Modeling and Molecular Dynamics of Membrane Proteins
|
|
|
- Ethel Collins
- 6 years ago
- Views:
Transcription
1 Modeling and Molecular Dynamics of Membrane Proteins Emad Tajkhorshid Department of Biochemistry, Center for Biophysics and Computational Biology, and Beckman Institute University of Illinois at Urbana-Champagin
2 Why Do Living Cells Need Membrane Channels (Proteins)? Living cells also need to exchange materials and information with the outside world however, in a highly selective manner. Extracellular (outside) Cytoplasm (inside)
3 Phospholipid Bilayers Are Excellent Materials For Cell Membranes Hydrophobic interaction is the driving force Self-assembly in water Tendency to close on themselves Self-sealing (a hole is unfavorable) Extensive: up to millimeters
4 Lipid Diffusion in a Membrane D lip = 10-8 cm 2.s -1 (50 Å in ~ 5 x 10-6 s) D wat = 2.5 x 10-5 cm 2.s -1 Modeling mixed lipid bilayers! Once in several hours! (~ 50 Å in ~ 10 4 s) ~9 orders of magnitude slower ensuring bilayer asymmetry
5 Fluid Mosaic Model of Membrane Lateral Diffusion Allowed Flip-flop Forbidden Ensuring the conservation of membrane asymmetric structure
6 Technical difficulties in Simulations of Biological Membranes Time scale Heterogeneity of biological membranes 60 x 60 Å Pure POPE 5 ns ~100,000 atoms
7 Coarse-grained modeling of lipids 150 particles 9 particles! Also, increasing the time step by orders of magnitude.
8 by: J. Siewert-Jan Marrink and Alan E. Mark, University of Groningen, The Netherlands
9 Protein/Lipid ratio Pure lipid: insulation (neuronal cells) Other membranes: on average 50% Energy transduction membranes (75%) Membranes of mitocondria and chloroplast Purple membrane of halobacteria Different functions = different protein composition
10 Protein / Lipid Composition The purple membrane of halobacteria
11 Gramicidin A Might be very sensitive to the lipid head group electrostatic and membrane potential
12 Central cavity
13 Analysis of Molecular Dynamics Simulations of Biomolecules A very complicated arrangement of hundreds of groups interacting with each other Where to start to look at? What to analyze? How much can we learn from simulations? It is very important to get acquainted with your system
14 Aquaporins Membrane water channels
15 Monomeric pores Water, glycerol, Tetrameric pore Perhaps ions???
16 Functionally Important Features Tetrameric architecture Amphipatic channel interior Water and glycerol transport Protons, and other ions are excluded Conserved asparagine-prolinealanine residues; NPA motif Characteristic half-membrane spanning structure N ~100% conserved -NPA- signature sequence E NPA E NPAR C
17 A Semi-hydrophobic channel
18 Molecular Dynamics Simulations Protein: ~ 15,000 atoms Lipids (POPE): ~ 40,000 atoms Water: ~ 51,000 atoms Total: ~ 106,000 atoms NAMD, CHARMM27, PME NpT ensemble at 310 K 1ns equilibration, 4ns production 10 days /ns 32-proc Linux cluster 3.5 days/ns O2000 CPUs 0.35 days/ns 512 LeMieux CPUs
19 Protein Embedding in Membrane Hydrophobic surface of the protein Ring of Tyr and Trp
20 Embedding GlpF in Membrane 77 A
21 112 A 122 A
22 A Recipe for Membrane Protein Simulations Align the protein along the z-axis (membrane normal): OPM, Orient. Decide on the lipid type and generate a large enough patch (MEMBRANE plugin in VMD, other sources). Size, area/lipid, shrinking. Overlay the protein with a hydrated lipid bilayer. Adjust the depth/ height to maximize hydrophobic overlap and matching of aromatic side chains (Trp/Tyr) with the interfacial region Remove lipids/water that overlap with the protein. Better to keep as many lipids as you can, so try to remove clashes if they are not too many by playing with the lipids. Add more water and ions to the two sides of the membrane (SOLVATE / AUTOIONIZE in VMD) Constrain (not FIX) the protein (we are still modeling, let s preserve the crystal structure; fix the lipid head groups and water/ion and minimize/simulate the lipid tails using a short simulation.
23 A Recipe for Membrane Protein Simulations Continue to constrain the protein (heavy atoms), but release everything else; minimize/simulate using a short constantpressure MD (NPT) to pack lipids and water against the protein and fill the gaps introduced after removal of protein-overlapping lipids. Watch water molecules; They normally stay out of the hydrophobic cleft. If necessary apply constraints to prevent them from penetrating into the open cleft between the lipids and the protein. Monitor the volume of your simulation box until the steep phase of the volume change is complete (.xst and.xsc files). Do not run the system for too long during this phase (over-shrinking; sometimes difficult to judge). Now release the protein, minimize the whole system, and start another short NPT simulation of the whole system. Switch to an NP n AT or an NVT simulation, when the system reaches a stable volume. Using the new CHARMM force field, you can stay with NPT.
24 Lipid-Protein Packing During the Initial NpT Simulation
25 Adjustment of Membrane Thickness to the Protein Hydrophobic Surface
26 Glycerol-Saturated GlpF
27
28 Description of full conduction pathway
29 Complete description of the conduction pathway Constriction region Selectivity filter
30 Channel Hydrogen Bonding Sites {set frame 0}{frame < 100}{incr frame}{ } animate goto $frame set donor [atomselect top name O N and within 2 of (resname GCL and name HO) ] lappend [$donor get index] list1 set acceptor [atomselect top resname GCL and name O and within 2 of (protein and name HN HO) ] lappend [$acceptor get index] list2
31 Channel Hydrogen Bonding Sites GLN 41 OE1 NE2 TRP 48 O NE1 GLY 64 O ALA 65 O HIS 66 O ND1 LEU 67 O ASN 68 ND2 ASP 130 OD1 GLY 133 O SER 136 O TYR 138 O PRO 139 O N ASN 140 OD1 ND2 HIS 142 ND1 THR 167 OG1 GLY 195 O PRO 196 O LEU 197 O THR 198 O GLY 199 O PHE 200 O ALA 201 O ASN 203 ND2 LYS 33 HZ1 HZ3 GLN 41 HE21 TRP 48 HE1 HIS 66 HD1 ASN 68 HD22 TYR 138 HN ASN 140 HN HD21 HD22 HIS 142 HD1 GLY 199 HN ASN 203 HN HD21HD22 ARG 206 HE HH21HH22
32 Channel Hydrogen Bonding Sites GLN 41 OE1 NE2 TRP 48 O NE1 GLY 64 O ALA 65 O HIS 66 O ND1 LEU 67 O ASN 68 ND2 ASP 130 OD1 GLY 133 O SER 136 O TYR 138 O PRO 139 O N ASN 140 OD1 ND2 HIS 142 ND1 THR 167 OG1 GLY 195 O PRO 196 O LEU 197 O THR 198 O GLY 199 O PHE 200 O ALA 201 O ASN 203 ND2 LYS 33 HZ1 HZ3 GLN 41 HE21 TRP 48 HE1 HIS 66 HD1 ASN 68 HD22 TYR 138 HN ASN 140 HN HD21 HD22 HIS 142 HD1 GLY 199 HN ASN 203 HN HD21HD22 ARG 206 HE HH21HH22
33 The Substrate Pathway is formed by C=O groups
34 The Substrate Pathway is formed by C=O groups Non-helical motifs are stabilized by two glutamate residues. N E NPA E NPAR C
35 Conservation of Glutamate Residue in Human Aquaporins
36 Glycerol water competition for hydrogen bonding sites
37 Revealing the Functional Role of Reentrant Loops Potassium channel
38 Both from E. coli AqpZ is a pure water channel GlpF is a glycerol channel AqpZ vs. GlpF We have high resolution structures for both channels
39 Steered Molecular Dynamics is a non-equilibrium method by nature A wide variety of events that are inaccessible to conventional molecular dynamics simulations can be probed. The system will be driven, however, away from equilibrium, resulting in problems in describing the energy landscape associated with the event of interest. Second law of thermodynamics W ΔG
40 Jarzynski s Equality Transition between two equilibrium states λ = λ i T λ = λ(t) T work W heat Q λ = λ f e -βw p(w) ΔG W W ΔG Δ G = G G f i p(w) C. Jarzynski, Phys. Rev. Lett., 78, 2690 (1997) C. Jarzynski, Phys. Rev. E, 56, 5018 (1997) e βw = e βδg In principle, it is possible to obtain free energy surfaces from repeated nonequilibrium experiments. β = 1 k T B
41 Steered Molecular Dynamics constant force (250 pn) constant velocity (30 Å/ns)
42 SMD Simulation of Glycerol Passage Trajectory of glycerol pulled by constant force
43 Constructing the Potential of Mean Force 4 trajectories v = 0.03, Å/ps k = 150 pn/å f ( t) = k[ z( t) z0 vt] W ( t) dtʹ vf( tʹ ) = t 0
44 Features of the Potential of Mean Force Captures major features of the channel The largest barrier 7.3 kcal/mol; exp.: 9.6±1.5 kcal/mol Jensen et al., PNAS, 99: , 2002.
