Cell Viability, Cytoskeleton Organization and Cytokines Secretion of RAW Macrophages Exposed to. Gram-Negative Bacterial Components
|
|
|
- Elmer Holmes
- 6 years ago
- Views:
Transcription
1 Cell Viability, Cytoskeleton Organization and Cytokines Secretion of RAW Macrophages Exposed to Gram-Negative Bacterial Components A thesis submitted in partial fulfillment of the requirements for the degree of Master of Science. By ALI AWADH ALSHEHRI B.S., King Abdulaziz University, Wright State University
2 WRIGHT STATE UNIVERSITY GRADUATE SCHOOL Dec 7,2016 I HEREBY RECOMMEND THAT THE THESIS PREPARED UNDER MY SUPERVISION BY Ali Awadh Alshehri ENTITLED Cell Viability, Cytoskeleton Organization and Cytokines Secretion of RAW Macrophages Exposed to Gram-Negative Bacterial Components BE ACCEPTED IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF Master of Science. Committee on Final Examination Nancy J. Bigley, Ph.D. Thesis Director Nancy J. Bigley, Ph.D. Professor of Microbiology and Immunology Barbara E. Hull, Ph.D. Director of Microbiology and Immunology Program, College of Science and Mathematics Barbara E. Hull, Ph.D. Professor of Biological Sciences Marjorie M. Markopoulos, M.S. Department of Environmental Health and Safety Robert E.W Fyffe, Ph.D. Vice President for Research and Dean of Graduate School
3 ABSTRACT Alshehri, Ali Awadh. M.S. Microbiology and Immunology Graduate Program, Wright State University, Cell Viability, Cytoskeleton Organization and Cytokines Expression of RAW Macrophages Exposed to Gram- Negative Bacterial Components. Macrophages play an important role in innate immunity by controlling cellular responses. In this study, the effects of gram-negative bacterial components (Flagellin, lipoprotein, lipopolysaccharides (LPS), outer membrane proteins-a (OMP-A) and peptidoglycan) were determined on cell viability, morphology, cytoskeletal filament and cytokines secretion of murine RAW macrophages at 24 hours. The effect of LPS, flagellin and peptidoglycan from gram negative bacteria on viability murine RAW macrophages were evaluated using different concentrations (1, 5 and 10 μg/ml). Cells stimulated with LPS displayed ~ 2-fold decrease (P 0.001) in cell viability compared to control cells at 24 hours whereas cells stimulated with flagellin displayed gradual and significant decrease (P 0.01; P 0.05) in cell viability with concentrations 5 and 10 μg/ml of this product. Effects of gram-negative bacteria components on the organization of F-actin and microtubule (tubulin) in murine RAW macrophages were monitored via immunofluorescent staining to study distribution of cytoskeleton such as elongation cell with cells stimulated with LPS. By using ImageJ analysis, the fluorescent intensity of immunofluorescent images were quantified to evaluate F-actin and tubulin rearrangement. Cells stimulated with Flagellin, lipoprotein, LPS, OMP-A or peptidoglycan show morphological changes as elongation and flatten with some components from gram negative bacteria with different concentrations (1, 5 and 10 μg/ml). LPS at all concentrations displayed a significant increase (p 0.001) in F-actin staining compared to control cells at 24 hours. The cells appeared flattened with irregular shapes compared to control cells appeared rounded. High level of TNF- α were secreted by murine RAW iii
4 264.7 macrophages exposed to LPS (2300 pg/ml) and flagellin (300 pg/ml) with 10 μg/ml concentration. These observations suggest that activation of TLRs leading to production of inflammatory cytokines such as TNF-α may account for our observed decreases in cell viability since LPS activates TLR4 and flagellin activates TLR5. iv
5 HYPOTHESIS Exposure of macrophage to bacterial substances from gram-negative bacteria will induce changes in cell viability, cell morphology, immunofluorescent intensity of microtubules (tubulin) staining and microfilaments (F-Actin) staining at 24 hours as well as differences a cytokines secretion. These characteristics will accompany the transition of unstimulated RAW macrophages to the proinflammatory phenotypes. v
6 TABLE OF CONTENTS LITERATURE REVIEW... 1 Macrophages... 1 Cytoskeleton of Macrophage... 2 Pathogen recognition receptors (PRRs)... 3 Toll-like receptors... 4 Signaling of toll-like receptors (TLRs)... 4 NOD-like receptors (NLRs)... 5 NOD1 and NOD2 receptors... 6 Signaling of NOD1 and NOD2 receptors... 7 Gram-negative bacteria... 7 Structure of gram-negative bacteria... 7 Cell wall... 7 Cell membranes... 8 Lipopolysaccarides (LPS)... 8 Peptidoglycan... 9 Lipoprotein... 9 OMP-A Flagellin MATERIALS AND METHODS Cell Line Cytokine Measurements Stimulated Cells with Gram-Negative Bacterial Components vi
7 Cell Viability Immunofluorescent Staining Image Analysis RESULTS Cell viability of RAW murine macrophage exposed to gram-negative components Cell morphology Morphological changes exposing to gram-negative bacterial compounds on RAW murine macrophage comparing to control cells Gram-negative bacterial components trigger tubulin cytoskeleton rearrangement in RAW murine macrophage Gram-negative bacterial components trigger F-actin cytoskeleton rearrangement in RAW murine macrophage Gram-negative bacterial components stimulate the production of cytokines by RAW murine macrophage DISCUSSION AND FUTURE STUDIES REFERENCES vii
8 LIST OF FIGURES Figure 1: Macrophage polarization... 2 Figure 2: Toll-like receptors (TLRs) pathway and activated signaling by PAMPs... 5 Figure 3: NOD1 and NOD2 signaling pathways and interaction partners by peptidoglycan of bacteria... 6 Figure 4: Structure of cell wall in gram-negative Figure 5: Trypan blue exclusion test Figure 6: Steps of Image analysis in image j Figure 7: Cell viability of RAW murine macrophage exposed to LPS from gram-negative bacteria in concentrations at 1, 5 and 10 μg/ml compared to control cells at 24 hours Figure 8: Cell viability of RAW murine macrophage exposed to flagellin from gramnegative bacteria in concentrations at 1, 5 and 10 μg/ml compared to control cells at 24 hours Figure 9: Cell viability of RAW murine macrophage exposed with peptidoglycan from gram-negative bacteria in concentrations at 1, 5 and 10 μg/ml compared to control cells at 24 hours Figure 10: Tubulin immunofluorescent images of RAW murine macrophages exposed to 1, 5 and 10 μg/ml of flagellin from gram-negative bacteria for 24 hours Figure 11: Tubulin immunofluorescent images of RAW murine macrophages exposed to 1, 5 and 10 μg/ml of lipoprotein from gram-negative bacteria for 24 hours Figure 12: Tubulin immunofluorescent images of RAW murine macrophages exposed to 1, 5 and 10 μg/ml of LPS from gram-negative bacteria for 24 hours viii
9 Figure 13: Tubulin immunofluorescent images of RAW murine macrophages show immunostaining exposed to 1, 5 and 10 μg/ml of OMP-A from gram-negative bacteria for 24 hours Figure 14: Tubulin immunofluorescent images of RAW murine macrophages show immunostaining exposed to 1, 5 and 10 μg/ml of peptidoglycan from gram-negative bacteria for 24 hours Figure 15: F-actin immunofluorescent images of RAW murine macrophages show immunostaining exposed to 1, 5 and 10 μg/ml of flagellin from gram-negative bacteria for 24 hours Figure 16: F-actin immunofluorescent images of RAW murine macrophages exposed to 1, 5 and 10 μg/ml of lipoprotein from gram-negative bacteria for 24 hours Figure 17: F-actin immunofluorescent images of RAW murine macrophages exposed to 1, 5 and 10 μg/ml of LPS from gram-negative bacteria for 24 hours Figure 18: F-actin immunofluorescent images of RAW murine macrophages exposed to 1, 5 and 10 μg/ml of OMP-A from gram-negative bacteria for 24 hours Figure 19: F-actin immunofluorescent images of RAW murine macrophages exposed to 1, 5 and 10 μg/ml of peptidoglycan from gram-negative bacteria for 24 hours Figure 20: RAW murine macrophages respond to gram-negative bacterial compounds (LPS, OMP-A, Flagellin, Lipoprotein, Peptidoglycan) by releasing IL-1b Figure 21: RAW murine macrophages respond to gram-negative bacterial compounds (LPS, OMP-A, Flagellin, Lipoprotein, Peptidoglycan) by releasing IL Figure 22: RAW murine macrophages respond to gram-negative bacterial compounds (LPS, OMP-A, Flagellin, Lipoprotein, Peptidoglycan) by releasing IL ix
10 Figure 23: RAW murine macrophages respond to gram-negative bacterial compounds (LPS, OMP-A, Flagellin, Lipoprotein, Peptidoglycan) by releasing TNF- α Figure 24: RAW murine macrophages respond to gram-negative bacterial compounds (OMP-A, Flagellin, Lipoprotein, Peptidoglycan) by releasing TNF- α Figure 25: RAW murine macrophage show morphological changes after interaction with LPS through TLR x
11 LIST OF TABLES Table 1: Cell viability calculations Table 2: Gram-negative components to which RAW cells have been exposed Table 3: The antibodies and the fluornresccent taps used in immunofluorescence experiments Table 4: Cytokines secretion of RAW murine macrophage after 24 hours xi
12 ACKNOWLDGEMENT I would like to express my special thanks of gratitude to Dr. Nancy Bigley who gave me the golden opportunity to do this research, and provided support throughout the duration of my project. Also, I would like to thank my program director, Dr. Barbara Hull for her suggestions and guidance. I would like to thank Mrs.Marjorie M. Markopoulos for serving as a member on my thesis committee and for her suggestions and contribution to my research. I would like to thank all my lab members for their assistance in my work. Finally, I would like to thank my family and my friends for supporting my decision to continue my education. xii
13 LITERATURE REVIEW Macrophage Tissue Macrophages are heterogeneous immune cells that play a major role in response to inflammation in both innate and adoptive immunities. Macrophages are mononuclear cells that are derived from blood monocytes which in turn develop from bone marrow stem cells, passing through proliferation and differentiation cascades (Murray and Wynn, 2011). Colony Stimulating Factor (CSF-1) is a critical growth factor that regulates the proliferation and differentiation of mononuclear phagocytic cells (P. Roth and Stanley 1992; Martinez et al. 2006). The main function of macrophages is to defend and clear pathogen- associated molecular patterns (PAMPs) and danger-associated molecular patterns (DAMPs) as well as to repair damage tissues (Mills, 2012). Macrophages recognize different PAMPs and DAMPs through recognition receptors such as toll like receptors TLR, nucleotide-binding oligomerization domain NOD-like receptors, retinoic acid-inducible gene I RIG-1 family, lectins and scavenger receptors that are distributed throughout the cell membrane and cytoplasmic organelles. Macrophages respond to PAMPs and DAMPs through signaling pathways that lead to the release of a variety of cytokines and chemokines. Macrophages also activate adaptive immune B and T cells to eliminate any pathogens (Small et al., 1994). Macrophages have been morphologically and functionally grouped into three classes. Depending on signals present in the microenvironment, resting macrophages M0 are polarized to pro-inflammatory M1 macrophages or antiinflammatory M2 macrophages. M0 are polarized to pro-inflammatory M1 in response to intracellular pathogens, microbial products such as LPS, or pro-inflammatory cytokines such as IFN-y and IL-1. Pro-inflammatory M1 cells respond to inflammation by releasing different cytokines, chemokines, and effector molecules such as TNF-α, IL-6, CCL3 and inos. IL-4, IL-10 or IL-13 polarize M0 to anti-inflammatory M2 macrophages. M2 cells play major roles in maintaining tissue hemostasis and in modulating inflammation through multiple mediators such as TGF-, IL4, IL10, and IL13. M2 elevates cellular metabolic activities, including phagocytosis, endocytosis, and tissue remodeling (Martinez et al. 1
14 2006). Macrophage subtypes are important in controlling immune response by keeping the balance between proinflammatory and anti-inflammatory macrophage populations (Junliang et al., 2010; Cheng et al., 2000). Figure 1: Macrophage polarization. M0 Resting cells are stimulated with PAMPS via TLR. LPS or IFN-γ polarized cells to M1 phenotype and secrete pro-inflammatory cytokines IL-1β, IL-6 and TNF-α. However, M2 antiinflammatory phenotype is polarized by IL-4, IL-10 and IL-13 and release IL-10 that down-regulates M1 functions. (Remodified from Nakagawa, Y., & Chiba, K. 2014). Cytoskeleton of Macrophage The cytoskeleton of a macrophage is organized in a complex networks of protein fibers that give the cell its shape, support the cell, and allow cell movement as a whole; these fibers also facilitate the organelles movements in an organized fashion. A macrophage s cytoskeleton is also a major player in the sliding movement toward target as well as in the endocytosis and phagocytosis processes that are required for these cells to achieve their proper function. The cytoskeleton provides an organized track for each cytokine and chemokine that is essential for the 2
