Retroviral Gag proteins are necessary and sufficient for the
|
|
|
- Alaina Washington
- 5 years ago
- Views:
Transcription
1 Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function Dimiter G. Demirov*, Akira Ono*, Jan M. Orenstein, and Eric O. Freed* *Laboratory of Molecular Microbiology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD ; and Department of Pathology, George Washington University Medical Center, Washington, DC Edited by John M. Coffin, Tufts University School of Medicine, Boston, MA, and approved December 5, 2001 (received for review September 27, 2001) Efficient budding of HIV-1 from the plasma membrane of infected cells requires the function of a 6-kDa protein known as p6. A highly conserved Pro-Thr-Ala-Pro (PTAP) motif (the late or L domain), is critical for the virus-budding activity of p6. Recently, it was demonstrated that the product of tumor susceptibility gene 101 (TSG101), which contains at its N terminus a domain highly related to ubiquitin-conjugating (E2) enzymes, binds HIV-1 Gag in a p6- dependent fashion. We examined the impact of overexpressing the N-terminal region of TSG101 on HIV-1 particle assembly and release. We observed that this domain (referred to as TSG-5 ) potently inhibits virus production. Examination of cells coexpressing HIV-1 Gag and TSG-5 by electron microscopy reveals a defect in virus budding reminiscent of that observed with p6 L domain mutants. In addition, the effect of TSG-5 depends on an intact p6 L domain; the assembly and release of virus-like particles produced by Gag mutants lacking a functional p6 PTAP motif is not significantly affected by TSG-5. Furthermore, assembly and release of murine leukemia virus and Mason Pfizer monkey virus are insensitive to TSG-5. TSG-5 is incorporated into virions, confirming the Gag TSG101 interaction in virus-producing cells. Mutations that inactivate the p6 L domain block TSG-5 incorporation. These data demonstrate a link between the E2-like domain of TSG101 and HIV-1 L domain function, and indicate that TSG101 derivatives can act as potent and specific inhibitors of HIV-1 replication by blocking virus budding. Retroviral Gag proteins are necessary and sufficient for the formation and release of virus-like particles from Gagexpressing cells. The Gag proteins of type C retroviruses and lentiviruses are synthesized as polyprotein precursors that are expressed in the cytosol and transported to the plasma membrane. At the membrane, Gag Gag interactions direct the assembly of virus particles that eventually bud off from the plasma membrane. Distinct domains in Gag provide the functions required for binding of Gag to membrane, Gag Gag multimerization, and virus release. In HIV-1, membranebinding determinants map to the N-terminal region of the matrix (MA or p17) domain; Gag interaction is mediated largely by a region spanning the C terminus of the capsid (CA or p24) domain, the p2 spacer peptide, and the N terminus of the nucleocapsid (NC or p7) domain; and the budding-off of particles from the plasma membrane is promoted by a PTAP motif located near the N terminus of p6 (1, 2). Domains have been defined in the Gag proteins of several retroviruses (2), and in the structural proteins of the filoviruses (3) and rhabdoviruses (4, 5) that encode a virus release function analogous to that provided by p6 of HIV-1. These domains are collectively referred to as late or L domains to reflect their role late in the budding process (6). One of the intriguing characteristics of these L domains is that they all contain highly conserved motifs known to play a role in mediating protein protein interactions among cellular proteins. For example, the L domain of HIV-1 p6 contains a PXXP motif (7, 8); L domain function of EIAV p9 is mediated by a YXXL motif (9); and PPXY motifs are responsible for the L domain activity of MLV p12 (10), Rous sarcoma virus p2b (11), pp16 of Mason Pfizer monkey virus (M-PMV) (12), and the rhabdovirus M protein (4, 5). The filovirus matrix protein, VP40, contains overlapping PTAP and PPXY motifs (3). PXXP, YXXL, and PPXY motifs present in cellular proteins mediate interactions with Srchomology region 3 (SH3) domains, clathrin-associated adapter protein complexes, and WW domains, respectively (13 15). In several instances, interactions or subcellular colocalization between L domain proteins and cellular proteins have been reported (3, 5, 16, 17). These observations suggest that retroviral L domains function by interacting with host factors. Although the physiological relevance of potential interactions between L domains and host factors remains to be established, a variety of lines of evidence suggest that L domains interact with the host ubiquitination machinery (reviewed in ref. 18): (i) The L domain protein of MLV (p12) and HIV-1 (p6) are themselves ubiquitinated (19). (ii) Depletion of free ubiquitin (Ub) in virus-producing cells with proteasome inhibitors impairs virus budding, producing a defect reminiscent of that observed with L domain mutations (20 22). (iii) The Ub ligase Nedd4 has been shown to interact with Rous sarcoma virus Gag, and the WW domain region from Nedd4 inhibits Rous sarcoma virus particle production (23). Finally, (iv) the cellular protein TSG101, which contains at its N terminus a domain related to Ub conjugating (E2) enzymes, binds Gag in a p6-dependent manner (24). The latter observation raises the possibility that TSG101 may play a physiologically relevant role in HIV-1 budding. TSG101 was originally identified in a random RNA knockout screen for genes whose homozygous disruption led to neoplasia (25). Subsequently, the TSG101 protein, and its yeast homologue Vps23, have been implicated in a number of cellular functions, including mitotic spindle formation (26), genome stability (26), transcriptional transactivation (27, 28), regulation of MDM2 and p53 levels (29); and endosomal sorting (30 33). TSG101 Vps23 contains several domains (Fig. 1A); as mentioned above, at its N terminus is a region that bears sequence and predicted structural similarity with E2 Ub-conjugating enzymes (34). E2 (also known as UBC) enzymes play a critical role, in conjunction with E3 Ub ligases, in attaching Ub to target proteins. E2s contain at their active sites a Cys residue to which Ub is directly conjugated via a high-energy thioester bond (35). The E2-like domain of TSG101 and other E2-like cellular proteins lacks the catalytic site Cys and is therefore incapable of directly conjugating Ub. TSG101 Vps23 has recently been shown to assemble into a high This paper was submitted directly (Track II) to the PNAS office. Abbreviations: TSG101, tumor susceptibility gene 101; MA, matrix; CA, capsid; NC, nucleocapsid; MLV, murine leukemia virus; M-PMV, Mason Pfizer monkey virus; L, late; Ub, ubiquitin; EM, electron microscopy; PR, protease; WT, wild type. To whom reprint requests should be addressed. EFreed@nih.gov. The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C solely to indicate this fact. MEDICAL SCIENCES cgi doi pnas PNAS January 22, 2002 vol. 99 no
2 molecular weight (350 kda) complex (termed ESCRT-1) that functions in endosomal sorting (31, 33). Despite the fact that the E2-like domain of TSG101 Vps23 cannot directly conjugate Ub, it seems to interact directly with ubiquitinated proteins and sort them into the multivesicular body pathway (33). To determine whether TSG101 plays a role in HIV-1 budding, we overexpressed the N-terminal region of the protein, containing the E2-like domain (TSG-5 ), with the full-length HIV-1 molecular clone pnl4 3. We observed that TSG-5 expression markedly inhibited HIV-1 budding from virus-producing cells. This effect was specific for HIV-1 and depended on an intact p6 L domain. Furthermore, we demonstrated that TSG-5 is incorporated into wild-type (WT) but not L-domain-deficient virus particles. We believe that these results have significant implications for our understanding of HIV-1 budding. Materials and Methods Cells and Plasmids. HeLa and 293T cells were passaged as reported previously (36). The full-length HIV-1 molecular clone pnl4 3 (37) and the p6-mutant derivatives p6 L1Term, p6 PTAP, and p6 S14Term have been described previously (8, 38), as have the protease (PR)-defective molecular clones pnl4 3 PR and pnl4 3 PR PTAP (8). WT MLV was expressed by using psv MLV-env (ref. 39; obtained through the National Institutes of Health AIDS Research and Reference Reagent Program from N. Landau). The M-PMV molecular clone psarm4 was generously provided by E. Hunter (Univ. of Alabama, Birmingham, AL). The TSG-5 expression vector (28) was obtained from Z. Sun (Stanford Univ., Stanford, CA). The 15A NC mutant (40) was a kind gift of J. Luban (Columbia Univ., New York). Transfections, Metabolic Labeling, Radioimmunoprecipitation, Western Blotting, and Electron Microscopy. Transfections and metabolic labeling were performed as described (41). In brief, HeLa cells were transfected in six-well dishes. In all transfections, DNA concentrations were held constant with vector plasmid. Two days after transfection, cells were metabolically labeled overnight in 500 Ci (1 Ci 37 GBq) per well [ 35 S]Met Cys. Cell and virion lysates were prepared and immunoprecipitated as described (41). Anti-HIV-1, MLV, and M-PMV immunoprecipitations were performed with anti-hiv-1 Ig (obtained through the NIH AIDS Research and Reference Reagent Program); goat anti- MLV Gag p30 (ViroMed Biosafety Laboratories, Camden, NJ), and goat anti-m-pmv Gag p27 (ViroMed Biosafety Laboratories), respectively. Anti-HA antibodies were obtained from Zymed, CLONTECH, or Sigma. Western blotting was performed as detailed (42). Quantitative Western blotting was performed with a fluorchem analyzer (Alpha Innotech, San Leandro, CA) by using the manufacturer s specifications. Virusexpressing HeLa cells were prepared and examined by transmission electron microscopy (EM) as described (43). Fig. 1. TSG-5 inhibits HIV-1 particle production. (A) Schematic organization of TSG101, depicting the major domains of the protein. The region of TSG101 expressed by the TSG-5 vector is indicated. (B) Radioimmunoprecipitation analysis of cell and viral lysates obtained from HeLa cells transfected with pnl4 3 alone or cotransfected with pnl4 3 TSG-5 at 1:1, 1:2, or 1:4 DNA ratios. The positions of the Env glycoproteins gp160 and gp120; the Gag precursor, Pr55 Gag ; the p41 Gag processing intermediate, and the mature CA protein are shown on the left. (C) Quantitative analysis of the effect of TSG-5 on the production of virion-associated Gag. Relative virus release efficiency was calculated as the amount of virion-associated p24(ca) as a fraction of total Gag [(cell-associated Pr55 Gag p41 Gag p24(ca) virion p24(ca)]. Data are averages from PhosphorImager analysis of at least four experiments, SE. Results TSG-5 Inhibits HIV-1 Particle Budding. If TSG101 plays a physiologically important role in HIV-1 assembly and release, expression of the N-terminal region of TSG101, which contains the E2-like domain, might interfere with HIV-1 particle production in a transdominant negative manner. To test this possibility, we used a vector that expresses the E2-like domain of TSG101 and a portion of the downstream Pro-rich motif [TSG-5 (28); Fig. 1A]. We cotransfected HeLa cells with pnl4 3 and TSG-5 at varying input DNA ratios (pnl4 3:TSG-5 ratios 1:1, 1:2, and 1:4). We then evaluated the effect on virus particle production by radioimmunoprecipitation analysis. The results indicated that TSG-5 expression induced a marked and dose-responsive inhibition of virus particle production. At a 1:1 DNA ratio, virus production was reduced by 60%; at a 1:4 ratio a 5-fold inhibition cgi doi pnas Demirov et al.
