Human Immunodeficiency Virus Type 1 Gag Engages the Bro1 Domain of ALIX/AIP1 through the Nucleocapsid
|
|
|
- Austen Hoover
- 5 years ago
- Views:
Transcription
1 JOURNAL OF VIROLOGY, Feb. 2008, p Vol. 82, No X/08/$ doi: /jvi Copyright 2008, American Society for Microbiology. All Rights Reserved. Human Immunodeficiency Virus Type 1 Gag Engages the Bro1 Domain of ALIX/AIP1 through the Nucleocapsid Sergei Popov, Elena Popova, Michio Inoue, and Heinrich G. Göttlinger* Program in Gene Function and Expression, Program in Molecular Medicine, University of Massachusetts Medical School, Worcester, Massachusetts Received 31 August 2007/Accepted 6 November 2007 Human immunodeficiency virus type 1 (HIV-1) and other retroviruses harbor short peptide motifs in Gag that promote the release of infectious virions. These motifs, known as late assembly (L) domains, recruit a cellular budding machinery that is required for the formation of multivesicular bodies (MVBs). The primary L domain of HIV-1 maps to a PTAP motif in the p6 region of Gag and engages the MVB pathway by binding to Tsg101. Additionally, HIV-1 p6 harbors an auxiliary L domain that binds to the V domain of ALIX, another component of the MVB pathway. We now show that ALIX also binds to the nucleocapsid (NC) domain of HIV-1 Gag and that ALIX and its isolated Bro1 domain can be specifically packaged into viral particles via NC. The interaction with ALIX depended on the zinc fingers of NC, which mediate the specific packaging of genomic viral RNA, but was not disrupted by nuclease treatment. We also observed that HIV-1 zinc finger mutants were defective for particle production and exhibited a similar defect in Gag processing as a PTAP deletion mutant. The effects of the zinc finger and PTAP mutations were not additive, suggesting a functional relationship between NC and p6. However, in contrast to the PTAP deletion mutant, the double mutants could not be rescued by overexpressing ALIX, further supporting the notion that NC plays a role in virus release. * Corresponding author. Mailing address: UMass Medical School, LRB 526, 364 Plantation Street, Worcester, MA Phone: (508) Fax: (508) heinrich.gottlinger@umassmed.edu. Published ahead of print on 21 November Retroviral Gag polyproteins harbor short peptide motifs that promote the detachment of assembled virions from the cell surface and from each other (16, 22, 24, 41, 49, 50, 52). These motifs, which are known as late assembly (L) domains, often remain functional even if moved to a different position within Gag and can be exchanged among unrelated viruses (39, 51). In the case of human immunodeficiency virus type 1 (HIV- 1), the primary L domain consists of a conserved P(T/S)AP motif near the beginning of the C-terminal p6 domain of the Gag polyprotein (22, 24). The PTAP motif serves as a binding site for the host protein Tsg101, which is required for the release of infectious virions (21, 31, 47). Tsg101 is a component of the endosomal sorting complex ESCRT-I, which functions in the sorting of ubiquitinated cargo into small vesicles that bud into the lumen of multivesicular bodies (MVBs) (3, 23, 27). This invagination event is topologically related to retrovirus budding and ultimately serves to deliver transmembrane proteins to lysosomal compartments for degradation. The components of this cellular budding pathway were initially identified in Saccharomyces cerevisiae, where a protein network that includes at least 18 different vacuolar protein-sorting (Vps) proteins is required for the formation of MVBs (3). The absence of any of these gene products causes the formation of an abnormal endosomal structure called the class E compartment (26). The MVB sorting pathway is conserved throughout eukaryotic evolution, and at least one homolog for each of the yeast class E Vps proteins exists in humans (48). Most of the class E Vps proteins participate in the formation of ESCRT-I or of two other high-molecularweight complexes called ESCRT-II and ESCRT-III, which function downstream of ESCRT-I (25). MVB sorting also depends on the activity of the AAA-type ATPase VPS4, another class E Vps protein that recycles the ESCRT machinery (4). Dominant-negative versions of various ESCRT-III subunits and of VPS4 potently inhibit the functions of all known types of viral L domains (21, 30, 43, 44, 48, 53), indicating that all act through components of the MVB sorting pathway. In addition to the PTAP motif, the p6 domain of HIV-1 Gag contains a LYPx n L motif near its C terminus that binds to ALIX/AIP1 (44), a homolog of the yeast class E Vps protein Bro1 that functions in endosomal trafficking (36). Like Tsg101, ALIX promotes viral egress, which requires the separation of the viral envelope from the plasma membrane, and is also involved in membrane fission events that are necessary for the completion of cytokinesis (10). ALIX has a modular organization, with a banana-shaped N-terminal Bro1 domain, a central domain that is shaped like a V and has therefore been termed the V domain, and a C-terminal proline-rich domain (PRD) that is thought to be unstructured (20, 29). ALIX interacts with the ESCRT-III component CHMP4 through its Bro1 domain (36), with LYPx n L-type viral L domains through its V domain (11, 33), and with Tsg101 and other endocytic proteins through its PRD (36). The few lentiviruses that are known to lack a PTAP motif all harbor a high-affinity binding site for ALIX in the C-terminal domain of Gag (6, 44). One of these is equine infectious anemia virus, which depends on ALIX for efficient virus release (20, 30, 41, 44, 48). However, in the case of HIV-1, the ALIX binding site in p6 has only an auxiliary role in virus release that becomes more evident in a minimal Gag context (20, 44). Nevertheless, exogenous ALIX can potently rescue the profound budding defect of HIV-1 PTAP mutants, provided that the ALIX binding site in p6 remains intact (20, 46). 1389
2 1390 POPOV ET AL. J. VIROL. We previously reported that the PTAP-type L domain in HIV-1 p6 does not function autonomously but depends on other Gag regions in order to function (45). Our results suggested that the PTAP motif needs to cooperate either with another determinant in p6, which is now known to bind ALIX, or with the nucleocapsid (NC)-p1 region, which immediately precedes the p6 domain in the context of the Gag precursor (45). The NC domain contains two highly conserved CCHC motifs that coordinate zinc and are required for the selective packaging of genomic viral RNA into progeny virions (5). NC also possesses a nonspecific nucleic acid binding activity that depends on its basic residues and is crucial for Gag polymerization and assembly (13). In contrast, disruption of the two CCHC motifs had no significant effect on Gag-Gag interactions (13). We now show that NC associates with the Bro1 domain of ALIX through its two CCHC motifs. We also show that the ability of ALIX to rescue a Tsg101 binding site mutant depends on intact CCHC motifs in NC. Disrupting both CCHC motifs led to defects in particle production and to an accumulation of the capsid (CA)-p2 and p41 Gag cleavage intermediates that is typically seen with L domain mutants (21, 22). However, the effects of disrupting the CCHC motifs and the PTAP motif in p6 were not additive, which supports the hypothesis that these motifs function in the same pathway during virus production. MATERIALS AND METHODS Proviral constructs. HXBH10, the parental proviral plasmid used in this study, is a vpu-positive version of the infectious HXB2 proviral clone of HIV-1. HXBH10-gag is unable to express Gag due to premature termination codons in the gag gene (19). The protease (PR)-negative variant HXBH10-PR, which was used to express Pr55 gag, and the HXBH10-based chimeric Gag constructs Z WT and Z WT -p6 have been described elsewhere (1). Z WT encodes an HIV-1 Gag precursor that has the NC-p1-p6 region precisely replaced by the GCN4 leucine zipper domain (1). Z WT -p6 has the HIV-1 p6 coding sequence directly fused to the 3 end of the GCN4 sequence (1). Pr55(Y 36 s) and Z WT -p6(y 36 s) are variants of HXBH10-PR and Z WT -p6, respectively, that harbor the previously described Y 36 s mutation (22), which introduces a premature termination codon in place of Tyr-36 of p6 without altering the pol frame. The codons for NC-p1 residues 56 to 71, 52 to 71, 36 to 71, and 15 to 71 were precisely deleted from HXBH10-Y 36 s without affecting any other Gag coding sequences. This was achieved by introducing silent mutations into the first two p6 codons to generate a PstI restriction site. All of these deletions remove the frameshift signal required for the expression of PR and thus prevent Gag processing. In contrast, the 15 39, 15 34/ C 36 S, 15 28, 30 31, C 28,49 S, and K 33,34 A mutations keep the frameshift signal intact and were therefore introduced into the PR-negative version of HXBH10- Y 36 s to prevent Gag processing. The 15 39, C 36 S, and mutants have two foreign codons specifying Leu-Gln inserted in place of the deleted NC codons. The C 28,49 S and mutations were also introduced into the PRpositive proviruses HXBH10 and PTAPP, a variant of HXBH10 with an inframe deletion of codons 7 through 11 of p6 (1). Expression vectors. The pbj5-based mammalian expression vectors for ALIX-HA and HA-ALIX have been described previously (44). The F 676 Dmutant of ALIX-HA was made using the QuikChange mutagenesis strategy (Stratagene). The expression vector for HA-ALIX Bro1 was obtained by inserting a PCR product encoding ALIX residues 1 to 362 with an N-terminal hemagglutinin (HA) tag between the NotI and EcoRI sites of pbj5. The coding sequence for APOBEC3G with a C-terminal FLAG tag was amplified from BC (Open Biosystems) and cloned between the HindIII and XhoI sites of pcdna3.1( ) (Invitrogen). In all mammalian expression vectors, the start codon is preceded by a Kozak consensus sequence for efficient translation initiation. Analysis of virus-like particles (VLP). 293T cells ( ) or HeLa cells ( ) were seeded into T80 flasks and cotransfected 24 h later with HIV-1 proviral DNA and ALIX expression vectors as indicated using a calcium phosphate precipitation technique. The total amount of transfected DNA was kept constant with carrier DNA (pbluescript). Twenty-four h (293T) or 48 h (HeLa) posttransfection, the cells were lysed in radioimmunoprecipitation assay buffer (140 mm NaCl, 8 mm Na 2 HPO 4,2mMNaH 2 PO 4, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.05% sodium dodecyl sulfate [SDS]), and the culture supernatants were clarified by low-speed centrifugation and passaged through m filters. Virions released into the medium were then pelleted through 20% sucrose cushions. Pelletable material and the cell lysates were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting as described elsewhere (1), using a rabbit anti-hiv CA serum (Advanced Biotechnologies) or the anti-hiv CA antibody 183-H12-5C (12) to detect Gag proteins. HA-tagged ALIX was detected with the anti-ha antibody HA.11 (Covance). GST pull-down assay. PCR fragments encoding the full-length NC (residues 1 to 55) and NC-p1 (residues 1 to 71) domains of HIV-1 HXB2 Gag were inserted in frame between the EcoRI and XhoI sites of pgex-4t-1 (GE Healthcare). Glutathione S-transferase (GST) fusion proteins were expressed in strain BL21 and immobilized on glutathione-sepharose beads (GE Healthcare). The beads were then incubated for 3 h at 4 Cwith hypotonic lysates of 293T cells transiently expressing epitope-tagged ALIX or APOBEC3G, followed by extensive washing in phosphate-buffered saline. Where indicated, the beads were incubated for another 30 min at 37 C in the presence or absence of 75 U (0.75 U/ l) benzonase nuclease (EMD Biosciences) in benzonase buffer (1.2 mm MgCl 2, 50 mm Tris- HCl [ph 8.0]). Bound proteins were eluted by boiling in SDS-PAGE sample buffer and resolved by SDS-PAGE. Epitope-tagged proteins were detected by Western blotting with anti-ha or anti-flag M2 antibody, and GST fusion proteins were visualized with colloidal Coomassie brilliant blue G-250. RESULTS NC-dependent incorporation of ALIX into HIV-1 virions. We previously examined the incorporation of ALIX into HIV-1 particles using the chimeric Gag constructs Z WT and Z WT -p6 (1, 44), which have NC-p1 replaced by the GCN4 leucine zipper domain (Fig. 1A). Because the GCN4 zipper domain rescues viral particle production by HIV-1 Gag in the absence of the NC-p1-p6 region (1), this approach allowed us to show that p6 mediates the specific incorporation of ALIX (44). Consistent with this finding, we and others have shown that both full-length ALIX and the isolated ALIX V domain bind to an LYPx n L motif near the C terminus of HIV-1 p6 (20, 29, 33, 44, 48). We therefore expected that the incorporation of ALIX into viral particles formed by authentic HIV-1 Gag would be abolished by the Y 36 s mutation (22), which converts the codon for Y 36 of p6 into a premature termination codon and prevents the synthesis of the LYPx n L motif. To test this notion, we coexpressed HA-tagged ALIX with HIV-1 proviral constructs expressing either no Gag, the wild-type (WT) Pr55 Gag polyprotein, or Pr55(Y 36 s) (Fig. 1A and B). As expected, HA-ALIX was readily detected in virions produced by WT HIV-1 Gag, and the incorporation of HA-ALIX was specific, because its appearance in the particulate fraction depended on the expression of Gag (Fig. 1B, lanes 1 and 2). However, we were surprised to find that the Y 36 s mutation reduced the uptake of HA-ALIX into viral particles only about twofold (Fig. 1B, lane 3), indicating that in an otherwise-wt HIV-1 Gag context the LYPx n L motif contributes to the recruitment of ALIX but is not essential. To determine whether the uptake of ALIX in the absence of the LYPx n L motif was mediated by the truncated p6 domain of the Y 36 s mutant, we introduced the Y 36 s mutation into Z WT -p6 (Fig. 1A). As previously reported (1, 44), the Z WT chimeric Gag molecule efficiently forms VLP that fail to incorporate coexpressed HA-ALIX (Fig. 1C, lane 1). In contrast, HA- ALIX was readily detectable in VLP produced by Z WT -p6 (Fig. 1C, lane 2), again consistent with previous results (44). Inter-
