Regulation of cholesterol metabolism: An IDOL-dependent pathway to degrade the LDL-receptor Sorrentino, V.
|
|
|
- Berniece Bradford
- 5 years ago
- Views:
Transcription
1 UvA-DARE (Digital Academic Repository) Regulation of cholesterol metabolism: An IDOL-dependent pathway to degrade the LDL-receptor Sorrentino, V. Link to publication Citation for published version (APA): Sorrentino, V. (2014). Regulation of cholesterol metabolism: An IDOL-dependent pathway to degrade the LDLreceptor General rights It is not permitted to download or to forward/distribute the text or part of it without the consent of the author(s) and/or copyright holder(s), other than for strictly personal, individual use, unless the work is under an open content license (like Creative Commons). Disclaimer/Complaints regulations If you believe that digital publication of certain material infringes any of your rights or (privacy) interests, please let the Library know, stating your reasons. In case of a legitimate complaint, the Library will make the material inaccessible and/or remove it from the website. Please Ask the Library: or a letter to: Library of the University of Amsterdam, Secretariat, Singel 425, 1012 WP Amsterdam, The Netherlands. You will be contacted as soon as possible. UvA-DARE is a service provided by the library of the University of Amsterdam ( Download date: 31 Oct 2018
2 CHAPTER 8 Summary And General Discussion
3
4 Summary and General Discussion Summary The central theme of this thesis is the functional and molecular characterization of the LXR- IDOL-LDLR nexus, a novel post-translational pathway to control abundance of the LDL receptor, complementary to the SREBP-driven transcriptional control of the LDLR. Genetic evidence is included to support a physiological role of IDOL in regulating whole-body cholesterol homeostasis in humans. Furthermore, work in this thesis identifies USP2 and the ESCRT machinery as new modulators of LDLR trafficking. The discovery and initial characterization of the LDLR pathway and cholesterol homeostasis mechanisms by Brown and Goldstein constitutes the groundwork of the research on steroldependent regulation of lipid metabolism, and is briefly reviewed in Chapter 1 from a historic perspective. Briefly, it reports a short description of the SREBP- and LXR-driven transcriptional programs controlling cellular cholesterol levels, and the astounding observations on feedback regulation of cholesterol synthesis by the LDL lipoprotein that led Brown and Goldstein to their famous hypothesis of a receptor-mediated pathway for cholesterol homeostasis 1. Chapter 2 provides a review on the LXR-inducible E3-ubiquitin ligase IDOL, a novel posttranslational regulator of the LDLR and the main subject of the research presented in this thesis. The review describes the initial steps in the characterization of IDOL and its function, from its discovery as an LXR-target gene to its role in lipid metabolism. In addition, the genetic association between the IDOL locus and variation in circulating levels of plasma LDL in humans is described, emphasizing the potential relevance of IDOL in LDL metabolism. Finally, the PCSK9-mediated lysosomal degradation of the LDLR is briefly introduced, and its integration with the LXR-IDOL pathway discussed. Chapter 3 reports the identification of two additional LDLR family members, VLDLR and ApoER2, as targets for ubiquitylation and degradation by IDOL. These two receptors bind multiple extracellular ligands, but they also share overlapping substrate specificity for the extracellular matrix protein Reelin. The Reelin-stimulated pathway is essential for proper neuronal positioning and brain development, and binding of this molecule to VLDLR and ApoER2 induces the first step in the Reelin pathway, phosphorylation of the adaptor protein Dab1. However, despite distinct substrate specificity the VLDLR and ApoER2 share a large sequence homology with the LDLR, and particularly their C-terminal cytoplasmic tail is highly conserved. Within this short, 50 amino acid domain, the highly conserved lysine residue that is targeted by IDOL on the LDLR is also conserved. As a consequence, activation of the LXR- IDOL pathway, or IDOL expression, leads to degradation of the VLDLR and APOER2. Functionally, this leads to attenuation of the Reelin signaling pathway in neurons. In this study we also studied the evolutionary conservation of the IDOL pathway. Remarkably, the LDLR and VLDLR are also ubiquitylation targets of DNR1, a distant homolog of IDOL from Drosophila melanogaster, the common fruit fly. Similarly, IDOL is able to degrade LpR, an ancient LDLR-related receptor from the migrating locust. Collectively, these data show that the IDOL pathway is an evolutionarily conserved mechanism to modulate lipoprotein uptake via degradation of LDLR-related lipoprotein receptors. Chapter 8 Chapter 4 presents a functional and biochemical characterization of IDOL s N-terminal FERM and C-terminal RING domains. The RING domain promotes formation of ubiquitin chains in vitro, formally establishing IDOL as a bona fide E3 ligase. Additionally, it promotes Lysine-63-specific ubiquitylation of the LDLR in vivo, marking the LDLR for lysosomal 107
5 Chapter 8 degradation. The FERM domain of IDOL is similar to the one found in other members of the ERM family of proteins, where this domain is involved in interactions between proteins and cell membranes. However, homology modeling indicates that IDOL s FERM also contains a phosphotyrosine-binding motif, and this chapter reports the initial characterization of the interaction interface between this element in the FERM and the intracellular tail of the LDLR. The FERM domain interacts with, and co-localizes, with the LDLR and VLDLR primarily at the plasma membrane, pointing towards the recycling LDLR pool as the target for IDOL-mediated degradation. As shown later in chapter 6 this indeed turns out to be the case, as activation of the LXR-IDOL axis rapidly removes LDLR from the plasma membrane and attenuates cellular LDL uptake. Chapter 5 describes the first genetic loss-of-function variant of IDOL in human individuals, potentially linked to low levels of circulating LDL-cholesterol found in the carriers of the IDOL mutation. The mutation was uncovered by a genetic screen for the identification of novel genetic loci associated with extreme LDL phenotypes in a large cohort of individuals from the Dutch population. The discovered mutation encodes a rare missense mutation in IDOL, introducing a premature stop codon in IDOL mrna, that if not subject to non-sense mediated RNA decay results in production of the truncated protein IDOL R266X. This variant represents a complete loss of IDOL function that is unable to promote ubiquitylation and degradation of the LDLR. Accordingly, this variant was identified in two related individuals with low circulating LDL-cholesterol. These findings reinforce the notion that IDOL contributes to variation in plasma LDL levels in humans and supports the idea of inhibiting IDOL activity as a therapeutic strategy to lower circulating LDL levels. Chapter 6 introduces a new layer of regulation to mechanisms controlling endocytosis and recycling of the LDLR, by reporting that the LXR-IDOL axis defines a novel internalization route of the LDLR that is independent of the classic clathrin endocytic pathway. Additionally, genetic, pharmacological and molecular approaches indicate that IDOL-dependent degradation of the receptor does not require the alternative dynamin-, caveolin-, or macroautophagy-dependent internalization pathways. Instead, IDOL activity seems to target a lipid raft-resident LDLR pool at the plasma membrane, and sorting of the ubiquitylated LDLR to the lysosomes requires the concerted action of the endocytic adaptor Epsin, the ESCRT complex and the deubiquitylase USP8. Chapter 7 reports the identification of the deubiquitylase USP2 as a novel IDOL-binding partner, and a positive modulator of the LDLR pathway. USP2 binds to, and deubiquitylates IDOL, leading to a marked stabilization of IDOL protein levels as a result of reduced proteasomal degradation of the ligase. Paradoxically, this results in a decreased IDOLdependent degradation of the LDLR and rescues LDL uptake. Conversely, loss of USP2 reduces LDLR protein in an IDOL-dependent manner and limits LDL uptake. Therefore, by controlling ubiquitylation-dependent activity of IDOL, USP2 could potentially act as a novel regulator of lipoprotein clearance and sterol metabolism. This chapter (Chapter 8) highlights findings presented in this thesis, discusses how they contribute to our understanding of the LXR-IDOL-LDLR nexus, proposes new directions for future research in this field, and presents a view on how these findings can be exploited in novel therapeutic strategies aimed at increasing plasma LDL clearance via upregulation of LDLR. 108
6 Summary and General Discussion General discussion Elevated levels of plasma LDL cholesterol have been established as a major risk factor for developing atherosclerosis and cardiovascular disease 2-4. Due to the central role of the LDLR in modulating circulating levels of LDL, understanding the mechanisms that regulate synthesis, endocytosis, recycling and degradation of this receptor is crucial to the development of therapeutic strategies for treating and preventing atherosclerosis and dyslipidemia. The discoveries of the Brown and Goldstein laboratory have largely contributed to the identification and characterization of the LDLR pathway. The synthesis of the LDLR is a well-defined event, elegantly regulated by the SREBP transcription factors as a means to maintain cellular cholesterol homeostasis 5. The LDLR-dependent uptake of LDL-cholesterol is the first and most known example of receptor-mediated endocytosis of ligands, and it is the key process by which LDL is internalized into the cell 6,7. Mutations in key components of this endocytic pathway, namely in the LDLR, APOB, or the endocytic LDLR adaptor ARH are known causes of FH autosomal disorders such as dominant hypercholesterolemia 8 and recessive hypercholesterolemia 9, respectively. More recently, post-transcriptional mechanisms of proteolysis of the LDLR in response to altered cellular sterol balance have been identified. These cholesterolresponsive mechanisms are based on the activity of PCSK9 10 and on the ubiquitylationdependent degradation of the LDLR, mediated by the E3-ubiquitin ligase IDOL 11. While the PCSK9 pathway is being thoroughly investigated and strategies for its inhibition recently tested in clinical trials 12-18, characterization and physiological relevance of the IDOL-mediated down-regulation of LDLR are less well understood. LXRs control LDL uptake through the E3 ubiquitin ligase IDOL In 2009, the E3-ubiquitin ligase IDOL was identified as a new post-transcriptional regulator of the LDLR, with its expression being regulated by the LXR transcription factors (Chapter 2 and Zelcer et al. 11 ). LXRs are master regulators of cholesterol homeostasis supporting primarily efflux of excess cellular cholesterol 19. In line with this, genetic loss of these nuclear receptors in mice results in marked susceptibility to developing atherosclerosis With hindsight, given that circulating LDL represents ~70% of plasma cholesterol and the essential function of the LDLR pathway it would seem reasonable to expect LXRs to regulate the LDLR pathway. If anything, it is surprising that IDOL was not discovered earlier, particularly as an early report by Witztum and colleagues reported IDOLlike activity in fibroblasts as early as One factor that may have contributed to this is the fact that most atherosclerosis studies are done in mice, which seem to have limited hepatic Idol activity as compared to humans. Nevertheless, some hints to a potential involvement of LXRs in LDL metabolism could be derived from experiments in which activation of LXRs with ligand was pursued in ApoE -/- mice challenged with a high-cholesterol diet 26, hamsters and primates 27 ; treatment of these animals with LXR agonists consistently led to an increase in plasma LDL-cholesterol. However, the mechanism underlying the ligand-dependent changes in lipid profiles was not addressed in the aforementioned studies. The identification of the LXR-IDOL-LDLR nexus introduces an elegant feedback loop for rapid sterol-dependent regulation of the LDLR independent of SREBP2, thereby allowing cells to tightly control uptake of LDL in response to intracellular sterol fluctuations. Importantly, by marking the LDLR for degradation through ubiquitylation, IDOL introduces a whole new layer of post-translational modulation in the LDLR pathway, the Ubiquitin- Proteasome System (UPS) (Figure 1). Chapter 8 109