3 Fig. 2. TSG-5 blocks HIV-1 budding. Cells transfected with pnl4 3 alone (A) or a 1:1 ratio of pnl4 3 TSG-5 (B D) were analyzed by EM. Note in B D the numerous tethered structures and doublet particles. (Bar 100 nm.) of virus production was observed (Fig. 1 B and C). Similar data were obtained in transfected 293T cells (data not shown). A severe defect in the kinetics of virus release in the presence of TSG-5 was also observed by pulse chase analysis (data not shown). To identify the step in virus production that is inhibited by TSG-5 expression, we performed EM on HeLa cells singly transfected with pnl4 3 or cotransfected with pnl4 3 TSG-5 (Fig. 2). In pnl4 3-transfected cells, numerous released mature virus particles were observed (Fig. 2A). In contrast, cells cotransfected with pnl4 3 and the TSG-5 expression vector displayed an abundance of particles tethered to the plasma membrane (Fig. 2 B D). Doublet particles were also frequently observed. This defect closely resembles that induced by mutations in the p6 L domain (7, 8, 38), strongly suggesting that TSG-5 inhibits budding by blocking L domain function. MEDICAL SCIENCES TSG-5 Does Not Inhibit the Release of L-Domain-Defective p6 Mutants. The EM data presented above suggest that TSG-5 disrupts p6 L domain function. Thus, the release of p6 mutants lacking a functional L domain should not be diminished further by TSG-5 expression. To test this prediction, we examined the impact of TSG-5 on the release of several Gag mutants (Fig. 3A): p6 L1Term, which contains a stop codon at p6 position 1 and therefore lacks all of p6 (8); p6 PTAP, which contains mutations in all four residues of the PTAP motif (8); and S14Term, which bears a stop codon at p6 position 14 but is release-competent (38). TSG-5 expression reduced p6 L1Term particle release only slightly (20%) and caused a small but reproducible increase in the efficiency of p6 PTAP release (Fig. 3 B and C). In contrast, release of the L-domain-competent p6 mutant S14Term was inhibited to an extent comparable with WT. Because the L1Term and PTAP p6 mutations severely impair virus release relative to WT, we wished to test another mutant that is similarly defective in virus production yet contains an intact L domain. Inhibition of the release of such a mutant by TSG-5 would validate the conclusion that L domain mutations and TSG-5 block the same step in the assembly release path- Fig. 3. The inhibition of virus budding by TSG-5 depends on the presence of an intact p6 L domain. (A) p6 mutations used in the study. The major Gag domains are depicted at Top, under which is shown the amino acid sequence of the N terminus of WT HIV-1 p6. The p6 L domain motif (PTAP) is shown in bold. Sequence of the L1Term, PTAP (8), and S14Term (38) mutants is provided. Asterisks denote stop codon; dashes denote sequence identity with WT. (B) Radioimmunoprecipitation analysis of cell and viral lysates obtained from HeLa cells transfected with the indicated p6 mutants alone or cotransfected with TSG-5 at a 1:1 DNA ratio. Positions of proteins are indicated as in Fig. 1 B and C. Quantitative analysis of the effect of TSG-5 on the production of virionassociated Gag, determined by PhosphorImager analysis as in Fig. 1C. The virus release efficiency in the absence of TSG-5 is set at 100% for each clone. Data are averages from at least three independent experiments, SE. Note that the virus release efficiency of the PTAP and L1Term molecular clones in the absence of TSG-5 is markedly lower than that of WT (8), whereas S14Term virus release efficiency is similar to that of WT (38). Demirov et al. PNAS January 22, 2002 vol. 99 no
4 Fig. 4. TSG-5 inhibits the release of the 15A NC mutant. Radioimmunoprecipitation analysis of cell and viral lysates obtained from HeLa cells transfected with the 15A molecular clone (40) or cotransfected with 15A and TSG-5 at a 1:1 DNA ratio. Positions of proteins are indicated as in Fig. 1B. In this experiment, TSG-5 expression caused a 60% reduction in virus release efficiency. way. For this purpose, we used the 15A NC mutant, in which all basic residues in NC were mutated to Ala (40). The 15A mutant displays a 10- to 20-fold defect in virus production relative to WT (40; data not shown), reportedly because of a defect in Gag assembly. In contrast to the insensitivity to TSG-5 observed with the L-domain-defective mutants, the release of 15A was inhibited to the same extent as WT (Fig. 4). Together, these results strongly suggest that TSG-5 disrupts virus release by specifically inhibiting p6 L domain function. TSG-5 Does Not Inhibit the Release of MLV or M-PMV Particles. If the virus-budding defect imposed by TSG-5 is specific for the HIV-1 p6 L domain, we would expect that the release of other retroviruses would be insensitive to TSG-5 expression. To test this expectation, we examined the impact of TSG-5 expression on MLV particle production. Unlike HIV-1, which contains a PTAP L domain, MLV contains an L domain (in p12-gag) of the PPPY-type (44). HeLa cells were transfected with the MLV expression vector psv MLV-env (39) or cotransfected with psv MLV-env TSG-5. Cell- and virion-associated proteins were detected by radioimmunoprecipitation analysis (Fig. 5). In contrast to results obtained with WT HIV-1, MLV particle production was not inhibited by TSG-5. We also tested the effect of TSG-5 expression on M-PMV particle release. Like MLV, M-PMV contains a PPPYtype L domain (12). We observed that M-PMV particle budding was unaffected by TSG-5 (data not shown). These results further support the conclusion that TSG-5 specifically inhibits HIV-1 p6 L domain function. TSG-5 Is Incorporated into Virions in a Manner That Depends on an Intact p6 L Domain. To probe the interaction between TSG-5 and Gag, we sought to determine whether TSG-5 is incorporated into HIV-1 virions. HeLa cells were transfected with pnl4 3 alone or pnl4 3 TSG-5. Transfected cells were metabolically labeled, and cell and virion lysates were immunoprecipitated with either anti-hiv-1 Ig or, because TSG-5 contains an N-terminal HA epitope tag (28), with anti-ha antiserum. A band of the expected size ( 30 kda) was clearly detected in virion lysates produced from cells cotransfected with pnl4 3 Fig. 5. TSG-5 does not inhibit the budding of MLV virions. HeLa cells were transfected with psv MLV-env (39) alone or at a 1:1 DNA ratio with TSG-5. (A) Radioimmunoprecipitation analysis of cell and viral lysates. The positions of the MLV Gag precursor Pr65 Gag and the mature CA protein p30 are indicated at the left. (B) Quantitative analysis of the effect of TSG-5 on the production of virion-associated Gag, determined by PhosphorImager analysis as in Fig. 1C. Data are averages from three independent experiments, SE. and TSG-5 but not in lysates of virions produced from cells expressing pnl4 3 alone (Fig. 6A). We demonstrated above (Fig. 3) that the release of p6 mutants lacking a functional L domain (e.g., L1Term and PTAP ) was not significantly inhibited by TSG-5 expression, whereas release of the S14Term mutant was markedly impaired by TSG-5. These results suggested that mutational inactivation of the p6 L domain might prevent the interaction of TSG-5 with Gag and the subsequent incorporation of TSG-5 into virions. To test this possibility, we evaluated the level of TSG-5 in L1Term, PTAP, and S14Term virions. To increase the sensitivity of the analysis, we used Western blotting to detect both Gag and TSG-5. The results (Fig. 6B) indicated that L1Term and PTAP mutant virions failed to incorporate detectable levels of TSG-5. In contrast, TSG-5 was readily detected in WT and S14Term virions. Cells transfected with TSG-5 alone did not release pelletable TSG-5 into the medium. The levels of virionassociated Gag were measured by quantitative Western blotting analysis (see Materials and Methods); the relative amounts of virion Gag in each sample are shown under each lane in Fig. 6B. We also examined TSG-5 incorporation into WT and p6-mutant virions by using the PR molecular clones pnl4 3 PR and pnl4 3 PR PTAP. The use of PR clones simplifies the detection of Gag, because only the Gag precursor is produced. Consistent with the data presented in Fig. 6B, virions produced upon expression of WT Gag readily incorporated TSG-5, whereas virions lacking the PTAP L domain motif failed to incorporate detectable levels of TSG-5 (Fig. 6C). Discussion In this study, we demonstrate that overexpression of the N- terminal region of TSG101 (TSG-5 ) markedly inhibits HIV-1 particle production by blocking efficient virus budding from the plasma membrane. The insensitivity of L-domain-defective mutants and heterologous retroviruses (MLV and M-PMV) to cgi doi pnas Demirov et al.