3 VOL. 82, 2008 ALIX BINDS TO HIV-1 NC 1391 FIG. 1. NC-mediated incorporation of ALIX into VLP. (A) Schematic illustration of the domain organizations of the HIV-1 Gag precursor (Pr55) and of mutant Gag molecules. A wavy line at the N terminus indicates the presence of a myristylation signal, and a gray box indicates the presence of a GCN4 zipper (Z) domain in place of NC-p1. (B) The ALIX binding site in p6 is not essential for the incorporation of ALIX into VLP. 293T cells were transfected with HIV-1 proviral DNA (5 g) expressing no Gag, WT Gag, or C-terminally truncated Gag, along with 5 g of a vector expressing ALIX-HA. VLP pellets and the cell lysates were analyzed by Western blotting to detect Gag and ALIX-HA as indicated. (C) The uptake of ALIX in the absence of the binding site in p6 depends on NC-p1. 293T cells were transfected with HIV-1 proviral DNAs (5 g) expressing Gag molecules with a leucine zipper in place of NC-p1, along with 5 g ofa vector expressing ALIX-HA. estingly, in the context of Z WT -p6, the Y 36 s mutation abolished the incorporation of HA-ALIX into VLP (Fig. 1C, lane 3). Of note, the Pr55(Y 36 s) and Z WT -p6(y 36 s) proviral constructs are precisely identical except for the region encoding NC-p1. We thus conclude that the ability of Pr55(Y 36 s) but not of Z WT - p6(y 36 s) Gag to direct the incorporation of ALIX is dependent on NC-p1. Role of NC zinc fingers in incorporation of ALIX. The NC domain of HIV-1 Gag harbors two copies of a CCHC-type zinc finger motif that are involved in the specific encapsidation of genomic viral RNA into nascent viral particles. NC also exhibits a nonspecific RNA binding activity that depends on clusters of basic residues and is thought to be critical for virus assembly (8, 13). However, NC deletion mutants are not drastically defective for viral particle production as long as the N-terminal basic NC region proximal to the first CCHC motif is retained (55). Furthermore, it has been shown that particle production by a mutant that lacks nearly all of NC can be rescued by a second site mutation that inactivates PR (38). To maintain FIG. 2. Effects of deletions in NC-p1 on the uptake of ALIX into VLP. (A) Schematic illustration of the Gag mutants examined. Crosshatched boxes indicate positions of the zinc fingers in NC. (B) p6- independent incorporation of ALIX depends on NC but not on p1. 293T cells were transfected with the indicated amounts of mutant HIV-1 proviral DNAs together with 3 g of a vector expressing ALIX- HA. VLP pellets and the cell lysates were analyzed by Western blotting to detect Gag and ALIX-HA as indicated. (C) Effects of a deletion that removes both zinc fingers of NC together with p1. viral particle production, we therefore used a PR-deficient provirus as the parental construct to generate a series of mutants with in-frame deletions in the NC-p1 coding region. Furthermore, all NC-p1 deletions were combined with the Y 36 s mutation to eliminate the ALIX binding site in the p6 domain of Gag (Fig. 2A). Since Gag multimerization is concentration dependent (40), we transfected relatively large amounts of these proviral constructs into 293T cells together with a vector expressing ALIX-
4 1392 POPOV ET AL. J. VIROL. FIG. 3. Role of NC zinc fingers in uptake of ALIX into VLP. (A) Schematic illustration of the Gag mutants examined. Cross-hatched boxes indicate the positions of the zinc fingers in NC, and arrowheads indicate the positions of residues that were substituted. (B) Effects of deletions that specifically target zinc fingers. 293T cells were transfected with the indicated amounts of mutant HIV-1 proviral DNAs, together with 5 g of a vector expressing ALIX-HA. VLP pellets and the cell lysates were analyzed by Western blotting to detect Gag and ALIX-HA as indicated. (C) Effect of disrupting both zinc fingers through point mutations. The C 28,49 S/Y 36 s mutant was examined in duplicate to verify reproducibility. (D) Effects of mutations in the region between the two zinc fingers. HA, in an effort to override potential Gag-Gag interaction defects. As a control for Gag-independent release of ALIX in cellular vesicles, the ALIX expression vector was cotransfected with a provirus that lacked the ability to express Gag due to the presence of premature termination codons in the gag gene. As shown in Fig. 2B, deleting the region between the second CCHC motif and p6 had no effect on particle production or on the incorporation of ALIX-HA (lanes 3 and 4). In contrast, a deletion which additionally removed the second CCHC motif ( 36 71) reduced the incorporation of ALIX-HA at least threefold (lane 5). Finally, ALIX-HA was no longer detectable in the particulate fraction if the first CCHC motif was also removed ( 15 71) (lane 6). In the experiment shown in Fig. 2B, the 15 71/Y 36 s mutant at 10 g produced somewhat fewer VLP than the parental proviral construct at 5 g. Therefore, the parental construct was titrated down to obtain equivalent levels of particle production. We found that VLP production by the 15 71/Y 36 s mutant transfected at 10 or 15 g matched or exceeded that by the parental Y 36 s provirus only when less than 1 g ofthe latter construct was used (Fig. 2C and data not shown). Importantly, this experiment confirmed that 15 71/Y 36 s VLP incorporate significantly less ALIX than Y 36 s VLP (Fig. 2C). Next, we selectively targeted the CCHC motifs in NC to determine their role in the LYPx n L-independent uptake of ALIX. All NC mutants depicted in Fig. 3A harbored the Y 36 s mutation to prevent the incorporation of ALIX via p6 and a mutation that inactivated PR to suppress potential defects in VLP production in the absence of NC sequences. As shown in Fig. 3B, a deletion that removed the first CCHC motif and a portion of the second ( 15 39) essentially abolished the ability of the Y 36 s mutant to incorporate ALIX (compare lanes 2 and 4). A similar effect was observed when a deletion that removed the first CCHC motif and a few flanking residues ( 15 34) was combined with a point mutation that changed the second CCHC motif to SCHC (lane 5). However, the ability to incorporate ALIX was partially preserved if only the first CCHC motif was deleted and the second was kept intact ( 15 28) (lane 6). We also note that the latter mutant produced more VLP than those which retained no intact CCHC motif (Fig. 3B). To specifically examine the role of zinc-coordinating residues in the incorporation of ALIX, we changed both CCHC
5 VOL. 82, 2008 ALIX BINDS TO HIV-1 NC 1393 motifs in NC to CCHS (Fig. 3A). As shown in Fig. 3C, VLP produced by the resulting C 28,49 S/Y 36 s mutant reproducibly incorporated less ALIX than those produced by the parental Y 36 s construct (compare lanes 1 and 2 to lanes 4 and 5). In contrast, mutations that targeted the region between the two CCHC motifs had no effect on the LYPx n L-independent incorporation of ALIX (Fig. 3D). Together, these results indicate that the LYPx n L-independent uptake of ALIX is mediated by the zinc fingers in NC. ALIX physically associates with HIV-1 NC in vitro. The packaging of ALIX into VLP in the absence of the LYPx n L motif in p6 suggested that ALIX may also bind to other Gag regions. To examine whether ALIX interacts with the NC-p1 region of HIV-1 Gag, GST-NC or GST-NCp1 was bound to glutathione-sepharose beads and incubated with extracts from 293T cells expressing FLAG- or HA-tagged ALIX. Epitopetagged ALIX was readily detected in association with both GST-NC and GST-NCp1, but not with GST alone (Fig. 4A and B). We also observed that the association of ALIX with GST- NCp1 was considerably reduced by the C 28,49 S mutation (Fig. 4B), consistent with the effect of this mutation on the packaging of ALIX into VLP (Fig. 3C). We previously showed that the C 28,49 S double mutation, which disrupts both CCHC motifs in NC, reduces the packaging of genomic viral RNA into HIV-1 virions more than 20- fold (18). We also showed that even the C 28 S change by itself reduced the binding of viral RNA to HIV-1 Gag 10-fold in a Northwestern blot assay (18). Since the C 28,49 S mutation also significantly reduced the interaction between NC-p1 and ALIX, we considered the possibility that the in vitro interaction depended on the presence of nucleic acid. To examine this possibility, HA-ALIX was pulled down with GST-NC-p1 bound to glutathione-sepharose, followed by incubation of the beads with benzonase, a promiscuous nuclease capable of degrading all forms of RNA and DNA. This treatment had no effect on the association of HA-ALIX with GST-NC-p1 (Fig. 4C, lanes 1 to 4). As a control, we examined whether the interaction between NC-p1 and APOPEC3G was affected by benzonase. It was shown previously that the ability of Gag- GST fusion proteins to bind APOBEC3G is inhibited by RNase A (54). Similarly, we observed that benzonase completely disrupted the interaction between GST-NCp1 and APOBEC3G-FLAG under the same experimental conditions in which it had no effect on the interaction with ALIX (Fig. 4C, lanes 5 to 7). Taken together, these results indicate that the interaction between NC or NC-p1 and ALIX is not simply a consequence of unspecific bridging by nucleic acids. NC engages the Bro1 domain of ALIX. ALIX possesses an N-terminal Bro1 domain and a central V domain that is named for its V-like shape (20, 29). The V domain is required for the binding of ALIX to HIV-1 p6 (44), and recent structural studies revealed that Phe-676 in a conserved hydrophobic pocket of the V domain plays a central role in ALIX-L domain interactions (20, 29). Mutating Phe-676 to Asp abolished the ability of the isolated V domain to inhibit HIV-1 particle release (29) and the ability of full-length ALIX to rescue viral particle production by PTAP HIV-1 (20). Consistent with these observations, the F 676 D mutation nearly eliminated the packaging of ALIX into VLP formed by Z WT -p6 (Fig. 5A, compare lanes 5 and 6), which depends on FIG. 4. In vitro interaction between ALIX and NC or NC-p1. (A) ALIX binds to GST-NC and GST-NC-p1. Glutathione-Sepharose beads decorated with bacterially expressed GST or GST fusion proteins were incubated with lysates from 293T cells expressing full-length ALIX with a C-terminal FLAG tag. GST proteins bound to the beads were detected by SDS-PAGE and Coomassie staining, and captured ALIX was detected by Western blotting with anti-flag. (B) A GST pull-down assay performed as for panel A, showing that the interaction between ALIX and GST-NC-p1 is impaired by the C 28,49 S mutation, which disrupts both CCHC motifs in NC. (C) ALIX remains bound to GST-NCp1 in the presence of nuclease, whereas the interaction between GST-NCp1 and APOBEC3G is disrupted. GST pull-down assays were performed as for panel A, followed by incubation of the beads with or without benzonase nuclease. Proteins that remained bound to the glutathione-sepharose beads were then detected as indicated. the LYPx n L motif in p6 (Fig. 1C). In contrast, the same mutation had only a moderate effect on the incorporation of ALIX into particles formed by the WT Pr55 Gag polyprotein (Fig. 5A, lanes 3 and 4). Furthermore, this effect was comparable to that of removing the LYPx n L motif in p6 on the incorporation of WT ALIX (Fig. 1B). Together, these results suggested that the packaging of ALIX via NC does not depend on the presence of an intact V domain. We therefore asked whether the isolated Bro1 domain of ALIX can be packaged into VLP. HA-ALIX Bro1 was readily detected in VLP produced by the Y 36 s mutant (Fig. 5B, lane 1), which expresses a Gag polyprotein that lacks the ALIX binding site in p6 but retains the WT NC domain (Fig. 1A). Indeed, HA-ALIX Bro1 was incorporated into Y 36 s mutant VLP at least as well as full-length ALIX (Fig. 5C). In marked contrast, HA-ALIX Bro1 was not at all packaged into VLP formed by Z WT -p6 (Fig. 5B, lane 2), which lacks NC but pos-