45 Periplasm Cytoplasm Features of the Potential of Mean Force Asymmetric Profile in the Vestibules Jensen et al., PNAS, 99: , 2002.
46 Artificial induction of glycerol conduction through AqpZ Y. Wang, K. Schulten, and E. Tajkhorshid Structure 13, 1107 (2005)
47 Three fold higher barriers periplasm cytoplasm SF NPA AqpZ 22.8 kcal/mol GlpF 7.3 kcal/mol Y. Wang, K. Schulten, and E. Tajkhorshid Structure 13, 1107 (2005)
48 Could it be simply the size? Y. Wang, K. Schulten, and E. Tajkhorshid Structure 13, 1107 (2005)
49 It is probably just the size that matters! Y. Wang, K. Schulten, and E. Tajkhorshid Structure 13, 1107 (2005)
50 Water permeation 5 nanosecond Simulation 18 water conducted In 4 monomers in 4 ns water/monomer/ns Exp. = ~ 1-2 /ns 7-8 water molecules in each channel
51
52 Correlated Motion of Water in the Channel Water pair correlation The single file of water molecules is maintained.
53 Diffusion of Water in the channel One dimensional diffusion: 2 Dt = ( z z ) Experimental value for AQP1: e-5 t 0 2
54 Diffusion of Water in the channel 2 Dt = ( z z ) t Time (ns) Improvement of statistics
55 Water Bipolar Configuration in Aquaporins
56 Water Bipolar Configuration in Aquaporins
57
58 R E M E M B E R: One of the most useful advantages of simulations over experiments is that you can modify the system as you wish: You can do modifications that are not even possible at all in reality! This is a powerful technique to test hypotheses developed during your simulations. Use it!
59 Electrostatic Stabilization of Water Bipolar Arrangement
60 Proton transfer through water H O H O H O H + H + H + H + H + H H H H + H + O H O H O H H + H + H H H H + H O H O H O H + H + H + H H H H +
61 Cl - channel K + channel Aquaporins
62
63 A Complex Electrostatic Interaction SF NPA Surprising and clearly not a hydrophobic channel M. Jensen, E. Tajkhorshid, K. Schulten, Biophys. J. 85, 2884 (2003)
64 A Repulsive Electrostatic Force at the Center of the Channel QM/MM MD of the behavior of an excessive proton
65 Combining all-atom and coarsegrained models to simulate transport across lipid bilayers
66 Peptide aggregation and Pore formation in lipid bilayers
67 Alamethicin 20-residue peptide No charge forming pores in the membrane 20 residue antimicrobial peptide
68 CG molecular systems allow for time scales of 3-4 orders of magnitude longer, because: Significant reduction of the degrees of freedom (or number of interacting particles/beads) Softer potentials allowing much longer time steps µm length scale and µs time scale Bilayer, micelle, and vesicle formation Fusion of bilayers and vesicles,
69 Alamethicin 20-residue peptide No charge forming pores in the membrane 20 residue antimicrobial peptide
70 Simulation Setting 49 peptides in 288 DMPC All-atom model equilibrated 1ns Converted to a CG model Simulated for 1µs 0.5 µs snapshot was reversecorarse-grained to an all atom model All atom model simulated for 20 ns
71 Coarse-Graining Four-to-one mapping P N C Q Employs CHARMM-like force fields parameterized using all-atom simulations
72 L. Thogersen, et al., Biophysical Journal (2008) Peptide Aggregation
73 g (R ) for peptides A g (R ) init t =1_s R / Å Number of clusters B Time / _s Av. number of neighbors C Time / _s MSD / Å D lipid peptide Time / _s
74 What brings the helices together?
75 L. Thogersen, et al., Biophysical Journal (2008) Peptide Insertion
76 Strong perturbation of the bilayer structure BUT no water permeation/pore formation!?
77 Hydration of the head group region in coarse-grained and all-atom models
78 Reverse Coarse-Graining X X
79 Reverse Coarse-Graining Mapping back CG beads to all-atom clusters Re-solvating the system 5000 steps of minimization Simulated annealing for 20 ps (T changing from 610K to 300K, ΔT = -10K) while constraining atoms to the position of the corresponding CG beads
80 Hydration of the head group region in coarse-grained and all-atom models
81 Alemethicin-induced Membrane Poration
82 Alemethicin-induced Membrane Poration
83 Alemethicin-induced Membrane Poration
Modeling and Molecular Dynamics of Membrane Proteins
 Modeling and Molecular Dynamics of Membrane Proteins Emad Tajkhorshid Department of Biochemistry, Center for Biophysics and Computational Biology, and Beckman Institute University of Illinois at Urbana-Champaign
Modeling and Molecular Dynamics of Membrane Proteins Emad Tajkhorshid Department of Biochemistry, Center for Biophysics and Computational Biology, and Beckman Institute University of Illinois at Urbana-Champaign
Lipid Bilayers Are Excellent For Cell Membranes
 Lipid Bilayers Are Excellent For Cell Membranes ydrophobic interaction is the driving force Self-assembly in water Tendency to close on themselves Self-sealing (a hole is unfavorable) Extensive: up to
Lipid Bilayers Are Excellent For Cell Membranes ydrophobic interaction is the driving force Self-assembly in water Tendency to close on themselves Self-sealing (a hole is unfavorable) Extensive: up to
Molecular Dynamics Simulation of Membrane Channels
 Molecular Dynamics Simulation of Membrane Channels Part II. Structure-Function Relationship and Transport in Aquaporins Emad Tajkhorshid Beckman Institute, UIUC Summer School on Theoretical and Computational
Molecular Dynamics Simulation of Membrane Channels Part II. Structure-Function Relationship and Transport in Aquaporins Emad Tajkhorshid Beckman Institute, UIUC Summer School on Theoretical and Computational
Analysis of Molecular Dynamics Simulations of Biomolecules
 Simulating Membrane Channels Part II. Structure-Function Relationship and Transport in Aquaporins Theoretical and Computational Biophysics Dec 2004, Boston, MA http://www.ks.uiuc.edu/training/ Analysis
Simulating Membrane Channels Part II. Structure-Function Relationship and Transport in Aquaporins Theoretical and Computational Biophysics Dec 2004, Boston, MA http://www.ks.uiuc.edu/training/ Analysis
Biological Membranes. Lipid Membranes. Bilayer Permeability. Common Features of Biological Membranes. A highly selective permeability barrier
 Biological Membranes Structure Function Composition Physicochemical properties Self-assembly Molecular models Lipid Membranes Receptors, detecting the signals from outside: Light Odorant Taste Chemicals
Biological Membranes Structure Function Composition Physicochemical properties Self-assembly Molecular models Lipid Membranes Receptors, detecting the signals from outside: Light Odorant Taste Chemicals
Arginine side chain interactions and the role of arginine as a mobile charge carrier in voltage sensitive ion channels. Supplementary Information
 Arginine side chain interactions and the role of arginine as a mobile charge carrier in voltage sensitive ion channels Craig T. Armstrong, Philip E. Mason, J. L. Ross Anderson and Christopher E. Dempsey
Arginine side chain interactions and the role of arginine as a mobile charge carrier in voltage sensitive ion channels Craig T. Armstrong, Philip E. Mason, J. L. Ross Anderson and Christopher E. Dempsey
Coarse grained simulations of Lipid Bilayer Membranes