5 Fig. 6. TSG-5 is incorporated into WT but not L-domain-deficient HIV-1 virions. (A) Radioimmunoprecipitation analysis of virions derived from cells transfected with pnl4 3 alone or pnl4 3 TSG-5 (1:1 DNA ratio). Immunoprecipitations were performed with either anti-hiv Ig or anti-ha (CLONTECH). Levels of cellassociated Gag expression were equivalent in each lysate (data not shown). (B) Western blot analysis of virions produced from HeLa cells transfected with TSG-5 alone, or a 1:1 ratio of TSG-5 pnl4 3, pnl4 3 PTAP, pnl4 3 L1Term, or pnl4 3 S14Term. Western blotting was performed with anti-ha antibody (Zymed) or anti-hiv Ig. The anti-hiv-1 Ig blots were subjected to quantitative fluorchem analysis (see Materials and Methods); the relative amounts of virionassociated Gag are indicated under each lane (Rel. Gag levels). Levels of cellassociated Gag and TSG-5 expression were comparable in each lysate transfected with an HIV-1 molecular clone (data not shown). (C) Western blot analysis of virions produced from cells cotransfected with a 1:1 DNA ratio of TSG-5 and either pnl4 3 PR or pnl4 3 PR PTAP (8). Western blotting was performed with anti-hiv-1 Ig or anti-ha antibody (Zymed). Levels of cell-associated Gag and TSG-5 were comparable in each lysate (data not shown). inhibition by TSG-5 strongly suggests that TSG-5 acts by blocking HIV-1 p6 L domain function. Virion incorporation data indicate that TSG-5 interacts with WT HIV-1 Gag but not Gag mutants lacking the PTAP L domain motif. The data presented here strongly support the hypothesis that TSG101 plays a physiologically relevant role in HIV-1 budding. Although precisely what role TSG101 plays in modulating HIV-1 release remains to be determined, consideration of the roles TSG101 and other E2-like proteins play in the cell (see the introduction) might offer some clues. Two general classes of models seem most likely: TSG101 could influence the ubiquitination of Gag itself or the ubiquitination of a host factor; or TSG101 could alter the localization or sorting of a ubiquitinated host protein. (i) The reported ability of TSG101 and other E2-like proteins to inhibit or modify the target specificity of ubiquitination (29, 45) raises the possibility that binding of TSG101 by p6 could alter the extent to which Gag becomes ubiquitinated. Whether ubiquitination of Gag itself plays a role in virus budding remains to be established. Direct fusion of Ub to Rous sarcoma virus Gag reverses the impact of proteasome inhibitors on particle release but does not eliminate the requirement for the L domain in budding (20). Removal of the sites of p6 ubiquitination (p6 residues Lys 27 and Lys 33 ) by amino acid substitution (46) or p6 truncation [e.g., S14Term (38)] has no effect on the efficiency of virus budding. Furthermore, S14Term remains sensitive to TSG-5 inhibition, and incorporates TSG-5 protein into virions, despite the fact that it lacks the sites of p6 ubiquitination. However, the latter observations do not exclude a possible role for Gag ubiquitination upstream of the p6 domain. Alternatively, ubiquitination of Gag itself may be an irrelevant consequence of the recruitment of ubiquitination machinery to the site of budding. Thus, the ubiquitination of unidentified host factor(s) could be the driving force in stimulating virus budding. Because ubiquitination plays a role both in endosomal sorting (33) and in the internalization of plasma membrane proteins (47), the modulation of host protein ubiquitination could alter the protein composition of the plasma membrane at the site of budding. (ii) Recent evidence (33) indicates that TSG101 plays a role in the sorting of ubiquitinated proteins into the endosomal pathway. Cellular machinery normally involved in budding reactions at intracellular sites (e.g., the multivesicular body) could be recruited to the site of budding at the plasma membrane through interactions with the L domain. The protein whose sorting is affected by TSG101 could be a host factor that directly or indirectly modulates budding. Our recent observation of a link between L domain function and rafts (48) suggests that the hypothetical host protein(s) whose expression or localization is affected by TSG101 may be raft localized. If TSG101 plays a central role in HIV-1 budding, how might TSG-5 inhibit particle release? TSG-5 could bind p6 and prevent it from interacting with its endogenous ligand (perhaps full-length TSG101). Alternatively, TSG-5 could interact with and disrupt cellular machinery, such as the high molecular weight ESCRT-1 complex in which TSG101 Vps23 is localized (ref. 33; see the introduction) thereby preventing it from functioning in HIV-1 particle release. Regardless of its mechanism of action, further investigations into the ability of TSG-5 to disrupt particle budding will likely provide new insights into this crucial step in the HIV-1 life cycle. The high degree of specificity displayed by TSG-5 in blocking HIV-1 budding raises the possibility that TSG-5, or other TSG101 derivatives, may be useful in gene therapy strategies to inhibit HIV-1 replication in vivo. Alternatively, small molecule inhibitors that disrupt the p6 TSG101 interaction may display antiviral activity. After submission of this manuscript, two articles were published that support the hypothesis that TSG101 plays a crucial role in HIV-1 budding. Garrus et al. (49) reported an interaction between HIV-1 p6 and TSG101; this interaction was blocked by mutations in the PTAP motif of p6. The functional relevance of the p6 TSG101 interaction was demonstrated by the finding that transient inhibition of TSG101 expression induced a budding defect similar to that observed with p6 L domain mutants. Martin-Serrano et al. (50) reported that both HIV-1 p6 and Ebola virus VP40 (which also contains a PTAP motif) interact with TSG101. Recruitment of TSG101 in trans to the site of HIV-1 budding could substitute for a functional L domain. Thus, with use of complementary approaches, these two studies, together with our report, highlight the importance of TSG101 in HIV-1 release. We thank Z. Sun, J. Luban, and E. Hunter for plasmids. The following reagents were obtained through the National Institutes of Health AIDS Research and Reference Reagent Program: psv MLV-env (from N. Landau) and anti HIV-Ig (from Nabi, Rockville, MD). MEDICAL SCIENCES 1. Swanstrom, R. & Wills, J. W. (1997) Synthesis, Assembly, and Processing of Viral Proteins (Cold Spring Harbor Lab. Press, Plainview, NY). 2. Freed, E. O. (1998) Virology 251, Harty, R. N., Brown, M. E., Wang, G., Huibregtse, J. & Hayes, F. P. (2000) Proc. Natl. Acad. Sci. USA 97, (First Published November 28, 2000; pnas ) Demirov et al. PNAS January 22, 2002 vol. 99 no
6 4. Craven, R. C., Harty, R. N., Paragas, J., Palese, P. & Wills, J. W. (1999) J. Virol. 73, Harty, R. N., Paragas, J., Sudol, M. & Palese, P. (1999) J. Virol. 73, Parent, L. J., Bennett, R. P., Craven, R. C., Nelle, T. D., Krishna, N. K., Bowzard, J. B., Wilson, C. B., Puffer, B. A., Montelaro, R. C. & Wills, J. W. (1995) J. Virol. 69, Gottlinger, H. G., Dorfman, T., Sodroski, J. G. & Haseltine, W. A. (1991) Proc. Natl. Acad. Sci. USA 88, Huang, M., Orenstein, J. M., Martin, M. A. & Freed, E. O. (1995) J. Virol. 69, Puffer, B. A., Parent, L. J., Wills, J. W. & Montelaro, R. C. (1997) J. Virol. 71, Yuan, B., Li, X. & Goff, S. P. (1999) EMBO J. 18, Xiang, Y., Cameron, C. E., Wills, J. W. & Leis, J. (1996) J. Virol. 70, Yasuda, J. & Hunter, E. (1998) J. Virol. 72, Mayer, B. J. & Eck, M. J. (1995) Curr. Biol. 5, Ohno, H., Stewart, J., Fournier, M. C., Bosshart, H., Rhee, I., Miyatake, S., Saito, T., Gallusser, A., Kirchhausen, T. & Bonifacino, J. S. (1995) Science 269, Sudol, M. (1996) Prog. Biophys. Mol. Biol. 65, Garnier, L., Wills, J. W., Verderame, M. F. & Sudol, M. (1996) Nature (London) 381, Puffer, B. A., Watkins, S. C. & Montelaro, R. C. (1998) J. Virol. 72, Vogt, V. M. (2000) Proc. Natl. Acad. Sci. USA 97, Ott, D. E., Coren, L. V., Copeland, T. D., Kane, B. P., Johnson, D. G., Sowder, R. C., 2nd, Yoshinaka, Y., Oroszlan, S., Arthur, L. O. & Henderson, L. E. (1998) J. Virol. 72, Patnaik, A., Chau, V. & Wills, J. W. (2000) Proc. Natl. Acad. Sci. USA 97, Schubert, U., Ott, D. E., Chertova, E. N., Welker, R., Tessmer, U., Princiotta, M. F., Bennink, J. R., Krausslich, H. G. & Yewdell, J. W. (2000) Proc. Natl. Acad. Sci. USA 97, Strack, B., Calistri, A., Accola, M. A., Palu, G. & Gottlinger, H. G. (2000) Proc. Natl. Acad. Sci. USA 97, Kikonyogo, A., Bouamr, F., Vana, M. L., Xiang, Y., Aiyar, A., Carter, C. & Leis, J. (2001) Proc. Natl. Acad. Sci. USA 98, (First Published September 18, 2001; pnas ) 24. VerPlank, L., Bouamr, F., LaGrassa, T. J., Agresta, B., Kikonyogo, A., Leis, J. & Carter, C. A. (2001) Proc. Natl. Acad. Sci. USA 98, (First Published June 26, 2001;, pnas ) 25. Li, L. & Cohen, S. N. (1996) Cell 85, Xie, W., Li, L. & Cohen, S. N. (1998) Proc. Natl. Acad. Sci. USA 95, Hittelman, A. B., Burakov, D., Iniguez-Lluhi, J. A., Freedman, L. P. & Garabedian, M. J. (1999) EMBO J. 18, Sun, Z., Pan, J., Hope, W. X., Cohen, S. N. & Balk, S. P. (1999) Cancer 86, Li, L., Liao, J., Ruland, J., Mak, T. W. & Cohen, S. N. (2001) Proc. Natl. Acad. Sci. USA 98, Li, Y., Kane, T., Tipper, C., Spatrick, P. & Jenness, D. D. (1999) Mol. Cell. Biol. 19, Babst, M., Odorizzi, G., Estepa, E. J. & Emr, S. D. (2000) Traffic 1, Bishop, N. & Woodman, P. (2001) J. Biol. Chem. 276, Katzmann, D. J., Babst, M. & Emr, S. D. (2001) Cell 106, Koonin, E. V. & Abagyan, R. A. (1997) Nat. Genet. 16, Varshavsky, A. (1997) Trends Biochem. Sci. 22, Murakami, T. & Freed, E. O. (2000) Proc. Natl. Acad. Sci. USA 97, Adachi, A., Gendelman, H. E., Koenig, S., Folks, T., Willey, R., Rabson, A. & Martin, M. A. (1986) J. Virol. 59, Demirov, D. G., Orenstein, J. M. & Freed, E. O. (2002) J. Virol. 76, Landau, N. R. & Littman, D. R. (1992) J. Virol. 66, Cimarelli, A. & Luban, J. (2000) J. Virol. 74, Freed, E. O. & Martin, M. A. (1994) J. Virol. 68, Kiernan, R. E., Ono, A., Englund, G. & Freed, E. O. (1998) J. Virol. 72, Freed, E. O., Orenstein, J. M., Buckler-White, A. J. & Martin, M. A. (1994) J. Virol. 68, Yuan, B., Campbell, S., Bacharach, E., Rein, A. & Goff, S. P. (2000) J. Virol. 74, Hofmann, R. M. & Pickart, C. M. (1999) Cell 96, Ott, D. E., Coren, L. V., Chertova, E. N., Gagliardi, T. D. & Schubert, U. (2000) Virology 278, Hicke, L. (2001) Nat. Rev. Mol. Cell. Biol. 2, Ono, A. & Freed, E. O. (2001) Proc. Natl. Acad. Sci. USA 98, Garrus, J. E., von Schwedler, U. K., Pornillos, O. W., Morham, S. G., Zavitz, K. H., Wang, H. E., Wettstein, D. A., Stray, K. M., Cote, M., Rich, R. L., et al. (2001) Cell 107, Martin-Serrano, J., Zang, T. & Bieniasz, P. D. (2001) Nat. Med. 7, cgi doi pnas Demirov et al.