7 Chapter 8 Figure 1 I The Ubiquitin system controls the LDLR through the E3-ubiquitin ligase IDOL. The ubiquitin system relies on the sequential activities of three components in order to tag proteins with the small protein ubiquitin: an E1- ubiquitin activating enzyme, an E2-ubiquitin loading enzyme and an E3-ubiquitin ligase. The substrate-specificity of the ubiquitin system is largely determined by the interaction of the E3 ligases with a restricted subset of protein targets. Acting as an E3 ligase, IDOL promotes ubiquitylation of the LDLR, thereby marking it for lysosomal degradation. Figure adapted from Shuai and Liu, Nature Reviews Immunology The IDOL-LDLR-UPS molecular network IDOL is an E3 ubiquitin ligase able to stimulate ubiquitylation of the LDLR 11. Ubiquitylation is a post-transcriptional protein modification best known for its involvement in proteostasis through degradation of ubiquitylated proteins in the proteasome 28. The UPS requires the concerted activity of an E1 activating enzyme to recruit and activate ubiquitin, an E2 component on which the ubiquitin molecule is loaded, and an E3 ligase that facilitates the transfer of ubiquitin to the protein target 29,30. There are two major families of E3 ligases: HECT domain and RING domain E3s. HECT domain E3 ligases mediate the direct transfer of ubiquitin by formation of a HECT-ubiquitin intermediate, whereas RING domain E3 ligases facilitate the direct transfer of ubiquitin from the E2 to the substrate 29,31,32. In addition to proteasomal-dependent protein turnover, the UPS is also involved in controlling signaling at the plasma membrane, endocytosis, and targeting of receptors for lysosomal degradation 33. Different polyubiquitylation labels on the targeted proteins distinguish proteasomal or lysosomal degradation events; a Lysine-48-linked ubiquitin chain is a canonical signal for protein turnover via the proteasome, while a Lysine-63 chain attached to membrane receptors can act as a sorting signal towards the lysosomes 33,34. IDOL is a RING-E3 ligase able to support Lysine-63 polyubiquitylation of the LDLR (Chapter 4), which is in agreement with lysosomal degradation of the receptor upon LXR activation or forced IDOL expression. Additionally, consistent with the self-degradation control mechanism of the majority of RING- E3 ligases 29,35, IDOL is also subject to auto-ubiquitylation and degradation in the proteasome (Chapter 2 and 7). The ability of IDOL to stimulate formation of differently linked ubiquitin chains is due to its partnering with specific E2 enzymes. In fact, the specificity of the ubiquitin chain linkage is controlled by the E2 components of the UPS 36. The human genome encodes approximately 40 E2s 37 ; these enzymes dictate the switch from ubiquitin chain initiation to elongation and establish the linkage of assembled chains, thereby determining the cellular fate of ubiquitylated proteins, in combination with their interacting E3s 38,39. Some E2s can catalyze formation of more polyubiquitylation signals, including Lysine-63 chains and proteasome-sensitive chains, based on their pairing with different E3 ligases 36,
8 Summary and General Discussion Work carried in the Tontonoz lab identified members of the UBE2D1-4 as potential E2 enzymes involved both in the ubiquitylation of the LDLR by IDOL and in the autoubiquitylation reaction of IDOL 41. Biochemical and structural characterization of the E2 IDOL complex demonstrated that disruption of UBE2D activity or the interaction interface between UBE2D and IDOL inhibits the degradation of the LDLR and IDOL autoubiquitylation. By using an inducible RNAi strategy to replace endogenous ubiquitin with mutants lacking Lysine-48 or Lysine-63 in U2OS cells, it emerged that IDOL autoubiquitylation and LDLR lysosomal targeting do not rely exclusively on Lysine-48- or Lysine-63-linked chains 42. Accordingly, UBE2D family proteins can catalyze the formation of different ubiquitin chains 40, supporting the conclusion that the same E2 can promote substrate and E3 ubiquitylation. However, the reported E2 was identified through a biased screening of only 19 candidate E2 enzymes, based on their previous characterization as preferentially interacting with the RING-type E3 ligases 38,39. In addition, the IDOL RING domain can support in vitro, and to a lesser extent in vivo, generation of lysine-63 polyubiquitin chains in complex with the E2 UBC13/UEV1a, which specifically promotes ubiquitin conjugation through Lysine-63 linkages in the presence of a compatible RING E3 43,44 (Chapter 4 and Zhang et al. 42 ). Therefore, it remains possible that additional E2 enzymes could also interact with, and regulate, IDOL and LDLR ubiquitylation and degradation in vivo. Regulation and reversibility of the IDOL-dependent ubiquitylation of the LDLR In addition to controlling the LDLR half-life, the involvement of the UPS system in the regulation in the LDLR pathway via IDOL introduces two further regulatory elements in the mechanisms of lipoprotein uptake, namely the Deubiquitylating enzymes (DUBs) USP2 and USP8, and the ESCRT system for the sorting of ubiquitylated membrane cargo (Chapter 6 and 7). Ubiquitylation is a reversible post-translational modification, and DUBs function in the UPS system by opposing the activity of E3 ligases 50,51. To date, about 100 DUBs have been identified and grouped in 5 enzyme families based on their catalytic domain signature 52. Most DUBs process ubiquitin chains into monoubiquitin, by binding two ubiquitins and cleaving the intermolecular isopeptide bond. Furthermore, DUBs can present high selectivity for specific ubiquitin-chain topologies, or show large substrate promiscuity, as in the case of USP family members 53. Therefore, similar to their E3 counterparts, DUBs are involved in a wide range of physiological processes, including protein degradation, DNA repair, endocytosis and signaling 51. USP2 is the only DUB currently known to play a role in lipid metabolism. In fact, one of its first recognized physiological substrates is the enzyme Fatty Acid Synthase (FAS), and depletion of USP2 by RNA interference results in increased proteasomal degradation of FAS 54,55. FAS is highly expressed in prostate cancer, suggesting a functional role for fatty acid synthesis in the growth or survival of cancer cells The identification of USP2 as a novel interacting partner and modulator of IDOL-mediated degradation of the LDLR (Chapter 7) further supports a role for this DUB in regulation of cellular lipid metabolism. The findings reported in Chapter 7 indicate that by deubiquitylating IDOL, USP2 is able to reduce IDOL-dependent degradation of the LDLR, leading to increased LDLR abundance and enhanced cellular LDL uptake. The proposed inactivation model introduces the possibility that IDOL could be ubiquitylated in trans by another E3 ligase, and that USP2 would target this activating ubiquitin signal. In essence, this alternative trans-ubiquitylation event would act as an on switch, converting IDOL to its active form that is able to promote LDLR degradation and auto-ubiquitylation. Under this model USP2 would act instead as the off switch, shifting IDOL to an inactive and stable form, characterized by decreased autoubiquitylation and reduced ability to target the LDLR. The described mechanism represents an unconventional facet of the DUB-E3 regulation network. In fact, Chapter 8 111
9 Chapter 8 deubiquitylation of E3 ligases impacts their autoubiquitylation or modification by exogenous E3s, generally leading to stabilization the E3 ligase and consequently, to enhanced ubiquitylation of its targets 59,60. A conceivable alternative to this model is that in addition to IDOL, USP2 could also recognize and deubiquitylate the LDLR, following its ubiquitylation by IDOL. Several in vitro-based findings suggest that USP2 is able to hydrolyze both Lysine-48 and lysine-63 ubiquitin chains 53,61,62, both signals used in vivo by IDOL for mediating its selfdegradation and for lysosomal turnover of the LDLR (Chapter 4 and Zhang et al. 42 ). Therefore, it is possible to envision a scenario in which USP2 could target in parallel both IDOL and the LDLR, and by doing so abolish IDOL-dependent degradation of the LDLR, in spite of accumulation of IDOL. Collectively, these findings underscore the importance of further investigating the role of USP2 and the ubiquitylation mechanisms regulating the activity and degradation of IDOL and LDLR, and more generally, of E3-ubiquitin ligases and their substrates. In addition to USP2 s substrate promiscuity and its widespread role in different cellular processes 61,63, USP2 expression has been found to be regulated by different stimuli related to nutrient sensing or hormonal signaling 54,64,65, but to be independent of SREBPs or LXRs (Chapter 7). Unveiling metabolism-dependent mechanisms governing expression of USP2 and consequently the IDOL-LDLR pathway would therefore increase the regulatory complexity of the LXR-IDOL-LDLR nexus, by potentially intertwining the LXRdependent sterol homeostasis with other metabolic networks and nutrient sensing signaling. The second DUB enzyme regulating the LDLR pathway is USP8 (Chapter 6). Different from USP2, USP8 does not interact with IDOL and its action range is determined by its association with the ESCRT system 66, placing USP8 downstream of IDOL-mediated ubiquitylation of the LDLR, regulating the ESCRT-dependent sorting of the ubiquitylated LDLR to the lysosomes, as it has been shown in the case of the EGFR 67. Complementary to the reduced LDLR degradation observed upon USP8 overexpression in the presence of IDOL, silencing of USP8 increases IDOL-dependent LDLR ubiquitylation, strongly suggesting that USP8 directly deubiquitylates the LDLR, a step used for salvaging ubiquitin prior to lysosomal destruction of the receptor 68. However, the USP8-dependent control of the LDLR ubiquitylation status could also be an indirect consequence of perturbing USP8 expression. In particular, two mechanisms could be plausible: (1) USP8 can also modify the ubiquitylation state of ESCRT components 66. Therefore, perturbing USP8 levels in the cell could result in ESCRT machinery functional impairment and blockage of ubiquitylated cargo transport to the lysosomes. (2) USP8 downregulation may result in increased accumulation of ubiquitylated proteins at the level of the early endosomes and consequent congestion of the ESCRT-mediated sorting. In support of these scenarios, both knockdown of overexpression of USP8 were found to inhibit IDOL-mediated degradation of the LDLR, instead of providing opposite phenotypes (Chapter 6 and Scotti et al. 68 ). Importantly, the discovery of ESCRT-USP8-based sorting of the LDLR following its ubiquitylation by IDOL adds an alternative internalization route for IDOL-sensitive lipoprotein receptors independent of the well-established clathrin-mediated endocytosis pathway, initially postulated and characterized by Brown and Goldstein (Chapter 1). Different lines of evidence illustrate that IDOL is able to recognize and ubiquitylate a lipid raft-localized LDLR pool in the plasma membrane, and through the engagement of the endocytic adaptor Epsin, initiate sorting of the LDLR through the ESCRT system, to route the receptor for degradation (Chapter 6 and Scotti et al. 68 ). The ESCRT is a conserved multi-complex machinery with a critical role in mediating sorting of ubiquitylated membrane proteins for lysosomal degradation 69. Ubiquitin-mediated degradation of membrane proteins is a general cellular quality control mechanism for misfolded receptors and for the attenuation of receptormediated signaling pathways 33,70,71. Internalization of these proteins by endocytosis leads to 112
10 Summary and General Discussion their localization in endosomes, which can form a platform for assembly of the ESCRT machinery on their cytosolic-facing membrane as a means to capture ubiquitylated cargo. This serves to prevent cargo recycling to the plasma membrane or retrograde trafficking 69. Subsequently, the ESCRT activity results in generation of endosomal invaginations where the cargo is sorted into, leading to the formation of Multi-Vesicular Bodies (MVBs) 72,73. In the last step of this sorting process, MVBs release their content into late endosomes/lysosomes eventually leading to degradation of the sorted cargo proteins 69,73. In agreement with this series of events, hampering the activity of different components of the ESCRT interferes with MVB-dependent lysosomal degradation of the LDLR (Chapter 6 and Scotti et al. 68 ). Recent evidence suggests that N-myc downstreamregulated gene 1 (NDRG1), a protein known to interact with components of the vesicular trafficking 74,75, is a novel regulator of MVBs biogenesis and IDOL-dependent endosomal trafficking of the LDLR, further emphasizing the role of IDOL in controlling LDLR trafficking and degradation through the ESCRT system. Consistent with the selective role of the ESCRT in recognition and trafficking of ubiquitylated membrane proteins, the PCSK9 route for LDLR degradation does not require ubiquitylation of the receptor and is independent of the ESCRT 76 ; rather, PCSK9 induces LDLR internalization in clathrin-coated pits, similar to the receptor-mediated lipoprotein uptake 77,78.Taken together, these findings confirm that IDOL and PCSK9 define independent but complementary pathways for regulating LDL uptake via lysosomal degradation of the LDLR. What is the substrate specificity of IDOL? While E2s, DUBs and the ESCRT are involved in the regulation of a broad spectrum of substrates through the UPS, E3 ligases determine the specificity of ubiquitylation by means of interaction with a restricted set of protein targets 30. The RING domain of IDOL is necessary and sufficient for establishing IDOL as a bona fide E3 ligase 45. Yet, it cannot promote ubiquitylation of the LDLR in vivo, indicating that the functional requirements for the interaction of IDOL with the LDLR are not embedded within the RING. Rather, the ability of IDOL to specifically ensure degradation of the LDLR through its E3 ligase activity depends on the presence the N-terminal FERM domain, critical for the interaction between IDOL and the intracellular tail of the LDLR and the two other lipoprotein receptors targeted by this E3 ligase, VLDLR and ApoER2 (Chapter 3 and Chapter 4). Currently, no protein structure of the intact IDOL, either in the absence or in the presence of its substrates, is available; therefore, all the structural analysis of the IDOL-lipoprotein receptor network relies on the generation of structural homology models, based on other known FERM structures or on the PTB domain of DAB1 binding to the ApoER2 cytosolic tail (Chapter4 and Calkin et al. 46 ). The findings presented in Chapter 3 and 4 illustrate two key features of the FERM-dependent recognition by IDOL 46 : (1) The critical role of conserved amino acid residues within the FERM s PTB motif in forming the interaction interface with the LDLR; this functional conservation is extended to the insect homologs of IDOL and LDLR, DNR1 and LpR respectively, as described in Chapter 3. (2) The identification of a conserved WxxKNxxSI/MxF sequence in the LDLR, VLDLR and ApoER2 cytosolic tails as an IDOL recognition motif. This sequence contains all the structural requirements for the interaction between these receptors and the FERM domain of IDOL, and it is located upstream of the key conserved Lysine targeted by IDOL for ubiquitin conjugation. These observations support the hypothesis of a direct interaction between the FERM and the LDLR, directly confirmed in vitro using fluorescence polarization assays to monitor binding of the FERM domain to a synthetic fluorescent LDLR peptide 46. The high FERM-dependent substrate specificity of IDOL further strengthens the concept that the IDOL-mediated turnover of these receptors is a conserved mechanism dedicated to the regulation of lipoprotein Chapter 8 113