6 1394 POPOV ET AL. J. VIROL. FIG. 5. NC engages the Bro1 domain of ALIX. (A) Effect of disrupting the ligand binding pocket in the V domain on incorporation of ALIX into VLP. 293T cells were transfected with HIV-1 proviral DNAs (7.5 g) expressing no Gag, WT Gag, or Gag with a leucine zipper in place of NC-p1, along with 5 g of a vector expressing WT or mutant ALIX-HA. VLP pellets and the cell lysates were analyzed by Western blotting to detect Gag and ALIX-HA as indicated. (B) NC-dependent incorporation of the isolated Bro1 domain of ALIX. 293T cells were transfected with HIV-1 proviral DNAs (7.5 g) expressing the indicated Gag molecules, along with 5 g of a vector expressing HA-ALIX Bro1. (C) NC-dependent incorporation of full-length HA-ALIX versus that of HA-ALIX Bro1. 293T cells were transfected with HIV-1 proviral DNA (7.5 g) expressing the Pr55(Y 36 s) Gag precursor, along with 5 g of vector expressing HA-ALIX or HA-ALIX Bro1. sesses an intact p6 domain (Fig. 1A). This result was not attributable to different levels of VLP production or to differences in the expression of HA-ALIX Bro1 (Fig. 5B). We thus conclude that the packaging of HA-ALIX Bro1 by the Y 36 s mutant is specific, and we infer that NC interacts with the Bro1 domain of ALIX. We previously described an HIV-1 mutant which encodes a minimal Gag protein called 8 87/ that lacks the globular domain of MA and the N-terminal domain of CA but produces WT levels of VLP (7). L domain function in this minimal Gag context was highly dependent on the presence of an intact ALIX binding site in p6 (44). Moreover, VLP production by ALIX binding site mutants could be rescued by overexpressing ALIX (44). This activity of ALIX depended on the presence of the PTAPP motif in p6, but not on the C- terminal PRD of ALIX (44), which is required for the rescue of PTAPP HIV-1 by ALIX (20, 46). Surprisingly, ALIX PRD markedly increased VLP production even by a variant of 8 87/ that harbors the Y 36 s mutation and thus lacks the entire ALIX binding site in p6 (44). We therefore examined whether the V domain of ALIX, which binds to p6 (20, 29), is required for the rescue of VLP production in this system. In both 293T and HeLa cells, HA-ALIX Bro1 increased VLP production by the Y 36 s version of the 8 87/ minimal Gag construct more than 10-fold without affecting Gag expression levels (Fig. 6A). As expected, HA-ALIX Bro1 did not increase the release of PTAPP HIV-1 (Fig. 6B, lanes 1 and 2). Furthermore, HA-ALIX Bro1 did not augment virion production by WT HIV-1 (Fig. 6B, lanes 3 and 4). Since the 8 87/ /Y 36 s minimal Gag construct retains the NC domain, these results raise the possibility that ALIX and the isolated Bro1 domain can act through NC. NC is required for the function of ALIX in HIV-1 budding. We and Sundquist and coworkers recently reported that the release defects of HIV-1 L domain mutants can be corrected by increasing the cellular expression levels of ALIX (20, 46). For instance, exogenous ALIX completely rescued particle production by a mutant termed PTAPP, which lacks the Tsg101 binding site in p6 (46). In this PTAPP rescue assay, the activity of ALIX was dependent on the presence of an intact ALIX binding site near the C terminus of p6 (20, 46). To determine whether NC also plays a role in the rescue by ALIX, we generated a variant of the PTAPP mutant that harbors the C 28,49 S mutation in NC, which interferes with the interaction between NC and ALIX (Fig. 3 and 4). Of note, in a previous study the C 28,49 S mutation had no effect on Gag- Gag interactions (13). Nevertheless, the C 28,49 S mutation by itself reduced viral particle production in 293T cells, although this defect was relatively mild compared to that caused by the PTAPP mutation (Fig. 7A). The C 28,49 S mutation also affected the pattern of cell-associated Gag proteins, which was altered in a manner similar to that seen with the PTAPP mutant. Specifically, both mutations altered the ratio of mature CA versus the CA-p2 processing intermediate and caused an accumulation of the p41 cleavage intermediate, as is typically seen if a late assembly step is defective (21, 22). Interestingly, the C 28,49 S/ PTAPP double mutant was no more defective for particle production than the original PTAPP mutant and produced a pattern of cell-associated Gag products similar to that seen with the two parental mutants (Fig. 7A). When an expression vector for ALIX was cotransfected, we observed a profound increase in viral particle production by the PTAPP mutant (Fig. 7B), as previously described (46). In contrast, particle production by the C 28,49 S/ PTAPP double mutant was only marginally improved (Fig. 7B). We also examined the effect of the deletion in the PTAPP rescue assay, since the deletion appeared to completely prevent the recruitment of ALIX via NC (Fig. 3B). In the presence of WT PR, the deletion in NC caused as profound a defect in viral particle production as the PTAPP L domain mutation and also had a comparable effect on the processing of cell-associated Gag (Fig. 8A, lanes 2 and 3). Remarkably, when the two mutations were combined, there again was no additive effect on particle production or on Gag
7 VOL. 82, 2008 ALIX BINDS TO HIV-1 NC 1395 FIG. 6. Effect of HA-ALIX Bro1 on particle production. (A) Rescue of VLP production by a minimal Gag construct that lacks the ALIX binding site in p6. 293T cells (lanes 1 to 4) or HeLa cells (lanes 5,6) were transfected with 1 g (lanes 1,2), 2 g (lanes 3,4), or 7.5 g (lanes 5,6) of proviral DNA expressing the Y 36 s version of the 8 87/ minimal Gag molecule (44), along with 2 g (lanes 1 and 2) or 5 g (lanes 3 to 6) of empty pbj5 or the HA-ALIX Bro1 expression vector. VLP production and Gag expression levels were examined by Western blotting with anti-ca antibody 183-H12-5C. The 8 87/ minimal Gag molecule lacks the globular domain of MA and the N-terminal domain of CA, and the Y 36 s truncation removes the ALIX binding site in p6. (B) HA-ALIX Bro1 does not increase virion production by PTAPP or WT HIV-1. HeLa cells were transfected with 7.5 g of PTAPP or WT HXBH10, along with 5 g of empty pbj5 or the HA-ALIX Bro1 expression vector, and virion production was examined as above. processing (Fig. 8A, lane 4). Moreover, particle production by the double mutant could not be rescued at all by exogenous ALIX (Fig. 8B). Taken together, these results indicate that the determinants in NC which mediate the interaction with ALIX also play a critical role in the function of ALIX in virus budding. DISCUSSION Several studies have shown that ALIX functionally interacts with the p6 domain of HIV-1 Gag (20, 29, 33, 44, 46, 48). We now report that ALIX also interacts with the NC-p1 region, which precedes the p6 domain in the context of the unprocessed Gag polyprotein. As a consequence, the specific incorporation of ALIX into HIV-1 virions is only moderately affected by removing its binding site in p6. While we initially identified ALIX based on its p6-dependent incorporation into HIV-1 VLP, this result is explained by the fact that the Gag constructs used in our previous study had NC-p1 replaced by the GCN4 leucine zipper (44). Indeed, the present study confirms that the incorporation of ALIX into VLP becomes completely dependent on the LYPx n L motif in p6 if NC-p1 is replaced by a heterologous dimerization domain. While the interaction with p6 is mediated by the V domain of ALIX (20, 29, 33), the association with NC was insensitive to FIG. 7. Effects of disrupting the two NC zinc fingers on particle production and on the ability of ALIX to rescue virus release. (A) Effects of the C 28,49 S mutation on particle production and Gag processing in the presence or absence of the PTAPP L domain. 293T cells were transfected with 1 g of PR-positive HIV-1 proviral DNAs, and virion production and the pattern of cell-associated Gag products were compared by Western blotting with anti-ca serum. (B) Effect of the C 28,49 S mutation on the ability of ALIX to rescue PTAPP HIV T cells were transfected with 1 g of PR-positive HIV-1 proviral DNAs and either empty pbj5 or a version expressing HA-ALIX (2 g).
8 1396 POPOV ET AL. J. VIROL. FIG. 8. Effects of removing the NC zinc fingers. (A) The deletion in NC, which removes all of the first zinc finger and part of the second, and the PTAPP deletion in p6 have comparable effects on particle production and Gag processing, and these effects are not additive. (B) The 15 39/ PTAPP double mutant is not rescued by ALIX. These experiments were performed as described in the legend for Fig. 7. a mutation in the ligand binding pocket of the V domain that disrupted binding to p6. Furthermore, the isolated Bro1 domain of ALIX was readily incorporated into viral particles in an NC-dependent manner, indicating that ALIX associates with NC through its N-terminal Bro1 domain. Thus, our results imply that ALIX can interact with HIV-1 Gag through both of its folded domains. The Bro1 domain of ALIX also interacts with ESCRT-III component CHMP4, and mutagenic analyses strongly suggest that this interaction is essential for the function of ALIX in virus budding (20, 46). For the interaction with NC to be functional, ALIX would thus have to be able to accommodate CHMP4 and NC simultaneously. In support of this notion, the overexpression of CHMP4B had no negative effect on the NC-dependent packaging of ALIX into HIV-1 virions (data not shown). The two CCHC boxes in NC are thought to mediate sequence-specific interactions with the genomic viral RNA by binding to exposed guanosine bases in the packaging signal (2, 15). In the present study, we found that the CCHC boxes also mediate the packaging of ALIX into virions. In particular, substituting zinc-coordinating residues in both CCHC boxes simultaneously nearly abolished the LYPx n L-independent incorporation of ALIX and the in vitro interaction between ALIX and NC-p1. The same modification has been shown to cause a dramatic defect in genomic RNA packaging (18) and to drastically reduce the incorporation of the RNA binding protein Staufen into HIV-1 particles (32). While these parallels point to a relationship between the packaging of ALIX and of the viral genome, there is no evidence that ALIX binds nucleic acid. Moreover, we observed that the in vitro interaction between ALIX and NC-p1 was insensitive to nuclease treatment. This finding indicates that nucleic acid is not required to maintain the interaction and that specific proteinprotein contacts are thus likely to be involved. The interaction of NC with RNA plays an important role in Gag multimerization and in virus assembly, apparently because RNA is needed as a scaffold to promote Gag-Gag interactions (9, 13, 35, 55). However, it is thought that this function of NC involves its nonspecific RNA binding activity, which is primarily mediated by basic residues and does not require intact zinc fingers (8, 13, 14, 17, 37, 42, 55). Nevertheless, we have observed that the C 28,49 S mutation, which disrupts both zinc fingers and interferes with ALIX binding, reduces particle production in HeLa cells about 10-fold (18). This observation contrasts with the finding that the C 28,49 S mutation had no effect on Gag-Gag interactions in an in vitro assay and only moderately affected the nonspecific RNA binding activity of NC (13). In the present study, the C 28,49 S mutation also reduced particle production in 293T cells, albeit to a lesser extent than what we had previously observed in HeLa cells (18). However, a deletion which affected both zinc fingers but left the rest of NC intact reduced particle production severely. Interestingly, these zinc finger mutations had little or no effect on the residual ability of the PTAPP mutant to produce viral particles, which suggests that the mutations in NC and in p6 affected the same step in virus assembly. Consistent with this possibility, the zinc finger mutations caused a similar Gagprocessing defect as the PTAPP mutant. These observations point to a role of HIV-1 NC in virus release, in addition to its well-documented function in virus assembly. Of note, a function of NC in virus release is also suggested by the late budding defect of certain NC mutants of the gammaretrovirus Moloney murine leukemia virus (34). Moreover, budding defects strikingly similar to those of L domain mutants have been observed upon deletion of either of the two CCHC motifs in the NC domain of an alpharetrovirus (28). We previously presented evidence indicating that in order to promote efficient virus release, the conserved PTAPP L domain core of HIV-1 needs to cooperate either with the ALIX binding site in p6 or with NC-p1 (45). Interestingly, the release defect of PTAPP mutants can be fully rescued by overexpressing ALIX, but only if the ALIX binding site in p6 remains intact (20, 46). In the present study, we observed that this effect of ALIX also depended on the zinc fingers in NC, and specifically on determinants in NC that mediate the interaction with ALIX. One interpretation of these results is that the interactions of ALIX with NC and with p6 are both involved in its function in virus budding. However, it also remains possible