Received 15 August 2002/Accepted 1 November 2002
 JOURNAL OF VIROLOGY, Feb. 2003, p. 1812 1819 Vol. 77, No. 3 0022-538X/03/$08.00 0 DOI: 10.1128/JVI.77.3.1812 1819.2003 Copyright 2003, American Society for Microbiology. All Rights Reserved. Overlapping
JOURNAL OF VIROLOGY, Feb. 2003, p. 1812 1819 Vol. 77, No. 3 0022-538X/03/$08.00 0 DOI: 10.1128/JVI.77.3.1812 1819.2003 Copyright 2003, American Society for Microbiology. All Rights Reserved. Overlapping
In Vivo Interference of Rous Sarcoma Virus Budding by cis Expression of a WW Domain
 JOURNAL OF VIROLOGY, Mar. 2002, p. 2789 2795 Vol. 76, No. 6 0022-538X/02/$04.00 0 DOI: 10.1128/JVI.76.6.2789 2795.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. In Vivo Interference
JOURNAL OF VIROLOGY, Mar. 2002, p. 2789 2795 Vol. 76, No. 6 0022-538X/02/$04.00 0 DOI: 10.1128/JVI.76.6.2789 2795.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. In Vivo Interference
Deletion of a Cys His motif from the Alpharetrovirus nucleocapsid domain reveals late domain mutant-like budding defects
 Virology 347 (2006) 226 233 www.elsevier.com/locate/yviro Deletion of a Cys His motif from the Alpharetrovirus nucleocapsid domain reveals late domain mutant-like budding defects Eun-Gyung Lee, Maxine
Virology 347 (2006) 226 233 www.elsevier.com/locate/yviro Deletion of a Cys His motif from the Alpharetrovirus nucleocapsid domain reveals late domain mutant-like budding defects Eun-Gyung Lee, Maxine
Cellular Factors Required for Lassa Virus Budding
 JOURNAL OF VIROLOGY, Apr. 2006, p. 4191 4195 Vol. 80, No. 8 0022-538X/06/$08.00 0 doi:10.1128/jvi.80.8.4191 4195.2006 Copyright 2006, American Society for Microbiology. All Rights Reserved. Cellular Factors
JOURNAL OF VIROLOGY, Apr. 2006, p. 4191 4195 Vol. 80, No. 8 0022-538X/06/$08.00 0 doi:10.1128/jvi.80.8.4191 4195.2006 Copyright 2006, American Society for Microbiology. All Rights Reserved. Cellular Factors
Role of ESCRT-I in Retroviral Budding
 JOURNAL OF VIROLOGY, Apr. 2003, p. 4794 4804 Vol. 77, No. 8 0022-538X/03/$08.00 0 DOI: 10.1128/JVI.77.8.4794 4804.2003 Copyright 2003, American Society for Microbiology. All Rights Reserved. Role of ESCRT-I
JOURNAL OF VIROLOGY, Apr. 2003, p. 4794 4804 Vol. 77, No. 8 0022-538X/03/$08.00 0 DOI: 10.1128/JVI.77.8.4794 4804.2003 Copyright 2003, American Society for Microbiology. All Rights Reserved. Role of ESCRT-I
Late Domain-Independent Rescue of a Release-Deficient Moloney Murine Leukemia Virus by the Ubiquitin Ligase Itch
 JOURNAL OF VIROLOGY, Jan. 2010, p. 704 715 Vol. 84, No. 2 0022-538X/10/$12.00 doi:10.1128/jvi.01319-09 Late Domain-Independent Rescue of a Release-Deficient Moloney Murine Leukemia Virus by the Ubiquitin
JOURNAL OF VIROLOGY, Jan. 2010, p. 704 715 Vol. 84, No. 2 0022-538X/10/$12.00 doi:10.1128/jvi.01319-09 Late Domain-Independent Rescue of a Release-Deficient Moloney Murine Leukemia Virus by the Ubiquitin
Late budding domains and host proteins in enveloped virus release
 Virology 344 (2006) 55 63 www.elsevier.com/locate/yviro Late budding domains and host proteins in enveloped virus release Paul D. Bieniasz Aaron Diamond AIDS Research Center and Laboratory of Retrovirology,
Virology 344 (2006) 55 63 www.elsevier.com/locate/yviro Late budding domains and host proteins in enveloped virus release Paul D. Bieniasz Aaron Diamond AIDS Research Center and Laboratory of Retrovirology,
Human Immunodeficiency Virus Type 1 Gag Engages the Bro1 Domain of ALIX/AIP1 through the Nucleocapsid
 JOURNAL OF VIROLOGY, Feb. 2008, p. 1389 1398 Vol. 82, No. 3 0022-538X/08/$08.00 0 doi:10.1128/jvi.01912-07 Copyright 2008, American Society for Microbiology. All Rights Reserved. Human Immunodeficiency
JOURNAL OF VIROLOGY, Feb. 2008, p. 1389 1398 Vol. 82, No. 3 0022-538X/08/$08.00 0 doi:10.1128/jvi.01912-07 Copyright 2008, American Society for Microbiology. All Rights Reserved. Human Immunodeficiency
Mutations of the Human Immunodeficiency Virus Type 1 p6 Gag Domain Result in Reduced Retention of Pol Proteins during Virus Assembly
 JOURNAL OF VIROLOGY, Apr. 1998, p. 3412 3417 Vol. 72, No. 4 0022-538X/98/$04.00 0 Copyright 1998, American Society for Microbiology Mutations of the Human Immunodeficiency Virus Type 1 p6 Gag Domain Result
JOURNAL OF VIROLOGY, Apr. 1998, p. 3412 3417 Vol. 72, No. 4 0022-538X/98/$04.00 0 Copyright 1998, American Society for Microbiology Mutations of the Human Immunodeficiency Virus Type 1 p6 Gag Domain Result
Identification of Domains in Gag Important for Prototypic Foamy Virus Egress
 JOURNAL OF VIROLOGY, May 2005, p. 6392 6399 Vol. 79, No. 10 0022-538X/05/$08.00 0 doi:10.1128/jvi.79.10.6392 6399.2005 Copyright 2005, American Society for Microbiology. All Rights Reserved. Identification
JOURNAL OF VIROLOGY, May 2005, p. 6392 6399 Vol. 79, No. 10 0022-538X/05/$08.00 0 doi:10.1128/jvi.79.10.6392 6399.2005 Copyright 2005, American Society for Microbiology. All Rights Reserved. Identification
The introduction of highly active antiretroviral therapy has led
 PA-457: A potent HIV inhibitor that disrupts core condensation by targeting a late step in Gag processing F. Li*, R. Goila-Gaur, K. Salzwedel*, N. R. Kilgore*, M. Reddick*, C. Matallana*, A. Castillo*,
PA-457: A potent HIV inhibitor that disrupts core condensation by targeting a late step in Gag processing F. Li*, R. Goila-Gaur, K. Salzwedel*, N. R. Kilgore*, M. Reddick*, C. Matallana*, A. Castillo*,
Differential Effects of Actin Cytoskeleton Dynamics on Equine Infectious Anemia Virus Particle Production
 JOURNAL OF VIROLOGY, Jan. 2004, p. 882 891 Vol. 78, No. 2 0022-538X/04/$08.00 0 DOI: 10.1128/JVI.78.2.882 891.2004 Copyright 2004, American Society for Microbiology. All Rights Reserved. Differential Effects
JOURNAL OF VIROLOGY, Jan. 2004, p. 882 891 Vol. 78, No. 2 0022-538X/04/$08.00 0 DOI: 10.1128/JVI.78.2.882 891.2004 Copyright 2004, American Society for Microbiology. All Rights Reserved. Differential Effects
Role of the C terminus Gag protein in human immunodefieieney virus type 1 virion assembly and maturation
 Journal of General Virology (1995), 76, 3171-3179. Printed in Great Britain 3171 Role of the C terminus Gag protein in human immunodefieieney virus type 1 virion assembly and maturation X.-F. Yu, ~ Z.
Journal of General Virology (1995), 76, 3171-3179. Printed in Great Britain 3171 Role of the C terminus Gag protein in human immunodefieieney virus type 1 virion assembly and maturation X.-F. Yu, ~ Z.