11 Chapter 8 metabolism. Yet, since most E3 ligases have multiple targets one can question whether IDOL has additional ubiquitylation targets beyond lipoprotein receptors? The myosin regulatory light chain protein (MRLC) has been also proposed as a potential IDOL target in neuronal cells 47 and cardiomyocytes 48 (Chapter 2). In fact, IDOL is also known as MYLIP, or MRLCinteracting protein, and it has been reported to inhibit nerve growth factor-driven neurite outgrowth in neuronal cells following ubiquitin-dependent degradation of MRLC 47,49. Alternatively, transcriptome analysis of 17 -estradiol-treated human myocardium identified IDOL/MYLIP as a sex-specific estrogen-responsive gene stimulating MRLC ubiquitylation and degradation in vivo 48. IDOL-dependent MRLC degradation has not been observed in the course of the identification of IDOL as a novel modulator of LDLR in non-neuronal cells 11, however it remains possible that IDOL could hold additional targets and functions in different tissues and cell types. Does IDOL have a physiological relevance in LDL metabolism? The functional and molecular characterization of the LXR-IDOL-LDLR nexus described in this thesis has deepened our understanding of the LDLR pathway and the complexity of its regulation. By modulating the levels of the LDLR, IDOL s activity ultimately controls cellular uptake of LDL-cholesterol. Activation of the LXR pathway, or exogenous expression of IDOL in the cell consistently results in substantial decrease of LDL internalization 11 (Chapters 5-7); opposite to this, silencing of IDOL 11, or attenuation of its activity by mutagenesis or by USP2 (Chapters 5 and Chapter 7), restores functional LDL uptake, due to decreased LDLR degradation. However, these functional studies are largely based on biochemical assays and in vitro observations made in model cell lines. Therefore, the physiological role of IDOL in LDL-cholesterol metabolism remains to be fully addressed. Gain-of-function experiments in mice show that IDOL is able to degrade the LDLR and decrease LDL clearance in vivo 11, while reduced IDOL expression due to genetic loss of LXRs leads a minor increase in hepatic LDLR content 11 (Chapter 2). Recently, the generation of Idol (-/-) mice has been reported 76 ; while their metabolic phenotype has not been described yet, Idol (-/-) embryonic stem cells were shown to have increased LDLR protein levels, enhanced LDL internalization, and to be insensitive to LXR-mediated degradation of the receptor 76. These effects were observed in spite of a functional SREBP pathway, in line with the fact that statins could still upregulate LDLR expression in Idol-null cells. Intriguingly, IDOL was amongst the few novel genes identified through GWAS studies as a modifier of circulating levels of LDL in humans 2,79-81 (Chapter 2). By screening individuals with extreme LDL phenotypes in the Dutch population, we recently reported the identification of the IDOL variant p.arg266x in individuals with low circulating levels of LDL, representing the first complete loss-of-function IDOL allele in humans (Chapter 5). In spite of our study design, our screening approach did not result in identification of gain-of-function mutations of IDOL. Given the limited number of individuals we screened it is possible that our approach lacked sufficient power to identify rare gain-of-function IDOL variants, which may be found through a larger sequencing effort. Alternatively, gain-of-function IDOL variants may not exist or be exceedingly rare, particularly taking into account the potency of IDOL activity in human cells, including hepatocytes. In spite of this, the genetic and molecular findings obtained so far on the LXR-IDOL-LDLR nexus support the notion of a role for IDOL in controlling the LDLR pathway and LDL-cholesterol metabolism in vivo. 114
12 Summary and General Discussion Future perspectives This thesis describes the molecular and functional characterization of IDOL, an LXR-inducible E3-ubiquitin ligase that marks the LDLR for lysosomal degradation by ubiquitylation. The ubiquitin system is involved in all cellular processes, and not surprisingly, forms an extended regulation network in sterol homeostasis by controlling the fate of HMGCR and other key components of the cholesterol biosynthesis pathway By controlling the LDLR pathway for cellular uptake of LDL-cholesterol, the LXR-IDOL-LDLR axis further establishes the ubiquitin system as a post-translational regulator of lipid metabolism. IDOL is a unique member within the E3-ubiquitin ligase family, being the only E3 combining the two functional domains FERM and RING. This allows IDOL to recognize with exquisite specificity a subset of lipoprotein receptors (Chapter 3 and Calkin et al. 46 ). Following the initial report on the role of IDOL in controlling LDLR abundance 11, a profound characterization of the IDOL s functional domains and of the LDLR has elucidated key structural patterns, interaction and catalytic mechanisms underlying the IDOL-LDLR network (Chapter 4 and Zhang et al. 41 ). These domain-based findings warrant further investigation aimed at solving the structure of the full-length IDOL, also in complex with its substrates, to achieve a better understanding of the degradation mechanism stimulated by IDOL. Importantly, the characterization of the LXR-IDOL-LDLR pathway has also led to the finding that IDOL s function can be regulated independently of transcriptional control by LXR. The identification of USP2 as a novel component of the IDOL-LDLR pathway (Chapter 7) indicates that IDOL can be regulated at the post-transcriptional level and that inhibition of its activity increases LDLR and cellular LDL uptake. Together with the ESCRT-USP8 dependent sorting of ubiquitylated LDLR (Chapter 6 and Scotti et al. 68 ), these findings substantiate the role of DUBs in controlling the LDLR pathway in conjunction with IDOL. Further investigation might unveil additional mechanisms that regulate the IDOL-LDLR nexus and their impact on LDL metabolism. In spite of this extended molecular understanding of the IDOL-LDLR pathway, whether IDOL physiologically contributes to maintenance of circulating levels of LDL is still unclear. A large body of evidence on LXR activation and IDOL gain- or loss-of-function in cells and mice indicates that IDOL modulates the LDLR pathway. This thesis presents, for the first time, evidence that loss-of-idol function in humans may impact LDL metabolism (Chapter 5). In this study we identified a mutation in IDOL resulting in complete loss of its E3-ubiquitin ligase activity in Dutch carriers with low circulating levels of LDL-cholesterol. Extending the search for genetic variation in loci associated with plasma LDL to larger cohorts and in different populations could potentially lead to the identification of novel mutations altering IDOL function, further establishing its physiological relevance. Given that elevated levels of plasma LDL increase the risk for developing atherosclerosis and cardiovascular disease, characterization and pharmacological targeting of cellular pathways controlling LDLR abundance and LDL clearance may allow development of novel strategies for treating dyslipidemia, complementary to statins 85. Inhibition of PCSK9 is being extensively investigated in clinical trials, and there are now numerous clinical studies demonstrating that blocking PCSK9-mediated LDLR degradation substantially decreases circulating LDL levels The research presented in this thesis supports investigating strategies to inhibit IDOL activity or expression in addition to PCSK9 and statin-based therapy. The findings of Chapter 5 and the observation that Idol (-/-) embryonic stem cells possess an enhanced, LXR-insensitive, LDLR pathway for uptake of LDL 76, suggest that RNAi technology could be used, for instance, to reduce expression of IDOL in the liver and increase LDLR levels, similar to PCSK9 repression 15. Alternatively, pharmacological inhibition of IDOL Chapter 8 115
13 Chapter 8 catalytic activity, or blocking the IDOL-E2 or IDOL-LDLR interactions could be used to achieve this goal. E3-ubiquitin ligases represent one of the largest protein families in the mammalian genome. Due to their exquisite substrate specificity they are the subject of heated investigations aimed at developing highly specific drugs that target their activity 86,87. In view of IDOL s unique role in controlling the LDLR pathway, the mechanistic understanding of IDOL function and regulation, and the seemingly narrow substrate specificity of IDOL, this E3 would be a prime candidate to target for lowering circulating levels of LDL cholesterol. Inhibition of hepatic IDOL activity is predicted to increase LDLR abundance in the liver and to enhance hepatic LDL-cholesterol clearance. Importantly, the experiments described in this thesis show that treatment of cells with statins does not impact the LXR-IDOL pathway, in agreement with the observation that loss of IDOL enhances the statin-mediated increase of LDLR and LDL uptake 11,76. This again illustrates the independent nature of SREBPs and LXRs in modulating the LDLR pathway and reinforces the concept that inhibition of IDOL could be complementary to current statin-based treatment of hypercholesterolemia. 116
14 Summary and General Discussion References 1. Brown, M. S., and Goldstein, J. L. A receptor-mediated pathway for cholesterol homeostasis. Science 232, (1986). 2. Teslovich, T. M. et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, (2010). 3. Chasman, D. I. et al. Genetic Determinants of Statin-Induced Low-Density Lipoprotein Cholesterol Reduction: The Justification for the Use of Statins in Prevention: An Intervention Trial Evaluating Rosuvastatin (JUPITER) Trial. Circulation: Cardiovascular Genetics 5, (2012). 4. Hobbs, H. H., Russell, D. W., Brown, M. S. & Goldstein, J. L. The LDL receptor locus in familial hypercholesterolemia: mutational analysis of a membrane protein. Annu. Rev. Genet. 24, (1989). 5. Horton, J. D., Goldstein, J. L. & Brown, M. S. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109, (2002). 6. Anderson, R. G. & Goldstein, J. L. Localization of low density lipoprotein receptors on plasma membrane of normal human fibroblasts and their absence in cells from a familial hypercholesterolemia homozygote. Proceedings of the National Academy of Sciences 73, (1976). 7. Goldstein, J. L. & Brown, M. S. The LDL Receptor. Arteriosclerosis, Thrombosis, and Vascular Biology 29, (2009). 8. Calandra, S. et al. Mechanisms and genetic determinants regulating sterol absorption, circulating LDL levels, and sterol elimination: implications for classification and disease risk. The Journal of Lipid Research 52, (2011). 9. Garcia, C. K. Autosomal Recessive Hypercholesterolemia Caused by Mutations in a Putative LDL Receptor Adaptor Protein. Science 292, (2001). 10. Horton, J. D., Cohen, J. C. & Hobbs, H. H. PCSK9: a convertase that coordinates LDL catabolism. The Journal of Lipid Research 50, S172 S177 (2008). 11. Zelcer, N., Hong, C., Boyadjian, R. & Tontonoz, P. LXR Regulates Cholesterol Uptake Through Idol-Dependent Ubiquitination of the LDL Receptor. Science 325, (2009). 12. Lindholm, M. W. et al. PCSK9 LNA Antisense Oligonucleotides Induce Sustained Reduction of LDL Cholesterol in Nonhuman Primates. Molecular Therapy 20, (2011). 13. Petrides, F. et al. The promises of PCSK9 inhibition. Current Opinion in Lipidology 24, (2013). 14. Stein, E. A. et al. Effect of a Monoclonal Antibody to PCSK9 on LDL Cholesterol. N Engl J Med 366, (2012). 15. Frank-Kamenetsky, M. et al. Therapeutic RNAi targeting PCSK9 acutely lowers plasma cholesterol in rodents and LDL cholesterol in nonhuman primates. Proceedings of the National Academy of Sciences 105, (2008). 16. Graham, M. J. et al. Antisense inhibition of proprotein convertase subtilisin/kexin type 9 reduces serum LDL in hyperlipidemic mice. The Journal of Lipid Research 48, (2007). 17. Chan, J. C. Y. et al. A proprotein convertase subtilisin/kexin type 9 neutralizing antibody reduces serum cholesterol in mice and nonhuman primates. Proceedings of the National Academy of Sciences 106, (2009). 18. Lambert, G., Sjouke, B., Choque, B., Kastelein, J. J. P. & Hovingh, G. K. The PCSK9 decade: Thematic Review Series: New Lipid and Lipoprotein Targets for the Treatment of Cardiometabolic Diseases. The Journal of Lipid Research 53, (2012). 19. Im, S. S. & Osborne, T. F. Liver X Receptors in Atherosclerosis and Inflammation. Circulation Research 108, (2011). 20. Calkin, A. C. & Tontonoz, P. Liver X Receptor Signaling Pathways and Atherosclerosis. Arteriosclerosis, Thrombosis, and Vascular Biology 30, (2010). Chapter 8 117