9 VOL. 82, 2008 ALIX BINDS TO HIV-1 NC 1397 that the role of ALIX in virus release strictly depends on the ability of NC to nucleate assembly and that zinc finger mutations affect this ability in a manner that has not been readily apparent in in vitro Gag multimerization assays (13). In either case, the involvement of NC in the activity of ALIX supports the notion that NC has a specific role in virus release. ACKNOWLEDGMENTS We thank Suman R. Das for making the APOBEC3G-FLAG expression vector and Yoshiko Usami for making the expression vector for HA-ALIX Bro1. The following reagent was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: HIV-1 p24 monoclonal antibody (183-H12-5C) from Bruce Chesebro and Kathy Wehrly. This work was supported by National Institutes of Health grant AI REFERENCES 1. Accola, M. A., B. Strack, and H. G. Gottlinger Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 74: Amarasinghe, G. K., R. N. De Guzman, R. B. Turner, K. J. Chancellor, Z. R. Wu, and M. F. Summers NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the -RNA packaging signal. Implications for genome recognition. fsj. Mol. Biol. 301: Babst, M A protein s final ESCRT. Traffic 6: Babst, M., B. Wendland, E. J. Estepa, and S. D. Emr The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17: Berkowitz, R., J. Fisher, and S. P. Goff RNA packaging. Curr. Top. Microbiol. Immunol. 214: Bibollet-Ruche, F., E. Bailes, F. Gao, X. Pourrut, K. L. Barlow, J. P. Clewley, J. M. Mwenda, D. K. Langat, G. K. Chege, H. M. McClure, E. Mpoudi- Ngole, E. Delaporte, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn New simian immunodeficiency virus infecting De Brazza s monkeys (Cercopithecus neglectus): evidence for a Cercopithecus monkey virus clade. J. Virol. 78: Borsetti, A., A. Ohagen, and H. G. Gottlinger The C-terminal half of the human immunodeficiency virus type 1 Gag precursor is sufficient for efficient particle assembly. J. Virol. 72: Bowzard, J. B., R. P. Bennett, N. K. Krishna, S. M. Ernst, A. Rein, and J. W. Wills Importance of basic residues in the nucleocapsid sequence for retrovirus Gag assembly and complementation rescue. J. Virol. 72: Burniston, M. T., A. Cimarelli, J. Colgan, S. P. Curtis, and J. Luban Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsiddimer interface and the basic region of matrix protein. J. Virol. 73: Carlton, J. G., and J. Martin-Serrano Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science 316: Chen, C., O. Vincent, J. Jin, O. A. Weisz, and R. C. Montelaro Functions of early (AP-2) and late (AIP1/ALIX) endocytic proteins in equine infectious anemia virus budding. J. Biol. Chem. 280: Chesebro, B., K. Wehrly, J. Nishio, and S. Perryman Macrophagetropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-celltropic isolates: definition of critical amino acids involved in cell tropism. J. Virol. 66: Cimarelli, A., S. Sandin, S. Hoglund, and J. Luban Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J. Virol. 74: Dawson, L., and X. F. Yu The role of nucleocapsid of HIV-1 in virus assembly. Virology 251: De Guzman, R. N., Z. R. Wu, C. C. Stalling, L. Pappalardo, P. N. Borer, and M. F. Summers Structure of the HIV-1 nucleocapsid protein bound to the SL3 -RNA recognition element. Science 279: Demirov, D. G., J. M. Orenstein, and E. O. Freed The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. J. Virol. 76: Derdowski, A., L. Ding, and P. Spearman A novel fluorescence resonance energy transfer assay demonstrates that the human immunodeficiency virus type 1 Pr55 Gag I domain mediates Gag-Gag interactions. J. Virol. 78: Dorfman, T., J. Luban, S. P. Goff, W. A. Haseltine, and H. G. Gottlinger Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 67: Dorfman, T., F. Mammano, W. A. Haseltine, and H. G. Gottlinger Role of the matrix protein in the virion association of the human immunodeficiency virus type 1 envelope glycoprotein. J. Virol. 68: Fisher, R. D., H. Y. Chung, Q. Zhai, H. Robinson, W. I. Sundquist, and C. P. Hill Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell 128: Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107: Gottlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 88: Gruenberg, J., and H. Stenmark The biogenesis of multivesicular endosomes. Nat. Rev. Mol. Cell Biol. 5: Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed p6 Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69: Hurley, J. H., and S. D. Emr The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 35: Katzmann, D. J., M. Babst, and S. D. Emr Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106: Katzmann, D. J., G. Odorizzi, and S. D. Emr Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 3: Lee, E. G., and M. L. Linial Deletion of a Cys-His motif from the alpharetrovirus nucleocapsid domain reveals late domain mutant-like budding defects. Virology 347: Lee, S., A. Joshi, K. Nagashima, E. O. Freed, and J. H. Hurley Structural basis for viral late-domain binding to Alix. Nat. Struct. Mol. Biol. 14: Martin-Serrano, J., A. Yarovoy, D. Perez-Caballero, and P. D. Bieniasz Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc. Natl. Acad. Sci. USA 100: Martin-Serrano, J., T. Zang, and P. D. Bieniasz HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7: Mouland, A. J., J. Mercier, M. Luo, L. Bernier, L. DesGroseillers, and E. A. Cohen The double-stranded RNA-binding protein Staufen is incorporated in human immunodeficiency virus type 1: evidence for a role in genomic RNA encapsidation. J. Virol. 74: Munshi, U. M., J. Kim, K. Nagashima, J. H. Hurley, and E. O. Freed An Alix fragment potently inhibits HIV-1 budding: characterization of binding to retroviral YPXL late domains. J. Biol. Chem. 282: Muriaux, D., S. Costes, K. Nagashima, J. Mirro, E. Cho, S. Lockett, and A. Rein Role of murine leukemia virus nucleocapsid protein in virus assembly. J. Virol. 78: Muriaux, D., J. Mirro, D. Harvin, and A. Rein RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. USA 98: Odorizzi, G The multiple personalities of Alix. J. Cell Sci. 119: Ono, A., A. A. Waheed, A. Joshi, and E. O. Freed Association of human immunodeficiency virus type 1 Gag with membrane does not require highly basic sequences in the nucleocapsid: use of a novel Gag multimerization assay. J. Virol. 79: Ott, D. E., L. V. Coren, E. N. Chertova, T. D. Gagliardi, K. Nagashima, R. C. Sowder II, D. T. Poon, and R. J. Gorelick Elimination of protease activity restores efficient virion production to a human immunodeficiency virus type 1 nucleocapsid deletion mutant. J. Virol. 77: Parent, L. J., R. P. Bennett, R. C. Craven, T. D. Nelle, N. K. Krishna, J. B. Bowzard, C. B. Wilson, B. A. Puffer, R. C. Montelaro, and J. W. Wills Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 69: Perez-Caballero, D., T. Hatziioannou, J. Martin-Serrano, and P. D. Bieniasz Human immunodeficiency virus type 1 matrix inhibits and confers cooperativity on Gag precursor-membrane interactions. J. Virol. 78: Puffer, B. A., L. J. Parent, J. W. Wills, and R. C. Montelaro Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 71: Sandefur, S., R. M. Smith, V. Varthakavi, and P. Spearman Mapping and characterization of the N-terminal I domain of human immunodeficiency virus type 1 Pr55 Gag. J. Virol. 74: Shim, S., L. A. Kimpler, and P. I. Hanson Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic 8: Strack, B., A. Calistri, S. Craig, E. Popova, and H. G. Gottlinger
10 1398 POPOV ET AL. J. VIROL. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114: Strack, B., A. Calistri, and H. G. Gottlinger Late assembly domain function can exhibit context dependence and involves ubiquitin residues implicated in endocytosis. J. Virol. 76: Usami, Y., S. Popov, and H. G. Gottlinger Potent rescue of human immunodeficiency virus type 1 late domain mutants by ALIX/AIP1 depends on its CHMP4 binding site. J. Virol. 81: VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc. Natl. Acad. Sci. USA 98: von Schwedler, U. K., M. Stuchell, B. Muller, D. M. Ward, H. Y. Chung, E. Morita, H. E. Wang, T. Davis, G. P. He, D. M. Cimbora, A. Scott, H. G. Krausslich, J. Kaplan, S. G. Morham, and W. I. Sundquist The protein network of HIV budding. Cell 114: Wills, J. W., C. E. Cameron, C. B. Wilson, Y. Xiang, R. P. Bennett, and J. Leis An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 68: Yasuda, J., and E. Hunter A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J. Virol. 72: Yuan, B., S. Campbell, E. Bacharach, A. Rein, and S. P. Goff Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J. Virol. 74: Yuan, B., X. Li, and S. P. Goff Mutations altering the Moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 18: Zamborlini, A., Y. Usami, S. R. Radoshitzky, E. Popova, G. Palu, and H. Gottlinger Release of autoinhibition converts ESCRT-III components into potent inhibitors of HIV-1 budding. Proc. Natl. Acad. Sci. USA 103: Zennou, V., D. Perez-Caballero, H. Gottlinger, and P. D. Bieniasz APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J. Virol. 78: Zhang, Y., and E. Barklis Effects of nucleocapsid mutations on human immunodeficiency virus assembly and RNA encapsidation. J. Virol. 71: Downloaded from on October 8, 2018 by guest
Retroviral Gag proteins are necessary and sufficient for the
 Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function Dimiter G. Demirov*, Akira Ono*, Jan M. Orenstein, and Eric O. Freed* *Laboratory of Molecular
Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function Dimiter G. Demirov*, Akira Ono*, Jan M. Orenstein, and Eric O. Freed* *Laboratory of Molecular
Late Domain-Independent Rescue of a Release-Deficient Moloney Murine Leukemia Virus by the Ubiquitin Ligase Itch
 JOURNAL OF VIROLOGY, Jan. 2010, p. 704 715 Vol. 84, No. 2 0022-538X/10/$12.00 doi:10.1128/jvi.01319-09 Late Domain-Independent Rescue of a Release-Deficient Moloney Murine Leukemia Virus by the Ubiquitin
JOURNAL OF VIROLOGY, Jan. 2010, p. 704 715 Vol. 84, No. 2 0022-538X/10/$12.00 doi:10.1128/jvi.01319-09 Late Domain-Independent Rescue of a Release-Deficient Moloney Murine Leukemia Virus by the Ubiquitin
Deletion of a Cys His motif from the Alpharetrovirus nucleocapsid domain reveals late domain mutant-like budding defects
 Virology 347 (2006) 226 233 www.elsevier.com/locate/yviro Deletion of a Cys His motif from the Alpharetrovirus nucleocapsid domain reveals late domain mutant-like budding defects Eun-Gyung Lee, Maxine
Virology 347 (2006) 226 233 www.elsevier.com/locate/yviro Deletion of a Cys His motif from the Alpharetrovirus nucleocapsid domain reveals late domain mutant-like budding defects Eun-Gyung Lee, Maxine
Role of ESCRT-I in Retroviral Budding
 JOURNAL OF VIROLOGY, Apr. 2003, p. 4794 4804 Vol. 77, No. 8 0022-538X/03/$08.00 0 DOI: 10.1128/JVI.77.8.4794 4804.2003 Copyright 2003, American Society for Microbiology. All Rights Reserved. Role of ESCRT-I
JOURNAL OF VIROLOGY, Apr. 2003, p. 4794 4804 Vol. 77, No. 8 0022-538X/03/$08.00 0 DOI: 10.1128/JVI.77.8.4794 4804.2003 Copyright 2003, American Society for Microbiology. All Rights Reserved. Role of ESCRT-I
Journal of Biology. BioMed Central
 Journal of Biology BioMed Central Open Access Research article Endophilins interact with Moloney murine leukemia virus Gag and modulate virion production Margaret Q Wang*, Wankee Kim, Guangxia Gao, Ted
Journal of Biology BioMed Central Open Access Research article Endophilins interact with Moloney murine leukemia virus Gag and modulate virion production Margaret Q Wang*, Wankee Kim, Guangxia Gao, Ted
AND JEREMY LUBAN 1,2 *
 JOURNAL OF VIROLOGY, Oct. 1999, p. 8527 8540 Vol. 73, No. 10 0022-538X/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. Human Immunodeficiency Virus Type 1 Gag Polyprotein
JOURNAL OF VIROLOGY, Oct. 1999, p. 8527 8540 Vol. 73, No. 10 0022-538X/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. Human Immunodeficiency Virus Type 1 Gag Polyprotein
JCB. After Hrs with HIV. The Journal of Cell Biology. Comment. Ali Amara 1 and Dan R. Littman 1,2
 JCB Comment After Hrs with HIV Ali Amara 1 and Dan R. Littman 1,2 1 Molecular Pathogenesis Program and 2 Howard Hughes Medical Institute, Skirball Institute of Biomolecular Medicine, New York University
JCB Comment After Hrs with HIV Ali Amara 1 and Dan R. Littman 1,2 1 Molecular Pathogenesis Program and 2 Howard Hughes Medical Institute, Skirball Institute of Biomolecular Medicine, New York University
Role of the C terminus Gag protein in human immunodefieieney virus type 1 virion assembly and maturation
 Journal of General Virology (1995), 76, 3171-3179. Printed in Great Britain 3171 Role of the C terminus Gag protein in human immunodefieieney virus type 1 virion assembly and maturation X.-F. Yu, ~ Z.