JCB. After Hrs with HIV. The Journal of Cell Biology. Comment. Ali Amara 1 and Dan R. Littman 1,2
 JCB Comment After Hrs with HIV Ali Amara 1 and Dan R. Littman 1,2 1 Molecular Pathogenesis Program and 2 Howard Hughes Medical Institute, Skirball Institute of Biomolecular Medicine, New York University
JCB Comment After Hrs with HIV Ali Amara 1 and Dan R. Littman 1,2 1 Molecular Pathogenesis Program and 2 Howard Hughes Medical Institute, Skirball Institute of Biomolecular Medicine, New York University
Packaging and Abnormal Particle Morphology
 JOURNAL OF VIROLOGY, OCt. 1990, p. 5230-5234 0022-538X/90/105230-05$02.00/0 Copyright 1990, American Society for Microbiology Vol. 64, No. 10 A Mutant of Human Immunodeficiency Virus with Reduced RNA Packaging
JOURNAL OF VIROLOGY, OCt. 1990, p. 5230-5234 0022-538X/90/105230-05$02.00/0 Copyright 1990, American Society for Microbiology Vol. 64, No. 10 A Mutant of Human Immunodeficiency Virus with Reduced RNA Packaging
VIROLOGY. Engineering Viral Genomes: Retrovirus Vectors
 VIROLOGY Engineering Viral Genomes: Retrovirus Vectors Viral vectors Retrovirus replicative cycle Most mammalian retroviruses use trna PRO, trna Lys3, trna Lys1,2 The partially unfolded trna is annealed
VIROLOGY Engineering Viral Genomes: Retrovirus Vectors Viral vectors Retrovirus replicative cycle Most mammalian retroviruses use trna PRO, trna Lys3, trna Lys1,2 The partially unfolded trna is annealed
Ubiquitination of Human Immunodeficiency Virus Type 1 Gag Is Highly Dependent on Gag Membrane Association
 JOURNAL OF VIROLOGY, Sept. 2007, p. 9193 9201 Vol. 81, No. 17 0022-538X/07/$08.00 0 doi:10.1128/jvi.00044-07 Copyright 2007, American Society for Microbiology. All Rights Reserved. Ubiquitination of Human
JOURNAL OF VIROLOGY, Sept. 2007, p. 9193 9201 Vol. 81, No. 17 0022-538X/07/$08.00 0 doi:10.1128/jvi.00044-07 Copyright 2007, American Society for Microbiology. All Rights Reserved. Ubiquitination of Human
Ebola Virus VP40 Late Domains Are Not Essential for Viral Replication in Cell Culture
 JOURNAL OF VIROLOGY, Aug. 2005, p. 10300 10307 Vol. 79, No. 16 0022-538X/05/$08.00 0 doi:10.1128/jvi.79.16.10300 10307.2005 Copyright 2005, American Society for Microbiology. All Rights Reserved. Ebola
JOURNAL OF VIROLOGY, Aug. 2005, p. 10300 10307 Vol. 79, No. 16 0022-538X/05/$08.00 0 doi:10.1128/jvi.79.16.10300 10307.2005 Copyright 2005, American Society for Microbiology. All Rights Reserved. Ebola
The assembly of type C retroviruses and lentiviruses occurs at
 Plasma membrane rafts play a critical role in HIV-1 assembly and release Akira Ono and Eric O. Freed* Laboratory of Molecular Microbiology, National Institute of Allergy and Infectious Diseases, National
Plasma membrane rafts play a critical role in HIV-1 assembly and release Akira Ono and Eric O. Freed* Laboratory of Molecular Microbiology, National Institute of Allergy and Infectious Diseases, National
Regulation of Marburg virus (MARV) budding by Nedd4.1: a different WW domain of Nedd4.1 is critical for binding to MARV and Ebola virus VP40
 Journal of General Virology (2010), 91, 228 234 DOI 10.1099/vir.0.015495-0 Regulation of Marburg virus (MARV) budding by Nedd4.1: a different WW domain of Nedd4.1 is critical for binding to MARV and Ebola
Journal of General Virology (2010), 91, 228 234 DOI 10.1099/vir.0.015495-0 Regulation of Marburg virus (MARV) budding by Nedd4.1: a different WW domain of Nedd4.1 is critical for binding to MARV and Ebola
Expression of Human Immunodeficiency Virus Type 1 Gag Modulates Ligand-Induced Downregulation of EGF Receptor
 JOURNAL OF VIROLOGY, Nov. 2004, p. 12386 12394 Vol. 78, No. 22 0022-538X/04/$08.00 0 DOI: 10.1128/JVI.78.22.12386 12394.2004 Copyright 2004, American Society for Microbiology. All Rights Reserved. Expression
JOURNAL OF VIROLOGY, Nov. 2004, p. 12386 12394 Vol. 78, No. 22 0022-538X/04/$08.00 0 DOI: 10.1128/JVI.78.22.12386 12394.2004 Copyright 2004, American Society for Microbiology. All Rights Reserved. Expression
Induction of APOBEC3G Ubiquitination and Degradation by an HIV-1 Vif-Cul5-SCF Complex
 Induction of APOBEC3G Ubiquitination and Degradation by an HIV-1 Vif-Cul5-SCF Complex Xianghui Yu, 1,2 * Yunkai Yu, 1 * Bindong Liu, 1 * Kun Luo, 1 Wei Kong, 2 Panyong Mao, 1 Xiao-Fang Yu 1,3 1 Department
Induction of APOBEC3G Ubiquitination and Degradation by an HIV-1 Vif-Cul5-SCF Complex Xianghui Yu, 1,2 * Yunkai Yu, 1 * Bindong Liu, 1 * Kun Luo, 1 Wei Kong, 2 Panyong Mao, 1 Xiao-Fang Yu 1,3 1 Department
Journal of Biology. BioMed Central
 Journal of Biology BioMed Central Open Access Research article Endophilins interact with Moloney murine leukemia virus Gag and modulate virion production Margaret Q Wang*, Wankee Kim, Guangxia Gao, Ted
Journal of Biology BioMed Central Open Access Research article Endophilins interact with Moloney murine leukemia virus Gag and modulate virion production Margaret Q Wang*, Wankee Kim, Guangxia Gao, Ted
Supplementary information. MARCH8 inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins
 Supplementary information inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins Takuya Tada, Yanzhao Zhang, Takayoshi Koyama, Minoru Tobiume, Yasuko Tsunetsugu-Yokota, Shoji
Supplementary information inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins Takuya Tada, Yanzhao Zhang, Takayoshi Koyama, Minoru Tobiume, Yasuko Tsunetsugu-Yokota, Shoji
JOURNAL OF VIROLOGY, July 1999, p Vol. 73, No. 7. Copyright 1999, American Society for Microbiology. All Rights Reserved.
 JOURNAL OF VIROLOGY, July 1999, p. 5654 5662 Vol. 73, No. 7 0022-538X/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. Formation of Virus Assembly Intermediate Complexes
JOURNAL OF VIROLOGY, July 1999, p. 5654 5662 Vol. 73, No. 7 0022-538X/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. Formation of Virus Assembly Intermediate Complexes
Effect of Mutations in Gag on Assembly of Immature Human Immunodeficiency Virus Type 1 Capsids in a Cell-Free System
 Virology 279, 257 270 (2001) doi:10.1006/viro.2000.0706, available online at http://www.idealibrary.com on Effect of Mutations in Gag on Assembly of Immature Human Immunodeficiency Virus Type 1 Capsids
Virology 279, 257 270 (2001) doi:10.1006/viro.2000.0706, available online at http://www.idealibrary.com on Effect of Mutations in Gag on Assembly of Immature Human Immunodeficiency Virus Type 1 Capsids
Specific Incorporation of Heat Shock Protein 70 Family Members into Primate Lentiviral Virions
 JOURNAL OF VIROLOGY, May 2002, p. 4666 4670 Vol. 76, No. 9 0022-538X/02/$04.00 0 DOI: 10.1128/JVI.76.9.4666 4670.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Specific Incorporation
JOURNAL OF VIROLOGY, May 2002, p. 4666 4670 Vol. 76, No. 9 0022-538X/02/$04.00 0 DOI: 10.1128/JVI.76.9.4666 4670.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Specific Incorporation
Cellular Motor Protein KIF-4 Associates with Retroviral Gag
 JOURNAL OF VIROLOGY, Dec. 1999, p. 10508 10513 Vol. 73, No. 12 0022-538X/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. Cellular Motor Protein KIF-4 Associates with
JOURNAL OF VIROLOGY, Dec. 1999, p. 10508 10513 Vol. 73, No. 12 0022-538X/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. Cellular Motor Protein KIF-4 Associates with
HIV-1 genome organization
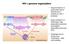 HIV-1 genome organization Gag encodes for a polyprotein that is cleaved into 4 proteins by HIV-1 protease. Gag is present in 2 transcripts: Gag polyprotein precursor and GagPol polyprotein precursor (5%).
HIV-1 genome organization Gag encodes for a polyprotein that is cleaved into 4 proteins by HIV-1 protease. Gag is present in 2 transcripts: Gag polyprotein precursor and GagPol polyprotein precursor (5%).
Virus Research 150 (2010) Contents lists available at ScienceDirect. Virus Research. journal homepage:
 Virus Research 150 (2010) 153 157 Contents lists available at ScienceDirect Virus Research journal homepage: www.elsevier.com/locate/virusres Short communication In vitro assembly of the feline immunodeficiency
Virus Research 150 (2010) 153 157 Contents lists available at ScienceDirect Virus Research journal homepage: www.elsevier.com/locate/virusres Short communication In vitro assembly of the feline immunodeficiency
Fayth K. Yoshimura, Ph.D. September 7, of 7 HIV - BASIC PROPERTIES
 1 of 7 I. Viral Origin. A. Retrovirus - animal lentiviruses. HIV - BASIC PROPERTIES 1. HIV is a member of the Retrovirus family and more specifically it is a member of the Lentivirus genus of this family.
1 of 7 I. Viral Origin. A. Retrovirus - animal lentiviruses. HIV - BASIC PROPERTIES 1. HIV is a member of the Retrovirus family and more specifically it is a member of the Lentivirus genus of this family.
The Interferon-Induced Gene ISG15 Blocks Retrovirus Release from Cells Late in the Budding Process
 JOURNAL OF VIROLOGY, May 2010, p. 4725 4736 Vol. 84, No. 9 0022-538X/10/$12.00 doi:10.1128/jvi.02478-09 Copyright 2010, American Society for Microbiology. All Rights Reserved. The Interferon-Induced Gene
JOURNAL OF VIROLOGY, May 2010, p. 4725 4736 Vol. 84, No. 9 0022-538X/10/$12.00 doi:10.1128/jvi.02478-09 Copyright 2010, American Society for Microbiology. All Rights Reserved. The Interferon-Induced Gene
Gag. Gag-Gag. TSG101 Nedd4. Two-Hybrid HIV Gag. Two-Hybrid. Two-Hybrid
 CytoTrap HIV Gag AnnexinA2 AUP1 AUP1 HIV AUP1 N Gag CytoTrap Gag-Gag TSG101 Nedd4 Two-Hybrid Two-Hybrid HIVGag Gag Two-Hybrid HIV Gag Gag HIV Two-Hybrid CytoTrap cdna Gag Gag CytoTrap Gag CytoTrap Gag-Gag
CytoTrap HIV Gag AnnexinA2 AUP1 AUP1 HIV AUP1 N Gag CytoTrap Gag-Gag TSG101 Nedd4 Two-Hybrid Two-Hybrid HIVGag Gag Two-Hybrid HIV Gag Gag HIV Two-Hybrid CytoTrap cdna Gag Gag CytoTrap Gag CytoTrap Gag-Gag
Retroviruses. ---The name retrovirus comes from the enzyme, reverse transcriptase.
 Retroviruses ---The name retrovirus comes from the enzyme, reverse transcriptase. ---Reverse transcriptase (RT) converts the RNA genome present in the virus particle into DNA. ---RT discovered in 1970.
Retroviruses ---The name retrovirus comes from the enzyme, reverse transcriptase. ---Reverse transcriptase (RT) converts the RNA genome present in the virus particle into DNA. ---RT discovered in 1970.