15 orchestrated macrophage function. The cytoskeleton is differentiated according to the size and roll of the filament; there are three types of filaments: microfilaments (about 7 nm in diameter), microtubules (about 25 nm in diameter), and intermediate filaments (about 10 nm in diameter) (Jaffe and Hall 2005). Gram-negative bacterial pathogens impact a macrophage s cytoskeleton by activating GTPases, which activate NF-kB; this in turn leads to increased gene expression of pro-inflammatory cytokines such as IL-1 and INF-y (Lee, 2000). Microtubules are hollow cylinders composed of polymeric proteins formed by α- and β-tubulin dimers. The microtubule organization center (MTOC) arranges and nucleates microtubules Hervé and Bourmeyster, 2015). Microtubule-associated Rac1 and RhoA play a role in the polymerization of tubulin subunits and the distribution of ATP levels inside cells. Activating Rac1 and RhoA signaling controls actin dynamics and causes the dynamic microtubules to be unstable. Rho activity is stimulated by the depolymerization of microtubules after Rho GTPase is activated at the plasma membrane (Jaffe and Hall 2005). Dynamics of microtubules are modulated via several bacteria, which can play an important role of cell movement (Yoshida and Sasakawa 2003). Microfilaments consist of linear polymers of double-stranded helical filamentous actin (F-actin). F-actin is formed by twisted subunits of a monomeric globular protein (G-actin). Microfilaments play a critical role in the dynamics of macrophages during infection with gram-negative bacteria. In response to infection, F-actin is involved in immune cell migration, chemotaxis, and phagocytosis. ATP molecules and Rho GTPases also play a role in the dynamics of microfilaments (Mattoo et al, 2007). Interactions between a pathogen with the cell membrane disturb the cytoskeleton of eukaryotic cells which is essential for phagocytosis and regulation of immune cell signaling. Rho GTPase is activated via many bacterial factors, which include virulence factors and toxins. This activation leads to the initiation of a cascade of immune cell signals (Jaffe and Hall 2005). Bacterial pathogens can survive the immune cell by manipulating Rho GTPase (Boquet and Lemichez, 2003). Pathogen recognition receptors (PRRs) The innate immune system recognizes and responds to pathogens by different immune surveillance mechanisms. Pathogen-associated molecular patterns (PAMPs) are identified by pathogen recognition receptors 3
16 (PRRs) on the plasma membrane of immune cells such as macrophages (Akira et al., 2006). Diverse PAMPs have been detected by various families of PRRs, such as C-type lectin receptors (CLRs), transmembrane receptors, Tolllike receptors (TLRs), RIG-I- like receptors (RLRs) and Nod-like receptors (NLRs) (Kumar et al., 2011). PRRs respond to PAMPs by creating an inflammatory response, in which chemokines, cytokines, and type-i interferons are released. These inflammatory cytokines attract other inflammatory cells, including innate and adaptive immune cells, to the site of infection (Takeuchi and Akira, 2010). Toll-like receptors Toll-like receptors (TLRs) play an important role in innate immunity. 13 TLRs detect certain PAMPs. TLRs induce the production of cytokines and chemokines and can also trigger NF-KB activation. TLRs are type-i transmembrane proteins that contain leucine-rich repeats (LRRs) on the extracellular domain and bind to corresponding PAMPs (Lemaitre et al., 1996). Some TLRs require co-receptors for ligand binding such as MD-2 for TLR4 when binding to LPS (Shimazu et al.,1999). The complex of TLR1/TLR2 is a heterodimer, while TLR5 has five loops that consist of 20 LRRs (Botos et al.,2011). Signaling of toll-like receptors (TLRs) In MyD88-dependent signaling pathway, interactions between TLRs on the cell surface and ligands of PAMPs activate a downstream signaling pathway. TIR-TIR domain interaction recruits one or more adaptor proteins. All TLRs, with the exception of TLR3, recruit the MyD88 adaptor protein that is essential for downstream signaling. IRAKs, TRAF6 and the TAK1 complex activate downstream TAB1, TAB2, TAK1 and MAP kinases. NEMO, IKKα and IKKβ make up the IKK complex that is activated by TAK1. NF-kB and IkBα are phosphorylated by IKKβ; phosphorylated NF-kB is then translocated to the nucleus of the cell. Finally, NF-kB and MAP kinases act as activators for pro-inflammatory cytokines IL-1 and INF-y. IKK complex is involved in the activation of ERK, P38, JNK, and MAP kinases (Mogensen, 2009). In the MyD88-independent/TRIF-dependent signaling pathway, IRF3 and IRF7 are interferon regulatory factors that increase the expression of type I interferons. TLR3 and TLR4 recruit an adaptor protein TRIF which 4
17 activates downstream IRF3 through TBK1 and IKK activation, leading to the production of pro-inflammatory cytokines (Mogensen, 2009). Figure 2: Toll-like receptor (TLR) pathways and activated signaling by PAMPs through MyD88 independent. phosphorylation is induced by IRAK after MyD88 recruit IRAK which associates with TRAF6 or TRAF3, leading to the activation of IKK complex or MKKs and resulting in the activation of NF-κB, CREB and AP1. (Remodified from O'Neill, et al 2013). NOD-like receptors (NLRs) NLRs consist of nucleotide-binding oligomerization domains of soluble proteins found in the cytoplasm and N-terminal effector domains. NLRs can be divided into four subfamilies: NLRA, NLRB, NLRC, and NLRP (Strober, 2006). CARDs, pyrin domains (PYDs), baculoviral inhibitor of apoptosis repeat (BIR) domains, or the transactivator domain (AD) are the effector domains in NLRs. PAMPs are detected by NLRs as well as TLRs, while DAMPs are 5
18 only detected by NLRs. Activation of NLR cytoplasmic domains leads to an inflammatory response, autophagy, or cell death. (Le Bourhis, 2007). NOD1 and NOD2 receptors NOD1 and NOD2 are intracellular receptors of innate immune cells. They are located in the cytosol of phagocytes. Peptidoglycan is recognized by NOD1 and NOD2 receptors expressed on epithelial cells and antigen presenting cells (APCs) such as macrophages. NOD1 and NOD2 consist of a C-terminal LRR domain and a central NOD which is a NACHT domain. They also contain an ATPase activity domain and an N-terminal domain such as CARDs or pyrin domain. In NOD1, the signal CARDs have C-terminal LRR, NACHT domain, and N-terminal effector domain. In NOD2, the two signal CARDs consist of C-terminal LRR, NACHT domain, and N-terminal effector domain. NOD1 has a high affinity for bacterial peptidoglycan and binds to D-γ-glutamyl-meso-DAP dipeptide (ie-dap). NOD2 recognizes the muramyl dipeptide (MDP) that is found in all bacteria (Le Bourhis, 2007). Figure 3: NOD1 and NOD2 signaling pathways and interaction partners by peptidoglycan of bacteria. In the cytosol, peptidoglycan is recognized by NOD1 or NOD2. NF- kb signaling is activated after NOD1 or NOD 2 bind RIP2 leading to the expression of pro-inflammatory genes to secrete IL-8 and TNF-α (Remodified from Claes, A et al, 2015). 6
19 Signaling of NOD1 and NOD2 receptors When PAMPs bind to NOD1 and NOD2 receptors, the CARD domain of NOD1 and NOD2 is activated; this leads to downstream activation of serine-threonine kinase RIP2. ciap1 and ciap2 that polyubiquitinate RIP2 and activate downstream signaling. As a result, IKK complex phosphorylates IkBα through activation of TRAF6 and TAK1 complex which have been recruited via RIP2. This leads NF-Kb to translocate to the nucleus. Activation of TAK1 also leads to activation of MAP kinase, ERK, p38 and JNK. This pathway leads to active the transcription factor AP-1 and the phosphorylation of histones H3, giving the transcription factor ERK access euchromatin. p38 works with both JNK and ERK to control the transcription factors. As a result of the translocation of NF-kB to the nucleus, there is increased gene expression of pro-inflammatory response cytokines and chemokines, including IL- 1, IL-6 and IL-8. This also leads to the release of an antimicrobial peptide that kills the invading bacteria (Strober, 2006; Le Bourhis, 2007). Gram-negative bacteria In 1884, Christian Gram discovered gram negative bacteria by their unique red color when using the gram staining technique (Silhavy et al, 2010). Some gram negative bacteria are saprophytic harmless and living in the bodies of many animal species, growing in the microflora of these species at the expense of harmful pathogens. Many gram negative bacteria are harmful and cause devastating conditions such as food poisoning, skin infection, gastrointestinal infection, urinary tract infection, and genital tract infection among others. These infections have a high impact on both medical and socioeconomic statuses in the USA and around the world (Bos et al, 2007). Structure of gram negative bacteria Cell wall Gram negative bacteria have cell walls 2-3nm thick containing 2-3 layers of interconnected peptidoglycan. Gram negative cell walls activate both innate and adaptive immune cells including macrophages in human. PAMPS from gram negative bacteria are recognized by pattern associated receptors of macrophages and other innate immune cells. This recognition triggers the release of many pro-inflammatory cytokines. Cell walls can also activate antibody 7
20 production from adoptive immune B cells. These antibodies are directed against cell wall antigens; the gram negative cell wall protects bacteria from osmotic lysis (Vomer, 2012). Cell membranes The outer membrane of gram negative bacteria is composed of a bilayer that is 7 nm thick and consists of phospholipids, lipoproteins, lipopolysaccharides (LPS), and proteins. The inner layer of the outer membrane is formed by phospholipids and lipoproteins and faces the peptidoglycan layer. The outer layer of membrane is formed by lipopolysaccharides. Lipid A is embedded in the lipid portion of the membrane while the polysaccharide portion extends outside the surface. The LPS portion of the outer membrane forms the endotoxin of the gram negative bacteria. The membrane contains many proteins that distinguish different gram negative bacteria from each other. This membrane also contains porins, which function as channels that allow transportation of nutrients into the bacteria. The periplasmic space separates the two layers and is formed from a gelatineous material that separates the outer membrane from the peptidoglycan. This space is around 15 nm thick and contains periplasmic binding proteins which help transport ATP- binding cassette (ABC system) as well as the transport of chemoreceptors for chemotaxis (Caroff and Karbian, 2003). Lipopolysaccarides (LPS) The main function of LPS is to protect gram-negative bacteria from environment attacks. LPS also contributes to stabilizing the outer membrane by keeping the negative charge on the outer membrane of the bacteria (Whitfield and Trent, 2014). Lipopolysaccharides consist of three different components in gram negative bacteria: a core of lipid A, an inner and outer core of polysaccharides, and O-antigen. Lipid A forms the hydrophobic endotoxin because of its high content of acyl chains (fatty acids); it also contains two glucosamines backbones that attach to the six acyl chains (Caroff and Karibian, 2003). The core oligosaccharide also includes sugars such as Kdo and heptose; Kdo is usually linked with lipid A. O-antigen is composed of repetitive subunits of glycosyl residues (2-8 monosaccharide moieties). The different composition of sugar moieties is the part of LPS that is targeted by host antibodies. Making O-antigen important for the activation of innate immunity (Erridge et al, 2002). LPS plays an important role in activating the innate immune system; LPS interacts with Toll-Like receptor 4 (TLR4) on the surface of macrophages. 8