Journal of General Virology (1995), 76, 3171-3179. Printed in Great Britain 3171 Role of the C terminus Gag protein in human immunodefieieney virus type 1 virion assembly and maturation X.-F. Yu, ~ Z.
Received 15 August 2002/Accepted 1 November 2002
 JOURNAL OF VIROLOGY, Feb. 2003, p. 1812 1819 Vol. 77, No. 3 0022-538X/03/$08.00 0 DOI: 10.1128/JVI.77.3.1812 1819.2003 Copyright 2003, American Society for Microbiology. All Rights Reserved. Overlapping
JOURNAL OF VIROLOGY, Feb. 2003, p. 1812 1819 Vol. 77, No. 3 0022-538X/03/$08.00 0 DOI: 10.1128/JVI.77.3.1812 1819.2003 Copyright 2003, American Society for Microbiology. All Rights Reserved. Overlapping
Mutations of the Human Immunodeficiency Virus Type 1 p6 Gag Domain Result in Reduced Retention of Pol Proteins during Virus Assembly
 JOURNAL OF VIROLOGY, Apr. 1998, p. 3412 3417 Vol. 72, No. 4 0022-538X/98/$04.00 0 Copyright 1998, American Society for Microbiology Mutations of the Human Immunodeficiency Virus Type 1 p6 Gag Domain Result
JOURNAL OF VIROLOGY, Apr. 1998, p. 3412 3417 Vol. 72, No. 4 0022-538X/98/$04.00 0 Copyright 1998, American Society for Microbiology Mutations of the Human Immunodeficiency Virus Type 1 p6 Gag Domain Result
Late budding domains and host proteins in enveloped virus release
 Virology 344 (2006) 55 63 www.elsevier.com/locate/yviro Late budding domains and host proteins in enveloped virus release Paul D. Bieniasz Aaron Diamond AIDS Research Center and Laboratory of Retrovirology,
Virology 344 (2006) 55 63 www.elsevier.com/locate/yviro Late budding domains and host proteins in enveloped virus release Paul D. Bieniasz Aaron Diamond AIDS Research Center and Laboratory of Retrovirology,
Identification of Domains in Gag Important for Prototypic Foamy Virus Egress
 JOURNAL OF VIROLOGY, May 2005, p. 6392 6399 Vol. 79, No. 10 0022-538X/05/$08.00 0 doi:10.1128/jvi.79.10.6392 6399.2005 Copyright 2005, American Society for Microbiology. All Rights Reserved. Identification
JOURNAL OF VIROLOGY, May 2005, p. 6392 6399 Vol. 79, No. 10 0022-538X/05/$08.00 0 doi:10.1128/jvi.79.10.6392 6399.2005 Copyright 2005, American Society for Microbiology. All Rights Reserved. Identification
Cellular Factors Required for Lassa Virus Budding
 JOURNAL OF VIROLOGY, Apr. 2006, p. 4191 4195 Vol. 80, No. 8 0022-538X/06/$08.00 0 doi:10.1128/jvi.80.8.4191 4195.2006 Copyright 2006, American Society for Microbiology. All Rights Reserved. Cellular Factors
JOURNAL OF VIROLOGY, Apr. 2006, p. 4191 4195 Vol. 80, No. 8 0022-538X/06/$08.00 0 doi:10.1128/jvi.80.8.4191 4195.2006 Copyright 2006, American Society for Microbiology. All Rights Reserved. Cellular Factors
Differential Effects of Actin Cytoskeleton Dynamics on Equine Infectious Anemia Virus Particle Production
 JOURNAL OF VIROLOGY, Jan. 2004, p. 882 891 Vol. 78, No. 2 0022-538X/04/$08.00 0 DOI: 10.1128/JVI.78.2.882 891.2004 Copyright 2004, American Society for Microbiology. All Rights Reserved. Differential Effects
JOURNAL OF VIROLOGY, Jan. 2004, p. 882 891 Vol. 78, No. 2 0022-538X/04/$08.00 0 DOI: 10.1128/JVI.78.2.882 891.2004 Copyright 2004, American Society for Microbiology. All Rights Reserved. Differential Effects
Effect of Mutations in Gag on Assembly of Immature Human Immunodeficiency Virus Type 1 Capsids in a Cell-Free System
 Virology 279, 257 270 (2001) doi:10.1006/viro.2000.0706, available online at http://www.idealibrary.com on Effect of Mutations in Gag on Assembly of Immature Human Immunodeficiency Virus Type 1 Capsids
Virology 279, 257 270 (2001) doi:10.1006/viro.2000.0706, available online at http://www.idealibrary.com on Effect of Mutations in Gag on Assembly of Immature Human Immunodeficiency Virus Type 1 Capsids
HIV-1 genome organization
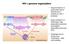 HIV-1 genome organization Gag encodes for a polyprotein that is cleaved into 4 proteins by HIV-1 protease. Gag is present in 2 transcripts: Gag polyprotein precursor and GagPol polyprotein precursor (5%).
HIV-1 genome organization Gag encodes for a polyprotein that is cleaved into 4 proteins by HIV-1 protease. Gag is present in 2 transcripts: Gag polyprotein precursor and GagPol polyprotein precursor (5%).
Charged Amino Acid Residues of Human Immunodeficiency Virus Type 1 Nucleocapsid p7 Protein Involved in RNA Packaging and Infectivity
 JOURNAL OF VIROLOGY, Oct. 1996, p. 6607 6616 Vol. 70, No. 10 0022-538X/96/$04.00 0 Copyright 1996, American Society for Microbiology Charged Amino Acid Residues of Human Immunodeficiency Virus Type 1 Nucleocapsid
JOURNAL OF VIROLOGY, Oct. 1996, p. 6607 6616 Vol. 70, No. 10 0022-538X/96/$04.00 0 Copyright 1996, American Society for Microbiology Charged Amino Acid Residues of Human Immunodeficiency Virus Type 1 Nucleocapsid
Recombinant Protein Expression Retroviral system
 Recombinant Protein Expression Retroviral system Viruses Contains genome DNA or RNA Genome encased in a protein coat or capsid. Some viruses have membrane covering protein coat enveloped virus Ø Essential
Recombinant Protein Expression Retroviral system Viruses Contains genome DNA or RNA Genome encased in a protein coat or capsid. Some viruses have membrane covering protein coat enveloped virus Ø Essential
Expression of Human Immunodeficiency Virus Type 1 Gag Modulates Ligand-Induced Downregulation of EGF Receptor
 JOURNAL OF VIROLOGY, Nov. 2004, p. 12386 12394 Vol. 78, No. 22 0022-538X/04/$08.00 0 DOI: 10.1128/JVI.78.22.12386 12394.2004 Copyright 2004, American Society for Microbiology. All Rights Reserved. Expression
JOURNAL OF VIROLOGY, Nov. 2004, p. 12386 12394 Vol. 78, No. 22 0022-538X/04/$08.00 0 DOI: 10.1128/JVI.78.22.12386 12394.2004 Copyright 2004, American Society for Microbiology. All Rights Reserved. Expression
Cellular Motor Protein KIF-4 Associates with Retroviral Gag
 JOURNAL OF VIROLOGY, Dec. 1999, p. 10508 10513 Vol. 73, No. 12 0022-538X/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. Cellular Motor Protein KIF-4 Associates with
JOURNAL OF VIROLOGY, Dec. 1999, p. 10508 10513 Vol. 73, No. 12 0022-538X/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. Cellular Motor Protein KIF-4 Associates with
Protein Trafficking in the Secretory and Endocytic Pathways
 Protein Trafficking in the Secretory and Endocytic Pathways The compartmentalization of eukaryotic cells has considerable functional advantages for the cell, but requires elaborate mechanisms to ensure
Protein Trafficking in the Secretory and Endocytic Pathways The compartmentalization of eukaryotic cells has considerable functional advantages for the cell, but requires elaborate mechanisms to ensure
Identification of the HIV-1 NC binding interface in Alix Bro1 reveals a role for RNA
 JVI Accepts, published online ahead of print on 15 August 2012 J. Virol. doi:10.1128/jvi.0126012 Copyright 2012, American Society for Microbiology. All Rights Reserved. 1 2 3 4 5 6 7 8 9 10 11 12 13 14
JVI Accepts, published online ahead of print on 15 August 2012 J. Virol. doi:10.1128/jvi.0126012 Copyright 2012, American Society for Microbiology. All Rights Reserved. 1 2 3 4 5 6 7 8 9 10 11 12 13 14
Fayth K. Yoshimura, Ph.D. September 7, of 7 HIV - BASIC PROPERTIES
 1 of 7 I. Viral Origin. A. Retrovirus - animal lentiviruses. HIV - BASIC PROPERTIES 1. HIV is a member of the Retrovirus family and more specifically it is a member of the Lentivirus genus of this family.
1 of 7 I. Viral Origin. A. Retrovirus - animal lentiviruses. HIV - BASIC PROPERTIES 1. HIV is a member of the Retrovirus family and more specifically it is a member of the Lentivirus genus of this family.
Materials and Methods , The two-hybrid principle.
 The enzymatic activity of an unknown protein which cleaves the phosphodiester bond between the tyrosine residue of a viral protein and the 5 terminus of the picornavirus RNA Introduction Every day there
The enzymatic activity of an unknown protein which cleaves the phosphodiester bond between the tyrosine residue of a viral protein and the 5 terminus of the picornavirus RNA Introduction Every day there
Packaging and Abnormal Particle Morphology
 JOURNAL OF VIROLOGY, OCt. 1990, p. 5230-5234 0022-538X/90/105230-05$02.00/0 Copyright 1990, American Society for Microbiology Vol. 64, No. 10 A Mutant of Human Immunodeficiency Virus with Reduced RNA Packaging
JOURNAL OF VIROLOGY, OCt. 1990, p. 5230-5234 0022-538X/90/105230-05$02.00/0 Copyright 1990, American Society for Microbiology Vol. 64, No. 10 A Mutant of Human Immunodeficiency Virus with Reduced RNA Packaging
Ubiquitination of Human Immunodeficiency Virus Type 1 Gag Is Highly Dependent on Gag Membrane Association
 JOURNAL OF VIROLOGY, Sept. 2007, p. 9193 9201 Vol. 81, No. 17 0022-538X/07/$08.00 0 doi:10.1128/jvi.00044-07 Copyright 2007, American Society for Microbiology. All Rights Reserved. Ubiquitination of Human
JOURNAL OF VIROLOGY, Sept. 2007, p. 9193 9201 Vol. 81, No. 17 0022-538X/07/$08.00 0 doi:10.1128/jvi.00044-07 Copyright 2007, American Society for Microbiology. All Rights Reserved. Ubiquitination of Human
The Interferon-Induced Gene ISG15 Blocks Retrovirus Release from Cells Late in the Budding Process
 JOURNAL OF VIROLOGY, May 2010, p. 4725 4736 Vol. 84, No. 9 0022-538X/10/$12.00 doi:10.1128/jvi.02478-09 Copyright 2010, American Society for Microbiology. All Rights Reserved. The Interferon-Induced Gene
JOURNAL OF VIROLOGY, May 2010, p. 4725 4736 Vol. 84, No. 9 0022-538X/10/$12.00 doi:10.1128/jvi.02478-09 Copyright 2010, American Society for Microbiology. All Rights Reserved. The Interferon-Induced Gene
There are approximately 30,000 proteasomes in a typical human cell Each proteasome is approximately 700 kda in size The proteasome is made up of 3
 Proteasomes Proteasomes Proteasomes are responsible for degrading proteins that have been damaged, assembled improperly, or that are of no profitable use to the cell. The unwanted protein is literally
Proteasomes Proteasomes Proteasomes are responsible for degrading proteins that have been damaged, assembled improperly, or that are of no profitable use to the cell. The unwanted protein is literally
Specific Incorporation of Heat Shock Protein 70 Family Members into Primate Lentiviral Virions
 JOURNAL OF VIROLOGY, May 2002, p. 4666 4670 Vol. 76, No. 9 0022-538X/02/$04.00 0 DOI: 10.1128/JVI.76.9.4666 4670.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Specific Incorporation
JOURNAL OF VIROLOGY, May 2002, p. 4666 4670 Vol. 76, No. 9 0022-538X/02/$04.00 0 DOI: 10.1128/JVI.76.9.4666 4670.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Specific Incorporation
Supplementary information. MARCH8 inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins
 Supplementary information inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins Takuya Tada, Yanzhao Zhang, Takayoshi Koyama, Minoru Tobiume, Yasuko Tsunetsugu-Yokota, Shoji
Supplementary information inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins Takuya Tada, Yanzhao Zhang, Takayoshi Koyama, Minoru Tobiume, Yasuko Tsunetsugu-Yokota, Shoji
Retroviruses. ---The name retrovirus comes from the enzyme, reverse transcriptase.