Tsg101 and the Vacuolar Protein Sorting Pathway Are Essential for HIV-1 Budding
 Cell, Vol. 107, 55 65, October 5, 2001, Copyright 2001 by Cell Press Tsg101 and the Vacuolar Protein Sorting Pathway Are Essential for HIV-1 Budding Jennifer E. Garrus, 1,5 Uta K. von Schwedler, 1,5 Owen
Cell, Vol. 107, 55 65, October 5, 2001, Copyright 2001 by Cell Press Tsg101 and the Vacuolar Protein Sorting Pathway Are Essential for HIV-1 Budding Jennifer E. Garrus, 1,5 Uta K. von Schwedler, 1,5 Owen
Reviewers' comments: Reviewer #1 (Remarks to the Author):
 Reviewers' comments: Reviewer #1 (Remarks to the Author): Nature Communications manuscript number NCOMMS-16-15882, by Miyakawa et al. presents an intriguing analysis of the effects of the tumor suppressor
Reviewers' comments: Reviewer #1 (Remarks to the Author): Nature Communications manuscript number NCOMMS-16-15882, by Miyakawa et al. presents an intriguing analysis of the effects of the tumor suppressor
Identification of late assembly domains of the human endogenous retrovirus-k(hml-2)
 Chudak et al. Retrovirology 2013, 10:140 RESEARCH Identification of late assembly domains of the human endogenous retrovirus-k(hml-2) Open Access Claudia Chudak 1, Nadine Beimforde 1, Maja George 2, Anja
Chudak et al. Retrovirology 2013, 10:140 RESEARCH Identification of late assembly domains of the human endogenous retrovirus-k(hml-2) Open Access Claudia Chudak 1, Nadine Beimforde 1, Maja George 2, Anja
Fayth K. Yoshimura, Ph.D. September 7, of 7 RETROVIRUSES. 2. HTLV-II causes hairy T-cell leukemia
 1 of 7 I. Diseases Caused by Retroviruses RETROVIRUSES A. Human retroviruses that cause cancers 1. HTLV-I causes adult T-cell leukemia and tropical spastic paraparesis 2. HTLV-II causes hairy T-cell leukemia
1 of 7 I. Diseases Caused by Retroviruses RETROVIRUSES A. Human retroviruses that cause cancers 1. HTLV-I causes adult T-cell leukemia and tropical spastic paraparesis 2. HTLV-II causes hairy T-cell leukemia
Mechanisms of enveloped RNA virus budding
 569 44 Brown, S.A. et al. (1998) Holoprosencephaly due to mutations in ZIC2, a homologue of Drosophila odd-paired. Nat. Genet. 20, 180 183 45 Aruga, J. et al. (2002) Zic2 controls cerebellar development
569 44 Brown, S.A. et al. (1998) Holoprosencephaly due to mutations in ZIC2, a homologue of Drosophila odd-paired. Nat. Genet. 20, 180 183 45 Aruga, J. et al. (2002) Zic2 controls cerebellar development
Charged Amino Acid Residues of Human Immunodeficiency Virus Type 1 Nucleocapsid p7 Protein Involved in RNA Packaging and Infectivity
 JOURNAL OF VIROLOGY, Oct. 1996, p. 6607 6616 Vol. 70, No. 10 0022-538X/96/$04.00 0 Copyright 1996, American Society for Microbiology Charged Amino Acid Residues of Human Immunodeficiency Virus Type 1 Nucleocapsid
JOURNAL OF VIROLOGY, Oct. 1996, p. 6607 6616 Vol. 70, No. 10 0022-538X/96/$04.00 0 Copyright 1996, American Society for Microbiology Charged Amino Acid Residues of Human Immunodeficiency Virus Type 1 Nucleocapsid
Supplementary Information. Supplementary Figure 1
 Supplementary Information Supplementary Figure 1 1 Supplementary Figure 1. Functional assay of the hcas9-2a-mcherry construct (a) Gene correction of a mutant EGFP reporter cell line mediated by hcas9 or
Supplementary Information Supplementary Figure 1 1 Supplementary Figure 1. Functional assay of the hcas9-2a-mcherry construct (a) Gene correction of a mutant EGFP reporter cell line mediated by hcas9 or
There are approximately 30,000 proteasomes in a typical human cell Each proteasome is approximately 700 kda in size The proteasome is made up of 3
 Proteasomes Proteasomes Proteasomes are responsible for degrading proteins that have been damaged, assembled improperly, or that are of no profitable use to the cell. The unwanted protein is literally
Proteasomes Proteasomes Proteasomes are responsible for degrading proteins that have been damaged, assembled improperly, or that are of no profitable use to the cell. The unwanted protein is literally
SUPPLEMENTARY INFORMATION
 sirna pool: Control Tetherin -HA-GFP HA-Tetherin -Tubulin Supplementary Figure S1. Knockdown of HA-tagged tetherin expression by tetherin specific sirnas. HeLa cells were cotransfected with plasmids expressing
sirna pool: Control Tetherin -HA-GFP HA-Tetherin -Tubulin Supplementary Figure S1. Knockdown of HA-tagged tetherin expression by tetherin specific sirnas. HeLa cells were cotransfected with plasmids expressing
Role of the Gag Matrix Domain in Targeting Human Immunodeficiency Virus Type 1 Assembly
 JOURNAL OF VIROLOGY, Mar. 2000, p. 2855 2866 Vol. 74, No. 6 0022-538X/00/$04.00 0 Copyright 2000, American Society for Microbiology. All Rights Reserved. Role of the Gag Matrix Domain in Targeting Human
JOURNAL OF VIROLOGY, Mar. 2000, p. 2855 2866 Vol. 74, No. 6 0022-538X/00/$04.00 0 Copyright 2000, American Society for Microbiology. All Rights Reserved. Role of the Gag Matrix Domain in Targeting Human
REGULATION OF TSG101 EXPRESSION BY THE STEADINESS BOX: A ROLE OF TSG101-ASSOCIATED LIGASE (TAL)
 REGULATION OF TSG101 EXPRESSION BY THE STEADINESS BOX: A ROLE OF TSG101-ASSOCIATED LIGASE (TAL) Bethan McDonald and Juan Martin-Serrano Department of Infectious Diseases; King's College London School of
REGULATION OF TSG101 EXPRESSION BY THE STEADINESS BOX: A ROLE OF TSG101-ASSOCIATED LIGASE (TAL) Bethan McDonald and Juan Martin-Serrano Department of Infectious Diseases; King's College London School of
letters Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein
 Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein Owen Pornillos 1,2, Steven L. Alam 1,2, Darrell R. Davis 2,3 and Wesley I. Sundquist 2 1 These authors contributed
Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein Owen Pornillos 1,2, Steven L. Alam 1,2, Darrell R. Davis 2,3 and Wesley I. Sundquist 2 1 These authors contributed
JOURNAL OF VIROLOGY, Jan. 1999, p Vol. 73, No. 1. Copyright 1999, American Society for Microbiology. All Rights Reserved.
 JOURNAL OF VIROLOGY, Jan. 1999, p. 19 28 Vol. 73, No. 1 0022-538X/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. Mutational Analysis of the Hydrophobic Tail of the
JOURNAL OF VIROLOGY, Jan. 1999, p. 19 28 Vol. 73, No. 1 0022-538X/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. Mutational Analysis of the Hydrophobic Tail of the
JOURNAL OF VIROLOGY, June 1999, p Vol. 73, No. 6. Received 29 December 1998/Accepted 12 March 1999
 JOURNAL OF VIROLOGY, June 1999, p. 4728 4737 Vol. 73, No. 6 0022-538X/99/$04.00 0 Reversion of a Human Immunodeficiency Virus Type 1 Matrix Mutation Affecting Gag Membrane Binding, Endogenous Reverse Transcriptase
JOURNAL OF VIROLOGY, June 1999, p. 4728 4737 Vol. 73, No. 6 0022-538X/99/$04.00 0 Reversion of a Human Immunodeficiency Virus Type 1 Matrix Mutation Affecting Gag Membrane Binding, Endogenous Reverse Transcriptase
Recombinant Protein Expression Retroviral system
 Recombinant Protein Expression Retroviral system Viruses Contains genome DNA or RNA Genome encased in a protein coat or capsid. Some viruses have membrane covering protein coat enveloped virus Ø Essential
Recombinant Protein Expression Retroviral system Viruses Contains genome DNA or RNA Genome encased in a protein coat or capsid. Some viruses have membrane covering protein coat enveloped virus Ø Essential
Role of Matrix in an Early Postentry Step in the Human Immunodeficiency Virus Type 1 Life Cycle
 JOURNAL OF VIROLOGY, May 1998, p. 4116 4126 Vol. 72, No. 5 0022-538X/98/$04.00 0 Copyright 1998, American Society for Microbiology Role of Matrix in an Early Postentry Step in the Human Immunodeficiency
JOURNAL OF VIROLOGY, May 1998, p. 4116 4126 Vol. 72, No. 5 0022-538X/98/$04.00 0 Copyright 1998, American Society for Microbiology Role of Matrix in an Early Postentry Step in the Human Immunodeficiency
Release of the enveloped HIV type 1 (HIV-1) in most infected
 The trans-golgi network-associated human ubiquitin-protein ligase POSH is essential for HIV type 1 production Iris Alroy*, Shmuel Tuvia*, Tsvika Greener*, Daphna Gordon*, Haim M. Barr*, Daniel Taglicht*,
The trans-golgi network-associated human ubiquitin-protein ligase POSH is essential for HIV type 1 production Iris Alroy*, Shmuel Tuvia*, Tsvika Greener*, Daphna Gordon*, Haim M. Barr*, Daniel Taglicht*,
Lecture Readings. Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell
 October 26, 2006 1 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
October 26, 2006 1 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
Vacuolar Protein Sorting Pathway Contributes to the Release of Marburg Virus
 JOURNAL OF VIROLOGY, Mar. 2009, p. 2327 2337 Vol. 83, No. 5 0022-538X/09/$08.00 0 doi:10.1128/jvi.02184-08 Copyright 2009, American Society for Microbiology. All Rights Reserved. Vacuolar Protein Sorting
JOURNAL OF VIROLOGY, Mar. 2009, p. 2327 2337 Vol. 83, No. 5 0022-538X/09/$08.00 0 doi:10.1128/jvi.02184-08 Copyright 2009, American Society for Microbiology. All Rights Reserved. Vacuolar Protein Sorting
Peptide hydrolysis uncatalyzed half-life = ~450 years HIV protease-catalyzed half-life = ~3 seconds
 Uncatalyzed half-life Peptide hydrolysis uncatalyzed half-life = ~450 years IV protease-catalyzed half-life = ~3 seconds Life Sciences 1a Lecture Slides Set 9 Fall 2006-2007 Prof. David R. Liu In the absence
Uncatalyzed half-life Peptide hydrolysis uncatalyzed half-life = ~450 years IV protease-catalyzed half-life = ~3 seconds Life Sciences 1a Lecture Slides Set 9 Fall 2006-2007 Prof. David R. Liu In the absence
AND JEREMY LUBAN 1,2 *
 JOURNAL OF VIROLOGY, Oct. 1999, p. 8527 8540 Vol. 73, No. 10 0022-538X/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. Human Immunodeficiency Virus Type 1 Gag Polyprotein
JOURNAL OF VIROLOGY, Oct. 1999, p. 8527 8540 Vol. 73, No. 10 0022-538X/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. Human Immunodeficiency Virus Type 1 Gag Polyprotein
Materials and Methods , The two-hybrid principle.