 Coarse grained simulations of Lipid Bilayer Membranes P. B. Sunil Kumar Department of Physics IIT Madras, Chennai 600036 sunil@iitm.ac.in Atomistic MD: time scales ~ 10 ns length scales ~100 nm 2 To study
Coarse grained simulations of Lipid Bilayer Membranes P. B. Sunil Kumar Department of Physics IIT Madras, Chennai 600036 sunil@iitm.ac.in Atomistic MD: time scales ~ 10 ns length scales ~100 nm 2 To study
Membranes & Membrane Proteins
 School on Biomolecular Simulations Membranes & Membrane Proteins Vani Vemparala The Institute of Mathematical Sciences Chennai November 13 2007 JNCASR, Bangalore Cellular Environment Plasma membrane extracellular
School on Biomolecular Simulations Membranes & Membrane Proteins Vani Vemparala The Institute of Mathematical Sciences Chennai November 13 2007 JNCASR, Bangalore Cellular Environment Plasma membrane extracellular
CS612 - Algorithms in Bioinformatics
 Spring 2016 Protein Structure February 7, 2016 Introduction to Protein Structure A protein is a linear chain of organic molecular building blocks called amino acids. Introduction to Protein Structure Amine
Spring 2016 Protein Structure February 7, 2016 Introduction to Protein Structure A protein is a linear chain of organic molecular building blocks called amino acids. Introduction to Protein Structure Amine
Cellular Neurophysiology I Membranes and Ion Channels
 Cellular Neurophysiology I Membranes and Ion Channels Reading: BCP Chapter 3 www.bioelectriclab All living cells maintain an electrical potential (voltage) across their membranes (V m ). Resting Potential
Cellular Neurophysiology I Membranes and Ion Channels Reading: BCP Chapter 3 www.bioelectriclab All living cells maintain an electrical potential (voltage) across their membranes (V m ). Resting Potential
Interaction of Functionalized C 60 Nanoparticles with Lipid Membranes
 Interaction of Functionalized C 60 Nanoparticles with Lipid Membranes Kevin Gasperich Advisor: Dmitry Kopelevich ABSTRACT The recent rapid development of applications for fullerenes has led to an increase
Interaction of Functionalized C 60 Nanoparticles with Lipid Membranes Kevin Gasperich Advisor: Dmitry Kopelevich ABSTRACT The recent rapid development of applications for fullerenes has led to an increase
During the last half century, much effort has been devoted
 Membranes serve as allosteric activators of phospholipase A 2, enabling it to extract, bind, and hydrolyze phospholipid substrates Varnavas D. Mouchlis a,b,1, Denis Bucher b, J. Andrew McCammon a,b,c,1,
Membranes serve as allosteric activators of phospholipase A 2, enabling it to extract, bind, and hydrolyze phospholipid substrates Varnavas D. Mouchlis a,b,1, Denis Bucher b, J. Andrew McCammon a,b,c,1,
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. (a) Uncropped version of Fig. 2a. RM indicates that the translation was done in the absence of rough mcirosomes. (b) LepB construct containing the GGPG-L6RL6-
Supplementary Figures Supplementary Figure 1. (a) Uncropped version of Fig. 2a. RM indicates that the translation was done in the absence of rough mcirosomes. (b) LepB construct containing the GGPG-L6RL6-
Biomolecules: amino acids
 Biomolecules: amino acids Amino acids Amino acids are the building blocks of proteins They are also part of hormones, neurotransmitters and metabolic intermediates There are 20 different amino acids in
Biomolecules: amino acids Amino acids Amino acids are the building blocks of proteins They are also part of hormones, neurotransmitters and metabolic intermediates There are 20 different amino acids in
Lecture 33 Membrane Proteins
 Lecture 33 Membrane Proteins Reading for today: Chapter 4, section D Required reading for next Wednesday: Chapter 14, sections A and 14.19 to the end Kuriyan, J., and Eisenberg, D. (2007) The origin of
Lecture 33 Membrane Proteins Reading for today: Chapter 4, section D Required reading for next Wednesday: Chapter 14, sections A and 14.19 to the end Kuriyan, J., and Eisenberg, D. (2007) The origin of
2. Which of the following amino acids is most likely to be found on the outer surface of a properly folded protein?
 Name: WHITE Student Number: Answer the following questions on the computer scoring sheet. 1 mark each 1. Which of the following amino acids would have the highest relative mobility R f in normal thin layer
Name: WHITE Student Number: Answer the following questions on the computer scoring sheet. 1 mark each 1. Which of the following amino acids would have the highest relative mobility R f in normal thin layer
This exam consists of two parts. Part I is multiple choice. Each of these 25 questions is worth 2 points.
 MBB 407/511 Molecular Biology and Biochemistry First Examination - October 1, 2002 Name Social Security Number This exam consists of two parts. Part I is multiple choice. Each of these 25 questions is
MBB 407/511 Molecular Biology and Biochemistry First Examination - October 1, 2002 Name Social Security Number This exam consists of two parts. Part I is multiple choice. Each of these 25 questions is
Electrostatic Tuning of Permeation and Selectivity in Aquaporin Water Channels
 2884 Biophysical Journal Volume 85 November 2003 2884 2899 Electrostatic Tuning of Permeation and Selectivity in Aquaporin Water Channels Morten Ø. Jensen, Emad Tajkhorshid, and Klaus Schulten Theoretical
2884 Biophysical Journal Volume 85 November 2003 2884 2899 Electrostatic Tuning of Permeation and Selectivity in Aquaporin Water Channels Morten Ø. Jensen, Emad Tajkhorshid, and Klaus Schulten Theoretical
Lipids: diverse group of hydrophobic molecules
 Lipids: diverse group of hydrophobic molecules Lipids only macromolecules that do not form polymers li3le or no affinity for water hydrophobic consist mostly of hydrocarbons nonpolar covalent bonds fats
Lipids: diverse group of hydrophobic molecules Lipids only macromolecules that do not form polymers li3le or no affinity for water hydrophobic consist mostly of hydrocarbons nonpolar covalent bonds fats
Chapter 12: Membranes. Voet & Voet: Pages
 Chapter 12: Membranes Voet & Voet: Pages 390-415 Slide 1 Membranes Essential components of all living cells (define boundry of cells) exclude toxic ions and compounds; accumulation of nutrients energy
Chapter 12: Membranes Voet & Voet: Pages 390-415 Slide 1 Membranes Essential components of all living cells (define boundry of cells) exclude toxic ions and compounds; accumulation of nutrients energy
The Basics: A general review of molecular biology:
 The Basics: A general review of molecular biology: DNA Transcription RNA Translation Proteins DNA (deoxy-ribonucleic acid) is the genetic material It is an informational super polymer -think of it as the
The Basics: A general review of molecular biology: DNA Transcription RNA Translation Proteins DNA (deoxy-ribonucleic acid) is the genetic material It is an informational super polymer -think of it as the
Chapter 1 Membrane Structure and Function
 Chapter 1 Membrane Structure and Function Architecture of Membranes Subcellular fractionation techniques can partially separate and purify several important biological membranes, including the plasma and