15 Chapter Zhang, Y. et al. Liver LXR expression is crucial for whole body cholesterol homeostasis and reverse cholesterol transport in mice. J. Clin. Invest. 122, (2012). 22. Zhao, C. & Dahlman-Wright, K. Liver X receptor in cholesterol metabolism. Journal of Endocrinology 204, (2010). 23. Zadelaar, S. et al. Mouse Models for Atherosclerosis and Pharmaceutical Modifiers. Arteriosclerosis, Thrombosis, and Vascular Biology 27, (2007). 24. Schuster, G. U. Accumulation of Foam Cells in Liver X Receptor-Deficient Mice. Circulation 106, (2002). 25. Sharkey, M. F., Miyanohara, A., Elam, R. L., Friedmann, T. & Witztum, J. L. Posttranscriptional regulation of retroviral vector-transduced low density lipoprotein receptor activity. The Journal of Lipid Research 31, (1990). 26. Verschuren, L., de Vries-van der Weij, J., Zadelaar, S., Kleemann, R. & Kooistra, T. LXR agonist suppresses atherosclerotic lesion growth and promotes lesion regression in apoe*3leiden mice: time course and mechanisms. The Journal of Lipid Research 50, (2008). 27. Groot, P. H. E. Synthetic LXR agonists increase LDL in CETP species. The Journal of Lipid Research 46, (2005). 28. Hershko, A. & Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 67, (1998). 29. Deshaies, R. J. & Joazeiro, C. A. P. RING Domain E3 Ubiquitin Ligases. Annu. Rev. Biochem. 78, (2009). 30. Pickart, C. M. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70, (2001). 31. Scheffner, M., Huibregtse, J. M., Vierstra, R. D. & Howley, P. M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75, (1993). 32. Deshaies, R. J. Scf And Cullin/Ring H2-Based Ubiquitin Ligases. Annu. Rev. Cell Dev. Biol. 15, (1999). 33. MacGurn, J. A., Hsu, P.-C. & Emr, S. D. Ubiquitin and Membrane Protein Turnover: From Cradle to Grave. Annu. Rev. Biochem. 81, (2012). 34. Thrower, J. S., Hoffman, L., Rechsteiner, M. & Pickart, C. M. Recognition of the polyubiquitin proteolytic signal. The EMBO Journal 19, (2000). 35. Yang, Y., Fang, S., Jensen, J. P., Weissman, A. M. & Ashwell, J. D. Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science 288, (2000). 36. Ye, Y. & Rape, M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol 10, (2009). 37. Bruford, E. A. et al. The HGNC Database in 2008: a resource for the human genome. Nucleic Acids Res. 36, D445 8 (2008). 38. van Wijk, S., de Vries, S. J. & Kemmeren, P. A comprehensive framework of E2 RING E3 interactions of the human ubiquitin proteasome system. Molecular systems Biology 5:295. doi: /msb (2009). 39. Markson, G. et al. Analysis of the human E2 ubiquitin conjugating enzyme protein interaction network. Genome Res. 19, (2009). 40. Kim, H. T. et al. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitinprotein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J. Biol. Chem. 282, (2007). 41. Zhang, L. et al. The IDOL-UBE2D complex mediates sterol-dependent degradation of the LDL receptor. Genes & Development 25, (2011). 42. Zhang, L., Xu, M., Scotti, E., Chen, Z. J. & Tontonoz, P. Both K63 and K48 ubiquitin linkages signal lysosomal degradation of the LDL receptor. The Journal of Lipid Research 54, (2013). 43. Nakada, S. et al. Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature 466, (2010). 44. Deng, L. et al. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103, (2000). 118
16 Summary and General Discussion 45. Sorrentino, V. et al. Distinct functional domains contribute to degradation of the low density lipoprotein receptor (LDLR) by the E3 ubiquitin ligase inducible Degrader of the LDLR (IDOL). Journal of Biological Chemistry 286, (2011). 46. Calkin, A. C. et al. FERM-dependent E3 ligase recognition is a conserved mechanism for targeted degradation of lipoprotein receptors. Proceedings of the National Academy of Sciences 108, (2011). 47. Olsson, P. A., Korhonen, L., Mercer, E. A. & Lindholm, D. MIR is a novel ERM-like protein that interacts with myosin regulatory light chain and inhibits neurite outgrowth. The Journal Of Biological Chemistry 274, (1999) 48. Kararigas, G. et al. Transcriptome characterization of estrogen-treated human myocardium identifies myosin regulatory light chain interacting protein as a sexspecific element influencing contractile function. J. Am. Coll. Cardiol. 59, (2012). 49. Bornhauser, B. C., Johansson, C. & Lindholm, D. Functional activities and cellular localization of the ezrin, radixin, moesin (ERM) and RING zinc finger domains in MIR. FEBS Letters 553, (2003). 50. Clague, M. J., Coulson, J. M. & Urbé, S. Cellular functions of the DUBs. Journal of Cell Science 124, (2012). 51. Reyes-Turcu, F. E., Ventii, K. H. & Wilkinson, K. D. Regulation and Cellular Roles of Ubiquitin-Specific Deubiquitinating Enzymes. Annu. Rev. Biochem. 78, (2009). 52. Komander, D., Clague, M. J. & Urbé, S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol 10, (2009). 53. Komander, D. et al. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep 10, (2009). 54. Graner, E., Tang, D., Rossi, S., Baron, A. & Migita, T. The isopeptidase USP2a regulates the stability of fatty acid synthase in prostate cancer. Cancer Cell 5, (2004). 55. Priolo, C. et al. The isopeptidase USP2a protects human prostate cancer from apoptosis. Cancer Research 66, (2006). 56. Dhanasekaran, S. M. et al. Delineation of prognostic biomarkers in prostate cancer. Nature 412, (2001). 57. Kuhajda, F. P. Fatty-acid synthase and human cancer: new perspectives on its role in tumor biology. Nutrition 16, (2000). 58. Rossi, S. et al. Fatty Acid Synthase Expression Defines Distinct Molecular Signatures in Prostate Cancer. Molecular Cancer Research 1, (2003). 59. Weissman, A. M., Shabek, N. & Ciechanover, A. The predator becomes the prey: regulating the ubiquitin system by ubiquitylation and degradation. Nat Rev Mol Cell Biol 12, (2011). 60. de Bie, P. & Ciechanover, A. Ubiquitination of E3 ligases: self-regulation of the ubiquitin system via proteolytic and non-proteolytic mechanisms. Cell Death and Differentiation 18, (2011). 61. Liu, Z. et al. The ubiquitin-specific protease USP2a prevents endocytosis-mediated EGFR degradation. Oncogene 32, (2012). 62. Virdee, S., Ye, Y., Nguyen, D. P., Komander, D. & Chin, J. W. Engineered diubiquitin synthesis reveals Lys29-isopeptide specificity of an OTU deubiquitinase. Nature Chemical Biology 6, (2010). 63. Shan, J., Zhao, W. & Gu, W. Suppression of Cancer Cell Growth by Promoting Cyclin D1 Degradation. Molecular Cell 36, (2009). 64. Molusky, M. M., Li, S., Ma, D., Yu, L. & Lin, J. D. Ubiquitin-specific protease 2 regulates hepatic gluconeogenesis and diurnal glucose metabolism through 11 hydroxysteroid dehydrogenase 1. Diabetes 61, (2012). 65. Calvisi, D. F. et al. Increased Lipogenesis, Induced by AKT-mTORC1-RPS6 Signaling, Promotes Development of Human Hepatocellular Carcinoma. Gastroenterology 140, e5 (2011). 66. Clague, M. J. & Urbé, S. Endocytosis: the DUB version. Trends in Cell Biology 16, (2006). 67. Mizuno, E. et al. Regulation of epidermal growth factor receptor down-regulation by Chapter 8 119
17 Chapter 8 UBPY-mediated deubiquitination at endosomes. Molecular Biology of the Cell 16, (2005). 68. Scotti, E. et al. IDOL stimulates clathrin-independent endocytosis and MVB-mediated lysosomal degradation of the LDLR. Molecular and Cellular Biology 33, (2013). 69. Raiborg, C. & Stenmark, H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 458, (2009). 70. Sigismund, S. et al. Clathrin-Mediated Internalization Is Essential for Sustained EGFR Signaling but Dispensable for Degradation. Developmental Cell 15, (2008). 71. Ettenberg, S. A., Katz, M., Tsygankov, A. Y. & Alroy, I. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-cbl/sli-1. Molecular Cell 4, (1999). 72. Wollert, T. et al. The ESCRT machinery at a glance. Journal of Cell Science 122, (2009). 73. Gruenberg, J. & Stenmark, H. Opinion: The biogenesis of multivesicular endosomes. Nat Rev Mol Cell Biol 5, (2004). 74. Hunter, M., Angelicheva, D., Tournev, I. & Ingley, E. NDRG1 interacts with APO AI and A-II and is a functional candidate for the HDL-C QTL on 8q24. Biochemical and Biophysical Research Communications 332, (2005). 75. Kachhap, S. K. et al. The N-Myc Down Regulated Gene1 (NDRG1) Is a Rab4a Effector Involved in Vesicular Recycling of E-Cadherin. PLoS ONE 2, e844 (2007). 76. Scotti, E. et al. Targeted Disruption of the Idol Gene Alters Cellular Regulation of the Low-Density Lipoprotein Receptor by Sterols and Liver X Receptor Agonists. Molecular and Cellular Biology 31, (2011). 77. Wang, Y., Huang, Y., Hobbs, H. H. & Cohen, J. C. Molecular characterization of proprotein convertase subtilisin/kexin type 9-mediated degradation of the LDLR. The Journal of Lipid Research 53, (2012). 78. McPherson, P. S., Attie, A. D., Prat, A. & Seidah, N. G. The cellular trafficking of the secretory proprotein convertase PCSK9 and its dependence on the LDLR Traffic 8, (2007). 79. Chasman, D. I. et al. Forty-Three Loci Associated with Plasma Lipoprotein Size, Concentration, and Cholesterol Content in Genome-Wide Analysis. PLoS Genet 5, e (2009). 80. Waterworth, D. M. et al. Genetic Variants Influencing Circulating Lipid Levels and Risk of Coronary Artery Disease. Arteriosclerosis, Thrombosis, and Vascular Biology 30, (2010). 81. Weissglas-Volkov, D. et al. The N342S MYLIP polymorphism is associated with high total cholesterol and increased LDL receptor degradation in humans. J. Clin. Invest. 121, (2011). 82. Gill, S., Stevenson, J., Kristiana, I. & Brown, A. J. Cholesterol-Dependent Degradation of Squalene Monooxygenase, a Control Point in Cholesterol Synthesis beyond HMG- CoA Reductase. Cell Metabolism 13, (2011). 83. Gong, Y. et al. Sterol-regulated ubiquitination and degradation of Insig-1 creates a convergent mechanism for feedback control of cholesterol synthesis and uptake. Cell Metabolism 3, (2006). 84. DeBose-Boyd, R. A. Feedback regulation of cholesterol synthesis: sterol-accelerated ubiquitination and degradation of HMG CoA reductase. Cell Res 18, (2008). 85. Brown, M. S., Goldstein J. L. Lowering Plasma Cholesterol by Raising LDL Receptors. Atherosclerosis Supplements 5, (1981). 86. Lydeard, J. R. & Harper, J. W. Inhibitors for E3 ubiquitin ligases. Nature Publishing Group 28, (2010). 87. Goldenberg, S. J., Marblestone, J. G., Mattern, M. R. & Nicholson, B. Strategies for the identification of ubiquitin ligase inhibitors. Biochem. Soc. Trans 38, 132 (2010). 120
Post-transcriptional regulation of lipoprotein receptors by the E3-ubiquitin ligase inducible degrader of the low-density lipoprotein receptor
 REVIEW C URRENT OPINION Post-transcriptional regulation of lipoprotein receptors by the E3-ubiquitin ligase inducible degrader of the low-density lipoprotein receptor Vincenzo Sorrentino and Noam Zelcer
REVIEW C URRENT OPINION Post-transcriptional regulation of lipoprotein receptors by the E3-ubiquitin ligase inducible degrader of the low-density lipoprotein receptor Vincenzo Sorrentino and Noam Zelcer
Citation for published version (APA): Von Eije, K. J. (2009). RNAi based gene therapy for HIV-1, from bench to bedside
 UvA-DARE (Digital Academic Repository) RNAi based gene therapy for HIV-1, from bench to bedside Von Eije, K.J. Link to publication Citation for published version (APA): Von Eije, K. J. (2009). RNAi based
UvA-DARE (Digital Academic Repository) RNAi based gene therapy for HIV-1, from bench to bedside Von Eije, K.J. Link to publication Citation for published version (APA): Von Eije, K. J. (2009). RNAi based
UvA-DARE (Digital Academic Repository) Marfan syndrome: Getting to the root of the problem Franken, Romy. Link to publication
 UvA-DARE (Digital Academic Repository) Marfan syndrome: Getting to the root of the problem Franken, Romy Link to publication Citation for published version (APA): Franken, R. (2016). Marfan syndrome: Getting
UvA-DARE (Digital Academic Repository) Marfan syndrome: Getting to the root of the problem Franken, Romy Link to publication Citation for published version (APA): Franken, R. (2016). Marfan syndrome: Getting
Cellular control of cholesterol. Peter Takizawa Department of Cell Biology
 Cellular control of cholesterol Peter Takizawa Department of Cell Biology Brief overview of cholesterol s biological role Regulation of cholesterol synthesis Dietary and cellular uptake of cholesterol
Cellular control of cholesterol Peter Takizawa Department of Cell Biology Brief overview of cholesterol s biological role Regulation of cholesterol synthesis Dietary and cellular uptake of cholesterol
UvA-DARE (Digital Academic Repository) Genetic basis of hypertrophic cardiomyopathy Bos, J.M. Link to publication
 UvA-DARE (Digital Academic Repository) Genetic basis of hypertrophic cardiomyopathy Bos, J.M. Link to publication Citation for published version (APA): Bos, J. M. (2010). Genetic basis of hypertrophic
UvA-DARE (Digital Academic Repository) Genetic basis of hypertrophic cardiomyopathy Bos, J.M. Link to publication Citation for published version (APA): Bos, J. M. (2010). Genetic basis of hypertrophic
Citation for published version (APA): van Munster, B. C. (2009). Pathophysiological studies in delirium : a focus on genetics.