21 LPS is expressed by gram negative bacteria bound to CD14 co-receptors on a macrophage surface. This complex activates TLR4-MD2 and leads to the downstream signaling activation that in turn releases the activated nuclear factor B (NF-B). The translocation of NF-kB to the nucleus leads to the release of pro inflammatory cytokines such as IL-1, IL-6, IL8 and TNF-α. Some of these cytokines polarize the cytoskeleton of the macrophage, leading to distribution in the morphology of the cell (Lu et al, 2008). Peptidoglycan Peptidoglycan is a protective layer in the bacterial membrane that covers the cytoplasmic membrane. It is composed of two alternating amino sugar residues, N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM), which form the backbone of the structure that is cross-linked by peptides. Because of its rigidity, the peptidoglycan layer gives the cell its shape. However, it is also flexible and porous, allowing cell growth and permitting compounds to enter and exit from the cell (Young, 2011). Peptidoglycan from gram negative bacteria interact with plasma intracellular receptors of the immune cell called Nod-Like Receptors (NLRs). NOD1 binds to N-acetylglucosamine- N-acetylmuramyl tripeptide (GM tripeptide) or the dipeptide D-glutamine- diaminopimelic acid (D-Glu DAP) found in the peptidoglycan of gram-negative bacteria. NOD2 reacts with peptidoglycan, which has the N-acetylmuramyl dipeptide (MDP), either in gram-negative bacteria or gram positive bacteria (Royet and Reichhart, 2003). Lipoprotein Lipoproteins of gram negative bacteria are formed from three fatty acids (N-Acyl-S-diacylglyceryl cysteine) which are modified by an unusual S-glycerylcysteine residue at the N-terminus in many lipoprotein Lp of Escherichia coli. N-terminus allows type II signal peptide sequences with a conserved lipid-modified cysteine residue from bacteria to attach onto the plasma membrane or outer membrane. Preprolipoprotein diacylglyceryl transferase (Lgt), prolipoprotein signal peptidase (Lsp) and apolipoprotein N-acyltransferase (Lnt) play important roles in the survival of enzymes required for the biosynthesis of bacterial lipoproteins in E. coli. A mature lipoprotein is localized within the cell wall of gram-negative bacteria by linking outer membrane with peptidoglycan (Nakayama et al, 2012). Bacterial lipoproteins play an important role in virulence and in inducing innate immunity via TLR2 in pattern 9
22 recognition receptors. TLR2 interacts with a bacterial lipoprotein that activates innate immune response through MHC-II in macrophages; this interaction is associated with an increase in production of cytokines such as IL-1beta, IL-12P40 and TNF-α by using MyD88-dependent signaling (Nakayama et al, 2012). OMP-A OmpA is complementary to outer membrane proteins in gram-negative bacteria. OMP-A is a highly conserved protein and contains two domains that are 325-residue, heat-modifiable proteins. In the outer membrane, the first domain of OMP-A contains an N-terminal transmembrane β-barrel domain, and in the periplasm, a C- terminal globular domain interacts with peptidoglycan in other domains (Wang, 2002). In many bacteria, the main function of OMP-A is to maintain in the structural integrity of the outer membrane along with murein lipoprotein and peptidoglycan-associated lipoprotein (Smith et al, 2007). During immune response, OmpA is a targeted adaptive immune defense and is a part of innate immunity. OMP-A plays an important role in activating antigen presenting cells (APCs) through TLR2. The acute-phase protein serum amyloid A optimizes OmpA, leading to enhanced phagocytosis by macrophages. (Confer and Ayalew, 2013). However, scavenger receptors, LOX-1 and SREC-I, mediate the ligands between OMP-A on gram-negative bacteria and TLR2 in macrophage (Egesten et al, 2008). Flagellin Flagellin is an extracellular locomotor and virulent organelle found on the bacterial surface of many gram negative bacteria. Flagella are motility systems that present on the surface of some gram negative bacteria. The flagellum is composed of 3 parts: a long hollow cylindrical helical flagellar filament, a short curved hook, and a basal body or flagellar motor which lies within the plasma membrane. The flagellum moves in a counter clockwise or clockwise direction and shifts bacterial movement utilizing the proton-motive force. The number of flagella range from 5-10 per bacterium, and length varies from 5 to 15 μm. Flagella are also used in gram negative bacteria as a sensory organ, helping the bacteria to attach to host cells such as immune cells. (Zhao et al, 2014). TLR5 on the surface of macrophages of the innate immunity system TLR5 allow the immune cells to recognize the flagellin protein forming the flagellum (Yoon et al, 2012). Flagellin ligands bind to TLR5, activating MyD88-dependent 10
23 signaling which leads to the activation of the pro-inflammatory transcription factor NF-κB in epithelial cells, monocytes, and dendritic cells. This activation lead to the release of pro-inflammatory cytokines such as TNF-α or IL-1B (Steiner, 2007). Figure 4: Structure of cell wall structure in gram-negative bacteria. Gram-negative bacteria contain two membranes which are outer membrane and inner membrane separated by peptidoglycan. Lipopolysaccharides is located in outer membrane and envelops the outer membrane. Lipoprotein is linked between outer membrane and peptidoglycan (Remodified from Lolis, and Bucala, 2003). 11
24 MATERIALS AND METHODS Cell Line The Cell line is RAW murine macrophage (purchased from American Type Culture Collection ATCC, Manassas, VA). Culture petri plates 100mm x 20mm (from BD Biosciences) are used to culture RAW murine macrophage with Dulbecco s Modified Eagle s Medium (DMEM; HyClone), Supplemented with 10% of fetal bovine serum (heat-inactivated) (from Fisher Scientific) and 1% of penicillin-streptomycin antibiotic (from MP Biomedical, LLC). At 37ºC, RAW murine macrophage were grown in a humidified 5%CO2 in incubator. Cells were splitting of the cells depends on confluency of the cell on the surface of petri plate by viewing cells under a microscope (approximately 2-3 times per a week). Cytokine Measurements After 24 hours, Exposure of gram-negative bacteria compounds onto RAW macrophage release cytokines concentrations that have been collected and measured via Luminex Multiplex Immunoassays. Stimulated Cells with Gram-Negative Bacterial Components RAW murine macrophages were grown in 12 well chambers (from Ibidi) to 70% confluency. RAW murine macrophages were stimulated with peptides (1,5 and 10 μg/ml) of LPS, peptidoglycan, lipoprotein, OMP-A and flagellin from gram negative bacteria. In each chamber, there were three wells for each concentration, and 3 wells with unstimulated cells as control. The chambers were incubated for 24 hours and the culture media were collected and frozen at -80 C. 12
25 Cell Viability At 70% confluency, the RAW cells were then stimulated with with peptide (1, 5 or 10 μg/ml) LPS, peptidoglycan, lipoprotein, OMP-A and Flagellin from gram negative bacteria. Unstimulated cells that served as control cells were incubated for 24 hours. After that, the cells, the control and each product (1,5 and 10 μg/ml), were grown on 12-well plates for for 24 hours were gathered. At 4 o C, using centrifuge on new supernatant at 1500 rpm was made for of 5 minutes. Then, solution of cell was aspirated and 1 ml of 10% DMEM medium was added to suspend the cell pellet. To determinant the cell viability, trypan blue stain was added the cells at a ratio of 1:2 the following equation was used to to classify the cell viability: % Cell Viability = [Total Viable Cells (Unstained) / Total Cells (Viable + Dead)] X 100 Figure 5: Trypan blue exclusion test (Hemocytometer method). Live cells appear colorless under microscope, whereas the dead cells stain blue. 13
26 Table 1: Cell viability calculations. Procedure Equation %Cell Viability [Total Viable cells (Unstained) / Total cells (Viable +Dead)] X 100 Table 2: Gram-negative components that have been exposed to RAW cell. Gram-negative components PRRs Concentration Company LPS TLR4 1, 5 and 10 μg/ml InvivoGen Flagellin TLR5 1, 5 and 10 μg/ml InvivoGen Peptidoglycan NOD1/2 1, 5 and 10 μg/ml InvivoGen Lipoprotein TLR2 1, 5 and 10 μg/ml InvivoGen OMP-A TLR2 1, 5 and 10 μg/ml Creative BioMart Table 3: The antibodies and the staining used in immunofluorescence experiments. Antibodies and stains Dilution Company A/B tubulin (Primary Antibody) 1:50 Cell Signaling Technology Texas Red-Phalloidin X(F-actin Florescence) 1:40 LifeSpan BioSciences Rabbit IgG Fab 2 Goat anti-rabbit Polycolonal 1:500 LifeSpan BioSciences Antibody (Secondary Antibody) 14
27 Immunofluorescent Staining On chambers, cells were washed with phosphate buffered saline (PBS) (3 times at 5 minutes each time). Next, (4%) paraformaldehyde was fixed the cells for 15 minutes. PBS was used to rinse the cells (3 times) (5 minutes). After that, 0.2% Triton diluted in PBS permeabilize the cells (5-10 minutes). One more time, PBS was used to rinse the cells (3 times at 5 minutes). The Blocking buffer (5% Goat serum, 3% BSA, and 0.05% tween) was used to block the cells for one hour preventing. Again, PBS was used to rinse the cells (3 times at 5 minutes). 3% BSA ( 1.5 mg of bovine serum albumin in 50 ml PBS) was used to dilute primary antibodies and cells were incubated at a temperature 4 o C overnight. Next day, (1%) BSA (0.5 mg of bovine serum albumin in 50 ml PBS) was used to rinse the cells (3 times at 5 minutes). In dark, Texas Red-Phalloidin X on secondary antibody were added for 1 2 hours. (1%) BSA (0.5 mg of bovine serum albumin in 50 ml PBS) was used to rinse the cells (3 times at 5 minutes). Vectashield (H- 1400) (from Vector Laboratories) was used to mount the stained cells onto the microscope slide by adding a drop in each well. The cells were evaluating on Olympus Epi- fluorescence microscope with a spot digital camera. Image Analysis Imaje J (National Institutes of Health, was used to qualify the Immunofluorescent of cells after exposure. Image J quantifies regions of interest (qroi) of the fluorescent via several steps. First of all, the type of color is 8-bit image by opening original image image type 8-bit image. After that, the background was subtracted to remove the pixel information from a continuous background. Also, the threshold 20 pixel values of black background for and 111 pixel values of bright saturated artifacts of image were adjusted. Finally, evaluating the image through analyze Particles by selecting analysis option then analyze the particles, so now the value total of the area and number of counted cells have been used to compare the images after stimulation via One-way ANOVA and T test in Sigma Plot 13.0 Software. 15
28 Figure 6: Steps of Image analysis in image j. 16
29 RESULTS Cell viability of RAW murine macrophage exposed to gram-negative components after 24 hours RAW murine macrophages (M0) were exposed to LPS, flagellin or Peptidoglycan in concentrations of 1, 5 or 10 μg/ml. The cell viability was measured after 24 hours using trypan blue staining assay. Macrophages exposed to LPS showed ~ 2-fold decrease (P 0.001) in cell viability at all concentrations compared to control cells as shown in Figure 7. Also, macrophages displayed a gradual decrease in variability with high concentrations of flagellin and showed a significant decrease with 10 μg/ml of flagellin (P 0.01) compared to control cells. Also, there was a significant decrease between M0 with 5 μg/ml flagellin (P 0.05) compared to control cells as shown in Figure 8. However, macrophages exposed to Peptidoglycan showed no differences in viability of the cells as shown in Figure 9. In general, RAW macrophages showed the highest response to LPS compared to flagellin or peptidoglycan of gram-negative bacteria. Cell morphology Morphological changes exposing to gram-negative bacterial compounds on RAW murine macrophage comparing to control cells M0 macrophages served as the control and appeared rounded after 24 hours. Macrophages were stimulated with gram-negative bacterial components, flagellin, lipoprotein, LPS, OMP-A and peptidoglycan, with different concentrations 1, 5 or 10 μg/ml. Morphologically, the cells appeared flattened and elongated with LPS from gram-negative bacterial. 17
30 Gram-negative bacterial components trigger tubulin cytoskeleton rearrangement in RAW murine macrophage At concentration of 5 and 10 μg/ml, the fllagellin caused dispersed staining of the microtubules throughout the cell cytoplasm of RAW macrophage as shown in Figure 10 A. The microtubule continued to show staining at lower concentrations with 5 and 1 μg/ml as compared to the control cells. At a concentration of 5 μg, bacterial lipoprotein, OMP-A, and peptidoglycan stimulated the macrophages to microtubule aggregation to one pole of the cell as shown in Figure 11A, 13A, 14A. In contrast, the 10 μg/ml concentration of lipoprotein caused the RAW264.7 cells to aggregate within 24 hours as shown figure11a. LPS expose to RAW264.7 cells at the three concentrations. Elongation of the cells morphology microtubule appeared throughout the cell as shown in Figure 12 A. Gram-negative bacterial components trigger F-actin cytoskeleton rearrangement in RAW murine macrophage Exposure of flagellin at concentration of 10 μg/ml, F-actin shows low staining whereas 1 and 5 μg/ml show higher level of staining on RAW macrophage as shown in Figure 15 A. At concentration of 5 and 10 μg/ml, bacterial lipoprotein caused aggregation one to one pole of the cell as shown Figure16 A. At the three concentrations, LPS caused flattened and extended irregular shapes of the cell morphology to express F-actin around the cells as shown in Figure 17 A. At concentration of 5 and 10 μg/ml, OMP-A displayed enlarged and flattened cells to express F-actin as shown in Figure 18 A. Peptidoglycan caused flattened cells at concentrations of 10 μg/ml for some cells as shown in Figure 19. Gram-negative bacterial components stimulate the production of cytokines by RAW murine macrophage Pro-inflammatory cytokine IL-1b production increased gradually when the cells were exposed to LPS, flagellin or peptidoglycan at all consternations 1, 5 and 10 μg/ml. Similarly, the macrophages stimulated with OMP- A at 5 μg/ml showed a slight increase in IL-1b compared to the control cells. Macrophages responded to lipoprotein 18