 The enzymatic activity of an unknown protein which cleaves the phosphodiester bond between the tyrosine residue of a viral protein and the 5 terminus of the picornavirus RNA Introduction Every day there
The enzymatic activity of an unknown protein which cleaves the phosphodiester bond between the tyrosine residue of a viral protein and the 5 terminus of the picornavirus RNA Introduction Every day there
Julianne Edwards. Retroviruses. Spring 2010
 Retroviruses Spring 2010 A retrovirus can simply be referred to as an infectious particle which replicates backwards even though there are many different types of retroviruses. More specifically, a retrovirus
Retroviruses Spring 2010 A retrovirus can simply be referred to as an infectious particle which replicates backwards even though there are many different types of retroviruses. More specifically, a retrovirus
Involvement of host cellular multivesicular body functions in hepatitis B virus budding
 Involvement of host cellular multivesicular body functions in hepatitis B virus budding Tokiko Watanabe*, Ericka M. Sorensen*, Akira Naito*, Meghan Schott*, Seungtaek Kim*, and Paul Ahlquist* *Institute
Involvement of host cellular multivesicular body functions in hepatitis B virus budding Tokiko Watanabe*, Ericka M. Sorensen*, Akira Naito*, Meghan Schott*, Seungtaek Kim*, and Paul Ahlquist* *Institute
Fine Mapping of a cis-acting Sequence Element in Yellow Fever Virus RNA That Is Required for RNA Replication and Cyclization
 JOURNAL OF VIROLOGY, Feb. 2003, p. 2265 2270 Vol. 77, No. 3 0022-538X/03/$08.00 0 DOI: 10.1128/JVI.77.3.2265 2270.2003 Copyright 2003, American Society for Microbiology. All Rights Reserved. Fine Mapping
JOURNAL OF VIROLOGY, Feb. 2003, p. 2265 2270 Vol. 77, No. 3 0022-538X/03/$08.00 0 DOI: 10.1128/JVI.77.3.2265 2270.2003 Copyright 2003, American Society for Microbiology. All Rights Reserved. Fine Mapping
Dr. Ahmed K. Ali Attachment and entry of viruses into cells
 Lec. 6 Dr. Ahmed K. Ali Attachment and entry of viruses into cells The aim of a virus is to replicate itself, and in order to achieve this aim it needs to enter a host cell, make copies of itself and
Lec. 6 Dr. Ahmed K. Ali Attachment and entry of viruses into cells The aim of a virus is to replicate itself, and in order to achieve this aim it needs to enter a host cell, make copies of itself and
Ubiquitination and deubiquitination of NP protein regulates influenza A virus RNA replication
 Manuscript EMBO-2010-74756 Ubiquitination and deubiquitination of NP protein regulates influenza A virus RNA replication Tsai-Ling Liao, Chung-Yi Wu, Wen-Chi Su, King-Song Jeng and Michael Lai Corresponding
Manuscript EMBO-2010-74756 Ubiquitination and deubiquitination of NP protein regulates influenza A virus RNA replication Tsai-Ling Liao, Chung-Yi Wu, Wen-Chi Su, King-Song Jeng and Michael Lai Corresponding
Herpesviruses. Virion. Genome. Genes and proteins. Viruses and hosts. Diseases. Distinctive characteristics
 Herpesviruses Virion Genome Genes and proteins Viruses and hosts Diseases Distinctive characteristics Virion Enveloped icosahedral capsid (T=16), diameter 125 nm Diameter of enveloped virion 200 nm Capsid
Herpesviruses Virion Genome Genes and proteins Viruses and hosts Diseases Distinctive characteristics Virion Enveloped icosahedral capsid (T=16), diameter 125 nm Diameter of enveloped virion 200 nm Capsid
Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid.
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
7.012 Quiz 3 Answers
 MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel Friday 11/12/04 7.012 Quiz 3 Answers A > 85 B 72-84
MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel Friday 11/12/04 7.012 Quiz 3 Answers A > 85 B 72-84
HIV-1 Virus-like Particle Budding Assay Nathan H Vande Burgt, Luis J Cocka * and Paul Bates
 HIV-1 Virus-like Particle Budding Assay Nathan H Vande Burgt, Luis J Cocka * and Paul Bates Department of Microbiology, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, USA
HIV-1 Virus-like Particle Budding Assay Nathan H Vande Burgt, Luis J Cocka * and Paul Bates Department of Microbiology, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, USA
A block in virus-like particle maturation following assembly of murine leukaemia virus in insect cells
 Available online at www.sciencedirect.com R Virology 314 (2003) 488 496 www.elsevier.com/locate/yviro A block in virus-like particle maturation following assembly of murine leukaemia virus in insect cells
Available online at www.sciencedirect.com R Virology 314 (2003) 488 496 www.elsevier.com/locate/yviro A block in virus-like particle maturation following assembly of murine leukaemia virus in insect cells
Structural vs. nonstructural proteins
 Why would you want to study proteins associated with viruses or virus infection? Receptors Mechanism of uncoating How is gene expression carried out, exclusively by viral enzymes? Gene expression phases?
Why would you want to study proteins associated with viruses or virus infection? Receptors Mechanism of uncoating How is gene expression carried out, exclusively by viral enzymes? Gene expression phases?
Polyomaviridae. Spring
 Polyomaviridae Spring 2002 331 Antibody Prevalence for BK & JC Viruses Spring 2002 332 Polyoma Viruses General characteristics Papovaviridae: PA - papilloma; PO - polyoma; VA - vacuolating agent a. 45nm
Polyomaviridae Spring 2002 331 Antibody Prevalence for BK & JC Viruses Spring 2002 332 Polyoma Viruses General characteristics Papovaviridae: PA - papilloma; PO - polyoma; VA - vacuolating agent a. 45nm
Chapter 13 Viruses, Viroids, and Prions. Biology 1009 Microbiology Johnson-Summer 2003
 Chapter 13 Viruses, Viroids, and Prions Biology 1009 Microbiology Johnson-Summer 2003 Viruses Virology-study of viruses Characteristics: acellular obligate intracellular parasites no ribosomes or means
Chapter 13 Viruses, Viroids, and Prions Biology 1009 Microbiology Johnson-Summer 2003 Viruses Virology-study of viruses Characteristics: acellular obligate intracellular parasites no ribosomes or means
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Krenn et al., http://www.jcb.org/cgi/content/full/jcb.201110013/dc1 Figure S1. Levels of expressed proteins and demonstration that C-terminal
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Krenn et al., http://www.jcb.org/cgi/content/full/jcb.201110013/dc1 Figure S1. Levels of expressed proteins and demonstration that C-terminal
Overview of virus life cycle
 Overview of virus life cycle cell recognition and internalization release from cells progeny virus assembly membrane breaching nucleus capsid disassembly and genome release replication and translation
Overview of virus life cycle cell recognition and internalization release from cells progeny virus assembly membrane breaching nucleus capsid disassembly and genome release replication and translation
Qin Yu and Casey D. Morrow 1. Department of Microbiology, University of Alabama at Birmingham, Birmingham, Alabama 35294
 Virology 254, 160 168 (1999) Article ID viro.1998.9542, available online at http://www.idealibrary.com on Complementarity between 3 Terminal Nucleotides of trna and Primer Binding Site Is a Major Determinant
Virology 254, 160 168 (1999) Article ID viro.1998.9542, available online at http://www.idealibrary.com on Complementarity between 3 Terminal Nucleotides of trna and Primer Binding Site Is a Major Determinant
The Ubiquitin-Vacuolar Protein Sorting System is Selectively Required During Entry of Influenza Virus into Host Cells
 Traffic 2003; 4: 857 868 Blackwell Munksgaard Copyright # Blackwell Munksgaard 2003 doi: 10.1046/j.1600-0854.2003.00140.x The Ubiquitin-Vacuolar Protein Sorting System is Selectively Required During Entry
Traffic 2003; 4: 857 868 Blackwell Munksgaard Copyright # Blackwell Munksgaard 2003 doi: 10.1046/j.1600-0854.2003.00140.x The Ubiquitin-Vacuolar Protein Sorting System is Selectively Required During Entry
Analysis of Rous Sarcoma Virus Gag Proteins by Mass Spectrometry Indicates Trimming by Host Exopeptidase
 JOURNAL OF VIROLOGY, May 1996, p. 3313 3318 Vol. 70, No. 5 0022-538X/96/$04.00 0 Copyright 1996, American Society for Microbiology Analysis of Rous Sarcoma Virus Gag Proteins by Mass Spectrometry Indicates
JOURNAL OF VIROLOGY, May 1996, p. 3313 3318 Vol. 70, No. 5 0022-538X/96/$04.00 0 Copyright 1996, American Society for Microbiology Analysis of Rous Sarcoma Virus Gag Proteins by Mass Spectrometry Indicates
October 26, Lecture Readings. Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell
 October 26, 2006 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
October 26, 2006 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
Retroviruses must gain access to the nucleus to replicate.
