKBOOK. How viruses make us sick: Viral Replication
 DEFINITIONS OF TERMS Eukaryotic: Non-bacterial cell type (bacteria are prokaryotes).. LESSON 4.4 WORKBOOK How viruses make us sick: Viral Replication This lesson extends the principles we learned in Unit
DEFINITIONS OF TERMS Eukaryotic: Non-bacterial cell type (bacteria are prokaryotes).. LESSON 4.4 WORKBOOK How viruses make us sick: Viral Replication This lesson extends the principles we learned in Unit
viruses ISSN
 Viruses 2013, 5, 1023-1041; doi:10.3390/v5041023 OPEN ACCESS viruses ISSN 1999-4915 www.mdpi.com/journal/viruses Review The Foamy Virus Gag Proteins: What Makes Them Different? Erik Müllers 1,2, * 1 2
Viruses 2013, 5, 1023-1041; doi:10.3390/v5041023 OPEN ACCESS viruses ISSN 1999-4915 www.mdpi.com/journal/viruses Review The Foamy Virus Gag Proteins: What Makes Them Different? Erik Müllers 1,2, * 1 2
Temperature-Sensitive Mutants Isolated from Hamster and
 JOURNAL OF VIROLOGY, Nov. 1975, p. 1332-1336 Copyright i 1975 American Society for Microbiology Vol. 16, No. 5 Printed in U.S.A. Temperature-Sensitive Mutants Isolated from Hamster and Canine Cell Lines
JOURNAL OF VIROLOGY, Nov. 1975, p. 1332-1336 Copyright i 1975 American Society for Microbiology Vol. 16, No. 5 Printed in U.S.A. Temperature-Sensitive Mutants Isolated from Hamster and Canine Cell Lines
Activation of Gene Expression by Human Herpes Virus 6
 Activation of Gene Expression by Human Herpes Virus 6 M. E. M. Campbell and S. McCorkindale 1 Introduction Human herpes virus type 6 (HHV-6) was first detected by Salahuddin et al. [6] and has been isolated
Activation of Gene Expression by Human Herpes Virus 6 M. E. M. Campbell and S. McCorkindale 1 Introduction Human herpes virus type 6 (HHV-6) was first detected by Salahuddin et al. [6] and has been isolated
Lecture 2: Virology. I. Background
 Lecture 2: Virology I. Background A. Properties 1. Simple biological systems a. Aggregates of nucleic acids and protein 2. Non-living a. Cannot reproduce or carry out metabolic activities outside of a
Lecture 2: Virology I. Background A. Properties 1. Simple biological systems a. Aggregates of nucleic acids and protein 2. Non-living a. Cannot reproduce or carry out metabolic activities outside of a
Homework Hanson section MCB Course, Fall 2014
 Homework Hanson section MCB Course, Fall 2014 (1) Antitrypsin, which inhibits certain proteases, is normally secreted into the bloodstream by liver cells. Antitrypsin is absent from the bloodstream of
Homework Hanson section MCB Course, Fall 2014 (1) Antitrypsin, which inhibits certain proteases, is normally secreted into the bloodstream by liver cells. Antitrypsin is absent from the bloodstream of
Last time we talked about the few steps in viral replication cycle and the un-coating stage:
 Zeina Al-Momani Last time we talked about the few steps in viral replication cycle and the un-coating stage: Un-coating: is a general term for the events which occur after penetration, we talked about
Zeina Al-Momani Last time we talked about the few steps in viral replication cycle and the un-coating stage: Un-coating: is a general term for the events which occur after penetration, we talked about
A Single Polymorphism in HIV-1 Subtype C SP1 Is Sufficient To Confer Natural Resistance to the Maturation Inhibitor Bevirimat
 ANTIMICROBIAL AGENTS AND CHEMOTHERAPY, July 2011, p. 3324 3329 Vol. 55, No. 7 0066-4804/11/$12.00 doi:10.1128/aac.01435-10 Copyright 2011, American Society for Microbiology. All Rights Reserved. A Single
ANTIMICROBIAL AGENTS AND CHEMOTHERAPY, July 2011, p. 3324 3329 Vol. 55, No. 7 0066-4804/11/$12.00 doi:10.1128/aac.01435-10 Copyright 2011, American Society for Microbiology. All Rights Reserved. A Single
human immunodeficiency virus, type 1; PI, protease inhibitor; LPV, lopinavir;
 THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 284, NO. 43, pp. 29692 29703, October 23, 2009 2009 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in the U.S.A. HIV-1 Gag Processing
THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 284, NO. 43, pp. 29692 29703, October 23, 2009 2009 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in the U.S.A. HIV-1 Gag Processing
Cellular Motor Protein KIF-4 Associates with Retroviral Gag
 JOURNAL OF VIROLOGY, Dec. 1999, p. 10508 10513 Vol. 73, No. 12 0022-538X/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. Cellular Motor Protein KIF-4 Associates with
JOURNAL OF VIROLOGY, Dec. 1999, p. 10508 10513 Vol. 73, No. 12 0022-538X/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. Cellular Motor Protein KIF-4 Associates with
NBP Protocol. Orders: Support: Web: NBP
 NBP2-29541 NBP2-29541 Protocol Orders: orders@novusbio.com Support: technical@novusbio.com Web: www.novusbio.com Protocols, Publications, Related Products, Reviews and more: www.novusbio.com/nbp2-29541
NBP2-29541 NBP2-29541 Protocol Orders: orders@novusbio.com Support: technical@novusbio.com Web: www.novusbio.com Protocols, Publications, Related Products, Reviews and more: www.novusbio.com/nbp2-29541
Wrapping up the bad news HIV assembly and release
 Meng and Lever Retrovirology 2013, 10:5 REVIEW Open Access Wrapping up the bad news HIV assembly and release Bo Meng and Andrew ML Lever * Abstract The late Nobel Laureate Sir Peter Medawar once memorably
Meng and Lever Retrovirology 2013, 10:5 REVIEW Open Access Wrapping up the bad news HIV assembly and release Bo Meng and Andrew ML Lever * Abstract The late Nobel Laureate Sir Peter Medawar once memorably
Under the Radar Screen: How Bugs Trick Our Immune Defenses
 Under the Radar Screen: How Bugs Trick Our Immune Defenses Session 7: Cytokines Marie-Eve Paquet and Gijsbert Grotenbreg Whitehead Institute for Biomedical Research HHV-8 Discovered in the 1980 s at the
Under the Radar Screen: How Bugs Trick Our Immune Defenses Session 7: Cytokines Marie-Eve Paquet and Gijsbert Grotenbreg Whitehead Institute for Biomedical Research HHV-8 Discovered in the 1980 s at the