 Retroviruses ---The name retrovirus comes from the enzyme, reverse transcriptase. ---Reverse transcriptase (RT) converts the RNA genome present in the virus particle into DNA. ---RT discovered in 1970.
Retroviruses ---The name retrovirus comes from the enzyme, reverse transcriptase. ---Reverse transcriptase (RT) converts the RNA genome present in the virus particle into DNA. ---RT discovered in 1970.
In Vivo Interference of Rous Sarcoma Virus Budding by cis Expression of a WW Domain
 JOURNAL OF VIROLOGY, Mar. 2002, p. 2789 2795 Vol. 76, No. 6 0022-538X/02/$04.00 0 DOI: 10.1128/JVI.76.6.2789 2795.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. In Vivo Interference
JOURNAL OF VIROLOGY, Mar. 2002, p. 2789 2795 Vol. 76, No. 6 0022-538X/02/$04.00 0 DOI: 10.1128/JVI.76.6.2789 2795.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. In Vivo Interference
Virus Research 150 (2010) Contents lists available at ScienceDirect. Virus Research. journal homepage:
 Virus Research 150 (2010) 153 157 Contents lists available at ScienceDirect Virus Research journal homepage: www.elsevier.com/locate/virusres Short communication In vitro assembly of the feline immunodeficiency
Virus Research 150 (2010) 153 157 Contents lists available at ScienceDirect Virus Research journal homepage: www.elsevier.com/locate/virusres Short communication In vitro assembly of the feline immunodeficiency
REGULATION OF TSG101 EXPRESSION BY THE STEADINESS BOX: A ROLE OF TSG101-ASSOCIATED LIGASE (TAL)
 REGULATION OF TSG101 EXPRESSION BY THE STEADINESS BOX: A ROLE OF TSG101-ASSOCIATED LIGASE (TAL) Bethan McDonald and Juan Martin-Serrano Department of Infectious Diseases; King's College London School of
REGULATION OF TSG101 EXPRESSION BY THE STEADINESS BOX: A ROLE OF TSG101-ASSOCIATED LIGASE (TAL) Bethan McDonald and Juan Martin-Serrano Department of Infectious Diseases; King's College London School of
FUNCTIONAL REDUNDANCY IN HIV-1 VIRUS PARTICLE ASSEMBLY. Running Title: Redundancy in HIV-1 Assembly. Frederick, MD 21702
 JVI Accepts, published online ahead of print on 19 September 2012 J. Virol. doi:10.1128/jvi.06287-11 Copyright 2012, American Society for Microbiology. All Rights Reserved. 1 2 FUNCTIONAL REDUNDANCY IN
JVI Accepts, published online ahead of print on 19 September 2012 J. Virol. doi:10.1128/jvi.06287-11 Copyright 2012, American Society for Microbiology. All Rights Reserved. 1 2 FUNCTIONAL REDUNDANCY IN
Introduction retroposon
 17.1 - Introduction A retrovirus is an RNA virus able to convert its sequence into DNA by reverse transcription A retroposon (retrotransposon) is a transposon that mobilizes via an RNA form; the DNA element
17.1 - Introduction A retrovirus is an RNA virus able to convert its sequence into DNA by reverse transcription A retroposon (retrotransposon) is a transposon that mobilizes via an RNA form; the DNA element
Supplementary Information. Supplementary Figure 1
 Supplementary Information Supplementary Figure 1 1 Supplementary Figure 1. Functional assay of the hcas9-2a-mcherry construct (a) Gene correction of a mutant EGFP reporter cell line mediated by hcas9 or
Supplementary Information Supplementary Figure 1 1 Supplementary Figure 1. Functional assay of the hcas9-2a-mcherry construct (a) Gene correction of a mutant EGFP reporter cell line mediated by hcas9 or
Basic Residues of the Retroviral Nucleocapsid Play Different Roles in Gag-Gag and Gag- RNA Interactions
 JOURNAL OF VIROLOGY, Aug. 2004, p. 8486 8495 Vol. 78, No. 16 0022-538X/04/$08.00 0 DOI: 10.1128/JVI.78.16.8486 8495.2004 Copyright 2004, American Society for Microbiology. All Rights Reserved. Basic Residues
JOURNAL OF VIROLOGY, Aug. 2004, p. 8486 8495 Vol. 78, No. 16 0022-538X/04/$08.00 0 DOI: 10.1128/JVI.78.16.8486 8495.2004 Copyright 2004, American Society for Microbiology. All Rights Reserved. Basic Residues
Fine Mapping of a cis-acting Sequence Element in Yellow Fever Virus RNA That Is Required for RNA Replication and Cyclization
 JOURNAL OF VIROLOGY, Feb. 2003, p. 2265 2270 Vol. 77, No. 3 0022-538X/03/$08.00 0 DOI: 10.1128/JVI.77.3.2265 2270.2003 Copyright 2003, American Society for Microbiology. All Rights Reserved. Fine Mapping
JOURNAL OF VIROLOGY, Feb. 2003, p. 2265 2270 Vol. 77, No. 3 0022-538X/03/$08.00 0 DOI: 10.1128/JVI.77.3.2265 2270.2003 Copyright 2003, American Society for Microbiology. All Rights Reserved. Fine Mapping
HIV-1 Virus-like Particle Budding Assay Nathan H Vande Burgt, Luis J Cocka * and Paul Bates
 HIV-1 Virus-like Particle Budding Assay Nathan H Vande Burgt, Luis J Cocka * and Paul Bates Department of Microbiology, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, USA
HIV-1 Virus-like Particle Budding Assay Nathan H Vande Burgt, Luis J Cocka * and Paul Bates Department of Microbiology, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, USA
Supplementary Material
 Supplementary Material Nuclear import of purified HIV-1 Integrase. Integrase remains associated to the RTC throughout the infection process until provirus integration occurs and is therefore one likely
Supplementary Material Nuclear import of purified HIV-1 Integrase. Integrase remains associated to the RTC throughout the infection process until provirus integration occurs and is therefore one likely
Fayth K. Yoshimura, Ph.D. September 7, of 7 RETROVIRUSES. 2. HTLV-II causes hairy T-cell leukemia
 1 of 7 I. Diseases Caused by Retroviruses RETROVIRUSES A. Human retroviruses that cause cancers 1. HTLV-I causes adult T-cell leukemia and tropical spastic paraparesis 2. HTLV-II causes hairy T-cell leukemia
1 of 7 I. Diseases Caused by Retroviruses RETROVIRUSES A. Human retroviruses that cause cancers 1. HTLV-I causes adult T-cell leukemia and tropical spastic paraparesis 2. HTLV-II causes hairy T-cell leukemia
Benchmarks Attenuated protein expression vectors for use in sirna rescue experiments
 Benchmarks Attenuated protein expression vectors for use in sirna rescue experiments Eiji Morita *, Jun Arii *, Devin Christensen, Jörg Votteler, and Wesley I. Sundquist Department of Biochemistry, University
Benchmarks Attenuated protein expression vectors for use in sirna rescue experiments Eiji Morita *, Jun Arii *, Devin Christensen, Jörg Votteler, and Wesley I. Sundquist Department of Biochemistry, University
Identification of late assembly domains of the human endogenous retrovirus-k(hml-2)
 Chudak et al. Retrovirology 2013, 10:140 RESEARCH Identification of late assembly domains of the human endogenous retrovirus-k(hml-2) Open Access Claudia Chudak 1, Nadine Beimforde 1, Maja George 2, Anja
Chudak et al. Retrovirology 2013, 10:140 RESEARCH Identification of late assembly domains of the human endogenous retrovirus-k(hml-2) Open Access Claudia Chudak 1, Nadine Beimforde 1, Maja George 2, Anja
Polyomaviridae. Spring
 Polyomaviridae Spring 2002 331 Antibody Prevalence for BK & JC Viruses Spring 2002 332 Polyoma Viruses General characteristics Papovaviridae: PA - papilloma; PO - polyoma; VA - vacuolating agent a. 45nm
Polyomaviridae Spring 2002 331 Antibody Prevalence for BK & JC Viruses Spring 2002 332 Polyoma Viruses General characteristics Papovaviridae: PA - papilloma; PO - polyoma; VA - vacuolating agent a. 45nm
Lecture 2: Virology. I. Background
 Lecture 2: Virology I. Background A. Properties 1. Simple biological systems a. Aggregates of nucleic acids and protein 2. Non-living a. Cannot reproduce or carry out metabolic activities outside of a
Lecture 2: Virology I. Background A. Properties 1. Simple biological systems a. Aggregates of nucleic acids and protein 2. Non-living a. Cannot reproduce or carry out metabolic activities outside of a
October 26, Lecture Readings. Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell
 October 26, 2006 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
October 26, 2006 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
Overview of virus life cycle
 Overview of virus life cycle cell recognition and internalization release from cells progeny virus assembly membrane breaching nucleus capsid disassembly and genome release replication and translation
Overview of virus life cycle cell recognition and internalization release from cells progeny virus assembly membrane breaching nucleus capsid disassembly and genome release replication and translation
Mechanisms of enveloped RNA virus budding
 569 44 Brown, S.A. et al. (1998) Holoprosencephaly due to mutations in ZIC2, a homologue of Drosophila odd-paired. Nat. Genet. 20, 180 183 45 Aruga, J. et al. (2002) Zic2 controls cerebellar development
569 44 Brown, S.A. et al. (1998) Holoprosencephaly due to mutations in ZIC2, a homologue of Drosophila odd-paired. Nat. Genet. 20, 180 183 45 Aruga, J. et al. (2002) Zic2 controls cerebellar development
JOURNAL OF VIROLOGY, July 1999, p Vol. 73, No. 7. Copyright 1999, American Society for Microbiology. All Rights Reserved.
 JOURNAL OF VIROLOGY, July 1999, p. 5654 5662 Vol. 73, No. 7 0022-538X/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. Formation of Virus Assembly Intermediate Complexes
JOURNAL OF VIROLOGY, July 1999, p. 5654 5662 Vol. 73, No. 7 0022-538X/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. Formation of Virus Assembly Intermediate Complexes
Ebola Virus VP40 Late Domains Are Not Essential for Viral Replication in Cell Culture
 JOURNAL OF VIROLOGY, Aug. 2005, p. 10300 10307 Vol. 79, No. 16 0022-538X/05/$08.00 0 doi:10.1128/jvi.79.16.10300 10307.2005 Copyright 2005, American Society for Microbiology. All Rights Reserved. Ebola
JOURNAL OF VIROLOGY, Aug. 2005, p. 10300 10307 Vol. 79, No. 16 0022-538X/05/$08.00 0 doi:10.1128/jvi.79.16.10300 10307.2005 Copyright 2005, American Society for Microbiology. All Rights Reserved. Ebola
Structural and functional studies of ALIX interactions with YPX n L late domains of HIV-1 and EIAV
 Structural and functional studies of ALIX interactions with YPX n L late domains of HIV-1 and EIAV Qianting Zhai 1,3, Robert D Fisher 1,3, Hyo-Young Chung 1, David G Myszka 2, Wesley I Sundquist 1 & Christopher
Structural and functional studies of ALIX interactions with YPX n L late domains of HIV-1 and EIAV Qianting Zhai 1,3, Robert D Fisher 1,3, Hyo-Young Chung 1, David G Myszka 2, Wesley I Sundquist 1 & Christopher
Section 6. Junaid Malek, M.D.