 UvA-DARE (Digital Academic Repository) Pathophysiological studies in delirium : a focus on genetics van Munster, B.C. Link to publication Citation for published version (APA): van Munster, B. C. (2009).
UvA-DARE (Digital Academic Repository) Pathophysiological studies in delirium : a focus on genetics van Munster, B.C. Link to publication Citation for published version (APA): van Munster, B. C. (2009).
From Biology to Therapy The biology of PCSK9 in humans Just LDL-cholesterol or more? May 24th. Dr. Gilles Lambert
 Dr. Gilles Lambert Associate Professor in Cell Biology University of Nantes Medical School Group Leader, Laboratory of Nutrition and Metabolism, University Hospital of Nantes From Biology to Therapy The
Dr. Gilles Lambert Associate Professor in Cell Biology University of Nantes Medical School Group Leader, Laboratory of Nutrition and Metabolism, University Hospital of Nantes From Biology to Therapy The
Gezinskenmerken: De constructie van de Vragenlijst Gezinskenmerken (VGK) Klijn, W.J.L.
 UvA-DARE (Digital Academic Repository) Gezinskenmerken: De constructie van de Vragenlijst Gezinskenmerken (VGK) Klijn, W.J.L. Link to publication Citation for published version (APA): Klijn, W. J. L. (2013).
UvA-DARE (Digital Academic Repository) Gezinskenmerken: De constructie van de Vragenlijst Gezinskenmerken (VGK) Klijn, W.J.L. Link to publication Citation for published version (APA): Klijn, W. J. L. (2013).
Citation for published version (APA): Sivapalaratnam, S. (2012). The molecular basis of early onset cardiovascular disease
 UvA-DARE (Digital Academic Repository) The molecular basis of early onset cardiovascular disease Sivapalaratnam, S. Link to publication Citation for published version (APA): Sivapalaratnam, S. (2012).
UvA-DARE (Digital Academic Repository) The molecular basis of early onset cardiovascular disease Sivapalaratnam, S. Link to publication Citation for published version (APA): Sivapalaratnam, S. (2012).
Lactase, sucrase-isomaltase, and carbamoyl phosphate synthase I expression in human intestine van Beers, E.H.
 UvA-DARE (Digital Academic Repository) Lactase, sucrase-isomaltase, and carbamoyl phosphate synthase I expression in human intestine van Beers, E.H. Link to publication Citation for published version (APA):
UvA-DARE (Digital Academic Repository) Lactase, sucrase-isomaltase, and carbamoyl phosphate synthase I expression in human intestine van Beers, E.H. Link to publication Citation for published version (APA):
Protein Trafficking in the Secretory and Endocytic Pathways
 Protein Trafficking in the Secretory and Endocytic Pathways The compartmentalization of eukaryotic cells has considerable functional advantages for the cell, but requires elaborate mechanisms to ensure
Protein Trafficking in the Secretory and Endocytic Pathways The compartmentalization of eukaryotic cells has considerable functional advantages for the cell, but requires elaborate mechanisms to ensure
Lipid Metabolism in Familial Hypercholesterolemia
 Lipid Metabolism in Familial Hypercholesterolemia Khalid Al-Rasadi, BSc, MD, FRCPC Head of Biochemistry Department, SQU Head of Lipid and LDL-Apheresis Unit, SQUH President of Oman society of Lipid & Atherosclerosis
Lipid Metabolism in Familial Hypercholesterolemia Khalid Al-Rasadi, BSc, MD, FRCPC Head of Biochemistry Department, SQU Head of Lipid and LDL-Apheresis Unit, SQUH President of Oman society of Lipid & Atherosclerosis
Study of different types of ubiquitination
 Study of different types of ubiquitination Rudi Beyaert (rudi.beyaert@irc.vib-ugent.be) VIB UGent Center for Inflammation Research Ghent, Belgium VIB Training Novel Proteomics Tools: Identifying PTMs October
Study of different types of ubiquitination Rudi Beyaert (rudi.beyaert@irc.vib-ugent.be) VIB UGent Center for Inflammation Research Ghent, Belgium VIB Training Novel Proteomics Tools: Identifying PTMs October
Cholesterol and its transport. Alice Skoumalová
 Cholesterol and its transport Alice Skoumalová 27 carbons Cholesterol - structure Cholesterol importance A stabilizing component of cell membranes A precursor of bile salts A precursor of steroid hormones
Cholesterol and its transport Alice Skoumalová 27 carbons Cholesterol - structure Cholesterol importance A stabilizing component of cell membranes A precursor of bile salts A precursor of steroid hormones
Cholesterol metabolism. Function Biosynthesis Transport in the organism Hypercholesterolemia
 Cholesterol metabolism Function Biosynthesis Transport in the organism Hypercholesterolemia - component of all cell membranes - precursor of bile acids steroid hormones vitamin D Cholesterol Sources: dietary
Cholesterol metabolism Function Biosynthesis Transport in the organism Hypercholesterolemia - component of all cell membranes - precursor of bile acids steroid hormones vitamin D Cholesterol Sources: dietary
Endocytosis and Intracellular Trafficking of Notch and Its Ligands
 CHA P T E R F IVE Endocytosis and Intracellular Trafficking of Notch and Its Ligands Shinya Yamamoto, *,1 Wu-Lin Charng, *,1 and Hugo J. Bellen *,,, Contents 1. Notch Signaling and its Regulation by Endocytosis
CHA P T E R F IVE Endocytosis and Intracellular Trafficking of Notch and Its Ligands Shinya Yamamoto, *,1 Wu-Lin Charng, *,1 and Hugo J. Bellen *,,, Contents 1. Notch Signaling and its Regulation by Endocytosis
UvA-DARE (Digital Academic Repository) Vascular factors in dementia and apathy Eurelings, Lisa. Link to publication
 UvA-DARE (Digital Academic Repository) Vascular factors in dementia and apathy Eurelings, Lisa Link to publication Citation for published version (APA): Eurelings, L. S. M. (2016). Vascular factors in
UvA-DARE (Digital Academic Repository) Vascular factors in dementia and apathy Eurelings, Lisa Link to publication Citation for published version (APA): Eurelings, L. S. M. (2016). Vascular factors in
ANSC/NUTR 618 LIPIDS & LIPID METABOLISM The LDL Receptor, LDL Uptake, and the Free Cholesterol Pool
 ANSC/NUTR 618 LIPIDS & LIPID METABOLISM The, LDL Uptake, and the Free Cholesterol Pool I. Michael Brown and Joseph Goldstein A. Studied families with familial hypercholesterolemia. B. Defined the relationship
ANSC/NUTR 618 LIPIDS & LIPID METABOLISM The, LDL Uptake, and the Free Cholesterol Pool I. Michael Brown and Joseph Goldstein A. Studied families with familial hypercholesterolemia. B. Defined the relationship
Molecular Graphics Perspective of Protein Structure and Function
 Molecular Graphics Perspective of Protein Structure and Function VMD Highlights > 20,000 registered Users Platforms: Unix (16 builds) Windows MacOS X Display of large biomolecules and simulation trajectories
Molecular Graphics Perspective of Protein Structure and Function VMD Highlights > 20,000 registered Users Platforms: Unix (16 builds) Windows MacOS X Display of large biomolecules and simulation trajectories
Cell Quality Control. Peter Takizawa Department of Cell Biology
 Cell Quality Control Peter Takizawa Department of Cell Biology Cellular quality control reduces production of defective proteins. Cells have many quality control systems to ensure that cell does not build
Cell Quality Control Peter Takizawa Department of Cell Biology Cellular quality control reduces production of defective proteins. Cells have many quality control systems to ensure that cell does not build
7.06 Cell Biology EXAM #3 April 24, 2003
 7.06 Spring 2003 Exam 3 Name 1 of 8 7.06 Cell Biology EXAM #3 April 24, 2003 This is an open book exam, and you are allowed access to books and notes. Please write your answers to the questions in the
7.06 Spring 2003 Exam 3 Name 1 of 8 7.06 Cell Biology EXAM #3 April 24, 2003 This is an open book exam, and you are allowed access to books and notes. Please write your answers to the questions in the
The Nobel Prize in Chemistry 2004
 The Nobel Prize in Chemistry 2004 Ubiquitous Quality Control of Life C S Karigar and K R Siddalinga Murthy The Nobel Prize in Chemistry for 2004 is shared by Aaron Ciechanover, Avram Hershko and Irwin
The Nobel Prize in Chemistry 2004 Ubiquitous Quality Control of Life C S Karigar and K R Siddalinga Murthy The Nobel Prize in Chemistry for 2004 is shared by Aaron Ciechanover, Avram Hershko and Irwin
Summary and concluding remarks
 Summary and concluding remarks This thesis is focused on the role and interaction of different cholesterol and phospholipid transporters. Cholesterol homeostasis is accomplished via a tightly regulated
Summary and concluding remarks This thesis is focused on the role and interaction of different cholesterol and phospholipid transporters. Cholesterol homeostasis is accomplished via a tightly regulated
PCSK9 and its Role in LDL Receptor Regulation Muscat, Oman - 9 February 2019
 PCSK9 and its Role in LDL Receptor Regulation Muscat, Oman - 9 February 2019 Professor Gilles Lambert, PhD LaboratoireInserm U1188 Universitéde la Réunion Faculté de Médecine Saint Denis de la Réunion,
PCSK9 and its Role in LDL Receptor Regulation Muscat, Oman - 9 February 2019 Professor Gilles Lambert, PhD LaboratoireInserm U1188 Universitéde la Réunion Faculté de Médecine Saint Denis de la Réunion,
Homework Hanson section MCB Course, Fall 2014
 Homework Hanson section MCB Course, Fall 2014 (1) Antitrypsin, which inhibits certain proteases, is normally secreted into the bloodstream by liver cells. Antitrypsin is absent from the bloodstream of
Homework Hanson section MCB Course, Fall 2014 (1) Antitrypsin, which inhibits certain proteases, is normally secreted into the bloodstream by liver cells. Antitrypsin is absent from the bloodstream of
UvA-DARE (Digital Academic Repository) Marfan syndrome: Getting to the root of the problem Franken, Romy. Link to publication
 UvA-DARE (Digital Academic Repository) Marfan syndrome: Getting to the root of the problem Franken, Romy Link to publication Citation for published version (APA): Franken, R. (2016). Marfan syndrome: Getting
UvA-DARE (Digital Academic Repository) Marfan syndrome: Getting to the root of the problem Franken, Romy Link to publication Citation for published version (APA): Franken, R. (2016). Marfan syndrome: Getting
Lysosomes and endocytic pathways 9/27/2012 Phyllis Hanson
 Lysosomes and endocytic pathways 9/27/2012 Phyllis Hanson General principles Properties of lysosomes Delivery of enzymes to lysosomes Endocytic uptake clathrin, others Endocytic pathways recycling vs.