 Nuclear entry and CRM1-dependent nuclear export of the Rous sarcoma virus Gag polyprotein Lisa Z. Scheifele*, Rachel A. Garbitt, Jonathan D. Rhoads, and Leslie J. Parent* *Cell and Molecular Biology Program
Nuclear entry and CRM1-dependent nuclear export of the Rous sarcoma virus Gag polyprotein Lisa Z. Scheifele*, Rachel A. Garbitt, Jonathan D. Rhoads, and Leslie J. Parent* *Cell and Molecular Biology Program
Effects of Gag Mutation and Processing on Retroviral Dimeric RNA Maturation
 JOURNAL OF VIROLOGY, Feb. 2006, p. 1242 1249 Vol. 80, No. 3 0022-538X/06/$08.00 0 doi:10.1128/jvi.80.3.1242 1249.2006 Copyright 2006, American Society for Microbiology. All Rights Reserved. Effects of
JOURNAL OF VIROLOGY, Feb. 2006, p. 1242 1249 Vol. 80, No. 3 0022-538X/06/$08.00 0 doi:10.1128/jvi.80.3.1242 1249.2006 Copyright 2006, American Society for Microbiology. All Rights Reserved. Effects of
125. Identification o f Proteins Specific to Friend Strain o f Spleen Focus forming Virus (SFFV)
 No. 101 Proc. Japan Acad., 54, Ser. B (1978) 651 125. Identification o f Proteins Specific to Friend Strain o f Spleen Focus forming Virus (SFFV) By Yoji IKAWA,*} Mitsuaki YOSHIDA,*) and Hiroshi YosHIKURA**>
No. 101 Proc. Japan Acad., 54, Ser. B (1978) 651 125. Identification o f Proteins Specific to Friend Strain o f Spleen Focus forming Virus (SFFV) By Yoji IKAWA,*} Mitsuaki YOSHIDA,*) and Hiroshi YosHIKURA**>
Human Immunodeficiency Virus
 Human Immunodeficiency Virus Virion Genome Genes and proteins Viruses and hosts Diseases Distinctive characteristics Viruses and hosts Lentivirus from Latin lentis (slow), for slow progression of disease
Human Immunodeficiency Virus Virion Genome Genes and proteins Viruses and hosts Diseases Distinctive characteristics Viruses and hosts Lentivirus from Latin lentis (slow), for slow progression of disease
JCB Article. HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein. The Journal of Cell Biology
 JCB Article HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein Owen Pornillos, 1 Daniel S. Higginson, 1 Kirsten M. Stray, 1 Robert D. Fisher, 1 Jennifer E. Garrus, 1 Marielle Payne,
JCB Article HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein Owen Pornillos, 1 Daniel S. Higginson, 1 Kirsten M. Stray, 1 Robert D. Fisher, 1 Jennifer E. Garrus, 1 Marielle Payne,
Supplemental Materials and Methods Plasmids and viruses Quantitative Reverse Transcription PCR Generation of molecular standard for quantitative PCR
 Supplemental Materials and Methods Plasmids and viruses To generate pseudotyped viruses, the previously described recombinant plasmids pnl4-3-δnef-gfp or pnl4-3-δ6-drgfp and a vector expressing HIV-1 X4
Supplemental Materials and Methods Plasmids and viruses To generate pseudotyped viruses, the previously described recombinant plasmids pnl4-3-δnef-gfp or pnl4-3-δ6-drgfp and a vector expressing HIV-1 X4
Section 6. Junaid Malek, M.D.
 Section 6 Junaid Malek, M.D. The Golgi and gp160 gp160 transported from ER to the Golgi in coated vesicles These coated vesicles fuse to the cis portion of the Golgi and deposit their cargo in the cisternae
Section 6 Junaid Malek, M.D. The Golgi and gp160 gp160 transported from ER to the Golgi in coated vesicles These coated vesicles fuse to the cis portion of the Golgi and deposit their cargo in the cisternae
Molecular Characterization of Feline Immunodeficiency Virus Budding
 JOURNAL OF VIROLOGY, Mar. 2008, p. 2106 2119 Vol. 82, No. 5 0022-538X/08/$08.00 0 doi:10.1128/jvi.02337-07 Molecular Characterization of Feline Immunodeficiency Virus Budding Benjamin G. Luttge, 1 Miranda
JOURNAL OF VIROLOGY, Mar. 2008, p. 2106 2119 Vol. 82, No. 5 0022-538X/08/$08.00 0 doi:10.1128/jvi.02337-07 Molecular Characterization of Feline Immunodeficiency Virus Budding Benjamin G. Luttge, 1 Miranda
The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein
 THESIS BOOK The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein Orsolya Buzás-Bereczki Supervisors: Dr. Éva Bálint Dr. Imre Miklós Boros University of
THESIS BOOK The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein Orsolya Buzás-Bereczki Supervisors: Dr. Éva Bálint Dr. Imre Miklós Boros University of
Translation. Host Cell Shutoff 1) Initiation of eukaryotic translation involves many initiation factors
 Translation Questions? 1) How does poliovirus shutoff eukaryotic translation? 2) If eukaryotic messages are not translated how can poliovirus get its message translated? Host Cell Shutoff 1) Initiation
Translation Questions? 1) How does poliovirus shutoff eukaryotic translation? 2) If eukaryotic messages are not translated how can poliovirus get its message translated? Host Cell Shutoff 1) Initiation
Multiple Functions for the Basic Amino Acids of the Human T-Cell Leukemia Virus Type 1 Matrix Protein in Viral Transmission
 JOURNAL OF VIROLOGY, Mar. 1999, p. 1860 1867 Vol. 73, No. 3 0022-538X/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. Multiple Functions for the Basic Amino Acids of
JOURNAL OF VIROLOGY, Mar. 1999, p. 1860 1867 Vol. 73, No. 3 0022-538X/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. Multiple Functions for the Basic Amino Acids of
The Clathrin Adaptor Complex AP-1 Binds HIV-1 and MLV Gag and Facilitates Their Budding D
 Vol. 18, 3193 3203, August 2007 The Clathrin Adaptor Complex AP-1 Binds HIV-1 and MLV Gag and Facilitates Their Budding D Grégory Camus,* Carolina Segura-Morales, Dorothee Molle, Sandra Lopez-Vergès,*
Vol. 18, 3193 3203, August 2007 The Clathrin Adaptor Complex AP-1 Binds HIV-1 and MLV Gag and Facilitates Their Budding D Grégory Camus,* Carolina Segura-Morales, Dorothee Molle, Sandra Lopez-Vergès,*
~Lentivirus production~
 ~Lentivirus production~ May 30, 2008 RNAi core R&D group member Lentivirus Production Session Lentivirus!!! Is it health threatening to lab technician? What s so good about this RNAi library? How to produce
~Lentivirus production~ May 30, 2008 RNAi core R&D group member Lentivirus Production Session Lentivirus!!! Is it health threatening to lab technician? What s so good about this RNAi library? How to produce
Viral structure م.م رنا مشعل
 Viral structure م.م رنا مشعل Viruses must reproduce (replicate) within cells, because they cannot generate energy or synthesize proteins. Because they can reproduce only within cells, viruses are obligate
Viral structure م.م رنا مشعل Viruses must reproduce (replicate) within cells, because they cannot generate energy or synthesize proteins. Because they can reproduce only within cells, viruses are obligate
Quantifying Lipid Contents in Enveloped Virus Particles with Plasmonic Nanoparticles
 Quantifying Lipid Contents in Enveloped Virus Particles with Plasmonic Nanoparticles Amin Feizpour Reinhard Lab Department of Chemistry and the Photonics Center, Boston University, Boston, MA May 2014
Quantifying Lipid Contents in Enveloped Virus Particles with Plasmonic Nanoparticles Amin Feizpour Reinhard Lab Department of Chemistry and the Photonics Center, Boston University, Boston, MA May 2014
Particle Size Determinants in the Human Immunodeficiency Virus Type 1 Gag Protein
 JOURNAL OF VIROLOGY, June 1998, p. 4667 4677 Vol. 72, No. 6 0022-538X/98/$04.00 0 Copyright 1998, American Society for Microbiology Particle Size Determinants in the Human Immunodeficiency Virus Type 1
JOURNAL OF VIROLOGY, June 1998, p. 4667 4677 Vol. 72, No. 6 0022-538X/98/$04.00 0 Copyright 1998, American Society for Microbiology Particle Size Determinants in the Human Immunodeficiency Virus Type 1
Human Ubc9 Contributes to Production of Fully Infectious Human Immunodeficiency Virus Type 1 Virions
 University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Virology Papers Virology, Nebraska Center for 2009 Human Ubc9 Contributes to Production of Fully Infectious Human Immunodeficiency
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Virology Papers Virology, Nebraska Center for 2009 Human Ubc9 Contributes to Production of Fully Infectious Human Immunodeficiency
A Single Polymorphism in HIV-1 Subtype C SP1 Is Sufficient To Confer Natural Resistance to the Maturation Inhibitor Bevirimat
 ANTIMICROBIAL AGENTS AND CHEMOTHERAPY, July 2011, p. 3324 3329 Vol. 55, No. 7 0066-4804/11/$12.00 doi:10.1128/aac.01435-10 Copyright 2011, American Society for Microbiology. All Rights Reserved. A Single
ANTIMICROBIAL AGENTS AND CHEMOTHERAPY, July 2011, p. 3324 3329 Vol. 55, No. 7 0066-4804/11/$12.00 doi:10.1128/aac.01435-10 Copyright 2011, American Society for Microbiology. All Rights Reserved. A Single
Interaction of influenza A virus matrix protein with RACK1 is required for virus release
 Cellular Microbiology (2012) 14(5), 774 789 doi:10.1111/j.1462-5822.2012.01759.x First published online 6 March 2012 Interaction of influenza A virus matrix protein with RACK1 is required for virus release
Cellular Microbiology (2012) 14(5), 774 789 doi:10.1111/j.1462-5822.2012.01759.x First published online 6 March 2012 Interaction of influenza A virus matrix protein with RACK1 is required for virus release
JOURNAL OF VIROLOGY, Apr. 1999, p Vol. 73, No. 4. Copyright 1999, American Society for Microbiology. All Rights Reserved.
 JOURNAL OF VIROLOGY, Apr. 1999, p. 2921 2929 Vol. 73, No. 4 0022-538X/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. A Proline-Rich Motif within the Matrix Protein
JOURNAL OF VIROLOGY, Apr. 1999, p. 2921 2929 Vol. 73, No. 4 0022-538X/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. A Proline-Rich Motif within the Matrix Protein
LESSON 4.4 WORKBOOK. How viruses make us sick: Viral Replication
 DEFINITIONS OF TERMS Eukaryotic: Non-bacterial cell type (bacteria are prokaryotes).. LESSON 4.4 WORKBOOK How viruses make us sick: Viral Replication This lesson extends the principles we learned in Unit
DEFINITIONS OF TERMS Eukaryotic: Non-bacterial cell type (bacteria are prokaryotes).. LESSON 4.4 WORKBOOK How viruses make us sick: Viral Replication This lesson extends the principles we learned in Unit
Gammaretroviral pol sequences act in cis to direct polysome loading and NXF1/NXT-dependent protein production by gag-encoded RNA
 University of Massachusetts Medical School escholarship@umms Open Access Articles Open Access Publications by UMMS Authors 9-12-2014 Gammaretroviral pol sequences act in cis to direct polysome loading
University of Massachusetts Medical School escholarship@umms Open Access Articles Open Access Publications by UMMS Authors 9-12-2014 Gammaretroviral pol sequences act in cis to direct polysome loading
RAPID COMMUNICATION. Canine Cyclin T1 Rescues Equine Infectious Anemia Virus Tat Trans-Activation in Human Cells
 Virology 268, 7 11 (2000) doi:10.1006/viro.1999.0141, available online at http://www.idealibrary.com on RAPID COMMUNICATION Canine Cyclin T1 Rescues Equine Infectious Anemia Virus Tat Trans-Activation
Virology 268, 7 11 (2000) doi:10.1006/viro.1999.0141, available online at http://www.idealibrary.com on RAPID COMMUNICATION Canine Cyclin T1 Rescues Equine Infectious Anemia Virus Tat Trans-Activation