 Section 6 Junaid Malek, M.D. The Golgi and gp160 gp160 transported from ER to the Golgi in coated vesicles These coated vesicles fuse to the cis portion of the Golgi and deposit their cargo in the cisternae
Section 6 Junaid Malek, M.D. The Golgi and gp160 gp160 transported from ER to the Golgi in coated vesicles These coated vesicles fuse to the cis portion of the Golgi and deposit their cargo in the cisternae
Lecture Readings. Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell
 October 26, 2006 1 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
October 26, 2006 1 Vesicular Trafficking, Secretory Pathway, HIV Assembly and Exit from Cell 1. Secretory pathway a. Formation of coated vesicles b. SNAREs and vesicle targeting 2. Membrane fusion a. SNAREs
CHEN LIANG, 1 LIWEI RONG, 1 YUDONG QUAN, 1 MICHAEL LAUGHREA, 1 LAWRENCE KLEIMAN, 1,2
 JOURNAL OF VIROLOGY, Aug. 1999, p. 7014 7020 Vol. 73, No. 8 0022-538X/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. Mutations within Four Distinct Gag Proteins Are
JOURNAL OF VIROLOGY, Aug. 1999, p. 7014 7020 Vol. 73, No. 8 0022-538X/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. Mutations within Four Distinct Gag Proteins Are
Structural vs. nonstructural proteins
 Why would you want to study proteins associated with viruses or virus infection? Receptors Mechanism of uncoating How is gene expression carried out, exclusively by viral enzymes? Gene expression phases?
Why would you want to study proteins associated with viruses or virus infection? Receptors Mechanism of uncoating How is gene expression carried out, exclusively by viral enzymes? Gene expression phases?
Wrapping up the bad news HIV assembly and release
 Meng and Lever Retrovirology 2013, 10:5 REVIEW Open Access Wrapping up the bad news HIV assembly and release Bo Meng and Andrew ML Lever * Abstract The late Nobel Laureate Sir Peter Medawar once memorably
Meng and Lever Retrovirology 2013, 10:5 REVIEW Open Access Wrapping up the bad news HIV assembly and release Bo Meng and Andrew ML Lever * Abstract The late Nobel Laureate Sir Peter Medawar once memorably
ALG-2 activates the MVB sorting function of ALIX through relieving its intramolecular interaction
 OPEN Citation: Cell Discovery (2015) 1, 15018; doi:10.1038/celldisc.2015.18 2015 SIBS, CAS All rights reserved 2056-5968/15 www.nature.com/celldisc ALG-2 activates the MVB sorting function of ALIX through
OPEN Citation: Cell Discovery (2015) 1, 15018; doi:10.1038/celldisc.2015.18 2015 SIBS, CAS All rights reserved 2056-5968/15 www.nature.com/celldisc ALG-2 activates the MVB sorting function of ALIX through
Julianne Edwards. Retroviruses. Spring 2010
 Retroviruses Spring 2010 A retrovirus can simply be referred to as an infectious particle which replicates backwards even though there are many different types of retroviruses. More specifically, a retrovirus
Retroviruses Spring 2010 A retrovirus can simply be referred to as an infectious particle which replicates backwards even though there are many different types of retroviruses. More specifically, a retrovirus
Sequences in the 5 and 3 R Elements of Human Immunodeficiency Virus Type 1 Critical for Efficient Reverse Transcription
 JOURNAL OF VIROLOGY, Sept. 2000, p. 8324 8334 Vol. 74, No. 18 0022-538X/00/$04.00 0 Copyright 2000, American Society for Microbiology. All Rights Reserved. Sequences in the 5 and 3 R Elements of Human
JOURNAL OF VIROLOGY, Sept. 2000, p. 8324 8334 Vol. 74, No. 18 0022-538X/00/$04.00 0 Copyright 2000, American Society for Microbiology. All Rights Reserved. Sequences in the 5 and 3 R Elements of Human
Homework Hanson section MCB Course, Fall 2014
 Homework Hanson section MCB Course, Fall 2014 (1) Antitrypsin, which inhibits certain proteases, is normally secreted into the bloodstream by liver cells. Antitrypsin is absent from the bloodstream of
Homework Hanson section MCB Course, Fall 2014 (1) Antitrypsin, which inhibits certain proteases, is normally secreted into the bloodstream by liver cells. Antitrypsin is absent from the bloodstream of
Gag. Gag-Gag. TSG101 Nedd4. Two-Hybrid HIV Gag. Two-Hybrid. Two-Hybrid
 CytoTrap HIV Gag AnnexinA2 AUP1 AUP1 HIV AUP1 N Gag CytoTrap Gag-Gag TSG101 Nedd4 Two-Hybrid Two-Hybrid HIVGag Gag Two-Hybrid HIV Gag Gag HIV Two-Hybrid CytoTrap cdna Gag Gag CytoTrap Gag CytoTrap Gag-Gag
CytoTrap HIV Gag AnnexinA2 AUP1 AUP1 HIV AUP1 N Gag CytoTrap Gag-Gag TSG101 Nedd4 Two-Hybrid Two-Hybrid HIVGag Gag Two-Hybrid HIV Gag Gag HIV Two-Hybrid CytoTrap cdna Gag Gag CytoTrap Gag CytoTrap Gag-Gag
Gammaretroviral pol sequences act in cis to direct polysome loading and NXF1/NXT-dependent protein production by gag-encoded RNA
 University of Massachusetts Medical School escholarship@umms Open Access Articles Open Access Publications by UMMS Authors 9-12-2014 Gammaretroviral pol sequences act in cis to direct polysome loading
University of Massachusetts Medical School escholarship@umms Open Access Articles Open Access Publications by UMMS Authors 9-12-2014 Gammaretroviral pol sequences act in cis to direct polysome loading
The Conserved Carboxy Terminus of the Capsid Domain of Human Immunodeficiency Virus Type 1 Gag Protein Is Important for Virion Assembly and Release
 JOURNAL OF VIROLOGY, Sept. 2004, p. 9675 9688 Vol. 78, No. 18 0022-538X/04/$08.00 0 DOI: 10.1128/JVI.78.18.9675 9688.2004 Copyright 2004, American Society for Microbiology. All Rights Reserved. The Conserved
JOURNAL OF VIROLOGY, Sept. 2004, p. 9675 9688 Vol. 78, No. 18 0022-538X/04/$08.00 0 DOI: 10.1128/JVI.78.18.9675 9688.2004 Copyright 2004, American Society for Microbiology. All Rights Reserved. The Conserved
NOTES. Activation of the Retroviral Budding Factor ALIX
 JOURNAL OF VIROLOGY, Sept. 2011, p. 9222 9226 Vol. 85, No. 17 0022-538X/11/$12.00 doi:10.1128/jvi.02653-10 Copyright 2011, American Society for Microbiology. All Rights Reserved. NOTES Activation of the
JOURNAL OF VIROLOGY, Sept. 2011, p. 9222 9226 Vol. 85, No. 17 0022-538X/11/$12.00 doi:10.1128/jvi.02653-10 Copyright 2011, American Society for Microbiology. All Rights Reserved. NOTES Activation of the
Dr. Ahmed K. Ali Attachment and entry of viruses into cells
 Lec. 6 Dr. Ahmed K. Ali Attachment and entry of viruses into cells The aim of a virus is to replicate itself, and in order to achieve this aim it needs to enter a host cell, make copies of itself and
Lec. 6 Dr. Ahmed K. Ali Attachment and entry of viruses into cells The aim of a virus is to replicate itself, and in order to achieve this aim it needs to enter a host cell, make copies of itself and
VIROLOGY. Engineering Viral Genomes: Retrovirus Vectors
 VIROLOGY Engineering Viral Genomes: Retrovirus Vectors Viral vectors Retrovirus replicative cycle Most mammalian retroviruses use trna PRO, trna Lys3, trna Lys1,2 The partially unfolded trna is annealed
VIROLOGY Engineering Viral Genomes: Retrovirus Vectors Viral vectors Retrovirus replicative cycle Most mammalian retroviruses use trna PRO, trna Lys3, trna Lys1,2 The partially unfolded trna is annealed
The Clathrin Adaptor Complex AP-1 Binds HIV-1 and MLV Gag and Facilitates Their Budding D
 Vol. 18, 3193 3203, August 2007 The Clathrin Adaptor Complex AP-1 Binds HIV-1 and MLV Gag and Facilitates Their Budding D Grégory Camus,* Carolina Segura-Morales, Dorothee Molle, Sandra Lopez-Vergès,*
Vol. 18, 3193 3203, August 2007 The Clathrin Adaptor Complex AP-1 Binds HIV-1 and MLV Gag and Facilitates Their Budding D Grégory Camus,* Carolina Segura-Morales, Dorothee Molle, Sandra Lopez-Vergès,*
Translation. Host Cell Shutoff 1) Initiation of eukaryotic translation involves many initiation factors
 Translation Questions? 1) How does poliovirus shutoff eukaryotic translation? 2) If eukaryotic messages are not translated how can poliovirus get its message translated? Host Cell Shutoff 1) Initiation
Translation Questions? 1) How does poliovirus shutoff eukaryotic translation? 2) If eukaryotic messages are not translated how can poliovirus get its message translated? Host Cell Shutoff 1) Initiation
Induction of APOBEC3G Ubiquitination and Degradation by an HIV-1 Vif-Cul5-SCF Complex
 Induction of APOBEC3G Ubiquitination and Degradation by an HIV-1 Vif-Cul5-SCF Complex Xianghui Yu, 1,2 * Yunkai Yu, 1 * Bindong Liu, 1 * Kun Luo, 1 Wei Kong, 2 Panyong Mao, 1 Xiao-Fang Yu 1,3 1 Department
Induction of APOBEC3G Ubiquitination and Degradation by an HIV-1 Vif-Cul5-SCF Complex Xianghui Yu, 1,2 * Yunkai Yu, 1 * Bindong Liu, 1 * Kun Luo, 1 Wei Kong, 2 Panyong Mao, 1 Xiao-Fang Yu 1,3 1 Department
Peptide hydrolysis uncatalyzed half-life = ~450 years HIV protease-catalyzed half-life = ~3 seconds
 Uncatalyzed half-life Peptide hydrolysis uncatalyzed half-life = ~450 years IV protease-catalyzed half-life = ~3 seconds Life Sciences 1a Lecture Slides Set 9 Fall 2006-2007 Prof. David R. Liu In the absence
Uncatalyzed half-life Peptide hydrolysis uncatalyzed half-life = ~450 years IV protease-catalyzed half-life = ~3 seconds Life Sciences 1a Lecture Slides Set 9 Fall 2006-2007 Prof. David R. Liu In the absence
Identification of Mutation(s) in. Associated with Neutralization Resistance. Miah Blomquist
 Identification of Mutation(s) in the HIV 1 gp41 Subunit Associated with Neutralization Resistance Miah Blomquist What is HIV 1? HIV-1 is an epidemic that affects over 34 million people worldwide. HIV-1
Identification of Mutation(s) in the HIV 1 gp41 Subunit Associated with Neutralization Resistance Miah Blomquist What is HIV 1? HIV-1 is an epidemic that affects over 34 million people worldwide. HIV-1
Choosing Between Lentivirus and Adeno-associated Virus For DNA Delivery
 Choosing Between Lentivirus and Adeno-associated Virus For DNA Delivery Presenter: April 12, 2017 Ed Davis, Ph.D. Senior Application Scientist GeneCopoeia, Inc. Outline Introduction to GeneCopoeia Lentiviral
Choosing Between Lentivirus and Adeno-associated Virus For DNA Delivery Presenter: April 12, 2017 Ed Davis, Ph.D. Senior Application Scientist GeneCopoeia, Inc. Outline Introduction to GeneCopoeia Lentiviral
Delineating Minimal Protein Domains and Promoter Elements for Transcriptional Activation by Lentivirus Tat Proteins
 JOURNAL OF VIROLOGY, Apr. 1995, p. 2605 2610 Vol. 69, No. 4 0022-538X/95/$04.00 0 Copyright 1995, American Society for Microbiology Delineating Minimal Protein Domains and Promoter Elements for Transcriptional
JOURNAL OF VIROLOGY, Apr. 1995, p. 2605 2610 Vol. 69, No. 4 0022-538X/95/$04.00 0 Copyright 1995, American Society for Microbiology Delineating Minimal Protein Domains and Promoter Elements for Transcriptional
Lysosomes and endocytic pathways 9/27/2012 Phyllis Hanson
 Lysosomes and endocytic pathways 9/27/2012 Phyllis Hanson General principles Properties of lysosomes Delivery of enzymes to lysosomes Endocytic uptake clathrin, others Endocytic pathways recycling vs.