Lysosomes and endocytic pathways 9/27/2012 Phyllis Hanson General principles Properties of lysosomes Delivery of enzymes to lysosomes Endocytic uptake clathrin, others Endocytic pathways recycling vs.
Plasma lipoproteins & atherosclerosis by. Prof.Dr. Maha M. Sallam
 Biochemistry Department Plasma lipoproteins & atherosclerosis by Prof.Dr. Maha M. Sallam 1 1. Recognize structures,types and role of lipoproteins in blood (Chylomicrons, VLDL, LDL and HDL). 2. Explain
Biochemistry Department Plasma lipoproteins & atherosclerosis by Prof.Dr. Maha M. Sallam 1 1. Recognize structures,types and role of lipoproteins in blood (Chylomicrons, VLDL, LDL and HDL). 2. Explain
UvA-DARE (Digital Academic Repository) Improving aspects of palliative care for children Jagt, C.T. Link to publication
 UvA-DARE (Digital Academic Repository) Improving aspects of palliative care for children Jagt, C.T. Link to publication Citation for published version (APA): Jagt, C. T. (2017). Improving aspects of palliative
UvA-DARE (Digital Academic Repository) Improving aspects of palliative care for children Jagt, C.T. Link to publication Citation for published version (APA): Jagt, C. T. (2017). Improving aspects of palliative
UvA-DARE (Digital Academic Repository) An electronic nose in respiratory disease Dragonieri, S. Link to publication
 UvA-DARE (Digital Academic Repository) An electronic nose in respiratory disease Dragonieri, S. Link to publication Citation for published version (APA): Dragonieri, S. (2012). An electronic nose in respiratory
UvA-DARE (Digital Academic Repository) An electronic nose in respiratory disease Dragonieri, S. Link to publication Citation for published version (APA): Dragonieri, S. (2012). An electronic nose in respiratory
INTERNAL MEDICINE - PEDIATRICS
 Rev. Med. Chir. Soc. Med. Nat., Iaşi 2016 vol. 120, no. 2 INTERNAL MEDICINE - PEDIATRICS UPDATES NEW CLASS OF DRUGS: THERAPEUTIC RNAi INHIBITION OF PCSK9 AS A SPECIFIC LDL-C LOWERING THERAPY A.L. Strat
Rev. Med. Chir. Soc. Med. Nat., Iaşi 2016 vol. 120, no. 2 INTERNAL MEDICINE - PEDIATRICS UPDATES NEW CLASS OF DRUGS: THERAPEUTIC RNAi INHIBITION OF PCSK9 AS A SPECIFIC LDL-C LOWERING THERAPY A.L. Strat
UNIVERSITA DI PISA CHIMICA E TECNOLOGIE FARMACEUTICHE FAMILIAL HYPERCHOLESTEROLEMIA DIPARTIMENTO DI FARMACIA GENETIC CAUSES AND THERAPY
 UNIVERSITA DI PISA DIPARTIMENTO DI FARMACIA CHIMICA E TECNOLOGIE FARMACEUTICHE CORSO DI BASI BIOCHIMICHE DELL AZIONE DEI FARMACI FAMILIAL HYPERCHOLESTEROLEMIA GENETIC CAUSES AND THERAPY ERIKA ROSARIA PACCIOLLA
UNIVERSITA DI PISA DIPARTIMENTO DI FARMACIA CHIMICA E TECNOLOGIE FARMACEUTICHE CORSO DI BASI BIOCHIMICHE DELL AZIONE DEI FARMACI FAMILIAL HYPERCHOLESTEROLEMIA GENETIC CAUSES AND THERAPY ERIKA ROSARIA PACCIOLLA
Cholesterol is essential for mammalian cells because it plays
 Brief Review Feedback Regulation of Cholesterol Uptake by the LXR IDOL LDLR Axis Li Zhang, Karen Reue, Loren G. Fong, Stephen G. Young, Peter Tontonoz Abstract Inducible degrader of the low-density lipoprotein
Brief Review Feedback Regulation of Cholesterol Uptake by the LXR IDOL LDLR Axis Li Zhang, Karen Reue, Loren G. Fong, Stephen G. Young, Peter Tontonoz Abstract Inducible degrader of the low-density lipoprotein
Acute lung injury in children : from viral infection and mechanical ventilation to inflammation and apoptosis Bern, R.A.
 UvA-DARE (Digital Academic Repository) Acute lung injury in children : from viral infection and mechanical ventilation to inflammation and apoptosis Bern, R.A. Link to publication Citation for published
UvA-DARE (Digital Academic Repository) Acute lung injury in children : from viral infection and mechanical ventilation to inflammation and apoptosis Bern, R.A. Link to publication Citation for published
Sorting and trafficking of proteins in oligodendrocytes during myelin membrane biogenesis Klunder, Lammert
 University of Groningen Sorting and trafficking of proteins in oligodendrocytes during myelin membrane biogenesis Klunder, Lammert IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's
University of Groningen Sorting and trafficking of proteins in oligodendrocytes during myelin membrane biogenesis Klunder, Lammert IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's
MCB130 Midterm. GSI s Name:
 1. Peroxisomes are small, membrane-enclosed organelles that function in the degradation of fatty acids and in the degradation of H 2 O 2. Peroxisomes are not part of the secretory pathway and peroxisomal
1. Peroxisomes are small, membrane-enclosed organelles that function in the degradation of fatty acids and in the degradation of H 2 O 2. Peroxisomes are not part of the secretory pathway and peroxisomal
Companion to Biosynthesis of Ketones & Cholesterols, Regulation of Lipid Metabolism Lecture Notes
 Companion to Biosynthesis of Ketones & Cholesterols, Regulation of Lipid Metabolism Lecture Notes The major site of acetoacetate and 3-hydorxybutyrate production is in the liver. 3-hydorxybutyrate is the
Companion to Biosynthesis of Ketones & Cholesterols, Regulation of Lipid Metabolism Lecture Notes The major site of acetoacetate and 3-hydorxybutyrate production is in the liver. 3-hydorxybutyrate is the
The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein
 THESIS BOOK The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein Orsolya Buzás-Bereczki Supervisors: Dr. Éva Bálint Dr. Imre Miklós Boros University of
THESIS BOOK The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein Orsolya Buzás-Bereczki Supervisors: Dr. Éva Bálint Dr. Imre Miklós Boros University of
TITLE: Effect of MUC1 Expression on EGFR Endocytosis and Degradation in Human Breast Cancer Cell Lines
 AD AWARD NUMBER: W81XWH-06-1-0464 TITLE: Effect of MUC1 Expression on EGFR Endocytosis and Degradation in Human Breast Cancer Cell Lines PRINCIPAL INVESTIGATOR: Rachid M El Bejjani CONTRACTING ORGANIZATION:
AD AWARD NUMBER: W81XWH-06-1-0464 TITLE: Effect of MUC1 Expression on EGFR Endocytosis and Degradation in Human Breast Cancer Cell Lines PRINCIPAL INVESTIGATOR: Rachid M El Bejjani CONTRACTING ORGANIZATION:
cholesterol structure Cholesterol FAQs Cholesterol promotes the liquid-ordered phase of membranes Friday, October 15, 2010
 cholesterol structure most plasma cholesterol is in the esterified form (not found in cells or membranes) cholesterol functions in all membranes (drives formation of lipid microdomains) cholesterol is
cholesterol structure most plasma cholesterol is in the esterified form (not found in cells or membranes) cholesterol functions in all membranes (drives formation of lipid microdomains) cholesterol is
Lipid metabolism in familial hypercholesterolemia
 Lipid metabolism in familial hypercholesterolemia Khalid Al-Rasadi, BSc, MD, FRCPC Head of Biochemistry Department, SQU Head of Lipid and LDL-Apheresis Unit, SQUH President of Oman society of Lipid & Atherosclerosis
Lipid metabolism in familial hypercholesterolemia Khalid Al-Rasadi, BSc, MD, FRCPC Head of Biochemistry Department, SQU Head of Lipid and LDL-Apheresis Unit, SQUH President of Oman society of Lipid & Atherosclerosis
UvA-DARE (Digital Academic Repository) The artificial pancreas Kropff, J. Link to publication
 UvA-DARE (Digital Academic Repository) The artificial pancreas Kropff, J. Link to publication Citation for published version (APA): Kropff, J. (2017). The artificial pancreas: From logic to life General
UvA-DARE (Digital Academic Repository) The artificial pancreas Kropff, J. Link to publication Citation for published version (APA): Kropff, J. (2017). The artificial pancreas: From logic to life General
Familial hypercholesterolemia in childhood: diagnostics, therapeutical options and risk stratification Rodenburg, J.
 UvADARE (Digital Academic Repository) Familial hypercholesterolemia in childhood: diagnostics, therapeutical options and risk stratification Rodenburg, J. Link to publication Citation for published version
UvADARE (Digital Academic Repository) Familial hypercholesterolemia in childhood: diagnostics, therapeutical options and risk stratification Rodenburg, J. Link to publication Citation for published version
Toll-like Receptors (TLRs): Biology, Pathology and Therapeutics
 Toll-like Receptors (TLRs): Biology, Pathology and Therapeutics Dr Sarah Sasson SydPATH Registrar 23 rd June 2014 TLRs: Introduction Discovered in 1990s Recognise conserved structures in pathogens Rely
Toll-like Receptors (TLRs): Biology, Pathology and Therapeutics Dr Sarah Sasson SydPATH Registrar 23 rd June 2014 TLRs: Introduction Discovered in 1990s Recognise conserved structures in pathogens Rely
Prediction of toxicity in concurrent chemoradiation for non-small cell lung cancer Uijterlinde, W.I.
 UvA-DARE (Digital Academic Repository) Prediction of toxicity in concurrent chemoradiation for non-small cell lung cancer Uijterlinde, W.I. Link to publication Citation for published version (APA): Uijterlinde,
UvA-DARE (Digital Academic Repository) Prediction of toxicity in concurrent chemoradiation for non-small cell lung cancer Uijterlinde, W.I. Link to publication Citation for published version (APA): Uijterlinde,
Antigen Presentation to T lymphocytes
 Antigen Presentation to T lymphocytes Immunology 441 Lectures 6 & 7 Chapter 6 October 10 & 12, 2016 Jessica Hamerman jhamerman@benaroyaresearch.org Office hours by arrangement Antibodies and T cell receptors
Antigen Presentation to T lymphocytes Immunology 441 Lectures 6 & 7 Chapter 6 October 10 & 12, 2016 Jessica Hamerman jhamerman@benaroyaresearch.org Office hours by arrangement Antibodies and T cell receptors
Introduction. Cancer Biology. Tumor-suppressor genes. Proto-oncogenes. DNA stability genes. Mechanisms of carcinogenesis.
 Cancer Biology Chapter 18 Eric J. Hall., Amato Giaccia, Radiobiology for the Radiologist Introduction Tissue homeostasis depends on the regulated cell division and self-elimination (programmed cell death)
Cancer Biology Chapter 18 Eric J. Hall., Amato Giaccia, Radiobiology for the Radiologist Introduction Tissue homeostasis depends on the regulated cell division and self-elimination (programmed cell death)
RAS Genes. The ras superfamily of genes encodes small GTP binding proteins that are responsible for the regulation of many cellular processes.
 ۱ RAS Genes The ras superfamily of genes encodes small GTP binding proteins that are responsible for the regulation of many cellular processes. Oncogenic ras genes in human cells include H ras, N ras,
۱ RAS Genes The ras superfamily of genes encodes small GTP binding proteins that are responsible for the regulation of many cellular processes. Oncogenic ras genes in human cells include H ras, N ras,
Cardiovascular Controversies: Emerging Therapies for Lowering Cardiovascular Risk
 Transcript Details This is a transcript of a continuing medical education (CME) activity accessible on the ReachMD network. Additional media formats for the activity and full activity details (including
Transcript Details This is a transcript of a continuing medical education (CME) activity accessible on the ReachMD network. Additional media formats for the activity and full activity details (including
Intracellular Vesicular Traffic Chapter 13, Alberts et al.