31 by releasing IL-1b, but there is no difference between levels of IL-1b in the three concentrations 1, 5 and 10 μg/ml as shown in Figure 7. Secretion of pro-inflammatory cytokine IL-10 decreased with cells that were exposed to LPS, OMP-A, flagellin, lipoprotein and peptidoglycan in all concentrations compared to IL-1b, IL-12 and TNF- α productions as shown in Figure 21. IL-12 production increased gradually with the cells that were exposed to LPS, which IL-12 increased slightly when macrophages were stimulated to other gram-negative bacterial compounds at concentrations 1, 5 and 10 μg/ml as shown in Figure 22. TNF-α increased highly with cells that were exposed to LPS, OMP-A, flagellin, lipoprotein and peptidoglycan in 1, 5 and 10 μg/ml as shown in Figure 24. However, stimulated -macrophage with LPS released the highest level of TNF-α compared to cells that exposed other gramnegative bacterial components as shown in Figure
32 Table 4: Cytokines secretion after exposure of gram-negative bacterial components to RAW murine macrophage after 24 hours. Bacterial compounds Cytokines Concentration LPS OMP-A Flagellin Lipoprotein Peptidoylycan IL-1b pg/ml IL-10 pg/ml IL-12 pg/ml TNF- pg/ml Control μg/ml μg/ml μg/ml Control μg/ml μg/ml μg/ml Control μg/ml μg/ml μg/ml Control μg/ml μg/ml μg/ml
33 Figure 7: Cell viability of RAW murine macrophage exposed to LPS from gram-negative bacteria in concentrations (1,5 and 10 μg/ml) compared to control cells at 24 hours. Each value represents the mean ± standard error (SE) of 3 separate experiments. ***, p< (n=3) Figure 8: Cell viability of RAW murine macrophage exposed to flagellin from gram-negative bacteria in concentrations (1,5 and 10 μg/ml) compared to control cells at 24 hours. Each value represents the mean ± standard error (SE) of 3 separate experiments. *, p<0.05; **, p< (n=3) 21
34 Figure 9: Cell viability of RAW murine macrophage exposed with peptidoglycan from gram-negative bacteria in concentrations (1,5 and 10 μg/ml) compared to control cells at 24 hours. Each value represents the mean ± standard error (SE) of 3 separate experiments. (n=3) 22
35 A) B) Figure 10: A) Tubulin immunofluorescent images of RAW murine macrophage. Macrophages cells show morphological changes and cytoskeleton immunostaining at 24 hours exposing to flagellin from gram-negative bacteria (Scale bar =20 μm). B) Tubulin immunofluorescent intensity for control and (1,5 and 10 μg/ml) of flagellin from gram-negative bacteria following 24 hours. Each value represents the mean ± standard error (SE) of 3 separate experiments. (n=3) 23
36 A) B) Figure 11: A) Tubulin immunofluorescent images of RAW murine macrophage. Macrophages cells show morphological changes and cytoskeleton immunostaining at 24 hours exposing to lipoprotein from gram-negative bacteria (Scale bar =20 μm). B) Tubulin immunofluorescent intensity for control and (1,5 and 10 μg/ml) of lipoprotein from gram-negative bacteria following 24 hours. Each value represents the mean ± standard error (SE) of 3 separate experiments. *, p<0.05; **, p< (n=3) 24
37 A) B) Figure 12: A) Tubulin immunofluorescent images of RAW murine macrophage. Macrophages cells show morphological changes and cytoskeleton immunostaining at 24 hours exposing to LPS from gram-negative bacteria (Scale bar =20 μm). B) Tubulin immunofluorescent intensity for control and (1,5 and 10 μg/ml) of LPS from gramnegative bacteria following 24 hours. Each value represents the mean ± standard error (SE) of 3 separate experiments. **, p<0.010; ***, p< (n=3) 25
38 A) B) Figure 13: A) Tubulin immunofluorescent images of RAW murine macrophage. Macrophages cells show morphological changes and cytoskeleton immunostaining at 24 hours exposing to OMP-A from gram-negative bacteria (Scale bar =20 μm). B) Tubulin immunofluorescent intensity for control and (1,5 and 10 μg/ml) of OMP-A from gram-negative bacteria following 24 hours.. Each value represents the mean ± standard error (SE) of 3 separate experiments. *, p<0.05. (n=3) 26
39 A) B) Figure 14: A) Tubulin immunofluorescent images of RAW murine macrophage. Macrophages cells show morphological changes and cytoskeleton immunostaining at 24 hours exposing to peptidoglycan from gram-negative bacteria (Scale bar =20 μm). B) Tubulin immunofluorescent intensity for control and (1,5 and 10 μg/ml) of peptidoglycan from gram-negative bacteria following 24 hours. Each value represents the mean ± standard error (SE) of 3 separate experiments. (n=3) 27
40 A) B) Figure 15: A) F-actin immunofluorescent images of RAW murine macrophage. Macrophages cells show morphological changes and cytoskeleton immunostaining at 24 hours exposing to Flagellin from gram-negative bacteria (Scale bar =20 μm). B) F-actin immunofluorescent intensity for control and (1,5 and 10 μg/ml) of flagellin from gram-negative bacteria following 24 hours. Each value represents the mean ± standard error (SE) of 3 separate experiments. (n=3) 28
41 A) B) Figure 16: A) F-actin immunofluorescent images of RAW murine macrophage. Macrophages cells show morphological changes and cytoskeleton immunostaining at 24 hours exposing to lipoprotein from gram-negative bacteria (Scale bar =20 μm). B) F-actin immunofluorescent intensity for control and (1,5 and 10 μg/ml) of lipoprotein from gram-negative bacteria following 24 hours. Each value represents the mean ± standard error (SE) of 3 separate experiments. **, p< (n=3) 29
42 A) B) Figure 17: A) F-actin immunofluorescent images of RAW murine macrophage. Macrophages cells show morphological changes and cytoskeleton immunostaining at 24 hours exposing to LPS from gram-negative bacteria (Scale bar =20 μm). B) F-actin immunofluorescent intensity for control and (1,5 and 10 μg/ml) of LPS from gramnegative bacteria following 24 hours. Each value represents the mean ± standard error (SE) of 3 separate experiments. ***, p< (n=3) 30
43 A) B) Figure 18: A) F-actin immunofluorescent images of RAW murine macrophage. Macrophages cells show morphological changes and cytoskeleton immunostaining at 24 hours exposing to OMP-A from gram-negative bacteria (Scale bar =20 μm). B) F-actin immunofluorescent intensity for control and (1,5 and 10 μg/ml) of OMP-A from gram-negative bacteria following 24 hours. Each value represents the mean ± standard error (SE) of 3 separate experiments. *, p<0.05. (n=3) 31
44 A) B) Figure 19: A) F-actin immunofluorescent images of RAW murine macrophage. Macrophages cells show morphological changes and cytoskeleton immunostaining at 24 hours exposing to peptidoglycan from gram-negative bacteria (Scale bar =20 μm). B) F-actin immunofluorescent intensity for control and (1,5 and 10 μg/ml) of peptidoglycan from gram-negative bacteria following 24 hours. Each value represents the mean ± standard error (SE) of 3 separate experiments. (n=3) 32
45 Figure 20: Graph shows RAW murine macrophage have been respond to gram-negative bacterial components (LPS, OMP-A, Flagellin, Lipoprotein, Peptidoglycan) by releasing IL-1b. Figure 21: Graph shows RAW murine macrophage have been respond to gram-negative bacterial compounds (LPS, OMP-A, Flagellin, Lipoprotein, Peptidoglycan) by releasing IL
46 Figure 22: Graph shows RAW murine macrophage have been respond to gram-negative bacterial compounds (LPS, OMP-A, Flagellin, Lipoprotein, Peptidoglycan) by releasing IL-12. Figure 23: Graph shows RAW murine macrophage have been respond to gram-negative bacterial compounds (LPS, OMP-A, Flagellin, Lipoprotein, Peptidoglycan) by releasing TNF-a. 34
47 Figure 24: Graph shows RAW murine macrophage have been respond to gram-negative bacterial compounds (OMP-A, Flagellin, Lipoprotein, Peptidoglycan) by releasing TNF-α. 35
48 DISCUSSION AND FUTURE STUDIES In this study, responses of murine RAW macrophages to substances displayed on the outer surfaces of gram-negative bacteria: LPS, lipoprotein, peptidoglycan, outer membrane protein A (OMP-A) and flagellin were studied. These materials did not cause a dramatic alteration in cell morphology except for LPS; LPS-stimulated cells appeared flattened and elongated (as shown in figure 25). Figure 25: RAW murine macrophage exhibited morphological changes after interaction with LPS through TLR4. The main objective of this study was to examine the cytoskeleton reorganization of RAW macrophages exposed to these gram-negative substances. The first response of LPS- stimulated macrophages is their migration to the sites of the infection (Duque and Descoteaux,2015). Tubulin filaments and F-actin were examined, and the amounts of fluorescent intensity staining of the tubulin and F-actin microfilaments were estimated by quantifying, using Image J analysis. LPS exerted the greatest effects on rearrangement of the cellular actin as the cells became flattened and irregularly shaped compared to the rounded morphology displayed by uninfected macrophages as 36
49 shown in Figure12A and 17A and the graphs show high level of tubulin and F-actin that are increased compared to the control as shown in Figure 12B and 17B. Also, OMP-A caused an effect on the cytoskeleton, as noted by the F- actin staining, at 5 μg/ml as shown in Figure 18 while the lipoprotein decreased staining of F-actin at 10 μg/ml compared to that seen in unstimulated control cells as shown in Figure 16. Upon exposure to LPS, innate immune system responds using the TLR4 pathway in macrophage. At downstream signaling, lipid A is a portion of LPS that generate the signaling to secrete pro-inflammatory cytokines such as TNF-α and IL1β (Katsuaki et al, 2016). Significant reductions in cell viability compared to unstimulated control cells were seen at 24 hours after exposure to 1, 5 and 10 μg/ml of LPS. Similar to the observation of Zhuang (1997), decreased cell viability was seen in cells stimulated with LPS compared to unstimulated cells after 24 hours. These observations suggest that activation of TLRs leading to production of inflammatory cytokines such as TNF-α may account for these decreases in cell viability; LPS activates TLR4 as shown in Figures 7 and 23. Hayashi et al. (2016) found that fagellin from Gram-negative bacteria stimulates RAW macrophages through TLR5. Triggering of the TLR5 receptor activates NF-kB and stimulates TNF-α production. In this study, none of the other substances tested caused decreases in cell viability except LPS and flagellin. Flagellin activates TLR5 as shown in Figures 8 and 24. Others have noted the decreased viability of cells expressing TNF-α (Wang et al., 2011). In future studies, one of our limitation on this study is the cell line which is RAW murine macrophage so it would be beneficial to study the effect of gram-negative bacterial components on another cell line such as J774A.1 macrophage phenotypes and human peripheral blood macrophages to compare the results of those cell line. It would be helpful to study the effects on viability, cytoskeleton filaments and cytokines expression on macrophages at 4, 8, 16 hours after interaction with gram-negative bacterial components with 1,5 and 10 μg/ml to understand the outcomes of the primary and immediate stages of infection. Also, it would be beneficial to expose RAW264.7 macrophages to flagellated and deflagellated E. coli to study the role of flagella on cell viability and secretion of cytokines from macrophage after exposing to those bacteria. Based on the results of this study, we predict that 37
Innate Immunity. Chapter 3. Connection Between Innate and Adaptive Immunity. Know Differences and Provide Examples. Antimicrobial peptide psoriasin
 Chapter Know Differences and Provide Examples Innate Immunity kin and Epithelial Barriers Antimicrobial peptide psoriasin -Activity against Gram (-) E. coli Connection Between Innate and Adaptive Immunity
Chapter Know Differences and Provide Examples Innate Immunity kin and Epithelial Barriers Antimicrobial peptide psoriasin -Activity against Gram (-) E. coli Connection Between Innate and Adaptive Immunity
Toll-like Receptor Signaling
 Toll-like Receptor Signaling 1 Professor of Medicine University of Massachusetts Medical School, Worcester, MA, USA Why do we need innate immunity? Pathogens multiply very fast We literally swim in viruses
Toll-like Receptor Signaling 1 Professor of Medicine University of Massachusetts Medical School, Worcester, MA, USA Why do we need innate immunity? Pathogens multiply very fast We literally swim in viruses
Chapter 3 The Induced Responses of Innate Immunity