Lysosomes and endocytic pathways 9/27/2012 Phyllis Hanson General principles Properties of lysosomes Delivery of enzymes to lysosomes Endocytic uptake clathrin, others Endocytic pathways recycling vs.
Virology Journal. Open Access. Abstract. BioMed Central
 Virology Journal BioMed Central Research Stimulation of poliovirus RNA synthesis and virus maturation in a HeLa cell-free in vitro translation-rna replication system by viral protein 3CD pro David Franco
Virology Journal BioMed Central Research Stimulation of poliovirus RNA synthesis and virus maturation in a HeLa cell-free in vitro translation-rna replication system by viral protein 3CD pro David Franco
Bioluminescence Resonance Energy Transfer (BRET)-based studies of receptor dynamics in living cells with Berthold s Mithras
 Bioluminescence Resonance Energy Transfer (BRET)-based studies of receptor dynamics in living cells with Berthold s Mithras Tarik Issad, Ralf Jockers and Stefano Marullo 1 Because they play a pivotal role
Bioluminescence Resonance Energy Transfer (BRET)-based studies of receptor dynamics in living cells with Berthold s Mithras Tarik Issad, Ralf Jockers and Stefano Marullo 1 Because they play a pivotal role
MedChem 401~ Retroviridae. Retroviridae
 MedChem 401~ Retroviridae Retroviruses plus-sense RNA genome (!8-10 kb) protein capsid lipid envelop envelope glycoproteins reverse transcriptase enzyme integrase enzyme protease enzyme Retroviridae The
MedChem 401~ Retroviridae Retroviruses plus-sense RNA genome (!8-10 kb) protein capsid lipid envelop envelope glycoproteins reverse transcriptase enzyme integrase enzyme protease enzyme Retroviridae The
numbe r Done by Corrected by Doctor
 numbe r 5 Done by Mustafa Khader Corrected by Mahdi Sharawi Doctor Ashraf Khasawneh Viral Replication Mechanisms: (Protein Synthesis) 1. Monocistronic Method: All human cells practice the monocistronic
numbe r 5 Done by Mustafa Khader Corrected by Mahdi Sharawi Doctor Ashraf Khasawneh Viral Replication Mechanisms: (Protein Synthesis) 1. Monocistronic Method: All human cells practice the monocistronic
Page 32 AP Biology: 2013 Exam Review CONCEPT 6 REGULATION
 Page 32 AP Biology: 2013 Exam Review CONCEPT 6 REGULATION 1. Feedback a. Negative feedback mechanisms maintain dynamic homeostasis for a particular condition (variable) by regulating physiological processes,
Page 32 AP Biology: 2013 Exam Review CONCEPT 6 REGULATION 1. Feedback a. Negative feedback mechanisms maintain dynamic homeostasis for a particular condition (variable) by regulating physiological processes,
Chapter 13B: Animal Viruses
 Chapter 13B: Animal Viruses 1. Overview of Animal Viruses 2. DNA Viruses 3. RNA Viruses 4. Prions 1. Overview of Animal Viruses Life Cycle of Animal Viruses The basic life cycle stages of animal viruses
Chapter 13B: Animal Viruses 1. Overview of Animal Viruses 2. DNA Viruses 3. RNA Viruses 4. Prions 1. Overview of Animal Viruses Life Cycle of Animal Viruses The basic life cycle stages of animal viruses
Hung-Hao Chu 1,2, Yu-Fen Chang 1,2 & Chin-Tien Wang 1,2, *
 Journal of Biomedical Science (2006)13:645 656 645 DOI 10.1007/s11373-006-9094-6 Mutations in the a-helix directly C-terminal to the major homology region of human immunodeficiency virus type 1 capsid
Journal of Biomedical Science (2006)13:645 656 645 DOI 10.1007/s11373-006-9094-6 Mutations in the a-helix directly C-terminal to the major homology region of human immunodeficiency virus type 1 capsid
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Stelter et al., http://www.jcb.org/cgi/content/full/jcb.201105042/dc1 S1 Figure S1. Dyn2 recruitment to edid-labeled FG domain Nups.
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Stelter et al., http://www.jcb.org/cgi/content/full/jcb.201105042/dc1 S1 Figure S1. Dyn2 recruitment to edid-labeled FG domain Nups.
Hsu-Chen Chiu, Szu-Yung Yao, and Chin-Tien Wang*
 JOURNAL OF VIROLOGY, Apr. 2002, p. 3221 3231 Vol. 76, No. 7 0022-538X/02/$04.00 0 DOI: 10.1128/JVI.76.7.3221 3231.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Coding Sequences
JOURNAL OF VIROLOGY, Apr. 2002, p. 3221 3231 Vol. 76, No. 7 0022-538X/02/$04.00 0 DOI: 10.1128/JVI.76.7.3221 3231.2002 Copyright 2002, American Society for Microbiology. All Rights Reserved. Coding Sequences
Hepatitis B Antiviral Drug Development Multi-Marker Screening Assay
 Hepatitis B Antiviral Drug Development Multi-Marker Screening Assay Background ImQuest BioSciences has developed and qualified a single-plate method to expedite the screening of antiviral agents against
Hepatitis B Antiviral Drug Development Multi-Marker Screening Assay Background ImQuest BioSciences has developed and qualified a single-plate method to expedite the screening of antiviral agents against
The Primary Nucleotide Sequence of the Bovine Leukemia Virus RNA Packaging Signal Can Influence Efficient RNA Packaging and Virus Replication
 Virology 301, 272 280 (2002) doi:10.1006/viro.2002.1578 The Primary Nucleotide Sequence of the Bovine Leukemia Virus RNA Packaging Signal Can Influence Efficient RNA Packaging and Virus Replication Louis
Virology 301, 272 280 (2002) doi:10.1006/viro.2002.1578 The Primary Nucleotide Sequence of the Bovine Leukemia Virus RNA Packaging Signal Can Influence Efficient RNA Packaging and Virus Replication Louis
JOURNAL OF VIROLOGY, Jan. 1999, p Vol. 73, No. 1. Copyright 1999, American Society for Microbiology. All Rights Reserved.
 JOURNAL OF VIROLOGY, Jan. 1999, p. 19 28 Vol. 73, No. 1 0022-538X/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. Mutational Analysis of the Hydrophobic Tail of the
JOURNAL OF VIROLOGY, Jan. 1999, p. 19 28 Vol. 73, No. 1 0022-538X/99/$04.00 0 Copyright 1999, American Society for Microbiology. All Rights Reserved. Mutational Analysis of the Hydrophobic Tail of the
Regulation of Marburg virus (MARV) budding by Nedd4.1: a different WW domain of Nedd4.1 is critical for binding to MARV and Ebola virus VP40
 Journal of General Virology (2010), 91, 228 234 DOI 10.1099/vir.0.015495-0 Regulation of Marburg virus (MARV) budding by Nedd4.1: a different WW domain of Nedd4.1 is critical for binding to MARV and Ebola
Journal of General Virology (2010), 91, 228 234 DOI 10.1099/vir.0.015495-0 Regulation of Marburg virus (MARV) budding by Nedd4.1: a different WW domain of Nedd4.1 is critical for binding to MARV and Ebola
The Role of Sorting Nexins in Antigen Presentation
 The Role of Sorting Nexins in Antigen Presentation Chng X.R.J 1 and Wong S.H. 2 Department of Microbiology Yong Loo Lin School of Medicine, National University of Singapore Block MD4, 5 Science Drive 2,
The Role of Sorting Nexins in Antigen Presentation Chng X.R.J 1 and Wong S.H. 2 Department of Microbiology Yong Loo Lin School of Medicine, National University of Singapore Block MD4, 5 Science Drive 2,
The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein
 THESIS BOOK The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein Orsolya Buzás-Bereczki Supervisors: Dr. Éva Bálint Dr. Imre Miklós Boros University of
THESIS BOOK The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein Orsolya Buzás-Bereczki Supervisors: Dr. Éva Bálint Dr. Imre Miklós Boros University of
SUPPLEMENTARY INFORMATION
 sirna pool: Control Tetherin -HA-GFP HA-Tetherin -Tubulin Supplementary Figure S1. Knockdown of HA-tagged tetherin expression by tetherin specific sirnas. HeLa cells were cotransfected with plasmids expressing
sirna pool: Control Tetherin -HA-GFP HA-Tetherin -Tubulin Supplementary Figure S1. Knockdown of HA-tagged tetherin expression by tetherin specific sirnas. HeLa cells were cotransfected with plasmids expressing
7.012 Problem Set 6 Solutions
 Name Section 7.012 Problem Set 6 Solutions Question 1 The viral family Orthomyxoviridae contains the influenza A, B and C viruses. These viruses have a (-)ss RNA genome surrounded by a capsid composed
Name Section 7.012 Problem Set 6 Solutions Question 1 The viral family Orthomyxoviridae contains the influenza A, B and C viruses. These viruses have a (-)ss RNA genome surrounded by a capsid composed
Viral Vectors In The Research Laboratory: Just How Safe Are They? Dawn P. Wooley, Ph.D., SM(NRM), RBP, CBSP
 Viral Vectors In The Research Laboratory: Just How Safe Are They? Dawn P. Wooley, Ph.D., SM(NRM), RBP, CBSP 1 Learning Objectives Recognize hazards associated with viral vectors in research and animal
Viral Vectors In The Research Laboratory: Just How Safe Are They? Dawn P. Wooley, Ph.D., SM(NRM), RBP, CBSP 1 Learning Objectives Recognize hazards associated with viral vectors in research and animal
Incorporation of Pol into Human Immunodeficiency Virus Type 1 Gag Virus-Like Particles Occurs Independently of the Upstream Gag Domain in Gag-Pol
 JOURNAL OF VIROLOGY, Jan. 2004, p. 1042 1049 Vol. 78, No. 2 0022-538X/04/$08.00 0 DOI: 10.1128/JVI.78.2.1042 1049.2004 Copyright 2004, American Society for Microbiology. All Rights Reserved. Incorporation
JOURNAL OF VIROLOGY, Jan. 2004, p. 1042 1049 Vol. 78, No. 2 0022-538X/04/$08.00 0 DOI: 10.1128/JVI.78.2.1042 1049.2004 Copyright 2004, American Society for Microbiology. All Rights Reserved. Incorporation
Removal of Arginine 332 Allows Human TRIM5 To Bind Human Immunodeficiency Virus Capsids and To Restrict Infection
 JOURNAL OF VIROLOGY, July 2006, p. 6738 6744 Vol. 80, No. 14 0022-538X/06/$08.00 0 doi:10.1128/jvi.00270-06 Copyright 2006, American Society for Microbiology. All Rights Reserved. Removal of Arginine 332
JOURNAL OF VIROLOGY, July 2006, p. 6738 6744 Vol. 80, No. 14 0022-538X/06/$08.00 0 doi:10.1128/jvi.00270-06 Copyright 2006, American Society for Microbiology. All Rights Reserved. Removal of Arginine 332
Received 6 August 2002/Accepted 16 September 2002
 JOURNAL OF VIROLOGY, Feb. 2003, p. 1772 1783 Vol. 77, No. 3 0022-538X/03/$08.00 0 DOI: 10.1128/JVI.77.3.1772 1783.2003 Copyright 2003, American Society for Microbiology. All Rights Reserved. A Structurally
JOURNAL OF VIROLOGY, Feb. 2003, p. 1772 1783 Vol. 77, No. 3 0022-538X/03/$08.00 0 DOI: 10.1128/JVI.77.3.1772 1783.2003 Copyright 2003, American Society for Microbiology. All Rights Reserved. A Structurally
JCB Article. HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein. The Journal of Cell Biology
 JCB Article HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein Owen Pornillos, 1 Daniel S. Higginson, 1 Kirsten M. Stray, 1 Robert D. Fisher, 1 Jennifer E. Garrus, 1 Marielle Payne,
JCB Article HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein Owen Pornillos, 1 Daniel S. Higginson, 1 Kirsten M. Stray, 1 Robert D. Fisher, 1 Jennifer E. Garrus, 1 Marielle Payne,