Chapter 1 Membrane Structure and Function Architecture of Membranes Subcellular fractionation techniques can partially separate and purify several important biological membranes, including the plasma and
Objective: You will be able to explain how the subcomponents of
 Objective: You will be able to explain how the subcomponents of nucleic acids determine the properties of that polymer. Do Now: Read the first two paragraphs from enduring understanding 4.A Essential knowledge:
Objective: You will be able to explain how the subcomponents of nucleic acids determine the properties of that polymer. Do Now: Read the first two paragraphs from enduring understanding 4.A Essential knowledge:
Biological systems interact, and these systems and their interactions possess complex properties. STOP at enduring understanding 4A
 Biological systems interact, and these systems and their interactions possess complex properties. STOP at enduring understanding 4A Homework Watch the Bozeman video called, Biological Molecules Objective:
Biological systems interact, and these systems and their interactions possess complex properties. STOP at enduring understanding 4A Homework Watch the Bozeman video called, Biological Molecules Objective:
Mechanisms of Selectivity in Channels and Enzymes Studied with Interactive Molecular Dynamics
 36 Biophysical Journal Volume 85 July 2003 36 48 Mechanisms of Selectivity in Channels and Enzymes Studied with Interactive Molecular Dynamics Paul Grayson, Emad Tajkhorshid, and Klaus Schulten Beckman
36 Biophysical Journal Volume 85 July 2003 36 48 Mechanisms of Selectivity in Channels and Enzymes Studied with Interactive Molecular Dynamics Paul Grayson, Emad Tajkhorshid, and Klaus Schulten Beckman
Coarse-Grained Molecular Dynamics Study of Cyclic Peptide Nanotube Insertion into a Lipid Bilayer
 4780 J. Phys. Chem. A 2009, 113, 4780 4787 Coarse-Grained Molecular Dynamics Study of Cyclic Peptide Nanotube Insertion into a Lipid Bilayer Hyonseok Hwang* Department of Chemistry and Institute for Molecular
4780 J. Phys. Chem. A 2009, 113, 4780 4787 Coarse-Grained Molecular Dynamics Study of Cyclic Peptide Nanotube Insertion into a Lipid Bilayer Hyonseok Hwang* Department of Chemistry and Institute for Molecular
Introduction to proteins and protein structure
 Introduction to proteins and protein structure The questions and answers below constitute an introduction to the fundamental principles of protein structure. They are all available at [link]. What are
Introduction to proteins and protein structure The questions and answers below constitute an introduction to the fundamental principles of protein structure. They are all available at [link]. What are
Cells. Variation and Function of Cells
 Cells Variation and Function of Cells Plasma Membrane= the skin of a cell, it protects and nourishes the cell while communicating with other cells at the same time. Lipid means fat and they are hydrophobic
Cells Variation and Function of Cells Plasma Membrane= the skin of a cell, it protects and nourishes the cell while communicating with other cells at the same time. Lipid means fat and they are hydrophobic
Short polymer. Dehydration removes a water molecule, forming a new bond. Longer polymer (a) Dehydration reaction in the synthesis of a polymer
 HO 1 2 3 H HO H Short polymer Dehydration removes a water molecule, forming a new bond Unlinked monomer H 2 O HO 1 2 3 4 H Longer polymer (a) Dehydration reaction in the synthesis of a polymer HO 1 2 3
HO 1 2 3 H HO H Short polymer Dehydration removes a water molecule, forming a new bond Unlinked monomer H 2 O HO 1 2 3 4 H Longer polymer (a) Dehydration reaction in the synthesis of a polymer HO 1 2 3
H 2 O. Liquid, solid, and vapor coexist in the same environment
 Water H 2 O Liquid, solid, and vapor coexist in the same environment WATER MOLECULES FORM HYDROGEN BONDS Water is a fundamental requirement for life, so it is important to understand the structural and
Water H 2 O Liquid, solid, and vapor coexist in the same environment WATER MOLECULES FORM HYDROGEN BONDS Water is a fundamental requirement for life, so it is important to understand the structural and
The Structure and Function of Macromolecules
 The Structure and Function of Macromolecules Macromolecules are polymers Polymer long molecule consisting of many similar building blocks. Monomer the small building block molecules. Carbohydrates, proteins
The Structure and Function of Macromolecules Macromolecules are polymers Polymer long molecule consisting of many similar building blocks. Monomer the small building block molecules. Carbohydrates, proteins
Chapter 7: Membranes
 Chapter 7: Membranes Roles of Biological Membranes The Lipid Bilayer and the Fluid Mosaic Model Transport and Transfer Across Cell Membranes Specialized contacts (junctions) between cells What are the
Chapter 7: Membranes Roles of Biological Membranes The Lipid Bilayer and the Fluid Mosaic Model Transport and Transfer Across Cell Membranes Specialized contacts (junctions) between cells What are the
Head. Tail. Carboxyl group. group. group. air water. Hydrocarbon chain. lecture 5-sa Seth Copen Goldstein 2.
 Lipids Some lipid structures Organic compounds Amphipathic Polar head group (hydrophilic) Non-polar tails (hydrophobic) Lots of uses Energy storage Membranes Hormones Vitamins HO O C H 2 C CH 2 H 2 C CH
Lipids Some lipid structures Organic compounds Amphipathic Polar head group (hydrophilic) Non-polar tails (hydrophobic) Lots of uses Energy storage Membranes Hormones Vitamins HO O C H 2 C CH 2 H 2 C CH
Research Article Interactions of Carbon Nanotube with Lipid Bilayer Membranes
 Nanomaterials Volume 211, Article ID 83436, 6 pages doi:1.1155/211/83436 Research Article Interactions of Carbon Nanotube with Lipid Bilayer Membranes Vamshi K. Gangupomu 1 and Franco M. Capaldi 2 1 Department
Nanomaterials Volume 211, Article ID 83436, 6 pages doi:1.1155/211/83436 Research Article Interactions of Carbon Nanotube with Lipid Bilayer Membranes Vamshi K. Gangupomu 1 and Franco M. Capaldi 2 1 Department
Distribution of Amino Acids in a Lipid Bilayer from Computer Simulations
 Biophysical Journal Volume 94 May 2008 3393 3404 3393 Distribution of Amino Acids in a Lipid Bilayer from Computer Simulations Justin L. MacCallum, W. F. Drew Bennett, and D. Peter Tieleman Department
Biophysical Journal Volume 94 May 2008 3393 3404 3393 Distribution of Amino Acids in a Lipid Bilayer from Computer Simulations Justin L. MacCallum, W. F. Drew Bennett, and D. Peter Tieleman Department
Lipids and Membranes
 Lipids and Membranes Presented by Dr. Mohammad Saadeh The requirements for the Pharmaceutical Biochemistry I Philadelphia University Faculty of pharmacy Biological membranes are composed of lipid bilayers
Lipids and Membranes Presented by Dr. Mohammad Saadeh The requirements for the Pharmaceutical Biochemistry I Philadelphia University Faculty of pharmacy Biological membranes are composed of lipid bilayers
Protein Structure Monday, March
 Michael Morales ffice: 204/202 ary; knock hard if you visit as my office is some door. Lab: 238 & 205 ary ffice hours Either make an appointment or just drop by Phone: 829-3965 E-Mail: moralesm@buffalo.edu