 Intracellular Vesicular Traffic Chapter 13, Alberts et al. The endocytic and biosynthetic-secretory pathways The intracellular compartments of the eucaryotic ell involved in the biosynthetic-secretory
Intracellular Vesicular Traffic Chapter 13, Alberts et al. The endocytic and biosynthetic-secretory pathways The intracellular compartments of the eucaryotic ell involved in the biosynthetic-secretory
Src-INACTIVE / Src-INACTIVE
 Biology 169 -- Exam 1 February 2003 Answer each question, noting carefully the instructions for each. Repeat- Read the instructions for each question before answering!!! Be as specific as possible in each
Biology 169 -- Exam 1 February 2003 Answer each question, noting carefully the instructions for each. Repeat- Read the instructions for each question before answering!!! Be as specific as possible in each
Enzyme-coupled Receptors. Cell-surface receptors 1. Ion-channel-coupled receptors 2. G-protein-coupled receptors 3. Enzyme-coupled receptors
 Enzyme-coupled Receptors Cell-surface receptors 1. Ion-channel-coupled receptors 2. G-protein-coupled receptors 3. Enzyme-coupled receptors Cell-surface receptors allow a flow of ions across the plasma
Enzyme-coupled Receptors Cell-surface receptors 1. Ion-channel-coupled receptors 2. G-protein-coupled receptors 3. Enzyme-coupled receptors Cell-surface receptors allow a flow of ions across the plasma
BIOL212 Biochemistry of Disease. Metabolic Disorders - Obesity
 BIOL212 Biochemistry of Disease Metabolic Disorders - Obesity Obesity Approx. 23% of adults are obese in the U.K. The number of obese children has tripled in 20 years. 10% of six year olds are obese, rising
BIOL212 Biochemistry of Disease Metabolic Disorders - Obesity Obesity Approx. 23% of adults are obese in the U.K. The number of obese children has tripled in 20 years. 10% of six year olds are obese, rising
Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression. Chromatin Array of nuc
 Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression Chromatin Array of nuc 1 Transcriptional control in Eukaryotes: Chromatin undergoes structural changes
Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression Chromatin Array of nuc 1 Transcriptional control in Eukaryotes: Chromatin undergoes structural changes
HIV-1 genome organization
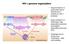 HIV-1 genome organization Gag encodes for a polyprotein that is cleaved into 4 proteins by HIV-1 protease. Gag is present in 2 transcripts: Gag polyprotein precursor and GagPol polyprotein precursor (5%).
HIV-1 genome organization Gag encodes for a polyprotein that is cleaved into 4 proteins by HIV-1 protease. Gag is present in 2 transcripts: Gag polyprotein precursor and GagPol polyprotein precursor (5%).
AperTO - Archivio Istituzionale Open Access dell'università di Torino
 AperTO - Archivio Istituzionale Open Access dell'università di Torino From the nucleus to the mitochondria and backthe odyssey of a multitask STAT3 This is the author's manuscript Original Citation: From
AperTO - Archivio Istituzionale Open Access dell'università di Torino From the nucleus to the mitochondria and backthe odyssey of a multitask STAT3 This is the author's manuscript Original Citation: From
PCSK9 Inhibition: From Genetics to Patients
 PCSK9 Inhibition: From Genetics to Patients John Chapman BSc, Ph.D., D.Sc., FESC Research Professor, University of Pierre and Marie Curie Director Emeritus, INSERM Dyslipidemia and Atherosclerosis Research
PCSK9 Inhibition: From Genetics to Patients John Chapman BSc, Ph.D., D.Sc., FESC Research Professor, University of Pierre and Marie Curie Director Emeritus, INSERM Dyslipidemia and Atherosclerosis Research
Cell Biology Lecture 9 Notes Basic Principles of cell signaling and GPCR system
 Cell Biology Lecture 9 Notes Basic Principles of cell signaling and GPCR system Basic Elements of cell signaling: Signal or signaling molecule (ligand, first messenger) o Small molecules (epinephrine,
Cell Biology Lecture 9 Notes Basic Principles of cell signaling and GPCR system Basic Elements of cell signaling: Signal or signaling molecule (ligand, first messenger) o Small molecules (epinephrine,
AMORE (Ablative surgery, MOulage technique brachytherapy and REconstruction) for childhood head and neck rhabdomyosarcoma Buwalda, J.
 UvA-DARE (Digital Academic Repository) AMORE (Ablative surgery, MOulage technique brachytherapy and REconstruction) for childhood head and neck rhabdomyosarcoma Buwalda, J. Link to publication Citation
UvA-DARE (Digital Academic Repository) AMORE (Ablative surgery, MOulage technique brachytherapy and REconstruction) for childhood head and neck rhabdomyosarcoma Buwalda, J. Link to publication Citation
CELLS. Cells. Basic unit of life (except virus)
 Basic unit of life (except virus) CELLS Prokaryotic, w/o nucleus, bacteria Eukaryotic, w/ nucleus Various cell types specialized for particular function. Differentiation. Over 200 human cell types 56%
Basic unit of life (except virus) CELLS Prokaryotic, w/o nucleus, bacteria Eukaryotic, w/ nucleus Various cell types specialized for particular function. Differentiation. Over 200 human cell types 56%
mirna Dr. S Hosseini-Asl
 mirna Dr. S Hosseini-Asl 1 2 MicroRNAs (mirnas) are small noncoding RNAs which enhance the cleavage or translational repression of specific mrna with recognition site(s) in the 3 - untranslated region
mirna Dr. S Hosseini-Asl 1 2 MicroRNAs (mirnas) are small noncoding RNAs which enhance the cleavage or translational repression of specific mrna with recognition site(s) in the 3 - untranslated region
General information. Cell mediated immunity. 455 LSA, Tuesday 11 to noon. Anytime after class.
 General information Cell mediated immunity 455 LSA, Tuesday 11 to noon Anytime after class T-cell precursors Thymus Naive T-cells (CD8 or CD4) email: lcoscoy@berkeley.edu edu Use MCB150 as subject line
General information Cell mediated immunity 455 LSA, Tuesday 11 to noon Anytime after class T-cell precursors Thymus Naive T-cells (CD8 or CD4) email: lcoscoy@berkeley.edu edu Use MCB150 as subject line
6.5 Enzymes. Enzyme Active Site and Substrate Specificity
 180 Chapter 6 Metabolism 6.5 Enzymes By the end of this section, you will be able to: Describe the role of enzymes in metabolic pathways Explain how enzymes function as molecular catalysts Discuss enzyme
180 Chapter 6 Metabolism 6.5 Enzymes By the end of this section, you will be able to: Describe the role of enzymes in metabolic pathways Explain how enzymes function as molecular catalysts Discuss enzyme
Citation for published version (APA): Oderkerk, A. E. (1999). De preliminaire fase van het rechtsvergelijkend onderzoek Nijmegen: Ars Aequi Libri
 UvA-DARE (Digital Academic Repository) De preliminaire fase van het rechtsvergelijkend onderzoek Oderkerk, A.E. Link to publication Citation for published version (APA): Oderkerk, A. E. (1999). De preliminaire
UvA-DARE (Digital Academic Repository) De preliminaire fase van het rechtsvergelijkend onderzoek Oderkerk, A.E. Link to publication Citation for published version (APA): Oderkerk, A. E. (1999). De preliminaire
Thyroid disease and haemostasis: a relationship with clinical implications? Squizzato, A.
 UvA-DARE (Digital Academic Repository) Thyroid disease and haemostasis: a relationship with clinical implications? Squizzato, A. Link to publication Citation for published version (APA): Squizzato, A.
UvA-DARE (Digital Academic Repository) Thyroid disease and haemostasis: a relationship with clinical implications? Squizzato, A. Link to publication Citation for published version (APA): Squizzato, A.
UvA-DARE (Digital Academic Repository) Workplace coaching: Processes and effects Theeboom, T. Link to publication
 UvA-DARE (Digital Academic Repository) Workplace coaching: Processes and effects Theeboom, T. Link to publication Citation for published version (APA): Theeboom, T. (2016). Workplace coaching: Processes
UvA-DARE (Digital Academic Repository) Workplace coaching: Processes and effects Theeboom, T. Link to publication Citation for published version (APA): Theeboom, T. (2016). Workplace coaching: Processes
Novel Reduction of PCSK9 Expression: Mechanistic Insights into the Anti-Atherosclerotic & Hypolipidemic Effects of Heat Shock Protein 27
 Novel Reduction of PCSK9 Expression: Mechanistic Insights into the Anti-Atherosclerotic & Hypolipidemic Effects of Heat Shock Protein 27 Ed O Brien, Jean-Claude Bakala-N Goma, Chunhua Shi Cumming School
Novel Reduction of PCSK9 Expression: Mechanistic Insights into the Anti-Atherosclerotic & Hypolipidemic Effects of Heat Shock Protein 27 Ed O Brien, Jean-Claude Bakala-N Goma, Chunhua Shi Cumming School
Citation for published version (APA): Parigger, E. M. (2012). Language and executive functioning in children with ADHD Den Bosch: Boxpress
 UvA-DARE (Digital Academic Repository) Language and executive functioning in children with ADHD Parigger, E.M. Link to publication Citation for published version (APA): Parigger, E. M. (2012). Language
UvA-DARE (Digital Academic Repository) Language and executive functioning in children with ADHD Parigger, E.M. Link to publication Citation for published version (APA): Parigger, E. M. (2012). Language
Insulin Resistance. Biol 405 Molecular Medicine
 Insulin Resistance Biol 405 Molecular Medicine Insulin resistance: a subnormal biological response to insulin. Defects of either insulin secretion or insulin action can cause diabetes mellitus. Insulin-dependent
Insulin Resistance Biol 405 Molecular Medicine Insulin resistance: a subnormal biological response to insulin. Defects of either insulin secretion or insulin action can cause diabetes mellitus. Insulin-dependent
Molecular Cell Biology 5068 In class Exam 1 October 2, Please print your name: Instructions:
 Molecular Cell Biology 5068 In class Exam 1 October 2, 2012 Exam Number: Please print your name: Instructions: Please write only on these pages, in the spaces allotted and not on the back. Write your number
Molecular Cell Biology 5068 In class Exam 1 October 2, 2012 Exam Number: Please print your name: Instructions: Please write only on these pages, in the spaces allotted and not on the back. Write your number
Unit IV Problem 3 Biochemistry: Cholesterol Metabolism and Lipoproteins
 Unit IV Problem 3 Biochemistry: Cholesterol Metabolism and Lipoproteins - Cholesterol: It is a sterol which is found in all eukaryotic cells and contains an oxygen (as a hydroxyl group OH) on Carbon number
Unit IV Problem 3 Biochemistry: Cholesterol Metabolism and Lipoproteins - Cholesterol: It is a sterol which is found in all eukaryotic cells and contains an oxygen (as a hydroxyl group OH) on Carbon number
endomembrane system internal membranes origins transport of proteins chapter 15 endomembrane system
 endo system chapter 15 internal s endo system functions as a coordinated unit divide cytoplasm into distinct compartments controls exocytosis and endocytosis movement of molecules which cannot pass through
endo system chapter 15 internal s endo system functions as a coordinated unit divide cytoplasm into distinct compartments controls exocytosis and endocytosis movement of molecules which cannot pass through
Identifying and evaluating patterns of prescription opioid use and associated risks in Ontario, Canada Gomes, T.
 UvA-DARE (Digital Academic Repository) Identifying and evaluating patterns of prescription opioid use and associated risks in Ontario, Canada Gomes, T. Link to publication Citation for published version
UvA-DARE (Digital Academic Repository) Identifying and evaluating patterns of prescription opioid use and associated risks in Ontario, Canada Gomes, T. Link to publication Citation for published version
Chapter VIII: Dr. Sameh Sarray Hlaoui
 Chapter VIII: Dr. Sameh Sarray Hlaoui Lipoproteins a Lipids are insoluble in plasma. In order to be transported they are combined with specific proteins to form lipoproteins: Clusters of proteins and lipids.
Chapter VIII: Dr. Sameh Sarray Hlaoui Lipoproteins a Lipids are insoluble in plasma. In order to be transported they are combined with specific proteins to form lipoproteins: Clusters of proteins and lipids.