 Chapter 3 The Induced Responses of Innate Immunity Pattern recognition by cells of the innate immune system Pattern recognition by cells of the innate immune system 4 main pattern recognition receptors
Chapter 3 The Induced Responses of Innate Immunity Pattern recognition by cells of the innate immune system Pattern recognition by cells of the innate immune system 4 main pattern recognition receptors
Newly Recognized Components of the Innate Immune System
 Newly Recognized Components of the Innate Immune System NOD Proteins: Intracellular Peptidoglycan Sensors NOD-1 NOD-2 Nod Protein LRR; Ligand Recognition CARD RICK I-κB p50 p65 NF-κB Polymorphisms in Nod-2
Newly Recognized Components of the Innate Immune System NOD Proteins: Intracellular Peptidoglycan Sensors NOD-1 NOD-2 Nod Protein LRR; Ligand Recognition CARD RICK I-κB p50 p65 NF-κB Polymorphisms in Nod-2
Innate Immunity & Inflammation
 Innate Immunity & Inflammation The innate immune system is an evolutionally conserved mechanism that provides an early and effective response against invading microbial pathogens. It relies on a limited
Innate Immunity & Inflammation The innate immune system is an evolutionally conserved mechanism that provides an early and effective response against invading microbial pathogens. It relies on a limited
Cell Structure. Morphology of Prokaryotic Cell. Cytoplasmic Membrane 4/6/2011. Chapter 3. Cytoplasmic membrane
 Cell Structure Chapter 3 Morphology of Prokaryotic Cell Cytoplasmic membrane Delicate thin fluid structure Surrounds cytoplasm of cell Defines boundary Defines boundary Serves as a selectively permeable
Cell Structure Chapter 3 Morphology of Prokaryotic Cell Cytoplasmic membrane Delicate thin fluid structure Surrounds cytoplasm of cell Defines boundary Defines boundary Serves as a selectively permeable
Role of Innate Immunity in Control of Adaptive Immunity
 Role of Innate Immunity in Control of Adaptive Immunity Innate Immunity The burden of pathogen sensing is placed on the innate immune system Danger hypothesis Missing Self Based on the detection of molecular
Role of Innate Immunity in Control of Adaptive Immunity Innate Immunity The burden of pathogen sensing is placed on the innate immune system Danger hypothesis Missing Self Based on the detection of molecular
Innate Immunity. Connection Between Innate and Adaptive Immunity. Know Differences and Provide Examples Chapter 3. Antimicrobial peptide psoriasin
 Know Differences and Provide Examples Chapter * Innate Immunity * kin and Epithelial Barriers * Antimicrobial peptide psoriasin -Activity against Gram (-) E. coli Connection Between Innate and Adaptive
Know Differences and Provide Examples Chapter * Innate Immunity * kin and Epithelial Barriers * Antimicrobial peptide psoriasin -Activity against Gram (-) E. coli Connection Between Innate and Adaptive
2. Innate immunity 2013
 1 Innate Immune Responses 3 Innate immunity Abul K. Abbas University of California San Francisco The initial responses to: 1. Microbes: essential early mechanisms to prevent, control, or eliminate infection;
1 Innate Immune Responses 3 Innate immunity Abul K. Abbas University of California San Francisco The initial responses to: 1. Microbes: essential early mechanisms to prevent, control, or eliminate infection;
Identification of Microbes
 Identification of Microbes Recognition by PRR (pattern recognition receptors) Recognize conserved molecular patterns on microbes called pathogen associated molecular patterns (PAMPs) which are not present
Identification of Microbes Recognition by PRR (pattern recognition receptors) Recognize conserved molecular patterns on microbes called pathogen associated molecular patterns (PAMPs) which are not present
Structure and Function of Antigen Recognition Molecules
 MICR2209 Structure and Function of Antigen Recognition Molecules Dr Allison Imrie allison.imrie@uwa.edu.au 1 Synopsis: In this lecture we will examine the major receptors used by cells of the innate and
MICR2209 Structure and Function of Antigen Recognition Molecules Dr Allison Imrie allison.imrie@uwa.edu.au 1 Synopsis: In this lecture we will examine the major receptors used by cells of the innate and
A.Kavitha Assistant professor Department of Botany RBVRR Womens college
 A.Kavitha Assistant professor Department of Botany RBVRR Womens college The Ultrastructure Of A Typical Bacterial Cell The Bacterial Cell This is a diagram of a typical bacterial cell, displaying all of
A.Kavitha Assistant professor Department of Botany RBVRR Womens college The Ultrastructure Of A Typical Bacterial Cell The Bacterial Cell This is a diagram of a typical bacterial cell, displaying all of
Innate Immunity. Hathairat Thananchai, DPhil Department of Microbiology Faculty of Medicine Chiang Mai University 2 August 2016
 Innate Immunity Hathairat Thananchai, DPhil Department of Microbiology Faculty of Medicine Chiang Mai University 2 August 2016 Objectives: Explain how innate immune system recognizes foreign substances
Innate Immunity Hathairat Thananchai, DPhil Department of Microbiology Faculty of Medicine Chiang Mai University 2 August 2016 Objectives: Explain how innate immune system recognizes foreign substances
Bacterial Structures. Capsule or Glycocalyx TYPES OF FLAGELLA FLAGELLA. Average size: µm 2-8 µm Basic shapes:
 PROKARYOTIC One circular chromosome, not in a membrane No histones No organelles Peptidoglycan cell walls Binary fission EUKARYOTIC Paired chromosomes, in nuclear membrane Histones Organelles Polysaccharide
PROKARYOTIC One circular chromosome, not in a membrane No histones No organelles Peptidoglycan cell walls Binary fission EUKARYOTIC Paired chromosomes, in nuclear membrane Histones Organelles Polysaccharide
Lecture on Innate Immunity and Inflammation
 Lecture on Innate Immunity and Inflammation Evolutionary View Epithelial barriers to infection Four main types of innate recognition molecules:tlrs, CLRs, NLRs, RLRs NF-κB, the master transcriptional regulator
Lecture on Innate Immunity and Inflammation Evolutionary View Epithelial barriers to infection Four main types of innate recognition molecules:tlrs, CLRs, NLRs, RLRs NF-κB, the master transcriptional regulator
The Innate Immune Response is Conserved Throughout Evolution and is Triggered by Pattern Recognition. Lipopolysaccharide = Lipid + Polysaccharide
 The Innate Immune Response is Conserved Throughout Evolution and is Triggered by Pattern Recognition Lipopolysaccharide = Lipid + Polysaccharide E.coli Cell wall organization Lipopolysaccharide Outer membrane
The Innate Immune Response is Conserved Throughout Evolution and is Triggered by Pattern Recognition Lipopolysaccharide = Lipid + Polysaccharide E.coli Cell wall organization Lipopolysaccharide Outer membrane
Immunology Part II. Innate Immunity. 18. April 2018, Ruhr-Universität Bochum Marcus Peters,
 Immunology Part II Innate Immunity 18. April 2018, Ruhr-Universität Bochum Marcus Peters, marcus.peters@rub.de Conserved structures of pathogens PAMPs are detected by Pattern Recognition Receptors PRRs
Immunology Part II Innate Immunity 18. April 2018, Ruhr-Universität Bochum Marcus Peters, marcus.peters@rub.de Conserved structures of pathogens PAMPs are detected by Pattern Recognition Receptors PRRs
Lecture on Innate Immunity and Inflammation. Innate Immunity: An Evolutionary View
 Lecture on Innate Immunity and Inflammation Evolutionary View Epithelial barriers to infection Four main types of innate recognition molecules:tlrs, CLRs, NLRs, RLRs NF-κB, the master transcriptional regulator
Lecture on Innate Immunity and Inflammation Evolutionary View Epithelial barriers to infection Four main types of innate recognition molecules:tlrs, CLRs, NLRs, RLRs NF-κB, the master transcriptional regulator
Innate immunity. Abul K. Abbas University of California San Francisco. FOCiS
 1 Innate immunity Abul K. Abbas University of California San Francisco FOCiS 2 Lecture outline Components of innate immunity Recognition of microbes and dead cells Toll Like Receptors NOD Like Receptors/Inflammasome
1 Innate immunity Abul K. Abbas University of California San Francisco FOCiS 2 Lecture outline Components of innate immunity Recognition of microbes and dead cells Toll Like Receptors NOD Like Receptors/Inflammasome
3/10/14. Ultrastructural organization. Gram Stain. Infection leads to production of inducers of inflammation. Gram negative.
 Infection leads to production of inducers of inflammation or dendritic cell Inflammatory mediators: Complex and many, but include: Lipids and Proteins (cytokines/chemokines) TNF Others Ultrastructural
Infection leads to production of inducers of inflammation or dendritic cell Inflammatory mediators: Complex and many, but include: Lipids and Proteins (cytokines/chemokines) TNF Others Ultrastructural
Prokaryotic Cell Structure
 Prokaryotic Cell Structure Chapter 3 Prokaryotes vs Eukaryotes DNA Prokaryotes Eukaryotes Organelles Size & Organization Kingdoms 1 Where do viruses fit in? Acellular microorganisms Cannot reproduce outside
Prokaryotic Cell Structure Chapter 3 Prokaryotes vs Eukaryotes DNA Prokaryotes Eukaryotes Organelles Size & Organization Kingdoms 1 Where do viruses fit in? Acellular microorganisms Cannot reproduce outside
Prokaryotic Cell Structure
 Prokaryotic Cell Structure Chapter 3 Prokaryotes vs Eukaryotes DNA Prokaryotes Eukaryotes Organelles Size & Organization Kingdoms Where do viruses fit in? Acellular microorganisms Cannot reproduce outside
Prokaryotic Cell Structure Chapter 3 Prokaryotes vs Eukaryotes DNA Prokaryotes Eukaryotes Organelles Size & Organization Kingdoms Where do viruses fit in? Acellular microorganisms Cannot reproduce outside
Chapter 4 Prokaryotic Profiles
 Chapter 4 Prokaryotic Profiles Topics: External Structures Cell Envelope Internal Structures Cell Shapes, Arrangement, and Sizes Prokaryotes are unicellular organisms Prokaryotes include two small groups
Chapter 4 Prokaryotic Profiles Topics: External Structures Cell Envelope Internal Structures Cell Shapes, Arrangement, and Sizes Prokaryotes are unicellular organisms Prokaryotes include two small groups
Journal club. Lama Nazzal
 Journal club Lama Nazzal Background Kidney stone disease affects about 12% of men and 5% of women during their lifetimes in the United States Intrarenal nephrocalcinosis is often asymptomatic, but can
Journal club Lama Nazzal Background Kidney stone disease affects about 12% of men and 5% of women during their lifetimes in the United States Intrarenal nephrocalcinosis is often asymptomatic, but can
Ch 4. Functional Anatomy of Prokaryotic and Eukaryotic Cells
 Ch 4 Functional Anatomy of Prokaryotic and Eukaryotic Cells Objectives Compare and contrast the overall cell structure of prokaryotes and eukaryotes. Identify the three basic shapes of bacteria. Describe
Ch 4 Functional Anatomy of Prokaryotic and Eukaryotic Cells Objectives Compare and contrast the overall cell structure of prokaryotes and eukaryotes. Identify the three basic shapes of bacteria. Describe
Glycosaminoglycans: Anionic polysaccharide chains made of repeating disaccharide units
 Glycosaminoglycans: Anionic polysaccharide chains made of repeating disaccharide units Glycosaminoglycans present on the animal cell surface and in the extracellular matrix. Glycoseaminoglycans (mucopolysaccharides)
Glycosaminoglycans: Anionic polysaccharide chains made of repeating disaccharide units Glycosaminoglycans present on the animal cell surface and in the extracellular matrix. Glycoseaminoglycans (mucopolysaccharides)
TOLL-LIKE RECEPTORS AND CYTOKINES IN SEPSIS
 TOLL-LIKE RECEPTORS AND CYTOKINES IN SEPSIS A/PROF WILLIAM SEWELL ST VINCENT S CLINICAL SCHOOL, UNSW SYDPATH, ST VINCENT S HOSPITAL SYDNEY GARVAN INSTITUTE INNATE VERSUS ADAPTIVE IMMUNE RESPONSES INNATE
TOLL-LIKE RECEPTORS AND CYTOKINES IN SEPSIS A/PROF WILLIAM SEWELL ST VINCENT S CLINICAL SCHOOL, UNSW SYDPATH, ST VINCENT S HOSPITAL SYDNEY GARVAN INSTITUTE INNATE VERSUS ADAPTIVE IMMUNE RESPONSES INNATE
10/13/11. Cell Theory. Cell Structure
 Cell Structure Grade 12 Biology Cell Theory All organisms are composed of one or more cells. Cells are the smallest living units of all living organisms. Cells arise only by division of a previously existing
Cell Structure Grade 12 Biology Cell Theory All organisms are composed of one or more cells. Cells are the smallest living units of all living organisms. Cells arise only by division of a previously existing
Microbiology for Environmental Health Officers. EHL0033 Prokaryotes 3
 Microbiology for Environmental Health Officers EHL0033 Prokaryotes 3 Mutualism: bacterial headlights. The glowing oval below the eye of the flashlight fish (Photoblepharon palpebratus) is an organ harboring
Microbiology for Environmental Health Officers EHL0033 Prokaryotes 3 Mutualism: bacterial headlights. The glowing oval below the eye of the flashlight fish (Photoblepharon palpebratus) is an organ harboring
D2 inhibits TLR2- initiated 12p40 transcription (-) TLR2 PGN MDP. MyD88 IRAK ECSIT TRAF6 NIK. Smallest unit of PGN muramyl dipeptide IKK.