Michael Morales ffice: 204/202 ary; knock hard if you visit as my office is some door. Lab: 238 & 205 ary ffice hours Either make an appointment or just drop by Phone: 829-3965 E-Mail: moralesm@buffalo.edu
Supplementary Information
 Coarse-Grained Molecular Dynamics Simulations of Photoswitchable Assembly and Disassembly Xiaoyan Zheng, Dong Wang,* Zhigang Shuai,* MOE Key Laboratory of Organic Optoelectronics and Molecular Engineering,
Coarse-Grained Molecular Dynamics Simulations of Photoswitchable Assembly and Disassembly Xiaoyan Zheng, Dong Wang,* Zhigang Shuai,* MOE Key Laboratory of Organic Optoelectronics and Molecular Engineering,
Transient β-hairpin Formation in α-synuclein Monomer Revealed by Coarse-grained Molecular Dynamics Simulation
 Transient β-hairpin Formation in α-synuclein Monomer Revealed by Coarse-grained Molecular Dynamics Simulation Hang Yu, 1, 2, a) Wei Han, 1, 3, b) Wen Ma, 1, 2 1, 2, 3, c) and Klaus Schulten 1) Beckman
Transient β-hairpin Formation in α-synuclein Monomer Revealed by Coarse-grained Molecular Dynamics Simulation Hang Yu, 1, 2, a) Wei Han, 1, 3, b) Wen Ma, 1, 2 1, 2, 3, c) and Klaus Schulten 1) Beckman
G protein coupled receptor interactions with cholesterol deep in the membrane
 G protein coupled receptor interactions with cholesterol deep in the membrane Samuel Genheden a,b, Jonathan W. Essex a,c, & Anthony G. Lee c,d * a School of Chemistry, University of Southampton, Southampton,
G protein coupled receptor interactions with cholesterol deep in the membrane Samuel Genheden a,b, Jonathan W. Essex a,c, & Anthony G. Lee c,d * a School of Chemistry, University of Southampton, Southampton,
Term Definition Example Amino Acids
 Name 1. What are some of the functions that proteins have in a living organism. 2. Define the following and list two amino acids that fit each description. Term Definition Example Amino Acids Hydrophobic
Name 1. What are some of the functions that proteins have in a living organism. 2. Define the following and list two amino acids that fit each description. Term Definition Example Amino Acids Hydrophobic
Structure and Dynamics of Model Pore Insertion into a Membrane
 Biophysical Journal Volume 88 May 2005 3083 3094 3083 Structure and Dynamics of Model Pore Insertion into a Membrane Carlos F. Lopez,* Steve O. Nielsen,* Bernd Ensing,* Preston B. Moore, y and Michael
Biophysical Journal Volume 88 May 2005 3083 3094 3083 Structure and Dynamics of Model Pore Insertion into a Membrane Carlos F. Lopez,* Steve O. Nielsen,* Bernd Ensing,* Preston B. Moore, y and Michael
Coarse Grained Molecular Dynamics Simulations of Transmembrane Protein-Lipid Systems
 Int. J. Mol. Sci. 2010, 11, 2393-2420; doi:10.3390/ijms11062393 OPEN ACCESS International Journal of Molecular Sciences ISSN 1422-0067 www.mdpi.com/journal/ijms Article Coarse Grained Molecular Dynamics
Int. J. Mol. Sci. 2010, 11, 2393-2420; doi:10.3390/ijms11062393 OPEN ACCESS International Journal of Molecular Sciences ISSN 1422-0067 www.mdpi.com/journal/ijms Article Coarse Grained Molecular Dynamics
Gold nanocrystals at DPPC bilayer. Bo Song, Huajun Yuan, Cynthia J. Jameson, Sohail Murad
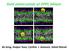 Gold nanocrystals at DPPC bilayer Bo Song, Huajun Yuan, Cynthia J. Jameson, Sohail Murad Coarse-grained mapping strategy of a DPPC lipid molecule. The structure of gold nanocrystals (bare gold nanoparticles)
Gold nanocrystals at DPPC bilayer Bo Song, Huajun Yuan, Cynthia J. Jameson, Sohail Murad Coarse-grained mapping strategy of a DPPC lipid molecule. The structure of gold nanocrystals (bare gold nanoparticles)
Lecture 15. Membrane Proteins I
 Lecture 15 Membrane Proteins I Introduction What are membrane proteins and where do they exist? Proteins consist of three main classes which are classified as globular, fibrous and membrane proteins. A
Lecture 15 Membrane Proteins I Introduction What are membrane proteins and where do they exist? Proteins consist of three main classes which are classified as globular, fibrous and membrane proteins. A
Proteins are sometimes only produced in one cell type or cell compartment (brain has 15,000 expressed proteins, gut has 2,000).
 Lecture 2: Principles of Protein Structure: Amino Acids Why study proteins? Proteins underpin every aspect of biological activity and therefore are targets for drug design and medicinal therapy, and in
Lecture 2: Principles of Protein Structure: Amino Acids Why study proteins? Proteins underpin every aspect of biological activity and therefore are targets for drug design and medicinal therapy, and in
Break Desired Compound into 3 Smaller Ones
 Break Desired Compound into 3 Smaller nes A B C Indole ydrazine Phenol When creating a covalent link between model compounds move the charge on the deleted into the carbon to maintain integer charge (i.e.
Break Desired Compound into 3 Smaller nes A B C Indole ydrazine Phenol When creating a covalent link between model compounds move the charge on the deleted into the carbon to maintain integer charge (i.e.
Water and Ion Permeation in baqp1 and GlpF Channels: A Kinetic Monte Carlo Study
 3690 Biophysical Journal Volume 87 December 2004 3690 3702 Water and Ion Permeation in baqp1 and GlpF Channels: A Kinetic Monte Carlo Study Gennady V. Miloshevsky and Peter C. Jordan Department of Chemistry,
3690 Biophysical Journal Volume 87 December 2004 3690 3702 Water and Ion Permeation in baqp1 and GlpF Channels: A Kinetic Monte Carlo Study Gennady V. Miloshevsky and Peter C. Jordan Department of Chemistry,
Amino Acids. Review I: Protein Structure. Amino Acids: Structures. Amino Acids (contd.) Rajan Munshi
 Review I: Protein Structure Rajan Munshi BBSI @ Pitt 2005 Department of Computational Biology University of Pittsburgh School of Medicine May 24, 2005 Amino Acids Building blocks of proteins 20 amino acids
Review I: Protein Structure Rajan Munshi BBSI @ Pitt 2005 Department of Computational Biology University of Pittsburgh School of Medicine May 24, 2005 Amino Acids Building blocks of proteins 20 amino acids
SUPPORTING INFORMATION FOR. A Computational Approach to Enzyme Design: Using Docking and MM- GBSA Scoring
 SUPPRTING INFRMATIN FR A Computational Approach to Enzyme Design: Predicting ω- Aminotransferase Catalytic Activity Using Docking and MM- GBSA Scoring Sarah Sirin, 1 Rajesh Kumar, 2 Carlos Martinez, 2
SUPPRTING INFRMATIN FR A Computational Approach to Enzyme Design: Predicting ω- Aminotransferase Catalytic Activity Using Docking and MM- GBSA Scoring Sarah Sirin, 1 Rajesh Kumar, 2 Carlos Martinez, 2
We parameterized a coarse-grained fullerene consistent with the MARTINI coarse-grained force field
 Parameterization of the fullerene coarse-grained model We parameterized a coarse-grained fullerene consistent with the MARTINI coarse-grained force field for lipids 1 and proteins 2. In the MARTINI force
Parameterization of the fullerene coarse-grained model We parameterized a coarse-grained fullerene consistent with the MARTINI coarse-grained force field for lipids 1 and proteins 2. In the MARTINI force
Supplementary Information A Hydrophobic Barrier Deep Within the Inner Pore of the TWIK-1 K2P Potassium Channel Aryal et al.