Proteasomes. When Death Comes a Knock n. Warren Gallagher Chem412, Spring 2001
 Proteasomes When Death Comes a Knock n Warren Gallagher Chem412, Spring 2001 I. Introduction Introduction The central dogma Genetic information is used to make proteins. DNA RNA Proteins Proteins are the
Proteasomes When Death Comes a Knock n Warren Gallagher Chem412, Spring 2001 I. Introduction Introduction The central dogma Genetic information is used to make proteins. DNA RNA Proteins Proteins are the
Inhibition of PCSK9: The Birth of a New Therapy
 Inhibition of PCSK9: The Birth of a New Therapy Prof. John J.P. Kastelein, MD PhD FESC Dept. of Vascular Medicine Academic Medical Center / University of Amsterdam The Netherlands Disclosures Dr. Kastelein
Inhibition of PCSK9: The Birth of a New Therapy Prof. John J.P. Kastelein, MD PhD FESC Dept. of Vascular Medicine Academic Medical Center / University of Amsterdam The Netherlands Disclosures Dr. Kastelein
UvA-DARE (Digital Academic Repository) Genetic variation in Helicobacter pylori Pan, Z. Link to publication
 UvA-DARE (Digital Academic Repository) Genetic variation in Helicobacter pylori Pan, Z. Link to publication Citation for published version (APA): Pan, Z. (1999). Genetic variation in Helicobacter pylori
UvA-DARE (Digital Academic Repository) Genetic variation in Helicobacter pylori Pan, Z. Link to publication Citation for published version (APA): Pan, Z. (1999). Genetic variation in Helicobacter pylori
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10643 Supplementary Table 1. Identification of hecw-1 coding polymorphisms at amino acid positions 322 and 325 in 162 strains of C. elegans. WWW.NATURE.COM/NATURE 1 Supplementary Figure
doi:10.1038/nature10643 Supplementary Table 1. Identification of hecw-1 coding polymorphisms at amino acid positions 322 and 325 in 162 strains of C. elegans. WWW.NATURE.COM/NATURE 1 Supplementary Figure
Chapter 13: Vesicular Traffic
 Chapter 13: Vesicular Traffic Know the terminology: ER, Golgi, vesicle, clathrin, COP-I, COP-II, BiP, glycosylation, KDEL, microtubule, SNAREs, dynamin, mannose-6-phosphate, M6P receptor, endocytosis,
Chapter 13: Vesicular Traffic Know the terminology: ER, Golgi, vesicle, clathrin, COP-I, COP-II, BiP, glycosylation, KDEL, microtubule, SNAREs, dynamin, mannose-6-phosphate, M6P receptor, endocytosis,
Genome of Hepatitis B Virus. VIRAL ONCOGENE Dr. Yahwardiah Siregar, PhD Dr. Sry Suryani Widjaja, Mkes Biochemistry Department
 Genome of Hepatitis B Virus VIRAL ONCOGENE Dr. Yahwardiah Siregar, PhD Dr. Sry Suryani Widjaja, Mkes Biochemistry Department Proto Oncogen and Oncogen Oncogen Proteins that possess the ability to cause
Genome of Hepatitis B Virus VIRAL ONCOGENE Dr. Yahwardiah Siregar, PhD Dr. Sry Suryani Widjaja, Mkes Biochemistry Department Proto Oncogen and Oncogen Oncogen Proteins that possess the ability to cause
Lipid Metabolism. Catabolism Overview
 Lipid Metabolism Pratt & Cornely, Chapter 17 Catabolism Overview Lipids as a fuel source from diet Beta oxidation Mechanism ATP production Ketone bodies as fuel 1 High energy More reduced Little water
Lipid Metabolism Pratt & Cornely, Chapter 17 Catabolism Overview Lipids as a fuel source from diet Beta oxidation Mechanism ATP production Ketone bodies as fuel 1 High energy More reduced Little water
Clinimetrics, clinical profile and prognosis in early Parkinson s disease Post, B.
 UvA-DARE (Digital Academic Repository) Clinimetrics, clinical profile and prognosis in early Parkinson s disease Post, B. Link to publication Citation for published version (APA): Post, B. (2009). Clinimetrics,
UvA-DARE (Digital Academic Repository) Clinimetrics, clinical profile and prognosis in early Parkinson s disease Post, B. Link to publication Citation for published version (APA): Post, B. (2009). Clinimetrics,
Citation for published version (APA): Martina-Mamber, C. E. (2014). GFAP as an understudy in adult neurogenesis. 's-hertogenbosch: Boxpress.
 UvA-DARE (Digital Academic Repository) GFAP as an understudy in adult neurogenesis Mamber, C.E. Link to publication Citation for published version (APA): Martina-Mamber, C. E. (2014). GFAP as an understudy
UvA-DARE (Digital Academic Repository) GFAP as an understudy in adult neurogenesis Mamber, C.E. Link to publication Citation for published version (APA): Martina-Mamber, C. E. (2014). GFAP as an understudy
Module 3 Lecture 7 Endocytosis and Exocytosis
 Module 3 Lecture 7 Endocytosis and Exocytosis Endocytosis: Endocytosis is the process by which cells absorb larger molecules and particles from the surrounding by engulfing them. It is used by most of
Module 3 Lecture 7 Endocytosis and Exocytosis Endocytosis: Endocytosis is the process by which cells absorb larger molecules and particles from the surrounding by engulfing them. It is used by most of
MBios 401/501: Lecture 12.1 Signaling IV. Slide 1
 MBios 401/501: Lecture 12.1 Signaling IV Slide 1 Pathways that require regulated proteolysis 1. Notch and Delta 2. Wnt/ b-catenin 3. Hedgehog 4. NFk-B Our last topic on cell signaling are pathways that
MBios 401/501: Lecture 12.1 Signaling IV Slide 1 Pathways that require regulated proteolysis 1. Notch and Delta 2. Wnt/ b-catenin 3. Hedgehog 4. NFk-B Our last topic on cell signaling are pathways that
UvA-DARE (Digital Academic Repository)
 UvA-DARE (Digital Academic Repository) Standaarden voor kerndoelen basisonderwijs : de ontwikkeling van standaarden voor kerndoelen basisonderwijs op basis van resultaten uit peilingsonderzoek van der
UvA-DARE (Digital Academic Repository) Standaarden voor kerndoelen basisonderwijs : de ontwikkeling van standaarden voor kerndoelen basisonderwijs op basis van resultaten uit peilingsonderzoek van der
Intracellular Compartments and Protein Sorting
 Intracellular Compartments and Protein Sorting Intracellular Compartments A eukaryotic cell is elaborately subdivided into functionally distinct, membrane-enclosed compartments. Each compartment, or organelle,
Intracellular Compartments and Protein Sorting Intracellular Compartments A eukaryotic cell is elaborately subdivided into functionally distinct, membrane-enclosed compartments. Each compartment, or organelle,
UvA-DARE (Digital Academic Repository) Obesity, ectopic lipids, and insulin resistance ter Horst, K.W. Link to publication
 UvA-DARE (Digital Academic Repository) Obesity, ectopic lipids, and insulin resistance ter Horst, K.W. Link to publication Citation for published version (APA): ter Horst, K. W. (2017). Obesity, ectopic
UvA-DARE (Digital Academic Repository) Obesity, ectopic lipids, and insulin resistance ter Horst, K.W. Link to publication Citation for published version (APA): ter Horst, K. W. (2017). Obesity, ectopic
Hypercholesterolemia: So much cholesterol, so many causes
 Hypercholesterolemia: So much cholesterol, so many causes Student group names kept anonymous Department of Biology, Lake Forest College, Lake Forest, IL 60045, USA Hypercholesterolemia is a disease characterized
Hypercholesterolemia: So much cholesterol, so many causes Student group names kept anonymous Department of Biology, Lake Forest College, Lake Forest, IL 60045, USA Hypercholesterolemia is a disease characterized
Overview: Conducting the Genetic Orchestra Prokaryotes and eukaryotes alter gene expression in response to their changing environment
 Overview: Conducting the Genetic Orchestra Prokaryotes and eukaryotes alter gene expression in response to their changing environment In multicellular eukaryotes, gene expression regulates development
Overview: Conducting the Genetic Orchestra Prokaryotes and eukaryotes alter gene expression in response to their changing environment In multicellular eukaryotes, gene expression regulates development
REGULATION OF ENZYME ACTIVITY. Medical Biochemistry, Lecture 25
 REGULATION OF ENZYME ACTIVITY Medical Biochemistry, Lecture 25 Lecture 25, Outline General properties of enzyme regulation Regulation of enzyme concentrations Allosteric enzymes and feedback inhibition
REGULATION OF ENZYME ACTIVITY Medical Biochemistry, Lecture 25 Lecture 25, Outline General properties of enzyme regulation Regulation of enzyme concentrations Allosteric enzymes and feedback inhibition
Focus on FH (Familial Hypercholesterolemia) Joshua W. Knowles, MD PhD for PCNA May, 2013
 Focus on FH (Familial Hypercholesterolemia) Joshua W. Knowles, MD PhD for PCNA May, 2013 Conflicts CMO for The FH Foundation Pre-talk quiz What is cascade screening? 1. screening all family members 2.
Focus on FH (Familial Hypercholesterolemia) Joshua W. Knowles, MD PhD for PCNA May, 2013 Conflicts CMO for The FH Foundation Pre-talk quiz What is cascade screening? 1. screening all family members 2.
Signaling. Dr. Sujata Persad Katz Group Centre for Pharmacy & Health research
 Signaling Dr. Sujata Persad 3-020 Katz Group Centre for Pharmacy & Health research E-mail:sujata.persad@ualberta.ca 1 Growth Factor Receptors and Other Signaling Pathways What we will cover today: How
Signaling Dr. Sujata Persad 3-020 Katz Group Centre for Pharmacy & Health research E-mail:sujata.persad@ualberta.ca 1 Growth Factor Receptors and Other Signaling Pathways What we will cover today: How
Chapter 6. Antigen Presentation to T lymphocytes
 Chapter 6 Antigen Presentation to T lymphocytes Generation of T-cell Receptor Ligands T cells only recognize Ags displayed on cell surfaces These Ags may be derived from pathogens that replicate within
Chapter 6 Antigen Presentation to T lymphocytes Generation of T-cell Receptor Ligands T cells only recognize Ags displayed on cell surfaces These Ags may be derived from pathogens that replicate within
Cellular functions of protein degradation
 Protein Degradation Cellular functions of protein degradation 1. Elimination of misfolded and damaged proteins: Environmental toxins, translation errors and genetic mutations can damage proteins. Misfolded
Protein Degradation Cellular functions of protein degradation 1. Elimination of misfolded and damaged proteins: Environmental toxins, translation errors and genetic mutations can damage proteins. Misfolded
Regulation of Gene Expression in Eukaryotes
 Ch. 19 Regulation of Gene Expression in Eukaryotes BIOL 222 Differential Gene Expression in Eukaryotes Signal Cells in a multicellular eukaryotic organism genetically identical differential gene expression
Ch. 19 Regulation of Gene Expression in Eukaryotes BIOL 222 Differential Gene Expression in Eukaryotes Signal Cells in a multicellular eukaryotic organism genetically identical differential gene expression
SUPPLEMENTARY INFORMATION
 Figure S1. Silver staining and immunoblotting of the purified TAK1 kinase complex. The TAK1 kinase complex was purified through tandem affinity methods (Protein A and FLAG), and aliquots of the purified
Figure S1. Silver staining and immunoblotting of the purified TAK1 kinase complex. The TAK1 kinase complex was purified through tandem affinity methods (Protein A and FLAG), and aliquots of the purified
Effects of biological response modifiers in psoriasis and psoriatic arthritis Goedkoop, A.Y.
 UvA-DARE (Digital Academic Repository) Effects of biological response modifiers in psoriasis and psoriatic arthritis Goedkoop, A.Y. Link to publication Citation for published version (APA): Goedkoop, A.
UvA-DARE (Digital Academic Repository) Effects of biological response modifiers in psoriasis and psoriatic arthritis Goedkoop, A.Y. Link to publication Citation for published version (APA): Goedkoop, A.
Ch. 18 Regulation of Gene Expression
 Ch. 18 Regulation of Gene Expression 1 Human genome has around 23,688 genes (Scientific American 2/2006) Essential Questions: How is transcription regulated? How are genes expressed? 2 Bacteria regulate
Ch. 18 Regulation of Gene Expression 1 Human genome has around 23,688 genes (Scientific American 2/2006) Essential Questions: How is transcription regulated? How are genes expressed? 2 Bacteria regulate
RNA Processing in Eukaryotes *
 OpenStax-CNX module: m44532 1 RNA Processing in Eukaryotes * OpenStax This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution License 3.0 By the end of this section, you
OpenStax-CNX module: m44532 1 RNA Processing in Eukaryotes * OpenStax This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution License 3.0 By the end of this section, you