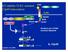 D2 inhibits TLR2- initiated 12p40 transcription CARD CARD NOD2 LRR RICK/Rip2 NIK MDP TRAF6 PGN TLR2 MyD88 IRAK ECSIT (-) IKK Smallest unit of PGN muramyl dipeptide IκB NF-κB atanabe et al, 2004 NF-κB IL-12p40
D2 inhibits TLR2- initiated 12p40 transcription CARD CARD NOD2 LRR RICK/Rip2 NIK MDP TRAF6 PGN TLR2 MyD88 IRAK ECSIT (-) IKK Smallest unit of PGN muramyl dipeptide IκB NF-κB atanabe et al, 2004 NF-κB IL-12p40
Fig. LPS in Gram negative bacteria
 Structure of bacterial cell Dentistry college - first class Medical biology- Lec.3 Lecturer D. Hanan S A- Cell wall ***Chemical composition of the cell wall Bacteria are divided into two separated groups
Structure of bacterial cell Dentistry college - first class Medical biology- Lec.3 Lecturer D. Hanan S A- Cell wall ***Chemical composition of the cell wall Bacteria are divided into two separated groups
T-cell activation T cells migrate to secondary lymphoid tissues where they interact with antigen, antigen-presenting cells, and other lymphocytes:
 Interactions between innate immunity & adaptive immunity What happens to T cells after they leave the thymus? Naïve T cells exit the thymus and enter the bloodstream. If they remain in the bloodstream,
Interactions between innate immunity & adaptive immunity What happens to T cells after they leave the thymus? Naïve T cells exit the thymus and enter the bloodstream. If they remain in the bloodstream,
T-cell activation T cells migrate to secondary lymphoid tissues where they interact with antigen, antigen-presenting cells, and other lymphocytes:
 Interactions between innate immunity & adaptive immunity What happens to T cells after they leave the thymus? Naïve T cells exit the thymus and enter the bloodstream. If they remain in the bloodstream,
Interactions between innate immunity & adaptive immunity What happens to T cells after they leave the thymus? Naïve T cells exit the thymus and enter the bloodstream. If they remain in the bloodstream,
Classification of Infectious Agents. Dr W. D. Colby
 Classification of Infectious Agents Dr W. D. Colby Nonliving Infectious Agents PRIONS: abnormally configured self-replicating protein templates VIRUSES: nucleic acid (DNA or RNA) genes packaged in protein
Classification of Infectious Agents Dr W. D. Colby Nonliving Infectious Agents PRIONS: abnormally configured self-replicating protein templates VIRUSES: nucleic acid (DNA or RNA) genes packaged in protein
CYTOKINE RECEPTORS AND SIGNAL TRANSDUCTION
 CYTOKINE RECEPTORS AND SIGNAL TRANSDUCTION What is Cytokine? Secreted popypeptide (protein) involved in cell-to-cell signaling. Acts in paracrine or autocrine fashion through specific cellular receptors.
CYTOKINE RECEPTORS AND SIGNAL TRANSDUCTION What is Cytokine? Secreted popypeptide (protein) involved in cell-to-cell signaling. Acts in paracrine or autocrine fashion through specific cellular receptors.
CORE Scholar. Wright State University. Mubarak Almutairi Wright State University
 Wright State University CORE Scholar Browse all Theses and Dissertations Theses and Dissertations 2016 The Impact of HSV-1 Infection, SOCS1 Peptide, and SOCS3 Peptide Mimetic on Cell Viability, Morphology,
Wright State University CORE Scholar Browse all Theses and Dissertations Theses and Dissertations 2016 The Impact of HSV-1 Infection, SOCS1 Peptide, and SOCS3 Peptide Mimetic on Cell Viability, Morphology,
CELLS. Cells. Basic unit of life (except virus)
 Basic unit of life (except virus) CELLS Prokaryotic, w/o nucleus, bacteria Eukaryotic, w/ nucleus Various cell types specialized for particular function. Differentiation. Over 200 human cell types 56%
Basic unit of life (except virus) CELLS Prokaryotic, w/o nucleus, bacteria Eukaryotic, w/ nucleus Various cell types specialized for particular function. Differentiation. Over 200 human cell types 56%
Structures in Cells. Cytoplasm. Lecture 5, EH1008: Biology for Public Health, Biomolecules
 Structures in Cells Lecture 5, EH1008: Biology for Public Health, Biomolecules Limian.zheng@ucc.ie 1 Cytoplasm Nucleus Centrioles Cytoskeleton Cilia Microvilli 2 Cytoplasm Cellular material outside nucleus
Structures in Cells Lecture 5, EH1008: Biology for Public Health, Biomolecules Limian.zheng@ucc.ie 1 Cytoplasm Nucleus Centrioles Cytoskeleton Cilia Microvilli 2 Cytoplasm Cellular material outside nucleus
The Effects of HSV-1 Challenge on Polarized Murine Macrophages: an In Vitro Model Using the J774A.1 Murine Macrophage Cell Line
 Wright State University CORE Scholar Browse all Theses and Dissertations Theses and Dissertations 2012 The Effects of HSV-1 Challenge on Polarized Murine Macrophages: an In Vitro Model Using the J774A.1
Wright State University CORE Scholar Browse all Theses and Dissertations Theses and Dissertations 2012 The Effects of HSV-1 Challenge on Polarized Murine Macrophages: an In Vitro Model Using the J774A.1
! gives mechanical strength to the cell and protects it from exploding due to osmotic lysis (shape and strength due to the peptidoglycan)
 Cell Wall! The cell wall is a rigid structure that surrounds the bacterial cell just outside of the plasma membrane Functions to:! gives the bacterium its shape! gives mechanical strength to the cell and
Cell Wall! The cell wall is a rigid structure that surrounds the bacterial cell just outside of the plasma membrane Functions to:! gives the bacterium its shape! gives mechanical strength to the cell and
Structures in Cells. Lecture 5, EH1008: Biology for Public Health, Biomolecules.
 Structures in Cells Lecture 5, EH1008: Biology for Public Health, Biomolecules Limian.zheng@ucc.ie 1 Cytoplasm Nucleus Centrioles Cytoskeleton Cilia Microvilli 2 Cytoplasm Cellular material outside nucleus
Structures in Cells Lecture 5, EH1008: Biology for Public Health, Biomolecules Limian.zheng@ucc.ie 1 Cytoplasm Nucleus Centrioles Cytoskeleton Cilia Microvilli 2 Cytoplasm Cellular material outside nucleus
Relative sizes of infectious agents
 Relative sizes of infectious agents Bacteria Protozoa Viruses RBC 0.005 0.01 0.03 01 03 05 1 3 5 10 30 50 100 300 Size in microns ( µm ) - log scale Immunity to Infection Principle 1 Every clinical infection
Relative sizes of infectious agents Bacteria Protozoa Viruses RBC 0.005 0.01 0.03 01 03 05 1 3 5 10 30 50 100 300 Size in microns ( µm ) - log scale Immunity to Infection Principle 1 Every clinical infection
Chapter 13: Cytokines
 Chapter 13: Cytokines Definition: secreted, low-molecular-weight proteins that regulate the nature, intensity and duration of the immune response by exerting a variety of effects on lymphocytes and/or
Chapter 13: Cytokines Definition: secreted, low-molecular-weight proteins that regulate the nature, intensity and duration of the immune response by exerting a variety of effects on lymphocytes and/or
The Cell Membrane (Ch. 7)
 The Cell Membrane (Ch. 7) Phospholipids Phosphate head hydrophilic Fatty acid tails hydrophobic Arranged as a bilayer Phosphate attracted to water Fatty acid repelled by water Aaaah, one of those structure
The Cell Membrane (Ch. 7) Phospholipids Phosphate head hydrophilic Fatty acid tails hydrophobic Arranged as a bilayer Phosphate attracted to water Fatty acid repelled by water Aaaah, one of those structure
Innate Immunity. Hathairat Thananchai, DPhil Department of Microbiology Faculty of Medicine Chiang Mai University 25 July 2017
 Innate Immunity Hathairat Thananchai, DPhil Department of Microbiology Faculty of Medicine Chiang Mai University 25 July 2017 Objectives: Explain how innate immune system recognizes foreign substances
Innate Immunity Hathairat Thananchai, DPhil Department of Microbiology Faculty of Medicine Chiang Mai University 25 July 2017 Objectives: Explain how innate immune system recognizes foreign substances
Eukaryotic cell. Premedical IV Biology
 Eukaryotic cell Premedical IV Biology The size range of organisms Light microscopes visible light is passed through the specimen and glass lenses the resolution is limited by the wavelength of the visible
Eukaryotic cell Premedical IV Biology The size range of organisms Light microscopes visible light is passed through the specimen and glass lenses the resolution is limited by the wavelength of the visible
Cell Structure and Function
 Cell Structure and Function Cell Structure and Function Topics External Structures Cell Envelope Internal Structures Cell Shapes, Arrangement, and Sizes Classification An Infectious Exam Patient with Tuberculosis
Cell Structure and Function Cell Structure and Function Topics External Structures Cell Envelope Internal Structures Cell Shapes, Arrangement, and Sizes Classification An Infectious Exam Patient with Tuberculosis
8/7/18. UNIT 2: Cells Chapter 3: Cell Structure and Function. I. Cell Theory (3.1) A. Early studies led to the development of the cell theory
 8/7/18 UNIT 2: Cells Chapter 3: Cell Structure and Function I. Cell Theory (3.1) A. Early studies led to the development of the cell theory 1. Discovery of Cells a. Robert Hooke (1665)-Used compound microscope
8/7/18 UNIT 2: Cells Chapter 3: Cell Structure and Function I. Cell Theory (3.1) A. Early studies led to the development of the cell theory 1. Discovery of Cells a. Robert Hooke (1665)-Used compound microscope
it selectively allows some molecules to pass into the organism
 Multiple Choice Quiz Procaryotic Cell Structure and Function Eucaryotic Cell Structure and Function Choose the best answer 1. The significance of the plasma membrane is that it selectively allows some
Multiple Choice Quiz Procaryotic Cell Structure and Function Eucaryotic Cell Structure and Function Choose the best answer 1. The significance of the plasma membrane is that it selectively allows some
JPEMS Nantes, Basic Immunology INNATE IMMUNITY
 JPEMS Nantes, 2014- Basic Immunology INNATE IMMUNITY Teacher: Pr. Régis Josien, Laboratoire d Immunologie and INSERM U1064, CHU Nantes Regis.Josien@univ-nantes.fr 1 Contents 1. General features and specificity
JPEMS Nantes, 2014- Basic Immunology INNATE IMMUNITY Teacher: Pr. Régis Josien, Laboratoire d Immunologie and INSERM U1064, CHU Nantes Regis.Josien@univ-nantes.fr 1 Contents 1. General features and specificity
FOR OPTIMAL GUT HEALTH KEMIN.COM/GUTHEALTH
 FOR OPTIMAL GUT HEALTH KEMIN.COM/GUTHEALTH ALETA A SOURCE OF 1,3-BETA GLUCANS Aleta is highly bioavailable, offering a concentration greater than 5% of 1,3-beta glucans. Aleta provides a consistent response
FOR OPTIMAL GUT HEALTH KEMIN.COM/GUTHEALTH ALETA A SOURCE OF 1,3-BETA GLUCANS Aleta is highly bioavailable, offering a concentration greater than 5% of 1,3-beta glucans. Aleta provides a consistent response
Innate Immunity: Nonspecific Defenses of the Host
 PowerPoint Lecture Presentations prepared by Bradley W. Christian, McLennan Community College C H A P T E R 16 Innate Immunity: Nonspecific Defenses of the Host Host Response to Disease Resistance- ability
PowerPoint Lecture Presentations prepared by Bradley W. Christian, McLennan Community College C H A P T E R 16 Innate Immunity: Nonspecific Defenses of the Host Host Response to Disease Resistance- ability
Overview of the Immune System
 Overview of the Immune System Immune System Innate (Nonspecific) Adaptive (Specific) Cellular Components Humoral Components Cell-Mediated Humoral (Ab) Antigens Definitions Immunogen Antigen (Ag) Hapten
Overview of the Immune System Immune System Innate (Nonspecific) Adaptive (Specific) Cellular Components Humoral Components Cell-Mediated Humoral (Ab) Antigens Definitions Immunogen Antigen (Ag) Hapten
number Done by Corrected by Doctor Dr. Hamed Al Zoubi
 number Done by Corrected by 46 2017/9/20 Doctor Dr. Hamed Al Zoubi 66 /8486535 مركز الرائد للخدمات الطالبية 66 /8486535 مركز الرائد للخدمات الطالبية 2 nd year Medical Students JU Bacterial Structure and
number Done by Corrected by 46 2017/9/20 Doctor Dr. Hamed Al Zoubi 66 /8486535 مركز الرائد للخدمات الطالبية 66 /8486535 مركز الرائد للخدمات الطالبية 2 nd year Medical Students JU Bacterial Structure and
Structure of Prokaryotic & Eukaryotic Cells
 Structure of Prokaryotic & Eukaryotic Cells Review of Prokaryotic & Eukaryotic Cells Nucleus vs nucleoid DNA : circular vs linear, presence of histones Membranous organelles Cell wall-peptidoglycan Cell
Structure of Prokaryotic & Eukaryotic Cells Review of Prokaryotic & Eukaryotic Cells Nucleus vs nucleoid DNA : circular vs linear, presence of histones Membranous organelles Cell wall-peptidoglycan Cell
CELL BIOLOGY - CLUTCH CH THE IMMUNE SYSTEM.