 Supplementary Information A Hydrophobic Barrier Deep Within the Inner Pore of the TWIK-1 K2P Potassium Channel Aryal et al. Supplementary Figure 1 TWIK-1 stability during MD simulations in a phospholipid
Supplementary Information A Hydrophobic Barrier Deep Within the Inner Pore of the TWIK-1 K2P Potassium Channel Aryal et al. Supplementary Figure 1 TWIK-1 stability during MD simulations in a phospholipid
Acta Crystallographica Section D
 Supporting information Acta Crystallographica Section D Volume 70 (2014) Supporting information for article: A conformational landscape for alginate secretion across the outer membrane of Pseudomonas aeruginosa
Supporting information Acta Crystallographica Section D Volume 70 (2014) Supporting information for article: A conformational landscape for alginate secretion across the outer membrane of Pseudomonas aeruginosa
I. Fluid Mosaic Model A. Biological membranes are lipid bilayers with associated proteins
 Lecture 6: Membranes and Cell Transport Biological Membranes I. Fluid Mosaic Model A. Biological membranes are lipid bilayers with associated proteins 1. Characteristics a. Phospholipids form bilayers
Lecture 6: Membranes and Cell Transport Biological Membranes I. Fluid Mosaic Model A. Biological membranes are lipid bilayers with associated proteins 1. Characteristics a. Phospholipids form bilayers
Conformational Flexibility of the Peptide Hormone Ghrelin in Solution and Lipid Membrane Bound: A Molecular Dynamics Study
 Open Access Article The authors, the publisher, and the right holders grant the right to use, reproduce, and disseminate the work in digital form to all users. Journal of Biomolecular Structure & Dynamics,
Open Access Article The authors, the publisher, and the right holders grant the right to use, reproduce, and disseminate the work in digital form to all users. Journal of Biomolecular Structure & Dynamics,
Role of Surface Ligands in Nanoparticle Permeation through a Model. Membrane: A Coarse grained Molecular Dynamics Simulations Study
 Role of Surface Ligands in Nanoparticle Permeation through a Model Membrane: A Coarse grained Molecular Dynamics Simulations Study Bo Song, Huajun Yuan, Cynthia J. Jameson, Sohail Murad * Department of
Role of Surface Ligands in Nanoparticle Permeation through a Model Membrane: A Coarse grained Molecular Dynamics Simulations Study Bo Song, Huajun Yuan, Cynthia J. Jameson, Sohail Murad * Department of
Chapter 9 - Biological Membranes. Membranes form a semi-permeable boundary between a cell and its environment.
 Chapter 9 - Biological Membranes www.gsbs.utmb.edu/ microbook/ch037.htmmycoplasma Membranes form a semi-permeable boundary between a cell and its environment. Membranes also permit subcellular organization
Chapter 9 - Biological Membranes www.gsbs.utmb.edu/ microbook/ch037.htmmycoplasma Membranes form a semi-permeable boundary between a cell and its environment. Membranes also permit subcellular organization
Biological Membranes & Transport
 Biological Membranes & Transport Life without membranes? Life without cells? 1. What are membranes made of? 2. Membranes, static or dynamic? 3. Transport across membranes? View The Inner Life of the Cell
Biological Membranes & Transport Life without membranes? Life without cells? 1. What are membranes made of? 2. Membranes, static or dynamic? 3. Transport across membranes? View The Inner Life of the Cell
Lecture Series 5 Cellular Membranes
 Lecture Series 5 Cellular Membranes Cellular Membranes A. Membrane Composition and Structure B. Animal Cell Adhesion C. Passive Processes of Membrane Transport D. Active Transport E. Endocytosis and Exocytosis
Lecture Series 5 Cellular Membranes Cellular Membranes A. Membrane Composition and Structure B. Animal Cell Adhesion C. Passive Processes of Membrane Transport D. Active Transport E. Endocytosis and Exocytosis
A. Membrane Composition and Structure. B. Animal Cell Adhesion. C. Passive Processes of Membrane Transport. D. Active Transport
 Cellular Membranes A. Membrane Composition and Structure Lecture Series 5 Cellular Membranes B. Animal Cell Adhesion E. Endocytosis and Exocytosis A. Membrane Composition and Structure The Fluid Mosaic
Cellular Membranes A. Membrane Composition and Structure Lecture Series 5 Cellular Membranes B. Animal Cell Adhesion E. Endocytosis and Exocytosis A. Membrane Composition and Structure The Fluid Mosaic
CHAPTER 21: Amino Acids, Proteins, & Enzymes. General, Organic, & Biological Chemistry Janice Gorzynski Smith
 CHAPTER 21: Amino Acids, Proteins, & Enzymes General, Organic, & Biological Chemistry Janice Gorzynski Smith CHAPTER 21: Amino Acids, Proteins, Enzymes Learning Objectives: q The 20 common, naturally occurring
CHAPTER 21: Amino Acids, Proteins, & Enzymes General, Organic, & Biological Chemistry Janice Gorzynski Smith CHAPTER 21: Amino Acids, Proteins, Enzymes Learning Objectives: q The 20 common, naturally occurring
Nanoparticle Permeation Induces Water Penetration, Ion Transport, and Lipid Flip-Flop
 pubs.acs.org/langmuir Nanoparticle Permeation Induces Water Penetration, Ion Transport, and Lipid Flip-Flop Bo Song, Huajun Yuan, Sydney V. Pham, Cynthia J. Jameson, and Sohail Murad* Department of Chemical
pubs.acs.org/langmuir Nanoparticle Permeation Induces Water Penetration, Ion Transport, and Lipid Flip-Flop Bo Song, Huajun Yuan, Sydney V. Pham, Cynthia J. Jameson, and Sohail Murad* Department of Chemical
Membrane Structure. Membrane Structure. Membrane Structure. Membranes
 Membrane Structure Membranes Chapter 5 The fluid mosaic model of membrane structure contends that membranes consist of: -phospholipids arranged in a bilayer -globular proteins inserted in the lipid bilayer
Membrane Structure Membranes Chapter 5 The fluid mosaic model of membrane structure contends that membranes consist of: -phospholipids arranged in a bilayer -globular proteins inserted in the lipid bilayer
Chemical Nature of the Amino Acids. Table of a-amino Acids Found in Proteins
 Chemical Nature of the Amino Acids All peptides and polypeptides are polymers of alpha-amino acids. There are 20 a- amino acids that are relevant to the make-up of mammalian proteins (see below). Several
Chemical Nature of the Amino Acids All peptides and polypeptides are polymers of alpha-amino acids. There are 20 a- amino acids that are relevant to the make-up of mammalian proteins (see below). Several
Reading from the NCBI
 Reading from the NCBI http://www.ncbi.nlm.nih.gov/books/bv.fcgi?highlight=thermodyn amics&rid=stryer.section.156#167 http://www.ncbi.nlm.nih.gov/books/bv.fcgi?highlight=stability,pr otein&rid=stryer.section.365#371
Reading from the NCBI http://www.ncbi.nlm.nih.gov/books/bv.fcgi?highlight=thermodyn amics&rid=stryer.section.156#167 http://www.ncbi.nlm.nih.gov/books/bv.fcgi?highlight=stability,pr otein&rid=stryer.section.365#371
Reading for lecture 6
 Reading for lecture 6 1. Lipids and Lipid Bilayers 2. Membrane Proteins Voet and Voet, Chapter 11 Alberts et al Chapter 6 Jones, R.A.L, Soft Condensed Matter 195pp Oxford University Press, ISBN 0-19-850590-6
Reading for lecture 6 1. Lipids and Lipid Bilayers 2. Membrane Proteins Voet and Voet, Chapter 11 Alberts et al Chapter 6 Jones, R.A.L, Soft Condensed Matter 195pp Oxford University Press, ISBN 0-19-850590-6
SDS-Assisted Protein Transport Through Solid-State Nanopores
 Supplementary Information for: SDS-Assisted Protein Transport Through Solid-State Nanopores Laura Restrepo-Pérez 1, Shalini John 2, Aleksei Aksimentiev 2 *, Chirlmin Joo 1 *, Cees Dekker 1 * 1 Department
Supplementary Information for: SDS-Assisted Protein Transport Through Solid-State Nanopores Laura Restrepo-Pérez 1, Shalini John 2, Aleksei Aksimentiev 2 *, Chirlmin Joo 1 *, Cees Dekker 1 * 1 Department
Chemical Mechanism of Enzymes
 Chemical Mechanism of Enzymes Enzyme Engineering 5.2 Definition of the mechanism 1. The sequence from substrate(s) to product(s) : Reaction steps 2. The rates at which the complex are interconverted 3.
Chemical Mechanism of Enzymes Enzyme Engineering 5.2 Definition of the mechanism 1. The sequence from substrate(s) to product(s) : Reaction steps 2. The rates at which the complex are interconverted 3.
9/6/2011. Amino Acids. C α. Nonpolar, aliphatic R groups
 Amino Acids Side chains (R groups) vary in: size shape charge hydrogen-bonding capacity hydrophobic character chemical reactivity C α Nonpolar, aliphatic R groups Glycine (Gly, G) Alanine (Ala, A) Valine
Amino Acids Side chains (R groups) vary in: size shape charge hydrogen-bonding capacity hydrophobic character chemical reactivity C α Nonpolar, aliphatic R groups Glycine (Gly, G) Alanine (Ala, A) Valine
Membrane Structure and Membrane Transport of Small Molecules. Assist. Prof. Pinar Tulay Faculty of Medicine
 Membrane Structure and Membrane Transport of Small Molecules Assist. Prof. Pinar Tulay Faculty of Medicine Introduction Cell membranes define compartments of different compositions. Membranes are composed
Membrane Structure and Membrane Transport of Small Molecules Assist. Prof. Pinar Tulay Faculty of Medicine Introduction Cell membranes define compartments of different compositions. Membranes are composed
Amino acids. Side chain. -Carbon atom. Carboxyl group. Amino group
 PROTEINS Amino acids Side chain -Carbon atom Amino group Carboxyl group Amino acids Primary structure Amino acid monomers Peptide bond Peptide bond Amino group Carboxyl group Peptide bond N-terminal (
PROTEINS Amino acids Side chain -Carbon atom Amino group Carboxyl group Amino acids Primary structure Amino acid monomers Peptide bond Peptide bond Amino group Carboxyl group Peptide bond N-terminal (
Supplementary Figure-1. SDS PAGE analysis of purified designed carbonic anhydrase enzymes. M1-M4 shown in lanes 1-4, respectively, with molecular
 Supplementary Figure-1. SDS PAGE analysis of purified designed carbonic anhydrase enzymes. M1-M4 shown in lanes 1-4, respectively, with molecular weight markers (M). Supplementary Figure-2. Overlay of
Supplementary Figure-1. SDS PAGE analysis of purified designed carbonic anhydrase enzymes. M1-M4 shown in lanes 1-4, respectively, with molecular weight markers (M). Supplementary Figure-2. Overlay of
Acid/Base chemistry. NESA Biochemistry Fall 2001 Review problems for the first exam. Complete the following sentences
 1 NESA Biochemistry Fall 2001 eview problems for the first exam Acid/Base chemistry 1. 2 3 is a weak acid. 2. The anion of a weak acid is a weak base 3. p is the measure of a solutions acidity. 4. 3 and
1 NESA Biochemistry Fall 2001 eview problems for the first exam Acid/Base chemistry 1. 2 3 is a weak acid. 2. The anion of a weak acid is a weak base 3. p is the measure of a solutions acidity. 4. 3 and
Molecular Biology. general transfer: occurs normally in cells. special transfer: occurs only in the laboratory in specific conditions.
 Chapter 9: Proteins Molecular Biology replication general transfer: occurs normally in cells transcription special transfer: occurs only in the laboratory in specific conditions translation unknown transfer:
Chapter 9: Proteins Molecular Biology replication general transfer: occurs normally in cells transcription special transfer: occurs only in the laboratory in specific conditions translation unknown transfer:
Supplementary Information
 Supplementary Information Two common structural motifs for TCR recognition by staphylococcal enterotoxins Karin Erica Johanna Rödström 1, Paulina Regenthal 1, Christopher Bahl 2, Alex Ford 2, David Baker
Supplementary Information Two common structural motifs for TCR recognition by staphylococcal enterotoxins Karin Erica Johanna Rödström 1, Paulina Regenthal 1, Christopher Bahl 2, Alex Ford 2, David Baker
Four Classes of Biological Macromolecules. Biological Macromolecules. Lipids
 Biological Macromolecules Much larger than other par4cles found in cells Made up of smaller subunits Found in all cells Great diversity of func4ons Four Classes of Biological Macromolecules Lipids Polysaccharides
Biological Macromolecules Much larger than other par4cles found in cells Made up of smaller subunits Found in all cells Great diversity of func4ons Four Classes of Biological Macromolecules Lipids Polysaccharides
Maha AbuAjamieh. Tamara Wahbeh. Mamoon Ahram
 12 Maha AbuAjamieh Tamara Wahbeh Mamoon Ahram - - Go to this sheet s last page for definitions of the words with an asterisk above them (*) - You should memorise the 3-letter abbreviations, of all the
12 Maha AbuAjamieh Tamara Wahbeh Mamoon Ahram - - Go to this sheet s last page for definitions of the words with an asterisk above them (*) - You should memorise the 3-letter abbreviations, of all the
paper and beads don t fall off. Then, place the beads in the following order on the pipe cleaner:
 Beady Pipe Cleaner Proteins Background: Proteins are the molecules that carry out most of the cell s dayto-day functions. While the DNA in the nucleus is "the boss" and controls the activities of the cell,
Beady Pipe Cleaner Proteins Background: Proteins are the molecules that carry out most of the cell s dayto-day functions. While the DNA in the nucleus is "the boss" and controls the activities of the cell,
Page 8/6: The cell. Where to start: Proteins (control a cell) (start/end products)
 Page 8/6: The cell Where to start: Proteins (control a cell) (start/end products) Page 11/10: Structural hierarchy Proteins Phenotype of organism 3 Dimensional structure Function by interaction THE PROTEIN
Page 8/6: The cell Where to start: Proteins (control a cell) (start/end products) Page 11/10: Structural hierarchy Proteins Phenotype of organism 3 Dimensional structure Function by interaction THE PROTEIN
University of Illinois at Urbana-Champaign NIH Resource for Macromolecular Modeling and Bioinformatics Beckman Institute.
 University of Illinois at Urbana-Champaign NIH Resource for Macromolecular Modeling and Bioinformatics Beckman Institute Aquaporins VMD Developers: John Stone Dan Wright John Eargle Fatemeh Khalili Elizabeth
University of Illinois at Urbana-Champaign NIH Resource for Macromolecular Modeling and Bioinformatics Beckman Institute Aquaporins VMD Developers: John Stone Dan Wright John Eargle Fatemeh Khalili Elizabeth
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/4/3/eaaq0762/dc1 Supplementary Materials for Structures of monomeric and oligomeric forms of the Toxoplasma gondii perforin-like protein 1 Tao Ni, Sophie I. Williams,
advances.sciencemag.org/cgi/content/full/4/3/eaaq0762/dc1 Supplementary Materials for Structures of monomeric and oligomeric forms of the Toxoplasma gondii perforin-like protein 1 Tao Ni, Sophie I. Williams,
Review II: The Molecules of Life
 Review II: The Molecules of Life Judy Wieber BBSI @ Pitt 2007 Department of Computational Biology University of Pittsburgh School of Medicine May 24, 2007 Outline Introduction Proteins Carbohydrates Lipids
Review II: The Molecules of Life Judy Wieber BBSI @ Pitt 2007 Department of Computational Biology University of Pittsburgh School of Medicine May 24, 2007 Outline Introduction Proteins Carbohydrates Lipids
Properties of amino acids in proteins
 Properties of amino acids in proteins one of the primary roles of DNA (but far from the only one!!!) is to code for proteins A typical bacterium builds thousands types of proteins, all from ~20 amino acids
Properties of amino acids in proteins one of the primary roles of DNA (but far from the only one!!!) is to code for proteins A typical bacterium builds thousands types of proteins, all from ~20 amino acids
SIMPLE BASIC METABOLISM
 SIMPLE BASIC METABOLISM When we eat food such as a tuna fish sandwich, the polysaccharides, lipids, and proteins are digested to smaller molecules that are absorbed into the cells of our body. As these
SIMPLE BASIC METABOLISM When we eat food such as a tuna fish sandwich, the polysaccharides, lipids, and proteins are digested to smaller molecules that are absorbed into the cells of our body. As these
Membranes. Chapter 5
 Membranes Chapter 5 Membrane Structure The fluid mosaic model of membrane structure contends that membranes consist of: -phospholipids arranged in a bilayer -globular proteins inserted in the lipid bilayer
Membranes Chapter 5 Membrane Structure The fluid mosaic model of membrane structure contends that membranes consist of: -phospholipids arranged in a bilayer -globular proteins inserted in the lipid bilayer
Chapter 12. Part II. Biological Membrane
 Chapter 12 Part II. Biological Membrane Single-tailed lipids tend to form micelles Critical micelle concentration (cmc): minimum concentration that forms micelles e.g.) cmc for SDS 1mM; cmc for phospholipids
Chapter 12 Part II. Biological Membrane Single-tailed lipids tend to form micelles Critical micelle concentration (cmc): minimum concentration that forms micelles e.g.) cmc for SDS 1mM; cmc for phospholipids
Part III - Bioinformatics Studies Using Multiseq in VMD. Aminoacyl trna Synthetases Aquaporins. San Francisco, 2005, Computational Biology Workshop
 Part III - Bioinformatics Studies Using Multiseq in VMD Aminoacyl trna Synthetases Aquaporins San Francisco, 2005, Computational Biology Workshop Multiple Sequence Alignments The aminoacyl-trna synthetases,
Part III - Bioinformatics Studies Using Multiseq in VMD Aminoacyl trna Synthetases Aquaporins San Francisco, 2005, Computational Biology Workshop Multiple Sequence Alignments The aminoacyl-trna synthetases,
Membrane transport. Pharmacy Dr. Szilvia Barkó
 Membrane transport Pharmacy 04.10.2017 Dr. Szilvia Barkó Cell Membranes Cell Membrane Functions Protection Communication Import and and export of molecules Movement of the cell General Structure A lipid
Membrane transport Pharmacy 04.10.2017 Dr. Szilvia Barkó Cell Membranes Cell Membrane Functions Protection Communication Import and and export of molecules Movement of the cell General Structure A lipid
Cell Membranes and Signaling
 5 Cell Membranes and Signaling Concept 5.1 Biological Membranes Have a Common Structure and Are Fluid A membrane s structure and functions are determined by its constituents: lipids, proteins, and carbohydrates.