 !! www.clutchprep.com CONCEPT: OVERVIEW OF HOST DEFENSES The human body contains three lines of against infectious agents (pathogens) 1. Mechanical and chemical boundaries (part of the innate immune system)
!! www.clutchprep.com CONCEPT: OVERVIEW OF HOST DEFENSES The human body contains three lines of against infectious agents (pathogens) 1. Mechanical and chemical boundaries (part of the innate immune system)
Innate immune regulation of T-helper (Th) cell homeostasis in the intestine
 Innate immune regulation of T-helper (Th) cell homeostasis in the intestine Masayuki Fukata, MD, Ph.D. Research Scientist II Division of Gastroenterology, Department of Medicine, F. Widjaja Foundation,
Innate immune regulation of T-helper (Th) cell homeostasis in the intestine Masayuki Fukata, MD, Ph.D. Research Scientist II Division of Gastroenterology, Department of Medicine, F. Widjaja Foundation,
Nucleic acids. Nucleic acids are information-rich polymers of nucleotides
 Nucleic acids Nucleic acids are information-rich polymers of nucleotides DNA and RNA Serve as the blueprints for proteins and thus control the life of a cell RNA and DNA are made up of very similar nucleotides.
Nucleic acids Nucleic acids are information-rich polymers of nucleotides DNA and RNA Serve as the blueprints for proteins and thus control the life of a cell RNA and DNA are made up of very similar nucleotides.
NOD1 contributes to mouse host defense against Helicobacter pylori via induction of type I IFN and activation of the ISGF3 signaling pathway
 Research article NOD1 contributes to mouse host defense against Helicobacter pylori via induction of type I IFN and activation of the ISGF3 signaling pathway Tomohiro Watanabe, 1,2 Naoki Asano, 1 Stefan
Research article NOD1 contributes to mouse host defense against Helicobacter pylori via induction of type I IFN and activation of the ISGF3 signaling pathway Tomohiro Watanabe, 1,2 Naoki Asano, 1 Stefan
The Cell. Biology 105 Lecture 4 Reading: Chapter 3 (pages 47 62)
 The Cell Biology 105 Lecture 4 Reading: Chapter 3 (pages 47 62) Outline I. Prokaryotic vs. Eukaryotic II. Eukaryotic A. Plasma membrane transport across B. Main features of animal cells and their functions
The Cell Biology 105 Lecture 4 Reading: Chapter 3 (pages 47 62) Outline I. Prokaryotic vs. Eukaryotic II. Eukaryotic A. Plasma membrane transport across B. Main features of animal cells and their functions
Immune System AP SBI4UP
 Immune System AP SBI4UP TYPES OF IMMUNITY INNATE IMMUNITY ACQUIRED IMMUNITY EXTERNAL DEFENCES INTERNAL DEFENCES HUMORAL RESPONSE Skin Phagocytic Cells CELL- MEDIATED RESPONSE Mucus layer Antimicrobial
Immune System AP SBI4UP TYPES OF IMMUNITY INNATE IMMUNITY ACQUIRED IMMUNITY EXTERNAL DEFENCES INTERNAL DEFENCES HUMORAL RESPONSE Skin Phagocytic Cells CELL- MEDIATED RESPONSE Mucus layer Antimicrobial
Intracellular MHC class II molecules promote TLR-triggered innate. immune responses by maintaining Btk activation
 Intracellular MHC class II molecules promote TLR-triggered innate immune responses by maintaining Btk activation Xingguang Liu, Zhenzhen Zhan, Dong Li, Li Xu, Feng Ma, Peng Zhang, Hangping Yao and Xuetao
Intracellular MHC class II molecules promote TLR-triggered innate immune responses by maintaining Btk activation Xingguang Liu, Zhenzhen Zhan, Dong Li, Li Xu, Feng Ma, Peng Zhang, Hangping Yao and Xuetao
Microbiology: A Systems Approach
 Microbiology: A Systems Approach First Edition Cowan & Talaro Chapter 4 Prokaryotic Profiles: the Bacteria and the Archaea Chapter 4 Fig. 4.1 3 3 parts flagella filament long, thin, helical structure composed
Microbiology: A Systems Approach First Edition Cowan & Talaro Chapter 4 Prokaryotic Profiles: the Bacteria and the Archaea Chapter 4 Fig. 4.1 3 3 parts flagella filament long, thin, helical structure composed
The Innate Immune Response is Conserved Throughout Evolution and is Triggered by Pattern Recognition. Lipopolysaccharide = Lipid + Polysaccharide
 The Innate Immune Response is onserved Throughout Evolution and is Triggered by Pattern Recognition Lipopolysaccharide = Lipid + Polysaccharide E.coli ell wall organization Lipopolysaccharide Outer membrane
The Innate Immune Response is onserved Throughout Evolution and is Triggered by Pattern Recognition Lipopolysaccharide = Lipid + Polysaccharide E.coli ell wall organization Lipopolysaccharide Outer membrane
Cells. Variation and Function of Cells
 Cells Variation and Function of Cells Cell Theory states that: 1. All living things are made of cells 2. Cells are the basic unit of structure and function in living things 3. New cells are produced from
Cells Variation and Function of Cells Cell Theory states that: 1. All living things are made of cells 2. Cells are the basic unit of structure and function in living things 3. New cells are produced from
Bio10 Cell Structure SRJC
 3.) Cell Structure and Function Structure of Cell Membranes Fluid mosaic model Mixed composition: Phospholipid bilayer Glycolipids Sterols Proteins Fluid Mosaic Model Phospholipids are not packed tightly
3.) Cell Structure and Function Structure of Cell Membranes Fluid mosaic model Mixed composition: Phospholipid bilayer Glycolipids Sterols Proteins Fluid Mosaic Model Phospholipids are not packed tightly
Innate Immunity. Natural or native immunity
 Innate Immunity 1 Innate Immunity Natural or native immunity 2 When microbes enter in the body 3 Secondly, it also stimulates the adaptive immune system 4 Immunologic memory 5 Components of Innate Immunity
Innate Immunity 1 Innate Immunity Natural or native immunity 2 When microbes enter in the body 3 Secondly, it also stimulates the adaptive immune system 4 Immunologic memory 5 Components of Innate Immunity
Topic 03 Prokaryotes (3.3)
 Topic 03 Prokaryotes (3.3) Topics Characteristics (comparison) External Structures Cell Envelope Internal Structures Cell Shapes, Arrangement, and Sizes Classification 1 Relative size of bacterial cell
Topic 03 Prokaryotes (3.3) Topics Characteristics (comparison) External Structures Cell Envelope Internal Structures Cell Shapes, Arrangement, and Sizes Classification 1 Relative size of bacterial cell
Innate Immunity. By Dr. Gouse Mohiddin Shaik
 Innate Immunity By Dr. Gouse Mohiddin Shaik Types of immunity Immunity Innate / inborn Non-specific Acquired / adaptive Specific 3rd line of defense Physical barriers Skin, Saliva, Mucous, Stomach acid,
Innate Immunity By Dr. Gouse Mohiddin Shaik Types of immunity Immunity Innate / inborn Non-specific Acquired / adaptive Specific 3rd line of defense Physical barriers Skin, Saliva, Mucous, Stomach acid,
Principles of Genetics and Molecular Biology
 Cell signaling Dr. Diala Abu-Hassan, DDS, PhD School of Medicine Dr.abuhassand@gmail.com Principles of Genetics and Molecular Biology www.cs.montana.edu Modes of cell signaling Direct interaction of a
Cell signaling Dr. Diala Abu-Hassan, DDS, PhD School of Medicine Dr.abuhassand@gmail.com Principles of Genetics and Molecular Biology www.cs.montana.edu Modes of cell signaling Direct interaction of a
ACTIVATION OF T LYMPHOCYTES AND CELL MEDIATED IMMUNITY
 ACTIVATION OF T LYMPHOCYTES AND CELL MEDIATED IMMUNITY The recognition of specific antigen by naïve T cell induces its own activation and effector phases. T helper cells recognize peptide antigens through
ACTIVATION OF T LYMPHOCYTES AND CELL MEDIATED IMMUNITY The recognition of specific antigen by naïve T cell induces its own activation and effector phases. T helper cells recognize peptide antigens through
Medical Virology Immunology. Dr. Sameer Naji, MB, BCh, PhD (UK) Head of Basic Medical Sciences Dept. Faculty of Medicine The Hashemite University
 Medical Virology Immunology Dr. Sameer Naji, MB, BCh, PhD (UK) Head of Basic Medical Sciences Dept. Faculty of Medicine The Hashemite University Human blood cells Phases of immune responses Microbe Naïve
Medical Virology Immunology Dr. Sameer Naji, MB, BCh, PhD (UK) Head of Basic Medical Sciences Dept. Faculty of Medicine The Hashemite University Human blood cells Phases of immune responses Microbe Naïve
Module 1. Introduction. Microbial Physiology
 Module 1 Introduction to Microbial Physiology Module 1 INTRODUCTION TO MICROBIAL PHYSIOLOGY Page 2 Last time * Discussed the central dogma of molecular biology * Look at, in detail, the process of replication,
Module 1 Introduction to Microbial Physiology Module 1 INTRODUCTION TO MICROBIAL PHYSIOLOGY Page 2 Last time * Discussed the central dogma of molecular biology * Look at, in detail, the process of replication,
Chapter 7. (7-1 and 7-2) A Tour of the Cell
 Chapter 7 (7-1 and 7-2) A Tour of the Cell Microscopes as Windows to the World of Cells Cells were first described in 1665 by Robert Hooke. By the mid-1800s, the accumulation of scientific evidence led
Chapter 7 (7-1 and 7-2) A Tour of the Cell Microscopes as Windows to the World of Cells Cells were first described in 1665 by Robert Hooke. By the mid-1800s, the accumulation of scientific evidence led
Macrophage Activation & Cytokine Release. Dendritic Cells & Antigen Presentation. Neutrophils & Innate Defense
 Macrophage Activation & Cytokine Release Dendritic Cells & Antigen Presentation Neutrophils & Innate Defense Neutrophils Polymorphonuclear cells (PMNs) are recruited to the site of infection where they
Macrophage Activation & Cytokine Release Dendritic Cells & Antigen Presentation Neutrophils & Innate Defense Neutrophils Polymorphonuclear cells (PMNs) are recruited to the site of infection where they
Introduction. Microbiology. Anas Abu-Humaidan M.D. Ph.D. Lecture 2
 Introduction to Microbiology Anas Abu-Humaidan M.D. Ph.D. Lecture 2 Cell structure / Eukaryotes 1. Nucleolus 2. Nucleus 3. Ribosome (80S) 4. Vesicle 5. Rough endoplasmic reticulum 6. Golgi apparatus (or
Introduction to Microbiology Anas Abu-Humaidan M.D. Ph.D. Lecture 2 Cell structure / Eukaryotes 1. Nucleolus 2. Nucleus 3. Ribosome (80S) 4. Vesicle 5. Rough endoplasmic reticulum 6. Golgi apparatus (or
UNIT 2: Cells Chapter 3: Cell Structure and Function
 UNIT 2: Cells Chapter 3: Cell Structure and Function I. Cell Theory (3.1) A. Early studies led to the development of the cell theory 1. Discovery of Cells a. Robert Hooke (1665)-Used compound microscope
UNIT 2: Cells Chapter 3: Cell Structure and Function I. Cell Theory (3.1) A. Early studies led to the development of the cell theory 1. Discovery of Cells a. Robert Hooke (1665)-Used compound microscope
BIOL 4374/BCHS 4313 Cell Biology Exam #2 March 22, 2001
 BIOL 4374/BCHS 4313 Cell Biology Exam #2 March 22, 2001 SS# Name This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses. Good luck! 1. (2) In the
BIOL 4374/BCHS 4313 Cell Biology Exam #2 March 22, 2001 SS# Name This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses. Good luck! 1. (2) In the
Unit 6: Adaptive Immunity. Adaptive Immunity (Humoral Immunity; Cell-Mediated Immunity; Immunodeficiency; Hypersensitivity)
 Unit 6: Adaptive Immunity Adaptive Immunity (Humoral Immunity; Cell-Mediated Immunity; Immunodeficiency; Hypersensitivity) : ADAPTIVE IMMUNITY: AN OVERVIEW OF INNATE AND ADAPTIVE IMMUNITY Adaptive Immunity
Unit 6: Adaptive Immunity Adaptive Immunity (Humoral Immunity; Cell-Mediated Immunity; Immunodeficiency; Hypersensitivity) : ADAPTIVE IMMUNITY: AN OVERVIEW OF INNATE AND ADAPTIVE IMMUNITY Adaptive Immunity
Chapter 4. Prokaryotic Cells. Prokaryotic cells. Prokaryotic vs. Eukaryotic Cells. Comparing Prokaryotic and. Eukaryotic cells.
 Chapter 4 Comparing Prokaryotic and Eukaryotic Cells Prokaryotic vs. Eukaryotic Cells Prokaryotic cells No nucleus No organelles Cell walls composed of peptidoglycan Reproduce asexually via binary fission
Chapter 4 Comparing Prokaryotic and Eukaryotic Cells Prokaryotic vs. Eukaryotic Cells Prokaryotic cells No nucleus No organelles Cell walls composed of peptidoglycan Reproduce asexually via binary fission
LIFE IS CELLULAR. Cell Theory. Cells Are Small. Prokaryotic Cell 10/4/15. Chapter 7 Cell Structure and Function
 Chapter 7 Cell Structure and Function The cell basic unit of life, all living things are made of a cell (unicellular) or more than one cell (multicellular). LIFE IS CELLULAR The invention of the microscope
Chapter 7 Cell Structure and Function The cell basic unit of life, all living things are made of a cell (unicellular) or more than one cell (multicellular). LIFE IS CELLULAR The invention of the microscope
11/25/2017. THE IMMUNE SYSTEM Chapter 43 IMMUNITY INNATE IMMUNITY EXAMPLE IN INSECTS BARRIER DEFENSES INNATE IMMUNITY OF VERTEBRATES
 THE IMMUNE SYSTEM Chapter 43 IMMUNITY INNATE IMMUNITY EXAMPLE IN INSECTS Exoskeleton made of chitin forms the first barrier to pathogens Digestive system is protected by a chitin-based barrier and lysozyme,
THE IMMUNE SYSTEM Chapter 43 IMMUNITY INNATE IMMUNITY EXAMPLE IN INSECTS Exoskeleton made of chitin forms the first barrier to pathogens Digestive system is protected by a chitin-based barrier and lysozyme,
Innate Immunity. Bởi: OpenStaxCollege
 Innate Immunity Bởi: OpenStaxCollege The vertebrate, including human, immune system is a complex multilayered system for defending against external and internal threats to the integrity of the body. The
Innate Immunity Bởi: OpenStaxCollege The vertebrate, including human, immune system is a complex multilayered system for defending against external and internal threats to the integrity of the body. The
Cell Quality Control. Peter Takizawa Department of Cell Biology
 Cell Quality Control Peter Takizawa Department of Cell Biology Cellular quality control reduces production of defective proteins. Cells have many quality control systems to ensure that cell does not build
Cell Quality Control Peter Takizawa Department of Cell Biology Cellular quality control reduces production of defective proteins. Cells have many quality control systems to ensure that cell does not build
Intrinsic cellular defenses against virus infection
 Intrinsic cellular defenses against virus infection Detection of virus infection Host cell response to virus infection Interferons: structure and synthesis Induction of antiviral activity Viral defenses
Intrinsic cellular defenses against virus infection Detection of virus infection Host cell response to virus infection Interferons: structure and synthesis Induction of antiviral activity Viral defenses
Cytokines modulate the functional activities of individual cells and tissues both under normal and pathologic conditions Interleukins,
 Cytokines http://highered.mcgraw-hill.com/sites/0072507470/student_view0/chapter22/animation the_immune_response.html Cytokines modulate the functional activities of individual cells and tissues both under
Cytokines http://highered.mcgraw-hill.com/sites/0072507470/student_view0/chapter22/animation the_immune_response.html Cytokines modulate the functional activities of individual cells and tissues both under
ORGANELLES OF THE ENDOMEMBRANE SYSTEM
 Membranes compartmentalize the interior of the cell and facilitate a variety of metabolic activities. Chloroplasts and a rigid cell wall are what distinguish a plant cell from an animal cell. A typical
Membranes compartmentalize the interior of the cell and facilitate a variety of metabolic activities. Chloroplasts and a rigid cell wall are what distinguish a plant cell from an animal cell. A typical
Modern Cell Theory. Plasma Membrane. Generalized Cell Structures. Cellular Form and Function. Three principle parts of a cell
 Cellular Form and Function Concepts of cellular structure Cell surface Membrane transport Cytoplasm Modern Cell Theory All living organisms are composed of cells. the simplest structural and functional
Cellular Form and Function Concepts of cellular structure Cell surface Membrane transport Cytoplasm Modern Cell Theory All living organisms are composed of cells. the simplest structural and functional
Chapter 7 Cell Structure and Function. Section Objectives: Relate advances in microscope technology to discoveries about cells and cell structure.
 Chapter 7 Cell Structure and Function Section Objectives: Relate advances in microscope technology to discoveries about cells and cell structure. Compare the operation of a microscope with that of an electron
Chapter 7 Cell Structure and Function Section Objectives: Relate advances in microscope technology to discoveries about cells and cell structure. Compare the operation of a microscope with that of an electron
CMB621: Cytoskeleton. Also known as How the cell plays with LEGOs to ensure order, not chaos, is temporally and spatially achieved
 CMB621: Cytoskeleton Also known as How the cell plays with LEGOs to ensure order, not chaos, is temporally and spatially achieved Lecture(s) Overview Lecture 1: What is the cytoskeleton? Membrane interaction
CMB621: Cytoskeleton Also known as How the cell plays with LEGOs to ensure order, not chaos, is temporally and spatially achieved Lecture(s) Overview Lecture 1: What is the cytoskeleton? Membrane interaction
BIOLOGY 103 Spring 2001 MIDTERM LAB SECTION
 BIOLOGY 103 Spring 2001 MIDTERM NAME KEY LAB SECTION ID# (last four digits of SS#) STUDENT PLEASE READ. Do not put yourself at a disadvantage by revealing the content of this exam to your classmates. Your
BIOLOGY 103 Spring 2001 MIDTERM NAME KEY LAB SECTION ID# (last four digits of SS#) STUDENT PLEASE READ. Do not put yourself at a disadvantage by revealing the content of this exam to your classmates. Your
What s in a Cell? From Ch. 4
 What s in a Cell? From Ch. 4 Plant cell walls Amit1b.files.wordpress.com genomebiology.com Figure 4.1 Arrangements of cocci. Plane of division Diplococci Streptococci Tetrad Sarcinae Staphylococci. Figure
What s in a Cell? From Ch. 4 Plant cell walls Amit1b.files.wordpress.com genomebiology.com Figure 4.1 Arrangements of cocci. Plane of division Diplococci Streptococci Tetrad Sarcinae Staphylococci. Figure
Ch 3 Cell Structure 10/1/2008. Cells Under the Microscope. Natural laws limit cell size. Biology Periods 2, 3, 4, & 6 Mrs.
 Ch 3 Cell Structure Cells Under the Microscope Electron microscopes have much higher magnifying and resolving powers than light microscopes. Biology Periods 2, 3, 4, & 6 Mrs. Stolipher Cell size and shape
Ch 3 Cell Structure Cells Under the Microscope Electron microscopes have much higher magnifying and resolving powers than light microscopes. Biology Periods 2, 3, 4, & 6 Mrs. Stolipher Cell size and shape
Innate Immunity. Jan 8 th Prof. dr. sc. Ivana Novak Nakir 1
 Innate Immunity Jan 8 th 2018. Prof. dr. sc. Ivana Novak Nakir 1 Adaptive Innate 2 Immune system overview 1 st line of defense skin (2m 2 ) and mucosal membranes (~400m 2 ): physical barrier, lymphoid
Innate Immunity Jan 8 th 2018. Prof. dr. sc. Ivana Novak Nakir 1 Adaptive Innate 2 Immune system overview 1 st line of defense skin (2m 2 ) and mucosal membranes (~400m 2 ): physical barrier, lymphoid
Test Bank for The Immune System 4th Edition by Parham
 Test Bank for The Immune System 4th Edition by Parham CHAPTER 3: INNATE IMMUNITY: THE INDUCED RESPONSE TO INFECTION 3 1 C-type lectins are so called because of the role of in facilitating receptor:ligand
Test Bank for The Immune System 4th Edition by Parham CHAPTER 3: INNATE IMMUNITY: THE INDUCED RESPONSE TO INFECTION 3 1 C-type lectins are so called because of the role of in facilitating receptor:ligand
Notes Chapter 7 Cell Structure and Function Hooke looked at cork under a simple microscope and found tiny chambers he named cells.
 Notes Chapter 7 Cell Structure and Function 7.1 Cell discovery and Theory 1665 Hooke looked at cork under a simple microscope and found tiny chambers he named cells. Cells are the basic structural and
Notes Chapter 7 Cell Structure and Function 7.1 Cell discovery and Theory 1665 Hooke looked at cork under a simple microscope and found tiny chambers he named cells. Cells are the basic structural and
Human height. Length of some nerve and muscle cells. Chicken egg. Frog egg. Most plant and animal cells Nucleus Most bacteria Mitochondrion
 10 m 1 m 0.1 m 1 cm Human height Length of some nerve and muscle cells Chicken egg Unaided eye 1 mm Frog egg 100 µm 10 µm 1 µm 100 nm 10 nm Most plant and animal cells Nucleus Most bacteria Mitochondrion
10 m 1 m 0.1 m 1 cm Human height Length of some nerve and muscle cells Chicken egg Unaided eye 1 mm Frog egg 100 µm 10 µm 1 µm 100 nm 10 nm Most plant and animal cells Nucleus Most bacteria Mitochondrion
TCR, MHC and coreceptors
 Cooperation In Immune Responses Antigen processing how peptides get into MHC Antigen processing involves the intracellular proteolytic generation of MHC binding proteins Protein antigens may be processed
Cooperation In Immune Responses Antigen processing how peptides get into MHC Antigen processing involves the intracellular proteolytic generation of MHC binding proteins Protein antigens may be processed
Pathogen Recognition and Inflammatory Signaling in Innate Immune Defenses
 CLINICAL MICROBIOLOGY REVIEWS, Apr. 2009, p. 240 273 Vol. 22, No. 2 0893-8512/09/$08.00 0 doi:10.1128/cmr.00046-08 Copyright 2009, American Society for Microbiology. All Rights Reserved. Pathogen Recognition
CLINICAL MICROBIOLOGY REVIEWS, Apr. 2009, p. 240 273 Vol. 22, No. 2 0893-8512/09/$08.00 0 doi:10.1128/cmr.00046-08 Copyright 2009, American Society for Microbiology. All Rights Reserved. Pathogen Recognition