5 Cell Membranes and Signaling Concept 5.1 Biological Membranes Have a Common Structure and Are Fluid A membrane s structure and functions are determined by its constituents: lipids, proteins, and carbohydrates.
Phenylketonuria (PKU) Structure of Phenylalanine Hydroxylase. Biol 405 Molecular Medicine
 Phenylketonuria (PKU) Structure of Phenylalanine Hydroxylase Biol 405 Molecular Medicine 1998 Crystal structure of phenylalanine hydroxylase solved. The polypeptide consists of three regions: Regulatory
Phenylketonuria (PKU) Structure of Phenylalanine Hydroxylase Biol 405 Molecular Medicine 1998 Crystal structure of phenylalanine hydroxylase solved. The polypeptide consists of three regions: Regulatory
BIOPHYSICS II. By Prof. Xiang Yang Liu Department of Physics,
 BIOPHYSICS II By Prof. Xiang Yang Liu Department of Physics, NUS 1 Hydrogen bond and the stability of macromolecular structure Membrane Model Amphiphilic molecule self-assembly at the surface and din the
BIOPHYSICS II By Prof. Xiang Yang Liu Department of Physics, NUS 1 Hydrogen bond and the stability of macromolecular structure Membrane Model Amphiphilic molecule self-assembly at the surface and din the
PROTEINS. Amino acids are the building blocks of proteins. Acid L-form * * Lecture 6 Macromolecules #2 O = N -C -C-O.
 Proteins: Linear polymers of amino acids workhorses of the cell tools, machines & scaffolds Lecture 6 Macromolecules #2 PRTEINS 1 Enzymes catalysts that mediate reactions, increase reaction rate Structural
Proteins: Linear polymers of amino acids workhorses of the cell tools, machines & scaffolds Lecture 6 Macromolecules #2 PRTEINS 1 Enzymes catalysts that mediate reactions, increase reaction rate Structural
Membrane Structure and Function
 BIOL1040 Page 1 Membrane Structure and Function Friday, 6 March 2015 2:58 PM Cellular Membranes Fluid mosaics of lipids and proteins Phospholipids - abundant Phospholipids are amphipathic molecules (has
BIOL1040 Page 1 Membrane Structure and Function Friday, 6 March 2015 2:58 PM Cellular Membranes Fluid mosaics of lipids and proteins Phospholipids - abundant Phospholipids are amphipathic molecules (has
Cryo EM structure of the ribosome SecYE complex in the
 Supplementary Information Cryo EM structure of the ribosome SecYE complex in the membrane environment Jens Frauenfeld,, James Gumbart, Eli O. van der Sluis,, Soledad Funes, Marco Gartmann,, Birgitta Beatrix,,
Supplementary Information Cryo EM structure of the ribosome SecYE complex in the membrane environment Jens Frauenfeld,, James Gumbart, Eli O. van der Sluis,, Soledad Funes, Marco Gartmann,, Birgitta Beatrix,,
Penetration of Gold Nanoparticle through Human Skin: Unraveling Its Mechanisms at the Molecular Scale
 Penetration of Gold Nanoparticle through Human Skin: Unraveling Its Mechanisms at the Molecular Scale Rakesh Gupta and Beena Rai* TATA Research Development & Design Centre, TCS Innovation labs, Pune India,
Penetration of Gold Nanoparticle through Human Skin: Unraveling Its Mechanisms at the Molecular Scale Rakesh Gupta and Beena Rai* TATA Research Development & Design Centre, TCS Innovation labs, Pune India,
Biomolecules Amino Acids & Protein Chemistry
 Biochemistry Department Date: 17/9/ 2017 Biomolecules Amino Acids & Protein Chemistry Prof.Dr./ FAYDA Elazazy Professor of Biochemistry and Molecular Biology Intended Learning Outcomes ILOs By the end
Biochemistry Department Date: 17/9/ 2017 Biomolecules Amino Acids & Protein Chemistry Prof.Dr./ FAYDA Elazazy Professor of Biochemistry and Molecular Biology Intended Learning Outcomes ILOs By the end
PAPER No. : 16, Bioorganic and biophysical chemistry MODULE No. : 22, Mechanism of enzyme catalyst reaction (I) Chymotrypsin
 Subject Paper No and Title 16 Bio-organic and Biophysical Module No and Title 22 Mechanism of Enzyme Catalyzed reactions I Module Tag CHE_P16_M22 Chymotrypsin TABLE OF CONTENTS 1. Learning outcomes 2.
Subject Paper No and Title 16 Bio-organic and Biophysical Module No and Title 22 Mechanism of Enzyme Catalyzed reactions I Module Tag CHE_P16_M22 Chymotrypsin TABLE OF CONTENTS 1. Learning outcomes 2.
The role of Ca² + ions in the complex assembling of protein Z and Z-dependent protease inhibitor: A structure and dynamics investigation
 www.bioinformation.net Hypothesis Volume 8(9) The role of Ca² + ions in the complex assembling of protein Z and Z-dependent protease inhibitor: A structure and dynamics investigation Zahra Karimi 1 *,
www.bioinformation.net Hypothesis Volume 8(9) The role of Ca² + ions in the complex assembling of protein Z and Z-dependent protease inhibitor: A structure and dynamics investigation Zahra Karimi 1 *,
Phospholipids. Extracellular fluid. Polar hydrophilic heads. Nonpolar hydrophobic tails. Polar hydrophilic heads. Intracellular fluid (cytosol)
 Module 2C Membranes and Cell Transport All cells are surrounded by a plasma membrane. Eukaryotic cells also contain internal membranes and membrane- bound organelles. In this module, we will examine the
Module 2C Membranes and Cell Transport All cells are surrounded by a plasma membrane. Eukaryotic cells also contain internal membranes and membrane- bound organelles. In this module, we will examine the
Biomembranes structure and function. B. Balen
 Biomembranes structure and function B. Balen All cells are surrounded by membranes Selective barrier But also important for: 1. Compartmentalization 2. Biochemical activities 3. Transport of dissolved
Biomembranes structure and function B. Balen All cells are surrounded by membranes Selective barrier But also important for: 1. Compartmentalization 2. Biochemical activities 3. Transport of dissolved
